
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Microbiol. , 22 September 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1034984
This article is a correction to:
Wildlife Is a Potential Source of Human Infections of Enterocytozoon bieneusi and Giardia duodenalis in Southeastern China
 Yan Zhang1
Yan Zhang1 Rongsheng Mi1
Rongsheng Mi1 Lijuan Yang1
Lijuan Yang1 Haiyan Gong1
Haiyan Gong1 Chunzhong Xu2
Chunzhong Xu2 Yongqi Feng2
Yongqi Feng2 Xinsheng Chen2
Xinsheng Chen2 Yan Huang1
Yan Huang1 Xiangan Han1
Xiangan Han1 Zhaoguo Chen1*
Zhaoguo Chen1*A corrigendum on
Wildlife is a potential source of human infections of Enterocytozoon bieneusi and Giardia duodenalis in southeastern China
by Zhang, Y., Mi, R., Yang, L., Gong, H., Xu, C., Feng, Y., Chen, X., Huang, Y., Han, X., and Chen, Z. (2021). Front. Microbiol. 12:692837. doi: 10.3389/fmicb.2021.692837
In the published article, there was an error in Figure 2 as published. The legend of Figure 2 is correct, but Figure 2 is exactly the same as Figure 3, which is wrong. The corrected Figure 2 and its caption appear below.
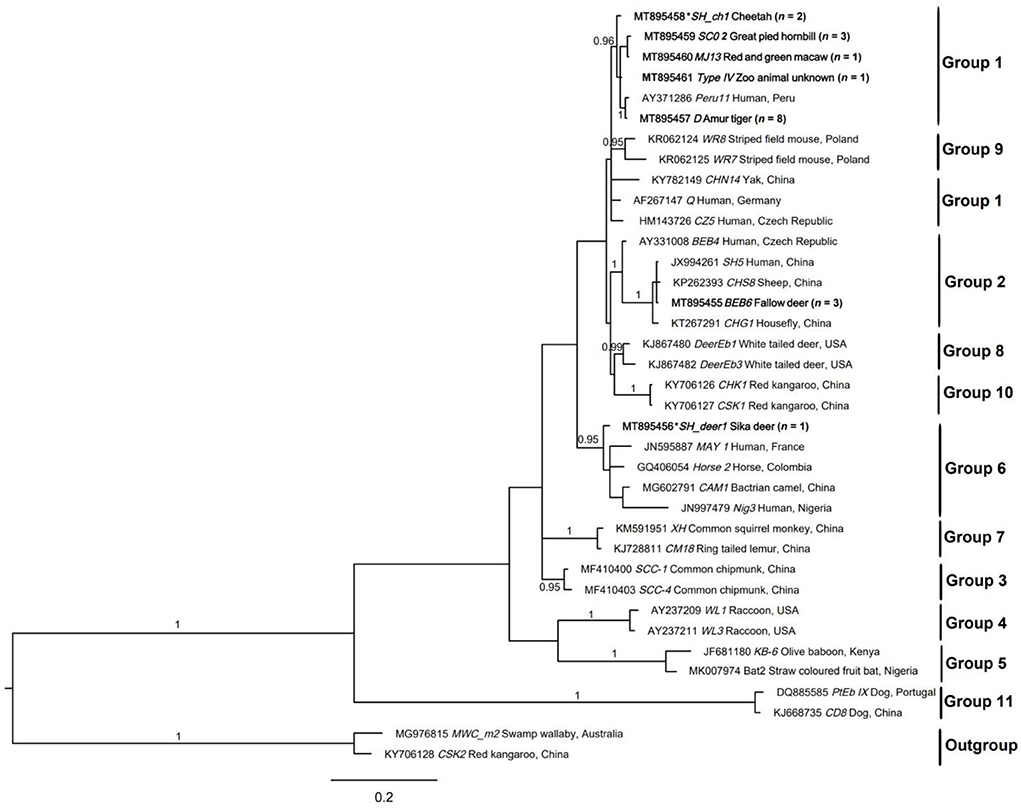
Figure 2. Relationships among the genotypes of Enterocytozoon bieneusi recorded in the wildlife in this study inferred from the phylogenetic analysis of sequence data for the internal transcribed spacer (ITS) of nuclear ribosomal DNA by Bayesian inference (BI). Statistically significant posterior probabilities (pps) are indicated on branches. Individual GenBank accession numbers precede genotype designation (in italics) followed by sample and locality descriptions. The Enterocytozoon bieneusi genotypes identified and characterized from fecal DNA samples in the present study are indicated in bold type. Clades were assigned group names based on the classification system established by Karim et al. (2015) and Li et al. (2019a). The scale bar represents the number of substitutions per site. The E. bieneusi genotypes PtEbIX (DQ885585) and CD8 (KJ668735) from dogs were used as outgroups. All the groups were strongly supported (pp = 0.96–1). pp < 0.95 were not shown.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Karim, M. R., Dong, H., Li, T., Yu, F., Li, D., Zhang, L., et al. (2015). Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: high genetic diversity and zoonotic significance. PLoS ONE 10:e0117991. doi: 10.1371/journal.pone.0117991
Keywords: Cryptosporidium, Enterocytozoon bieneusi, Giardia duodenalis, genotypes, wildlife, prevalence, zoonotic potential
Citation: Zhang Y, Mi R, Yang L, Gong H, Xu C, Feng Y, Chen X, Huang Y, Han X and Chen Z (2022) Corrigendum: Wildlife is a potential source of human infections of Enterocytozoon bieneusi and Giardia duodenalis in southeastern China. Front. Microbiol. 13:1034984. doi: 10.3389/fmicb.2022.1034984
Received: 02 September 2022; Accepted: 05 September 2022;
Published: 22 September 2022.
Copyright © 2022 Zhang, Mi, Yang, Gong, Xu, Feng, Chen, Huang, Han and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoguo Chen, emhhb2d1b2NoZW5Ac2h2cmkuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.