
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 December 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1071530
This article is part of the Research TopicCommunity Series in Antimicrobial Peptides: Molecular Design, Structure Function Relationship and Biosynthesis OptimizationView all 22 articles
Burkholderia arboris, which belongs to the Burkholderia cepacia complex, has been shown to possess antifungal activity against several plant fungal pathogens; however, the antifungal compounds are yet to be identified. Here, we identified the antifungal compounds produced by B. arboris using genetic and metabolomic approaches. We generated a Tn5 transposon mutation library of 3,000 B. arboris mutants and isolated three mutants with reduced antifungal activity against the plant fungal pathogen Fusarium oxysporum. Among the mutants, the M464 mutant exhibited the weakest antifungal activity. In the M464 genome, the transposon was inserted into the cobA gene, encoding uroporphyrin-III methyltransferase. Deletion of the cobA gene also resulted in reduced antifungal activity, indicating that the cobA gene contributed to the antifungal activity of B. arboris. Furthermore, a comparison of the differential metabolites between wild type B. arboris and the ∆cobA mutant showed a significantly decreased level of tetrapeptide His-Ala-Phe-Lys (Hafk) in the ∆cobA mutant. Therefore, a Hafk peptide with D-amino acid residues was synthesized and its antifungal activity was evaluated. Notably, the Hafk peptide displayed significant antifungal activity against F. oxysporum and Botrytis cinerea, two plant pathogens that cause destructive fungal diseases. Overall, a novel antifungal compound (Hafk) that can be used for the biocontrol of fungal diseases in plants was identified in B. arboris.
Protection against crop losses caused by pathogens, especially fungi, is important to ensure sustainable crop production to meet the demand of the growing global population (Nazarov et al., 2020). Although chemical control using synthetic fungicides is effective in reducing crop loss caused by fungal diseases (Russell, 2005; Goldenhar and Hausbeck, 2019), there has been an increased concern on the potential environmental (Latin and Ou, 2018; Fraaije et al., 2020; Krichilsky et al., 2021) and health risks of indiscriminate fungicides use, as well as the rising threat of fungicide resistance in plant pathogens (Canal-Raffin et al., 2008; van den Bosch and Gilligan, 2008). Therefore, biological control using microbes or microbial compounds has been suggested as a sustainable alternative for plant protection against fungal diseases (Joo and Hussein, 2022). Since Bacillus was used as a biocontrol agent in 1940, large efforts have been made to identify effective biocontrol bacteria, most of which belong to the Bacillus and Pseudomonas genera (Poveda et al., 2021). Biocontrol bacteria antagonize pathogens mainly by producing bioactive compounds, including antibiotics, siderophores, peptides, lipopeptides, polyketides, bacteriocins, and enzymes (Mercado-Blanco and Bakker, 2007; Daungfu et al., 2019; Chen et al., 2020). Antagonistic compounds are primarily produced by non-ribosomal peptide synthase (NRPS) and polyketide synthases (PKS). NRPS are multi-domain and multi-enzymatic mega-synthases responsible for the synthesis of non-ribosomal peptides, sometimes with modifications, such as oxidation, acylation, methylation, or glycosylation (Marahiel, 2009; Thomson and Dennis, 2012). PKS is involved in the synthesis of polyketides, similar to fatty acid synthase (Staunton and Weissman, 2001; Hertweck, 2009). Among these antagonistic compounds, antimicrobe peptides, including lipopeptides, glycans, and cyclic peptides, have received substantial attention owing to their ecofriendly characteristics (Fira et al., 2018). The genome sequence of B. subtilis revealed that approximately 5% of its genome is dedicated to the production of antimicrobe compounds (Caulier et al., 2019; Tran et al., 2022). Among 2,700 antimicrobial peptides in Antimicrobial Peptide Database, 1,000 of them have antifungal activity, and most are produced by Bacillus, Paenibacillus, Pseudomonas, and Aspergillus (Li et al., 2021). Antifungal peptides are usually referred to as peptides containing 10–100 amino acid residues (Zhang et al., 2020); however, more and more short chain polypeptides have been found to process antifungal ability. His (2-aryl)-Trp-Arg has been reported to antagonize Cryptococcus neoformans (Sharma et al., 2021). In B. subtilis, 57 peptides with less than five amino acids were found to have antifungal activity, among which two tetrapeptides AWYW and HWWY strongly suppressed the growth of Phytophthora capsici and Penicillin chrysogenum (Qiao et al., 2020).
Burkholderia is a genus comprising over 60 species of gram-negative bacteria, and are widely present in soil around the rhizosphere zone, water, plants, and animals (Mahenthiralingam et al., 2008). Burkholderia was firstly defined in 1942 by Burkholder and initially named Pseudomonas caryophyll until 1992 when seven species with similar genetic characters were transferred from the Pseudomonas genus to establish the genus Burkholderia (Burkholder, 1942; Yabuuchi et al., 1992). Burkholderia species have been utilized as natural sources of biotechnological agents in agricultural production owing to their ability to produce several biomolecules, including antibiotics; extracellular hydrolytic enzymes, such as chitinase, glucanase, and protease (Elshafie and Camele, 2021); and bioactive secondary metabolites, such as alkaloids, lipopeptides, and polypeptides (El-Banna and Winkelmann, 1998; Sultan et al., 2008). Burkholderia cepacia complex (BCC) is a group of antibiotic resistant bacteria, including Burkholderia arboris, capable of producing bioactive compounds via NRPS and PKS (Thomson and Dennis, 2012). Burkholderia arboris has been widely isolated from the rhizosphere zone of the soil, and has been shown to possess antifungal activity (Barrera-Galicia et al., 2021); however, the antifungal compound is yet to be identified.
Here, we identified the antifungal compound in B. arboris using molecular and metabolomic techniques and evaluated the antifungal activity against selected plant fungi. It is anticipated that the antifungal agent identified in B. arboris would be effective as a biocontrol agent against phytopathogenic fungi causing Fusarium wilt and gray mold disease.
The bacterial and fungal strains used in this study are listed in Supplementary Table 1. Burkholderia arboris 1 (CGMCC No.16905), a strain isolated from Vernicia fordii (tung oil tree) in China, Botrytis cinerea B05.10, and Fusarium oxysporum f. sp. lycopersici were used in this study. Burkholderia arboris was cultured in nutrient agar (NA) medium at 28°C (Edwards et al., 1965), and F. oxysporum and B. cinerea were cultured in potato dextrose agar (PDA) medium at 28°C (Robbins, 1924). Escherichia coli were cultured in Luria-Bertani (LB) medium at 37°C (Elder, 1983). Kanamycin (Kan) was added to the medium at a final concentration of 50 μg/ml when screening for clones positive for E. coli, and at 500 μg/ml when screening for clones positive for B. arboris.
The Tn5 transposon mutant library was constructed as previously described (He et al., 2021). Briefly, a fragment of the kanamycin resistance gene (Kan) was amplified from plasmid pBBR1MCS-2 by PCR using primers MEKANA-F/R containing Tn5 mosaic end (ME) sequences. The PCR product was ligated into the pCE2 vector using a TA/Blunt-Zero Cloning Kit (Vazyme Biotech, Nanjing, China). After confirmation by sequencing, the Kan-ME fragment was transformed into B. arboris using an EZ-Tn5™ Transposase Kit (Lucigen, Middleton, WI, United States). Putative mutants were selected using 500 μg/ml kanamycin.
The plasmids used in this study are listed in Supplementary Table 1. The primer sequences used for plasmid construction are listed in Supplementary Table 2. Genomic DNA of B. arboris was extracted using a TIANamp Bacteria DNA Kit (Tiangen Biotech, Beijing, China). To knockout cobA, a fragment of the cobA gene in B. arboris genome was amplified by PCR. The PCR product was digested with BamHI and EcoRI restriction enzymes and then ligated into the suicide plasmid pJP5603 (Riedel et al., 2013), which was digested with the same enzymes. The ligation product was transferred into E. coli S17-1 λpir cells by heat shock. Positive clones were selected with 50 mg/ml kanamycin resistance and verified by sequencing the cobA insertion in the pJP5603-cobA plasmid. The resulting pJP5603-cobA plasmid was transferred into B. arboris competent cells with electroporation. Putative mutants were selected with 500 mg/ml kanamycin and verified by PCR with primers from 16S rDNA of B. arboris and the conserved R6K origin of replication (Riedel et al., 2013). The transcript levels of cobA in mutants were determined by qRT-PCR.
Tn5 insertion sites were identified using thermal asymmetric interlaced PCR (TAIL-PCR; Liu and Whittier, 1995). Arbitrary degenerated primers served as forward primers for all three rounds of PCR, and specific primers, including SP1, SP2, and SP3, were used as the reverse primers for primary, secondary, and tertiary rounds of PCR, respectively. The primer sequences are listed in Supplementary Table 2. The primary-round PCR product was 100-fold diluted and used as a DNA template for the secondary PCR reaction, which was 100-fold diluted and used as the template for the tertiary PCR reaction. Cycling conditions were as previously described.
Growth curves were generated as previously described (Xian et al., 2020). Bacterial suspension was centrifuged at 5,000 rpm for 5 min after overnight culture at 28°C. The pellet was resuspended in ddH2O and adjusted to an OD600 of 0.005 using fresh medium. Absorbance at OD600 was measured with a microplate spectrophotometer (Multiskan™ FC, Thermo Fisher Scientific, Waltham, MA, United States) every 30 min for 36 h to obtain the growth curves. The growth rate, the doubling time and the lagging time were calculated.
Swimming ability was determined as previously described (Bahar et al., 2009). Bacterial strains were cultured overnight at 28°C until OD600 reached 0.6. Thereafter, the bacteria suspension (2 μl) was point-inoculated on the center of a plate containing semi-solid LB broth with 0.3% agar, and the diameter of the swimming zone was measured 2 days after inoculation.
Biofilm formation assay was conducted using the crystal violet staining method (Holmberg et al., 2009). Bacterial strains were cultured overnight until the OD600 reached 0.5, and then transferred to 96-well cell plate at 100 μl per well and cultured at 28°C for 72 h without shaking. Thereafter, the wells were washed with sterile water twice and stained with 125 μl 0.1% crystal violet (w/v) for 30 min to quantitatively measure the attached biofilms. After removing the dye, wells were thoroughly dried at 37°C, and 1% sodium dodecyl sulfate (SDS) was added to dissolve the biofilm at 200 μl per well. The quantity of biofilm formed was determined at an absorbance of 570 nm using a Multiskan™ FC microplate spectrophotometer (Thermo Fisher Scientific).
Bacterial culture (300 ml) was centrifuged at 4,000 rpm for 20 min after culturing overnight, and the supernatant was mixed with an equal volume of ethyl acetate. The solution from the organic phase was transferred to a conical flask containing anhydrous Na2SO4 to remove residual moisture, and was finally filtered through a triangular funnel with cotton. Thereafter, the organic phase filtrate was transferred to a pre-weighed collecting bottle, and the ethyl acetate was evaporated with an IKA® RV 10 digital rotary evaporator at 42°C, 80 rpm under control of IKA® MVP 10 basic compact vacuum pump (IKA, Staufen, Germany). The crude extract was dissolved in 1 ml ethyl acetate.
For determination of antifungal activity of B. arboris bacteria, fungal mycelial disks and fungal spore suspension were used as described previously with slight modifications (Kumar et al., 2018). Fungal disks and B. arboris were co-cultivated on PDA medium with a distance of 30 mm from each other. The inhibitory zone was measured 6 days after co-cultivation. To prepare the fungal spore suspension, the fungal mycelia and spores on the agar plate were washed off with sterile ddH2O and then filtered through two layers of sterile gauzes. The concentration of spore suspension was determined with a hemocytometer. For determination of antifungal activity of B. arboris extracts, fungal spore suspension (1.0 × 105 CFU/ml, 500 μl) was evenly spread on PDA medium, and then 8 mm round holes were removed from the solid medium using a sterile punch, followed by the addition of 20 μl of melting PDA medium to each hole to seal the bottom. After cooling, 150 μl of crude B. arboris extract was added to the holes, and the diameters of the inhibition zones were measured 6 days later.
Hafk peptides with D-amino acid residues were synthesized by ChenPeptide Biotech (Nanjing, China). LPE (CAS # 53862–35-4) was purchased from Aladdin Ltd. (Shanghai, China). The antifungal activity of Hafk peptides and LPE was determined as described previously with slight modifications (Shi et al., 2010). Briefly, a stock solution of the Hafk peptides (100 mM) and LPE (100 mM) was prepared using ddH2O and ethyl acetate, respectively. PDA medium was added to a 12-well cell plate at 2 ml per well, and then 50 μl of different concentrations of the peptide or LPE solution were evenly spread on the surface of the medium. Thereafter, fungal mycelial disks were placed at the center of the dried medium, and the diameters of fungal mycelial growth were measured 3 days later.
Untargeted metabolomic analysis of the bacterial sample was performed at Metware Biotechnology Co., Ltd.1 as previously described (Oh et al., 2021). Bacterial samples were collected by centrifugation, and then a 500 μl solution (methanol: water = 4: 1, v/v) containing an internal standard was added to each bacterial sample and vortexed for 3 min. Samples were placed in liquid nitrogen for 5 min, dried on ice for 5 min, thawed on ice, and vortexed for 2 min. This freeze–thaw cycle was repeated thrice. Then they were centrifuged at 12,000 rpm for 10 min at 4°C, and 300 μl of supernatant was collected and kept at −20°C for 30 min, followed by further centrifugation at 12,000 rpm for 3 min at 4°C. Thereafter, 200 μl aliquot of the supernatant was collected for UPLC-QTOF/MS analysis. The analytical conditions were as follows: column, Waters ACQUITY UPLC HSS T3 C18 (1.8 μm, 2.1 mm × 100 mm); column temperature, 4°C; flow rate, 0.4 ml/min; injection volume, 2 μl; solvent system, water (0.1% formic acid), and acetonitrile (0.1% formic acid); gradient program, 95: 5 V/V at 0 min, 10: 90 V/V at 11.0 min, 10: 90 V/V at 12.0 min, 95: 5 V/V at 12.1 min, and 95:5 V/V at 14.0 min. Mass spectrometric analysis was performed using a quadrupole time-of-flight mass spectrometer in the positive and negative ion modes. The gas flow rate was set at 8 L/min; gas and sheath temperatures were 325°C; the fragmetor was 135 V; and the nebulizer was 40 V. The ESI voltage was 2,500 V in the positive-ion mode and 1,500 V in the negative-ion mode.
The original data file acquired by UPLC-QTOF/MS was converted into the mzML format using ProteoWizard software. Peak extraction, alignment, and retention time corrections were performed using the XCMS program. The “SVR” method was used to correct the peak area. Peaks with detection rate lower than 50% in each group samples were discarded. Subsequently, metabolic identification information was obtained by searching the laboratory’s self-built database, integrated public database, AI database, and metDNA. Differential metabolites between wild type and mutant B. arboris were determined at variable importance in projection (VIP) ≥ 1, p < 0.05 (Student’s t-test), and absolute log2 fold change (FC) ≥ 1.0. VIP values extracted from the OPLS-DA results, which also contained score plots and permutation plots, were generated using MetaboAnalystR package in R software. The data were log transformed (log2) and mean centered before OPLS-DA. A permutation test (200 permutations) was performed to avoid overfitting.
Hafk peptide in the bacterial supernatant was identified by LC–MS analysis. The bacterial supernatant (100 ml) was freeze-dried, dissolved in 4 ml 40% acetonitrile, and then subjected to LC–MS (6400 Series Triple Quadrupole LC–MS by Agilent). The standard sample is the synthesized Hafk peptide (Purity: 81.98%).
Burkholderia spp. are highly resistant to several antibiotics. Therefore, the sensitivity of B. arboris to common antibiotics was examined to facilitate genetic analysis of the species. The growth of B. arboris was suppressed by 500 μg/ml of Kan, 10-fold higher than the requirement for most bacteria; in contrast, streptomycin (Strep), tetracyclines (Tet), spectinomycin (Spe), and chloroamphenicol (Chl) did not inhibit the growth of B. arboris (Supplementary Figure 1). Subsequently, plasmids carrying Kan-resistant and mCherry fluorescent genes were transferred into competent B. arboris cells using electroporation (Supplementary Figure 2). Positive clones were selected using NA media containing 500 μg/ml of Kan, and all selected clones displayed pink color under bright field and fluorescent conditions (Figure 1). The pink color did not decrease after 20 generations of subculture. Overall, these results indicated that 500 μg/ml Kan can be used as a selective antibiotic for B. arboris. Transposon random mutagenesis technology was used to generate B. arboris mutant library. The Kan gene fragment (795 bp) was amplified from a plasmid (Supplementary Figure 3) by PCR, ligated to adapters containing a Tn5 mosaic end (ME, 19 bp), and the resulting Kan-ME fragments (833 bp) were introduced into B. arboris along with transposases (Figure 2A). Overall, 3,000 putative transposon insertion mutants were isolated from Kan-resistant plates.
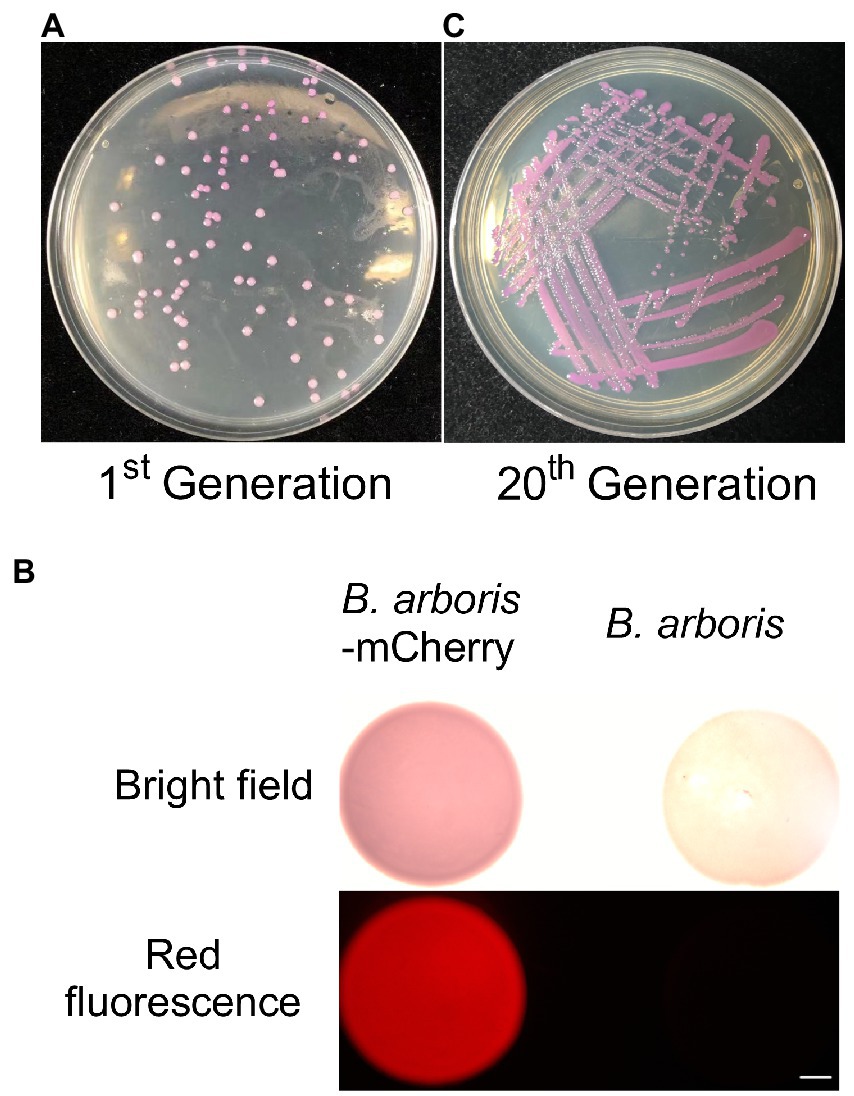
Figure 1. Kanamycin can be used as selection antibiotic for Burkholderia arboris. (A) Generation of B. arboris-mCherry using kanamycin resistance. Plasmids containing mCherry and kanamycin resistance genes were transferred to B. arboris, and positive clones were selected on the NA agar plate (90 mm) with 500 μg/ml of kanamycin 48 h after cultivation at 28°C. (B) B. arboris-mCherry colonies under fluorescent microscopy. Bars, 1,000 mm. (C) The morphology of B. arboris-mCherry colonies after 20 generations subculturing.
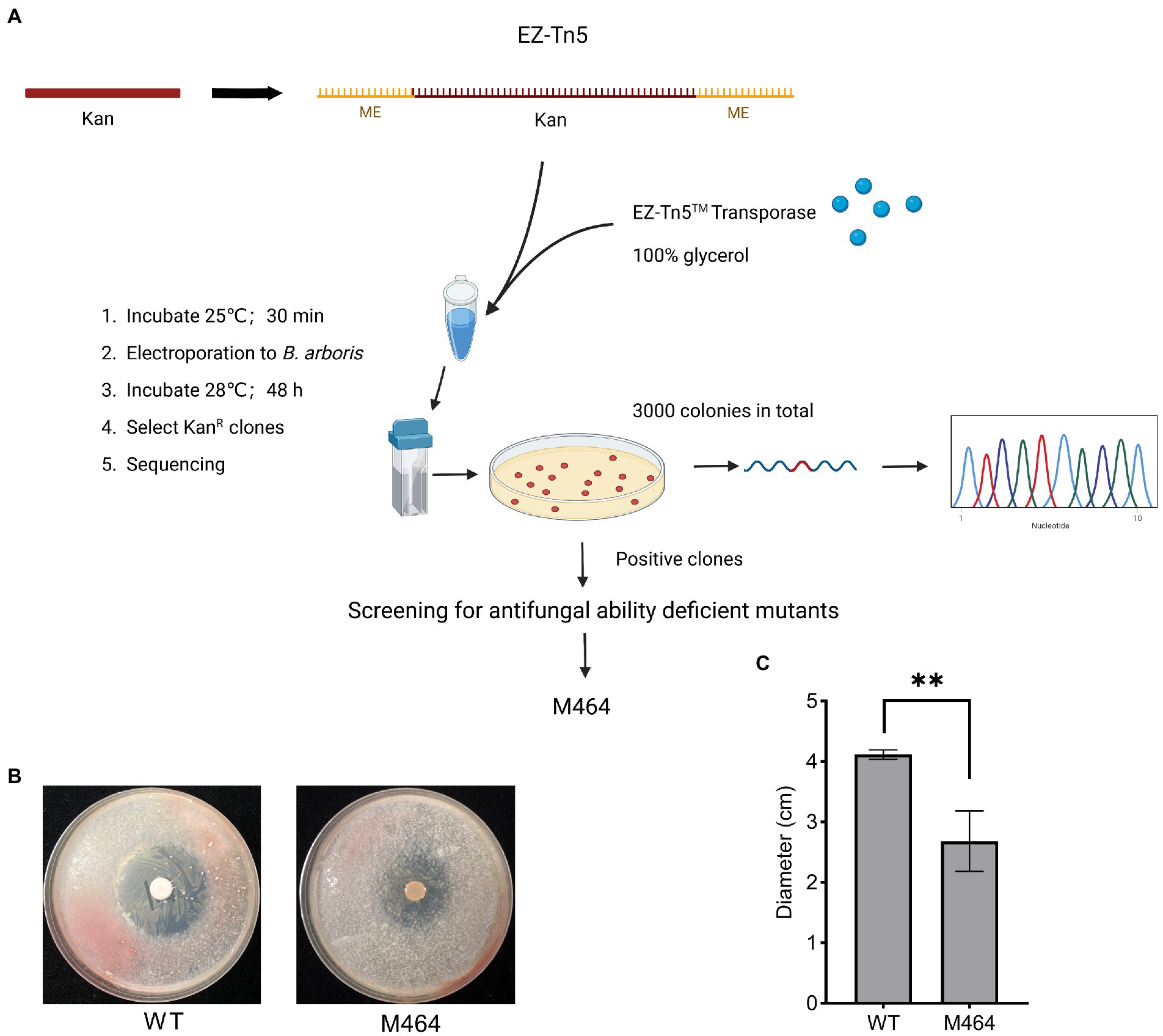
Figure 2. Isolation of Burkholderia arboris M464 mutant with reduced antifungal activity. (A) Diagram of construction of B. arboris Tn5 insertion mutants. (B) Confrontation plate experiment of the antifungal activity of B. arboris against Fusarium oxysporum. Bacterial solution (10 μl at OD600 of 1.0) from wild type (WT) and M464 mutant were dropped in the center of 90 mm PDA plates, and fungi (1.0 × 105 spores/ml, 500 μl) were evenly sprayed on each plate 48 h after the cultivation of B. arboris. Plates were incubated at 28°C and images were taken 6 days after the fungal spray. (C) The M464 mutant showed reduced antifungal activity, as evidenced by the smaller diameter of the inhibition zone against F. oxysporum. Data are presented as mean ± SD (n = 3). Asterisks indicate significant differences between the wild type and M464 (**p < 0.01, Student’s t-test). The experiments were repeated twice, and similar results were obtained.
A plate confrontation experiment was performed to identify B. arboris mutants with weak antifungal activity against Fusarium oxysporum. Wild type B. arboris exhibited high antagonistic activity against F. oxysporum (Supplementary Figure 4), whereas three out of 3,000 putative transposon insertion mutants showed reduced antagonistic activity (Supplementary Figure 5). Among the three, M464, with a little yellow color, displayed the weakest inhibitory effect against F. oxysporum, as evidenced by the small diameters of the inhibition zones (Figures 2B,C); therefore, it was selected for further analysis.
The Kan fragment from the M464 mutant was amplified by PCR, and the resulting PCR product was electrophoresed on an agarose gel. As expected, 833 bp of PCR fragments were observed in the gel (Supplementary Figure 6), confirming that the Kan sequence were inserted into the M464 genome. Thermal asymmetrical interlaced PCR (TAIL-PCR) was performed to amplify the flanking sequence adjacent to the border of Kan gene to identify the insertion site of Kan sequence in the M464 genome. Three specific primers with high annealing temperatures were designed from the Kan gene: SP1, SP2, and SP3. TAIL-PCR was performed using nested specific primers in three subsequent rounds of reactions, together with six arbitrary degenerate (AD) primers (Figure 3A). A specific band of approximately 1,000 bp was obtained in the secondary rounds of PCR with the SP2 primer when the M464 template was amplified, but not the wild type B. arboris genomic DNA (Figure 3B). Two bands were obtained during the tertiary rounds of PCR using the SP3 primer (Figure 3B). Since the PCR products from the tertiary rounds of PCR are supposed to be smaller than those from the secondary round, we purified the small band (around 800 bp) and sequenced it using the primer SP3. After alignment with the B. arboris genome sequence using the nucleotide Basic Local Alignment Search Tool (BLAST), we found that Kan sequence was inserted in the cobA gene in B. arboris chromosome (Figure 3C). Subsequently, the insertion site of Kan was confirmed by PCR using specific primers (Figure 3D). Overall, these results suggest that the cobA gene is mutated in M464 mutants.
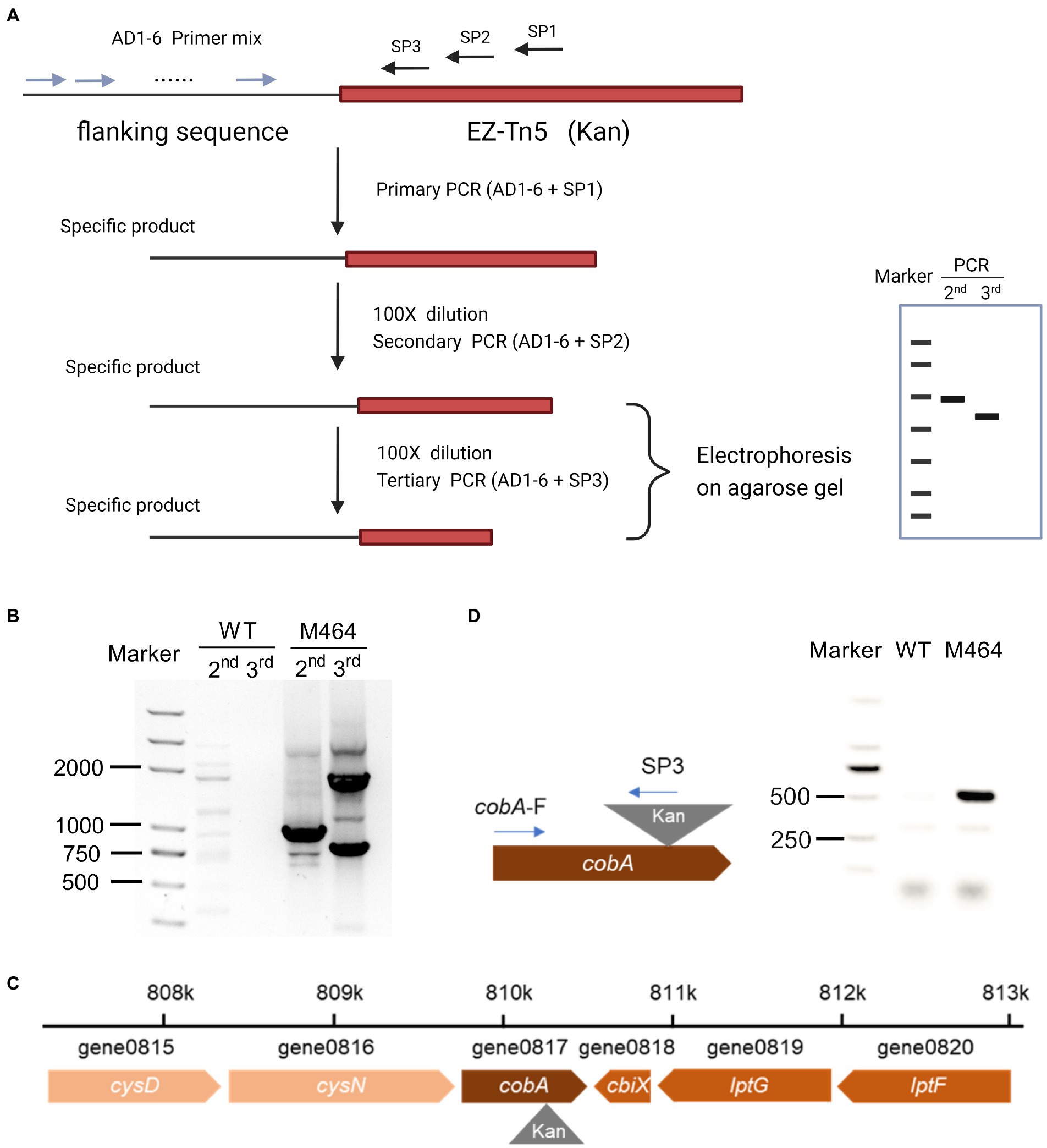
Figure 3. The cobA gene was mutated in M464 mutant of Burkholderia arboris. (A) Diagram of thermal asymmetric interlaced PCR (TAIL-PCR). (B) Gel image of TAIL-PCR products. The second and third round of PCR products from wild type (WT) and M464 mutants were run on agarose gel electrophoresis. (C) Diagram of the insertion site of the kanamycin resistance gene (Kan) in the M464 mutant. The cobA gene was mutated in the M464 B. arboris mutant. (D) Fragments of PCR products were detected on agarose gel with specific primers from cobA and Kan sequences.
A cobA knockdown mutant was generated by site-specific mutagenesis to verify that cobA is important for the antifungal activity of B. arboris against F. oxysporum. To achieve this, cobA sequence was amplified from the B. arboris genome and ligated to the enzyme-digested mobilizable suicide plasmid pJP5603. Knockdown mutants were generated by homologous recombination and screened for resistance to high concentrations of Kan. Once the ∆cobA mutant had been verified, a confrontation plate test was performed to evaluate the antifungal activity of ∆cobA against F. oxysporum. The ∆cobA mutants showed significantly lower antifungal activity against F. oxysporum compared to the wild type, but higher than that of the M464 mutant (Figures 4A,B). Additionally, unlike M464, ∆cobA mutants did not show a yellow color, probably because of other mutations in M464. Nevertheless, deletion of cobA impaired the antifungal activity of B. arboris.
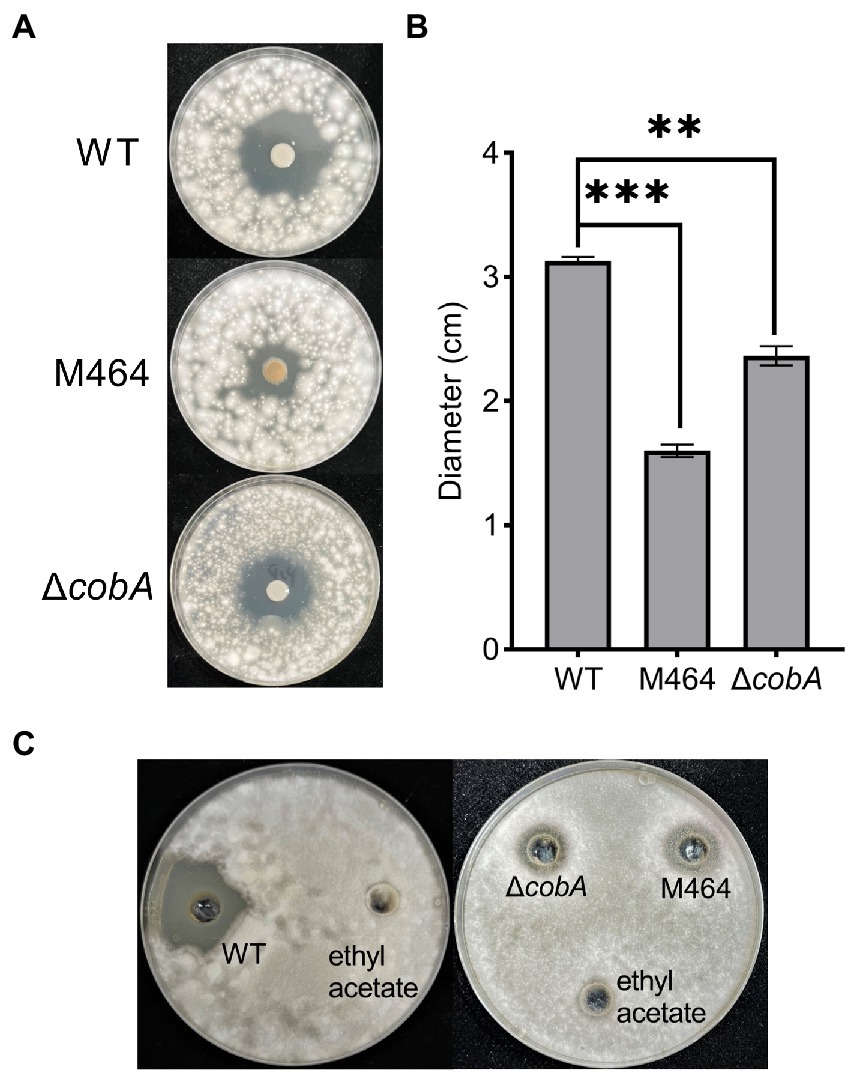
Figure 4. The ΔcobA mutant exhibited reduced antifungal activity. (A) Confrontation plate experiment of the antifungal activity of Burkholderia arboris against Fusarium oxysporum. Bacterial solutions (10 μl at OD600 of 1.0) from wild type (WT), ΔcobA, and M464 mutant were dropped in the center of the 90 mm PDA plate, and fungi (1.0 × 105 spores/ml, 500 μl) were evenly sprayed on each plate 48 h after the cultivation of B. arboris. Plates were incubated at 28°C and images were taken 6 days after the fungal spray. (B) The ΔcobA mutant showed reduced antifungal activity, as evidenced by the small diameter of the inhibition zone. Data are presented mean ± SD (n = 3). Asterisks indicate significant differences between the wild type and mutants (**p < 0.01, ***p < 0.001, Student’s t-test). The experiments were repeated twice, and similar results were obtained. (C) Crude extract of B.arboris inhibits the growth of F. oxysporum. Crude metabolites were extracted from the wild-type (WT), ΔcobA, and M464 mutants and dissolved in ethyl acetate. The antifungal ability against F. oxysporum was determined on the PDA plates (90 mm).
Furthermore, the potential mechanisms by which cobA mutation suppressed the antifungal activity of B. arboris were examined. First, we determined whether the cobA mutation affected the growth of B. arboris by comparing the growth curves of the wild type and ΔcobA mutant for 36 h, and found that the growth of ΔcobA mutant, including the doubling time and lagging time, had no substantial differences from that of the wild type (Supplementary Figure 7 A; Supplementary Table 3). Additionally, biofilm formation assay was performed, and it was observed that cobA mutation did not alter biofilm formation by B. arboris (Supplementary Figure 7 B). Moreover, swimming ability assay showed that the ΔcobA mutant had significantly higher swimming motility than wild type B. arboris (Supplementary Figure 7 C). Since ΔcobA mutant displayed reduced antifungal activity, it is unlikely that the increased swimming motility of the mutant was the major cause of the impaired antifungal activity. Overall, these results suggest that cobA mutation did not affect bacterial growth and physiological characteristics. Therefore, we hypothesized that the cobA may be involved in the production of antifungal compounds by B. arboris. To verify this hypothesis, the antifungal activities of metabolic crude extracts of the wild type and mutant B. arboris were evaluated. The crude extract from wild-type B. arboris significantly inhibited the growth of F. oxysporum, whereas that of ΔcobA mutant did not (Figure 4C), confirming that cobA is important for the production of antifungal compounds.
Untargeted metabolomics was performed to identify the antifungal compounds in B. arboris, using UPLC-QTOF/MS, and differential metabolites between B. arboris and the ΔcobA mutant were identified (Supplementary Table 4). Compared with wild type B. arboris, 17 compounds were identified in the ΔcobA mutant, among which three were downregulated (Table 1). The cobA gene, also known as cysG (Spencer et al., 1993; Woodcock et al., 1998), has been shown to be involved in sulfur and selenium metabolic pathways (Storbeck et al., 2009; Park et al., 2011; Moore et al., 2012). Accordingly, it was observed that several of the differentially expressed metabolites between B. arboris and the ΔcobA mutant contained these elements, indicating that cobA mutation affected the synthesis of the metabolites. The cobA gene also contributes to the biosynthesis of siroheme and vitamin B12 (Woodcock et al., 1998; Rodionov et al., 2003); however, the ΔcobA mutant did not show significantly reduced levels of heme and vitamin B12, which may be due to the detection sensitivity of the equipment or redundancy of cobA gene in B. arboris. Therefore, the antifungal activities of the three downregulated compounds in the ΔcobA mutant were evaluated.
The three downregulated compounds were dimethyl diselenide, lysophosphatidylethanolamine (LPE or lysoPE), and His-Ala-Phe-Lys (Hafk peptide). Dimethyl diselenide is volatile with a low boiling point of 57°C at 760 mmHg (Lide, 2007). However, we found that B. arboris did not inhibit the growth of F. oxysporum in a petri dish with separate compartments, suggesting that the antifungal compounds are not volatile (Supplementary Figure 8). Similarly, 10 and 100 mM of LPE did not inhibit the growth of F. oxysporum on PDA media (Supplementary Figure 9), indicating that both dimethyl diselenide and LPE were not the antifungal compounds produced by B. arboris. Overall, these results indicated that Hafk peptide may be the compound with antifungal activity.
To investigate whether the short peptide Hafk possesses antifungal activity, we first determined the stability of crude extracts from B. arboris suspensions. The crude extracts were heated for 30 min at 40, 50, and 60°C respectively, followed by a plate confrontation experiment to verify the antifungal activity against F. oxysporum (Supplementary Figure 10 A). Preheating at high temperatures did not reduce the antifungal activity of B. arboris. Additionally, the crude extract exhibited antifungal activity after treatment with proteinase K, which catalyzes the degradation of L-isoforms of amino acids (Supplementary Figure 10 B), indicating that the antifungal compounds produced by B. arboris are not peptides or amino acids with L-isoforms (Kannan et al., 2020). Therefore, we synthesized a Hafk peptide containing all the D-amino acids, and its antifungal ability against F. oxysporum was assessed in a 12-well cell plate. PDA media (2 ml) was added at the bottom of each well, and then 50 μl of different concentrations of Hafk peptide solution was spread evenly on the surface. A F. oxysporum mycelium disk was placed at the center of each well and the mycelial growth zone was measured 72 h after placement. Fusarium oxysporum growth was significantly inhibited by the addition of 10 and 100 mM Hafk peptide compared with the control group; however, concentrations lower than 10 mM did not significantly inhibit fungal growth (Figures 5A,B). To detect whether Hafk peptide was secreted by B. arboris, the crude extracts from bacterial supernatant were subjected to LC–MS analysis. Synthesized Hafk peptide was used as a standard. The chromatogram in standard solution was overlapped with that in the supernatant of B. arboris, suggesting that Hafk peptide is present in the supernatant (Supplementary Figure 11). To test whether the Hafk peptide antagonizes other phytopathogenic fungi, its antifungal activity against B. cinerea was evaluated. Consistent with the findings in F. oxysporum, 10 mM of Hafk peptide significantly inhibited the growth of B. cinerea (Figures 5C,D). Similarly, B. arboris inhibited the growth of B. cinerea in the confrontation assay (Supplementary Figure 12). Overall, these results indicated that Hafk peptide possesses antifungal activity.
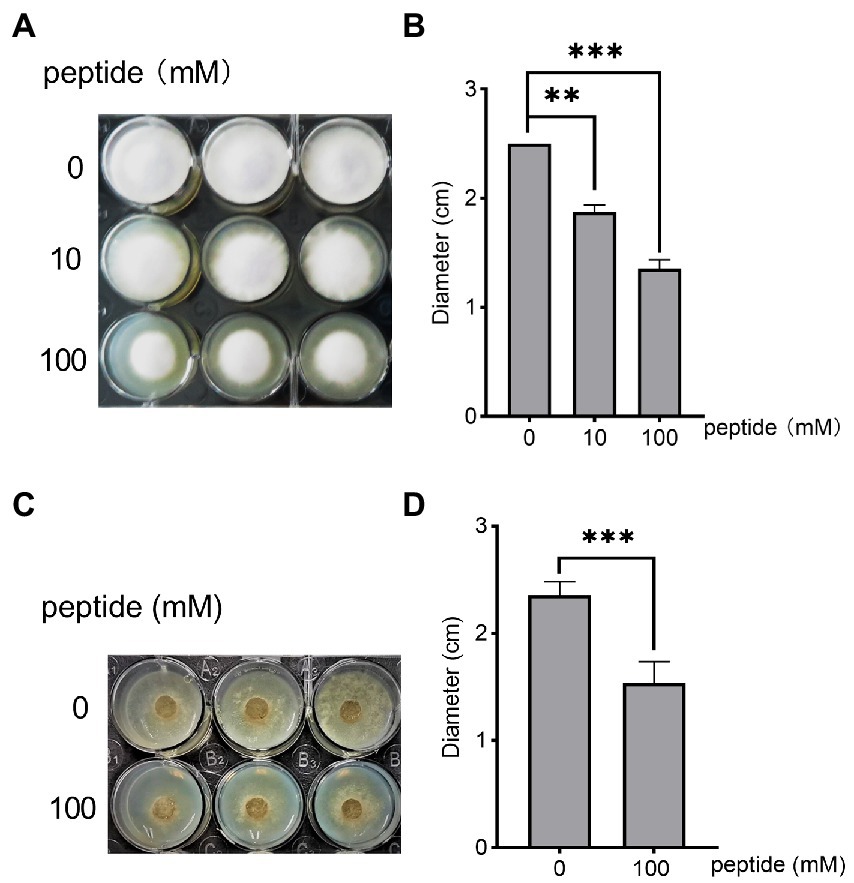
Figure 5. The peptide His-Ala-Phe-Lys (Hafk) antagonizes Fusarium oxysporum and Botrytis cinerea. (A) Growth inhibition assay of Hafk peptide against F. oxysporum. Different concentrations of Hafk peptide (50 μl) were evenly spread on the surface of the agar media in a 12-well-plate (diameter of each well, 25 mm), and then a fungal disk (diameter, 5 mm) was placed in the center of each well and cultured at 28°C. Images were taken 3 days after placement. A representative image is presented. (B) Hafk peptide inhibited the growth of F. oxysporum, as evidenced by the diameter of fungal mycelial growth. Data are presented as mean ± SD (n = 3). Asterisks indicate significant differences with or without the addition of the peptide (**p < 0.01, ***p < 0.001, Student’s t-test). The experiments were repeated with similar results. (C) Growth inhibition assay of Hafk peptide against Botrytis cinerea. The experimental conditions were the same as those in (A), except that B. cinerea was used. (D) Hafk peptide inhibited the growth of B. cinerea, as evidenced by the diameter of fungal mycelial growth. Data are presented as mean ± SD (n = 3). Asterisks indicate significant differences with or without the addition of peptides (***p < 0.001, Student’s t-test). The experiments were repeated with similar results.
The identification of novel antifungal peptides has attracted much attention because it possesses considerable potential as alternatives to chemical pesticides or antibiotics in the control of fungal diseases in agriculture (Whetstone and Hammock, 2007; Fira et al., 2018; Zhao et al., 2019; Niu et al., 2020). In this study, a new short peptide (Hafk) consisting of four amino acids residues (His-Ala-Phe-Lys) was identified in B. arboris. The Hafk peptide with D-amino acids significantly inhibited the growth of F. oxysporum and B. cinerea, two phytopathogens that cause wilt and gray mold diseases. Overall, Hafk peptides could serve as a reference for the development of antifungal agents with broad-spectrum applications.
Antifungal peptides are produced by several organisms, including insects, plants, fungi, and bacteria (Maskey et al., 2003; Wen et al., 2014; Al Souhail et al., 2016; Khani et al., 2019). Most antifungal peptides are composed of 10–100 amino acid residues, and are classified as lipopeptides and linear, cyclic, and glucan peptides (Donzelli et al., 2001; Ortiz-Lopez et al., 2015; Park et al., 2018). Several mechanisms through which antifungal peptides inhibit fungal growth have been elucidated, which include the blocking of the biosynthesis of chitin and b-glucan, two major fungal cell wall components (Donzelli et al., 2001; Pushpanathan et al., 2012; Moghaddam et al., 2017; Tong et al., 2020). Additionally, some antifungal peptides can disrupt the fungal membrane, while others can pass through cell membranes to target intracellular DNA and RNA (Han et al., 2019; Khani et al., 2020; Bansal et al., 2022; Ding et al., 2022). Moreover, accumulating evidence indicates that short peptides containing 2–10 amino acids also exhibit antifungal activity (Velivelli et al., 2018; Gong et al., 2022). Compared to longer peptides, short antifungal peptides possess several advantages, including higher stability, lower toxicity to the host, and lower cost of synthesis (Apostolopoulos et al., 2021). However, the antifungal mechanisms underlying short antifungal peptides require further study. The Hafk peptide represents a new class of antifungal compounds; thus, it could serve as a model short peptide to elucidate the mechanism of antifungal activity.
Amino acids exist in two isomeric forms, D- and L-amino acids, with the exception of glycine (Du et al., 2019). Although L-amino acids are the most common form produced through chemical and enzymatic syntheses, D-amino acids are also involved in several biological processes. D-Ala and D-Glu were found to be the most common D-amino acids in bacterial extracellular or periplasmic polymers (Radkov and Moe, 2014). Several D-amino acids can synthesize biological peptides through the NRPS pathway, including antibiotics and toxins, such as daptomycin containing D-Ala (Baltz, 2009), which can inhibit spore growth (Shrestha et al., 2017). D-amino acids have also been reported to antagonize pathogens, such as Agrobacterium tumefaciens, Mycobacterium smegmatis, and Saccharomyces cerevisiae (Fox et al., 1944; Bopp, 1965; Yabu and Huempfner, 1974; Ruiz et al., 2021). In addition to their potential antimicrobial ability, D-amino acids are more stable than L- amino acids (Zhao et al., 2016). Therefore, D-amino acids could serve as experimental materials to elucidate the antimicrobial ability of peptides (Kapil and Sharma, 2021). In the present study, Hafk (His-Ala-Phe-Lys) peptide was identified in the wild-type using UPLC-QTOF/MS, while ∆cobA mutant showed reduced levels. Although ribosome cobA might not directly regulate the synthesis of D-amino acids, it is possible that ribosome cobA might control the production of a precursor substance which is required for the formation of mature Hafk peptide. Considering the cost, peptides with all 12 possible combinations of D and L isoforms were not synthesized, but instead assessed the antifungal activity of peptides with D-isoforms. In the present study, the Hafk peptide (D-isoform) significantly inhibited fungal growth; however, the concentration required is still high. Therefore, the antifungal activity of all combinations needs to be examined in future studies to improve the efficiency and reduce the cost.
CobA is homologous to the C-terminal module of CysG, which is a multiple functional enzyme involved in the metabolism of sulfur and selenium (Warren et al., 1994; Stroupe et al., 2003; Dereven'kov et al., 2017). In the present study, untargeted metabolomics between wild type and ∆cobA mutant identified compounds containing sulfur, selenium, and aromatic compounds. Since these elements are important for amino acid metabolism, it is possible that the cobA gene contributes to the biosynthesis of Hafk peptides. Moreover, cobA is important for the synthesis of cobalamin (vitamin B12) and siro heme (Storbeck et al., 2009; Park et al., 2011; Moore et al., 2012); however, both cobalamin and siroheme were not detected in the wild type and ∆cobA mutant, which could be attributed to low detection efficiency of the equipment or redundancy of cobA in B. arboris. Moreover, cobalamin and siroheme have not been reported to exhibit antifungal activity, indicating that both cobalamin and siro heme are not involved in antifungal activity in B. arboris. However, further studies are necessary to elucidate how cobA regulates the biosynthesis of Hafk peptide.
In summary, B. arboris exerts antifungal activity by producing Hafk peptide under the regulation of cobA gene. The Hafk peptide possesses considerable potential as a biocontrol agent for crop fungal diseases.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
HZ and CX conducted most of the experiments and analyzed the data. YC isolated Burkholderia arboris strain and initiated this project. HZ and YL wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Key Research and Development Program of Zhejiang Province (2021C02009, 2022C02016, and 2021C02064-7).
The authors thank Prof. Mengcen Wang from Zhejiang University for his assistance with the preparation of crude extracts, Xiaodan Wu and Mei Fang from Analysis Center of Agrobiology and Environmental Science of Zhejiang University for assistance with LC–MS analysis, and Xiao-xiao Feng from Agricultural Experiment Station of Zhejiang University for her assistance with plant pathogens.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1071530/full#supplementary-material
Al Souhail, Q., Hiromasa, Y., Rahnamaeian, M., Giraldo, M. C., Takahashi, D., Valent, B., et al. (2016). Characterization and regulation of expression of an antifungal peptide from hemolymph of an insect, Manduca sexta. Dev. Comp. Immunol. 61, 258–268. doi: 10.1016/j.dci.2016.03.006
Apostolopoulos, V., Bojarska, J., Chai, T. T., Elnagdy, S., Kaczmarek, K., Matsoukas, J., et al. (2021). A global review on short peptides: frontiers and perspectives. Molecules 26:430. doi: 10.3390/molecules26020430
Bahar, O., Goffer, T., and Burdman, S. (2009). Type IV pili are required for virulence, twitching motility, and biofilm formation of Acidovorax avenae subsp. citrulli. Mol. Plant-Microbe Interact. 22, 909–920. doi: 10.1094/MPMI-22-8-0909
Baltz, R. H. (2009). Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. Curr. Opin. Chem. Biol. 13, 144–151. doi: 10.1016/j.cbpa.2009.02.031
Bansal, S., Vu, K., Liu, R. W., Ajena, Y., Xiao, W. W., Menon, S. M., et al. (2022). Discovery and characterization of a potent antifungal peptide through one-bead, one-compound combinatorial library screening. ACS Infect. Dis. 8, 1291–1302. doi: 10.1021/acsinfecdis.2c00019
Barrera-Galicia, G. C., Peniche-Pavia, H. A., Pena-Cabriales, J. J., Covarrubias, S. A., Vera-Nunez, J. A., and Delano-Frier, J. P. (2021). Metabolic footprints of Burkholderia sensu lato rhizosphere bacteria active against maize Fusarium pathogens. Microorganisms 9:2061. doi: 10.3390/microorganisms9102061
Bopp, M. (1965). Inhibition of agrobacterium tumefaciens by d-amino acids. Z. Naturforsch. B 20, 899–905. doi: 10.1515/znb-1965-0914
Burkholder, W. H. (1942). Three bacterial plant pathogens: Pathogenss caryophylli sp n. Phytomonas alliicola sp.n., and Phytomonas manihotis (Arthaud-Berthet et Bondar) Viégas. Phytopathology 32, 141–149.
Canal-Raffin, M., L'azou, B., Jorly, J., Hurtier, A., Cambar, J., and Brochard, P. (2008). Cytotoxicity of folpet fungicide on human bronchial epithelial cells. Toxicology 249, 160–166. doi: 10.1016/j.tox.2008.05.003
Caulier, S., Nannan, C., Gillis, A., Licciardi, F., Bragard, C., and Mahillon, J. (2019). Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 10:302. doi: 10.3389/fmicb.2019.00302
Chen, K., Tian, Z. H., He, H., Long, C. A., and Jiang, F. T. (2020). Bacillus species as potential biocontrol agents against citrus diseases. Biol. Control 151:104419. doi: 10.1016/j.biocontrol.2020.104419
Daungfu, O., Youpensuk, S., and Lumyong, S. (2019). Endophytic bacteria isolated from citrus plants for biological control of citrus canker in lime plants. Trop. Life Sci. Res. 30, 73–88. doi: 10.21315/tlsr2019.30.1.5
Dereven'kov, I. A., Salnikov, D. S., and Makarov, S. V. (2017). Interaction between super-reduced cobalamin and selenite. Russ. J. Phys. Chem. A 91, 2404–2408. doi: 10.1134/S003602441711005X
Ding, K., Shen, P., Xie, Z., Wang, L., and Dang, X. (2022). In vitro and in vivo antifungal activity of two peptides with the same composition and different distribution. Comp. Biochem. Physiol. C 252:109243. doi: 10.1016/j.cbpc.2021.109243
Donzelli, B. G., Lorito, M., Scala, F., and Harman, G. E. (2001). Cloning, sequence and structure of a gene encoding an antifungal glucan 1,3-beta-glucosidase from Trichoderma atroviride (T. harzianum). Gene 277, 199–208. doi: 10.1016/S0378-1119(01)00681-3
Du, S., Wang, Y., Alatrash, N., Weatherly, C. A., Roy, D., Macdonnell, F. M., et al. (2019). Altered profiles and metabolism of l- and d-amino acids in cultured human breast cancer cells vs. non-tumorigenic human breast epithelial cells. J. Pharm. Biomed. Anal. 164, 421–429. doi: 10.1016/j.jpba.2018.10.047
Edwards, J. L. Jr., Busta, F. F., and Speck, M. L. (1965). Thermal inactivation characteristics of Bacillus subtilis spores at ultra high temperatures. Appl. Microbiol. 13, 851–857. doi: 10.1128/am.13.6.851-857.1965
El-Banna, N., and Winkelmann, G. (1998). Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J. Appl. Microbiol. 85, 69–78. doi: 10.1046/j.1365-2672.1998.00473.x
Elder, R. T. (1983). Molecular cloning a laboratory manual. Bioscience 33, 721–722. doi: 10.2307/1309366
Elshafie, H. S., and Camele, I. (2021). An overview of metabolic activity, beneficial and pathogenic aspects of Burkholderia Spp. Meta 11:321. doi: 10.3390/metabo11050321
Fira, D., Dimkic, I., Beric, T., Lozo, J., and Stankovic, S. (2018). Biological control of plant pathogens by bacillus species. J. Biotechnol. 285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044
Fox, S. W., Fling, M., and Bollenback, G. N. (1944). Inhibition of bacterial growth by d-leucine. J. Biol. Chem. 155, 465–468. doi: 10.1016/S0021-9258(18)51176-5
Fraaije, B., Atkins, S., Hanley, S., Macdonald, A., and Lucas, J. (2020). The multi-fungicide resistance status of Aspergillus fumigatus populations in arable soils and the wider european environment. Front. Microbiol. 11:599233. doi: 10.3389/fmicb.2020.599233
Goldenhar, K. E., and Hausbeck, M. K. (2019). Fungicides for control of downy mildew on pickling cucumber in Michigan. Plant Hlth. Prog. 20, 165–169. doi: 10.1094/PHP-04-19-0025-RS
Gong, Y., Li, H., Wu, F., Li, Y., and Zhang, S. (2022). Fungicidal activity of AP10W, a short peptide derived from AP-2 complex subunit mu-a, in vitro and in vivo. Biomol. Ther. 12:965. doi: 10.3390/biom12070965
Han, Y., Zhao, J., Zhang, B., Shen, Q., Shang, Q., and Li, P. (2019). Effect of a novel antifungal peptide P852 on cell morphology and membrane permeability of Fusarium oxysporum. Biochim. Biophys. Acta Biomembr. 1861, 532–539. doi: 10.1016/j.bbamem.2018.10.018
He, Y. Z., Xu, Y., Sun, J., Gao, B. L., Li, G., Zhou, Y. F., et al. (2021). Novel plasmid-borne fimbriae-associated gene cluster participates in biofilm formation in Escherichia coli. Microb. Drug Resist. 27, 1624–1632. doi: 10.1089/mdr.2020.0512
Hertweck, C. (2009). The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Eng. 48, 4688–4716. doi: 10.1002/anie.200806121
Holmberg, A., Lood, R., Morgelin, M., Soderquist, B., Holst, E., Collin, M., et al. (2009). Biofilm formation by Propionibacterium acnes is a characteristic of invasive isolates. Clin. Microbiol. Infect. 15, 787–795. doi: 10.1111/j.1469-0691.2009.02747.x
Joo, J. H., and Hussein, K. A. (2022). Biological control and plant growth promotion properties of volatile organic compound-producing antagonistic Trichoderma spp. Front. Plant Sci. 13:897668. doi: 10.3389/fpls.2022.897668
Kannan, S., Aronica, P. G. A., Ng, S., Gek Lian, D. T., Frosi, Y., Chee, S., et al. (2020). Macrocyclization of an all-d linear alpha-helical peptide imparts cellular permeability. Chem. Sci. 11, 5577–5591. doi: 10.1039/C9SC06383H
Kapil, S., and Sharma, V. (2021). D-amino acids in antimicrobial peptides: a potential approach to treat and combat antimicrobial resistance. Can. J. Microbiol. 67, 119–137. doi: 10.1139/cjm-2020-0142
Khani, S., Seyedjavadi, S. S., Hosseini, H. M., Goudarzi, M., Valadbeigi, S., Khatami, S., et al. (2020). Effects of the antifungal peptide Skh-AMP1 derived from Satureja khuzistanica on cell membrane permeability, ROS production, and cell morphology of conidia and hyphae of Aspergillus fumigatus. Peptides 123:170195. doi: 10.1016/j.peptides.2019.170195
Khani, S., Seyedjavadi, S. S., Zare-Zardini, H., Hosseini, H. M., Goudarzi, M., Khatami, S., et al. (2019). Isolation and functional characterization of an antifungal hydrophilic peptide, Skh-AMP1, derived from Satureja khuzistanica leaves. Phytochemistry 164, 136–143. doi: 10.1016/j.phytochem.2019.05.011
Krichilsky, E., Centrella, M., Eitzer, B., Danforth, B., Poveda, K., and Grab, H. (2021). Landscape composition and fungicide exposure influence host-pathogen dynamics in a solitary bee. Environ. Entomol. 50, 107–116. doi: 10.1093/ee/nvaa138
Kumar, R., Swain, D. M., Yadav, S. K., Tyagi, I., Kumar, R., Das, J., et al. (2018). Bacteria-fungal confrontation and fungal growth prevention assay. Bio. Protoc. 8:e2694. doi: 10.21769/BioProtoc.2694
Latin, R., and Ou, L. (2018). Influence of irrigation and wetting agent on fungicide residues in creeping bentgrass. Plant Dis. 102, 2352–2360. doi: 10.1094/PDIS-11-17-1844-RE
Li, T., Li, L., Du, F., Sun, L., Shi, J., Long, M., et al. (2021). Activity and mechanism of action of antifungal peptides from microorganisms: a review. Molecules 26:3438. doi: 10.3390/molecules26113438
Lide, D.R. (2007). CRC Handbook of Chemistry and Physics. Boca Raton, FL: CRC Press, Taylor & Francis.
Liu, Y. G., and Whittier, R. F. (1995). Thermal asymmetric interlaced PCR - automatable amplification and sequencing of insert end fragments from P1 and Yac clones for chromosome walking. Genomics 25, 674–681. doi: 10.1016/0888-7543(95)80010-J
Mahenthiralingam, E., Baldwin, A., and Dowson, C. G. (2008). Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104, 1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x
Marahiel, M. A. (2009). Working outside the protein-synthesis rules: insights into non-ribosomal peptide synthesis. J. Pept. Sci. 15, 799–807. doi: 10.1002/psc.1183
Maskey, R. P., Li, F., Qin, S., Fiebig, H. H., and Laatsch, H. (2003). Chandrananimycins A-C: production of novel anticancer antibiotics from a marine Actinomadura sp. isolate M048 by variation of medium composition and growth conditions. J. Antibiot. 56, 622–629. doi: 10.7164/antibiotics.56.622
Mercado-Blanco, J., and Bakker, P. A. H. M. (2007). Interactions between plants and beneficial pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie Van Leeuwenhoek 92, 367–389. doi: 10.1007/s10482-007-9167-1
Moghaddam, M. R. B., Vilcinskas, A., and Rahnamaeian, M. (2017). The insect-derived antimicrobial peptide metchnikowin targets Fusarium graminearum beta(1,3)-glucanosyltransferase Gel1, which is required for the maintenance of cell wall integrity. Biol. Chem. 398, 491–498. doi: 10.1515/hsz-2016-0295
Moore, T. C., Newmister, S. A., Rayment, I., and Escalante-Semerena, J. C. (2012). Structural insights into the mechanism of four-coordinate cob(II)alamin formation in the active site of the salmonella enterica ATP: co(I)rrinoid adenosyltransferase enzyme: critical role of residues Phe91 and Trp93. Biochemistry 51, 9647–9657. doi: 10.1021/bi301378d
Nazarov, P. A., Baleev, D. N., Ivanova, M. I., Sokolova, L. M., and Karakozova, M. V. (2020). Infectious plant diseases: etiology, current status, problems and prospects in plant protection. Acta Nat. 12, 46–59. doi: 10.32607/actanaturae.11026
Niu, X., Thaochan, N., and Hu, Q. (2020). Diversity of linear non-ribosomal peptide in biocontrol fungi. J. Fungi 6:61. doi: 10.3390/jof6020061
Oh, M., Park, S., Kim, H., Choi, G. J., and Kim, S. H. (2021). Application of UPLC-QTOF-MS based untargeted metabolomics in identification of metabolites induced in pathogen-infected rice. Plants 10:213. doi: 10.3390/plants10020213
Ortiz-Lopez, F. J., Monteiro, M. C., Gonzalez-Menendez, V., Tormo, J. R., Genilloud, O., Bills, G. F., et al. (2015). Cyclic colisporifungin and linear cavinafungins, antifungal lipopeptides isolated from Colispora cavincola. J. Nat. Prod. 78, 468–475. doi: 10.1021/np500854j
Park, A. K., Chi, Y. M., and Moon, J. H. (2011). Crystal structure of PduO-type ATP: cob(I)alamin adenosyltransferase from Bacillus cereus in a complex with ATP. Biochem. Biophys. Res. Commun. 408, 417–421. doi: 10.1016/j.bbrc.2011.04.036
Park, S. C., Kim, J. Y., Kim, E. J., Cheong, G. W., Lee, Y., Choi, W., et al. (2018). Hydrophilic linear peptide with histidine and lysine residues as a key factor affecting antifungal activity. Int. J. Mol. Sci. 19:3781. doi: 10.3390/ijms19123781
Poveda, J., Roeschlin, R. A., Marano, M. R., and Favaro, M. A. (2021). Microorganisms as biocontrol agents against bacterial citrus diseases. Biol. Control 158:104602. doi: 10.1016/j.biocontrol.2021.104602
Pushpanathan, M., Rajendhran, J., Jayashree, S., Sundarakrishnan, B., Jayachandran, S., and Gunasekaran, P. (2012). Identification of a novel antifungal peptide with chitin-binding property from marine metagenome. Protein Pept. Lett. 19, 1289–1296. doi: 10.2174/092986612803521620
Qiao, H., Zhang, B., Chen, X., Su, L., Jiao, C., Chen, S., et al. (2020). Short peptides secreted by Bacillus subtilis inhibit the growth of mold on fresh-cut pumpkin (Cucurbita pepo). J. Sci. Food Agric. 100, 936–944. doi: 10.1002/jsfa.10021
Radkov, A. D., and Moe, L. A. (2014). Bacterial synthesis of d-amino acids. Appl. Microbiol. Biotechnol. 98, 5363–5374. doi: 10.1007/s00253-014-5726-3
Riedel, T., Rohlfs, M., Buchholz, I., Wagner-Dobler, I., and Reck, M. (2013). Complete sequence of the suicide vector pJP5603. Plasmid 69, 104–107. doi: 10.1016/j.plasmid.2012.07.005
Robbins, W. J. (1924). Isoelectric points for the mycelium of fungi. J. Gen. Physiol. 6, 259–271. doi: 10.1085/jgp.6.3.259
Rodionov, D. A., Vitreschak, A. G., Mironov, A. A., and Gelfand, M. S. (2003). Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278, 41148–41159. doi: 10.1074/jbc.M305837200
Ruiz, S. J., Van't Klooster, J. S., Bianchi, F., and Poolman, B. (2021). Growth inhibition by amino acids in Saccharomyces cerevisiae. Microorganisms 9:7. doi: 10.3390/microorganisms9010007
Russell, P. E. (2005). A century of fungicide evolution. J. Agr. Sci. 143, 11–25. doi: 10.1017/S0021859605004971
Sharma, K. K., Ravi, R., Maurya, I. K., Kapadia, A., Khan, S. I., Kumar, V., et al. (2021). Modified histidine containing amphipathic ultrashort antifungal peptide, his[2-p-(n-butyl)phenyl]-Trp-Arg-OMe exhibits potent anticryptococcal activity. Eur. J. Med. Chem. 223:113635. doi: 10.1016/j.ejmech.2021.113635
Shi, P., Yao, G., Yang, P., Li, N., Luo, H., Bai, Y., et al. (2010). Cloning, characterization, and antifungal activity of an endo-1,3-beta-D: -glucanase from Streptomyces sp. S27. Appl. Microbiol. Biotechnol. 85, 1483–1490. doi: 10.1007/s00253-009-2187-1
Shrestha, R., Lockless, S. W., and Sorg, J. A. (2017). A Clostridium difficile alanine racemase affects spore germination and accommodates serine as a substrate. J. Biol. Chem. 292, 10735–10742. doi: 10.1074/jbc.M117.791749
Spencer, J. B., Stolowich, N. J., Roessner, C. A., and Scott, A. I. (1993). The Escherichia coli cysG gene encodes the multifunctional protein, siroheme synthase. FEBS Lett. 335, 57–60. doi: 10.1016/0014-5793(93)80438-Z
Staunton, J., and Weissman, K. J. (2001). Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18, 380–416. doi: 10.1039/a909079g
Storbeck, S., Walther, J., Muller, J., Parmar, V., Schiebel, H. M., Kemken, D., et al. (2009). The Pseudomonas aeruginosa nirE gene encodes the S-adenosyl-L-methionine-dependent uroporphyrinogen III methyltransferase required for heme d1biosynthesis. FEBS J. 276, 5973–5982. doi: 10.1111/j.1742-4658.2009.07306.x
Stroupe, M. E., Leech, H. K., Daniels, D. S., Warren, M. J., and Getzoff, E. D. (2003). CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis. Nat. Struct. Biol. 10, 1064–1073. doi: 10.1038/nsb1007
Sultan, M. Z., Park, K., Lee, S. Y., Park, J. K., Varughese, T., and Moon, S. S. (2008). Novel oxidized derivatives of antifungal pyrrolnitrin from the bacterium Burkholderia cepacia K87. J. Antibiot. 61, 420–425. doi: 10.1038/ja.2008.58
Thomson, E. L. S., and Dennis, J. J. (2012). A Burkholderia cepacia complex non-ribosomal peptide-synthesized toxin is hemolytic and required for full virulence. Virulence 3, 286–298. doi: 10.4161/viru.19355
Tong, S., Li, M., Keyhani, N. O., Liu, Y., Yuan, M., Lin, D., et al. (2020). Characterization of a fungal competition factor: production of a conidial cell-wall associated antifungal peptide. PLoS Pathog. 16:e1008518. doi: 10.1371/journal.ppat.1008518
Tran, C., Cock, I. E., Chen, X. J., and Feng, Y. J. (2022). Antimicrobial bacillus: metabolites and their mode of action. Antibiotics 11:88. doi: 10.3390/antibiotics11010088
Van Den Bosch, F., and Gilligan, C. A. (2008). Models of fungicide resistance dynamics. Annu. Rev. Phytopathol. 46, 123–147. doi: 10.1146/annurev.phyto.011108.135838
Velivelli, S. L. S., Islam, K. T., Hobson, E., and Shah, D. M. (2018). Modes of action of a bi-domain plant defensin MtDef5 against a bacterial pathogen Xanthomonas campestris. Front. Microbiol. 9:934. doi: 10.3389/fmicb.2018.00934
Warren, M. J., Bolt, E. L., Roessner, C. A., Scott, A. I., Spencer, J. B., and Woodcock, S. C. (1994). Gene dissection demonstrates that the Escherichia coli cysG gene encodes a multifunctional protein. Biochem. J. 302, 837–844. doi: 10.1042/bj3020837
Wen, C., Guo, W., and Chen, X. (2014). Purification and identification of a novel antifungal protein secreted by Penicillium citrinum from the southwest indian ocean. J. Microbiol. Biotechnol. 24, 1337–1345. doi: 10.4014/jmb.1405.05008
Whetstone, P. A., and Hammock, B. D. (2007). Delivery methods for peptide and protein toxins in insect control. Toxicon 49, 576–596. doi: 10.1016/j.toxicon.2006.11.009
Woodcock, S. C., Raux, E., Levillayer, F., Thermes, C., Rambach, A., and Warren, M. J. (1998). Effect of mutations in the transmethylase and dehydrogenase/chelatase domains of sirohaem synthase (CysG) on sirohaem and cobalamin biosynthesis. Biochem. J. 330, 121–129. doi: 10.1042/bj3300121
Xian, L., Yu, G., Wei, Y., Rufian, J. S., Li, Y., Zhuang, H., et al. (2020). A bacterial effector protein hijacks plant metabolism to support pathogen nutrition. Cell Host Microbe 28, 548–557.e7. doi: 10.1016/j.chom.2020.07.003
Yabu, K., and Huempfner, H. R. (1974). Inhibition of growth of mycobacterium smegmatis and of cell-wall synthesis by d-serine. Antimicrob. Agents Chem. 6, 1–10. doi: 10.1128/AAC.6.1.1
Yabuuchi, E., Kosako, Y., Oyaizu, H., Yano, I., Hotta, H., Hashimoto, Y., et al. (1992). Proposal of Burkholderia gen. Nov. and transfer of seven species of the genus pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36, 1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x
Zhang, D., Lu, Y. L., Chen, H. C., Wu, C. T., Zhang, H., Chen, L. Y., et al. (2020). Antifungal peptides produced by actinomycetes and their biological activities against plant diseases. J. Antibiot. 73, 265–282. doi: 10.1038/s41429-020-0287-4
Zhao, P., Xue, Y., Li, J., Li, X., Zu, X., Zhao, Z., et al. (2019). Non-lipopeptide fungi-derived peptide antibiotics developed since 2000. Biotechnol. Lett. 41, 651–673. doi: 10.1007/s10529-019-02677-3
Keywords: Burkholderia arboris, antifungal ability, Tn5, untargeted metabolomics, peptide
Citation: Zhu H, Xu C, Chen Y and Liang Y (2022) His-Ala-Phe-Lys peptide from Burkholderia arboris possesses antifungal activity. Front. Microbiol. 13:1071530. doi: 10.3389/fmicb.2022.1071530
Received: 16 October 2022; Accepted: 21 November 2022;
Published: 06 December 2022.
Edited by:
Jianhua Wang, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Bijit Bhowmik, Croda Inc., United StatesCopyright © 2022 Zhu, Xu, Chen and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Liang, eWFubGlhbmdAemp1LmVkdS5jbg==; Yicun Chen, eWljdW5fY2hlbkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.