
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 07 November 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1026887
This article is part of the Research Topic Innovative Antiviral Strategy against Immune Evasion View all 8 articles
Viruses are strictly intracellular parasites requiring host cellular functions to complete their reproduction cycle involving virus infection of host cell, viral genome replication, viral protein translation, and virion release. Ribosomes are protein synthesis factories in cells, and viruses need to manipulate ribosomes to complete their protein synthesis. Viruses use translation initiation factors through their own RNA structures or cap structures, thereby inducing ribosomes to synthesize viral proteins. Viruses also affect ribosome production and the assembly of mature ribosomes, and regulate the recognition of mRNA by ribosomes, thereby promoting viral protein synthesis and inhibiting the synthesis of host antiviral immune proteins. Here, we review the remarkable mechanisms used by RNA viruses to regulate ribosomes, in particular, the mechanisms by which RNA viruses induce the formation of specific heterogeneous ribosomes required for viral protein translation. This review provides valuable insights into the control of viral infection and diseases from the perspective of viral protein synthesis.
Viruses are intracellular parasitic organisms with infectious potential and are divided into two categories, namely DNA viruses and RNA viruses, according to their genomic characteristics. In recent years, RNA viruses have attracted the increasing attention of researchers because of their high adaptability, high variability, and strong pathogenicity (Colson et al., 2022; Liu et al., 2022; Tomar et al., 2022). Depending on their genomic characteristics, RNA viruses can be classified as double-stranded RNA viruses, positive-stranded RNA viruses and negative-stranded RNA viruses. Of course, the life cycle of different RNA viruses in the cell also differs. However, viruses invade the host cell by binding to receptors on the cell membrane, releasing the viral genome into the cell, subsequently initiating the translation of viral proteins, and performing genome replication, viral assembly, and viral budding and release, thereby completing the viral replication cycle. Viruses complete each step of their life cycle through an intricate interaction with the host cells. Because the viral genome is small and does not have a translation system for coded proteins, all viruses use the host protein synthetic machinery to translate viral mRNAs. To form mature ribosomes and initiate the translation of viral proteins, viruses recruit the host’s protein translation initiation factors through genomic RNA 5′-UTR-specific structures or covalently bound viral proteins to generate translation initiation complexes that subsequently bind to both the small and large subunits of the ribosomes (Lopez-Lastra et al., 2010). With the evolution of viruses, different viral species have adopted different mechanisms for recruiting translation initiation factors such as the classical cap-dependent and cap-independent types (Internal Ribosome Entry Site [IRES], etc.) (Vallejos et al., 2010; Filbin et al., 2013; Olivares et al., 2014). In addition, during the life cycle which are as follows. Viruses usurp the host translation machinery, encode multifunctional proteins to regulate gene expression, use overlapping open reading frames to store viral genetic information, or use a single messenger RNA to synthesize multiple proteins, all of these processes require a series of sophisticated strategies to evade host immune surveillance (Meyers, 2003; Gaucherand et al., 2019; Liu et al., 2019). Presently, the mechanism of viral protein translation initiation is well understood. However, the mechanism by which viral mRNA forms the translation initiation complex and then recruits ribosomal subunits to form mature ribosomes required for viral protein translation is unclear.
The ribosome is the host protein synthesis factory, and it is formed by the assembly of rRNA, the 40S ribosomal subunit, and the 60S ribosomal subunit. There are more than 80 known ribosomal proteins (RPs), the 40S subunit comprises about 30 proteins, while the 60S subunit contains 50 proteins, but it has been found that not all RPs are involved in the formation of mature ribosomes. However, not all RPs are involved in the formation of mature ribosomes (Hamidi et al., 2018). Therefore, the size and composition of ribosomes in different microenvironments may vary correspondingly. Around the 1980s, it was found that different mRNAs are translated by specific ribosomes (Hui and de Boer, 1987; Ferretti et al., 2017). Ribosomal heterogeneity refers to changes in the stoichiometry of rRNAs and RPs during ribosome generation as well as post-transcriptional rRNA and post-translational RP modifications that result in the incorrect expression of some genes (Genuth and Barna, 2018; Cheng et al., 2019; Emmott et al., 2019). Defects or accumulation of substances required for body metabolism can lead to ribosomopathies such as Diamond–Blackfan anemia (DBA), some cancers, some neurodevelopmental disorders such as autism spectrum disorders and microcephaly are associated with heterogeneous ribosomes or mutated RPs (Segev and Gerst, 2018). Recent evidence suggests that viral infection induces heterogeneous ribosome production, leading to changes in ribosome conformation and composition (DiGiuseppe et al., 2020).
In this review, we focus on the regulation of the host ribosome following viral invasion of the host cell. Including how viruses shut down ribosomal translation of the host genome and how viruses use the host ribosome to preferentially translate the viral genome.
Viral infection is a dynamic process in which viruses and organisms compete with each other. Depending on the mechanism of infection, viruses can be classified as those that cause cell-killing, stable, and integrated infections (Maree et al., 2016). Compared to viruses that cause stable and integrated infection of the host, cell-killing viruses encode early viral proteins that capture the synthesis machinery of the host cell for their use to inhibit or even shut down the synthesis of host RNA and proteins (Curran and Kolakofsky, 1989; Vidal et al., 1990; Boeck et al., 1992; Chenik et al., 1995). Through this approach, viruses can disrupt normal cellular metabolism and ultimately lead to cell death. Hence, this section focuses on two aspects of RNA viruses that induce ribosomes to shut down the translation of host proteins: inhibition of host mRNA recognition and prevention of ribosome subunit assembly.
During viral infection, the virus regulates the expression of host proteins through the control of ribosomes. The main pathways include remodeling the host mRNA pool and inhibiting the recognition of host mRNA by ribosomes. Currently, some viruses can remodel the host mRNA library by encoding viral proteins that interfere with host pre-mRNA synthesis, induce host mRNA degradation, or block host mRNA nuclear export pathway. Among these approaches, interference with host mRNA synthesis is mainly achieved by strategies such as inhibition of pre-mRNA splicing by the incorporation of poly(A) or direct inhibition of pre-mRNA polyadenylation. For example, the Vpr protein of human immunodeficiency virus (HIV) was identified to inhibit pre-mRNA splicing, and the NS1 protein of avian influenza virus (AIV) inhibits pre-mRNA splicing and polyadenylation (Nemeroff et al., 1998; Hashizume et al., 2007; Nacken et al., 2021). Moreover, the NSP1 protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the polymerase of feline calicivirus (FCV), and the PA-X protein of AIV were confirmed to degrade host mRNA (DuBois et al., 2012; Khaperskyy et al., 2016; Finkel et al., 2021; Wu et al., 2021). The inhibition of nuclear export of host mRNA mainly occurs by encoding viral proteins that interfere with the normal biological role of nuclear pore proteins, thereby inhibiting the recognition of host mRNA by the ribosome and facilitating the expression and inheritance of viral proteins such as the L protein of Theiler’s murine encephalomyelitis virus (TMEV), 2A protein of poliovirus (PV), M protein of vesicular stomatitis virus (VSV), and 3C protein of human rhinovirus (HRV) (von Kobbe et al., 2000; Castello et al., 2009; Walker et al., 2013; Masaki et al., 2019).
In recent years, some viruses were found to prevent ribosomes from recognizing host mRNA, resulting in the inhibition of normal cellular physiological functions and providing a more supportive environment for virus proliferation. In studies of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which causes the worldwide coronavirus disease 2019 (COVID-19) pandemic affecting millions of people, it was found that the viral non-structural protein 1 (NSP1) encoding the virus early in infection binds with high affinity to the ribosomal mRNA entry channel, preventing recognition of host and viral mRNA by the ribosome (Tidu et al., 2020). The interaction between NSP1 and RPs RPS3, RPS2 and rRNA h18 relies mainly on electrostatic and hydrophobic forces (Yuan et al., 2020). The viral genome hijacking ribosome mainly uses 5′ end stem-loop structure to interact with NSP1, regulating the binding between NSP1 and 40S and exposing the mRNA entry channel to complete the translation of the viral genome (Tidu et al., 2020). Such fine-grained regulatory roles between the host ribosome and SARS-CoV-2 should leave much to be investigated. Another example is about the level of host protein expression was significantly suppressed in the late stages of HIV-1 infection due to the high expression of host RNA decapping enzyme (DDX3) (Labaronne et al., 2022). DDX3 has been found to be hijacked by a variety of RNA viruses, facilitating viral transcription, nuclear export, translation and assembly of viral particles, while threatening the innate immune response (Valiente-Echeverria et al., 2015). It follows that different strategies can be used to inhibit translation of host proteins by the ribosome, regardless of the type of viral infection.
Ribosome biogenesis is primarily in the nucleolus, where rRNA precursors (pre-rRNA) are processed into mature molecules with mature RPs that assemble into 40S ribosomal subunits and 60S ribosomal subunits for release into the cytoplasm. The 40S ribosomal subunit forms a 48S pre-initiation complex with the translation initiation complex and mRNA, which subsequently recruits the 60S ribosomal subunit to eventually form the translationally functional 80S ribosome. Viruses cannot carry their protein synthesis machinery and need to manipulate ribosomes to complete their protein translation. Hence, the viral genome competitively utilizes ribosomes with host mRNA. Viruses have evolved mechanisms to influence the assembly and maturation of ribosomes.
Some viral proteins interact directly with the host 40S or 60S, thereby preventing the formation of the 43S initiation complex and the assembly of the 60S and 40S subunits. For example, SARS-CoV NSP1 protein interacts with the 40S ribosomal subunit to inhibit the formation of the 80S ribosome (Kamitani et al., 2009; Lokugamage et al., 2012). Notably, the effects of NSP1 encoded by different coronaviruses on the ribosome are different. For example, the NSP1 protein encoded by Middle East respiratory syndrome coronavirus (MERS-CoV) binds with different stability and intracellular localization to the 40S ribosomal subunit as compared to the NSP1 protein encoded by SARS-CoV-2 (Lokugamage et al., 2015). This suggests that the degree of selective inhibition of host protein translation through the strategy of blocking ribosomal mRNA entry into the channel by the binding of NSP1 to the 40S subunit varies with different coronaviruses. Discovery of similar mechanisms is relatively rare, mainly due to the fact that translation of the viral genome requires an intact ribosome to synthesize viral proteins.
The modification and splicing of eukaryotic translation initiation factors required for ribosome assembly by viruses lead to ribosome-mediated translation barriers. Most RNA viruses inhibit host protein translation by preventing the binding of ribosomal subunits through strategies such as promoting phosphorylation or dephosphorylation modification of translation initiation factors, cleaving eukaryotic translation initiation factors, or cleaving poly(A)-PABP (Lamphear et al., 1995; Gingras et al., 1996; Gradi et al., 1998; Kerekatte et al., 1999; Kuyumcu-Martinez et al., 2004; Willcocks et al., 2004; de Breyne et al., 2008; Rojas et al., 2010). The coat proteins encoded by neuro necrosis virus (NNV) manipulate PABP from the cytoplasm of the host cell into the nucleus to degrade it by ubiquitination (Cheng et al., 2021). Thus, viruses inhibit the expression of the host protein but do not affect the expression of the viral protein.
In contrast, viruses can use the diversity of genomes and the expression of viral proteins to promote the assembly of ribosomal subunits and facilitate the formation of heterogeneous ribosomes to selectively translate the viral genome (Lago et al., 2017). The ribosomal protein RPL13 interacts with the IRES element of the Seneca Valley virus (SVV) and classical swine fever virus (CSFV) to manipulate ribosomes to translate viral proteins (Han et al., 2020). At present, it has been confirmed that the IRES structure of Foot and Mouth Disease Virus (FMDV) utilizes the interaction between RPL13 and helicase DDX3 to promote the assembly of 80S ribosomes and then translate viral protein (Han et al., 2020). This approach was also found in other picornaviruses such as coxsackievirus (CBV). However, DDX3 cannot contribute to the translation of other RNA viruses that do not contain IRES elements, such as VSV and measles virus (MV) (Quaranta et al., 2020). Together, these findings demonstrate the IRES element of the viral genome could manipulate the host’s antiviral factor to inhibit the host natural immune response and thereby facilitate viral protein translation.
In summary, RNA viruses manipulate the functional ribosome to translate the viral mRNA. However, the biosynthesis and assembly mechanism of ribosomes are very complex and elusive. As shown in Figures 1, 2, RNA viruses use different strategies to control the ribosome, such as inhibiting of ribosome recognizing host genome and preventing the assembly of large and small ribosomal subunits. The mechanisms by which RNA viruses affect ribosome assembly remain unclear. For example, it is unknown why viruses affect the expression of some host proteins but not all. Some studies have shown that the ribosomal components are not fully involved in the assembly of ribosomes, and the ribosome structures in different cell regions are significantly different (Shi and Barna, 2015; Luan et al., 2022). These findings indicate that the ribosome is composed of a core region and various genetically adapted RPs (Ngoveni et al., 2019; Patino-Galindo et al., 2021). Therefore, the effect of heterogeneous ribosomes on viral protein translation deserves further study.
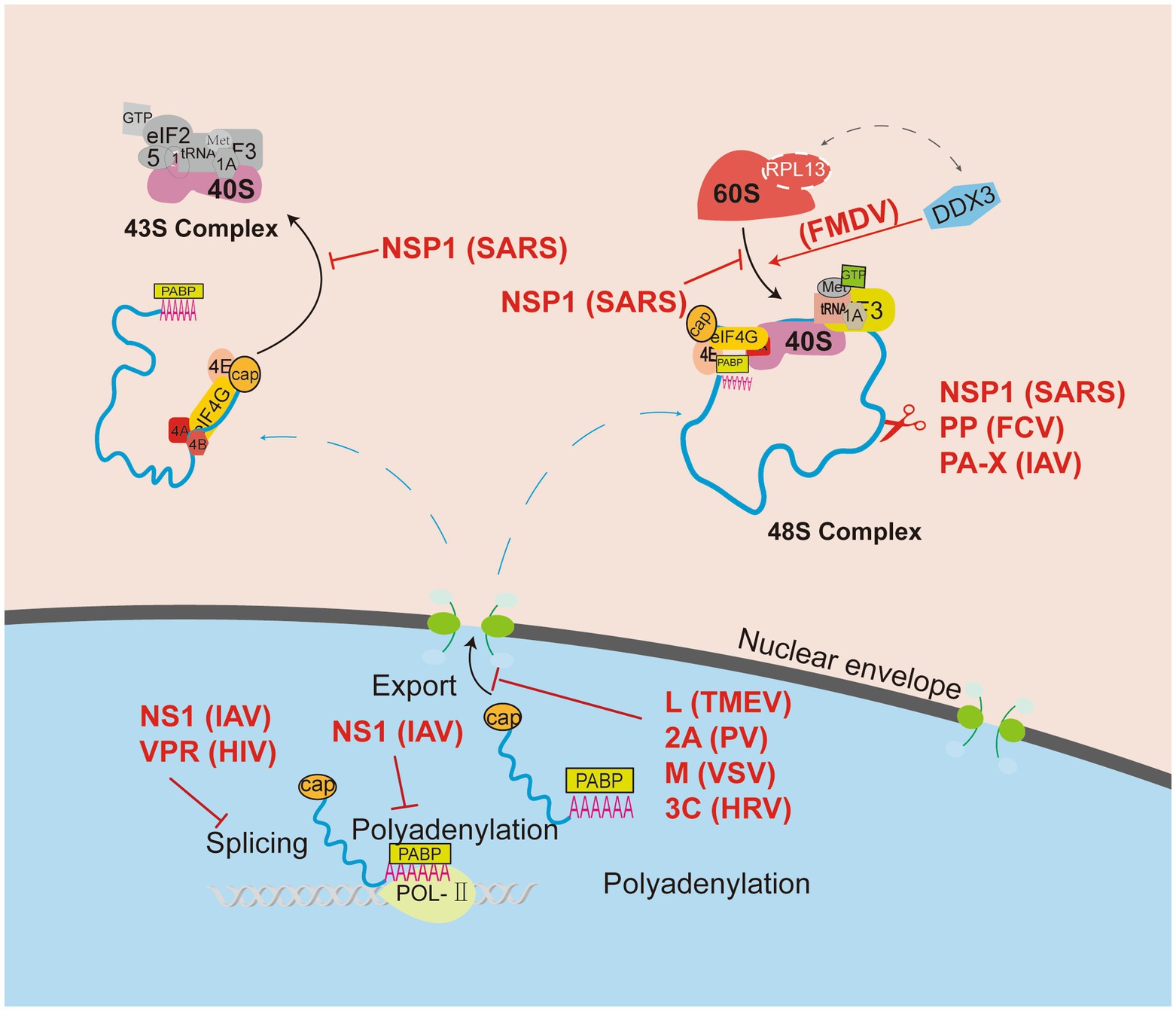
Figure 1. Viruses manipulate ribosomes to shut down host protein translation. Cellular mRNA biogenesis begins in the nucleus, and viruses use various mechanisms to block the translation of host mRNA, including remodeling of the host mRNA library, inhibiting host mRNA transport from the nucleus, and blocking ribosomal recruitment of eukaryotic translation initiation factor (eIF). Consequently, viruses can block ribosomal mRNA entry channels or inhibit the binding of ribosomal size subunits to prevent the translation of host proteins.
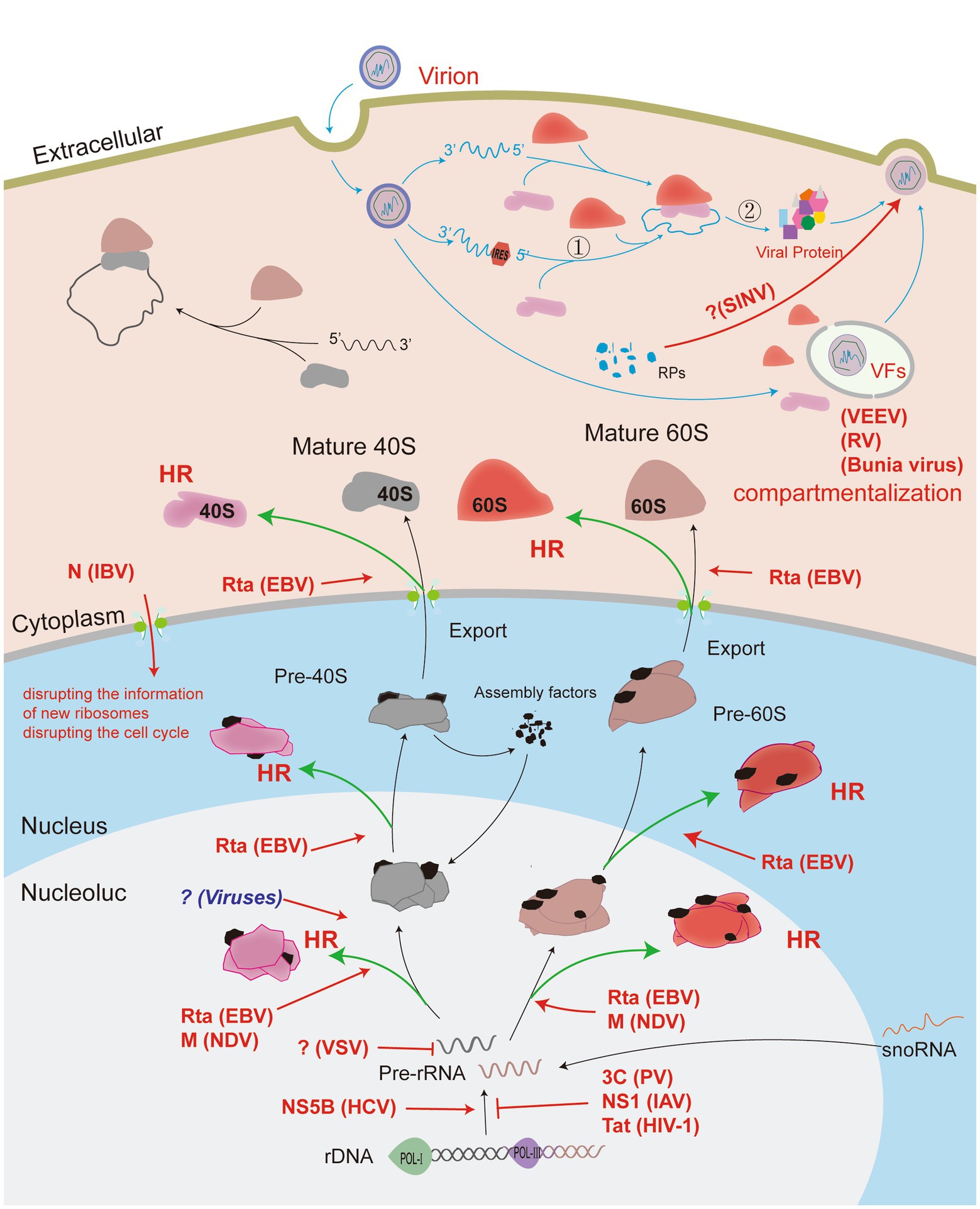
Figure 2. Viruses manipulate ribosomes to complete the translation of viral proteins. Viruses invading host cells can manipulate ribosomes to initiate the translation of viral proteins by modulating their own genome structure. The viral genome can also regulate the distribution of host ribosomal compartments or manipulate host ribosomal components to assemble viral particles that facilitate viral protein translation. Ribosome biogenesis begins in the nucleolus, and RNA virus infection causes nucleolus stress, with viral proteins acting on the different functional regions of the nucleolus to induce heterogeneous ribosome biosynthesis (HR). The effect of the virus on the host (red arrow); normal host physiological processes (black arrow); the process of virus proliferation in the cell (blue arrow); the virus-mediated production of heterogeneous ribosomes (green arrow). Process ① represents the recruitment of ribosomes by viruses using a cap-independent structure. Process ② indicates the translation process wherein viruses can use the specific structure of their own mRNA to reinitiate downstream viral proteins.
Viral infection induces a comprehensive stress response in cells that stimulates the phosphorylation of eIF2α and inhibits viral proliferation (Huang et al., 2017). To defend against host antiviral responses, some viruses have evolved cap-dependent translation initiation mechanisms. For example, some viruses adopt a strategy that is independent of the translation initiation factor eIF2 during the translation process (Garaigorta and Chisari, 2009; Roth et al., 2017). Regardless of this mechanism, the ribosome is also the key machine for viral protein translation. Here, we review strategies for the binding of viral RNA to ribosomes to initiate protein translation and the effects of viral infection on the assembly and conformation of the ribosomal size subunit (as shown in Figure 2).
Mutation or recombination of the viral genome can increase its diversity, thereby allowing viruses to easily adapt to a new host and to mediate immune escape and exhibit resistance to gene therapy (Pilipenko et al., 1992; Miras et al., 2014; Xiao et al., 2016; Ngoveni et al., 2019; Patino-Galindo et al., 2021). This evolutionary pattern makes the viral genome more prone to prey on ribosomes. Viruses have evolved various strategies to initiate viral protein translation, such as cap-dependent, cap-independent, and IRES-dependent approaches.
Discovered in 1988 by Jang and Pelletier et al., IRES is a secondary structure of the untranslated region (UTR) at the 5′ end of PV mRNA that binds directly to 40S and remodels the ribosome structure to form a heterogeneous ribosome that preferentially translates the viral genome (Jang et al., 1988; Pelletier and Sonenberg, 1988; Jobe et al., 2019). Notably, it has been demonstrated that the HCV IRES structure can directly hijack the translating 80S ribosome, but the subsequent mechanism of action needs further investigation (Yokoyama et al., 2019). Currently, IRES structures have been identified in picornaviruses, flaviviruses, retroviruses, and some cellular mRNAs, but IRES elements in these RNA viruses lack conserved features (Martinez-Salas et al., 2017). According to the secondary structure of IRES, it is mainly divided into five types (I–V). Each type of IRES requires different translation initiation factors to initiate translation, and the pattern of binding of each type of IRES to ribosomes is also different (as shown in Table 1; Martinez-Salas et al., 2017).
IGR IRES of double-stranded viruses represent the simplest class of IRES (Roberts and Groppelli, 2009; Kerr et al., 2015; Gross et al., 2017). Each ORF encoded by the virus has unique IRES structure (Roberts and Groppelli, 2009; Kerr et al., 2015; Gross et al., 2017). However, many other IRES structures are challenging to categorize because of their diversity, such as IRES structures in SARS-CoV-2, alphavirus, infectious pancreatic necrosis virus (IPNV), drosophila C virus (DCV), dengue fever virus (DENV), and calicivirus (Kamrud et al., 2007; Landry et al., 2009; Rivas-Aravena et al., 2017; Fernandez-Garcia et al., 2020; Arhab et al., 2022; Slobodin et al., 2022). Notably, viral IRES elements are expressed in heterologous viral genomes in hosts with mixed infectious viruses (Arhab et al., 2020). In the case of avian calicivirus and picornavirus co-infection, the 5’-UTR of avian calicivirus was found to contain a conformation consistent with the 5’-UTR secondary structure of picornavirus (IRES type V) (Arhab et al., 2022). These discoveries of IRES structure may be due to genetic recombination with other viruses and reflects the importance of analyzing the interaction of the ribosome with the viral RNA genome to resolve the mechanism of viral protein synthesis.
Eukaryotic mRNAs are usually monocistronic, while some viral mRNAs are polycistronic. Thus, in order to survive and reproduce in the host cell, viruses have a superior means of solving the problems of time, space and quality of genetic information expression than cells. The strategies include leaky scanning, translation termination re-initiation, ribosomal shunting, ribosome hopping, and frameshifting. As summarized in Table 2, these re-initiation strategies vary in their approach and characteristics.
Viral genomes take advantage of diverse translation strategies not only to facilitate the production of two or more proteins but also to regulate the ratio of viral proteins generated. Viruses regulate gene expression by modulating the arrangement of nucleotides on mRNA, for example by forming specific Kozak sequences or relatively complex secondary structures such as DPL sequences, TURBS sequences and 2A sequences, which affect the rate and mode of translation of the ribosome. As found in both Sendai and parainfluenza viruses, the viral genome can cleverly hijack ribosomal initiation for translation by using different initiation codons through nucleotide changes at specific sites of mRNA (Curran and Kolakofsky, 1989; Vidal et al., 1990; Boeck et al., 1992). Notably, the ribosomal “skipping” occurs inside the ribosome with little influence from external factors. The nucleotides of the viral protein are very delicately arranged and facilitate the stable expression of viral genes by regulating the movement of ribosomes on mRNA. Therefore, identifying the specific roles of ribosome components in the movement of RNA virus genomes is particularly important for the assembly mechanism of ribosomes required for viral protein translation.
Viral infection induces ribosome aggregation. RNA viruses induce ribosomal localization to recruit viral mRNA. In eukaryotes, some transcripts could be assembled into specific RNA condensates to form ribonucleoprotein (RNP) particles (Kachaev et al., 2021). These particles are transported by motor proteins to sites where translation is active, thus resulting in areas of liquid–liquid phase separation in the cytoplasm (Kachaev et al., 2021). Some RNA viruses, such as Zika virus (ZIKV), DENV, and hepatitis C virus (HCV) infect host cells to stimulate specific intracellular structures to form virus factories (VFs) (Paul et al., 2014; Reid et al., 2018; Mohd Ropidi et al., 2020). VFs could facilitate the replication and assembly of viral genomes, but additional studies are required to determine whether VFs recruit ribosomes to mediate the generation of heterogeneous ribosomes (Lashkevich and Dmitriev, 2021). As early as the 1970s, ribosomal aggregates in the VF formed early in Venezuelan equine encephalomyelitis virus (VEEV) infection were identified by electron microscopy (Bykovsky et al., 1969). It is worth mentioning that ribosomes and translation initiation factors are clustered around VFs formed in the bunyavirus and reovirus infection (Matsumoto et al., 2012; Desmet et al., 2014). In addition, Reoviruses infect cells mainly by regulating the distribution of translation initiation factors, but not by promoting the production of translation initiation factors. Taken together, these studies suggest that viral infection induces intracellular compartmentalization of the host translational machinery and replicative components.
Ribosome-associated proteins are involved in viral particle assembly. Different from the machines by viral genome, Sindbis virus (SINV) infects hosts with viral particles that contain host-derived ribosomal components, and SINVHeavy is more infectious than SINVLight because of its ribosomal component (Sokoloski et al., 2013; Mackenzie-Liu et al., 2018). SINVHeavy particles are found only in vertebrates, whereas SINVLight particles are found in invertebrate blood. The intermediate host mosquito can absorb only SINVLight particles, but the virus particles undergo proliferative replication in the mosquito to produce SINVHeavy virus particles to reinfect vertebrates (Sokoloski et al., 2013; Mackenzie-Liu et al., 2018). This mechanism suggests that the virus manipulates the ribosomal component of the host as it proliferates in the host, but whether this mechanism contributes to the assembly of ribosomal subunits during the next round of viral genome translation needs to be further explored.
With the development of cryo-electron microscopy, the delicate structure of the ribosome is gradually being revealed. Numerous studies have found that viral infection affects ribosome biogenesis, modification of ribosomal components, and changes in ribosome conformation.
The biosynthesis of ribosomes occurs mainly in the nucleolus, and disturbances in the process at any point can potentially lead to nucleolar stress, is also known ribosomal stress (Bianco and Mohr, 2019; Iarovaia et al., 2021; Luan et al., 2022). To manage this stressful state, cells cease the transcription of ribosomal genes (Iarovaia et al., 2021). Hence, the intensity of ribosome biogenesis is directly dependent on cellular demand and the strength of cell growth (Iarovaia et al., 2021).
Some RNA virus infections are accompanied by viral proteins that need to be transported to the nucleolus and possibly affect ribosome biogenesis. The HCV NS5B protein and core protein upregulate the expression of rRNA, thereby mediating the production of heterogeneous ribosomes and promoting viral protein translation (Goh et al., 2004). It is well known that HCV is one of the causes of hepatic cancer, which may be related to abnormal ribosome function leading to uncontrolled protein synthesis and increased demand for ribosome biosynthesis. A previous study reported that the matrix protein (M) encoded by Newcastle disease virus (NDV) at the beginning of infection is mainly localized in the nucleolus, and it may affect ribosome biogenesis and inhibit the synthesis of host proteins (Peeples et al., 1992). Presently, RPs that interact with the swine acute diarrhea syndrome coronavirus (SADS-CoV) M protein are quantified by liquid chromatography-mass spectrometry (LC–MS/MS) methods (Xu et al., 2022). This is consistent with the argument of Duan et al. on how M protein entry into the nucleus promotes RPL18 production found in NDV (Duan et al., 2022). Mouse L cells infected by VSV show inhibition of the synthesis of 45S rRNA precursors and their processing into 28S and 18S rRNA, which prevents ribosome assembly and RP synthesis (Jaye et al., 1980; Zan et al., 1990). With this similarity, the HIV Tat protein is known to be localized primarily in the nucleus and nucleolus. Interestingly, as Tat expression increases, cytoplasmic ribosome production decreases due to Tat’s interaction with the pro-fibronectin-U3 snoRNA complex, which interferes with the maturation of nucleolus precursor rRNA (Ponti et al., 2008). By using this mechanism, HIV regulates the host response, leading to apoptosis and shutting down protein synthesis in uninfected HIV cells (Ponti et al., 2008). In recent studies, researchers have found that influenza A virus (IAV) NS1 and the poliovirus 3C protease inhibit the transcription of rDNA in the nucleolus, while the infectious bronchitis virus (IBV) N protein can enter the nucleolus together with RPs to prevent the formation of new ribosomes and disrupt the cell cycle to interfere with translation in the host cell (Hiscox et al., 2001; You et al., 2005). However, the inhibition of ribosome biogenesis by this strategy is as adverse to virus proliferation as preventing the assembly of ribosomal subunits. Hence, the precise mechanism of these viruses needs to be further explored.
The host is a whole interconnected organism, and viral infections can affect the metabolic functions of the organism in various ways. Viruses and host cells are in a constant state of battle, and an increasing number of mechanisms and countermeasures are gradually being revealed that play an important role in maintaining an effective infection process and the stability of the internal environment of the host cell.
rRNA and RPs are the main components of ribosomes. Presently, we know that viral infection induces post-translational modifications of RPs, but it remains unknown whether rRNA changes occur due to viral infection. rRNAs are the main effectors of ribosome dynamics during mRNA decoding, peptide bond formation, and translation (Erales et al., 2017). In addition to being components of ribosomes, RPs play an important role in ribosome biogenesis, and they regulate many fundamental life processes, including cell cycle, cell proliferation, apoptosis, tumor development, genome integrity and development, and so on (Li, 2019; Miller et al., 2021).
Mutations, amplicon changes, and post-transcriptional modifications of rRNA alleles not only affect the fidelity of ribosomal translational mRNA, but are also a major aspect that contributes to ribosomal heterogeneity (Sloan et al., 2017; Fujii et al., 2018; Poitevin et al., 2020; Jansson et al., 2021; Gay et al., 2022). However, the effects of viral infection-induced changes in rRNA need further study. As mentioned above, IRES structures encoded by the viral genome bind to the 40S subunit to facilitate the translation of viral proteins. There is abundant evidence that encephalomyocarditis virus (EMCV), FMDV, hepatitis A virus (HAV), and HCV control the ribosome through the interaction of the IRES structure of genomic RNA with 18S rRNA in the 40S ribosomal subunit to translate viral proteins (Le et al., 1993, 1995). By using chemical detection and cryogenic electron microscopy (cryo-EM), the genome of HCV-infected cells was found to be complementary to the amplified segment 7 (ES7) base sequence of 18S rRNA (Malygin et al., 2013; Quade et al., 2015). Epstein–Barr virus (EBV), one of the DNA viruses, encodes the transcription factor Rta, which binds to rRNA in the 40S, 60S and 80S ribosomal subunits of nucleosomes and promotes the formation of heterogeneous ribosomes to preferentially translate the viral genome (Hwang et al., 2020). Such binding mechanisms remain to be explored in RNA virus-infected cells. Eukaryotic rRNAs include 18S rRNA, 28S rRNA, 5.8S rRNA, and 5S rRNA. Further studies are required to determine whether virus infection causes changes in rRNA mutation and modification.
Differences in post-translational modifications of RPs, such as phosphorylation, acylation, methylation, or glycosylation, contribute to ribosomal heterogeneity. Recent studies have shown that the deletion of RPs that constitute the subunits of the ribosome alters the level of translation of cellular proteins (Emmott et al., 2019; Luan et al., 2022). Different viral infections lead to various phosphorylation modifications of the ribosomal protein; this facilitates the expression of viral proteins (DiGiuseppe et al., 2020). Regarding phosphorylation of RACK1, researchers suggest that mutation of the phosphorylation site in the RACK1 protein to a negatively charged amino acid not only results in a conformational change in the 80S ribosomal subunit and a change in the conformation of the 40S ribosomal head, but it also promotes viral protein translation (Rollins et al., 2021). In summary, viral infection and modification of ribosomal components lead to heterogeneity of ribosomal function and promote the expression of viral proteins. However, the mechanisms by which heterogeneous ribosomes selectively translate viral proteins need to be further investigated.
Currently, it is known that the different translation strategies used by the viral genome to manipulate the ribosome cause changes in ribosomal conformation (Poitevin et al., 2020). However, it is not sufficient to conduct quantitative comparisons of ribosome structures, and comparisons of ribosome composition and studies of ribosome conformation are required (Poitevin et al., 2020). RPL40 promotes translation initiation in VSV, MV, and rabies virus (Lee et al., 2013). Moreover, one cannot help but suspect that during VSV infection, the conformation of the 60S subunit may change in response to the interaction of viral mRNA with RPL40 to allow the ribosome to preferentially translate the viral genome. By using cryo-EM, the spatial structure of the interaction between alphavirus mRNA and host ribosomal 18S rRNA was revealed, and this interaction resulted in a conformational change of rRNA extended segment 6 (ES6S) that is necessary for alphavirus translation (Toribio et al., 2016, 2018). In addition, it was found in HCV IRES translation initiation that the viral genome utilizes eIF2 or eIF5B to form a specific 48S complex conformation, which in turn recruits the 60S ribosomal large subunit to initiate protein translation (Brown et al., 2022). Thus, it was demonstrated that viral infection manipulates the ribosome and induces conformational changes in the ribosome to selectively translate viral mRNA.
To date, there are no reports that identify viral-induced heterogeneous ribosomal components. Therefore, it is crucial to conduct more studies on heterogeneous ribosomes to clarify the translation mechanism of viral proteins.
Viruses antagonize the antiviral immune response of the host and promote virus replication through complex interactions with the host. At present, several studies have shown that ribosomes have non-ribosomal functions in addition to translation after virus infection.
Ribosomal proteins positively regulate viral proliferation, mainly by promoting the replication, transcription, and assembly of viral genomes or by serving as viral receptors. The lymphocytic choriomeningitis virus (LCMV) Z protein interacts with the ribosomal protein RPLP0 and plays an important role in virion assembly (Pedersen, 1973; Borden et al., 1998). Moreover, the LCMV replication-transcription complex (RTC) contains the RPs eL10a and eS6, and these RPs facilitate replication of the viral genome. By interacting with RPs, the LCMV nucleoprotein (N) can inhibit the antiviral immune response of the host (Baird et al., 2012). Similarly, the 3′-extremity region of rabbit hemorrhagic disease virus (RHDV) interacts with RPS5 to promote replication of viral genes (Guo et al., 2020). Infectious bursal disease virus (IBDV) VP3 protein interacts with RPs to form ribosomal protein assembly complexes to promote viral genome replication (Ma et al., 2021). HIV-1 encodes Gag proteins that interact with RPL7 to facilitate the assembly of viral particles (Karnib et al., 2020). The efficient particle assembly of mouse mammary tumor virus (MMTV) depends on the interaction of Gag and RPL9 in the nucleolus of infected cells (Beyer et al., 2013). RPS2 (RPSA), as a ribosomal binding receptor, binds to the envelope glycoprotein E of DENV and yellow fever virus to promote viral infection (Zidane et al., 2012). After Semliki Forest virus (SFV) invades host cells, it needs the large ribosomal subunit to uncoat and release the viral genome (Wengler et al., 1992). Existing studies have shown that RPs can not only play an important role in the translation of the viral genome as a major component of the protein translation factory, but they can also contribute to viral life cycle progression.
Some RPs have antiviral functions. RPs interact with viral proteins to directly inhibit viral transcription or translation. During CSFV infection, the viral protein Npro interacts with uS10 and induces Npro degradation by the proteasome (Lv et al., 2017). Because TLR3 overexpression or uS10 knockdown does not promote CSFV replication, it was found that uS10 may indirectly regulate CSFV replication through TLR3 (Lv et al., 2017). Some RPs also activate antiviral signaling pathways. The ribosomal protein RPSA interacts with FMDV structural proteins to inhibit the activation of the mitogen-activated protein kinase (MAPK) pathway, thereby inhibiting viral genome replication (Zhu et al., 2020). Finally, RPS3, as an example, not only plays a role in apoptosis and DNA repair, but it also acts as a novel antiviral factor to inhibit CSFV proliferation by increasing the secretion of antiviral cytokines (Zhao et al., 2022).
In summary, the biological significance of many RPs in the infection of different viruses is gradually being revealed. In recent years, the interaction between viral and host proteins has been analyzed by high-throughput quantitative mass spectrometry where over 50 RPs interact with SFV capsid proteins (Contu et al., 2021). However, the specific functions need further study. Similarly, a proteomic approach has been used to explore the interactions between the encoded viral proteins and host proteins following IBDV infection (Ma et al., 2021). Therefore, the above finding demonstrates the feasibility of this approach for determining the role of viral proteins following viral invasion of the host. Currently, two-dimensional gel electrophoresis and mass spectrometry are commonly used in proteomics, and the abovementioned studies also suggest that the use of proteomic techniques, ribosome analysis, and bioinformatics to analyze ribosomal components has far-reaching potential.
Ribosomes are protein synthesis factories not only for eukaryotic mRNA but also for viral mRNA. After the virus infects the host cell, it needs to manipulate the ribosome to complete the translation of viral protein. The control of the ribosome by viruses involves two main aspects. First, viruses shut down the ribosome to translate host mRNA. For example, some viral proteins regulate the compartmentalization of organelles or recruit translation initiation factors to facilitate the assembly of the translation machinery and viral protein translation. To inhibit host mRNA translation by the ribosome, viruses also promote the assembly of large/small ribosomal subunits by regulating the secondary structure of viral mRNAs. Second, viruses manipulate the ribosome to complete viral protein translation. The viral mRNA secondary structure not only influences the assembly rate of the large and small subunits of the ribosome, but it also regulates how ribosomes move on the viral mRNA during different translation stages. Viral infection also induces changes in the composition and conformation of ribosomes, resulting in the formation of heterogeneous ribosomes. Therefore, analyzing the mechanism of viral control of ribosomes and studying the properties and assembly mechanisms of heterogeneous ribosomes required for viral protein translation will help us to fully understand the mechanism of viral protein translation.
With the development of science and technology, especially proteomics and cryo-EM, the structure of eukaryotic ribosomes has been gradually revealed. The crystal structure of the human 80S ribosome and the crystal structure of the yeast 90S ribosomal precursor has been reported, thereby opening a new door for designing small-molecule drugs targeting the ribosome (Khatter et al., 2015; Sun et al., 2017). Some recent studies have shown that Alzheimer’s disease, Parkinson’s disease, tumorigenesis, cancer metastasis, and other diseases are associated with the abnormal ribosomal function (Ebright et al., 2020; Elhamamsy et al., 2022; Stein et al., 2022). The necessity for correct assembly of ribosomes for hematopoietic stem cell regeneration and the involvement of the ribosomal protein RPL39L in spermatogenesis also reflect the importance of ribosomes for maintaining body homeostasis and genetic stability (Lv et al., 2021; Zou and Qi, 2021). Because these diseases are dependent on abnormal ribosomal function, we wondered whether this “butterfly effect” could be cured by developing drugs that target ribosomes. The antibiotic tetracenomycin blocks bacterial protein synthesis by targeting bacterial ribosomes, and it is precise because of this targeting that the emergence of bacterial resistance can be effectively avoided (Osterman et al., 2020). This discovery suggests a promising future for the development of drugs targeting heterogeneous ribosomes. The heterogeneity of ribosomal components in Kaposi’s sarcoma-associated herpesvirus infected cells has been analyzed using quantitative proteomics (Murphy et al., 2022). The deletion of ribosomal biogenesis factor BUD23 was found to significantly inhibit the replication of viral particles (Murphy et al., 2022). This suggests that the inhibition of new heterogeneous ribosome production after viral infection of host cells can effectively kill viral particles in the organism. Therefore, the identification and structural analysis of the heterogeneous ribosomal components required for viral protein translation will contribute to a comprehensive understanding of the mechanism of viral protein synthesis and open a new door for developing antiviral drugs in the future. It also provides new strategies for optimizing vaccines and raises new questions for RNA vaccines or drugs, that is, whether related drugs or vaccines after entering the body will have corresponding adverse reactions on ribosomes.
GL and JZ: conceptualization and review and editing. XW and JZ: writing original draft preparation. GL: supervision and project administration. DZ and XW: visualization. All authors have read and agreed to the published version of the manuscript.
This study was sponsored by the Natural Science Foundation of Shanghai (22ZR1476300), the National Natural Science Foundation of China (32000109 and 32172832), the Shanghai Sailing Program (20YF1457700), Shanghai Municipal Science and Technology Major Project (ZD2021CY001), and the Central Public-interest Scientific Institution Basal Research Fund (2022JB01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arhab, Y., Bulakhov, A. G., Pestova, T. V., and Hellen, C. U. T. (2020). Dissemination of internal ribosomal entry sites (IRES) between viruses by horizontal gene transfer. Viruses 12:612. doi: 10.3390/v12060612
Arhab, Y., Miscicka, A., Pestova, T. V., and Hellen, C. U. T. (2022). Horizontal gene transfer as a mechanism for the promiscuous acquisition of distinct classes of IRES by avian caliciviruses. Nucleic Acids Res. 50, 1052–1068. doi: 10.1093/nar/gkab1243
Baird, N. L., York, J., and Nunberg, J. H. (2012). Arenavirus infection induces discrete cytosolic structures for RNA replication. J. Virol. 86, 11301–11310. doi: 10.1128/JVI.01635-12
Bakhshesh, M., Groppelli, E., Willcocks, M. M., Royall, E., Belsham, G. J., and Roberts, L. O. (2008). The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J. Virol. 82, 1993–2003. doi: 10.1128/JVI.01957-07
Beckham, S. A., Matak, M. Y., Belousoff, M. J., Venugopal, H., Shah, N., Vankadari, N., et al. (2020). Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES. Nucleic Acids Res. 48, 8006–8021. doi: 10.1093/nar/gkaa519
Beyer, A. R., Bann, D. V., Rice, B., Pultz, I. S., Kane, M., Goff, S. P., et al. (2013). Nucleolar trafficking of the mouse mammary tumor virus gag protein induced by interaction with ribosomal protein L9. J. Virol. 87, 1069–1082. doi: 10.1128/JVI.02463-12
Bhatt, P. R., Scaiola, A., Loughran, G., Leibundgut, M., Kratzel, A., Meurs, R., et al. (2021). Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 372, 1306–1313. doi: 10.1126/science.abf3546
Bhattacharyya, S., Verma, B., Pandey, G., and Das, S. (2008). The structure and function of a cis-acting element located upstream of the IRES that influences Coxsackievirus B3 RNA translation. Virology 377, 345–354. doi: 10.1016/j.virol.2008.04.019
Bianco, C., and Mohr, I. (2019). Ribosome biogenesis restricts innate immune responses to virus infection and DNA. elife 8:e49551. doi: 10.7554/eLife.49551
Boeck, R., Curran, J., Matsuoka, Y., Compans, R., and Kolakofsky, D. (1992). The parainfluenza virus type 1 P/C gene uses a very efficient GUG codon to start its C' protein. J. Virol. 66, 1765–1768. doi: 10.1128/JVI.66.3.1765-1768.1992
Borden, K. L., Campbelldwyer, E. J., Carlile, G. W., Djavani, M., and Salvato, M. S. (1998). Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J. Virol. 72, 3819–3826. doi: 10.1128/JVI.72.5.3819-3826.1998
Brown, Z. P., Abaeva, I. S., De, S., Hellen, C. U. T., Pestova, T. V., and Frank, J. (2022). Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES. EMBO J. 41:e110581. doi: 10.15252/embj.2022110581
Bykovsky, A. F., Yershov, F. I., and Zhdanov, V. M. (1969). Morphogenesis of Venezuelan equine encephalomyelitis virus. J. Virol. 4, 496–504. doi: 10.1128/JVI.4.4.496-504.1969
Caceres, C. J., Angulo, J., Contreras, N., Pino, K., Vera-Otarola, J., and Lopez-Lastra, M. (2016). Targeting deoxyhypusine hydroxylase activity impairs cap-independent translation initiation driven by the 5'untranslated region of the HIV-1, HTLV-1, and MMTV mRNAs. Antivir. Res. 134, 192–206. doi: 10.1016/j.antiviral.2016.09.006
Cao, F., and Tavis, J. E. (2011). RNA elements directing translation of the duck hepatitis B virus polymerase via ribosomal shunting. J. Virol. 85, 6343–6352. doi: 10.1128/JVI.00101-11
Carvajal, F., Vallejos, M., Walters, B., Contreras, N., Hertz, M. I., Olivares, E., et al. (2016). Structural domains within the HIV-1 mRNA and the ribosomal protein S25 influence cap-independent translation initiation. FEBS J. 283, 2508–2527. doi: 10.1111/febs.13756
Castello, A., Izquierdo, J. M., Welnowska, E., and Carrasco, L. (2009). RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J. Cell Sci. 122, 3799–3809. doi: 10.1242/jcs.055988
Chamond, N., Deforges, J., Ulryck, N., and Sargueil, B. (2014). 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of type II IRESs. Nucleic Acids Res. 42, 10373–10384. doi: 10.1093/nar/gku720
Cheng, C. A., Luo, J. M., Chiang, M. H., Fang, K. Y., Li, C. H., Chen, C. W., et al. (2021). Nervous necrosis virus coat protein mediates host translation shutoff through nuclear translocalization and degradation of polyadenylate binding protein. J. Virol. 95:e0236420. doi: 10.1128/JVI.02364-20
Cheng, Z., Mugler, C. F., Keskin, A., Hodapp, S., Chan, L. Y., Weis, K., et al. (2019). Small and large ribosomal subunit deficiencies Lead to distinct gene expression signatures that reflect cellular growth rate. Mol. Cell 73, 36–47.e10. doi: 10.1016/j.molcel.2018.10.032
Chenik, M., Chebli, K., and Blondel, D. (1995). Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J. Virol. 69, 707–712. doi: 10.1128/JVI.69.2.707-712.1995
Colson, P., Gautret, P., Delerce, J., Chaudet, H., Pontarotti, P., Forterre, P., et al. (2022). The emergence, spread and vanishing of a French SARS-CoV-2 variant exemplifies the fate of RNA virus epidemics and obeys the Mistigri rule. J. Med. Virol. doi: 10.1002/jmv.28102 [Epub ahead of print].
Contu, L., Balistreri, G., Domanski, M., Uldry, A.-C., and Mühlemann, O. (2021). Characterization of the Semliki Forest virus-host cell interactome reveals the viral capsid protein as an inhibitor of non-sense-mediated mRNA decay. PLoS Pathog. 17:e1009603. doi: 10.1101/2020.10.12.335497
Curran, J., and Kolakofsky, D. (1989). Scanning independent ribosomal initiation of the Sendai virus Y proteins in vitro and in vivo. EMBO J. 8, 521–526. doi: 10.1002/j.1460-2075.1989.tb03406.x
Davies, M. V., and Kaufman, R. J. (1992). The sequence context of the initiation codon in the encephalomyocarditis virus leader modulates efficiency of internal translation initiation. J. Virol. 66, 1924–1932. doi: 10.1128/JVI.66.4.1924-1932.1992
de Breyne, S., Bonderoff, J. M., Chumakov, K. M., Lloyd, R. E., and Hellen, C. U. (2008). Cleavage of eukaryotic initiation factor eIF5B by enterovirus 3C proteases. Virology 378, 118–122. doi: 10.1016/j.virol.2008.05.019
de Breyne, S., Simonet, V., Pelet, T., and Curran, J. (2003). Identification of a cis-acting element required for shunt-mediated translational initiation of the Sendai virus Y proteins. Nucleic Acids Res. 31, 608–618. doi: 10.1093/nar/gkg143
Desmet, E. A., Anguish, L. J., and Parker, J. S. (2014). Virus-mediated compartmentalization of the host translational machinery. MBio 5, e01463–e01414. doi: 10.1128/mBio.01463-14
DiGiuseppe, S., Rollins, M. G., Astar, H., Khalatyan, N., Savas, J. N., and Walsh, D. (2020). Proteomic and mechanistic dissection of the poxvirus-customized ribosome. J. Cell Sci. 134:jcs246603. doi: 10.1242/jcs.246603
Duan, Z., Tang, H., Wang, Y., Zhao, C., Zhou, L., and Han, Y. (2022). The association of ribosomal protein L18 with Newcastle disease virus matrix protein enhances viral translation and replication. Avian Pathol. 51, 129–140. doi: 10.1080/03079457.2021.2013435
DuBois, R. M., Slavish, P. J., Baughman, B. M., Yun, M. K., Bao, J., Webby, R. J., et al. (2012). Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog. 8:e1002830. doi: 10.1371/journal.ppat.1002830
Ebright, R. Y., Lee, S., Wittner, B. S., Niederhoffer, K. L., Nicholson, B. T., Bardia, A., et al. (2020). Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 367, 1468–1473. doi: 10.1126/science.aay0939
Elhamamsy, A. R., Metge, B. J., Alsheikh, H. A., Shevde, L. A., and Samant, R. S. (2022). Ribosome biogenesis: a central player in cancer metastasis and therapeutic resistance. Cancer Res. 82, 2344–2353. doi: 10.1158/0008-5472.CAN-21-4087
Emmott, E., Jovanovic, M., and Slavov, N. (2019). Ribosome stoichiometry: from form to function. Trends Biochem. Sci. 44, 95–109. doi: 10.1016/j.tibs.2018.10.009
Erales, J., Marchand, V., Panthu, B., Gillot, S., Belin, S., Ghayad, S. E., et al. (2017). Evidence for rRNA 2'-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. U. S. A. 114, 12934–12939. doi: 10.1073/pnas.1707674114
Fernandez-Garcia, L., Angulo, J., Ramos, H., Barrera, A., Pino, K., Vera-Otarola, J., et al. (2020). The internal ribosome entry site of the dengue virus mRNA is active when cap-dependent translation initiation is inhibited. J. Virol. 95:e01998-20. doi: 10.1128/JVI.01998-20
Ferretti, M. B., Ghalei, H., Ward, E. A., Potts, E. L., and Karbstein, K. (2017). Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements. Nat. Struct. Mol. Biol. 24, 700–707. doi: 10.1038/nsmb.3442
Filbin, M. E., Vollmar, B. S., Shi, D., Gonen, T., and Kieft, J. S. (2013). HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat. Struct. Mol. Biol. 20, 150–158. doi: 10.1038/nsmb.2465
Finkel, Y., Gluck, A., Nachshon, A., Winkler, R., Fisher, T., Rozman, B., et al. (2021). SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 594, 240–245. doi: 10.1038/s41586-021-03610-3
Fujii, K., Susanto, T. T., Saurabh, S., and Barna, M. (2018). Decoding the function of expansion segments in ribosomes. Mol. Cell 72, 1013–1020.e6. doi: 10.1016/j.molcel.2018.11.023
Garaigorta, U., and Chisari, F. V. (2009). Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 6, 513–522. doi: 10.1016/j.chom.2009.11.004
Gaucherand, L., Porter, B. K., Levene, R. E., Price, E. L., Schmaling, S. K., Rycroft, C. H., et al. (2019). The influenza a virus endoribonuclease PA-X usurps host mRNA processing machinery to limit host gene expression. Cell Rep. 27, 776–792.e7. doi: 10.1016/j.celrep.2019.03.063
Gay, D. M., Lund, A. H., and Jansson, M. D. (2022). Translational control through ribosome heterogeneity and functional specialization. Trends Biochem. Sci. 47, 66–81. doi: 10.1016/j.tibs.2021.07.001
Genuth, N. R., and Barna, M. (2018). The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell 71, 364–374. doi: 10.1016/j.molcel.2018.07.018
Gingras, A. C., Svitkin, Y., Belsham, G. J., Pause, A., and Sonenberg, N. (1996). Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. U. S. A. 93, 5578–5583. doi: 10.1073/pnas.93.11.5578
Goh, P. Y., Tan, Y. J., Lim, S. P., Tan, Y. H., Lim, S. G., Fuller-Pace, F., et al. (2004). Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J. Virol. 78, 5288–5298. doi: 10.1128/jvi.78.10.5288-5298.2004
Gradi, A., Svitkin, Y. V., Imataka, H., and Sonenberg, N. (1998). Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. U. S. A. 95, 11089–11094. doi: 10.1073/pnas.95.19.11089
Gross, L., Vicens, Q., Einhorn, E., Noireterre, A., Schaeffer, L., Kuhn, L., et al. (2017). The IRES5'UTR of the dicistrovirus cricket paralysis virus is a type III IRES containing an essential pseudoknot structure. Nucleic Acids Res. 45, 8993–9004. doi: 10.1093/nar/gkx622
Guo, H., Zhu, J., Miao, Q., Qi, R., Tang, A., Liu, C., et al. (2020). RPS5 interacts with the rabbit hemorrhagic disease virus 3′ extremities region and plays a role in virus replication. Vet. Microbiol. 249:108858. doi: 10.1016/j.vetmic.2020.108858
Habeta, M., Luttermann, C., and Meyers, G. (2014). Feline calicivirus can tolerate gross changes of its minor capsid protein expression levels induced by changing translation reinitiation frequency or use of a separate VP2-coding mRNA. PLoS One 9:e102254. doi: 10.1371/journal.pone.0102254
Hamidi, T., Singh, A. K., Veland, N., Vemulapalli, V., Chen, J., Hardikar, S., et al. (2018). Identification of Rpl29 as a major substrate of the lysine methyltransferase Set7/9. J. Biol. Chem. 293, 12770–12780. doi: 10.1074/jbc.RA118.002890
Han, S., Sun, S., Li, P., Liu, Q., Zhang, Z., Dong, H., et al. (2020). Ribosomal protein L13 promotes IRES-driven translation of foot-and-mouth disease virus in a helicase DDX3-dependent manner. J. Virol. 94:e01679-19. doi: 10.1128/JVI.01679-19
Hashizume, C., Kuramitsu, M., Zhang, X., Kurosawa, T., Kamata, M., and Aida, Y. (2007). Human immunodeficiency virus type 1 Vpr interacts with spliceosomal protein SAP145 to mediate cellular pre-mRNA splicing inhibition. Microbes Infect. 9, 490–497. doi: 10.1016/j.micinf.2007.01.013
Hinton, T. M., and Crabb, B. S. (2001). The novel picornavirus equine rhinitis B virus contains a strong type II internal ribosomal entry site which functions similarly to that of Encephalomyocarditis virus. J. Gen. Virol. 82, 2257–2269. doi: 10.1099/0022-1317-82-9-2257
Hiscox, J. A., Wurm, T., Wilson, L., Britton, P., Cavanagh, D., and Brooks, G. (2001). The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 75, 506–512. doi: 10.1128/JVI.75.1.506-512.2001
Huang, C., Kolokoltsova, O. A., Mateer, E. J., Koma, T., and Paessler, S. (2017). Highly pathogenic New World arenavirus infection activates the pattern recognition receptor protein kinase R without attenuating virus replication in human cells. J. Virol. 91:e01090-17. doi: 10.1128/JVI.01090-17
Hui, A., and de Boer, H. A. (1987). Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 84, 4762–4766. doi: 10.1073/pnas.84.14.4762
Hwang, S. P., Huang, L. C., Wang, W. H., Lin, M. H., Kuo, C. W., Huang, H. H., et al. (2020). Expression of Rta in B lymphocytes during Epstein-Barr virus latency. J. Mol. Biol. 432, 5227–5243. doi: 10.1016/j.jmb.2020.07.011
Iarovaia, O. V., Ioudinkova, E. S., Velichko, A. K., and Razin, S. V. (2021). Manipulation of cellular processes via nucleolus Hijaking in the course of viral infection in mammals. Cells 10:1597. doi: 10.3390/cells10071597
Jang, S. K., Krausslich, H. G., Nicklin, M. J., Duke, G. M., Palmenberg, A. C., and Wimmer, E. (1988). A segment of the 5′ non-translated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62, 2636–2643. doi: 10.1128/JVI.62.8.2636-2643.1988
Jansson, M. D., Hafner, S. J., Altinel, K., Tehler, D., Krogh, N., Jakobsen, E., et al. (2021). Regulation of translation by site-specific ribosomal RNA methylation. Nat. Struct. Mol. Biol. 28, 889–899. doi: 10.1038/s41594-021-00669-4
Jaye, M., Wu, F. S., and Lucas-Lenard, J. M. (1980). Inhibition of synthesis of ribosomal proteins and of ribosome assembly after infection of L cells with vesicular stomatitis virus. Biochim. Biophys. Acta 606, 1–12. doi: 10.1016/0005-2787(80)90092-1
Jobe, A., Liu, Z., Gutierrez-Vargas, C., and Frank, J. (2019). New insights into ribosome structure and function. Cold Spring Harb. Perspect. Biol. 11:a032615. doi: 10.1101/cshperspect.a032615
Kachaev, Z. M., Ivashchenko, S. D., Kozlov, E. N., Lebedeva, L. A., and Shidlovskii, Y. V. (2021). Localization and functional roles of components of the translation apparatus in the eukaryotic cell nucleus. Cells 10:3239. doi: 10.3390/cells10113239
Kamitani, W., Huang, C., Narayanan, K., Lokugamage, K. G., and Makino, S. (2009). A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 16, 1134–1140. doi: 10.1038/nsmb.1680
Kamrud, K. I., Custer, M., Dudek, J. M., Owens, G., Alterson, K. D., Lee, J. S., et al. (2007). Alphavirus replicon approach to promoterless analysis of IRES elements. Virology 360, 376–387. doi: 10.1016/j.virol.2006.10.049
Karnib, H., Nadeem, M. F., Humbert, N., Sharma, K. K., Grytsyk, N., Tisne, C., et al. (2020). The nucleic acid chaperone activity of the HIV-1 gag polyprotein is boosted by its cellular partner RPL7: a kinetic study. Nucleic Acids Res. 48, 9218–9234. doi: 10.1093/nar/gkaa659
Kerekatte, V., Keiper, B. D., Badorff, C., Cai, A., Knowlton, K. U., and Rhoads, R. E. (1999). Cleavage of poly(a)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 73, 709–717. doi: 10.1128/JVI.73.1.709-717.1999
Kerr, C. H., Wang, Q. S., Keatings, K., Khong, A., Allan, D., Yip, C. K., et al. (2015). The 5′ untranslated region of a novel infectious molecular clone of the dicistrovirus cricket paralysis virus modulates infection. J. Virol. 89, 5919–5934. doi: 10.1128/JVI.00463-15
Khaperskyy, D. A., Schmaling, S., Larkins-Ford, J., McCormick, C., and Gaglia, M. M. (2016). Selective degradation of host RNA polymerase II transcripts by influenza a virus PA-X host shutoff protein. PLoS Pathog. 12:e1005427. doi: 10.1371/journal.ppat.1005427
Khatter, H., Myasnikov, A. G., Natchiar, S. K., and Klaholz, B. P. (2015). Structure of the human 80S ribosome. Nature 520, 640–645. doi: 10.1038/nature14427
Kjaer, J., and Belsham, G. J. (2018). Modifications to the foot-and-mouth disease virus 2A peptide: influence on polyprotein processing and virus replication. J. Virol. 92:e02218-17. doi: 10.1128/JVI.02218-17
Kuyumcu-Martinez, M., Belliot, G., Sosnovtsev, S. V., Chang, K. O., Green, K. Y., and Lloyd, R. E. (2004). Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(a)-binding protein. J. Virol. 78, 8172–8182. doi: 10.1128/JVI.78.15.8172-8182.2004
Labaronne, E., Décimo, D., Bertrand, L., Guiguettaz, L., Sohier, T. J. M., Cluet, D., et al. (2022). Extensive uORF translation from HIV-1 transcripts conditions DDX3 dependency for expression of main ORFs and elicits specific T cell immune responses in infected individuals. bioRxiv [Preprint]. doi: 10.1101/2022.04.29.489990
LaFontaine, E., Miller, C. M., Permaul, N., Martin, E. T., and Fuchs, G. (2020). Ribosomal protein RACK1 enhances translation of poliovirus and other viral IRESs. Virology 545, 53–62. doi: 10.1016/j.virol.2020.03.004
Lago, M., Bandin, I., Olveira, J. G., and Dopazo, C. P. (2017). In vitro reassortment between infectious pancreatic necrosis virus (IPNV) strains: the mechanisms involved and its effect on virulence. Virology 501, 1–11. doi: 10.1016/j.virol.2016.11.003
Lamphear, B. J., Kirchweger, R., Skern, T., and Rhoads, R. E. (1995). Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270, 21975–21983. doi: 10.1074/jbc.270.37.21975
Landry, D. M., Hertz, M. I., and Thompson, S. R. (2009). RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 23, 2753–2764. doi: 10.1101/gad.1832209
Lashkevich, K. A., and Dmitriev, S. E. (2021). mRNA targeting, transport and local translation in eukaryotic cells: from the classical view to a diversity of new concepts. Mol. Biol. 55, 507–537. doi: 10.1134/S0026893321030080
Le, S. Y., Chen, J. H., Sonenberg, N., and Maizel, J. V. Jr. (1993). Conserved tertiary structural elements in the 5′ non-translated region of cardiovirus, aphthovirus and hepatitis a virus RNAs. Nucleic Acids Res. 21, 2445–2451. doi: 10.1093/nar/21.10.2445
Le, S. Y., Sonenberg, N., and Maizel, J. V. Jr. (1995). Unusual folding regions and ribosome landing pad within hepatitis C virus and pestivirus RNAs. Gene 154, 137–143. doi: 10.1016/0378-1119(94)00859-q
Lee, A. S., Burdeinick-Kerr, R., and Whelan, S. P. (2013). A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc. Natl. Acad. Sci. U. S. A. 110, 324–329. doi: 10.1073/pnas.1216454109
Li, S. (2019). Regulation of ribosomal proteins on viral infection. Cells 8:508. doi: 10.3390/cells8050508
Li, Y., Firth, A. E., Brierley, I., Cai, Y., Napthine, S., Wang, T., et al. (2019). Programmed-2/−1 ribosomal frameshifting in Simarteriviruses: an evolutionarily conserved mechanism. J. Virol. 93:e00370-19. doi: 10.1128/JVI.00370-19
Liu, Y., Chin, J. M., Choo, E. L., and Phua, K. K. L. (2019). Messenger RNA translation enhancement by immune evasion proteins: a comparative study between EKB (vaccinia virus) and NS1 (influenza a virus). Sci. Rep. 9:11972. doi: 10.1038/s41598-019-48559-6
Liu, T., Wang, Y., Tan, T. J. C., Wu, N. C., and Brooke, C. B. (2022). The evolutionary potential of influenza a virus hemagglutinin is highly constrained by epistatic interactions with neuraminidase. Cell Host Microbe 30, 1363–1369.e4. doi: 10.1016/j.chom.2022.09.003
Lokugamage, K. G., Narayanan, K., Huang, C., and Makino, S. (2012). Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 86, 13598–13608. doi: 10.1128/JVI.01958-12
Lokugamage, K. G., Narayanan, K., Nakagawa, K., Terasaki, K., Ramirez, S. I., Tseng, C. T., et al. (2015). Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 89, 10970–10981. doi: 10.1128/JVI.01352-15
Lopez-Lastra, M., Ramdohr, P., Letelier, A., Vallejos, M., Vera-Otarola, J., and Valiente-Echeverria, F. (2010). Translation initiation of viral mRNAs. Rev. Med. Virol. 20, 177–195. doi: 10.1002/rmv.649
Luan, Y., Tang, N., Yang, J., Liu, S., Cheng, C., Wang, Y., et al. (2022). Deficiency of ribosomal proteins reshapes the transcriptional and translational landscape in human cells. Nucleic Acids Res. 50, 6601–6617. doi: 10.1093/nar/gkac053
Luke, G. A., de Felipe, P., Lukashev, A., Kallioinen, S. E., Bruno, E. A., and Ryan, M. D. (2008). Occurrence, function and evolutionary origins of '2A-like' sequences in virus genomes. J. Gen. Virol. 89, 1036–1042. doi: 10.1099/vir.0.83428-0
Luttermann, C., and Meyers, G. (2014). Two alternative ways of start site selection in human norovirus reinitiation of translation. J. Biol. Chem. 289, 11739–11754. doi: 10.1074/jbc.M114.554030
Lv, H., Dong, W., Qian, G., Wang, J., Li, X., Cao, Z., et al. (2017). uS10, a novel Npro-interacting protein, inhibits classical swine fever virus replication. J. Gen. Virol. 98, 1679–1692. doi: 10.1099/jgv.0.000867
Lv, K., Gong, C., Antony, C., Han, X., Ren, J. G., Donaghy, R., et al. (2021). HectD1 controls hematopoietic stem cell regeneration by coordinating ribosome assembly and protein synthesis. Cell Stem Cell 28, 1275–1290.e9. doi: 10.1016/j.stem.2021.02.008
Ma, S. T., Wang, Y. S., Wang, X. L., Xia, X. X., Bi, Z. W., Wang, J. Y., et al. (2021). Mass spectrometry-based proteomic analysis of potential infectious bursal disease virus VP3-interacting proteins in chicken embryo fibroblasts cells. Virus Genes 57, 194–204. doi: 10.1007/s11262-021-01828-x
Mackenzie-Liu, D., Sokoloski, K. J., Purdy, S., and Hardy, R. W. (2018). Encapsidated host factors in alphavirus particles influence midgut infection of Aedes aegypti. Viruses 10:263. doi: 10.3390/v10050263
Malygin, A. A., Kossinova, O. A., Shatsky, I. N., and Karpova, G. G. (2013). HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 41, 8706–8714. doi: 10.1093/nar/gkt632
Maree, F., de Klerk-Lorist, L. M., Gubbins, S., Zhang, F., Seago, J., Perez-Martin, E., et al. (2016). Differential persistence of foot-and-mouth disease virus in African Buffalo is related to virus virulence. J. Virol. 90, 5132–5140. doi: 10.1128/JVI.00166-16
Martinez-Salas, E., Francisco-Velilla, R., Fernandez-Chamorro, J., and Embarek, A. M. (2017). Insights into structural and mechanistic features of viral IRES elements. Front. Microbiol. 8:2629. doi: 10.3389/fmicb.2017.02629
Masaki, K., Sonobe, Y., Ghadge, G., Pytel, P., and Roos, R. P. (2019). TDP-43 proteinopathy in Theiler's murine encephalomyelitis virus infection. PLoS Pathog. 15:e1007574. doi: 10.1371/journal.ppat.1007574
Matsumoto, Y., Hayashi, Y., Omori, H., Honda, T., Daito, T., Horie, M., et al. (2012). Bornavirus closely associates and segregates with host chromosomes to ensure persistent intranuclear infection. Cell Host Microbe 11, 492–503. doi: 10.1016/j.chom.2012.04.009
McKnight, K. L., and Lemon, S. M. (2018). Hepatitis a virus genome organization and replication strategy. Cold Spring Harb. Perspect. Med. 8:a033480. doi: 10.1101/cshperspect.a033480
Meyers, G. (2003). Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J. Biol. Chem. 278, 34051–34060. doi: 10.1074/jbc.M304874200
Meyers, G. (2007). Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J. Virol. 81, 9623–9632. doi: 10.1128/JVI.00771-07
Miller, C. M., Selvam, S., and Fuchs, G. (2021). Fatal attraction: the roles of ribosomal proteins in the viral life cycle. Wiley Interdiscip Rev. RNA 12:e1613. doi: 10.1002/wrna.1613
Minskaia, E., and Ryan, M. D. (2013). Protein coexpression using FMDV 2A: effect of "linker" residues. Biomed. Res. Int. 2013:291730. doi: 10.1155/2013/291730
Miras, M., Sempere, R. N., Kraft, J. J., Miller, W. A., Aranda, M. A., and Truniger, V. (2014). Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 202, 233–246. doi: 10.1111/nph.12650
Mohd Ropidi, M. I., Khazali, A. S., Nor Rashid, N., and Yusof, R. (2020). Endoplasmic reticulum: a focal point of Zika virus infection. J. Biomed. Sci. 27:27. doi: 10.1186/s12929-020-0618-6
Murphy, J. C., Schumann, S., Harrington, E., Vasconcelos, E. J. R., Mottram, T. J., Aspden, J. L., et al. (2022). Kaposi’s sarcoma-associated herpesvirus induces specialized ribosomes to efficiently translate viral lytic mRNAs. bioRxiv [Preprint]. doi: 10.1101/2022.03.11.483946
Nacken, W., Schreiber, A., Masemann, D., and Ludwig, S. (2021). The effector domain of the influenza a virus non-structural protein NS1 triggers host shutoff by mediating inhibition and global deregulation of host transcription when associated with specific structures in the nucleus. MBio 12:e0219621. doi: 10.1128/mBio.02196-21
Namy, O., Moran, S. J., Stuart, D. I., Gilbert, R. J., and Brierley, I. (2006). A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441, 244–247. doi: 10.1038/nature04735
Napthine, S., Hill, C. H., Nugent, H. C. M., and Brierley, I. (2021). Modulation of viral programmed ribosomal frameshifting and stop codon read through by the host restriction factor shiftless. Viruses 13:1230. doi: 10.3390/v13071230
Nemeroff, M. E., Barabino, S. M., Li, Y., Keller, W., and Krug, R. M. (1998). Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3'end formation of cellular pre-mRNAs. Mol. Cell 1, 991–1000. doi: 10.1016/s1097-2765(00)80099-4
Ngoveni, H. G., van Schalkwyk, A., and Koekemoer, J. J. O. (2019). Evidence of intragenic recombination in African horse sickness virus. Viruses 11:654. doi: 10.3390/v11070654
Olivares, E., Landry, D. M., Caceres, C. J., Pino, K., Rossi, F., Navarrete, C., et al. (2014). The 5′ untranslated region of the human T-cell lymphotropic virus type 1 mRNA enables cap-independent translation initiation. J. Virol. 88, 5936–5955. doi: 10.1128/JVI.00279-14
Osterman, I. A., Wieland, M., Maviza, T. P., Lashkevich, K. A., Lukianov, D. A., Komarova, E. S., et al. (2020). Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel. Nat. Chem. Biol. 16, 1071–1077. doi: 10.1038/s41589-020-0578-x
Patino-Galindo, J. A., Filip, I., and Rabadan, R. (2021). Global patterns of recombination across human viruses. Mol. Biol. Evol. 38, 2520–2531. doi: 10.1093/molbev/msab046
Paul, D., Madan, V., and Bartenschlager, R. (2014). Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe 16, 569–579. doi: 10.1016/j.chom.2014.10.008
Pedersen, I. R. (1973). Different classes of ribonucleic acid isolated from lymphocytic choriomeningitis virus. J. Virol. 11, 416–423. doi: 10.1128/JVI.11.3.416-423.1973
Peeples, M. E., Wang, C., Gupta, K. C., and Coleman, N. (1992). Nuclear entry and nucleolar localization of the Newcastle disease virus (NDV) matrix protein occur early in infection and do not require other NDV proteins. J. Virol. 66, 3263–3269. doi: 10.1128/JVI.66.5.3263-3269.1992
Pelletier, J., and Sonenberg, N. (1988). Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325. doi: 10.1038/334320a0
Pilipenko, E. V., Gmyl, A. P., Maslova, S. V., Svitkin, Y. V., Sinyakov, A. N., and Agol, V. I. (1992). Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cells 68, 119–131. doi: 10.1016/0092-8674(92)90211-t
Poitevin, F., Kushner, A., Li, X., and Dao Duc, K. (2020). Structural heterogeneities of the ribosome: new Frontiers and opportunities for Cryo-EM. Molecules 25:4262. doi: 10.3390/molecules25184262
Ponti, D., Troiano, M., Bellenchi, G. C., Battaglia, P. A., and Gigliani, F. (2008). The HIV tat protein affects processing of ribosomal RNA precursor. BMC Cell Biol. 9:32. doi: 10.1186/1471-2121-9-32
Quade, N., Boehringer, D., Leibundgut, M., van den Heuvel, J., and Ban, N. (2015). Cryo-EM structure of hepatitis C virus IRES bound to the human ribosome at 3.9-a resolution. Nat. Commun. 6:7646. doi: 10.1038/ncomms8646
Quaranta, P., Lottini, G., Chesi, G., Contrafatto, F., Russotto, R., Macera, L., et al. (2020). DDX3 inhibitors show antiviral activity against positive-sense single-stranded RNA viruses but not against negative-sense single-stranded RNA viruses: the coxsackie B model. Antivir. Res. 178:104750. doi: 10.1016/j.antiviral.2020.104750
Racine, T., and Duncan, R. (2010). Facilitated leaky scanning and atypical ribosome shunting direct downstream translation initiation on the tricistronic S1 mRNA of avian reovirus. Nucleic Acids Res. 38, 7260–7272. doi: 10.1093/nar/gkq611
Reid, D. W., Campos, R. K., Child, J. R., Zheng, T., Chan, K. W. K., Bradrick, S. S., et al. (2018). Dengue virus selectively annexes endoplasmic reticulum-associated translation machinery as a strategy for co-opting host cell protein synthesis. J. Virol. 92:e01766-17. doi: 10.1128/JVI.01766-17
Rethwilm, A., and Bodem, J. (2013). Evolution of foamy viruses: the most ancient of all retroviruses. Viruses 5, 2349–2374. doi: 10.3390/v5102349
Rivas-Aravena, A., Munoz, P., Jorquera, P., Diaz, A., Reinoso, C., Gonzalez-Catrilelbun, S., et al. (2017). Study of RNA-A initiation translation of the infectious pancreatic necrosis virus. Virus Res. 240, 121–129. doi: 10.1016/j.virusres.2017.07.014
Roberts, L. O., and Groppelli, E. (2009). An atypical IRES within the 5' UTR of a dicistrovirus genome. Virus Res. 139, 157–165. doi: 10.1016/j.virusres.2008.07.017
Rojas, M., Arias, C. F., and Lopez, S. (2010). Protein kinase R is responsible for the phosphorylation of eIF2alpha in rotavirus infection. J. Virol. 84, 10457–10466. doi: 10.1128/JVI.00625-10
Rollins, M. G., Shasmal, M., Meade, N., Astar, H., Shen, P. S., and Walsh, D. (2021). Negative charge in the RACK1 loop broadens the translational capacity of the human ribosome. Cell Rep. 36:109663. doi: 10.1016/j.celrep.2021.109663
Roth, H., Magg, V., Uch, F., Mutz, P., Klein, P., Haneke, K., et al. (2017). Flavivirus infection uncouples translation suppression from cellular stress responses. mBio 8:e02150-16. doi: 10.1128/mBio.02150-16
Schepetilnikov, M., Schott, G., Katsarou, K., Thiebeauld, O., Keller, M., and Ryabova, L. A. (2009). Molecular dissection of the prototype foamy virus (PFV) RNA 5'-UTR identifies essential elements of a ribosomal shunt. Nucleic Acids Res. 37, 5838–5847. doi: 10.1093/nar/gkp609
Sean, P., Nguyen, J. H., and Semler, B. L. (2008). The linker domain of poly(rC) binding protein 2 is a major determinant in poliovirus cap-independent translation. Virology 378, 243–253. doi: 10.1016/j.virol.2008.05.007
Segev, N., and Gerst, J. E. (2018). Specialized ribosomes and specific ribosomal protein paralogs control translation of mitochondrial proteins. J. Cell Biol. 217, 117–126. doi: 10.1083/jcb.201706059
Senanayake, S. D., and Brian, D. A. (1997). Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 48, 101–105. doi: 10.1016/s0168-1702(96)01423-2
Shi, Z., and Barna, M. (2015). Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 31, 31–54. doi: 10.1146/annurev-cellbio-100814-125346
Sloan, K. E., Warda, A. S., Sharma, S., Entian, K. D., Lafontaine, D. L. J., and Bohnsack, M. T. (2017). Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152. doi: 10.1080/15476286.2016.1259781
Slobodin, B., Sehrawat, U., Lev, A., Hayat, D., Zuckerman, B., Fraticelli, D., et al. (2022). Cap-independent translation and a precisely located RNA sequence enable SARS-CoV-2 to control host translation and escape anti-viral response. Nucleic Acids Res. 50, 8080–8092. doi: 10.1093/nar/gkac615
Smirnova, E., Firth, A. E., Miller, W. A., Scheidecker, D., Brault, V., Reinbold, C., et al. (2015). Discovery of a small non-AUG-initiated ORF in Poleroviruses and Luteoviruses that is required for long-distance movement. PLoS Pathog. 11:e1004868. doi: 10.1371/journal.ppat.1004868
Sokoloski, K. J., Snyder, A. J., Liu, N. H., Hayes, C. A., Mukhopadhyay, S., and Hardy, R. W. (2013). Encapsidation of host-derived factors correlates with enhanced infectivity of Sindbis virus. J. Virol. 87, 12216–12226. doi: 10.1128/JVI.02437-13
Stein, K. C., Morales-Polanco, F., van der Lienden, J., Rainbolt, T. K., and Frydman, J. (2022). Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis. Nature 601, 637–642. doi: 10.1038/s41586-021-04295-4
Sun, Q., Zhu, X., Qi, J., An, W., Lan, P., Tan, D., et al. (2017). Molecular architecture of the 90S small subunit pre-ribosome. elife 6:e22086. doi: 10.7554/eLife.22086
Sweeney, T. R., Dhote, V., Yu, Y., and Hellen, C. U. (2012). A distinct class of internal ribosomal entry site in members of the kobuvirus and proposed salivirus and paraturdivirus genera of the picornaviridae. J. Virol. 86, 1468–1486. doi: 10.1128/JVI.05862-11
Thompson, S. R., and Sarnow, P. (2003). Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology 315, 259–266. doi: 10.1016/s0042-6822(03)00544-0
Tidu, A., Janvier, A., Schaeffer, L., Sosnowski, P., Kuhn, L., Hammann, P., et al. (2020). The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA 27, 253–264. doi: 10.1261/rna.078121.120
Tolbert, M., Morgan, C. E., Pollum, M., Crespo-Hernandez, C. E., Li, M. L., Brewer, G., et al. (2017). HnRNP A1 alters the structure of a conserved enterovirus IRES domain to stimulate viral translation. J. Mol. Biol. 429, 2841–2858. doi: 10.1016/j.jmb.2017.06.007
Tomar, P. P. S., Krugliak, M., Singh, A., and Arkin, I. T. (2022). Zika M-A potential viroporin: mutational study and drug repurposing. Biomedicine 10:641. doi: 10.3390/biomedicines10030641
Toribio, R., Diaz-Lopez, I., Boskovic, J., and Ventoso, I. (2016). An RNA trapping mechanism in alphavirus mRNA promotes ribosome stalling and translation initiation. Nucleic Acids Res. 44, 4368–4380. doi: 10.1093/nar/gkw172
Toribio, R., Diaz-Lopez, I., Boskovic, J., and Ventoso, I. (2018). Translation initiation of alphavirus mRNA reveals new insights into the topology of the 48S initiation complex. Nucleic Acids Res. 46, 4176–4187. doi: 10.1093/nar/gky071
Ullah, H., Hou, W., Dakshanamurthy, S., and Tang, Q. (2019). Host targeted antiviral (HTA): functional inhibitor compounds of scaffold protein RACK1 inhibit herpes simplex virus proliferation. Oncotarget 10, 3209–3226. doi: 10.18632/oncotarget.26907
Valiente-Echeverria, F., Hermoso, M. A., and Soto-Rifo, R. (2015). RNA helicase DDX3: at the crossroad of viral replication and antiviral immunity. Rev. Med. Virol. 25, 286–299. doi: 10.1002/rmv.1845
Vallejos, M., Ramdohr, P., Valiente-Echeverria, F., Tapia, K., Rodriguez, F. E., Lowy, F., et al. (2010). The 5′-untranslated region of the mouse mammary tumor virus mRNA exhibits cap-independent translation initiation. Nucleic Acids Res. 38, 618–632. doi: 10.1093/nar/gkp890
Vidal, S., Curran, J., and Kolakofsky, D. (1990). Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J. Virol. 64, 239–246. doi: 10.1128/JVI.64.1.239-246.1990
von Kobbe, C., van Deursen, J. M., Rodrigues, J. P., Sitterlin, D., Bachi, A., Wu, X., et al. (2000). Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell 6, 1243–1252. doi: 10.1016/s1097-2765(00)00120-9
Walker, E. J., Younessi, P., Fulcher, A. J., McCuaig, R., Thomas, B. J., Bardin, P. G., et al. (2013). Rhinovirus 3C protease facilitates specific nucleoporin cleavage and mislocalization of nuclear proteins in infected host cells. PLoS One 8:e71316. doi: 10.1371/journal.pone.0071316
Walter, B. L., Parsley, T. B., Ehrenfeld, E., and Semler, B. L. (2002). Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76, 12008–12022. doi: 10.1128/jvi.76.23.12008-12022.2002
Walters, B., Axhemi, A., Jankowsky, E., and Thompson, S. R. (2020). Binding of a viral IRES to the 40S subunit occurs in two successive steps mediated by eS25. Nucleic Acids Res. 48, 8063–8073. doi: 10.1093/nar/gkaa547
Wengler, G., Wurkner, D., and Wengler, G. (1992). Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology 191, 880–888. doi: 10.1016/0042-6822(92)90263-o
Wennesz, R., Luttermann, C., Kreher, F., and Meyers, G. (2019). Structure-function relationship in the 'termination upstream ribosomal binding site' of the calicivirus rabbit hemorrhagic disease virus. Nucleic Acids Res. 47, 1920–1934. doi: 10.1093/nar/gkz021
Willcocks, M. M., Carter, M. J., and Roberts, L. O. (2004). Cleavage of eukaryotic initiation factor eIF4G and inhibition of host-cell protein synthesis during feline calicivirus infection. J. Gen. Virol. 85, 1125–1130. doi: 10.1099/vir.0.19564-0
Willcocks, M. M., Zaini, S., Chamond, N., Ulryck, N., Allouche, D., Rajagopalan, N., et al. (2017). Distinct roles for the IIId2 sub-domain in pestivirus and picornavirus internal ribosome entry sites. Nucleic Acids Res. 45, 13016–13028. doi: 10.1093/nar/gkx991
Wilson, J. E., Powell, M. J., Hoover, S. E., and Sarnow, P. (2000). Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 20, 4990–4999. doi: 10.1128/MCB.20.14.4990-4999.2000
Wu, H., Huang, J., Liu, Y., Pan, Y., Li, Y., Miao, Q., et al. (2021). Feline Calicivirus proteinase-polymerase protein degrades mRNAs to inhibit host gene expression. J. Virol. 95:e0033621. doi: 10.1128/JVI.00336-21
Xiao, Y., Rouzine, I. M., Bianco, S., Acevedo, A., Goldstein, E. F., Farkov, M., et al. (2016). RNA recombination enhances adaptability and is required for virus spread and virulence. Cell Host Microbe 19, 493–503. doi: 10.1016/j.chom.2016.03.009
Xu, J., Cao, Z., Ji, C., Zhou, L., Yan, X., Sun, Y., et al. (2022). Analysis of interaction network between host protein and M protein of swine acute diarrhea syndrome coronavirus. Front. Microbiol. 13:858460. doi: 10.3389/fmicb.2022.858460
Yamamoto, H., Collier, M., Loerke, J., Ismer, J., Schmidt, A., Hilal, T., et al. (2015). Molecular architecture of the ribosome-bound hepatitis C virus internal ribosomal entry site RNA. EMBO J. 34, 3042–3058. doi: 10.15252/embj.201592469
Yang, L., Toh, D. K., Krishna, M. S., Zhong, Z., Liu, Y., Wang, S., et al. (2020). Tertiary Base triple formation in the SRV-1 frameshifting pseudoknot stabilizes secondary structure components. Biochemistry 59, 4429–4438. doi: 10.1021/acs.biochem.0c00611
Yang, X., Zeng, Q., Wang, M., Cheng, A., Pan, K., Zhu, D., et al. (2018). DHAV-1 2A1 peptide - a newly discovered co-expression tool that mediates the ribosomal "skipping" function. Front. Microbiol. 9:2727. doi: 10.3389/fmicb.2018.02727
Yokoyama, T., Machida, K., Iwasaki, W., Shigeta, T., Nishimoto, M., Takahashi, M., et al. (2019). HCV IRES captures an actively translating 80S ribosome. Mol. Cell 74:e1208, 1205–1214.e8. doi: 10.1016/j.molcel.2019.04.022
You, J., Dove, B. K., Enjuanes, L., DeDiego, M. L., Alvarez, E., Howell, G., et al. (2005). Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Gen. Virol. 86, 3303–3310. doi: 10.1099/vir.0.81076-0
Yu, Y., Ji, H., Doudna, J. A., and Leary, J. A. (2005). Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci. 14, 1438–1446. doi: 10.1110/ps.041293005
Yu, Y., Sweeney, T. R., Kafasla, P., Jackson, R. J., Pestova, T. V., and Hellen, C. U. (2011). The mechanism of translation initiation on aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 30, 4423–4436. doi: 10.1038/emboj.2011.306
Yuan, S., Peng, L., Park, J. J., Hu, Y., Devarkar, S. C., Dong, M. B., et al. (2020). Non-structural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol. Cell 80:e1056, 1055–1066.e6. doi: 10.1016/j.molcel.2020.10.034
Zan, M., Evans, P., and Lucas-Lenard, J. (1990). The inhibition of mouse L-cell 45 S ribosomal RNA processing is a highly uv-resistant property of vesicular stomatitis virus. Virology 177, 75–84. doi: 10.1016/0042-6822(90)90461-y
Zhao, D., Zhang, L., Song, M., and Mail, Y. Z. (2022). RPS3-induced antiviral cytokines inhibit the proliferation of classical swine fever virus. Acta Virol. 66, 55–64. doi: 10.4149/av_2022_107
Zhu, Z., Li, W., Zhang, X., Wang, C., Gao, L., Yang, F., et al. (2020). Foot-and-mouth disease virus capsid protein VP1 interacts with host ribosomal protein SA to maintain activation of the MAPK signal pathway and promote virus replication. J. Virol. 94:e01350-19. doi: 10.1128/JVI.01350-19
Zidane, N., Ould-Abeih, M. B., Petit-Topin, I., and Bedouelle, H. (2012). The folded and disordered domains of human ribosomal protein SA have both idiosyncratic and shared functions as membrane receptors. Biosci. Rep. 33, 113–124. doi: 10.1042/BSR20120103
Zinoviev, A., Hellen, C. U. T., and Pestova, T. V. (2015). Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol. Cell 57, 1059–1073. doi: 10.1016/j.molcel.2015.01.039
Keywords: ribosome, translation, RNA viruses, heterogeneous ribosomes, protein
Citation: Wang X, Zhu J, Zhang D and Liu G (2022) Ribosomal control in RNA virus-infected cells. Front. Microbiol. 13:1026887. doi: 10.3389/fmicb.2022.1026887
Received: 24 August 2022; Accepted: 19 October 2022;
Published: 07 November 2022.
Edited by:
Fang He, Zhejiang University, ChinaReviewed by:
Shanhui Ren, Lanzhou Veterinary Research Institute (CAAS), ChinaCopyright © 2022 Wang, Zhu, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangqing Liu, bGl1Z3FAc2h2cmkuYWMuY24=; Jie Zhu, emoxMjFAc2h2cmkuYWMuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.