
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 13 January 2023
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1037733
 Kai Zhou1†
Kai Zhou1† Bingjie Hu1†
Bingjie Hu1† Xinzhuan Zhao1†
Xinzhuan Zhao1† Hongbo Chi1
Hongbo Chi1 Juan Pan1
Juan Pan1 Yufen Zheng1
Yufen Zheng1 Xiaojie Bi1
Xiaojie Bi1 Mengyuan Chen1
Mengyuan Chen1 Jicheng Xie1
Jicheng Xie1 Jiaqin Xu1
Jiaqin Xu1 Tao-Hsin Tung2
Tao-Hsin Tung2 Bo Shen1*
Bo Shen1* Hongguo Zhu1*
Hongguo Zhu1*Objective: In 2022, a new coronavirus variant (Omicron) infection epidemic broke out in Shanghai, China. However, it is unclear whether the duration of this omicron variant is different from that of the prototype strain.
Methods: We retrospectively analyzed 157 cases of Omicron variant infection in Taizhou Public Health Center from March 29, 2022, to April 18, 2022, and observed the dynamics of nucleic acid Ct values during the admission and discharge of patients. Clinical and laboratory indicators of these patients were also obtained.
Results: Compared to the prototype strain, the Omicron variant showed a broad population susceptibility in infected individuals (regardless of age and presence of underlying disease) and had slight damage to the immune system and renal function; the viral loads peaked was 2-3 days from disease onset; the median duration of omicron variant was 15-18 days; the nucleic acid Ct value of nasopharyngeal swabs of infected patients is lower than that of throat swabs, and the Ct value of oropharyngeal swabs is unstable during the recovery period.
Conclusion: Therefore, we found that the time to peak viral load of this Omicron variant was 2-3 days after the onset of the disease, and the duration was 15-18 days; symptomatic patients had higher viral load and longer hospitalization time. This finding will provide a basis for understanding omicron variants and formulating the national prevention and control strategy.
The first case of SARS-Cov-2 infection emerged in December 2019 and became a global public health event in 2020 following Severe Acute Respiratory Syndrome pneumonia (Mahase, 2020). In addition, new variants of the virus continue to emerge, with the currently predominantly prevalent SARS-CoV-2 Omicron variant first identified in South Africa, which has a more mutated genome than the wild type, accelerated transmission (Fan et al., 2022) and increased infectivity (Luo et al., 2021). There were 57,438 confirmed cases and 580 cumulative deaths in Shanghai, where a wave of infections started mid-to-late February (Shanghai Municipal Health Commission News release, 2022).
Studies have reported that Omicron variants have higher viral loads (Luo et al., 2021). Omicron’s exponential growth and increased infectivity are mainly attributed to immune escape due to altered spike-in antigens (Carreño et al., 2022; Dejnirattisai et al., 2022). However, increased viral load has also been found to contribute to the increased infectivity of previously emerged variants and to be associated with symptomatic versus asymptomatic infections (Teyssou et al., 2021). By assessing viral load based on reverse transcriptase PCR (RT-PCR) cycling threshold (Ct), Salvatore et al. found that prototype strain infections had lower Ct values in symptomatic individuals compared to asymptomatic ones and were negatively correlated with duration of illness (Salvatore et al., 2021). Luo et al. (2021) compared Ct values in respiratory specimens between the Alpha, B.1.2, and Delta variants. They found that Delta Ct values were lower than those of the Alpha variant (20.08 vs. 21.74, p < 0.05) but not statistically different from those of the B.1.2 variant.
The omicron cohort in this study was homozygous for the Shanghai COVID-19 patients infected in the same period. Phylogenetic features of SARS-CoV-2 viral genomes in this period, and inferring their relationship with those available on the GISAID database, indicated that all of the new viral genomes in Shanghai were clustered into the SARS-CoV-2 BA.2.2 sub-lineage (Zhang et al., 2022). There are few longitudinal studies on the viral load after Omicron infection and its relationship with clinical symptoms. The viral load correlates with the presence or absence of symptoms and has a specific impact on the course of the disease and patients’ infectiousness. Therefore, this paper further discusses the above aspects to guide clinical practice.
A retrospective analysis was made on 301 cases of patients with COVID-19 who were admitted to Taizhou Public Health Center from January 17, 2020, to March 10, 2020, and from March 30, 2022, to April 18, 2022. The infected SAS-cov-2 virus strain divided the patients into 157 cases infected with the Omicron variant and 144 cases infected with the wild-type strain (prototype strain) (Figure 1).
Data on epidemiological, clinical, and laboratory characteristics were obtained through data collection forms in the hospital’s electronic medical records and reviewed by a team of trained physicians. Clinical information included personal characteristics (gender, age, and BMI), comorbidities, symptom onset date, and length of hospital stay. Underlying conditions had hypertension, diabetes, cardiovascular disease, lung disease, kidney disease, liver disease, and a history of prior malignancy. We considered that the first symptoms began with a fever, cough, sore throat, headache, sputum production, nasal congestion, muscle aches, and diarrhea. Laboratory characteristic data included complete blood count, blood gases, electrolytes, coagulation indicators, and organ function characteristics. The chest CT scan was performed using a combined 40-row UCT530 scanner (United imaging, Shanghai, China). The acquired CT images were sent to the GE-PACS workstation and read by two senior physicians in a double-blind manner.
The nucleic acid test Ct values of patients infected with Omicron variants or prototype strains were collected from throat swabs, nasopharyngeal swabs, and sputum and fecal specimens from admission to discharge.
RNA assay of the prototype strain was performed with throat swabs, nasopharyngeal swabs, sputum, and stool as test specimens. The sample nucleic acids were extracted by nucleic acid extraction reagent (Zhijiang Biological, Shanghai, China). The target gene was detected with the novel coronavirus 2019-nCoV nucleic acid detection kit (fluorescence PCR method) (Reagent No: SFDA: 20203400057, Zhijiang Bio, Shanghai, China). This kit uses RT-PCR (one-step method for reversing transcription-polymerase chain reaction) combined with Taqman technology to detect the novel coronavirus’s RdRP gene, N gene, and E gene. If the RdRp gene is positive (Ct ≤ 43) with E or/and N gene also being positive (Ct ≤ 43), the result is determined as positive.
RNA detection of the Omicron variants was performed mostly with throat and nasopharyngeal swabs as specimens. Nucleic acid was extracted by the novel coronavirus 2019-nCoV nucleic acid extraction kit (magnetic bead method) (Mingde, Wuhan, China) detection kit (fluorescence PCR) (Reagent No: SFDA: 20203400212, Wuhan Mingde Bio, Wuhan, China) for testing. This kit uses a one-step RT-PCR combined with Taqman technology to detect the novel coronavirus’s ORF1ab gene and N gene. If both the ORF1ab gene and the N gene are positive (Ct < 38), the test result is determined to be positive.
Continuous variables were expressed as a median and interquartile range, and categorical variables were expressed as counts and percentages. Mann–Whitney U test was used for comparison between the two groups for continuous variables, the Kruskal-Wallis test for multiple comparisons, and χ2 and Fisher test for categorical variables. All statistical analyses were performed using IBM SPSS 22.0 statistical software, and graphs were plotted using GraphPad 9.0. p ≤ 0.05 was defined as a statistically significant difference.
A total of 157 cases of Omicron variants infection and 144 cases of prototype strain infection were included in this study. Table 1 summarizes the clinical characteristics of the subjects, including gender, age, BMI, symptoms, and underlying diseases. There were 81 males (51.6%) and 76 females (48.4%) in the omicron group, 77 males (53.5%) and 76 females (46.5%) in the prototype strain group. The median age of patients infected with the omicron variant is younger than that of the prototype strain (35.0 vs. 47.0, p < 0.001). Patients infected with the omicron variant had a lower median BMI than the prototype strain (22.5 vs. 24.4, p = 0.015). Hypertension (6.3%) and diabetes mellitus (1.9%) were the most common underlying diseases for omicron variant infections, and most infected patients had no specific underlying diseases.
These results indicate a general population susceptibility to Omicron variants; about 50% of patients with Omicron variants infection had first symptoms of cough, fever, sore throat, and ground glass/splatter showed by the CT image; compared with patients with prototype strain infection, the proportion of patients with lung solid lesions was lower. Laboratory parameters showed higher lymphocyte counts and eGFR levels in Omicron variants-infected patients compared to prototype strain-infected patients, which also indicated that Omicron variants caused less damage to the immune system and renal function of infected patients. The laboratory test results are detailed in the Supplementary material.
Notably, the median time from onset to hospitalization (2 vs. 6) days and the median length of hospitalization (15 vs. 22) days of Omicron variants infected patients were shorter than those of the prototype strain.
We compared the changes of Ct values longitudinally among patients after Omicron variants and prototype strain infections, and the data showed that patients had the lowest Ct values at the point of admission (highest viral nucleic acid load) (Figure 2A); Omicron strain infected patients had the lowest Ct values at a median time of 2 (2–3) days from the onset to admission, while prototype strain group had the lowest Ct values at a median time of 6 (4–10) days from the onset to hospital admission (Figure 2A); the duration of Omicron variant infection was 15 (13–18) days, while that of patients infected with the prototype strain was 22 (13–28) days (Table 1). The above results suggest that the viral load of Omicron variants peaks 2–3 days after the onset, and the time for the test result to turn negative in Omicron variants infection is 6–10 days shorter than that in prototype strain infection. Further study found that the Omicron variant infection group had lower nucleic acid Ct values in nasopharyngeal swabs than in throat swabs, and the Ct values reached the same at 12 days.
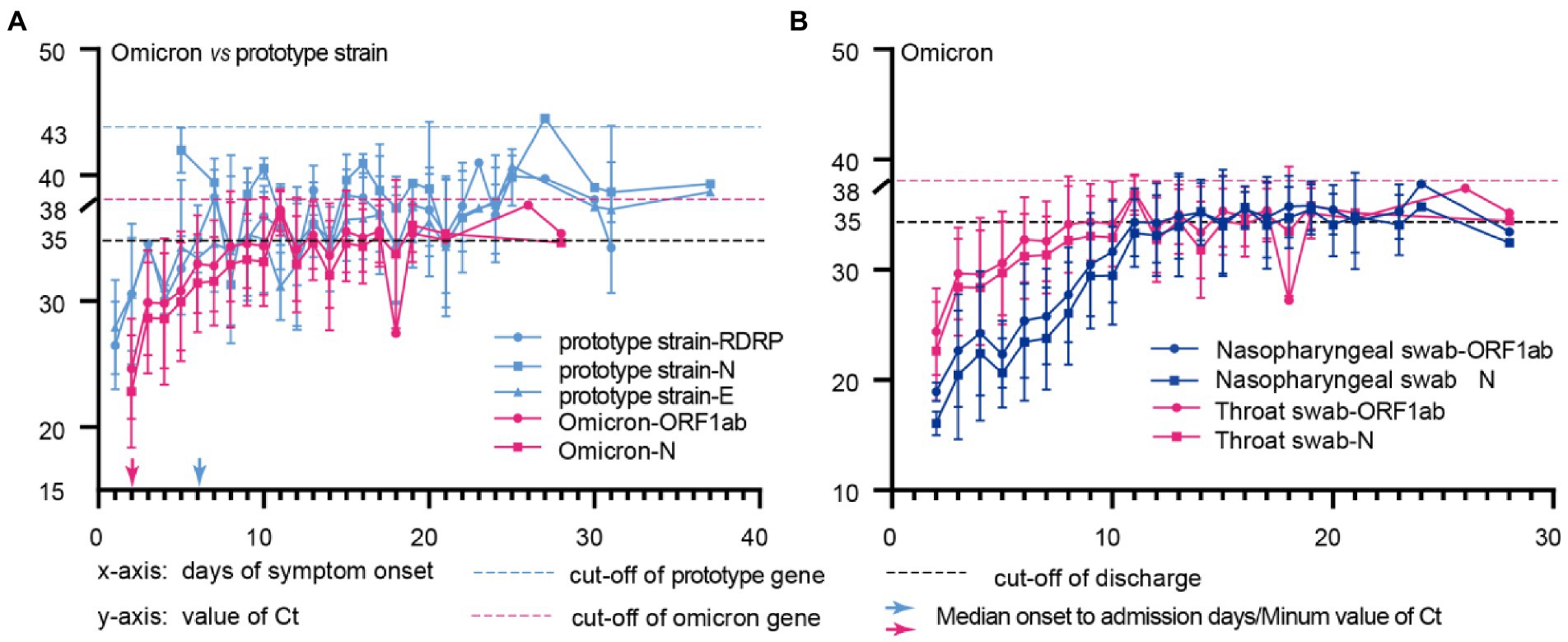
Figure 2. Comparison of the dynamic changes in Ct values of Omicron and prototype strain genes in different sample types. (A), Throat swab from patients with omicron or prototype strain infection; (B), Throat swabs and Nasopharyngeal swabs of Omicron infected patients; The x-axis represents the time of disease course since symptom onset, and the y-axis shows the Ct values for each feature of prototype strain(blue) and Omicron(red) in different sample types. The blue and red arrows represent the median days from onset to admission for omicron and prototype strain-infected patients.
We compared Ct values between patients’ nasopharyngeal and throat swabs at admission. The Ct values of nasopharyngeal swabs were lower than those of throat swabs (ORF1ab gene 21.72 vs. 29.67, p < 0.001; N gene 19.5 vs. 28.59, p < 0.001), and the latters fluctuated during the recovery period (Figure 2B). The above results suggest that the viral load of nasopharyngeal swabs is higher and stable in the early stage of viral infection. Therefore, nasopharyngeal swabs should be the primary samples for early detection. And Omicron variants should be diagnosed and quarantined as soon as symptoms appear to cut off the source of infection in the shortest possible time.
Approximately 90% of patients with domestic epidemic Omicron variant infections in China are asymptomatic (Zhang et al., 2022). We further investigated the dynamics of Ct values in symptomatic and asymptomatic individuals. In the Omicron variant cohort, 75 subjects provided the vaccination information. 2 (2.7%) patients were not vaccinated against the COVID-19 vaccine, 2(2.7%) patients were vaccinated with one dose, 27 (36.0%) patients were vaccinated with two doses, and 44 (58.7%) patients received a booster vaccination. The patient has no record of reinfection. We found that symptomatic ones with lower Ct values at admission early in the infection (ORF1ab gene, p = 0.02; N gene, p = 0.007; Figures 3A, 4A) had a longer length of stay than asymptomatic ones (15 vs. 13) days (p = 0.006; Figure 3; Table 1).
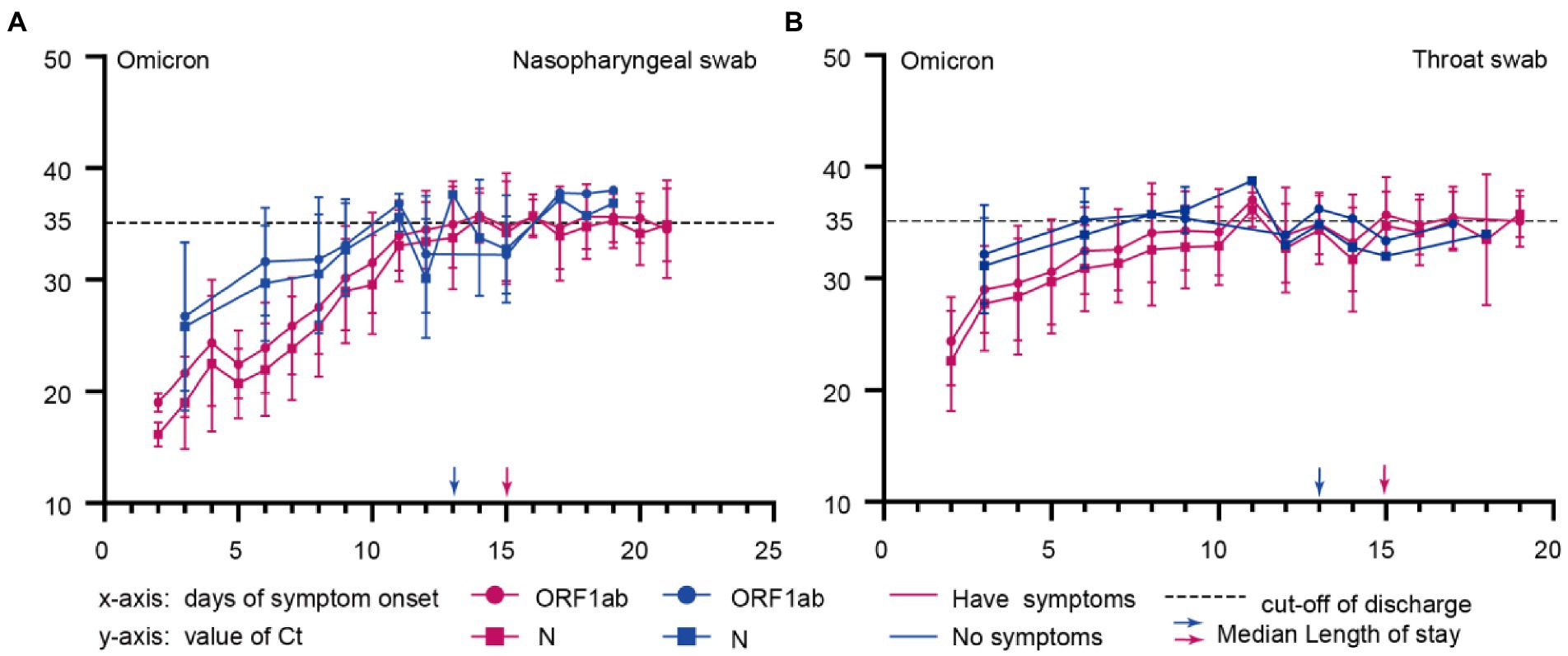
Figure 3. Comparison of Ct dynamic changes in omicron-infected patients with symptoms or no symptoms. (A), Nasopharyngeal swabs of Omicron infected patients with symptoms or no symptoms; (B), Throat swabs of Omicron infected patients with symptoms or no symptoms; The x-axis represents the time of disease course since symptom onset, and the y-axis shows the Ct values for Omicron infected patients with symptoms or without symptoms. Blue and red represent asymptomatic and symptomatic infected persons, respectively.
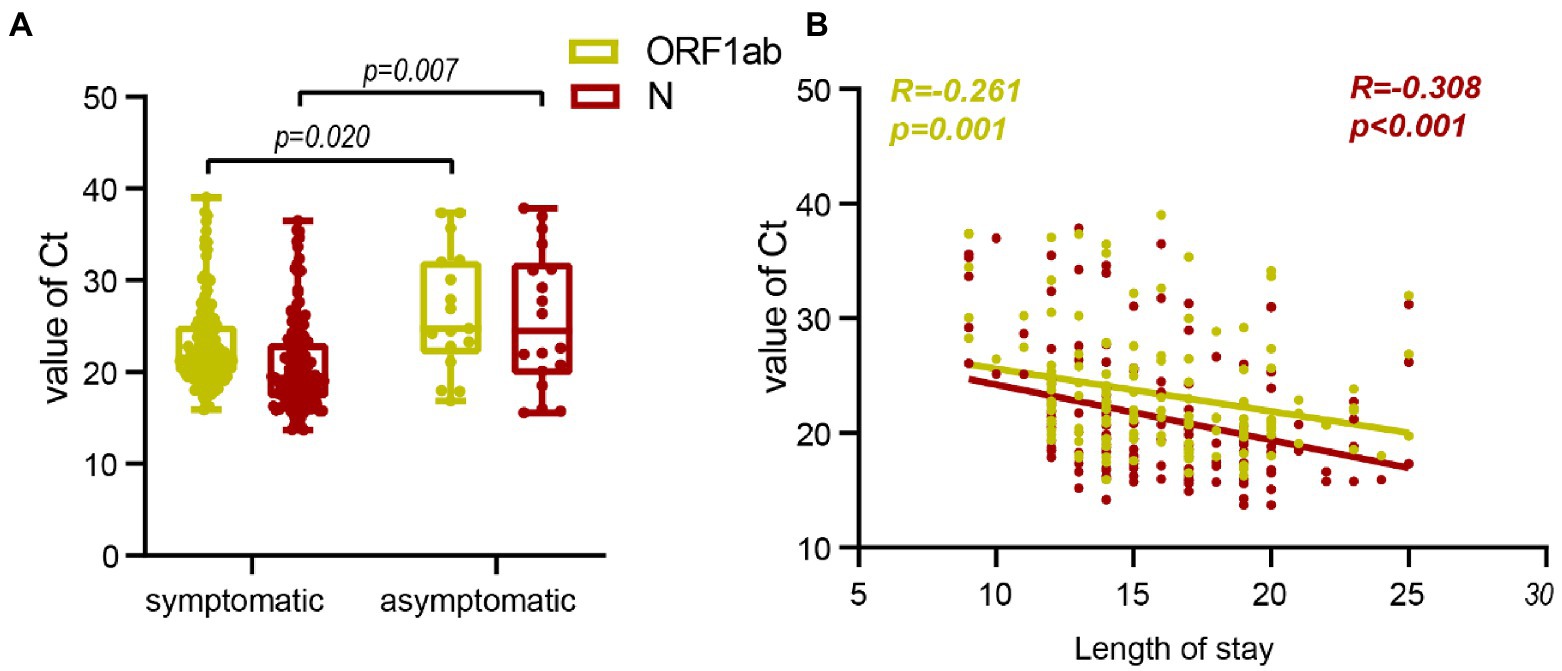
Figure 4. Correlation between Ct value and symptoms onset and length of stay. (A), Comparison of admission Ct values between patients with symptoms and asymptomatic patients. (B), Correlation between admission Ct value and length of stay.
Correlation analysis was conducted to further assess the difference between symptomatic and asymptomatic clinical indicators of Ct after Omicron variants infection. It showed a negative correlation between lower admission Ct values and longer length of stay (Figures 3, 4B).
We systematically described the clinical characteristics of 157 Omicron variants-infected patients and the dynamic changes of Omicron variants virus loads and disease progression in 1,723 multi-type samples. The dynamic changes of the viral loads after prototype strain infection were also compared. Our study found that the median time of peak viral replication in respiratory samples from Omicron variant-infected patients was 2 days after onset and the lowest point of nucleic acid Ct values. The viral load decreased as the disease progressed, with a viral duration of 15 (13–18) days in Omicron variants. And prototype strain infection, the peak time of viral replication was 6–10 days after the onset, which is consistent with a previous study by Zheng et al. (Zheng et al., 2020; Salvatore et al., 2021), the duration of viral shedding was 22 (13–28) days. Boucau et al. (2022) reported that the viral load started to decay from the first PCR positive or symptom onset. However, the duration of viral shedding in the Omicron variant in this study failed to be characterized as well.
Since Omicron variants replicate better in the upper respiratory tract than previous variants, it has been proposed that saliva or oral specimens may be more sensitive than nasopharyngeal samples in detecting Omicron (Marais et al., 2021; University of Hong Kong, 2021). Our study found that in the early stages of Omicron variants infection, the nucleic acid Ct value of nasopharyngeal swabs was lower than that of throat swabs. Both of them reached the same Ct value in 12–15 days of the disease, while Ct values of throat swabs were more likely to fluctuate during the recovery period, which may lead to false negatives in nucleic acid detection. This is detrimental to the prevention and control of the epidemic because of sampling errors and instability of throat swabs. Ct values of Nasopharyngeal swabs have been stable during the recovery period, and therefore nasopharyngeal swabs still have high analytical sensitivity and stability for RT-PCR (Matic et al., 2022). However, oral specimens are still an option, although attention should be paid to the lower viral load, especially when there may be difficulties in obtaining nasopharyngeal swabs (Williams et al., 2020).
The asymptomatic carrier rate of Omicron variants is much higher than that of other variants. In the outbreak of Omicron variants in Shanghai, 90.8% of infected patients were asymptomatic (Zhang et al., 2022). This high rate of asymptomatic infection may be a significant factor in this variant’s widespread and rapid global spread (Garrett et al., 2022; Zhang et al., 2022). The mean Ct value at 30.1 in asymptomatic Omicron patients, higher than that in symptomatic ones at 25.9, increased with the number of days of symptoms (Pollock et al., 2021; Schrom et al., 2022). We studied the admission Ct value of asymptomatic individuals and found that the admission Ct value was negatively correlated with the length of stay. This will help to strengthen the management of asymptomatic infection patients and shorten the time for nucleic acid transformation.
Several limitations should be considered when interpreting this study’s results. First, we only selected the subjects infected with the Omicron variant in one city as the study population. However, there may be differences between cities and regions. In addition, the sample size for our analysis is not large enough, which may affect the statistical power. Therefore, further generalization and external validity studies are needed to clarify the intrinsic mechanism.
In conclusion, the viral load of Omicron variants reached its peak value 2–3 days after the onset and the duration of virus was 15–18 days; The viral load of symptomatic patients was higher, and the overall length of hospitalization was longer, and the two indicators were negatively correlated; In the early stage of Omicron variants infection, nasopharyngeal swab test is still the first choice and shows better stability in the later stage. Supplementary material showed higher lymphocyte counts and eGFR levels in those infected with the Omicron variant compared to those infected with the prototype strain, suggesting that the Omicron variant causes less damage to the immune system and renal system function in those infected. This study provides a basis for the development of prevention and control strategies. Once symptoms appear, diagnosis and quarantine should be carried out as soon as possible, the source of infection should be cut off in the shortest time, and closed-loop management measures.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
This study was approved by the Ethics Committee of Taizhou Hospital, Zhejiang Province (K20210218). Informed consent was obtained from each subject. Minors’ participation in the study was approved by their parents and/or legal guardians.
HZ and BS supervised the project. BS, KZ, and T-HT designed the study. KZ organized the data and wrote the manuscript. KZ, BH, XZ, HC, JP, YZ, XB, MC, and JXu collected clinical data and provided clinical supervision. All authors contributed to the article and approved the submitted version.
This work is supported by the Zhejiang Provincial Medical and Health Science and Technology Program (2019RC089), Zhejiang Provincial Natural Science Foundation of China (Grant No. LTGY23H290007), Sichuan Provincial Natural Science Foundation of China (Grant No. 23NSFSC1861), and Scientific Research Fund of Zhejiang Provincial Education Department (Y202146073).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1037733/full#supplementary-material
Boucau, J., Marino, C., Regan, J., Uddin, R., Choudhary, M. C., Flynn, J. P., et al. (2022). Duration of viable virus shedding in SARS-CoV-2 omicron variant infection. medRxiv. doi: 10.1101/2022.03.01.22271582
Carreño, J. M., Alshammary, H., Tcheou, J., Singh, G., Raskin, A. J., Kawabata, H., et al. (2022). Activity of convalescent and vaccine serum against SARS-CoV-2 omicron. Nature 602, 682–688. doi: 10.1038/s41586-022-04399-5
Dejnirattisai, W., Huo, J., Zhou, D., Zahradník, J., Supasa, P., Liu, C., et al. (2022). SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cells 185, 467–484. doi: 10.1016/j.cell.2021.12.046
Fan, Y., Li, X., Zhang, L., Wan, S., Zhang, L., and Zhou, F. (2022). SARS-CoV-2 Omicron variant: recent progress and future perspectives. Signal Transduct. Target. Ther. 7:141. doi: 10.1038/s41392-022-00997-x
Garrett, N., Tapley, A., Andriesen, J., Seocharan, I., Fisher, L. H., Bunts, L., et al. (2022). High rate of asymptomatic carriage associated with variant strain omicron. medRxiv. doi: 10.1101/2021.12.20.21268130
Luo, C. H., Morris, C. P., Sachithanandham, J., Amadi, A., Gaston, D., Li, M., et al. (2021). Infection with the SARS-CoV-2 Delta variant is associated with higher infectious virus loads compared to the alpha variant in both unvaccinated and vaccinated individuals. MedRxiv. doi: 10.1101/2021.08.15.21262077
Mahase, E. (2020). China coronavirus: WHO declares international emergency as death toll exceeds 200. BMJ 368:m408. doi: 10.1136/bmj.m408
Marais, G., Iranzadeh, N. H. A., Doolabh, D., and Enoch, A. (2021). Saliva swabs are the preferred sample for Omicron detection. medRxiv. doi: 10.1101/2021.12.22.21268246
Matic, N., Lowe, C. F., Ritchie, G., Young, M., Lawson, T., and Romney, M. G. (2022). Omicron (B.1.1.529) SARS-CoV-2 viral load among nasopharyngeal and oral samples compared to other variants of concern and impact on diagnostic testing strategy. Clin. Microbiol. Infect. 28, 1302–1303. doi: 10.1016/j.cmi.2022.04.022
Pollock, N. R., Jacobs, J. R., Tran, K., Cranston, A. E., Smith, S., O'kane, C. Y., et al. (2021). Performance and implementation evaluation of the Abbott BinaxNOW rapid antigen test in a high-throughput drive-through community testing site in Massachusetts. J. Clin. Microbiol. 59:e00083-21. doi: 10.1128/JCM.00083-21
Salvatore, P. P., Dawson, P., Wadhwa, A., Rabold, E. M., Buono, S., Dietrich, E. A., et al. (2021). Epidemiological correlates of polymerase chain reaction cycle threshold values in the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 72, e761–e767. doi: 10.1093/cid/ciaa1469
Schrom, J., Marquez, C., Pilarowski, G., Wang, C. Y., Mitchell, A., Puccinelli, R., et al. (2022). Comparison of SARS-CoV-2 reverse transcriptase polymerase chain reaction and BinaxNOW rapid antigen tests at a community site during an omicron surge: a cross-sectional study. Ann. Intern. Med. 175, 682–690. doi: 10.7326/M22-0202
Shanghai Municipal Health Commission News release (2022). Available at: https://wsjkw.sh.gov.cn/xwfb/index.html
Teyssou, E., Delagrèverie, H., Visseaux, B., Lambert-Niclot, S., Brichler, S., Ferre, V., et al. (2021). The Delta SARS-CoV-2 variant has a higher viral load than the Beta and the historical variants in nasopharyngeal samples from newly diagnosed COVID-19 patients. J. Infect. 83, e1–e3. doi: 10.1016/j.jinf.2021.08.027
University of Hong Kong (2021). HKUMed finds omicron SARS-CoV-2 can infect faster and better than Delta in human bronchus but with less severe infection in lung, [online]. Available at: www.med.hku.hk/en/news/press/20211215-omicron-saes-cov-2-infection
Williams, E., Bond, K., Zhang, B., Putland, M., and Williamson, D. A. (2020). Saliva as a noninvasive specimen for detection of SARS-CoV-2. J. Clin. Microbiol. 58:e00776-20. doi: 10.1128/JCM.00776-20
Zhang, X., Zhang, W., and Chen, S. (2022). Shanghai's life-saving efforts against the current omicron wave of the COVID-19 pandemic. Lancet 399, 2011–2012. doi: 10.1016/S0140-6736(22)00838-8
Keywords: Omicron variant, nucleic acid Ct value, time to peak viral load, duration of this Omicron variant, asymptomatic infected patients
Citation: Zhou K, Hu B, Zhao X, Chi H, Pan J, Zheng Y, Bi X, Chen M, Xie J, Xu J, Tung T-H, Shen B and Zhu H (2023) Longitudinal observation of viral load in patients infected with Omicron variant and its relationship with clinical symptoms. Front. Microbiol. 13:1037733. doi: 10.3389/fmicb.2022.1037733
Received: 06 September 2022; Accepted: 28 December 2022;
Published: 13 January 2023.
Edited by:
Chenjian Gu, Fudan University, ChinaReviewed by:
Yili Fang, Shanghai General Hospital, ChinaCopyright © 2023 Zhou, Hu, Zhao, Chi, Pan, Zheng, Bi, Chen, Xie, Xu, Tung, Shen and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Shen, ✉ c2hlbmJAZW56ZW1lZC5jb20=; Hongguo Zhu, ✉ emh1aGdAZW56ZW1lZC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.