
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 22 December 2022
Sec. Systems Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1074398
This article is part of the Research TopicUntangle the Broad Connections and Tight Interactions Between Human Microbiota and Complex Diseases Through Data-Driven ApproachesView all 13 articles
 Tingting Niu1†
Tingting Niu1† Yi Cui1†
Yi Cui1† Xu Shan1
Xu Shan1 Shuzhen Qin1
Shuzhen Qin1 Xuejie Zhou1
Xuejie Zhou1 Rui Wang1
Rui Wang1 Alan Chang2
Alan Chang2 Nan Ma3*
Nan Ma3* Jingjing Jing4*
Jingjing Jing4* Jianwei He1*
Jianwei He1*Introduction: The industrial yeast Pichia pastoris is widely used as a cell factory to produce proteins, chemicals and advanced biofuels. We have previously constructed P. pastoris strains that overexpress protein disulfide isomerase (PDI), which is a kind of molecular chaperone that can improve the expression of an exogenous protein when they are co-expressed. Chicken cystatin (cC) is a highly thermostable cysteine protease inhibitor and a homologous protein of human cystatin C (HCC). Wild-type cC and the two mutants, I66Q and ΔW (a truncated cC lacking the á-helix 2) represent proteins with different degrees of stability.
Methods: Wild-type cC, I66Q and ΔW were each overexpressed in P. pastoris without and with the coexpression of PDI and their extracellular levels were determined and compared. Transcriptomic profiling was performed to compare the changes in the main signaling pathways and cell components (other than endoplasmic reticulum quality control system represented by molecular chaperones) in P. pastoris in response to intracellular folding stress caused by the expression of exogenous proteins with different stabilities. Finally, hub genes hunting was also performed.
Results and discussion: The coexpression of PDI was able to increase the extracellular levels of both wild-type cC and the two mutants, indicating that overexpression of PDI could prevent the misfolding of unstable proteins or promote the degradation of the misfolded proteins to some extent. For P. pastoris cells that expressed the I66Q or ΔW mutant, GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) analyses of the common DEGs in these cells revealed a significant upregulation of the genes involved in protein processing, but a significant downregulation of the genes enriched in the Ribosome, TCA and Glycolysis/Gluconeogenesis pathways. Hub genes hunting indicated that the most downregulated ribosome protein, C4QXU7 in this case, might be an important target protein that could be manipulated to increase the expression of foreign proteins, especially proteins with a certain degree of instability.
Conclusion: These findings should shed new light on our understanding of the regulatory mechanism in yeast cells that responds to intracellular folding stress, providing valuable information for the development of a convenient platform that could improve the efficiency of heterologous protein expression in P. pastoris.
Human cystatin C (HCC) is a papain-like cysteine protease inhibitor that belongs to the cystatin superfamily, and it is also one of the most extensively studied endogenous inhibitors as well as an important biomarker of renal function (Dubin, 2005). Abnormal changes in the expression and secretion of HCC in the brain have been described for various neurological disorders such as amyotrophic lateral sclerosis (ALS), rare heritable neurodegenerative disorders, ischemia, some forms of epilepsy, Alzheimer’s disease (AD) (Mathews and Levy, 2016) and recurrent hemorrhagic stroke (Merz et al., 1997; Zhou et al., 2015).
Previous studies have reported that the fatal amyloid disease, hereditary cystatin amyloid angiopathy (HCCAA), found in young Icelanders is mainly caused by the HCC hereditary amyloidogenic mutant L68Q, which has a high dimerization potential that can lead to self-aggregation and hyper-amyloidosis (Janowski et al., 2001; Palsdottir et al., 2006). The instability of the soluble HCC monomer has constrained any structural studies on its physicochemical properties. Meanwhile, chicken cystatin (cC) has a number of characteristics similar to HCC, and both proteins share about 44% sequence homology. Thus, cC is considered an ideal model for studying protein domain exchange and amyloid-related diseases (Bode et al., 1988; Yu et al., 2010). Residue 66 in cC corresponds to residue 68 in HCC, and the I66Q mutant of cC has similar amyloidogenic properties to L68Q of HCC under physiological conditions (Bjarnadottir et al., 2001).
The AS (appending structure) region of cC contains α-helix 2, which is crucial for the stability of cC and is considered to be the biggest difference between HCC and cC (Grubb et al., 1984). Therefore, the α-helix 2-truncated mutant (ΔW) with a deletion at residues 77–85 was constructed as the unstable cC model protein. Based on our previous results, the secreted amount of ΔW is much lower than that of WT cC or I66Q when expressed in P. pastoris, indicating that the absence of α-helix 2 in the AS region may be one of the factors contributing to the structural instability of HCC (Zhou et al., 2019).
Pichia pastoris (reclassified as Komagataella phaffii/pastoris) is a methylotrophic yeast and a highly successful system for producing recombinant proteins in the pharmaceutical and biofuel industries (Yu et al., 2017). The ability of P. pastoris to express recombinant proteins is facilitated by the strong promoter of its alcohol oxidase 1 (AOX1) gene. The activity of the AOX1 promoter is tightly regulated by the carbon source. Thus, recombinant proteins expressed from the AOX1 promoter in P. pastoris cells can be induced with methanol once cell growth has reached high densities to obtain a high level of expression for the proteins. For example, a His-Qtagged lipase A from Beauveria bassiana has been successfully produced in P. pastoris and shown to have potential use for biodiesel production via ethanolysis (Vici et al., 2015). Another example is the expression of α-L-arabinofuranosidase (ARA) in P. pastoris, which can be improved 5.5-fold by codon optimization. The recombinant ARA has significant potential in the catalytic conversion of corn stover to fermentable sugars during biofuel production (Vici et al., 2015). However, overexpression of recombinant proteins may lead to more misfolded proteins and trigger endoplasmic reticulum (ER) stress (Jolly and Morimoto, 2000; Oakes and Papa, 2015). The cells might then respond to ER stress by increasing the expression of some molecular chaperones, including PDI, HSP90, and HSP72 (Vogl and Glieder, 2013; Delic et al., 2014; Gu et al., 2015). To prevent protein misfolding and aggregation, the newly synthesized molecular chaperones would increase folding efficiency by capturing the folded intermediates and promoting refolding or degradation. Co-expression of PDI has been used to improve the expression of heterologous proteins in P. pastoris by overcoming the burden of protein folding and secretion (Inan et al., 2006; Navone et al., 2021a). In our previous study, the overexpression of PDI in P. pastoris GS115 strains was found to significantly increase the expression of cC (Zhou et al., 2019). On this basis, three representative proteins with different stabilities (WT cC and its two mutants I66Q and ΔW) (Figure 1) were used as model proteins to screen for factors other than the ER quality control system represented by the molecular chaperone PDI that could influence the expression of foreign proteins that may not be properly folded in P. pastoris. Subsequently, transcriptomic profiling was performed to identify the transcriptomic changes and pathways involved in the molecular network and changes in the dynamic mechanism of the foreign protein secretion pathway in P. pastoris.
Pichia Pastoris GS115 strain was provided by Dr. Shutao Liu at Fuzhou University. The plasmids pPIC3.5K and pPICZαA were purchased from Invitrogen. GS115 strain and the previously constructed GS115 PDI-overexpressing strain were used as starting strains for the construction of the PDI and cC co-overexpressing strains. The yeast cells were first cultured at 30°C in Yeast Peptone Dextrose medium (YPD) (1% yeast extract, 2% peptone, and 2% glucose) to logarithmic growth phase (OD600 = 5.0) followed by methanol induction in Yeast Extract Peptone Medium (YPM) [1% yeast extract, 2% peptone, 0.5% Methanol (v/v)] for 72 h to induce the expression of PDI and cC.
GS115 competent cells were transformed with the linearized plasmid pPICZαA-cC, pPICZαA-I66Q, and pPICZαA-ΔW by electroporation to generate GS115-cC, GS115-I66Q, GS115-ΔW recombinant strains, respectively. Similarly, GS115-PDI-cC, GS115-PDI-I66Q, GS115-PDI-ΔW strains were obtained by transforming GS115 PDI with pPICZαA-cC, pPICZαA-I66Q, and pPICZαA-ΔW, respectively.
Extracellular protein samples were obtained as previously described (Zhou et al., 2019). Intracellular protein samples were extracted from yeast cells after treatment with Yeast Protein Extraction Reagent (Takara, Dalian, China). SDS-PAGE and western blotting were carried out following the procedure described previously (Zhou et al., 2019). The protein bands in one gel were visualized by staining the gel with PAGE Gel Silver Staining Kit (Takara, Beijing, China) whereas the protein bands in the other gel were transferred to a PVDF membrane (Millipore, MA, USA) for western blot analysis. After protein transfer, the PVDF membrane was incubated in a blocking buffer containing TBST plus 5% skimmed milk powder for 2 h. This was followed by three 10 min washes in TBST buffer, and 1 h of incubation in rabbit anti-cC antiserum (1:2000) at room temperature. After that, the membrane was again washed three times in TBST, with each wash lasting for 10 min. Finally, the blot was incubated with anti-rabbit peroxidase conjugate (1:10000) for 1 h at RT, and then subjected to detection using the eECL Reagent (Beyotime, Shanghai, China).
Total RNA was extracted from the different GS115 strains using Yeast RNAiso Kit (Takara, Dalian, China). The extraction was performed according to the manufacturer’s protocol. The mRNA fraction was purified from the total RNA using MicroPoly Purist kit (Takara, Dalian, China) according to the manufacturer’s protocol. The concentration and integrity of the mRNA were measured using a NanoDrop 2000 (Thermo Fisher Scientific, MA, USA) and the Agilent 2100 LabChip system (Agilent Technologies, CA, USA). The RNA was sheared, and reverse transcribed using random primers to obtain the cDNA, which was then used for the construction of a cDNA library. Illumina RNA sequencing (RNA-Seq) libraries were subsequently performed using VAHTS Universal V6 RNA-seq Library P the SMARTer Stranded RNA Seq Kit (Vazyme Biotech Co. Ltd.) according to the manufacturer’s instructions. Finally, RNA-Seq data were generated in Fastq format. The sequencing data have been submitted to National Center for Biotechnology Information (NCBI) under accession PRJNA8928871.
The differentially expressed genes (DEGs) for WT vs. I66Q, PDI-WT vs. PDI-I66Q, WT vs. ΔW and PDI-WT vs. PDI-ΔW were identified by the BioMarker cloud platform with adjusted p-value <0.01 and log2 fold change (FC) >2. Moreover, the common DEGs between different groups have been identified by the same method.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed on the BioMarker cloud platform with a p-value <0.05. ClueGo plug-in in Cytoscape software (3.8 version) was used for showing the ClueGO network diagram.
Common DEGs were used to construct the protein–protein interaction (PPI) network by using the SRTING online database with a confidence score of more than 0.7. Hub genes of the PPI network were identified using a degree algorithm from cytoHubba, a plugin in Cytoscape, and visualized using Cytoscape (v3.8.0).
To investigate the intracellular distribution pattern and retention level of recombinant cC as well as its extracellular secretion in P. pastoris, WT cC, I66Q and ΔW were expressed in both P. pastoris GS115 strains without and with the overexpression of PDI. The extracellular secretion of wild-type cC and its two mutants was detected by silver staining (Figure 2A). Overexpression of PDI (57 kDa) can significantly enhance the expression of WT cC (14 kDa) and the I66Q mutant. In the case of ΔW, the protein was only secreted when it was co-expressed with PDI (Figure 2B). As shown in Figure 2B, almost no I66Q and ΔW were detected as an intracellular form in both PDI-overexpressing GS115 and wild-type GS115 strains. Interestingly, for WT cC, no significant difference in intracellular level was observed between the two yeast strains. This might suggest that when the yeast expressed WT cC, most of the proteins were capable of folding into the native form and were subsequently secreted out of the cell, with little misfolded protein being produced and residing in the ER despite an increase in the amount of the newly synthesized protein entering the ER. For the mutant I66Q, its intracellular level was proportional to its extracellular level in either yeast strain because of its amyloidogenic properties. Consequently, the three cC-overexpressing GS115 strains and the three cC-overexpressing GS115 strains that also co-expressed PDI were used for the following transcriptomic studies.
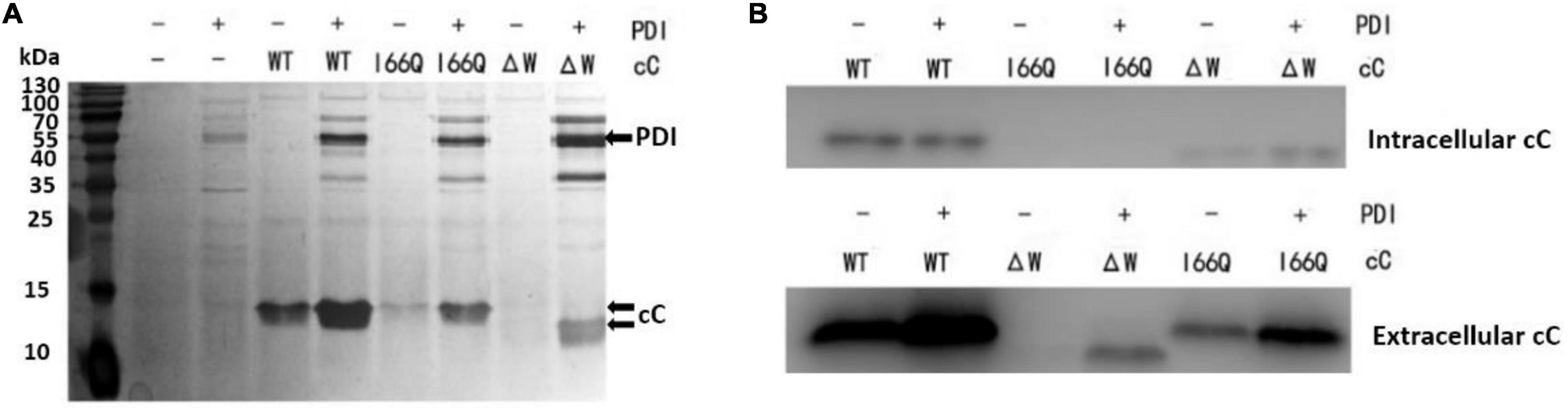
Figure 2. Analysis of the expression of the different versions of chicken cystatin (cC) (MW: 13 kDa) in both GS115 strains that did and did not overexpress PDI (MW: 57 kDa). (A) Secretion of the WT cC, I66Q and ΔW as detected in the culture supernatant after centrifugation as shown by SDS-PAGE. The gel was visualized by silver staining. (B) Expression of WT cC, I66Q and ΔW in GS115 as analyzed by western blot. Intracellular refers to the soluble fraction of cell lysate, and Extracellular refers to the culture supernatant. “+” and “–” indicates the cells were transfected with and without the corresponding cC-coding gene, respectively.
It has been proven that genes are being expressed at different levels in different individual organisms as a result of biological variability (Robasky et al., 2014). Therefore, biological replicates were included to ensure the validity of the following experiments. The Pearson Correlation Coefficient r refers to the biological assessment of repeated samples and it was used to analyze the correlation between every two samples (Schulze et al., 2012). The closer r2 is to 1, the stronger the correlation between the two replicates (Figure 3). Subsequently, 24 samples were subjected to the transcriptomic analysis after RNA-sequencing was completed, with three replicates included for each strain and the data are summarized in Table 1. The clean reads of each sample were compared with the designated P. pastoris GS115 genome. The mapped data obtained after alignment were used to evaluate the quality of the library such as randcheck, insert size, and saturation test. Typically, FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) was used as an indicator to measure the expression level of a gene (Zhao et al., 2021). Identification of DEGs was carried out according to the gene expression levels in different samples.
Different numbers of up-and down-regulated DEGs were obtained by comparing each of the two sample groups as shown in Figure 4. Interestingly, when the WT cC-overexpressing strain was compared with either the I66Q- or ΔW-overexpressing strain, the DEGs, especially the up-regulated genes, were significantly increased. Nevertheless, this trend was not observed in the comparison of PDI-WT vs. PDI-I66Q/ΔW, implying a healthy intracellular cell condition in GS115-PDI-I66Q and GS115-PDI-ΔW afforded by the overexpression of PDI. From this point, it became important to investigate the common DEGs identified from the comparison of both WT vs. I66Q/ΔW and PDI-WT vs. PDI-I66Q/ΔW, since these common DEGs are vital for the overexpression of foreign proteins that are less stable. The total number of up-regulated and down-regulated genes in each comparison group was depicted in a Venn diagram (Figure 4B). Among all the compared genes, 203 and 210 common DEGs indicated in Figure 4B were selected for subsequent KEGG analysis, GO annotation, and hub gene identification.
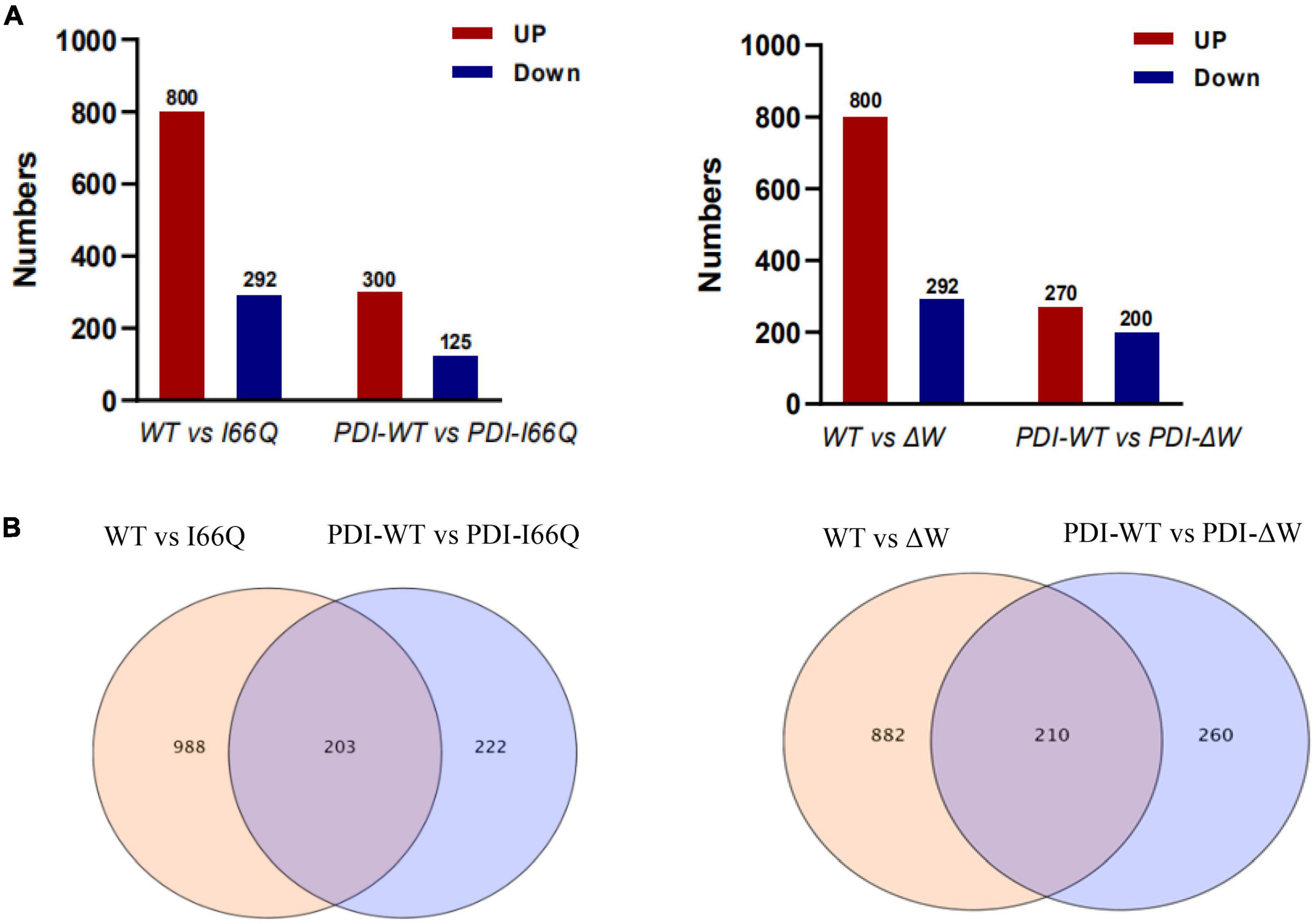
Figure 4. Count of differentially expressed genes (DEGs) in WT vs. I66Q/ΔW and PDI-WT vs. PDI-I66Q/ΔW comparisons. (A) Total number of up and downregulated DEGs in the four pairs: WT vs. I66Q, WT vs. ΔW, PDI-WT vs. PDI-I66Q and PDI-WT vs. PDI-ΔW. (B) Venn diagram depicting the total number of proteins, including the up and down-regulated genes in the four groups.
Kyoto encyclopedia of genes and genomes analysis was applied to explore the potential molecular functions and molecular mechanisms associated with the functions of the common DEGs. As shown in Figure 5A, for WT vs. I66Q and PDI-WT vs. PDI-I66Q comparisons, several signaling pathways were significantly enriched, including the pathways for the biosynthesis of amino acids, citrate cycle, and Tricarboxylic Acid cycle (TCA cycle), and the metabolic pathways. In the comparison of WT vs. ΔW and of PDI-WT vs. PDI-ΔW, in addition to the pathways that were enriched in both WT vs. I66Q and PDI-WT vs. PDI-I66Q, the DNA replication and ribosome pathways were also indicated, suggesting that the activation of genes in different KEGG pathways may be due to the different physicochemical properties of the I66Q and ΔW cC mutants (Figure 5B).
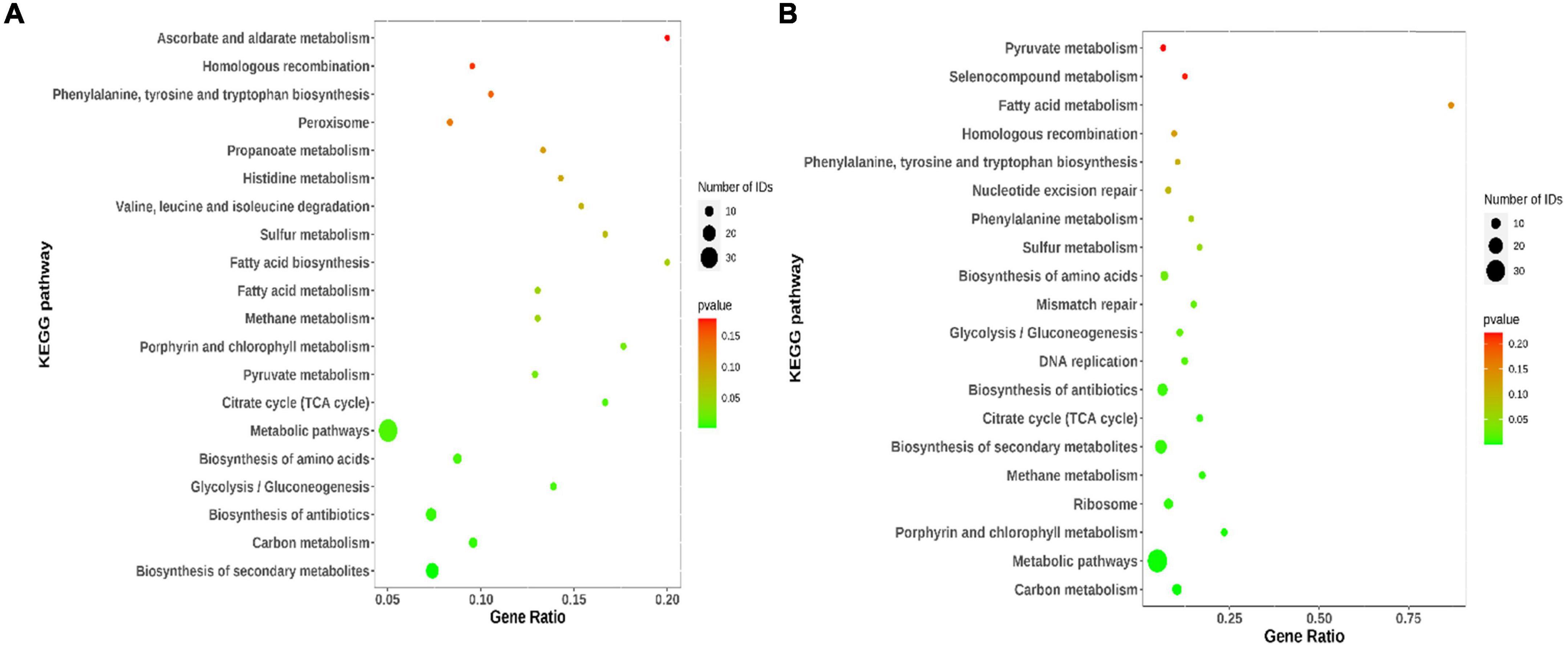
Figure 5. Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis of common differentially expressed genes (DEGs). (A) KEGG pathway enrichment of 203 common DEGs identified from the WT vs. I66Q and PDI-WT vs. PDI-I66Q comparisons. (B) KEGG pathway enrichment of 210 common DEGs from the WT vs. ΔW and PDI-WT vs. PDI-ΔW comparisons.
Enrichment analysis was performed on the 203 and 210 common DEGs using the ClueGO v2.5.4 plugin. After setting the P-value as <0.05 and the Kappa Score Threshold as 0.4, 25, and 24 GO terms were identified from the group WT vs. I66Q and PDI-WT vs. PDI-I66Q, WT vs. ΔW and PDI-WT vs. PDI-ΔW, respectively. For the comparison of WT vs. I66Q and PDI-WT vs. PDI-I66Q, the enriched genes were mainly involved in protein folding, translation, and the acetyl-CoA metabolic and monocarboxylic acid biosynthetic processes (Figure 6A). For WT vs. ΔW and PDI-WT vs. PDI-ΔW, the enriched genes were not only distributed in the above pathways but also in the DNA metabolic process and porphyrin-containing compound biosynthetic process (Figure 6B). The result obtained from ClueGO enrichment analysis provided a global understanding of the scenario when proteins with different stabilities were expressed in P. pastoris. The common DEGs enriched in both the metabolic and protein processing pathways were quite noticeable in the comparison of WT vs. I66Q and that of WT vs. ΔW, suggesting that when amyloid mutants and unstable exogenous proteins are expressed in the P. pastoris, there might be a need to adjust the basic metabolic speed/efficiency and reproduction speed of the cells.
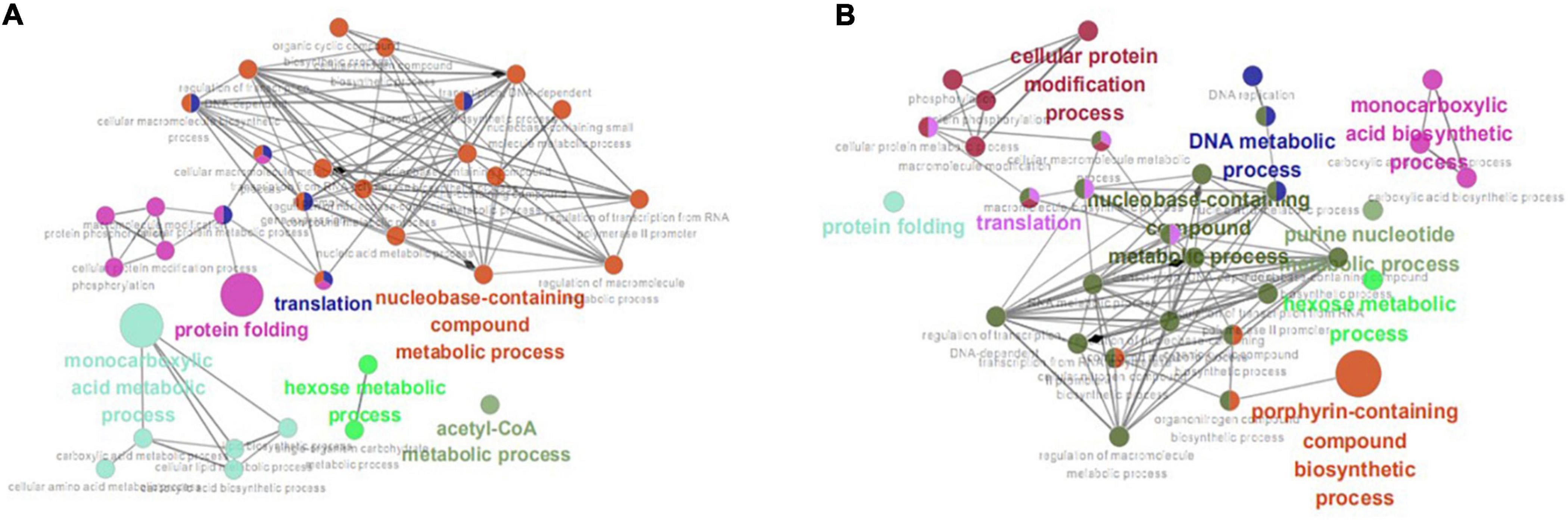
Figure 6. ClueGO enrichment analysis. (A) Significantly enriched gene ontology (GO) terms of the common differentially expressed genes (DEGs) identified from the WT vs. I66Q and PDI-WT vs. PDI-I66Q comparisons. (B) Significantly enriched GO terms of the common DEGs from the WT vs. ΔW and PDI-WT vs. PDI-ΔW comparisons.
To further investigate the key cellular components and biological processes in the wild-type GS115 (I66Q/ΔW) and PDI-overexpressing GS115 strains (I66Q/ΔW), both of which were found to have common enriched DEGs, the STRING (Search Tool for the Retrieval of Interacting Genes) online tool was used to construct a PPI network of common DEGs. A combined score of >0.7 was set as the cut-off criterion for statistical significance. Next, the PPI network was downloaded and visualized as shown in Figure 7. A total of 132 nodes and 101 edges were identified for the common genes belonging to the comparison of WT vs. I66Q and of PDI-WT vs. PDI-I66Q (Figure 7A). According to the Degree sores in the cytoHubba, the top ten highest-scored genes were selected as the hub genes, including the common DEGs that encode the proteins C4QXU7, C4QZL4, C4R7T6, C4R447, C4R196, C4R0F8, C4R7T7, C4R7D3, C4R596 (Figure 7B). As for the comparison of WT vs. ΔW and of PDI-WT vs. PDI-ΔW, 140 nodes and 118 edges were found in the PPI network (Figure 7C), and the Hub genes included the common DEGs that encode C4QXU7, C4QVA2, C4R0T8, C4R447, C4QZE7, C4R7A1, C4R8M3, C4QWG3, C4R7T6, C4R7T6, C4QWY6 (Figure 7D).
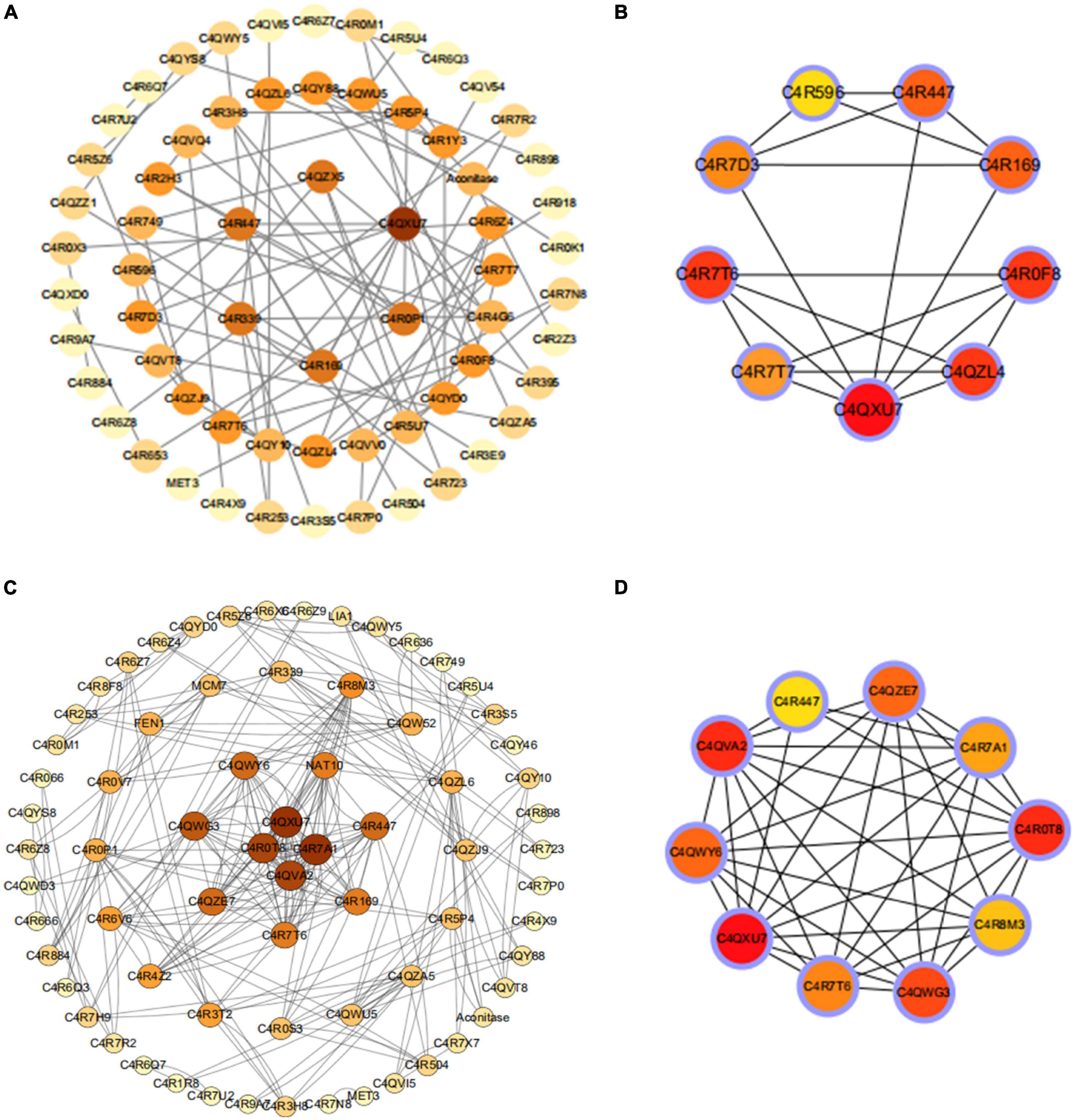
Figure 7. Protein–protein interaction (PPI) network of the proteins encoded by the common differentially expressed genes (DEGs) and selection of hub genes. (A,B) PPI network and the top 10 common DEGs between WT and I66Q and between PDI-WT and PDI-I66Q. (C,D) PPI network and the top 10 common DEGs between the group WT and ΔW and between PDI-WT and PDI-ΔW.
C4QXU7, C4R7T6, and C4R0F8 were the top three key proteins identified in the comparison of WT vs. I66Q and of PDI-WT vs. PDI-I66Q. C4QXU7 is a small subunit of ribosomal protein S28e, whereas C4R7T6 is the large subunit of the ribosomal protein LP1, and C4R0F8 is a small subunit of the ribosomal protein S26e. In the case of WT vs. ΔW and PDI-WT vs. PDI-ΔW comparisons, the top three key proteins included C4QXU7, C4QVA2, and C4R0T8. C4QVA2 is a small subunit of ribosomal protein S3e, and C4R0T8 is a small subunit of ribosomal protein S15A. All of the above-mentioned proteins are responsible for the structural integrity of the ribosome and the protein translation rate. Interestingly, all the top three hub genes belong to the components of ribosomal subunits and they were found to be downregulated. Moreover, they were all found to be enriched in the translation pathway. This observation underlined the importance of the rate of protein synthesis by ribosomes and suggested that protein synthesis by ribosomes may be the ultimate goal that yeast cells need to adjust when an exogenous unstable protein is expressed in P. pastoris.
It is generally believed that the amount of secreted amyloid mutant proteins and unstable mutant proteins is usually lower than that of their wild-type counterparts when eukaryotic expression systems are used (Whyteside et al., 2011; Bou Ali et al., 2013; He et al., 2019). When yeast cells express foreign proteins, the misfolded proteins may become toxic to the cells, and therefore, the co-overexpression of molecular chaperones may promote the folding and posttranslational modifications of the expressed proteins, thereby facilitating the secretion of the foreign proteins. Our results showed that co-expression of PDI with wild-type cC or mutated cC (I66Q or ΔW) could improve the secretion of cC to different extents. However, the distinct client-recruiting system of molecular chaperones may have a limit in improving the secretion of foreign proteins that may not be properly folded (Yan et al., 2021). This limitation could prevent P. pastoris from becoming an ideal protein production platform that can accommodate a variety of production requirements. Although modifications of specific transcription factors have been adopted to alter the regulation mode, it is still difficult to markedly increase the expression of foreign proteins in wild-type P. pastoris (Nusse and Lindau, 1988; Wang et al., 2017; Vogl et al., 2018). In this context, it is essential to pursue other regulatory genes and related cellular pathways that are capable of increasing the expression of foreign proteins in P. pastoris. Accordingly, WT, I66Q and ΔW were designed to be secreted by P. pastoris cells that did not overexpress PDI and those that did overexpress PDI, since both cell types could achieve different levels of protein expression in response to the various stabilities of WT cC and the two cC mutants. Our results, along with previously reported results, have shown that following their expression in P. pastoris, improperly folded foreign proteins may be found in lower levels compared with those that are properly folded, such as the improperly folded cC mutants versus their wide-type counterpart. Interestingly, the two cC mutants had different stabilities, which directly impacted their extracellular and intracellular levels. Consequently, the order of decreasing stability for WT cC, I66Q and ΔW turned out to be well-suited for establishing P. pastoris strains that could be used to investigate the key regulatory genes that the host cell would modulate when expressing foreign proteins that may not be properly folded.
Both P. pastoris GS115 which did not overexpress PDI and P. pastoris GS115 overexpressed PDI were used as the starting strains to express different variants of cC having different stabilities (WT cC, I66Q and ΔW). RNA-seq analysis was then performed in order to screen for the common DEGs between the WT cC-expressing strain and I66Q-expressing strain or ΔW-expressing strain. For the comparison of WT vs. I66Q with PDI-WT vs. PDI-I66Q, GO analysis indicated the consumption of energy by the host cells as an important factor in the case of foreign protein expression since protein synthesis is an energy-costing process. Glycolysis and the TCA cycle are processes of energy acquisition, so reducing energy acquisition is a way to force the slow-down of protein synthesis because, without ATP and GTP, protein synthesis cannot occur. Glycolysis also leads to the pentose phosphate pathway, which provides nucleotides for nucleic acid synthesis. Thus, lowering glycolysis also affects RNA synthesis, and hence protein synthesis. The result of KEGG pathway analysis showed that in both groups, the common DEGs were enriched in the ribosome and TCA cycle, as well as in carbon metabolism and glycolysis/gluconeogenesis, consistent with the results of GO analysis. The basal metabolic level of P. pastoris is closely related to the protein expression level (Liang et al., 2012; Renuse et al., 2014). When P. pastoris expressed I66Q and ΔW during the methanol induction stage, the common DEGs enriched in the TCA and glycolytic pathways were significantly decreased compared with those in the WT cC-expressing strain. At the same time, the common DEGs enriched in the protein translation and protein folding processes were up-regulated, indicating an initial increased expression of protein synthesis-related genes, and even glycolysis and the TCA cycle-related genes in the case of the I66Q- and ΔW-expressing strains. At a later time, the burden caused by these misfolded proteins started to affect the cells and resulted in an adjustment to reduce the expression of the foreign proteins. However, the selective pressure of methanol induction was still ongoing, so protein synthesis could only be slowed down by reducing the flow of energy to protein synthesis (Sola et al., 2007; Orman et al., 2009). These findings indicated that when foreign proteins that may not be properly folded are expressed in P. pastoris, the cells need to adjust their own metabolic states in order to maintain intracellular homeostasis, based on the degree of protein instability.
Hub genes are considered to be key genes that play vital roles in biological processes and can affect the regulation of other genes in a related pathway (Liu et al., 2022; Shu et al., 2022). It is of significance to note that the key gene C4QXU7, a small subunit of the ribosomal protein S28e, was identified by both the comparisons of WT vs. I66Q/ΔW and PDI-WT vs. PDI-I66Q/ΔW (Figure 4B). Yeast ribosomal proteins play important roles in the biogenesis and function of the ribosome (Aw et al., 2017). Deletion of a particular ribosome protein can delay or impair the subunit assembly, indicating that the decelerated elongation stage of translation might promote the co-translational folding rate of heterologous proteins (Liao et al., 2019). In P. pastoris, overexpression of xylanase A (a foreign protein) can lead to a significant down-regulation of numerous ribosomal proteins, resulting in decelerated translation elongation and enhanced folding efficiency for xylanase A (Navone et al., 2021b). Meanwhile, studies have shown that knocking out the C4QXU7 gene in P. pastoris does not affect its growth, but can lead to a significant increase in the secretion levels of exogenous proteins (e.g., Pfu and Phytase), indicating that the decelerated elongation rate caused by the loss of C4QXU7 might promote the co-translational folding rate of heterologous proteins, increasing the expression of Pfu and Phytase (Liao et al., 2019). Together with our data on protein expression in P. pastoris, these observations could collectively indicate that knocking out the C4QXU7 gene may promote the expression of foreign proteins that are not easily folded in P. pastoris.
On the other hand, because P. pastoris is widely used for the expression of foreign proteins in industrial protein production, how to design and develop a new P. pastoris expression system capable of yielding a high expression level and flexible regulation characteristics is one of the key problems and important goals faced by bioengineering and synthetic biotechnology. In this study, the protein expression levels of three model proteins with the order of decreasing stability WT > I66Q > ΔW were significantly increased in P. pastoris GS115 that simultaneously overexpressed PDI. In addition to molecular chaperones, our data also revealed that some ribosomal proteins, e.g., C4QXU7, may also be important targets that can be modulated to increase the expression of foreign proteins. The modulation of key P. pastoris ribosomal protein genes will expand its application potential in a broader scenario. From this viewpoint, our research has provided valuable information for developing a convenient platform to improve the efficiency of heterologous protein expression in P. pastoris, which may also contribute to the application of synthetic biology in a special field, such as in the field of biofuel production.
The data presented in this study are deposited in the BioSample database repository of National Center for Biotechnology Information (NCBI), accession number PRJNA892887.
JH and JJ: conceived and designed the research. TN, JH, and YC: wrote the manuscript. SQ and XZ: performed the experiments. TN, YC, and NM: performed the data analyses. XS and RW: provided experiment assistance, data curation, and validation. JH and NM: funding acquisition. AC: writing—review and editing. All authors contributed to the article and approved the submitted version.
This work was supported by grants from National Natural Science Foundation of China (No. 31670103).
We thank Hailong Li and Defu Liu for their kind assistance in the transcriptomic analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aw, R., Barton, G. R., and Leak, D. J. (2017). Insights into the prevalence and underlying causes of clonal variation through transcriptomic analysis in Pichia pastoris. Appl. Microbiol. Biotechnol. 101, 5045–5058. doi: 10.1007/s00253-017-8317-2
Bjarnadottir, M., Nilsson, C., Lindstrom, V., Westman, A., Davidsson, P., Thormodsson, F., et al. (2001). The cerebral hemorrhage-producing cystatin C variant (L68Q) in extracellular fluids. Amyloid 8, 1–10. doi: 10.3109/13506120108993809
Bode, W., Engh, R., Musil, D., Thiele, U., Huber, R., Karshikov, A., et al. (1988). The 2.0 A X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J. 7, 2593–2599. doi: 10.1002/j.1460-2075.1988.tb03109.x
Bou Ali, M., Karray, A., Gargouri, Y., and Ben Ali, Y. (2013). N-terminal domain of turkey pancreatic lipase is active on long chain triacylglycerols and stabilized by colipase. PLoS One 8:e71605. doi: 10.1371/journal.pone.0071605
Delic, M., Gongrich, R., Mattanovich, D., and Gasser, B. (2014). Engineering of protein folding and secretion-strategies to overcome bottlenecks for efficient production of recombinant proteins. Antioxid Redox Signal. 21, 414–437. doi: 10.1089/ars.2014.5844
Dubin, G. (2005). Proteinaceous cysteine protease inhibitors. Cell. Mol. Life Sci. 62, 653–669. doi: 10.1007/s00018-004-4445-9
Grubb, A., Jensson, O., Gudmundsson, G., Arnason, A., Lofberg, H., and Malm, J. (1984). Abnormal metabolism of gamma-trace alkaline microprotein. The basic defect in hereditary cerebral hemorrhage with amyloidosis. N. Engl. J. Med. 311, 1547–1549. doi: 10.1056/NEJM198412133112406
Gu, L., Zhang, J., Du, G., and Chen, J. (2015). Multivariate modular engineering of the protein secretory pathway for production of heterologous glucose oxidase in Pichia pastoris. Enzyme Microb. Technol. 68, 33–42. doi: 10.1016/j.enzmictec.2014.10.006
He, J., Tang, F., Chen, D., Yu, B., Luo, Y., Zheng, P., et al. (2019). Design, expression and functional characterization of a thermostable xylanase from Trichoderma reesei. PLoS One 14:e0210548. doi: 10.1371/journal.pone.0210548
Inan, M., Aryasomayajula, D., Sinha, J., and Meagher, M. M. (2006). Enhancement of protein secretion in Pichia pastoris by overexpression of protein disulfide isomerase. Biotechnol. Bioeng. 93, 771–778. doi: 10.1002/bit.20762
Janowski, R., Kozak, M., Jankowska, E., Grzonka, Z., Grubb, A., Abrahamson, M., et al. (2001). Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat. Struct. Biol. 8, 316–320. doi: 10.1038/86188
Jolly, C., and Morimoto, R. I. (2000). Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 92, 1564–1572. doi: 10.1093/jnci/92.19.1564
Liang, S., Wang, B., Pan, L., Ye, Y., He, M., Han, S., et al. (2012). Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genom. 13:738. doi: 10.1186/1471-2164-13-738
Liao, X., Zhao, J., Liang, S., Jin, J., Li, C., Xiao, R., et al. (2019). Enhancing co-translational folding of heterologous protein by deleting non-essential ribosomal proteins in Pichia pastoris. Biotechnol. Biofuels 12:38. doi: 10.1186/s13068-019-1377-z
Liu, W., Jiang, Y., Peng, L., Sun, X., Gan, W., Zhao, Q., et al. (2022). Inferring gene regulatory networks using the improved markov blanket discovery algorithm. Interdiscip. Sci. 14, 168–181. doi: 10.1007/s12539-021-00478-9
Mathews, P. M., and Levy, E. (2016). Cystatin C in aging and in alzheimer’s disease. Ageing Res. Rev. 32, 38–50. doi: 10.1016/j.arr.2016.06.003
Merz, G. S., Benedikz, E., Schwenk, V., Johansen, T. E., Vogel, L. K., Rushbrook, J. I., et al. (1997). Human cystatin C forms an inactive dimer during intracellular trafficking in transfected CHO cells. J. Cell. Physiol. 173, 423–432. doi: 10.1002/(SICI)1097-4652(199712)173:3<423::AID-JCP15>3.0.CO;2-C
Navone, L., Vogl, T., Luangthongkam, P., Blinco, J. A., Luna-Flores, C. H., Chen, X., et al. (2021a). Disulfide bond engineering of AppA phytase for increased thermostability requires co-expression of protein disulfide isomerase in Pichia pastoris. Biotechnol. Biofuels 14:80. doi: 10.1186/s13068-021-01936-8
Navone, L., Vogl, T., Luangthongkam, P., Blinco, J. A., Luna-Flores, C., Chen, X., et al. (2021b). Synergistic optimisation of expression, folding, and secretion improves E. coli AppA phytase production in Pichia pastoris. Microb. Cell Fact. 20:8. doi: 10.1186/s12934-020-01499-7
Nusse, O., and Lindau, M. (1988). The dynamics of exocytosis in human neutrophils. J. Cell Biol. 107, 2117–2123. doi: 10.1083/jcb.107.6.2117
Oakes, S. A., and Papa, F. R. (2015). The role of endoplasmic reticulum stress in human pathology. Annu. Rev. Pathol. 10, 173–194. doi: 10.1146/annurev-pathol-012513-104649
Orman, M. A., Calik, P., and Ozdamar, T. H. (2009). The influence of carbon sources on recombinant-human- growth-hormone production by Pichia pastoris is dependent on phenotype: A comparison of muts and mut+ strains. Biotechnol. Appl. Biochem. 52, 245–255. doi: 10.1042/BA20080057
Palsdottir, A., Snorradottir, A. O., and Thorsteinsson, L. (2006). Hereditary cystatin C amyloid angiopathy: Genetic, clinical, and pathological aspects. Brain Pathol. 16, 55–59. doi: 10.1111/j.1750-3639.2006.tb00561.x
Renuse, S., Madugundu, A. K., Kumar, P., Nair, B. G., Gowda, H., Prasad, T. S., et al. (2014). Proteomic analysis and genome annotation of Pichia pastoris, a recombinant protein expression host. Proteomics 14, 2769–2779. doi: 10.1002/pmic.201400267
Robasky, K., Lewis, N. E., and Church, G. M. (2014). The role of replicates for error mitigation in next-generation sequencing. Nat. Rev. Genet. 15, 56–62. doi: 10.1038/nrg3655
Schulze, S. K., Kanwar, R., Golzenleuchter, M., Therneau, T. M., and Beutler, A. S. (2012). SERE: Single-parameter quality control and sample comparison for RNA-Seq. BMC Genom. 13:524. doi: 10.1186/1471-2164-13-524
Shu, J., Wei, W., and Zhang, L. (2022). Identification of molecular signatures and candidate drugs in vascular dementia by bioinformatics analyses. Front. Mol. Neurosci. 15:751044. doi: 10.3389/fnmol.2022.751044
Sola, A., Jouhten, P., Maaheimo, H., Sanchez-Ferrando, F., Szyperski, T., and Ferrer, P. (2007). Metabolic flux profiling of Pichia pastoris grown on glycerol/methanol mixtures in chemostat cultures at low and high dilution rates. Microbiology 153, 281–290. doi: 10.1099/mic.0.29263-0
Vici, A. C., da Cruz, A. F., Facchini, F. D., de Carvalho, C. C., Pereira, M. G., Fonseca-Maldonado, R., et al. (2015). Beauveria bassiana lipase A expressed in Komagataella (Pichia) pastoris with potential for biodiesel catalysis. Front. Microbiol. 6:1083. doi: 10.3389/fmicb.2015.01083
Vogl, T., and Glieder, A. (2013). Regulation of Pichia pastoris promoters and its consequences for protein production. N. Biotechnol. 30, 385–404. doi: 10.1016/j.nbt.2012.11.010
Vogl, T., Sturmberger, L., Fauland, P. C., Hyden, P., Fischer, J. E., Schmid, C., et al. (2018). Methanol independent induction in Pichia pastoris by simple derepressed overexpression of single transcription factors. Biotechnol. Bioeng. 115, 1037–1050. doi: 10.1002/bit.26529
Wang, J., Wang, X., Shi, L., Qi, F., Zhang, P., Zhang, Y., et al. (2017). Methanol-independent protein expression by AOX1 promoter with trans-acting elements engineering and glucose-glycerol-shift induction in Pichia pastoris. Sci. Rep. 7:41850. doi: 10.1038/srep41850
Whyteside, G., Alcocer, M. J., Kumita, J. R., Dobson, C. M., Lazarou, M., Pleass, R. J., et al. (2011). Native-state stability determines the extent of degradation relative to secretion of protein variants from Pichia pastoris. PLoS One 6:e22692. doi: 10.1371/journal.pone.0022692
Yan, P., Zou, Z., Zhang, S., Wang, R., Niu, T., Zhang, X., et al. (2021). Defining the mechanism of PDI interaction with disulfide-free amyloidogenic proteins: Implications for exogenous protein expression and neurodegenerative disease. Int. J. Biol. Macromol. 174, 175–184. doi: 10.1016/j.ijbiomac.2021.01.172
Yu, X. W., Sun, W. H., Wang, Y. Z., and Xu, Y. (2017). Identification of novel factors enhancing recombinant protein production in multi-copy Komagataella phaffii based on transcriptomic analysis of overexpression effects. Sci. Rep. 7:16249. doi: 10.1038/s41598-017-16577-x
Yu, Y., Wang, Y., He, J., Liu, Y., Li, H., Zhang, H., et al. (2010). Structural and dynamic properties of a new amyloidogenic chicken cystatin mutant I108T. J. Biomol. Struct. Dyn. 27, 641–649. doi: 10.1080/07391102.2010.10508578
Zhao, Y., Li, M. C., Konate, M. M., Chen, L., Das, B., Karlovich, C., et al. (2021). TPM, FPKM, or normalized counts? a comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J. Transl. Med. 19:269. doi: 10.1186/s12967-021-02936-w
Zhou, X., Lu, X., Qin, S., Xu, L., Chong, X., Liu, J., et al. (2019). Is the absence of alpha-helix 2 in the appendant structure region the major contributor to structural instability of human cystatin C? J. Biomol. Struct. Dyn. 37, 4522–4527. doi: 10.1080/07391102.2018.1552625
Keywords: cystatin, transcriptomic, amyloid disease, pichia pastoris, biofuel
Citation: Niu T, Cui Y, Shan X, Qin S, Zhou X, Wang R, Chang A, Ma N, Jing J and He J (2022) Comparative transcriptomic analysis-based identification of the regulation of foreign proteins with different stabilities expressed in Pichia pastoris. Front. Microbiol. 13:1074398. doi: 10.3389/fmicb.2022.1074398
Received: 19 October 2022; Accepted: 28 November 2022;
Published: 22 December 2022.
Edited by:
Qi Zhao, University of Science and Technology Liaoning, ChinaReviewed by:
Luís Fernando Parizi, Federal University of Rio Grande do Sul, BrazilCopyright © 2022 Niu, Cui, Shan, Qin, Zhou, Wang, Chang, Ma, Jing and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianwei He, andoZUBsbnUuZWR1LmNu; Nan Ma, bWFuYW5AbW90Y2F0cy5hYy5jbg==; Jingjing Jing, aGVsbG9qamppbmdAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.