- 1State Key Laboratory for Managing Biotic and Chemical Treats to the Quality and Safety of Agro-products, Institute of Plant Protection and Microbiology, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2College of Plant Protection, Southwest University, Chongqing, China
Introduction: The insect gut harbors numerous microorganisms that may have functions in development and reproduction, digestion, immunity and protection, and detoxification. Recently, the influence factors on gut microbiota were evaluated in the rice leaffolder Cnaphalocrocis medinalis, a widespread insect pest in paddy fields. However, the relationship between gut microbiota composition and geography is poorly understood in C. medinalis.
Methods: To reveal the patterns of C. medinalis gut bacterial communities across geographic sources and the ecological processes driving the patterns, C. medinalis were sampled from six geographic sources in China, Thailand, and Vietnam in 2016, followed by gut bacterial 16S ribosomal RNA gene sequencing.
Results: A total of 22 bacterial phyla, 56 classes, 84 orders, 138 families, 228 genera, and 299 species were generated in C. medinalis from six geographic sources. All alpha diversity indices differed among the samples from different geographic sources. Analysis of similarity (ANOSIM) and permutational multivariate analysis of variance (PERMANOVA) both revealed significant differences in the gut microbiota of C. medinalis from six geographic sources. A total of 94 different taxa were screened as indicators for the gut microbiota of C. medinalis from six geographic sources by linear discriminant analysis effect size (LEfSe). The gene ontology (GO) pathways of the gut microbiota in C. medinalis differed among geographic sources. In total, the bacterial communities within geographic sources were mainly determined by stochastic processes, and those between geographic sources were mainly determined by deterministic processes.
Discussion: This study elucidates that geography plays a crucial role in shaping the gut microbiota of C. medinalis. Thus, it enriches our knowledge of gut bacteria in C. medinalis and sheds light on the mechanisms underlying C. medinalis gut microbial shifts across geography.
Introduction
The insect gut is colonized by numerous microorganisms (Douglas, 2015), and these microorganisms have been reported to contribute to many important functions, including development and reproduction, nutrient metabolism, pathogen and insecticidal resistance, immunity, digestion, and detoxification (Dillon and Dillon, 2004; Anand et al., 2010; Engel and Moran, 2013; Beran and Gershenzon, 2016; Cheng et al., 2017; Voirol et al., 2018; Blanton and Peterson, 2020; Pyszko et al., 2020; Li et al., 2021; Moore and Aparicio, 2022). In the fruit fly (Drosophila melanogaster), microbiome interactions were reported to shape host fitness (Gould et al., 2018). Core gut symbiotic bacteria—Lactococcus, Enterococcus, Serratia, and Dysgonomonas—isolated from the bamboo snout beetle Cyrtotrachelus buqueti have the capability of degrading lignocellulose (Luo et al., 2019). The gut microbiota is crucial in regulating the interconnections between both the host and gut pathogens, and the interaction of the gut microbiota and pathogenic fungi could result in the acceleration of mosquito mortality (Wei et al., 2017). However, the gut microbiota facilitates the weight gain of honeybees through bacterial metabolism and hormonal signaling, and increases the fitness of the pine weevil by degrading conifer diterpenes (Berasategui et al., 2017; Zheng et al., 2017). Gut symbionts could enhance resistance to trichlorphon, an organophosphate insecticide in the oriental fruit fly Bactrocera dorsalis (Hendel) (Cheng et al., 2017). Likewise, gut bacteria were proven to increase the resistance of the silkworm Bombyx mori against chlorpyrifos, another organophosphate insecticide (Chen et al., 2020). The honeybee (Apis mellifera) intestinal microbiota could promote host endogenous detoxification ability by regulating P450 gene expression in the digestive tract (Wu et al., 2020). In a Plutella xylostella population susceptible to Cry1Ac protoxin, the gut microbiota is correlated with the host immunological response (Li et al., 2021). Considering the importance of insect gut microbiota, application in gut symbiont-driven pest control may be a novel concept to make insect pests more susceptible to wide population management approaches by targeting microbial symbionts (Berasategui et al., 2016). Exploiting the functions of gut microorganisms could advance knowledge of gut microbiota-insect interactions and facilitate the application of gut symbiont-driven pest management (Frago et al., 2012; Berasategui et al., 2016; Kyritsis et al., 2017).
Many factors can influence the composition and diversity of gut microbiota including development stages, diet, pH, temperature and humidity, and geographic source (Schmitt-Wagner et al., 2003; Behar et al., 2008; Mikaelyan et al., 2015; Jones et al., 2018; Yang et al., 2020; Jaramillo and Castañeda, 2021). Bosmans et al. (2018) found that land-use change could cause differences in the gut microbial communities in bumblebees (Bombus terrestris). Jones et al. (2019) suggested that the host plant is the main motivation to shape intestinal microbiota, while variations in insect physiology, intestinal tract, and local external conditions could be responsible for differences in microbiota. The relative abundances of anaerobes were significantly different in insects, and the gut bacterial diversity was impacted by host environmental location, food, growth status, and phylogeny (Yun et al., 2014). Liu et al. (2021) reported that larval bacterial communities differed between laboratory and wild populations of ghost moths (Thitarodes sp.). Yu et al. (2021) compared the microbiota in Leptinotarsa decemlineata (Say), Colorado potato beetles, derived from distinct geographic locations in China, and found that order and genus were proper taxonomic categories to discriminate the geographical locations of Colorado potato beetles. Honey bee colonies from different habitats could harbor significantly different gut microbiota (Jones et al., 2018). Similarly, the composition and diversity of gut microbiota differed in large black chafer (Holotrichia parallela) populations from various geographical sources (Huang and Zhang, 2013).
The rice leaffolder Cnaphalocrocis medinalis (Guenée; Lepidoptera: Crambidae), which mainly feeds on rice plants (Oryza sativa L.), is a significant insect pest in Asia and might result in tremendous financial losses to rice production (Barrion et al., 1991; Cheng, 1996; Yang et al., 2015). This insect is an important long-distance migratory insect pest and cannot overwinter in temperate climates (Cheng, 1996). The C. medinalis population migrates northward beginning in March every year from the southern areas, expanding their regional distributions to cover the rice production zones in China, the Korean Peninsula, and Japan, and their direction becomes predominantly southbound from September onward (Chang et al., 1980; Riley et al., 1995; Wang et al., 2017). In 2015, C. medinalis devastated 15.5 million hectares of rice plants, leading to a yield reduction of 0.47 million tons in China (Yang et al., 2015). A growing number of environmentally friendly alternatives and suggestions for controlling C. medinalis have been researched and put forth, despite the fact that the main method of management for this insect pest relied on chemicals (Yang et al., 2015; Kalaivani et al., 2018; Senthil-Nathan, 2019; Ramasubramanian et al., 2022). Several microorganisms including Bacillus thuringiensis, Bacillus subtilis, Metarhizium anisopliae, and Beauveria bassiana were evaluated their capacity to suppress C. medinalis (Kandibane et al., 2010; Kirubakaran et al., 2014; Shakir et al., 2015; Ramasubramanian et al., 2022). Given the importance of microbes, investigation on the gut bacteria of C. medinalis will contribute to the understanding of insect ecology, and assist the development of innovative pest management approaches based on the application of microorganisms. The gut microbiota in C. medinalis larvae was characterized by traditional isolation and culture methods and Illumina MiSeq technology (Yang, 2012; Liu et al., 2016). The structure and composition of the gut microbiota in C. medinalis were analyzed across developmental stages (Yang et al., 2020). In this study, to assess the patterns of C. medinalis gut microbial communities across geographic sources and the ecological processes generating the patterns, C. medinalis were sampled from six geographic sources in China, Thailand, and Vietnam, followed by bacterial 16S ribosomal RNA gene sequencing with the Illumina MiSeq technology. The results of this study will add to our knowledge of the gut microbiota of C. medinalis, and provide fresh insights into C. medinalis population features.
Materials and methods
Sample preparation
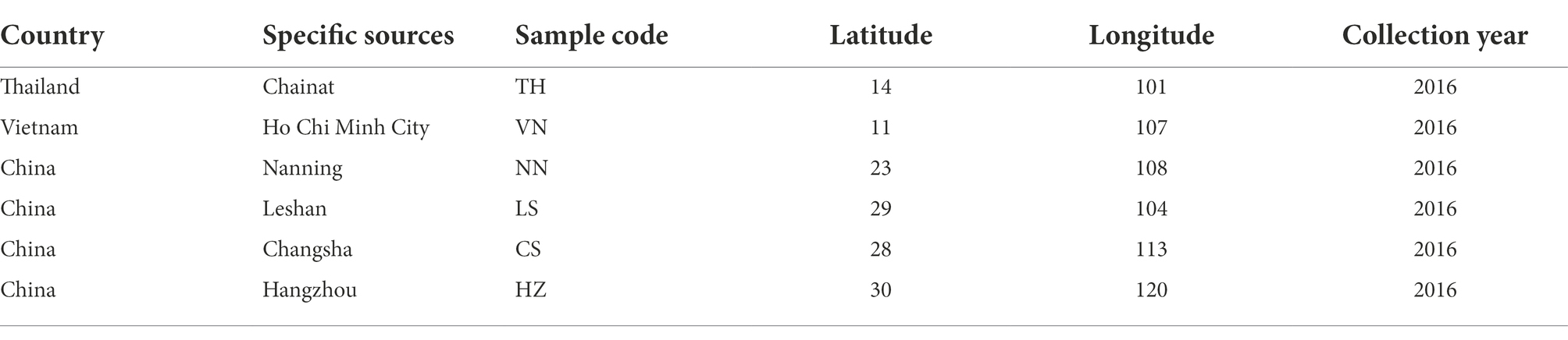
Cnaphalocrocis medinalis adults were collected from paddy fields from four sites in China, one site in Thailand, and one site in Vietnam in 2016. Detailed information on the collected populations is listed in Table 1. This study was conducted based on the populations collected in 2016, and the fifth-instar larvae derived from the eggs laid by the collected moths were used in the study. The collected adults were cultivated with 10% honey solution in the laboratory under controlled circumstances of 26 ± 1°C temperature, 70 ± 10% relative humidity, and a photoperiod of 16:8 (L:D) h. The larvae derived from each geographic source were fed rice leaves. Rice plants were potted in the greenhouse. Rice plant leaves were washed with sterile ddH2O, then air dried before being fed to C. medinalis larvae, and adequate rice leaves were prepared for the C. medinalis larvae.
One-day fifth instar larvae of C. medinalis with similar size were chosen at random to dissect intestinal tract from each geographic source for investigation. A biological sample was created by pooling 20 guts, and three duplicates were established for each geographic source. The larvae of C. medinalis were washed with sterile ddH2O before being disinfected with 75% ethanol for 90s, and then rinsed again with sterile ddH2O. The intestines were collected after dissection and stored at −80°C in 1.5 ml sterile tubes until use.
DNA extraction, PCR, and sequencing
The dissected intestines of C. medinalis were homogenized with 0.5 ml of phosphate buffer saline (PBS buffer, pH 7.5) by shaking with sterile glass beads (0.5 mm diameter) in a sterile tube for 15 min on a vortex. Total DNA were isolated from the samples using the bacterial DNA extraction kit (E.Z.N.A.®, OMEGA, USA) according to the manufacturer’s instructions. Amplicons were generated in a 20 μl reaction system with 5 × FastPfu Buffer (4 μl), 2.5 mM dNTPs (2 μl), each primer (5 μM, 0.8 μL), FastPfu Polymerase (0.4 μl), and sample DNA(10 ng) by PCR (95°C for 2 min, followed by 25 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 5 min) of V4-V5 regions of the bacterial 16S ribosomal RNA gene using the primers 515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′). Amplicons were extracted and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.). Then, they were measured using QuantiFluor™-ST (Promega, U.S.), pooled in equimolar amounts, and paired-end sequenced (2 × 250) using standard methods on an Illumina MiSeq platform.
Data analyses
Raw sequencing data with FASTQ format were demultiplexed and quality-filtered using QIIME (version 1.17).1 Based on the sequence similarity, effective sequences were classified into multiple operational taxonomic units (OTUs) at a similarity level of 97% with UPARSE (version 7.1),2 and then chimeric sequences were identified and removed with UCHIME.3 All of the sequences were annotated and blasted against the Silva (SSU115) 16S ribosomal RNA database with a confidence level of 70% for each 16S ribosomal RNA gene sequence assessed through RDP Classifier.4
To assess alpha diversity, five parameters were used, including OTU number, ACE, Chao1, Shannon, and Simpson’s index. The alpha diversity indices and relative abundance data were subjected to one-way analysis of variance (ANOVA) with SPSS 26.0 (IBM SPSS Statistics), and multiple comparisons were assessed with Tukey’s test. Rarefaction curves were graphed with R software.5 Principal coordinate analysis (PCoA) was performed among all the samples from different geographic sources based on pairwise weighted UniFrac distances and Bray–Curtis distances. Nonmetric multidimensional scaling (NMDS) plots were drawn based on Bray–Curtis distance metrics. Additionally, an analysis of similarity (ANOSIM) was performed to assess the differences in the composition of microbiota across samples from six geographic sources. Individual repetitions were included in the model as a random effect, and 999 permutations were used to construct the permutational multivariate analysis of variance (PERMANOVA). R software6 was used to analyze or plot the data for PCoA, NMDS, ANOSIM, and PERMANOVA. Linear discriminant analysis (LDA) was applied to assess the biomarkers for significant variations among samples from six geographic sources with LDA scores larger than two. Galaxy7 was used to generate a cladogram to reveal the distribution of the significant biomarkers at various taxonomic levels. Functional predictions were performed for the gut microbiota through annotating gene ontology (GO) pathways of OTUs against the Ref99NR database employing the Tax4Fun2 package in R software.
Ecological processing of the microbial communities and phylogenetic turnover within the system was conducted following the framework of Stegen et al. (2013). In this framework, two distinct metrics were utilized to evaluate the contributions of selection, dispersal, and drift that drive an observed assembly of the bacterial community. First, the strength of selection was evaluated by assessing variations in phylogenetic distance based on beta-mean nearest taxon distance (βMNTD) and the β-nearest taxon index (βNTI) for all pairwise comparisons within the samples. Following the confirmation of the roles of selective processes, pairwise comparisons without significant βNTI metrics were evaluated through the Raup-Crick bray (RCbray) index metric to reflect the strength of dispersal on assembly. βMNTD and βNTI were performed using R software with the codes provided by Stegen et al. (2013).8 RCbray index was performed through R software with the codes provided by Stegen et al. (2013).9 Supplementary Table S1 provides a thorough explanation of ecological processing (Stegen et al., 2015).
Results
Sequence analysis and taxa generation
We sequenced the gut microbiota of C. medinalis larvae derived from six geographic sources (Table 1) and retrieved a total of 1,000,338 trimmed paired reads (Supplementary Table S2). Among these reads, 558 OTUs were identified. The high values of Good’s coverage index in all the samples from geographic sources reflected that the sequencing depth was adequate to profile the microbial communities (Supplementary Table S2). Rarefaction curves based on 16S ribosomal RNA gene sequences indicated that the majority of the OTUs were detected (Supplementary Figure S1). A total of 22 bacterial phyla, 56 classes, 84 orders, 138 families, 228 genera, and 299 species were generated in C. medinalis from six geographic sources (Table 2).
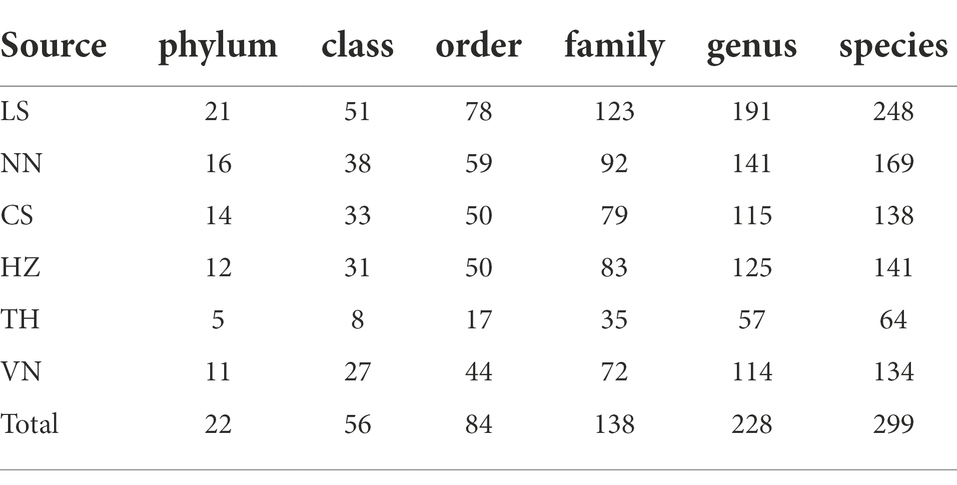
Table 2. Number of gut bacterial taxonomic categories in Cnaphalocrocis medinalis from different geographic sources.
Composition of the gut microbiota in Cnaphalocrocis medinalis from different geographic sources
At the phylum level, Firmicutes and Proteobacteria are the dominant taxa in the gut microbiota of C. medinalis from different geographic sources (Figure 1A). The relative abundance of Firmicutes varied from 57.31 to 93.08%, and the highest relative abundance was observed in the HZ population, followed by the NN, TH, VN, CS, and LS populations. The relative abundance of Proteobacteria varied from 4.94 to 28.66%, and the highest relative abundance was observed in the LS population, followed by the NN, TH, VN, CS, and HZ populations.
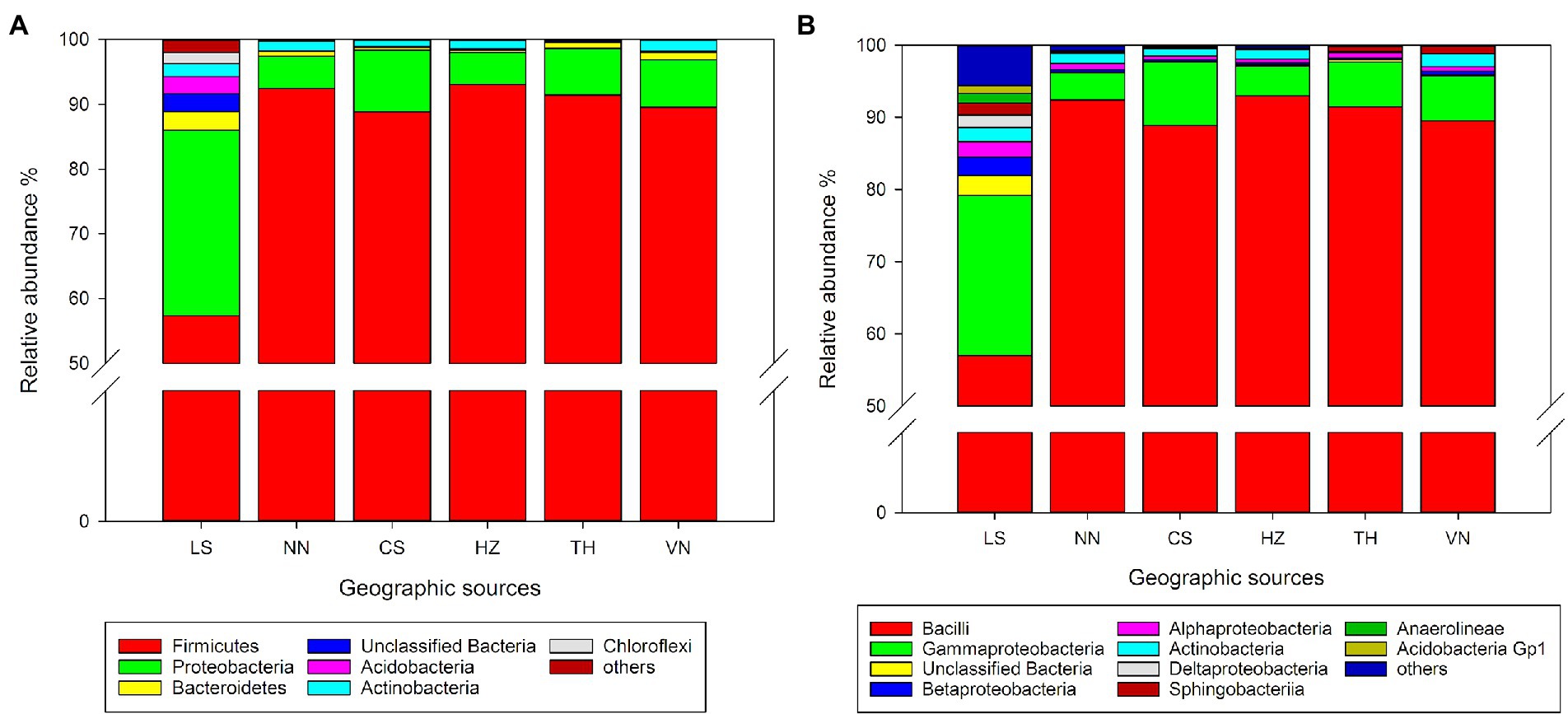
Figure 1. Relative abundance of the gut microbiota in C. medinalis from different geographic sources at the phylum (A) and class (B) levels.
At the class level, Bacilli and Gammaproteobacteria were the dominant taxa in the gut microbiota of C. medinalis from six geographic sources (Figure 1B). Actinobacteria, Alphaproteobacteria, Bacilli, Betaproteobacteria, Gammaproteobacteria, Sphingobacteriia, and unclassified Bacteria occupied the top 10 taxa in the samples from all six geographic sources. The relative abundance of Bacilli varied from 56.99 to 92.98%, and the highest relative abundance was observed in the HZ population, followed by the NN, TH, VN, CS, and LS populations. The relative abundance of Gammaproteobacteria varied from 3.78 to 22.17%, and the highest relative abundance was observed in the LS population, followed by the CS, VN, TH, HZ, and NN populations. The relative abundance of Betaproteobacteria varied from 0.21 to 2.55%, and the highest relative abundance was observed in the LS population, followed by the VN, NN, HZ, CS, and TH populations.
At the genus level, Enterococcus and unclassified Enterobacteriaceae were the dominant taxa in the gut microbiota of C. medinalis from six geographic sources (Figure 2). The relative abundance of Enterococcus varied from 56.20 to 92.92%, and the highest relative abundance was observed in the HZ population, followed by the NN, TH, VN, CS, and LS populations. The relative abundance of unclassified Enterobacteriaceae varied from 3.06 to 19.36%, and the highest relative abundance was observed in the LS population, followed by the CS, VN, TH, HZ, and NN populations.
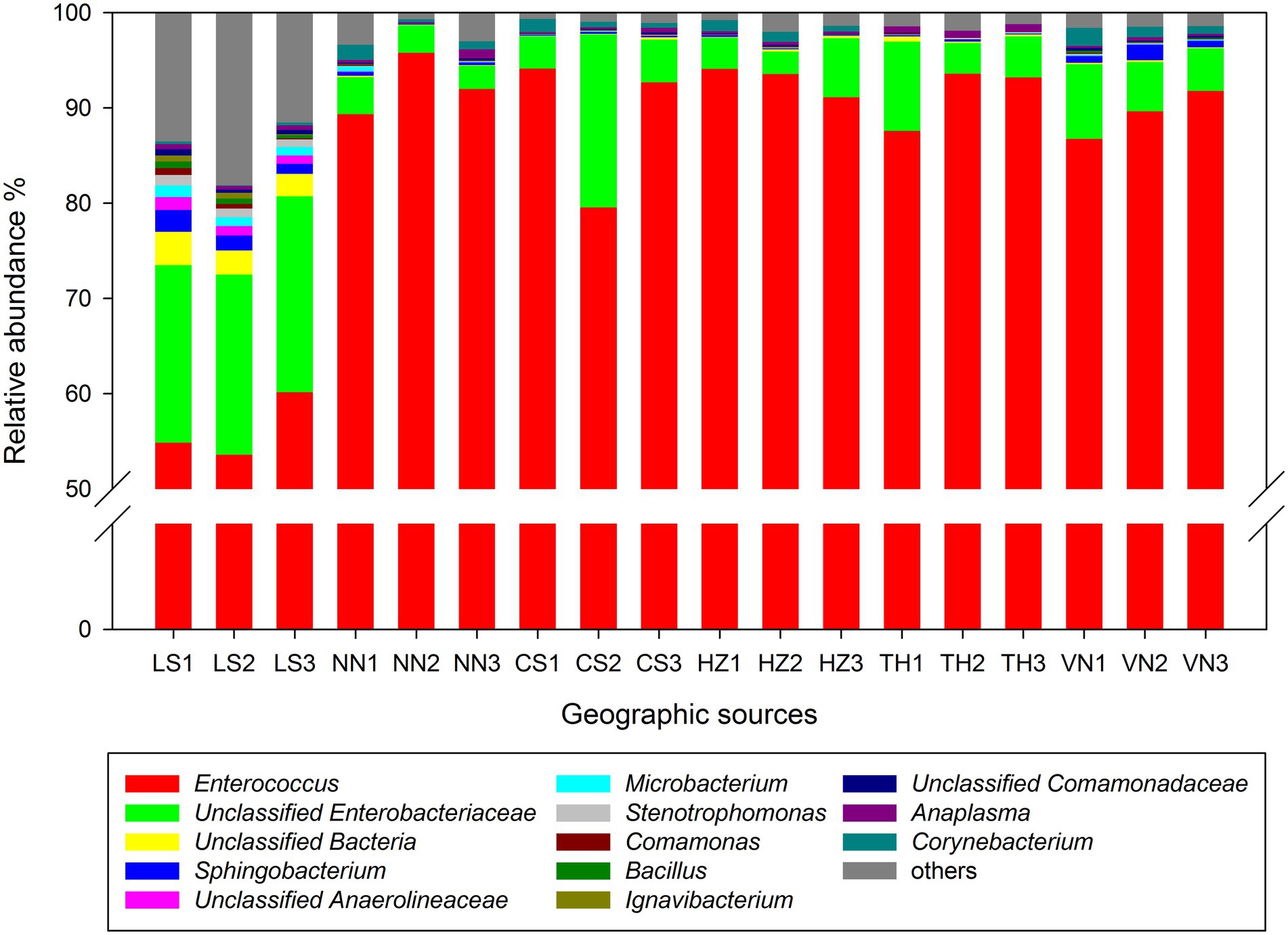
Figure 2. Relative abundance of the gut microbiota in C. medinalis from different geographic sources at the genus level.
Alpha diversity of the gut microbiota in Cnaphalocrocis medinalis from different geographic sources
Alpha diversity indices were analyzed to clarify the differences in the richness and diversity of the bacterial communities of C. medinalis from different geographic sources. The OTU numbers of C. medinalis samples from six geographic sources ranged from 54 to 311, the Ace index ranged from 58.50 to 322.67, the Chao1 index ranged from 61.59 to 319.94, the Shannon index ranged from 0.36 to 3.59, and the Simpson index ranged from 0.32 to 0.92 (Supplementary Table S3). The OTU number and the Shannon index were considerably greater in the LS population than in the other populations, Ace was higher in the LS population than in the HZ and TH populations, the Chao1 index was higher in the LS population than in the CS, HZ, TH, and VN populations, and the Simpson index was lower in the LS population than in the other populations (Table 3). Analysis of variance (ANOVA) revealed that all alpha diversity indices exhibited significant differences across samples from distinct geographic sources (OTU numbers: F = 14.969, p = 8.45E-05; Ace: F = 6.050, p = 0.005; Chao1: F = 5.396, p = 0.008; Shannon: F = 54.464, p = 7.84E-08; Simpson: F = 27.618, p = 3.44E-06; Supplementary Table S4).
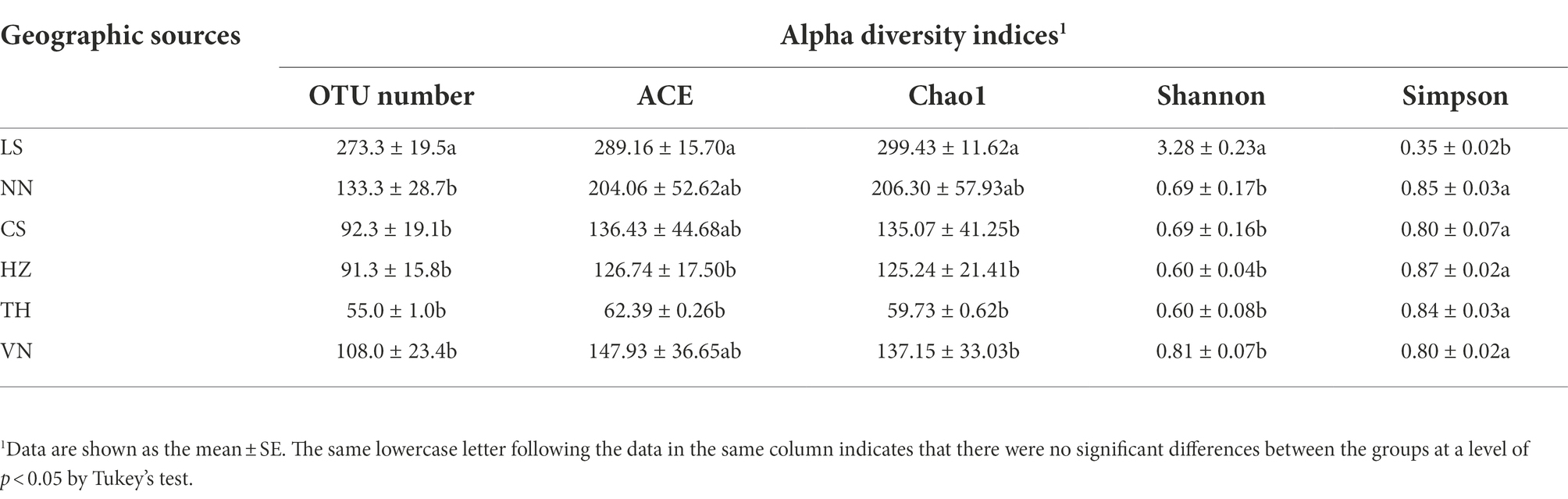
Table 3. Alpha diversity indices of gut bacterial communities in Cnaphalocrocis medinalis from different geographic sources.
Beta diversity of the gut microbiota in Cnaphalocrocis medinalis from different geographic sources
Principal coordinates analysis (PCoA) based on the weighted UniFrac distance and Bray–Curtis distance was conducted to present the bacterial community similarities between samples from different geographic sources. The abscissa and ordinate of the PCoA scatter plot represented the two characteristic values contributing the most to variations between the samples from six geographic sources, with effect degrees of 34.51 and 9.97% based on weighted UniFrac distance (Figure 3A) and 65.96 and 28.39% based on Bray–Curtis distance (Figure 3B), respectively. Significant differences in the gut microbiota of C. medinalis from different geographic sources were observed based on permutational multivariate analysis of variance (PERMANOVA; R2 = 0.25, p = 0.009). Nonmetric multidimensional scaling (NMDS) analysis demonstrated that the gut microbiota of C. medinalis from different geographic sources differed significantly (Figure 4). Analysis of similarity (ANOSIM) revealed significant differences in the gut microbiota of C. medinalis from different geographic sources (R = 0.544, p = 0.002).
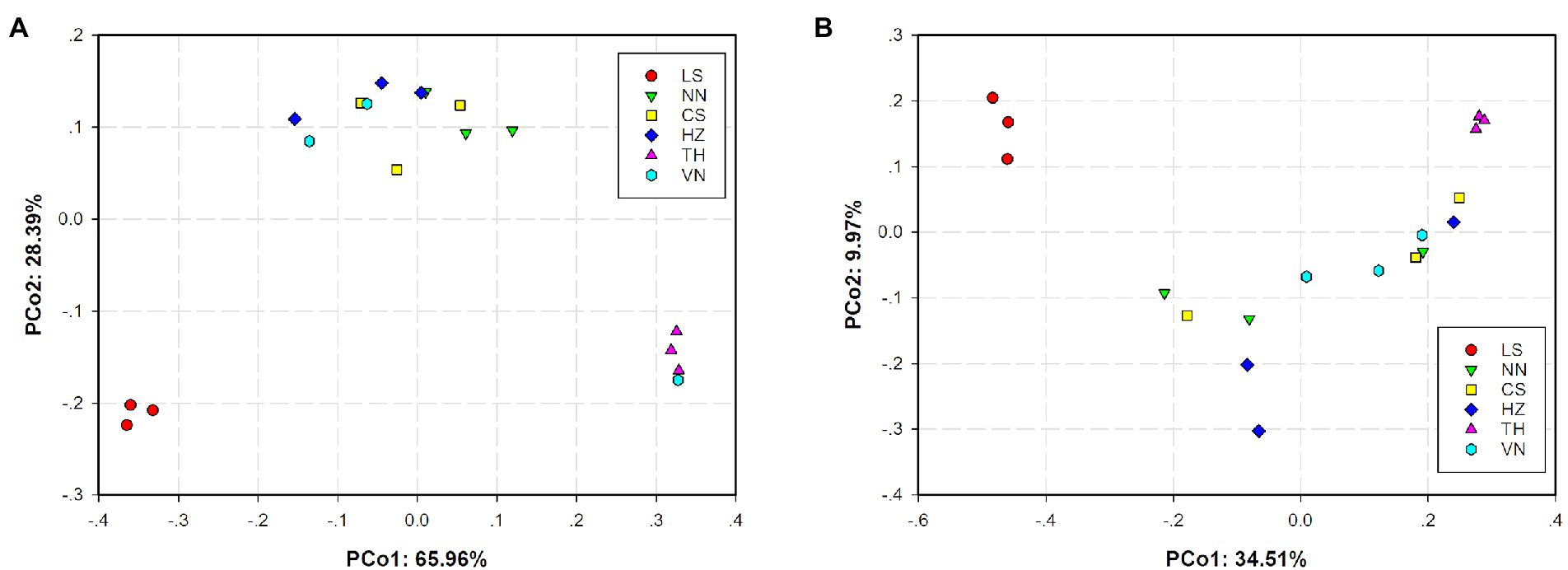
Figure 3. Principal coordinate analysis (PCoA) of bacterial communities in C. medinalis from different geographic sources based on weighted UniFrac (A) and Bray–Curtis (B) distances.
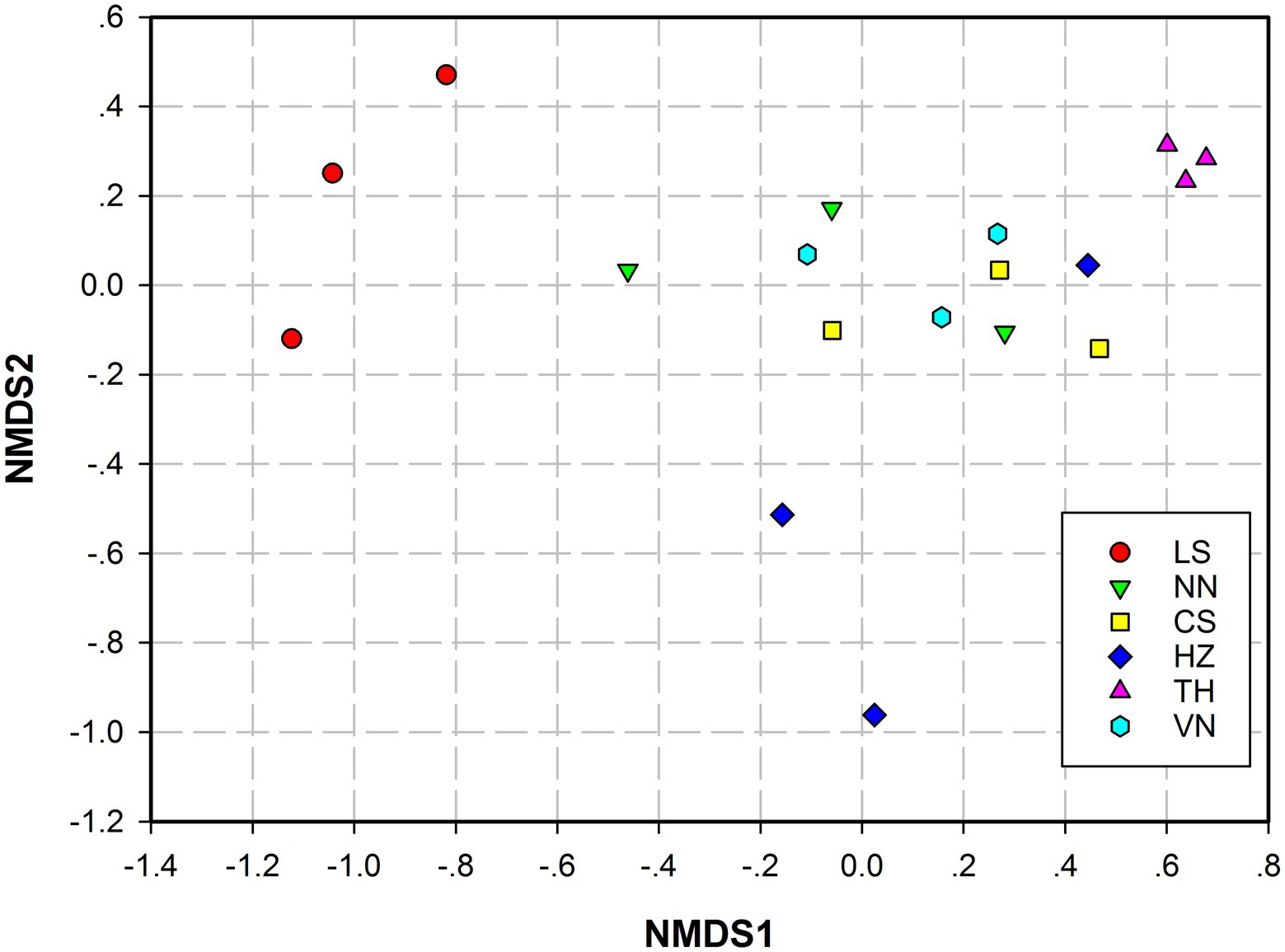
Figure 4. Nonmetric multidimensional scaling (NMDS) analysis of the bacterial communities in different C. medinalis geographic populations. NMDS plots were constructed using Bray–Curtis. Stress values (stress = 0.079) indicate a good fit.
Linear discriminant analysis effect size on the gut microbiota in Cnaphalocrocis medinalis from different geographic sources
To discover significant biomarkers among samples from different geographic sources, linear discriminant analysis (LDA) effect size (LEfSe) was applied to screen out distinct taxa from kingdom to species levels among samples from six geographic sources based on a standard of LDA > 2 (Figure 5). Simultaneously, a cladogram was drawn to overall understand the distribution of these various taxa at different levels from phylum to species (Figure 6). There were 94 different taxa mainly belonging to Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and other Bacteria in the gut microbiota of C. medinalis from different geographic sources (LDA > 2). Most of the different taxa (LDA > 2) were found in the gut microbiota of the LS population. Six taxa belonging to Proteobacteria, two taxa belonging to Bacteroidetes, and three taxa belonging to Actinobacteria were found in the gut microbiota of the VN population (LDA > 2). Two taxa belonging to Bacteroidetes were found in the gut microbiota of the TH population (LDA > 2). Six taxa belonging to Proteobacteria were found in the gut microbiota of the NN population (LDA > 2). A total of 39 taxa belonging to Proteobacteria, eight taxa belonging to Bacteroidetes, 17 taxa belonging to Actinobacteria, four taxa belonging to Firmicutes, and six taxa belonging to other Bacteria were found in the gut microbiota of the LS population (LDA > 2). One taxon belonging to Actinobacteria was found in the gut microbiota of the HZ population (LDA > 2). There were no different taxa found in the gut microbiota of the CS population (LDA > 2).
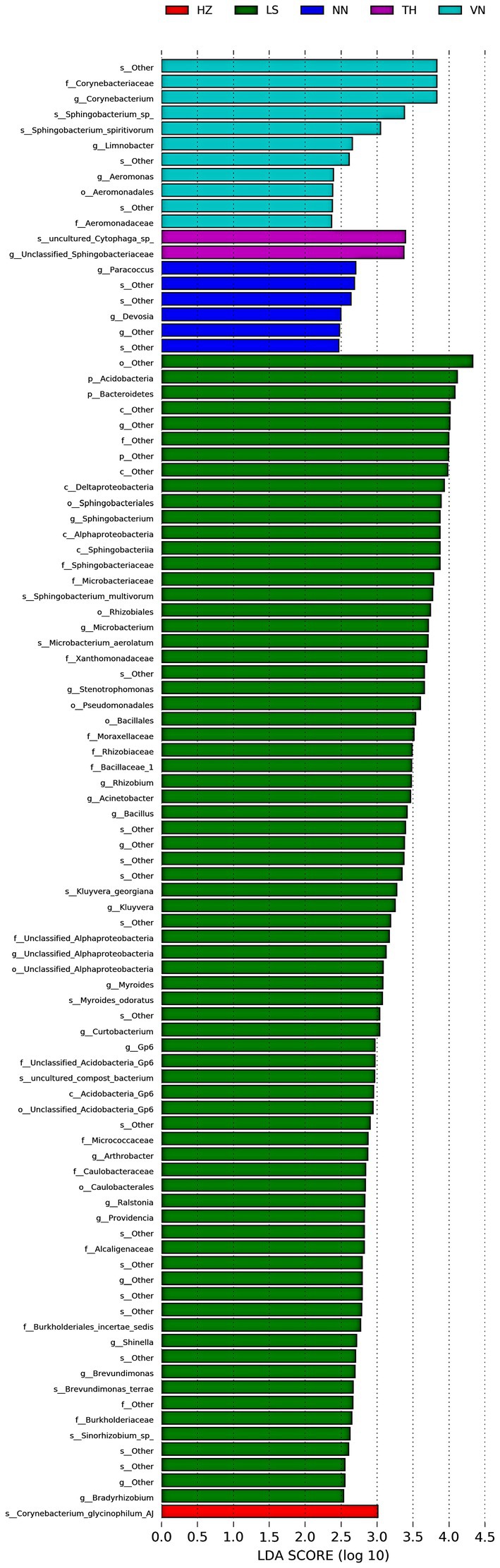
Figure 5. Bacterial taxa with linear discriminant analysis (LDA) scores greater than two in the gut microbiota of C. medinalis from different geographic sources.
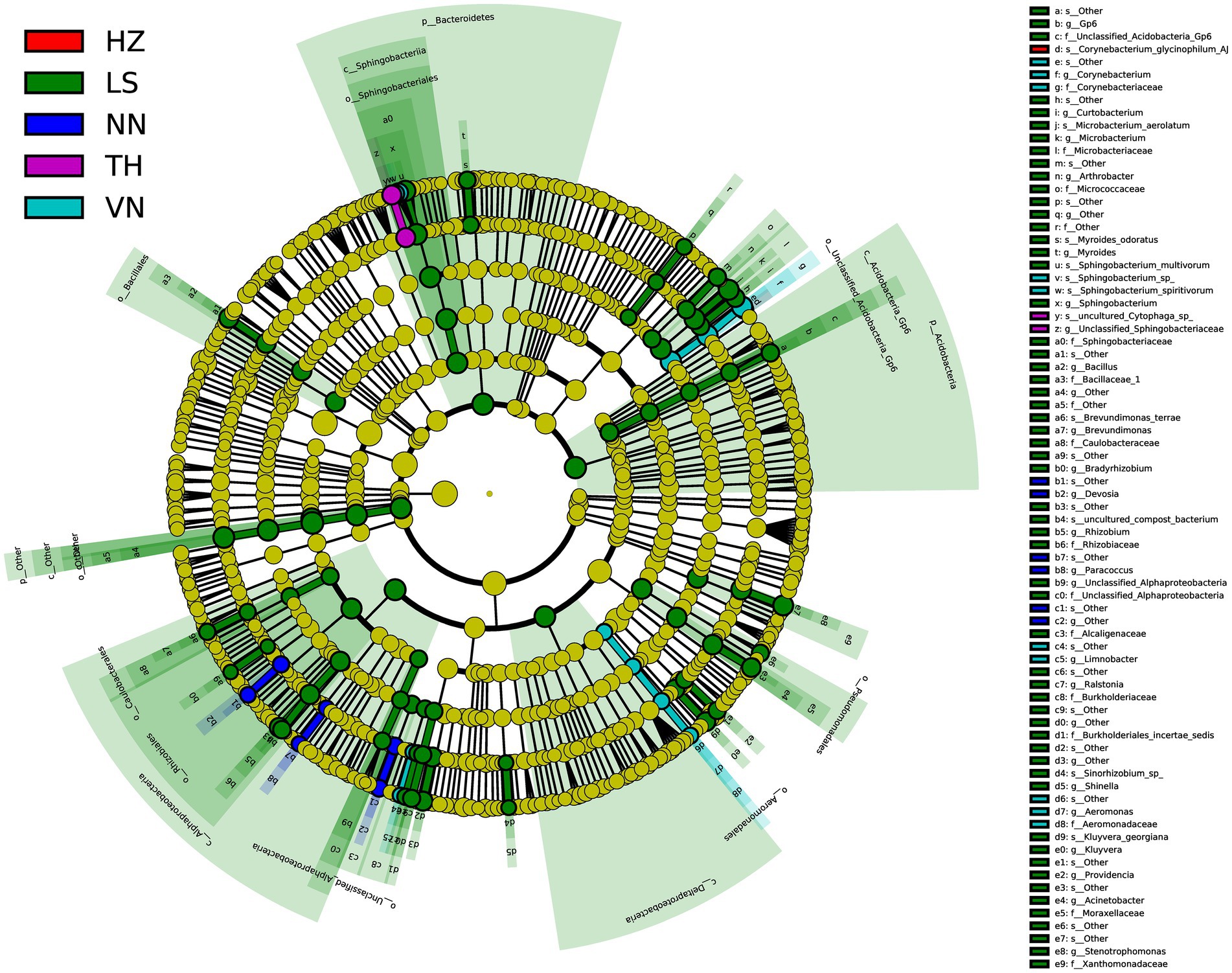
Figure 6. Cladogram of bacterial biomarkers, from the phylum (innermost ring) to species (outermost ring) level, with an LDA score > 2. Differential bacterial taxa are marked by lowercase letters. Each small circle at different taxonomic levels represents a taxon at that level, and the diameter of the circle is proportional to the relative abundance. The coloring principle is to color the taxa with no significant difference as yellow and the other different taxa as the group with the highest abundance of the species. Different colors represent different groups, and nodes with different colors represent the communities that play an important role in the group represented by the color.
There were 52 different taxa among samples from different geographic sources on a standard of LDA > 3, including two taxa belonging to Bacteroidetes and three taxa belonging to Actinobacteria in the gut microbiota of the VN population, two taxa belonging to Bacteroidetes in the gut microbiota of the TH population, 20 taxa belonging to Proteobacteria, eight taxa belonging to Bacteroidetes, five taxa belonging to Actinobacteria, four taxa belonging to Firmicutes, one taxon belonging to Acidobacteria, and six taxa belonging to other Bacteria in the gut microbiota of the LS population, and one taxon belonging to Actinobacteria in the gut microbiota of the HZ population.
Five different taxa were found in the gut microbiota among samples from different geographic sources on a standard of LDA > 4, including one taxon belonging to Actinobacteria, one taxon belonging to Bacteroidetes, and three taxa belonging to other Bacteria in the gut microbiota of the LS population.
Functional prediction of the gut microbiota in Cnaphalocrocis medinalis from different geographic sources
To reveal the significant role of the gut microbiota of C. medinalis from six geographic sources, the function of the gut microbiota in C. medinalis from different geographic sources was predicted using R software with Tax4Fun2 based on 16S ribosomal RNA gene sequencing data and the Ref99NR database (Figure 7). Functional prediction demonstrated that metabolism (70.22–70.53%) was the most functional category, followed by environmental information processing (15.79–17.21%), cellular process (5.29–6.12%), genetic information processing (3.90–4.06%), human diseases (2.32–2.72%), and organismal systems (0.82–0.94%).
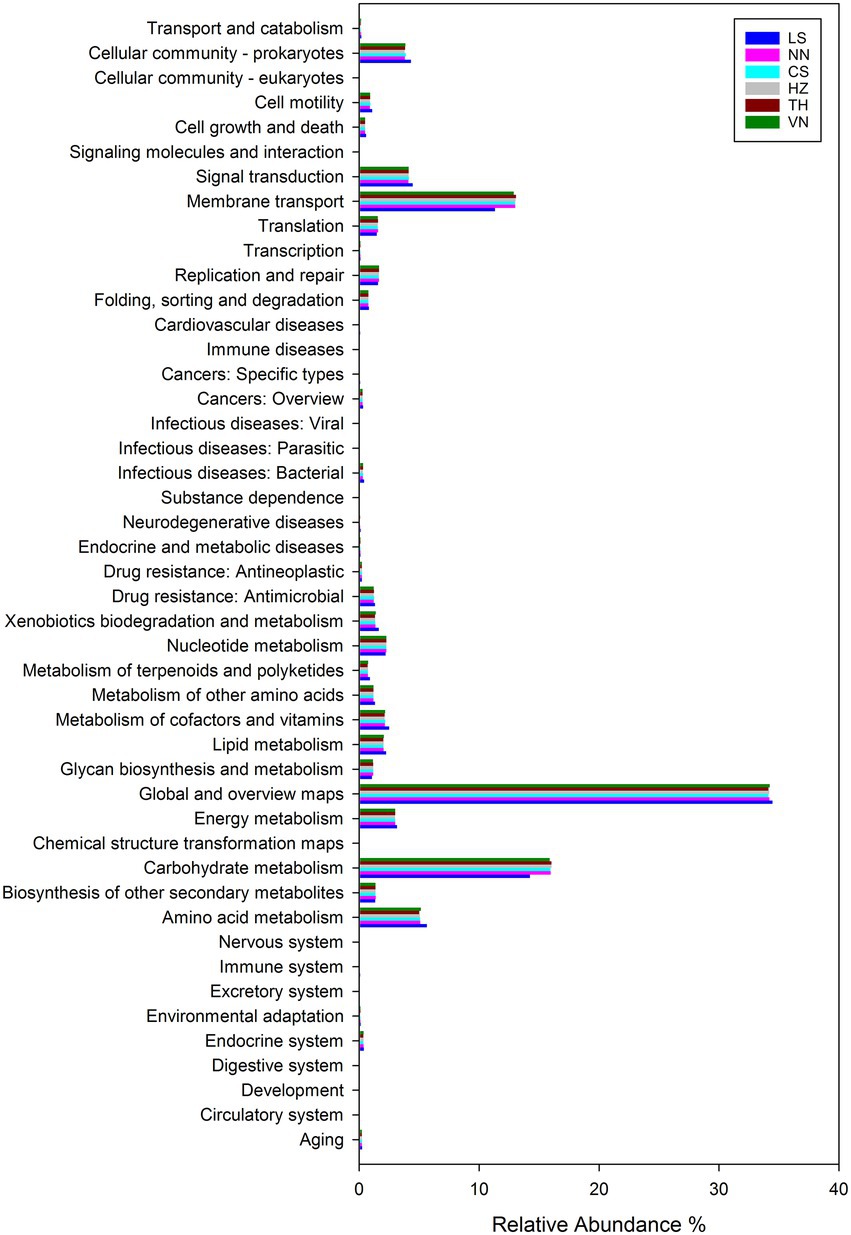
Figure 7. Comparison of predicted GO functions of the gut bacteria in C. medinalis from different geographic sources.
The highest abundance was found in gut microbiota functioning in global and overview maps (34.12–34.45%) among the categories of metabolism, followed by those functioning in carbohydrate metabolism (14.23–16.04%), amino acid metabolism (4.99–5.62%), energy metabolism (2.98–3.13%), nucleotide metabolism (2.18–2.26%), metabolism of cofactors and vitamins (2.11–2.49%), lipid metabolism (2.00–2.24%), xenobiotics biodegradation and metabolism (1.31–1.62%), other secondary metabolites biosynthesis (1.33–1.36%), other amino acids metabolism (1.17–1.30%), glycan biosynthesis and metabolism (1.06–1.15%), terpenoids and polyketides metabolism(0.70–0.88%), and chemical structure transformation maps (0.01–0.02%). The highest abundance was found in gut microbiota functioning in membrane transport (11.33–13.07%) among the categories of environmental information processing, followed by those functioning in signal transduction (4.10–4.45%) and signaling molecules and interactions (0.01–0.02%). The highest abundance was found in gut microbiota functioning in the cellular community (3.79–4.31%) among the categories of cellular processes, followed by those functioning in cell motility (0.88–1.07%), cell growth and death (0.48–0.57%), and transport and catabolism (0.14–0.17%). The highest abundance was found in gut microbiota functioning in replication and repair (1.55–1.65%) among the categories of genetic information processing, followed by those functioning in translation (1.46–1.55%), folding, sorting, and degradation (0.75–0.80%), and transcription (0.09–0.10%). The highest abundance was found in gut microbiota functioning in drug resistance: antimicrobial (1.19–1.29%) among the categories of human diseases, followed by those functioning in infectious diseases: bacterial (0.30–0.40%), cancers: overview (0.26–0.31%), drug resistance: antineoplastic (0.21%), endocrine and metabolic diseases (0.10–0.11%), neurodegenerative diseases (0.07–0.11%), cardiovascular diseases (0.06–0.09%), cancers: specific types (0.05–0.08%), infectious diseases: viral (0.04–0.05%), and immune diseases (0.03%). The highest abundance was found in gut microbiota functioning in the endocrine system (0.35–0.37%) among the categories of organismal systems, followed by those functioning in aging (0.19–0.22%), environmental adaptation (0.10–0.12%), the immune system (0.07–0.08%), the nervous system (0.06–0.07%), and the digestive system (0.05%).
At level 1, the top 20 GO functional pathways of the gut microbiota of C. medinalis from six geographic sources had relative abundances of 63.96–67.91% (Table 4). The relative abundances of the gut microbiota functioning in microbial metabolism in diverse environments, two-component systems, and quorum sensing were all considerably greater in the LS population than in the other geographic populations. The relative abundances of the gut microbiota functioning in ABC transporters, the phosphotransferase system (PTS), starch and sucrose metabolism, amino acids biosynthesis, amino sugar and nucleotide sugar metabolism, glycolysis/gluconeogenesis, fructose and mannose metabolism, purine metabolism, pyruvate metabolism, ribosome, pyrimidine metabolism, and cysteine and methionine metabolism in the LS population of C. medinalis were significantly lower than those in other geographic populations. PERMANOVA demonstrated significant variations in the GO pathways of the gut microbiota of C. medinalis from different geographic sources (R2 = 0.25, p = 0.009). ANOSIM results revealed substantial changes in the GO pathways of the gut microbiota of C. medinalis from different geographic sources (R = 0.3169, p = 0.007).
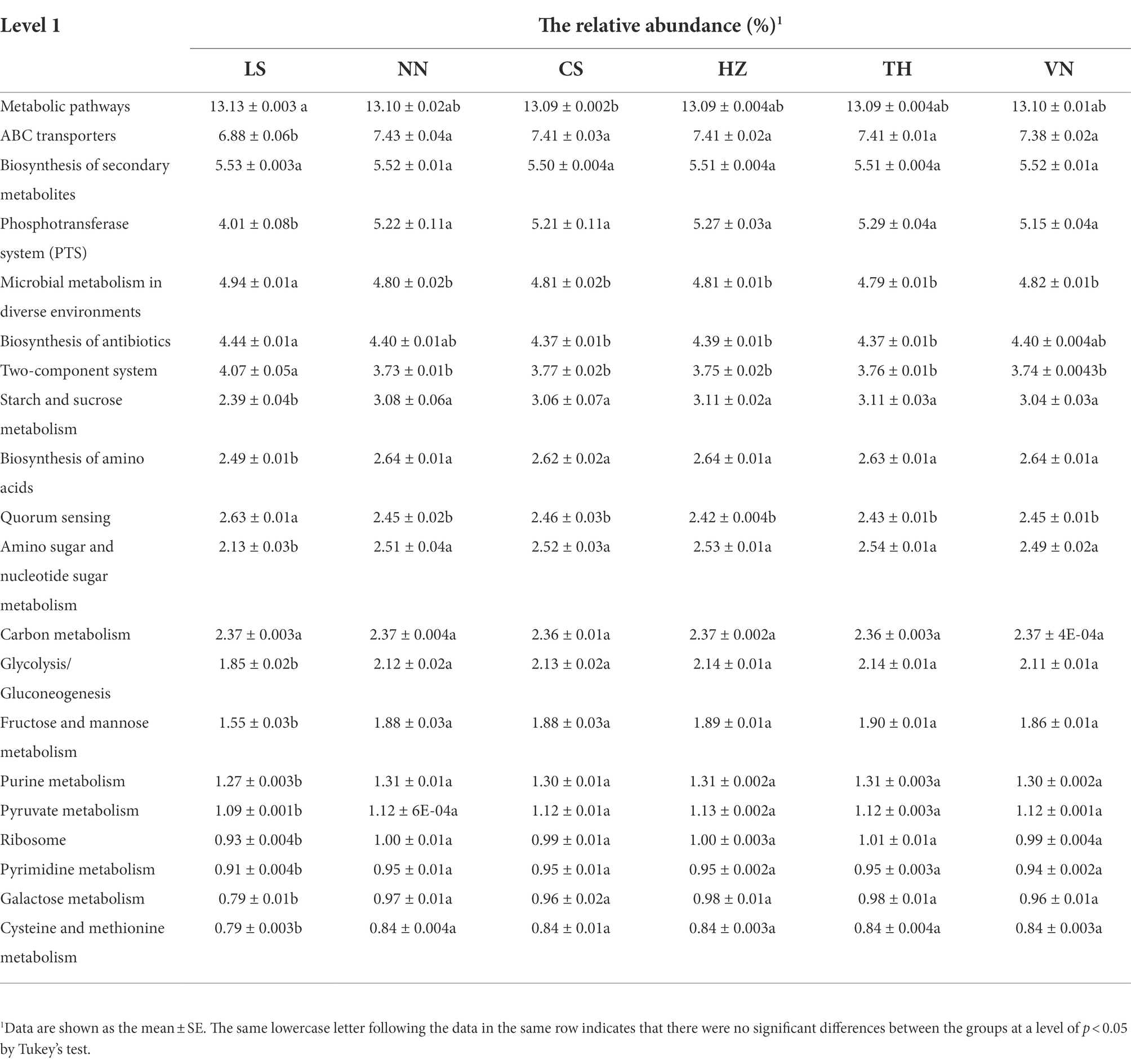
Table 4. The relative abundance of the top 20 GO pathway functions (Level 1) of the gut microbiota of Cnaphalocrocis medinalis from geographic sources.
Ecological processing
The assembly processes of the microbial communities were evaluated within geographic sources (Figures 8A,B) and between geographic sources (Figures 8C,D). Within geographic sources, dispersal processes and selection processes accounted for 61.11 and 38.89% of the microbial community turnover observed between samples within geographic sources, respectively (Figure 9). Variable selection plays an important role within the NN population, and homogeneous selection plays an important role within the TH population (Figures 8A,B). In pairwise comparisons within geographic sources, βNTI mean and median values close to 0 suggested that dispersal (stochastic) processes were the dominant ecological processes within the other geographic sources (Figure 8A). Overall, between geographic sources, selection processes, comprising variable selection (30.90%) and homogeneous selection (24.31%), were the dominant processes, and dispersal processes, including dispersal limitation (2.78%) and homogenizing dispersal (35.76%), accounted for 38.54% of the total (Figure 9). Variable selection strongly influenced pairwise comparisons between the LS population (77.08%) and other geographic sources. Homogenizing dispersal was the dominant ecological process affecting pairwise comparisons between the NN (39.58%), CS(45.83%), HZ(47.92%), TH(43.75%), and VN (37.50%) populations and other geographic sources (Supplementary Table S5; Figures 8C,D).
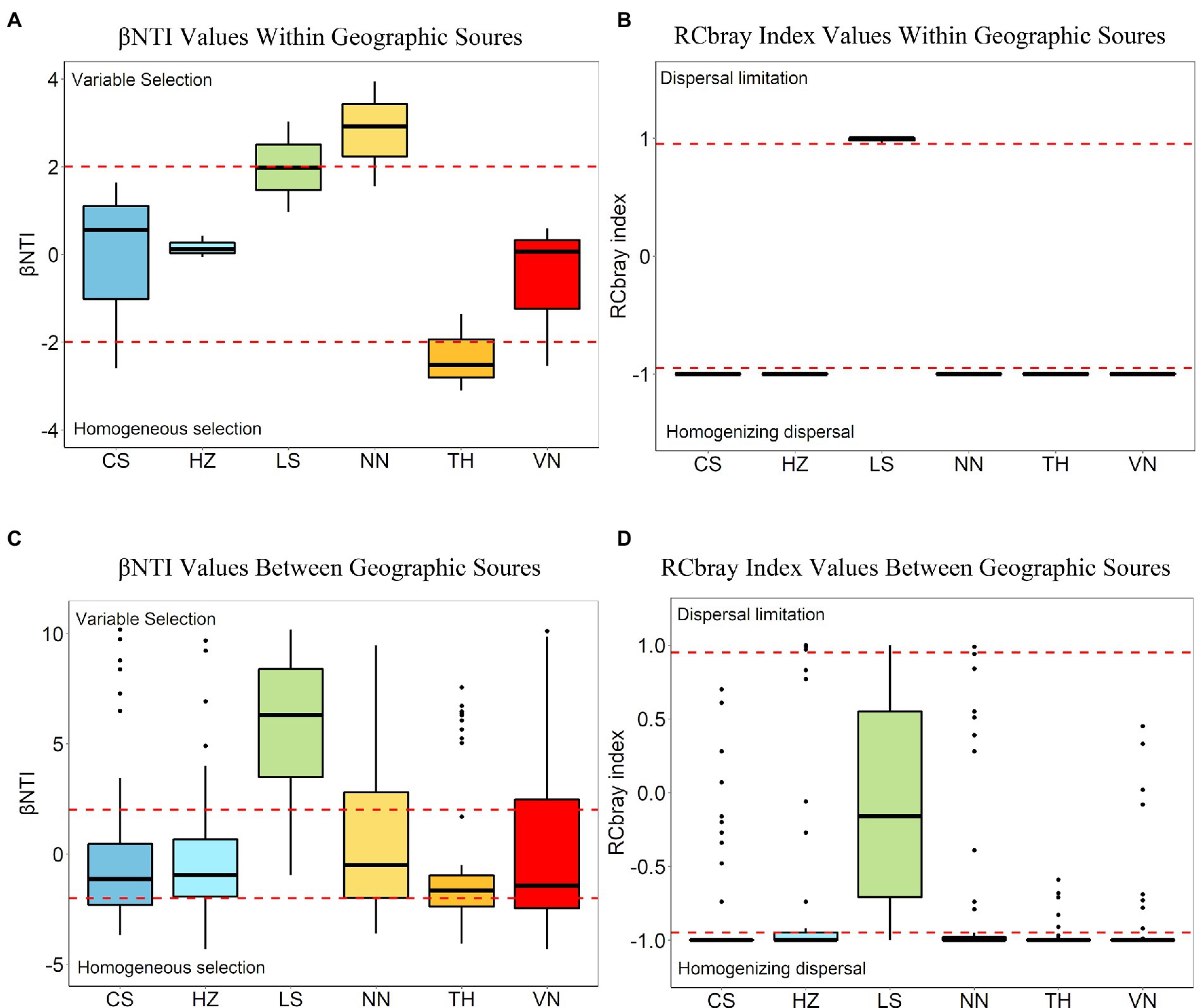
Figure 8. Boxplots showing the distribution of βNTI (A) and RCbray (B) values within geographic sources and the distribution of βNTI (C) and RCbray (D) values between geographic sources. Boxplots represent pairwise comparisons within geographic sources (A,B) and pairwise comparisons between geographic sources (C,D). The dashed black line in the plots represents the null expectation. Dashed red lines in each plot represent significance thresholds for both the βNTI and RCbray metrics. Plotted RCbray values (B,D) do not include values from pairwise comparisons that have significant βNTI values. Boxplots display the distribution of the data, whereas the main colored box displays the interquartile range (25th to 75th percentile) of the data. Solid lines within the boxes represent the median of the data. Whiskers on the plot display the maximum and minimum expected values of the data distribution, and black-colored points represent outlier data points with respect to the plotted distribution.
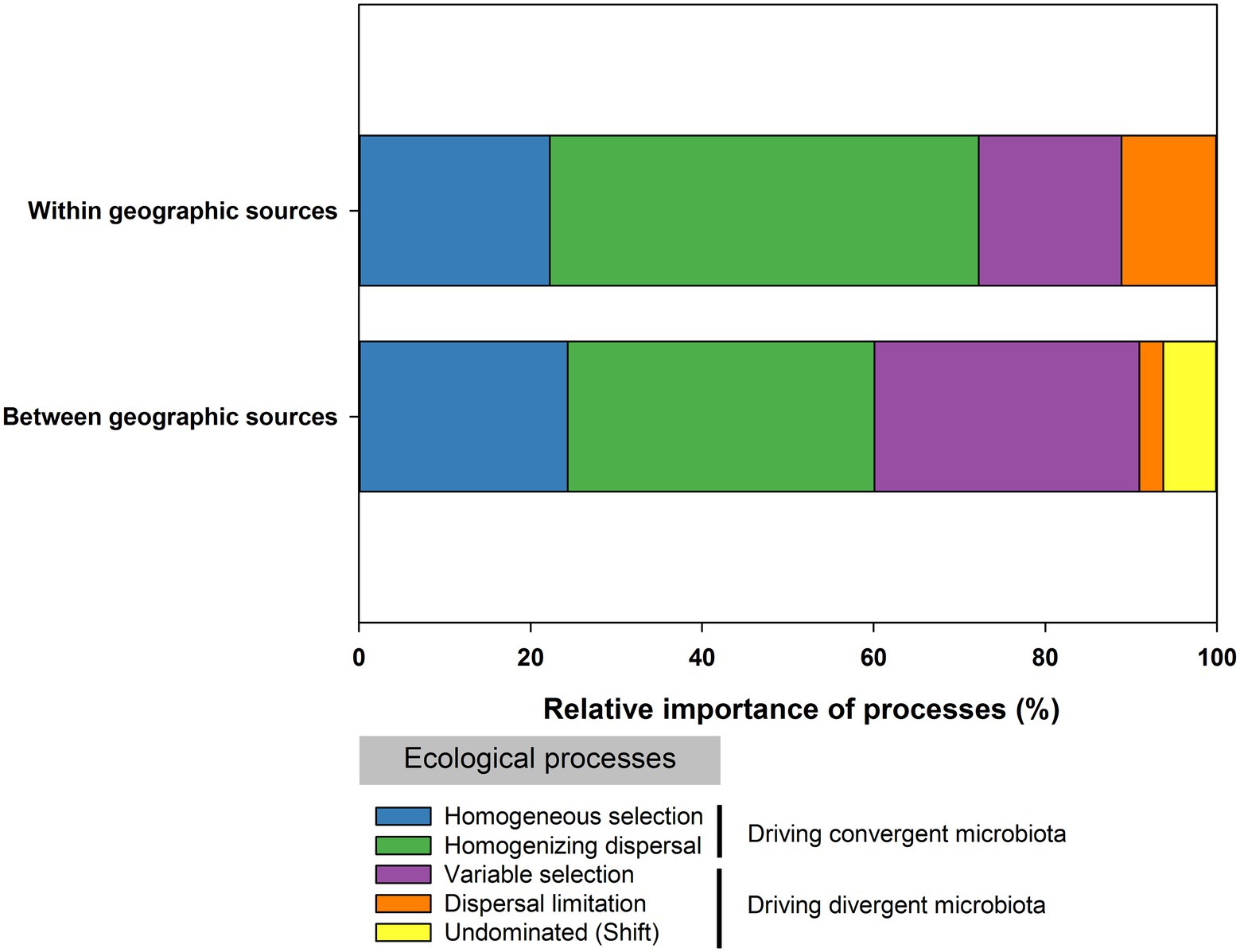
Figure 9. The relative importance of ecological processes in governing the gut bacterial communities of C. medinalis within or between geographical sources.
Discussion
This study profiled the gut microbiota of C. medinalis from six geographic sources. More information on the C. medinalis gut microbiota could be obtained through MiSeq sequencing than through traditional methods of isolation and culturing (Yang, 2012; Liu et al., 2016). The structure and diversity of the gut microbiota in C. medinalis vary across the life cycle of C. medinalis (Yang et al., 2020). Recently, we characterized the bacterial structures of C. medinalis fed rice or maize for three generations and found that host plant and host plant × generation might have a substantial effect on C. medinalis intestinal bacteria based on the PERMANOVA (Yang et al., 2022). In this study, 22 bacterial phyla, 56 classes, 84 orders, 138 families, 228 genera, and 299 species were generated in C. medinalis from different geographic sources, including four sites in China, one in Thailand, and one in Vietnam. In the gut microbiota of C. medinalis from six geographic sources, the phyla Proteobacteria and Firmicutes were also the most common bacterial taxa. Alpha diversity indices suggested that bacterial community richness and diversity could be affected by geographic sources. Our findings in this study add to our understanding of the connection between the C. medinalis populations and their intestinal microbiota, and will deepen the cognition of the population characteristics of C. medinalis and promote the investigation of the C. medinalis gut microbiota.
Insect gut microbiota could be shaped by a range of complex factors such as development stages and host plants. In addition, the geographic source was considered as an important factor in many studies on gut microbiota. We analyzed the bacterial configuration of C. medinalis from six geographic sources and found that geographic sources could influence the composition of the C. medinalis gut microbiota in this work. Firmicutes (57.31–93.08% relative abundance) and Proteobacteria (4.94–28.66% relative abundance) were the dominant taxa at the phylum level in the gut microbiota of C. medinalis from different geographic sources. ANOSIM and PERMANOVA indicated that geographic sources could significantly affect the C. medinalis gut microbiota. Similarly, bacterial communities of Dendroctonus valens vary with geographic sites in the United States of America, and the bacterial community of the house fly Musca domestica varies with geographical location and environment (Adams et al., 2010; Park et al., 2019). Microbiota variations were observed in a vector mosquito Culex nigripalpus from various geographic sources (Duguma et al., 2019). Gut microbial communities in indigenous honey bees (Apis mellifera jemenitica) from two distinct ecological locations in Saudi Arabia were investigated using 16S ribosomal RNA gene sequencing and culture-dependent methods, and the investigation showed that the relative abundance of the isolated bacteria in native honey bees was high in Al-Baha, with 15 bacterial species (45.5%) specific, and 12 bacterial species (36.4%) were specific to Riyadh (Khan et al., 2017). The discrepancy in the gut microbial diversity of honey bees may be influenced by ontogenetic stage, age, and geographic source (Hroncova et al., 2015). However, Ramalho et al. (2017) discovered that host phylogeny can affect the total bacterial community of spiny ants, while geographic source had no effect. Similar bacterial communities were observed among different populations of Tuta absoluta (Meyrick) (Wang et al., 2022). The gut microbial community structures in the bark beetles D. valens and Dendroctonus mexicanus are obviously stable across geographical sources, and the ANOSIM test indicated no significant changes among the metabolic pathways of the microbial communities in D. valens and D. mexicanus across geographical sources (Hernández-García et al., 2018). In fact, the geographic source is a complex factor incorporated with biotic and abiotic factors such as host plant, temperature, humidity, and environmental microbes. Coincidentally, these factors with geographic sources assist the insect population with geographic characteristics. In our study, we fed larvae the same rice variety to rule out the interference of the host plant on the variation in gut microbiota of samples from distinct locations. Hu et al. (2020) found that environmental conditions significantly affect the diversity and composition of the Triatoma rubrofasciata gut microbiota, and Staphylococcus was more common in the laboratory-reared population, while Enterococcus was dominant in the wild population. Ge et al. (2021) suggested that host species and geographic location affected the gut microbial communities of the honeybee by altering the relative contribution of assembly processes of the microbial community. Bascuñán et al. (2018) found that the developmental stage of mosquitoes, followed by geographical source, is a more crucial determinant of gut bacterial structure than mosquito species or adult feeding status.
The gut microbiota always has an essential function in the host insect. Essential nutrient provisioning is the primary function of gut bacteria in Cryptorhynchus lapathi (L.), followed by digestion and detoxification (Jing et al., 2020). Functional prediction of the gut microbiota revealed that metabolism (70.22–70.53%) was found to be the most functional category in this study, followed by environmental information processing (15.79–17.21%), cellular process (5.29–6.12%), genetic information processing (3.90–4.06%), human diseases (2.32–2.72%), and organismal systems (0.82–0.94%). Likewise, intestinal bacteria in C. medinalis fed different host plants perform metabolism most commonly, followed by environmental information processing, cellular process, and genetic information processing according to functional prediction (Yang et al., 2022). Among the top 20 GO functional pathways, the relative abundances of three pathways in the LS population were significantly higher, and 11 pathways in the LS population were significantly lower than those in the other geographic populations. Variation in the relative abundance of the function of the gut microbiota may affect the population characteristics of insects. PERMANOVA and ANOSIM both showed that there were significant variations among the GO pathways of the gut microbiota of C. medinalis from different geographic sources. Global and overview maps, carbohydrate metabolism, membrane transport, amino acid metabolism, signal transduction, cellular community—prokaryotes, energy metabolism, nucleotide metabolism, cofactors and vitamins metabolism, and lipid metabolism were the top10 GO functional pathways in the roles of the gut microbiota in C. medinalis from six geographic sources. As members of the Bacilli class, Enterococcus and unclassified Enterobacteriaceae are the dominant taxa at the genus level in the gut microbiota of C. medinalis from six geographic sources. Many microbes are present in all developmental stages of C. medinalis, and Enterococcus and unclassified Enterobacteriaceae occupy the majority of the bacterial community in larvae, pupae, and adults (Yang et al., 2020). Enterococcus and unclassified Enterobacteriaceae may be important in the life processes of C. medinalis, especially in metabolism. Enterococcus has also been reported to function in protecting insects against pathogens, fixing toxins from plants, increasing insect fitness, and tolerating toxic diets (Shao et al., 2011; Johnston and Rolff, 2015; Vilanova et al., 2016; Shao et al., 2017). Additionally, Enterobacteriaceae has been documented to be associated with insect metabolism, insect resistance or susceptibility to pathogens and insecticides, and insect adaptation and reproduction (Shi et al., 2012; Wang et al., 2020; Álvarez-Lagazzi et al., 2021; Pers and Hansen, 2021; Polenogova et al., 2021). Host-specific Enterococcus could produce dramatically different interactions, and while Enterococcus isolated from Spodoptera frugiperda facilitates the utilization of a poor diet substrate by S. frugiperda, it was obviously antagonistic to Spodoptera exigua under the same conditions (Mason et al., 2022). The variation in the relative abundance of pathways of the gut microbiota may be a potentially critical factor in understanding differences in the population characteristics of C. medinalis. As important resources, some microbes, such as B. thuringiensis, B. subtilis, M. anisopliae, and B. bassiana, have been used in the management of C. medinalis (Kandibane et al., 2010; Kirubakaran et al., 2014; Shakir et al., 2015; Ramasubramanian et al., 2022). Gut microbes could be utilized to manage C. medinalis by disrupting the makeup of gut microbes, weakening their natural functions, or combining with other control measures. Furthermore, many experiments should be carried out to estimate the effect of the variation in gut microbiota on the population characteristics and validate the function of these microbes, and then more and more functional microorganisms may be incorporated into the strategy of C. medinalis management in the future.
In an effort to thoroughly understand the underlying mechanisms of the geographic patterns of the C. medinalis gut microbiota, we assessed the phylogenetic signals of ecological processes that could drive microbial communities to diverge (variable selection and dispersal limitation), converge (homogeneous selection and homogeneous dispersal), and drift (the undominated process) (Stegen et al., 2013, 2015). The results indicated that variable selection possesses a significant role within the NN population, and homogeneous selection possesses a significant role within the TH population. Deterministic processes (variable selection and homogeneous selection) and stochastic processes (homogenizing dispersal and dispersal limitation) accounted for 61.11 and 38.89% of the bacterial community turnover between samples within geographic sources, respectively. Variable selection forcefully influenced paired comparisons between the LS population (77.08%) and other geographic sources, and homogenizing dispersal was the dominant ecological process affecting paired comparisons between the NN (39.58%), CS (45.83%), HZ (47.92%), TH (43.75%), and VN (37.50%) populations and other geographic sources. Within or between geographic sources, variable selection and homogeneous dispersal drive the convergence of C. medinalis gut bacterial communities, and variable selection, dispersal limitation, and undominated processes drive C. medinalis gut bacterial communities to diverge. Homogeneous dispersal probably demonstrated the transfer of gut bacteria between C. medinalis individuals within the same geographic population or exposed to the identical environmental microbial pool. Homogeneous selection likely demonstrated the selection of microbiota under the identical inner intestinal environment or external environment. Geography also influences the relative importance of distinct ecological processes in structuring the gut microbial communities of honeybees (Ge et al., 2021). As an important migratory insect pest, the C. medinalis population migrates northward from March every year and then southward from September onward (Chang et al., 1980; Riley et al., 1995; Wang et al., 2017). Southeast Asia is considered as the main source area of the C. medinalis populations in China (Yan, 2021). Wang et al. (2017) reported that the Nanning population could migrate to the middle and lower reaches of the Yangtze River, including Changsha and Hangzhou. In addition, different locations may have particular weather, and weather could influence the activity of C. medinalis (Khan and Ramamurthy, 2004). The LS population harbored a particular geography in Leshan, which is located in the southwestern Sichuan Basin with particular weather surrounding high mountains in southwestern China. The special geography may constrain the communication and migration of C. medinalis, making the sources of the LS population complicated. Our study indicated that the ecological processing of gut microbiota in the LS population differed from those in other populations. The LS population, with a special geographic environment, may cause variations in the ecological processing of the gut microbiota with other populations. Although microbes differ among the developmental stages of C. medinalis, some microbes, including Enterococcus, can be found across all the C. medinalis stages (Yang et al., 2020). A recent study indicated that the host plant also affects the microbiota of C. medinalis along with the insect generation (Yang et al., 2022). Communication between populations and geography may be the crucial factor affecting the ecological processing of the microbiota in C. medinalis. In total, the bacterial communities within geographic sources of C. medinalis were mainly determined by stochastic processes, and the bacterial communities between geographic sources were mainly determined by deterministic processes. In the Helicoverpa armigera caterpillars, the structure of the gut microbiota is mainly determined by stochastic processes (Li et al., 2022). Therefore, considering the importance of the microbiota, gut microbial shifts in C. medinalis across geography may influence the characteristics of the C. medinalis populations. Nevertheless, more experiments should be carried out to clarify the assembly and function of the C. medinalis gut microbiota.
Conclusion
Geography may be quite important to determining the C. medinalis gut microbiota. The alpha diversity indices of C. medinalis gut microorganisms could be influenced by geographic source. PERMANOVA and ANOSIM indicated that the C. medinalis gut bacteria could be significantly affected by geographic sources. The most dominant function of the C. medinalis intestinal microbiota is metabolism, followed by environmental information processing, cellular process, genetic information processing, human diseases, and organismal systems. PERMANOVA and ANOSIM both revealed significant differences among the GO pathways of the gut microbiota of C. medinalis from different geographic sources. In total, the bacterial communities within geographic sources were mainly determined by stochastic processes, and the bacterial communities between geographic sources were mainly determined by deterministic processes. Furthermore, more research should be carried out to elucidate the role of these microbes, which will facilitate the screening of novel targets for the C. medinalis management. The findings in this work contribute to a better knowledge of the microbial diversity associated with migratory pests, enrich the character of C. medinalis populations, lay a theoretical groundwork for future research on C. medinalis gut microbes, and shed light on the population relationship and the shaping of the gut microbiota in C. medinalis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA838393, SRR19216016-SRR19216021.
Author contributions
YY, YL, and ZL contributed to conceptualization of the study. YY, JG, and ZL contributed to funding acquisition. XL investigated the study. YY, HX, and ZL contributed to the methodology. YL and ZL contributed to supervision. YY, JG, and XL visualized the study. YY wrote the original draft and contributed to writing, reviewing, and editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the earmarked fund for China Agriculture Research System, grant number CARS-01, and Zhejiang Provincial Natural Science Foundation of China, grant numbers LY20C140004 and LQ22C140006.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1035644/full#supplementary-material
Supplementary Figure S1 | Rarefaction curves of the bacterial community of Cnaphalocrocis medinalis from different geographic sources based on Illumina MiSeq sequencing data.
Footnotes
3. ^http://www.drive5.com/uchime/uchime_download.html
5. ^https://www.r-project.org/
6. ^https://www.r-project.org/
7. ^http://huttenhower.sph.harvard.edu/galaxy/ (accessed on 22 March 2022).
8. ^https://github.com/stegen/Stegen_etal_ISME_2013/blob/master/bNTI_Local_Machine.r
9. ^https://github.com/stegen/Stegen_etal_ISME_2013/blob/master/Raup_Crick_Abundance.r
References
Adams, A. S., Adams, S. M., Currie, C. R., Gillette, N. E., and Raffa, K. F. (2010). Geographic variation in bacterial communities associated with the red turpentine beetle (Coleoptera: Curculionidae). Environ. Entomol. 39, 406–414. doi: 10.1603/EN09221
Álvarez-Lagazzi, A. P., Cabrera, N., Francis, F., and Ramírez, C. C. (2021). Bacillus subtilis (Bacillales, Bacillaceae) spores affect survival and population growth in the grain aphid Sitobion avenae (Hemiptera, Aphididae) in relation to the presence of the facultative bacterial endosymbiont Regiella insecticola (Enterobacteriales, Enterobacteriaceae). J. Econ. Entomol. 114, 2043–2050. doi: 10.1093/jee/toab164
Anand, A. A., Vennison, S. J., Sankar, S. G., Prabhu, D. I. G., Vasan, P. T., Raghuraman, T., et al. (2010). Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J. Insect Sci. 10, 1–20. doi: 10.1673/031.010.10701
Barrion, A. T., Litsinger, J. A., Medina, E. B., Aguda, R. M., Bandong, J. P., Pantua, P. C. Jr., et al. (1991). The rice Cnaphalocrocis and Marasmia (Lepidoptera: Pyralidae) leaffolder complex in the Philippines: taxonomy, bionomics, and control. Phil. Entomol. 8, 987–1074.
Bascuñán, P., Niño-Garcia, J. P., Galeano-Castañeda, Y., Serre, D., and Correa, M. M. (2018). Factors shaping the gut bacterial community assembly in two main Colombian malaria vectors. Microbiome 6:148. doi: 10.1186/s40168-018-0528-y
Behar, A., Yuval, B., and Jurkevitch, E. (2008). Community structure of the mediterranean fruit fly microbiota: seasonal and spatial sources of variation. Isr. J. Ecol. Evol. 54, 181–191. doi: 10.1080/15659801.2008.10639612
Beran, F., and Gershenzon, J. (2016). Microbes matter: herbivore gut endosymbionts play a role in breakdown of host plant toxins. Environ. Microbiol. 18, 1306–1307. doi: 10.1111/1462-2920.13258
Berasategui, A., Salem, H., Paetz, C., Santoro, M., Gershenzon, J., Kaltenpoth, M., et al. (2017). Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Mol. Ecol. 26, 4099–4110. doi: 10.1111/mec.14186
Berasategui, A., Shukla, S., Salem, H., and Kaltenpoth, M. (2016). Potential applications of insect symbionts in biotechnology. Appl. Microbiol. Biotechnol. 100, 1567–1577. doi: 10.1007/s00253-015-7186-9
Blanton, A. G., and Peterson, B. F. (2020). Symbiont-mediated insecticide detoxification as an emerging problem in insect pests. Front. Microbiol. 11:547108. doi: 10.3389/fmicb.2020.547108
Bosmans, L., Pozo, M. I., Verreth, C., Crauwels, S., Wilberts, L., Sobhy, I. S., et al. (2018). Habitat specific variation in gut microbial communities and pathogen prevalence in bumblebee queens (Bombus terrestris). PLoS One 13:e0204612. doi: 10.1371/journal.pone.0204612
Chang, S. S., Lo, Z. C., Keng, C. G., Li, G. Z., Chen, X. L., Wu, X. W., et al. (1980). Studies on the migration of rice leaf roller, Cnaphalocrocis medinalis Guenée. Acta Entomol. Sin. 23, 130–140.
Chen, B., Zhang, N., Xie, S., Zhang, X., He, J., Muhammad, A., et al. (2020). Gut bacteria of the silkworm Bombyx mori facilitate host resistance against the toxic effects of organophosphate insecticides. Environ. Int. 143:105886. doi: 10.1016/j.envint.2020.105886
Cheng, D., Guo, Z., Riegler, M., Xi, Z., Liang, G., and Xu, Y. (2017). Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 5:13. doi: 10.1186/s40168-017-0236-z
Dillon, R. J., and Dillon, V. M. (2004). The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49, 71–92. doi: 10.1146/annurev.ento.49.061802.123416
Douglas, A. E. (2015). Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. doi: 10.1146/annurev-ento-010814-020822
Duguma, D., Hall, M. W., Smartt, C. T., Debboun, M., and Neufeld, J. D. (2019). Microbiota variations in Culex nigripalpus disease vector mosquito of West Nile virus and Saint Louis encephalitis from different geographic origins. Peer 6:e6168. doi: 10.7717/peerj.6168
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Frago, E., Dicke, M., and Godfray, H. C. J. (2012). Insect symbionts as hidden players in insect–plant interactions. Trends Ecol. Evol. 27, 705–711. doi: 10.1016/j.tree.2012.08.013
Ge, Y., Jing, Z., Diao, Q., He, J. Z., and Liu, Y. J. (2021). Host species and geography differentiate honeybee gut bacterial communities by changing the relative contribution of community assembly processes. MBio 12, e00751–e00721. doi: 10.1128/mBio.00751-21
Gould, A. L., Zhang, V., Lamberti, L., Jones, E. W., Obadia, B., Korasidis, N., et al. (2018). Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. U. S. A. 115, E11951–E11960. doi: 10.1073/pnas.1809349115
Hernández-García, J. A., Gonzalez-Escobedo, R., Briones-Roblero, C. I., Cano-Ramírez, C., Rivera-Orduña, F. N., and Zúñiga, G. (2018). Gut bacterial communities of Dendroctonus valens and D. mexicanus (Curculionidae: Scolytinae): a metagenomic analysis across different geographical locations in Mexico. Int. J. Mol. Sci. 19:2578. doi: 10.3390/ijms19092578
Hroncova, Z., Havlik, J., Killer, J., Doskocil, I., Tyl, J., Kamler, M., et al. (2015). Variation in honey bee gut microbial diversity affected by ontogenetic stage, age and geographic location. PLoS One 10:e0118707. doi: 10.1371/journal.pone.0118707
Hu, Y., Xie, H., Gao, M., Huang, P., Zhou, H., Ma, Y., et al. (2020). Dynamic of composition and diversity of gut microbiota in Triatoma rubrofasciata in different developmental stages and environmental conditions. Front. Cell. Infect. Microbiol. 10:587708. doi: 10.3389/fcimb.2020.587708
Huang, S. W., and Zhang, H. Y. (2013). The impact of environmental heterogeneity and life stage on the hindgut microbiota of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). PLoS One 8:e57169. doi: 10.1371/journal.pone.0057169
Jaramillo, A., and Castañeda, L. E. (2021). Gut microbiota of Drosophila subobscura contributes to its heat tolerance and is sensitive to transient thermal stress. Front. Microbiol. 12:654108. doi: 10.3389/fmicb.2021.654108
Jing, T. Z., Qi, F. H., and Wang, Z. Y. (2020). Most dominant roles of insect gut bacteria: digestion, detoxification, or essential nutrient provision? Microbiome 8:38. doi: 10.1186/s40168-020-00823-y
Johnston, P. R., and Rolff, J. (2015). Host and symbiont jointly control gut microbiota during complete metamorphosis. PLoS Pathog. 11:e1005246. doi: 10.1371/journal.ppat.1005246
Jones, J. C., Fruciano, C., Hildebrand, F., Toufalilia, H. A., Balfour, N. J., Bork, P., et al. (2018). Gut microbiota composition is associated with environmental landscape in honey bees. Ecol. Evol. 8, 441–451. doi: 10.1002/ece3.3597
Jones, A. G., Mason, C. J., Felton, G. W., and Hoover, K. (2019). Host plant and population source drive diversity of microbial gut communities in two polyphagous insects. Sci. Rep. 9:2792. doi: 10.1038/s41598-019-39163-9
Kalaivani, K., Kalaiselvi, M. M., and Senthil-Nathan, S. (2018). Effect of methyl salicylate (MeSA) induced changes in rice plant (Oryza sativa) that affect growth and development of the rice leaf folder, Cnaphalocrocis medinalis. Physiol. Mol. Plant Pathol. 101, 116–126. doi: 10.1016/j.pmpp.2017.07.001
Kandibane, M., Kumar, K., and Adiroubane, D. (2010). Effect of bacillus thuringiensis Berliner formulation against the rice leaf folder Cnaphalocrocis medinalis Guenee (Pyralidae: Lepidoptera). J. Biopest. 3, 445–447.
Khan, K. A., Ansari, M. J., Al-Ghamdi, A., Nuru, A., Harakeh, S., and Iqbal, J. (2017). Investigation of gut microbial communities associated with indigenous honey bee (Apis mellifera jemenitica) from two different eco-regions of Saudi Arabia. Saudi J. Biol. Sci. 24, 1061–1068. doi: 10.1016/j.sjbs.2017.01.055
Khan, Z. H., and Ramamurthy, V. V. (2004). Influence of weather factors on the activity of rice leaf folder, Cnaphalocrocis medinalis (Guenee). Ann. Plant Prot. Sci. 12, 263–266.
Kirubakaran, S. A., Sathish-Narayanan, S., Revathi, K., Chandrasekaran, R., and Senthil-Nathan, S. (2014). Effect of oil-formulated Metarhizium anisopliae and Beauveria bassiana against the rice leaffolder Cnaphalocrocis medinalis Guenée (Lepidoptera: Pyralidae). Arch. Phytopathol. Plant Prot. 47, 977–992. doi: 10.1080/03235408.2013.828388
Kyritsis, G. A., Augustinos, A. A., Caceres, C., and Bourtzis, K. (2017). Medfly gut microbiota and enhancement of the sterile insect technique: similarities and differences of Klebsiella oxytoca and Enterobacter sp. AA26 probiotics during the larval and adult stages of the VIENNA 8(D53+) genetic sexing strain. Front. Microbiol. 8:2064. doi: 10.3389/fmicb.2017.02064
Li, S. L., Tang, R., Yi, H., Cao, Z. C., Sun, S. L., Liu, T. X., et al. (2022). Neutral processes provide an insight into the structure and function of gut microbiota in the cotton bollworm. Front. Microbiol. 13:849637. doi: 10.3389/fmicb.2022.849637
Li, S., Xu, X., Mandal, S. D., Shakeel, M., Hua, Y., Shoukat, R. F., et al. (2021). Gut microbiota mediate Plutella xylostella susceptibility to Bt cry 1Ac protoxin is associated with host immune response. Environ. Pollut. 271:116271. doi: 10.1016/j.envpol.2020.116271
Liu, X. G., Yang, Y. J., Liao, Q. J., Xu, H. X., Liu, Y. H., and Lu, Z. X. (2016). Analysis of the bacterial community structure and diversity in the intestine of Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). Acta Entomol. Sin. 59, 965–976. doi: 10.16380/j.kcxb.2016.09.006
Liu, G., Zheng, X., Long, H., Rao, Z., Cao, L., and Han, R. (2021). Gut bacterial and fungal communities of the wild and laboratory-reared Thitarodes larvae, host of the Chinese medicinal fungus Ophiocordyceps sinensis on tibetan plateau. Insects 12:327. doi: 10.3390/insects12040327
Luo, C., Li, Y., Chen, Y., Fu, C., Long, W., Xiao, X., et al. (2019). Bamboo lignocellulose degradation by gut symbiotic microbiota of the bamboo snout beetle Cyrtotrachelus buqueti. Biotechnol. Biofuels 12:70. doi: 10.1186/s13068-019-1411-1
Mason, C. J., Peiffer, M., Chen, B., Hoover, K., and Felton, G. W. (2022). Opposing growth responses of lepidopteran larvae to the establishment of gut microbiota. Microbiol. Spectr. 10:e01941-22. doi: 10.1128/spectrum.01941-22
Mikaelyan, A., Dietrich, C., Köhler, T., Poulsen, M., Sillam-Dussès, D., and Brune, A. (2015). Diet is the primary determinant of bacterial community structure in the guts of higher termites. Mol. Ecol. 24, 5284–5295. doi: 10.1111/mec.13376
Moore, J. M., and Aparicio, J. C. M. (2022). “Enhancing pathogen resistance: the gut microbiota and malaria,” in Comprehensive Gut Microbiota. ed. M. Glibetic (Amsterdam: Elsevier), 143–167.
Park, R., Dzialo, M. C., Spaepen, S., Nsabimana, D., Gielens, K., Devriese, H., et al. (2019). Microbial communities of the house fly Musca domestica vary with geographical location and habitat. Microbiome 7:147. doi: 10.1186/s40168-019-0748-9
Pers, D., and Hansen, A. K. (2021). The boom and bust of the aphid's essential amino acid metabolism across nymphal development. G3 (Bethesda) 11:jkab115. doi: 10.1038/srep39853
Polenogova, O. V., Noskov, Y. A., Yaroslavtseva, O. N., Kryukova, N. A., Alikina, T., Klementeva, T. N., et al. (2021). Influence of Bacillus thuringiensis and avermectins on gut physiology and microbiota in Colorado potato beetle: impact of enterobacteria on susceptibility to insecticides. PLoS One 16:e0248704. doi: 10.1371/journal.pone.0248704
Pyszko, P., Višňovská, D., Drgová, M., Šigut, M., and Drozd, P. (2020). Effect of bacterial and fungal microbiota removal on the survival and development of bryophagous beetles. Environ. Entomol. 49, 902–911. doi: 10.1093/ee/nvaa060
Ramalho, M. O., Bueno, O. C., and Moreau, C. S. (2017). Microbial composition of spiny ants (Hymenoptera: Formicidae: Polyrhachis) across their geographic range. BMC Evol. Biol. 17:96. doi: 10.1186/s12862-017-0945-8
Ramasubramanian, R., Karthi, S., Senthil-Nathan, S., Sivanesh, H., Shyam Sundar, N., Stanley-Raja, V., et al. (2022). Effect of bacterial toxin identified from the Bacillus subtilis against the Cnaphalocrocis medinalis Guenée (Lepidoptera: Crambidae). Toxin Rev. doi: 10.1080/15569543.2022.2111444
Riley, J. R., Reynolds, D. R., Smith, A. D., Edwards, A. S., Zhang, X. X., and Cheng, X. N. (1995). Observations of the autumn migration of the rice leaf roller, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae) and other moths in eastern China. Bull. Entomol. Res. 85, 397–414. doi: 10.1017/S0007485300036130
Schmitt-Wagner, D., Friedrich, M. W., Wagner, B., and Brune, A. (2003). Axial dynamics, stability, and interspecies similarity of bacterial community structure in the highly compartmentalized gut of soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69, 6018–6024. doi: 10.1128/AEM.69.10.6018-6024.2003
Senthil-Nathan, S. (2019). Effect of methyl jasmonate (MeJA)-induced defenses in rice against the rice leaffolder Cnaphalocrocis medinalis (Guenèe) (Lepidoptera: Pyralidae). Pest Manag. Sci. 75, 460–465. doi: 10.1002/ps.5139
Shakir, H. U., Saeed, M., Anjum, N., Farid, A., Khan, I. A., Liaquat, M., et al. (2015). Combined effect of entomopathogenic fungus Beauveria bassiana, imidacloprid and potassium silicate against Cnaphalocrocis medinalis Guenée (Lepidoptera: Pyralidae) in rice crop. J. Entomol. Zool. Stud. 3, 173–177.
Shao, Y. Q., Chen, B. S., Sun, C., Ishida, K., Hertweck, C., and Boland, W. (2017). Symbiont-derived antimicrobials contribute to the control of the lepidopteran gut microbiota. Cell Chem. Biol. 24, 66–75. doi: 10.1016/j.chembiol.2016.11.015
Shao, Y., Spiteller, D., Tang, X., Ping, L., Colesie, C., Munchberg, U., et al. (2011). Crystallization of alpha- and beta-carotene in the foregut of Spodoptera larvae feeding on a toxic food plant. Insect Biochem. Mol. Biol. 41, 273–281. doi: 10.1016/j.ibmb.2011.01.004
Shi, Z., Wang, L., and Zhang, H. (2012). Low diversity bacterial community and the trapping activity of metabolites from cultivable bacteria species in the female reproductive system of the oriental fruit fly, Bactrocera dorsalis Hendel (Diptera: Tephritidae). Int. J. Mol. Sci. 13, 6266–6278. doi: 10.3390/ijms13056266
Stegen, J. C., Lin, X., Fredrickson, J. K., Chen, X., Kennedy, D. W., Murray, C. J., et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME 7, 2069–2079. doi: 10.1038/ismej.2013.93
Stegen, J. C., Lin, X., Fredrickson, J. K., and Konopka, A. E. (2015). Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 6:370. doi: 10.3389/fmicb.2015.00370
Vilanova, C., Baixeras, J., Latorre, A., and Porcar, M. (2016). The generalist inside the specialist: gut bacterial communities of two insect species feeding on toxic plants are dominated by Enterococcus sp. Front. Microbiol. 7:1005. doi: 10.3389/fmicb.2016.01005
Voirol, L. R. P., Frago, E., Kaltenpoth, M., Hilker, M., and Fatouros, N. E. (2018). Bacterial symbionts in Lepidoptera: their diversity, transmission, and impact on the host. Front. Microbiol. 9:556. doi: 10.3389/fmicb.2018.00556
Wang, H., Xian, X., Gu, Y., Castañé, C., Arnó, J., Wu, S., et al. (2022). Similar bacterial communities among different populations of a newly emerging invasive species, Tuta absoluta (Meyrick). Insects 13:252. doi: 10.3390/insects13030252
Wang, F. Y., Yang, F., Lu, M. H., Luo, S. Y., Zhai, B. P., Lim, K. S., et al. (2017). Determining the migration duration of rice leaf folder (Cnaphalocrocis medinalis (Guenée)) moths using a trajectory analytical approach. Sci. Rep. 7:39853. doi: 10.1038/srep39853
Wang, Q. Y., Yuan, E. L., Ling, X. Y., Zhu-Salzman, K., Guo, H. J., Ge, F., et al. (2020). An aphid facultative symbiont suppresses plant defence by manipulating aphid gene expression in salivary glands. Plant Cell Environ. 43, 2311–2322. doi: 10.1111/pce.13836
Wei, G., Lai, Y., Wang, G., Chen, H., Li, F., and Wang, S. (2017). Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc. Natl. Acad. Sci. U. S. A. 114, 5994–5999. doi: 10.1073/pnas.1703546114
Wu, Y., Zheng, Y., Chen, Y., Wang, S., Chen, Y., Hu, F., et al. (2020). Honey bee (Apis mellifera) gut microbiota promotes host endogenous detoxification capability via regulation of P 450 gene expression in the digestive tract. Microb. Biotechnol. 13, 1201–1212. doi: 10.1111/1751-7915.13579
Yan, R. (2021). Study on the monitoring methods of population migration in Cnaphalocrocis medinalis (Lepidoptera: Pyrididae). master's thesis. Beijing: Chinese Academy of Agricultural Sciences.
Yang, H. (2012). Diversity of gut bacteria in larval of four lepidopteran insect species. master's thesis. Nanjing: Nanjing Agricultural University.
Yang, Y., Liu, X., Xu, H., Liu, Y., Ali, P., Bodlah, M. A., et al. (2020). The abundance and diversity of gut bacteria of rice leaffolder Cnaphalocrocis medinalis (Guenée) across life stages. J. Asia Pacific Entomol. 23, 430–438. doi: 10.1016/j.aspen.2020.03.006
Yang, Y. J., Liu, X. G., Xu, H. X., Liu, Y. H., and Lu, Z. X. (2022). Effects of host plant and insect generation on shaping of the gut microbiota in the rice leaffolder, Cnaphalocrocis medinalis. Front. Microbiol. 13:824224. doi: 10.3389/fmicb.2022.824224
Yang, Y. J., Xu, H. X., Zheng, X. S., Tian, J. C., Lu, Y. H., and Lu, Z. X. (2015). Progresses in management technology of rice leaffolders in China. J. Plant Prot. 42, 691–701. doi: 10.13802/j.cnki.zwbhxb.2015.05.002
Yu, Y., Wang, Y., Li, H., Yu, X., Shi, W., and Zhai, J. (2021). Comparison of microbial communities in Colorado potato beetles (Leptinotarsa decemlineata say) collected from different sources in China. Front. Microbiol. 12:639913. doi: 10.3389/fmicb.2021.639913
Yun, J. H., Roh, S. W., Whon, T. W., Jung, M. J., Kim, M. S., Park, D. S., et al. (2014). Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80, 5254–5264. doi: 10.1128/AEM.01226-14
Keywords: rice leaffolder, gut microbiota, function, geography, selection, ecological processes
Citation: Yang Y, Liu X, Guo J, Xu H, Liu Y and Lu Z (2022) Gut bacterial communities and their assembly processing in Cnaphalocrocis medinalis from different geographic sources. Front. Microbiol. 13:1035644. doi: 10.3389/fmicb.2022.1035644
Edited by:
Senthil-Nathan Sengottayan, Manonmaniam Sundaranar University, IndiaReviewed by:
Habib Ali, Khwaja Fareed University of Engineering and Information Technology (KFUEIT), PakistanAnandham Rangasamy, Tamil Nadu Agricultural University, India
Copyright © 2022 Yang, Liu, Guo, Xu, Liu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinghong Liu, eWhsaXVAc3d1LmVkdS5jbg==; Zhongxian Lu, bHV6eG1oQDE2My5jb20=
 Yajun Yang
Yajun Yang Xiaogai Liu
Xiaogai Liu Jiawen Guo
Jiawen Guo Hongxing Xu1
Hongxing Xu1 Zhongxian Lu
Zhongxian Lu