- 1Department of Pathogenic Biology, College of Basic Medicine, Chengde Medical University, Chengde, Hebei, China
- 2Laboratory of Microbiology Detection, Chengde Center for Disease Control and Prevention, Chengde, China
- 3Yancheng Center for Disease Control and Prevention, Yancheng, China
Rodents are the primary natural reservoirs of Bartonella spp., and some of which are zoonotic causative agents. Hence, surveillance of Bartonella sp. infection in rodents is very important for the prevention of human bartonellosis caused by them. In this study, rodents were captured, and their spleen samples were collected for Bartonella sp. DNA detection and identification by amplifying the 16S rRNA, gltA, and ftsz genes using semi-nested polymerase chain reaction (PCR). The results indicated that Bartonella sp. DNA was detected in seven Rattus norvegicus individuals with a detection rate of 6.7% in Chengde City and bacterial DNA in 31 Apodemus agrarius individuals with a detection rate of 28.4% in Handan City. The DNA detection rate across the genders and ages of rodents was not found to be statistically significant. Furthermore, sequence analysis of the above-mentioned three genes demonstrated that at least eight Bartonella species were circulating in Hebei Province, of which three, including Bartonella rattimassiliensis, Bartonella grahamii, and Bartonella tribocorum, are human pathogens, thus suggesting the existence of a major public health risk. Overall, these results revealed the detection rate and genetic diversity of Bartonella species infection in rodents in Hebei Province, which could be potentially helpful for the prevention of bartonellosis caused by rodent-associated Bartonella species. This study highlights the urgent need for the surveillance of Bartonella infections in rodents and ectoparasites that affect both rodents and humans and can cause fever of unknown origin or endocarditis.
Introduction
Bartonella spp. belong to the genus Bartonella within the family Bartonellaceae and are Gram-negative facultative intracellular alphaproteobacteria (Okaro et al., 2017). Before the reclassification of Bartonella in 1993, only one species, B. bacilliformis, was recorded. After that, the number of Bartonella spp. has continued to increase rapidly over the past 30 years, with currently more than 50 validated species and more than 10 candidate species (Okaro et al., 2017; Krügel et al., 2022). In addition, some complete genome sequences representing potential novel species are likely to further expand the number of species in the genus Bartonella (Lin et al., 2008; Sato et al., 2012). Bartonella spp. can infect a wide range of different domestic and wild animals, including cats, dogs, rodents, cattle, sheep, and bats (Okaro et al., 2017). Moreover, an increasing variety of animals have been confirmed as hosts of Bartonella spp., such as the beluga whale (Maggi et al., 2008), kangaroo (Fournier et al., 2007), camel (Rasis et al., 2014), and wild carnivores (Marciano et al., 2016; López-Pérez et al., 2017). Bartonella spp. are zoonotic bacteria and can be transmitted from animals to humans indirectly by blood-sucking arthropods (Deng et al., 2012), as well as through direct contact through the scratch of an infected cat or with the infected feces of a vector (Krügel et al., 2022). Bartonella henselae and Bartonella quintana, which mainly cause cat scratch disease (CSD) and trench fever in humans, respectively, have attracted more attention due to more patients being attributed as having the infections (Okaro et al., 2017). More seriously, at least another 18 species are considered to be human pathogens or have been identified in humans with the clinical symptom of fever (Maggi et al., 2009; Chomel and Kasten, 2010; Vayssier-Taussat et al., 2016; Okaro et al., 2017; Krügel et al., 2022).
Rodents are the natural reservoir of many human pathogens, including viruses, bacteria, and protozoans (Meerburg et al., 2009). There is no doubt that rodents play an important role in the maintenance and transmission of Bartonella spp., and at least 40 Bartonella species have been identified in a great diversity of rodents (Okaro et al., 2017; do Amaral et al., 2022; Krügel et al., 2022). Among them, eight have been proven to be pathogenic to humans and are hosted by rodents. These include Bartonella doshiae (Vayssier-Taussat et al., 2016), Bartonella elizabethae (Daly et al., 1993), Bartonella grahamii (Kerkhoff et al., 1999), Bartonella rattimassiliensis (Kosoy et al., 2010), Bartonella rochalimae (Eremeeva et al., 2007), Bartonella tribocorum (Kosoy et al., 2010), Bartonella vinsonii (Roux et al., 2000), and Bartonella washoensis (Kosoy et al., 2003). In addition, B. henselae, mainly hosted by cats, has also been detected in several rodent species, although their role in the maintenance and transmission of B. henselae remains unclear (Helan et al., 2018; Divari et al., 2020; Böge et al., 2021). In China, at least 22 species, including eight human pathogens, have been reported in rodents, with infection rates ranging from 6.4% to 57.7% to date (Saisongkorh et al., 2009; Li et al., 2015; Xia et al., 2015; Qin et al., 2019; Rao et al., 2021; Krügel et al., 2022; Yao et al., 2022). Therefore, we speculate that the Chinese population could be at risk of being affected by rodent-associated Bartonella spp., although human infections have not yet been identified.
Rodent-associated Bartonella spp. have been found to be widely distributed around the world, as well as in China, and a number of previous studies have mainly focused on the field environment (Okaro et al., 2017; Krügel et al., 2022). Hebei Province has a complex and varied terrain consisting of plateaus, mountains, and plains, which are beneficial to rodent colonization and survival. Moreover, closer contact between humans and rodents in residential areas, as well as diverse human activities that continue to invade the wild habitat of rodents, can significantly increase the transmission risk of Bartonella spp. from rodents to humans. Bartonellosis, which is caused by the rodent-associated Bartonella spp., is a natural focal disease, and thus investigations of the epidemiology and genetic diversity of Bartonella spp. infection in rodents can be of great significance for the prevention of human bartonellosis. Therefore, in this study, rodent samples were collected from Hebei Province to analyze the Bartonella sp. infection in the rodent populations, especially those pathogenic to humans. The results of this study will not only benefit the prevention and control of human bartonellosis in the local population but will also be helpful for the accurate diagnosis of diseases with fever of unknown origin (FUO).
Materials and methods
Rodent sample collection and deoxyribonucleic acid extraction
During 2020, rodents were captured using live capture traps baited with peanuts in residential areas of Chengde City and field areas of Handan City in Hebei Province, and two sampling sites were established in each city (Figure 1). The species, age, and sex of all the captured rodents were identified and recorded (Chen, 1987; Wang, 2003). All the captured rodents were euthanized, and the spleen specimens were aseptically collected.
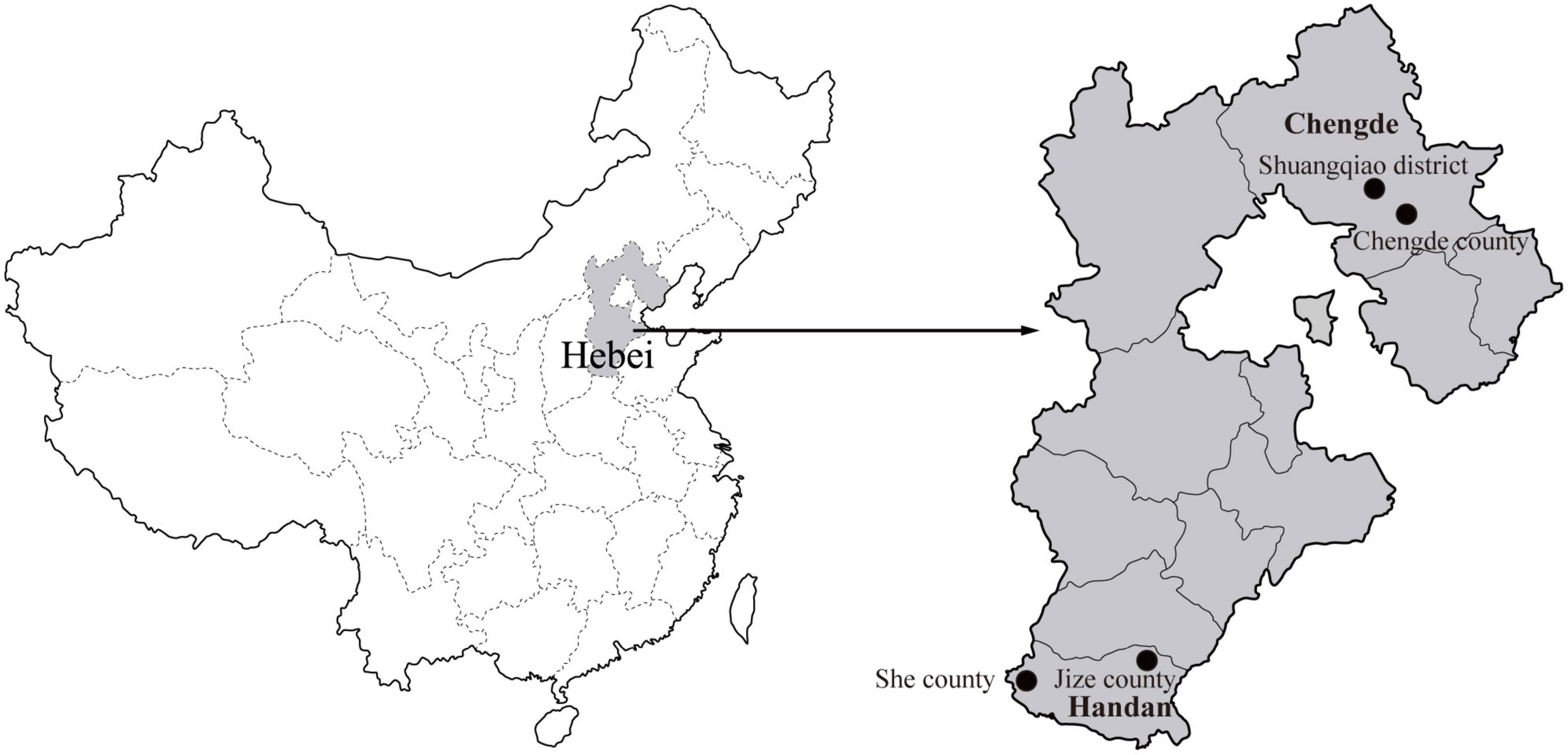
Figure 1. Geographical distribution of the rodent sampling sites (black dots) in Hebei Province, China. The map of China was obtained freely from http://english.freemap.jp/item/asia/china.html. The map of Hebei Province was generated using ArcGIS.
Total DNA was extracted from the spleen samples using a DNA extraction kit (OMEGA, Doraville, CA, USA) in a fume hood in a separate room. The extracted DNA was eluted with 50 μl of double-distilled water and stored at−20°C before analysis. In addition, the identification of the rodent species was further confirmed by sequence analysis of the mt-cyt b gene obtained by PCR using the extracted DNA as the template (Guo et al., 2013).
Detection and molecular characterization of Bartonella spp.
The presence of Bartonella spp. in rodents was detected by amplifying part of the 16S rRNA gene using a semi-nested polymerase chain reaction (PCR) and subsequently confirmed by further sequencing. The primer pairs Bar-SF1/Bar-SR and Bar-SF2/Bar-SR designed in this study were used for the first and second rounds of PCR to amplify an 813-bp 16S rRNA gene fragment.
In addition, the nearly complete 16S rRNA, the partial citrate synthase gene (gltA), and the cell division protein gene (ftsZ) were obtained from the Bartonella sp. DNA-detected samples to better determine and characterize the Bartonella species. Briefly, the primer pairs Bar-SF/Bar-SR1 and Bar-SF/Bar-SR2 were used to obtain the rest of the 16S rRNA with a length of 712 bp. The partial gltA gene (1,036 bp) was amplified with the primer pairs Bar-gltA-F/Bar-gltA-R1 and Bar-gltA-F/Bar-gltA-R2 for the primary and secondary rounds of the semi-nested PCR, respectively. The partial ftsz gene (860 bp) was amplified with the primer pairs Bar-ftsz-F1/Bar-ftsz-R and Bar-ftsz-F2/Bar-ftsz-R for the primary and secondary rounds of the semi-nested PCR, respectively. Alternatively, Bar-gltA-FM and Bar-ftsz-RM, instead of Bar-gltA-F and Bar-ftsz-R, were employed to amplify the shorter gltA (476 bp) and ftsz (575 bp) genes, respectively. All the primer sequences used in the present study are shown in Table 1.
The PCR reaction for the first round of the nested PCR was performed in a 20 μl reaction volume, containing 10 μl Premix Taq (Takara, Dalian, China), 1.5 μl extracted DNA, 0.8 μl of each primer (10 pmol), and 6.9 μl water. For the second round, the PCR mixture contained 25 μl Premix Taq (Takara, Dalian, China), 3 μl of the first-round PCR products, 2 μl of each primer (10 pmol), and 18 μl water for a final volume of 50 μl. The same thermal cycling condition was used for both rounds, including a denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 40 s, annealing at 56°C for 40 s, and elongation at 72°C for 1 min, followed by a final extension at 72°C for 7 min. In addition, to prevent contamination, the PCR mixture preparation, template addition, and agarose gel electrophoresis were performed in a fume hood in three separate rooms, and filter tips were also used in each assay. Furthermore, ddH2O was used as a negative control.
The PCR product was electrophoresed on a 1.0% agarose gel, and the amplicon of the expected size was purified using the Takara MiniBEST Agarose Gel DNA Extraction Kit Version 4.0 (Takara, Dalian, China). The purified PCR product was subjected to bidirectional sequencing with the PCR primers. Alternatively, the purified PCR product was ligated into the pGEM-T vector for sequencing when it produced complex sequence chromatograms in direct sequencing. The PCR products were sequenced using the ABI-PRISM Dye Termination Sequencing Kit and the ABI 3730 genetic analyzer. All the newly generated sequences in this study were submitted to GenBank under the accession numbers OP363479-OP363516 for the rrs gene, OP382327-OP382454 for the gltA gene, and OP382391-OP382426 for the ftsZ gene.
Sequence analysis
The nearly complete 16S rRNA gene was assembled by partially overlapping oligonucleotides using BioEdit version 7.1.11 (Hall, 1999). The nucleotide identities of the 16S rRNA, gltA, and ftsZ genes were calculated using the MegAlign program within the DNASTAR Lasergene software (Burland, 2000). Phylogenetic trees based on the 16S rRNA, gltA, and ftsZ gene sequences were reconstructed using the maximum-likelihood (ML) method within PhyML version 3.2, and the reliability of branches in the tree was evaluated by calculating the bootstrap values with 1,000 replicates (Guindon et al., 2010). The general time-reversible (GTR) substitution model with a gamma (Γ) model of rate heterogeneity and a proportion of invariable sites (GTR + Γ + I) determined by MEGA 7.0 was found to be the best fit model for the phylogenetic analysis (Kumar et al., 2016). The ML tree was rooted at its midpoint using MEGA 7.0.
Statistical analyses
The P-value with the chi-square test was calculated using SPSS 21.0 software for the statistical analyses to determine the association between the detection rate and the gender and age of the rodents. A P-value of less than 0.05 was considered to be statistically significant.
Ethics statement
This study was approved by the ethics committee of the Chengde Medical University (No. 202004). All the rodent experiments were performed in strict accordance with the Guidance for Experimental Animal Welfare and Ethical Treatment by the Ministry of Science and Technology of China. In addition, the rodents were anesthetized with ether and dissected to collect spleen samples for the detection of Bartonella sp. DNA, and analgesics were administered to minimize the suffering of the captured rodents.
Results
Sample collection and detection of Bartonella sp. deoxyribonucleic acid in rodents
A total of 223 rodents were captured from four sampling sites in Chengde and Handan cities in Hebei Province, China (Table 2). The collected rodents were identified into five distinct species comprising Rattus norvegicus, R. tanezumi, Mus musculus, Apodemus agrarius, and Cricetulus triton. In Chengde City, the rodents were sampled in residential areas, and R. norvegicus was found to be the most abundant species, while A. agrarius was the predominant species in Handan, where it was captured in the field.
Based on the amplification of the 16S rRNA gene, PCR products of the expected sizes were detected in 38 spleen samples collected from the 223 rodents. After sequencing of the PCR products, all the obtained 16S rRNA gene sequences were blasted against the GenBank nucleotide database, and the results showed that all of them shared more than 99.4% nucleotide identity with the most closely related 16S rRNA gene sequence of a Bartonella species. Hence, the DNA detection rate was found to be 17.0%. Specifically, seven of the Bartonella sp. DNA-detected samples were identified as being from R. norvegicus in Chengde with a detection rate of 6.7% and 31 from A. agrarius in Handan with a detection rate of 28.4%. Furthermore, the infection rates in female and male rodents were observed to be 16.7% (95% CI: 4.3–29.1%) and 17.3% (95% CI: 6.0–27.4%), respectively, which were not statistically significant (χ2 = 0.808, P = 0.567). Similarly, no significant difference (χ2 = 0.887, P = 0.538) was noted in the infection rates between juvenile (12.4%, 95% CI: 5.5–26.8%) and adult rodents (20.6%, 95% CI: 6.5–30.1%).
Molecular characterization of the Bartonella species
To better identify and characterize the Bartonella species in the current study, 37 nearly complete 16S rRNA gene sequences (with a length of approximately 1,400 bp), 28 partial gltA gene sequences (16 with a length of approximately 1,000 bp and 12 others with a length of approximately 440 bp), and 36 partial ftsZ gene sequences (31 with a length of approximately 830 bp and 5 others with a length of approximately 550 bp) were obtained from all the samples with Bartonella sp.-DNA detection. The sequencing and BLAST analyses based on three gene sequences indicated that all the Bartonella strains identified in this study were classified into at least eight different Bartonella species, namely, B. japonica, Bartonella sp. 1-1C (a B. rochalimae-like species), B. rattimassiliensis, B. grahamii, B. taylorii, B. tribocorum, B. mastomydis, and B. kosoyi. In addition, a 770-bp 16S rRNA gene sequence of Shexian-Aa46 from A. agrarius in Handan shared the highest nucleotide identity with B. vinsonii (99.5%), while its ftsZ gene exhibited the highest nucleotide identity with that of Bartonella sp. 1-1C (99.6%), which might have been caused by coinfection with two different Bartonella species. Due to a failure to obtain the gltA and ftsZ gene sequences resembling those of B. vinsonii from Shexian-Aa46, B. vinsonii or B. vinsonii-like bacteria could have been circulating in A. agrarius in local areas.
Specifically, five distinct Bartonella species, including B. japonica (n = 1), Bartonella sp. 1-1C (n = 2), B. rattimassiliensis (n = 2), B. grahamii (n = 1) and B. taylorii (n = 1), were detected in R. norvegicus in Chengde City. In Handan City, Bartonella sp. 1-1C, B. tribocorum, B. mastomydis, and B. kosoyi were identified from 6, 10, 1, and 12 A. agrarius, respectively. For Jize-Aa8 belonging to B. mastomydis in A. agrarius, only an approximate 1,000-bp gltA gene sequence was obtained, which shared the highest nucleotide identity of 99.4% with that of the B. mastomydis strain 008 (KY555066), while its 1,400-bp 16S rRNA belonged to B. tribocorum, which also may have been potentially caused by co-infection with B. mastomydis and B. tribocorum. Of the eight Bartonella species, only one, Bartonella sp. 1-1C, was detected in both R. norvegicus in Chengde City and A. agrarius in Handan City.
The strain Chengde-Rn16 shared the highest nucleotide identities of 99.6% and 97.9% with the B. japonica strain Fuji 18-1 for the 16S rRNA and gltA genes, respectively. The strain Chengde-Rn85 shared the highest nucleotide identities of 100%, 99.6%, and 99.6% with the B. grahamii strain as4aup for the 16S rRNA, gltA, and ftsz genes, respectively. The strain Chengde-Rn40 showed the highest nucleotide identity of 100% with the B. taylorii strain IBS296 for the 16S rRNA, 100% with the B. taylorii strain Far East I for the gltA, and 98.9% with the B. taylorii strain M1 for the ftsz genes. The two B. rattimassiliensis strains, Shuangqiao-Rn48 and Chengde-Rn25, displayed the highest nucleotide identities of 99.9% and 99.9%, respectively, with the B. rattimassiliensis strain 15908 for the 16S rRNA and ftsz genes, and 99.1% and 98.9%, respectively, with the B. rattimassiliensis strain SD-10 for the gltA gene. The seven Bartonella sp. 1-1C strains identified here shared 99.8–100%, 98.9–100%, and 97.6–100% nucleotide identities with each other and the highest nucleotide identities of 99.9–100%, 99.3–100%, and 97.0–100% with the Bartonella sp. 1-1C for the 16S rRNA, gltA, and ftsz genes, respectively. The 10 B. tribocorum strains identified here shared 99.9–100%, 99.3–100%, and 99.5–100% nucleotide identities with each other and the highest nucleotide identities of 99.9%, 99.6–99.0%, and 99.6–100% with the known B. tribocorum strains for the 16S rRNA, gltA, and ftsz genes, respectively. The 13 B. kosoyi strains identified here shared 99.6–100%, 99.5–99.8%, and 99.8–100% nucleotide identities with each other and the highest nucleotide identities of 99.7–100%, 99.6–99.8%, and 99.8–100% with the B. kosoyi strain Tel Aviv for the 16S rRNA, gltA, and ftsz genes, respectively.
Phylogenetic trees based on the 16S rRNA, gltA, and ftsz gene sequences were reconstructed, and these three trees presented similar topologies. Consistent with the genetic analysis, all sequences in the three trees were divided into eight different groups that corresponded to B. japonica, Bartonella sp. 1-1C, B. rattimassiliensis, B. grahamii, B. taylorii, B. tribocorum, B. mastomydis, and B. kosoyi (Figures 2-4). Moreover, the 16S rRNA gene sequence of Shexian-Aa46 clustered with that of B. vinsonii in the 16S rRNA tree (Figure 2). However, in the phylogenetic tree of ftsz, all sequences obtained in this study were classified into six different groups due to the absence of the ftsz gene sequence in the B. japonica strain Chengde-Rn16 and the B. mastomydis strain Handan-Aa8.
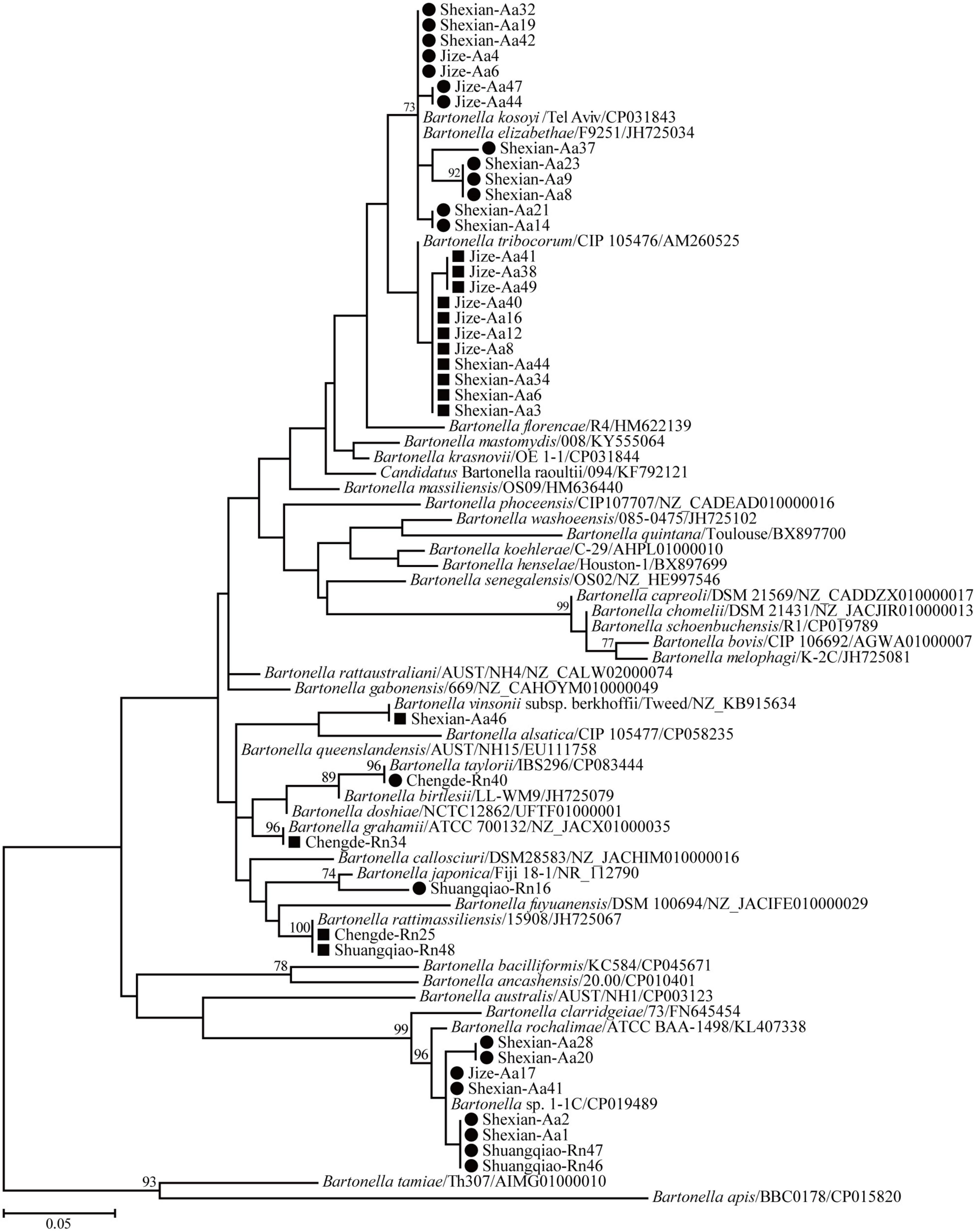
Figure 2. Maximum-likelihood (ML) tree reconstructed based on the 16S rRNA gene sequences of Bartonella species. Bootstrap values were calculated with 100 replicates and only > 70% are shown. Sequences of human-pathogenic Bartonella species determined herein are marked as black squares and others as black circles.
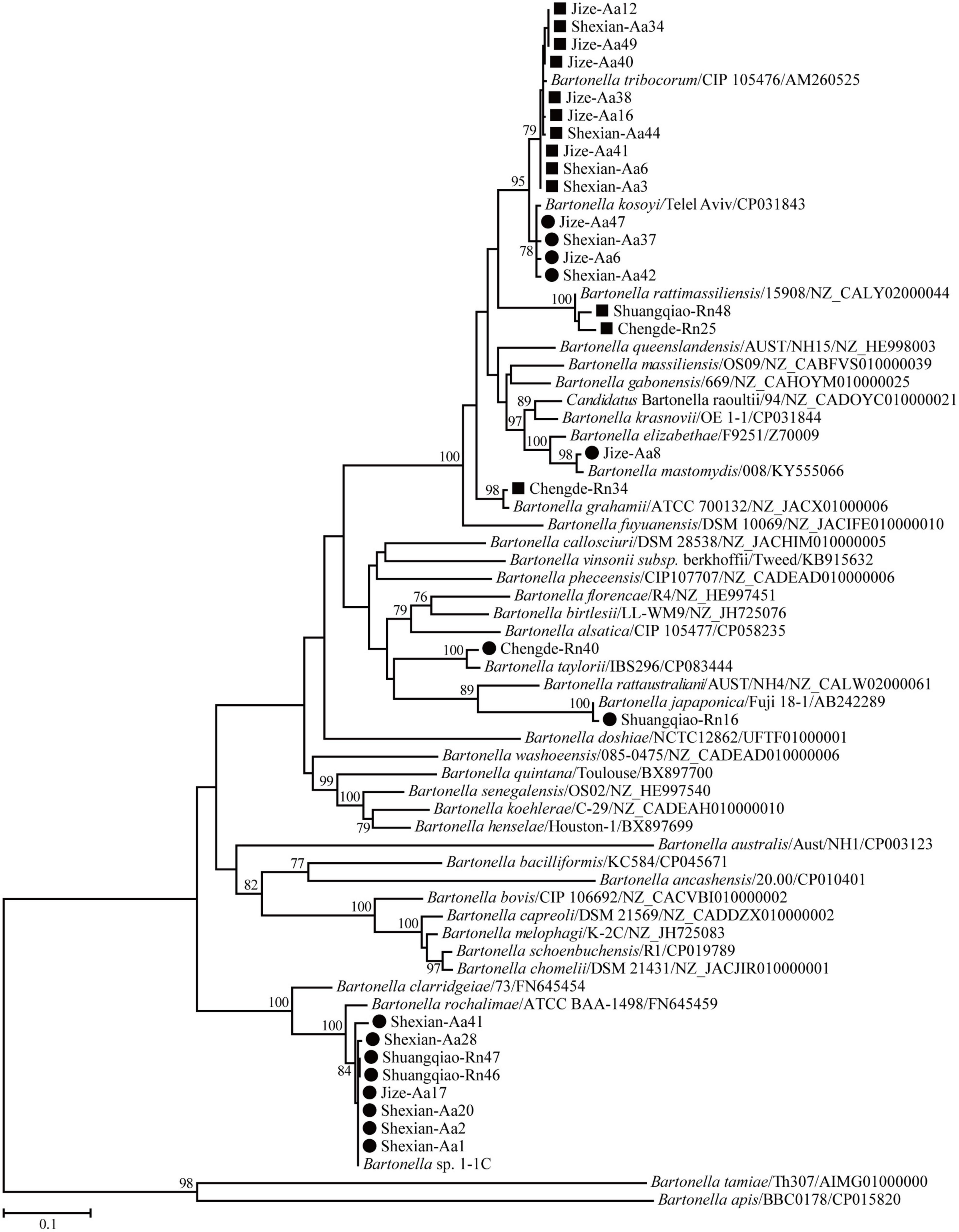
Figure 3. Maximum-likelihood (ML) tree reconstructed based on the gltA gene sequences of Bartonella species. The legend follows that of Figure 2.
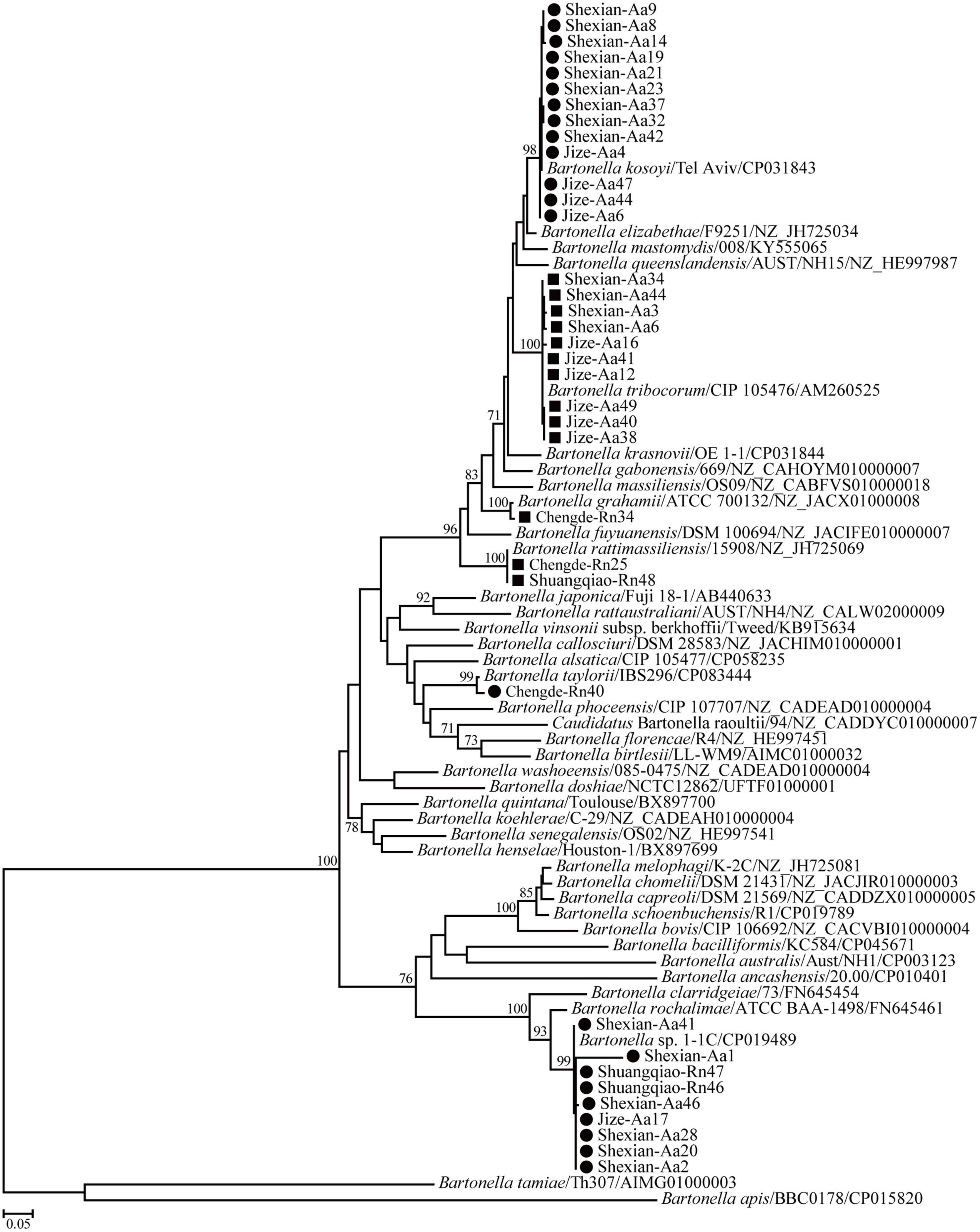
Figure 4. Maximum-likelihood (ML) tree reconstructed based on the ftsz gene sequences of Bartonella species. The legend follows that of Figure 2.
Discussion
Over the past 30 years, an increasing number of rodent-borne Bartonella species and associated patients with fever or endocarditis have been reported, suggesting more common human exposures to these agents than previously suspected (Daly et al., 1993; Kerkhoff et al., 1999; Roux et al., 2000; Kosoy et al., 2003, 2010; Eremeeva et al., 2007; Vayssier-Taussat et al., 2016; Okaro et al., 2017; Krügel et al., 2022). In addition, some species that were not previously thought to infect humans have now been confirmed to be human pathogens (Eremeeva et al., 2007; Vayssier-Taussat et al., 2016; Krügel et al., 2022). Hence, human-pathogenic Bartonella spp. are being considered emerging zoonotic causative agents, and more attention should be paid to their infection in reservoirs, vectors, and humans for the better prevention of bartonellosis.
Rodents are considered to be the primary natural hosts of Bartonella spp., and most known species have been detected in a wide variety of rodents. In China, more than half of the rodent-associated Bartonella species have been identified from rodents in residential and field areas (Saisongkorh et al., 2009; Xia et al., 2015; Rao et al., 2021; Krügel et al., 2022). R. norvegicus and A. agrarius are the predominant species found in residential areas and the wild, respectively, and they can also act as the hosts of Bartonella spp. In previous studies, at least seven validated Bartonella species, namely, B. elizabethae, B. grahamii, B. rattimassiliensis, B. rochalimae, B. tribocorum, B. mastomydis, and B. queenslandensis, were identified in R. norvegicus (Qin et al., 2019; An et al., 2020; Liu et al., 2022; Yao et al., 2022; Yu et al., 2022c), and six species, namely, B. fuyuanensis, B. grahamii, B. phoceensis, B. japonica, B. taylorii, and B. coopersplainsensis, were identified in A. agrarius (Li et al., 2015; Qin et al., 2019; Lu et al., 2022; Yu et al., 2022b). In the present study, eight official or candidate Bartonella species, in addition to B. vinsonii-like species, were identified in rodents. The findings indicated that genetically diverse Bartonella species are circulating in rodent populations in Hebei Province, China. In addition, this was the first identification of B. kosoyi in China.
Consistent with the previous study, no significant difference in the detection rate was associated with either the gender or the age of the rodents (Yao et al., 2022). In this study, it was found that the overall detection rate of Bartonella sp. infection in rodents was 17.0%, lower than that in rodents from the Qinghai-Tibetan Plateau (30.1%, Yu et al., 2022a), the Shangdang Basin (37.4%, Yu et al., 2022b), the Qaidam Basin (38.6%, Rao et al., 2021), Heixiazi Island (57.7%, Li et al., 2015), Shaanxi (26.1%, An et al., 2020), Zhongtiao Mountain (49.5%, Yu et al., 2022c), Zhejiang (31.4%, Liu et al., 2010), and South China (43.5%, Ying et al., 2002), while higher than that in rodents from eastern China (8.4%, Qin et al., 2019) and Guangzhou (6.4%, Yao et al., 2022). In addition, similar detection rates to those found in the present study were observed in rodents from Guizhou (16.1%; Lu et al., 2022) and southeastern China (14.9%; Liu et al., 2022). The discrepancy might have been due to the rodent species, habitats, and arthropod vector populations (Meheretu et al., 2013). In addition, compared to rodents collected in residential areas, a higher detection rate was observed in rodents collected in the wild (Yu et al., 2022b). Similarly, in this study, a Bartonella sp. DNA detection rate of 28.4% was observed in R. norvegicus in residential areas, while a 6.7% rate was found in A. agrarius in the field. However, R. norvegicus and A. agrarius were captured from different areas; thus, it was not clear whether the rodent species or the habitat was the factor that affected the detection rate, which is a limitation of this study.
In China, all eight rodent-related Bartonella species pathogenic to humans have been identified, mostly in the past 10 years (Li et al., 2015; Su et al., 2020; Yao et al., 2022). In this study, three human-pathogenic Bartonella species were detected, including B. rattimassiliensis and B. grahamii in Chengde and B. tribocorum in Handan. More importantly, the presence of B. tribocorum should be of concern due to the high detection rate of A. agrarius. In addition, attention should be paid to B. rattimassiliensis and B. grahamii due to the close association between their host, R. norvegicus, and humans, though they displayed a relatively low detection rate in rodents. Bartonella spp. are mainly transmitted from rodents to humans by blood-sucking arthropod vectors (Deng et al., 2012). In China, diverse Bartonella spp. have been identified in ectoparasites such as ticks, lice, and fleas (Li et al., 2007, 2013; Hao et al., 2020). Unfortunately, we failed to collect ectoparasites from the sampled rodents, which is another limitation of our study. Therefore, ectoparasites should be collected and analyzed in future studies. Furthermore, the presence of Bartonella spp. should be monitored and investigated in humans with fever of unknown origin or endocarditis in the local population.
As the natural reservoir hosts of many different types of microorganisms, rodents are considered to have a long-term co-divergence or co-speciation with the pathogens they carry, such as hantaviruses (Guo et al., 2013) and arenaviruses (Gonzalez et al., 2007). In terms of Bartonella species, Kosoy et al. also considered that they present potential co-evolution or co-speciation with their rodent hosts according to the host specificity (Kosoy et al., 2000). In addition, some previous studies have also demonstrated the host specificity of Bartonella sp. infection in rodents (Telfer et al., 2007; Abreu-Yanes et al., 2018; Qin et al., 2019; Divari et al., 2021). However, an increasing amount of evidence has suggested a lack of host specificity because the same Bartonella species can be identified from a diversity of rodent hosts, even from those belonging to different families (Billeter et al., 2014; Krügel et al., 2022). In the current study, B. japonica, B. rattimassiliensis, B. grahamii, and B. taylorii only in R. norvegicus and B. tribocorum, B. mastomydis, and B. kosoyi only in A. agrarius appear to support the view that Bartonella infection in rodents can exhibit host specificity. However, the R. norvegicus and A. agrarius in this study were sampled from different cities. In addition, the Bartonella species identified in this study were found to be carried by various rodents, as shown in previous studies. Hence, the apparent host specificity observed in this study might have been caused by the different habitats of the rodents.
Conclusion
At least eight Bartonella species, including three human causative agents, were identified in two rodent species in Hebei Province, China. In addition, Bartonella sp. DNA detection rates of 28.4% and 6.7% were observed in A. agrarius in Handan City and in R. norvegicus in Chengde City, respectively. Our results also indicated a lack of host specificity for Bartonella sp. infection in rodents. Overall, our results indicate that more attention should be paid to the surveillance of rodent-associated Bartonella species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
This animal study was reviewed and approved by Ethics Committee of the Chengde Medical University.
Author contributions
W-PG conceived and designed the experiments and contributed to writing, reviewing, and editing the manuscript. RJ, QR, JX, G-CX, G-QC, and LD performed the experiments and analyzed the data. W-PG and JW helped to collect the samples. W-PG and RJ participated in writing the original draft. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Young Talent Program of Higher School in Hebei Province (No. BJ2020024), the Natural Science Foundation of Hebei Province (No. C2022406003), the Scientific Research Foundation for High-level Talents of Chengde Medical University (No. 202001), and key discipline construction of colleges and universities in Hebei Province (Ji Jiao Gao No. [2013]4).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1039665/full#supplementary-material
References
Abreu-Yanes, E., Martin-Alonso, A., Martin-Carrillo, N., Livia, K. G., Marrero-Gagliardi, A., Valladares, B., et al. (2018). Bartonella in rodents and ectoparasites in the Canary Islands, Spain: New insights into host-vector-pathogen relationships. Microb. Ecol. 75, 264–273. doi: 10.1007/s00248-017-1022-y
An, C. H., Chen, B. B., Lyu, W., Nie, S. M., Li, S. Z., Fan, S. P., et al. (2020). Bartonella species investigated among rodents from Shaanxi province of China. Biomed. Environ. Sci. 33, 201–205. doi: 10.3967/bes2020.028
Billeter, S. A., Borchert, J. N., Atiku, L. A., Mpanga, J. T., Gage, K. L., and Kosoy, M. Y. (2014). Bartonella species in invasive rats and indigenous rodents from Uganda. Vector Borne Zoonotic Dis. 14, 182–188. doi: 10.1089/vbz.2013.1375
Böge, I., Pfeffer, M., Htwe, N. M., Maw, P. P., Sarathchandra, S. R., Sluydts, V., et al. (2021). First detection of Bartonella spp. in small mammals from rice storage and processing facilities in Myanmar and Sri Lanka. Microorganisms 9:658. doi: 10.3390/microorganisms9030658
Burland, T. G. (2000). DNASTAR’s lasergene sequence analysis software. Methods Mol. Biol. 132, 71–91.
Chen, H. X. (1987). Classification and identification of medical animals. Beijing: The Institute of Epidemiology and Microbiology, Chinese Academy of Preventive Medicine, 123–125. (in Chinese).
Chomel, B. B., and Kasten, R. W. (2010). Bartonellosis, an increasingly recognized zoonosis. J. Appl. Microbiol. 109, 743–750. doi: 10.1111/j.1365-2672.2010.04679.x
Daly, J. S., Worthington, M. G., Brenner, D. J., Moss, C. W., Hollis, D. G., Weyant, R. S., et al. (1993). Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 31, 872–881. doi: 10.1128/jcm.31.4.872-881.1993
Deng, H., Le Rhun, D., Buffet, J. P., Cotté, V., Read, A., Birtles, R. J., et al. (2012). Strategies of exploitation of mammalian reservoirs by Bartonella species. Vet. Res. 43:15. doi: 10.1186/1297-9716-43-15
Divari, S., Danelli, M., Pregel, P., Ghielmetti, G., Borel, N., and Bollo, E. (2021). Biomolecular investigation of Bartonella spp. in wild rodents of two swiss regions. Pathogens 10:1331. doi: 10.3390/pathogens10101331
Divari, S., Pregel, P., Zanet, S., Ferroglio, E., Giannini, F., Scaglione, F. E., et al. (2020). Molecular evidence of Bartonella spp. in rodents: A study in Pianosa Island, Italy. Animals (Basel) 10:2070. doi: 10.3390/ani10112070
do Amaral, R. B., Cardozo, M. V., Varani, A. M., Gonçalves, L. R., Furquim, M. E. C., Dias, C. M., et al. (2022). Bartonella machadoae sp. nov. isolated from wild rodents in the Pantanal wetland. Acta Trop. 229:106368. doi: 10.1016/j.actatropica.2022.106368
Eremeeva, M. E., Gerns, H. L., Lydy, S. L., Goo, J. S., Ryan, E. T., Mathew, S. S., et al. (2007). Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N. Engl. J. Med. 356, 2381–2387. doi: 10.1056/NEJMoa065987
Fournier, P. E., Taylor, C., Rolain, J. M., Barrassi, L., Smith, G., and Raoult, D. (2007). Bartonella australis sp. nov. from kangaroos, Australia. Emerg. Infect. Dis. 13, 1961–1962. doi: 10.3201/eid1312.060559
Gonzalez, J. P., Emonet, S., de Lamballerie, X., and Charrel, R. (2007). Arenaviruses. Curr. Top. Microbiol. Immunol. 315, 253–288. doi: 10.1007/978-3-540-70962-6_11
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guo, W. P., Lin, X. D., Wang, W., Tian, J. H., Cong, M. L., Zhang, H. L., et al. (2013). Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 9:e1003159. doi: 10.1371/journal.ppat.1003159
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis. Nuclc. Acids Symp. 41, 95–98.
Hao, L., Yuan, D., Guo, L., Hou, W., Mo, X., Yin, J., et al. (2020). Molecular detection of Bartonella in ixodid ticks collected from yaks and plateau pikas (Ochotona curzoniae) in Shiqu County, China. BMC Vet. Res. 16:235. doi: 10.1186/s12917-020-02452-x
Helan, J. V. G., Grinberg, A., Gedye, K., Potter, M. A., and Harrus, S. (2018). Molecular detection of Bartonella coopersplainsensis and B. henselae in rats from New Zealand. N. Z. Vet. J. 66, 257–260. doi: 10.1080/00480169.2018.1483781
Kerkhoff, F. T., Bergmans, A. M., van Der Zee, A., and Rothova, A. (1999). Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J. Clin. Microbiol. 37, 4034–4038. doi: 10.1128/JCM.37.12.4034-4038.1999
Kosoy, M., Bai, Y., Sheff, K., Morway, C., Baggett, H., Maloney, S. A., et al. (2010). Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am. J. Trop. Med. Hyg. 82, 1140–1145. doi: 10.4269/ajtmh.2010.09-0778
Kosoy, M., Murray, M., Gilmore, R. D. Jr., Bai, Y., and Gage, K. L. (2003). Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J. Clin. Microbiol. 41, 645–650. doi: 10.1128/JCM.41.2.645-650.2003
Kosoy, M. Y., Saito, E. K., Green, D., Marston, E. L., Jones, D. C., and Childs, J. E. (2000). Experimental evidence of host specificity of Bartonella infection in rodents. Comp. Immunol. Microbiol. Infect. Dis. 23, 221–238. doi: 10.1016/s0147-9571(99)00075-2
Krügel, M., Król, N., Kempf, V. A. J., Pfeffer, M., and Obiegala, A. (2022). Emerging rodent-associated Bartonella: A threat for human health? Parasit. Vectors 15:113. doi: 10.1186/s13071-022-05162-5
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, D. M., Hou, Y., Song, X. P., Fu, Y. Q., Li, G. C., Li, M., et al. (2015). High prevalence and genetic heterogeneity of rodent-borne Bartonella species on Heixiazi Island, China. Appl. Environ. Microbiol. 81, 7981–7992. doi: 10.1128/AEM.02041-15
Li, D. M., Liu, Q. Y., Yu, D. Z., Zhang, J. Z., Gong, Z. D., and Song, X. P. (2007). Phylogenetic analysis of Bartonella detected in rodent fleas in Yunnan, China. J. Wildl. Dis. 43, 609–617. doi: 10.7589/0090-3558-43.4.609
Li, H., Liu, W., Zhang, G. Z., Sun, Z. Z., Bai, J. Y., Jiang, B. G., et al. (2013). Transmission and maintenance cycle of Bartonella quintana among rhesus macaques, China. Emerg. Infect. Dis. 19, 297–300. doi: 10.3201/eid1902.120816
Lin, J. W., Chen, C. Y., Chen, W. C., Chomel, B. B., and Chang, C. C. (2008). Isolation of Bartonella species from rodents in Taiwan including a strain closely related to ‘Bartonella rochalimae’ from Rattus norvegicus. J. Med. Microbiol. 57, 1496–1501. doi: 10.1099/jmm.0.2008/004671-0
Liu, H., Han, T., Liu, W., Xu, G., Zheng, K., and Xiao, F. (2022). Epidemiological characteristics and genetic diversity of Bartonella species in rodents from southeastern China. Zoonoses Public Health 69, 224–234. doi: 10.1111/zph.12912
Liu, Q., Sun, J., Lu, L., Fu, G., Ding, G., Song, X., et al. (2010). Detection of Bartonella species in small mammals from Zhejiang province. China J. Wildl. Dis. 46, 179–185. doi: 10.7589/0090-3558-46.1.179
López-Pérez, A. M., Osikowicz, L., Bai, Y., Montenieri, J., Rubio, A., Moreno, K., et al. (2017). Prevalence and phylogenetic analysis of Bartonella species of wild carnivores and their fleas in northwestern Mexico. Ecohealth 14, 116–129. doi: 10.1007/s10393-017-1216-2
Lu, M., Tang, G., Ren, Z., Zhang, J., Wang, W., Qin, X., et al. (2022). Ehrlichia, Coxiella and Bartonella infections in rodents from Guizhou province, Southwest China. Ticks Tick Borne Dis. 13:101974. doi: 10.1016/j.ttbdis.2022.101974
Maggi, R. G., Kosoy, M., Mintzer, M., and Breitschwerdt, E. B. (2009). Isolation of Candidatus Bartonella melophagi from human blood. Emerg. Infect. Dis. 15, 66–68. doi: 10.3201/eid1501.081080
Maggi, R. G., Raverty, S. A., Lester, S. J., Huff, D. G., Haulena, M., Ford, S. L., et al. (2008). Bartonella henselae in captive and hunter-harvested beluga (Delphinapterus leucas). J. Wildl. Dis. 44, 871–877. doi: 10.7589/0090-3558-44.4.871
Marciano, O., Gutiérrez, R., Morick, D., King, R., Nachum-Biala, Y., Baneth, G., et al. (2016). Detection of Bartonella spp. in wild carnivores, hyraxes, hedgehog and rodents from Israel. Parasitology 143, 1232–1242. doi: 10.1017/S0031182016000603
Meerburg, B. G., Singleton, G. R., and Kijlstra, A. (2009). Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 35, 221–270. doi: 10.1080/10408410902989837
Meheretu, Y., Leirs, H., Welegerima, K., Breno, M., Tomas, Z., Kidane, D., et al. (2013). Bartonella prevalence and genetic diversity in small mammals from Ethiopia. Vector Borne Zoonotic Dis. 13, 164–175. doi: 10.1089/vbz.2012.1004
Okaro, U., Addisu, A., Casanas, B., and Anderson, B. (2017). Bartonella species, an emerging cause of blood-culture-negative endocarditis. Clin. Microbiol. Rev. 30, 709–746. doi: 10.1128/CMR.00013-17
Qin, X. R., Liu, J. W., Yu, H., and Yu, X. J. (2019). Bartonella species detected in rodents from eastern China. Vector Borne Zoonotic Dis. 19, 810–814. doi: 10.1089/vbz.2018.2410
Rao, H., Li, S., Lu, L., Wang, R., Song, X., Sun, K., et al. (2021). Genetic diversity of Bartonella species in small mammals in the Qaidam Basin, western China. Sci. Rep. 11:1735. doi: 10.1038/s41598-021-81508-w
Rasis, M., Rudoler, N., Schwartz, D., and Giladi, M. (2014). Bartonella dromedarii sp. nov. isolated from domesticated camels (Camelus dromedarius) in Israel. Vector Borne Zoonotic Dis. 14, 775–782. doi: 10.1089/vbz.2014.1663
Roux, V., Eykyn, S. J., Wyllie, S., and Raoult, D. (2000). Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J. Clin. Microbiol. 38, 1698–1700. doi: 10.1128/JCM.38.4.1698-1700.2000
Saisongkorh, W., Rolain, J. M., Suputtamongkol, Y., and Raoult, D. (2009). Emerging Bartonella in humans and animals in Asia and Australia. J. Med. Assoc. Thai. 92, 707–731.
Sato, S., Kabeya, H., Yamazaki, M., Takeno, S., Suzuki, K., Kobayashi, S., et al. (2012). Prevalence and genetic diversity of Bartonella species in sika deer (Cervus nippon) in Japan. Comp. Immunol. Microbiol. Infect. Dis. 35, 575–581. doi: 10.1016/j.cimid.2012.07.001
Su, Q., Chen, Y., Wang, B., Huang, C., Han, S., Yuan, G., et al. (2020). Epidemiology and genetic diversity of zoonotic pathogens in urban rats (Rattus spp.) from a subtropical city, Guangzhou, southern China. Zoonoses Public Health 67, 534–545. doi: 10.1111/zph.12717
Telfer, S., Clough, H. E., Birtles, L. R., Bennett, M., Carslake, D., Helyar, S., et al. (2007). Ecological differences and coexistence in a guild of microparasites: Bartonella in wild rodents. Ecology 88, 1841–1849. doi: 10.1890/06-1004.1
Vayssier-Taussat, M., Moutailler, S., Féménia, F., Raymond, P., Croce, O., La Scola, B., et al. (2016). Identification of novel zoonotic activity of Bartonella spp., France. Emerg. Infect. Dis. 22, 457–462. doi: 10.3201/eid2203.150269
Wang, Y. X. (2003). A complete checklist of mammal species and subspecies in China - a taxonomic and geographic reference. Beijing: China Forestry Publishing House, 27–59. (in Chinese).
Xia, X., Liu, S., Zhang, L., Dai, M., Zhang, J., and Zhu, H. (2015). Tick infestation and tick-borne pathogen infection in human and ticks in a military camp and nearby area in Xiaogan, Hubei. Int. J. Med. Parasit. Dis. 42, 337–340. (in Chinese), doi: 10.3760/cma.j.issn.1673-4122.2015.06.007
Yao, X. Y., Liu, H., Sun, J., Zhang, Y. Q., Lv, Z. H., Zhang, X. L., et al. (2022). Epidemiology and genetic diversity of Bartonella in rodents in urban areas of Guangzhou, southern China. Front. Microbiol. 13:942587. doi: 10.3389/fmicb.2022.942587
Ying, B., Kosoy, M. Y., Maupin, G. O., Tsuchiya, K. R., and Gage, K. L. (2002). Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am. J. Trop. Med. Hyg. 66, 622–627. doi: 10.4269/ajtmh.2002.66.622
Yu, J., Zhang, X. Y., Chen, Y. X., Cheng, H. B., Li, D. M., and Rao, H. X. (2022c). Molecular detection and genetic characterization of small rodents associated Bartonella species in Zhongtiao Mountain, China. PLoS One. 17:e0264591. doi: 10.1371/journal.pone.0264591
Yu, J., Xie, B., Bi, G. Y., Zuo, H. H., Du, X. Y., Bi, L. F., et al. (2022b). Prevalence and diversity of small rodent-associated Bartonella species in Shangdang Basin, China. PLoS Negl. Trop. Dis. 16:e0010446. doi: 10.1371/journal.pntd.0010446
Keywords: Bartonella, epidemiology, genetic diversity, human-pathogenic, rodents
Citation: Jian R, Ren Q, Xue J, Xie G-C, Wang J, Chen G-Q, Du L and Guo W-P (2022) Genetic diversity of Bartonella infection in residential and field rodents in Hebei, China. Front. Microbiol. 13:1039665. doi: 10.3389/fmicb.2022.1039665
Received: 08 September 2022; Accepted: 07 November 2022;
Published: 25 November 2022.
Edited by:
Hongliang Chai, Northeast Forestry University, ChinaReviewed by:
Ryan Oliver Marino Rego, Academy of Sciences of the Czech Republic (ASCR), CzechiaPaulo Eduardo Velho, State University of Campinas, Brazil
Jana Radzijevskaja, Vytautas Magnus University, Lithuania
Copyright © 2022 Jian, Ren, Xue, Xie, Wang, Chen, Du and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Ping Guo, Z3Vvd2VucGluZ0Bud3N1YWYuZWR1LmNu
†These authors have contributed equally to this work
 Rui Jian1†
Rui Jian1† Wen-Ping Guo
Wen-Ping Guo