- College of Animal Science and Technology, Yangzhou University, Yangzhou, China
Enterobacter cloacae is widely distributed in the aquatic environment, and has been determined as a novel pathogen of various aquatic animals recently. Our previous studies have indicated E. cloacae caused repeated infections in Macrobrachium rosenbergii, suggesting a high survival ability of the bacteria, and rpoS gene has been known to regulate stress response and virulence of many bacteria. In this study, the E. cloacae-rpoS RNAi strain was constructed by RNAi technology, and the regulation role of rpoS in stress resistance and virulence of E. cloacae was explored by transcriptomic and phenotype analysis. The transcriptome analysis showed a total of 488 differentially expressed genes (DEGs) were identified between rpoS-RNAi and wild-type strains, including 30 up-regulated genes and 458 down-regulated genes, and these down-regulated DEGs were mainly related to environmental response, biofilm formation, bacterial type II secretory system, flagellin, fimbrillin, and chemotactic protein which associated with bacterial survival and virulence. The phenotype changes also showed the E. cloacae-rpoS RNAi strain exhibited significantly decreasing abilities of survival in environmental stresses (starvation, salinity, low pH, and oxidative stress), biofilm production, movement, adhesion to cells, pathogenicity, and colonization to M. rosenbergii. These results reveal that rpoS plays an important regulatory role in environmental stress adaptation and virulence of E. cloacae.
Introduction
Enterobacter cloacae an enteric bacterium that belongs to Enterobacteriaceae, is widely distributed in various aquatic and terrestrial environments (Krzymińska et al., 2010), and has been considered as an opportunist pathogen causing variety of infections both in human and animals (Lee et al., 2017). Furthermore, the control of E. cloacae mainly relies on antibiotics, which leads the emergence of multi-resistant E. cloacae strains (Band et al., 2016). Recently, E. cloacae has been recognized as an aquatic pathogen to several aquatic animals, e.g., Macrobrachium rosenbergii (Gao et al., 2019, 2020, 2021), Procambarus clarkia (Dong et al., 2020), Mugil cephalus (Thillai et al., 2008), and Coryphaena hippurus (Hansen et al., 1990). Our previous studies also showed the E. cloacae caused repeated infections in M. rosenbergii (Gao et al., 2019, 2021), suggested that E. cloacae has developed survival strategies to adapt various stresses in the aquatic environment. Pathogenic bacteria of aquatic animals usually encounter various stresses in the aquatic and host environments, such as nutrient starvation, oxidative stress, changes in pH, temperature and osmolality, etc. Hence, the bacteria overcome these environmental stresses require self-regulation of major genes expression that commonly controlled by sigma factors (Qiu et al., 2013).
Sigma factors are considered as important regulators involved in transcription and regulation of various genes, which regulate many cellular activities, such as growth, environmental stresses adaptation, biofilm formation, and virulence (Feng et al., 2021). Among these, the alternative sigma factor (RpoS) has been widely considered as a key regulatory factor, which is responsible for regulating stress response in many bacteria (Landini et al., 2014). Previous studies showed that RpoS plays important roles in tolerance of high temperature, salt stress, acid stress and many other environmental stresses, e.g., the rpoS mutant strains of the bacteria such as Vibrio alginolyticus, Edwardsiella tarda, Escherichia coli, Shewanella baltica, Serratia plymuthica, and Yersinia pseudotuberculosis exhibited significantly decreasing abilities of survival in various stresses compared to the wild type (Battesti et al., 2011; Liu et al., 2014, 2016; Guan et al., 2015; Huang et al., 2019; Zhang et al., 2021).
In addition to its role in the stress response, RpoS also plays important regulatory roles in virulence of pathogenic bacteria (Yin et al., 2018). RpoS is responsible for regulating virulence by increasing resistance to host environment stresses and regulating expression of virulence-related genes in serval bacteria (Dong and Schellhorn, 2010). Hülsmann et al. (2003) reported that RpoS could positively regulate many extracellular enzymes activities, which may be required for invasion to host. Similarly, Ma et al. (2009) reported that the rpoS mutant of Vibrio anguillarum decreased markedly production of extracellular enzymes and virulence to zebra fish. RpoS also controls the expression of various virulence genes, e.g., RpoS could positively regulate the expression of spvR and spvABCD virulence genes, which are responsible for causing disease in mice and humans (Fang et al., 1992; Norel et al., 1992).
Although RpoS has been recognized as an important regulator in stress resistance and virulence of many bacteria, its regulatory mechanism in E. cloacae is still scarce. In this study, the knockdown of rpoS in E. cloacae was constructed by RNA interference (RNAi), growth curves, stress response, biofilm formation, adhesion ability, virulence, and transcriptomic changes between rpoS-RNAi and wild strains, were investigated. Our results will contribute to reveal the regulatory mechanisms of rpoS in stress response and virulence of E. cloacae, and provide theoretical support for prevention and control the diseases caused by E. cloacae in aquatic animals.
Materials and methods
Bacterial strains and growth conditions
The E. cloacae XL3-1 strain was isolated from the diseased M. rosenbergii in Gaoyou County of Jiangsu Province (Gao et al., 2019), and incubated in LB medium at 28°C with shaking at 180 rpm for 18 h. E. coli DH5α (TransGen Biotech, Beijing, China) incubated in LB medium at 37°C with shaking at 180 rpm for 18 h.
Stable gene silence
The E. cloacae-rpoS RNAi strain was performed following previously described methods (Luo et al., 2019). Five short hairpin RNA sequences, respectively, targeting the rpoS (Genbank accession number: Y13230.1) were designed by Invitrogrn Block-iT RNAi Designer and synthetized by Shanghai Generay Biotech Co., Ltd. (Shanghai China). The designed synthetic shRNA was annealed, and then ligated to the pCM130/tac plasmid after digestion with the enzymes NsiI and BsrGI (New England Biolabs) to construct pCM130/tac-rpoS. The recombinant plasmid was transformed into E. coli DH5α by heat shock, and then extracted and introduced into E. cloacae by electroporation. The rpoS-RNAi E. cloacae was screened by tetracycline (10 μg/mL) and qRT-PCR.
Transcriptome sequencing and data analysis
Total RNA was extracted from the wild-type and rpoS-RNAi E. cloacae (n = 3) for cDNA preparations, and the contaminating genomic DNA was removed by RNase-free DNase I. Further, the RNA quantity and quality were assessed using Agilent 2,100 Bioanalyzer (Agilent technologies, United States) and NanoDrop 2000 (Thermo Fisher Scientific, United States). Then, rRNA from the total RNA of each sample was removed, followed by RNA fragmentation. The first strand cDNA was synthesized with random primers and M-MuLV reverse transcriptase. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Prior to final amplification, the synthesized cDNA fragments were purified, end repaired, ligated to sequencing adapters, and selectively degraded with USER enzyme. After that, sequencing was carried out on the Hiseq2000 (Beijing Novogene Co., Ltd., Beijing China). Then the clean reads were obtained from raw reads by removing adapters, higher N rate reads, and low quality reads. Then the sequencing reads were compared with the reference genome (GenBank accession number: CP012162.1) using Bowtie software. The gene expression levels were calculated by using FPKM (Fragments Per Kilobase of transcript per Million mapped reads) method. The criteria of differentially expressed genes (DEGs) was log2(FoldChange)| > 0 and padj <0.05. The enrichment analysis of DEGs were conducted by using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Conesa et al., 2005; Kanehisa et al., 2014).
Quantitative real-time PCR analysis
The qRT-PCR was carried out using Thermofisher QuantStudio Real-Time PCR System PCR System with ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing Co., Ltd., China), and the primer sequences are displayed in Supplementary Table S1. The 16S rRNA was chosen as an internal control, and relative mRNA expression was calculated by the 2−ΔΔCt method.
Growth assay
Overnight cultures of wild-type and rpoS-RNAi E. cloacae were adjusted to OD600 = 0.5, diluted 1:100 into LB medium, and then incubated at 28 h with shaking for 24 h. The values of OD600 were recorded at 2-h intervals, the growth curves were conducted both the wild-type and rpoS-RNAi strains.
Motility assay
The concentration of overnight cultured wild-type and rpoS-RNAi E. cloacae was adjusted to OD600 = 0.3. 1 μl of the bacterial suspension was spotted into LB medium with 0.4% agar, incubated at 28°C for 24 h, and then the diameters of the colonies surrounding the punctured portion of the agar media were measured (Wang et al., 2019).
Biofilm formation
The biofilm formation assay for E. cloacae was conducted as described by Huang et al. (2019). The overnight cultures of wild-type and rpoS-RNAi E. cloacae were adjusted OD600 = 0.2. Then 100 μl of bacterial suspension was added to a 96-well plate and incubated at 28°C for 24 h. After incubation, the plate was rinsed three times with sterile PBS (pH 7.4), stained with 200 μl 1% crystal violet for 15 min, and then the plate was rinsed again and air-dried. Finally, 200 μl of 95% ethanol solution was added to solubilize the stained biofilm, which was measured at OD590 nm using a microplate reader. The above results were normalized by the bacterial numbers and conducted for six replicates.
Adhesion assays
Overnight cultures of wild-type and rpoS-RNAi E. cloacae were adjusted to OD600 = 1, the bacteria were 10-fold serially diluted using sterilized PBS (pH 7.4), and then then bacterial colonies were counted by plating 100 μl of bacteria solution on LB agar. For adhesion assay, the human renal epithelial cells (293 T) were subsequently infected at a multiplicity of infection (MOI) of 10 for 2 h. The infected cells were rinsed with PBS three times to remove non-adherent bacteria. Adherent bacteria were isolated from 293 T cells using 0.1% Triton-X 100. The serial dilution of collected bacterial suspension was coated onto LB plates, and then bacterial colonies were counted. The percentage of adhesion was calculated as adherent CFU × 100/total inoculum CFU. The assay was performed in triplicate.
Stress survival assay
The wild-type and rpoS-RNAi E. cloacae were grown in LB medium at 28°C to stationary phase. For osmotic stress assay, the cultures were diluted 50-fold into containing 4% NaCl. For acid stress assay, the cultures were diluted 50-fold into LB medium which were adjusted to pH 4.0. For H2O2 stress assay, the cultures were diluted 50-fold into LB medium with 1.5 mM H2O2. In addition, the cultures of wild-type and rpoS-RNAi E. cloacae were also diluted into normal LB medium as control groups. After incubating at 28°C for 1 h, the cultures were serially diluted and plated onto LB agar plates, colonies were counted after 24 h incubation. Additionally, the starvation stress assay was performed as described by Gao et al. (2022), the starved cells were sampled at day 7, 14, 21, and 28 for colonies counts. The survival rate was calculated as follows: (CFU of stressed cells/CFU of untreated cells) × 100%. The assay was performed in triplicate.
Virulence test
To investigate pathogenicity of wild-type and rpoS-RNAi E. cloacae, the 2 months after hatching old M. rosenbergii (0.23 ± 0.05 g) were obtained from a local farm in Gaoyou county and acclimated for a week. These prawns were divided into test and control groups, the test groups were infected by exposure to 2.4 × 107 CFU/mL of wild-type and rpoS-RNAi E. cloacae, respectively, while the control group prawns were cultured in fresh water without any bacteria inoculation. The experimental M. rosenbergii were kept at 28 ± 2°C, monitored for 96 h without feeding and water change. The mortalities of prawns were monitored at 6, 12, 24, 48, 72 and 96 h, the dead M. rosenbergii were immediately removed and re-isolated to confirm the mortality caused by E. cloacae. In addition, the bacterial burden in the prawns was detected at 12, 24, and 48 h. 100 mg of prawns was homogenized in 1 ml of sterile PBS (pH 7.4), and the serial diluted homogenates were plated onto LB agar plates for bacterial colonies counting.
Statistical analysis
Statistically significant differences among wild-type and rpoS-RNAi E. cloacae groups were analyzed one-way ANOVA using SPSS 16.0 software. The results data were expressed as means ± standard deviation (SD), and p < 0.05 was considered as significant difference.
Results
Construction of the rpoS-RNAi strain
The expression of rpoS in the wild-type and rpoS-RNAi E. cloacae was compared. All of the five shRNAs significantly decreased the expression of rpoS with different efficiency, and the reduction in the expression level of rpoS-shRNA-99, rpoS-shRNA-121, rpoS-shRNA-546, rpoS-shRNA-585 and rpoS-shRNA-750 was 37.30, 68.06, 82.62, 51.09, and 74.21%, respectively (Figure 1A). Therefore, the rpoS-shRNA-546 was selected for further studies. In addition, the growth curves showed that the growth rate of rpoS silenced strain was lower than that of the wild-type (Figure 1B).
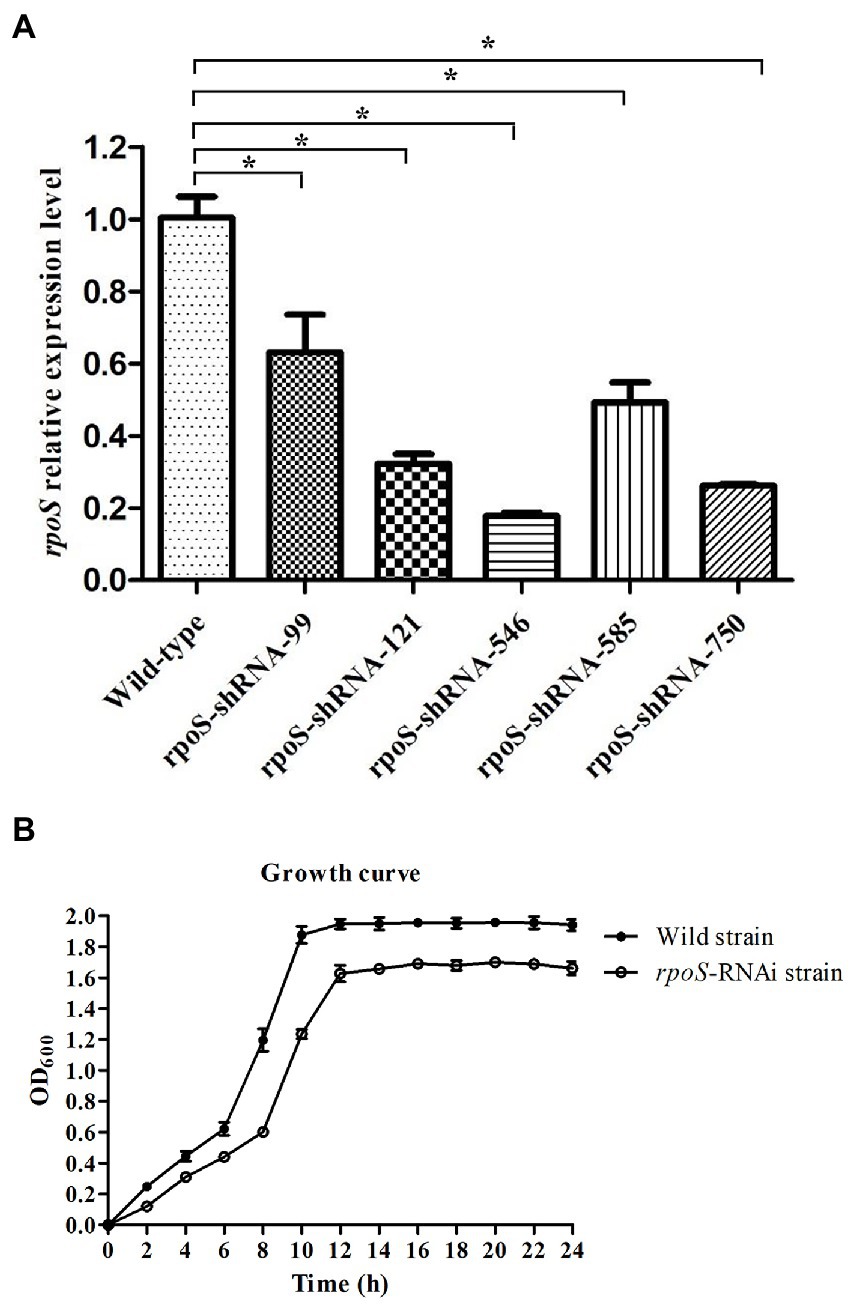
Figure 1. Construction of RNAi strains of Enterobacter cloacae. (A) Gene expression levels of rpoS after stable gene silencing, the asterisk indicated significant differences between the RNAi and wild E. cloacae, p < 0.05. (B) Growth curve of wild-type and rpoS-RNAi strains.
Transcriptome changes induced by knockdown of rpoS
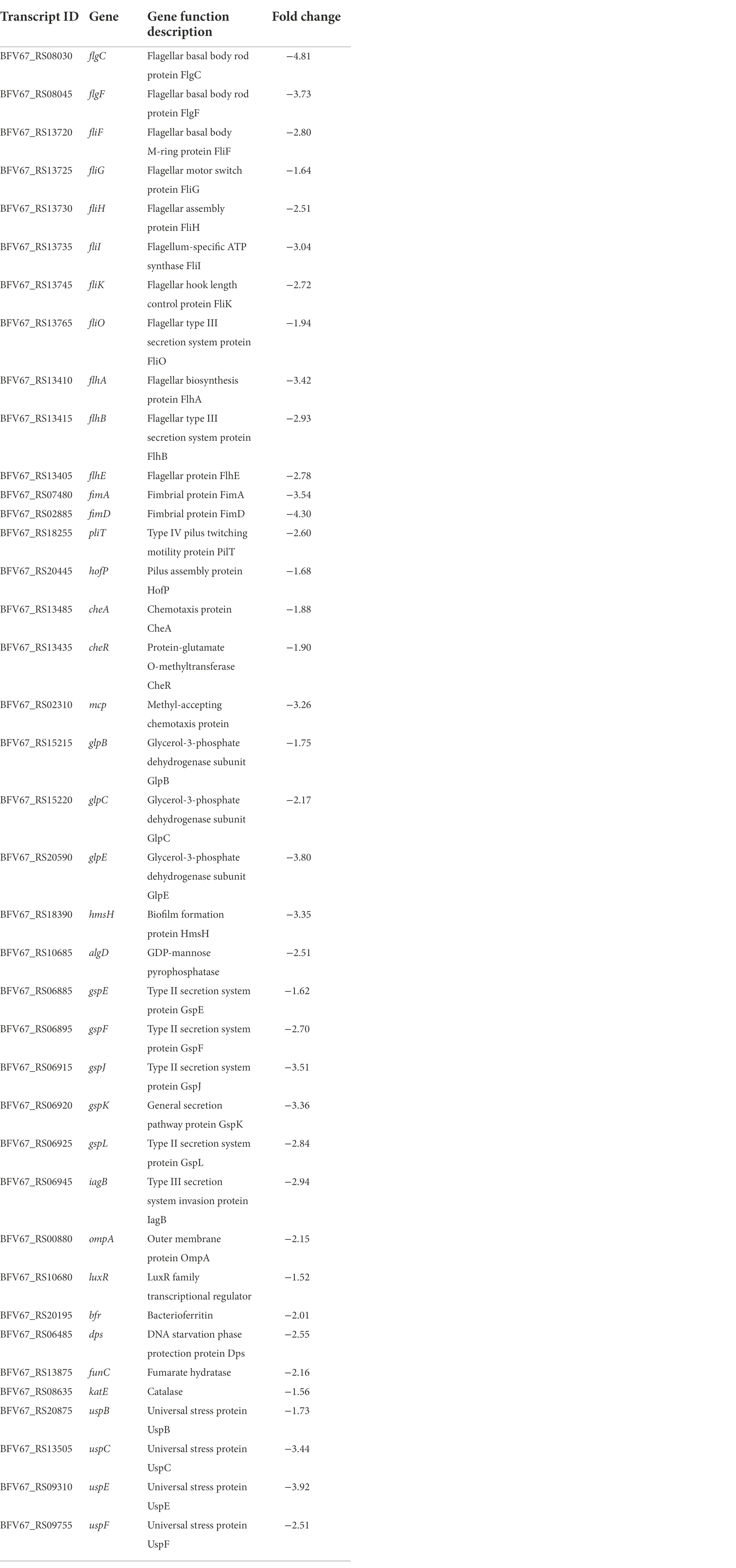
To investigate the regulatory role of rpoS in E. cloacae, transcriptome profiles of the rpoS-RNAi and wild-type E. cloacae were analyzed using RNA sequencing. After filtering through the raw reads, a total of 11,583,012, 10,789,068, 10,939,872 clean reads from rpoS-RNAi strains, 11,649,402, 9,878,616, 14,998,822 clean reads from wild-type strains were obtained, respectively (Supplementary Table S2). A total of 488 DEGs in rpoS-RNAi were identified in comparison with the wild-type strain, including 458 downregulated genes and 30 upregulated genes (Figure 2A). Among DEGs, genes responsible for resistance to environmental stress (e.g., bfr, dps, funC, katE、uspB, uspC, uspE, and uspF), flagellar assembly (flgC, flgF, fliF, fliG, fliH, fliI, fliK, fliO, flhA, flhB, and fimC), fimbria protein (fimA, fimD, pliT, and hofP), chemotaxis (cheA, cheR, and mcp), biofilm formation (glpB, glpC, glpE, hmsH, and algD), type II secretion system (gspE, gspF, gspJ, gspK, and gspL) were downregulated significantly in the rpoS-RNAi strain (Tables 1, 2), which suggested that rpoS was involved in stress response and virulence regulation of E. cloacae.
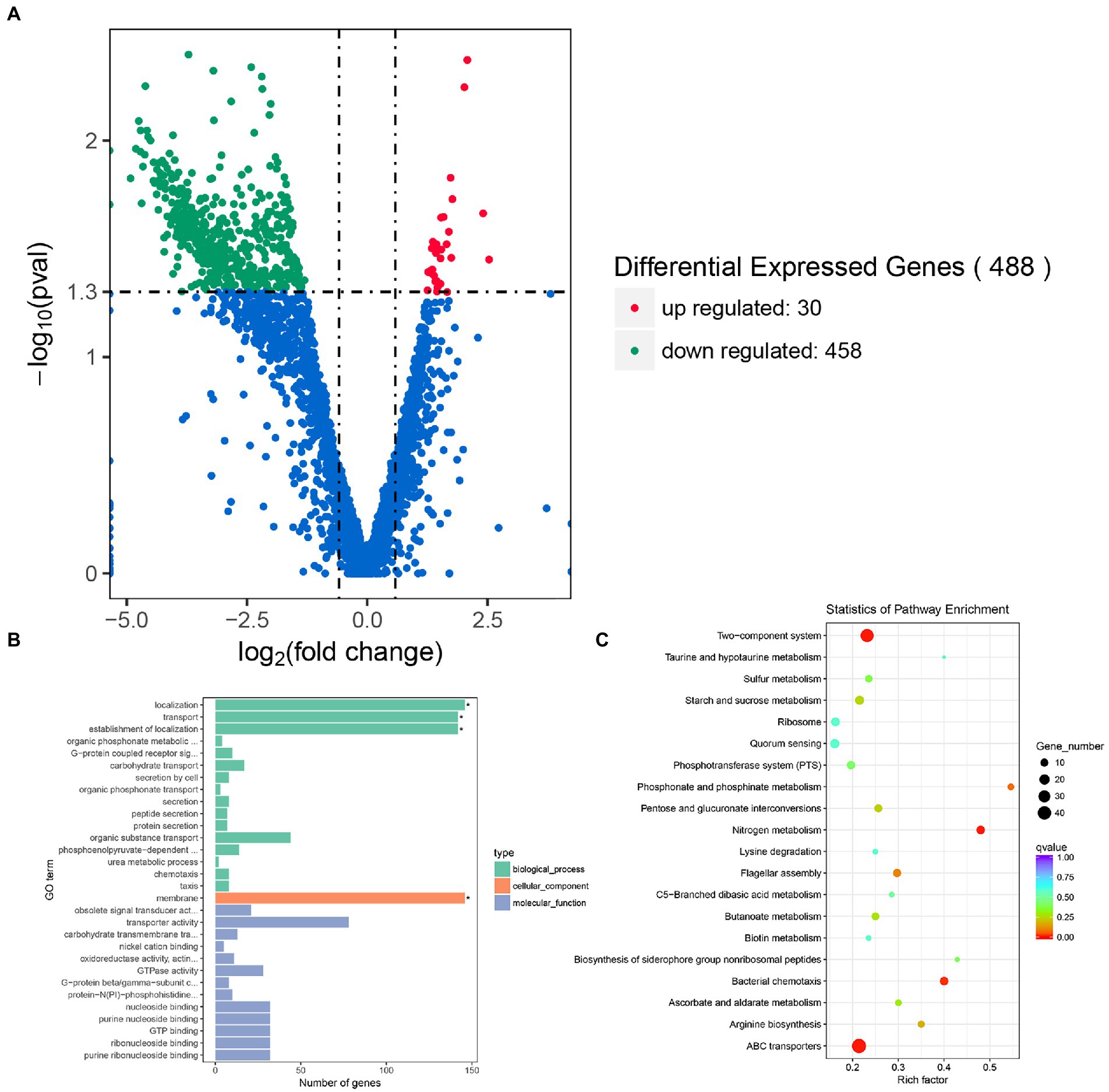
Figure 2. Transcriptomic analysis of E. cloacae between rpoS-RNAi and wild-type strains. (A) The volcano plot of DEGs, the red dots were up-regulated genes and the green dots were down-regulated genes. (B) GO analysis of differential expression genes of rpoS-RNAi and wild-type strains. (C) The Top20 KEGG enrichment pathways for DEGs, the rich factor refers to the ratio of the number of DEGs in the pathway and the number of all annotated genes in the pathway.
The functions of the 458 DEGs between the rpoS-RNAi and wild-type E. cloacae were analyzed by Go tools and categorized into different enriched functional groups, among which the important functions included localization, transport, establishment of localization, membrane, transporter activity, active transmembrane transporter activity and GTPase activity (Figure 2B). The 458 DEGs were also performed KEGG pathway enrichment analysis and enriched in 67 KEGG pathways, including two-component system, ABC transporters, flagellar assembly, bacterial chemotaxis, quorum sensing and so on (Figures 2B,C).
Effect of rpoS on the survival of Enterobacter cloacae under environmental stresses
To investigate the effect of rpoS on the survival rate of E. cloacae under environmental stress conditions, the survival rates of the rpoS-RNAi and wild-type E. cloacae under environmental stress conditions (starvation, 4% NaCl, 1.5 mM H2O2 or pH 4) were measured. As shown in Figure 3, the survival rates of the rpoS-RNAi strain under these stress conditions decreased compared with the wild-type strain. After 28 d of starvation, the survival rate of the wild-type strain was 3.50-fold higher than the rpoS-RNAi strain (Figure 3A). Similarly, After1 h of exposure to 4% NaCl, 1.5 mM H2O2, and pH 4, survival rates of the WT were 1.32, 1.42 and 1.62-fold higher than the rpoS-RNAi strain, respectively (Figures 3B–D). The results of transcriptome analysis and qPCR verification also suggested that after the expression the expression of rpoS was inhibited, the resistance to environmental stress related genes including bacterioferritin (bfr), DNA starvation phase protection protein (dps), fumarate hydratase (fumC), catalase (cat), universal stress proteins (uspB, uspC, uspE, and uspF) were significantly down-regulated (Figure 3E).
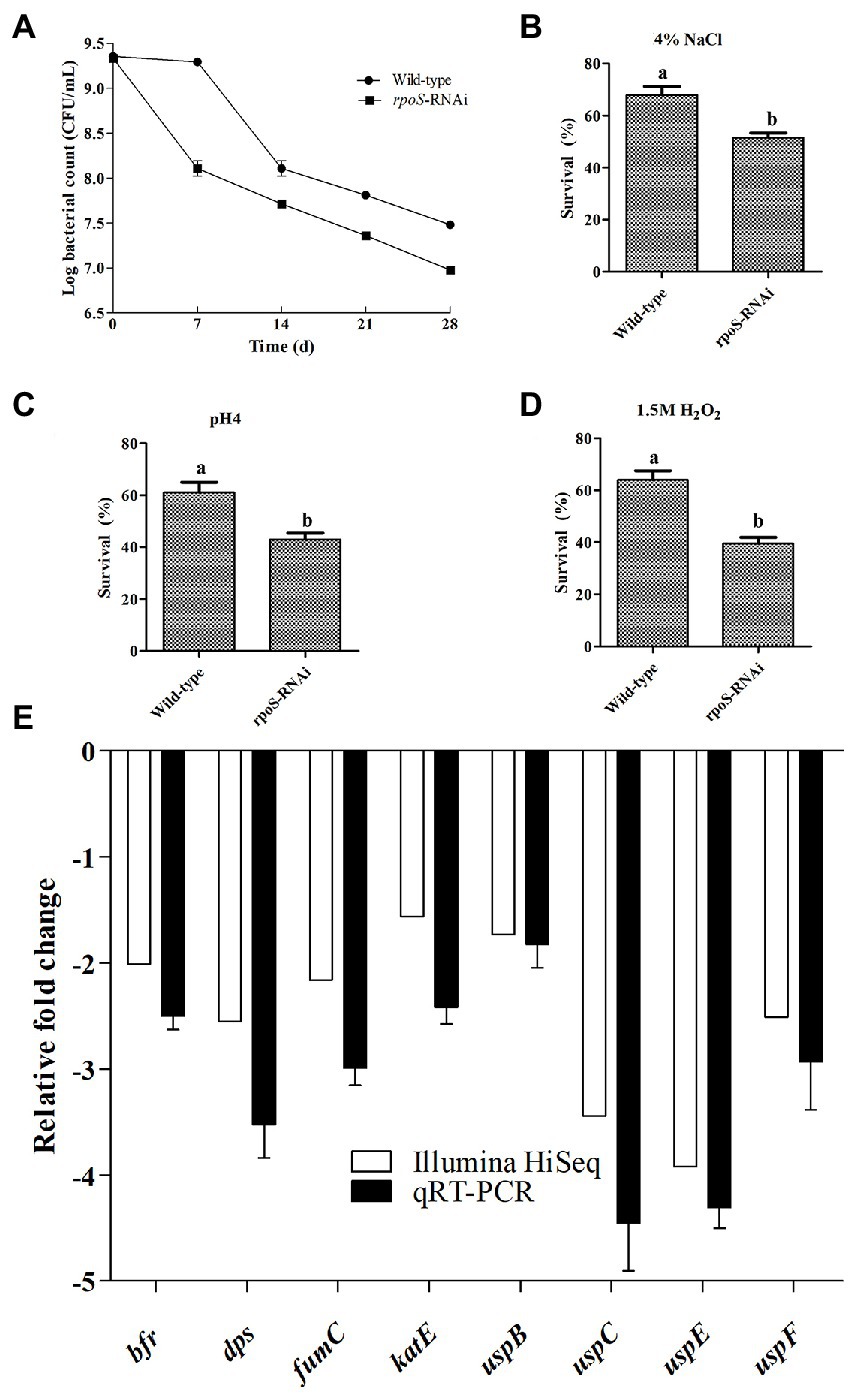
Figure 3. Effects of rpoS knockdown on stress resistance in Enterobacter cloacae. (A) Survival of wild-type and rpoS-RNAi strains under starvation. (B) Survival of wild-type and rpoS-RNAi strains in 4% NaCl. (C) Survival of wild-type and rpoS-RNAi strains in pH 4. (D) Survival of wild-type and rpoS-RNAi strains in 1.5 mmol/l H2O2. (E) Effects of rpoS on stress related genes expression in E. cloacae. The values marked with different letters (a, b) are significantly different (p < 0.05).
Effect of rpoS on biofilm formation ability of Enterobacter cloacae
The biofilm formation ability compared between the rpoS-RNAi and wild-type strains was shown in Figure 4. The ability to form biofilms was decreased significantly in the rpoS-RNAi strain compared to the wild-type strain (Figure 4A). The results of transcriptome analysis and qPCR verification also showed the biofilm formation-related genes including glpB, glpC, glpE, hmsH, and algD, were significantly down-regulated (Figure 4B), which suggested that that rpoS positively regulated the expression of the biofilm formation-related genes.
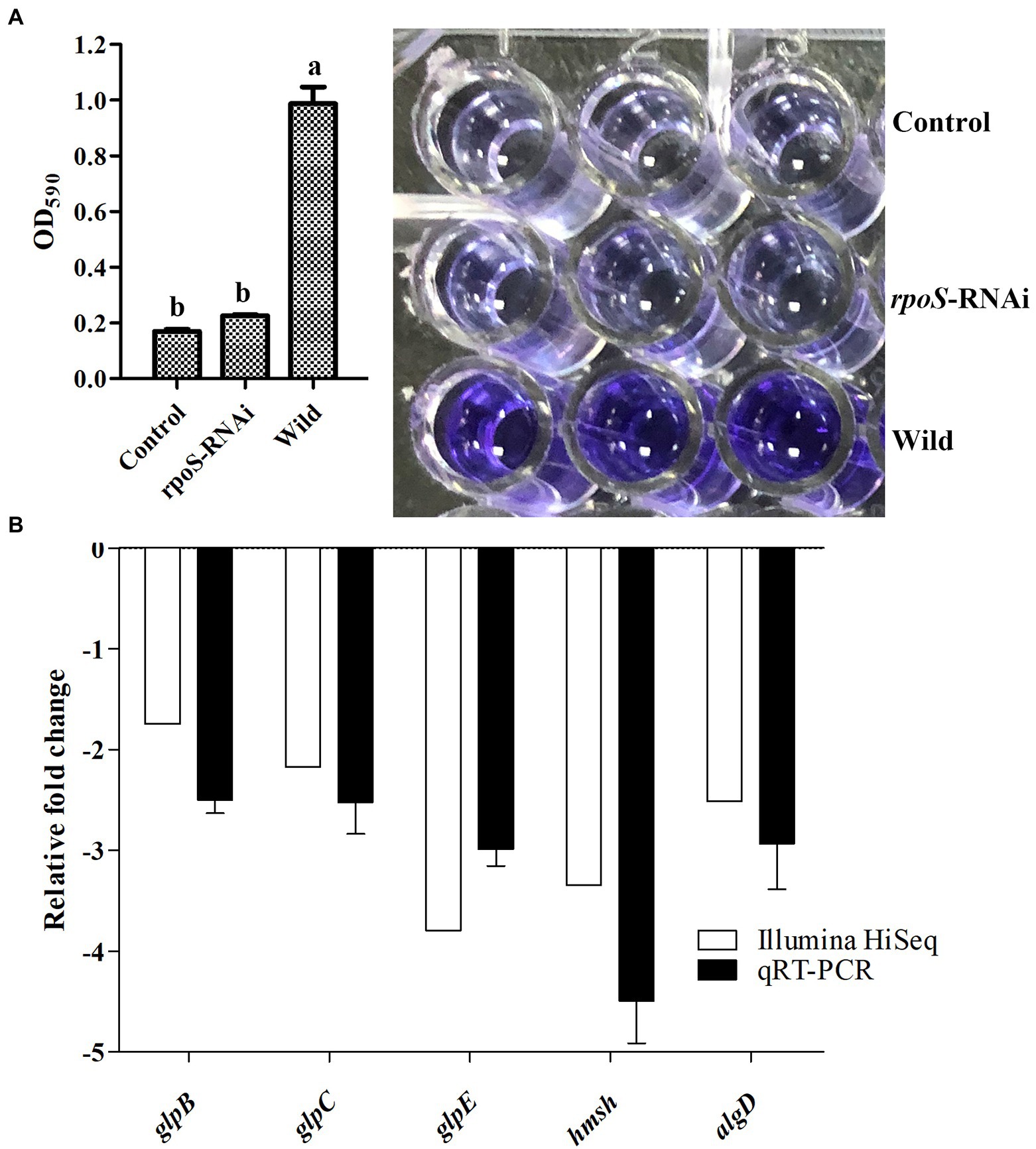
Figure 4. Effect of rpoS on biofilm formation in E. cloacae. (A) Biofilm formation ability of wild-type and rpoS-RNAi strains. (B) Expressions of biofilm formation related genes in rpoS-RNAi strain; The values marked with different letters (a, b) are significantly different (p < 0.05).
Effect of rpoS on motility of Enterobacter cloacae
The effect of rpoS expression on the motility of E. cloacae are shown in Figure 5. The rpoS-RNAi strain was motile, but the diameters of the colonies (12.0 ± 0.8 mm) after 24 h of culture were significantly smaller than that of the wild-type strain (18.7 ± 0.5 mm) (Figure 5A). In addition, the genes related to flagellar assembly (flgC, flgF, fliF, fliG, fliH, fliI, fliK, fliO, flhA, flhB, and fimC) were significantly down-regulated (Figure 5B). These results confirmed that the reduction in E. cloacae motility may be due to knockdown of rpoS.
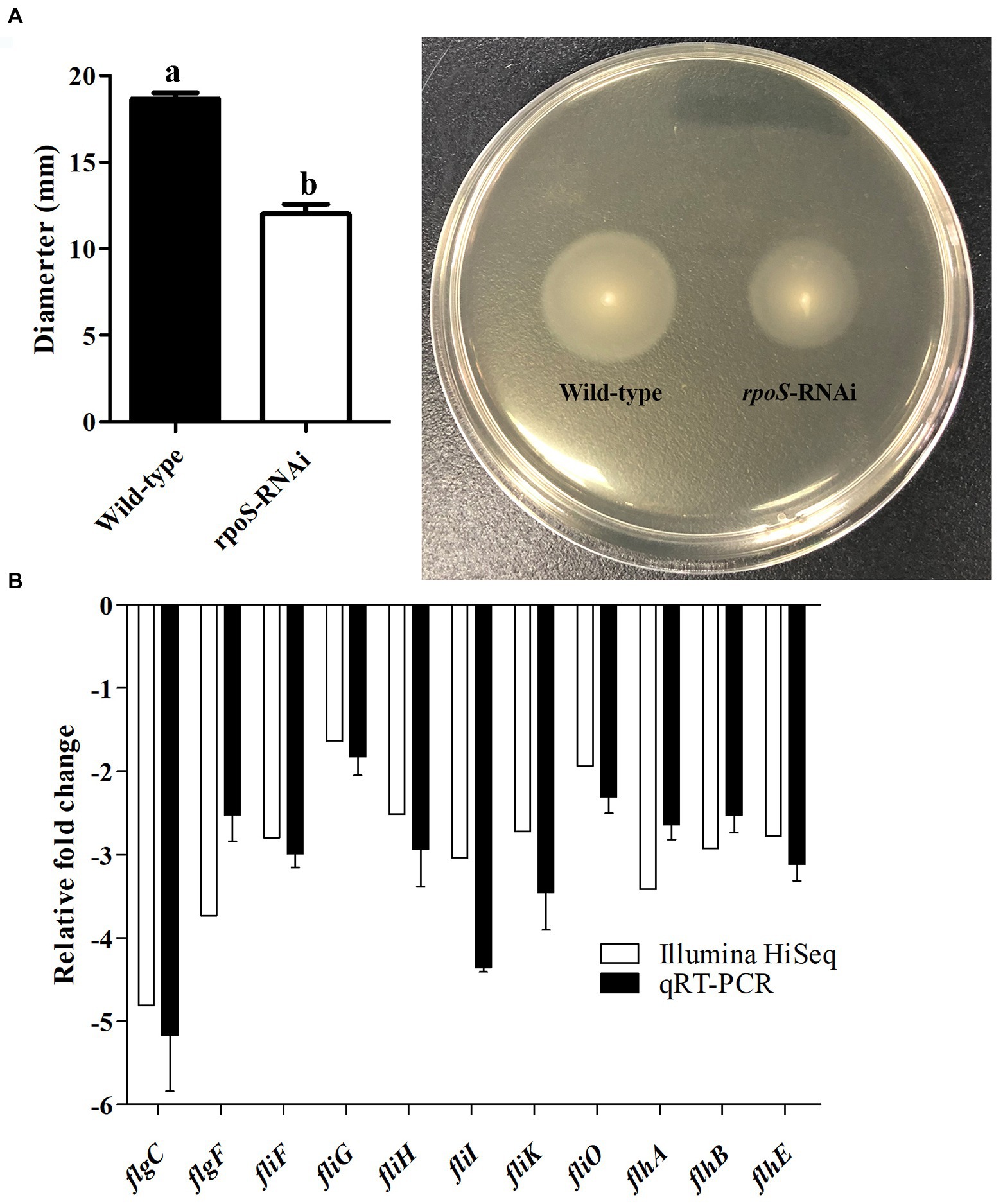
Figure 5. Effects of rpoS on motility of Enterobacter cloacae. (A) Detection of motility of E. cloacae wild-type and rpoS-RNAi strains; (B) Expressions of motility-related genes in rpoS-RNAi strain; The values marked with different letters (a, b) are significantly different (p < 0.05).
Knockdown of rpoS reduced adhesion ability of Enterobacter cloacae
Prior to studying the effects of rpoS on adhesion ability of E. cloacae, we tested the adhesion ability of the rpoS-RNAi and wild-type E. cloacae to live 293 T cells. The adhesion rate of the rpoS-RNAi strain into 293 T cells was 14.95%, while the adhesion rate of the wild-type strain into 293 T cells was 44.35% (Figure 6A). In addition, the results of transcriptome analysis and qPCR verification also showed some genes related to pilus biogenesis (fimA, fimD, pliT, hofP), bacterial chemotaxis (cheA, cheR, mcp) were significantly down-regulated after the expression inhibition of rpoS (Figure 6B). These results suggested that rpoS involved in the process of E. cloacae adhesion.
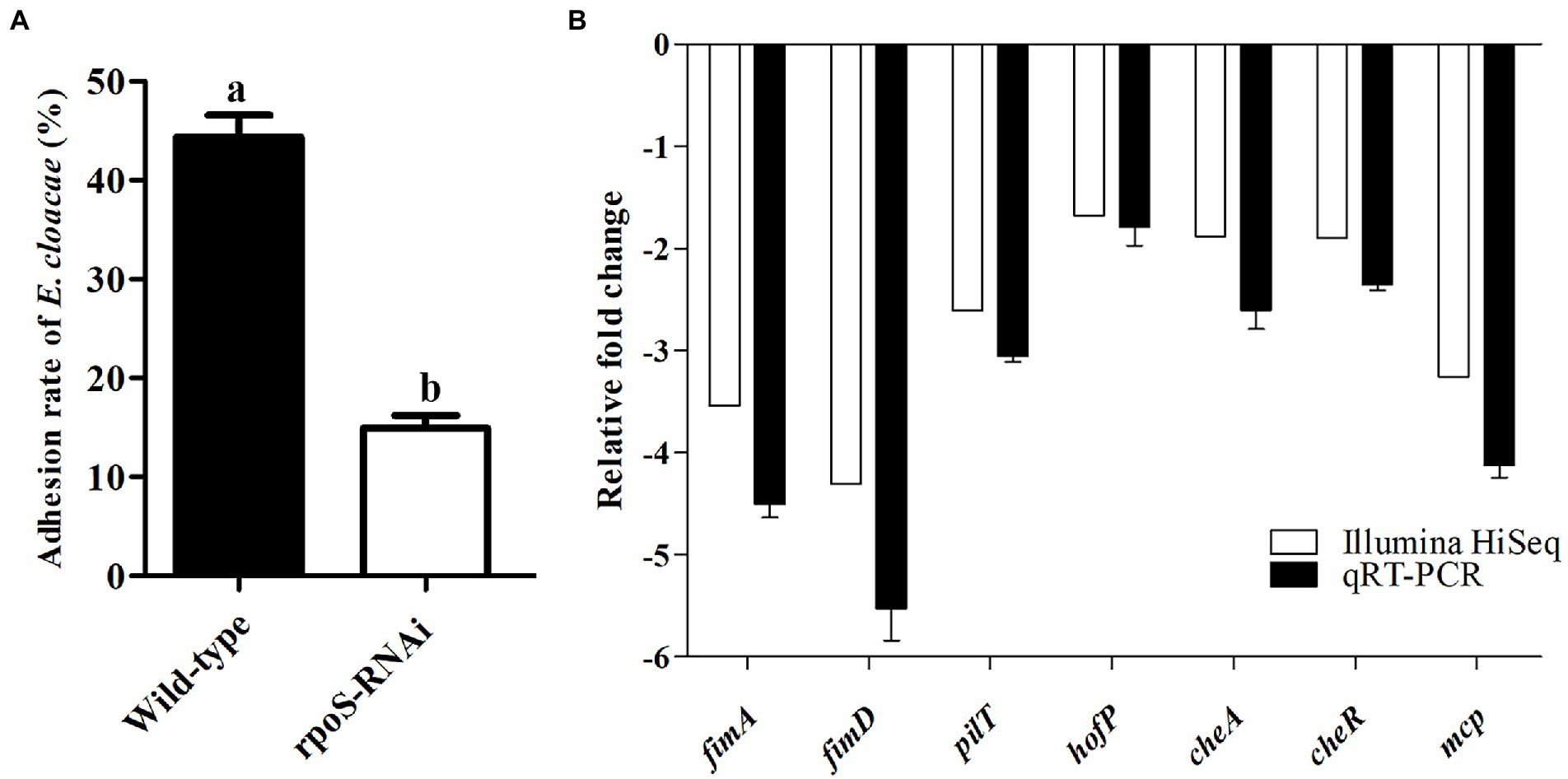
Figure 6. Effects of rpoS on adhesion of Enterobacter cloacae. (A) The adhesion rate of E. cloacae wild-type and rpoS-RNAi strains to 293 T cells; (B) Expressions of adhesion-related genes in rpoS-RNAi strain; The values marked with different letters (a, b) are significantly different (p < 0.05).
Effect of rpoS on virulence of Enterobacter cloacae
Comparison of pathogenicity between the rpoS-RNAi and wild-type E. cloacae were showed in Figure 7. The virulence test showed the M. rosenbergii injected with the wild-type E. cloacae showed 58% survival rate at 96 hpi, while the M. rosenbergii injected with the rpoS-RNAi E. cloacae showed 20% survival rate (Figure 7A). Besides, the bacterial burden of the rpoS-RNAi E. cloacae in M. rosenbergii (4.29 Log10 CFU/g) was significantly lower than that of the wild-type E. cloacae (4.65 Log10 CFU/g) at 48 hpi (Figure 7B). Additionally, the expression of multiple virulence genes, such as gspE, gspF, gspJ, gspK, gspL, iagB, luxR, and ompA were significantly down-regulated in the rpoS-RNAi strain (Figures 7B,C). These results suggested that rpoS was likely involved in the regulation of bacterial virulence.
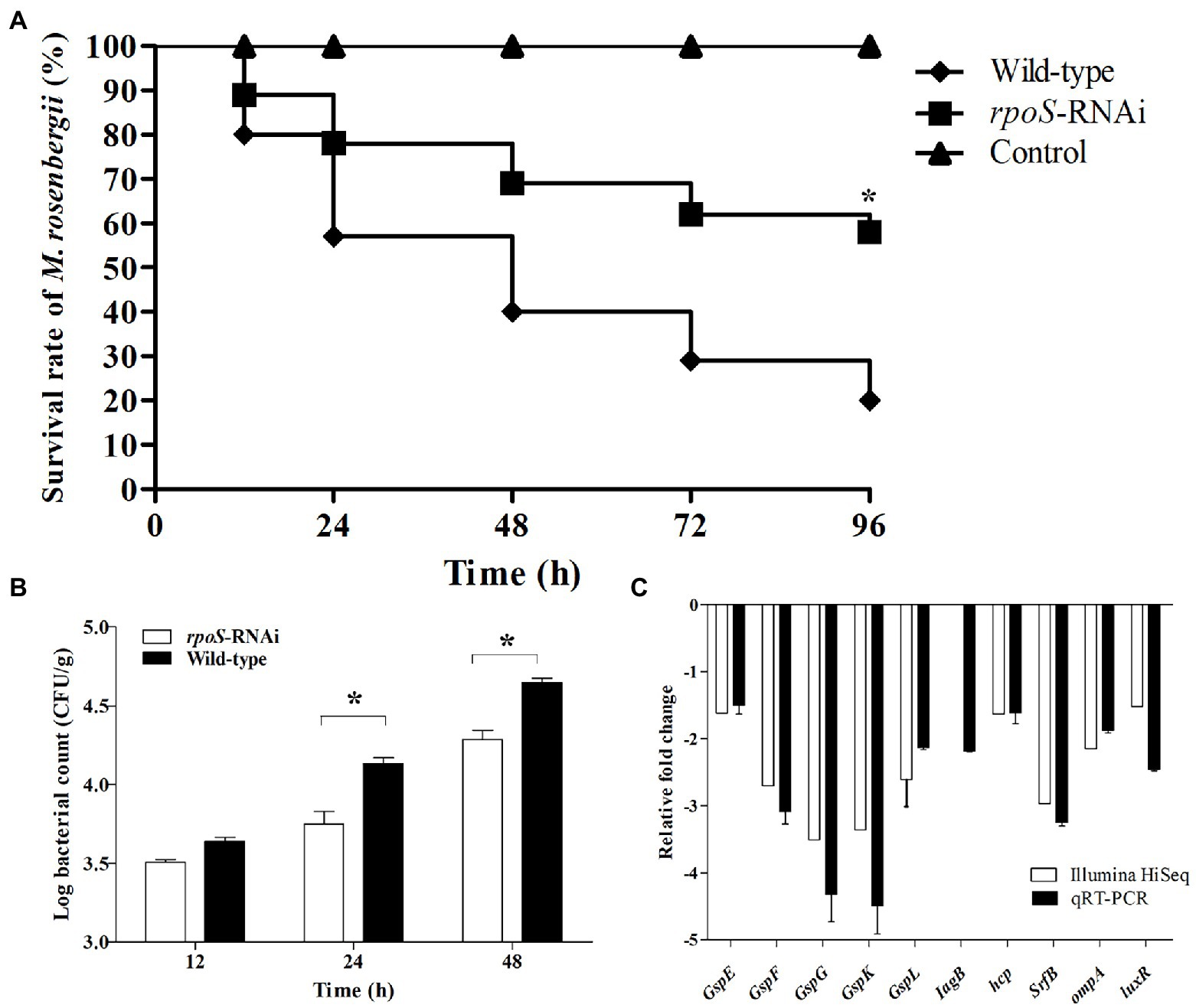
Figure 7. Effects of rpoS on virulence of Enterobacter cloacae. (A) Survival of M. rosenbergii after infected with E. cloacae wild-type and rpoS-RNAi strains; (B) Bacterial burden of M. rosenbergii after infected with E. cloacae wild-type and rpoS-RNAi strains; (C) Expressions of virulence-related genes in rpoS-RNAi strain; The asterisk indicated significant differences between the E. cloacae wild-type and rpoS-RNAi infected groups (p < 0.05).
Discussion
It is well known that aquatic pathogenic bacteria are frequently subjected to a variety of stresses in the aquatic environments or host systems, and must adapt to survive in these environmental stresses, especially during infection (Sun et al., 2016; Awan et al., 2018; Gao et al., 2018, 2022). As a result, to investigate the environmental adaptation mechanism of bacterial pathogens, many studies focused to identify the key stress resistance related genes (Kamareddine et al., 2018; Zhan et al., 2021). Among these, RpoS has been characterized as an alternative sigma factor responsible for the regulation of stress response genes in many bacteria (Guo et al., 2019; Fernández-Gómez et al., 2020). The rpoS gene encodes RpoS, and has been proved to be critical for adaptation to environment stresses many bacteria, but limited information is available regarding the role of rpoS in E. cloacae. To address this issue, the phenotypes and transcriptome of wild-type and rpoS-RNAi strains were compared to clarify the role of rpoS in the E. cloacae.
In this study, the transcriptome analysis generated a novel dataset describing the functional response of E. cloacae after knockdown of rpoS. Our results suggested that the rpoS may regulate the expression of at least 488 genes, including 458 downregulated genes and 30 upregulated genes. These DEGs were mainly were involved into stress resistance, flagellar assembly, bacterial chemotaxis, biofilm formation, two-component system, quorum sensing, adherence, and secretion, etc., which suggested that RpoS is a critical regulator of stress resistance and virulence in E. cloacae. The RpoS can positively regulated a lot of stress resistance related genes in our study as previously reported in other bacterial, e.g., Weber et al. (2005) reported that RpoS controls the expression of approximately 10–23% of all genes in E. coli, and these genes were involved in responses to involved in responses to varying environmental conditions. Meanwhile, RpoS has been also shown to regulate the expression of many virulence genes in several pathogens (Chang and Lee, 2018; Yin et al., 2018). For example, RpoS could activate the expression of the plasmid-borne spvR and spvABCD related virulence in Salmonella typhimurium (Waterman and Small, 2003). Additionally, the analysis of KEGG enrichment may help us to understand the molecular regulation mechanism of rpoS in E. cloacae. KEGG analysis in our study showed that the DEGs of the rpoS-RNAi and wild-type strain were mainly enriched in two-component system, ABC transporters, flagellar assembly, bacterial chemotaxis and quorum sensing KEGG pathways, which associated with bacterial survival and virulence (Higgins, 2001; Joelsson et al., 2007; Reading et al., 2009; Wang et al., 2015). These results indicated that rpoS was involved in stress response and virulence regulation of E. cloacae.
To further determine the functional roles of rpoS in survival of E. cloacae, the effects of rpoS on survival under stress conditions and the biofilm formation were investigated. In this study, we found that in E. cloacae, knockdown of rpoS led to decreased survival rates after starvation, acid, osmotic, and oxidative stresses. Similar results were also observed in Yersinia pseudotuberculosis, rpoS mutation led to decreased survival rates after oxidative, acid, osmotic and 42°C heat shock stresses (Guan et al., 2015). Zhan et al. (2021) also reported that membrane formation, acid tolerance and drying tolerance ability was decreased significantly in the mutant strain ΔrpoS of Cronobacter sakazakii. As expected, knockdown of rpoS in E. cloacae also resulted in decreased expression of the majority of RpoS-dependent stress resistance related genes (bfr, dps, funC, katE、uspB, uspC, uspE, uspF). Furthermore, the previous studies have showed that rpoS is involved in biofilm production in many bacteria, and biofilm formation could enhance the resistance to natural environment stresses and host immune killing (Kulikalova et al., 2014; Beshiru and Igbinosa, 2018; Olwal et al., 2018). Our studies also showed that knockdown of rpoS could decrease the biofilm production in E. cloacae, which demonstrated that rpoS plays an important role in E. cloacae biofilm production. Additionally, this study suggested that rpoS can positively regulate the expression of biofilm formation related genes, such as glpB, glpC, glpE, hmsH, and algD. The results of the present study indicated that rpoS has an important effect on the regulation of stress response genes in E. cloacae, and thus allows the bacteria to survive in adverse environments.
In addition to regulating environmental resistance functions, RpoS is essential for virulence regulation in several pathogens (Fang et al., 1992; Dong and Schellhorn, 2010; Huang et al., 2019). In this study, the effects of rpoS on E. cloacae virulence were evaluated by motility test, adhesion test and virulence test. It is well known that motility is important for colonization, adhesion and causing disease of bacteria. Our studies showed that RNAi-treated bacteria exhibited significantly impaired abilities of motility, consistent with the findings in V. vulnificus, P. aeruginosa, and Y. pseudotuberculosis (Dong and Schellhorn, 2010; Guan et al., 2015; Huang et al., 2019). Meanwhile, we demonstrated that rpoS can positively regulate the flagellum expression to control motility in E. cloacae, consistent with previous findings that rpoS is necessary for motility (Wang et al., 2012). Furthermore, adhesion of pathogenic bacteria to the mucus of host animals is often the initial step of infection, and rpoS have been proved to regulate adhesion in other bacteria (Adnan et al., 2017). In the present study, the knockdown of rpoS can significantly reduce the adhesion, consistent with that rpoS can positively regulate adhesion in other bacteria (Huang et al., 2019). Finally, the virulence test showed that the rpoS-RNAi E. cloacae exhibited attenuated pathogenicity and colonization to M. rosenbergii. Similar results were also observed in Vibrio anguillarum, σS mutation led to reduced production of hemolysin, catalase and phospholipase. rpoS mutation led to was reduced production of hemolysin, catalase and phospholipase, as well as decreased virulence to zebra fish (Ma et al., 2009). These findings revealed that rpoS was involved in the regulation of E. cloacae virulence.
In conclusion, these findings suggest the rpoS gene not only contributes survival under adverse environments, but also is crucial for the virulence regulation in E. cloacae. The results will provide a better understanding in environmental adaptation and pathogenesis mechanism of E. cloacae, and provides reference for the prevention of E. cloacae caused diseases in aquaculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in NCBI with BioProject ID PRJNA857757.
Ethics statement
All treatments of prawns in this study were strictly in accordance with the guidelines of Animal Experiment Ethics Committee of Yangzhou University. The protocol was approved by Animal Experiment Ethics Committee of Yangzhou University.
Author contributions
XG conceived, designed, and carried out the experiments, analyzed the data, and wrote the manuscript. QQ, YZ, ZC, JX, WX, QJ, and JW participated in performing experiments, and analyzing the data. XZ participated in designing the experiments and revising the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Fund (Nos. 31972830 and 32202982), Natural Science Foundation of Jiangsu Province (no. BK20220584), earmarked fund for Jiangsu Agricultural Industry Technology System (JATS [2022] 501, JATS [2022] 504), “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS [2021] 120), Project of Major Agricultural Technology Cooperative Promotion in Jiangsu Province (no. 2021-ZYXT-05-4).
Acknowledgments
We would like to express our great appreciation to Qingpi Yan for providing the pCM130/tac plasmid.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1030955/full#supplementary-material
References
Adnan, M., Sousa, A. M., Machado, I., Pereira, M. O., Khan, S., Morton, G., et al. (2017). Role of bolA and rpoS genes in biofilm formation and adherence pattern by Escherichia coli K-12 MG1655 on polypropylene, stainless steel, and silicone surfaces. Acta Microbiol. Imm. H. 64, 179–189. doi: 10.1556/030.63.2016.018
Awan, F., Dong, Y., Wang, N., Liu, J., Ma, K., and Liu, Y. (2018). The fight for invincibility: environmental stress response mechanisms and Aeromonas hydrophila. Microb. Pathogenesis 116, 135–145. doi: 10.1016/j.micpath.2018.01.023
Band, V. I., Crispell, E. K., Napier, B. A., Herrera, C. M., Tharp, G. K., Vavikolanu, K., et al. (2016). Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat. Microbiol. 1:16053. doi: 10.1038/nmicrobiol.2016.53
Battesti, A., Majdalani, N., and Gottesman, S. (2011). The Rpos-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213. doi: 10.1146/annurev-micro-090110-102946
Beshiru, A., and Igbinosa, E. O. (2018). Characterization of extracellular virulence properties and biofilm-formation capacity of vibrio species recovered from ready-to-eat (RTE) shrimps. Microb. Pathogenesis 119, 93–102. doi: 10.1016/j.micpath.2018.04.015
Chang, S. C., and Lee, C. Y. (2018). OpaR and RpoS are positive regulators of a virulence factor PrtA in Vibrio parahaemolyticus. Microbiology 164, 221–231. doi: 10.1099/mic.0.000591
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Dong, T., and Schellhorn, H. E. (2010). Role of RpoS in virulence of pathogens. Infect. Immun. 78, 887–897. doi: 10.1128/IAI.00882-09
Dong, J., Zhang, L., Zhou, S., Xu, N., Yang, Q., Liu, Y., et al. (2020). Identification of a multi-resistant Enterobacter cloacae strain from diseased crayfish (Procambarus clarkii). Aquacult. Rep. 17:100405. doi: 10.1016/j.aqrep.2020.100405
Fang, F. C., Libby, S. J., Buchmeier, N. A., Loewen, P. C., Switala, J., Harwood, J., et al. (1992). The alternative sigma factor katF (rpoS) regulates salmonella virulence. P. Natl. Acad. Sci. U. S. A. 89, 11978–11982. doi: 10.1073/pnas.89.24.11978
Feng, L., Bi, W., Chen, S., Zhu, J., and Liu, X. (2021). Regulatory function of sigma factors RpoS/RpoN in adaptation and spoilage potential of Shewanella baltica. Food Microbiol. 97:103755. doi: 10.1016/j.fm.2021.103755
Fernández-Gómez, P., López, M., Prieto, M., González-Raurich, M., and Alvarez-Ordóñez, A. (2020). The role of the general stress response regulator RpoS in Cronobacter sakazakii biofilm formation. Food Res. Int. 136:109508. doi: 10.1016/j.foodres.2020.109508
Gao, X., Jiang, Z., Zhang, S., Chen, Q., Tong, S., Liu, X., et al. (2020). Transcriptome analysis and immune-related genes expression reveals the immune responses of Macrobrachium rosenbergii infected by Enterobacter cloacae. Fish Shellfish Immun. 101, 66–77. doi: 10.1016/j.fsi.2020.03.042
Gao, X., Pi, D., Chen, N., Li, X., Liu, X., Yang, H., et al. (2018). Survival, virulent characteristics, and transcriptomic analyses of the pathogenic vibrio anguillarum under starvation stress. Front. Cell. Infect. Microbiol. 8:389. doi: 10.3389/fcimb.2018.00389
Gao, X., Zhang, H., Jiang, Q., Chen, N., Li, X., Liu, X., et al. (2019). Enterobacter cloacae associated with mass mortality in zoea of giant freshwater prawns Macrobrachium rosenbergii and control with specific chicken egg yolk immunoglobulins (IgY). Aquaculture 501, 331–337. doi: 10.1016/j.aquaculture.2018.11.050
Gao, X., Zhang, Z., Qian, Q., Chen, Q., Gu, S., Li, J., et al. (2022). Survival characteristics and transcriptomic analyses reveal the adaptive response of the aquatic pathogen non-O1/O139 vibrio cholerae to starvation stress. Microbiol. Spectr. 10, e01939–e01921. doi: 10.1128/spectrum.01939-21
Gao, X., Zhou, Y., Zhu, X., Tang, H., Li, X., Jiang, Q., et al. (2021). Enterobacter cloacae: a probable etiological agent associated with slow growth in the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 530:735826. doi: 10.1016/j.aquaculture.2020.735826
Guan, J., Xiao, X., Xu, S., Gao, F., Wang, J., Wang, T., et al. (2015). Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J. Microbiol. 53, 633–642. doi: 10.1007/s12275-015-0099-6
Guo, Y., Li, Y., Zhan, W., Wood, T. K., and Wang, X. (2019). Resistance to oxidative stress by inner membrane protein ElaB is regulated by OxyR and RpoS. Microb. Biotechnol. 12, 392–404. doi: 10.1111/1751-7915.13369
Hansen, G. H., Raa, J., and Olafsen, J. A. (1990). Isolation of Enterobacter agglomerans from dolphin fish, Coryphaena hippurus L. J. Fish Dis. 13, 93–96. doi: 10.1111/j.1365-2761.1990.tb00761.x
Higgins, C. F. (2001). ABC transporters: physiology, structure and mechanism-an overview. Res. Microbiol. 152, 205–210. doi: 10.1016/S0923-2508(01)01193-7
Huang, L., Guo, L., Xu, X., Qin, Y., Zhao, L., Su, Y., et al. (2019). The role of rpoS in the regulation of vibrio alginolyticus virulence and the response to diverse stresses. J. Fish Dis. 42, 703–712. doi: 10.1111/jfd.12972
Hülsmann, A., Rosche, T. M., Kong, I. S., Hassan, H. M., Beam, D. M., and Oliver, J. D. (2003). RpoS-dependent stress response and exoenzyme production in Vibrio vulnificus. Appl. Environ. Microb. 69, 6114–6120. doi: 10.1128/AEM.69.10.6114-6120.2003
Joelsson, A., Kan, B., and Zhu, J. (2007). Quorum sensing enhances the stress response in vibrio cholerae. Appl. Environ. Microb. 73, 3742–3746. doi: 10.1128/AEM.02804-06
Kamareddine, L., Wong, A. C., Vanhove, A. S., Hang, S., Purdy, A. E., Kierek-Pearson, K., et al. (2018). Activation of vibrio cholerae quorum sensing promotes survival of an arthropod host. Nat. Microbiol. 3, 243–252. doi: 10.1038/s41564-017-0065-7
Kanehisa, M., Goto, S., Sato, Y., Kawashima, M., Furumichi, M., and Mao, T. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. doi: 10.1093/nar/gkt1076
Krzymińska, S., Koczura, R., Mokracka, J., Puton, T., and Kaznowski, A. (2010). Isolates of the Enterobacter cloacae complex induce apoptosis of human intestinal epithelial cells. Microb. Pathogenesis 49, 83–89. doi: 10.1016/j.micpath.2010.04.003
Kulikalova, E., Sappo, S., Urbanovich, L. Y., Markov, E. Y., Mironova, L., and Balakhonov, S. (2014). Model of vibrio cholerae biofilm as a mechanism of its survival in surface water reservoirs. Contemp. Probl. Ecol. 7, 12–18. doi: 10.1134/S1995425514010089
Landini, P., Egli, T., Wolf, J., and Lacour, S. (2014). sigmaS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Env. Microbiol. Rep. 6, 1–13. doi: 10.1111/1758-2229.12112
Lee, J. H., Bae, I. K., Lee, C. H., and Jeong, S. (2017). Molecular characteristics of first IMP-4-producing Enterobacter cloacae sequence type 74 and 194 in Korea. Front. Microbiol. 8:2343. doi: 10.3389/fmicb.2017.02343
Liu, X., Wu, Y., Chen, Y., Xu, F., Halliday, N., Gao, K., et al. (2016). RpoS differentially affects the general stress response and biofilm formation in the endophytic Serratia plymuthica G3. Res. Microbiol. 167, 168–177. doi: 10.1016/j.resmic.2015.11.003
Liu, E., Ye, J., Song, S., Wang, K., Zhang, Y., and Zhang, H. (2014). Impact of co-deficiency of RpoN and RpoS on stress tolerance, virulence and gene regulation in Edwardsiella tarda. J Basic Microb. 54, 678–687. doi: 10.1002/jobm.201300622
Luo, G., Xu, X., Zhao, L., Qin, Y., Huang, L., Su, Y., et al. (2019). clpV is a key virulence gene during in vivo pseudomonas plecoglossicida infection. J. Fish Dis. 42, 991–1000. doi: 10.1111/jfd.13001
Ma, L., Chen, J., Liu, R., Zhang, X. H., and Jiang, Y. A. (2009). Mutation of rpoS gene decreased resistance to environmental stresses, synthesis of extracellular products and virulence of vibrio anguillarum. FEMS Microbiol. Ecol. 70, 286–292. doi: 10.1111/j.1574-6941.2009.00713.x
Norel, F., Robbe-Saule, V., Popoff, M. Y., and Coynault, C. (1992). The putative sigma factor KatF (RpoS) is required for the transcription of the salmonella typhimurium virulence gene spvB in Escherichia coli. FEMS Microbiol. Lett. 99, 271–276. doi: 10.1111/j.1574-6968.1992.tb05580.x
Olwal, C. O., Angienda, P. O., Onyango, D. M., and Ochiel, D. O. (2018). Susceptibility patterns and the role of extracellular DNA in Staphylococcus epidermidis biofilm resistance to physico-chemical stress exposure. BMC Microbiol. 18, 1–13. doi: 10.1186/s12866-018-1183-y
Qiu, Y., Nagarajan, H., Embree, M., Shieu, W., Abate, E., Juarez, K., et al. (2013). Characterizing the interplay between multiple levels of organization within bacterial sigma factor regulatory networks. Nat. Commun. 4:1755. doi: 10.1038/ncomms2743
Reading, N. C., Rasko, D. A., Torres, A. G., and Sperandio, V. (2009). The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. P. Natl. Acad. Sci. U. S. A. 106, 5889–5894. doi: 10.1073/pnas.0811409106
Sun, J., Gao, X., Qun, J., Du, X., Bi, K., Zhang, X., et al. (2016). Comparative analysis of the survival and gene expression of pathogenic strains Vibrio harveyi after starvation. FEMS Microbiol. Lett. 363:fnw250. doi: 10.1093/femsle/fnw250
Thillai, S. V., Santiago, T. C., Vijayan, K. K., Alavandi, S. V., Stalin, R. V., Rajan, J. J. S., et al. (2008). Involvement of Enterobacter cloacae in the mortality of the fish, Mugil cephalus. Lett. Appl. Microbiol. 46, 667–672. doi: 10.1111/j.1472-765X.2008.02365.x
Wang, H., Ayala, J. C., Benitez, J. A., and Silva, A. J. (2012). Interaction of the histone-like nucleoid structuring protein and the general stress response regulator RpoS at vibrio cholerae promoters that regulate motility and hemagglutinin/protease expression. J. Bacteriol. 194, 1205–1215. doi: 10.1128/JB.05900-11
Wang, L., Huang, L., Su, Y., Qin, Y., Kong, W., Ma, Y., et al. (2015). Involvement of the flagellar assembly pathway in vibrio alginolyticus adhesion under environmental stresses. Front. Cell. Infect. Microbiol. 5:59. doi: 10.3389/fcimb.2015.00059
Wang, S., Yan, Q., Zhang, M., Huang, L., Mao, L., Zhang, M., et al. (2019). The role and mechanism of icmF in Aeromonas hydrophila survival in fish macrophages. J. Fish Dis. 42, 895–904. doi: 10.1111/jfd.12991
Waterman, S. R., and Small, P. (2003). Transcriptional expression of Escherichia coli glutamate-dependent acid resistance genes gadA and gadBC in an hns rpoS mutant. J. Bacteriol. 185, 4644–4647. doi: 10.1128/JB.185.15.4644-4647.2003
Weber, H., Polen, T., Heuveling, J., Wendisch, V. F., and Hengge, R. (2005). Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187, 1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005
Yin, K., Guan, Y., Ma, R., Wei, L., Liu, B., Liu, X., et al. (2018). Critical role for a promoter discriminator in RpoS control of virulence in Edwardsiella piscicida. PLoS Pathog. 14:e1007272. doi: 10.1371/journal.ppat.1007272
Zhan, J., Qiao, J., and Wang, X. (2021). Role of sigma factor RpoS in Cronobacter sakazakii environmental stress tolerance. Bioengineered 12, 2791–2809. doi: 10.1080/21655979.2021.1938499
Keywords: Enterobacter cloacae, rpoS, transcriptomic analysis, stress resistance, virulence
Citation: Gao X, Qian Q, Zhu Y, Chen Z, Xu J, Xu W, Jiang Q, Wang J and Zhang X (2022) Transcriptomic and phenotype analysis revealed the role of rpoS in stress resistance and virulence of pathogenic Enterobacter cloacae from Macrobrachium rosenbergii. Front. Microbiol. 13:1030955. doi: 10.3389/fmicb.2022.1030955
Edited by:
Nityananda Chowdhury, Medical University of South Carolina, United StatesReviewed by:
Pande Gde Sasmita Julyantoro, Udayana University, IndonesiaXiuzhen Sheng, Ocean University of China, China
Copyright © 2022 Gao, Qian, Zhu, Chen, Xu, Xu, Jiang, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Zhang, enhqOTMwN0AxNjMuY29t
 Xiaojian Gao
Xiaojian Gao Qieqi Qian
Qieqi Qian Qun Jiang
Qun Jiang Xiaojun Zhang
Xiaojun Zhang