
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 21 October 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1020220
This article is part of the Research Topic Virus Induced Innate Immune Response and Inflammation View all 8 articles
Early growth response 1 (EGR1) is a multifunctional mammalian transcription factor capable of both enhancing and/or inhibiting gene expression. EGR1 can be activated by a wide array of stimuli such as exposure to growth factors, cytokines, apoptosis, and various cellular stress states including viral infections by both DNA and RNA viruses. Following induction, EGR1 functions as a convergence point for numerous specialized signaling cascades and couples short-term extracellular signals to influence transcriptional regulation of genes required to initiate the appropriate biological response. The role of EGR1 has been extensively studied in both physiological and pathological conditions of the adult nervous system where it is readily expressed in various regions of the brain and is critical for neuronal plasticity and the formation of memories. In addition to its involvement in neuropsychiatric disorders, EGR1 has also been widely examined in the field of cancer where it plays paradoxical roles as a tumor suppressor gene or oncogene. EGR1 is also associated with multiple viral infections such as Venezuelan equine encephalitis virus (VEEV), Kaposi’s sarcoma-associated herpesvirus (KSHV), herpes simplex virus 1 (HSV-1), human polyomavirus JC virus (JCV), human immunodeficiency virus (HIV), and Epstein–Barr virus (EBV). In this review, we examine EGR1 and its role(s) during viral infections. First, we provide an overview of EGR1 in terms of its structure, other family members, and a brief overview of its roles in non-viral disease states. We also review upstream regulators of EGR1 and downstream factors impacted by EGR1. Then, we extensively examine EGR1 and its roles, both direct and indirect, in regulating replication of DNA and RNA viruses.
Early growth response 1 (EGR1) is a transcription factor (TF) that was simultaneously identified by numerous research groups in the 1980s and as such there are several alternate names in the literature including: EGR1 (Sukhatme et al., 1988), NGFI-A (Milbrandt, 1987), Krox-24 (Lemaire et al., 1988), TIS8 (Lim et al., 1987, 1989), and Zif268 (Christy et al., 1988). The EGR1 gene itself is highly conserved between numerous species including mouse, rat, chicken, zebrafish, chimpanzee, dog, cow, and human (Havis and Duprez, 2020). EGR1 is expressed in numerous cell types and is activated transiently and rapidly by a wide array of stimuli such as growth factors, cytokines, mitogens, apoptosis, oxygen deprivation, oxidative stress, shear stress, and tissue injury (Cao et al., 1990; Ouellette et al., 1990; Hirata et al., 1998; Yan et al., 1999; Nishi et al., 2002; Pines et al., 2005; Banerji and Saroj, 2021; Khachigian, 2021). EGR1 has a broad range of physiological functions and associates with numerous cellular signaling cascades and binding partners in response to stimulation. EGR1’s primary DNA binding domain consists of three zinc finger motifs that preferentially bind to the GC-rich promoter sequence 5’-GCG(T/G)GGGCG-3′ (Christy and Nathans, 1989; Cao et al., 1990; Lemaire et al., 1990). Once induced, EGR1 is capable of transmitting activation signals downstream to influence transcriptional regulation of specific target genes required to initiate the appropriate biological response (McMahon and Monroe, 1996).
EGR1 is most notably known for its roles in the adult nervous system where it regulates critical processes underlying neuronal activity, from neurotransmission and synaptic plasticity, to higher order processes such as learning and memory, as well as reward and stress responses (Duclot and Kabbaj, 2017). EGR1 is readily expressed in various regions of the brain such as the medial prefrontal cortex, striatum, hippocampus, and amygdala and baseline expression levels of EGR1 are maintained through normal ongoing neuronal activity (Milbrandt, 1987; Herdegen et al., 1990; Mack et al., 1990; Waters et al., 1990; Aguilar et al., 2011; Duclot and Kabbaj, 2017). EGR1 is involved in the maintenance of long-term potentiation which is critical for the formation of memories, both long-term memory and for reconsolidation of memory after reactivation during retrieval (Jones et al., 2001). Altered EGR1 expression patterns have been documented in humans with neuropsychiatric disorders, like schizophrenia, where decreased EGR1 expression in the dorsal lateral prefrontal cortex is thought to contribute to patient symptoms. Furthermore, these patients also had increased EGR1 mRNA transcripts in the bloodstream (Gallo et al., 2018).
Expression of EGR1 is altered in numerous cancers where EGR1 can play paradoxical roles as either a tumor suppressor or oncogene, depending on the type of cancer and other relevant physiological circumstances. In prostate and gastric cancers, EGR1 expression is often overexpressed in the tumor as compared to surrounding tissue (Myung et al., 2013; Gabriel et al., 2016; Li et al., 2019; Ma et al., 2021). Meanwhile, EGR1 also has tumor suppressor properties through regulation of tumor suppressor genes such as transforming growth factor beta 1 (TGF1β), PTEN, and p53 (Baron et al., 2006). In gliomas and melanocytomas, EGR1 upregulates tumor suppressor gene p21Waf1/Cip1 which ultimately contributes to tumor cell apoptosis (Calogero et al., 2001; Schmidt et al., 2019). Low expression levels of EGR1 are also found in other tumor types, including breast, lung, and fibrosarcoma (Aliperti et al., 2019).
Numerous other disease states including cardiovascular disease, ischemia–reperfusion injury, acute lung injury, atopic dermatitis, and sepsis, among others, have been associated with EGR1 dysregulation and are reviewed elsewhere (Nishi et al., 2002; Ngiam et al., 2007; Chen et al., 2019; Khachigian, 2021; Yeo et al., 2021). Finally, EGR1 has been implicated to play a role in bacterial and viral infections; however, EGR1 and its role in viral infections have not previously been extensively reviewed. Thus, the purpose of this review is to elucidate the roles of EGR1 during viral infections.
EGR1 is a zinc-finger transcription factor encoded by two exons mapped to human chromosome 5. As a modular structure, EGR1 has an extended strong activation domain on the N-terminus [amino acid (aa) 1 to 281] (Figure 1). The strong activation domain is followed by an inhibitory domain which consists of about 35 amino acids (aa281 to 315). Next is the highly conserved DNA-binding domain which is comprised of three Cys2-His2 type zinc-fingers (aa338 to 418). Within the DNA-binding domain is the nuclear localization domain which also includes bipartite nuclear localization signals (aa315 to 419). Lastly, is the weak activation domain on the C-terminus (aa420 to 543; Gashler and Sukhatme, 1995).
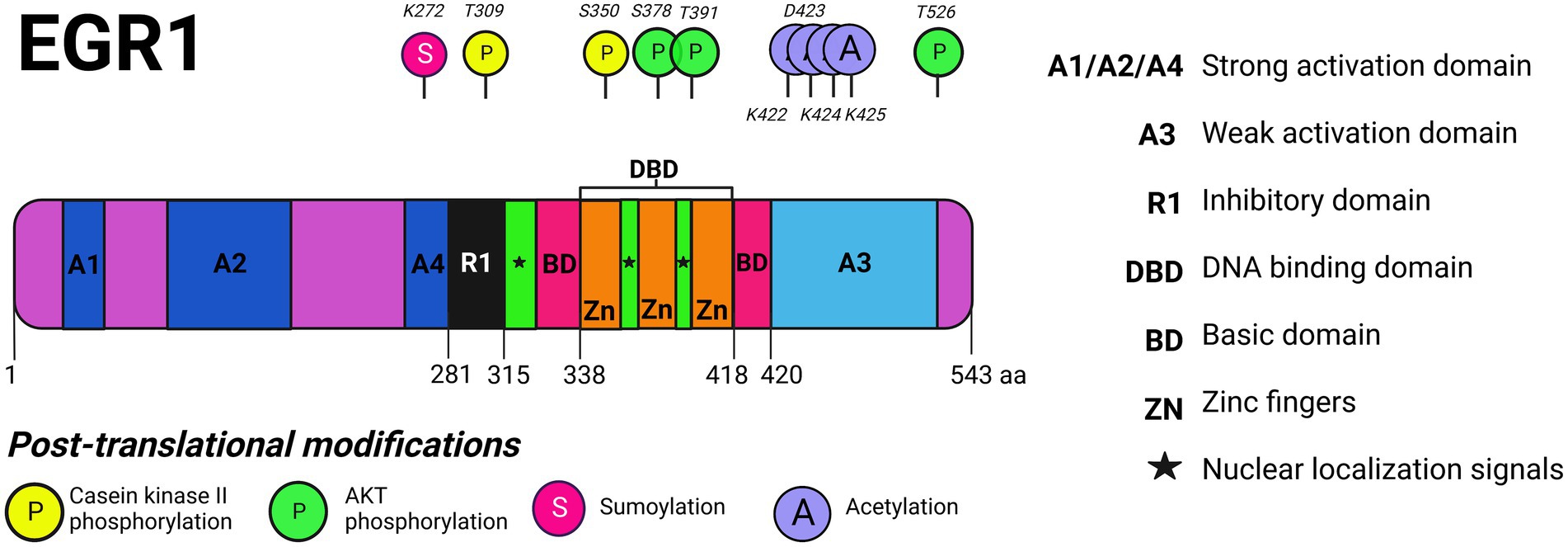
Figure 1. EGR1 protein domains and post-translational modifications. The multiple protein domains of EGR1 are depicted including its strong activation domain, weak activation domain, inhibitory domain, DNA binding domain, and basic domain as well as its zinc finger motifs and nuclear localization signals (NLS). EGR1 is phosphorylated by AKT on S378, T391, and T526 and by casein kinase II (CKII) on T309 and S350. EGR1 is sumoylated on K272 and acetylated on K422, D423, K424, and K425. Created with BioRender.com.
The inhibitory domain functions as a binding site for two transcriptional co-repressors known as NGFI-A binding proteins 1 and 2 (NAB1, NAB2) both of which are capable of suspending the biological activity of EGR1 (Russo et al., 1995; Svaren et al., 1996; Bhattacharyya et al., 2013). The prevalence of NAB1 and NAB2 in a particular cell is critical for EGR1 functionality because induction of EGR1 as a transcriptional regulator may be neutralized by NAB1 or NAB2 (Srinivasan et al., 2006). Furthermore, EGR1 directly regulates NAB2, indicating that EGR1 controls its own biological activity in a negative feedback loop via the synthesis of NAB2 (Havis and Duprez, 2020). Moreover, the binding of EGR1 to its own promoter leads to transcriptional repression or activation, depending on the cell type (Cao et al., 1993). For example, EGR1 regulation of itself leads to transcriptional activation in macrophages whereas in fibroblasts it leads to transcriptional repression (McMahon and Monroe, 1996). The C′ terminal weak activation domain is rich in proline, serine, and threonine residues with weak transactivation capabilities as compared to the strong N′ terminal activation domain which is rich in serine and threonine residues with strong transactivation properties (DeLigio and Zorio, 2009). Current literature lacks insight as to the structural differences between the strong and inhibitory binding domains, other than the N′ terminal being predominantly active as compared to the C′ terminal weak activation domain.
A major aspect of protein regulation in eukaryotic cells is the trafficking of proteins between the cytoplasm and the nucleus through the nuclear pore complex (NPC). EGR1 expression levels are poised at baseline in both the nucleus and the cytoplasm (albeit to a lesser extent) and increased expression in either location can be induced after exposure to various stimuli. Two nuclear localization signals (NLSs) have been identified for EGR1: one positioned at amino acids 315–330 and another between amino acids 406–417 (Chen et al., 2011). Another NLS, named SPS (a motif of three amino acids – Ser/Thr-Pro-Ser/Thr), directs target proteins to the nucleus through direct interaction with importin-7 (Imp-7) and EGR1 contains an SPS on the C′ terminus downstream of its zinc fingers. In response to growth factors, such as serum, EGR1 was found to form a complex with Imp-7 and Imp-7 was required for the nuclear translocation of EGR1. The SPS in EGR1 can serve as an independent NLS and stimulation may induce SPS to facilitate increased nuclear import of EGR1 to promote enhanced transcriptional activity in response to extracellular stimulation (Chen et al., 2011).
In terms of post-translational modifications, phosphorylation events of EGR1’s domains are controlled by protein kinases and phosphatases (Cao et al., 1992). There are at least five known phosphorylation sites that are regulated by casein kinase II phosphorylation (S378, T391, T526) or AKT phosphorylation (T309, S350) as depicted in Figure 1. Phosphorylation of the different EGR1 domains can either enhance or suspend the transcriptional activity of EGR1. Following UV irradiation in a fibrosarcoma model, EGR1 is phosphorylated by both protein kinase C (PKC) and tyrosine kinases and protects cells from apoptosis (Huang et al., 1998). Meanwhile, phosphorylation by casein kinase II in fibrosarcoma cells decreases the transcriptional activity of EGR1 as well as its DNA binding activity (Jain et al., 1996). In terms of acetylation, the cyclic AMP responsive element binding protein (CREB)-binding protein (CBP)/p300 complex can acetylate EGR1 within its weak activation domain (K422, D423, K424, K425; Yu et al., 2004). EGR1 can also undergo sumoylation (K272) and ubiquitination following translation which has been reviewed elsewhere (Havis and Duprez, 2020). EGR1’s functionality as an activator or repressor of target genes is dependent on its post-translational modifications which modulate its activity.
EGR1 belongs to the EGR family of C2H2-type zinc finger proteins which also include EGR2, EGR3, and EGR4 (Beckmann and Wilce, 1997). EGR family members are rapidly induced in response to environmental and cellular stimuli and are also transcriptional regulators capable of activating specialized signaling cascades. All four EGR family members contain three cystine2-histidine2 zinc fingers and are highly homologous both within and between species suggesting possible target and functional overlap (Duclot and Kabbaj, 2017). Furthermore, alignment of the DNA binding domains of the EGR family members from humans, rats, and mice suggests that differences between family members are greater within species than between species which suggests that the characteristics of each EGR member are evolutionarily conserved (Duclot and Kabbaj, 2017). However, with the exception of their shared DNA-binding domain, the sequences between the family members are much less homologous suggesting specificities in protein–protein interactions allowing for differences in gene regulation, reactivity, and transcriptional control (Poirier et al., 2008). EGR2 is most notably associated with the onset of myelination within the peripheral nervous system as well as hindbrain segmentation (Warner et al., 1998; Taefehshokr et al., 2017). Defects in EGR2 expression can clinically manifest as various forms of neuropathies such as Charcot–Marie–Tooth disease. Meanwhile, EGR3 is associated with muscle-spindle development, lymphocyte development, endothelial cell growth and migration, and neuronal development. EGR3 dysfunction has been implicated in psychiatric disorders such as schizophrenia and bipolar disorder (Pfaffenseller et al., 2018). EGR4 is a paralog of EGR2 and has been implicated in posterior hindbrain development and also plays a critical role in spermatogenesis in male murine fertility (Bae et al., 2015; Taefehshokr et al., 2017). In alignment with the other EGR family members, EGR4 defects are associated with psychiatric disorders such as schizophrenia as well as neuropathies. Outside of the nervous system, EGR1, EGR2, and EGR3 have been implicated to be critical for response to external stimuli and to direct lineage differentiation within the immune system (Sun et al., 2019).
The promoter region of the EGR1 gene contains transcription factor binding sites such as cyclic adenosine 3′, 5′-monophosphate (cAMP) response elements (CRE), an activator protein-1 (AP-1) binding site, an EGR binding site (EBS), specificity protein 1 (Sp1) elements, nuclear factor kappa B (NF-κB) binding site, and serum response elements (SREs; Figure 2). The CREs region can be occupied by members of the CREB protein family of transcription factors (Banerji and Saroj, 2021). The SREs function as binding sites for serum response factors (SRFs) and ternary complex factors (TCFs), such as Elk-1. As a bivalent regulator of EGR1, Elk-1 can induce transcription of EGR1 through recruitment of histone acetyltransferases, such as p300/CBP (Li et al., 2003), or function to repress EGR1 transcription via recruitment of histone deacetylases, such as mSin3A-HDAC (Yang et al., 2001). The EGR1 promoter also contains two CpG islands that are susceptible to DNA methylation. Promoters with methylated CpG regions generally correspond with repressed gene expression. However, due to its structural plasticity EGR1 is capable of binding to its target DNA sequence regardless of methylation state and is capable of binding to target regions that are completely methylated by adapting its conformational structure to not only recognize the target DNA but also maintain its affinity for the target sequence (Zandarashvili et al., 2015).
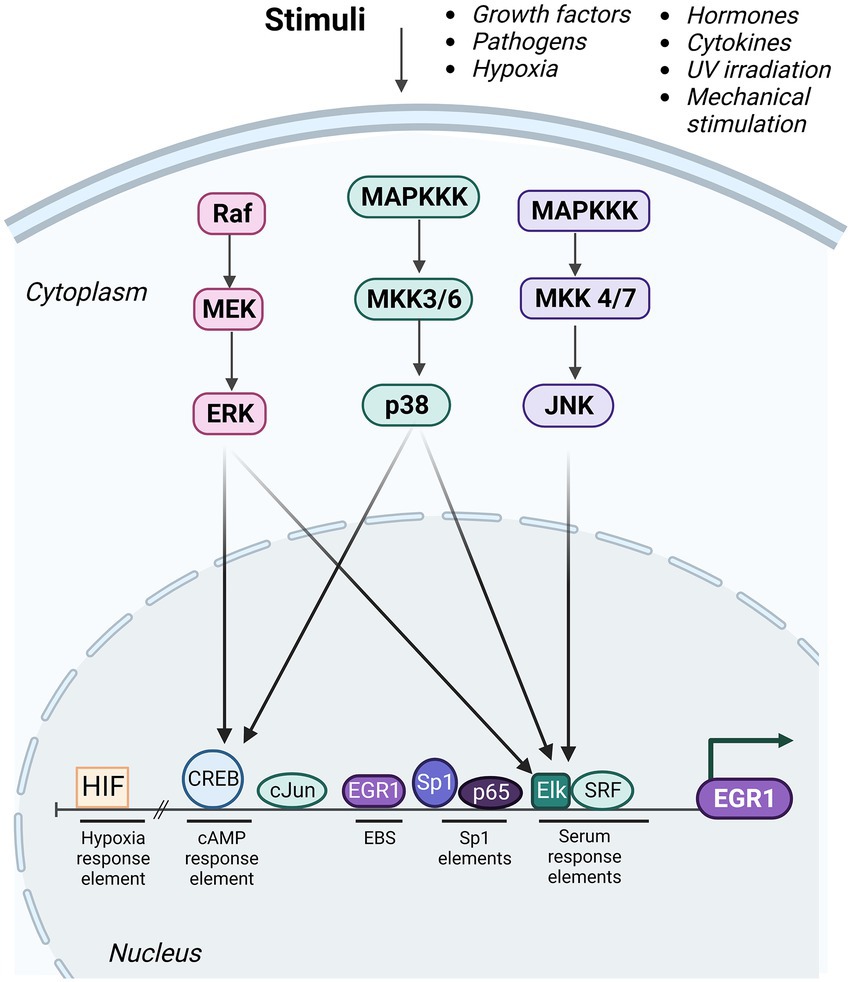
Figure 2. Upstream regulators of EGR1. The various regulatory elements of the EGR1 promoter are depicted including the hypoxic response element (HIF), cyclic adenosine 3′, 5′-monophosphate (cAMP) response elements (CRE), EGR1 binding sites (EBS), specificity protein 1 (Sp1) elements, and serum response elements (SREs). Multiple transcription factors are capable of binding to these elements to regulated EGR1 expression including HIF, cAMP response element binding protein (CREB), cJun, EGR1, Sp1, nuclear factor kappa B (NF-κB)/p65, Elk, and serum response factor (SRF). Multiple stimuli induce activation of ERK, JNK, and p38 MAPK signaling cascades, which results in activation of Elk and/or CREB transcription factors and induction of EGR1 gene expression. Created with BioRender.com.
Activation of EGR1 gene expression is induced by numerous upstream signal transduction pathways that are activated via external stimuli and subsequently these pathways direct molecules to the EGR1 promoter (de Klerk et al., 2017; Figure 2). Mitogen-activated protein kinases (MAPKs), protein kinase A (PKA), protein kinase B (PKB)/AKT, PKC, and NF-κB signaling factors have all been shown to regulate EGR1 gene expression (Havis and Duprez, 2020; Banerji and Saroj, 2021; Khachigian, 2021). The most well-characterized signal transduction cascade that impacts EGR1 transcriptional regulation is the MAPK pathway. MAPKs are ubiquitously expressed phosphorylated protein kinases that are known to regulate a variety of cellular activities such as stress response, apoptosis, and genes required for immune response induction. The MAPK pathway is part of an extensively integrated signaling network and MAPK signaling regulates many physiological responses that vary according to stimulus and cell type (Johnson, 2011). All MAPK pathways are composed of three core signaling modules including the Extracellular Signal-Related Kinase (ERK1/2), the c-Jun NH2-terminal kinase (JNK), and p38 MAPK (Kyriakis and Avruch, 2001). ERK1/2 is activated in response to growth factors, hormones, and various pro-inflammatory stimuli while cellular and environmental stressors activate JNK1/2/3 and p38 MAPK pathways (Banerji and Saroj, 2021). Previous studies have shown that EGR1 can be transcriptionally regulated via ERK-dependent phosphorylation of Elk-1, which as mentioned above can result in either target gene activation or repression, depending on the epigenetic modifications of the EGR1 promoter. Genome-wide microarray analysis revealed EGR1 as the most upregulated gene following JNK activation and c-Jun as an essential effector in EGR1 transcriptional regulation (Hoffmann et al., 2008). c-Jun directly binds the AP-1 element and is required for EGR1 promoter activation through the three distal SREs (Hoffmann et al., 2008). Activated p38 can directly induce transcription factors such as CREB to bind to its respective binding site on the EGR1 promoter and thus induce EGR1 upregulation (Bachstetter and Van Eldik, 2010).
Transcriptional regulation of EGR1 is a complex and dynamic process especially considering its many interactions between transcription factor binding, cofactor recruitment, and chromatin modifications including histone methylation, acetylation, phosphorylation, and nucleosome remodeling (Duclot and Kabbaj, 2017). Exposure to phorbol esters rapidly induces EGR1 expression within minutes but returns to baseline transcriptional levels by 180 min after exposure. Riffo-Campos et al. propose a model in which Elk1, CREB and EGR1 co-localize to rapidly induce EGR1 transcription (Riffo-Campos et al., 2015). Three components of histone deacetylase (HDAC) complexes are found on the EGR1 promoter prior to exposure to phorbol esters. However, CREB, Elk-1, SRF and RNA polymerase II can also be found at the EGR1 promoter prior to its induction thus corroborating the understanding that EGR1 is poised at baseline. This can be partially attributed to favorable nucleosome positioning. Phorbol ester exposure triggers classical nucleosome repositioning and triggers partial eviction of the +1 and − 1 nucleosomes as well as sliding of the – 2 nucleosome at 15 min post-exposure. Simultaneously, CREB and Elk1, both bound to the EGR1 promoter, are phosphorylated in a p38- and MEK1/2- dependent manner (Tur et al., 2010) which triggers an increase in phosphorylation (pS10) and acetylation (AcK14) of histone H3 at the +1 nucleosome (Riffo-Campos et al., 2015) and is likely mediated by the transcriptional co-factor CBP. Altogether, EGR1 gene regulation is dependent on a variety of factors such as binding partners bound to its promoter and upstream signaling pathways, including the MAPK cascades (Banerji and Saroj, 2021; Figure 2).
There are many potential downstream targets of EGR1 and it was initially assumed that genes containing EGR1 binding sites were thought to be directly regulated by EGR1 (Swirnoff and Milbrandt, 1995). However, experimental evidence found that EGR1 can also regulate gene expression indirectly through interacting with other transcription factors such as c/EBPβ, Fos, or Jun, thereby further expanding the range of potential EGR1 targets and biological processes regulated by EGR1 (Duclot and Kabbaj, 2017). Initially, identification of EGR1 dependent genes was investigated on a single-gene basis. The development of genomic wide screening technologies enabled the identification of EGR1 regulated genes on a global scale. However, it is important to keep in mind that many of the genes identified to be regulated by EGR1 are likely specific to the cell type and insult encountered prior to samples undergoing sequencing. Results gained from the Encyclopedia of DNA Elements (ENCODE) project identified approximately 8,552 of 15,872 genes annotated (~54%) contain at least one EGR1 binding region within 3 kb of their transcriptional start sites (Consortium EP, 2012). These studies further corroborated previous speculation that EGR1 can bind a large number of genes across several human cell types and thus exert transcriptional regulation for a wide variety of biological functions. Functional analysis of genes identified in the ENCODE project revealed enrichment of pathways and processes related to growth factor signaling and intracellular signaling cascades. The molecular functions of EGR1-bound genes were found to range from chromatin and transcription factor activity to guanyl-nucleotide exchange factor activity through serine/threonine kinase activity. Moreover, EGR1-bound target genes were localized from the chromatin to the cell membrane (Duclot and Kabbaj, 2017). Similarly, EGR1 ChIP-seq studies of mouse brains identified enrichment of biological processes and pathways related to protein trafficking, synaptic vesicle transport, endocytosis, protein phosphorylation, and intracellular signaling cascades (Koldamova et al., 2014).
Numerous tumor suppressor genes have been identified to be under the transcriptional regulation of EGR1 (Figure 3). TGFβ1, a protein critically involved in regulation of cell proliferation, differentiation, and growth, is directly regulated by EGR1 in human fibrosarcoma cells (Liu et al., 1996; De Belle et al., 2000) and monkey kidney cells (Dey et al., 1994). Furthermore, fibronectin, a downstream target of TGFβ1/Smad signaling pathway, was also found to be a direct target of EGR1, with two closely spaced EBS located within its proximal promoter (Liu et al., 2000). Phosphatase and tensin homolog (PTEN), a proapoptotic factor that is altered in numerous cancers, is also directly regulated by EGR1. In vivo studies of wild-type mice exposed to irradiation indicated a strong upregulation of EGR1 and PTEN mRNA. However, EGR1 knock-out mice exposed to irradiation did not result in an upregulation of PTEN, indicating that EGR1 directly regulates PTEN in vivo (Virolle et al., 2001). Additionally, an extensive review of clinical cases of human non-small-cell lung cancer found that EGR1 expression levels are predictive of both PTEN expression levels and survival outcome with a high degree of significance (Baron et al., 2006). EGR1 has also been identified as a regulator of p53 in both humans and mice, both of which contain EBS within their promoter regions. Loss of EGR1 in mouse embryonic fibroblasts (MEFs) suppresses p53-dependent growth regulatory functions (Krones-Herzig et al., 2003) and ChIP and EMSA assays confirmed EGR1 binding to the p53 promoter both in vitro and in vivo (Krones-Herzig et al., 2005).
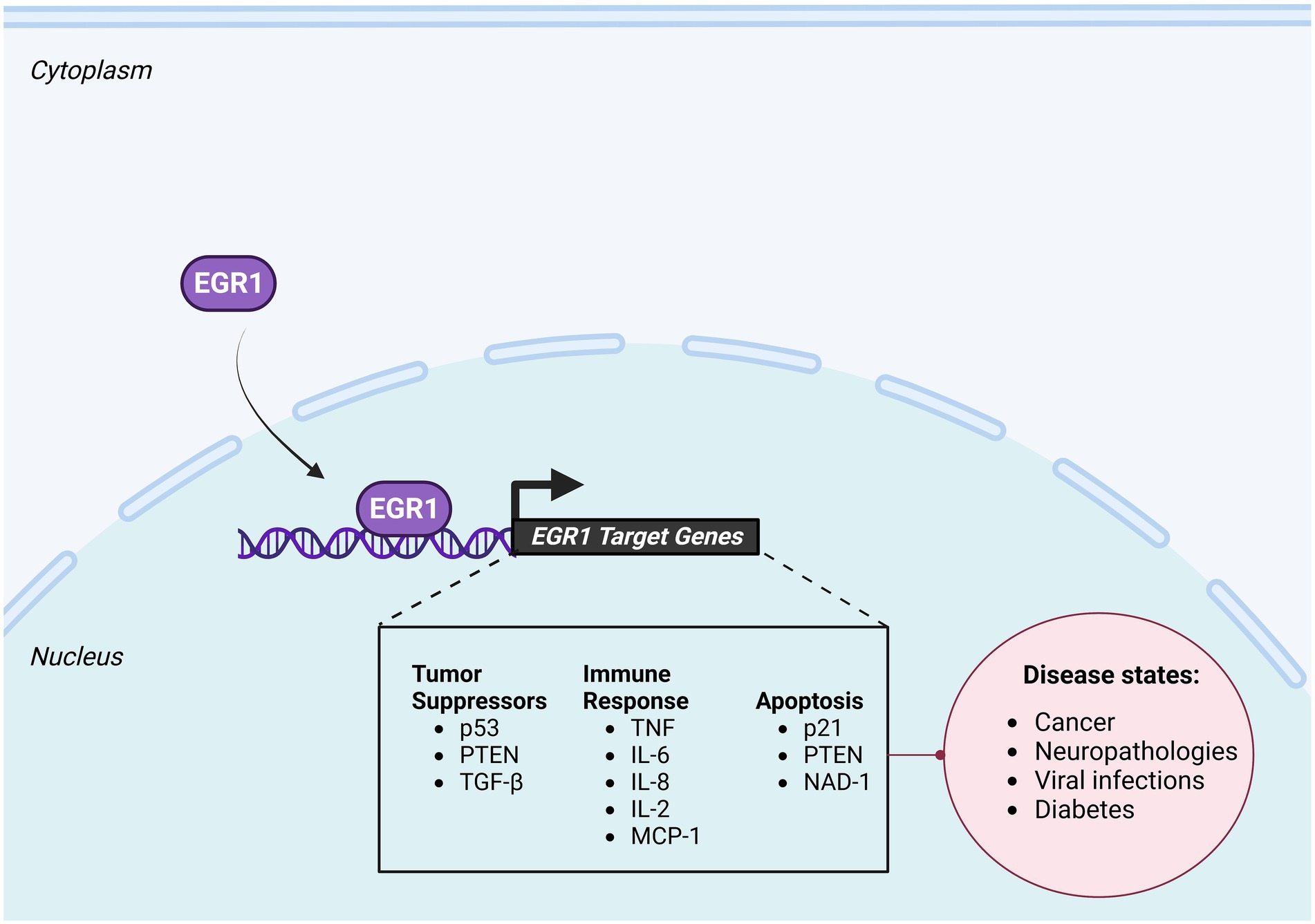
Figure 3. Downstream Targets of EGR1. EGR1 induces transcription of a large repertoire of genes, including tumor suppressors, immune response genes, and apoptotic factors. Changes in EGR1 dependent gene expression have been correlated with multiple disease states including cancer, diabetes, neuropathologies, and viral infections. Created with BioRender.com.
EGR1 is known to target immunologically significant genes, such as tumor necrosis factor (TNF; Figure 3). As a cytokine, TNF plays an important role in the early phases of the immune response by promoting inflammation. As such, the promoter region of TNF is GC-rich and contains a binding site for EGR1 (McMahon and Monroe, 1996). Additionally, previous studies in RAW 264.7 cells showed that following LPS-induced ERK1/2 activation, TNFα mRNA is upregulated in an EGR1 dependent manner and that EGR1 binds to the TNFα promoter (Shi et al., 2002). Moreover, the promoter of the inflammatory gene IL-2 was found to have an overlapping Sp1 and EBS and EGR1, in combination with NFAT, was found to upregulate the expression of IL-2 following LPS stimulation (Decker et al., 1998). In vivo ChIP analysis revealed EGR1 regulates additional proinflammatory mediators such as IL-8, IL-6, and monocyte chemoattractant protein 1 (CCL2/MCP-1; Hoffmann et al., 2008).
EGR1 has also been implicated in apoptosis (Figure 3), most notably in cancer studies where EGR1 expression is significantly reduced in developing tumors (Huang et al., 1997). Upon exposure to damaging ionizing radiation EGR1 binds to the 5’ UTR of the PTEN gene, which contains an EBS, a tumor suppressor and pro-apoptotic gene, to induce apoptosis (Virolle et al., 2001). Additionally, EGR1 was shown to activate transcription of p21Waf1/Cip1, a gene involved in DNA damage response, differentiation, senescence, and apoptosis, independently of p53 in response to curcumin treatment in human astrocytoma cells (Choi et al., 2008). Another pro-apoptotic gene, NSAID-activated gene 1 (NAD-1), is regulated by EGR1 and has been shown to facilitate apoptosis in colon carcinoma cells, lung cancer cells, and hepatocellular carcinoma cells (Chen et al., 2007; Shin et al., 2008).
Host-pathogen interactions are dynamic and complex. Throughout their co-evolution, hosts have acquired ways to overcome viral infection and likewise viruses have developed countermeasures to overcome the host’s antiviral cellular state. Viruses, with their relatively compact genomes and high replication rates, are prone to mutations and thus continuously present the host with ever-evolving challenges. To combat the host antiviral response, viruses encode mechanisms to control interferon signaling as well as disrupt cross-talk between cellular pathways that are known to induce apoptosis and inflammation. By utilizing numerous evasion strategies viruses are also able to subvert host cellular processes such as cell cycle regulation, major histocompatibility complex restricted antigen presentation, intracellular protein transport, apoptosis, interferon signaling, and humoral immune responses (Fan et al., 2018). A primary evasion strategy by viruses is direct interaction of viral proteins with host proteins to inhibit their functionality and by doing this the virus can hijack the cells biosynthetic machinery in order to enhance viral replication in a variety of ways such as inducing posttranslational modifications and interfering with host restriction factors to regulate their function and/or degradation. Emergence of functional genomic, proteomic, and bioinformatic technologies has allowed for more-thorough studies to aid in the quest of identifying host factors critical for viral replication and potential novel targets that can be exploited for therapeutic development. EGR1 has been associated with multiple viral infections such as VEEV (Venezuelan equine encephalitis virus), KSHV (Kaposi’s sarcoma-associated herpesvirus), HSV-1 (herpes simplex 1), JCV (human polyomavirus JC virus), HIV (human immunodeficiency virus) and EBV (Epstein–Barr virus). During viral infections, EGR1 expression can facilitate either an anti-viral or pro-viral cellular state, depending on the virus and the transcriptional events regulated by EGR1. This information is summarized in Tables 1, 2, including the cell types where the findings were discovered.
JC virus (JCV), also known as human polyomavirus 2, is the causative agent of the fatal demyelinating disease known as progressive multifocal leukoencephalopathy (PML). EGR1 is upregulated in human astrocytoma cells infected with JCV and binds to the JCV late promoter to enhance viral transcription (Romagnoli et al., 2008). Moreover, mutation of the EBS impaired the ability of EGR1 to bind to the viral promoter and resulted in significant reduction in late gene expression and DNA replication during the course of infection (Romagnoli et al., 2008). Importantly, EGR1 was found to be upregulated in PML clinical samples and immunohistochemistry staining revealed the strong presence of EGR1 in the nuclei of oligodendrocytes, the primary site of JCV infection and replication (Romagnoli et al., 2008). These results suggest that EGR1 induction following infection with JCV may play an important role in JCV replication and PML pathogenesis.
EGR1 upregulation has been documented in several herpesviruses including HSV-1, bovine herpesvirus 1 (BHV-1), KSHV, EBV, and human cytomegalovirus (CMV). HSV-1 is a neurotropic virus and initial infection, which is usually mild or asymptomatic in nature, typically occurs during childhood. HSV-1 infections of epithelial cells initiates lytic replication upon which virions infect neurons in the trigeminal ganglia ultimately resulting in lifelong latency (Taylor et al., 2002). An EBS was previously found in the intron of HSV-1 viral gene ICP22 and EGR1 was found to regulate viral genes ICP4 and ICP22 (Bedadala et al., 2007). More recently, EGR1 has been shown to be upregulated at both the mRNA and protein level following infection with HSV-1 in Vero cells (Bedadala et al., 2011). Importantly, EGR1 was shown to enhance viral gene expression, viral replication, and release of infectious particles. Mechanistic studies involving ChIP assays identified NF-κB and phosphorylated CREB (pCREB) as being bound to the EGR1 promoter following HSV-1 infection in rabbit corneal (SIRC) cells, but not HEK-293 T cells, thus suggesting NFκB and pCREB may play a role in regulating EGR1 upregulation during HSV-1 infections (Bedadala et al., 2011). Clinical infections of HSV-1 can progress to herpes encephalitis and induce permanent neurological damage. Given the many roles EGR1 plays within the brain in terms of neuronal plasticity as well as memory and learning, it would be interesting to further examine the role of EGR1 in the development of HSV-1 induced neurological sequalae.
BHV-1 is considered an economically important pathogen to the cattle industry worldwide. Clinical infections of BHV-1 manifest in a variety of pathologies including respiratory symptoms, gastrointestinal symptoms, genital disorders, and abortions. RNA sequencing of BHV-1 infected MDBK cells identified EGR1 as being differentially expressed and upregulated following infection and EGR1 induction was confirmed at both the mRNA and protein level (Hou et al., 2019). Importantly, EGR1 was found to enhance viral replication in BHV-1 infected MDBK cells, thus suggesting EGR1 plays a pro-viral role during BHV-1 infections. Additionally, EGR1 was found to bind to an EBS within the promoter region of the viral gene UL46 (Hou et al., 2019), but not ICP22, which differs from previous findings that EGR1 binds and regulates ICP22 in HSV-1, however this is not entirely unexpected considering they are different viruses with distinct differences in tropism and biology (Bedadala et al., 2011). Moreover, EGR1 was found to be a direct target of cellular micro-RNA bta-miR-2,361, which was also found to inhibit viral replication through downregulation of EGR1 (Hou et al., 2019). These results suggest a potential therapeutic role for bta-miR-2,361 in mitigating BHV-1 infections by directly targeting EGR1.
EGR1 has also been found to regulate the replication and transcription activator (RTA) gene, which regulates the switch from latent to lytic replication during KSHV infection (Dyson et al., 2010, 2012). Loss of EGR1 via small interfering RNA (siRNA) resulted in decreased virion production following lytic reactivation (Sarkar and Verma, 2017) suggesting a pro-viral role for EGR1 during KSHV infections. Previous studies suggested a role for CBP/p300 in modulating the RTA promoter activity (Gwack et al., 2001) and since EGR1 has a specific binding site for CBP/p300 it was speculated that EGR1 and CBP/p300 could be working cooperatively to regulate RTA activity. EGR1 was not essential to activate the RTA promoter however, expression of EGR1 significantly enhanced activation of the RTA promoter thus suggesting that EGR1 can regulate the transcriptional activities of CBP/p300 (Sarkar and Verma, 2017). These results suggest a potential role for EGR1 as a key regulator for KSHV lytic reactivation. In line with these results, there are also studies suggesting that EGR1 plays a role in regulating Epstein–Barr virus (EBV) lytic cycle activation (Chang et al., 2006; Vockerodt et al., 2013). EBV latent membrane protein 2A (LMP2A) maintains EBV latency (Miller et al., 1994, 1995) and EGR1 is significantly upregulated in primary human germinal center B cells transfected with LMP2A (Fujii et al., 1991). However, EGR1 upregulation was not observed following LMP2A expression in Hodgkin’s lymphoma cells lacking a functional B cell receptor, which is correlated with a lack of viral lytic replication in these cells (Vockerodt et al., 2013). Expression of EBV latent membrane protein 1 (LMP1), which is required for EBV B cell immortalization (Young and Rickinson, 2004), also induces EGR1 upregulation in a NF-κB dependent manner (Kim et al., 2007). EGR1 is upregulated in tumor samples from patients with extranodal natural killer T-cell lymphoma (ENKTL), which is an aggressive cancer associated with EBV infection (Lee et al., 2021). EGR1 expression was found to have prognostic value, with EGR1 being upregulated in the ENKTL low risk group. Moreover, EGR1 has been shown to regulate UL138, the latency gene, in CMV infections. CMV interferes with host cell signaling and downregulates epidermal growth factor receptor (EGFR) and its downstream pathways during early stages of infection in order to establish latency (Kim et al., 2017). During CMV infection, EGR1 is induced downstream of EGFR signaling via the MEK/ERK pathway (Buehler et al., 2016). EGR1 binds to the CMV latency gene, UL138, which stimulates its expression and consequently reinforces a latent infection state. CMV mutant viruses lacking EGR1 binding sites fail to establish latency. Thus, antagonizing the EGFR/EGR1 signaling enables CMV to maintain a productive infection (Buehler et al., 2019) and suggests EGR1 serves a pro-viral role during CMV infections.
Infection with vaccinia virus (VACV), a member of the Poxviridae family, induces MAPK pathway activation which subsequently activates EGR1. Activation of MEK/ERK/EGR1 pathway enhances VACV replication in certain cell lines, including fibroblasts. Loss of EGR1 via siRNA targeting EGR1 or knock-out cells results in a ~ 1 log10 reduction of VACV replication in serum starved cells (de Oliveira et al., 2018). Furthermore, virions derived from infected EGR1 null MEFs were shown to have reduced infectivity as compared to virions derived from infected wild-type MEFs. Taken together, these results suggest a pro-viral role for EGR1 during VACV infection.
Enterovirus 71 (EV71) infections in young children can manifest clinically from asymptomatic, to mild hand, foot and mouth disease, to severe neurological disease. EGR1 is upregulated following infection with EV71 in RD and SK-N-SH cells and was found to facilitate viral replication (Song et al., 2015). Moreover, EGR1 directly interacts with the 5’UTR of EV71 in the cytoplasm of infected cells to facilitate viral replication in a miR-141-independent manner. Previous studies suggested that EGR1 facilitates EV71 virus production through activation of miR-141 and suppression of eIF4E (Ho et al., 2011). However, Song et al. identified an additional mechanism in that EGR1 binds to both the cloverleaf and step-loop IV secondary structures of the EV71 IRES, which contribute to viral replication and viral protein translation, respectively (Song et al., 2015). Thus, EGR1 seemingly plays a multi-functional pro-viral role in EV71 infections.
Conversely, EGR1 appears to have an antiviral effect during other picornavirus infections. EGR1 was found to be activated irrespective of IFN-β treatment following infection with foot and mouth disease virus (FMDV) in PK-15 and HEK-293 T cells (Zhu et al., 2018). Knockdown of EGR1 via siRNA considerably enhanced expression of the viral protein VP1, a protein involved in humoral immune response and the regulation of interferon production (Zhu et al., 2018; Peng et al., 2020). Moreover, loss of EGR1 resulted in increased viral mRNA which correlated with an increase in viral titers, thus suggesting an antiviral role for EGR1 during FMDV infection (Zhu et al., 2018). Mechanistic studies determined that EGR1 enhanced type I IFN signaling and suppressed viral replication. Moreover, EGR1 enhanced type I IFN signaling and suppressed viral replication in Seneca Valley virus (SVV), another picornavirus, which further corroborates an antiviral role for EGR1 for these viruses under these conditions (Zhu et al., 2018).
In the early 1990’s, EGR1 was found to be upregulated in human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2 infected T cells (Mallett et al., 2012). Further, the viral protein Tax was found to modulate cellular gene expression via CREB/activating transcription factors (ATF)-, SRF- and NF-κB-associated pathways (Matsuoka and Jeang, 2007). More recent studies have shown that HTLV-1 infection, and Tax more specifically, upregulates EGR1 expression in infected T-cells (Huang et al., 2017). Moreover, using a luciferase reporter plasmid system, it was shown that both the SRE element and NF-κB binding sites within the EGR1 promoter are essential for regulating EGR1 transcription and Tax directly binds to the NF-κB binding site within the EGR1 promoter (Huang et al., 2017). Consequently, HTLV-1 infection and dysregulation of EGR1 in infected cells was found to be regulated by Tax-induced persistent activation of the NF-κB pathway (Huang et al., 2017). Following Tax binding to the NF-κB binding site within the EGR1 promoter, upregulation of EGR1 results in NF-κB activation, which enhances EGR1 transcription via a positive feedback loop. The establishment of this positive feedback loop further enhances Tax-induced constitutive NF-κB activation in HTLV-1 infected cells, promoting T cell transformation and ultimately adult T-cell leukemia/lymphoma pathogenesis (Huang et al., 2017).
EGR1 is also upregulated in human immunodeficiency virus-1 (HIV-1) infected U937 monocytic cells (Dron et al., 1999). A common therapeutic strategy in treating HIV-1 infections is to “kick and kill” whereby latency reversal agonists, including protein kinase C agonists (PKCa), are employed to reactivate latent HIV-1 reservoirs so that they can be cleared by viral cytopathic effect or immune-mediated clearance. Using a Jurkat T-cell model of HIV latency, EGR1 was found to be robustly upregulated following treatment with various PKCa and a linear regression analysis of EGR1 induction compared to HIV reactivation revealed that EGR1 is highly correlated with PKCa induced HIV-1 reactivation (Wong et al., 2022). Moreover, EGR1 was shown to directly interact with the HIV-1 promoter to induce HIV-1 transcription and/or latency reversal, however further investigation is required to tease out the underlying molecular mechanisms (Wong et al., 2022). Altogether, these studies suggest a pro-viral role for EGR1 in retroviral infections which makes EGR1 an attractive target to exploit in terms of therapeutic development.
EGR1 appears to play paradoxical roles when it comes to coronavirus infections. There are numerous studies involving coronaviruses where activated EGR1 seemingly plays a pro-viral role. Mouse hepatitis virus (MHV) is a coronavirus capable of inducing significant damage to the CNS and is often used as an animal model for modeling multiple sclerosis in humans due to the similarities in demyelination pathology (Lampert et al., 1973; Herndon et al., 1977). EGR1 is upregulated following MHV infection of DBT cells which are mouse primary astrocyte cells (Cai et al., 2006). EGR1 was found to be activated via ERK1/2 signaling and loss of EGR1 via siRNA resulted in a significant decrease in viral replication (~2 log10) suggesting EGR1 may play a role in viral replication (Cai et al., 2006). More recent studies found EGR1 mRNA and protein levels are upregulated following infection with infectious bronchitis virus (IBV) in H1299, Vero, and DF1 cells (Yuan et al., 2022). Other coronaviruses examined, including porcine epidemic diarrhea virus (PEDV), human coronavirus (HCoV)-229E, and HCoV-OC43, were also found to activate EGR1 at both the mRNA and protein level following infection in both H1299 and Vero cells (Yuan et al., 2022). Following siRNA-mediated knock-down of EGR1, it was shown that loss of EGR1 has no significant impact on IBV replication but knock-down cells had significantly higher percentages of PARP cleavage, an indicator of cell death, which suggests that EGR1 may function as a survival factor during IBV infection of H1299 cells (Yuan et al., 2022). Knock-down of EGR1 resulted in slightly, but not significantly, reduced replication of PEDV and increased PARP cleavage, similar to results observed with IBV. Moreover, the effects of EGR1-knockdown on the expression of cFos and cJun, both AP-1 family members, was also examined and loss of EGR1 resulted in drastically reduced induction of both cFos and cJun, albeit more so at later timepoints (Yuan et al., 2022). The observation that IBV infection induces a mutual regulatory effect on both EGR1 and AP-1 genes propelled the authors to determine the upstream kinases that potentially regulate EGR1 expression (Yuan et al., 2022). To this end, small molecule inhibitors targeting ERK1/2, JNK, or p38 pathways were utilized and results indicated that ERK1/2 may function as an upstream regulator of EGR1 during IBV, PEDV, and HCoV-229E coronavirus infections (Yuan et al., 2022). Overall, these results suggest that EGR1 may serve a pro-viral function during coronavirus infections through suppression of the host antiviral response and potentially as a survival factor in virally induced apoptosis in PEDV and IBV infections.
EGR1 has been implicated in multiple pandemic viruses including SARS-CoV and SARS-CoV-2. Studies have also shown that EGR1 upregulates TGF-β1 via activation of ROS/p38 MAPK/STAT3 pathway in human lung epithelial cells and mouse models infected with SARS-CoV (Li et al., 2016). Additionally, EGR1 is dysregulated in COVID-19 patient lung samples as identified via transcriptomic and bioinformatic analysis (Islam and Khan, 2020; Alexander et al., 2021; Xie et al., 2021).
Although, EGR1 has been shown to act in a proviral capacity for multiple coronaviruses (discussed above), there is also evidence that EGR1 has an antiviral role during HCoV-229E and PEDV infections. Yuan et al. found that EGR1 was upregulated following infection of H1299 cells with HCoV-229E. Knockdown of EGR1 in H1299 cells followed by infection with HCoV-229E resulted in significantly reduced VP-1 protein, viral mRNA, and PARP cleavage (Yuan et al., 2022). Additionally, in the section above the pro-viral role for EGR1 in PEDV infections was described. However, there appears to be conflicting evidence as to the role of EGR1 in these infections, some of these differences are likely due to the cells and conditions under which the role of EGR1 was examined. Wang et al. recently described an antiviral role for EGR1 following PEDV infection of LLC-PK1 cells, which are normal pig kidney cells and represent a more physiologically relevant model for studying PEDV. EGR1 was found to be upregulated at both the mRNA and protein level following infection with PEDV in LLC-PK1 cells (Wang et al., 2021). Furthermore, overexpression of EGR1 significantly decreased PEDV N protein as well as viral mRNA in Vero cells and these results were further confirmed using siRNA targeting EGR1 in which similar results were achieved (Wang et al., 2021). Additionally, mechanistic experiments involving ChIP assays revealed that EGR1 inhibits PEDV replication through direct binding of EGR1 to the IFN-regulated antiviral (IRAV) promoter to induce IRAV expression (Wang et al., 2021). Moreover, siRNA mediated knockdown of IRAV increased viral replication and microscopy studies revealed that IRAV colocalizes with PEDV N protein to facilitate its subsequent degradation and IRAV was found to be essential for EGR1 suppression of PEDV replication (Wang et al., 2021). Altogether, these data provide a more robust argument for EGR1 functioning in an antiviral capacity during PEDV infections.
Viral and host transcriptomic studies identified EGR1 as being highly upregulated after infection with encephalitic Venezuelan equine encephalitis virus (VEEV) in U87MG cells, a human astrocytoma cell line (Baer et al., 2016). Next generation RNA sequencing (RNA-Seq) data was leveraged to elucidate clinically relevant alterations in the mRNA transcriptome of VEEV infected astrocytes. EGR1 was found to be differentially expressed after VEEV infection and results suggested that EGR1 may serve as a potential link between the innate immune response and unfolded protein response (UPR) pathways, two pathways intimately associated with apoptosis. Interestingly, loss of EGR1 inhibited VEEV-induced apoptosis but did not alter viral replication kinetics. Neuronal cell death is a hallmark of VEEV infection and VEEV induced neurological sequalae is common in survivors (Ronca et al., 2016). Additional studies indicated that EGR1 expression is upregulated in primary human astrocytes during VEEV infection and that EGR1 induction is dependent on ERK1/2 and protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK; Dahal et al., 2020). More recent studies identified EGR1 dependent genes associated with inflammation, apoptosis, and/or encephalitis in VEEV infected astrocytes including ATF3, FOS, JUN, KLF4, EGR2, and EGR4 transcription factors and CXCL3, CXCL8, CXCL10, TNF, and prostaglandin synthase 2 (PTGS2) inflammatory mediators. Targeting of PTGS2 with the small molecule inhibitor celecoxib resulted in increased cell viability as compared to vehicle-treated infected cells (Lehman et al., 2022). Interestingly, EGR1 gene expression is also induced in U87MG cells infected with other alphaviruses, including eastern equine encephalitis virus (EEEV), chikungunya virus (CHIKV), and Sindbis virus (SINV; Lehman et al., 2022). EEEV infected cells displayed EGR1 dependent gene expression changes of ATF3, JUN, CXCL3, CXCL8, CXCL10, TNF, and PTGS2, similar to the results observed with VEEV. Additional studies are needed to further elucidate the underlying molecular mechanisms as to how EGR1 contributes to neuronal cell death following infection with VEEV.
EGR1 was identified as an activated host gene in ex vivo mouse brain samples from mice that were infected with Japanese encephalitis virus (JEV; Saha and Rangarajan, 2003). Following infection of astrocytes with Zika virus (ZIKV), a flavivirus associated with microcephaly and Guillain–Barré syndrome (Musso et al., 2019), EGR1 mRNA is upregulated approximately 2-fold (Lehman et al., 2022). Loss of function studies indicated that EGR1 at least partially regulates the expression of transcription factor KLF4 and inflammatory mediator TNF-α following ZIKV infection of astrocytes. However, to date, the full functional significance of EGR1 in JEV or ZIKV infections has yet to be determined.
Rift Valley fever virus (RVFV) is a hemorrhagic fever virus known to induce significant hepatitis and encephalitis (Gaudreault et al., 2019). Infection of astrocytes with RVFV induced a robust induction of EGR1 mRNA (~70-fold over mock; Lehman et al., 2022). Further, loss of function studies indicated that EGR1 at least partially regulates the transcription of KLF4 and CXCL10 (Lehman et al., 2022). Given the sex differences in EGR1 expression and the abortion storms as a result of RVFV infection, it would be interesting to investigate if EGR1 plays any role in the manifestation of that disease phenotype.
As reviewed above, induction of EGR1 gene expression is a common theme amongst a wide variety of viruses, spanning from large DNA viruses to small RNA viruses. This is likely due to dsRNA and multiple cytokines, including IFNα, INFβ, IFNλ, and TNFα, being capable of increasing EGR1 transcription (Ramana and Das, 2020). However, there is a dearth of information on the impact of EGR1 on viral pathogenesis in vivo. This is somewhat surprising given the availability of EGR1 knockout mice (Lee et al., 1995), which have been used to elucidate the importance of EGR1 for other infectious diseases, including bacterial infections (Pang et al., 2019). Recent studies using Assay for Transposase-Accessible Chromatin (ATAC) sequencing have characterized EGR1 as a transcriptional regulator of inflammatory genes, suppressing the inflammatory response of macrophages (Trizzino et al., 2021). Complete loss of EGR1 using knockout mouse models has its limitations, and conditional knockout mice and/or therapeutics targeting EGR1 should be used as tools to more fully understand the impact of EGR1 on viral pathogenesis. To date there are no FDA approved small molecule inhibitors of EGR1 and only one research grade small molecule inhibitor of EGR1, AB1711, has been developed which blocks the ability of EGR1 to bind DNA (Yeo et al., 2021). Treatment of mice with AB1711 was able to prevent skin inflammation in a mouse model of atopic dermatitis. Future work should focus on elucidating the impact of EGR1 on viral pathogenesis especially as it relates to immune modulation and inflammatory processes.
Given the role of EGR1 in learning and memory (Duclot and Kabbaj, 2017; Gallo et al., 2018), the potential impacts of EGR1 on the development of neurological sequelae following viral infections is especially of interest. Particularly, the capability of EGR1 binding to methylated DNA and then mediating TET1 recruitment to demethylate target sequences may result in long-term epigenetic changes and altered cellular functions (Sun et al., 2019). Since epigenetic changes are reversible, it would be extremely valuable to the scientific community to precisely define the epigenetic aberrations associated with viral infections and further explore them as potential therapeutic targets. However, at the cellular level, biological systems are heterogeneous and viral infections may induce cell-type specific responses in individual cells. Growing evidence indicates that EGR1 may have distinct sets of target genes across tissues and thus play diverse roles in different types of cells. Although we have confirmed that EGR1 is able to bind to methylated DNA, it remains unclear the prerequisite(s) for EGR1 to achieve cell-type specific bindings. Mechanistic studies will aid in determining the causal links between EGR1 binding, histone modifications, and chromatin accessibility. Over the past decade, rapid development of single cell techniques have enabled the generation of multi-omics data simultaneously. In addition, recent advances in spatial sequencing technology enables obtainment of a high-resolution view of multicellular responses to viral infections in a three-dimensional manner without the loss of spatial information. Future single-cell plus spatial sequencing technologies will provide a better understanding of EGR1 functions within the highly complex tissue of the brain in response to viral infections and unlock novel EGR1-dependent epigenetic markers as therapeutic targets for drug development.
CW and KK-H: conceptualization and writing—review and editing. CW: writing—original draft preparation. KK-H: funding acquisition. All authors contributed to the article and approved the submitted version.
This work was funded through the Defense Threat Reduction Agency (DTRA) grant HDTRA1-21-1-0008 to KK-H: Funders do not have any role in the design of the study and collection, analysis, and interpretation of data and nor in writing the manuscript.
We thank Hehuang Xie and Cody Swilley, both from Virginia Tech, for helpful discussions, insights, and editing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilar, P. V., Estrada-Franco, J. G., Navarro-Lopez, R., Ferro, C., Haddow, A. D., and Weaver, S. C. (2011). Endemic Venezuelan equine encephalitis in the Americas: hidden under the dengue umbrella. Future Virol. 6, 721–740. doi: 10.2217/fvl.11.50
Alexander, M. R., Brice, A. M., Jansen van Vuren, P., Rootes, C. L., Tribolet, L., Cowled, C., et al. (2021). Ribosome-profiling reveals restricted Post transcriptional expression of antiviral cytokines and transcription factors during SARS-CoV-2 infection. Int. J. Mol. Sci. 22:3392. doi: 10.3390/ijms22073392
Aliperti, V., Sgueglia, G., Aniello, F., Vitale, E., Fucci, L., and Donizetti, A. (2019). Identification, characterization, and regulatory mechanisms of a novel EGR1 splicing isoform. Int. J. Mol. Sci. 20. doi: 10.3390/ijms20071548
Bachstetter, A. D., and Van Eldik, L. J. (2010). The p38 MAP kinase family as regulators of Proinflammatory cytokine production in degenerative diseases of the CNS. Aging Dis. 1, 199–211.
Bae, C. J., Jeong, J., and Saint-Jeannet, J. P. (2015). A novel function for Egr4 in posterior hindbrain development. Sci. Rep. 5:7750. doi: 10.1038/srep07750
Baer, A., Lundberg, L., Swales, D., Waybright, N., Pinkham, C., Dinman, J. D., et al. (2016). Venezuelan equine encephalitis virus induces apoptosis through the unfolded protein response activation of EGR1. J. Virol. 90, 3558–3572. doi: 10.1128/JVI.02827-15
Banerji, R., and Saroj, S. D. (2021). Early growth response 1 (EGR1) activation in initial stages of host-pathogen interactions. Mol. Biol. Rep. 48, 2935–2943. doi: 10.1007/s11033-021-06305-0
Baron, V., Adamson, E. D., Calogero, A., Ragona, G., and Mercola, D. (2006). The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 13, 115–124. doi: 10.1038/sj.cgt.7700896
Beckmann, A. M., and Wilce, P. A. (1997). Egr transcription factors in the nervous system. Neurochem. Int. 31, 477–510; discussion 7-6. doi: 10.1016/S0197-0186(96)00136-2
Bedadala, G. R., Palem, J. R., Graham, L., Hill, J. M., McFerrin, H. E., and Hsia, S. C. (2011). Lytic HSV-1 infection induces the multifunctional transcription factor early growth Response-1 (EGR-1) in rabbit corneal cells. Virol. J. 8:262. doi: 10.1186/1743-422X-8-262
Bedadala, G. R., Pinnoji, R. C., and Hsia, S. C. (2007). Early growth response gene 1 (Egr-1) regulates HSV-1 ICP4 and ICP22 gene expression. Cell Res. 17, 546–555. doi: 10.1038/cr.2007.44
Bhattacharyya, S., Fang, F., Tourtellotte, W., and Varga, J. (2013). Egr-1: new conductor for the tissue repair orchestra directs harmony (regeneration) or cacophony (fibrosis). J. Pathol. 229, 286–297. doi: 10.1002/path.4131
Buehler, J., Carpenter, E., Zeltzer, S., Igarashi, S., Rak, M., Mikell, I., et al. (2019). Host signaling and EGR1 transcriptional control of human cytomegalovirus replication and latency. PLoS Pathog. 15:e1008037. doi: 10.1371/journal.ppat.1008037
Buehler, J., Zeltzer, S., Reitsma, J., Petrucelli, A., Umashankar, M., Rak, M., et al. (2016). Opposing regulation of the EGF receptor: a molecular switch controlling cytomegalovirus latency and replication. PLoS Pathog. 12:e1005655. doi: 10.1371/journal.ppat.1005655
Cai, Y., Liu, Y., and Zhang, X. (2006). Induction of transcription factor Egr-1 gene expression in astrocytoma cells by murine coronavirus infection. Virology 355, 152–163. doi: 10.1016/j.virol.2006.07.012
Calogero, A., Arcella, A., De Gregorio, G., Porcellini, A., Mercola, D., Liu, C., et al. (2001). The early growth response gene EGR-1 behaves as a suppressor gene that is down-regulated independent of ARF/Mdm2 but not p53 alterations in fresh human gliomas. Clin. Cancer Res. 7, 2788–2796.
Cao, X. M., Koski, R. A., Gashler, A., McKiernan, M., Morris, C. F., Gaffney, R., et al. (1990). Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol. Cell. Biol. 10, 1931–1939.
Cao, X., Mahendran, R., Guy, G. R., and Tan, Y. H. (1992). Protein phosphatase inhibitors induce the sustained expression of the Egr-1 gene and the hyperphosphorylation of its gene product. J. Biol. Chem. 267, 12991–12997. doi: 10.1016/S0021-9258(18)42372-1
Cao, X., Mahendran, R., Guy, G. R., and Tan, Y. H. (1993). Detection and characterization of cellular EGR-1 binding to its recognition site. J. Biol. Chem. 268, 16949–16957. doi: 10.1016/S0021-9258(19)85286-9
Chang, Y., Lee, H. H., Chen, Y. T., Lu, J., Wu, S. Y., Chen, C. W., et al. (2006). Induction of the early growth response 1 gene by Epstein-Barr virus lytic transactivator Zta. J. Virol. 80, 7748–7755. doi: 10.1128/JVI.02608-05
Chen, Y. L., Lin, P. C., Chen, S. P., Lin, C. C., Tsai, N. M., Cheng, Y. L., et al. (2007). Activation of nonsteroidal anti-inflammatory drug-activated gene-1 via extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase revealed a isochaihulactone-triggered apoptotic pathway in human lung cancer A549 cells. J. Pharmacol. Exp. Ther. 323, 746–756. doi: 10.1124/jpet.107.126193
Chen, J., Liu, M. Y., Parish, C. R., Chong, B. H., and Khachigian, L. (2011). Nuclear import of early growth response-1 involves importin-7 and the novel nuclear localization signal serine-proline-serine. Int. J. Biochem. Cell Biol. 43, 905–912. doi: 10.1016/j.biocel.2011.03.004
Chen, F., Wang, Y., Zhang, W., Cai, Y., Zhao, T., Mai, H., et al. (2019). A functional polymorphism-mediated disruption of EGR1/ADAM10 pathway confers the risk of sepsis progression. MBio 10. doi: 10.1128/mBio.01663-19
Choi, B. H., Kim, C. G., Bae, Y. S., Lim, Y., Lee, Y. H., and Shin, S. Y. (2008). p21 Waf1/Cip1 expression by curcumin in U-87MG human glioma cells: role of early growth response-1 expression. Cancer Res. 68, 1369–1377. doi: 10.1158/0008-5472.CAN-07-5222
Christy, B. A., Lau, L. F., and Nathans, D. (1988). A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with "zinc finger" sequences. Proc. Natl. Acad. Sci. U. S. A. 85, 7857–7861.
Christy, B., and Nathans, D. (1989). DNA binding site of the growth factor-inducible protein Zif268. Proc. Natl. Acad. Sci. U. S. A. 86, 8737–8741. doi: 10.1073/pnas.86.22.8737
Consortium EP (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. doi: 10.1038/nature11247
Dahal, B., Lin, S. C., Carey, B. D., Jacobs, J. L., Dinman, J. D., van Hoek, M. L., et al. (2020). EGR1 upregulation following Venezuelan equine encephalitis virus infection is regulated by ERK and PERK pathways contributing to cell death. Virology 539, 121–128. doi: 10.1016/j.virol.2019.10.016
De Belle, I., Mercola, D., and Adamson, E. D. (2000). Method for cloning in vivo targets of the Egr-1 transcription factor. BioTechniques 29, 162–169. doi: 10.2144/00291rr03
de Klerk, N., Saroj, S. D., Wassing, G. M., Maudsdotter, L., and Jonsson, A. B. (2017). The host cell transcription factor EGR1 is induced by bacteria through the EGFR-ERK1/2 pathway. Front. Cell. Infect. Microbiol. 7:16. doi: 10.3389/fcimb.2017.00016
de Oliveira, L. C., Brasil, B., Unger, B., Trindade, G. S., Abrahao, J. S., Kroon, E. G., et al. (2018). The host factor early growth response gene (EGR-1) regulates Vaccinia virus infectivity during infection of starved mouse cells. Viruses 10. doi: 10.3390/v10040140
Decker, E. L., Skerka, C., and Zipfel, P. F. (1998). The early growth response protein (EGR-1) regulates interleukin-2 transcription by synergistic interaction with the nuclear factor of activated T cells. J. Biol. Chem. 273, 26923–26930.
DeLigio, J. T., and Zorio, D. A. (2009). Early growth response 1 (EGR1): a gene with as many names as biological functions. Cancer Biol. Ther. 8, 1889–1892. doi: 10.4161/cbt.8.20.9804
Dey, B. R., Sukhatme, V. P., Roberts, A. B., Sporn, M. B., Rauscher, F. J., and Kim, S. J. (1994). Repression of the transforming growth factor-beta 1 gene by the Wilms' tumor suppressor WT1 gene product. Mol. Endocrinol. 8, 595–602.
Djavani, M. M., Crasta, O. R., Zapata, J. C., Fei, Z., Folkerts, O., Sobral, B., et al. (2007). Early blood profiles of virus infection in a monkey model for Lassa fever. J. Virol. 81, 7960–7973. doi: 10.1128/JVI.00536-07
Domdom, M. A., Brest, P., Grosjean, I., Romeo, B., Landi, M. T., Gal, J., et al. (2020). A multifactorial score including autophagy for prognosis and care of COVID-19 patients. Autophagy 16, 2276–2281. doi: 10.1080/15548627.2020.1844433
Dron, M., Hameau, L., Benboudjema, L., Guymarho, J., Cajean-Feroldi, C., Rizza, P., et al. (1999). Cloning of a long HIV-1 readthrough transcript and detection of an increased level of early growth response protein-1 (Egr-1) mRNA in chronically infected U937 cells. Arch. Virol. 144, 19–28. doi: 10.1007/s007050050482
Duclot, F., and Kabbaj, M. (2017). The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front. Behav. Neurosci. 11:35. doi: 10.3389/fnbeh.2017.00035
Dyson, O. F., Traylen, C. M., and Akula, S. M. (2010). Cell membrane-bound Kaposi's sarcoma-associated herpesvirus-encoded glycoprotein B promotes virus latency by regulating expression of cellular Egr-1. J. Biol. Chem. 285, 37491–37502. doi: 10.1074/jbc.M110.159103
Dyson, O. F., Walker, L. R., Whitehouse, A., Cook, P. P., and Akula, S. M. (2012). Resveratrol inhibits KSHV reactivation by lowering the levels of cellular EGR-1. PLoS One 7:e33364. doi: 10.1371/journal.pone.0033364
Fan, Y., Sanyal, S., and Bruzzone, R. (2018). Breaking bad: how viruses subvert the cell cycle. Front. Cell. Infect. Microbiol. 8:396. doi: 10.3389/fcimb.2018.00396
Fu, Z. F., Weihe, E., Zheng, Y. M., Schafer, M. K., Sheng, H., Corisdeo, S., et al. (1993). Differential effects of rabies and Borna disease viruses on immediate-early- and late-response gene expression in brain tissues. J. Virol. 67, 6674–6681. doi: 10.1128/jvi.67.11.6674-6681.1993
Fujii, M., Niki, T., Mori, T., Matsuda, T., Matsui, M., Nomura, N., et al. (1991). HTLV-1 tax induces expression of various immediate early serum responsive genes. Oncogene 6, 1023–1029.
Gabriel, K. N., Jones, A. C., Nguyen, J. P., Antillon, K. S., Janos, S. N., Overton, H. N., et al. (2016). Association and regulation of protein factors of field effect in prostate tissues. Int. J. Oncol. 49, 1541–1552. doi: 10.3892/ijo.2016.3666
Gallo, F. T., Katche, C., Morici, J. F., Medina, J. H., and Weisstaub, N. V. (2018). Immediate early genes, memory and psychiatric disorders: focus on c-Fos, Egr1 and arc. Front. Behav. Neurosci. 12:79. doi: 10.3389/fnbeh.2018.00079
Gashler, A., and Sukhatme, V. P. (1995). Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog. Nucleic Acid Res. Mol. Biol. 50, 191–224. doi: 10.1016/S0079-6603(08)60815-6
Gaudreault, N. N., Indran, S. V., Balaraman, V., Wilson, W. C., and Richt, J. A. (2019). Molecular aspects of Rift Valley fever virus and the emergence of reassortants. Virus Genes 55, 1–11. doi: 10.1007/s11262-018-1611-y
Gwack, Y., Byun, H., Hwang, S., Lim, C., and Choe, J. (2001). CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75, 1909–1917. doi: 10.1128/JVI.75.4.1909-1917.2001
Havis, E., and Duprez, D. (2020). EGR1 transcription factor is a multifaceted regulator of matrix production in tendons and other connective tissues. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21051664
Herdegen, T., Walker, T., Leah, J. D., Bravo, R., and Zimmermann, M. (1990). The KROX-24 protein, a new transcription regulating factor: expression in the rat central nervous system following afferent somatosensory stimulation. Neurosci. Lett. 120, 21–24.
Herndon, R. M., Price, D. L., and Weiner, L. P. (1977). Regeneration of oligodendroglia during recovery from demyelinating disease. Science 195, 693–694. doi: 10.1126/science.190678
Hirata, H., Asanuma, M., and Cadet, J. L. (1998). Superoxide radicals are mediators of the effects of methamphetamine on Zif268 (Egr-1, NGFI-A) in the brain: evidence from using CuZn superoxide dismutase transgenic mice. Brain Res. Mol. Brain Res. 58, 209–216. doi: 10.1016/S0169-328X(98)00055-2
Ho, B. C., Yu, S. L., Chen, J. J., Chang, S. Y., Yan, B. S., Hong, Q. S., et al. (2011). Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9, 58–69. doi: 10.1016/j.chom.2010.12.001
Hoffmann, E., Ashouri, J., Wolter, S., Doerrie, A., Dittrich-Breiholz, O., Schneider, H., et al. (2008). Transcriptional regulation of EGR-1 by the Interleukin-1-JNK-MKK7-c-Jun pathway. J. Biol. Chem. 283, 12120–12128. doi: 10.1074/jbc.M800583200
Hou, P., Zhao, M., He, W., He, H., and Wang, H. (2019). Cellular microRNA bta-miR-2361 inhibits bovine herpesvirus 1 replication by directly targeting EGR1 gene. Vet. Microbiol. 233, 174–183. doi: 10.1016/j.vetmic.2019.05.004
Hsia, S. C., Graham, L. P., Bedadala, G. R., Balish, M. B., Chen, F., and Figliozzi, R. W. (2013). Induction of transcription factor early growth response protein 1 during HSV-1 infection promotes viral replication in corneal cells. Br. Microbiol Res. J. 3, 706–723. doi: 10.9734/BMRJ/2013/4817
Huang, R. P., Fan, Y., de Belle, I., Niemeyer, C., Gottardis, M. M., Mercola, D., et al. (1997). Decreased Egr-1 expression in human, mouse and rat mammary cells and tissues correlates with tumor formation. Int. J. Cancer 72, 102–109.
Huang, R. P., Fan, Y., deBelle, I., Ni, Z., Matheny, W., and Adamson, E. D. (1998). Egr-1 inhibits apoptosis during the UV response: correlation of cell survival with Egr-1 phosphorylation. Cell Death Differ. 5, 96–106. doi: 10.1038/sj.cdd.4400322
Huang, Q., Niu, Z., Han, J., Liu, X., Lv, Z., Li, H., et al. (2017). HTLV-1 tax upregulates early growth response protein 1 through nuclear factor-kappaB signaling. Oncotarget 8, 51123–51133. doi: 10.18632/oncotarget.17699
Islam, A., and Khan, M. A. (2020). Lung transcriptome of a COVID-19 patient and systems biology predictions suggest impaired surfactant production which may be druggable by surfactant therapy. Sci. Rep. 10:19395. doi: 10.1038/s41598-020-76404-8
Jain, N., Mahendran, R., Philp, R., Guy, G. R., Tan, Y. H., and Cao, X. (1996). Casein kinase II associates with Egr-1 and acts as a negative modulator of its DNA binding and transcription activities in NIH 3T3 cells. J. Biol. Chem. 271, 13530–13536.
Jones, M. W., Errington, M. L., French, P. J., Fine, A., Bliss, T. V., Garel, S., et al. (2001). A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat. Neurosci. 4, 289–296. doi: 10.1038/85138
Khachigian, L. M. (2021). Early growth Response-1, an integrative sensor in cardiovascular and inflammatory disease. J. Am. Heart Assoc. 10:e023539. doi: 10.1161/JAHA.121.023539
Kim, J. H., Collins-McMillen, D., Buehler, J. C., Goodrum, F. D., and Yurochko, A. D. (2017). Human cytomegalovirus requires epidermal growth factor receptor signaling to enter and initiate the early steps in the establishment of latency in CD34(+) human progenitor cells. J. Virol. 91. doi: 10.1128/JVI.01206-16
Kim, J. H., Kim, W. S., Kang, J. H., Lim, H. Y., Ko, Y. H., and Park, C. (2007). Egr-1, a new downstream molecule of Epstein-Barr virus latent membrane protein 1. FEBS Lett. 581, 623–628. doi: 10.1016/j.febslet.2007.01.020
Koldamova, R., Schug, J., Lefterova, M., Cronican, A. A., Fitz, N. F., Davenport, F. A., et al. (2014). Genome-wide approaches reveal EGR1-controlled regulatory networks associated with neurodegeneration. Neurobiol. Dis. 63, 107–114. doi: 10.1016/j.nbd.2013.11.005
Krones-Herzig, A., Adamson, E., and Mercola, D. (2003). Early growth response 1 protein, an upstream gatekeeper of the p53 tumor suppressor, controls replicative senescence. Proc. Natl. Acad. Sci. U. S. A. 100, 3233–3238. doi: 10.1073/pnas.2628034100
Krones-Herzig, A., Mittal, S., Yule, K., Liang, H., English, C., Urcis, R., et al. (2005). Early growth response 1 acts as a tumor suppressor in vivo and in vitro via regulation of p53. Cancer Res. 65, 5133–5143. doi: 10.1158/0008-5472.CAN-04-3742
Kyriakis, J. M., and Avruch, J. (2001). Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81, 807–869. doi: 10.1152/physrev.2001.81.2.807
Lampert, P. W., Sims, J. K., and Kniazeff, A. J. (1973). Mechanism of demyelination in JHM virus encephalomyelitis. Elec. Microsc. Studi. Acta Neuropathol. 24, 76–85. doi: 10.1007/BF00691421
Lee, J. Y., Kim, J. H., Bang, H., Cho, J., Ko, Y. H., Kim, S. J., et al. (2021). EGR1 as a potential marker of prognosis in extranodal NK/T-cell lymphoma. Sci Rep. 11. doi: 10.1038/s41598-021-89754-8
Lee, S. L., Tourtellotte, L. C., Wesselschmidt, R. L., and Milbrandt, J. (1995). Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J. Biol. Chem. 270, 9971–9977. doi: 10.1074/jbc.270.17.9971
Lehman, C. W., Smith, A., Kelly, J., Jacobs, J. L., Dinman, J. D., and Kehn-Hall, K. (2022). EGR1 Upregulation during encephalitic viral infections contributes to inflammation and cell death. Viruses 14:1210. doi: 10.3390/v14061210
Lemaire, P., Revelant, O., Bravo, R., and Charnay, P. (1988). Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc. Natl. Acad. Sci. U. S. A. 85, 4691–4695.
Lemaire, P., Vesque, C., Schmitt, J., Stunnenberg, H., Frank, R., and Charnay, P. (1990). The serum-inducible mouse gene Krox-24 encodes a sequence-specific transcriptional activator. Mol. Cell. Biol. 10, 3456–3467.
Li, L., Ameri, A. H., Wang, S., Jansson, K. H., Casey, O. M., Yang, Q., et al. (2019). EGR1 regulates angiogenic and osteoclastogenic factors in prostate cancer and promotes metastasis. Oncogene 38, 6241–6255. doi: 10.1038/s41388-019-0873-8
Li, S. W., Wang, C. Y., Jou, Y. J., Yang, T. C., Huang, S. H., Wan, L., et al. (2016). SARS coronavirus papain-like protease induces Egr-1-dependent up-regulation of TGF-beta1 via ROS/p38 MAPK/STAT3 pathway. Sci. Rep. 6:25754. doi: 10.1038/srep25754
Li, Q. J., Yang, S. H., Maeda, Y., Sladek, F. M., Sharrocks, A. D., and Martins-Green, M. (2003). MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 22, 281–291. doi: 10.1093/emboj/cdg028
Lim, R. W., Varnum, B. C., and Herschman, H. R. (1987). Cloning of tetradecanoyl phorbol ester-induced 'primary response' sequences and their expression in density-arrested Swiss 3T3 cells and a TPA non-proliferative variant. Oncogene 1, 263–270.
Lim, R. W., Varnum, B. C., O'Brien, T. G., and Herschman, H. R. (1989). Induction of tumor promotor-inducible genes in murine 3T3 cell lines and tetradecanoyl phorbol acetate-nonproliferative 3T3 variants can occur through protein kinase C-dependent and -independent pathways. Mol. Cell. Biol. 9, 1790–1793.
Liu, C., Adamson, E., and Mercola, D. (1996). Transcription factor EGR-1 suppresses the growth and transformation of human HT-1080 fibrosarcoma cells by induction of transforming growth factor beta 1. Proc. Natl. Acad. Sci. U. S. A. 93, 11831–11836. doi: 10.1073/pnas.93.21.11831
Liu, C., Yao, J., Mercola, D., and Adamson, E. (2000). The transcription factor EGR-1 directly transactivates the fibronectin gene and enhances attachment of human glioblastoma cell line U251. J. Biol. Chem. 275, 20315–20323. doi: 10.1074/jbc.M909046199
Ma, Z., Gao, X., Shuai, Y., Wu, X., Yan, Y., Xing, X., et al. (2021). EGR1-mediated linc01503 promotes cell cycle progression and tumorigenesis in gastric cancer. Cell Prolif. 54:e12922. doi: 10.1111/cpr.12922
Mack, K., Day, M., Milbrandt, J., and Gottlieb, D. I. (1990). Localization of the NGFI-A protein in the rat brain. Brain Res. Mol. Brain Res. 8, 177–180. doi: 10.1016/0169-328X(90)90062-I
Mallett, K. A., Turrisi, R., Billingsley, E., Comer, C. D., Read, A., Varvil-Weld, L., et al. (2012). Enhancing patients' satisfaction and sun-protective behaviors using the ABC method of physician-patient communication. Arch. Dermatol. 148, 1087–1089. doi: 10.1001/archdermatol.2012.1659
Matsuoka, M., and Jeang, K. T. (2007). Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 7, 270–280. doi: 10.1038/nrc2111
McMahon, S. B., and Monroe, J. G. (1996). The role of early growth response gene 1 (egr-1) in regulation of the immune response. J. Leukoc. Biol. 60, 159–166. doi: 10.1002/jlb.60.2.159
Milbrandt, J. (1987). A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science 238, 797–799. doi: 10.1126/science.3672127
Miller, C. L., Burkhardt, A. L., Lee, J. H., Stealey, B., Longnecker, R., Bolen, J. B., et al. (1995). Integral membrane-protein 2 of Epstein-Barr-virus regulates reactivation from latency through dominant-negative effects on protein-tyrosine kinases. Immunity 2, 155–166. doi: 10.1016/S1074-7613(95)80040-9
Miller, C. L., Lee, J. H., Kieff, E., and Longnecker, R. (1994). An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc. Natl. Acad. Sci. U. S. A. 91, 772–776. doi: 10.1073/pnas.91.2.772
Musso, D., Ko, A. I., and Baud, D. (2019). Zika virus infection - after the pandemic. N. Engl. J. Med. 381, 1444–1457. doi: 10.1056/NEJMra1808246
Myung, E., Park, Y. L., Kim, N., Chung, C. Y., Park, H. B., Park, H. C., et al. (2013). Expression of early growth response-1 in human gastric cancer and its relationship with tumor cell behaviors and prognosis. Pathol. Res. Pract. 209, 692–699. doi: 10.1016/j.prp.2013.08.001
Ngiam, N., Post, M., and Kavanagh, B. P. (2007). Early growth response factor-1 in acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 293, L1089–L1091. doi: 10.1152/ajplung.00265.2007
Nishi, H., Nishi, K. H., and Johnson, A. C. (2002). Early growth Response-1 gene mediates up-regulation of epidermal growth factor receptor expression during hypoxia. Cancer Res. 62, 827–834.
Ouellette, A. J., Malt, R. A., Sukhatme, V. P., and Bonventre, J. V. (1990). Expression of two "immediate early" genes, Egr-1 and c-fos, in response to renal ischemia and during compensatory renal hypertrophy in mice. J. Clin. Invest. 85, 766–771. doi: 10.1172/JCI114502
Pang, Z., Raudonis, R., McCormick, C., and Cheng, Z. (2019). Early growth response 1 deficiency protects the host against Pseudomonas aeruginosa lung infection. Infect. Immun. 88. doi: 10.1128/IAI.00678-19
Peng, J., Yi, J., Yang, W., Ren, J., Wen, Y., Zheng, H., et al. (2020). Advances in foot-and-mouth disease virus proteins regulating host innate immunity. Front. Microbiol. 11:2046. doi: 10.3389/fmicb.2020.02046
Pfaffenseller, B., Kapczinski, F., Gallitano, A. L., and Klamt, F. (2018). EGR3 immediate early gene and the brain-derived Neurotrophic factor in bipolar disorder. Front. Behav. Neurosci. 12:15. doi: 10.3389/fnbeh.2018.00015
Pines, A., Bivi, N., Romanello, M., Damante, G., Kelley, M. R., Adamson, E. D., et al. (2005). Cross-regulation between Egr-1 and APE/Ref-1 during early response to oxidative stress in the human osteoblastic HOBIT cell line: evidence for an autoregulatory loop. Free Radic. Res. 39, 269–281. doi: 10.1080/10715760400028423
Poirier, R., Cheval, H., Mailhes, C., Garel, S., Charnay, P., Davis, S., et al. (2008). Distinct functions of egr gene family members in cognitive processes. Front. Neurosci. 2, 47–55. doi: 10.3389/neuro.01.002.2008
Ramana, C. V., and Das, B. (2020). Regulation of early growth response-1 (Egr-1) gene expression by Stat1-independent type I interferon signaling and respiratory viruses. Comput. Math. Biophys. 9, 289–303. doi: 10.1101/2020.08.14.244897
Riffo-Campos, A. L., Castillo, J., Tur, G., Gonzalez-Figueroa, P., Georgieva, E. I., Rodriguez, J. L., et al. (2015). Nucleosome-specific, time-dependent changes in histone modifications during activation of the early growth response 1 (Egr1) gene. J. Biol. Chem. 290, 197–208. doi: 10.1074/jbc.M114.579292
Romagnoli, L., Sariyer, I. K., Tung, J., Feliciano, M., Sawaya, B. E., Del Valle, L., et al. (2008). Early growth response-1 protein is induced by JC virus infection and binds and regulates the JC virus promoter. Virology 375, 331–341. doi: 10.1016/j.virol.2008.02.021
Ronca, S. E., Dineley, K. T., and Paessler, S. (2016). Neurological Sequelae resulting from encephalitic Alphavirus infection. Front. Microbiol. 7:959. doi: 10.3389/fmicb.2016.00959
Russo, M. W., Sevetson, B. R., and Milbrandt, J. (1995). Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc. Natl. Acad. Sci. U. S. A. 92, 6873–6877. doi: 10.1073/pnas.92.15.6873
Saha, S., and Rangarajan, P. N. (2003). Common host genes are activated in mouse brain by Japanese encephalitis and rabies viruses. J. Gen. Virol. 84, 1729–1735. doi: 10.1099/vir.0.18826-0
Sarkar, R., and Verma, S. C. (2017). Egr-1 regulates RTA transcription through a cooperative involvement of transcriptional regulators. Oncotarget 8, 91425–91444. doi: 10.18632/oncotarget.20648
Schmidt, K., Carroll, J. S., Yee, E., Thomas, D. D., Wert-Lamas, L., Neier, S. C., et al. (2019). The lncRNA SLNCR recruits the androgen receptor to EGR1-bound genes in melanoma and inhibits expression of tumor suppressor p21. Cell Rep. 27, 2493–2507.e4. doi: 10.1016/j.celrep.2019.04.101
Shi, L., Kishore, R., McMullen, M. R., and Nagy, L. E. (2002). Lipopolysaccharide stimulation of ERK1/2 increases TNF-alpha production via Egr-1. Am. J. Physiol. Cell Physiol. 282, C1205–C1211. doi: 10.1152/ajpcell.00511.2001
Shin, D. Y., Kim, G. Y., Kim, N. D., Jung, J. H., Kim, S. K., Kang, H. S., et al. (2008). Induction of apoptosis by pectenotoxin-2 is mediated with the induction of DR4/DR5, Egr-1 and NAG-1, activation of caspases and modulation of the Bcl-2 family in p53-deficient Hep3B hepatocellular carcinoma cells. Oncol. Rep. 19, 517–526. doi: 10.3892/or.19.2.517
Si, J., Tang, X., Xu, L., Fu, H., Li, H., He, Y., et al. (2021). High throughput sequencing of whole transcriptome and construct of ceRNA regulatory network in RD cells infected with enterovirus D68. Virol. J. 18:216. doi: 10.1186/s12985-021-01686-x
Song, Y., Cheng, X., Yang, X., Zhao, R., Wang, P., Han, Y., et al. (2015). Early growth response-1 facilitates enterovirus 71 replication by direct binding to the viral genome RNA. Int. J. Biochem. Cell Biol. 62, 36–46. doi: 10.1016/j.biocel.2015.02.012
Srinivasan, R., Mager, G. M., Ward, R. M., Mayer, J., and Svaren, J. (2006). NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex. J. Biol. Chem. 281, 15129–15137. doi: 10.1074/jbc.M600775200
Sukhatme, V. P., Cao, X. M., Chang, L. C., Tsai-Morris, C. H., Stamenkovich, D., Ferreira, P. C., et al. (1988). A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cells 53, 37–43.
Sun, Z., Xu, X., He, J., Murray, A., Sun, M. A., Wei, X., et al. (2019). EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat. Commun. 10:3892. doi: 10.1038/s41467-019-11905-3
Svaren, J., Sevetson, B. R., Apel, E. D., Zimonjic, D. B., Popescu, N. C., and Milbrandt, J. (1996). NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol. Cell. Biol. 16, 3545–3553. doi: 10.1128/MCB.16.7.3545
Swirnoff, A. H., and Milbrandt, J. (1995). DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol. Cell. Biol. 15, 2275–2287. doi: 10.1128/MCB.15.4.2275
Taefehshokr, S., Key, Y. A., Khakpour, M., Dadebighlu, P., and Oveisi, A. (2017). Early growth response 2 and Egr3 are unique regulators in immune system. Cent Eur J Immunol. 42, 205–209. doi: 10.5114/ceji.2017.69363
Tatebe, K., Zeytun, A., Ribeiro, R. M., Hoffmann, R., Harrod, K. S., and Forst, C. V. (2010). Response network analysis of differential gene expression in human epithelial lung cells during avian influenza infections. BMC Bioinform. 11:170. doi: 10.1186/1471-2105-11-170
Taylor, T. J., Brockman, M. A., McNamee, E. E., and Knipe, D. M. (2002). Herpes simplex virus. Front. Biosci. 7, d752–d764. doi: 10.2741/taylor
Trizzino, M., Zucco, A., Deliard, S., Wang, F., Barbieri, E., Veglia, F., et al. (2021). EGR1 is a gatekeeper of inflammatory enhancers in human macrophages. Sci. Adv. 7. doi: 10.1126/sciadv.aaz8836
Tur, G., Georgieva, E. I., Gagete, A., Lopez-Rodas, G., Rodriguez, J. L., and Franco, L. (2010). Factor binding and chromatin modification in the promoter of murine Egr1 gene upon induction. Cell. Mol. Life Sci. 67, 4065–4077. doi: 10.1007/s00018-010-0426-3
Virolle, T., Adamson, E. D., Baron, V., Birle, D., Mercola, D., Mustelin, T., et al. (2001). The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat. Cell Biol. 3, 1124–1128. doi: 10.1038/ncb1201-1124
Vockerodt, M., Wei, W., Nagy, E., Prouzova, Z., Schrader, A., Kube, D., et al. (2013). Suppression of the LMP2A target gene, EGR-1, protects Hodgkin's lymphoma cells from entry to the EBV lytic cycle. J. Pathol. 230, 399–409. doi: 10.1002/path.4198
Wagner, A., Doerks, A., Aboud, M., Alonso, A., Tokino, T., Flugel, R. M., et al. (2000). Induction of cellular genes is mediated by the Bel1 transactivator in foamy virus-infected human cells. J. Virol. 74, 4441–4447. doi: 10.1128/JVI.74.10.4441-4447.2000
Wang, H., Kong, N., Jiao, Y., Dong, S., Sun, D., Chen, X., et al. (2021). EGR1 suppresses porcine epidemic diarrhea virus replication by regulating IRAV to degrade viral Nucleocapsid protein. J. Virol. 95:e0064521. doi: 10.1128/JVI.00645-21
Wang, X. P., Wen, B., Zhang, X. J., Ma, L., Liang, X. L., and Zhang, M. L. (2022). Transcriptome analysis of genes responding to infection of Leghorn male hepatocellular cells with fowl adenovirus serotype 4. Front Vet Sci. 9:871038. doi: 10.3389/fvets.2022.871038
Warner, L. E., Mancias, P., Butler, I. J., McDonald, C. M., Keppen, L., Koob, K. G., et al. (1998). Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat. Genet. 18, 382–384. doi: 10.1038/ng0498-382
Waters, C. M., Hancock, D. C., and Evan, G. I. (1990). Identification and characterisation of the egr-1 gene product as an inducible, short-lived, nuclear phosphoprotein. Oncogene 5, 669–674.
Wong, L. M., Li, D., Tang, Y., Mendez-Lagares, G., Thompson, G. R., Hartigan-O'Connor, D. J., et al. (2022). Human immunodeficiency Virus-1 latency reversal via the induction of early growth response protein 1 to bypass protein kinase C agonist-associated immune activation. Front. Microbiol. 13:836831. doi: 10.3389/fmicb.2022.836831
Xie, T. A., He, Z. J., Liang, C., Dong, H. N., Zhou, J., Fan, S. J., et al. (2021). An integrative bioinformatics analysis for identifying hub genes associated with infection of lung samples in patients infected with SARS-CoV-2. Eur. J. Med. Res. 26:146. doi: 10.1186/s40001-021-00609-4
Yan, S. F., Mackman, N., Kisiel, W., Stern, D. M., and Pinsky, D. J. (1999). Hypoxia/hypoxemia-induced activation of the procoagulant pathways and the pathogenesis of ischemia-associated thrombosis. Arterioscler. Thromb. Vasc. Biol. 19, 2029–2035. doi: 10.1161/01.ATV.19.9.2029
Yang, S. H., Vickers, E., Brehm, A., Kouzarides, T., and Sharrocks, A. D. (2001). Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 21, 2802–2814. doi: 10.1128/MCB.21.8.2802-2814.2001
Yeo, H., Ahn, S. S., Lee, J. Y., Jung, E., Jeong, M., Kang, G. S., et al. (2021). Disrupting the DNA binding of EGR-1 with a small-molecule inhibitor ameliorates 2,4-Dinitrochlorobenzene-induced skin inflammation. J. Invest. Dermatol. 141, 1851–1855. doi: 10.1016/j.jid.2020.12.029
Young, L. S., and Rickinson, A. B. (2004). Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4, 757–768. doi: 10.1038/nrc1452
Yu, J., de Belle, I., Liang, H., and Adamson, E. D. (2004). Coactivating factors p300 and CBP are transcriptionally crossregulated by Egr1 in prostate cells, leading to divergent responses. Mol. Cell 15, 83–94. doi: 10.1016/j.molcel.2004.06.030
Yuan, L., Fung, T. S., He, J., Chen, R. A., and Liu, D. X. (2022). Modulation of viral replication, apoptosis and antiviral response by induction and mutual regulation of EGR and AP-1 family genes during coronavirus infection. Emerg. Microbes Infect. 11, 1717–1729. doi: 10.1080/22221751.2022.2093133
Zandarashvili, L., White, M. A., Esadze, A., and Iwahara, J. (2015). Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1. FEBS Lett. 589, 1748–1753. doi: 10.1016/j.febslet.2015.05.022
Keywords: early growth response protein 1, transcription, virus, herpesvirus, inflammation, Venezuelan equine encephalitis virus, alphavirus
Citation: Woodson CM and Kehn-Hall K (2022) Examining the role of EGR1 during viral infections. Front. Microbiol. 13:1020220. doi: 10.3389/fmicb.2022.1020220
Received: 16 August 2022; Accepted: 26 September 2022;
Published: 21 October 2022.
Edited by:
Bin Su, Capital Medical University, ChinaReviewed by:
Dinesh Verma, The University of Utah, United StatesCopyright © 2022 Woodson and Kehn-Hall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kylene Kehn-Hall, a2tlaG5oYWxsQHZ0LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.