- 1Department of Laboratory Medicine, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Graduate School, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 3Beijing Key Laboratory for Mechanisms Research and Precision Diagnosis of Invasive Fungal Diseases (BZ0447), Beijing, China
- 4Department of Laboratory Medicine, Peking University Third Hospital, Beijing, China
- 5Medical Research Center, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 6Department of Laboratory Medicine, No.908 Hospital of Joint Logistics Support Force, Nanchang, Jiangxi, China
- 7Department of Laboratory Medicine, The Second Xiangya Hospital, Central South University, Changsha, Hubei, China
- 8Department of Infectious Diseases and Clinical Microbiology, Beijing Institute of Respiratory Medicine, Beijing Chao-Yang Hospital, Capital Medical University, Beijing, China
Candida haemulonii var. vulnera is a rare variant of C. haemulonii, which has been previously reported to cause human infections. Owing to the close kinship between C. haemulonii sensu stricto and C. haemulonii var. vulnera, accurate identification of C. haemulonii var. vulnera relied on DNA sequencing assay targeting, for example, rDNA internal transcribed spacer (ITS) region. In this work, two strains of C. haemulonii var. vulnera were collected from the China Hospital Invasive Fungal Surveillance Net (CHIF-NET). The identification capacity of three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and VITEK 2 YST ID biochemical methods were evaluated against ITS sequencing. In addition, antifungal susceptibility testing was performed using Sensititre YeastOne. Moreover, we comprehensively screened drug-resistant related genes by whole-genome sequencing. The two strains were not correctly identified to species variant level using MALDI-TOF MS and YST ID cards. Both strains were resistant to amphotericin B (minimum inhibitory concentration [MIC] > 2 μg/ml). Moreover, strain F4564 and F4584 exhibited high MIC to fluconazole (>256 μg/ml) and 5-flucytosine (>64 μg/ml), respectively, which were supposed to result from key amino acid substitutions Y132F and G307A in Erg11p and V58fs and G60K substitutions in Fur1p. The rare species C. haemulonii var. vulnera has emerged in China, and such drug-resistant fungal species that can cause invasive diseases require further close attention.
Introduction
Candida haemulonii var. vulnera belongs to the C. haemulonii species complex, along with C. haemulonii sensu stricto and C. duobushaemulonii (Gade et al., 2020; Rodrigues et al., 2021; Ramos et al., 2022). Generally, C. haemulonii species complex isolates display high multidrug-resistant rates and transmission properties, which have attracted increased attention (Gade et al., 2020).
In 2012, C. haemulonii var. vulnera was reported for the first time by Cendejas-Bueno et al. They found four strains with low rDNA internal transcribed spacer (ITS) sequence identity to C. haemulonii sensu stricto type strain (∼96%) (Cendejas-Bueno et al., 2012). Infections caused by C. haemulonii var. vulnera were later reported in Brazil, India, Argentina, and Peru (de Almeida et al., 2016; Kumar et al., 2016; Isla et al., 2017; Pérez-Lazo et al., 2021; Ramos et al., 2022). In addition, by retesting a set of previously collected strains, Rodrigues et al. found a C. haemulonii var. vulnera strain isolated in 2009, which became the earliest strain of the species variant discovered till now and its genome sequence was elucidated (Rodrigues et al., 2020). Moreover, antifungal resistance to azoles, echinocandins, and amphotericin B has been reported in C. haemulonii var. vulnera isolates (Cendejas-Bueno et al., 2012; Isla et al., 2017; Ramos et al., 2022).
In this study, we reported two fungemia cases caused by Candida haemulonii var. vulnera found from China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study. Clinical characters, identification capacity of Vitek YST card and three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems, isolates’ antifungal susceptibility phenotypes, and potential resistant mechanisms were illustrated. To our best knowledge, these were the first Candida haemulonii var. vulnera infection cases reported in China, including the first 5-flucytocine resistant strain discovered globally.
Materials and methods
Ethics statement
This study was approved by the Human Research Ethics Committee of the Peking Union Medical College Hospital (No. S-263). Written informed consent was obtained from all patients who participated in this study, which aimed to culture and study the isolates obtained from the patients.
Microorganisms and identification
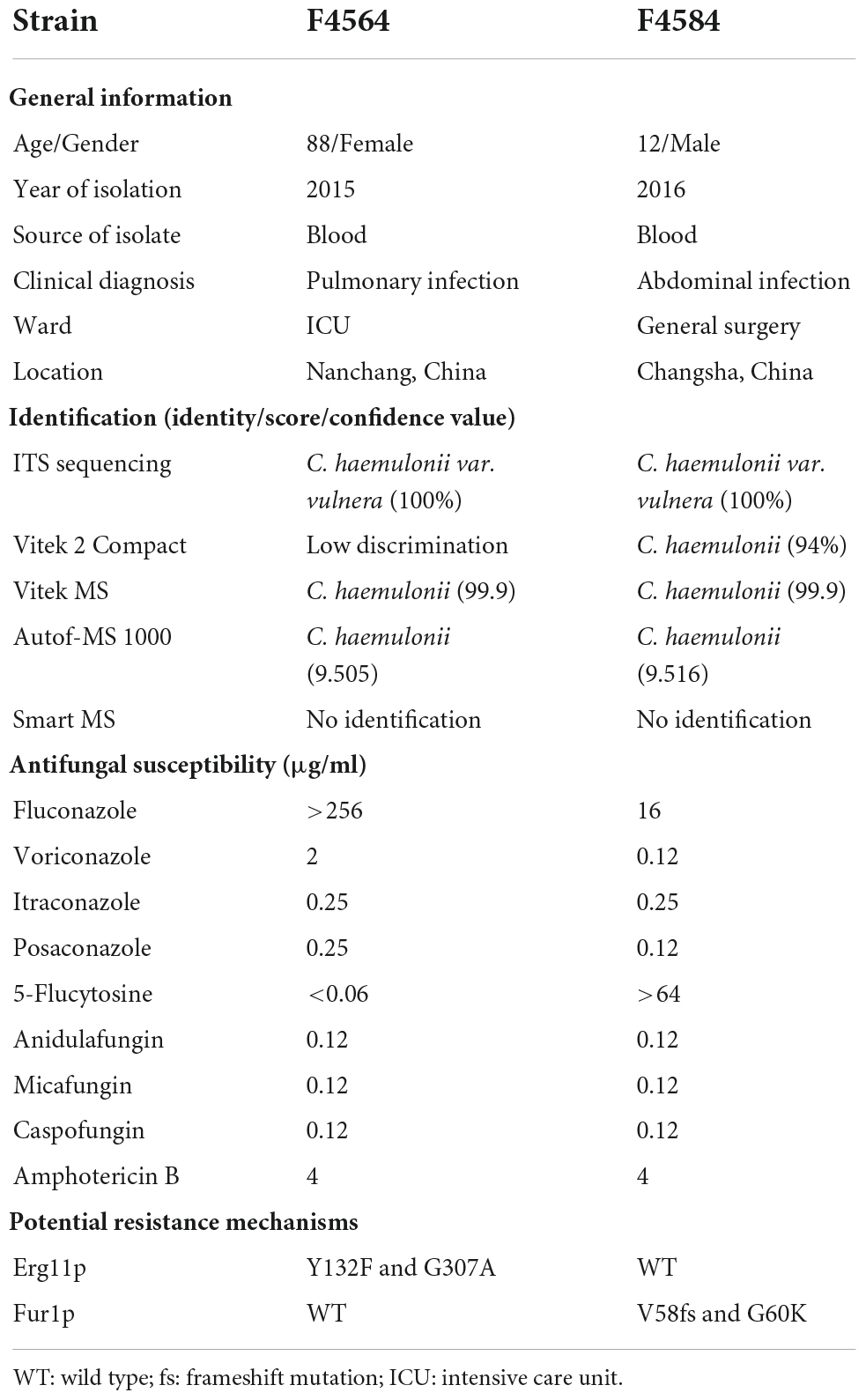
From 2010 to 2017 (Table 1), two C. haemulonii var. vulnera isolates were collected from different hospitals in two provinces from the CHIF-NET study. The colony morphology of two strains is smooth, moist, and circular, which is the typical appearance of Candida species. Gram-stained microscopy showed budding yeast cells (Figure 1). The isolates were identified using Autof MS 1000 (Autobio, Zhengzhou, China), Smart MS (DL, Zhuhai, China), and Vitek MS (bioMérieux, Marcy l’Étoile, France) MALDI-TOF MS systems, in addition to Vitek 2 YST ID Card using VITEK 2 (9.02 version, bioMérieux, Marcy-l’Etoile, France) following the manufacturer’s instructions. Primers ITS1 and ITS4 (Hou et al., 2016) were used for ITS amplification and sequencing, and Sanger sequencing was performed using ABI 3730XL DNA analyzer (Thermo Fisher Scientific, Cleveland, OH, USA). A phylogenetic tree of the ITS sequences was constructed using Mega X based on 1000 bootstrap replicates using the maximum likelihood method (Kumar et al., 2018).
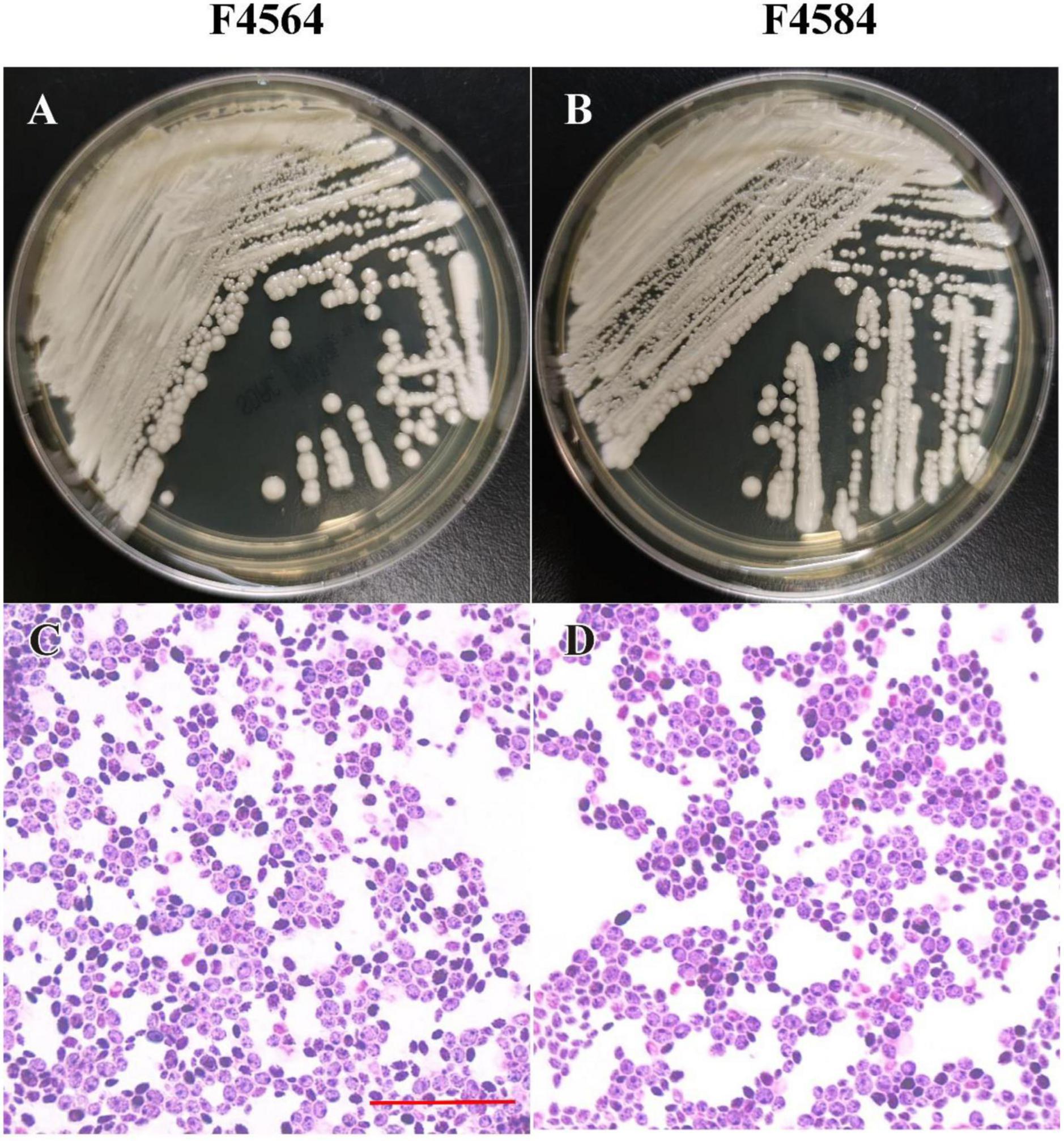
Figure 1. Phenotypic characteristics of two C. haemulonii var. vulnera isolates on Sabouraud dextrose agar. The plate contains colonies with typical appearance of circular, smooth, and moist (A,B). Microscopic examination showed budding yeast cells with Gram’s staining (C,D). Scale bar = 10 μm (C).
DNA extraction and whole-genome sequencing
The whole genomic DNA of C. haemulonii var. vulnera was extracted as previously reported (Huang et al., 2022). The 350-bp DNA library was constructed using NEBNext® Ultra™, following the manufacturer’s instructions. Library integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Sequencing was performed on an Illumina NovaSeq using the PE150 strategy (Beijing Novogene Bioinformatics Technology Co., Ltd.). The Illumina reads generated in this study were obtained from the National Center for Biotechnology Information (NCBI) under BioProject PRJNA890168.
Genome analysis
The C. haemulonii var. vulnera K1 (GenBank accession number GCA_012184645.1) genome was concatenated and used as a reference genome for read mapping. BWA 0.5.9, SAMtools, and bcftools 0.1.19 (Li and Durbin, 2009; Li et al., 2009) were used for single-nucleotide polymorphism (SNP) and insertion/deletion (indel) analysis, and snpEff 4.3 was used for SNP and indel function annotations (Cingolani et al., 2012). Mutations on antifungal-resistant related genes, including ERG11 and TAC1b for azoles, FUR1 for 5-flucytosine, FKS1 and FKS2 for echinocandins, ERG3 and ERG6 for amphotericin B, and ERG4 for other ergosterol pathway genes, were analyzed in detail (Arendrup and Patterson, 2017; Berkow and Lockhart, 2017).
Antifungal susceptibility testing
Antifungal susceptibility testing was performed using Sensititre YeastOne YO10 methodology (Thermo Scientific, Cleveland, OH, USA). Candida krusei ATCC 6258 and Candida parapsilosis ATCC 22019 were used as quality controls. As clinical breakpoints or epidemiological cutoff values against C. haemulonii var. vulnera have not been established, we used interpretation criteria for Candida species referring to CLSI M27-S3 guidelines (CLSI, 2008), including MIC value of ≥32 μg/mL as resistance to fluconazole ≥32 μg/mL as resistance to 5-flucytosine. In addition, MIC of ≥2 μg/mL was used for interpreting “resistance” to amphotericin B (Pfaller et al., 2012).
Review of Candida haemulonii var. vulnera cases reported
A literature review of previously reported C. haemulonii var. vulnera infections was done. A literature search was performed on 28 August 2022, using the following three databases: PubMed1, Web of Science2, and Embase3. The terms “Candida haemulonii var. vulnera” were entered in the category of “Title/Abstract” in the PubMed Advanced Search Builder, and “TS (Candida haemulonii var. vulnera)” was entered into the Web of Science database. The search in Embase was conducted in the advanced search area, including the terms “Candida haemulonii var. vulnera’: ab,ti.”. All hits were further screened manually to find out all infection cases with antifungal susceptibility reports.
Results
Isolate information
The first case was an 88-year-old male patient who was admitted to the intensive care unit (ICU) with clinical diagnosis of pulmonary infection. F4564 was isolated from peripheral blood of this patient and initially misidentified as “Candida krusei” using CHROMagar chromogenic medium at local laboratory (Table 1). The second case was a 12-year-old boy who was admitted to the general surgical ward, and clinical diagnosis was abdominal infection. F4584 was isolated from peripheral blood of this patient and identified as “Candida spp.” using CHROMagar chromogenic medium initially (Table 1).
Identification of Candida. haemulonii var. vulnera using ITS sequencing, MALDI-TOF MS, and Vitek 2
The ITS sequences of the isolates exhibited 100% identity with the corresponding ITS sequences of the reference C. haemulonii var. vulnera CBS12439T isolates (GenBank accession number: JX459686.1). Furthermore, both clinical isolates were identified as C. haemulonii by the Autof MS 1000 and Vitek MS, but with “no identification” results by Smart MS. While using the Vitek 2 Compact system, one strain was identified with low discrimination and the other was identified as C. haemulonii (score = 94%) (Table 1). In phylogenetic tree generated by C. haemulonii complex, C. auris, and C. pseudohaemulonii ITS sequences, F4564 and F4584 were in the branch of C. haemulonii var. vulnera (Figure 2).
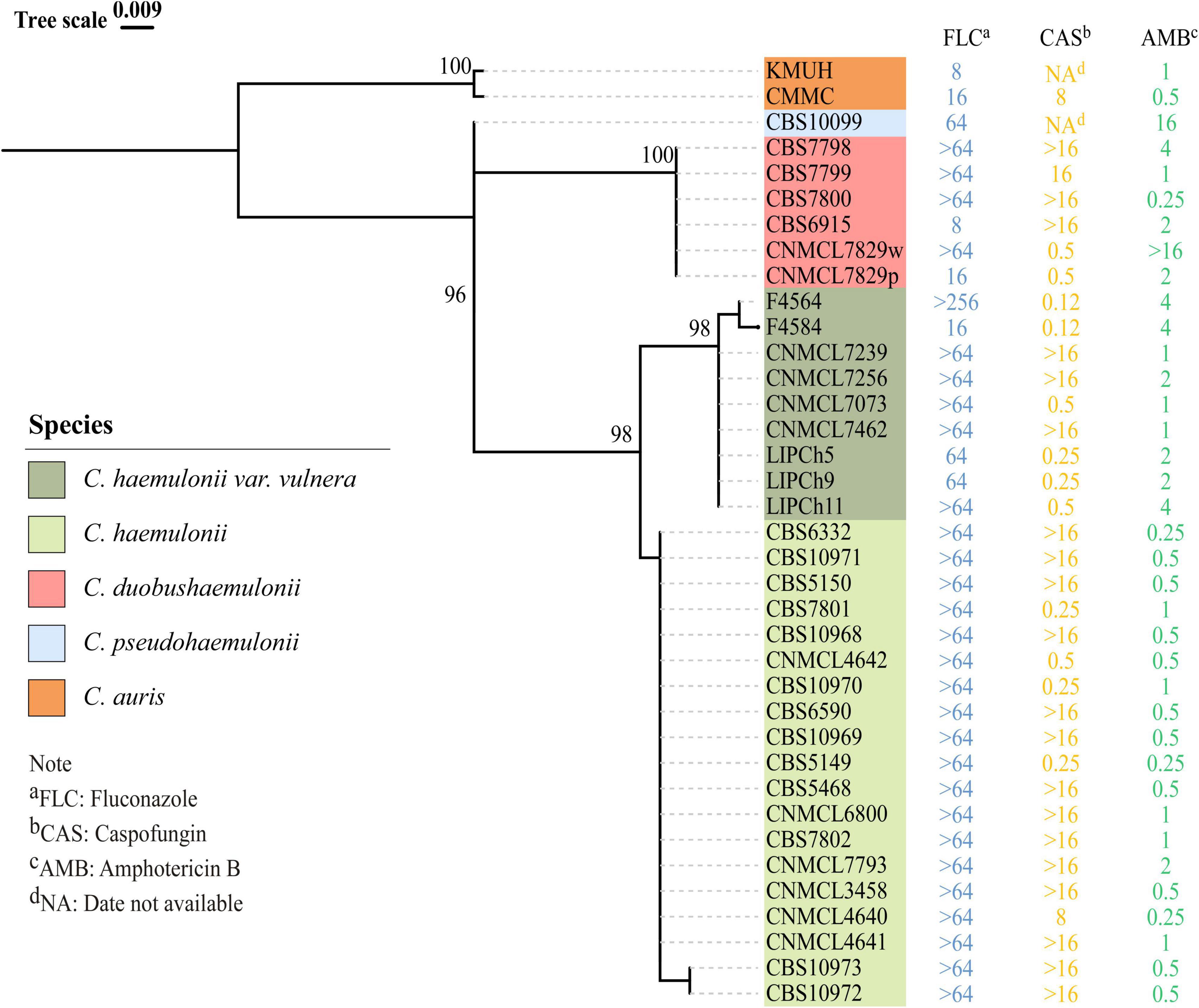
Figure 2. Phylogenetic tree for rDNA ITS region of previously reported Candida haemulonii, C. duobushaemulonii, C. pseudohaemulonii, C. auris, and C. haemulonii var. vulnera, including two C. haemulonii var. vulnera clinical strains identified in this study. Phylogenetic tree was constructed using Mega X with 1000 bootstrap replicates based on the maximum likelihood method.
Genome analysis
We sequenced genomes of F4564 and F4584, and their total genome sizes were 13.51 Mb and 13.26 Mb, respectively, with an average GC content of 45% (N50 was 346,464 bp and 283,647 bp, and a number of assembled contigs were 270 and 262, comprising 95.7% and 95.6% of data according to BUSCO analysis, respectively).
Antifungal susceptibility
MIC values obtained for the nine antifungal agents against C. haemulonii var. vulnera isolates are shown in Table 1. F4564 was resistant to fluconazole with an MIC of >256 μg/ml. In addition, F4564 was classified as susceptible dose-dependent to voriconazole (2 μg/ml) and itraconazole (0.25 μg/ml). F4684 was resistant to 5-flucytosine (>64 μg/ml) and susceptible dose-dependent to itraconazole. In addition, both isolates had high MIC values of 4 μg/ml for amphotericin B, while neither strain was non-susceptible to posaconazole, caspofungin, anidulafungin, and micafungin (Table 1).
Potential genomic variations contributed to antifungal resistance
Compared with the reference genome K1, which is from an isolate susceptible to all antifungals except for amphotericin B, we found that F4564 has two previously reported key amino acid substitutions (Y132F and G307A) in Erg11p. Interestingly, we discovered two novel mutations in Fur1p (V58fs and G60K) in strain F4584 with high 5-flucytosine MIC. However, we did not find any key variation in sterol metabolism-related genes like ERG2, ERG3, and ERG6 that may result in amphotericin B resistance. In addition, all strains were susceptible to echinocandins and did not carry substitutions in Fks1p or Fks2p.
Literature review
We found eight articles in all reported antifungal susceptibility of C. haemulonii var. vulnera isolates. A number of strains exhibit high MICs to fluconazole or even all azoles (Cendejas-Bueno et al., 2012), but there was not any investigation on related resistance mechanisms. In addition, one study reported the emergence of pan-echinocandin-resistant C. haemulonii var. vulnera strains (Cendejas-Bueno et al., 2012). To date, there have not been any reports describing 5-flucytosine-resistant C. haemulonii var. vulnera strain (Table 2).
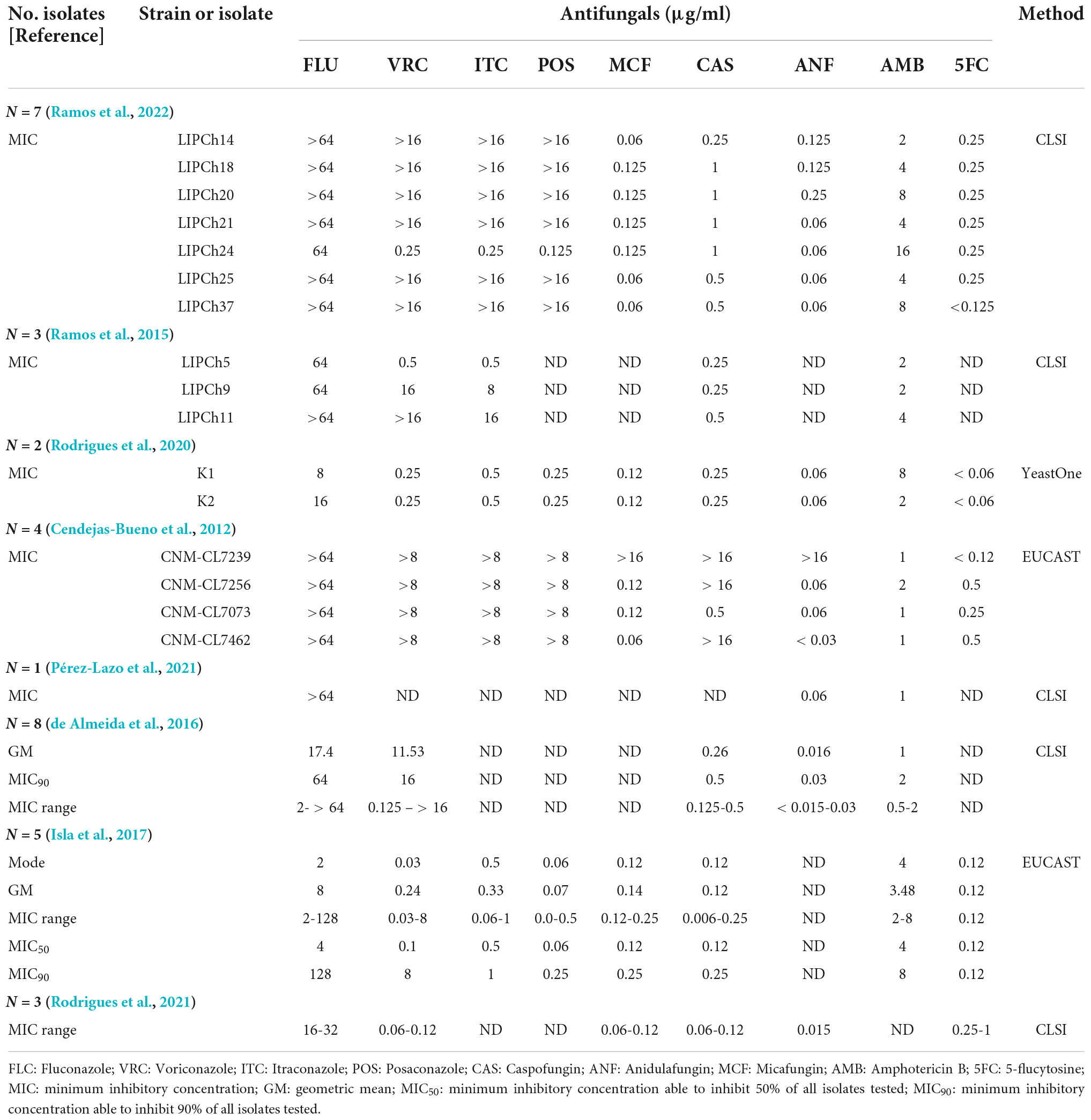
Table 2. Overview of published reports on antifungal susceptibility profiles of Candida haemulonii var. vulnera.
Discussion
Candida haemulonii var. vulnera was identified ever-first by ITS sequencing (Cendejas-Bueno et al., 2012). Although it has been involved in identification database of latest Vitek 2 YST ID system, the results from the current study and Rodrigues et al. (2020) still found YST ID card was not able to achieve a reliable identification between C. haemulonii sensu stricto and C. haemulonii var. vulnera. Furthermore, none of the current available MALDI-TOF MS systems could accurately identify C. haemulonii var. vulnera (Kathuria et al., 2015; Grenfell et al., 2016; Rodrigues et al., 2020), though Grenfell et al. described that some discriminatory protein peaks may be able to differentiate C. haemulonii sensu stricto and C. haemulonii var. vulnera in using FlexAnalysis and ClinProTools to analysis MALDI-TOF MS results (Grenfell et al., 2016). Therefore, DNA sequencing remained the only applicable methods for the identification of C. haemulonii var. vulnera. Interestingly, Cendejas-Bueno reported that C. haemulonii sensu stricto and C. haemulonii var. vulnera can be separated by ITS sequences, but these two species have identical RPB1, RPB2, and D1/D2 sequences. Therefore, these strategies could not be used for the identification of these two species (Cendejas-Bueno et al., 2012). To date, isolation of C. haemulonii var. vulnera has only been reported in Brazil, Argentina, and Peru. Due to the above-mentioned limitations of commercial identification systems, we reidentified all C. haemulonii complex isolates collected in CHIF-NET study by ITS sequencing and finally found two C. haemulonii var. vulnera strains among >80 cases.
Previously, fluconazole- and amphotericin B-resistant C. haemulonii var. vulnera have been discovered (Cendejas-Bueno et al., 2012; Ramos et al., 2015; de Almeida et al., 2016). Like findings in previous reports, one of the isolates we discovered was also resistant to fluconazole and amphotericin B. However, we also found a 5-flucytosine-resistant strain, and it is also cross-resistant to amphotericin B. Although 5-flucytosine resistance has been recognized in C. haemulonii sensu stricto and C. duobushaemulonii in China and other regions (Kathuria et al., 2015; Hou et al., 2016), this is the first 5-flucytosine-resistant C. haemulonii var. vulnera case characterized to date. Of note, both strains we discovered remained susceptible to all echinocandins.
Compared with C. haemulonii sensu stricto and C. duobushaemulonii that have been well characterized, there are very few studies on resistant mechanisms of C. haemulonii var. vulnera. Key amino acid substitutions in Erg11p and Fur1p have been noted as major reasons contributing to azole and 5-flucytosine resistance, respectively, in other C. haemulonii complex species (Gade et al., 2020) and Chen et al. (2022). In this study, it was found that the fluconazole-resistant C. haemulonii var. vulnera isolate carried Y132F and G307A mutations in Erg11p. Erg11p Y132F substitutions have been reported in a broad range of fluconazole-resistant Candida species including C. albicans, C. tropicalis, and C. haemulonii (Fan et al., 2019; Warrilow et al., 2019; Gade et al., 2020), while G307A has been reported in C. parapsilosis (Arastehfar et al., 2020; Binder et al., 2020). While in our 5-flucytosine-resistant C. haemulonii var. vulnera strain, a frameshift V58fs and a mutation G60K were found in Fur1p, which has not been recovered in any other Candida strains to our best knowledge. Literature review showed a high proportion of C. haemulonii var. vulnera isolates (13/17, 76.5%) were with reduced susceptibility to amphotericin B (≥2 μg/ml). However, resistant mechanisms to amphotericin B were not well understood, and in this study, we also failed to found any mutations in key genes may potentially contributed to amphotericin B resistance. As a major limitation, our study did not collect detailed medical records of these two cases; therefore, we were not able to further analyze the source of infections or assessing clinical risk factors of this species.
In conclusion, there is a potential threat posed by C. haemulonii var. vulnera, a highly antifungal-resistant fungal species. Resistance to fluconazole and 5-flucytosine in C. haemulonii var. vulnera was supposed to be resulted from variations in Erg11p and Fur1p, respectively. Although invasive infection with C. haemulonii var. vulnera remained rare, further monitoring of this specie is still warranted.
Data availability statement
The datasets presented in this study can be found in the online repositories. The names of the repository/repositories and accession numbers(s) can be found in the manuscript.
Author contributions
X-FC, XH, HZ, and X-MJ conceived and designed the experiments. L-PN and WC provided the isolates. X-FC, HZ, X-MJ, XF, XH, J-JH, W-HY, GZ, J-JZ, and WK performed experiments. X-FC and HZ analyzed the data and wrote the manuscript. MX, XH, and Y-CX revised the manuscript. All authors contributed to the manuscript and approved the submitted version.
Funding
This study was supported by the National Key Research and Development Program of China (2022YFC2300017), National Natural Science Foundation of China (82002178), National High Level Hospital Clinical Research Funding (2022-PUMCH-B-074), Tsinghua University-Peking Union Medical College Hospital Initiative Scientific Research Program (Grant No. 20191080604), Beijing Key Clinical Specialty for Laboratory Medicine-Excellent Project (No. ZK201000), and CAMS Innovation Fund for Medical Sciences (2021-I2M-1-038 and 2021-I2M-1-044).
Acknowledgments
The authors thank all Hospitals involved in the CHIF-NET study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Arastehfar, A., Daneshnia, F., Hilmioglu-Polat, S., Fang, W., Yasar, M., Polat, F., et al. (2020). First report of candidemia clonal outbreak caused by emerging fluconazole-resistant Candida parapsilosis isolates harboring Y132F and/or Y132F+K143R in Turkey. Antimicrob. Agents Chemother. 64, e01001-20. doi: 10.1128/AAC.01001-20
Arendrup, M. C., and Patterson, T. F. (2017). Multidrug-resistant candida: Epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 216, S445–S451. doi: 10.1093/infdis/jix131
Berkow, E. L., and Lockhart, S. R. (2017). Fluconazole resistance in Candida species: A current perspective. Infect. Drug Resist. 10, 237–245. doi: 10.2147/IDR.S118892
Binder, U., Arastehfar, A., Schnegg, L., Hortnagl, C., Hilmioglu-Polat, S., Perlin, D. S., et al. (2020). Efficacy of LAMB against emerging azole- and multidrug-resistant Candida parapsilosis isolates in the Galleria mellonella model. J. Fungi (Basel, Switzerland) 6:377. doi: 10.3390/jof6040377
Cendejas-Bueno, E., Kolecka, A., Alastruey-Izquierdo, A., Theelen, B., Groenewald, M., Kostrzewa, M., et al. (2012). Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: Three multiresistant human pathogenic yeasts. J. Clin. Microbiol. 50, 3641–3651. doi: 10.1128/jcm.02248-12
Chen, X.-F., Zhang, H., Jia, X.-M., Cao, J., Li, L., Hu, X.-L., et al. (2022). Antifungal susceptibility profiles and drug resistance mechanisms of clinical Candida duobushaemulonii isolates from China. Front. Microbiol. doi: 10.3389/fmicb.2022.1001845
Cingolani, P., Platts, A., Wang le, L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92. doi: 10.4161/fly.19695
CLSI (2008). Reference method for broth dilution anti fungal susceptibility testing of yeasts, third informational supplement. CLSI document M27-S3, 3rd Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
de Almeida, J. N. Jr., Assy, J. G., Levin, A. S., Del Negro, G. M., Giudice, M. C., Tringoni, M. P., et al. (2016). Candida haemulonii Complex Species, Brazil, January 2010-March 2015. Emerg. Infect. Dis. 22, 561–563. doi: 10.3201/eid2203.151610
Fan, X., Xiao, M., Zhang, D., Huang, J. J., Wang, H., Hou, X., et al. (2019). Molecular mechanisms of azole resistance in Candida tropicalis isolates causing invasive candidiasis in China. Clin. Microbiol. Infect. 25, 885–891. doi: 10.1016/j.cmi.2018.11.007
Gade, L., Munoz, J. F., Sheth, M., Wagner, D., Berkow, E. L., Forsberg, K., et al. (2020). Understanding the emergence of multidrug-resistant Candida: Using whole-genome sequencing to describe the population structure of Candida haemulonii species complex. Front. Genet. 11:554. doi: 10.3389/fgene.2020.00554
Grenfell, R. C., Da Silva Junior, A. R., Del Negro, G. M. B., Munhoz, R. B., Gimenes, V. M. F., Assis, D. M., et al. (2016). Identification of Candida haemulonii complex species: Use of ClinProToolsTM to overcome limitations of the bruker BiotyperTM, VITEK MSTM IVD, and VITEK MSTM RUO databases. Front. Microbiol. 7:940. doi: 10.3389/fmicb.2016.00940
Hou, X., Xiao, M., Chen, S. C., Wang, H., Cheng, J. W., Chen, X. X., et al. (2016). Identification and antifungal susceptibility profiles of Candida haemulonii species complex clinical isolates from a multicenter study in China. J. Clin. Microbiol. 54, 2676–2680. doi: 10.1128/JCM.01492-16
Huang, J. J., Chen, X. F., Tsui, C. K. M., Pang, C. J., Hu, Z. D., Shi, Y., et al. (2022). Persistence of an epidemic cluster of Rhodotorula mucilaginosa in multiple geographic regions in China and the emergence of a 5-flucytosine resistant clone. Emerg. Microbes Infect. 11, 1079–1089. doi: 10.1080/22221751.2022.2059402
Isla, G., Taverna, C. G., Szusz, W., Vivot, W., García-Effron, G., and Davel, G. (2017). Candida haemulonii sensu lato: Update of the determination of susceptibility profile in Argentina and literature review. Curr. Fungal Infect. Rep. 11, 203–208. doi: 10.1007/s12281-017-0300-y
Kathuria, S., Singh, P. K., Sharma, C., Prakash, A., Masih, A., Kumar, A., et al. (2015). Multidrug-resistant Candida auris misidentified as Candida haemulonii: Characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and etest method. J. Clin. Microbiol. 53, 1823–1830. doi: 10.1128/jcm.00367-15
Kumar, A., Prakash, A., Singh, A., Kumar, H., Hagen, F., Meis, J. F., et al. (2016). Candida haemulonii species complex: An emerging species in India and its genetic diversity assessed with multilocus sequence and amplified fragment-length polymorphism analyses. Emerg. Microbes Infect. 5:e49. doi: 10.1038/emi.2016.49
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Pérez-Lazo, G., Morales-Moreno, A., Soto-Febres, F., Hidalgo, J. A., Neyra, E., and Bustamante, B. (2021). Liver abscess caused by Candida haemulonii var. vulnera. First case report in Peru. Rev. Iberoam Micol. 38, 138–140. doi: 10.1016/j.riam.2020.12.001
Pfaller, M. A., Espinel-Ingroff, A., Canton, E., Castanheira, M., Cuenca-Estrella, M., Diekema, D. J., et al. (2012). Wild-type MIC distributions and epidemiological cutoff values for amphotericin B, flucytosine, and itraconazole and Candida spp. as determined by CLSI broth microdilution. J. Clin. Microbiol. 50, 2040–2046. doi: 10.1128/JCM.00248-12
Ramos, L. S., Figueiredo-Carvalho, M. H. G., Barbedo, L. S., Ziccardi, M., Chaves, A. L. S., Zancopé-Oliveira, R. M., et al. (2015). Candida haemulonii complex: Species identification and antifungal susceptibility profiles of clinical isolates from Brazil. J. Antimicrob. Chemother. 70, 111–115. doi: 10.1093/jac/dku321
Ramos, L. S., Figueiredo-Carvalho, M. H. G., Silva, L. N., Siqueira, N. L. M., Lima, J. C., Oliveira, S. S., et al. (2022). The threat called Candida haemulonii species complex in Rio de Janeiro State, Brazil: Focus on antifungal resistance and virulence attributes. J. Fungi (Basel, Switzerland) 8:574. doi: 10.3390/jof8060574
Rodrigues, D. K. B., Bonfietti, L. X., Garcia, R. A., Araujo, M. R., Rodrigues, J. S., Gimenes, V. M. F., et al. (2021). Antifungal susceptibility profile of Candida clinical isolates from 22 hospitals of São Paulo State, Brazil. Braz. J. Med. Biol. Res. 54:e10928. doi: 10.1590/1414-431X2020e10928
Rodrigues, L. S., Gazara, R. K., Passarelli-Araujo, H., Valengo, A. E., Pontes, P. V. M., Nunes-da-Fonseca, R., et al. (2020). First genome sequences of two multidrug-resistant Candida haemulonii var. vulnera isolates from pediatric patients with candidemia. Front. Microbiol. 11:1535. doi: 10.3389/fmicb.2020.01535
Warrilow, A. G., Nishimoto, A. T., Parker, J. E., Price, C. L., Flowers, S. A., Kelly, D. E., et al. (2019). The evolution of azole resistance in Candida albicans sterol 14alpha-demethylase (CYP51) through incremental amino acid substitutions. Antimicrob. Agents Chemother. 63:e02586-18. doi: 10.1128/AAC.02586-18
Keywords: Candida haemulonii var. vulnera, antifungal susceptibility, ERG11, FUR1, whole-genome sequence, drug resistant mechanisms
Citation: Chen X-F, Hou X, Zhang H, Jia X-M, Ning L-P, Cao W, Fan X, Huang J-J, Yang W-H, Zhang G, Zhang J-J, Kang W, Xiao M and Xu Y-C (2022) First two fungemia cases caused by Candida haemulonii var. vulnera in China with emerged antifungal resistance. Front. Microbiol. 13:1036351. doi: 10.3389/fmicb.2022.1036351
Received: 04 September 2022; Accepted: 17 October 2022;
Published: 10 November 2022.
Edited by:
Wenjie Fang, Shanghai Changzheng Hospital, ChinaReviewed by:
Macit Ilkit, Çukurova University, TurkeyLászló Majoros, University of Debrecen, Hungary
Copyright © 2022 Chen, Hou, Zhang, Jia, Ning, Cao, Fan, Huang, Yang, Zhang, Zhang, Kang, Xiao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Xiao, Y2p0Y3hpYW9tZW5nQGFsaXl1bi5jb20=; Ying-Chun Xu, eHljcHVtY2hAMTM5LmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Xin-Fei Chen
Xin-Fei Chen Xin Hou
Xin Hou Han Zhang
Han Zhang Xin-Miao Jia
Xin-Miao Jia Li-Ping Ning6
Li-Ping Ning6 Xin Fan
Xin Fan Jing-Jing Huang
Jing-Jing Huang Meng Xiao
Meng Xiao Ying-Chun Xu
Ying-Chun Xu