
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 December 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1026991
This article is part of the Research Topic Rhizospheric Microbiota-Plant Interactions: A Bioremediation Strategy for Inorganic Pollutants View all 5 articles
Morocco holds the vast majority of the world’s phosphate reserves, but due to the processes involved in extracting and commercializing these reserves, large quantities of de-structured, nutritionally deficient mine phosphate wastes are produced each year. In a semi-arid climate, these wastes severely hamper plant growth and development leading to huge unvegetated areas. Soil indigenous Plant Growth-Promoting Bacteria (PGPB) play a pivotal role in restauration of these phosphate mining wastes by revegetation, by increasing plants development, soil functioning, and nutrient cycling. The development of a vegetative cover above the degraded phosphate wastes, could stabilize and reintegrate these wastes in the surrounding environment. The current study’s objectives were to isolate, characterize, and identify indigenous bacterial strains, and test their PGP activity in vitro and, for the best-performing strains in planta, in order to assess their potential for acting as biofertilizers. A quantitative test for the synthesis of auxin and the production of siderophores as well as a qualitative test for the solubilization of phosphate were performed on all isolated bacterial strains. The production of hydrogen cyanide (HCN), exopolysaccharides (EPS), and enzymes were also examined. Three bacteria, selected among the best PGPB of this study, were tested in planta to determine whether such indigenous bacteria could aid plant growth in this de-structured and nutrient-poor mining soil. Using 16S rRNA gene sequencing, 41 bacterial strains were isolated and 11 genera were identified: Acinetobacter, Agrococcus, Bacillus, Brevibacterium, Microbacterium, Neobacillus, Paenibacillus, Peribacillus, Pseudarthrobacter, Stenotrophomonas, and Raoultella. Among the three best performing bacteria (related to Bacillus paramycoides, Brevibacterium anseongense, and Stenotrophomonas rhizophila), only Stenotrophomonas rhizophila and Brevibacterium anseongense were able to significantly enhance Lupinus albus L. growth. The best inoculation results were obtained using the strain related to Stenotrophomonas rhizophila, improving the plant’s root dry weight and chlorophyll content. This is also, to our knowledge, the first study to show a PGP activity of Brevibacterium anseongense.
Phosphate mining is one of the most important sectors of the Moroccan economy. Morocco has around 50 billion metric tons of rock phosphate reserves making it by far, the country with the largest reserves, and the 2nd biggest phosphate producer in the world, with annual production reaching 37 million metric tons in 2020 (USGS, 2020). During open-pit mining, huge volumes of wastes (hundreds of millions of tons per year) are removed to access the phosphate rocks. Wastes result also from the long process to generate the final product, implicating crushing, screening, and washing of phosphate rocks, followed by attacks with chemicals. All these represent approximately 18,000 tons of wastes every day, stockpiled on huge surface areas and exposed to atmospheric phenomena (Trifi et al., 2020; Mobaligh et al., 2021). These unstructured wastes, poor in nutrients, lacking organic matter, and contaminated by excessive phosphorus and sometimes hazardous contaminants such as fluoride, strontium, chromium, or cadmium, strongly limit the natural formation of a vegetative cover. These unvegetated wastes, damaging the esthetics of the landscape, are subjected to erosion and transport of small particles by wind and water and represent a potential threat to the environment and the local population’s health (Zine et al., 2020, 2021). The semi-arid climate, characterized by strong winds and short but intense rainfall events (annual precipitation not exceeding 250 mm), makes the establishment of a vegetative cover even more challenging. Furthermore, the salt concentrations at the surface of soils are generally high in semi-arid area due to low water infiltration and high evaporation rate (Mendez and Maier, 2008). Revegetation of phosphate wastes represents an effective and economic way to stabilize the ground, limit erosion, and allow the reintegration of the site in the surrounding ecosystem.
Due to their major role in soil functioning and plant development, indigenous plant growth-promoting bacteria could be of great help (Hassani et al., 2018). Numerous studies have been conducted on the use of indigenous bacteria to remediate contaminated sites (Wu et al., 2019; Jalali et al., 2020; Khairina et al., 2021). These bacteria act either through direct or indirect mechanisms (Mahmud et al., 2021). The direct mechanisms englobe (i) biological phytohormones production such as IAA (indole 3-acetic-acid) which plays an important role in root development and root hair formation, and hence in plant growth (Duca et al., 2014); (ii) some nutrients solubilization such as phosphorus or siderophores production important for iron captation (Upadhyay and Chauhan, 2022). Iron is a fundamental cofactor in cell machinery (Ma et al., 2016), and is highly abundant in soils but generally unavailable to microorganisms and plants. Siderophores are secondary metabolites that bind to iron and form an assimilable complex that provides plants with iron (Gray and Smith, 2005). Microbial siderophores usually have a higher affinity for iron than phytosiderophores (Pattnaik et al., 2019). The indirect mechanisms include the production of biocontrol agents such as hydrogen cyanide (HCN) which is known for its ability to prevent the development of pathogens; enzymes like cellulase, which can play an important role in restoring damaged soils by boosting seed germination or enhancing root development (Kuhad et al., 2011); and catalase which enhances plant defense against reactive oxygen species by decomposing hydrogen peroxide at an exceptionally rapid rate (Sharma and Ahmad, 2014; Nandi et al., 2017; Upadhyay and Chauhan, 2022). Exopolysaccharides (EPS) are metabolites produced by bacteria to protect themselves from harsh environmental conditions such as metals, drought or salinity (by sequestrating metals, or binding cations like Na+ alleviating salt stress, or enabling water conservation by maintaining a hydrated microenvironment around the cells) (Naseem and Bano, 2014; Sandhya and Ali, 2015). The EPS can also increase soil aggregation being adsorbed by some particles such as clay thus forming micro- and macroaggregates (Sandhya and Ali, 2015). Bacteria tolerating high concentrations of salinity can also be employed to counteract the negative effects of salt stress on plants, by enhancing physiological functions thus facilitating the installation and development of plants (Sharma et al., 2021).
To help establish a vegetation cover on these phosphate wastes, indigenous bacteria could represent an economical and eco-friendly method, acting as biofertilizer due to the properties described above. However, the documentation on plant growth promoting activities of indigenous bacteria from phosphate mine waste is poor. Therefore, the specific objectives of this study were to isolate and identify indigenous bacterial strains from phosphate mine wastes and characterize their PGP activities in vitro and for the best performing bacteria, in planta, in order to assess their use as biofertilizers. Three bacterial strains were selected for further biocontrol and metal tolerance tests and were used in single and consortium inoculation to evaluate their effect on the Lupinus albus L. growth, thus validating the results of in vitro PGP activities tests. This legume is an indigenous hyper-accumulator plant known for its capacity to adapt to harsh conditions such as drought stress, poor nutrients, and toxic metal contaminated soil, especially chromium, manganese, mercury, and lead-contaminated soils (Gawronski et al., 2011). This plant can also thrive on soils with very limited nutrient availability and a variability of pH from acid to alkaline (Esteban et al., 2003). Lupinus albus L. is also valued for its ability to improve soils by extracting toxic metals (Garau et al., 2021; González et al., 2021). The choice of Lupinus albus L. was based on (i) its ecophysiological traits allowing this indigenous legume to easily adapt to contaminated phosphate mining soils in Morocco, to (ii) its robust root system that facilitate the absorption of soil components (González et al., 2021), and to (iii) its ability to help recover de-structured soils via its capacity to provide nitrogen (Esteban et al., 2003).
Samples of phosphate tailings were collected in October 2017 from one of the experimental cells established at the abandoned pyrrhotite Kettara mine near Marrakech in Morocco (31.8748, –8.1760). A rehabilitation scenario for this mine includes covering acidic tailings with alkaline phosphate mine wastes to limit water infiltration and hence acid mine drainage (Ouakibi et al., 2013; Bossé et al., 2015). Five subsamples were collected from different random locations, then were processed in the laboratory within a week of sampling. The physicochemical properties of these phosphate tailings were identified and are detailed in Mghazli et al. (2021) and Supplementary Table 1.
A soil solution was prepared using 10 g of phosphate tailings that were dissolved in 90 ml of sterilized physiological water (0.9% NaCl) and stirred for 1 h. A series of 10–1 to 10–5 dilutions were made from the soil solution, and 100 μL aliquots from each dilution were spread on a modified yeast starch agar medium (YSA) from Kadmiri et al. (2010). The modified-YSA medium contains glucose and starch as source of carbon instead of only starch, and it contains per liter 4 g of yeast extract, 2 g of starch, 8 g of glucose, 0.5 g of MgSO4 7H2O, 0.5 g K2HPO4, and 15 g of agar. To select the maximum culturable bacteria, four different pH media ranging from 6 to 9 were used. The bacterial strains were transferred many times until a pure isolate was obtained.
Bacterial DNA was extracted using a standard phenol-chloroform protocol (Köchl et al., 2005), and 16S rRNA gene was amplified using MyTaq™ DNA Polymerase (Bioline, Meridian Bioscience, London, UK) and the universal primers 27f/1492r (Frank et al., 2008) or fD1/rD1 (Weisburg et al., 1991). The amplification was carried out as described in the following protocol: initial denaturation at 95°C for 1 min followed by 30 cycles of denaturation at 95°C for 15 s, annealing at 56°C for 15 s and extension at 72°C for 10 s, then a final extension step at 72°C for 1 min. The amplified products were subjected to Sanger paired-end sequencing and the resulting sequences were cleaned and processed using DNADragon and Mega X programs. The sequences were then BLASTed against the NCBI database to identify the isolated strains. All sequences were deposited in NCBI database and their accession numbers are shown in Figure 1 and Supplementary Table 2.
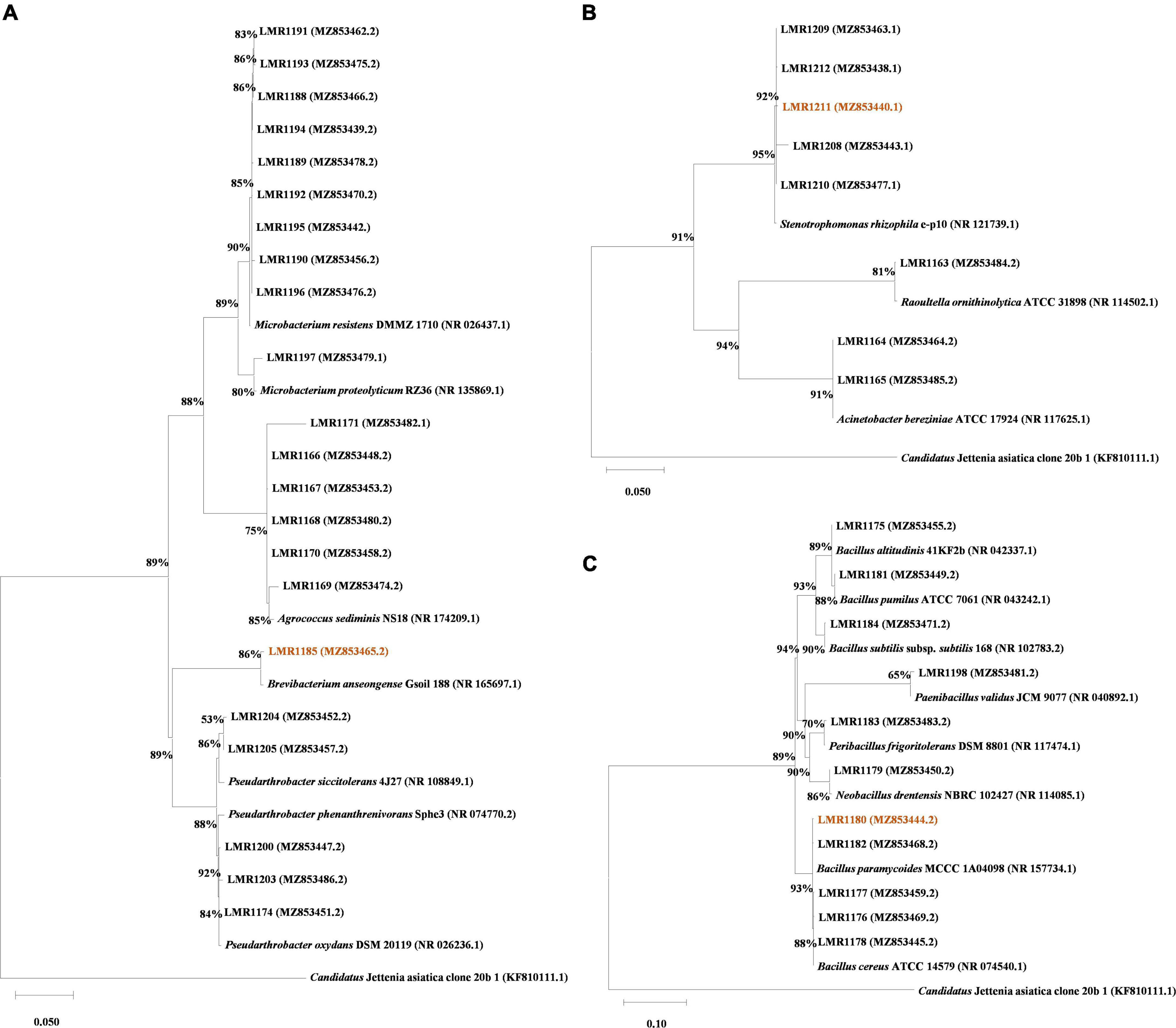
Figure 1. Phylogenetic trees of all isolated strains (indicated in bold) based on ClustalW aligned 16S rRNA gene. A total of 1,617 nt were used for Actinobacteria phylum alignment (A), 1,526 nt were used for the Proteobacteria phylum alignment (B), and 1,622 nt were used for the Firmicutes phylum alignment (C). The Tamura-Nei model (Tamura and Nei, 1993) with a discrete Gamma distribution was used to construct the phylogenetic trees. Accession numbers are given between parenthesis. The bootstrapping replicates was 1,000 for all trees and the percentage of replicate trees is shown beside the branches. Candidatus Jettenia asiatica clone 20b_1 was used as outgroup, and the strains used for in planta test are highlighted.
The phylogenetic tree was constructed using Mega X (Kumar et al., 2018). All the isolated sequences were aligned alongside reference sequences from NCBI database and an external reference sequence [Candidatus Jettenia asiatica clone 20b_1 (KF810111.1)]. The Maximum Likelihood method was used. The evolutionary distances were computed using the Tamura-Nei model (Tamura and Nei, 1993) with a discrete Gamma distribution with 5 rates categories.
A pre-culture bacterial cell suspension was prepared in YSA medium and was used for all the following phenotypic tests. YSA medium with a pH ranging from 4 to 12 using KH2PO4/K2HPO4 and HCl/Tris buffer solutions, were used to study the ability of the isolated strain to tolerate different pH. While for the salinity test, YSA medium was supplemented with different concentrations of NaCl ranging from 3 to 9%. For these two tests, the bacteria were dot spotted in Petri dishes and incubated at 28°C for at least 48 h. The capacity of our strains to tolerate high temperatures was assessed in Petri dishes containing YSA medium incubated at different temperatures ranging from 35 to 60°C for at least 48 h. All tests were repeated three times.
Pre-cultures were prepared and inoculated on a yeast mannitol medium (Sigma-Aldrich), supplemented with 0.5 g L-Tryptophane. After 48 h at 28°C, the cultures were centrifuged at 10,000 rpm for 20 min, then filtered through a 0.22 μm filter to eliminate any remaining bacterial cells. One mL of the filtrate was added to 1 mL of Salkowski reagent and incubated at room temperature in the dark for 30 min (Gordon and Weber, 1951). The optical density was then measured at 540 nm to quantify auxin production. The results were compared to a standard curve prepared using a pure IAA solution.
A volume of the pre-cultures was inoculated on Luria–Bertani (LB) medium (Sigma-Aldrich). The tubes were incubated at 28°C for 7 days, then centrifuged at 10,000 rpm for 20 min and filtered through a 0.22 μm filter. The pH of the filtrate was then adjusted to neutral and 1 mL of the suspension was added to 1 mL of CAS reagent (Alexander and Zuberer, 1991) in hemolysis tubes pre-treated with HCl. The mixture was incubated at room temperature in the dark for 30 min, before optical density was measured at 630 nm. The semi-quantitative results were calculated based on the following equation:
Where %SU, % of siderophore unit; Ar, absorbance of the control; As, absorbance of the strains.
Ten microliters of the strain’s pre-culture were deposited in Pikovskaya’s medium (PVK) (Pikovskaya, 1948) containing tricalcium phosphate (TCP) as a source of phosphate. The Petri dishes were incubated at 28°C for at least 7 days. The presence of a transparent halo beside the colonies indicates TCP solubilization. The phosphate solubilization index (PSI) was calculated based on the following equation (Premono et al., 1996):
The production of hydrogen cyanide (HCN) was determined using the protocol proposed by Castellano-Hinojosa and Bedmar (2017). For instance, bacterial strains were grown for 48 h in YSA medium supplemented with 0.4% glycine, then a sterile disc of 90 mm was dipped in 0.5% picric acid supplemented with 2% Na2CO3 and placed on the lid of each Petri dish, the plates were sealed and incubated at 30°C for at least 48 h. A change in the color of the bacteria to orange or brown signaled the production of HCN.
To detect strains able to produce cellulase a carboxymethylcellulose (CMC) agar medium (Kasana et al., 2008) was used. Bacterial strains were prepared in liquid medium and incubated overnight, then 5 μL of each bacterial suspension were spot plated. The Petri dishes were incubated at 28°C for 48 h, then were flooded for 30–40 min with 1% hexadecyltrimethyl ammonium bromide. The appearance of an unstained area around the colony signaled cellulase production (Kasana et al., 2008).
A simple method suggested by Reiner (2010) was used. In brief, overnight grown bacterial strains were rinsed and resuspended in 1 mL 0.9% NaCl solution. Ten microliters of bacterial strains were spotted on a sterile and empty Petri dish, and a drop of 3% H2O2 solution was spotted on top of the bacterial strain. The strains were considered catalase positive if bubbles formed.
To assess the ability of our bacterial strains to produce exopolysaccharides (EPS) the protocol proposed by Fusconi and Godinho (2002) was used. Bacteria liquid cultures (0.8 OD at 600 nm) were dot spotter on a mineral salt medium with 10% saccharose and incubated at 28°C for at least 48 h, the bacteria were considered EPS-positive if they are mucoid.
In a previous study (Mghazli et al., 2021), the bacterial diversity of phosphate wastes was characterized using Illumina MiSeq sequencing. The V4 region of the 16S rRNA gene sequences was targeted and revealed a huge number of sequences, totaling 8 million raw reads. In the present study, the TaxASS (Rohwer et al., 2018) workflow was used to BLAST the metabarcoding sequences against a customized database containing the sequences of the isolated strains. The Silva 132 database was chosen as a general reference database.
The bacterial strains used for the inoculation assay were subjected to antagonism tests to prevent any further inhibition effect in the in planta tests. Two methods were used, the first consisted of streaking two bacterial strains side-by-side on the same Petri dish containing YSA medium to test their diffusible compound effect (Jain et al., 2012). The second method consisted of spreading 100 μL of each bacterial strain on a Petri dish and place two dishes on top of each other to test their volatile compounds’ effect. The Petri plates were hermetically sealed with parafilm and incubated at 28°C for at least 3 days.
The antifungal activity through volatile compounds test was used as described by Ebadzadsahrai et al. (2020). In brief, bacterial strains were streaked on a YSA medium while a 6 mm diameter disc of the fungus (Botrytis cinerea and Fusarium oxysporum) was placed in the center of a Petri dish containing Potato Dextrose Agar (PDA) medium. The two plates were placed on top of each other and sealed, then incubated in the dark at 25°C for at least 7 days. The diffusible compounds antagonism test was also used as described by Hernández-León et al. (2015). To this end, bacterial strains were grown on solid LB medium for 24 h, before being filtered through a 0.22 μm filter, and 100 μL of the filtrate was spread on a Petri dish containing PDA medium. The plates were placed at 4°C for 2 h to help the diffusion of the bacterial filtrate through the medium, then a 6 mm fungal disc was placed in the center of each Petri dish and the plates were incubated in the dark at 25°C for 7 days. The percentage of inhibition of radial growth (PIRG) was calculated following the formula (Stracquadanio et al., 2020):
Where Rc is the radius of the fungal control and Rs is the radius of the fungal in presence of the bacterial strain.
Tryptone Yeast (TY) medium (containing per liter: 5 g tryptone, 3 g yeast, and 0.8 g CaCl2) was supplemented with 200–900 mg/mL FeCl2.4H2O, 100–550 mg/L NaAsO2 [As (III)], 50–300 mg/L HgCl2, 10–300 mg/L CdCl2.4H2O, and 50–500 mg/L ZnCl2 each time. The plates were dot spotted with bacterial strains and incubated at 28°C for 10 days before final reading.
Lupinus albus L. seeds were disinfected with 40% commercial bleach (12%) for 4 min and washed many times using sterile water. The seeds were then imbibed in sterile water for 4 h and germinated on a 0.6% water-agar medium. After 4 days, when the radicles start growing, the plantlets were placed in sieved non-sterile phosphate waste rock that served to isolate the bacterial strains.
Four treatments with five replicates were evaluated, three of which consisted of single inoculation, and one consisted of a consortium of the different strains. Non-inoculated plants were used as a control. Plants were grown in a greenhouse for 90 days. Fresh and dry weights and length of root and shoot parts were measured. Chlorophyll content was estimated following the protocol of Sharma et al. (2012). In brief, 0.5 g of fresh leaves was suspended in 25 mL of 80% acetone and 0.1% w/v CaCO3. The mixture was well-digested and mixed using the homogenizer, then centrifuged at 3000 rpm for 10 min at 4°C. The optical density of the supernatant was measured at 663, 645, and 470 nm. To determine the chlorophyll and the carotenoid contents, the following formula was used (Lichtenthaler and Wellburn, 1983):
Where Chl a is the chlorophyll a content, Chl b is the chlorophyll b content, Car is the carotenoid content.
All tests were repeated three times and data was analyzed using R (V 4.3) using variance analysis (ANOVA) and Tukey test (p < 0.05). Graphs visualization was done using ggplot2 package.
A total of 41 bacterial colonies were isolated from the phosphate mine wastes. Their sequences were BLASTed within NCBI database, and similar type strain sequences (with different similarity percentages) were then aligned with our strains using Mega X (Figure 1). Actinobacteria was the dominant phylum with 53.7% bacterial strains (22 out of 41), while 26.8% of the bacterial strains (11 out of 41) were affiliated to Firmicutes phylum, and only 19.5% (8 out of 41) was affiliated to Proteobacteria phylum. The most abundant genus was Microbacterium with 10 representatives, followed by Bacillus with 8 representatives. The remaining strains were affiliated to Acinetobacter, Agrococcus, Brevibacterium, Neobacillus, Paenibacillus, Peribacilllus, Pseudarthrobacter, Stenotrophomonas, and Raoultella genera, with 2, 6, 1, 1, 1, 1, 5, 5, and 1 representatives, respectively (Supplementary Table 2). The comparison between culture-dependent and culture-independent bacteria using TaxASS software revealed that all isolated bacteria were already present in the metabarcoding sequence data (Supplementary Figure 1).
All isolates were able to tolerate up to 5% of NaCl; 38 strains were able to tolerate 7% of NaCl; 26 strains tolerated 8% of NaCl while only 9 strains were able to grow in the presence of 9% of NaCl but none of the isolated strains was able to grow above this concentration (Figure 2).
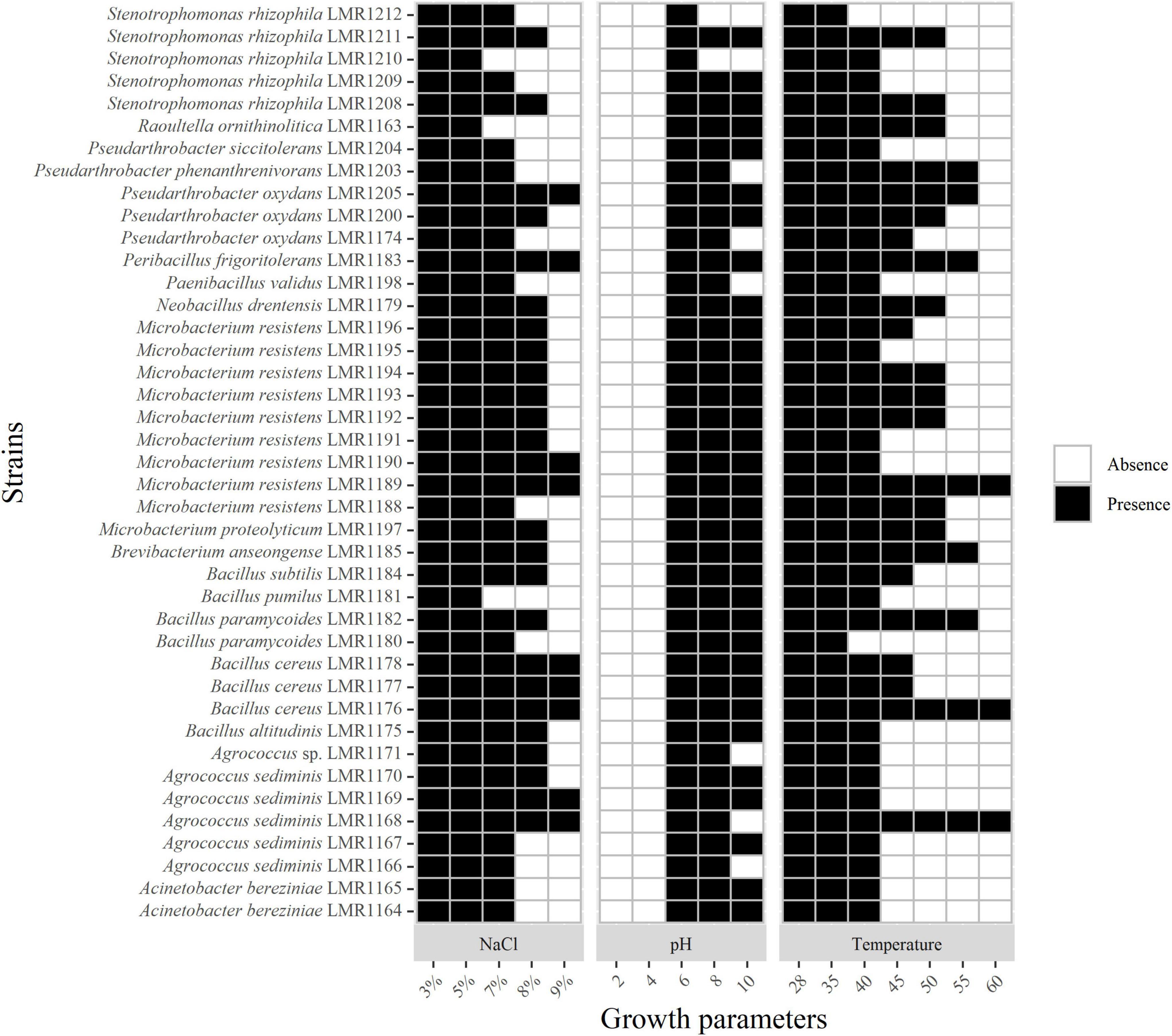
Figure 2. Heatmap of the capacity of the 41 isolated strains to tolerate different concentrations of NaCl and different pH and temperature. Presence and absence indicate the presence or the absence of growth.
As mentioned in Supplementary Table 1, the pH of phosphate mining wastes in this study is alkaline (8.1). None of the 41 bacterial strains was able to grow at a pH below 4; the optimal pH of all strains was 7. All representatives of Acinetobacter, Bacillus, Microbacterium and the strains similar to Brevibacterium, Peribacillus, and Raoultella genera were able to grow at pH 10, whereas none of the isolated bacteria was able to grow above this pH (Figure 2).
All strains were able to tolerate temperatures up to 40°C except Stenotrophomonas rhizophila LMR1212 and Bacillus paramycoides LMR1180 (Figure 2). Only three strains were able to grow at 60°C: Agrococcus sediminis LMR1168, Bacillus cereus LMR1176, and Microbacterium resistens LMR1189.
Twenty-two isolates were positive for catalase with different intensities (Figure 3). Bacillus paramycoides LMR1180 showed the best catalase activity, followed by the two strains of Acinetobacter bereziniae (LMR1164 and LMR1165), and by Brevibacterium anseongense LMR1185, whereas the remaining 18 strains showed quite low catalase activity.
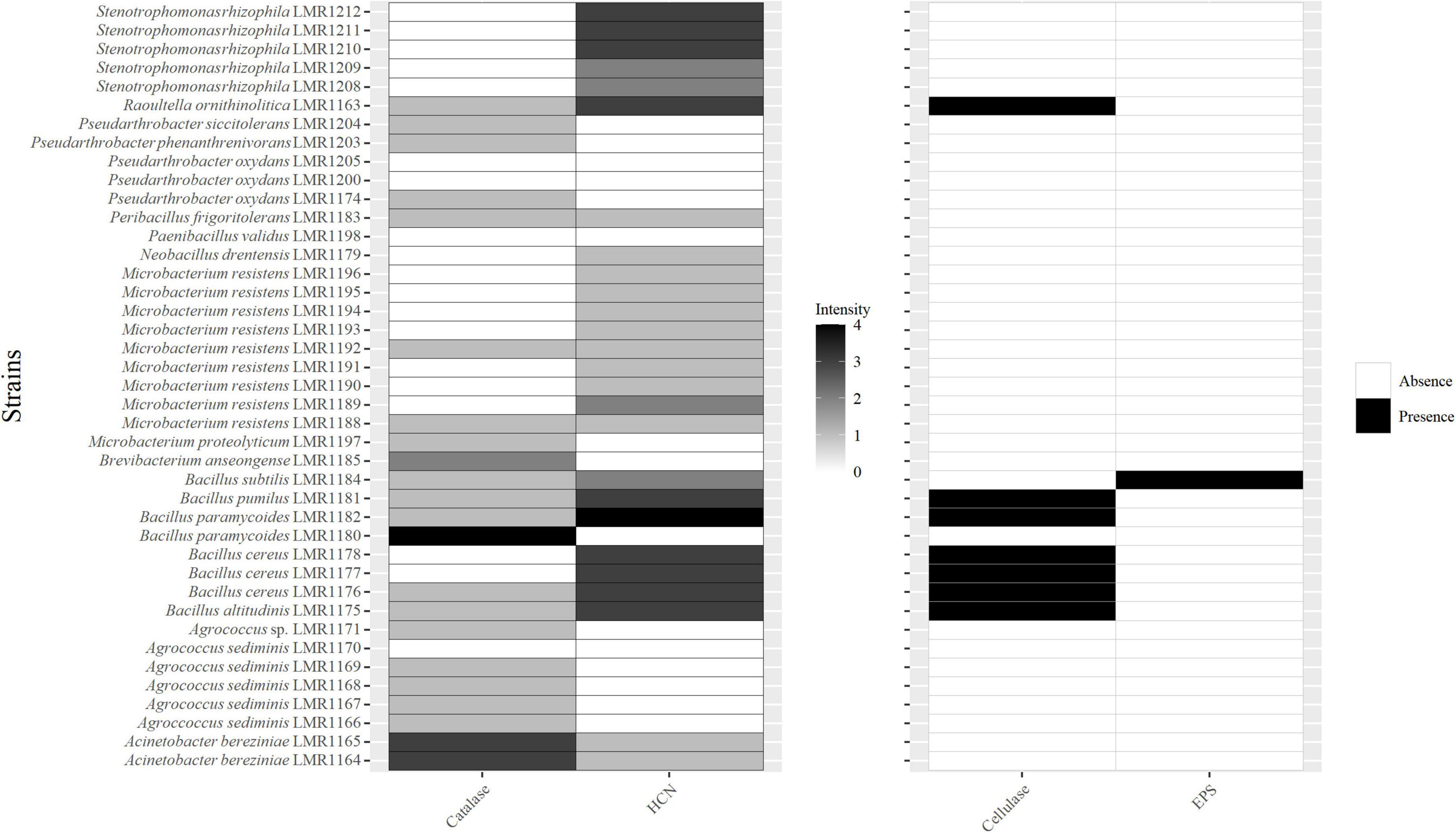
Figure 3. Heatmap of the capacity of the 41 isolated bacterial strains to produce catalase; hydrogen cyanide (HCN); cellulase and exopolysaccharides. For catalase and HCN; different intensities are presented, 0 for no activity, 1 for low activity, 2 for medium activity, 3 for high activity, and 4 for very high activity.
Twenty-six strains were positive for HCN (Figure 3). The highest activity was observed with Bacillus paramycoides LMR1182, followed by five strains of Bacillus (Bacillus altitudinis LMR1175, Bacillus cereus LMR1176; LMR1177 and LMR1178, and Bacillus pumilus LMR1181), Raoultella ornithinolytica LMR1163, and 3 strains of Stenotrophomonas rhizophila (LMR1210; LMR1211; and LMR1212). Bacillus subtilis LMR1184 and Microbacterium resistens LMR1189 showed medium production, while the remaining 14 strains showed poor HCN production.
Only strains of Raoultella (Raoultella ornithinolytica LMR1163) and Bacillus (Bacillus paramycoides LMR1182; Bacillus pumilus LMR1181; Bacillus cereus LMR1176-1178 and Bacillus altitudinis LMR1175) showed a positive reaction to the cellulase activity (Figure 3), while only Bacillus subtilis LMR1184 was EPS positive.
The majority of our bacterial strains tested positive for IAA production (Figure 4). IAA production ranged from 0.005 to 34.68 μg/mL. Among representatives of Actinobacteria phylum, all strains except Agrococcus sediminis LMR1169 were able to produce different quantities of IAA, while among Firmicutes representatives, Peribacillus frigoritolerans LMR1183 produced 9.3 μg/mL of IAA, and as for Proteobacteria phylum, Raoultella ornithinolytica LMR1163 produced the highest concentration of IAA, i.e., exceeding 13 μg/mL. The highest IAA production was recorded in Brevibacterium anseongense LMR1185 (34.68 μg/mL), followed by Microbacterium resistens LMR1196 (28.05 μg/mL) and Microbacterium proteolyticum LMR1197 (11.76 μg/mL).
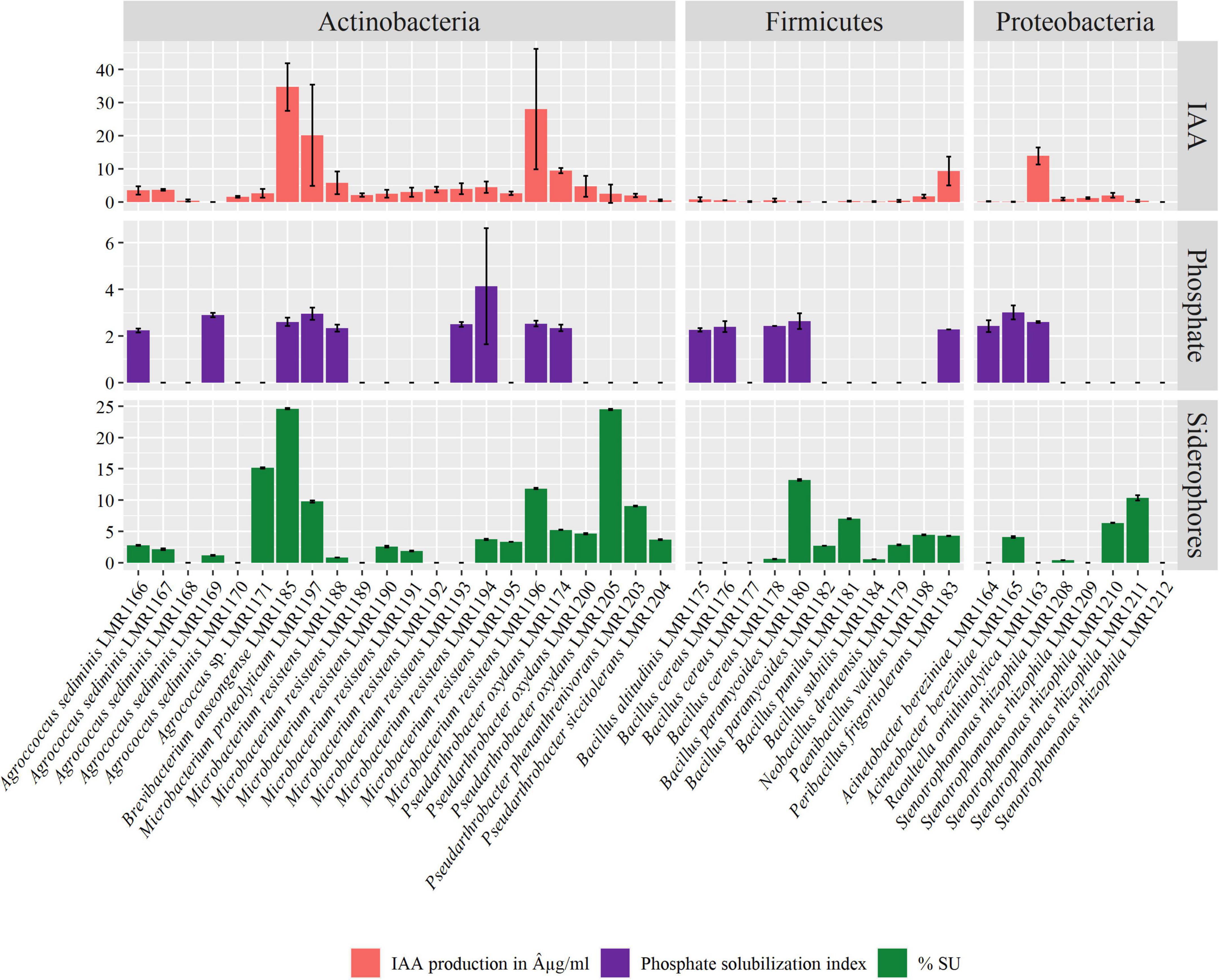
Figure 4. Plant growth promoting activities of the 41 isolated strains. Indole-3-acetic acid (IAA) production in μg/mL, phosphate solubilization index, and siderophore production in percent siderophore unit (%SU).
Seventeen strains were able to solubilize phosphate with a solubilization index ranging from 2.24 for Agrococcus sediminis LMR1166 to 4.13 for Microbacterium resistens LMR1194 (Figure 4). Nine strains of Actinobacteria phylum, five of Firmicutes phylum, and three of Proteobacteria phylum were able to solubilize different amounts of TCP.
Finally, 29 strains were able to produce siderophores, with production ranging from 0.38 to 24.6% SU (Figure 4). Brevibacterium anseongense LMR1185 produced the largest quantity of siderophores, followed by Pseudarthrobacter oxydans LMR1205 (24.5% SU). Four other strains (Agrococcus sp. LMR1171, Bacillus paramycoides LMR1180, Microbacterium resistens LMR1196, and Stenotrophomonas rhizophila LMR1211) produced more than 10% SU.
Based on all above results, three phylogenetically diversified strains, Bacillus paramycoides LMR1180, Brevibacterium anseongense LMR1185, and Stenotrophomonas rhizophila LMR1211, were chosen for the plant inoculation assay. The selection was based on all tests with a focus on PGP activities and discarding all strains with a potential pathogenic activity on human. These chosen bacteria were tested for other activities (antifungal, metal tolerance, and compatibility tests; Supplementary Table 3). The three selected bacteria were tested for their antagonism effects to eliminate any potential antagonism between strains that may affect the growth of the plant. No growth inhibition effect was detected between strains, neither via diffusible compounds nor through volatile compounds.
All the selected strains were able to inhibit the growth of the two fungi (Fusarium oxysporum and Botrytis cinerea) through volatile and diffusible compounds. Bacillus paramycoides LMR1180 showed the best effect against both fungi via diffusible compounds, but presents also quite good effects via volatile compounds. Brevibacterium anseongense LMR1185 and Stenotrophomonas rhizophila LMR1211 had the best effects against Fusarium oxysporum and Botrytis cinerea, respectively, via volatile compounds (Table 1).
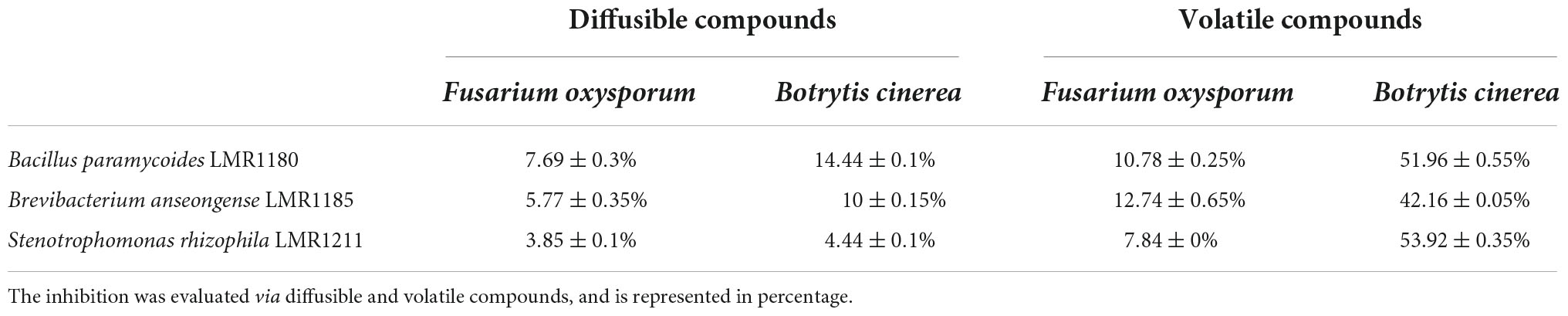
Table 1. Effects of the three selected strains (Bacillus paramycoides LMR1180, Brevibacterium anseongense LMR1185, and Stenotrophomonas rhizophila LMR1211) on the radial growth of two fungi (Botrytis cinerea and Fusarium oxysporum).
The three selected strains showed a quite high metal tolerance to all tested metals (NaAsO2, FeCl2, and HgCl2) except for CdCl2 where Bacillus paramycoides LMR1180 and Brevibacterium anseongense LMR1185 could not tolerate more than 20 and 10 mg/L, respectively, or for ZnCl2 where Bacillus paramycoides LMR1180 could not tolerate more than 50 mg/L (Table 2).
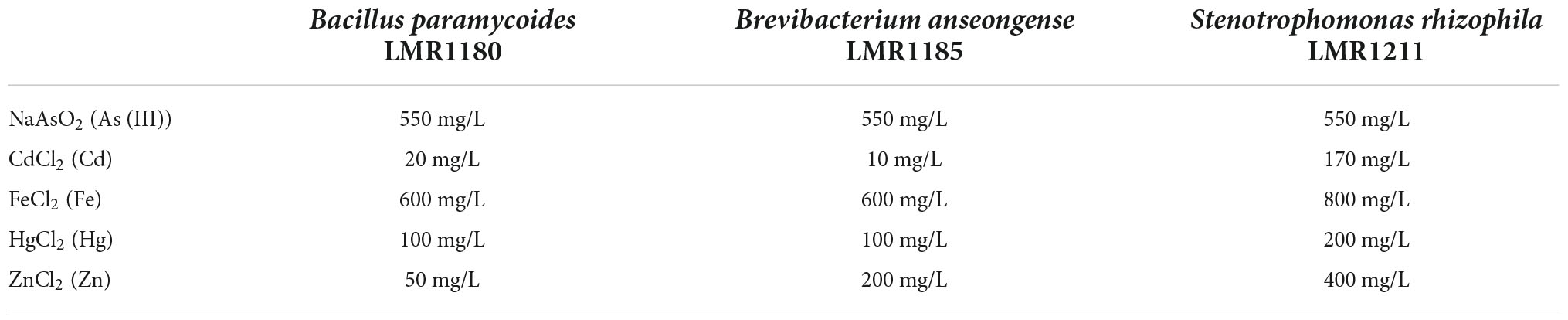
Table 2. Table representing the highest concentrations of toxic metals tolerated by the three selected strains (Bacillus paramycoides LMR1180, Brevibacterium anseongense LMR1185, and Stenotrophomonas rhizophila LMR1211).
Non-sterilized phosphate waste rocks were used for the inoculation test. Five germinated plantlets were used per test, and along the inoculation assay, the plantlets were sprinkled with distilled water every other day. After 90 days, different plant parameters were measured.
The shoot weights of plants treated with Brevibacterium anseongense LMR1185, and Stenotrophomonas rhizophila LMR1211 were similar and significantly higher than non-inoculated plants (Figure 5). No significant effects were observed for plants treated with Bacillus paramycoides LMR1180 or with the consortium compared to the control. Only plants treated with Stenotrophomonas rhizophila LMR1211, showed also significant differences in the root dry weight compared to the non-inoculated plants. Similarly, plants treated with Stenotrophomonas rhizophila LMR1211 and Brevibacterium anseongense LMR1185 had a significantly higher shoots than the control (Figure 5).
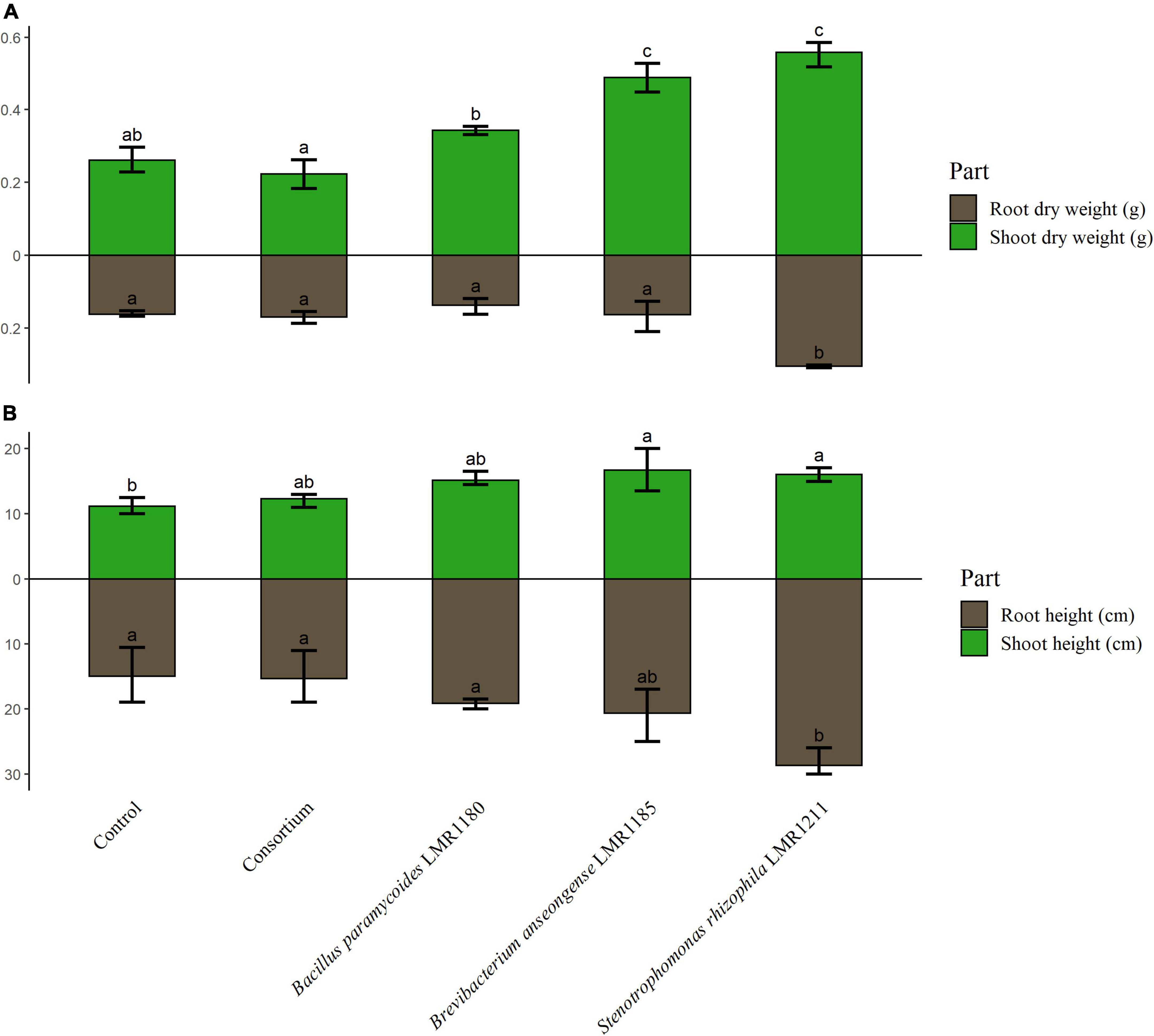
Figure 5. The measurement of control and inoculated Lupinus albus plants using the three selected strains (Bacillus paramycoides LMR1180, Brevibacterium anseongense LMR1185, and Stenotrophomonas rhizophila LMR1211). The dry weight of shoot and root parts (A) is represented in g, and the height of shoot and root parts (B) in cm after 90 days under greenhouse conditions. The letters (a, b, and c) present the compact display letters (CDL). Values in each bar followed by the same letter are not significantly different at 0.05% (Tukey’s).
The total chlorophyll content was only significantly higher for plants treated with Stenotrophomonas rhizophila LMR1211 (5.16 ± 0.12 mg/g) compared to the control, while no significant difference was observed in the carotenoid content from Stenotrophomonas rhizophila LMR1211 and Brevibacterium anseongense LM1185 compared to the control (Figure 6). We also note that Bacillus paramycoides LMR1180 and the consortium showed the lowest chlorophyll and carotenoid contents.
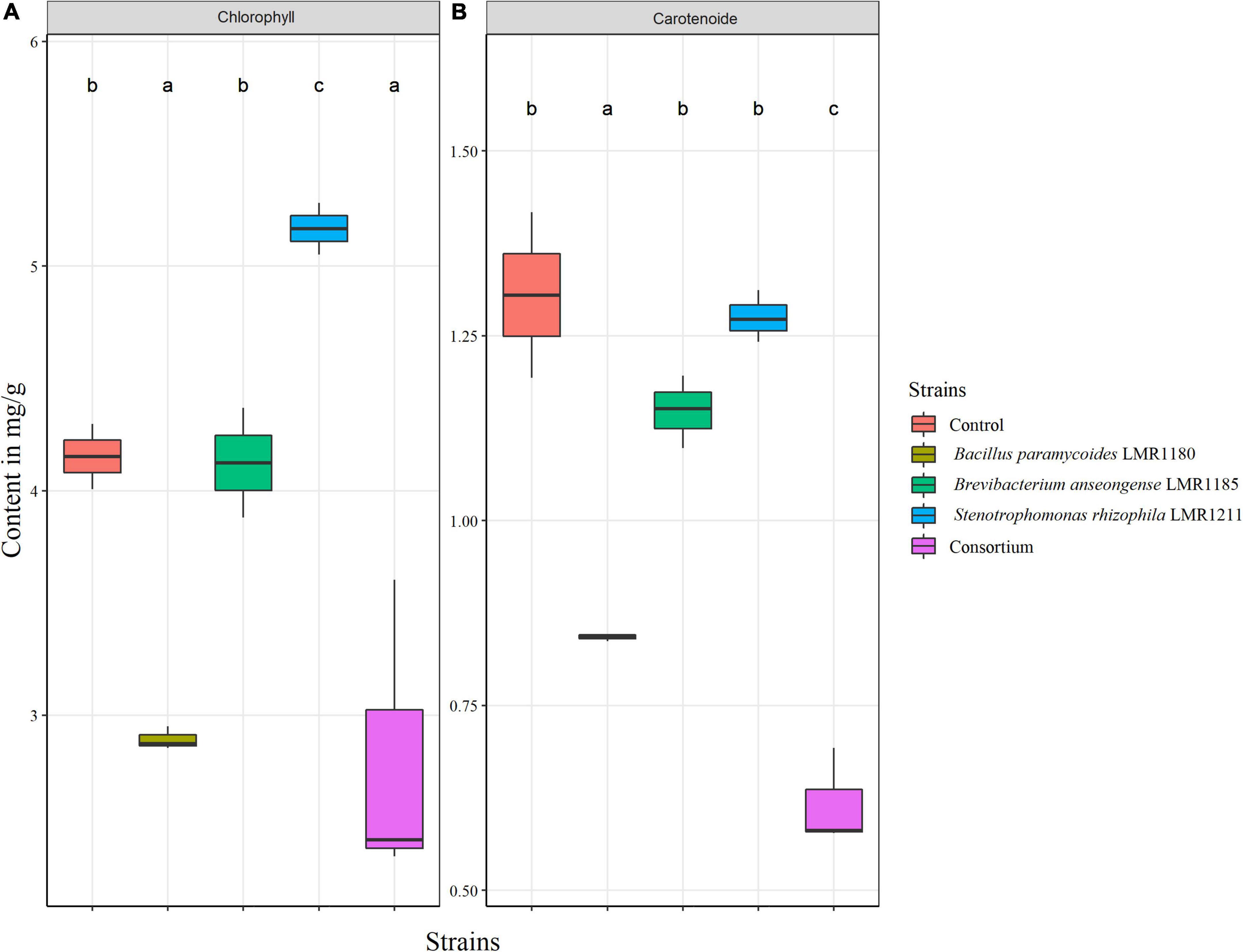
Figure 6. The estimation of total chlorophyll (A) and carotenoid (B) content in control and inoculated Lupinus albus plants using the three selected strains (Bacillus paramycoides LMR1180, Brevibacterium anseongense LMR1185, and Stenotrophomonas rhizophila LMR1211) in mg/g of biomass after 90 days under greenhouse conditions. The letters (a, b, and c) present the compact display letters (CDL). Values in each bar followed by the same letter are not significantly different at 0.05% (Tukey’s).
In Morocco, the development of vegetation in the harsh conditions of phosphate mine wastes in a semi-arid climate (requiring drought-, heat-, metal-, and salt-tolerant plants adapted to high-velocity winds) is key to the bioremediation by revegetation of these wastes dumps. As bacterial communities play a key role in the development of soil and plants, understanding bacterial structure, diversity, and their potential plant growth activities is the first step toward revegetation of phosphate tailings (Nannipieri et al., 2003; Luo et al., 2018). The objectives were to isolate and characterize the potential indigenous PGPB from phosphate wastes and select the most efficient strains for the growth of plants. To this end, forty-one bacterial strains were isolated, identified, and screened for their PGP activities in vitro. The best performing bacteria were also tested in planta. A very limited number of studies have focused on isolating the bacterial community from phosphate wastes and they mainly studied the phosphate solubilizing activity in bacteria for their use as phosphate biofertilizers (Hamdali et al., 2008, 2012; Yang et al., 2012; Aliyat et al., 2020; Benbrik et al., 2020; Qarni et al., 2021). To our knowledge, this is one of the first studies on the effect of indigenous bacteria on plant growth for revegetation purposes of phosphate wastes.
Among the 11 bacterial genera isolated in this study, only 4 were retrieved in precedent works in phosphate mine wastes: Acinetobacter, Bacillus, Brevibacterium, and Raoultella (Yang et al., 2012; Benbrik et al., 2020; Qarni et al., 2021). However, other genera such as Micromonospora, Streptomyces, Pseudomonas, Sphingobacterium, Staphylococcus, and Serratia were not identified in the present study (Hamdali et al., 2008, 2012; Yang et al., 2012; Benbrik et al., 2020; Qarni et al., 2021). This could be due to the isolation medium used or to the type of phosphate, and process implicated in generating these wastes.
The majority of the isolated strains were able to tolerate high temperatures, substrate salinity and alkalinity. This can be due to the presence of arid to semi-arid climate of study site, which results in a high evaporation rate causing serious drought stress and salinity in the surface soil.
Among the isolated strains, six Bacillus strains and one Raoultella were able to produce cellulase, an enzyme that could help in the bioremediation of de-structured soils by increasing seeds germination and/or by playing a key role in roots development (Kuhad et al., 2011). While only Bacillus subtilis LMR1184 was able to produce EPS. This is in accordance with the results of Moghannem et al. (2018) showing that different species of Bacillus genus were able to produce EPS. These metabolites, produced by bacteria, could help the development of plants in harsh condition by segregating metals, alleviating salinity, improving humidity of soil and soil aggregation (Sandhya and Ali, 2015).
Many of the isolated strains were able to produce HCN and catalase. Bacillus paramycoides LMR1182 produced the most HCN quantity, while Bacillus paramycoides LMR1180 exhibited the highest catalase production. In general, Bacillus spp. are well-known for their HCN (Walia et al., 2014; Brzezinska et al., 2020) and catalase production (Fibriarti et al., 2021). HCN is known for its capacity to counter the development and growth of plant pathogens (Nandi et al., 2017). Besides their biocontrol capacity, HCN-producing bacteria can enhance plant development by increasing the quantity of bioavailable phosphate in soil (Rijavec and Lapanje, 2016). Whereas catalase-producing bacteria can protect themselves and the surrounding soil, including plants from hydrogen peroxide and reactive oxygen species, responsible for cell wall degradation when present in excess (Reiner, 2010). Stress suppressing catalase can break down H2O2 molecules, thereby providing more oxygen to the soil (Singh and Kumar, 2019).
More than 95% of the isolated strains (39 strains) were able to produce IAA, which enhances plant growth by improving seed germination, shoot elongation, and the formation of root hairs which increases water and nutrient uptake (Small and Degenhardt, 2018; Ullah and Bano, 2019). In the present work, IAA production ranged from 0.005 μg/mL for Bacillus paramycoides LMR1180 to 34.7 μg/mL for Brevibacterium anseongense LMR1185 with 12 strains exceeding 10 μg/mL. This is in accordance with findings reported by Aliyat et al. (2020) where the production of IAA by strains isolated from a Moroccan phosphate sludge ranged from 1.37 to 43.8 μg/mL, with 5 strains out of 27 exceeding 10 μg/mL.
Alkaline soils are more subject to iron deficiency, hence affecting the growth of plants in these soils (Balparda et al., 2021). Iron solubility in soil is proportional to soil pH, the higher the pH is, the less soluble iron is (Mann et al., 2017; Singh et al., 2022). Siderophore producing bacteria alleviate iron deficiency in plants and minimize negative effects of saline soils such as chlorosis, as well as increase chlorophyll content (Ferreira et al., 2019; Silambarasan et al., 2020; Singh et al., 2022). In the present study, 29 strains were able to produce siderophores. Bacillus paramycoides LMR1180, Brevibacterium anseongense LMR1185, and Stenotrophomonas rhizophila LMR1211, produced the most siderophores in their respective phyla with 13.2, 24.7, and 10.8% SU, respectively. This is in agreement with research by El-Sersawy et al. (2021) and Liu et al. (2022) showing, respectively, that Bacillus paramycoides and Stenotrophomonas rhizophila can produce siderophores.
As phosphorus predominates in phosphate mine wastes, our bacteria do not generally display the need to solubilize additional phosphate for their cell machinery. As described by Michas et al. (2021), the phosphate solubilizing bacteria community changes in parallel with variations in soil composition, especially the carbon to phosphorus (C:P) ratio. These factors could explain the small number of bacterial isolates able to solubilize tricalcium phosphate in the present study.
The preliminary testing revealed that the majority of the isolated strains displayed good PGP activities. This is also the first time the recently described species Brevibacterium anseongense (Jung et al., 2018) and Agrococcus sediminis (Qu et al., 2020) were studied for their PGP activities.
To go further in this study, three phylogenetically diversified strains with excellent performance in in vitro analyses were subjected to metal resistance, biocontrol activities and a further pot experiment. The three selected strains, LMR1180, LMR1185, and LMR1211, showed high similarity to Bacillus paramycoides MCCC 1A04098, Brevibacterium anseongense Gsoil 188, and Stenotrophomonas rhizophila e-p10, respectively (Supplementary Table 2). The present works’ additional in planta test findings aided in determining how well the chosen strains can promote plant growth in stressed environments, contaminated with excess concentration of phosphate associated with nutrient-poor soils.
As phosphate mining wastes in Morocco may contain toxic metallic elements (Hakkou et al., 2009; Ouakibi et al., 2013), the resistance of the strains chosen for pot experiments were tested for some metals and metalloids. All these strains performed similarly in As(III), with a tolerance reaching 550 mg/L of NaAsO2. For Cd, Stenotrophomonas rhizophila LMR1211 was the most tolerant strain (until 170 mg/L of CdCl2), exceeding the concentration of Gao et al. (2020). Stenotrophomonas rhizophila LMR1211 showed also the most tolerance capacity for Fe, Hg, and Zn with concentrations reaching 800, 200, and 400 mg/L, respectively. This is in agreement with results of Ryan et al. (2009) describing Stenotrophomonas rhizophila as a potent multi-metal resistant strain.
Another mechanism that aids in plant establishment under stressed conditions, is the capacity of some bacteria to act as biocontrol agents, which could limit the negative effects of numerous pathogenic agents (Glick, 2012). Indeed, Mahmud et al. (2021) demonstrated the positive effect that biocontrol agents play in plant-growth promotion through their antagonistic relationship with pathogenic microorganisms in general, and fungi specifically. In the present study, Bacillus paramycoides LMR1180 performed the best in mycelial growth inhibition via diffusible compounds, and was able to inhibit the growth of Fusarium oxysporum and Botrytis cinerea by 7.7 and 14.4%, respectively. A similar study has shown that Bacillus paramycoides was able to limit F. oxysporum growth by more than 52% (El-Sersawy et al., 2021). As for the volatile compounds, Stenotrophomonas rhizophila LMR1211 inhibited Botrytis cinerea by 53.9%. Ryan et al. (2009) reported that Stenotrophomonas rhizophila inhibited also more than 9% of Rhizoctonia solani mycelium growth. Brevibacterium anseongense LMR1185 limited the growth of F. oxysporum by 12.7%. This is, to our knowledge, the first time Brevibacterium anseongense was tested for its biocontrol activity.
The in-pot experiments were performed with the white lupin (Lupinus albus L.), a Mediterranean annual nitrogen fixing legume (Huyghe, 1997). This plant, was shown to be hyperaccumulator in contaminated sites (González et al., 2021; Quiñones et al., 2021) making it a good candidate for bioremediation schemes and the establishment of a vegetative cover on top of phosphate wastes. In the current work, Stenotrophomonas rhizophila LMR1211 enhanced significantly the shoot and root dry weight as well as the root height. This is in line with the reported results by Ryan et al. (2009) suggesting that Stenotrophomonas rhizophila entertains a beneficial microorganism-plant interaction. Additionally, the inoculation with Brevibacterium anseongense LMR1185 showed a significant increase in shoot dry weight and height, and in root height. This is in accordance with the findings of González et al. (2021) showing that the inoculation of Lupinus albus L. with a species of Brevibacterium (Brevibacterium frigoritolerans) had a significant effect on shoots and roots parts parameters. Whilst neither the inoculation with Bacillus paramycoides LMR1180 nor with the consortium showed any improvement on Lupinus albus L. dry weight or height over the non-inoculated control. The use of a consortium, suggested by different previous study, was conducted in the hope to cumulate the PGP effects of each bacterium (Adams et al., 2014; Zhang et al., 2020). However, this assumption was not correct in the present study.
Biologists recognized the relevance of measuring the chlorophyll content from plant leaves. It has been shown that the content of leaves chlorophyll is correlated with the plant photosynthetic capacity (Buttery and Buzzell, 1977) thus helping plant growth and development (Rojas-Tapias et al., 2012). Stenotrophomonas rhizophila LMR1211 increased significantly the chlorophyll content in Lupinus albus leaves, which may explain the great development observed in this study, especially dry weight and length. The results observed in the consortium could be explained by the PGP activities antagonism. As a hypothesis, we wanted to check if the PGP activities of the three strains were kept when in consortium, and it has been shown that both Stenotrophomonas rhizophila LMR1211 and Brevibacterium anseongense LMR1185 inhibited the production of IAA by Bacillus paramycoides LMR1180, which may explain the poor development of the consortium root system, impacting the shoots development especially and the plant growth in general. While none of the two other activities (siderophore production or phosphate solubilization) were significantly affected in the consortium.
To help the establishment of a vegetative cover on phosphate mining wastes, this study explored the plant growth activity of indigenous bacteria. Forty-one bacterial strains were isolated and identified and the three best-performing strains (Bacillus paramycoides LMR1180, Brevibacterium anseongense LMR1185, and Stenotrophomonas rhizophila LMR1211) were chosen for further in vitro and in planta analyses. Stenotrophomonas rhizophila LMR1211 exhibited the best performance in promoting white lupin growth, a Mediterranean annual legume, followed by Brevibacterium anseongense LMR1185. These two strains should be put forward for further analyses to prove their positive impact on plant growth under stressed conditions and ultimately be used as a potential biofertilizer. To our knowledge, this is the first time Brevibacterium anseongense was reported to have good PGP activities and the ability to promote plant growth in pot experiments. The use of these strains as biofertilizers could represent an economical and eco-friendly method for the rehabilitation of phosphate mining waste in Morocco.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, MZ853464.2; MZ853485.2; MZ853448.2; MZ853453.2; MZ853480.2; MZ853474.2; MZ853458.2; MZ853482.1; MZ853455.2; MZ853469.2; MZ853459.2; MZ853445.2; MZ853444.2; MZ853449.2; MZ853468.2; MZ853483.2; MZ853471.2; MZ853465.2; MZ853466.2; MZ853478.2; MZ853456.2; MZ853462.2; MZ853470.2; MZ853475.2; MZ853439.2; MZ853442.2; MZ853476.2; MZ853479.1; MZ853450.2; MZ853481.2; MZ853451.2; MZ853447.2; MZ853486.2; MZ853452.2; MZ853457.2; MZ853484.2; MZ853443.1; MZ853463.1; MZ853477.1; MZ853440.1; MZ853438.1.
NM, OB, RH, and LS designed and contributed to the conception of the work. NM performed the sampling along with the experiments and wrote the first draft of the manuscript. RZ contributed to the experiments. NM and LS analyzed the data. OB contributed to the data interpretation. All authors reviewed and approved the final version of this manuscript.
The authors would like to thank Razana Zegrari for proofreading the final version of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1026991/full#supplementary-material
Adams, G. O., Tawari-Fufeyin, P., Igelenyah, E., and Odukoya, E. (2014). Assessment of heavy metals bioremediation potential of microbial consortia from poultry litter and spent oil contaminated site. Int. J. Environ. Bioremed. Biodegrad. 2, 84–92. doi: 10.12691/ijebb-2-2-6
Alexander, D. B., and Zuberer, D. A. (1991). Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 12, 39–45. doi: 10.1007/BF00369386
Aliyat, F. Z., Maldani, M., El Guilli, M., Nassiri, L., and Ibijbijen, J. (2020). Isolation and characterization of phosphate solubilizing bacteria from phosphate solid sludge of the moroccan phosphate mines. Open Agric. J. 14, 16–24.
Balparda, M., Armas, A. M., Gomez-Casati, D. F., and Pagani, M. A. (2021). PAP/SAL1 retrograde signaling pathway modulates iron deficiency response in alkaline soils. Plant Sci. 304:110808. doi: 10.1016/j.plantsci.2020.110808
Benbrik, B., Elabed, A., El Modafar, C., Douira, A., Amir, S., Filali-Maltouf, A., et al. (2020). Reusing phosphate sludge enriched by phosphate solubilizing bacteria as biofertilizer: Growth promotion of Zea mays. Biocatal. Agric. Biotechnol. 30:101825. doi: 10.1016/j.bcab.2020.101825
Bossé, B., Bussière, B., Hakkou, R., Maqsoud, A., and Benzaazoua, M. (2015). Field experimental cells to assess hydrogeological behavior of store-and-release covers made with phosphate mine waste. Can. Geotech. J. 52, 1255–1269. doi: 10.1139/cgj-2014-0263
Brzezinska, M. S., Kalwasińska, A., Świątczak, J., Żero, K., and Jankiewicz, U. (2020). Exploring the properties of chitinolytic Bacillus isolates for the pathogens biological control. Microb. Pathog. 148:104462. doi: 10.1016/j.micpath.2020.104462
Buttery, B. R., and Buzzell, R. I. (1977). The relationship between chlorophyll content and rate of photosynthesis in soybeans. Can. J. Plant Sci. 57, 1–5. doi: 10.4141/cjps77-001
Castellano-Hinojosa, A., and Bedmar, E. J. (2017). “Methods for evaluating plant growth promoting rhizobacteria traits,” in En: Advances in PGPR research, eds H. Bahadur Singh, B. Sarma, and C. Keswani (Wallingford: CABI Academic), 255–274. doi: 10.1079/9781786390325.0255
Duca, D., Lorv, J., Patten, C. L., Rose, D., and Glick, B. R. (2014). Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek 106, 85–125. doi: 10.1007/s10482-013-0095-y
Ebadzadsahrai, G., Higgins Keppler, E. A., Soby, S. D., and Bean, H. D. (2020). Inhibition of fungal growth and induction of a novel volatilome in response to Chromobacterium vaccinii volatile organic compounds. Front. Microbiol. 11:1035. doi: 10.3389/fmicb.2020.01035
El-Sersawy, M. M., Hassan, S. E. D., El-Ghamry, A. A., Abd El-Gwad, A. M., and Fouda, A. (2021). Implication of plant growth-promoting rhizobacteria of Bacillus spp. as biocontrol agents against wilt disease caused by Fusarium oxysporum Schlecht. in Vicia faba L. Biomol. Concepts 12, 197–214. doi: 10.1515/bmc-2021-0020
Esteban, E., Carpena, R. O., and Meharg, A. A. (2003). High-affinity phosphate/arsenate transport in white lupin (Lupinus albus) is relatively insensitive to phosphate status. N. Phytol. 158, 165–173.
Ferreira, C. M. H., Vilas, B. A., Sousa, C. A., Soares, H. M. V. M., and Soares, E. V. (2019). Comparison of five bacterial strains producing siderophores with ability to chelate iron under alkaline conditions. AMB Express 9, 1–12. doi: 10.1186/s13568-019-0796-3
Fibriarti, B. L., Feliatra, F., Amin, B., and Darwis, D. (2021). Biodegradation of LDPE plastic by local strain of Bacillus sp. Isolated from dump soil of Pekanbaru, Indonesia. Biodiversitas J. Biol. Diversity 22, 5484–5490. doi: 10.13057/biodiv/d221232
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Fusconi, R., and Godinho, M. J. L. (2002). Screening for exopolysaccharide-producing bacteria from sub-tropical polluted groundwater. Braz. J. Biol. 62, 363–369. doi: 10.1590/S1519-69842002000200020
Gao, J., Wu, S., Liu, Y., Wu, S., Jiang, C., Li, X., et al. (2020). Characterization and transcriptomic analysis of a highly Cr (VI)-resistant and-reductive plant-growth-promoting rhizobacterium Stenotrophomonas rhizophila DSM14405T. Environ. Pollut. 263:114622. doi: 10.1016/j.envpol.2020.114622
Garau, M., Castaldi, P., Diquattro, S., Pinna, M. V., Senette, C., Roggero, P. P., et al. (2021). Combining grass and legume species with compost for assisted phytostabilization of contaminated soils. Environ. Technol. Innov. 22:101387. doi: 10.1016/j.eti.2021.101387
Gawronski, S. W., Greger, M., and Gawronska, H. (2011). “Plant taxonomy and metal phytoremediation,” in Detoxification of heavy metals (Berlin: Springer), 91–109. doi: 10.1007/978-3-642-21408-0_5
Glick, B. R. (2012). Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012:963401. doi: 10.6064/2012/963401
González, D., Blanco, C., Probanza, A., Jiménez, P. A., and Robas, M. (2021). Evaluation of the PGPR capacity of four bacterial strains and their mixtures, tested on Lupinus albus var. Dorado seedlings, for the bioremediation of mercury-polluted soils. Processes 9:1293. doi: 10.3390/pr9081293
Gordon, S. A., and Weber, R. P. (1951). Colorimetric estimation of indoleacetic acid. Plant Physiol. 26:192. doi: 10.1104/pp.26.1.192
Gray, E. J., and Smith, D. L. (2005). Intracellular and extracellular PGPR: Commonalities and distinctions in the plant-bacterium signaling processes. Soil Biol. Biochem. 37, 395–412. doi: 10.1016/j.soilbio.2004.08.030
Hakkou, R., Benzaazoua, M., and Bussiere, B. (2009). Laboratory evaluation of the use of alkaline phosphate wastes for the control of acidic mine drainage. Mine Water Environ. 28, 206–218.
Hamdali, H., Bouizgarne, B., Hafidi, M., Lebrihi, A., Virolle, M. J., and Ouhdouch, Y. (2008). Screening for rock phosphate solubilizing actinomycetes from moroccan phosphate mines. Appl. Soil Ecol. 38, 12–19. doi: 10.1016/j.apsoil.2007.08.007
Hamdali, H., Moursalou, K., Tchangbedji, G., Ouhdouch, Y., and Hafidi, M. (2012). Isolation and characterization of rock phosphate solubilizing actinobacteria from a Togolese phosphate mine. Afr. J. Biotechnol. 11, 312–320. doi: 10.5897/AJB11.774
Hassani, M. A., Durán, P., and Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6, 1–17. doi: 10.1186/s40168-018-0445-0
Hernández-León, R., Rojas-Solís, D., Contreras-Pérez, M., del Carmen Orozco-Mosqueda, M., Macías-Rodríguez, L. I., and Reyes-de la Cruz, H. (2015). Characterization of the antifungal and plant growth-promoting effects of diffusible and volatile organic compounds produced by Pseudomonas fluorescens strains. Biol. Control 81, 83–92. doi: 10.1016/j.biocontrol.2014.11.011
Huyghe, C. (1997). White lupin (Lupinus albus L.). Field Crops Res. 53, 147–160. doi: 10.1016/S0378-4290(97)00028-2
Jain, A., Singh, S., Kumar Sarma, B., and Bahadur Singh, H. (2012). Microbial consortium–mediated reprogramming of defence network in pea to enhance tolerance against Sclerotinia sclerotiorum. J. Appl. Microbiol. 112, 537–550. doi: 10.1111/j.1365-2672.2011.05220.x
Jalali, J., Gaudin, P., Capiaux, H., Ammar, E., and Lebeau, T. (2020). Isolation and screening of indigenous bacteria from phosphogypsum-contaminated soils for their potential in promoting plant growth and trace elements mobilization. J. Environ. Manag. 260:110063. doi: 10.1016/j.jenvman.2020.110063
Jung, M. S., Quan, X. T., Siddiqi, M. Z., Liu, Q., Kim, S. Y., Wee, J. H., et al. (2018). Brevibacterium anseongense sp. nov., isolated from soil of ginseng field. J. Microbiol. 56, 706–712. doi: 10.1007/s12275-018-8181-5
Kadmiri, I. M., Amahdar, L., Sarkodi, A., Amghar, S., and Hilali, A. (2010). Isolation and identification of bacterial strains from “EL HALASSA” phosphate deposit (Morocco). Vice Editor in Chief/Vice Rédacteur en chef 12:30.
Kasana, R. C., Salwan, R., Dhar, H., Dutt, S., and Gulati, A. (2008). A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr. Microbiol. 57, 503–507. doi: 10.1007/s00284-008-9276-8
Khairina, Y., Jog, R., Boonmak, C., Toyama, T., Oyama, T., and Morikawa, M. (2021). Indigenous bacteria, an excellent reservoir of functional plant growth promoters for enhancing duckweed biomass yield on site. Chemosphere 268:129247. doi: 10.1016/j.chemosphere.2020.129247
Köchl, S., Niederstätter, H., and Parson, W. (2005). DNA extraction and quantitation of forensic samples using the phenol-chloroform method and real-time PCR. Forensic DNA Typing Protoc. 297, 13–30. doi: 10.1385/1-59259-867-6:013
Kuhad, R. C., Gupta, R., and Singh, A. (2011). Microbial cellulases and their industrial applications. Enzyme Res. 2011:280696. doi: 10.4061/2011/280696
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35:1547. doi: 10.1093/molbev/msy096
Lichtenthaler, H. K., and Wellburn, A. R. (1983). Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 603, 591–592. doi: 10.1042/bst0110591
Liu, Y., Gao, J., Wang, N., Li, X., Fang, N., and Zhuang, X. (2022). Diffusible signal factor enhances the saline-alkaline resistance and rhizosphere colonization of Stenotrophomonas rhizophila by coordinating optimal metabolism. Sci. Total Environ. 834:155403. doi: 10.1016/j.scitotenv.2022.155403
Luo, Y., Wu, Y., Wang, H., Xing, R., Zheng, Z., Qiu, J., et al. (2018). Bacterial community structure and diversity responses to the direct revegetation of an artisanal zinc smelting slag after 5 years. Environ. Sci. Pollut. Res. 25, 14773–14788. doi: 10.1007/s11356-018-1573-6
Ma, Y., Rajkumar, M., Zhang, C., and Freitas, H. (2016). Beneficial role of bacterial endophytes in heavy metal phytoremediation. J. Environ. Manag. 174, 14–25. doi: 10.1016/j.jenvman.2016.02.047
Mahmud, A. A., Upadhyay, S. K., Srivastava, A. K., and Bhojiya, A. A. (2021). Biofertilizers: A Nexus between soil fertility and crop productivity under abiotic stress. Curr. Res. Environ. Sustain. 3:100063. doi: 10.1016/j.crsust.2021.100063
Mann, A., Singh, A. L., Oza, S., Goswami, N., Mehta, D., and Chaudhari, V. (2017). Effect of iron source on iron deficiency induced chlorosis in groundnut. Legume Res. 40, 241–249. doi: 10.18805/lr.v0iOF.6849
Mendez, M. O., and Maier, R. M. (2008). Phytoremediation of mine tailings in temperate and arid environments. Rev. Environ. Sci. Bio Technol. 7, 47–59. doi: 10.1007/s11157-007-9125-4
Mghazli, N., Sbabou, L., Hakkou, R., Ouhammou, A., Adnani, M. E., and Bruneel, O. (2021). Description of microbial communities of phosphate mine wastes in Morocco, a semi-arid climate, using high-throughput sequencing and functional prediction. Front. Microbiol. 12:1767. doi: 10.3389/fmicb.2021.666936
Michas, A., Pastore, G., Chiba, A., Grafe, M., Clausing, S., Polle, A., et al. (2021). Phosphorus availability alters the effect of tree girdling on the diversity of phosphorus solubilizing soil bacterial communities in temperate beech forests. Front. Forests Glob. Change 4:79. doi: 10.3389/ffgc.2021.696983
Mobaligh, M., Meddich, A., Imziln, B., and Fares, K. (2021). The use of phosphate washing sludge to recover by composting the leachate from the controlled landfill. Processes 9:1735. doi: 10.3390/pr9101735
Moghannem, S. A., Farag, M., Shehab, A. M., and Azab, M. S. (2018). Exopolysaccharide production from Bacillus velezensis KY471306 using statistical experimental design. Braz. J. Microbiol. 49, 452–462. doi: 10.1016/j.bjm.2017.05.012
Nandi, M., Selin, C., Brawerman, G., Fernando, W. D., and de Kievit, T. (2017). Hydrogen cyanide, which contributes to Pseudomonas chlororaphis strain PA23 biocontrol, is upregulated in the presence of glycine. Biol. Control 108, 47–54. doi: 10.1016/j.biocontrol.2017.02.008
Nannipieri, P., Ascher, J., Ceccherini, M., Landi, L., Pietramellara, G., and Renella, G. (2003). Microbial diversity and soil functions. Eur. J. Soil Sci. 54, 655–670. doi: 10.1046/j.1351-0754.2003.0556.x
Naseem, H., and Bano, A. (2014). Role of plant growth-promoting rhizobacteria and their exopolysaccharide in drought tolerance of maize. J. Plant Interact. 9, 689–701. doi: 10.1080/17429145.2014.902125
Ouakibi, O., Loqman, S., Hakkou, R., and Benzaazoua, M. (2013). The potential use of phosphatic limestone wastes in the passive treatment of AMD: A laboratory study. Mine Water Environ. 32, 266–277. doi: 10.1007/s10230-013-0226-8
Pattnaik, S., Mohapatra, B., Kumar, U., Pattnaik, M., and Samantaray, D. (2019). “Microbe-mediated plant growth promotion: A mechanistic overview on cultivable plant growth-promoting members,” in Biofertilizers for sustainable agriculture and environment, Vol. 55, eds B. Giri, R. Prasad, Q. S. Wu, and A. Varma (Cham: Springer), 435–463. doi: 10.1007/978-3-030-18933-4_20
Pikovskaya, R. I. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17, 362–370.
Premono, M. E., Moawad, A. M., and Vleck, P. L. G. (1996). Effect of phosphate solubilizing Pseudomonas putida on the growth of maize and its survival in the rhizosphere. Indones. J. Crop Sci. 11, 13–23.
Qarni, A., Billah, M., Hussain, K., Shah, S. H., Ahmed, W., Alam, S., et al. (2021). Isolation and characterization of phosphate solubilizing microbes from rock phosphate mines and their potential effect for sustainable agriculture. Sustainability 13:2151. doi: 10.3390/su13042151
Qu, J. H., Yue, Y. F., Tian, H. L., Zhou, J., Yang, B. B., Deng, L., et al. (2020). Agrococcus sediminis sp. nov., an actinobacterium isolated from lake sediment. Int. J. Syst. Evol. Microbiol. 70, 4986–4992. doi: 10.1099/ijsem.0.004371
Quiñones, M. A., Fajardo, S., Fernández-Pascual, M., Lucas, M. M., and Pueyo, J. J. (2021). Nodulated white lupin plants growing in contaminated soils accumulate unusually high mercury concentrations in their nodules, roots and especially cluster roots. Horticulturae 7:302. doi: 10.3390/horticulturae7090302
Rijavec, T., and Lapanje, A. (2016). Hydrogen cyanide in the rhizosphere: Not suppressing plant pathogens, but rather regulating availability of phosphate. Front. Microbiol. 7:1785. doi: 10.3389/fmicb.2016.01785
Rohwer, R. R., Hamilton, J. J., Newton, R. J., and McMahon, K. D. (2018). TaxAss: Leveraging a custom freshwater database achieves fine-scale taxonomic resolution. Msphere 3, e327–e318. doi: 10.1128/mSphere.00327-18
Rojas-Tapias, D., Moreno-Galván, A., Pardo-Díaz, S., Obando, M., Rivera, D., and Bonilla, R. (2012). Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Appl. Soil Ecol. 61, 264–272. doi: 10.1016/j.apsoil.2012.01.006
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. doi: 10.1038/nrmicro2163
Sandhya, V., and Ali, S. Z. (2015). The production of exopolysaccharide by Pseudomonas putida GAP-P45 under various abiotic stress conditions and its role in soil aggregation. Microbiology 84, 512–519. doi: 10.1134/S0026261715040153
Sharma, A., Kumar, A., and Dhaka, T. S. (2012). Impact of sugar factory effluent on chlorophyll and protein contents of Cicer arietinum and Tigonella foenum-gracecum. Curr. Adv. Agric. Sci. 4, 62–63.
Sharma, I., and Ahmad, P. (2014). “Catalase: A versatile antioxidant in plants,” in Oxidative damage to plants (Cambridge, MA: Academic Press), 131–148. doi: 10.1016/B978-0-12-799963-0.00004-6
Silambarasan, S., Logeswari, P., Valentine, A., Cornejo, P., and Kannan, V. R. (2020). Pseudomonas citronellolis strain SLP6 enhances the phytoremediation efficiency of Helianthus annuus in copper contaminated soils under salinity stress. Plant Soil 457, 241–253. doi: 10.1007/s11104-020-04734-7
Singh, P., Chauhan, P. K., Upadhyay, S. K., Singh, R. K., Dwivedi, P., Wang, J., et al. (2022). Mechanistic insights and potential use of siderophores producing microbes in rhizosphere for mitigation of stress in plants grown in degraded land. Front. Microbiol. 13:898979. doi: 10.3389/fmicb.2022.898979
Singh, P., and Kumar, S. (2019). “Microbial enzyme in food biotechnology,” in Enzymes in food biotechnology (Cambridge, MA: Academic Press), 19–28. doi: 10.1016/B978-0-12-813280-7.00002-5
Sharma, P., Ngo, H. H., Khanal, S., Larroche, C., Kim, S. H., and Pandey, A. (2021). Efficiency of transporter genes and proteins in hyperaccumulator plants for metals tolerance in wastewater treatment: Sustainable technique for metal detoxification. Environ. Technol. Innov. 23:101725. doi: 10.1016/j.eti.2021.101725
Small, C. C., and Degenhardt, D. (2018). Plant growth regulators for enhancing revegetation success in reclamation: A review. Ecol. Eng. 118, 43–51. doi: 10.1016/j.ecoleng.2018.04.010
Stracquadanio, C., Quiles, J. M., Meca, G., and Cacciola, S. O. (2020). Antifungal activity of bioactive metabolites produced by Trichoderma asperellum and Trichoderma atroviride in liquid medium. J. Fungi 6:263. doi: 10.3390/jof6040263
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. doi: 10.1093/oxfordjournals.molbev.a040023
Trifi, H., Najjari, A., Achouak, W., Barakat, M., Ghedira, K., Mrad, F., et al. (2020). Metataxonomics of Tunisian phosphogypsum based on five bioinformatics pipelines: Insights for bioremediation. Genomics 112, 981–989. doi: 10.1016/j.ygeno.2019.06.014
Ullah, A., and Bano, A. (2019). Role of PGPR in the reclamation and revegetation of saline land. Pak. J. Bot. 51, 27–35. doi: 10.30848/PJB2019-1(43)
Upadhyay, S. K., and Chauhan, P. K. (2022). Optimization of eco-friendly amendments as sustainable asset for salt-tolerant plant growth-promoting bacteria mediated maize (Zea Mays L.) plant growth, Na uptake reduction and saline soil restoration. Environ. Res. 211:113081. doi: 10.1016/j.envres.2022.113081
USGS (2020). U.S. geological survey, mineral resources program (MRP). Available online at: https://www.statista.com/statistics/681617/phosphate-rock-production-by-country/ (accessed July 9, 2021).
Walia, A., Mehta, P., Chauhan, A., and Shirkot, C. K. (2014). Effect of Bacillus subtilis strain CKT1 as inoculum on growth of tomato seedlings under net house conditions. Proc. Natl. Acad. Sci. India Section B Biol. Sci. 84, 145–155. doi: 10.1007/s40011-013-0189-3
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Wu, Z., Kong, Z., Lu, S., Huang, C., Huang, S., He, Y., et al. (2019). Isolation, characterization and the effect of indigenous heavy metal-resistant plant growth-promoting bacteria on sorghum grown in acid mine drainage polluted soils. J. Gen. Appl. Microbiol. 65, 2018–2011. doi: 10.2323/jgam.2018.11.004
Yang, P. X., Ma, L., Chen, M. H., Wi, J. Q., He, F., Duan, C. Q., et al. (2012). Phosphate solubilizing ability and phylogenetic diversity of bacteria from P-rich soils around Dianchi Lake drainage area of China. Pedosphere 22, 707–716. doi: 10.1016/S1002-0160(12)60056-3
Zhang, C., Wu, D., and Ren, H. (2020). Bioremediation of oil contaminated soil using agricultural wastes via microbial consortium. Sci. Rep. 10, 1–8. doi: 10.1038/s41598-020-66169-5
Zine, H., Hakkou, R., El Adnani, M., Lamnai Kamal, K., Elgadi, S., Rachid Ait, B., et al. (2020). Phytostabilization of store-and-release cover made with phosphate mine wastes in arid and semiarid climate using wild local plants. Remediation 31, 105–122. doi: 10.1002/rem.21662
Keywords: phosphate waste rocks, semi-arid climate, PGPB, revegetation, bioremediation
Citation: Mghazli N, Bruneel O, Zouagui R, Hakkou R and Sbabou L (2022) Characterization of plant growth promoting activities of indigenous bacteria of phosphate mine wastes, a first step toward revegetation. Front. Microbiol. 13:1026991. doi: 10.3389/fmicb.2022.1026991
Received: 24 August 2022; Accepted: 28 November 2022;
Published: 15 December 2022.
Edited by:
Eloisa Pajuelo, University of Seville, SpainReviewed by:
Sudhir K. Upadhyay, Veer Bahadur Singh Purvanchal University, IndiaCopyright © 2022 Mghazli, Bruneel, Zouagui, Hakkou and Sbabou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laila Sbabou, bC5zYmFib3VAdW01ci5hYy5tYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.