
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 21 December 2022
Sec. Aquatic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1048619
 Diwen Liang1,2,3†
Diwen Liang1,2,3† Hailin Luo1
Hailin Luo1 Chunrong Huang1
Chunrong Huang1 Zhen Ye1
Zhen Ye1 Shuangshuang Sun1
Shuangshuang Sun1 Jiahua Dong1
Jiahua Dong1 Mingyi Liang1
Mingyi Liang1 Senjie Lin3*
Senjie Lin3* Yufeng Yang2*
Yufeng Yang2*Knowledge of in situ diet of widespread rotifers is crucial for accurately understanding the trophic position, ecological function, and adaptability to environmental changes in aquatic ecosystems. However, it is challenging to achieve the in situ diet information due to the lack of efficient and comprehensive methods. Here, we investigated the diet composition of Polyarthra in a subtropical lake using high-throughput sequencing (HTS) of a rRNA metabarcode for Polyarthra and ambient water samples. After eliminating Polyarthra sequences, a total of 159 operational taxonomic units (OTUs) from taxa in 15 phyla were detected from Polyarthra gut content samples. Most of the OTUs belong to Chlorophyta, followed by unclassified Fungi, Chrysophyta, Dinoflagellata, Ciliophora, Bacillariophyta, Cryptophyta, Arthropoda, Cercozoa, Mollusca, Apicomplexa, Haptophyta, Amoebozoa, Chordata and other eukaryotes. Our results showed that Polyarthra mainly grazed on Chlorophyta, which may result from the high relative abundance of Chlorophyta in ambient waters. In contrast, Chrysophyceae and Synurophyceae were enriched in Polyarthra’s gut, indicating that this rotifer prefers these taxa as food. Moreover, correlation analysis showed that total nitrogen, transparency, depth, Chlorophyll-a and total phosphorus were key factors for the variation of the eukaryotic community in the Polyarthra gut contents. When the concentration of nutrients in the water environment decreased, Polyarthra shifted from herbivorous feeding to more carnivorous feeding. Thus, Polyarthra is generally omnivorous but preference for Chrysophytes and Synurophytes, and it responds to the environmental changes by adopting a flexible feeding strategy. This could partly explain why the widespread rotifers have apparently wide tolerance toward spatial and environmental changes.
Rotifers, a group of essential zooplankton in aquatic ecosystems, are sensitive to changes of the environment, acting as reliable indicators of the trophic status (Sládeček, 1983; Liang et al., 2020). Rotifers connect primary producers and secondary consumers, playing an important role not only in the food chain but also the micro-food web (Arndt, 1993; Galir Balkić, 2019). Polyarthra dolichoptera and Polyarthra vulgaris, cosmopolitan rotifer species, are more tolerant to seasonal changes than other rotifers and widely dominant not only in freshwaters but also brackish waters (Liang et al., 2022). However, it is still unclear why these rotifers have a strong adaptability to environmental changes. On the one hand, their higher genetic diversity may be the reason for them to adapt to specific habitats (Obertegger et al., 2014). On the other hand, the adaptation for survival of widespread species could be attributed to their tolerances to harsh environment (e.g., thermal and toxins stress), as well as their wide food spectrum (Boersma et al., 2016; Liang et al., 2017; Lin et al., 2018).
Studying the diets of widespread species is an important approach to clarify their trophic position and ecological function in aquatic ecosystems, understanding exchanges of material and energy transfer through food webs affected by environmental changes (Boersma et al., 2016; Hu et al., 2018). The diets of primary consumers may reflect both environmental quality and their survival status. Therefore, the variations of diet composition may help us to understand their survival strategies. Many studies demonstrated that Polyarthra not only grazed on phytoplankton but also consumed ciliates (Arndt, 1993; Joaquim-Justo et al., 2004). However, its feeding preference among different prey species still needs further investigation.
Previous dietary studies on zooplankton have mainly been based on traditional methods, such as morphological identification on gut contents, feeding behavioral observation under microscope and laboratory experiments (Arndt, 1993; Drzewicki et al., 2015; Sarma et al., 2019). These approaches do not provide complete information on food spectrum of rotifers in natural waters. Although natural dietary information can be obtained from feeding behavioral observation of wild-caught zooplankton, the currently available microtechnique is challenging because some zooplanktons with strong escaping ability are hard to follow and their fragmental prey can be extremely difficult to identify (Hu et al., 2014). While information from incubation experiments has constituted the backbone of aquatic planktonic trophic ecology, it falls short in documenting the full range of the trophic linkages among the complex components in the natural environment (Hu et al., 2015). In addition, pigment analysis has been used to study the prey diversity, but it is limited to phytoplankton and are of low taxonomic resolution (Turner, 2004). The approach using fluorescent microparticles successfully estimated the predation of rotifers on ciliates but only under semi-in situ conditions (Joaquim-Justo et al., 2004). Also, stable isotope analysis of fatty acids is reliable in tracing sources of nitrogen or carbon and provide information of trophic level, but still could not depict the whole picture of diet composition (Gonçalves et al., 2012; Boersma et al., 2016).
In recent years, DNA-based methods have been widely applied to study the feeding ecology of microzooplankton due to its sensitivity, specificity and rapidness over traditional methods. The 18S ribosomal RNA gene (18S rDNA) is widely used in PCR as a reliable taxonomic marker because it consists of multiple copies in the genomes of eukaryotic organisms and the hypervariable region can be used to distinguish genetic differences among species (Hu et al., 2014). Universal 18S rDNA primers (targeting the hypervariable V4 region) have been proven as an effective and efficient method in studying the eukaryotic diversity (Cheung et al., 2010). Although clone-based sequencing has successfully explored the in situ diet of copepod, the data generated by this approach only provide general information on the structure of eukaryote communities but are not sufficient for meaningful comparisons among libraries (Cheung et al., 2010). On the contrary, high-throughput sequencing (HTS) can provide semi-quantitative information on the contribution of different prey to the diet of a consumer and detect rare prey species in gut contents (Ho et al., 2017). A weak but positive relationship between relative read abundance (eDNA) and biomass (traditional approach) has been found in recent studies of zooplankton (Ershova et al., 2021), benthic invertebrate (Klunder et al., 2022) and fish (Stoeckle et al., 2021). HTS has been successfully used to detect the food composition of invertebrates, mollusca, fish and marine mammal (Ho et al., 2017; Lin et al., 2018). Thus, it is a promising method for dietary studies of microorganisms such as rotifers.
The aims of this study were: (1) to provide more detailed information on the diet spectrum of rotifer Polyarthra, exploring the food resources of this widespread rotifer; (2) to characterize the in situ diet composition of Polyarthra and the feeding responses to environmental changes; (3) to understand the feeding strategy that enables widespread rotifers to survive and thrive under different trophic conditions.
A total of 24 samples, including 12 rotifer samples and 12 ambient water samples were collected from Lake Liuye, Changde, China (Supplementary Figure S1; Supplementary Table S1). The average interval between sampling sites was set to about four kilometers. Samples were taken on a boat at every site quarterly from December 2017 to September 2018. The ambient water samples (1 L) were collected with polyethylene bottles from the surface and subsurface layer. The water samples were filtered immediately on a 0.2 μm polycarbonate membrane (EMD Millipore GTPP04700, USA) and stored at −80°C. Rotifer samples were collected by towing a plankton net (mesh size 30 μm) horizontally at surface and subsurface depths for 5 min. To prevent changes in rotifer gut contents in the sampling process, rotifers samples were fixed on site immediately in neutral Lugol’s solution at 2% final concentration and transported in a cooler before storing at −20°C.
Water temperature (Temp), dissolved oxygen (DO), pH, and salinity, were measured using a calibrated multiprobe (YSI-Plus, USA). Water depth (Dep) was measured using a fathometer (SM-5, USA). Water transparency (SD) was measured with a Secchi disc. Chlorophyll-a (Chl-a), chemical oxygen demand (CODMn), total phosphorus (TP), ammonium nitrogen (NH4-N) and total nitrogen (TN) were determined in the laboratory following standard analytical methods (MEE, 2002).
In the laboratory, Polyathra dolichoptera and P. vulgaris were identified and 50–100 individuals of similar size were isolated with sterilized micropipettes and Sedgewick-Rafter chambers under the stereo microscope (Supplementary Figure S2). The isolated individuals were serially rinsed four times thoroughly with double distilled water under the stereo microscope, examined to ensure no attachment of other visible organisms on the body surface and appendages. All washed individuals were transferred into a 1.5 ml Eppendorf tube for DNA extraction.
The Polyarthra rotifers in the 1.5 ml EP tube were homogenized using a sterilized disposable grinding rod. Total DNA in each rotifer sample was then extracted following the HotSHOT protocol (Montero-Pau et al., 2008). Total DNA in each ambient water sample was extracted from the polycarbonate membrane using DNA Lysis buffer + protein K + CTAB + Clean & Concentrator kit (Zymo Research, USA) methods following the improved protocol (Yuan et al., 2015). The concentrations and purities of DNA were measured on a NanoDrop 2000 Spectrophotometer.
The V4 region of the 18S rRNA gene was amplified to detect the eukaryotic communities in both Polyarthra gut contents and ambient waters using universal primers 528F (5′-GCGGTAATTCCAGCTCCAA-3′) and 706R (5′-AATCCRAGAATTTCACCTCT-3; Cheung et al., 2010). Thermal cycling conditions of amplification were: 3 min of denaturation at 94°C, followed by 27 cycles of 30 s at 94°C, 30 s at 55°C, 30 s at 72°C and a final extension at 72°C for 5 min. PCR reactions were performed in triplicate 20 μl mixture containing 4 μl of 5 × FastPfu Buffer, 2 μl of 2.5 mM dNTPs, 0.8 μl of each primer (5 mM), 0.4 μl of FastPfu Polymerase and 10 ng of template DNA. The PCR products were extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluorTM-ST (Promega, USA) according to the manufacturer’s protocol. Purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina Miseq platform (Illumina, San Diego, USA) using NEXTFLEX Rapid DNA-Seq Kit. Sequencing was conducted in the Shanghai Meiji Sequencing Centre.
Raw FASTQ files were demultiplexed, quality-filtered by fastp and merged by FLASH to minimize the effects of random sequencing errors (Magoč and Salzberg, 2011). Briefly, sequences were removed if: (1) they contained ambiguous base calls, (2) their lengths were shorter than 240 bps or the average quality score were <20. After sequence screening, operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff using UPARSE (version 7.11) and chimeric sequences were identified and removed using UCHIME (Edgar, 2013). The taxonomy of each OTU representative sequence was analyzed using RDP Classifier algorithm2 against the NCBI nucleotide sequence database NT (NT_v20200327/18S_eukaryota) using confidence threshold of 0.7. Finally, the corresponding species information of each OUT was obtained.
Since this study was aimed at analyzing the composition of eukaryotes in the Polyarthra gut contents, it was necessary to remove the sequences of Polyarthra themselves from the samples before analysis. Polyarthra reads from its gut contents samples were subtracted before relative abundance (based on reads abundance) calculation. Rarefaction curves were plotted on OUT level by using Mothur for each sample (Schloss et al., 2009). Venn diagrams were generated with online tools to show the common and unique species in ambient water and Polyarthra samples.3
To calculate the differences of the eukaryotic communities between the ambient water and Polyarthra samples, the dissimilarity tests between groups were performed by the analysis of similarities (ANOSIM). In general, R > 0.75 means large difference; 0.75 > R > 0.5 means medium difference, 0.5 > R > 0.25 means small difference. Food selection of Polyarthra was estimated using the selectivity index E (Ivlev, 1961; Cotonnec et al., 2001), given by E = (ri – pi)/(ri + pi). ri is the relative abundance of one eukaryotic genus in the diet of Polyarthra sample. While pi is the relative abundance of the same eukaryotic genus in the ambient water sample. In general, E > + 0.25 indicates a preference; E < − 0.25 indicates discrimination against particular prey items; − 0.25 < E < + 0.25 indicates non-selective feeding.
A heatmap was also generated using the R ‘vegan’ package. Canonical correspondence analysis (CCA) or Redundancy analysis (RDA) was used to identify the effects of environmental factors on the in situ diet composition of Polyarthra. Whether CCA or RDA model was used was determined based on the community composition by Detrended Correspondence Analysis (DCA). If the longest gradient was >4, the unimodal method (CCA) was applied. If that value was <3, the linear method (RDA) was a better choice. All the above analyses were conducted with R (version 4.1.14) using ‘vegan’, ‘graphics’, ‘maptools’ and ‘ggplot2’ packages. The relative abundance difference and the significance level of species between ambient water and Polyarthra samples were compared by Wilcoxon ranksum test using ‘stats’ packages. The calculated value of p was gone through FDR Correction, taking FDR ≤ 0.05 as a threshold.
A phylogenetic tree was constructed based on sequences of dominant OTUs in Polyarthra samples using a Maximum Likelihood method with a bootstrap value of 1,000 displayed by MEGA X and the plot was generated on iTOL (Letunic and Bork, 2011).
Rarefaction curves for all the samples were nearly saturated (Supplementary Figure S3), suggesting sufficient sequencing depth for this study. High-throughput sequencing of 18S rRNA V4 region yielded 715,818 clean metabarcode sequences from all 12 ambient water samples. After blasting against NCBI using BLASTN, a total of 1,348 OTUs in ambient water samples were divided into five groups. The highest numbers of OTUs was observed as unclassified eukaryotes (704 OTUs), followed by phytoplankton (including dinoflagellates; 389 OTUs), fungi (156 OTUs), protists (excluding dinoflagellates; 63 OTUs), and zooplankton (36 OTUs; Figure 1A).
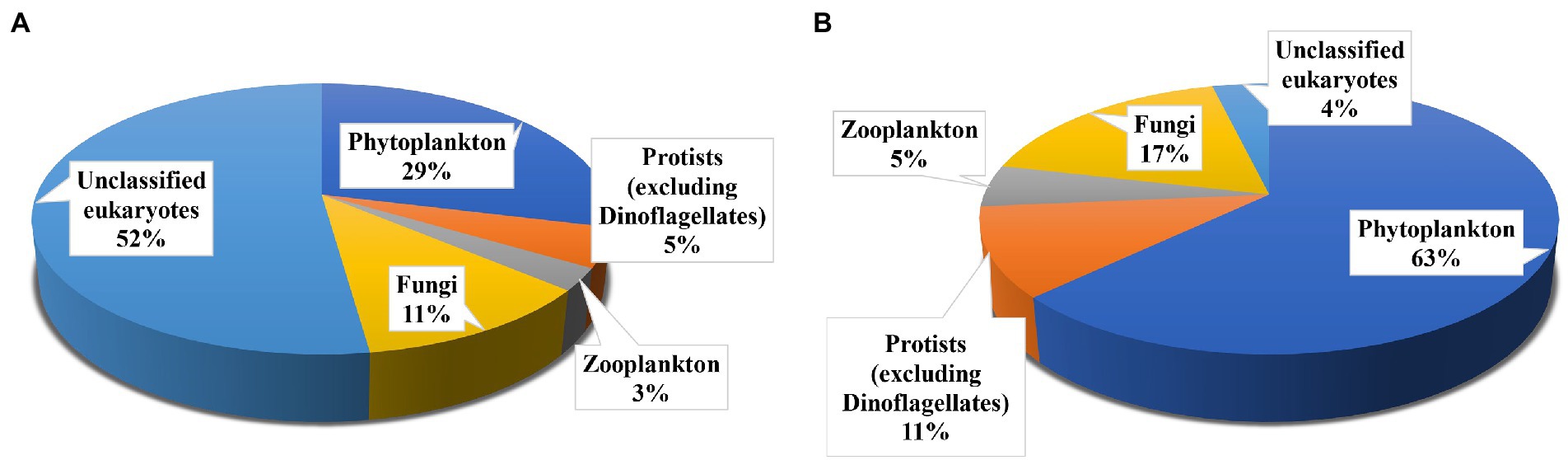
Figure 1. Operational taxonomic unit (OTU) richness of main groups in the ambient waters (A), and Polyarthra gut contents samples (B).
After eliminating Polyarthra sequences, 6,789 sequences were obtained for all 12 Polyarthra gut contents samples. A total of 159 OTUs were observed in Polyarthra gut contents. Most of the observed OTUs belonged to phytoplankton (100 OTUs), followed by fungi (28 OTUs), protozoa (17 OTUs), zooplankton (8 OTUs) and unclassified eukaryotes (6 OTUs; Figure 1B). Moreover, 129 OTUs were shared between the ambient waters and the Polyarthra gut contents samples (Supplementary Figure S4).
Phylogenetic tree of OTUs from Polyarthra gut contents samples based on Maximum Likelihood showed that the 159 OTUs sequences could be classified into 15 Phyla and 39 Classes (Figure 1B). A large number of phytoplankton taxa (100 OTUs) was detected in Polyarthra gut contents samples. Most of the observed OTUs belonged to Chlorophyta (54 OTUs), followed by unclassified Fungi (28 OTUs), Chrysophyta (17 OTUs), Dinoflagellata (10 OTUs), Ciliophora (10 OTUs). Bacillariophyta (9 OTUs), Cryptophyta (9 OTUs), Arthropoda (8 OTUs), Cercozoa (5 OTUs), Mollusca (3 OTUs), Apicomplexa (1 OTUs), Haptophyta (1 OTUs), Amoebozoa (1 OTUs), Chordata (1 OTUs) and other eukaryotes (2 OTUs).
A small number of sequences of animal and macroalgal were also detected, including fishes of Cyprinus, molluscs of Tubificidae, Arionoidea, large zooplankton of Sinocalanus, Pseudodiaptomus, Eucyclops, Thermocyclops, and algae from the genus Ulva (Figure 2).
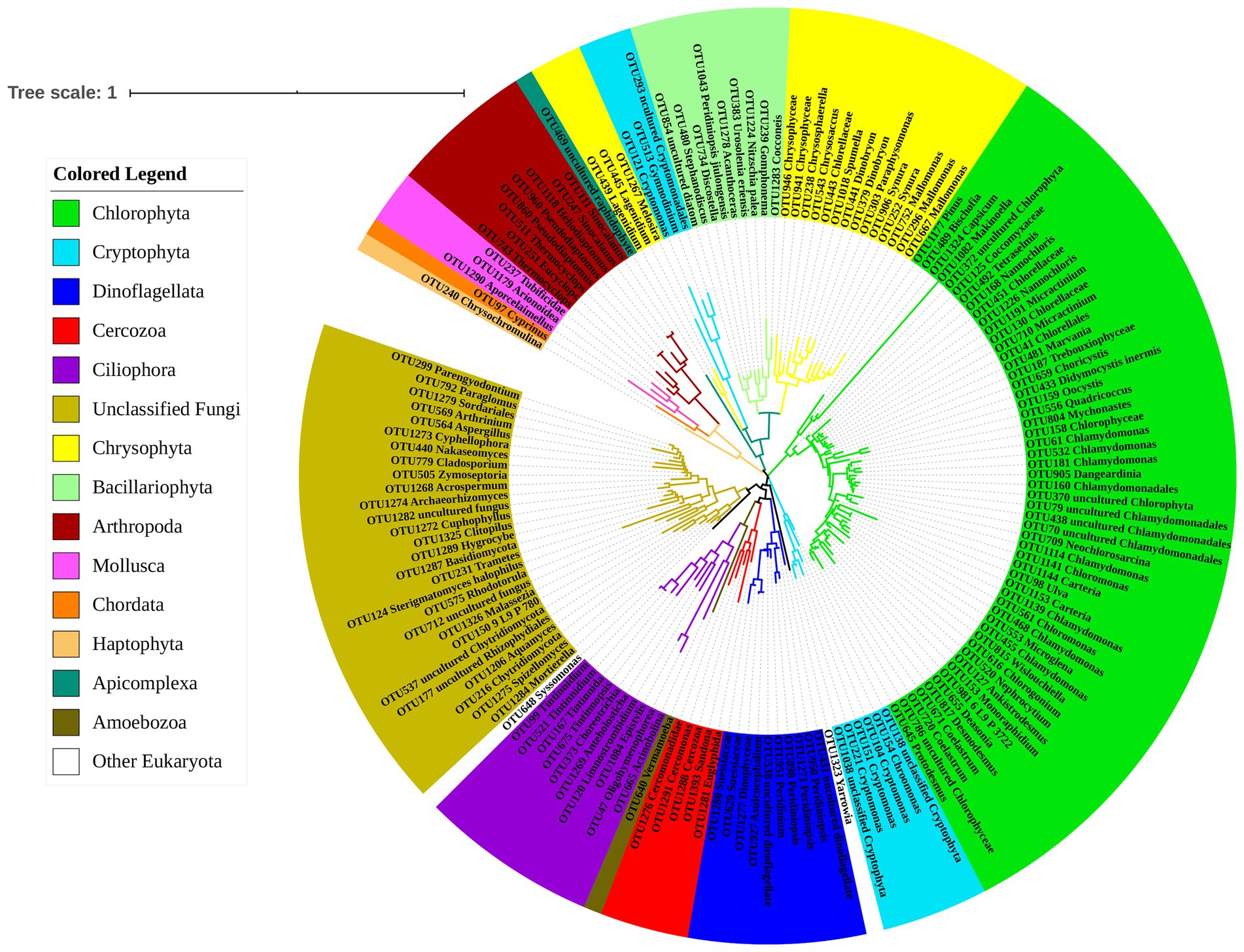
Figure 2. Phylogenetic tree of OTUs from the samples of Polyarthra gut contents based on Maximum Likelihood. The color coding of OTUs represents different phyla.
For ambient water samples, the most abundant class was Chlorophyceae, with the average relative abundance of 28.1%. The next Class was Cryptophyceae, with an average relative abundance of 19.8%, followed by Trebouxiophyceae (10.8%), Maxillopoda (9.1%), Dinophyceae (5.4%) and Bacillariophyceae (4.1%). For Polyarthra gut contents samples, the most abundant class was also Chlorophyceae, with the average relative abundance of 16%. The next Class was Oligohymenophorea, with an average relative abundance of 14.6%. In addition, Dinophyceae (11.8%), Spirotrichea (11.4%), Cryptophyceae (10.2%) were also abundant (Figure 3).
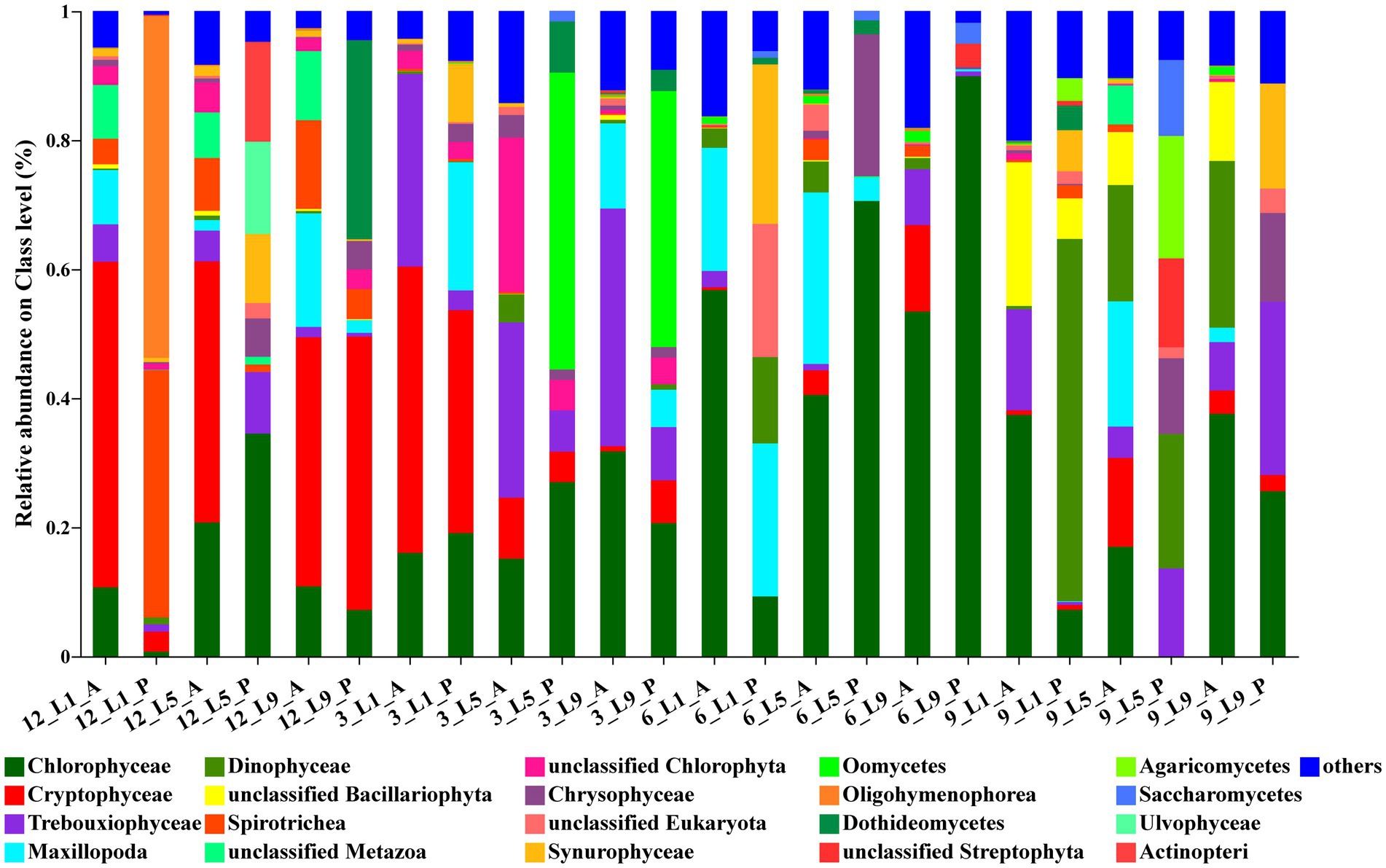
Figure 3. Relative abundance (based on reads abundance) of the ambient water samples and the Polyarthra samples on class level (top 20). Polyarthra reads from the 12 gut contents samples were removed before calculation. A: the ambient water samples; P: the Polyarthra gut contents samples; 12: December; 3: March; 6: June; 9: September.
The seasonal variations of eukaryotic composition of water environment at the three sites were consistent (Figure 3). Cryptophyceae dominated in the ambient water in December 2017, with the relative abundances reaching 39%–51%. The relative abundances of Trebouxiophyceae (27%–37%) increased in March 2018. While in June, Chlorophyceae dominated in the ambient water, with the relative abundances reaching 41%–57%. In September, the relative abundances of Chlorophyceae decreased to 17%–38%, and those of Bacillariophyceae increased to 8%–22%. The composition of eukaryotic phytoplankton community determined by metabarcoding was consistent with the results based on the morphological approach (Supplementary Figure S5).
The average relative abundance of Chlorophyta was the highest in the Polyarthra gut content samples, reaching 21.8%. However, the variations of eukaryotic composition of the Polyarthra gut contents were different among the three sites. In general, the relative abundance of Cryptophyceae and Oomycetes in the Polyarthra gut contents in December and March were greater than those in June and September. While Chlorophyceae dominated the Polyarthra gut contents in all seasons, and the relative abundance of Dinophyceae, Trebouxiophyceae and Chrysophyceae in the Polyarthra gut contents increased in September (Figure 3).
The results of the community similarity analysis (ANOSIM) on the class level showed that there was small difference of eukaryotic communities between the ambient waters and the Polyarthra gut contents (R = 0.35, p < 0.01). The samples collected from hot season (Jun and Sep) mostly stayed together, while the ambient water samples and the Polyarthra gut contents samples were mostly together (Figure 4). Moreover, phytoplankton such as Chlorophyceae, Trebouxiophyceae, Crytophyta, and Dinophyceae dominated not only in the ambient waters but also in the Polyarthra gut contents. These results indicated that the eukaryotic community in the Polyarthra gut contents varied with the plankton community in the ambient waters.
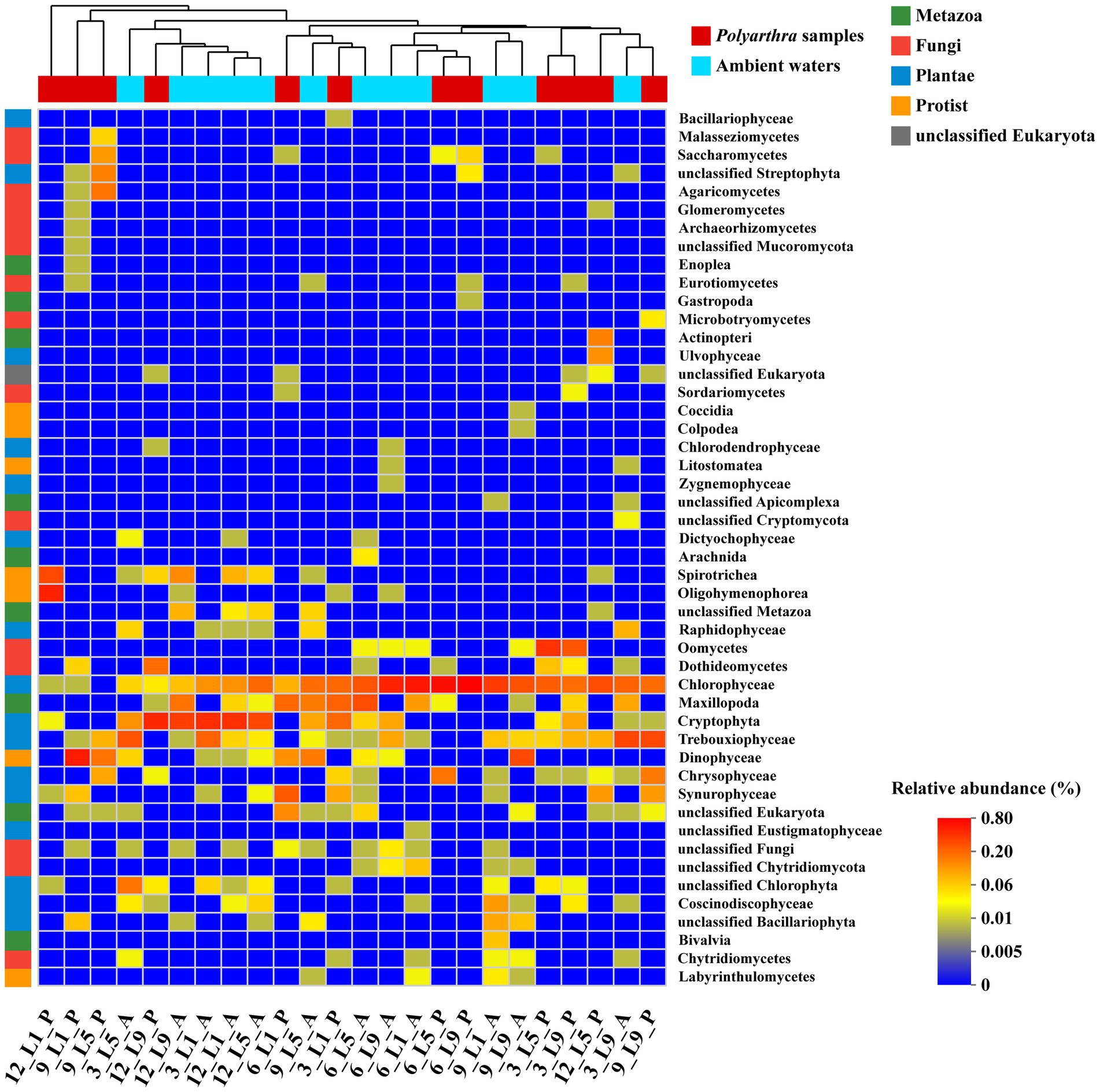
Figure 4. Heatmap analysis of eukaryotic communities between ambient water (A) and Polyarthra (P) based on Bray - Curtis distance (Top 50 abundant OTUs on Class level). Samples and taxa were automatically organized by hierarchical clustering. A: the ambient water samples; P: the Polyarthra gut contents samples; 12: December; 3: March; 6: June; 9: September.
The Wilcoxon rank-sum test was used to analyze the significant difference of species composition between the Polyarthra gut contents and the ambient water samples (Figure 5). Chlorophyceae, Cryptophyceae, Trebouxiophyceae, Maxillopoda, Dinophyceae dominated in both the ambient waters and the Polyarthra gut contents. The higher relative abundances of Dinophyceae, Oomycetes, Spirotrichea, Oligohymenophorea, Chrysophyceae and Synurophyceae were found in the Polyarthra gut contents than in the ambient water samples. In addition, the relative abundances of Chrysophyceae and Synurophyceae in the samples of the Polyarthra gut contents were significantly greater than those in ambient waters (p < 0.05; Figure 5).
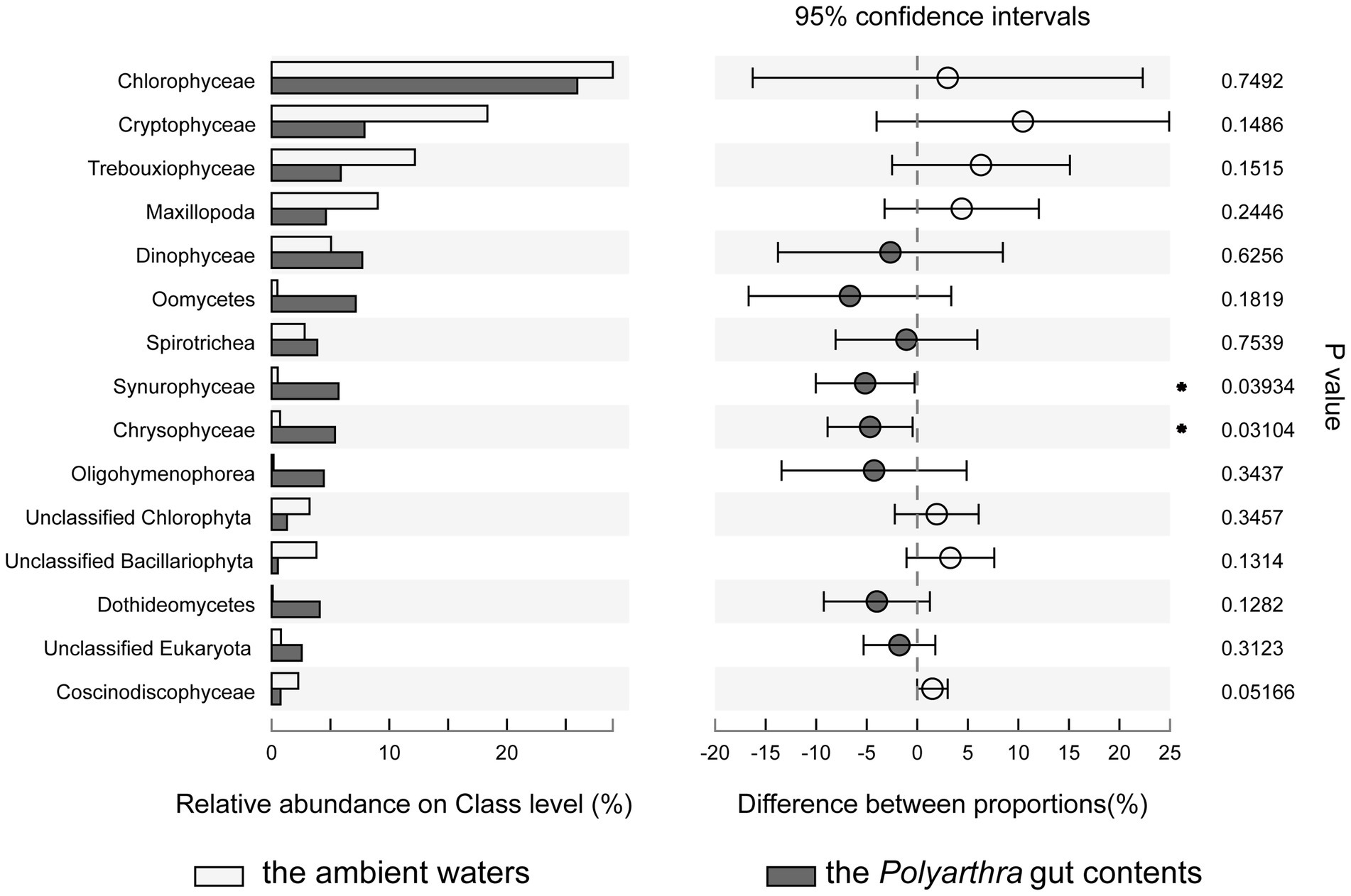
Figure 5. Comparison of the differences in the average relative abundance of eukaryotes on class level (top 15) between the Polyarthra gut contents and the ambient water samples (*p < 0.05; Wilcoxon rank-sum test).
The E values (Table 1) and the significant differences of eukaryotic compositions between the Polyarthra gut contents and the ambient water samples (Figure 5) indicated a selective feeding on Chrysophyceae and Synurophyceae. Whereas the Chrysophyceae and Synurophyceae were in relatively low concentrations in ambient waters, they were enriched in Polyarthra diets. By contrast with Chrysophyceae and Synurophyceae, the selectivity index showed a non-selective feeding on Chlorophyceae, Cryptophyceae and Trebouxiophyceae for Polyarthra, as they were dominant phytoplankton in both the Polyarthra gut contents and the ambient waters (Figure 5; Table 1).
As the longest gradient of DCA for the ambient water samples was 1.6, a RDA model was used to estimate the relationship between eukaryotic communities of the ambient waters and the environmental factors. Since the longest gradient of DCA for Polyarthra samples was 4.4, CCA was a better choice. For RDA, the first two ordinate axes explained 67% of the eukaryotes-environment variability in the ordination of environmental variables (Figure 6A; Supplementary Table 2). After forward selection with Monte Carlo permutation tests, only temperature, NH4-N, transparency, Chlorophyll-a, depth and total phosphorus were significant contributors to the variation of the eukaryotic community in the ambient waters.
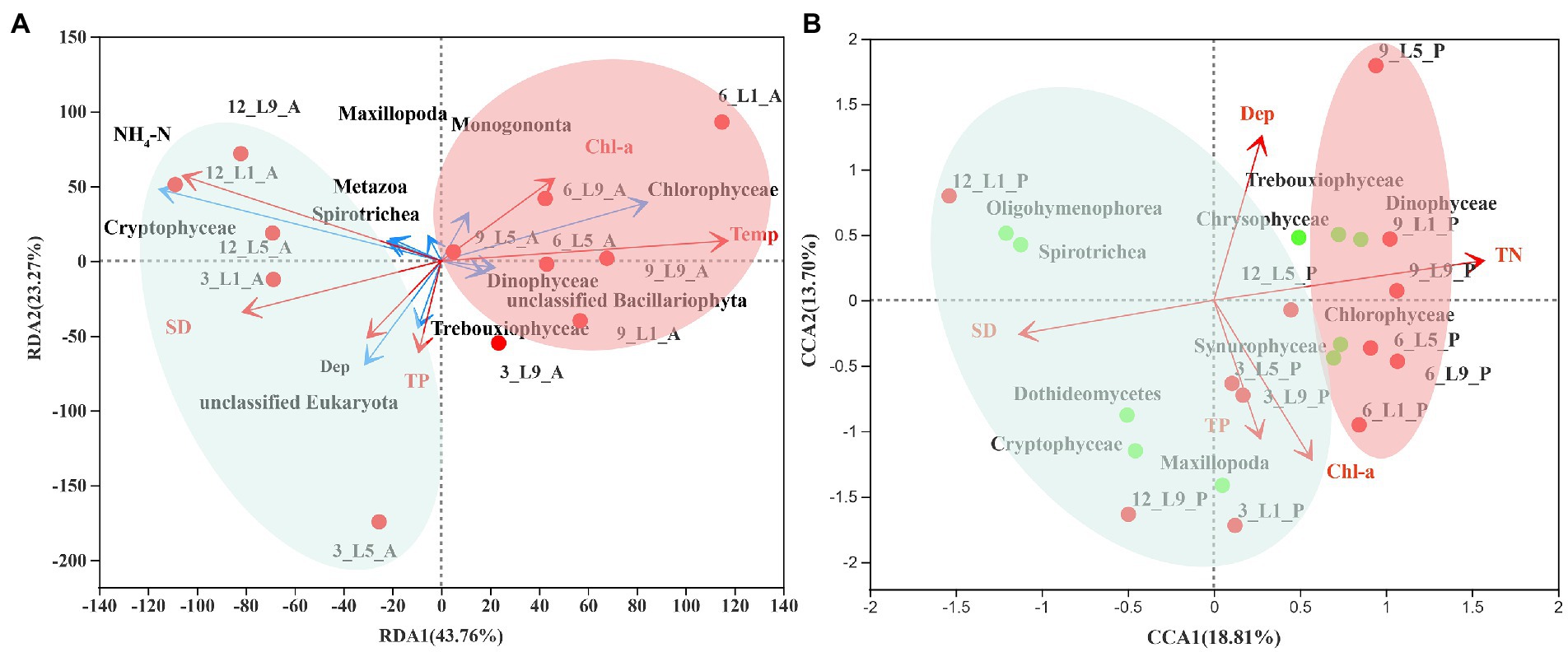
Figure 6. Restrictive ordination analysis of relationship between eukaryotic communities and environmental factors. (A) RDA of ambient water samples: Relative abundance of top 10 eukaryotes (blue arrows), environmental variables (red arrows). (B) CCA of Polyarthra samples: Relative abundance of top 10 eukaryotes. 12 and 3: December and March (cyan circle); 6 and 9: June and September (red circle).
For CCA, the first two ordinate axes explained 33% of the eukaryotes-environment variability in the ordination of environmental variables (Figure 6B; Supplementary Table S3). After forward selection with Monte Carlo permutation tests, only total nitrogen, transparency, depth, Chlorophyll-a and total phosphorus were significant contributors to the variation of the eukaryotic community in the Polyarthra gut contents.
A total of 159 OTUs of eukaryotes belonging to phytoplankton, metazoans, protozoans, fungi and other unclassified eukaryotes were observed in Polyarthra gut contents by HTS. The in situ diets confirmed the omnivorous feeding of Polyarthra, in accordance with previous laboratory experiments (Arndt, 1993; Joaquim-Justo et al., 2004; Espinosa-Rodríguez et al., 2021). Arndt (1993) summarized rotifers diets using traditional observation and found that phytoplankton (e.g., Chlorophyta, Cryptophyta), protozoa (e.g., Ciliates, Flagellates, Amoebas, Rhizopos), fungi (e.g., yeasts), bacteria (e.g., Aerobacter) and organic detritus were food resources for rotifers. Laboratory investigations showed that Polyarthra mainly feed on edible algae, especially Chlamydomonas (Gilbert and Bogdan, 1981; Wen et al., 2011). Also, it took advantage of autotrophic and heterotrophic flagellates such as Chilomonas, Euglena, Bodo, and Cyathomonas (Buikema et al., 1977; Pourriot, 1977). In the present study, 11 OTUs of different Chlamydomonas and 17 OTUs of protozoa (e.g., Vermamoeba, Tintinnina, Raphidophyte) were detected by HTS in the gut content of Polyarthra. This molecular evidence supported the notion that protozoa are important food sources for rotifers. This implies that rotifers are not only micrograzers but also predators of micro-food web and that, as proposed in the 20th-century (Arndt, 1993).
In the view of functional groups, Polyarthra is considered as raptorial and macrofilter-feeder, since it shows active grasping and piercing actions to catch single food items (Obertegger et al., 2011). However, there are few reports about rotifers feeding on crustaceans, mollusks, macrophytes and so on. Many unexpected prey taxa that were overlooked by microscopic observation could be detected from gut contents through HTS (Lin et al., 2018). A small number of sequences of fishes as Cyprinus, molluscs as Tubificidae, Arionoidea, arthropods as Sinocalanus, Pseudodiaptomus, Eucyclops, Thermocyclops, macroalgaes as Ulva were detected in the Polyarthra gut contents in the present study. It was also reported that copepods, the macrozooplankton, feed on metazoans that larger than itself such as Hydrozoans, Ctenophores, Arrow worms, Echinoderms, and Tunicates in the form of detritus and eggs (Ho et al., 2017; Yi et al., 2017; Xu et al., 2020). Since Polyarthra is a micro-zooplankton with the body length ranges from 90 to 245 μm, it is more likely to feed on the eggs and gametes than adults of arthropods, fishes and macroalgae. Besides the form of eggs or gametes, metazoans could also be ingested in the form of organic particles/detritus (Yi et al., 2017). The energy supplied by a metazoan diet with high levels of lipids and proteins, may be more accessible and easily utilized by zooplankton than plant food items (Hu et al., 2018). Additionally, Polyarthra usually shows significantly positive correlation with CODMn and thrives in habitats with higher nutrient content and lower transparency, supporting that it inclines to consume detritus particles (Ji et al., 2013; Liang et al., 2020).
Macroalgae is one of the potential food sources for Polyarthra. Microscopic observation and molecular analysis have shown that heterotrophic ciliates can acquire Ulva sequence by ingesting their gametes, chloroplasts and detritus (McManus et al., 2004). Also, copepods could use plant detritus as supplementary food sources when phytoplankton production is limited (Hu et al., 2015). It is possible that Polyarthra could take advantage of the gametes or detritus of macroalgae by adopting an “opportunistic feeding.”
Twenty eight OTUs of fungi, belonging to Chytridiomycota, Ascomycota, Basidiomycota and Mucoromycota were detected in the Polyarthra samples. Fungi were also found in the diet of European eel larvae and in the diet of calanoid copepods through molecular approach (Riemann et al., 2010; Xu et al., 2020). As an important component of microorganisms in aquatic ecosystems, most fungi are either parasitic or symbiotic to microorganisms including zooplankton, or attach to nutrient-rich sediments. Only a minority are free-living (Richards et al., 2012). So far, the relationship between fungi and zooplankton is still unclear (Xu et al., 2020). Thus, the fungi detected in the Polyarthra samples may be either parasites or constituted the diet of Polyarthra. Further studies combining scanning electron microscopy and fluorescence tracing are needed to confirm the trophic relationship between fungi and Polyarthra. In any case, this study documented the dietary information of natural rotifer Polyarthra and expanded the knowledge of their in situ food resources in detail.
Our results showed that Polyarthra had a high ingestion on Chlorophyta in this river–lake ecosystem. It seems that Chlorophyceae is the staple diet for rotifers. Chlorella, Scenedesmus and Chlamydomonas have been considered the most suitable food for the laboratory cultured rotifers (Gilbert and Bogdan, 1981; Geng and Xie, 2008; Sun et al., 2020). However, the E values and the differences of average relative abundances in the present study suggests that Polyarthra mainly grazed on Chlorophyta, which may result from its “opportunistic feeding” strategy. As Chlorophyceae, Cryptophyceae, Trebouxiophyceae, Dinophyceae dominated in the ambient waters, they became more readily accessible to Polyarthra than other food items. On the contrary, Polyarthra exhibited a high preference for Chrysophyceae and Synurophyceae.
The prey preference for macro-zooplankton could be related to the nutritional state of the food items (Yi et al., 2017). Dinoflagellates were more abundant in the copepod diet compared to diatoms, because diatoms were of lower nutritional value than dinoflagellates (Jones and Flynn, 2005). Isochrysis and Mallomonas, belonging to Chrysophyceae, contain higher levels of unsaturated fatty acid (e.g., DHA, EPA) than many other phytoplanktons, so they have been used as food for laboratory cultured Scallops and zooplankton (Seychelles et al., 2009; Bi et al., 2014; Freites et al., 2016). Mallomonas, the high-quality Chrysophyceae, can support high population growth rates and offspring production in zooplankton (Taipale et al., 2019). Since zooplankton are known to preferentially feed on high nutritional quality algae (Taipale et al., 2019), the prey preference of Polyarthra can be explained by the nutritional value of the food items.
Both active and passive feeding behavior of zooplankton could be related to the prey size-selectivity (Chen et al., 2021). Polyarthra showed preference toward food items with a particle size ranging from 1 to 40 μm (Arndt, 1993). Most Chlorophyceae in the in situ diet of Polyarthra are well within nano particle size, such as Chlorella (5–20 μm), Chlamydomonas (3–10 μm) and Monoraphidium (7–20 μm). In addition, Synura (2–40 μm), Chromulina (3–20 μm), Mallomonas (2–16 μm) Paraphysomonas (8–17 μm) belonging to Chrysophyta and Chroomonas (4–20 μm) belonging to Cryptophyta are also in micro to nano particle size (Hu and Wei, 2006), which are suitable for direct ingestion by Polyarthra.
Copepods can regulate their food intake according to their energy demands when exposed to warming environments. Their feeding preference shift with temperature, with a change from a diatom-dominated diet at 29°C to a metazoan-dominated diet at 35°C (Hu et al., 2018). However, our study indicates that Polyarthra adjusted their feeding strategy according to food availability by applying an “opportunistic feeding” strategy. RDA analysis showed that the eukaryotic community of the ambient waters presented a seasonal differentiation pattern and was controlled by temperature and trophic status, which was consistent with the previous studies of zooplankton and phytoplankton communities in Lake Liuye (Liu, 2019; Liang et al., 2020). CCA analysis showed that the eukaryotic community of the Polyarthra gut contents also exhibited a distinct pattern of seasonal differentiation, which was consistent with that of the ambient waters. The dietary composition of Polyarthra seems to be regulated by environmental factors related to trophic status.
The “opportunistic feeding” strategy is also common for macro-zooplankton as copepod in food-limited conditions (Hu et al., 2020). Dominant species can better their survival by widening the choice of potential food resources (Xu et al., 2020). They are capable to regulate their feeding, by exhibiting a rhythm of herbivorous feeding in midday and carnivorous feeding in morning and night, to better coordinate with other competitors for utilization of food resources (Hu et al., 2020). For example, the genus Centropages was more omnivorous-carnivorous and consistently displayed selection for large motile prey when the phytoplankton density was low in the outfall region (Xu et al., 2020). In addition, they can use terrigenous detritus as supplementary food sources when phytoplankton production is limited (Hu et al., 2015). Polyarthra mainly grazed on phytoplankton and consumed the animal-derived prey (metazoan detritus or protozoan) as supplementary food sources. In the present study, the spatial–temporal pattern of the eukaryotic community in the Polyarthra gut contents varied along an increasing gradient of the trophic status (including TN, TP, Chl-a, and SD) in ambient waters (Figure 6B). Moreover, Polyarthra mainly feed on Chlorophyceae Trebouxiophyceae, Dinophyceae, Maxillopoda, Chrysophyceae and Synurophyceae when the trophic status was relatively high. To adopt to the relatively lower trophic status, Polyarthra had probably shifted from herbivorous feeding to relatively carnivorous (Oligohymenophorea and Spirotrichea) feeding. Our study indicates that Polyarthra is omnivorous with preference and responds to the environmental changes by adopting a flexible feeding strategy. This could partly explain why the cosmopolitan rotifers have apparently wide tolerance toward spatial and environmental changes.
Carcasses of metazoan will release a continuum of particulate organic matter (POM) and dissolved organic matter (DOM), a complex suite of polymers and molecules, if they are not eaten by predators. These organic matters are further decomposed by bacteria and archaea and converted into inorganic nutrients (e.g., C, N, P, Si and Fe) which can be used by primary producers (Worden et al., 2015). Utilization of DOM and POM by zooplankton might increase the efficiency of the upward trophic transfer of matter and energy flow through secondary consumer under food-limited conditions, and thereby promotes the efficiency of aquatic ecosystem cycling (Hu et al., 2018).
Changes of the diet composition of zooplankton play a key role in the transfer of materials and energy along the food chain (Liu et al., 2006). Phytoplankton density being affected by a feedback mechanism of zooplankton consumer grazing is termed consumer-driven nutrient recycling (Sterner et al., 1995; Trommer et al., 2012). The composition of zooplankton and their excretions can regulate and stimulate the phytoplankton growth and biomass accumulation under N or P limitations (Trommer et al., 2012). Zooplankton feeding on protozoa and metazoan detritus could obtain higher carbon than herbivorous feeding alone. If the POM content of phytoplankton in the water environment is insufficient to satisfy their daily carbon requirements (19 μg), the food intake of protozoa and organic detritus increases (David et al., 2006). Thus, when phytoplankton density was low in Lake Liuye, the high relative abundance of protozoans in the Polyarthra gut contents may be related to their carbon requirements. The carnivorous and detritus feeding of zooplankton ensures that secondary consumer can still contribute to the pathways of aquatic carbon flow, through the food chain while under low primary production.
Because feeding strategy of most zooplankton adaptively adjusts with the environment, knowledge of the in situ diet composition is crucial for accurately clarifying their ecological function in the aquatic ecosystem, understanding the prey diversity and the ingestion rate (Lin et al., 2018). Our study indicates that HTS is a promising approach that provides us a more comprehensive perspective and more details on the diet composition of microorganism than by microscopic observations. However, to further explore the role and mechanism of zooplankton in biogeochemical cycles, both molecular evidence and microscopy should be combined to investigate the dietary assimilation, release, and physiological turnover rates of C, N and P in zooplanktons under different conditions.
1. Our HTS data has revealed the dietary information of natural rotifer Polyarthra and expanded the knowledge of their in situ food resources. A total of 159 OTUs of eukaryotes belonging to phytoplankton, metazoans, protozoans, fungi and other unclassified eukaryotes were detected in Polyarthra gut contents.
2. Diet composition indicated that Polyarthra is omnivorous with preference and responds to the environmental changes by adopting a flexible feeding strategy. Polyarthra mainly grazed on Chlorophyta, which may result from its “opportunistic feeding” strategy. On the contrary, it exhibited a high preference for Chrysophyceae and Synurophyceae.
3 When the concentration of nutrients in the water environment decreased, Polyarthra may shift from herbivorous feeding to relatively carnivorous feeding. This could partly explain why the cosmopolitan rotifers have apparently wide tolerance toward spatial and environmental changes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found at: https://www.ncbi.nlm.nih.gov/, BioProject No. PRJNA611635.
DL conceived the ideas and led the experiments, data analysis, and paper writing. SL guided paper revising, data analysis, and interpretation. YY were project administrator and supported the research. HL conducted the graphic production. Other authors supported the paper revising and editing. DL, HL, CH, ZY, SS, JD, ML, SL, and YY were critically involved in writing the manuscript. All authors contributed to the article and approved the submitted version.
This work received funding from the Guangzhou Municipal Science and Technology Project (No. 202201010592 and 202201010045), the National Natural Science Foundation of China (41673080), and the Fundamental Research Funds for the Central Public Welfare Research Institutes (PM-zx703-202207-267 and PM-zx703-202207-268) for financial support.
The paper benefited greatly from the comments on revision by Dajun Qiu, Qing Wang, and Xuejia He. Special thanks to the reviewers for their helpful comments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1048619/full#supplementary-material
Arndt, H. (1993). Rotifers as predators on components of the microbial web (bacteria, heterotrophic flagellates, ciliates)—a review. Hydrobiologia 255–256, 231–246. doi: 10.1007/BF00025844
Bi, R., Arndt, C., and Sommer, U. (2014). Linking elements to biochemicals: effects of nutrient supply ratios and growth rates on fatty acid composition of phytoplankton species. J. Phycol. 50, 117–130. doi: 10.1111/jpy.12140
Boersma, M., Mathew, K. A., Niehoff, B., Schoo, K. L., Franco-Santos, R. M., and Meunier, C. L. (2016). Temperature driven changes in the diet preference of omnivorous copepods: no more meat when it’s hot? Ecol. Lett. 19, 45–53. doi: 10.1111/ele.12541
Buikema, A., Cairns, J., Edmunds, P., and Krakauer, T. (1977). Culturing and Ecology Studies of the rotifer Polyarthra vulgaris. Washington, DC: Environmental Protection Agency.
Chen, M., Si, Y., Han, L., Liu, X., Huang, B., and Kang, C. (2021). Effect of prey selectivity and trophic cascades induced by mesozooplankton on the dynamics of phytoplankton. Mar. Ecol. Prog. Ser. 662, 35–51. doi: 10.3354/meps13627
Cheung, M. K., Au, C. H., Chu, K. H., Kwan, H. S., and Wong, C. K. (2010). Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing. ISME J. 4, 1053–1059. doi: 10.1038/ismej.2010.26
Cotonnec, G., Brunet, C., Sautour, B., and Thoumelin, G. (2001). Nutritive value and selection of food particles by copepods during a spring bloom of Phaeocystis sp. in the English Channel, as determined by pigment and fatty acid analyses. J. Plankton Res. 23, 693–703. doi: 10.1093/plankt/23.7.693
David, V., Sautour, B., Galois, R., and Chardy, P. (2006). The paradox high zooplankton biomass–low vegetal particulate organic matter in high turbidity zones: what way for energy transfer? J. Exp. Mar. Biol. Ecol. 333, 202–218. doi: 10.1016/j.jembe.2005.12.045
Drzewicki, A., Kowalska, E., Pajdak-Stós, A., Fiałkowska, E., Kocerba-Soroka, W., Sobczyk, Ł., et al. (2015). Experimental attempt at using Lecane inermis rotifers to control filamentous bacteria Eikelboom type 0092 in activated sludge. Water Environ. Res. 87, 205–210. doi: 10.2175/106143015X14212658613037
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Ershova, E. A., Wangensteen, O. S., Descoteaux, R., Barth-Jensen, C., and Præbel, K. (2021). Metabarcoding as a quantitative tool for estimating biodiversity and relative biomass of marine zooplankton. ICES J. Mar. Sci. 78, 3342–3355. doi: 10.1093/icesjms/fsab171
Espinosa-Rodríguez, C. A., Sarma, S. S. S., Sarma, N., and Wallace, R. L. (2021). Substratum selection and feeding responses influence the demography of the sessile rotifer Cupelopagis vorax (Collothecacea: Atrochidae). Int. Rev. Hydrobiol. 106, 18–28. doi: 10.1002/iroh.202002051
Freites, L., Lodeiros, C., Guevara, M., Jordan, N., Maeda-Martínez, A. N., and Himmelman, J. H. (2016). Fatty acids profiles as selecting criteria of microalgal diets used for hatchery-rearing of the tropical scallop Euvola ziczac. Aquac. Res. 47, 2670–2676. doi: 10.1111/are.12695
Galir Balkić, A. (2019). The importance of environmental differences in the structuring of rotifer functional diversity. J. Limnol. 78, 284–295. doi: 10.4081/jlimnol.2019.1903
Geng, H., and Xie, P. (2008). Experimental studies on the effects of toxic Microcystis aeruginosa PCC7820 on the survival and reproduction of two freshwater rotifers Brachionus calyciflorus and Brachionus rubens. Ecotoxicology 17, 709–715. doi: 10.1007/s10646-008-0219-8
Gilbert, J. J., and Bogdan, K. G. (1981). Selectivity of Polyarthra and Keratella for flagellate and aflagellate cells. SIL Proc. 21, 1515–1521. doi: 10.1080/03680770.1980.11897224
Gonçalves, A. M. M., Azeiteiro, U. M., Pardal, M. A., and De Troch, M. (2012). Fatty acid profiling reveals seasonal and spatial shifts in zooplankton diet in a temperate estuary. Estuar. Coast. Shelf Sci. 109, 70–80. doi: 10.1016/j.ecss.2012.05.020
Ho, T. W., Hwang, J.-S., Cheung, M. K., Kwan, H. S., and Wong, C. K. (2017). DNA-based study of the diet of the marine calanoid copepod Calanus sinicus. J. Exp. Mar. Biol. Ecol. 494, 1–9. doi: 10.1016/j.jembe.2017.04.004
Hu, S., Guo, Z., Li, T., Carpenter, E. J., Liu, S., and Lin, S. (2014). Detecting in situ copepod diet diversity using molecular technique: development of a copepod/symbiotic ciliate-excluding eukaryote-inclusive PCR protocol. PLoS One 9:e103528. doi: 10.1371/journal.pone.0103528
Hu, S., Guo, Z., Li, T., Xu, C., Huang, H., Liu, S., et al. (2015). Molecular analysis of in situ diets of coral reef copepods: evidence of terrestrial plant detritus as a food source in Sanya Bay, China. J. Plankton Res. 37, 363–371. doi: 10.1093/plankt/fbv014
Hu, S., Li, T., Liu, S., and Huang, H. (2020). Dietary separation between co-occurring copepods in a food-limited tropical coral reef of the Sanya Bay. Acta Oceanol. Sin. 39, 65–72. doi: 10.1007/s13131-020-1583-3
Hu, S., Liu, S., Wang, L., Li, T., and Huang, H. (2018). Feeding response of the tropical copepod Acartia erythraea to short-term thermal stress: more animal-derived food was consumed. PeerJ 6:e6129. doi: 10.7717/peerj.6129
Hu, H. J., and Wei, X. Y. (2006). The Freshwater Algae of China—Systematics, Taxonomy and Ecology. Beijing: Science Press.
Ivlev, V. S. (1961). Experimental ecology of the feeding of fishes (Scott D, trans.) New Haven, CT: Yale University Press.
Ji, G., Wang, X., and Wang, L. (2013). Planktonic rotifers in a subtropical shallow Lake: succession, relationship to environmental factors, and use as bioindicators. Sci. World J. 2013, 1–14. doi: 10.1155/2013/702942
Joaquim-Justo, C., Detry, C., Caufman, F., and Thomé, J.-P. (2004). Feeding of planktonic rotifers on ciliates: a method using natural ciliate assemblages labelled with fluorescent microparticles. J. Plankton Res. 26, 1289–1299. doi: 10.1093/plankt/fbh120
Jones, R. H., and Flynn, K. J. (2005). Nutritional status and diet composition affect the value of diatoms as copepod prey. Science 307, 1457–1459. doi: 10.1126/science.1107767
Klunder, L., Bleijswijk, J. D. L., Kleine Schaars, L., Veer, H. W., Luttikhuizen, P. C., and Bijleveld, A. I. (2022). Quantification of marine benthic communities with metabarcoding. Mol. Ecol. Resour. 22, 1043–1054. doi: 10.1111/1755-0998.13536
Letunic, I., and Bork, P. (2011). Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
Liang, D., McManus, G. B., Wang, Q., Sun, X., Liu, Z., Lin, S., et al. (2022). Genetic differentiation and phylogeography of rotifer Polyarthra dolichoptera and P. vulgaris populations between southeastern China and eastern North America: high intercontinental differences. Ecol. Evol. 12:e8912. doi: 10.1002/ece3.8912
Liang, Y., Ouyang, K., Chen, X., Su, Y., and Yang, J. (2017). Life strategy and grazing intensity responses of Brachionus calyciflorus fed on different concentrations of microcystin-producing and microcystin-free Microcystis aeruginosa. Sci. Rep. 7:43127. doi: 10.1038/srep43127
Liang, D., Wang, Q., Wei, N., Tang, C., Sun, X., and Yang, Y. (2020). Biological indicators of ecological quality in typical urban river-lake ecosystems: the planktonic rotifer community and its response to environmental factors. Ecol. Indic. 112:106127. doi: 10.1016/j.ecolind.2020.106127
Lin, X., Hu, S., Liu, S., and Huang, H. (2018). Unexpected prey of juvenile spotted scat (Scatophagus argus) near a wharf: the prevalence of fouling organisms in stomach contents. Ecol. Evol. 8, 8547–8554. doi: 10.1002/ece3.4380
Liu, C. (2019). Characteristics of phytoplankton community structure and water environment in Liuye Lake and its adjacent waters in Changde (in Chinese). Jinan University, Master’s thesis. Available at: https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=CMFD202101&filename=1020604815.nh
Liu, S., Wang, W., and Huang, L. (2006). Phosphorus dietary assimilation and efflux in the marine copepod Acartia erythraea. Mar. Ecol. Prog. Ser. 321, 193–202. doi: 10.3354/meps321193
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
McManus, G. B., Zhang, H., and Lin, S. (2004). Marine planktonic ciliates that prey on macroalgae and enslave their chloroplasts. Limnol. Oceanogr. 49, 308–313. doi: 10.4319/lo.2004.49.1.0308
MEE (2002). Environmental Quality Standards for Surface Water (GB3838-2002). Beijing, China: The Ministry of Ecology and Environmental Protection of the People’s Republic of China.
Montero-Pau, J., Gómez, A., and Muñoz, J. (2008). Application of an inexpensive and high-throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs: rapid DNA extraction for diapausing eggs. Limnol. Oceanogr. Methods 6, 218–222. doi: 10.4319/lom.2008.6.218
Obertegger, U., Flaim, G., and Fontaneto, D. (2014). Cryptic diversity within the rotifer Polyarthra dolichoptera along an altitudinal gradient. Freshw. Biol. 59, 2413–2427. doi: 10.1111/fwb.12447
Obertegger, U., Smith, H. A., Flaim, G., and Wallace, R. L. (2011). Using the guild ratio to characterize pelagic rotifer communities. Hydrobiologia 662, 157–162. doi: 10.1007/s10750-010-0491-5
Pourriot, R. (1977). Food and feeding habits of Rotifera. Archiv für Hydrobiologie-Beiheft Ergebnisse der Limnologie 8, 243–260.
Richards, T. A., Jones, M. D. M., Leonard, G., and Bass, D. (2012). Marine fungi: their ecology and molecular diversity. Annu. Rev. Mar. Sci. 4, 495–522. doi: 10.1146/annurev-marine-120710-100802
Riemann, L., Alfredsson, H., Hansen, M. M., Als, T. D., Nielsen, T. G., Munk, P., et al. (2010). Qualitative assessment of the diet of European eel larvae in the Sargasso Sea resolved by DNA barcoding. Biol. Lett. 6, 819–822. doi: 10.1098/rsbl.2010.0411
Sarma, S. S. S., Miracle, M. R., Nandini, S., and Vicente, E. (2019). Predation by Acanthocyclops americanus (Copepoda: Cyclopoida) in the hypertrophic shallow waterbody, Lake Albufera (Spain): field and laboratory observations. Hydrobiologia 829, 5–17. doi: 10.1007/s10750-018-3546-7
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Seychelles, L. H., Audet, C., Tremblay, R., Fournier, R., and Pernet, F. (2009). Essential fatty acid enrichment of cultured rotifers (Brachionus plicatilis, Müller) using frozen-concentrated microalgae. Aquac. Nutr. 15, 431–439. doi: 10.1111/j.1365-2095.2008.00608.x
Sládeček, V. (1983). Rotifers as indicators of water quality. Hydrobiologia 100, 169–201. doi: 10.1007/BF00027429
Sterner, R. W., Chrzanowski, T. H., Elser, J. J., and George, N. B. (1995). Sources of nitrogen and phosphorus supporting the growth of bacteria and phytoplankton in an oligotrophic Canadian shield lake. Limnol. Oceanogr. 40, 242–249. doi: 10.4319/lo.1995.40.2.0242
Stoeckle, M. Y., Adolf, J., Charlop-Powers, Z., Dunton, K. J., Hinks, G., and Van Morter, S. M. (2021). Trawl and eDNA assessment of marine fish diversity, seasonality, and relative abundance in coastal New Jersey, USA. ICES J. Mar. Sci. 78, 293–304. doi: 10.1093/icesjms/fsaa225
Sun, X., Liang, D., Luo, H., and Yang, Y. (2020). Effects of selenium supplementation on the antioxidation response in the rotifer (Brachionus plicatilis). Aquac. Nutr. 26, 1636–1646. doi: 10.1111/anu.13108
Taipale, S. J., Aalto, S. L., Galloway, A. W. E., Kuoppamäki, K., Nzobeuh, P., and Peltomaa, E. (2019). Eutrophication and browning influence daphnia nutritional ecology. Inland Waters 9, 374–394. doi: 10.1080/20442041.2019.1574177
Trommer, G., Pondaven, P., Siccha, M., and Stibor, H. (2012). Zooplankton-mediated nutrient limitation patterns in marine phytoplankton: an experimental approach with natural communities. Mar. Ecol. Prog. Ser. 449, 83–94. doi: 10.3354/meps09508
Turner, J. T. (2004). The importance of small planktonic copepods and their roles in pelagic marine food webs. Zool. Stud. 43, 255–266.
Wen, X. L., Xi, Y. L., Qian, F. P., Zhang, G., and Xiang, X. L. (2011). Comparative analysis of rotifer community structure in five subtropical shallow lakes in East China: role of physical and chemical conditions. Hydrobiologia 661, 303–316. doi: 10.1007/s10750-010-0539-6
Worden, A. Z., Follows, M. J., Giovannoni, S. J., Wilken, S., Zimmerman, A. E., and Keeling, P. J. (2015). Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347:1257594. doi: 10.1126/science.1257594
Xu, C., Hu, S., Guo, Z., Li, T., Huang, H., Chan, L. L., et al. (2020). Flexible feeding patterns of copepod Centropages tenuiremis in fluctuating conditions: a possible survival strategy to cope with disturbance. Acta Oceanol. Sin. 39, 59–68. doi: 10.1007/s13131-020-1553-9
Yi, X., Huang, Y., Zhuang, Y., Chen, H., Yang, F., Wang, W., et al. (2017). In situ diet of the copepod Calanus sinicus in coastal waters of the South Yellow Sea and the Bohai Sea. Acta Oceanol. Sin. 36, 68–79. doi: 10.1007/s13131-017-0974-6
Keywords: zooplankton, 18S rDNA, food web, feeding response, food spectrum, dietary analysis
Citation: Liang D, Luo H, Huang C, Ye Z, Sun S, Dong J, Liang M, Lin S and Yang Y (2022) High-throughput sequencing reveals omnivorous and preferential diets of the rotifer Polyarthra in situ. Front. Microbiol. 13:1048619. doi: 10.3389/fmicb.2022.1048619
Received: 19 September 2022; Accepted: 29 November 2022;
Published: 21 December 2022.
Edited by:
Francisco J. A. Nascimento, Stockholm University, SwedenReviewed by:
Wuchang Zhang, Institute of Oceanology (CAS), ChinaCopyright © 2022 Liang, Luo, Huang, Ye, Sun, Dong, Liang, Lin and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Senjie Lin, c2VuamllLmxpbkB1Y29ubi5lZHU=; Yufeng Yang, dHl5ZkBqbnUuZWR1LmNu
†ORCID: Diwen Liang https://orcid.org/0000-0003-3415-393X
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.