- 1Department of Microbiology, Yongchuan Hospital of Chongqing Medical University, Chongqing, China
- 2Department of Pathogenic Biology, Jiamusi University School of Basic Medicine, Jiamusi, China
- 3Department of Microbiology, The First Affiliated Hospital of Jiamusi University, Jiamusi, China
- 4Department of Microbiology, Shenzhen University General Hospital, Shenzhen, China
- 5Department of Microbiology, Affiliated Hangzhou Xixi Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 6Department of Life Science and Technology, Mudanjiang Normal University, Mudanjiang, China
Carbapenem-resistant Enterobacter cloacae complex (CRECC) has increasingly emerged as a major cause of healthcare-associated infections, with colistin being one of the last-resort antibiotics of treatment. Mobile colistin resistance (mcr)-9 is a member of a growing family of mcr genes and has been reported to be an inducible gene encoding an acquired phosphoethanolamine transferase. Here, we collected 24 ECC strains from Chongqing, China from 2018 to 2021. Subsequently, antibiotic resistance genes and the transmission dynamics of the strains were determined by PCR, whole-genome sequencing, and bioinformatic analysis. The mcr-9 was identified in IncHI2/2A or IncHI2/2A + IncN plasmids from six CRECC strains and was co-located with blaNDM-1 or blaIMP-4 in 2/6 plasmids. The genetic environment of mcr-9.1 was composed of IS903B-mcr-9.1-wbuC-IS26 in the five mcr-9.1-harboring-plasmid, but IS1B was located downstream of mcr-9.2 in the pECL414-1 sequence. We also found that the pNDM-068001 plasmid carrying mcr-9.1 could be a hybrid plasmid, formed by a Tn6360-like blaNDM-1 region inserted into an mcr-9.1-positive IncHI2/2A plasmid. A conjugation assay showed that plasmids mediated the co-dissemination of mcr-9 and metallo-β-lactamase (MBL) genes. In addition, we performed induction assays with sub-inhibitory concentrations of colistin and found an increase in the relative expression levels of the mcr-9.2, qseC, and qseB genes, as well as an increase in the minimum inhibitory concentration values of colistin in the CRECC414 strain. These findings provide a basis for studying the regulatory mechanisms of mcr-9 expression and highlight the importance of effective monitoring to assess the prevalence of MBL and mcr-9 co-existing plasmids.
Introduction
The dramatic increase in the prevalence and clinical impact of infections caused by Enterobacterales producing carbapenemases, such as Klebsiella pneumoniae carbapenemase-2 (KPC-2) and New Delhi metallo-β-lactamase-1 (NDM-1), has triggered a global health crisis (Kumarasamy et al., 2010; Munoz-Price et al., 2013). Colistin belongs to the family of polymyxins, cationic polypeptides. Owing to its nephrotoxicity, the clinical use of colistin was discontinued in the 1980s; however, it was reintroduced in the 1990s because of the lack of effective antimicrobial agents for the treatment of multidrug-resistant Gram-negative pathogens (Falagas and Kasiakou, 2005). Previously reported mechanisms of colistin resistance are chromosomally mediated and involve the regulation of two regulatory systems (pmrAB, phoPQ, and its negative regulator mgrB in K. pneumoniae), resulting in the modification of lipid A by phosphoethanolamine or 4-amino-4-arabinose (Gunn, 2008; Cannatelli et al., 2013). However, a plasmid-mediated mobilized colistin resistance (mcr) gene mcr-1 was first reported in China in late 2015 (Liu et al., 2016), which largely threatens the use of colistin in clinical settings, and nine mcr homologs (mcr-2 to -10) have since been detected (Nang et al., 2019). Although the transmission of most mcr genes to carbapenem-resistant Enterobacterales (CRE) has been limited (Carroll et al., 2019; Kieffer et al., 2019; Nang et al., 2019), mcr-9, first identified in Salmonella enterica clinically isolated in the United States in 2019 (Carroll et al., 2019), has been identified in several CRE backgrounds (Chavda et al., 2019; Lalaoui et al., 2019; Liu et al., 2021). In addition, the first identified strain that carried mcr-9 was sensitive to colistin, but mcr-9 confers resistance to colistin after cloning and overexpression in the laboratory (Carroll et al., 2019). Further studies have revealed that mcr-9 expression is inducible in the presence of colistin when mcr-9 is located upstream of the qseBC two-component system (Kieffer et al., 2019). However, other studies have suggested that mcr-9 and colistin resistance may not be related (Tyson et al., 2020). These results suggest that there is uncertainty as to when mcr-9 confers elevated minimum inhibitory concentration (MIC) values for colistin, which may be related to the genetic background or differences in host and other unidentified regulatory genes (Chavda et al., 2019; Simoni et al., 2021).
In the present study, we investigated the antibiotic resistance genes (ARGs) and transmission dynamics of 24 ECC isolates collected from 2018 to 2021 using PCR, whole-genome sequencing (WGS), and bioinformatic analysis. We analyzed WGS data and compared them with publicly available data to determine the genome structure of strains carrying the mcr-9 gene. We determined the phenotypic impact of carbapenem-resistant Enterobacter cloacae complex (CRECC) strains carrying mcr-9 under the sub-inhibitory concentrations of colistin induction. Moreover, plasmid-mediated co-transmission of the mcr-9 and MBL genes was determined using a conjugation assay.
Materials and methods
Bacterial isolates, susceptibility testing, and clinical data collection
Enterobacter cloacae complex (ECC) strains that were resistant to meropenem, imipenem, or ertapenem or produced carbapenemase isolated from 2018 to 2021 at Yongchuan Affiliated Hospital of Chongqing Medical University, China, were collected, in addition to one carbapenem-sensitive Enterobacter cloacae complex (CSECC) strain carrying mcr-9, identified in the screening, which was also included in this study. All 24 ECC strains were identified using a VITEK-2 Compact automatic microbiology analyzer and MALDI-TOF MS (Bruker, Billerica, MA, United States). Antimicrobial susceptibility was evaluated for all isolates following the Clinical and Laboratory Standards Institute (CLSI) guidelines (M100-S30; CLSI, 2020), and the results were interpreted according to these guidelines (Clinical and Laboratory Standards Institute (CLSI), 2020), except that colistin resistance was defined according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST; version 10.0) clinical breakpoints (The European Committee on Antimicrobial Susceptibility Testing, 2020). Metadata including patients’ departments, dates of specimen collection, and specimen types were recorded.
PCR and sequencing
Total DNA was extracted from each strain of the 24 ECC strains using the boiling method and used as a template in PCR experiments (Gong et al., 2018). Resistance genes were detected by PCR, including colistin-resistant genes (mcr-1 and mcr-9), carbapenemase genes (blaNDM, blaKPC, blaOXA-48, blaVIM-1, blaVIM-2, blaIMP-4, and blaIMP-8), AmpC β-lactamase enzyme genes (blaACC and blaDHA), ESBL genes (blaTEM, blaSHV, blaCTX-M-1, and blaCTX-M-9), and quinolone resistance genes (qnrA, qnrB, qnrS, qepA, and aac(6′)Ib-cr). All primers have been used in previous studies (Park et al., 2006; Ma et al., 2009; Gong et al., 2018). Positive amplification products were subjected to Sanger sequencing, and the obtained sequences were subsequently submitted to Basic Local Alignment Search Tool (BLAST).
Whole-genome sequencing and bioinformatic analysis
WGS was performed using an Illumina HiSeq PE150 (Illumina, San Diego, CA, United States) on the 24 ECC isolates, with six CRECC isolates carrying mcr-9 proceeding to further long-read sequencing (Oxford Nanopore, Oxford, United Kingdom). Long-read sequencing assembly was performed using the unicycler v0.4.8 (Wick et al., 2017), and the correction was performed using the pilon v1.22 (Walker et al., 2014). In silico multi-locus sequence typing (MLST) was performed using PubMLST1 (Jolley et al., 2018). The plasmid replicon type and plasmid MLST (pMLST) were identified using PlasmidFinder 2.1,2 and pMLST 2.03 (Carattoli et al., 2014). ResFinder 4.14 was employed for the identification of ARGs (Bortolaia et al., 2020). Complete plasmid sequence alignments were performed using BLAST and visualized using the BRIG tool. Sequence alignments among mcr-9-carrying or qseC-qseB-carrying plasmids or chromosomes were performed using BLAST and visualized using Easyfig v 2.2.5.
A total of 2,337 complete genomes of ECC strains were retrieved from the NCBI database (accessed on 31 December 2021) and were subjected to comparative alignments of mcr-9 positive plasmid genomes isolated from patients using BLAST and visualized using Easyfig v 2.2.5.
Core genome MLST and cg-SNP
All strains contained 3,048 core genomes according to the reference genome (GenBank accession no. CP010377). The results of core genome MLST (cg-MLST) were visualized using PHYLOViZ, based on the goeBURST algorithm (Ribeiro-Goncalves et al., 2016). The results of core genome single nucleotide polymorphism (cg-SNP) were visualized using ggtree after building the evolutionary tree with FastMe (Lefort et al., 2015).
Conjugation experiment
We selected six isolates of CRECC carrying the mcr-9 and carbapenemase genes and used them as donors in a conjugation experiment, with rifampicin-resistant Escherichia coli EC600 as the recipient. Conjugation experiments were performed using the membrane bonding method according to previously described method (Gong et al., 2018). Briefly, donor (CRECC strain) and recipient cultures (E. coli EC600) were mixed at a 1:3 ratio in Luria-Bertani (LB) broth. The mixtures were placed on a membrane and subsequently incubated at 35°C for 24 h. Mueller–Hinton agar plates were supplemented with meropenem (1 mg/l) and rifampicin (600 mg/l) to select transconjugants. The VITEK-2 Compact system and 16S rRNA sequence were performed to confirm the transconjugants and PCR was determined to the presence of resistance genes.
Colistin induction assays
Colistin induction assays were performed as previously described with some modifications (Kieffer et al., 2019; Liu et al., 2021). Briefly, the CRECC414 strain was inoculated in 4 ml LB broth with shaking at 37°C for 2 h and then supplemented with colistin (1 and 2 mg/l) or without at a final bacterial suspension of 1.0 McFarland turbidity standard. After shaking at 37°C for 6 h, bacterial suspensions were used for mRNA extraction.
mRNA extraction and RT-qPCR
Total RNA was extracted using the PureLinkTIM RNA Mini Kit (Thermo Fisher Scientific), as specified by the manufacturer. Each RNA sample was reverse-transcribed using the PrimeScript™ RT reagent kit (TaKaRa, Japan) according to the manufacturer’s instructions. The relative expression level of mcr-9 was determined by quantitative real-time PCR (RT-qPCR) using SYBR Green detection reagents on the CFX96 RealTime PCR system. The primer sequences used are listed in Supplementary Table 1. Relative expression levels were calculated using the 2-ΔΔCT method, with rpoB as a reference gene for comparison with those of samples that were not induced. Three independent replicates were performed with and without induction.
Nucleotide sequence accession number
WGS data were submitted to NCBI with accession numbers in Supplementary Table 2.
Results
Isolate characteristics
We collected 24 non-repetitive clinical ECC isolates, 23 of which were CRECC strains and 1 CSECC strain (Supplementary Table 2). In total, 24 ECC isolates were identified to five species: Enterobacter hormaechei (n = 19), Enterobacter kobei (n = 2), Enterobacter cloacae (n = 1), Enterobacter asburiae (n = 1), and Enterobacter mori (n = 1; Figure 1). Regarding the source of the strains, sputum accounted for the highest proportion (54.2%; 13/24), followed by urine (12.5%; 3/24), blood (12.5%; 3/24), and bronchoalveolar lavage fluid (12.5%; 3/24). Notably, at least nine isolates (37.5%; 9/24) were recovered from sterile site specimens of patients (Supplementary Table 2). All 23 CRECC strains carried the MBL genes (blaNDM-1, blaNDM-5, or blaIMP-4, except for the CRECC410 strain carrying both blaNDM-5 and blaIMP-4), the ESBL gene (blaTEM-1), and the quinolone resistance gene (aac(6’)Ib-cr; Figure 1). Among them, six CRECC strains (CRECC68, CRECC78, CRECC404, CRECC405, CRECC411, and CRECC414) carried mcr-9 on IncHI2-ST1 plasmids and carried qseC-qseB-like two-component regulatory system on the chromosomes. The six CRECC strains belonged to three species: E. hormaechei (66.6%; 4/6), E. kobei (16.7%, 1/6), and E. cloacae (16.7%, 1/6; Figure 1). According to the MLST analysis, 24 ECC strains showed that the main ST types were ST177 (CRECC72, CRECC73, CRECC75, CRECC76, CRECC67, CRECC66, and CRECC79) and ST171 (CRECC406, CRECC408, CRECC409, and CRECC112; Figure 1). Based on the cg-MLST results, there were 1–115 allelic differences between the ST177 strains, as well as an average nucleotide identity (ANI) of more than 99.9% (Figure 2 and Supplementary Figure 1), which further illustrates the similarity between strains. In addition, the ANI among ST171 strains was more than 99.9%, but the ANI results between CRECC112 and other ST171 strains ranged from 99.82 to 99.88% (Supplementary Figure 1). Phylogenetic analysis of cg-SNP also showed similarities between CRECC406, CRECC408, and CRECC409 (Supplementary Figure 2). Therefore, this may represent the clonal dissemination of ST177 strains between the PICU, neonatal ward, respiratory medicine department, and pediatric department of our institution from October 21, 2018, to June 6, 2019 (Figures 1, 2; Supplementary Table 2).
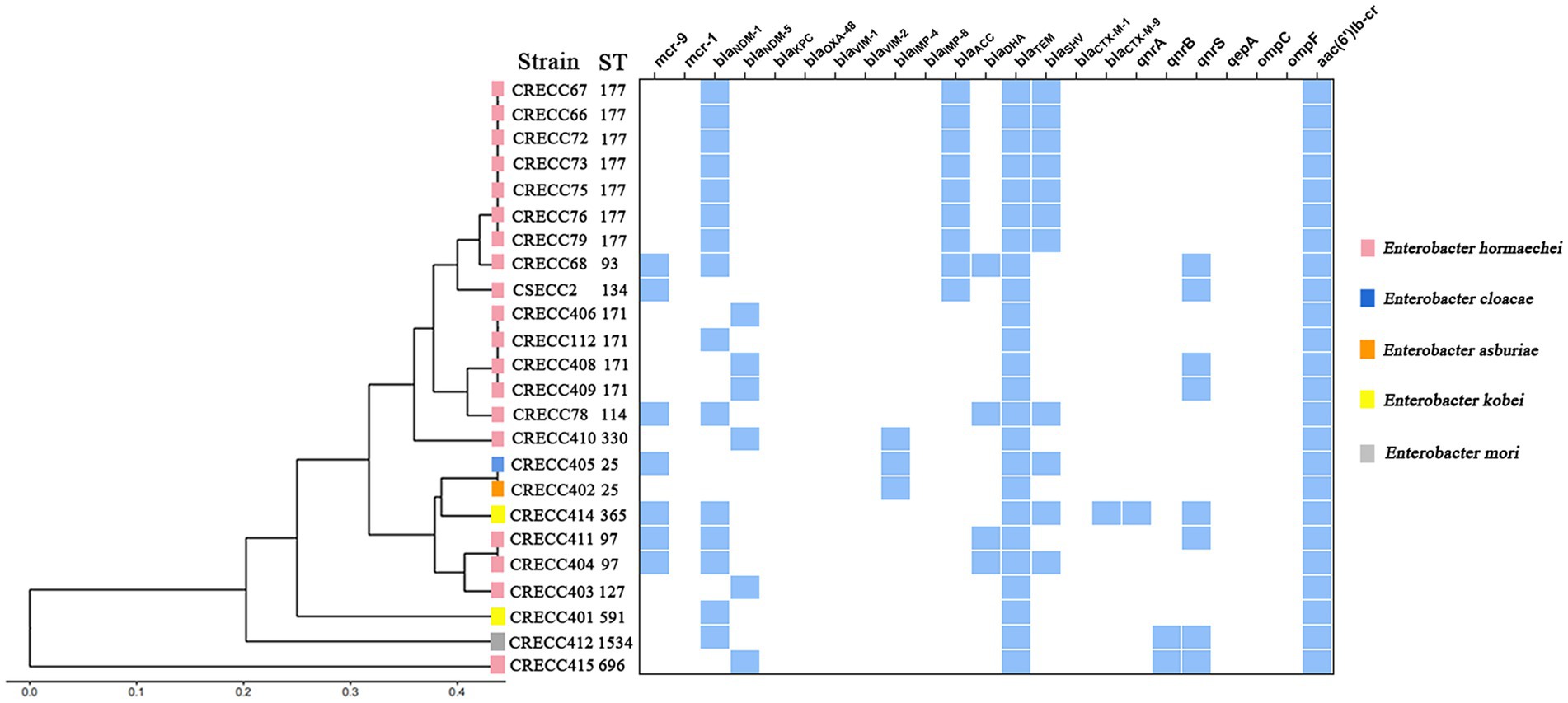
Figure 1. Clustering analysis and resistance determinants of 24 ECC strains from our institution. The resulting structure reflected the similarity between the sequences, and resistance determinants present (in blue) in each strain are shown on the right.
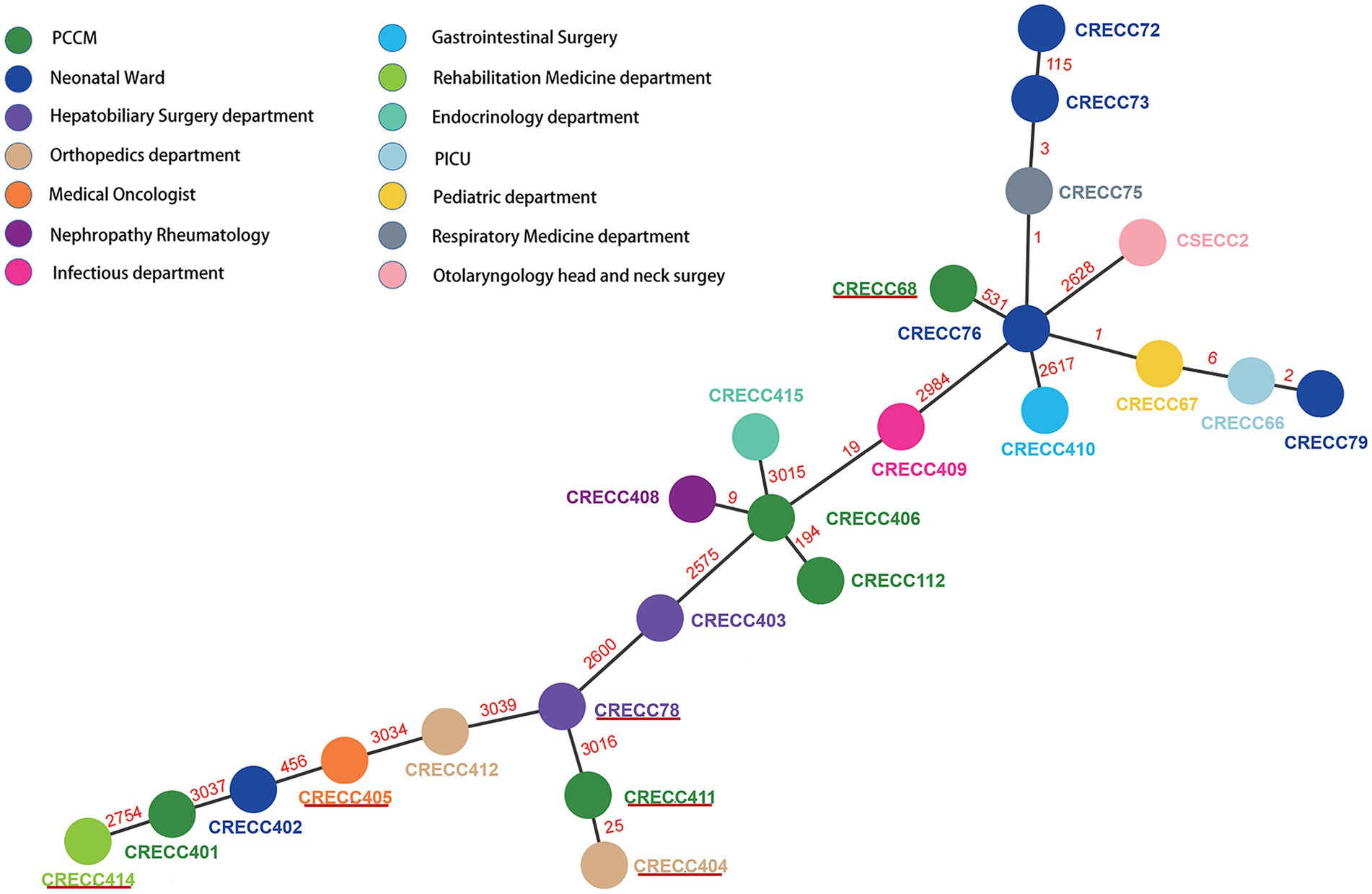
Figure 2. Minimum spanning tree for multilocus sequence typing of the core genome based on 3,084 targets. Allelic distances between isolates are represented numerically. The resulting structure reflected the similarity between strains isolated by different departments. Solid red lines represent CRECC strains carrying mcr-9.
Transferability of mcr-9-carrying plasmids
The mcr-9 genes of CRECC68 and CRECC405 strains co-occurred on the same plasmids as blaNDM-1 and blaIMP-4, respectively. However, mcr-9 and blaNDM-1 of other CRECC strains (CRECC78, CRECC404, CRECC411, and CRECC414) were located on plasmids of different sizes (Supplementary Table 3). Meropenem was used for screening and six E. coli EC600 transconjugants were identified using VITEK-2, 16S rRNA, and PCR. The six transconjugants were found to contain the MBL genes (blaNDM-1 or blaIMP-4) and the colistin-resistant genes (mcr-9; Supplementary Table 3), and were resistant to meropenem and rifampicin. This suggests that using meropenem may promote the co-transmission of mcr-9 and MBL genes, which could lead to a more serious public health crisis.
Genetic environment of mcr-9-carrying isolates
We identified 26 mcr-9-containing complete plasmid genomes isolated from patients in the NCBI database (Supplementary Figure 3). We found that most (24/26) of the mcr-9 genetic environments showed 100% homology to the backbone composed of rcnR-rcnA-pcoE-pcoS-IS903B-mcr-9. Among them, more than half (19/24) were found to have wbuC-IS26 downstream of mcr-9, and two (2/24) plasmid sequences were found to have qseC-qseB-IS1 downstream of mcr-9. Another two (2/24) plasmid sequences were found where pcoS was interrupted by ISVsa5. It is suggested that mcr-9 may be transferred as a gene cassette (rcnR-rcnA-pcoE-pcoS-IS903B-mcr-9-wbuC-IS26) among the plasmids. In the present study, mcr-9 was located in an almost conserved region in the five mcr-9-harboring plasmids (pNDM-068001, pMCR-078001, pECL404-1, pECL405-1, and pECL411-1). wbuC was downstream of mcr-9.1 and mcr-9.1-wbuC was surrounded upstream by an IS903B and downstream by an IS26; however, comparative alignments of the genetic environments found that IS1B was located downstream of mcr-9.2 in the pECL414-1 sequence (Figure 3).
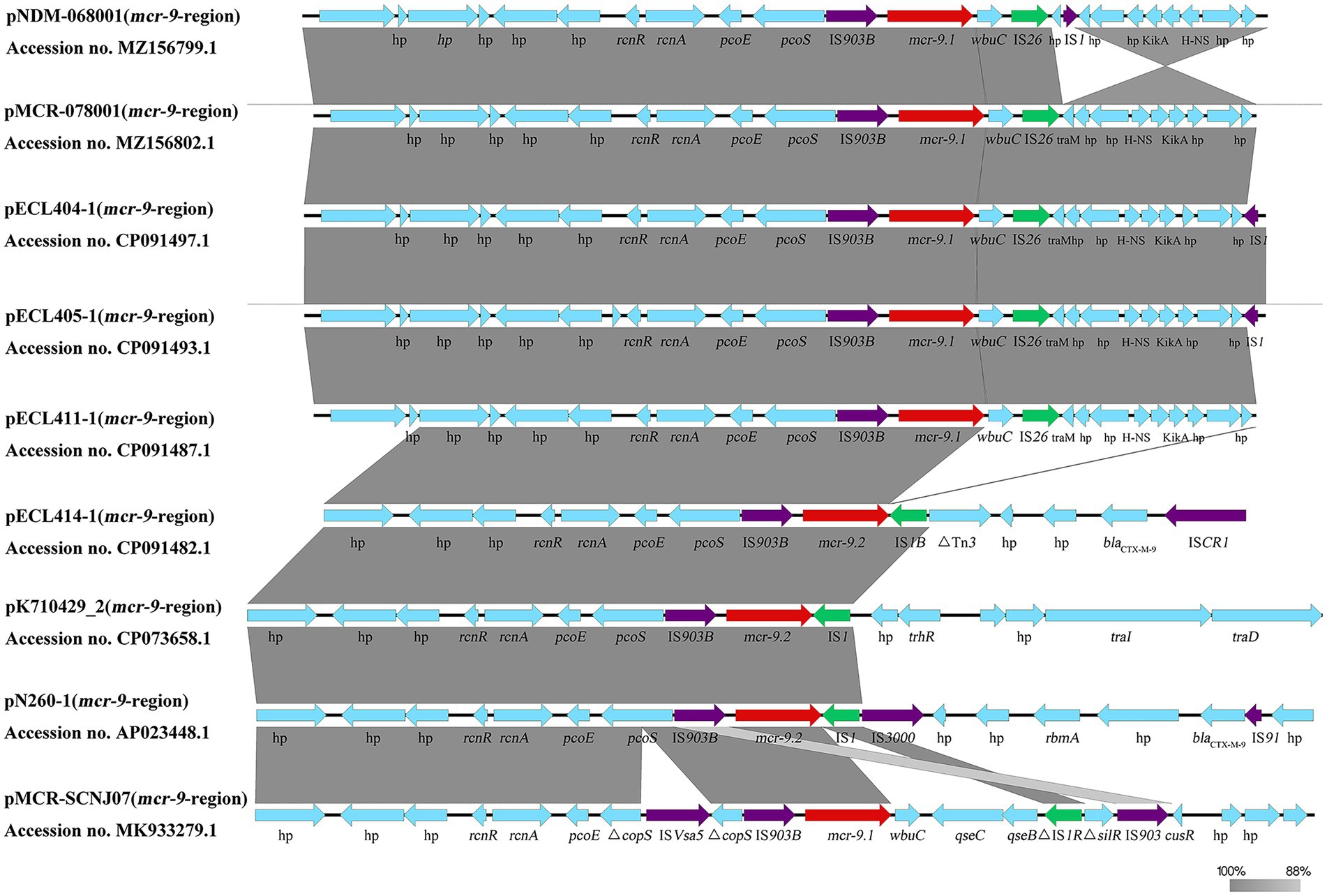
Figure 3. Different genetic structure surrounding the mcr-9 gene. Comparison of mcr-9 regions detected in pNDM-068001, pMCR-078001, pECL404-1, pECL405-1, pECL411-1, pECL414-1, pN260-1, pKPN7104292-2, and pMCR-SCNJ07. Gray shading denotes regions of shared homology. Arrows indicate the direction of gene transcription.
Analysis of mcr-9-harboring plasmids from our institution
In our setting, mcr-9 was found to be located on one IncHI2/2A + IncN plasmid and five different IncHI2/IncHI2A plasmids, named pNDM-068001, pMCR-078001, pECL404-1, pECL405-1, pECL411-1, and pECL414-1, with sizes of 444,489, 342,942, 319,000, 362,923, 316,592 and 294,412 bp, respectively. pECL414-1 shared 99.97% identity and 84% coverage with plasmid pMCR-SCNJ07 carried by E. hormaechei in Neijiang, Sichuan, which was identified in 2019, and pECL414-1 shared 100% identity and 97% coverage with plasmid pK710429 carried by K. pneumoniae in Anhui, Sichuan, which was identified in 2021. Comparison of the six mcr-9-carrying plasmids revealed that with a coverage range of 81–100%, they shared at least 99% identity, whereas the genome sequence of pNDM-068001 to pECL414-1 showed a lower coverage of 71% and nucleotide identity of 99.52%. This showed that they shared a similar backbone that mostly included regions essential for plasmid replication, maintenance, and conjugative transference (Figure 4). Except for mcr-9, multiple ARGs located on mcr-9-harboring plasmids were mainly responsible for the multidrug resistance phenotype of the isolates (Supplementary Tables 2, 4). These included blaNDM-1, blaIMP-4, blaDHA-1, blaSHV-12, blaCTX-M-9, and blaTEM-1 for β-lactam resistance; strAB, aph(3′)-Ia, aac(6′)-llc, and aac(6′)-lb for aminoglycoside resistance; dfrA19, dfrA16, and dfrA14 for trimethoprim resistance; sul1 for sulfonamide resistance; tet(D) and tet(A) for tetracycline resistance; qnrS1, qnrA1, and qnrB4 for quinolone resistance; and ere(A) and mph(A) for macrolide resistance (Supplementary Table 4). This prompts that mcr-9 was frequently present on mobile genetic elements with other ARGs, implying that the use of these types of antibiotics has the potential to co-select for the continued presence of mcr-9.
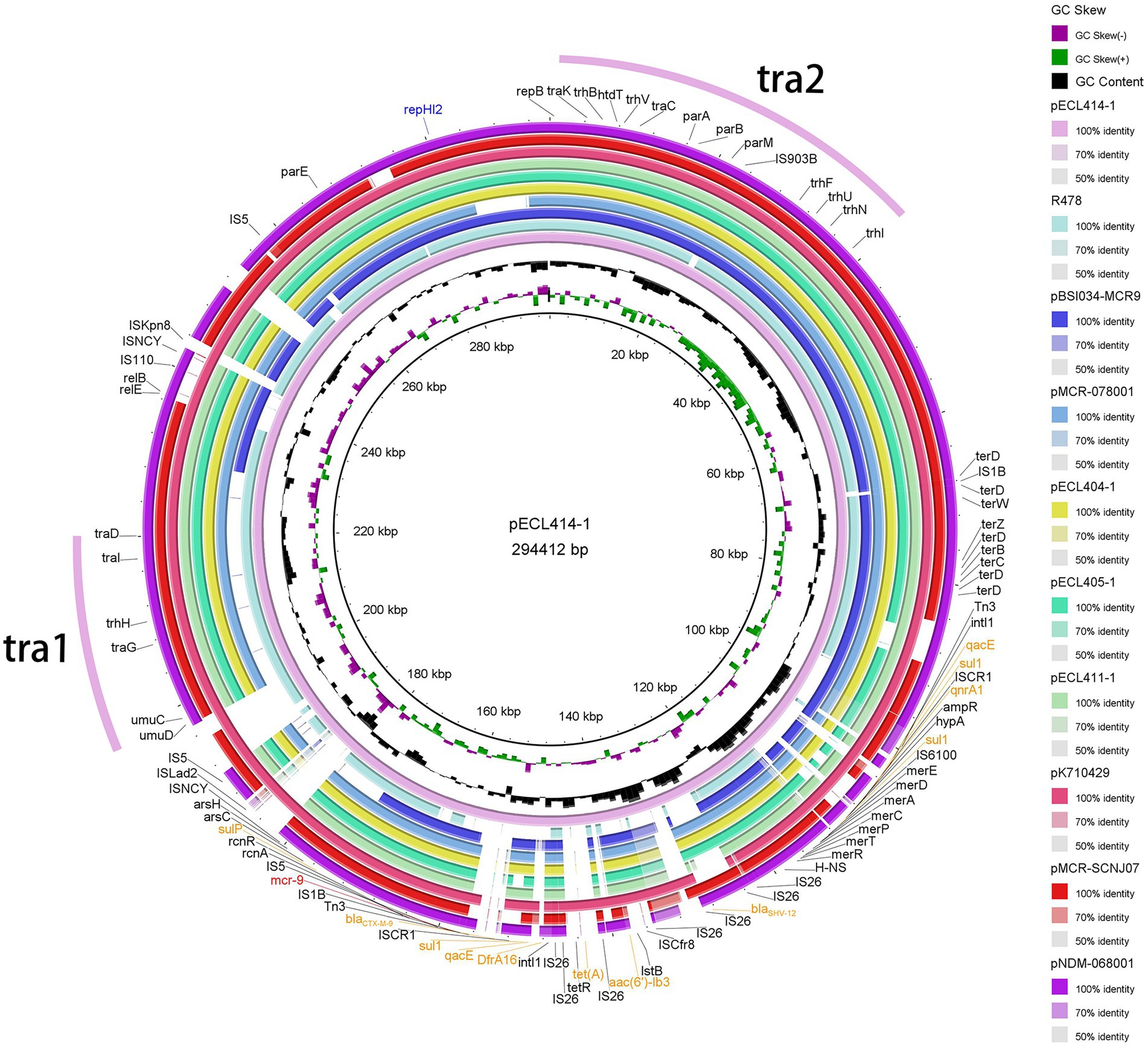
Figure 4. Plasmid structure of mcr-9. Sequence alignment of mcr-9-harboring plasmids pNDM-068001, pMCR-078001, pECL404-1, pECL405-1, pECL411-1, pECL414-1, pR478, pK710429, pMCR-SCNJ07, and pBSI034-MCR9. The complete plasmid pECL414-1 sequence was used as the reference, and the white and colored regions of the circles indicate absence and presence, respectively. Antibiotic resistance genes are shown in orange (mcr-9 is shown in red), and the tra1 and tra2 regions for conjugative transfer are indicated by pink curves on the outer circle.
Colistin induction assays and colistin susceptibility testing
The strains were exposed to sub-inhibitory concentrations of colistin, and the CRECC414 strain was used as the template for mRNA extraction. Notably, a 2.6-fold increase in the plasmid-located mcr-9 mRNA was detected once the culture was induced with 1 mg/l colistin; however, there was no statistically significant difference in the relative expression levels of the qseB and qseC genes located on chromosomes (Figure 5). After induction with 2 mg/l colistin, the relative expression levels of mcr-9, qseC, and qseB mRNA increased by 8.5-, 2.6-, and 2.7-fold, respectively (Figure 5). In parallel, the MIC value of colistin for the CRECC414 strain was 4 mg/l, and the MIC values were 64 and 128 mg/l after induction with 1 or 2 mg/l colistin, respectively.
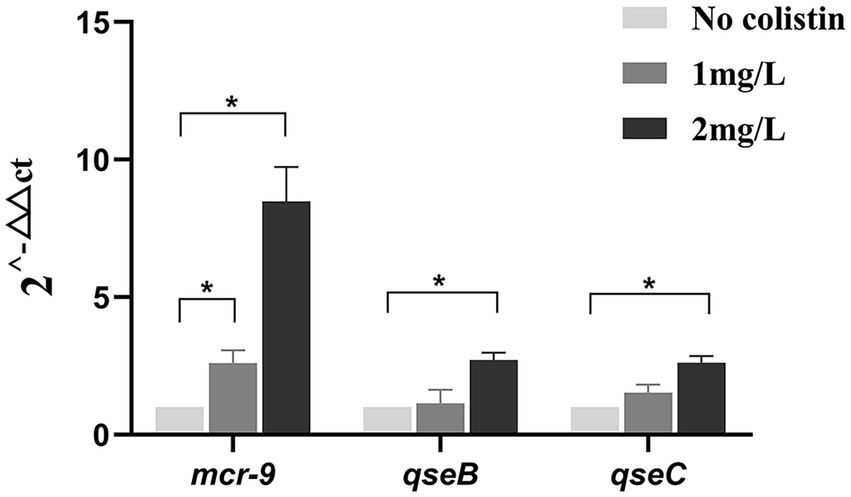
Figure 5. Expression of genes in clinical isolate CRECC414 with and without colistin exposure. The genes are the plasmid-located mcr-9 and the chromosome-located qseB and qseC. Fold changes in mRNA levels were determined by RT-qPCR. The data have been normalized to values for reference rpoB gene. Three independent replicates were performed and the data shown represent the means. The asterisks indicate statistical significance at different levels by Student’s t-tests: *p ≤ 0.05.
Discussion
The IncHI2-ST1 plasmid is the main replicon type carrying mcr-9, which is a member of a growing family of mobile colistin resistance genes. The mcr-9 is predominantly present in MBL-producing strains at our institution; therefore, novel beta-lactam/beta-lactamase inhibitor combinations have no activity against them. The use of colistin in CRE infections remains a priority. However, the present study found an increase in the MIC value of strains mcr-9-carrying for colistin under antibiotic selection pressure, which may raise concerns regarding the use of colistin.
We performed phylogenetic analysis of plasmids carrying mcr-9 from published studies and our institution. The results showed that 26 plasmids had the IncHI2 replicon alone, one had IncHI2 + IncR, one had IncHI2 + IncM1, and one had IncHI2 + IncN (Supplementary Figure 4). This suggests that different replicon-type plasmids (IncHI2, IncHI2 + IncR, and IncHI2 + IncN) may represent important vehicles for mediating the dissemination of mcr-9 in China. In addition, IncHI2 and IncHI2 + IncN were the main replicon-type in our setting, which may be frequently involved in the spread of multiple ARGs (Chowdhury et al., 2019; Borjesson et al., 2020; Supplementary Tables 3, 4). The IncHI2-IncN hybrid plasmid, pNDM-068001 (accession no. MZ156799.1), clustered with several IncHI2 plasmids (accession no. CP080514.1, MN937241.1, CP059887.1, and AP024500.1), implying that they may have a common ancestor and a subsequent genetic recombination event, leading to the formation of hybrid plasmids. The results of the conjugation assay showed that mcr-9-encoding plasmids can be horizontally transmitted between different species of Enterobacterales. Phylogenetic analysis also showed that mcr-9-encoding plasmids have been identified in several types of strains, with ECC isolates being the main isolates that disseminate mcr-9 (Li et al., 2020; Supplementary Figure 4). Notably, mcr-9-harbouring plasmids recovered from human isolates were interspersed with plasmids from animals and food (Supplementary Figure 4), and it is possible that the use of colistin in food animals largely contributes to the prevalence of mcr genes in humans and the environment (Sun et al., 2018). Interestingly, we found that mcr-9 genetic environments were major composed of IS903B-mcr-9-wbuC-IS26 in our setting (Figure 3), and this genetic environment has been reported previously (Wu et al., 2022). In addition, few reports have described mcr-9 integration into the chromosome of E. hormaechei (Wu et al., 2022), Citrobacter (Ribeiro et al., 2021), and Salmonella isolates (Tyson et al., 2020; Pan et al., 2021) with a genetic context similar to that observed in mcr-9-harboring plasmid sequences, suggesting that mcr-9 may be transferred between plasmids and chromosomes in the form of a gene cassette.
According to MLST typing, six CRECC isolates carrying mcr-9 were ST97 (33.3%, 2/6), ST93, ST114, ST25, and ST365 (Figure 1), which are most likely horizontal transmission rather than clonal transmission in our hospital. Interestingly, molecular epidemiology revealed that ST93 is the predominant sequence type of the CRECC strain, followed by ST171 and ST145 in China (Chen et al., 2021); and ST114 is a common global CRECC strain clone (Peirano et al., 2018). The results of the current study showed that the CRECC68 and CRECC78 strains carrying mcr-9 belonged to ST93 or ST114, indicating that mcr-9 may have co-evolved with the host, which carries a high risk of transmission.
Studies have been conducted on the phenotypic effects of strains under antibiotic selection pressure. Kieffer et al. (2019) found that the expression of mcr-9 is inducible by subinhibitory concentrations of colistin and is related to the presence of qseC and qseB genes downstream of mcr-9. However, the MIC values of a few isolates in the presence of the qseB/qseC-like two-component system downstream of mcr-9 remained unchanged after colistin exposure (Kim et al., 2021; Macesic et al., 2021; Simoni et al., 2021). It was also found that the absence of qseC and qseB genes downstream of mcr-9 also does not change the MIC values of the strains after colistin exposure (Chavda et al., 2019; Bitar et al., 2020; Wu et al., 2022). The different results could be related to differences in the genetic background, host, and other undiscovered regulatory genes (Chavda et al., 2019; Simoni et al., 2021). In the present study, six CRECC strains carrying mcr-9 were isolated from six patients from different departments, all of whom had different diseases, but none of them had been treated with colistin (Supplementary Table 5). First, we found that the colistin resistance rate of six CRECC strains carrying mcr-9 was 50% (Supplementary Table 2). Nevertheless, it has been reported that few isolates carrying mcr-9 display colistin resistance at baseline (Carroll et al., 2019; Kieffer et al., 2019; Tyson et al., 2020; Macesic et al., 2021). Alignment revealed no possible mutations in chromosomal resistance genes in our strains. Second, the relative expression levels of mcr-9.2 and MIC values of the CRECC414 strain increased after colistin exposure, which is consistent with the results of K. pneumoniae (Liu et al., 2021). Although qseC and qseB are not located downstream of mcr-9.2 on the plasmid, our experiments showed an increase in the relative expression levels of qseC and qseB after colistin induction. Therefore, we hypothesized that once mcr-9 is integrated into the chromosome, it may be regulated by the qseB/qseC-like two-component of the lysM-mdaB-qseC-qseB-parC-parF-sufl genetic background (Supplementary Figure 5), ultimately leading to a possible increase in the relative expression levels of mcr-9 and the MIC value of colistin in the CRECC strain under antibiotic selection pressure. In addition, comparative alignments of the genetic environments revealed that IS1B was located downstream of mcr-9.2 in both the CRECC414 and K. pneumoniae strains (Figure 3). Similarly, IS1 was found downstream of mcr-9 in both pN260-1 and pMCR-SCNJ07 sequences, and these strains displayed colistin resistance at baseline (Yuan et al., 2019; Umeda et al., 2021; Figure 3). We hypothesized that the induced expression of mcr-9 is related to the presence of downstream IS1. Therefore, further validation of these possible regulatory mechanisms will be the focus of future research.
The strength of the present study was the extensive use of Illumina and Nanopore read sequencing to construct high-quality hybrid plasmid assemblies. Notably, two mcr-9-harboring plasmids also carried blaIMP-4 (pECL405-1) or blaNDM-1 (pNDM-068001), respectively (Supplementary Table 3). We found that blaNDM-1 and mcr-9 were located on the IncHI2/2A + IncN plasmid, and the blaNDM-1 cassette was similar to the recently reported cassette of Tn6360 (carrying blaNDM-1 and located on the IncN1 plasmid; Zhao et al., 2017), suggesting that pNDM-068001 could be a hybrid plasmid, formed by a Tn6360-like blaNDM-1 region inserted into an mcr-9-positive IncHI2/HI2A plasmid. In addition, blaIMP-4 and mcr-9 were located on the IncHI2/2A plasmid, and blaIMP-4 was characterized by the following structure: IS6-dfrA19-IntI1-blaIMP-4-ltrA-SMR-sul1-GNAT-IS6 (IS6100), which may mediate the transfer of blaIMP-4. This indicated that horizontal gene transfer events played a significant role in plasmid evolution. Notably, the results of our study showed a co-transmission characteristic of the mcr-9 and MBL genes, and colistin for CRE infections remains an important priority. We also found that mcr-9 was inducible in the CRECC strains; therefore, this may pose some challenges for the use of colistin in the clinical environment. The current study has a minor limitation; the boiling method extracts a small amount of DNA with many impurities, and DNA breakage may occur, leading to errors. However, PCR and sequencing of the extracted nucleic acids for the primary screening of ARGs in strains still have some value.
In conclusion, the spread of mcr-9 is primarily driven by the IncHI2/2A plasmid. Our results alert physicians to the circulation of plasmid-mediated co-transmission of mcr-9 and MBL genes already in the hospital setting, which could lead to a serious public health crisis; therefore, effective monitoring is urgently needed to assess the prevalence of MBL and mcr-9 co-existing plasmids and to find an effective measure to control their spread. We found that mcr-9 showed increased expression when induced with colistin in the CRECC strain, which may be related to IS downstream of mcr-9 or qseB/qseC-like two-component systems on chromosomes. Further clarification of their regulatory mechanisms for mcr-9 gene expression will be the focus of our future research.
Data availability statement
The data presented in the study are deposited at GenBank, accession number(s) can be found in the article/Supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
XZ, CL, SJ, and XW conceived and designed the study. SJ and XW wrote the manuscript and participated in the whole experiment process. XL and KH helped with the experimental process. SJ, XW, HY, JZ, JW, and JL analyzed and interpreted the data. JW, JL, XGong, XGou, YY, SJ, and XW collected the isolates and the clinical data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by General projects of Chongqing Natural Science Foundation (cstc2020jcyj-msxm0067), Yongchuan Natural Science Foundation (2021yc-jckx20053), Talent introduction project of Yongchuan Hospital of Chongqing Medical University (YJYJ202004 and YJYJ202005), and Program for Youth Innovation in Future Medicine, Chongqing Medical University (W0113).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1032833/full#supplementary-material
Footnotes
References
Bitar, I., Papagiannitsis, C. C., Kraftova, L., Chudejova, K., Mattioni Marchetti, V., and Hrabak, J. (2020). Detection of five mcr-9-carrying Enterobacterales isolates in four Czech hospitals. mSphere 5:e01008-20. doi: 10.1128/mSphere.01008-20
Borjesson, S., Greko, C., Myrenas, M., Landen, A., Nilsson, O., and Pedersen, K. (2020). A link between the newly described colistin resistance gene mcr-9 and clinical Enterobacteriaceae isolates carrying blaSHV-12 from horses in Sweden. J. Glob. Antimicrob. Resist. 20, 285–289. doi: 10.1016/j.jgar.2019.08.007
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Cannatelli, A., D'Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible salmonella enterica serotype typhimurium isolate. MBio 10:e00853-19. doi: 10.1128/mBio.00853-19
Chavda, K. D., Westblade, L. F., Satlin, M. J., Hemmert, A. C., Castanheira, M., Jenkins, S. G., et al. (2019). First report of blaVIM-4-and mcr-9-coharboring Enterobacter species isolated from a pediatric patient. mSphere 4:e00629-19. doi: 10.1128/mSphere
Chen, J., Tian, S., Nian, H., Wang, R., Li, F., Jiang, N., et al. (2021). Carbapenem-resistant Enterobacter cloacae complex in a tertiary hospital in Northeast China, 2010-2019. BMC Infect. Dis. 21:611. doi: 10.1186/s12879-021-06250-0
Chowdhury, P., Fourment, M., DeMaere, M. Z., Monahan, L., Merlino, J., Gottlieb, T., et al. (2019). Identification of a novel lineage of plasmids within phylogenetically diverse subclades of IncHI2-ST1 plasmids. Plasmid 102, 56–61. doi: 10.1016/j.plasmid.2019.03.002
Clinical and Laboratory Standards Institute (CLSI) (2020). Performance Standards for Antimicrobial Susceptibility Testing. CLIS DOCUMENT M100, (30th Edn.). Wayne, PA: Clinical and Laboratory Standards Institute.
Falagas, M. E., and Kasiakou, S. K. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40, 1333–1341. doi: 10.1086/429323
Gong, X., Zhang, J., Su, S., Fu, Y., Bao, M., Wang, Y., et al. (2018). Molecular characterization and epidemiology of carbapenem non-susceptible Enterobacteriaceae isolated from the eastern region of Heilongjiang Province. China. BMC Infect Dis. 18:417. doi: 10.1186/s12879-018-3294-3
Gunn, J. S. (2008). The Salmonella Pmr AB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16, 284–290. doi: 10.1016/j.tim.2008.03.007
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Kieffer, N., Royer, G., Decousser, J. W., Bourrel, A. S., Palmieri, M., Ortiz De La Rosa, J. M., et al. (2019). Mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob. Agents Chemother. 63, e00965–e00919. doi: 10.1128/AAC.00965-19
Kim, J. S., Kwon, M. J., Jeon, S. J., Park, S. H., Han, S., Park, S. H., et al. (2021). Identification of a carbapenem-resistant Enterobacter kobei clinical strain co-harbouring mcr-4.3 and mcr-9 in Republic of Korea. J Glob Antimicrob. Resist. 26, 114–116. doi: 10.1016/j.jgar.2021.05.008
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/s1473-3099(10)70143-2
Lalaoui, R., Djukovic, A., Bakour, S., Hadjadj, L., Sanz, J., Salavert, M., et al. (2019). Genomic characterization of Citrobacter freundii strains coproducing OXA-48 and VIM-1 carbapenemase enzymes isolated in leukemic patient in Spain. Antimicrob. Resist. Infect. Control 8:167. doi: 10.1186/s13756-019-0630-3
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Li, Y., Dai, X., Zeng, J., Gao, Y., Zhang, Z., and Zhang, L. (2020). Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci. Rep. 10:8113. doi: 10.1038/s41598-020-65106-w
Liu, Z., Hang, X., Xiao, X., Chu, W., Li, X., Liu, Y., et al. (2021). Co-occurrence of blaNDM-1 and mcr-9 in a conjugative IncHI2/HI2A plasmid from a bloodstream infection-causing carbapenem-resistant Klebsiella pneumoniae. Front. Microbiol. 12:756201. doi: 10.3389/fmicb.2021.756201
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Ma, J., Zeng, Z., Chen, Z., Xu, X., Wang, X., Deng, Y., et al. (2009). High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob. Agents Chemother. 53, 519–524. doi: 10.1128/AAC.00886-08
Macesic, N., Blakeway, L. V., Stewart, J. D., Hawkey, J., Wyres, K. L., Judd, L. M., et al. (2021). Silent spread of mobile colistin resistance gene mcr-9.1 on IncHI2 'superplasmids' in clinical carbapenem-resistant Enterobacterales. Clin. Microbiol. Infect. 27, 1856.e7–1856.e13. doi: 10.1016/j.cmi.2021.04.020
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/s1473-3099(13)70190-7
Nang, S. C., Li, J., and Velkov, T. (2019). The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841X.2018.1492902
Pan, Y., Fang, Y., Song, X., Lyu, N., Chen, L., Feng, Y., et al. (2021). Co-occurrence of mcr-9, extended spectrum beta-lactamase (ESBL) and AmpC genes in a conjugative IncHI2A plasmid from a multidrug-resistant clinical isolate of salmonella diarizonae. J. Infect. 82, 84–123. doi: 10.1016/j.jinf.2020.11.008
Park, C. H., Robicsek, A., Jacoby, G. A., Sahm, D., and Hooper, D. C. (2006). Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50, 3953–3955. doi: 10.1128/AAC.00915-06
Peirano, G., Matsumura, Y., Adams, M. D., Bradford, P., Motyl, M., Chen, L., et al. (2018). Genomic epidemiology of global Carbapenemase-producing Enterobacter spp., 2008-2014. Emerg. Infect. Dis. 24, 1010–1019. doi: 10.3201/eid2406.171648
Ribeiro, T. G., Izdebski, R., Urbanowicz, P., Carmeli, Y., Gniadkowski, M., and Peixe, L. (2021). Citrobacter telavivum sp. nov. with chromosomal mcr-9 from hospitalized patients. Eur. J. Clin. Microbiol. Infect. Dis. 40, 123–131. doi: 10.1007/s10096-020-04003-6
Ribeiro-Goncalves, B., Francisco, A. P., Vaz, C., Ramirez, M., and Carrico, J. A. (2016). PHYLOViZ online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 44, W246–W251. doi: 10.1093/nar/gkw359
Simoni, S., Mingoia, M., Brenciani, A., Carelli, M., Lleo, M. M., Malerba, G., et al. (2021). First IncHI2 plasmid carrying mcr-9.1, blaVIM-1, and double copies of blaKPC-3 in a multidrug-resistant Escherichia coli human isolate. mSphere 6:e0030221. doi: 10.1128/mSphere.00302-21
Sun, J., Zhang, H., Liu, Y. H., and Feng, Y. (2018). Towards understanding MCR-like colistin resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/j.tim.2018.02.006
The European Committee on Antimicrobial Susceptibility Testing. (2020). Breakpoint tables for interpretation of MICs and zone diameters, version 10.0.
Tyson, G. H., Li, C., Hsu, C. H., Ayers, S., Borenstein, S., Mukherjee, S., et al. (2020). The mcr-9 gene of salmonella and Escherichia coli is not associated with colistin resistance in the United States. Antimicrob. Agents Chemother. 64:e00573-20. doi: 10.1128/AAC.00573-20
Umeda, K., Nakamura, H., Fukuda, A., Matsumoto, Y., Motooka, D., Nakamura, S., et al. (2021). Genomic characterization of clinical Enterobacter roggenkampii co-harbouring blaIMP-1-and blaGES-5-encoding IncP6 and mcr-9-encoding IncHI2 plasmids isolated in Japan. J Glob Antimicrob. Resist. 24, 220–227. doi: 10.1016/j.jgar.2020.11.028
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, L., Xu, T., Ji, Y., Song, J., Wang, F., Huang, J., et al. (2022). Occurrence and characteristics of Mcrs among gram-negative bacteria causing bloodstream infections of infant inpatients between 2006 and 2019 in China. Microbiol Spectr. 10:e0193821. doi: 10.1128/spectrum.01938-21
Yuan, Y., Li, Y., Wang, G., Li, C., Xiang, L., She, J., et al. (2019). Coproduction of MCR-9 and NDM-1 by colistin-resistant Enterobacter hormaechei isolated from bloodstream infection. Infect Drug Resist. 12, 2979–2985. doi: 10.2147/IDR.S217168
Keywords: carbapenem-resistant Enterobacter cloacae complex, mcr-9, metallo-β-lactamase genes, whole-genome sequencing, IncHI2/2A, colistin
Citation: Jiang S, Wang X, Yu H, Zhang J, Wang J, Li J, Li X, Hu K, Gong X, Gou X, Yang Y, Li C and Zhang X (2022) Molecular antibiotic resistance mechanisms and co-transmission of the mcr-9 and metallo-β-lactamase genes in carbapenem-resistant Enterobacter cloacae complex. Front. Microbiol. 13:1032833. doi: 10.3389/fmicb.2022.1032833
Edited by:
Mona I. Shaaban, Mansoura Universiy, EgyptReviewed by:
Nayeem Ahmad, All India Institute of Medical Sciences, IndiaPatrick Rik Butaye, Ghent University, Belgium
Rabaab Zahra, Quaid-i-Azam University, Pakistan
Copyright © 2022 Jiang, Wang, Yu, Zhang, Wang, Li, Li, Hu, Gong, Gou, Yang, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Zhang, am1zenhsMTIzQDE2My5jb20=; Chunjiang Li, bWRqbGNqMDdAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Shan Jiang1,2†
Shan Jiang1,2† Xiaoli Zhang
Xiaoli Zhang