
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 November 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1031688
This article is part of the Research Topic Emerging Multidrug-Resistant Bacterial Pathogens "superbugs": A Rising Public Health Threat View all 7 articles
 Muhammad Shafiq1
Muhammad Shafiq1 Mi Zeng1
Mi Zeng1 Budi Permana2
Budi Permana2 Hazrat Bilal3
Hazrat Bilal3 Jinhu Huang4
Jinhu Huang4 Fen Yao5
Fen Yao5 Abdelazeem Mohamed Algammal6
Abdelazeem Mohamed Algammal6 Xin Li1
Xin Li1 Yumeng Yuan1
Yumeng Yuan1 Xiaoyang Jiao1*
Xiaoyang Jiao1*The emergence of pathogens is conferring resistance to last-resort therapies such as tigecycline, colistin, and carbapenems, limiting the therapeutic options, and raising concerns about the emergence of new “superbugs.” This study reports the first incident of a blaNDM–5 and tet(X4) co-harboring Escherichia coli with resistance to carbapenem and tigecycline recovered as the causative agent of a urinary tract infection in a 94-year-old patient. The E. coli strain ECCL209 carries multiple resistance genes [i.e., blaTEM–1B, blaNDM–5, blaCMY–2, aadA22, florR, erm(B), mph(A), erm(42), lnuG, qnrS1, and sul2] and exhibits resistance to almost all clinically used antibiotics. MLST analysis found that the strain belongs to ST648, considered a worldwide high-risk pandemic clone. Moreover, multiple plasmid incompatibility types were detected, i.e., IncHI1A, IncHI1B, IncFII, IncFIA, IncFIB, IncQ1, Col, and IncX4. Genetic analysis revealed that blaNDM–5 and tet(X4) genes were localized on two hybrid plasmids with multiple replicons. Continuous monitoring studies are suggested to quantify the antimicrobial resistance and assess the dissemination of such superbugs into a human healthcare setting.
Antimicrobial resistance (AMR) has been an emerging and increasing threat to global health (World Health Organization [WHO], 2014; Su et al., 2017). A report from 2016 predicted that global fatalities from infectious diseases caused by AMR will rise from 0.7 to 10 million by 2050, with a vast estimated inaction cost of US$100 trillion between 2016 and 2050 (O’Neill, 2016). The antibiotic-resistant bacteria (ARB) of particular interest are multidrug-resistant (MDR), extensively drug-resistant (XDR), and pandrug-resistant (PDR) (Basak et al., 2016). These ARBs are called superbugs, and they can cause severe bacterial infections due to their acquired and intrinsic resistance mechanisms and render the efficacy of many existing antibiotics (Potter et al., 2016; Acolatse et al., 2022).
Of particular concern is AMR among Gram-negative bacterial species, especially the carbapenem-resistant Escherichia coli (CRE), which is the leading cause of urinary tract infections (UTIs) and is challenging to treat with last-resort carbapenem antibiotics. Carbapenems were developed to tackle bacteria producing extended-spectrum β-lactamases (ESBLs). However, Gram-negative bacteria have become resistant to this group of drugs by developing and/or acquiring bla genes encoding carbapenem hydrolyzing enzymes, named carbapenemases (Codjoe and Donkor, 2017; Nordmann and Poirel, 2019). Among the newly emerging carbapenemases, New Delhi Metallo-β-lactamase (NDM) is very important due to its widespread dissemination and allelic variations (Suay-García and Pérez-Gracia, 2021). The pathogens harboring these genes resist almost all β-lactam antibiotics (Wu et al., 2019). Tigecycline and colistin were relatively effective and used as the last-resort treatments to treat such infections caused by MDR and XDR bacteria (He et al., 2019). However, the recent discoveries of plasmid-mediated colistin resistance genes (mcr-1 to mcr-10) and/or the tigecycline resistance genes tet(X1) to tet(X15) among Enterobacteriaceae, especially in CRE, predict a return to the pre-antibiotic era and pose a severe threat to public health (He et al., 2019; Hussein et al., 2021). Furthermore, the co-occurrence of tet(X4) and mcr-1 as well as the combination of tet(X4) and blaNDM–5 genes in tigecycline- colistin- and carbapenem-resistant E. coli strains recovered from animals in China, posing a significant threat to public health, which requires urgent monitoring in terms of its prevalence (He et al., 2020; Sun et al., 2021; Lu et al., 2022).
To the best of our knowledge, herein, we identified the first case of XDR E. coli isolate co-harboring plasmid-mediated blaNDM–5 and tet(X4) genes from a clinical sample from a human patient.
During a routine surveillance project on AMR, an E. coli isolate ECCL209 was recovered from a 94-year-old man, admitted for >6 months in the respiratory and critical care department at Shantou Hospital, Guangdong Province, China. The patient was diagnosed with UTI. The E. coli strain ECCL209 was identified by automated mass spectrometry systems (VitekMS, bioMerieux, Marcy I’Etoile, France) and further confirmed by PCR utilizing the primers specific to the uidA gene as reported previously (Shafiq et al., 2019).
Antimicrobial susceptibility testing was accomplished by Vitek 2 COMPACT (bioMerieux, Marcy I’Etoile, France) with AST-N334 cards for the following antimicrobial agents: amikacin (AMK), cefoperazone/sulbactam (SCF), cefepime (FEP), cefoxitin (FOX), cefotaxime (CTX), ertapenem (ETP), imipenem (IMP), amoxicillin/clavulanic acid (AMC), cefuroxime (CXM), ceftriaxone (CRO), ceftazidime (CAZ), piperacillin/tazobactam (TZP), ticarcillin/clavulanic (TCC), ceftazidime-avibactam (CZA), ciprofloxacin (CIP), doxycycline (DOX), tigecycline (TIG), aztreonam (ATM), minocycline (MIC), tobramycin (TOB), trimethoprim/sulfamethoxazole (SXT), and colistin (COL). Antibiotic susceptibility for levofloxacin (LEV) was determined using Levofloxacin Susceptibility Test Paper (Thermo Scientific™ Oxoid™, Leicestershire, United Kingdom). Results for all antibiotics were interpreted following the standard of the Clinical and Laboratory Standard Institute (CLSI M100; 31st edition) guidelines, except imipenem, ertapenem, amoxicillin-clavulanic acid, and ceftazidime/avibactam for which the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints were considered.1
Detection of common ESBL genes (i.e., blaTEM, blaCTX–M, and blaSHV), carbapenemases (blaNDM, blaKPC, blaIMP, blaVIM, and blaOXA), and tigecycline-resistant genes tet (X3 and X4) was performed using PCR to identify resistance genes. All the primers used in this study are summarized in Supplementary Table 1.
Conjugation experiments were performed according to a previously described method (Shafiq et al., 2019). The donor strain [blaNDM and tet(X4)-positive E. coli] was diluted to the 0.5 McFarland standard and mixed with rifampicin-resistant recipient strain (E. coli C600) at a ratio of 1:1, respectively, on the microporous membrane. After cultures were incubated at 37°C for 12–14 h, the mixtures were collected and streaked on freshly made Luria-Bertani (LB) agar plates containing tigecycline (2 mg/L), meropenem (2 mg/L), and rifampicin (300 mg/L). The presence of blaNDM and tet(X4) in transconjugants was confirmed by PCR and corresponding resistance phenotyping. The number of positive transconjugants per recipient calculated the transfer frequency of conjugation.
To determine the genomic background, the ECCL209 E. coli strain was subjected to whole-genome sequencing (WGS) on the Illumina Miseq and Oxford Nanopore MinION platforms. The total DNA of E. coli strain ECCL209 was collected from fresh overnight cultures using a DNA kit (QIAamp® DNA Mini Kit, Germany) according to the manufacturer’s guidelines. The quality and quantity of extracted genomic DNA were measured and confirmed using a Nanodrop OD-1000 spectrophotometer (Thermo-Scientific®). DNA libraries were constructed using NEBNext® UltraTM DNA Library Prep Kit for Illumina (NEB, USA) and sequenced using an Illumina MiSeq sequencer (Illumina, San Diego, CA, USA). For the Nanopore platform, a Rapid Barcoding Sequencing Kit was used to construct the libraries and sequenced with a mini device (MinION), as previously reported (Maestri et al., 2019). Guppy base-calling software version 2.2 was used to generate fast5 files harboring the 1D DNA sequence from fast5 files. The quality of raw data from paired-end sequencing was checked using FastQC (version 0.11.6). Fastp (version 0.23.2) (Chen et al., 2018) was performed for the quality filtering to remove the low-quality reads, adapters, and poly-G tails. De novo assembly was accomplished using SPAdes (version 3.15.3) and Flye (version 2.8.3) with default parameters.
The assembled genomes were subjected to determine the resistome, virulome, MLST, serotype, mobile genetic elements (MGEs), and plasmidome using online search tools such as ResFinder 4.0; VirulenceFinder 2.0, MLST 2.0, SerotypeFinder 2.0, MobileElementFinder, and PlasmidFinder 2.0, at the Center for Genomic Epidemiology (CGE).2 Genome annotation and visualization were performed using Prokka (version 1.14.6) and Proksee.3 Plasmid replicons were identified using Abricate (version 1.0.1)4 from the assemblies. EasyFig (version 2.2.2) was used to compare and visualize the region of interest between similar sequences. The sequence similarity search was performed using BLAST against the NCBI nucleotide database. The significant hits were investigated, and related information, including the source organisms and hosts, was visualized along the BLAST result tree using ggtree version 3.4.
The E. coli strain exhibited resistance against 19 antimicrobial agents, an XDR phenotype (Magiorakos et al., 2012), including SCF, FEP, FOX, CTX, ETP, IMP, AMC, LEV, TZP, CXM, CRO, CAZ, TCC, CZA, CIP, DOX, TIG, ATM, and MIC, while susceptible to AMK, TOB, SXT, and COL. The susceptibility data are shown in Supplementary Table 2. The E. coli isolate ECCL209 was resistant to carbapenems and tigecycline and harbored blaTEM, blaNDM, and tet(X4) genes, amplified by PCR and subsequently confirmed by Sanger sequencing.
To determine the transmissibility of blaNDM and tet(X4) genes, we performed conjugation experiments with a recipient E. coli strain C600. The outcomes of conjugation proved that the blaNDM and tet(X4) genes in donor E. coli isolate ECCL209, with their corresponding resistance against imipenem and tigecycline, were successfully moved to the recipient strain C600, suggesting that blaNDM and tet(X4) genes were located on conjugative plasmids. The cotransfer of blaNDM and tet(X4) was at a frequency of (1.67 ± 0.2) × 10–1 to (3.12 ± 0.1) × 10–3 cells per recipient.
The main comprehensive results from the WGS analysis of Illumina and Nanopore are summarized in Table 1. The ECCL209 isolate was assigned as serotype O83:H42 using SerotypeFinder 2.0,5 which is an extraintestinal pathogenic E. coli (ExPEC) primarily found in samples from animals, indicating their possible transmission from animal to humans (Abreu-Salinas et al., 2020; Shafiq et al., 2021a,b, 2022). MLST analysis revealed that E. coli isolate ECCL209 in this study belonged to sequence type (ST648), which had been previously reported to carry blaCTX–M–, blaCMY–2–, blaNDM–, blaOXA–48–, and mcr-1 encoding genes and caused a significant proportion of infections in humans (Hornsey et al., 2011; Poirel et al., 2018; Chowdhury et al., 2022). This clonal lineage has emerged as a pandemic high-risk clone, being globally reported in humans, animals, and the environment (Hornsey et al., 2011; Fernandes et al., 2018; Furlan et al., 2020; Chowdhury et al., 2022; Landolsi et al., 2022). To the best of our knowledge, this is the first report of ST648, with blaNDM and tet(X4).
Our resistome results confirmed aminoglycosides (aadA22), amphenicols (floR), β-lactams (blaTEM–1B, blaNDM–5, and blaCMY–2), sulfonamides (sul2), macrolides [ermB, mphA, erm(42), and lnuG], quinolones (qnrS1), and tetracycline-resistant genes [tet(X4) and tetM]. Moreover, we found chromosomal mutations in parE (p. S458A), parC (p. S801), and gyrA (p. S83L, p. D87N), which encodes high-level resistance to fluoroquinolones (Mohsin et al., 2019). Multiple plasmids were detected in the E. coli ECCL209 strain, including, IncHI1A, IncHI1B, IncFII, IncFIA, IncFIB, IncQ1, Col, and IncX4. Detection of multiple plasmid types reflects the strains’ severity because all these replicons identified have the ability of horizontal transfer and play a vital role in spreading AMR genes (Rodríguez-Beltrán et al., 2021).
Regarding virulence genes, the presence of iutA (ferric aerobactin receptor), terC (tellurium ion resistance protein), IpfA (long polar fimbriae), traT (outer membrane protein complements resistance), air (enteroaggregative immunoglobulin repeat protein), sitA (iron transport protein), hra (heat resistance agglutinin), yfcV (fimbrial protein), iucC (aerobactin synthetase), eilA (Salmonella HilA homolog), and chuA (outer membrane hemin receptor) were identified in E. coli strain ECCL209. These virulence genes could enhance bacterial pathogenicity, and a recent study also described their direct interaction with ARGs in terms of bacterial survivability, which need to be disclosed in future studies (Zhang et al., 2019).
To further understand the genetic contexts of blaNDM–5 and tet(X4), we carried out long-read sequencing of E. coli ECCL209 isolate with the Oxford Nanopore MinION platform to obtain complete genome sequences. This assembled genome had four contigs, with a total length of 5,411,927 bp and an average G + C content of 50.31%. Bioinformatic analysis revealed that isolate ECCL209 harbored a chromosome and three circular plasmids comprising pECCL209-tetX4-190-kb, pECCL209-blaNDM5-157-kb, and pECCL209-blaCMY2-36-kb.
pECCL209-tetX4 was a 190,682-bp plasmid co-fused with IncHI1A, IncHI1B, and IncFIA, forming multiple replicon plasmids. Similarly, a fusion plasmid has been previously reported from China recently, where a tet(X4) gene was located in Enterobacter cloacae on a hybrid plasmid (∼190 kb) with IncFIA, IncHI1A, and IncHI1B replicons (Wu et al., 2022). This high homology of plasmids from animal and human origin suggests that tet(X4)-carrying plasmids could be conjugated from E. cloacae to E. coli. The BLASTn search was performed against the NCBI database to examine the sequence similarity of pECCL209-tetX4-190-kb and pECCL209-blaNDM5-157-kb. Phylogenetic analysis revealed that the pECCL209-tetX4 plasmid was similar to other bacterial strains with ≥90% query coverage. Most of the plasmid sequences matched with pECCL209-tetX4 were from animal origins, while this is the first human-origin E. coli plasmid harboring tet(X4) resistant gene (Figure 1A). The result showed that the pECCL209-tetX4-like plasmid might have been widely spread in different species of Enterobacteriaceae. pECCL209-tetX4-190-kb displayed a mosaic structure harboring five AMR genes, including flor (phenicol resistance), qnrS1 (quinolone resistance), blaTEM–1B (β-lactam resistance), aadA22 (aminoglycosides resistance), and lnu(G) (lincosamide resistance), and the MGEs found in the MDR region were IS26, ISVsa3, IS6, and IS91 (Figure 1B). The backbone of plasmid pECCL209-tetX4-25-kb from this study harboring a tet(X4) gene showed >99% nucleotide homology and 100% query coverage to several other tet(X4) carrying plasmids in Klebsiella pneumoniae and E. coli reported from animal origins, including plasmid p3Z-5L-2-X4 (GeneBank accession no. CP072517.1) in Klebsiella quasipneumoniae and pTKPN_3-186k-tetX4 (GeneBank accession no. MZ773211.1) in K. pneumoniae, while plasmid pYUGZP1-tetX (GeneBank accession no. MW439255.1) in E. coli of unknown animal origin (Figure 1C). This high similarity and dissemination of this type of plasmid suggest that plasmids harboring tet(X4) had been widely propagated in animals (He et al., 2020; Sun et al., 2021; Lu et al., 2022). Moreover, the tet(X4) genetic context in plasmid pECCL209-tetX4 of this study showed a resemblance with the above three plasmids from the NCBI database, showing that the tet(X4) gene tends to be adjacent to the upstream rdmC gene and flanked by a complete IS1R element.
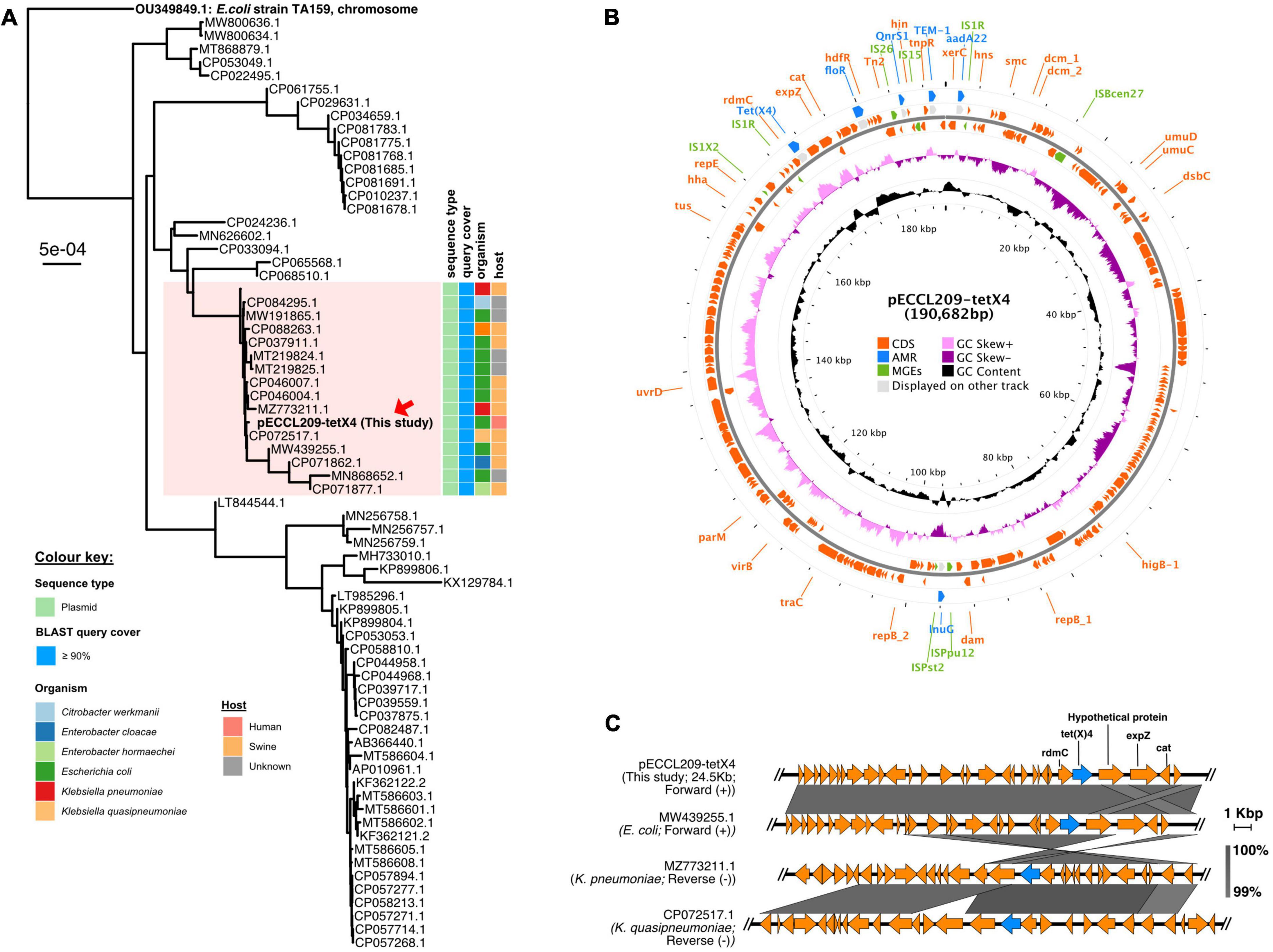
Figure 1. Structure of the tet(X4)-carrying plasmid and comparison of the genetic context of tet(X4). (A) BLAST tree comparison of plasmid pECCL209-tetX4 with other homologous plasmids available in the NCBI database. (B) Structure of the tet(X4)-carrying plasmid pECCL209. (C) Sequence comparison of the genetic context of a plasmid carrying tet(X4) gene from different sources. The arrows showed the direction of the transcription. Regions of >99% of homology are displayed by gray shading.
Similarly, pECCL209-blaNDM5 was a 157,741-bp hybrid plasmid with three replicon types, i.e., IncFII, IncFIA, and IncFIB. This plasmid showed ≥90% sequence identity with other blaNDM–5-carrying plasmids in K. pneumoniae plasmid pEH13_2 (GeneBank accession no. CP089099.1) and E. coli plasmid pYSP8-1-CTX-M-14 (GeneBank accession no. CP037912.1) of human and animal origin, respectively, suggesting that blaNDM–5-carrying plasmids had widely disseminated in China (Figure 2A). The blaNDM–5 gene resided in a complex region of the plasmid pECCL209-blaNDM5-157,741-bp. The plasmid carried other resistance genes, including aadA22 (aminoglycosides resistance), erm(B), erm(42), mph(A) (macrolides resistance), tet(M) (tetracycline resistance), and sul2 (sulfonamide resistance), and MGEs found in the MDR region, including IS26, ISVsa3, IS5, ISEc9, ISKox3, and IS91 (Figure 2B). The blaNDM–5 gene was located within a 10.8-kb region, which was highly similar (99% identity) to E. coli plasmid pGZ3_NDM5 (GeneBank accession no. CP017981.1) obtained from patient urine with intra-abdominal infections and Salmonella enterica plasmid unnamed2 (GeneBank accession no. CP019444.1) collected from a patient stool in China. In the plasmid backbone of pECCL209-blaNDM5-10kb, IS5 was inserted with ISAba125 upstream of blaNDM–5, and the bleMBL trpF, dsbD, and IS26 were located downstream from blaNDM–5 as shown Figure 2C. Interestingly, on sequence alignment of our plasmid pECCL209-blaNDM5 with other identical plasmids ≥60% BLAST query coverage found that the blaNDM–5 region in other strains was mostly missing as shown in Figure 3, suggesting that blaNDM–5 in pECCL209-blaNDM5 was captured from other mobile elements.
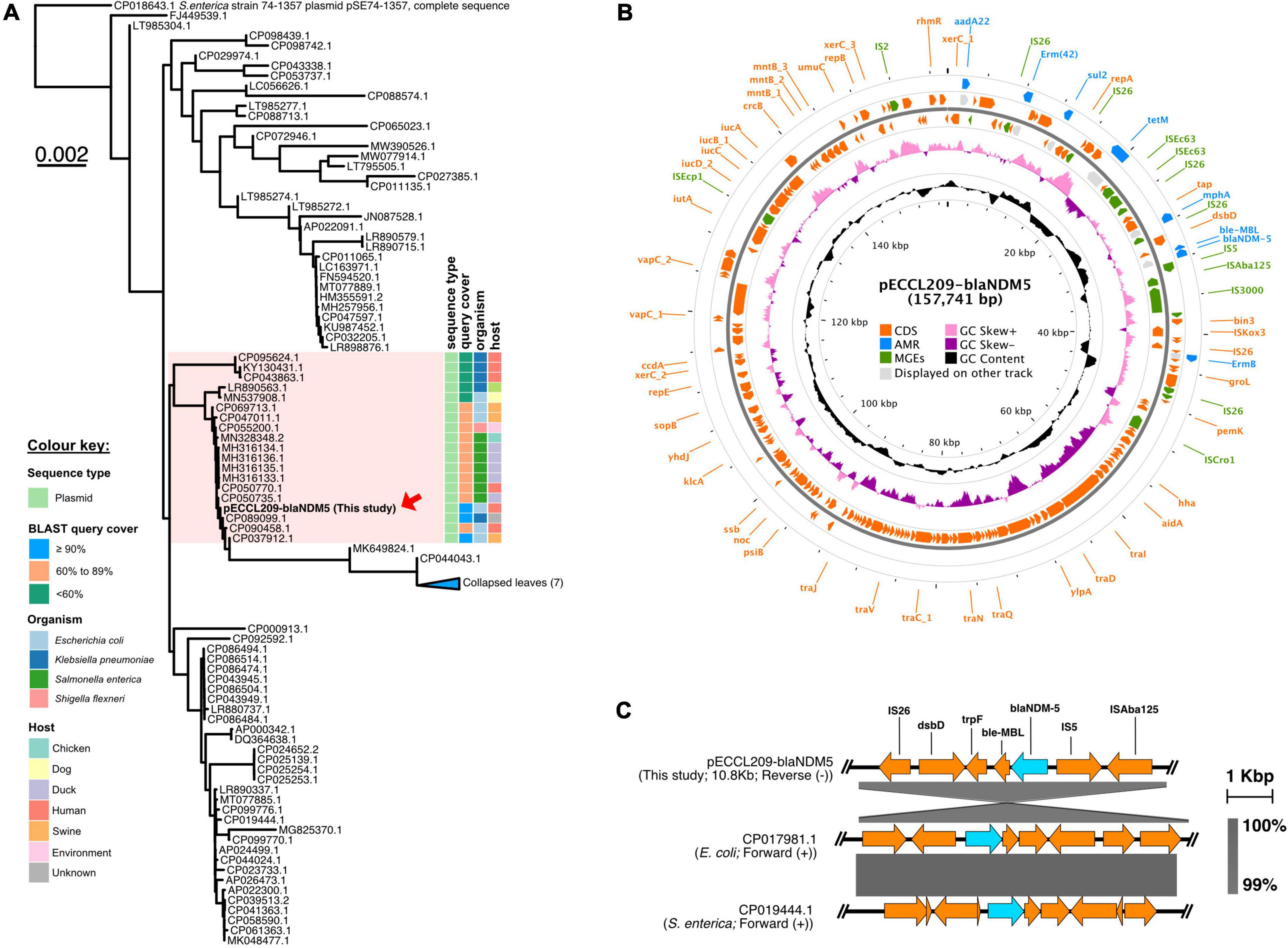
Figure 2. Structure of the blaNDM–5-carrying plasmid and comparison of the genetic context of blaNDM–5. (A) BLAST tree comparison of plasmid pECCL209-blaNDM5 with other homologous plasmids available in the NCBI database. (B) Structure of the blaNDM–5-carrying plasmid pECCL209. (C) Sequence comparison of the genetic context of a plasmid carrying blaNDM–5 gene from different sources. The arrows showed the direction of the transcription. Regions of >99% of homology are displayed by gray shading.
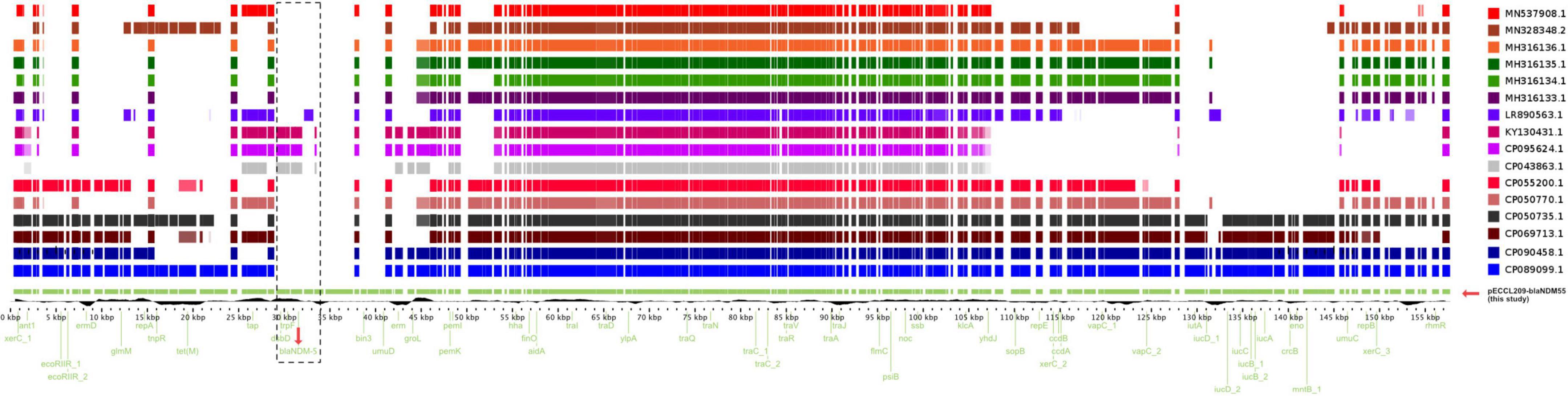
Figure 3. Linear alignment of the selected blaNDM gene comparison with other homologous plasmids available in the NCBI database.
As far as we know, this is the first report that emphasized the emergence of high-risk E. coli clone ST648 of a human origin, which carries the mobile carbapenem and tigecycline resistance determinants blaNDM–5 and tet(X4), respectively. Regardless of their low prevalence rate in humans and animal-associated sources, the mobile plasmid-mediated resistance genes in such superbugs can pose a significant threat to public health. Therefore, continuous monitoring of such MDR and XDR bacteria in humans, animals, and the environment should be considered under the aegis of the One Health approach and to guide the deployment of public health interventions before clinical cases increase.
The sequence data mentioned in this present study were deposited to the GenBank NCBI database under the BioProject PRJNA850111 with accession number: SRR19844396.
Ethical approval was provided by the Human Research Ethics Committee of Shantou Central Hospital and Shantou University Medical College (Ref 047 and SUMC-2021-51, respectively). Consent forms from the patients were waived by the Ethical Committee as all the clinical samples were obtained from the hospital laboratory.
MS and XJ designed the experiments. MS, MZ, and XL performed the experiments. MS wrote the original manuscript. BP and YY helped in the analysis. JH, HB, FY, and AA reviewed and edited the manuscript. All authors read and approved the final manuscript.
This study was graciously supported by the National Natural Science Foundation of China for International Young Scientists (No. 42150410383) and the 2020 Li Ka Shing Foundation Cross-Disciplinary Research Grant (project number: 2020LKSFG03E).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1031688/full#supplementary-material
Supplementary Table 1 | List of primers used in this study.
Supplementary Table 2 | Antibiotic susceptibility profile of Escherichia coli strain ECCL209.
Abreu-Salinas, F., Díaz-Jiménez, D., García-Meniño, I., Lumbreras, P., López-Beceiro, A. M., Fidalgo, L. E., et al. (2020). High prevalence and diversity of cephalosporin-resistant Enterobacteriaceae including extraintestinal pathogenic E. coli CC648 lineage in rural and urban dogs in Northwest Spain. Antibiotics 9:468. doi: 10.3390/antibiotics9080468
Acolatse, J. E. E., Portal, E. A., Boostrom, I., Akafity, G., Dakroah, M. P., Chalker, V. J., et al. (2022). Environmental surveillance of ESBL and carbapenemase-producing gram-negative bacteria in a Ghanaian Tertiary Hospital. Antimicrob. Resist. Infect. Control 11:49. doi: 10.1186/s13756-022-01090-2
Basak, S., Singh, P., and Rajurkar, M. (2016). Multidrug resistant and extensively drug resistant bacteria: A study. J. Pathog. 2016:4065603. doi: 10.1155/2016/4065603
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chowdhury, G., Ramamurthy, T., Das, B., Ghosh, D., Okamoto, K., Miyoshi, S. I., et al. (2022). Characterization of NDM-5 carbapenemase-encoding gene (bla (NDM-5)) – positive multidrug resistant commensal Escherichia coli from diarrheal patients. Infect. Drug Resist. 15, 3631–3642. doi: 10.2147/IDR.S364526
Codjoe, F. S., and Donkor, E. S. (2017). Carbapenem resistance: A review. Med. Sci. 6:1. doi: 10.3390/medsci6010001
Fernandes, M. R., Sellera, F. P., Moura, Q., Gaspar, V. C., Cerdeira, L., and Lincopan, N. (2018). International high-risk clonal lineages of CTX-M-producing Escherichia coli F-ST648 in free-roaming cats, South America. Infect. Genet. Evol. 66, 48–51. doi: 10.1016/j.meegid.2018.09.009
Furlan, J. P. R., Savazzi, E. A., and Stehling, E. G. (2020). Widespread high-risk clones of multidrug-resistant extended-spectrum β-lactamase-producing Escherichia coli B2-ST131 and F-ST648 in public aquatic environments. Int. J. Antimicrob. Agents 56:106040. doi: 10.1016/j.ijantimicag.2020.106040
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
He, T., Wei, R., Li, R., Zhang, L., Sun, L., Bao, H., et al. (2020). Co-existence of tet(X4) and mcr-1 in two porcine Escherichia coli isolates. J. Antimicrob. Chemother. 75, 764–766. doi: 10.1093/jac/dkz510
Hornsey, M., Phee, L., and Wareham, D. W. (2011). A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55, 5952–5954. doi: 10.1128/AAC.05108-11
Hussein, N. H., Al-Kadmy, I., Taha, B. M., and Hussein, J. D. (2021). Mobilized colistin resistance (mcr) genes from 1 to 10: A comprehensive review. Mol. Biol. Rep. 48, 2897–2907. doi: 10.1007/s11033-021-06307-y
Landolsi, S., Selmi, R., Hadjadj, L., Ben Haj Yahia, A., Ben Romdhane, K., Messadi, L., et al. (2022). First report of extended-spectrum β-lactamase (bla(CTX-M1)) and colistin resistance gene mcr-1 in E. coli of lineage ST648 from cockroaches in Tunisia. Microbiol. Spectr. 10:e0003621. doi: 10.1128/spectrum.00036-21
Lu, X., Du, Y., Peng, K., Zhang, W., Li, J., Wang, Z., et al. (2022). Coexistence of tet(X4), mcr-1, and bla(NDM-5) in ST6775 Escherichia coli isolates of animal origin in China. Microbiol. Spectr. 10:e0019622. doi: 10.1128/spectrum.00196-22
Maestri, S., Cosentino, E., Paterno, M., Freitag, H., Garces, J. M., Marcolungo, L., et al. (2019). A rapid and accurate MinION-based workflow for tracking species biodiversity in the field. Genes 10:468. doi: 10.3390/genes10060468
Magiorakos, A.-P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M., Giske, C., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mohsin, M., Azam, M., Ur Rahman, S., Esposito, F., Sellera, F. P., Monte, D. F., et al. (2019). Genomic background of a colistin-resistant and highly virulent MCR-1-positive Escherichia coli ST6395 from a broiler chicken in Pakistan. Pathog. Dis. 77:ftz064. doi: 10.1093/femspd/ftz064
Nordmann, P., and Poirel, L. (2019). Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin. Infect. Dis. 69, S521–S528. doi: 10.1093/cid/ciz824
O’Neill, J. (2016). Tackling drug-resistant infections globally: Final report and recommendations, ed. A. Ro (London: HM Government and Wellcome Trust).
Poirel, L., Madec, J. Y., Lupo, A., Schink, A. K., Kieffer, N., Nordmann, P., et al. (2018). Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 6:14. doi: 10.1128/microbiolspec.ARBA-0026-2017
Potter, R. F., D’souza, A. W., and Dantas, G. (2016). The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Updates 29, 30–46. doi: 10.1016/j.drup.2016.09.002
Rodríguez-Beltrán, J., Delafuente, J., Leon-Sampedro, R., Maclean, R. C., and San Millan, A. (2021). Beyond horizontal gene transfer: The role of plasmids in bacterial evolution. Nat. Rev. Microbiol. 19, 347–359. doi: 10.1038/s41579-020-00497-1
Shafiq, M., Huang, J., Rahman, S. U., Shah, J. M., Chen, L., Gao, Y., et al. (2019). High incidence of multidrug-resistant Escherichia coli coharboring mcr-1 and blaCTX-M-15 recovered from pigs. Infect. Drug Resist. 12, 2135–2149. doi: 10.2147/IDR.S209473
Shafiq, M., Huang, J., Shah, J. M., Ali, I., Rahman, S. U., and Wang, L. (2021a). Characterization and resistant determinants linked to mobile elements of ESBL-producing and mcr-1-positive Escherichia coli recovered from the chicken origin. Microb. Pathog. 150:104722. doi: 10.1016/j.micpath.2020.104722
Shafiq, M., Huang, J., Shah, J. M., Wang, X., Rahman, S., Ali, I., et al. (2021b). Characterization and virulence factors distribution of bla CTX-M and mcr-1 carrying Escherichia coli isolates from bovine mastitis. J. Appl. Microbiol. 131, 634–646. doi: 10.1111/jam.14994
Shafiq, M., Rahman, S. U., Bilal, H., Ullah, A., Noman, S. M., Zeng, M., et al. (2022). Incidence and molecular characterization of ESBL-producing and colistin-resistant Escherichia coli isolates recovered from healthy food-producing animals in Pakistan. J. Appl. Microbiol. 133, 1169–1182. doi: 10.1111/jam.15469
Su, J.-Q., An, X.-L., Li, B., Chen, Q.-L., Gillings, M. R., Chen, H., et al. (2017). Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome 5:84. doi: 10.1186/s40168-017-0298-y
Suay-García, B., and Pérez-Gracia, M. T. (2021). “Present and future of carbapenem-resistant Enterobacteriaceae infections,” in Advances in clinical immunology, medical microbiology, COVID-19, and big data, ed. R. Bawa (New York, NY: Jenny Stanford Publishing), 435–456.
Sun, H., Zhai, W., Fu, Y., Li, R., Du, P., and Bai, L. (2021). Co-occurrence of plasmid-mediated resistance genes tet (X4) and bla NDM-5 in a multidrug-resistant Escherichia coli isolate recovered from chicken in China. J. Glob. Antimicrob. Resist. 24, 415–417. doi: 10.1016/j.jgar.2021.02.010
World Health Organization [WHO] (2014). Antimicrobial resistance global report on surveillance: 2014 summary. Geneva: World Health Organization.
Wu, W., Feng, Y., Tang, G., Qiao, F., Mcnally, A., and Zong, Z. (2019). NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32, e00115–e00118. doi: 10.1128/CMR.00115-18
Wu, Y., He, R., Qin, M., Yang, Y., Chen, J., Feng, Y., et al. (2022). Identification of plasmid-mediated tigecycline-resistant gene tet(X4) in Enterobacter cloacae from pigs in China. Microbiol. Spectr. 10:e0206421. doi: 10.1128/spectrum.02064-21
Keywords: antimicrobial resistance, Escherichia coli, blaNDM–5, tet(X4), coexistence, superbugs, hybrid plasmids
Citation: Shafiq M, Zeng M, Permana B, Bilal H, Huang J, Yao F, Algammal AM, Li X, Yuan Y and Jiao X (2022) Coexistence of blaNDM–5 and tet(X4) in international high-risk Escherichia coli clone ST648 of human origin in China. Front. Microbiol. 13:1031688. doi: 10.3389/fmicb.2022.1031688
Received: 30 August 2022; Accepted: 13 October 2022;
Published: 10 November 2022.
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Jiyun Li, Hunan Agricultural University, ChinaCopyright © 2022 Shafiq, Zeng, Permana, Bilal, Huang, Yao, Algammal, Li, Yuan and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyang Jiao, eHlqaWFvQHN0dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.