- 1Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 2Department of Laboratory Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 3Department of Infectious Diseases, The First Affiliated Hospital, College of Clinical Medicine, Henan University of Science and Technology, Luoyang, China
- 4Department of Structure and Morphology, Jinan Microecological Biomedicine Shandong Laboratory, Jinan, Shandong, China
- 5Research Units of Infectious Diseases and Microecology, Chinese Academy of Medical Sciences, Beijing, China
The worldwide spread of carbapenem-resistant Enterobacteriaceae (CRE) has led to a major challenge to human health. In this case, colistin is often used to treat the infection caused by CRE. However, the coexistence of genes conferring resistance to carbapenem and colistin is of great concern. In this work, we reported the coexistence of blaOXA-181, blaCTX-M-55, and mcr-8 in an ST273 Klebsiella pneumoniae isolate for the first time. The species identification was performed using MALDI-TOF MS, and the presence of various antimicrobial resistance genes (ARGs) and virulence genes were detected by PCR and whole-genome sequencing. Antimicrobial susceptibility testing showed that K. pneumoniae 5589 was resistant to aztreonam, imipenem, meropenem, ceftriaxone, cefotaxime, ceftazidime, levofloxacin, ciprofloxacin, gentamicin, piperacillin-tazobactam, cefepime, and polymyxin B, but sensitive to amikacin. S1-pulsed-field gel electrophoresis (PFGE) and Southern blotting revealed the mcr-8 gene was carried on a ~ 138 kb plasmid with a conserved structure (IS903B-ymoA-inhA-mcr-8-copR-baeS-dgkA-ampC). In addition, blaOXA-181 was found on another ~51 kb plasmid with a composite transposon flanked by insertion sequence IS26. The in vitro conjugation experiments and plasmid sequence probe indicated that the plasmid p5589-OXA-181 and the p5589-mcr-8 were conjugative, which may contribute to the propagation of ARGs. Relevant detection and investigation measures should be taken to control the prevalence of pathogens coharboring blaOXA-181, blaCTX-M-55 and mcr-8.
Introduction
As one of the significant challenges to global public health, bacterial resistance has attracted much attention in clinical treatment (Xiao et al., 2016; Lai et al., 2021). Especially the infection caused by carbapenem-resistant Enterobacteriaceae (CRE) puts pressure on the health care system in China (Zheng et al., 2018, 2019a; Tompkins and van Duin, 2021).
OXA-48, one of the most common carbapenemases, was first reported in a K. pneumoniae isolated from a patient in Turkey (Mairi et al., 2018). OXA-48, unlike the other major carbapenemases, is an ambler class D enzyme that shows low activity against carbapenems and spares extended-spectrum cephalosporins (Stewart et al., 2018). Therefore, it is challenging to detect blaOXA-48-habouring bacteria clinically. Till now, OXA-48 has more than 10 variants, and OXA-181 is currently the second most common global derivative, which differs from OXA-48 by four amino acid substitutions (Messaoudi et al., 2021). Unlike the prevalence of KPC, NDM and IMP, OXA-181 mainly occurs in India, Europe and the South-East Mediterranean region (Nigg et al., 2019; Shanthini et al., 2019). The emergence of blaOXA-181 in China has aroused concern extensively.
Extended-spectrum β-lactamases (ESBLs) are a class of enzymes that mainly confer resistance to beta-lactam antibiotics, including SHV, TEM, CTX-M and PER. Among them, the CTX-M has been reported to be the predominant type of ESBLs in various Enterobacteriaceae. Since the CTX-M-55 first appeared in India, it has been found in countries worldwide through the transmission of many mobile genetic elements. Recently, considering the increasing detection rate of blaCTX-M-55 in China, many researches were performed about its characteristics.
Currently, colistin is widely used in clinical practice, mainly for treating infections caused by CRE (Durante-Mangoni et al., 2019). However, the mobile colistin resistance gene mcr-8 significantly affects the therapeutic efficacy of colistin and the prognosis of patients with associated infections (Phetburom et al., 2021). In 2016, mcr-8 was first identified in K. pneumoniae (Wang et al., 2018). Several mcr-8 variants have been reported in K. pneumoniae, Klebsiella quasipneumoniae, Raoultella ornithinolytica, and Enterobacter cloacae, including mcr-8.1 mcr-8.4 (Wang et al., 2019, 2022; Yang et al., 2019).
The spread of the mcr genes into CRE, which has been reported globally, is of great clinical concern, leading to the emergence of true pan-drug-resistant pathogens (Mediavilla et al., 2016; Zheng et al., 2017; Han S. et al., 2020; Chen et al., 2022). Meanwhile, the isolation and culture of such pathogen coharboring mcr and carbapenemase-encoding gene from the blood sample is rare. Accordingly, our work aims to describe the antimicrobial susceptibility, plasmid characteristics and genomic features of a K. pneumoniae strain co-producing OXA-181, CTX-M-55, and MCR-8 from China for the first time.
Materials and methods
Species identification and antimicrobial susceptibility testing
Isolates were collected from a tertiary hospital in Zhengzhou, Henan province, China, during our routine surveillance of CRE. Species identification was performed by matrix-assist laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (Bruker Daltonik GmbH, Bremen, Germany). The mobile colistin resistance genes mcr-1 to mcr-8 and the major carbapenemase genes, such as blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP, were identified using PCR, as described previously (Zheng et al., 2019b; Liang et al., 2021).
The susceptibility of K. pneumoniae 5589 and its transconjugants to antibiotics was tested using the agar dilution method, except for the polymyxins, which was performed using the broth microdilution method (Liu et al., 2021). The results were interpreted based on the Clinical and Laboratory Standards Institute (CLSI) and the European Committee for Antimicrobial Susceptibility Testing (EUCAST) guidelines. K. pneumoniae ATCC700603 and Escherichia coli ATCC25922 were used as the quality control.
Plasmid analysis and conjugation assay
The number and size of plasmids in K. pneumoniae 5589 were detected by S1-PFGE of total DNA (Chi et al., 2020). The locations of plasmids harboring the blaOXA-181 and mcr-8 were determined by Southern blotting and hybridization with digoxigenin-labeled specific probes. Furthermore, rifampin-resistant P. aeruginosa PAO1Ri was used as a recipient bacterium in transformation conjugation experiments to investigate whether the plasmids can transfer (Liu et al., 2021). The transconjugants which showed growth on Mueller-Hinton medium simultaneously containing 300 mg/L rifampicin and 2 mg/L meropenem were identified by MALDI-TOF/MS. The existence of blaOXA-181 and mcr-8 in transconjugants was detected by PCR and the antimicrobial susceptibility testing of transconjugants to confirm whether the plasmids carrying target genes were successfully transferred.
Whole-genome sequencing and analysis
The genome of K. pneumoniae 5589 was extracted using a specific bacterial DNA Kit (QIAGEN, Hilden, Germany). To better understand the genetic features, DNA sequencing was performed on the Illumina NovaSeq 6000 (Illumina, San Diego, CA, United States) and the Oxford Nanopore (Oxford Nanopore Technologies, Oxford, United Kingdom) platform (Bao et al., 2022). Then, the whole genome was annotated with Prokka. Additionally, the acquired ARGs were detected by ResFinder 4.11, and the plasmid replicon type was identified by PlasmidFinder 2.1.2 The transposon and insertion sequence were detected using the ISFinder database.3 Finally, the circular comparison images of multiplex plasmids were generated by BLAST Ring Image Generator (BRIG). The linear comparison figures of multiple genomic loci surrounding the blaOXA-181 and mcr-8 were generated by Easyfig 2.0 software (Sullivan et al., 2011).
Results
Isolation of Klebsiella pneumoniae 5589 and antimicrobial susceptibility testing
Carbapenem-resistant K. pneumoniae 5589 was isolated from a blood sample of a patient who was hospitalized for myelodysplastic syndrome (MDS). During his hospitalization, the patient developed thrombocytopenia, high fever and groin infection. Subsequently, the patient’s condition was controlled with a normal body temperature after the biapenem and tigecycline treatment.
The antimicrobial susceptibility profiles of K. pneumoniae 5589 and transconjugants were demonstrated in Table 1. K. pneumoniae 5589 was resistant to multiple antibiotics such as aztreonam, imipenem, meropenem, ceftriaxone, cefotaxime, ceftazidime, levofloxacin, ciprofloxacin, gentamicin, piperacillin-tazobactam, cefepime and polymyxin B, but remained susceptible to amikacin. Moreover, the transconjugants 5589-PAO1Ri showed a similarity antibiotic resistance profile to K. pneumoniae 5589 but was intermediate to imipenem and sensitive to ciprofloxacin and gentamicin.
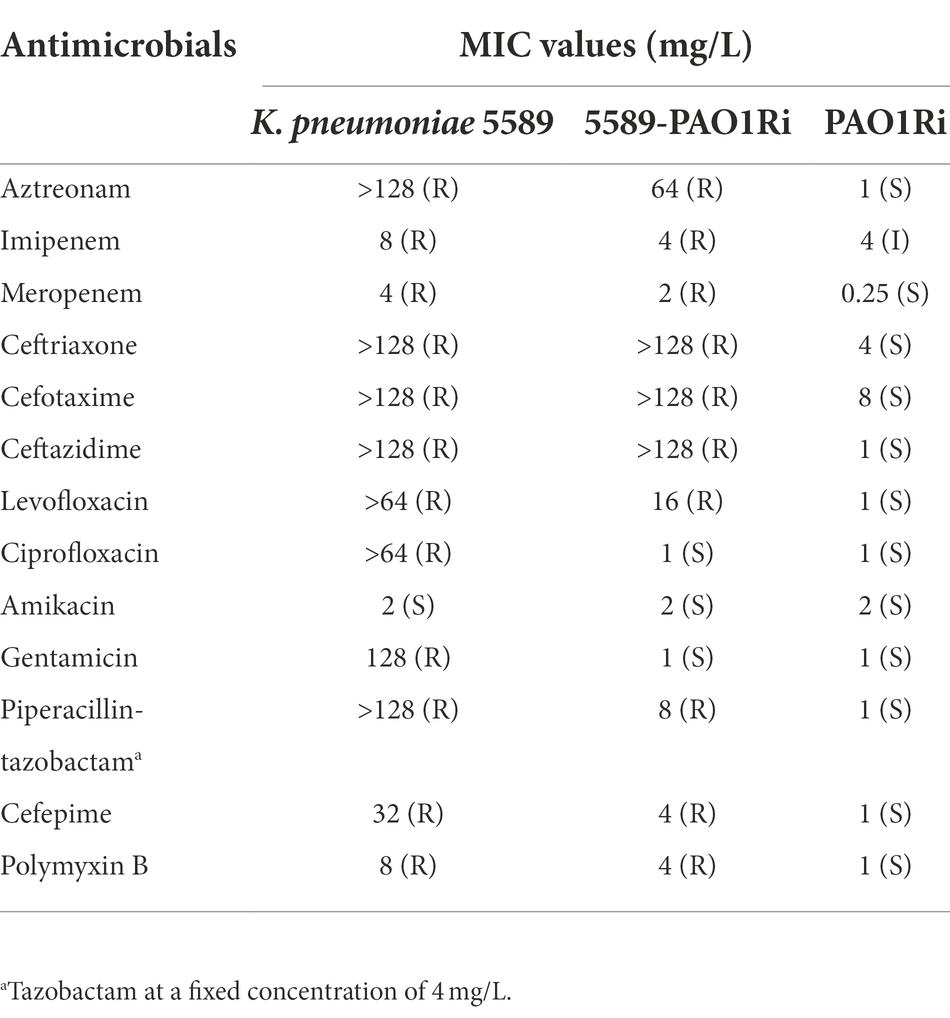
Table 1. MIC values of antimicrobials for Klebsiella pneumoniae 5589, transconjugant 5589-PAO1Ri, and recipient strain PAO1Ri.
Genomics features of Klebsiella pneumoniae 5589
The K. pneumoniae 5589 genome contains a 5,279,178 bp circular chromosome with an average GC content of 57.5% and three plasmids of different sizes from 51,479 bp to 290,720 bp (Supplementary Table S1). WGS revealed that K. pneumoniae 5589 was identified as ST273, which belongs to the clonal group 147. By researching the ARGs on ResFinder, 48 acquired resistance genes were detected (Supplementary Table S1). The chromosome of strain K. pneumoniae 5589 was found to harbor ARGs which confer resistance to beta-lactams (blaSHV-11, blaSHV-67), fosfomycin (fosA), chloramphenicol (OqxA, OqxB). Moreover, it carried multiple virulence genes such as coding for outer membrane receptor (fepA), transcriptional regulator (fimK), regulator protein (ykgK) and transcriptional activator (mrkH).
Characterization of plasmid bearing blaOXA-181
S1-PFGE and southern blot results revealed that the resistance gene blaOXA-181 was located on a 51,479 kb plasmid (p5589-OXA-181), which belongs to IncX3-ColKP3 with a GC content of 46% (Supplementary Figure S1). The plasmid carrying blaOXA − 181 was successfully transferred to a P. aeruginosa PAO1Ri recipient strain. The plasmid p5589-OXA-181 carries not only the blaOXA-181 but also qnrS1, which enables the strain to be resistant to ciprofloxacin. According to the result of the BLAST search, plasmid p5589-OXA-181 was almost identical to pNIPH17_0036_1 (accession no: LC483179), pKBN10P04869C (accession no: CP026476), pABC264-OXA-181 (accession no: MK412917) and pBC947-OXA-181 (accession no: MK412920) with the similarity between 99% and 100% (Figure 1A). The genetic environment analysis showed blaOXA-181 was located on a composite transposon surrounded by two copies of insertion sequence IS26. A similar region can be seen in E. coli plasmid pKBN10P04869C (accession no: CP026476), and E. coli plasmid pABC264-OXA-181 (accession no: MK412917). In plasmid p5589-OXA-181, the gene repA1 is responsible for encoding ColKP3-type replication initiation protein, and the ISKpn19 fragment is located downstream of blaOXA-181, while IS3000 is located on the upstream (Supplementary Figure S2A). Many other functional genes such as encoding DNA topoisomerase (topB), proteasome-associated ATPase (mpa), type IV secretion system protein (ptlH, virB9, virB8), transcription antitermination protein (rfaH) are distributed on the backbone.
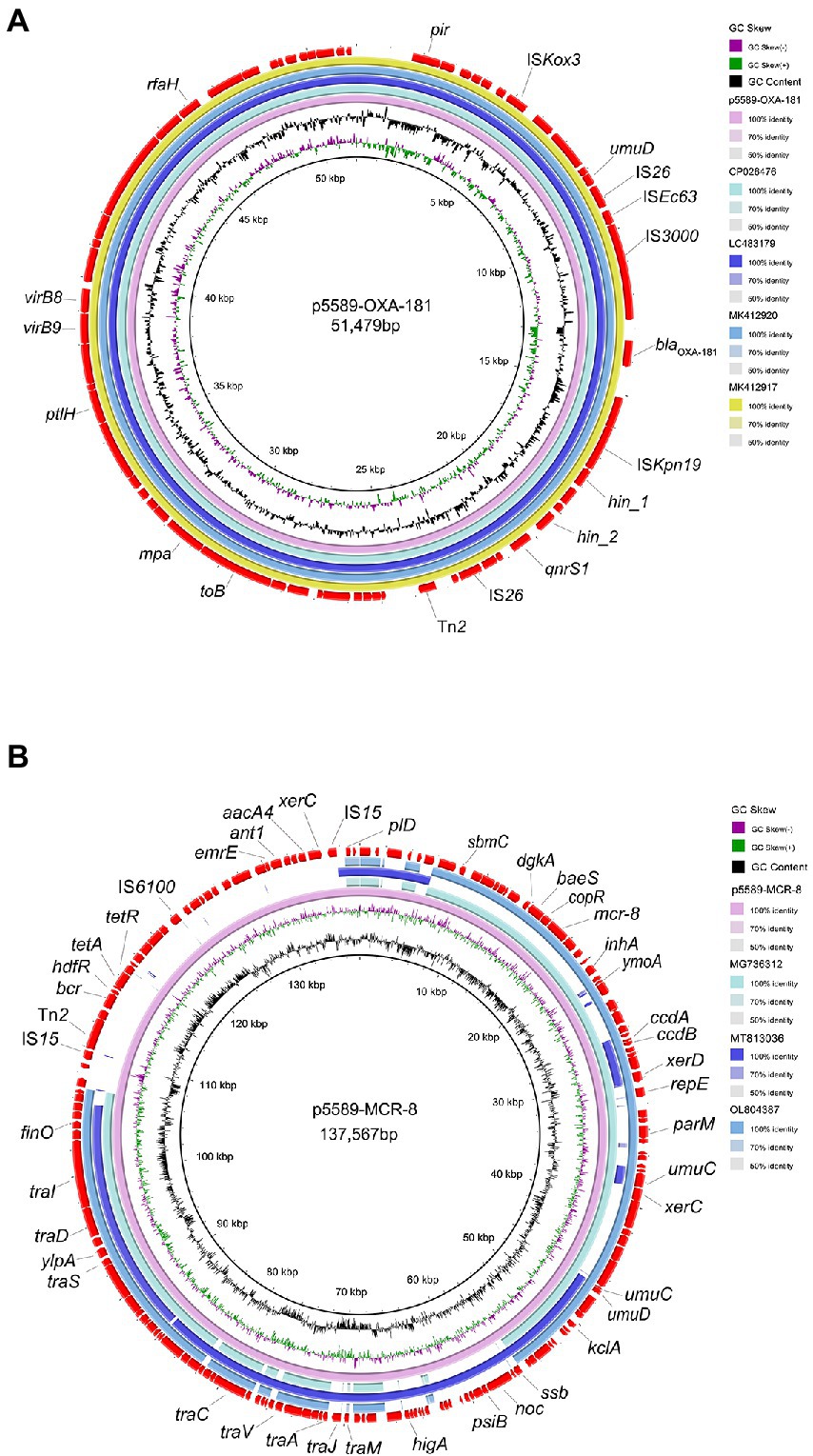
Figure 1. Comparative analysis of plasmids p5589-OXA-181 and p5589-mcr-8 detected in Klebsiella pneumoniae 5589. (A) Comparison of blaOXA-181 bearing plasmid p5589-OXA-181 with pKBN10P04869C (GenBank accession no. CP026476), pNIPH17_0036_1 (GenBank accession no. LC483179), pBC947-OXA-181(GenBank accession no. MK412920) and pABC264-OXA-181(GenBank accession no. MK412917). (B) Comparison of mcr-8-carrying plasmid p5589-MCR-8 with pKP91(GenBank accession no. MG736312), pKQBSI104-1(GenBank accession no. MT813036) and pKP3(GenBank accession no. OL804387).
Characterization of plasmid bearing mcr-8
The mcr-8 gene was carried by another plasmid of size 137,567 kb (p5589-mcr-8) with the replicon type of IncFIA-FII (Supplementary Figure S1). Meanwhile, we obtained the transconjugant harboring mcr-8 successfully. The plasmid p5589-mcr-8 contained additional genes that make strain exhibit resistance to multiple antibiotics, such as bleomycin (bleO), spectinomycin (aadA16), ciprofloxacin (aac (6′)-Ib-cr, qnrB91), rifampicin (arr-3), azithromycin (mph(A)), trimethoprim (dfrA27), sulfamethoxazole (sul1), tetracycline (tet(A)), chlorhexidine (qacE), chloramphenicol (floR). On the other hand, the plasmid p5589_MCR-8 showed great similarity to pKP91(65%coverage and 99.70%identity; accession no: MG736312), pKQBSI104-1(52%coverage and 99.47%identity; accession no: MT813036), pKP3(65%coverage and 99.86%identity; accession no: OL804387; Figure 1B).
The inspection of the genetic regions revealed that mcr-8 in this work was similar to that of the mcr-8.1 gene in plasmid pK91 and pKP3. The upstream was the IS903B and other functional genes (inhA, YmoA). At the same time, the downstream of mcr-8 were the transfer or transcription-associated genes (copR, sasA dgkA) and encoding β-lactamase gene (ampc) (Supplementary Figure S2B). The p5589-mcr-8 backbone carried regions responsible for toxin-antitoxin (TA) systems (ccdA, ccdB, higA, ylpA), replication (repE), mobilization (tra, xerD, xerC, klcA, finO) and stability (parM, umuC, umuD, ssb, noc, psiB). Other genes encode enzymes associated with DNA replication (sbmC, ant1, aacA4, pld), and proteins associated with the resistance and transport of multidrug (bcr, tetA, tetR, emrE) also can be found.
Discussion
OXA-181-producing Enterobacteriaceae have been reported in several countries, including Portugal, South Africa, and Singapore, but have rarely been described in China, where Klebsiella pneumoniae carbapenemase (KPC) is the major carbapenemase (Han R. et al., 2020; Chew et al., 2021). To our knowledge, OXA-181 has not emerged in China until 2015, and the report of blaOXA-181 in China is still uncommon (Qin et al., 2018). Infections caused by OXA-181 in nonendemic areas were often associated with the travelling of patients to endemic areas (Chudejova et al., 2021). However, the identification of blaOXA-181 in this work was from a patient without a history of foreign residence, which indicated its wider dissemination than previously anticipated.
So far, several studies have reported plasmids carrying blaOXA-181 with different replicons, such as IncX3, IncA/C, ColE, IncT, IncN, IncFIIK and ColKP3 (Villa et al., 2013; Naha et al., 2021). But the most common plasmid in China is the IncX3 type which has a similar genetic environment to others (Liu et al., 2020). The similar blaOXA-181 bearing IncX3 plasmid further highlights the role of IncX3-type plasmid as an irreplaceable vector of ARGs (Santos Tufic-Garutti et al., 2022). In p5589-OXA-181, blaOXA-181 was found on a composite transposon which was considered to facilitate its horizontal transmission (Supplementary Figure S2A). Therefore, the absence of upstream mobile element ISEcp1 was detected in p5589-OXA-181, which was consistent with other studies (Naha et al., 2021). Generally, ISEcp1 plays an essential role in the transmission of ARGs; its absence may affect transposase activity and the maintenance of resistance genes on a plasmid (Potron et al., 2013; Naha et al., 2021).
The colistin resistance mechanism in Enterobacteriaceae is complicated and has not been wholly investigated (El-Sayed Ahmed et al., 2020). In general, the resistance to colistin can be acquired by intrinsic mutation or adaptation mechanisms and the horizontal transfer of mcr gene and its variants (Moffatt et al., 2019). Since the initial report of mcr-8, this gene has been discovered in Enterobacteriaceae isolates from humans, animals and various environments worldwide (Anyanwu et al., 2020; Ngbede et al., 2020).
There have already been several studies about the genetic context analysis of mcr-8 (Wu et al., 2020). The mcr-8 gene was firstly recognized on a typical IncFII-type plasmid pKP91, with a conservative region flanked by IS903B, but in p5589-mcr-8, the IS903B located on the upstream was absent (Supplementary Figure S2B; Wang et al., 2018). The mcr-8-carrying plasmid in Raoultella ornithinolytica also harbored only one copy of IS903B located upstream (Wang et al., 2019). In addition, Farzana has reported the upstream IS903B of mcr-8 in ST15 K. pneumoniae was replaced by ISKpn21 (Farzana et al., 2020). Thus, it was reasonable to speculate that the upstream IS903B surrounding mcr-8 is unstable and replaceable. However, additional studies focusing on this are warranted.
K. pneumoniae 5589 in this work also carried blaCTX-M-55, which was frequently found in E. coli (Zhang et al., 2014; Birgy et al., 2018; Feng et al., 2019). Hence, we could also pay more attention on blaCTX − M − 55-positive K. pneumoniae to better understand the molecular epidemiology of CTX-M-55 in China. Simultaneously, K. pneumoniae 5589 was assigned to ST273, which was recognized as the reservoir of many carbapenemase genes, including blaKPC, blaVIM, blaNDM and blaIMP (Chou et al., 2016; Liu et al., 2018). ST273 was divided into the specific clonal group 147, which had a high epidemic potential (Rodrigues et al., 2022). The original detection of ST273 was in Europe and had been gradually identified in Italy, Norway, and Russia, even causing the outbreak in Southeast Asia. Furthermore, whether the ST273 could influence the epidemiology of blaOXA-181 and mcr-8 remains unknown, and necessary attention should be paid to these sequence types to avoid epidemic outbreaks.
Previous studies have reported the co-producing of OXA-181 and other carbapenemases, such as co-harboring blaOXA-181 and blaNDM-5 in Nepal, blaOXA-181 and blaNDM-1 in French, and blaOXA-181 and blaKPC-121 in Italy (Sherchan et al., 2020; Gaibani et al., 2022). Remarkably, the spread of mcr gene into CRE resulting the accumulation of multidrug resistance genes. Nevertheless, the study about the co-carriage of blaOXA-181 and mcr-8 is limited, except for a report of an Escherichia coli co-producing OXA-181 and MCR-1 (Pulss et al., 2017). Identifying the isolate co-carrying blaOXA-181 and mcr-8 in K. pneumoniae reminds us that persistence detection and further exploration are needed to prevent the emergence and evolution of such MDR isolates.
Conclusion
In summary, our work firstly described the co-occurrence of OXA-181, CTX-M-55, and MCR-8 in K. pneumoniae. Our study also characterized the blaOXA-181 and mcr-8-carrying plasmids, which contribute to exploring the transmission mechanism. The appearance of such clinical isolates producing carbapenemases and MCR narrows the therapeutic options and reveals the severe situation of antimicrobial resistance. Continuous observation and exploration are essential to control its spread.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
XG and BZ conceived and designed the experiments. HG, JQ, RC, CL, and JZ collected samples and performed the experiments. HX, RL, and XH analyzed the data. HG wrote the manuscript. BZ reviewed and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by research grants from Henan Science and Technology Department (192102310059), the National Natural Science Foundation of China (82072314), the Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022011B), the Fundamental Research Funds for the Central Universities (2022ZFJH003), CAMS Innovation Fund for Medical Sciences (2019-I2M-5-045), Henan Province Medical Science and Technology Research Project Joint Construction Project (LHGJ20190232), and Zhejiang Provincial Natural Science Foundation of China (LQ20H200003).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1020500/full#supplementary-material
Footnotes
References
Anyanwu, M. U., Jaja, I. F., and Nwobi, O. C. (2020). Occurrence and characteristics of Mobile Colistin resistance (mcr) gene-containing isolates from the environment: a review. Int. J. Environ. Res. Public Health 17:E1028. doi: 10.3390/ijerph17031028
Bao, D., Xu, X., Wang, Y., and Zhu, F. (2022). Emergence of a multidrug-resistant Escherichia coli co-carrying a new mcr-1.33 variant and Bla (NDM-5) genes recovered from a urinary tract infection. Infect. Drug Resist. 15, 1499–1503. doi: 10.2147/idr.S358566
Birgy, A., Madhi, F., Hogan, J., Doit, C., Gaschignard, J., Caseris, M., et al. (2018). CTX-M-55-, MCR-1-, and FosA-producing multidrug-resistant Escherichia coli infection in a child in France. Antimicrob. Agents Chemother. 62:e00127-18. doi: 10.1128/aac.00127-18
Chen, C., Xu, H., Liu, R., Hu, X., Han, J., Wu, L., et al. (2022). Emergence of neonatal sepsis caused by MCR-9- and NDM-1-co-producing Enterobacter hormaechei in China. Front. Cell. Infect. Microbiol. 12:879409. doi: 10.3389/fcimb.2022.879409
Chew, K. L., Octavia, S., Lai, D., Lin, R. T. P., and Teo, J. W. P. (2021). Genomic characterization of Klebsiella quasipneumoniae from clinical specimens in Singapore. Antimicrob. Agents Chemother. 65:e0041221. doi: 10.1128/aac.00412-21
Chi, X., Zhang, J., Xu, H., Yu, X., Shen, P., Ji, J., et al. (2020). Emergence of KPC-2-producing Raoultella ornithinolytica isolated from a hospital wastewater treatment plant. Antimicrob. Agents Chemother. 64:e01983-19. doi: 10.1128/aac.01983-19
Chou, A., Roa, M., Evangelista, M. A., Sulit, A. K., Lagamayo, E., Torres, B. C., et al. (2016). Emergence of Klebsiella pneumoniae ST273 carrying Bla(NDM-7) and ST656 carrying Bla(NDM-1) in Manila, Philippines. Microb. Drug Resist. 22, 585–588. doi: 10.1089/mdr.2015.0205
Chudejova, K., Kraftova, L., Mattioni Marchetti, V., Hrabak, J., Papagiannitsis, C. C., and Bitar, I. (2021). Genetic plurality of OXA/NDM-encoding features characterized from Enterobacterales recovered from Czech hospitals. Front. Microbiol. 12:641415. doi: 10.3389/fmicb.2021.641415
Durante-Mangoni, E., Andini, R., and Zampino, R. (2019). Management of carbapenem-resistant Enterobacteriaceae infections. Clin. Microbiol. Infect. 25, 943–950. doi: 10.1016/j.cmi.2019.04.013
El-Sayed Ahmed, M. A. E., Zhong, L. L., Shen, C., Yang, Y., Doi, Y., and Tian, G. B. (2020). Colistin and its role in the era of antibiotic resistance: an extended review (2000-2019). Emerg. Microbes Infect. 9, 868–885. doi: 10.1080/22221751.2020.1754133
Farzana, R., Jones, L. S., Barratt, A., Rahman, M. A., Sands, K., Portal, E., et al. (2020). Emergence of Mobile Colistin resistance (mcr-8) in a highly successful Klebsiella pneumoniae sequence type 15 clone from clinical infections in Bangladesh. mSphere 5:e00023-20. doi: 10.1128/mSphere.00023-20
Feng, C., Wen, P., Xu, H., Chi, X., Li, S., Yu, X., et al. (2019). Emergence and comparative genomics analysis of extended-Spectrum-beta-lactamase-producing Escherichia coli carrying mcr-1 in fennec fox imported from Sudan to China. mSphere 4:e00732-19. doi: 10.1128/mSphere.00732-19
Gaibani, P., Amadesi, S., Lazzarotto, T., and Ambretti, S. (2022). Genome characterization of a Klebsiella pneumoniae co-producing OXA-181 and KPC-121 resistant to ceftazidime/avibactam, meropenem/vaborbactam, imipenem/relebactam and cefiderocol isolated from a critically ill patient. J. Glob. Antimicrob. Resist. 30, 262–264. doi: 10.1016/j.jgar.2022.06.021
Han, R., Shi, Q., Wu, S., Yin, D., Peng, M., Dong, D., et al. (2020). Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among Carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front. Cell. Infect. Microbiol. 10:314. doi: 10.3389/fcimb.2020.00314
Han, S., Kim, J. S., Hong, C. K., Park, S. H., Kim, H. S., Yu, J. K., et al. (2020). Identification of an extensively drug-resistant Escherichia coli clinical strain harboring mcr-1 and Bla(NDM-1) in Korea. J. Antibiot. (Tokyo) 73, 852–858. doi: 10.1038/s41429-020-0350-1
Lai, C. C., Chen, S. Y., Ko, W. C., and Hsueh, P. R. (2021). Increased antimicrobial resistance during the COVID-19 pandemic. Int. J. Antimicrob. Agents 57:106324. doi: 10.1016/j.ijantimicag.2021.106324
Liang, G., Rao, Y., Wang, S., Chi, X., Xu, H., and Shen, Y. (2021). Co-occurrence of NDM-9 and MCR-1 in a human gut colonized Escherichia coli ST1011. Infect. Drug Resist. 14, 3011–3017. doi: 10.2147/idr.S321732
Liu, C., Fang, Y., Zeng, Y., Lu, J., Sun, Q., Zhou, H., et al. (2020). First report of OXA-181-producing Klebsiella pneumoniae in China. Infect. Drug Resist. 13, 995–998. doi: 10.2147/idr.S237793
Liu, L., Feng, Y., Long, H., McNally, A., and Zong, Z. (2018). Sequence type 273 Carbapenem-resistant Klebsiella pneumoniae carrying Bla(NDM-1) and Bla(IMP-4). Antimicrob. Agents Chemother. 62:e00160-18. doi: 10.1128/aac.00160-18
Liu, S., Xu, H., Guo, X., Li, S., Wang, Q., Li, Y., et al. (2021). Emergence and genetic characterization of plasmid-encoded VIM-2-producing pseudomonas stutzeri with novel Integron In1998 isolated from cerebrospinal fluid. Infect. Drug Resist. 14, 3415–3424. doi: 10.2147/idr.S320294
Mairi, A., Pantel, A., Sotto, A., Lavigne, J. P., and Touati, A. (2018). OXA-48-like carbapenemases producing Enterobacteriaceae in different niches. Eur. J. Clin. Microbiol. Infect. Dis. 37, 587–604. doi: 10.1007/s10096-017-3112-7
Mediavilla, J. R., Patrawalla, A., Chen, L., Chavda, K. D., Mathema, B., Vinnard, C., et al. (2016). Colistin- and Carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio 7:e01191-16. doi: 10.1128/mBio.01191-16
Messaoudi, A., Mansour, W., Tilouche, L., Châtre, P., Drapeau, A., Chaouch, C., et al. (2021). First report of carbapenemase OXA-181-producing Serratia marcescens. J. Glob. Antimicrob. Resist. 26, 205–206. doi: 10.1016/j.jgar.2021.06.004
Moffatt, J. H., Harper, M., and Boyce, J. D. (2019). Mechanisms of Polymyxin resistance. Adv. Exp. Med. Biol. 1145, 55–71. doi: 10.1007/978-3-030-16373-0_5
Naha, S., Sands, K., Mukherjee, S., Saha, B., Dutta, S., and Basu, S. (2021). OXA-181-like Carbapenemases in Klebsiella pneumoniae ST14, ST15, ST23, ST48, and ST231 from septicemic neonates: coexistence with NDM-5, resistome, transmissibility, and genome diversity. mSphere 6:e01156-20. doi: 10.1128/mSphere.01156-20
Ngbede, E. O., Poudel, A., Kalalah, A., Yang, Y., Adekanmbi, F., Adikwu, A. A., et al. (2020). Identification of mobile colistin resistance genes (mcr-1.1, mcr-5 and mcr-8.1) in Enterobacteriaceae and Alcaligenes faecalis of human and animal origin, Nigeria. Int. J. Antimicrob. Agents 56:106108. doi: 10.1016/j.ijantimicag.2020.106108
Nigg, A., Brilhante, M., Dazio, V., Clément, M., Collaud, A., Gobeli Brawand, S., et al. (2019). Shedding of OXA-181 carbapenemase-producing Escherichia coli from companion animals after hospitalisation in Switzerland: an outbreak in 2018. Eurosurveillance 24:1900071. doi: 10.2807/1560-7917.Es.2019.24.39.1900071
Phetburom, N., Boueroy, P., Chopjitt, P., Hatrongjit, R., Akeda, Y., Hamada, S., et al. (2021). Klebsiella pneumoniae Complex harboring mcr-1, mcr-7, and mcr-8 isolates from slaughtered pigs in Thailand. Microorganisms 9:2436. doi: 10.3390/microorganisms9122436
Potron, A., Rondinaud, E., Poirel, L., Belmonte, O., Boyer, S., Camiade, S., et al. (2013). Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae. Int. J. Antimicrob. Agents 41, 325–329. doi: 10.1016/j.ijantimicag.2012.11.007
Pulss, S., Semmler, T., Prenger-Berninghoff, E., Bauerfeind, R., and Ewers, C. (2017). First report of an Escherichia coli strain from swine carrying an OXA-181 carbapenemase and the colistin resistance determinant MCR-1. Int. J. Antimicrob. Agents 50, 232–236. doi: 10.1016/j.ijantimicag.2017.03.014
Qin, S., Cheng, J., Wang, P., Feng, X., and Liu, H. M. (2018). Early emergence of OXA-181-producing Escherichia coli ST410 in China. J. Glob. Antimicrob. Resist. 15, 215–218. doi: 10.1016/j.jgar.2018.06.017
Rodrigues, C., Desai, S., Passet, V., Gajjar, D., and Brisse, S. (2022). Genomic evolution of the globally disseminated multidrug-resistant Klebsiella pneumoniae clonal group 147. Microb. Genom. 8:000737. doi: 10.1099/mgen.0.000737
Santos Tufic-Garutti, S. D., de Araújo Longo, L. G., Fontana, H., Garutti, L. H. G., de Carvalho Girão, V. B., Fuga, B., et al. (2022). OXA-181 carbapenemase carried on an IncX3 plasmid in high-risk Escherichia coli ST167 isolated from a traveler returning from sub-Saharan Africa to Brazil. Diagn. Microbiol. Infect. Dis. 102:115570. doi: 10.1016/j.diagmicrobio.2021.115570
Shanthini, T., Manohar, P., Samna, S., Srividya, R., Bozdogan, B., Rameshpathy, M., et al. (2019). Emergence of plasmid-borne Bla (oxa-181) gene in Ochrobactrum intermedium: first report from India. Access Microbiol. 1:e000024. doi: 10.1099/acmi.0.000024
Sherchan, J. B., Tada, T., Shrestha, S., Uchida, H., Hishinuma, T., Morioka, S., et al. (2020). Emergence of clinical isolates of highly carbapenem-resistant Klebsiella pneumoniae co-harboring Bla(NDM-5) and Bla(OXA-181 or −232) in Nepal. Int. J. Infect. Dis. 92, 247–252. doi: 10.1016/j.ijid.2020.01.040
Stewart, A., Harris, P., Henderson, A., and Paterson, D. (2018). Treatment of infections by OXA-48-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 62:e01195-18. doi: 10.1128/aac.01195-18
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tompkins, K., and van Duin, D. (2021). Treatment for carbapenem-resistant Enterobacterales infections: recent advances and future directions. Eur. J. Clin. Microbiol. Infect. Dis. 40, 2053–2068. doi: 10.1007/s10096-021-04296-1
Villa, L., Carattoli, A., Nordmann, P., Carta, C., and Poirel, L. (2013). Complete sequence of the IncT-type plasmid pT-OXA-181 carrying the blaOXA-181 carbapenemase gene from Citrobacter freundii. Antimicrob. Agents Chemother. 57, 1965–1967. doi: 10.1128/aac.01297-12
Wang, S., Ju, X., Dong, N., Li, R., Li, Y., Zhang, R., et al. (2022). Emergence of mobilized Colistin resistance gene mcr-8.2 in multidrug-resistant Enterobacter cloacae isolated from a patient in China. Microbiol. Spectr. 10:e0121722. doi: 10.1128/spectrum.01217-22
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8 in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7:122. doi: 10.1038/s41426-018-0124-z
Wang, X., Wang, Y., Zhou, Y., Wang, Z., Wang, Y., Zhang, S., et al. (2019). Emergence of Colistin resistance gene mcr-8 and its variant in Raoultella ornithinolytica. Front. Microbiol. 10:228. doi: 10.3389/fmicb.2019.00228
Wu, B., Wang, Y., Ling, Z., Yu, Z., Shen, Z., Zhang, S., et al. (2020). Heterogeneity and diversity of mcr-8 genetic context in chicken-associated Klebsiella pneumoniae. Antimicrob. Agents Chemother. 65:e01872-20. doi: 10.1128/aac.01872-20
Xiao, Y., Wang, J., Shen, P., Zheng, B., Zheng, Y., and Li, L. (2016). Retrospective survey of the efficacy of mandatory implementation of the essential medicine policy in the primary healthcare setting in China: failure to promote the rational use of antibiotics in clinics. Int. J. Antimicrob. Agents 48, 409–414. doi: 10.1016/j.ijantimicag.2016.06.017
Yang, X., Liu, L., Wang, Z., Bai, L., and Li, R. (2019). Emergence of mcr-8.2-bearing Klebsiella quasipneumoniae of animal origin. J. Antimicrob. Chemother. 74, 2814–2817. doi: 10.1093/jac/dkz213
Zhang, J., Zheng, B., Zhao, L., Wei, Z., Ji, J., Li, L., et al. (2014). Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14:659. doi: 10.1186/s12879-014-0659-0
Zheng, B., Chen, Y., Violetta, L., Xiao, Y., and Li, L. (2019a). Bloodstream infections caused by Entero-bacteriaceae in China. Lancet Infect. Dis. 19, 810–811. doi: 10.1016/S1473-3099(19)30352-4
Zheng, B., Feng, C., Xu, H., Yu, X., Guo, L., Jiang, X., et al. (2019b). Detection and characterization of ESBL-producing Escherichia coli expressing mcr-1 from dairy cows in China. J. Antimicrob. Chemother. 74, 321–325. doi: 10.1093/jac/dky446
Zheng, B., Lv, T., Xu, H., Yu, X., Chen, Y., Li, J., et al. (2018). Discovery and characterisation of an escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China. Int. J. Antimicrob. Agents 51, 273–275. doi: 10.1016/j.ijantimicag.2017.09.005
Keywords: Klebsiella pneumoniae, OXA-181, CTX-M-55, MCR-8, bacteremia
Citation: Ge H, Qiao J, Xu H, Liu R, Chen R, Li C, Hu X, Zhou J, Guo X and Zheng B (2022) First report of Klebsiella pneumoniae co-producing OXA-181, CTX-M-55, and MCR-8 isolated from the patient with bacteremia. Front. Microbiol. 13:1020500. doi: 10.3389/fmicb.2022.1020500
Edited by:
Biao Tang, Zhejiang Academy of Agricultural Sciences, ChinaReviewed by:
Tieli Zhou, First Affiliated Hospital of Wenzhou Medical University, ChinaSun Chengtao, China Agricultural University, China
Wang Shanmei, Henan Provincial People's Hospital, China
Copyright © 2022 Ge, Qiao, Xu, Liu, Chen, Li, Hu, Zhou, Guo and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beiwen Zheng, emhlbmdid0B6anUuZWR1LmNu; Xiaobing Guo, Z3hiaW5nOTI4QHp6dS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Haoyu Ge
Haoyu Ge Jie Qiao
Jie Qiao Hao Xu
Hao Xu Ruishan Liu
Ruishan Liu Ruyan Chen
Ruyan Chen Chenyu Li
Chenyu Li Xinjun Hu
Xinjun Hu Jiawei Zhou
Jiawei Zhou Xiaobing Guo
Xiaobing Guo Beiwen Zheng
Beiwen Zheng