- 1College of Pharmacy, Duksung Women’s University, Seoul, South Korea
- 2Innovative Drug Center, Duksung Women’s University, Seoul, South Korea
Caffeine, a methylxanthine derivative, affects various physiological conditions such as cell growth, proliferation, and energy metabolism. A genome-wide screening for genes required for caffeine resistance in Schizosaccharomyces pombe revealed several candidates, including Pap1 and downstream target genes involved in caffeine efflux. We found that Yap1, a budding yeast AP-1 homolog required for oxidative stress response, has a caffeine tolerance function. Although the Yap1 mutant is not sensitive to caffeine, overexpression of Yap1 renders cells resistant to high concentrations of caffeine. Caffeine sensitivity of mutants lacking two multidrug transporters, Pdr5 or Snq2, is completely recovered by Yap1 overexpression. Among Yap1-dependent target genes, FLR1, a fluconazole-resistant gene, is necessary but not sufficient for caffeine tolerance. Low concentrations of hydrogen peroxide induce Yap1 activation, which restores cell viability against caffeine toxicity. Intriguingly, oxidative stress-mediated cellular adaptation to caffeine toxicity requires Yap1, but not Flr1. Moreover, caffeine is involved in reduction of intracellular reactive oxygen species (ROS), as well as mutation rate and Rad52 foci formation. Altogether, we identified novel reciprocal crosstalk between ROS signaling and caffeine resistance.
Introduction
Caffeine is a natural purine analog and one of the most frequently ingested methylxanthine derivatives in beverages and medications. It has a wide variety of biological effects with multiple cellular targets in fungi, plants, and humans. In higher animals, caffeine plays beneficial roles in neuronal stimulation accompanied by antidepressant and diuretic effects (López-Cruz et al., 2018). Caffeine is a relatively non-selective drug that affects multiple cellular processes such as cell growth, proliferation, and energy metabolism by triggering calcium mobilization and inhibiting the target of rapamycin (TOR) signaling as well as the DNA damage repair system (Combettes et al., 1994; Ruta and Farcasanu, 2020).
Several studies have reported that in yeast, caffeine threatens cell wall integrity and causes cytotoxicity by perturbing several steps in DNA damage repair pathways and cell cycle checkpoint signaling (Martin et al., 2000; Moser et al., 2000; Kuranda et al., 2006). Caffeine inhibits the repair of UV-induced pyrimidine dimers by interfering with DNA photolyase and UvrA. It also disrupts the recruitment of Sae2 and Dna2 nucleases required for break end resection, one of the early steps in homologous recombination (HR), and Rad51, a strand exchange protein that detects repair homology, leading to failure in DNA double-strand break (DSB) repair (Selby and Sancar, 1990; Zelensky et al., 2013; Tsabar et al., 2015a, 2015b).
A genome-scale screen for caffeine-sensitive mutants in the fission yeast Schizosaccharomyces pombe has revealed that mutants lacking RAD3, RAD51, RAD54, and SSB3, which are responsible for HR and damage checkpoint activation, are all highly sensitive to caffeine (Calvo et al., 2009). Caffeine disturbs cell cycle progression and exerts mutagenic effects through the suppression of Tel1 and Mec1, yeast homologs of ataxia-telangiectasia mutated (ATM) and ATM-related (ATR) kinases for checkpoint signaling (Saiardi et al., 2005).
Interestingly in S. pombe, caffeine sensitivity and toxicity are also involved in the level of intracellular reactive oxygen species (ROS) and the ensuing activation of the oxidative stress response. Significant caffeine tolerance has been observed in mutants with increased activity of Pap1, an AP-1-like transcription factor in S. pombe, that induces the expression of various antioxidant enzymes against oxidative stress (Benko et al., 2004). Cells lacking Pap1 display a highly sensitive phenotype to caffeine, and in this case, caffeine resistance is largely due to Hba2, a Pap1-mediated drug efflux pump (Calvo et al., 2009). The lack of Trr1, a thioredoxin reductase, causes an elevation of intracellular ROS leading to constitutive activation of Pap1 and caffeine tolerance (Vivancos et al., 2004; Calvo et al., 2009). These observations indicate that cellular responses to caffeine might be linked to the oxidative stress response pathway or the ROS signaling process (Chung, 2021).
Yap1, a Pap1 homolog in Saccharomyces cerevisiae, is crucial for the oxidative stress response (Rodrigues-Pousada et al., 2019). Yap1 shuttles between the cytoplasm and the nucleus mediated by Crm1, an exportin, and by Pse1, an importin (Yan et al., 1998; Isoyama et al., 2001). Unlike S. pombe counterparts that are dispensable for survival, both Crm1 and Pse1 are essential in S. cerevisiae, indicating that the localization or cellular distribution of Yap1 is critical to maintaining cellular homeostasis. Overexpression of Yap1 renders cells resistant to several toxic chemicals, such as diazaborine and cadmium, mediated by the transcriptional induction of Flr1 and Ycf1, two multidrug transporters, in a Yap1-dependent manner (Wemmie et al., 1994; Jungwirth et al., 2000).
In this study, we assessed whether the oxidative stress pathway is involved in caffeine tolerance in S. cerevisiae. Strong caffeine resistance was observed when Yap1 is overexpressed, whereas the lack of Yap1 made no difference in caffeine sensitivity. We discovered that Flr1, a fluconazole-resistant transporter, is mainly responsible for caffeine tolerance under Yap1 activation. While intracellular localization and/or activation of Yap1 is not affected by caffeine, Yap1-mediated oxidative stress response confers resistance to caffeine toxicity. Based on the results that caffeine reduces the level of intracellular ROS, spontaneous mutation frequency, as well as DNA damage-induced foci formation, we suggest a significant reciprocal crosstalk between ROS signaling and caffeine resistance, which provides a cooperative mechanism for the maintenance of cell integrity.
Materials and methods
Strains, plasmids, and growth media
All yeast strains used in this study are isogenic derivatives of S. cerevisiae BY4741 (MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) obtained from Yeast Knockout (YKO) collection (YSC1053 glycerol stock, Thermo Fisher Scientific). The genotypes of all strains used in this study are listed in Supplementary Table 1. The strains with C-terminally GFP-fused proteins, Yap1-GFP and Rad52-GFP, were constructed by oligonucleotide-directed in-frame tagging method as previously described (Huh et al., 2003). Yeast cell cultures and treatments with caffeine, hydrogen peroxide (H2O2) or phleomycin were performed in YEPD (1% yeast extract, 2% peptone, and 2% dextrose) or minimal SD media supplemented with the required amino acids. Overexpression plasmids including YAP1, SKN7, SOD1, TSA1, or FLR1 were created using high copy number shuttle vector pRS426 with the wild-type ADH1 promoter (Mumberg et al., 1995).
Drug sensitivity analysis
The sensitivity of yeast cells to drugs was determined by spot plating assay. A small part of the overnight-grown cell culture was taken and grown again in fresh liquid media to reach mid-log phase (3–4 × 107 cells/ml). Cells were then diluted 10-fold serially and spotted in rows onto YEPD or synthetic dropout (SD) plates containing H2O2 and/or caffeine. After spotting, cells were incubated for 3 days at 30°C and then photographed.
Fluorescence microscopy
Fluorescence microscopic pictures were acquired with Leica DMi8 automated microsystems. LAS X software was used for the image analysis. To detect localization of Yap1-GFP to the nucleus, green fluorescent cells merged with DAPI stain signals were quantified after H2O2 or caffeine treatment. To measure the amount of DSB lesions and genomic instability, the percentage of cells with subnuclear Rad52-GFP foci formation in the population was determined (Lisby et al., 2001). At least 300 individual cells for each experiment were counted and analyzed manually.
RNA isolation and reverse transcription-PCR (RT-PCR) analysis
Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The quality of the isolated RNA was assessed by Agilent 2,100 bioanalyzer using the RNA 6000 Nano Chip (Agilent Technologies). The concentration of RNA was determined by measuring the UV absorbance at 260 nm in a spectrophotometer, and highly purified samples with comparable A260/A280 ratios (1.8 ~ 2.1) were used for RT-PCR analysis. The sequences of the PCR primer pairs for the amplification of target genes were shown in Supplementary Table 2. PCR was performed with T100 thermal cycler (BIO-RAD). All amplification set-up went through 22 ~ 28 cycles unless noted otherwise.
The QuantSeq mRNA sequencing and data analysis
The 3’ mRNA-Seq libraries were prepared using QuantSeq 3’ mRNA-Seq Library Prep Kit (Lexogen) following the manufacturer’s instructions. Each 500 ng total RNA was hybridized to the oligo-dT primer containing an Illumina-compatible sequence at its 5′ end. After reverse-transcription and degradation of the RNA template, second strand synthesis was initiated by a random primer containing an Illumina-compatible linker sequence at its 5′ end and was purified. The library was amplified to add the complete adapter sequences required for cluster generation. High-throughput sequencing was determined using the single-end 75 sequencing platform of NextSeq 500 (Illumina). QuantSeq 3′ sequencing reads were aligned using Bowtie2 software (Langmead and Salzberg, 2012). Differential gene expression was evaluated based on counts from unique and multiple alignments using coverage in Bedtools and was determined using EdgeR within R (R development Core Team, 2016). Gene classification was based on searches done by DAVID1 and Medline databases.2 Student’s t-test was used to determine significant differences (value of p < 0.05). All of raw sequencing data is deposited at NCBI GEO site.3
Measurement of intracellular ROS level
To measure intracellular ROS level in yeast, cells were grown to exponential phase and diluted to an OD600 of ~0.2. H2DCFDA (2′,7′-dichlorodihydrofluorescein diacetate, Thermo Fisher Scientific), a cell-permeant indicator for ROS detection, was added at a final concentration of 5 μg/ml, and then cells were incubated with shaking for 2 h at 30°C. Intracellular ROS levels were determined using a BD FACS Canto II flow cytometer (Becton Dickinson) as previously described (Madeo et al., 1999). A baseline of zero (background level of fluorescence) was set based on the maximum value of control sample without the ROS indicator. The cells with higher ROS level than background were counted and converted into a percentage. All values represent the average of at least three independent experiments.
Measurement of cell viability and mutation frequency
Yeast cells were inoculated into 5 ml of YEPD media and grown overnight at 30°C. The next day, cells were diluted into 5 ml of fresh media to an OD600 of ~0.2 and incubated with shaking for 6 h. More than 100 cells were counted and plated onto three YEPD plates including H2O2 and/or caffeine at the indicated final concentrations. The plates were incubated at 30°C for 3 days and then colonies were counted for a colony forming unit (CFU) assay. The rate of spontaneous or induced mutations as a result of genome instability was measured by a forward-mutation assay that detects mutations in the CAN1 gene (Motegi and Myung, 2007). Yeast cells were treated with H2O2 and/or caffeine for 2 h at the indicated concentrations and then were grown on plates with or without 60 μg/ml canavanine. Spontaneous mutation rates were determined by counting CFUs after incubation for 3–4 days at 30°C. All rates represent the average of three independent experiments and error bars indicate the standard deviation.
Results
Cells lacking Yap1 are not sensitive to caffeine whereas overexpression of Yap1 confers resistance
The screening analysis of several caffeine-resistant mutants using fission yeast S. pombe strains has revealed that all the caffeine-tolerant phenotypes identified are related to an elevated expression of Pap1 or its constitutive localization in the nucleus (Kumada et al., 1996; Benko et al., 2004). In S. pombe, there are two alternative oxidative stress responses by which different signaling pathways are triggered. One is mediated by Pap1, an AP-1-like transcription factor, responding to moderate concentrations of H2O2, and the other is the Sty1 MAP kinase pathway activated not only by oxidative stress but also by general stress conditions such as heat shock and osmotic stress (Toone et al., 1998; Vivancos et al., 2004). Overexpression of Pap1, but not Sty1, makes cells resistant to high doses of caffeine, and Pap1- and Sty1-deficient cells are both highly sensitive to caffeine (Benko et al., 2004; Calvo et al., 2009).
Based on these observations, we examined the caffeine sensitivity of cells lacking Yap1 or Skn7, two specialized transcriptional regulators for oxidative stress response in S. cerevisiae. Skn7 collaborates with Yap1 on several promoters such as TRX2 and AHP1 to induce transcription in response to oxidative stress (Morgan et al., 1997; Lee et al., 1999). In contrast with the homologs in S. pombe, we could not observe any caffeine sensitivity in yap1 or skn7 null mutants (Figure 1A). The expression of Sod1 and Tsa1, two representative antioxidant enzymes, is mainly under the control of Yap1 and/or Skn7. When caffeine sensitivity was examined in cells lacking Sod1 or Tsa1, no difference in sensitivity was shown. Next we overexpressed YAP1, SKN7, SOD1, or TSA1 in WT yeast cells using the multicopy plasmid pRS426ADH, and found that only the expression of Yap1 confers yeast cells with significantly enhanced resistance to high doses of caffeine (Figure 1B).
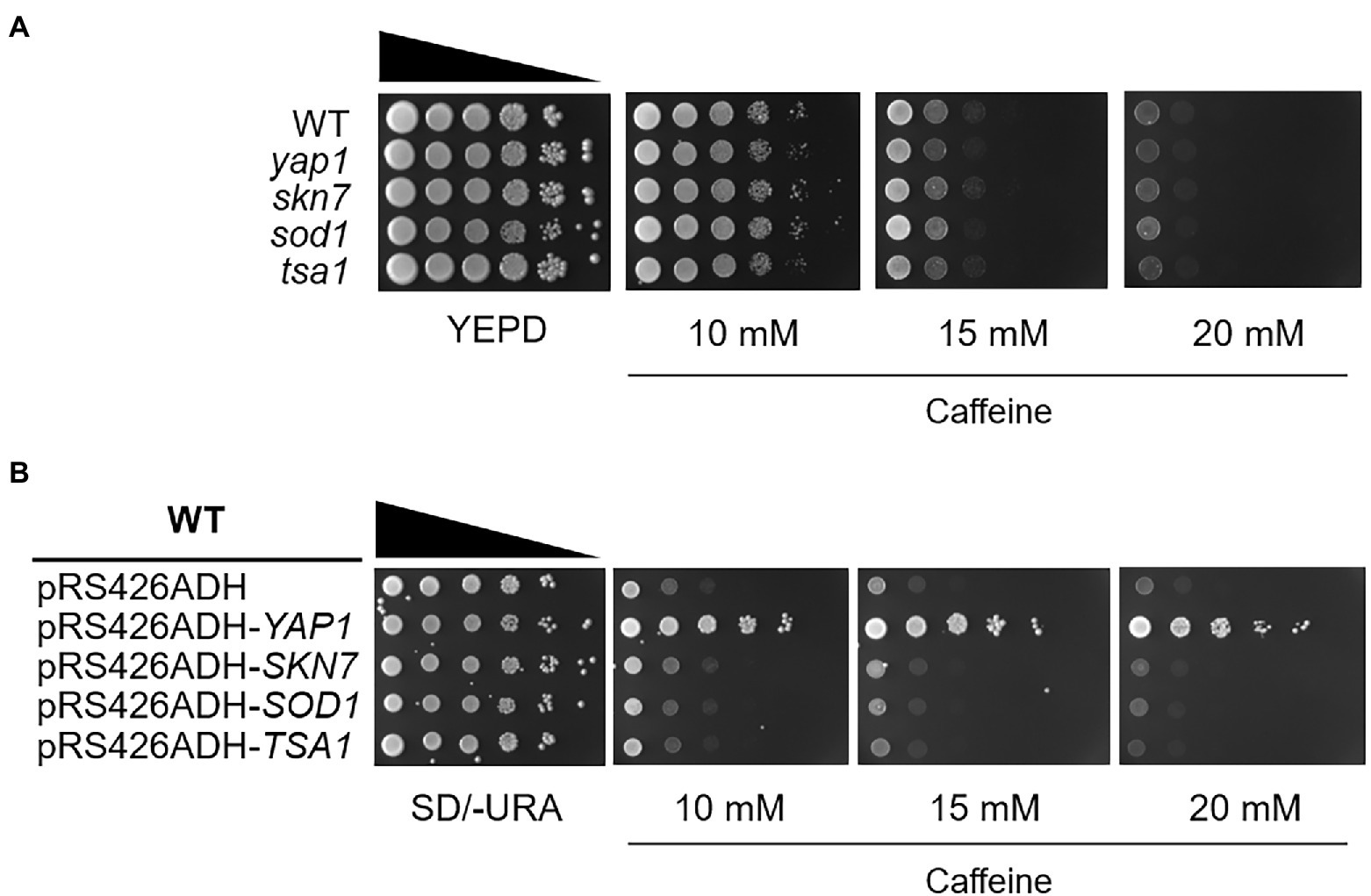
Figure 1. The yap1 null mutant is not sensitive to caffeine while overexpression of Yap1 confers significant resistance. (A) 10-fold serial dilutions of yeast mutant strains were spotted on YEPD media in the presence of caffeine and incubated for 3 days at 30°C. (B) 10-fold serial dilutions of yeast overexpression strains were spotted on SD/-URA media containing caffeine and incubated for 3 days at 30°C.
Caffeine sensitivity of the flr1 null mutant is not overcome by Yap1 overexpression
Previously, in multicopy vector-based genomic library screening, two ATP-binding cassette (ABC) transporters, Pdr5, and Snq2, have been identified as major caffeine efflux pumps in S. cerevisiae (Kolaczkowski et al., 1996; Tsujimoto et al., 2015). We evaluated whether caffeine tolerance mediated by Yap1 is involved in the function of these multidrug resistance transporters. Consistent with the previous report that Snq2 is more functional in caffeine efflux than Pdr5, the snq2 mutant was slightly more sensitive to caffeine than the pdr5 mutant, and the pdr5 snq2 double mutant showed even more sensitivity than each of the single mutants (Figure 2A). However, we observed that the overexpression of Yap1 almost completely surmounts caffeine sensitivity in the pdr5 snq2 double mutant as well as each of the single mutants. These results suggest that caffeine tolerance is associated with elevated levels of Yap1 and is not derived from an improvement in caffeine efflux mediated by either Pdr5 or Snq2 (Figure 2A).
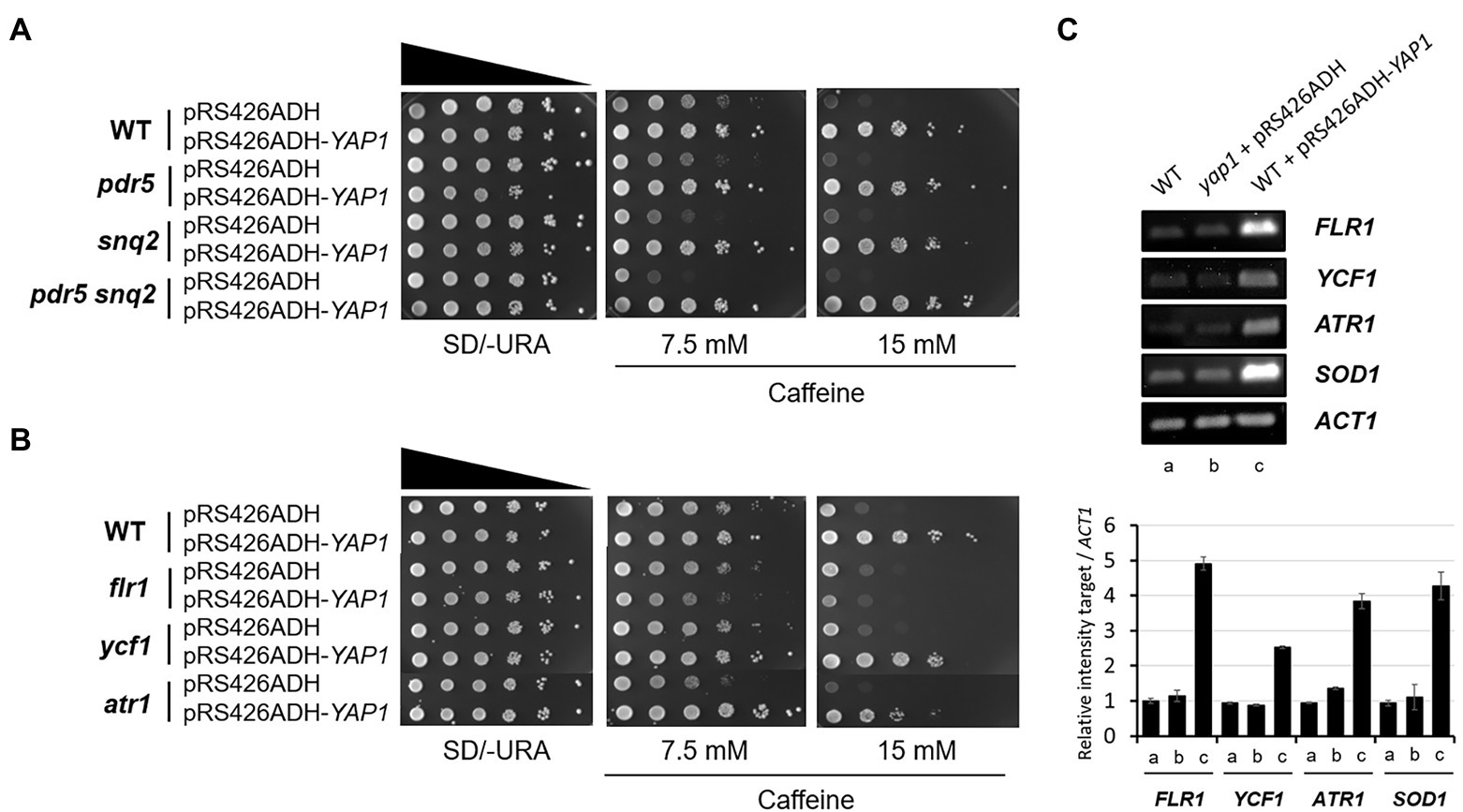
Figure 2. Caffeine sensitivity of flr1 mutant is not recovered by Yap1 overexpression. (A,B) 10-fold serial dilutions of yeast mutant strains with YAP1 overexpression plasmids were spotted on SD/-URA media containing caffeine and incubated for 3 days at 30°C. (C) RT-PCR analysis of Yap1-dependent target genes expressed in WT, yap1 and YAP1 overexpression strains. ACT1 as a loading control. The products were analyzed by electrophoresis on a 1.5% agarose gel. The numbers of PCR cycles used are 22 (FLR1 and SOD1), 28 (YCF1 and ATR1) and 30 (ACT1), respectively.
Flr1, Ycf1 and Atr1, three efflux pumps in S. cerevisiae, confer resistance to several drugs under the control of Yap1. Ycf1 is required for cadmium tolerance and arsenite detoxification (Wemmie et al., 1994; Ghosh et al., 1999). Flr1 is essential for Yap1-mediated resistance to fluconazole, cycloheximide, aminotriazole, and 4-nitroquinoline-N-oxide (4-NQO; Alarco et al., 1997), and both Ycf1 and Flr1 are involved in Yap1-mediated defense against cerulenin and diazaborine toxicity (Oskouian and Saba, 1999; Jungwirth et al., 2000). In addition, Atr1 is responsible for resistance to 4-NQO in both Yap1- and Gcn4-dependent manners (Coleman et al., 1997).
To examine whether these membrane-associated transporters are involved in the acquisition of caffeine resistance in a Yap1-related manner, each deletion mutant was spotted onto caffeine-infused plates with or without Yap1 overexpression. Except for a slightly sensitive atr1 mutant, neither the flr1 nor the ycf1 mutant was found to be sensitive to caffeine compared to the WT strain under normal conditions (Figure 2B). Interestingly, however, caffeine resistance of the flr1 mutant is not enhanced by Yap1 overexpression at all. The caffeine resistance of ycr1 and atr1 mutants is just slightly less than that of WT strain. These results suggest that FLR1 is a major target gene regulated by Yap1 and is responsible for a specific Yap1-dependent caffeine tolerance. Analysis with reverse transcription-PCR (RT-PCR) revealed that the expression level of Flr1 is not almost altered in yap1 mutants compared to WT and increases significantly in cells that overexpress Yap1 (Figure 2C). When Yap1 is overexpressed, the amount of transcriptional induction in FLR1 is higher than that of YCF1 or ATR1. This accounts for our previous results that there is no difference in caffeine sensitivity between WT and the yap1 mutant but Yap1 overexpression increases resistance (Figure 1).
Flr1 is required but not sufficient for the increase of Yap1-dependent caffeine tolerance
Contrary to mutant cells carrying deletions in both PDR5 and SNQ2 that have considerable caffeine sensitivity, the flr1 mutant is as sensitive to caffeine as the WT strain, and the pdr5 snq2 flr1 triple mutant is as sensitive as the pdr5 snq2 double mutant. This suggests that the Flr1 transporter alone is not significantly involved in caffeine efflux when compared to Pdr5 and Snq2 under no Yap1 overexpression conditions (Figure 3A).
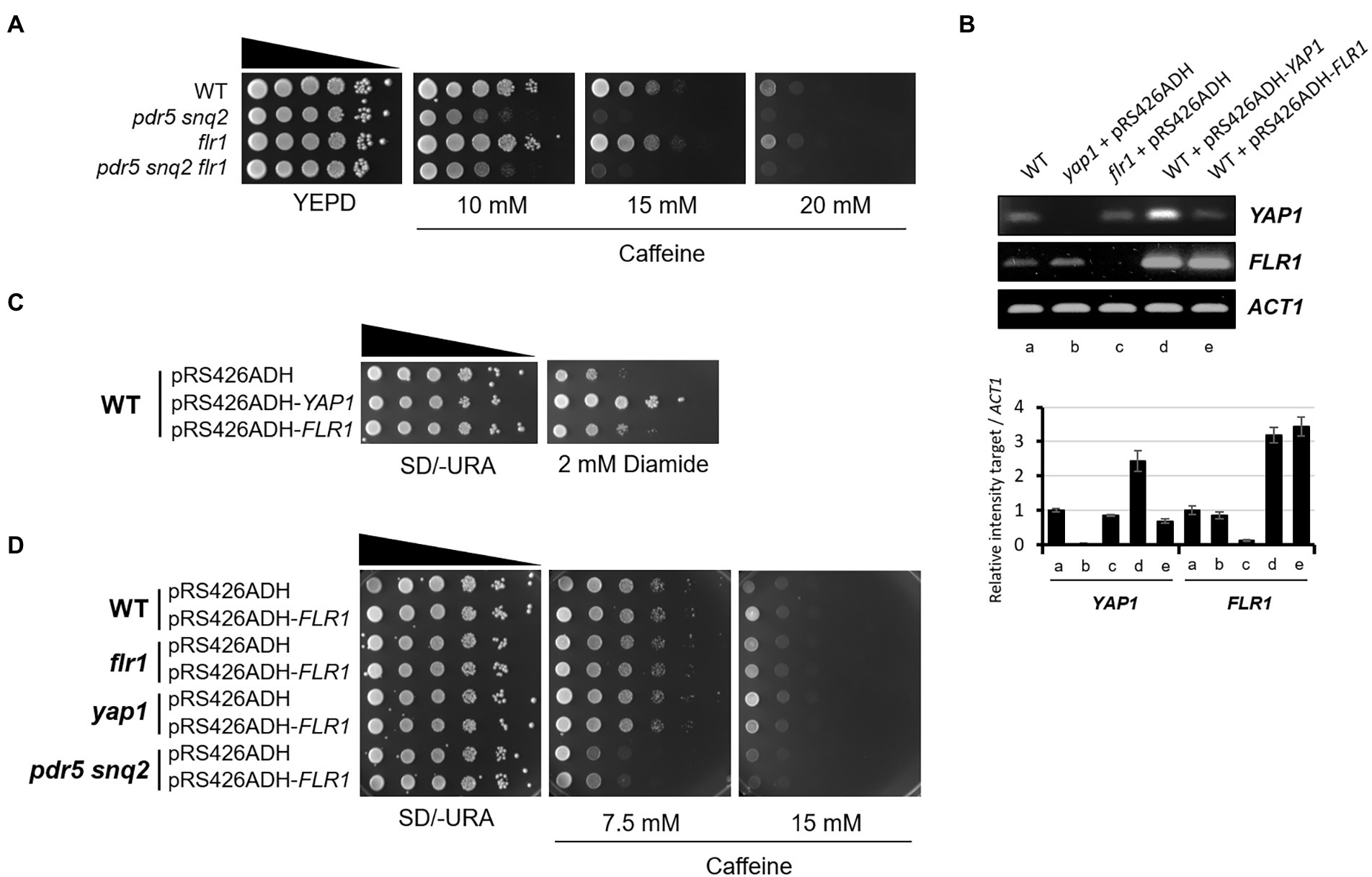
Figure 3. Flr1 is required but not sufficient for the increase of Yap1-dependent caffeine tolerance. (A) 10-fold serial dilutions of yeast mutant strains were spotted on YEPD media containing caffeine and incubated for 3 days at 30°C. (B) RT-PCR analysis of YAP1 and FLR1 expressed in WT, yap1, flr1, and strains overexpressing YAP1 or FLR1. ACT1 as a loading control. The numbers of PCR cycles used are 28 (YAP1), 22 (FLR1) and 25 (ACT1), respectively. (C) 10-fold serial dilution of yeast strains with YAP1 or FLR1 overexpression were spotted on SD/-URA media containing diamide and incubated for 3 days at 30°C. (D) 10-fold serial dilutions of yeast mutant strains with FLR1 overexpression plasmids were spotted on SD/-URA media containing caffeine and incubated for 3 days at 30°C.
To investigate whether Flr1 could functionally substitute Pdr5 or Snq2 in caffeine export, we overexpressed FLR1 in a pdr5 snq2 double mutant and tested its caffeine resistance. FLR1 was overexpressed under the control of the ADH1 promoter and its transcription level was compatible with that of Yap1 overexpression strain (Figure 3B). Consistent with the previous report by Nguyên et al. (2001), the amount of Flr1 produced was also enough to show resistance to diamide, a thiol oxidizing agent (Figure 3C). However, we found that the caffeine sensitivity of the pdr5 snq2 double mutant was not sufficiently recovered by Flr1 overexpression, indicating that Flr1 is not functionally redundant with Pdr5 or Snq2 in caffeine tolerance (Figure 3D). This also implies that Flr1 alone is not enough to endure a caffeine threat or to show strong caffeine resistance and cells still need to employ more unknown target(s) mediated by activated Yap1 besides the Flr1 transporter.
Neither Yap1 activation nor Flr1 expression is induced by caffeine
Previous studies have shown that Yap1 is activated and accumulates in the nucleus specifically under oxidative stress conditions and its absence causes hypersensitivity to several oxidants such as hydroperoxides and diamide (Kuge and Jones, 1994; Rodrigues-Pousada et al., 2019). We examined whether Yap1 and the transcription of its downstream target genes are activated by caffeine. Yap1 is activated and is relocated to the nucleus within 30 min of hydrogen peroxide treatment, whereas it is largely cytoplasmic with no changes after caffeine treatment even at high concentrations (Figure 4A).
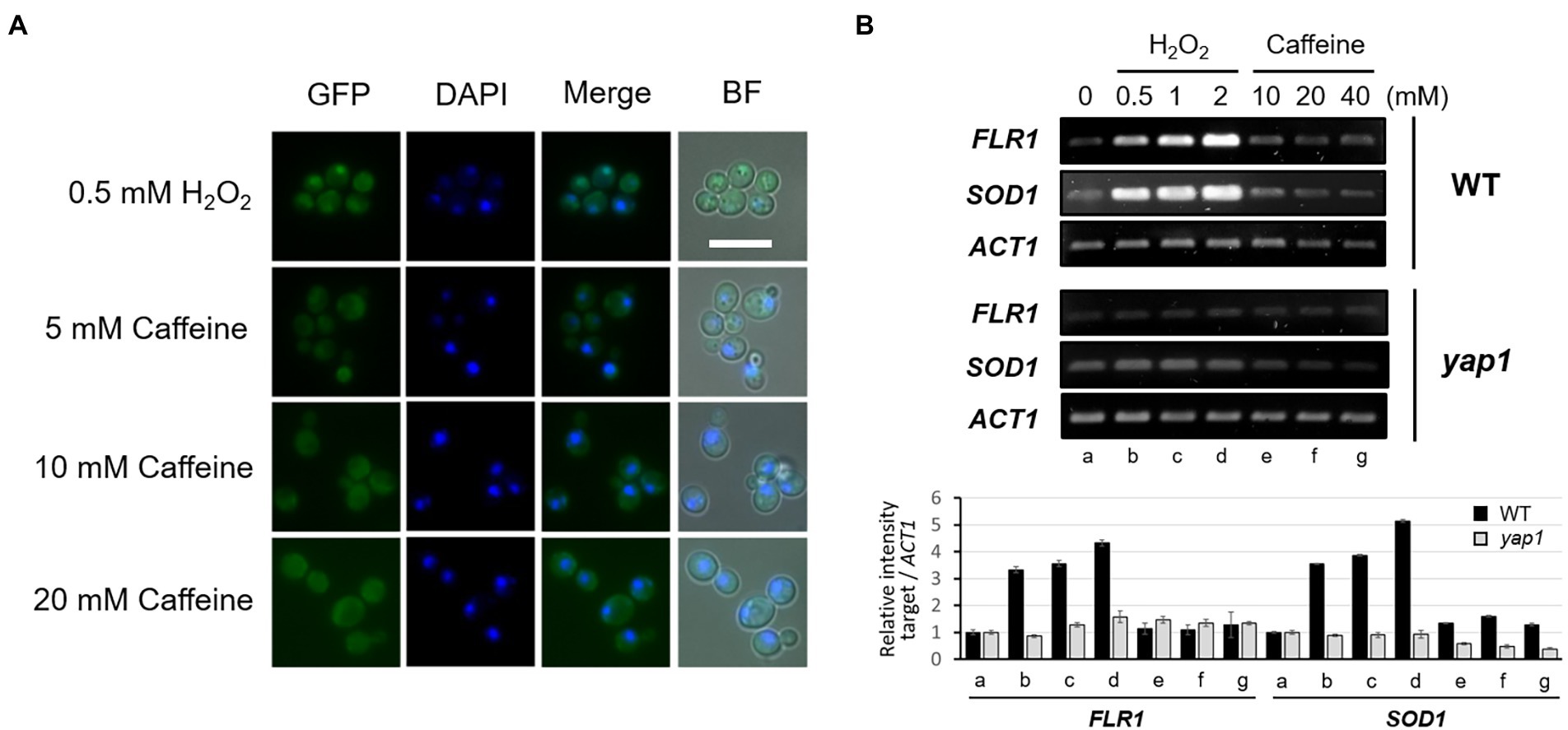
Figure 4. Activation of Yap1 and its intracellular localization are not affected by caffeine treatment. (A) Fluorescent microscopy of the spontaneous or induced nuclear localization of Yap1-GFP in WT with H2O2 or caffeine treatment. White scale bar corresponds to 10 μm. (B) RT-PCR analysis of FLR1 and SOD1 expression in WT and yap1 mutant. ACT1 as a loading control. Cells were incubated at 30°C for 30 min with H2O2 or caffeine treatment. The numbers of PCR cycles used are 25 (SOD1) and 28 (FLR1 and ACT1), respectively.
We next performed RT-PCR analysis to inquire about a possible alteration in FLR1 expression level after H2O2 or caffeine treatment. As shown in Figure 4B, transcription of FLR1 is induced by H2O2, but not by caffeine, only in the presence of functional Yap1, and its induction pattern is quite similar to that of SOD1, another target of Yap1 induction. Caffeine tolerance rendered by Yap1 overexpression in the presence of Flr1 was induced by the oxidative stress response, but not by caffeine itself. These results imply that intracellular caffeine response and ROS signaling may have a cooperative linkage to maintain cell integrity.
Flr1 expression is regulated mainly by Yap1 and is important for caffeine tolerance
Caffeine may alter the transcriptional profiles of multiple genes that are responsible for the transport of various metabolites and proteins. Kuranda et al. (2006) reported that the expression of at least 50 genes implicated in cellular transport was elicited by 20 mM caffeine treatment. DNA microarray studies have revealed that the expression of three multidrug transporter genes, SNQ2, QDR3, and MDL2, was upregulated by caffeine (Kuranda et al., 2006).
To more thoroughly screen for unknown genes whose expression is induced by caffeine, and to investigate whether the induction is involved in the activation of Yap1, we performed quantitative analysis of transcriptomes in three groups of strains: 50 mM caffeine-treated WT, yap1 mutant, and Yap1 overexpressed cells (Supplementary Figure 1). Out of 7,125 genes that were analyzed, the transcription of 2,285 genes (32%) was upregulated with a fold change of > 1.5 by 50 mM caffeine. This number is about three times higher than that in the yap1 mutant (9%) or Yap1 overexpression strain (11%). Among these genes, transcription levels of at least seven encoding multidrug transporters that were implicated in pleiotropic drug resistance, including PDR5 and SNQ2, were upregulated by over 1.5-fold after 1 h of treatment with 50 mM caffeine, and the expression of PDR5 was two times higher than that of SNQ2 (Figures 5A,B; Supplementary Figure 2). This result is consistent with a previous report showing that Pdr5 and Snq2 are two major caffeine efflux pumps in S. cerevisiae (Tsujimoto et al., 2015). Consistent with transcriptome results, we also found that in quantitative PCR analysis transcriptional induction of PDR5 is higher than that of SNQ2 by various concentrations of caffeine, but not by H2O2 (Figure 5C).
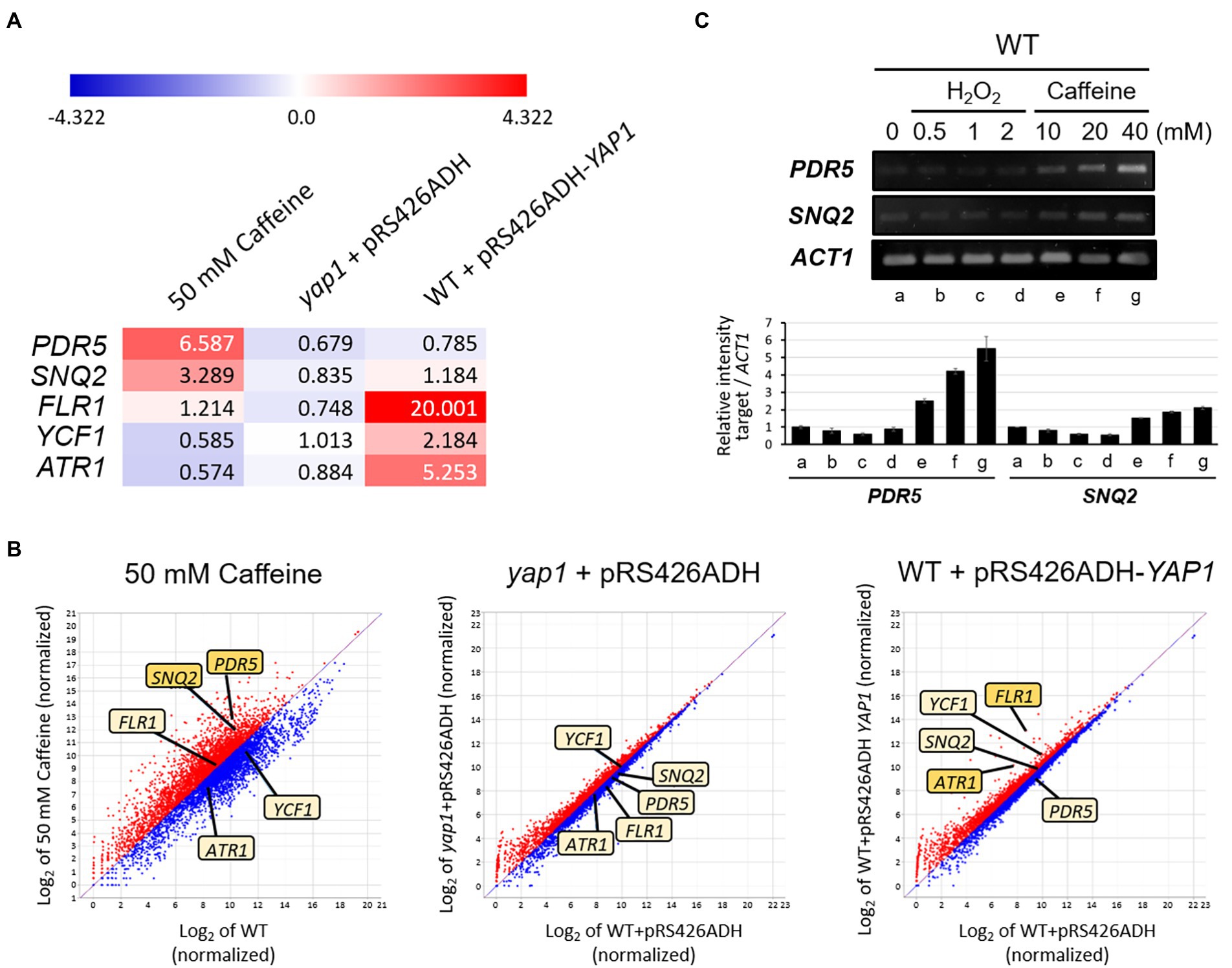
Figure 5. QuantSeq analysis to screen Yap1-dependent target genes for caffeine resistance. (A) Heat map depiction with numerical values of induction and (B) Scatter plot show differentially expressed mRNA levels of several transporter genes in caffeine-treated WT, yap1 and YAP1 overexpression strains. Red color represents upregulation and blue represents downregulation of mRNA expression. The degrees of darkness in colors represent the proportions of the expressive level. The 7,125 mRNAs were standardized and log2-transformed to show on a scatter plot by ExDEGA software. The X-axis displays the expression level of genes from WT, whereas the Y-axis shows the expression level of genes (C) RT-PCR analysis of PDR5 and SNQ2 expression in cells with H2O2 or caffeine treatment. Cells were incubated at 30°C for 30 min with H2O2 or caffeine treatment. The numbers of PCR cycles used are all 28.
However, the transcription of FLR1, YCF1, and ATR1 did not increase in response to caffeine. Instead, mRNA levels of these three genes were elevated under the condition of Yap1 overexpression, as expected for Yap1-dependent genes, and this upregulation disappeared completely in the absence of Yap1 (Figure 5A). Specifically, the expression of FLR1 increases significantly up to 20-fold, the highest among transporter-encoding genes, and ATR1 with over 5-fold increase is the next. These results are consistent with our previous observation that the caffeine-resistant phenotype of Yap1 overexpression strain completely disappeared in an flr1 mutant background and was slightly impaired in an atr1 mutant (Figure 2B).
Reactive oxygen species signaling modulates cellular adaptation to caffeine in a Yap1-dependent manner
An observation that the cellular process to acquire caffeine tolerance involves the activation of the oxidative stress response pathway aroused our curiosity as to whether ROS signaling modulates cellular adaptation to caffeine toxicity. We examined the caffeine sensitivity of cells with or without oxidative stress by using spotting analysis. While cell growth is severely impaired on minimal media with 10 mM caffeine as shown earlier in Figure 1B, the impaired growth of WT cells is significantly restored when cells are grown on caffeine media and treated with 0.5 mM H2O2, and even more with 1 mM H2O2 (Figure 6A). However, the recovery of caffeine resistance completely disappeared in the absence of Yap1. These results give a strong implication that the oxidative stress response is mediated by Yap1 in adaptation to caffeine toxicity. The concentration of H2O2 used for this analysis is not sufficient to inhibit cell growth solely, but enough to generate caffeine tolerance when treated together with caffeine.
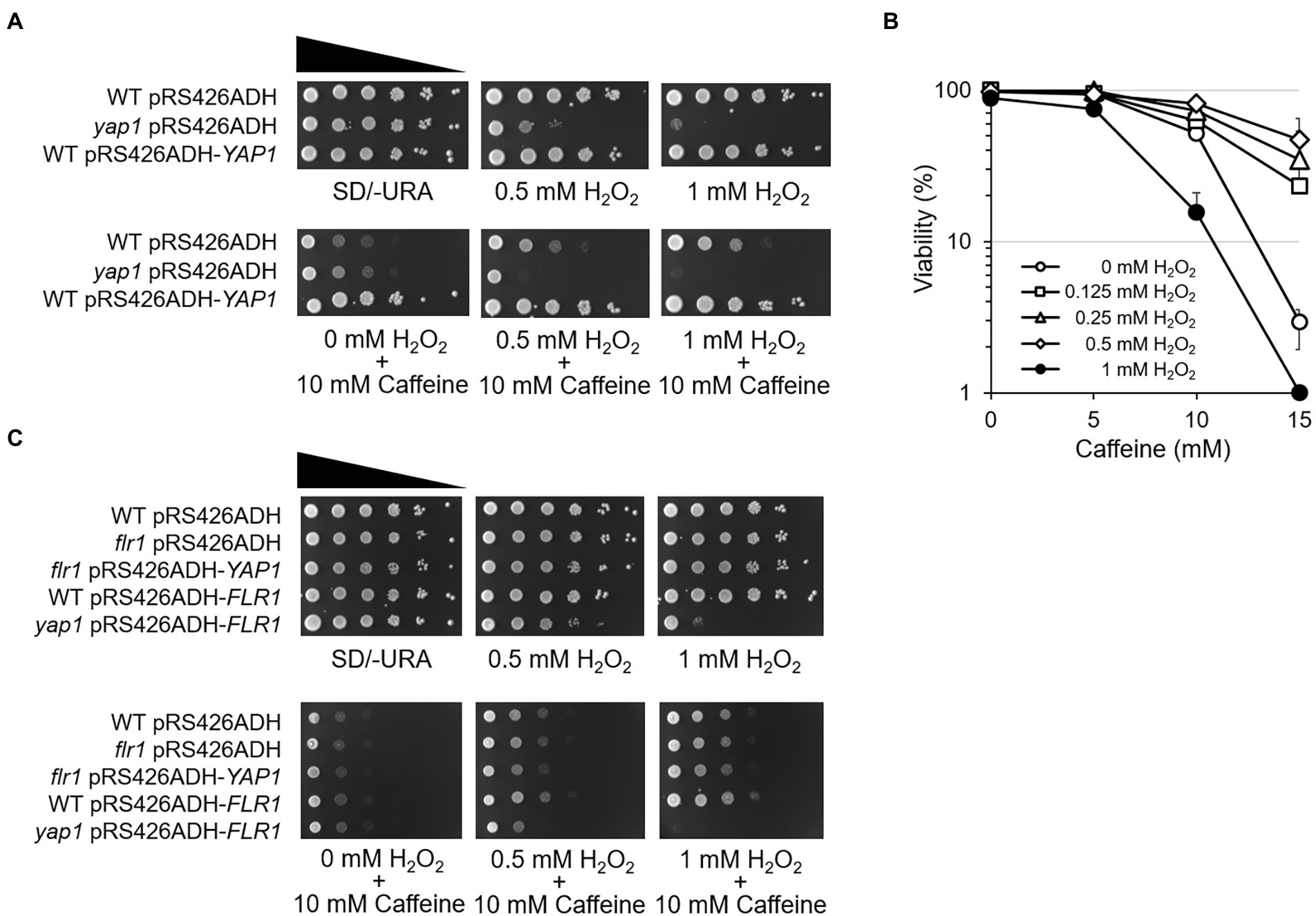
Figure 6. ROS signaling modulates cellular adaptation to caffeine. (A) 10-fold serial dilutions of yeast mutant strains were spotted on SD/-URA media in the presence of caffeine with or without H2O2 and incubated for 3 days at 30°C. (B) Cell viability in the presence of caffeine was measured by counting CFUs on YEPD plates with or without H2O2. (C) 10-fold serial dilutions of yeast mutant strains were spotted on SD/-URA media in the presence of the caffeine with or without H2O2 and incubated for 3 days at 30°C.
To consolidate the notion that ROS signaling renders cell adaptation to caffeine, cell viability was analyzed using a CFU counting assay. When 100 of WT cells were spread onto 15 mM caffeine-containing plates, only ~ 2% were viable (Figure 6B). Interestingly, the viability increases as higher concentrations of H2O2 are treated with caffeine, and reached up to ~ 60% with 0.5 mM H2O2. However, cells are no longer viable with 1 mM H2O2 on plates containing 15 mM caffeine.
Next, spotting analysis was carried out with flr1 mutants and Flr1 overexpression strains to address whether Flr1 is involved in ROS-mediated caffeine adaptation. Surprisingly, we found that H2O2-induced caffeine adaptation still occurs in the absence of FLR1 (Figure 6C). Yap1 overexpression does not make flr1 mutants more resistant to caffeine when treated with H2O2 compared to WT or flr1 mutant cells, although flr1 mutant survives caffeine toxicity better with H2O2 than without H2O2. However, yap1 mutants display no adaptation even with Flr1 overexpression. These results suggest that Yap1, but not Flr1, is essential for oxidative stress-mediated cellular adaptation to caffeine toxicity.
Reciprocal crosstalk between ROS signaling and caffeine resistance
Caffeine has potential antioxidant activity, protecting cells against oxygen radicals and repairing some oxidative damages (Devasagayam et al., 1996; Vieira et al., 2020). Since several toxic drugs are known to trigger ROS production in the cell and many transporters responsible for multidrug resistance belong to oxidative stress-responsive regulons, we next investigated the possible roles of caffeine in the regulation of oxidative stress and the accompanying DNA damages and mutagenesis (Chung, 2021).
When the intracellular ROS level was measured using flow cytometry with H2DCFDA, a cell-permeant indicator for general oxidative stress, we found that the amount of ROS in WT cells increases significantly when treated with higher concentrations of H2O2 (Figure 7A). However, co-treatment of 25 mM caffeine with 6 mM H2O2 dramatically reduces ROS levels from 82.4 to 37.6%, although caffeine on its own shows almost no change in ROS level (1.9%).
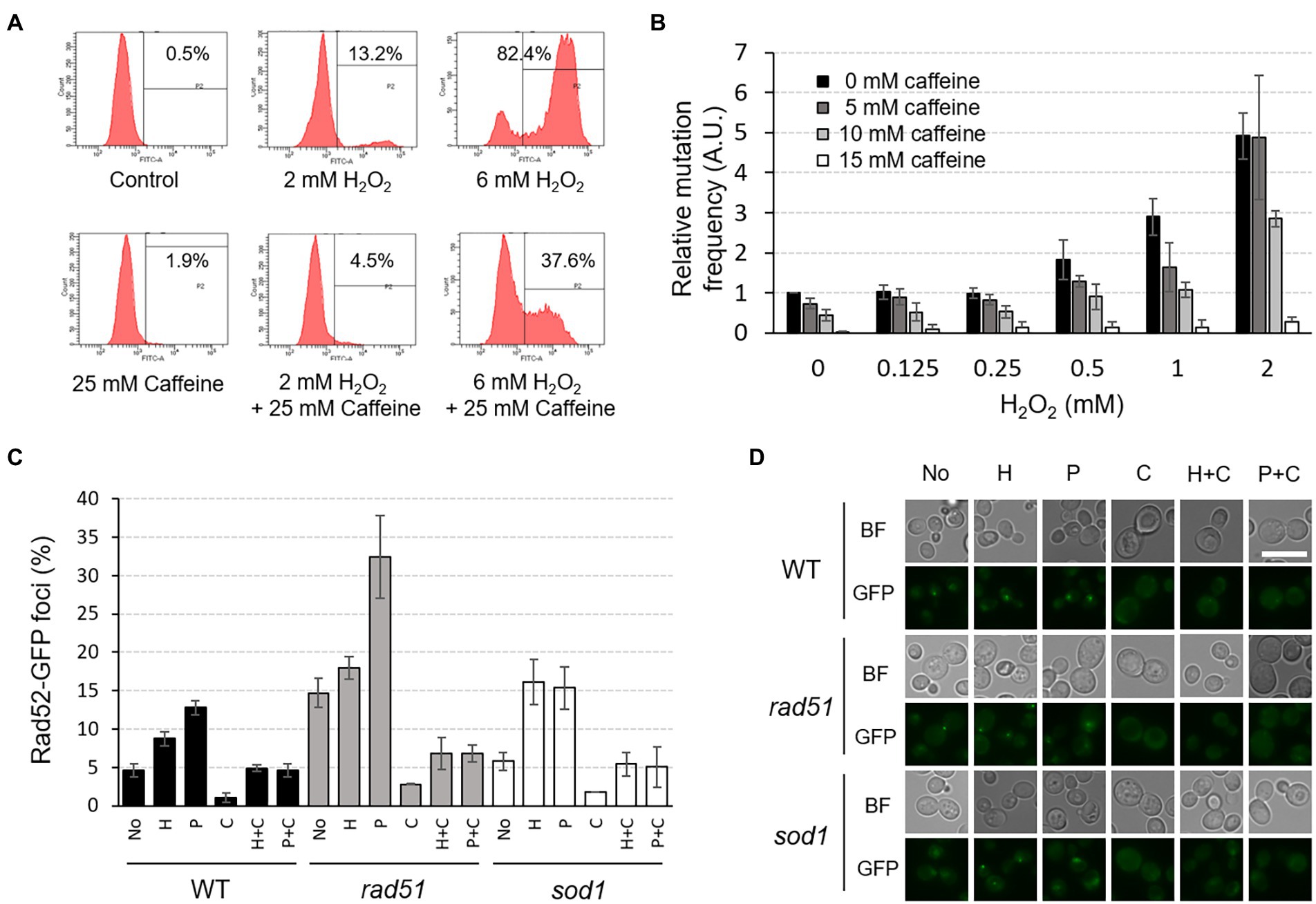
Figure 7. Caffeine reduces intracellular ROS level, mutation rates, and Rad52-GFP foci formation induced by H2O2 or phleomycin treatment. (A) The intracellular ROS level was measured by FACS analysis under the indicated conditions. Cells were treated with H2O2 and/or caffeine for 2 h prior to ROS detection. P2 percentage indicates the proportion of cells with an increased production of ROS, emitting above the background levels of DCF fluorescence. Averages from three independent experiments are presented. (B) Relative mutation frequency was measured by CAN1 forward-mutation assay in response to various concentrations of H2O2 with or without caffeine treatment. (C) The proportion of cells containing Rad52-GFP foci was determined under the indicated conditions of H2O2 (H), phleomycin (P), and caffeine (C) in WT, rad51, and sod1 mutant strains. (D) Fluorescent microscopy of Rad52-GFP foci analyzed in (C). White scale bar corresponds to 10 μm. BF, bright field.
To test whether caffeine directly affects genome stability, we measured spontaneous mutation frequency using CAN1 forward-mutation assay with or without H2O2 and caffeine. Consistent with our previous observations, mutation rates of WT cells start to increase from 0.5 mM concentration of H2O2 and reached up to five times at 2 mM H2O2 treatment compared to those with no treatment (Yi et al., 2016; Figure 7B). However, when cells are treated with caffeine together with H2O2, the mutation rates decrease in a concentration-dependent manner, and the rate with 15 mM caffeine is about 100-fold lower than that of the untreated WT. The increased mutation rate with 2 mM H2O2 decreased significantly to ~ 95% with 15 mM caffeine compared with no caffeine treatment. Caffeine seems to remove ROS and successfully mask its harmful effects on mutagenesis.
The number of Rad52-GFP foci in the nucleus, an indicator of accumulated DNA damage including DSBs, also decreases significantly when cells are treated with caffeine (Figures 7C,D). The foci formation induced by H2O2 or phleomycin is inhibited efficiently when treated together with caffeine, and similar results appeared in cells lacking Rad51 or Sod1, showing increased intracellular DSBs or ROS, respectively. All these observations reveal that caffeine acts as a strong antioxidant and protects cells from potential genome instability and mutagenesis by inhibiting the generation of DNA lesions.
Taken together, our results coherently indicate reciprocal crosstalk between oxidative stress response and caffeine metabolism (Figure 8). Yap1 activation by elevated ROS levels and the subsequent induction of its downstream target genes, undoubtedly including Flr1, confer cells resistance to caffeine toxicity, and inversely, appropriate doses of caffeine play important roles in the removal of deleterious oxidants from cells and the protection of chromosomes from ROS-mediated damage and accompanying mutagenesis.
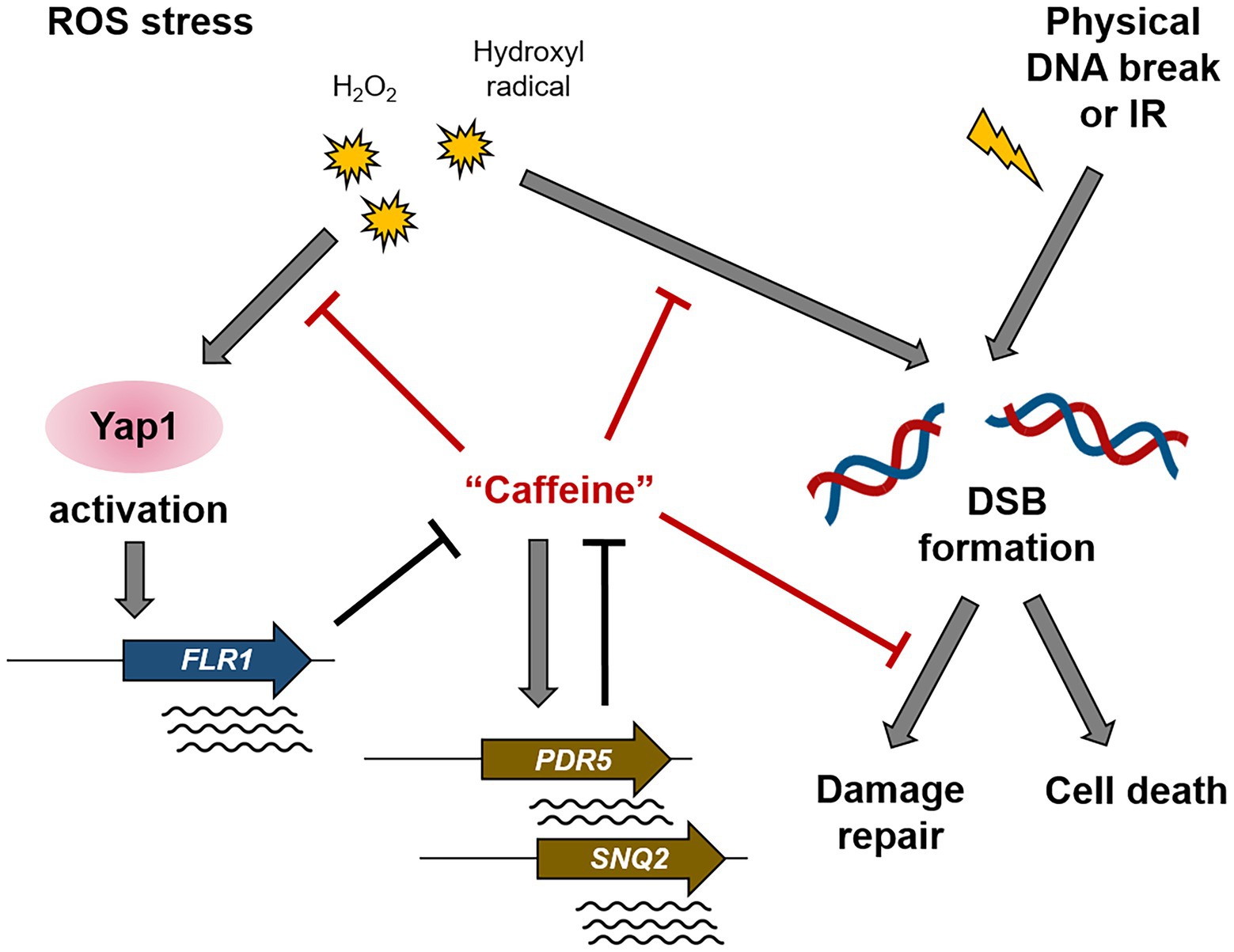
Figure 8. A schematic diagram illustrating the mechanism of action of caffeine in Yap1-dependent ROS signaling and DNA damage response. The ROS-induced Yap1 activation protects cells from caffeine toxicity, and caffeine reduces intracellular ROS level, DNA damage, and mutation rates. Controversially, caffeine disturbs several steps of DSB repair pathway.
Discussion
Caffeine elicits well-documented cytotoxic effects to bacteria and various eukaryotic cells including yeast and humans, inactivating a lot of important cellular activities. Caffeine abolishes DNA damage-induced cell cycle arrest by inhibiting DNA damage response (DDR) signaling pathway orchestrated by the sensor kinases, ATM and ATR, and perturbs multiple steps in DNA damage repair pathways (Blasina et al., 1999; Sarkaria et al., 1999; Tsabar et al., 2015b). In this study, however, we addressed several possible beneficial roles of caffeine that maintains chromosomal fidelity by reducing intracellular ROS level and inhibiting DSB formation and mutagenesis. We showed that Yap1 and its target gene FLR1 are the key mediators of regulatory crosstalk between ROS signaling and cellular resistance to caffeine.
Although the export of a wide range of structurally and functionally unrelated drugs out of the cell by a single transporter is still open to debate, our results demonstrate that Flr1, previously known as an efflux pump of fluconazole, diazaborine, and methotrexate, is also highly involved in caffeine detoxification. The exact mechanism of its involvement in caffeine tolerance is not clarified, but the process might be related to the regulation of physiological conditions such as cellular redox state or metal concentrations since Yap1, a highly conserved transcription factor for oxidative stress response and cadmium resistance, is required for the induction of FLR1 expression (Wemmie et al., 1994; Alarco et al., 1997; Kuge et al., 2001; Figure 2).
Flr1 is a yeast representative of the major facilitator superfamily (MFS) of multidrug resistance. The promoter region of FLR1 contains three Yap1-responsive elements (YREs), each with differential binding activity to Yap1 (Nguyên et al., 2001). Therefore, the transcription of FLR1 is induced by oxidizing agents such as H2O2, diamide, diethylmaleate, and even the alkylating agent methyl methanesulfonate (MMS), which stalls replication forks leading to DNA DSBs.
In fact, a number of oxidative stress regulons are responsible for the expression of the multidrug resistance system including ABC transporters. A total of 22 genes encode ABC transporters in the S. cerevisiae genome and many of them are crucial for adaptation to environmental stresses (Paumi et al., 2009). Among them, two major caffeine efflux pumps, Pdr5 and Snq2, and a Yap1-dependent cadmium transporter, Ycf1, have been reported to be commonly involved in both multidrug resistance and the oxidative stress response (Buechel and Pinkett, 2020). Miyahara et al. (1996) have shown that both PDR5 and SNQ2 have one YRE sequence in their promoter regions. This is plausible because many toxic drugs could trigger ROS accumulation in the cell.
However, our QuantSeq analysis and several studies with mutants revealed that the expression of neither PDR5 nor SNQ2 seems to be regulated by Yap1, and FLR1 is the only strong target of Yap1 activation among the potential candidates responsible for caffeine resistance (Figures 2, 5). In S. cerevisiae, 9 proteins, including Pdr5, and Snq2, have been identified as pleiotropic drug resistance (PDR) protein subfamily members, but Flr1 does not belong to that subfamily (Prasad and Goffeau, 2012). It is possible that the transcriptional induction of PDR5 and SNQ2 is also dependent on Yap2, a functional homolog of Yap1 with overlaps in oxidative and metal stress response, but with gene targets that are distinct from Yap1, especially during oxidative stress (Stephen et al., 1995; Buechel and Pinkett, 2020).
The reason why Pap1-impaired S. pombe cells are sensitive to caffeine is that the expression of the efflux pump Hba2 is significantly reduced in the absence of Pap1 (Calvo et al., 2009). Excessive production of ABC transporter Caf5 also enhances caffeine resistance in a Pap1-dependent manner. What can be the functional counterparts of Hba2 or Caf5 in S. cerevisiae? We presented several RT-PCR and QuantSeq data showing that the transcription of FLR1, ATR1, and YCF1 is clearly induced by Yap1, but none of them are actually required for caffeine resistance (Figures 2, 5A). Overexpression of FLR1 fails to confer cells with as much resistance to caffeine as overexpression of Yap1 (Figures 2B, 3B). Most importantly, however, overexpression of Yap1 shows an obvious caffeine resistant effect only in the presence of Flr1, indicating that other unknown Yap1 target proteins could work for caffeine tolerance together with Flr1 (Figure 2B). Interestingly, overexpression of Yap1 also confers resistance to cerulenin, a potent inhibitor of fatty acid synthase through the Flr1 transporter, and this observation again demonstrates that the Yap1-Flr1 stress response system has a wide spectrum of detoxifying capability (Oskouian and Saba, 1999).
It has been reported that the QDR3, encoding another member of multidrug transporters in the MFS, is induced by 20 mM caffeine treatment and is also crucial for cellular resistance to cisplatin and bleomycin, DNA DSB-inducing reagents (Tenreiro et al., 2005; Kuranda et al., 2006). High-throughput microscopic screening by Tkach et al. (2012) showed that the level of spontaneous Rad52 foci is elevated in atr1 null mutant. These observations suggest that the efficacy of caffeine efflux pumps is also highly involved in the protection or repair of deleterious DNA damages as well as protection against oxidative stress.
In this research, we provided strong evidence that a relatively moderate concentration of caffeine plays a role as an antioxidant, scavenging harmful ROS out of the cell, and pretreatment with H2O2 could provide adaptation to the cytotoxic effects of caffeine in a Yap1-dependent manner (Figure 6). Importantly, while Yap1 overexpression-mediated caffeine resistance required FLR1, H2O2-induced cellular adaptation to caffeine toxicity relied on the presence of Yap1, but not FLR1 (Figures 2, 6). This implies that ROS-induced Yap1 activation could be somehow different from the overexpression of Yap1 in the transcriptional induction of Yap1 target genes, or it is probably because increased H2O2 might alter broader range of transcriptional profiles involved in caffeine tolerance than just a Yap1-related group of genes.
Consistent with the recent report by Czachor et al. (2020) that coffee infusions reduce Rad52-GFP foci formation almost by half, our results demonstrated that caffeine itself is effective in decreasing both DNA DSB damages and spontaneously occurring mutations (Figure 7). Caffeine inhibits the repair of damaged DNA by disturbing the correct operation of repair proteins and damage checkpoint signaling, but at the same time, caffeine attenuates cytotoxic effects by efficiently reducing intracellular ROS level in the earlier steps as well as damage-induced mutagenesis (Saiardi et al., 2005; Tsabar et al., 2015a, 2015b; Figure 8). Taken together, we conclude that caffeine regulates a variety of cellular metabolic processes as a double-edged sword; cells maintain well-balanced growth condition by reciprocal crosstalk between the effects of caffeine and the ROS signaling pathway, especially in a Yap1-dependent manner.
Data availability statement
The data presented in the study are deposited in the NCBI GEO repository, accession number GSE214902.
Author contributions
JC designed and performed almost all experiments. S-HH performed a part of experiments. W-HC designed the entire research and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2020R1A2C1011370 and 2016R1A6A1A03007648).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1026780/full#supplementary-material
Abbreviations
WT, Wild type, ROS, Reactive oxygen species; 4-NQO, 4-Nitroquinoline-N-oxide; GFP, Green fluorescent protein; H2DCFDA, 2′,7′-dichlorodihydrofluorescein diacetate; TOR, Target of rapamycin; HR, Homologous recombination; DSB, Double-strand break; ATM, Ataxia-telangiectasia mutated; ATR, ATM-related; PDR, Pleiotropic drug resistance; ABC transporter, ATP-binding cassette transporter; CFU, Colony forming unit.
Footnotes
1. ^http://david.abcc.ncifcrf.gov/
2. ^http://www.ncbi.nlm.nih.gov/
3. ^https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214902
References
Alarco, A. M., Balan, I., Talibi, D., Mainville, N., and Raymond, M. (1997). AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 272, 19304–19313. doi: 10.1074/jbc.272.31.19304
Benko, Z., Fenyvesvolgyi, C., Pesti, M., and Sipiczki, M. (2004). The transcription factor Pap1/Caf3 plays a central role in the determination of caffeine resistance in Schizosaccharomyces pombe. Mol. Gen. Genomics. 271, 161–170. doi: 10.1007/s00438-003-0967-3
Blasina, A., Price, B. D., Turenne, G. A., and McGowan, C. H. (1999). Caffeine inhibits the checkpoint kinase ATM. Curr. Biol. 9, 1135–1138. doi: 10.1016/S0960-9822(99)80486-2
Buechel, E. R., and Pinkett, H. W. (2020). Transcription factors and ABC transporters: from pleiotropic drug resistance to cellular signaling in yeast. FEBS Lett. 594, 3943–3964. doi: 10.1002/1873-3468.13964
Calvo, I. A., Gabrielli, N., Iglesias-Baena, I., García-Santamarina, S., and Hoe, K. L. (2009). Genome-wide screen of genes required for caffeine tolerance in fission yeast. PLoS One 4:e6619.
Chung, W. H. (2021). Pleiotropic effects of caffeine leading to chromosome instability and cytotoxicity in eukaryotic microorganisms. J. Microbiol. Biotechnol. 31, 171–180. doi: 10.4014/jmb.2011.11042
Coleman, S. T., Tseng, E., and Moye-Rowley, W. S. (1997). Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J. Biol. Chem. 272, 23224–23230. doi: 10.1074/jbc.272.37.23224
Combettes, L., Berthon, B., and Claret, M. (1994). Caffeine inhibits cytosolic calcium oscillations induced by noradrenaline and vasopressin in rat hepatocytes. Biochem. J. 301, 737–744. doi: 10.1042/bj3010737
Czachor, J., Miłek, M., Galiniak, S., Stępień, K., Dżugan, M., and Mołoń, M. (2020). Coffee extends yeast chronological lifespan through antioxidant properties. Int. J. Mol. Sci. 21:9510. doi: 10.3390/ijms21249510
Devasagayam, T. P., Kamat, J. P., Mohan, H., and Kesavan, P. C. (1996). Caffeine as an antioxidant: inhibition of lipid peroxidation induced by reactive oxygen species. Biochim. Biophys. Acta 1282, 63–70. doi: 10.1016/0005-2736(96)00040-5
Ghosh, M., Shen, J., and Rosen, B. P. (1999). Pathways of as(III) detoxification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 96, 5001–5006. doi: 10.1073/pnas.96.9.5001
Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., et al. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. doi: 10.1038/nature02026
Isoyama, T., Murayama, A., Nomoto, A., and Kuge, S. (2001). Nuclear import of the yeast AP-1-like transcription factor Yap1p is mediated by transport receptor Pse1p, and this import step is not affected by oxidative stress. J. Biol. Chem. 276, 21863–21869. doi: 10.1074/jbc.M009258200
Jungwirth, H., Wendler, F., Platzer, B., Bergler, H., and Högenauer, G. (2000). Diazaborine resistance in yeast involves the efflux pumps Ycf1p and Flr1p and is enhanced by a gain-of-function allele of gene YAP1. Eur. J. Biochem. 267, 4809–4816. doi: 10.1046/j.1432-1327.2000.01537.x
Kolaczkowski, M., van der Rest, M. E., Cybularz-Kolaczkowska, A., Soumillion, J.-P., Konings, W. N., and Goffeau, A. (1996). Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J. Biol. Chem. 271, 31543–31548. doi: 10.1074/jbc.271.49.31543
Kuge, S., Arita, M., Murayama, A., Maeta, K., Izawa, S., Inoue, Y., et al. (2001). Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21, 6139–6150. doi: 10.1128/MCB.21.18.6139-6150.2001
Kuge, S., and Jones, N. (1994). YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13, 655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x
Kumada, K., Yanagida, M., and Toda, T. (1996). Caffeine-resistance in fission yeast is caused by mutations in a single essential gene, crm1+. Mol. Gen. Genet. 250, 59–68.
Kuranda, K., Leberre, V., Sokol, S., Palamarczyk, G., and François, J. (2006). Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 61, 1147–1166. doi: 10.1111/j.1365-2958.2006.05300.x
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee, J., Godon, C., Lagniel, G., Spector, D., Garin, J., Labarre, J., et al. (1999). Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274, 16040–16046. doi: 10.1074/jbc.274.23.16040
Lisby, M., Rothstein, R., and Mortensen, U. H. (2001). Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. U. S. A. 98, 8276–8282. doi: 10.1073/pnas.121006298
López-Cruz, L., Salamone, J. D., and Correa, M. (2018). Caffeine and selective adenosine receptor antagonists as new therapeutic tools for the motivational symptoms of depression. Front. Pharmacol. 9:526. doi: 10.3389/fphar.2018.00526
Madeo, F., Fröhlich, E., Ligr, M., Grey, M., Sigrist, S. J., Wolf, D. H., et al. (1999). Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145, 757–767. doi: 10.1083/jcb.145.4.757
Martin, H., Rodriguez-Pachon, J. M., Ruiz, C., Nombela, C., and Molina, M. (2000). Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275, 1511–1519. doi: 10.1074/jbc.275.2.1511
Miyahara, K., Hirata, D., and Miyakawa, T. (1996). yAP-1- and yAP-2-mediated, heat shock-induced transcriptional activation of the multidrug resistance ABC transporter genes in Saccharomyces cerevisiae. Curr. Genet. 29, 103–105. doi: 10.1007/BF02221572
Morgan, B. A., Banks, G. R., Toone, W. M., Raitt, D., Kuge, S., and Johnston, L. H. (1997). The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16, 1035–1044. doi: 10.1093/emboj/16.5.1035
Moser, B. A., Brondello, J. M., Baber-Furnari, B., and Russell, P. (2000). Mechanism of caffeine-induced checkpoint override in fission yeast. Mol. Cell. Biol. 20, 4288–4294. doi: 10.1128/MCB.20.12.4288-4294.2000
Motegi, A., and Myung, K. (2007). Measuring the rate of gross chromosomal rearrangements in Saccharomyces cerevisiae: a practical approach to study genomic rearrangements observed in cancer. Methods 41, 168–176. doi: 10.1016/j.ymeth.2006.07.025
Mumberg, D., Muller, R., and Funk, M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122. doi: 10.1016/0378-1119(95)00037-7
Nguyên, D. T., Alarco, A. M., and Raymond, M. (2001). Multiple Yap1p-binding sites mediate induction of the yeast major facilitator FLR1 gene in response to drugs, oxidants, and alkylating agents. J. Biol. Chem. 276, 1138–1145. doi: 10.1074/jbc.M008377200
Oskouian, B., and Saba, J. D. (1999). YAP1 confers resistance to the fatty acid synthase inhibitor cerulenin through the transporter Flr1p in Saccharomyces cerevisiae. Mol. Gen. Genet. 261, 346–353. doi: 10.1007/s004380050975
Paumi, C. M., Chuk, M., Snider, J., Stagljar, I., and Michaelis, S. (2009). ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily. Microbiol. Mol. Biol. Rev. 73, 577–593. doi: 10.1128/MMBR.00020-09
Prasad, R., and Goffeau, A. (2012). Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu. Rev. Microbiol. 66, 39–63. doi: 10.1146/annurev-micro-092611-150111
Rodrigues-Pousada, C., Devaux, F., Caetano, S. M., Pimentel, C., da Silva, S., Cordeiro, A. C., et al. (2019). Yeast AP-1 like transcription factors (yap) and stress response: a current overview. Microb. Cell 6, 267–285. doi: 10.15698/mic2019.06.679
Ruta, L. L., and Farcasanu, I. C. (2020). Saccharomyces cerevisiae and caffeine implications on the eukaryotic cell. Nutrients 12:2440. doi: 10.3390/nu12082440
Saiardi, A., Resnick, A. C., Snowman, A. M., Wendland, B., and Snyder, S. H. (2005). Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc. Natl. Acad. Sci. U. S. A. 102, 1911–1914. doi: 10.1073/pnas.0409322102
Sarkaria, J. N., Busby, E. C., Tibbetts, R. S., Roos, P., Taya, Y., Karnitz, L. M., et al. (1999). Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59, 4375–4382.
Selby, C. P., and Sancar, A. (1990). Molecular mechanisms of DNA repair inhibition by caffeine. Proc. Natl. Acad. Sci. U. S. A. 87, 3522–3525. doi: 10.1073/pnas.87.9.3522
Stephen, D. W., Rivers, S. L., and Jamieson, D. J. (1995). The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol. Microbiol. 16, 415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x
Tenreiro, S., Vargas, R. C., Teixeira, M. C., Magnani, C., and Sá-Correia, I. (2005). The yeast multidrug transporter Qdr3 (Ybr043c): localization and role as a determinant of resistance to quinidine, barban, cisplatin, and bleomycin. Biochem. Biophys. Res. Commun. 327, 952–959. doi: 10.1016/j.bbrc.2004.12.097
Tkach, J. M., Yimit, A., Lee, A. Y., Riffle, M., Costanzo, M., Jaschob, D., et al. (2012). Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 14, 966–976. doi: 10.1038/ncb2549
Toone, W. M., Kuge, S., Samuels, M., Morgan, B. A., Toda, T., and Jones, N. (1998). Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 12, 1453–1463. doi: 10.1101/gad.12.10.1453
Tsabar, M., Eapen, V. V., Mason, J. M., Memisoglu, G., Waterman, D. P., Long, M. J., et al. (2015a). Caffeine impairs resection during DNA break repair by reducing the levels of nucleases Sae2 and Dna2. Nucleic Acids Res. 43, 6889–6901. doi: 10.1093/nar/gkv520
Tsabar, M., Mason, J. M., Chan, Y. L., Bishop, D. K., and Haber, J. E. (2015b). Caffeine inhibits gene conversion by displacing Rad51 from ssDNA. Nucleic Acids Res. 43, 6902–6918. doi: 10.1093/nar/gkv525
Tsujimoto, Y., Shimizu, Y., Otake, K., Nakamura, T., Okada, R., Miyazaki, T., et al. (2015). Multidrug resistance transporters Snq2p and Pdr5p mediate caffeine efflux in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 79, 1103–1110. doi: 10.1080/09168451.2015.1010476
Vieira, A. J. S. C., Gaspar, E. M., and Santos, P. M. P. (2020). Mechanisms of potential antioxidant activity of caffeine. Radiat. Phys. Chem. 174:108968. doi: 10.1016/j.radphyschem.2020.108968
Vivancos, A. P., Castillo, E. A., Jones, N., Ayte, J., and Hidalgo, E. (2004). Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol. Microbiol. 52, 1427–1435. doi: 10.1111/j.1365-2958.2004.04065.x
Wemmie, J. A., Szczypka, M. S., Thiele, D. J., and Moye-Rowley, W. S. (1994). Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J. Biol. Chem. 269, 32592–32597. doi: 10.1016/S0021-9258(18)31675-2
Yan, C., Lee, L. H., and Davis, L. I. (1998). Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17, 7416–7429. doi: 10.1093/emboj/17.24.7416
Yi, D. G., Kim, M. J., Choi, J. E., Lee, J., Jung, J., Huh, W. K., et al. (2016). Yap1 and Skn7 genetically interact with Rad51 in response to oxidative stress and DNA double-strand break in Saccharomyces cerevisiae. Free Radic. Biol. Med. 101, 424–433. doi: 10.1016/j.freeradbiomed.2016.11.005
Keywords: oxidative stress response, caffeine resistance, Yap1, Flr1, DNA damage, adaptation, Saccharomyces cerevisiae
Citation: Choi JE, Heo S-H and Chung W-H (2022) Yap1-mediated Flr1 expression reveals crosstalk between oxidative stress signaling and caffeine resistance in Saccharomyces cerevisiae. Front. Microbiol. 13:1026780. doi: 10.3389/fmicb.2022.1026780
Edited by:
You-Hee Cho, CHA University, South KoreaReviewed by:
Keun Kim, Chung-Ang University, South KoreaPer Sunnerhagen, University of Gothenburg, Sweden
Xuefeng Chen, Wuhan University, China
Copyright © 2022 Choi, Heo and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woo-Hyun Chung, d2hjaHVuZzIzQGR1a3N1bmcuYWMua3I=
 Ji Eun Choi1,2
Ji Eun Choi1,2 Woo-Hyun Chung
Woo-Hyun Chung