
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 November 2022
Sec. Microbiological Chemistry and Geomicrobiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1028969
This article is part of the Research Topic Phosphorus Along the Soil-Freshwater-Ocean Continuum, volume II View all 7 articles
 Jin Liu1*†
Jin Liu1*† Yang Li1†
Yang Li1† Chaoqun Han1
Chaoqun Han1 Dongling Yang1
Dongling Yang1 Jianjun Yang2
Jianjun Yang2 Barbara J. Cade-Menun3
Barbara J. Cade-Menun3 Yuanquan Chen1
Yuanquan Chen1 Peng Sui1
Peng Sui1Intercropping often substantially increases phosphorus (P) availability to plants compared with monocropping, which could be an effective strategy for soil legacy P recovery and agricultural production. However, the biogeochemical interactions among plants, microbes, and soil that mobilize P remain largely unknown in intercropping systems. Pot experiments with maize-soybean intercropping in a calcareous soil were conducted to investigate the potential chemical and biological transformation mechanisms of inorganic P (Pi) and organic P (Po) using sequential extraction and Illumina MiSeq sequencing. Compared to monocropping of each crop, maize-soybean intercropping significantly enhanced total P uptake of the two crops by mobilizing Ca2-Pi [extracted by bicarbonate (NaHCO3)], Al-Pi/Po [extracted by ammonium fluoride (NH4F)] and Fe-Pi [extracted by sodium hydroxide and sodium carbonate (NaOH-Na2CO3)] fractions. Furthermore, there were significant increases in the organic carbon content and alkaline phosphomonoesterase (ALP) and phosphodiesterase (PDE) activities as well as the abundances of Microvirga, Lysobacter, Microlunatus and Sphingomonas under maize-soybean intercropping relative to monocropping. In contrast, compared to monocroppping, no significant change in the soil pH was observed under maize-soybean intercropping. Therefore, the enhanced P uptake of the maize-soybean intercropping probably resulted from a synergistic effect of rhizosphere organic carbon deposit, increased activities of ALP and PDE, together with the bacteria (Microvirga, Lysobacter, Microlunatus and Sphingomonas) which showed correlation with soil P forms, while the generally recognized rhizosphere acidification was excluded in this investigated calcareous soil. Moreover, the selected bacterial genera exhibited a closer network in the rhizosphere of soybean compared to maize, suggesting enhanced interactions among bacteria in the soybean rhizosphere. These results provide theoretical bases for the recovery of soil legacy P by maize-soybean intercropping.
Phosphorus (P) limits crop yield on more than 30% of the world’s arable land (Vance et al., 2003). Widespread use of chemical phosphate fertilizer has substantially depleted phosphate rock resources, and also has led to environmental pollution due to loss of fertilizer P from agricultural land to water bodies (Sattari et al., 2012). Traditional fertilization practices generally result in the accumulation of high P concentrations in soil. It is estimated that 15% of applied P is absorbed by plants, but 60% remains in the topsoil (up to 15 cm depth) in forms that are not readily available to plants (Wagar et al., 1986). There is a growing recognition that the recovery of this residual or legacy soil P could reduce the reliance on chemical fertilizers and their negative impacts on environment (Sattari et al., 2012; Liu et al., 2015; Withers et al., 2019).
Crop intercropping, growing more than one crop simultaneously, has been shown to increase crop yields and nutrient acquisition (Li et al., 2007; Tian et al., 2020). The intercropping of cereals with legumes, widely recognized to enhance soil nitrogen (N) fertility, could facilitate the recovery of soil legacy P (Li et al., 2007). For example, previous studies indicated intercropping with legumes mobilized more soil P compared to single-cropped maize and wheat (Liao et al., 2020; Messaoudi et al., 2020). Since the bioavailability of soil P compounds depends on their chemical forms (Liu et al., 2013), the recovery of soil P by intercropping was mainly through root-induced transformation of specific soil P pools or fractions. Soil P speciation greatly depends on soil type and land use (Negassa and Leinweber, 2009; Liu et al., 2018), and previous studies have shown that different types of crops access different soil P pools. For example, maize could readily deplete the NaOH-extractable inorganic P (NaOH-Pi) fraction determined by the Hedley fractionation method (Cabeza et al., 2017), while in a monocropping study, soybean enriched the NaHCO3-extracted organic P (Po) fraction, but had less depletion of the NaOH-Pi fraction compared with maize (Rubio et al., 2012). In intercropping studies of maize and faba bean, more residual-P fraction was depleted under intercropping compared to monocropping after a 4-year study withholding nitrogen (N) and P fertilization (Liao et al., 2020). However, the transformation of soil P fractions under maize-soybean intercropping remains largely unknown.
Multiple strategies to mobilize soil P through the transformation of different legacy P fractions in agricultural soils exist in cereal-legume intercropping systems, including root-induced pH changes, root exudates, and enhanced phosphatase and microbial activities (Hinsinger, 2001; Roohi et al., 2020). Tang et al. (2016) found microbial biomass P significantly increased in the rhizosphere of faba bean when intercropped with durum wheat, which highlighted the potential involvement of microbes in rhizospheric P cycling. Bargaz et al. (2017) reported that the soil microbial community shaped by the rhizosphere effect of wheat under wheat-soybean intercropping could promote P uptake of wheat. Recently, Roohi et al. (2020) demonstrated that cowpea-maize intercropping could deplete HCl-extracted Pi fractions relative to monocropping in semi-arid soils under convention NPK fertilization, which probably resulted from a combination of microbial and rhizosphere processes. However, the biochemical driving mechanisms involved in maize-soybean intercropping remain unclear, thus deserving further investigation.
The Huang-Huai-Hai Plain is one of the main crop producing area in China. The soil in this area is generally rich in carbonates and clay minerals, which strongly react with P compounds through sorption and precipitation, resulting in low P use efficiency of applied phosphate fertilizer (Liu et al., 2019). Therefore, the objectives of this study were to: (1) Identify the transformation of P fractions; (2) investigate chemical and biological factors driving the transformation of specific P fractions, and (3) explore the bacterial community composition under maize-soybean intercropping.
A pot experiment was conducted in a greenhouse where one maize (Zea mays L.) and one soybean [Glycine max (L.) Merr.] were planted in rhizoboxes. Three treatments of root interactions were set up with three replicates: maize-soybean intercropping with no barrier (NB); with nylon net isolation (mesh barrier, MB, pore size = 40 μm) to prevent root interactions between different crops, but allow root exudate exchange; and plastic plate isolation (solid barrier, SB) to simulate monocropping. The soil used for the pot experiment is classified as Inceptisols according to the USDA Soil Taxonomy (USDA, 1999). The rhizoboxes (28 × 14 × 18 cm) were filled with 6 kg of air-dried soil and moistened to 50% of the maximum water holding capacity. The crops were sown directly into each rhizobox and no extra nutrients were applied during the whole growth period.
Plants were harvested after growing for 3 months. The shoots were oven-dried at 105°C for 0.5 h and dried to constant weight at 60°C, then weighed for biomass determination and finely ground for total P measurement by inductively coupled plasma optical emission spectroscopy (ICP-OES, Agilent 5110) after nitric acid-perchloric acid digestion (Smith and Joynson, 1974). The residual plants with roots were manually removed from the soil, and then shaken gently to remove loosely adhering soil from the roots. The rhizosphere soil was collected by vigorously shaking and gently brushing the root system (Yang et al., 2010). Soil from an unplanted treatment was also sampled as the bulk soil (BS). A portion of each soil sample was air-dried and sieved (<2 mm) for the measurements of soil properties and sequential P fractionation. Sub-samples were stored at 4°C for enzyme assays, and –80°C for DNA sequencing.
Soil pH was measured in deionized water (1:2.5 w/v; Lu, 1999). Total carbon (C) and total N (TN) were determined by element analyzer (Vario Max). Prior to organic C (OC) measurement, soil samples were pre-treated with hydrochloric acid to remove inorganic C (IC). Soil IC was calculated as the difference between TC and OC. Total P was determined by H2SO4-HClO4 digestion, Po was determined by ignition, both analyzed colorimetrically using the molybdate blue method (Murphy and Riley, 1962). The alkaline phosphomonoesterase (ALP) and phosphodiesterase (PDE) activities were measured by p-nitrophenyl phosphate as substrate with the buffer pH 11.0, and bis-p-nitrophenyl phosphate as substrate with the buffer pH 8.0, respectively (Tabatabai, 1994).
Soil P fractions were extracted by the fractionation scheme recommended by Jiang and Gu (1989). Soil samples was sequentially extracted by (1) 0.25 mol L–1 NaHCO3 at pH 7.5 [calcium (Ca)2-P]; (2) 0.5 mol L–1 NH4Ac at pH 4.2 (Ca8-P); (3) 0.5 mol L–1 NH4F [aluminum (Al)-P]; (4) 0.1 mol L–1 NaOH-Na2CO3 at pH 8.2 [iron (Fe)-P]; (5) 0.3 mol L–1 citrate solution and Na2S2O4 kept at 40°C [occluded (O)-P]; (6) 0.25 mol L–1 H2SO4 (Ca10-P). Ca2-P extracted by bicarbonate is categorized as labile. The identifications of each fraction (“Ca2-P,” “Al-P,” etc.) are those indicated in the fractionation protocol; we acknowledge that these fractions were operationally defined, and may likely contain more than one P compound. The concentration of molybdate-reactive P, considered here to be Pi, in each extract was determined by colorimetric analysis within 24 h (Murphy and Riley, 1962), and the total P concentration in each extract was analyzed by ICP-OES. Molybdate-unreactive P concentrations were calculated as the difference between total P and Pi, and for this study is considered to be Po.
Soil DNA was extracted using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to manufacturer’s protocol, and quantified with a NanoDrop 2000 spectrophotometer. The primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify the V3-V4 region of the 16S rRNA genes by an ABI GeneAmp® 9700 PCR thermocycler. The amplification was performed in triplicates as follows: 3 min initial denaturation at 95°C, followed by 27 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, with a final 10 min extension at 72°C, and stored at 4°C. The PCR mixtures contain 4 μL of 5 × TransStart FastPfu buffer, 2 μL 2.5 mM dNTPs, 0.8 μL forward primer (5 μM), 0.8 μL reverse primer (5 μM), 0.4 μL TransStart FastPfu DNA Polymerase, 10 ng template DNA, and finally ddH2O up to 20 μL. The PCR product was extracted from a 2% agarose gel, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA), and quantified using Quantus Fluorometer (Promega, USA). The purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
Raw sequence reads were processed using the Quantitative Insights into Microbial Ecology (QIIME2) pipeline (Caporaso et al., 2010). The adaptor sequences, barcodes and low-quality bases at the end of each read were removed. The raw paired-end reads were quality-filtered by fastp (version 0.19.6) and merged by FLASH (version 1.2.7; Magoc and Salzberg, 2011), with the following criteria: (i) The maximum mismatch ratio of overlap region is 0.2. Reads that could not be assembled were discarded; (ii) only overlapping sequences longer than 10 bp were assembled according to their overlapped sequence. The operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE version 7.1 (Edgar, 2013). Totally, there were 941,784 optimized sequences (392,730,025 bp) in 21 samples. A representative sequence of each OTU was analyzed by RDP Classifier version 2.2 (Wang, 2007) against the 16S rRNA gene database (Silva v138) using confidence threshold of 0.7. To avoid potential bias caused by differences in sequences depth, the number of sequences in each sample were rarefied to 28,570 sequences, which still yielded an average Good’s coverage of 97.61%, respectively. The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA782812).
The P fractionation data were centered log-ratio transformed before the statistical analysis (Liu et al., 2015). For the data of soil properties, P fractionation and plant biomass as well as P uptake, one-way analysis of variance (ANOVA) was conducted using SPSS 25 software (SPSS, Inc.), followed by a least significant difference (LSD) test with α = 0.05. Bioinformatic analysis of the soil was conducted by Majorbio Cloud1. Mothur v1.30.2 to identify them, using “classify.otu.” Rarefaction curves (Supplementary Figure 2), Chao richness, Shannon and Simpson indices were calculated at the 0.03 cut off level to assess the alpha diversity (Cole et al., 2009). The “Stats” package in R and “Scipy” in python were used to test the differences in the relative abundance of OTUs or genera between treatments using the Benjamini-Hochberg P-value correction. And the differences were analyzed with a one-way ANOVA followed by Tukey’s honestly significant difference (HSD) test. The data means were considered significantly different at P < 0.05. “Pheatmap” package in R was conducted to calculate the correlation coefficient between environmental factors and the selected species. Both hierarchical cluster method of environmental factors and species were average. The co-occurrence networks were constructed to explore the internal community relationships across the samples (Barberan et al., 2012). The network was constructed by Spearman correlation among genera with the absolute value of that correlation coefficient ≥ 0.5 and P < 0.05, which was analyzed by Networkx (python package Hagberg et al., 2008). We reconstructed a Maximum likelihood (ML) phylogeny based on the aligned OTU sequences of Silva collections using the FastTree (version 2.1.3)2. The ML phylogeny was visualized using the R (version 3.3.1).
Maize-soybean intercropping generally enhanced aboveground biomass and total P uptake of crops compared to monocropping (Table 1). For maize, plant biomass and P uptake were highest in the NB treatment followed by the MB treatment, both of which were higher than the SB treatment. For soybean, plant biomass and P uptake were highest in the MB treatment and lowest in the NB treatment. For the sum of two crops, total biomass and P uptake were significantly higher for the MB and NB treatments compared to the SB treatment.
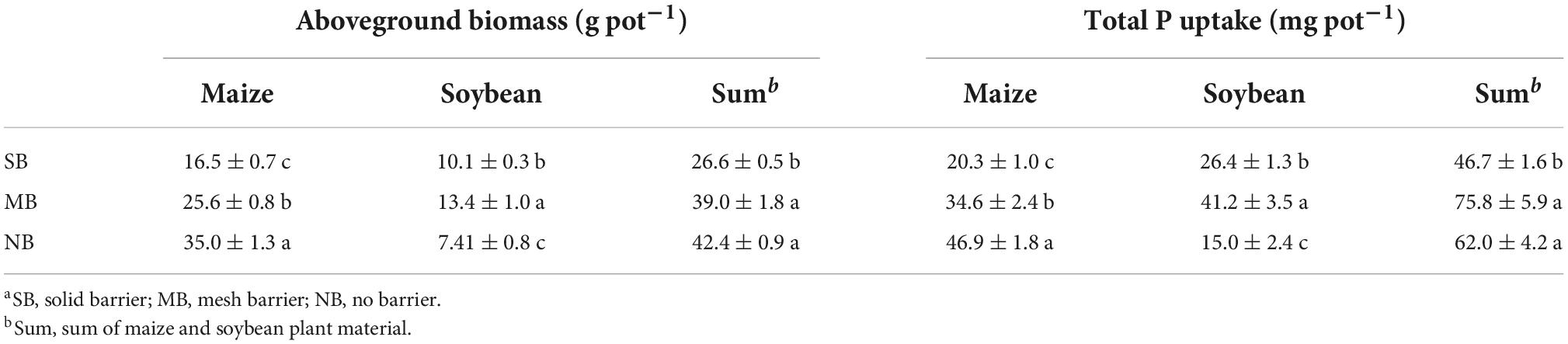
Table 1. Aboveground biomass and total phosphorus (P) uptake of maize and soybean under the different separation treatmentsa (means ± standard errors, n = 3).
For the BS, pH was 8.5 and TN, TC, OC, and IC concentrations were 0.07, 1.74, 0.59, and 1.14%, respectively (Table 2). Total P was 773.4 mg kg–1 in the BS where a small concentration of 83.5 mg kg–1 was Po. The ALP and PDE activities in the BS were 176.3 and 14.6 μg g–1h–1 (Table 2). Maize and soybean monocropping and intercropping significantly affected soil chemical and biological properties. Compared to the BS, maize monocropping (SB) significantly reduced rhizosphere soil pH but increased Po contents and PDE activities (Table 2). Similar changes were also observed for soybean monocropping (SB) relative to the BS (Table 2). Compared to the SB, the NB treatment increased TN, TC, OC, Po, as well as ALP and PDE activities, but decreased IC contents in the rhizosphere soils for both maize and soybean (Table 2). For both crops, higher ALP and PDE activities occurred in the MB treatment than the NB treatment (Table 2). A larger concentration of Po in the MB treatment was shown compared to the NB treatments for soybean, while this change was insignificant for maize (Table 2).
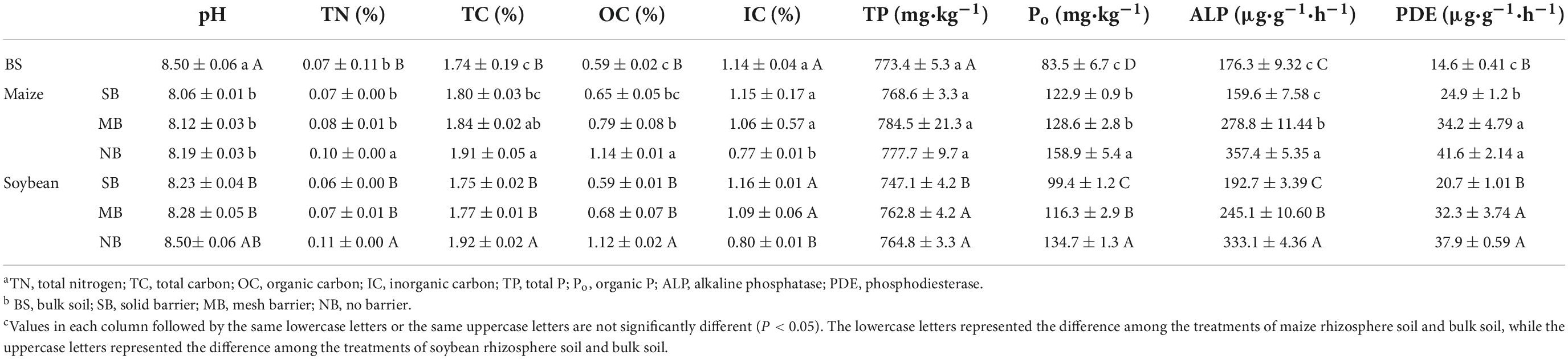
Table 2. Chemical propertiesa of the bulk and rhizospheric soils of the maize and soybeans under the different separation treatmentsb (means ± standard errors, n = 3)c.
Sequential fractionation recovered 71.7–75.3% of Pi (Table 3). The majority of soil Pi was extracted by H2SO4 (Ca10-Pi), followed by NH4Ac (Ca8-Pi). Phosphorus transformation was induced in the rhizosphere of both crops by monocropping and intercropping. In the monocropping (SB) treatment, Al-Pi and O-Pi significantly decreased in the maize rhizosphere soil, and Ca8-Pi and Fe-Pi significantly decreased in the soybean rhizosphere soil, respectively, compared to the BS; there were no significant changes in the other Pi fractions (Table 3). Under the intercropping treatment with no barrier, there were significant decreases of Ca2-Pi, Al-Pi, Fe-Pi, but a significant increase of Ca8-Pi in the rhizosphere soils of the two crops compared to the SB (Table 3). Similarly, this effect was observed in the MB treatment with higher Ca8-Pi, and O-Pi for maize and higher Ca8-Pi and lower O-Pi for soybean than the SB soil.
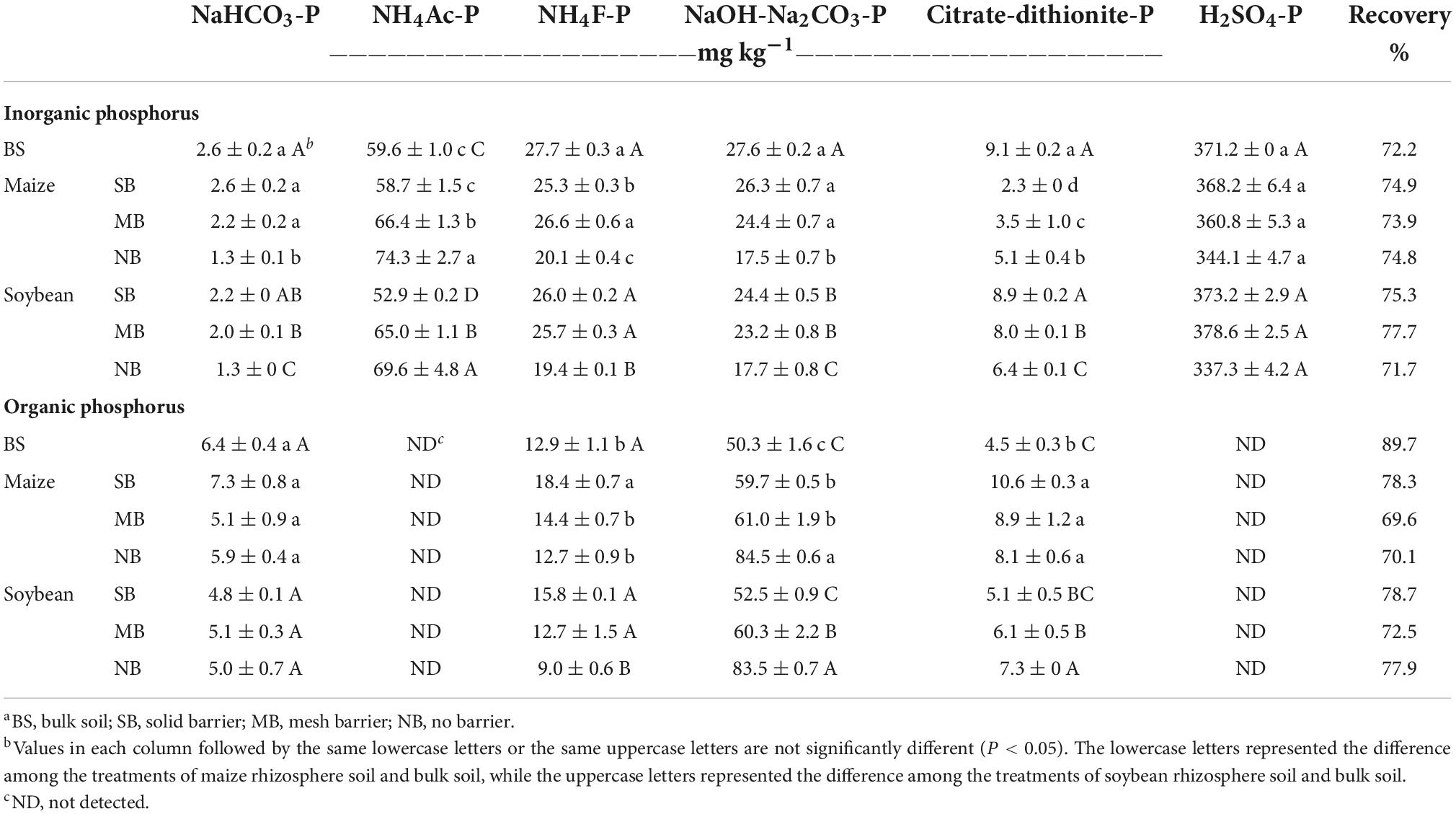
Table 3. Phosphorus (P) fractions in the bulk and rhizospheric soils of the maize and soybean under the different separation treatmentsa (mean ± standard errors, n = 3)b.
Fractionation recovered 69.6–89.7% of Po, most of which was Fe-Po (Table 2). For the SB treatment, there were significant increases in the Al-Po, Fe-Po and O-Po contents, but no significant changes in the Ca2-Po content in the maize rhizosphere relative to the BS treatment (Table 3). In contrast, there were no significant differences in the detected Po fractions in the soybean rhizosphere soil under the SB treatment compared to the BS (Table 3). Compared to the SB, there were significant decreases in Al-Po and increases in the Fe-Po fractions in the rhizosphere soils of both crops under the NB treatment, with no significant changes in the Ca2-Po content (Table 3). Consistently, the decreased Al-Po for maize and increased Fe-Po for soybean occurred under the MB treatment compared to the SB (Table 3).
For both maize and soybean, soil bacterial richness (Chao) was significantly decreased in the intercropping (NB vs. SB treatment) rather than the monocropping system (SB vs. BS treatment; Supplementary Table 1). In contrast, the Shannon index was not affected by intercropping or monocropping (except for maize; Supplementary Table 1). Moreover, Actinobacteriota, Proteobacteria, Chloroflexi, and Acidobacteriota were the main phyla for all soil treatments of the two crops (Supplementary Figure 1).
Specific bacteria showed significant differences in relative abundances at the genus level in the rhizosphere soils of the two crops under different separation treatments. Compare to the BS, the SB treatment significantly reduced the abundance of Pontibacter in the rhizosphere of the two crops, and increased the abundance of Arthrobacter in the maize rhizosphere (Figures 1A, 2A). In contrast, there was a significant decrease in the abundance of Arthrobacter but significant increases in the abundances of Microvirga, Lysobacter and Microlunatus in the rhizosphere soils of the two crops under the NB treatment relative to the SB treatment (Figures 1A, 2A). Similarly, the abundance of Sphingomonas in the soybean rhizosphere soil significantly increased under the NB treatment compared to the SB treatment (Figure 2A). For the MB treatment, the abundance of each selected bacterial genus (Arthrobacter, Sphingomonas, Microvirga, Lysobacter, Microlunatus, Skemanella, Haliangium) except Pontibacter, showed a transition trend compared to the NB and SB treatments (Figures 1A, 2A). Additionally, certain non-ranked bacteria phyla were also found under maize-soybean intercropping, which had close genetic relationships with the aforementioned bacteria related to P cycling (Supplementary Figure 3). For all bacteria involved in P cycling, including the non-ranked bacteria, a tighter engagement in the network of these bacteria was observed in the rhizosphere of soybean than maize (Figure 3).
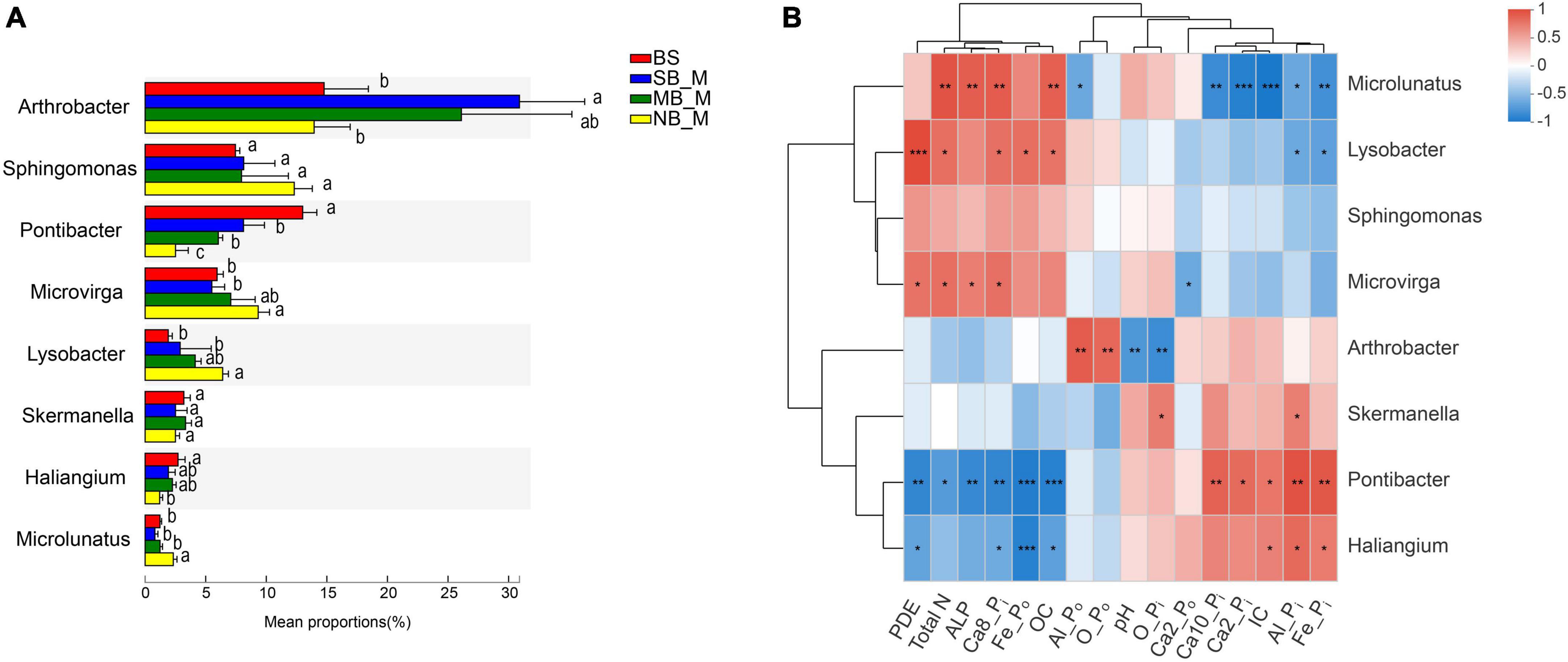
Figure 1. Abundances of the selected bacteria in the bulk and maize rhizosphere soils at the genus level under different separation treatments (A) and their correlation with selected P fractions and soil properties (B). BS, bulk soil; SB, solid barrier; MB, mesh barrier; NB, no barrier. Values marked by the same lowercase letters in Figure 1A are not significantly different among different separation treatments (P < 0.05). *P < 0.05, **P < 0.01, ***P < 0.001.
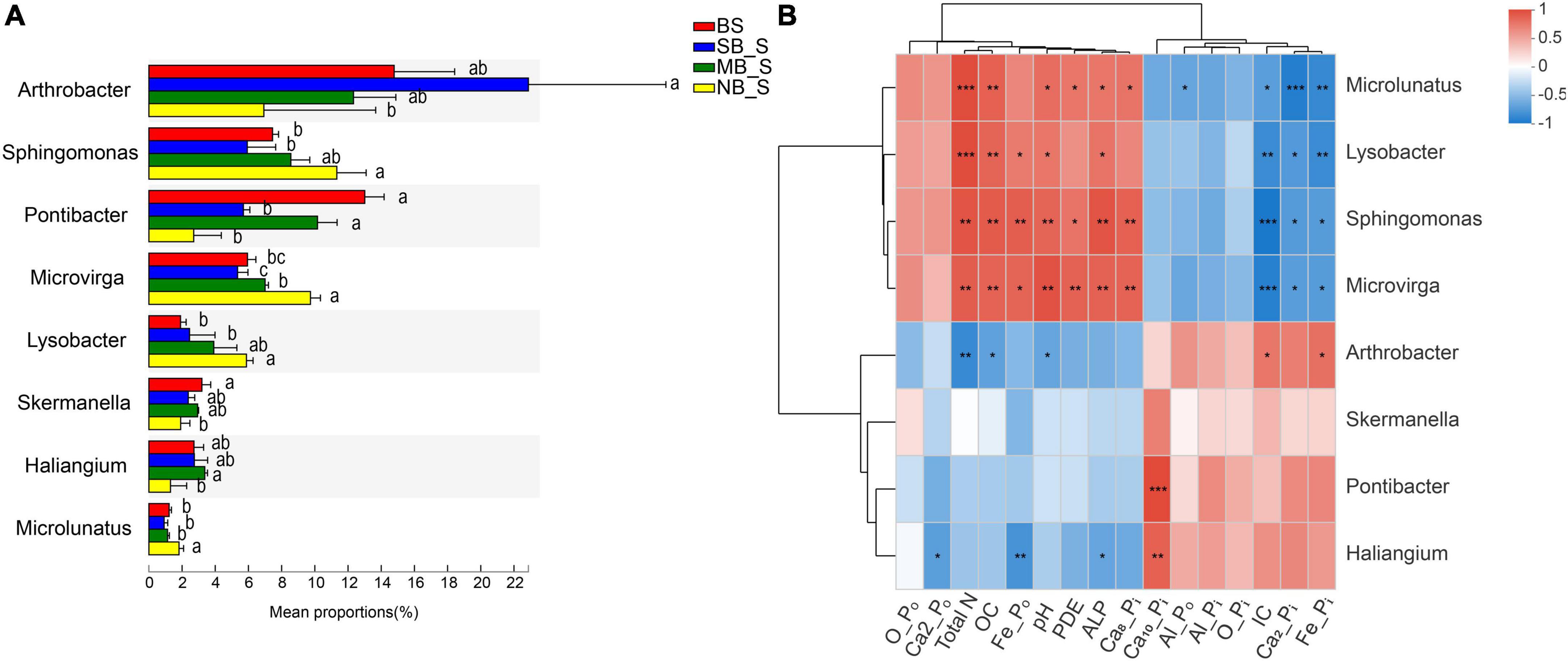
Figure 2. Abundances of the selected bacteria in the bulk and soybean rhizosphere soils at the genus level under different separation treatments (A) and their correlation with selected P fractions and soil properties (B). BS, bulk soil; SB, solid barrier; MB, mesh barrier; NB, no barrier. Values marked by the same lowercase letters in Figure 2A are not significantly different among different separation treatments (P < 0.05). *P < 0.05, **P < 0.01, ***P < 0.001.
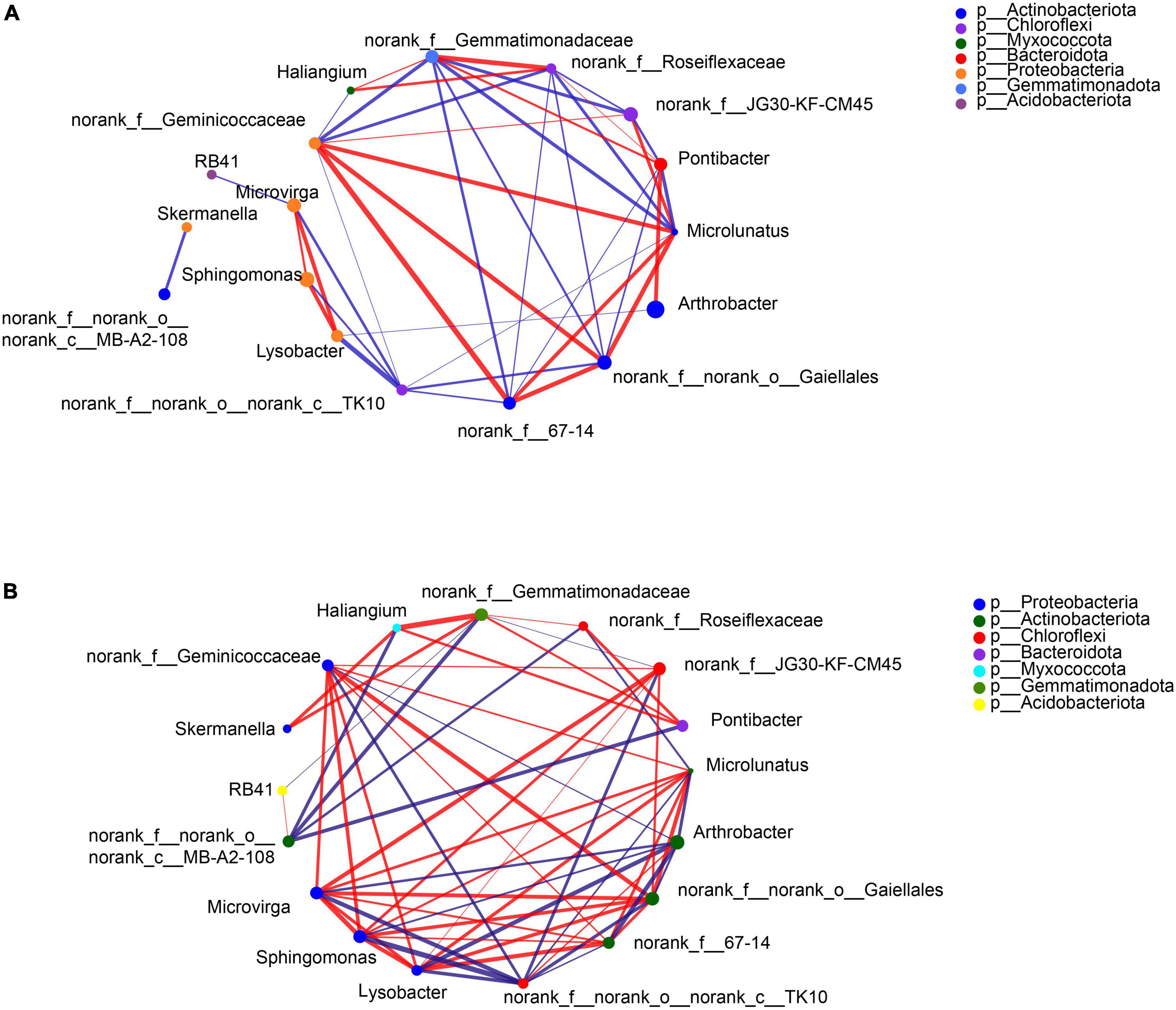
Figure 3. Correlation network of the selected bacteria at the genus level in the maize (A) and soybean (B) rhizosphere soils under intercropping.
The correlation analyses of P fractions, soil biochemical properties and the abundances of selected bacteria genera were conducted for the rhizosphere soils of the two crops. Abundance of Pontibacter was positively correlated with concentrations of Al-Pi, Ca2-Pi, Fe-Pi, Ca10-Pi and IC, but negatively correlated with concentrations of OC, Ca8-Pi, TN, ALP, and Fe-Po in the maize rhizosphere soils (Figure 1B). In contrast, the abundance of Pontibacter was positively correlated only with the Ca10-Pi concentration in the soybean rhizosphere soils (Figure 2B). Furthermore, the abundance of Arthrobacter exhibited positive correlations with Al-Po and O-Po concentrations, but negative correlations with the O-Pi concentration and pH in the maize rhizosphere soils (Figure 1B), and positive correlations with IC and Fe-Pi concentrations, but negative correlations with TN and OC concentrations and pH in the soybean rhizosphere soils (Figure 2B). Moreover, Microvirga and Lysobacter both only had positive correlations with TN, PDE and Ca8-Pi concentrations in the maize rhizosphere soil (Figure 1B), but were positively correlated with TN, OC, Fe-Po, concentration, pH, and ALP activity but were negatively correlated with IC, Ca2-Pi, Fe-Pi concentrations in the soybean rhizosphere soil (Figure 2B). For both crops, the abundance of Microlunatus positively correlated with TN, OC, ALP and Ca8-Pi concentrations, but negatively correlated with Al-Po, IC, Ca2-Pi, Fe-Pi concentrations (Figures 1B, 2B). In addition, the abundance of Sphingornonas showed no significant correlations with any soil P fractions and biochemical properties in the maize rhizosphere soils (Figure 1B). However, it was positively correlated with TN, pH, OC, Ca8-Pi and Fe-Po concentrations as well as ALP and PDE activities, and was negatively correlated with IC, Ca2-Pi and Fe-Pi concentrations in the soybean rhizosphere soils (Figure 2B).
Maize-soybean intercropping significantly promoted crop growth and P uptake, as indicated by the increase of the total above-ground biomass and P uptake of the two crops (maize and soybean combined, Table 1). These results agreed with previous reports showing that maize-legume intercropping benefited crop production and P accumulation (Betencourt et al., 2012; Liao et al., 2020). The lowest biomass and P uptake of soybean but highest of maize under the NB treatment relative to the SB treatment (Table 1) resulted from the competition for soil nutrients between maize and soybean, and maize could absorb more nutrients than soybeans (Zhang et al., 2015). Generally, most cereal/legume polycultures are asymmetric and thus lead to imbalanced nutrient absorption by crops (Zhang et al., 2016).
Besides enhancing crop P uptake, maize-soybean intercropping induced significant transformation of P fractions in this calcareous soil. This was indicated by the significant decrease in concentrations of Ca2-Pi, Al-Pi/Po and Fe-Pi, but significant increases in Ca8-Pi and Fe-Po concentrations in the rhizospheres of maize and soybeans under intercropping (NB) relative to monocropping (SB, Table 3). However, compared to the bulk soil, only concentrations of Al-Pi and O-Pi in the maize rhizosphere soil and Ca8-Pi and Fe-Pi in the soybean rhizosphere soil significantly decreased, while Al-Po, Fe-Po and O-Po concentrations significantly increased in the maize rhizosphere soil under monocropping (SB, Table 3). These results indicate that maize monoculture improved the recovery of P from Al-Pi and O-Pi fractions, while soybean monoculture could mobilize P from Ca8-Pi and Fe-Pi pools. In contrast, maize-soybean intercropping could further utilize the Ca2-Pi fraction, in addition to Al-Pi and Fe-Pi fractions. Generally, the Ca2-Pi fraction, as the most bioavailable P fraction to crops, could be supplied from the diffusion of labile P from the bulk soil P pools to the rhizosphere (Hinsinger, 2001). The depletion of rhizospheric Ca2-Pi fraction under maize-soybean intercropping rather than maize or soybean monoculture highlighted the stronger recovery capacity of soil legacy P under intercropping relative to monoculture. Moreover, the significant decrease of Al-Pi and Fe-Pi fractions in the rhizosphere of both crops under the NB treatment relative to the SB treatment further highlights that maize-soybean intercropping facilitates the recovery of P sorbed to Al/Fe oxides. This probably resulted from the rhizosphere effects of maize-soybeans, which could mobilize micronutrients such as Fe in calcareous soils (Dragicevic et al., 2015). Although reports on the influence of maize-soybean intercropping on soil P transformations are scarce in the literature, one field study of 4-year maize-legume (i.e., faba bean) intercropping without P fertilization suggests intercropping could significantly deplete NaOH-extracted Pi fraction, generally assigned as Pi associated with Fe/Al oxides (Liu et al., 2013), in calcareous soil compared to monocropping (Liao et al., 2020). Additionally, maize-soybean intercropping also resulted in the rhizospheric accumulation of Fe-Po and decrease of Al-Po fractions for the two crops (Table 3). These results indicated maize-soybean intercropping facilitated the transformation of Pi fractions (Ca2-Pi, Al-Pi, and Fe-Pi) into Fe-Po fractions in the calcareous soil, which agreed with the increased rhizospheric Po contents for the two crops under the NB treatment (Table 2).
Maize-soybean intercropping could increase soil P availability through multiple mechanisms including root exudates and proton-induced dissolution of P-containing minerals, increased phosphatase activity, and functional microbe-mediated Pi mobilization and Po mineralization (Hinsinger, 2001; Betencourt et al., 2012; Amadou et al., 2021; Park et al., 2022). In this study, rhizospheric OC content significantly increased for both crops under the NB treatment relative to the SB treatment (Table 2), which reflected the accumulation of rhizosphere OC under the NB treatment. Rhizosphere OC deposition has multiple sources including root exudates, microbial turnover and litter decomposition (Hinsinger et al., 2009), and root exudates were generally considered as the major driving force to induce the dissolution of solid phase associated Pi fractions including Ca2-Pi, Al-Pi and Fe-Pi (Table 3; Hinsinger, 2001). Generally, all three aforementioned Pi fractions are sensitive to low molecular-weight organic acids, which, as one of the major components in root exudates, could dissolve brushite, Al and Fe oxides through ligand-induced complexation and dissolution (Qin et al., 2013, 2018). Furthermore, phytosiderophores and phenolic acid were widely reported to be present in the root exudates of maize and legume, respectively (Bekkara et al., 1998; Bernards et al., 2002; Wang et al., 2014), which could destroy Fe oxides minerals and mobilized their associated Pi (Hunt et al., 2007; Reichard et al., 2007). This probably also accounted for the decrease of Fe-Pi fraction in the rhizosphere soils of both crops (Table 3). Moreover, the enhanced rhizospheric TN contents caused by soybean-mediated N fixation processes in the maize-soybean intercropping would also be beneficial to soil P mobilization (Tables 2, 3). There were several potential driving factors derived from the soybeans, including the production of root exudates and phosphatases and enhanced proton release during N2-fixation (Betencourt et al., 2012). In this study, rhizospheric acidification was excluded as the major driving factor for the observed transformation of P fractions under the maize-soybean intercropping, given the lack of significant changes in the rhizospheric pH for both crops among the three separation treatments (Table 2). The high pH buffer capacity of the investigated calcareous soil probably resulted in the resistance of rhizospheric acidification in this soil under the intercropping (Andersson et al., 2015). Therefore, the accumulation of rhizosphere OC deposit and phosphatases probably contributed to the recovery of soil Pi fractions, which agreed with the enhanced OC contents and phosphatase activities under the maize-soybean intercropping (Table 2).
For the cycling of Po pools, enzymes serve as one of the major driving factors to mineralize Po into the available Pi pool for crop uptake (Richardson et al., 2009; Park et al., 2022; Yu et al., 2022). Generally, the ALP in soils is considered as microbial origin, while the PDE could be from plant roots and microbes (Amadou et al., 2021). Compared to the SB treatment, the enhanced ALP and PDE activities in the rhizosphere soils of both crops under the NB treatment (Table 2) implies increased mineralization of soil Po fractions. However, the net accumulation of Po contents under the NB treatment indicated that mineralization rates of soil Po fractions were less than the accumulation rates of Po fractions under the maize-soybean intercropping. The depletion of the Al-Po fraction in the rhizosphere soils of both crops under the NB treatment (Table 3) was less likely related to enhanced enzyme-mediated Po mineralization; instead, it probably resulted from ligand-induced dissolution of Al oxides by root exudates considering the enhanced rhizospheric OC content of the two crops under the NB treatment relative to the SB treatment (Table 2). Additionally, the enrichment of the Fe-Po fraction in the rhizosphere soils of both crops under maize-soybean intercropping probably resulted from the rhizospheric transformation of crystalline Fe oxides to amorphous phase, which have relatively high surface area and in turn enhance Po immobilization and accumulation (Liu et al., 2015). A previous long-term field study also found increased P concentration in the Fe-Po fractions of soil under long-term maize monocropping (Liu et al., 2017).
Maize-soybean intercropping selectively enriched specific bacterial phyla in the rhizosphere (Figures 1A, 2A). The enrichment of Microvirga, Lysobacter, and Microlunatus in the rhizosphere of both crops under the NB treatment suggests that maize and soybean could harbor certain bacterial genera in their rhizosphere soils under intercropping. Microvirga (Rhizobiales), as an N-fixing bacteria (Pathan et al., 2020; Zhang et al., 2021), could contribute to increased soil N concentrations and subsequent plant growth, which probably indirectly affected the mobilization of soil P as a result of enhanced release of root exudates (Betencourt et al., 2012). This agreed with the positive correlation of the abundance of Microvirga with the TN content in the rhizosphere soils of both crops (Figures 1B, 2B). Furthermore, the abundance of Microvirga was negatively correlated to the Ca2-Po fraction but positively related to PDE and ALP activities. Legume roots, generally benefiting the colonization of N-fixing bacteria, also release high amounts of phosphatase (Betencourt et al., 2012), which probably contributed to the negative correlation between the Microvirga and Ca2-Po fraction. Moreover, Lysobacter could release acid and alkaline phosphatases to degrade Po (Chen et al., 2019). The enrichment of Lysobacter in the rhizosphere soils of the two crops under the NB treatment may have contributed to the enhanced PDE and ALP activity in the maize and soybean rhizospheres, respectively (Table 2). This implies that bacteria-mediated Po cycling was enhanced in the rhizosphere of these crops, although the total Po contents increased for both crops under the NB treatment (Table 2). In addition, Microlunatus facilitates the formation and accumulation of polyphosphate (Zhong et al., 2019). The enriched Microlunatus positively correlated with the Ca8-Pi fraction, but negatively correlated with the Ca2-Pi, Al-Pi and Fe-Pi fractions in both maize and soybean rhizospheres (Figures 1B, 2B), suggesting that Microlunatus may be involved in transformation of Ca2-Pi, Al-Pi and Fe-Pi fractions into the Ca8-Pi fraction through the formation of polyphosphate by Microlunatus and the subsequent hydrolysis of polyphosphate to replenish Ca2-Pi, Al-Pi and Fe-Pi pools. A previous study also showed that the application of polyphosphate fertilizers increased the NaOH-extracted soil P fraction as a result of the relatively high solubility of polyphosphate (Shah and Chu, 2021). Since all the selected representative strains of these taxa related to P cycling were obtained from Illumina MiSeq sequencing analysis, future investigations on the isolation of representative strains of these taxa to test their functions are needed.
The presence of specific microorganisms also depended critically on the crop species among the root barrier treatments. There was a tighter interaction of bacteria, reported to be involved in P-cycling, in the rhizosphere of soybean than maize, which demonstrated the enhanced interactions of bacteria in the soybean rhizosphere (Figure 3). Furthermore, Arthrobacter was significantly enriched in maize monocropping (i.e., SB treatment) but was inhibited in the maize-soybean intercropping (i.e., NB treatment). The C–P lyase, detected in the Arthrobacter sp. (Kertesz et al., 1991), could contribute to the degradation of phosphonates by cleaving the C–P bond. This may have accounted for the higher Po content in the maize-soybean intercropping relative to the maize monocropping. Sphingomonas, being able to utilize phytate (Sanangelantoni et al., 2018), accumulated in the rhizosphere of soybean but not for maize, which could account for the positive relationship of Sphingomonas with the Fe-Po fraction enriched with orthophosphate monoester containing high abundance of phytate (Liu et al., 2013).
This study provides new insights into the transformations of specific soil P fractions under maize-soybean intercropping and associated chemical and biological driving mechanisms. Maize-soybean intercropping facilitated the recovery of soil legacy P and induced the release of P from the Ca2-Pi, Al-Pi/Po and Fe-Pi fractions, which probably resulted from synergetic effects of rhizosphere OC deposit, the enhanced enzymes (ALP and PDE) activities and the bacteria (Microvirga, Lysobacter, Microlunatus, and Sphingomonas) which exhibited correlation with soil P pools. The new finding that a tighter engagement in the network of the bacteria in the rhizosphere of soybean than that of maize suggests the potential application of soybean intercropping with cereal crops to recover soil legacy P resources. This facilitated the maintenance of soil fertility with low-carbon economics compared to conventional chemical fertilization, although future field scale studies are needed to confirm these results.
The data presented in this study are deposited in the Sequence Read Archive (SRA) of NCBI (https://www.ncbi.nlm.nih.gov/) (accession no. PRJNA782812).
JL designed the study. CH and JL conducted the pot experiment. CH, YL, JL, and DY performed the data analyses. JL and YL wrote the manuscript with inputs from JY, BC-M, YC, and PS. All authors read and approved the manuscript.
This work was financially supported by the National Natural Science Foundation of China (No. 41977091).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1028969/full#supplementary-material
Amadou, I., Houben, D., and Faucon, M.-P. (2021). Unravelling the role of rhizosphere microbiome and root traits in organic phosphorus mobilization for sustainable phosphorus fertilization. A review. Agronomy 11:2267. doi: 10.3390/agronomy11112267
Andersson, K. O., Tighe, M. K., Guppy, C. N., Milham, P. J., and McLaren, T. I. (2015). Incremental acidification reveals phosphorus release dynamics in alkaline vertic soils. Geoderma 259, 35–44. doi: 10.1016/j.geoderma.2015.05.001
Barberan, A., Bates, S. T., Casamayor, E. O., and Fierer, N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. doi: 10.1038/ismej.2011.119
Bargaz, A., Noyce, G. L., Fulthorpe, R., Carlsson, G., Furze, J. R., Jensen, E. S., et al. (2017). Species interactions enhance root allocation, microbial diversity and P acquisition in intercropped wheat and soybean under P deficiency. Appl. Soil Ecol. 120, 179–188. doi: 10.1016/j.apsoil.2017.08.011
Bekkara, F., Jay, M., Viricel, M. R., and Rome, S. (1998). Distribution of phenolic compounds within seed and seedlings of two Vicia faba cvs differing in their seed tannin content, and study of their seed and root phenolic exudations. Plant Soil 203, 27–36. doi: 10.1023/A:1004365913726
Bernards, M. L., Jolley, V. D., Stevens, W. B., and Hergert, G. W. (2002). Phytosiderophore release from nodal, primary, and complete root systems in maize. Plant Soil 241, 105–113. doi: 10.1023/A:1016084023377
Betencourt, E., Duputel, M., Colomb, B., Desclaux, D., and Hinsinger, P. (2012). Intercropping promotes the ability of durum wheat and chickpea to increase rhizosphere phosphorus availability in a low P soil. Soil Biol. Biochem. 46, 181–190. doi: 10.1016/j.soilbio.2011.11.015
Cabeza, R. A., Myint, K., Steingrobe, B., Stritsis, C., Schulze, J., and Claassen, N. (2017). Phosphorus fractions depletion in the rhizosphere of young and adult maize and oilseed rape plants. J. Soil Sci. Plant Nutr. 17, 824–838. doi: 10.4067/S0718-95162017000300020
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, D. M., Huang, J. G., and Yuan, L. (2019). A new function of the biocontrol bacterium Lysobacter enzymogenes LE16 in the mineralization of soil organic phosphorus. Plant Soil 442, 299–309. doi: 10.1007/s11104-019-04175-x
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The ribosomal database project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37(Suppl._1), D141–D145. doi: 10.1093/nar/gkn879
Dragicevic, V., Oljaca, S., Stojiljkovic, M., Simic, M., Dolijanovic, Z., and Kravic, N. (2015). Effect of the maize-soybean intercropping system on the potential bioavailability of magnesium, iron and zinc. Crop Pasture Sci. 66, 1118–1127. doi: 10.1071/CP14211
Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Hagberg, A. A., Swart, P. J., and Schult, D. A. (2008). “Exploring network structure, dynamics, and function using NetworkX,” in Proceedings of the 7th Python in science conference, eds G. Varoquaux, T. Vaught, and J. Millman (Pasadena, CA), 11–15.
Hinsinger, P. (2001). Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: A review. Plant Soil 237, 173–195. doi: 10.1023/A:1013351617532
Hinsinger, P., Bengough, A. G., Vetterlein, D., and Young, I. M. (2009). Rhizosphere: Biophysics, biogeochemistry and ecological relevance. Plant Soil 321, 117–152. doi: 10.1007/s11104-008-9885-9
Hunt, J. F., Ohno, T., He, Z. Q., Honeycutt, C. W., and Dail, D. B. (2007). Inhibition of phosphorus sorption to goethite, gibbsite, and kaolin by fresh and decomposed organic matter. Biol. Fertil. Soils 44, 277–288. doi: 10.1007/s00374-007-0202-1
Jiang, B., and Gu, Y. (1989). A suggested fractionation scheme of inorganic phosphorus in calcareous soils. Fertil. Res. 20, 159–165. doi: 10.1007/BF01054551
Kertesz, M., Elgorriaga, A., and Amrhein, N. (1991). Evidence for two distinct phosphonate-degrading enzymes (C-P lyases) in Arthrobacter sp, GLP-1. Biodegradation 2, 53–59. doi: 10.1007/BF00122425
Li, L., Li, S., Sun, J., Zhou, L., Bao, X., Zhang, H., et al. (2007). Diversity enhances agricultural productivity via rhizosphere phosphorus facilitation on phosphorus-deficient soils. Proc. Natl. Acad. Sci. U.S.A. 104, 11192–11196. doi: 10.1073/pnas.0704591104
Liao, D., Zhang, C., Li, H., Lambers, H., and Zhang, F. (2020). Changes in soil phosphorus fractions following sole cropped and intercropped maize and faba bean grown on calcareous soil. Plant Soil 448, 587–601. doi: 10.1007/s11104-020-04460-0
Liu, J., Cade-Menun, B. J., Yang, J., Hu, Y., Liu, C. W., Tremblay, J., et al. (2018). Long-term land use affects phosphorus speciation and the composition of phosphorus cycling genes in agricultural soils. Front. Microbiol. 9:1643. doi: 10.3389/fmicb.2018.01643
Liu, J., Hu, Y., Yang, J., Abdi, D., and Cade-Menun, B. J. (2015). Investigation of soil legacy phosphorus transformation in long-term agricultural fields using sequential fractionation, P K-edge XANES and solution P-NMR spectroscopy. Environ. Sci. Technol. 49, 168–176. doi: 10.1021/es504420n
Liu, J., Sui, P., Cade-Menun, B. J., Hu, Y., Yang, J., Huang, S., et al. (2019). Molecular-level understanding of phosphorus transformation with long-term phosphorus addition and depletion in an alkaline soil. Geoderma 353, 116–124. doi: 10.1016/j.geoderma.2019.06.024
Liu, J., Yang, J., Cade-Menun, B. J., Hu, Y., Li, J., Peng, C., et al. (2017). Molecular speciation and transformation of soil legacy phosphorus with and without long-term phosphorus fertilization: Insights from bulk and microprobe spectroscopy. Sci. Rep. 7:15354. doi: 10.1038/s41598-017-13498-7
Liu, J., Yang, J., Cade-Menun, B. J., Liang, X., Hu, Y., Liu, C. W., et al. (2013). Complementary phosphorus speciation in agricultural soils by sequential fractionation, solution P nuclear magnetic resonance, and P K-edge X-ray absorption near-edge structure spectroscopy. J. Environ. Qual. 42, 1763–1770. doi: 10.2134/jeq2013.04.0127
Lu, R. (1999). Analytical methods of soil and agricultural chemistry. Beijing: China Agricultural Science and Technology Press.
Magoc, T., and Salzberg, S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Messaoudi, H., Gerard, F., Dokukin, P., Djamai, H., Rebouh, N. Y., and Latati, M. (2020). Effects of intercropping on field-scale phosphorus acquisition processes in a calcareous soil. Plant Soil 449, 331–341. doi: 10.1007/s11104-020-04491-7
Murphy, J., and Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 27, 31–36. doi: 10.1016/S0003-2670(00)88444-5
Negassa, W., and Leinweber, P. (2009). How does the Hedley sequential phosphorus fractionation reflect impacts of land use and management on soil phosphorus: A review. J. Plant Nutr. Soil Sci. 172, 305–325. doi: 10.1002/jpln.200800223
Park, Y., Solhtalab, M., Thongsomboon, W., and Aristilde, L. (2022). Strategies of organic phosphorus recycling by soil bacteria: Acquisition, metabolism, and regulation. Environ. Microbiol. Rep. 14, 3–24. doi: 10.1111/1758-2229.13040
Pathan, S. I., Scibetta, S., Grassi, C., Pietramellara, G., Orlandini, S., Ceccherini, M. T., et al. (2020). Response of soil bacterial community to application of organic and inorganic phosphate based fertilizers under Vicia faba L. cultivation at two different phenological stages. Sustainability 12:9706. doi: 10.3390/su12229706
Qin, J., Enya, O., and Lin, C. (2018). Dynamics of Fe, Mn, and Al liberated from contaminated soil by low-molecular-weight organic acids and their effects on the release of soil-borne trace elements. Appl. Sci. 8:2444. doi: 10.3390/app8122444
Qin, L., Zhang, W., Lu, J., Stack, A. G., and Wang, L. (2013). Direct imaging of nanoscale dissolution of dicalcium phosphate dihydrate by an organic ligand: Concentration matters. Environ. Sci. Technol. 47, 13365–13374. doi: 10.1021/es402748t
Reichard, P. U., Kretzschmar, R., and Kraemer, S. M. (2007). Dissolution mechanisms of goethite in the presence of siderophores and organic acids. Geochim. Cosmochim. Acta 71, 5635–5650. doi: 10.1016/j.gca.2006.12.022
Richardson, A. E., Barea, J.-M., McNeill, A. M., and Prigent-Combaret, C. (2009). Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321, 305–339. doi: 10.1007/s11104-009-9895-2
Roohi, M., Arif, M. S., Yasmeen, T., Riaz, M., Rizwan, M., Shahzad, S. M., et al. (2020). Effects of cropping system and fertilization regime on soil phosphorous are mediated by rhizosphere-microbial processes in a semi-arid agroecosystem. J. Environ. Manag. 271:111033. doi: 10.1016/j.jenvman.2020.111033
Rubio, G., Faggioli, V., Scheiner, J. D., and Gutierrez-Boem, F. H. (2012). Rhizosphere phosphorus depletion by three crops differing in their phosphorus critical levels. J. Plant Nutr. Soil Sci. 175, 810–871. doi: 10.1002/jpln.201200307
Sanangelantoni, A. M., Malatrasi, M., Trivelloni, E., Visioli, G., and Agrimonti, C. (2018). A novel beta-propeller phytase from the dioxin-degrading bacterium Sphingomonas wittichii RW-1. Appl. Microbiol. Biotechnol. 102, 8351–8358. doi: 10.1007/s00253-018-9248-2
Sattari, S. Z., Bouwman, A. F., Giller, K. E., and van Ittersum, M. K. (2012). Residual soil phosphorus as the missing piece in the global phosphorus crisis puzzle. Proc. Natl. Acad. Sci. U.S.A. 109, 6348–6353. doi: 10.1073/pnas.1113675109
Shah, J. A., and Chu, G. (2021). Short-chain soluble polyphosphate fertilizers increased soil P availability and mobility by reducing P fixation in two contrasting calcareous soils. Peerj 9:e11493. doi: 10.7717/peerj.11493
Smith, F. W., and Joynson, A. D. (1974). Rapid semi-micro digestion technique for major element analysis of plant material. Lab. Pract. 23, 51–52.
Tabatabai, M. A. (1994). “Soil enzymes,” in Methods of soil analysis. Part2: Microbiological and biochemical properties, eds S. H. Mickelson and J. M. Bifham (Madison, WI: Soil Science Society of America, Inc), 775–833.
Tang, X., Placella, S. A., Dayde, F., Bernard, L., Robin, A., Journet, E.-P., et al. (2016). Phosphorus availability and microbial community in the rhizosphere of intercropped cereal and legume along a P-fertilizer gradient. Plant Soil 407, 119–134. doi: 10.1007/s11104-016-2949-3
Tian, J., Tang, M., Xu, X., Luo, S., Condron, L. M., Lambers, H., et al. (2020). Soybean (Glycine max (L.) Merrill) intercropping with reduced nitrogen input influences rhizosphere phosphorus dynamics and phosphorus acquisition of sugarcane (Saccharum officinarum). Biol. Fertil. Soils 56, 1063–1075. doi: 10.1007/s00374-020-01484-7
USDA (1999). Soil taxonomy: A basic system of soil classification for making and interpreting soil surveys. Washington, DC: U.S. Government Printing Office, 489.
Vance, C. P., Uhde-Stone, C., and Allan, D. L. (2003). Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Wagar, B. I., Stewart, J. W. B., and Moir, J. O. (1986). Changes with time in the form and availability of residual fertilizer phosphorus on Chernozemic soils. Can. J. Soil Sci. 66, 105–119. doi: 10.4141/cjss86-011
Wang, Q. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, Z. R., Straub, D., Yang, H. Y., Kania, A., Shen, J. B., Ludewig, U., et al. (2014). The regulatory network of cluster-root function and development in phosphate-deficient white lupin (Lupinus albus) identified by transcriptome sequencing. Physiol. Plant. 151, 323–338. doi: 10.1111/ppl.12187
Withers, P. J. A., Vadas, P. A., Uusitalo, R., Forber, K. J., Hart, M., Foy, R. H., et al. (2019). A global perspective on integrated strategies to manage soil phosphorus status for eutrophication control without limiting land productivity. J. Environ. Qual. 48, 1234–1246. doi: 10.2134/jeq2019.03.0131
Yang, J., Hu, S., Chen, X., Yu, M., Liu, J., Li, H., et al. (2010). Transformation of lead solid fraction in the rhizosphere of Elsholtzia splendens: The importance of organic matter. Water Air Soil Poll. 205, 333–342. doi: 10.1007/s11270-009-0077-x
Yu, X., Tong, D., Wang, S., Wang, D., Xu, L, Li, B., et al. (2022). Long-term rice cultivation promoted microbial mineralization of organic P in a black soil. Soil Sci. Soc. Am. J. 86, 540–551. doi: 10.1002/saj2.20384
Zhang, D., Zhang, C., Tang, X., Li, H., Zhang, F., Rengel, Z., et al. (2016). Increased soil phosphorus availability induced by faba bean root exudation stimulates root growth and phosphorus uptake in neighbouring maize. New Phytol. 209, 823–831. doi: 10.1111/nph.13613
Zhang, H., Han, L., Jiang, B., and Long, C. (2021). Identification of a phosphorus-solubilizing Tsukamurella tyrosinosolvens strain and its effect on the bacterial diversity of the rhizosphere soil of peanuts growth-promoting. World J. Microbiol. Biotechnol. 37:109. doi: 10.1007/s11274-021-03078-3
Zhang, L., Tang, L., and Zheng, Y. (2015). Phosphorus aborption of crops affected by root interaction in maize and soybean intercropping system. J. Plant Nutr. Fertil. 21, 1142–1149. doi: 10.11674/zwyf.2015.0506
Keywords: legacy P, mobilization, rhizosphere effect, phosphatase, P-solubilizing bacteria
Citation: Liu J, Li Y, Han C, Yang D, Yang J, Cade-Menun BJ, Chen Y and Sui P (2022) Maize-soybean intercropping facilitates chemical and microbial transformations of phosphorus fractions in a calcareous soil. Front. Microbiol. 13:1028969. doi: 10.3389/fmicb.2022.1028969
Received: 26 August 2022; Accepted: 10 October 2022;
Published: 16 November 2022.
Edited by:
Junli Hu, Institute of Soil Science (CAS), ChinaReviewed by:
Jihui Tian, South China Agricultural University, ChinaCopyright © 2022 Jin Liu, Yang Li, Chaoqun Han, Dongling Yang, Jianjun Yang, Yuanquan Chen, Peng Sui His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Barbara J. Cade-Menun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Liu, amxpdTIwN0BjYXUuZWR1LmNu
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.