
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 October 2022
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1036042
This article is part of the Research Topic The Potential Role of Gut Microbiome in Animal Gut-Linked Diseases View all 16 articles
 Xiushuang Chen1,2†
Xiushuang Chen1,2† Qinghui Kong3†
Qinghui Kong3† Xiaoxiao Zhao1,2
Xiaoxiao Zhao1,2 Chenxi Zhao1,2
Chenxi Zhao1,2 Pin Hao1,2
Pin Hao1,2 Irfan Irshad4
Irfan Irshad4 Hongjun Lei1,2
Hongjun Lei1,2 Muhammad Fakhar-e-Alam Kulyar5,6
Muhammad Fakhar-e-Alam Kulyar5,6 Zeeshan Ahmad Bhutta7
Zeeshan Ahmad Bhutta7 Hassan Ashfaq4
Hassan Ashfaq4 Qiang Sha8
Qiang Sha8 Kun Li1,2*
Kun Li1,2* Yi Wu1,2*
Yi Wu1,2*Diarrhea is a word-widely severe disease coupled with gastrointestinal dysfunction, especially in cattle causing huge economic losses. However, the effects of currently implemented measures are still not enough to prevent diarrhea. Previously we found that dropped short-chain fatty acids in diarrhea yaks, and butyrate is commonly known to be related to the epithelial barrier function and intestinal inflammation. However, it is still unknown whether sodium acetate/sodium butyrate could alleviate diarrhea in animals. The present study is carried out to explore the potential effects of sodium acetate/sodium butyrate on lipopolysaccharide-induced diarrhea in mice. Fifty ICR mice were randomly divided into control (C), LPS-induced (L), and sodium acetate/sodium butyrate (D, B, A)-treated groups. Serum and intestine samples were collected to examine inflammatory cytokines, antioxidant levels, relative gene expressions via real-time PCR assay, and gut microbiota changes through high-throughput sequencing. Results indicated that LPS decreased the villus height (p < 0.0001), increased the crypt depth (p < 0.05), and lowered the villus height to crypt depth ratio (p < 0.0001), while sodium acetate/sodium butyrate supplementation caused a significant increase in the villus height (p < 0.001), decrease in the crypt depth (p < 0.01), and increase in the villus height to crypt depth ratio (p < 0.001), especially. In mice treated with LPS, it was found that the serum level of IL-1β, TNF-α (p < 0.001), and MDA (p < 0.01) was significantly higher; however, sodium acetate/sodium butyrate supplementation significantly reduced IL-1β (p < 0.001), TNF-α (p < 0.01), and MDA (p < 0.01), respectively. A total of 19 genera were detected among mouse groups; LPS challenge decreased the abundance of Lactobacillus, unidentified F16, unidentified_S24-7, Adlercreutzia, Ruminococcus, unclassified Pseudomonadales, [Ruminococcus], Acetobacter, cc 1, Rhodococcus, unclassified Comamonadaceae, Faecalibacterium, and Cupriavidus, while increased Shigella, Rhodococcus, unclassified Comamonadaceae, and unclassified Pseudomonadales in group L. Interestingly, sodium acetate/sodium butyrate supplementation increased Lactobacillus, unidentified F16, Adlercreutzia, Ruminococcus, [Ruminococcus], unidentified F16, cc 115, Acetobacter, Faecalibacterium, and Cupriavidus, while decreased Shigella, unclassified Enterobacteriaceae, unclassified Pseudomonadales, Rhodococcus, and unclassified Comamonadaceae. LPS treatment upregulated the expressions of ZO-1 (p < 0.01) and NLRP3 (p < 0.0001) genes in mice; however, sodium acetate/sodium butyrate solution supplementation downregulated the expressions of ZO-1 (p < 0.05) and NLRP3 (p < 0.05) genes in treated mice. Also, the LPS challenge clearly downregulated the expression of Occludin (p < 0.001), Claudin (p < 0.0001), and Caspase-1 (p < 0.0001) genes, while sodium acetate/sodium butyrate solution supplementation upregulated those gene expressions in treated groups. The present study revealed that sodium acetate/sodium butyrate supplementation alleviated LPS-induced diarrhea in mice via enriching beneficial bacterium and decreasing pathogens, which could regulate oxidative damages and inflammatory responses via NLRP3/Caspase-1 signaling. The current results may give insights into the prevention and treatment of diarrhea.
Diarrhea is a severe disease coupled with gastrointestinal dysfunction that has a global impact on fertility rate, milk production, and immunity in livestock (Coura et al., 2015; Li et al., 2022). Nowadays diarrhea in dairy cows and yak is very serious. It has a high incidence rate, especially neonatal calf diarrhea is found usually with high morbidity and mortality, causing considerable economic damage to the industry due to the heavy treatment expenses and impairments in the growth of animal (Coura et al., 2015; Schmoeller et al., 2021). Despite measures such as improved hygiene and scientific feeding management with the use of extensive drugs, this disease, i.e., diarrhea, remains serious (Li et al., 2022). The imbalance in gut microbiota was commonly recognized as the primary cause of diarrhea (Schmoeller et al., 2021), and many studies found changed intestine microbiota in diarrhea of cattle (Chuang et al., 2022; Coelho et al., 2022; Li et al., 2022; Liu J. et al., 2022).
Gut microflora is composed of millions of microorganisms that contribute remarkably to physiological processes, i.e., functions of nutrition absorption, metabolism, and immunity of the host by producing various metabolites (Wei et al., 2020). The anaerobic bacterial fermented short-chain fatty acids (SCFAs) are six carbon-containing fatty acids in the gut (Du et al., 2021). Acetate and butyrate are mainly produced through bacterial catabolism of dietary fibers in the host colon (Ezzine et al., 2022), which are a primary source of energy for colonic epithelial cells (Fu et al., 2019). Previous studies found that acetate could promote small intestinal barrier function in mice (Yosi et al., 2022) and regulate IgA reactivity (Takeuchi et al., 2021). As an important short-chain fatty acid, butyrate can not only provide energy for enterocyte regeneration but also modulate the intestine microbial community and contribute to the host’s health (Jiminez et al., 2017). Some of the studies reported that butyrate has an important role in the proper functioning of the immune system, nervous system, and energy metabolism (Koh et al., 2016; Fu et al., 2019). It was observed in the previous study that intestinal disease like ulcerative colitis is highly related to inadequate use of butyrate (Leonel and Alvarez-Leite, 2012). Butyrate could enhance epithelial barrier function, promote goblet cells mucus secretion, and reduce intestinal inflammation by reducing pro-inflammatory cytokines’ levels (Hamer et al., 2008; Gaudier et al., 2009; Guilloteau et al., 2010).
In our previous study, we found a significant decrease in concentrations of SCFAs, especially acetic acid and butyric acid in yaks (Li et al., 2022). We hypothesized that sodium acetate/sodium butyrate supplementation could alleviate diarrhea in animals, similar to how Lactobacillus plantarum alleviated diarrhea in a previous study by balancing gut microbiota and regulating SCFAs (Yue et al., 2020). The widely known lipopolysaccharide is an important membrane component of gram-negative bacteria, causing the inflammatory reaction, oxidative damage, and gut dysbiosis in hosts (Xu et al., 2021). Previous studies found that ROS were an important second messenger of the atypical domain (NOD)-like receptor containing pyrin domain 3 inflammasomes, and Caspase-1 was activated by NLRP3 and then cause inflammation reaction (Dashdorj et al., 2013; Sho and Xu, 2019; He et al., 2022). Hence, we conducted this study to explore the alleviation effect and potential mechanism of sodium acetate/sodium butyrate supplementation on LPS-induced diarrhea in mice via NLRP3/Caspase-1 signaling.
A total of 50, four weeks of age, ICR mice with an equal number of male and female animals (average weight of 18 ± 2 g) were purchased from Qing Long Shan Dong Wu Fan Zhi (Nanjing, China). After 30 days of rearing, mice were randomly divided into five groups, namely control (C), LPS (L), and treatment groups (A, B, and D). Group D (400: 200), B (300:300), and A (200: 400) were treated with 600 mg/kg sodium acetate/sodium butyrate solution via gavage for 18 days, while mice in C and L were treated with equal volume of normal saline. On day 19, mice in groups L, A, B, and D were treated with 20 mg/kg LPS (Solarbio life science, China), and after 24 hours mice were euthanized to collect serum, small intestine, and rectum samples (Figure 1). All animals were given normal water and feeds, and kept in the laboratory animal center of Nanjing Agricultural University. The body weights and diarrhea were documented.
Duodenum, jejunum, and ileum samples from all the groups were preserved in 4% paraformaldehyde for at least 48 hrs and then processed for commercial H&E staining (Wuhan Pinuofei Biological Technology Co., Ltd., China). On an Olympus CX23 microscope with an integrated digital imaging analysis system, histological slices were examined (Olympus Co., Japan). The villus height and crypt depth were measured as depicted in the previous study (Xu et al., 2021).
Blood samples of mice were centrifuged at 4,000 g for 10 min and stored at –20°C for future analysis. Antioxidant capacity was examined by detecting the levels of superoxide dismutase (SOD), glutathione peroxidase (GSH-px), total anti-oxidation capacity (T-AOC), and malondialdehyde (MDA) by utilizing commercial assay kits (Nanjing Jiancheng Bioengineering Institute, China). Meanwhile, the NO concentrations were determined via commercial assay kits (Nanjing Jiancheng Bioengineering Institute, China). The concentration of cytokines including interleukin 1 beta (IL-1β), interleukin 6 (IL-6), interleukin 10 (IL-10), and tumor necrosis factor-alpha (TNF-α) was measured by using commercial ELISA kits (Solarbio life science, China).
The total microbial genomic DNA was extracted from the rectum contents of each mouse. The samples from groups C (n = 4), L (n = 4), and A (n = 4) were extracted utilizing the fast DNA Stool Mini Kit (Qiagen, German) according to the manufacturer’s specifications. The quantity and quality of all extracted DNA samples were examined by using NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, USA) and agarose gel electrophoresis, respectively. Gene amplification of bacterial 16S rRNA gene was performed using the V3–V4 regions primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Then all amplicon products were purified by employing Vazyme VAHTSTM DNA Clean Beads (Vazyme, China) and quantified using the QuantiFluor™-ST (Promega, USA). At last, all samples were sequenced by using the Illumina MiSeq platform (Bioyi Biotechnology Co., Ltd. China) with MiSeq Reagent Kit v3.
All of the achieved sequencing raw data were cleaned using the DADA2 (Callahan et al., 2016) and Vsearch (Rognes et al., 2016) to generate accurate and reliable results for microbiome bioinformatic analysis through QIIME2 (2019.4)1 (Bokulich et al., 2018). Phylogenetic trees were constructed via mafft (Katoh et al., 2002) and FastTree (Price et al., 2010). Alpha-diversity metrics of Chao1 (Chao, 1984), observed species, Shannon (Shannon, 1948), Simpson (Simpson, 1997), Faith’s PD (Faith, 1992), Pielou’s evenness (Pielou, 1966), and Good’s coverage (Good, 1953) were estimated among samples. Beta diversity metrics of principal coordinate analysis (PCoA) (Alban, 2010), non-metric multidimensional scaling (NMDS) (Legendre and Montréal, 2003), and unweighted pair-group method with arithmetic means (UPGMA) were estimated among samples. Taxonomy was assigned to non-singleton amplicon sequence variants (ASVs) using the classify-sklearn naïve Bayes taxonomy classifier in the feature-classifier plugin (Bokulich et al., 2018) against the SILVA Release 132 Database2 (Quast et al., 2012). Tree diagram of classification levels and GraPhlAn evolutionary was generated via ggtree3. Krona species composition map was generated via KronaTools (v2.7)4 (Ondov et al., 2011). Significant difference analyses among different mouse groups were performed via PERMANOVA and Adonis in QIIME2 (2019.4). Venn5, heatmap, metagenomeSeq, LEFSe (Mahadevan et al., 2008; Segata et al., 2011), OPLS-DA (Mahadevan et al., 2008), and random forest analysis were carried to explore the significant difference in species. Network analysis (Faust and Raes, 2012) was performed to find potential keystone. The functional potential prediction was carried through the phylogenetic investigation of communities by the reconstruction of unobserved states (PICRUSt2) (Gavin et al., 2019) using MetaCyc6 and KEGG7 databases.
Intestinal tissue RNA extraction from all mouse groups was performed by utilizing TRIzol reagent (Life Technologies, USA). All of the RNA samples were examined via denaturing formaldehyde gel electrophoresis and NanoDrop 2000 analyzer (Thermo Fisher Scientific, China) to validate their integrity and concentrations, respectively. Then commercial SuperScript™IV first strand cDNA synthesis kits (Invitrogen™, Thermo Fisher Scientific, USA) were used for translating RNA samples into cDNA under the guidance of the manufacturer’s specifications. Finally, qRT-PCR for all groups was carried out by using 25 uL of reaction mixtures consisting of 2 uL of intestinal tissues cDNA, 12.5 uL of Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (Yeasen, China), 2uL of primers, and 8.5 uL nuclease-free water, then the procedure was performed in the StepOnePlus™ Real-Time PCR System (Applied Biosystems, USA). All sample reactions were repeated three times and the method of 2–ΔΔCT was utilized for calculating gene relative quantification. All primer pairs used in the present study were synthesized by Sangon Biotech (China) and are shown in Table 1.
All the generated data were evaluated via ANOVA, Student’s t-test, Kruskal–Wallis, and Dunn’s test via IBM SPSS (22.0) software. Data presented as means ± SD and statistically significant are considered when P < 0.05.
The mice were weighed on a daily basis and the weight of the mice in group A was slightly higher than mice in other groups (Figure 2A). The diarrhea was found in the mouse of group L (induced by LPS), whereas sodium acetate/sodium butyrate supplementation alleviated diarrhea, especially in mice of group A. The intestines’ morphology was examined via H&E staining and found that LPS caused a decrease in the villus height (p < 0.0001), an increase in the crypt depth (p < 0.05), and it also lowered the villus height to crypt depth ratio (p < 0.0001). Whereas, sodium acetate/sodium butyrate supplementation resulted in a significant increase in the villus height (p < 0.001), decrease in the crypt depth (p < 0.01), and increase in the villus height to crypt depth ratio (p < 0.001), especially in the mice of groups B and A (Figure 2).
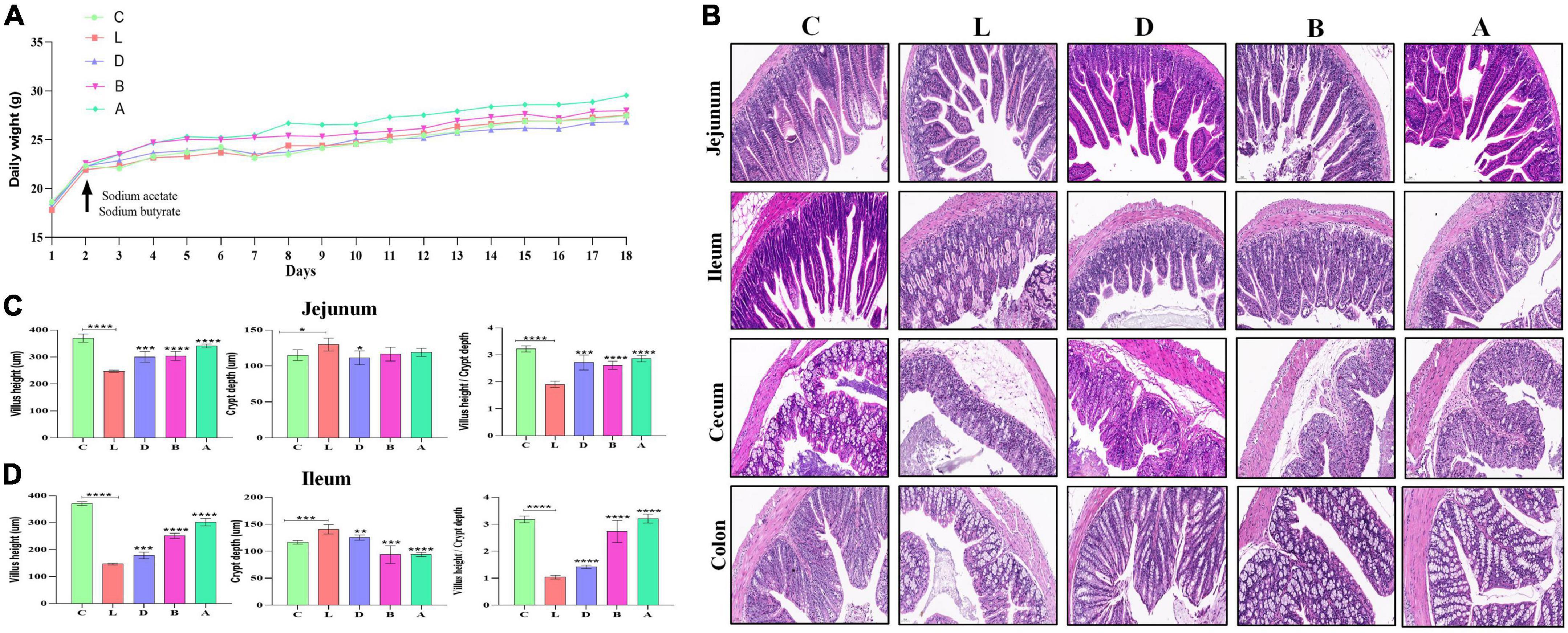
Figure 2. Effects of sodium acetate/sodium butyrate supplementation on body weight, diarrhea score, and intestinal damage induced by LPS. (A) Mouse daily weights, (B) H&E staining analysis of small intestine of mouse, (C) villus height, crypt depth, and villus height/crypt depth ratio of Jejunum, and (D) villus height, crypt depth, and villus height/crypt depth ratio of Ileum. Scale bar 50 μm. Significance is presented as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; data are presented as the mean ± SEM (n = 3).
In mice serum, no obvious difference was found in IL-6, IL-10, NO, GSH-px, and SOD levels among the control group and LPS-induced groups, respectively. T-AOC in group L was significantly low than in group C (p < 0.05), while there was no marked difference between group L and treated groups D, B, and A, respectively. In group C, the serum level of mice induced by LPS was found prominently high for IL-1β (p < 0.001), TNF-α (p < 0.001), and MDA (p < 0.01), respectively. However, in the serum of groups D, B, and A, sodium acetate/sodium butyrate supplementation caused a remarkable decrease in IL-1β (p < 0.001), TNF-α (p < 0.01), and MDA (p < 0.01), respectively (Figure 3).
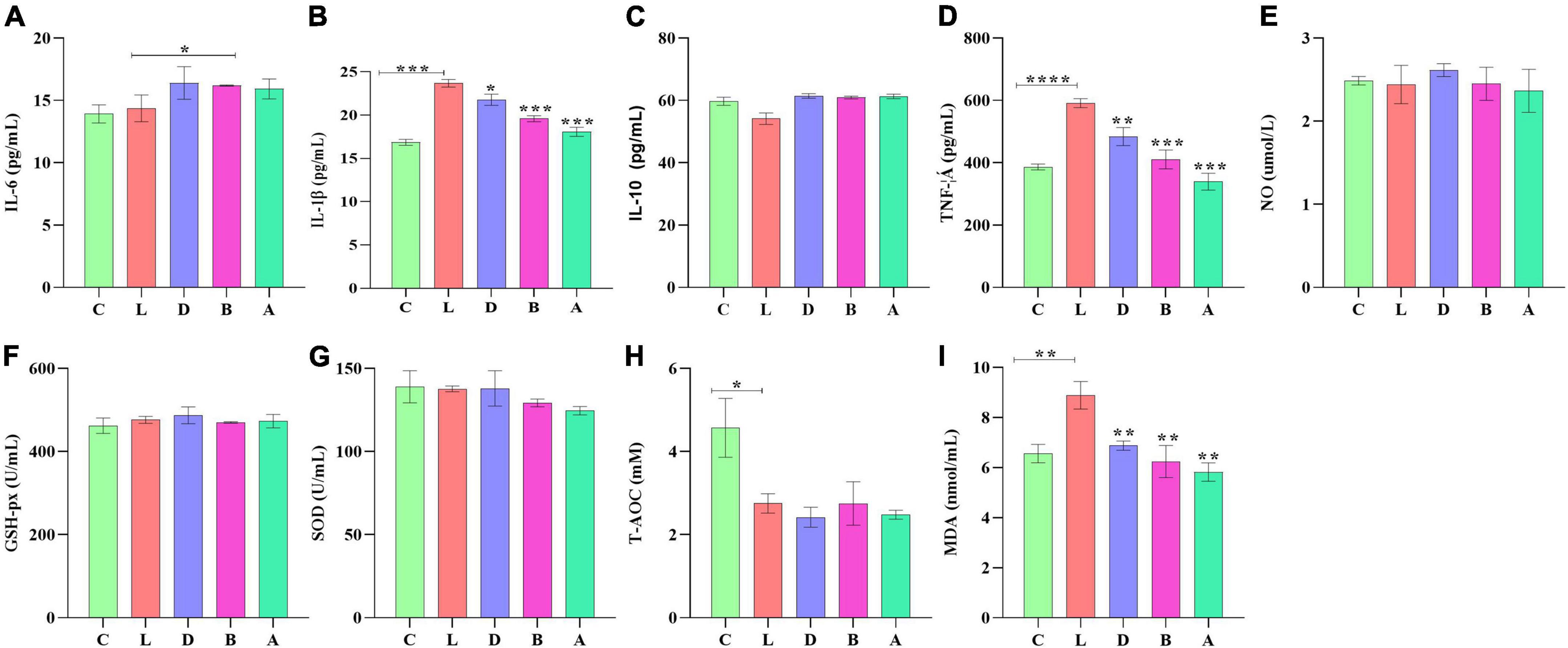
Figure 3. Sodium acetate/sodium butyrate supplementation improved the inflammation response and oxidative stress of mouse induced by LPS. The concentrations of inflammatory cytokines IL-6 (A), IL-1β (B), IL-10 (C), TNF-α (D), and NO (E) in serums. Oxidative status indices levels of GSH-px (F), SOD (G), T-AOC (H), and MDA (I) in serums. Significance is presented as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; data are presented as the mean ± SEM (n = 3).
In the current study, over 110 000, 95 000, 90 000, 94 000, 69 000, and 69 000 of input, filtered, denoised, merged, non-chimeric, and non-singleton data were achieved, respectively, in different mouse samples (Table 2). A significant difference in non-chimeric (p < 0.05) and non-singleton (p < 0.05) was found between groups C and L (Figure 4A). The majority of sequence lengths were around 430 bp (Figure 4B). As shown in Figure 4C, flatness broken lines were present in all samples, which reflected the evenness of OTUs composition in samples. Alpha-diversity index of Chao1, Simpson, Shannon, Pielou’s evenness, observed species, Faith’s PD, and Goods coverage is shown in Table 3. There was a significant difference in only Pielou’s evenness between groups C and A (p < 0.05) (Figure 4D). Beta diversity analysis indicated a far distance of points in samples of group L, whereas relatively near points were found in group A via PCoA and NMDS analyses, respectively (Figure 4E). UPGMA analysis found that the branch length in group C was relatively shorter in group A compared with samples in group L (Figure 4E). Significant differences between groups were found in groups A and C through PERMANOVA (p < 0.05), ANOSIM (p < 0.05), and PERMDISP (p < 0.05), respectively (Figure 4F).
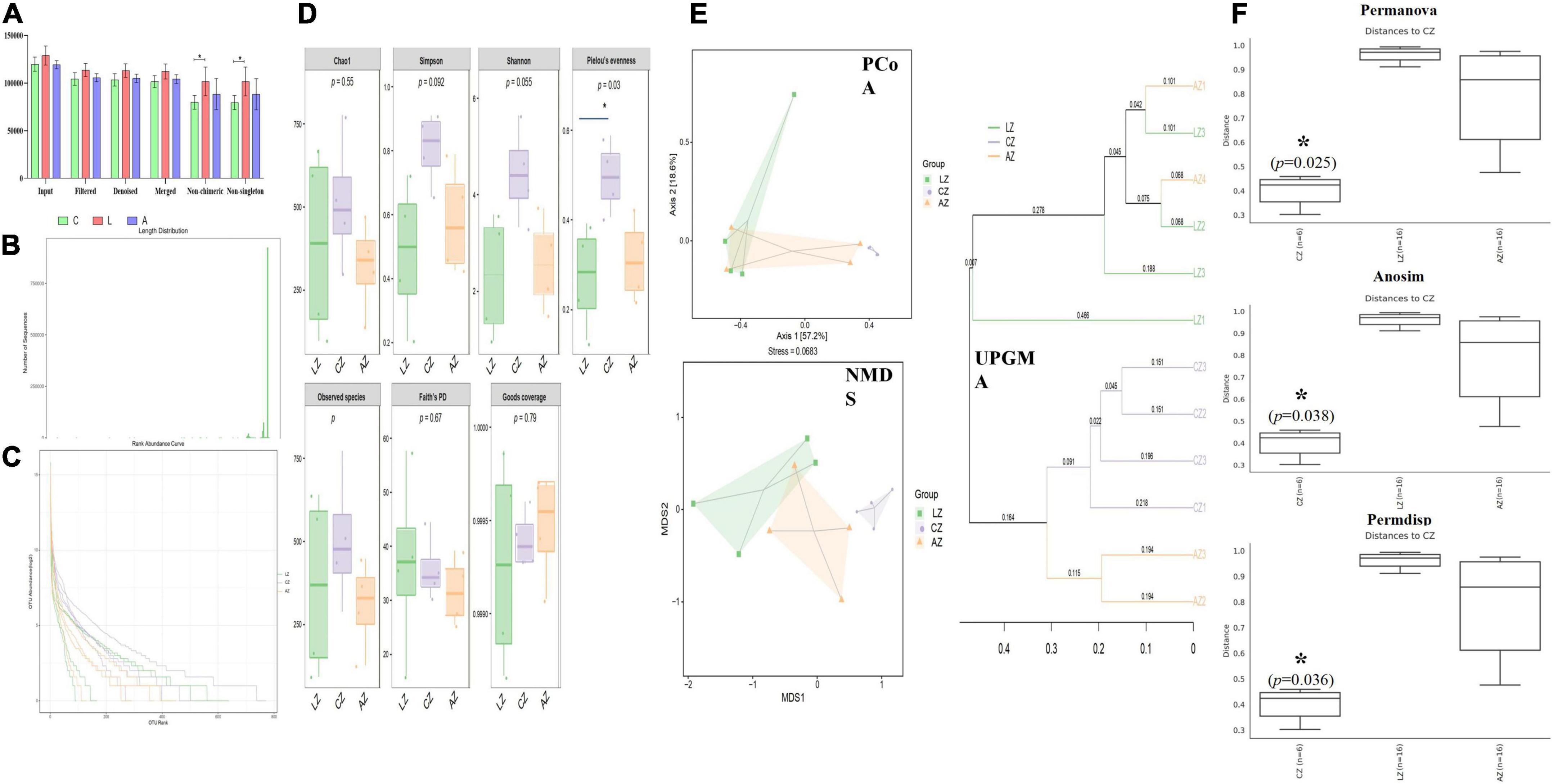
Figure 4. Sodium acetate/sodium butyrate supplementation restored the structure and diversity of mouse gut microbiota affected by LPS. (A) Sequencing data statistical analysis, (B) length distribution of sequencing data, (C) rank abundance curve, (D) alpha-diversity index analysis, (E) beta diversity analysis, and (F) group difference analysis. Significance is presented as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; data are presented as the mean ± SEM (n = 4).
The number of taxa contained in different levels of phylum, class, order, family, genus, and species is shown in Figure 5. At the phylum level, the dominating phyla in group L were Proteobacteria (83.02%) and Bacteroidetes (13.00%). While Firmicutes (83.36%), Bacteroidetes (12.80%), Firmicutes (45.35%), and Proteobacteria (46.88%) were the main phyla in groups C and A. At the Class level, the main classes were Gammaproteobacteria (81.28%) and Bacteroidia (12.99%) in group L, while Bacilli (74.72%), Bacteroidia (12.80%), Clostridia (8.08%), Gammaproteobacteria (45.90%), Bacilli (44.04%), and Bacteroidia (6.80%) were the dominant classes in groups C and A, respectively. At the order level, the primary classes were Enterobacteriales (55.60%), Pseudomonadales (25.65%), and Bacteroidales (12.99%), while Lactobacillales (72.68%), Bacteroidales (12.80%), Clostridiales (8.08%), Lactobacillales (43.52%), Enterobacteriales (41.07%), and Bacteroidales (6.80%) were the staple orders in groups C and A, respectively. At the family level, the dominating families in group L were Enterobacteriaceae (55.59%), Pseudomonadaceae (25.47%), and Bacteroidaceae (9.09%), while the main families were Lactobacillales (72.47%) and S24-7 (12.50%) in group C, whereas Lactobacillales (43.35), Enterobacteriaceae (41.07%), and Bacteroidaceae (4.84%) in group A, respectively. At the genus level, the major genera in group L were Shigella (54.62%), Pseudomonas (25.42%), and Bacteroides (9.09%), while Lactobacillus (72.45%), unidentified_S24-7 (12.50), Lactobacillus (43.34%), Shigella (40.71), and Bacteroides (4.84%) were the dominating genera in groups C and A (Figure 5).
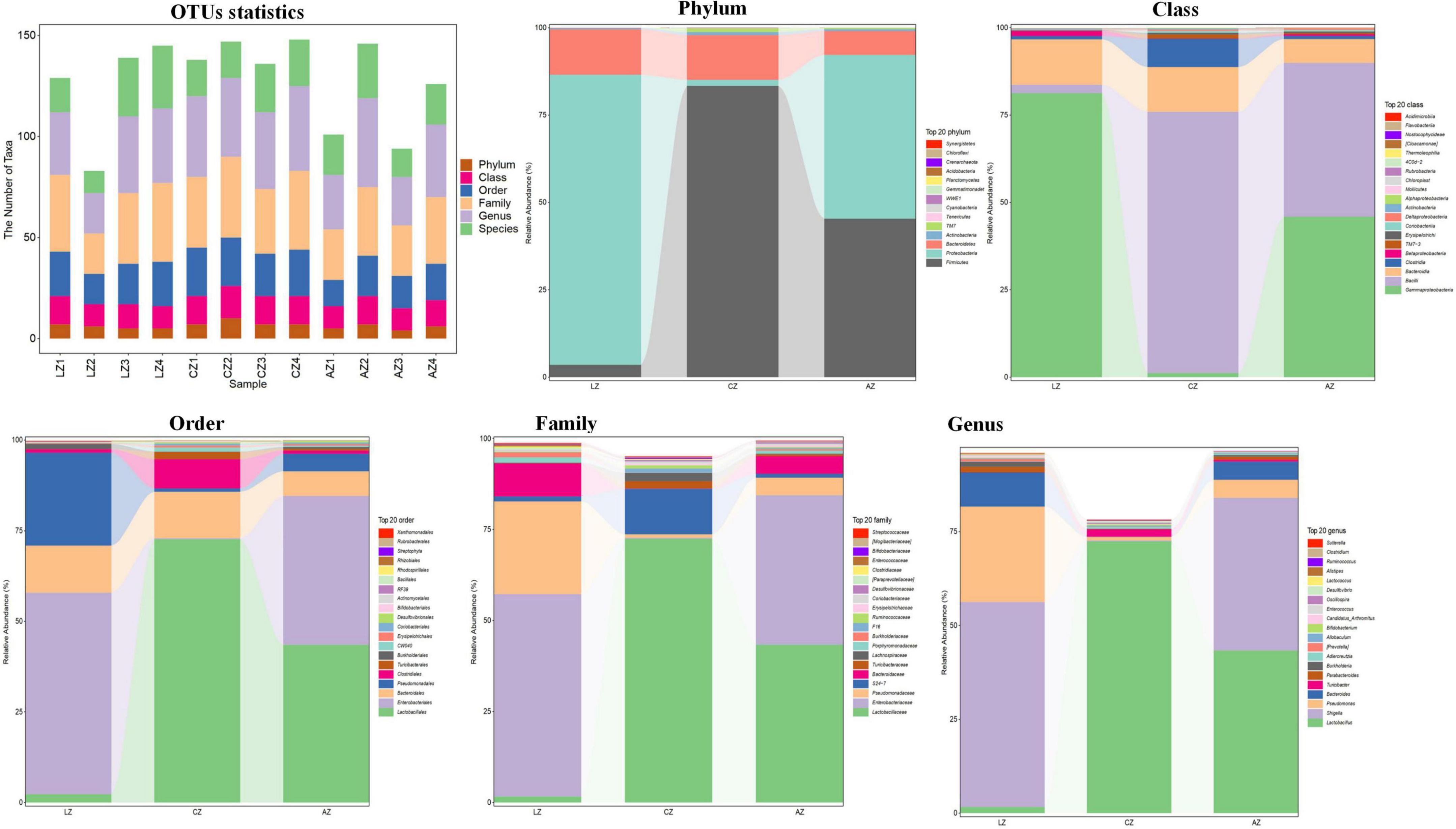
Figure 5. Effect of dietary sodium acetate/sodium butyrate supplementation on the relative abundance of gut microbiota in different taxa levels.
The classification levels tree diagram showed that larger sectors of Ruminococcus, Oscillospira, Lactobacillus vaginalis, Lactobacillus helveticus, Lactobacillus hamsteri, Turicibacter, Allobaculum, Adlercreutzia, and F16 with light orange and lavender color in groups A and C, respectively, while larger sectors of Candidatus Arthromitus, Lactococcus, Enterococcus, Bacteroides uniformis, Bacteroides caccae, Bacteroides acidifaciens, Parabacteroides gordonii, Parabacteroides distasonis, Alistipes finegoldii, Prevotella, Shigella, Pseudomonas syringae, and Pseudomonas Pseudomonas with green color were found in group L (Figure 6A). GraPhlAn evolutionary tree diagram showed that the abundance of Lactobacillus, Shigella, Pseudomonas Pseudomonas, Bacteroides, Turicibacter, Parabacteroides, Burkholderia, Adlercreutzia, Prevotella, and Enterobacteriaceae Pseudomonas depicted with various colors were found significantly different among different mouse groups (Figure 6B). Krona species composition diagram indicated that the main genera were Shigella (72%), Bacteroides (9%), and Pseudomonas (8%) in group L, while Shigella (56%), Pseudomonas (26%), and Bacteroides (9%) in group A, and unidentified S34-7 (45%), unidentified Clostridia (15%), Lachnospiraceae (7%), and Turicibacter (7%) in group C, respectively (Figure 6C).
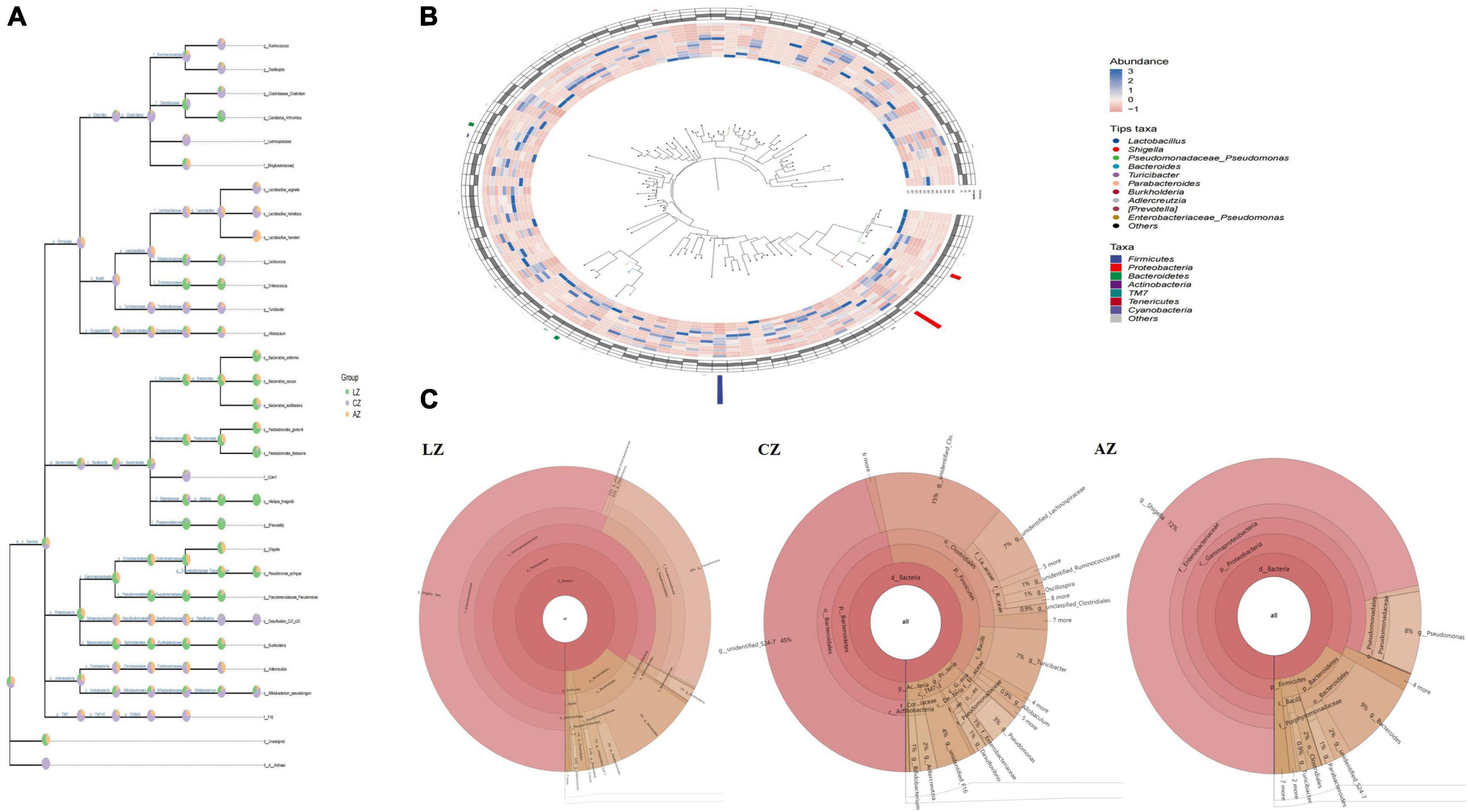
Figure 6. Species composition analysis of mouse gut microbiota. (A) Classification levels tree diagram, (B) GraPhlAn evolutionary tree diagram, and (C) Krona species composition diagram.
To find different species and markers, species in mouse microbiota induced by LPS, we performed the Venn diagram, bar chart of ASV/OTU numbers in different regions of the Venn diagram, bar graphs of ASV/OTU abundance in different regions of the Venn diagram, Genera composition heatmap, PCA, and OPLS-DA analysis. Results showed that 216 (7.71%) OTUs were shared in groups C and L, while 335 (11.96%) OTUs were shared in groups C and A (Figure 7A). Then ASV/OTU abundance was explored in different regions of the Venn diagram. The results showed that at the phylum level, groups C and A shared Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and TM7 Phyla, while groups C and L shared Firmicutes, Proteobacteria, and Bacteroidetes Phyla. At the genus level, groups C and L shared Shigella, Lactobacillus, Pseudomonas, Bacteroides, Parabacteroides, Oscillospira, Allobaculum, and Ruminococcus Genera, while groups C and A shared Lactobacillus, Adlercreutzia, Oscillospira, Ruminococcus, and Allobaculum genera (Figure 7B). ASV/OTU number analysis found that at the Phylum level, groups C and L shared Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Cyanobacteria, while groups C and A shared Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, TM7, and Tenericutes Phyla. At the genus level, groups C and L shared Lactobacillus, Bacteroides, Shigella, Oscillospira, Parabacteroides, Pseudomonas, Adlercreutzia, Ruminococcus, and Enterococcus, while groups C and A shared Lactobacillus, Bacteroides, Oscillospira, Pseudomonas, Adlercreutzia, Ruminococcus, Bifidobacterium, and Enterococcus genera (Figure 7C). It is depicted in the heatmap that the abundance of Bacteroides, Parabacteroides, Shigella, Prevotella, Phenylobacterium, Candidatus Arthromitus, Prevotella, Rhodococcus, Burkholderia, Blautia, Dorea, Enterococcus, Pseudomonas, Melissococcus, Rubrobacter, Acinetobacter, Butyricimonas, Prauserella, Anaerotruncus, Alistipes, and Subdoligranulum shown in red color in group L were obviously higher than in groups C and A, while Lactobacillus, Adlercreutzia, Ruminococcus, Coprobacillus, and Faecalibacterium shown in blue color in group L were significantly lower than in groups C and A (Figure 7D). PCA analysis represents that the main genera among mouse groups were Lactobacillus, Shigella, and Pseudomonas. The distance between points of group A (green) projected on the coordinate axis was clearly farther than groups A (light orange) and C (lavender), which revealed a difference between group L, and groups A and C, respectively (Figure 7E). Also, OPLS-DA analysis revealed similar results to PCA analysis (Figure 7F).

Figure 7. Different species and marker species analysis in mouse microbiota induced by LPS. (A) Venn diagram, (B) bar chart of ASV/OTU numbers in different regions of Venn diagram, (C) bar graphs of ASV/OTU abundance in different regions of the Venn diagram, (D) genera composition heatmap, (E) PCA, and (F) OPLS-DA.
For further investigation, MetagenomeSeq analysis was performed and different genera like Bacteroidales and Lactobacillales were found significantly on the upside of the broken lines with decreased ASV 102 (p < 0.05), ASV 23 (p < 0.05), ASV 14 (p < 0.05), and ASV 53 (p < 0.05), increased ASV 73 (p < 0.05), ASV 1 (p < 0.01), ASV 194 (p < 0.05), ASV 50 (p < 0.05), ASV 18 (p < 0.01), and ASV 5 (p < 0.001) between groups C and A. Different genera like Bacteroidales, Lactobacillales, Clostridiales, Burkholderiales, Desulfovibrionales, Enterobacteriales, and Pseudomonadales were found significantly in groups C and L, with 107 decreased ASV and 86 increased ASV (Figure 8A). Biomarker bacteria in mouse groups were uncovered by using LEfSe analysis, which were from the class Gammaproteobacteria, phylum Proteobacteria, genus Shigella, family Enterobacteriaceae, order Enterobacteriales, Rhizobiales, order Burkholderiales, class Betaproteobacteria, and genus Pseudomonas in group L (green color), family S24-7, order Clostridiales, class Clostridia, genus Gemella, order Gemellales, family Gemellaceae, phylum Tenericutes, class Mollicutes, family Lachnospiraceae, order RF39, family F16, class TMT-3, order CW040, phylum TM7, and genus Ruminococcus in group C (lavender), genus Faecalibacterium, family Acetobacteraceae, and order Rhodospirillales in group A (light orange) (Figure 8B).
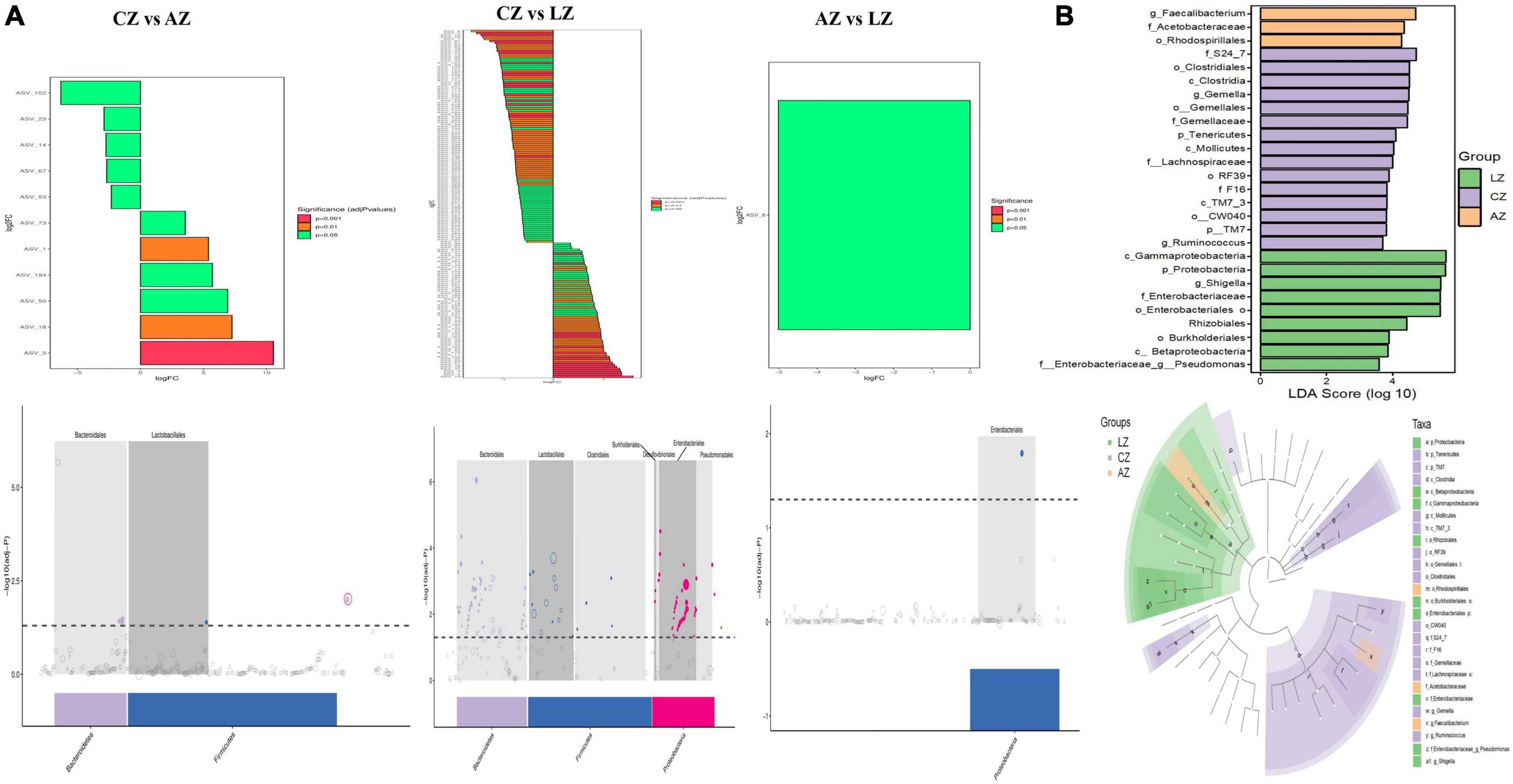
Figure 8. Gut microbiota difference analysis between mouse groups. (A) MetagenomeSeq analysis and (B) LEfSe analysis.
Random forest analysis was performed with an accuracy ratio of 2 (Figure 9A). Important genera among mouse groups are shown in Figure 9B including Clostridium, Ruminococcus, Lactococcus, Gemella, etc. Network analysis revealed that there were more edges between groups A and C than between groups L and C, which inferred a higher similarity between groups A and C (Figure 9C). The dominant genera in the network were unidentified F16, Lactobacillus, Adlercreutzia, unidentified S24-7, and Turicibacter between groups A and C, while Lactococcus and Allobaculum were the main genera in the network between groups L and C (Figure 9D).
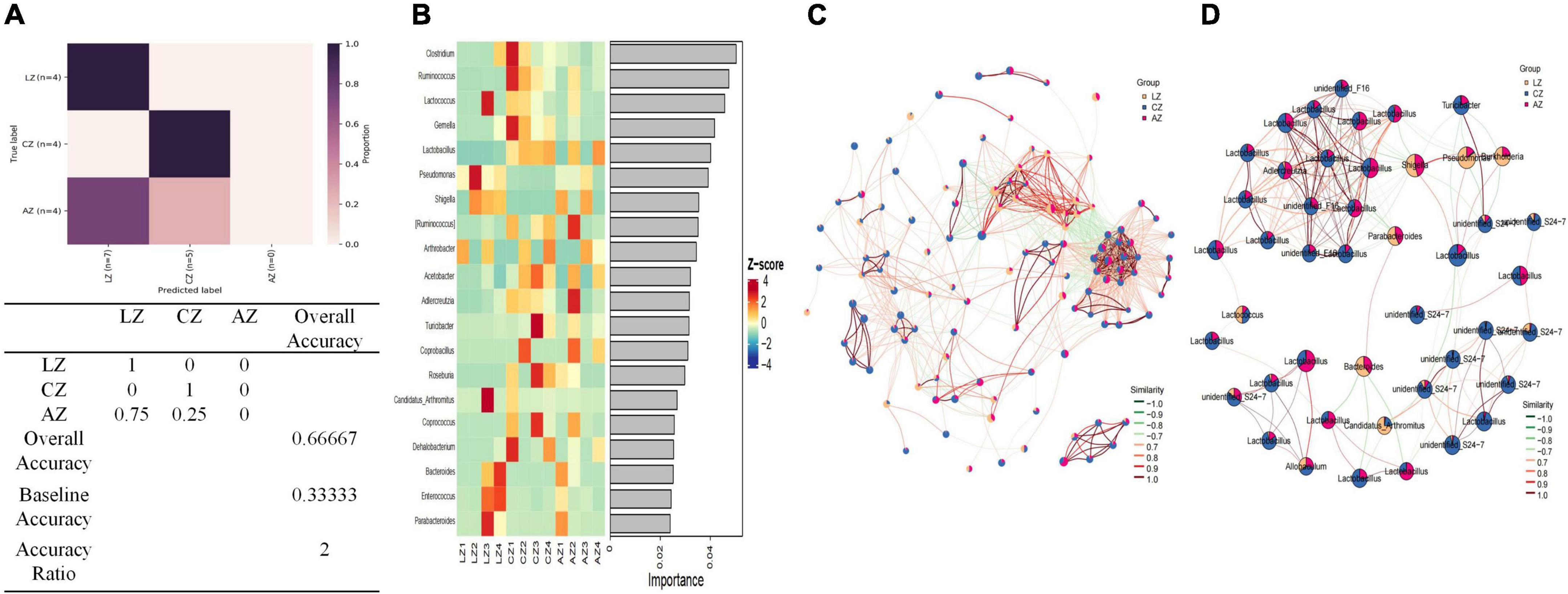
Figure 9. Random Forests and network analysis. (A) Model accuracy, (B) ASV taxa heatmap, (C) network analysis, and (D) subnetwork analysis of dominant genera.
By comparing the abundance of genera of mouse microbiota, 19 genera were detected among mouse groups. The abundance of Lactobacillus (p < 0.0001), unidentified F16 (p < 0.0001), Adlercreutzia (p < 0.01), Ruminococcus (p < 0.05), [Ruminococcus] (p < 0.05), Acetobacter (p < 0.05), cc 115 (p < 0.05), and Cupriavidus (p < 0.05) in group L was obviously lower than group C, respectively. While Shigella (p < 0.05) was prominently higher in group L than in group C. Unidentified_S24-7 and unclassified RF39 were significantly higher in group C than in groups A (p < 0.05) and L (p < 0.05), respectively, while unclassified Enterobacteriaceae was significantly lower in group C than in groups A (p < 0.05) and L (p < 0.05), respectively. The abundance of unclassified Pseudomonadales (p < 0.05), Rhodococcus (p < 0.05), unclassified Comamonadaceae (p < 0.05), and Lysobacter (p < 0.05) in group L was conspicuously higher than group A, respectively. The abundance of Faecalibacterium in group L was clearly lower than groups A (p < 0.05) and L (p < 0.01), respectively. The abundance of Gluconacetobacter in group A was prominently higher than groups L (p < 0.05) and C (p < 0.01), respectively. The abundance of unclassified Bradyrhizobiaceae in group C was clearly higher than in group A (p < 0.05) (Figure 10).
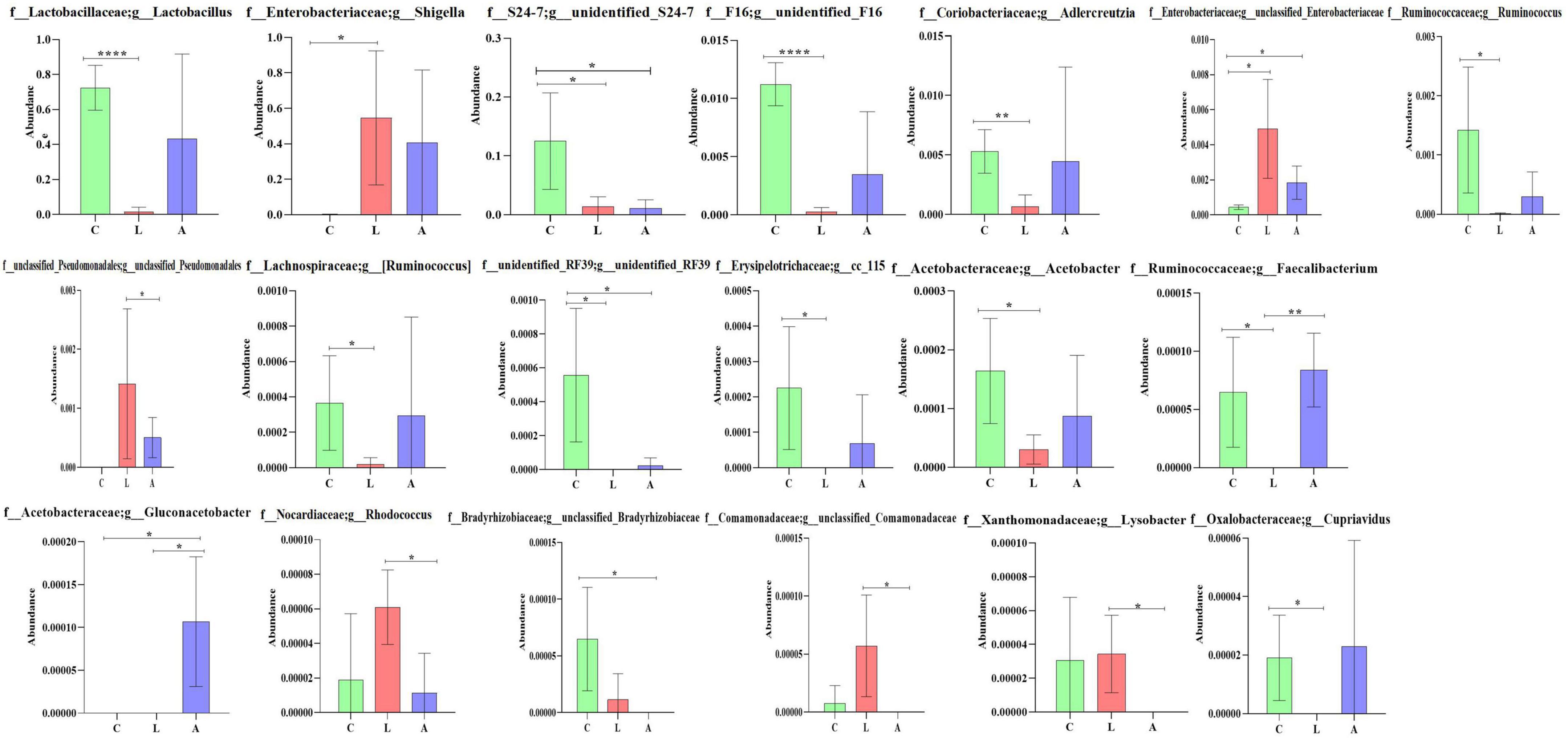
Figure 10. Comparing genera difference of mouse microbiota. Significance is presented as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; data are presented as the mean ± SEM (n = 4).
In summary, different analysis methods include relative abundance taxa, classification levels tree diagram, GraPhlAn evolutionary tree diagram, Krona species composition diagram, Venn Diagram, Genera composition heatmap, PCA, OPLS-DA, metagenomeSeq analysis, LEfSe analysis, and random forests and network analyses demonstrated that LPS challenge changed microbiota composing in mice; however, sodium acetate/sodium butyrate supplementation could partly restore the gut microbiota in animals.
PICRUSt2 was utilized for potential function prediction analysis of mouse microbiota. PCoA analysis found that closer points are projected on the coordinate axis in groups A and L in both functional units of KO and EC (Figure 11A), which revealed a more similar functional composition in these groups. KEGG analysis showed that the main pathways were related to metabolism (Figure 11B). MetaCyc analysis showed that the main pathways were related to biosynthesis, degradation/utilization/assimilation, and generation of precursor metabolite and energy (Figure 11C). MetaCyc metabolic pathways comparing analysis found 44 (p < 0.05), 9 (p < 0.01), and 20 (p < 0.001) significant different pathways between groups C and L, 37 (p < 0.05), 27 (p < 0.01), and 53 (p < 0.001) significant different pathways between groups A and C, respectively (Figure 11D). KEGG metabolic pathways comparing analysis revealed that 15 (p < 0.05), 15 (p < 0.01), and 51 (p < 0.001) significant different pathways between groups C and L, while one (p < 0.001) significant different pathway between groups A and C was examined (Figure 11E).
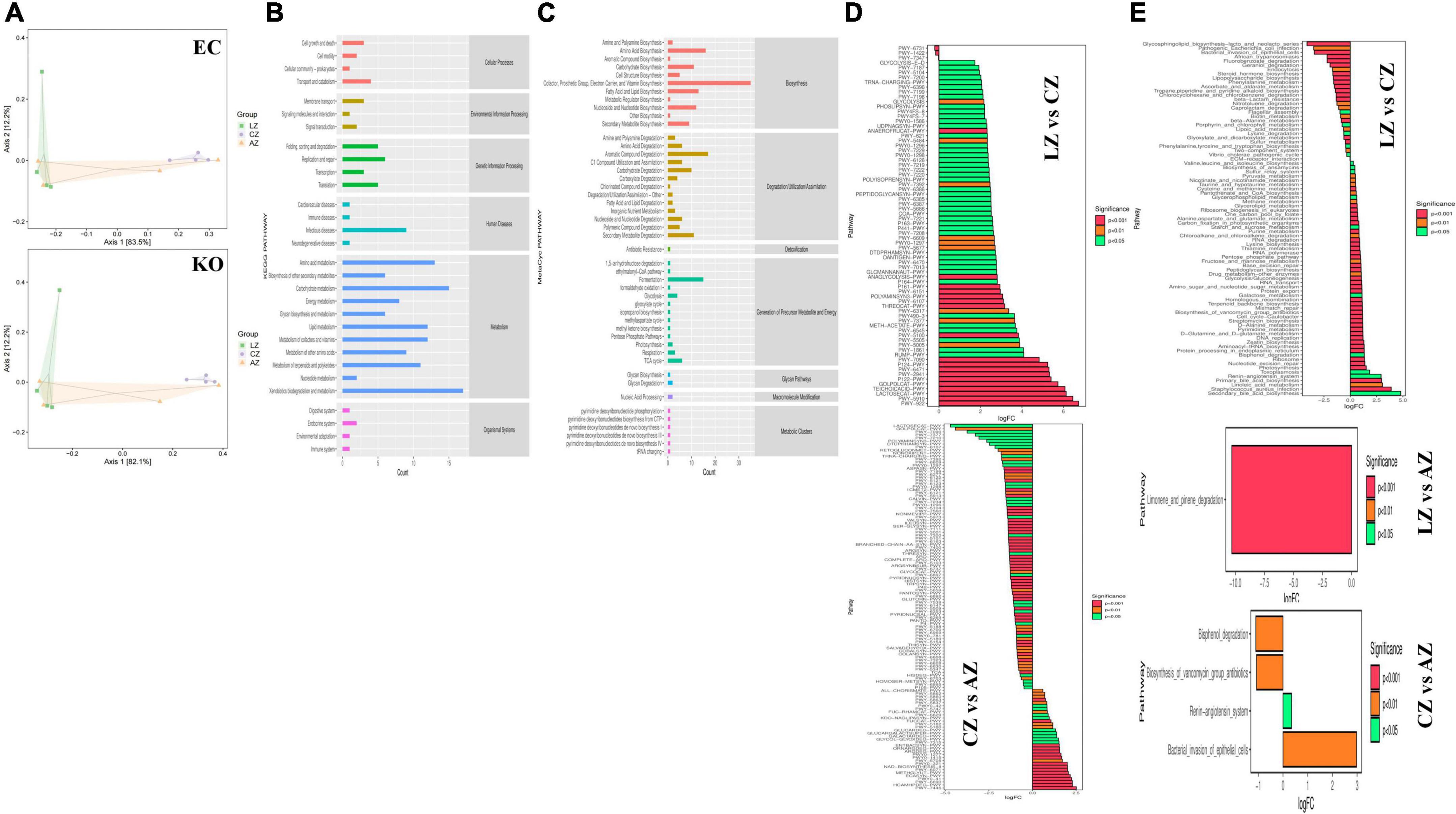
Figure 11. Potential function prediction analysis of mouse microbiota. (A) PCoA, (B) KEGG, (C) MetaCyc, (D) MetaCyc metabolic pathways, and (E) KEGG metabolic pathways.
Relative gene expression of Zonula occludens 1 (ZO-1), Occludin, Claudin, Caspase-1, and NLRP3 were detected by employing qRT-PCR. LPS induction prominently upregulated the expression of ZO-1 (p < 0.01) and NLRP3 (p < 0.0001) in mice of group L; however, sodium acetate/sodium butyrate solution supplementation downregulated the expression of ZO-1 (p < 0.05) and NLRP3 (p < 0.05) genes in treated mice. Furthermore, change in LPS clearly downregulated the expression of Occludin (p < 0.001), Claudin (p < 0.0001), and Caspase-1 (p < 0.0001) genes in group L, while sodium acetate/sodium butyrate solution supplementation upregulated those gene expressions in treated groups of D, B, and A (Figure 11).
Correlation assessment between differential bacteria (abundance of top 20 genera), intestine morphology indices, inflammatory cytokines, oxidative indices, and gene expressions was performed through Statistical Analysis System. Results showed that Lactobacillus, Turicibacter, Roseburia, Allobaculum, Bifidobacterium, Faecalibacterium, Ruminococcus, Coprococcus, cc_15, Gemella, Pediococcus, Butyricicoccus, and Cupriavidus were positively related to villus height and villus height to crypt depth ratio, while Rhodococcus, Phenylobacterium, Lysobacter, and W22 were positively related to villus height. Lactobacillus, Allobaculum, Rhodococcus, Butyricicoccus, and Ralstonia were found positively related to antioxidant ability. Pseudomonas, Faecalibacterium, Faecalibacterium, Candidatus Arthromitus, Ruminococcus, Roseburia, Gemella, Faecalibacterium, Gluconacetobacter, Rhodococcus, Anaeroplasma, (Clostridium), Phenylobacterium, and Aminobacter were positively related to inflammatory cytokines. Pseudomonas, Faecalibacterium, Gluconacetobacter, Rhodococcus, (Clostridium), and Gluconobacter were positively related to tight junction proteins, while Adlercreutzia, Candidatus Arthromitus, Lactococcus, Ruminococcus, Coprococcus, Acinetobacter, cc_115, Gemella, Anaeroplasma, Phenylobacterium, Lysobacter, and W22 were negatively related to tight junction proteins. Adlercreutzia, Ruminococcus, Coprococcus, cc_115, Roseburia, Gemella, and Anaeroplasma were negatively related to expressions of Caspase-1 and NLRP3 (Figure 12).
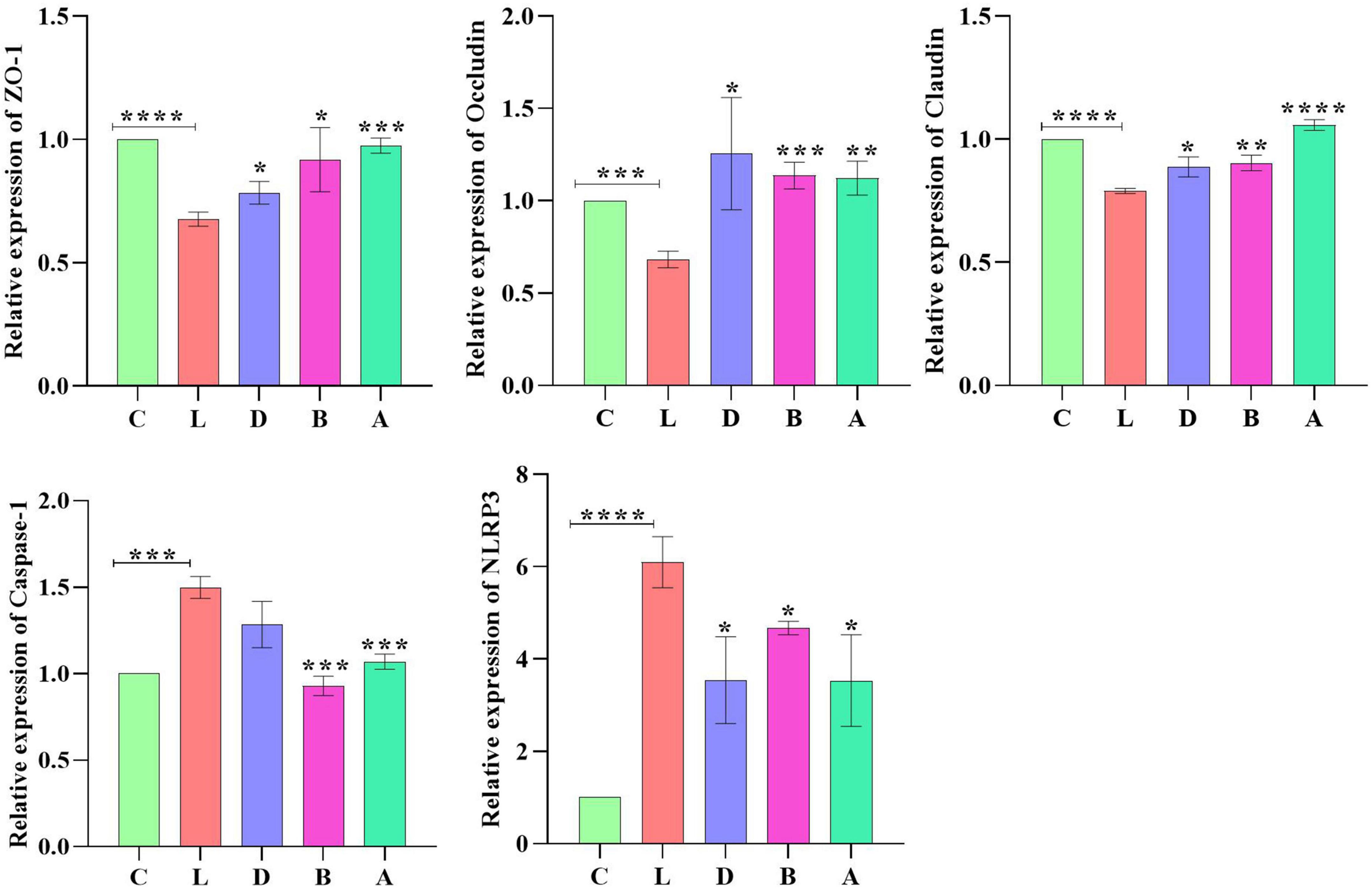
Figure 12. Relative expression analysis of ZO-1, Occludin, Claudin, Caspase-1, and NLRP3 via qRT-PCR. Significance is presented as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; data are presented as the mean ± SEM (n = 3).
Though various kinds of measures have been implemented so far to fight against diarrhea, there is still a long way to go (Cho et al., 2013). A recent review found that higher antimicrobial resistance was associated with the drugs being used for the treatment of diarrhea (Zhang H. et al., 2022; Zhang X. et al., 2022;) therefore, establishing a method to positively regulate the gut microbiota to ameliorate diarrhea is of great importance and meaningful (Coelho et al., 2022).
In the present study, mice were supplemented with sodium acetate/sodium butyrate before inducing diarrhea by utilizing LPS. Serve damage to intestines particularly jejunum, ileum, cecum, and colon was examined (Figure 1B), which was in accordance with previous results (He et al., 2022). An obvious decrease in villus height, increase in crypt depth, and lowered villus height to crypt depth ratio were uncovered in group C, while interestingly, sodium acetate/sodium butyrate supplementation could significantly increase the villus height, decrease the crypt depth, and increase the villus height to crypt depth ratio in both jejunum and ileum, especially in groups B and A (Figures 1C,D). The current results demonstrated that sodium acetate/sodium butyrate could improve intestinal integrity deteriorated by LPS, as intestinal barrier integrity is considered an important indicator of intestinal health (Liu H. et al., 2022). The current results may reveal that sodium acetate/sodium butyrate supplementation is a hopeful and effective method to alleviate diarrhea in animals.
To explore the potential mechanisms, we detected the gene expressions of tight junction proteins in intestines. Among them Occludin and Claudins were recognized as important components of intestinal permeability (Chen et al., 2015). The expression of Occludin in group L was significantly lower than in group C, while expression levels of Occludin increased in sodium acetate/sodium butyrate-treated groups (Figure 11). The current results were in line with a study on inflammatory bowel disease in humans with the downregulation of Occludin (Chen et al., 2015). The expression of Claudin in group L was significantly lower, which is in line with previous results (He et al., 2022); however, with the treatment of sodium acetate/sodium butyrate, the expression of Claudin upregulated significantly. The plaque protein ZO1 is an adaptor connecting trans-membrane protein with the peri-junctional actomyosin ring (Ulluwishewa et al., 2011). Like previous results found lower expressions of ZO-1 (Cao et al., 2022; He et al., 2022), the expression of ZO-1 in group L was significantly higher than in group C, while expression levels of ZO-1 decreased in sodium acetate/sodium butyrate-treated groups. The current findings suggested that acetate/sodium butyrate could improve intestinal barrier function by modulating tight junction gene expressions.
Reactive oxygen species are widely known for their important role in inflammatory diseases like colitis. The intestinal tissue injury is mediated through the administration of the antioxidants (Dashdorj et al., 2013; Aziz et al., 2021; Hassan et al., 2021; Liu et al., 2021; Murtaza et al., 2021). Oxidative agents of SOD, T-AOC, GSH-Px, and MDA are commonly known important enzymes related to oxidative stress, which may cause intestinal damage (Mehmood et al., 2019; Liu J. et al., 2022). In mice serums, no obvious difference was found in GSH-px and SOD between the control group and LPS-induced groups, respectively, while a prominently higher level of MDA (p < 0.01) was detected in group L; however, there was a significant decrease in MDA (p < 0.01) levels in sodium acetate/sodium butyrate supplemented groups D, B, and A, respectively (Figure 3), suggesting that sodium acetate/sodium butyrate improves intestinal oxidative damage by reducing MDA contents. Previous studies found that the activation of Caspase-1 by NLRP3 inflammasome could cause inflammation reaction by promoting the maturation of IL-1β, and ROS was a generally accepted second key messenger of NLRP3 inflammasome (Dashdorj et al., 2013; Sho and Xu, 2019; He et al., 2022). LPS inducing obviously upregulated the expression of NLRP3 (p < 0.0001) in mice in group L; however, sodium acetate/sodium butyrate solution supplementation downregulated the expression of NLRP3 (p < 0.05) genes in treated mice. Also, the LPS challenge clearly upregulated the Caspase-1 (p < 0.0001) gene in group L, while sodium acetate/sodium butyrate solution supplementation downregulated it in treated groups D, B, and A (Figure 11). TNF-α is an important cytokine in inflammation (Zelová and Hošek, 2013), Significantly higher IL-1β and TNF-α (p < 0.001) were found in group C, which was consistent with previous results found higher inflammatory factors and upregulation of NLRP3 in HUVEC cells (Liu et al., 2021). However, sodium acetate/sodium butyrate supplementation significantly decreased IL-1β and TNF-α in groups D, B, and A, respectively (Figure 3), demonstrating that sodium acetate/sodium butyrate could reduce intestine inflammation caused by LPS by alleviating oxidative damage via the downregulation of NLRP3 and Caspase-1.
Previous study found that LPS-induced ROS caused gut microbiota dysbiosis in piglets (Xu et al., 2021). Similar results were found in the current results in mice. The structure and diversity of mouse gut microbiota were significantly altered in LPS-induced mice, as determined by alpha and beta diversity analysis (Figure 13), taxa analysis at different levels (Figure 5), Classification levels tree diagram, and GraPhlAn evolutionary tree diagram analysis (Figures 6A,C). Potential function prediction analysis by PICRUSt2 and KEGG analysis found that LPS significantly changed the main pathways of mice (Figure 11). In consistent with previous studies (Yue et al., 2020; Xu et al., 2021; Chang et al., 2022), sodium acetate/sodium butyrate supplementation restored the structure, diversity, and partly function of mouse gut microbiota. Venn diagram, heatmap, PCA, OPLS-DA (Figure 7), metagenomeSeq (Figure 8), and random forests and network analysis (Figure 9) analyses were performed to reveal different species and their markers in mice microbiota induced by LPS. A total of 19 genera were detected among mouse groups (Figure 10). LPS challenge decreased the abundance of Lactobacillus, unidentified F16, unidentified_S24-7, Adlercreutzia, Ruminococcus, unclassified Pseudomonadales, (Ruminococcus), Acetobacter, cc 1, Rhodococcus, unclassified Comamonadaceae, Faecalibacterium, and Cupriavidus, while increased Shigella, Rhodococcus, unclassified Comamonadaceae, and unclassified Pseudomonadales in group L. Interestingly sodium acetate/sodium butyrate supplementation increased Lactobacillus, unidentified F16, Adlercreutzia, Ruminococcus, (Ruminococcus), unidentified F16, cc 115, Acetobacter, Faecalibacterium, and Cupriavidus, while decreased Shigella, unclassified Enterobacteriaceae, unclassified Pseudomonadales, Rhodococcus, and unclassified Comamonadaceae. Lactobacillus genus bacteria are probiotic microorganisms that have beneficial effects on to host (Rajab et al., 2020), and previous studies demonstrated that Lactobacillus could improve diarrhea in infants and piglets (Hu et al., 2019; Wang et al., 2020). A previous study reported that the genus Adlercreutzia was related to body weight loss (Liu Z. et al., 2022), the abundance of Ruminococcus was significantly decreased in diarrhea piglets (Liu C. et al., 2019; Liu G. et al., 2019). The changes of Adlercreutzia and Ruminococcus in the current study were consistent with previous findings. Bacteria from the Acetobacter genus are related to lignin degradation and intestine metabolism (Wu et al., 2022), and the decreasing of this genus may affect the metabolism of animals. Also, these bacteria can generate acetate (Zhang H. et al., 2022), which confirmed that diarrhea was related to the inefficiency of acetate. Bacteria of Rhodococcus are from natural environments (Alvarez et al., 2021), which may contrite little to diarrhea. A previous study reported that unclassified Comamonadaceae were related to the degradation of organics (Zhang et al., 2020), which may infer that LPS inducing could affect organics metabolism. The lower abundance of Faecalibacterium was consistent with the previous study observed in patients with colitis and Crohn’s disease (Ponziani et al., 2020). Faecalibacterium could cause inflammation through decreased SCFAs (Nishiwaki et al., 2020), which showed that LPS-induced diarrhea may attribute to the absence of SCFAs. Bacteria from Cupriavidus genus are related to biodegradation and biodetoxifcation, which may indicate that LPS could reduce the degradation (Al-Nussairawi et al., 2020) and detoxification ability of mouse. Shigella spp. are well-known pathogens causing bacterial dysentery (Liu C. et al., 2019), which should reveal the reason of diarrhea caused by LPS. Bacteria from the top 20 abundance genera related to intestine damage, oxidation resistance, inflammatory factor, and gene expressions are shown in Figure 12, which demonstrated that sodium acetate/sodium butyrate could regulate microbiota to improve diarrhea induced by LPS in mice.
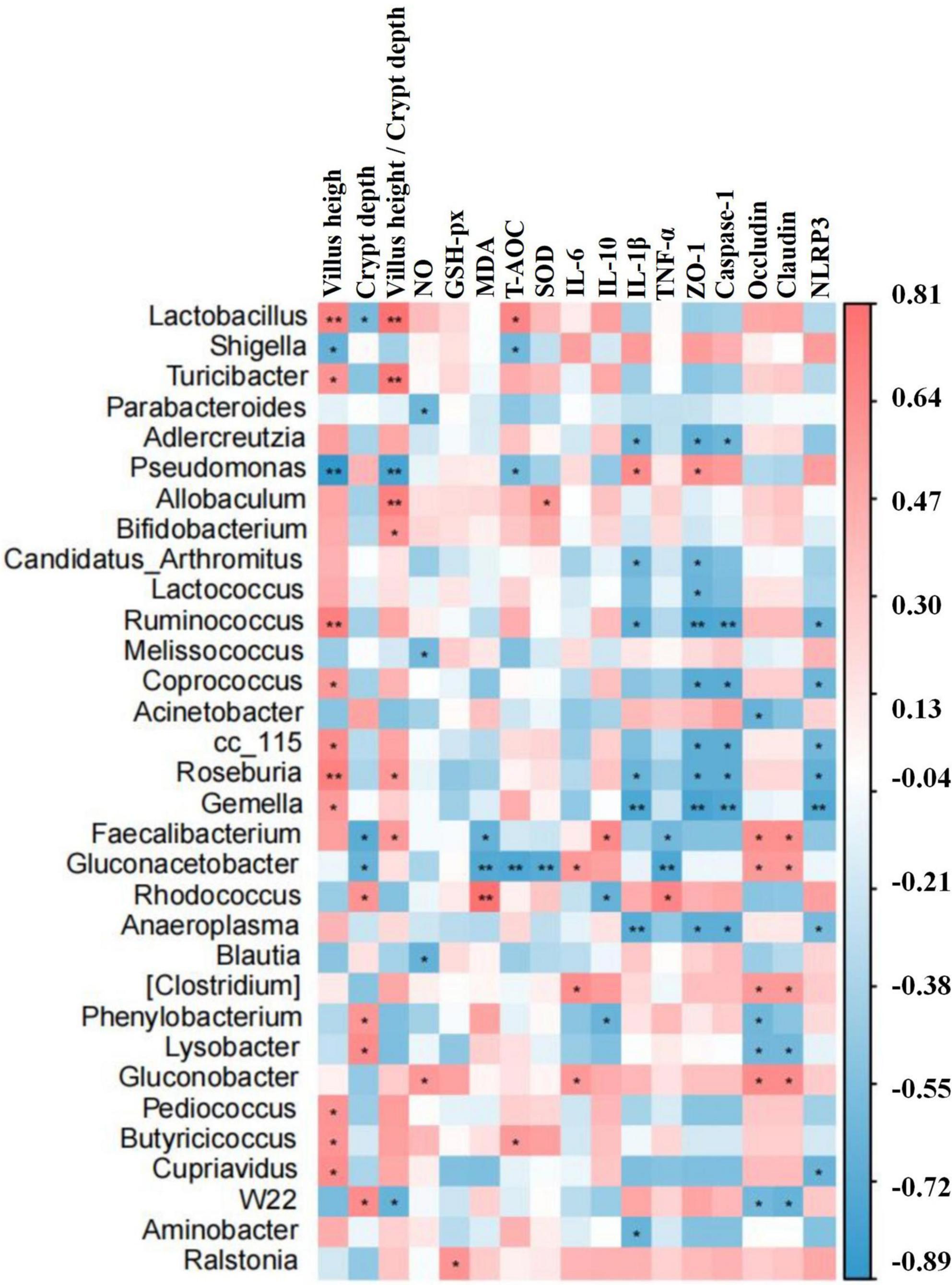
Figure 13. The differential bacteria of mouse were notably corrected with the indices of inflammatory cytokines, oxidative status, intestine morphology, and gene expressions by the statistical analysis system. Significance is presented as *p < 0.05 and **p < 0.01; data are presented as the mean ± SEM (n = 4).
In conclusion, we revealed that sodium acetate/sodium butyrate could alleviate LPS-induced diarrhea in mice by increasing beneficial bacteria and decreasing pathogens, which could regulate oxidative damage and inflammatory responses via NLRP3/Caspase-1 signaling (Figure 14). The current results may give insights into the prevention and treatment of diarrhea.
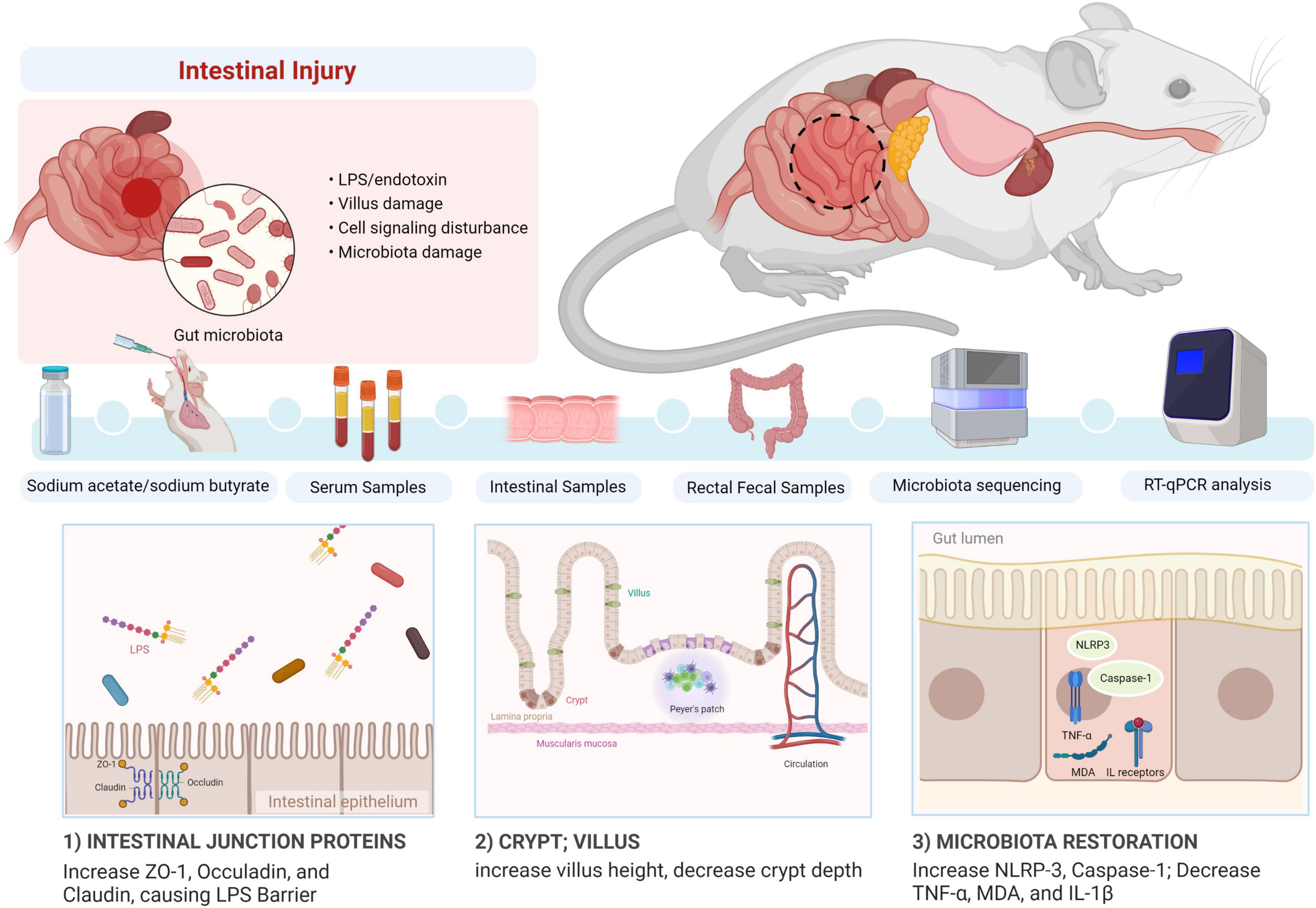
Figure 14. Summary of the potential mechanism of acetate/sodium butyrate alleviates LPS-induced diarrhea in mice.
The data presented in this study are deposited in the https://www.ncbi.nlm.nih.gov/ repository, accession numbers: PRJNA872847 and SAMN30471364-SAMN30471375.
This animal study was reviewed and approved by Laboratory Animals Research Centre of Jiangsu, China and the Ethics Committee of Nanjing Agricultural University (NJAU.No20220520108).
KL and QK: research idea, methodology, visualization, and supervision. XC, QK, XZ, CZ, PH, and HL: reagents, materials, and analysis tools. KL: writing—original draft and preparation. MK, ZB, II, HA, QS, and KL: writing—review and editing. All authors contributed to the article and approved the submitted version.
This study was supported by the Basic Research Funds for Central Universities (Tibet Joint Fund) (KYYZ2022003) and the National Natural Science Foundation of China (Nos. 31872514 and 32172900).
We thank Bioyi Biotechnology Co., Ltd., (Wuhan, China) for providing sequencing help in our research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alban, R. (2010). Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62, 142–160. doi: 10.1111/j.1574-6941.2007.00375.x
Al-Nussairawi, M., Risa, A., Garai, E., Varga, E., Szabó, I., Csenki-Bakos, Z., et al. (2020). Mycotoxin biodegradation ability of the Cupriavidus Genus. Curr. Microbiol. 77, 2430–2440. doi: 10.1007/s00284-020-02063-7
Alvarez, H. M., Hernández, M. A., Lanfranconi, M. P., Silva, R. A., and Villalba, M. S. (2021). Rhodococcus as biofactories for microbial oil production. Molecules 26:4871. doi: 10.3390/molecules26164871
Aziz, S., Abdullah, S., Anwar, H., Latif, F., and Mustfa, W. (2021). Effect of engineered nickel oxide nano particles on antioxidant enzymes in fresh water fish, Labeo rohita. Pak. Vet. J. 41, 424–428. doi: 10.29261/pakvetj/2021.044
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cao, Z., Gao, J., Huang, W., Yan, J., Shan, A., and Gao, X. (2022). Curcumin mitigates deoxynivalenol-induced intestinal epithelial barrier disruption by regulating Nrf2/p53 and NF-κB/MLCK signaling in mice. Food Chem. Toxicol. 167:113281. doi: 10.1016/j.fct.2022.113281
Chang, M., Wang, F., Ma, F., Jin, Y., and Sun, P. (2022). Supplementation with galacto-oligosaccharides in early life persistently facilitates the microbial colonization of the rumen and promotes growth of preweaning Holstein dairy calves. Anim. Nutr. 10, 223–233. doi: 10.1016/j.aninu.2022.04.009
Chao, A. (1984). Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270.
Chen, J., Tellez, G., Richards, J. D., and Escobar, J. (2015). Identification of potential biomarkers for gut barrier failure in broiler chickens. Front. Vet. Sci. 2:14. doi: 10.3389/fvets.2015.00014
Cho, Y., Han, J., Wang, C., Cooper, V., Schwartz, K., Engelken, T., et al. (2013). Case-control study of microbiological etiology associated with calf diarrhea. Vet. Microbiol. 166, 375–385. doi: 10.1016/j.vetmic.2013.07.001
Chuang, S., Chen, C., Hsieh, J., Li, K., Ho, S., and Chen, M. (2022). Development of next-generation probiotics by investigating the interrelationships between gastrointestinal Microbiota and Diarrhea in Preruminant Holstein Calves. Animals 12:695. doi: 10.3390/ani12060695
Coelho, M. G., Virgínio Júnior, G. F., Tomaluski, C. R., de Toledo, A. F., Reis, M. E., Dondé, S. C., et al. (2022). Comparative study of different liquid diets for dairy calves and the impact on performance and the bacterial community during diarrhea. Sci. Rep. 12:13394. doi: 10.1038/s41598-022-17613-1
Coura, F. M., Freitas, M. D., Ribeiro, J., de Leme, R. A., de Souza, C., Alfieri, A. A., et al. (2015). Longitudinal study of Salmonella spp., diarrheagenic Escherichia coli, Rotavirus, and Coronavirus isolated from healthy and diarrheic calves in a Brazilian dairy herd. Trop. Anim. Health Prod. 47, 3–11. doi: 10.1007/s11250-014-0675-5
Dashdorj, A., Jyothi, K. R., Lim, S., Jo, A., Nguyen, M. N., Ha, J., et al. (2013). Mitochondria-targeted antioxidant MitoQ ameliorates experimental mouse colitis by suppressing NLRP3 inflammasome-mediated inflammatory cytokines. BMC Med. 11:178. doi: 10.1186/1741-7015-11-178
Du, K., Bereswill, S., and Heimesaat, M. M. (2021). A literature survey on antimicrobial and immune-modulatory effects of butyrate revealing non-antibiotic approaches to tackle bacterial infections. Eur. J. Microbiol. Immunol. 11, 1–9. doi: 10.1556/1886.2021.00001
Ezzine, C., Loison, L., Montbrion, N., Bole-Feysot, C., Dechelotte, P., Coeffier, M., et al. (2022). Fatty acids produced by the gut microbiota dampen host inflammatory responses by modulating intestinal SUMOylation. Gut Microbes 14:2108280. doi: 10.1080/19490976.2022.2108280
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Faust, K., and Raes, J. (2012). Microbial interactions: From networks to models. Nat. Rev. Microbiol. 10, 538–550. doi: 10.1038/nrmicro2832
Fu, X., Liu, Z., Zhu, C., Mou, H., and Kong, Q. (2019). Nondigestible carbohydrates, butyrate, and butyrate-producing bacteria. Crit. Rev. Food Sci. 59, S130–S152. doi: 10.1080/10408398.2018.1542587
Gaudier, E., Rival, M., Buisine, M., Robineau, I., and Hoebler, C. (2009). Butyrate enemas upregulate Muc genes expression but decrease adherent mucus thickness in mice colon. Physiol. Res. 58, 111–119. doi: 10.33549/physiolres.931271
Gavin, M. D., Vincent, J. M., Jesse, Z., Svetlana, N. Y., James, R. B., Christopher, M. T., et al. (2019). PICRUSt2: An improved and customizable approach for metagenome inference. bioRxiv [Preprint]. doi: 10.1101/672295
Good, I. J. (1953). The population frequencies of species and the estimation of population parametes. Biometrika 40, 237–264. doi: 10.1093/biomet/40.3-4.237
Guilloteau, P., Martin, L., Eeckhaut, V., Ducatelle, R., Zabielski, R., and Van Immerseel, F. (2010). From the gut to the peripheral tissues: The multiple effects of butyrate. Nutr. Res. Rev. 23, 366–384. doi: 10.1017/S0954422410000247
Hamer, H. M., Jonkers, D., Venema, K., Vanhoutvin, S., Troost, F. J., and Brummer, R. J. (2008). Review article: The role of butyrate on colonic function. Aliment. Pharm. Ther. 27, 104–119. doi: 10.1111/j.1365-2036.2007.03562.x
Hassan, M., Ali, A., Wajid, M., Ahmad, A., Saleemi, M. K., Sarwar, Y., et al. (2021). Purification and antigenic detection of lipopolysaccharides of Salmonella enterica serovar Typhimurium isolate from Faisalabad, Pakistan. Pak. Vet. J. 41, 434–438.
He, Y., Li, Z., Xu, T., Luo, D., Chi, Q., Zhang, Y., et al. (2022). Polystyrene nanoplastics deteriorate LPS-modulated duodenal permeability and inflammation in mice via ROS drived-NF-κB/NLRP3 pathway. Chemosphere 307:135662. doi: 10.1016/j.chemosphere.2022.135662
Hu, P., Zhao, F., Zhu, W., and Wang, J. (2019). Effects of early-life lactoferrin intervention on growth performance, small intestinal function and gut microbiota in suckling piglets. Food Funct. 10, 5361–5373. doi: 10.1039/C9FO00676A
Jiminez, J. A., Uwiera, T. C., Abbott, D. W., Uwiera, R. R. E., and Inglis, G. D. (2017). Butyrate supplementation at high concentrations alters enteric bacterial communities and reduces intestinal inflammation in mice infected with Citrobacter rodentium. mSphere 2:e00243-17. doi: 10.1128/mSphere.00243-17
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., and Bäckhed, F. (2016). From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345. doi: 10.1016/j.cell.2016.05.041
Legendre, P., and Montréal, U. D. (2003). Numerical ecology, 2nd English Edn. Developments in environmental modelling Vol. 20. Amsterdam: Elsevier.
Leonel, A., and Alvarez-Leite, J. (2012). Butyrate implications for intestinal function. Curr. Opin. Clin. Nutr. Metab. Care 15, 474–479. doi: 10.1097/MCO.0b013e32835665fa
Li, K., Zeng, Z., Liu, J., Pei, L., Wang, Y., Li, A., et al. (2022). Effects of short-chain fatty acid modulation on potentially Diarrhea-causing pathogens in Yaks Through Metagenomic Sequencing. Front. Cell. Infect. Microbiol. 12:805481. doi: 10.3389/fcimb.2022.805481
Liu, C., Liang, X., Wei, X., Jin, Z., Chen, F., Tang, Q., et al. (2019). Gegen Qinlian decoction treats Diarrhea in piglets by modulating gut microbiota and short-chain fatty Acids. Front. Microbiol. 10:825. doi: 10.3389/fmicb.2019.00825
Liu, G., Pilla, G., and Tang, C. M. (2019). Shigellahost: Pathogen interactions: Keeping bacteria in the loop. Cell. Microbiol. 21:e13062. doi: 10.1111/cmi.13062
Liu, H., Li, X., Shi, S., Zhou, Y., Zhang, K., Wang, Y., et al. (2022). Chlorogenic acid improves growth performance and intestinal health through autophagy-mediated nuclear factor erythroid 2-related factor 2 pathway in oxidatively stressed broilers induced by dexamethasone. Poult. Sci. 101:102036. doi: 10.1016/j.psj.2022.102036
Liu, J., Wang, X., Zhang, W., Kulyar, M. F., Ullah, K., Han, Z., et al. (2022). Comparative analysis of gut microbiota in healthy and diarrheic yaks. Microb. Cell Fact. 21:111. doi: 10.1186/s12934-022-01836-y
Liu, X., Lu, B., Fu, J., Zhu, X., Song, E., and Song, Y. (2021). Amorphous silica nanoparticles induce inflammation via activation of NLRP3 inflammasome and HMGB1/TLR4/MYD88/NF-kb signaling pathway in HUVEC cells. J. Hazard. Mater. 404:124050. doi: 10.1016/j.jhazmat.2020.124050
Liu, Z., Peng, Y., Zhao, L., and Li, X. (2022). MFE40—the active fraction of Mume Fructus alcohol extract—alleviates Crohn’s disease and its complications. J. Ethnopharmacol. 296:115465. doi: 10.1016/j.jep.2022.115465
Mahadevan, S., Shah, S. L., Marrie, T. J., and Slupsky, C. M. (2008). Analysis of metabolomic data using support vector machines. Anal. Chem. 80, 7562–7570. doi: 10.1021/ac800954c
Mehmood, K., Zhang, H., Yao, W., Jiang, X., Waqas, M., Li, A., et al. (2019). Protective effect of Astragaloside IV to inhibit thiram-induced tibial dyschondroplasia. Environ. Sci. Pollut. Res. 26, 16210–16219. doi: 10.1007/s11356-019-05032-1
Murtaza, S., Khan, J. A., Aslam, B., and Faisal, M. N. (2021). Pomegranate peel extract and quercetin possess antioxidant and hepatoprotective activity against concanavalin A-induced liver injury in mice. Pak. Vet. J. 41, 197–202.
Nishiwaki, H., Ito, M., Ishida, T., Hamaguchi, T., Maeda, T., Kashihara, K., et al. (2020). Meta-Analysis of Gut Dysbiosis in Parkinson’s disease. Mov. Disord. 35, 1626–1635. doi: 10.1002/mds.28119
Ondov, B. D., Bergman, N. H., and Phillippy, A. M. (2011). Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12:385. doi: 10.1186/1471-2105-12-385
Pielou, E. C. J. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Ponziani, F. R., Scaldaferri, F., Siena, M., Mangiola, F., Matteo, M., Pecere, S., et al. (2020). Increased Faecalibacterium abundance is associated with clinical improvement in patients receiving rifaximin treatment. Benef. Microbes 11, 519–525. doi: 10.3920/BM2019.0171
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rajab, S., Tabandeh, F., Shahraky, M. K., and Alahyaribeik, S. (2020). The effect of lactobacillus cell size on its probiotic characteristics. Anaerobe 62:102103. doi: 10.1016/j.anaerobe.2019.102103
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: A versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Schmoeller, E., Matos, A. D. C. D., Rahal, N. M., Feijo, J. O., Brauner, C. C., Pino, F. A. B. D., et al. (2021). Diarrhea duration and performance outcomes of pre-weaned dairy calves supplemented with bacteriophage. Can. J. Anim. Sci. 102, 165–174. doi: 10.1139/cjas-2021-0074
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shannon, C. E. (1948). A mathematical theory of communication, 1948. Bell Syst. Tech. J. 27, 3–55. doi: 10.1002/j.1538-7305.1948.tb01338.x
Sho, T., and Xu, J. (2019). Role and mechanism of ROS scavengers in alleviating NLRP3-mediated inflammation. Biotechnol. Appl. Biochem. 66, 4–13. doi: 10.1002/bab.1700
Takeuchi, T., Miyauchi, E., Kanaya, T., Kato, T., Nakanishi, Y., Watanabe, T., et al. (2021). Acetate differentially regulates IgA reactivity to commensal bacteria. Nature 595, 560–564. doi: 10.1038/s41586-021-03727-5
Ulluwishewa, D., Anderson, R. C., Mcnabb, W. C., Moughan, P. J., Wells, J. M., and Roy, N. C. (2011). Regulation of tight junction permeability by intestinal bacteria and dietary. J. Nutr. 141, 769–776. doi: 10.3945/jn.110.135657
Wang, X., Zhang, M., Wang, W., Lv, H., Zhang, H., Liu, Y., et al. (2020). The in vitro effects of the probiotic strain, Lactobacillus casei ZX633 on gut microbiota composition in infants with Diarrhea. Front. Cell. Infect. Microbiol. 10:576185. doi: 10.3389/fcimb.2020.576185
Wei, H., Li, X., Tang, L., Yao, H., Ren, Z., Wang, C., et al. (2020). 16S rRNA gene sequencing reveals the relationship between gut microbiota and ovarian development in the swimming crab Portunus trituberculatus. Chemosphere 254:126891. doi: 10.1016/j.chemosphere.2020.126891
Wu, X., Wang, J., Amanze, C., Yu, R., Li, J., Wu, X., et al. (2022). Exploring the dynamic of microbial community and metabolic function in food waste composting amended with traditional Chinese medicine residues. J. Environ. Manage. 319:115765. doi: 10.1016/j.jenvman.2022.115765
Xu, B., Yan, Y., Yin, B., Zhang, L., Qin, W., Niu, Y., et al. (2021). Dietary glycyl-glutamine supplementation ameliorates intestinal integrity, inflammatory response, and oxidative status in association with the gut microbiota in LPS-challenged piglets. Food Funct. 12, 3539–3551. doi: 10.1039/D0FO03080E
Yosi, F., Sharma, S., Sener-Aydemir, A., Koger, S., Baskara, A. P., and Metzler-Zebeli, B. U. (2022). Short-chain fatty acids promote jejunal barrier function and caecal muscle contractibility in laying hens ex vivo. Br. Poult. Sci. 63, 406–413. doi: 10.1080/00071668.2021.2008312
Yue, Y., He, Z., Zhou, Y., Ross, R. P., Stanton, C., Zhao, J., et al. (2020). Lactobacillus plantarum relieves diarrhea caused by enterotoxin-producing Escherichia coli through inflammation modulation and gut microbiota regulation. Food Funct. 11, 10362–10374. doi: 10.1039/D0FO02670K
Zelová, H., and Hošek, J. (2013). TNF-α signalling and inflammation: Interactions between old acquaintances. Inflamm. Res. 62, 641–651. doi: 10.1007/s00011-013-0633-0
Zhang, B., Yue, J., Guo, Y., Liu, T., Zhou, M., Yang, Y., et al. (2020). Effects of bioporous carriers on the performance and microbial community structure in side-stream anaerobic membrane bioreactors. Can. J. Microbiol. 66, 475–489. doi: 10.1139/cjm-2019-0632
Zhang, H., Elolimy, A. A., Akbar, H., Thanh, L. P., Yang, Z., and Loor, J. J. (2022). Association of residual feed intake with peripartal ruminal microbiome and milk fatty acid composition during early lactation in Holstein dairy cows. J. Dairy Sci. 105, 4971–4986. doi: 10.3168/jds.2021-21454
Keywords: sodium acetate, sodium butyrate, LPS, diarrhea, microbiota
Citation: Chen X, Kong Q, Zhao X, Zhao C, Hao P, Irshad I, Lei H, Kulyar MF-e-A, Bhutta ZA, Ashfaq H, Sha Q, Li K and Wu Y (2022) Sodium acetate/sodium butyrate alleviates lipopolysaccharide-induced diarrhea in mice via regulating the gut microbiota, inflammatory cytokines, antioxidant levels, and NLRP3/Caspase-1 signaling. Front. Microbiol. 13:1036042. doi: 10.3389/fmicb.2022.1036042
Received: 03 September 2022; Accepted: 29 September 2022;
Published: 28 October 2022.
Edited by:
Tang Zhaoxin, South China Agricultural University, ChinaCopyright © 2022 Chen, Kong, Zhao, Zhao, Hao, Irshad, Lei, Kulyar, Bhutta, Ashfaq, Sha, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Wu, d3V5aTIwMDFjbkAxNjMuY29t; Kun Li, bGszMDA1QG5qYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.