- 1National Engineering Laboratory of Biological Feed Safety and Pollution Prevention and Control, Key Laboratory of Molecular Nutrition, Ministry of Education, Key Laboratory of Animal Nutrition and Feed, Ministry of Agriculture and Rural Affairs, Key Laboratory of Animal Nutrition and Feed Science of Zhejiang Province, Institute of Feed Science, Zhejiang University, Hangzhou, China
- 2Key Laboratory of Forage Cultivation, Processing and High Efficient Utilization, Ministry of Agriculture, Resources and Environment, Inner Mongolia Agricultural University, Hohhot, China
- 3Institute of Grassland Research, Chinese Academy of Agricultural Sciences, Hohhot, China
- 4Inner Mongolia Yihelvjin Agricultural Development Co., Ltd., Chifeng, China
Bioaugmentation of native grass ensiling with Lactobacillus plantarum or Lactobacillus buchneri or Pediococcus pentosaceus on the ensiling performance and bacterial community was investigated after 30 days of the fermentation process. The native grass was inoculated with distilled water, Lactobacillus plantarum, Lactobacillus plantarum, and Lactobacillus buchneri, and Lactobacillus plantarum, Lactobacillus buchneri, and Pediococcus pentosaceus as the CON treatment, T1 treatment, T2 treatment, and T3 treatment, respectively. The addition of lactic acid bacteria was added at a total of 1 × 106 colony-forming unit/g of fresh weight. As expected, the markedly (p < 0.05) lower water-soluble carbohydrate content was tested in the T2 and T3 treatments compared to the CON and T1 treatments. Compared to the CON and T1 treatment, significantly (p < 0.05) higher crude protein content, and lower acid detergent fiber and neutral detergent fiber contents were found in the T2 and T3 treatments. Compared to the CON treatment, the pH significantly (p < 0.05) decreased in the lactic acid bacteria (LAB) inoculated silage, and the lowest pH was measured in the T3 treatment. Similarly, significantly higher lactic acid and acetic acid contents were also found in the T3 treatment compared to those in other treatments. After 30 days of ensiling, the Shannon and Chao1 indexes in silages decreased compared to that in the fresh materials (FMs). The principal coordinate analysis indicated that both FM and silage were distinctly separated in each treatment with no interactions on the confidence ellipse (R = 0.8933, p = 0.001). At the phylum level, the dominant phylum was shifted from Proteobacteria to Firmicutes after the fermentation process. Interestingly, Weissella dominated the fermentation in the CON treatment and Lactobacillus dominated the fermentation in all inoculated LAB silages at the genus level. Results of functional prediction analyses showed that the metabolism of amino acid, cofactors, and vitamins, and membrane transport was reduced, while the metabolism of nucleotide and majority carbohydrates was increased after ensiling. The complex LAB (Lactobacillus plantarum, Lactobacillus buchneri, and Pediococcus pentosaceus) exhibited the potential possibility to decrease pH and enhance the relative abundance of LAB in response to obtaining high-quality silage by the synergistic effects. These results suggested that the complex LAB could improve the ensiling performance of native grass silage, and lay a theoretical basis for inoculant application in native grass.
Introduction
The availability of feed in terms of quality, quantity, and continuity directly influences livestock productivity (Tahuk et al., 2021). Native grass provides feed for a large number of ruminants to meet the demand for meat and milk for consumers. Nevertheless, the quality and quantity of native grass are volatile because of seasonal changes (Tahuk et al., 2021; Du et al., 2022). The native grass production will increase during the summer, which leads to a positive effect on increasing biomass production, whereas, in the winter, low quality and feed deficiency constrained the continuous development of animal husbandry (Zhou et al., 2022).
Ensiling has received increased attention as a traditional, important, and reliable forages and grass preservation technique to provide a continuous supply for animal husbandry development throughout the year, particularly in developed countries (Zhou et al., 2019; Lin et al., 2021). Ensiling is a complex biochemistry process that was determined by several factors, including temperature, moisture, raw materials nutritional compositions, harvest time, raw materials length, pack density, the microbiome in raw materials, and others (Puntillo et al., 2022). In particular, the preservation of silage from forages and grasses depends on the microbial ecological diversity and the epiphytic lactic acid bacteria (LAB) play a determining role in the conservation of silage with high quality. The epiphytic LAB can be found ranging from 101 to 107 cfu/g−1 of raw material and the abundance may lead to undesirable fermentation when the epiphytic LAB was lower than 105 cfu/g−1 of raw material (Cai et al., 1999). Different additives have been used to improve ensiling performance and preserve the nutrients in silage, among which LAB is one of the most current alternatives because it could improve the fermentation quality and prolong the storage of raw materials (Puntillo et al., 2022). The LAB inoculants could rapidly dominate and overcome the complex microbiome in the raw materials and novel strains have been isolated and identified from various materials and silages as new silage inoculants (Carvalho et al., 2021). It is difficult to directly produce high-quality native grass silage because of the lower moisture and water-soluble carbohydrate (WSC) contents, and lower LAB population (You et al., 2021a). A previous study indicated that the LAB could improve the fermentation quality of native grass silage, especially Lactobacillus plantarum (You et al., 2021a). Previously reports also have indicated that the homofermentative LAB could accelerate the fermentation process by utilizing the glucose to produce lactic acid (LA) through the Embden–Meyerhof pathway, such as the certain strains Lactobacillus plantarum, Pediococcus acidilactici, and Pediococcus pentosaceus, whereas, the heterofermentative LAB were also widely used for producing a mixture of LA, acetic acid (AA), and ethanol via the phosphoketolase pathway by improving the aerobic stability, such as the certain strains Lactobacillus buchneri (Gallagher et al., 2018; Alhaag et al., 2019; Cao et al., 2021; Silva et al., 2022). However, due to the diversity of, not all LAB inoculants could significantly improve silage, making it impossible to create universal LAB-based products (Fijalkowska et al., 2020; Cao et al., 2021).
Previous reports also found that the synergistic effects determine the final community structure and functions when the first species arrive (Cheong et al., 2021; Debray et al., 2021). Additionally, within specific niches, microbe–microbe interactions can play a critical role in driving community structures and functional properties (Cheong et al., 2021). Nevertheless, there is less research on how priority effects influence bacterial community structure and functional profiles in native grass inoculated with various LAB inoculants. As a result, the present study aimed to determine the following: the bioaugmentation efficacy of various LAB additives on ensiling performance in terms of fermentation characteristics, fermentation characteristics, and bacterial community, the correlations between ensiling performance and bacterial community in native grass silage and the functional profiles.
Materials and methods
Substrate and silage preparation
The native grass was collected in the typical steppe flora of Bairin Left Banner, Inner Mongolian Plateau, China. Stipa gigantea L. and Leymus chinensis (Trin.) Tzvel. as the dominant species in this grassland. To get high-quality native grass and silage, the native grass was harvested at the 5 cm cutting height at the milk stage of the dominant species (You et al., 2021b). The native grass was inoculated with Lactobacillus plantarum (Chikuso-1, Snow Brand Seed Co., Ltd, Sapporo, Japan) at 1 × 106 colony-forming unit/g fresh matter as the T1 treatment; the native grass was inoculated with a mixture of Lactobacillus plantarum and Lactobacillus buchneri (Sci-plus Biotech. Co, Ltd, Inner Mongolia, China) at 1 × 106 colony-forming unit/g fresh matter as the T2 treatment and the native grass was inoculated with a mixture of Lactobacillus plantarum, Lactobacillus buchneri, and Pediococcus pentosaceus (Lactosan GmbH and Co, KG, Austria) at 1 × 106 colony-forming unit/g fresh matter as the T3 treatment, the native grass ensiling directly with the same volume deionized water as the control group (CON). The native grass was chopped into 30 mm size by a forage cuter (Fulida Tool Co., Ltd., Linyi, China) and then taken into the laboratory immediately. The native grass treated without or with LAB inoculants was transferred into vacuum-sealing polyethylene plastic bags (260 mm × 180 mm) and vacuum-sealed. Each group was ensiled with three replicates with about 250 g of forage per bag. The native grass silage bags were stored at room temperature (25~27°C) and sampled after 30 days of the fermentation period. The chemical compositions, fermentation quality, and bacterial community were analyzed after the silage bags ensiling for 30 days.
Fermentation characteristics and chemical compositions analyses
For chemical composition parameters, the fresh weight (FW) and native grass silage were blended uniformly before sampling and the samples were put into envelopes. The samples were measured for dry matter (DM) content in an oven for 72 h at 65°C and then passed a 1-mm screen (FW100, Taisite Instrument Co., Ltd., Tianjin, China) for subsequent analysis. The anthrone method was used to determine the WSC content (Thomas, 1977). The ash and crude protein (CP) contents were analyzed based on the methods of the Association of Official Analytical Chemists (methods: 2001.11; AOAC, 2005). The acid detergent (ADF) and neutral fiber (NDF) contents were tested with an ANKOM A200i Fiber Analyzer (ANKOM Technology, Macedon, NY, USA) and were described exclusively as residual ash (Van Soest et al., 1991; Li et al., 2020). Ten grams of silage samples were mixed with 90 ml of deionized water to derive extract at 4°C fridge for 24 h. Then, the extracts underwent filtration through a four-layer cheesecloth for analyzing fermentation characteristics. The pH value of the filtrate was tested by a glass-electrode pH meter. The ammonia nitrogen (NH3-N) content was analyzed with the previous report (Broderick and Kang, 1980). Organic acid contents of the filtrate were analyzed by high-performance liquid chromatography (HPLC) with a UV detector (210 nm) and 3 mmol/L of HClO4 was the mobile phase at a flow rate of 1.0 mL min−1 at 50°C (You et al., 2021b). The microbial population in the fresh materials (FMs) was counted by the plate count method and expressed on colony-forming units (cfu)/g of FW. The numbers of LAB and coliform bacteria were counted on de Man, Rogosa, Sharpe agar (Difco Laboratories, Detroit, MI, USA) and blue light broth agar (Nissui Ltd., Tokyo, Japan) incubated at 30°C for 48 h, the numbers of mold and yeast, and aerobic bacteria were counted on potato dextrose agar (Nissui Ltd., Tokyo, Japan) and nutrient agar Nissui Ltd., Tokyo, Japan) incubated at 30°C for 24 h, respectively (You et al., 2021b).
DNA extraction, PCR amplicon, and sequencing
Before analyzing the microbiome of the native grass silage, all samples were stored at −80°C before the extraction of DNA. The microbial DNA of FM and silage was extracted following the manufacturer's protocols with the HiPure Stool DNA Kits (Magen, Guangzhou, China). Amplicons spanning the V3–V4 hypervariable regions of the 16S rRNA gene were produced using primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) to determine the bacterial community (Logue et al., 2016). The sequences of these samples have been uploaded into the public database with the accession number PRJNA871931.
Microbial community analyses
The Flash (v1.2.8) was used to assign, truncate, and merge the paired-end reads (Tanja and Salzberg, 2011). Operational taxonomic units (OTUs) with a 97% similarity cutoff (Liu et al., 2019) were clustered by UPARSE (version 7.1, http://drive5.com/uparse/), and the high-quality sequences higher than 97% similarity were put into the same OTU (Rognes et al., 2016). The OTUs were classified using the Silva database (https://www.arbsilva.de/) with a confidence threshold of 70%, and the false discovery rate-adjusted Kruskal–Wallis multiple comparisons (q < 0.05) were used to detect bacterial community at the phylum and genus levels (Omontese et al., 2022), and permutational multivariate analysis of variance test was used to analyze the significant difference (Chambers and Hastie, 1992). The Venn diagram was constructed by R (v 1.6.2) according to the unique and common OTUs. The alpha diversity and Good's coverage were calculated in QIIME (version 1.9.1; Caporaso et al., 2010). The principal coordinates analysis (PCoA) was generated based on Bray–Curtis in the R package (2.5.3). The bacterial abundance and community composition were performed with Krona (version 2.6) and R package (version 2.2.1), respectively (Ondov et al., 2011; Wickham, 2011). The pheatmap package was used to display the heatmap of genus abundance (version 1.0.12; Kolde and Kolde, 2015). Spearman correlation analysis of species was calculated in the R package (version 1.8.4; Revelle and Revelle, 2015). The linear discriminant analysis (LDA) effect size (LEfSe) analyses were conducted via an online tool (https://www.omicstudio.cn/tool/60), and the LDA score > 4 and p < 0.05 were selected as the threshold.
Statistical analysis
The analyzed data of chemical compositions (DM, CP, WSC, ADF, and NDF contents) and fermentation characteristics (pH, LA, AA, PA, BA, and NH3-N parameters) were expressed on the mean ± standard error of means of three replicates. The effects of additives on the silage quality were evaluated with SAS 9.0 (SAS Institute, 2007 Cary, NC, USA). All the measured data were analyzed with the additive effect by the general linear models (GLMs): Yij = μ + αi + εij, where, μ is the overall mean, αi is the additive effect, and εij is the residual error (Ren et al., 2021).
Results
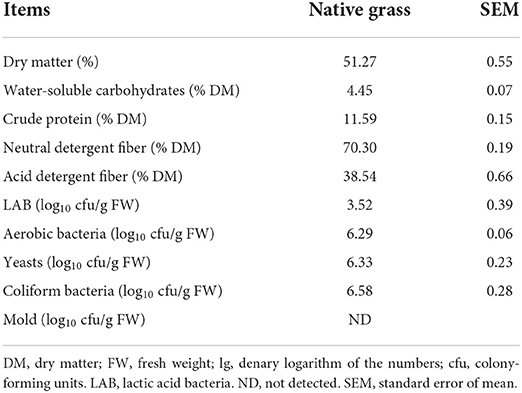
Chemical and microbial compositions of native grass
The chemical compositions and microbial population of the FM before ensiling are shown in Table 1. The DM content was 51.27% of the raw materials. The concentrations of WSC, CP, NDF, and ADF of native grass were 4.45, 11.29, 70.30, and 38.54% of DM, respectively. The native grass contained low desirable LAB (3.52 log cfu/g of FW) and high coliform bacteria (6.58 log cfu/g of FW) counts. The numbers of aerobic bacteria and yeasts were 6.29 and 6.33 log cfu/g of FW, respectively. Mold was not detected.
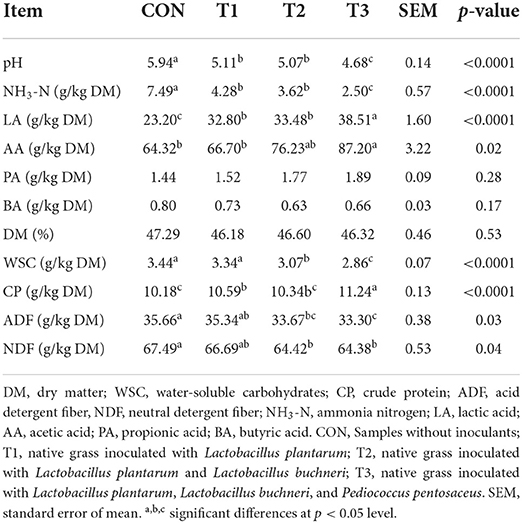
Ensiling performance of native grass without or with LAB inoculation
The fermentation quality and nutritional profiles of native grass ensiled with various LAB are shown in Table 2. As expected, the markedly (p < 0.05) lower WSC content was tested in the T2 and T3 treatments compared to the CON and T1 treatments, and no significant difference was found between the CON and T1 treatments. Interestingly, no significant difference was found in the DM content among these treatments. Additionally, compared to the CON and T1 treatments, significantly (p < 0.05) higher CP content, and lower ADF and NDF contents were found in the T2 and T3 treatments. After 30 days of ensiling, compared to the CON treatment, the pH was decreased in the LAB-inoculated silage, and the lowest pH was measured in the T3 treatment. Similarly, the highest LA and AA contents were also found in the T3 treatment compared to those in other treatments. No significant difference was analyzed in the propionic acid (PA) and butyric acid (BA) concentrations among these treatments.
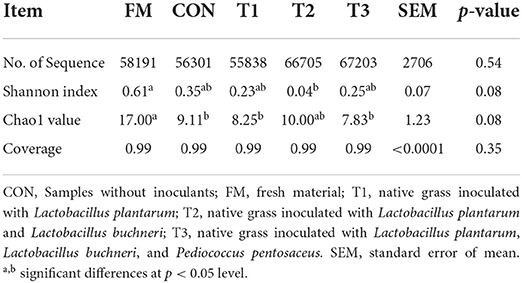
Bacterial diversity of native grass inoculated without or with LAB
The diversity of bacteria in native grass silage is shown in Table 3. According to the 16S rRNA amplicon sequencing of native grass materials and silage bacteria, an average of 60, 847 sequence numbers was obtained from each sample (data are not shown) and no significant difference was found in sequence numbers among these samples. After 30 days of ensiling, the diversity (Shannon) and richness (Chao1) in silages were decreased compared to that in the FM. A Venn diagram was constructed to depict similar and overlapping OTUs among all raw materials and silage (Figure 1A). As displayed, 51 OTUs were shared by the FM, CON, T1, T2, and T3 treatments, and the unique OTUs among these treatments were 598, 16, 14, 8, and 26, respectively. As indicated in the PCoA plot (Figure 1B; R = 0.8933, p = 0.001), both fresh native grass and silage were separated in each treatment with no interactions on the confidence ellipse. Figure 1C shows the abundance of the phyla in the raw materials and silages after 30 days of ensiling. Proteobacteria dominated the community at the phylum level in the fresh native grass. After the fermentation process, Firmicutes dramatically increased and became the most abundant phylum in native grass silage. Figure 1C shows the community structures in the FM and silages after 30 days of ensiling at the genus level. In the FM, the most abundant genus was Pantoea, followed by Enterobacter and Curtobacterium. Interestingly, the bacterial community structures in native grass inoculated without or with LAB were diverse. The genus, Weissella plays the important role in the CON group, and the genus Lactobacillus dominated the fermentation in all LAB-inoculated silage. Differences of bacterial taxa in native grass silage with different treatments were performed by LEfSe analyses (Figure 1E). As described, the phylum Proteobacteria and genus Pantoea were significantly (p < 0.05) enriched in the FM compared to these in the silage, and the phylum Firmicutes was markedly (p < 0.05) enriched in the T3 treatment than that in the fresh native grass and other treatments. The genus Weissella was significantly (p < 0.05) concentrated in the CON and T1 treatments compared to that in the T2 and T3 treatments. Additionally, compared to the CON and T1 treatments, the genus Lactobacillus was dramatically (p < 0.05) enriched in the T2 and T3 treatments.
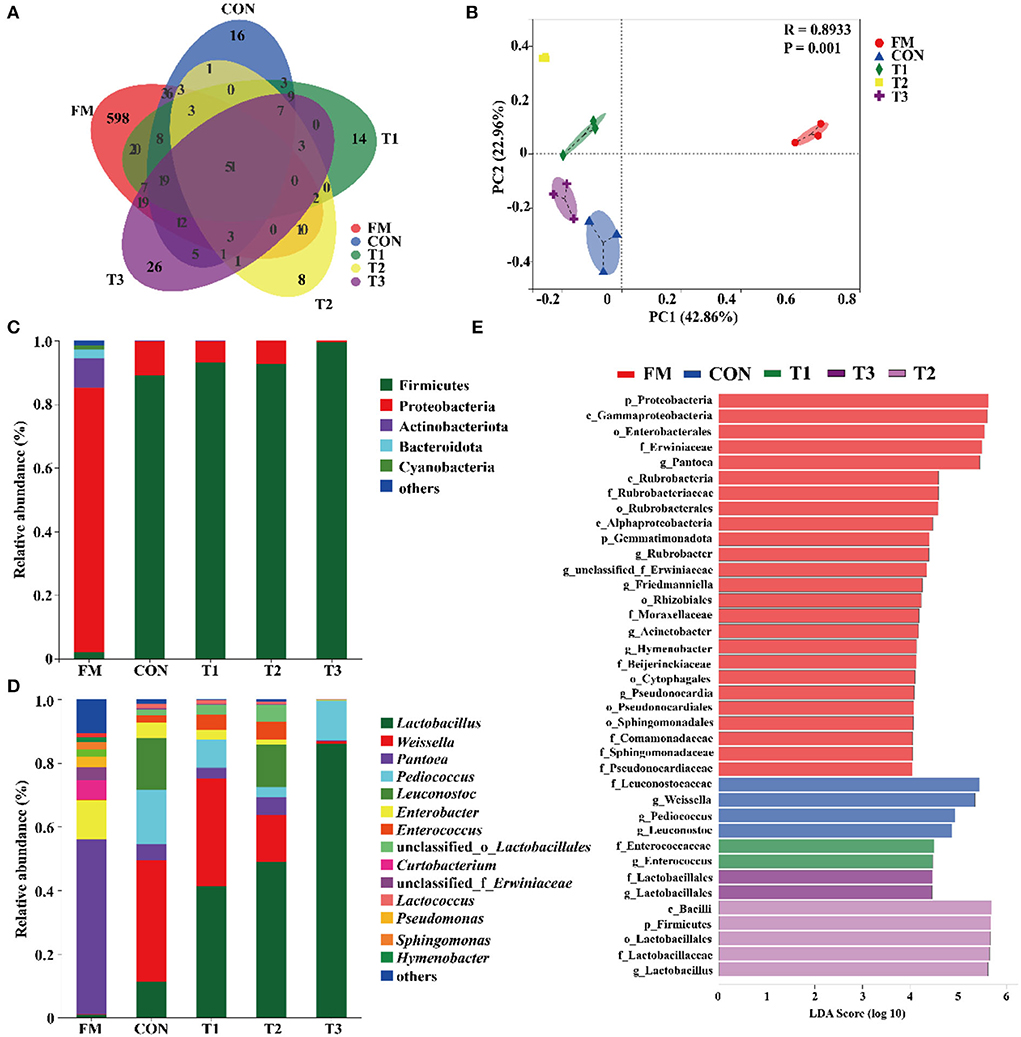
Figure 1. Bacterial community diversities and dissimilarities of native grass silage. FM, fresh material; CON, Samples without inoculants; T1, native grass inoculated with Lactobacillus plantarum; T2, native grass inoculated with Lactobacillus plantarum and Lactobacillus buchneri; T3, native grass inoculated with Lactobacillus plantarum, Lactobacillus buchneri, and Pediococcus pentosaceus. (A) Venn diagram representing the unique and common OTUs. (B) The bacterial community dissimilarities in different treatments were calculated by Bray–Curtis with coordinates calculated by principal coordinates analysis. (C,D) Relative abundances of native grass silage bacterial phylum and genus across different treatments. (E) Differences of bacterial taxa in native grass silage with different treatments.
Correlations between the bacterial community and ensiling performance in native grass inoculated without or with LAB
In the present study, the heatmap was used to evaluate the correlations between the bacterial genus (Top 10) and chemical constituents/fermentation profile based on the Spearman analysis in Figure 2. The BA, NDF, NH3-N and WSC contents, and pH were significantly associated with Lactobacillus (BA: rho = −0.631, p = 0.028; NDF: rho = −0.650, p = 0.022; NH3-N: rho = −0.713, p < 0.01; WSC: rho = −0.592, p = 0.04; pH: rho = −0.630; p = 0.03), whereas, the genus Pediococcus was positively associated with CP content (rho = 0.681; p = 0.02). The NDF, NH3-N, and WSC contents were also observed to have significantly positive connections with the genus Enterobacter (NDF: rho = 0.584, p = 0.05; NH3-N: rho = 0.674, p = 0.02; WSC: rho = 0.652, p = 0.02) and Weissella (NDF: rho = 0.625, p = 0.03; NH3-N: rho = 0.608, p = 0.04; WSC: rho = 0.627, p = 0.03).
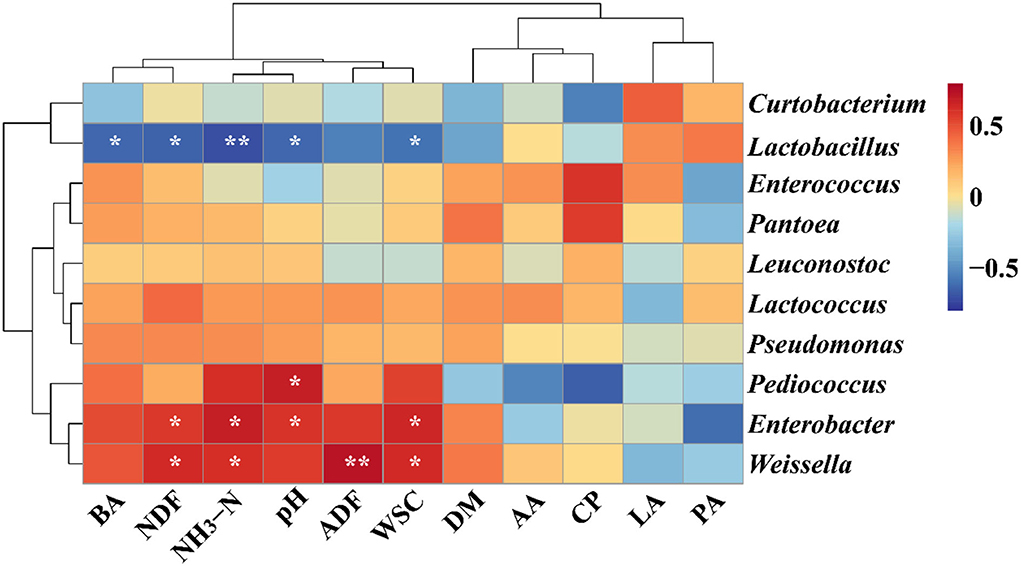
Figure 2. Correlation analysis between genera (top 10 significant genera) and ensiling performance of native grass silage inoculated without or with lactic acid bacteria. *Significant correlation at p < 0.05 level, **Significant correlation at p < 0.01 level. DM, dry matter; WSC, water-soluble carbohydrates; CP, crude protein; ADF, acid detergent fiber; NDF, neutral detergent fiber; NH3-N, ammonia nitrogen; LA, lactic acid; AA, acetic acid; PA, propionic acid; BA, butyric acid.
Predicted functions and pathways of bacterial community in native grass silage
In the current study, the predicted function of bacterial communities was performed by the PICRUSt (Figure 3). As shown in Figure 3A, the abundance of “Metabolism” was more than 40% and was markedly higher than other pathways, followed by environmental information processing and genetic information processing. The top 20 with significant differences in metabolic function are shown in Figure 3B. The proportions of membrane transport, carbohydrate metabolism, and amino acid metabolism were much higher than the other pathways. The carbohydrate metabolism was significantly (p < 0.05) inhibited in LAB-inoculated silage, especially in the T3 treatment, whereas the proportions of amino acid metabolism were significantly (p < 0.05) increased in the T1 treatment compared to the other treatments. Interestingly, compared to the CON group, the proportion of membrane transport was markedly in the LAB-inoculated silage. As shown in Figure 3C, the metabolic functions of the bacterial community were diverse in FM and silage. Results of functional prediction analyses showed that the metabolism of amino acid, cofactors, vitamins, and membrane transport was reduced, while the metabolism of nucleotide and the majority of carbohydrates was increased after ensiling. The carbohydrate metabolism (TCA cycle, starch and sucrose metabolism, and fructose and mannose metabolism) and amino acid metabolism (cysteine and methionine metabolism) were enriched in the T3 treatment compared to that in the CON group.
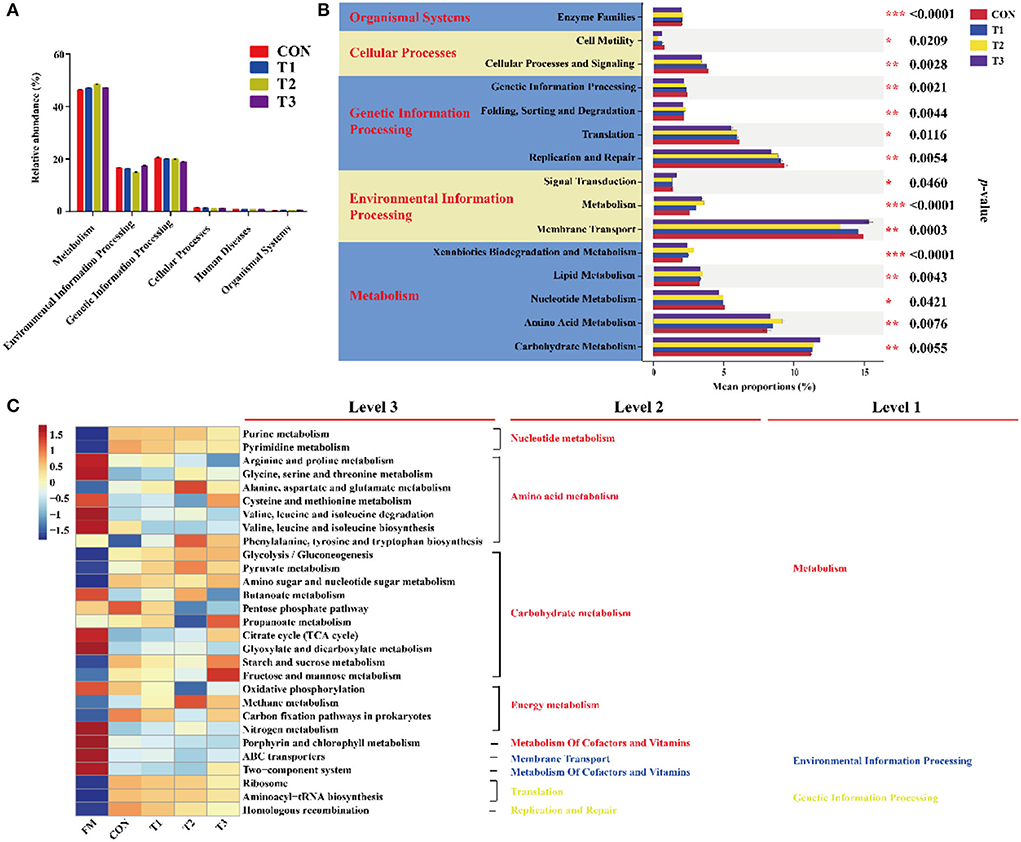
Figure 3. Dynamics of bacterial functional profiles in different treatments analyzed by PICRUSt (n = 3). (A) Level 1 metabolic pathway. (B) Level 2 KEGG ortholog functional predictions of the relative abundances with significant differences in the top 20 metabolic functions. (C) Level 3 KEGG ortholog functional predictions of the relative abundances of the top 30 metabolic functions. FM, fresh materials; CON, Samples without inoculants; FM, fresh material; T1, native grass inoculated with Lactobacillus plantarum; T2, native grass inoculated with Lactobacillus plantarum and Lactobacillus buchneri; T3, native grass inoculated with Lactobacillus plantarum, Lactobacillus buchneri, and Pediococcus pentosaceus.de. * Indicates significant at p < 0.05, ** indicates significant at p < 0.01 and *** indicates significant at p < 0.001.
Discussion
Ensiling is a useful method to prolong the supply of feed to ruminants. The LAB inoculants, including L. buchneri, L. plantarum, and P. pentosaceus are widely used worldwide in silages. On the one hand, the chemical compositions in the forages and grass can be effectively preserved by anaerobic fermentation; on the other hand, the microorganisms also produce organic acid (mainly LA and AA) that can improve the nutritional structure during the fermentation process (Lin et al., 2021). Unfortunately, the effective LAB inoculant is limited to improving the ensiling performance on native grass, and the complex LAB on native grass silage remains largely unknown. Here, the multiple fermentation characteristic analyses were combined with the16S rRNA sequencing to reveal the changes in native grass inoculated without or with various LAB inoculants during anaerobic fermentation. This is the first tentative model for the integrative analysis of the bacteria to respond to the anaerobic bioaugmentation of native grass ensiling with various LAB inoculants.
Chemical and microbial compositions of native grass
In the present study, the ADF and NDF contents were higher, and the CP content was lower compared to the previous report by You et al. (2021b), which could be contributed to the internal and external factors, including native grass community composition, harvest time and environment (Wang et al., 2020). The requirement of WSC content of good-quality silages is more than 5% DM (Amer et al., 2012), and the WSC content (4.45%) in the present study might meet the requirement. Moreover, the fermentation process was determined by the number of LAB, and the minimum requirement for the number of LAB in fresh materials should be higher than 5.0 log cfu/g FW (Cai et al., 1999). In the present study, the lower LAB (3.52 log cfu/g FW) counts and higher harmful microorganisms (more than 5.0 log cfu/g FW) were also found, which led to undesirable fermentation and end products. Therefore, it is necessary to add and reveal the role of LAB how to determine the silage fermentation and the shift of bacteria.
Ensiling performance of native grass silage
The LA content plays a determining role in dropping pH in silage and the desirable pH is ~3.8–4.2 for well-preserved silage. Nevertheless, the pH in all native grass silages was higher than 4.20 after 30 days of the fermentation process, which is in disagreement with the previous report that the pH (< 4.20) of native grass with 37.72% DM content ensiled with LAB additives after 30 days of fermentation. These differences could be explained by the DM content. Compared to the CON treatment, the LAB-inoculated silage decreased the pH value and the lowest pH was found in the T3 treatment, which could be contributed to the LAB additives improving the accumulation of LA and AA by Embden–Meyerhof pathway, phosphoketolase pathway, and pentose phosphate pathway for homofermentative LAB and pentose phosphate pathway for the homofermentative LAB by utilizing the WSC content (Ganzle, 2015; Muck et al., 2018; Valk et al., 2020; Lee et al., 2021). Moreover, the T3 treatment inoculated with three species of LAB could produce an acidic environment that was more beneficial for the LAB by the synergistic effects, and the WSC content was continuously utilized by the species of LAB (Cai et al., 1999; Svoboda et al., 2018). Therefore, lower DM and pH, and higher LA and AA contents were found in the T3 treatment, especially, the lowest WSC content was observed in the T3 treatment, followed by the T2 and T1 treatments. The PA and BA contents are undesirable fermentation end products in silages because the production pathway is an energy-waste metabolism (Dong et al., 2022). The little production of PA and BA contents indicated that extensive secondary fermentation did not occur throughout the whole fermentation process (Dong et al., 2022). The NH3-N content is negatively associated with the CP content. The plant and microbial enzymes degraded the protein into non-protein fractions throughout the fermentation process, including NH3, NH3-N, free amino acids, and peptides, by the proteolysis (Dong et al., 2022), reflecting the protein degradation throughout the ensiling period (Kung et al., 2018). Furthermore, the lower NH3-N and higher CP contents in the T3 treatment indicated that the forage protein was well-preserved after the fermentation process, which could have contributed to the lower pH, which could have inhibited the growth and metabolism of undesirable microorganisms, such as Clostridium (Kung et al., 2018; You et al., 2021b). At the same time, the digestible cell wall was broken down under hydrolytic activities, including microbial activities, enzymatic, and acidolysis when the silage is made (Zhao et al., 2018). Consequently, lower ADF and NDF contents were observed in the T3 treatment.
Bacterial diversity of native grass silage
The bacterial diversity and compositions in native grass silage were revealed by 16S rRNA sequencing. The variances of the bacterial community were performed by alpha diversity. The coverage in all samples was more than 0.99 (Table 3), suggesting that the depth of 16S rRNA sequencing had adequately reasonable to reflect the profile of the bacterial community (Ren et al., 2021). In the current study, the OTUs diversity and richness were decreased after ensiling compared to that in the FM (Table 3 and Figure 1A), which is in agreement with the previous reports that the decreased alpha diversity was observed because the undesirable microorganisms were inhibited by pH and gradually replaced by LAB (Xu et al., 2020; Ren et al., 2021). The PCoA plot clearly illustrates the variance of the bacterial community structures by the fermentation-additives-based separation of the treatments, suggesting that the additives had remarkable effects on the bacterial community structures of native grass silage. The predominant bacterial phylum, Proteobacteria, was found in FM, and the, most dominant bacterial phylum, Firmicutes, was found across all silage samples. In the current study, the dominant phylum was Proteobacteria and the primary genus was Pantoea, Pseudomonas, and Sphingomonas in FM, which is similar to the previous results that Pantoea, Pseudomonas, and Sphingomonas were the dominant genus in the raw materials (Ogunade et al., 2018; Romero et al., 2021; Long et al., 2022). Moreover, Firmicutes become the predominant phylum after the fermentation process (Yuan et al., 2020; Long et al., 2022). The shift from Proteobacteria to Firmicutes could be contributed to the microorganisms belonging to Firmicutes can thrive under low pH and anaerobic conditions (Wang et al., 2018; Dong et al., 2019). After ensiling, the abundance of Panotea markedly decreased in this study. The abundance of Panotea might be inhibited in an acidic environment (pH < 5.40) during anaerobic conditions (McGarvey et al., 2013; Sun et al., 2021). After the fermentation process, the bacterial community structures were diverse in native grass inoculated without or with LAB. The genus Weissella dominated the fermentation in the CON group, followed by Pediococcus and Leuconostoc; whereas, the abundance of Lactobacillus was increased and the abundance of Weissella was dropped with the increase of LAB species inoculated in the native grass. The previous report indicated that Lactococcus and Pediococcus initiated the fermentation at the early stage (Cai et al., 1998), and they were replaced by Lactobacillus with more acid-tolerant characteristics (Graf et al., 2016). Moreover, the increase of LAB species inoculated in the native grass produced a complex environment and enriched the bacterial diversity. As a consequence, a higher level of Lactobacillus was observed in the T3 treatment.
Correlation analysis of bacterial community and ensiling performance in native grass silage
The variations in fermentation characteristics could be characterized by the diversity of the microbial community (Ni et al., 2017). Moreover, the additives had significant effects on silage quality and microbial community (Wang et al., 2021). In the present study, Lactobacillus was negatively associated with pH and NH3-N, which is similar to the previous report (Fang et al., 2022). Nevertheless, Lactobacillus was also negatively associated with the WSC content, which is in disagreement with the previous study that the genus Lactobacillus was positively associated with the WSC content (Fang et al., 2022). In the whole-plant quinoa, the WSC content was higher than 30% of DM and the DM content in the present study was lower than the requirement of 5% of DM. The sufficient WSC content was beneficial for the growth of Lactobacillus and the abundance of Lactobacillus was inhibited, while the WSC content is limited. Therefore, the genus Lactobacillus was negatively associated with the WSC content. These results provide sufficient data to support that Lactobacillus has played a crucial role in improving the fermentation quality and preserving the native grass silage. Enterobacter was positively connected with pH and NH3-N. Additionally, the genus Enterococcus belongs to cocci LAB and could be inhibited in a low pH environment (McGarvey et al., 2013). Therefore, a lower abundance of Enterobacter and NH3-N content was observed in the T3 treatment.
Metabolic profile in native grass silage affected by LAB
The predicted functional profiles of the bacterial community were evaluated based on the Kyoto Encyclopedia of Genes and Genomes databases by PICRUSt to assess the metabolic pathways of native grass silage. The first-level directory and second-level directory indicated the first and second metabolic pathway levels, respectively (Wang et al., 2022). A previous report also indicated that multiple sub-directories were involved with various related signal pathways (Ogata et al., 1999). As shown in Figure 3A, metabolism was the primary metabolic pathway, indicating that the bacteria could convert fermentable substrates to various metabolites by the bacterial activities in metabolism pathways. The metabolic pathways with significant differences were including amino acid, carbohydrate, and nucleotide (Figure 3B), which is in accordance with the previous report (Bai et al., 2021). As shown in Figure 3C, carbohydrate metabolism mainly contained gluconeogenesis and glycolysis metabolism, TCA cycles, pentose phosphate pathway, and other pathways (Kanehisa and Goto, 2000). In this study, the carbohydrate metabolism pathway of native grass silage inoculated with LAB was stronger than that of the CON treatment, especially in the T3 treatment. These results indicated that the addition of LAB in native grass silage has a higher capacity to metabolize WSC than the epiphytic bacteria in the control treatment by the addition of LAB which could enhance the competitive strength through the synergistic effects (Svoboda et al., 2018). It was consistent with the previous results that higher LA and AA concentrations were found in the T3 treatment. Additionally, amino acid metabolism is necessary to promote primary metabolism and plant protein synthesis in plants (Wang et al., 2021). In the present study, some of the amino acid metabolism was suppressed after the fermentation process, which could be contributed to the diverse pH environment after ensiling and some undesirable microorganisms that directly influenced the amino acid metabolism were inhibited by the acidic environment (Flythe and Russell, 2004; Wang et al., 2021).
Conclusion
In conclusion, the present study shows the influences of various LAB on the ensiling performance of native grass by integrating the 16S rRNA gene sequences combined and multiple fermentation parameters. These results showed that LAB could directly affect chemical compositions and fermentation quality by modulating the bacterial community of native grass silage. The complex LAB (Lactobacillus plantarum, Lactobacillus buchneri, and Pediococcus pentosaceus) exhibited the potential possibility to decrease pH and enhance the relative abundance of LAB in response to obtaining high-quality silage by the synergistic effects. These results suggested that the complex LAB could improve the ensiling performance of native grass silage, and lay a theoretical basis for inoculant application in native grass.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA871931.
Author contributions
SD and SY designed the study and analyzed the data. SD wrote the manuscript. SD, SY, XJ, YL, and RW performed the experiments. SD, SY, GG, and YJ reviewed and edited the manuscript. YJ funded and supervised the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key Program of the National Dairy Innovation Center, China (2021-National Dairy Innovation Center-1).
Conflict of interest
Author RW is employed by the Inner Mongolia Yihelvjin Agricultural Development Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alhaag, H., Yuan, X., Mala, A., and Bai, J., Shao, T. (2019). Fermentation characteristics of Lactobacillus plantarum and Pediococcus species isolated from sweet sorghum silage and their application as silage inoculants. Appl. Sci. 9, 1247. doi: 10.3390/app9061247
Amer, S., Hassanat, F., Berthiaume, R., Seguin, P., and Mustafa, A. F. (2012). Effects of water soluble carbohydrate content on ensiling characteristics, chemical composition and in vitro gas production of forage millet and forage sorghum silages. Anim. Feed Sci. Tech. 177, 23–29. doi: 10.1016/j.anifeedsci.2012.07.024
Bai, J., Ding, Z., Ke, W., Xu, D., Wang, M., Huang, W., et al. (2021). Different lactic acid bacteria and their combinations regulated the fermentation process of ensiled alfalfa: ensiling characteristics, dynamics of bacterial community and their functional shifts. Microb. Biotechnol. 14, 1171–1182. doi: 10.1111/1751-7915.13785
Broderick, G. A., and Kang, J. H. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 63, 64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
Cai, Y., Benno, Y., Ogawa, M., Ohmomo, S., Kumai, S., Nakase, T., et al. (1998). Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl. Environ. Microbiol. 64, 2982–2987. doi: 10.1128/AEM.64.8.2982-2987.1998
Cai, Y., Suyanandana, P., Saman, P., and Benno, Y. (1999). Classification and characterization of lactic acid bacteria isolated from the intestines of common carp and fresh water prawns. J. Gen. Appl. Microbiol. 45, 177–184. doi: 10.2323/jgam.45.177
Cao, C. X., Bao, W. C., Li, W. C., Zhao, F. Y., Kwok, L. Y., Zhang, W. Y., et al. (2021). Changes in physico-chemical characteristics and viable bacterial communities during fermentation of alfalfa silages inoculated with Lactobacillus plantarum. World J. Microb. Biot. 37, 127. doi: 10.1007/s11274-021-03095-2
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high throughput community sequencing data. Nat. Methods. 7, 335–336. doi: 10.1038/nmeth.f.303
Carvalho, B. F., Sales, G. F. C., Schwan, R. F., and Ávila, C. L. S. (2021). Criteria for lactic acid bacteria screening to enhance silage quality. J. Appl. Microbiol. 130, 341–355. doi: 10.1111/jam.14833
Chambers, J. M., and Hastie, T. J. (1992). Statistical Models. Pacific Grove, CA: Wadsworth and Brooks/Cole.
Cheong, J. Z. A., Johnson, C. J., Wan, H. X., Liu, A. P., Kernien, J. F., Gibson, A. L. F., et al. (2021). Priority effects dictate community structure and alter virulence of fungal-bacterial biofilms. ISME J. 15, 2012–2027. doi: 10.1038/s41396-021-00901-5
Debray, R., Herbert, R. A., Jaffe, A. L., Crits-Christoph, A., Power, M. E., Koskella, B., et al. (2021). Priority effects in microbiome assembly. Nat. Rev. Microbiol. 20, 109–121. doi: 10.1038/s41579-021-00604-w
Dong, Z., Li, J., Chen, L., Wang, S., and Shao, T. (2019). Effects of freeze-thaw event on microbial community dynamics during red clover ensiling. Front. Microbiol. 10, 1559. doi: 10.3389/fmicb.2019.01559
Dong, Z. H., Li, J. F., Wang, S. R., Dong, D., and Shao, T. (2022). Time of day for harvest affects the fermentation parameters, bacterial community, and metabolic characteristics of sorghum-sudangrass hybrid silage. mSphere. 7, e00168-22. doi: 10.1128/msphere.00168-22
Du, S., You, S., Jiang, X., Li, Y., and Jia, Y. (2022). Longitudinal investigation of the native grass hay from storage to market reveals mycotoxin-associated fungi. Microorganisms. 10, 1154. doi: 10.3390/microorganisms10061154
Fang, D., Dong, Z. H., Wang, D. L., Li, B., Shi, P. B., Yan, J., et al. (2022). Evaluating the fermentation quality and bacterial community of high-moisture whole plant quinoa silage ensiled with different additives. J. Appl. Microbiol. 132, 3578–3589. doi: 10.1111/jam.15506
Fijalkowska, M., Przemieniecki, S. W., Purwin, C., Lipinski, K., Kurowski, T. P., Karwowska, A., et al. (2020). The effect of an additive containing three Lactobacillus species on the fermentation pattern and microbiological status of silage. J. Sci. Food Agric. 100, 1174–1184. doi: 10.1002/jsfa.10126
Flythe, M. D., and Russell, J. B. (2004). The effect of pH and a bacteriocin (bovicin HC5) on Clostridium sporogenes MD1, a bacterium that has the ability to degrade amino acids in ensiled plant materials. FEMS Microbiol. Ecol. 47, 215–222. doi: 10.1016/S0168-6496(03)00259-9
Gallagher, D., Parker, D., Allen, D. J., and Tsesmetzis, N. (2018). Dynamic bacterial and fungal microbiomes during sweet sorghum ensiling impact bioethanol production. Bioresource Technol. 264, 163–173. doi: 10.1016/j.biortech.2018.05.053
Ganzle, M. G. (2015). Lactic metabolism revisited: metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr. Opin. Food Sci. 2, 106–117. doi: 10.1016/j.cofs.2015.03.001
Graf, K., Ulrich, A., Idler, C., and Klocke, M. (2016). Bacterial community dynamics during ensiling of perennial ryegrass at two compaction levels monitored by terminal restriction fragment length polymorphism. J. Appl. Microbiol. 120, 1479–1491. doi: 10.1111/jam.13114
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kung, L., Shaver, R. D., Grant, R. J., and Schmidt, R. J. (2018). Silage review: interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 101, 4020–4033. doi: 10.3168/jds.2017-13909
Lee, Y., Cho, H. J., Choi, J. I., and Woo, H. M. (2021). Hybrid Embden–Meyerhof–Parnas pathway for reducing CO2 loss and increasing the Acetyl-CoA levels during microbial fermentation. ACS Sustain. Chem. Eng. 9, 12394–12405. doi: 10.1021/acssuschemeng.1c04812
Li, Y. Q., Wang, Z. X., Li, T., Jiang, S. H., Sun, Z. Y., Jiang, H., et al. (2020). Changes in microbial community and methanogenesis during high solid anaerobic digestion of ensiled corn stover. J. Clean. Prod. 242, 118479. doi: 10.1016/j.jclepro.2019.118479
Lin, H. Y., Lin, S. Q., Awasthi, M. K., Wang, Y. F., and Xu, P. (2021). Exploring the bacterial community and fermentation characteristics during silage fermentation of abandoned fresh tea leaves. Chemosphere. 283, 131234. doi: 10.1016/j.chemosphere.2021.131234
Liu, B., Huan, H., Gu, H., Xu, N., Shen, Q., Ding, C., et al. (2019). Dynamics of a microbial community during ensiling and upon aerobic exposure in lactic acid bacteria inoculation-treated and untreated barley silages. Bioresource Technol. 273, 212–219. doi: 10.1016/j.biortech.2018.10.041
Logue, J. B., Stedmon, C. A., Kellerman, A. M., Nielsen, N. J., Andersson, A. F., Laudon, H., LindstrÖm, E. S., et al. (2016). Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J. 10, 533–545. doi: 10.1038/ismej.2015.131
Long, S. H., Li, X. F., Yuan, X. J., Su, R. A., Pan, J. X., Chang, Y., et al. (2022). The effect of early and delayed harvest on dynamics of fermentation profile, chemical composition, and bacterial community of King grass silage. Front. Microbiol. 13, 864649. doi: 10.3389/fmicb.2022.864649
McGarvey, J. A., Franco, R. B., Palumbo, J. D., Hnasko, R., Stanker, L., Mitloehner, F. M., et al. (2013). Bacterial population dynamics during the ensiling of Medicago sativa (alfalfa) and subsequent exposure to air. J. Appl. Microbiol. 114, 1661–1670. doi: 10.1111/jam.12179
Muck, R. E., Nadeau, E. M. G., McAllister, T. A., Contreras-Govea, F. E., Santos, M. C., Kung, L., et al. (2018). Silage review: recent advances and future uses of silage additives. J. Dairy Sci. 101, 3980–4000. doi: 10.3168/jds.2017-13839
Ni, K., Wang, F., Zhu, B., Yang, J., Zhou, G., Pan, Y. I., Tao, Y., et al. (2017). Effects of lactic acid bacteria and molasses additives on the microbial community and fermentation quality of soybean silage. Bioresource Technol. 238, 706–715. doi: 10.1016/j.biortech.2017.04.055
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., Kanehisa, M., et al. (1999). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. doi: 10.1093/nar/27.1.29
Ogunade, I. M., Jiang, Y., Cervantes, A. P., Kim, D. H., Oliveira, A. S., Vyas, D., et al. (2018). Bacterial diversity and composition of alfalfa silage as analyzed by Illumina MiSeq sequencing: Effects of Escherichia coli O157: H7 and silage additives. J. Dairy Sci. 101, 2048–2059. doi: 10.3168/jds.2017-12876
Omontese, B. O., Sharma, A. K., Davison, S., Jacobson, E., DiConstanzo, A., Webb, M. J., et al. (2022). Microbiome network traits in the rumen predict average daily gain in beef cattle under different backgrounding systems. Anim Microbiome. 4, 25. doi: 10.1186/s42523-022-00175-y
Ondov, B. D., Bergman, N. H., and Phillippy, A. M. (2011). Interactive metagenomic visualization in a Web browser. BMC Bioinform. 12, 385. doi: 10.1186/1471-2105-12-385
Puntillo, M., Peralta, G. H., Burgi, M. D. M., Huber, P., Gaggiotti, M., Binetti, A. G., et al. (2022). Metaprofiling of the bacterial community in sorghum silages inoculated with lactic acid bacteria. J. Appl. Microbiol. 00, 1–15. doi: 10.1111/jam.15698
Ren, H. W., Sun, W. L., Yan, Z. H., Zhang, Y. C., Wang, Z. Y., Song, B., et al. (2021). Bioaugmentation of sweet sorghum ensiling with rumen fluid: Fermentation characteristics, chemical composition, microbial community, and enzymatic digestibility of silages. J. Clean. Prod. 294, 126308. doi: 10.1016/j.jclepro.2021.126308
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mah,é, F. (2016). Vsearch: a versatile open source tool for metagenomics. Peer J. 4, e2584. doi: 10.7717/peerj.2584
Romero, J. J., Park, J., Joo, Y., Zhao, Y. C., Killerby, M., Reyes, D. C., et al. (2021). A combination of Lactobacillus buchneri and Pediococcus pentosaceus extended the aerobic stability of conventional and brown midrib mutants–corn hybrids ensiled at low dry matter concentrations by causing a major shift in their bacterial and fungal community. J. Anim. Sci. 99, skab141. doi: 10.1093/jas/skab141
Silva, D. A., Liu, E. B., Caitlin, X. J., Tanya, M., Jason, G. F. S. D., et al. (2022). Effect of dry matter content on the microbial community and on the effectiveness of a microbial inoculant to improve the aerobic stability of corn silage. J. Dairy Sci. 105, 5024–5043. doi: 10.3168/jds.2021-21515
Sun, L., Bai, C. S., Xu, H. W., Na, N., Jiang, Y., Yin, G. M., et al. (2021). Succession of bacterial community during the initial aerobic, intense fermentation, and stable phases of whole-plant corn silages treated with lactic acid bacteria suspensions prepared from other silages. Front. Microbiol. 12, 655095. doi: 10.3389/fmicb.2021.655095
Svoboda, P., Lindstrom, E. S., Osman, O. A., and Langenheder, S. (2018). Dispersal timing determines the importance of priority effects in bacterial communities. ISME J.12, 644–646. doi: 10.1038/ismej.2017.180
Tahuk, P. K., Bira, G. F., and Taga, H. (2021). Physical characteristics analysis of complete silage made of sorghum forage, king grass and natural grass. IOP Conf Series Earth Environ. Sci. 465, 012022. doi: 10.1088/1755-1315/465/1/012022
Tanja, M., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Thomas, T. A. (1977). An automated procedure for the determination of soluble carbohydrates in herbage. J. Sci. Food Agric. 28, 639–642. doi: 10.1002/jsfa.2740280711
Valk, L. C., Luttik, M. A. H., Ram, D. E., Pabst, C., Van den Broek, M., Van Loosdrecht, M., et al. J. T. (2020). A novel D-galacturonate fermentation pathway in Lactobacillus suebicus links initial reactions of the Galacturonate-Isomerase route with the phosphoketolase pathway. Front. Microbiol. 10, 3027. doi: 10.3389/fmicb.2019.03027
Van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Wang, S., Zhang, G., Zhang, P., Ma, X., Li, F., Zhang, H., et al. (2018). Rumen fluid fermentation for enhancement of hydrolysis and acidification of grass clipping. J. Environ. Manag. 220, 142. doi: 10.1016/j.jenvman.2018.05.027
Wang, S. R., Li, J. F., Zhao, J., Dong, Z. H., Dong, D., Shao, T., et al. (2021). Effect of epiphytic microbiota from napiergrass and Sudan grass on fermentation characteristics and bacterial community in oat silage. J. Appl. Microbiol. 132, 919–932. doi: 10.1111/jam.15293
Wang, S. R., Li, J. F., Zhao, J., Dong, Z. H., Dong, D., Shao, T., et al. (2022). Dynamics of the bacterial communities and predicted functional profiles in wilted alfalfa silage. J. Appl. Microbiol. 132, 2613–2624. doi: 10.1111/jam.15417
Wang, S. R., Zhao, J., Dong, Z. H., Li, J. F., and Kaka, N. A., Shao, T. (2020). Sequencing and microbiota transplantation to determine the role of microbiota on the fermentation type of oat silage. Biores Technol. 309, 123371. doi: 10.1016/j.biortech.2020.123371
Xu, D. M., Wang, N., Rinne, M., Ke, W. C., Weinberg, Z. G., Da, M., et al. (2020). The bacterial community and metabolome dynamics and their interactions modulate fermentation process of whole crop corn silage prepared with or without inoculants. J. Microbial. Biotechnol. 14, 561–576. doi: 10.1111/1751-7915.13623
You, S. H., Du, S., Ge, G. T., Wan, T., and Jia, Y. S. (2021a). Selection of lactic acid bacteria from native grass silage and its effects as inoculant on silage fermentation. Agron. J. 113, 3169–3177. doi: 10.1002/agj2.20720
You, S. H., Du, S., Ge, G. T., Wan, T., and Jia, Y. S. (2021b). Microbial community and fermentation characteristics of native grass prepared without or with isolated lactic acid bacteria on the Mongolian Plateau, Front. Microbiol. 12, 731770. doi: 10.3389/fmicb.2021.731770
Yuan, X. J., Li, J. F., Dong, Z. H., and Shao, T. (2020). The reconstitution mechanism of napier grass microiota during the ensiling of alfalfa and their contributions to fermentation quality of silage. Bioresour. Technol. 297, 122391. doi: 10.1016/j.biortech.2019.122391
Zhao, J., Dong, Z. H., Li, J. F., Chen, L., Bai, Y. F., Jia, Y. S., et al. (2018). Ensiling as pretreatment of rice straw: the effect of hemicellulase and Lactobacillus plantarum on hemicellulose degradation and cellulose conversion. Bioresour. Technol. 266, 158–165. doi: 10.1016/j.biortech.2018.06.058
Zhou, Y. L., Sun, L., Cheng, Q. M., Li, Y. C., Chen, J. X., Zhao, B., et al. (2022). Effect of pelleted alfalfa or native grass total mixed ration on the rumen bacterial community and growth performance of lambs on the Mongolian Plateau. Small Ruminant Res. 207, 106610. doi: 10.1016/j.smallrumres.2021.106610
Keywords: lactic acid bacteria, fermentation quality, bacterial community, silage, native grass
Citation: Du S, You S, Jiang X, Li Y, Wang R, Ge G and Jia Y (2022) Evaluating the fermentation characteristics, bacterial community, and predicted functional profiles of native grass ensiled with different additives. Front. Microbiol. 13:1025536. doi: 10.3389/fmicb.2022.1025536
Received: 23 August 2022; Accepted: 30 September 2022;
Published: 18 October 2022.
Edited by:
Siran Wang, Nanjing Agricultural University, ChinaReviewed by:
Qing Zhang, South China Agricultural University, ChinaJie Zhao, Nanjing Agricultural University, China
Copyright © 2022 Du, You, Jiang, Li, Wang, Ge and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuai Du, ZHVzaHVhaV9ubUBzaW5hLmNvbQ==; Yushan Jia, anlzX25tQHNpbmEuY29t
 Shuai Du
Shuai Du Sihan You
Sihan You Xiaowei Jiang3
Xiaowei Jiang3 Yushan Jia
Yushan Jia