- 1Department of Oral Microbiology and Immunology, and Dental Research Institute, School of Dentistry, Seoul National University, Seoul, South Korea
- 23BIGS Co., Ltd., Hwaseong, South Korea
- 3Department of Agricultural Biotechnology, Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul, South Korea
- 4Institutes of Green Bio Science and Technology, Seoul National University, Pyeongchang, South Korea
Staphylococcus aureus is a pathogen that causes a variety of infectious diseases such as pneumonia, endocarditis, and septic shock. Methicillin-resistant S. aureus (MRSA) evades virtually all available treatments, creating the need for an alternative control strategy. Although we previously demonstrated the inhibitory effect of sodium propionate (NaP) on MRSA, the regulatory mechanism of this effect remains unclear. In this study, we investigated the regulatory mechanism responsible for the inhibitory effect of NaP on MRSA using RNA-Seq analysis. Total RNAs were isolated from non-treated and 50 mM NaP-treated S. aureus USA300 for 3 h and transcriptional profiling was conducted by RNA-Seq analysis. A total of 171 differentially expressed genes (DEGs) with log2 fold change ≥2 and p < 0.05 was identified in the NaP treatment group compared with the control group. Among the 171 genes, 131 were up-regulated and 40 were down-regulated. Upon gene ontology (GO) annotation analysis, total 26 specific GO terms in “Biological process,” “Molecular function,” and “Cellular component” were identified in MRSA treated with NaP for 3 h. “Purine metabolism”; “riboflavin metabolism”; and “glycine, serine, and threonine metabolism” were identified as major altered metabolic pathways among the eight significantly enriched KEGG pathways in MRSA treated with NaP. Furthermore, the MRSA strains deficient in purF, ilvA, ribE, or ribA, which were the up-regulated DEGs in the metabolic pathways, were more susceptible to NaP than wild-type MRSA. Collectively, these results demonstrate that NaP attenuates MRSA growth by altering its metabolic pathways, suggesting that NaP can be used as a potential bacteriostatic agent for prevention of MRSA infection.
Introduction
Staphylococcus aureus is one of the most frequently isolated human Gram-positive pathogens that causes a variety of human diseases such as pneumonia, gastroenteritis, endocarditis, and septic shock (Lowy, 1998; Alberti et al., 2002). Since S. aureus can readily develop or obtain resistance to antibiotics, emergence of antibiotic-resistant S. aureus is a growing problem in human healthcare as treatment failures are associated with not only risk of the life but also enormous medical cost (Graves et al., 2010; Vestergaard et al., 2019). Methicillin-resistant S. aureus (MRSA) is the most prevalent such pathogen, with persistently high morbidity and mortality (Turner et al., 2019). In the United States, total of 119,247 cases of S. aureus infections, including MRSA, with 19,832 deaths had been reported in 2017 (Kourtis et al., 2019). Community-acquired MRSA (CA-MRSA) has rapidly increased the global incidence of S. aureus infection and S. aureus USA300 is known as one of the most predominant CA-MRSA strain worldwide (Planet, 2017). Furthermore, S. aureus USA300 is considered as an epidemic strain leading to severe infections and outbreaks due to its relatively higher virulence and invasive capacity compared with other CA-MRSA strains (Selan et al., 2018). Vancomycin and daptomycin are used as first-line treatment options for MRSA infection (Liu et al., 2011), but these antibiotic treatments have limitations including low tissue absorption, slow bactericidal efficacy, and antibiotic resistance (Kullar et al., 2013; Moise et al., 2013; Ortwine and Bhavan, 2018). Moreover, since an S. aureus vaccine is not currently available for preventing infection (Clegg et al., 2021), a novel strategy to effectively control MRSA infection, especially USA300 strain, is urgently needed.
Short-chain fatty acids (SCFAs) are small organic monocarboxylic acids that are mainly produced by commensal bacteria in the colon through fermentation of dietary fibers and carbohydrates (Silva et al., 2020). Although roughly 500–600 mM of SCFAs are produced in the human gut per day, their amount and distribution can change depending on the fiber content in the diet and microbiota composition (Macfarlane and Macfarlane, 2003). Acetate (C2), propionate (C3), and butyrate (C4) are representative SCFAs accounting for more than 95% of all SCFAs; and their molar ratio in the human colon is about 60:20:20 (Cummings et al., 1987; Fernandes et al., 2014; Luu et al., 2019). SCFAs are utilized as energy sources by certain bacteria. For instance, Treponema species in dental plaque utilize butyrate produced by Prevotella and Porphyromonas species as their energy source (Hojo et al., 2009). Furthermore, SCFAs can regulate host immune responses in various ways. For example, butyrate suppresses S. aureus lipoprotein-induced nitric oxide production in macrophages (Park et al., 2019). In addition, butyrate and propionate inhibit the activation of antigen-specific CD8+ T lymphocytes by suppressing IL-12 production in dendritic cells (Nastasi et al., 2017).
Previous studies have reported antimicrobial effects of SCFAs on various pathogenic bacteria. For instance, butyrate inhibited the growth of Helicobacter pylori by impairing integrity of the cell envelope (Yonezawa et al., 2012) while acetate suppressed the growth of Escherichia coli by interfering with its methionine biosynthesis (Roe et al., 2002) or perturbing acetyl phosphate concentration needed for sugar uptake (Pinhal et al., 2019). In addition, propionate decreased the growth of clinically isolated Enterococcus faecalis (Jeong et al., 2019b). Consistent with these observations, our previous study demonstrated a bacteriostatic effect of propionate on MRSA (Jeong et al., 2019a). In that study, propionate was more effective at attenuating bacterial growth than acetate and butyrate. Moreover, propionate effectively alleviated MRSA infection by inhibiting bacterial growth in a murine skin infection model. Although these findings suggest propionate as an alternative strategy to treat MRSA infection, underlying mechanisms have not been fully characterized.
Recently, development of transcriptomics based on next-generation sequencing allows quantitative analysis of the expression of numerous genes simultaneously and consequential comparative analysis (Westermann and Vogel, 2021). Among transcriptomics methods, RNA sequencing (RNA-Seq) permits meticulous evaluation of transcription levels compared with other methods (Westermann and Vogel, 2021). Thus, we conducted transcriptomic analysis to characterize gene alterations to identify the underlying mechanisms of the bacteriostatic effect of propionate on MRSA.
Materials and methods
Reagent and chemicals
Sodium propionate (NaP) was purchased from Sigma-Aldrich Inc. (St. Louis, MO, United States). NaP was dissolved in endotoxin-free distilled water and filtered using a syringe filter (0.2 μm pore size; Corning, Corning, NY, United States). Trypticase soy broth (TSB) and Bacto agar were purchased from BD Biosciences (Franklin Lakes, NJ, United States).
Bacterial growth
Staphylococcus aureus USA300 was used as a representative MRSA strain, and its wild-type and mutant strains deficient for the purF (∆purF), ilvA (∆ilvA), ribE (∆ribE), or ribA (∆ribA) gene were obtained from the Nebraska Transposon Mutant Library (Omaha, NE, United States). A single colony of each strain of S. aureus USA300 grown on a TSB agar plate (TSB broth containing 1.5% Bacto agar) was cultured in TSB broth at 37°C overnight, and 1% of the culture was inoculated into fresh TSB broth in the presence or absence of 50 mM NaP on a flat-bottom 96-well plate (Thermo Scientific, Waltham, MA, United States). The bacteria were cultured at 37°C under aerobic shaking (180 rpm/min) culture conditions, and the bacterial growth was estimated by measuring optical density at 600 nm every 1 or 2 h up to 12 h using a microplate reader (SPARK 10 M, Tecan, Zurich, Swiss).
Minimum inhibitory concentration and minimum bactericidal concentration test
The minimum inhibitory concentration (MIC) test was performed as previously described (Jeong et al., 2019a). Briefly, 1% of the overnight cultured S. aureus USA300 was inoculated into fresh TSB broth containing serially diluted (0, 3.9, 7.8, 15.6, 31.3, 62.5, 125, 250, or 500 mM) NaP on a flat-bottom 96-well plate. The bacteria were cultured at 37°C for 24 h under aerobic shaking culture conditions, and optical density at 600 nm was measured using a microplate reader (SPARK, Tecan). MIC was defined as the minimum concentration of NaP needed for non-visible growth of S. aureus USA300 at 24 h. To determine minimum bactericidal concentration (MBC), the bacterial cultures showing low or no bacterial growth on the MIC test were inoculated into fresh TSB broth and incubated at 37°C for 24 h under aerobic shaking culture conditions. Bacterial growth was examined by measuring optical density at 600 nm (SPARK).
Extraction and purification of RNA
One percent of the overnight cultured S. aureus USA300 was inoculated into fresh TSB broth containing 50 mM NaP in 50 ml conical tubes (SPL Life Science, Gyeonggi-Do, Republic of Korea) and cultured at 37°C for 3 h under aerobic shaking culture conditions. The bacteria were harvested by centrifugation at 8,000 × g for 10 min, and bacterial pellets were suspended in phosphate-buffered saline (PBS) containing 100 μg/ml of lysostaphin (Sigma-Aldrich Inc.). After incubation at 37°C for 15 min, the pellets were collected by centrifugation at 8,000 × g for 10 min; and total RNA was isolated and purified using a RNeasy® Mini kit (Qiagen, Gaithersburg, MD, United States) according to the manufacturer’s instructions. The purified total RNA was used to construct complementary DNA (cDNA) libraries for RNA-Seq.
Construction of a cDNA library and RNA-Seq
The construction of cDNA library and RNA-Seq data were performed by Macrogen (Seoul, Republic of Korea). The mRNA in the prepared total RNA was initially converted into a cDNA library using the Illumina® TruSeq™ stranded mRNA sample prep kit (Illumina Inc., San Diego, CA, United States) according to the manufacturer’s guidance. In brief, bacterial rRNA was removed using the NEBNext® rRNA depletion kit (NEB, Ipswich, MA, United States). The purified mRNA was fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were transcribed into first-strand cDNA using SuperScript II reverse transcriptase (Invitrogen, Waltham, MA, United States) and random primers. Then, the cDNA was synthesized to second-strand cDNA using DNA polymerase I, RNase H, and dUTP. Finally, single adenine bases were added to the cDNA fragments using a 3′ to 5′ exonuclease, and these adenylated cDNA fragments were ligated with adapters. The products were purified and enriched by PCR to create the cDNA library. The cDNA libraries were quantified using a KAPA library quantification kit (KAPA Biosystems, Wilmington, MA, United States) according to the qPCR quantification protocol guide (KAPA Biosystems) and quantified using the Agilent D1000 ScreenTape System (Agilent Technologies, Santa Clara, CA, United States). The resulting cDNA libraries (three libraries per treatment group) were sequenced on the NovaSeq 6,000 platform (Illumina Inc.).
Identification and annotation of differentially expressed genes
The quality of raw data was checked by the FastQC.1 Trimmomatic (v.0.38) was then applied to examine adaptor contamination and to remove low-quality reads (Bolger et al., 2014). The clean reads were aligned onto the reference genome of S. aureus subsp. aureus USA300_FPR3757 (NCBI Reference Sequence: NC_007793.1) using HISAT2 (2.1.0) aligner (Kim et al., 2015). In addition, the abundance of mapped reads was counted and measured by fragments per kilobase of exon per million fragments mapped (FPKM; Anders et al., 2015). For analysis of differentially expressed genes (DEGs), the log2 values (FPKM) were initially calculated and normalized by quantile normalization to reduce systematic bias. Statistical analysis between treatment groups was conducted by independent t-test with the mean FPKM value for each gene, and genes with a fold-change ≥2 or ≤−2 with p < 0.05 were assigned as DEGs. All statistical analysis for DEGs was performed using the DESeq2 R package (v.1.26.0; Anders and Huber, 2010). For functional analysis of DEGs, Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis was conducted using a KEGG orthology-based annotation system (KOBAS; Bu et al., 2021), and gene ontology (GO) annotation analysis was performed using BLAST search against UniProt database (Ashburner et al., 2000). A p-value of <0.05 calculated by Fisher’s exact test was set as the cutoff criteria for the KEGG pathway enrichment analysis.
Real-time PCR validation
To validate the RNA-Seq data, real-time PCR analysis was conducted to quantify the mRNA transcripts of eight randomly selected DEGs as previously described (Song et al., 2021) with minor modifications. Briefly, one microgram of total RNA isolated from S. aureus USA300 under identical processing conditions with those of the RNA-Seq samples was subjected to cDNA synthesis using random hexamers and reverse transcriptase (Promega, Madison, WI, United States). The mRNA expression was determined using the StepOnePlus real-time system (Applied Biosystems, Waltham, MA, United States) under these reaction conditions: initial denaturation at 95°C for 10 min, followed by amplification by 40 cycles of 95°C for 15 s, and 60°C for 1 min. In addition, 16S rRNA was used as an internal control to normalize the target gene mRNA expression. The information from 10 randomly selected DEGs and details of primers used in the current study are listed in Supplementary Table S1.
Results
Bacteriostatic effect of NaP on methicillin-resistant Staphylococcus aureus
To confirm the growth inhibitory effect of NaP on MRSA as previously reported (Jeong et al., 2019a), growth of S. aureus USA300 in the presence or absence of 50 mM NaP was initially examined at the indicated time points under aerobic shaking culture conditions. As shown in Figure 1A, the growth inhibitory property of NaP was potent up to 12 h. Since the growth inhibition peaked at 6 h, RNA for RNA-Seq analysis was extracted 3 h post-treatment when initial growth inhibition by NaP was observed. To confirm that the inhibitory property of NaP on MRSA growth is bacteriostatic and not bactericidal, MIC and MBC tests were conducted. Since bacterial growth of S. aureus USA300 was not observed under 250 mM NaP treatment, this concentration was determined as the MIC of NaP (Figure 1B). Furthermore, an MBC test was conducted by examining the growth of bacterial cultures having low or no growth in the MIC test. Since bacterial growth was observed for all tested bacterial cultures even at 500 mM NaP, there was no MBC for NaP (Figure 1C). These results suggest that the inhibitory effect of NaP on MRSA growth is mediated by its bacteriostatic effect.
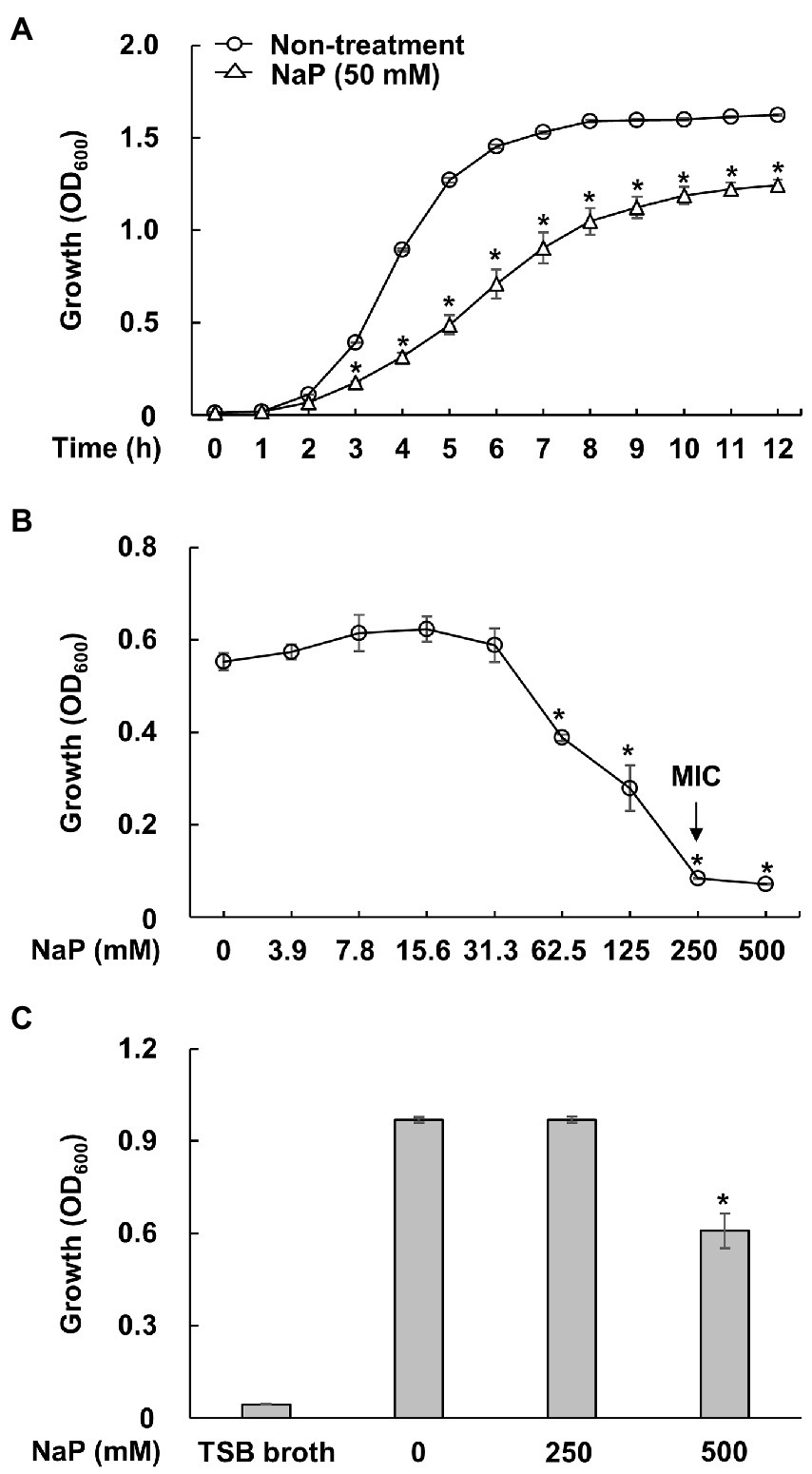
Figure 1. Bacteriostatic effect of NaP on MRSA. (A) S. aureus USA300 was cultured in the presence or absence of 50 mM NaP at 37°C for 12 h under aerobic shaking culture conditions. Bacterial growth was examined by measuring optical density at 600 nm hourly up to 12 h. Data shown are the mean value ± SEM of triplicate samples. *p < 0.05 compared with the non-treatment group at each time point. (B) S. aureus USA300 was incubated in the presence of various concentrations of NaP at 37°C for 24 h under aerobic shaking culture conditions. After incubation, optical density at 600 nm was measured; the black arrows in the figure indicate the MIC of NaP. Data shown are the mean ± SEM of triplicate samples. *p < 0.05 compared with the control group. (C) The bacteria cultures showing low or no bacterial growth on the MIC test were inoculated in fresh TSB broth and incubated at 37°C for 24 h under aerobic shaking culture conditions. The bacterial growth was then examined by measuring optical density at 600 nm. Data shown are the mean ± SEM of triplicate samples. *p < 0.05 compared with the non-treatment group.
Differentially expressed genes and their validation
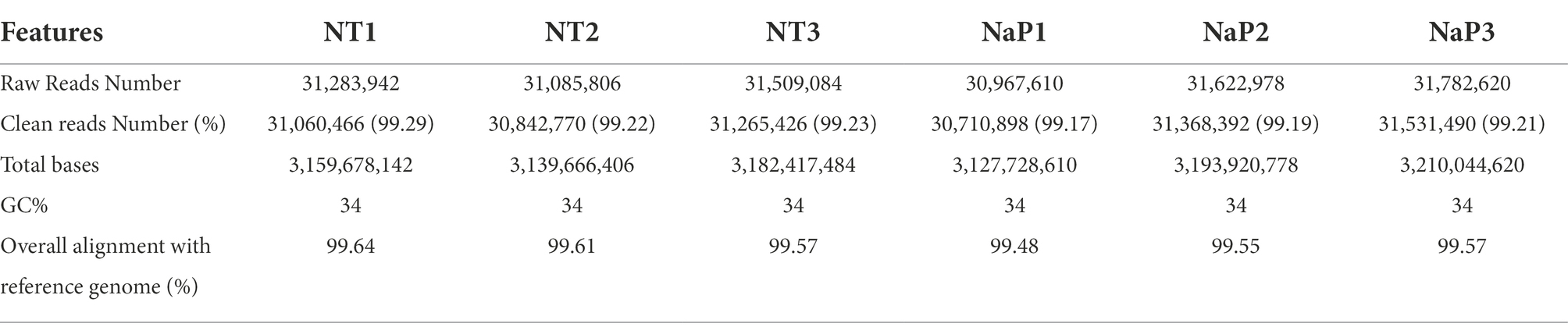
To obtain a comprehensive analysis of the bacteriostatic effect of NaP on MRSA, six cDNA libraries were constructed from the NaP-treated and control S. aureus USA300, and each library was sequenced with Illumina NovaSeq 6,000 platform. RNA-Seq generated 31,085,806-31,782,620 raw reads per each cDNA library (a total of 188,252,040 from six libraries). For quality control, raw reads were confirmed using FastQC software (data not shown). Among them, 30,710,898-31,060,466 clean reads (99.17–99.29%) were generated from each sample after removing contaminated adaptor and low-quality sequences using Trimmomatic software (Table 1). Furthermore, we found that 99.48–99.64% of the clean reads were successfully aligned to the reference genome of S. aureus subsp. aureus USA300_FPR3757 (NCBI Reference Sequence: NC_007793.1). The NaP treatment group was compared with the control group (NT) to estimate the transcription profile of MRSA under NaP treatment. The selection criteria for DEGs were log2 fold change ≥2 or ≤−2 and p < 0.05. The expression of 171 genes was significantly different in the NaP treatment group compared with its control group. Among these, 131 genes were up-regulated, and 40 genes were down-regulated (Figure 2A). To examine a DEG profile under NaP treatment conditions, DEGs with similar metabolic functions were clustered by hierarchical clustering analysis using FPKM values. The clustering analysis showed that DEG expression patterns of MRSA were distinctly altered by NaP treatment (Figure 2B). The up- and down-regulated genes from the NaP treatment are listed in Supplementary Tables S2, S3, respectively. Furthermore, real-time PCR was conducted to validate the transcriptional profiles of RNA-Seq for the eight randomly selected DEGs adh, purN, purF, purM, tpiA, pgm, gap, and pgk. As shown in Figure 3, the DEG validation results were similar to those obtained from RNA-Seq analysis, suggesting that the DEGs were successfully identified by RNA-Seq.
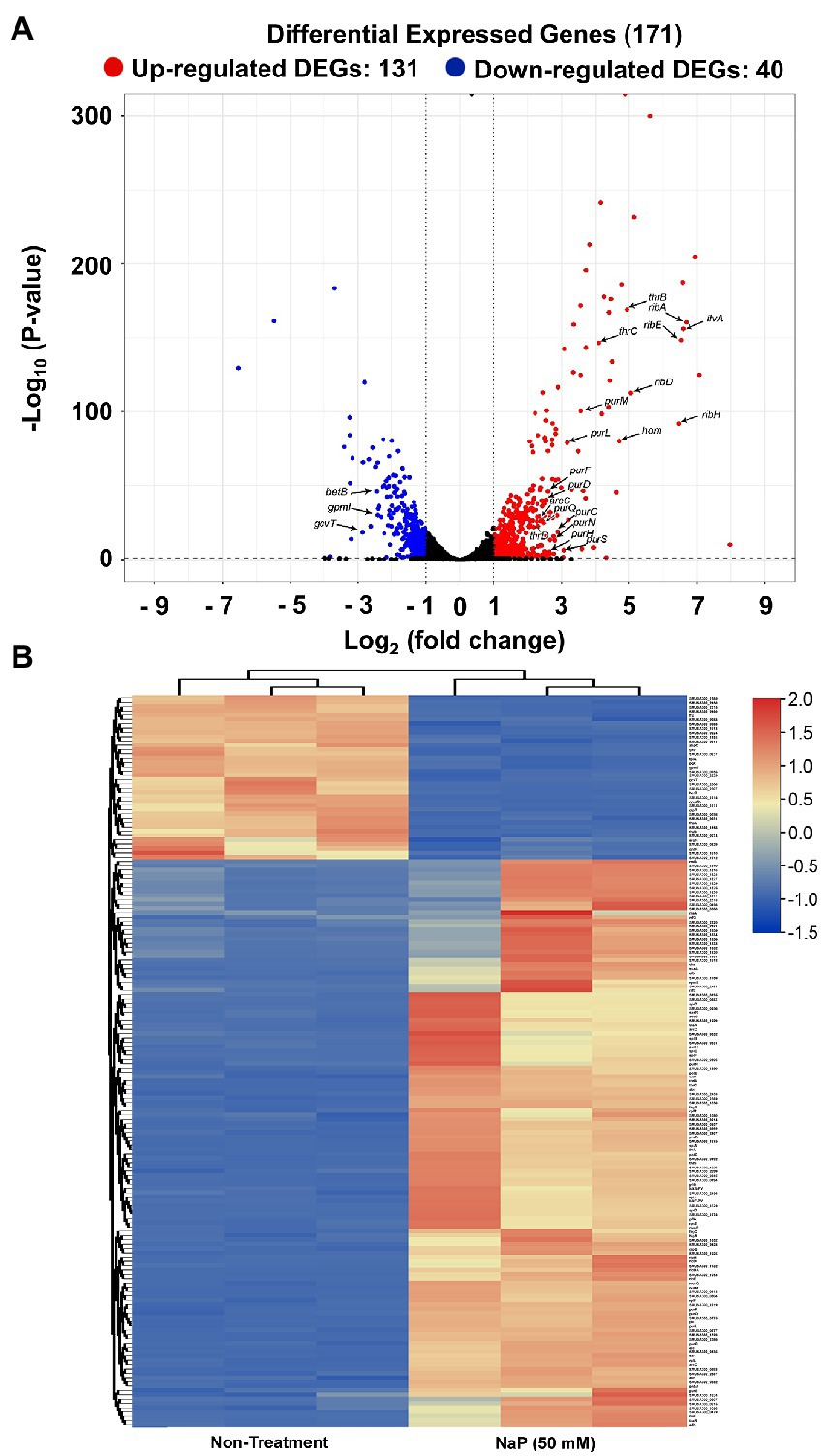
Figure 2. Differentially expressed genes in MRSA treated with NaP. The cDNA libraries were constructed from S. aureus USA300 incubated with or without 50 mM NaP for 3 h under aerobic shaking culture conditions. The libraries were subjected to RNA-Seq as described in the Materials and methods section. (A) Volcano plot of DEGs in the NaP-treated MRSA. Among the genes with log2 fold change ≥2 or ≤−2, up- and down-regulated genes at p < 0.05 in the NaP treatment group compared to non-treatment group were classified into DEGs and are represented by red and blue dots, respectively. (B) Heat map of the hierarchical clustering based on 171 DEGs (log2 fold change ≥2 or ≤−2, p < 0.05). Red and blue indicate up- and down-regulated DEGs in the NaP treatment group compared to the non-treatment group, respectively.
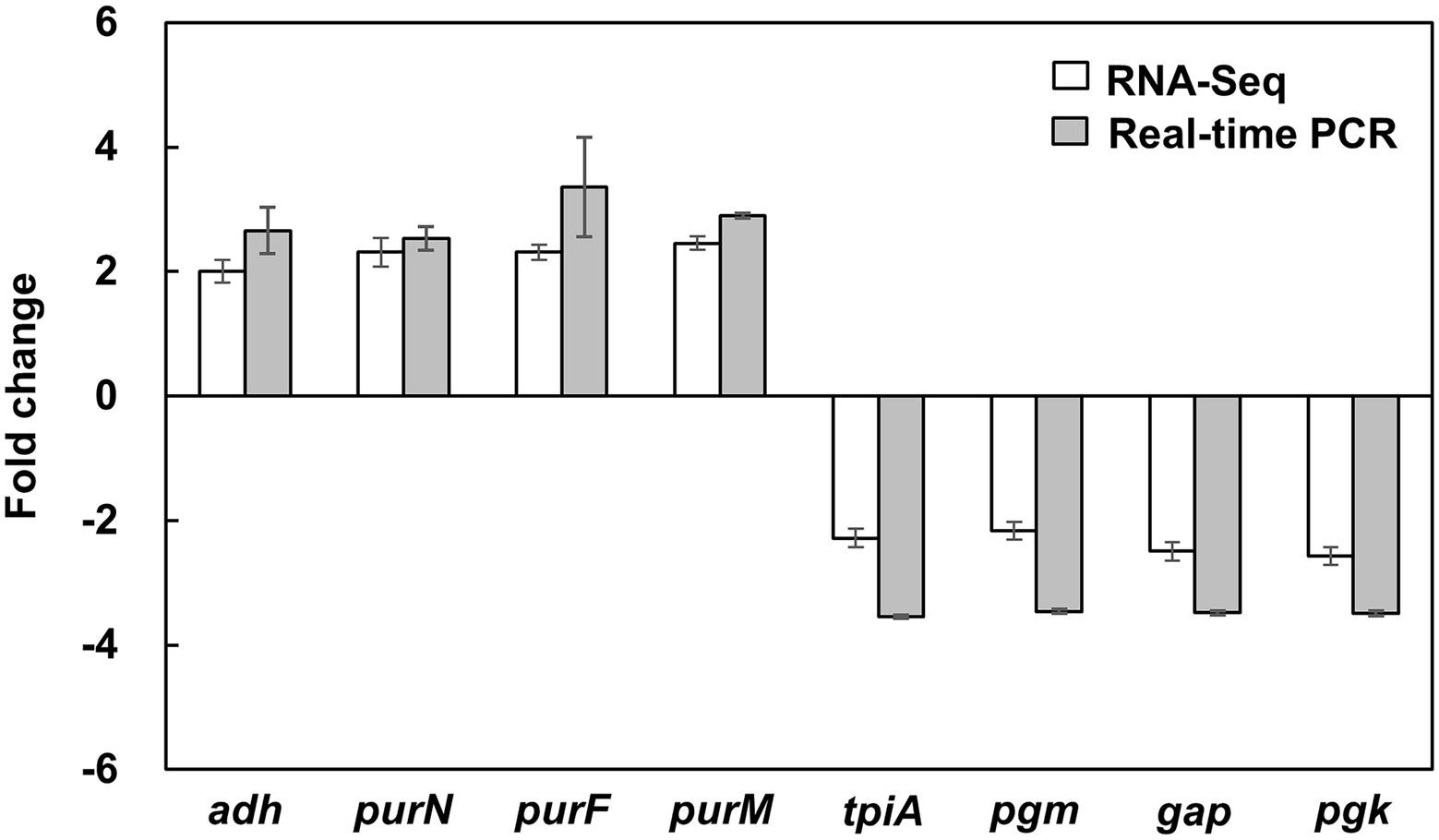
Figure 3. Validation of DEGs in MRSA treated with NaP using real-time PCR. S. aureus USA300 was cultured in the presence or absence of 50 mM NaP at 37°C for 3 h under aerobic shaking culture conditions. After incubation, total RNA was isolated and subjected to real-time PCR analysis for the eight randomly selected DEGs shown in Supplementary Table S1 as described in the Materials and methods. Data shown are the mean ± SEM of triplicate samples after normalization with 16S rRNA for each target gene.
Gene ontology annotation analysis
To understand the function of the identified DEGs in the bacteriostatic effect of NaP on MRSA, GO annotation analysis was conducted for the 171 DEGs. For this, the DEGs were assigned to one or more GO terms and mainly categorized into three categories, including “cellular components,” “molecular function,” and “biological process” comprising 200, 129, and 93 subcategories, respectively. The total 26 GO terms at subcategory levels belonging to the three categories were identified and illustrated in Figure 4A. In the cellular components’ category, nine specific GO terms were identified in MRSA treated with NaP, including “organelle part,” “extracellular region part,” “membrane part,” “organelle,” “extracellular region,” “membrane,” “protein-containing complex,” “cell,” and “cell part.” In addition, six specific GO terms belonging to category of molecular function were annotated, such as “transcription regulation activity,” “structural molecule activity,” “antioxidant activity,” “binding,” “transporter activity,” and “catalytic activity.” On the other hand, total 10 specific GO terms composed of biological process were annotated, including “cell killing,” “regulation of biological process,” “signaling,” “biological adhesion,” “response to stimulus,” “multi-organism process,” “biological regulation,” “component organization and biogenesis,” “cellular process,” and “localization.” Details of the identified GO terms at subcategory levels together with a list of DEGs belonging to each GO term are presented in Supplementary Table S4.
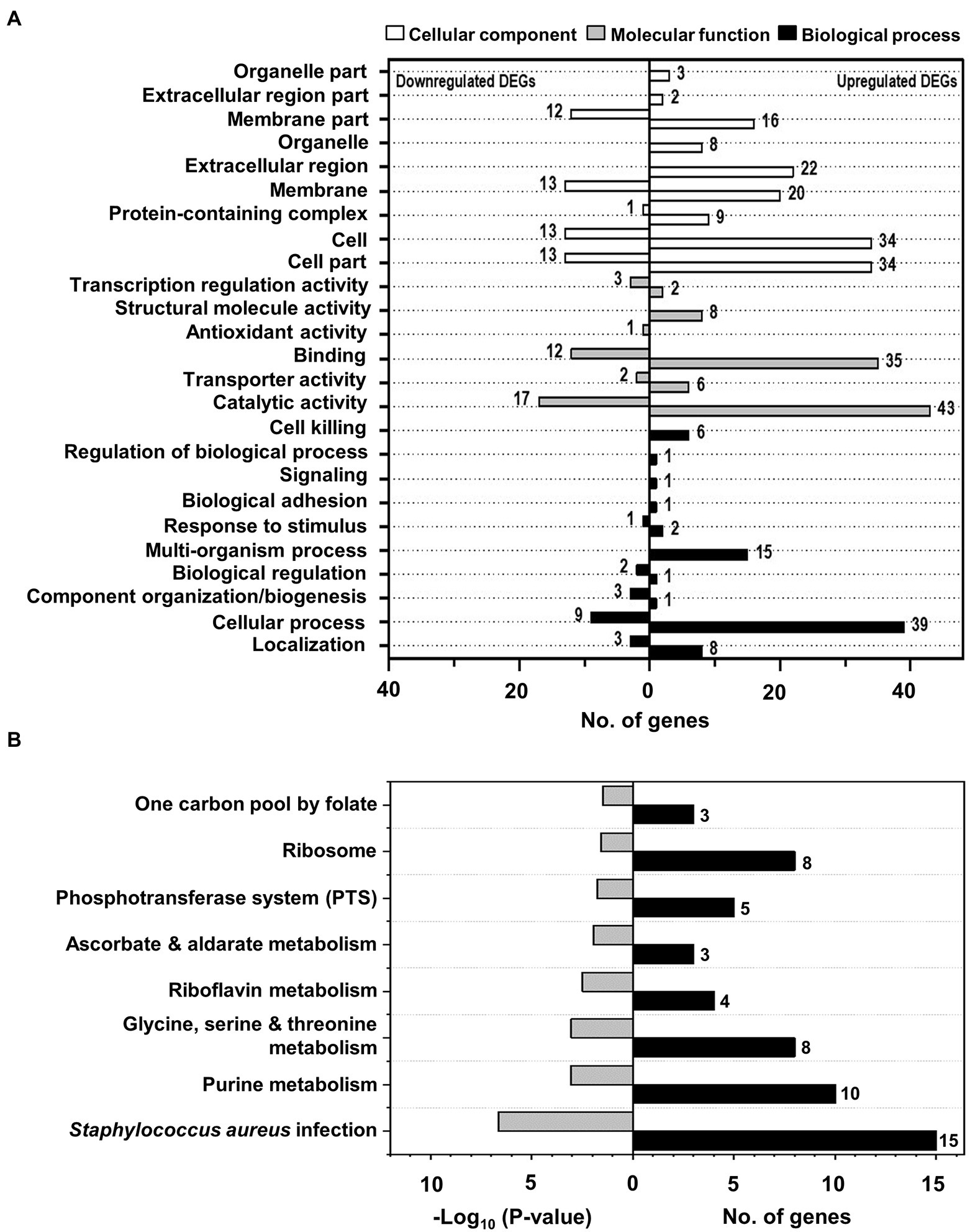
Figure 4. The identified GO terms and significantly enriched KEGG pathways of DEGs in MRSA treated with NaP. (A) GO annotation analysis for 171 DEGs was conducted using the UniProt GO database. The figure shows the identified 26 GO terms at subcategory levels of the three categories, including “cellular components,” “molecular function,” and “biological process,” with the numbers of up- and down-regulated DEGs belonging to each identified GO term. (B) KEGG pathway enrichment for 171 DEGs was performed using KOBAS, and significantly enriched pathways were identified by hypergeometric distribution at p < 0.01. The figure shows the eight most significantly enriched KEGG pathways with their p-values and the number of DEGs belonging to each enriched pathway.
Kyoto encyclopedia of genes and genomes pathway enrichment
To further identify the involved pathways in the inhibition of MRSA growth by NaP treatment, DEGs were mapped to the KEGG database, and KEGG pathway enrichment analysis was performed. A total of eight pathways was significantly enriched: “one carbon pool by folate”; “ribosome”; “phosphotransferase system (PTS)”; “ascorbate and aldarate metabolism”; “riboflavin metabolism”; “glycine, serine, and threonine metabolism”; “purine metabolism”; and “Staphylococcus aureus infection” (Figure 4B; Supplementary Table S5; Supplementary Figure S1–S4). Since bacteriostatic agents mainly affect the metabolic state of the bacterium (Lobritz et al., 2015), we selected “purine metabolism”; “riboflavin metabolism”; and “glycine, serine, and threonine metabolism” as potent metabolic pathways responsible for the bacteriostatic effect of NaP on MRSA. Since it has been reported that purine metabolism plays a key role in cell wall synthesis which is closely related to changes in bacterial morphology (Sobral et al., 2007), we previously examined the effects of NaP on morphology of S. aureus USA300 by using scanning electron microscopy. The morphological changes, including bacterial size and membrane integrity, were not observed by NaP treatment when compared to those of non-treatment group (Jeong et al., 2019a). Furthermore, we examined the expression of genes in the selected pathways (Supplementary Figure S1). In the purine metabolic pathway, 9 of 10 genes encoding enzymes involved in purine biosynthesis were up-regulated by NaP treatment, including phosphoribosylamino imidazole-succinocarboxamide synthase (purC; EC: 6.3.2.6), phosphoribosylformyl glycinamidine synthase (purS; EC: 6.3.5.3), IMP cyclohydrolase (purH; EC: 2.1.2.3; 3.5.4.10), phosphoribosylglycinamide formyltransferase (purN; EC: 2.1.2.2), phosphoribosylamine-glycine ligase (purD; EC: 6.3.4.13), phosphoribosylformyl glycinamidine synthase II (purL; EC: 6.3.5.3), phosphoribosylformylglycinamidine synthase I (purQ; EC: 6.3.5.3), phosphoribosylaminoimidazole synthetase (purM; EC: 6.3.3.1), and amidophosphoribosyl transferase (purF; EC: 2.4.2.14). A gene encoding carbamate kinase (arcC; EC: 2.7.2.2) was not up-regulated by NaP. In the riboflavin metabolic pathway, four genes encoding enzymes and other proteins involved in riboflavin synthesis were significantly up-regulated by NaP, such as riboflavin synthase subunit alpha (ribE; EC: 2.5.1.9), riboflavin biosynthesis protein (ribD; EC: 3.5.4.26; 1.1.1.193), 6,7-dimethyl-8-ribityllumazine synthase (ribH; EC: 2.5.1.78), and riboflavin biosynthesis protein (ribA; EC: 3.5.4.25; 4.199.12). In the glycine, serine, and threonine metabolic pathway, five genes encoding enzymes involved in threonine synthesis and down-stream metabolism were up-regulated, including threonine dehydratase (ilvA; EC: 4.3.1.19), homoserine kinase (thrB; EC: 2.7.1.39), homoserine dehydrogenase (hom; EC: 1.1.1.3), threonine synthase (thrC; EC: 4.2.3.1), and aspartate kinase (thrD; EC: 2.7.2.4). However, three genes were down-regulated in this metabolic pathway, such as phosphoglyceromutase (gpmI; EC: 5.4.2.12), glycine cleavage system aminomethyltransferase T (gcvT; EC: 2.1.2.10), and glycine betaine aldehyde dehydrogenase (betB; EC: 1.2.1.8).
Function of identified pathways in growth of methicillin-resistant Staphylococcus aureus
To further investigate the role of the selected pathways in the bacteriostatic effect of NaP, bacterial growth of S. aureus USA300 strains that lack the up-regulated DEGs in the selected metabolic pathways was evaluated in the presence of NaP and compared with that of wild-type. Among various up-regulated DEGs in the selected pathways, the four DEG-deficient strains purF (∆purF) for purine metabolism; ilvA (∆ilvA) for glycine, serine, and threonine metabolism; and ribE (∆ribE) and ribA (∆ribA) for riboflavin metabolism were adapted for growth analysis. As shown in Figures 5A–D, all tested mutant strains were more susceptible to NaP compared with wild-type after 4 h. These results suggest that “purine metabolism”; “riboflavin metabolism”; and “glycine, serine, and threonine metabolism” are critical metabolic pathways responsible for the bacteriostatic effect of NaP on MRSA.
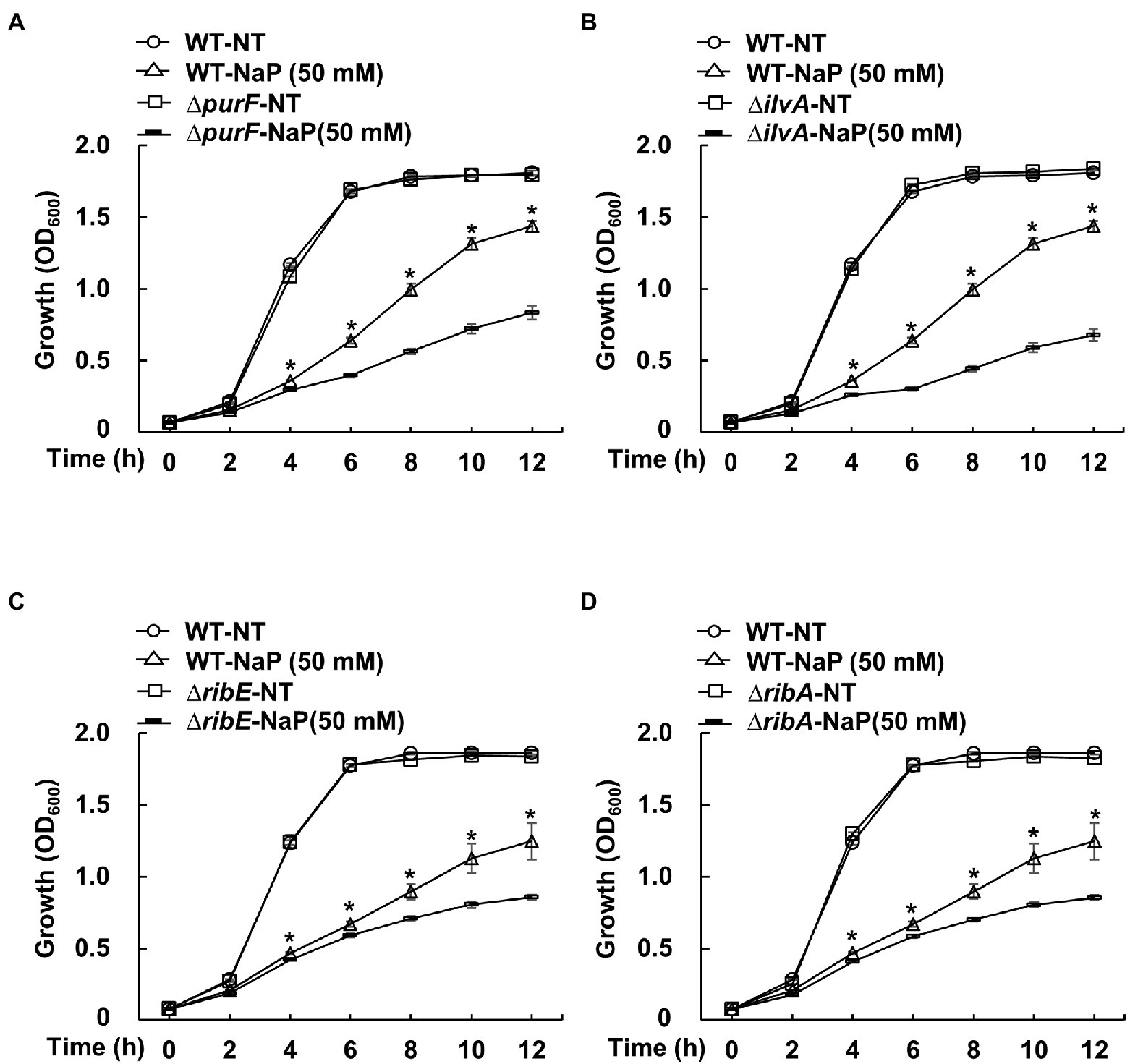
Figure 5. The effect of NaP on growth of MRSA mutant strains deficient for the up-regulated DEGs in the selected metabolic pathways. S. aureus USA300 wild-type and mutant strains that lack the up-regulated DEGs of the metabolism pathways including (A) purF (∆purF) for purine metabolism; (B) ilvA (∆ilvA) for glycine, serine, and threonine metabolism; and (C) ribE (∆ribE) and (D) ribA (∆ribA) for riboflavin metabolism were incubated with 50 mM NaP for 12 h under aerobic shaking culture conditions. Bacterial growth was examined by measuring optical density at 600 nm every 2 h up to 12 h. Data shown are the mean ± SEM of triplicate samples. *p < 0.05 compared with the wild-type treated with 50 mM NaP (WT-NaP) at each time point.
Discussion
MRSA is a serious clinical threat with persistently high morbidity and mortality, suggesting an urgent need for alternative therapeutic strategies. We have previously demonstrated potential utilization of NaP as an effective bacteriostatic agent against MRSA infection (Jeong et al., 2019a). However, the underlying mechanisms were unclear. In the current study, we found genetic evidence for the bacteriostatic effect of NaP on MRSA by transcriptomic analysis using RNA-Seq. In NaP-treated MRSA, 131 and 40 genes were significantly up-regulated and down-regulated, respectively. Moreover, KEGG pathway enrichment analysis showed that “purine metabolism”; “riboflavin metabolism”; and “glycine, serine, and threonine metabolism” were major metabolic pathways altered by NaP. Furthermore, the growth of MRSA strains deficient for purF, ilvA, ribE, or ribA, which are up-regulated genes in these metabolic pathways, was more susceptible to NaP compared with wild-type, suggesting that the bacteriostatic effect of NaP on MRSA is mediated through alteration of these pathways. Based on the KEGG pathway enrichment analysis, purine metabolism seems to function as an up-stream metabolic pathway regulating both riboflavin metabolism and glycine, serine, and threonine metabolism (Supplementary Figure S1A). In fact, since purine metabolic pathway produces guanosine triphosphate and glycine, which are essential precursor metabolites for synthesis of riboflavin, and serine and threonine, respectively, it can directly regulate these two metabolic pathways (Averianova et al., 2020; Shimizu and Matsuoka, 2022). Therefore, we concluded that purine metabolism is a central metabolic pathway responsible for NaP-induced bacteriostatic effect in MRSA growth.
In the current study, we found differentially expressed purine metabolic pathway with eight up-regulated DEGs in NaP-treated S. aureus USA300. Since nine of 10 up-regulated genes, purC, purS, purH, purN, purD, purL, purQ, purM, and purF, belong to the pur purine biosynthetic operon, NaP probably up-regulates purine biosynthesis of MRSA. Similar to our results, enhanced purine synthesis was observed in E. coli under stress conditions induced by various antibiotics (Yang et al., 2019). Although purine biosynthesis is critical for bacterial growth via DNA and RNA syntheses and ATP energy supply (Zhang et al., 2008), the mechanism by which increased purine biosynthesis during stress conditions is associated with bacterial growth is unclear. Based on recent studies, this can be explained by enhanced DNA damage following increased purine biosynthesis under stress conditions (Yee et al., 2015; Yang et al., 2019; Lopatkin and Yang, 2021). Thus, stress conditions, such as those occurring with antibiotic treatment, induced disruption of the nucleotide pool, causing adenine deficiency and triggering purine biosynthesis. Also, activated central carbon metabolism resulting from increased purine biosynthesis subsequently enhanced reactive oxygen species (ROS) production. Finally, ROS-damaged DNA leads to inhibited growth or increased death of bacteria. Therefore, the bacteriostatic effects of NaP may be affected by up-regulated purine biosynthesis leading to enhanced bacterial DNA damage. This possibility is supported by the higher susceptibility of MRSA mutants involved in purine biosynthesis, such as purM, to various stress conditions, including antibiotics, heat, and low pH (Yee et al., 2015).
The riboflavin metabolic pathway was also differentially expressed in MRSA when treated with NaP. In the pathway, the four genes ribE, ribD, ribH, and ribA existing in the rib riboflavin biosynthetic operon were up-regulated, suggesting that NaP probably enhances MRSA riboflavin biosynthesis. Moreover, since riboflavin was mainly synthesized from guanosine-5′-triphosphate derived from the purine biosynthetic pathway (Garcia-Angulo, 2017), the NaP-induced purine biosynthesis may cause increased riboflavin biosynthesis. The role of NaP-induced riboflavin biosynthesis in the bacteriostatic effect of NaP can be explained. In bacteria, riboflavin plays important roles in bacterial growth through various bacterial metabolic pathways, such as fatty acid metabolism, tricarboxylic acid cycle function, and electron transport chain progression, via its derivatives, including flavin mononucleotide and flavin adenine dinucleotide (FAD; Massey, 2000; Lin et al., 2014). However, enhanced riboflavin synthesis can result in excessive ROS production by promoting autoxidation of FAD through FADH2 oxidation, leading to ROS-induced DNA damage (Imlay, 2013). A previous study reported that stress conditions induced by an anti-bacterial reagent in Edwardsiella tarda up-regulated riboflavin biosynthesis and subsequent ROS production, leading to ROS-mediated DNA damage and bacterial growth inhibition (Ye et al., 2018). Therefore, excessive ROS production through enhanced riboflavin biosynthesis by NaP may be a potential mechanism responsible for the bacteriostatic effect of NaP on MRSA.
The differentially expressed glycine, serine, and threonine metabolic pathway caused by NaP administration can also inhibit MRSA growth. Up-regulation of four genes involved in threonine biosynthesis from L-aspartate, hom, thrB, thrC, and thrD, may involve enhanced threonine biosynthesis in MRSA under NaP treatment. Synthesized threonine can then be converted into glycine and serine and, subsequently, pyruvate for energy or into isoleucine (Fang et al., 2021). During the conversion, ilvA is responsible for conversion of serine to pyruvate and, together with sdaAB, the conversion of threonine into 2-oxobutyrate (2-OBA), a precursor metabolite of isoleucine. However, NaP did not affect the gene expression of enzymes involved in conversion of threonine into glycine or of glycine into serine, suggesting that NaP-induced ilvA expression is involved in 2-OBA generation. Of note, 2-OBA is a toxic metabolic intermediate leading to the arrest of cell growth in most microorganisms (LaRossa et al., 1987; Fang et al., 2021). Thus, NaP-induced ilvA expression following up-regulated threonine biosynthesis may suppress MRSA growth by excessive 2-OBA production. However, we also observed down-regulation of gpmI, gcvT, and betB in this metabolic pathway; alteration of the gcvT gene by NaP is another possible explanation for the inhibitory effect of NaP on MRSA growth. Although glycine is an essential amino acid involved in bacterial growth as a precursor of pyruvate for energy metabolism (Fang et al., 2021), excessive glycine can inhibit growth of various bacteria, including Bacillus subtilis, Streptococcus lactis, Streptomyces griseus, and S. aureus (Tezuka and Ohnishi, 2014). To counteract this phenomenon, most bacteria possess a glycine cleavage system as a detoxification mechanism to mediate the oxidative cleavage of glycine into CO2, NH4+, and a methylene group (Kikuchi et al., 2008). Therefore, NaP-induced down-regulation of gcvT, which is a key gene of the glycine cleavage system in S. aureus, may lead to suppressed MRSA growth by inhibiting this system.
Based on our results, the bacteriostatic effect of NaP against MRSA seems to be mediated by different mechanism compared with other bacterial growth inhibitory substances. According to the previous studies, propionate effectively inhibited bacterial growth of S. aureus and Salmonella typhimurium by disrupting their intracellular pH homeostasis (Wang et al., 2014; Jacobson et al., 2018). On the other hand, inorganic acid and alkaline shock suppressed S. aureus growth by inducing alteration of gene expression for adaptation to pH stress (Anderson et al., 2010). Thus, we have examined the pH changes in the extracellular culture medium of S. aureus USA300 in the presence of NaP. However, NaP did not affect pH of the culture medium throughout the experiment (data not shown). Therefore, bacteriostatic effect of NaP might be mediated by altered metabolic pathways, including purine metabolism, riboflavin metabolism, and glycine, serine, and threonine metabolism, but not by pH stress.
Among various antibiotics resistant to S. aureus, MRSA is the most prevalent form, with persistently high morbidity and mortality (Turner et al., 2019). Although vancomycin and daptomycin are currently approved anti-bacterials for treatment of MRSA infection (Liu et al., 2011), reports on treatment failure with vancomycin and decreased susceptibility to daptomycin necessitate the development of alternative strategies (Ortwine and Bhavan, 2018). Recently, semisynthetic lipoglycopeptides, including telavancin, dalbavancin and oritavancin, have been suggested as alternative anti-bacterials with reliable bactericidal activity against MRSA. However, these often accompany various adverse effects such as hypersensitivity, anaphylaxis, and osteomyelitis (Ortwine and Bhavan, 2018). In our previous study, bacterial load and dermonecrosis in the co-treatment group of S. aureus USA300 and 50 mM NaP were effectively reduced compared to S. aureus USA300 single-treatment group in a murine MRSA skin infection model (Jeong et al., 2019a). Furthermore, the same concentration of NaP (50 mM) did not induce pathology or inflammation in the murine MRSA skin infection model, suggesting a non-toxic effect of NaP at 50 mM. In addition, the non-toxic effect of high concentration of NaP even at 200 mM has been demonstrated in a mouse with a month of NaP oral administration (Ciarlo et al., 2016). Moreover, since propionate is present in various sites of the human body, such as liver, lung, and colon, its utilization as a bacteriostatic agent against MRSA suggests biocompatibility. The clinical applications of propionate as a potential antimicrobial agent would be appropriate as a bacteriostatic agent for the prevention of MRSA infection and/or its disease progression rather than a bactericidal agent for MRSA treatment. Since the underlying mechanisms responsible for the bacteriostatic effect of propionate have remained unclear so far, the current findings might contribute to the development of an effective and safe antimicrobial agent using propionate against MRSA infection.
Data availability statement
The raw data associated with this study are accessible through the NCBI Sequence Read Archive (SRA) database under BioProject ID PRJNA887926.
Author contributions
JI, DL, O-JP, and SH designed the research. JI, DL, and, SN carried out the experiments. JI, DL, O-JP, SN, JP, C-HY, and SH analyzed and interpreted the data. JI, DL, O-JP, SN, JP, C-HY, and SH prepared and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2018R1A5A2024418, NRF-2019R1A2C2007041, NRF-2022M3A9F3082330, and RS-2022-00164722).
Conflict of interest
SN and JP were employed by 3BIGS Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1063650/full#supplementary-material
Footnotes
References
Alberti, C., Brun-Buisson, C., Burchardi, H., Martin, C., Goodman, S., Artigas, A., et al. (2002). Epidemiology of sepsis and infection in ICU patients from an international multicentre cohort study. Intensive Care Med. 28, 108–121. doi: 10.1007/s00134-001-1143-z
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-A python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Anderson, K. L., Roux, C. M., Olson, M. W., Luong, T. T., Lee, C. Y., Olson, R., et al. (2010). Characterizing the effects of inorganic acid and alkaline shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol. Med. Microbiol. 60, 208–250. doi: 10.1111/j.1574-695X.2010.00736.x
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Averianova, L. A., Balabanova, L. A., Son, O. M., Podvolotskaya, A. B., and Tekutyeva, L. A. (2020). Production of vitamin B2 (riboflavin) by microorganisms: an overview. Front. Bioeng. Biotechnol. 8:570828. doi: 10.3389/fbioe.2020.570828
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., He, Z., et al. (2021). KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, W317–W325. doi: 10.1093/nar/gkab447
Ciarlo, E., Heinonen, T., Herderschee, J., Fenwick, C., Mombelli, M., Le Roy, D., et al. (2016). Impact of the microbial derived short chain fatty acid propionate on host susceptibility to bacterial and fungal infections in vivo. Sci. Rep. 6:37944. doi: 10.1038/srep37944
Clegg, J., Soldaini, E., McLoughlin, R. M., Rittenhouse, S., Bagnoli, F., and Phogat, S. (2021). Staphylococcus aureus vaccine research and development: the past, present and future, including novel therapeutic strategies. Front. Immunol. 12:705360. doi: 10.3389/fimmu.2021.705360
Cummings, J. H., Pomare, E. W., Branch, W. J., Naylor, C. P., and Macfarlane, G. T. (1987). Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28, 1221–1227. doi: 10.1136/gut.28.10.1221
Fang, Y., Zhang, S., Wang, J., Yin, L., Zhang, H., Wang, Z., et al. (2021). Metabolic detoxification of 2-oxobutyrate by remodeling Escherichia coli acetate bypass. Meta 11:30. doi: 10.3390/metabo11010030
Fernandes, J., Su, W., Rahat-Rozenbloom, S., Wolever, T. M., and Comelli, E. M. (2014). Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr. Diabetes 4:e121. doi: 10.1038/nutd.2014.23
Garcia-Angulo, V. A. (2017). Overlapping riboflavin supply pathways in bacteria. Crit. Rev. Microbiol. 43, 196–209. doi: 10.1080/1040841X.2016.1192578
Graves, S. F., Kobayashi, S. D., and DeLeo, F. R. (2010). Community-associated methicillin-resistant Staphylococcus aureus immune evasion and virulence. J. Mol. Med. (Berl) 88, 109–114. doi: 10.1007/s00109-009-0573-x
Hojo, K., Nagaoka, S., Ohshima, T., and Maeda, N. (2009). Bacterial interactions in dental biofilm development. J. Dent. Res. 88, 982–990. doi: 10.1177/0022034509346811
Imlay, J. A. (2013). The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454. doi: 10.1038/nrmicro3032
Jacobson, A., Lam, L., Rajendram, M., Tamburini, F., Honeycutt, J., Pham, T., et al. (2018). A gut commensal-produced metabolite mediates colonization resistance to salmonella infection. Cell Host Microbe 24, 296–307.e7. doi: 10.1016/j.chom.2018.07.002
Jeong, S., Kim, H. Y., Kim, A. R., Yun, C. H., and Han, S. H. (2019a). Propionate ameliorates Staphylococcus aureus skin infection by attenuating bacterial growth. Front. Microbiol. 10:1363. doi: 10.3389/fmicb.2019.01363
Jeong, S., Lee, Y., Yun, C. H., Park, O. J., and Han, S. H. (2019b). Propionate, together with triple antibiotics, inhibits the growth of enterococci. J. Microbiol. 57, 1019–1024. doi: 10.1007/s12275-019-9434-7
Kikuchi, G., Motokawa, Y., Yoshida, T., and Hiraga, K. (2008). Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 84, 246–263. doi: 10.2183/pjab.84.246
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kourtis, A. P., Hatfield, K., Baggs, J., Mu, Y., See, I., Epson, E., et al. (2019). Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections - United States. MMWR Morb. Mortal. Wkly Rep. 68, 214–219. doi: 10.15585/mmwr.mm6809e1
Kullar, R., Casapao, A. M., Davis, S. L., Levine, D. P., Zhao, J. J., Crank, C. W., et al. (2013). A multicentre evaluation of the effectiveness and safety of high-dose daptomycin for the treatment of infective endocarditis. J. Antimicrob. Chemother. 68, 2921–2926. doi: 10.1093/jac/dkt294
LaRossa, R. A., Van Dyk, T. K., and Smulski, D. R. (1987). Toxic accumulation of alpha-ketobutyrate caused by inhibition of the branched-chain amino acid biosynthetic enzyme acetolactate synthase in salmonella typhimurium. J. Bacteriol. 169, 1372–1378. doi: 10.1128/jb.169.4.1372-1378.1987
Lin, Z., Xu, Z., Li, Y., Wang, Z., Chen, T., and Zhao, X. (2014). Metabolic engineering of Escherichia coli for the production of riboflavin. Microb. Cell Factories 13:104. doi: 10.1186/s12934-014-0104-5
Liu, C., Bayer, A., Cosgrove, S. E., Daum, R. S., Fridkin, S. K., Gorwitz, R. J., et al. (2011). Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin. Infect. Dis. 52, 285–292. doi: 10.1093/cid/cir034
Lobritz, M. A., Belenky, P., Porter, C. B., Gutierrez, A., Yang, J. H., Schwarz, E. G., et al. (2015). Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. U. S. A. 112, 8173–8180. doi: 10.1073/pnas.1509743112
Lopatkin, A. J., and Yang, J. H. (2021). Digital insights into nucleotide metabolism and antibiotic treatment failure. Front. Digit Health 3:583468. doi: 10.3389/fdgth.2021.583468
Lowy, F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Luu, M., Pautz, S., Kohl, V., Singh, R., Romero, R., Lucas, S., et al. (2019). The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun. 10:760. doi: 10.1038/s41467-019-08711-2
Macfarlane, S., and Macfarlane, G. T. (2003). Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 62, 67–72. doi: 10.1079/PNS2002207
Massey, V. (2000). The chemical and biological versatility of riboflavin. Biochem. Soc. Trans. 28, 283–296. doi: 10.1042/bst0280283
Moise, P. A., Amodio-Groton, M., Rashid, M., Lamp, K. C., Hoffman-Roberts, H. L., Sakoulas, G., et al. (2013). Multicenter evaluation of the clinical outcomes of daptomycin with and without concomitant beta-lactams in patients with Staphylococcus aureus bacteremia and mild to moderate renal impairment. Antimicrob. Agents Chemother. 57, 1192–1200. doi: 10.1128/AAC.02192-12
Nastasi, C., Fredholm, S., Willerslev-Olsen, A., Hansen, M., Bonefeld, C. M., Geisler, C., et al. (2017). Butyrate and propionate inhibit antigen-specific CD8(+) T cell activation by suppressing IL-12 production by antigen-presenting cells. Sci. Rep. 7:14516. doi: 10.1038/s41598-017-15099-w
Ortwine, J. K., and Bhavan, K. (2018). Morbidity, mortality, and management of methicillin-resistant S. aureus bacteremia in the USA: update on antibacterial choices and understanding. Hosp. Pract. (1995) 46, 64–72. doi: 10.1080/21548331.2018.1435128
Park, J. W., Kim, H. Y., Kim, M. G., Jeong, S., Yun, C. H., and Han, S. H. (2019). Short-chain fatty acids inhibit staphylococcal lipoprotein-induced nitric oxide production in murine macrophages. Immune Netw. 19:e9. doi: 10.4110/in.2019.19.e9
Pinhal, S., Ropers, D., Geiselmann, J., and de Jong, H. (2019). Acetate metabolism and the inhibition of bacterial growth by acetate. J. Bacteriol. 201, e00147–e00119. doi: 10.1128/JB.00147-19
Planet, P. J. (2017). Life after USA300: the rise and fall of a superbug. J. Infect. Dis. 215, S71–S77. doi: 10.1093/infdis/jiw444
Roe, A. J., O’Byrne, C., McLaggan, D., and Booth, I. R. (2002). Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology (Reading) 148, 2215–2222. doi: 10.1099/00221287-148-7-2215
Selan, L., Vrenna, G., Ettorre, E., Papa, R., and Artini, M. (2018). Virulence of MRSA USA300 is enhanced by sub-inhibitory concentration of two different classes of antibiotics. J. Chemother. 30, 384–388. doi: 10.1080/1120009X.2018.1533085
Shimizu, K., and Matsuoka, Y. (2022). Feedback regulation and coordination of the main metabolism for bacterial growth and metabolic engineering for amino acid fermentation. Biotechnol. Adv. 55:107887. doi: 10.1016/j.biotechadv.2021.107887
Silva, Y. P., Bernardi, A., and Frozza, R. L. (2020). The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. (Lausanne) 11:25. doi: 10.3389/fendo.2020.00025
Sobral, R. G., Jones, A. E., Des Etages, S. G., Dougherty, T. J., Peitzsch, R. M., Gaasterland, T., et al. (2007). Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 189, 2376–2391. doi: 10.1128/JB.01439-06
Song, J., Jung, K. J., Yang, M. J., Han, S. C., and Lee, K. (2021). Assessment of acute and repeated pulmonary toxicities of oligo 2- 2-ethoxy ethoxyethyl guanidium chloride in mice. Toxicol. Res. 37, 99–113. doi: 10.1007/s43188-020-00058-x
Tezuka, T., and Ohnishi, Y. (2014). Two glycine riboswitches activate the glycine cleavage system essential for glycine detoxification in Streptomyces griseus. J. Bacteriol. 196, 1369–1376. doi: 10.1128/JB.01480-13
Turner, N. A., Sharma-Kuinkel, B. K., Maskarinec, S. A., Eichenberger, E. M., Shah, P. P., Carugati, M., et al. (2019). Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol. 17, 203–218. doi: 10.1038/s41579-018-0147-4
Vestergaard, M., Frees, D., and Ingmer, H. (2019). Antibiotic resistance and the MRSA problem. Microbiol. Spectr. 7:GPP3-0057-2018. doi: 10.1128/microbiolspec.GPP3-0057-2018
Wang, Y., Dai, A., Huang, S., Kuo, S., Shu, M., Tapia, C. P., et al. (2014). Propionic acid and its esterified derivative suppress the growth of methicillin-resistant Staphylococcus aureus USA300. Benef. Microbes 5, 161–168. doi: 10.3920/BM2013.0031
Westermann, A. J., and Vogel, J. (2021). Cross-species RNA-seq for deciphering host-microbe interactions. Nat. Rev. Genet. 22, 361–378. doi: 10.1038/s41576-021-00326-y
Yang, J. H., Wright, S. N., Hamblin, M., McCloskey, D., Alcantar, M. A., Schrubbers, L., et al. (2019). A white-box machine learning approach for revealing antibiotic mechanisms of action. Cells 177, 1649–1661.e9. doi: 10.1016/j.cell.2019.04.016
Ye, J. Z., Su, Y. B., Lin, X. M., Lai, S. S., Li, W. X., Ali, F., et al. (2018). Alanine enhances aminoglycosides-induced ROS production as revealed by proteomic analysis. Front. Microbiol. 9:29. doi: 10.3389/fmicb.2018.00029
Yee, R., Cui, P., Shi, W., Feng, J., and Zhang, Y. (2015). Genetic screen reveals the role of purine metabolism in Staphylococcus aureus persistence to rifampicin. Antibiotics (Basel) 4, 627–642. doi: 10.3390/antibiotics4040627
Yonezawa, H., Osaki, T., Hanawa, T., Kurata, S., Zaman, C., Woo, T. D. H., et al. (2012). Destructive effects of butyrate on the cell envelope of helicobacter pylori. J. Med. Microbiol. 61, 582–589. doi: 10.1099/jmm.0.039040-0
Keywords: short-chain fatty acids, propionate, bacteriostatic, RNA-Seq, metabolic pathway
Citation: Im J, Lee D, Park O-J, Natarajan S, Park J, Yun C-H and Han SH (2022) RNA-Seq-based transcriptome analysis of methicillin-resistant Staphylococcus aureus growth inhibition by propionate. Front. Microbiol. 13:1063650. doi: 10.3389/fmicb.2022.1063650
Edited by:
Michal Letek, Universidad de León, SpainReviewed by:
Heather Bennett Miller, High Point University, United StatesVolker Behrends, University of Roehampton London, United Kingdom
Jie Hui, Harvard Medical School, United States
Copyright © 2022 Im, Lee, Park, Natarajan, Park, Yun and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seung Hyun Han, c2hoYW4tbWlAc251LmFjLmty
 Jintaek Im
Jintaek Im Dongwook Lee
Dongwook Lee Ok-Jin Park
Ok-Jin Park Sathishkumar Natarajan
Sathishkumar Natarajan Junhyung Park
Junhyung Park Cheol-Heui Yun
Cheol-Heui Yun Seung Hyun Han
Seung Hyun Han