- 1Winogradsky Institute of Microbiology, Federal Research Centre of Biotechnology, Russian Academy of Sciences, Moscow, Russia
- 2Department of Biotechnology, Delft University of Technology, Delft, Netherlands
- 3Institute of Bioengineering, Federal Research Centre of Biotechnology, Russian Academy of Sciences, Moscow, Russia
- 4Faculty of Biology, Lomonosov Moscow State University, Moscow, Russia
Extremely halophilic archaea (haloarchaea) of the class Halobacteria is a dominant group of aerobic heterotrophic prokaryotic communities in salt-saturated habitats, such as salt lakes and solar salterns. Most of the pure cultures of haloarchaea were enriched, isolated, and cultivated on rich soluble substrates such as amino acids, peptides or simple sugars. So far, the evidences on the capability of haloarchaea to use different polysaccharides as growth substrates remained scarce. However, it is becoming increasingly obvious that these archaea can also actively participate in mineralization of complex biopolymers, in particular cellulose and chitin–two dominant biomass polysaccharides on the planet. Here we used an array of commercially available homo- and heteropolysaccharides to enrich hydrolytic haloarchaea from hypersaline salt lakes with neutral pH and from alkaline soda lakes. This resulted in isolation of a range of halo- and natrono-archaea, respectively, belonging to already described taxa as well as several new genus-level lineages. In some cases, the isolates enriched with different polysaccharides happened to be closely related, thus representing generalistic ecotype, while the others were narrow specialists. In general, soda lakes yielded a broader range of polysaccharide-utilizing specialists in comparison to neutral salt lakes. The results demonstrated a significant diversity of halo(natrono)archaea with a previously unrecognized potential for utilization of a broad range of natural polysaccharides in hypersaline habitats.
Introduction
Hypersaline lakes and solar salterns at its final evaporation stage represent unique salt-saturated habitats dominated by extremely halophilic microbial communities among which the extremely halophilic archaea of the class Halobacteria is the particularly successful group (Cui and Dyall-Smith, 2021). These archaea (at least those known in culture) are mostly aerobic organoheterotrophs, utilizing simple soluble organic compounds, such as amino acids and sugars (Andrei et al., 2012; Oren, 2013, 2015; Grant and Jones, 2016). Haloarchaea typically have very high cell density that gives the characteristic reddish color to hypersaline brines in intracontinental athalassic lakes and thalassic endeavaporite pools of the marine solar salt concentrators. Only handful of cultivated haloarchaeal species can grow with polymeric substances, such as starch, proteins or olive oil (Bhatnagar et al., 2005; Enache and Kamekura, 2010; Moshfegh et al., 2013; Selim et al., 2014; Amoozegar et al., 2017). Recently, this spectrum has been expanded by recalcitrant insoluble polysaccharides, such as cellulose and chitin as well as some other partially soluble polysaccharides, such as galactomannan and xylan. Utilization of native insoluble forms of cellulose has recently been shown for the neutrophilic genera Halococcoides, Halomicrobium, and Halosimplex (Sorokin et al., 2015, 2019a,2020a) and for two genera of natronoarchaea from soda lakes–Natronolimnobius and Natronobiforma (Sorokin et al., 2015, 2018, 2019b). The ability to use chitin as the growth substrate has been proven for the neutrophilic genera Halomicrobium and Salinarchaeum and for the natronoarchael genus Natrarchaeobius (Sorokin et al., 2015, 2019c,2020b; Minegishi et al., 2017). Finally, growth with locust bean galacto-beta-1,4-mannan was shown for the neutrophilic genera Natronoarchaeum and Haloarcula (Shimane et al., 2010; Enomoto et al., 2020).
The potential of haloarchaea to utilize various recalcitrant polysaccharides produced mostly by plants and algae is of significant interest both for fundamental understanding of their functional importance for the organic matter mineralization in hypersaline environments and also by regarding them as a source of extremely halo(alkali)stable extracellular hydrolases which have important application potential in production of biofuel from lignocellulosic wastes because this process often starts with a decrystallization pretreatment step, performed either with alkali or ionic liquids (Kaar and Holtzapple, 2000; Zavrel et al., 2010; Begemann et al., 2011).
In this work, the search for polysaccharide-utilizing haloarchaea was extended beyond the most abundant cellulose and chitin. For this, a range of commercially available polysaccharides of plant and microbial origin was used for selective enrichment and further isolation in pure cultures of halo(natrono)archaea able to utilize these polymers as growth substrate. The de novo sequenced genomes of these strains allowed to establish their phylogenies as well as to detect the genes, encoding enzymes responsible for their polysacharidolytic capacities. The results demonstrated significant diversity of polysaccharide-specialized haloarchaea belonging to already described genera and species (mostly for salt lakes) and several new genera (mostly among natronoarchaea), all of which have enzymatic repertoire sufficient for decomposition of the respective polysaccharides.
Experimental procedures
Samples
Sediment (top 3 cm) and brine samples were obtained from five hypersaline chloride-sulfate lakes with neutral pH in Kulunda Steppe (Altai, Russia) and from hypersaline alkaline (soda) lakes in Kulunda Steppe (three lakes), northeastern Mongolia (two lakes) and North America (California, two lakes) (Sorokin et al., 2015). Two “master mixes,” one for each type of lakes, were created by mixing equal parts of sediments and brines from each lake and used at 5% (v/v) for primary enrichments.
Enrichment and growth conditions
The neutrophilic haloarchaea originated from salt lakes were enriched, purified and further cultivated in a neutral base medium 1 with the following composition (g l–1): 230 NaCl, 5 KCl, 0.2 NH4Cl, 2.5 K2HPO4, pH 6.8. After sterilization, the base was supplemented with vitamin and trace metal mix (Pfennig and Lippert, 1966) (1 ml l–1 each) and 2 mM MgSO4. For the soda lake enrichments and further cultivation of alkaliphilic natronoarchaea, a sodium carbonate/bicarbonate-based medium 2 containing 4 M total Na+ [(g l–1): 190 Na2CO3, 30 NaHCO3, 16 NaCl, 5 KCl and 1 K2HPO4, final pH 10 after sterilization] was supplemented with the same additions as for the medium 1, except that the amount of Mg was two times lower and that 4 mM NH4Cl was added after sterilization. This alkaline medium was mixed 1:3 with the neutral medium 1, resulting in the final pH of 9.6.
Polysaccharides (Sigma-Aldrich and Megazyme; Supplementary Table 1) were either added from suspensions in sterile distilled water (when heat sterilization was not possible) or from 5% heat-sterilized (110°C for 20 min) stocks to a final concentration of 0.5 g l–1. At the stage of initial enrichments and further 1:100 transfers, a mixture of streptomycin and kanamycin (final concentration 100 mg l–1) was added to suppress bacterial development. Cultivation was performed in 30 ml bottles sealed with gray-rubber septa (to prevent evaporation) containing 10 ml medium at 35°C on a rotary shaker at 150 rpm. Solid media were prepared by 3:2 (v:v) mixing of the fully prepared liquid media with 4% washed agar at 50°C. Solid NaCl was added to the portions of liquid media before heating to bring the salinity back to 4 M of total Na+ after agar addition. For isolation of pure cultures, the initial positive enrichments were passed 2 times into new media containing target polysaccharides at 1:100 dilution to obtain sediment-free cultures, followed with dilution to extinction series and finally plating the maximal positive dilutions onto solid media with the same composition. The dominant colony types or those showed visible signs of polymer degradation (where possible) were picked up with sterile Pasteur capillary with pooled tips under control of binocular and placed into liquid media. Only those cultures which showed vigorous growth in liquid media were further purified by repeating the colony formation procedure. The purity of isolates were confirmed by microscopy, 16S-rRNA gene sequencing (Sanger and amplicon profiling) and in several cases by full genome sequencing.
Polysaccharide utilization activity
The main indication of polysaccharide utilization was consistent microbial growth in liquid culture whereby the polysaccharide in question served as the sole carbon and energy source. In addition, whenever it was possible, the hydrolytic activities of spot-colonies were also visualized on agar plates, either by formation of clearance zones (amorphous cellulose, beta-mannan) or reagent-developed hydrolysis zone: Lugol solution for starch, pullulan and pectin and Congo Red/1 M NaCl for xylan, xyloglucan, arabonoxylan, and glucomannan.
Genomic sequencing and phylogenetic analysis
Genomic DNA isolation, DNA library preparation, sequencing as well as genome assembly were performed as described earlier (Sorokin et al., 2022a).
Phylogenomic analysis based on the “ar122” set of conserved single copy archaeal proteins (Rinke et al., 2021) was performed as follows: the protein sequences were identified and aligned in in silico proteomes of the type species of all genera within Halobacteria class (non-type species were taken for Halalkalicoccus and Natronoarchaeum genera because the genomes of the type species were not available) using the GTDB-tk v.1.7.0 with reference data v.202 (Chaumeil et al., 2019). The maximum likelihood tree was inferred using the RAxML v.8.2.12 (Stamatakis, 2014) with the PROTGAMMAILG model of amino acid substitution; support values were calculated from the 1000 rapid bootstrap replications. The phylogenetic tree was polished using iTOL v.6.5.8 (Letunic and Bork, 2019).
Genome analysis
The genomes were annotated with NCBI Prokaryotic Genome Annotation Pipeline (Tatusova et al., 2016). Carbohydrate-active enzymes (CAZymes) including glycosidases, polysaccharide lyases, carbohydrate esterases, glycosyl transferases, and carbohydrate oxidases genes were predicted using dbCAN2 script v2.0.11 (Zhang et al., 2018) with HMMER v3.3 (Mistry et al., 2013) with default thresholds. The most probable activities of identified CAZymes (excluding glycosyl transferases, for which the manual verification of the predictions was not performed) were predicted using BLAST search against Swiss-Prot database (Boutet et al., 2016).
Genbank accession numbers
The 16S rRNA gene sequences generated in this study were deposited in the GenBank under accession numbers ON787970-ON788000. The whole genome sequence are available in GenBank under the following accession numbers: JAOPJY000000000, JAOPJZ000000000, JAOPKA000000000, JAOPKB000000000, JAOPKC000000000, JAOPKD000000000, and JAOPKE000000000.
Results
Polysaccharide-utilizing haloarchaea from hypersaline lakes with neutral pH
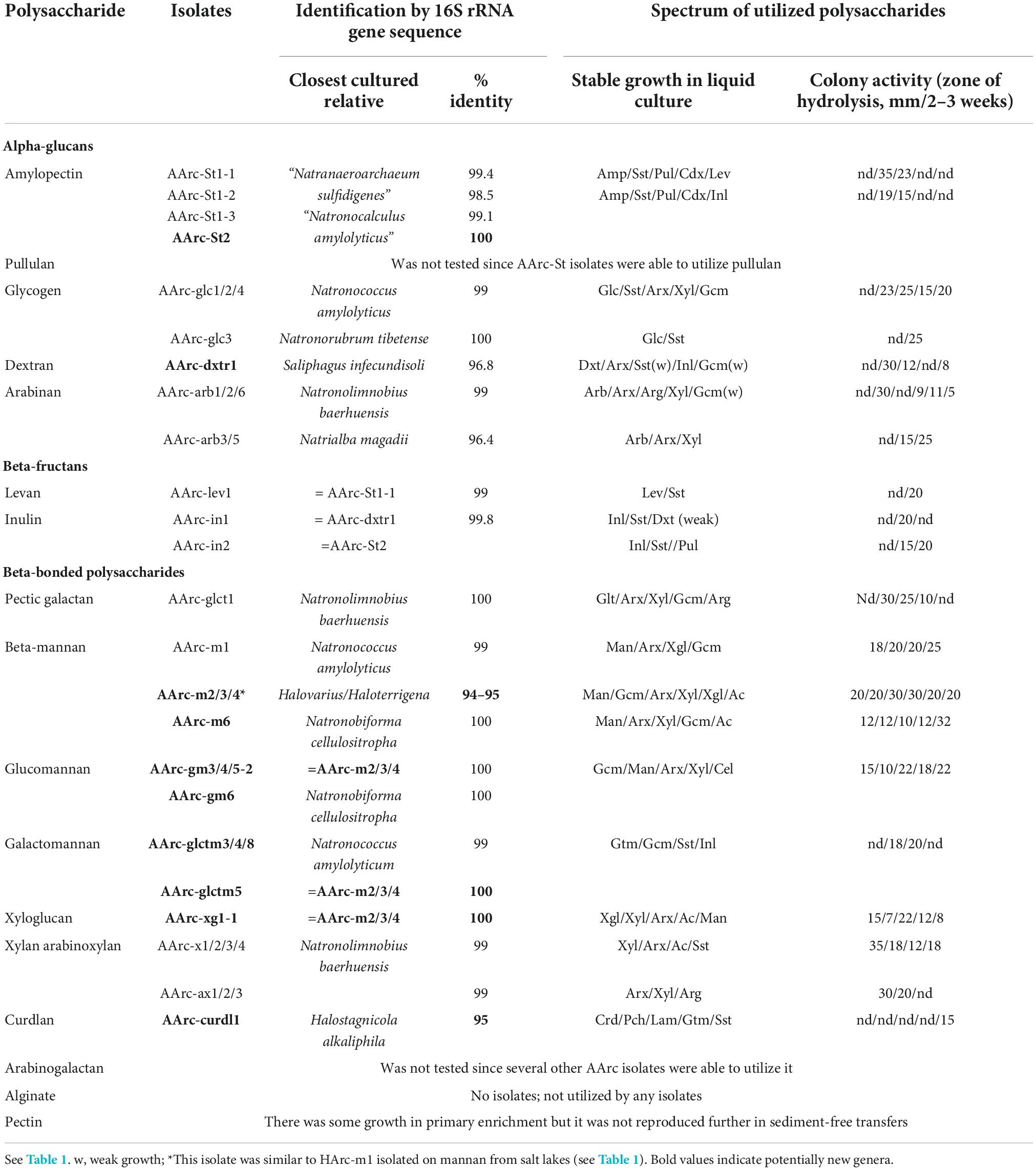
Positive enrichment cultures from salt lakes were obtained with 13 out of 18 polysaccharide compounds tested. The fastest development (maximal growth yield was achieved after 1 week) was observed with starch-like compounds (amylopectin, pullulan, and glycogen), while the slowest (up to a month)–with insoluble beta-linked polysaccharides (beta-mannan and curdlan). Other positive cultures showed growth in between 2 and 3 weeks. All positive primary enrichments were transferred into a sediment-free stage after 2–3 consecutive 1:100 (v:v) transfers on the same synthetic medium and acquired pink coloration with a domination of polymorphic flat cells characteristic of haloarchaea accompanied by visible degradation of substrates in case of insoluble polysaccharides. Further dilutions to extinction were performed without antibiotics and were generally positive up to 10–8–10–9. Final purification was achieved by isolation of individual colonies on solid media. Pure cultures of polysacharidolytic haloarchaea were obtained from those colony morphotype(s) (not always dominant ones) which consistently grew back in liquid medium with the target polysaccharide used in the enrichment. The list of isolates is given in Table 1.
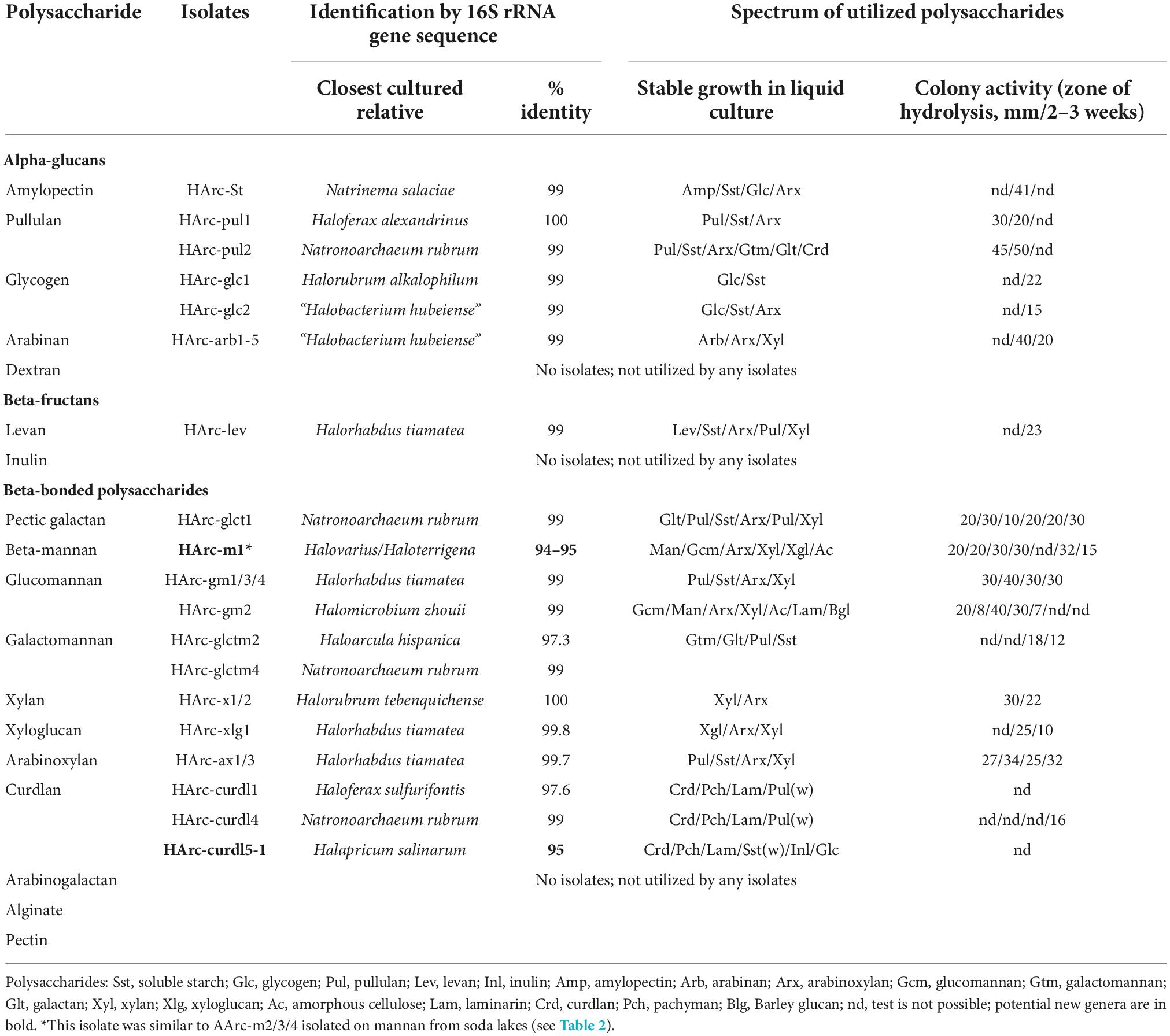
Table 1. Polysaccharide-utilizing neutrophilic haloarchaea enriched and isolated from hypersaline salt lakes with neutral pH.
All isolates selected on alpha-bonded polysaccharides belonged to the well-characterized genera of haloarchaea, for some of which utilization of starch is known. However, to our knowledge, the capacity to utilize arabinan and arabinoxylan has never been shown/tested for any cultivated species of haloarchaea. This is also true for Halorhabdus tiamateia, a broadly-specialized polysaccharidolytic haloarchaeon, according to our current results (Table 1) and previous studies (Wainø and Ingvorsen, 2003; Werner et al., 2014). However, its ability to grow on levan (beta-fructan) was not known before. Isolates belonging to the genus Natronoarchaeum were second after Halorhabdus by the number of isolated strains and were selected with four polysaccharides, including pullulan, beta-galactan, galactomannan, and curdlan. While utilization of galactomannan has already been reported for N. mannanilyticum (Shimane et al., 2010), growth with beta-1,4-galactan and beta-1,3-glucans, such as curdlan, have never been observed in any pure cultures of haloarchaea. Same is true for other two beta-1,4-bonded polysaccharides, including xyloglucan and beta-mannan. Looking from the taxonomical perspective, only two insoluble polysaccharides, mannan (consisted of beta-1,4-linked mannose) and curdlan (consisted of beta-1,3-linked glucose) (Table 1) resulted in selection of new genus-level isolates from the neutral salt lakes.
The cross-specificity for various polysaccharides supporting growth of the neutrophilic haloarchaea selected with a single polymer is shown in Figure 1. All five species enriched on alpha-glucans utilized soluble starch and five out of the six alpha-glucan specialists also grew with arabinoxylan which has alpha-bonded arabinose residues in the side chains. In turn, a single case of cross-specificity was also observed in Halobacterium strains which were selected with both glycogen (HArc-glc2) and arabinan (HArc-arb1-5) (Figure 1A).
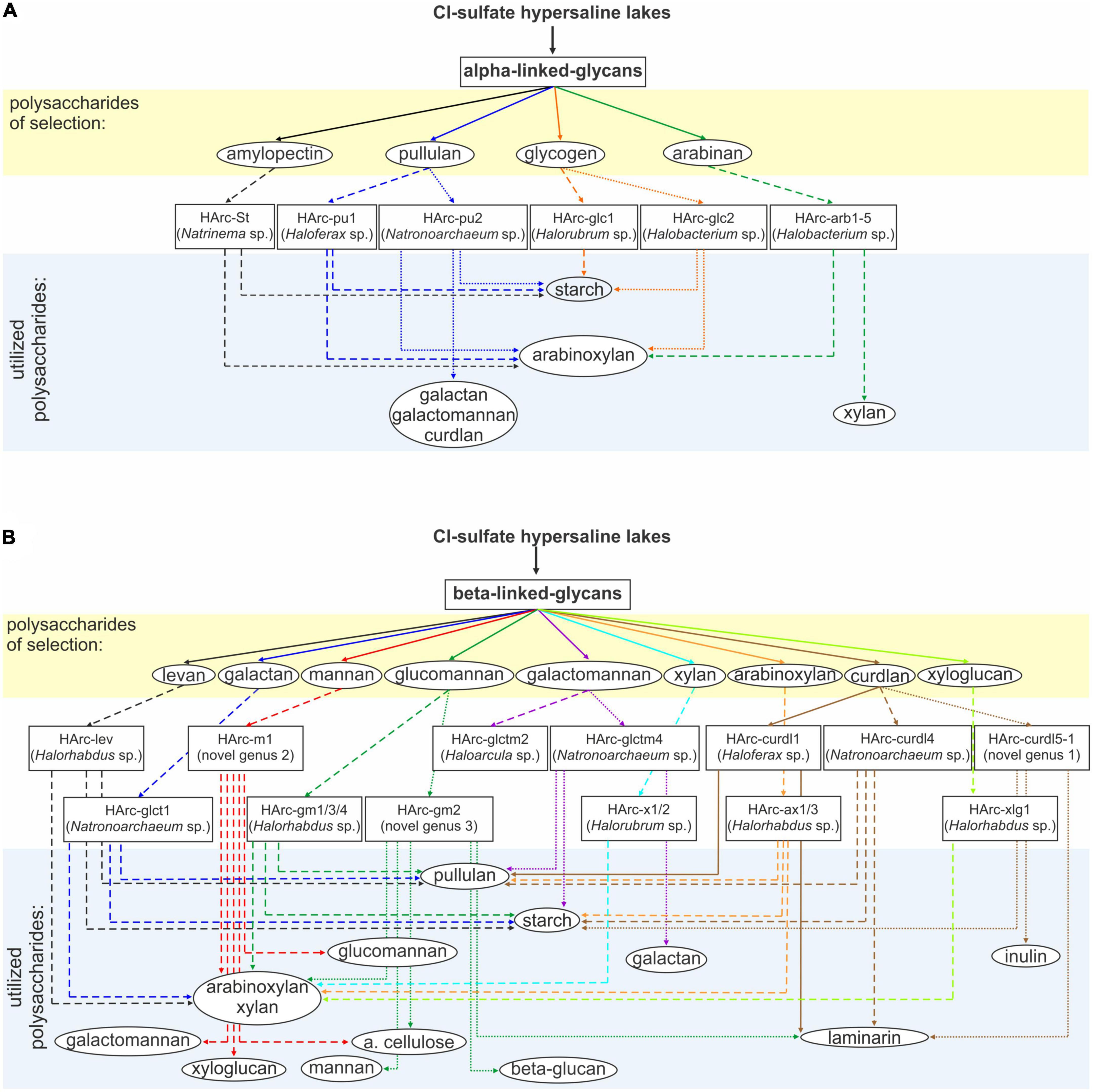
Figure 1. Schematic representation of selective enrichments of haloarchaea from hypersaline salt lakes on (A) alpha-bonded and (B) beta-bonded polysaccharides. Assignment to the novel genus was based on protein sequence-based phylogenomic analysis and 16S rRNA gene sequence identity values.
Among the haloarchaeal strains selected with various beta-bonded polysaccharides the most common cross-substrates were xylan and arabinoxylan, while alpha-glucans (starch and pullulan) were only utilized by a few beta-glucan specialists (Figure 1B). Two out of the three isolates enriched with either mannan (HArc-m1) or glucomannan (HArc-gm2) were able to grow with amorphous cellulose indicating related selectivity of these beta-1,4 backbone polysaccharides. The Halomicrobium strain HArc-gm2 selected with glucomannan was identical in its 16S rRNA gene sequence to Halomicrobium sp. HArcel3–the cellulose-enriched haloarchaeon most closely related to H. zhouii (Sorokin et al., 2015). We tested the type strain H. zhouii JCM 17095 and it appeared to be able to grow with amorphous cellulose as well.
Strain HArc-m1 was identical (according to the 16S rRNA gene sequence analysis) to several natronoarchaeal isolates enriched from soda lakes with beta-1,4 mannan backbone polysaccharides (see below) confirming a link between the cellulose and the beta-mannan selectivity. This pattern has already been observed in cellulotrophic Natronobiforma cellulositropha which was enriched on cellulose but can also grow with beta-mannan (Sorokin et al., 2018). Furthermore, HArc-m1 is the only strain enriched from the neutral salt lakes being closely related to isolates from soda lakes representing quite a rare example in our long-term work with hypersaline lakes. It is also worth to notice that, in this particular case, the substrate selectivity (mannan) overruled the dominant selective factor – the nature of sodium salt (chloride vs. carbonate) and, therefore, the considerable difference in the pH-osmotic pressure combination. Growth experiments confirmed that strain HArc-m1 can indeed grow both at neutral pH and up to pH 9.5, thus being a facultative alkaliphile.
Polysaccharide-utilizing natronoarchaea from hypersaline soda lakes
Positive stable enrichment cultures from soda lakes were obtained with 14 out of 16 polysaccharides tested (pullulan and arabinogalactan were not tested). Similar for salt lakes, alginate and pectin enrichments were negative. Another similarity was the fastest growth (1 week) of the starch-like alpha-glucans (amylopectin and glycogen) and the slowest growth (up to a month) of the insoluble beta-linked-glycans (beta-mannan and curdlan) and α-1,6-glucan dextran (which was negative in case of salt lake samples) utilizing enrichment cultures. However, in contrast to salt lakes further dilution to extinction from the soda lake enrichments had still to be done in the presence of antibiotics since in their absence the cultures were rapidly overrun by bacteria. It was even necessary to add antibiotics to the solid media at the final stage of pure culture isolation. The contaminating bacteria mostly belonged to the genus Halomonas which were unable to grow on the target polysaccharide but most probably scavenged the hydrolysis products. The Halomonas colonies were easily distinguished from the pink-orange colonies of natronoarchaea which helped to purify the latter, although, in most cases only 1-2 types of such colonies grew back in the liquid medium with the target polysaccharide. The list of isolated polysaccharide-utilizing natronoarchaea is given in Table 2.
Four different alpha-bonded glucans and fructans resulted in selection of seven isolates, of which, all except two belonged to know genera. Two isolates enriched and isolated either on amylopectin or inulin were identical in their 16S rRNA gene sequences and represented a novel genus and species Natronocalculus amylovorans (Sorokin et al., 2022a). The other three amylopectin-utilizing isolates were closely related to each other and to a facultively anaerobic sulfur-respiring amylolytic Natranaeroarchaeum sulfidigenes (Sorokin et al., 2022b). Together with the isolate AArc-lev selected with beta-fructan–levan (Table 2) those four strains have recently been described as a new species Natranaeroarchaeum aerophilus (Sorokin et al., 2022b). So, there seems to be a connection in starch-like alpha-glucans and beta-fructans selectivity among natronoarchaea.
Interestingly, although glycogen is structurally similar to amylopectin (both are branched alpha-glucans), the two glycogen-selected natronoarchaeal isolates from the genera Natronococcus and Natronorubrum were not related to above-mentioned new taxa. But all four were able to grow with soluble starch and pullulan, similar to the members of the genus Natronococcus which are well-known for their ability to utilize starch and to produce alkalistable amylases (Kobayashi et al., 1992; Kanal et al., 1995).
To our knowledge, dextrans have never been shown or even suspected to support growth of any known Halobacteria species, while it was mentioned among positive substrates in anaerobic hyperthermophilic archaea Desulfurcococcus fermentas (Perevalova et al., 2005), D. kamchatkensis (Kublanov et al., 2009), Thermoccoccus sibiricus (Mardanov et al., 2009), and Thermococcus sp. strain 2319 × 1 (Gavrilov et al., 2016). While starch-like polysaccharides can have side branches with alpha-bonded glucose other than α-1,4, none but dextrans have the α-1,6 backbone, which probably makes them difficult substrates for hydrolytic archaea. From two forms of the cyanobacterial dextran tested in this work (19.5 and 200 kDa), only the low molecular weight variety resulted in a positive enrichment and isolation of a single natronoarchaeal strain AArc-dxtr1 representing a distant novel species in the genus Saliphagus (Table 2).
Finally, an alpha-1,5-arabinan enrichment from soda lakes yielded a stable binary culture impossible to separate by serial dilutions. Plating showed two distinctive types of colonies: a dominant type with small red colonies and less abundant larger and nearly colorless colonies. Both grew back in liquid pure cultures with arabinan. Interestingly, the arabinan utilization in the liquid culture inoculated with the colorless colonies resulted in a formation of soluble yellow-brownish product, while the red colonies culture supernatant remained colorless. The isolate AArc-arb3/5 with colorless colonies was identified as a novel Natrialba species (with the highest 16S rRNA sequence identity of 97% to “N. wudunaoensis”), while the second isolate AArc-arb1/2/6 with red colonies was closely related to the known species Natronolimnobius baerhuensis.
The natronoarchaea selected from soda lakes on various beta-bonded polysaccharides can be divided into two major groups: preferably xylanolytic and cellulo-/mannanolytic (Table 2). The xylanolytics selected on either xylan, arabinoxylan and galactan belonged to the known genus Natronolimnobius. Xyloglucan, galactomannan, and mannan enrichments were all dominated by a novel genus-level lineage to which a facultatively alkaliphilic strain HArc-m1 (see above; Table 2) also belonged. Furthermore, a less abundant component in the galactomannan enrichment was identified as a member of the genus Natronococcus. A dominant organism in a glucomannan enrichment was identical to the cellulose/mannan-specialized Natronobiforma cellulositropha (Sorokin et al., 2018). This is similar to the selectivity of glucomannan in salt lakes resulted in isolation of a cellulolytic haloarchaeon HArc-gm2 closely related to cellulotrophic Halomicrobium HArcel3 dominating in cellulose enrichments from hypersaline lakes (Sorokin et al., 2015).
Curdlan, a beta-1,3-glucan homopolysacharide, selected a single natronoarchael strain representing a new genus lineage of the polysaccharide-utilizing archaea. The enrichment culture was very slow in development resulting in degradation of antibiotics and massive development of bacteria belonging to Halomonas. Several repeated attempts with sequential addition of antibiotics resulted in the sufficient enrichment of the archaeal component appropriate for further purification on a solid medium. Despite its general chemical similarity to cellulose, curdlan molecules have a different physical structure (helical in contrast to flat ribbon cellulose fibrils) (Deslandes et al., 1980). Altogether, its structural characteristics, both primary and secondary, as well as low occurrence in nature makes it highly selective substrate in comparison with the more common beta-1,4 glucans. We did not manage to find any published data on curdlan utilization in haloarchaea. On the other hand, two out of ten CAZymes families (GH81 and GH16) including members with the endo-β-1,3-glucanase activities (EC 3.2.1.39) have archaeal representatives. While archaeal representatives of the GH81 family are known exclusively by the presence of the respective genes in their genomes, a single archaeal GH16 glycosidase from Pyrococcus furiosus (Gueguen et al., 1997) was characterized as a laminarinase able to hydrolyze laminarin, lichenan, and barley β-glucan. However, no information of its capability to hydrolyze curdlan was provided. It should be noted that the halo- and natronoarchaea enriched and isolated on curdlan in the course of this work were capable to grow on laminarin, a soluble beta-1,3/1,6 glucan.
The growth cross-specificity for various polysaccharides among the natronoarchaeal isolates is shown in Figure 2. From the strains isolated on alpha-bonded polysaccharides, the amylolytics were most restricted in their polymer-utilizing profiles with only beta-fructans as the alternative substrates. The only exception was Natronococcus AArc-glc1/2/4, isolated on glycogen and able to grow with a few beta-1,4 bonded polysaccharides. On the other hand, strains selected with other alpha-bonded polysaccharides, such as dextran and arabinan, were all able to utilize xylan and arabinoxylan and one of them also grew with arabinogalactan (Figure 2A).
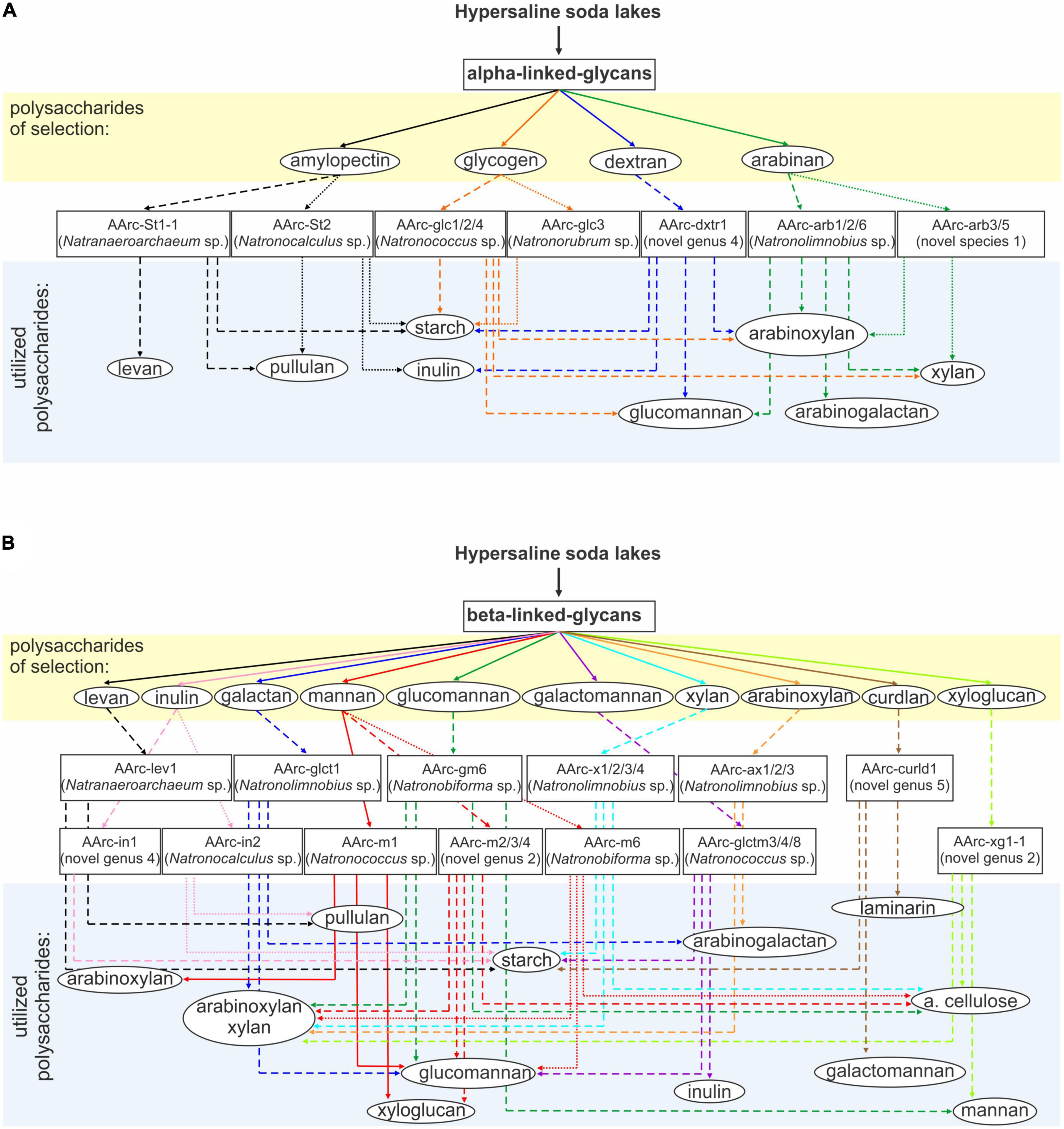
Figure 2. Schematic representation of selective enrichments of natronoarchaea from hypersaline soda lakes on (A) alpha-bonded and (B) beta-bonded polysaccharides. Assignment to the novel genus was based on protein sequence-based phylogenomic analysis and 16S rRNA gene sequence identity values (for strain AArc-arb3/5 only 16S rRNA gene sequence identity values were used).
Among the strains selected with beta-bonded polysaccharides, the most narrowly specialized was strain AArc-curdl1 isolated on curdlan: it only grew with two other polysaccharides with the beta-1,3-backbone (pachyman and laminarin) and on starch (Figure 2B). In contrast, the natronoarchaea enriched with various beta-1,4-bonded polysaccharides had a broader substrate range with xylan and arabonoxylan being the most common cross-substrates among them. Similar to the salt lake isolates, the soda lake strains selected with beta-1,4 mannan and glucomannan were also capable of growth on native celluloses. On the other hand, in contrast to salt lakes, the same taxa were also selected on galactomannan. This difference is significant, taking into account importance of cellulose for natural habitats but the reason for this is not clear yet.
Phylogenomic and functional genomic analysis
Genomes of seven stains were de novo sequenced and assembled. Quality check of the assemblies revealed high completeness (99–100%) and low contamination (0-1.87%) levels what makes them suitable for both phylogenomic and functional analyses (Supplementary Table 2). Genome sizes varied from 2.81 to 5.59 Mbp while the G + C contents for different genomes were 59.2–66%.
Phylogenomic analysis showed that the novel polysaccharidolytic strains are uniformly dispersed within the Halobacteria tree (Figure 3). Strain AArc-St1-1 belonged to the genus Natranaeroarchaeum and is recently described as a new species N. aerophilus (Sorokin et al., 2022b), while strain AArc-St2 is described as a novel genus and species Natronocalculus amylovorans (Sorokin et al., 2022a). Strain HArc-gm2 was most closely related to the members of the genus Halosiccatus. The neutrophilic curdlan-utilizing haloarchaea strains HArc-curdl5-1 and HArc-curdl7 are most closely related to the genus Halapricum. The nearest relatives of AArc-dxtr1 are among the Halostagnicola species, while AArc-xg1-1, AArc-m2/3/4 and AArc-cudrl1 are related to the cellulolytic Natronobiforma. Establishing the exact taxonomic rank (novel species or genus) of these novel haloa(natrono)archaea will need a more in-depth phenotypic and chemotaxonomical characterization.
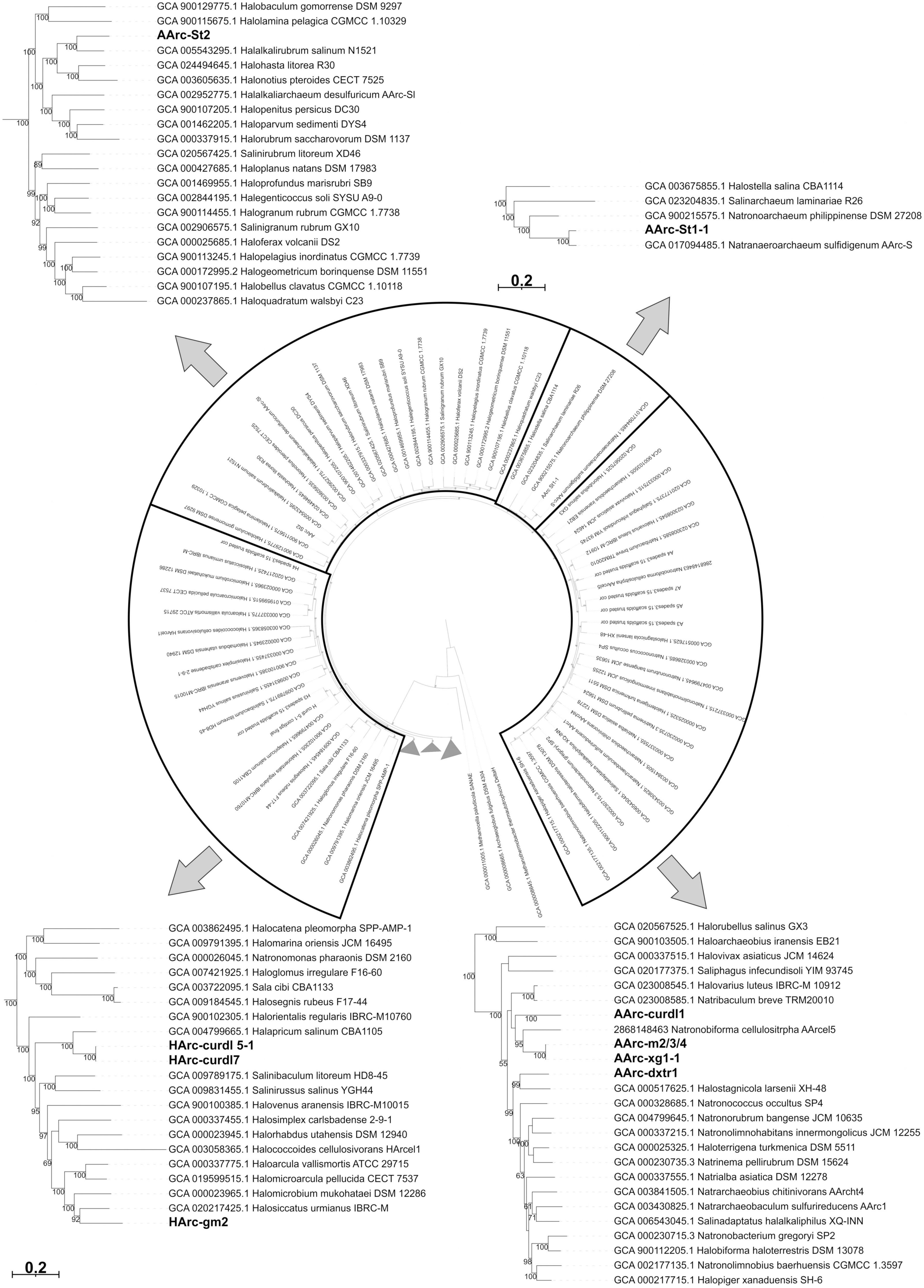
Figure 3. Maximum likelihood phylogenomic tree showing position of polysaccharide-utilizing halo(natrono)archaea enriched from hypersaline lakes within the class Halobacteria. Sequences of 122 conserved archaeal proteins were used to infer the tree.
Polysaccharide-utilizing haloarchaea must have a set of carbohydrate active enzymes (CAZymes), as a prerequisite for successful decomposition of insoluble and soluble poly- and oligosaccharides. Indeed, the sequenced genomes encoded all types of the CAZymes: glycosidases (GH), polysaccharide lyases (PL), glycosyl transferases (GT), carbohydrate esterases (CE), carbohydrate oxidases (AA) as well as carbohydrate-binding modules (CBM). The detailed analysis was focused on GHs and PLs (Figure 4; Supplementary Table 3) due to their major role in polysaccharide depolymerization.
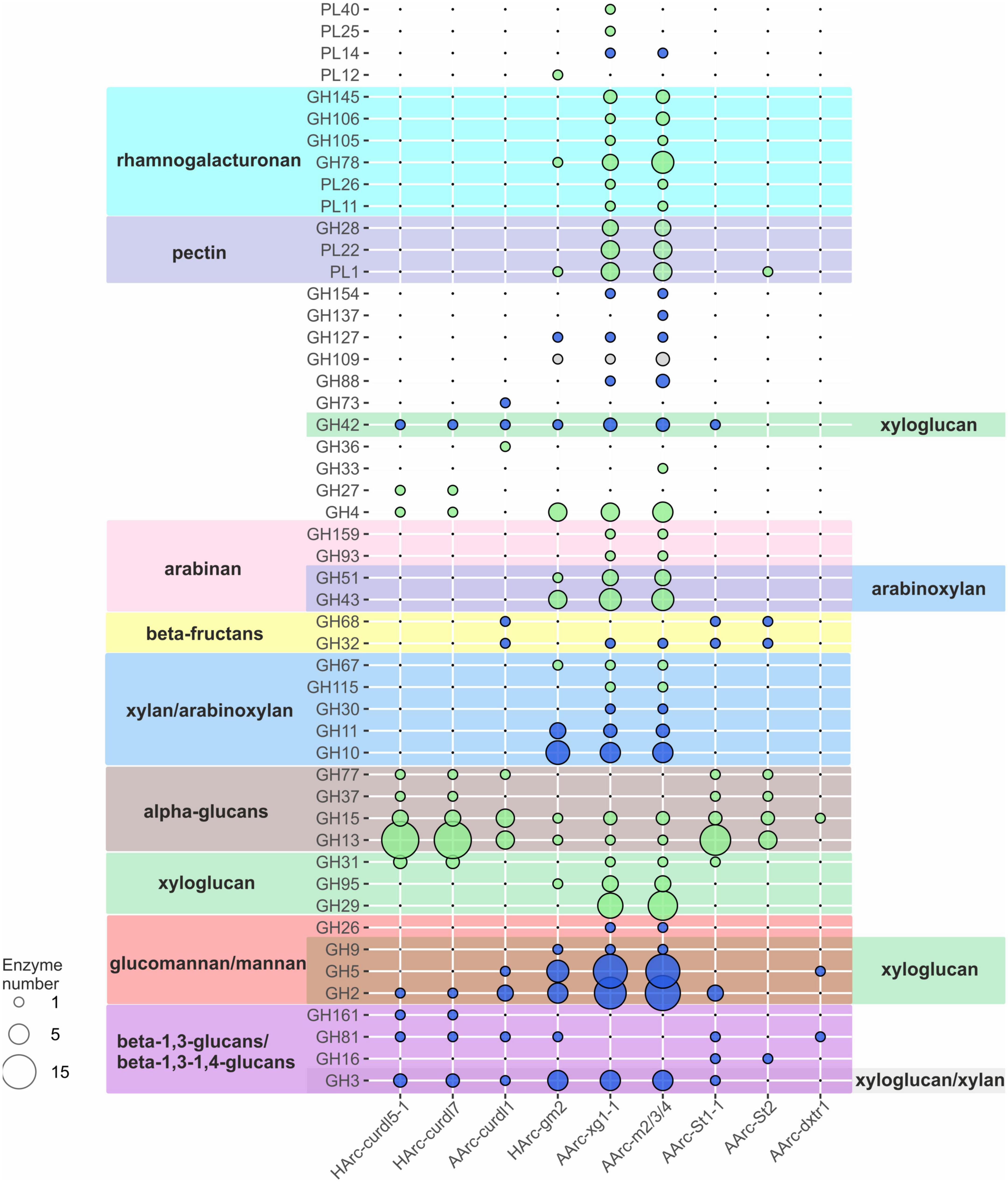
Figure 4. Sets of GH and PL genes found in the genomes of novel polysaccharidolytic haloarchaea. Size of bubbles indicate number of enzymes; the color of bubbles indicate the prevailing type of glycosidic bonds hydrolyzed by the enzymes (alpha-bonds—green, beta-bonds—blue, both—gray). Color of background indicates these CAZymes families contains the characterized enzyme(s) capable of depolymerization of respective polysaccharides. No color means the activities, characteristic to these families are not directly linked to the substrates used in this work.
Closely related neutrophilic strains HArc-curld5-1 and HArc-curdl7 enriched on curdlan had identical CAZyme repertoires. Their capability to degrade curdlan as well as pachyman and laminarin is due to the action of endo-beta-1,3(4)-glucanase (GH81), beta-1,3-glucan phosphorylase (GH161) and beta-glucosidase (GH3). Both strains also grew on starch by means of fourteen alpha-amylases (GH13), three oligo-1,6-glucosidases (GH13), two glucoamylases (GH15), and a 4-alphaglucanotransferase encoded in their genomes. The genome of alkaliphilic strain AArc-curld1 also isolated on curdlan had a smaller set of genes for respective enzymes yet it included an essential endo-beta-1,3(4)-glucanase (GH81) and a beta-glucosidase (GH3). A neopullulanase (GH13), three alpha-amylases (GH13), a 4-alphaglucanotransferase (GH77), and two glucoamylases (GH15) apparently allowed the strain to utilize soluble starch.
The neutrophilic strain HArcl-gm2 isolated on glucomannan has a machinery for its decomposition including endo-beta-1,4-mannosidase (GH5), several endoglucanases (four from the GH5 and one from the GH9 families), two beta-mannosidases (GH2) and a beta-glucosidase (GH3). The strain also can utilize laminarin and beta-glucan due to the presence of the endo-beta-1,3(4)-glucanase (GH81); xylan and arabinoxylan by means of endoxylanases (seven enzymes from GH10 and three from GH11), beta-xylosidases (GH3), and arabinosidases (GH43 and GH51) responsible for hydrolysis of side chains of arabinoxylan.
Comparative genomic analysis of closely related natronarchaeal strains AArc-xg1-1 (isolated on xyloglucan) and AArc-m2/3/4 (isolated on beta-1,4-mannan) showed a nearly identical and the largest CAZyme sets among the studied haloarchaea. In particular, the genes encoding several endo-beta-1,4-mannosidases (five enzymes from GH5 family and one from GH26), two beta-mannosidases (GH2), endoglucanases (10 proteins from GH5 and one enzyme from GH9), three beta-glucosidases (GH3 family), endo-beta-1,4-xylanases (5 enzymes from GH10 and two enzymes from GH11), beta-xylosidase (GH3) were found. Moreover, the genes of enzymes known to degrade galacturonate-containing polymers (polygalacturonate, pectin, rhamnogalacturonan) were also detected: three polygalacturonases (GH28), putative pectate lyases (four proteins from PL1 family and four from PL22), two rhamnogalacturonan lyases (PL11, PL26) as well as arabinan-hydrolyzing enzymes (six proteins from GH43, three GH51 enzymes and one enzyme from GH93). However, none of these isolates grew with either pectins, polygalacturonate, rhamnogalacturonan, or arabinan. This example clearly demonstrates that functional conclusions based solely on the genomic evidence should be considered only as preliminary.
The genome of the natronarchaeal amylolytic strain AArc-St1-1 (Natranaeroarcheum aerophilus, Sorokin et al., 2022a) contains a large number of genes coding for alpha-bond degrading glycosidases, including ten alpha-amylases (GH13 family), two oligo-1,6-glucosidases (GH13), 4-alpha-glucanotransferase (GH77), glucoamylase (GH15), and trehalase (GH37). Finally, two beta-fructosidases (GH32 and GH68) were also found, apparently allowing the strain to grow on levan. Also, two beta-1,3(4)-glucanases (GH16, GH81), beta-glucosidase/beta-xylosidase (GH3), beta-mannosidase (GH2), alpha-xylosidase (GH31), and several beta-galactosidases (GH2 and GH42 families) genes were present. Another amylolytic strain AArc-St2 (Natronocalculus amylovorans) possessed similar CAZymes set (alpha-glucan-specific enzymes from GH13, GH15 and GH77 families as well beta-fructosidases) but the number of alpha-amylases and oligo-1,6-glucosidases was significantly lower (Sorokin et al., 2020a). Despite the presence of genes for putative GH16 beta-glucanase and PL1 pectate lyase, no growth was observed on their specific substrates including beta-glucan, lichenan, curdlan, pachyman, or pectin.
Only three GH genes and no PL genes were found in the genome of dextran-utilizing natronoarchaeon AArc-dxtr1: trehalase (GH15), endoglucanase (GH5), and beta-1,3(4)-glucanase (GH81). The typical dextran-degrading enzymes from GH13, GH49, GH66, or GH70 families a were not encoded and it might be only speculated that dextran is hydrolyzed as a result of side activity of a GH15 enzyme since this family is known to contain glucodextranases (Mizuno et al., 2004). An endoglucanase and a beta-1,3(4)-glucanase could be responsible for hydrolysis of glucomannan and arabinoxylan which this organism can also utilize.
Altogether, the repertoire of CAZymes within the in silico proteomes of the nine polysacharidolytic halo/natronoarchaea analyzed in this work (summarized on Figure 4) was mostly consistent with their polysaccharide untilization spectrum. In total, the number of CAZyme genes found in the studied HArcel/AArcel strains greatly varied from 30 (strain AArc-St2) to 160 (strain AArc-m2/3/4). Closely related strains isolated with the same (HArc-curld5-1 and HArc-curdl7) or different (AArc-xg1-1 and AArc-m2/3/4) substrates may have identical or similar CAZymes gene sets indicating that a selection substrate will not necessarily lead to isolation of a distinctive phylogenetic lineage. At the same time, despite that the phylogenetically distant strains enriched and isolated with the same polysaccharide have different CAZymes sets, they all had similar patterns of particular CAZymes responsible for the hydrolysis of selective substrate. Surprisingly, the genomes of several isolates including AArc-xg1-1, AArc-m2/3/4, AArc-St2, and HArc-gm2 also contained polysaccharide lyases genes which are nearly unknown within the Archaeal kingdom. However, the growth experiments revealed that none of those strains can grow on alginate, pectin, polygalacturonate or rhamnogalacturonan indicating that these enzymes might have an unknown activity in halophilic archaea. This is also substantiated by the fact that all our attempts to enrich pectin- or alginate-utilizing haloarchaea failed so far. Among the possible reasons might be that these uronic acids-based polysaccharides are hardly present in hypersaline habitats or that hypersalinity changes the chemical properties and enzyme accessibility of the polymers. On the other hand, the high diversity of hydrolytic haloarchaea with the potential to utilize various polysacchirides with different types of glycosidic bonds indicate that such substrates might be available in hypersaline habitats. One of the most probable source of these polymers are external terrestrial plants growing in the area surrounding hypersaline lakes.
Conclusion
The obtained results allow to significantly extend the knowledge on polysaccharide-utilizing capabilities of haloarchaea and archaea in general. Selective enrichment approach leaded to recover the so called “best-fit” organisms specialized on a narrow-specialized conversion of a particular substrate. In case of polysaccharides, however, most of the haloarchaeal isolates enriched with a certain substrate were still able to utilize several other polymers. It was also found that polymers with the alpha-1,4 or beta-1,4 linkage backbones more often resulted in positive enrichments than with other types of linkage, such as the alpha-1,6- or beta-1,3 bonding. Finally, no haloarchaea, growing on uronic acid-based polysaccharides (pectin and alginate), commonly utilized by bacteria, were isolated.
In the course of this work, the first haloarchaea able to grow on such recalcitrant polysaccharides as dextran, curdlan, xyloglucan, and beta-mannan were isolated. The enlarged variety of polysaccharidolytic halo(natrono)archaea recovered from hypersaline lakes with novel substrate utilization specificities is offering a good opportunity for further studies of their extremely halo(alkali)stable hydrolases, both in fundamental enzymology research and prospective application.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
DS and TVKh were responsible for microbiology work. AE and IK analyzed the genomes and run phylogenetic analysis. TVKo was responsible for 16S rRNA gene sequencing and identification of the isolates. DS, AE, and IK wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Russian Science Foundation (grant 20-14-00250) and by the Russian Ministry of Science and Higher Education (field and primary enrichment work).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1059347/full#supplementary-material
References
Amoozegar, M. A., Siroosi, M., Atashgahi, S., Smidt, H., and Ventosa, A. (2017). Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiology 163, 623–645. doi: 10.1099/mic.0.000463
Andrei, A. Ş, Banciu, H. L., and Oren, A. (2012). Living with salt: Metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol. Lett. 330, 1–9. doi: 10.1111/j.1574-6968.2012.02526.x
Begemann, M. B., Mormile, M. R., Paul, V. G., and Vidt, D. J. (2011). “Potential enhancement of biofuel production through enzymatic biomass degradation activity and biodiesel production by halophilic microorganisms,” in Halophiles and hypersaline environments: Current research and future trends, eds A. Ventosa, A. Oren, and Y. Ma (Heidelberg: Springer), 341–357.
Bhatnagar, T., Boutaiba, S., Hacene, H., Cayol, J.-L., Fardeau, M.-L., Ollivier, B., et al. (2005). Lipolytic activity from halobacteria: Screening and hydrolase production. FEMS Microbiol. Lett. 248, 133–140.
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the manually annotated section of the uniprot knowledgebase: How to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2019). GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Cui, H. L., and Dyall-Smith, M. L. (2021). Cultivation of halophilic archaea (class Halobacteria) from thalassohaline and athalassohaline environments. Mar. Life Sci. Technol. 3, 243–251.
Deslandes, Y., Marchessault, R. H., and Sarko, A. (1980). Triple-helical structure of (1/3)-b-D-glucan. Macromolecules 13, 1466–1471.
Enache, M., and Kamekura, M. (2010). Hydrolytic enzymes of halophilic microorganisms and their economic values. Rom. J. Biochem. 47, 47–59. doi: 10.1002/bit.27639
Enomoto, S., Shimane, Y., Ihara, K., Kamekura, M., Itoh, T., Ohkuma, M., et al. (2020). Haloarcula mannanilytica sp. nov., a galactomannan-degrading haloarchaeon isolated from commercial salt. Int. J. Syst. Evol. Microbiol. 70, 6331–6337. doi: 10.1099/ijsem.0.004535
Gavrilov, S. N., Stracke, C., Jensen, K., Menzel, P., Kallnik, V., Slesarev, A., et al. (2016). Isolation and characterization of the first xylanolytic hyperthermophilic euryarchaeon Thermococcus sp. Strain 2319x1 and its unusual multidomain glycosidase. Front. Microbiol. 7:552. doi: 10.3389/fmicb.2016.00552
Grant, W. D., and Jones, B. E. (2016). “Bacteria, Archaea and viruses of soda lakes,” in Soda Lakes of East Africa, ed. M. Schagerl (Berlin: Springer International Publishing), 97–147.
Gueguen, Y., Voorhorst, W. G. B., van der Oost, J., and de Vos, W. M. (1997). Molecular and biochemical characterization of an endo-β-1,3-glucanase of the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 272, 31258–31264.
Kaar, W. E., and Holtzapple, M. T. (2000). Using lime pretreatment to facilitate the enzymic hydrolysis of corn stover. Biomass Bioenergy 18, 189–199.
Kanal, H., Kobayashi, T., Aono, R., and Kudo, T. (1995). Natronococcus amylolyticus sp. nov., a haloalkaliphilic archaeon. Int. J. Syst. Bacteriol. 45, 762–766. doi: 10.1099/00207713-45-4-762
Kobayashi, T., Kanai, H., Hayashi, T., Akiba, T., Akaboshi, R., and Horikoshi, K. (1992). Haloalkaliphilic maltotriose-forming α-amylase from the archaebacterium Natronococcus sp. strain Ah-36. J. Bacteriol. 174, 3439–3444. doi: 10.1128/jb.174.11.3439-3444.1992
Kublanov, I. V., Bidjieva, S. K. H., Mardanov, A. V., and Bonch-Osmolovskaya, E. A. (2009). Desulfurococcus kamchatkensis sp. nov., a novel hyperthermophilic protein-degrading archaeon isolated from a Kamchatka hot spring. Int. J. Syst. Evol. Microbiol. 59, 1743–1747.
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47:W256–W259. doi: 10.1093/nar/gkz239
Mardanov, A. V., Ravin, N. V., Svetlitchnyi, V. A., Beletsky, A. V., Miroshnichenko, M. L., Bonch-Osmolovskaya, E. A., et al. (2009). Metabolic versatility and indigenous origin of the archaeon Thermococcus sibiricus, isolated from a Siberian oil reservoir, as revealed by genome analysis. Appl. Environ. Microbiol. 75, 4580–4588. doi: 10.1128/AEM.00718-09
Minegishi, H., Enomoto, S., Echigo, A., Shimane, Y., Kondo, Y., Inoma, A., et al. (2017). Salinarchaeum chitinilyticum sp. nov., a chitin-degrading haloarchaeon isolated from commercial salt. Int. J. Syst. Evol. Microbiol. 67, 2274–2278. doi: 10.1099/ijsem.0.001941
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., and Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41:e121. doi: 10.1093/nar/gkt263
Mizuno, M., Tonozuka, T., Suzuki, S., Uotsu-Tomita, R., Kamitori, S., Nishikawa, A., et al. (2004). Structural insights into substrate specificity and function of glucodextranase. J. Biol. Chem. 279, 10575–10583. doi: 10.1074/jbc.M310771200
Moshfegh, M., Shahverdi, A. R., Zarrini, G., and Faramarzi, M. A. (2013). Biochemical characterization of an extracellular polyextremophilic α-amylase from the halophilic archaeon Halorubrum xinjiangense. Extremophiles 17, 677–687. doi: 10.1007/s00792-013-0551-7
Oren, A. (2013). “Life at high salt concentrations,” in The Prokaryotes: Prokaryotic Communities and Ecophysiology, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer Berlin Heidelberg), 421–440.
Oren, A. (2015). Halophilic microbial communities and their environments. Curr. Opin. Biotechnol. 33, 119–124.
Perevalova, A. A., Svetlichny, V. A., Kublanov, I. V., Chernyh, N. A., Kostrikina, N. A., Tourova, T. P., et al. (2005). Desulfurococcus fermentans sp. nov., a novel hyperthermophilic archaeon from a Kamchatka hot spring, and emended description of the genus Desulfurococcus. Int. J. Syst. Evol. Microbiol. 55, 995–999. doi: 10.1099/ijs.0.63378-0
Pfennig, N., and Lippert, K. D. (1966). Über das vitamin B12-bedürfnis phototropher schwefelbakterien. Arch. Microbiol. 55, 245–256.
Rinke, C., Chuvochina, M., Mussig, A. J., Chaumeil, P.-A., Davín, A. A., Waite, D. W., et al. (2021). A standardized archaeal taxonomy for the genome taxonomy database. Nat. Microbiol. 6, 946–959. doi: 10.1038/s41564-021-00918-8
Selim, S., Hagagy, N., Aziz, M. A., El-Meleigy, E.-S., and Pessione, E. (2014). Thermostable alkaline halophilic-protease production by Natronolimnobius innermongolicus WN18. Nat. Prod. Res. 28, 1476–1479.
Shimane, Y., Hatada, Y., Minegishi, H., Mizuki, T., and Echigo, A. (2010). Natronoarchaeum mannanilyticum gen. nov., sp. nov., an aerobic, extremely halophilic archaeon isolated from commercial salt. Int. J. Syst. Evol. Microbiol. 60, 2529–2534. doi: 10.1099/ijs.0.016600-0
Sorokin, D. Y., Elcheninov, A. G., Khizhniak, T. V., Koenen, M., Bale, N. J., Damsté, J. S. S., et al. (2022a). Natronocalculus amylovorans gen. nov., sp. nov., and Natranaeroarchaeum aerophilus sp. nov., dominant culturable amylolytic natronoarchaea from hypersaline soda lakes in southwestern Siberia. Syst. Appl. Microbiol. 45:126336. doi: 10.1016/j.syapm.2022.126336
Sorokin, D. Y., Yakimov, M. M., Messina, E., Merkel, A. Y., Koenen, M., Bale, N. J., et al. (2022b). Natranaeroarchaeum sulfidigenes gen. nov., sp. nov., carbohydrate-utilizing sulfur-respiring Haloarchaeon from Hypersaline lakes, a member of a new family Natronoarchaeaceae fam. nov. in the order Halobacteriales. Syst. Appl. Microbiol. 45:126356. doi: 10.1016/j.syapm.2022.126356
Sorokin, D. Y., Khijniak, T. V., Kostrikina, N. A., Elcheninov, A. G., Toshchakov, S. V., Bale, N. J., et al. (2018). Natronobiforma cellulositropha gen. nov., sp. nov., a novel haloalkaliphilic member of the family Natrialbaceae (class Halobacteria) from hypersaline alkaline lakes. Syst. Appl. Microbiol. 41, 355–362. doi: 10.1016/j.syapm.2018.04.002
Sorokin, D. Y., Khijniak, T. V., Kostrikina, N. A., Elcheninov, A. G., Toshchakov, S. V., Bale, N. J., et al. (2019a). Halococcoides cellulosivorans gen. nov., sp. nov., an extremely halophilic cellulose-utilizing haloarchaeon from Hypersaline lakes. Int. J. Syst. Evol. Microbiol. 69, 1327–1335. doi: 10.1099/ijsem.0.003312
Sorokin, D. Y., Kublanov, I. V., Elcheninov, A. G., and Oren, A. (2019b). Natronobiforma. In: Bergey’s Manual of Systematics of Archaea and Bacteria. Hoboken: John Wiley & Sons, Inc, doi: 10.1002/9781118960608.gbm01838
Sorokin, D. Y., Elcheninov, A. G., Toshchakov, S. V., Bale, N. J., Sinninghe Damsté, J. S., Khijniak, T. V., et al. (2019c). Natrarchaeobius chitinivorans gen. nov., sp. nov., and Natrarchaeobius halalkaliphilus sp. nov., alkaliphilic, chitin-utilizing haloarchaea from hypersaline alkaline lakes. Syst. Appl. Microbiol. 42, 309–318. doi: 10.1016/j.syapm.2019.01.001
Sorokin, D. Y., Merkel, A. Y., Kublanov, I. V., and Oren, A. (2020a). Halococcoides. In: Bergey’s Manual of Systematics of Archaea and Bacteria. Hoboken: John Wiley & Sons, Inc, doi: 10.1002/9781118960608.gbm01944
Sorokin, D. Y., Merkel, A. Y., Kublanov, I. V., and Oren, A. (2020b). Natrarchaeobius. In: Bergey’s Manual of Systematics of Archaea and Bacteria. Hoboken: John Wiley & Sons, Inc, doi: 10.1002/9781118960608.gbm01945
Sorokin, D. Y., Toshchakov, S. V., Kolganova, T. V., and Kublanov, I. V. (2015). Halo(natrono)archaea isolated from hypersaline lakes utilize cellulose and chitin as growth substrates. Front. Microbiol. 6:942. doi: 10.3389/fmicb.2015.00942
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Wainø, M., and Ingvorsen, K. (2003). Production of β-xylanase and β-xylosidase by the extremely halophilic archaeon Halorhabdus utahensis. Extremophiles 7, 87–93.
Werner, J., Ferrer, M., Michel, G., Mann, A. J., Huang, S., and Juarez, S. (2014). Halorhabdus tiamatea: Proteogenomics and glycosidase activity measurements identify the first cultivated euryarchaeon from a deep-sea anoxic brine lake as potential polysaccharide degrader. Environ. Microbiol. 16, 2525–2537. doi: 10.1111/1462-2920.12393
Zavrel, M., Bross, D., Funke, M., Büchs, J., and Spiess, A. C. (2010). High-throughput screening for ionic liquids dissolving (ligno-)cellulose. Bioresour. Technol. 100, 2580–2587.
Keywords: halo(natrono)archaea, hypersaline lakes, soda lakes, polysaccharides, hydrolytic
Citation: Sorokin DY, Elcheninov AG, Khijniak TV, Kolganova TV and Kublanov IV (2022) Selective enrichment on a wide polysaccharide spectrum allowed isolation of novel metabolic and taxonomic groups of haloarchaea from hypersaline lakes. Front. Microbiol. 13:1059347. doi: 10.3389/fmicb.2022.1059347
Received: 01 October 2022; Accepted: 07 November 2022;
Published: 23 November 2022.
Edited by:
Rosa María Martínez-Espinosa, University of Alicante, SpainCopyright © 2022 Sorokin, Elcheninov, Khijniak, Kolganova and Kublanov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dimitry Y. Sorokin, c29yb2NAaW5taS5ydQ==; ZC5zb3Jva2luQHR1ZGVsZnQubmw=
 Dimitry Y. Sorokin
Dimitry Y. Sorokin Alexander G. Elcheninov
Alexander G. Elcheninov Tatiana V. Khijniak1
Tatiana V. Khijniak1 Tatiana V. Kolganova
Tatiana V. Kolganova Ilya V. Kublanov
Ilya V. Kublanov