- College of Horticulture, Shenyang Agricultural University, Shenyang, China
Heterotrophic nitrification is a process of organic nitrogen degradation completed by the participation of heterotrophic nitrifying microorganisms, which can accelerate the nitrogen transformation process. However, the current research mainly focuses on heterotrophic nitrifying bacteria and their ammonium degradation capacities. And there is little accumulation of research on fungi, the main force of heterotrophic nitrification, and their capacities to transform organic nitrogen. In this study, novel heterotrophic nitrifying fungus (XTY1) and bacterium (GS2) were screened and isolated from upland soil, and the strains were identified and registered through GenBank comparison. After 24 h single nitrogen source tests and 15N labeling tests, we compared and preliminarily determined the heterotrophic nitrification capacities and pathways of the two strains. The results showed that XTY1 and GS2 had different transformation capacities to different nitrogen substrates and could efficiently transform organic nitrogen. However, the transformation capacity of XTY1 to ammonium was much lower than that of GS2. The two strains did not pass through NH2OH and NO2− during the heterotrophic nitrification of organic nitrogen, and mainly generated intracellular nitrogen and low N2O. Other novel organic nitrogen metabolism pathways may be existed, but they remain to be further validated.
Highlights
•Two strains of heterotrophic nitrifying microorganisms were screened and isolated from soil. The fungus was named Pichia kudriavzevii XTY1, and the bacterium was named Enterobacter asburiae GS2. The gene sequences were 471 bp and 1,385 bp in length, and the GenBank registration numbers were MW132885 and MW126534, respectively.
•XTY1 and GS2 could efficiently utilize organic nitrogen for heterotrophic nitrification with transformation rates of 83.6 and 82.8%, respectively. However, the transformation capacities of inorganic N were different, and the transformation rate of ammonium by XTY1 (41.3%) was much lower than that of GS2 (82.0%).
•During the heterotrophic nitrification of organic nitrogen by XTY1 and GS2, no hydroxylamine and nitrite were produced, more than 60% of the organic nitrogen was generated into intracellular nitrogen, and less than 0.3% was generated into N2O. Unlike previous findings, there may be novel pathways.
Introduction
Heterotrophic nitrifying microorganisms transform soil organic nitrogen (Norg) directly to nitrate (NO3−) without ammonification, thereby accelerating the process of nitrogen transformation. Heterotrophic nitrification is considered to be a new pathway in the nitrogen transformation process, which launched the research on heterotrophic nitrifying microorganisms (Robertson and Kuenen, 1983; Serna and Pomares, 1992; Brierley and Wood, 2001). Over the last three decades, many researchers have identified a variety of heterotrophic nitrifying microorganisms in various habitats, and described their ammonium (NH4+) transformation capacities and metabolite characteristics (Pedersen et al., 1999; Joo et al., 2005; Schmidt et al., 2011; Zhang et al., 2014; Bai et al., 2018; Yu et al., 2020; Zhang et al., 2020), but little is known about the Norg transformation pathways and characteristics.
At the same time, the studies on fungi (Boer and Kowalchuk, 2001; Liu et al., 2016), the main undertaker of nitrification, are far fewer than that on heterotrophic nitrifying bacteria. These studies have only focused on the screening of fungi, and did not explore their nitrogen (N) transformation in depth. Therefore, studying heterotrophic nitrifying fungi and their capacities to transform Norg are beneficial to deepen researchers’ understanding of heterotrophic nitrifying microorganisms.
Different heterotrophic nitrifying microorganisms have different nitrogen transformation pathways. For example, Alcaligenes faecalis No. 4 and Acinetobacter sp. JR1 transformed NH4+ first to hydroxylamine (NH2OH), then to nitrite (NO2−), and finally to NO3− via nitrification (Joo et al., 2005; Yang et al., 2019). By comparison, Alcaligenes faecalis NR transformed NH4+ to NH2OH and then directly to nitrous oxide (N2O) and nitrogen gas (N2) without passing through NO2− and NO3− (Zhao et al., 2012). Agrobacterium sp. LAD9 isolated by Chen and Ni (2012) and Alcaligenes ammonioxydans sp. HO-1 isolated by Wu et al. (2021) denitrified to N2 after transformation of NH4+ to NO2−. In contrast, Pseudomonas putida Y-9 directly transformed NH4+ to nitric oxide (NO) and then to N2O, and no other intermediates were found (Huang et al., 2019). Therefore, we also need to track the metabolites and pathways of the isolated strains to understand the nitrogen transformation method of each microorganism.
Under the background of global changes such as atmospheric nitrogen deposition and atmospheric warming, screening and isolating heterotrophic nitrifying strains from soil and understanding the transformation capacities of strains to organic nitrogen are of great significance for preventing soil pollution and reducing greenhouse gas emissions, etc. Subsequently, our research group screened and isolated fungus and heterotrophic nitrifying bacterium with high organic nitrogen transformation capacities in the soil of the upland experimental area, and identified their species after comparing with the GenBank. We inoculated the strains with Norg (glycine), NH4+, NO3−, potential intermediate products (NH2OH and NO2−) as the sole nitrogen sources of the medium for 24 h incubation tests, respectively, and determined the heterotrophic nitrification and aerobic denitrification capacities and intermediate products of the two strains. We then inoculated the strains with Norg (glycine), NH4+, NO3− as the sole nitrogen sources of the medium, combined with 15N labeling method to determine the N2O and N2 emissions of heterotrophic nitrification and aerobic denitrification processes, to preliminarily understand and analyze the Norg transformation pathways of two strains. Finally, it provided potential strains for heterotrophic nitrification and further enriched the nitrogen transformation pathways of heterotrophic nitrifying microorganisms. We hypothesized that: (i) Do heterotrophic nitrifying microorganisms have both autotrophic nitrification and heterotrophic nitrification capabilities? Is there a difference between the two? (ii) Are the intermediates of heterotrophic nitrification processes the same as those of conventional autotrophic nitrification processes?
Materials and methods
Media
The fungal Screening medium (Bengal Red) contained the following components (per liter): 5.0 g peptone, 10.0 g glucose, 1.0 g KH2PO4, 0.5 g MgSO4·7H2O, 0.03 g Bengal Red, and 0.1 g chloramphenicol. The bacterial enrichment medium (LB) contained the following components (per liter): 3.0 g beef paste, 10.0 g peptone, 5.0 g NaCl. The bacterial isolation and purification medium contained the following components (per liter): 0.5 g glycine, 4.0 g trisodium citrate, 1.6 g NaCl, 8.2 g KH2PO4, 0.5 g MgSO4·7H2O, 0.01 g FeSO4·7H2O, 0.5 g KCl. The basal medium contained the following components (per liter): 1.0 g KH2PO4, 0.5 g MgSO4·7H2O, 0.03 g FeSO4·7H2O, 0.12 g NaCl, 0.01 g CaCl2, and 0.1 g chloramphenicol (Bacterial medium was not added). Different nitrogen and carbon sources were added to the basal medium depending on the experiment (Tables 1 and 2). The initial pH of the medium was adjusted to 7.2 ± 0.2 using 0.1 M HCl or 0.1 M NaOH. The solid medium was supplemented with 1.3–1.5% agar. All media were autoclaved at 121°C for 20 min before use. All chemicals were of analytical grade and were weighed on an electronic scale (precision 1 × 10−4).
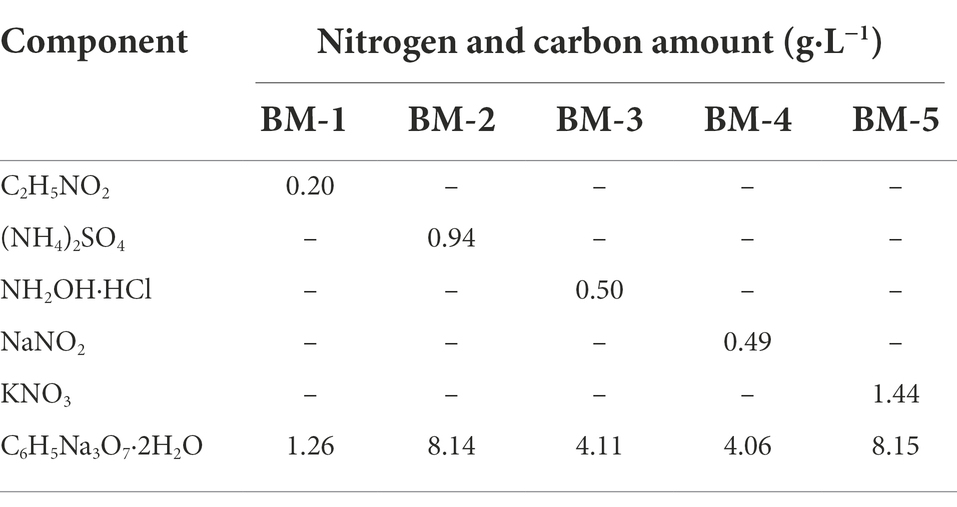
Table 1. List of carbon and nitrogen sources added to basal medium (BM) for nitrification and denitrification by XTY1.
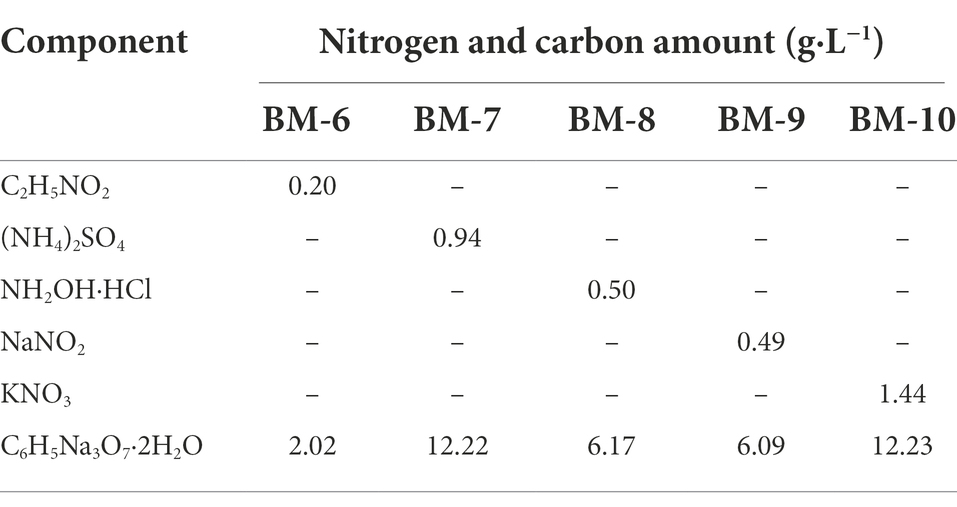
Table 2. List of carbon and nitrogen sources added to basal medium (BM) for nitrification and denitrification by GS2.
Screening and identification
Soil samples were collected from upland at the Baicao Garden Teaching and Research Base of Shenyang Agricultural University, Shenyang, Liaoning Province, China (41.83° N, 123.57° E). The site was a perennial grassland planted with Poa annua L. (species of excellence) without fertilization treatment and belongs to a temperate semi-humid continental climate zone, with an annual average precipitation of 580 mm and average temperate of 24.8°C from April to October. The brown soil texture is 27.2% sand, 50.2% silt and 22.6% clay. The soil pH was 5.74, total carbon (C) was 32.11 g·kg−1, total phosphorus was 1.42 g·kg−1, total potassium was 30.29 g·kg−1, total N was 1.22 g·kg−1, NH4+ was 13.70 mg·kg−1 and NO3− was 5.53 mg·kg−1, available phosphorus was 13.40 mg·kg−1, available potassium was 132.34 mg·kg−1.
Three sites with good growth and no baldness were searched for in the sampling area to take 0–10 cm soil samples using the three-point method. Ten gram of the soil sample was placed in a 250-mL conical flask containing 90 ml sterile water and incubated in a rotary shaker at 25°C and 150 rpm for 1 h. After standing for 2 h, 2 ml of the suspension was added to 250-mL conical flasks containing 98 ml fungal screening medium and bacterial enrichment medium and incubated at 35°C and 150 rpm for 2 d. For bacterial screening, an additional 2 ml suspension of enrichment medium was added to a 250-mL conical flask containing 98 ml of bacterial screening medium and incubated at 35°C at 150 rpm for 1 d. The above culture medium inoculation processes were repeated three times.
We applied 200 μl of the suspensions on the respective solid screening medium, and the strains were isolated by plate streaking until a single colony could be picked and inoculated in 250-mL conical flasks containing 100 ml BM-1, 6 medium (Tables 1, 2). These cultures were shaken at a constant temperature for 24 h. The glycine concentration in the medium was measured, and the fungal and bacterial strains with the highest transformation rate were selected for further experiments. The strains were inoculated in 250-mL conical flasks containing 98 ml respective screening medium and shaken at 35°C until the culture reached logarithmic phase, whereby it was mixed 1:1 with 30% glycerol and placed at −80°C for long-term storage.
Scanning electron microscopy and transmission electron microscopy (bacteria only) were used to observe the fungal and bacterial cell morphology. ITS rRNA and 16S rDNA were determined by polymerase chain reaction (PCR) amplification. Fungal primers ITS1 (TCCGTAGGTGAACCTGCGG) and ITS4 (TCCTCCGTTATTGATATATGC), bacterial universal primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5’-CTACGGCTACCTTGTTACGA-3′), these primer synthesis and culture identification processes were completed by Sangon Biotech (Shanghai, China). The PCR thermal program of the fungus was conducted according to the following cycle profile: pre-denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min and 30 s in a thermal cycler. After the last cycle, incubate for another 7 min at 72°C to allow the extension to complete. The PCR thermal program for bacteria was the same as above, except for 1 min at 72°C in a thermal cycler. Finally, BLAST analysis of the National Center for Biotechnology Information was used for sequence analysis and homology comparison to determine fungi and bacterial genera.
Heterotrophic nitrification and aerobic denitrification capacity
Firstly, the optimal conditions of heterotrophic nitrifying strains were screened by orthogonal experiment. The results showed that the fungal transformation rate was highest with trisodium citrate as the carbon source, 200 mg·L−1 initial glycine, and the ratio of total carbon to total nitrogen in the culture medium (C/N ratio) of 10, 35°C, and 130 rpm rotating speed. The bacterial transformation rate was highest under culture conditions with trisodium citrate as the carbon source, 200 mg·L−1 initial glycine, and C/N ratio of 15, 35°C, and 160 rpm. Therefore, subsequent experiments were conducted under these conditions. Researches with soil revealed that free amino acids were an important contributor to the soil soluble Norg pool, with glycine, aspartic acid, and glutamic acid accounting for the largest fraction of Norg (Paul and Williams, 2005; Streeter et al., 2015). Hence, glycine was chosen as the organic nitrogen source for heterotrophic nitrification in this study.
Heterotrophic nitrification and aerobic denitrification of the candidate fungal and bacterial strains were determined using Norg (glycine), (NH4)2SO4, and KNO3 as sole nitrogen sources. Two milliliters of fungal and bacterial sample solutions stored at −80°C were inoculated in 250-mL conical flasks containing 98 ml BM-1, 6 medium and incubated at 35°C and 130 (160) rpm for 24 h to activate the strains. The pre-cultures were centrifuged at 4000 G and 4°C for 2 min, and then washed three times with sterile water. Next, 2 ml of the sterile water-rinsed solutions was inoculated in 250-mL conical flasks containing 98 ml BM-1, 2, 5 and BM-6, 7, 10 medium (Tables 1, 2), and incubated at 130 (160) rpm and 35°C for 24 h. Each sample was assayed in triplicates, and medium without inoculum was used as the control. Samples were taken at 2 h intervals, centrifuged at 11000 G for 10 min and the supernatants were taken to determine glycine, NH4+, NO2−, NH2OH, NO3−, intracellular nitrogen, and total nitrogen concentrations. as well as cell growth by measuring optical density at a 600 nm wavelength (OD600) without centrifuging the samples.
Utilization of hydroxylamine and nitrite
Pre-treatment of the activated sample solution was as described in section 2.3. The sterile water-rinsed fungal and bacterial solutions (2 ml) were inoculated into 250-mL conical flasks containing 98 ml BM-3, 4 and BM-8, 9 medium (Tables 1, 2) with NH2OḤHCl and NaNO2 as sole nitrogen sources, and incubated at 130 (160) rpm and 35°C for 24 h. Three replicates were performed. Samples were taken every 4 h to determine NH4+, NO2−, NH2OH, NO3−, intracellular nitrogen, and total nitrogen concentrations, as well as OD600.
Gaseous product determination
N2O and N2 produced during heterotrophic nitrification and aerobic denitrification were determined using a single 15N-labeled nitrogen source (99 atom%). The activated sample solutions (2 ml) were inoculated in 250-mL airtight silk-mouthed bottles containing 98 ml BM-1, 2, 5 and BM-6, 7, 10 media labeled with 15N. The space above the liquid was filled with ambient air and flasks were shaken at 35°C and 130 (160) rpm for 24 h. Then, 2 ml of the gas above each sample was extracted for the determination of N2 and N2O content. Three replicates were performed. These analyses were conducted at the Shenyang Institute of Applied Ecology, Chinese Academy of Sciences.
Analytical methods and calculations
Cell growth was determined by measuring OD600 on an ultraviolet (UV) spectrophotometer. Glycine was determined using the ninhydrin colorimetric method, NH4+ using the indophenol blue colorimetric method, NH2OH by the Frear and Burrell (1955) method, and NO2− by the N-(1-naphthyl)-ethylenediamine method. Nitrate and total nitrogen were determined by UV spectrophotometry (Clescerl, 1998). Intracellular nitrogen was measured by differential subtraction, whereby the total nitrogen concentration after centrifugation was subtracted from that before centrifugation (Rout et al., 2017). N2O and N2 were determined by mass spectrometry.
The MEGA 5.0 software was used to build the phylogenetic tree. Data analysis was performed using Microsoft Excel and the SPSS 22 software. Graphs were plotted using SigmaPlot 10.0, and results were presented as means ± standard errors.
Results and discussion
Screening and identification
Three strains of heterotrophic nitrifying fungi were screened on solid Bengal Red medium, revealing glycine transformation rates of 30.1, 50.2, and 83.8%. The fungus with the strongest transformation capacity was named XTY1 and was selected for subsequent experiments. Its colonies appeared round, creamy white, with a smooth and moist surface, whereas cells were elongated and (2.4–2.5) μm × 0.8 μm in length (Figures 1A,C). To identify the genus of the strain, a phylogenetic tree constructed after PCR amplification of a 471 bp fragment revealed that the candidate strain had 99% homology with Pichia kudriavzevii (Figure 2A). Hence, the strain was named Pichia kudriavzevii XTY1, referred to herein as XTY1. The sequence was deposited in the GenBank database under registration number MW132885.
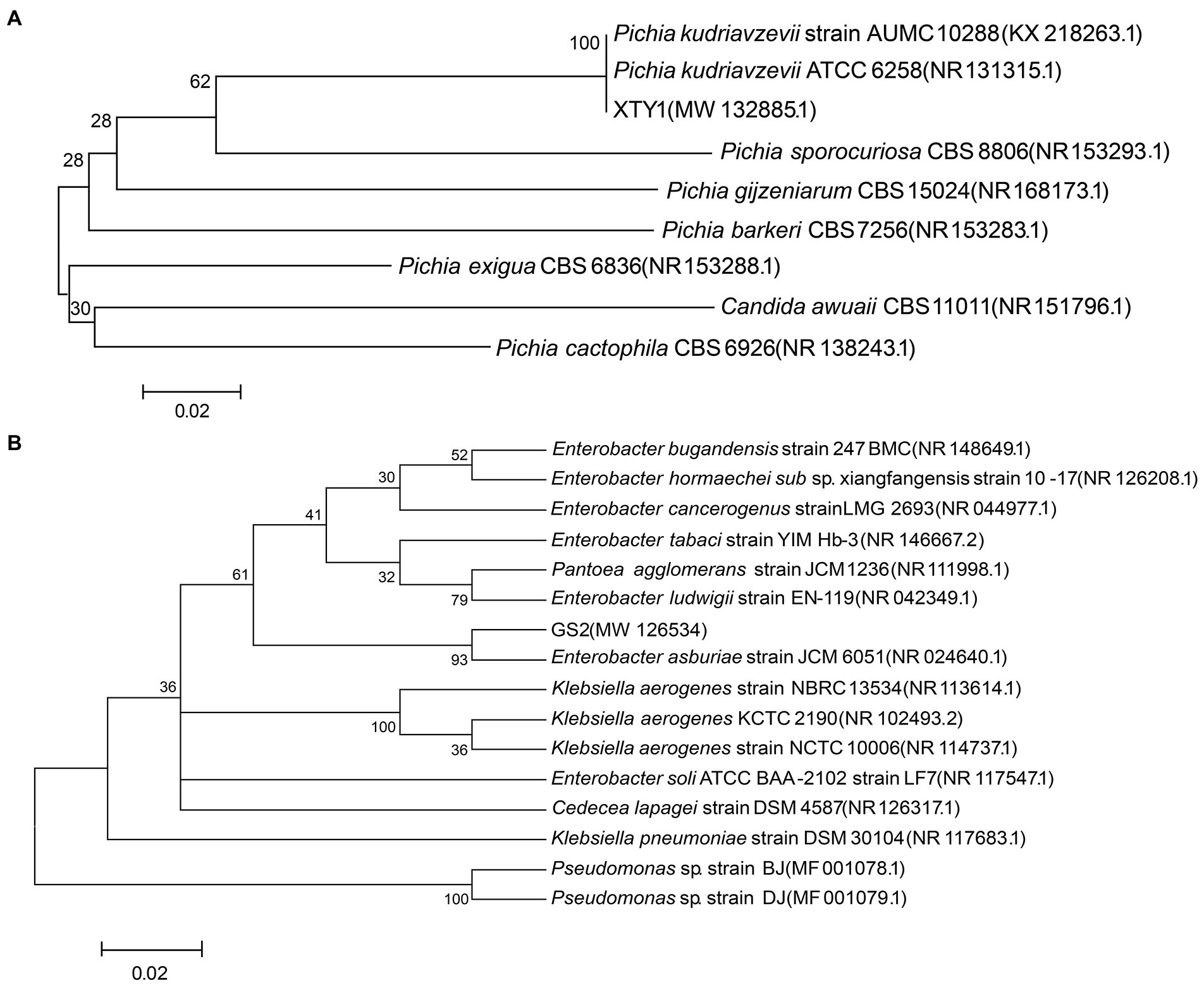
Figure 2. Phylogenetic tree of XTY1 (A) and GS2 (B). Node support values are presented, the scale bar defines the branch length.
Two strains of heterotrophic nitrifying bacteria were also screened, with glycine transformation rates of 57.0 and 82.7%. The second bacterium with stronger transformation capacity was named GS2 and used for the follow-up experiment. The colonies were round, off-white and translucent with smooth and moist surface and regular edges. The cells were short rod-shaped with flagella, and the size was about (1.3–1.6) μm × (0.6–0.8) μm (Figures 1B,D). A fragment of 1,385 bp was amplified by PCR, and the comparison revealed that the candidate strain had 99% homology with Enterobacter asburiae (Figure 2B). The strain was finally named Enterobacter asburiae GS2 (GS2 for short) and was registered in the GenBank database under registration number MW126534.
Heterotrophic nitrification and aerobic denitrification capacity
As shown in Figure 3A, fungus XTY1 grown on glycine displayed a lag phase of 4 h, and started the logarithmic phase after 12 h, reaching maximum growth (D600 = 0.42) after 20 h and entered senescence thereafter. At the same time, glycine decreased rapidly after 12 h, reached the maximum transformation rate of 29.60 mg·L−1·h−1 at 14 h, and stabilized after 20 h. This decreasing trend was consistent with the growth trend of XTY1, indicating that the strain could utilize glycine for growth. After 20 h, the cell growth decreased slightly, while the NH4+ concentration increased, which can be explained by cellular transformation, possibly due to the release of NH4+ by senescent death of the strain (Chen et al., 2014; Li et al., 2015). with the fungus XTY1 and the bacterium GS2 (Figure 3B) were not significantly different in their capacity to transform glycine, and the concentrations of nitrogen-containing intermediates NH4+, NH2OH, NO2−, and NO3− did not change significantly during the 24 h incubation period.
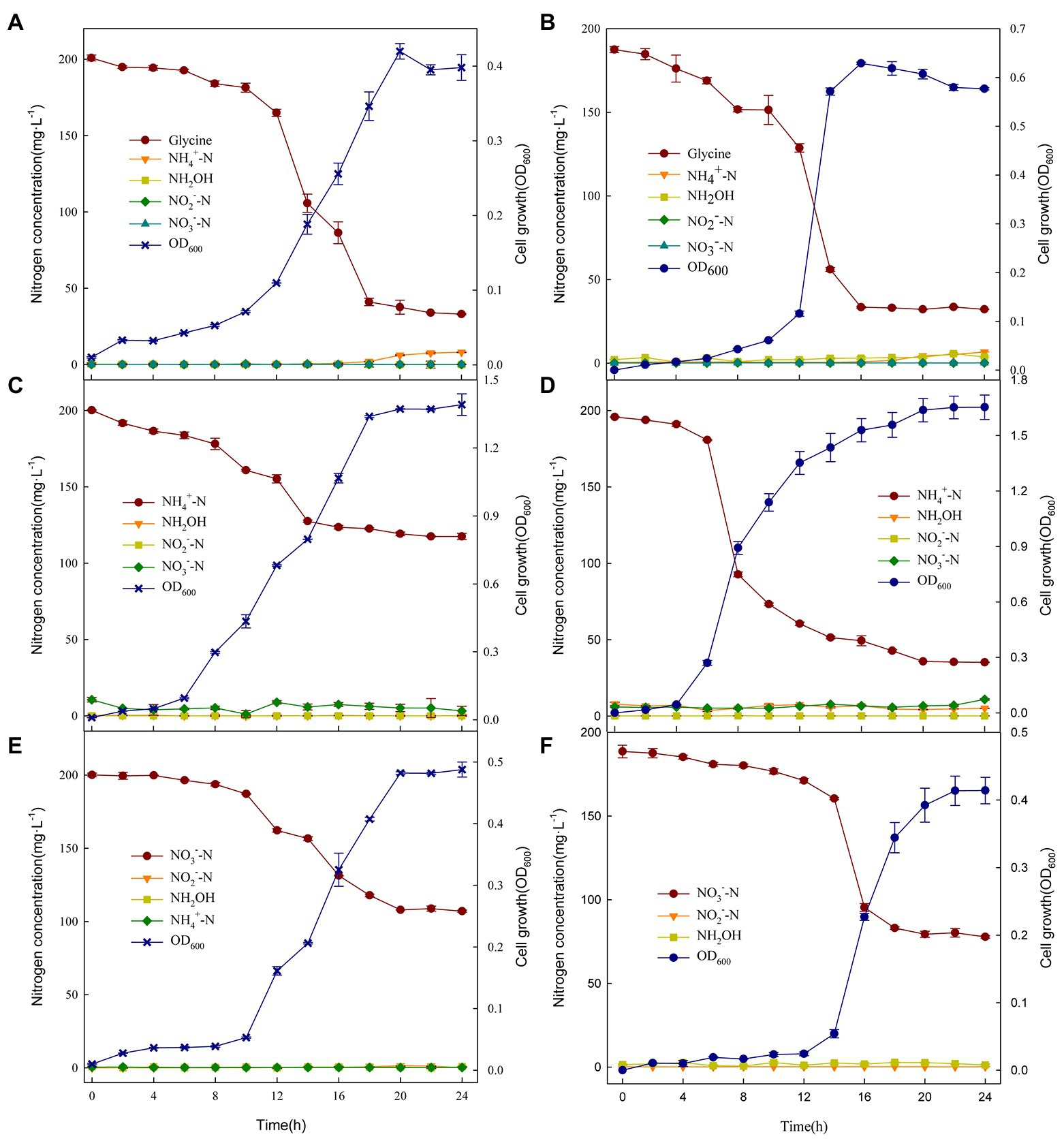
Figure 3. Changes in nitrogen compounds and growth of XTY1 and GS2 when glycine (A, B), (NH4)2SO4 (C, D), or KNO3 (E, F) were used as sole nitrogen sources under aerobic conditions.
As shown in Figure 3C, growth of XTY1 on (NH4)2SO4 as the nitrogen source led to no lag phase and stabilization after 18 h, with a maximum OD600 of 1.39. The NH4+ concentration decreased from 200.15 mg·L−1 to 117.56 mg·L−1, with a maximum transformation rate of 13.88 mg·L−1·h−1, indicating that NH4+ could be used for both growth and nitrogen transformation. At 0 h, a small accumulation of NO3− was observed, probably due to oxidation of NH4+ followed by its gradual reduction during the subsequent incubation (Li et al., 2015). Compared to GS2 (Figure 3D), the fungal heterotrophic nitrification of ammonium was very low. No significant changes in other intermediates were detected, suggesting that even though the fungi play a major role in heterotrophic nitrification (Zhang et al., 2011), the heterotrophic nitrifying fungi were much less capable of transformation than the bacteria. There were small fluctuations in NO3− throughout the incubation period of both strains, probably due to the transformation of NH4+ to NO3− by the strains.
Incubation with KNO3 as the sole nitrogen source resulted in a long lag phase for XTY1, which entered the logarithmic phase only after 8 h, when the concentration of NO3− decreased sharply, dropping to 107.07 mg·L−1 at 24 h (Figure 3E). This suggests that the strain could use NO3− for aerobic denitrification. Compared with GS2 (Figure 3F), XTY1 showed similar aerobic denitrification of nitrate, with no significant changes in the content of other nitrogenous substances throughout the incubation.
Study on heterotrophic nitrification intermediates and pathways of strains
At present, researchers believe that hydroxylamine and nitrite are the most likely intermediates for heterotrophic nitrifying strains to transform nitrogen sources, such as Pseudomonas bauzanensis DN13-1 (Zhang et al., 2020) and Ochrobactrum anthropi HND19 (Ren et al., 2021). Therefore, NH2OḤHCl (Figures 4A,B) and NaNO2 (Figures 4C,D) were used as the sole nitrogen sources for XTY1 and GS2 culture experiments. However, the experiments showed that the two strains did not grow and the nitrogen concentration did not change over 24 h. This result confirmed that NH2OH and NO2− were not detected during the incubation process as a result of non-production and not by transformation. There might be new metabolites in the nitrogen transformation process. Therefore, the metabolites of XTY1 and GS2 with glycine as nitrogen source (BM-1, 6 medium) for 16 h were preliminarily detected. We compared the nitrogen-containing substances in the culture medium with glycine transformation products retrieved from Glycine, serine and threonine metabolism1 in KEGG official website, such as Glyoxylic acid (C00048), S-Amino-methyldihydrolipoylprotein (C01242), Lipoylprotein (C02051), 5, 10-methodology-THF (C00143), etc. These substances were not found in the culture medium. On the contrary, pantothenic acid, anthranilic acid and other nitrogen-containing substances had obvious up-regulation, but whether these up-regulation nitrogen-containing substances were related to glycine metabolism pathways of strains needs further follow-up.
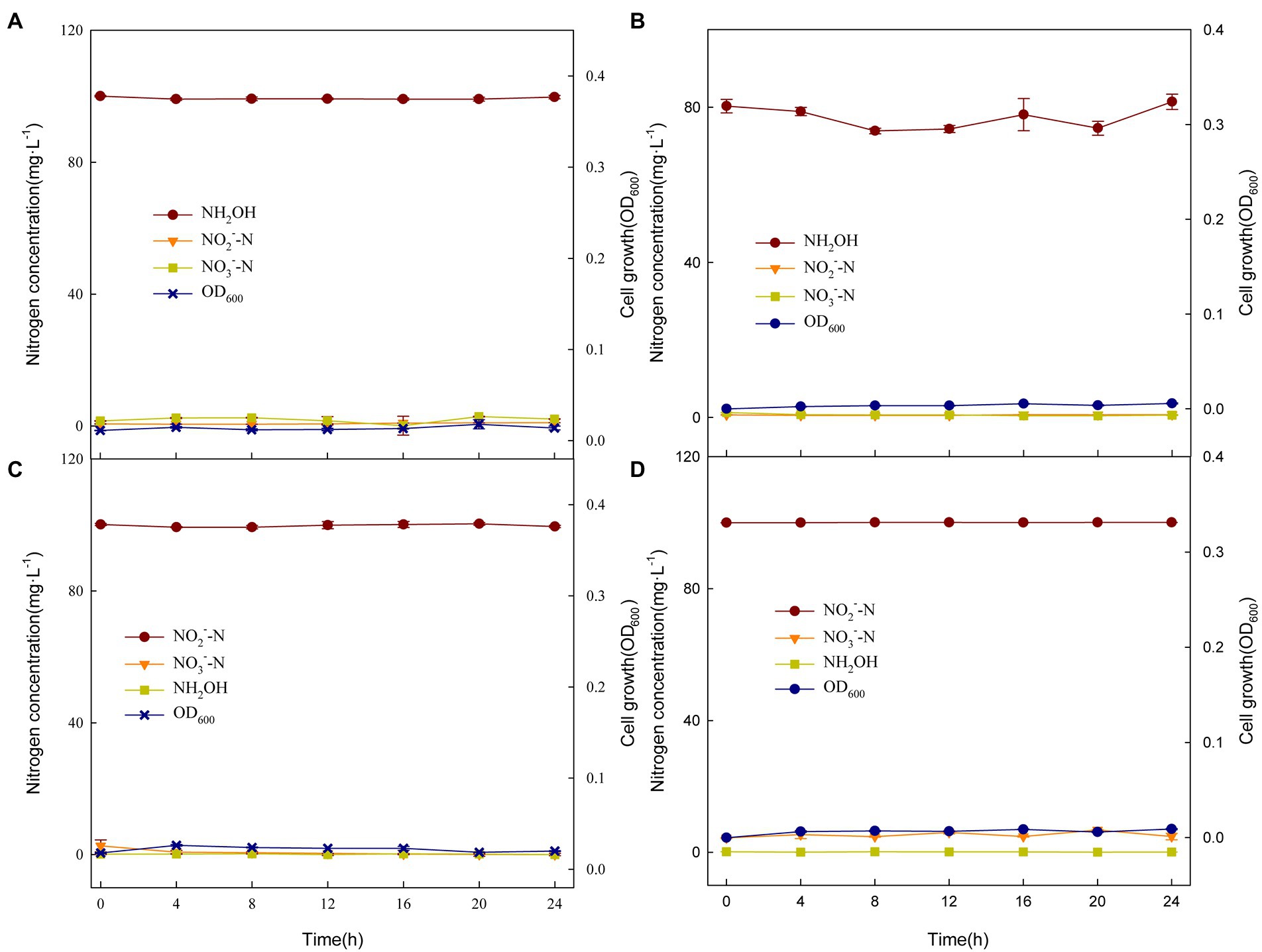
Figure 4. Changes in nitrogen compounds and growth of XTY1 and GS2 when NH2OḤHCl (A, B) or NaNO2 (C, D) were used as sole nitrogen sources under aerobic conditions.
Nitrogen balance by heterotrophic nitrification and aerobic denitrification
The nitrogen balance calculation (Table 3) revealed that both the fungus XTY1 and the bacterium GS2 were able to transform glycine (83.6 and 82.8%), ammonium (41.3 and 82.0%) and nitrate (46.5 and 58.7%). As shown in Table 3, both strains produced and released N2O and N2 during nitrification and denitrification. However, the amount of N2O produced during heterotrophic nitrification of Norg was much lower than that produced by aerobic denitrification (36.2 and 30.6%), indicating that N2O was produced by NO3− via denitrification.
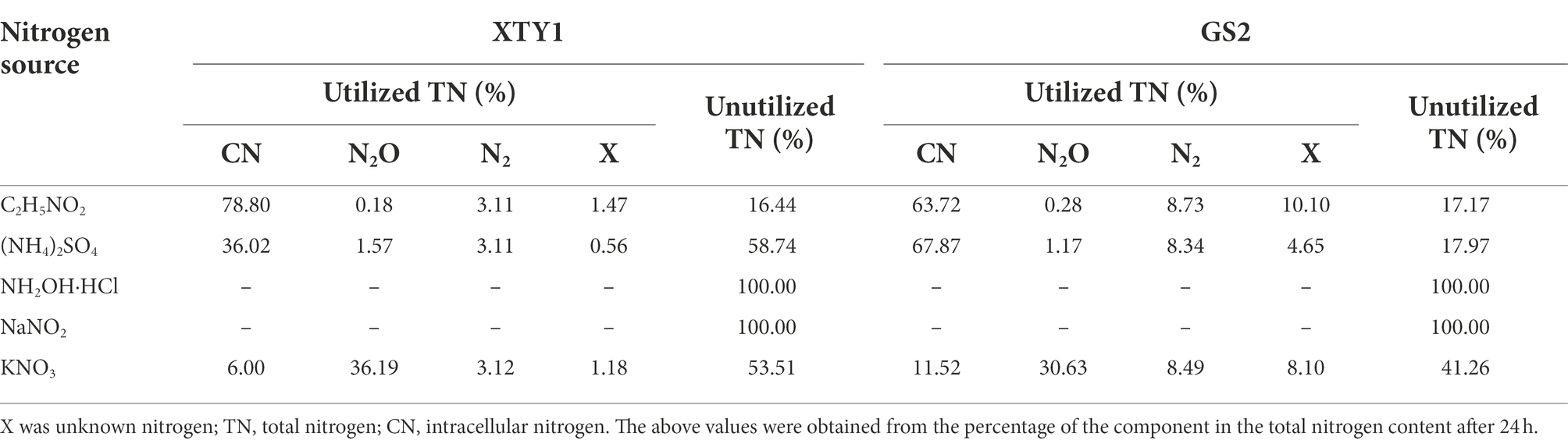
Table 3. Nitrogen balance after 24 h of heterotrophic nitrification and denitrification processes by XTY1 and GS2.
During heterotrophic nitrification of Norg, the fungus XTY1 and the bacterium GS2 produced 94.3 and 63.7% of the total nitrogen concentration of intracellular nitrogen (CN), respectively. This result indicated that both strains transformed most of Norg into CN by assimilation during heterotrophic nitrification, which is consistent with Acinetobacter sp. JR1 transforming part of the nitrogen source into CN (Yang et al., 2019). This pathway was also demonstrated in a study by Joo et al. (2005) on the denitrification efficiency of Alcaligenes faecalis No. 4 under high ammonium concentrations. In that study, almost all ammonium was removed after 93 h of inoculating the strain when the initial ammonium mass concentration was up to 1,200 mg·L−1. Fifty percent of this was transformed to intracellular nitrogenous material, 3% was transformed to nitrification products, and 40 to 50% underwent denitrification. In a study by Chen and Ni (2012) on the nitrogen balance of Agrobacterium LAD9 during heterotrophic nitrification and aerobic denitrification, the initial mass concentration of total nitrogen (TN) was 7.2 mg·L−1. After 24 h of inoculation with this bacterium, 50.1% of TN was transformed to gaseous nitrogen compounds and 40.8% was also transformed to CN.
The remaining small fraction of Norg was eventually transformed to N2 via aerobic denitrification, but the specific intermediates were still unclear because they did not pass through NH2OH and NO2−. Further experiments using methods such as 15N labeling and metabolomics are needed to determine the specific metabolic pathway used by the strains.
Conclusion
Our results with fungus XTY1 and heterotrophic nitrifying bacterium GS2 revealed that:
1. XTY1 and GS2 had different transformation capacities to different nitrogen sources, and the heterotrophic nitrification capacities of organic nitrogen were stronger (the transformation rates were more than 80%), but the nitrification capacity of XTY1 to ammonium nitrogen (41.3%) was lower.
2. The organic nitrogen transformation intermediates of the two strains were free of hydroxylamine and nitrite. More than 60% of the total nitrogen were transformed to intracellular nitrogen, and less than 0.3% to N2O.
3. Compared with the founded heterotrophic nitrifying strains, the two strains screened in this study may have novel metabolic pathways.
4. In future studies, methods such as 15N labeling and metabolomics could be used to track the metabolite characteristics of the two strains and explore their Norg metabolism pathways.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, MW132885 https://www.ncbi.nlm.nih.gov/genbank/, MW126534.
Author contributions
TX, LB, and ZP contributed to conceptualization and design of the study. TX and SS performed the lab work and analyzed the data. TX wrote the first draft of the manuscript. LB, BR, JY, and JL contributed to writing, review and editing. LB contributed to funding acquisition. All authors have read and approved to the published version of the manuscript.
Funding
This research was financially supported by Liaoning Provincial Science and Technology Plan Projects (2020JH1/10300006) in China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Bai, S. N., Xu, T. Y., Liu, Y., Wang, X. H., and Bai, L. (2018). Isolation and screening of heterotrophic nitrifying bacteria from lawn soil and characterization of nitrification products. J. Hortic. 45, 1338–1346. doi: 10.16420/j.issn.0513-353x.2017-0773
Boer, W. D., and Kowalchuk, G. A. (2001). Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol. Biochem. 33, 853–866. doi: 10.1016/S0038-0717(00)00247-9
Brierley, E. D. R., and Wood, M. (2001). Heterotrophic nitrification in an acid forest soil: isolation and characterisation of a nitrifying bacterium. Soil Biol. Biochem. 33, 1403–1409. doi: 10.1016/S0038-0717(01)00045-1
Chen, Q., and Ni, J. R. (2012). Ammonium removal by agrobacterium sp. LAD9 capable of heterotrophic nitrification-aerobic denitrification. J. Biosci. Bioeng. 113, 619–623. doi: 10.1016/j.jbiosc.2011.12.012
Chen, M. X., Wang, W. C., Feng, Y., Zhu, X. H., Zhou, H. Z., Tan, Z. L., et al. (2014). Impact resistance of different factors on ammonia removal by heterotrophic nitrification-aerobic denitrification bacterium Aeromonas sp. HN-02. Bioresour. Technol. 167, 456–461. doi: 10.1016/j.biortech.2014.06.001
Clescerl, L. S. (1998). Standard Methods for the Examination of Water and Wastewater 20th. Washington: APHA.
Frear, D. S., and Burrell, R. C. (1955). Spectrophotometric method for determining hydroxylamine reductase activity in higher plants. Anal. Chem. 27, 1664–1665. doi: 10.1021/ac60106a054
Huang, X. J., Xu, Y., He, T. X., Jia, H. J., Feng, M., Xiang, S. D., et al. (2019). Ammonium transformed into nitrous oxide via nitric oxide by pseudomonas putida Y-9 under aerobic conditions without hydroxylamine as intermediate. Bioresour. Technol. 277, 87–93. doi: 10.1016/j.biortech.2019.01.040
Joo, H. S., Hirai, M., and Shoda, M. (2005). Characteristics of ammonium removal by heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis no. 4. J. Biosci. Bioeng. 100, 184–191. doi: 10.1263/jbb.100.184
Li, C. E., Yang, J. S., Wang, X., Wang, E. T., Li, B. Z., He, R. X., et al. (2015). Removal of nitrogen by heterotrophic nitrification–aerobic denitrification of a phosphate accumulating bacterium pseudomonas stutzeri YG-24. Bioresour. Technol. 182, 18–25. doi: 10.1016/j.biortech.2015.01.100
Liu, Y. X., Hu, T. T., Jing, Z., Lv, Y. K., and Ren, R. P. (2016). Simultaneous removal of carbon and nitrogen by mycelial pellets of a heterotrophic nitrifying fungus-Penicillium sp. L1. J. Biosci. Bioeng. 123, 223–229. doi: 10.1016/j.jbiosc.2016.08.009
Paul, J. P., and Williams, B. L. (2005). Contribution of α-amino N to extractable organic nitrogen (DON) in three soil types from the Scottish uplands. Soil Biol. Biochem. 37, 801–803. doi: 10.1016/j.soilbio.2004.09.011
Pedersen, H., Dunkin, K. A., and Firestone, M. K. (1999). The relative importance of autotrophic and heterotrophic nitrification in a conifer forest soil as measured by 15N tracer and pool dilution techniques. Biogeochemistry 44, 135–150. doi: 10.1007/BF00992975
Ren, J. L., Bai, X. Y., Liu, Y. C., and Huang, X. (2021). Simultaneous nitrification and aerobic denitrification by a novel isolated Ochrobactrum anthropi HND19. Bioresour. Technol. 340:125582. doi: 10.1016/j.biortech.2021.125582
Robertson, L. A., and Kuenen, J. G. (1983). Thiosphaera pantotropha gen. Nov. sp. nov., a Facultatively anaerobic, Facultatively autotrophic Sulphur bacterium. J. Gen. Microbiol. 129, 2847–2855. doi: 10.1099/00221287-129-9-2847
Rout, P. R., Bhunia, P., and Dash, R. R. (2017). Simultaneous removal of nitrogen and phosphorous from domestic wastewater using Bacillus cereus GS-5 strain exhibiting heterotrophic nitrification, aerobic denitrification and denitrifying phosphorous removal. Bioresour. Technol. 244, 484–495. doi: 10.1016/j.biortech.2017.07.186
Schmidt, C. S., Richardson, D. J., and Baggs, E. M. (2011). Constraining the conditions conducive to dissimilatory nitrate reduction to ammonium in temperate arable soils. Soil Biol. Biochem. 43, 1607–1611. doi: 10.1016/j.soilbio.2011.02.015
Serna, M. D., and Pomares, F. (1992). Evaluation of chemical indices of soil organic nitrogen availcapacity in calcareous soils. Soil Sci. Soc. Am. J. 56, 1486–1491. doi: 10.2136/sssaj1992.03615995005600050025x
Streeter, T. C., Bol, R., and Bardgett, R. D. (2015). Amino acids as a nitrogen source in temperate upland grasslands: the use of dual labelled (13C, 15N) glycine to test for direct uptake by dominant grasses. Rapid Commun. Mass Spectrom. 14, 1351–1355. doi: 10.1002/1097-0231(20000815)14:15<1351::AID-RCM23>3.0.CO;2-9
Wu, M. R., Hou, T. T., Liu, Y., Miao, L. L., Ai, G. M., Ma, L., et al. (2021). Novel alcaligenes ammonioxydans sp. nov. from wastewater treatment sludge oxidizes ammonia to N2 with a previously unknown pathway. Environ. Microbiol. 23, 6965–6980. doi: 10.1111/1462-2920.15751
Yang, J. R., Wang, Y., Chen, H., and Lyu, Y. K. (2019). Ammonium removal characteristics of an acid-resistant bacterium Acinetobacter sp. JR1 from pharmaceutical wastewater capable of heterotrophic nitrification-aerobic denitrification. Bioresour. Technol. 274, 56–64. doi: 10.1016/j.biortech.2018.10.052
Yu, X., Li, D. J., Li, D., Zhang, G. H., Zhou, H. S., Li, S., et al. (2020). Enhanced wet deposition of water-soluble organic nitrogen during the harvest season: influence of biomass burning and in-cloud scavenging. J. Geophys. Res. Atmos. 125:e2020JD032699. doi: 10.1029/2020jd032699
Zhang, M. X., Li, A. Z., Yao, Q., Wu, Q. P., and Zhu, H. H. (2020). Nitrogen removal characteristics of a versatile heterotrophic nitrifying-aerobic denitrifying bacterium, pseudomonas bauzanensis DN13-1, isolated from deep-sea sediment. Bioresour. Technol. 305:122626. doi: 10.1016/j.biortech.2019.122626
Zhang, J. B., Müller, C., Zhu, T. B., Cheng, Y., and Cai, Z. C. (2011). Heterotrophic nitrification is the predominant NO3− production mechanism in coniferous but not broad-leaf acid forest soil in subtropical China. Biol. Fertil. Soils 47, 533–542. doi: 10.1007/s00374-011-0567-z
Zhang, J. B., Sun, W. J., Zhong, W. H., and Cai, Z. C. (2014). The substrate is an important factor in controlling the significance of heterotrophic nitrification in acidic forest soils. Soil Biol. Biochem. 76, 143–148. doi: 10.1016/j.soilbio.2014.05.001
Keywords: organic nitrogen transformation, fungi, heterotrophic nitrifying bacteria, heterotrophic nitrification, 15N labeling
Citation: Xu T, Song S, Ren B, Li J, Yang J, Bai L and Piao Z (2022) Fungus Pichia kudriavzevii XTY1 and heterotrophic nitrifying bacterium Enterobacter asburiae GS2 cannot efficiently transform organic nitrogen via hydroxylamine and nitrite. Front. Microbiol. 13:1038599. doi: 10.3389/fmicb.2022.1038599
Edited by:
Jincai Ma, College of New Energy and Environment/Jilin Unversity, ChinaReviewed by:
Man Lang, Nanjing University of Information Science and Technology, ChinaHaifang Zhang, Agro-Environmental Protection Institute (CAAS), China
Copyright © 2022 Xu, Song, Ren, Li, Yang, Bai and Piao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Long Bai, YmFpbG9uZzIwMThAc3lhdS5lZHUuY24=; Zhongyun Piao, enlwaWFvQHN5YXUuZWR1LmNu
 Tianyue Xu
Tianyue Xu Simeng Song
Simeng Song Baihui Ren
Baihui Ren Jiahuan Li
Jiahuan Li Jiyun Yang
Jiyun Yang Long Bai
Long Bai Zhongyun Piao
Zhongyun Piao