- 1MARA Key Laboratory of Sustainable Crop Production in the Middle Reaches of the Yangtze River (Co-construction by Ministry and Province), College of Agriculture, Yangtze University, Jingzhou, China
- 2Guizhou Provincial Academician Workstation of Microbiology and Health, Guizhou Academy of Tobacco Science, Guiyang, Guizhou, China
- 3School of Food Science and Technology & School of Chemical Engineering, Hubei University of Arts and Science, Xiangyang, China
- 4China Tobacco Sichuan Industrial Corporation Technical Centre, Chengdu, China
- 5Guizhou Institute of Plant Protection, Guizhou Academy of Agricultural Sciences, Guiyang, Guizhou, China
Rhizopus oryzae is a destructive pathogen that frequently causes tobacco pole rot in curing chambers. Phenotypic characterization of the pathogen was conducted to provide basic biological and pathological information using Biolog Phenotype MicroArray (PM). In addition, the Y5 strain of R. oryzae was sequenced using Illumina HiSeq and Pacific Biosciences (PacBio) technologies. Using PM plates 1–8, 758 growth conditions were tested. Results indicated that R. oryzae could metabolize 54.21% of tested carbon sources, 86.84% of nitrogen sources, 100% of sulfur sources, and 98.31% of phosphorus sources. About 37 carbon compounds, including D-xylose, N-acetyl-D-glucosamine, D-sorbitol, β-methyl-D-glucoside, D-galactose, L-arabinose, and D-cellobiose, significantly supported the growth of the pathogen. PM 3 indicated the active nitrogen sources, including Gly-Asn, Ala-Asp., Ala-Gln, and uric acid. PM 6–8 showed 285 different nitrogen pathways, indicating that different combinations of different amino acids support the growth of the pathogen. Genome sequencing results showed that the R. oryzae Y5 strain had raw data assembled into 2,271 Mbp with an N50 value of 10,563 bp. A genome sequence of 50.3 Mb was polished and assembled into 53 contigs with an N50 length of 1,785,794 bp, maximum contig length of 3,223,184 bp, and a sum of contig lengths of 51,182,778 bp. A total of 12,680 protein-coding genes were predicted using the Nonredundant, Gene Ontology, Clusters of Orthologous Groups, Kyoto Encyclopedia of Genes and Genomes, and SWISS-PROT databases. The genome sequence and annotation resources of R. oryzae provided a reference for studying its biological characteristics, trait-specific genes, pathogen-host interaction, pathogen evolution, and population genetic diversity. The phenomics and genome of R. oryzae will provide insights into microfungal biology, pathogen evolution, and the genetic diversity of epidemics.
Introduction
Tobacco (Nicotiana tabacum L.) is a leafy, annual, solanaceous plant grown commercially for its leaves (Chen et al., 2020). It is one of the most widely grown commercial non-food crops in the world (Liu et al., 2017). Tobacco pole rot has been described as a postharvest pathogen (Chen et al., 2020) as it frequently infects tobacco leaves when that temperature was above 35°C, which occurs in the curing chamber, and had not been shown to infect tobacco leaves in the field. Yearly losses due to tobacco pole rot are immense. In the last 5 years, it became a potentially serious disease of flue-cured tobacco that led to the complete loss of the harvest in southwest China, especially in Guizhou, the second largest tobacco production province. Under high humidity and warm temperatures during curing, leaf rot can damage the whole leaf within the first 2 days in the curing chamber. The first symptoms are white fuzzy spots on the petiole and a watery brown soft rot. Afterward, dark fruiting bodies are formed. These fruiting bodies are filled with spores easily released by the wind (Kortekamp et al., 2003). It frequently occurs during the curing stage, and the disease incidence rate can reach 100% (Surhone et al., 2010).
The fungus of Rhizopus oryzae is widely studied, it commonly used for production of some oriental traditional foods, it is mainly recognized as a good producer of lactic acid (Londoño-Hernández et al., 2017). Meanwhile, the pathogen is also the primary cause of mucormycosis, an emerging, life-threatening infection characterized by rapid angioinvasive growth with an overall mortality rate that exceeds 50% (Ma et al., 2009). R. oryzae, the causative agent of tobacco pole rot, has also received much attention in recent years. Many studies on the biological characteristics of R. oryzae have been conducted. It has a wide range of temperature adaptability, ranging from 25–45°C, with an optimum temperature of 38°C (Chen et al., 2021). Early studies showed that R. oryzae was a heat-resistant pathogenic fungus, exhibiting higher growth rates at 25–37°C, and much lower rates were observed at temperatures higher than 40°C (Gayed et al., 1972). Previous studies showed that tobacco pole rot usually occurred during the flue-cured stage, and the termination temperature was about 45°C (Su et al., 2018; Zhang et al., 2018). The pathogen mainly infected petioles and leaves, the pathogenicity varied among different tissues, and the petiole was more conducive to disease (Chen et al., 2021). Carbons such as ribitol, D-arabitol, and ß-cyclodextrin (Wang et al., 2018a), as well as pH (Chen et al., 2021) affect the growth of R. oryzae. However, the metabolic basis for the absence of host specificity by R. oryzae is unknown. This includes the absorption and utilization of carbon, nitrogen, phosphorus, and sulfur and whether there is a significant difference in R. oryzae infection of tobacco under adverse conditions. Knowing the metabolic phenotype of R. oryzae will be valuable to understanding its biochemical properties. It may also help develop potential measures to decrease the overall effect of tobacco pole rot.
Genome sequencing is an important tool for studying the pathogenicity mechanism of plant pathogens. Genomic data are a useful resource to understand the mechanism of plant-pathogen interaction and are used in the phylogenetic analyses of the species (Ailloud et al., 2015). Beyond the isolate of R. oryzae from tobacco, many strains of R. oryzae were isolated from sweet potato, mulberry, lily (Holmes et al., 2002) and the human body (Nguyen et al., 2020). Information obtained from the National Center for Biotechnology Information (NCBI) could be learned that a total of 43 strains of R. oryzae had been sequenced, and there were less differences of genomic data with different R. oryzae strains. The genome size ranged from 37.46–55.79 Mb except for strain GL39, with a size of 72.36 Mb. The GC content of most strains was 34%. However, R. oryzae has a wide range of hosts. Therefore, more genomic sequences are required to analyze the entire species, especially those isolates from tobacco. To better understand the functions of pathogenicity determinants and the traits of aggressiveness of R. oryzae under different ecological environments, the entire genome of the pathogen isolated from tobacco must be sequenced.
Therefore, the objectives of this study were to (i) characterize the metabolic phenotype of R. oryzae and (ii) sequence the genome of R. oryzae. The genome combined with the metabolic phenotype of the pathogen could provide a reference for the study of fungal biological characteristics, trait-specific genes determination, pathogen-host interaction, pathogen evolution, and population genetic diversity. The data provided by this study will be valuable in expanding our knowledge of the biochemical and metabolic phenomics of R. oryzae. It would aid in developing more effective control measures for tobacco pole rot.
Materials and methods
Fungal strain and culture conditions
One isolate of R. oryzae strain Y5 (Wang et al., 2016) (GenBank Accession Nos. KT390187) from tobacco was chosen randomly among the isolates of the pathogen (Chen et al., 2020) for analysis from the laboratory of Guizhou Academy of Tobacco Science. It was also conserved at the China Typical Microorganism Conservation Center with conservation No. CCTCC M2015720. The isolate was maintained on potato dextrose agar (PDA), in a controlled climate cabinet at 25°C in darkness. After 5 days of incubation on PDA, conidia were produced (Figure 1), based on the observed colony attributes, the presence of pale brown sporangiospores (5 to 8 μm in diameter) with bluish stripes (Watanabe, 2002). Sterile cotton swabs were moistened with sterile FF Inoculating Fluid (FF-IF), spores were collected by rotating the cotton swabs on the surface of the colony, and then the swabs were mixed into 12 ml FF-IF inoculation solution. The suspension was filtered through a double-layer of sterile cheesecloth (Grade # 40: 24 × 20 threads per inch) to remove mycelial fragments, and the resulting conidia suspension was diluted to a final concentration of 1 × 105 spores mL−1.
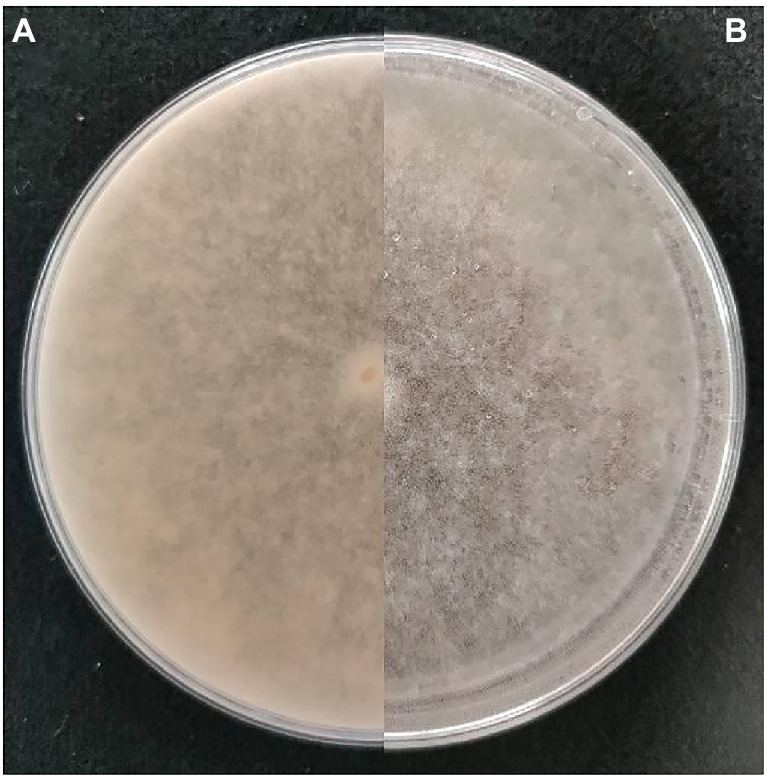
Figure 1. Morphological characteristics of colony of Rhizopus oryzae Y5. (A,B). Colony on PDA after 5 days’ incubation at 25°C in the dark (front and reverse).
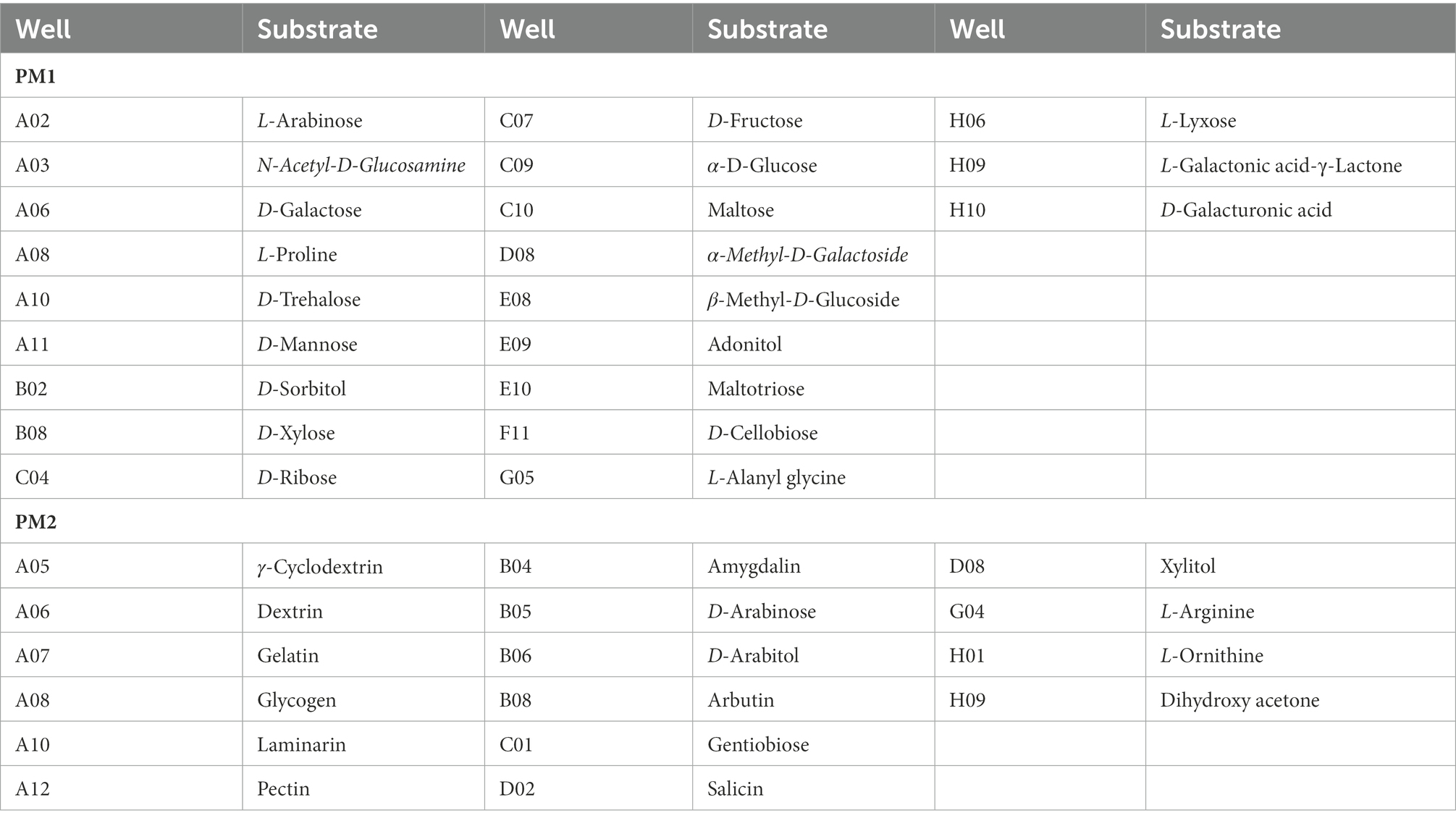
Table 1. Substrates in PM 1–2 carbon source Micro plates significantly supported the growth of Rhizopus oryzae.
Phenotypic characterization
The metabolic capacity of R. oryzae was tested using the Phenotype MicroArray (PM) system (Biolog, Hayward, CA, United States) to determine its phenotype (Bochner et al., 2001; Zhou et al., 2003; Von Eiff et al., 2006). The PM system involved 758 different growth conditions, including 190 diverse carbon sources (PM 1–2), 95 nitrogen sources (PM 3), 285 nitrogen pathways (PM 6–8), 59 phosphorus sources (PM 4), 35 sulfur sources (PM 4) and 94 biosynthetic pathways (PM 5). All materials, media, and reagents for the PM system were purchased from Biolog corporation. In total, 8 PM plates were used in this study. Carbon, nitrogen, phosphorus, sulfur, and biosynthetic pathways were tested for catabolic pathways in Plates 1–8. R. oryzae conidia suspension was prepared as detailed above and suspended in a suitable medium containing sterile FF-IF. The turbidity of conidial suspension was tested and was modified to a density of 62% T (transmittance). FF-IF was used for PM plates 1 and 2, FF-IF plus 100 mM D-glucose, 5 mM potassium phosphate (pH 6.0), and 2 mM sodium sulfate was used for PM plates 3 and 5–8. FF-IF plus 100 mM D-glucose was used for PM plate 4 (Wang M. S. et al., 2015). Plates containing 100 μl of the specified mixture were incubated in an OmniLog at 28°C for 1 week, and readings were taken every 15 min. Incubation and recording of phenotypic data were performed in the OmniLog station by capturing digital images of microarrays and storing turbidity values. Kinetic and Parametric software (Biolog, Hayward, CA, United States) was used to analyze the data. The phenotype was estimated according to the area of each well under the staining formation kinetics curve. The experiment was repeated twice.
Phylogenetic analyses of the sequences of genomic strains of R. oryzae
Reference sequences (Supplementary Table S1) from Nguyen et al. (2020) were downloaded from GenBank, the evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987), evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016).
Whole genome sequencing of R. oryzae strain of Y5
Genome sequencing
Extraction of genome DNA
Genomic DNA was extracted with the SDS method (Lim et al., 2016). The harvested DNA was detected by the agarose gel electrophoresis and quantified by Qubit® 2.0 Fluorometer (Thermo Scientific).
Library construction
Illumina NovaSeq platform
A total amount of 1 μg DNA per sample was used as input material for the DNA sample preparations. Sequencing libraries were generated using NEBNext® Ultra™ DNA Library Prep Kit for Illumina (NEB, United States) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, the DNA sample was fragmented by sonication to a size of 350 bp, then DNA fragments were end-polished, A-tailed, and ligated with the full-length adaptor for Illumina sequencing with further PCR amplification. At last, PCR products were purified (AMPure XP system) and libraries were analyzed for size distribution by Agilent2100 Bioanalyzer and quantified using real-time PCR.
PacBio sequel platform
Libraries for single-molecule real-time (SMRT) sequencing was constructed with an insert size of 20 kb using the SMRT bell TM Template kit, version 1.0. Briefly, the process was that fragment and concentrate DNA, repair DNA damage and ends, prepare blunt ligation reaction, purify SMRTbell Templates with 0.45X AMPure PB Beads, size-selection using the BluePippin System, repair DNA damage after size-selection. At last, the library quality was assessed on the Qubit® 2.0 Fluorometer (Thermo Scientific) and detected the insert fragment size by Agilent 2,100 (Agilent Technologies).
Sequencing
The whole genome of strain Y5 was sequenced using PacBio Sequel platform and Illumina NovaSeq PE150 at the Beijing Novogene Bioinformatics Technology Co., Ltd. The Illumina reads was only used to polish the assembly generated by PacBio reads and helped to reduce gaps and merge contigs.
Genome assembly
Preliminary assembly with SMRT Link v5.0.1 (Ardui et al., 2018; Reiner et al., 2018).
In order to ensure the accuracy of the subsequent analysis results, the low-quality reads were filtered (less than 500 bp) to obtain clean data. Using the automatic error correction function of SMRT portal, the long reads were selected (more than 6,000 bp) as the seed sequence, and the other shorter reads were aligned to the seed sequence by Blasr, so that the accuracy of the seed sequence could be improved further. After assembling we got an initial result.
Correct the results of the preliminary assembly
By the variant Caller module of the SMRT Link software, the arrow algorithm was used to correct and count the variant sites in the preliminary assembly results.
Genome component prediction
Genome component prediction included the prediction of the coding gene, repetitive sequences and non-coding RNA. The available steps were proceeded as follows:
For Fungi, by default, the Augustus (Stanke et al., 2008) 2.7 program to retrieve the related coding gene was used. Homology reference gene sequences and transcript sequencing data were provided, a complete annotation pipeline, PASA, as implemented at the Broad Institute, involves the following steps: (A) ab initio gene finding using a selection of the following software tools: GeneMarkHMM, FGENESH, Augustus, and SNAP, GlimmerHMM. (B) protein homology detection and intron resolution using the GeneWise software and the uniref90 non-redundant protein database. (C) alignment of known ESTs, full-length cDNAs, and most recently, Trinity RNA-Seq assemblies to the genome. (D) PASA alignment assemblies based on overlapping transcript alignments from step (C). (E) use of EVidenceModeler (EVM) to compute weighted consensus gene structure annotations based on the above (A, B, C, D). (F) use of PASA to update the EVM consensus predictions, adding UTR annotations and models for alternatively spliced isoforms (leveraging D and E).
The interspersed repetitive sequences were predicted using the RepeatMasker (Saha et al., 2008).1 The tandem Repeats were analyzed by the TRF (Tandem repeats finder; Benson, 1999).
Transfer RNA (tRNA) genes were predicted by the tRNAscan-SE (Lowe and Eddy, 1997). Ribosome RNA (rRNA) genes were analyzed by the rRNAmmer (Lagesen et al., 2007). sRNA, snRNA and miRNA were predicted by BLAST against the Rfam database (Gardner et al., 2009; Nawrocki and Kolbe, 2009).
Gene function prediction
Seven databases were used to predict gene functions. They were respective GO (Gene Ontology; Ashburner et al., 2000), KEGG (Kyoto Encyclopedia of Genes and Genomes; Kanehisa et al., 2004, 2006), KOG (Clusters of Orthologous Groups), NR (Non-Redundant Protein Database databases; Li et al., 2002), TCDB (Transporter Classification Database; Milton et al., 2014), P450 (Crešnar and Petrič, 2011) and Swiss-Prot (Amos and Rolf, 2000). A whole genome Blast search (E-value less than 1e-5, minimal alignment length percentage larger than 40%) was performed against above seven databases. The secretory proteins were predicted by the Signal P database (Petersen et al., 2011). Meanwhile, we analyzed the secondary metabolism gene clusters by the antiSMASH (Medema et al., 2011). For pathogenic fungi, we added the pathogenicity and drug resistance analyses. We used the PHI (Martin et al., 2015; Pathogen Host Interactions), DFVF (database of fungal virulence factors) to perform the above analyses. Carbohydrate-Active enzymes were predicted by the Carbohydrate-Active enZYmes Database (Cantarel et al., 2009).
Results
Phenotypic characterization of R. oryzae strain Y5
Using the OmniLog PM system, a kinetic response curve which parallels microbial growth can be generated for each well, allowing growth to be compared between samples through multiple parameters such as lag, slope, and area under the curve (Figure 2). The isolate Y5 of R. oryzae tested presented a representative phenotypic fingerprint. The fungus was able to metabolize 54.21% of tested carbon sources (51/95 in plate PM1 and 52/95 in plate PM2), 86.84% of nitrogen sources (61/95 in plate PM3, 93/95 in plate PM6, 94/95 in plate PM7, and 82/95 in plate PM8), 100% of sulfur sources (35/35 in plate PM4, wells F02-H12) and 98.31% of phosphorus sources (58/59 in plate PM4, wells A02-E12). The efficient metabolism of carbon sources rate was 19.27%, and the opportune metabolism of carbon sources rate was 17.19%. The efficient metabolism of nitrogen sources rate was 53.91%, and the opportune metabolism of carbon sources rate was 27.34%. The efficient metabolism of sulfur sources rate was 97.14%. The efficient metabolism of phosphorus sources rate was 76.27%, and the opportune metabolism of carbon sources rate was 16.49%. The pathogen presented 94 different biosynthetic pathways (94/94 tested, plate PM5, wells A3-H12).

Figure 2. Data for biology phenotype microarray PM 1–8 plates of the pathogen Rhizopus oryzae. Utilization of the isolate Y5 of R. oryzae from tobacco was indicated by green areas in the growth curve for each substrate, the larger the green area, the higher the utilization. A kinetic response curve which parallels microbial growth can be generated for each well, allowing growth to be compared between samples through multiple parameters such as lag, slope, and area under the curve.
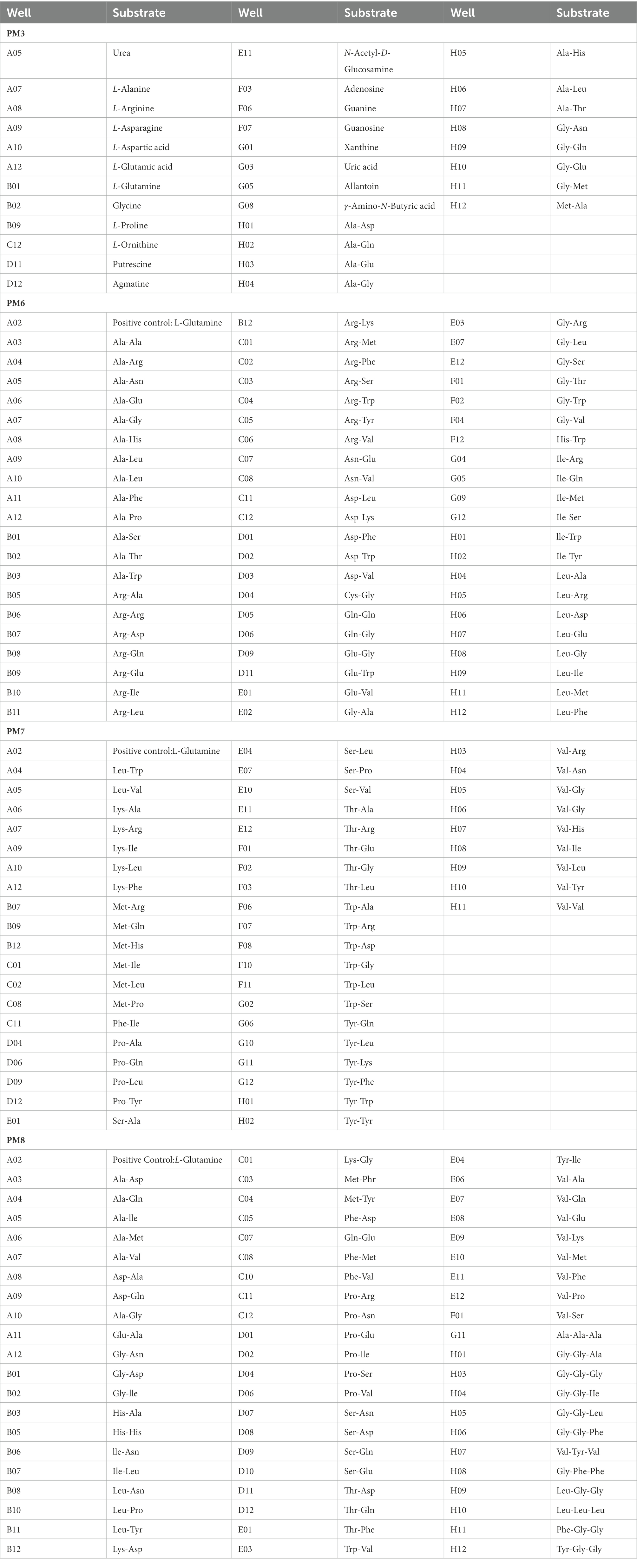
Table 2. Substrates in PM 3, 6–8 nitrogen source Micro plates significantly supported the growth of Rhizopus oryzae.
Carbon source utilization characteristics of strain Y5
Based on data from PM1 and PM2 (carbon sources), the isolate of R. oryzae from tobacco could use 103 different carbon sources and about 37 compounds (Table 1), including D-xylose, N-acetyl-D-glucosamine, D-sorbitol, β-methyl-D-glucoside, D-galactose, L-arabinose, D-cellobiose, D-Mannose, D-Galacturonic acid, maltotriose, D-trehalose, L-alanyl glycine, D-fructose, maltose, α-D-glucose, L-proline, D-ribose, adonitol, L-lyxose, L-galactonic acid-γ-lactone, α-methyl-D-galactoside, γ-cyclodextrin, pectin, dextrin, laminarin, gentiobiose, salicin, D-arabinose, D-arabitol, dihydroxy acetone, arbutin, glycogen, L-ornithine, xylitol, amygdalin, gelatin, and L-arginine significantly supported the growth of the pathogen. In comparison, around 87 compounds significantly inhibited the growth of the pathogen. Therefore, the utilization rate of carbon was lower than that of other sources.
Nitrogen sources utilization characteristics of strain Y5
Based on data from the PM3 plate, the isolate was tested for its ability to grow on 95 different nitrogen sources (amino acids). Sixty one compounds supported the growth of the pathogen, typical compounds included Gly-Asn, Ala-Asp., Ala-Gln, uric acid, Ala-Gly, Gly-Gln, Met-Ala, Gly-Glu, L-arginine, L-glutamine, L-ornithine, Ala-Glu, L-asparagine, Ala-Thr, Ala-Leu, allantoin, Gly-Met, agmatine, urea, guanine, Ala-His, guanosine, putrescine, xanthine, L-glutamic acid, L-aspartic acid, adenosine, L-proline, L-alanine, N-acetyl-D-glucosamine, glycine, and γ-amino-N-butyric acid. In comparison, 34 out of 95 nitrogen sources supported growth in the negative control, indicating that R. oryzae cannot metabolize these compounds. Based on PM6 to PM8 (nitrogen pathway) data, R. oryzae showed 285 different nitrogen pathways, indicating that different combinations of different amino acids support the growth of the pathogen. The result showed that 260 nitrogen pathways supported the growth of the pathogen, and more than 175 efficient nitrogen pathways, including L-glutamine, Arg-Arg, Ala-Arg, Arg-Lys, Ala-Asn, Arg-Met, Arg-Gln, Arg-Ala, Arg-Ile, Arg-Val, Arg-Asp., Ile-Arg, Arg-Glu, Leu-Phe, Arg-Leu, Arg-Ser, Ala-Pro, Arg-Trp, Leu-Arg, Ile-Gln, Gly-Arg, Asn-Glu, Asn-Val, Arg-Phe, Arg-Tyr, Ala-Gly, Ile-Ser, Ala-Trp, Asp-Lys, Ala-Ala, and lle-Trp. In comparison, around 25 compounds significantly inhibited the growth of the pathogen (Table 2). Therefore, the utilization rate of nitrogen sources was higher than that of carbon sources.
Phosphorus and sulfur sources utilization characteristics of strain Y5
The pathogen presented unapparent growth in the negative control without any phosphorus source (plate PM4, Well A01). Meanwhile, the pathogen could assimilate all S-containing compounds tested (35/35 tested, plate PM4, Wells F1-H12) (Table 3). Typical compounds included tetramethylene sulfone, butane sulfonic acid, L-methionine sulfone, hypotaurine, methane sulfonic acid, thiourea, L-cysteine sulfinic acid, D, L-lipoamide, L-cysteic acid, ρ-amino benzene sulfonic acid, 1-thio-β-D-glucose, glycyl-L-methionine, taurine, L-cysteinylglycine, 2-hydroxyethane sulfonic acid, cysteamine, thiosulfate, and cystathionine. The pathogen presented 94 different biosynthetic pathways (94/94 tested, plate PM5, wells A3-H12). Typical biosynthetic pathways included Tween 80, caprylic acid, myo-inositol, Tween 20, 2′-deoxy inosine, D, L-carnitine, 2′-deoxy adenosine, 2′-deoxy uridine, D, L-α-lipoic acid (oxidized form), Tween 60, thymidine, butyric acid, and L-glutamine (Table 4).
Phylogenetic analyses
The optimal tree with the sum of branch length = 31.57 was shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) were shown next to the branches (Felsenstein, 1985). The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and were in the units of the number of base substitutions per site. The analysis involved 37 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated. There was a total of 276 positions in the final dataset (Supplementary Figure S1).
Whole genome sequencing and statistical analysis
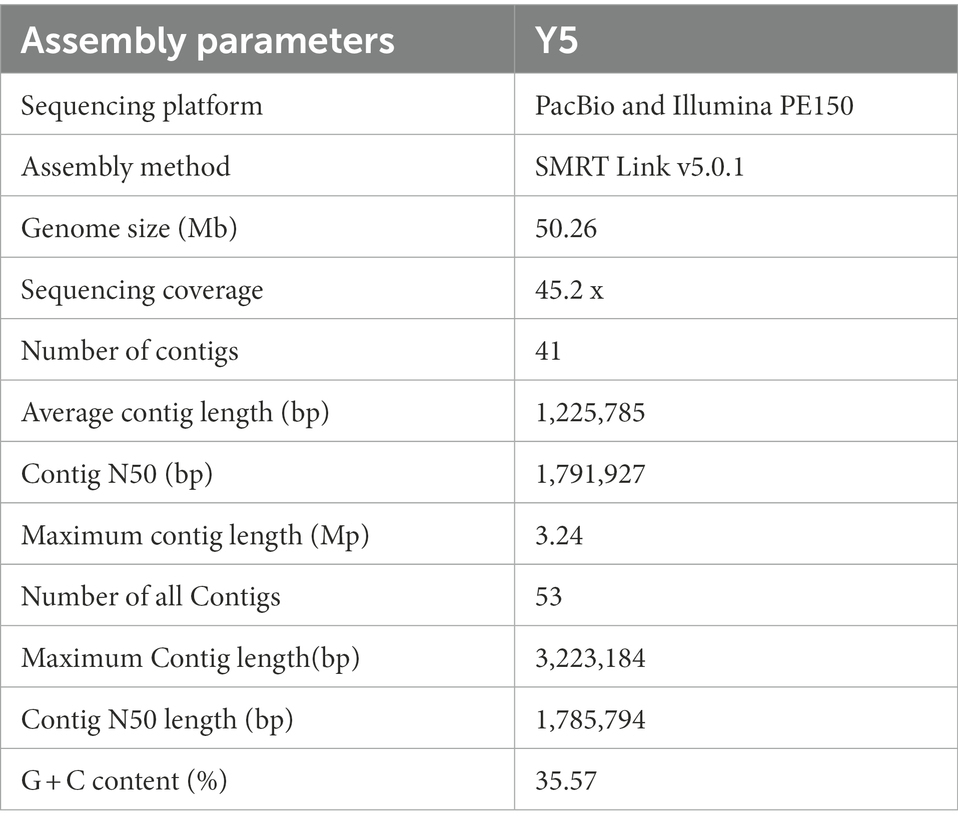
Whole genome sequencing was performed using single-molecule real-time (SMRT) sequencing on the PacBio RS II (Eid et al., 2009) and HiSeq PE150 platforms. PacBio RS II platform yielded 283,154 reads encompassing 2,129,349,468 bp, with an N50 value of 10,563 bp, using SMRT Link v5.1.0 software2 (Ardui et al., 2018; Reiner et al., 2018) for genome assembly to obtain the preliminary result which can reflect the basic condition of the sample genome. A genome sequence of 50.3 Mb was polished and assembled into 53 contigs with an N50 length of 1,785,794 bp, maximum contig length of 3,223,184 bp, and a total contig length of 51,182,778 bp. After the quality control, the number of contigs was 41 with an N50 length of 1,791,927 bp, maximum contig length of 3,235,075 bp, and total length of 50,257,186 bp (~45× genome average coverage; Table 5), and GC content of R. oryzae genome was found to be 35.57% which is similar to other forty-three sequenced R. oryzae (Figure 3; Supplementary Table S1).
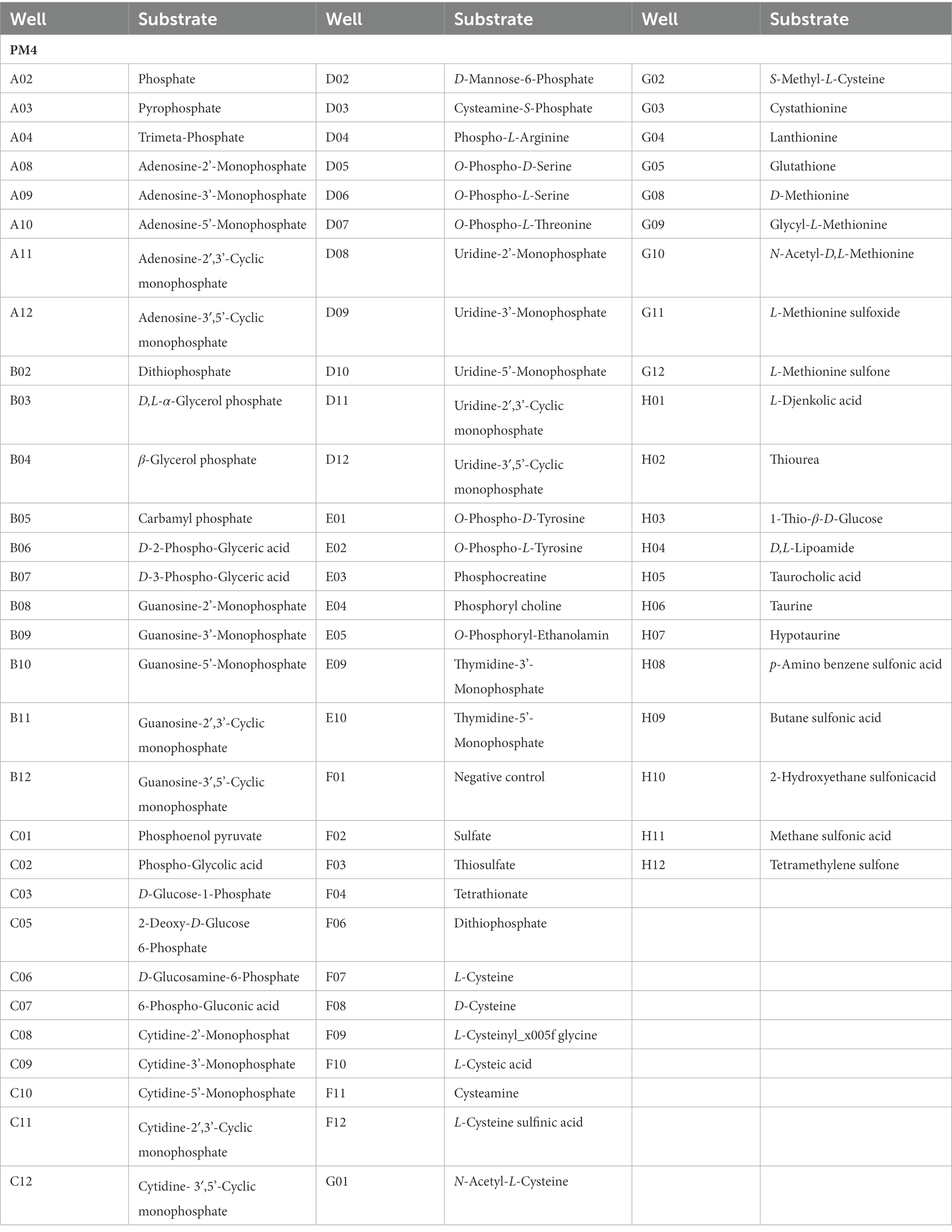
Table 3. Substrates in PM 4 phosphorus and sulfur source Micro plates significantly supported the growth of Rhizopus oryzae.
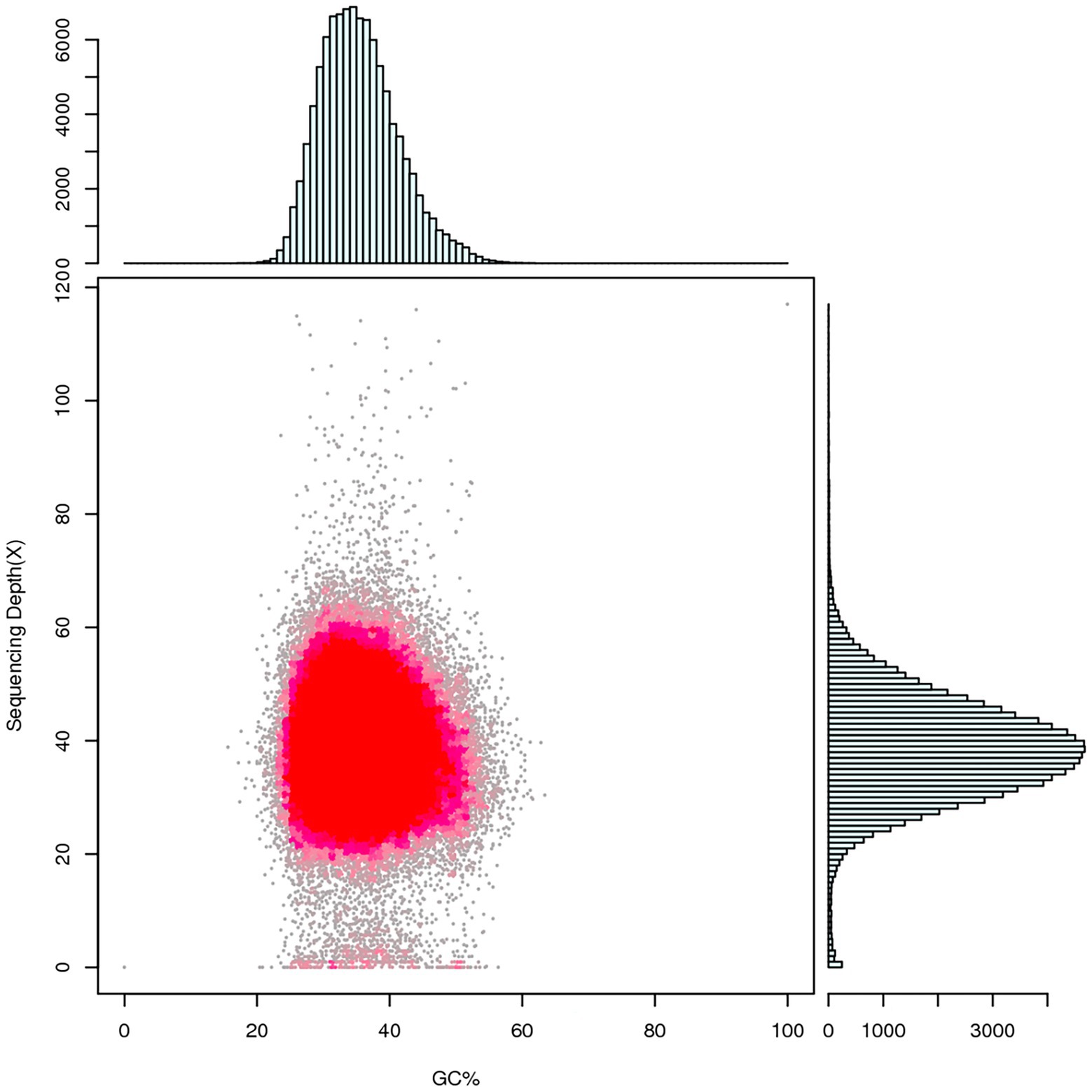
Figure 3. Statistical diagram of correlation analysis between GC content and sequencing depth. Summarize the GC bias and repetitive sequences of the genome by counting the GC content and reads coverage depth of assembled sequences. The horizontal coordinate indicates the GC content, the vertical coordinate indicates the sequencing depth, the right side is the sequencing depth distribution, and the upper side is the GC content distribution.
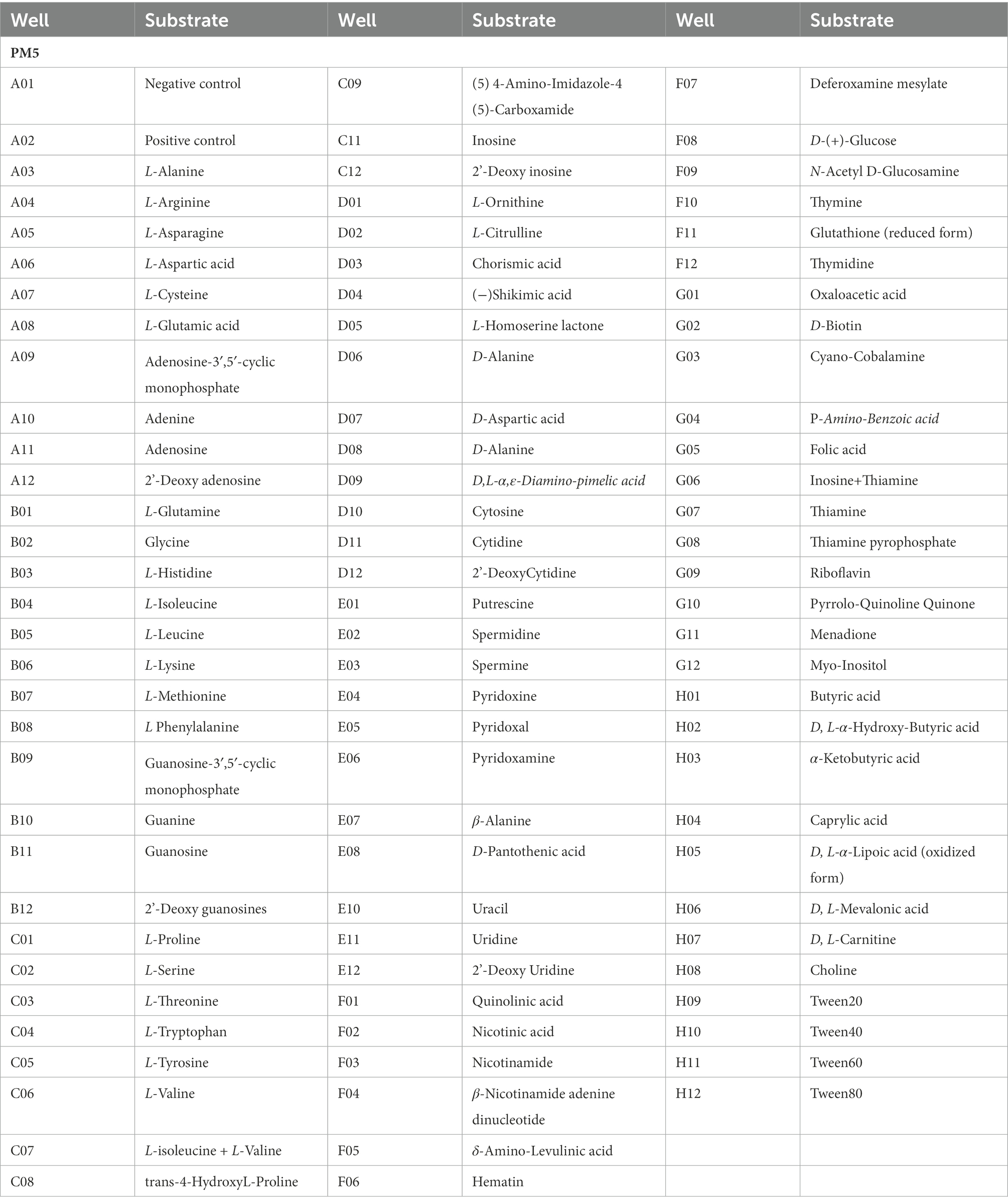
Table 4. Substrates in PM 5 nutrient supplements Micro plates significantly supported the growth of Rhizopus oryzae.
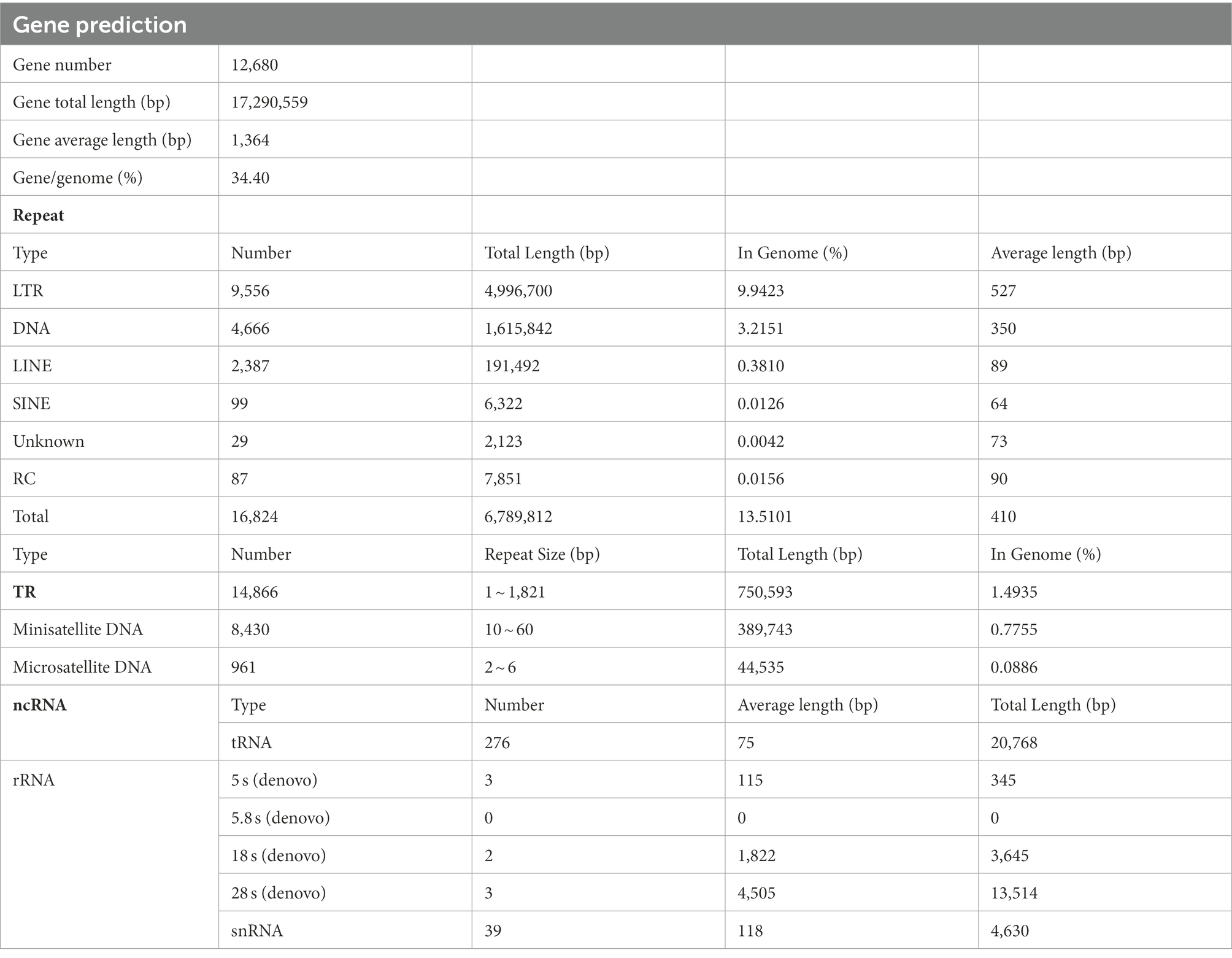
Table 6. Genomic component statistics and transposable element repeat class analysis of Rhizopus oryzae strain Y5.
Genome component analysis of coding genes, repeat sequence and ncRNA
The gene prediction was annotated, resulting in 12,680 genes. The total length of genes was 17,290,559 bp, and the average length was 1,364 bp, accounting for 34.4% of the genome. The TEs (transposable elements) represented 13.51% of the genome assembly with a total length of 6,787,689 bp. The total number of TE families analyzed with RepeatMasker in the genome assembly was 16,795 of which 16,708 (99.0%) belonged to the known TEs, including 12,042 retrotransposons (Class I) and 4,666 DNA transposons (Class II; Table 6). Class I retrotransposons could be mainly divided into three groups of TEs, including LINE, LTR, and SINE. Owing to the redundancy of repeats, these duplications were more tolerant of mutations, such as transposon insertions and sequence rearrangements, and might therefore act as a hotspot for genome expansion (Lippman et al., 2004). Tandem Repeat (TR) units with species composition specificity could be used as genetic traits of species for the study of evolutionary relationships. The TR represented 1.49% of the genome assembly with a total length of 750,593 bp. Non-coding RNAs (ncRNA) resembled mRNA in structure and function, they could regulate the transcription and translation of mRNAs in close proximity to them, except that unlike mRNAs that could be translated into proteins. The total number of ncRNA was 323, with a total length of 42,902 bp.; this suggested that ncRNA formed only a small proportion of the overall genome size (Table 6).
Genome function analysis of pathogenicity related genes
The annotation result statistics of the encoded genes were shown in the Supplementary Table S2, and 9.4% (3,531 genes) could be annotated and classified into different functional categories using COG (Cluster of Orthologous Groups of proteins; Figure 4), and 3,531 predicted genes could be assigned to 23 COG families. Except for the genes predicted to have general (382 genes) or unknown functions (133 genes), the largest group of genes were involved in “translation, ribosomal structure, and biogenesis” (525 genes, 8.93%). In addition, 5,408 predicted genes had KEGG orthologs, and 4,013 predicted genes had Swiss-Prot orthologs. A total of 12,167 predicted genes had NR orthologs, the largest proportion. The PHI database (Martin et al., 2015), mainly derived from fungi, oomycetes, and bacterial pathogens, indicated that the infected hosts include animals, plants, fungi, and insects. Complete proteome of R. oryzae was aligned to PHI database to reveal the pathogenicity related proteins. We observed a total of 1,147 (9.05%) PHI genes were classified into different classes such as “chemistry target: resistance to chemical” “effector (plant a virulence determinant)” “increased virulence (hypervirulence)” “lethal” “loss of pathogenicity” “no data found” “reduced virulence” and “unaffected pathogenicity.” As shown in Figure 5, we observed 10 genes associated with chemistry target about resistance to chemical. Furthermore, 115 genes belonged to lethal and 25 to increased virulence class. Remaining three classes viz., loss of pathogenicity, reduced virulence and unaffected pathogenicity were having 152, 589 and 253 genes, respectively. Pathogenicity related genes identified in this study have high relevance in future fungicide designing. The database is important for finding target genes for drug interventions, and it also includes antifungal compounds and corresponding target genes. The whole-genome sequence and annotation of Rhizopus oryzae isolate Y5 have been deposited at NCBI3 with accession PRJNA814049; BioSample SAMN26535981.
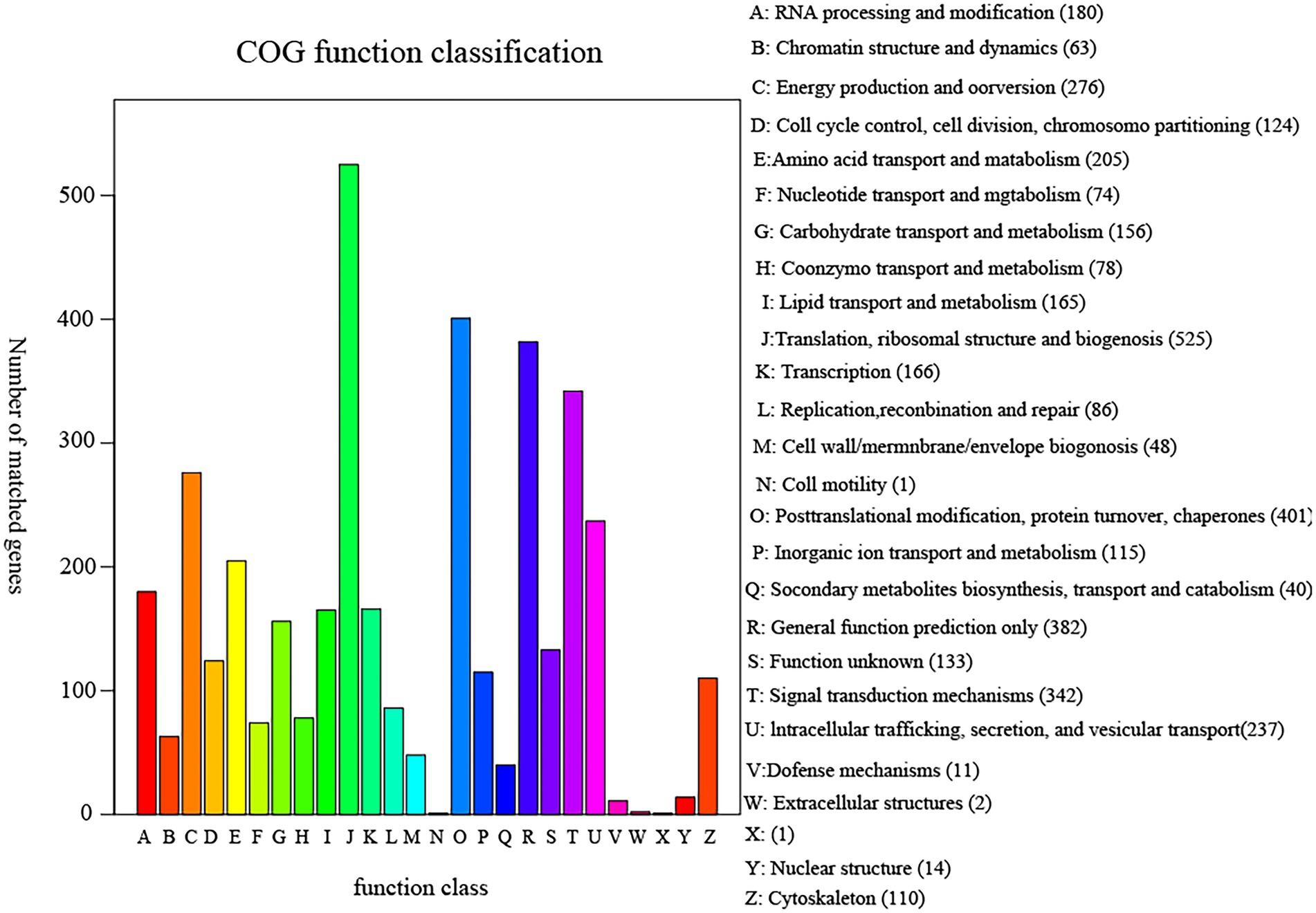
Figure 4. COG functional classification diagram of sample Y5 gene function annotation. COG (Cluster of Orthologous Groups of proteins) is constructed based on the classification of phylogenetic relationships of proteins encoded in complete genomes of bacteria, algae and eukaryotes. By comparison, a protein sequence can be annotated to a particular COG, and each cluster of COGs consists of direct homologous sequences, allowing the function of that sequence to be inferred. The abscissa represents the type of COG function and the ordinate represents the number of genes on the annotation.
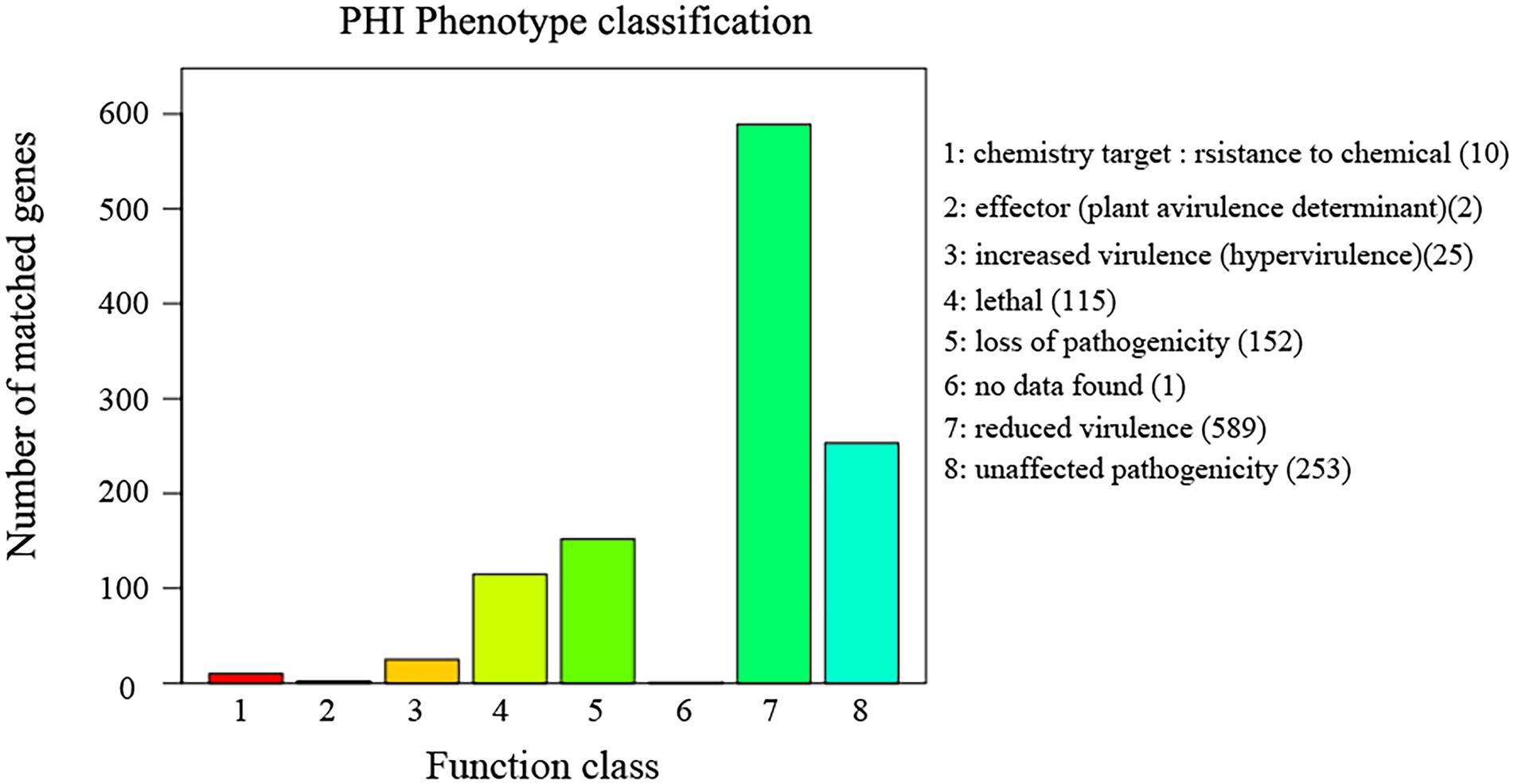
Figure 5. Distribution diagram of PHI phenotypic mutation type in the sample Y5 gene function annotation. PHI (Pathogen Host Interactions Database), pathogen-host interactions database, which is mainly derived from fungal, oomycete and bacterial pathogens and infects hosts including animals, plants, fungi as well as insects. This database is important for finding target genes for drug intervention studies, and it also includes antifungal compounds and their corresponding target genes. The horizontal coordinate indicates the type of phenotypic mutation and the vertical coordinate indicates the number of genes on the annotation.
Discussion
Rhizopus oryzae is a destructive fungal pathogen during flue-cured tobacco. The pathogen’s extensive host range includes Rosaceae, Cucurbitaceae, Solanaceae, Brassicaceae, and Umbelliferae (Xu et al., 2020). Many molecular biology, genetic, and genomic studies have been conducted on R. oryzae (Ellis, 1985; Ma et al., 2009). PMs have been used to analyze many microorganisms’ phenotypes, including Botrytis cinerea (Wang et al., 2018b), Phytophthora parasitica (Wang M. S. et al., 2015), and Alternaria alternata (Wang H. C. et al., 2015), comparisons were made with these pathogens. In this study, the metabolic ability of a R. oryzae isolate obtained from flue-cured tobacco was systematically studied using PMs, and important metabolic diversity information was obtained. So far, many Zygomycetes genomes have been sequenced and are publicly available, including R. oryzae from different hosts. We sequenced the genome of strain Y5 from flue-cured tobacco using Illumina HiSeq and Pacific Biosciences (PacBio) technologies. The genome combined with the metabolic phenotype of the pathogen could provide a reference for the study of fungal biological characteristics and population genetic diversity. Genome sequencing can reveal role of each individual gene and their networks responsible for plant pathogen interaction, growth, evolutionary relationship and genes for pathogenicity (Iquebal et al., 2017). Whole genome sequencing of R. oryzae is imperative not only to study the host-pathogen (HP) interaction but such knowledge discovery may lead to more effective disease combating strategy. Annotated genes/ predicted proteins can be directly used as new targets in fungicides development using computational approach (Acero et al., 2011).
Our study revealed that R. oryzae had a comparatively narrow utilization range of carbon sources. In comparison to carbon, nitrogen utilization efficiency was comparatively higher, which explained the R. oryzae infection mechanism for tobacco. Higher levels of an unutilized carbon source or less accessible nitrogen sources may affect pathogen infection and subsequently restrain disease. The PM 4 and PM 6–8 plates showed high levels of metabolic activity; this is comparable to findings from other researchers studying R. oryzae (Wang et al., 2018a). In our study, for the carbon or nitrogen substrates, carbohydrates or amino acids and peptides were greatly utilized by R. oryzae. The utilized substrates may sustain the continued existence of R. oryzae in different hosts and thus affect the pathogenicity of the pathogen. Compared with other pathogens, A. alternata had a comparatively small range of accessible carbon compounds, and most nitrogen, sulfur, and phosphorus sources are metabolized. Pseudomonas syringae had a comparatively small range of accessible carbon compounds, and most nitrogen, sulfur, and phosphorus sources could not be metabolized. B. cinerea had a small range of accessible carbon compounds with different crops, and most nitrogen, sulfur, and phosphorus sources were metabolized comparatively. Most pathogens made full use of nitrogen sources and used carbon sources to a larger or smaller extent. The prevention and control of tobacco pole rot through exploring the absorption and utilization of different nutritional elements by the pathogenic fungus, combined with the nutritional elements absorbed by tobacco, are discussed further.
We report the genomic analysis of R. oryzae, one of the most widely used extracellular enzyme producers, which could also cause disease in plants and humans. A genome sequence of 50.3 Mb was assembled into 41 contigs with an N50 length of 1,791,927 bp, maximum contig length of 3,223,184 bp, and total contig length of 50,257,186 bp. Genome information of all R. oryzae strains is supplied in Supplementary Table S1. Small differences were recorded in the genome sizes of the R. oryzae strains, ranging from 37.5–55.8 Mb except for strain GL39 (72.36 Mb), and the average GC content of the genomes was 34.7%, which was lower than that of strain Y5 of R. oryzae GC content. The genome host of the R. oryzae strains were “Homo sapiens,” “urine,” and “unknown.” Most of the strains were derived from the medical environment, for instance, “lung transplant,” “sinus,” “tracheal biopsy,” “nasal cavity,” “bone marrow,” “ethmoid sinus of diabetic” and “bronchial wash.” The Y5 strain isolated from tobacco was infrequently mentioned in genomic. In this study, the total number of predicted protein-coding genes was 12,680, compared to approximately 14,000 as the highest number from other strains, with a total length of 17,290,559 bp, average gene length of 1,364 bp, and gene length/genome (%) of 34.4%. Pathogenicity related genes identified in this study have high relevance in future fungicide designing using PHI database (Cools and Hammond-Kosack, 2013). Our enlisted lethal and virulence genes can be used in future research of fungal disease management, especially by designing of new generation genomic based fungicide (Seringhaus et al., 2006), the number of lethal genes in our study was found to be 115. In other ascomycotina, for example in S. cerevisiae, it was found to have 900 lethal genes (Miklos and Rubin, 1996).
The characteristics of each strain depend on which genes had changed and how that affected the phenotype. Although numerous studies had investigated the genome-wide properties of R. oryzae and the differences between multiple strains, few reports had compared the strain’s genome with its metabolic phenotype, despite advances in genetic techniques. Further studies of the effects of such metabolism in strains using gene knockout methods are needed. The deluge of data generated by genome sequencing has led to an increasing reliance on bioinformatic predictions, since the traditional experimental approach of characterizing gene function one at a time cannot possibly keep pace with the sequence-based discovery of novel genes (Johnson et al., 2008). One common application of the PM system is to detect phenotypic changes associated with gene knockouts, and this strategy has been used to study gene function and to assess and improve genome annotation (Loh et al., 2006). We have further explored the linkage between metabolic phenotypes and genomes in the next study. Aspergillus luchuensis (Hong et al., 2013) could grow on carbon sources other than glucose. Further RNA sequencing studies are needed to provide quantitative evidence to support our results and elucidate expression levels under stress conditions. In addition, Orthologous Groups (OGs) containing multiple genes also affect the traits of the lines; thus, these OGs should be further studied. Further Studies to know the relationship between metabolic activities under stress conditions are needed. This study demonstrated the power of whole genome analysis to reveal novel gene sequences between different strains. Whole genome analysis of R. oryzae should reveal the unique genes and non-coding regions of each strain. Therefore, the genome sequences of R. oryzae provided a unique resource for studying pathogenicity determinants in this pathogen, understanding host adaptation mechanisms, and designing specific disease management control strategies.
Conclusion
This study demonstrates that phenotypic characterization of a strain of the pathogen R. oryzae isolated from tobacco was conducted to provide basic biological and pathological information using Biolog Phenotype MicroArray (PM). In addition, the Y5 strain of R. oryzae was sequenced using Illumina HiSeq and Pacific Biosciences (PacBio) technologies. It indicated that R. oryzae could metabolize 54.21% of tested carbon sources, 86.84% of nitrogen sources, 100% of sulfur sources, and 98.31% of phosphorus sources. And also, genome sequencing results showed that the R. oryzae Y5 strain had raw data assembled into of 2,271 Mbp with an N50 value of 10,563 bp. A genome sequence of 50.3 Mb was polished and assembled into 53 contigs with an N50 length of 1,785,794 bp, maximum contig length of 3,223,184 bp, and a sum of contig lengths of 51,182,778 bp. A total of 12,680 protein-coding genes were predicted. This study demonstrated the power of whole genome analysis to reveal novel gene sequences between different strains. Whole genome analysis of R. oryzae should reveal the unique genes and non-coding regions of each strain. Therefore, the genome sequences of R. oryzae provided a unique resource for studying pathogenicity determinants in this pathogen, understanding host adaptation mechanisms, and designing specific disease management control strategies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA814049.
Author contributions
ZL, C-hS, YH, H-cW, W-hL and L-tC contributed to conceptualize and design of the study. ZL organized the database and performed the statistical analysis. ZL and H-cW wrote the first draft of the manuscript. H-cW and YH revised the manuscript and wrote some sections. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the China National Tobacco Corporation [110202101048(LS-08), 110202001035(LS-04)], the National Natural Science Foundation of China (31960550, 32160522), the Hundred Level Innovative Talent Foundation of Guizhou Province [GCC(2022)028–1], the Guizhou Science Technology Foundation (ZK[2021]Key036), the International Science and Technology Cooperation Base ([2020]4102), Guizhou Provincial Academician Workstation of Microbiology and Health([2020]4004), and the Guizhou Tobacco Company (2020XM22, 2020XM03). The authors declare that this study received funding from the China National Tobacco Corporation and Guizhou Tobacco Company. The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. All authors declare no other competing interests.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1031023/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Evolutionary relationships of Rhizopus oryzae. Phylogenetic tree showing the phylogenetic relationship amongst difference belonging to the species Rhizopus oryzae (Rhizopus oryzae, also known as Rhizopus arrhizus), is a filamentous fungus that is the most common cause of mucormycosis, also referred to as zygomycosis. An opportunistic pathogen, R. oryzae causes disease primarily in immunocompromised people, such as those with diabetes mellitus, cancer, or AIDS. R. oryzae is found in soil, decaying fruit and vegetables, old bread, and animal dung. It is used in the preparation of fermented foods and alcoholic beverages in Asia. R. oryzae is also a destructive pathogen that frequently causes tobacco pole rot in curing chambers.
Footnotes
References
Acero, F. J. F., Carbú, M., El-Akhal, M. R., Garrido, C., González-Rodríguez, V. E., and Cantoral, J. M. (2011). Development of proteomics-based fungicides: new strategies for environmentally friendly control of fungal plant diseases. Int. J. Mol. Sci. 12, 795–816. doi: 10.3390/ijms12010795
Ailloud, F., Lowe, T., Cellier, G., Roche, D., Allen, C., and Prior, P. (2015). Comparative genomic analysis of Ralstonia solanacearum reveals candidate genes for host specificity. BMC Genomics 16:270. doi: 10.1186/s12864-015-1474-8
Amos, B., and Rolf, A. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. NAR. 28, 45–48.
Ardui, S., Ameur, A., Vermeesch, J. R., and Hestand, M. S. (2018). Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 46, 2159–2168. doi: 10.1093/nar/gky066
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. NAR. 27, 573–580. doi: 10.1093/nar/27.2.573
Bochner, B. R., Gadzinski, P., Panomitros, E., Gadzinski, P., and Panomitros, E. (2001). Phenotype microarrays for high-throughput phenotypictesting and assay of gene function. Genome Res. 11, 1246–1255. doi: 10.1101/gr.186501
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. NAR 37, D233–D238. doi: 10.1093/nar/gkn663
Chen, Q. L., Cai, L., Wang, H. C., Cai, L. T., Goodwin, P., Ma, J., et al. (2020). Fungal composition and diversity of the tobacco leaf phyllosphere during curing of leaves. Front. Microbiol. 11:2136. doi: 10.3389/fmicb.2020.554051
Chen, Q. L., Wang, H. C., Huang, Y., et al. (2021). Pathogenicity and metabolic phenotypic characteristics of Rhizopus oryzae in tobacco under different osmotic pressure and pH environments. Tobacco Science & Technology. 2022, 16–23. doi: 10.16135/j.issn1002-0861.2021.0081
Cools, H. J., and Hammond-Kosack, K. E. (2013). Exploitation of genomics in fungicide research: current status and future perspectives. Mol. Plant Pathol. 14, 197–210. doi: 10.1111/mpp.12001
Crešnar, B., and Petrič, S. (2011). Cytochrome P450 enzymes in the fungal kingdom. Biochim. Biophys. Acta 1814, 29–35. doi: 10.1016/j.bbapap.2010.06.020
Eid, J., Fehr, A., Gray, J., Luong, K., Lyle, J., Otto, G., et al. (2009). Real-timeDNA sequencing from single polymerase molecules. Science 323, 133–138. doi: 10.1126/science.1162986
Ellis, J. J. (1985). Species and varieties in the Rhizopus arrhizus-Rhizopus oryzae group as indicated by their dna complementarity. Mycologia 77, 243–247. doi: 10.1080/00275514.1985.12025091
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Gardner, P. P., Daub, J., Tate, J. G., Nawrocki, E. P., Kolbe, D. L., Lindgreen, S., et al. (2009). Rfam: updates to the RNA families database. NAR. 37, D136–D140. doi: 10.1093/nar/gkn766
Gayed, S. K., et al. (1972). Rhizopus arrhizus causing pole rot of flue-cured tobacco in Ontario. Can. J. Plant Sci. 52, 103–106. doi: 10.4141/cjps72-013
Holmes, G. J., Stange, R. R., et al. (2002). Influence of wound type and storage duration on susceptibility of sweet potatoes to Rhizopus soft rot. Plant Dis. 86, 345–348. doi: 10.1094/PDIS.2002.86.4.345
Hong, S. B., Lee, M., Kim, D. H., Varga, J., Frisvad, J. C., Perrone, G., et al. (2013). Aspergillus luchuensis, an industrially important black Aspergillus in East Asia. PLoS One 8:e63769. doi: 10.1371/journal.pone.0063769
Iquebal, M. A., Tomar, R. S., Parakhia, M. V., Singla, D., Jaiswal, S., Rathod, V. M., et al. (2017). Draft whole genome sequence of groundnut stem rot fungus Athelia rolfsii revealing genetic architect of its pathogenicity and virulence. Sci. Rep. 7, 1–10. doi: 10.1038/s41598-017-05478-8
Johnson, D. A., Tetu, S. G., Phillippy, K., Chen, J., Ren, Q., and Paulsen, I. T. (2008). High-throughput phenotypic characterization of Pseudomonas aeruginosa membrane transport genes. PLoS Genet. 4:e1000211. doi: 10.1371/journal.pgen.1000211
Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F., Itoh, M., Kawashima, S., et al. (2006). From genomics to chemical genomics: new developments in KEGG. NAR. 34, D354–D357. doi: 10.1093/nar/gkj102
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. NAR. 32, 277D–2280D. doi: 10.1093/nar/gkh063
Kortekamp, A., Schmidtke, M., Serr, A., et al. (2003). Infection and decay of tobacco caused by Rhizopus oryzae/die Infektion und Fäulnis von Tabak verursacht durch Rhizopus oryzae. J. Plant. Dis. Protect. 110, 535–543.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lagesen, K., Hallin, P., Rødland, E. A., Stærfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. NAR. 35, 3100–3108. doi: 10.1093/nar/gkm160
Li, W., Jaroszewski, L., and Godzik, A. (2002). Tolerating some redundancy significantly speeds up clustering of large protein databases. CCF TCBI. 18, 77–82. doi: 10.1093/bioinformatics/18.1.77
Lim, H. J., Lee, E. H., Yoon, Y., Chua, B., and Son, A. (2016). Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J. Appl. Microbiol. 120, 379–387. doi: 10.1111/jam.13011
Lippman, Z., Gendrel, A. V., Black, M., Vaughn, M. W., Dedhia, N., Richard McCombie, W., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476. doi: 10.1038/nature02651
Liu, L. P., Qu, J. W., Yi, X. Q., Huang, H. H., et al. (2017). Genome-wide identification, classification and expression analysis of the mildew resistance locus o (MLO) gene family in sweet orange (Citrus sinensis). Braz. Arch. Biol.Technol. 60, 1678–4324. doi: 10.1590/1678-4324-2017160474
Loh, K. D., Gyaneshwar, P., Markenscoff Papadimitriou, E., Fong, R., Kim, K. S., Parales, R., et al. (2006). A previously undescribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. U. S. A. 103, 5114–5119. doi: 10.1073/pnas.0600521103
Londoño-Hernández, L., Ramírez-Toro, C., Ruiz, H. A., Ascacio-Valdés, J. A., Aguilar-Gonzalez, M. A., Rodríguez-Herrera, R., et al. (2017). Rhizopus oryzae–ancient microbial resource with importance in modern food industry. Int. J. Food Microbiol. 257, 110–127. doi: 10.1016/j.ijfoodmicro.2017.06.012
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. NAR. 25, 955–964. doi: 10.1093/nar/25.5.955
Ma, L. J., Ibrahim, A. S., Skory, C., Grabherr, M. G., Burger, G., Butler, M., et al. (2009). Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 5:e1000549. doi: 10.1371/journal.pgen.1000549
Martin, U., Rashmi, P., Arathi, R., et al. (2015). “The pathogen-host interactions database (phi-base): additions and future developments,” in Nucleic Acids Research (NAR). Oxford, England: Oxford University Press.
Medema, M. H., Blin, K., Cimermancic, P., de Jager, V., Zakrzewski, P., Fischbach, M. A., et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. NAR. 39, W339–W346. doi: 10.1093/nar/gkr466
Miklos, G. L. G., and Rubin, G. M. (1996). The role of the genome project in determining gene function: insights from model organisms. Cells 86, 521–529. doi: 10.1016/S0092-8674(00)80126-9
Milton, S. J., Vamsee, S. R., Dorjee, G. T., et al. (2014). “The transporter classification database,” in Nucleic Acids Research (NAR). Oxford, England: Oxford University Press.
Nawrocki, E. P., and Kolbe, D. L. (2009). Eddy SR: infernal 1.0: inference of RNA alignments. CCF TCBI. 25, 1335–1337. doi: 10.1093/bioinformatics/btp157
Nguyen, M. H., Kaul, D., Muto, C., Cheng, S. J., Richter, R. A., Bruno, V. M., et al. (2020). Genetic diversity of clinical and environmental Mucorales isolates obtained from an investigation of mucormycosis cases among solid organ transplant recipients. Microb. Genomics. 6:e000473. doi: 10.1099/mgen.0.000473
Petersen, T. N., Brunak, S., Von Heijne, G., et al. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Reiner, J., Pisani, L., Qiao, W., Singh, R., Yang, Y., Shi, L., et al. (2018). Cytogenomic identification and long-read single molecule real-time (SMRT) sequencing of aBardet–Biedl syndrome 9(BBS9). NPJ Genom. Med. 3, 1–5. doi: 10.1038/s41525-017-0042-3
Saha, S., Bridges, S., Magbanua, Z. V., and Peterson, D. G. (2008). Empirical comparison of ab initio repeat finding programs. NAR 36, 2284–2294. doi: 10.1093/nar/gkn064
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Seringhaus, M., Paccanaro, A., Borneman, A., Snyder, M., and Gerstein, M. (2006). Predicting essential genes in fungal genomes. Genome Res. 16, 1126–1135. doi: 10.1101/gr.5144106
Stanke, M., Diekhans, M., Baertsch, R., and Haussler, D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. CCF TCBI. 24, 637–644. doi: 10.1093/bioinformatics/btn013
Su, J. E., Pan, F. L., Ma, M., et al. (2018). Effects of different curing process on tobacco mildew disease. Tianjin Agricultural Sciences. 24, 51–55. doi: 10.3969/j.issn.1006–6500.2018.02.013
Surhone, L. M., Timpledon, M. T., Marseken, S. F., et al. (2010). Quicksand: hydrogel, granular matter, mercury (element), water circulation, non-newtonian fluid, viscosity. United Kingdom: Betascript Publishing.
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. PNAS 101, 11030–11035. doi: 10.1073/pnas.0404206101
Von Eiff, C., McNamara, P., Becker, K., Bates, D., Lei, X. H., et al. (2006). Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J. Bacteriol. 188, 687–693. doi: 10.1128/JB.188.2.687-693.2006
Wang, H. C., Huang, Y. F., Tang, X. G., Cai, L. T., Chen, Q. Y., Wang, M. S., et al. (2016). Leaf and stem rot of tobacco (Nicotiana tabacum) caused by Rhizopus oryzae in closed curing barns in Guizhou Province of China. Plant Dis. 100, 536–536. doi: 10.1094/PDIS-08-15-0935-PDN
Wang, H. C., Huang, Y. F., Xia, H. Q., Wang, J., Wang, M. S., Zhang, C. Q., et al. (2015). Phenotypic analysis of Alternaria alternata, the causal agent of tobacco Brown spot. Plant Pathology J. 14, 79–85. doi: 10.3923/ppj.2015.79.85
Wang, H. C., Li, L. C., Cai, B., Cai, L. T., Chen, X. J., Yu, Z. H., et al. (2018b). Metabolic phenotype characterization of Botrytis cinerea, the causal agent of Gray Mold. Front. Microbiol. 13, 9–470. doi: 10.3389/fmicb.2018.00470
Wang, M. S., Wang, H. C., Huang, Y. F., et al. (2015). Phenotypic analysis of Phytophthora parasitica by using high throughput phenotypic microarray. Acta Microbiol Sin. 55, 1356–1363. doi: 10.13343/j.cnki.wsxb.20150036
Wang, H. C., Zhang, M., Zhang, C. Q., et al. (2018a). Pathogen detection, carbon metabolic phenotype analysis of Rhizopus oryzae from tobacco and its sensitivity to seven fungicides. J. Pestic. Sci. 20, 743–748. doi: 10.16801/j.issn.1008-7303.2018.0095
Watanabe, T. (2002). Pictorial atlas of soil and seed fungi: Morphologies of cultured fungi and key to species. United States: CRC press.
Xu, D., Li, R. G., Xiao, S. J., et al. (2020). Host range of the lily pathogen Rhizopus oryzae. J. yi chun University. 42, 95–99.
Zhang, Y. Q., Li, Q. S., Wang, C. Y., et al. (2018). Study on color parameters and curing characteristics of flue-cured tobacco leaves in process ofmaturity. Southwest China J Agric Sci. 31, 62–67. doi: 10.16213/j.cnki.scjas.2018.1.011
Keywords: Rhizopus oryzae, whole genome sequence, tobacco pole rot, biolog phenotype MicroArray, metabolic fingerprint
Citation: Li Z, Shi C-h, Huang Y, Wang H-c, Li W-h and Cai L-t (2023) Phenotypic analysis and genome sequence of Rhizopus oryzae strain Y5, the causal agent of tobacco pole rot. Front. Microbiol. 13:1031023. doi: 10.3389/fmicb.2022.1031023
Edited by:
Stepan Toshchakov, Kurchatov Institute, RussiaReviewed by:
Feng Zhang, Nanjing Agricultural University, ChinaAlexander N. Ignatov, Peoples' Friendship University of Russia, Russia
Copyright © 2023 Li, Shi, Huang, Wang, Li and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cai-hua Shi, ✉ c2hpY2FpaHVhMTk4MEAxMjYuY29t; Han-cheng Wang, ✉ eGlhb2JhaXlhbmcxMjZAaG90bWFpbC5jb20=
 Zhen Li
Zhen Li Cai-hua Shi1,3*
Cai-hua Shi1,3*