- Laboratory of Bacterial Drug Resistance, Institute of Biochemistry and Biophysics Polish Academy of Sciences, Warsaw, Poland
Streptococcus anginosus together with S. constellatus and S. intermedius constitute the Streptococcus anginosus group (SAG), until recently considered to be benign commensals of the human mucosa isolated predominantly from oral cavity, but also from upper respiratory, intestinal, and urogenital tracts. For years the virulence potential of SAG was underestimated, mainly due to complications in correct species identification and their assignment to the physiological microbiota. Still, SAG representatives have been associated with purulent infections at oral and non-oral sites resulting in abscesses formation and empyema. Also, life threatening blood infections caused by SAG have been reported. However, the understanding of SAG as potential pathogen is only fragmentary, albeit certain aspects of SAG infection seem sufficiently well described to deserve a systematic overview. In this review we summarize the current state of knowledge of the S. anginosus pathogenicity factors and their mechanisms of action.
Introduction
In the genus Streptococcus the species S. pyogenes, S. agalactiae, S. pneumoniae, and S. dysgalactiae subsp. equisimilis (SDSE) are most frequently responsible for morbidity and mortality in humans (Carapetis et al., 2005; O’Brien et al., 2009; Rantala, 2014; Lawn et al., 2017). However, other streptococci, previously considered just human commensals, are recognised as a cause of human diseases, e.g., oral streptococci S. sanguinis (Vogkou et al., 2016; Zhu et al., 2018), S. intermedius (Sadykov et al., 2003; Mishra and Fournier, 2013; Issa et al., 2020), S. constellatus (Akuzawa et al., 2017), and S. anginosus (Clarridge et al., 2001; Asam and Spellerberg, 2014). Clinical data indicate that also S. suis and S. iniae, porcine and fish pathogens, respectively, can infect humans (Weinstein et al., 1997; Lun et al., 2007; Baiano and Barnes, 2009). Distinguishing between pathogenic and non-pathogenic strains was historically based on their ability to lyse erythrocytes on blood-agar plates: those forming a clear zone of hemolysis (β-hemolysis), were considered potentially pathogenic. A further differentiation was proposed in 1933 by Rebecca Lancefield, who identified different versions of the major cell wall polysaccharide of streptococci, later called the group polysaccharide or the Lancefield antigen. According to this classification, the human pathogens S. pyogenes and S. agalactiae belong to group A (GAS) and group B (GBS) streptococci, respectively. Other streptococci are not homogenous in expressing the Lancefield antigens, e.g., S. anginosus strains can bear group F, C, G, or even group A antigens, or none (Obszańska et al., 2016; Baracco, 2019; Figure 1). Rather than basing on the antigens, the currently accepted classification of streptococci reflects their phylogenetic relationships. Accordingly, they are classed into seven groups: pyogenic, anginosus, mitis, salivarius, bovis, mutans, and species of unknown position, as for example S. suis (Köhler, 2007). S. anginosus, S. constellatus and S. intermedius constitute the Streptococus anginosus group (SAG, the Anginosus group). Formerly, these three species were regarded by European and Japanese microbiologists as a single species “Streptococcus milleri.” However, they were distinguished by microbiologists from North America. [reviewed in (Coykendall, 1989) and recently in (Pilarczyk-Zurek et al., 2022)]. Previously, SAG was classed in a diverse group of streptococci called the viridans group which was poorly characterized(Coykendall, 1989). The name “viridans” comes from the Latin “viridis” meaning “green” and was given to the bacteria due to a greenish halo around their colonies on blood agar. The green or brown colouring results from the oxidation of hemoglobin to methemoglobin and is called α-hemolysis. However, the name viridans was misleading, as not all S. anginosus strains present this type of hemolysis: some are β-hemolytic and some are even nonhemolytic, in other words – they present γ-hemolysis, a confusing term referring to a lack of hemolysis (Coykendall, 1989; Whiley et al., 1990; Fox et al., 1993; Broyles et al., 2009; Fischetti and Ryan, 2015; Whiley and Hardie, 2015; Figure 1). In 2013, a further differentiation of SAG was proposed on the basis of the nucleotide sequences of seven housekeeping genes: S. constellatus was divided into three subspecies, constellatus, pharyngis, and viborgensis, and S. anginosus into two subspecies, anginosus and whileyi; S. intermedius remained as a single species (Jensen et al., 2013). The type strain S. anginosus NCTC 10713 ( = ATCC 33397, ATCC 12395), a β-hemolytic strain with the Lancefield group G antigen isolated from human throat, is classified as S. anginosus subsp. anginosus. It should be noted here that detection of Lancefield antigen and biochemical tests used in laboratory practice for SAG identification, and even mass spectrometry (MALDI-TOF-MS), do not allow a reliable identification at species level and the results should be viewed with caution (Whiley et al., 1990; Wenzler et al., 2015). The precise SAG species identification can be achieved with the use of the molecular biology-based methods; the SAG identification issues have been recently reviewed in (Pilarczyk-Zurek et al., 2022).
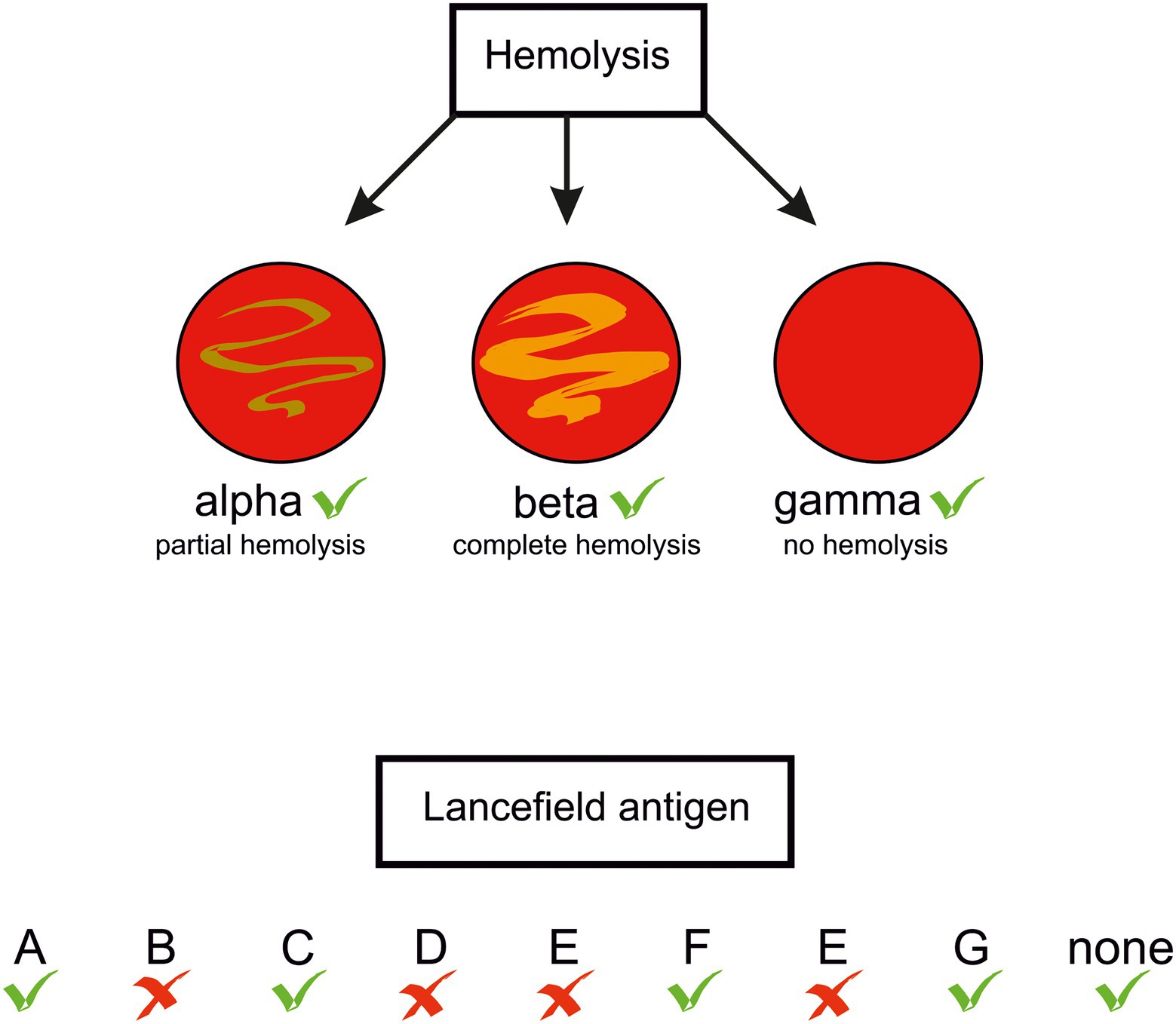
Figure 1. Types of hemolysis and Lancefield antigens presented by S. anginosus strains. Green tick –feature present in some S. anginosus strains, red cross – absent.
In the past all SAG species were considered to be human commensals, part of the healthy human microbiota, commonly residing on mucosal membranes of the oral cavity, preferentially found in dental plaques, but also on mucosal membranes of the gastrointestinal, upper respiratory, and urogenital tracts (Hamada and Slade, 1980; Whiley et al., 1992). However, more and more data link SAG members with health problems in humans, especially in immunocompromised, cancer, or cystic fibrosis patients [reviewed in (Pilarczyk-Zurek et al., 2022)]. According to observations, the clinical syndromes differed depending on the SAG species involved: S. constellatus was associated with odontogenic, soft tissue, pleuropulmonary, and intra-abdominal abscesses, S. intermedius with pleuropulmonary infections, abscesses of brain, and deep soft tissues, whereas S. anginosus was rarely responsible for abscesses and more frequently than two other SAGs was isolated from blood, infected soft tissues, and urine (Whiley et al., 1992; Clarridge et al., 2001; Junckerstorff et al., 2014; Issa et al., 2020; Jiang et al., 2020). Nevertheless, there are several reports of isolation of S. anginosus not only from dental abscesses, but also from infective endocarditis, abscesses of brain, liver, spleen, corpus cavernosum, probably as a result of hematogenous spread (Fisher and Russell, 1993; Dugdale et al., 2013; Junckerstorff et al., 2014; Esplin et al., 2017; Finn et al., 2018; Wu and Zheng, 2020). A correlation between S. anginosus infections in oncological patients with esophageal cancer have been suggested (Morita et al., 2003; Rawla et al., 2017). S. anginosus was frequently isolated from the sputum of cystic fibrosis (CF) patients together with Pseudomonas aeruginosa and Staphylococcus aureus, the principal CF pathogens, and presumably was responsible for exacerbation of pneumonia (Waite et al., 2012; Whiley et al., 2014). In vitro S. anginosus was shown to enhance P. aeruginosa pathogenicity (Whiley et al., 2014; Waite et al., 2017). S. anginosus was also recovered from patients in co-infections with other species, e.g., Mycobacterium tuberculosis, and Eikenella corrodens (Rabuñal et al., 2009; Patel et al., 2020). All these data strongly suggest that S. anginosus should be viewed as an emerging opportunistic pathogen of substantial clinical importance (Reißmann et al., 2010; Pilarczyk-Zurek et al., 2022).
Streptococci use diverse strategies to adhere to, invade, and finally colonize host cells or tissues to acquire nutrition, and also to evade the defence mechanisms by suppressing the immune response (Zachary, 2017; Sitkiewicz, 2018). Numerous streptococcal proteins responsible for the adherence to eukaryotic cells have been identified. They interact with components of the connective tissue and the extracellular matrix (ECM), such as collagen, fibrinogen, fibronectin, and laminin, and also bind to salivary proteins (Nobbs et al., 2009). The attachment to host surfaces is mediated by adhesins, e.g., lectin-like adhesins, and extracellular appendages such as pili and fibrils (Nobbs et al., 2009; Schüler et al., 2012). To avoid being eliminated, streptococci trick the host immune system by hiding their antigens under thick capsules, by surviving inside macrophages, or by inducing macrophage apoptosis with pore-forming enzymes from the hemolysin group (Dinkla et al., 2007; Timmer et al., 2009; Cumley et al., 2012). Using nuclease A or DNases (Chang et al., 2011; de Buhr et al., 2014), streptococci dismantle neutrophil extracellular traps (NETs) built of chromosomal DNA and bactericidal proteins, which are released by neutrophils upon stimulation with, i.e., interleukin (IL)-8 or hydrogen peroxide (Von Köckritz-Blickwede and Nizet, 2009). Other virulence factors, such as hyaluronidase and chondroitin sulphatase, are involved in degradation of the host ECM (Asam and Spellerberg, 2014). SRRP1 (serine-rich repeat protein 1) and enolase aid the crossing of the blood – brain barrier (Sorge et al., 2009; Sun et al., 2016). Individual streptococcal species produce different repertoires of virulence factors, not all of equal importance.
Our understanding of the repertoire of virulence factors of SAG, and especially of their regulation, is still rather limited, although some mechanisms and their role in different stages of SAG infections seem firmly establish. Because of the progress in this field in recent years, the two brilliant reviews by Olson (Olson et al., 2013) on streptococcal virulence factors identified in SAG genomes in silico, and by Asam and Spellerberg (2014) on molecular pathogenicity of SAG species, published, respectively, in 2013 and 2014, are no longer up to date. Therefore, in the present review we will update the information on S. anginosus, its virulence factors and their prevalence in strains isolated from humans.
Streptococcal virulence factors
The term “virulence factor” itself refers to a component or structure of a microorganism that helps in the establishing of an infection or in eliciting a disease. During an infection, the virulence factors of bacteria battle the protection mechanisms of the host. A streptococcal infection proceeds in three main stages: adhesion to, invasion, and colonization of the host tissues (Nobbs et al., 2009). In the genus Streptococcus, 189 genes coding for known and putative virulence factors have been identified (Olson et al., 2013). An analysis of 18 clinical SAG strains has revealed the presence of 55 of such factors in the SAG pan-genome, however, an individual S. anginosus carried only 30–34 virulence genes, of which 16 genes were found in all the SAGs analysed (Olson et al., 2013). In this review, factors with an experimentally-determined role in different aspects of S. anginosus virulence will be presented in detail, while the putative ones will only be mentioned briefly.
Adhesion
The most important step of infection is adhesion of a pathogenic bacteria to the host tissue. This step determinates whether the bacterium will continue invasion and colonisation or not. Several host-tissue components can be exploited as targets of adhesion: fibronectin, fibrinogen, collagen, laminin and other proteins of the ECM.
Fibronectin-binding proteins
Fibronectin, a protein found in the ECM of connective tissue, is an important ligand used by Gram-positive cocci to adhere to host tissues. Fibronectin binding proteins (FPBs), are also called “microbial surface components recognizing adhesive matrix molecules,” or MSCRAMMs (Schwarz-Linek et al., 2004). FBPs are anchored in the bacterial cell wall, and typically comprise an N-terminal signal peptide for secretion, an LPXTG membrane anchorage motif, and a fibronectin-binding domain (Hanski et al., 1992). However, several atypical FBPs of streptococci are also known which lack a conventional secretion signal, an anchorage motif, or a typical fibronectin-binding domain, e.g., Fbp54 of S. pyogenes (Courtney et al., 1996). A gene encoding an atypical FBP homologue was also identified in S. anginosus NCTC 10713 type strain (Kodama et al., 2018). The fbp62 gene codes for a 62.8-kDa cell-wall localized protein (Fbp62) without a signal peptide and a membrane anchorage motif. While S. anginosus NCTC 10713 was able to bind to immobilized fibronectin and epithelial cells of the HEp-2 (human laryngeal carcinoma) and DOK (human dysplastic keratinocyte) lines, its knock-out mutant Δfbp62 was not. Moreover, the fbp62 mutation lowered mortality and abscess formation in a mouse infection model (Kodama et al., 2018). These results imply that Fbp62 is a bona fide virulence factor of S. anginosus. Earlier studies have revealed that all clinical SAG isolates recovered from abscesses, chest infection or nephritis can bind rat fibronectin, but with efficiencies varying 10-fold, especially among the S. anginosus strains (Willcox, 1995).
Also glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of S. pyogenes or S. suis, a glycolytic enzyme, found at the cell surface of these bacteria, binds fibronectin. By showing these two unrelated activities it can be classed as a moonlighting protein [for review (Henderson and Martin, 2011)]. In addition to fibronectin, S. pyogenes GAPDH binds lysozyme, myosin, and actin (Pancholi and Fischetti, 1992). The fibronectin-binding function of GAPDH in S. anginosus has not been tested. Nevertheless, a fraction of this enzyme was detected on the cell surface and among plasminogen-binding proteins (Kinnby et al., 2008; see below). In another survey the gapC gene, coding for a monomer of cell-surface located GAPDH, was detected by PCR in all 22 S. anginosus clinical strains tested, indicating that GAPDH could act as a virulence factor also in S. anginosus (Chang and Lo, 2013).
Fibrinogen-binding proteins
Fibrinogen (Fg) is a blood plasma glycoprotein composed of two trimers of non-identical α, β, and γ chains. It plays a crucial role in hemostasis and in the innate immune system by forming insoluble fibrin clots (Herrick et al., 1999; Rubel et al., 2001). Numerous proteins of diverse Gram-positive bacteria, including streptococci can interact with Fg by distinct mechanisms [for review (Rivera et al., 2007)]. In S. pyogenes the M protein, its main virulence factor, binds Fg and plays an anti-phagocytic role thus enhancing bacterial survival in the host (Courtney et al., 2006). Besides Fg, the M protein can also interact with other serum proteins, such as plasminogen and immunoglobulins, and with collagen (Courtney et al., 2002). In S. suis muramidase-released protein (MRP), a 136-kDa cell wall-anchored surface protein, is a major fibrinogen-binding protein. A recent study showed that binding of MRP with human fibrinogen improves the viability of S. suis in the human blood and also favours the development of meningitis (Pian et al., 2015). Two fibrinogen-binding proteins in S. agalactiae, FbsA and FbsB, were shown to bind human fibrinogen and form a semi-flexible polymer-like network effectively preventing phagocytosis (Tenenbaum et al., 2005). Ability to bind human Fg was also shown for clinical S. anginosus strains, although the exact mechanism of Fg binding by S. anginosus is not yet clear (Willcox, 1995). Moreover, a whole-genome comparative analysis of seven other clinical S. anginosus strains identified eight proteins out of 58 with the LPXTG motif as fibrinogen-binding ones (Olson et al., 2013).
Laminin-binding protein
Laminins are glycoproteins of the ECM and a major component of the basement membrane; laminin binding facilitates adhesion of streptococci. The clinical S. anginosus strain SLEH753 isolated from infective endocarditis was shown to adhere strongly to an exposed basement membrane of human and porcine valves (mimicking native endovascular surface) in a laminin- as well as fibronectin-dependent manner (Allen et al., 2002). An 80-kDa surface lipoprotein was isolated from this strain as a putative laminin binding protein (PLBP), albeit direct binding of the purified PLBP to laminins was not shown (Allen and Höök, 2002).
Putative adhesion factors
A/ Serum opacity factor (SOF), an extracellular lipoproteinase, is another major fibronectin-binding protein of streptococci expressed by ca. 50% of invasive S. pyogenes strains (Courtney and Pownall, 2010). Its main function is disrupting the organization of high-density lipoproteins, which results in the formation of large water-insoluble lipid vesicles making the serum opaque. Apart from fibronectin, SOF binds other host molecules involved in bacterial adherence, such as fibrinogen and fibulin-1 (Courtney and Pownall, 2010). In S. pyogenes SOF is encoded by the sof gene. Its homologue was detected in several clinical isolates of various streptoccoci, including two of five analysed strains of SAG of group F (Reitmeyer et al., 1989).
B/ Pneumococcal surface adhesin A (PsaA) is a 37-kDa protein encoded by the psaA gene reported in a S. pneumoniae strain serotype 6B (Jado et al., 2001). This protein is likely to be involved in pneumococcal virulence, as immunization of mice with PsaA reduced the nasal load of pneumococci (Briles et al., 2000). The psaA homologue identical in 95% or 94% to the pneumococcal one was found in S. mitis and S. oralis, respectively (Jado et al., 2001). In S. anginosus type strain NCTC 10713 the psaA homologue is 90% identical with pneumococcal psaA. However, its role in SAG virulence has not been studied.
C/ Antigen I/II – the major adhesins of oral cavity streptococci are Antigens I/II (Ag I/II), cell surface-anchored proteins of ca. 180–210 kDa (Moisset et al., 1994). Proteins of the Ag I/II family are multifunctional, with functions varying among bacterial species and the niches occupied in the host [for review (Manzer et al., 2020)]. The first Ag I/II was characterized in S. mutans where it plays a crucial role in teeth colonisation. It is encoded by the spaP gene (also called pac; Russell et al., 1980). Another S. mutans Ag I/II-related protein, SR (salivary receptor), interacts with salivary glycoproteins adsorbed on the tooth surface (Hajishengallis et al., 1992). When present in the bloodstream, the Ag I/II proteins can modulate the immune response and induce a proinflammatory reactions (for review (Manzer et al., 2020)). Sequences coding for homologues of the Ag I/II and SR antigen were detected in two α-hemolytic S. anginosus strains NMH10 and PC4890 by Southern hybridization with the spaP gene fragments (Ma et al., 1991). Further studies are needed to confirm the presence of the Ag I/II-like antigens on the cell surface of SAGs.
D/ Pneumococcal adherence and virulence factor B (PavB), encoded by the pavB gene, is a surface-exposed adhesin of S. pneumoniae. A homologue of pavB has been detected in some S. anginosus strains. It codes for a protein 62% identical to pneumococal PavB on 56% of its length (Olson et al., 2013).
F/ Internalin A is a major invasion protein of Listeria monocytogenes, an intercellular pathogen, involved in the attachment to hepatocyte, epithelial, and endothelial cells and in the invasion of their vacuoles; it is encoded by inlA (Bierne et al., 2018). An internalin A homologue, the Slr (Streptococcus leucine-rich) protein, is produced by S. pyogenes (Reid et al., 2003). A gene homologous to inlA was also found in two of seven genomes of S. anginosus strains analysed. It codes for a protein 47% identical to internalin on 89% of its length (Sawyer et al., 1996; Olson et al., 2013).
G/ Fimbriae (Pili). Bacterial adhesion to surfaces and the resulting virulence is also dependent on hair-like structures located on the bacterial cell surface. In contrast to those of Gram-negative bacteria, in Gram-positive bacteria they are not well described [for review (Telford et al., 2006)]. Two types of surface appendages have been detected in Gram-positive bacteria: fimbriae (also called pili) – up to 3 μm long, flexible, thick rods (3–10 nm in diameter), and fibrils – short, thin rods (1–2 nm in diameter) extending for 70–500 nm from the bacterial surface. Historically, pili and fimbriae were discovered independently and named by two research groups. Although nowadays their names are considered synonymous (Telford et al., 2006), fimbriae seem to be related mainly to adhesion, while pili also to DNA uptake (Ottow, 1975). The pili of Gram-positive bacteria are composed of three covalently bound protein subunits carrying LPXTG sorting motif and also covalently bound to the peptidoglycan. The amino acid sequences of the pili of invasive streptococcal strains are similar to the MSCRAMM proteins involved in interaction with components of the ECM. Genes encoding pili subunits and a sortase, as well as their regulators are located in the pilus pathogenicity island organised similarly in all main groups of streptococci such as GAS, GBS, and pneumococci (Telford et al., 2006). Fibrils produced by streptococci help to bind fibronectin and better adhere to the host tissue (Telford et al., 2006). They have been identified in, e.g., S. agalactiae, S. pneumoniae, S. pyogenes, S. mutans, and in two SAG species: S. intermedius, and S. constellatus [(see Nobbs et al., 2009)].
In S. intermedius fimbriae are involved in saliva-mediated aggregation and associated with adherence. They are encoded by the saf operon, where saf3 encodes the main pilus subunit (Yamaguchi et al., 2009). An analysis of 16 clinical strains of SAG, including six S. anginosus strains, identified the saf3 gene in only four S. intermedius strains (Yamaguchi et al., 2009). In an earlier study the saliva-mediated aggregation was observed for 17 S. intermedius and two S. constellatus strains and for none of the six S. anginosus strains (Yamaguchi and Matsunoshita, 2004). It is therefore likely that S. anginosus does not produce fimbriae.
Invasion and colonisation
Factors enabling bacteria to evade the host immune system and to improve the niche for living are considered important for the colonisation and invasion of host tissues.
Capsule
One of the first streptococcal virulence factors identified was the capsule. It is built from capsular polysaccharide (CPS) and has been detected in numerous streptococcal strains of various species: S. pyogenes, S. pneumoniae, S. mitis, S. gordonii, S. agalactiae, S. suis, and SAGs (Smith et al., 1999; Rukke et al., 2012; Toniolo et al., 2015; Skov Sørensen et al., 2016). The biochemical composition of the capsule differs between species. For example, in S. pyogenes capsular polysaccharide is hyaluronic acid [for review (Wessels, 2019)], and the simplest CPS of S. pneumoniae is a linear polymer with a repeat unit comprising two or more monosaccharides [for review (Paton and Trappetti, 2019)]. A typical cps cluster encoding enzymes of the CPS synthesis pathway in S. pneumoniae is located in the chromosome between the dexB and aliA genes, and comprises 12 to 20 genes. The capsule not only helps S. pneumoniae to defend its own cells against the host immune system, phagocytosis and binding by NETs, but it also contributes to pathogenesis in many ways and is absolutely required in systemic infections (Wartha et al., 2007; Hyams et al., 2010; Paton and Trappetti, 2019).
A cps locus similar to that of S. pneumoniae is often harboured by oral streptococci belonging to the groups, salivarius (S. salivarius), mitis (S. mitis and S. oralis), sanguinis (S. sanguinis and S. gordonii), and anginosus (S. anginosus and S. intermedius; Skov Sørensen et al., 2016). The main role of the capsule of oral streptococci is adherence to salivary components, biofilm formation, and interaction with other bacterial species; in the latter case CPS acts as a receptor for lectin-like adhesins (Cisar et al., 1995).
The structure of the locus associated with CPS synthesis has been studied in detail in type strain S. anginosus ATCC 33397 (NCTC 10713; Tsunashima et al., 2012; Figure 2). The region consists of 24 ORFs with 14 central genes, cpsA – cpsN, directly associated with CPS synthesis. The first four, cpsA – cpsD, are predicted to code for, respectively, a membrane-bound transcription factor for the cps operon, phosphotyrosine-protein phosphatase, a regulatory protein, and a tyrosine protein kinase. Based on the homology of the subsequent cps genes of S. anginosus to cps genes of other streptococci, they are inferred to encode transferases for undecaprenyl-phosphate glucose-1-phosphate (cpsE), rhamnose (cpsF), N-acetylgalactosamine (cpsG), O-acetylserine (cpsH), acetylgalactosamine (cpsI), rhamnose (cpsJ), and galactofuranose (cpsK). The following genes code for: the polysaccharide polymerase Wzy (cpsL) which joins the repeating units to form polysaccharide, the repeating unit transporter (flippase) Wzx (cpsM) engaged in transporting of the repeating units to the outer surface of the membrane and UDP-galactopyranose mutase (cpsN). The importance of cpsE for CPS production in S. anginosus has been verified experimentally (Tsunashima et al., 2012).

Figure 2. Organization of the cps locus of S. anginosus. Arrows depict genes coding for: transferases (green), regulatory proteins (yellow), polymerase and flippase (orange), mutase (dark green), other genes are in grey. Details in the text.
The genes upstream of cpsA code for: nrdD – ribonucleoside-triphosphate reductase, orfW, orfX and orfY – acetyltransferases, nrdG – anaerobic ribonucleoside triphosphate reductase activator protein. Downstream of cpsN lie genes encoding: orfO – a phosphoglycerate mutase family protein, orfP – a cell wall surface anchor family protein, orfQ – a DNA-binding response regulator, orfR – a sensor histidine kinase, and polI – DNA polymerase I (Tsunashima et al., 2012). The cps loci of S. gordonii and S. sanguinis have an organization and chromosomal localization similar to that of S. anginosus (Tsunashima et al., 2012).
Plasminogen-binding proteins: α-enolase and glyceraldehyde-3-phosphate dehydrogenase
α-Enolase is a glycolytic enzyme, but it can also be found on the surface of bacterial cells where it acts as a receptor for human plasminogen to aid invasion (Kornblatt et al., 2011). Similarly to GAPDH, α-enolase is therefore a moonlighting protein [for review (Henderson and Martin, 2014)]. α-Enolase in its receptor function has been found in S. pneumoniae, S. pyogenes, S. suis, S. mutans, and S. iniae (Pancholi and Fischetti, 1998; Bergmann et al., 2001; Ge et al., 2004; Esgleas et al., 2008; Membrebe et al., 2016). In S. suis α-enolase can also bind fibronectin and fibrinogen (Esgleas et al., 2008; Pian et al., 2015), thereby improving an antiphagocytic effect.
The streptococcal α-enolases are homo-octamers of 45-kDa monomers, each with two potential plasminogen binding sites (Cork et al., 2009; Kornblatt et al., 2011). In S. pneumoniae α-enolase the plasminogen binding depends on two C-terminal lysine residues and an internal plasminogen binding motif (IPM; Itzek et al., 2010). An analysis of 56 clinical strains of oral streptococci, including eight SAG strains, has shown that IPM is conserved in their α-enolases (Itzek et al., 2010). Plasminogen binding was analysed in detail in three S. anginosus strains of human oral origin. In one S. anginosus strain the plasminogen-binding proteins were identified as α-enolase and phosphoglycerate mutase; the second produced 11 plasminogen-binding proteins, of which five were identified as the glycolytic enzymes α-enolase, phosphoglycerate kinase, GAPDH, phosphoglycerate mutase, and triosephosphate isomerase; and the third – 14 proteins identified as standard glycolytic enzymes or their isoforms (Kinnby et al., 2008).
Streptococcal hemolysins
Hemolysins are well-described streptococcal virulence factors. Different types of hemolysins have been identified and named after the species or genus in which they were detected. Some hemolysins are produced by several species, like streptolysin S (SLS), an oxygen stable cytolytic toxin of S. pyogenes, S. iniae, and S. constellatus, and streptolysin O, an oxygen labile one, produced by S. pyogenes, S. canis, and S. dysgalactiae subsp. equisimilis (SDSE). Both of them lyse erythrocytes, leukocytes and platelets, forming holes in their membranes. Streptolysin S is a small peptide (2.7 kDa) synthesised ribosomally and extensively modified post-translationally (Nizet et al., 2000). The latter feature makes it related to the post-translationally modified bacteriocins (Molloy et al., 2011). These modifications result in the formation of aromatic thiazole and (methyl-)oxazole heterocycles prior to SLS export (Lee et al., 2008). Proteins involved in SLS production are encoded by the sag operon. Typically, as in S. pyogenes, it comprises nine genes sagA–I, with sagA encoding an SLS precursor. The sagB product is a 36.0-kDa protein 22% identical to a dehydrogenase that modifies McbA, the precursor of the E. coli bacteriocin microcin B17, one of the thiazole/oxazole-modified microcins (TOMMs). SagC (40.3 kDa) is 13% identical to the McbA cyclodehydratase, and SagD (51.6 kDa) is a component of the SagBCD complex and probably regulates its enzymatic activity (Nizet et al., 2000; Lee et al., 2008). sagE encodes a 25.4-kDa immunity protein localized in the membrane, similarly to the 26.2-kDa product of sagF. The sagGHI-encoded proteins are putative ABC transporters, important in extracellular transport (Nizet et al., 2000).
Streptolysin O (63,65 kDa) is a cholesterol-dependent cytolysin composed of 571 amino acids, with a 33-residues N-terminal secretion signal peptide (Kehoe et al., 1987). Other hemolysins seem to be unique to single bacterial species only, e.g., intermedilysin to S. intermedius (Nagamune et al., 1996), suilysin to S. suis (Xu et al., 2010), and pneumolysin to S. pneumoniae (Shumway and Klebanoff, 1971). Although most hemolysins show the expected hemolytic activity, there are exceptions – some pneumolysins have no hemolytic activity (Nadeem Khan et al., 2014). Pneumolysins were originally believed to be released only upon S. pneumoniae death, but later data showed that their active forms are in fact bound to the bacterial cell surface (Price and Camilli, 2009).
Notably, recent data have revealed that various streptococcal hemolysins have more sophisticated functions than simple lysis of erythrocytes. Streptolysin O suppresses the neutrophil oxidative burst, a rapid production of reactive oxygen species in response to pathogen invasion, and also helps to avoid the bacteria killing by neutrophil by blocking their degranulation, interleukin-8, and elastase secretion, which in turn suppresses the formation of NETs (Timmer et al., 2009; Uchiyama et al., 2015). Some hemolysins also affect the invasion of eukaryotic cells, as for example the cell-bound fraction of intermedilysin during invasion of human liver cell line HepG2 by S. intermedius (Sukeno et al., 2005). Also pneumolysins are involved in the S. pneumoniae pathogenicity regardless of their hemolytic activity, as they are engaged in biofilm formation (Kirkham et al., 2006; Shak et al., 2013).
Hemolysins of Streptococcus anginosus
As mentioned earlier, few of the S. anginosus strains isolated from infections are β-hemolytic. The β-hemolysin of S. anginosus has been identified as SLS (Asam et al., 2013; Tabata et al., 2013), no streptolysin O producing strains have been reported. SLS is synthesised as a precursor comprising a 23-amino acid signal peptide and a 30-amino acid structural peptide which is further modified post-translationally, similarly to other members of the SLS-like group of peptides (Bernheimer, 1967; Molloy et al., 2011). SLS is also classified as a member of the TOMM family. Similarly to gene clusters coding for other TOMM bacteriocins, the sagA gene encoding the SLS is followed by sagB, sagC and sagD, products of which introduce aromatic thiazole and (methyl-)oxazole heterocycles onto the SLS (Lee et al., 2008; Molloy et al., 2011). In β-hemolytic S. anginosus strains, the sag operon is composed of 10 genes sagA-I, including two copies of sagA, sagA1 and sagA2, encoding SLS (Tabata et al., 2013). The genes responsible for the β-hemolytic phenotype were first identified in S. anginosus type strain NCTC 10713 (Tabata et al., 2013). A PCR screen of 125 clinical S. anginosus strains for the sagA1 and sagA2 genes showed that all β-hemolytic strains (18.4%; n = 23) carried the both genes, as did the type strain NCTC 10713, and all nonhemolytic strains (81.6%; n = 102) carried neither (Tabata et al., 2013). A mutant of NCTC 10713 depleted of both sagA genes was nonhemolytic, and introduction of either sagA1 or sagA2 restored the β-hemolytic activity. Also both single NCTC 10713 mutants, ΔsagA1 and ΔsagA2, were still β-hemolytic (Tabata et al., 2013).
SLS of S. anginosus is a broad-range β-hemolysin causing lysis of red blood cells of humans, horse, sheep, cattle, rabbit and even chicken (Asam et al., 2015). SLS shows no cytotoxic activity towards THP-1 (acute monocytic leukemia) cells or human granulocytes after 2 h of treatment (Asam et al., 2015). However, after 24 h morphological changes, such as a flattened morphology and bleb formation are observed in the THP-1 and HSC-2 (human oral squamous cell carcinoma line) cell lines, presumably due to the leakage of the cellular contents following disruption of the cell membrane (Tabata et al., 2019). Those changes were shown to be SLS-depended: the β-hemolytic strain NCTC 10713 markedly decreased the HSC-2 cells survival, while NCTC 10713 depleted of both sagAs was not cytotoxic, demonstrating that SLS of the β-hemolytic S. anginosus NCTC 10713 is responsible for cytotoxicity (Tabata et al., 2019).
Prevalence of β-hemolytic Streptococcus anginosus strains
The prevalence of β-hemolytic strains of S. anginosus among natural isolates is not entirely clear. While the survey of 125 clinical strains mentioned earlier (Tabata et al., 2013) found as much as 18.4% (n = 23) of those to be β-hemolytic, this proportion was markedly lower in two other studies. An analysis of 164 strains of S. anginosus isolated from throats of 1,480 US students with pharyngitis reported only 1.2% β-hemolytic ones (n = 2; Fox et al., 1993), and a survey of 29 S. anginosus strains isolated from patients with skin or soft tissue infections found only 7% (n = 2; Summanen et al., 2009). Notably, in the latter analysis all the other strains were α-hemolytic (Summanen et al., 2009). It is worth mentioning that the analysis described by Fox et al. (1993) was performed before the molecular biology-based methods of SAG identification were elaborated. The prevalence of β-hemolytic strains among isolates from apparently healthy subjects is even smaller: among SAG strains isolated from throat swabs collected from 3,416 healthy children in India only ca. 0.18% (n = 6) isolates were β-hemolytic (Nayak et al., 2019). Similarly no β-hemolytic strains were found among 25 samples collected from non-symptomatic students in the US study (Fox et al., 1993). β-hemolysis of S. anginosus is overall rare, albeit more frequent among clinical strains isolated from infections. Still, since a vast majority of the clinical strains are non-β-hemolytic one may conclude that, while being a cytotoxin, SLS, is not a critical virulence factor of S. anginosus.
DNases
Historically, four different types of extracellular DNases, A, B, C, and D, produced by GAS have been distinguished serologically, and each strain produces at least one of them (Wannamaker, 1958; Wannamaker et al., 1967; Wannamaker and Yasmineh, 1967). DNase B, known as mitogenic factor (MF) or streptococcal pyrogenic exotoxin F (SPE-F), is chromosomally encoded by the spd (also known as mf, speF or spdB) gene and is specific for GAS (Sriskandan et al., 2000; Das et al., 2017). DNase C and DNase D are encoded on prophages by genes spd3 and sdaD2, respectively (Sriskandan et al., 2000; Sumby et al., 2005). Currently, DNase A remains known only serologically and as a protein. Up till now, still other GAS DNases have been identified [for review (Remmington and Turner, 2018)], including cell-wall-anchored nuclease A encoded by spnA (Hasegawa et al., 2010; Chang et al., 2011). It took over 60 years from discovery of the GAS-produced extracellular DNases to the conclusion about their importance in the pathogen invasion (Sumby et al., 2005). The DNases protect S. pyogenes from extracellular killing by degrading DNA scaffolding of the NETs (Beiter et al., 2006; Buchanan et al., 2006; de Buhr et al., 2014). In turn, by degrading streptococcal DNA, DNases enable evading the host innate immune system by avoidance of Toll-like receptor 9 (TLR9)-mediated recognition of unmethylated CpG-rich motifs in bacterial DNA, followed by cytokine overproduction (Uchiyama et al., 2012). Extracellular DNases have been identified in S. suis, S. pyogenes, S. pneumoniae, and S. agalactiae (Hasegawa et al., 2002a; Beiter et al., 2006; Korczynska et al., 2012; Derré-Bobillot et al., 2013; de Buhr et al., 2015). Moreover, GAS, similarly to SDSE, secretes streptodornase, encoded by the sda gene (sdc in SDSE; Wolinowska et al., 1991; Hasegawa et al., 2002b).
The DNase activity was tested for 518 SAG strains from clinical specimens (Jacobs and Stobberingh, 1995). Among 307 S. anginosus isolates the majority (63%) were positive for DNase activity, and the corresponding figures for S. constellatus and S. intermedius strains were 58 and 78%, respectively. It is worth noting here that species identification was based on antigenic and biochemical tests. Different results were obtained in a survey of 128 SAG strains from sputum of CF patients, where among 45 isolates recognised as S. anginosus all but one (98%) were DNase producers (Grinwis et al., 2010).
Hyaluronidase
Hyaluronic acid is the major component of the ECM of the host connective tissue. Hyaluronan is a mediator of inflammation, however its role depends of its size: small fragments are regarded pro-inflammatory while large ones serve to suppress the inflammatory response [for review (Petrey and de la Motte, 2014)]. Hyaluronidase (hyaluronate lyase) is secreted by many streptococcal strains, including SAGs; it is an important factor facilitating the spreading of bacteria through the host tissues (Günther et al., 1996). Accordingly, secretion of hyaluronidase by S. agalactiae (GBS) has been shown to suppress the immune response and increase the intracellular survival of the bacteria in macrophages (Wang et al., 2014; Kolar et al., 2015). In GAS, GBS, and clinical strains of S. pneumoniae hyaluronidase also enables the use of hyaluronic acid produced by the host, or even that derived from capsules of other streptococci, for example GAS, as an alternative carbon source for growth (Rivera Starr and Cary, 2006; Marion et al., 2012). Moreover, hyaluronidase is involved in the spreading of S. intermedius by facilitating their detachment from a biofilm (Pecharki et al., 2008). The extracellular hyaluronidase produced by S. pyogenes is a protein of 99.6 kDa encoded by the hylA gene (Hynes et al., 2000, 2009).
Some light on the prevalence of hyaluronidase in diverse serological groups of streptococci has been shed by a large research on 614 GAS, 247 GBS, 225 group C streptococci (GCS) and 143 group G (GGS) strains (Günther et al., 1996). Among the GAS only 12.5% produced hyaluronidase, whereas in the other groups hyaluronidase positive strains were more frequent: GBS – 72.1%, GCS – 84%, and GGS – 85.5%. A survey of SAG strains (no species identified) isolated from clinical cases (n = 165) and from healthy people (n = 97) showed that on average 41% of the strains were hyaluronidase-positive, but this rate was twice as high among the clinical strains (ca. 50%) compared with those from a normal flora (ca. 25%; Unsworth, 1989). The frequency of hyaluronidase production also differs markedly for the three SAG species. In a study of 518 SAG strains recovered from clinical swabs a hyaluronidase activity was detected in only 8% (n = 25) of all S. anginosus strains (n = 307), both related to infections (11 of 163, 7%) and of unknown clinical importance (14 of 144, 10%; Jacobs and Stobberingh, 1995). In stark contrast, hyaluronidase producers represented 96 and 83% of S. constellatus and S. intermedius strains, respectively. Consistently, in a study of the SAG strains isolated from patients with acute dento-alveolar abscesses, none of 15 S. anginosus strains identified produced hyaluronidase (Fisher and Russell, 1993). A similar outcome was also obtained in an analysis of 157 SAG strains (64 dental plaque isolates and 91 from various other clinical sources), among which an average of 67% produced hyaluronidase (Whiley et al., 1990). However, of the 47 S. anginosus strains only 4% (n = 2) did, while 98% of S. intermedius and 88% of S. constellatus strains were hyaluronidase-positive (Whiley et al., 1990). Thus, hyaluronidase is produced by strikingly few S. anginosus strains and, therefore, appears not to be as important in their virulence as in other streptococci. However, it should be noted that in surveys mentioned above (Whiley et al., 1990; Fisher and Russell, 1993; Jacobs and Stobberingh, 1995) the SAG species were identified with antigenic and biochemical tests, prior the molecular biology-based methods have been introduced.
Hydrogen sulfide production
Hydrogen sulfide (H2S) is toxic for mammals at high concentrations. Its major effect is covalent modification of hemoglobin and its release from erythrocytes (Beauchamp et al., 1984). H2S is produced from L-cysteine in an α, β-elimination reaction catalysed by L-cysteine desulfhydrase (βC-S lyase, Lcd; Guarneros and Ortega, 1970; Kurzban et al., 1999). An involvement of H2S in progression of periodontal diseases has been proposed, especially for oral Gram-negative bacteria, the major H2S producers (Ratcliff and Johnson, 1999). Notably, the crude extracts from S. anginosus have a much higher capacity to produce hydrogen sulfide than those from other oral streptococci (Yoshida et al., 2003). The lcd gene encoding L-cysteine desulfhydrase from the laboratory S. anginosus strain FW73 was cloned in E. coli and the enzyme purified (Yoshida et al., 2002); exposition of sheep erythrocytes to L-cysteine and βC-S lyase resulted in the release of hemoglobin and its modification. Moreover, in BALB/c mice dental abscesses were formed after dorsal subcutaneous injection of the FW73 strain (Takahashi et al., 2011). When the mice were injected S. anginosus together with L-cysteine, the mean diameter of the abscesses was larger and also the level of lcd expression in the pus was over 15-fold higher than in mice injected with S. anginosus without L-cysteine. This indicated that H2S produced by L-cysteine desulfhydrase was responsible for the formation of the odontogenic abscesses in mouse (Takahashi et al., 2011).
Superantigens
Bacterial superantigens are toxins acting as T lymphocytes mitogens. They bypass antigen-presenting cells and directly stimulate the massive polyclonal proliferation of T cells, what results in the systemic release of pro-inflammatory cytokines (“cytokine storm”), including interferon-gamma (IFN-γ), tumour necrosis factor-alpha (TNFα) interleukin-beta (IL-1β), and IL-6, which in turn lead to a high fever and a toxic shock (Fraser and Proft, 2008; Wilde et al., 2021). In S. pyogenes the superantigens comprise streptococcal pyrogenic toxins (SPEs), streptococcal superantigen (SSA), and streptococcal mitogenic exotoxin Zn (SMEZn; Spaulding et al., 2013; Commons et al., 2014). The SPEs are divided into serotypes named A, C and G to O. SPEG and SMEZn are encoded chromosomally, in contrast to the other superantigens which are encoded by bacteriophages.
A collection of 124 β-hemolytic streptococcal strains isolated from 1,040 patients with diagnosed acute pharyngitis was tested by PCR for the presence of 11 known S. pyogenes superantigen-encoding genes (Anand et al., 2012). With the use of antigenic and biochemical tests 67 isolates (54%) were identified as S. anginosus, 38 (31%) as SDSE, and 19 (15%) as S. pyogenes. The speC, speG, and smeZ genes were detected in 100, 97.1, and 77.1% of the strains, respectively. The largest set comprising speC, speG, speA, speH, speI, and smeZ was detected in seven strains including one S. anginosus. Moreover, every S. anginosus strain harboured at least one gene encoding a superantigen. However, their nucleotide sequences were not determined (Anand et al., 2012). Recently, in a study of 59 β-hemolytic S. anginosus strains collected from endocarditis cases, the speC and speG genes as well as sdc and sdaD coding for DNases were detected in 1–8% of strains (Babbar et al., 2017). The genes were sequenced showing 100% identity with the respective S. pyogenes genes.
SAA, a novel Streptococcus anginosus antigen
Nitric oxide (NO) produced in response to bacterial infection has bactericidal effects [for review (Vatansever et al., 2013)]. Components of the S. mutans cell wall, rhamnose glucose polymers, and lipoteichoic acid from S. sanguis and S. mutans have been shown to induce NO production in macrophages (English et al., 1996; Martin et al., 1997). A similar feature has been observed for S. anginosus – induction of the production of both NO and inflammatory cytokines by murine peritoneal exudate cells (PEC; Sasaki et al., 2001). The agent responsible for that was purified from a culture supernatant of S. anginosus NCTC 10713 type strain and named S. anginosus antigen (SAA). SAA was then shown to stimulate NO production by PEC and accumulation of induced NO synthetase mRNA in a dose dependent manner in vitro. Furthermore, SAA also induced the accumulation of mRNA of TNF-α, IL–1β, and IL–6 (Sasaki et al., 2001). SAA was recognized as a tyrosine tRNA synthetase (TyrRS) belonging to the aminoacyl-tRNA synthetase family (Shimoyama et al., 2020). TyrRS could be isolated from whole-cell lysates of eight oral streptococci type strains (S. anginosus, S. mutans, S. intermedius, S. mitis, S. sobrinus, S. gordonii, S. sanguinus, and S. constellatus) but not from S. salivarius or S. oralis. It was also detected in culture supernatants of six out of seven clinical strains of S. anginosus from dental plaques but not in the supernatants of nine other oral streptococci. Interestingly, TyrRS has been found extracellularly uniquely in S. anginosus. The secretory system exporting TyrRS of S. anginosus outside the cell is yet to be elucidated as the protein lacks a typical N-terminal signal peptide recognised by the Sec secretion system (Shimoyama et al., 2020).
Signalling mediated by LuxS orthologues
Biofilm is the predominant type of growth for most bacterial species as it gives their communities a much higher resistance to changing environmental factors or antimicrobials compared to free-floating planktonic cells (Mah and O’Toole, 2001). In a process called quorum sensing numerous opportunistic pathogens produce autoinducers in response to extracellular signals to coordinate population behaviour (Miller and Bassler, 2001). Inter-species communication, frequent in both Gram-positive and Gram-negative bacteria, is based on the universal signalling molecule called autoinducer–2 (AI–2) whose synthesis requires the activity of LuxS (Vendeville et al., 2005). LuxS-mediated signalling has been identified in diverse species, including S. mitis, S. gordonii, S. pneumoniae, and S. agalactiae, therefore an attempt has been made to detect the luxS gene also in the S. anginosus type strain NCTC 10713 (Petersen et al., 2006). A PCR-amplified internal fragment of putative luxS showed approx. 80% nucleotide sequence identity with the luxS sequences of S. pneumoniae, S. gordonii, and S. mutans. Also, a culture supernatant of S. anginosus NCTC 10713 induced bioluminescence in Vibrio harveyi BB170, an AI-2 sensor, 5-fold more efficiently than that of its isogenic ΔluxS mutant. In accordance with the expected role of the putative LuxS, the luxS deletion decreased the ability to form biofilm between 2- and 5-fold, depending on medium used. Interestingly, under anaerobic conditions S. anginosus formed biofilm less efficiently than in a 5% CO2 aerobic atmosphere, regardless of its luxS status (Petersen et al., 2006). Moreover, the luxS mutation increased susceptibility to erythromycin and ampicillin (Ahmed et al., 2007), suggesting that the communication among bacteria could be taken advantage of as a possible target for designing a new antibacterials (Ahmed et al., 2007).
Other putative factors
A/ UDP-glucose pyrophosphorylase encoded by hasC is part of the chromosomal hasABC operon, essential for the synthesis of the hyaluronic acid capsule of group A streptococci (Crater et al., 1995). A homologue of hasC encoding a protein 86% identical to that of S. pyogenes has been found in all 17 SAG genomes tested, including seven S. anginosus strains (Olson et al., 2013); however, its role in SAG remains to be determined.
B/ Pullulanase is an enzyme degrading pullulan, a polysaccharide consisting of maltotriose units joined through α-D-1-6 glycosidic bonds. In S. pneumoniae pullulanase (SpuA) is a cell wall-anchored protein of 143 kDa shown to be necessary for full virulence in a mouse-lung model of infection (Bongaerts et al., 2000; Hava and Camilli, 2002). Also in S. pyogenes pullulanase (PulA) is a surface enzyme (129 kDa) involved in adhesion to the nasopharyngeal epithelium (Hytönen et al., 2003); it may contribute to the S. pyogenes virulence by its glycoprotein-binding activity and also by its potential to supply energy by carbohydrate hydrolysis. In all seven genomes of S. anginosus strains isolated from pulmonary and blood infections, analysed by Olsen et al. (Olson et al., 2013), a pulA homologue was detected coding for a protein 81% identical to the S. pyogenes protein along 59% of its length. However, the putative pullulanase of S. anginosus has not been studied yet.
C/ Streptococcal invasion locus. Another factor involved in invasion and evasion of streptococci in the host is the sil locus (streptococcus invasion locus). In S. pneumoniae it comprises five genes, silA-silE, which work as a quorum-sensing competence regulon. In GAS the sil locus has been shown to markedly increase invasiveness and fatal infections in an animal model (Hidalgo-Grass et al., 2002). Homologues of the silA to silE genes have also been identified in S. anginosus. The encoded proteins are identical in 52, 41, 41, 54, and 80% to those of S. pyogenes, respectively, and cover 98, 84, 55, 99, and 99.7% of their length (Olson et al., 2013).
D/ Two-component global regulatory system – the CsrR/CsrS regulon. In GAS the two-component system CsrR/CsrS represses several genes, e.g., those enabling the synthesis of the hyaluronic acid capsule, SLS and other toxins (Engleberg et al., 2001). The importance of CsrR/CsrS in the pathogenesis of GBS has also been reported, showing that inactivation of csrR increased the expression of a cluster of cyl genes and enhanced the hemolytic activity (Spellerberg et al., 2000; Lamy et al., 2004; Jiang et al., 2005). In SAG a csrR homologue encoding a protein 46% identical to CsrR of S. pyogenes along 99% of its length has been identified (Olson et al., 2013).
Additionally, several loci coding for other putative virulence factors have been detected in individual S. anginosus strains, basing on their homology to the Streptococcus virulence gene database (Olson et al., 2013). As already stated, S. anginosus is the most diverse species of the SAG group and the repertoire of its virulence factors differs substantially among strains (Olson et al., 2013).
Conclusion
Due to the increasing number of reports on the association of S. anginosus with a wide variety of infections ranging from oral infections, through empyema, abscesses and pulmonary infections to systemic blood infections, especially of immunocompromised and critically ill patients, it is now considered an emerging pathogen. However, in contrast to other streptococcal pathogens, the virulence factors and their regulation in S. anginosus are still poorly understood. One cannot exclude that its repertoire of factors facilitating infection is much broader than currently appreciated – those listed here do not exhaust the arsenal of proteins binding the host extracellular matrix and allowing the infection caused by these bacteria to spread. We hope that this review will systematize the present knowledge on the molecular basis of S. anginosus virulence and attract interest to this fascinating but somewhat neglected species. Further studies on the role of these factors in the infections are essential and should contribute to a better understanding of the transmission mechanisms of S. anginosus. This knowledge will in turn help to develop more effective means of preventing and dealing with the wide range of infections caused by this species.
Author contributions
AK, MS, and IK-Z created conception of the study and wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Science Centre under Grant 2018/29/B/NZ6/00624.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, N. A. A. M., Petersen, F. C., and Scheie, A. A. (2007). AI-2 quorum sensing affects antibiotic susceptibility in Streptococcus anginosus. J. Antimicrob. Chemother. 60, 49–53. doi: 10.1093/jac/dkm124
Akuzawa, N., Hatori, T., Kitahara, Y., and Kurabayashi, M. (2017). Multiple liver abscesses and bacteremia caused by Streptococcus constellatus infection: a case report. Clin. Case Rep. 5, 69–74. doi: 10.1002/ccr3.774
Allen, B. L., and Höök, M. (2002). Isolation of a putative laminin binding protein from Streptococcus anginosus. Microb. Pathog. 33, 23–31. doi: 10.1006/mpat.2002.0510
Allen, B. L., Katz, B., and Höök, M. (2002). Streptococcus anginosus adheres to vascular endothelium basement membrane and purified extracellular matrix proteins. Microb. Pathog. 32, 191–204. doi: 10.1006/mpat.2002.0496
Anand, T. D., Rajesh, T., Rajendhran, J., and Gunasekaran, P. (2012). Superantigen profiles of emm and emm-like typeable and nontypeable pharyngeal streptococcal isolates of South India. Ann. Clin. Microbiol. Antimicrob. 11:3. doi: 10.1186/1476-0711-11-3
Asam, D., Mauerer, S., and Spellerberg, B. (2015). Streptolysin S of Streptococcus anginosus exhibits broad-range hemolytic activity. Med. Microbiol. Immunol. 204, 227–237. doi: 10.1007/s00430-014-0363-0
Asam, D., Mauerer, S., Walheim, E., and Spellerberg, B. (2013). Identification of β-haemolysin-encoding genes in Streptococcus anginosus. Mol Oral Microbiol 28, 302–315. doi: 10.1111/omi.12026
Asam, D., and Spellerberg, B. (2014). Molecular pathogenicity of Streptococcus anginosus. Mol Oral Microbiol 29, 145–155. doi: 10.1111/omi.12056
Babbar, A., Kumar, V. N., Bergmann, R., Barrantes, I., Pieper, D. H., Itzek, A., et al. (2017). Members of a new subgroup of Streptococcus anginosus harbor virulence related genes previously observed in streptococcus pyogenes. Int. J. Med. Microbiol. 307, 174–181. doi: 10.1016/j.ijmm.2017.02.002
Baiano, J. C. F., and Barnes, A. C. (2009). Towards control of streptococcus iniae. Emerg. Infect. Dis. 15, 1891–1896. doi: 10.3201/eid1512.090232
Baracco, G. J. (2019). Infections caused by group C and G Streptococcus (streptococcus dysgalactiae subsp. equisimilis and others): epidemiological and clinical aspects. Microbiol Spectr 7. doi: 10.1128/microbiolspec.GPP3-0016-2018
Beauchamp, R. O., Bus, J. S., Popp, J. A., Boreiko, C. J., Andjelkovich, D. A., and Leber, P. (1984). A critical review of the literature on hydrogen sulfide toxicity. Crit. Rev. Toxicol. 13, 25–97. doi: 10.3109/10408448409029321
Beiter, K., Wartha, F., Albiger, B., Normark, S., Zychlinsky, A., and Henriques-Normark, B. (2006). An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 16, 401–407. doi: 10.1016/j.cub.2006.01.056
Bergmann, S., Rohde, M., Chhatwal, G. S., and Hammerschmidt, S. (2001). α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40, 1273–1287. doi: 10.1046/j.1365-2958.2001.02448.x
Bernheimer, A. W. (1967). Physical behavior of streptolysin S. J. Bacteriol. 93, 2024–2025. doi: 10.1128/jb.93.6.2024-2025.1967
Bierne, H., Milohanic, E., and Kortebi, M. (2018). To be cytosolic or vacuolar: the double life of listeria monocytogenes. Front. Cell. Infect. Microbiol. 8:136. doi: 10.3389/fcimb.2018.00136
Bongaerts, R. J. M., Heinz, H. P., Hadding, U., and Zysk, G. (2000). Antigenicity, expression, and molecular characterization of surface-located pullulanase of Streptococcus pneumoniae. Infect. Immun. 68, 7141–7143. doi: 10.1128/IAI.68.12.7141-7143.2000
Briles, D. E., Ades, E., Paton, J. C., Sampson, J. S., Carlone, G. M., Huebner, R. C., et al. (2000). Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect. Immun. 68, 796–800. doi: 10.1128/IAI.68.2.796-800.2000
Broyles, L. N., Van Beneden, C., Beall, B., Facklam, R., Lynn Shewmaker, P., Malpiedi, P., et al. (2009). Population-based study of invasive disease due to β-hemolytic streptococci of groups other than A and B. Clin. Infect. Dis. 48, 706–712. doi: 10.1086/597035
Buchanan, J. T., Simpson, A. J., Aziz, R. K., Liu, G. Y., Kristian, S. A., Kotb, M., et al. (2006). DNase expression allows the pathogen group a streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16, 396–400. doi: 10.1016/j.cub.2005.12.039
Carapetis, J. R., Steer, A. C., Mulholland, E. K., and Weber, M. (2005). The global burden of group a streptococcal diseases. Lancet Infect. Dis. 5, 685–694. doi: 10.1016/S1473-3099(05)70267-X
Chang, A., Khemlani, A., Kang, H., and Proft, T. (2011). Functional analysis of streptococcus pyogenes nuclease a (SpnA), a novel group a streptococcal virulence factor. Mol. Microbiol. 79, 1629–1642. doi: 10.1111/j.1365-2958.2011.07550.x
Chang, Y. C., and Lo, H. H. (2013). Identification, clinical aspects, susceptibility pattern, and molecular epidemiology of β-haemolytic group G Streptococcus anginosus group isolates from Central Taiwan. Diagn. Microbiol. Infect. Dis. 76, 262–265. doi: 10.1016/j.diagmicrobio.2013.03.021
Cisar, J. O., Sandberg, A. L., Abeygunawardana, C., Reddy, G. P., and Bush, C. A. (1995). Lectin recognition of host-like saccharide motifs in streptococcal cell wall polysaccharides. Glycobiology 5, 655–662. doi: 10.1093/glycob/5.7.655
Clarridge, J. E., Attorri, S., Musher, D. M., Hebert, J., and Dunbar, S. (2001). Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (“Streptococcus milleri group”) are of different clinical importance and are not equally associated with abscess. Clin. Infect. Dis. 32, 1511–1515. doi: 10.1086/320163
Commons, R. J., Smeesters, P. R., Proft, T., Fraser, J. D., Robins-Browne, R., and Curtis, N. (2014). Streptococcal superantigens: categorization and clinical associations. Trends Mol. Med. 20, 48–62. doi: 10.1016/j.molmed.2013.10.004
Cork, A. J., Jergic, S., Hammerschmidt, S., Kobe, B., Pancholi, V., Benesch, J. L. P., et al. (2009). Defining the structural basis of human plasminogen binding by streptococcal surface enolase. J. Biol. Chem. 284, 17129–17137. doi: 10.1074/jbc.M109.004317
Courtney, H. S., Dale, J. B., and Hasty, D. L. (1996). Differential effects of the streptococcal fibronectin-binding protein, FBP54, on adhesion of group a streptococci to human buccal cells and HEp-2 tissue culture cells. Infect. Immun. 64, 2415–2419. doi: 10.1128/iai.64.7.2415-2419.1996
Courtney, H. S., Hasty, D. L., and Dale, J. B. (2002). Molecular mechanisms of adhesion, colonization, and invasion of group a streptococci. Ann. Med. 34, 77–87. doi: 10.1080/07853890252953464
Courtney, H. S., Hasty, D. L., and Dale, J. B. (2006). Anti-phagocytic mechanisms of Streptococcus pyogenes: binding of fibrinogen to M-related protein. Mol. Microbiol. 59, 936–947. doi: 10.1111/j.1365-2958.2005.04977.x
Courtney, H. S., and Pownall, H. J. (2010). The structure and function of serum opacity factor: a unique streptococcal virulence determinant that targets high-density lipoproteins. J. Biomed. Biotechnol. 2010:956071. doi: 10.1155/2010/956071
Coykendall, A. L. (1989). Classification and identification of the viridans streptococci. Clin. Microbiol. Rev. 2, 315–328. doi: 10.1128/CMR.2.3.31527
Crater, D. L., Dougherty, B. A., and Van de Rijn, I. (1995). Molecular characterization of hasC from an operon required for hyaluronic acid synthesis in group a streptococci. Demonstration of UDP-glucose pyrophosphorylase activity. J. Biol. Chem. 270, 28676–28680. doi: 10.1074/jbc.270.48.28676
Cumley, N. J., Smith, L. M., Anthony, M., and May, R. C. (2012). The CovS/CovR acid response regulator is required for intracellular survival of group B streptococcus in macrophages. Infect. Immun. 80, 1650–1661. doi: 10.1128/IAI.05443-11
Das, S., Dileepan, T., Johnson, D. R., Kaplan, E. L., and Patrick, C. P. (2017). Enzyme-linked immunosorbent assay for group a streptococcal anti-DNase B in human sera, using recombinant proteins – comparison to the DNA methyl green micromethod. J. Immunol. Methods 451, 111–117. doi: 10.1016/j.jim.2017.09.006
de Buhr, N., Neumann, A., Jerjomiceva, N., von Köckritz-Blickwede, M., and Baums, C. G. (2014). Streptococcus suis DNase SsnA contributes to degradation of neutrophil extracellular traps (NETs) and evasion of NET-mediated antimicrobial activity. Microbiol 160, 385–395. doi: 10.1099/mic.0.072199-0
de Buhr, N., Stehr, M., Neumann, A., Naim, H. Y., Valentin-Weigand, P., von Köckritz-Blickwede, M., et al. (2015). Identification of a novel DNase of Streptococcus suis (EndAsuis) important for neutrophil extracellular trap degradation during exponential growth. Microbiology 161, 838–850. doi: 10.1099/mic.0.000040
Derré-Bobillot, A., Cortes-Perez, N. G., Yamamoto, Y., Kharrat, P., Couvé, E., Da Cunha, V., et al. (2013). Nuclease a (Gbs0661), an extracellular nuclease of Streptococcus agalactiae, attacks the neutrophil extracellular traps and is needed for full virulence. Mol. Microbiol. 89, 518–531. doi: 10.1111/mmi.12295
Dinkla, K., Sastalla, I., Godehardt, A. W., Janze, N., Chhatwal, G. S., Rohde, M., et al. (2007). Upregulation of capsule enables streptococcus pyogenes to evade immune recognition by antigen-specific antibodies directed to the G-related α2-macroglobulin-binding protein GRAB located on the bacterial surface. Microb. Infect. 9, 922–931. doi: 10.1016/j.micinf.2007.03.011
Dugdale, C. M., Tompkins, A. J., Reece, R. M., and Gardner, A. F. (2013). Cavernosal abscess due to Streptococcus anginosus: a case report and comprehensive review of the literature. Curr. Urol. 7, 51–56. doi: 10.1159/000343555
Engleberg, N. C., Heath, A., Miller, A., Rivera, C., and DiRita, V. J. (2001). Spontaneous mutations in the CsrRS two-component regulatory system of streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J. Infect. Dis. 183, 1043–1054. doi: 10.1086/319291
English, B. K., Patrick, C. C., Orlicek, S. L., McCordic, R., and Shenep, J. L. (1996). Lipoteichoic acid from viridans streptococci induces the production of tumor necrosis factor and nitric oxide by murine macrophages. J. Infect. Dis. 174, 1348–1351. doi: 10.1093/infdis/174.6.1348
Esgleas, M., Li, Y., Hancock, M. A., Harel, J., Dubreuil, J. D., and Gottschalk, M. (2008). Isolation and characterization of α-enolase, a novel fibronectin-binding protein from Streptococcus suis. Microbiology 154, 2668–2679. doi: 10.1099/mic.0.2008/017145-0
Esplin, N., Stelzer, J. W., All, S., Kumar, S., Ghaffar, E., and Ali, S. (2017). A case of Streptococcus anginosus brain abscess caused by contiguous spread from sinusitis in an immunocompetent patient. Cureus 9:e1745. doi: 10.7759/cureus.1745
Finn, T., Schattner, A., Dubin, I., and Cohen, R. (2018). Streptococcus anginosus endocarditis and multiple liver abscesses in a splenectomised patient. BMJ Case Rep. 2018:bcr-2018-224266. doi: 10.1136/bcr-2018-224266
Fischetti, V. A., and Ryan, P. (2015). “Streptococcus,” in Practical Handbook of Microbiology. 3rd Edn. (Boca Raton: CRC Press).
Fisher, L. E., and Russell, R. R. B. (1993). The isolation and characterization of milleri group streptococci from dental periapical abscesses. J. Dent. Res. 72, 1191–1193. doi: 10.1177/00220345930720080501
Fox, K., Turner, J., and Fox, A. (1993). Role of β-hemolytic group C streptococci in pharyngitis: incidence and biochemical characteristics of Streptococcus equisimilis and Streptococcus anginosus in patients and healthy controls. J. Clin. Microbiol. 31, 804–807. doi: 10.1128/jcm.31.4.804-807.1993
Fraser, J. D., and Proft, T. (2008). The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 225, 226–243. doi: 10.1111/j.1600-065X.2008.00681.x
Ge, J., Catt, D. M., and Gregory, R. L. (2004). Streptococcus mutans surface α-enolase binds salivary mucin MG2 and human plasminogen. Infect. Immun. 72, 6748–6752. doi: 10.1128/IAI.72.11.6748-6752.2004
Grinwis, M. E., Sibley, C. D., Parkins, M. D., Eshaghurshan, C. S., Rabin, H. R., and Surette, M. G. (2010). Characterization of Streptococcus milleri group isolates from expectorated sputum of adult patients with cystic fibrosis. J. Clin. Microbiol. 48, 395–401. doi: 10.1128/JCM.01807-09
Guarneros, G., and Ortega, M. V. (1970). Cysteine desulfhydrase activities of salmonella typhimurium and Escherichia coli. Biochim. Biophys. Acta Enzymol. 198, 132–142. doi: 10.1016/0005-2744(70)90041-0
Günther, E., Ozegowski, J. H., and Köhler, W. (1996). Occurrence of extracellular hyaluronic acid and hyaluronatlyase in streptococci of groups a, B, C, and G. Zentralblatt fur Bakteriol 285, 64–73. doi: 10.1016/s0934-8840(96)80023-5
Hajishengallis, G., Nikolova, E., and Russell, M. W. (1992). Inhibition of Streptococcus mutans adherence to saliva-coated hydroxyapatite by human secretory immunoglobulin a (S-IgA) antibodies to cell surface protein antigen I/II: reversal by IgA1 protease cleavage. Infect. Immun. 60, 5057–5064. doi: 10.1128/iai.60.12.5057-5064.1992
Hamada, S., and Slade, H. D. (1980). Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44, 331–384. doi: 10.1128/mr.44.2.331-384.1980
Hanski, E., Horwitz, P. A., and Caparon, M. G. (1992). Expression of protein F, the fibronectin-binding protein of streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 60, 5119–5125. doi: 10.1128/iai.60.12.5119-5125.1992
Hasegawa, T., Minami, M., Okamoto, A., Tatsuno, I., Isaka, M., and Ohta, M. (2010). Characterization of a virulence-associated and cell-wall-located DNase of streptococcus pyogenes. Microbiology 156, 184–190. doi: 10.1099/mic.0.031955-0
Hasegawa, T., Torii, K., Hashikawa, S., Iinuma, Y., and Ohta, M. (2002a). Cloning and characterization of two novel DNases from streptococcus pyogenes. Arch. Microbiol. 177, 451–456. doi: 10.1007/s00203-002-0412-8
Hasegawa, T., Torii, K., Hashikawa, S., Iinuma, Y., and Ohta, M. (2002b). Cloning and characterization of the deoxyribonuclease sdα gene from streptococcus pyogenes. Curr. Microbiol. 45, 13–17. doi: 10.1007/s00284-001-0042-4
Hava, D. L., and Camilli, A. (2002). Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406. doi: 10.1046/j.1365-2958.2002.03106.x
Henderson, B., and Martin, A. (2011). Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79, 3476–3491. doi: 10.1128/IAI.00179-11
Henderson, B., and Martin, A. C. R. (2014). The biological and biomedical consequences of protein moonlighting. Biochem. Soc. Trans. 42, 1671–1678. doi: 10.1042/BST20140273
Herrick, S., Blanc-Brude, O., Gray, A., and Laurent, G. (1999). Fibrinogen. Int. J. Biochem. Cell Biol. 31, 741–746. doi: 10.1016/s1357-2725(99)00032-1
Hidalgo-Grass, C., Ravins, M., Dan-Goor, M., Jaffe, J., Moses, A. E., and Hanski, E. (2002). A locus of group a streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 46, 87–99. doi: 10.1046/j.1365-2958.2002.03127.x
Hyams, C., Camberlein, E., Cohen, J. M., Bax, K., and Brown, J. S. (2010). The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect. Immun. 78, 704–715. doi: 10.1128/IAI.00881-09
Hynes, W. L., Dixon, A. R., Walton, S. L., and Aridgides, L. J. (2000). The extracellular hyaluronidase gene (hylA) of streptococcus pyogenes. FEMS Microbiol. Lett. 184, 109–112. doi: 10.1111/j.1574-6968.2000.tb08999.x
Hynes, W., Johnson, C., and Stokes, M. (2009). A single nucleotide mutation results in loss of enzymatic activity in the hyaluronate lyase gene of Streptococcus pyogenes. Microb. Pathog. 47, 308–313. doi: 10.1016/j.micpath.2009.09.008
Hytönen, J., Haataja, S., and Finne, J. (2003). Streptococcus pyogenes glycoprotein-binding strepadhesin activity is mediated by a surface-associated carbohydrate-degrading enzyme, pullulanase. Infect. Immun. 71, 784–793. doi: 10.1128/IAI.71.2.784-793.2003
Issa, E., Salloum, T., and Tokajian, S. (2020). From normal flora to brain abscesses: a review of Streptococcus intermedius. Front. Microbiol. 11:826. doi: 10.3389/fmicb.2020.00826
Itzek, A., Gillen, C. M., Fulde, M., Friedrichs, C., Rodloff, A. C., Chhatwal, G. S., et al. (2010). Contribution of plasminogen activation towards the pathogenic potential of oral streptococci. PLoS One 5:e13826. doi: 10.1371/journal.pone.0013826
Jacobs, J. A., and Stobberingh, E. E. (1995). Hydrolytic enzymes of Streptococcus anginosus, Streptococcus constellatus and Streptococcus intermedius in relation to infection. Eur. J. Clin. Microbiol. Infect. Dis. 14, 818–820. doi: 10.1007/BF01691002
Jado, I., Fenoll, A., Casal, J., and Pérez, A. (2001). Identification of the psaA gene, coding for pneumococcal surface adhesin a, in viridans group streptococci other than Streptococcus pneumoniae. Clin. Diagn. Lab. Immunol. 8, 895–898. doi: 10.1128/CDLI.8.5.895-898.2001
Jensen, A., Hoshino, T., and Kilian, M. (2013). Taxonomy of the Anginosus group of the genus streptococcus and description of Streptococcus anginosus subsp. whileyi subsp. nov. and Streptococcus constellatus subsp. viborgensis subsp. nov. Int. J. Syst. Evol. Microbiol. 63, 2506–2519. doi: 10.1099/ijs.0.043232-0
Jiang, S. M., Cieslewicz, M. J., Kasper, D. L., and Wessels, M. R. (2005). Regulation of virulence by a two-component system in group B streptococcus. J. Bacteriol. 187, 1105–1113. doi: 10.1128/JB.187.3.1105-1113.2005
Jiang, S., Li, M., Fu, T., Shan, F., Jiang, L., and Shao, Z. (2020). Clinical characteristics of infections caused by Streptococcus anginosus group. Sci. Rep. 10:9032. doi: 10.1038/s41598-020-65977-z
Junckerstorff, R. K., Robinson, J. O., and Murray, R. J. (2014). Invasive Streptococcus anginosus group infection-does the species predict the outcome? Int. J. Infect. Dis. 18, 38–40. doi: 10.1016/j.ijid.2013.09.003
Kehoe, M. A., Miller, L., Walker, J. A., and Boulnois, G. J. (1987). Nucleotide sequence of the streptolysin O (SLO) gene: structural homologies between SLO and other membrane-damaging, thiol-activated toxins. Infect. Immun. 55, 3228–3232. doi: 10.1128/iai.55.12.3228-3232.1987
Kinnby, B., Booth, N. A., and Svensäter, G. (2008). Plasminogen binding by oral streptococci from dental plaque and inflammatory lesions. Microbiology 154, 924–931. doi: 10.1099/mic.0.2007/013235-0
Kirkham, L.-A. S., Jefferies, J. M. C., Kerr, A. R., Jing, Y., Clarke, S. C., Smith, A., et al. (2006). Identification of invasive serotype 1 pneumococcal isolates that express nonhemolytic pneumolysin. J. Clin. Microbiol. 44, 151–159. doi: 10.1128/JCM.44.1.151-159.2006
Kodama, Y., Ishikawa, T., Shimoyama, Y., Sasaki, D., Kimura, S., and Sasaki, M. (2018). The fibronectin-binding protein homologue Fbp62 of Streptococcus anginosus is a potent virulence factor. Microbiol. Immunol. 62, 624–634. doi: 10.1111/1348-0421.12646
Köhler, W. (2007). The present state of species within the genera streptococcus and enterococcus. Int. J. Med. Microbiol. 297, 133–150. doi: 10.1016/j.ijmm.2006.11.008
Kolar, S. L., Kyme, P., Tseng, C. W., Soliman, A., Kaplan, A., Liang, J., et al. (2015). Group B streptococcus evades host immunity by degrading hyaluronan. Cell Host Microb. 18, 694–704. doi: 10.1016/j.chom.2015.11.001
Korczynska, J. E., Turkenburg, J. P., and Taylor, E. J. (2012). The structural characterization of a prophage-encoded extracellular DNase from streptococcus pyogenes. Nucleic Acids Res. 40, 928–938. doi: 10.1093/nar/gkr789
Kornblatt, M. J., Kornblatt, J. A., and Hancock, M. A. (2011). The interaction of canine plasminogen with streptococcus pyogenes enolase: they bind to one another but what is the nature of the structures involved? PLoS One 6:e28481. doi: 10.1371/journal.pone.0028481
Kurzban, G. P., Chu, L., Ebersole, J. L., and Holt, S. C. (1999). Sulfhemoglobin formation in human erythrocytes by cystalysin, an L-cysteine desulfhydrase from Treponema denticola. Oral Microbiol. Immunol. 14, 153–164. doi: 10.1034/j.1399-302x.1999.140303.x
Lamy, M. C., Zouine, M., Fert, J., Vergassola, M., Couve, E., Pellegrini, E., et al. (2004). CovS/CovR of group B streptococcus: a two-component global regulatory system involved in virulence. Mol. Microbiol. 54, 1250–1268. doi: 10.1111/j.1365-2958.2004.04365.x.
Lawn, J. E., Bianchi-Jassir, F., Russell, N. J., Kohli-Lynch, M., Tann, C. J., Hall, J., et al. (2017). Group B streptococcal disease worldwide for pregnant women, stillbirths, and children: why, what, and how to undertake estimates? Clin. Infect. Dis. 65, S89–S99. doi: 10.1093/cid/cix653
Lee, S. W., Mitchell, D. A., Markley, A. L., Hensler, M. E., Gonzalez, D., Wohlrab, A., et al. (2008). Discovery of a widely distributed toxin biosynthetic gene cluster. Proc. Natl. Acad. Sci. U. S. A. 105, 5879–5884. doi: 10.1073/pnas.0801338105
Lun, Z. R., Wang, Q. P., Chen, X. G., Li, A. X., and Zhu, X. Q. (2007). Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect. Dis. 7, 201–209. doi: 10.1016/S1473-3099(07)70001-4
Ma, J. K. C., Kelly, C. G., Munro, G., Whiley, R. A., and Lehner, T. (1991). Conservation of the gene encoding streptococcal antigen I/II in oral streptococci. Infect. Immun. 59, 2686–2694. doi: 10.1128/iai.59.8.2686-2694.1991
Mah, T. F. C., and O’Toole, G. A. (2001). Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39. doi: 10.1016/s0966-842x(00)01913-2
Manzer, H. S., Nobbs, A. H., and Doran, K. S. (2020). The multifaceted nature of streptococcal antigen I/II proteins in colonization and disease pathogenesis. Front. Microbiol. 11:602305. doi: 10.3389/fmicb.2020.602305
Marion, C., Stewart, J. M., Tazi, M. F., Burnaugh, A. M., Linke, C. M., Woodiga, S. A., et al. (2012). Streptococcus pneumoniae can utilize multiple sources of hyaluronic acid for growth. Infect. Immun. 80, 1390–1398. doi: 10.1128/IAI.05756-11
Martin, V., Kleschyov, A. L., Klein, J. P., and Beretz, A. (1997). Induction of nitric oxide production by polyosides from the cell walls of streptococcus mutants OMZ 175, a gram-positive bacterium, in the rat aorta. Infect. Immun. 65, 2074–2079. doi: 10.1128/iai.65.6.2074-2079.1997
Membrebe, J. D., Yoon, N. K., Hong, M., Lee, J., Lee, H., Park, K., et al. (2016). Protective efficacy of streptococcus iniae derived enolase against streptococcal infection in a zebrafish model. Vet. Immunol. Immunopathol. 170, 25–29. doi: 10.1016/j.vetimm.2016.01.004
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Mishra, A. K., and Fournier, P.-E. (2013). The role of Streptococcus intermedius in brain abscess. Eur. J. Clin. Microbiol. Infect. Dis. 32, 477–483. doi: 10.1007/s10096-012-1782-8
Moisset, A., Schatz, N., Lepoivre, Y., Amadio, S., Wachsmann, D., Scholler, M., et al. (1994). Conservation of salivary glycoprotein-interacting and human immunoglobulin G-cross-reactive domains of antigen I/II in oral streptococci. Infect. Immun. 62, 184–193. doi: 10.1128/iai.62.1.184-193.1994
Molloy, E. M., Cotter, P. D., Hill, C., Mitchell, D. A., and Ross, R. P. (2011). Streptolysin S-like virulence factors: the continuing sagA. Nat. Rev. Microbiol. 9, 670–681. doi: 10.1038/nrmicro2624
Morita, E., Narikiyo, M., Yano, A., Nishimura, E., Igaki, H., Sasaki, H., et al. (2003). Different frequencies of Streptococcus anginosus infection in oral cancer and esophageal cancer. Cancer Sci. 94, 492–496. doi: 10.1111/j.1349-7006.2003.tb01471.x
Nadeem Khan, M., Coleman, J. R., Vernatter, J., Varshney, A. K., Dufaud, C., and Pirofski, L. A. (2014). An ahemolytic pneumolysin of Streptococcus pneumoniae manipulates human innate and CD4+ T-cell responses and reduces resistance to colonization in mice in a serotype-independent manner. J. Infect. Dis. 210, 1658–1669. doi: 10.1093/infdis/jiu321
Nagamune, H., Ohnishi, C., Katsuura, A., Fushitani, K., Whiley, R. A., Tsuji, A., et al. (1996). Intermedilysin, a novel cytotoxin specific for human cells secreted by Streptococcus intermedius UNS46 isolated from a human liver abscess. Infect. Immun. 64, 3093–3100. doi: 10.1128/iai.64.8.3093-3100.1996
Nayak, K., Shobha, K. L., and Prasad, V. (2019). Streptococcus anginosus throat colonization in healthy school going children. Indian J. Public Heal. Res. Dev. 10, 167–172. doi: 10.5958/0976-5506.2019.00990.2
Nizet, V., Beall, B., Bast, D. J., Datta, V., Kilburn, L., Low, D. E., et al. (2000). Genetic locus for streptolysin S production by group a streptococcus. Infect. Immun. 68, 4245–4254. doi: 10.1128/IAI.68.7.4245-4254.2000
Nobbs, A. H., Lamont, R. J., and Jenkinson, H. F. (2009). Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450. doi: 10.1128/MMBR.00014-09
O’Brien, K. L., Wolfson, L. J., Watt, J. P., Henkle, E., Deloria-Knoll, M., McCall, N., et al. (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902. doi: 10.1016/S0140-6736(09)61204-6
Obszańska, K., Kern-Zdanowicz, I., Kozińska, A., Machura, K., Stefaniuk, E., Hryniewicz, W., et al. (2016). Streptococcus anginosus (milleri) group strains isolated in Poland (1996-2012) and their antibiotic resistance patterns. Polish. J. Microbiol. 65, 33–41. doi: 10.5604/17331331.1197323
Olson, A. B., Kent, H., Sibley, C. D., Grinwis, M. E., Mabon, P., Ouellette, C., et al. (2013). Phylogenetic relationship and virulence inference of Streptococcus anginosus group: curated annotation and whole-genome comparative analysis support distinct species designation. BMC Genomics 14:895. doi: 10.1186/1471-2164-14-895
Ottow, J. C. (1975). Ecology, physiology, and genetics of fimbriae and pili. Annu. Rev. Microbiol. 29, 79–108. doi: 10.1146/annurev.mi.29.100175.000455
Pancholi, V., and Fischetti, V. A. (1992). A major surface protein on group a streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176, 415–426. doi: 10.1084/jem.176.2.415
Pancholi, V., and Fischetti, V. A. (1998). α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273, 14503–14515. doi: 10.1074/jbc.273.23.14503
Patel, K., Park, C., Memon, Z., and Ilyas, N. (2020). Eikenella corrodens and Streptococcus anginosus co-infection causing skull base osteomyelitis and internal carotid artery lesion. IDCases 20:e00740. doi: 10.1016/j.idcr.2020.e00740
Paton, J. C., and Trappetti, C. (2019). Streptococcus pneumoniae capsular polysaccharide. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.GPP3-0050-2018
Pecharki, D., Petersen, F. C., and Scheie, A. A. (2008). Role of hyaluronidase in Streptococcus intermedius biofilm. Microbiology 154, 932–938. doi: 10.1099/mic.0.2007/012393-0
Petersen, F. C., Ahmed, N. A. A. M., Naemi, A., and Scheie, A. A. (2006). LuxS-mediated signalling in Streptococcus anginosus and its role in biofilm formation. Antonie Van Leeuwenhoek 90, 109–121. doi: 10.1007/s10482-006-9065-y
Petrey, A. C., and de la Motte, C. A. (2014). Hyaluronan, a crucial regulator of inflammation. Front. Immunol. 5:101. doi: 10.3389/fimmu.2014.00101
Pian, Y., Wang, P., Liu, P., Zheng, Y., Zhu, L., Wang, H., et al. (2015). Proteomics identification of novel fibrinogen-binding proteins of Streptococcus suis contributing to antiphagocytosis. Front. Cell. Infect. Microbiol. 5:19. doi: 10.3389/fcimb.2015.00019
Pilarczyk-Zurek, M., Sitkiewicz, I., and Koziel, J. (2022). The clinical view on Streptococcus anginosus group – opportunistic pathogens coming out of hiding. Front. Microbiol. 13:956677. doi: 10.3389/fmicb.2022.956677
Price, K. E., and Camilli, A. (2009). Pneumolysin localizes to the cell wall of Streptococcus pneumoniae. J. Bacteriol. 191, 2163–2168. doi: 10.1128/JB.01489-08
Rabuñal, R., Corredoira, J., Monte, R., and Coira, A. (2009). Co-infection by Streptococcus anginosus and mycobacterium tuberculosis: three case reports. J. Med. Case Rep. 3:37. doi: 10.1186/1752-1947-3-37
Rantala, S. (2014). Streptococcus dysgalactiae subsp. equisimilis bacteremia: an emerging infection. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1303–1310. doi: 10.1007/s10096-014-2092-0
Ratcliff, P. A., and Johnson, P. W. (1999). The relationship between oral malodor, gingivitis, and periodontitis. A review. J. Periodontol. 70, 485–489. doi: 10.1902/jop.1999.70.5.485
Rawla, P., Vellipuram, A. R., Bandaru, S. S., and Pradeep, R. (2017). Colon carcinoma presenting as Streptococcus anginosus bacteremia and liver abscess. Gastroenterol. Res. 10, 376–379. doi: 10.14740/gr884w
Reid, S. D., Montgomery, A. G., Voyich, J. M., DeLeo, F. R., Lei, B., Ireland, R. M., et al. (2003). Characterization of an extracellular virulence factor made by group a streptococcus with homology to the listeria monocytogenes internalin family of proteins. Infect. Immun. 71, 7043–7052. doi: 10.1128/IAI.71.12.7043-7052.2003
Reißmann, S., Friedrichs, C., Rajkumari, R., Itzek, A., Fulde, M., Rodloff, A. C., et al. (2010). Contribution of Streptococcus anginosus to infections caused by groups C and G streptococci, southern India. Emerg. Infect. Dis. 16, 656–663. doi: 10.3201/eid1604.090448
Reitmeyer, J., Guthrie, R., and Steele, J. (1989). The occurrence of nicotinamide adenine dinucleotide glycohydrolase and the serum opacity reaction in groups B, C, F and G streptococci. Microbios 58, 183–187.
Remmington, A., and Turner, C. E. (2018). The DNases of pathogenic Lancefield streptococci. Microbiology 164, 242–250. doi: 10.1099/mic.0.000612
Rivera Starr, C., and Cary, E. N. (2006). Role of hyaluronidase in subcutaneous spread and growth of group a streptococcus. Infect. Immun. 74, 40–48. doi: 10.1128/IAI.74.1.40-48.2006
Rivera, J., Vannakambadi, G., and Höök, M. S. P. (2007). Fibrinogen-binding proteins of gram-positive bacteria. Thromb. Haemost. 98, 503–511. doi: 10.1160/TH07-03-0233
Rubel, C., Fernández, G. C., Dran, G., Bompadre, M. B., Isturiz, M. A., and Palermo, M. S. (2001). Fibrinogen promotes neutrophil activation and delays apoptosis. J. Immunol. 166, 2002–2010. doi: 10.4049/jimmunol.166.3.2002
Rukke, H. V., Hegna, I. K., and Petersen, F. C. (2012). Identification of a functional capsule locus in Streptococcus mitis. Mol. Oral. Microbiol. 27, 95–108. doi: 10.1111/j.2041-1014.2011.00635.x
Russell, M. W., Bergmeier, L. A., Zanders, E. D., and Lehner, T. (1980). Protein antigens of Streptococcus mutans: purification and properties of a double antigen and its protease-resistant component. Infect. Immun. 28, 486–493. doi: 10.1128/iai.28.2.486-493.1980
Sadykov, M., Asami, Y., Niki, H., Handa, N., Itaya, M., Tanokura, M., et al. (2003). Multiplication of a restriction-modification gene complex. Mol. Microbiol. 48, 417–427. doi: 10.1046/j.1365-2958.2003.03464.x
Sasaki, M., Ohara-Nemoto, Y., Tajika, S., Kobayashi, M., Yamaura, C., and Kimura, S. (2001). Antigenic characterisation of a novel Streptococcus anginosus antigen that induces nitric oxide synthesis by murine peritoneal exudate cells. J. Med. Microbiol. 50, 952–958. doi: 10.1099/0022-1317-50-11-952
Sawyer, R. T., Drevets, D. A., Campbell, P. A., and Potter, T. A. (1996). Internalin a can mediate phagocytosis of listeria monocytogenes by mouse macrophage cell lines. J. Leukoc. Biol. 60, 603–610. doi: 10.1002/jlb.60.5.603
Schüler, V., Lussi, A., Kage, A., and Seemann, R. (2012). Glycan-binding specificities of Streptococcus mutans and Streptococcus sobrinus lectin-like adhesins. Clin. Oral Investig. 16, 789–796. doi: 10.1007/s00784-011-0568-1
Schwarz-Linek, U., Höök, M., and Potts, J. R. (2004). The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52, 631–641. doi: 10.1111/j.1365-2958.2004.04027.x
Shak, J. R., Ludewick, H. P., Howery, K. E., Sakai, F., Yi, H., Harvey, R. M., et al. (2013). Novel role for the Streptococcus pneumoniae toxin pneumolysin in the assembly of biofilms. MBio 4:e00655, –e00613. doi: 10.1128/mBio.00655-13
Shimoyama, Y., Ishikawa, T., Kodama, Y., Kimura, S., and Sasaki, M. (2020). Tyrosine tRNA synthetase as a novel extracellular immunomodulatory protein in Streptococcus anginosus. FEMS Microbiol. Lett. 367:fnaa153. doi: 10.1093/femsle/fnaa153
Shumway, C. N., and Klebanoff, S. J. (1971). Purification of pneumolysin. Infect. Immun. 4, 388–392. doi: 10.1128/iai.4.4.388-392.1971
Sitkiewicz, I. (2018). How to become a killer, or is it all accidental? Virulence strategies in oral streptococci. Mol Oral Microbiol 33, 1–12. doi: 10.1111/omi.12192
Skov Sørensen, U. B., Yao, K., Yang, Y., Tettelin, H., and Kilian, M. (2016). Capsular polysaccharide expression in commensal streptococcus species: genetic and antigenic similarities to Streptococcus pneumoniae. MBio 7:e01844. doi: 10.1128/mBio.01844-16
Smith, H. E., Damman, M., van der Velde, J., Wagenaar, F., Wisselink, H. J., Stockhofe-Zurwieden, N., et al. (1999). Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67, 1750–1756. doi: 10.1128/IAI.67.4.1750-1756.1999
Sorge, N. M. V., Quach, D., Gurney, M. A., Sullam, P. M., Nizet, V., and Doran, K. S. (2009). The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J. Infect. Dis. 199, 1479–1487. doi: 10.1086/598217
Spaulding, A. R., Salgado-Pabón, W., Kohler, P. L., Horswill, A. R., Leung, D. Y. M., and Schlievert, P. M. (2013). Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 26, 422–447. doi: 10.1128/CMR.00104-12
Spellerberg, B., Martin, S., Brandt, C., and Lütticken, R. (2000). The cyl genes of Streptococcus agalactiae are involved in the production of pigment. FEMS Microbiol. Lett. 188, 125–128. doi: 10.1111/j.1574-6968.2000.tb09182.x
Sriskandan, S., Unnikrishnan, M., Krausz, T., and Cohen, J. (2000). Mitogenic factor (MF) is the major DNase of serotype M89 streptococcus pyogenes. Microbiology 146, 2785–2792. doi: 10.1099/00221287-146-11-2785
Sukeno, A., Nagamune, H., Whiley, R. A., Jafar, S. I., Aduse-Opoku, J., Ohkura, K., et al. (2005). Intermedilysin is essential for the invasion of hepatoma HepG2 cells by Streptococcus intermedius. Microbiol. Immunol. 49, 681–694. doi: 10.1111/j.1348-0421.2005.tb03647.x
Sumby, P., Barbian, K. D., Gardner, D. J., Whitney, A. R., Welty, D. M., Long, R. D., et al. (2005). Extracellular deoxyribonuclease made by group a streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc. Natl. Acad. Sci. U. S. A. 102, 1679–1684. doi: 10.1073/pnas.0406641102
Summanen, P. H., Rowlinson, M. C., Wooton, J., and Finegold, S. M. (2009). Evaluation of genotypic and phenotypic methods for differentiation of the members of the Anginosus group streptococci. Eur. J. Clin. Microbiol. Infect. Dis. 28, 1123–1128. doi: 10.1007/s10096-009-0758-9
Sun, Y., Li, N., Zhang, J., Liu, H., Liu, J., Xia, X., et al. (2016). Enolase of Streptococcus suis serotype 2 enhances blood–brain barrier permeability by inducing IL-8 release. Inflammation 39, 718–726. doi: 10.1007/s10753-015-0298-7
Tabata, A., Nakano, K., Ohkura, K., Tomoyasu, T., Kikuchi, K., Whiley, R. A., et al. (2013). Novel twin streptolysin S-like peptides encoded in the sag operon homologue of β-hemolytic Streptococcus anginosus. J. Bacteriol. 195, 1090–1099. doi: 10.1128/JB.01344-12
Tabata, A., Yamada, T., Ohtani, H., Ohkura, K., Tomoyasu, T., and Nagamune, H. (2019). β-Hemolytic Streptococcus anginosus subsp. anginosus causes streptolysin S-dependent cytotoxicity to human cell culture lines in vitro. J. Oral Microbiol. 11:1609839. doi: 10.1080/20002297.2019.1609839
Takahashi, Y., Yoshida, A., Nagata, E., Hoshino, T., Oho, T., Awano, S., et al. (2011). Streptococcus anginosus L-cysteine desulfhydrase gene expression is associated with abscess formation in BALB/c mice. Mol Oral Microbiol 26, 221–227. doi: 10.1111/j.2041-1014.2010.00599.x
Telford, J. L., Barocchi, M. A., Margarit, I., Rappuoli, R., and Grandi, G. (2006). Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4, 509–519. doi: 10.1038/nrmicro1443
Tenenbaum, T., Bloier, C., Adam, R., Reinscheid, D. J., and Schroten, H. (2005). Adherence to and invasion of human brain microvascular endothelial cells are promoted by fibrinogen-binding protein FbsA of Streptococcus agalactiae. Infect. Immun. 73, 4404–4409. doi: 10.1128/IAI.73.7.4404-4409.2005
Timmer, A. M., Timmer, J. C., Pence, M. A., Hsu, L. C., Ghochani, M., Frey, T. G., et al. (2009). Streptolysin O promotes group a streptococcus immune evasion by accelerated macrophage apoptosis. J. Biol. Chem. 284, 862–871. doi: 10.1074/jbc.M804632200
Toniolo, C., Balducci, E., Romano, M. R., Proietti, D., Ferlenghi, I., Grandi, G., et al. (2015). Streptococcus agalactiae capsule polymer length and attachment is determined by the proteins CpsABCD. J. Biol. Chem. 290, 9521–9532. doi: 10.1074/jbc.M114.631499
Tsunashima, H., Miyake, K., Motono, M., and Iijima, S. (2012). Organization of the capsule biosynthesis gene locus of the oral streptococcus Streptococcus anginosus. J. Biosci. Bioeng. 113, 271–278. doi: 10.1016/j.jbiosc.2011.10.013
Uchiyama, S., Andreoni, F., Schuepbach, R. A., Nizet, V., and Zinkernagel, A. S. (2012). DNase Sda1 allows invasive M1T1 group a streptococcus to prevent TLR9-dependent recognition. PLoS Pathog. 8:e1002736. doi: 10.1371/journal.ppat.1002736
Uchiyama, S., Döhrmann, S., Timmer, A. M., Dixit, N., Ghochani, M., Bhandari, T., et al. (2015). Streptolysin O rapidly impairs neutrophil oxidative burst and antibacterial responses to group a streptococcus. Front. Immunol. 6:581. doi: 10.3389/fimmu.2015.00581
Unsworth, P. F. (1989). Hyaluronidase production in Streptococcus milleri in relation to infection. J. Clin. Pathol. 42, 506–510. doi: 10.1136/jcp.42.5.506
Vatansever, F., de Melo, W. C. M. A., Avci, P., Vecchio, D., Sadasivam, M., Gupta, A., et al. (2013). Antimicrobial strategies centered around reactive oxygen species – bactericidal antibiotics, photodynamic therapy, and beyond. FEMS Microbiol. Rev. 37, 955–989. doi: 10.1111/1574-6976.12026
Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M., and Hardie, K. R. (2005). Making “sense” of metabolism: Autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3, 383–396. doi: 10.1038/nrmicro1146
Vogkou, C. T., Vlachogiannis, N. I., Palaiodimos, L., and Kousoulis, A. A. (2016). The causative agents in infective endocarditis: a systematic review comprising 33,214 cases. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1227–1245. doi: 10.1007/s10096-016-2660-6
Von Köckritz-Blickwede, M., and Nizet, V. (2009). Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J. Mol. Med. 87, 775–783. doi: 10.1007/s00109-009-0481-0
Waite, R. D., Qureshi, M. R., and Whiley, R. A. (2017). Modulation of behaviour and virulence of a high alginate expressing Pseudomonas aeruginosa strain from cystic fibrosis by oral commensal bacterium Streptococcus anginosus. PLoS One 12:e0173741. doi: 10.1371/journal.pone.0173741
Waite, R. D., Wareham, D. W., Gardiner, S., and Whiley, R. A. (2012). A simple, semiselective medium for anaerobic isolation of anginosus group streptococci from patients with chronic lung disease. J. Clin. Microbiol. 50, 1430–1432. doi: 10.1128/JCM.06184-11
Wang, Z., Guo, C., Xu, Y., Liu, G., Lu, C., and Liu, Y. (2014). Two novel functions of hyaluronidase from Streptococcus agalactiae are enhanced intracellular survival and inhibition of proinflammatory cytokine expression. Infect. Immun. 82, 2615–2625. doi: 10.1128/IAI.00022-14
Wannamaker, L. W. (1958). The differentiation of three distinct desoxyribonucleases of group a streptococci. J. Exp. Med. 107, 797–812. doi: 10.1084/jem.107.6.797
Wannamaker, L. W., Hayes, B., and Yasmineh, W. (1967). Streptococcal nucleases. II. Characterization of DNAse D. J. Exp. Med. 126, 497–508. doi: 10.1084/jem.126.3.497
Wannamaker, L. W., and Yasmineh, W. (1967). Streptococcal nucleases. I. Further studies on the a, B, and C enzymes. J. Exp. Med. 126, 475–496. doi: 10.1084/jem.126.3.475
Wartha, F., Beiter, K., Albiger, B., Fernebro, J., Zychlinsky, A., Normark, S., et al. (2007). Capsule and d-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell. Microbiol. 9, 1162–1171. doi: 10.1111/j.1462-5822.2006.00857.x
Weinstein, M. R., Litt, M., Kertesz, D. A., Wyper, P., Rose, D., Coulter, M., et al. (1997). Invasive infections due to a fish pathogen, streptococcus iniae. S. Iniae study group. N. Engl. J. Med. 337, 589–594. doi: 10.1056/NEJM199708283370902
Wenzler, E., Chandrasekaran, V., Salvador, P., Anwar, M., Pancholi, P., and McGwire, B. S. (2015). Clinical and microbiological outcomes in patients with Streptococcus anginosus group bacteraemia identified through use of a rapid microarray assay. J. Med. Microbiol. 64, 1369–1374. doi: 10.1099/jmm.0.000176
Wessels, M. R. (2019). Capsular polysaccharide of group a streptococcus. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.GPP3-0050-2018
Whiley, R. A., Beighton, D., Winstanley, T. G., Fraser, H. Y., and Hardie, J. M. (1992). Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (the Streptococcus milleri group): association with different body sites and clinical infections. J. Clin. Microbiol. 30, 243–244. doi: 10.1128/jcm.30.1.243-244.1992
Whiley, R. A., Fraser, H., Hardie, J. M., and Beighton, D. (1990). Phenotypic differentiation of Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus strains within the “Streptococcus milleri group”. J. Clin. Microbiol. 28, 1497–1501. doi: 10.1128/jcm.28.7.1497-1501.1990
Whiley, R. A., and Hardie, J. M. (2015). Streptococcus. Bergey’s Manu. System. Archaea Bacteria, eds. M.E. Trujillo, S. Dedysh, P. DeVos, B. Hedlund, P. Kämpfer, F.A. Rainey and W.B. Whitman, doi: 10.1002/9781118960608.gbm00612
Whiley, R. A., Sheikh, N. P., Mushtaq, N., Hagi-Pavli, E., Personne, Y., Javaid, D., et al. (2014). Differential potentiation of the virulence of the Pseudomonas aeruginosa cystic fibrosis Liverpool epidemic strain by oral commensal streptococci. J. Infect. Dis. 209, 769–780. doi: 10.1093/infdis/jit568
Wilde, S., Johnson, A. F., and LaRock, C. N. (2021). Playing with fire: Proinflammatory virulence mechanisms of group a streptococcus. Front. Cell. Infect. Microbiol. 11:704099. doi: 10.3389/fcimb.2021.704099
Willcox, M. D. P. (1995). Potential pathogenic properties of members of the “Streptococcus milleri” group in relation to the production of endocarditis and abscesses. J. Med. Microbiol. 43, 405–410. doi: 10.1099/00222615-43-6-405
Wolinowska, R., Cegłowski, P., Kok, J., and Venema, G. (1991). Isolation, sequence and expression in Escherichia coli, Bacillus subtilis and Lactococcus lactis of the DNase (streptodornase)-encoding gene from streptococcus equisimilis H46A. Gene 106, 115–119. doi: 10.1016/0378-1119(91)90574-u
Wu, H., and Zheng, R. (2020). Splenic abscess caused by Streptococcus anginosus bacteremia secondary to urinary tract infection: a case report and literature review. Open Med. 15, 997–1002. doi: 10.1515/med-2020-0117
Xu, L., Huang, B., Du, H., Zhang, X. C., Xu, J., Li, X., et al. (2010). Crystal structure of cytotoxin protein suilysin from Streptococcus suis. Protein Cell 1, 96–105. doi: 10.1007/s13238-010-0012-3
Yamaguchi, T., Matsumoto, M., Sugimoto, Y., Soutome, S., and Oho, T. (2009). Gene cloning and characterization of Streptococcus intermedius fimbriae involved in saliva-mediated aggregation. Res. Microbiol. 160, 809–816. doi: 10.1016/j.resmic.2009.09.014
Yamaguchi, T., and Matsunoshita, N. (2004). Isolation and some properties of fimbriae of oral Streptococcus intermedius. Curr. Microbiol. 49, 59–65. doi: 10.1007/s00284-004-4281-z
Yoshida, Y., Nakano, Y., Amano, A., Yoshimura, M., Fukamachi, H., Oho, T., et al. (2002). Lcd from streptococcus encodes a C-S lyase with α,β-elimination activity that degrades L-cysteine. Microbiology 148, 3961–3970. doi: 10.1099/00221287-148-12-3961
Yoshida, Y., Negishi, M., Amano, A., Oho, T., and Nakano, Y. (2003). Differences in the βC-S lyase activities of viridans group streptococci. Biochem. Biophys. Res. Commun. 300, 55–60. doi: 10.1016/s0006-291x(02)02803-6
Zachary, J. F. (2017). “Mechanisms of microbial infections,” in Pathologic Basis of Veterinary Disease Expert Consult (Amsterdam: Elsevier Inc)
Keywords: SAG, Streptococcus milleri, commensal, opportunistic infections, invasive infections
Citation: Kuryłek A, Stasiak M and Kern-Zdanowicz I (2022) Virulence factors of Streptococcus anginosus – a molecular perspective. Front. Microbiol. 13:1025136. doi: 10.3389/fmicb.2022.1025136
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Mogens Kilian, Aarhus University, DenmarkClemence Massip, Université Toulouse III Paul Sabatier, France
Copyright © 2022 Kuryłek, Stasiak and Kern-Zdanowicz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Izabela Kern-Zdanowicz, aXphQGliYi53YXcucGw=
†These authors have contributed equally to this work
 Aleksandra Kuryłek†
Aleksandra Kuryłek† Monika Stasiak
Monika Stasiak Izabela Kern-Zdanowicz
Izabela Kern-Zdanowicz