
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 21 December 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1066641
This article is part of the Research TopicExploring the Role of Microorganisms in Silages: Species, Communities, Interactions, and Functional CharacteristicsView all 35 articles
Objective: The objective of this study was to determine the effect of Lactobacillus plantarum (L. plantarum) and propionic acid (PA) on the microbial community and fermentation performance of high-moisture amaranth silage.
Methods: Amaranth silages were rown without addition (AhGCK) as a control and with L. plantarum JYLP-002 (AhGLP) or propionic acid (AhGPA) and then were opened after 60 days of ensiling to determine the microbial community and fermentation quality.
Results: Crude protein (CP) content, lactic acid (LA) content, and lactic acid bacteria (LAB) counts were significantly higher in AhGLP and AhGPA compared with those in AhGCK (p < 0.05). In contrast, pH, acetic acid (AA) content, and yeast and aerobic bacteria counts were significantly lower in AhGLP and AhGPA compared with those in AhGCK (p < 0.05). In addition, propionic acid (PA) levels were markedly higher in AhGPA (p < 0.05). In terms of microbial communities, the silage in the additive groups showed an increased relative abundance of Lactiplantibacillus plantarum and Lentilactobacillus buchneri and a reduced relative abundance of Enterobacter cloacae and Clostridium tyrobutyricum. The abundance of Xanthomonas oryzae was significantly increased in AhGPA, but completely inhibited in the silage supplemented with L. plantarum. Spearman’s correlation analysis revealed that Lentilactobacillus buchneri and Levilactobacillus brevis were positively associated with LA and negatively associated with pH. Conversely, Clostridium tyrobutyricum and Enterobacter cloacae were negatively associated with LA, but positively associated with pH and AA content. AA content was inversely correlated with Lentilactobacillus buchneri. Functional prediction analysis showed that LAB dominated the three groups of silage and the silages containing additives had improved carbohydrate and amino acid metabolism compared with the control silage; in particular, the AhGLP group had more heterotypic fermentation processes and a richer metabolic pathway. Furthermore, the epiphytic Lactiplantibacillus plantarum and Lentilactobacillus buchneri could inhibit the reproductive activity of undesirable microorganisms to a certain extent, thus slowing the spoilage process of the silage.
Conclusion: In conclusion, L. plantarum can improve fermentation characteristics by modulating the microbial community attached to high-moisture amaranth silage and will prove useful for preserving high-moisture silage.
Modern livestock systems have an enormous impact on the environment. Designing sustainable and intensive livestock production systems is a promising way to effectively reduce feed costs and achieve local access to local feed resources (Herrero et al., 2015). Silage storage technology is widely used to increase the productivity of animals and improve the efficiency of the livestock system when forage supplies are insufficient to maintain the productive performance of ruminants (Wilkinson et al., 2020). The main principle of silage fermentation is the production of lactic acid (LA) by fresh crop-attached lactic acid bacteria (LAB) under anaerobic conditions using water-soluble carbohydrates (WSC) as a fermentation substrate, which in turn creates an acidic environment and inhibits spoilage microorganisms (Dong et al., 2019). In China, the predominant limiting factor for livestock production is the shortage of fodder, which primarily occurs during the dry season. Consequently, Consequently, it is crucial that new, readily available, feed resources are developed to deal with such feed shortages (Du et al., 2021). Amaranth (Amaranthus hypochondriaus), an annual herb that has been cultivated for 8,000 years, may be a potential protein-feed resource. In recent years, some amaranth species have been used as forage crops owing their strong environmental adaptability (Rezaei et al., 2014; Peiretti, 2018). A. hypochondriaus is a C4 plant capable of producing 16.7 t/ha of dry matter (DM). In addition, it is one of the preferred feeds for sheep and beef and dairy cattle (Rahjerdi et al., 2015). Fresh grass yields of 86.4 t/ha and hay yields of 13.2 t/ha has been reported for amaranth (Abbasi et al., 2018). In addition, amaranth had a higher CP content (285 g/kg DM) and lower lignin content (40 g/kg DM) and lower nitrate and oxalate acid contents compared with those of maize (Rezaei et al., 2009; Rahjerdi et al., 2015). However, the high moisture and protein content of amaranth make it prone to spoilage. Exploring effective ways of preserving high-moisture amaranth is therefore crucial to develop this plant for use as forage in the livestock industry.
Silage quality can be directly improved by the administration of organic acids. Previous studies have compared the effects of formic acid (FA) and propionic acid (PA) on maize silage, with PA additives retaining more nutrients in maize silage and being able to be sprayed onto the forage, thus proving more beneficial than FA (Gheller et al., 2021). The PA additive inhibits yeasts and molds and can improve the aerobic stability of silage without additives (Kung et al., 2003; Coblentz and Akins, 2021). Lactobacillus inoculants have also been widely used in the silage fermentation process to produce high-quality silage. The inoculants were classified as homogeneous and heterogeneous fermenters based on their fermentation characteristics (Muck et al., 2018). Homogeneous LAB, represented by L. plantarum, grow and multiply rapidly. In the early stages of fermentation, they quickly produce LA, which lowers the pH of the system. This swift formation of an acidic environment in the fermentation system inhibits the growth and reproduction of harmful micro-organisms and effectively maintains feed nutrition (Ranjit and Kung, 2000). Researchers have inoculated L. plantarum in whole crop corn silage and Moringa oleifera leaf silage (Wang Y. et al., 2018), and sugarcane silage (Rabelo et al., 2019) and found that sugarcane and Moringa oleifera leaves exhibited better fermentation was achieved without the addition of L. plantarum. However, L. plantarum was highly effective as an inoculant for a high moisture amaranth and straw mixed silage. Compared with mixed silage without additives, L. plantarum increased the relative abundance of Lactobacillus, decreased the relative abundance of Weissella, Pediococcus and Lactococcus, decreased pH, and acetic acid (AA) and NH3-N concentrations, and increased concentration of LA (Mu et al., 2020). All types of microbial communities and biochemical reactions are involved in the ensiling process with fermentation quality predominantly dependent on the microbial community and its dynamic succession and fermentation metabolites (Yang et al., 2019). Consequently, obtaining detailed knowledge of the microbial community in silage is crucial to improve the quality of silage, especially in conditions of high moisture. In recent years, methods have been developed that allow the analysis of microbial communities in a culture-independent manner to circumvent the limitations of traditional culture methods (Ercolini, 2004). Next-generation sequencing (NGS) technology was employed to explore silage bacterial communities in the current study.
We hypothesized that inoculation of L. plantarum and PA additives during silage preparation might alter the bacterial community and succession patterns and affect fermentation performance in high-moisture seed amaranth silage. The effect of L. plantarum and PA on silage was investigated in terms of chemical composition, fermentation characteristics and bacterial community in conjunction with NGS techniques.
The test material was red-fruited amaranth (Amaranthus hypochondriaus), which was planted on June 9, 2021 at the forage trial site of Inner Mongolia Agricultural University (111°430 E, 40°480 N, altitude 1,056 m above sea level) Hohhot, Inner Mongolia. Three 1-m2 plots were selected for harvesting (mature stage) on August 10. The collected red fruit amaranth was placed on a clean plastic sheet and was cut into lengths of approximately 3 cm using a hand-held guillotine (Mode-8,200; Minghong Business, Shandong, China).
For inoculant preparation, Lactobacillus plantarum (L. plantarum) JYLP-002 was supplied by Shandong Zhongke Jiayi Biological Engineering Company (Shandong, China). PA was manufactured by Shanghai McLean Biochemical Technology Company (Shanghai, China). Three treatment groups were established with the prepared amaranth: (1) no additive control (AhGCK); (2) L. plantarum JYLP-002 (AhGLP), applied into the fresh forage at a rate of 1 × 109 cfu L. plantarum/g fresh matter (FM); (3) PA (AhGPA), added at 4 g/kg. The two additives were dissolved in deionized water, and 10 mL of solution per kg of amaranth forage was applied using a manual sprayer. The control silage was sprayed with an equivalent volume of deionized water. The amaranth (500 g) were packed in polyethylene plastic bags (size: 300 mm × 400 mm; Shenyang Huasheng Plastic Packaging Products Co., Ltd., Shenyang, China), which were then vacuum sealed (DZ400/2D vacuum sealer; Wenzhou Dafeng Machinery Co., Ltd., Wenzhou, China) and ensiled at ambient temperature (24–30°C) for 60 days. Chemical and fermentation characteristics, microbial populations, and microbial community of the control (AhGCK), L. plantarum (AhGLP), and PA (AhGPA) silages were then measured.
Each sample of fresh amaranth silage was replicated. Dry matter (DM) content was measured following the method of Zhang et al. (2016). The crude protein (CP) content was measured according to the method of Patrica (1997), utilizing on a Kjeldahl nitrogen tester (Gehart Vapodest 50 s, Germany). Neutral detergent fiber (NDF) and acid detergent fiber (ADF) content were measured using an Ankom A2000i fiber analyzer (Ankom Technology, Macedon, NY, USA) and the method of Van Soest (Van Soest et al., 1991). The WSC content was measured following the method of Chen et al. (2015).
A sample of silage (10 g) was mixed with 90 g deionized water following the description of Cai (2004) and stored in a refrigerator at 4°C for 24 h. The leachate was filtered through four layers of gauze and filter paper and the pH, ammonia nitrogen (NH3-N), and organic acids of the leachate were measured. pH was measured using a glass electrode pH meter (Leici pH S-3C, Shanghai, China). LA, AA, PA, and butyric acid (BA) contents of the silage were determined by high-performance liquid chromatography (HPLC; Model: Waters e2695, Milford, CT, USA) according to the method of Cheng et al. (2021). The method of Broderick and Kang (1980) was used to determine ammoniacal nitrogen (NH3-N) concentrations. Microbial populations (LAB, yeasts, mold, anerobic bacteria, and coliform bacteria) in the FM were assessed as described in a previous report (You et al., 2021).
The bacterial community compositions of ensiled high-moisture amaranth fermented for 60 days were analyzed by using 16S rRNA gene sequencing. Total DNA was extracted from the amaranth silage samples according to Liu et al. (2019). The procedures of metagenomic DNA extraction and PCR amplification of the bacterial 16S ribosomal RNA gene were performed according to Guo et al. (2021). Briefly, DNA was amplified with primers 27F (5′-AGRGTTTGATYNTGGCTCAG-3′) and 1492R (5′-TASGGHTACCTTGTTASGACTT-3′). PCR conditions were an initial denaturation at 98°C for 2 min, 30 cycles of denaturation at 98°C for 30 s, annealing at 50°C for 30 s, and elongation at 72°C for 60 s, followed by a final extension at 72°C for 5 min. The PCR products were purified for sequencing and analysis. Each treatment was performed in triplicate. DNA was amplified with primers 27F (5′-AGRGTTTGATYNTGGCTCAG-3′) and 1492R (5′-TASGGHTACCTTGTTASGACTT-3′). PCR conditions were an initial denaturation at 98°C for 2 min, 30 cycles of denaturation at 98°C for 30 s, annealing at 50°C for 30 s, and elongation at 72°C for 60 s, followed by a final extension at 72°C for 5 min. The PCR products were purified for sequencing and analysis. Each treatment was performed in triplicate.
Next-generation sequencing (NGS) sequencing was performed by Biomarker Technologies (Beijing, China) on a Pacbio_SMRT platform (Pacbio Sequel II, CA, USA). Coverage of alpha-diversity indicators Chao1 and Good was calculated using QIIME v1.9.1 (Caporaso et al., 2010). Principal coordinate analysis (PCoA) was performed using the R program (version 3.2.5) on the basis of beta-diversity unweighted or weighted unifrac distances. Operational taxonomic units (OTUs) were classified using Ribosome Database Project (RDP) Classifier (version 2.2) against the SILVA (Release 128) 16S rRNA database with a minimum confidence cutoff of 0.7, and were then denominated at the phylum, genus, and species levels. Mothur (version v.1.30) software was used to evaluate the alpha-diversity indices (ACE, Chao 1, Simpson, and Shannon) of the samples. A heat map of correlation analyses was produced using a R-based statistics tool. LEfSe [Linear Discriminant Analysis (LDA) effect size] was able to find biomarkers that are statistically different between groups (Segata et al., 2011). It was performed using a free online platform.1 Phylogenetic Investigation of Communities by Reconstruction of Unobserved States II (PICRUSt2) software was used to predict microbial functions from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The PICRUSt2 software was used to analyze functional differences between different samples or subgroups by annotating the characteristic sequences to be predicted with the species in the phylogenetic tree available in the software and using the IMG microbial genomic data to output functional information and thus extrapolate the functional gene composition of the samples (Ward et al., 2017).
One-way analyses of variance (ANOVA) based on SAS’ General Linear Model (GLM) program (version 9.3; SAS Institute Inc., Cary, NC, United States) were conducted on fermentation and nutritional characteristics and microbial counts of fresh and silage amaranth. Effects were considered significant when p < 0.05. The graphs of microbial community data were created using the BMK Cloud online platform and GraphPad prism 8.
The chemical composition and microbial counts of the raw material are presented in Table 1. The raw material had a low DM content (21.00%) and a CP content of 6.94% DM, which was lower than that reported by Mu et al. (2020), where the CP content of fresh amaranth was 91.9 g/kg at 136 g/kg DM. The low DM content might affect silage production, as high moisture content previously led to nutrient losses and markedly increased clostridial fermentation (He et al., 2020b). The NDF and ADF contents of the feedstock were 46.37% DM and 33.22% DM, respectively. The WSC content was only 1.18% DM. This might be due to the fact that amaranth is a thermophilic plant and the amaranth variety in this study was grown in a different environment with a large diurnal temperature difference, which affected its growth and nutrient accumulation. The WSC content and epiphytic LAB count of the material are another two determinants of fermentation quality (Dong et al., 2020). Cai et al. (1999) indicated that a minimum of 105 cfu/g of LAB was required for the storage of well-preserved silage. Zhang et al. (2016) suggested that the minimum WSC level for successful fermentation is 60 g/kg DM. The amount of LAB in the fresh material was 3.84 log10 cfu/g FM in the current study. Based on the criterion, fresh amaranth failed to meet the requirement. However, the high number of coliforms (2.73 log10 cfu/g FM), yeasts (4.55 log10 cfu/g FM), and molds (2.00 log10 cfu/g FM) in the fresh material were lower than the bacterial counts of harmful microorganisms attached to rice straw, Leymus chinensis, and paper mulberry ingredients by previous studies (He et al., 2020a; Wu B. et al., 2022; Wu C. et al., 2022). Thus, silage additives like LAB were subsequently found to be indispensable in the preparation of silage.
The chemical and fermentation characteristics and microbial populations of the silage at 60 days are shown in Table 2. A significantly higher CP content was found in AhGPA compared with that in AhGLP and AhGCK (p < 0.05). The addition of PA reduced the degradation of CP in silage and avoided nutrient losses during ensiling. The additive groups had lower levels of NDF and ADF compared with the AhGCK group (p > 0.05). The growth of objectionable microorganisms and the accumulation of NH3-N illustrated by Dong et al. (2020) were predominantly manifested by the low CP content and high NH3-N content of AhGCK, while the additive groups performed well. This was coincident with low pH inhibiting the growth and activity of undesirable microorganisms (Arriola et al., 2011; Heinritz et al., 2012).
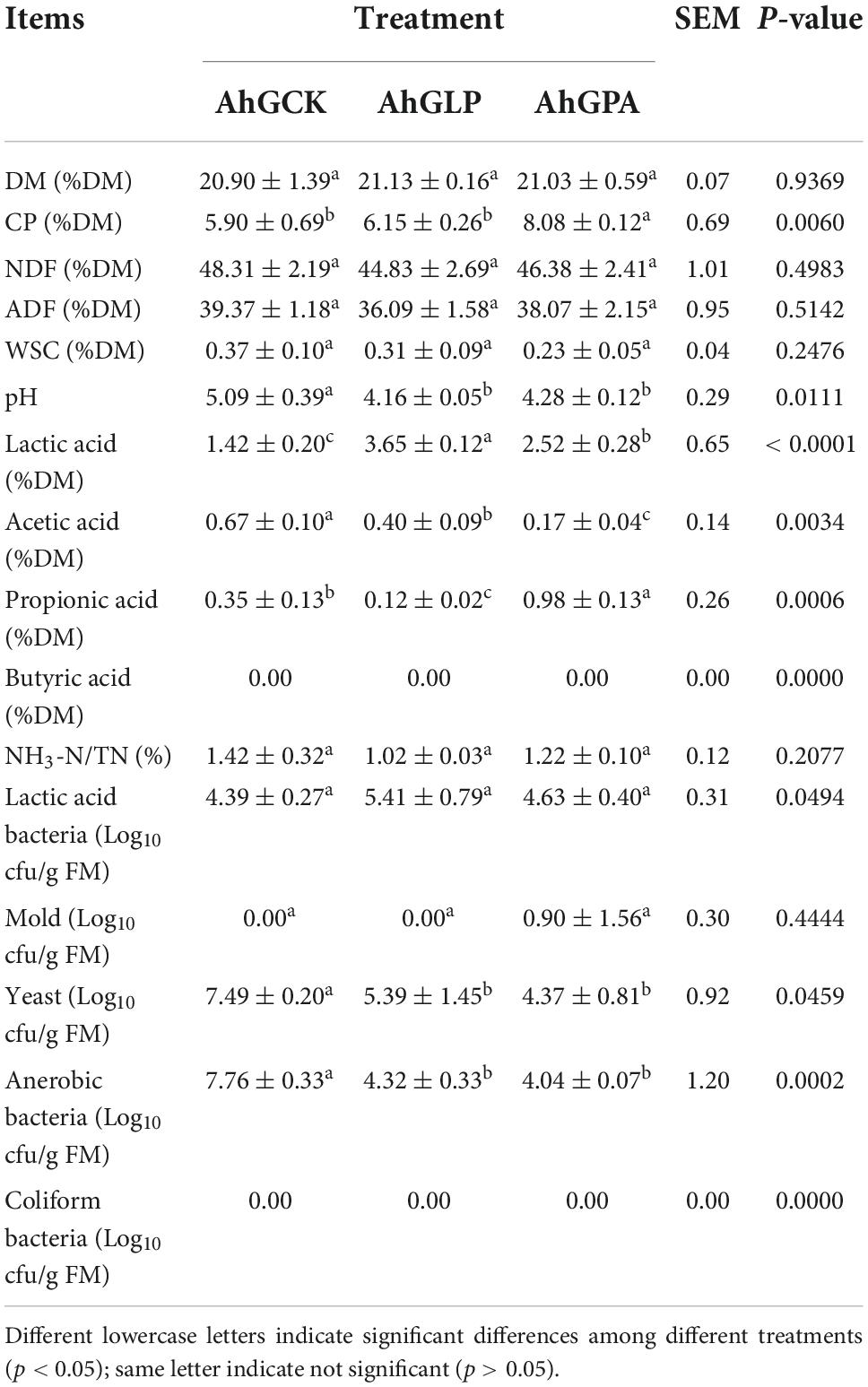
Table 2. Chemical compositions, fermentation characteristics, and microbial populations on 60 days of ensiling.
Compared with the control silage, the inoculated silage had a lower WSC content, which was congruent with the findings of Kleinschmit and Kung (2006). During the fermentation process, in response to LAB fermentation, WSC is converted to organic acids, ethanol, and carbon dioxide (Muck et al., 2018; Wang S. et al., 2018). A high moisture content was previously reported to be detrimental to natural fermentation and led to a decrease in organic acid concentration and an increase in pH, which adversely affected the fermentation process of alfalfa silage (Cai et al., 1998; Zheng et al., 2017). Both AhGLP and AhGPA had a significantly lower pH compared with AhGCK (p < 0.05); in particular, the pH of AhGLP was lower than 4.20. These results were congruent with the conclusions of Wang et al. (2019), who considered that LAB accelerated the accumulation of LA and lowered the pH of silage. Yang et al. (2019) also concluded that the growth of harmful bacteria was effectively inhibited when the pH was below 4.20, ensuring quality fermentation, and that the addition of LAB accelerated the LA fermentation process of alfalfa silage. In contrast, the pH of AhGCK in our study was as high as 5.09. This can potentially be attributed to the high metabolism of LA and other nutrients consumed by yeasts, molds, and other aerobic microorganisms, which results in a rise in pH (Woolford, 1990). Escherichia coli and molds were undetectable after 60 days of fermentation in high-moisture amaranth silages, congruent with the report of Dong et al. (2020). This was due to the low pH suppressing the activities of adverse microorganisms (Lima et al., 2010; Reich and Kung, 2010).
Lactobacillus plantarum JYLP-002 (AhGLP) had significantly higher LA content compared with AhGCK and AhGPA, but AhGPA had significantly lower and higher AA and PA contents, respectively, compared with AhGLP and AhGCK (p < 0.05). PA and BA are indicators of silage quality and consume some of the metabolic energy during production. It was previously reported that conversion of LA to BA resulted in the loss of more than half of the DM content and some total energy (Muck, 2010), and this impacted the feed intake of livestock (Dong et al., 2020). The BA content of the different silages in the present study was 0.00. Some microorganisms that were detrimental to silage fermentation (for example, yeast, molds, Clostridium, and Enterobacter) often attached to silage or poor quality silage at different times of fermentation (Driehuis et al., 2018; Zhang et al., 2019; Guo et al., 2020). As fermentation proceeds, these microorganisms were gradually inactivated or disappear. In silage, Bacillus, Clostridium, and Enterobacter and yeasts are the microorganisms that play a central role in BA formation by excreting amino acid decarboxylases to produce BA (Jia et al., 2021). The low abundance of these harmful microorganisms in this study may also be one of the reasons why BA was not detected. AhGPA had a higher PA content compared with AhGLP and AhGCK, and this might be attributed to the increase in exogenously added PA and silage fermentation products. Fermentation characteristics were positively affected by inoculation with L. plantarum; this LAB not only decreased the pH but also promoted the accumulation of LA.
The third-generation Pacific Biosciences (PacBio) SMRT has proved outstanding in its ability to boost the sensitivity and accuracy of microbial community classification in ensiling (Xu et al., 2019). Consequently, the bacterial communities of high-moisture amaranth silage in the current study were further investigated by sequencing using SMRT.
Table 3 shows the bacterial alpha diversity of the amaranth silages. Ensiling is a bacteria-driven process, and the type of bacteria and their abundance in the fermentation process can directly affect the quality of the fermentation (Guan et al., 2018; He et al., 2020a). Sequencing coverage values for all samples were greater than 99%, indicating that the sequencing depth was sufficient for effective bacterial community characterization. Alpha diversity reflects bacterial diversity and species richness in individual samples. Shannon and Simpson indices indicate species diversity, while Chao1 and ACE indices represent species richness. The Simpson index of AhGCK was higher compared with those of AhGLP and AhGPA, but not by a significant margin (p > 0.05). The two additive groups of silage had higher OTU, ACE, Chao1, and Shannon indices compared with those of AhGCK. Among them, the ACE and Chao1 indices were higher in AhGLP compared with those in AhGCK. These results were in accordance with the findings of Du et al. (2021) who detected an increase in Chao1 and Ace indices in paper mulberry silage after inoculation with LAB compared with the control group. This suggests that the anaerobic and acidic environment displaces a large number of aerobic microorganisms in the silage by beneficial microorganisms (e.g., LAB), which in turn affects the overall micro-ecological environment. Collectively, the above results indicate that the screened LAB in the current study can rapidly reduce the pH of amaranth silage, thereby inhibiting harmful microorganisms and diminishing the alpha diversity of bacteria (Li et al., 2021). Once LAB became the prevailing species, bacterial diversity decreased, which was similar to the observations of Ni et al. (2017). Fermentation with exogenous L. plantarum and PA positively impacted the production of LA. The results of current study further demonstrated that exogenous L. plantarum exhibits a strong competitive advantage during the fermentation of high-moisture amaranth silage. Therefore, there is a need to study the dominant species of bacterial communities in amaranth silage.
Principal coordinate analysis (PCoA) clearly reflected the variations within the microbial community (Figure 1). There was a marked division of bacterial communities in the control and additive groups. The addition of additives highlighted the variability of bacterial communities in the silage samples of the different treatments, indicating that the composition and structure of the bacterial communities were significantly altered in AhGLP and AhGPA.
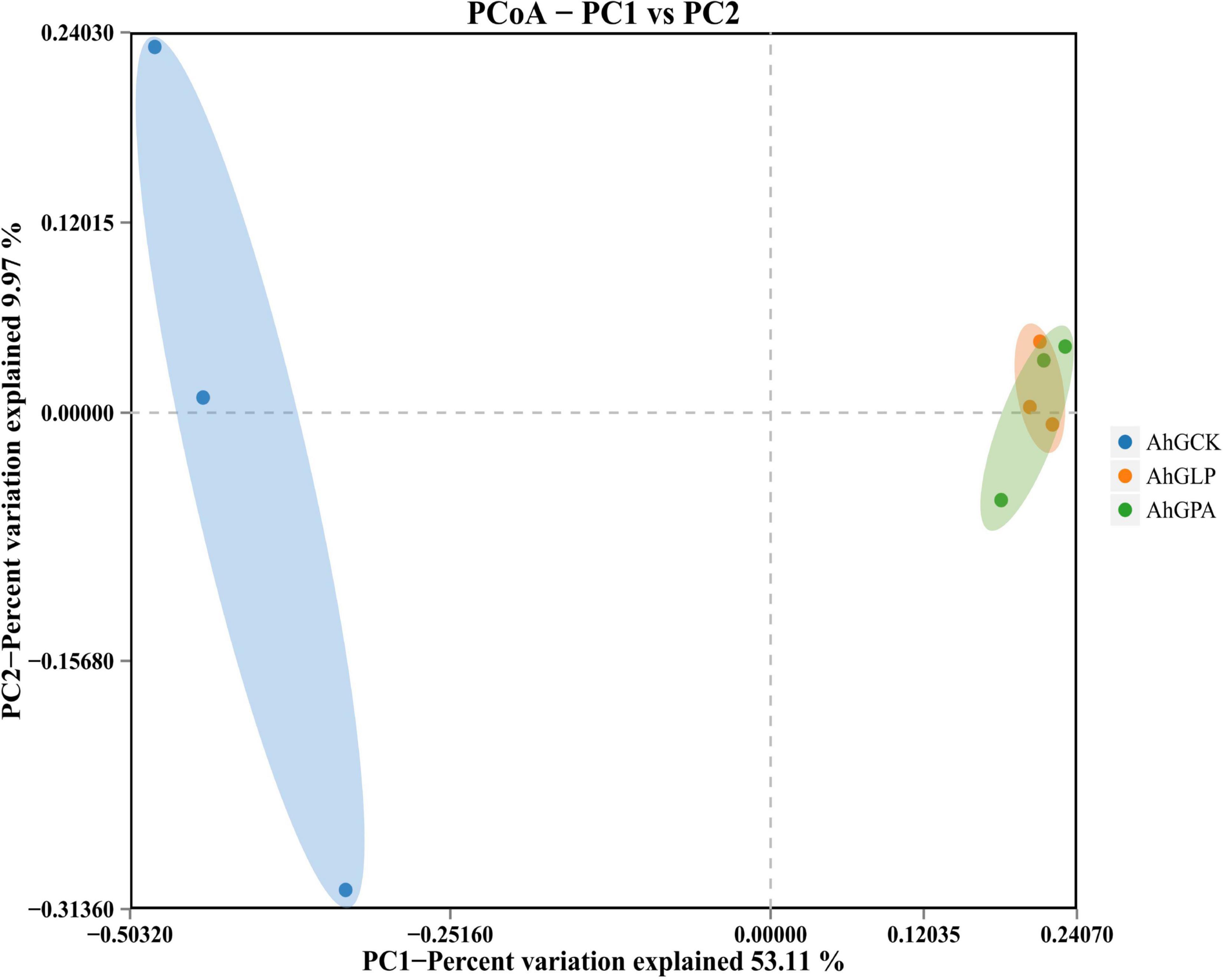
Figure 1. Bacterial β-diversity of high-moisture amaranth silage with Lactobacillus plantarum and propionic acid additives. AhGCK, control; AhGLP, L. plantarum; AhGPA, propionic acid.
Of the numerous elements that influence the silage fermentation process, the predominant microbial species often determine the quality of the silage (Ennahar et al., 2003; Ni et al., 2017). Therefore, it is essential to analyze changes in fermentation characteristics and microbial composition during silage to facilitate understanding of the silage process and enhance fermentation quality (Namihira et al., 2010). The relative abundance of bacteria in amaranth forage is shown in Figure 2, with the bacterial community at the phylum level presented in Figure 2A. Firmicutes, Proteobacteria, and Bacteroidota were the dominant phyla in all three groups after 60 days of ensiling. There was a significant increase and decrease in the relative abundance of Firmicutes and Proteobacteria, respectively, in AhGLP and AhGPA compared with AhGCK. The relative abundance of Firmicutes was more than 60% in AhGLP and AhGPA, while the relative abundance of Proteobacteria was lower in these two groups compared with that in AhGCK. Firmicutes were the main clade in the silage in the present study, which was consistent with the findings of Bao et al. (2016). This might be due to the low pH or anaerobic conditions favoring the growth of members of the phylum Firmicutes during silage (Keshri et al., 2018). Figure 2B shows the significant differences among groups at the phylum level. It indicated that there were significant differences in Actinobacteriota between three groups (p < 0.05).
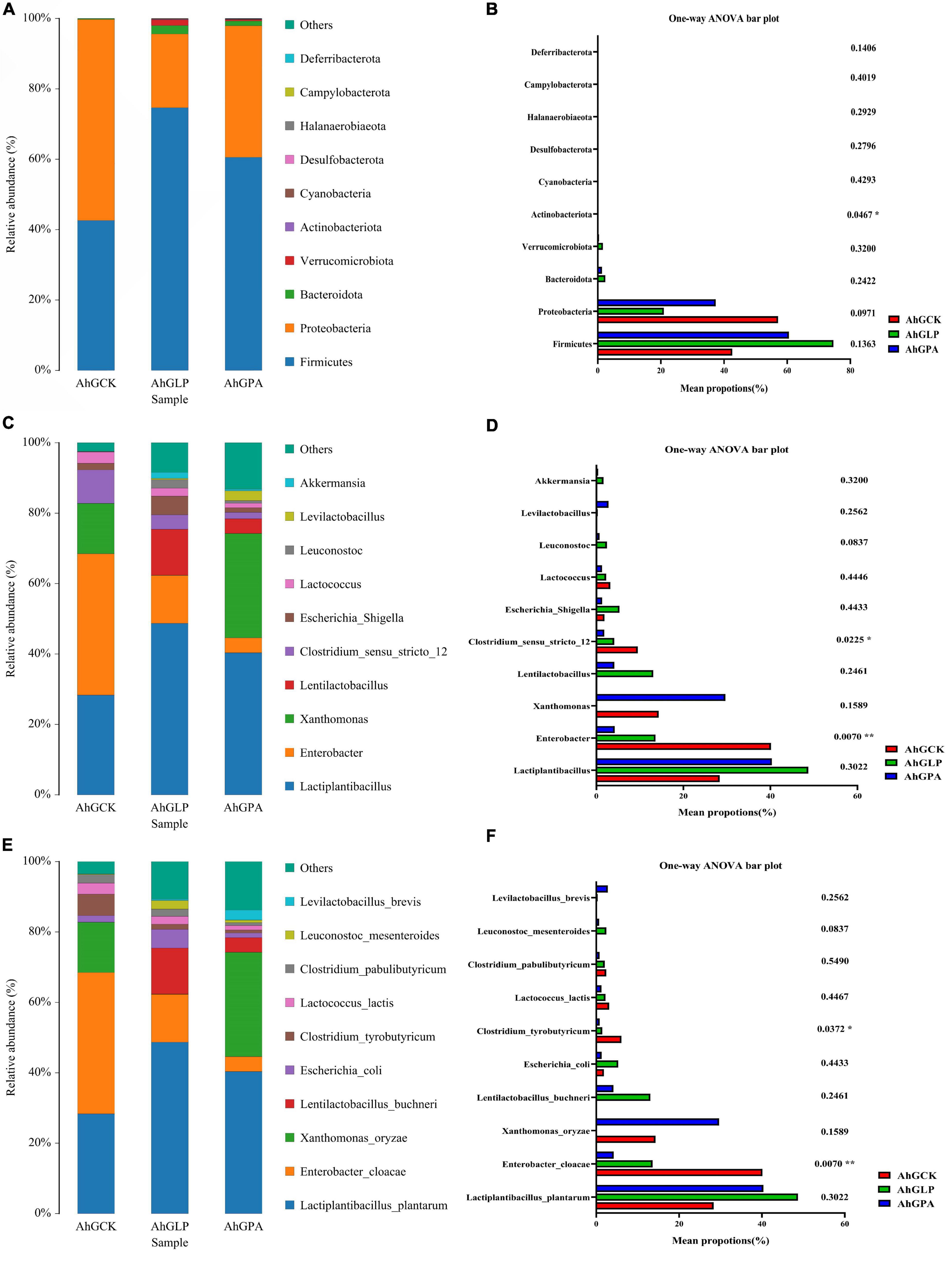
Figure 2. Relative abundance of bacteria at the phylum (A), genus (C), and species (E) levels in high-moisture amaranth silage with Lactobacillus plantarum and propionic acid additives. The extended error bar plot displaying the significant differences among groups at the phylum (B), genus (D), and species (F) levels. AhGCK, control; AhGLP, L. plantarum; AhGPA, propionic acid.
Relative abundances of bacteria at the genus level are presented in Figure 2C. The dominant genus in AhGCK was Enterobacter, and the subdominant genera were Lactiplantibacillus and Xanthomonas. The dominant genus in AhGLP and AhGPA was Lactiplantibacillus, while the subdominant genera in AhGLP were Enterobacter, Lentilactobacillus, and Escherichia Shigella, and those in AhGPA were Enterobacter and Xanthomonas. The relative abundances of Lactobacillus and Enterobacter in the additive groups were significantly higher and lower, respectively, compared with those in AhGCK, which could explain the high LAB populations in the additive groups. In a previous study, competition between Enterobacter and LAB for available WSC in silage led to the production of more AA, succinic acid, 2,3-butanediol, some endotoxins, and ammonia nitrogen and reduced nutritional value and palatability (Muck, 2010). Therefore, Enterobacter are an undesirable genus in silage. The dominant genus in AhGCK was Enterobacter, and this was confirmed by its poor silage quality. In AhGLP and AhGPA, Enterobacter was the second dominant genus after Lactiplantibacillus, but the addition of L. plantarum and PA reduced the abundance of Enterobacter. Clostridium sensu stricto 12 and Enterobacter had significant and highly significant differences among three groups (p < 0.01 and p < 0.05) (Figure 2D). We concluded that the intervention of additives in the silage fermentation process plays an important role in the fermentation of silage.
Figure 2E illustrates the relative abundance of the top 10 bacterial species in amaranth silage. LAB play an essential role in fermentation during the ensiling process. In this study, Lactiplantibacillus plantarum, Enterobacter cloacae, and Xanthomonas oryzae were the dominant species in AhGCK and AhGPA. PA is often used to improve the aerobic stability of silage. In the present study, the addition of PA enhanced the relative abundance of Lactiplantibacillus plantarum and Lentibacillus buchneri. Exogenous addition of PA lowers the pH owing to the acidity of the PA itself, thereby inhibiting acid-intolerant bacteria, creating a favorable anaerobic environment, encouraging acid-tolerant LAB fermentation (e.g., Lactiplantibacillus plantarum and Lentibacillus buchneri), and reducing undesirable fermentation and protein hydrolysis. Papendick and Singh-Verma (1972) and Jia et al. (2021) also produced similar results. The dominant species of AhGLP in the current study were Lactiplantibacillus plantarum, Enterobacter cloacae, and Lentilactobacillus buchneri, among which, L. plantarum is known to play a key role in silage fermentation. Enterobacter cloacae and Clostridium tyrobutyricum exhibited significant and highly significant differences among three groups (p < 0.01 and p < 0.05), and were the common harmful microorganisms found in high-moisture silage (Figure 2F). Enterobacter cloacae, Clostridium tyrobutyricum, and some yeasts produce biogenic amines (an endogenous metabolic component of cells in plants) by excreting amino acid decarboxylases (Halász et al., 1994). X. oryzae was the third-ranked dominant species in the silage of AhGCK and AhGPA. Members of the genus Xanthomonas are pathogenic for more than 300 plant species (Boch and Bonas, 2010; Büttner and Bonas, 2010). X. oryzae is obligatorily aerobic, does not form spores, grows optimally between 25 and 30°C, is peroxidase-positive, cannot reduce nitrate, and is a weak producer of acid from carbohydrates (Bradbury, 1984). Xanthomonas differs from non-pathogenic bacteria in that it relies on a variety of potent substances secreted through different kinds of protein secretion systems, predominantly the type III secretion system, to suppress host immunity and to obtain nutrients from plants. Amaranth was considered a promising crop in Argentina (Noelting et al., 2019), but was found to be a host of Xanthomonas euvesicatoria (Santos et al., 2020), Curtobacterium flaccumfaciens pv. flaccumfaciens (Nascimento et al., 2020), and Xanthomonas citri subsp. citri (Ebrahimi et al., 2020). It also demonstrates that X. oryzae accounts for a large proportion of amaranth silage. Our results show that L. plantarum might inhibit the growth and multiplication of pathogens. As a fermentation inhibitor, PA inhibited aerobic bacteria, mold, and Bacillus bacteria, but X. oryzae exhibited the opposite trend in the amaranth silage with PA (i.e., X. oryzae increased in abundance) compared with the amaranth silage with L. plantarum added (i.e., X. oryzae decreased in abundance). In addition, X. oryzae could not produce LA and AA with soluble sugar as the fermentation substrate, which also explains the poor fermentation quality in AhGPA compared with AhGLP. Thus, L. plantarum can affect the bacterial diversity and community structure of high moisture amaranth silage by inhibiting the colonization of undesirable microorganisms, such as Enterobacter cloacae and Clostridium tyrobutyricum.
The LEfSe method was used to evaluate differences in microbial communities between the three groups of silage and to explore specific bacterial species in each group [linear discriminant analysis (LDA) score, > 4.0]. L. plantarum markedly affected the microbial community in the silage (Figure 3). In AhGCK, a group of 8 bacteria were significantly enriched, with Enterobacter cloacae (LDA score, 5.60) having the highest LDA score. In AhGLP, a group of 11 bacteria were significantly enriched, with Lentilactobacillus buchneri (LDA score, 5.12) having the highest LDA score. In AhGPA, three subgroups of bacteria were significantly enriched, with Levilactobacillus brevis (LDA score, 4.42) having the highest LDA score. These results suggested that there was some variation in species abundance in specific communities of high-moisture amaranth silage with different additives. The reason of why the Lactiplantibacillus plantarum has no difference in the LEfSe after the addition of Lactobacillus plantarum may be due to the fact that Lactiplantibacillus plantarum was not the most dominant strain in all groups.
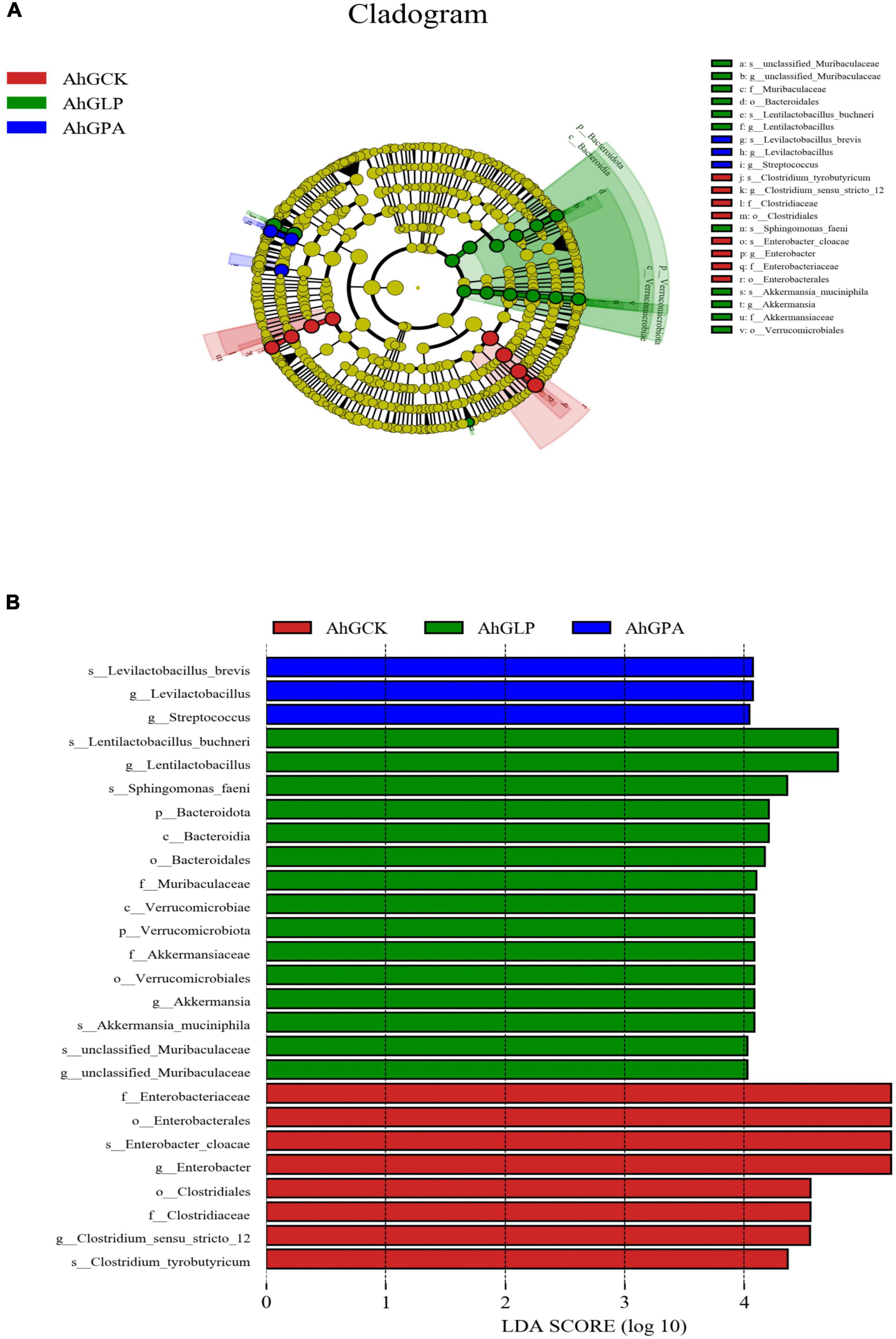
Figure 3. Comparison of microbial variations using the LEfSe online tool for high-moisture amaranth silage with Lactobacillus plantarum and propionic acid additives. AhGCK, control; AhGLP, L. plantarum; AhGPA, propionic acid. (A) Cladogram for taxonomic representation of significantly differences. (B) Indicator bacteria with an linear discriminant analysis (LDA) score of 4 or more in the silage bacterial community under different treatments.
Spearman correlation clustering was used to explore correlation between microbial communities and fermentation characteristics (species level; Figure 4). Lentilactobacillus buchneri and Levilactobacillus brevis were positively associated with LA (R2 = 0.590 and R2 = 0.577, respectively; p < 0.05 for both), which suggested that the generation of LA was largely attributable to these two species. In addition, these two species were negatively associated with pH (R2 = –0.662 and R2 = –0.602 for Lentilactobacillus buchneri and Levilactobacillus brevis, respectively; p < 0.01 for both). Clostridium tyrobutyricum and Enterobacter cloacae were negatively associated with LA (R2 = –0.585, p < 0.05; R2 = –0.672, p < 0.01), but positively correlated with pH (R2 = 0.668 and R2 = 0.678, respectively; p < 0.05 for both) and AA (R2 = 0.589 and R2 = 0.485, respectively; p < 0.05 for both). AA was also inversely correlated with Lentilactobacillus buchneri (R2 = –0.654, p < 0.01).
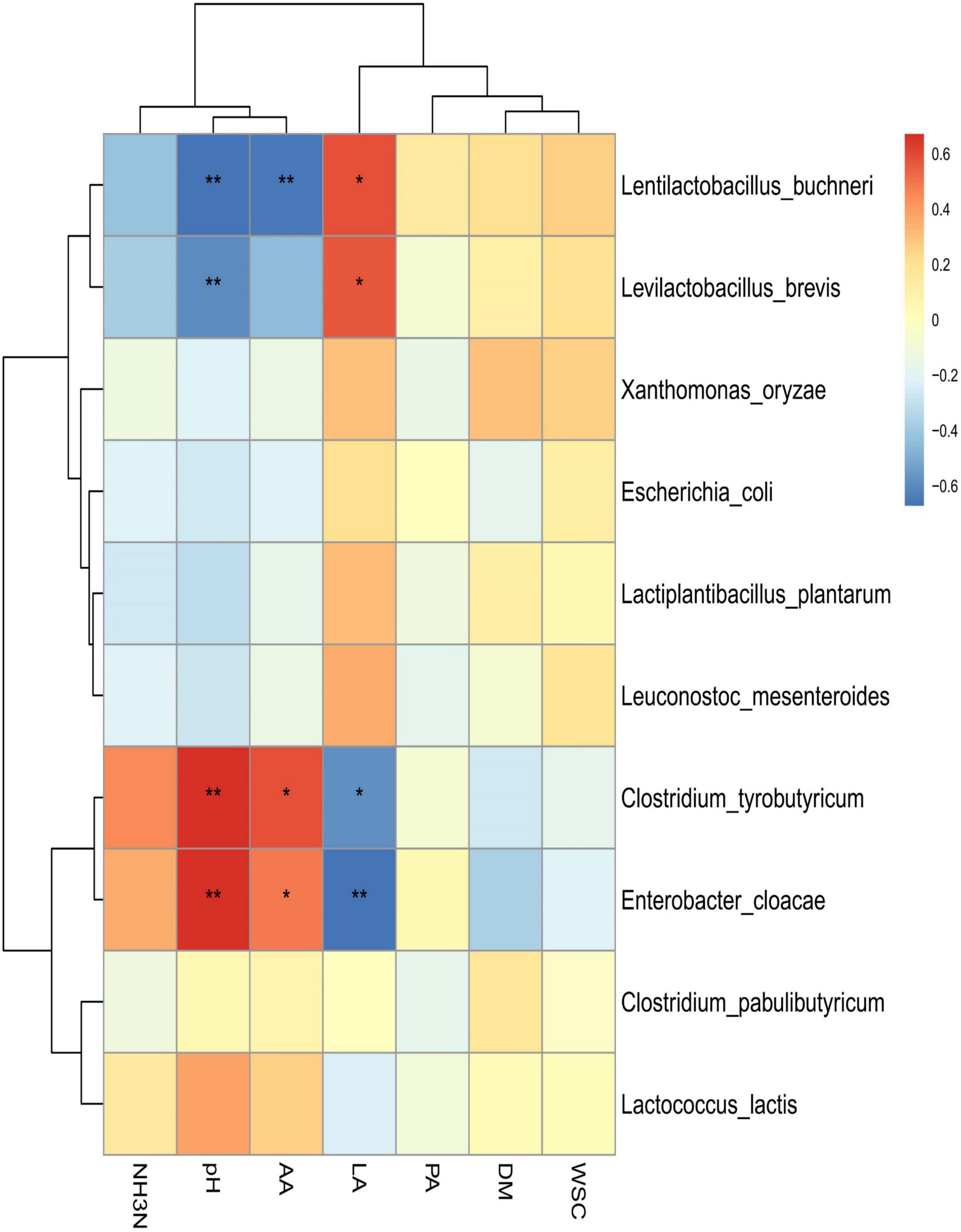
Figure 4. Spearman correlation heatmap of abundance of microbial community and fermentation properties in high-moisture amaranth silage with Lactobacillus plantarum and propionic acid additives. AhGCK, control; AhGLP, L. plantarum; AhGPA, propionic acid; AA, acetic acid; DM, dry matter; LA, lactic acid; PA, propionic acid; WSC, water soluble carbohydrate. *p < 0.05; **p < 0.01.
Lentilactobacillus buchneri and Levilactobacillus brevis were previously found to be major producers of LA and were instrumental in lowering pH during ensiling (Cai et al., 1998; Wang et al., 2020; Yang et al., 2021). The low AA content indicated that heterofermentative LAB produce PA and AA through fermentation to enhance aerobic stability (Naiara et al., 2019). Fermentation products are the result of the joint action of the resident microorganisms, and eventually, more PA and a small amount of AA were produced during fermentation because ofthe different metabolic pathways of the microorganisms (Xu et al., 2019). Consequently, Lentilactobacillus buchneri may produce more products other than AA when fermenting through the HMP pathway (Muck et al., 2018). Clostridium tyrobutyricum and Enterobacter cloacae were the predominant contributors to the reduction in pH and increase in AA production and thus have a negative impact on the production of an acidic environment during the silage process. This may be the main reason for the growth and multiplication of undesirable bacteria attached to the silage and consequently increases nutrient losses.
In summary, this analysis demonstrated that L. plantarum and PA enhanced the fermentation characteristics of high-moisture amaranth by enhancing the relative abundance of Lactiplantibacillus plantarum and Lentilactobacillus buchneri and decreasing the relative abundance of Enterobacter cloacae.
Kyoto encyclopedia of genes and genomes (KEGG) is a bioinformatics tool for understanding the capacities and utilities of cells and living beings at both high-level and genomic points of view. The prediction of useful shifts in bacterial communities facilitates the assessment of the impact of organisms on energy changes in silage quality. KEGG pathway analysis was therefore used to predict the function of the microbial community of high-moisture amaranth silage. A total of 364 predicted functions were generated, some of which were significantly different between the three silage groups (Figure 5). The main predicted functional genes at level 1 during fermentation were distributed among the functions of metabolism, environmental information processing, genetic information processing, and cellular processes (Figure 5A). Compared with the control (AhGCK), metabolism and genetic information processing were significantly increased in AhGLP and AhGPA, while the functions of environmental information processing and cellular processes were significantly reduced. This may be because the growth of LAB in silage without additives is much lower compared with that in the additive silage, thus the growth and metabolic activity of other microorganisms is inhibited, along with the biosynthesis of secondary metabolites and antibiotics. KEGG pathway levels 2 and 3 of bacterial gene function are shown in Figures 5B, C. In the metabolism function, there were significant differences or highly significant differences between the control and additive-treated silage (p < 0.05 or p < 0.01) in the KEGG pathway terms of biosynthesis of secondary metabolites; biosynthesis of antibiotics; biosynthesis of amino acids, carbon metabolism, purine metabolism, pentose phosphate pathway; alanine, aspartate, and glutamate metabolism; methane metabolism; carbon fixation pathways in prokaryotes; and sulfur metabolism. In contrast, aminoacyl-tRNA biosynthesis and ribosome in the genetic information processing function showed significant or highly significant differences (p < 0.05 or p < 0.01) between the control and additive-treated silage. Sulfur metabolism, methane metabolism, and carbon metabolism were lower in the additive groups compared with the control group. All KEGG pathways at level 3 were enriched in the silage containing additives compared with the control. The specific fermentation characteristics of the different silage groups showed that the exogenous microbiota of amaranth improved the quality of the fermentation by altering the cellular characteristics, inhibiting membrane transport and signal transduction of undesirable bacteria, and accelerating the rate of multiplication and metabolic levels of beneficial bacteria such as LAB species.
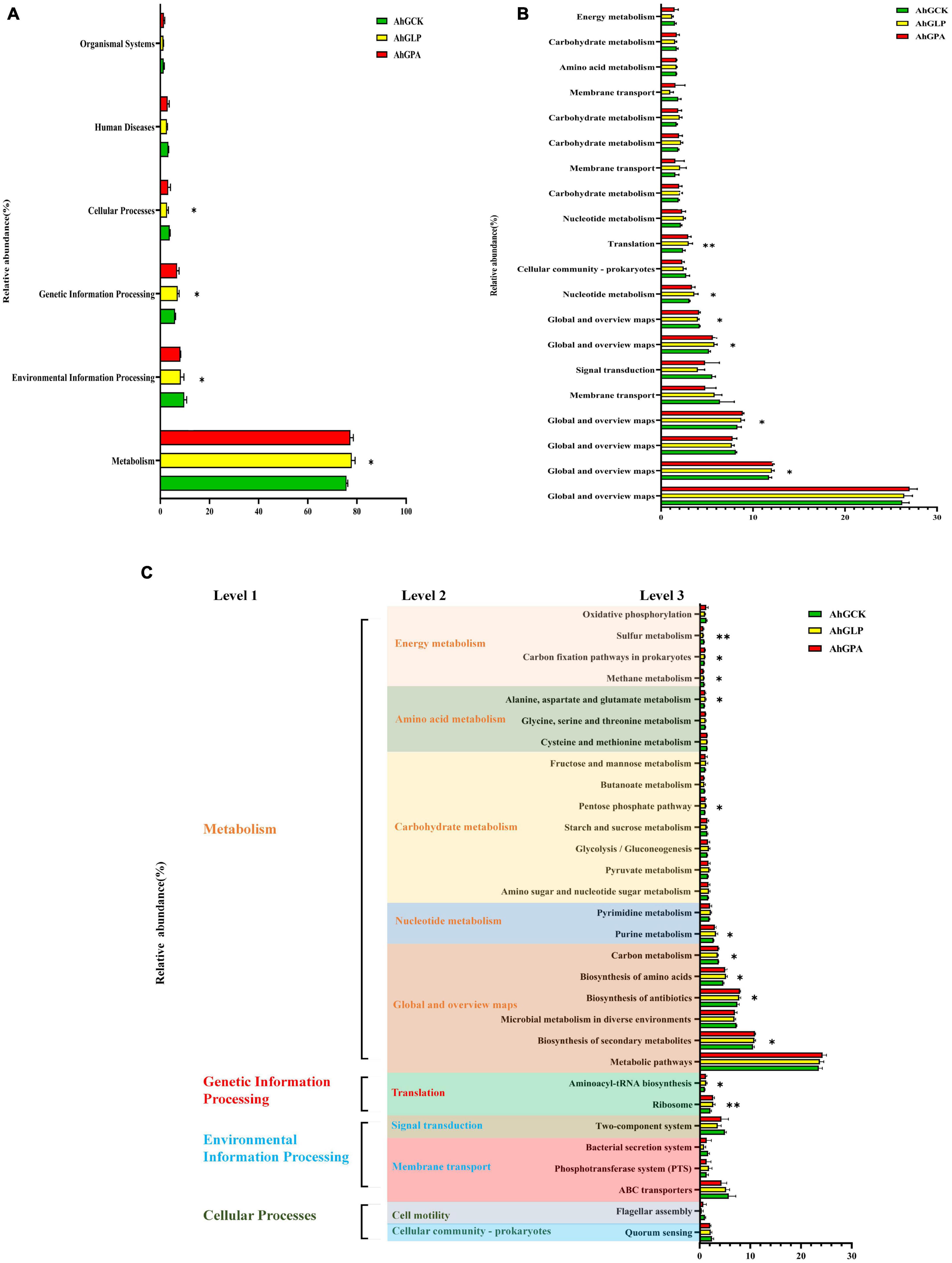
Figure 5. Functional predictions for silage microbiota with significantly different Kyoto encyclopedia of genes and genomes (KEGG) pathways (p < 0.05) among the three groups of silage (AhGCK, AhGLP, and AhGPA). KEGG pathways at level 1 (A), level 2 (B), and level 3 (C) are represented. AhGCK, control; AhGLP, L. plantarum; AhGPA, propionic acid. *p < 0.05; **p < 0.01.
The fermentation process of silage comprises microbial activity to degrade substrates or convert metabolites through complex metabolic pathways. The additives facilitated the metabolic pathways of carbohydrates in silage in this study, which was in line with the metabolic pathways associated with silage fermentation that were identified by Xu et al. (2021)—namely, the metabolism of carbohydrates, amino acids, energy and cofactors, and vitamins. This indicates that silage microorganisms (Lactiplantibacillus plantarum and Lentilactobacillus buchneri) complemented with L. plantarum and PA had a greater ability to metabolize WSC. The expression of carbohydrate metabolic pathways was correlated with the relative abundance of total LAB in the bacterial community. A relatively high abundance of total LAB and a poor relative abundance of carbohydrate metabolism in the bacterial community was observed in AhGLP. According to Pessione et al. (2010), the three main energy metabolism pathways in Lactobacillus were amino acid decarboxylation, malate decarboxylation, and arginine IX decarboxylation, and these pathways contributed to the accumulation of LA during fermentation. This differed from our study, possibly due to the low carbohydrate content of amaranth and its high consumption as a fermentation substrate during the silage process. Homofermentative LAB have been classified as obligate homofermentative and facultative heterofermentative (Orla-Jensen, 1919; Kandler, 1983; Axelsson, 2004). Obligate homofermentative LAB mainly metabolize hexose through the glycolytic pathway and then convert it all to pyruvate, which is reduced to lactic acid by lactate dehydrogenase. Facultative heterofermentative LAB metabolize pentose via the pentose phosphate pathway to produce LA and AA. Interestingly, obligate homofermentative LAB do not metabolize pentose or gluconate. The carbohydrate metabolism pathway of level 3 suggested that the L. plantarum added to our study may belong to the facultative heterofermentative. Silages with different additives showed variability in the pentose phosphate pathway, with some heterogeneously fermented LAB producing AA via this pathway. This indirectly explains the higher relative abundance of Lactilantibacillus and Lentilactobacillus in AhGLP. At the same time, this suggests that AhGLP has more heterotypic fermentation processes and a richer metabolic pathway than AhGCK and AhGPA. Furthermore, the epiphytic Lactiplantibacillus plantarum and Lentilactobacillus buchneri could inhibit the reproductive activity of undesirable microorganisms to a certain extent, thus slowing the spoilage process of the silage.
Amino acids are the basic building blocks of proteins. While the highest CP content was found in silage inoculated with PA, the highest metabolic abundance of alanine, aspartate, and glutamate metabolism was found in silage inoculated with L. plantarum. This was consistent with the lowest pH and NH3-N detected in silage inoculated with L. plantarum. The relative abundance of X. oryzae was higher in AhGPA compared with that in AhGCK and AhGLP, which may explain why AhGPA had the highest CP content but differed from the silage supplemented with L. plantarum in terms of metabolic rate and silage quality. The low metabolism might reflect the capacity of the initial microbial populations, in the silage, to synthesize amino acid de novo. In contrast, as LAB do not synthesize all their essential amino acid, they rely on proteolytic systems to provide essential amino acids for their growth (Novik and Savic, 2020). Therefore, the dynamics of amino acid metabolism observed in AhGCK and additive-treated silages may reflect the metabolism of the dominant populations throughout the ensiling process. Above all, the low pH of silage inoculated with L. plantarum inhibited amino acid metabolism induced by the undesirable microorganisms of the silage such as Enterobacter cloacae and X. oryzae (Flythe and Russell, 2004; Yuan et al., 2020). Despite these observations, the reliability of KEGG gene function predictions is limited, and we will therefore combine the KEGG analysis with metabolomics for multi-omics validation in future work.
Inoculation of the epiphytic microbiota with L. plantarum and PA significantly altered the fermentation products and bacterial community composition and predicted metabolic pathways of high-moisture amaranth silage. The dominant bacterial species of AhGLP were Lactiplantibacillus plantarum, Enterobacter cloacae, and Lentilactobacillus buchneri. The predominant bacterial species of AhGLP were Lactiplantibacillus plantarum, Enterobacter cloacae, and X. oryzae. LAB dominated both additive silages. Furthermore, L. plantarum markedly inhibited the reproductive activity of X. oryzae and reduced the depletion of nutrients despite amaranth being a known host of phytopathogenic X. oryzae. However, the effect of exogenous PA on the bacterial community of the silage was limited. In summary, our results confirm the feasibility of regulating high-moisture amaranth silage by inoculation with L. plantarum.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA888204.
MZ: conceptualization, investigation, visualization, methodology, formal analysis, and writing—original draft. ZW: supervision and writing—review and editing. SD, LS, JB, and JH: conceptualization, investigation, and formal analysis. GG: conceptualization, methodology, validation, investigation, writing—review and editing, and funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the Key Laboratory of Forage Cultivation and the Processing and Highly Efficient Utilization of the Ministry of Agriculture, the Key Laboratory of Grassland Resources of the Ministry of Education, and funded by the National Technical System of Forage Industry for Dry Grass Storage (CARS-34) and Research and Demonstration of Key Technology for Processing, Utilization and Storage of “Double Increase and Double Extraction” of Alfalfa (2021GG0109).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbasi, M., Rouzbehan, Y., Rezaei, J., and Jacobsen, S. E. (2018). The effect of lactic acid bacteria inoculation, molasses, or wilting on the fermentation quality and nutritive value of amaranth (Amaranthus hypochondriaus) silage. J. Anim. Sci. 96, 3983–3992. doi: 10.1093/jas/sky257
Arriola, K. G., Kim, S. C., and Adesogan, A. T. (2011). Effect of applying inoculants with heterolactic or homolactic and heterolactic bacteria on the fermentation and quality of corn silage. J. Dairy Sci. 94, 1511–1516. doi: 10.3168/jds.2010-3807
Axelsson, L. (2004). Lactic acid bacteria: classification and physiology. Food Sci. Technology-New York-Marcel Dekker 139, 1–66. doi: 10.1201/9780824752033.ch1
Bao, W., Mi, Z., Xu, H., Zheng, Y., Kwok, L. Y., Zhang, H., et al. (2016). Assessing quality of Medicago sativa silage by monitoring bacterial composition with single molecule, real-time sequencing technology and various physiological parameters. Sci. Rep. 6:28358. doi: 10.1038/srep28358
Boch, J., and Bonas, U. (2010). Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. doi: 10.1146/annurev-phyto-080508-081936
Bradbury, J. F. (1984). “Genus II. xanthomonas dowson,” in Bergey’s Manual of Systematic Bacteriology, eds N. R. Krieg and J. G. Holt (Baltimore, MD: Williams and Wilkins), 199–210.
Broderick, G. A., and Kang, J. H. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 63, 64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
Büttner, D., and Bonas, U. (2010). Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. doi: 10.1111/j.1574-6976.2009.00192.x
Cai, Y. (2004). “Analysis method for silage,” in Japanese Society of Grassland Science, ed. Field and laboratory methods for grassland science (Tokyo: Thsho Printing Co., Ltd), 279–282.
Cai, Y., Benno, Y., Ogawa, M., Ohmomo, S., Kumai, S., and Nakase, T. (1998). Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl. Environ. Microbiol. 64, 2982–2987. doi: 10.1128/AEM.64.8.2982-2987.1998
Cai, Y., Suyanandana, P., Saman, P., and Benno, Y. (1999). Classification and characterization of lactic acid bacteria isolated from the intestines of common carp and fresh water prawns. J. Gen. Appl. Microbiol. 45, 177–184. doi: 10.2323/jgam.45.177
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, L., Guo, G., Yu, C., Zhang, J., Shimojo, M., and Shao, T. (2015). The effects of replacement of whole-plant corn with oat and common vetch on the fermentation quality, chemical composition and aerobic stability of total mixed ration silage in Tibet. Animal Sci. J. 86, 69–76. doi: 10.1111/asj.12245
Cheng, Q., Li, P., Xiao, B., Yang, F., Li, D., Ge, G., et al. (2021). Effects of LAB inoculant and cellulase on the fermentation quality and chemical composition of forage soybean silage prepared with corn stover. Grassland Sci. 67, 83–90. doi: 10.1111/grs.12289
Coblentz, W. K., and Akins, M. S. (2021). Nutritive value, silage fermentation characteristics, and aerobic stability of round-baled, alfalfa–grass forages ensiled at 2 moisture concentrations with or without a propionic-acid-based preservative. Appl. Animal Sci. 37, 89–105. doi: 10.15232/aas.2020-02128
Dong, L., Liu, J., Zhong, Z., Wang, S., Wang, H., Huo, Y., et al. (2019). Dietary tea tree oil supplementation improves the intestinal mucosal immunity of weanling piglets. Animal Feed Sci. Technol. 255:114209. doi: 10.1016/j.anifeedsci.2019.114209
Dong, L., Zhang, H., Gao, Y., and Diao, Q. (2020). Dynamic profiles of fermentation characteristics and bacterial community composition of Broussonetia papyrifera ensiled with perennial ryegrass. Bioresour. Technol. 310:123396. doi: 10.1016/j.biortech.2020.123396
Driehuis, F., Wilkinson, J. M., Jiang, Y., Ogunade, I., and Adesogan, A. T. (2018). Silage review: animal and human health risks from silage. J. Dairy Sci. 101, 4093–4110. doi: 10.3168/jds.2017-13836
Du, Z., Sun, L., Chen, C., Lin, J., Yang, F., and Cai, Y. (2021). Exploring microbial community structure and metabolic gene clusters during silage fermentation of paper mulberry, a high-protein woody plant. Animal Feed Sci. Technol. 275:114766. doi: 10.1016/j.anifeedsci.2020.114766
Ebrahimi, Z., Rezaei, R., Masoumi-Asl, A., and Charehgani, H. (2020). Variation in the aggressiveness of Xanthomonas citrisubsp. citri pathotypes A and A* on three Citrus species, and epiphytic growth on some citrus weeds. Crop Pasture Sci. 71, 260–267. doi: 10.1071/CP19448
Ennahar, S., Cai, Y., and Fujita, Y. (2003). Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 69, 444–451. doi: 10.1128/AEM.69.1.444-451.2003
Ercolini, D. (2004). PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56, 297–314. doi: 10.1016/j.mimet.2003.11.006
Flythe, M. D., and Russell, J. B. (2004). The effect of pH and a bacteriocin (bovicin HC5) on Clostridium sporogenes MD1, a bacterium that has the ability to degrade amino acids in ensiled plant materials. FEMS Microbiol. Ecol. 47, 215–222. doi: 10.1016/S0168-6496(03)00259-9
Gheller, L. S., Ghizzi, L. G., Takiya, C. S., Grigoletto, N. T., Silva, T. B., Marques, J. A., et al. (2021). Different organic acid preparations on fermentation and microbiological profile, chemical composition, and aerobic stability of whole-plant corn silage. Animal Feed Sci. Technol. 281:115083. doi: 10.1016/j.anifeedsci.2021.115083
Guan, H., Yan, Y., Li, X., Li, X., Shuai, Y., Feng, G., et al. (2018). Microbial communities and natural fermentation of corn silages prepared with farm bunker-silo in Southwest China. Bioresour. Technol. 265, 282–290. doi: 10.1016/j.biortech.2018.06.018
Guo, L., Wang, X., Lin, Y., Yang, X., Ni, K., and Yang, F. (2021). Microorganisms that are critical for the fermentation quality of paper mulberry silage. Food Energy Security 10:e304. doi: 10.1002/fes3.304
Guo, L., Yao, D., Li, D., Lin, Y., Bureenok, S., Ni, K., et al. (2020). Effects of lactic acid bacteria isolated from rumen fluid and feces of dairy cows on fermentation quality, microbial community, and in vitro digestibility of alfalfa silage. Front. Microbiol. 10:2998. doi: 10.3389/fmicb.2019.02998
Halász, A., Barath, A., Simon-Sarkadi, L., and Holzapfel, W. (1994). Biogenic amines and their production by microorganisms in food. Trends Food Sci. Technol. 5, 42–49. doi: 10.1016/0924-2244(94)90070-1
He, L., Wang, C., Xing, Y., Zhou, W., Pian, R., Chen, X., et al. (2020a). Ensiling characteristics, proteolysis and bacterial community of high-moisture corn stalk and stylo silage prepared with Bauhinia variegate flower. Bioresour. Technol. 296:122336. doi: 10.1016/j.biortech.2019.122336
He, L., Zhou, W., Xing, Y., Pian, R., Chen, X., and Zhang, Q. (2020b). Improving the quality of rice straw silage with moringa oleifera leaves andpropionic acid: fermentation, nutrition, aerobic stability and microbial communities. Bioresour. Technol. 299:122579. doi: 10.1016/j.biortech.2019.122579
Heinritz, S. N., Martens, S. D., Avila, P., and Hoedtke, S. (2012). The effect of inoculant and sucrose addition on the silage quality of tropical forage legumes with varying ensilability. Animal Feed Sci. Technol. 174, 201–210. doi: 10.1016/j.anifeedsci.2012.03.017
Herrero, M., Wirsenius, S., Henderson, B., Rigolot, C., Thornton, P., Havlík, P., et al. (2015). Livestock and the environment: what have we learned in the past decade? Annual Rev. Environ. Resources 40, 177–202. doi: 10.1146/annurev-environ-031113-093503
Jia, T., Yun, Y., and Yu, Z. (2021). Propionic acid and sodium benzoate affected biogenic amine formation, microbial community, and quality of oat silage. Front. Microbiol. 12:750920. doi: 10.3389/fmicb.2021.750920
Kandler, O. (1983). Carbohydrate metabolism in lactic acid bacteria. Antonie van Leeuwenhoek 49, 209–224. doi: 10.1007/BF00399499
Keshri, J., Chen, Y., Pinto, R., Kroupitski, Y., and Weinberg, Z. G. (2018). Microbiome dynamics during ensiling of corn with and without Lactobacillus plantarum inoculant. Appl. Microbiol. Biotechnol. 102, 4025–4037. doi: 10.1007/s00253-018-8903-y
Kleinschmit, D. H., and Kung, L. Jr. (2006). A meta-analysis of the effects of Lactobacillus buchneri on the fermentation and aerobic stability of corn and grass and small-grain silages. J. Dairy Sci. 89, 4005–4013. doi: 10.3168/jds.S0022-0302(06)72444-4
Kung, L., Stokes, M. R., and Lin, C. J. (2003). Silage Additives: Silage Science and Technology. Madison: American Society of Agronomy.
Li, P., Lu, Y., Zhao, M., Chen, L., Zhang, C., Cheng, Q., et al. (2021). Effects of phenyllactic acid, lactic acid bacteria, and their mixture on fermentation characteristics and microbial community composition of timothy silage. Front. Microbiol. 12:743433. doi: 10.3389/fmicb.2021.743433
Lima, R., Lourenco, M., Diaz, R., Castro, A., and Fievez, V. (2010). Effect of combined ensiling of sorghum and soybean with or without molasses and Lactobacilli on silage quality and in vitro rumen fermentation. Animal Feed Sci. Technol. 155, 122–131. doi: 10.1016/j.anifeedsci.2009.10.008
Liu, B., Huan, H., Gu, H., Xu, N., Shen, Q., and Ding, C. (2019). Dynamics of a microbial community during ensiling and upon aerobic exposure in lactic acid bacteria inoculation-treated and untreated barley silages. Bioresour. Technol. 273, 212–219. doi: 10.1016/j.biortech.2018.10.041
Mu, L., Xie, Z., Hu, L., Chen, G., and Zhang, Z. (2020). Cellulase interacts with Lactobacillus plantarum to affect chemical composition, bacterial communities, and aerobic stability in mixed silage of high-moisture amaranth and rice straw. Bioresour. Technol. 315:123772. doi: 10.1016/j.biortech.2020.123772
Muck, R. E. (2010). Silage microbiology and its control through additives. Rev. Brasileira Zootecnia 39, 183–191. doi: 10.1590/S1516-35982010001300021
Muck, R. E., Nadeau, E. M. G., McAllister, T. A., Contreras-Govea, F. E., Santos, M. C., and Kung, L. Jr. (2018). Silage review: recent advances and future uses of silage additives. J. Dairy Sci. 101, 3980–4000. doi: 10.3168/jds.2017-13839
Naiara, C., Nascimento, C. F., Campos, V. M. A., Alves, M. A. P., Resende, F. D., Daniel, J. L. P., et al. (2019). Influence of storage length and inoculation with Lactobacillus buchneri on the fermentation, aerobic stability, and ruminal degradability of high-moisture corn and rehydrated corn grain silage. Anim. Feed Sci. Tech. 251, 124–133. doi: 10.1016/j.anifeedsci.2019.03.003
Namihira, T., Shinzato, N., Akamine, H., Maekawa, H., and Matsui, T. (2010). Influence of nitrogen fertilization on tropical-grass silage assessed by ensiling process monitoring using chemical and microbial community analyses. J. Appl. Microbiol. 108, 1954–1965. doi: 10.1111/j.1365-2672.2009.04591.x
Nascimento, D. M., Oliveira, L. R., Melo, L. L., Silva, J. C., Soman, J. M., Girotto, K. T., et al. (2020). Survival of Curtobacterium flaccumfaciens pv. flaccumfaciens in weeds. Plant Pathol. 69, 1357–1367. doi: 10.1111/ppa.13206
Ni, K., Wang, F., Zhu, B., Yang, J., Zhou, G., Pan, Y. I., et al. (2017). Effects of lactic acid bacteria and molasses additives on the microbial community and fermentation quality of soybean silage. Bioresour. Technol. 238, 706–715. doi: 10.1016/j.biortech.2017.04.055
Noelting, M. C., Ferreira, J., Galvão, S. R., Greizerstein, E. J., Molina, M. D. C., López, C. G., et al. (2019). Amaranthus caudatus subsp. mantegazzianus: a new host of ‘Candidatus Phytoplasma hispanicum’(subgroup 16Sr XIII-A). J. Phytopathol. 167, 618–623. doi: 10.1111/jph.12852
Novik, G., and Savich, V. (2020). Beneficial microbiota. Probiotics and pharmaceutical products in functional nutrition and medicine. Microbes Infect. 22, 8–18. doi: 10.1016/j.micinf.2019.06.004
Papendick, K., and Singh-Verma, S. B. (1972). The effect of propionic acid and formic acid as silage additives. Das Wirtschaft. Futter 18, 293–304.
Patrica, C. (1997). Official Method of Analysis of AOAC International, 16th Edn. Washington, DC: AOAC International.
Pessione, A., Lamberti, C., and Pessione, E. (2010). Proteomics as a tool for studying energy metabolism in lactic acid bacteria. Mol. BioSystems 6, 1419–1430. doi: 10.1039/c001948h
Rabelo, C. H. S., Härter, C. J., Ávila, C. L. D. S., and Reis, R. A. (2019). Meta-analysis of the effects of Lactobacillus plantarum and Lactobacillus buchneri on fermentation, chemical composition and aerobic stability of sugarcane silage. Grassland Sci. 65, 3–12. doi: 10.1111/grs.12215
Rahjerdi, N. K., Rouzbehan, Y., Fazaeli, H., and Rezaei, J. (2015). Chemical composition, fermentation characteristics, digestibility, and degradability of silages from two amaranth varieties (Kharkovskiy and Sem), corn, and an amaranth–corn combination. J. Anim. Sci. 93, 5781–5790. doi: 10.2527/jas.2015-9494
Ranjit, N. K., and Kung, L. Jr. (2000). The effect of Lactobacillus buchneri, Lactobacillus plantarum, or a chemical preservative on the fermentation and aerobic stability of corn silage. J. Dairy Sci. 83, 526–535. doi: 10.3168/jds.S0022-0302(00)74912-5
Reich, L. J., and Kung, L. Jr. (2010). Effects of combining Lactobacillus buchneri 40788 with various lactic acid bacteria on the fermentation and aerobic stability of corn silage. Animal Feed Sci. Technol. 159, 105–109. doi: 10.1016/j.anifeedsci.2010.06.002
Rezaei, J., Rouzbehan, Y., and Fazaeli, H. (2009). Nutritive value of fresh and ensiled amaranth (Amaranthus hypochondriacus) treated with different levels of molasses. Animal Feed Sci. Technol. 151, 153–160. doi: 10.1016/j.anifeedsci.2008.12.001
Rezaei, J., Rouzbehan, Y., Fazaeli, H., and Zahedifar, M. (2014). Effects of substituting amaranth silage for corn silage on intake, growth performance, diet digestibility, microbial protein, nitrogen retention and ruminal fermentation in fattening lambs. Animal Feed Sci. Technol. 192, 29–38. doi: 10.1016/j.anifeedsci.2014.03.005
Santos, L. V., Melo, E. A., Silva, A. M., Félix, K., Quezado-Duval, A. M., Albuquerque, G. M., et al. (2020). Weeds as alternate hosts of Xanthomonas euvesicatoria pv. euvesicatoria and X. campestris pv. campestris in vegetable-growing fields in the state of Pernambuco. Brazil. Trop. Plant Pathol. 45, 484–492. doi: 10.1007/s40858-020-00350-z
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, 1–18. doi: 10.1186/gb-2011-12-6-r60
Van Soest, P. V., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Wang, S., Li, J., Dong, Z., Chen, L., Yuan, X., and Shao, T. (2018). The effects of lactic acid bacteria strains isolated from various substrates on the fermentation quality of common vetch (Vicia sativa L.) in Tibet. Grass Forage Sci. 73, 639–647. doi: 10.1111/gfs.12363
Wang, S., Sun, Y., Zhao, J., Dong, Z., Li, J., Nazar, M., et al. (2020). Assessment of inoculating various epiphytic microbiota on fermentative profile and microbial community dynamics in sterile Italian ryegrass. J. Appl. Microbiol. 129, 509–520. doi: 10.1111/jam.14636
Wang, Y., He, L., Xing, Y., Zhou, W., Pian, R., Yang, F., et al. (2019). Bacterial diversity and fermentation quality of Moringa oleifera leaves silage prepared with lactic acid bacteria inoculants and stored at different temperatures. Bioresour. Technol. 284, 349–358. doi: 10.1016/j.biortech.2019.03.139
Wang, Y., Wang, C., Zhou, W., Yang, F. Y., Chen, X. Y., and Zhang, Q. (2018). Effects of wilting and Lactobacillus plantarum addition on the fermentation quality and microbial community of Moringa oleifera leaf silage. Front. Microbiol. 9:1817. doi: 10.3389/fmicb.2018.01817
Ward, T., Larson, J., Meulemans, J., Hillmann, B., Lynch, J., Sidiropoulos, D., et al. (2017). BugBase predicts organism-level microbiome phenotypes. BioRxiv [preprint] doi: 10.1101/133462
Wilkinson, J. M., Lee, M. R., Rivero, M. J., and Chamberlain, A. T. (2020). Some challenges and opportunities for grazing dairy cows on temperate pastures. Grass Forage Sci. 75, 1–17. doi: 10.1111/gfs.12458
Woolford, M. K. (1990). The detrimental effects of air on silage. J. Appl. Microbiol. 68, 101–116. doi: 10.1111/j.1365-2672.1990.tb02554.x
Wu, B., Hu, Z., Wei, M., Yong, M., and Niu, H. (2022). Effects of inoculation of Lactiplantibacillus plantarum and Lentilactobacillus buchneri on fermentation quality, aerobic stability, and microbial community dynamics of wilted Leymus chinensis silage. Front. Microbiol. 13:928731. doi: 10.3389/fmicb.2022.928731
Wu, C., Sun, W., Huang, Y., Dai, S., Peng, C., Zheng, Y., et al. (2022). Effects of different additives on the bacterial community and fermentation mode of whole-plant paper mulberry silage. Front. Microbiol. 13:904193. doi: 10.3389/fmicb.2022.904193
Xu, D., Ding, W., Ke, W., Li, F., Zhang, P., and Guo, X. (2019). Modulation of metabolome and bacterial community in whole crop corn silage by inoculating homofermentative Lactobacillus plantarum and heterofermentative Lactobacillus buchneri. Front. Microbiol. 9:3299. doi: 10.3389/fmicb.2018.03299
Xu, D. M., Wang, N., Rinne, M., Ke, W. C., Weinberg, Z. G., Da, M., et al. (2021). The bacterial community and metabolome dynamics and their interactions modulate fermentation process of whole crop corn silage prepared with or without inoculants. Microb. Biotechnol. 14, 561–576. doi: 10.1111/1751-7915.13623
Yang, F., Zhao, S., Wang, Y., Fan, X., Wang, Y., and Feng, C. (2021). Assessment of bacterial community composition and dynamics in alfalfa silages with and without Lactobacillus plantarum inoculation using absolute quantification 16S rRNA sequencing. Front. Microbiol. 11:629894. doi: 10.3389/fmicb.2020.629894
Yang, L. L., Yuan, X. J., Li, J. F., Dong, Z. H., and Shao, T. (2019). Dynamics of microbial community and fermentation quality during ensiling of sterile and nonsterile alfalfa with or without Lactobacillus plantarum inoculant. Bioresource. Technol. 275, 280–287. doi: 10.1016/j.biortech.2018.12.067
You, S., Du, S., Ge, G., Wan, T., and Jia, Y. (2021). Microbial community and fermentation characteristics of native grass prepared without or with isolated lactic acid bacteria on the Mongolian Plateau. Front. Microbiol. 12:731770. doi: 10.3389/fmicb.2021.731770
Yuan, X. J., Li, J. F., Dong, Z. H., and Shao, T. (2020). The reconstitution mechanism of napier grass microiota during the ensiling of alfalfa and their contributions to fermentation quality of silage. Bioresour. Technol. 297:122391. doi: 10.1016/j.biortech.2019.122391
Zhang, Q., Wu, B., Nishino, N., Wang, X., and Yu, Z. (2016). Fermentation and microbial population dynamics during the ensiling of native grass and subsequent exposure to air. Animal Sci. J. 87, 389–397. doi: 10.1111/asj.12427
Zhang, Y. C., Li, D. X., Wang, X. K., Lin, Y. L., Zhang, Q., Chen, X. Y., et al. (2019). Fermentation dynamics and diversity of bacterial community in four typical woody forages. Annals Microbiol. 69, 233–240. doi: 10.1007/s13213-018-1398-z
Keywords: amaranth, fermentation quality, Lactobacillus plantarum, microbial community, propionic acid, silage
Citation: Zhao M, Wang Z, Du S, Sun L, Bao J, Hao J and Ge G (2022) Lactobacillus plantarum and propionic acid improve the fermentation quality of high-moisture amaranth silage by altering the microbial community composition. Front. Microbiol. 13:1066641. doi: 10.3389/fmicb.2022.1066641
Received: 11 October 2022; Accepted: 28 November 2022;
Published: 21 December 2022.
Edited by:
Yimin Cai, Japan International Research Center for Agricultural Sciences, JapanReviewed by:
Ping Li, Guizhou University, ChinaCopyright © 2022 Zhao, Wang, Du, Sun, Bao, Hao and Ge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gentu Ge, Z2VnZW50dUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.