
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 05 October 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1029426
This article is part of the Research TopicAnimal Emerging and Reemerging DiseasesView all 24 articles
 Feng Liu1†
Feng Liu1† Qing Yao1†
Qing Yao1† Jing Huang2
Jing Huang2 Jiajia Wan1
Jiajia Wan1 Tingting Xie1
Tingting Xie1 Xuejun Gao1
Xuejun Gao1 Diangang Sun1
Diangang Sun1 Fuxian Zhang1
Fuxian Zhang1 Weicheng Bei3*
Weicheng Bei3* Liancheng Lei1,4*
Liancheng Lei1,4*Actinobacillus pleuropneumoniae, a major bacterial porcine respiratory tract pathogen causing pig pleuropneumonia, has resulted in high economic losses worldwide. The mutation of the two-component system CpxAR strongly impacted the virulence of A. pleuropneumoniae, but the underlying regulatory mechanism remained unclear. Here, we found that CpxAR positively regulated the cpxDCBA gene cluster involved in polysaccharide capsule export. A capsular layer was confirmed in wild-type cells by transmission electron microscopy, whereas cpxAR and cpxD mutants were non-capsulated. The mutants for polysaccharide capsule export gene cpxD exhibited non-capsulated and were strongly impaired in virulence for mice, indicating a major role of CPS export system in virulence. We then demonstrated that CpxR directly regulated the transcription of the CPS export gene cluster cpxDCBA. Taken together, our data suggested that CpxAR is a key modulator of capsule export that facilitates A. pleuropneumoniae survival in the host.
Actinobacillus pleuropneumoniae is a Gram-negative facultative anaerobic bacterium belonging to the Pasteurellaceae family (Sassu et al., 2018). This pathogen is the aetiological agent of porcine pleuropneumonia in pigs of all ages, which is a highly contagious and often deadly respiratory disease causing substantial economic losses in the swine industry worldwide (Cohen et al., 2021). A. pleuropneumoniae colonizes the tonsils and nasal cavities of infected pigs, and transmission from pig to pig occurs mainly by respiratory droplets or direct contact (Bosse et al., 2002). It is currently classified into 19 serovars based on the antigenic properties of their capsular polysaccharides and lipopolysaccharides (Stringer et al., 2021). Previous studies have reported that many virulence factors play an important role in the pathogenicity of A. pleuropneumoniae, such as lipopolysaccharide, Apx toxins, polysaccharide capsule, and iron acquisition proteins (Loera-Muro et al., 2021).
Capsule polysaccharide (CPS) is encoded by the genes of the CPS biosynthetic locus cpsABCD and the CPS export locus cpxDCBA in A. pleuropneumoniae (Hathroubi et al., 2016). Unlike the former, the CPS export genes, cpxDCBA, are conserved in different A. pleuropneumoniae serovars (Bandara et al., 2003). The four genes encode an outer membrane lipoprotein (CpxD), a cytoplasmic membrane protein (CpxC), an integral membrane protein (CpxB), and an ATP-binding protein (CpxA) (Ward and Inzana, 1997).
Two-component signaling systems (TCSs), usually comprising a sensor histidine kinase (HK) and a cytoplasmic response regulator (RR), are involved in bacterial colonization and virulence through sensing environmental stimulus and responding accordingly (West and Stock, 2001; Li et al., 2018). Once it senses a physical or chemical signal, the histidine kinase will lead to autophosphorylation. The phosphoryl group is then transferred to the response regulator, which subsequently binds the target DNA promoters to alter gene expression (Gu et al., 2020). The genome of A. pleuropneumoniae encodes 5 paired of two-component systems, such as CpxAR, QseBC, ArcAB, PhoBR, and NarPQ (Xu et al., 2008).
The well-known TCS CpxAR, including the sensor histidine kinase CpxA, the cytopalsmic response regulator CpxR and the accessory protein CpxP, is required for sensing and coordinating the response to the envelope stress in E. coli (Raivio and Silhavy, 1997; Xu et al., 2014). In A. pleuropneumoniae, the Cpx system is composed of the histidine kinase CpxA and response regulator CpxR, and their genes constitute the cpxRA gene cluster (Li et al., 2018). Previous studies showed that the CpxAR system is a pleiotropic TCS in A. pleuropneumoniae, and involved in biofilm formation, oxidative stress, osmotic stress, heat stress, and virulence (Li et al., 2018; Yan et al., 2020). Li et al. found that CpxAR affects biofilm formation by regulating the expression of the pgaABCD operon through rpoE in A. pleuropneumoniae (Li et al., 2018). CpxAR has been reported to contribute to the virulence of A. pleuropneumoniae by altering O-antigen repeating unit biosynthesis (Yan et al., 2020). However, more underlying mechanisms of the CpxAR-mediated pathogensis of A. pleuropneumoniae remain to be elucidated. Here, the principal objective of this study is to reveal that the regulatory mechanism employed by CpxAR contributes to A. pleuropneumoniae virulence.
For this study, we used A. pleuropneumoniae strain S4074 as a representative strain. A. pleuropneumoniae strain was routinely grown at 37°C with shaking (180 rpm) in tryptic soy broth (TSB) medium (Solarbio, Beijing, China) supplemented with 10 μg/ml Nicotinamide Adenine Dinucleotide (NAD; Biofroxx, Einhausen, Germany) and 10% (vol/vol) newborn bovine serum (FBS; EVERY GREEN, Hangzhou, China). Escherichia coli strain BL21 was grown at 37°C with shaking (180 rpm) in Luria-Bertani (LB) medium. E. coli β2155 was grown in LB medium supplemented with 50 μg/ml diaminopimelic acid (Sigma-Aldrich, St. Louis, United States). We added 5 μg/ml chloramphenicol, or 50 μg/ml kanamycin as required. All strains and plasmids used in this study are listed in Table 1, and primers (Sangon Biotech Co., Ltd., Shanghai, China) are shown in Table 2.
The pEMOC2 suicide plasmid was used to construct the mutant strain ΔcpxD following an allelic exchange methodology, as described earlier (Liu et al., 2018). The complementation strain CΔcpxD was generated using the shuttle plasmid pJFF224-XN as previously described (Liu et al., 2018). The ΔcpxD mutant and complementation strain CΔcpxD were verified by PCR (Supplementary Figure 1) and sequencing.
RNA extraction and qRT-PCR assays were performed as described earlier, with some modifications (Huang et al., 2018). The wild-type and ∆cpxAR mutant strains were grown in 5 ml of TSB overnight, normalized to an optical density of 600 nm (OD600) of 0.05, and incubated at 37°C with shaking (180 rpm) to an OD600 of 0.6. After the cells have grown to an OD600 of 0.6, cells were centrifuged at 4°C for 5 min at 10, 000 g, treated with the Bacteria Total RNA Isolation Kit (Sangon Biotech, Shanghai, China), and stored at −80°C until analysis. Reverse transcription was performed using the HiScript II first-strand cDNA synthesis kit (Vazyme, Nanjing, China). qRT-PCR was performed using the ViiA-7 Real-Time PCR System (Applied Biosystems, Waltham, United States) and SYBR qPCR Mix (Vazyme, Nanjing, China). Fold change data were normalized according to the housekeeping gene 16S rRNA, and analyzed using the 2−ΔΔCt method (Livak and Schmittgen, 2001). RT-PCR acrossing the cps2A-cpxD, cpxD-cpxC, cpxC-cpxB, and cpxB-cpxA junctions was conducted as described previously (Xiong et al., 2019; Cheng et al., 2021). Amplified RT-PCR products were electrophoresed and photographed using a Gel Image Analyzing JS-1800 system (Peiqing, China).
The PCR-amplified cpxR gene from A. pleuropneumoniae strain S4074 was cloned into the pET30a vector by digesting with Nde I and Xho I. Then, the CpxR expression plasmid was transformed into E. coli BL21(DE3), and their expression was induced by the addition of 0.5 mM IPTG and incubated at 16°C overnight. The cells were disrupted using a Ultrasonic Homogenizer (SCIENTZ, Ningbo, China), and centrifuged at 12,000 g for 20 min to remove cellular debris. The supernatant was extracted with Ni-nitrilotriacetic acid (Ni-NTA) resin affinity chromatography following the manufacturer’s instructions. The purified protein concentration was determined by a BCA protein assay kit (Beyotime, Shanghai, China), and the purity was checked by 12% SDS-PAGE.
To study the binding of CpxR to the DNA probes, the electrophoretic mobility shift assays (EMSAs) were performed using a Chemiluminescent EMSA Kit (Beyotime, Shanghai, China) as previously described (Cheng et al., 2021). The DNA probes were generated by PCR amplification from 1 to 196 bp upstream of the start codon of cpxD gene, purified using a Gel Extraction Kit (Omega, Norcross, United States), and labeled using a EMSA Probe Biotin Labeling Kit (Beyotime, Shanghai, China). The recombinant protein CpxR was phosphorylated in vitro by 50 mM acetyl phosphate (Sigma, St. Louis, United States) (Pogliano et al., 1997). Increasing amounts of phosphorylated CpxR protein (0 to 4 pmol) were incubated with the labeled probe (1 μM) in binding buffer (50 mM Tris (pH 8.0), 100 mM KCl, 2.5 mM MgCl2, 0.2 mM dithiothreitol (DTT), 2 μg salmon sperm DNA, 10% glycerol) for 20 min at room temperature. The reaction mixtures were directly subjected to 4% non-denaturing polyacrylamide electrophoresis. The gel was transferred to a nylon membrane (Beyotime, Shanghai, China), and imaging was performed using JS-1070 fluorescent chemiluminescence gel imaging system (Peiqing, Shanghai, China).
For preparation of fluorescent 6-carboxyfluorescein (FAM) labeled probe, the cpxD promoter was amplified from 1 to 196 bp upstream of the start codon by PCR using the plasmid pEASY-cpxD as a template and primers of M13F (FAM) and M13R. For each assay, the 6-FAM-labeled probe (400 ng) was mixed with different amounts of phosphorylated CpxR protein in a 40 μl reaction volume for 30 min at 25°C. Subsequently, the mixture was incubated with 0.015 unit DNase I (Promega, Madison, United States) at 37°C for 1 min. The reaction was terminated by adding 140 μl DNase I stop solution (200 mM unbuffered sodium acetate, 30 mM EDTA and 0.15% SDS). The samples were extracted by phenol/chloroform, and the pellets containing DNA were dissolved in 30 μl water. The results were analyzed using 3130xl DNA analyzer and Peak Scanner software v1.0 (Applied Biosystems, Waltham, United States).
For transmission electron microscope (TEM), A. pleuropneumoniae strain was grown on TSA plates overnight at 37°C, harvested by centrifugation (5,000 g, 4°C, 5 min), and then incubated in FBS (EVERY GREEN, Hangzhou, China) for 30 min at 37°C. The samples were fixed for 2 h with 2.5% glutaraldehyde and placed onto 200 mesh copper grids. Subsequently, the copper grids were air-dried and observed with a 120KV biological transmission electron microscope (HITACHI, Tokyo, Japan).
In all experiments, Kunming (KM) mice (female, 6 weeks old) purchased from CTGU University Laboratory Animal Center (Quality Certificate No. 42010200007283) were used. All animal experiments were approved by the Animal Ethics Committee of the Yangtze University. To investigate the survival curves of the mice, 1 × 107 CFUs of WT S4074, ∆cpxAR, ∆cpxD, C∆cpxAR or C∆cpxD was injected via the abdominal cavity of each mouse (8 mice per group). The survival rates were recorded daily in 1 week after challenge.
In addition, to evaluate the colonization ability, another 5 groups (8 mice/group) were given intraperitoneal injections with 1 × 107 CFUs of WT S4074, ∆cpxAR, ∆cpxD, C∆cpxAR or C∆cpxD. At 8 h after injection, one half of the lung and liver from each mice was collected aseptically, weighed, homogenized, diluted serially, and plated to determine bacterial counts. The remaining lungs were fixed in tissuse’s fixative (Biosharp, Beijing, China) at 4°C for histo pathological analysis as described earlier (Guo et al., 2018).
The promoter and the transcriptional start site of the cpxD gene were, respectively, predicated by using BPROM1 and BDGP.2 Two-tailed Student’s t tests were used to analyze the significance between various mutants and WT strain using GraphPad Prism version 7.0 (GraphPad, La Jolla, United States). Values of p < 0.05 was considered statistically significant.
The growth traits of the WT S4074, ∆cpxAR, C∆cpxAR, ∆cpxD, and C∆cpxD strains were investigated. As shown in Figure 1A, only the ∆cpxAR mutant exhibited growth defects, which was consistent with previous studies (Li et al., 2018). To elucidate the mechanism by which CpxAR impacts the pathogenicity of A. pleuropneumoniae, qRT-PCR was performed to identify the genes regulated by CpxAR. The qRT-PCR analysis showed that the relative transcript levels of the four CPS export genes, cpxD, cpxC, cpxB, and cpxA (capsular polysaccharide export gene), were significantly decreased in the ∆cpxAR mutant strain (Figure 1B). These data indicated that CpxAR regulate the expression of the four CPS export genes in A. pleuropneumoniae.
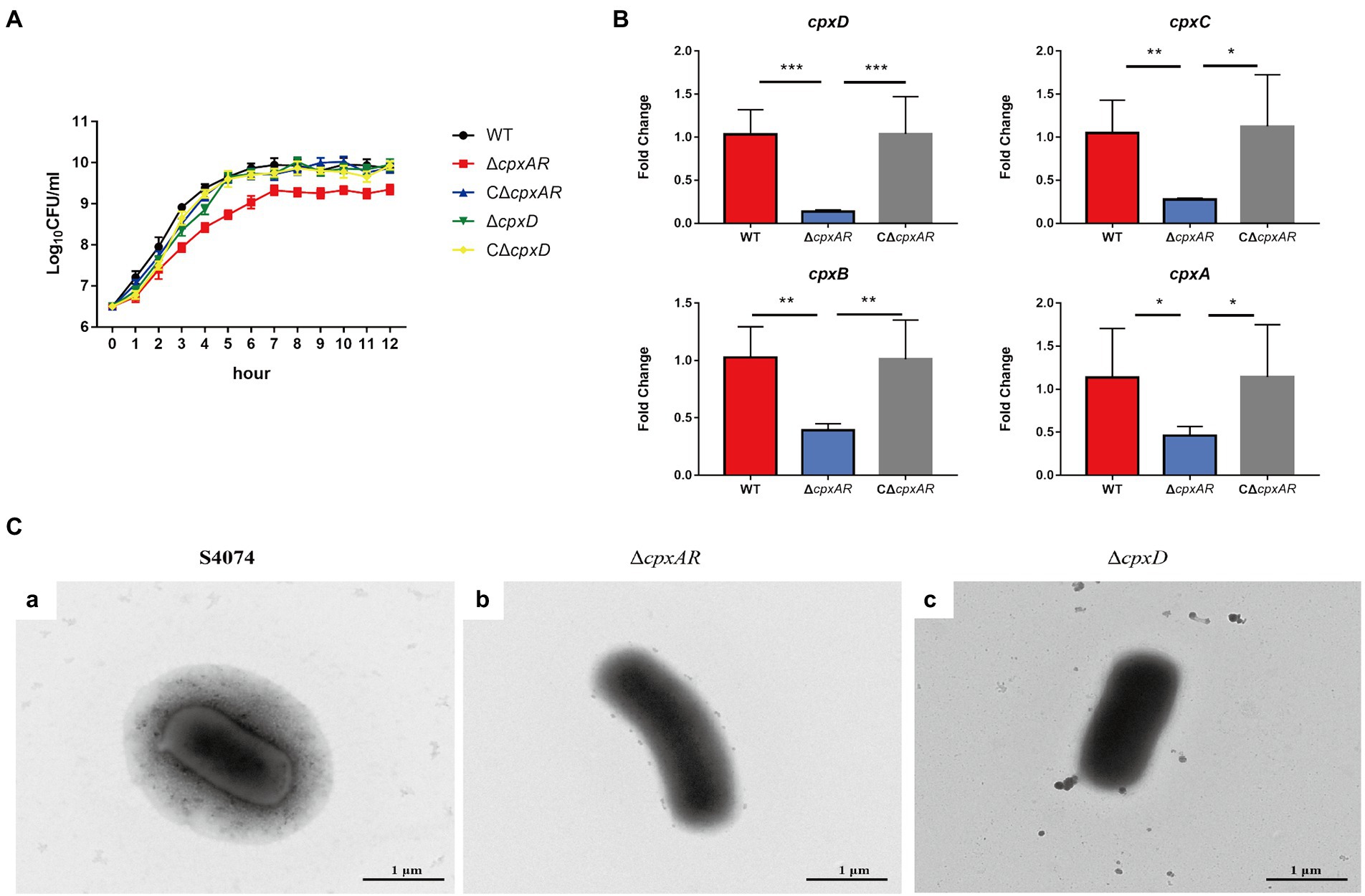
Figure 1. CpxAR impacts the capsule production. (A) The growth analysis of the WT S4074, ∆cpxAR, C∆cpxAR, ∆cpxD, and C∆cpxD strains. (B) The relative mRNA levels of cpxD, cpxC, cpxB, and cpxA (capsular polysaccharide export gene) in the ΔcpxAR mutant compared to the parent strain determined by RT-qPCR. *p < 0.05, **p < 0.01, ***p < 0.001. (C) Observation of the capsule layer of the WT S4074 (A), ∆cpxAR (B), ∆cpxD (C), strains by TEM.
Previous work on A. pleuropneumoniae found that the WT S4074 strain forms a extensive layer of capsular material covering the cells (Jacques et al., 1988). Here, we observed an extensive layer of capsular material covering the WT S4074 as expected, whereas ∆cpxAR and ∆cpxD mutant strains were not found layer of capsular material (Figure 1C). These data indicated that CpxAR contributes to capsule synthesis in A. pleuropneumoniae.
These four capsule export genes, including cpxD, cpxC, cpxB, and cpxA (capsular polysaccharide export gene), are in the chromosome, and such cluster is adjacent to the gene cps2A (Figures 2A,B). To verify whether these four genes are controlled by one promoter, we performed RT-PCR across the cps2A-cpxD, cpxD-cpxC, cpxC-cpxB, and cpxB-cpxA junctions. The RT-PCR analysis indicated that the cpxDCBA gene cluster is a single operon (Figure 2C).
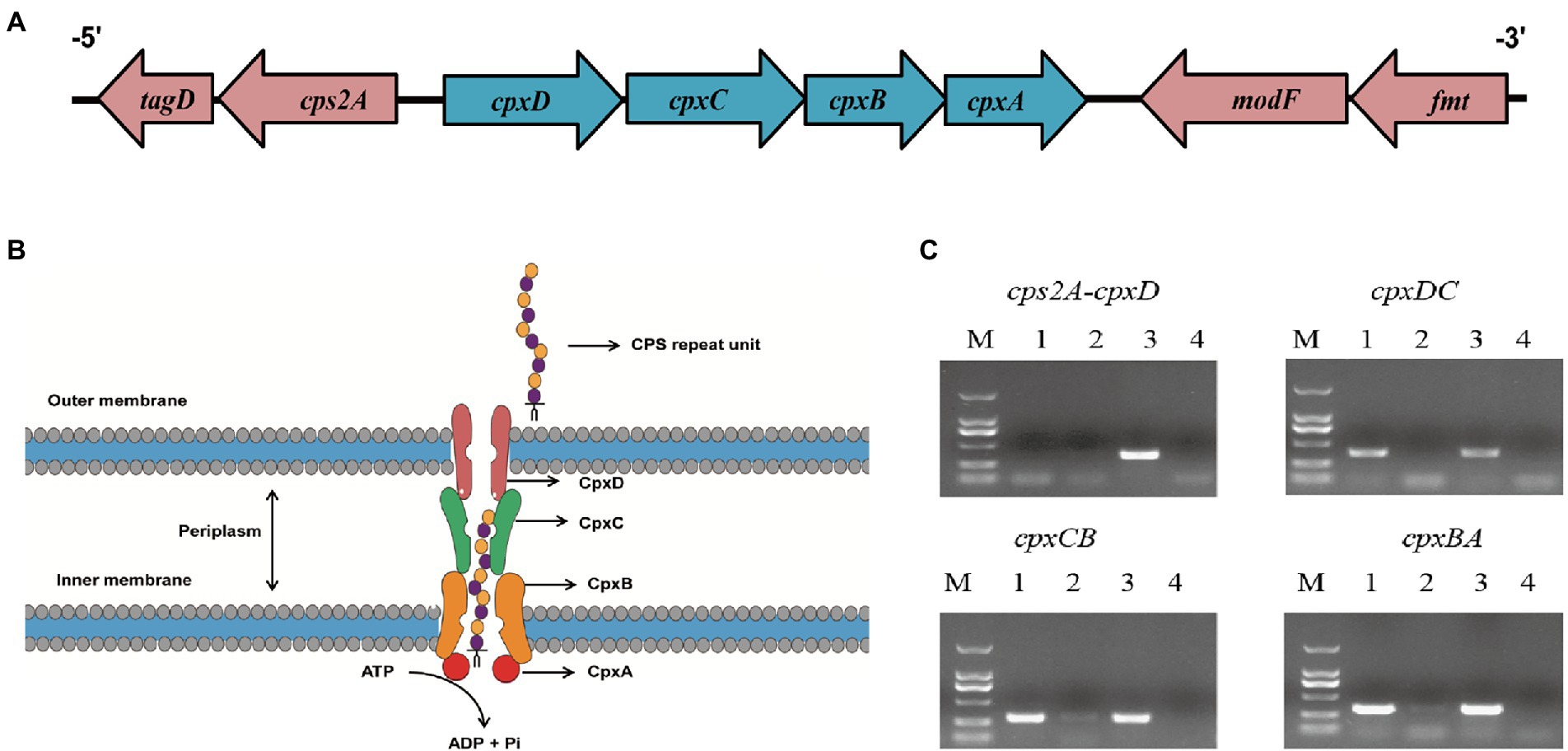
Figure 2. The cpxD, cpxC, cpxB, and cpxA genes form a single operon. (A) Schematic presentation of the capsule export locus. (B) Diagram showing the role of CpxD, CpxC, CpxB, and CpxA (capsular polysaccharide export protein) in CPS synthesis. (C) RT-PCR analysis confirmed that the cpxD, cpxC, cpxB, and cpxA (capsular polysaccharide export gene) genes form an operon. The Lane 1–4 were cDNA, total RNA, genomic DNA, and no-template control, respectively.
To investigate the mechanism of CpxAR-mediated transcriptional regulation of the cpxDCBA operon, we analyzed the binding site for CpxR in the upstream region of cpxDCBA operon using EMSA. EMSAs showed that the CpxR protein could bind to the promoter region of the cpxDCBA operon (Figure 3A). To further analyze the CpxR-cpxD interaction, DNase I footprinting was used to map the precise binding site. Two CpxR-binding sites were found 92 to 121 bp and 146 to 170 bp upstream of the start codon, and the sequences were 5’-TCTATTTACTTTCTTTACAAATGAT-3′ and 5’-TTTTGTAAATTTTTTATATTTAATTTCTCT-3′ respectively (Figure 3B). Collectively, these findings indicated that CpxR directly regulates the expression of cpxDCBA operon.
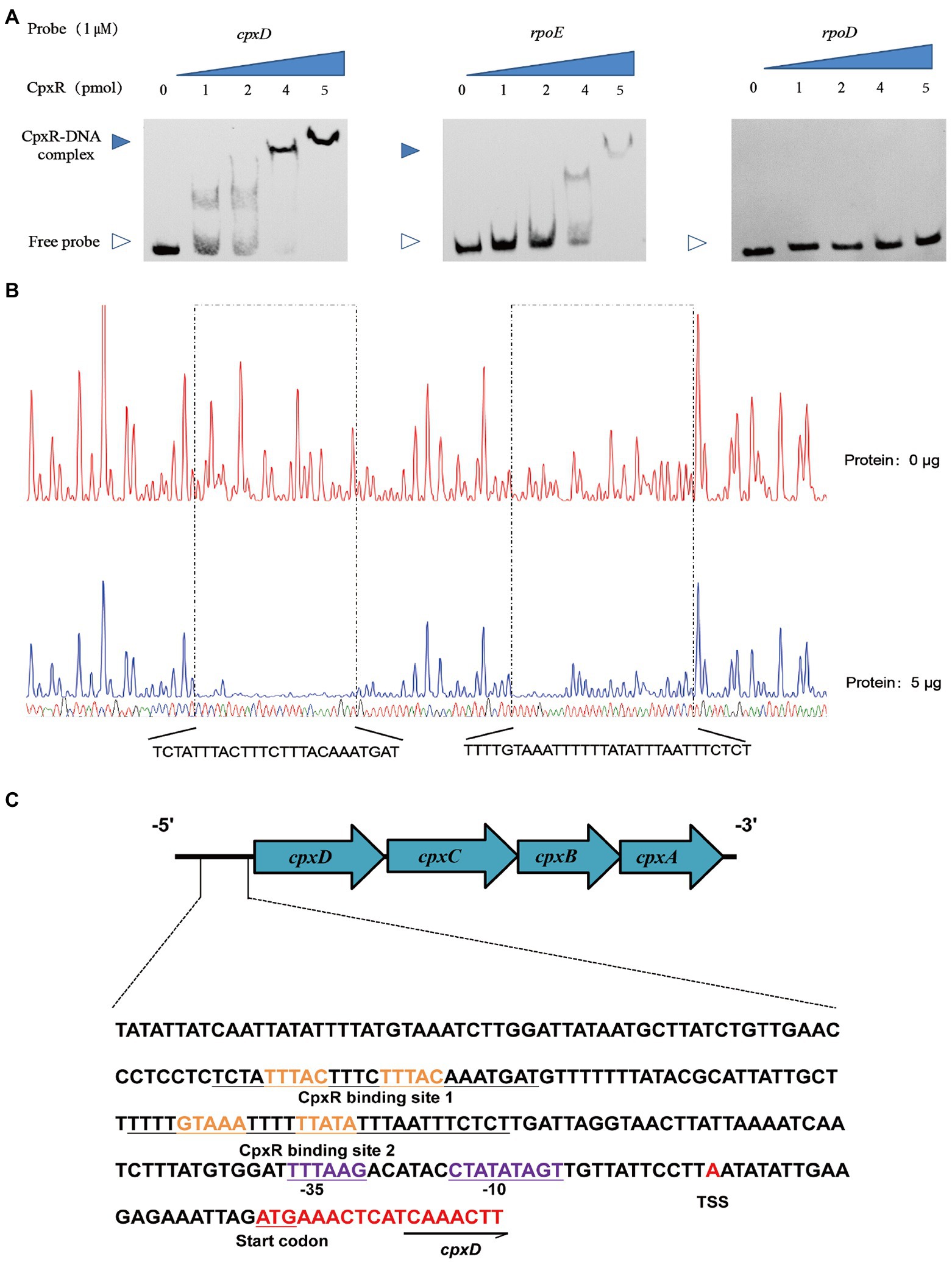
Figure 3. CpxR binds to the cpxD promoter region. (A) EMSAs performed with various concentrations of phosphorylated CpxR (0–4 pmol) and the promoter regions of cpxD, rpoE (positive control) and rpoD (negative control). (B) Mapping the CpxR binding sites in the cpxD promoter by DNase I footprinting. Protected regions were shown below the footprinting results. (C) Nucleotide sequences of cpxD promoter. CpxR-binding site was shown in blue nucleotides boxed in black, and − 35 box, −10 box were underlined and shown in green. Start codon of cpxD was underlined and shown in red, and the TSS was shown in red.
To obtain a more detailed picture of the CpxR binding site, the promoter and the transcription start site of the cpxD gene were, respectively, predicted by BPROM and BDGP. The cpxD transcriptional start site, designated as TSS, was detected 21-bp upstream of the start codon, and determined as A (Figure 3C). In addition, we performed a bioinformatic search in the promoter region of cpxD and identified a putative ˗10 CTATATAGT box and a putative ˗35 TTTAAG box, respectively, located 33 bp and 48 bp upstream of the start codon (Figure 3C).
To verify whether the CpxAR-cpxDCBA pathway plays an important role in the virulence of A. pleuropneumoniae, the WT S4074, ∆cpxAR, C∆cpxAR, ∆cpxD, and C∆cpxD strains were further compared through survival and colonization assays in vivo in KM mice. As shown in Figure 4A, the mice infected by the ∆cpxAR and ∆cpxD strain showed significantly higher survival rate of 83 and 100% respectively; whereas, the mice infected by the WT, C∆cpxAR and C∆cpxD strains showed the survival rate of 0, 17 and 0%, respectively. As shown in Figure 4B, the bacterial loads in the lungs and livers of ∆cpxAR- and ∆cpxD-infected mice were significantly lower than that of WT-, C∆cpxAR- and C∆cpxD-infected mice.
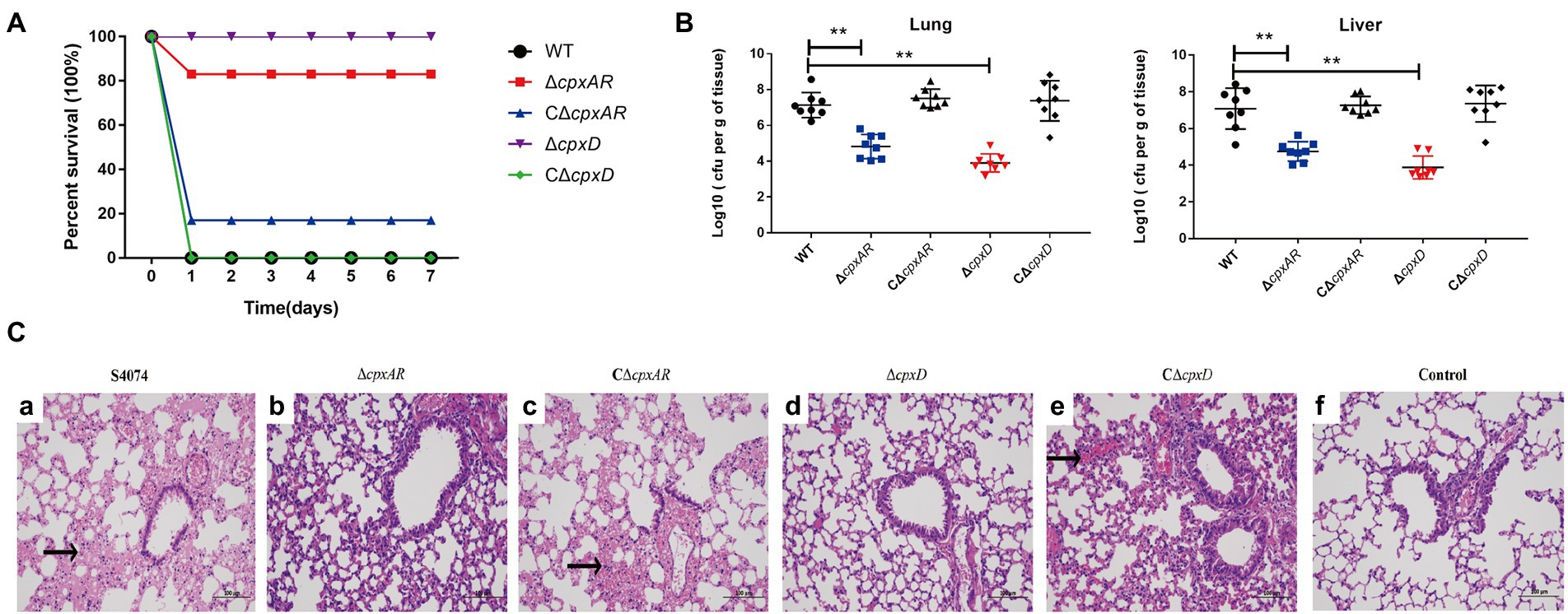
Figure 4. CpxAR and CpxD contribute to the virulence and colonization of A. pleuropneumoniae. The survival rates (A), bacterial loads in the lungs and livers (B), and histopathology of the lung tissues (C) of Kunming mice challenged with the WT S4074 (a), ΔcpxAR (b), ΔcpxD (c), CΔcpxAR (d), and CΔcpxD (e) strains at a dose of 1 × 107 CFU/mouse. The control group (f) was only injected with 100 µl of PBS **p < 0.01.
Histologic examination of the lung tissue sections from all of the mice infected with WT, C∆cpxAR and C∆cpxD strains showed the classic features of pneumonia, such as hyperemia, swelling, hemorrhage, edema, consolidation, but this was not evident in ∆cpxAR- and ∆cpxD-infected mice (Figure 4C). Taken together, these observations indicated that the CpxAR-cpxDCBA pathway contributs to the pathogenesis of A. pleuropneumoniae.
Two-component system is one of the key bacterial mechanisms that enables bacteria to sense and respond to host stimulation, which is critical for the pathogenic process (Matanza et al., 2021). The CpxAR system is the principal determinant of many biological processes in A. pleuropneumoniae, such as biofilm formation, heat stress and O-antigen repeating unit biosynthesis (Li et al., 2018; Yan et al., 2020). Previous studies have found that CpxAR is required for virulence in A. pleuropneumoniae (Li et al., 2018; Yan et al., 2020). However, the crucial role of CpxAR in the pathogenesis of A. pleuropneumoniae requires further investigation. In this study, we explored whether CpxAR-regulated genes are critical to the successful infection of A. pleuropneumoniae.
CPS is one of the major virulence factors of A. pleuropneumoniae, which can protect the bacteria from the host’s immune response, such as phagocytic uptake and complement-mediated bacteriolysis (Budde et al., 2020). The thickness of the capsule is related to the virulence of A. pleuropneumoniae, and generally, the thicker the capsule, the more virulent the strain (Dubreuil et al., 2000). However, the regulation of capsule production in A. pleuropneumoniae is still unknown. In this study, TEM confirmed that the capsule layer of the ΔcpxAR strain was difficult to observe. These observations identified that CpxAR is a capsule regulator, which has been implicated in controlling the gene expression and production of CPS in A. pleuropneumoniae.
Previous studies showed that the sequence GTAAA-(N)4–8-GTAAA, or TTTAC-(N)4–8-TTTAC is the binding consensus sequence of CpxR (Keilwagen et al., 2009; Srinivasan et al., 2012; Feldheim et al., 2016; Tian et al., 2016). Here, two putative CpxR-binding sequence (TTTAC-N4-TTTAC and GTAAA-N4-TTATA) were, respectively, located 39–68 bp and 94–118 bp upstream of the promoter −35 region of the cpxD gene. Furthermore, we found that CpxR could directly bind to the cpxD promoter region by EMSA, and identified two CpxR-binding sites by DNase I footprinting which were consistent with the two putative sequences. In general, the CpxR-binding site is located upstream of the promoter region and activates their transcription (Raffa and Raivio, 2002). Previous work identified only one CpxR-binding site, but we found two in this study. The reason and the function of the two sites is worthy of further investigation.
In the present study, we showed that CpxAR positively regulates the CPS export operon cpxDCBA using qRT-PCR. Furthermore, EMSA and DNase I footprinting demonstrated that CpxR directly regulated the cpx operon by binding to the cpxD promoter region. In addition, these animal challenge tests showed that CpxD plays an important role in the virulence of A. pleuropneumoniae. Previous studies have shown that an insertion mutant in cpxC, encoding a cytoplasmic membrane protein which is essential for polysaccharide transport, caused less mortality in pigs compared to the parent strain (Rioux et al., 2000). These findings indicated that CpxAR contributes to the virulence of A. pleuropneumoniae by directly regulating the CPS export locus, cpxDCBA (Figure 5).
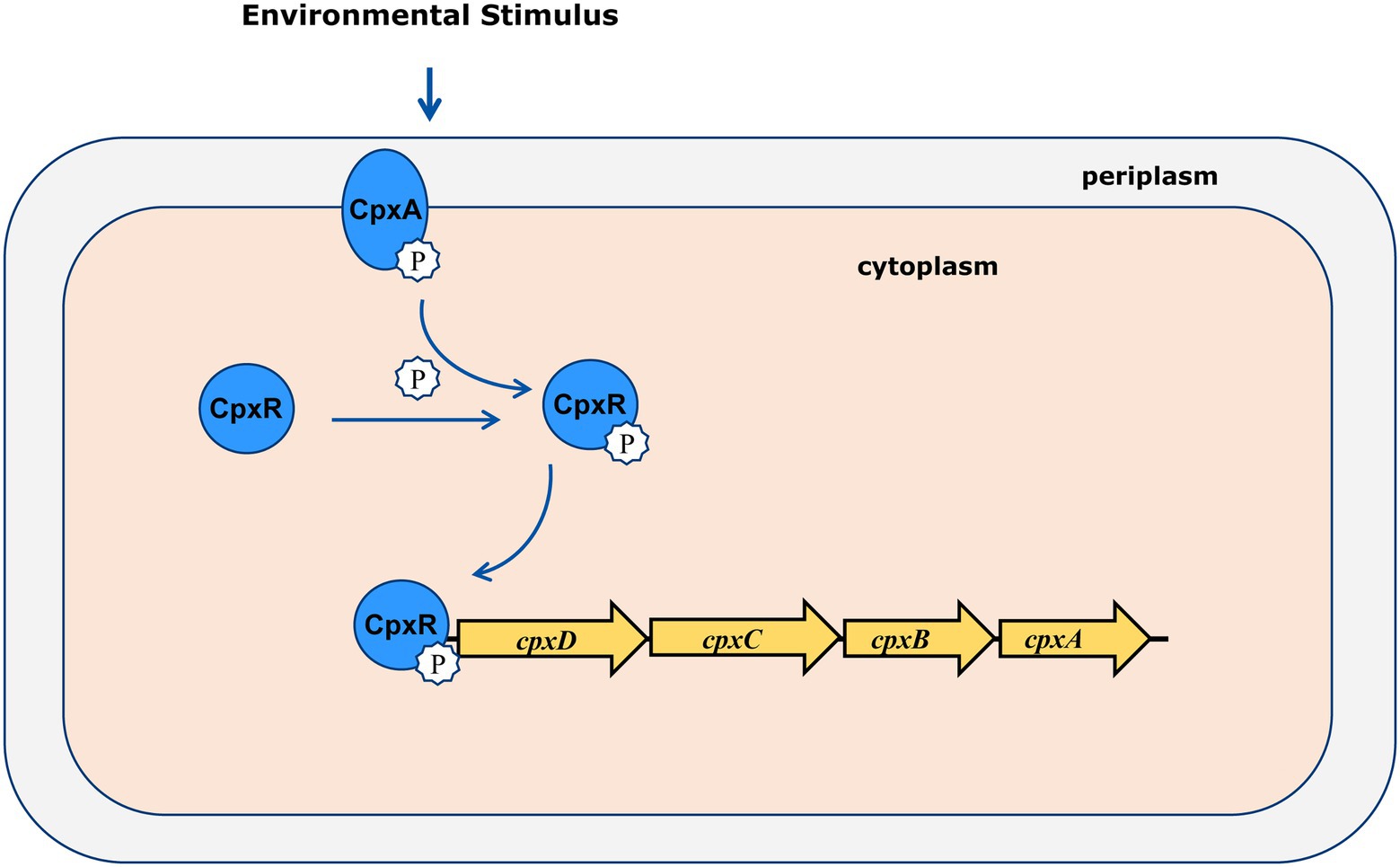
Figure 5. Molecular model of virulence regulation by CpxAR. CpxDCBA transcription is activated by CpxR-P, which contributes to the virulence in A. pleuropneumoniae.
Our results showed that CpxAR plays a contributing role in virulence by affecting capsule synthesis through directly regulating the expression of the cpxDCBA operon. The CpxAR and CPS are present in many bacteria (Vogt and Raivio, 2012; Whitfield et al., 2020). Therefore, our findings may not only contribute to the understanding of the pathogenesis of A. pleuropneumoniae, but also to other bacteria. Future studies will focus on identifying more virulence factors regulated by CpxAR in A. pleuropneumoniae.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by the Animal Ethics Committee of the Yangtze University.
FL, WB, and LL: conceived and designed the experiments. QY, JW, and TX: performed the experiments. FL and QY: analyzed the data. XG, DS, and FZ: contributed reagents, materials, and analysis tools. JH: polished the language. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This research was supported by the National Natural Science Foundation of China (32002252).
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1029426/full#supplementary-material
Bandara, A. B., Lawrence, M. L., Veit, H. P., and Inzana, T. J. (2003). Association of Actinobacillus pleuropneumoniae capsular polysaccharide with virulence in pigs. Infect. Immun. 71, 3320–3328. doi: 10.1128/IAI.71.6.3320-3328.2003
Bosse, J. T., Janson, H., Sheehan, B. J., Beddek, A. J., Rycroft, A. N., Kroll, J. S., et al. (2002). Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 4, 225–235. doi: 10.1016/s1286-4579(01)01534-9
Budde, I., Litschko, C., Fuhring, J. I., Gerardy-Schahn, R., Schubert, M., and Fiebig, T. (2020). An enzyme-based protocol for cell-free synthesis of nature-identical capsular oligosaccharides from Actinobacillus pleuropneumoniae serotype 1. J. Biol. Chem. 295, 5771–5784. doi: 10.1074/jbc.RA120.012961
Cheng, C., Liu, F., Jin, H., Xu, X., Xu, J., Deng, S., et al. (2021). The DegU orphan response regulator contributes to heat stress resistance in listeria monocytogenes. Front. Cell. Infect. Microbiol. 11:761335. doi: 10.3389/fcimb.2021.761335
Cohen, L. M., Bosse, J. T., Stegger, M., Li, Y., Langford, P. R., Kielland, C., et al. (2021). Comparative genome sequence analysis of Actinobacillus pleuropneumoniae Serovar 8 isolates from Norway, Denmark, and the United Kingdom indicates distinct phylogenetic lineages and differences in distribution of antimicrobial resistance genes. Front. Microbiol. 12:729637. doi: 10.3389/fmicb.2021.729637
Dubreuil, J. D., Jacques, M., Mittal, K. R., and Gottschalk, M. (2000). Actinobacillus pleuropneumoniae surface polysaccharides: their role in diagnosis and immunogenicity. Anim. Health Res. Rev. 1, 73–93. doi: 10.1017/s1466252300000074
Feldheim, Y. S., Zusman, T., Speiser, Y., and Segal, G. (2016). The legionella pneumophila CpxRA two-component regulatory system: new insights into CpxR's function as a dual regulator and its connection to the effectors regulatory network. Mol. Microbiol. 99, 1059–1079. doi: 10.1111/mmi.13290
Gu, D., Zhang, Y., Wang, Q., and Zhou, X. (2020). S-nitrosylation-mediated activation of a histidine kinase represses the type 3 secretion system and promotes virulence of an enteric pathogen. Nat. Commun. 11:5777. doi: 10.1038/s41467-020-19506-1
Guo, Y., Xiao, Z., Wang, Y., Yao, W., Liao, S., Yu, B., et al. (2018). Sodium butyrate ameliorates Streptozotocin-induced type 1 diabetes in mice by inhibiting the HMGB1 expression. Front. Endocrinol. 9:630. doi: 10.3389/fendo.2018.00630
Hathroubi, S., Hancock, M. A., Bosse, J. T., Langford, P. R., Tremblay, Y. D., Labrie, J., et al. (2016). Surface polysaccharide mutants reveal that absence of O antigen reduces biofilm formation of Actinobacillus pleuropneumoniae. Infect. Immun. 84, 127–137. doi: 10.1128/IAI.00912-15
Huang, Y., Wang, H., and Yang, Y. (2018). Expression of fibroblast growth factor 5 (FGF5) and its influence on survival of breast cancer patients. Med. Sci. Monit. 24, 3524–3530. doi: 10.12659/MSM.907798
Jacques, M., Foiry, B., Higgins, R., and Mittal, K. R. (1988). Electron microscopic examination of capsular material from various serotypes of Actinobacillus pleuropneumoniae. J. Bacteriol. 170, 3314–3318. doi: 10.1128/jb.170.7.3314-3318.1988
Keilwagen, J., Baumbach, J., Kohl, T. A., and Grosse, I. (2009). MotifAdjuster: a tool for computational reassessment of transcription factor binding site annotations. Genome Biol. 10:R46. doi: 10.1186/gb-2009-10-5-r46
Li, H., Liu, F., Peng, W., Yan, K., Zhao, H., Liu, T., et al. (2018). The CpxA/CpxR two-component system affects biofilm formation and virulence in Actinobacillus pleuropneumoniae. Front. Cell. Infect. Microbiol. 8:72. doi: 10.3389/fcimb.2018.00072
Liu, F., Peng, W., Liu, T., Zhao, H., Yan, K., Yuan, F., et al. (2018). Biological role of Actinobacillus pleuropneumoniae type IV pilus proteins encoded by the apf and pil operons. Vet. Microbiol. 224, 17–22. doi: 10.1016/j.vetmic.2018.08.006
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loera-Muro, A., Ramirez-Castillo, F. Y., Moreno-Flores, A. C., Martin, E. M., Avelar-Gonzalez, F. J., and Guerrero-Barrera, A. L. (2021). Actinobacillus pleuropneumoniae surviving on environmental multi-species biofilms in swine farms. Front Vet Sci 8:722683. doi: 10.3389/fvets.2021.722683
Matanza, X. M., Lopez-Suarez, L., do Vale, A., and Osorio, C. R. (2021). The two-component system RstAB regulates production of a polysaccharide capsule with a role in virulence in the marine pathogen Photobacterium damselae subsp. damselae. Environ. Microbiol. 23, 4859–4880. doi: 10.1111/1462-2920.15731
Pogliano, J., Lynch, A. S., Belin, D., Lin, E. C., and Beckwith, J. (1997). Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11, 1169–1182. doi: 10.1101/gad.11.9.1169
Raffa, R. G., and Raivio, T. L. (2002). A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45, 1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x
Raivio, T. L., and Silhavy, T. J. (1997). Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179, 7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997
Rioux, S., Galarneau, C., Harel, J., Kobisch, M., Frey, J., Gottschalk, M., et al. (2000). Isolation and characterization of a capsule-deficient mutant of Actinobacillus pleuropneumoniae serotype 1. Microb. Pathog. 28, 279–289. doi: 10.1006/mpat.1999.0347
Sassu, E. L., Bosse, J. T., Tobias, T. J., Gottschalk, M., Langford, P. R., and Hennig-Pauka, I. (2018). Update on Actinobacillus pleuropneumoniae-knowledge, gaps and challenges. Transbound. Emerg. Dis. 65, 72–90. doi: 10.1111/tbed.12739
Srinivasan, V. B., Vaidyanathan, V., Mondal, A., and Rajamohan, G. (2012). Role of the two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. PLoS One 7:e33777. doi: 10.1371/journal.pone.0033777
Stringer, O. W., Bosse, J. T., Lacouture, S., Gottschalk, M., Fodor, L., Angen, O., et al. (2021). Proposal of Actinobacillus pleuropneumoniae serovar 19, and reformulation of previous multiplex PCRs for capsule-specific typing of all known serovars. Vet. Microbiol. 255:109021. doi: 10.1016/j.vetmic.2021.109021
Tian, Z. X., Yi, X. X., Cho, A., O'Gara, F., and Wang, Y. P. (2016). CpxR activates MexAB-OprM efflux pump expression and enhances antibiotic resistance in both laboratory and clinical nalB-type isolates of Pseudomonas aeruginosa. PLoS Pathog. 12:e1005932. doi: 10.1371/journal.ppat.1005932
Vogt, S. L., and Raivio, T. L. (2012). Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol. Lett. 326, 2–11. doi: 10.1111/j.1574-6968.2011.02406.x
Ward, C. K., and Inzana, T. J. (1997). Identification and characterization of a DNA region involved in the export of capsular polysaccharide by Actinobacillus pleuropneumoniae serotype 5a. Infect. Immun. 65, 2491–2496. doi: 10.1128/iai.65.6.2491-2496.1997
West, A. H., and Stock, A. M. (2001). Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26, 369–376. doi: 10.1016/s0968-0004(01)01852-7
Whitfield, C., Wear, S. S., and Sande, C. (2020). Assembly of bacterial capsular polysaccharides and exopolysaccharides. Annu. Rev. Microbiol. 74, 521–543. doi: 10.1146/annurev-micro-011420-075607
Xiong, Y., Huang, H., Chen, S., Dai, H., and Zhang, L. (2019). ERK5regulated RERG expression promotes cancer progression in prostatic carcinoma. Oncol. Rep. 41, 1160–1168. doi: 10.3892/or.2018.6852
Xu, L., Zhou, X., and He, X. (2014). Cpx two-component regulatory system in gram-negative bacteria--a review. Wei Sheng Wu Xue Bao 54, 269–275.
Xu, Z., Zhou, Y., Li, L., Zhou, R., Xiao, S., Wan, Y., et al. (2008). Genome biology of Actinobacillus pleuropneumoniae JL03, an isolate of serotype 3 prevalent in China. PLoS One 3:e1450. doi: 10.1371/journal.pone.0001450
Keywords: Actinobacillus pleuropneumoniae, two-component system, CpxAR, polysaccharide capsule export, cpxDCBA
Citation: Liu F, Yao Q, Huang J, Wan J, Xie T, Gao X, Sun D, Zhang F, Bei W and Lei L (2022) The two-component system CpxA/CpxR is critical for full virulence in Actinobacillus pleuropneumoniae. Front. Microbiol. 13:1029426. doi: 10.3389/fmicb.2022.1029426
Received: 27 August 2022; Accepted: 21 September 2022;
Published: 05 October 2022.
Edited by:
Qing Pan, Qingdao Agricultural University, ChinaReviewed by:
Yuexiu Zhang, The Ohio State University, United StatesCopyright © 2022 Liu, Yao, Huang, Wan, Xie, Gao, Sun, Zhang, Bei and Lei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weicheng Bei, YmVpd2NAbWFpbC5oemF1LmVkdS5jbg==; Liancheng Lei, bGVpbGlhbmNoZW5nQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.