- 1Department of Internal Medicine, Chi Mei Medical Center, Tainan, Taiwan
- 2Department of Medical Research, Chi Mei Medical Center, Tainan, Taiwan
- 3Department of Food Science, National Chiayi University, Chiayi, Taiwan
- 4Division of Hospital Medicine, Department of Internal Medicine, Chi Mei Medical Center, Tainan, Taiwan
- 5Department of Intensive Care Medicine, Chi Mei Medical Center, Liouying, Tainan, Taiwan
Objectives: This study investigated the inhibitory effect of Lactobacillus spp. with prebiotics against Klebsiella pneumoniae carbapenemase-2 (KPC-2)-producing Klebsiella pneumoniae using both in vitro experiments and animal models.
Methods: Thirty-three Lactobacillus spp. strains were confirmed by 16S rDNA sequencing, and four different PFGE genotyped KPC-2-producing K. pneumoniae strains were selected for investigation. In vitro studies, including broth microdilution assays, changes in pH values in lactobacilli cultures with different prebiotics, time-kill tests of Lactobacillus spp. against KPC-2-producing K. pneumoniae and further in vivo Lactobacillus alone or in combination with prebiotics against KPC-2-producing K. pneumoniae in an animal model, were performed.
Results: The lower pH value of the cell-free supernatant was associated with a lower minimal inhibitory percentage of the Lactobacillus strain against KPC-2-producing K. pneumoniae. Furthermore, lactulose/isomalto-oligosaccharide/inulin and fructo-oligosaccharide can enhance the inhibitory effect of all 107 CFU/ml Lactobacillus strains against KPC001. Three Lactobacillus strains (LYC1154, LYC1322, and LYC1511) that could be persistently detected in the stool were tested for their ability to reduce the amount of KPC001 in the feces individually or in combination. A significantly better effect in reducing the amount of KPC001 was observed for the combination of three different Lactobacillus species than for each of them alone. Furthermore, their inhibitory effect was enhanced after adding lactulose or isomalto-oligosaccharide (both p < 0.05).
Conclusion: This study demonstrates the inhibitory effect of probiotic Lactobacillus, including LYC1154, LYC1322, and LYC1511, with prebiotics such as lactulose or isomalto-oligosaccharide against the colonization of KPC-2-producing K. pneumoniae.
Introduction
Probiotics are living bacteria or fungi that are consumed, and prebiotics are nondigestible compounds that are selectively fermented by commensal microbiota in the human gut (Gibson and Roberfroid, 1995; Newman and Arshad, 2020). Both probiotics and prebiotics can provide health benefits to the host and promote human health. Probiotics as well as prebiotics exert their effect through the production of short-chain fatty acids from metabolic precursors, leading to the downstream effects of immune modulation and increased mucosal barrier function (Patel and DuPont, 2015). Moreover, probiotics with or without prebiotics can exhibit the additional effect of producing antimicrobial compounds and restoring the enteric microbiome (Patel and DuPont, 2015; Newman and Arshad, 2020). In fact, several commercial probiotics have been used to manage Clostridioides difficile infections, traveler’s diarrhea, and irritable bowel syndrome (Sniffen et al., 2018). Moreover, the use of prebiotics, probiotics and synbiotics shows promising potential in the manipulation of the microbiome and resistome and helps combat MDROs (Newman and Arshad, 2020).
Antibiotics are essential in the treatment of acute bacterial infections; however, overuse or misuse of antibiotics in humans, agriculture, and animal husbandry has resulted in the emergence of a wide range of multidrug-resistant organisms (MDROs) (Dadgostar, 2019). Among MDROs, carbapenem-resistant gram-negative bacteria, including carbapenem-resistant Enterobacterales (CREs), Acinetobacter baumannii and Pseudomonas aeruginosa, have become the common cause of health care-associated infections and pose a great threat to global health (Dadgostar, 2019; Lai and Yu, 2021; Wozniak et al., 2022; Jean et al., 2022b). Both the World Health Organization and Centers for Disease Control and Prevention have designated that CRE, such as Klebsiella species, Escherichia coli and Enterobacter species, are the most crucial emerging resistance threats worldwide (Hetzler et al., 2022; Lodise et al., 2022; Oka et al., 2022; Sy et al., 2022; Jean et al., 2022a). The most common resistance mechanism of carbapenem resistance in Enterobacterales is the synthesis of carbapenemase enzymes, which include class A Klebsiella pneumoniae carbapenemase (KPC), class B metallo-β-lactamases, and class D OXA β-lactamases (Durante-Mangoni et al., 2019). Because these bacteria are difficult to treat due to high levels of antibiotic resistance and the decreased focus of pharmaceutical industries on research and development of newer effective antibiotics to fight these MDROs, novel strategies such as nanoparticles, phage therapy, antimicrobial peptides, and fecal microbiota transplantation are urgently needed to fight these MDROs (Sannathimmappa et al., 2021).
In our previous in vitro studies using agar well diffusion and broth microdilution assays and time-kill tests, we demonstrated the potent activity of Lactobacillus spp. against CRE and carbapenemase-producing Enterobacterales (Chen et al., 2019, 2021). Furthermore, we found that lactic acid produced by Lactobacillus strains is the major antimicrobial mechanism. However, we considered whether adding prebiotics with Lactobacillus spp. would help enhance their activity against these MDROs. Therefore, we conducted this study to assess the inhibitory effect of Lactobacillus spp. with prebiotics against KPC-2-producing K. pneumoniae using both in vitro experiments and animal models.
Materials and methods
Bacterial strains
Thirty-three Lactobacillus spp. strains were isolated from Chinese sauerkraut as confirmed by 16S rDNA sequencing. The 16S rRNA gene was amplified with primers F27 (AGAGTTTGATCM TGGCTCAG) and R1492 (TACGGYTACCTTGTTACGACTT) as previously reported (Cruciani et al., 2015). The sequences were searched with the NCBI BLAST web service (blast.ncbi.nlm.nih.gov) to confirm the taxonomic identification at the species level. Strains including 13 L. plantarum, 8 L. paracasei, 7 L. fermentum, 4 L. rhamnosus, and 1 L. brevis were selected. The basic growth medium for Lactobacillus spp. was Man-Rogosa-Sharpe (MRS; Oxoid Inc., Ogdensburg, NY, United States). Four different pulse field gel electrophoresis (PFGE)-genotyped KPC-2-producing K. pneumonia strains were selected with a CHEF DR II apparatus (Bio-Rad Laboratories, Hercules, CA, United States) (Tang et al., 2019).
Cell-free supernatant preparation
The culture supernatants of lactobacilli were grown in MRS broth (pH adjusted to 6.5) at 37°C for 24 h. The culture broths were then centrifuged at 10,000g at 4°C for 30 min. The supernatants were sterilized by filtration through a 0.22 μm cellulose acetate filter (Millipore, Billerica, MA, United States) and stored at −80°C until use.
Broth microdilution assay
A broth microdilution assay was conducted as previously described with modifications (Śliżewska and Chlebicz-Wójcik, 2020). Overnight cultures of KPC-2-producing K. pneumoniae were inoculated into fresh Mueller-Hinton broth (MHB) media and seeded into 96-well plates (BD Discovery Labware, Bedford, MA, United States). The CFSs were diluted with MRS broth (pH = 6.5) and used at different percentages (i.e., 6.25%, 12.5%, 25%, and 50%) in the final 200 μl volume. The minimum inhibitory percentage (MIP), defined as the lowest percentage of supernatant that can inhibit the growth of pathogens, was monitored by measuring the optical density (OD600 nm). All tests were performed in triplicate.
The effect on pH value by Lactobacillus and different prebiotics
The effect of Lactobacillus spp. strains after adding prebiotics, including inulin (IN), fructooligosaccharide (FOS), and lactulose (LU) (Sigma–Aldrich, St. Louis, MO, United States), isomalto-oligosaccharide (IMO) (FUJIFILM, Chuo-Ku, Osaka, Japan), and xylo-oligosaccharide (XOS) (Americanway Bio-Technology, Tainan, Taiwan), on pH value was analyzed. Lactobacillus spp. were added as 104 CFU/ml inoculum to MRS broths in which glucose was substituted with 2% prebiotic or sucrose (Sigma–Aldrich, St. Louis, MO, United States; Lu et al., 2018; Śliżewska and Chlebicz-Wójcik, 2020; Chen et al., 2021). The cultures were incubated for 24 h at 37°C. The pH values were measured using a pH meter. All tests were performed in triplicate.
Time-kill test of Lactobacillus spp. against KPC-2-producing Klebsiella pneumoniae
First, KPC-2-producing K. pneumoniae and Lactobacillus strains were individually cultured in their own broth medium at 37°C for 24 h. The cultures were centrifuged at 6,000 rpm and 22°C for 10 min to collect the cell pellet. Second, KPC-2-producing K. pneumoniae were inoculated at 1 × 106 CFU/ml and cocultured with 1 × 105, 1 × 106, or 1 × 107 CFU/ml lactobacilli in tubes containing 10 ml of MRS-MH broth (1:1) with 2% different probiotics, sucrose as a control or no carbohydrate at 37°C for 48 h (Drago et al., 1997). Samples were collected at 0, 3, 6, 24, and 48 h for the determination of viable cell count and pH measurements. A 1 ml aliquot of each sample was used to prepare serial dilutions that were poured onto the appropriate agar plates; MRS agar (pH 5.5) was used for Lactobacillus spp., while EMB agar (Oxoid Inc., Ogdensburg, NY, United States) + 16 μg/ml ampicillin (Sigma–Aldrich, St. Louis, MO, United States) was used for pathogen plates that were incubated at 37°C for 24 h, and colonies were counted. All tests were performed in triplicate.
Antimicrobial treatment protocol to eradicate the intestinal pathogen in mice
This animal study was approved by the Institutional Animal Care and Use Committee of Chi Mei Medical Center (IACUC Approval No 108101803). Animals were housed under controlled temperatures and 12 h/12 h light/dark cycles with ad libitum access to food and water. Before the study, the mice were allowed to adapt for the last 5 days. The humane endpoint was set to a loss of 20% body weight compared with the starting weight in each experiment. Female inbred BALB/c mice (Animal Center, National Science Council, Taipei, Taiwan) weighing 18–20 g (6–8 weeks old) were used in this study. For every animal, antimicrobials started with 3 days of 0.1 mg/ml amphotericin-B for 0.1 ml by gavage every 12 h. In addition, water flasks were supplemented with 1 g/L ampicillin (Bristol Meyers Squibb, New York City, NY), 5 mg/ml vancomycin, 10 mg/ml neomycin, and 10 mg/ml metronidazole. All drugs were purchased from Sigma–Aldrich (Sigma–Aldrich, St. Louis, MO, United States). Fresh antibiotics were added daily into drinking water for three consecutive days (Reikvam et al., 2011). After 3 days of antibiotic treatment, the fresh feces of each animal were collected directly into a preweighed 1.5 ml capped microtube. Tubes with fecal pellets were kept on ice and weighed, and the weight of the pellets was calculated. Fecal pellets were resuspended in 1 ml PBS by vortexing. The fecal suspension was then plated on blood agar, anaerobic blood agar, and yeast agar (Sabouraud agar) with 100 μl suspension on each plate. Blood agar and Sabouraud agar plates were incubated aerobically at 37°C with 5% CO2 for 72 h, while anaerobic blood agar plates were incubated at 37°C in anaerobic conditions for 72 h. At the end of incubation, the numbers of colonies on the plates were counted to confirm whether the fecal bacteria were eradicated. The detection limit of the assay was defined as 1 cfu/mg feces (Reikvam et al., 2011; Jeong et al., 2017).
Lactobacillus colonization test
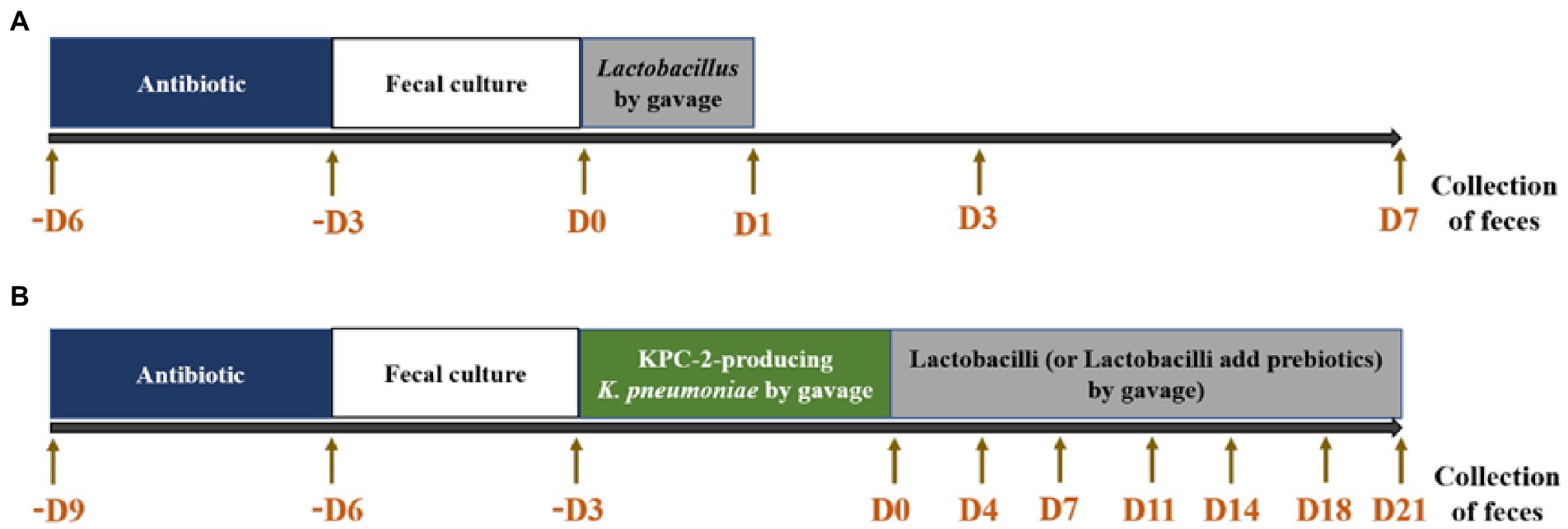
After 3 days of antimicrobial treatment, no microbes were detected in the animal feces. A total of 2 × 109 CFU/animal of each Lactobacillus strain, including 6 L. plantarum, 3 L. paracasei, and 3 L. rhamnosus, resuspended in PBS was added individually to the gavage tube. On Days 1, 3, and 7 after adding lactobacilli, fresh feces were collected. The fecal suspensions were serially diluted 10-fold, plated on MRS agar and incubated at 37°C for 48 h. After incubation, the numbers of colonies on the plates (MRS pH 5.0 + 32 μg/ml vancomycin) were counted, and the number of Lactobacillus spp. per g of feces was calculated (Figure 1A).
Lactobacillus combined with prebiotics against KPC-2-producing Klebsiella pneumoniae in an animal model
After 3 days of antimicrobial treatment and animal feces without detectable microbes, 3 × 108 CFU of KPC-2-producing K. pneumoniae was added by oral gavage for 3 days for every animal. On the day after the last pathogen treatment, LYC1154, LYC1322, and LYC1511 (2 × 109 CFU each Lactobacillus/animal/200 μl) combined with 20% LU, IMO, or PBS alone were added daily until the end of the study. Feces were collected on Days 0 (1 h before Lactobacillus treatment), 4, 7, 11, 14, 18, and 21 (Figure 1B). Lactobacillus was detected by MRS agar (pH = 5.0) + 32 μg/ml vancomycin, and KPC-2-producing K. pneumoniae was detected by EMB agar with 16 μg/ml eartapenem + 64 μg/ml ampicillin + 16 μg/ml cefotaxime.
Lactobacillus alone or in combination with three strains against KPC-2-producing Klebsiella pneumoniae in mice
LYC1154, LYC1322, and LYC1511 were treated alone at 6 × 109 CFU in PBS. The combination of LYC1154, LYC1322, and LYC1511 included 2 × 109 CFU per strain. Antibiotic treatment and KPC-2-producing K. pneumoniae infection were performed as described above. LYC1154, LYC1322, and LYC1511 were treated alone at 6 × 109 CFU in PBS. Alternatively, LYC1154, LYC1322, and LYC1511 were combined (2 × 109 CFU per strain). The fecal treatments are described above.
Statistical analysis
When appropriate, data are presented as the mean and standard deviation (mean ± SD). The two-tailed t-test was used for statistical analysis. The p-value for statistical significance for all analyses was defined as p < 0.05.
Results
The minimal inhibition percentage against the CPE and pH of lactobacilli CFS
The pH of Lactobacillus strain CFS varied according to different Lactobacillus species (Table 1). The resulting pH value was lowest in the CFS of L. plantarum, followed by L. paracasei, L. fermentum, and L rhamnosus. Furthermore, the lower pH value of CFS was associated with a lower MIP of the Lactobacillus strain against KPC-2-producing K. pneumoniae, and this finding was consistent across 4 KPC-2-producing K. pneumoniae strains (Table 1 and Figure 2). Then, 28 Lactobacillus strains with MIP ≤ 50% were selected for the following tests with prebiotics.
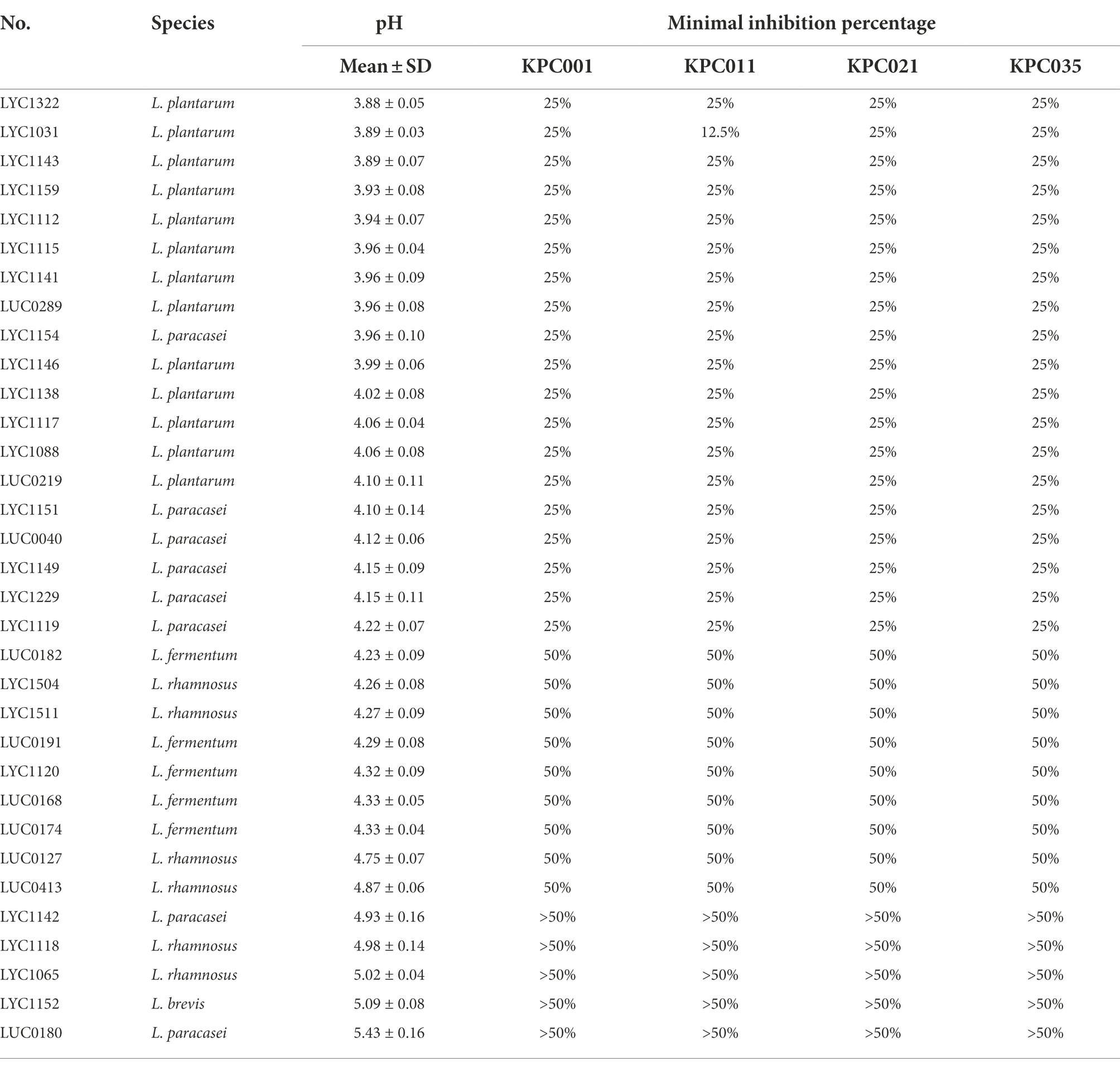
Table 1. The resulting pH and MIPs of Lactobacillus strain cell-free supernatants (%) against KPC-2-producing Klebsiella pneumoniae.
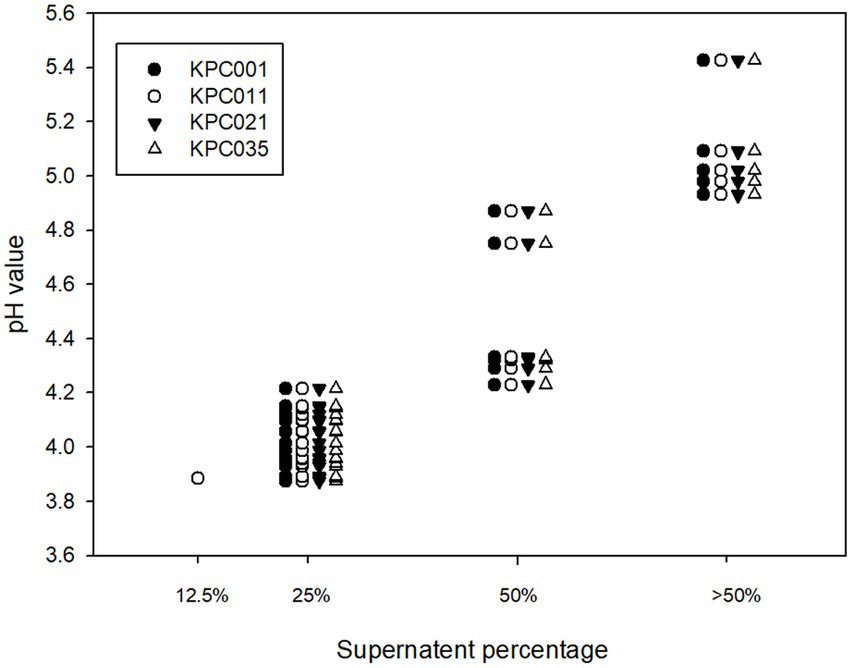
Figure 2. Correlation of the pH of cell-free supernatants and MIP against carbapenemase-producing Klebsiella pneumoniae.
pH value in Lactobacillus cultures with different prebiotics
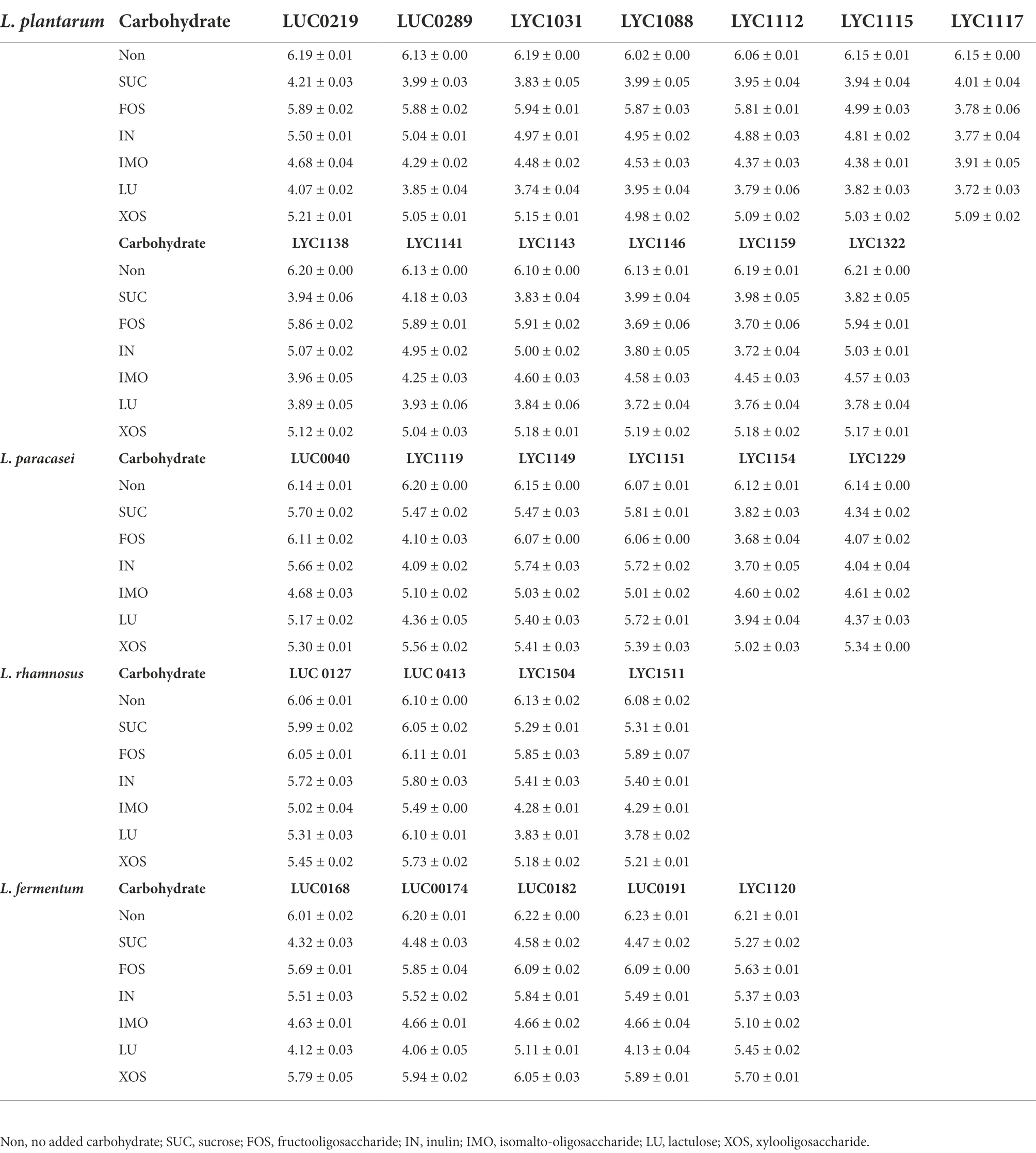
Overall, the pH value in the 19/28 Lactobacillus strains cultured with LU was lower than that in those cultured with other prebiotics. Among 13 L. plantarum strains, LYC1031, L1112, L1117, LYC1146, LYC1159, and LYC1322 could be associated with lower pH after adding LU than other strains (Table 2). Among the 6 L. paracasei strains, LYC1119, LYC1154, and LYC1229 had lower pH values with LU. Among the 4 L. rhamnosus strains, the pH was lower in the coculture with LYC1504 and LYC15111. However, a lower pH in the coculture with prebiotics was not observed for the L. fermentum strains. Therefore, 11 Lactobacillus strains, including LYC1031, L1112, L1117, LYC1146, LYC1159, LYC1322, LYC1119, LYC1154, LYC1229, LYC1504, and LYC1511, which could produce lower pH values with prebiotics, were selected for testing with prebiotics against KPC-2-producing K. pneumoniae.
Time-kill test of Lactobacillus spp. with prebiotics against KPC-2-producing Klebsiella pneumoniae
For all 11 Lactobacillus strains tested, LU enhanced the inhibitory effect against KPC001 (Table 3). IMO helped enhance the inhibitory effect of 10 Lactobacillus strains, including LYC1031, LYC1112, LYC1117, LYC1146, LYC1159, LYC1322, LYC1154, LYC1229, LYC1504, and LYC1511. IN and FOSs were found to enhance the inhibitory effect of 7 Lactobacillus strains, including LYC1117, LYC1146, LYC1159, LYC1119, LYC1154, LYC1229, and LYC1511. However, no inhibitory effect was observed for Lactobacillus strains with xylo-oligosaccharide.
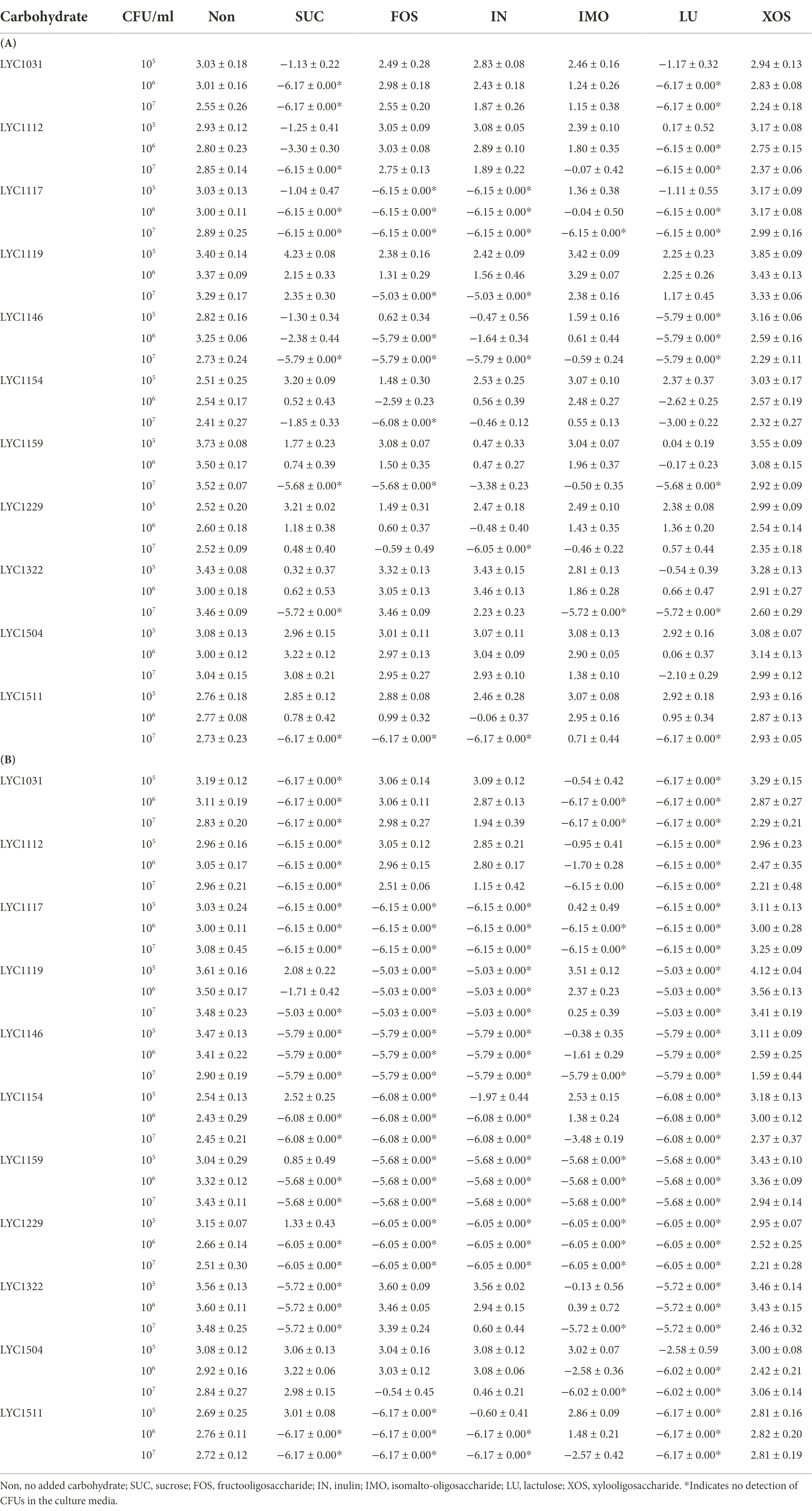
Table 3. The change in log10 CFU/ml of KPC001 cocultured with Lactobacillus strains in MRS-MH broth with different carbohydrates at 24 (A) and 48 (B) hours.
Animal model
For 3 L. paracasei strains (LYC119, LYC1154, and LYC1229), all of them could be persistently detected, and their amount remained at ~106 CFU/g in the stool on Day 7 (Figure 3A). Among 6 L. plantarum strains (LYC1031, LYC1112, LYC1117, LYC1146, LYC1150, and LYC1322), 4 became undetectable on Day 7, and only 3 (LYC1322, LYC1031, and LYC1159) remained detectable with amounts >104 CFU/g (Figure 3B). For 2 L. rhamnosus strains (LYC1504 and LYC1511), both were undetectable on Day 7 (Figure 3C). Thereafter, three strains, LYC1154, 1322, and 1511, were selected for further tests against KPC-2-producing K. pneumoniae.
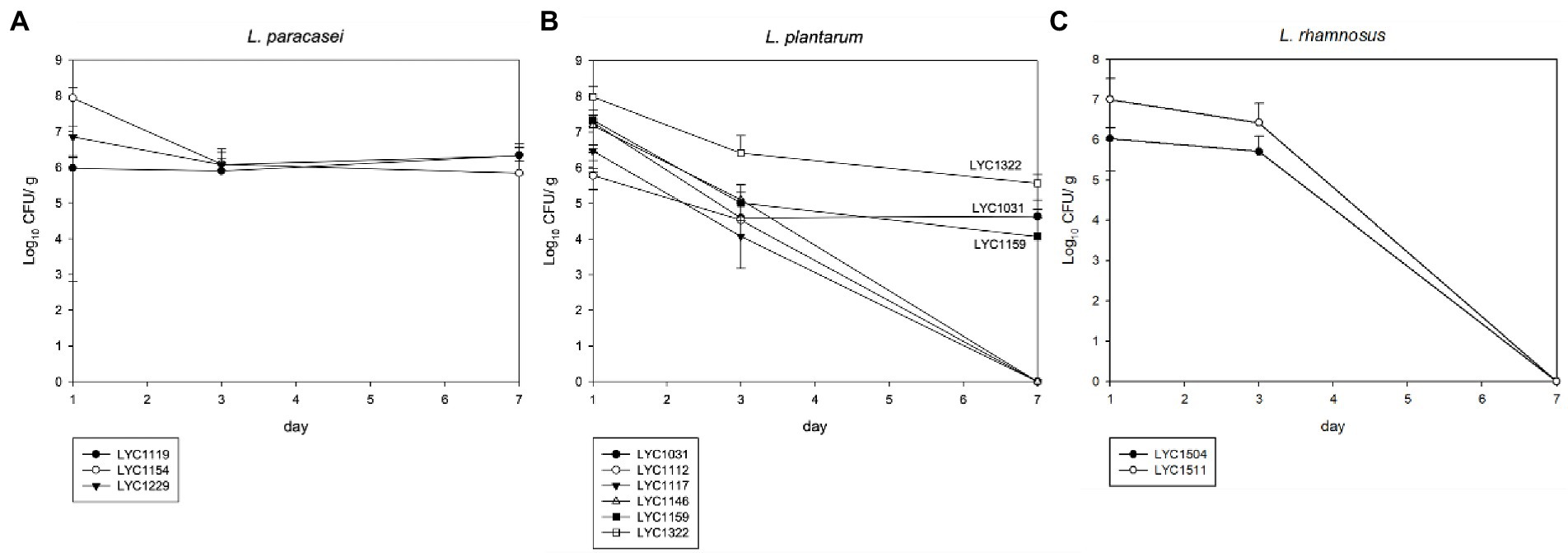
Figure 3. The Lactobacillus number in feces after each oral treatment. (A) L. paracasei, (B) L. plantarum, and (C) L. rhamnosus. (The Lactobacillus strains in stool are shown in CFUs/g as the mean ± SD.)
Three Lactobacillus strains (LYC1154, LYC1322, and LYC1511) were tested for their ability to reduce the amount of KPC001 in the feces individually or in combination (Figure 4). A significantly better effect in reducing the amount of KPC001 was observed for the combination of 3 different Lactobacillus species than for each species alone (Figure 4). Furthermore, their inhibitory effect was enhanced after adding LU or IMO (both p < 0.05, Figure 5).
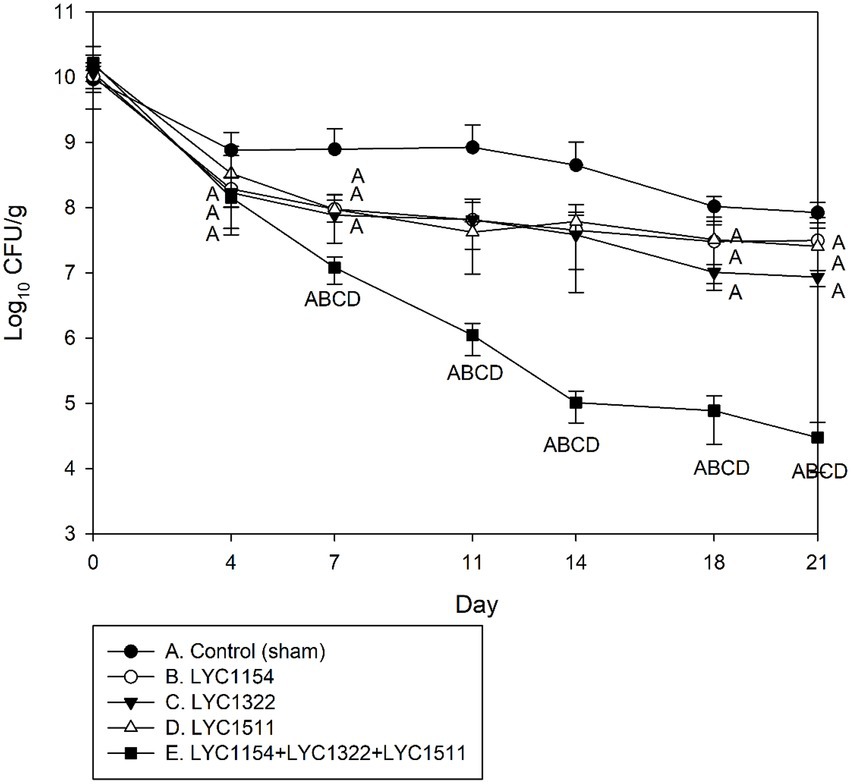
Figure 4. Kinetics of KPC001 from feces in treatments with one of three Lactobacillus strains or the three lactobacilli combined. Each symbol represents the mean ± SD of colony-forming units per gram (CFU/g) in feces from different treatment mice. (Group A: no Lactobacillus treatment, B: LYC1154, C: LYC1322, D: LYC1511, and E: LYC1154 + LYC1322 + LYC1511. The letter indicates a significant difference compared with the indicated group.)
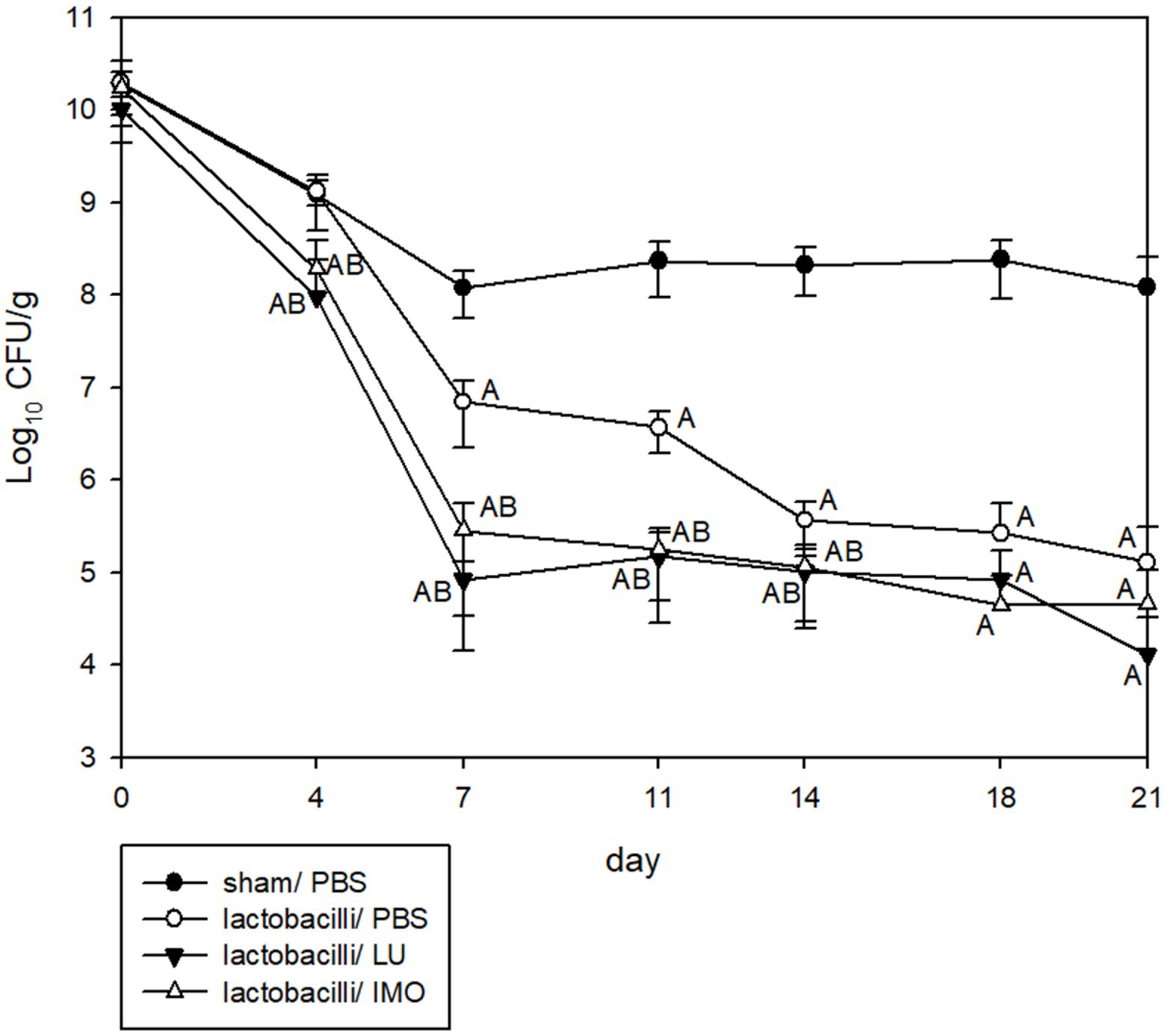
Figure 5. Kinetics of KPC001 from feces in three lactobacilli combined with PBS, lactulose or isomalto-oligosaccharide. Each symbol represents the mean ± SD of colony-forming units per gram (CFU/g) in feces from different treatment mice. (Group A: no lactobacilli or prebiotics, B: lactobacilli only, C: LYC1322, LYC1154, and LYC1511 with added LU, D: LYC1322, LYC1154, and LYC1511 with added IMO. LU: lactulose; IMO: isomalto-oligosaccharide. The letter indicates a significant difference compared with the indicated group.)
Discussion
This is the first study to investigate the antibacterial effect of Lactobacillus spp. with prebiotics against KPC-2-producing K. pneumoniae. From the serial tests of 33 Lactobacillus strains and five prebiotics, including FOS, IN, IMO, LU, and xylooligosaccharide, we identified three Lactobacillus strains (LYC1154, LYC1322, and LYC1511), and LU or IMO exhibited the greatest potency against KPC-2-producing K. pneumonia.
This finding was supported by the following evidence. First, the CSFs of the three Lactobacillus strains exhibited low pH values and low MIPs against the four KPC-2-producing K. pneumoniae strains tested (Table 1). Second, the pH values of all three strains with LU or IMO were lower than those with other prebiotics (Table 2). Third, the time-killing methods showed that both LU and IMO can help enhance the inhibitory effect of these three strains against KPC-2-producing K. pneumoniae—KPC001 (Table 3). Fourth, the animal model demonstrated that each of them can reduce the intestinal colonization of KPC001, and the combination of all three strains exhibited a better anti-colonization effect than each of them alone (Figure 4). Finally, after adding LU or IMO, the anti-colonization effect of the Lactobacillus strain against KPC001 was further enhanced in the animal experiment (Figure 5). All of these findings based on in vitro studies and animal models indicated the potent anti-KPC-2-producing K. pneumoniae of three Lactobacillus strains (LYC1154, LYC1322, and LYC1511) with the prebiotics LU and IMO. Moreover, Abramov et al. demonstrated that Limosilactobacillus fermentum strain 3872 and Actigen prebiotic (Alltech Inc., Nicholasville, KY, United States) could exhibit synergistic anti-adhesive activity against gram-negative pathogens (Abramov et al., 2022). Overall, these findings suggest the promising role of synbiotics (probiotics + prebiotics) as a new strategy for fighting MDROs.
In addition to KPC-2-producing K. pneumoniae, several studies (Asahara et al., 2001; Wieërs et al., 2020; Hai and Huang, 2021; Scillato et al., 2021) have reported the effect of probiotics against MDROs. Hai et al. demonstrated that the growth of MDR Salmonella enteritidis SE05 decreased over time by coculturing with L. reuteri Lb11 (isolated from the chicken intestinal tract) in vitro, and the pH value significantly decreased (Hai and Huang, 2021). Scillato et al. showed that the CFS of L. gasseri 1A-TV, L. fermentum 18A-TV, and L. crispatus 35A-TV (isolated from the vaginal microbiota of healthy premenopausal women) and their combination revealed a strong bactericidal effect on uropathogens, such as S. agalactiae, E. coli, K. pneumoniae, S. aureus, P. aeruginosa, P. vulgaris, and P. mirabilis, and MDROs, such as KPC-3-producing K. pneumoniae and vancomycin-resistant enterococcus (Scillato et al., 2021). Moreover, a randomized clinical trial reported that treatment with a probiotic mixture containing Saccharomyces boulardii, Lactobacillus acidophilus NCFM, Lactobacillus paracasei Lpc-37, Bifidobacterium lactis Bl-04, and Bifidobacterium lactis Bi-07 could significantly reduce gastrointestinal colonization with P. aeruginosa and AmpC-producing enterobacteria after amoxicillin-clavulanate pretreatment (p = 0.041). Similarly, Asaharathe et al., using an opportunistic antibiotic-induced murine infection model, showed that an increase in the concentration of organic acids and a lowered pH in the intestine due to bifidobacterial colonization were correlated with anti-infectious activity (Asahara et al., 2001). Although our findings were in line with these studies (Asahara et al., 2001; Wieërs et al., 2020; Hai and Huang, 2021; Scillato et al., 2021) and confirmed the inhibitory effect of probiotics against MDROs, our finding further indicated that multiple probiotics in combination with prebiotics would have greater activity.
However, one RCT found that the administration of a synbiotic product twice a day for 7 days via the oral/enteral route was not effective for decolonizing hospitalized patients harboring MDR gram-negative bacilli (Salomão et al., 2016). In this single-center study of 116 patients, no significant difference was observed in the negative recovery rate of rectal swabs for MDR gram-negative bacilli after treatment between the synbiotic group and the placebo group (16.7% [8/48] vs. 20.7% [11/53], p = 0.60) (Salomão et al., 2016). The possible explanation for these conflicting findings between the RCT and the present study could be the different study designs—in vitro or in vivo vs. human study. In addition, we identified three Lactobacillus strains (LYC1154, LYC1322, and LYC1511) and two prebiotics, LU and IMO, as exhibiting the greatest activity, but the RCT used the synbiotics Lactobacillus bulgaricus, Lactobacillus rhamnosus, and FOSs. Therefore, further study is needed to clarify our findings.
This study has two limitations. First, as we only focused on KPC-2-producing K. pneumoniae, our findings cannot be generalized to other MDROs. Second, the composition and percentage of the three lactobacilli and prebiotics were not clearly defined in the study. Further in vivo studies are warranted.
In conclusion, our study strengthens the concept of using probiotic Lactobacillus with prebiotics to protect the host against the MDR pathogen KPC-2-producing K. pneumoniae, and these results support novel therapeutic strategies as new synbiotics for the prevention and treatment of MDRO colonization or infections.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
H-JT, C-CC, C-CL, and C-MC contributed to conception and design of the study. H-JT, C-CC, Y-CL, H-LH, H-JC, Y-CC, C-CL investigation, analysis, or interpretation of data for the work. C-CC, Y-CL, H-LH, H-JC, performed the statistical analysis. H-JT, C-CL, Y-CC, C-MC wrote the first draft of the manuscript. H-JT and C-MC critical review. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was supported by a research grant (MOST 111-2314-B-384-008) from the Ministry of Science and Technology and the Chi-Mei Medical Center Research Foundation (CMFHR11067 and CMFHT11001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abramov, V. M., Kosarev, I. V., Machulin, A. V., Priputnevich, T. V., Chikileva, I. O., Deryusheva, E. I., et al. (2022). Limosilactobacillus fermentum strain 3872: antibacterial and Immunoregulatory properties and synergy with prebiotics against socially significant antibiotic-resistant infections of animals and humans. Antibiotics (Basel) 11:1437. doi: 10.3390/antibiotics11101437
Asahara, T., Nomoto, K., Shimizu, K., Watanuki, M., and Tanaka, R. (2001). Increased resistance of mice to salmonella enterica serovar typhimurium infection by synbiotic administration of Bifidobacteria and transgalactosylated oligosaccharides. J. Appl. Microbiol. 91, 985–996. doi: 10.1046/j.1365-2672.2001.01461.x
Chen, C. C., Lai, C. C., Huang, H. L., Huang, W. Y., Toh, H. S., Weng, T. C., et al. (2019). Antimicrobial activity of lactobacillus species against Carbapenem-resistant Enterobacteriaceae. Front. Microbiol. 10:789. doi: 10.3389/fmicb.2019.00789
Chen, C. C., Lai, C. C., Huang, H. L., Su, Y. T., Chiu, Y. H., Toh, H. S., et al. (2021). Antimicrobial ability and mechanism analysis of lactobacillus species against carbapenemase-producing Enterobacteriaceae. J. Microbiol. Immunol. Infect. 54, 447–456. doi: 10.1016/j.jmii.2020.01.005
Cruciani, F., Biagi, E., Severgnini, M., Consolandi, C., Calanni, F., Donders, G., et al. (2015). Development of a microarray-based tool to characterize vaginal bacterial fluctuations and application to a novel antibiotic treatment for bacterial vaginosis. Antimicrob. Agents Chemother. 59, 2825–2834. doi: 10.1128/AAC.00225-15
Dadgostar, P. (2019). Antimicrobial resistance: implications and costs. Infect. Drug Resist. 12, 3903–3910. doi: 10.2147/IDR.S234610
Drago, L., Gismondo, M. R., Lombardi, A., De Haën, C., and Gozzini, L. (1997). Inhibition of in vitro growth of enteropathogens by new lactobacillus isolates of human intestinal origin. FEMS Microbiol. Lett. 153, 455–463. doi: 10.1111/j.1574-6968.1997.tb12610.x
Durante-Mangoni, E., Andini, R., and Zampino, R. (2019). Management of carbapenem-resistant Enterobacteriaceae infections. Clin. Microbiol. Infect. 25, 943–950. doi: 10.1016/j.cmi.2019.04.013
Gibson, G. R., and Roberfroid, M. B. (1995). Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125, 1401–1412. doi: 10.1093/jn/125.6.1401
Hai, D., and Huang, X. (2021). Protective effect of lactobacillus reuteri Lb11 from chicken intestinal tract against salmonella Enteritidis SE05 in vitro. Antonie Van Leeuwenhoek 114, 1745–1757. doi: 10.1007/s10482-021-01625-4
Hetzler, L., Kollef, M. H., Yuenger, V., Micek, S. T., and Betthauser, K. D. (2022). New antimicrobial treatment options for severe gram-negative infections. Curr. Opin. Crit. Care 28, 522–533. doi: 10.1097/MCC.0000000000000968
Jean, S. S., Harnod, D., and Hsueh, P. R. (2022a). Global threat of Carbapenem-resistant gram-negative bacteria. Front. Cell. Infect. Microbiol. 12:823684. doi: 10.3389/fcimb.2022.823684
Jean, S. S., Lee, Y. L., Liu, P. Y., Lu, M. C., Ko, W. C., and Hsueh, P. R. (2022b). Multicenter surveillance of antimicrobial susceptibilities and resistance mechanisms among Enterobacterales species and non-fermenting gram-negative bacteria from different infection sources in Taiwan from 2016 to 2018. J. Microbiol. Immunol. Infect. 55, 463–473. doi: 10.1016/j.jmii.2021.07.015
Jeong, D., Kim, D. H., Kang, I. B., Kim, H., Song, K. Y., Kim, H. S., et al. (2017). Modulation of gut microbiota and increase in fecal water content in mice induced by administration of lactobacillus kefiranofaciens DN1. Food Funct. 8, 680–686. doi: 10.1039/C6FO01559J
Lai, C. C., and Yu, W. L. (2021). Klebsiella pneumoniae harboring Carbapenemase genes in Taiwan: its evolution over 20 years, 1998-2019. Int. J. Antimicrob. Agents 58:106354. doi: 10.1016/j.ijantimicag.2021.106354
Lodise, T. P., Bassetti, M., Ferrer, R., Naas, T., Niki, Y., Paterson, D. L., et al. (2022). All-cause mortality rates in adults with carbapenem-resistant gram-negative bacterial infections: a comprehensive review of pathogen-focused, prospective, randomized, interventional clinical studies. Expert Rev. Anti-Infect. Ther. 20, 707–719. doi: 10.1080/14787210.2022.2020099
Lu, E., Yeung, M., Yeung, C. K. J. F., and Sciences, N. (2018). Comparative analysis of lactulose and fructooligosaccharide on growth kinetics, fermentation, and antioxidant activity of common probiotics. Food Nutr. Sci. 09, 161–178. doi: 10.4236/fns.2018.93013
Newman, A. M., and Arshad, M. (2020). The role of probiotics, prebiotics and Synbiotics in combating multidrug-resistant organisms. Clin. Ther. 42, 1637–1648. doi: 10.1016/j.clinthera.2020.06.011
Oka, K., Matsumoto, A., Tetsuka, N., Morioka, H., Iguchi, M., Ishiguro, N., et al. (2022). Clinical characteristics and treatment outcomes of carbapenem-resistant Enterobacterales infections in Japan. J. Glob. Antimicrob. Resist. 29, 247–252. doi: 10.1016/j.jgar.2022.04.004
Patel, R., and Dupont, H. L. (2015). New approaches for bacteriotherapy: prebiotics, new-generation probiotics, and synbiotics. Clin. Infect. Dis. 60, S108–S121. doi: 10.1093/cid/civ177
Reikvam, D. H., Erofeev, A., Sandvik, A., Grcic, V., Jahnsen, F. L., Gaustad, P., et al. (2011). Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One 6:e17996. doi: 10.1371/journal.pone.0017996
Salomão, M. C., Heluany-Filho, M. A., Menegueti, M. G., Kraker, M. E., Martinez, R., and Bellissimo-Rodrigues, F. (2016). A randomized clinical trial on the effectiveness of a symbiotic product to decolonize patients harboring multidrug-resistant gram-negative bacilli. Rev. Soc. Bras. Med. Trop. 49, 559–566. doi: 10.1590/0037-8682-0233-2016
Sannathimmappa, M. B., Nambiar, V., and Aravindakshan, R. (2021). Antibiotics at the crossroads – do we have any therapeutic alternatives to control the emergence and spread of antimicrobial resistance? J. Educ. Health Promot. 10:438.
Scillato, M., Spitale, A., Mongelli, G., Privitera, G. F., Mangano, K., Cianci, A., et al. (2021). Antimicrobial properties of lactobacillus cell-free supernatants against multidrug-resistant urogenital pathogens. Microbiology 10:e1173. doi: 10.1002/mbo3.1173
Śliżewska, K., and Chlebicz-Wójcik, A. (2020). The in vitro analysis of prebiotics to be used as a component of a Synbiotic preparation. Nutrients 12:1272. doi: 10.3390/nu12051272
Sniffen, J. C., Mcfarland, L. V., Evans, C. T., and Goldstein, E. J. C. (2018). Choosing an appropriate probiotic product for your patient: an evidence-based practical guide. PLoS One 13:e0209205. doi: 10.1371/journal.pone.0209205
Sy, C. L., Chen, P. Y., Cheng, C. W., Huang, L. J., Wang, C. H., Chang, T. H., et al. (2022). Recommendations and guidelines for the treatment of infections due to multidrug resistant organisms. J. Microbiol. Immunol. Infect. 55, 359–386. doi: 10.1016/j.jmii.2022.02.001
Tang, H.-J., Lai, C.-C., Chen, C.-C., Zhang, C.-C., Weng, T.-C., Chiu, Y.-H., et al. (2019). Colistin-sparing regimens against Klebsiella pneumoniae carbapenemase-producing K. pneumoniae isolates: Combination of tigecycline or doxycycline and gentamicin or amikacin. J. Microbiol. Immunol. Infect. 52, 273–281. doi: 10.1016/j.jmii.2016.03.003
Wieërs, G., Verbelen, V., Van Den Driessche, M., Melnik, E., Vanheule, G., Marot, J. C., et al. (2020). Do probiotics during in-hospital antibiotic treatment prevent colonization of gut microbiota with multi-drug-resistant bacteria? A randomized placebo-controlled trial comparing saccharomyces to a mixture of lactobacillus, Bifidobacterium, and saccharomyces. Front. Public Health 8:578089.doi: 10.3389/fpubh.2020.578089
Keywords: KPC, Klebsiella pneumoniae, probiotic, prebiotic, synbiotic
Citation: Tang H-J, Chen C-C, Lu Y-C, Huang H-L, Chen H-J, Chuang Y-C, Lai C-C and Chao C-M (2022) The effect of Lactobacillus with prebiotics on KPC-2-producing Klebsiella pneumoniae. Front. Microbiol. 13:1050247. doi: 10.3389/fmicb.2022.1050247
Edited by:
Ning Dong, Soochow University, ChinaReviewed by:
Noble K. Kurian, Chandigarh University, IndiaGamal Mohamed El-Said El-Sherbiny, Al-Azhar University, Egypt
Jingpeng Yang, Nanjing Normal University, China
Qipeng Cheng, Anhui Normal University, China
Copyright © 2022 Tang, Chen, Lu, Huang, Chen, Chuang, Lai and Chao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chien-Ming Chao, Y2NtODcwOTU4QHlhaG9vLmNvbS50dw==
 Hung-Jen Tang
Hung-Jen Tang Chi-Chung Chen2
Chi-Chung Chen2 Hung-Jui Chen
Hung-Jui Chen Chih-Cheng Lai
Chih-Cheng Lai