
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 25 October 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1030564
This article is part of the Research TopicPrevalence, Transmission and Control of Clinically Important Antimicrobial-Resistant Bacteria/Genes within One Health FrameworkView all 22 articles
The aquatic environment is an important medium for the accumulation and dissemination of antibiotic-resistant bacteria as it is often closely related to human activities. Previous studies paid little attention to the prevalence and mechanism of polymyxin-resistant bacteria in the aquatic environment. As a Gram-negative opportunistic pathogen widely distributed in aquatic ecosystems, the antibiotic-resistant profile of Aeromonas spp. deserves much attention. In this study, we identified 61 Aeromonas spp. isolates from water samples in the section of the Yangtze River. The total polymyxin B (PMB) resistance rate of these strains was 49.18% (30/61), showing a high level of polymyxin resistance in Aeromonas spp. The MIC50 and MIC90 for PMB exhibited a significant discrepancy among different species (p < 0.001). The MIC50 and MIC90 for PMB in the Aeromonas hydrophila were 128 mg/L and above 128 mg/L while in Aeromonas caviae and Aeromonas veronii, the MIC50 and MIC90 value were both 2 mg/L. Only two A. veronii strains (MIC = 2 mg/L) and one A. caviae strain (MIC = 0.5 mg/L) were identified as carrying mobilized polymyxin resistant gene mcr-3.42, and mcr-3.16. All mcr genes were located in the chromosome. This is the first report that the downstream region of mcr-3.42 was the truncated mcr-3-like gene separated by the insertion sequences of ISAs20 (1,674 bp) and ISAs2 (1,084 bp). Analysis of epidemiology of mcr-positive Aeromonas genomes from GenBank database showed that the genus Aeromonas and the aquatic environment might be the potential container and reservoir of mcr-3. By the whole-genome sequencing and qRT-PCR, we inferred that the sequence differences in the AAA domain of MlaF protein and its expression level among these three species might be involved in the development of polymyxin resistance. Our study provided evidences of the possible mechanism for the variety of polymyxin susceptibility in different species of the genus Aeromonas and a theoretical basis for the surveillance of the aquatic environment.
Antimicrobial resistance (AMR) has been a major challenge to global public health with the use of antibiotics over the decades. Antimicrobial resistance genes (ARGs) are considered as environmental pollutants as the result of the continuous release of residual antibiotics by human activities into the environment such as hospitals, livestock, and sewage treatment plant, which increased occurrence of antimicrobial resistant bacteria (ARB) (Pruden et al., 2006; Zurfluh et al., 2017). Moreover, the aquatic environment itself can serve as the sites of the spreading of AMR and the reservoir of ARGs, like qnrA, qnrB, qnrS, mef, and so on (Pei et al., 2006; Aedo et al., 2014; Nonaka et al., 2015). There are a large number of bacteria (> 100 million/liter of seawater) in the aquatic environment, from which bacteria such as Aeromonas spp., Vibrio, and Pseudomonas sp. are often isolated (Noonburg, 2005). Especially, Aeromonas spp., consisting of 30 species, is widely distributed in various kinds of natural water ecosystems (Chen et al., 2021). Aeromonas spp. are gram-negative, rod-shaped aerobes belonging to the Aeromonadaceae family. Among the members of Aeromonas spp., Aeromonas hydrophila, Aeromonas veronii, and Aeromonas caviae are the common zoonotic pathogens both for human and aquatic animals. In recent years, Aeromonas species have become the third most common enteric bacterial pathogens, with a clinical isolation rate of 56.73% (Yuwono et al., 2021). The antimicrobial susceptibility of Aeromonas has been reported in many studies, showing that Aeromonas were resistant to ampicillin, penicillin, and cephalosporins, tetracycline (Scarano et al., 2018; Jia et al., 2020; Sun et al., 2021).
Polymyxins, including polymyxin B (PMB) and polymyxin E (colistin), are a group of cationic polypeptides that disrupts membrane integrity by replacing cations like Mg2+ and Ca2+ in the outer membrane, which leads to the cell lysis. Due to the emergence of bacteria resistant to the most common-use antibiotic and the shortage of new antimicrobial agents to resist them, polymyxins have become the last line of defense against multiple drug-resistant gram-negative bacteria infections (Falagas and Rafailidis, 2008; Trimble et al., 2016). The most common mechanism of colistin resistance is the chromosomal mutations in genes involved in the synthesis and modification of lipid A of lipopolysaccharide (LPS). These genes include the arnBCADTEF operon which is regulated by the two-component system (TCS) PhoP/Q, PmrA/B, ParR/S, CprR/S, and ColR/S (Olaitan et al., 2014). In addition to that, the horizontal transfer of the plasmid-carrying gene mcr encoding phosphoethanolamine (PEtN) transferase which modifies the lipid A by addition of phosphoethanolamine has become another important cause of colistin resistance. Since the first plasmid-mediated colistin resistance gene mcr-1 was identified in 2015, a total of 10 mcr gene variants (mcr-1 to mcr-10) have been reported, mainly among the Enterobacterales (Hussein et al., 2021).
In Aeromonas species, mcr-2, mcr-3, mcr-5, and mcr-7 have been reported, especially the variants of mcr-3, which were widely detected and frequently disseminated between Enterobacteriaceae and Aeromonas (Ma et al., 2018; Sun et al., 2018; Ragupathi et al., 2020; Wang et al., 2021). Beyond that, the mcr-positive Aeromonas strains showed the discrepancy of polymyxin-resistant phenotype, manifesting as a wide span of and the minimum inhibitory concentration (MIC) of polymyxin ranging from <0.5 mg/L to >128 mg/L. But the exact mechanism of polymyxin resistance in Aeromonas and reasons for the differences of polymyxin susceptibility in different Aeromonas species remains poorly understood and less attention has been paid to aquatic environments (Ling et al., 2017; Tuo et al., 2018; Ragupathi et al., 2020).
Here, we determined the polymyxin susceptibility in different Aeromonas species isolated from the section of the Yangtze River, investigated the prevalence of the mcr gene, and explored the connections between gene expression levels in the TCS involved in the synthesis and modification of lipid A and the polymyxin resistance. A total of 61 Aeromonas strains were detected and 30 strains showed polymyxin resistance, showing a high level of polymyxin resistance rate of 49.18% (30/61). Among different species in the genus Aeromonas, the polymyxin resistance rate showed a distinct discrepancy (p < 0.001). Two A. veronii strains were identified as mcr-3.42 positive strains, and one A. caviae strain was identified as mcr-3.16 while the rest strains remained negative of mcr-1 to mcr-10. We inferred that the existence of arnBCADTEF and mutations in MlaF protein among different Aeromonas species could affect the function of phospholipid transportation and contribute to the resistance to polymyxins.
To investigate the dissemination of mcr in the genus Aeromonas from the aquatic environment, 16 water samples were collected in sterile 1-L bottles from A section of the Yangtze River. All samples were stored at 4°C after collection and analyzed within 12 h. Luria-Bertani (LB), Mueller-Hinton (MH), and Salmonella Shigella (SS) culture mediums were used to perform the culture and isolation of bacteria. The whole isolates were screened for the presence of mcr-1 to mcr-10 by colony PCR (Woodman et al., 2016). The positive PCR products were verified by Sanger sequencing. Primer pairs used in this study were listed in Supplementary Table S1. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS identification and 16S rRNA sequencing were adopted for bacterial identification.
Antimicrobial susceptibility testing (AST) was conducted by the broth microdilution method and agar dilution method. Testing was performed according to Clinical and Laboratory Standards Institute guidelines (CLSI) (2021). The minimum inhibitory concentration (MIC) of PMB (Sigma, Shanghai, China), tigecycline (TGC), and tetracycline (TCY) (Solarbio, Shanghai, China) were determined by the broth microdilution method, and the MIC of common drugs for clinical therapy including amikacin, cefepime, ciprofloxacin, meropenem, aztreonam et al. (Solarbio, Shanghai, China) were determined by agar dilution method. The breakpoint of TGC was interpreted according to the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) version 10.0.1 The other antimicrobial agents were interpreted according to the guidelines of the CLSI M100 (2021 version, 31st edition) and M45-A2 (2015 version, 3rd edition) documents. Escherichia coli ATCC 25922 was used as the quality control strain for antimicrobial susceptibility testing.
The conjugation experiment was carried out to investigate the transferability of mcr gene. Procedures were performed according to the method described previously (Hadjadj et al., 2019). Sodium azide-resistant E. coli J53 (MIC of 200 mg/L) was used as recipient strain. The transconjugants were selected on the medium containing azide at 200 mg/L and PMB, at 1 mg/L was consistent with the MIC of the donor. Resultant colonies were screened for the presence of mcr by PCR.
S1-PFGE analysis of mcr-positive isolates was used to estimate the sizes of mcr-positive plasmids. XbaI-restricted DNA of Salmonella enterica serovar Braenderup H9812 was used as a DNA marker. Shortly, agarose plugs were made using the colonies in the medium of a single colony inoculated and digested with S1 nuclease (TaKaRa, Dalian, China). The DNA was separated using the CHEF-MAPPER PFGE system (Bio-Rad) under the following conditions: 14°C, 6 V/cm, and a 120° pulse angle for 16 h, with the initial and final pulses conducted for 2.16 s and 63.8 s, respectively. Then the dyed gel was visualized with the imaging system.
Genomic DNA of mcr-positive isolates were extracted using Gentra Puregene Yeast/Bact. Kit (Qiagen, CA, United States), according to the manufacturer’s instructions. Illumina Hiseq PE150 platform was used for the whole-genome sequencing (WGS). After read quality control, the reads from Illumina Hiseq platform were assembled using SPAdes 3.10.18 which is based on De-Bruijn algorithm and takes the appropriate k-mer values. Genetic predictions of assembly results were using the web platforms of the National Center for Biotechnological Information (NCBI) and ORFfinder.2 The sequences were annotated using the RAST annotation server3 and prokka 1.14.6.4 Assembled contigs were analyzed via the Center for Genomic Epidemiology website5 to screen for the presence of acquired AMR genes, plasmid incompatibility types, and Multi Locus Sequence Typing (MLST). Linear comparisons of mcr were generated using Easyfig. 2. Multiple alignments of nucleotide sequences were performed by using The European Bioinformatics Institute (EMBL-EBI) tools.6 To investigate the distribution of mcr-positive strains of the genus Aeromonas, we retrieved the isolates in sequences deposited in GenBank by Genomes Browser,7 (accessed 26 April 2022) and Isolates Browser8 (accessed 26 April 2022).
Total RNA was extracted from mid-long phase (OD600 ~ 0.6) bacterial cultures using the Qiagen RNeasy Mini kit (QIAGEN, Alameda, United States), according to the manufacturer’s instructions. Complementary DNA (cDNA) was generated from total RNA using the RT Premix. Supplementary Table S2 shows the sequences of transcript-specific primers used for qPCR. All samples were performed in triplicate, and the data were normalized to 16S rRNA levels and analyzed using the 2−ΔΔCT method to calculate the fold-change relative to the control. The difference between groups was analyzed by Welch’s t-test.
Multiple alignments of amino acid sequences were performed by using The European Bioinformatics Institute (EMBL-EBI) tools9 and Jalview 2.11.1.4 was used to perform the multiple sequence alignment editing, visualization, and analysis. PROVEAN Protein tools10 were calculated to evaluate whether amino acid alterations in PhoP/PhoQ, OmpR/EnvZ, and MlaF affected biological function. Moreover, the protein domains of PhoP/PhoQ, OmpR/EnvZ, and MlaF were subjected to SMART analysis.11 The secondary structure and complex structure of MlaF protein were predicted by PSIPRED12 and SWISS-MODEL,13 respectively. The images of biomolecular structures of MlaF protein were generated by PyMoL (Seeliger and de Groot, 2010).
The whole-genome nucleotide sequences of 15 Aeromonas strains have been submitted to DDBJ/ENA/GenBank under the accession numbers JANKLR000000000-JANKLZ000000000, JANKMA000000000-JANKME000000000, and JANPYL000000000 and were in a processing queue (BioProject: PRJNA864564). The accession numbers were listed in Table 1.
In order to investigate the distribution of polymyxin resistance in the genus Aeromonas bacteria isolated from the aquatic environment, we collected a comprehensive set of samples from the section of the Yangtze River, in Hubei province. In total, 163 gram-negative bacteria strains were isolated and identified by MALDI-TOF MS. Among them, 37.42% (61/163) of the isolates were identified as belonging to Aeromonas spp., namely Aeromonas hydrophila (n = 29), Aeromonas caviae (n = 21), and Aeromonas veronii (n = 11). Antimicrobial susceptibility testing was performed to investigate the drug resistance characteristics of 61 Aeromonas spp. isolates against fifteen common clinical drugs. 100% of A. hydrophila isolates exhibited resistance to PMB while 100 and 90.91% (10/11) of A. caviae and A. veronii isolates exhibited susceptibility to PMB. 96.72% (59/61), 93.44% (57/61), 90.16% (55/61), 93.44% (57/61), 55.74% (34/61), 65.57% (40/61), 73.77% (45/61), 96.72% (59/61), 50.82% (31/61), 77.05% (47/61), 75.41% (46/61), 83.61% (51/61) of Aeromonas spp. strains showed susceptibility to clinically important antibiotic drugs meropenem, imipenem, tigecycline, tazobactam/piperacillin, ciprofloxacin, levofloxacin, aztreonam, amikacin, gentamycin, cefepime, ceftazidime, cefoperazone/sulbactam, respectively. But for the sulfamethoxazole/trimethoprim and tetracycline, the majority of Aeromonas spp. remained resistant, with a resistance rate of 67.21% (41/61) and 60.66% (37/61) respectively (Supplementary Table S3). The results of the comparator antimicrobial drugs tested against 61 Aeromonas spp. isolates were listed in Table 2. And the MIC50 and MIC90 for PMB were 128 mg/L and above 128 mg/L in the A. hydrophila group. However, both the MIC50 and MIC90 were 2 mg/L in the A. caviae and A. veronii groups (Figure 1A). This result shows that the characteristics of PMB resistance in the genus Aeromonas vary in different species (p < 0.001).
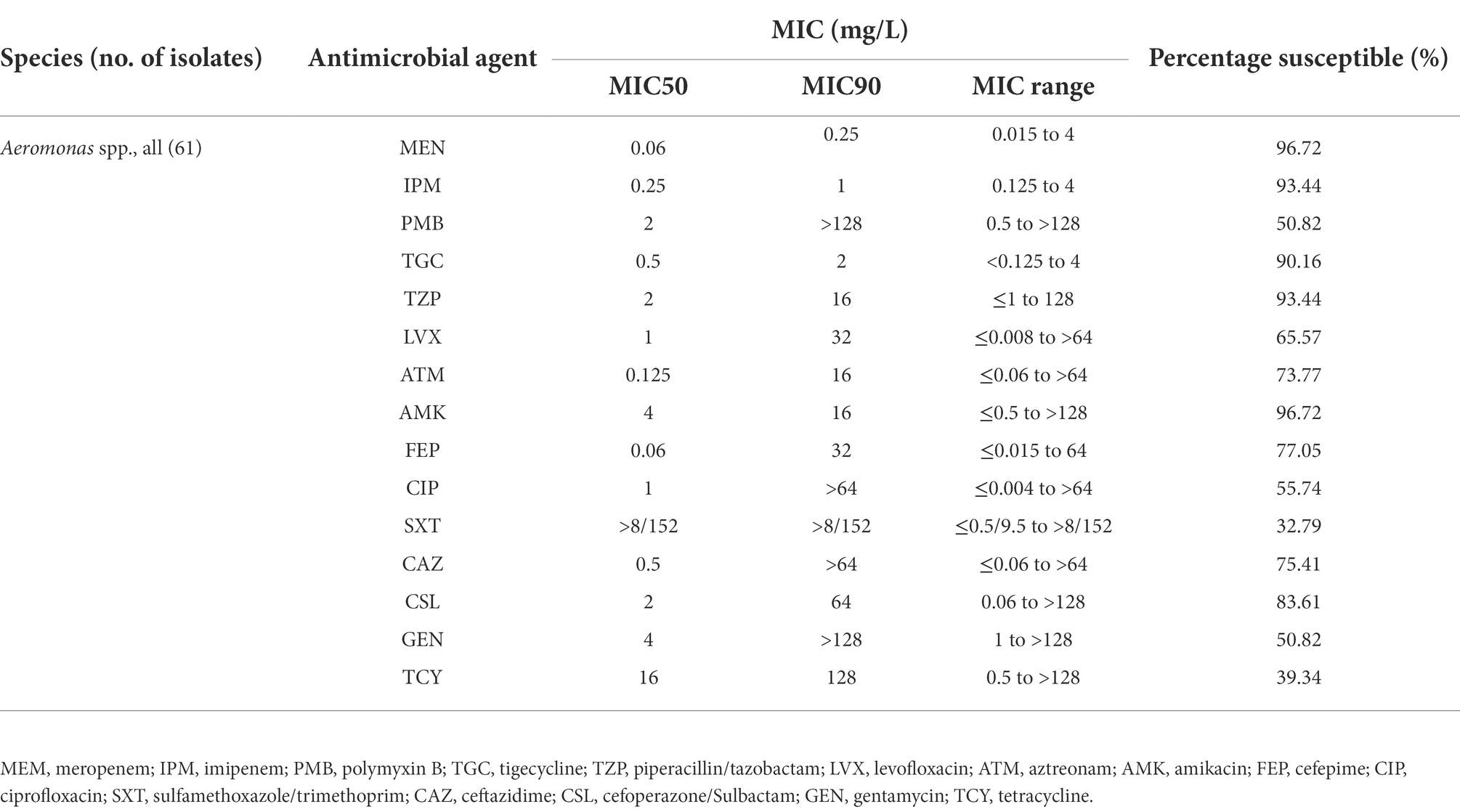
Table 2. Comparator antimicrobial agents tested against 61 isolates of Aeromonas spp. collected in a section of Yangtze River.
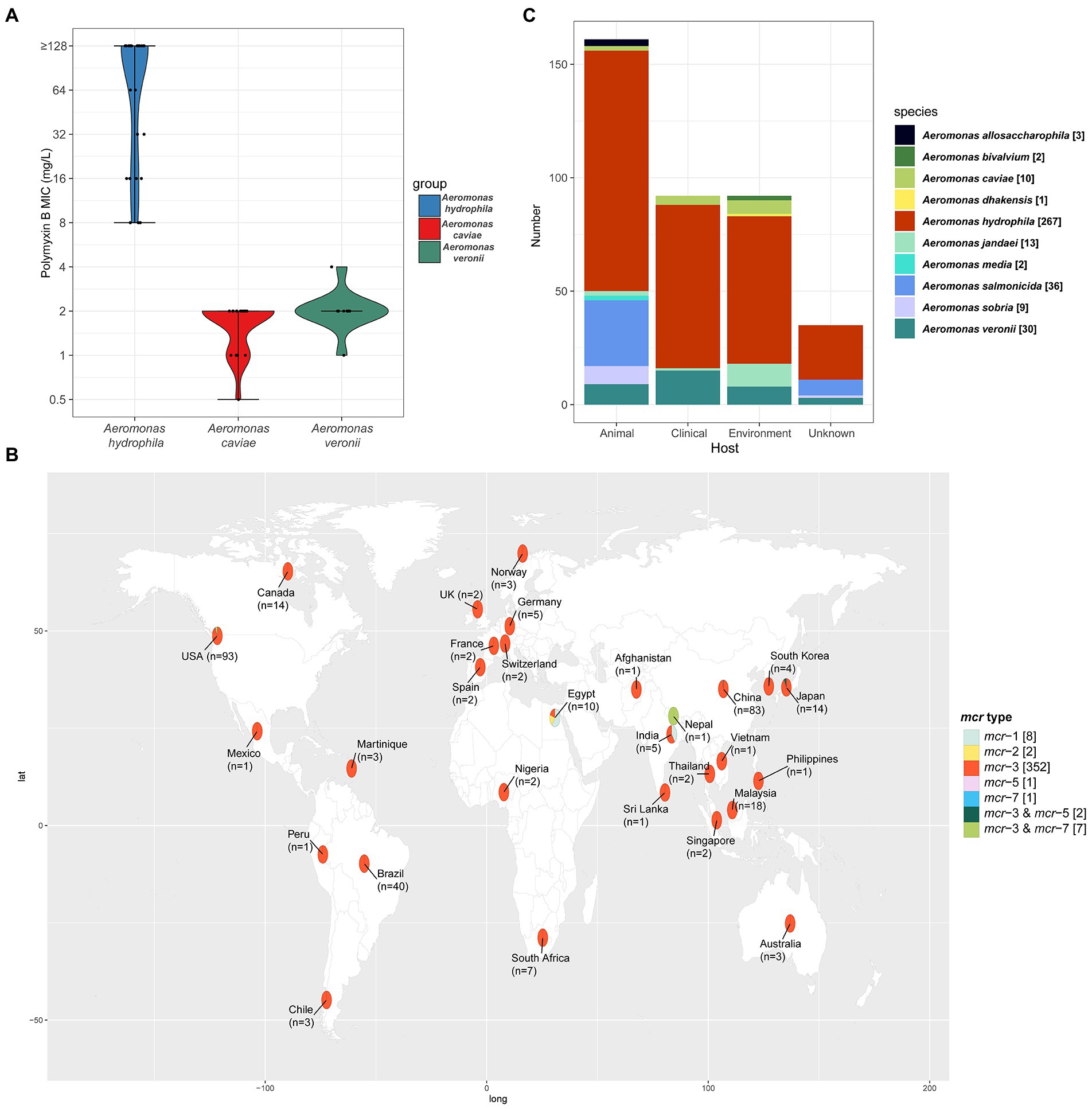
Figure 1. (A) The Violin-plot. The MIC of PMB in the three species of the genus Aeromonas. The black dot represents the dispersion of MIC value. And the shape of the violin represents the distribution of MICs in the corresponding group. Blue, red, and green indicated the species of A. hydrophila, A. caviae, and A. veronii. (B) Geographical distribution of the 326 isolates with definitive location, among all 373 mcr-positive Aeromonas isolates from the NCBI database. Different mcr types of the isolates were labeled with different colors and the number of each mcr type is given in square brackets. The number of all the mcr-positive isolates in each country was labeled on the world map under the country name. In addition, the location of the 47 mcr-positive isolates was still unknown. The world map was created using the corresponding map data with R package ggplot2 v3.3.5 (https://github.com/tidyverse/ggplot2). (C) The species and number of mcr-positive Aeromonas isolates from different sources. In total, 338 mcr-positive Aeromonas isolates were isolated and identified from the specific host and the sample sources of 35 isolates were still unknown. Different colors indicated the different species of the genus Aeromonas.
To investigate the prevalence of mcr in the genus Aeromonas bacteria isolated from the aquatic environment, colony PCR was conducted to screen mcr genes using primers as described in materials and methods. Only two mcr-3 positive A. veronii strains (M683 and M694, MIC = 2 mg/L) and one mcr-3 positive A. caviae S611 (MIC = 0.5 mg/L) were identified, while the rest of the 58 isolates were all negative of mcr-1 to mcr-10, indicating that the prevalence of mcr in Aeromonas from the section of the Yangtze River is 4.92% (3/61). However, none of the three mcr-positive Aeromonas strains showed resistance to PMB (MIC <4 mg/L). Conjugation experiments showed that these three mcr-positive isolates were not able to transfer the polymyxin resistance phenotype to receipt strain E. coli J53. S1-PFGE and DNA hybridization were conducted to separate the plasmid of different sizes and locate the genetic position of mcr-3. No plasmid was found in the A. caviae S611. But a plasmid fragment of approximately 5.47 × 104 bp was found in A. veronii strains (M683 and M694). Southern blotting result suggested that the mcr-3 was located in the chromosome (Supplementary Figure S1).
To investigate the characteristics of the epidemiology of mcr-positive Aeromonas strains, we retrieved all the details of isolates deposited in GenBank as described in materials and methods. The mcr-positive Aeromonas strains in the GenBank were reported to carry mcr-1, mcr-2, mcr-3, mcr-5 as well as mcr-7, and mcr-3 was the predominant type in these mcr-positive Aeromonas strains (47.70%, 352/738). Most of the mcr-positive Aeromonas were isolated from the United States (24.93%, 93/373) and China (22.25%, 83/373), and mcr-3 was the main type (94.37%, 352/373) (Figure 1B). Nearly half of the mcr-positive Aeromonas were isolated from animals (42.63%, 159/373), while 24.40% (91/373) and 23.59% (88/373) of the positive samples were from the environment and clinic (Figure 1C). By the phylogenetic tree analysis, we found that the evolution of A. caviae S611 isolated from Yangtze River was closely related to A. caviae SCAc2001 (WUTZ00000000.1) isolated from clinical samples in China. The evolution distance of A. veronii M683 and M694 isolated from the aquatic environment was quite approximated and closely related to A. veronii MS-17-88 (GCA_003611985.1) isolated from animal samples in the United States (Figure 2).
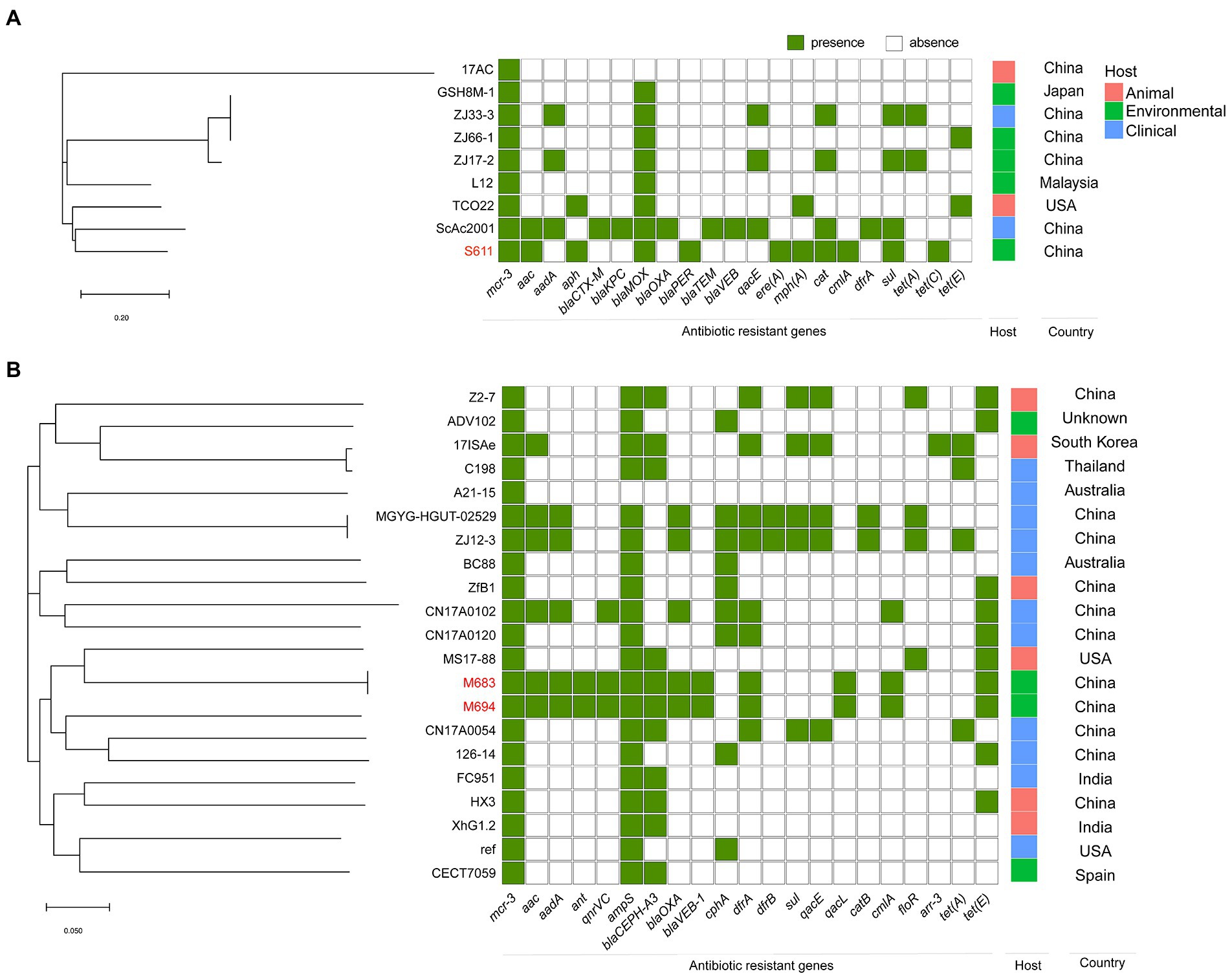
Figure 2. Phylogenetic tree generated from the core-genome sequences of the mcr-3-positive isolates identified in this study and other mcr-3-positive isolates within the same species randomly selected from GenBank database. Significant antimicrobial resistance genes, country, and host were shown. (A) Phylogenetic tree of mcr-3-positive A. caviae. (B) Phylogenetic tree of mcr-3-positive A. veronii.
To fully understand the genetic context of mcr-positive Aeromonas strains isolated from the section of Yangtze River, we perform WGS with three mcr-3 positive isolates in this study (A. veronii M683, M694, and A. caviae S611) and the contigs were properly assembled.
In the A. veronii strains M683 and M694, ResFinder was used to reveal the antibiotic resistance genes, such as the aminoglycoside and fluoroquinolone resistance genes (aac, aadA, ant, and qnrVC), beta-lactam (ampS, blaCEPH-A3, blaOXA, and blaVEB-1), folate pathway antagonist (dfrA), quaternary ammonium compound (qacL), amphenicol (cmlA), and tetracycline antibiotic genes (tet(E)). In the A. caviae S611, the following antibiotic resistance genes were predicted, aac, aph, blaMOX, blaPER, sul, macrolide resistance genes (mph(A) and ere(A)), cat, cmlA, and tet(C) (Figure 2).
The nucleotide sequence of mcr-3 in the A. caviae S611 showed 100% identity with mcr-3.16 (NG_060517.1). A 1626-bp mcr-3-like gene lay downstream of the mcr-3. Its nucleotide sequence showed 83.03 and 67.32% identity to mcr-3.16 and lipid A phosphoethanolamine transferase gene (eptA) in the A. caviae S611. Downstream region of the mcr-3-like gene lay dgkA encoding diacylglycerol kinase, a gene arrangement (mcr-3-mcr-3-like) that is often observed in many mcr-3 positive Aeromonas strains. But A. veronii strains M683 and M694 were the exception. Their mcr-3-mcr-3-like segment was separated by the insertion sequence of ISAs20 (1,674 bp) and ISAs2 (1,084 bp) (Figure 3). This is the first report of the truncated mcr-3-mcr-3-like segment in the mcr-3 positive A. veronii strains. Both of the two 1,623-bp mcr-3 variant genes of M683 and M694 exhibited 99.94% nucleotide sequence identity to the mcr-3.32 (NG_070770.1), and the corresponding proteins exhibited 99.81% amino acid sequence identity to MCR-3.32 (WP_188331891.1). The two novel mcr-3 variant genes of A. veronii M683 and M694 were termed as mcr-3.42. And the accession numbers were OP297669 and OP297670, respectively. The discovery of mcr-3.42 genes in A. veronii was the first report about the mcr-3 positive A. veronii strains isolated from the aquatic environment in China.
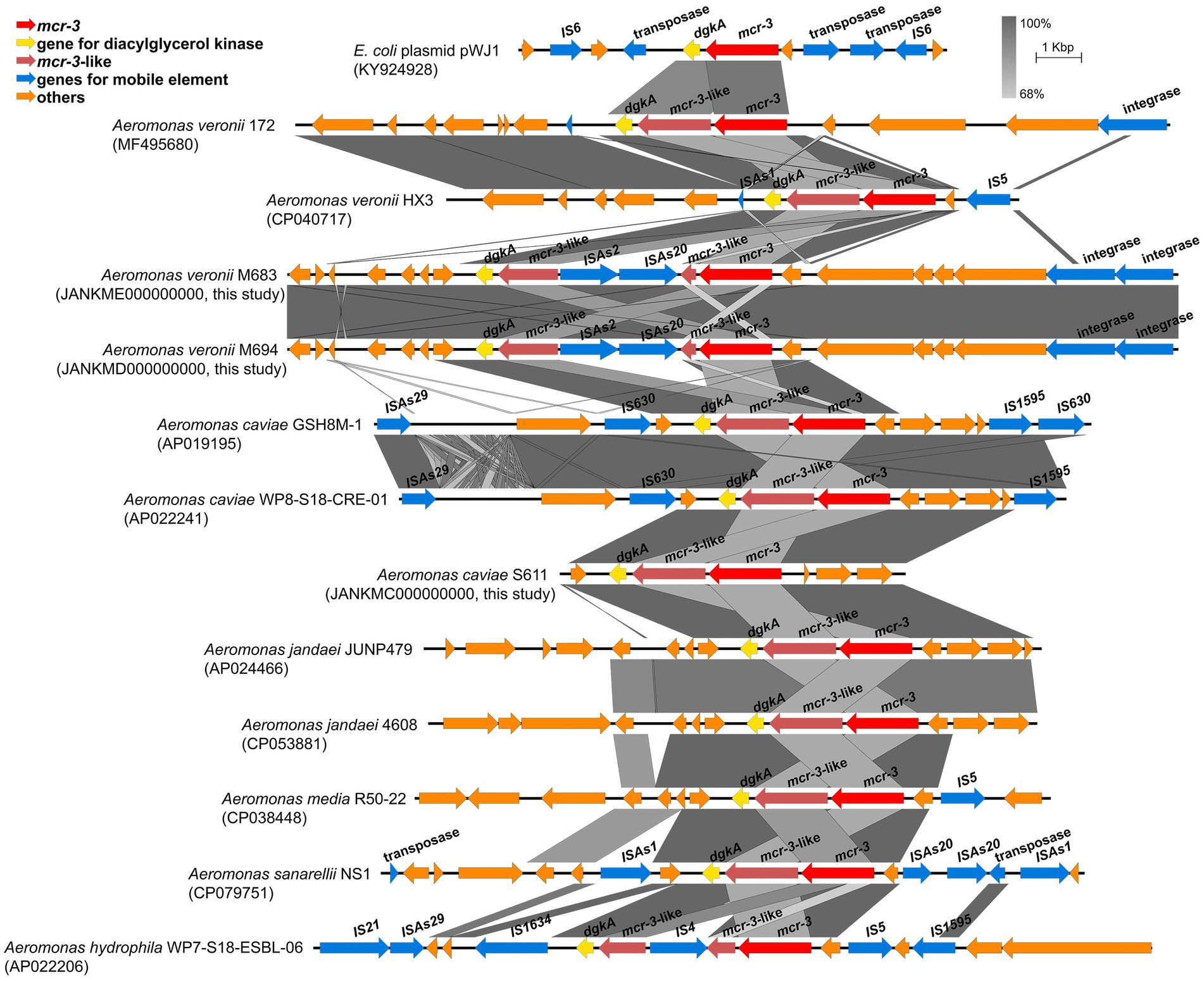
Figure 3. Schematic representation and comparison of the genetic environment of the mcr-3-flanking region in each genomic backbone type. Arrows indicate the direction of transcription of each gene, and different genes are shown in different colors. Regions of above 68% nucleotide sequence identify are shaded grey.
Although mcr-3 gene were detected in some Aeromonas strains, these bacteria did not show high colistin resistance. And other Aeromonas strains with high colistin resistance did not possess mcr gene. Therefore, we suspected that the mcr gene might not be the primary cause for the colistin resistance diversity among Aeromonas. In order to investigate the possible reason for that, we screened 12 mcr-negative Aeromonas strains in three species with different levels of PMB sensibility (MIC of 4 A. hydrophila isolates was ≥128 mg/L, MIC of 4 A. caviae and 4 A. veronii isolates were ≤ 2 mg/L). Whole genome sequencing was performed to analyze the differences in the gene sequences of TCS such as PhoP/PhoQ, EnvZ/OmpR TCS, and MlaF in these 12 Aeromonas strains. And amino acids analysis and protein structure prediction was performed as described in materials and methods.
We compared the genome sequences of A. hydrophila with A. caviae and A. veronii, colistin resistance gene arnBCADTEF was only found in A. hydrophila, implying that arnBCADTEF operon might be the possible cause for the variety of colistin sensitivity among these species. We also screened the nucleic acids differences in the TCS PhoP/PhoQ, EnvZ/OmpR, and MlaF. Many base mutations were found in these genes in A. hydrophila, but most of the mutations did not change the amino acid sequences. For example, compared with A. caviae genome sequence, A. hydrophila possessed 12 site mutations in phoP, 53 site mutations in phoQ, 7 site mutations in ompR, 32 site mutations in envZ, and 32 site mutations in mlaF. While compared with A. veronii, there were 12, 93, 10, 55, and 5 site mutations in these genes (Supplementary Table S4). In order to evaluate whether these mutations would impact the protein function, PROVEAN software was used to predict the result of these mutations (Table 3). For A. hydrophila, PhoP amino acid substitution in Q39G, R69S/T, V153T/A, PhoQ in P82R, Y233F, OmpR in F30V, EnvZ in G108K, Q198L, Q302Y, L339Q, P369A, H407Y, and MlaF in S63F, A231E/D were supposed to cause disfunction. But secondary structure alignment showed that the AAA domain in MlaF of A. hydrophila (174aa) was shorter than that of A. veronii and A. caviae (198aa) (Figure 4). And protein structures predictions of MlaF based on the model proteins showed that MlaF of A. hydrophila lacked the C-terminal helix structure that exists in both MlaF of A. veronii and A. caviae (Figure 5).
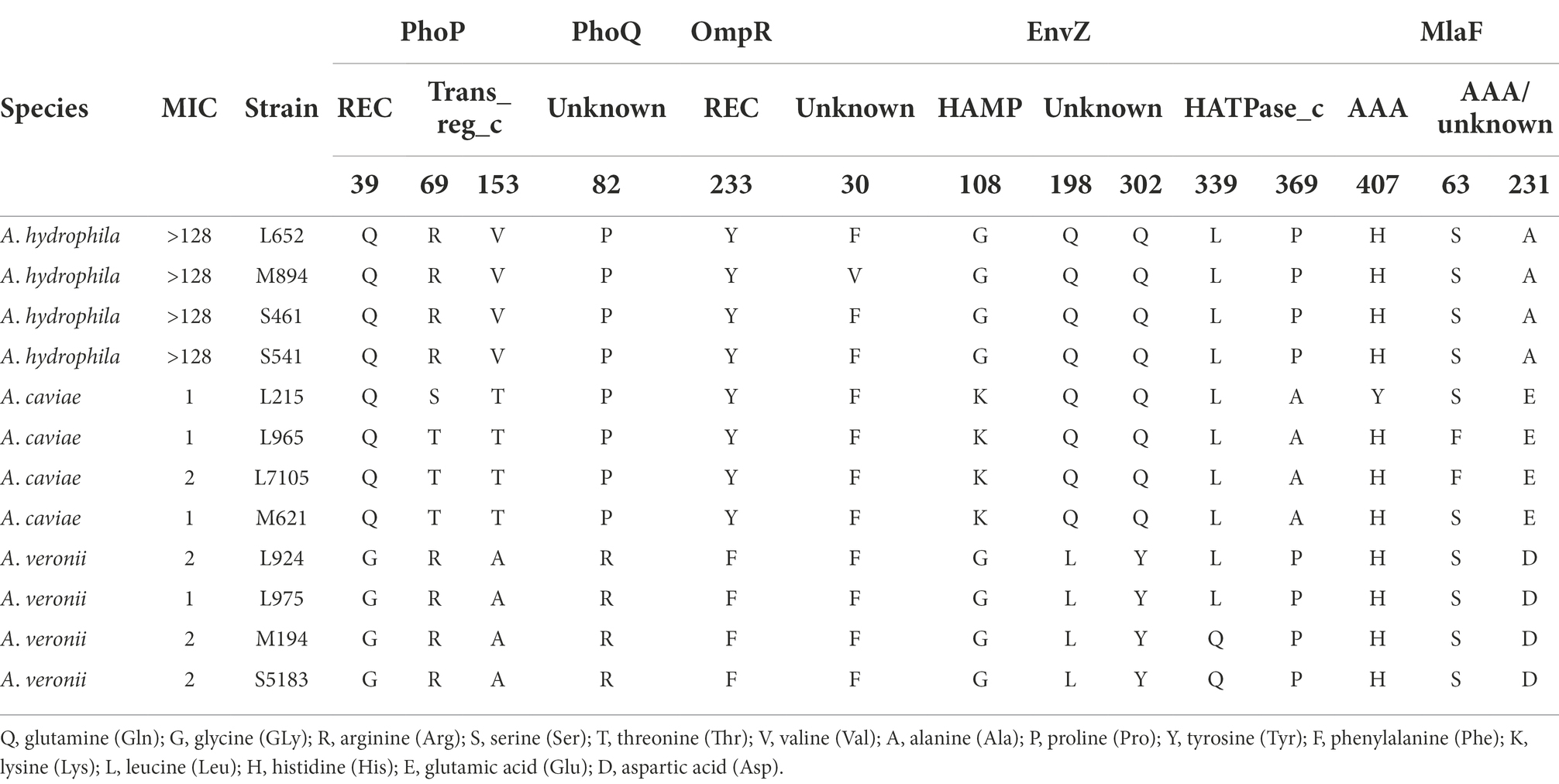
Table 3. Amino acid variations in functional domains of PhoP/PhoQ, EnvZ/OmpR, and MlaF in Aeromonas.
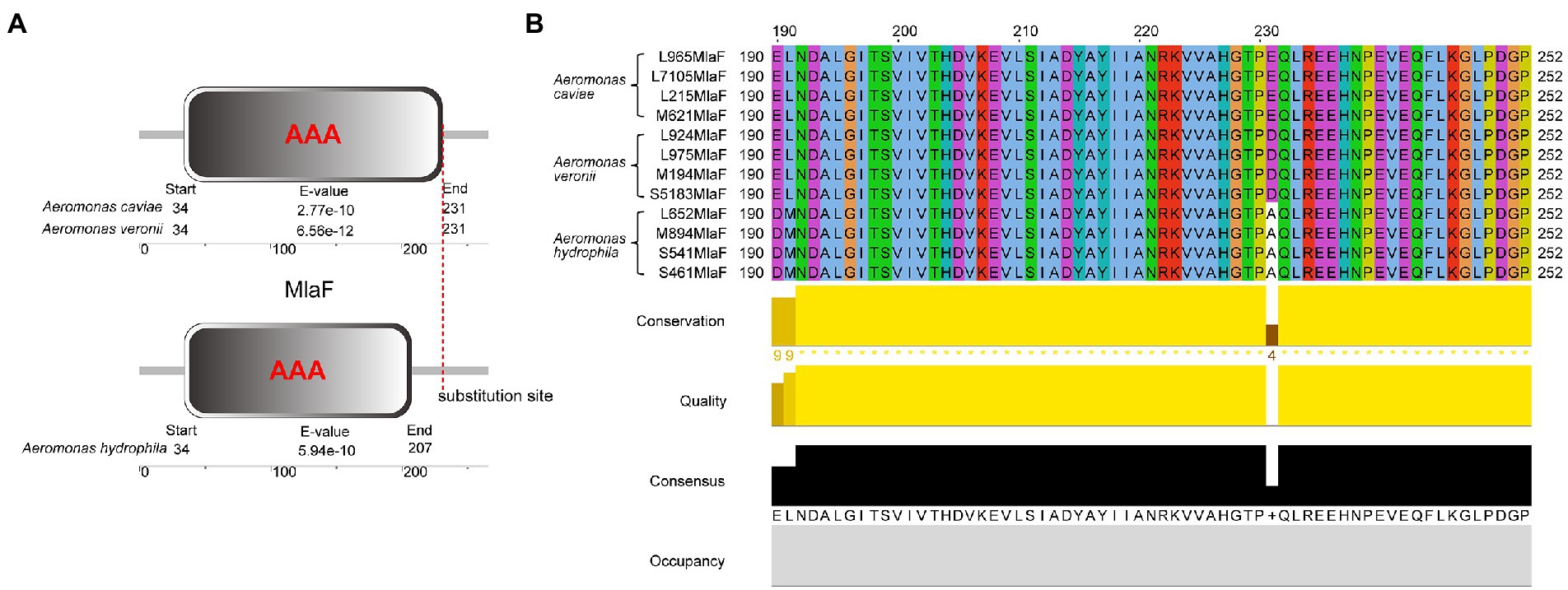
Figure 4. The structure and multiple sequence alignment of MlaF protein. (A) The structure of MlaF domain. (B) The multiple sequence alignment of MlaF from 225 position to the end in three species of the genus Aeromonas. The beautification of the image was used the Jalview 2.11.1.7 and the amino acid color was used Clustalx.
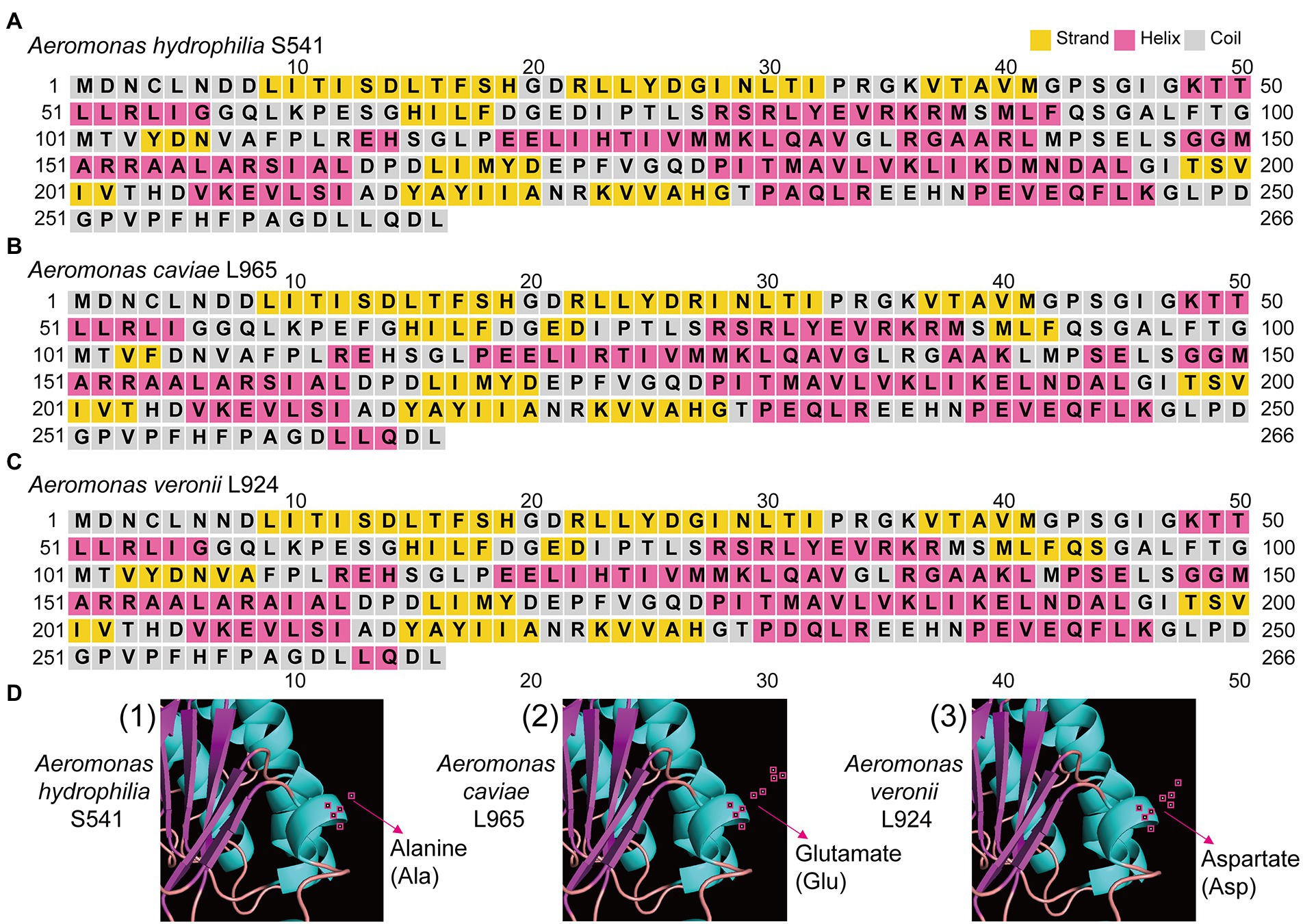
Figure 5. The structure of MlaF protein in three Aeromonas species. (A) The predicted secondary structure of MlaF protein in A. hydrophilia S541. (B) The predicted secondary structure of MlaF protein in A. caviae L965. (C) The predicted secondary structure of MlaF protein in A. veronii L924. (D) The predicted three-dimensional structure of MlaF protein in corresponding strains. The amino acid of 231 position of MlaF was pointed by red arrow.
In order to investigate whether expression levels of these TCS were involved with discrepancy of polymyxin resistance, we selected one strain as a representative strain from each of the three species with the fewest intraspecific mutations based on multiple alignments of TCS genes to perform qRT-PCR. Transcriptional levels of phoP/Q, envZ/ompR, and mlaF in A. hydrophila S541 (PMB MIC>128 mg/L), A. caviae L965 (PMB MIC = 1 mg/L), and A. veronii L924 (PMB MIC = 2 mg/L) were detected by qRT-PCR. A. hydrophila S541 was acted as the control. As shown in Figure 6, compared with A. hydrophila S541, transcriptional levels of phoP, envZ, ompR, and mlaF decreased in A. veronii L924, while only expression levels of phoP/Q decreased in A. caviae L965. We inferred that upregulated TCS PhoPQ in the colistin-resistant strain A. hydrophila S541 might participate in the diversity of colistin resistance among the genus Aeromonas.
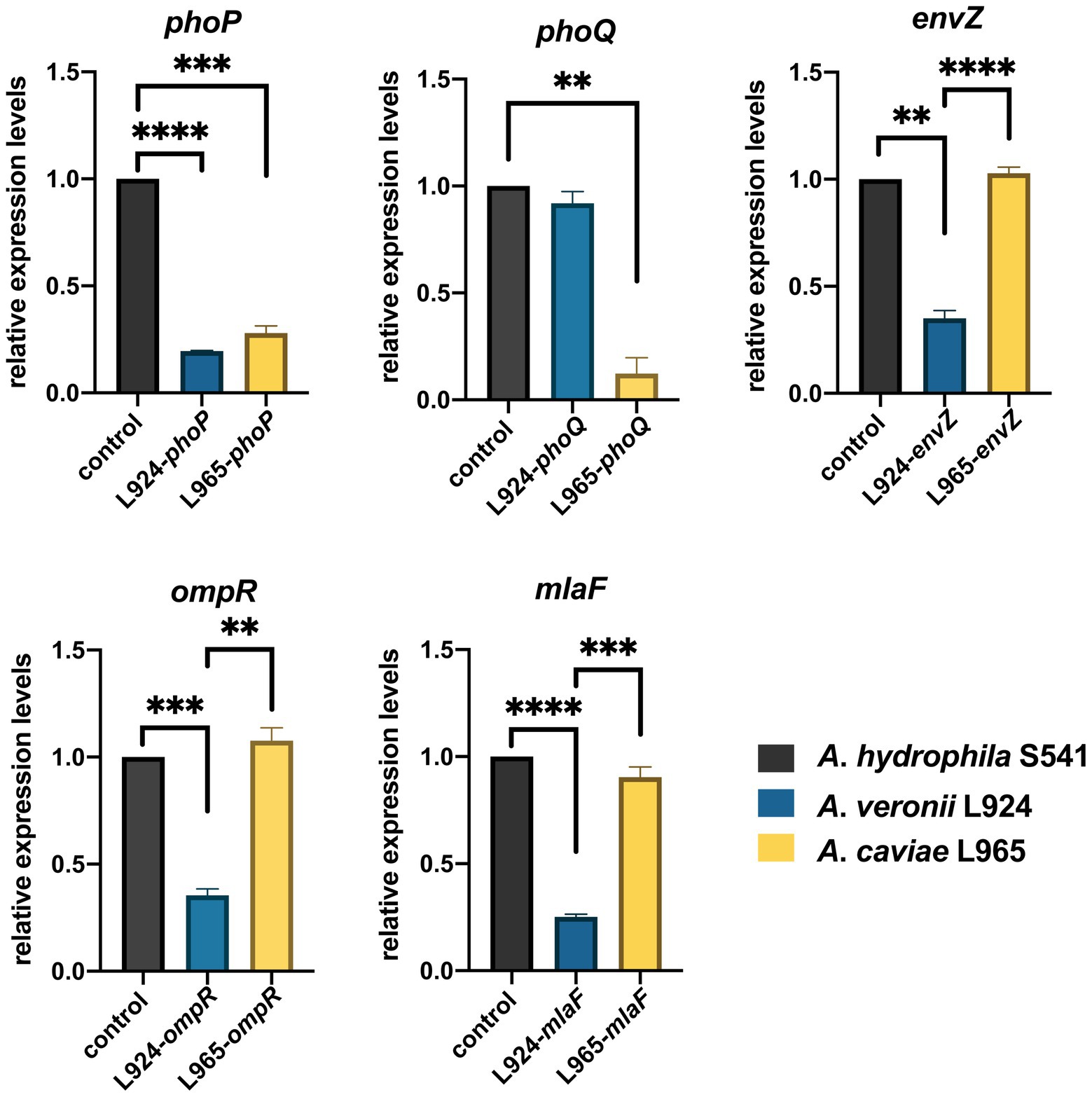
Figure 6. Gene expression of A. hydrophila S541, A. veronii L924, A. caviae L965. Relative expression levels of genes were determined using the 2−ΔΔct method. Error bars represent the standard deviation of three biological replicates. **p < 0.01; ***p < 0.001, ****p < 0.0001.
Water, especially the sewage regeneration system is the medium for the spread of Aeromonas spp. (McLellan et al., 2010; Pablos et al., 2011). The epidemiological study of Aeromonas spp. and sewage metagenomic studies have reported that A. caviae strains could be isolated from drinking water samples and diarrhea feces. Chen et al confirmed that horizontal gene transfer (HGT) was the dominant way to spread drug resistance in the water phase (Chen et al., 2019). The aquatic environment could be a reservoir for the transmission of polymyxin-resistant bacteria. For example, the aquatic bacterium Shewanella and its associated aquaculture are the reservoir for the dissemination of mcr-4 (Zhang et al., 2019a,b). It has been reported that Aeromonas may be the container for mcr-3 and mcr-7, although it is not the inherent host of mcr-3 (Parker and Shaw, 2011; Eichhorn et al., 2018; Shen et al., 2018). According to available research reports, it reveals that variants of mcr-1 to mcr-10 exhibit selectivity to some specific species. For example, mcr-3 and mcr-7 genes have been reported to locate on the chromosomes in the genus Aeromonas such as A. hydrophila, A. veronii, A. caviae, A. jandaei (Parker and Shaw, 2011; Ling et al., 2017). Our study proved this and showed that the genus Aeromonas and aquatic environment might be the potential container and reservoir of mcr-3.
Compared with abundant studies focused on polymyxin-resistant bacteria from clinics, less work has been carried out in the aquatic environment in China (Yang et al., 2017; Tuo et al., 2018; Pan et al., 2022). By phylogenetic tree analysis, we found that three mcr-3 positive Aeromonas strains isolated from Yangtze River have a close evolution distance with clinical samples from China and animal samples from the United States, showing a wide spread of the mcr-3 in Aeromonas strains. In this study, susceptibility of PMB between different species in the genus Aeromonas exhibited a distinct discrepancy according to the results of MIC50 and MIC90 (p < 0.001). No mcr gene was found in A. hydrophila, but all the strains showed high-level resistance to PMB (8 mg/L ≤ MIC ≤128 mg/L). Three mcr-3 positive strains were found in the A. caviae and A. veronii strains while most A. caviae and A. veronii isolates were susceptible to PMB (0.5 mg/L ≤ MIC ≤4 mg/L). Gene mcr-3 in this study were all located on chromosomes as other studies have reported (Ling et al., 2017; Liu et al., 2020; Wang et al., 2021). This is the first report about the identification of mcr-3.42 positive A. veronii strains isolated from the aquatic environment in China and was also the first report of the variant of polymyxin resistance phenotype in different species of the genus Aeromonas.
Genetic analysis in the surrounding gene–environment of the mcr-3 positive isolates found that the mcr-3-like was truncated by ISAs20 (1,674 bp) and ISAs2 (1,084 bp) in two A. veronii isolates, leading to the separation of mcr-3-mcr-3-like segment. The truncated mcr-3-mcr-3-like segment is seldomly observed in the genus Aeromonas. Likewise, the mcr-3-like gene was also divided into two fragments by IS4 in the A. hydrophila WP7-S18-ESBL-06. It indicated that the mcr-3-mcr-3-like segment was not so conserved that it could be divided by IS. In the upstream region of mcr-3, there also lied an IS. Wang et al have reported a novel transposon Tn6518 in the A. veronii w55, composing the genetic element ISAs2-ISAhy2-ISAs20-mcr-3.6-mcr-3-like-dgkA-ISAs2 (Wang et al., 2020). In A. veronii FC951, mcr-3.19 was inserted by an ISAs18 and become inactive, making the A. veronii FC951 susceptible to colistin (Ragupathi et al., 2020). Some IS family members, including IS3, IS30, IS110, IS26, and ISCR1 elements, utilize circular DNA intermediates containing accessory genes to undergo gene translocation via copy-and-paste mechanisms. We supposed that the IS-gene-mcr-3-mcr-3-like-truncated-IS would be conducive to the spread of colistin resistance in different strains and leads to the dissemination of mcr-3 in the aquatic environment. The three mcr-positive strains isolated from the Yangtze River were all susceptible to PMB. Among the reported mcr-3 positive Aeromonas, most of the A. veronii and A. caviae were sensitive to polymyxins, while A. hydrophila were highly resistant to polymyxin (Ling et al., 2017; Shen et al., 2018; Hatrongjit et al., 2020; Huang et al., 2020; Wang et al., 2021). Therefore, we speculated that mcr-3 might not be the main factor leading to the resistance to polymyxin among Aeromonas. One possible reason could be that the expression of mcr-3 might not be activated due to the loss of expression elements during the process of the insertion of ISAs20 and ISAs2.
Genome analysis of the isolates discovered that arnBCADTEF operon only existed in A. hydrophila genomes but not in A. caviae and A. veronii genomes, which might be a reason for the different phenotypes of polymyxin susceptibility among these Aeromonas strains. It has been reported that A. hydrophila could thrive in a complex colistin environment with the help of EnvZ/OmpR TCS and MlaF (Liu et al., 2021). We used these as the entry point and tried to investigate the possible cause for diversity in the colistin resistance in the Aeromonas genus by comparing gene sequences of PhoP/PhoQ TCS, EnvZ/OmpR TCS, and MlaF. Though some site mutations were found in PhoP/PhoQ and EnvZ/OmpR TCS, this nucleic acid substitution did not change the conformation structure of these proteins. Only nucleic acid changes in MlaF were predicted to impact the length of its domain. MlaF was one of the proteins of the Mla system that has played an essential role in phospholipid (PL) transport and constituted the inner membrane ABC transporter complex, MlaFEDB (Coudray et al., 2020). It is reported that MlaF was involved in the antibiotic resistance in A. hydrophila and the mutation in mlaF (mlaFD173A) confers high-level colistin resistance via upregulation of the Mla system (Liu et al., 2021; Zhou et al., 2021). In this study, the substitution at the 231 position of the MlaF was predicated to be deleterious and this substitution may affect the biological function when the acidic amino acid that is E or D in the susceptible strains (A. caviae and A. veronii) is substituted for the neutral amino acid that is A in the resistant strains (A. hydrophila). We infer that the substitution at 231 position in MlaF in the susceptible strains (A. caviae and A. veronii) may alter the steric hindrance and may be related to the polymyxin resistance in different species. In addition, MlaF subunit possesses a unique ~25 aa C-terminal extension (CTE) forming a domain-swapped reciprocal ‘handshake’ that interacts with the adjacent MlaB to fulfill the function (Kolich et al., 2020). And results of the secondary structure alignment of MlaF showed that helix existed in the susceptible strains (A. caviae and A. veronii, Figures 5B,C), not in the resistant strains (A. hydrophila, Figure 5A), which might have an influence on the interaction with MlaB, hence affecting the Mla system. In addition, the expression level of mlaF in A. hydrophila S541 and A. caviae L965 was much higher than that in A. veronii L924 (Figure 6). We supposed that the higher expression of mlaF might cause an increased level of PL and maintain the membrane homeostasis so as to withstand the polymyxin. Thus, we inferred that the amino acids differences in MlaF protein and its expression levels among different Aeromonas species could affect the function of transportation in the Mla system which is involved in the synthesis of membrane components and contribute to the resistance to polymyxins. In general, we speculated that the existence of arnBCADTEF and sequence differences in MlaF might contribute to the variety of polymyxin susceptibility in different species of the genus Aeromonas.
In sum, this is the first report describing the variety in the phenotype of polymyxin susceptibility in different species of the genus Aeromonas. And our study also identified two novel mcr-3.42 positive A. veronii strains and one mcr-3.16 positive A. caviae strain in the aquatic environment in China for the first time. We provided pieces of evidence of the possible mechanism for the different polymyxin susceptibility in different species of the genus Aeromonas, but the exact mechanism deserved further research.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
QL conceived and designed the experiments. LX, HF, and JF performed the experiments. YY, QL, and HF contributed to the reagents, materials, and analysis tools. LX wrote the original draft. QL and FW checked and revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the National Key R&D Program of China (2021YFC2300302), National Natural Science Foundation of China (31900021), National Science Foundation of Zhejiang Province, China (LY20H190002), and Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022031C).
We would like to thank the First Affiliated Hospital of Zhejiang University for providing the equipment required for the experiment. We sincerely thank Xiao for providing the ATCC 25922 and E. coli J53.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1030564/full#supplementary-material
1. ^https://eucast.org/clinical_breakpoints/
2. ^https://ncbi.nlm.nih.gov/orffinder/
4. ^http://github.com/tseemann/prokka
5. ^http://genomicepidemiology.org/
6. ^https://www.ebi.ac.uk/Tools/msa/clustalo/
7. ^https://www.ncbi.nlm.nih.gov/data-hub/genome/
8. ^https://www.ncbi.nlm.nih.gov/pathogens/isolates/
9. ^https://www.ebi.ac.uk/Tools/msa/clustalo/
10. ^http://provean.jcvi.org/index.php
Aedo, S., Ivanova, L., Tomova, A., and Cabello, F. C. (2014). Plasmid-related quinolone resistance determinants in epidemic Vibrio parahaemolyticus, uropathogenic Escherichia coli, and marine bacteria from an aquaculture area in Chile. Microb. Ecol. 68, 324–328. doi: 10.1007/s00248-014-0409-2
Chen, J. S., Hsu, G. J., Hsu, B. M., Yang, P. Y., Kuo, Y. J., Wang, J. L., et al. (2021). Prevalence, virulence-gene profiles, antimicrobial resistance, and genetic diversity of human pathogenic Aeromonas spp. from shellfish and aquatic environments. Environ. Pollut. 287:117361. doi: 10.1016/j.envpol.2021.117361
Chen, Y., Li, P., Huang, Y., Yu, K., Chen, H., Cui, K., et al. (2019). Environmental media exert a bottleneck in driving the dynamics of antibiotic resistance genes in modern aquatic environment. Water Res. 162, 127–138. doi: 10.1016/j.watres.2019.06.047
Coudray, N., Isom, G. L., MacRae, M. R., Saiduddin, M. N., Bhabha, G., and Ekiert, D. C. (2020). Structure of bacterial phospholipid transporter MlaFEDB with substrate bound. elife 9:e62518. doi: 10.7554/eLife.62518
Eichhorn, I., Feudi, C., Wang, Y., Kaspar, H., Feßler, A. T., Lübke-Becker, A., et al. (2018). Identification of novel variants of the colistin resistance gene mcr-3 in Aeromonas spp. from the national resistance monitoring programme GERM-vet and from diagnostic submissions. J. Antimicrob. Chemother. 73, 1217–1221. doi: 10.1093/jac/dkx538
Falagas, M. E., and Rafailidis, P. I. (2008). Re-emergence of colistin in today's world of multidrug-resistant organisms: personal perspectives. Expert Opin. Investig. Drugs 17, 973–981. doi: 10.1517/13543784.17.7.973
Hadjadj, L., Baron, S. A., Olaitan, A. O., Morand, S., and Rolain, J. M. (2019). Co-occurrence of variants of mcr-3 and mcr-8 genes in a Klebsiella pneumoniae isolate from Laos. Front. Microbiol. 10:2720. doi: 10.3389/fmicb.2019.02720
Hatrongjit, R., Kerdsin, A., Takeuchi, D., Wongsurawat, T., Jenjaroenpun, P., Chopjitt, P., et al. (2020). Genomic analysis of Aeromonas veronii C198, a novel Mcr-3.41-harboring isolate from a patient with septicemia in Thailand. Pathogens 9:1031. doi: 10.3390/pathogens9121031
Huang, Z., Dong, N., Chen, S., Shu, L., Sun, Q., Zhou, H., et al. (2020). Detection and genetic characterization of the colistin resistance gene mcr-3.3 in an Aeromonas veronii strain isolated from alligator faeces. J. Glob. Antimicrob. Resis. 22, 860–861. doi: 10.1016/j.jgar.2020.07.003
Hussein, N. H., Al-Kadmy, I. M. S., Taha, B. M., and Hussein, J. D. (2021). Mobilized colistin resistance (mcr) genes from 1 to 10: a comprehensive review. Mol. Biol. Rep. 48, 2897–2907. doi: 10.1007/s11033-021-06307-y
Jia, J., Cheng, M., Xue, X., Guan, Y., and Wang, Z. (2020). Characterization of tetracycline effects on microbial community, antibiotic resistance genes and antibiotic resistance of Aeromonas spp. in gut of goldfish Carassius auratus Linnaeus. Ecotoxicol. Environ. Saf. 191:110182. doi: 10.1016/j.ecoenv.2020.110182
Kolich, L. R., Chang, Y. T., Coudray, N., Giacometti, S. I., MacRae, M. R., Isom, G. L., et al. (2020). Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation. elife 9:60030. doi: 10.7554/eLife.60030
Ling, Z., Yin, W., Li, H., Zhang, Q., Wang, X., Wang, Z., et al. (2017). Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob. Agents Chemother. 61:e01272. doi: 10.1128/AAC.01272-17
Liu, D., Song, H., Ke, Y., Xia, J., Shen, Y., Ou, Y., et al. (2020). Co-existence of two novel phosphoethanolamine transferase gene variants in Aeromonas jandaei from retail fish. Int. J. Antimicrob. Agents 55:105856. doi: 10.1016/j.ijantimicag.2019.11.013
Liu, J., Xiao, G., Zhou, W., Yang, J., Wang, Y., Wu, Y., et al. (2021). Various novel Colistin resistance mechanisms interact to facilitate adaptation of Aeromonas hydrophila to complex Colistin environments. Antimicrob. Agents Chemother. 65:e0007121. doi: 10.1128/aac.00071-21
Ma, S., Sun, C., Hulth, A., Li, J., Nilsson, L. E., Zhou, Y., et al. (2018). Mobile colistin resistance gene mcr-5 in porcine Aeromonas hydrophila. J. Antimicrob. Chemother. 73, 1777–1780. doi: 10.1093/jac/dky110
McLellan, S. L., Huse, S. M., Mueller-Spitz, S. R., Andreishcheva, E. N., and Sogin, M. L. (2010). Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ. Microbiol. 12, 378–392. doi: 10.1111/j.1462-2920.2009.02075.x
Nonaka, L., Maruyama, F., Suzuki, S., and Masuda, M. (2015). Novel macrolide-resistance genes, mef(C) and mph(G), carried by plasmids from vibrio and Photobacterium isolated from sediment and seawater of a coastal aquaculture site. Lett. Appl. Microbiol. 61, 1–6. doi: 10.1111/lam.12414
Noonburg, G. E. (2005). Management of extremity trauma and related infections occurring in the aquatic environment. J. Am. Acad. Orthop. Surg. 13, 243–253. doi: 10.5435/00124635-200507000-00004
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Pablos, M., Huys, G., Cnockaert, M., Rodriguez-Calleja, J. M., Otero, A., Santos, J. A., et al. (2011). Identification and epidemiological relationships of Aeromonas isolates from patients with diarrhea, drinking water and foods. Int. J. Food Microbiol. 147, 203–210. doi: 10.1016/j.ijfoodmicro.2011.04.006
Pan, Y., Zeng, Z., Niu, H., Huang, L., Hu, J., Li, G., et al. (2022). Whole-genome epidemiology and characterisation of mcr-1-encoding Escherichia coli in aquatic bird farms from the Pearl River Delta, China, 2019–2020. Int. J. Antimicrob. Agents 59:106478. doi: 10.1016/j.ijantimicag.2021.106478
Parker, J. L., and Shaw, J. G. (2011). Aeromonas spp. clinical microbiology and disease. J. Infect. 62, 109–118. doi: 10.1016/j.jinf.2010.12.003
Pei, R., Kim, S. C., Carlson, K. H., and Pruden, A. (2006). Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 40, 2427–2435. doi: 10.1016/j.watres.2006.04.017
Pruden, A., Pei, R., Storteboom, H., and Carlson, K. H. (2006). Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ. Sci. Technol. 40, 7445–7450. doi: 10.1021/es060413l
Ragupathi, N. K. D., Sethuvel, D. P. M., Anandan, S., Murugan, D., Asokan, K., Neethi Mohan, R. G., et al. (2020). First hybrid complete genome of Aeromonas veronii reveals chromosome-mediated novel structural variant mcr-3.30 from a human clinical sample. Access Microbiol. 2:acmi000103. doi: 10.1099/acmi.0.000103
Scarano, C., Piras, F., Virdis, S., Ziino, G., Nuvoloni, R., Dalmasso, A., et al. (2018). Antibiotic resistance of Aeromonas ssp. strains isolated from Sparus aurata reared in Italian mariculture farms. Int. J. Food Microbiol. 284, 91–97. doi: 10.1016/j.ijfoodmicro.2018.07.033
Seeliger, D., and de Groot, B. L. (2010). Ligand docking and binding site analysis with PyMOL and autodock/Vina. J. Comput. Aided Mol. Des. 24, 417–422. doi: 10.1007/s10822-010-9352-6
Shen, Y., Xu, C., Sun, Q., Schwarz, S., Ou, Y., Yang, L., et al. (2018). Prevalence and genetic analysis of mcr-3-positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob. Agents Chemother. 62:e00404. doi: 10.1128/AAC.00404-18
Sun, Q., Hu, Y., Zhou, H., Shu, L., Wang, H., Huang, Z., et al. (2018). Alkaline peptone water-based enrichment method for mcr-3 from acute diarrheic outpatient gut samples. Front. Med. 5:99. doi: 10.3389/fmed.2018.00099
Sun, Y., Zhao, Y., Xu, W., Fang, R., Wu, Q., He, H., et al. (2021). Taxonomy, virulence determinants and antimicrobial susceptibility of Aeromonas spp. isolated from bacteremia in southeastern China. Antimicrob. Resist. Infect. Control 10:43. doi: 10.1186/s13756-021-00911-0
Trimble, M. J., Mlynárčik, P., Kolář, M., and Hancock, R. E. (2016). Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 6:a025288. doi: 10.1101/cshperspect.a025288
Tuo, H., Yang, Y., Tao, X., Liu, D., Li, Y., Xie, X., et al. (2018). The prevalence of Colistin resistant strains and antibiotic resistance gene profiles in Funan River China. Front. Microbiol. 9:3094. doi: 10.3389/fmicb.2018.03094
Wang, X., Zhai, W., Wang, S., Shen, Z., Wang, Y., and Zhang, Q. (2020). A novel transposon, Tn6518, mediated transfer of mcr-3 variant in ESBL-producing Aeromonas veronii. Infect. Drug Res. 13, 893–899. doi: 10.2147/idr.S239865
Wang, Y., Hou, N., Rasooly, R., Gu, Y., and He, X. (2021). Prevalence and genetic analysis of chromosomal mcr-3/7 in Aeromonas from US animal-derived samples. Front. Microbiol. 12:667406. doi: 10.3389/fmicb.2021.667406
Woodman, M. E., Savage, C. R., Arnold, W. K., and Stevenson, B. (2016). Direct PCR of intact bacteria (Colony PCR). Curr. Protoc. Microbiol. 42, A.3d.1–a.3d.7. doi: 10.1002/cpmc.14
Yang, D., Qiu, Z., Shen, Z., Zhao, H., Jin, M., Li, H., et al. (2017). The occurrence of the Colistin resistance gene mcr-1 in the Haihe River (China). Int. J. Environ. Res. Public Health 14:576. doi: 10.3390/ijerph14060576
Yuwono, C., Wehrhahn, M. C., Liu, F., Riordan, S. M., and Zhang, L. (2021). The isolation of Aeromonas species and other common enteric bacterial pathogens from patients with gastroenteritis in an Australian population. Microorganisms 9:1440. doi: 10.3390/microorganisms9071440
Zhang, H., Hou, M., Xu, Y., Srinivas, S., Huang, M., Liu, L., et al. (2019a). Action and mechanism of the colistin resistance enzyme MCR-4. Commun. Biol. 2:36. doi: 10.1038/s42003-018-0278-1
Zhang, H., Wei, W., Huang, M., Umar, Z., and Feng, Y. (2019b). Definition of a family of nonmobile Colistin resistance (NMCR-1) determinants suggests aquatic reservoirs for MCR-4. Adv. Sci. 6:1900038. doi: 10.1002/advs.201900038
Zhou, C., Shi, H., Zhang, M., Zhou, L., Xiao, L., Feng, S., et al. (2021). Structural insight into phospholipid transport by the MlaFEBD complex from P. aeruginosa. J. Mol. Biol. 433:166986. doi: 10.1016/j.jmb.2021.166986
Keywords: aquatic environment, polymyxin resistance, mcr, MlaF, Aeromonas
Citation: Xu L, Fan J, Fu H, Yang Y, Luo Q and Wan F (2022) The variants of polymyxin susceptibility in different species of genus Aeromonas. Front. Microbiol. 13:1030564. doi: 10.3389/fmicb.2022.1030564
Received: 29 August 2022; Accepted: 07 October 2022;
Published: 25 October 2022.
Edited by:
Chang-Wei Lei, Sichuan University, ChinaReviewed by:
Jun Li, Zhejiang University of Technology, ChinaCopyright © 2022 Xu, Fan, Fu, Yang, Luo and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fen Wan, ZmVudzA4MThAMTYzLmNvbQ==; Qixia Luo, cWl4aWFfbHVvQHpqdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.