
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 November 2022
Sec. Extreme Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1047580
This article is part of the Research Topic Thermophilic and Halophilic Extremophiles in Eurasian Environments, Volume II View all 9 articles
 Maria A. Khomyakova1†
Maria A. Khomyakova1† Daria G. Zavarzina1†
Daria G. Zavarzina1† Alexander Y. Merkel1
Alexander Y. Merkel1 Alexandra A. Klyukina1
Alexandra A. Klyukina1 Valeria A. Pikhtereva1,2
Valeria A. Pikhtereva1,2 Sergey N. Gavrilov1*
Sergey N. Gavrilov1* Alexander I. Slobodkin1
Alexander I. Slobodkin1The continental subsurface harbors microbial populations highly enriched in uncultured taxa. OPB41 is an uncultured order-level phylogenetic lineage within the actinobacterial class Coriobacteriia. OPB41 bacteria have a wide geographical distribution, but the physiology and metabolic traits of this cosmopolitan group remain elusive. From two contrasting subsurface environments, a terrestrial mud volcano and a deep subsurface aquifer, located in the central part of Eurasia, within the Caucasus petroleum region, we have isolated two pure cultures of anaerobic actinobacteria belonging to OPB41. The cells of both strains are small non-motile rods forming numerous pili-like appendages. Strain M08DHBT is mesophilic, while strain Es71-Z0120T is a true thermophile having a broad temperature range for growth (25–77°C). Strain M08DHBT anaerobically reduces sulfur compounds and utilizes an aromatic compound 3,4-dihydroxybenzoic acid. Strain Es71-Z0120T is an obligate dissimilatory Fe(III) reducer that is unable to utilize aromatic compounds. Both isolates grow lithotrophically and consume molecular hydrogen or formate using either thiosulfate, elemental sulfur, or Fe(III) as an electron acceptor. Genomes of the strains encode the putative reductive glycine pathway for autotrophic CO2 fixation, Ni-Fe hydrogenases, putative thiosulfate/polysulfide reductases, and multiheme c-type cytochromes presumably involved in dissimilatory Fe(III) reduction. We propose to assign the isolated strains to the novel taxa of the species–order levels and describe strain M08DHBT as Anaerosoma tenue gen. nov., sp. nov., and strain Es71-Z0120T as Parvivirga hydrogeniphila gen. nov., sp. nov., being members of Anaerosomatales ord. nov. This work expands the knowledge of the diversity, metabolic functions, and ecological role of the phylum Actinomycetota.
One of the least explored environments on Earth, the continental subsurface, harbors microbial populations highly enriched in uncultured taxa. Understanding the ecological roles, adaptation strategies and interspecies interactions of microorganisms whose existence has only been traced by metagenomic studies is hampered without obtaining cultivated laboratory cultures. OPB41 is an uncultured phylogenetic lineage within the phylum Actinomycetota. Designation ‘OPB41’ refers to the 16S rRNA gene sequence found by Hugenholtz et al. in Obsidian Pool, a Yellowstone National Park hot spring (Hugenholtz et al., 1998). The SILVA rRNA database places OPB41-related sequences in a separate order-level group within the class Coriobacteriia, which currently contains about 2000 entries (Quast et al., 2013).1 According to Genome Taxonomy Database2, OPB41 also represents a separate order (o_OPB41) within the class Coriobacteriia (Parks et al., 2022). No members of OPB41 have been isolated in pure culture or even enriched in selective medium so far. Around 50 complete genomes of OPB41 are publicly available; but studies of the metabolic potential of this bacterial group based on cultivation-independent data are scarce (Bird et al., 2019).
Actinobacteria play important roles in biogeochemical cycling of elements (Goodfellow and Williams, 1983; Holmalahti et al., 1994; Hill et al., 2011). Several actinobacterial species, e.g., from Nocardia and Streptomyces genera, are capable of aerobic chemolithoautotrophic growth (Aggag and Schlegel, 1973; Gadkari et al., 1990). Anaerobic hydrogenotrophy is a rarer feature of actinobacteria and had only been reported for Denitrobacterium detoxificans, a member of Coriobacteriaceae family (Anderson et al., 2000). Recently, the genes for key enzymes of the Wood-Ljungdahl pathway for CO2 fixation have been identified in MAGs and SAGs of several actinobacteria, including those of Coriobacteriaceae family and OPB41 group (Vavourakis et al., 2018; Liu et al., 2020; Merino et al., 2020; Jiao et al., 2021). All of the analyzed genomic data were retrieved from metagenomes of extreme environments, such as serpentinite-hosted systems or hot springs, no putative mesophilic lithotrophs have been proposed so far among OPB41 representatives.
Environmental distribution of ribosomal gene sequences shows that OPB41 is a cosmopolitan group of prokaryotes. We have noticed the presence of OPB41 in the samples from two contrasting types of environments related to deep subsurface – terrestrial mud volcanoes (TMVs) and mineral water aquifers. Our studied sites were located in the central part of Eurasia, within the Caucasus petroleum region, which is subdivided into several petroleum provinces based on their geographic location, shared tectonostratigraphic architecture and petroleum systems elements (Tari et al., 2021).
Terrestrial mud volcanoes are the structures located on the Earth’s surface but geologically connected to deep petroleum fields, and thus, to the subsurface biosphere. Mud volcanism is one of the most exciting geological phenomena with significant implications for hydrocarbon exploration, seismicity, and the atmospheric budget of methane (Mazzini and Etiope, 2017). TMVs can provide direct way to recover subsurface microbial communities due to emission of mud, breccias, liquids, and gases from deep reservoirs to the day surface through fracture networks extending to a depth of several kilometers. TMV fluids generate local physicochemical gradients that allow the proliferation of microorganisms with various metabolic patterns (Khomyakova et al., 2020; Slobodkina et al., 2020; Frolova et al., 2021). Representatives of the phylum Actinomycetota are not a major component of TMV’s microbial communities, where methane-oxidizing and sulfur-metabolizing prokaryotes usually predominate (Wang et al., 2014; Lin et al., 2018; Mardanov et al., 2020; Merkel et al., 2021; Tu et al., 2022). However, in several TMVs the relative abundance of Actinomycetota reaches 12–15% (Yang et al., 2012; Khomyakova et al., 2022).
Yessentukskoye mineral water deposit (YMWD) located on the territory of the Stavropol Upland petroleum province, is famous for balneologically valuable waters extracted from a kilometer deep Upper Cretaceous (K2s-m) aquifer. The YMWD is characterized by the proximity of the crystalline basement and heating zones to its aquifers, the presence of igneous intrusions (laccoliths) embedded in the whole thickness of water-bearing sedimentary rocks, and tectonic faults serving as well-permeable conduits between the basement and the sedimentary cover. This geological complexity leads to the formation of highly diverse mineral waters with salinity ranging from 0.5 to 14.0 g l−1, temperatures ranging from 10 to ~70°С, and variation of predominant ions (carbonate, bicarbonate, chloride or sulfate; Abramov and Vavichkin, 2010; Filimonova et al., 2020; Lavrushin et al., 2020). We have previously detected the representatives of the phylum Actinomycetota in waters extracted from two production wells of YMWD. The wells penetrated K2s-m aquifer where the abundance of uncultured actinobacterial groups reached 30% (Gavrilov et al., 2022).
Here we report the physiological and genomic characterization of two pure cultures of anaerobic bacteria belonging to OPB41 group, isolated from subsurface environments of the Caucasus petroleum region, where they constitute significant parts of two different microbial communities. The isolates are physiologically dissimilar from each other, yet share a common metabolic feature of anaerobic hydrogenotrophic or formatotrophic lithotrophy.
Strain M08DHBT was isolated from a mixed sample of sediment and water named B03 and collected in September 2018 from a salsa lake of terrestrial mud volcano Karabetova Gora (45.202 N, 36.783 E), Taman Peninsula, Krasnodar kray, Russia. Temperature and pH at the sampling site were 15°C and 8.0–8.5, respectively, salinity was 6.0 g l−1. The water contained 24.3 mM chloride and 1.2 mM sulfate. Dissolved sulfide, nitrate, and nitrite were under the detection level (<0.01 mM). The water did not contain soluble iron ions, but the sediment had high HCl-extractable iron content (81.9 mM Fe(II) and 2.7 mM Fe(III)), due to the presence of iron-bearing minerals.
Enrichment and isolation were performed in the liquid medium of the following composition (per liter distilled water): KH2PO4, NH4Cl, KCl, MgCl2·6H2O (0.33 g each), CaCl2·6H2O, 0.033 g, NaHCO3, 2.0 g, Na2S·9H2O, 0.5 g, 1 ml trace element solution (Slobodkin et al., 2012), 1 ml vitamin solution (Wolin et al., 1963) and 1 ml resazurin solution (up to 0.001 g l−1 in the medium). The medium was prepared by boiling and cooling it under N2:CO2 (80:20) flow, afterwards, NaHCO3, vitamins, and Na2S·9H2O were added. Sodium sulfide was used as a reducing agent. Resazurin was added as a redox indicator. The medium was dispensed in 10 ml aliquots into 17 ml Hungate tubes; the headspace was filled with N2:CO2 (80:20, high-purity grade). The medium was autoclaved at 1 atm, 121°C for 20 min. The pH of the sterile medium was adjusted to 7.0–7.5 at 25°C with 10% sterile anaerobic NaOH solution using anaerobic technique. 3,4-dihydroxybenzoic (protocatechuic) acid and MgSO4.7H2O from sterile anoxic stock solutions were added before inoculation to a final concentration of 10 mM each.
Strain Es71-Z0120T was isolated from water sampled in September 2020 from the well 71 of YMWD that is utilized for industrial extraction of Yessentuki no. 4 type mineral water. This well (44.189 N, 42.942 E) has a maximal depth of 999 m with open boreholes in the interval of 676.0–998.9 m. Temperature and pH values of water at the wellhead were 40.5°C and 6.8–6.9, respectively, salinity was 7.7 g l−1. The water contained 71.8 mM bicarbonate and 47.4 mM chloride, as well as 10.4 ppb dissolved iron ions. Dissolved sulfate, sulfide, nitrate, and nitrite were under the detection level (<0.01 mM). Water-bearing sediments of the aquifer penetrated by the well 71 contained the iron minerals siderite and glauconite, according to the local geological survey data.
Sterile 17 ml Hungate anaerobic culture tubes, pre-filled with 100% CO2 gas, synthesized ferrihydrite (SF), and acetate were used to obtain enrichment cultures. 10 ml of fresh water, sampled from the wellhead, were injected into the tubes with sterile syringes and needles, and the tubes were further incubated in the dark at 47°C, the average temperature observed during well operation in tap mode. Thus, water samples served as both the basic mineral medium and the inoculum for primary enrichments. The initial content of Fe(III) and acetate in the primary enrichments comprised 10 mM each. The basic cation-anion composition of the medium used for pure culture isolation corresponded to that previously determined for Yessentuki no. 4 type mineral water (Zavarzina et al., 2022). The medium contained (per liter distilled water): KH2PO4, NH4Cl, KCl, MgCl2·6H2O (0.33 g each), CaCl2·6H2O, 0.033 g, NaCl, 2.00 g, NaHCO3, 6.00 g, Na2S·9H2O, 0.01 g, 1 ml trace mineral solution (Kevbrin and Zavarzin, 1992), 1 ml vitamin solution (Wolin et al., 1963). Sodium formate 1 g l−1, or H2 (10% v/v) was used as the electron donor and SF as the electron acceptor. SF was prepared as described previously (Zavarzina et al., 2006) and added to the culture medium up to the final Fe(III) content of 10 mM, before sterilization. The medium was prepared by boiling and cooling it under CO2 flow, afterwards, NaHCO3, vitamins, and Na2S·9H2O were added. The medium was dispensed in 10 ml aliquots into 17 ml Hungate tubes, the headspace was filled with extra pure CO2. The medium was autoclaved at 1 atm, 121°C for 20 min. The pH of the medium after sterilization was 6.8 at room temperature.
Growth of the isolates was monitored by direct cell counting using a phase-contrast microscope (Olympus CX-43) for the cultures of strain M08DHBT, and a fluorescence microscope Axio Lab.A1 (Zeiss, Germany) for the cultures of strain Es71-Z0120T whose subsamples were pre-stained with acridine orange dye for DNA. Transmission electron microscopy was performed to determine cell morphology using JEM-100 and JEM-1400 electron microscopes (JEOL, Japan) at the UNIQEM Collection Core Facility, FRC Biotechnology of the Russian Academy of Science. Morphology of the cultures with iron minerals was examined by scanning electron microscopy (SEM) using TESCAN VEGA 3 LMU device with an INCA Energy 350/X-max 80 energy-dispersive analysis system (OXFORD Instruments NanoAnalysis, United Kingdom). Specimens were pre-fixed with a carbon double-sized scotch tape and triply coated with Au.
All the cultivation experiments were performed in duplicate using Hungate tubes, 1 ml inoculum volume was used for the transfers of the strain M08DHBT, and 0.5 ml inoculum volume was used for the strain Es71-Z0120T. Temperature (from 10 to 60°C) and pH growth ranges of the strain M08DHBT were determined using the same medium as for the strain isolation, with formate and thiosulfate serving as the electron donor and acceptor, respectively. The pH of the medium was adjusted to different values in the range of 5.0–10.0 with sterile anaerobic solutions of HCl (2 M) or NaOH (1.25 M). The NaCl requirement for growth was determined in a medium of similar mineral composition, but lacking NaCl and containing 0.33 g l−1 MgCl2·6H2O. Varying amounts of NaCl (0–100 g l−1) were added directly into Hungate tubes from pre-sterilized anaerobic stock solutions.
To determine optimal growth conditions of strain Es71-Z0120T, temperatures ranging from 10 to 78°C, NaCl concentrations up to 25g l−1, sodium bicarbonate concentrations up to 30 g l−1, and pH ranging from 5.5 to 8.15 were tested with formate and Fe(III)-citrate (10 mM) as the electron donor and acceptor, respectively. In these cases, sodium sulfide was omitted from the medium. Cell counts were performed within 5–20 days of incubation. Optimal NaCl concentrations for growth were determined on a modified medium in which all chlorides, other than NaCl, were replaced with equimolar concentrations of sulfates. pH optimum of strain Es71-Z0120T was determined on a medium containing 3.0 g l−1 NaHCO3, prepared under extra pure CO2 gas flow. 6 M HCl solution was used for pH adjustment to the values below 6.1, while different pH values in the range of 6.1–7.0 were reached by the gradual replacement of CO2 with N2 in the gas phase. Solution of Na2CO3 (100 g l−1) and pure N2 in the gas phase were used to sustain pH values above 7.0.
For both strains, aerobic and microaerobic growth was tested using the media lacking the reducing agent, under 2, 4, or 10% O2 (v/v in CO2) or 100% air in the gas phase. Catalase activity was tested by the bubble production assay with 3% (v/v) solution of H2O2. Oxidase activity was determined with 10 g l−1 tetramethyl-p-phenylenediamine (Cappuccino and Sherman, 2002). Electron donors and acceptors were added from sterile anaerobic stock solutions before inoculation. All the organic substrates (peptides, carbohydrates, alcohols, and organic acids) were filter-sterilized using 0.2 μm pore size syringe filters (Millipore) and added to a final concentration of 3 g l−1 or 0.3% (v/v). Growth with molecular hydrogen (100% or 10% in the gas phase) as the electron donor and utilization of non-fermentable substrates were tested in the presence of an optimal electron acceptor. Formate concentration was monitored on a Stayer HPLC (Aquilon) equipped with a refractometric detector (Knauer) and an Aminex HPX-87H column (Bio-Rad), operated isocratically, with 5 mM H2SO4 as an eluent at 0.6 ml min−1. Subsamples of the cultures for chromatographic analysis were prepared by centrifugation at 12100 g for 3 min and further pH adjustment of the supernatants to 2.0 with 5 mM H2SO4. Molecular hydrogen consumption and the formation of gaseous metabolites were monitored by gas chromatography (GC) on a HayeSep N 80/100 mesh column at 40°C with argon as the carrier gas at 20 ml min−1 flow rate. Sulfide concentration was determined colorimetrically with dimethyl-p-phenylenediamine (Trüper and Schlegel, 1964). Fe(II) production during the growth with Fe(III) compounds was monitored with ferrozine (Stookey, 1970), Fe(II) from SF and magnetite was preliminary extracted with 0.6 M HCl. 9,10-anthraquinone-2,6-disulfonate (AQDS) reduction was monitored by the change in the medium color.
Chemotaxonomic analysis was performed with the cells grown on formate with thiosulfate (for strain M08DHBT) or ferric citrate (for strain Es71-Z0120T) serving as the electron acceptors. For fatty acid analysis, the isolates were cultivated at optimal physicochemical growth conditions. Biomass was collected in the late exponential growth phase. Cellular fatty acid profiles were determined by GC–MS using methyl ester derivatives prepared from 5 mg of freeze-dried biomass treated with anhydrous HCl/MeOH. The determination was based on the retention time, reference equivalent chain length values (Hartig, 2008), and mass spectra using Supelco standards and NIST MS Search 2.0 program provided with the GC–MS setup. Cellular fatty acids content was determined as percentages of the total ion current peak area. Quinones were analyzed as described by Collins et al. (Collins and Jones, 1981; Collins, 1985) using Finnigan 123 LCQ Advantage MAX APSI/MS ion trap mass spectrometer.
Genomic DNA isolation and 16S rRNA gene amplification and sequencing were performed as described previously (Slobodkina et al., 2016). The GenBank accession numbers for 16S rRNA gene sequences of strains M08DHBT and Es71-Z0120T are ON668121 and OP389241, respectively. The 16S rRNA gene sequences of the isolates were compared with other sequences in GenBank (Benson et al., 1999) by using the BLAST program (Altschul et al., 1990) and by means of the EzBioCloud server3 (Yoon et al., 2017) to identify their closest relatives. Sequences were aligned by MAFFT v7.427 (G-INS-i strategy; Nakamura et al., 2018) for 16S rRNA gene-based phylogenetic analyses.
Complete genome sequencing of both strains was carried out on a MiSeq system (Illumina, San Diego, California, United States) using the reagent kit providing for 2 × 250 bp readings. Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JALNTY010000000 for M08DHBT and JAMCCO010000000 for Es71-Z0120T. Gene search and annotation were performed by BLAST (Altschul et al., 1990) and IMG (Chen et al., 2021) services. AAI values were calculated using an online tool developed by the Kostas group at the Georgia Institute of Technology (Rodriguez-R and Konstantinidis, 2016). For genome-based phylogenetic reconstructions, 120 bacterial single-copy conservative marker genes were used as described previously (Parks et al., 2020). The trees were built using the IQ-TREE 2 program (Minh et al., 2020) with fast model selection via ModelFinder (Kalyaanamoorthy et al., 2017) and ultrafast approximation for phylogenetic bootstrap (Hoang et al., 2018), as well as approximate likelihood-ratio test for branches (Anisimova and Gascuel, 2006).
Analysis of the prokaryotic diversity in salsa lake sediments of Karabetova Gora mud volcano by high-throughput sequencing of 16S rRNA gene amplicons showed that representatives of OPB41 group constituted 13–14% of all reads (Khomyakova et al., 2022). An enrichment culture was initiated by 10% (w/v) inoculation of a mud sample into anaerobic sterile medium with 3,4-dihydroxybenzoic (protocatechuic) acid and sulfate. Protocatechuic acid was chosen as a biodegradable substrate, modelling the aromatic hydrocarbons present in mud volcanoes (Remizovschi et al., 2020). After 3 weeks of incubation at 30°C in the dark the growth of small thin rods was observed. After three subsequent 10% (v/v) transfers, the enrichment was partially purified (up to 60%) by serial 10-fold dilutions in the same liquid medium (Supplementary Figure S1). Well-separated brownish round colonies (1–2 mm in diameter) appeared on Gelrite blocks after 14–17 days of further incubation on 3,4-dihydroxybenzoic acid. When cultivated with formate and thiosulfate, the isolate formed whitish microcolonies, invisible to the naked eye. Individual colony from the 10−6 dilution was transferred to a test tube with liquid medium, after 4–7 days of incubation, microbial growth was observed, and this culture, designated strain M08DHBT was used for further studies.
16S rRNA gene sequence profiling of different microbial communities of YMWD mineral waters retrieved relative abundance of the representatives of OPB41 group in the range of 3–9% of all prokaryotic reads (Supplementary Figure S1). An enrichment culture from well 71 was initiated by incubation of a freshly taken water sample into a test-tube supplemented with acetate and ferrihydrite under 100% CO2 in the gas phase. Active microbial growth was observed after 4 days of incubation being accompanied by the transformation of brown-colored precipitate of SF into a black magnetic mineral, presumably magnetite. Microscopic observations of acridine orange-stained samples of the primary enrichment revealed the predominance of vibrio-like cells in the culture and lower representation of very small rods. Both morphotypes were closely associated with iron mineral particles. 16S rRNA gene fragment profiling of the primary enrichment revealed the predominance of novel representatives of the order Deferribacterales (79% relative abundance), and a small share of OPB41 group uncultured actinobacteria (3%). Acetate replacement with formate led to the substitution of the predominating morphotype in the enrichment with small rods. 16S rRNA gene fragment profiling of the novel culture indicated increased representation of an OPB41-related phylotype (58%). This bacterium was isolated into a pure culture by serial ten-fold dilutions. The culture in the last positive dilution (10−7) contained morphologically homogeneous non-motile small rod-shaped cells and was designated strain Es71-Z0120T. The purity of the culture was assessed by routine microscopic examination and confirmed by complete 16S rRNA gene and complete genome sequencing.
According to the results of our phylogenomic analysis based on concatenated partial amino acid sequences of 120 bacterial single-copy conserved marker genes (Figure 1), both new strains belong to the phylogenetic lineage OPB41 of the order rank, and represent separate genera in a single family-level lineage, which is designated as “UBA2279” in the Genome Taxonomy DataBase, and for which we propose the name ‘Anaerosomataceae’ (see below). Division of M08DHBT and Es71-Z0120T isolates into two genera is strongly supported by AAI comparison, which retrieved 54.4–57.7% identity of one-way AAI and 62.5% identity of two-way AAI, as well as by 94.3% 16S rRNA gene sequence identity. On a 16S rRNA-based tree, M08DHBT and Es71-Z0120T isolates also form a separate group within class Coriobacteriia, although statistical support for this separation is not strong (Supplementary Figure S2).
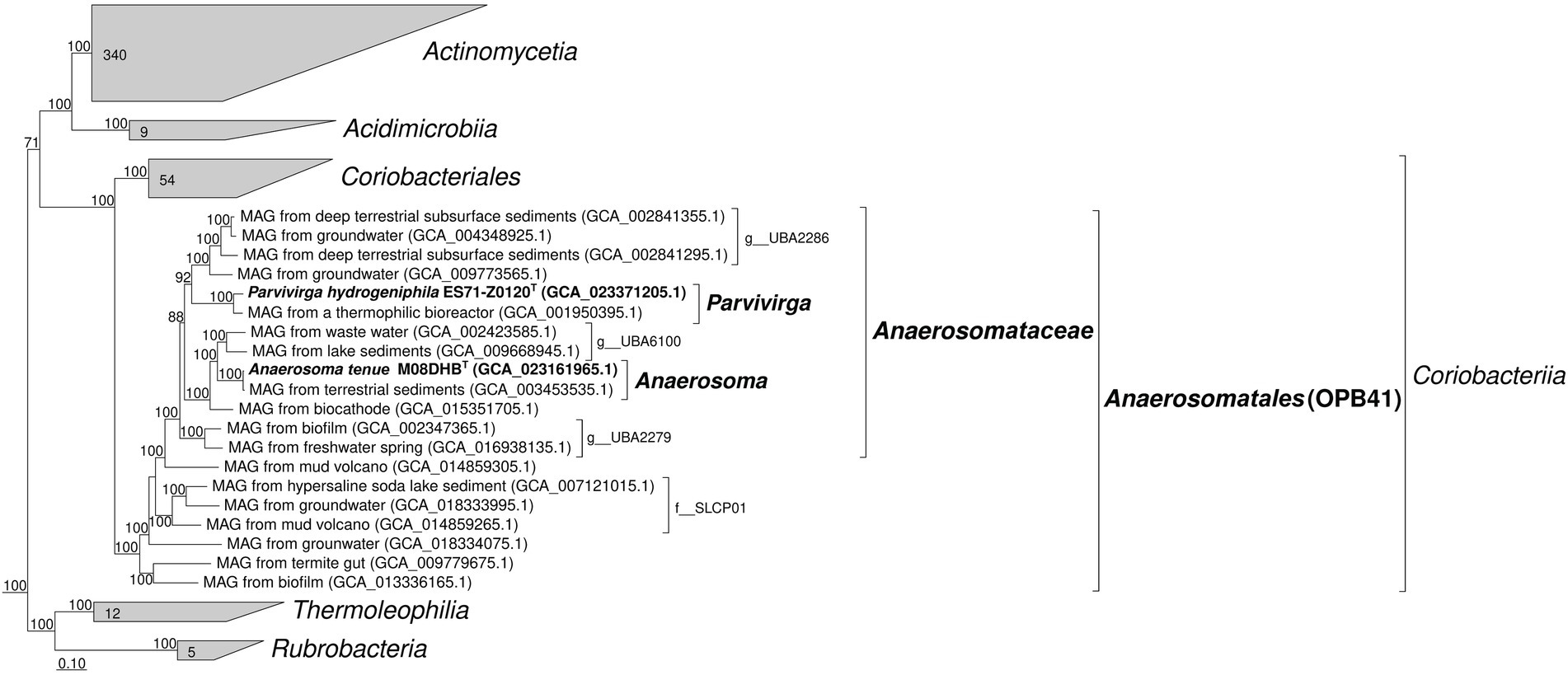
Figure 1. Phylogenomic placement of strains M08DHBT and Es71-Z0120T within the Actinomycetota phylum based on concatenated partial amino acid sequences of 120 bacterial single copy conserved marker genes with taxonomic designations according to the Genome Taxonomy DataBase. Bootstrap consensus tree is shown with values above 70%, placed at the nodes. Bar, 0.1 changes per position.
The survey of the environmental distribution of publicly available 16S rRNA gene sequences, related to the family-level lineage ‘Anaerosomataceae’ (Figure 1), shows that similar shares of sequences were retrieved from soils and different oil-bearing environments, such as petroleum crude oil, oil sands, oil wells, oil-contaminated wastewaters and tailings. A lesser share of this group sequences was retrieved from various sedimentary and bottom water environments, including the sediments of pristine waterbodies, soda lakes and marine shallow waters (Supplementary Figure S3 A). The majority of MAGs related to ‘Anaerosomataceae’ were retrieved from oil-bearing ecosystems (Supplementary Figure S3 B). Somewhat dissimilar result was obtained when all the OPB41-related MAGs were considered for the analysis. The number of MAGs retrieved from bottom waters and sediments was almost twice as much as the number of those assembled from the samples of oil-containing environments or subsurface habitats, not associated with oil and gas deposits (19 v/s 10 and 12 sequences, respectively, Supplementary Figure S4). Totally, the majority of publicly available OPB41-related phylotypes were detected in oil-bearing, sedimentary and subsurface environments.
Morphological, physiological and chemotaxonomic characteristics of strains M08DHBT and Es71-Z0120T are summarized in Table 1. The cells of the isolates are small, non-motile rods with Gram-positive cell wall type (Figures 2, 3). Both strains have pili-like appendages that cover the entire cell surface. The pili of strain Es71-Z0120T are 0.8–0.9 nm in diameter and appear only in the presence of ferrihydrite, but not in the presence of soluble Fe(III)-citrate (Figures 3B,C). The cells of strain Es71-Z0120T usually form dense clusters strongly associated with ferrihydrite particles (Figure 3A; Supplementary Figure S5 A). In the late stationary growth phase or during long storage, cells of strain Es71-Z0120T form dense extracellular matrix. This matrix stains green with acridine orange dye and is visualized under a fluorescent microscope as large bubble-like structures interconnected by long strands from which small bubbles of various diameters bud off (Supplementary Figures S5 B,D). Energy dispersive X-ray spectroscopy (EDS) revealed that the ‘bubbles’ observed in the late stationary growth phase consist mainly of oxygen and carbon but are enriched with silicon and iron comparing to the cells themselves (Supplementary Figures S5 D, S6B). Scanning electron microscopy revealed that siderite is likely to be the main mineral product of ferrihydrite reduction (Supplementary Figures S5 C, S6A) by the culture of Es71-Z0120T.
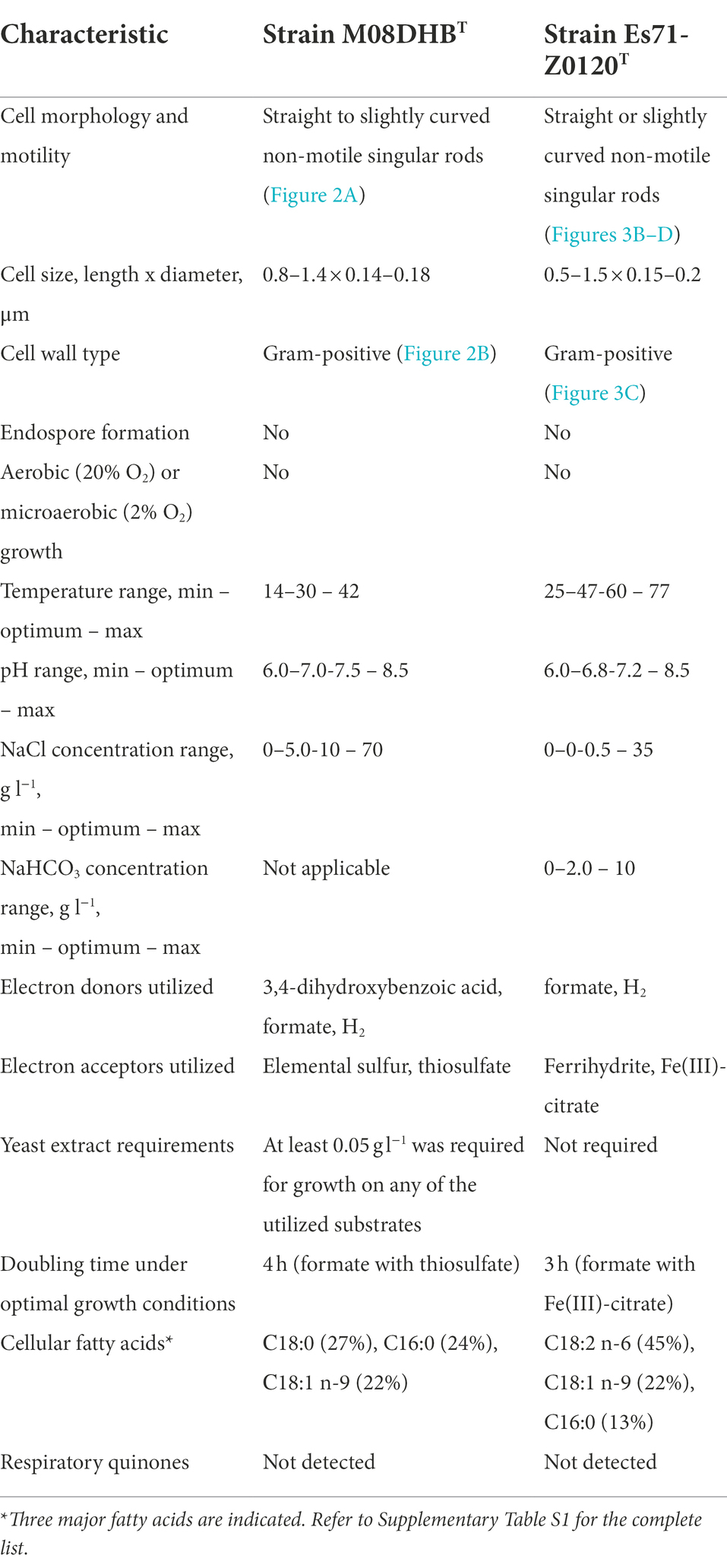
Table 1. Morphological, physiological and chemotaxonomic characteristics of strains M08DHBT and Es71-Z0120T.
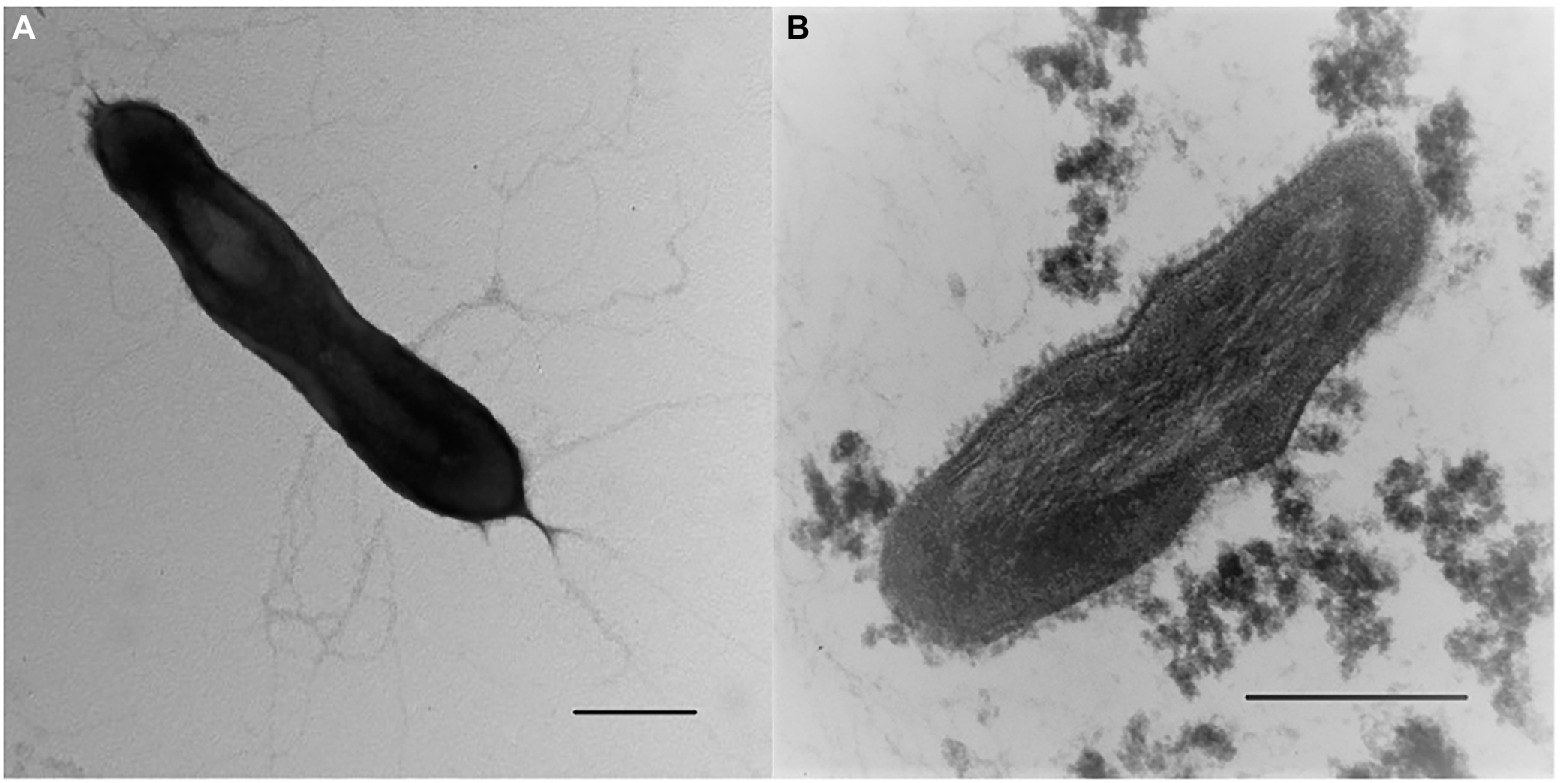
Figure 2. Cellular morphology of strain M08DHBT. (A) Electron micrograph of negatively stained cells showing an overall cell morphology and localization of filamentous appendages around a cell. Bar, 0.2 μm. (B) Ultrathin section of a cell, showing thick bilayered cell-wall structure. Bar, 0.2 μm.
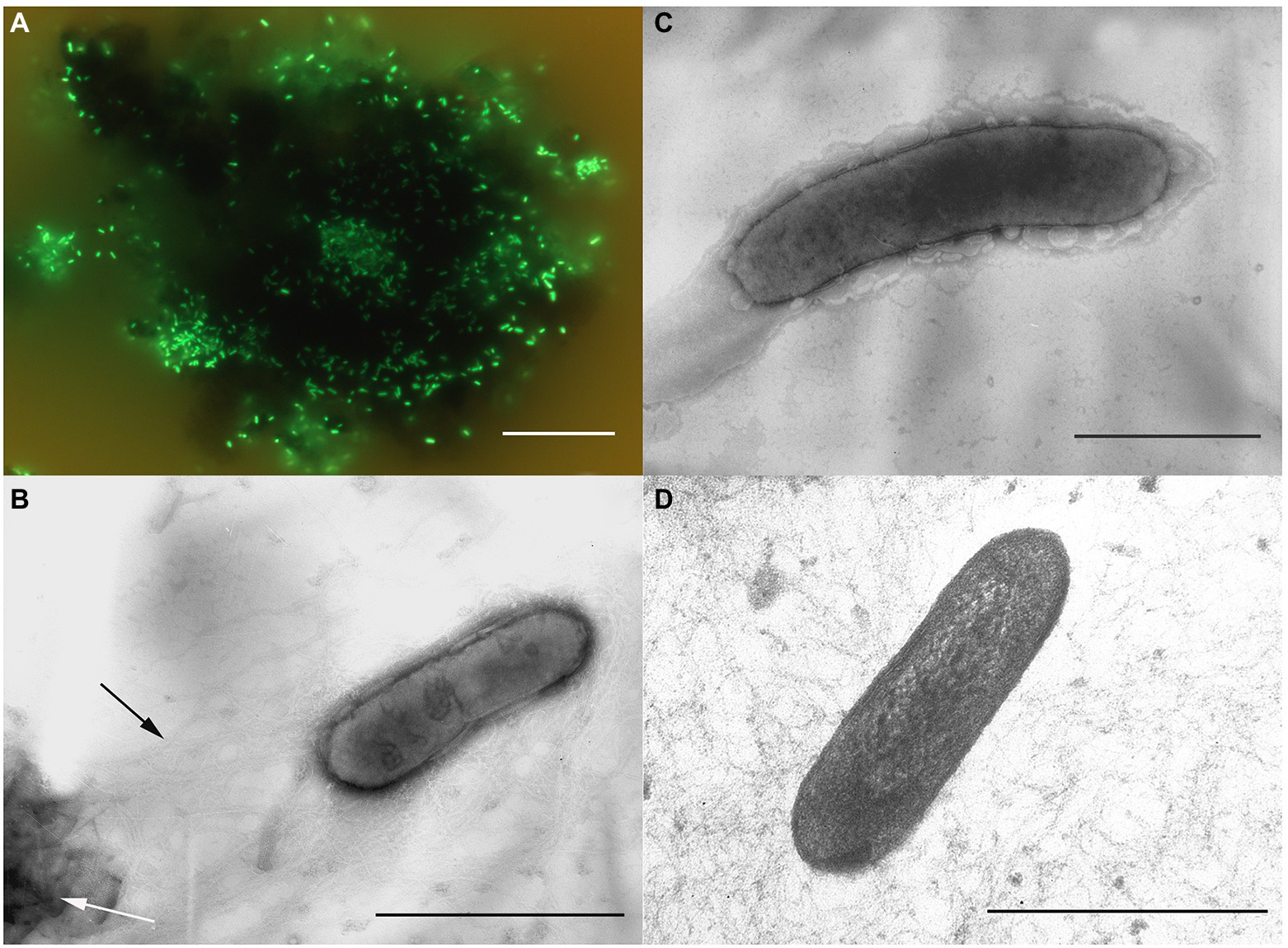
Figure 3. Cellular morphology of strain Es71-Z0120T. (A) Acridine orange-stained cells colonizing an SF particle. (B) Electron micrograph of a negatively stained cell grown with SF. Black arrow indicates a network of pili-like appendages, white arrow points at a mineral particle of SF. (C) Electron micrograph of a negatively stained cell grown with soluble Fe(III)-citrate. (D) Ultrathin section of a cell. Bars: a = 10 μm, b–d = 0.3 μm.
Both strains are obligate anaerobes, no growth was observed at 2% or 20% O2 in the gas phase, but the strains were positive for catalase and oxidase tests. Strain M08DHBT is mesophilic, while strain Es71-Z0120T is thermophilic and can grow in a wide temperature range from 25 to 77°C, with broad temperature optimum of 47–60°C. Strains M08DHBT and Es71-Z0120T use a limited number of electron donors and acceptors for their growth. The common features of both isolates are hydrogenotrophy and formatotrophy. The strains are capable of lithotropic growth with molecular hydrogen and an inorganic electron acceptor (Figure 4). Strain M08DHBT grows with molecular hydrogen in the presence of thiosulfate and requires 50 mg l−1 of yeast extract and vitamins for its lithotrophic growth (Figure 4A). Strain Es71-Z0120T grows lithoautotrophically with ferrihydrite in the presence of vitamins and in the absence of yeast extract (Figure 4B). Under these growth conditions, the highest cell density of strain Es71-Z0120T was observed at low hydrogen content (5% v/v in the gas phase) with 10 mM Fe(III) supplied as ferrihydrite mineral (SF). The growth was accompanied by stoichiometric H2 consumption and Fe(II) production in the form of siderite (Figure 4B). Both strains consume ca. 1.5 mM H2 within 14 days of lithotrophic growth at optimal conditions. In the presence of thiosulfate or S0 strain М08DHBT oxidizes formate to CO2 and produces sulfide. Acetate production was not detected. In contrast, the growth of strain Es71-Z0120TT on 10 mM formate under ferrihydrite-reducing conditions is accompanied by the production of acetate in trace amounts. Both strains do not grow on formate or hydrogen without an electron acceptor, as well as on vitamins or yeast extract in the absence of hydrogen or formate. However, strain M08DHBT utilizes 3,4-dihydroxybenzoic acid in the absence of electron acceptors but in the presence of 50 g l−1 yeast extract. Metabolic products of growth on this substrate are acetate, CO2 and traces of H2. Strain Es71-Z0120TT does not grow on 3,4-dihydroxybenzoic acid in the presence or absence of yeast extract or electron acceptors.
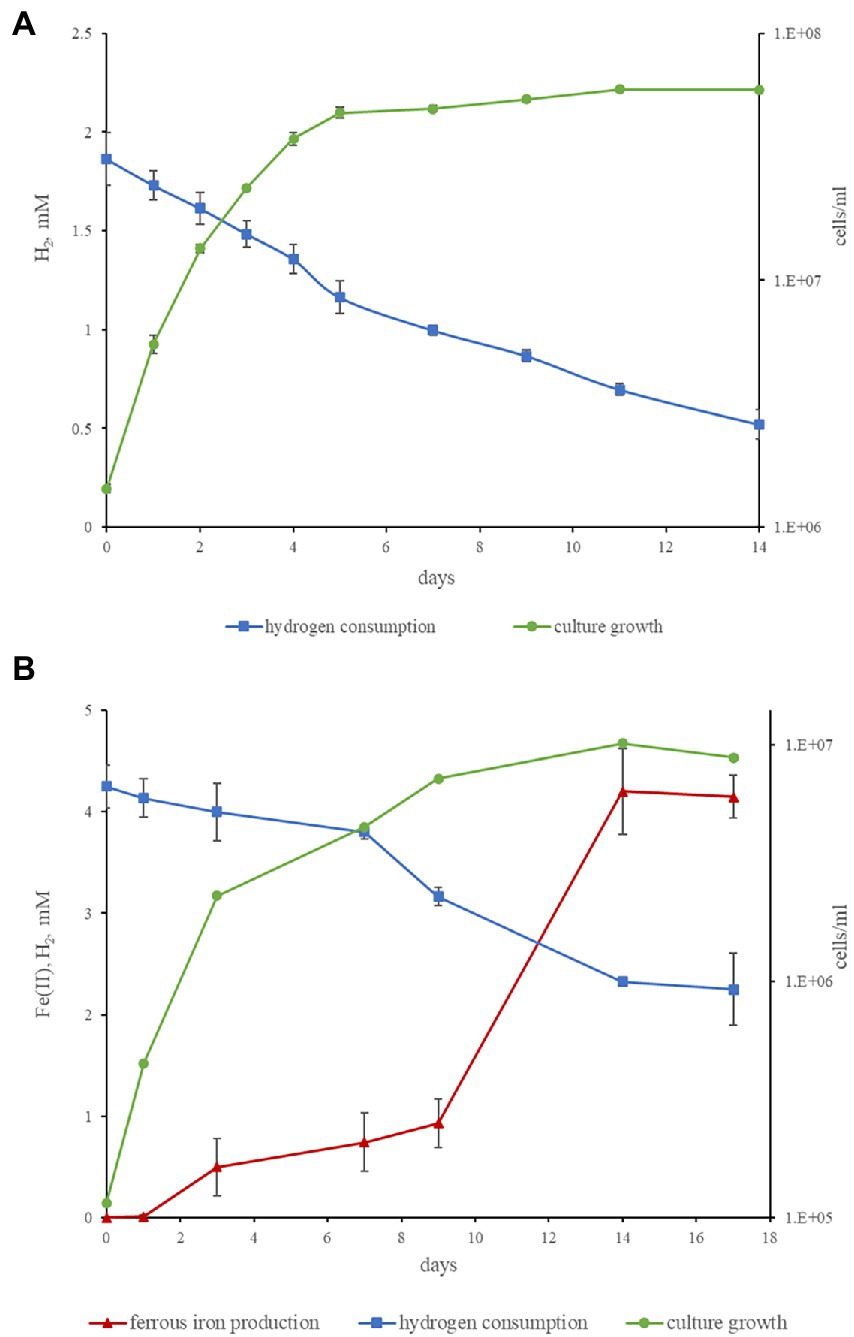
Figure 4. Kinetics of hydrogenotrophic growth of strains M08DHBT and Es71-Z0120T. (A) Growth curve of strain M08DHBT on the medium with hydrogen and thiosulfate in the presence of yeast extract, hydrogen consumption by the strain. (B) Growth curve of strain Es71-Z0120T on the medium with hydrogen and SF, hydrogen consumption and Fe(III) reduction. Note different cell yields of the strain M08DHBT growing lithoheterotrophically with yeast extract and the strain Es71-Z0120T growing lithoautotrophically with insoluble electron acceptor ferrihydrite.
The draft genome of strain M08DHBT was assembled into 6 contigs with genome size of 2,107,022 bp and the DNA G + C content of 66.58%. The genome was predicted to contain 2,100 protein-coding sequences and 53 RNA genes including 3 rRNA and 46 tRNA. NCBI PGAP genome annotation tools identified 2038 genes and 9 pseudogenes. The draft genome of strain Es71-Z0120T was assembled into 9 contigs with genome size of 1,840,996 bp and the DNA G + C content of 67.65%. The genome contained 1,808 protein-coding sequences and 54 RNAs genes, including a single copy of 16S rRNA gene. Detailed genomes statistics can be found in Supplementary Table S2. We have analyzed the genomic data in relation to central carbon and energy metabolism of the novel isolates.
Genes of the proton-translocating type I NADH-dehydrogenase are present in both strains but in differentially organized loci. Strain M08DHBT possesses the genes encoding 10 subunits of the respiratory complex I in a locus MCK8114634-44 in the order nuoNMLKJIHDCBA. Strain Es71-Z0120T possesses two loci, MCL4079026-34 encoding 8 subunits NuoACDHJKLM of the complex I, and MCL4079286-96 encoding 10 subunits NuoNMLKJIHDCBA. Both strains lack the genes encoding the NuoEFG subunits known to form the domain involved in NADH binding and oxidation (Sazanov, 2007). Thus, the respiratory complexes I of these organisms are likely to accept electrons from alternative donors besides NADH (e.g., reduced ferredoxin). Genes for the respiratory complex II, and canonical or alternative complexes III are absent from the genomes of both strains. Oxidative phosphorylation in these organisms is determined by canonical F0F1-type ATP-synthases encoded in the loci MCK8114601-8 and MCL4078772-9 in the strains M08DHBT and Es71-Z0120T, respectively. Alternatively, both organisms could synthetize ATP via bacterial A/V-type ATPases which are supposed to be bifunctional (Stewart et al., 2014). These enzyme complexes are encoded in the loci MCK8114895-903 (atpDBAFCEKI genes) and MCK8115595-602 (atpDBAFCEL) in the strain M08DHBT. Similar order of genes encoding A/V-type ATPases is observed in the loci MCL4079475-83 and MCL4078579-85 of Es71-Z0120T strain genome.
The inability of strains M08DHBT and Es71-Z0120T to utilize sugars, which are typical energy and carbon sources for actinobacteria, is obviously caused by the absence of glucose or other sugar transporter genes in both genomes. In addition, both strains lack the genes of phosphofructokinase or glucokinase but possess fructose-1,6-bisphosphatase genes instead, which provide for the irreversible transformation of fructose-1,6-bisphosphate to fructose-6-phosphate in gluconeogenesis direction of glucose metabolism. All the other genes determining the glycolysis/gluconeogenesis pathway are present in the genomes of strains M08DHBT and Es71-Z0120T, starting from pyruvate:ferredoxin oxidoreductases encoded in the loci MCK8115834-35 and MCL4079529-30, respectively, and ending with phosphoglucomutases encoded by MCK8114681 and MCL4079339. Both these proteins share considerable homology (32% amino acid sequence identity at an e-value 10−36 and complete query sequence coverage) with structurally characterized archaeal phosphoglucomutase of Thermococcus kodakaraensis. Interestingly, the interconversion of 2-and 3-phosphoglycerate is catalyzed in the strains M08DHBT and Es71-Z0120T by 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (iPGAM) which is a metalloenzyme distantly related to archaeal iPGAM and distinct from the unrelated cofactor-dependent bacterial dPGAM (Jedrzejas et al., 2000). This enzyme is encoded by MCK8113924 in strain M08DHBT and by MCL4078879 in strain Es71-Z0120T. In both our isolates, the products of gluconeogenesis could enter the metabolic network as glucose-1-phosphate via uridylyltransferase reaction and further transformation of UDP-glucose, or as glucose-6-phosphate via the reactions of incomplete pentose phosphate pathway. Both isolates possess incomplete gene sets determining the TCA cycle. The genomes encode citrate synthetases, aconitate hydratases, isocitrate dehydrogenases, 2-oxoglutarate dehydrogenases, succinyl-CoA synthetases, fumarate hydratases. Genes encoding succinate dehydrogenases/fumarate reductases are absent from both genomes. Strain M08DHBT also lacks malate dehydrogenase genes but malate could be formed from pyruvate by the NADP-dependent malic enzyme encoded by MCK8114652.
In both strains, acetyl-CoA, formed within autotrophic CO2 fixation, can enter the gluconeogenesis pathway via pyruvate:ferredoxin oxidoreductase, and the TCA cycle via the citrate synthetase. Oxaloacetate can be produced from pyruvate by the pyruvate carboxylase encoded in the locus MCK8114401-2 in strain M08DHBT, and in the locus MCL4079139-40 in strain Es71-Z0120T. Alternatively, acetyl-CoA could be converted to acetate by acetate-CoA ligase encoded in the loci MCK8114182 and MCL4079557.
In the genomes of strains M08DHBT and Es71-Z0120T, we did not find genes encoding the key enzymes of six microbial carbon fixation pathways, viz. ribulose 1,5-bisphosphate carboxylase (Calvin-Benson cycle), carbon monoxide dehydrogenase/acetyl-CoA synthase complex (reductive acetyl-CoA pathway), ATP-citrate lyase and citryl-CoA lyase (two variants of the reductive tricarboxylic acid cycle), 4-hydroxybutyryl-CoA dehydratase (3-hydroxypropionate/4-hydroxybutyrate and dicarboxylate/4-hydroxybutyrate cycles), and malonyl-CoA reductase (3-hydroxypropionate bi-cycle). The absence of succinate dehydrogenase genes from both genomes indicates the inability of our isolates to use the reversed oxidative tricarboxylic acid cycle for CO2 assimilation (Mall et al., 2018; Nunoura et al., 2018).
We assume that CO2 fixation in strains M08DHBT and Es71-Z0120T proceeds via the reductive glycine pathway (Sánchez-Andrea et al., 2020). Genomes of both strains contain a similar set of genes providing autotrophic carbon assimilation through glycine formation. The molybdenum-containing formate dehydrogenase (MCK8114746-7; MCL4079382-3) reduces CO2 to formate. Formate-tetrahydrofolate ligase (MCK8114519; MCL4079206) catalyzes the condensation of formate with tetrahydrofolate (THF) and, together with the methylene-THF dehydrogenase/cyclohydrolase (MCK8114455; MCL4079205), converts formate to methylene-THF. Glycine cleavage system (MCK8114290-3; MCL4079664-7), operating in the reductive carboxylation direction, produces glycine from CO2, methylene-THF and ammonia. Glycine could be further transformed to acetyl-phosphate with participation of thioredoxin-disulfide reductase (MCK8115865-6; MCL4078363-4) or to serine by serine hydroxymethyltransferase (MCK8114613; MCL4078767). Acetyl-phosphate can be converted to acetyl-CoA by phosphoacetyltransferase (MCK8114184; MCL4079559) or to acetate by acylphosphatase (MCK8115765; MCL4078211). Serine could be assimilated into the central metabolism employing L-serine dehydratase (MCK8115376-7; MCL4078168-9) to form pyruvate. Ammonia, necessary for the formation of glycine, is delivered to the cytoplasm of strain M08DHBT using specific transporter (MCK8115391). Strain Es71-Z0120T lacks any homologs of MCK8115391 ammonium transporter protein. Its function could be performed by a less selective transporter protein MCL4078770 sharing weak but considerable homology (30% sequence identity at an e-value 0.0001) with AmtB protein of Azospirillum brasilense belonging to 1.A.11 ammonium channel transporters family (according to Transporter Classification Database4, Saier et al., 2021).
The genomes of strains M08DHBT and Es71-Z0120T contain a gene cluster encoding membrane-bound hydrogenase belonging to the Group 1i respiratory H2-uptake [NiFe] hydrogenases of Coriobacteriia-type, according to the HydDB classifier5. This gene cluster encodes large and small hydrogenase subunits, cytochrome b subunit and a hydrogenase maturation protease (MCK8113990-2; MCL4078930-2). [NiFe] 1i hydrogenases have been found exclusively in Actinomycetota of the order Coriobacteriales and likely mediate hydrogenotrophic respiration using cytochrome b and quinone with an unresolved electron acceptor (Søndergaard et al., 2016). The genomes of both strains also encode several hydrogenase-related proteins HypABCDEF involved in metallocenter assembly (MCK8114656-7; MCK8115855-9; MCL4078352-4; MCL4079307-8). In addition, only the genome of strain M08DHBT encode a [NiFe] hydrogenase belonging to the Group 3b. Some complexes of this cytoplasmic bidirectional NADP-coupled enzyme are proposed to harbor sulfhydrogenase activity, reducing elemental sulfur with molecular hydrogen (Ma et al., 1993). We identified a gene cluster (MCK8114995-8) encoding alpha and delta hydrogenase subunits and beta and gamma sulfur reductase subunits of a sulfhydrogenase complex. Beta and gamma subunits of the sulfur reductase from strain M08DHBT have 32 and 40% amino acid sequence identity with HydB and HydG of the archaeon Pyrococcus furiosus, respectively. MCK8114997 protein contains FAD/NAD-binding pocket, and MCK8114998 one contains 4Fe-4S dicluster domain. Thus, the ability of strain M08DHBT to grow hydrogenotrophically with S0 as an electron acceptor may be provided by the action of the sulfhydrogenase.
Both isolates possess wide repertoire of multiheme c-type cytochromes with predicted membrane bound or cell surface-associated localizations. The genomes of strains M08DHBT and Es71-Z0120T encode 18 and 17 multihemes, respectively. Their genes are scattered along the genomes, although some clusters could be emphasized. One of these clusters in the strain M08DHBT (loci MCK8114672-75) encodes homologs of outer membrane-associated Fe(III)-reducing multihemes MtrA of Shewanella oneidensis (White et al., 2016) and OmcH of Geobacter sulfurreducens (Aklujkar et al., 2013), as well as a homolog of putative terminal Fe(III) reductase CFE_2239 identified in Carboxydocella thermautotrophica (Toshchakov et al., 2018). Another gene cluster of the strain M08DHBT encode weak homologs of MtrA and OmcH together with cytochrome c biogenesis proteins (loci MCK8114945-47), the homologs of MtrA, putative terminal Fe(III) reductase OcwA of ‘Thermincola potens’ (Costa et al., 2019), and a rubrerythrin (redoxin) protein (loci MCK8115166-68). An additional cluster encodes two putative quinol oxidizing multihemes related to Ga0395992_02_135863_137242 of Carboxydothermus ferrireducens (Gavrilov et al., 2021) together with an MtrA homolog (loci MCK8115470-72). Interestingly, only the last gene cluster is reproduced in the genome of strain Es71-Z0120T (loci MCL4078710-12), while other multiheme genes of this Fe(III) reducing strain are combined in two peculiar clusters. The first cluster (MCL4079323-32) encodes homologs of MtrA, OmcH and a quinol oxidizing multiheme of C. ferrireducens together with cytochrome c biogenesis proteins and a Rex-type redox-sensing suppressor which genes are absent from M08DHBT genome. The second cluster (MCL4079376-90) encodes homologs of the same quinol oxidizing multiheme and the CFE_2239 multiheme, as well as a molybdopterin oxidoreductase and a weak homolog of SmhB cytochrome, involved in soluble ferric citrate reduction in C. ferrireducens (Gavrilov et al., 2021). Simultaneous involvement of membrane-bound multiheme cytochrome c and molybdopterin enzyme complexes in Fe(III) oxide reduction has been recently proposed for the archaeon Pyrodictium delaneyi (Kashyap and Holden, 2021). Probably, peculiar organization of the cluster MCL4079376-90 in the strain Es71-Z0120T is the main feature determining the Fe(III) reducing ability in contrast to its non-Fe(III) reducing counterpart.
Strain M08DHBT was isolated under anaerobic conditions with 3,4-dihydroxybenzoic acid and sulfate. Further experiments demonstrated its inability to metabolize sulfate as the electron acceptor, indicating protocatechuic acid involvement in energy and carbon metabolism of the organism. Genome analysis revealed the absence of key enzymes (protocatechuate-3,4-dioxygenase and protocatechuate-4,5-dioxygenase) for aerobic catabolism of protocatechuate and catechol in strains M08DHBT and Es71-Z0120T. We also did not detect any of the genes encoding 3-hydroxybenzoate-CoA ligase and 4-hydroxybenzoyl-CoA reductase of peripheral pathways for anaerobic degradation of aromatic compounds in both strains. The ABC subunits of benzoyl-CoA reductase (BcrABCD) from Thauera aromatica show 54–67% similarity to the two-component acyl-CoA dehydratase activase/2-hydroxyacyl-CoA dehydratase (HAD; MCK8115292-3) of strain M08DHBT. HAD belongs to the same radical enzyme family as Bcr-type benzoyl-CoA reductases and is typical for glutamate-fermenting Bacillota and Fusobacteriota (Buckel et al., 2014). Since M08DHB is not able to grow on glutamate, we suppose that the protein MCK8115292-93 can be involved in the degradation of aromatics. In addition, three genes from strain M08DHBT genome are similar to bamB, bamC and bamD genes of a distinct benzoyl-CoA reductase from Geobacter metallireducens (Wischgoll et al., 2005). However, these genes are not co-localized in M08DHBT.
Besides the sulfhydrogenase complex (see above), the genome of strain M08DHBT encodes three subunits of a complex iron–sulfur molybdoenzyme (CISM, Rothery et al., 2008), namely: a molybdopterin cofactor-containing catalytic subunit (MCK8114746), a four-cluster protein subunit that contains 4 [Fe-S] clusters (MCK8114747), and a membrane anchor protein, containing a diheme b-binding PF01292 domain. Amino acid sequence analysis of these three proteins revealed their highest similarity with the subunits of predicted FdnGHI-type formate dehydrogenases (max. 52% sequence identity). These enzymes are distantly related to thiosulfate/polysulfide reductases of PhsA/PsrA group, and further phylogenetic analysis of the proteins from strain M08DHBT is needed to correctly predict the exact function of its CISM family complex. However, characteristic features of the membrane anchor protein MCK8114748 (4 predicted transmembrane helices and the presence of the diheme b-binding conserved domain) indicate similarity of the oxidoreductase complex with thiosulfate/polysulfide reductases (Rothery et al., 2008). This fact together with the absence of other determinants of thiosulfate reduction in the genome of M08DHBT allows us to propose the key role of the MCK8114746-8 protein complex in thiosulfate, and probably sulfur, respiration. Genome of strain Es71-Z0120T encodes only a molybdopterin-containing catalytic domain of CISM family oxidoreductases (MCL4079383) presumed to be a part of the formate dehydrogenase. The other genes typical for CISM-family complexes are absent in the organism that is consistent with its inability to reduce sulfur or thiosulfate.
We have isolated in a pure culture two strains of anaerobic bacteria belonging to so far uncultured actinobacterial OPB41 group. The habitats of both isolates are related to deep subsurface. The mud volcanoes of the Taman Peninsula and aquifers of the YMWD are located 500 km apart from each other and were formed during the Alpine geosyncline (Tveritinova et al., 2015). A common feature of these environments is the presence of a gas-water fluid which, however, has different origins. In mud volcanoes of the Taman Peninsula, feeder channels penetrate the Cenozoic sediments and approach the Cretaceous and Jurassic sedimentary rock strata, where the gas-water fluid is formed (Kholodov, 2019). The source of gas (methane) is the buried organic matter transformed at high pressure and temperature. In YMWD, the gas-water fluid flows from the crystalline basement through vertical fractures and cracks, and the major gas component is carbon dioxide with traces of molecular hydrogen of juvenile origin (Lavrushin et al., 2020). Both regions are characterized by oil and gas-bearing manifestations (Tari et al., 2021). The survey of the environmental distribution of OPB41-related phylotypes, and in particular, of the phylotypes related to the proposed ‘Anaerosomataceae’ lineage within the OPB41 group (Figure 1), shows that these actinobacteria are generally cosmopolitan. However, they are wider represented in oil-bearing environments, subsurface and various sedimentary ecosystems (Supplementary Figures S3, S4). Only 2 of 106 OPB41-related 16S rRNA sequences and MAGs, considered for our analysis, were retrieved from endosymbiotic microbial communities (from a termite gut and a sphagnum moss) indicating preferably free-living lifestyle of these actinobacteria.
OPB41 group belongs to the class Coriobacteriia which is currently subdivided into two orders, Coriobacteriales and Eggerthellales. Majority of Coriobacteriia species are the components of normal enteric microbiomes. The only and the type species of the genus Coriobacterium, C. glomerans, is an endosymbiont of pyrrhocorid bugs. Eggerthella lenta is a member of the normal human intestinal microbiome and has been most commonly associated with infections from a gastrointestinal tract. In contrast, our isolates are free-living bacteria, and the majority of OPB41-related phylotypes are also likely to represent free-living organisms. Our isolates have only several features widespread among Coriobacteriia: they are Gram-positive non-motile strictly anaerobic non spore-forming neutrophilic rods (Table 2). The common physiological characteristic of strains M08DHBT and Es71-Z0120T is their ability to grow lithotrophically on molecular hydrogen. This capability indicates that OPB41 bacteria are likely involved in primary organic matter production. Such ecological role is unusual for Coriobacteriia, among which fermentative species predominate. Only one member of this class, Denitrobacterium detoxificans, has been reported to oxidize H2 with trimethylamine oxide or dimethyl sulfoxide as an electron donor in the presence of ruminal fluid (Anderson et al., 2000). Our genomic analysis suggests that during hydrogenotrophic growth, strains M08DHBT and Es71-Z0120T could assimilate CO2 via reductive glycine pathway. This seventh metabolic route of inorganic carbon fixation in prokaryotes has been poorly investigated, and so far, has been proposed or experimentally confirmed only in a limited number of microorganisms belonging to Pseudomonadota (of the former class Deltaproteobacteria) and Bacillota (Figueroa et al., 2017; Sánchez-Andrea et al., 2020; Song et al., 2020).
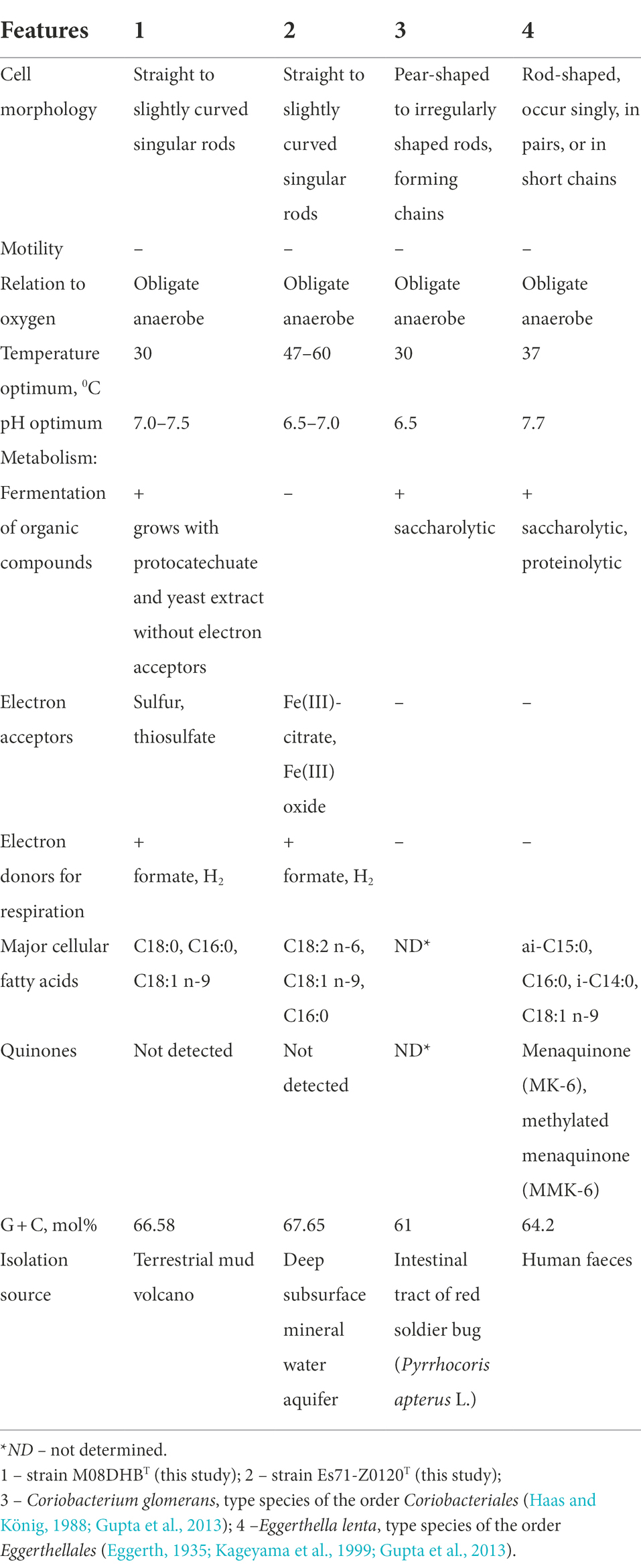
Table 2. Comparative characteristics of novel isolates and type representatives of the сlass Coriobacteriia.
The main physiological differences between strains M08DHBT and Es71-Z0120T include thermophily, the ability to grow on aromatic compounds and the spectrum of utilized electron acceptors. Strain M08DHBT is mesophilic, while strain Es71-Z0120T is a true thermophile with a broad temperature range for growth (25–77°C). Thus, different representatives of OPB41 group can proliferate at moderate as well as at elevated temperatures, which is consistent with molecular data on their environmental distribution (Supplementary Figures S3, S4).
Only strain M08DHBT can grow on 3,4-dihydroxybenzoic acid. This compound, with a trivial name protocatechuate, could be formed during aerobic or anaerobic degradation of lignin-associated phenolic compounds (Philipp et al., 2002; Linger et al., 2014; Upadhyay and Lali, 2021). About 10 to 20% of petroleum is composed of hydrocarbons originating either from biosynthetic processes or from organic matter transformation during diagenesis, which gives grounds to consider plant lignin as the plausible source of petroleum (Libes, 2009). All the currently reported lignin-degrading bacteria belong to aerobic Actinomycetota, Pseudomonadota, or Bacillota phyla (Bugg et al., 2011). Information about the organisms anaerobically degrading lignin-associated or any other aromatic compounds is scarce, and no data on anaerobic degradation of aromatics by Actinomycetota has been reported so far. Analysis of strain M08DHBT genome did not reveal any complete aerobic or anaerobic pathways of aromatic compounds degradation, but some crucial determinants of protocatechuate oxidation has been identified. Further investigations are needed to uncover biochemical routes of aromatic compounds transformation in the strain M08DHBT and Coriobacteriia, in general.
Peculiar metabolic feature of strain Es71-Z0120T is its obligate dependence on Fe(III) as the electron acceptor for growth, that is a rare case among dissimilatory iron-reducing microorganisms. In contrast, strain M08DHBT does not use Fe(III) as the electron acceptor. However, both isolates possess similar set of 17 genes encoding multiheme c-type cytochromes, which are regarded to be the major determinants of extracellular electron transfer to Fe(III) compounds in various prokaryotes (Shi et al., 2016). These genes are differentially clustered with each other and regulatory regions in the genomes of two strains. Imperfect gene clustering, i.e., caused by random mutations, could be the main reason for the inability of strain M08DHBT to reduce Fe(III). Alternatively, the cytochromes of M08DHBT isolate might have high specificity to a particular Fe(III) form, different from ferric oxides or soluble complexes, e.g., to Fe(III) silicate minerals. Genomic background of the inability of Es71-Z0120T strain to reduce sulfur or thiosulfate is clearer. This strain, in contrast to M08DHBT, lacks the genes of iron–sulfur and membrane anchor subunits of CISM family enzyme complexes to which thiosulfate/polysulfide reductases belong. The specificity of our isolates to electron acceptors correlates with different representation of sulfur and iron compounds in their natural environments. Soluble iron but not sulfur compounds were detected in mineral waters used for the isolation of the Fe(III)-reducing strain Es71-Z0120T, while no iron but 1.2 mM sulfate was detected in the samples taken for the isolation of the non-iron-reducing strain M08DHBT, capable of sulfur and thiosulfate respiration. Accordingly, the availability of inorganic electron acceptors seems to be an important selective factor driving the evolution of metabolic capacities within the OPB41 group actinobacteria. This fact and the capability for lithotrophic growth with formate or hydrogen are novel cases for the organisms of Coriobacteriia class which has been previously represented mainly by endosymbiotic organotrophs. This work expands the knowledge of the diversity, metabolic functions and ecological role of the phylum Actinomycetota. Based on phylogenetic position, phenotypic, physiological and genomic properties of strains M08DHBT and Es71-Z0120T, we propose to assign them to the novel taxa of the species–order level.
Anaerosoma (An.ae.ro.so’ma. Gr. pref. an-without; Gr. masc. n. aer – air; Gr. neut. n. soma - body; N.L. neut. n. Anaerosoma, anaerobic body).
Non-motile, slightly curved to rod-shaped bacterium. Mesophilic, obligate anaerobe, catalase- and oxidase-positive. Non spore-forming. Gram-positive. Elemental sulfur or thiosulfate are used as the electron acceptors and reduced to hydrogen sulfide. Molecular hydrogen and formate can be used as electron donors. Unable to ferment carbohydrates and proteins. The major fatty acids are С18:03, C16:0 and С18:1 n-9. Quinones were not detected. Member of the family Anaerosomataceae, order Anaerosomatales, class Coriobacteriia. The type species is Anaerosoma tenue.
Anaerosoma tenue (te’nu.e. L. neut. Adj. tenue – small, slender).
Nonmotile, rod-shaped bacterium with pili-like appendages, 0.8–1.4 μm in length and 0.14–0.18 μm in diameter. Strictly anaerobic, but catalase- and oxidase-positive. Non-spore-forming. Gram-positive type of the cell wall. Grows at 14–42°C (optimum 30°C), at pH 6.0–8.5 (optimum 7.0–7.5) and at NaCl concentrations of 0–70 g l−1 (optimum 5.0–10 g l−1). Yeast extract is necessary for growth. Grows on 3,4-dihydroxybenzoic acid only in the presence of yeast extract, producing acetate, CO2, and traces of H2. Is able to reduce elemental sulfur or thiosulfate as the electron acceptor with formate or molecular hydrogen as the electron donor. Products of chemolithotrophic growth are carbon dioxide and hydrogen sulfide. No growth is observed on D-glucose, D-fructose, D-mannose, D-ribose, D-sucrose, lactose, cellobiose, succinate, pyruvate, malate, fumarate, lactate, galactose, arabinose, citrate, acetate, yeast extract, tryptone, peptone, methanol, ethanol, trimethylamine, phenol, lactate, butyrate, glycerol, glutamate, benzoate, 2-methoxyphenol, 2-methoxybenzoate, 3,4-dimethoxybenzoate, vanillate, 2-methoxycinnamic acid, 2-hydroxybenzoate, 3-hydroxybenzoate, 2,5-dihydroxybenzoate, 2-hydroxycinnamic acid, 4-aminobenzoate, 2-chlorobenzoate. Ferrihydrite (poorly crystalline Fe(III) oxide), sulfate, nitrate, nitrite, sulfite, AQDS, dimethylsulfoxide, arsenate, selenate, selenite, crotonate are not reduced and do not support growth. The genome of the type strain is characterized by the size of 2.11 Mb and a G + C content of 66.6%. The type strain, M08DHBT (=VKM B-3570T = JCM 39237T = KCTC 25380T = UQM 41472T), was isolated from terrestrial mud volcano, Taman Peninsula, Russia.
Parvivirga (Par.vi.vir’ga. L. masc. Adj. parvus, tiny, small; L. fem. n. virga, rod; N.L. fem. n. Parvivirga, a tiny rod).
Non-motile, slightly curved to rod-shaped bacterium. Non spore-forming. Gram-positive. Obligate anaerobe, positive for catalase and oxidase. Neutrophile and thermophile. Chemolithotrophic growth is possible with molecular hydrogen or formate as the electron donors. Capable of dissimilatory Fe(III)-reduction. Unable to ferment carbohydrates and proteins. The major fatty acids are C18:2 n-6, C18:1 n-9, C16:0. Respiratory quinones were not detected. Member of the family Anaerosomataceae, order Anaerosomatales, class Coriobacteriia. The type species is Parvivirga hydrogeniphila.
Parvivirga hydrogeniphila (hy.dro.ge.ni’phi.la. N.L. neut. n. hydrogenum, hydrogen; N.L. masc. Adj. philus (from Gr. masc. adj. philos), loving; N.L. fem. Adj. hydrogeniphila, hydrogen-liking, referring to its ability to grow lithotrophically on molecular hydrogen).
Non-motile or slightly curved small single rods, 0.5–1.0 μm in length and 0.12–0.18 μm in width. In the presence of ferrihydrite, cells form exocellular, long pili-like appendages, 0.8–0.9 nm in diameter. Non spore-forming. Gram-positive type of the cell wall. Strictly anaerobic, but catalase- and oxidase-positive. Thermophile, grows at 25–70°C (optimum 47–60°C) at pH 6.0–8.5 (optimum 6.8–7.2), at NaCl concentrations of 0–35 g l−1 (optimum at 0–0.5 g l−1) and at NaHCO3 concentrations of 0–10 g l−1 (optimum 2.0 g l−1). Vitamins, but not yeast extract are necessary for growth. Chemolithotroph, capable of growth using molecular hydrogen or formate as the only electron donor with synthetic ferrihydrite (SF) or Fe(III)-citrate as the electron acceptors. The products of chemolithotrophic growth are carbon dioxide and ferrous iron in the form of siderite or magnetite. Unable to oxidize methanol, ethanol, n-propanol, butanol, acetate, lactate, pyruvate, succinate, malate, citrate, microcrystalline- or carboxymethylcellulose, N-acetyl-D-glucosamine in the presence of SF or Fe(III)-citrate as the electron acceptors. Cannot ferment peptone, yeast extract, beef extract, tryptone, D-glucose, D-sucrose, cellobiose. Does not grow on 3,4-dihydroxybenzoic acid, whether in the presence or absence of yeast extract. Sulfate, sulfite, thiosulfate, elemental sulfur, dimethylsulfoxide, nitrate, nitrite, arsenate, selenate, selenite, crotonate, fumarate, Fe(III)-EDTA, AQDS are not reduced and do not support growth. The genome of the type strain is characterized by the size of 1.84 Mb and a G + C content of 67.7%. The type strain, Es71-Z0120T (=VKM B-3556T = JCM 39246T), was isolated from subsurface mineral water of Yessentukskoye mineral water deposit (production well 71), Stavropol Krai, Russia.
Anaerosomataceae (An.ae.ro.so.ma.ta.ce’ae. N.L. neut. n. Anaerosoma, a bacterial genus; −aceae ending to denote a family; N.L. fem. pl. n. Anaerosomataceae, the Anaerosoma family).
Cells are Gram-stain positive, thin rods, non spore-forming. Cells are nonmotile, possess pili-like appendages. Mesophilic and thermophilic. Chemolithotrophic and chemoorganotrophic, asaccharolytic. Respiratory quinones were not detected. The type genus is Anaerosoma. The family belongs to the order Anaerosomatales of the class Coriobacteriia.
Anaerosomatales (An.ae.ro.so.ma.ta’les. N.L. neut. n. Anaerosoma, a bacterial genus; −ales ending to denote an order; N.L. fem. pl. n. Anaerosomatales, the Anaerosoma order).
Cells are Gram-stain positive, rod-shaped, anaerobic. The type genus is Anaerosoma. The order contains the single family Anaerosomataceae and belongs to the class Coriobacteriia.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, ON668121, OP389241, JALNTY010000000, JAMCCO010000000.
MK, DZ, AM, AK, and VP: experimental work. AS, MK, AM, and SG: genome annotation and analysis. MK, DZ, and SG: writing – original draft preparation. AS, MK, and SG: writing –review and editing. MK, DZ, and AM: visualization. AS and SG: funding acquisition. All authors contributed to the article and approved the submitted version.
This research was partially funded by the Russian Science Foundation, grant number 22–14-00011 (MK, AM, AS, isolation and characterization of strain M08DHB), and number 21–14-00333 (DZ, SG, VP, isolation, sequencing and characterization of strain Es71-Z0120), and by the Ministry of Science and Higher Education of the Russian Federation (M08DHB genome sequencing and annotation).
We are grateful to M. Chernov for performing scanning microscopy using the equipment acquired with the funding of Moscow State University Development Program. We are grateful to Holding Aqua LLC for providing the access to the production wells of YMWD.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1047580/full#supplementary-material
Abramov, V. J., and Vavichkin, A. J. (2010). Features of formation of the thermal gas chemical composition of mineral waters of the Yessentuki basin. Prospect and protection of mineral resources [in Russian] 10, 27–32.
Aggag, M., and Schlegel, H. G. (1973). Studies on a gram-positive hydrogen bacterium, Nocardia opaca strain lb. I. Description and physiological characterization. Arch. Microbiol. 88, 299–318. doi: 10.1007/BF00409942
Aklujkar, M., Coppi, M. V., Leang, C., Kim, B. C., Chavan, M. A., Perpetua, L. A., et al. (2013). Proteins involved in electron transfer to Fe(III) and Mn(IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology 159, 515–535. doi: 10.1099/mic.0.064089-0
Altschul, S., Gish, W., Miller, W., Myers, E., and Lipman, D. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anderson, R. C., Rasmussen, M. A., Jensen, N. S., and Allison, M. J. (2000). Denitrobacterium detoxificans gen. nov., sp. nov., a ruminal bacterium that respires on nitrocompounds Int. J. Syst. Evol. Microbiol. 50, 633–638. doi: 10.1099/00207713-50-2-633
Anisimova, M., and Gascuel, O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. doi: 10.1080/10635150600755453
Benson, D., Boguski, M., Lipman, D., Ostell, J., Ouellette, B., Rapp, B. A., et al. (1999). GenBank. Nucleic Acids Res. 46, D41–D47. doi: 10.1093/nar/gkx1094
Bird, J. T., Tague, E. D., Zinke, L., Schmidt, J. M., Steen, A. D., Reese, B., et al. (2019). Uncultured microbial phyla suggest mechanisms for multi-thousand-year subsistence in Baltic Sea sediments. MBio 10, e02376–e02318. doi: 10.1128/mBio.02376-18
Buckel, W., Kung, J. W., and Boll, M. (2014). The benzoyl-coenzyme a reductase and 2-hydroxyacyl-coenzyme a dehydratase radical enzyme family. Chem Bio Chem 15, 2188–2194. doi: 10.1002/cbic.201402270
Bugg, T. D., Ahmad, M., Hardiman, E. M., and Singh, R. (2011). The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22, 394–400. doi: 10.1016/j.copbio.2010.10.009
Cappuccino, J. G., and Sherman, N. (2002). Microbiology: A laboratory manual, 6th edn., Pearson Education Inc., San Francisco, 15–224.
Chen, I. A., Chu, K., Palaniappan, K., Ratner, A., Huang, J., Huntemann, M., et al. (2021). The IMG/M data management and analysis system vol 6.0: new tools and advanced capabilities. Nucleic Acids Res. 49, D751–D763. doi: 10.1093/nar/gkaa939
Collins, M. D. (1985). Analysis of isoprenoid quinones. Methods Microbiol. 18, 329–366. doi: 10.1016/S0580-9517(08)70480-X
Collins, M. D., and Jones, D. (1981). Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol. Rev. 45, 481–354. doi: 10.1128/mr.45.3.481-481.1981
Costa, N. L., Hermann, B., Fourmond, V., Faustino, M. M., Teixeira, M., Einsle, O., et al. (2019). How thermophilic gram-positive organisms perform extracellular electron transfer: characterization of the cell surface terminal reductase OcwA. mBio 10, e01210–e01219. doi: 10.1128/mBio.01210-19
Eggerth, A. H. (1935). The gram-positive non-spore-bearing anaerobic bacilli of human feces. J. Bacteriol. 30, 277–299. doi: 10.1128/jb.30.3.277-299.1935
Figueroa, I. A., Barnum, T. P., Somasekhar, P. Y., Carlström, C. I., Engelbrektson, A. L., and Coates, J. D. (2017). Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proc. Natl. Acad. Sci. 115, E92–E101. doi: 10.1073/pnas.1715549114
Filimonova, E., Kharitonova, N., Sartykov, A., Maximova, E., Baranovskaya, E., Korzun, A., et al. (2020). Hydrogeology and hydrogeochemistry of mineral sparkling groundwater within Yessentuki area (Caucasian mineral water region). Environ. Earth Sci. 79, 1–12. doi: 10.1007/s12665-019-8721-2
Frolova, A. A., Merkel, A. Y., Kuchierskaya, A. A., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I. (2021). Pseudodesulfovibrio alkaliphilus, sp. nov., an alkaliphilic sulfate-reducing bacterium isolated from a terrestrial mud volcano. Antonie Van Leeuwenhoek 114, 1387–1397. doi: 10.1007/s10482-021-01608-5
Gadkari, D., Schricker, K., Acker, G., Kroppenstedt, R. M., and Meyer, O. (1990). Streptomyces thermoautotrophicus sp. nov., a thermophilic CO-and H2-oxidizing obligate chemolithoautotroph. Appl. Environ. Microbiol. 56, 3727–3734. doi: 10.1128/aem.56.12.3727-3734.1990
Gavrilov, S. N., Potapov, E. G., Prokof’eva, M. I., Klyukina, A. A., Merkel, A. Y., Maslov, A. A., et al. (2022). Diversity of novel uncultured prokaryotes in microbial communities of the Yessentukskoye underground mineral water deposit. Microbiology (Moscow) 91, 28–44. doi: 10.1134/S0026261722010039
Gavrilov, S. N., Zavarzina, D. G., Elizarov, I. M., Tikhonova, T. V., Dergousova, N. I., Popov, V. O., et al. (2021). Novel extracellular electron transfer channels in a gram-positive thermophilic bacterium. Front. Microbiol. 11:597818. doi: 10.3389/fmicb.2020.597818
Goodfellow, M., and Williams, S. T. (1983). Ecology of actinomycetes. Ann. Rev. Microbiol. 37, 189–216. doi: 10.1146/annurev.mi.37.100183.001201
Gupta, R. S., Chen, W. J., Adeolu, M., and Chai, Y. (2013). Molecular signatures for the class Coriobacteriia and its different clades; proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam. Nov., and Eggerthellales Ord. Nov., containing the family Eggerthellaceae fam. Nov. Int. J. Syst. Evol. Microbiol. 63, 3379–3397. doi: 10.1099/ijs.0.048371-0
Haas, F., and König, H. (1988). Coriobacterium glomerans gen. Nov., sp. nov. from the intestinal tract of the red soldier bug. Int. J. Syst. Evol. Microbiol. 38, 382–384. doi: 10.1099/00207713-38-4-382
Hartig, C. (2008). Rapid identification of fatty acid methyl esters using a multidimensional gas chromatography-mass spectrometry database. J. Chromatogr. 1177, 159–169. doi: 10.1016/j.chroma.2007.10.089
Hill, P., Krištůfek, V., Dijkhuizen, L., Boddy, C., Kroetsch, D., and van Elsas, J. D. (2011). Land use intensity controls actinobacterial community structure. Microb. Ecol. 61, 286–302. doi: 10.1007/s00248-010-9752-0
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Holmalahti, J., Wright, A., and Raatikainen, O. (1994). Variations in the spectra of biological activities of actinomycetes isolated from different soils. Lett. Appl. Microbiol. 18, 144–146. doi: 10.1111/j.1472-765X.1994.tb00829.x
Hugenholtz, P., Pitulle, C., Hershberger, K. L., and Pace, N. R. (1998). Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180, 366–376. doi: 10.1128/JB.180.2.366-376.1998
Jedrzejas, M. J., Chander, M., Setlow, P., and Krishnasamy, G. (2000). Mechanism of catalysis of the cofactor-independent phosphoglycerate mutase from Bacillus stearothermophilus. Crystal structure of the complex with 2-phosphoglycerate. J. Biol. Chem. 275, 23146–23153. doi: 10.1074/jbc.M002544200
Jiao, J. Y., Fu, L., Hua, Z. S., Liu, L., Salam, N., Liu, P. F., et al. (2021). Insight into the function and evolution of the wood-Ljungdahl pathway in Actinobacteria. ISME J. 15, 3005–3018. doi: 10.1038/s41396-021-00935-9
Kageyama, A., Benno, Y., and Nakase, T. (1999). Phylogenetic evidence for the transfer of Eubacterium lentum to the genus Eggerthella as Eggerthella lenta gen. Nov., comb. nov. Int. J. Syst. Bacteriol. 49, 1725–1732. doi: 10.1099/00207713-49-4-1725
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). Model finder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kashyap, S., and Holden, J. F. (2021). Microbe-mineral interaction and novel proteins for iron oxide mineral reduction in the hyperthermophilic crenarchaeon Pyrodictium delaneyi. Appl. Environ. Microbiol. 87, e02330–e02320. doi: 10.1128/AEM.02330-20
Kevbrin, V. V., and Zavarzin, G. A. (1992). The influence of sulfur compounds on the growth of halophilic homoacetic bacterium Acetohalobium arabaticum. Microbiology (Moscow) 61, 563–571.
Kholodov, V. N. (2019). The origin of mud volcanoes. Geology and Mineral Resources of World Ocean 15, 57–80. doi: 10.15407/gpimo2019.04.057
Khomyakova, M. A., Merkel, A. Y., Kopitsyn, D. S., and Slobodkin, A. I. (2022). Pelovirga terrestris gen. Nov., sp. nov., anaerobic, alkaliphilic, fumarate-, arsenate-, Fe(III)- and sulfur-reducing bacterium isolated from a terrestrial mud volcano. Syst. Appl. Microbiol. 45:126304. doi: 10.1016/j.syapm.2022.126304
Khomyakova, M. A., Merkel, A. Y., Petrova, D. A., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I. (2020). Alkalibaculum sporogenes sp. nov., isolated from a terrestrial mud volcano and emended description of the genus Alkalibaculum. Int. J. Syst. Evol. Microbiol. 70, 4914–4919. doi: 10.1099/ijsem.0.004361
Lavrushin, V. Y., Lisenkov, A. B., and Aidarkozhina, A. S. (2020). Genesis of the Yessentuki carbon dioxide field (northern Caucasus). Geochemistry [in Russian] 65, 75–91. doi: 10.31857/S0016752520010082
Libes, S. M. (2009). “The origin of petroleum in the marine environment” in Introduction to marine biogeochemistry (Amsterdam: Elsevier Science), 1–33.
Lin, Y. T., Tu, T. H., Wei, C. L., Rumble, D., Lin, L. H., and Wang, P. L. (2018). Steep redox gradient and biogeochemical cycling driven by deeply sourced fluids and gases in a terrestrial mud volcano. FEMS Microbiol. Ecol. 94:3796. doi: 10.1093/femsec/fiy171
Linger, J. G., Vardon, D. R., Guarneri, M. T., Karp, E. M., Hunsinger, G. B., Franden, M. A., et al. (2014). Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. U. S. A. 111, 12013–12018. doi: 10.1073/pnas.1410657111
Liu, Y. F., Chen, J., Liu, Z. L., Shou, L. B., Lin, D. D., Zhou, L., et al. (2020). Anaerobic degradation of paraffins by thermophilic Actinobacteria under methanogenic conditions. Environ. Sci. Technol. 54, 10610–10620. doi: 10.1021/acs.est.0c02071
Ma, K., Schicho, R. N., Kelly, R. M., and Adams, M. W. (1993). Hydrogenase of the hyperthermophile Pyrococcus furiosus is an elemental sulfur reductase or sulfhydrogenase: evidence for a sulfur-reducing hydrogenase ancestor. Proc. Natl. Acad. Sci. U. S. A. 90, 5341–5344. doi: 10.1073/pnas.90.11.5341
Mall, A., Sobotta, J., Huber, C., Tschirner, C., Kowarschik, S., Bačnik, K., et al. (2018). Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science 359, 563–567. doi: 10.1126/science.aao2410
Mardanov, A. V., Kadnikov, V. V., Beletsky, A. V., and Ravin, N. V. (2020). Sulfur and methane-oxidizing microbial community in a terrestrial mud volcano revealed by metagenomics. Microorganisms. 8:1333. doi: 10.3390/microorganisms8091333
Mazzini, A., and Etiope, G. (2017). Mud volcanism: an updated review. Earth-Sci. Rev. 168, 81–112. doi: 10.1016/j.earscirev.2017.03.001
Merino, N., Kawai, M., Boyd, E. S., Colman, D. R., McGlynn, S. E., Nealson, K. H., et al. (2020). Single-cell genomics of novel Actinobacteria with the wood–Ljungdahl pathway discovered in a serpentinizing system. Front. Microbiol. 11:1031. doi: 10.3389/fmicb.2020.01031
Merkel, A. Y., Chernyh, N. A., Pimenov, N. V., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I. (2021). Diversity and metabolic potential of the terrestrial mud volcano microbial community with a high abundance of archaea mediating the anaerobic oxidation of methane. Life (Basel). 11:953. doi: 10.3390/life11090953
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Nakamura, T., Yamada, K. D., Tomii, K., and Katoh, K. (2018). Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34, 2490–2492. doi: 10.1093/bioinformatics/bty121
Nunoura, T., Chikaraishi, Y., Izaki, R., Suwa, T., Sato, T., Harada, T., et al. (2018). A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science 359, 559–563. doi: 10.1126/science.aao3407
Parks, D. H., Chuvochina, M., Chaumeil, P. A., Rinke, C., Mussig, A. J., and Hugenholtz, P. (2020). A complete domain-to-species taxonomy for bacteria and archaea. Nat. Biotechnol. 38, 1079–1086. doi: 10.1038/s41587-020-0501-8
Parks, D. H., Chuvochina, M., Rinke, C., Mussig, A. J., Chaumeil, P. A., and Hugenholtz, P. (2022). GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794. doi: 10.1093/nar/gkab776
Philipp, B., Kemmler, D., Hellstern, J., Gorny, N., Caballero, A., and Schink, B. (2002). Anaerobic degradation of protocatechuate (3,4-dihydroxybenzoate) by Thauera aromatica strain AR-1. FEMS Microbiol. Lett. 212, 139–143. doi: 10.1111/j.1574-6968.2002.tb11257.x
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Opens external link in new window. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Remizovschi, A., Carpa, R., Forray, F. L., Chiriac, C., Roba, C. A., Beldean-Galea, S., et al. (2020). Mud volcanoes and the presence of PAHs. Sci. Rep. 10:1253. doi: 10.1038/s41598-020-58282-2
Rodriguez-R, L. M., and Konstantinidis, K. (2016). The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. Peer. J. Prepr. doi: 10.7287/peerj.preprints.1900
Rothery, R. A., Workun, G. J., and Weiner, J. H. (2008). The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim. Biophys. Acta 1778, 1897–1929. doi: 10.1016/j.bbamem.2007.09.002
Saier, M. H., Reddy, V. S., Moreno-Hagelsieb, G., Hendargo, K. J., Zhang, Y., Iddamsetty, V., et al. (2021). The transporter classification database (TCDB): 2021 update. Nucleic Acids Res. 49, D461–D467. doi: 10.1093/nar/gkaa1004
Sánchez-Andrea, I., Guedes, I. A., Hornung, B., Boeren, S., Lawson, C. E., Sousa, D. Z., et al. (2020). The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans. Nat. Commun. 11:5090. doi: 10.1038/s41467-020-18906-7
Sazanov, L. A. (2007). Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry 46, 2275–2288. doi: 10.1021/bi602508x
Shi, L., Dong, H., Reguera, G., Beyenal, H., Lu, A., Liu, J., et al. (2016). Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 14, 651–662. doi: 10.1038/nrmicro.2016.93
Slobodkin, A. I., Reysenbach, A. L., Slobodkina, G. B., Baslerov, R. V., Kostrikina, N. A., Wagner, I. D., et al. (2012). Thermosulfurimonas dismutans gen. Nov., sp. nov., an extremely thermophilic sulfur disproportionating bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 62, 2565–2571. doi: 10.1099/ijs.0.034397-0
Slobodkina, G. B., Baslerov, R. V., Novikov, A. A., Viryasov, M. B., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I. (2016). Inmirania thermothiophila gen. nov., sp. nov., a thermophilic, facultatively autotrophic, sulfur-oxidizing gammaproteobacterium isolated from a shallow-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol 66, 701–706. doi: 10.1099/ijsem.0.000773
Slobodkina, G. B., Merkel, A. Y., Novikov, A. A., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I. (2020).Pelomicrobium methylotrophicum gen. nov., sp. nov.a moderately thermophilic, facultatively anaerobic, lithoautotrophic and methylotrophic bacterium isolated from a terrestrial mud volcano. Extremophiles 24, 177–185. doi: 10.1007/s00792-019-01145-0
Søndergaard, D., Pedersen, C. N. S., and Greening, C. (2016). Hyd DB: a web tool for hydrogenase classification and analysis. Sci. Rep. 6:34212. doi: 10.1038/srep34212
Song, Y., Lee, J. S., Shin, J., Lee, G. M., Jin, S., Kang, S., et al. (2020). Functional cooperation of the glycine synthase/reductase and wood-Ljungdahl pathways for autotrophic growth of Clostridium drakei. Proc. Natl. Acad. Sci. U. S. A. 117, 7516–7523. doi: 10.1073/pnas.1912289117
Stewart, A. G., Laming, E. M., Sobti, M., and Stock, D. (2014). Rotary ATPases-dynamic molecular machines. Curr. Opin. Struct. Biol. 25, 40–48. doi: 10.1016/j.sbi.2013.11.013
Stookey, L. L. (1970). Ferrozine-a new spectrophotometric reagent for iron. Anal. Chem. 42, 779–781. doi: 10.1021/ac60289a016
Tari, G., Blackbourn, G., Boote, D. R. D., Sachsenhofer, R. F., and Yukler, A. (2021). Exploration plays in the Caucasus region. J. Pet. Geol. 44, 213–236. doi: 10.1111/jpg.12791
Toshchakov, S. V., Lebedinsky, A. V., Sokolova, T. G., Zavarzina, D. G., Korzhenkov, A. A., Teplyuk, A. V., et al. (2018). Genomic insights into energy metabolism of Carboxydocella thermautotrophica coupling hydrogenogenic CO oxidation with the reduction of Fe(III) minerals. Front. Microbiol. 9:1759. doi: 10.3389/fmicb.2018.01759
Trüper, H. G., and Schlegel, H. G. (1964). Sulphur metabolism in Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium okenii. Antonie Van Leeuwenhoek 30, 225–238. doi: 10.1007/BF02046728
Tu, T. H., Chen, L. L., Chiu, Y. P., Lin, L. H., Wu, L. W., Italiano, F., et al. (2022). The biogeographic pattern of microbial communities inhabiting terrestrial mud volcanoes across the Eurasian continent. Biogeosciences 19, 831–843. doi: 10.5194/bg-19-831-2022
Tveritinova, T. Y., Sobisevich, A. L., Sobisevich, L. Y., and Likhodeev, D. V. (2015). [Strukturnaya pozitsiya i osobennosti stroeniya gryazevogo vulkana gory Karabetovoy]. Structural position and features of the structure of Karabetova Gora mud volcano. Geologiya i poleznye iskopaemye mirovogo okeana [in Russian] 2, 106–122.
Upadhyay, P., and Lali, A. (2021). Protocatechuic acid production from lignin-associated phenolics. Prep. Biochem. Biotechnol. 51, 979–984. doi: 10.1080/10826068.2021.1881908
Vavourakis, C. D., Andrei, A. S., Mehrshad, M., Ghai, R., Sorokin, D. Y., and Muyzer, G. (2018). A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments. Microbiome 6, 168–118. doi: 10.1186/s40168-018-0548-7
Wang, P. L., Chiu, Y. P., Cheng, T. W., Chang, Y. H., Tu, W. X., and Lin, L. H. (2014). Spatial variations of community structures and methane cycling across a transect of lei-gong-Hou mud volcanoes in eastern Taiwan. Front. Microbiol. 5:2946. doi: 10.3389/fmicb.2014.00121
White, G. F., Edwards, M. J., Gomez-Perez, L., Richardson, D. J., Butt, J. N., and Clarke, T. A. (2016). “Mechanisms of bacterial extracellular electron exchange” in Advances in microbial physiology. Advances in bacterial electron transport systems and their regulation. ed. R. K. Poole (Cambridge, MA: Academic Press), 87–138.
Wischgoll, S., Heintz, D., Peters, F., Erxleben, A., Sarnighausen, E., Reski, R., et al. (2005). Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol. Microbiol. 58, 1238–1252. doi: 10.1111/j.1365-2958.2005.04909.x
Wolin, E. A., Wolin, M. J., and Wolfe, R. S. (1963). Formation of methane by bacterial extracts. J. Biol. Chem. 238, 2882–2886. doi: 10.1016/S0021-9258(18)67912-8
Yang, H. M., Lou, K., Sun, J., Zhang, T., and Ma, X. L. (2012). Prokaryotic diversity of an active mud volcano in the Usu City of Xinjiang. China. J. Basic Microbiol. 52, 79–85. doi: 10.1002/jobm.201100074
Yoon, S. H., Ha, S. M., Kwon, S. J., Lim, J. M., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Zavarzina, D. G., Kolganova, T. V., Boulygina, E. S., Kostrikina, N. A., Tourova, T. P., and Zavarzin, G. A. (2006). Geoalkalibacter ferrihydriticus gen. Nov. sp. nov., the first alkaliphilic representative of the family Geobacteraceae, isolated from a soda lake. Microbiology (Moscow) 75, 673–682. doi: 10.1134/S0026261706060099
Zavarzina, D. G., Prokofeva, M. I., Pikhtereva, V. A., Klyukina, A. A., Maslov, A. A., Merkel, A. Y., et al. (2022). Deferrivibrio essentukiensis sp. nov., gen. Nov., a representative of Deferrivibrionaceae fam. Nov., isolated from the subsurface aquifer of Caucasian mineral drinking waters. Microbiology (Moscow) 91, 143–159. doi: 10.1134/S0026261722020114
Keywords: mud volcano, mineral waters, subsurface biosphere, lithotrophy, thermophile, Fe(III) reduction, protocatechuate, Coriobacteriia
Citation: Khomyakova MA, Zavarzina DG, Merkel AY, Klyukina AA, Pikhtereva VA, Gavrilov SN and Slobodkin AI (2022) The first cultivated representatives of the actinobacterial lineage OPB41 isolated from subsurface environments constitute a novel order Anaerosomatales. Front. Microbiol. 13:1047580. doi: 10.3389/fmicb.2022.1047580
Received: 18 September 2022; Accepted: 24 October 2022;
Published: 10 November 2022.
Edited by:
Hongchen Jiang, China University of Geosciences Wuhan, ChinaReviewed by:
Suresh Kaushik, Indian Agricultural Research Institute (ICAR), IndiaCopyright © 2022 Khomyakova, Zavarzina, Merkel, Klyukina, Pikhtereva, Gavrilov and Slobodkin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sergey N. Gavrilov, c25nYXZyaWxvdkBnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.