- 1Aix-Marseille Université, IRD, APHM, MEPHI, IHU–Méditerranée Infection, Marseille, France
- 2Center National de la Recherche Scientifique, Marseille, France
- 3Laboratoire d’Hématologie, Hôpital de La Timone, APHM, Boulevard Jean-Moulin, Marseille, France
It has been known for many years that the angiotensin-converting enzyme 2 (ACE2) is a cell surface enzyme involved in the regulation of blood pressure. More recently, it was proven that the severe acute respiratory syndrome coronavirus (SARS-CoV-2) interacts with ACE2 to enter susceptible human cells. This functional duality of ACE2 tends to explain why this molecule plays such an important role in the clinical manifestations of coronavirus disease 2019 (COVID-19). At the very start of the pandemic, a publication from our Institute (entitled “ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome”), was one of the first reviews linking COVID-19 to the duplicitous nature of ACE2. However, even given that COVID-19 pathophysiology may be driven by an imbalance in the renin-angiotensin system (RAS), we were still far from understanding the complexity of the mechanisms which are controlled by ACE2 in different cell types. To gain insight into the physiopathology of SARS-CoV-2 infection, it is essential to consider the polymorphism and expression levels of the ACE2 gene (including its alternative isoforms). Over the past 2 years, an impressive amount of new results have come to shed light on the role of ACE2 in the pathophysiology of COVID-19, requiring us to update our analysis. Genetic linkage studies have been reported that highlight a relationship between ACE2 genetic variants and the risk of developing hypertension. Currently, many research efforts are being undertaken to understand the links between ACE2 polymorphism and the severity of COVID-19. In this review, we update the state of knowledge on the polymorphism of ACE2 and its consequences on the susceptibility of individuals to SARS-CoV-2. We also discuss the link between the increase of angiotensin II levels among SARS-CoV-2-infected patients and the development of a cytokine storm associated microvascular injury and obstructive thrombo-inflammatory syndrome, which represent the primary causes of severe forms of COVID-19 and lethality. Finally, we summarize the therapeutic strategies aimed at preventing the severe forms of COVID-19 that target ACE2. Changing paradigms may help improve patients’ therapy.
Introduction
Present in a large number of tissues, including endothelial cells of the arteries, arterioles, and venules of the heart and kidney, angiotensin-converting enzyme 2 (ACE2) is a fascinating molecule which plays a crucial role in maintaining blood pressure homeostasis. ACE2 is only one of the actors in a complex biological network known as the renin-angiotensin system (RAS). ACE2 mainly exerts its functions by regulating the ratio of two major mediators: angiotensin II (Ang II) and angiotensin-[1–7; Ang-(1–7)]. Ang II synthesis is catalyzed by angiotensin-converting enzyme (ACE) while Ang-(1–7) is obtained after hydrolysis of Ang II by ACE2. Ang-(1–7) can also be generated from Ang-(1–9) formed after the action of ACE2 on Ang I by the action of ACE itself. Despite their contrasting physiological functions, the ACE2 is considered to have evolved through ACE gene duplication and exhibits 42% amino acid homology with ACE (Donoghue et al., 2000; Turner and Hooper, 2002; Towler et al., 2004).
Besides being widely studied in cardiology, ACE2 became attractive for other fields of medical sciences and, particularly, virology (Devaux et al., 2020). In 2003 a novel coronavirus infecting humans, the severe acute respiratory syndrome coronavirus (SARS-CoV, provisionally renamed SARS-CoV-1) emerged in Asia, causing an outbreak of severe pneumopathy (Ksiazek et al., 2003; Marra et al., 2003; Rota et al., 2003). ACE2 was demonstrated to be the cellular receptor for SARS-CoV-1, as it had been previously reported for another coronaviruses infecting humans, HCoV-NL63, a coronavirus causing the common winter cold (Hofmann et al., 2005; Li et al., 2007; Ge et al., 2013; Graham et al., 2013). In 2019, new cases of severe pneumopathy were reported in China, with the disease being characterized by a multiple organ dysfunction syndrome (MODS) as well as acute respiratory distress syndrome (ARDS) sometimes requiring the need for ventilation or extracorporeal membrane oxygenation (ECMO). The severe forms of the disease lead to death in ∼ 0.5–2.5% of cases, with a high fatality risk increasing with age and the existence of underlying comorbidities (Huang et al., 2020; Zhou et al., 2020; Zhu et al., 2020). Under chest computerized tomography (CT) scans, the majority of patients show bilateral ground glass-like opacities and subsegmental areas of consolidation indicative of pneumonia. This disease was later defined as COVID-19, the aetiological agent of which was found to be a new human coronavirus named severe acute respiratory syndrome coronavirus (SARS-CoV-2). Although not highly symptomatic for the majority of those infected, the virus has spread worlwide causing more than 6 million deaths for ∼603 million reported cases of infections (World Health Organization COVID-19 Dashboard on 6 September 2022; https://covid19.who.int/). SARS-CoV-2 shares 79.5% nucleotide identity with SARS-CoV-1, and both these Sarbecoviruses isolated from humans are genetically close to coronaviruses circulating in wildlife (Ge et al., 2013; Afelt et al., 2018; Wang et al., 2020; Zhou et al., 2020; Frutos et al., 2021). Once SARS-CoV-2 was characterized, the search for its cellular receptor became a priority. Due to the sequence similarity between SARS-CoV-1 and SARS-CoV-2, studies quickly focused on ACE2 and the role of this molecule as a viral entry receptor was demonstrated (Qiu et al., 2020; Yan et al., 2020).
Due to the central role played by ACE2 in maintaining blood pressure homeostasis, the objective of this work is to review the state of knowledge regarding the possible imbalance of the RAS in the context of a SARS-CoV-2 infection and to highlight the role of ACE2 in SARS-CoV-2 infection and replication, as well as its contribution in the severity of COVID-19.
The renin-angiotensin system: A molecular network which regulates blood pressure homeostasis and ion-fluid balance
In humans and other mammals, intravascular RAS plays a key role in maintaining blood pressure homeostasis as well as fluid and salt balance, while tissue RAS is mainly involved in the pathogenesis of inflammatory diseases (Paul et al., 2006; Greenberg, 2008; de Kloet et al., 2010). The kidneys, as a sensor of ion fluid balance and producer of renin, play a fundamental role in the long-term control of arterial pressure (Tigerstedt and Bergman, 1898; Phillips and Schmidt-Ott, 1999; Yim and Yoo, 2008; Prieto et al., 2011; Gonzalez et al., 2017). Active renin is secreted into the blood circulation in response to hypotension or hypernatremia. Upon activation of the juxtaglomerular apparatus of the kidneys’ afferent arterioles, proteases (proconvertase 1, cathepsin B) catalyze the removal of the 20-amino-acid terminal prosegment of prorenin to produce a polypeptide composed of 297 amino-acids (Davis and Freeman, 1976; Hadman et al., 1984; Cohen-Haguenauer et al., 1989; Sealey and Rubattu, 1989; Neves et al., 1996; Muller et al., 1999). The active form of renin cleaves the alpha-globulin angiotensinogen (formerly angiotonin, a 118-amino-acid-long polypeptide), giving rise to angiotensin I (Ang I), the N-terminal decapeptide of angiotensinogen (Goldblatt et al., 1934; Page and Helmer, 1940; James and Sielecki, 1985). The conversion of Ang I (Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His-Leu) to the octapeptide Ang II (Asp-Arg-Val-Tyr-Ile-His-Pro-Phe), requires the cleavage of its C-terminal dipeptide catalyzed by ACE (provisionally named ACE1) expressed at the endothelial surface of the blood vessels, epithelium of the lungs and upper respiratory system (Skeggs et al., 1956; Crisan and Carr, 2000; Wakahara et al., 2007). The vasoconstrictor octapeptide Ang II was evidenced to be a substrate for ACE2, which acts as an essential factor in the RAS pathway homeostasis. By removing a single residue phenylalanine (Phe) from Ang II, the membrane form of ACE2 (mACE2) plays a central role in the synthesis of the cardiovascular protective heptapeptide Ang-(1–7) that acts by limiting the adverse vasoconstrictor and profibrotic effects of Ang II and reduces the oxidative stress of Ang II on endothelial arteries (Crackower et al., 2002; Pena Silva et al., 2012). ACE2 can also catalyze the conversion of Ang I to Ang-(1–9) by removing the C-terminal leucine (Leu) residue of Ang I, but with a catalytic efficiency ∼ 400-fold lower than the hydrolysis of Ang II to produce Ang-(1–7). Besides Ang II and Ang I, ACE2 can cleave several other substrates including des-Arg9-bradykinin (DABK), apelin-13, and dynorphin A-(1–13; Skidgel and Erdos, 1987; Ferrario et al., 1997; Vickers et al., 2002; Oudit et al., 2003) In addition to its membrane form, ACE2 can be found in a soluble form (sACE2) and increasing sACE2 has been reported in patients with cardiomyopathies and heart failure (Epelman et al., 2008). In patients with aortic stenosis, increasing levels of sACE2 associated with reduced myocardial ACE2 gene expression and severe myocardial fibrosis is considered as a death risk biomaker (Rajagopal et al., 2010). Thus, increased sACE2 plasma levels have been associated with heart failure, cardiovascular disease, and cardiac remodeling (Epelman et al., 2009; Sama et al., 2020; Garcia-Escobar et al., 2021). Using animal models, it was shown that knocking out (KO) of the ACE2 gene results in increased levels of Ang II, followed by vasoconstriction reducing coronary blood flow and leading to cardiac dysfunction (Danilczyk et al., 2003). The expression of mACE2 in the kidneys and heart is influenced by salt rich and/or glucose-rich diets, and can be correlated with pathological disorders (Reich et al., 2008; Lavrentyev and Malik, 2009; Bernardi et al., 2012; Wysocki et al., 2013). In the respiratory tract, DABK is a substrate of mACE2 and a decrease in ACE2 could lead to an increase in vascular permeability and fluid extravasation (Chung et al., 2020). Using a mouse animal model, it was found that loss of ACE2 led to activation of the DABK/braddykinin receptor B1 (BKB1R) axis associated with release of proinflammatory chemokines (e.g., CXCL5, MIP2, and TNFα) and increase in neutrophil infiltration (Sodhi et al., 2017).
Resulting from the cleavage of Ang II by the mACE2 protease, Ang-(1–7) exhibits vasodilatory, anti-proliferative, anti-inflammatory, and antifibrotic effects via the G protein-coupled receptor (GPGR) known as Mas 1 (Santos et al., 2003, 2018; Simoes e Silva et al., 2013; Patel et al., 2016; Karnik et al., 2017; Bader et al., 2018). However, biochemical studies have failed to demonstrate a direct interaction between Ang-(1–7) and Mas1 (Gaidarov et al., 2018). In addition to mACE2, several peptidases, including vascular endothelium prolyl peptidases, neprilysin (NEP), and smooth muscle thimet oligopeptidase, can produce Ang-(1–7; Chappell, 2019). NEP and thimet oligopeptides produce Ang-(1–7) directly from Ang I. Ang-(1–7) has been shown to potentiate bradykinin (BK 1–9), a potent vasodilator of the kinin system which mediates its effects through the B2 receptor (BKB2R) abundant in vascular tissue (Jackman et al., 2002). ACE2 overexpression and Ang-(1–7) infusion have beneficial effects on atherosclerosis, whereas ACE2 deficiency accentuates vascular atherosclerosis in animal models (Dong et al., 2008; Thomas et al., 2010; Yang et al., 2013). The up-regulation of the ACE2/Ang-(1–7)/MasR axis promotes the expression of E-cadherin (E-cad) adhesion molecules by suppressing the PAK1/NF-κB/Snail1 pathway (Yu et al., 2016). Moreover, Ang-(1–7) can exert cerebroprotective functions in endothelin-1-induced ischaemic stroke (Mecca et al., 2011).
For many years, it has been known that there is cross-talk between insulin and the RAS, providing possible links between hypertension, obesity, and diabetes (Alderman et al., 1991; Frederich et al., 1992; Velloso et al., 1996; Boustany et al., 2004; Schmieder et al., 2007). Moreover, a low expression of ACE2 mRNA or protein is associated with an increase in AngII levels, hypertension, diabetes and heart disease (Crackower et al., 2002; Diez-Freire et al., 2006; Tikellis et al., 2012; Velkoska et al., 2016).Interestingly, these diseases are the major comorbidities in the severe forms of COVID-19 (Bavishi et al., 2020). The occurrence of specific comorbidities associated with an RAS imbalance could be decisive for the clinical outcome of COVID-19 (Devaux et al., 2020; Rysz et al., 2021).
RAS imbalance and overproduction of harmful Ang II
Clinical investigations have provided convincing evidence that RAS imbalance is capable of stimulating atherosclerosis, which ultimately lead to the rupture of atherosclerosis plaques and thrombosis (Schmidt-Ott et al., 2000; Jacoby and Rader, 2003; Verdecchia et al., 2008). Ang II is the main harmful effector molecule synthesized in excess in situations of RAS imbalance. Ang II, inactivates the vasodilator bradykinin and can control the ion-fluid balance by acting on the adrenal cortex to stimulate the release of aldosterone, leading to sodium and water retention (Jaspard et al., 1993; Brewster and Perazella, 2004; Xue et al., 2012; Aroor et al., 2016; Nishimura, 2017). The action of Ang II (proximal tubule) and aldosterone (collecting duct) are complementary to influence sodium reabsorption across the nephron (Gurley et al., 2011). Thereby, Ang II functions as a powerful regulator of vascular tone and intravascular volume. Increased circulating levels of Ang II is associated with vasoconstriction and hypertension and accelerates thrombosis in arterioles by activating the coagulation cascade and the platelet-derived growth factor (PDGF; Gustafsson and Holstein-Rathlou, 1999; Heeneman et al., 2000; Senchenkova et al., 2010, 2014; Singh and Karnik, 2016; Samavati and Uhal, 2020). It also induces hypertrophy of vascular smooth muscle cells (Berk et al., 1989; Griendling et al., 1997; Funakoshi et al., 2002). Ang II can also exert tissue-specific actions, such as neurotransmission inducing adipocytes growth in adipose tissues (Li and Ferguson, 1993; Massiéra et al., 2001).
These multiple effects of Ang II are obtained through its ability to bind to Ang II type I and type II receptors (AT1R and AT2R, respectively) expressed in arterioles and several organs including the kidney, pancreas, heart, and the brain. The AT1R, a 359-amino-acids protein spanning cell membrane, and AT2R have a 34% nucleic acid sequence homology (Arendse et al., 2019). Ang II can bind to both to AT1R and AT2R, which are receptors with opposite effects (i.e., AT1R mediates vasoconstriction, inflammation and fibrosis while AT2R mediates opposite effects). AT2R is poorly expressed compared to AT1R, which causes the Ang II to primarily exhibit an effect through AT1R (Murphy et al., 1991; de Gasparo et al., 2000; Forrester et al., 2018; Furuhashi et al., 2020). The activation of AT1R by Ang II is transient and associated with the phosphorylation of the receptor by kinases, including PKC and GRKs. The phosphorylated AT1R is internalized through a mechanism that involves β-arrestin 2, the adaptor protein complex 2 (APC2), clathrin, and intersectin 2 (AbdAlla et al., 2000; de Gasparo et al., 2000; Gáborik et al., 2001). These AT1R-mediated signals lead to overexpression of the prorenin receptor (PRR), thereby increasing renin activity and contributing to the local accumulation of Ang II, fibrosis, and hypertension (Nguyen et al., 2002; Advani et al., 2009; Peng et al., 2013; Wang et al., 2014; Xu et al., 2016; Ichihara and Yatabe, 2019). At the opposite, Ang II also exerts a negative feedback signaling on juxtaglomerular cells that reduces the REN gene transcription and renal renin secretion (Naftilan and Oparil, 1978).
The interaction of Ang II with AT1R functions as a pluripotent mediator to enhance oxidative injury by reactive oxygen species (ROS), and endothelial injury by inhibiting nitric oxide (NO) synthesis. Ang II is a potent activator of NADPH oxidase and an inducer of ROS (Garrido and Griendling, 2009). Interestingly, CHOP−/− mice are protected from Ang II-induced NADPH oxidase activation, hypertension, and cardiovascular disease (Kassan et al., 2016). This is consistent with the observation that Ang II increases the transcription of the CHOP and ATF4 genes (Kassan et al., 2012; Spitler and Webb, 2014; Takayanagi et al., 2015). Activation of AT1R by Ang II also induces various signaling pathways, including G-protein-coupled receptors, PKC, serine/threonine kinase, serine tyrosine kinases, ERK/JNK activation, leading to proinflammatory responses characterized by the synthesis of IL-6, TNFα, and other cytokines (Sadoshima et al., 1995; Han et al., 1999; Nataraj et al., 1999; Ruiz-Ortega et al., 2001; Watanabe et al., 2005; Luther et al., 2006; Rushworth et al., 2008; Dikalov and Nazarewicz, 2013). Furthermore, Ang II activates the flow of neutrophils and macrophages to the affected tissues and inhibits the production of NO, leading to vascular injury (Nabah et al., 2004).
ACE2 tissue distribution in human
Angiotensin-converting enzyme 2 is expressed in virtually all organs with higher levels in capillary rich organs such as the lungs, heart, or kidneys (Donoghue et al., 2000; Tipnis et al., 2000; Ferrario and Varagic, 2010; Tikellis and Thomas, 2012; Figure 1). A study of ACE2 mRNA and protein in more than 150 cell types concluded that ACE2 is mainly observed in enterocytes, renal tubules, the gallbladder, cardiomyocytes, male reproductive cells, placental trophoblasts, ductal cells, eyes, and the vasculature. In the respiratory system, its expression was limited to a subset of cells (Hikmet et al., 2020).
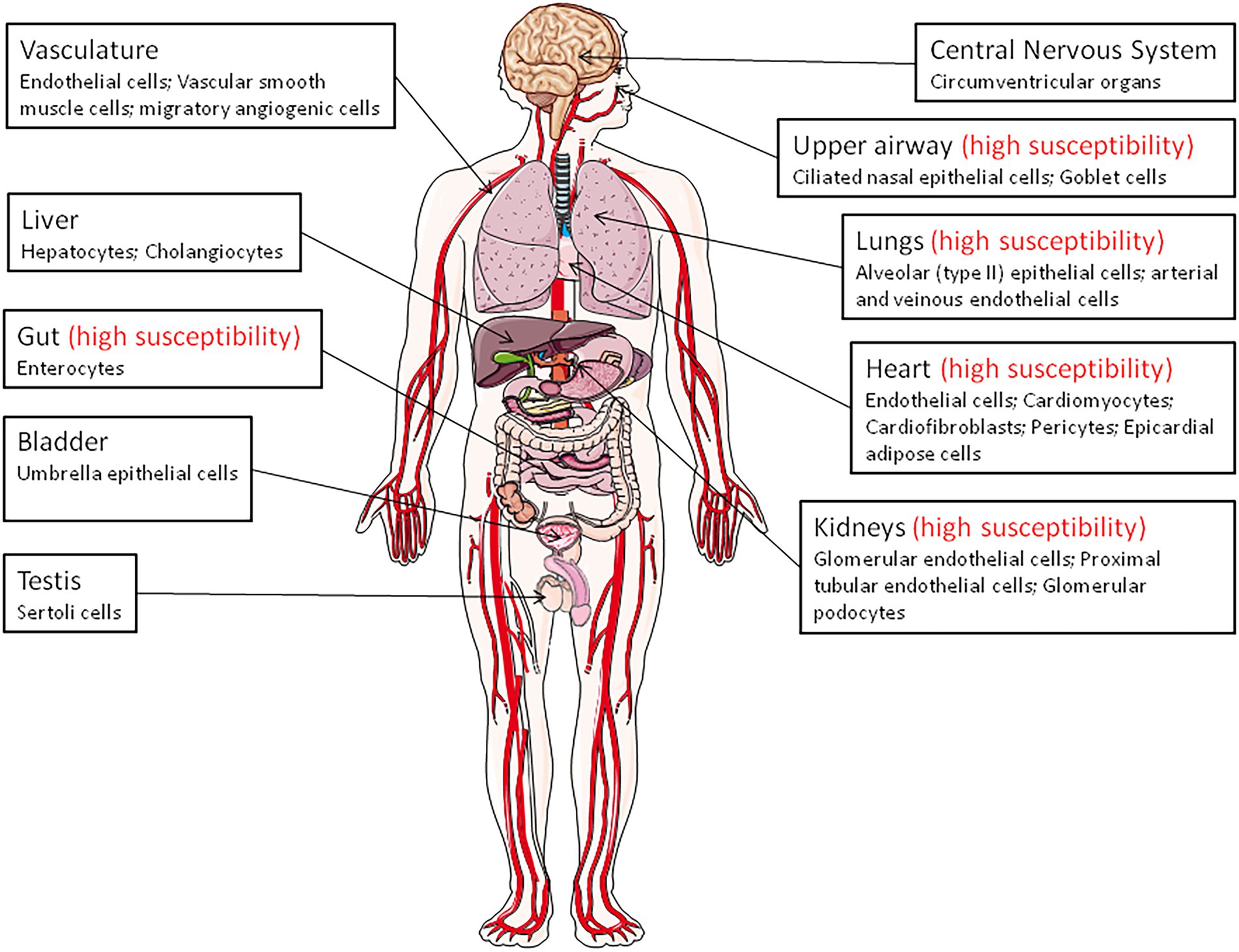
Figure 1. Angiotensin-converting enzyme 2 (ACE2) expression throughout the body (the main ACE2+ target cells are indicated). The organs vulnerability to SARS-CoV-2 infection is also indicated (high susceptibility).
Remarkably, in the upper airway, goblet and ciliated cells show the highest expression of ACE2 and are thought to play a major role in human infection with SARS-CoV-2. The expression of the mACE2 protein is highest within regions of the sinonasal cavity and pulmonary alveoli and in the lung parenchyma (Descamps et al., 2020; Ortiz et al., 2020). In normal human lungs, the mACE2 protein is found on a very small subset of alveolar type II epithelial lung cells (Ortiz et al., 2020; Delorey et al., 2021). Alveolar epithelial type II cells (which represent ∼5% of the alveoli and serves at stem cells to generate type I alveolar epithelial cells), are thought to be a main target for SARS-CoV-2 in the respiratory tract and, consequently, can be destroyed during viral replication (Barkauskas et al., 2013). However, ACE2-positive cells are more abundant in the nasal mucosa than in the bronchus (Hikmet et al., 2020). Moreover, the mACE2 peptidase is also expressed in the arterial and venous endothelial cells present in abundance in the lungs and arterial smooth muscles (Hamming et al., 2004). Expression of ACE2 was found to be drastically increased in airway epithelial cells 24 h after SARS-CoV-1 infection (Li et al., 2020). In COVID-19 related ARDS, ACE2 was found to be upregulated in endothelial cells, but not in type II alveolar epithelial lung cells (Gerard et al., 2021).
The expression of ACE2 in the heart is higher than in the lungs and ACE2 is found in the endothelial cells of coronary arteries, arterioles, venules, and capillaries (Danilczyk et al., 2003; Robinson et al., 2020). The mACE2 is strongly expressed in cardiomyocytes, endothelial cells, cardiac fibroblasts, vascular smooth muscle cells, and was also found in cardiac pericytes, which play crucial role in the microvasculature and may be the target for SARS-CoV-2 (Chen et al., 2020; Hikmet et al., 2020). Patients with heart failure show a significant increase in ACE2 mRNA expression (Goulter et al., 2004), suggesting that ACE2 gene overexpression may explain why heart dysfunction is found within the list of COVID-19 comorbidities. In a rat model of diabetic cardiomyopathy, the overexpression of ACE2 attenuates cardiac hypertrophy, myocardial fibrosis, and dysfunction induced by diabetes (Dong et al., 2012). Post-mortem examinations of endomyocardial biopsies from COVID-19 patients highlighted the presence of SARS-CoV-2 in the myocardium (Lindner et al., 2020; Marchiano et al., 2021).
In the kidneys, ACE2 is expressed in the proximal tubule cells, epithelial cells of the Bowman’s capsule, endothelial cells, mesengial cells (glomerulus central area), glomerular podocytes, proximal cell brush border, and cells from the collecting ducts (Aragao et al., 2011; Hikmet et al., 2020; Martinez-Rojas et al., 2020). Patients with diabetic or hypertensive nephropathy had lower glomerular ACE2 expression compared to healthy controls (Mizuiri et al., 2008; Wysocki et al., 2013). Between 3 and 10% of COVID-19 patients have abnormal renal function (diagnosed with elevated creatinine or urea nitrogen), and 7% experienced acute renal injury (Fan et al., 2021). In the pancreas, ACE2 plays a major glycemia-protective role (Pedersen et al., 2013). In testis, the Sertoli cells, which protect germ cells by forming blood-testis barrier, have a high expression of mACE2, suggesting that SARS-CoV-2 might cause reproductive disorders in infected patients (Shen et al., 2020; Fan et al., 2021).
A high expression of ACE2 was reported in the epithelial cells of the oral mucosa. This is rarely seen in esophageal mucosa (mainly composed of squamous epithelial cells) and is abundantly expressed in the glandular cells of the gastric, duodenal, and rectal epithelia, possibly contributing to the oral transmission of SARS-CoV-2 and then to viral spreading into the gastrointestinal tract, a major target for the virus (Lamers et al., 2020; Xu et al., 2020; Devaux et al., 2021a; Osman et al., 2022). mACE2 is highly expressed thorough the ileum where it may cleave circulating Ang II in the mesenteric arterial blood into Ang-(1–7), which is destined for portal circulation and the liver. The mACE2 also exerts RAS-independent functions in the gastrointestinal tract through cleaving carboxy-terminal amino acids from nutrient proteins and by acting as a chaperon for the expression of the B0AT1 amino acid transporter (Crackower et al., 2002; Camargo et al., 2009; Singer and Camargo, 2011; Fairweather et al., 2012; Hashimoto et al., 2012; Vuille-Dit-Bille et al., 2015; Wang et al., 2015). The mACE2 regulates the gut homeostasis, microbiota composition, the expression of antimicrobial peptides (Reg3γ, α-defensin, such as HD5 and HD6, β-defensin, and lysozyme; Singer et al., 2012; Perlot and Penninger, 2013; Ferrand et al., 2019). This probably explains the diarrhea that is sometimes observed in SARSCoV-2 patients, and supports the use of antibiotic treatment in COVID-19 patients. In addition, it was reported that HD5 secreted by intestinal Paneth cells, interacts with ACE2 (Wang et al., 2020), suggesting that the presence of HD5 in abundance in the ileal fluid may compete with SARS-CoV-2 to bind to ACE2. The infection of Caco2 cells by SARS-CoV-2 was found to be significantly reduced when cultured in the presence of HD5 and this effect was confirmed on intestinal and lung epithelial cells and for different SARS-CoV-2 variants (Wang et al., 2020; Xu et al., 2021). Although the ACE2 regulation of gut homeostasis was considered to be RAS-independent, α-defensins expression has also been associated with atherosclerosis, being involved in the lipoprotein metabolism in the vessel wall and inhibiting fibrinolysis (Kougias et al., 2005; Nassar et al., 2007; Abdeen et al., 2021).
Structure of the human ACE2 protein
The ACE2 gene encodes a type I transmembrane glycoprotein of ∼ 100 kDa composed of 805 amino acids(Figure 2; Marian, 2013; Gheblawi et al., 2020), including six amino acids (Asn53, Asn90, Asn103, Asn322, Asn432, and Asn546), which can potentially be N-glycosylated (Lubbe et al., 2020). This metalloprotease resembles a chimera molecule composed of a single ACE-like catalytic ectodomain (41.8% sequence homology with the amino domain of ACE) fused to a collectrin-like domain (48% homology with collectrin; Donoghue et al., 2000; Zhang et al., 2001). The functional domains of ACE2 include: (i) a N-terminal signal peptide region of 17 amino acid residues; (ii) a peptidase domain (PD; amino acids 19–615) with its zinc binding metalloprotease motif (catalytic domain; amino acids 374–378); (iii) a C-terminal collectrin-like domain (CLD; amino acids 616–768 acting as a regulator of renal amino acid transport and insulin exocytosis), containing a ferredoxin-like fold “neck” domain (amino acids 615–726); and (iv) an hydrophobic transmembrane hydrophobic helix region of 22 amino acids followed by an intracellular cytoplasmic tail of 43 amino acids (Donoghue et al., 2000; Zhang et al., 2001; Cerdà-Costa and Gomis-Rüth, 2014). The C-terminal segment of mACE2 contains a PDZ-binding motif (amino acids 803–805) Thr803-Ser804-Phe805 (TSFCOOH) targeting protein-interacting domains from proteins (SNX27, SHANK3, MAST2, and NHERF2) involved in protein trafficking (Caillet-Saguy and Wolf, 2021; Kliche et al., 2021).
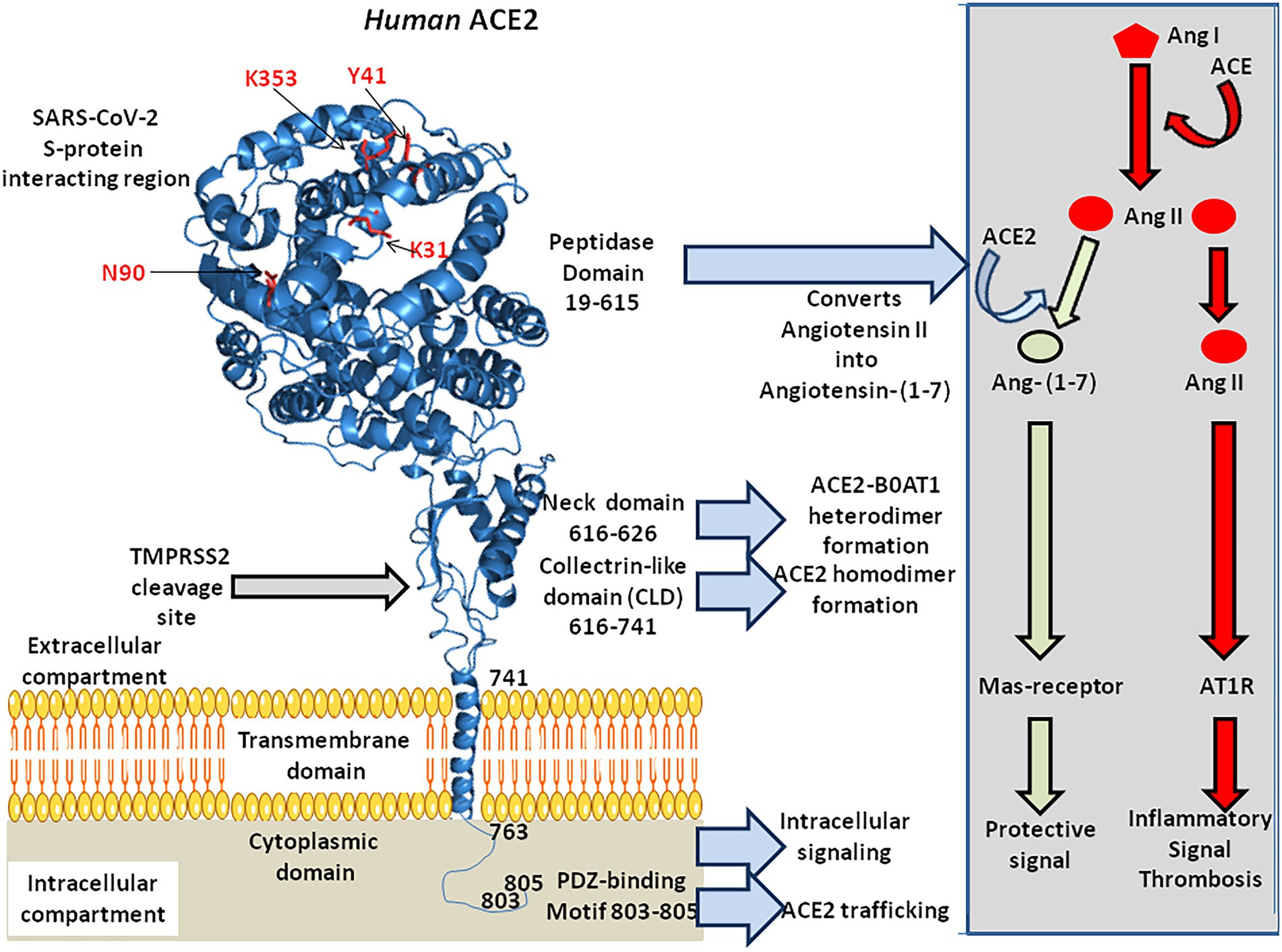
Figure 2. 3-D model of ACE2 protein structure according to the published data PDB: 6M1D by Yan and colleagues in 2020. ACE2 is composed of 805 amino acids and characterized by an N-terminal signal peptide of 17 amino acid residues, a peptidase domain (PD; residues 19–615) with its HEXXH zinc binding metalloprotease motif, a C-terminal collectrin-like domain (CLD; residues 616–741) that includes a ferredoxin-like fold “neck” domain (616–626), that end with a hydrophobic transmembrane hydrophobic helix region (741–763) followed by an intracellular segment of 43 amino acid residues. No information is available regarding the 3-D structure of the ACE2 cytoplasmic tail (763–805), which was drawn to appear on the diagram. Some of the amino acids which are considered important for SARS-CoV-2 interaction are located in the 3-D model (amino acids in red). Arg652, Arg708, and Arg710 (not shown) are the active residues for ACE2-TMPRSS2 docking. The function of each domain is indicated on the middle right side of the figure. The biochemical pathway of the RAS and the beneficial ACE2/Ang-(1–7) arm of RAS are summarized in the right box.
The mACE2 functions predominantly as a monocarboxypeptidase, with a substrate preference for hydrolysis between a proline and a hydrophobic or basic C-terminal residue (Turner and Hooper, 2002). The catalytic domain of mACE2 consists of two subdomains (subdomains 1 and 2) forming the two sides of a long deep cleft bridged together by a hinge region. Upon substrate binding, the two catalytic subdomains undergo a hinge-bending movement and form a binding cavity required to initiate substrate hydrolysis (Towler et al., 2004). The His-Glu-X-X-His motif (or HEXXH motif where X is any amino acid), coordinates a catalytic zinc ion, characteristic of zinc-dependent metalloproteases. The zinc is co-ordinated by His374, His378, Glu402, and one water molecule in the subdomain 1, whereas a chloride ion is co-ordinated by Arg169, Trp477, and Lys481 in the subdomain 2. The Arg514 of mACE2 is considered as a residue critical for substrate selectivity (Luther et al., 2006).
Both the PD and neck domains of mACE2 contribute to dimerization, whereas each B0AT1 interacts with the neck and TM helix in the adjacent mACE2 (Yan et al., 2020). Complexes of mACE2/B0AT1 heterodimers have been evidenced at the intestinal apical membrane but did not occur in lung pneumocytes. Steric hindrance to the B0AT1 binding site on mACE2 or down-regulation of mACE2 due to the presence of SARS-CoV-2 is likely to display impaired intestinal tryptophan uptake (Devaux et al., 2021a).
Finally, the Arg652 of ACE2 is a target for the catalytic site of proteases ADAM17 and TMPRSS2, which leads to the shedding of a soluble form of ACE2 (sACE2; Heurich et al., 2014; Lanjanian et al., 2021).
The human ACE2 gene variant mRNAs
The prototype human ACE2 cDNA (or ACHE for angiotensin-converting enzyme homolog) was cloned more than 2 decades ago from a human cardiac left ventricle cDNA library and a lymphoma cDNA library (Donoghue et al., 2000; Ferrario and Varagic, 2010). The ACE2 gene, which contains 20 introns and 19 exons maps to chromosome Xp22 and spans 39.98 kb of genomic DNA (Turner and Hooper, 2002). Two isoforms of ACE2 with 18 or 19 exons (v1 and v2) that encode the same protein (805 amino acids) have been described, as well as three other smaller variants: x1–x3 (Chen et al., 2020; Khayat et al., 2020). ACE2 shows similarities with the ACE gene located at chromosome 17q23 (Hubert et al., 1991). Although ACE2 is one of the genes escaping X chromosome inactivation, there is evidence of sex bias (Tukiainen et al., 2017; Cai, 2020; Gay et al., 2021). Indeed, there is a plausible mechanism of androgen-induced expression of ACE2 that contributes to increased susceptibility or severity of COVID-19 in males (Baratchian et al., 2021). The tissue levels of mACE2 represent equilibrium between transcription/translation of mACE2 and shedding rate of sACE2. It was reported that a positive relationship exists between renin and sACE2 levels in male and female subjects, and between sACE2 levels and body mass index (BMI) in males, with possible implication for COVID-19 (Jehpsson et al., 2021). Variations in mACE2 with age were first demonstrated using animal models (Xie et al., 2006). A negative association between age and sACE2 plasma concentrations in people above the age of 55 year-old, was reported (AlGhatrif et al., 2021). The mACE2 deficiency is considered to be linked to cardiovascular disease and diabetes, suggesting that mACE2 deficiency may increase the risk of developing severe COVID-19 (Oudit and Pfeffer, 2020; Verdecchia et al., 2020; Wang et al., 2020).
The transcription of full-length ACE2 (2,721 bp mRNA) is initiated from either a proximal or a distal promoter with tissue-specific differences in their usage (Itoyama et al., 2005; Fan et al., 2021). The proximal site contains a TATA box motif at position-110/−96 of the transcription start site and a GATA motif and two HNF1 binding sites at position-165/−131. The distal site contains YY1/COUP, C/EBPβ, and STAT/FOXA motifs. Site-directed mutagenesis of the human ACE2 promoter region from position −2069 to +20, has enabled the identification of an activating domain in the −516 to −481 region (Kuan et al., 2011) and a potential binding site, ATTTGGA, homologous to that of an Ikaros-like binding domain which can be regulated by the levels of Ang II. It has also been reported that the NAD + -dependent deacetylase silent information regulator T1 (SIRT1 known for its ability to deacetylate proteins such as p53 and forkhead box O), binds to the ACE2 promoter and regulates ACE2 gene expression under condition of energy stress which increase AMP-activated protein kinase, while IL-1β treatment decreased the binding of SIRT1 to the ACE2 promoter (Clarke et al., 2014). In addition, there is a cAMP-responsive element (CREB)-binding site within an upstream region of the start site containing both p300 (a CREB co-activator that relaxes the chromatin and recruits RNA polymerase II) and the CREB site (Figure 3).
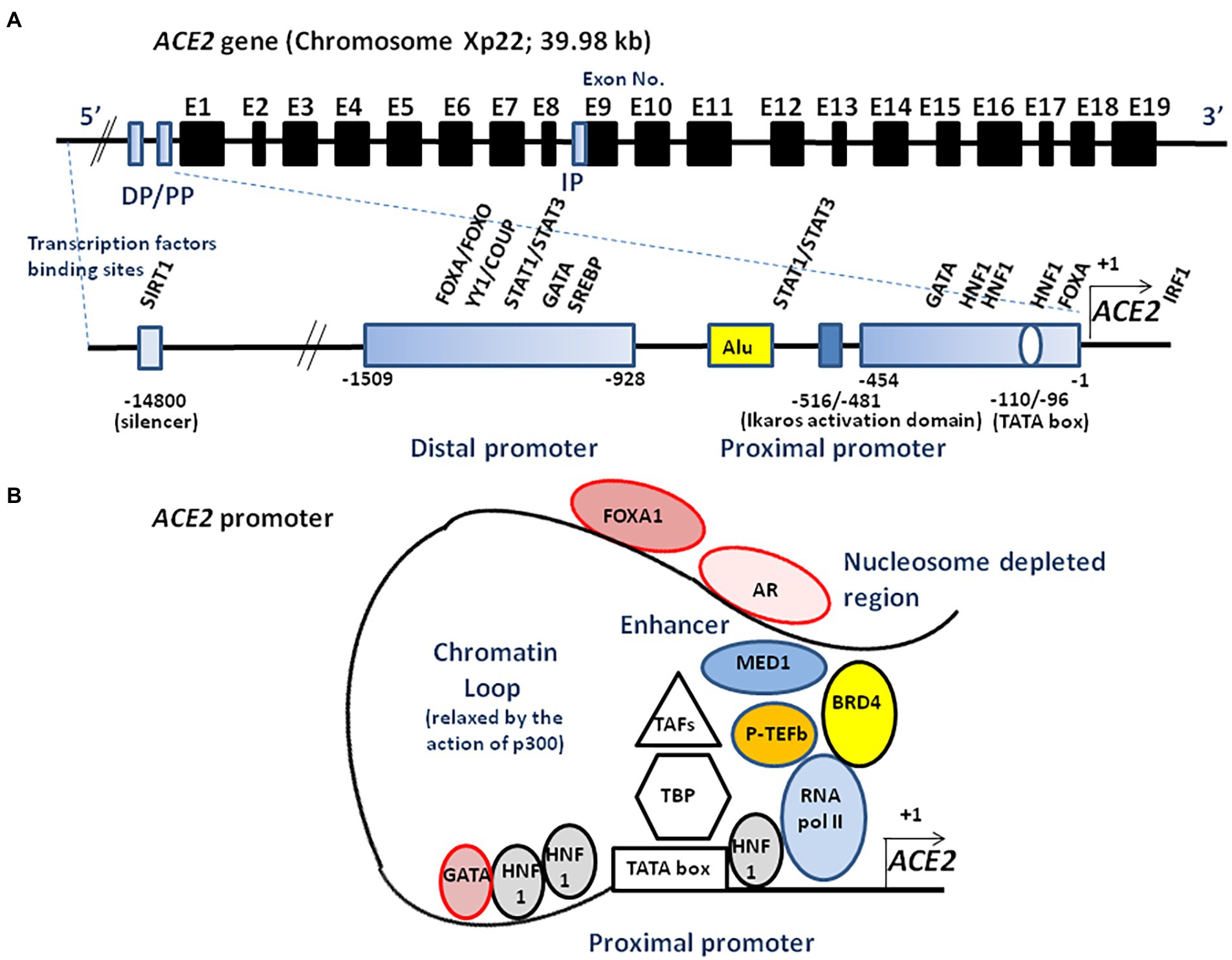
Figure 3. Schematic illustration of ACE2 transcriptional regulation. (A) A schematic diagram of the ACE2 gene structure (upper panel). The known exons (E1–E19) are depicted as black boxes. The location of the distal promotor (DP) and proximal promoter (PP) are depicted as blue boxes. The ACE2 gene can encode several transcript leading to several isoforms. An internal promoter (IP) is thought to activate the transcription of an mRNA encoding a short isoform of ACE2 which lacks the SARS-CoV-2 binding site. The 5′ region upstream of the ACE2 gene contains two promoters (proximal and distal) separated by a repetitive Alu element (lower panel). The transcription of full-length ACE2 is initiated from either the proximal or distal promoter with tissue-specific differences in their usage. Transcription factors binding to the proximal and the distal upstream promoter regions are indicated. Ang II is likely to regulate the ACE2 expression through the Ikaros activation domain. Truncated ACE2 forms (e.g., dACE2) can also be expressed. (B) ACE2 transcriptome. AR binds to the enhancer element of the ACE2 gene, connecting the regulatory circuit between the enhanceosome complex (comprising MED1, BRD4, etc.) and the promoter-bound RNA polymerase machinery to activate gene expression. P-TEFb, positive transcription elongation factor; TBP, TATA-binding protein; TAFs, TBP-associated factors; FOXA1, forkhead box A1; BRD4, bromodomain-containing protein 4; MED1, mediator complex subunit 1; SREBP, sterol regulatory element binding protein; and SIRT1, silent information regulator T1.
The in silico study of candidate binding sites within the 400 bp upstream of the transcription start site identified putative sites for various DNA-binding molecules, with different tissue expression such as CDX2 in the lungs, colon, and terminal ileum; HNF1A in the colon, kidneys, and terminal ileum; FOXA1 in the cervix, colon and terminal ileum; SOX11 in the kidneys, and TCF7/LEF1 in the lungs (Barker and Parkkila, 2020). The ACE2 promoter also contains an androgen receptor (AR) binding site, and AR antagonists (e.g., enzalutamide, apalutamide) have been reported as being able to decrease SARS-CoV-2 infection (Qiao et al., 2021). Moreover, forkhead box A1 (FOXA1; also known as HNF3α) involved in AR signaling, and bromodomain-containing protein 4 (BRD4) binding sites, overlap with open chromatin regions. Bromodomain and extra terminal domain (BET) antagonists (e.g., JQ1, OTX015), inhibit BRD4, a factor able to interact with positive elongation factor (P-TEFb) cyclin-dependent kinase required for transcription elongation through RNA polymerase II (RNA pol II), also decrease SARS-CoV-2 infection through the inhibition of BRD4. The distal-less homeobox 2 (DLX2) and CCAAT/enhancer binding protein epsilon (CEBPE) are more represented in ACE2-expressing cells (Sherman and Emmer, 2021). Evidence for additional transcription factor binding sites (e.g., SP1, CEBP, GATA3, HNF4A, USF1, etc.) has also been reported (Beacon et al., 2021).
Putative binding sites for signal transducer and activator of transcription, STATs (−662 to −647 region and −911 to −897 region), and interferon-regulatory factors, IRFs, have also been demonstrated (Ziegler et al., 2020). Indeed, interferon modulates ACE2 expression and can lead to the transcription of a truncated form of ACE2, designated as deltaACE2 (dACE2) which lacks 356 amino-terminal amino acids and fails to bind to SARS-CoV-2 (Onabajo et al., 2020). The transcription of such a truncated form of ACE2 involves the activation of a promoter located downstream of the transcription start site with a splicing event introducing a new ATG start codon. The analysis of this region identified ISGF-3-, AP-1-, and NF-κB-binding sites (Blume et al., 2021). Treating cells with IFNβ significantly induces the dominant expression of dACE2 over ACE2 (Onabajo et al., 2020). In addition, the possible role of alternatively spliced isoforms of ACE2 in SARS-CoV-2 homing, infectivity, and influence on COVID-19 evolution, should be investigated (Heyman et al., 2021; Nikiforuk et al., 2021). Polymorphisms in ACE2 gene 5′ upstream regions might influence ACE2 expression. Differences greater than 1% of minor allele frequency (MAF) in the 10 Kb region upstream to ACE2 analyzed using data from the 1,000 Genomes project, found 57 polymorphisms (Lanjanian et al., 2021). A single nucleotide polymorphism (SNP), rs5934250, with a change from G to T at approximately 5,700 bp upstream of the start codon of the ACE2 gene, presented a penetration difference among populations. This allele is almost absent in the East Asian population, while it has a MAF in almost half of Europeans (East Asians: 1%; Africans: 10%; South Asians: 22%; Americans: 29%; and Europeans: 47%). Another SNP, rs2097723, also shows a very heterogeneous distribution among populations (Africans: 7%; South Asians: 22%; Europeans: 28%; Americans: 32%; and East Asians: 42%).
Human ACE2 polymorphism
Exploration of the ACE2 genetic polymorphism was conducted to define SNPs associated with hypertension and heart diseases. Special attention was drawn to 14 SNP (rs2285666, rs1978124, rs2074192, rs2106809, rs4830542, rs4240157, rs879922, rs2158083, rs233574, rs1514282, rs1514283, rs4646155, rs4646176, and rs4646188). The best characterized SNP is a splice region variant (rs2285666, G > A, Intron 3/4), known to be associated with hypertension, coronary heart disease, and diabetes (Yang et al., 2015; Pinheiro et al., 2019; Bosso et al., 2020). A number of SNPs, including genotypes of rs2048683, rs233575, rs2158083, rs2074192, rs2106809, rs4240157, rs4646155, and rs4830542 were linked with moderate risks of hypertension, while rs4646188 and rs879922 were linked to high hypertension risks (Yi et al., 2006; Fan et al., 2009; Patnaik et al., 2014; Dai et al., 2015; Meng et al., 2015; Chen et al., 2016; Liu et al., 2018; Luo et al., 2019), and the rs2074192 and rs2106809 were associated with left ventricular hypertrophy in hypertensive patients (Fan et al., 2019). The ACE2 A1075G allele found in China was associated with hypertension and the ACE2 G8790A allele is associated with susceptibility to hypertension, type 2 diabetes, and increased plasma concentration of sACE2 (Niu et al., 2007; Wu et al., 2017; Pinheiro et al., 2019). An allele frequency heterogeneity for the rs2285666 (East Asians: 17%; South Asians: 23%; Americans: 37%; Africans: 48%; Europeans 48%; and with the highest frequency in Indians: 71%) has been reported (Khayat et al., 2020) while the rs4646140 has a MAF ranging from zero in Indians to 13% in Africans. Polymorphisms, including rs233574, rs2074192, and rs4646188 with MAF of 16, 36, and 6%, respectively, were able to induce a significant RNA secondary structure change (Pouladi and Abdolahi, 2021). These alterations may lead to dysregulations in ACE2 transcription/translation or its protein stability. Indeed, in the case of the mutated alleles, the splicing regulatory molecule ETR-3 is unable to bind to the pre-mRNA. Similarly, in the case of the mutated forms of rs2158083 and rs2285666, the binding of YB-1 and hnRNP DL, respectively, are impaired, resulting in exon retention. In the case of the mutated form of rs1514283, the SF2/ASF, and SRp40 proteins bind and lead to the creation of a new intron splicing enhancer and exon inclusion. In the case of the mutated form of rs879922, there is a possibility of interaction with the SC35 and DAZAP1 proteins that leads to exon inclusion. In addition, the binding of proteins of the hnRNP A1, A0, A2/B1, D, and DL family creates a new intronic splice silencer and intron exclusion. In the case of the mutated form of rs4646155, the NOVA-1 protein induces an exon inclusion, while SLM-2 and Sam68 lead to intron exclusion. In the case of the mutated form of rs2106809, the hnRNP H proteins lead to an intron exclusion.
As COVID-19 emerged, it was postulated that SNPs in the ACE2 gene could affect susceptibility for SARS-CoV-2 infection (Darbani, 2020; Devaux et al., 2020; Hou et al., 2020). Particular attention was paid to the impact of the G8790A mutation on the severity of COVID-19, although its role in this disease remains controversial (Gómez et al., 2020; Möhlendick et al., 2021). About 77% of GG genotype, 13% of GA genotype and 9% of AA genotype were found in Caucasian SARS-CoV-2-positive patients and 70% of GG genotype, 14% of GA genotype and 16% of AA genotype carriers in SARS-CoV-2-negative people, respectively. A meta-analysis concluded that the ACE2 variant rs190509934:C (a rare variant) characterized by a lower ACE2 expression in individuals carrying the C allele, reduces the risk of SARS-CoV-2 infection (Horowitz et al., 2021).
Analysis of inter-individual ACE2 polymorphism, based on broad genomic databases reveal a link with the susceptibility to SARS-CoV-2 and the severity of COVID-19 (Brest et al., 2020; Cao et al., 2020). The pioneering work by Cao and colleagues identified 15 unique expression quantitative trait loci variants (14 SNPs and 1 InDel) with a higher frequency of minor alleles in the Asian population than in the European population. For example, the rs143695310 variant among East Asian populations was found to be associated with elevated expression of ACE2. Moreover, it was reported that Asian men have a higher ACE2 mRNA expression in their lungs than women, and that Asian people express higher amount of ACE2 than Caucasian and African American populations according to single-cell RNA-seq analysis (Zhao et al., 2020; Figure 4). Similar data were obtained using expression quantitative trait loci (eQTL), indicating a higher expression of ACE2 in South Asian and East Asian populations compared to Europeans, while the lowest expression levels were observed for Africans (Ortiz-Fernández and Sawalha, 2020). Dozen of human ACE2 variants were identified, which could impact on protein stability (e.g., Lys26Arg, Gly211Arg, and Asn720Asp variants) or internalization (e.g., Leu351Val and Pro389His variants; Benetti et al., 2020; Cao et al., 2020; Othman et al., 2020). The rs41303171 C polymorphism, which is practically exclusive to Europeans (MAF 1.8%), is a missense SNP causing an Asn720Asp replacement, which can trigger a conformational disorder in ACE2 changing viral interactions (Khayat et al., 2020). The Pro389His variant occurs in Latino American population with an allele frequency of 0.015%. Only African Americans carry Met383Thr and Asp427Tyr variants with allele frequencies of 0.003 and 0.01%, respectively. The Arg514Gly occurs in African Americans with an allele frequency of 0.003% (Hou et al., 2020). The European population with Arg708Trp, Arg710Cys, Arg710His, or Arg716Cys variants in mACE2 may have mild symptom of COVID-19 as ACE2 lose the cleavage site by TMPRSS2 (Hou et al., 2020; Lanjanian et al., 2021). The Ser19Pro variant (rs73635825 genotype) common in African populations, may protect against COVID-19 while the Lys26Arg variant (rs75548401 genotype) might predispose to severe forms of COVID-19 (Calcagnile et al., 2021). Recently, Suryamohan and colleagues found 298 unique ACE2 variants (Suryamohan et al., 2021). Among these variants they predicted that the Lys31Arg polymorphism breaks an interaction with Gln493 in the viral RBD and destabilizes the charge-neutralizing interaction with the virus and that the Glu37Lys polymorphism disrupts the critical interactions with ACE2 Lys353 by removing the polar intramolecular interaction that stabilizes contacts with the SARS-CoV-2 RBD. Similarly, the His34Arg was predicted to result in a loss of interface polar contact. Thus, individuals carrying these variants are predicted to be less susceptible to SARS-CoV-2 infection. Fourteen human ACE2 variants (Ile21Val, Glu23Lys, Lys26Arg, Asn64Lys, Thr92Ile, Gln102Pro, Asp206Gly, Gly211Arg, Arg219Cys, Glu329Gly, His378Arg, Val447Phe, Ala501Thr, and Asn720Asp) which could enhance susceptibility to SARS-CoV-2 were found to have an higher allele frequencies in European populations than East Asian populations, while two additional ACE2 variants (Glu35Lys and Phe72Val) possibly conferring resistance to the virus, have higher allele frequencies in East Asian populations, while they are low or not expressed in European populations (Chen et al., 2021). Recently, a total of 570 genetic variations (SNP and InDel) on the ACE2 gene were reported in the Iranian population (Lanjanian et al., 2021).
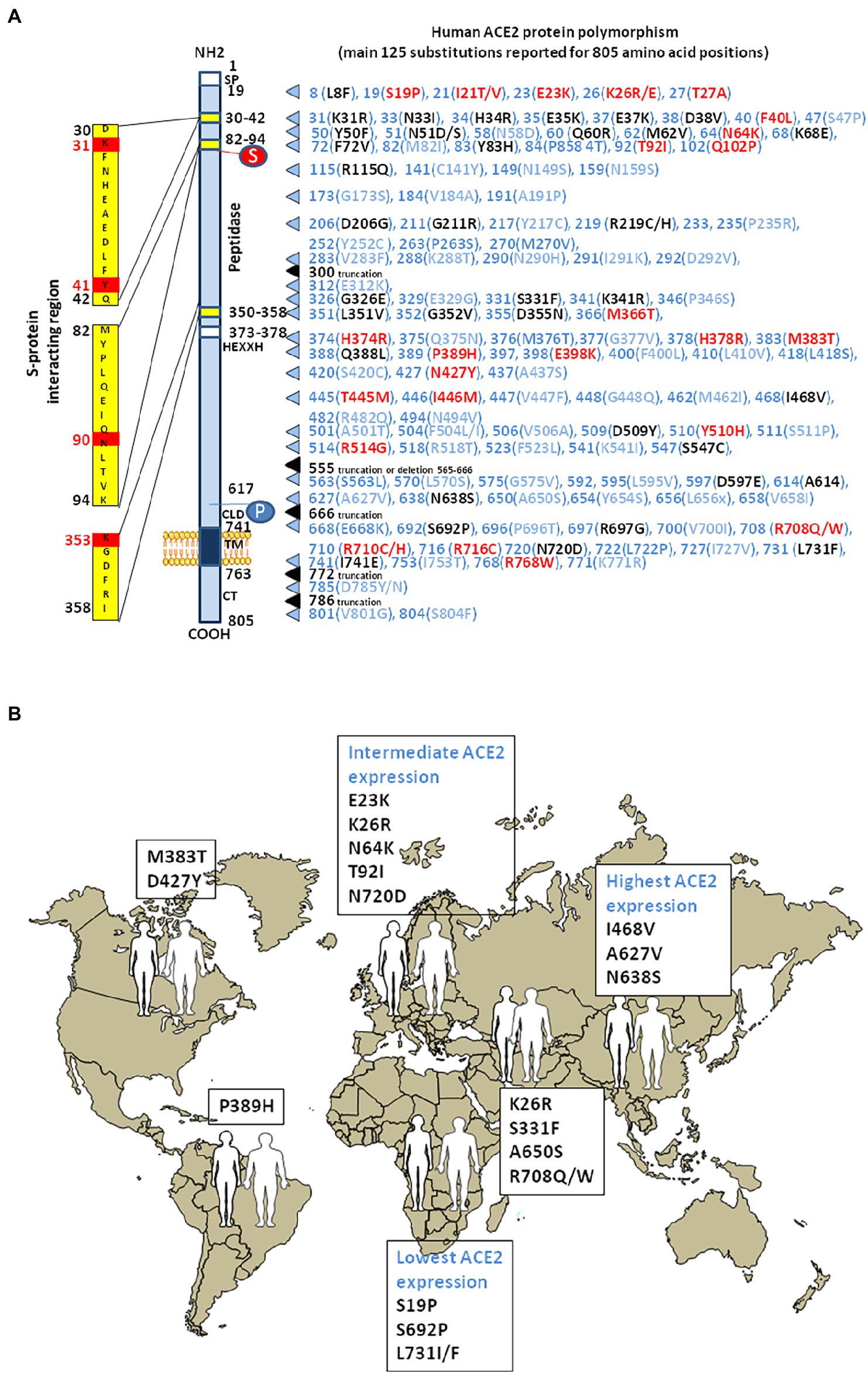
Figure 4. Human ACE2 polymorphism. (A) Schematic representation of the cell surface of the human ACE2 molecule and its major domains is drawn on left side of the figure. The amino acid positions are in black. Some of the amino acids considered to be important for viral tropism are marked in red. S, sugar; P, phosphorylation. The right part of the figure is a compilation of the main substitutions described in the literature. To simplify the figure, we used the single letter amino acids code instead of multiple letters code. The ACE2 substitutions in blue are considered neutral. The ACE2 substitutions in red are predicted to increase cell susceptibility to SARS-CoV-2. The ACE2 substitutions in black are predicted to decrease cell susceptibility to SARS-CoV-2. Polymorphisms in intronic regions might modify ACE2 regulation. Polymorphisms were able to induce a significant RNA secondary structure change. These alterations may lead to dysregulations in ACE2 transcription/translation or its protein stability. (B) The main geographical distribution of ACE2 protein polymorphisms in human populations. Representative substitutions in the human mACE2 per geographic areas.
ACE2 production and regulation inside human cells
Angiotensin-converting enzyme 2 surface abundance differ among cell types, indicating a complex epigenetic regulation of the ACE2 gene. The interaction between tissue or cell type specific enhancer/repressor is required for gene expression (Andersson et al., 2014). The ACE2 gene expression is also increased in individuals with pulmonary arterial hypertension, chronic obstructive pulmonary disease, obesity, diabetes, and older people (Muus et al., 2020; Pinto et al., 2020). In patients with hypertensive cardiopathy a marked ACE upregulation and ACE2 downregulation associated with Ang II/AT1R induced activation of the ERK1/2 and p38 MAP kinase, was reported (Koka et al., 2008). DNA methylation (5mC) was found to be involved in the silencing of ACE2 gene expression and CpG methylation was greater in patients with hypertension compared to healthy controls (Fan et al., 2017; Chlamydas et al., 2020; Cardenas et al., 2021). In contrast, enhanced ACE2 expression might also be protective in COVID-19 if it increases the peptidase activity of ACE2 thereby reducing Ang II concentration. Hypomethylation of specific sites in the ACE2 promoter was reported to correlate with increased ACE2 gene expression (Corley and Ndhlovu, 2020). Three CpGs (cg04013915, cg08559914, and cg03536816) at the ACE2 gene were reported as having lower methylation in lung epithelial cells compared to the other tissues (Beacon et al., 2021). The search for ACE2 topologically associating domains (TADs) with active histone markers, including H3 acetylated at K27 (H3K27ac) and H3 trimethylated at K4 (H3K4me3) or repressive histone markers (H3K27me3), revealed the presence of H3K4me3 at the promoter and after the first exon of ACE2, and the presence of H3K27ac in human kidneys (Beacon et al., 2021). The association of H3K4me3 correlates with ACE2 gene expression in the kidneys, heart, and small intestine. In contrast, H3K4me3 peaks are not detected in lung tissues.
MicroRNAs (miRNAs) are non-coding RNAs which can bind the 3′-untranslated regions (3’-UTRs) of target mRNAs, thereby regulating gene expression at a post-transcriptional level. Lysine-specific demethylase 5B, JARID1B, is responsible for the downregulation of several miRNAs that target ACE2 (Henzinger et al., 2020). Putative miRNA-binding sites were identified in the 3′-UTR of the ACE2 transcript thereby repressing translation. Both the miR-421, an miRNA implicated in the development of thrombosis and the miR-200c-3p were found to downregulate the ACE2 mRNA expression (Hirano and Murakami, 2020). In contrast the increases ACE2 mRNA expression (Sato et al., 2013; Siddiquee et al., 2013; Zhang et al., 2017). Other miRNAs predicted to bind to ACE2 mRNA 3’-UTR, such as miR-9-5p and miR-218-5p, were found to be differentially expressed in different cell types (Pierce et al., 2020). Moreover, the repression of the Xu and Li, 2021; Figure 5).An in silico studies aimed at predicting miRNAs that regulate ACE2-related networks with a possible impact on COVID-19 outcome, suggests that the top miRNAs regulating ACE2 networks are miR-27a-3p, miR-26b-5p, miR-10b-5p, miR-302c-5p, hsa-miR-587, hsa-miR-1305, hsa-miR-200b-3p, hsa-miR-124-3p, and hsa-miR-16-5p (Wicik et al., 2020). sACE2 shed into systemic circulation maintains its ability to generate Ang-(1–7). This process is fine-tuned by ADAM17 (also known as TACE), the metalloprotease ADAM10, and the transmembrane protease serine 2 (TMPRSS2), but only TMPRSS2 increases the entry of both SARS-CoV-1 and SARS-CoV-2 into susceptible cells (Lambert et al., 2005; Heurich et al., 2014; Hoffmann et al., 2020; Qiao et al., 2021). The ADAM17 and ADAM10 sheddases can trigger ACE2 ectodomain shedding by cleavage between amino acids 716 and 741 near the predicted transmembrane domain (Xiao et al., 2014), while TMPRSS2 trigger cleavage between amino acids 697 and 716 (Lanjanian et al., 2021). Phorbol ester and ionomycin as well as the proinflammatory cytokines IL-1β and TNF-alpha, can induce cellular proteases to catalyze sACE2 shedding (Jia et al., 2009). A study of plasma samples from 534 subjects indicated that up to 67% of the phenotypic variation in sACE2 shedding could be accounted for by genetic factors (Rice et al., 2006). mACE2 also interacts with several PDZ-binding proteins such as NHERF, involved in the internalization and recycling of mACE2 (Zhang et al., 2021). The in silico study of proteins belonging to the ACE2 interactome and which could be affected by SARS-CoV-2 infection, highlighted that the most affected interactions were associated with microtubule-associated serine and threonine kinase 2 (MAST2), and [Calmodulin 1 (CALM1; Wicik et al., 2020]. It was previously reported that CALM1 inhibitors increase sACE2 shedding by preventing calmodulin binding to the cytoplasmic tail of mACE2 (Lambert et al., 2008).
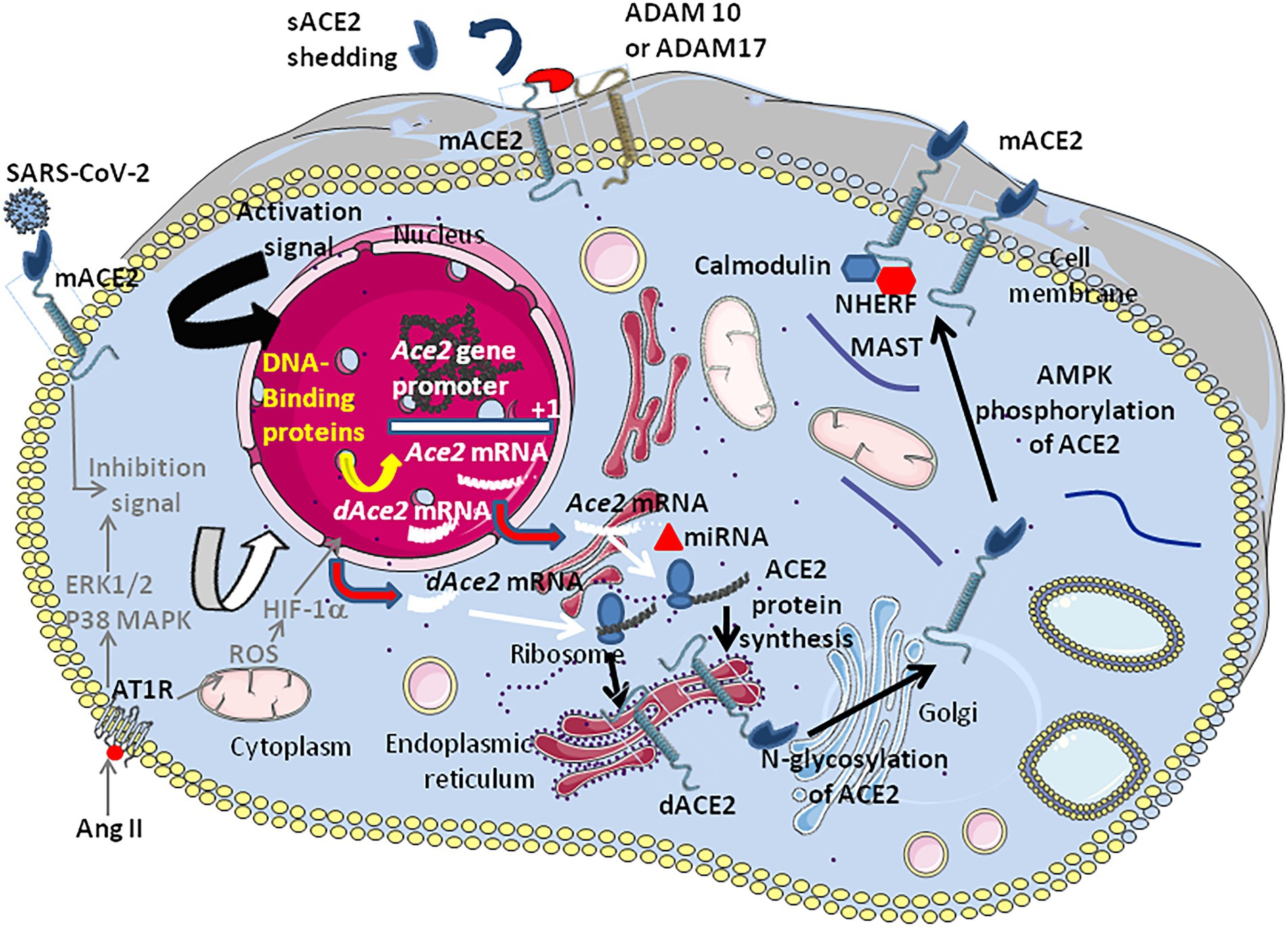
Figure 5. Schematic representation of the regulation of ACE2 expression. The transcription of the Ace2 gene is under control of several DNA-binding proteins that bind the Ace2 gene promotor (see Figure 3). In patients with hypertension (hypertensive cardiopathy and hypertensive nephropathy) a down-regulation ACE2 is observed. Angiotensin II was shown to down-regulate ACE2. The inhibition of ACE2 expression was shown to be associated with the activation of ERK and p38 MAP kinases (however this signaling pathway also activates NF-κB which is an activator of ACE2, suggesting a complex regulation of positive and negative signals which remains to be characterized). The binding of AngII to AT1R induces the hypoxia-inducible factor (HIF)-1α, which regulates several genes involved in the RAS (e.g., ACE1). Post-transcriptional regulation by miRNA (e.g., miRNA143 and miRNA421) could occur. Following translation the newly synthesized ACE2 proteins are target of post-transcriptional modifications such as phosphorylation of Ser680 by AMPK that enhances the stability of ACE2, and N-glycosylations (seven potential N-glycosylation sites). Once expressed at the cell membrane the mACE2 protein can be regulated by sheddases (e.g., ADAM10 and ADAM17) that cleave the ACE2 extracellular domain and release a circulating soluble form, sACE2. IFNβ is likely to induce dominant expression of dACE2 over ACE2.
ACE2 through the ages
Structural comparisons of genes indicated that ACE2 and ACE arose by duplication from a common ancestor (Riordan, 2003). Although the evolutionary tree of ACE2 genes from 36 representative vertebrates is consistent with the species evolutionary tree, certain differences found in coelacanths and frogs may suggest a very slow evolutionary rate in the initial evolution of ACE2 in vertebrates (Lv et al., 2018; Damas et al., 2020; Lam et al., 2020; Luan et al., 2020; Lubbe et al., 2020; Liu et al., 2021). Orthologs of ACE2 and ACE also exist in bacteria, chordates and tunicates, suggesting an early origin of the RAS (Fournier et al., 2012). Although intriguing, the observation that the ACE2-like carbopeptidase from Paenibacillus sp. B38 catalyzes the conversion of Ang II to Ang-(1–7) and can suppress Ang II-induced hypertension, cardiac hypertrophy, and fibrosis in mice does not necessarily mean that the origin of the RAS goes back to bacteria but that a molecule with an ACE2-like carbopeptidase activity was maintained during speciation (Minato et al., 2020). ACE2-ancestors may then have acquired important new functions in tissues during speciation, as evidenced in humans. Beside the ACE2-like carbopeptidase, bacteria also express the neutral amino acid transporter SLC6A19, the homologous of B0AT1 in human, suggesting that SLC6A19 and the bacterial ACE2 ortholog may have already been molecular partners in bacteria (Gallucio et al., 2020). It is remarkable to note that an ACE-like bacterial protein named XcACE from Xanthomonas axonopodis pv. citri, hydrolyses Ang I into Ang II (Rivière et al., 2007). Other bacteria belonging to Lactococcus (L. lactis, L. helveticus, L. acidophilus, and L. casei) and Bifidobacterium species, release peptides with in vitro ACE-inhibitory activity (Fuglsang et al., 2003; Donkor et al., 2007).
The Ance genes from Drosophila melanogaster shares similarities with the human ACE2 (Burnham et al., 2005). In Acyrthrosiphon pisum, expression of the insect ACE2-ortholog is inducible upon feeding (Wang et al., 2015). The simultaneous KO of A. pisum ACE2 and ACE resulted in enhanced feeding and increased aphid mortality. It was also reported that the challenging of Anopheles gambiae with Staphylococcus aureus and Staphylococcus typhimurium upregulated the transcription of the Anopheles homolog of ACE, named AnoACE (Aguilar et al., 2005). Moreover, it was reported that treatment of A. gambiae with an ACE inhibitor resulted in larval death (Abu Hasan et al., 2017).
While searching for the zoonotic origin of SARS-CoV-2, special attention has been drawn to bats, minks and hamsters ACE2 molecules, as they might serve as viral receptors. Using multiple sequence alignments, we found that the bat ACE2 protein polymorphism grouped in the dendrogram according to the 18 subspecies of bats studied (Devaux et al., 2021c). The ACE2 from Rhinolophus bats appeared to be an appropriate candidate for interacting with SARS-CoV-2-related viruses, despite species polymorphism (i.e., R. sinicus with Lys31, Tyr41His, Asn82, Asn90, and Lys353). The Lys31Asp variant found in R. ferrumequinum may possibly alter the binding of the SARS-CoV-2 spike to the bat mACE2 receptor. The mACE2 sequences from other bat species showed increasing amino acid substitutions at positions considered to be required for SARS-CoV-2 spike binding (e.g., D. rotundus with Lys31Asn, Tyr41, Asn82Thr, Asn90Asp., and Lys353Asn). The mACE2 proteins from Myotis bats examined were characterized by Lys31Asn, Tyr41His, Asn82Thr, Asn90, and Lys353, including substitutions incompatible with SARS-CoV-2-like viruses binding. Regarding the ACE2 from minks we found that the mink ACE2 sequences from Neovison vison and Mustela lutreola displayed 99.51% similarity to one another, but shared only 83.73 and 83.48% amino acid identity with the human ACE2, respectively (Devaux et al., 2021b). The similarity between human ACE2 and mink ACE2 dropped to 63.34% in the region described to be involved in the interaction with the SARS-CoV-2 spike protein (regions 30–41, 82–93, and 353–358). Despite the fact that more than 130 substitutions out of 805 amino acids were observed between the human ACE2 and mink ACE2 (e.g., 131 substitutions and 133 substitutions for N. vison ACE2 and M. lutreola ACE2, respectively), including an Asn90Asp substitution possibly impacting the affinity of mink ACE2 for the virus, the Lys31, Tyr41, and Lys353 amino acids required for human ACE2 interaction with the SARS-CoV-2 spike protein are conserved in minks mACE2. This amino acids triad is also conserved in hamsters. The Figure 6A, illustrates a comparison of ACE2 amino acid sequences from humans, mink, hamsters, mice and bats.
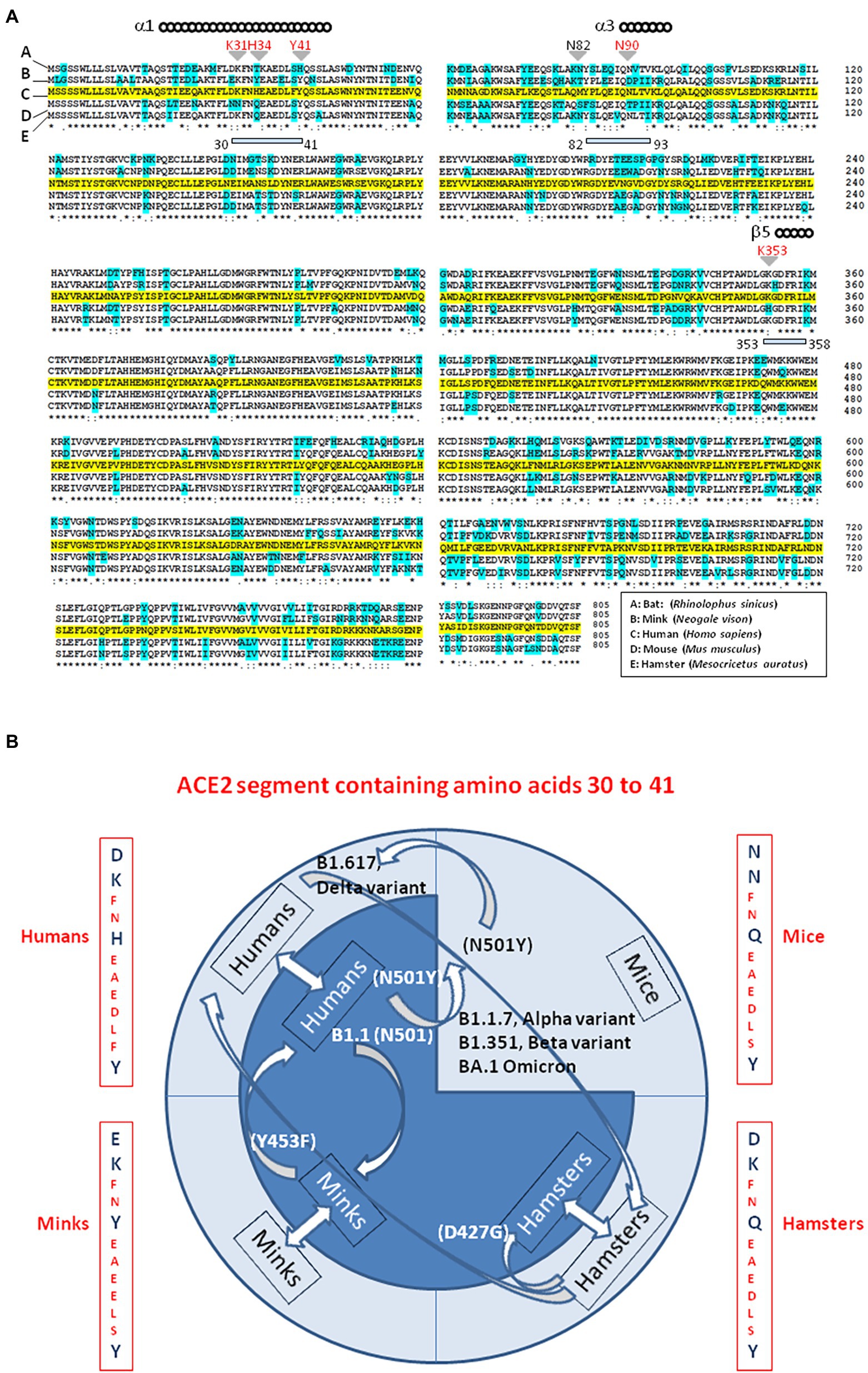
Figure 6. Interspecies viral circulation. (A) ACE2 multiple sequence alignment. The consensus ACE2 sequence from humans (Homo sapiens; GeneBank BAB40370.1) was compared to ACE2 sequences from mink (Neogale vison NCBI ref. sequence XP_044091953.1), hamsters (Mesocricetus auratus; NCBI ref. sequence XP_005074266.1), mice (Mus musculus; NCBI ref. sequence NP_081562.2), and bats (Rhinolophus sinicus; GeneBank: AGZ48803.1), using the Clustal Omega multiple sequence alignment (EMBL-EBI bioinformatic tool; Copyright © EMBL 2020; https://www.ebi.ac.uk/Tools/msa/clustalo/). The human ACE2 sequence is highlighted in yellow. Amino acids that differ from the human ACE2 sequence in ACE2 from other species are highlighted in cyan. The (*) symbol indicates sequence identity between the ACE2 of the five species. Some of the amino acids found to be important for viral tropism in previous studies (in particular amino acid residues 31, 34, 41, 90, and 353 are important for viral spike binding). (B) SARS-CoV-2 is spreading on their ability to recognize a receptor and circumvent the host immune defenses. This principle accounts for the circulation of SARS-CoV-2 between species. Species living in various ecosystem show different amino acid substitutions at positions considered to be required for SARS-CoV-2 spike binding to ACE2. The ACE2 from minks shares 83% amino acid identity with the human ACE2 (63% in the region described to be involved in the interaction with the SARS-CoV-2 spike protein). Despite more than 130 substitutions out of 805 amino acids the interspecies transmission of SARS-CoV-2 from humans to minks and back to humans is possible and generates specific amino acid substitutions in each species, which improved the affinity for the ACE2 receptor as observed in Denmark’ farms. The same applies in the case of the hamster-adapted Delta variant recently described in Hong Kong. SARS-CoV-2 (Wuhan-HU1 strain) cannot use mouse ACE2. It was reported that the B1.1.7 (20I/501Y.V1; UK variant), (B) 1.351 (20H/501Y.V2; South Africa variant) and P1 (20J/501Y.V3, Brazilian variant) SARS-CoV-2 variants and other N501Y-carrying variants exhibit extended host range to mice. Moreover, it has been postulated that the new lineage SARS-CoV-2 Omicron (BA.1, BA.2) could have a murine origin. Omicron variants (e.g., BA.5) are the SARS-CoV-2 lineages that currently cause the most cases of human infections. The amino acid differences in residues 30–41 of the N-terminal region of the ACE2 of humans, minks, hamsters, and mice, are indicated. Arrows indicate interspecies circulation of SARS-CoV-2 strains.
ACE2 as SARS-CoV-2 receptor
Severe acute respiratory syndrome coronavirus is an enveloped single-stranded positive-sense RNA virus (its genome contains ∼32 kb). The SARS-CoV-2 viral envelope consists of a lipid bilayer, where the viral membrane (M), envelope (E), and spike (S) structural proteins are anchored. The S proteins surrounding the viral particles consist of two subunits, S1 and S2. This S protein determines the cellular tropism of the virus. In 2020, ACE2 was identified as the main entry receptor for the SARS-CoV-2 virus (Zhao et al., 2020; Zhuang et al., 2020; Baggen et al., 2021). SARS-CoV-2 is the third human coronavirus after SARS-CoV-1 and HCoV-NL63 which use the human mACE2 as a cellular receptor (Li et al., 2003, 2007). A unique feature of SARS-CoV-2 compared with SARS-CoV-1 is the presence of a polybasic motif (RRAR) at the S1/S2 boundary, which can be cleaved by furin (Walls et al., 2020), resulting in a C-terminally exposed RRAR peptide. Two independent studies showed that this peptide directly binds to neuropilin-1 (NRP1) and that NRP1 promotes SARS-CoV-2 infection (Cantuti-Castelvetri et al., 2020; Daly et al., 2020).
A critical step in the SARS-CoV-2 infection cycle is the binding of the homotrimeric viral spike protein through RBD to the peptidase domain of mACE2 (Lan et al., 2020; Shang et al., 2020; Yan et al., 2020). Despite high similarity between the RBD of SARS-CoV-1 and SARS-CoV-2, several amino acid variations in the binding domain of SARS-CoV-2, increase its affinity for ACE2 (Lan et al., 2020; Yan et al., 2020). The interaction is driven by two domains in the S1 subunit of the molecule, namely the RBD and the N-terminal domain (NTD). The NTD displays a flat electropositive ganglioside binding site enabling the virus to interact with lipid rafts of the cell membrane (Fantini et al., 2021). At the N terminus of the viral spike, Gln498, Thr500, and Asn501 of the RBD form a network of H-bonds with Tyr41, Gln42, Lys353, and Arg357 of the human mACE2. In addition, in the middle of the bridge, Lys417 and Tyr453 of the RBD interact with Asp30 and His34 of ACE2, respectively. Moreover, Gln474 of the RBD is H-bonded to Gln24 of ACE2, whereas Phe486 of the RBD interacts with Met82 of ACE2 through van der Waals forces (Yan et al., 2020). Binding of S1 to the mACE2 receptor triggers an ACE2 ectodomain cleavage by ADAM17 (Lambert et al., 2005; Heurich et al., 2014; Oarhe et al., 2015). The ACE2 cleavages by ADAM17 and a serine protease (TMPRSS2 or TMPRSS4) induce the shedding of cellular ACE2 and systemic release of S1/sACE2 complex, and primes for cellular viral entry (Hoffmann et al., 2020). When S1 binds to mACE2, another site on S2 is exposed and cleaved by host proteases. S2 does not interact with mACE2 but harbors the functional elements which guides membrane fusion. So, SARS-CoV-2 can therefore utilize two pathways to infected ACE2 positive cells: the virus can either fuse at the plasma membrane (early pathway) or, it can fuse at the endosomal membrane (late pathway). The privileged pathway is determined by the proteases present at the cell membrane (Wicik et al., 2020; Caillet-Saguy and Wolf, 2021). When the fusion occurs at the cell membrane, this process is followed by the formation of a funnel like structure built by two heptad repeats in the S2 protein in an antiparallel six-helix bundle, facilitating the fusion and release of the viral genome into the cytoplasm. When the protease is absent, SARS-CoV-2 can be endocytosed via clathrin-and non-clathrin-mediated internalization and the virion is then activated in endosomal vesicles by the action of low pH-dependant protease Cathepsin L (Tang et al., 2020). Thus, the expression and polymorphism of both ACE2 and TMPRSS2 are likely to dictate SARS-CoV-2 tissue tropism (Hou et al., 2020; Zou et al., 2020). Whether overexpression of mACE2 would facilitate infection (increasing the number of receptors available for the virus) or restrict the risks of developing the most severe forms of the disease, has long been a source of controversy (Vaduganathan et al., 2020). Once bound to mACE2, SARS-CoV-2 down-regulates the cellular expression of the ACE2 gene and mACE2 protein and the unopposed action of Ang II was deemed responsible for worsening the outcome of COVID-19 (Hendren et al., 2020).
The ACE2 key residues at the ACE2/S-protein-RBD interface include Ser19, Gln24, Thr27, Phe28, Asp30, Lys31, His34, Glu35, Glu37, Asp38, Tyr41, Gln42, Leu45, Leu79, Met82, Tyr83, Thr324, Gln325, Gly326, Glu329, Asn330, Lys353, Gly354, Asp355, Arg357, Pro389, and Arg393 (Suryamohan et al., 2021). The Lys31 and Lys353 residues in human mACE2 form hydrogen bonds with the main chain of Asn501 and Gln493 in the RBD. ACE2 variants Ser19Pro, Ile21Val, Glu23Lys, and Lys26Arg (which stabilizes core ACE2 α-helical interactions), Thr27Ala (which removes interactions between Thr27 and Glu30), Asn64Lys, Thr92Ile, Gln102Pro and His378Arg were predicted to increase cell susceptibility to SARS-CoV-2. In contrast, ACE2 variants Lys31Arg (which breaks an interaction with Gln493 in the SARS-CoV-2 spike RBD), Asn33Ile, His34Arg (which results in a loss polar contact at the interface with SARS-CoV-2 spike RBD), Glu35Lys (which affects the critical polar contact with SARS-CoV-2 spike Gln493), Glu37Lys, Asp38Val (which compromises the Asp38-Lys353 interaction), Tyr50Phe, Asn51Ser, Met62Val, Lys68Glu, Phe72Val, and Tyr83His (which prevents insertion of SARS-CoV-2 spike residue Phe486 into an hydrophobic pocket driven by residue Tyr83), Gly326Glu, Gly352Val, Asp355Asn, Gln388Leu, and Asp509Tyr were predicted to be less sensitive to SARS-CoV-2 (Procko, 2020; Suryamohan et al., 2021). When considering ACE2 variants, high mACE2 cell-surface expression can mask the effects of impaired binding while low cell surface expression reveals a range of infection efficiencies across variants, supporting a major role for binding avidity during viral entry (Shukla et al., 2021). Using an in vitro model of infection of cells expressing suboptimal surface ACE2, it was found that the mACE2 variants Asp355Asn, Arg357Ala, and Arg357Thr abrogated entry of SARS-CoV-2 while Tyr41Ala showed only a slight effect on SARS-CoV-2 entry although it inhibited SARS-CoV-1. The NTD and RBD domains in the viral S protein act synergistically to insure virus adhesion (Fantini et al., 2021). Moreover, an inverse correlation was established between ACE2 expression and COVID-19 severity (Chen et al., 2021).
Particular attention was drawn to polymorphism of ACE2 in bat (considered to be a reservoir of SARS-CoV-related virus; Zhou et al., 2020; Wacharapluesadee et al., 2021) this species, and in minks (because they have been shown to be susceptible to infection by SARS-CoV-2 from humans and then to be a source of the virus being able to reinfect humans; Boklund et al., 2021; Oude Munnink et al., 2021; Shuai et al., 2021). It was found that when SARS-CoV-2 of human origin become host-adapted to mink, a Tyr453Phe substitution located in the RBD was selected. This process is driven by the fact that mink mACE2 has a Tyr34 instead of the H34 found in human mACE2 and that the Tyr453Phe substitution improves the virus binding to the mink mACE2 (Ren et al., 2021). The hamster is another species of interest for ACE2, because hamster-adapted SARS-CoV-2 Delta variants were isolated in Hong Kong, and the virus was transmitted back to human and further human-to-human transmission was then demonstrated (Kok et al., 2022; Yen et al., 2022). We found that once adapted to the hamster ACE2, the variant virus show mutations (e.g., Asp427Gly) that could make this virus more efficient at infecting humans (Fantini et al., 2022). Although a large number of animal species were considered to be susceptible to infection by SARS-CoV-2 (Stawiski et al., 2020), SARS-CoV-2 (Wuhan-HU1 strain) cannot use mouse ACE2 (Zhou et al., 2020). The presence of Asn30 (instead of Asp30) and Asn31 (instead of Lys31) in mouse ACE2 is likely to cause the lack of salt bridges and the critical H-bond at the mouseACE2-SARS-COV-2 RBD interface. In addition, the presence of His353 (instead of Lys353), leads to unfavorable interactions with the SARS-CoV-2 S protein RBD (Brooke and Prischi, 2020; Gao and Zhang, 2020). However, this does not rule out the possibility of low efficiency mouse infection through an alternative receptor. It was reported that the expression of human basigin/CD147 in mice, enabled SARS-CoV-2 infection with detectable viral loads in the lungs (Wang et al., 2020). However, this model remains controversial (Shilts et al., 2021). It has been reported that the B1.1.7 (20I/501Y.V1; United Kingdom variant), B.1.351 (20H/501Y.V2; South Africa variant), and P1 (20J/501Y.V3; Brazilian variant) SARS-CoV-2 variants and other N501Y-carrying variants exhibit extended host ranges to mice (Montagutelli et al., 2021; Shuai et al., 2021). Moreover, it has been postulated that the new lineage SARS-CoV-2 Omicron variant (BA.1, BA.2), has a murine origin (Wei et al., 2021). Indeed, the interspecies conservation of ACE2 turns out to be sufficient to allow viruses that use this receptor to circulate between animal hosts and humans. Viruses do not spread based on species but based on their ability to recognize a receptor and circumvent the host immune defenses. We have proposed that this general principle accounts for the circulation of SARS-CoV-2 between species (Frutos et al., 2021, 2022; Figure 6B).
Immune response against SARS-CoV-2 and auto-antibodies against ACE2 in COVID-19 patients
Infection with SARS-CoV-2 initiates an antiviral immunoglobulin (Ig)M and IgA response, detectable during the first week of symptoms, whereas IgG are found later. The antibody titres reaches a plateau within 6 days after seroconversion (Guo et al., 2020; Kellam and Barclay, 2020; Long et al., 2020; Zhao et al., 2020). The serum level of SARS-CoV-2 specific IgA is positively correlated with the severity of COVID-19 (Ma et al., 2020; Yu et al., 2020). The state of hyperstimulation of the immune system that occurs in severely ill patients contributes to autoimmune manifestations and is associated with an increased need for oxygen therapy (Gagiannis et al., 2020). Moreover, it was recently reported that Ang II induces ROS release from monocytes able to induce DNA damages and apoptosis in neighboring T-cells leading to lymphopenia in certain patients with severe forms of COVID-19 (Kundura et al., 2022). It is neither the purpose of this paragraph to discuss the complex pattern of immune response in COVID-19 (e.g., a decrease in the total number of CD4+ and CD8+ T cells, B cells, and NK and a recruitment of neutrophils; a massive increase in the release of inflammatory cytokines or ‘cytokine storm’, and chemokines such as IL-2, IL6, IL-7, IL-8, IL-10, TNF, IFN; Amor et al., 2020; Campbell and Kahwash, 2020; Han et al., 2020; Luo et al., 2020; Mehta et al., 2020; Tay et al., 2020; Vitte et al., 2020; Zheng et al., 2020), nor is it to review the abnormal expression of Ag II in COVID-19 patients that could stimulate proinflammatory processes (Naftilan and Oparil, 1978; Moore et al., 2015; Varanat et al., 2017; Silva et al., 2020; Raghavan et al., 2021; Vandestienne et al., 2021; Yamamoto et al., 2021), but rather to briefly summarize the contribution of anti-ACE2 and anti-AT1R auto-antibodies in COVID-19, since these molecules could play an important role in the immunological puzzle of clinical variability of the disease.
What was intriguing in SARS-CoV-2 infected patients with respect to the RAS, was the report of the development of ACE2 auto-antibodies. Among 53 patients who had detectable anti-SARS-CoV-2 RBD, 40 (75%) had anti-ACE2 antibodies (Arthur et al., 2021). Among them, 26 (81%) belonged to the convalescent group and 14 (15; 93%) were patients hospitalized for symptoms of COVID-19. Healthy controls with no history of SARS-CoV-2 were all negative for anti-ACE2 antibodies. The median activity of sACE2 in patients with ACE2 auto-antibodies was 263 pmol/min/ml compared to 1,056 pmol/min/ml for those who did not develop an anti-ACE2 immune response. The binding of anti-ACE2 antibodies to ACE2 in normal cells could have the potential to mediate profound pathophysiological effects long after the original antigen itself has disappeared, particularly in the long term COVID-19 patients (e.g., possibly inducing myocarditis or neurological illnesses; Figure 7).
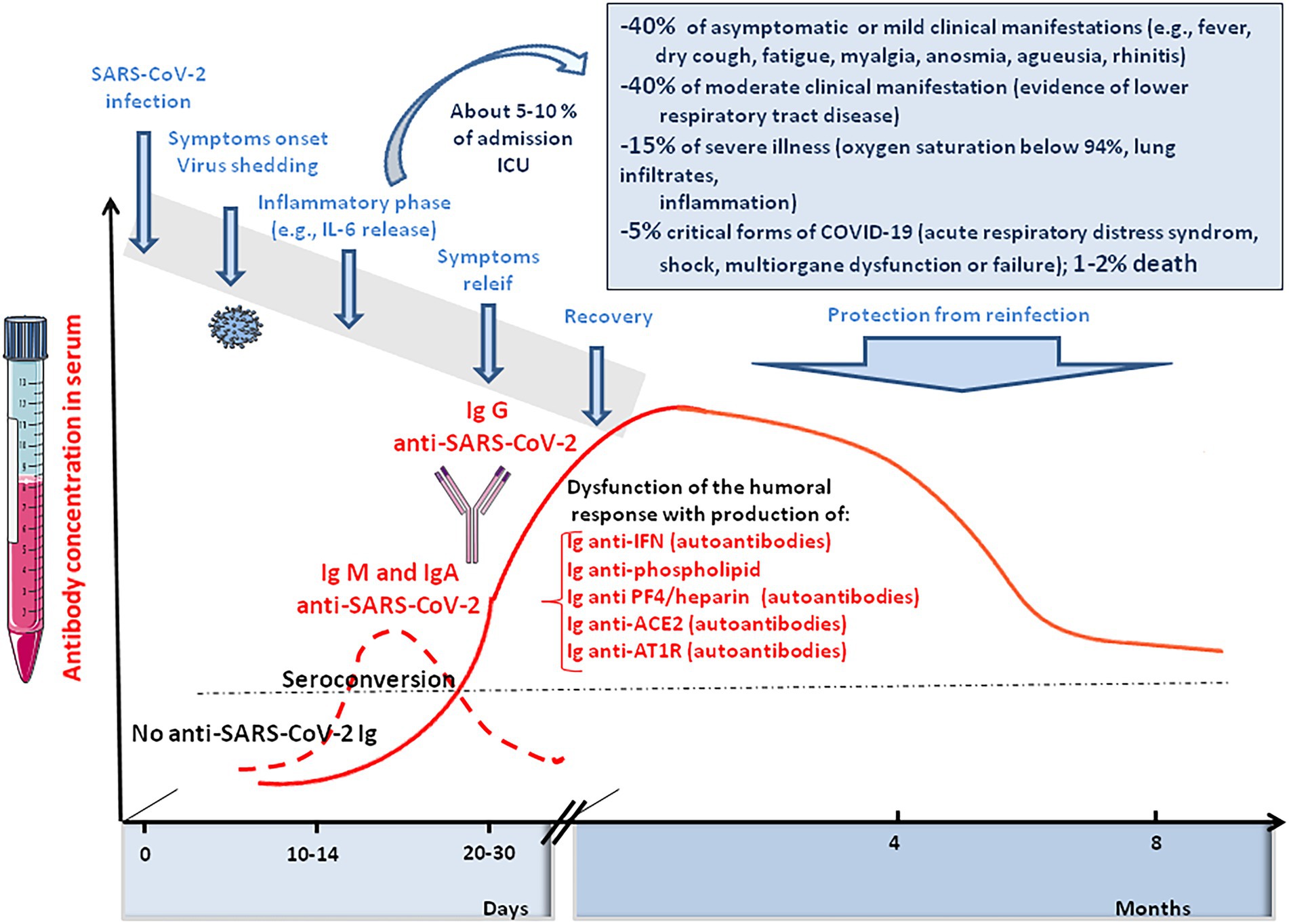
Figure 7. Schematic representation of the clinical course of SARS-CoV-2 infection and COVID-19. It illustrates the immune response of SARS-CoV-2 infected people, including anti-SARS-CoV-2 IgM, IgA, and IgG responses and the induction of auto-immune Ig (anti-phospholipid A, anti-PF4/heparin, anti-ACE2, and anti-AT1R).
Considering the similarities between vasculopathy in severe COVID-19 and antibody-mediated rejection after lung transplantation induced by auto-antibodies against AT1R (Cozzi et al., 2017), the presence of AT1R auto-antibodies in COVID-19 patients was investigated and compared to patients with a favorable disease course. A significant increase (42%) of anti-AT1R Ig was found in COVID-19 patients with an unfavorable disease course (Miedema et al., 2021). These AT1R auto-antibodies are expected to mimick the proinflammatory effect of Ang II, as previously reported (Dragun et al., 2005). Tissue transglutaminase (TG2)-mediated modification of AT1R contributes to AT1R auto-antibody production and hypertension associated with preeclampsia; the post-translational modification of Gln187 in the second extracellular loop of the AT1R loop creates a neo-epitope that induces the production of an autoantibody that can activate the receptor (Liu et al., 2015). Endothelin receptor type A (ETAR) auto-antibodies were also more frequent in severe COVID-19 patients (Miedema et al., 2021). These antibodies are known to stimulate chemotactic activity and neutrophils trafficking (Cabral-Marques et al., 2018). Both anti-AT1R and anti-ETAR antibodies could be associated with cardiovascular disease and hypertension in severe COVID-19 patients (Philogene et al., 2019).
Among other auto-antibodies found in COVID-19 patients, anti-interferon Ig was found in patients with severe COVID-19 while no such auto-antibodies were found in patients with mild disease (Bastard et al., 2020). Anti-phospholipid antibodies have also been observed as being associated with thrombotic events in COVID-19 cases (Bertin et al., 2020; Daviet et al., 2020; Harzallah et al., 2020; Helms et al., 2020; Manne et al., 2020; Siguret et al., 2020; Tan et al., 2020; Xiao et al., 2020; Zhang Y. et al., 2020; Zuo et al., 2020; Brodard et al., 2021).
SARS-CoV-2 triggers a vascular and coagulation disease
Venous thromboembolism is a relatively common side effect of SARS-CoV-2 infection. It is characterized by an acute pulmonary embolism or intravascular coagulopathy that predisposes the patients to thrombotic events (Faggiano et al., 2020; Leonard-Lorant et al., 2020; Middeldorp et al., 2020). After the first month of infection, individuals with COVID-19 are at an increased risk of cardiovascular disease, including cerebrovascular disorders, dysrhythmias, ischemic and non-ischemic heart disease, pericarditis, myocarditis, heart failure, and thromboembolic disease (Xie et al., 2022). A nationwide cohort found an increased risk of a deep vein thrombosis up to 3 months after COVID-19, pulmonary embolisms up to 6 months, and bleeding events up to 2 months, with the risk of pulmonary embolism being especially high (Katsoularis et al., 2022). Elevated D-dimers (which reflects the degradation of fibrin and a process of hypercoagulation) upon admission of patients is a marker of hypercoagulation and pulmonary embolism and is associated with increased mortality in severe COVID-19 patients (Lippi and Favaloro, 2020; Sakka et al., 2020; Stefely et al., 2020; Smadja et al., 2021). High levels of D-dimers are found in ∼ 20–40% of critically ill COVID-19 patients (Poissy et al., 2020; Zhang Y. et al., 2020; Zhang S. et al., 2020; Xie et al., 2022). Usual thrombosis prophylaxis is often not sufficient to prevent thrombotic coagulopathy in patients with severe forms of COVID-19 (Berthelot et al., 2020). These lesions usually start with intimal proliferation, followed by fragmented and discontinuous internal elastic lamina (Carvelli et al., 2020; Hofman et al., 2021). Perivascular inflammation was reported to be patchy and scattered, composed mainly of lymphocytes, with thrombi in the branches of the pulmonary artery and focal areas of congestion in the alveolar septal capillaries, as well as septal capillary lesions with wall and luminal fibrin deposition (Deshmukh et al., 2020).
The pathological manifestation of COVID-19 has a strong vascular component, with exacerbated effects on the microvasculature comprising the arterioles, capillaries, venules, and microthrombosis events. The increased occurrence of microvascular thrombi provides a good explanation for the sometimes sudden development of hypoxemia in COVID-19 patients, since the thrombi prevent gas exchange in the oxygenated areas of tissues. Beside the formation of fibrin thrombi, ARDS is characterized by increased alveolar capillary permeability and exudation into the alveoli, where inflammatory cells are present in abundance, as well as coagulation factors including fibrinogen. Regarding COVID-19, it was suggested to name severe pulmonary COVID-19 as “MicroCLOTs” for “microvascular COVID-19 lung vessels obstructive thromboinflammatory syndrome” (Ciceri et al., 2020). The analysis of autopsy lung specimens from COVID-19 patients has shown inflammatory perivascular lymphocyte infiltration, the presence of microvascular thrombi containing platelets, fibrin and numerous neutrophil extracellular traps (NETs) releasing (Carsana et al., 2020; Hofman et al., 2021). Deposits of complement components C3, C4d and C5b-9 were found in the microvasculature of the lungs (Magro et al., 2020). Patients diagnosed with elevated D-dimer and thrombosis during severe forms of COVID-19 have higher blood levels of markers of NETs and calprotectin (Zuo et al., 2020). The formation of NETs in turn, perpetuates complement activation. When activated by proinflammatory cytokines, or NETs, the vascular endothelial cells produce von Willebrand factor (vWF) that retains platelets and leucocytes to the vessel wall and activates coagulation leading to the repair of local damage. Finally, microangiopathic vessel occlusions and endothelium damage has been described in the kidneys (Goshua et al., 2020).
Among the mechanisms implicated in this thrombo-inflammation, AngII seems to have pleiotropic effects. Indeed, regarding the central role played by ACE2 as the viral entry receptor, and its role in the regulation of Ang II blood levels, the balance between ACE2 expression and the accumulation of Ang II in the blood stream may contribute to explain the immunothrombosis. The analysis of RAS dysfunction and Ang II side effects is critical for the understanding of the pathophysiological changes due to SARS-CoV-2 infection. Ang II has a significant effect on the platelet and coagulation/fibrinolytic system and causes mild activation of the coagulation cascade with increases in plasma levels of the thrombin–antithrombin complex and prothrombin (Brown and Vaughan, 2000; Larsson et al., 2000; Fletcher-Sandersjöö and Bellander, 2020; Gando and Wada, 2021). The platelet activation described after COVID-19 is thought to be due in part to the binding of AngII to AT1R. Moreover, SARS-CoV-2 can directly activate platelets by binding to platelet ACE2 (Zhang S. et al., 2020). Through binding to AT1R, Ang II stimulates the expression of Tissue Factor (TF), which triggers coagulation cascade (Nemerson, 1988; Nishimura et al., 1997; Muller et al., 2000; Felmeden et al., 2003; He et al., 2006; Brambilla et al., 2018). Ang II also induces expression of plasminogen activator inhibitor-1(PAI-1), the main inhibitor of tissue plasminogen activator and urokinase-type plasminogen activator, in cultured endothelial cells (Fogari et al., 2011). Increased levels of PAI-1 can occur locally upon SARS-CoV-2 infection, leading to the formation of plugs in the body.
The binding of SARS-Cov2 to ACE2 at the surface of endothelial cells (ECs) of blood and lymph vessels, leads to activation of the complement system, promoting a pro-coagulative state, leukocyte infiltration, vascular dysfunction, and thrombosis (Jin et al., 2020). In severe COVID-19 patients, the plasma levels intercellular adhesion molecule 1 (I-CAM-1), vascular cell adhesion molecule-1 (VCAM-1), and vascular adhesion protein-1 (VAP-1), are elevated (Escher et al., 2020; Tong et al., 2020), indicating that the endothelial barrier is damaged consecutive to viral infection. Soluble E-selectin, soluble ICAM-1, and soluble platelet endothelial adhesion molecule 1 (sPECAM-1) correlate with disease severity (Li et al., 2021; Vassiliou et al., 2021). ICAM-1 promotes fibrin adhesion and leukocyte transmigration and increased thrombus formation. Disruption of the vascular barrier is associated with the inhibition of protein C, a major anticoagulant (Dahlbäck and Villoutreix, 2005). The plasma levels of vWF, angiopoietin-2, Fms-related tyrosine kinase 3 ligand (FLT-3L), and PAI-1 are significantly elevated in patients with COVID-19 (Liu and Zhang, 2021; Figure 8). Moreover, a decrease ADAMTS13, which ensure vWF hemostatic function, has been reported in severe forms of COVID-19 (Bazzan et al., 2020; Rodriguez Rodriguez et al., 2021). It was also reported that the SARS-CoV-2 main protease Mpro causes microvascular brain pathology by cleaving NEMO (an essential modulator of NF-κB) in infected brain ECs (Wenzel et al., 2021).
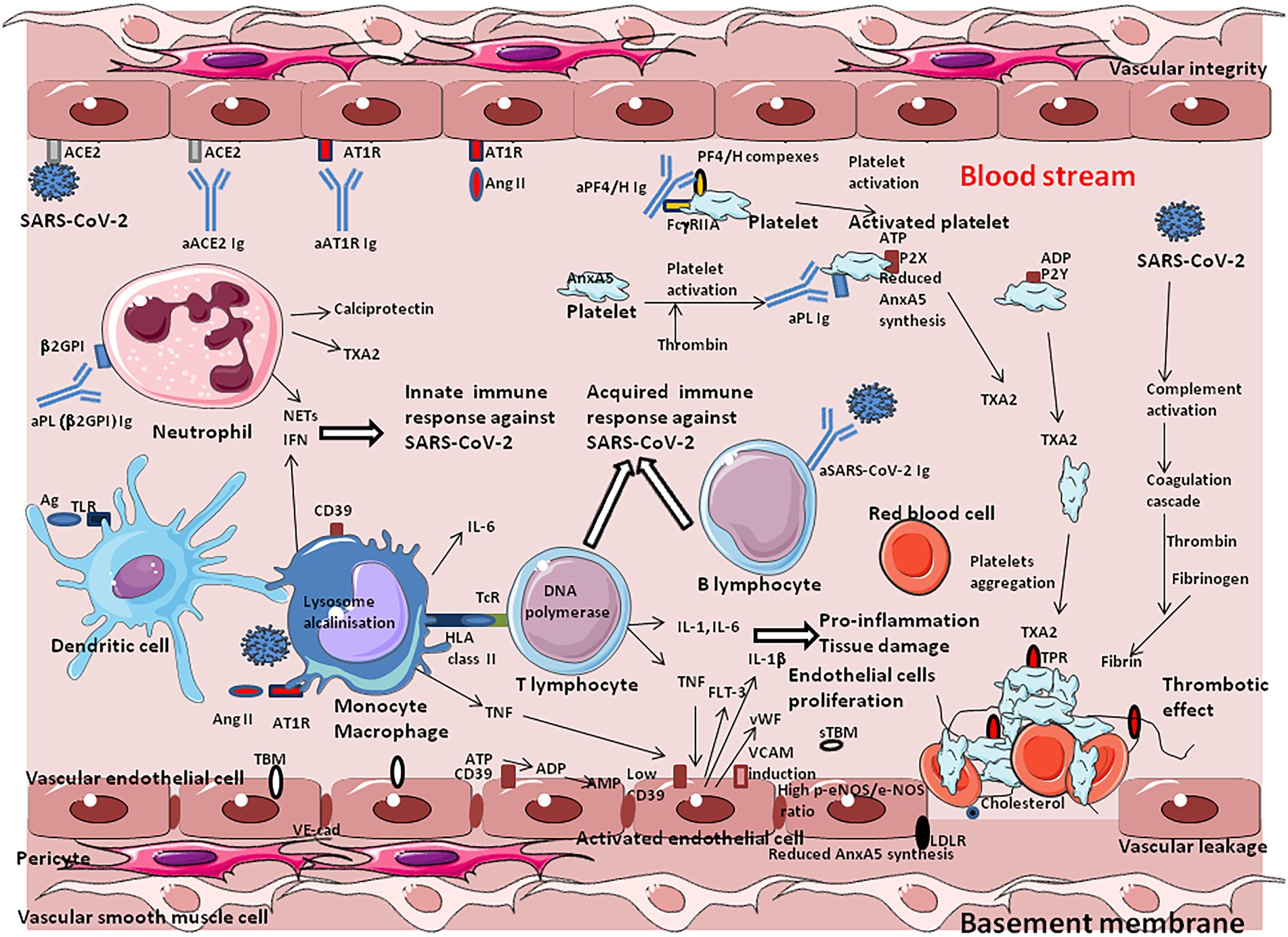
Figure 8. Under physiological conditions, the vascular endothelium, composed of vascular endothelial cells (VECs), functions as an integral barrier with intercellular junctions ensured by adhesion molecules such VE-cadherin. It maintains blood fluidity by acting as an anticoagulant through the suppression of platelet activation and the induction of fibrinolysis, a mechanism including heparan sulfate proteoglycans and CD39. During SARS-CoV-2 infection, innate and acquired immune defense mechanisms are activated with the overproduction of cytokines such as IL-1, IL-2, IL-6, IL-8, IL-17, and TNFα (a phenomenon known as the “cytokine storm”), which disrupts blood vessel walls and provokes tissue damage in the lung parenchyma and the immediately adjacent bronchial alveolar lymphoid tissue. The endothelial cells express the ACE2 molecule that acts as a cell-surface-receptor, facilitating SARS-CoV-2 entry into these cells. The SARS-CoV-2 induced increased concentrations of Ang II have mild platelet-activating effects, thereby enhancing coagulation and are also associated with monocyte and macrophage accumulation, which produces proinflammatory cytokines and worsen hypertension. When blood vessels are strained due to high blood pressure, the endothelial cells of the blood vessels are damaged, and the function of the endothelium at preventing arteriosclerosis is lost. When activated by proinflammatory cytokines, or neutrophil extracellular traps, endothelial cells produce von Willebrand factor, which retains platelets and leucocytes to the vessel wall and activates coagulation systems resulting in the rapid activation of mechanisms leading to the repair of local damage, the accumulation of immune cells to prevent infection, and the aggregation of platelets for primary and secondary hemostasis. Anti-PF4/polyanion (heparin) complex immunoglobulins directly activate platelets via their Fc gamma type 2 receptor A (FcγRIIA). The hyper-reaction set up in response to vascular damage, can influence a propensity toward local vascular micro-thrombosis. COVID-19 patients suffer from prominent alveolar oedema, intra-alveolar proteinosis, cell infiltration (including lymphocytes), apoptosis of virally-infected pneumocytes, and fibrin deposition. aPL, antiphospholipids immunoglobulins; aACE2 Ig, anti-ACE2 immunoglobulins; aAT1R Ig, anti-AT1R immunoglobulins; aPF4/H Ig, anti-PF4/heparin immunoglobulins; AnxA5, annexinA5 (or annexin V or anchorin CII; anticoagulant, interact with phospholipids); Ag, antigen; Ang II, angiotensin II; AT1R, angiotensin II receptor type 1; IFN, interferon; CD39/ENTPD1, ectonucleoside triphosphate diphosphohydrolase-1 (also known as P2 receptors: P2X receptors are ion channels that open upon binding of ATP; P2Y receptors mediate cellular response to purine and pyrimidine, such as ATP, ADP, and UTP; in physiological conditions CD39 catalyzes the reduction of ATP and ADP pool to AMP and CD73 transform AMP to adenosine whereas nucleotides released during cell activation/injury bind to P2 receptors to activate thrombo-inflammatory programs); IL-6, interleukin-6; LDLR, low density lipoprotein receptor (bind LDL/cholesterol); NETs, neutrophil extracellular traps; TcR, T-cell receptor; TLR, toll like receptor; TNF, tumor necrosis factor; TXA2, Thromboxane A2 (induce platelets aggregation); TPR, thromboxane A2 prostanoid receptor: VE-cad, VE-cadherin; TBM, thrombomodulin prevents thrombosis; upon endothelial cell activation a soluble form of TBM (sTBM) is released in plasma further promoting procoagulant mechanisms. VWF, von Willebrand factor; Fibrin, fibrin is formed from blood plasma fibrinogen (produced in the liver) by the action of thrombin; red thrombus is composed of erythrocytes enmeshed in a fibrin network.
In response to COVID-19, the activation of ECs was also associated with the overexpression of proangiogenic factors, such as vascular endothelial growth factor (VEGF), basic fibroblast growth factor (FGF-2), and placental growth factors (PlGF; Smadja et al., 2021). Soluble Flt-1 (sFlt-1), a circulating truncated form of the VEGF-A receptor, was markedly increased in severe forms of COVID-19 (Rovas et al., 2020). Damage to ACE2+ pericytes and ECs leads to vascular permeability in severe COVID-19 (Cardot-Leccia et al., 2020; Afzali et al., 2021). The viral Spike induces oxidative stress, ERK1/2 activation through the CD147 receptor and NF-κB nuclear translocation in pericytes, thereby prompting dysfunction of the vascular pericytes (Avolio et al., 2021; Khaddaj-Mallat et al., 2021). CD147, considered to have a potential proatherosclerotic effect (Wang et al., 2015), is upregulated in COVID-19 patients and can act as a receptor for SARS-CoV-2 in cells expressing low ACE2 (Radzikowska et al., 2020). Interestingly, statins, the action of which partly relies on CD147 downregulation, have been recommended in the therapeutic arsenal against COVID-19 (Zhang X.J. et al., 2020).
Modulation of ACE2 and other actors of the RAS in COVID-19 patients
Coronavirus disease 2019 is a systemic disease characterized by a cytokine storm associated with high levels of C reactive protein (CRP), high fibrinogen, high fibrin degradation to D-Dimers, microvascular injury, and obstructive thrombo-inflammatory syndrome. As knowledge grows, the need for a deeper understanding of the molecular cross-talk leading to thrombosis appears as a research priority in order to gain a better understanding of ARDS and MODS associated with severe COVID-19.
In a pioneer study it was demonstrated that SARS-CoV-1 infection was associated with ACE2 downregulation and impaired degradation of Ang II (Kuba et al., 2005). Since both SARS-CoV-1 and SAR-CoV-2 enter cells through ACE2 and induce simimlar diseases, attention rapidely focused on the consequences of virus-ACE2 interaction on the dysregulation of RAS. This is complexified by the fact that SARS-CoV-2 infection triggers IFN activation, which in turn can upregulate ACE2 (Garvin et al., 2020). Another element of complexity resides in the fact that a greater number of ACE2+ cells seems to circulate in the lungs of patients with severe COVID-19 (Ackermann et al., 2020). Using a swine animal model it was demonstrated that blocking ACE2 (or infusing Ang II) leads to increased pulmonary artery pressure, reduced blood oxygenation, increased coagulation, diffuse alveolar damage, and acute tubular necrosis (Aroor et al., 2016).
Despite efforts that have been made to quantify the compound of the RAS in COVID-19 patients, this exploration remained incomplete and debatable. An early study found no difference in the Ang II/Ang I ratio in the plasma sample of 31 COVID-19 patients, but reported that the plasma sACE2 activity was increased in patients treated with an ACE inhibitor (Kintscher et al., 2020). Another study reported increased plasma levels of Ang II in 12 patients with severe COVID-19 pneumonia (Liu et al., 2020). Furthermore, no alteration of RAS was found in a cohort of nonsevere COVID-19 patients (Rieder et al., 2021). More recently, a sevenfold ACE2 increase was found in patients with COVID-19 and Ang II as well as Ang-(1–7) concentrations was significantly higher in patients with severe COVID-19 (Reindl-Schwaighofer et al., 2021). Another investigation in a cohort of 306 COVID-19 patients revealed that elevated plasma sACE2 from COVID-19 patients was significantly associated with severe forms of disease, particularly in hospitalized patients intubated at the time of sample collection (Kragstrup et al., 2021). ARDS in patients with COVID-19 was found associated with an increase in blood pressure and decrease in serum potassium concentration (Vicenzi et al., 2020). Surprisingly, another recent report suggests a significant reduction of Ang II concentration and increased Ang-(1–7) in COVID-19 patients (Martins et al., 2021). The divergent results reported concerning the variation of RAS molecules in plasma from SARS-CoV-2-positive patients could either be explained by the differing severity of COVID-19 in the groups of patients tested and/or by the method used for quantification of the molecules (Figure 9).
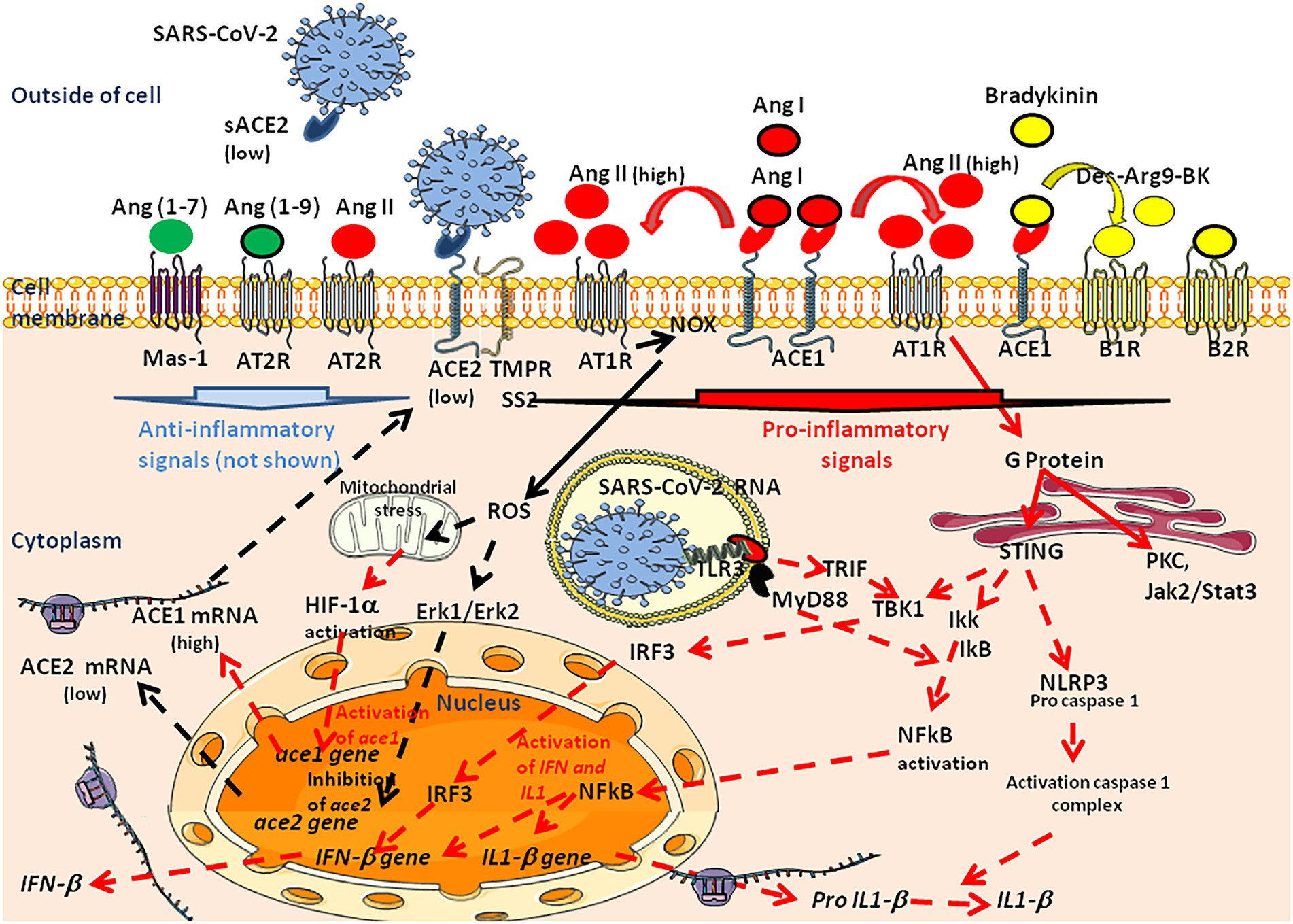
Figure 9. Signaling pathways that are activated during SARS-CoV-2 infection and replication. SARS-CoV-2 infects human cells expressing the ACE2 receptor and the serine protease TMPRSS2. This process results in the downregulation of ACE2 mRNA expression, the reduced expression of mACE2, dysfunction of the RAS and increasing levels of AngII in the circulation. High Ang II levels trigger signaling through AT1R. This activates a number of signaling pathways, such as G protein–mediated (Gq and Gi), Janus kinase/signal transducers and activators of transcription, extracellular signal-regulated kinase (ERK), IFN regulatory factor (IRF)3, NF-κB, NLRP3 procaspase 1 pathways leading to induction of IFNs and pro-inflammatory cytokines, and the HIF-1α pathway. In addition, G protein–independent signaling takes place through the adapter proteins β-arrestin 1 and β-arrestin 2 that can have distinct functional and physiological consequences (not shown). Crosstalk between AT1R and AT2R was evidenced and stimulation of one receptor modulates the expression of the other. Ang II can bind both to AT1R and AT2R, which are receptors with opposite effects but the low expression of AT2R compared to AT1R, account for a privilegied effect of Ang II through AT1R when the plasma levels of Ang II increase. The single AT1R gene in humans encodes a 359-amino-acid protein and AT1R is widely expressed and well conserved between species The MAS receptor can interact with AT1R, explaining that it is the physiological antagonist of Ang II signaling. Mas1 activation induces the second-messenger cAMP, phospholipase A2 pathway, and the phosphoinositide 3-kinase/AKT pathway, and mediates antiapoptotic, anti-inflammatory, vasodilatory, and antithrombotic effects. Excess of Ang II results in organ damage, hypertension, thrombotic microangiopathy, progression to fibrosis, and cardiovascular remodeling.
Recently, by exploring different biomarkers in a cohort of COVID-19 patients (30 prolonged viral shedders and 14 short viral shedders) we found that circulating blood cells (in particular monocytes) from COVID-19 patients expressed less ACE2 mRNA than cells from healthy volunteers (Osman et al., 2021). Moreover, although we found the expression of sACE2 to be heterogenous among individuals from each group, the sACE2 plasma concentrations were found to be lower in prolonged viral shedders than in healthy controls, while the concentration of sACE2 returned to normal levels in short viral shedders. In the plasma of prolonged viral shedders, we also found higher concentrations of Ang II and Ang I. However, the plasma levels of Ang-(1–7) were found to be almost stable in prolonged viral shedders, but seemed insufficient to prevent the adverse effects of Ang II accumulation, strongly suggesting that increased levels of Ang II contribute to thrombotic events associated with the severe forms of COVID-19.
Targeting ACE2 for the therapeutic prevention of severe forms of COVID-19
In COVID-19 patients, the downregulation of ACE2 and the reduced capacity to counteract the detrimental effects of Ang II are likely to play a critical role in the development of severe forms of the disease. In experimental animal models, ACE2 KO mice experienced more severe forms of acute lung injury than wild type mice, highlighting the protective role of ACE2 (Imai et al., 2005). The loss of ACE2 resulted in enhanced vascular permeability, neutrophils accumulation and increased lung edema. Both angiotensinogen-specific antisense oligonucleotides and small interfering RNA (siRNA) lowered blood pressure in rat models of hypertension (Mullick et al., 2017; Uijl et al., 2019). In humans with weight excess and hypertension, renin inhibitors (e.g., aliskiren) and ACEi (e.g., ramipril), improve renal and systemic hemodynamics and reduce arterial pressure (Kwakernaak et al., 2017). Thus, therapeutic solutions for reducing COVID-19 severity could be found in the pharmacopeia used by cardiologists to intervene on the RAS.
All FDA approved drugs for treatment of patients with high blood pressure (renin inhibitors, ACEi, and ARBs) are primarily designed to block or reduce the detrimental effects of Ang II (Wright, 2000; Mentz et al., 2013; Arendse et al., 2019; Figure 10A). Reducing the formation of Ang II by ACEi or antagonizing its effect by blocking the AT1R through ARBs may be a suitable strategy for reducing symptoms of COVID-19 patients (Schiffrin et al., 2020). ARBs (e.g., losartan) were found to protect against acute lung injury through the reduction of Ang II/AT1R stimulation (Shen et al., 2009; Meng et al., 2020). Hypertensive patients taking ARBs presented a lower risk of severe COVID-19 (Sarzani et al., 2020). However, the interpretation of the benefits and harmful effects of ACEi and ARBs may be premature due to the multiple effects of such molecules on the RAS (Kai et al., 2021; Tereshchenko et al., 2022). Indeed, it has been reported that ACEi and ARBs increase ACE2 (Ferrario et al., 2005; Furuhashi et al., 2015), which could also increase the binding of SARS-CoV-2. However, it has been reported that ACE2 is not increased by ACEi or ARBS in the respiratory cilia (Lee et al., 2020). We recently reported that in vitro treatment of SARS-CoV-2 permissive ACE2+/AT1R+ Vero E6 cells with various ARBs resulted into ∼50% increase in SARS-CoV-2 production correlated with the ARBs-induced up-regulation of ACE2 expression (Pires de Souza et al., 2022). However, we also observed a downregulation of AT1R, suggesting that Ang II harmful effects should be strongly reduced (Pires de Souza et al., 2022). The upregulation of ACE2 can have opposed effects on SARS-CoV-2 infection and organ pathophysiology (Devaux, 2020). The study of large cohorts support the beneficial effects of RAS inhibitors in patients with COVID-19 (Bean et al., 2020; Zhang P. et al., 2020). In patients, ARBs is preferred over ACEi for first line hypertension treatment and discontinuing treatment is not required (Abbasi, 2021; Lopez et al., 2021). Since, the activation of AT1R by Ang II induces ROS through the NADPH oxidase pathway and activates the hypoxia-inducible factor (HIF)-1α leading to the synthesis of the transient receptor potential channel ankyrin repeat (TRPA1) which controls intracellular calcium increase and potentially contributes to pulmonary inflammation, it was also suggested to use calcium channel blokers as an alternative to ACEi and ARBS (Fang and Karakiulakis, 2020; Tignarelli et al., 2020; Devaux and Raoult, 2022; Zhang et al., 2022). Moreover, acting on HIF-1α, may improve the outcome of COVID-19 by decreasing hypoxia (Devaux and Raoult, 2022).
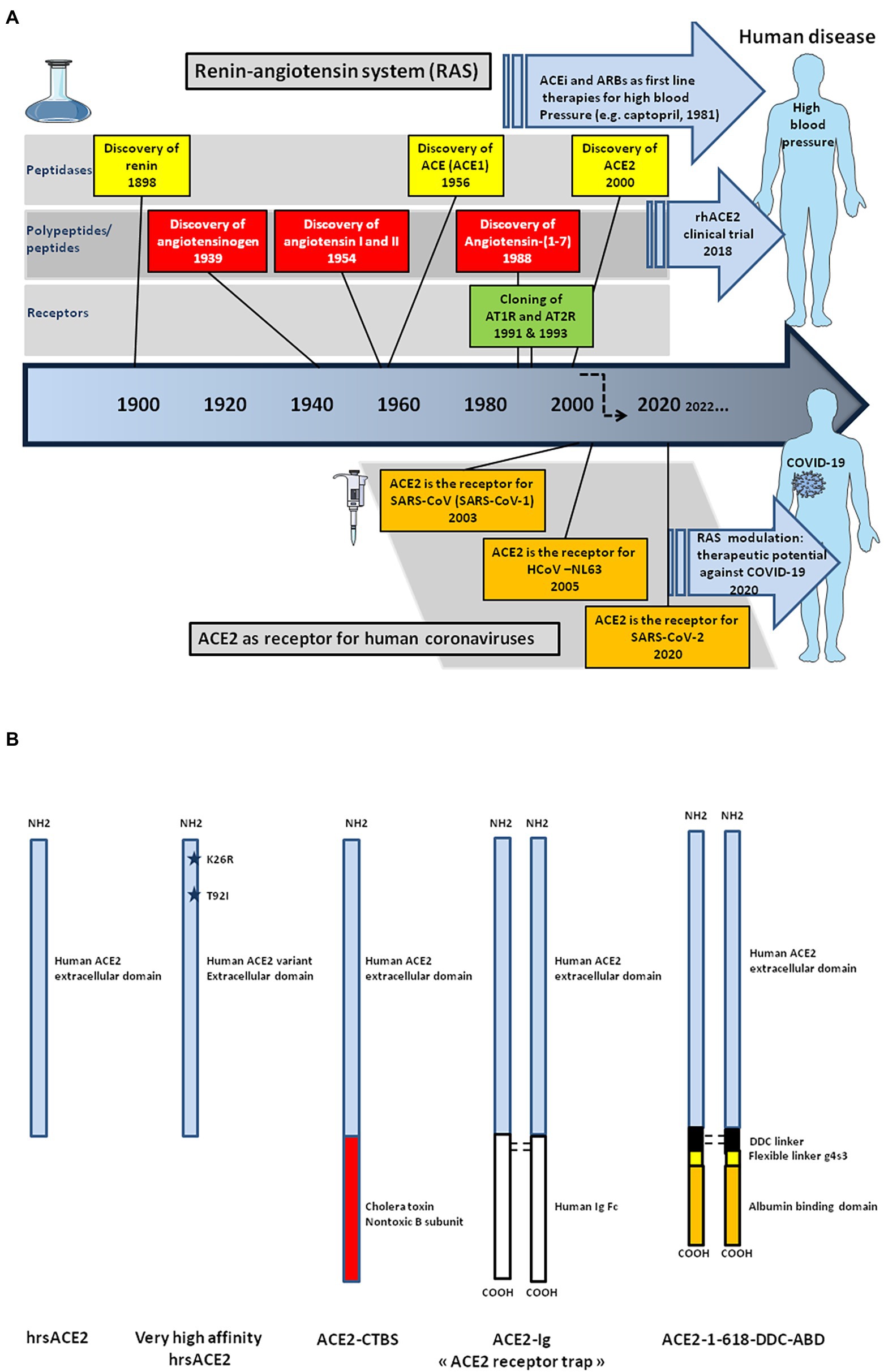
Figure 10. Schematic representation of the historical discovery of the main components of the renin-angiotensin system (RAS) and ACE2 candidate therapeutic molecules. (A) In 1898, renin was the first component of the RAS to be discovered. Vasoconstriction of the renal artery was then shown to lead to high blood pressure, thus driving the discovery of hypertensin and angiotonin (a compound later termed angiotensin). Angiotensin was subsequently characterized, as well as two downstream conpounds, the Ang I and Ang II, respectively. The ACE peptidase responsible for processing of Ang I into Ang II was subsequently characterized in 1956. The first orally active angiotensin-converting enzyme inhibitor, captopril, was used as antihypertensive therapy in patients with high blood pressure from the early 80s. The AT1R receptor was cloned in 1991, followed by the cloning of AT2R. Then, the counter-regulatory pathway of RAS was described in 2000, with the discovery of ACE2 by two independent research groups and identification of the Ang-(1–7)/Mas receptor interacting partners was achieved two decades later. The cardioprotective effects of ACE2 were discovered as was its ability to process Ang II into Ang-(1–7). Finally, studies have identified the ACE2 protease domain as the receptor for severe acute respiratory syndrome-coronavirus (SARS-CoV-1) in 2003, HCoV-NL63 in 2005, and, more recently (2020), SARS-CoV-2. (B) The ACE2 peptidase extracellular domain known to bind the SARS-CoV-2 Spike can be produced as a recombinant soluble molecule able to neutralize SARS-CoV-2. Various amino acid substitutions can be introduced in the sequence of the ACE2 extracellular domain by genetic engeneering to change the affinity of the recombinant molecule for the viral spike. However, hrsACE2 appears to have a short half-life the efficiency of which could be improved by engineering fusion proteins. The fusion of the rhACE2 extracellular domain with the nontoxic subunit B of cholera toxin (ACE2-CTBS) improves transmucosal transport. The recombinant ACE2-Ig fusion protein consists of a homodimer of the ACE2 extracellular domain linked to an Fc domain of human IgG increasing the stability of the molecule but could also act as cargo for the virus through the Fc-Tag to attach cells like macrophages that express high levels of the Fc receptor. The addition of an albumin binding domain in fusion with ACE2 extend the duration of ACE2 action.
In silico methods of molecular docking have been used to develop allosteric activators of ACE2 such as xanthenone (XNT), the antiprotozoal dimiazene aceturate (DIZE) drug, and resorcinolnaphthalein (Hernández Prada et al., 2008; Gjymishka et al., 2010). Activation of ACE2 by XNT, prevented elevated right ventricular systolic pressure, ventricular hypertrophy, and increased pulmonary vessel wall thickness (Ferreira et al., 2009; Fraga-Silva Rodrigo et al., 2013; Cole-Jeffrey et al., 2015). DIZE was found to reduce the severity of hyperoxic lungs injury by inhibiting the inflammatory response and oxidative stress, to attenuate the myocardial infarction, and to prevent atherosclerosis by increasing ACE2 mRNA expression (Kulemina and Ostrov, 2011; Qi et al., 2013; Shenoy et al., 2013; Haber et al., 2014; Qiu et al., 2014; Goru et al., 2017; Fang et al., 2019; Qaradakhi and Gadanec, 2020). Various FDA-approved molecules are under study for evaluating their ability to reduce COVID-19 severity (Albini et al., 2020; Lubbe et al., 2020; Qaradakhi and Gadanec, 2020). For example, the virtual screening of 2,456 approved drugs as inhibitors of SARS-CoV-2 spike-ACE2 interaction highlighted the properties of riboflavin (a vitamin), fenoterol (a bronchodilator), vidaranine (an anti-neoplastic agent), and cangrelor (an anti-platelet agent; Prajapat et al., 2020).
A better understanding of the role of the ACE2, encouraged the use of human recombinant soluble ACE2 (hrsACE2) in medicine (Kuba et al., 2005; Oudit et al., 2010; Johnson et al., 2011). In a tolerability study in healthy volunteers, doses up to 1.2 mg/kg hrsACE2 were administered intraveinously and the plasma half-life of the molecule was in the range of 10 h (Haschke et al., 2013). Despite a fast clearance rate, the administration of hrsACE2 alleviated the severity of influenza A H7N9 and respiratory syncytial virus (RSV)-induced lung injury (Yang et al., 2014; Gu et al., 2016). This hrsACE2 was also found to reduce IL-6 when given to healthy volunteers suffering from ARDS (Khan et al., 2017; Zhang and Baker, 2017). Through its binding to the viral S protein, sACE2 could act as a decoy receptor and could reduce the harmful effect of Ang II by making mACE2 available for the conversion of Ang II (Patel et al., 2014; Devaux et al., 2020; Issa et al., 2021; Krishnamurthy et al., 2021). The infusion of a single dose of hrsACE2 (GSK2586881 at 0.4 mg/kg i.v.), was found to be well-tolerated and to have potential haemodynamic benefits in pulmonary arterial hypertension (Hemnes et al., 2018). A hrsACE2 clinical trial is ongoing (NCT00886353) for the treatment of cardiovascular diseases (Ghatage et al., 2021). It has been reported that ACE2 S680D gain-function knock-in mice are protected against hypoxia-induced pulmonary hypertension (Zhang et al., 2018). This has also opened the way to modified ACE2 for gene transfer (Guignabert et al., 2018). It was recently demonstrated that recombinant ACE2 is effective for treating SARS-CoV-2 RBD protein-aggravated LPS-induced acute lung injury in a mouse experimental model and that the protection occurs by acting on the ACE2-AngII-AT1R-NOX1/2 axis that is otherwise overactivated by the SARS-CoV-2 infection (Zhang et al., 2022).
A proof-of-concept of the efficiency of the hrsACE2 therapeutic approach in COVID-19 was described in a case report of a 45-year-old woman infected by SARS-CoV-2 who was admitted to hospital with a 7-day history of severe symptoms. Two days after hospital admission, she was treated with 0.4 mg/kg of hrsACE2 intravenous infusion twice daily. Surprisingly after the first injection the patient became afebrile, the biological investigation indicated a marked reduction of Ang II, an increase of sACE2 in plasma, and her clinical condition improved gradually (Zoufaly et al., 2020). A large phase II clinical trial has been initiated by the Austrian pharmaceutical company APEIRON to treat COVID-19 patients with APN01-rhACE2. More recently, hrsACE2, in combination with sub-toxic remdesivir, was found to reduce viral load by 60% in a model of SARS-CoV-2 infected kidney organoids (Monteil et al., 2021).
Given such encouraging results, it seemed important to design molecules with improved activity against SARS-CoV-2 (Maiti, 2021; Figure 10B) A fusion molecule consisting of murine rACE2 with a Fc fragment (rACE2-Fc), demonstrated a long-lasting ability to protect organs in mice models of Ang II-dependent hypertension (Liu et al., 2018). It was also reported that Lactobacillus paracasi probiotic expressing a hrACE2 extracellular domain in fusion with the nontoxic subunit B of cholera toxin, resulted in increased ACE2 activities in serum of mice treated with this compound (Verma et al., 2019). Another molecule, rACE2 extracellular domain fused to the FC region of the IgG1, has been shown to neutralize viruses pseudotyped with SARS-CoV-2 spike proteins in vitro (Lei et al., 2020). A new set of molecules named “ACE2 receptor trap” that contain the extracellular domain residues 18–614 (including the SARS-CoV-2 RBD), and collectrin domain of ACE2 fused to human IgG1 Fc fragment for increased stabilization and avidity, were designed (Glasgow et al., 2020). In silico, ACE2 variants Lys26Arg and Thr92Ile were predicted to have increased affinity for the viral S protein when compared to wildtype ACE2. Consistent with this, soluble ACE2 Lys26Arg and Thr92Ile were more effective in blocking the entry of the SARS-CoV-2 S protein pseudotyped virus (Suryamohan et al., 2021). Another type of therapeutic molecules containging the hrsACE2 fused with a 5 kD albumin binding domain and bridged via a dimerization hinge-like peptide motif (termed ACE2 1-618-DDC-ABD) was first tested in an animal model prevented mortality in the treated group while untreated animals became severely hill and were found to have extensive pulmonary hemorrhage and mononuclear infiltrates (Hassler et al., 2021). The very rapid accumulation of three-dimensional structural data is likely to greatly accelerate the development of molecules aimed at treating COVID-19 patients (Sorokina et al., 2020).
Discussion
For virologists, ACE2 is the receptor for Sarbecoviruses. But to see ACE2 as a simple receptor necessary to initiate the replication cycle of the virus would be to ignore the essential role of ACE2 in the pathophysiology of COVID-19. ACE2 was not maintained during species evolution to wait for an unlikely meeting with a spike of Sarbecovirus. The regulatory function of the RAS pathway naturally devolved to ACE2 is the key element to be considered. The imbalance of the RAS pathway followed by the uncontrolled elevation of Ang II levels in SARS-CoV-2 infected patients and signaling through AT1R is the triggering event that can lead to severe forms of COVID-19. Thereby, COVID-19 is primarily a vascular rather than a respiratory disease and Ang II/AT1R blockade might attenuate progression to COVID-19.
The global COVID-19 Host Genetics Initiative (HGI) was set up to bring together international experts in human genetics and epidemiology to explore the genetic determinants of COVID-19 susceptibility, who have shed light on several host factors including ACE2, ACE, TMPRSS2, several chemokine receptors, the IL-6 receptor, IFN, and HLA, which are likely at the forefront of parameters affecting the disease severity (Correale et al., 2020; Ellinghaus et al., 2020; Initiative, 2020; Karaderi et al., 2020; Lorente et al., 2020; Nguyen et al., 2020; Strafella et al., 2020; Zhang Q. et al., 2020; Fricke-Galindo, 2021; Goujon et al., 2021; Martin-Sanchos et al., 2021; Pairo-Castineira et al., 2021). The list of genes possibly involved in the severity of COVID-19 continues to grow and indicates that the predisposition to severe COVID-19 is multifactorial. Understanding these pathways may help identifying targets for COVID-19 therapy and prophylaxis. Although the implication of a multiplicity of genes in the severity of COVID-19 is oubvious when considering the heterogenity in patient’s cases, we consider members of RAS as the main actors of the pathophysiological process. Besides ACE2 and Ang II, RAS can also be regulated by insertion/deletion (I/D) polymorphism of the ACE gene increasing the risk of severe forms of COVID-19 (Marshall et al., 2002; Harrap et al., 2003; Sayed-Tabatabaei et al., 2006; Gupta et al., 2009; Yamamoto et al., 2021). Association between the I/D polymorphism and blood pressure status has been reported (Jeunemaitre et al., 1992; Schmidt et al., 1993; Duru et al., 1994; Kario et al., 1999; Giner et al., 2000; Martinez et al., 2000; Agachan et al., 2003). Infusion of Ang I into normosensitive men was followed by higher venous levels of Ang II and increase in blood pressure in D/D carriers compared with I/I carriers (Ueda et al., 1995). Moreover, it has been reported that higher plasma IL-6 levels can be detected in ST segment elevation myocardial infarction patients, when the D allele is present (Dai et al., 2019). The possible association between the ACE genotype and the severity of COVID-19 should be further explored (Celik et al., 2021; Verma et al., 2021). One report indicates that the prevalence of D/D polymorphism is higher in COVID-19 patients with pulmonary embolism (PE) than patients without PE (Calabrese et al., 2021).
Our review highlights that the most important factors associated with severe COVID-19 outcome are related to the RAS and the regulation of blood pressure and coagulation. By focusing our attention to ACE2, we have come to the conclusion that this molecule may potentially play contrasting roles at different stages of the disease, with its ability to enable viral entry into the cell at early stages of infection thereby increasing disease susceptibility and later by decreasing Ang II/AT1R signaling thereby reducing the severity of the disease. Revisiting the structure and function of this molecule highlights the crucial role of ACE2 in the pathophysiology of sarbecoviruses-induced diseases, particularly in the context of inflammation and thrombosis. A low expression of ACE2 in the respiratory tract (e.g., epithelial cells, arterial and venous endothelial cells present in abundance in the lungs, and arterial smooth muscles) is associated with increased circulating levels of Ang II. The interaction of Ang II with AT1R and activation of various AT1R-dependent signaling pathways induce ROS release from monocytes able to trigger DNA damages and apoptosis in neighboring T-cells leading to lymphopenia, and endothelial injury by inhibiting NO synthesis. It is associated with vasoconstriction, hypertension, vascular permeability, fluid extravasation, and accelerated thrombosis in arterioles by activating hemostasis and the complement system. This process is accompanied by a recruitment of neutrophils and macrophages to the affected tissues leading to the “cytokine storm” (e.g., IL-6, MIP2, TNFα, and IFN responses). Perivascular inflammation is composed mainly of lymphocytes, with thrombi in the branches of the pulmonary artery and focal areas of congestion in the alveolar septal capillaries, as well as septal capillary lesions with wall and luminal fibrin deposition. Taken as a whole, these observations lead us to assume that instead of considering COVID-19 as respiratory tract diseases, we should rather see this disease as a clinical picture of hypercoagulopathy, microvascular immunothrombosis, and hyperinflammation. The loss of ACE2 function after the binding of SARS-CoV-2 is driven by mACE2 receptor endocytosis, activation of proteolytic cleavage of mACE2 and ACE2 gene transcriptional downregulation. Accurate quantification of RAS biomarkers should be added to the collection of tools aimed at monitoring COVID-19 infection both at pre-clinical and clinical levels.
According to the literature and our own observations, ACE2 and Ang II are the most relevant host factors in later stages of the disease and ACE2 should be seen as an ally in the global fight against COVID-19 and should be considered when designing appropriated drugs for COVID-19 therapy. It now appears that we can see the direction in wich work to deal with this disease should head. It requires treatment consisting in maintaining the homeostasis of the RAS pathway by preventing the elevation of the circulating levels of Ang II through sufficient biodisponibility of ACE2 to hydrolyze Ang II (including the use of human recombinant soluble ACE2), by inhibiting the Ang II/AT1R axis using ARBs which decrease the surface expression of AT1R, and/or by using calcium channel blokers as an alternative to ACEi and ARBS.
Author contributions
CD conceived the manuscript and wrote the first draft. LC-J participated in the correction of the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by the French Government under the “Investissements d’avenir” (Investments for the Future) program managed by the Agence Nationale de la Recherche (French ANR: National Agency for Research; reference: Méditerranée Infection 10-IAHU-03 to Professor Didier Raoult), and annual funds from Aix-Marseille university and IRD to the MEPHI research unit (Director: Professor Jean-Christophe Lagier). Other funding sources were limited to the salaries of the authors (Center National de la Recherche Scientifique for CAD, Assistance Publique Hôpitaux de Marseille for LCJ), with no other role or involvement.
Acknowledgments
We thank JC Lagier and D. Raoult for stimulating discussions and their unwavering support. Figures were designed using the Servier Medical Art supply of images available under a Creative Commons CC BY 3.0 license.
Conflict of interest
CD declares a link of interest with the Sanofi and Merck pharmaceutical companies.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbasi, J. (2021). Choose ARBs over ACE inhibitors for first-line hypertension treatment, large new analysis suggests. JAMA 326, 1244–1245. doi: 10.1001/jama.2021.14017
AbdAlla, S., Lother, H., and Quitterer, U. (2000). AT1-receptor heterodimers show enhanced G-protein activation and altered receptor sequestration. Nature 407, 94–98. doi: 10.1038/35024095
Abdeen, S., Bdeir, K., Abu-Fanne, R., Maraga, E., Higazi, M., Khurram, N., et al. (2021). Alpha-defensins: risk factor for thrombosis in COVID-19 infection. Brit. J. Haematol. 194, 44–52. doi: 10.1111/bjh.17503
Abu Hasan, Z., Williams, H., Ismail, N. M., Othman, H., Cozier, G. E., Acharya, K. R., et al. (2017). The toxicity of angiotensin converting enzyme inhibitors to larvae of the disease vectors Aedes aegypti and Anopheles gambiae. Sci. Rep. 7:45409. doi: 10.1038/srep45409
Ackermann, M., Verleden, S. E., Kuehnel, M., Haverich, A., Welte, T., Laenger, F., et al. (2020). Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 383, 120–128. doi: 10.1056/NEJMoa2015432
Advani, A., Kelly, D. J., Cox, A. J., White, K. E., Advani, S. L., Thai, K., et al. (2009). The (pro)renin receptor: site-specific and functional linkage to the vacuolar H+-ATPase in the kidney. Hypertension 54, 261–269. doi: 10.1161/HYPERTENSIONAHA.109.128645
Afelt, A., Frutos, R., and Devaux, C. (2018). Bats, coronaviruses, and deforestation: toward the emergence of novel infectious diseases? Front. Microbiol. 9:702. doi: 10.3389/fmicb.2018.00702
Afzali, B., Noris, M., Lambrecht, B. N., and Kemper, C. (2021). The state of complement in COVID-19. Nat. Rev. Immunol. 22, 77–84. doi: 10.1038/s41577-021-00665-1
Agachan, B., Isbir, T., Yilmaz, H., and Akoglu, E. (2003). Angiotensin converting enzyme I/D, angiotensinogen T174M-M235T and angiotensin II type 1 receptor A1166C gene polymorphisms in Turkish hypertensive patients. Exp. Mol. Med. 35, 545–549. doi: 10.1038/emm.2003.71
Aguilar, R., Jedlicka, A. E., Mintz, M., Mahairaki, V., Scott, A. L., and Dimopoulos, G. (2005). Global gene expression analysis of Anopheles gambiae responses to microbial challenge. Insect Biochem. Mol. Biol. 35, 709–719. doi: 10.1016/j.ibmb.2005.02.019
Albini, A., Di Guardo, G., McClain Noonan, D., and Lombardo, M. (2020). The SARS-CoV-2 receptor, ACE-2, is expressed on many diferent cell types: implications for ACE-inhibitor-and angiotensin II receptor blocker-based cardiovascular therapies. Intern. Emerg. Med. 15, 759–766. doi: 10.1007/s11739-020-02364-6
Alderman, M. H., Madhavan, S., Ooi, W. L., Cohen, H., Sealey, J. E., and Laragh, J. H. (1991). Association of the renin sodium profile with the risk of myocardial-infarction in patients with hypertension. New Engl. J. Med. 324, 1098–1104. doi: 10.1056/NEJM199104183241605
AlGhatrif, M., Tanaka, T., Moore, A. Z., Bandinelli, S., Lakatta, E. G., and Ferrucci, L. (2021). Age-associated difference in circulating ACE2, the gateway for SARS-COV-2, in humans: results from the InCHIANTI study. GeroScience 43, 619–627. doi: 10.1007/s11357-020-00314-w
Amor, S., Fernandez Blanco, L., and Baker, D. (2020). Innate immunity during SARS-CoV-2: evasion strategies and activation trigger hypoxia and vascular damage. Clin. Exp. Immunol. 202, 193–209. doi: 10.1111/cei.13523
Andersson, R., Gebhard, C., Miguel-Escalada, I., Hoof, I., Bornholdt, J., Boyd, M., et al. (2014). An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. doi: 10.1038/nature12787
Aragao, D. S., Cunha, T. S., Arita, D. Y., Andrade, M. C. C., Fernandes, A. B., Watanabe, I. K. M., et al. (2011). Purification and characterization of angiotensin converting enzyme 2 (ACE2) from murine model of mesangial cell in culture. Int. J. Biol. Macromol. 49, 79–84. doi: 10.1016/j.ijbiomac.2011.03.018
Arendse, L. B., Danser, A. H. J., Poglitsch, M., Touyz, R. M., Burnett, J. C., Llorens-Cortes, C., et al. (2019). Novel therapeutic approaches targeting the renin-angiotensin system and associated peptides in hypertension and heart failure. Pharmacol. Rev. 71, 539–570. doi: 10.1124/pr.118.017129
Aroor, A., Zuberek, M., Duta, C., Meuth, A., Sowers, J. R., Whaley-Connell, A., et al. (2016). Angiotensin II stimulation of DPP4 activity regulates megalin in the proximal tubules. Int. J. Mol. Sci. 17:780. doi: 10.3390/ijms17050780
Arthur, J. M., Forrest, J. C., Boehme, K. W., Kennedy, J. L., Owens, S., Herzog, C., et al. (2021). Development of ACE2 autoantibodies after SARSCoV-2 infection. PLoS One 16:e0257016. doi: 10.1371/journal.pone.025701
Avolio, E., Carrabba, M., Milligan, R., Kavanagh Williamson, M., Beltrami, A. P., Gupta, K., et al. (2021). The SARS-CoV-2 spike protein disrupts human cardiac pericytes function through CD147-receptor-mediated signalling: a potential non-infective mechanism of COVID-19 microvascular disease. Clin. Sci. 135, 2667–2689. doi: 10.1042/CS20210735
Bader, M., Alenina, N., Young, D., Santos, R. A. S., and Touyz, R. M. (2018). The meaning of Mas. Hypertension 72, 1072–1075. doi: 10.1161/HYPERTENSIONAHA.118.10918
Baggen, J., Vanstreels, E., Jansen, S., and Daelemans, D. (2021). Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 6, 1219–1232. doi: 10.1038/s41564-021-00958-0
Baratchian, M., McManus, J. M., Berk, M. P., Nakamura, F., Mukhopadhyay, S., Xu, W., et al. (2021). Androgen regulation of pulmonary AR, TMPRSS2 and ACE2 with implications for sex-discordant COVID-19 outcomes. Sci. Rep. 11:11130. doi: 10.1038/s41598-021-90491-1
Barkauskas, C. E., Cronce, M. J., Rackley, C. R., Bowie, E. J., Keene, D. R., Stripp, B. R., et al. (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036. doi: 10.1172/JCI68782
Barker, H., and Parkkila, S. (2020). Bioinformatic characterization of angiotensin-converting enzyme 2, the entry receptor for SARS-CoV-2. PLoS One 15:e0240647. doi: 10.1371/journal.pone.0240647
Bastard, P., Rosen, L. B., Zhang, Q., Michailidis, E., Hoffmann, H. H., Zhang, Y., et al. (2020). Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370:eabd4585. doi: 10.1126/science.abd4585
Bavishi, C., Maddox, T. M., and Messerli, F. H. (2020). Coronavirus disease 2019 (COVID-19) infection and renin angiotensin system blockers. JAMA Cardiol. 5, 745–747. doi: 10.1001/jamacardio.2020.1282
Bazzan, M., Montaruli, B., Sciascia, S., Cosseddu, D., Norbiato, C., and Roccatello, D. (2020). Low ADAMTS 13 plasma levels are predictors of mortality in COVID-19 patients. Intern. Emerg. Med. 15, 861–863. doi: 10.1007/s11739-020-02394-0
Beacon, T. H., Delcuve, G. P., and Davie, J. R. (2021). Epigenetic regulation of ACE2, the receptor of the SARS-CoV-2 virus. Genome 64, 386–399. doi: 10.1139/gen-2020-0124
Bean, D. M., Kraljevic, Z., Searle, T., Bendayan, R., Kevin, O., Pickles, A., et al. (2020). Angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers are not associated with severe COVID-19 infection in a multi-site UK acute hospital trust. Eur. J. Heart Fail. 22, 967–974. doi: 10.1002/ejhf.1924
Benetti, E., Tita, R., Spiga, O., Ciolfi, A., Birolo, G., Bruselles, A., et al. (2020). ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 28, 1602–1614. doi: 10.1038/s41431-020-0691-z
Berk, B. C., Vekshtein, V., Gordon, H. M., and Tsuda, T. (1989). Angiotensin II-stimulated protein synthesis in cultured vascular smooth muscle cells. Hypertension 13, 305–314. doi: 10.1161/01.HYP.13.4.305
Bernardi, S., Toffoli, B., Zennaro, C., Tikellis, C., Monticone, S., Losurdo, P., et al. (2012). High-salt diet increases glomerular ACE/ACE2 ratio leading to oxidative stress and kidney damage. Nephrol. Dial. Transplant. 27, 1793–1800. doi: 10.1093/ndt/gfr600
Berthelot, J. M., Drouet, L., and Lioté, F. (2020). Kawasaki-like diseases and thrombotic coagulopathy in COVID-19: delayed over-activation of the STING pathway? Emerg. Microb. Infect. 9, 1514–1522. doi: 10.1080/22221751.2020.1785336
Bertin, D., Brodovitch, A., Beziane, A., Hug, S., Bouamri, A., Mege, J. L., et al. (2020). Anticardiolipin IgG autoantibody level is an independent risk factor for COVID-19 severity. Arthritis Rheum. 72, 1953–1955. doi: 10.1002/art.41409
Blume, C., Jackson, C. L., Spalluto, C. M., Legebeke, J., Nazlamova, L., Conforti, F., et al. (2021). A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat. Genet. 53, 205–214. doi: 10.1038/s41588-020-00759
Boklund, A., Hammer, A. S., Quaade, M. L., Rasmussen, T. B., Lohse, L., Strandbygaard, B., et al. (2021). SARS-CoV-2 in danish mink farms: course of the epidemic and a descriptive analysis of the outbreaks in 2020. Animals 11:164. doi: 10.3390/ani11010164
Bosso, M., Thanaraj, T. A., Abu-Farha, M., Alanbaei, M., Abubaker, J., and Fahd, A.-M. (2020). The two faces of ACE2: the role of ACE2 receptor and its polymorphisms in hypertension and COVID-19. Mol. Ther. Meth. Clin. Dev. 18, 321–327. doi: 10.1016/j.omtm.2020.06.017
Boustany, C. M., Bharadwaj, K., Daugherty, A., Brown, D. R., Randall, D. C., and Cassis, L. A. (2004). Activation of the systemic and adipose renin-angiotensin system in rats with diet-induced obesity and hypertension. Am. J. Physiol. Regul. Integrat. Comparat. Physiol. 287, R943–R949. doi: 10.1152/ajpregu.00265.2004
Brambilla, M., Gelosa, P., Rossetti, L., Castiglioni, L., Zara, C., Canzano, P., et al. (2018). Impact of angiotensin-converting enzyme inhibition on platelet tissue factor expression in stroke-prone rats. J. Hypertens. 36, 1360–1371. doi: 10.1097/HJH.0000000000001702
Brest, P., Refae, S., Mograbi, B., Hofman, P., and Milano, G. (2020). Host polymorphisms may impact SARS-CoV-2 infectivity. Trends Genet. 36, 813–815. doi: 10.1016/j.tig.2020.08.003
Brewster, U. C., and Perazella, M. A. (2004). The renin-angiotensinaldosterone system and the kidney disease. Am. J. Med. 116, 263–272. doi: 10.1016/j.amjmed.2003.09.034
Brodard, J., Kremer Hovinga, J. A., Fontana, P., Studt, J. D., Gruel, Y., and Greinacher, A. (2021). COVID-19 patients often show high-titer non-platelet-activating anti-PF4/heparin IgG antibodies. J. Thromb. Haemost. 19, 1294–1298. doi: 10.1111/jth.15262
Brooke, G. N., and Prischi, F. (2020). Structural and functional modelling of SARS-CoV-2 entry in animal models. Sci. Rep. 10:15917. doi: 10.1038/s41598-020-72528-z
Brown, N. J., and Vaughan, D. E. (2000). Prothrombotic effects of angiotensin. Adv. Intern. Med. 45, 419–429.
Burnham, S., Smith, J. A., Lee, A. J., Isaac, R. E., and Shirras, A. D. (2005). The angiotensin-converting enzyme (ACE) gene family of Anopheles gambiae. BMC Genomics 6:172. doi: 10.1186/1471-2164-6-172
Cabral-Marques, O., Marques, A., Giil, L. M., De Vito, R., Rademacher, J., Gunther, J., et al. (2018). GPCR-specific autoantibody signatures are associated with physiological and pathological immune homeostasis. Nat. Commun. 9:5224. doi: 10.1038/s41467-018-07598-9
Cai, H. (2020). Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir. Med. 8:e20. doi: 10.1016/S2213-2600(20)30117-X.P
Caillet-Saguy, C., and Wolf, N. (2021). PDZ-containing proteins targeted by the ACE2 receptor. Viruses 13:2281. doi: 10.3390/v13112281
Calabrese, C., Annunziata, A., Coppola, A., Pafundi, P. C., Guarino, S., Di Spirito, V., et al. (2021). ACE gene I/D polymorphism and acute pulmonary embolism in COVID19 pneumonia: a potential predisposing role. Front. Med. 7:631148. doi: 10.3389/fmed.2020.631148
Calcagnile, M., Forgez, P., Iannelli, A., Bucci, C., Alifano, M., and Alifano, P. (2021). Molecular docking simulation reveals ACE2 polymorphisms that may increase the affinity of ACE2 with the SARS-CoV-2 spike protein. Biochimie 180, 143–148. doi: 10.1016/j.biochi.2020.11.004
Camargo, S. M., Singer, D., Makrides, V., Huggel, K., Pos, K. M., Wagner, C. A., et al. (2009). Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology 136, 872–882.e3. doi: 10.1053/j.gastro.2008.10.055
Campbell, C. M., and Kahwash, R. (2020). Will complement inhibition be the new target in treating COVID-19 related systemic thrombosis? Circulation 141, 1739–1741. doi: 10.1161/CIRCULATIONAHA.120.047419
Cantuti-Castelvetri, L., Ojha, R., Pedro, L. D., Djannatian, M., Franz, J., Kuivanen, S., et al. (2020). Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860. doi: 10.1126/science.abd2985
Cao, Y., Li, L., Feng, Z., Wan, S., Huang, P., Sun, X., et al. (2020). Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 6, 4–7. doi: 10.1038/s41421-020-0147-1
Cardenas, A., Rifas-Shiman, S. L., Sordillo, J. E., DeMeo, D. L., and Baccarelli, A. A. (2021). DNA methylation architecture of the ACE2 gene in nasal cells of children. Sci. Rep. 11:7107. doi: 10.1038/s41598-021-86494-7
Cardot-Leccia, N., Hubiche, T., Dellamonica, J., and Burel-Vandenbos, F. (2020). Pericyte alteration shed light on micro-vasculopathy in COVID-19 infection. Intensive Care Med. 46, 1777–1778. doi: 10.1007/s00134-020-06147-7
Carsana, L., Sanzagni, A., Nasr, A., Rossi, R. S., Pellegrinelli, A., Zerbi, P., et al. (2020). Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-Centre descriptive study. Lancet 20, 1135–1140. doi: 10.1016/S1473-3099(20)30434-5
Carvelli, J., Demaria, O., Vély, F., Batista, L., Chouaki Benmansour, N., Fares, J., et al. (2020). Association of COVID-19 inflammation with activation of the C5a–C5aR1 axis. Nature 588, 146–150. doi: 10.1038/s41586-020-2600-6
Celik, S. K., Genç, G. C., Piskin, N., Açikgöz, B., Altinsoy, B., Issiz, B. K., et al. (2021). Polymorphisms of ACE (I/D) and ACE2 receptor gene (Rs2106809, Rs2285666) are not related to the clinical course of COVID-19: a case study. J. Med. Virol. 93, 5947–5952. doi: 10.1002/jmv.27160
Cerdà-Costa, N., and Gomis-Rüth, F. X. (2014). Architecture and function of metallopeptidase catalytic domains. Protein Sci. 23, 123–144. doi: 10.1002/pro.2400
Chappell, M. C. (2019). The angiotensin-(1-7) Axis: formation and metabolism pathways. Springer nature Switzerland. Angiotensin 22, 1–26. doi: 10.1007/978-3-030-22696-1_1
Chen, J., Jiang, Q., Xia, X., Liu, K., Yu, Z., Tao, W., et al. (2020). Individual variation of the SARS-CoV-2 receptor ACE2 gene expression and regulation. Aging Cell 19:e13168. doi: 10.1111/acel.13168
Chen, L., Li, X., Chen, M., Feng, Y., and Xiong, C. (2020). The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 116, 1097–1100. doi: 10.1093/cvr/cvaa078
Chen, Y. Y., Liu, D., Zhang, P., and Zhong, J. C. (2016). Impact of ACE2 gene polymorphism on antihypertensive efficacy of ACE inhibitors. J. Hum. Hypertens. 30, 766–771. doi: 10.1038/jhh.2016.24
Chen, F., Zhang, Y., Li, X., Li, W., Liu, X., and Xue, X. (2021). The impact of ACE2 polymorphisms on COVID-19 disease: susceptibility, severity, and therapy. Front. Cell. Infect. Microbiol. 11:753721. doi: 10.3389/fcimb.2021.753721
Chlamydas, S., Papavassiliou, A. G., and Piperi, C. (2020). Epigenetic mechanisms regulating COVID-19 infection. Epigenetics 16, 263–270. doi: 10.1080/15592294.2020.1796896
Chung, M. K., Karnik, S., Saef, J., Bergmann, C., Barnard, J., Lederman, M. M., et al. (2020). SARS-CoV-2 and ACE2: the biology and clinical data settling the ARB and ACEI controversy. EBioMed. 58:102907. doi: 10.1016/j.ebiom.2020.102907
Ciceri, F., Beretta, L., Mara Scandroglio, A., Colombo, S., Landoni, G., Ruggeri, A., et al. (2020). Microvascular COVID-19 lung vessels obstructive thromboinflammatory syndrome (MicroCLOTS): an atypical acute respiratory distress syndrome working hypothesis. Crit. Care Resusc. 22, 95–97. doi: 10.51893/2020.2.pov2
Clarke, N. E., Belyaev, N. D., Lambert, D. W., and Turner, A. J. (2014). Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin. Sci. 126, 507–516. doi: 10.1042/CS20130291
Cohen-Haguenauer, O., Soubrier, F., Van Cong, N., Serero, S., Turleau, C., Jegou, C., et al. (1989). Regional mapping of the human renin gene to 1q32 by in situ hybridisation. Ann. Genet. 32, 16–20.
Cole-Jeffrey, C. T., Liu, M., Katovich, M. J., Raizada, M. K., and Shenoy, V. (2015). ACE2 and microbiota: emerging targets for cardiopulmonary disease therapy. J. Cardiovasc. Pharmacol. 66, 540–550. doi: 10.1097/FJC.0000000000000307
Corley, MJ, and Ndhlovu, LC. DNA methylation analysis of the COVID-19 host cell receptor, angiotensin I converting enzyme 2 gene (ACE2) in the respiratory system reveal age and gender differences. (2020). Preprints [Preprint]. doi: 10.20944/preprints202003.0295.v1
Correale, P., Mutti, L., Pentimalli, F., Baglio, G., Saladino, R. E., Sileri, P., et al. (2020). HLAB * 44 and C * 01 prevalence correlates with Covid19 spreading across Italy. Int. J. Mol. Sci. 21:5205. doi: 10.3390/ijms21155205
Cozzi, E., Calabrese, F., Schiavon, M., Feltracco, P., Seveso, M., Carollo, C., et al. (2017). Immediate and catastrophic antibody-mediated rejection in a lung transplant recipient with anti-angiotensin ii receptor type 1 and AntiEndothelin-1 receptor type a antibodies. Am. J. Transplant. 17, 557–564. doi: 10.1111/ajt.14053
Crackower, M. A., Sarao, R., Oudit, G. Y., Yagil, C., Kozieradzki, I., Scanga, S. E., et al. (2002). Angiotensinconverting enzyme 2 is an essential regulator of heart function. Nature 417, 822–828. doi: 10.1038/nature00786
Crisan, D., and Carr, J. (2000). Angiotensin I-converting enzyme. J. Mol. Diagn. 2, 105–115. doi: 10.1016/S1525-1578(10)60624-1
Dahlbäck, B., and Villoutreix, B. O. (2005). Regulation of blood coagulation by the protein C anticoagulant pathway: novel insights into structure-function relationships and molecular recognition Arterioscler. Thromb. Vasc. Biol. 25, 1311–1320. doi: 10.1161/01.ATV.0000168421.13467.82
Dai, S., Ding, M., Liang, N., Li, Z., Guan, L., and Liu, H. (2019). Associations of ACE I/D polymorphism with the levels of ACE, kallikrein, angiotensin II and interleukin-6 in STEMI patients. Sci. Rep. 9:19719. doi: 10.1038/s41598-019-56263-8
Dai, C. F., Xie, X., Ma, Y. T., Yang, Y. N., Li, X. M., Fu, Z. Y., et al. (2015). Relationship between CYP17A1 genetic polymorphism and essential hypertension in a Chinese population. Aging Dis. 6, 486–498. doi: 10.14336/AD.2015.0505
Daly, J. L., Simonetti, B., Klein, K., Chen, K. E., Williamson, M. K., Anton-Plagaro, C., et al. (2020). Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370, 861–865. doi: 10.1126/science.abd3072
Damas, J., Hughes, G. M., Keough, K. C., Painter, C. A., Persky, N. S., Corbo, M., et al. (2020). Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. U. S. A. 117, 22311–22322. doi: 10.1073/pnas.2010146117
Danilczyk, U., Eriksson, U., Crackower, M. A., and Penninger, J. M. (2003). A story of two ACEs. J. Mol. Med. 81, 227–234. doi: 10.1007/s00109-003-0419-x
Darbani, B. (2020). The expression and polymorphism of entry machinery for COVID-19 in human: juxtaposing population groups, gender, and different tissues. Int. J. Environ. Res. Public Health 17:3433. doi: 10.3390/ijerph17103433
Daviet, F., Guervilly, C., Baldesi, O., Bernard-Guervilly, F., Pilarczyk, E., Genin, A., et al. (2020). Heparin-induced thromcytopenia in severe COVID-19. Circulation 142, 1875–1877. doi: 10.1161/CIRCULATIONAHA.120.049015
Davis, J. O., and Freeman, R. H. (1976). Mechanisms regulating renin release. Physiol. Rev. 56, 1–56. doi: 10.1152/physrev.1976.56.1.1
de Gasparo, M., Catt, K. J., Inagami, T., Wright, J. W., and Unger, T., International Union of Pharmacology. XXIII (2000). The angiotensin II receptors. Pharmacol. Rev. 52, 415–472.
de Kloet, A. D., Krause, E. G., and Woods, S. C. (2010). The renin angiotensin system and the metabolic syndrome. Physiol. Behav. 100, 525–534. doi: 10.1016/j.physbeh.2010.03.018
Delorey, T. M., Ziegler, C. G. K., Heimberg, G., Normand, R., Yang, Y., Segerstolpe, A., et al. (2021). COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 595, 107–113. doi: 10.1038/s41586-021-03570-8
Descamps, G., Verset, L., Trelcat, A., Hopkins, C., Lechien, J. R., Journe, F., et al. (2020). ACE2 protein landscape in the head and neck region: the conundrum of SARS-CoV-2 infection. Biology 9:235. doi: 10.3390/biology9080235
Deshmukh, V., Motwani, R., Kumar, A., Kurnari, C., and Raza, K. (2020). Histopathological observations in COVID-19: a systematic review. J. Clin. Pathol. 74, 76–83. doi: 10.1136/jclinpath-2020-206995
Devaux, C. A. (2020). Are ACE inhibitors and ARBs more beneficials than harmful in the treatment of severe COVID-19 disease? Cardiovasc. Med. Cardiol. 7, 101–103. doi: 10.17352/2455-2976.000122
Devaux, C. A., Lagier, J.-C., and Raoult, D. (2021a). New insights into the physiopathology of COVID-19: SARS-CoV-2-associated gastrointestinal illness. Front. Med. 8:640073. doi: 10.3389/fmed.2021.640073
Devaux, C. A., Pinault, L., Delerce, J., Raoult, D., Levasseur, A., and Frutos, R. (2021b). Spread of mink SARS-CoV-2 variants in humans: a model of Sarbecovirus interspecies evolution. Front. Microbiol. 12:675528. doi: 10.3389/fmicb.2021.675528
Devaux, C. A., Pinault, L., Osman, I. O., and Raoult, D. (2021c). Can ACE2 receptor polymorphism predict species susceptibility to SARS-CoV-2? Front. Public Health 8:608765. doi: 10.3389/fpubh.2020.608765
Devaux, C. A., and Raoult, D. (2022). The impact of COVID-19 on populations living at high altitude: role of hypoxia-inducible factors (HIFs) signaling pathway in SARS-CoV-2 infection and replication. Front. Physiol. 13:960308. doi: 10.3389/fphys.2022.960308
Devaux, C. A., Rolain, J. M., and Raoult, D. (2020). ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immnol. Inf. 53, 425–435. doi: 10.1016/j.jmii.2020.04.015
Diez-Freire, C., Vazquez, J., Correa de Adjounian, M. F., Ferrari, M. F. R., Yuan, L., Silver, X., et al. (2006). ACE2 gene transfer attenuates hypertension-linked pathophysiological changes in the SHR. Physiol. Genomics 27, 12–19. doi: 10.1152/physiolgenomics.00312.2005
Dikalov, S. I., and Nazarewicz, R. R. (2013). Angiotensin II-induced production of mitochondrial reactive oxygen species: potential mechanisms and relevance for cardiovascular disease. Antioxid. Redox Signal. 19, 1085–1094. doi: 10.1089/ars.2012.4604
Dong, B., Yu, Q. T., Dai, H. Y., Gao, Y. Y., Zhou, Z. L., Zhang, L., et al. (2012). Angiotensin-converting enzyme-2 overexpression improves left ventricular remodeling and function in a rat model of diabetic cardiomyopathy. J. Am. Coll. Cardiol. 59, 739–747. doi: 10.1016/j.jacc.2011.09.071
Dong, B., Zhang, C., Feng, J. B., Zhao, Y. X., Li, S. Y., Yang, Y. P., et al. (2008). Overexpression of ACE2 enhances plaque stability in a rabbit model of atherosclerosis. Arteriosc. Thromb. Vascular Biol. 28, 1270–1276. doi: 10.1161/ATVBAHA.108.164715
Donkor, O. N., Henriksson, A., Vasiljevic, T., and Shah, N. P. (2007). Proteolytic activity of dairy lactic acid bacteria and probiotics as determinant of growth and in vitro angiotensin-converting enzyme inhibitory activity in fermented milk. Lait 86, 21–38. doi: 10.1051/lait:2006023
Donoghue, M., Hsieh, F., and Baronas, E. (2000). A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 87, E1–E9. doi: 10.1161/01.RES.87.5.e1
Dragun, D., Muller, D. N., Brasen, J. H., Fritsche, L., Nieminen-Kelha, M., Dechend, R., et al. (2005). Angiotensin II type 1-receptor activating antibodies in renal allograft rejection. N. Engl. J. Med. 352, 558–569. doi: 10.1056/NEJMoa035717
Duru, K., Farrow, S., Wang, J. M., Lockette, W., and Kurtz, T. (1994). Frequency of a deletion polymorphism in the gene for angiotensin converting enzyme is increased in african-Americans with hypertension. Am. J. Hypertens. 7, 759–762. doi: 10.1093/ajh/7.8.759
Ellinghaus, D., Degenhardt, F., Bujanda, L., Buti, M., Albillos, A., Invernizzi, P., et al. (2020). Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 383, 1522–1534. doi: 10.1056/NEJMoa2020283
Epelman, S., Shrestha, K., Troughton, R. W., Francis, G. S., Sen, S., Klein, A. L., et al. (2009). Soluble angiotensin-converting enzyme 2 in human heart failure: relation with myocardial function and clinical outcomes. J. Card. Fail. 15, 565–571. doi: 10.1016/j.cardfail.2009.01.014
Epelman, S., Tang, W. H., Chen, S. Y., Lente, F. V., Francis, G. S., Sen, S., et al. (2008). Detection of soluble angiotensin-converting enzyme 2 in heart failure: insights into the endogenous counter-regulatory pathway of the renin-angiotensin-aldosterone system. J. Am. Coll. Cardiol. 52, 750–754. doi: 10.1016/j.jacc.2008.02.088
Escher, R., Breakey, N., and Lammle, B. (2020). Severe COVID-19 infection associated with endothelial activation. Thromb. Res. 190:62. doi: 10.1016/j.thromres.2020.04.014
Faggiano, P., Bonelli, A., Paris, S., Milesi, G., Bisegna, S., Bernardi, N., et al. (2020). Acute pulmonary embolism in COVID-19 disease: preliminary report on seven patients. Int. J. Cardiol. 313, 129–131. doi: 10.1016/j.ijcard.2020.04.028
Fairweather, S. J., Broer, A., O’Mara, M. L., and Broer, S. (2012). Intestinal peptidases from functional complexes with neutral amino acid transporter B0AT1. Biochem. J. 446, 135–148. doi: 10.1042/BJ20120307
Fan, C., Lu, W., Li, K., Ding, Y., and Wang, J. (2021). ACE2 expression in kidney and testis may cause kidney and testis infection in COVID-19 patients. Front. Med. 7:563893. doi: 10.3389/fmed.2020.563893
Fan, R., Mao, S. Q., Gu, T. L., Zhong, F. D., Gong, M. L., Hao, L. M., et al. (2017). Preliminary analysis of the association between methylation of the ACE2 promoter and essential hypertension. Mol. Med. Rep. 15, 3905–3911. doi: 10.3892/mmr.2017.6460
Fan, Z., Wu, G., Yue, M., Ye, J., Chen, Y., Xu, B., et al. (2019). Hypertension and hypertensive left ventricular hypertrophy are associated with ACE2 genetic polymorphism. Life Sci. 225, 39–45. doi: 10.1016/j.lfs.2019.03.059
Fan, X., Yb, W., Wang, H., Sun, K., Zhang, W.-l., Song, X., et al. (2009). Polymorphisms of angiotensinconverting enzyme (ACE) and ACE2 are not associated with orthostatic blood pressure dysregulation in hypertensive patients. Acta Pharmacol. Sin. 30, 1237–1244. doi: 10.1038/aps.2009.110
Fang, Y., Gao, F., and Liu, Z. (2019). Angiotensin-converting enzyme 2 attenuates inflammatory response and oxidative stress in hyperoxic lung injury by regulating NF-κB and Nrf2 pathways. QJM 112, 914–924. doi: 10.1093/qjmed/hcz206
Fang, L., and Karakiulakis, R. M. (2020). Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 8:e21. doi: 10.1016/S2213-2600(20)30116-8
Fantini, J., Chahinian, H., and Yahi, N. (2021). Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19. Biochem. Biophys. Res. Commun. 538, 132–136. doi: 10.1016/j.bbrc.2020.10.015
Fantini, J., Devaux, C. A., Yahi, N., and Frutos, R. (2022). The novel hamster-adapted SARS-CoV-2 Delta variant may be selectively advantaged in humans. J. Inf. Secur. 84, e53–e54. doi: 10.1016/j.jinf.2022.03.001
Felmeden, D. C., Spencer, C. G. C., Chung, N. A. Y., Belgore, F. M., Blann, A. D., Beevers, D. G., et al. (2003). Relation of thrombogenesis in systemic hypertension to angiogenesis and endothelial damage/dysfunction (a substudy of the Anglo-Scandinavian cardiac outcomes trial [ASCOT]). Am. J. Cardiol. 92, 400–405. doi: 10.1016/s0002-9149(03)00657-x
Ferrand, A., Nabhani, Z. A., Tapias, N. S., Mas, E., Hugoy, J. P., and Barreau, F. (2019). NOD2 expression in intestinal epithelial cells protects toward the development of inflammation and associated carcinogenesis. Cell. Mol. Gastroenterol. Hepatol. 7, 357–369. doi: 10.1016/j.jcmgh.2018.10.009
Ferrario, C. M., Chappell, M. C., Tallant, E. A., Brosnihan, K. B., and Diz, D. I. (1997). Counterregulatory actions of angiotensin-(1-7). Hypertension 30, 535–541. doi: 10.1161/01.hyp.30.3.535
Ferrario, C. M., Jessup, J., Chappell, M. C., Averill, D. B., Brosnihan, K. B., Tallant, E. A., et al. (2005). Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation 111, 2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461
Ferrario, C. M., and Varagic, J. (2010). The ANG-(1-7)/ACE2/mas axis in the regulation of nephron function. Am. J. Physiol. Ren. Physiol. 298, F1297–F1305. doi: 10.1152/ajprenal.00110.2010
Ferreira, A. J., Shenoy, V., Yamazato, Y., Sriramula, S., Francis, J., Yuan, L., et al. (2009). Evidence for angiotensin-converting enzyme 2 as a therapeutic target for the prevention of pulmonary hypertension. Am. J. Respir. Crit. Care Med. 179, 1048–1054. doi: 10.1164/rccm.200811-1678OC
Fletcher-Sandersjöö, A., and Bellander, B. M. (2020). Is COVID-19 associated thrombosis caused by overactivation of the complement cascade? Thromb. Res. 194, 36–41. doi: 10.1016/j.thromres.2020.06.027
Fogari, R., Zoppi, A., Mugellini, A., Maffioli, P., Lazzari, P., and Derosa, G. (2011). Role of angiotensin II in plasma PAI-1 changes induced by imidapril or candesartan in hypertensive patients with metabolic syndrome. Hypertens. Res. 34, 1321–1326. doi: 10.1038/hr.2011.137
Forrester, S. J., Booz, G. W., Sigmund, C. D., Coffman, T. M., Kawai, T., Rizzo, V., et al. (2018). Angiotensin II signal transduction: an update on mechanisms of physiology and pathophysiology. Physiol. Rev. 98, 1627–1738. doi: 10.1152/physrev.00038.2017
Fournier, D., Luft, F. C., Bader, M., Ganten, D., and Andrade-Navarro, M. A. (2012). Emergence and evolution of the renin–angiotensin–aldosterone system. J. Mol. Med. 90, 495–508. doi: 10.1007/s00109-012-0894-z
Fraga-Silva Rodrigo, A., Costa-Fraga Fabiana, P., Murca Tatiane, M., Moraes Patrıcia, L., Martins Lima, A., Lautner Roberto, Q., et al. (2013). Angiotensin-converting enzyme 2 activation improves endothelial function. Hypertension 61, 1233–1238. doi: 10.1161/HYPERTENSIONAHA.111.00627
Frederich, R. C. Jr., Kahn, B. B., Peach, M. J., and Flier, J. S. (1992). Tissue-specific nutritional regulation of angiotensinogen in adipose tissue. Hypertension 19, 339–344. doi: 10.1161/01.hyp.19.4.339
Fricke-Galindo, I. (2021). Falfa´ n-Valencia R genetics insight for COVID-19 susceptibility and severity: a review. Front. Immunol. 12:622176. doi: 10.3389/fimmu.2021.622176
Frutos, R., Gavotte, L., and Devaux, C. A. (2021). Understanding the origin of COVID-19 requires to change the paradigm on zoonotic emergence from the spillover to the circulation model. Infect. Genet. Evol. 95:104812. doi: 10.1016/j.meegid.2021.104812
Frutos, R., Serra-Cobo, J., Pinault, L., Lopez Roig, M., and Devaux, C. A. (2021). Emergence of bat-related Betacoronaviruses: Hazard and risks. Front. Microbiol. 12:591535. doi: 10.3389/fmicb.2021.591535
Frutos, R., Yahi, N., Gavotte, L., Fantini, J., and Devaux, C. A. (2022). Role of spike compensatory mutations in the interspecies transmission of SARS-CoV-2. One Health 15:100429. doi: 10.1016/j.onehlt.2022.100429
Fuglsang, A., Rattray, F. P., Nilsson, D., and Nyborg, N. C. B. (2003). Lactic acid bacteria: inhibition of angiotensin converting enzyme in vitro and in vivo. Antonie Van Leeuwenhoek 83, 27–34. doi: 10.1023/A:1022993905778
Funakoshi, Y., Ichiki, T., Takeda, K., Tokuno, T., Lino, N., and Takeshita, A. (2002). Critical role of cAMP-response element-binding protein for angiotensin II-induced hypertrophy of vascular smooth muscle cells. J. Biol. Chem. 277, 18710–18717. doi: 10.1074/jbc.M110430200
Furuhashi, M., Moniwa, N., Mita, T., Fuseya, T., Ishimura, S., Ohno, K., et al. (2015). Urinary angiotensin-converting enzyme 2 in hypertensive patients may be increased by olmesartan, an angiotensin II receptor blocker. Am. J. Hypertens. 28, 15–21. doi: 10.1093/ajh/hpu086
Furuhashi, M., Moniwa, N., Takizawa, H., Ura, N., and Shimamoto, K. (2020). Potential differential effects of renin-angiotensin system inhibitors on SARS-CoV-2 infection and lung injury in COVID-19. Hypertens. Res. 43, 837–840. doi: 10.1038/s41440-020-0478-1
Gáborik, Z., Szaszák, M., Szidonya, L., Balla, B., Paku, S., Catt, K. J., et al. (2001). Beta-arrestin-and dynamin-dependent endocytosis of the AT1 angiotensin receptor. Mol. Pharmacol. 59, 239–247. doi: 10.1124/mol.59.2.239
Gagiannis, D., Steinestel, J., Hackenbroch, C., Schreiner, B., Hannemann, M., Bloch, W., et al. (2020). Clinical, serological, and histopathological similarities between severe COVID-19 and acute exacerbation of connective tissue disease-associated interstitial lung disease (CTD-ILD). Front. Immunol. 11:587517. doi: 10.3389/fimmu.2020.587517
Gaidarov, I., Adams, J., Frazer, J., Anthony, T., Chen, X., Gatlin, J., et al. (2018). Angiotensin (1-7) does not interact directly with MAS1, but can potently antagonize signaling from the AT1 receptor. Cell. Signal. 50, 9–24. doi: 10.1016/j.cellsig.2018.06.007
Gallucio, M., Pantanella, M., Gludice, D., Brescia, S., and Indiveri, C. (2020). Low temperature bacterial expression of the neutral amino acid transporters SLC1A5 (ASCT2), and SLC6A19 (B0AT1). Mol. Biol. Rep. 47, 7283–7289. doi: 10.1007/s11033-020-05717-8
Gando, S., and Wada, T. (2021). Thromboplasminflammation in COVID-19 coagulopathy: three viewpoints for diagnostic and therapeutic strategies. Front. Immunol. 12:649122. doi: 10.3389/fimmu.2021.649122
Gao, S., and Zhang, L. (2020). ACE2 partially dictates the host range and tropism of SARS-CoV-2. Comput. Struct. Biotechnol. J. 18, 4040–4047. doi: 10.1016/j.csbj.2020.11.032
Garcia-Escobar, A., Jimenez-Valero, S., Galeote, G., Jurado-Roman, A., Garcia-Rodriguez, J., and Moreno, R. (2021). The soluble catalytic ectodomain of ACE2 a biomarker of cardiac remodelling: new insights for heart failure and COVID19. Heart Fail. Rev. 26, 961–971. doi: 10.1007/s10741-020-10066-6
Garrido, A. M., and Griendling, K. K. (2009). NADPH oxidases and angiotensin II receptor signaling. Mol. Cell. Endocrinol. 302, 148–158. doi: 10.1016/j.mce.2008.11.003
Garvin, M. R., Alvarez, C., Miller, J. I., Prates, E. T., Walker, A. M., Amos, B. K., et al. (2020). A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. elife 9:e59177. doi: 10.7554/eLife.59177
Gay, L., Melenotte, C., Lakbar, I., Mezouar, S., Devaux, C., Raoult, D., et al. (2021). Sexual dimorphism and gender in infectious diseases. Front. Immunol. 12:698121. doi: 10.3389/fimmu.2021.698121
Ge, X. Y., Li, J. L., Yang, X. L., Chmura, A. A., Zhu, G., Epstein, J. H., et al. (2013). Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503, 535–538. doi: 10.1038/nature12711
Gerard, L., Lecocq, M., Bouzin, C., Hoton, D., Schmit, G., Pereira, J. P., et al. (2021). Increased angiotensin-converting enzyme 2 and loss of alveolar type II cells in COVID-19 related ARDS. Am. J. Respir. Crit. Care Med. 204, 1024–1034. doi: 10.1164/rccm.202012-4461OC
Ghatage, T., Gopal Goyal, S., Dhar, A., and Bhat, A. (2021). Novel therapeutics for the treatment of hypertension and its associated complications: peptide-and nonpeptide-based strategies. Hypertens. Res. 44, 740–755. doi: 10.1038/s41440-021-00643-z
Gheblawi, M., Wang, K., Viveiros, A., Nguyen, Q., Zhong, J. C., Turner, A. J., et al. (2020). Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 126, 1456–1474. doi: 10.1161/CIRCRESAHA.120.317015
Giner, V., Poch, E., Bragulat, E., Oriola, J., Gonzalez, D., Coca, A., et al. (2000). Renin-angiotensin system genetic polymorphisms and salt sensitivity in essential hypertension. Hypertension 35, 512–517. doi: 10.1161/01.hyp.35.1.512
Gjymishka, A., Kulemina, L. V., Shenoy, V., Katovich, M. J., Ostrov, D. A., and Raizada, M. K. (2010). Diminazene aceturate is an ACE2 activator and a novel antihypertensive drug. FASEB J. 24:1032. doi: 10.1096/fasebj.24.1_supplement.1032.3
Glasgow, A., Glasgow, J., Limonta, D., Solomon, P., Lui, I., Zhang, Y., et al. (2020). Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 117, 28046–28055. doi: 10.1073/pnas.2016093117
Goldblatt, H., Lynch, J., Hanzal, R. F., and Summerville, W. W. (1934). Studies on experimental hypertension: I. the production of persistent elevation of systolic blood pressure by means of renal ischemia. J. Exp. Med. 59, 347–379. doi: 10.1084/jem.59.3.347
Gómez, J., Albaiceta, G. M., García-Clemente, M., López-Larrea, C., Amado Rodríguez, L., Lopez-Alonso, I., et al. (2020). Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene 762:145102. doi: 10.1016/j.gene.2020.145102
Gonzalez, A. A., Lara, L. S., and Prieto, M. C. (2017). Role of collecting duct renin in the pathogenesis of hypertension. Curr. Hypertens. Rep. 19:62. doi: 10.1007/s11906-017-0763-9
Goru, S. K., Kadakol, A., Malek, V., Pandey, A., and Gaikwad, S. N. A. B. (2017). Diminazene aceturate prevents nephropathy by increasing glomerular ACE2 and AT(2) receptor expression in a rat model of type1 diabetes. Br. J. Pharmacol. 174, 3118–3130. doi: 10.1111/bph.13946
Goshua, G., Pine, A. B., Meizlish, M. L., Chang, C. H., Zhang, H., Bahel, P., et al. (2020). Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-Centre, cross-sectional study. Lancet Heamatol. 7, e575–e582. doi: 10.1016/S2352-3026(20)30216-7
Goujon, C, Rebendenne, A, Roy, P, Bonaventure, B, Valadao, AC, Desmarets, L, et al. (2021). Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs. Research Square [Preprint]. doi: 10.21203/rs.3.rs-555275/v1
Goulter, A. B., Goddard, M. J., Allen, J. C., and Clark, K. L. (2004). ACE2 gene expression is up-regulated in the human failing heart. BMC Med. 2:19. doi: 10.1186/1741-7015-2-19
Graham, R. L., Donaldson, E. F., and Baric, R. S. (2013). A decade after SARS: strategies to control emerging coronaviruses. Nat. Rev. Microbiol. 11, 836–848. doi: 10.1038/nrmicro3143
Greenberg, B. (2008). An ACE in the hole alternative pathways of the renin angiotensin system and their potential role in cardiac remodeling. J. Am. Coll. Cardiol. 52, 755–757. doi: 10.1016/j.jacc.2008.04.059
Griendling, K. K., Ushio-Fukai, M., Lassègue, B., and Alexander, R. W. (1997). Angiotensin II signaling in vascular smooth muscle. Hypertension 29, 366–370. doi: 10.1161/01.HYP.29.1.366
Gu, H., Xie, Z., Li, T., Zhang, S., Lai, C., Zhu, P., et al. (2016). Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci. Rep. 6:19840. doi: 10.1038/srep19840
Guignabert, C., de Man, F., and Lombes, M. (2018). ACE2 as therapy for pulmonary arterial hypertension: the good outweighs the bad. Eur. Respir. J. 51:1800848. doi: 10.1183/13993003.00848-2018
Guo, L., Ren, L., Yang, S., Xiao, M., Chang, D., Yang, F., et al. (2020). Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin. Infect. Dis. 71, 778–785. doi: 10.1093/cid/ciaa310
Gupta, S., Aqrawal, B. K., Goel, R. K., and Sehajpal, P. K. (2009). Angiotensin-converting enzyme gene polymorphism in hypertensive rural population of Haryana, India. J. Emerg. Trauma Shock. 2, 150–154. doi: 10.4103/0974-2700.55323
Gurley, S. B., Riquier-Brison, A. D. M., Schnermann, J., Sparks, M. A., Allen, A. M., Haase, V. H., et al. (2011). AT1A angiotensin receptors in the renal proximal tubule regulate blood pressure. Cell Metab. 13, 469–475. doi: 10.1016/j.cmet.2011.03.001
Gustafsson, F., and Holstein-Rathlou, N. H. (1999). Angiotensin II modulates conducted vasoconstriction to norepinephrine and local electrical stimulation in rat mesenteric arterioles. Cardiovasc. Res. 44, 176–184. doi: 10.1016/S0008-6363(99)00174-1
Haber, P. K., Ye, M., Wysocki, J., Maier, C., Haque, S. K., and Battle, D. (2014). Angiotensin-converting enzyme 2–independent action of presumed angiotensin-converting enzyme 2 activators. Hypertension 63, 774–782. doi: 10.1161/HYPERTENSIONAHA.113.02856
Hadman, J. A., Mort, Y. J., Catanzaro, D. F., Tallam, J. T., Baxter, J. D., Morris, B. J., et al. (1984). Primary structure of human renin gene. DNA 3, 457–468. doi: 10.1089/dna.1.1984.3.457
Hamming, I., Timens, W., Bulthuis, M., Lely, T., Navis, G., and van Goor, H. (2004). Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. J. Pathol. 203, 631–637. doi: 10.1002/path.1570
Han, H., Ma, Q., Li, C., Liu, R., Zhao, L., Wang, W., et al. (2020). Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microb. Infect. 9, 1123–1130. doi: 10.1080/22221751.2020.1770129
Han, Y., Runge, M. S., and Brasier, A. R. (1999). Angiotensin II induces Interleukin-6 transcription in vascular smooth muscle cells through pleiotropic activation of nuclear factor-kB transcription factors. Circ. Res. 84, 695–703. doi: 10.1161/01.RES.84.6.695
Harrap, S. B., Tzourio, C., Cambien, F., Poirier, O., Raoux, S., Chalmers, J., et al. (2003). The ACE gene I/D polymorphism is not associated with the blood pressure and cardiovascular benefits of ACE inhibition. Hypertension 42, 297–303. doi: 10.1161/01.HYP.0000088322.85804.96
Harzallah, I., Debliquis, A., and Drenou, B. (2020). Lupus anticoagulant is frequent in patients with Covid-19. J. Thromb. Haemost. 18, 2064–2065. doi: 10.1111/jth.14867
Haschke, M., Schuster, M., Poglitsch, M., Loiner, H., Salzberg, M., Bruggisser, M., et al. (2013). Pharmacokinetics and pharmacodynamics of recombinant human angiotensin-converting enzyme 2 in healthy human subjects. Clin. Pharmacokinet. 52, 783–792. doi: 10.1007/s40262-013-0072-7
Hashimoto, T., Perlot, T., Rehman, A., Trichereau, J., Ishiguro, H., Paolino, M., et al. (2012). ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487, 477–481. doi: 10.1038/nature11228
Hassler, L, Wysocki, J, Gelarden, I, Tomatsidou, A, Gula, H, Nicoleascu, V, et al. (2021). A novel soluble ACE2 protein totally protects from lethal disease caused by SARS-CoV-2 infection. bioRxiv [Preprint] doi: 10.1101/2021.03.12.435191
He, M., He, X., Xie, Q., Chen, F., and He, S. (2006). Angiotensin II induces the expression of tissue factor and its mechanism in human monocytes. Thromb. Res. 117, 579–590. doi: 10.1016/j.thromres.2005.04.033
Heeneman, S., Haendeler, J., Saito, Y., Ishida, M., and Berk, B. C. (2000). Angiotensin II induces transactivation of two different populations of the platelet-derived growth factor beta receptor. Key role for the p66 adaptor protein Shc. J. Biol. Chem. 275, 15926–15932. doi: 10.1074/jbc.M909616199
Helms, J., Tacquard, C., Severac, F., Leonard-Lorant, I., Ohana, M., Delabranche, X., et al. (2020). High risk of thrombosis in patients with severe SARS-CoV-2 infection: a multicenter prospective cohort study. Intensive Care Med. 46, 1089–1098. doi: 10.1007/s00134-020-06062-x
Hemnes, A. R., Rathinasabapathy, A., Austin, E. A., Brittain, E. L., Carrier, E. J., Chen, X., et al. (2018). A potential therapeutic role for angiotensin-converting enzyme 2 in human pulmonary arterial hypertension. Eur. Respir. J. 51:1702638. doi: 10.1183/13993003.02638-2017
Hendren, N. S., Drazner, M. H., Bozkurt, B., and Cooper, L. T. (2020). Description and proposed management of the acute COVID-19 cardiovascular syndrome. Circulation 141, 1903–1914. doi: 10.1161/CIRCULATIONAHA.120.047349
Henzinger, H., Barth, D. A., Klec, C., and Pichler, M. (2020). Non-Coding RNAs and SARS-related coronaviruses. Viruses 12:1374. doi: 10.3390/v12121374
Hernández Prada, J. A., Ferreira, A. J., Katovich, M. J., Shenoy, V., Qi, Y., Santos, R. A., et al. (2008). Structure-based identification of small-molecule angiotensin-converting enzyme 2 activators as novel antihypertensive agents. Hypertension 51, 1312–1317. doi: 10.1161/HYPERTENSIONAHA.107.108944
Heurich, A., Hofmann-Winkler, H., Gierer, S., Liepold, T., Jahn, O., and Pöhlmann, S. (2014). TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 88, 1293–1307. doi: 10.1128/JVI.02202-13
Heyman, S. N., Kinaneh, S., and Abassi, Z. (2021). The duplicitous nature of ACE2 in COVID-19 disease. EBioMed. 67:103356. doi: 10.1016/j.ebiom.2021.103356
Hikmet, F., Méar, L., Edvinsson, A., Micke, P., Uhlén, M., and Lindskog, C. (2020). The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 16:e9610. doi: 10.15252/msb.20209610
Hirano, T., and Murakami, M. (2020). COVID-19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity 52, 731–733. doi: 10.1016/j.immuni.2020.04.003
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Kruger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cells 181, 271–280.e8. doi: 10.1016/j.cell.2020.02.052
Hofman, P., Copin, M. C., Tauziede-Espariat, A., Adle-Biassette, H., Fortarezza, F., Passeron, T., et al. (2021). Histopathological features due to the SARS-CoV-2 [in French]. Ann. Pathol. 41, 9–22. doi: 10.1016/j.annpat.2020.12.009
Hofmann, H., Pyrc, K., van der Hoek, L., Geier, M., Berkhout, B., and Pöhlmann, S. (2005). Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 102, 7988–7993. doi: 10.1073/pnas.0409465102
Horowitz, JE, Kosmicki, JA, Damask, A, Sharma, D, Roberts, GHL, Justice, AE, et al. Genome-wide analysis in 756, 646 individuals provides first genetic evidence that ACE2 expression influences COVID-19 risk and yields genetic risk scores predictive of severe disease [Internet]. medRxiv [Preprint]. (2021). doi: 10.1101/2020.12.14.20248176
Hou, Y., Zhao, J., Martin, W., Kallianpur, A., Chung, M. K., Jehi, L., et al. (2020). New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 18:216. doi: 10.1186/s12916-020-01673-z
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. doi: 10.1016/S0140-6736(20)30183-5
Hubert, C., Houot, A. M., Corvol, P., and Soubrier, F. (1991). Structure of the angiotensin I-converting enzyme gene. Two alternate promoters correspond to evolutionary steps of a duplicated gene. J. Biol. Chem. 266, 15377–15383. doi: 10.1016/S0021-9258(18)98626-6
Ichihara, A., and Yatabe, M. S. (2019). The (pro)renin receptor in health and disease. Nat. Rev. Nephrol. 15, 693–712. doi: 10.1038/s41581-019-0160-5
Imai, Y., Kub, K., Rao, S., Huan, Y., Guo, F., Guan, B., et al. (2005). Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436, 112–116. doi: 10.1038/nature03712
Initiative, C.-H. G. (2020). The COVID-19 host genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARSCoV-2 virus pandemic. Eur. J. Hum. Genet. 28, 715–718. doi: 10.1038/s41431-020-0636-6
Issa, H., Eid, A. H., Berry, B., Takhviji, V., Khosravi, A., Mantash, S., et al. (2021). Combination of angiotensin (1-7) agonists and convalescent plasma as a new strategy to overcome angiotensin converting enzyme 2 (ACE2) inhibition for the treatment of COVID-19. Front. Med. 8:620990. doi: 10.3389/fmed.2021.620990
Itoyama, S., Keicho, N., Hijikata, M., Quy, T., Chi Phi, N., Long, H. T., et al. (2005). Identifcation of an alternative 5’-untranslated exon and new polymorphisms of angiotensin-converting enzyme 2 gene: lack of association with SARS in the Vietnamese population. Am. J. Med. Genet. 136, 52–57. doi: 10.1002/ajmg.a.30779
Jackman, H. L., Massad, M. G., Sekosan, M., Tan, F., Brovkovych, V., Marcic, B. M., et al. (2002). Angiotensin 1-9 and 1-7 release in human heart: role of cathepsin a. Hypertension 39, 976–981. doi: 10.1161/01.HYP.0000017283.67962.02
Jacoby, D. S., and Rader, D. J. (2003). Renin-angiotensin system and atherothrombotic disease: from genes to treatment. Arch. Intern. Med. 163, 1155–1164. doi: 10.1001/archinte.163.10.1155
James, M. N., and Sielecki, A. R. (1985). Stereochemical analysis of peptide bond hydrolysis catalyzed by the aspartic proteinase penicillopepsin. Biochemistry 24, 3701–3713. doi: 10.1021/bi00335a045
Jaspard, E., Wei, I., and Alhenc-Gelas, F. (1993). Differences in the properties and enzymatic specifities of the two active sites of angiotensin I-converting enzyme (kininase II). Studies with bradykinin and other natural peptides. J. Biol. Chem. 268, 9496–9503. doi: 10.1016/S0021-9258(18)98378-X
Jehpsson, L., Sun, J., Nilsson, P. M., Edsfeldt, A., and Swärd, P. (2021). Serum renin levels increase with age in boys resulting in higher renin levels in Young men compared to Young women, and soluble angiotensin-converting enzyme 2 correlates with renin and body mass index. Front. Physiol. 11:622179. doi: 10.3389/fphys.2020.622179
Jeunemaitre, X., Lifton, R. P., Hunt, S. C., Williams, R. R., and Lalouel, J. M. (1992). Absence of linkage between the angiotensin converting enzyme locus and human essential hypertension. Nat. Genet. 1, 72–75. doi: 10.1038/ng0492-72
Jia, H. P., Look, D. C., and Tan, P. (2009). Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am. J. Phys. Lung Cell. Mol. Phys. 297, L84–L96. doi: 10.1152/ajplung.00071.2009
Jin, Y., Ji, W., Yang, H., Chen, S., Zhang, W., and Duan, G. (2020). Endothelial activation and dysfunction in COVID-19: from basic mechanisms to potential therapeutic approaches. Signal Transduct. Target. Ther. 5:293. doi: 10.1038/s41392-020-00454-7
Johnson, J. A., West, J., Maynard, K. B., and Hemnes, A. R. (2011). ACE2 improves right ventricular function in a pressure overload model. PLoS One 6:e20828. doi: 10.1371/journal.pone.0020828
Kai, H., Kai, M., Niiyama, H., Okina, N., Sasaki, M., Maeda, T., et al. (2021). Overexpression of angiotensin-converting enzyme 2 by renin-angiotensin system inhibitors. Truth or myth? A systematic review of animal studies. Hypertens. Res. 44, 955–968. doi: 10.1038/s41440-021-00641-1
Karaderi, T., Bareke, H., Kunter, I., Seytanoglu, A., Cagnan, I., Balci, D., et al. (2020). Host genetics at the intersection of autoimmunity and COVID-19: a potential key for heterogeneous COVID-19 severity. Front. Immunol. 11:586111. doi: 10.3389/fimmu.2020.586111
Kario, K., Hoshide, S., Umeda, Y., Sato, Y., Ikeda, U., Nishiuma, S., et al. (1999). Angiotensinogen and angiotensin-converting enzyme genotypes, and day and night blood pressures in elderly Japanese hypertensives. Hypertens. Res. 22, 95–103. doi: 10.1291/hypres.22.95
Karnik, S. S., Singh, K. D., Tirupula, K., and Unal, H. (2017). Significance of angiotensin 1-7 coupling with MAS1 receptor and other GPGRs to the renin-angiotensin system: IUPHAR review 22. Br. J. Pharmacol. 174, 737–753. doi: 10.1111/bph.13742
Kassan, M., Ait-Aissa, K., Radwan, E., Mali, V., Haddox, S., Gabani, M., et al. (2016). Essential role of smooth muscle STIM1 in hypertension and cardiovascular dysfunction. Arterioscler. Thromb. Vasc. Biol. 36, 1900–1909. doi: 10.1161/ATVBAHA.116.307869
Kassan, M., Galán, M., Partyka, M., Saifudeen, Z., Henrion, D., Trebak, M., et al. (2012). Endoplasmic reticulum stress is involved in cardiac damage and vascular endothelial dysfunction in hypertensive mice. Arterioscler. Thromb. Vasc. Biol. 32, 1652–1661. doi: 10.1161/ATVBAHA.112.249318
Katsoularis, I., Fonseca-Rodriguez, O., Farrington, P., Jerndal, H., Häggström Lundevaller, E., Sund, M., et al. (2022). Risks of deep vein thrombosis, pulmonary embolism, and bleeding after covid-19: nationwide self-controlled cases series and matched cohort study. BMJ 377:e069590. doi: 10.1136/bmj-2021-069590
Kellam, P., and Barclay, W. (2020). The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J. Gen. Virol. 101, 791–797. doi: 10.1099/jgv.0.001439
Khaddaj-Mallat, R., Aldib, N., Bernard, M., Paquette, A. S., Ferreira, A., Lecordier, S., et al. (2021). SARS-CoV-2 deregulates the vascular and immune functions of brain pericytes via spike protein. Neurobiol. Dis. 161:105561. doi: 10.1016/j.nbd.2021.105561
Khan, A., Benthin, C., Zeno, B., Albertson, T. E., Boyd, J., Christie, J. D., et al. (2017). A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit. Care 21:234. doi: 10.1186/s13054-017-1823-x
Khayat, AS, de Assumpc, PP, Meireles Khayat, BC, Thomaz Arau Jo, TM, Batista-Gomes, JA, Imbiriba, L.C., et al. (2020). ACE2 polymorphisms as potential players in COVID-19 outcome. PLoS One 15:e0243887. doi: 10.1371/journal.pone.024388
Kintscher, U., Slagman, A., Domenig, O., Röhle, R., Konietschke, F., Poglitsch, M., et al. (2020). Plasma angiotensin peptide profiling and ACE (angiotensin-converting enzyme)-2 activity in COVID-19 patients treated with pharmacological blockers of the renin-angiotensin system. Hypertension 76, e34–e36. doi: 10.1161/HYPERTENSIONAHA.120.15841
Kliche, J., Kuss, H., Ali, M., and Ivarsson, Y. (2021). Cytoplasmic short linear motifs in ACE2 and integrin B3 link SARS-CoV-2 host cell receptors to mediators of endocytosis and autophagy. Sci. Signal. 14:ebaf1117. doi: 10.1126/scisignal.abf1117
Kok, K. H., Wong, S. C., Chan, W. M., Lei, W., Chu, A. W. H., Ip, J. D., et al. (2022). Cocirculation of two SARSCoV-2 variant strains within imported pet hamsters in Hong Kong. Emerg. Microb. Infect. 11, 689–698. doi: 10.1080/22221751.2022.2040922
Koka, V., Huang, X. R., Chung, A. C. K., Wang, W., Truong, L. D., and Lan, H. Y. (2008). Angiotensin II up-regulates angiotensin I converting enzyme (ACE), but Down-regulates ACE2 via the AT1-ERK/p38 MAP kinase pathway. Am. J. Pathol. 172, 1174–1183. doi: 10.2353/ajpath.2008.070762
Kougias, P., Chai, H., Lin, P. H., Yao, Q., Lumsden, A. B., and Chen, C. (2005). Defensins and cathelicidins: neutrophil peptides with roles in inflammation, hyperlipidemia and atherosclerosis. J. Cell. Mol. Med. 9, 3–10. doi: 10.1111/j.1582-4934.2005.tb00332.x
Kragstrup, T. W., Sogaard Singh, H., Grundberg, I., Langkilde-Lauesen Nielsen, A., Rivellese, F., Mehta, A., et al. (2021). Plasma ACE2 predicts outcome of COVID-19 in hospitalized patients. PLoS One 16:e0252799. doi: 10.1371/journal.pone.0252799
Krishnamurthy, S., Lockey, R. F., and Kolliputi, N. (2021). Soluble ACE2 as a potential therapy for COVID-19. Am. J. Phys. Cell Physiol. 320, C279–C281. doi: 10.1152/ajpcell.00478.2020
Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., et al. (2003). A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966. doi: 10.1056/NEJMoa030781
Kuan, T. C., Yang, T. H., Wen, C. H., Chen, M. Y., Lee, I. L., and Lin, C. S. (2011). Identifying the regulatory element for human angiotensin-converting enzyme 2 (ACE2) expression in human cardiofibroblasts. Peptides 32, 1832–1839. doi: 10.1016/j.peptides.2011.08.009
Kuba, K., Imai, Y., Rao, S., Gao, H., Guo, F., Guan, B., et al. (2005). A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11, 875–879. doi: 10.1038/nm1267
Kulemina, L. V., and Ostrov, D. A. (2011). Prediction of off-target effects on angiotensin-converting enzyme 2. J. Biomol. Screen. 16, 878–885. doi: 10.1177/1087057111413919
Kundura, L., Gimenez, S., Cezar, R., André, S., Younas, M., Lin, Y. L., et al. (2022). Angiotensin II induces reactive oxygen species, DNA damage, and T-cell apoptosis in severe COVID-19. J. Allergy Clin. Immunol. 150, 594–603.e2. doi: 10.1016/j.jaci.2022.06.020
Kwakernaak, A. J., Roksnoer, L. C., Lambers Heerspink, H. J., Ven den Berg-Garrelds, I., Lochorn, G. A., Embden, J. H., et al. (2017). Effects of direct renin blockade on renal & systemic hemodynamics and on RAAS activity, in weight excess and hypertension: a randomized clinical trial. PLoS One 12:e0169258. doi: 10.1371/journal.pone.0169258
Lam, S. D., Bordin, N., Waman, V. P., Scholes, H. M., Ashford, P., Sen, N., et al. (2020). SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Sci. Rep. 10:16471. doi: 10.1038/s41598-020-71936-5
Lambert, D. W., Clarke, N. E., Hooper, N. M., and Turner, A. J. (2008). Calmodulin interacts with angiotensin-converting enzyme-2 (ACE2) and inhibits shedding of its ectodomain. FEBS Lett. 582, 385–390. doi: 10.1016/j.febslet.2007.11.085
Lambert, D. W., Yarski, M., and Warner, F. J. (2005). Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J. Biol. Chem. 280, 30113–33019. doi: 10.1074/jbc.M505111200
Lamers, M. M., Beumer, J., van der Vaart, J., Knoops, K., Puschhof, J., Breugem, T. I., et al. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54. doi: 10.1126/science.abc1669
Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., et al. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220. doi: 10.1038/s41586-020-2180-5
Lanjanian, H., Moazzam-Jazi, M., Hedayati, M., Akbarzadeh, M., Guity, K., Sedaghati-Khayat, B., et al. (2021). SARS-CoV-2 infection susceptibility infuenced by ACE2 genetic polymorphisms: insights from Tehran cardio-metabolic genetic study. Sci. Rep. 11:1529. doi: 10.1038/s41598-020-80325-x
Larsson, P. T., Schwieler, J. H., and Wallen, N. H. (2000). Platelet activation during angiotensin II infusion in healthy volunteers. Blood Coag. Fibrinolysis 11, 61–69. doi: 10.1097/00001721-200011010-00007
Lavrentyev, E. N., and Malik, K. U. (2009). High glucose-induced Nox1-derived superoxides downregulate PKC-βII, which subsequently decreases ACE2 expression and ANG(1-7) formation in rat VSMCs. Am. J. Physiol. Circ. Physiol. 296, H106–H118. doi: 10.1152/ajpheart.00239.2008
Lee, I. I., Nakayama, T., Wu, C. T., Goltsey, Y., Jiang, S., Gall, P. A., et al. (2020). ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat. Commun. 11:5453. doi: 10.1038/s41467-020-19145-6
Lei, C., Qian, K., Li, T., Zhang, S., Fu, W., Ding, M., et al. (2020). Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 11:2070. doi: 10.1038/s41467-020-16048-4
Leonard-Lorant, I., Delabranche, X., Severac, F., Helms, J., Pauzet, C., Collange, O., et al. (2020). Acute pulmonary embolism in COVID-19 patients on CT angiography and relationship to D-dimer levels. Radiology 296, E189–E191. doi: 10.1148/radiol.2020201561
Li, Z., and Ferguson, A. V. (1993). Subfornical organ efferents to paraventricular nucleus utilize angiotensin as a neurotransmitter. Am. J. Phys. Regul. Integr. Comp. Phys. 265, R302–R309. doi: 10.1152/ajpregu.1993.265.2.R302
Li, G., He, X., Zhang, L., Ran, Q., Wang, J., Xiong, A., et al. (2020). Assessing ACE2 expression patterns in lung tissues in the pathogenesis of COVID-19. J. Autoimmun. 112:102463. doi: 10.1016/j.jaut.2020.102463
Li, L., Huang, M., Shen, J., Wang, Y., Wang, R., Yuan, C., et al. (2021). Serum levels of soluble platelet endothelial cell adhesion molecule 1 in COVID-19 patients are associated with disease severity. J. Infect. Dis. 223, 178–179. doi: 10.1093/infdis/jiaa642
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454. doi: 10.1038/nature02145
Li, W., Sui, J., Huang, I. C., Kuhn, J. H., Radoshitzky, S. R., Marasco, W. A., et al. (2007). The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ace2. Virology 367, 367–374. doi: 10.1016/j.virol.2007.04.035
Lindner, D., Fitzek, A., Bräuninger, H., Aleshcheva, G., Edler, C., Meissner, K., et al. (2020). Association of cardiac infection with SARS-CoV-2 in confirmed COVID-19 autopsy cases. JAMA Cardiol. 5, 1281–1285. doi: 10.1001/jamacardio.2020.3551
Lippi, G., and Favaloro, E. J. (2020). D-dimer is associated with severity of coronavirus disease 2019: a pooled analysis. Thromb. Haemost. 120, 876–878. doi: 10.1055/s-0040-1709650
Liu, Y., Hu, G., Wang, Y., Ren, W., Zhao, X., Ji, F., et al. (2021). Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 118:e2025373118. doi: 10.1073/pnas.2025373118
Liu, C., Li, Y., Guan, T., Lai, Y., Shen, Y., Zeyaweiding, A., et al. (2018). ACE2 polymorphisms associated with cardiovascular risk in Uygurs with type 2 diabetes mellitus. Cardiovasc. Diabetol. 17:127. doi: 10.1186/s12933-018-0771-3
Liu, C., Luo, R., Elliott, S. E., Wang, W., Parchim, N. F., Iriyama, T., et al. (2015). Elevated transglutaminase activity triggers angiotensin receptor activating autoantibody production and pathophysiology of preeclampsia. J. Am. Heart Assoc. 4:e002323. doi: 10.1161/JAHA.115.002323
Liu, P., Wysocki, J., Souma, T., Ye, M., Ramirez, V., Zhou, B., et al. (2018). Novel ACE2-fc chimeric fusion provides long-lasting hypertension control and organ protection in mouse models of systemic renin angiotensin system activation. Kidney Int. 94, 114–125. doi: 10.1016/j.kint.2018.01.029
Liu, Y., Yang, Y., Zhang, C., Huang, F., Wang, F., Yuan, J., et al. (2020). Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 63, 364–374. doi: 10.1007/s11427-020-1643-8
Liu, Y., and Zhang, H. G. (2021). Vigilance on new-onset atherosclerosis following SARS-CoV-2 infection. Front. Med. 7:629413. doi: 10.3389/fmed.2020.629413
Long, Q. X., Liu, B. Z., Deng, H. J., Wu, G. C., Deng, K., Chen, Y. K., et al. (2020). Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 26, 845–848. doi: 10.1038/s41591-020-0897-1
Lopez, R. D., Macedo, A. V. S., de Baros, E., Silva, P. G. M., Moll-Bernardes, J., dos Santos, T., et al. (2021). Continuing angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers on days alive and out of the hospital in patients admitted with COVID-19. A randomized clinical trial. JAMA 325, 254–264. doi: 10.1001/jama.2020.25864
Lorente, L., Martın, M. M., Franco, A., Barrios, Y., Caceres, J. J., Solé-Violán, J., et al. (2020). HLA genetic polymorphisms and prognosis of patients with COVID-19. Med. Int. 45, 96–103. doi: 10.1016/j.medin.2020.08.004
Luan, J., Lu, Y., Jin, X., and Zhang, L. (2020). Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARSCoV-2 infection. Biochem. Biophys. Res. Commun. 526, 165–169. doi: 10.1016/j.bbrc.2020.03.047
Lubbe, L., Cozier, G. E., Dosthuizen, D., Acharya, K. R., and Sturrock, E. D. (2020). ACE2 and ACE: structure-based insights into mechanism, regulation and receptor recognition by SARS-CoV. Clin. Sci. 134, 2851–2871. doi: 10.1042/CS20200899
Luo, Y., Liu, C., Guan, T., Li, Y., Lai, Y., Li, F., et al. (2019). Association of ACE2 genetic polymorphisms with hypertension-related target organ damages in South Xinjiang. Hypertens. Res. 42, 681–689. doi: 10.1038/s41440-018-0166-6
Luo, Y., Mao, L., Yan, X., Xue, Y., Lin, Q., Tang, G., et al. (2020). Predicted model based on the combination of cytokines and lymphocytes subsets for prognosis of SARS-CoV-2 infection. J. Clin. Immunol. 40, 960–969. doi: 10.1007/s10875-020-00821-7
Luther, J. M., Gainer, J. V., Murphey, L. J., Yu, C., Vaughan, D. E., Morrow, J. D., et al. (2006). Hypertension 48, 1050–1057. doi: 10.1161/01.HYP.0000248135.97380.76
Lv, Y., Li, Y., Yi, Y., Zhang, L., Shi, Q., and Yang, J. (2018). A genomic survey of angiotensin-converting enzymes provides novel insights into their molecular evolution in vertebrates. Moelcules 23:2923. doi: 10.3390/molecules23112923
Ma, H., Zeng, W., He, H., Zhao, D., Jiang, D., Zhou, P., et al. (2020). Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 17, 773–775. doi: 10.1038/s41423-020-0474-z
Magro, C., Mulvey, J. J., Berlin, D., Nuovo, G., Salvatore, S., Harp, J., et al. (2020). Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 220, 1–13. doi: 10.1016/j.trsl.2020.04.007
Maiti, B. K. (2021). Bioengineered angiotensin-converting-enzyme-2: a potential therapeutic option against SARS-CoV-2 infection. J. Hum. Hypertens. 36, 488–492. doi: 10.1038/s41371-021-00636-y
Manne, B. K., Denorme, F., Middleton, E. A., Portier, I., Rowley, J. W., Stubben, C. J., et al. (2020). Platelet gene expression and function in COVID-19 patients. Blood 136, 1317–1329. doi: 10.1182/-blood.2020007214
Marchiano, S., Hsiang, T. Y., Khanna, A., Higashi, T., Whitmore, L. S., Bargehr, J., et al. (2021). SARS-CoV-2 infects human pluripotent stem cell-derived cardiomyocytes, impairing electrical and mechanical function. Stem Cell Rep. 16, 478–492. doi: 10.1016/j.stemcr.2021.02.008
Marian, A. J. (2013). The discovery of the ACE2 gene. Circ. Res. 112, 1307–1309. doi: 10.1161/CIRCRESAHA.113.301271
Marra, M. A., Jones, S. J., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S., et al. (2003). The genome sequence of the SARS-associated coronavirus. Science 300, 1399–1404. doi: 10.1126/science.1085953
Marshall, R. P., Webb, S., Bellingan, G. J., Montgomery, H. E., Chaudhari, B., Mc Arthur, R. J., et al. (2002). Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 166, 646–650. doi: 10.1164/rccm.2108086
Martinez, E., Pura, A., Escribano, J., Sanchis, C., Carrion, L., Artigao, M., et al. (2000). Angiotensin-converting enzyme (ACE) gene polymorphisms, serum ACE activity and blood pressure in a Spanish-Mediterranean population. J. Hum. Hypertens. 14, 131–135. doi: 10.1038/sj.jhh.1000958
Martinez-Rojas, M. A., Vega-Vega, O., and Bobadilla, X. N. A. (2020). Is the kidney a target of SARS-CoV-2? Am. J. Physiol. Ren. Physiol. 318, F1454–F1462. doi: 10.1152/AJPRENAL.00160.2
Martins, A. L. V., da Silva, F. A., Bolais-Ramos, L., Capanema de Oliveira, G., Cunha Ribeiro, R., Alves Pereira, D. A., et al. (2021). Increased circulating levels of angiotensin-(1–7) in severely ill COVID-19 patients. ERJ Open Res. 7, 00114–02021. doi: 10.1183/23120541.00114-2021
Martin-Sanchos, L., Lewinski, M. K., Pache, L., Stoneham, C. A., Yin, X., Becker, M., et al. (2021). Functional landscape of SARS-CoV-2 cellular restriction. Mol. Cell 81, 2656–2668.e8. doi: 10.1016/j.molcel.2021.04.008
Massiéra, F., Bloch-Faure, M., Ceiler, D., Murakami, K., Fukamizu, A., Gasc, J.-M., et al. (2001). Adipose angiotensinogen is involved in adipose tissue growth and blood pressure regulation. FASEB J. 15, 2727–2729. doi: 10.1096/fj.01-0457fje
Mecca, A. P., Regenhardt, R. W., O'Connor, T. E., Jospeh, J. P., Raizada, M. K., Katovich, M. J., et al. (2011). Cerebroprotection by angiotensin-(1-7) in endothelin-1-induced ischaemic stroke. Exp. Physiol. 96, 1084–1096. doi: 10.1113/expphysiol.2011.058578
Mehta, P., Mc Autley, D. F., Brown, M., Sanchez, E., Tattersall, R. S., Manson, J. J., et al. (2020). COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034. doi: 10.1016/S0140-6736(20)30628-0
Meng, J., Xiao, G., Zhang, J., He, X., Ou, M., Bi, J., et al. (2020). Renin-angiotensin system inhibitors improve the clinical outcomes of COVID-19 patients with hypertension. Emerg. Microb. Infect. 9, 757–760. doi: 10.1080/22221751.2020.1746200
Meng, N., Zhang, Y., Ma, J., Li, H., Zhou, F., and Qu, Y. (2015). Association of polymorphisms of angiotensin I converting enzyme 2 with retinopathy in type 2 diabetes mellitus among Chinese individuals. Eye 29, 266–271. doi: 10.1038/eye.2014.254
Mentz, R. J., Bakris, G. L., Waeber, B., McMurray, J. J., Gheorghiade, M., Ruilope, L. M., et al. (2013). The past, present and future of renin-angiotensin aldosterone system inhibition. Int. J. Cardiol. 167, 1677–1687. doi: 10.1016/j.ijcard.2012.10.007
Middeldorp, S., Coppens, M., van Haaps, T. F., Foppen, M., Vlaar, A. P., Miller, M. C. A., et al. (2020). Incidence of venous thromboembolism in hospitalized patients with COVID-19. J. Thromb. Haemost. 18, 1995–2002. doi: 10.1111/jth.14888
Miedema, J., Schreurs, M., van der Sarvan der Brugge, S., Paats, M., Baart, S., Bakker, M., et al. (2021). Antibodies against angiotensin II receptor type 1 and endothelin a receptor are associated with an unfavorable COVID19 disease course. Front. Immunol. 12:684142. doi: 10.3389/fimmu.2021.684142
Minato, T., Nirasawa, S., Sato, T., Yamaguchi, T., Hoshizaki, M., Inagaki, T., et al. (2020). B38-CAP is a bacteria-derived ACE2-like enzyme that suppresses hypertension and cardiac dysfunction. Nat. Commun. 11:1058. doi: 10.1038/s41467-020-14867-z
Mizuiri, S., Hemmi, H., Arita, M., Ohashi, Y., Tanaka, Y., Miyagi, M., et al. (2008). Expression of ACE and ACE2 in individuals with diabetic kidney disease and healthy controls. Am. J. Kidney Dis. 51, 613–623. doi: 10.1053/j.ajkd.2007.11.022
Möhlendick, B., Schönfelder, K., Breuckmann, K., Elsner, C., Babel, N., Balfanz, P., et al. (2021). ACE2 polymorphism and susceptibility for SARS-CoV-2 infection and severity of COVID-19. Pharmacogenet. Genomics 31, 165–171. doi: 10.1097/FPC.0000000000000436
Montagutelli, X, Prot, M, Levillayer, L, Baquero SAlazar, E, Jouvion, G, Conquet, L., et al. The B1.351 and P. 1 variants extend SARS-CoV-2 host range to mice. bioRxiv [Preprint]. (2021). doi: 10.1101/2021.03.18.436013
Monteil, V., Dyczynski, M., Lauschke, V. M., Kwon, H., Wirnsberger, G., Youhanna, S., et al. (2021). Human soluble ACE2 improves the effect of remdesivir in SARS-CoV-2 infection. EMBO Mol. Med. 13:e13426. doi: 10.15252/emmm.202013426
Moore, J. P., Vinh, A., Tuck, K., Sakkal, S., Krishnan, S. M., Chan, C. T., et al. (2015). M2 macrophage accumulation in the aortic wall during angiotensin II infusion in mice is associated with fibrosis, elastin loss, and elevated blood pressure. Am. J. Physiol. Circ. Physiol. 309, H906–H917. doi: 10.1152/ajpheart.00821.2014
Muller, D. N., Hilgers, K. F., Mathews, S., Breu, V., Fischli, W., Uhlmann, R., et al. (1999). Effects of human prorenin in rats transgenic for human angiotensinogen. Hypertension 33, 312–317. doi: 10.1161/01.HYP.33.1.312
Muller, D. N., Mervaala, E. M. A., Dechend, R., Fiebeler, A., Park, J. K., Schmidt, F., et al. (2000). Angiotensin II (AT1) receptor blockade reduces vascular tissue factor in angiotensin II-induced cardiac vasculopathy. Am. J. Pathol. 157, 111–122. doi: 10.1016/S0002-9440(10)64523-3
Mullick, A. E., Yeh, S. T., Graham, M. J., Engelhardt, J. A., Prakash, T. P., and Crooke, R. M. (2017). Blood pressure lowering and safety improvements with liver angiotensinogen inhibition in models of hypertension and kidney injury. Hypertension 70, 566–576. doi: 10.1161/HYPERTENSIONAHA.117.09755
Murphy, T. J., Alexander, R. W., Griendling, K. K., Runge, M. S., and Bernstein, K. E. (1991). Isolation of a cDNA encoding the vascular type-1 angiotensin II receptor. Nature 351, 233–236. doi: 10.1038/351233a0
Muus, C, Luecken, M, Eraslan, G, Waghray, A, Heimberg, G, Sikkema, L, et al. (2020). Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. bioRxiv [Preprint]. doi: 10.1101/2020.04.19.049254
Nabah, Y. N. A., Mateo, T., Estellés, R., Mata, M., Zagorski, J., Sarau, H., et al. (2004). Angiotensin II induces neutrophil accumulation in vivo through generation and release of CXC chemokines. Circulation 110, 3581–3586. doi: 10.1161/01.CIR.0000148824.93600.F3
Naftilan, A. J., and Oparil, S. (1978). Inhibition of renin release from rat kidney slices by the angiotensins. Am. J. Phys. 235, F62–F68. doi: 10.1152/ajprenal.1978.235.1.F62
Nassar, H., Lavi, E., Akkawi, S., Bdeir, K., Heyman, S. M., Raghunath, P. N., et al. (2007). Alpha-Defensin: link between inflammation and atherosclerosis. Atherosclerosis 194, 452–457. doi: 10.1016/j.atherosclerosis.2006.08.046
Nataraj, C., Oliverio, M. I., Mannon, R. B., Mannon, P. J., Audoly, L. P., Amuchastegui, C. S., et al. (1999). Angiotensin II regulates cellular immune responses through a calcineurin-dependent pathway. J. Clin. Invest. 104, 1693–1701. doi: 10.1172/JCI7451
Neves, F. A., Duncan, K. G., and Baxter, J. D. (1996). Cathepsin B is a prorenin processing enzyme. Hypertension 27, 514–517. doi: 10.1161/01.hyp.27.3.514
Nguyen, A., David, J. K., Maden, S. K., Wood, M. A., Weeder, B. R., Nellore, A., et al. (2020). Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 94, e00510–e00520. doi: 10.1128/JVI.00510-20
Nguyen, G., Delarue, F., Burcklé, C., Bouzhir, L., Giller, T., and Sraer, J. D. (2002). Pivotal role of the renin/prorenin receptor in angiotensin II production and cellular responses to renin. J. Clin. Invest. 109, 1417–1427. doi: 10.1172/JCI14276
Nikiforuk, A. M., Kuchinski, K. S., Twa, D. D. W., Lukac, C. D., Sbihi, H., Basham, C. A., et al. (2021). The contrasting role of nasopharyngeal angiotensin converting enzyme 2 (ACE2) expression in SARS-CoV-2 infection: a cross-sectional study of people tested for COVID-19 in British Columbia. EBio Med. 66:103316. doi: 10.1016/j.ebiom.2021.103316
Nishimura, H. (2017). Renin-angiotensin system in vertebrates: phylogenetic view of structure and function. Anat. Sci. Int. 92, 215–247. doi: 10.1007/s12565-016-0372-8
Nishimura, H., Tsuji, H., Masuada, H., Nakagawa, K., Nakahara, Y., Kitamura, H., et al. (1997). Angiotensin II increases plasminogen activator Inhibitor-1 and tissue factor mRNA expression without changing that of tissue type plasminogen activator or tissue factor pathway inhibitor in cultured rat aortic endothelial cells. Thromb. Haemost. 77, 1189–1195. doi: 10.1055/s-0038-1656136
Niu, W., Qi, Y., Hou, S., Zhou, W., and Qiu, C. (2007). Correlation of angiotensin-converting enzyme 2 gene polymorphisms with stage 2 hypertension in Han Chinese. Transl. Res. 150, 374–380. doi: 10.1016/j.trsl.2007.06.002
Oarhe, C. I., Dang, V., Dang, M. T., Nguyen, H., Gopallawa, I., Gewolb, I. H., et al. (2015). Hyperoxia downregulates angiotensin-converting enzyme-2 in human fetal lung fibroblasts. Pediatr. Res. 77, 656–662. doi: 10.1038/pr.2015.27
Onabajo, O. O., Banday, A. R., Stanifer, M. L., Yan, W., Obajemu, A., Santer, D. M., et al. (2020). Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 52, 1283–1293. doi: 10.1038/s41588-020-00731-9
Ortiz, M. E., Thurman, A., Pezzulo, A. A., Leidinger, M. R., Klesney-Tait, J. A., Karp, P. H., et al. (2020). Heterogeneous expression of the SARS Coronavirus-2 receptor ACE2 in the human respiratory tract. EBioMed. 60:102976. doi: 10.1016/j.ebiom.2020.102976
Ortiz-Fernández, L., and Sawalha, A. H. (2020). Genetic variability in the expression of the SARS-CoV-2 host cell entry factors across populations. Genes Immun. 21, 269–272. doi: 10.1038/s41435-020-0107-7
Osman, I. O., Garrec, C., de Souza, G. A. P., Zarubica, A., Belhaouari, D. B., Baudoin, J.-P., et al. (2022). Control of CDH1/E-cadherin gene expression and release of a soluble form of E-cadherin in SARS-CoV-2 infected Caco-2 intestinal cells: Physiopathological consequences for the intestinal forms of COVID-19. Front. Cell. Infect. Microbiol. 12:798767. doi: 10.3389/fcimb.2022.798767
Osman, I. O., Melenotte, C., Brouqui, P., Million, M., Lagier, J.-C., Parola, P., et al. (2021). Expression of ACE2, soluble ACE2, angiotensin I, angiotensin II and angiotensin-(1-7) is modulated in COVID-19 patients. Front. Immunol. 12:625732. doi: 10.3389/fimmu.2021.625732
Othman, H., Bouslama, Z., Brandenburg, J. T., da Rocha, J., Hamdi, Y., Ghedira, K., et al. (2020). Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem. Biophys. Res. Commun. 527, 702–708. doi: 10.1016/j.bbrc.2020.05.028
Oude Munnink, B. B., Sikkema, R. S., Nieuwenhuijse, D. F., Molenaar, R. J., Munger, E., Molenkamp, R., et al. (2021). Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371, 172–177. doi: 10.1126/science.abe5901
Oudit, G. Y., Crackower, M. A., Backx, P. H., and Penninger, J. M. (2003). The role of ACE2 in cardiovascular physiology. Trends Cardiovasc. Med. 13, 93–101. doi: 10.1016/s1050-1738(02)00233-5
Oudit, G. Y., Liu, G. C., Zhong, J., Basu, R., Chow, F. L., Zhou, J., et al. (2010). Human recombinant ACE2 reduces the progression of diabetic nephropathy. Diabetes 59, 529–538. doi: 10.2337/db09-1218
Oudit, G. Y., and Pfeffer, M. A. (2020). Plasma angiotensin-converting enzyme 2: novel biomarker in heart failure with implications for COVID-19. Eur. Heart J. 41, 1818–1820. doi: 10.1093/eurheartj/ehaa414
Page, I. H., and Helmer, O. M. (1940). Angiotonin-activator, renin-and angiotonin-inhibitor, and the mechanism of angiotonin tachyphylaxis in normal, hypertensive, and nephrectomized animals. J. Exp. Med. 71, 495–519. doi: 10.1084/jem.71.4.495
Pairo-Castineira, E., Clohisey, S., Klaric, L., Bretherick, A., Rawlik, K., Parkinson, N., et al. (2021). Genetic mechanisms of critical illness in Covid-19. Nature 591, 92–98. doi: 10.1038/s41586-020-03065-y
Patel, S. K., Velkoska, E., Freeman, M., Wai, B., Lancefield, T. F., and Burrell, L. M. (2014). From gene to protein - experimental and clinical studies of ACE2 in blood pressure control and arterial hypertension. Front. Physiol. 5:227. doi: 10.3389/fphys.2014.00227
Patel, V. B., Zhong, J. C., Grant, M. B., and Oudit, G. Y. (2016). Role of the ACE2/angiotensin 1-7 axis of the renin-angiotensin system in heart failure. Circ. Res. 118, 1313–1326. doi: 10.1161/CIRCRESAHA.116.307708
Patnaik, M., Pati, P., Swain, S. N., Mohapatra, M. K., Dwibedi, B., Kar, S. K., et al. (2014). Association of angiotensinconverting enzyme and angiotensin-converting enzyme-2 gene polymorphisms with essential hypertension in the population of Odisha, India. Ann. Hum. Biol. 41, 145–152. doi: 10.3109/03014460.2013.837195
Paul, M., Poyan Mehr, A., and Kreutz, R. (2006). Physiology of local renin-angiotensin systems. Physiol. Rev. 86, 747–803. doi: 10.1152/physrev.00036.2005
Pedersen, K. B., Chhabra, K. H., Nguyen, V. K., Xia, H., and Lazartigues, E. (2013). The transcription factor HNF1alpha induces expression of angiotensin-converting enzyme 2 (ACE2) in pancreatic islets from evolutionarily conserved promoter motifs. Biochim. Biophys. Acta 1829, 1225–1235. doi: 10.1016/j.bbagrm.2013.09.007
Pena Silva, R. A., Chu, Y., Miller, J. D., Mitchell, I. J., Penninger, J. M., Faraci, F. M., et al. (2012). Impact of ACE2 deficiency and oxidative stress on cerebrovascular function with aging. Stroke 43, 3358–3363. doi: 10.1161/STROKEAHA.112.667063
Peng, H., Li, W., Seth, D. M., Nair, A. R., Francis, J., and Feng, Y. (2013). (pro)renin receptor mediates both angiotensin II-dependent and-independent oxidative stress in neuronal cells. PLoS One 8:e58339. doi: 10.1371/journal.pone.0058339
Perlot, T., and Penninger, J. M. (2013). ACE2-from the renin-angiotensin system to gut microbiota and malnutrition. Microbes Infect. 15, 866–873. doi: 10.1016/j.micinf.2013.08.003
Phillips, M. I., and Schmidt-Ott, K. M. (1999). The discovery of renin 100 years ago. News Physiol. Sci. 14, 271–274. doi: 10.1152/physiologyonline.1999.14.6.271
Philogene, M. C., Johnson, T., Vaught, A. J., Zakaria, S., and Fedarko, N. (2019). Antibodies against angiotensin II type 1 and endothelin a receptors: relevance and pathogenicity. Hum. Immunol. 80, 561–567. doi: 10.1016/j.humimm.2019.04.012
Pierce, J. B., Simion, V., Icli, B., Perez-Cremades, D., Cheng, H. S., and Feinberg, M. W. (2020). Computational analysis of targeting SARS-CoV-2, viral entry proteins ACE2 and TMPRSS2, and interferon genes by host MicroRNAs. Genes (Basel) 11:1354. doi: 10.3390/genes11111354
Pinheiro, D. S., Santos, R. S., Jardim, P. C. B. V., Silva, E. G., Reis, A. A. S., Pedrino, G. R., et al. (2019). The combination of ACE I/D and ACE2 G8790A polymorphisms reveals susceptibility to hypertension: a genetic association study in Brazilian patients. PLoS One 14:e0221248. doi: 10.1371/journal.pone.0221248
Pinto, B. G. G., Oliveira, A. E. R., Singh, Y., Jimenez, L., Gonçalves, A. N. A., Ogava, R. L. T., et al. (2020). ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. J. Infect. Dis. 222, 556–563. doi: 10.1093/infdis/jiaa332
Pires de Souza, G. A., Osman, I. O., Le Bideau, M., Baudoin, J.-P., Jaafar, R., Devaux, C., et al. (2022). Angiotensin II receptor blockers (ARBs antihypertensive agents) increase replication of SARS-CoV-2 in Vero E6 cells. Front. Cell. Infect. Microbiol. 11:639177. doi: 10.3389/fcimb.2021.639177
Poissy, J., Goutay, J., Caplan, M., Parmentier, E., Duburcq, T., Lassale, F., et al. (2020). Pulmonary embolism in patients with COVID19. Circulation 142, 184–186. doi: 10.1161/CIRCULATIONAHA.120.047430
Pouladi, N., and Abdolahi, S. (2021). Investigating the ACE2 polymorphisms in COVID-19 susceptibility: An in silico analysis. Mol. Genet. Genom. Med. 9:e1672. doi: 10.1002/mgg3.1672
Prajapat, M., Shekhar, N., Sarma, P., Avti, P., Singh, S., Kaur, H., et al. (2020). Virtual screening and molecular dynamics study of approved drugs as inhibitors of spike protein S1 domain and ACE2 interaction in SARS-CoV-2. J. Mol. Graph. Model. 101:107716. doi: 10.1016/j.jmgm.2020.107716
Prieto, M. C., Gonzalez-Villalobos, R. A., Botros, F. T., Martin, V. L., Pagan, J., Satou, R., et al. (2011). Reciprocal changes in renal ACE/ANG II and ACE2/ANG 1-7 are associated with enhanced collecting duct renin in Goldblatt hypertensive rats. Am. J. Physiol. Ren. Physiol. 300, F749–F755. doi: 10.1152/ajprenal.00383.2009
Procko, E. The sequence of human ACE2 is suboptimal for binding the S spike protein of SARS coronavirus 2. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.1103.1116.994236
Qaradakhi, T., and Gadanec, L. (2020). Could DIZE be the answer to COVID-19? Maturitas 140, 83–84. doi: 10.1016/j.maturitas.2020.07.002
Qi, Y. F., Zhang, J., Cole-Jeffrey, C. T., Shenoy, V., Espejo, A., Hanna, M., et al. (2013). Diminazene Aceturate enhances angiotensin-converting enzyme 2 activity and attenuates ischemia-induced cardiac pathophysiology. Hypertension 62, 746–752. doi: 10.1161/HYPERTENSIONAHA.113.01337
Qiao, Y., Wang, X. M., Mannan, R., Pitchiaya, S., Zhang, Y., Wotring, J. W., et al. (2021). Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc. Natl. Acad. Sci. U. S. A. 118:e2021450118. doi: 10.1073/pnas.2021450118
Qiu, Y., Shil, P. K., Zhu, P., Yang, H., Verma, A., Lei, B., et al. (2014). Angiotensin-converting enzyme 2 (ACE2) activator Diminazene Aceturate ameliorates endotoxin-induced uveitis in mice. Invest. Ophthalmol. Vis. Sci. 55, 3809–3818. doi: 10.1167/iovs.14-13883
Qiu, Y., Zhao, Y. B., Wang, Q., Li, J. Y., Zhou, Z. J., Liao, C. H., et al. (2020). Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microbes Infect. 22, 221–225. doi: 10.1016/j.micinf.2020.03.003
Radzikowska, U., Ding, M., Tan, G., Zhakparov, D., Peng, Y., Wawrzyniak, P., et al. (2020). Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 75, 2829–2845. doi: 10.1111/all.14429
Raghavan, S., Kenchappa, D. B., and Leo, M. D. (2021). SARS-CoV-2 spike protein induces degradation of junctional proteins that maintain endothelial barrier integrity. Front. Cardiovasc. Med. 8:687783. doi: 10.3389/fcvm.2021.687783
Rajagopal, S., Rajagopal, K., and Lefkowitz, R. J. (2010). Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat. Rev. Drug Discov. 9, 373–386. doi: 10.1038/nrd3024
Reich, H. N., Oudit, G. Y., Penninger, J. M., Scholey, J. W., and Herzenberg, A. M. (2008). Decreased glomerular and tubular expression of ACE2 in patients with type 2 diabetes and kidney disease. Kidney Int. 74, 1610–1616. doi: 10.1038/ki.2008.497
Reindl-Schwaighofer, R., Hödlmoser, S., Eskandary, F., Poglitsch, M., Bonderman, D., Strassl, R., et al. (2021). ACE2 elevation in severe COVID-19. Am. J. Resp. Critic. Care Med. 203, 1191–1196. doi: 10.1164/rccm.202101-0142LE
Ren, W., Lan, J., Ju, X., Gong, M., Long, Q., Zhu, Z., et al. (2021). Mutation Y453F in the spike protein of SARS-CoV-2 enhances interaction with the mink ACE2 receptor for host adaption. PLoS Pathog. 17:e1010053. doi: 10.1371/journal.ppat.1010053
Rice, G. I., Jones, A. L., Grant, P. J., Carter, A. M., Turner, A. J., and Hooper, N. M. (2006). Circulating activities of angiotensin-converting enzyme, its homolog, angiotensin-converting enzyme 2, and neprilysin in a family study. Hypertension 48, 914–920. doi: 10.1161/01.HYP.0000244543.91937.79
Rieder, M., Wirth, L., Pollmeier, L., Jeserich, M., Goller, I., Baldus, N., et al. (2021). Serum ACE2, angiotensin II, and aldosterone levels are unchanged in patients with COVID-19. Am. J. Hypertens. 34, 278–281. doi: 10.1093/ajh/hpaa169
Riordan, J. F. (2003). Angiotensin-I-converting enzyme and its relatives. Genome Biol. 4:225. doi: 10.1186/gb-2003-4-8-225
Rivière, G., Michaud, A., Corradi, H. R., Sturrock, E. D., Acharya, K. R., Coqez, V., et al. (2007). Characterization of the first angiotensin-converting like enzyme in bacteria: ancestor ACE is already active. Gene 399, 81–91. doi: 10.1016/j.gene.2007.05.010
Robinson, F. A., Mihealsick, R. P., Wagener, B. M., Hanna, P., Poston, M. D., Efimov, I. R., et al. (2020). Role of angiotensin-converting enzyme 2 and pericytes in cardiac complications of COVID-19 infection. Am. J. Physiol. Heart Circ. Physiol. 319, H1059–H1068. doi: 10.1152/ajpheart.00681.2020
Rodriguez Rodriguez, M., Castro Quismondo, N., Zafra Torres, D., Gil Alos, D., Ayala, R., and Martinez-Lopez, J. (2021). Increased von Willebrand factor antigen and low ADAMTS13 activity are related to poor prognosis in covid-19 patients. Int. J. Lab. Hematol. 43, O152–O155. doi: 10.1111/ijlh.13476
Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., et al. (2003). Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300, 1394–1399. doi: 10.1126/science.1085952
Rovas, A., Osiaevi, I., Buscher, K., Sackarnd, J., Tepasse, P. R., Fobker, M., et al. (2020). Microvascular dysfunction in COVID-19: the MYSTIC study. Angiogenesis 24, 145–157. doi: 10.1007/s10456-020-09753-7
Ruiz-Ortega, M., Lorenzo, O., Suzuki, Y., Rupe’rez, M., and Egido, J. (2001). Proinflammatory actions of angiotensins. Curr. Opin. Nephrol. Hypertens. 10, 321–329. doi: 10.1097/00041552-200105000-00005
Rushworth, C. A., Guy, J. L., and Turner, A. J. (2008). Residues affecting the chloride regulation and substrate selectivity of the angiotensin-converting enzymes (ACE and ACE2) identified by site-directed mutagenesis. FEBS J. 275, 6033–6042. doi: 10.1111/j.1742-4658.2008.06733.x
Rysz, S., Al-Saadi, J., Sjöström, A., Farm, M., Campoccia Jalde, F., Plattén, M., et al. (2021). COVID-19 pathophysiology may be driven by an imbalance in the renin-angiotensin-aldosterone system. Nat. Commun. 12:2417. doi: 10.1038/s41467-021-22713-z|
Sadoshima, J., Qiu, Z. H., Morgan, J. P., and Izumo, S. (1995). Angiotensin-II and other hypertrophic stimuli mediated by G-protein-coupled receptors activate tyrosine kinase, mitogen-activated protein-kinase, and 90-Kd S6 kinase in cardiac myocytes - the critical role of Ca2+−dependent signaling. Circ. Res. 76, 1–15. doi: 10.1161/01.res.76.1.1
Sakka, M., Connors, J. M., He Kimian, G., Martin-Toutain, I., Crichi, B., Colmegna, I., et al. (2020). Association between D-dimer levels and mortality in patients with coronavirus disease 2019 COVID19: a systematic review and pooled analysis. J. Med. Vasc. 45, 268–274. doi: 10.1016/j.jdmv.2020.05.003
Sama, I. E., Ravera, A., Santema, B. T., van Goor, H., ter Maaten, J. M., Cleland, J. G. F., et al. (2020). Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin–angiotensin–aldosterone inhibitors. Eur. Heart J. 41, 1810–1817. doi: 10.1093/eurheartj/ehaa373
Samavati, L., and Uhal, B. D. (2020). ACE2, much more than just a receptor for SARS-COV-2. Front. Cell. Infect. Microbiol. 10:317. doi: 10.3389/fcimb.2020.00317
Santos, R. A. S., Sampaio, W. O., Alzamora, A. C., Motta-Santos, D., Alenina, N., Bader, M., et al. (2018). The ACE2/angiotensin-(1–7)/MAS axis of the renin-angiotensin system: focus on angiotensin-(1–7). Physiol. Rev. 98, 505–553. doi: 10.1152/physrev.00023.2016
Santos, R. A., Silva AC, S. E., Maric, C., DMR, S., Machado, R. P., de Buhr, I., et al. (2003). Angiotensin-(1-7) is an endogenous ligand for the G protein-coupled receptor Mas. Proc. Natl. Acad. Sci. U. S. A. 100, 8258–8263. doi: 10.1073/pnas.1432869100
Sarzani, R., Giulietti, F., Di Pentima, C., Filipponi, A., and Spannella, F. (2020). Antagonizing the renin-angiotensin-aldosterone system in the era of COVID-19. Intern. Emerg. Med., 15, 885–887. doi: 10.1007/s11739-020-02365-5
Sato, T., Suzuki, T., Watanabe, H., Kadowaki, A., Fukamizu, A., Liu, P. P., et al. (2013). Apelin is a positive regulator of ACE2 in failing hearts. J. Clin. Invest. 123, 5203–5211. doi: 10.1172/JCI69608
Sayed-Tabatabaei, F. A., Oostra, B. A., Isaacs, A., van Duijn, C. M., and Witteman, J. C. M. (2006). ACE polymorphism. Circ. Res. 98, 1123–1133. doi: 10.1161/01.RES.0000223145.74217.e7
Schiffrin, E. L., Flack, J. M., Ito, S., Muntner, P., and Webb, R. C. (2020). Hypertension and COVID-19. Am. J. Hypertens. 33, 373–374. doi: 10.1093/ajh/hpaa057
Schmidt, S., van Hooft, I. M., Grobbee, D. E., Ganten, D., and Ritz, E. (1993). Polymorphism of the angiotensin I converting enzyme gene is apparently not related to high blood pressure: Dutch hypertension and offspring study. J. Hypertens. 11, 345–348. doi: 10.1097/00004872-199304000-00003
Schmidt-Ott, K. M., Kagiyama, S., and Philips, M. I. (2000). The multiple actions of angiotensin II in atherosclerosis. Regul. Pept. 93, 65–77. doi: 10.1016/s0167-0115(00)00178-6
Schmieder, R. E., Hilgers, K. F., Schlaich, M. P., and Schmidt, B. M. W. (2007). Renin-angiotensin system and cardiovascular risk. Lancet 369, 1208–1219. doi: 10.1016/S0140-6736(07)60242-6
Sealey, J. E., and Rubattu, S. (1989). Prorenin and renin as separate mediators of tissue and circulating systems am. J. Hypertens. 2, 358–366. doi: 10.1093/ajh/2.5.358
Senchenkova, E. Y., Russell, J., Almeida-Paula, L. D., Harding, J. W., and Granger, D. N. (2010). Angiotensin II-mediated microvascular thrombosis. Hypertension 56, 1089–1095. doi: 10.1161/HYPERTENSIONAHA.110.158220
Senchenkova, E. Y., Russell, J., Esmon, C. T., and Granger, D. N. (2014). Roles of coagulation and fibrinolysis in angiotensin II enhanced microvascular thrombosis. Microcirculation 21, 401–407. doi: 10.1111/micc.12120
Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., et al. (2020). Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224. doi: 10.1038/s41586-020-2179-y
Shen, L., Mo, H., Cai, L., Kong, T., Zheng, W., Ye, J., et al. (2009). Losartan prevents sepsis-induced acute lung injury and decreases activation of nuclear factor kappaB and mitogen-activated protein kinases. Shock 31, 500–506. doi: 10.1097/SHK.0b013e318189017a
Shen, Q., Xiao, X., Aierken, A., Yue, W., Wu, X., Liao, M., et al. (2020). The ACE2 expression in Sertoli cells and germ cells may cause male reproductive disorder after SARS-CoV-2 infection. J. Cell. Mol. Med. 24, 9472–9477. doi: 10.1111/jcmm.15541
Shenoy, V., Gjymishka, A., Jarajapu, Y. P., Qi, Y., Afzal, A., Rigatto, K., et al. (2013). Diminazene attenuates pulmonary hypertension and improves angiogenic progenitor cell functions in experimental models. Am. J. Respir. Crit. Care Med. 187, 648–657. doi: 10.1164/rccm.201205-0880OC
Sherman, E. J., and Emmer, B. T. (2021). ACE2 protein expression within isogenic cell lines is heterogeneous and associated with distinct transcriptomes. Sci. Rep. 11:15900. doi: 10.1038/s41598-021-95308-9
Shilts, J., Crozier, T. W. M., Greenwood, E. J. D., Lehner, P. J., and Wright, G. J. (2021). No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 11:413. doi: 10.1038/s41598-020-80464-1
Shuai, H., Chan, J. F. W., Yuen, T. T. T., Yoon, C., Hu, J. C., Wen, L., et al. (2021). Emerging SARS-CoV-2 variants expand species tropism to murines. eBioMed. 73:103643. doi: 10.1016/j.ebiom.2021.103643
Shukla, N., Roelle, S. M., Suzart, V. G., Bruchez, A. M., and Matreyek, K. A. (2021). Mutants of human ACE2 differentially promote SARS-CoV and SARS-CoV-2 spike mediated infection. PLoS Pathog. 17:e1009715. doi: 10.1371/journal.ppat.1009715
Siddiquee, K., Hampton, J., McAnally, D., May, L., and Smith, L. (2013). The apelin receptor inhibits the angiotensin II type 1 receptor via allosteric trans-inhibition. Br. J. Pharmacol. 168, 1104–1117. doi: 10.1111/j.1476-5381.2012.02192.x
Siguret, V., Voicu, S., Neuwirth, M., Delrue, M., Gayat, E., Stépanian, A., et al. (2020). Are antiphospholipid antibodies associated with thrombotic complications in critically ill COVID-19 patients? Thromb. Res. 195, 74–76. doi: 10.1016/j.thromres.2020.07.016
Silva, G. M., França-Falcão, M. S., Calzerra, N. T. M., Luz, M. S., Gadelha, D. D. A., Balarini, C. M., et al. (2020). Role of renin angiotensin system components in atherosclerosis: focus on Ang-II, ACE2, and Ang-1–7. Front. Physiol. 11:1067. doi: 10.3389/fphys.2020.01067
Simoes e Silva, A. C., Silveira, K. D., Ferreira, A. J., and Teixeira, M. M. (2013). ACE2, angiotensin-(1-7) and Mas receptor axis in inflammation and fibrosis. Br. J. Pharmacol. 169, 477–492. doi: 10.1111/bph.12159
Singer, D., and Camargo, S. M. R. (2011). Collectrin and ACE2 in renal and intestinal amino acid transport. Channels 5, 410–423. doi: 10.4161/chan.5.5.16470
Singer, D., Camargo, S. M. R., Ramadam, T., Schäfer, M., Mariotta, L., Herzog, B., et al. (2012). Defective intestinal amino acidabsorption in ACE2 null mice. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G686–G695. doi: 10.1152/ajpgi.00140.2012
Singh, K. D., and Karnik, S. S. (2016). Angiotensin receptors: structure, function, signaling and clinical applications. J. Cell Signal. 1:111. doi: 10.4172/jcs.1000111
Skeggs, L. T. Jr., Kahn, J. R., and Shumway, N. P. (1956). The preparation and function of the hypertensin-converting enzyme. J. Exp. Med. 103, 295–299. doi: 10.1084/jem.103.3.295
Skidgel, R. A., and Erdos, E. G. (1987). The broad substrate specificity of human angiotensin I converting enzyme. Clin. Exp. Hypertens. A 9, 243–259. doi: 10.3109/10641968709164184
Smadja, D. M., Mentzer, S., Fontenay, M., Laffan, M. A., Ackermann, M., Helms, J., et al. (2021). COVID-19 is a systemic vascular hemopathy: insight for mechanistic and clinical aspects. Angiogenesis 24, 755–788. doi: 10.1007/s10456-021-09805-6
Sodhi, C. P., Wohlford-Lenane, C., Yamaguchi, Y., Prindle, T., Fulton, W. B., Wang, S., et al. (2017). Attenuation of pulmonary ACE2 activity impairs inactivation of des-Arg9 bradykinin/BKB1R axis and facilitates LPS-induced neutrophil infiltration. AJP Lung Cell. Mol. Physiol. 314, L17–L31. doi: 10.1152/ajplung.00498.2016
Sorokina, M., Teixeira, J. M. C., Barrera-Vilarmau, S., Paschkle, R., Papasotiriou, I., Rodrigues, J. P. L. M., et al. (2020). Structural models of human ACE2 variants with SARS-CoV-2 spike protein for structure-based drug design. Sci. Data 7:309. doi: 10.1038/s41597-020-00652-6
Spitler, K. M., and Webb, R. C. (2014). Endoplasmic reticulum stress contributes to aortic stiffening via proapoptotic and fibrotic signaling mechanisms. Hypertension 63, e40–e45. doi: 10.1161/HYPERTENSIONAHA.113.02558
Stawiski, EW, Diwanji, D, Suryamohan, K, Gupta, R., Fellouse, FA, Sathirapongsasuti, JF, et al. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. bioRxiv [Preprint] (2020). doi: 10.1101/2020.04.07.024752
Stefely, J. A., Christensen, B. B., Gogakos, T., Conne Sullivan, J. K., Montgomery, G. G., Barranco, J. P., et al. (2020). Marked factor V activity elevation in severe COVID-19 is associated with venous thromboembolism. Am. J. Hematol. 95, 1522–1530. doi: 10.1002/ajh.25979
Strafella, C., Caputo, V., Termine, A., Barati, S., Caltagirone, C., Giardina, E., et al. (2020). Investigation of genetic variations of IL6 and IL6r as potential prognostic and pharmacogenetics biomarkers: implications for covid-19 and neuroinflammatory disorders. Lifestyles 10, 1–10. doi: 10.3390/life10120351
Suryamohan, K., Diwanji, D., Stawiski, E. W., Gupta, R., Miersch, S., Liu, J., et al. (2021). Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Comm. Biol. 4:475. doi: 10.1038/s42003-021-02030-3
Takayanagi, T., Kawai, T., Forrester, S. J., Obama, T., Tsuji, T., Fukuda, Y., et al. (2015). Role of epidermal growth factor receptor and endoplasmic reticulum stress in vascular remodeling induced by angiotensin II. Hypertension 65, 1349–1355. doi: 10.1161/HYPERTENSIONAHA.115.05344
Tan, Y. K., Goh, C., Leow, A. S. T., Tambyah, P. A., Ang, A., Yap, E. S., et al. (2020). COVID-19 and ischemic stroke: a systematic review and meta-summary of the literature. J. Thromb. Thrombolysis 50, 587–595. doi: 10.1007/s11239-020-02228-y
Tang, T., Bidon, M., Jaimes, J. A., Whittaker, G. R., and Daniel, S. (2020). Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 178:104792. doi: 10.1016/j.antiviral.2020.104792
Tay, M. Z., Poh, C. M., Renia, L., MacAry, P. A., and Ng, L. F. P. (2020). The trinity of COVID19: immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374. doi: 10.1038/s41577-020-0311-8
Tereshchenko, L. G., Johnson, K., Khayyat-Kholghi, M., and Johnson, B. (2022). Rate of angiotensin-converting enzyme inhibitors and angiotensin receptor blockers use and the number of COVID-19–confirmed cases and deaths. Am. J. Cardiol. 165, 101–108. doi: 10.1016/j.amjcard.2021.10.050
Thomas, M. C., Pickering, R. J., Tsorotes, D., Koitka, A., Sheehy, K., Bernardi, S., et al. (2010). Genetic Ace2 deficiency accentuates vascular inflammation and atherosclerosis in the ApoE knockout mouse. Circ. Res. 107, 888–897. doi: 10.1161/CIRCRESAHA.110.219279
Tigerstedt, R., and Bergman, P. G. (1898). Niere und kreislauf. Skand. Arch. Physiol. 8, 223–271. doi: 10.1111/j.1748-1716.1898.tb00272.x
Tignarelli, C. J., Ingraham, N. E., Sparks, M. A., Reilkoff, R., Bezdicek, T., Benson, B., et al. (2020). Antihypertensive drugs and risk of COVID-19? Lancet Respir. Med. 8, e30–e31. doi: 10.1016/S2213-2600(20)30153-3
Tikellis, C., Pickering, R., Tsorotes, D., Du, X. J., Kiriazis, H., Nguyen-Huu, T. P., et al. (2012). Interaction of diabetes and ACE2 in the pathogenesis of cardiovascular disease in experimental diabetes. Clin. Sci. (Lond.) 123, 519–529. doi: 10.1042/CS20110668
Tikellis, C., and Thomas, M. C. (2012). Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int. J. Pept. Actions 2012:256294. doi: 10.1155/2012/256294
Tipnis, S. R., Hooper, N. M., and Hyde, R. (2000). A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 275, 33238–33243. doi: 10.1074/jbc.M002615200
Tong, M., Jiang, Y., Xia, D., Xiong, Y., Zheng, Q., Chen, F., et al. (2020). Elevated serum endothelial cell adhesion molecules expression in COVID-19 patients. J. Infect. Dis. 222, 894–898. doi: 10.1093/infdis/jiaa349
Towler, P., Staker, B., Prasad, S. G., Menon, S., Tang, J., Parsons, T., et al. (2004). ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 279, 17996–18007. doi: 10.1074/jbc.M311191200
Tukiainen, T., Villani, A. C., Yen, A., Rivas, M. A., Marshall, J. L., Satija, R., et al. (2017). Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248. doi: 10.1038/nature24265
Turner, A. J., and Hooper, N. M. (2002). The angiotensin-converting enzyme gene family: genomics and pharmacology. Trends Pharmacol. Sci. 23, 177–183. doi: 10.1016/s0165-6147(00)01994-5
Ueda, S., Elliott, H. L., Morton, J. J., and Connell, J. M. (1995). Enhanced pressor response to angiotensin I in normotensive men with the deletion genotype (DD) for angiotensin-converting enzyme. Hypertension 25, 1266–1269. doi: 10.1161/01.hyp.25.6.1266
Uijl, E., Mirabito Colafella, K. M., Sun, Y., Ren, L., van Veghel, R., Garrelds, I. M., et al. (2019). Strong and sustained antihypertensive effect of small interfering RNA targeting liver angiotensinogen. Hypertension 73, 1249–1257. doi: 10.1161/HYPERTENSIONAHA.119.12703
Vaduganathan, M., Varden, O., Michel, T., McMurray, J. J. V., Pfeffer, M. A., and Solomon, S. D. (2020). Renin–angiotensin–aldosterone system inhibitors in patients with Covid-19. N. Engl. J. Med. 382, 1653–1659. doi: 10.1056/NEJMsr2005760
Vandestienne, M., Zhang, Y., Santos-Zas, I., Al-Rifai, R., Joffre, J., Giraud, A., et al. (2021). TREM-1 orchestrates angiotensin II-induced monocyte trafficking and promotes experimental abdominal aortic aneurysm. J. Clin. Invest. 131:e142468. doi: 10.1172/JCI142468
Varanat, M., Haase, E. M., Kay, J. G., and Scannapieco, F. A. (2017). Activation of the TREM-1 pathway in human monocytes by periodontal pathogens and oral commensal bacteria. Mol Oral Microbiol 32, 257–287. doi: 10.1111/omi.12169
Vassiliou, A. G., Keskinidou, C., Jahaj, E., Gallos, P., Dimopoulou, I., Kotanidou, A., et al. (2021). ICU admission levels of endothelial biomarkers as predictors of mortality in critically ill COVID-19 patients. Cells 10:186. doi: 10.3390/cells10010186
Velkoska, E., Patel, S. K., and Burrell, L. M. (2016). Angiotensin converting enzyme 2 and diminazene: role in cardiovascular and blood pressure regulation. Curr. Opin. Nephrol. Hypertens. 25, 384–395. doi: 10.1097/MNH.0000000000000254
Velloso, L. A., Folli, F., Sun, X. J., White, M. F., Saad, M. J. A., and Kahn, C. R. (1996). Cross-talk between the insulin and angiotensin signaling systems. Proc. Natl. Acad. Sci. U. S. A. 93, 12490–12495. doi: 10.1073/pnas.93.22.12490
Verdecchia, P., Angeli, F., Mazzotta, G., Gentile, G., and Reboldi, G. (2008). The renin angiotensin system in the development of cardiovascular disease: role of aliskiren in risk reduction. Vasc. Health Risk Manag. 4, 971–981. doi: 10.2147/vhrm.s3215
Verdecchia, P., Cavallini, C., Spanevello, A., and Angeli, F. (2020). The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 76, 14–20. doi: 10.1016/j.ejim.2020.04.037
Verma, S., Abbas, M., Verma, S., Khan, F. H., Raza, S. T., Siddigi, Z., et al. (2021). Impact of I/D polymorphism of angiotensin-converting enzyme 1 (ACE1) gene on the severity of COVID-19 patients. Infect. Genet. Evol. 91:104801. doi: 10.1016/j.meegid.2021.104801
Verma, A., Xu, K., Du, T., Zhu, P., Liang, Z., Liao, S., et al. (2019). Expression of human ACE2 in lactobacillus and beneficial effects in diabetic retinopathy in mice. Mol. Ther. Methods Clin. Dev. 14, 161–170. doi: 10.1016/j.omtm.2019.06.007
Vicenzi, M., Di Cosola, R., Ruscica, M., Ratti, A., Rota, I., Rota, F., et al. (2020). The liaison between respiratory failure and high blood pressure: evidence from COVID-19 patients. Eur. Respir. J. 56:2001157. doi: 10.1183/13993003.01157-2020
Vickers, C., Hales, P., Kaushik, V., Dick, L., Gavin, J., Tang, J., et al. (2002). Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 277, 14838–14843. doi: 10.1074/jbc.M200581200
Vitte, J., Diallo, A. B., Boumaza, A., Lopez, A., Michel, M., Allardet-Servent, J., et al. (2020). A granulocytic signature identifies COVID-19 and its severity. J. Infect. Dis. 222, 1985–1996. doi: 10.1093/infdis/jiaa591
Vuille-Dit-Bille, R. N., Camargo, S. M., Emmenegger, L., Sasse, T., Kummer, E., Jando, J., et al. (2015). Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids 47, 693–705. doi: 10.1007/s00726-014-1889-6
Wacharapluesadee, S., Tan, C. W., Maneeorn, P., Duengkae, P., Zhu, F., Joyjinda, Y., et al. (2021). Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 12:972. doi: 10.1038/s41467-021-21240-1
Wakahara, S., Konoshita, T., Mizuno, S., Motomura, M., Aoyama, C., Makino, Y., et al. (2007). Synergistic expression of angiotensin-converting enzyme (ACE) and ACE2 in human renal tissue and confounding effects of hypertension on the ACE to ACE2 ratio. Endocrinology 148, 2453–2457. doi: 10.1210/en.2006-1287
Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., and Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cells 181, 281–292. doi: 10.1016/j.cell.2020.02.058
Wang, K., Chen, W., Zhang, Z., Deng, Y., Lian, J. Q., Peng, D., et al. (2020). CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 5:283. doi: 10.1038/s41392-020-00426-x
Wang, H., Ji, Y., Wu, G., Sun, K., Sun, Y., Li, W., et al. (2015). L-tryptophan activates mammalian target of rapamycin and enhances expression of tight junction proteins in intestinal porcine epithelial cells. J. Nutr. 145, 1156–1162. doi: 10.3945/jn.114.209817
Wang, C., Jin, R., Zhu, X., Yan, J., and Li, G. (2015). Function of CD147 in atherosclerosis and atherothrombosis. J. Cardiovasc. Transl. Res. 8, 59–66. doi: 10.1007/s12265-015-9608-6
Wang, F., Lu, X., Peng, K., Zhou, L., Li, C., Wang, W., et al. (2014). COX-2 mediates angiotensin II-induced (pro)renin receptor expression in the rat renal medulla. Am. J. Physiol. Ren. Physiol. 307, F25–F32. doi: 10.1152/ajprenal.00548.2013
Wang, W., Luo, L., Lu, H., Chen, S., Kang, L., and Cui, F. (2015). Angiotensin-converting enzymes modulate aphid–plant interactions. Sci. Rep. 5:8885. doi: 10.1038/srep08885
Wang, C., Wang, S., Li, D., Wei, D. Q., Zhao, J., and Wang, J. (2020). Human intestinal defensin 5 inhibits SARS-CoV-2 invasion by cloaking ACE2. Gastroenterology 159, 1145–1147.e4. doi: 10.1053/j.gastro.2020.05.015
Wang, Q., Zhang, Y., Wu, L., Niu, S., Song, C., Zhang, Z., et al. (2020). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cells 181, 894–904.e9. doi: 10.1016/j.cell.2020.03.045
Wang, M. Y., Zhao, R., Gao, L. J., Gao, X. F., Wang, D. P., and Cao, J. M. (2020). SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front. Cell. Infect. Microbiol. 10:587269. doi: 10.3389/fcimb.2020.587269
Watanabe, T., Barker, T. A., and Berk, B. C. (2005). Angiotensin II and the endothelium: diverse signals and effects. Hypertension 45, 163–169. doi: 10.1161/01.HYP.0000153321.13792.b9
Wei, C., Shan, K. J., Wang, W., Zhang, S., Huan, Q., and Qian, W. (2021). Evidence for a mouse origin of the SARS-CoV-2 omicron variant. J. Genet. Genom. 48, 1111–1121. doi: 10.1016/j.jgg.2021.12.003
Wenzel, J., Lampe, J., Müller-Fielitz, H., Schuster, R., Zille, M., Müller, K., et al. (2021). The SARS-CoV-2 main protease Mpro causes microvascular brain pathology by cleaving NEMO in brain endothelial cells. Nat. Neurosci. 24, 1522–1533. doi: 10.1038/s41593-021-00926-1
Wicik, Z., Eyileten, C., Jakubik, D., Simoes, S. N., Martins, D. C. Jr., Pavao, R., et al. (2020). ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors. J. Clin. Med. 9:3743. doi: 10.3390/jcm9113743
Wright, J. M. (2000). Choosing a first-line drug in the management of elevated blood pressure: what is the evidence? 3: angiotensin-converting-enzyme inhibitors. CMAJ 163, 293–296.
Wu, Y.-H., Li, J.-Y., Wang, C., Zhang, L.-M., and Qiao, H. (2017). The ACE2 G8790A polymorphism: involvement in type 2 diabetes mellitus combined with cerebral stroke. J. Clin. Lab. Anal. 31:e22033. doi: 10.1002/jcla.22033
Wysocki, J., Garcia-Halpin, L., Ye, M., Maier, C., Sowers, K., Burns, K. D., et al. (2013). Regulation of urinary ACE2 in diabetic mice. Am. J. Physiol. Physiol. 305, F600–F611. doi: 10.1152/ajprenal.00600.2012
Xiao, M., Zhang, Y., Zhang, S., Qin, X., Xia, P., Cao, W., et al. (2020). Anti-phospholipid antibodies in critically ill patients with coronavirus disease 2019 (COVID-19). Arthritis Rheum. 72, 1998–2004. doi: 10.1002/art.41425
Xiao, F., Zimpelmann, J., Agaybi, S., Gurley, S. B., Puente, L., and Burns, K. D. (2014). Characterization of angiotensin-converting enzyme 2 ectodomain shedding from mouse proximal tubular cells. PLoS One 9:e85958. doi: 10.1371/journal.pone.0085958
Xie, X., Chen, J., Wang, X., Zhang, F., and Liu, Y. (2006). Age-and genderrelated difference of ACE2 expression in rat lung. Life Sci. 78, 2166–2171. doi: 10.1016/j.lfs.2005.09.038
Xie, Y., Xu, E., Bowe, B., and Al-Aly, Z. (2022). Long-term cardiovascular outcomes of COVID-19. Nat. Med. 28, 583–590. doi: 10.1038/s41591-022-01689-3
Xu, Q., Jensen, D. D., Peng, H., and Feng, Y. (2016). The critical role of the central nervous system (pro)renin receptor in regulating systemic blood pressure. Pharmacol. Ther. 164, 126–134. doi: 10.1016/j.pharmthera.2016.04.006
Xu, Y., and Li, Y. (2021). MicroRNA-28-3p inhibits angiotensin-converting enzyme 2 ectodomain shedding in 293T cells treated with the spike protein of severe acute respiratory syndrome coronavirus 2 by targeting a disintegrin and metalloproteinase 17. Int. J. Mol. Med. 48:189. doi: 10.3892/ijmm.2021.5022
Xu, C., Wang, A., Marin, M., Honnen, W., Ramasamy, S., Porter, E., et al. (2021). Human Defensins inhibit SARS-CoV-2 infection by blocking viral entry. Viruses 13:1246. doi: 10.3390/v13071246
Xu, H., Zhong, L., Deng, J., Peng, J., Dan, H., Zeng, X., et al. (2020). High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 12:8. doi: 10.1038/s41368-020-0074-x
Xue, B., Zhang, Z., Roncari, C. F., Guo, F., and Johnson, A. K. (2012). Aldosterone acting through the central nervous system sensitizes angiotensin II-induced hypertension. Hypertension 60, 1023–1030. doi: 10.1161/HYPERTENSIONAHA.112.196576
Yamamoto, N., Nishida, N., Yamamoto, R., Gojobori, T., Shimotohno, K., Mizokami, M., et al. (2021). Angiotensin–converting enzyme (ACE) 1 gene polymorphism and phenotypic expression of COVID-19 symptoms. Gene 12:1572. doi: 10.3390/genes12101572
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., Zhou, Q., et al. (2020). Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448. doi: 10.1126/science.abb2762
Yang, J. M., Dong, M., Meng, X., Zhao, Y. X., Yang, X. Y., Liu, X. L., et al. (2013). Angiotensin-(1-7) dose-dependently inhibits atherosclerotic lesion formation and enhances plaque stability by targeting vascular cells. Arteriosc. Thromb. Vascular Biol. 33, 1978–1985. doi: 10.1161/ATVBAHA.113.301320
Yang, P., Gu, H., Zhao, Z., Wang, W., Cao, B., Lai, C., et al. (2014). Angiotensin-converting enzyme 2 (ACE2) mediates influenza H7N9 virus-induced acute lung injury. Sci. Rep. 4:7027. doi: 10.1038/srep07027
Yang, M., Zhao, J., Xing, L., and Shi, L. (2015). The association between angiotensinconverting enzyme 2 polymorphisms and essential hypertension risk: a meta-analysis involving 14, 122 patients. J. Renin-Angiotensin-Aldosterone Syst. 16, 1240–1244. doi: 10.1177/1470320314549221
Yen, H. L., Sit, T. H. C., Brackman, C. J., Chuk, S. S. Y., Gu, H., Tam, K. W. S., et al. (2022). Transmission of SARS-CoV-2 delta variant (AY.127) from pet hamsters to humans leading to onward human-to-human transmission: a case study. Lancet 399, 1070–1078. doi: 10.1016/S0140-6736(22)00326-9
Yi, L., Gu, Y. H., Wang, X. L., An, L. Z., Xie, X. D., Shao, W., et al. (2006). Association of ACE, ACE2 and UTS2 polymorphisms with essential hypertension in Han and Dongxiang populations from North-Western China. J. Int. Med. Res. 34, 272–283. doi: 10.1177/147323000603400306
Yim, H. E., and Yoo, K. H. (2008). Renin-angiotensin system-considerations for hypertension and kidney. Electrol. Blood Press. 6, 42–50. doi: 10.5049/EBP.2008.6.1.42
Yu, H. Q., Sun, B. Q., Fanf, Z. F., Zhao, J. C., Liu, X. Y., Li, Y. M., et al. (2020). Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur. Respir. J. 56:2001526. doi: 10.1183/13993003.01526-2020
Yu, C., Tang, W., Wang, Y., Shen, Q., Wang, B., Cai, C., et al. (2016). Downregulation of ACE2/Ang-(1-7)/Mas axis promotes breast cancer metastasis by enhancing store-operated calcium entry. Cancer Lett. 376, 268–277. doi: 10.1016/j.canlet.2016.04.006
Zhang, H., and Baker, A. (2017). Recombinant human ACE2: acing out angiotensin II in ARDS therapy. Crit. Care 21:305. doi: 10.1186/s13054-017-1882-z
Zhang, Q., Bastard, P., Liu, Z., Le Pen, J., Moncada-Velez, M., Chen, J., et al. (2020). Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370:eabd4570. doi: 10.1126/science.abd4570
Zhang, J., Dong, J., Martin, M., He, M., Gongol, B., Marin, T. L., et al. (2018). AMP-activated protein kinase phosphorylation of ACE2 in endothelium mitigates pulmonary hypertension. Am. J. Respir. Crit. Care Med. 198, 509–520. doi: 10.1164/rccm.201712-2570OC
Zhang, Q., Gefter, J., Sneddon, W. B., Mamonava, T., and Friedman, P. A. (2021). ACE2 interaction with cytoplasmic PDZ protein enhances SARS-CoV-2 invasion. iScience 24:102770. doi: 10.1016/j.isci.2021.102770
Zhang, S., Liu, Y., Wang, X., Yang, L., Li, H., Wang, Y., et al. (2020). SARS-CoV-2 binds platelet ACE2 to enhance thrombosis in COVID-19. J. Hematol. Oncol. 13:120. doi: 10.1186/s13045-020-00954-7
Zhang, X. J., Qin, J. J., Cheng, X., Shen, L., Zhao, Y. C., Yuan, Y., et al. (2020). In-hospital use of statins is associated with a reduced risk of mortality among individuals with COVID-19. Cell Metab. 32, 176–187. doi: 10.1016/j.cmet.2020.06.015.e4
Zhang, C., Shen, L., Le, K. J., Pan, M. M., Kong, L. C., Gu, Z. C., et al. (2020). Incidence of venous thromboembolism in hospitalized coronavirus disease 2019 patients: a systematic review and meta-analysis. Front. Cardiovasc. Med. 7:151. doi: 10.3389/fcvm.2020.00151
Zhang, H., Wada, J., Hida, K., Tsuchiyama, Y., Hiragushi, K., Shikata, K., et al. (2001). Collectrin, a collecting duct-specific transmembrane glycoprotein, is a novel homolog of ACE2 and is developmentally regulated in embryonic kidneys. J. Biol. Chem. 276, 17132–17139. doi: 10.1074/jbc.M006723200
Zhang, Z. Z., Wang, W., Jin, H. Y., Chen, X., Cheng, Y. W., Xu, Y. L., et al. (2017). Apelin is a negative regulator of angiotensin II-mediated adverse myocardial remodeling and dysfunction. Hypertension 70, 1165–1175. doi: 10.1161/HYPERTENSIONAHA.117.10156
Zhang, Y., Xiao, M., Zhang, S., Xia, P., Cao, W., Jiang, W., et al. (2020). Coagulopathy and antiphospholipid antibodies in patients with Covid-19. N. Engl. J. Med. 382:e38. doi: 10.1056/NEJMc2007575
Zhang, L., Zhang, Y., Qin, X., Jiang, X., Zhang, J., Mao, L., et al. (2022). Recombinant ACE2 protein protects against acute lung injury induced by SARS-CoV-2 spike RBD protein. Crit. Care 26:171. doi: 10.1186/s13054-022-04034-9
Zhang, P., Zhu, L., Cai, J., Lei, F., Qin, J. J., Xie, J., et al. (2020). Association of inpatient use of angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers with mortality among patients with hypertension hospitalized with COVID-19. Circ. Res. 126, 1671–1681. doi: 10.1161/CIRCRESAHA.120.317242
Zhao, J., Yuan, Q., Wang, H., Liu, W., Liao, X., Su, Y., et al. (2020). Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 71, 2027–2034. doi: 10.1093/cid/ciaa344
Zhao, Y., Zhao, Z., Wang, Y., Zhou, Y., Ma, Y., and Zuo, W. (2020). Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 202, 756–759. doi: 10.1164/rccm.202001-0179LE
Zheng, M., Gao, Y., Wang, G., Song, G., Liu, S., Sun, D., et al. (2020). Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 17, 533–535. doi: 10.1038/s41423-020-0402-2
Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. doi: 10.1038/s41586-020-2012-7
Zhou, F., Yu, T., Du, R., Fan, G., Liu, Y., Liu, Z., et al. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395, 1054–1062. doi: 10.1016/S0140-6736(20)30566-3
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020). A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733. doi: 10.1056/NEJMoa2001017
Zhuang, M. W., Cheng, Y., Zhang, J., Jiang, X. M., Wang, L., Deng, J., et al. (2020). Increasing host cellular receptor-angiotensin-converting enzyme 2 (ACE2) expression by coronavirus may facilitate 2019-nCoV (or SARS-CoV2) infection. J. Med. Virol. 92, 2693–2701. doi: 10.1002/jmv.26139
Ziegler, C. G. K., Allon, S. J., Nyquist, S. K., Mbano, I. M., Miao, V. N., Tzouanas, C. N., et al. (2020). SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cells 181, 1016–1035.e19. doi: 10.1016/j.cell.2020.04.035
Zou, X., Chen, K., Zou, J., Han, P., Hao, J., and Han, Z. (2020). Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 14, 185–192. doi: 10.1007/s11684-020-0754-0
Zoufaly, A., Poglitsch, M., Aberle, J. H., Hoepler, W., Seitz, T., Traugott, M., et al. (2020). Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 8, 115–120. doi: 10.1016/S2213-2600(20)30418-5
Keywords: ACE2, renin-angiotensin system, hypertension, coagulation, coronavirus—COVID-19, therapy
Citation: Devaux CA and Camoin-Jau L (2022) An update on angiotensin-converting enzyme 2 structure/functions, polymorphism, and duplicitous nature in the pathophysiology of coronavirus disease 2019: Implications for vascular and coagulation disease associated with severe acute respiratory syndrome coronavirus infection. Front. Microbiol. 13:1042200. doi: 10.3389/fmicb.2022.1042200
Edited by:
Jian Shang, Zhengzhou University, ChinaReviewed by:
Kirsty Pringle, The University of Newcastle, AustraliaDulce Elena Casarini, Federal University of São Paulo, Brazil
Copyright © 2022 Devaux and Camoin-Jau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian A. Devaux, Y2hyaXN0aWFuLmRldmF1eEBtZWRpdGVycmFuZWUtaW5mZWN0aW9uLmNvbQ==
 Christian A. Devaux
Christian A. Devaux Laurence Camoin-Jau
Laurence Camoin-Jau