- 1College of Horticulture and Plant Protection, Inner Mongolia Agricultural University, Hohhot, China
- 2Institute of Agro-Food Technology, Jilin Academy of Agricultural Sciences, Changchun, China
- 3Jilin Provincial Key Laboratory of Agricultural Biotechnology, Jilin Academy of Agricultural Sciences, Changchun, China
Introduction: Endophytes are non-pathogenic inhabitants of healthy plant tissues and have been found to promote plant growth and health. The endophytic bacterial strain Q2H1 was isolated from the roots of the potato and was identified to exhibit growth-promoting effects in potato plants.
Methods: Whole-genome sequencing was performed to reveal the mechanism underlying its growth-promoting effect. The obtained sequencing data of approximately 5.65 MB encompassed 5,533 coding sequences. Of note, nine secondary metabolite gene clusters, including siderophore gene clusters, closely associated with plant growth promotion (PGP) were predicted by antiSMASH software. Comparative genomic analysis revealed that Q2H1 belongs to the genus Peribacillus. By gene function annotation, those genes related to plant growth-promoting activities, including indole-3-acetic acid (IAA) synthesis in tryptophan metabolism, siderophore biosynthetic activity, phosphate solubilization, nitrogen fixation, and related genes, were summarized. IAA (14.4 μg/ml) was presumptively produced by Q2H1 using the Salkowski colorimetric method. A total of five genes, namely, phoU, pstB, pstA1, pstC, and pstS, were annotated for phosphate solubilization, which is associated with the ability of the Q2H1 strain to solubilize phosphate under in vitro conditions.
Results: It is revealed that genes in the Q2H1 genome associated with nitrogen fixation belonged to three groups, namely, nitrogen fixation (nifU, sufU, salA, and nifS), nitrogen metabolism (nirA, nrtB, and nasA), and glutamate synthesis (glnA, gltB, gltD, and gudB), supported by evidence that Q2H1 grew on medium without nitrogen. We have also identified a siderophore gene cluster located on the chromosome of Q2H1, including seven genes (viz., rbsR, rhbf, rhbE, rhbD, rhbC, rhbA, ddc, and an unknown gene). In the in vitro assay, a prominent brown circle around the colony was produced on the chrome azurol S medium at 48 and 72 h post-inoculation, indicating that the siderophore gene cluster in Q2H1 harbored the ability to produce siderophores.
Conclusion: In summary, these findings implied that identifying strain-specific genes for their metabolic pathways in bacterial endophytes may reveal a variety of significant functions of plant growth-promoting mechanisms.
Introduction
Endophytes are microbes that live in plants but do not cause plant diseases (Gouda et al., 2016). Cavalcante and Dobereiner (1988) first identified and isolated an endophytic nitrogen-fixing bacterium from sugarcane tissue and named it Acetobacter diazotrophicus because of its ability to oxidize ethanol to acetic acid and fix nitrogen under acidic conditions. Endophytes have been demonstrated to play crucial roles in plant disease resistance, secondary metabolite synthesis, plant growth promotion (PGP), and environmental stress resistance (Patel and Archana, 2017; Afzal et al., 2019; Ouyabe et al., 2020; Lopes Antunes et al., 2022).
Several studies have shown that endophytes regulate plant growth by nitrogen fixation, phosphate solubilization, siderophore production, 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity, and indole-3-acetic acid (IAA) synthesis (Yan et al., 2018). Endophytic bacteria from rice plants (Oryza sativa) were the earliest investigated plant endophytes, which showed versatile PGP activities, such as phosphate solubilization, siderophore production, and IAA synthesis (Etesami et al., 2014). Endophytes and symbiotic nitrogen-fixing bacteria in the nodules of leguminous plants have been demonstrated to improve the pathogen resistance of host plants (Zgadzaj et al., 2015). In sugarcane, endophytic bacteria, including Enterobacter sp. and Klebsiella sp., were found to significantly promote host plant growth by nitrogen fixation (Lin et al., 2015; Taule et al., 2016), which was also reported to assist in stubborn plant growth on barren dunes (Zhang and White, 2021). Jatropha curcas L., a fuel plant that grows on barren sandy soil, contains a rich population of growth-promoting and nitrogen-fixing bacteria (Methylobacterium strain L2-4), which significantly increases host plant growth and seed yield (Madhaiyan et al., 2014). Endophytes isolated from potato roots exhibited the characteristics of nitrogen fixation, phytohormone production, and resistance against pathogenic bacteria. It is also demonstrated that inoculation of endophytic bacteria in potatoes increased the shoot and root biomass of the plant and reduced the pathogenic bacterial biomass (Pageni et al., 2014).
Endophytes colonize plants, stably protecting them from environmental stresses; however, they independently divide, reproduce, and transmit inside plants (Adeleke et al., 2021). Azorhizobium caulinodans is known to enter the root system of cereals via intercellular invasion between epidermal cells and to internally colonize the plant, including the xylem (Dreyfus et al., 1988). Many endophytic bacteria have been reported to successfully colonize non-leguminous plants, such as rice, wheat, maize, and sugarcane, forming symbiotic relationships and fixing nitrogen (Santoyo et al., 2016). It was reported that plant height, fresh weight, and chlorophyll content of sugarcane significantly increased when inoculated with the endophytic nitrogen-fixing bacteria Klebsiella sp. DX120E was observed by GFP-labeled DX120E, colonization in root hairs, new lateral roots, wounds, and leaves (Gu et al., 2018). The colonization of endophytic nitrogen-fixing bacteria in plants has an obvious advantage to plant growth over rhizosphere and symbiotic nitrogen-fixing bacteria.
In most cases, the mechanism of PGP by endophytes may result from either better uptake of soil nutrients or endophytic nitrogen fixation (Santoyo et al., 2016). Furthermore, endophytes can also secrete phytohormones such as IAA, gibberellin, and cytokinin that stimulate plant growth (Orozco-Mosqueda et al., 2020). IAA synthesis plays a vital role in promoting plant growth. Bacillus subtilis LK14 was reported to produce IAA and ACC deaminase in vitro. After the inoculation of endophytic Bacillus subtilis LK14 in tomato plants, the biomass of the shoot and root and the chlorophyll content significantly increased (Khan et al., 2016). Various IAA biosynthetic pathways have been proposed for endophytes, including tryptophan-dependent and tryptophan-independent pathways. Tryptophan-dependent pathways include indole-3-pyruvate (IPyA), indole acetamide synthesis (IAM), tryptamine (TAM), indole-3-acetonitrile (IAN), and tryptophan side chain oxidase (TSO) pathways (Liu et al., 2008). The TAM pathway is extensively studied in bacteria. A previous study found that Azospirillum can convert exogenous tryptamine into IAA (Normanly, 2010). It was reported that Pantoea agglomerans contain both IAM and IPyA genes to promote IAA synthesis. Also, the bacterial pathogen Pseudomonas syringae is reported to catalyze the formation of IAA from indole-3-acetaldehyde (IAAld) by indole-3-acetaldehyde dehydrogenase (AldA) (Zhao, 2012).
Siderophores are produced by endophytes. They usually combine with iron to form complexes, contributing to iron uptake directly from the complexes or by exchanging ligands (Garzón-Posse et al., 2019). With the increase in iron uptake from bacterial siderophores, plants increase their development and growth, for example, in rice, wheat, corn, and other crops (Santoyo et al., 2016; Afzal et al., 2019). Siderophores can form steady complexes with heavy metals, such as Cd, Cu, Pb, Zn, or radionuclide-U, reducing phytotoxicity in heavy metal-contaminated areas and improving plant growth (Grobelak and Hiller, 2017).
In addition to promoting plant growth, endophytes enhance the tolerance of host plants to abiotic and biotic stresses. Compared with the control, inoculation with the endophyte Neotyphodium spp. enhanced the tolerance of grasses to drought stress (Malinowski and Belesky, 2000). Inoculation with the bacterial endophyte Burkholderia phytofirmans strain PsJN can promote the growth of grapes, maintain physiological activity at low temperatures, and improve its ability to resist cold stress (Redman et al., 2002; Barka et al., 2006). Endophytes also play a significant role in salt stress and phytoremediation of contaminated soil and water (Mei and Flinn, 2010). Endophytes can also decrease biotic stress in plants. An isolate of Burkholderia sp. KJ006 from rice soil has a broad spectrum of antifungal properties (Cho et al., 2007). Bacillus subtilis (Lu144) significantly reduced the incidence of bacterial wilt in mulberry before infection by the pathogen. In tomato plants infected with potato virus Y (PVY), potato virus X (PVX), endophytic Bacillus subtilis 26D, and Ttl2 were reported to induce systemic resistance (ISR) by activating the transcription of salicylic acid- and jasmonic acid-related genes and regulating hormone levels, which resulted in reduced virus accumulation in plants (Veselova et al., 2022).
Whole-genome analysis of endophytes can be used to classify genes associated with PGP activities while providing insights into their molecular and functional mechanisms (Kamada et al., 2015; Lo et al., 2018; Lin et al., 2019; Franco-Sierra et al., 2020; Guo et al., 2020). Whole-genome sequencing of the Enterobacter roggenkampii ED5 strain, an endophytic strain of sugarcane, revealed a series of genes associated with PGP activity, including nitrogen fixation, production of plant hormones, biotic and abiotic stresses, induction of resistance, and root colonization (Guo et al., 2020). Based on the whole-genome sequencing data of Bacillus subtilis EA-CB0575, possible mechanisms of PGP were proposed by analyzing metabolic pathways (Franco-Sierra et al., 2020). Whole-genome sequencing of endophytic Bacillus toyonensis BAC3151 revealed its potential role in anti-microbial development associated with controlling crop diseases after analyzing secondary metabolites (Lopes et al., 2017).
In our previous study, we isolated a bacterial strain Q2H1 from potato roots treated at high temperatures and reported that it had an apparent growth-promoting effect on the potato; however, its underlying mechanism was unknown. In the current study, we determined the PGP activity of Q2H1 in potatoes and explored the possible PGP mechanisms of Q2H1 by whole-genome sequencing analysis. We analyzed the genomic characteristics of Q2H1 and compared them with those of other species. Gene clusters that were possibly related to IAA production, siderophore production, phosphate solubilization, and nitrogen fixation were analyzed. Our findings investigated the fundamental knowledge of endophytic strain Q2H1 and possible mechanisms of PGP, which will be helpful for its potential future application in enhancing potato production.
Materials and methods
Q2H1 strain and culture conditions
The endophytic strain Q2H1 was isolated from potato (Atlantic) roots and stored at –80°C with glycerol (50%). The cells were cultured in Luria–Bertani (LB) medium (tryptone: 10 g, yeast extract: 5 g, NaCl: 10 g, agar: 15 g in 1,000 ml of distilled water) at 28°C in an electrothermal constant-temperature incubator.
Genomic DNA was extracted from a 12-h logarithmic growth period LB liquid cell suspension of endophytic strain Q2H1 using a bacterial genomic DNA extraction kit (Tiangen Biotech, Beijing, China). DNA quality and concentration were estimated using NanoDrop One (Thermo Scientific). The 16S rRNA gene of Q2H1 was amplified by polymerase chain reaction (PCR) using a pair of primers (7F and 1540R). PCR amplification was performed using genomic DNA as the template (Supplementary Table 1). The PCR product was purified and sequenced by BGI (Shenzhen, China). The sequencing results were compared with the National Center for Biotechnology Information (NCBI) GenBank database. A phylogenetic tree was created using Molecular Evolutionary Genetic Analysis (MEGA, version 7.0, Mega Limited, Auckland, New Zealand).
Plant growth-promoting assay of Q2H1 in potatoes in a greenhouse
The Q2H1 strain was stored at –80°C, and the inoculation loop was dipped in the bacterial solution and streaked onto a newly prepared LB solid medium plate at 28°C until the growth of the first colony. Q2H1 colonies were picked using an inoculation loop and then were inoculated into 100 ml of liquid LB medium at 28°C for 24 h at 180 rpm. The resultant liquid was then centrifuged at 4,000 rpm for 10 min, and an equal volume of sterile water was used to suspend, dilute to OD600 = 1, and then set aside. For this study, the Atlantic potato variety was used. The soil and nutrient soil were mixed in a 1:1 ratio by volume, watered, and mixed thoroughly. The exact size of potato sprouting tubers was sown in pots, with one piece each. When four or more leaves were grown from the tubers, 30 ml of the aforementioned bacterial suspension was inoculated with root irrigation, and 30 ml of sterile water was added as the negative control. The plants were grown under greenhouse conditions (22°C, 16-h light, and 8-h dark). To examine the effects of Q2H1 on potato growth, plant height, fresh weight, root weight, and chlorophyll content were measured 30 days post-inoculation. The experiment was repeated three times.
Sample preparation of Q2H1 and genome sequencing
A single colony of Q2H1 from the culture plate was inoculated into 20 ml of liquid LB medium and cultured at 28°C with shaking at 180 rpm for 12 h. Bacterial liquid (2 ml) was taken in a centrifuge tube and centrifuged at 14,000 × g for 1 min at 25°C; the medium was discarded; and the bacteria were quickly transferred to liquid nitrogen for 1–3 h and then transferred to –80°C for storage.
Q2H1 samples were sent to Biomarker Technologies (Beijing, China) for genome sequencing with PacBio platform single-molecule real-time (SMRT) sequencing. The experimental procedure was carried out following the standard protocol provided by PacBio and included sample quality detection, library construction, library quality detection, and library sequencing. PacBio sequencing technology uses an SMRT chip as a sequencing carrier. DNA polymerase was bound to the template in the nanopore inside the SMRT chip, and the dNTP mix was labeled with four-color fluorescence. In the base pairing stage, different bases labeled with different lights are determined according to the wavelength and peak value of the light. SMRT Link version 8.0 software was used to process the off-machine PB data to obtain circular consensus sequencing (CCS) data. After further filtering the reads of short fragments (length < 2,000 bp), the total dataset was obtained. Hifiasm v0.12 (r304) software was used to assemble the filtered CCS data, and Pilon version 1.22 software was used for further error correction using second-generation data to obtain genomes with higher accuracy for subsequent analysis.
Genome component analysis and functional annotation
Genomic component analyses include coding gene prediction (Hyatt et al., 2010), repeat sequence prediction, clustered regularly interspaced palindromic repeats (CRISPR) sequence prediction, pseudogene prediction, gene island prediction, prophage prediction, gene cluster prediction, promoter prediction, and paralogous gene prediction (Bland et al., 2007; Bertelli and Brinkman, 2018; Blin et al., 2019). Biosynthetic gene clusters (BGCs) in bacterial genome sequences were identified and analyzed using antiSMASH v5.0.0 software and combined with the results of NCBI BLAST alignment analysis. The sequencing results were compared with seven databases, namely, the non-redundant protein database (Nr), gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), eggNOG, Pfam, SwissProt, and TrEMBL for gene function annotation and the annotation information of each database. Using the predicted genomic data, a genome map of Q2H1 was created using Circos v0.66 software (Krzywinski et al., 2009). From the KEGG database, the genes related to IAA production, phosphate solubilization, and nitrogen fixation were analyzed. Gene clusters from the secondary metabolites of Q2H1 were investigated.
Comparative genomics analysis of Q2H1
Among average nucleotide identity (ANI) software calculations and tools, Jspecies is one of the most commonly used. We used the online website of JspeciesWS1 to determine whether two or more genomes belonged to the same species by calculating the ANI value (Richter et al., 2016). The genomic data of eight closely related strains were downloaded from the NCBI database. ANI results were analyzed using TBtools software (version v1.098765), and the results were displayed as heat maps (Chen et al., 2020).
Based on the ANI analysis results of the Q2H1 genome, two strains (Peribacillus frigoritolerans strain Ant232 and Peribacillus simplex strain NBRC 15720) with the closest genetic relationship to Q2H1 were used for comparative genomic analysis. The genome sequences of P. frigoritolerans strain Ant232 (GenBank accession number: CP084539.1) and P. simplex strain NBRC 15720 (GenBank accession number: CP017704.1) were downloaded from the NCBI database. To conduct the gene family clustering analysis, OrthoMCL version 2.0 software (Li et al., 2003) was used to perform family clustering on the protein sequences predicted by the sequencing strains and the protein sequences of the reference genome. We then analyzed the gene families, including strain-specific gene families and common gene families, in different strains. Venn diagrams or petal diagrams were constructed to identify gene families. Functional annotations based on the Pfam database were used in this study. For genome collinearity analysis using the Q2H1 genome as the reference genome, the protein sequences of Q2H1 were compared with those of the reference genome using BLAST (Wang et al., 2012). Then, according to the position of the homologous gene in the genome sequence, a collinear relationship was obtained at the nucleotide level.
Detection of siderophore from Q2H1 by chrome azurol S assay
To confirm the expression of the siderophore gene cluster, we performed an in vitro assay in chrome azurol S (CAS) assay medium (sucrose: 2 g, casein acid hydrolyzed: 3 g, 1 mM CaCl2: 1 ml, 1 mM MgSO4⋅7H2O: 20 ml, agar: 20 g, distilled water: 1,000 ml. The mixture was sterilized in an autoclave at 121°C for 30 min) and waited until the medium temperature dropped approximately to 60°C. Subsequently, 5 ml of phosphate buffer and 5 ml of CAS staining solution were added per 100 ml of medium. The CAS staining solution was dissolved in water to form a blue complex with hexadecyl trimethyl ammonium bromide (CTAB) (Chen, 2020). To culture Q2H1 in LB solid medium, a single colony was picked with a sterile toothpick and inoculated on the CAS detection medium. When the complex was degraded, it produced a yellow or brown halo around the colony. The experiment was repeated three times, the aperture diameter was measured at 48 and 72 h, and the siderophore ability was analyzed.
Nitrogen-fixing and phosphate solubilization of Q2H1
Phosphate solubilization experiments were validated on organic phosphate medium [glucose: 10.0 g, (NH4)2SO4: 0.5 g, NaCl: 0.3 g, KCl: 0.3 g, FeSO4⋅7H2O: 0.03 g, MgSO4⋅7H2O: 0.3 g, MnSO4⋅4H2O: 0.03 g, Ca3(PO4)2: 5 g, agar: 15 g, distilled water: 1,000 ml, pH 7.0] and inorganic phosphate medium (NaCl: 5.0 g, beef extract: 5.0 g, peptone: 10.0 g, agar: 15 g, distilled water: 1,000 ml, pH 7–7.5, add 3 ml of fresh egg yolk liquid to each 50 ml for temporary use) (Gao et al., 2015). A single colony of Q2H1 was picked using a sterile toothpick and inoculated to the inorganic phosphate medium and organic phosphate medium. The cells were incubated at 28°C for 7 days to observe the development of a halo. Nitrogen-fixing ability was measured using Ashby nitrogen-free medium (mannitol: 10 g, KH2PO4: 0.2 g, MgSO4⋅7H2O: 0.2 g, NaCl: 0.2 g, CaSO4⋅2H2O: 0.1 g, CaCO3: 5 g, agar: 15 g, distilled water: 1,000 ml) (Singh et al., 2020). Q2H1 was inoculated into the Ashby medium at 28°C for 7 days to determine its viability.
Indole-3-acetic acid production assay
Indole-3-acetic acid production was estimated using the Salkowski colorimetric method in the presence of tryptophan (Patten and Glick, 2002). Strains were grown overnight in the DF medium (Zhang et al., 2016), and 200 μl was transferred to the DF medium containing 0.1% L-tryptophan. The strains were cultured at 28°C for 7 days and then centrifuged at 8,000 rpm for 10 min. In total, 1 ml of the fermentation supernatant was mixed with 2 ml of Fe-H2SO4 solution (1 ml of 0.5 M FeCl3⋅6H2O in 75 ml of 6.13 M H2SO4) and placed in a dark room for 45 min. The IAA concentration was presumed by measuring the absorbance of the samples at 450 nm.
Antagonism assay against phytopathogenic fungi
In the in vitro antifungal activity test of the strain Q2H1, five pathogenic fungi, namely Fusarium oxysporum, Fusarium commune, Fusarium graminearum, Rhizoctonia solani, and Stemphylium solani, were tested on the PDA medium. These pathogenic fungi are from the College of Horticulture and Plant Protection, Inner Mongolia Agricultural University, China. A 5 mm diameter of the culture medium from each pathogenic fungi was cut and placed on the PDA plate together with the Q2H1 strain. The plate containing only pathogenic fungi was used as the control. Then, the plates were incubated at 28°C for 3–5 days, the fungus radii were measured, and the antifungal rates were calculated.
Statistical analysis
All genomic analyses were performed following the manufacturer’s instructions. All PGP trials were performed in triplicate, and data were analyzed using analysis of variance and Duncan’s multiple range test.
Results
Plant growth promotion of endophytic strain Q2H1 in potato
We isolated the endophytic strain Q2H1 from potato roots (Figure 1A). The Q2H1 growth promotion assay was performed on potato plants grown in a greenhouse (Figure 1B). Compared with the control (treated with sterile water), potato plants treated with Q2H1 bacterial suspension showed an average increase in fresh weight of 6.83 g (Figure 1C) and an average increase in root weight of 1.5 g (Figure 1D), with significant differences. Plant height increased by 1.11 cm, chlorophyll content increased by 3.89, and the difference was not significant compared with the control.
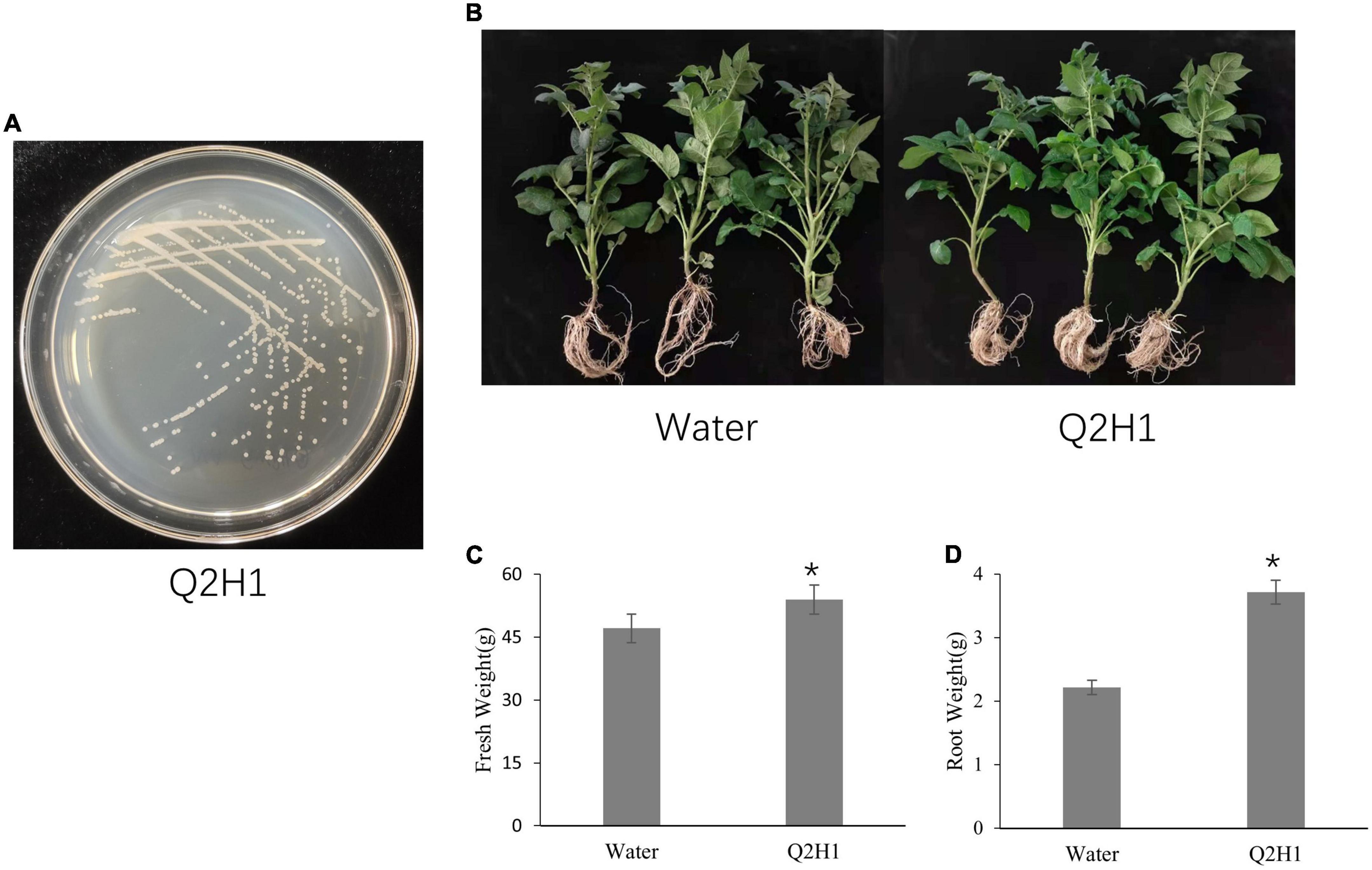
Figure 1. Plant growth promotion of endophytic strain Q2H1 in potato: (A) Colony morphology of strain Q2H1 on LB medium; (B) photographs of potato plants at 30 days post-inoculation with Q2H1; (C,D) fresh weight and root weight of potato plants measured at 30 days post-inoculation with Q2H1; *show the significant differences (P < 0.05) according to multiple ranges of LSD.
Genomic properties of endophytic strain Q2H1
The Q2H1 strain was identified by 16S rRNA gene sequencing, and the obtained sequences were matched with the nucleotide sequences of the National Center for Biotechnology Information (NCBI) GenBank database using the Basic Local Alignment Search Tool (BlastN) program. We constructed a phylogenetic tree using the BLAST results for Q2H1, and the results are shown in Supplementary Figure 1. Q2H1 was preliminarily identified as Peribacillus.
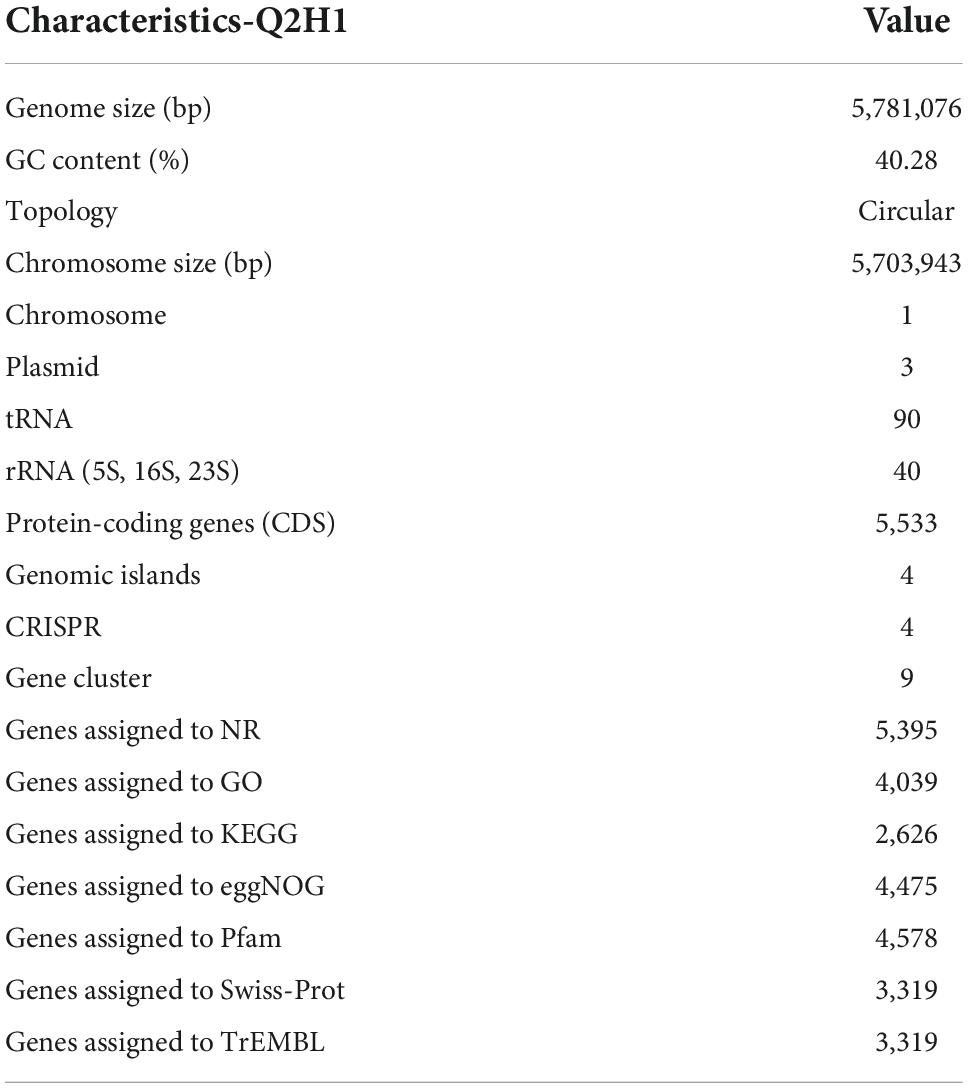
To explore the genomic properties of the endophytic strain Q2H1, we performed whole-genome sequencing using single-molecule real-time (SMRT) sequencing (Biomarker Technologies Beijing China). The general characteristics of the Q2H1 genome are listed in Table 1. The Q2H1 genome contains a chromosome of 5,703,943 bp and three plasmids with an average genome-wide GC content of 40.28%. The genome was predicted to contain 5,533 coding sequences (CDSs). Using the CRT, antiSMASH, and IslandPath-DIMOB software, we predicted four clustered regularly interspaced palindromic repeat (CRISPR) sequences, nine gene clusters, and four gene islands. Moreover, the genome of Q2H1 includes 90 tRNA and 40 rRNA genes (13 5S rRNA, 14 16S rRNA, and 13 23S rRNA) (Kalvari et al., 2018; Chan and Lowe, 2019). The number of CDSs assigned to the KEGG, eggNOG, and GO databases was 2,626, 4,475, and 4,039, respectively. The results of plasmid genome annotation are shown in Supplementary Table 2 and Supplementary Figure 2. Circos visualizes the genome, which could more clearly explore the relationship between genomic components or locations (Figure 2). The different colors in Figure 2 represent different functions of genes annotated in the COG database, including amino acid transport metabolism, general function prediction, secondary metabolism biosynthesis, transport, and catabolism. A complete genome sequence of this strain has been submitted at GenBank with accession numbers CP110132-CP110135.
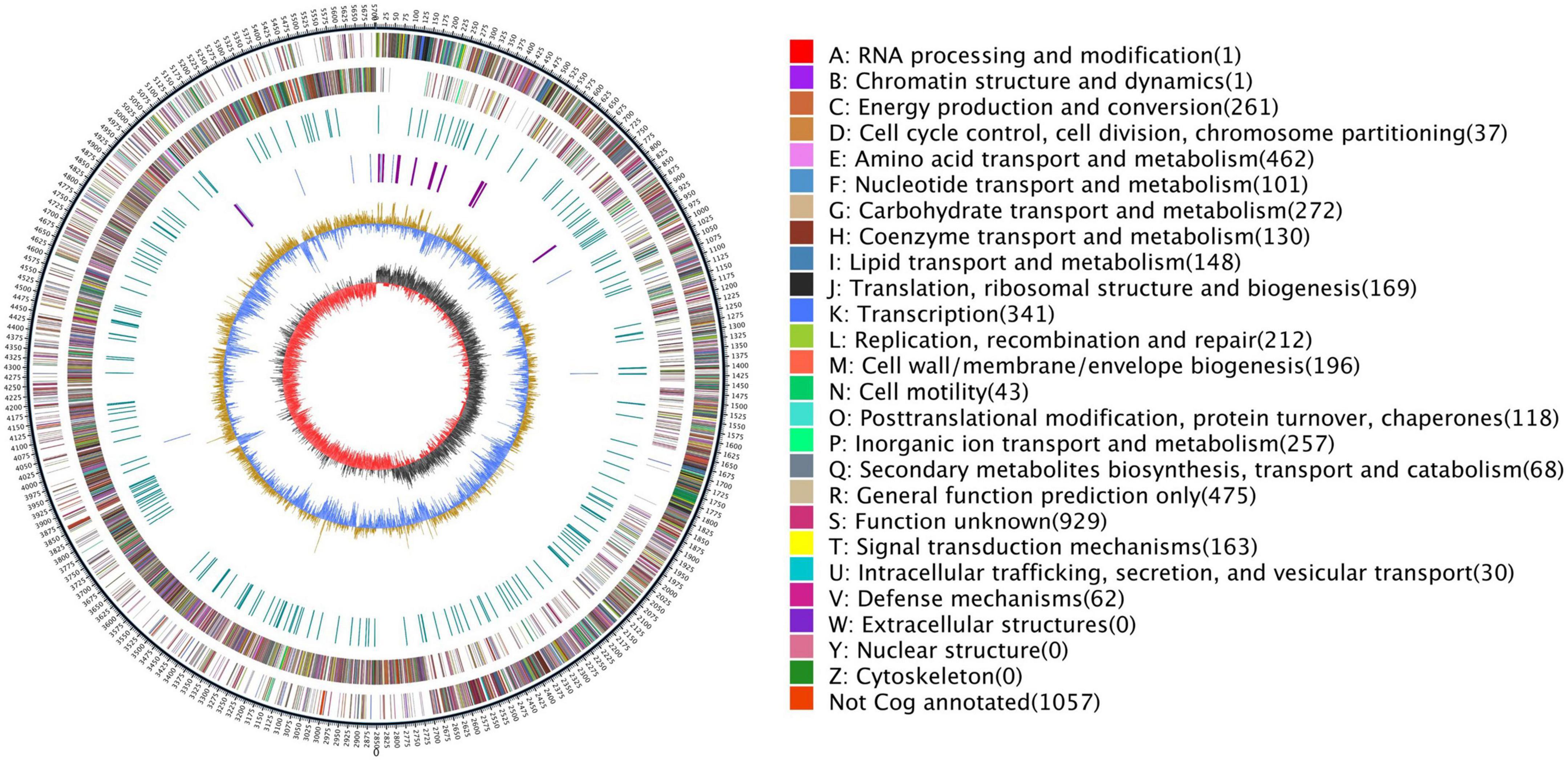
Figure 2. Genome map of Q2H1: The outermost circle is the indication of the genome size; the second and third circles are genes on the positive and negative strands of the genome, respectively; and different colors represent different COG functional classifications; the fourth circle is the repeat sequence; the fifth circle is tRNA and rRNA, blue is tRNA, and purple is rRNA; the sixth circle is the GC content; and the innermost circle is GC skew. A–Z, respectively, show the functional classification of the CDS genes in the chromosome.
Comparative genomics analysis of Q2H1 strain
To analyze the similarity of the Q2H1 strain with other closely related species, phylogenetic and comparative genomic analyses were performed. Heat maps of average nucleotide identity (ANI) between the strain Q2H1 and the eight closest phylogenetically related species were created by TBtools software. ANI analysis showed that the ANI similarity between the Q2H1 genome and P. frigoritolerans strain FJAT-2396 was 97.34% and that between Q2H1 genome and P. frigoritolerans strain Ant232 was 96.53%. Genome similarity between the Q2H1 genome and P. simplex NBRC 15720 was 93.08%. These ANI results indicated that Q2H1 belonged to P. frigoritolerans (Figure 3A).
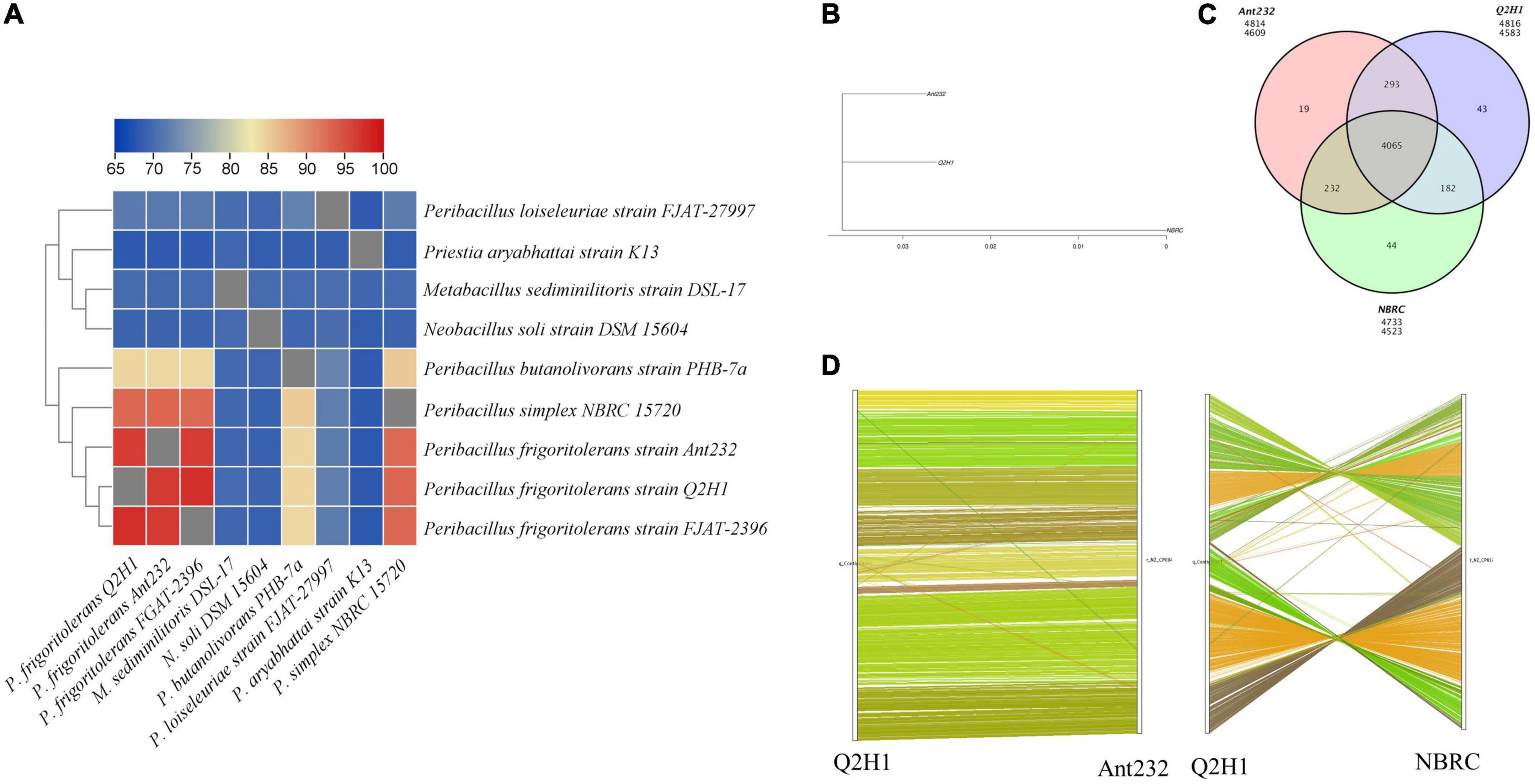
Figure 3. Comparative genomics analysis of Q2H1 strain: (A) Heat maps of average nucleotide identity (ANI) between strain Q2H1 and phylogenetically eight closely related species. (B) Evolutionary relationship between Q2H1 and two species. (C) Venn diagram of gene family statistics. (D) Genome collinearity of Q2H1/Ant232 and Q2H1/NBRC.
Analysis of the 16S rRNA gene and ANI showed that P. frigoritolerans strain FJAT-2396 was most similar to Q2H1, but the NCBI genome database retrieved only one scaffold for the P. frigoritolerans strain FJAT-2396 genome, whereas P. frigoritolerans strain Ant232 (named as Ant232) and P. simplex strain NBRC 15720 (named as NBRC) had complete genome information. Therefore, Ant232 and NBRC were selected for comparative genomic analysis. To analyze the difference between the Q2H1 strain and the selected Ant232 and NBRC strains, a comparative genomics analysis was performed. An evolutionary tree showing the evolutionary relationship between species was created by PhyML software (Guindon et al., 2010; Figure 3B). These three strains belonged to the same clade, and the evolutionary relationship between Q2H1 and Ant232 was closer than that between Q2H1 and NBRC. Q2H1 is most similar to the P. frigoritolerans strain Ant232. Gene family cluster analysis revealed 4,583 gene families in the Q2H1 strain, 4,609 in the Ant232 strain, and 4,523 in the NBRC strain. The number of gene families shared by the three strains was 4,065, and 43 unique genes were identified in the Q2H1 strain, 19 in the Ant232 strain, and 44 in the NBRC strain (Figure 3C).
To analyze the positional and evolutionary relationships of homologous genes on chromosomes between Q2H1 and closely related species, genome collinearity analysis was performed to compare the protein sequence of Q2H1 with the protein sequence of each reference genome using BLAST. We then obtained collinearity at the nucleic acid level according to the position information of the homologous gene on the genome sequence. The results showed that Q2H1 and Ant232 had good collinearity at the nucleotide level, and only translocations of the individual genes occurred (Figure 3D). However, in the collinear map of Q2H1 and NBRC, it can be seen that there is only a localized region of collinearity, and a large number of gene inversion, translocation, and genome rearrangement events occur in NBRC. The accumulated variation between Q2H1 and Ant232 genomes was lower. More features were retained from the ancestors, and the order of the genes was relatively consistent, indicating that the two have a shorter differentiation time. The difference between Q2H1 and NBRC may be due to the longer differentiation time, which leads to the accumulation of inter-species variation, the reduction of shared features, and the acquisition of fewer collinear fragments.
Predictive gene clusters involved in the synthesis of secondary metabolites in Q2H1
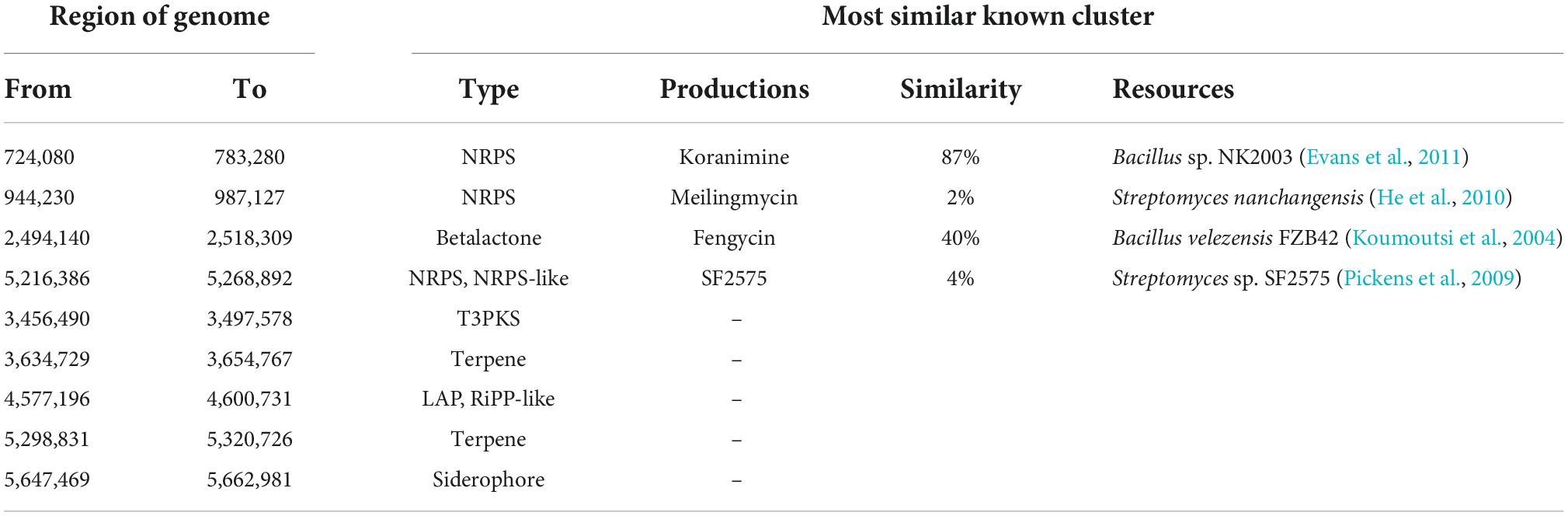
To identify the gene clusters involved in the synthesis of secondary metabolites in Q2H1, antiSMASH software prediction and NCBI BLAST comparison analyses were performed. The results showed that the Q2H1 strain encodes nine gene clusters involved in secondary metabolite synthesis. Among them, there were three non-ribosomal peptide synthases (NRPSs), including NRPS-type koranimine, meilingmycin, SF2575, one betalactone (betalactone-type fengycin), one type III PKS (polyketide synthase), two terpenes, one linear azol(in)e-containing peptides (LAPs), and one siderophore cluster (Table 2). In comparison to the known gene clusters, the similarity with the koranimine-encoded gene cluster reached 87%, and the similarity of the fengycin-encoded gene cluster was 40%. However, the similarity of the other two, meilingmycin and SF2575, is quite low (2 and 4%, respectively), indicating that some secondary metabolites may be specific to Q2H1. In addition, five unknown metabolites were encoded, which were initially predicted to be T3PKS, terpene, LAP RiPP-like, terpene, and siderophore, which requires further study.
Expression of plant growth-promoting traits found in Q2H1 genome in vitro
Through whole-genome sequencing, we found a series of genes possibly related to growth promotion, IAA production (Supplementary Figures 3, 4), phosphate solubilization (Supplementary Figure 5), siderophore production, and nitrogen fixation (Supplementary Figure 6) in the Q2H1 genome. This helped us better understand the PGP mechanisms of Q2H1.
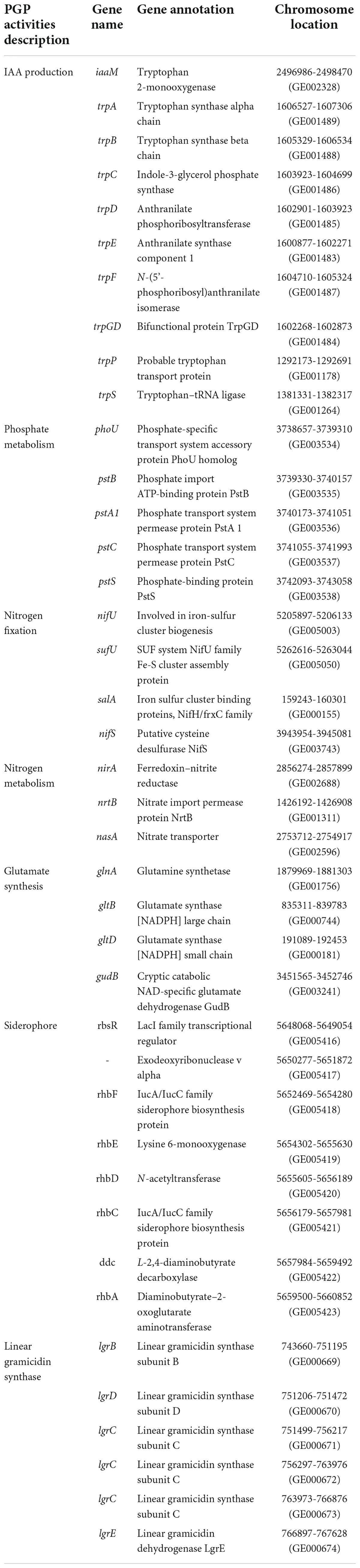
The key IAA-producing gene iaaM and a series of related genes were found in the genome, which underlined the IAA-producing ability of Q2H1. Tryptophan is a precursor of bacterial IAA synthesis. We identified the tryptamine pathway (TAM pathway) in the tryptophan metabolism KEGG pathway map (Supplementary Figure 3). In addition, a set of tryptophan biosynthesis genes, trpABCDEFGDPS, was found in the tryptophan biosynthesis pathway map (Supplementary Figure 4) and was closely related to IAA synthesis (Guo et al., 2020; Table 3). The Salkowski colorimetric method was used for the in vitro presumptive determination of IAA, and Q2H1 produced IAA (14.4 μg/ml).
We also identified a cluster of genes associated with phosphate metabolism in the Q2H1 genome (Table 3 and Figure 4A). Through KEGG analysis, phosphate transporters encoded by pstS, pstC, pstA, and pstB were found in the phosphate metabolism pathway (Supplementary Figure 5). To determine whether these genes could be expressed in vitro, a phosphate solubilization assay was performed. The Q2H1 culture was dotted on organic and inorganic phosphate mediums grown for 7 days. The results showed obvious halos around the dots on the organic phosphate medium (Figure 4B). No halos were observed in the inorganic phosphate medium. This indicated that Q2H1 could efficiently dissolve organic phosphate in the medium.
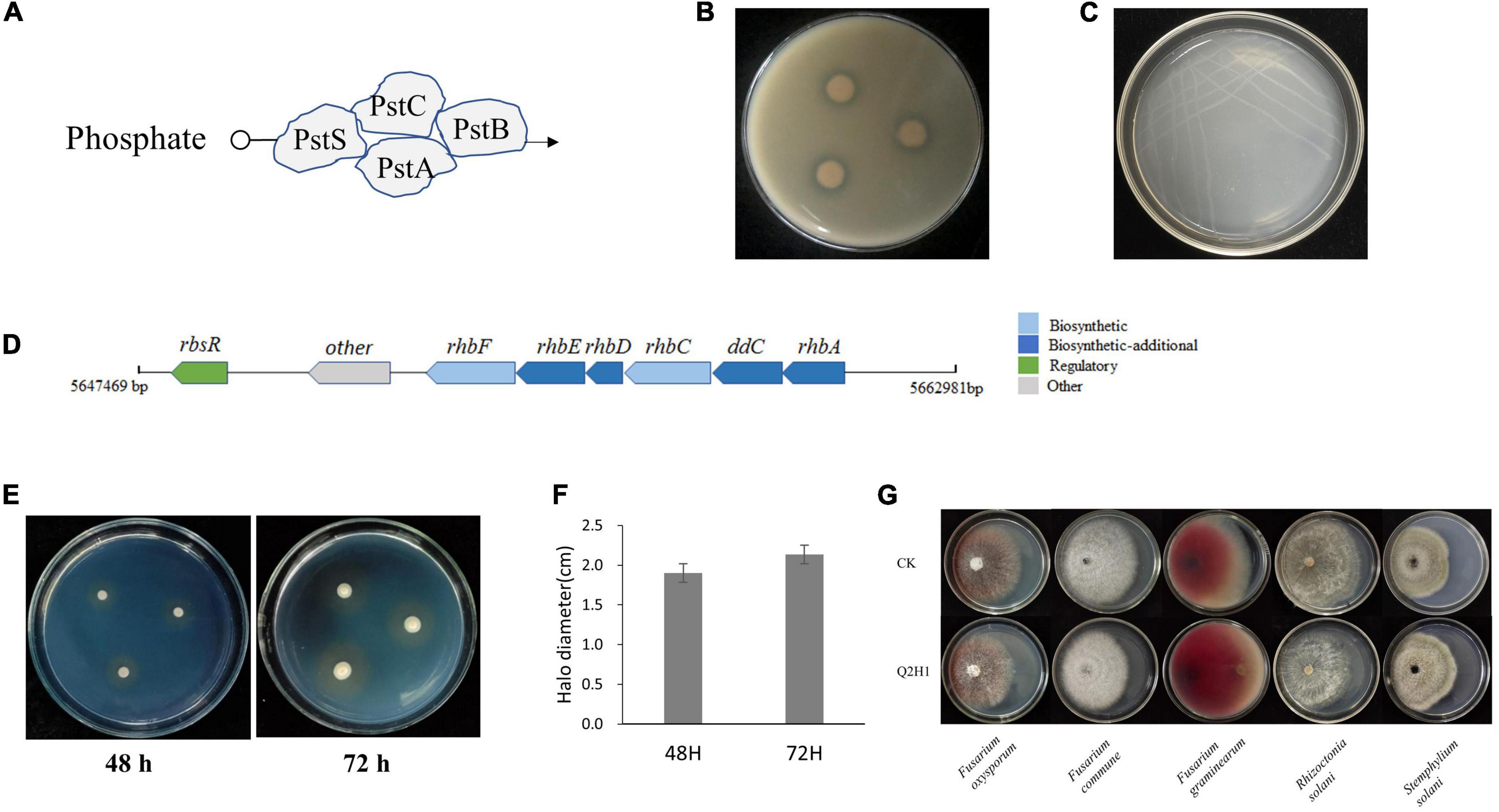
Figure 4. In vitro assay of PGP activity: (A) Phosphate transport pattern diagram. (B) Phosphate solubilization experiment in organic phosphate medium. (C) Ashby nitrogen-free medium. (D) Siderophore gene cluster arrangement. (E) Siderophore CAS detection medium. (F) Diameter of the halo in CAS medium at 48 h and 72 h. (G) Antagonism of Q2H1 against pathogenic fungi.
We also identified a cluster of genes associated with nitrogen fixation in Q2H1 (Table 3). Unfortunately, we did not identify a complete nitrogenase gene cluster. Only nifU, nifS, and sufU genes were present in the strain genome. Annotation revealed that these genes were related to nitrogenase activity and were responsible for biological nitrogen fixation. The regulatory genes nirA, nasA, and nrtB are found in the nitrogen metabolism pathway. The protein encoded by nrtB transports extracellular nitrate into the cell, which is reduced to nitrate under the control of nasA, and nitrate produces ammonia under the action of nirA (Supplementary Figure 6). The glutamine synthase gene glnA and the glutamate synthase genes gltB and gltD were found in the nitrogen metabolism pathway map (Supplementary Figure 6). To test the nitrogen fixation ability of strain Q2H1, we performed an assay to grow Q2H1 in a medium without nitrogen. We found that Q2H1 could grow in a medium without nitrogen (Figure 4C).
We sorted the genes encoding siderophore clusters in the genome sequence of Q2H1. The siderophore synthesis pathway and related genes were also analyzed. The annotation results are shown in Table 3, which lays the foundation for an in-depth study of siderophores and reveals the mechanism of PGP. We found that the siderophore gene cluster was located on chromosome (5647469-5662981). A schematic representation of the siderophore gene cluster arrangement is shown in Figure 4D. To verify whether the siderophore gene cluster can be expressed in vitro, a siderophore test was carried out using the CAS detection medium. The results showed an apparent brown circle around the Q2H1 colony on the plate 48 h post-inoculation (Figure 4E). The halo expanded further after 72 h (Figure 4F), indicating that Q2H1 can produce siderophores.
However, we found the linear gramicidin synthase-related genes lgrB, lgrC, lgrD, and lgrE from SwissProt annotation, which may be associated with disease resistance (Table 3). According to the results of the antagonistic test of Q2H1 against five pathogenic fungi, we found that QH21 had no antagonistic activity (Figure 4G).
Discussion
Bacteria in plant roots and rhizospheres benefit from root exudates. Some bacteria can enter plants as endophytes, which can establish a mutualistic association (Gaiero et al., 2013). It has been reported that the promotion of plant growth by endophytes may be due to nitrogen fixation (Lowman et al., 2016), phytohormone production, biological control of phytopathogens (through the production of antifungal or microbial agents), siderophore production, nutrient competition, induction of systemic acquired host resistance, or increasing availability of minerals (Ali et al., 2014; Santoyo et al., 2016; Mukherjee et al., 2017; Lata et al., 2018). Exploring the mechanisms of PGP will aid in the application of endophytic bacteria to enhance plant production.
In this study, we isolated an endophytic bacterial strain, Q2H1, from potato roots, which plays a growth-promoting role in potato plants. The fresh and root weights of potato plants were significantly increased. The average nucleotide identity (ANI) is an indicator for comparing the relationship between two genomes at the nucleotide level. ANI is defined as the average base similarity between the homologous fragments of two microbial genomes. It is characterized by a high degree of discrimination between closely related species, and it is generally considered that an ANI ≥ 95% indicates the same species (Goris et al., 2007; Yoon et al., 2017). Based on the 16S rRNA gene and ANI analysis, Q2H1 is closely related to P. frigoritolerans.
We performed whole-genome sequencing of Q2H1 to explore the possible mechanisms of PGP. Through comparative genome analysis, we found that evolutionary relationships suggest that these three strains belong to the same clade (Figure 3B); more specifically, Q2H1 and Ant232 are closer than Q2H1 and NBRC. Gene family cluster analysis of Q2H1 and both Ant232 and NBRC revealed that the three strains shared 4,065 genes, and Q2H1 had 43 unique genes. A considerable number of gene inversion, translocation, and genome rearrangement events occurred in NBRC, as can be seen in the collinear map of Q2H1 and NBRC. There was less variation in accumulation between the genomes of Q2H1 and Ant232. The fact that most ancestor-derived traits were retained and the gene order was largely constant suggests that the two strains took less time for differentiation. Good collinearity between the genomes of Q2H1 and Ant232 indicated relatively consistent gene sequences.
Next, we focused on the genes in the Q2H1 genome that were predictably associated with PGP. Studies have shown that secondary metabolites of plant endophytes can improve the resistance of plants to biotic and abiotic stresses (Gouda et al., 2016; Bharadwaj et al., 2020). A total of nine secondary metabolite gene clusters were predicted in the genome, including NRPS-type koranimine, meilingmycin, SF2575, betalactone-type fengycin, five unknown products, one type III PKS (polyketide synthase), two terpenes, one linear azol(in)e-containing peptides (LAPs), and one siderophore cluster (Koumoutsi et al., 2004; Pickens et al., 2009; He et al., 2010; Evans et al., 2011).
We found genes and enzymes involved in tryptophan metabolism in the KEGG metabolic pathway, which is closely related to the synthesis of indoleacetic acid. In addition, genes iaaM, trpABCDEFPS, and trpGD in the tryptophan synthesis pathway were also found. iaaM is a key gene in the IAM pathway, and trpABCDFEPS and trpGD are involved in tryptophan synthesis (Liu et al., 2008). This is consistent with the results of the IAA production assay in vitro. Similar to our findings, it was previously recognized that the presence of tryptophan-related genes in bacterial genomes is associated with IAA production, as described for Enterobacter strain 638 (Taghavi et al., 2010) and Enterobacter cloacae UW5 (Coulson and Patten, 2015). Asaf et al. (2018) identified a tryptophan biosynthesis gene (trpABD) involved in IAA production in Sphingomonas sp. LK11 genome.
A group of phosphate metabolism genes phoU, pstA, pstB, pstC, and pstS are presumed to have phosphate solubilization capabilities. Through functional annotation of Q2H1, we learned that these genes (phoU, pstA, pstB, pstC, and pstS) encode phosphate transporters, which together constitute the phosphate transport system. We also demonstrated that Q2H1 could solubilize organic phosphate in vitro. Similar to our results, other endophytes, such as Bacillus subtilis RS10 (Iqbal et al., 2021), E. roggenkampii ED5 (Guo et al., 2020), and B. subtilis EA-CB0575 (Franco-Sierra et al., 2020), have also been reported as phosphate solubilizers. Genes related to phosphate transport (pstACS) were found in the genome of B. subtilis strain RS10, which was confirmed in vitro and had favorable plant growth-promoting traits. These genes are also present in E. roggenkampii ED5 and have been confirmed in vitro.
Regarding nitrogen fixation, only individual nitrogen fixation-related genes nifU, nifS, sufU, and salA were found. The presence of nifU and nifS, which are required for the components of the enzymatic module encoding nitrogenase, was determined (Li et al., 2016). The nifU protein plays a major role in Fe-S cluster aggregation, which is necessary for nitrogen fixation (Smith et al., 2005). This suggests the possibility of the strain fixing environmental nitrogen, which was experimentally determined by P. frigoritolerans Q2H1 growth on the Ashby medium. Correspondingly, we found that Q2H1 was able to grow on a medium without nitrogen. The regulatory genes nirA, nasA, and nrtB, which can convert nitrate to ammonia, are found in the nitrogen metabolism pathway. Glutamine plays an important role in nitrogen metabolism and regulates nitrogenase activity in some nitrogen-fixing photosynthetic bacteria and Azospirillum spp. Glutamine was first shown to regulate nitrogenase activity in Rhodospirillum rubrum (Nordlund and Högbom, 2013). The glutamate synthesis-related genes glnA, gltB, gltD, and gudB were also annotated. The presence of nifU, glnA, and gltBD in the genome of the sugarcane endophyte E. roggenkampii ED5 confirms the nitrogen fixation effect of ED5 and can significantly promote sugarcane growth (Guo et al., 2020).
When plants grow in an iron-deficient environment, siderophore production by microorganisms can chelate Fe3+ ions that can be difficult for plants to absorb directly for its utilization. Siderophores are relatively low-molecular weight (500–1,000) organic chelators that bind to insoluble iron in the environment and form Fe3+–siderophore complexes. Siderophore-producing bacteria change the availability of iron in the soil through the chelation of siderophores, thereby improving the iron availability in the rhizosphere of plants and meeting the nutritional needs of iron for plant growth (Garzón-Posse et al., 2019). In this study, we found a siderophore gene cluster in the Q2H1 genome; however, the siderophore type was not determined. The siderophore gene cluster was evaluated and confirmed in vitro. Consistent with the present study, siderophore production has been detected in Enterobacter cloacae SBP8 (Singh et al., 2017) and Pseudomonas sp. UW4 (Duan et al., 2013), with multiple PGP properties.
Conclusion
Using whole-genome sequencing, we obtained the sequence of P. frigoritolerans Q2H1. Several genes and gene clusters related to growth-promoting effects have also been identified. The PGP characteristics of Q2H1, including IAA production, phosphate solubilization, nitrogen fixation, and siderophore production, were annotated to genes in the genome, which were verified in vitro. Our results are important for investigating the mechanism of PGP and setting the groundwork for future plant growth applications.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
MZ, YW, QZ, and XY wrote the manuscript. MZ, HYZ, and HLZ designed the experiments. YW, ZS, YL, HH, and QZ performed the experiments. YW, DW, and BD analyzed the sequencing data. YZ, QZ, XY, and HYZ advised on the experimental design and English language editing. All authors read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Jilin Provincial Agricultural Science & Technology Innovation Project (grant no. KYJF2021ZR005), Natural Science Foundation of Inner Mongolia (grant no. 2021ZD06), National Natural Science Foundation of China (grant no. 31960533), and China Agriculture Research System of MOF and MARA (grant no. CARS-07-C-3).
Acknowledgments
We thank Jianying Yue, Haijuan Wang, and Zhiying Wang for critical discussions on the project design and technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1035901/full#supplementary-material
Footnotes
References
Adeleke, B. S., Babalola, O. O., and Glick, B. R. (2021). Plant growth-promoting root-colonizing bacterial endophytes. Rhizosphere 20:100433. doi: 10.1016/j.rhisph.2021.100433
Afzal, I., Shinwari, Z. K., Sikandar, S., and Shahzad, S. (2019). Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221, 36–49. doi: 10.1016/j.micres.2019.02.001
Ali, S., Charles, T. C., and Glick, B. R. (2014). Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol. Biochem. 80, 160–167. doi: 10.1016/j.plaphy.2014.04.003
Asaf, S., Khan, A. L., Khan, M. A., Al-Harrasi, A., and Lee, I. J. (2018). Complete genome sequencing and analysis of endophytic Sphingomonas sp. LK11 and its potential in plant growth. 3 Biotech. 8:389. doi: 10.1007/s13205-018-1403-z
Barka, E. A., Nowak, J., and Clement, C. (2006). Enhancement of chilling resistance of inoculated grapevine plantlets with a plant growth-promoting rhizobacterium, Burkholderia phytofirmans strain PsJN. Appl. Environ. Microb. 72, 7246–7252. doi: 10.1128/AEM.01047-06
Bertelli, C., and Brinkman, F. S. L. (2018). Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics 34, 2161–2167. doi: 10.1093/bioinformatics/bty095
Bharadwaj, R., Jagadeesan, H., Kumar, S. R., and Ramalingam, S. (2020). Molecular mechanisms in grass-Epichloe interactions: Towards endophyte driven farming to improve plant fitness and immunity. World J. Microbiol. Biotechnol. 36:92. doi: 10.1007/s11274-020-02868-5
Bland, C., Ramsey, T. L., Sabree, F., Lowe, M., Brown, K., Kyrpides, N. C., et al. (2007). CRISPR Recognition Tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8:209. doi: 10.1186/1471-2105-8-209
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Cavalcante, V. A., and Dobereiner, J. (1988). A new acid-tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil 108, 23–31.
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Gene Predict. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y. (2020). Screening of endophytic siderophore-producing bacteria of bamboo and complete genome analysis of high-activity strains. Ph.D thesis. Jiangxi, IL: Jiangxi Agricultural University, doi: 10.27177/d.cnki.gjxnu.2020.000209
Cho, H. S., Park, S. Y., Ryu, C. M., Kim, J. F., Kim, J. G., and Park, S. H. (2007). Interference of quorum sensing and virulence of the rice pathogen Burkholderia glumae by an engineered endophytic bacterium. Fems Microbiol. Ecol. 60, 14–23. doi: 10.1111/j.1574-6941.2007.00280.x
Coulson, T. J. D., and Patten, C. L. (2015). Complete Genome Sequence of Enterobacter cloacae UW5, a Rhizobacterium Capable of High Levels of Indole-3-Acetic Acid Production. Genome Announc. 3, e00843–15. doi: 10.1128/genomeA.00843-15
Dreyfus, B., Garcia, J. L., and Gillis, M. (1988). Characterization of Azorhizobium caulinodans gen. nov., sp. nov., a stem-nodulating nitrogen-fixing bacterium isolated from Sesbania rostrata. Int. J. Syst. Evol. Microbiol. 38, 89–98. doi: 10.1099/00207713-38-1-89
Duan, J., Jiang, W., Cheng, Z., Heikkila, J. J., and Glick, B. R. (2013). The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. PLoS One 8:e58640. doi: 10.1371/journal.pone.0058640
Etesami, H., Hosseini, H. M., and Alikhani, H. A. (2014). In planta selection of plant growth promoting endophytic bacteria for rice (Oryza sativa L.). J. Soil Sci. Plant Nutr. 14, 491–503.
Evans, B. S., Ntai, I., Chen, Y., Robinson, S. J., and Kelleher, N. L. (2011). Proteomics-Based Discovery of Koranimine, a Cyclic Imine Natural Product. J. Am. Chem. Soc. 133, 7316–7319. doi: 10.1021/ja2015795
Franco-Sierra, N. D., Posada, L. F., Santa-Maria, G., Romero-Tabarez, M., Villegas-Escobar, V., and Alvarez, J. C. (2020). Bacillus subtilis EA-CB0575 genome reveals clues for plant growth promotion and potential for sustainable agriculture. Funct. Integr. Genom. 20, 575–589. doi: 10.1007/s10142-020-00736-x
Gaiero, J. R., McCall, C. A., Thompson, K. A., Day, N. J., Best, A. S., and Dunfield, K. E. (2013). Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 100, 1738–1750. doi: 10.3732/ajb.1200572
Gao, M., Zhou, J. J., Wang, E. T., Chen, Q., Xu, J., and Sun, J. G. (2015). Multiphasic characterization of a plant growth promoting bacterial strain, Burkholderia sp. 7016 and its effect on tomato growth in the field. J. Integr. Agric. 14, 1855–1863. doi: 10.1016/S2095-3119(14)60932-1
Garzón-Posse, F., Quevedo-Acosta, Y., Mahecha-Mahecha, C., and Acosta-Guzmán, P. (2019). Recent Progress in the Synthesis of Naturally Occurring siderophores. Eur. J. Org. Chem. 2019, 7747–7769. doi: 10.1002/ejoc.201901257
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Gouda, S., Das, G., Sen, S. K., Shin, H. S., and Patra, J. K. (2016). Endophytes: A Treasure House of Bioactive Compounds of Medicinal Importance. Front. Microbiol. 7:1538. doi: 10.3389/fmicb.2016.01538
Grobelak, A., and Hiller, J. (2017). Bacterial siderophores promote plant growth: Screening of catechol and hydroxamate siderophores. Int. J. Phytoremediation 19, 825–833. doi: 10.1080/15226514.2017.1290581
Gu, C. C., Wang, Z., Wang, L. R., Mao, L. Y., Song, Q. Q., Yang, L. T., et al. (2018). gfp maker of nitrogen-fixing bacteria DX120E and inoculation effects on different sugarcane varieties. J. South. Agric. 49, 1075–1081. doi: 10.3969/j.issn.2095-1191.2018.06.05
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guo, D. J., Singh, R. K., Singh, P., Li, D. P., Sharma, A., Xing, Y. X., et al. (2020). Complete Genome Sequence of Enterobacter roggenkampii ED5, a Nitrogen Fixing Plant Growth Promoting Endophytic Bacterium With Biocontrol and Stress Tolerance Properties, Isolated From Sugarcane Root. Front. Microbiol. 11:580081. doi: 10.3389/fmicb.2020.580081
He, Y., Sun, Y., Liu, T., Zhou, X., Bai, L., and Deng, Z. (2010). Cloning of Separate Meilingmycin Biosynthesis Gene Clusters by Use of Acyltransferase-Ketoreductase Didomain PCR Amplification. Appl. Environ. Microb. 76, 3283–3292. doi: 10.1128/AEM.02262-09
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Iqbal, S., Ullah, N., and Janjua, H. A. (2021). In Vitro Evaluation and Genome Mining of Bacillus subtilis Strain RS10 Reveals Its Biocontrol and Plant Growth-Promoting Potential. Agriculture 11:1273. doi: 10.3390/agriculture11121273
Kalvari, I., Argasinska, J., Quinones-Olvera, N., Nawrocki, E. P., Rivas, E., Eddy, S. R., et al. (2018). Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46, D335–D342. doi: 10.1093/nar/gkx1038
Kamada, M., Hase, S., Fujii, K., Miyake, M., Sato, K., Kimura, K., et al. (2015). Whole-Genome Sequencing and Comparative Genome Analysis of Bacillus subtilis Strains Isolated from Non-Salted Fermented Soybean Foods. PLoS One 10:e0141369. doi: 10.1371/journal.pone.0141369
Khan, A. L., Halo, B. A., Elyassi, A., Ali, S., Al-Hosni, K., Hussain, J., et al. (2016). Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanum lycopersicum. Electron. J. Biotechn. 21, 58–64. doi: 10.1016/j.ejbt.2016.02.001
Koumoutsi, A., Chen, X. H., Henne, A., Liesegang, H., Hitzeroth, G., Franke, P., et al. (2004). Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186, 1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lata, R., Chowdhury, S., Gond, S. K., and White, J. F. Jr. (2018). Induction of abiotic stress tolerance in plants by endophytic microbes. Lett. Appl. Microbiol. 66, 268–276. doi: 10.1111/lam.12855
Li, L., Stoeckert, C. J., and Roos, D. S. (2003). OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Li, X. X., Liu, Q., Liu, X. M., Shi, H. W., and Chen, S. F. (2016). Using synthetic biology to increase nitrogenase activity. Microb. Cell Fact. 15:43. doi: 10.1186/s12934-016-0442-6
Lin, B., Song, Z., Jia, Y., Zhang, Y., Wang, L., Fan, J., et al. (2019). Biological characteristics and genome-wide sequence analysis of endophytic nitrogen-fixing bacteria Klebsiella variicola GN02. Biotechnol. Biotech. Equip. 33, 108–117. doi: 10.1080/13102818.2018.1555010
Lin, L., Wei, C., Chen, M., Wang, H., Li, Y., Li, Y., et al. (2015). Complete genome sequence of endophytic nitrogen-fixing Klebsiella variicola strain DX120E. Stand. Genomic Sci. 10:22. doi: 10.1186/s40793-015-0004-2
Liu, D., Li, Q., and Li, G. (2008). Research progress on the key gene iaaM in IAA synthesis. Biotechnology 02, 87–90. doi: 10.16519/j.cnki.1004-311x.2008.02.019
Lo, K. J., Lin, S. S., Lu, C. W., Kuo, C. H., and Liu, C. T. (2018). Whole-genome sequencing and comparative analysis of two plant-associated strains of Rhodopseudomonas palustris (PS3 and YSC3). Sci. Rep. 8:12769. doi: 10.1038/s41598-018-31128-8
Lopes, R., Cerdeira, L., Tavares, G. S., Ruiz, J. C., Blom, J., Horacio, E. C. A., et al. (2017). Genome analysis reveals insights of the endophytic Bacillus toyonensis BAC3151 as a potentially novel agent for biocontrol of plant pathogens. World J. Microbiol. Biotechnol. 33:185. doi: 10.1007/s11274-017-2347-x
Lopes Antunes, J. E., Santiago de Freitas, A. D., Bonifacio, A., de Souza Oliveira, L. M., de Rosalia e Silva Santos, C. E., Catanho Pereira de Lyra, M. D. C., et al. (2022). Isolation and Characterization of Plant Growth-Promotion Diazotrophic Endophytic Bacteria Associated to Sugarcane (Saccharum officinarum L.) Grown in Paraiba, Brazil. Braz. Arch. Biol. Techn. 65:e22200439. doi: 10.1590/1678-4324-2022200439
Lowman, S., Kim-Dura, S., Mei, C., and Nowak, J. (2016). Strategies for enhancement of switchgrass (Panicum virgatum L.) performance under limited nitrogen supply based on utilization of N-fixing bacterial endophytes. Plant Soil 405, 47–63. doi: 10.1007/s11104-015-2640-0
Madhaiyan, M., Chan, K. L., and Ji, L. (2014). Draft Genome Sequence of Methylobacterium sp. Strain L2-4, a Leaf-Associated Endophytic N-Fixing Bacterium Isolated from Jatropha curcas L. Genome Announc. 2, e1306–e1314. doi: 10.1128/genomeA.01306-14
Malinowski, D. P., and Belesky, D. P. (2000). Adaptations of endophyte-infected cool-season grasses to environmental stresses: Mechanisms of drought and mineral stress tolerance. Crop Sci. 40, 923–940. doi: 10.2135/cropsci2000.404923x
Mei, C., and Flinn, B. S. (2010). The use of beneficial microbial endophytes for plant biomass and stress tolerance improvement. Recent Patents Biotechnol. 4, 81–95.
Mukherjee, A., Bhattacharjee, P., Das, R., Pal, A., and Paul, A. K. (2017). Endophytic bacteria with plant growth promoting abilities from Ophioglossum reticulatum L. AIMS Microbiol. 3, 596–612. doi: 10.3934/microbiol.2017.3.596
Nordlund, S., and Högbom, M. (2013). ADP-ribosylation, a mechanism regulating nitrogenase activity. FEBS J. 280, 3484–3490.
Normanly, J. (2010). Approaching Cellular and Molecular Resolution of Auxin Biosynthesis and Metabolism. Cold Spring Harb. Perspect. Biol. 2:a001594. doi: 10.1101/cshperspect.a001594
Orozco-Mosqueda, M. D. C., Glick, B. R., and Santoyo, G. (2020). ACC deaminase in plant growth-promoting bacteria (PGPB): An efficient mechanism to counter salt stress in crops. Microbiol. Res. 235:126439. doi: 10.1016/j.micres.2020.126439
Ouyabe, M., Irie, K., Tanaka, N., Kikuno, H., Pachakkil, B., and Shiwachi, H. (2020). Response of Upland Rice (Oryza sativa L.) Inoculated with Non-Native Plant Growth-Promoting Bacteria. Agronomy 10:903. doi: 10.3390/agronomy10060903
Pageni, B. B., Lupwayi, N. Z., Akter, Z., Larney, F. J., Kawchuk, L. M., and Gan, Y. (2014). Plant growth-promoting and phytopathogen-antagonistic properties of bacterial endophytes from potato (Solanum tuberosum L.) cropping systems. Can. J. Plant Sci. 94, 835–844. doi: 10.4141/CJPS2013-356
Patel, J. K., and Archana, G. (2017). Diverse culturable diazotrophic endophytic bacteria from Poaceae plants show cross-colonization and plant growth promotion in wheat. Plant Soil 417, 99–116. doi: 10.1007/s11104-017-3244-7
Patten, C. L., and Glick, B. R. (2002). Regulation of indoleacetic acid production in Pseudomonas putida GR12-2 by tryptophan and the stationary-phase sigma factor RpoS. Can. J. Plant Sci. 48, 635–642. doi: 10.1139/W02-053
Pickens, L. B., Kim, W., Wang, P., Zhou, H., Watanabe, K., Gomi, S., et al. (2009). Biochemical Analysis of the Biosynthetic Pathway of an Anticancer Tetracycline SF2575. J. Am. Chem. Soc. 131, 17677–17689. doi: 10.1021/ja907852c
Redman, R. S., Sheehan, K. B., Stout, R. G., Rodriguez, R. J., and Henson, J. M. (2002). Thermotolerance generated by plant/fungal symbiosis. Science 298, 1581–1581. doi: 10.1126/science.1072191
Richter, M., Rossello-Mora, R., Oliver Glockner, F., and Peplies, J. (2016). JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Santoyo, G., Moreno-Hagelsieb, G., Orozco-Mosqueda Mdel, C., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Singh, R. K., Singh, P., Li, H. B., Guo, D. J., Song, Q. Q., Yang, L. T., et al. (2020). Plant-PGPR interaction study of plant growth-promoting diazotrophs Kosakonia radicincitans BA1 and Stenotrophomonas maltophilia COA2 to enhance growth and stress-related gene expression in Saccharum spp. J. Plant Interact. 15, 427–445. doi: 10.1080/17429145.2020.1857857
Singh, R. P., Nalwaya, S., and Jha, P. N. (2017). The draft genome sequence of the plant growth promoting rhizospheric bacterium Enterobacter cloacae SBP-8. Genom. Data 12, 81–83. doi: 10.1016/j.gdata.2017.03.006
Smith, A. D., Jameson, G. N. L., Santos, P. C. D., Agar, J. N., Naik, S., Krebs, C., et al. (2005). NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry 44, 12955–12969. doi: 10.1021/bi051257i
Taghavi, S., Van der Lelie, D., Hoffman, D. A., Zhang, Y. B., Walla, M. D., Vangronsveld, J., et al. (2010). Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 6:e1000943. doi: 10.1371/journal.pgen.1000943
Taule, C., Castillo, A., Villar, S., Olivares, F., and Battistoni, F. (2016). Endophytic colonization of sugarcane (Saccharum officinarum) by the novel diazotrophs Shinella sp UYSO24 and Enterobacter sp UYSO10. Plant Soil 403, 403–418. doi: 10.1007/s11104-016-2813-5
Veselova, S. V., Sorokan, A. V., Burkhanova, G. F., Rumyantsev, S. D., Cherepanova, E. A., Alekseev, V. Y., et al. (2022). By Modulating the Hormonal Balance and Ribonuclease Activity of Tomato Plants Bacillus subtilis Induces Defense Response against Potato Virus X and Potato Virus Y. Biomolecules 12:288. doi: 10.3390/biom12020288
Wang, Y. P., Tang, H. B., DeBarry, J. D., Tan, X., Li, J. P., Wang, X. Y., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Yan, X. M., Wang, Z., Mei, Y., Wang, L. Q., Wang, X., Xu, Q. S., et al. (2018). Isolation, Diversity, and Growth-Promoting Activities of Endophytic Bacteria From Tea Cultivars of Zijuan and Yunkang-10. Front. Microbiol. 9:1848. doi: 10.3389/fmicb.2018.01848
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Zgadzaj, R., James, E. K., Kelly, S., Kawaharada, Y., de Jonge, N., Jensen, D. B., et al. (2015). A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria. PLoS Genet. 11:e1005280. doi: 10.1371/journal.pgen.1005280
Zhang, L., Yuan, M., Sun, J. G., Fan, M. S., Gao, M., and Zheng, H. L. (2016). Isolation and functional characterizations of potato endogenous nitrogen-fixing bacteria. Soil Fertilizer Sci. China 06, 139–145. doi: 10.11838/sfsc.20160623
Zhang, Q., and White, J. F. (2021). Bioprospecting Desert Plants for Endophytic and Biostimulant Microbes: A Strategy for Enhancing Agricultural Production in a Hotter, Drier Future. Biol. Basel 10:961. doi: 10.3390/biology10100961
Keywords: bacterial endophytes, Q2H1, potato, plant growth-promoting, whole-genome sequencing
Citation: Wang Y, Zhao Q, Sun Z, Li Y, He H, Zhang Y, Yang X, Wang D, Dong B, Zhou H, Zhao M and Zheng H (2022) Whole-genome analysis revealed the growth-promoting mechanism of endophytic bacterial strain Q2H1 in potato plants. Front. Microbiol. 13:1035901. doi: 10.3389/fmicb.2022.1035901
Received: 03 September 2022; Accepted: 03 November 2022;
Published: 01 December 2022.
Edited by:
José David Flores Félix, Universidade da Beira Interior, PortugalReviewed by:
Rocio Roca Couso, University of Salamanca, SpainMukesh Kumar Malviya, Guangxi Academy of Agricultural Sciences, China
Copyright © 2022 Wang, Zhao, Sun, Li, He, Zhang, Yang, Wang, Dong, Zhou, Zhao and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingmin Zhao, bWluZ21pbnpoQDE2My5jb20=; Hongli Zheng, emhsZmN5NjZAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Yuhu Wang
Yuhu Wang Qianqian Zhao2†
Qianqian Zhao2† Mingmin Zhao
Mingmin Zhao