
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 08 December 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1060947
The clustered regularly interspaced short palindromic repeats (CRISPR) system is a natural adaptive immune system of prokaryotes. The CRISPR-Cas system is currently divided into two classes and six types: types I, III, and IV in class 1 systems and types II, V, and VI in class 2 systems. Among the CRISPR-Cas type VI systems, the CRISPR/Cas13a system has been the most widely characterized for its application in molecular diagnostics, gene therapy, gene editing, and RNA imaging. Moreover, because of the trans-cleavage activity of Cas13a and the high specificity of its CRISPR RNA, the CRISPR/Cas13a system has enormous potential in the field of molecular diagnostics. Herein, we summarize the applications of the CRISPR/Cas13a system in the detection of pathogens, including viruses, bacteria, parasites, chlamydia, and fungus; biomarkers, such as microRNAs, lncRNAs, and circRNAs; and some non-nucleic acid targets, including proteins, ions, and methyl groups. Meanwhile, we highlight the working principles of some novel Cas13a-based detection methods, including the Specific High-Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) and its improved versions, Cas13a-based nucleic acid amplification-free biosensors, and Cas13a-based biosensors for non-nucleic acid target detection. Finally, we focus on some issues that need to be solved and the development prospects of the CRISPR/Cas13a system.
The clustered regularly interspaced short palindromic repeats (CRISPR) system, which exists in almost half of bacteria (40%) and almost all archaea (90%) (Grissa et al., 2007; Kunin et al., 2007), is the natural adaptive immune system for prokaryotes to defend against extraneous invasion by phages and transformation of plasmids (Sorek et al., 2008; Garneau et al., 2010). The CRISPR array, which comprises short direct repeats separated by spacer sequences acquired from foreign nucleic acids (Mojica et al., 2005; Hochstrasser and Doudna, 2015), was first discovered in 1987 (Ishino et al., 1987). Additional elements of the CRISPR system include the leader sequence and CRISPR-associated (Cas) genes (Pougach et al., 2010; Yosef et al., 2012; Makarova et al., 2015). The mechanism of adaptive immunity driven by the CRISPR system includes three common stages (van der Oost et al., 2014; Amitai and Sorek, 2016; Sternberg et al., 2016; Hille et al., 2018; Wang et al., 2019): (1) the adaption stage, in which the CRISPR system recognizes foreign nucleic acids and integrates them into spacer sequences in the CRISPR array to form immunologic memory; (2) the CRISPR-RNA (crRNA) expression and maturation stage, in which the CRISPR array is transcribed into precursor CRISPR-RNA (pre-crRNA) that is subsequently processed to mature crRNA; (3) the interference stage, in which Cas protein–crRNA complex targets and cleaves the identified foreign DNAs and RNAs (Marraffini and Sontheimer, 2008; Hale et al., 2009).
The currently discovered CRISPR-Cas system is generally divided into two classes, six types, and 33 subtypes after multiple refinements (Makarova et al., 2011, 2015, 2020). The class 1 systems, including types I, III, and IV, are represented by multi-protein effecter complexes, while the class 2 systems, containing types II, V, and VI, utilize a single, large, multi-domain Cas protein as the effecter module (Mohanraju et al., 2016; Koonin et al., 2017). The CRISPR VI systems have four subtypes: VI-A, VI-B, VI-C, and VI-D, which are also termed Cas13a, Cas13b, Cas13c, and Cas13d systems, respectively (Makarova et al., 2020). Cas13a (also known as C2c2) in type VI systems is an RNA-guided RNase, which contains two conserved HEPN (Higher Eukaryotes and Prokaryotes Nucleotide-binding) domains (Abudayyeh et al., 2016). Cas13a comprises two lobes termed the crRNA-recognition (REC) lobe and the nuclease (NUC) lobe. The Helical-1 domain and the N-terminal domain (NTD) constitute the REC lobe, whereas the NUC lobe contains the HEPN1 domain, HEPN2 domain, Helical-2 domain, and a Linker between two HEPN domains (Figure 1A; Liu et al., 2017). The mature crRNA contains a 3′ spacer and a 5′ handle, which is divided into 5′ flank, 5′ stem, Loop, 3′ stem, and 3′ flank (Figure 1B; Knott et al., 2017). Cas13a processes pre-crRNA into mature crRNA independently and combines with its crRNA to form the surveillance complex that recognizes the foreign target RNA and cleaves the target and surrounding RNAs (Figure 1C; Knott et al., 2017).
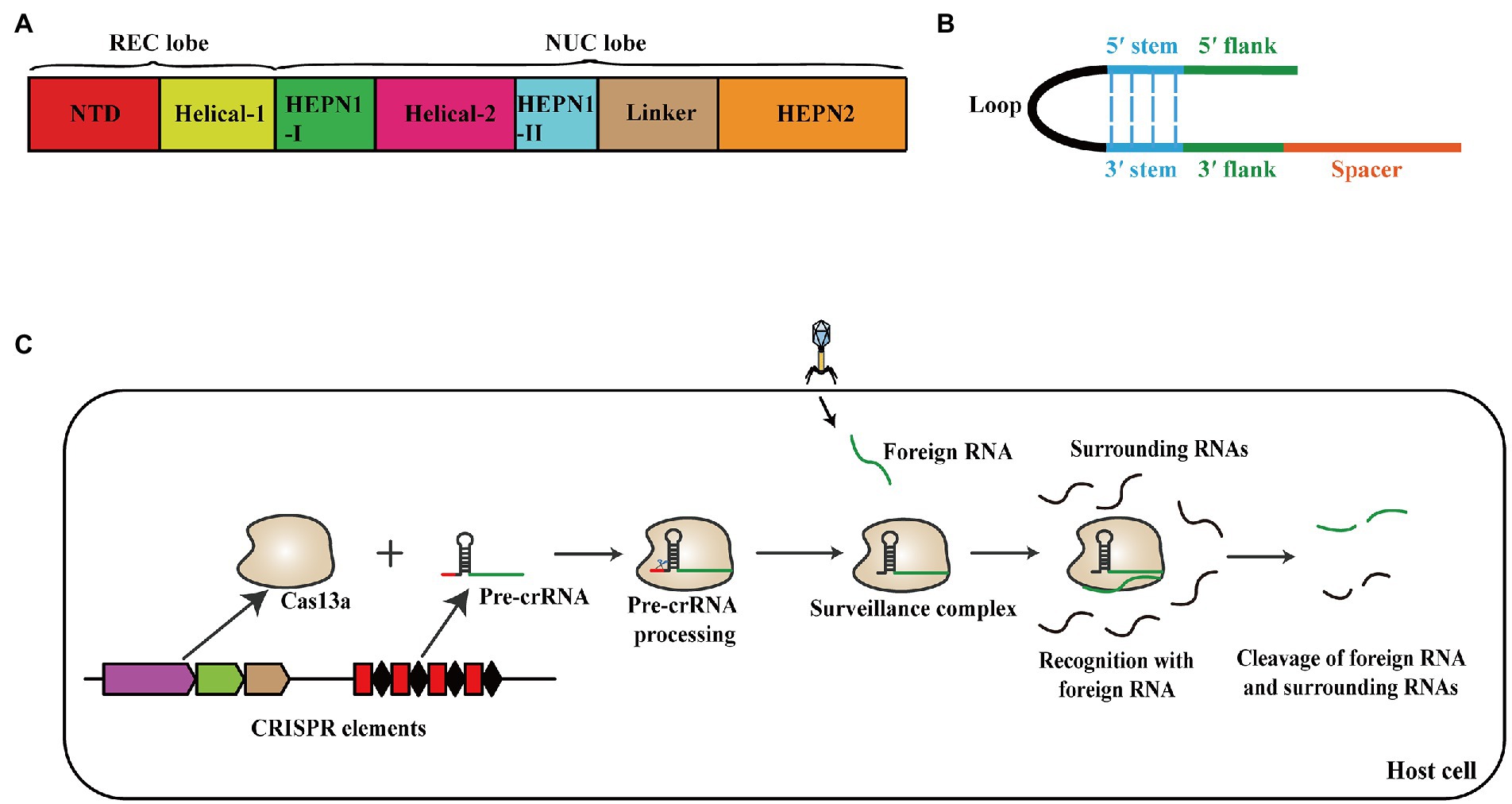
Figure 1. (A) Domain organization schematic of Cas13a. Cas13a comprises REC lobe and NUC lobe. The Helical-1 domain and the N-terminal domain constitute REC lobe, the NUC lobe contains the HEPN1 domain, HEPN2 domain, Helical-2 domain, and a Linker. (B) Schematic representation of the mature crRNA structure. The mature crRNA contains 3′ spacer and 5′ handle, which consists of 5′ flank, 5′ stem, Loop, 3′ stem, and 3′ flank. (C) Schematic of the effect mechanism of Cas13a system against foreign nucleic acid. Cas13a processes pre-crRNA into mature crRNA independently and combines with its crRNA to form the surveillance complex that recognizes the foreign target RNA and cleaves the target and surrounding RNAs.
Cas13a possesses two RNase activities, one for pre-crRNA processing and the other for cis-cleavage of the target RNA and trans-cleavage of non-specific RNAs. The pre-crRNA is cleaved in a metal-independent and acid–base manner by conserved residues in the Helical-1 and HEPN2 domains (Knott et al., 2017). The position of processing generally occurs 2–5 nucleotides upstream of the repeat sequences of crRNA (East-Seletsky et al., 2016). The two nucleotides adjacent to the broken site are essential for processing activity. Notably, the processing of pre-crRNA is not necessary for target cleavage because effecter complexes with pre-crRNA still have cleavage capacity (East-Seletsky et al., 2017). In addition to the common target RNA cleavage mechanism (cis-cleavage), Cas13a also exhibits non-specific collateral cleavage activity of the surrounding non-target RNAs; this collateral activity can achieve robust signal amplification with 104 turnover ability (East-Seletsky et al., 2016). The cis- and trans-cleavage activities of Cas13a are dominated by two HEPN domains in a divalent cation-dependent manner (Nalefski et al., 2021). Different Cas13a orthologs exhibit A or U preferences for the cleavage site (East-Seletsky et al., 2017). The length of the spacer and the number of and position of mismatches between the spacer and target RNA are all vital for activating the RNase activity of Cas13a. The spacer length is required to be as short as 20 nt to retain the cleavage ability of Cas13a (Abudayyeh et al., 2017). A single mismatch between the crRNA and the target has a minor effect on the cleavage activity of Cas13a, and double mismatches greatly reduce the cleavage activity. Meanwhile, the “central seed region” of spacer-target duplex is more sensitive to mismatches (Abudayyeh et al., 2016). Intriguingly, Tambe et al. have demonstrated that the target binding affinity of Cas13a was uncoupled with the activation of RNase activity in LbuCas13a, and they found a “HEPN-nuclease switch region,” in which mismatches result in unaltered target binding affinity but lead to inactivation of the HEPN-nuclease (Tambe et al., 2018).
As the most widely characterized system of CRISPR, Cas13a system has been applied for molecular diagnostics, gene therapy, gene editing, and RNA imaging (Abudayyeh et al., 2017; Cox et al., 2017; Gootenberg et al., 2017; Li et al., 2020). Among the numerous applications of the Cas13a system, we focused on its application in molecular diagnostics since various studies have been reported in this field and Cas13a-based biosensors have made great progress. The activated collateral cleavage activity of Cas13a and the simple programmability of crRNA provide tremendous potential for Cas13a-based molecular diagnostics. East et al. first detected the bacteriophage λ gene by harnessing the RNA-guided trans-cleavage activity of LbuCas13a, in which activated LbuCas13a, followed by specific crRNA-target recognition, can cleave fluorophore quencher-labeled reporter RNA to cause increased fluorescence (East-Seletsky et al., 2016). Since then, CRISPR/Cas13a-based biosensors have become the spotlight in the field of molecular diagnostics and have been applied for the detection of various kinds of targets. The CRISPR/Cas13a system has been developed as an effective tool for molecular diagnostics, with expectations to improve the sensitivity, specificity, operability, portability and cost performance of molecular diagnostics tools (such as antigen–antibody reaction, PCR-based techniques, genome sequencing, etc.). In this review, we summarize the previously reported applications of the CRISPR/Cas13a system in molecular diagnostics for detecting pathogens, including viruses, bacteria, parasites, chlamydia, and fungus; biomarkers, such as microRNAs, lncRNAs, and circRNAs; proteins; ions; and methyl groups (Figure 2). The specific detection methods, targets, amplification methods, readout, sensitivity, and running time-based CRISPR/Cas13a detection technology are listed in Table 1. We also detail the working principles of some CRISPR/Cas13a-based detection methods, including the Specific High-Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) and its improved versions, Cas13a-based nucleic acid amplification-free biosensors, and Cas13a-based biosensors for non-nucleic acid target detection. Finally, we point out some issues associated with CRISPR/Cas13a-based biosensors that need to be solved and focus on the future development of Cas13a-based biosensors.
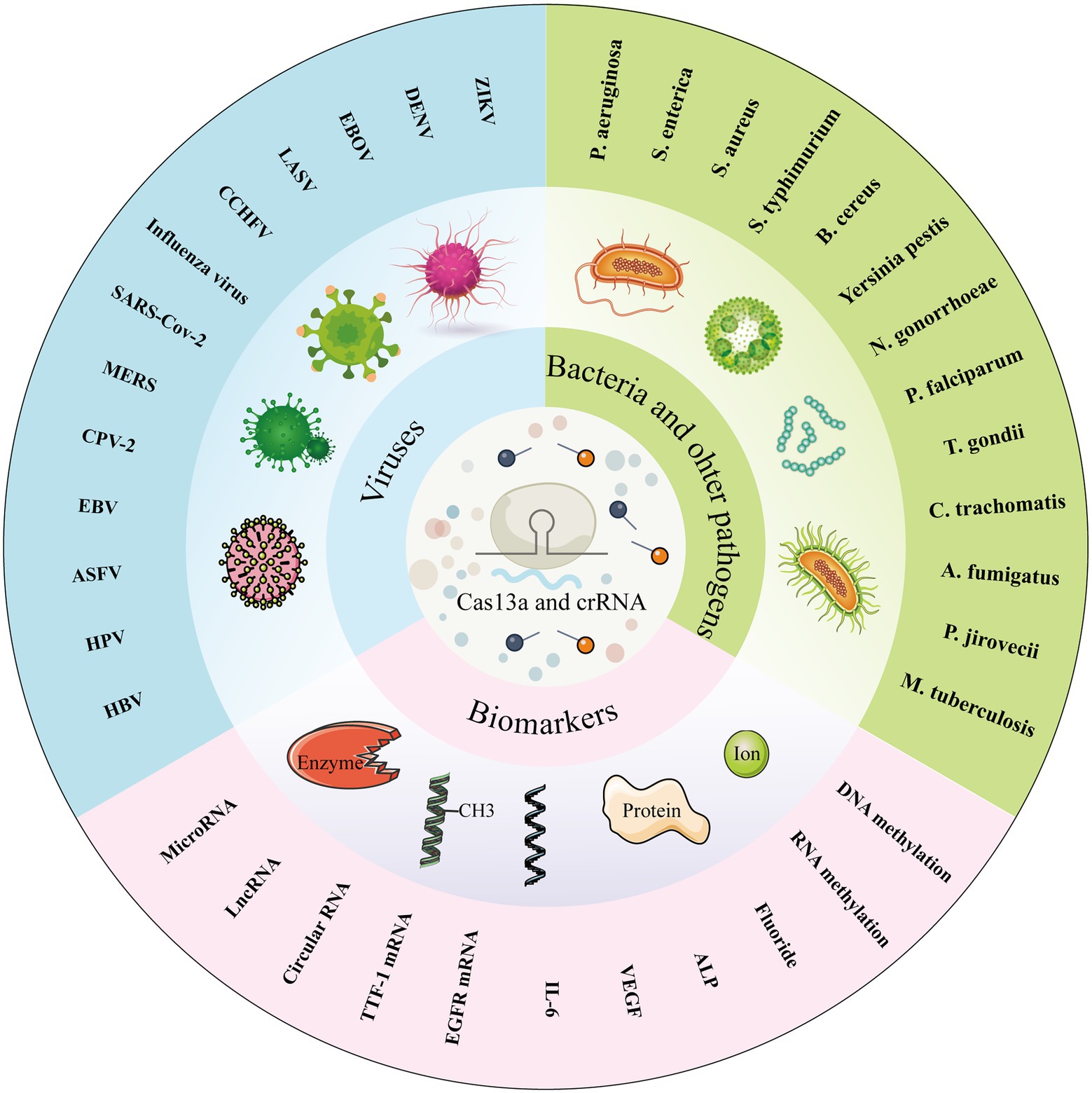
Figure 2. Various targets detected by Cas13a-based biosensors. Cas13a-based biosensors are able to detect pathogens, including viruses, bacteria, parasites, chlamydia, and fungus; biomarkers, such as microRNAs, IncRNAs, and circRNAs; and some non-nucleic acid targets, including proteins, ions, and methyl groups.
The global pandemic of flaviviruses, including Dengue virus (DENV), Zika virus (ZIKV), West Nile virus (WNV), and Yellow fever virus (YFV), has placed a huge burden on global health for the last several decades. Among these viruses, DENV causes nearly 400 million infections each year, and the Dengue fever epidemic has affected a quarter of the world’s population (Pierson and Diamond, 2020). In addition, the spread of ZIKV in recent years also led to numerous infection cases in the American region (Pierson and Diamond, 2018). The serious problems caused by flaviviruses have made rapid and accurate diagnosis of the viruses essential. A novel CRISPR-based nucleic acid detection platform has been established, called the SHERLOCK, which combines isothermal amplification that comprises recombinase polymerase amplification (RPA) and T7 transcription with the collateral cleavage effect of Cas13a for virus detection (Figure 3). Because of the isothermal amplification of the target RNA and the specificity of crRNA-guided recognition, the SHERLOCK assay achieved attomolar sensitivity and single-base specificity for ZIKV and DENV detection. Furthermore, SHERLOCK is also available in paper spotting and lyophilization forms, which significantly improve its flexibility and portability (Gootenberg et al., 2017; Kellner et al., 2019). Before long, Gootenberg et al. developed the SHERLOCK version 2 (SHERLOCKv2) by refining the original method. This method combined CRISPR/Cas13a-based detection with four multiplexing channels and achieved instrument-free detection of DENV or ZIKV ssRNA within 90 min, with a sensitivity of 2 aM. In addition to fluorescence readout, the lateral-flow test was employed for SHERLOCKv2 as a visual readout for field applications (Figure 3). Furthermore, the use of Csm6, an auxiliary CRISPR-associated enzyme, significantly increased the sensitivity of RPA-SHERLOCK (Gootenberg et al., 2018). In general, field-deployable tests can detect clinical samples without extracting nucleic acids. Therefore, an extraction-free method was reported, called heating unextracted diagnostic samples to obliterate nucleases (HUDSON), in which samples are directly added to the RPA process after HUDSON (37°C–50°C, 5–20 min for nuclease inactivation and 64°C–95°C, 5 min for viral inactivation) treatment without purification or dilution (Figure 3). The HUDSON method combined with RPA-SHERLOCK was used to directly detect DENV in patient samples within 2 h, and four serotypes of DENV were distinguished using this method. ZIKV and viral single-nucleotide polymorphisms (SNP) were also confirmed. Furthermore, this extraction-free assay still achieved attomolar detection sensitivity (Myhrvold et al., 2018). Recently, one research reported a novel electrochemical method in which the Cas13a system was combined with catalytic hairpin assembly (CHA) signal amplification. In this method, the activated Cas13a cleaves the reporter RNA hybridized to the swing arm-DNA walker (DW), promoting a series of downstream strand displacement reactions to produce electrochemical signals. Using this assay, they could detect DENV-1 at a level as low as 0.78 fM (Wang J. et al., 2021).
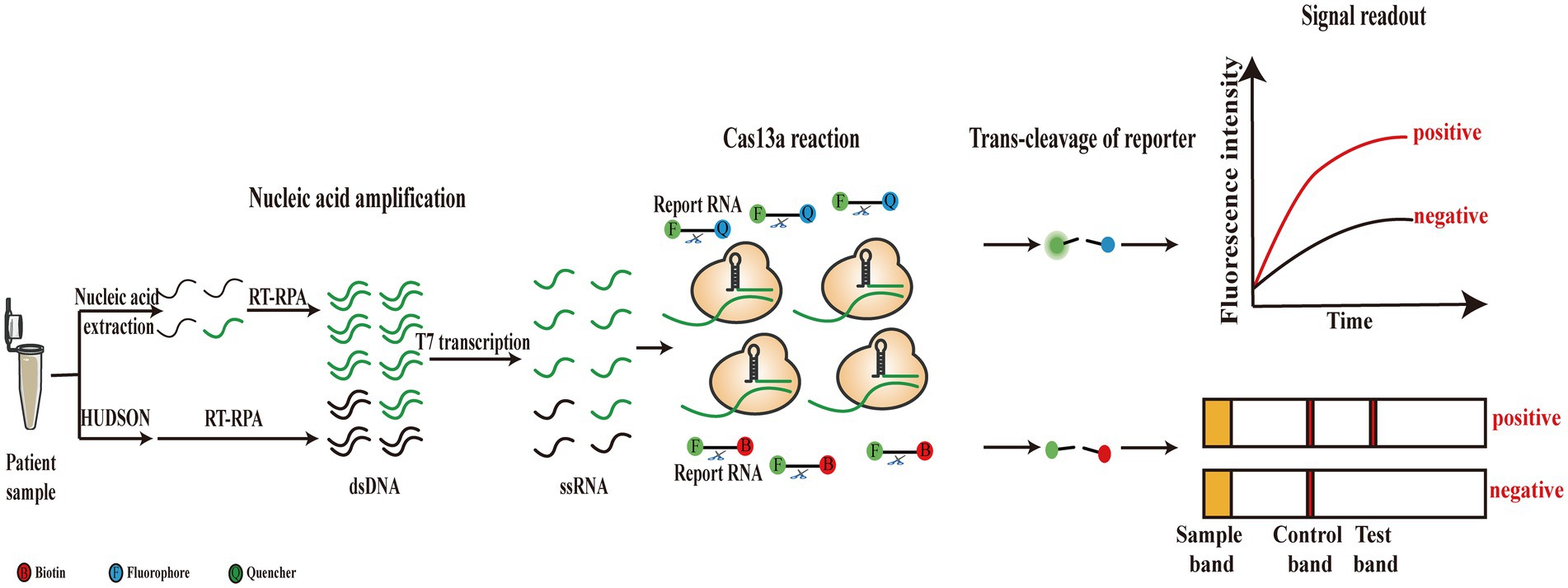
Figure 3. Schematic of original SHERLOCK and its improved versions. Target RNAs are extracted from patient sample and then amplified through RT-RPA and T7 transcription. Subsequently, amplification products are recognized and trans-cleaved by Cas13a system with fluorescence or lateral flow readout. Patient sample can also be directly treated by HUDSON method without the extraction step.
The Ebola virus (EBOV) is a member of the Filoviridae and can result in the fulminating infectious disease, Ebola hemorrhagic fever, which is the deadliest viral hemorrhagic fever. The outbreak of Ebola in 2014 resulted in over 28,000 infected cases and over 11,000 deaths (Cenciarelli et al., 2015). Similarly, the Lassa virus (LASV), a single-stranded negative-strand RNA virus of the family Arenavirus, generally causes severe Lassa fever, which leads to approximately 5,000–10,000 deaths per year (Happi et al., 2019). Both the two hemorrhagic fever viruses constitute serious global health threats. However, the diagnosis of these two diseases is challenging owing to non-specific symptoms. Therefore, sensitive and specific methods for point-of-care detection of the Ebola and Lassa virus are required. Qin et al. reported an automated microfluidic system combined with the collateral activity of Cas13a for the detection of EBOV. Using a sensitive custom integrated fluorometer for reading fluorescence signals, they achieved a limit of detection (LOD) of 5. 45 × 104 copies/μl for EBOV oligos without target RNA amplification and improved point-of-care testing for the EBOV (Qin et al., 2019). Moreover, the previously reported HUDSON method combined with RPA-SHERLOCK was employed to detect the Ebola and Lassa viruses, with an LOD of as low as 10 copies/μl, in which the fluorescent readout of LASV-II was 100% consistent with the results of reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR). They also detected EBOV in the whole blood, urine, and saliva samples using this approach (Barnes et al., 2020). Recently, a novel method called the Cas-Roller was reported, which combines the Cas13a reaction with a DNA roller reaction to detect Ebola RNA and achieved an LOD of 291 aM within 40 min (Hang et al., 2022). In addition, the RPA-SHERLOCK method was employed to detect the Crimean-Congo hemorrhagic fever virus (CCHFV), another severe hemorrhagic fever virus (Bente et al., 2013), realizing an LOD of 1 copy/μl within 30–40 min and discriminating CCHFV from a series of CCHFV-related viruses (Li et al., 2022a).
Influenza viruses have spread worldwide for decades, and each outbreak has caused severe destruction to human society. In 2013, a new influenza virus H7N9 was found in China; since then, 1,568 laboratory-confirmed H7N9 infected cases have been reported with the mortality rate up to 39.2% (Gao et al., 2013; Liu W. J. et al., 2021). Therefore, establishing a sensitive and rapid method for the detection of the H7N9 virus is required. A previous study applied Cas13-based detection for H7N9 virus via targeting its hemagglutinin (HA) and neuraminidase (NA) genes. They could detect 1 nM of NA ssRNA within 5 min without amplification, and the method could test 1 fM of HA ssRNA in 50 min when combined with the RPA-SHERLOCK assay (Liu et al., 2019).
The outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to millions of deaths worldwide. Although most patients experience minor symptoms, people with low-level infections, especially asymptomatic carriers, may accelerate the spread of the virus; thus, an applicable detection approach is required for timely and accurate detection of SARS-CoV-2 (Bai et al., 2020). RPA-SHERLOCK has been utilized to detect SARS-CoV-2 by fluorescence and lateral-flow readout, with an LOD of 42 copies per reaction by targeting the S gene of SARS-CoV-2. Compared to RT-qPCR, the RPA-SHERLOCK assay achieved a sensitivity and specificity of 96 and 100%, respectively, using fluorescence readout, and 88% and 100%, respectively, using lateral-flow readout for detecting the S gene of SARS-CoV-2 in patient samples. In addition, the RPA-SHERLOCK method was optimized by adding an RNase contamination detection method on the lateral-flow test strip, thereby avoiding false positives or negatives resulting from exogenous RNase contamination (Patchsung et al., 2020). Furthermore, Zhang et al. developed a refined Cas13a-based test strip assay combining quantum dot microspheres with a Cas13a-based test strip for SARS-CoV-2 detection. In this assay, an LOD of 1 copy/ml was achieved in 1 h, with 100% specificity and 100% sensitivity compared to RT-PCR (Zhang Q. et al., 2022). Recently, SHERLOCK coupled with reverse transcriptase loop-mediated amplification (RT-LAMP) was used to detect 60 clinical samples of SARS-CoV-2, with 100% detection accuracy (Khan et al., 2021). Furthermore, several studies for extraction-free Cas13a detection of SARS-CoV-2 have also been reported. One study employed SHERLOCK and HUDSON integration to navigate epidemics (SHINE; the previously reported HUDSON-SHERLOCK method) for SARS-CoV-2 detection without RNA extraction; they also integrated RPA and the T7 transcription steps of SHERLOCK into one step to simplify the operating process and deployed an in-tube fluorescent readout, which could reduce the risk of contamination. The LOD of SHINE was as low as 10 copies/μl by testing HUDSON-treated saliva with an in-tube fluorescent readout in 50 min (Arizti-Sanz et al., 2020). Azmi et al. developed Cas13 assisted saliva-based and smartphone-integrated testing (CASSPIT), which further optimizes the HUDSON-SHERLOCK method by refining the chemical treatment and heat inactivation processes to detect saliva samples of SARS-CoV-2 with lateral-flow readout; the method achieved an LOD of 200 copies per reaction (Azmi et al., 2021). Notably, a recent study reported the RT-LAMP-CRISPR-Cas13a method based on SHERLOCK. This novel RNA extraction-free assay combined RT-LAMP, Cas13a reaction, proteinase K-heat inactivation (PK-HID), and exhibited 83% sensitivity and 100% specificity for the detection of SARS-CoV-2 in the nasopharyngeal samples compared to RT-qPCR. Meanwhile, positive predictive value (PPV) of 100% and negative predictive values (NPVs) of 81% for cycle threshold (CT) values of <20 were also verified using this method (Ortiz-Cartagena et al., 2022). Moreover, nucleic acid amplification-free methods for Cas13a-based detection of SARS-CoV-2 have made significant progress. A previous study reported a novel nucleic acid amplification-free method in which two or three crRNAs targeting distinct regions of the SARS-CoV-2 genome were combined to significantly increase the sensitivity of Cas13a-based detection (Figure 4A). This assay could achieve an LOD of 100 copies/μl within 30 min; furthermore, triple combinations of crRNAs enabled sensitive detection of as low as 31 copies/μl (Fozouni et al., 2021). Tian et al. recently established a neoteric method for nucleic acid amplification-free detection, called the ultralocalized Cas13a assay, which harnesses the bioinspired confinement effect by separating the reaction mixture of Cas13a, crRNA, target RNA, and RNA reporters into tens of thousands of monodisperse picoliter-sized droplets. This assay greatly increased the local concentration of the reaction and subsequently caused a more than 10,000-fold enhancement in sensitivity without nucleic acid amplification (Figure 4B). Using this method, they could detect as low as 6 copies/μl of the SARS-CoV-2 N gene, and absolute digital quantification was achieved (Tian et al., 2021). Furthermore, a novel nucleic acid amplification-free platform was developed recently, termed the automated platform on CRISPR-based nucleic acid amplification-free digital RNA detection (opn-SATORI), which combines the Cas13a reaction, microchamber technology, and magnetic beads technology. In this method, biotin-labeled Cas13a complexes are captured by streptavidin-labeled magnetic beads, which are concentrated by magnetic force into microchambers and are activated in the presence of target RNAs (Figure 4C). Using this assay, the authors detected as low as 3.9 copies/μl of SARS-CoV-2 RNA within 9 min and distinguished three variants of SARS-CoV-2: alpha, delta, and omicron (Shinoda et al., 2022). Furthermore, Heo et al. (2022) developed an electrochemical biosensor in which activated Cas13a cleaves the report RNA on the surface of the electrode to release methylene blue, resulting in an altered peak current. This method achieved an LOD of 4.4 × 10−2 and 8.1 × 10−2 fg/ml for the ORF and S genes of SARS-CoV-2, respectively. Recently, a Cas13a-based biosensor was established that employs the Cas13a reaction and graphene field-effect transistor (gFET) to test SARS-CoV-2 and respiratory syncytial virus, with an LOD of 1 aM. In this assay, activated Cas13a cleaved PolyU20 on gFET, leading to reduced electron transfer (Li et al., 2022b). In addition, a novel detection assay called CRISPR/CAS-based colorimetric nucleic acid detection (CASCADE) was reported, which combines the Cas13a system with gold nanoparticles (AuNPs) to allow naked-eye detection of SARS-CoV-2. LODs of 3 fM and 40 aM were obtained when the method was coupled with RPA and nucleic acid sequence-based amplification (NASBA), respectively. (López-Valls et al., 2022). PCR technology was also integrated with the Cas13a reaction: Niu et al. (2022) developed a PCR-CRISPR method to detect the HV69-70del mutant of SARS-CoV-2, with an LOD of 1 copy/μl, and utilized this approach to successfully distinguish various mutants of SARS-CoV-2. To avoid false replication in the reverse transcription process, a light-up CRISPR-Cas13 transcription amplification method was reported, in which transcription amplification without the reverse transcription process is initiated after specific recognition and ligation of the probe pairs, presubstrate A and presubstrate B; then, the specifically activated Cas13a digests an RNA aptamer, leading to a reduction in the fluorescence signal produced by the RNA aptamer-DFHBI-1 T complex. They detected MERS, SARS, SARS-CoV-2, and influenza viruses using this light-up detection method and identified a gene mutation in SARS-CoV-2 variants. Notably, the dual amplification and recognition of the light-up CRISPR-Cas13 transcription amplification method ensures its high sensitivity and specificity of detection (Wang Y. et al., 2021). Furthermore, Wang et al. (2022) recently established a CRISPR-Cas13a cascade-based viral RNA (Cas13C) assay, in which activated Cas13a cleaves the pre-primer into mature primer to promote the downstream transcription amplification to produce light-up RNA aptamers for generating fluorescence. Using this assay, they detected SARS-CoV-2 with an LOD of 0.216 fM per reaction and distinguished SARS-CoV-2 from its N501Y variant. In addition, a novel enzyme-free Cas13a-based detection technique coupled with hybridization chain reaction amplification, a type of toehold-mediated strand displacement, was employed to detect SARS-CoV-2; the method achieved LODs of 10 aM (6 copies/μl), 100 aM (60 copies/μl), and 10 aM (6 copies/μl) for the S, N, and Orf1ab genes of SARS-CoV-2, respectively, within 1 h (Yang et al., 2021).
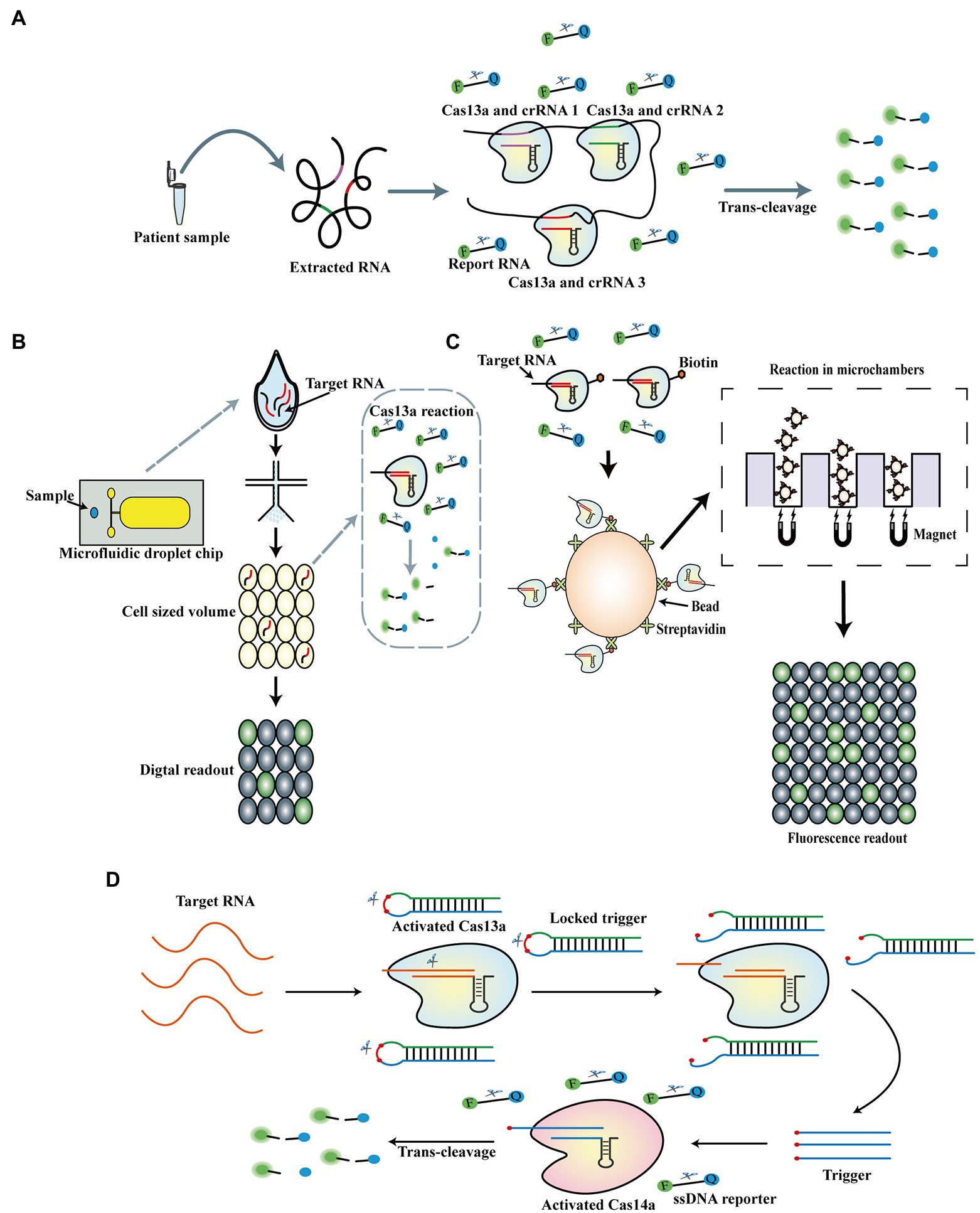
Figure 4. Cas13a-based nucleic acid amplification-free biosensors. (A) Schematic of Cas13a-based biosensor using combination of crRNAs. Two or three crRNAs targeting distinct regions of SARS-CoV-2 genome are mixed to increase the sensitivity of Cas13a-based detection. (B) Schematic of ultralocalized Cas13a assay. The Cas13a reaction is confined in cell-like-sized reactor to enhance the local reaction concentration. (C) Schematic of opn-SATORI. In this detection, biotin-labeled Cas13a complexes are captured by streptavidin-labeled magnetic beads, which are concentrated by the magnetic force into microchambers and are activated in the presence of target RNAs. (D) Schematic of casCRISPR. The trans-cleavage activities of both Cas14a and Cas13a reactions are orderly combined to detect targets.
SHERLOCK is not only for the detection of RNA viruses, but also confers the capability of Cas13a-based detection assay for diagnosis of DNA viruses through its RPA and T7 transcription processes. Several studies have verified the capacity of SHERLOCK for DNA virus detection (Khan et al., 2019; Wu et al., 2019; Wei et al., 2022). Canine parvovirus type 2 (CPV-2), a type of dog DNA virus, was detected by the RPA-SHERLOCK assay with an LOD of 100 amol/L of CPV-2 DNA within 30 min (Khan et al., 2019). Wu et al. (Wu et al., 2019) optimized RPA primers to make SHERLOCK work at 25°C and used it to detect DNA of the Epstein–Barr virus (EBV); this is the first report of room temperature detection using the SHERLOCK platform. The African swine fever virus (ASFV) was also detected by recombinase aided amplification SHERLOCK (RAA-SHERLOCK) with lateral-flow readout, achieving an LOD of 10 copies/μl within 1 h. This assay also exhibited significant specificity for ASFV with no cross-reactivity with other swine viruses and a 100% concordance rate with PCR results (Wei et al., 2022). Additionally, one research group established a rapid one-tube SHERLOCK detection system for ASFV by performing the RPA reaction and Cas13a reaction in one tube; the products of the RPA reaction in the inner tube enter the outer tube by centrifugation via two hydrophobic holes and then contact the Cas13a reaction system. This method could accomplish rapid virus detection within 25 min, with an LOD of 3 copies/μl (Hu et al., 2022). Besides, Cui et al. (2022) performed a picoinjection-aided digital reaction unlocking (PADLOCK) assay, which combines a droplet generator with the addition of the reaction initiator MgOAc through a droplet microfluidic device to avoid premature amplification, allowing detection of HPV16 at as low as 5 copies/μl. Recently, Zhang X. et al. (2022) established an RCA-PCR-CRISPR assay that combines the Cas13a reaction with dual amplification—rolling circle amplification (RCA) and PCR—to detect covalently closed circular DNA (cccDNA) of the hepatitis B virus (HBV). Using this approach, they could detect 1 copy/μL HBV cccDNA, with a higher positive detection rate than those of real-time PCR (qPCR), droplet digital PCR (ddPCR), RCA-qPCR, and PCR-CRISPR.
Food contamination and some severe diseases caused by bacteria and their toxins have led to an enormous burden on public health (Martinovic et al., 2016). Therefore, there is an urgent need to apply the appropriate methods for the detection of bacteria, especially in food safety inspection and infectious disease diagnosis (Zhou J. et al., 2020). The CRISPR/Cas13a system, as a sensitive, rapid, and specific technique, is being increasingly applied for the detection of bacteria. Some studies have focused on pathogenic bacteria of foodborne diseases. An et al. (2021) established a one-tube SHERLOCK method in which the RPA reaction and Cas13a reaction are performed in combination in a single tube and achieved detection of Salmonella spp. at as low as 102 copies/μl within 20 min. Shen et al. (2020) developed a novel allosteric probe-initiated catalysis and CRISPR/Cas13a (APC-Cas) method to diagnose S. enteritidis. In the method, the presence of the target pathogen activates the allosteric probe-initiated amplification, and then, the ssRNA products promote crRNA-guided Cas13a collateral cleavage of the reporter probes to produce fluorescence signals. The APC-Cas assay could detect as low as 1 CFU/ml of S. enteritidis and exhibited superior specificity because of the dual recognition and triple amplification processes. Moreover, detection of viable bacteria is essential for bacterial virulence; therefore, one study developed a CRISPR/Cas13-based NASBA assay (called the cNASBA assay) that couples NASBA with the Cas13a reaction to detect and quantify viable S. enterica within 2.5 h, with an LOD of 1.5 CFU/ml of viable bacteria (Xue et al., 2022). In addition, Cas13a-based detection combined with PCR amplification was performed to detect S. aureus, another pathogen of foodborne diseases, exhibiting an LOD of 1 CFU/ml within 4 h (Zhou J. et al., 2020). In concordance with the detection of S. aureus, another study applied this PCR-Cas13a-based technique to detect S. typhimurium and achieved an LOD of as low as 1 CFU/ml (Gao et al., 2021). Furthermore, Zhang et al. (2021) deployed a Cas13a-based biosensor integrated with the light-up RNA aptamer technique to detect as low as 10 CFU/ml of B. cereus. They also confirmed the ability of this method for estimating the trend of food spoilage by quantifying the percentage of viable pathogenic bacteria. Diagnosis of infectious diseases caused by bacteria is also necessary. The ultralocalized Cas13a assay mentioned above was used to detect P. aeruginosa with an LOD of as low as 6 × 103 copies/μl (Tian et al., 2021). Additionally, Yersinia pestis, the main pathogen causing plague, was detected by the RPA-SHERLOCK method with an LOD of as low as 700 zM (420 copies/ml; Schultzhaus et al., 2021). Using the RPA-SHERLOCK assay, Luo et al. (2021) detected 10 copies/μl of the porA gene of Neisseria gonorrhoeae and successfully identified two azithromycin resistance mutations.
In addition to detecting viruses and bacteria, the CRISPR/Cas13a system was also used for the detection of parasites, chlamydia, and fungus. Cunningham et al. (2021) employed RPA-SHERLOCK to achieve attomolar sensitivity for Plasmodium falciparum detection and achieved a desirable specificity for diverse plasmodium species. The Cas13a reaction coupled with RAA was also introduced to detect as low as 1 × 10−6 ng/μl of Toxoplasma gondii (Zhao et al., 2022). Additionally, a one-pot CRISPR/Cas13a assay was developed to detect dsDNA of Chlamydia trachomatis with an LOD of 10 fM (Huang et al., 2021). Recently, RPA-SHERLOCK was used to detect Aspergillus fumigatus, with an LOD of 3 copies/L within a total time of 70 min (Li et al., 2021). In addition, Zhan et al. combined transcription-mediated amplification (TMA) with Cas13a detection system and used this method to detect mitochondrial large subunit ribosomal RNA of Pneumocystis jirovecii, with an LOD of 2 copies/μl (Zhan et al., 2022).
Antimicrobial resistance (AMR) of pathogens has become a growing global public health concern during the past decade (Dean et al., 2022). The social and financial burden are estimated to be $100 trillion if this issue is not resolved (Piddock, 2016). Therefore, the detection of antimicrobial-resistant pathogens and subsequent rational drug use are vital for the containment of AMR. Cas13a-based detection methods could recognize SNP in target gene via artificially adding mismatch between crRNA and target RNA, exhibiting the potential to detect AMR mutants. Following to this mechanism, several studies have achieved detection of AMR mutants in different pathogens (Kiga et al., 2020; Cunningham et al., 2021; Luo et al., 2021; Bai et al., 2022). Cunningham et al. demonstrated the capability of RPA-SHERLOCK assay for recognizing the A581G mutant in P. falciparum associated with sulfadoxine resistance (Cunningham et al., 2021). Similarly, another group utilized RPA-SHERLOCK assay to identify the A2059G and C2611T mutants in Neisseria gonorrhoeae associated with azithromycin resistance, in accordance with sequencing (Luo et al., 2021). In addition, Bai et al. used Cas13a system with synthetic mismatch to significantly distinguish the wild type strains of Mycobacterium tuberculosis from fluroquinolone resistance mutant strains (G88A, A90V, S91P, D94N, D94H, D94T, and D94G; Bai et al., 2022). Moreover, another study established a novel method for detection of antimicrobial resistant Escherichia coli, termed CRISPR-Cas13a-based antibacterial nucleocapsids (CapsidCas13a). In this study, researchers constructed a bacteriophage capsid which contains programmed Cas13a system targeting AMR gene of Escherichia coli. Similar to drug sensitive test, Escherichia coli carrying AMR gene is unable to grow in the plate with the treatment of CapsidCas13a. Using this assay, they achieved detection of several AMR gene including blaIMP-1, blaOXA-48, blaNDM-1, and blaVIM-2 (Kiga et al., 2020).
Aberrant expression of microRNAs (miRNAs) is known to be associated with various tumors and has been confirmed as biomarker for initial cancer diagnosis (Chen et al., 2018). The detection of miRNA remains challenging given its low cellular abundance, small size, and high homology (Pritchard et al., 2012). Thus, the CRISPR/Cas13a system has been developed as a novel diagnostic tool to detect microRNA. Shan et al. (2019) first utilized the Cas13a system to directly detect as low as 4.5 amol of miR-17 and demonstrated its high specificity to distinguish SNPs in miRNAs. Subsequently, several studies refined the original CRISPR/Cas13a detection platform to detect miR-17. One study introduced a gold nanoparticle-based colorimetric assay integrated with the Cas13a system, allowing naked-eye detection of as low as 500 fM of miR-17 and promoting point-of-care detection based on the Cas13a assay (Yuan et al., 2020). Furthermore, a CRISPR/Cas13a-powered portable electrochemiluminescence chip, called the PECL-CRISPR, was established and could detect miR-17 at levels as low as 1 fM. In PECL-CRISPR, the activated Cas13a cleaves the pretrigger (PT) into mature trigger and then promotes downstream DNA amplification. Subsequently, the amplified dsDNA products interact with the light switch [Ru(phen)2dppz]2+ to produce the luminance signal. The PECL-CRISPR approach provided a single-nucleotide resolution for miR-17 together with its highly homologous family members and successfully analyzed the expression of miR-17 in human breast adenocarcinoma and human hepatocellular carcinoma cells (Zhou T. et al., 2020). Notably, instead of harnessing conventional methods for signal amplification, Sha et al. (2021) introduced a cascade CRISPR/Cas system (casCRISPR), which combines trans-cleavage activities of both Cas14a and Cas13a to detect miR-17 in an nucleic acid amplification-free manner (Figure 4D), with an LOD of 1.33 fM and high specificity for homologous miRNAs. As mentioned above, the ultralocalized Cas13a assay was also employed to detect and accurately quantify miRNA-17 in four different cell lines (Tian et al., 2021). In addition, other pivotal miRNAs have also been detected in previous studies. Bruch et al. exhibited a Cas13a-powered microfluidic electrochemical biosensor platform to quantify miR19b and miR20a, achieving an LOD of 10 pM within 4 h and confirming its feasibility for miR19b detection in serum samples of children suffering from medulloblastoma (Bruch et al., 2019). An electrochemical biosensor combining CHA with the Cas13a system was introduced to detect miR-21 and achieved an LOD of 2.6 fM using dual signal amplification of the Cas13a reaction and CHA amplification (Cui et al., 2021). Intriguingly, a novel magnetic relaxation switching (MRS)-based strategy was combined with the Cas13a system for the detection of as low as 0.22 pM of miR-21 with a unique T2 signal readout (Guo et al., 2022). Furthermore, Zhou et al. (2021) developed a Cas13a-powered rolling circle amplified DNAzyme colorimetric detection system, in which miRNA-activated Cas13a cleaves the pre-primer to initiate downstream DNA polymerase-mediated RCA. The resulting products catalyze the substrates to produce a color change. They could detect miR-10b with an LOD of as low as 1 fM using this method. In addition to miRNAs, lncRNAs are also considered significant biomarkers of cancers; thus, one group recently reported a plasmonically enhanced CRISPR-powered fluoroimmunoassay (p-FLISA), which integrates plasmonic-fluor as a fluorescent nanolabel with the Cas13a system. In this assay, p-FLISA was utilized to detect lncRNA H19 with an LOD of as low as 6 pM and could quantitatively detect H19 in human ovarian cancer cells and breast cancer cells. Furthermore, the feasibility of quantitative detection of tumor tissues obtained from a human ovarian cancer xenograft mouse model was confirmed (Liu L. et al., 2021). Recently, Song et al. (2022) reported a Cas13a-induced exponential amplification assay in which activated Cas13a cleaves stem-loop DNA primers to produce massive double stem-loop DNAs and promote loop-mediated isothermal amplification. Using this assay, they could detect circRNA sponge for miR-7 (ciRS-7) with an LOD of as low as 1 fM within 30 min and successfully discriminated circRNA from linear RNA. Notably, the diagnosis of clinical diseases by detecting a single specific biomarker is not specific; thus, it is indispensable to detect a set of biomarkers associated with a certain disease for accurate diagnosis. Bruch et al. (2021) refined a previously-developed Cas13a-powered microfluidic electrochemical biosensor platform into a multiplexed version, called CRISPR-Biosensor X, and realized synchronous quantification of a maximum of eight miRNAs. In another study, a Cas-CHDC-powered electrochemical RNA sensing technology (COMET) was developed, which combines the catalytic hairpin DNA circuit (CHDC) with the Cas13a system to detect and analyze six RNAs associated with non-small-cell lung carcinoma (NSCLC): miR-17, miR-155, miR-19b, miR-210, TTF-1 mRNA, and EGFR mRNA. This method could successfully discriminate between patients with early-stage NSCLC and patients with benign lung disease through combinatory analysis of multiple RNA biomarkers (Sheng et al., 2021).
Most researchers pay more attention to Cas13a-based detection of nucleic acids; however, the Cas13a system can also be employed for detecting non-nucleic acid targets. Chen et al. (2020) established a CRISPR/Cas13a signal amplification linked immunosorbent assay (CLISA), in which conventional horseradish peroxidase is replaced by dsDNA containing a T7 promoter sequence to activate the downstream Cas13a reaction system (Figure 5A). In this assay, two biomarkers, human IL-6 and VEGF, were detected with LODs of 2.29 fM and 0.81 fM, respectively. Wang et al. (2021a) developed a Cas13a-based detection assay that combines the target-induced transcription amplification with the trans-cleavage activity of Cas13a (called TITAC-Cas) and employs the dephosphorylation of alkaline phosphatase (ALP) as a switch to protect template dsDNA from digestion by λ exonuclease and activate the subsequent T7 transcription and trans-cleavage activity of Cas13a (Figure 5B). This assay allowed ultrasensitive detection of ALP activity (6 ± 0.52 mU/L). Another study established a Cas13a-based method for the detection of fluoride with an LOD of 1.7 μM, in which the presence of fluoride promotes the fluoride riboswitch to regulate transcription and produce full-length RNA, which then activates the collateral cleavage of downstream Cas13a to generate a fluorescence signal (Figure 5C; Ma et al., 2021). Furthermore, a high throughput analytical method called SHERLOCK-based profiling of in vitro transcription was reported, which combines ligand-dependent transcription activation of allosteric transcription factor and riboswitch with the Cas13a reaction. Because diverse ligands can be employed, it could quantify various compounds containing cofactors, nucleotides, metabolites of amino acids, tetracycline, and monatomic ions simultaneously based on ligand-dependent transcription (Iwasaki and Batey, 2020). Moreover, the Cas13a system was used to test the presence of methylation (Figure 5D). A study reported that the Cas13a system could be utilized to analyze methylation by artificially designed mismatches in crRNA based on the disability of methylated bases to pair with other bases. Based on this principle, two studies detected the presence of RNA methylation, including N1-methyladenosine (m1A) and N6-methyladenosine (m6A) and quantitated the percentage of methylated RNA in RNA samples (Figure 5E; Chen et al., 2019; Yang et al., 2020). Furthermore, Wang et al. (2021b) introduced a dual methylation-sensitive restriction endonuclease coupling with an RPA-assisted CRISPR/Cas13a System (DESCS), in which methylated DNA cannot be digested by two methylation-sensitive restriction endonucleases (BstUI/HhaI) and then initiates the downstream RPA, T7 transcription, and the collateral cleavage activity of Cas13a (Figure 5E). This approach could detect C5 methylation in the SEPT9 gene, with LODs of 86.4 aM and 200 aM using fluorescence and lateral-flow readout, respectively.
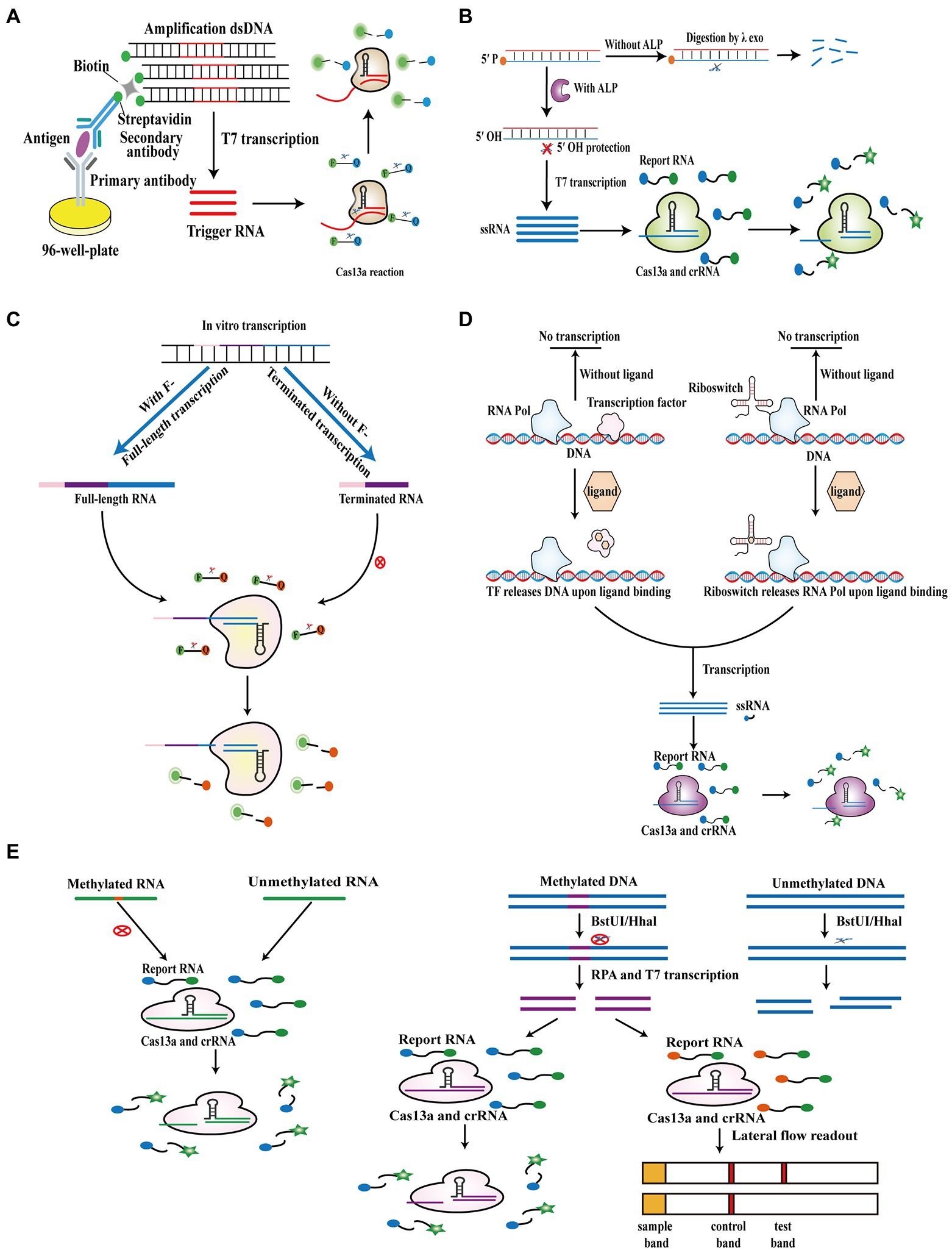
Figure 5. Cas13a-based biosensors for non-nucleic acid target detection. (A) Schematic of CLISA. Horseradish peroxidase in ELISA is replaced by biotinylated dsDNA containing a T7 promoter sequence to achieve Cas13a-based detection of antigen. (B) Schematic of TITAC-Cas. In the presence of ALP, 5′ P-dsDNA is converted into 5′ OH-dsDNA, avoiding digestion by λexo and activating the downstream T7 transcription and the Cas13a reaction. (C) Schematic of Cas13a-based biosensor for fluoride detection. In the absence of fluoride, template DNA is transcribed into terminated RNA, which cannot be recognized by the Cas13a system. Instead, in the presence of fluoride, template DNA is transcribed into full-length RNA, which can activate the downstream Cas13a reaction. (D) Schematic of SPRINT. The presence of ligands leads to the release of DNA by transcription factor or the release of RNA pol by riboswitch. Subsequently, the transcription products are recognized by the Cas13a system and activate the trans-cleavage activity of Cas13a. (E) Schematic of Cas13a-based biosensor for RNA and DNA methylation detection. Methylated RNA is not recognized by the Cas13a system due to the mismatch between methylated and unmethylated bases (above at left). Methylated DNA cannot be digested by BstUI/HhaI and then initiates the downstream RPA, T7 transcription, and the collateral cleavage activity of Cas13a (above at right).
So far, scientists have discovered two classes of the CRISPR-Cas system, among which class II effectors are expected to be widely employed because of their simplicity and high efficiency. Although some Cas9-based detection methods have been developed, the application of the CRISPR-Cas system as a biosensor only came under the spotlight when the collateral activity of Cas13a was reported. Currently, mainstream detection methods include PCR-based technologies, antigen–antibody reaction, genome sequencing, etc. (Yin et al., 2021). PCR-based methods are currently considered the gold standard of detection; however, PCR-based methods usually require expensive instruments, complete laboratory settings, and professionals, thereby limiting their deployment in low-resource regions, such as Africa (Kevadiya et al., 2021). Antigen–antibody reaction exists disadvantages of cross-reactivity and low sensitivity. Genome sequencing is time-consuming and highly expensive. Compared to these traditional diagnostic tools, CRISPR/Cas13a based detection methods are able to simultaneously achieve high sensitivity, high specificity, low cost and low time-consuming, by combining Cas13a system with isothermal amplification technique, visual readout, extraction-free method, etc. The SHERLOCK platform reported by Gootenberg et al. (2017) combined RPA nucleic acid amplification with the collateral activity of Cas13a; the dual signal amplification achieved by this method allowed attomolar sensitivity. Compared to RT-qPCR, the SHERLOCK assay is based on isothermal amplification (37–42°C) and does not require an expensive thermal cycler; furthermore, the SHERLOCK paper test just costs $0.61 per test. Subsequently, in SHERLOCKv2, an auxiliary enzyme was introduced, which significantly improved the detection sensitivity. Moreover, a lateral-flow strip readout was also added to SHERLOCK for point-of-care detection. Inspiringly, the SHERLOCK platform has gained approval for the detection of SARS-CoV-2 under emergencies by the Food and Drug Administration.
Since the SHERLOCK assay was developed, numerous studies have refined Cas13a-based biosensors by improving the SHERLOCK method or coupling the Cas13a reaction with advanced technologies. The multiple steps of SHERLOCK may cause aerosol pollution, operation complexity, and loss of sample; additionally, the amplification steps might be accompanied by amplification bias. Accordingly, a range of refined methods have been developed to simplify the complicated steps and reduce the testing time. Some studies utilized nucleic acid amplification-free methods, such as multiple combinations of crRNAs, ultralocalized Cas13a-based detection, opn-SATORI, and cascaded collateral activities of two Cas effectors (Fozouni et al., 2021; Sha et al., 2021; Tian et al., 2021; Shinoda et al., 2022). One extraction-free approach called HUDSON was also reported (Myhrvold et al., 2018). Furthermore, some studies combined the RPA and Cas13a reactions in a single tube, avoiding sample contamination during the transition of different steps and significantly reducing the total test time (Arizti-Sanz et al., 2020; An et al., 2021; Hu et al., 2022).
The programmability of crRNA makes the Cas13a system capable of detecting nearly all nucleic acid targets. Currently, Cas13a-based detection has been deployed to test pathogens, such as viruses, bacteria, parasites, chlamydia, and fungi; and biomarkers, such as microRNAs, lncRNAs, and circRNAs. The Cas13a system can effectively perform DNA detection because of transcription after amplification. Moreover, some non-nucleic acid targets, including proteins and ions, have also been detected by combining novel methods with the Cas13a reaction. Additionally, methylation and SNPs were successfully detected based on the mismatches between the guide and target. However, for the detection of methylation, the current methods can only test known methylation sites for supplementary validation. Furthermore, different readout methods, such as fluorescence readout, lateral-flow readout, colorimetric readout, and electrochemical readout, have been combined with the Cas13a reaction for detection.
The ideal detection approach is expected to be rapid, sensitive, specific, inexpensive, easy, and deployable. The majority of Cas13a-based biosensors still require multiple steps, including nucleic acid extraction, amplification, and the Cas13a reaction, which prolong the detection time. One-pot or tube methods shorten the detection time but they always give up high sensitivity. Combinations of crRNAs significantly increase the sensitivity of detection, whereas the capacity of this method to detect SNP remains to be verified (Fozouni et al., 2021). The sensitivity of a single Cas13a reaction is low, and nucleic acid amplification is still necessary to improve sensitivity. In addition, the issues of amplification bias and false-positive results caused by contamination during amplification remain to be solved (Yin et al., 2021; Li et al., 2022b). The prospective development of Cas13a-based detection is supposed to focus on improving the multiplexing capability by deploying a microfluidic system or through the orthogonal utilization of different Cas effectors. Ackerman et al., 2020 established a combinatorial arrayed reactions for multiplexed evaluation of nucleic acids (CARMEN) platform that can test more than 4,500 guide-target pairs on a single array and successfully detected 169 viruses. Since the casCRISPR system was developed, more different combinations of Cas proteins are expected to be further exploited. Furthermore, balancing sensitivity, availability, and operation time is also crucial for the future application of Cas13a-based biosensors, which remains to be studied further.
Effectively designing crRNA and clarifying the effect of the Cas13a system are vital for improving the detection efficiency. Tambe et al. (2018) demonstrated that there are two important functional regions in crRNA and target RNA: the seed region and the HEPN-nuclease switch region. Mismatches in the former lead to a significant reduction in the binding affinity, whereas mismatches in the latter inactivate the HEPN-nuclease without affecting the binding affinity. A previous study confirmed a 3′ protospacer flanking sequence called the anti-tag, which is complementary to the 5′ flanking region of the crRNA spacer, also termed as crRNA tag. This extra complementarity between the anti-tag and crRNA tag prevents the Cas13a-mediated cleavage of crRNA (Meeske and Marraffini, 2018). A subsequent structure analysis study demonstrated that the tag: anti-tag duplex results in conformational inactivation of the Cas13a protein (Wang B. et al., 2021). Intriguingly, (Li et al., 2020) found that targeting the NS3 region of DENV by the Cas13a system results in the deletion of the NS2B region, and the detection of NS3-crRNA alone without the Cas13a protein also provided an anti-viral effect. Thus, it is expected to clarify the specific effect mechanism of the Cas13a system to rationally select and design crRNAs and the Cas13a protein for accurate diagnosis.
In summary, the development and application of the CRISPR/Cas13a system may herald the emergence of a next-generation diagnostic tool, which will provide novel insights for the detection of diseases. Despite many advantages, there are still some challenges to be resolved. The ideal future development of Cas13a-based biosensors includes: (1) Improving the sensitivity of detection without relying on nucleic acid amplification methods (such as combinations of different Cas proteins, combinations of crRNAs, etc). The high-fidelity of Cas13a protein is often ignored as the efficient nuclease to recognize the amplification products (Zhou T. et al., 2020). (2) Combining sample treatment, amplification and Cas13a reaction into a real one-pot reaction that does not need additional manual operation and requires less time. (Soh et al., 2022). (3) Combining multiplexing technique with variable programmed ability of Cas13a system to achieve high throughput detection of various targets Ackerman et al. (2020). (4) Combining visual readout with mobile device to reduce artificial bias (Tian et al., 2022). Nevertheless, it is conceivable that the CRISPR/Cas13a system will become a promising diagnostic tool in future.
LZ was involved in conceptualization, writing original draft, writing—review and editing, and visualization. MQ was involved in conceptualization and writing—review and editing. XL and JY were involved in writing—review and editing. JL was involved in conceptualization, writing—review and editing, and supervision. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (grant number 81570497) and the Science Foundation of AMU (grant number 21QNPY004).
The authors wish to thank the National Natural Science Foundation of China (grant number 81570497) and the Science Foundation of AMU (grant number 21QNPY004) for financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., et al. (2017). RNA targeting with CRISPR-Cas13. Nature 550, 280–284. doi: 10.1038/nature24049
Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B., et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353:aaf5573. doi: 10.1126/science.aaf5573
Ackerman, C. M., Myhrvold, C., Thakku, S. G., Freije, C. A., Metsky, H. C., Yang, D. K., et al. (2020). Massively multiplexed nucleic acid detection with Cas13. Nature 582, 277–282. doi: 10.1038/s41586-020-2279-8
Amitai, G., and Sorek, R. (2016). CRISPR-Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 14, 67–76. doi: 10.1038/nrmicro.2015.14
An, B., Zhang, H., Su, X., Guo, Y., Wu, T., Ge, Y., et al. (2021). Rapid and sensitive detection of salmonella spp. using CRISPR-Cas13a combined with recombinase polymerase amplification. Front. Microbiol. 12:732426. doi: 10.3389/fmicb.2021.732426
Arizti-Sanz, J., Freije, C. A., Stanton, A. C., Boehm, C. K., Petros, B. A., Siddiqui, S., et al. (2020). Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. bioRxiv [Preprint].
Azmi, I., Faizan, M. I., Kumar, R., Raj Yadav, S., Chaudhary, N., Kumar Singh, D., et al. (2021). A saliva-based RNA extraction-free workflow integrated with Cas13a for SARS-CoV-2 detection. Front. Cell. Infect. Microbiol. 11:632646. doi: 10.3389/fcimb.2021.632646
Bai, X., Gao, P., Qian, K., Yang, J., Deng, H., Fu, T., et al. (2022). A highly sensitive and specific detection method for mycobacterium tuberculosis fluoroquinolone resistance mutations utilizing the CRISPR-Cas13a system. Front. Microbiol. 13:847373. doi: 10.3389/fmicb.2022.847373
Bai, Y., Yao, L., Wei, T., Tian, F., Jin, D. Y., Chen, L., et al. (2020). Presumed asymptomatic carrier transmission of COVID-19. JAMA 323, 1406–1407. doi: 10.1001/jama.2020.2565
Barnes, K. G., Lachenauer, A. E., Nitido, A., Siddiqui, S., Gross, R., Beitzel, B., et al. (2020). Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time. Nat. Commun. 11:4131. doi: 10.1038/s41467-020-17994-9
Bente, D. A., Forrester, N. L., Watts, D. M., McAuley, A. J., Whitehouse, C. A., and Bray, M. (2013). Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res. 100, 159–189. doi: 10.1016/j.antiviral.2013.07.006
Bruch, R., Baaske, J., Chatelle, C., Meirich, M., Madlener, S., Weber, W., et al. (2019). CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. 31:e1905311. doi: 10.1002/adma.201905311
Bruch, R., Johnston, M., Kling, A., Mattmuller, T., Baaske, J., Partel, S., et al. (2021). CRISPR-powered electrochemical microfluidic multiplexed biosensor for target amplification-free miRNA diagnostics. Biosens. Bioelectron. 177:112887. doi: 10.1016/j.bios.2020.112887
Cenciarelli, O., Gabbarini, V., Pietropaoli, S., Malizia, A., Tamburrini, A., Ludovici, G. M., et al. (2015). Viral bioterrorism: learning the lesson of Ebola virus in West Africa 2013-2015. Virus Res. 210, 318–326. doi: 10.1016/j.virusres.2015.09.002
Chen, Y. X., Huang, K. J., and Niu, K. X. (2018). Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review. Biosens. Bioelectron. 99, 612–624. doi: 10.1016/j.bios.2017.08.036
Chen, Q., Tian, T., Xiong, E., Wang, P., and Zhou, X. (2020). CRISPR/Cas13a signal amplification linked immunosorbent assay for Femtomolar protein detection. Anal. Chem. 92, 573–577. doi: 10.1021/acs.analchem.9b04403
Chen, Y., Yang, S., Peng, S., Li, W., Wu, F., Yao, Q., et al. (2019). N1-Methyladenosine detection with CRISPR-Cas13a/C2c2. Chem. Sci. 10, 2975–2979. doi: 10.1039/c8sc03408g
Cox, D. B. T., Gootenberg, J. S., Abudayyeh, O. O., Franklin, B., Kellner, M. J., Joung, J., et al. (2017). RNA editing with CRISPR-Cas13. Science 358, 1019–1027. doi: 10.1126/science.aaq0180
Cui, Y., Fan, S., Yuan, Z., Song, M., Hu, J., Qian, D., et al. (2021). Ultrasensitive electrochemical assay for microRNA-21 based on CRISPR/Cas13a-assisted catalytic hairpin assembly. Talanta 224:121878. doi: 10.1016/j.talanta.2020.121878
Cui, J. Q., Liu, F. X., Park, H., Chan, K. W., Leung, T., Tang, B. Z., et al. (2022). Droplet digital recombinase polymerase amplification (ddRPA) reaction unlocking via picoinjection. Biosens. Bioelectron. 202:114019. doi: 10.1016/j.bios.2022.114019
Cunningham, C. H., Hennelly, C. M., Lin, J. T., Ubalee, R., Boyce, R. M., Mulogo, E. M., et al. (2021). A novel CRISPR-based malaria diagnostic capable of plasmodium detection, species differentiation, and drug-resistance genotyping. EBioMedicine 68:103415. doi: 10.1016/j.ebiom.2021.103415
Dean, A. S., Tosas Auguet, O., Glaziou, P., Zignol, M., Ismail, N., Kasaeva, T., et al. (2022). 25 years of surveillance of drug-resistant tuberculosis: achievements, challenges, and way forward. Lancet Infect. Dis. 22, e191–e196. doi: 10.1016/S1473-3099(21)00808-2
East-Seletsky, A., O’Connell, M. R., Burstein, D., Knott, G. J., and Doudna, J. A. (2017). RNA targeting by functionally orthogonal type VI-A CRISPR-Cas enzymes. Mol. Cell 66, 373–383.e3. doi: 10.1016/j.molcel.2017.04.008
East-Seletsky, A., O’Connell, M. R., Knight, S. C., Burstein, D., Cate, J. H., Tjian, R., et al. (2016). Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273. doi: 10.1038/nature19802
Fozouni, P., Son, S., de Leon Derby, M. D., Knott, G. J., Gray, C. N., Ambrosio, M. V. D., et al. (2021). Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cells 184, 323–333 e9. doi: 10.1016/j.cell.2020.12.001
Gao, R., Cao, B., Hu, Y., Feng, Z., Wang, D., Hu, W., et al. (2013). Human infection with a novel avian-origin influenza a (H7N9) virus. N. Engl. J. Med. 368, 1888–1897. doi: 10.1056/NEJMoa1304459
Gao, S., Liu, J., Li, Z., Ma, Y., and Wang, J. (2021). Sensitive detection of foodborne pathogens based on CRISPR-Cas13a. J. Food Sci. 86, 2615–2625. doi: 10.1111/1750-3841.15745
Garneau, J. E., Dupuis, M. E., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. doi: 10.1038/nature09523
Gootenberg, J. S., Abudayyeh, O. O., Kellner, M. J., Joung, J., Collins, J. J., and Zhang, F. (2018). Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444. doi: 10.1126/science.aaq0179
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442. doi: 10.1126/science.aam9321
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172
Guo, X., Tian, T., Deng, X., Song, Y., Zhou, X., and Song, E. (2022). CRISPR/Cas13a assisted amplification of magnetic relaxation switching sensing for accurate detection of miRNA-21 in human serum. Anal. Chim. Acta 1209:339853. doi: 10.1016/j.aca.2022.339853
Hale, C. R., Zhao, P., Olson, S., Duff, M. O., Graveley, B. R., Wells, L., et al. (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cells 139, 945–956. doi: 10.1016/j.cell.2009.07.040
Hang, X. M., Liu, P. F., Tian, S., Wang, H. Y., Zhao, K. R., and Wang, L. (2022). Rapid and sensitive detection of Ebola RNA in an unamplified sample based on CRISPR-Cas13a and DNA roller machine. Biosens. Bioelectron. 211:114393. doi: 10.1016/j.bios.2022.114393
Happi, A. N., Happi, C. T., and Schoepp, R. J. (2019). Lassa fever diagnostics: past, present, and future. Curr. Opin. Virol. 37, 132–138. doi: 10.1016/j.coviro.2019.08.002
Heo, W., Lee, K., Park, S., Hyun, K. A., and Jung, H. I. (2022). Electrochemical biosensor for nucleic acid amplification-free and sensitive detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA via CRISPR/Cas13a trans-cleavage reaction. Biosens. Bioelectron. 201:113960. doi: 10.1016/j.bios.2021.113960
Hille, F., Richter, H., Wong, S. P., Bratovic, M., Ressel, S., and Charpentier, E. (2018). The biology of CRISPR-Cas: backward and forward. Cells 172, 1239–1259. doi: 10.1016/j.cell.2017.11.032
Hochstrasser, M. L., and Doudna, J. A. (2015). Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem. Sci. 40, 58–66. doi: 10.1016/j.tibs.2014.10.007
Hu, F., Liu, Y., Zhao, S., Zhang, Z., Li, X., Peng, N., et al. (2022). A one-pot CRISPR/Cas13a-based contamination-free biosensor for low-cost and rapid nucleic acid diagnostics. Biosens. Bioelectron. 202:113994. doi: 10.1016/j.bios.2022.113994
Huang, K., Yu, H., Chen, Z., Lin, G., Zhang, Z., Zhang, X., et al. (2021). CRISPR-Cas13a-based diagnostic method for chlamydia trachomatis from nongonococcal urethritis. Bioanalysis 13, 901–912. doi: 10.4155/bio-2021-0022
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987
Iwasaki, R. S., and Batey, R. T. (2020). SPRINT: a Cas13a-based platform for detection of small molecules. Nucleic Acids Res. 48:e101. doi: 10.1093/nar/gkaa673
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O., and Zhang, F. (2019). SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 14, 2986–3012. doi: 10.1038/s41596-019-0210-2
Kevadiya, B. D., Machhi, J., Herskovitz, J., Oleynikov, M. D., Blomberg, W. R., Bajwa, N., et al. (2021). Diagnostics for SARS-CoV-2 infections. Nat. Mater. 20, 593–605. doi: 10.1038/s41563-020-00906-z
Khan, W. A., Barney, R. E., and Tsongalis, G. J. (2021). CRISPR-cas13 enzymology rapidly detects SARS-CoV-2 fragments in a clinical setting. J. Clin. Virol. 145:105019. doi: 10.1016/j.jcv.2021.105019
Khan, H., Khan, A., Liu, Y., Wang, S., Bibi, S., Xu, H., et al. (2019). CRISPR-Cas13a mediated nanosystem for attomolar detection of canine parvovirus type 2. Chin. Chem. Lett. 30, 2201–2204. doi: 10.1016/j.cclet.2019.10.032
Kiga, K., Tan, X. E., Ibarra-Chávez, R., Watanabe, S., Aiba, Y., Sato’o, Y., et al. (2020). Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat. Commun. 11:2934. doi: 10.1038/s41467-020-16731-6
Knott, G. J., East-Seletsky, A., Cofsky, J. C., Holton, J. M., Charles, E., O’Connell, M. R., et al. (2017). Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat. Struct. Mol. Biol. 24, 825–833. doi: 10.1038/nsmb.3466
Koonin, E. V., Makarova, K. S., and Zhang, F. (2017). Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78. doi: 10.1016/j.mib.2017.05.008
Kunin, V., Sorek, R., and Hugenholtz, P. (2007). Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8:R61. doi: 10.1186/gb-2007-8-4-r61
Li, H., Bello, A., Smith, G., Kielich, D. M. S., Strong, J. E., and Pickering, B. S. (2022a). Degenerate sequence-based CRISPR diagnostic for Crimean-Congo hemorrhagic fever virus. PLoS Negl. Trop. Dis. 16:e0010285. doi: 10.1371/journal.pntd.0010285
Li, H., Wang, S., Dong, X., Li, Q., Li, M., Li, J., et al. (2020). CRISPR-Cas13a cleavage of dengue virus NS3 gene efficiently inhibits viral replication. Mol. Ther. Nucleic Acids 19, 1460–1469. doi: 10.1016/j.omtn.2020.01.028
Li, Z., Wang, M., Xu, T., Zhan, Y., Chen, F., Lin, Y., et al. (2021). Development and clinical implications of a novel CRISPR-based diagnostic test for pulmonary aspergillus fumigatus infection. J. Microbiol. Immunol. Infect. 55, 749–756. doi: 10.1016/j.jmii.2021.11.008
Li, H., Yang, J., Wu, G., Weng, Z., Song, Y., Zhang, Y., et al. (2022b). Amplification-free detection of SARS-CoV-2 and respiratory syncytial virus using CRISPR Cas13a and graphene field-effect transistors. Angew. Chem. Int. Ed. Engl. 61:e202203826. doi: 10.1002/anie.202203826
Liu, L., Li, X., Wang, J., Wang, M., Chen, P., Yin, M., et al. (2017). Two distant catalytic sites are responsible for C2c2 RNase activities. Cells 168, 121–134.e12. doi: 10.1016/j.cell.2016.12.031
Liu, L., Wang, Z., Wang, Y., Luan, J., Morrissey, J. J., Naik, R. R., et al. (2021). Plasmonically enhanced CRISPR/Cas13a-based bioassay for amplification-free detection of cancer-associated RNA. Adv. Healthc. Mater. 10:e2100956. doi: 10.1002/adhm.202100956
Liu, W. J., Xiao, H., Dai, L., Liu, D., Chen, J., Qi, X., et al. (2021). Avian influenza a (H7N9) virus: from low pathogenic to highly pathogenic. Front. Med. 15, 507–527. doi: 10.1007/s11684-020-0814-5
Liu, Y., Xu, H., Liu, C., Peng, L., Khan, H., Cui, L., et al. (2019). CRISPR-Cas13a Nanomachine based simple Technology for Avian Influenza a (H7N9) virus on-site detection. J. Biomed. Nanotechnol. 15, 790–798. doi: 10.1166/jbn.2019.2742
López-Valls, M., Escalona-Noguero, C., Rodríguez-Díaz, C., Pardo, D., Castellanos, M., Milán-Rois, P., et al. (2022). CASCADE: naked eye-detection of SARS-CoV-2 using Cas13a and gold nanoparticles. Anal. Chim. Acta 1205:339749. doi: 10.1016/j.aca.2022.339749
Luo, H., Chen, W., Mai, Z., Yang, J., Lin, X., Zeng, L., et al. (2021). Development and application of Cas13a-based diagnostic assay for Neisseria gonorrhoeae detection and azithromycin resistance identification. J. Antimicrob. Chemother. 77, 656–664. doi: 10.1093/jac/dkab447
Ma, Y., Mou, Q., Yan, P., Yang, Z., Xiong, Y., Yan, D., et al. (2021). A highly sensitive and selective fluoride sensor based on a riboswitch-regulated transcription coupled with CRISPR-Cas13a tandem reaction. Chem. Sci. 12, 11740–11747. doi: 10.1039/d1sc03508h
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477. doi: 10.1038/nrmicro2577
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83. doi: 10.1038/s41579-019-0299-x
Marraffini, L. A., and Sontheimer, E. J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845. doi: 10.1126/science.1165771
Martinovic, T., Andjelkovic, U., Gajdosik, M. S., Resetar, D., and Josic, D. (2016). Foodborne pathogens and their toxins. J. Proteomics 147, 226–235. doi: 10.1016/j.jprot.2016.04.029
Meeske, A. J., and Marraffini, L. A. (2018). RNA guide complementarity prevents self-targeting in type VI CRISPR systems. Mol. Cell 71, 791–801.e3. doi: 10.1016/j.molcel.2018.07.013
Mohanraju, P., Makarova, K. S., Zetsche, B., Zhang, F., Koonin, E. V., and van der Oost, J. (2016). Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353:aad5147. doi: 10.1126/science.aad5147
Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3
Myhrvold, C., Freije, C. A., Gootenberg, J. S., Abudayyeh, O. O., Metsky, H. C., Durbin, A. F., et al. (2018). Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444–448. doi: 10.1126/science.aas8836
Nalefski, E. A., Patel, N., Leung, P. J. Y., Islam, Z., Kooistra, R. M., Parikh, I., et al. (2021). Kinetic analysis of Cas12a and Cas13a RNA-guided nucleases for development of improved CRISPR-based diagnostics. iScience 24:102996. doi: 10.1016/j.isci.2021.102996
Niu, M., Han, Y., Dong, X., Yang, L., Li, F., Zhang, Y., et al. (2022). Highly sensitive detection method for HV69-70del in SARS-CoV-2 alpha and omicron variants based on CRISPR/Cas13a. Front. Bioeng. Biotechnol. 10:831332. doi: 10.3389/fbioe.2022.831332
Ortiz-Cartagena, C., Fernandez-Garcia, L., Blasco, L., Pacios, O., Bleriot, I., Lopez, M., et al. (2022). Reverse transcription-loop-mediated isothermal amplification-CRISPR-Cas13a technology as a promising diagnostic tool for SARS-CoV-2. Microbiol. Spectr. 10:e0239822. doi: 10.1128/spectrum.02398-22
Patchsung, M., Jantarug, K., Pattama, A., Aphicho, K., Suraritdechachai, S., Meesawat, P., et al. (2020). Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 4, 1140–1149. doi: 10.1038/s41551-020-00603-x
Piddock, L. J. V. (2016). Reflecting on the final report of the O’Neill review on antimicrobial resistance. Lancet Infect. Dis. 16, 767–768. doi: 10.1016/S1473-3099(16)30127-X
Pierson, T. C., and Diamond, M. S. (2018). The emergence of Zika virus and its new clinical syndromes. Nature 560, 573–581. doi: 10.1038/s41586-018-0446-y
Pierson, T. C., and Diamond, M. S. (2020). The continued threat of emerging flaviviruses. Nat. Microbiol. 5, 796–812. doi: 10.1038/s41564-020-0714-0
Pougach, K., Semenova, E., Bogdanova, E., Datsenko, K. A., Djordjevic, M., Wanner, B. L., et al. (2010). Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 77, 1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x
Pritchard, C. C., Cheng, H. H., and Tewari, M. (2012). MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 13, 358–369. doi: 10.1038/nrg3198
Qin, P., Park, M., Alfson, K. J., Tamhankar, M., Carrion, R., Patterson, J. L., et al. (2019). Rapid and fully microfluidic Ebola virus detection with CRISPR-Cas13a. ACS Sens 4, 1048–1054. doi: 10.1021/acssensors.9b00239
Schultzhaus, Z., Wang, Z., and Stenger, D. (2021). Systematic analysis, identification, and use of CRISPR/Cas13a-associated crRNAs for sensitive and specific detection of the lcrV gene of Yersinia pestis. Diagn. Microbiol. Infect. Dis. 99:115275. doi: 10.1016/j.diagmicrobio.2020.115275
Sha, Y., Huang, R., Huang, M., Yue, H., Shan, Y., Hu, J., et al. (2021). Cascade CRISPR/cas enables amplification-free microRNA sensing with fM-sensitivity and single-base-specificity. Chem. Commun. 57, 247–250. doi: 10.1039/d0cc06412b
Shan, Y., Zhou, X., Huang, R., and Xing, D. (2019). High-Fidelity and rapid quantification of miRNA combining crRNA programmability and CRISPR/Cas13a trans-cleavage activity. Anal. Chem. 91, 5278–5285. doi: 10.1021/acs.analchem.9b00073
Shen, J., Zhou, X., Shan, Y., Yue, H., Huang, R., Hu, J., et al. (2020). Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat. Commun. 11:267. doi: 10.1038/s41467-019-14135-9
Sheng, Y., Zhang, T., Zhang, S., Johnston, M., Zheng, X., Shan, Y., et al. (2021). A CRISPR/Cas13a-powered catalytic electrochemical biosensor for successive and highly sensitive RNA diagnostics. Biosens. Bioelectron. 178:113027. doi: 10.1016/j.bios.2021.113027
Shinoda, H., Iida, T., Makino, A., Yoshimura, M., Ishikawa, J., Ando, J., et al. (2022). Automated amplification-free digital RNA detection platform for rapid and sensitive SARS-CoV-2 diagnosis. Commun. Biol. 5:473. doi: 10.1038/s42003-022-03433-6
Soh, J. H., Balleza, E., Abdul Rahim, M. N., Chan, H. M., Mohd Ali, S., Chuah, J. K. C., et al. (2022). CRISPR-based systems for sensitive and rapid on-site COVID-19 diagnostics. Trends Biotechnol. 40, 1346–1360. doi: 10.1016/j.tibtech.2022.06.002
Song, P., Zhang, P., Qin, K., Su, F., Gao, K., Liu, X., et al. (2022). CRISPR/Cas13a induced exponential amplification for highly sensitive and specific detection of circular RNA. Talanta 246:123521. doi: 10.1016/j.talanta.2022.123521
Sorek, R., Kunin, V., and Hugenholtz, P. (2008). CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6, 181–186. doi: 10.1038/nrmicro1793
Sternberg, S. H., Richter, H., Charpentier, E., and Qimron, U. (2016). Adaptation in CRISPR-Cas systems. Mol. Cell 61, 797–808. doi: 10.1016/j.molcel.2016.01.030
Tambe, A., East-Seletsky, A., Knott, G. J., Doudna, J. A., and O’Connell, M. R. (2018). RNA binding and HEPN-nuclease activation are decoupled in CRISPR-Cas13a. Cell Rep. 24, 1025–1036. doi: 10.1016/j.celrep.2018.06.105
Tian, T., Qiu, Z., Jiang, Y., Zhu, D., and Zhou, X. (2022). Exploiting the orthogonal CRISPR-Cas12a/Cas13a trans-cleavage for dual-gene virus detection using a handheld device. Biosens. Bioelectron. 196:113701. doi: 10.1016/j.bios.2021.113701
Tian, T., Shu, B., Jiang, Y., Ye, M., Liu, L., Guo, Z., et al. (2021). An Ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. ACS Nano 15, 1167–1178. doi: 10.1021/acsnano.0c08165
van der Oost, J., Westra, E. R., Jackson, R. N., and Wiedenheft, B. (2014). Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 12, 479–492. doi: 10.1038/nrmicro3279
Wang, F., Wang, L., Zou, X., Duan, S., Li, Z., Deng, Z., et al. (2019). Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnol. Adv. 37, 708–729. doi: 10.1016/j.biotechadv.2019.03.016
Wang, J., Xia, Q., Wu, J., Lin, Y., and Ju, H. (2021). A sensitive electrochemical method for rapid detection of dengue virus by CRISPR/Cas13a-assisted catalytic hairpin assembly. Anal. Chim. Acta 1187:339131. doi: 10.1016/j.aca.2021.339131
Wang, Y., Xue, T., Wang, M., Ledesma-Amaro, R., Lu, Y., Hu, X., et al. (2022). CRISPR-Cas13a cascade-based viral RNA assay for detecting SARS-CoV-2 and its mutations in clinical samples. Sens. Actuators B 362:131765. doi: 10.1016/j.snb.2022.131765
Wang, Y., Zhang, Y., Chen, J., Wang, M., Zhang, T., Luo, W., et al. (2021). Detection of SARS-CoV-2 and its mutated variants via CRISPR-Cas13-based transcription amplification. Anal. Chem. 93, 3393–3402. doi: 10.1021/acs.analchem.0c04303
Wang, B., Zhang, T., Yin, J., Yu, Y., Xu, W., Ding, J., et al. (2021). Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems. Mol. Cell 81, 1100–1115 e5. doi: 10.1016/j.molcel.2020.12.033
Wang, X., Zhou, S., Chu, C., Yang, M., Huo, D., and Hou, C. (2021a). Target-induced transcription amplification to trigger the trans-cleavage activity of CRISPR/Cas13a (TITAC-Cas) for detection of alkaline phosphatase. Biosens. Bioelectron. 185:113281. doi: 10.1016/j.bios.2021.113281
Wang, X., Zhou, S., Chu, C., Yang, M., Huo, D., and Hou, C. (2021b). Dual methylation-sensitive restriction endonucleases coupling with an RPA-assisted CRISPR/Cas13a system (DESCS) for highly sensitive analysis of DNA methylation and its application for point-of-care detection. ACS Sens. 6, 2419–2428. doi: 10.1021/acssensors.1c00674
Wei, N., Zheng, B., Niu, J., Chen, T., Ye, J., Si, Y., et al. (2022). Rapid detection of genotype II African swine fever virus using CRISPR Cas13a-based lateral flow strip. Viruses 14:179. doi: 10.3390/v14020179
Wu, Y., Liu, S. X., Wang, F., and Zeng, M. S. (2019). Room temperature detection of plasma Epstein-Barr virus DNA with CRISPR-Cas13. Clin. Chem. 65, 591–592. doi: 10.1373/clinchem.2018.299347
Xue, T., Lu, Y., Yang, H., Hu, X., Zhang, K., Ren, Y., et al. (2022). Isothermal RNA amplification for the detection of viable pathogenic bacteria to estimate the salmonella virulence for causing enteritis. J. Agric. Food Chem. 70, 1670–1678. doi: 10.1021/acs.jafc.1c07182
Yang, Y., Liu, J., and Zhou, X. (2021). A CRISPR-based and post-amplification coupled SARS-CoV-2 detection with a portable evanescent wave biosensor. Biosens. Bioelectron. 190:113418. doi: 10.1016/j.bios.2021.113418
Yang, S., Wu, F., Peng, S., Wang, F., Chen, Y., Yuan, Y., et al. (2020). A m(6) a sensing method by its impact on the stability of RNA double helix. Chem. Biodivers. 17:e2000050. doi: 10.1002/cbdv.202000050
Yin, L., Man, S., Ye, S., Liu, G., and Ma, L. (2021). CRISPR-Cas based virus detection: recent advances and perspectives. Biosens. Bioelectron. 193:113541. doi: 10.1016/j.bios.2021.113541
Yosef, I., Goren, M. G., and Qimron, U. (2012). Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576. doi: 10.1093/nar/gks216
Yuan, C., Tian, T., Sun, J., Hu, M., Wang, X., Xiong, E., et al. (2020). Universal and naked-eye gene detection platform based on the clustered regularly interspaced short palindromic repeats/Cas12a/13a system. Anal. Chem. 92, 4029–4037. doi: 10.1021/acs.analchem.9b05597
Zhan, Y., Gao, X., Li, S., Si, Y., Li, Y., Han, X., et al. (2022). Development and evaluation of rapid and accurate CRISPR/Cas13-based RNA diagnostics for pneumocystis jirovecii pneumonia. Front. Cell. Infect. Microbiol. 12:904485. doi: 10.3389/fcimb.2022.904485
Zhang, Q., Li, J., Li, Y., Tan, G., Sun, M., Shan, Y., et al. (2022). SARS-CoV-2 detection using quantum dot fluorescence immunochromatography combined with isothermal amplification and CRISPR/Cas13a. Biosens. Bioelectron. 202:113978. doi: 10.1016/j.bios.2022.113978
Zhang, X., Tian, Y., Xu, L., Fan, Z., Cao, Y., Ma, Y., et al. (2022). CRISPR/Cas13-assisted hepatitis B virus covalently closed circular DNA detection. Hepatol. Int. 16, 306–315. doi: 10.1007/s12072-022-10311-0
Zhang, T., Zhou, W., Lin, X., Khan, M. R., Deng, S., Zhou, M., et al. (2021). Light-up RNA aptamer signaling-CRISPR-Cas13a-based mix-and-read assays for profiling viable pathogenic bacteria. Biosens. Bioelectron. 176:112906. doi: 10.1016/j.bios.2020.112906
Zhao, J., Li, Y., Xue, Q., Zhu, Z., Zou, M., and Fang, F. (2022). A novel rapid visual detection assay for toxoplasma gondii combining recombinase-aided amplification and lateral flow dipstick coupled with CRISPR-Cas13a fluorescence (RAA-Cas13a-LFD). Parasite 29:21. doi: 10.1051/parasite/2022021
Zhou, T., Huang, R., Huang, M., Shen, J., Shan, Y., and Xing, D. (2020). CRISPR/Cas13a powered portable Electrochemiluminescence Chip for ultrasensitive and specific MiRNA detection. Adv. Sci. 7:1903661. doi: 10.1002/advs.201903661
Zhou, T., Huang, M., Lin, J., Huang, R., and Xing, D. (2021). High-Fidelity CRISPR/Cas13a trans-cleavage-triggered rolling circle amplified DNAzyme for visual profiling of MicroRNA. Anal. Chem. 93, 2038–2044. doi: 10.1021/acs.analchem.0c03708
Keywords: CRISPR-Cas, CRISPR/Cas13a, biosensor, SHERLOCK, molecular diagnostics
Citation: Zhao L, Qiu M, Li X, Yang J and Li J (2022) CRISPR-Cas13a system: A novel tool for molecular diagnostics. Front. Microbiol. 13:1060947. doi: 10.3389/fmicb.2022.1060947
Received: 04 October 2022; Accepted: 09 November 2022;
Published: 08 December 2022.
Edited by:
Michael Köpke, LanzaTech, United StatesReviewed by:
Maria Tomas, A Coruña University Hospital Complex (CHUAC), SpainCopyright © 2022 Zhao, Qiu, Li, Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jintao Li, bGp0cW1zQHRtbXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.