
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 05 January 2023
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1068251
This article is part of the Research TopicRising Stars in Microbial Symbioses: 2022View all 8 articles
 Chao-Chin Liu1
Chao-Chin Liu1 Mei-Hui Lin1,2,3*
Mei-Hui Lin1,2,3*Staphylococcus aureus, which lacks pili and flagella, is nonmotile. However, it hitchhikes motile bacteria, such as Pseudomonas aeruginosa, to migrate in the environment. This study demonstrated that the hitchhiking motility of S. aureus SA113 was reduced after the tagO, which encodes an enzyme for wall teichoic acids (WTA) synthesis, was deleted. The hitchhiking motility was restored after the mutation was complemented by transforming a plasmid expressing TagO into the mutant. We also showed that adding purified lipopolysaccharide (LPS) to a culture that contains S. aureus SA113 and P. aeruginosa PAO1, reduced the movement of S. aureus, showing that WTA and LPS are involved in the hitchhiking motility of S. aureus. This study also found that P. aeruginosa promoted the movement of S. aureus in the digestive tract of Caenorhabditis elegans and in mice. In conclusion, this study reveals how S. aureus hitchhikes P. aeruginosa for translocation in an ecosystem. The results from this study improve our understanding on how a nonmotile pathogen moves in the environment and spreads in animals.
Microorganisms in an ecosystem often compete for resources to improve their survival (Ghoul and Mitri, 2016). One of the well-known examples occurs in a cystic fibrosis infection model where Pseudomonas aeruginosa changes the physiology and inhibits the growth of Staphylococcus aureus (Mashburn et al., 2005; Pernet et al., 2014); nevertheless, S. aureus prevents growth inhibition by P. aeruginosa through changes in metabolic pathways and growth characteristics (Hoffman et al., 2006; Biswas et al., 2009; Briaud et al., 2019). Another example regarding interspecies interaction is that Candida albicans enhances biofilm formation, antibiotic resistance and virulence of S. aureus through activating the staphylococcal agr quorum sensing system (Todd et al., 2019). Additionally, the expression of C. albicans drug resistant gene is upregulated during co-culture with S. aureus (Hu et al., 2021).
Motility is crucial for bacteria to escape dangers and search for nutrients in the environment, which are important to their virulence and pathogenicity (Matilla and Krell, 2018). Recent studies have demonstrated that nonmotile bacteria are capable of attaching motile bacteria to move and relocate to favorable niches (Hagai et al., 2014; Samad et al., 2017). For instance, Xanthomonas perforans, a weak swarmer, produces substances allowing the organism to attach to a motile bacterium, Paenibacillus vortex, to swarm efficiently on the surface of host plants (Hagai et al., 2014). Another example is that swarming P. vortex carries antibiotic-resistant Escherichia coli to help the organism against antibiotic stress (Finkelshtein et al., 2015). These studies indicate that microbes in a community often using hitchhiking strategy to migrate and overcome stresses in the environment.
Staphylococcus aureus is nonmotile due to the lack of flagella and pili (Pollitt and Diggle, 2017). However, this organism does move on soft agar surface via spreading (Kaito and Sekimizu, 2007; Lin et al., 2016). Its movement is mediated through the accumulation of water in a colony and the production of biosurfactants termed phenol soluble modulins (PSMs; Kaito and Sekimizu, 2007; Tsompanidou et al., 2013; Lin et al., 2016), which weakens the surface tension of the water, allowing water to flood on the agar surface and spreading of the organism (Lin et al., 2016). Meanwhile, in an ecosystem, S. aureus has opportunities to interact with neighboring microbes. Staphylococcus aureus and P. aeruginosa are two human pathogens often sharing the same niche (Pastar et al., 2013; DeLeon et al., 2014). A recent study demonstrated that swimming P. aeruginosa promotes dispersion of S. aureus (Samad et al., 2017). However, the mechanism underlying the interaction between S. aureus and P. aeruginosa remains unclear. This study demonstrated that S. aureus hitchhikes P. aeruginosa via interaction between wall teichoic acids (WTA) of S. aureus and lipopolysaccharide (LPS) of P. aeruginosa. The hitchhiking motility of S. aureus prompted by P. aeruginosa was also observed in vivo in a Caenorhabditis elegans model and a mouse model.
Staphylococcus aureus SA113 (ATCC35556) was used for the study of hitchhiking motility (Herbert et al., 2010; Tsompanidou et al., 2011). A ΔtagO mutant of S. aureus SA113 contains a deletion in tagO and does not produce WTA (Weidenmaier et al., 2004). For complementing the ΔtagO mutation, the strain was transformed with a TagO-expressing plasmid, pHY-tagO. Pseudomonas aeruginosa PAO1 and its isogenic mutant ΔfliA, a flagellum-defective mutant (Lo et al., 2016) were kindly provided by Professor Hwan-You Chang. Escherichia coli EPI300 (Epicentre Technologies, Wisconsin, United States) and S. aureus RN4220, a restriction-deficient strain (Nair et al., 2011), were used as hosts for cloning. Staphylococcus aureus strains were transformed with pCtuf-ppmch (Mauthe et al., 2012), which encodes mCherry and exhibits red fluorescence. Staphylococcus aureus CGL1190, a bioluminescent strain that constitutively expresses a luciferase reporter. Plasmid pHY-tagO was constructed by inserting a PCR-amplified DNA fragment containing the tagO from S. aureus SA113. The fragment was amplified with primers 5′-CGGTCTAGATAGCACTTGTTACTGCAGCA and 5′-TTACCCGGGATCCCATACAGCTATGCTTT and digested with XbaI and SmaI and finally inserted into the same sites in pHY300pLK (TaKaRa Bio) to generate pHY-tagO. Bacteria were cultured in tryptic soy broth (TSB) and tryptic soy agar (TSA; Oxoid). Mannitol salt agar (BD Difco) was used for differentiating and enumerating S. aureus colonies. Antibiotic-resistant colonies were selected on media that contained ampicillin (100 μg/ml), chloramphenicol (10 μg/ml), kanamycin (50 μg/ml), and tetracycline (5 μg/ml).
To generate a bioluminescent S. aureus strain, a temperature-sensitive plasmid pRP1190 (Plaut et al., 2013), which contains a modified luxBADCE operon from Photorhabdus luminescens, was transformed into S. aureus SA113. Integration of plasmids into bacterial chromosomes by homologous recombination was performed according to a method described previously (Plaut et al., 2013). The plates were imaged using an in vivo Imaging System (IVIS Lumina III, PerkinElmer, Waltham, Massachusetts, United States). Luminescent colonies were selected, and the integration site was confirmed by PCR.
Hitchhiking motility was assayed according to a method described elsewhere (Samad et al., 2017) but with modifications. Staphylococcus aureus and Pseudomonas aeruginosa strains were cultured to the mid-log phase and then mixed at a ratio of 1:100 in TSB. Bacterial mixture (200 μl) that contained 106 colony forming unit (CFU) of S. aureus and 108 CFU of P. aeruginosa was transferred into the wells in a Calgary Biofilm Device (CBD; Innovotech, Edmonton, Alberta, Canada). The plate was placed at room temperature for 1 h. Subsequently, the pegs on a CBD lid was immersed in the wells for 30 s to allow attachment of the bacteria. The pegs on the lid were then washed by immersing the pegs in 200 μl PBS in each well of a 96-well microtiter plate with gentle shaking using a Thermomixer (Eppendorf, Hamburg, Germany). After washing, the lid was transferred and immersed in a new 96-well plate containing 200 μl PBS and 0.1 mm-diameter glass beads (Biospec Products, Bartlesville, Oklahoma, United States). The plate was vortexed gently using a Thermomixer (Eppendorf, Hamburg, Germany) to remove the bacteria from the pegs. The bacterial suspension was then serially diluted and plated on mannitol salt agar. The number of S. aureus SA113 on the pegs was enumerated by viable cell count.
The pegs on the lid of CBD were removed and fixed overnight in 2% glutaraldehyde. The bacteria were examined under a field emission scanning electron microscope (FE-SEM, Hitachi SU8220, Hitachi High-Tech, Tokyo, Japan) according to a method described elsewhere (Samad et al., 2017). Images were colored using Photoshop CS3 (Adobe Systems, San Jose, California, United States).
TSA plates prepared with 0.4% agarose (TSA-0.4; Seakem, Lonza Rockland, Maine, United States) were used for swarming assay. Bacterial culture (2 μl) that contained 1×104 CFU of S. aureus and 1 × 106 CFU of P. aeruginosa was spotted at the center of a TSA-0.4 plate. The plate was subsequently incubated at 37°C for 4 h to allow P. aeruginosa to swarm. The bacteria at the center and the edge of a swarm were picked with a toothpick, which was inserted vertically into the bottom of the plate. The bacteria adhered to the toothpick were dissolved in 1 ml PBS. After serial dilution, bacteria were plated on mannitol salt agar. The number of S. aureus in the bacterial suspension was enumerated by viable cell count.
To detect the binding of LPS from P. aeruginosa to S. aureus, FITC-LPS (Sigma-Aldrich, United States) and free LPS (Sigma-Aldrich, United States) were mixed with 105 CFU of S. aureus. The fluorescence intensity was detected, and the number of S. aureus SA113 bound to FITC-LPS was enumerated with a Guava easyCyte flow cytometer (Merck Millipore). To assess the binding of WTA of S. aureus or free lipoteichoic acids (LTA; Sigma-Aldrich, United States) to P. aeruginosa, mid-log phase S. aureus and P. aeruginosa were stained with hexidium iodide (Thermo Fisher Scientific, Waltham, MA, United States) and SYTO 9 (Invitrogen; Thermo Fisher Scientific) according to the instruction provided by the vendors. Labeled S. aureus and P. aeruginosa were mixed at a ratio of 1:1 and 105 CFU/ml of the bacteria was used for flow cytometry analysis. Five thousand fluorescent-stained cells were counted in a gate set in a hexidium iodide versus SYTO 9 dot plot.
Caenorhabditis elegans N2 was cultured and maintained according to a method described previously (Brenner, 1974). Staphylococcus aureus strains harboring pCtuf-ppmch and SYTO 9-labeled P. aeruginosa were used in this study. Staphylococcus aureus and P. aeruginosa were subcultured for 4 h. Then, bacterial pellets were collected, mixed, and suspended in 200 μl PBS. The bacterial suspension was inoculated on TSA plates, and the inoculum was allowed to dry at room temperature. Caenorhabditis elegans at the L4 stage were placed onto bacteria-containing TSA plates and incubated at 20°C for 30 min. The worms were then transferred and mounted onto 2% agarose pads on a slide covered with 1% levamisole (Sigma-Aldrich, United States) to paralyze the worms. Bacteria in the digestive tracts of C. elegans were observed and imaged under a confocal laser-scanning microscope at 0 and 60 min after feeding, worms were imaged under a Olympus FV10i microscope. Meanwhile, the images were captured at 30 s interval for 5 min and converted into a time-lapse video. The distance of bacterial migration was measured and analyzed using FV10-ASW 4.2 software (Olympus).
Six- to eight-week-old BALB/c mice were used in this study. The animal experiments were approved by the Chang Gung University Animal Care Committee (approval no. CGU108-191) and performed in accordance with the approved animal care guidelines and protocols. To improve the imaging quality, the backs of the mice were shaved. Mice were anesthetized by isoflurane inhalation and 50 μl of bacterial suspension, which contained 1 × 107 CFU of luminescent S. aureus CGL1190 and 5 × 107 CFU of P. aeruginosa PAO1, was injected subcutaneously into the dorsal region of the animals. The mice were then imaged with an IVIS instrument at 0 and 6 h post-injection. After observation, the mice were sacrificed, and the skin tissues at the injection site and the areas 2 cm away from the injection site were collected using a single hole puncher. The skin tissues were homogenized in 1 ml sterile PBS followed by centrifugation at 500 ×g for 5 min. The supernatant was serially diluted and plated on mannitol salt agar plates. The number of luminescent S. aureus was determined by viable cell count.
The between-group differences were determined using two-tailed Student’s t test and one-way analysis of variance (ANOVA) test. According to the Shapiro–Wilk’s test, the results of C. elegans studies did not exhibit a normal distribution; the Kruskal-Wallis test was further used for statistical analysis of the C. elegans studies. All statistical analysis were performed using GraphPad Prism software version 8.0 (GraphPad Software, San Diego, California, United States).
An earlier study demonstrated that S. aureus achieves movement in a liquid environment by interacting with a swimming bacterium (Samad et al., 2017). A hitchhiking motility assay (Samad et al., 2017) was modified and used in this study to measure the ability of P. aeruginosa to carry S. aureus during swimming. We added a mixture of S. aureus and P. aeruginosa into the wells of a CBD. After the bacterial mixture was stratified into two different layers with motile bacteria on the top and nonmotile bacteria at the bottom in the wells, we collected the bacteria on the top using the pegs on the lid of the CBD for CFU enumeration (Figure 1A). The results showed that when S. aureus SA113 was incubated in wells alone, the average CFU of S. aureus SA113 that adhered to a peg was 173. The CFU of bacteria on the pegs increased 17-fold to 2,966 CFU when S. aureus was incubated with P. aeruginosa PAO1 (Figure 1B). We also used a ΔfliA mutant of P. aeruginosa PAO1, which lacks flagellum. Compared with flagellated P. aeruginosa PAO1, the mutant carried 100 times less S. aureus SA113 to the pegs (Figure 1C), indicating that adherence of S. aureus to the pegs depends on the motility of P. aeruginosa. These results were consistent with an earlier finding that S. aureus is carried by P. aeruginosa to migrate (Samad et al., 2017). The attachment of P. aeruginosa PAO1 and S. aureus SA113 to the pegs was also verified by SEM (Figure 1A).
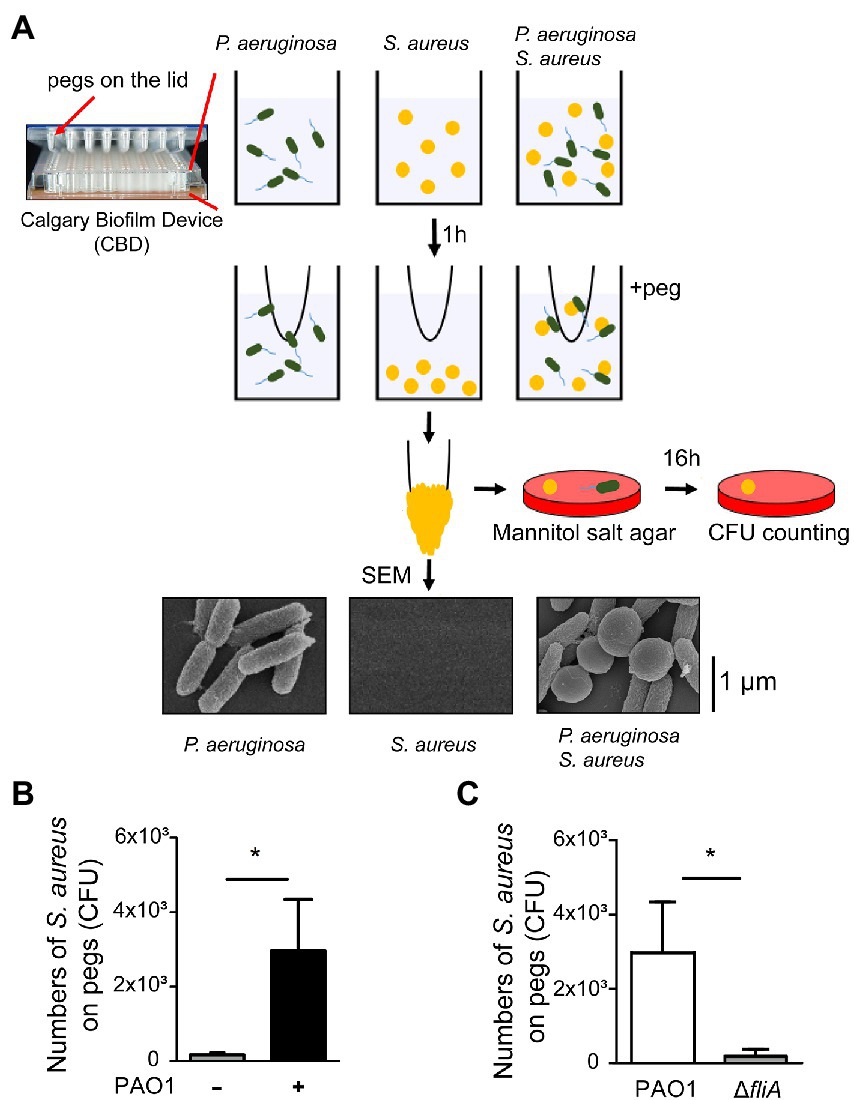
Figure 1. Promotion of Staphylococcus aureus movement by motile Pseudomonas aeruginosa. (A) Assay of S. aureus SA113 movement prompted by swimming P. aeruginosa PAO1. Bacteria were cultured in wells of CBD plates. After incubation for 1 h, the pegs on the lid were immersed into the culture for 30 s. Staphylococcus aureus adhered on the pegs was enumerated by viable cell count. Bacteria on pegs were also examined by SEM. (B,C) Wells were seeded with S. aureus SA113 only or mixed with P. aeruginosa PAO1 or its ΔfliA mutant strain. The number of S. aureus SA113 adhered on pegs was determined. Data are presented as the mean of the results from more than three independent experiments and were analyzed statistically using Student’s t test. Error bars indicate standard deviations. Significant differences (p < 0.05) are denoted with *.
The interaction between S. aureus and P. aeruginosa suggested that surface components of S. aureus were involved in the hitchhiking motility. Our study found that the number of S. aureus that adhered to a peg was reduced after deleting tagO from strain SA113, which is required for the synthesis of wall teichoic acids (WTA; Weidenmaier et al., 2004). The results showed that when cocultured with P. aeruginosa PAO1, the average CFU of S. aureus SA113 that adhered to a peg was 3.1 × 103 (Figure 2A). However, after tagO was deleted, the average CFU of bacteria adhered to a peg reduced 4.3-fold to 7 × 102 (Figure 2A). The number increased to 3.8 × 103 CFU per peg when the mutant strain was transformed with pHY-tagO, a plasmid expressing TagO (Figure 2A), showing the importance of WTA in the hitchhiking motility. Similar results were also observed by SEM (Figure 2A, lower panel).
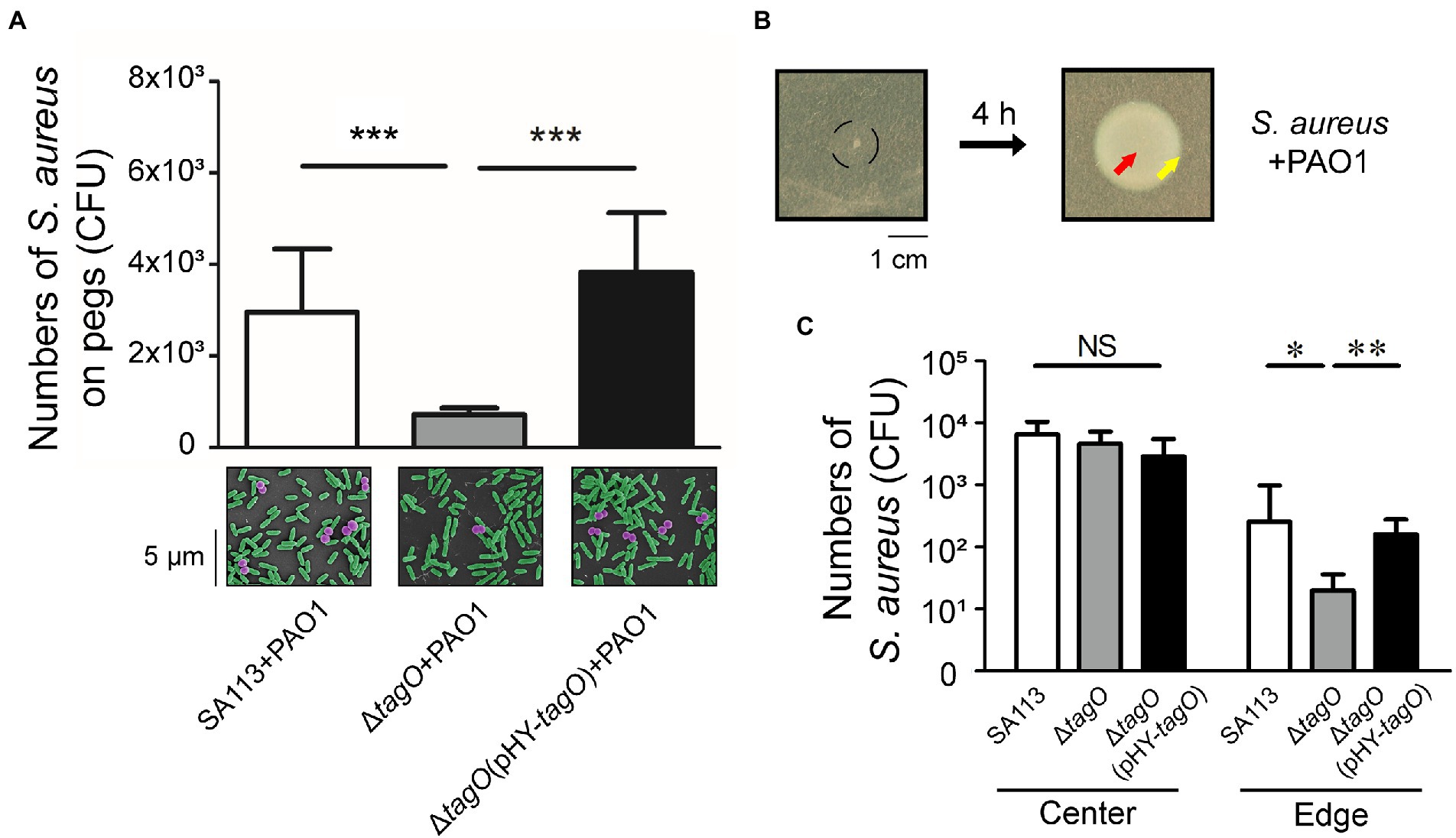
Figure 2. Influence of WTA in the hitchhiking motility of Staphylococcus aureus. (A) Staphylococcus aureus SA113, ΔtagO mutant of S. aureus SA113, and the mutant transformed with pHY-tagO were mixed with Pseudomonas aeruginosa PAO1 and added into wells of CBD plates. The number of S. aureus adhered on pegs was determined. The lower panel shows SEM images of S. aureus (purple) associated with P. aeruginosa (green) on pegs. The images shown are representative of three independent experiments. (B) Staphylococcus aureus was mixed with P. aeruginosa and inoculated on TSA-0.4 plates. The black circle indicates the area of inoculation (left panel). The plates were incubated for 4 h to allow bacteria to swarm. (C) Staphylococcus aureus located at the center (red arrow) and the edge (yellow arrow) of the swarm were collected using a toothpick. Bacteria on the toothpick were suspended in PBS and plated on mannitol salt agar for viable cell counts. Data are presented as the mean of the results from more than three independent experiments. Data were analyzed statistically using One-way ANOVA. Error bars indicate the standard deviations. Significant differences are denoted as follows: * indicates p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.001. NS, no significance.
The study further examined whether P. aeruginosa carried S. aureus during swarming. It is known that S. aureus spreads on soft agar surfaces, a movement resembles swarming (Kaito and Sekimizu, 2007). As S. aureus SA113 does not spread due to its agr-quorum sensing system is defective (Herbert et al., 2010; Tsompanidou et al., 2011), the movement of S. aureus SA113 during P. aeruginosa swarming could not be attributed to the spreading of S. aureus. Two microliter bacterial culture that contained 1 × 104 CFU of S. aureus and 1 × 106 CFU of P. aeruginosa was spotted at the center of a TSA-0.4 plate. Four hours after inoculation on agar medium with a mixture containing S. aureus SA113 and P. aeruginosa PAO1, the bacteria at the center and the edge were picked with a toothpick (Figure 2B). The results showed that the average CFU of S. aureus SA113, ΔtagO mutant of SA113, and the ΔtagO mutant of SA113 transformed with pHY-tagO at the center were similar (Figure 2C). However, at the edge of the swarm, average CFU of S. aureus SA113, the ΔtagO mutant, and the mutant harboring pHY-tagO were 251, 20 and 158 CFU, respectively (Figure 2C). The results indicated that S. aureus SA113 does not migrate to the edge of a swarm efficiently after tagO is deleted. According to the results from Figures 1, 2A, P. aeruginosa interacted with WTA of S. aureus and carried S. aureus to move around in the liquid environment, which facilitated S. aureus up to top layer and adhered to pegs. The phenomenon of WTA dependent carriage between P. aeruginosa and S. aureus is also observed on surface. Meanwhile, the whole swarm colony was harvested for CFU counting. The CFU of S. aureus increased from 1 × 104 to 1 × 105, indicating that during co-incubation with P. aeruginosa for 4 h, S. aureus still multiplied.
An earlier study showed that Bifidobacterium interacts with the LPS of E. coli to neutralize its toxicity (Park et al., 2007), implying that LPS is a potential candidate involved in associations with Gram-positive bacteria. To investigate the components of P. aeruginosa that interacted with WTA of S. aureus, free LPS from P. aeruginosa was used in the hitchhiking motility assay according to the method described in Figure 1A. The results showed that when free LPS was mixed with S. aureus before coincubation with P. aeruginosa, the number of S. aureus that adhered to a peg decreased in a dose-dependent manner (Figure 3A). Average CFU of S. aureus that adhered to the pegs was 3.6 × 103. The CFU decreased to 2.4 × 103, 1.1 × 103, and 0.4 × 103 when the bacteria were cultured in TSB containing 0.1, 1, and 10 μg/ml free LPS, respectively (Figure 3A). The results indicated that adding LPS competes for the binding of S. aureus to P. aeruginosa. Flow cytometry analysis also verified the binding of FITC-LPS to S. aureus (Figure 3B). The results demonstrated that the mean fluorescence intensity (MFI) of S. aureus SA113 alone was 19. The MFI increased to 253 when S. aureus SA113 was incubated with FITC-LPS. When unlabeled LPS was added, the MFI of FITC-LPS bound S. aureus decreased to 186 (Figure 3C). To assess whether WTA of S. aureus was required for the binding to LPS of P. aeruginosa, free LPS, hexidium iodide-labeled S. aureus SA113, ΔtagO mutant of SA113, and the mutant transformed with pHY-tagO and SYTO 9-labeled P. aeruginosa were used in flow cytometry analysis. The results demonstrated that when S. aureus SA113 was mixed with P. aeruginosa at a ratio of 1:1, the average percentage of P. aeruginosa associated with S. aureus SA113 was 32 (Figure 3E). When tagO was deleted from strain SA113, the average percentage of P. aeruginosa associated with the mutant reduced to 21.8 (Figure 3E); but increased to 34.7 when the mutant strain was transformed with pHY-tagO (Figure 3E). We also added free LPS to the mixture to determine whether LPS competes for the binding of S. aureus to P. aeruginosa. The results showed that compared with the strains untreated with LPS, the addition of free LPS decreased the binding percentage of P. aeruginosa to S. aureus SA113, the SA113 ΔtagO mutant and the mutant transformed with pHY-tagO from 32, 21.8, and 34.7 to 16, 6.3, and 18, respectively (Figure 3E). Furthermore, we examined whether lipoteichoic acids (LTA), which is a teichoic acid that anchored to the membrane lipid, is involved in the interaction between S. aureus and P. aeruginosa. When free LTA from S. aureus was used in the hitchhiking motility assay, the similar results were obtained as that of LPS (Figure 3D). The results showed that adding free LTA competes for the binding of S. aureus to P. aeruginosa in a dose-dependent manner (Figure 3D). Flow cytometry analysis also confirmed the percentage reductions after adding LTA. The results demonstrated that adding free LTA decreased the average percentage of the binding of P. aeruginosa to S. aureus SA113, the SA113 ΔtagO mutant, and the mutant transformed with pHY-tagO from 32, 21.8, and 34.7 to 10.4, 7.9, and 10.8 (Figure 3E), verifying that P. aeruginosa interacts with S. aureus via the interaction of LPS with WTA or LTA (Figure 3E). However, in comparison with S. aureus SA113, addition of free LTA to the ΔtagO mutant of SA113 or the mutant transformed with pHY-tagO, the differences of the binding of P. aeruginosa to S. aureus is not statistically significant between the groups (Figure 3E). The results suggested that WTA and LTA target different components on the P. aeruginosa surface for interaction.
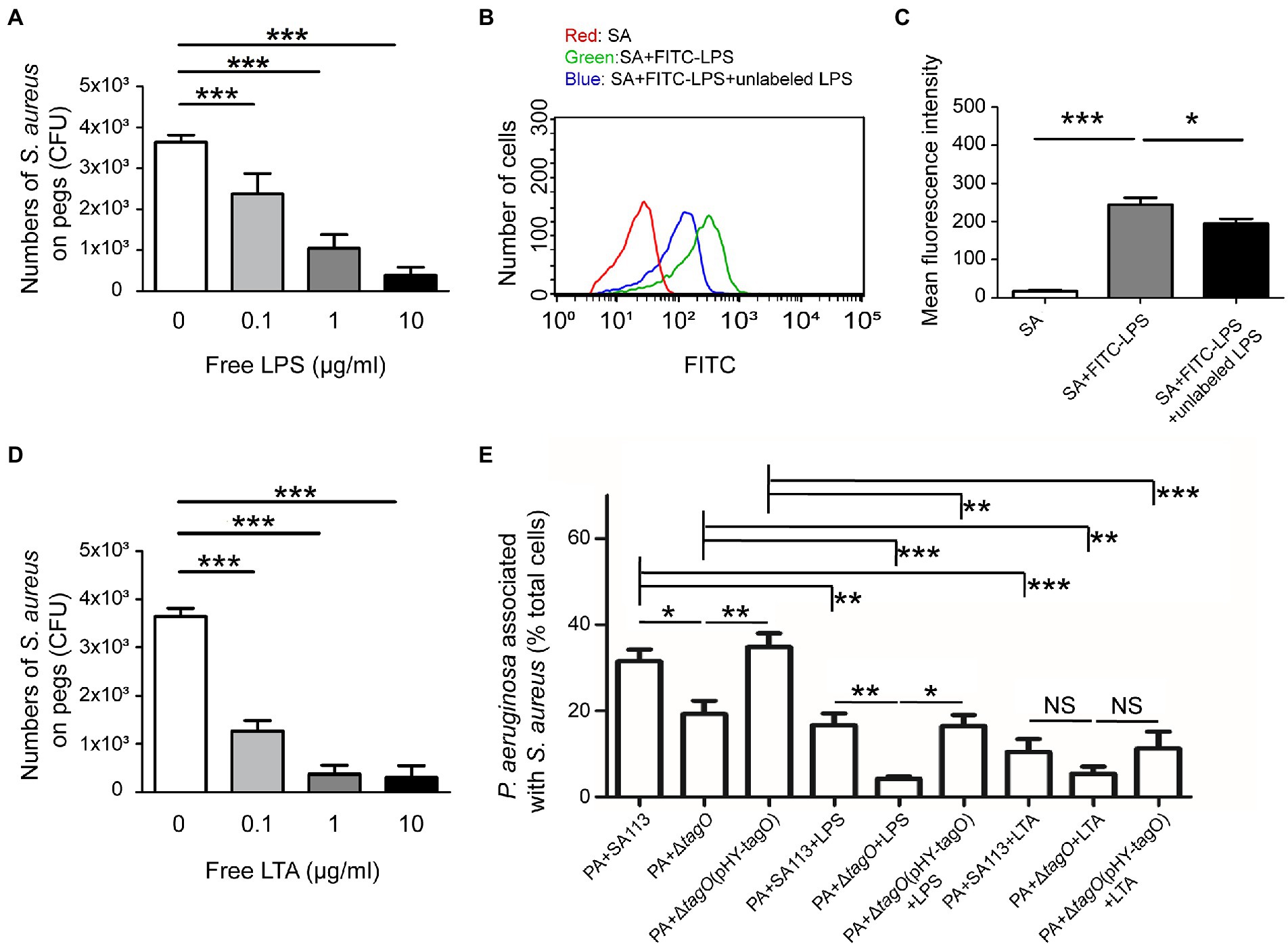
Figure 3. Interaction between WTA of Staphylococcus aureus and LPS of Pseudomonas aeruginosa. Staphylococcus aureus SA113 was mixed with various concentrations of free LPS (A) or LTA (D) for 10 min, and then mixed with P. aeruginosa PAO1. The mixtures were seeded into the wells of CBD plates. The number of S. aureus on pegs was determined. (B) Staphylococcus aureus SA113 (SA) was incubated alone (red) or with FITC-LPS in the absence (green) or presence (blue) of unlabeled LPS. The binding of FITC-LPS to S. aureus SA113 was analyzed by flow cytometry. The representative histogram of flow cytometry was shown. (C) Mean of fluorescence intensity of FITC-LPS bound S. aureus was measured and analyzed using Guavasoft 3.3. (E) Pseudomonas aeruginosa and S. aureus strains were labeled with SYTO 9 and hexidium iodide, respectively. Pseudomonas aeruginosa was incubated with S. aureus in the absence or presence of free LPS or LTA. After incubation, the cells were analyzed by flow cytometry. The data shown are the average percentage of P. aeruginosa associated with S. aureus in total analyzed P. aeruginosa. Bar presents as the mean of the results from more than three independent experiments and analyzed statistically using One-way ANOVA. Error bars indicate the standard deviations. Significant differences are denoted as follows: * indicates p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.001.
After demonstrating that P. aeruginosa PAO1 promotes the movement of S. aureus SA113 on surface, this study further investigated whether P. aeruginosa facilitates the movement of S. aureus in a C. elegans model. In this study, S. aureus SA113, the SA113 ΔtagO mutant and the mutant with pHY-tagO were transformed with pCtuf-ppmch to express mCherry (Mauthe et al., 2012). Meanwhile, P. aeruginosa PAO1 was stained with SYTO 9 to exhibit green fluorescence.
Caenorhabditis elegans was fed with bacteria and the movement of the bacteria in the digestive track was followed and imaged. The images revealed that when the worms were fed with S. aureus SA113, little movement of the bacteria was observed; the bacteria at the same location during an hour; the bacteria moved only 0.3 μm within an hour (Figures 4Aa–f, C). When the worms were fed a mixture of S. aureus SA113 and P. aeruginosa PAO1, S. aureus SA113 moved along with P. aeruginosa PAO1 with a distance of 6.0 μm (Figures 4Ba–f, C). When the worms were fed with a bacterial mixture containing the ΔtagO mutant of SA113 and P. aeruginosa PAO1, little movement of the mutant was observed; the mutant moved only 0.9 μm (Figures 4Bg–l, C). However, the mutant was found to co-migrate with P. aeruginosa PAO1 and reached a distance of 4.3 μm at 1 h after feeding if the mutant was transformed with pHY-tagO to complement the mutation (Figures 4Bm–r, C). Time-lapse video showed that S. aureus (red) moved together with P. aeruginosa (green; Supplementary Videos S1, S2). However, due to prolonged exposure to laser damaged the digestive tract and was lethal to the worm, this study was unable to collect the results beyond the 1 h period.
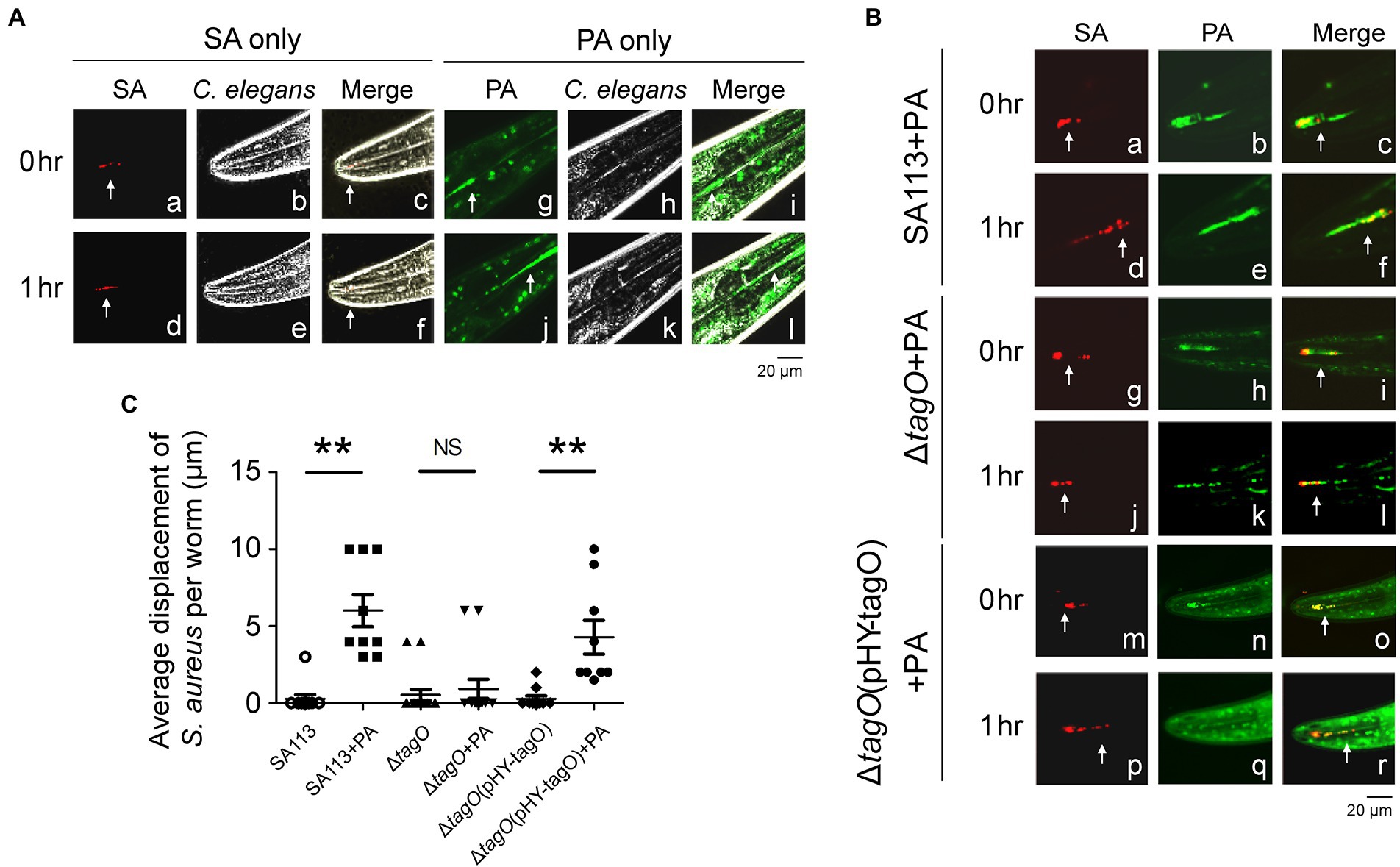
Figure 4. Pseudomonas aeruginosa promoted the movement of Staphylococcus aureus in Caenorhabditis elegans. (A) Caenorhabditis elegans were placed on TSA plates that had been inoculated with S. aureus SA113(pCtuf-ppmch; SA) or CYTO 9-labeled P. aeruginosa PAO1 (PA). (B) CYTO 9-labeled P. aeruginosa PAO1 (PA) was mixed with S. aureus SA113(pCtuf-ppmch) (a–f), ΔtagO(pCtuf-ppmch) (g–l) and ΔtagO(pHY-tagO, pCtuf-ppmch) (m–r) and inoculated on TSA plates. The worms were placed on the bacteria-containing agar, and fluorescence (white arrow) in the digestive tracts of C. elegans was imaged under a confocal laser-scanning microscope. Another image was obtained at the same location of the worm 1 h later. Autofluorescence (green) was observed in the C. elegans background. Phase-contrast microscopy images showed the shape of the worm. Merged images showed that bacteria were located inside C. elegans. The images shown are representative of each group (n ≥ 7 per group). (C) The distance of bacterial migration was measured using FV10-ASW 4.2 software. Data are presented as the mean of the results from each group and were analyzed statistically using Kruskal–Wallis test. Error bars indicate standard deviations. Significant differences (p < 0.01) are denoted as **. NS, no significance.
Staphylococcus aureus and P. aeruginosa are commonly associated with wound infection (DeLeon et al., 2014). To clarify whether P. aeruginosa promotes spreading of S. aureus in skin tissues, we used IVIS and observed the movement of S. aureus CGL1190, a strain exhibiting bioluminescence, in mice. The results showed that bioluminescence signals were present in the injection site, indicating that the bioluminescent S. aureus was useful in a mouse skin model (Figures 5Aa,c). Compared to inoculations with S. aureus alone, S. aureus spread farther when co-inoculated with P. aeruginosa (Figures 5Ab,d). To quantify the number of S. aureus moved away from the original injection site, the skin tissues at the inoculated areas and the areas 2 cm away were removed from the mice to enumerate the bacteria in the regions. The results showed that at 6 h after inoculation with S. aureus SA113 alone, the tissue in the area 2 cm away from the inoculation site contained only a few S. aureus, the average CFU was 3 (Figure 5B). In contrast, when inoculated with the bacterial mixture containing S. aureus SA113 and P. aeruginosa PAO1, the average CFU of S. aureus in the skin tissue 2 cm away from the inoculation site was 1.9 × 104 (Figure 5B). However, the average CFU of S. aureus was 1.5 × 103 CFU when inoculated with a mixture containing the ΔtagO mutant and P. aeruginosa PAO1 (Figure 5B). The results indicated that P. aeruginosa promotes the movement of S. aureus in the mouse skin model. The results also showed that WTA is required for hitchhiking P. aeruginosa by S. aureus SA113 in vivo. Furthermore, we plated 50 μl blood collected from the mice at 6 h post-inoculation, on mannitol salt agar plates and did not detected any S. aureus and P. aeruginosa in the blood. Additionally, our IVIS study revealed that the bioluminescence was largely confined near the tail, rather than the whole body suggesting that bacteria movement does not involve the dispersion of the bacteria via the circulatory system. The results excluded the possibility that the bacterial dissemination was due to the entry of the bacteria into the bloodstream.
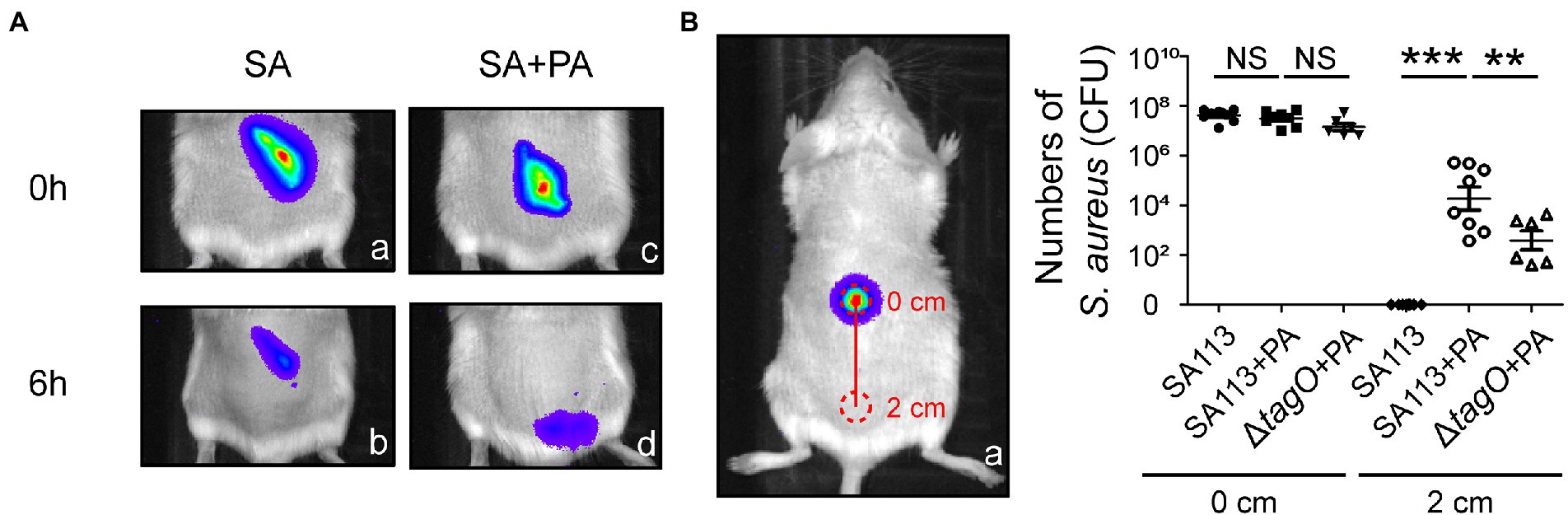
Figure 5. Movement of Staphylococcus aureus mediated by Pseudomonas aeruginosa in mice. (A) Mice were injected subcutaneously with bioluminescent S. aureus CGL1190 (SA) alone (a,b) or SA mixed with P. aeruginosa PAO1 (SA + PA) (c,d). Bioluminescence of SA in mice was observed and imaged at 0 and 6 h post-inoculation using an IVIS instrument. The images shown are representative of each group (n = 8 per group). (B) After observation, the mice were sacrificed, and skin tissue at areas of 0 and 2 cm (red circles) was removed. The number of S. aureus recovered from the skin tissues was determined. Data are presented as the mean of the results from each group and were analyzed statistically using One-way ANOVA. Error bars indicate the standard deviations. Significant differences are denoted as follows: ** indicates p < 0.01, and *** indicates p < 0.001. NS, no significance.
Motility is crucial for bacteria to travel in the environment. Due to the lack of pili and flagellum, S. aureus is nonmotile. Although S. aureus does not move actively, the bacterium uses a mechanism similar to that of B. subtilis swarming to extract water from agar medium and expresses biosurfactants, such as PSMs, to weaken the water surface tension, thereby facilitating colony spreading (Ke et al., 2015; Lin et al., 2016). Additionally, like B. subtilis, an S. aureus spreading colony often forms dendrites during spreading (Pollitt et al., 2015). An earlier study demonstrated that S. aureus associates with flagellated bacteria, such as P. aeruginosa, to facilitate hitchhiking motility (Samad et al., 2017). This study investigates how S. aureus hitchhikes P. aeruginosa for movement.
TagO is an enzyme required for the synthesis of WTA, which is a major surface component of Gram-positive bacteria (Brown et al., 2013). WTA of S. aureus is important to biofilm formation (Vergara-Irigaray et al., 2008) and colonizing the human skin (Burian et al., 2021). Moreover, a receptor for the binding of staphylococcal WTA has been identified on human nose epithelial cells (Baur et al., 2014). WTA also serves as key factors for recognition by bacteriophages (Eugster and Loessner, 2012). These studies indicated that WTA of S. aureus has a strong binding ability to different molecules. Indeed, our study found that tagO mutation decreased the hitchhiking motility of S. aureus prompted by P. aeruginosa (Figures 2, 3). In addition to WTA, we found that LTA, another important cell wall polymer found in Gram-positive bacteria, is also involved in interaction between P. aeruginosa and S. aureus (Figures 3D, E). However, whether LTA binds to LPS of P. aeruginosa remaining unclear. Nevertheless, involvement of LTA in binding to P. aeruginosa may explain why WTA deficiency alone does not completely abolish the binding of P. aeruginosa to S. aureus (Figure 2). We also examined whether factors other than WTA and LTA in S. aureus are involved in the interaction with P. aeruginosa. We found that a mutant strain of SA113 that is defective in fibronectin-binding protein (FnBP) synthesis (unpublished) interacts with P. aeruginosa PAO1 at a level comparable to the parental strain, showing that FnBP is uninvolved in the interaction (data not shown).
Both WTA and LTA of S. aureus are polyanionic polymers, modification of the backbone of the polymer with D-alanine, mono- or oligosaccharides has been shown to change surface properties of S. aureus, which provides structures for binding or interacting with a variety of receptors (van Dalen et al., 2020). D-alanylation is known to increase the positive charge of the polymers and confers them the zwitterionic properties, which are important for interacting with their binding partners (Collins et al., 2002; Weidenmaier et al., 2010). In addition to D-alanylation, glycosylation alters physiological properties of the polymers and influences their binding with extracellular molecules (Winstel et al., 2015; Gerlach et al., 2018). According to the results from flow cytometry analysis (Figure 3E), although the binding ability of S. aureus SA113 to P. aeruginosa PAO1 reduces after tagO mutation, the binding is restored if the mutation is complemented. Furthermore, adding free LTA decreases the binding between P. aeruginosa PAO1 and the tagO mutant (Figure 3E). The level of decrease is comparable to the reduction observed between P. aeruginosa PAO1 and S. aureus SA113 after adding LTA (Figure 3E), suggesting that WTA and LTA are two independent factors affecting the binding of S. aureus to P. aeruginosa. Therefore, we propose that D-alanylation increases the positive charge of WTA thus reducing the repulsion between WTA and LPS, which is also a polyanionic polymer on surface of Gram-negative bacteria. When WTA is near LPS, the interaction between glycan modification of WTA and LPS occurs.
LPS is a structure that functions similarly to WTA (Huszczynski et al., 2019). In addition to being a virulence factor, LPS is an important ligand for bacterial adhesion (Walker et al., 2004; Huszczynski et al., 2019). An earlier study showed that Bifidobacterium binds to LPS in E. coli (Park et al., 2007). In the present study, LPS supplementation competes for the binding of S. aureus to P. aeruginosa. Flow cytometry analysis also showed that WTA of S. aureus is required for binding to LPS. The results suggested that the binding of WTA of S. aureus to LPS of P. aeruginosa is the underlying mechanism for the hitchhiking motility of S. aureus. Although WTA and LPS are two major components involved in the interaction between S. aureus and P. aeruginosa, this study found that free LPS cannot completely abolish the binding of S. aureus to P. aeruginosa (Figure 3E), indicating that factors other than LPS on P. aeruginosa may be involved in interacting with S. aureus.
This study found that in addition to P. aeruginosa, other motile bacteria, such as E. coli, and even motile Gram-positive bacteria, such as B. subtilis and Listeria monocytogenes, also promote the hitchhiking motility of S. aureus, although less efficiently than P. aeruginosa (Supplementary Figure S1). Teichoic acids or related polymers are known to bind to cell wall proteins, S-layers, or mycolic acids in other Gram-positive bacteria (Jonquieres et al., 1999; Zoll et al., 2012). It is plausible that the interactions between Gram-positive bacteria involve these factors. However, adding free LTA into the cultures containing S. aureus and L. monocytogenes does not impair the hitchhiking motility of S. aureus (data not shown), indicating that LTA is unlikely involved in the interaction between S. aureus and L. monocytogenes. Furthermore, this study finds that these motile bacteria although support the hitchhiking motility of S. aureus SA113, do not fully assist the movement of the ΔtagO mutant of S. aureus (Supplementary Figure S1), indicating that WTA of S. aureus is a crucial factor associated with either Gram-negative or Gram-positive bacteria. The results suggested that S. aureus using WTA to hitchhike on motile bacteria is a general strategy for its dispersal.
Coinfection with P. aeruginosa and S. aureus is common in chronic lung infection and wound infections (Yung et al., 2021). A recent study showed that P. aeruginosa is capable of sensing S. aureus from a distance and initiates single-cell movement to invade the colony and eventually inhibits the growth of S. aureus. The study demonstrated that secreted factors controlled by the Agr quorum sensing system of S. aureus trigger the exploratory motility of P. aeruginosa toward S. aureus (Limoli et al., 2019). Due to S. aureus SA113 lacks the Agr quorum sensing system, the strain is unlikely to influence the movement of P. aeruginosa in our study. Furthermore, another single-cell study showed different results, which indicated that S. aureus are highly competitive against P. aeruginosa on surfaces (Niggli et al., 2021). The study demonstrated that when P. aeruginosa contacts with S. aureus, the contact enhances the growth of P. aeruginosa and activates the quorum sensing system of P. aeruginosa to inhibit S. aureus (Niggli et al., 2021). However, in our swarming and mouse studies, we did not observe the inhibition of S. aureus SA113 growth by P. aeruginosa PAO1 (Figures 2, 5), suggesting that interspecies interactions under co-culture conditions may not reflect the phenomena observed in the single-cell studies. However, several co-culture studies also showed that P. aeruginosa produces anti-staphylococcal compounds such as siderophores, rhamnolipids, and phenazines to inhibit the growth of S. aureus (Haba et al., 2003; Mashburn et al., 2005; Bharali et al., 2013; Borrero et al., 2014; Filkins et al., 2015); the mucoid P. aeruginosa produces exogenous alginate, which downregulates the expression of antistaphylococcal compounds, leading to protection of S. aureus from killing by P. aeruginosa (Limoli et al., 2017; Price et al., 2020). A transcriptomic study revealed that coexistence with P. aeruginosa alters the S. aureus transcriptome and its virulence (Briaud et al., 2019). These studies indicate that the interspecies interactions and coexisting mechanisms of P. aeruginosa and S. aureus are very complicate. The microorganisms change their moving behavior, gene expression and even lifestyle to achieve a competitive niche. Base on used bacterial strains, different culture conditions and study models, the conclusions derived from the studies may not be consistent. Despite the inconsistency, the studies, nevertheless, provide useful information on how bacteria use the interaction to survive in an ecosystem.
In this study, we showed that S. aureus hitchhikes flagellated P. aeruginosa to spread by interaction between WTA of S. aureus and LPS of P. aeruginosa. The hitchhiking motility promotes the migration of S. aureus in a C. elegans model as well as in mice, showing that the motility occurs not only in culture but also in animals. The overall results demonstrated how a mobile bacterium carries another motionless pathogen to spread in the environment and hosts in which provide useful information for developing strategy to treatment of chronic polymicrobial infections.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Chang Gung University Animal Care Committee.
M-HL and C-CL conceived and designed the study, performed the experiments, analyzed the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from Chang Gung Memorial Hospital (CMRPD1F0061-3 and CMRPD1J0271-3) and the Ministry of Science and Technology (MOST), Taiwan (MOST 108-2320-B-182-028).
We thank Dr. Friedrich Götz for providing the Staphylococcus aureus strains SA113 and SA113ΔtagO, Dr. Hwan-You Chang for providing the Pseudomonas aeruginosa strain ΔfliA, and Roger D. Plaut for providing the pRP1190 plasmid. The authors would like to thank the Microscopy Core Laboratory at the Linkou Chang Gung Memorial Hospital (CGMH), Taiwan, for microscopy support. We also thank Dr. Po-Hsiang Chen for the technical assistance in the Caenorhabditis elegans experiments. We would also like to thank Shih-Tung Liu for his critiques.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1068251/full#supplementary-material
Baur, S., Rautenberg, M., Faulstich, M., Grau, T., Severin, Y., Unger, C., et al. (2014). A nasal epithelial receptor for Staphylococcus aureus WTA governs adhesion to epithelial cells and modulates nasal colonization. PLoS Pathog. 10:e1004089. doi: 10.1371/journal.ppat.1004089
Bharali, P., Saikia, J. P., Ray, A., and Konwar, B. K. (2013). Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: a novel chemotaxis and antibacterial agent. Colloids Surf. B Biointerfaces 103, 502–509. doi: 10.1016/j.colsurfb.2012.10.064
Biswas, L., Biswas, R., Schlag, M., Bertram, R., and Gotz, F. (2009). Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75, 6910–6912. doi: 10.1128/AEM.01211-09
Borrero, N. V., Bai, F., Perez, C., Duong, B. Q., Rocca, J. R., Jin, S., et al. (2014). Phenazine antibiotic inspired discovery of potent bromophenazine antibacterial agents against Staphylococcus aureus and Staphylococcus epidermidis. Org. Biomol. Chem. 12, 881–886. doi: 10.1039/c3ob42416b
Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. doi: 10.1093/genetics/77.1.71
Briaud, P., Camus, L., Bastien, S., Doleans-Jordheim, A., Vandenesch, F., and Moreau, K. (2019). Coexistence with Pseudomonas aeruginosa alters Staphylococcus aureus transcriptome, antibiotic resistance and internalization into epithelial cells. Sci. Rep. 9:16564. doi: 10.1038/s41598-019-52975-z
Brown, S., Santa Maria, J. P. Jr., and Walker, S. (2013). Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336. doi: 10.1146/annurev-micro-092412-155620
Burian, M., Plange, J., Schmitt, L., Kaschke, A., Marquardt, Y., Huth, L., et al. (2021). Adaptation of Staphylococcus aureus to the human skin environment identified using an ex vivo tissue model. Front. Microbiol. 12:728989. doi: 10.3389/fmicb.2021.728989
Collins, L. V., Kristian, S. A., Weidenmaier, C., Faigle, M., Van Kessel, K. P., Van Strijp, J. A., et al. (2002). Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J Infect Dis 186, 214–219. doi: 10.1086/341454
DeLeon, S., Clinton, A., Fowler, H., Everett, J., Horswill, A. R., and Rumbaugh, K. P. (2014). Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 82, 4718–4728. doi: 10.1128/IAI.02198-14
Eugster, M. R., and Loessner, M. J. (2012). Wall teichoic acids restrict access of bacteriophage endolysin Ply118, Ply511, and PlyP40 cell wall binding domains to the listeria monocytogenes peptidoglycan. J. Bacteriol. 194, 6498–6506. doi: 10.1128/JB.00808-12
Filkins, L. M., Graber, J. A., Olson, D. G., Dolben, E. L., Lynd, L. R., Bhuju, S., et al. (2015). Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J. Bacteriol. 197, 2252–2264. doi: 10.1128/JB.00059-15
Finkelshtein, A., Roth, D., Ben Jacob, E., and Ingham, C. J. (2015). Bacterial swarms recruit cargo bacteria to pave the way in toxic environments. MBio 6, e00074–e00015. doi: 10.1128/mBio.00074-15
Gerlach, D., Guo, Y., De Castro, C., Kim, S. H., Schlatterer, K., Xu, F. F., et al. (2018). Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity. Nature 563, 705–709. doi: 10.1038/s41586-018-0730-x
Ghoul, M., and Mitri, S. (2016). The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845. doi: 10.1016/j.tim.2016.06.011
Haba, E., Pinazo, A., Jauregui, O., Espuny, M. J., Infante, M. R., and Manresa, A. (2003). Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol. Bioeng. 81, 316–322. doi: 10.1002/bit.10474
Hagai, E., Dvora, R., Havkin-Blank, T., Zelinger, E., Porat, Z., Schulz, S., et al. (2014). Surface-motility induction, attraction and hitchhiking between bacterial species promote dispersal on solid surfaces. ISME J. 8, 1147–1151. doi: 10.1038/ismej.2013.218
Herbert, S., Ziebandt, A. K., Ohlsen, K., Schafer, T., Hecker, M., Albrecht, D., et al. (2010). Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 78, 2877–2889. doi: 10.1128/IAI.00088-10
Hoffman, L. R., Deziel, E., D'Argenio, D. A., Lepine, F., Emerson, J., McNamara, S., et al. (2006). Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 103, 19890–19895. doi: 10.1073/pnas.0606756104
Hu, Y., Niu, Y., Ye, X., Zhu, C., Tong, T., Zhou, Y., et al. (2021). Staphylococcus aureus synergized with Candida albicans to increase the pathogenesis and drug resistance in cutaneous abscess and peritonitis murine models. Pathogens 10:1036. doi: 10.3390/pathogens10081036
Huszczynski, S. M., Lam, J. S., and Khursigara, C. M. (2019). The role of Pseudomonas aeruginosa lipopolysaccharide in bacterial pathogenesis and physiology. Pathogens 9:6. doi: 10.3390/pathogens9010006
Jonquieres, R., Bierne, H., Fiedler, F., Gounon, P., and Cossart, P. (1999). Interaction between the protein InlB of listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol. 34, 902–914. doi: 10.1046/j.1365-2958.1999.01652.x
Kaito, C., and Sekimizu, K. (2007). Colony spreading in Staphylococcus aureus. J. Bacteriol. 189, 2553–2557. doi: 10.1128/JB.01635-06
Ke, W. J., Hsueh, Y. H., Cheng, Y. C., Wu, C. C., and Liu, S. T. (2015). Water surface tension modulates the swarming mechanics of Bacillus subtilis. Front. Microbiol. 6:1017. doi: 10.3389/fmicb.2015.01017
Limoli, D. H., Warren, E. A., Yarrington, K. D., Donegan, N. P., Cheung, A. L., and O'Toole, G. A. (2019). Interspecies interactions induce exploratory motility in Pseudomonas aeruginosa. Elife 8:e47365. doi: 10.7554/eLife.47365
Limoli, D. H., Whitfield, G. B., Kitao, T., Ivey, M. L., Davis, M. R. Jr., Grahl, N., et al. (2017). Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. MBio 8:e00186-17. doi: 10.1128/mBio.00186-17
Lin, M. H., Ke, W. J., Liu, C. C., and Yang, M. W. (2016). Modulation of Staphylococcus aureus spreading by water. Sci. Rep. 6:25233. doi: 10.1038/srep25233
Lo, Y. L., Shen, L., Chang, C. H., Bhuwan, M., Chiu, C. H., and Chang, H. Y. (2016). Regulation of motility and phenazine pigment production by FliA is cyclic-di-GMP dependent in Pseudomonas aeruginosa PAO1. PLoS One 11:e0155397. doi: 10.1371/journal.pone.0155397
Mashburn, L. M., Jett, A. M., Akins, D. R., and Whiteley, M. (2005). Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187, 554–566. doi: 10.1128/JB.187.2.554-566.2005
Matilla, M. A., and Krell, T. (2018). The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol. Rev. 42, 40–67. doi: 10.1093/femsre/fux052
Mauthe, M., Yu, W., Krut, O., Kronke, M., Gotz, F., Robenek, H., et al. (2012). WIPI-1 positive autophagosome-like vesicles entrap pathogenic Staphylococcus aureus for lysosomal degradation. Int. J. Cell Biol. 2012:179207. doi: 10.1155/2012/179207
Nair, D., Memmi, G., Hernandez, D., Bard, J., Beaume, M., Gill, S., et al. (2011). Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J. Bacteriol. 193, 2332–2335. doi: 10.1128/JB.00027-11
Niggli, S., Wechsler, T., and Kummerli, R. (2021). Single-cell imaging reveals that Staphylococcus aureus is highly competitive against Pseudomonas aeruginosa on surfaces. Front. Cell. Infect. Microbiol. 11:733991. doi: 10.3389/fcimb.2021.733991
Park, M. S., Kim, M. J., and Ji, G. E. (2007). Assessment of lipopolysaccharide-binding activity of Bifidobacterium and its relationship with cell surface hydrophobicity, autoaggregation, and inhibition of interleukin-8 production. J. Microbiol. Biotechnol. 17, 1120–1126.
Pastar, I., Nusbaum, A. G., Gil, J., Patel, S. B., Chen, J., Valdes, J., et al. (2013). Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8:e56846. doi: 10.1371/journal.pone.0056846
Pernet, E., Guillemot, L., Burgel, P. R., Martin, C., Lambeau, G., Sermet-Gaudelus, I., et al. (2014). Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Nat. Commun. 5:5105. doi: 10.1038/ncomms6105
Plaut, R. D., Mocca, C. P., Prabhakara, R., Merkel, T. J., and Stibitz, S. (2013). Stably luminescent Staphylococcus aureus clinical strains for use in bioluminescent imaging. PLoS One 8:e59232. doi: 10.1371/journal.pone.0059232
Pollitt, E. J., Crusz, S. A., and Diggle, S. P. (2015). Staphylococcus aureus forms spreading dendrites that have characteristics of active motility. Sci. Rep. 5:17698. doi: 10.1038/srep17698
Pollitt, E. J. G., and Diggle, S. P. (2017). Defining motility in the staphylococci. Cell. Mol. Life Sci. 74, 2943–2958. doi: 10.1007/s00018-017-2507-z
Price, C. E., Brown, D. G., Limoli, D. H., Phelan, V. V., and O'Toole, G. A. (2020). Exogenous alginate protects Staphylococcus aureus from killing by Pseudomonas aeruginosa. J. Bacteriol. 202:e00559-19. doi: 10.1128/JB.00559-19
Samad, T., Billings, N., Birjiniuk, A., Crouzier, T., Doyle, P. S., and Ribbeck, K. (2017). Swimming bacteria promote dispersal of non-motile staphylococcal species. ISME J. 11, 1933–1937. doi: 10.1038/ismej.2017.23
Todd, O. A., Fidel, P. L. Jr., Harro, J. M., Hilliard, J. J., Tkaczyk, C., Sellman, B. R., et al. (2019). Candida albicans augments Staphylococcus aureus virulence by engaging the staphylococcal agr quorum sensing system. MBio 10:e00910-19. doi: 10.1128/mBio.00910-19
Tsompanidou, E., Denham, E. L., Becher, D., de Jong, A., Buist, G., van Oosten, M., et al. (2013). Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces. Appl. Environ. Microbiol. 79, 886–895. doi: 10.1128/AEM.03157-12
Tsompanidou, E., Sibbald, M. J., Chlebowicz, M. A., Dreisbach, A., Back, J. W., van Dijl, J. M., et al. (2011). Requirement of the agr locus for colony spreading of Staphylococcus aureus. J. Bacteriol. 193, 1267–1272. doi: 10.1128/JB.01276-10
van Dalen, R., Peschel, A., and van Sorge, N. M. (2020). Wall teichoic acid in Staphylococcus aureus host interaction. Trends Microbiol. 28, 985–998. doi: 10.1016/j.tim.2020.05.017
Vergara-Irigaray, M., Maira-Litran, T., Merino, N., Pier, G. B., Penades, J. R., and Lasa, I. (2008). Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology 154, 865–877. doi: 10.1099/mic.0.2007/013292-0
Walker, S. L., Redman, J. A., and Elimelech, M. (2004). Role of cell surface lipopolysaccharides in Escherichia coli K12 adhesion and transport. Langmuir 20, 7736–7746. doi: 10.1021/la049511f
Weidenmaier, C., Kokai-Kun, J. F., Kristian, S. A., Chanturiya, T., Kalbacher, H., Gross, M., et al. (2004). Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10, 243–245. doi: 10.1038/nm991
Weidenmaier, C., McLoughlin, R. M., and Lee, J. C. (2010). The zwitterionic cell wall teichoic acid of Staphylococcus aureus provokes skin abscesses in mice by a novel CD4+ T-cell-dependent mechanism. PLoS One 5:e13227. doi: 10.1371/journal.pone.0013227
Winstel, V., Kuhner, P., Salomon, F., Larsen, J., Skov, R., Hoffmann, W., et al. (2015). Wall teichoic acid glycosylation governs Staphylococcus aureus nasal colonization. MBio 6:e00632. doi: 10.1128/mBio.00632-15
Yung, D. B. Y., Sircombe, K. J., and Pletzer, D. (2021). Friends or enemies? The complicated relationship between Pseudomonas aeruginosa and Staphylococcus aureus. Mol. Microbiol. 116, 1–15. doi: 10.1111/mmi.14699
Keywords: Staphylococcus aureus, Pseudomonas aeruginosa, TagO, teichoic acids, lipopolysaccharide, hitchhiking motility
Citation: Liu C-C and Lin M-H (2023) Hitchhiking motility of Staphylococcus aureus involves the interaction between its wall teichoic acids and lipopolysaccharide of Pseudomonas aeruginosa. Front. Microbiol. 13:1068251. doi: 10.3389/fmicb.2022.1068251
Received: 12 October 2022; Accepted: 13 December 2022;
Published: 05 January 2023.
Edited by:
Chih-Horng Kuo, Academia Sinica, TaiwanReviewed by:
Rolf Kümmerli, University of Zurich, SwitzerlandCopyright © 2023 Liu and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei-Hui Lin, ✉ dGhlYUBtYWlsLmNndS5lZHUudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.