- 1Laboratory of Clinical Virology, WHO Reference Laboratory for Poliomyelitis and Measles in the Eastern Mediterranean Region, Pasteur Institute of Tunis, University Tunis El Manar (UTM), Tunis, Tunisia
- 2Research Laboratory “Virus, Vectors and Hosts: One Health Approach and Technological Innovation for a Better Health,” LR20IPT02, Pasteur Institute of Tunis, University Tunis El Manar (UTM), Tunis, Tunisia
- 3Laboratory of Biomedical Genomics and Oncogenetics (LR16IPT05), Pasteur Institute of Tunis, University Tunis El Manar (UTM), Tunis, Tunisia,
- 4Department of Gastroenterology, Polyclinic of CNSS, Sousse, Tunisia
- 5Department of Gastroenterology, Hospital of Tahar Maamouri, Nabeul, Tunisia
Hepatitis B virus (HBV) infection remains a serious public health concern worldwide despite the availability of an efficient vaccine and the major improvements in antiviral treatments. The aim of the present study is to analyze the mutational profile of the HBV whole genome in ETV non-responder chronic HBV patients, in order to investigate antiviral drug resistance, immune escape, and liver disease progression to Liver Cirrhosis (LC) or Hepatocellular Carcinoma (HCC). Blood samples were collected from five chronic hepatitis B patients. For each patient, two plasma samples were collected, before and during the treatment. Whole genome sequencing was performed using Sanger technology. Phylogenetic analysis comparing the studied sequences with reference ones was used for genotyping. The mutational profile was analyzed by comparison with the reference sequence M32138. Genotyping showed that the studied strains belong to subgenotypes D1, D7, and D8. The mutational analysis showed high genetic variability. In the RT region of the polymerase gene, 28 amino acid (aa) mutations were detected. The most significant mutations were the pattern rtL180M + rtS202G + rtM204V, which confer treatment resistance. In the S gene, 35 mutations were detected namely sP120T, sT126S, sG130R, sY134F, sS193L, sI195M, and sL216stop were previously described to lead to vaccine, immunotherapy, and/or diagnosis escape. In the C gene, 34 mutations were found. In particular, cG1764A, cC1766G/T, cT1768A, and cC1773T in the BCP; cG1896A and cG1899A in the precore region and cT12S, cE64D, cA80T, and cP130Q in the core region were associated with disease progression to LC and/or HCC. Other mutations were associated with viral replication increase including cT1753V, cG1764A/T, cC1766G/T, cT1768A, and cC1788G in the BCP as well as cG1896A and cG1899A in the precore region. In the X gene, 30 aa substitutions were detected, of which substitutions xT36D, xP46S, xA47T, xI88F, xA102V, xI127T, xK130M, xV131I, and xF132Y were previously described to lead to LC and/or HCC disease progression. In conclusion, our results show high genetic variability in the long-term treatment of chronic HBV patients causing several effects. This could contribute to guiding national efforts to optimize relevant HBV treatment management in order to achieve the global hepatitis elimination goal by 2030.
Introduction
Hepatitis B virus (HBV) infection remains a serious public health concern worldwide despite the availability of an efficient vaccine and the major improvements in antiviral treatments. The World Health Organization (WHO) estimates that, in 2021, approximately 296 million persons are chronic HBV carriers. Among them, 820,000 represent a high risk of mortality caused by developing progressive liver diseases including hepatocellular carcinoma (HCC) and liver cirrhosis (LC) (WHO, 2021).
The genome of HBV is a circular DNA partially double-stranded of 3.2 kb and classified into 10 genotypes from A to J (Sunbul, 2014). It is organized into four main open overlapped reading frames (ORFs; pre-S1/pre-S2/S, pre-C/C, P, and X), encoding several proteins including the surface proteins S, M, and L holding the HBs antigen (HBsAg), the precore/core proteins holding HBeAg and HBcAg antigens, the polymerase (P), and the X protein holding the antigen HBxAg. Thus, mutations that occur in one gene can result in significant changes in the other overlapping genes.
Long-term treatment of HBV chronic patients with the available antiviral molecules can lead to the emergence of mutations throughout the whole genome. Mutations that occur within the reverse transcriptase (RT) domain of the P gene, target of antiviral treatment, may lead to treatment failure (Locarnini and Mason, 2006). Potential resistance-related mutations are grouped into 4 categories, primary mutations (category 1) could reduce antiviral susceptibility and HBV replication fitness. Secondary/compensatory mutations (category 2) developed subsequently and could restore functional defects in the RT activity of HBV caused by primary mutations. Putative antiviral resistance mutations (category 3) were reported as possible drug-resistant mutations but not verified experimentally and may be related to prolonged treatment or replication compensation. Pre-treatment mutations (category 4) could be found among treatment-naive patients but their role in antiviral treatment resistance has not been elucidated (Liu et al., 2010; Ciftci et al., 2014).
Moreover, mutations that emerge throughout a prolonged therapy could affect not only the RT region (Locarnini and Mason, 2006) but also the different overlapping genes. Therefore, such variations might result in hepatitis B immunoglobulin (HBIG) therapy escape, vaccine escape, misdiagnosis, and immune escape. They also could enhance viral replication capacity and viral persistence leading to the progression of severe liver diseases such as HCC or LC (Sheldon et al., 2006; Sheldon, 2008; Rajoriya et al., 2017).
On the other hand, it has been found that the presence of pre-existing naturally occurring mutations in treatment-naive patients may influence the efficacy of antiviral treatments. Therefore, knowledge of the mutational profile by whole genome sequencing of the HBV genome, for chronically infected patients, is of great interest for a complete diagnosis toward an efficient therapy scheme.
For HBV chronic patients in Tunisia, the national therapeutic schema is based on Entecavir (ETV) as a first-line of HBV treatment, and it is fully covered by the National Health Insurance Fund (NHIF), in case of resistance, Tenofovir disoproxil fumarate (TDF), is recommended alone or combined to ETV. However, the TDF is not covered by the NHIF.
The aim of the present study is to analyze the mutational profile through the HBV whole genome in ETV non-responder chronic HBV patients, in order to investigate antiviral drug resistance, immune escape, and liver disease progression to LC or HCC.
Materials and methods
Patients and samples
HBV chronic patients with quantifiable viral load and suspected to be ETV non-responders after viral breakthrough were included in the study. Blood samples were collected from five chronic hepatitis B patients investigated during the routine diagnostic activity of the Laboratory of Clinical Virology in Pasteur Institute of Tunis. For each included patient two plasma samples were collected: one before treatment as part of the pre-treatment diagnostic, and one during the treatment upon request of the treating physician. The period separating the second sample from the date of treatment beginning ranging between 14 and 72 months depending on the time of the viral breakthrough for each patient. Virological and clinical data are shown in Table 1.
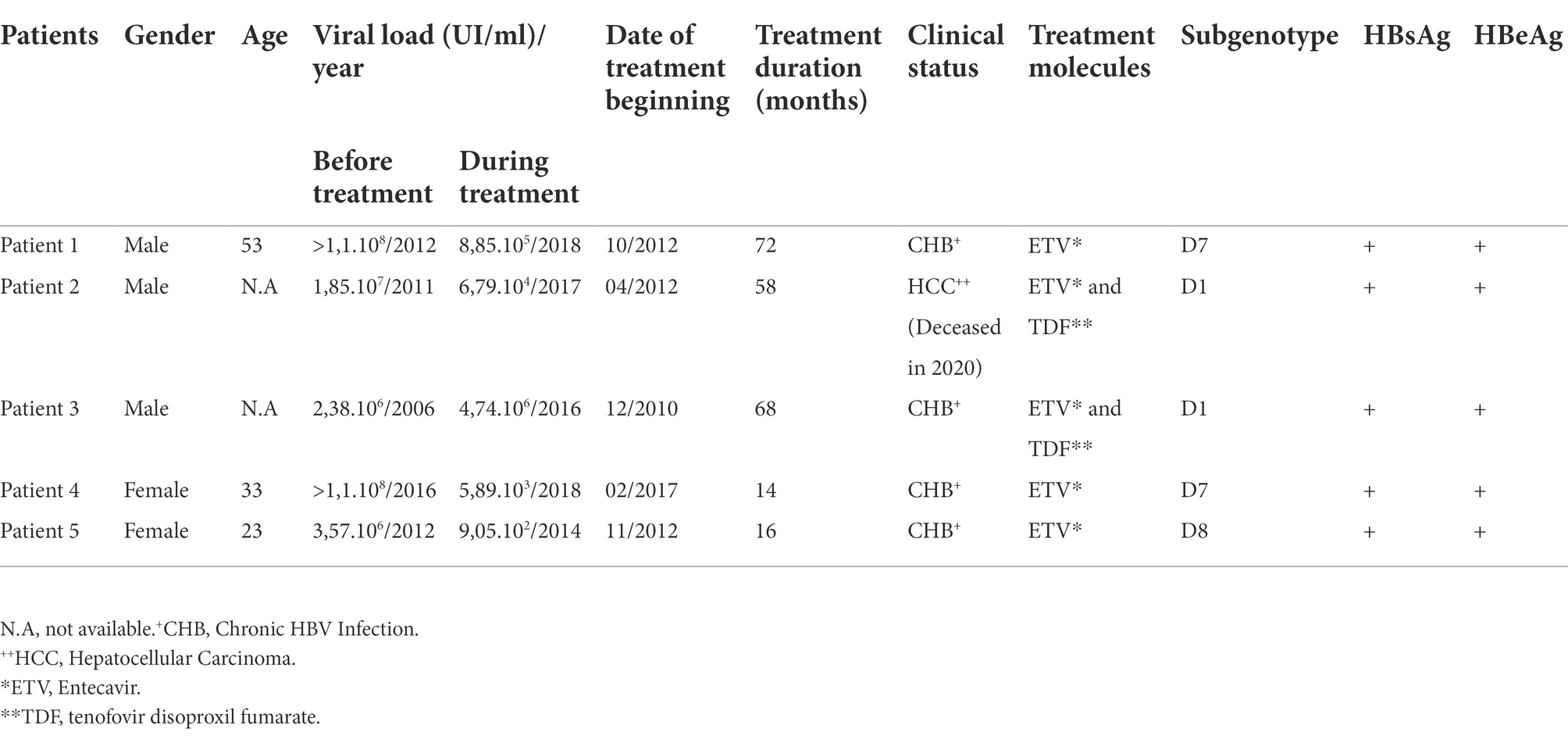
Table 1. Virological, treatment molecules, and treatment duration data for the five Tunisian chronic HBV infected patients before and during therapy.
Methods
DNA extraction, amplification, and sequencing
DNA was extracted from 200 μl of plasma using the Qiagen QIAamp® DNA extraction kit (QIAGEN® Inc., Hilden, Germany) according to the manufacturer’s instructions. Three pairs of primers previously described by Chekaraou et al. (2010) were used to amplify 3 overlapping amplicons of 1,228-bp (nt 2,817–863), 1,253 bp (nt 448–1,701), and 1,653 bp (nt 1,609–80) covering the whole HBV genome as shown in Figure 1.
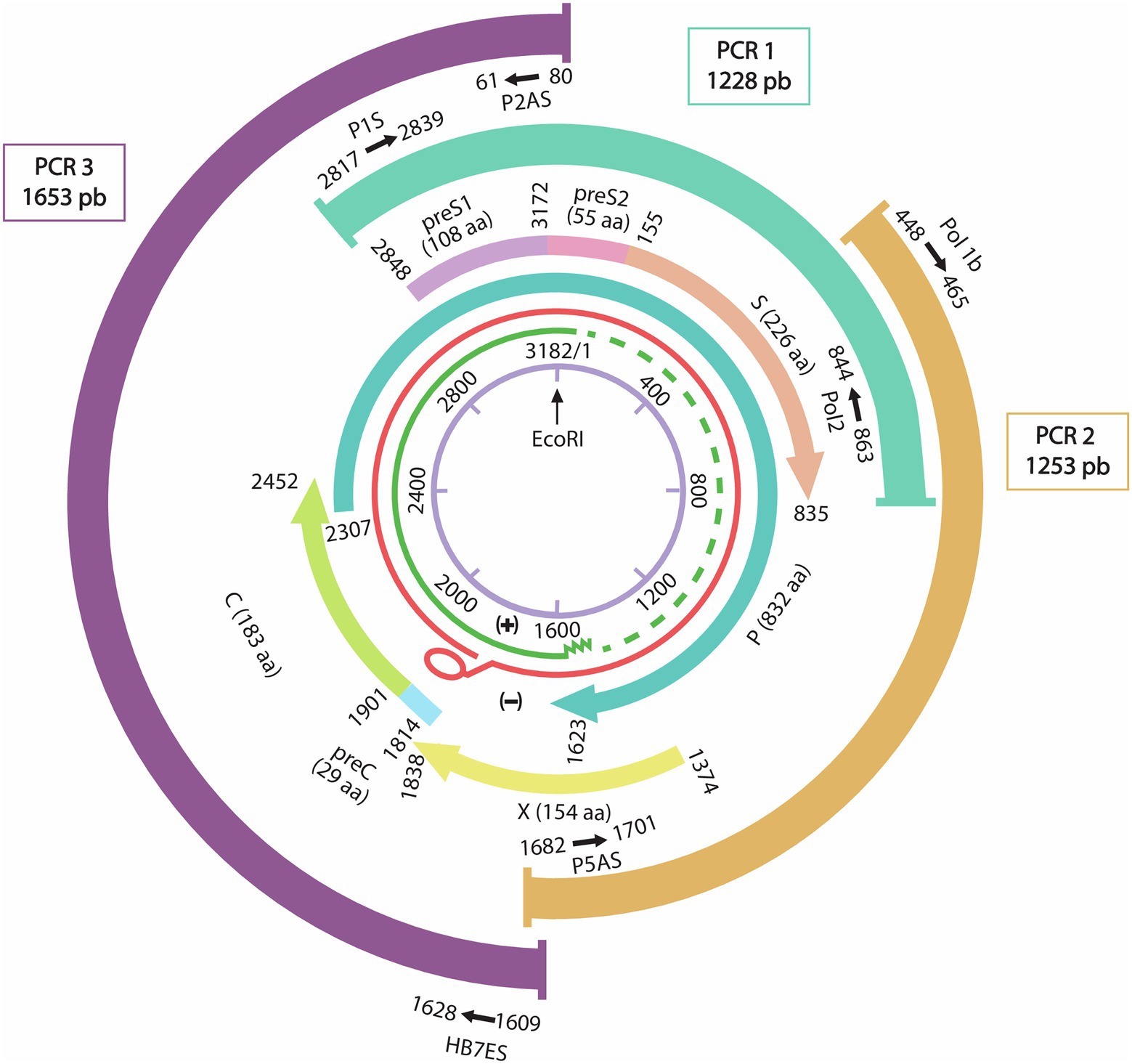
Figure 1. Representation of the HBV whole genome with the three overlapping PCR products and their primers sets positions. From inside to outside: schematic representation of the HBV genome with the EcoR1 restriction enzyme site representing nucleotide number 1. In green the incomplete positive strand, in Red the complete negative strand of the HBV genome. The overlapping genes P: Polymerase gene, S: Surface gene, C: capsid gene and X gene. Three overlapping fragments covering the whole genome are represented with the used primers P1S-Pol2 for PCR1 (1,228 nt), Pol1b-P5as for PCR2 (1,253 bp), and P2AS-HB7ES for PCR3 (1,653 bp).
PCR reactions were performed in 50 μl of reaction mixture containing 1X polymerase buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 μM of each primer, 1.25 U of Taq Core MP® (Applied Biosystems) and nuclease-free water. The amount of DNA extract added varied between 10 to 35 μl depending on the viral load. PCR cycling was as follows: 94°C for 5 min, 40 cycles (94°C for 1 min, 56°C/57°C/62.5°C for regions 1, 2, and 3, respectively, 72°C for 1 min) with a final extension step at 72°C for 10 min. PCR products were analyzed by electrophoresis on 1% agarose gels stained with 1,25X of Red gel™ dye Nucleic Acid (Biotium®) and visualized by UV transilluminator.
The purified template DNA was sequenced using a BigDye Terminator Ready Reaction Cycle Sequencing Kit (Applied Biosystems) using the same primers pairs on an ABI Prism 3130 Genetic Analyzer (Applied Biosystems).
HBV genome assembly, genotyping, and subtyping
The obtained overlapping sequences were then assembled using BLAST multiple sequences software by comparison with a reference sequence (M32138).1 The generated final sequences were submitted to Genbank under accession numbers: MT591274-MT591281 and OP121186.
Sequence alignment was performed with MAFFT online server using default parameters2 by comparing the obtained genomic sequences with 58 reference sequences representing the 10 HBV genotypes (A–J) and their corresponding subgenotypes. The resulting alignment was used to build a maximum likelihood phylogenetic tree using the IQ-TREE web server, supported by 1,000 bootstrap replicates.3 The phylogenetic tree was then visualized using Figtree software.4 The tree was rooted using the midpoint rooting method. Genotypes were also confirmed by the National Center for Biotechnology Information’s (NCBI) E-genotype online software.5 HBV subtypes were inferred from sequences of the S gene by identifying amino acids (aa) at positions 122, 160, 127, 140, and 159 according to an algorithm previously described (Purdy et al., 2006).
Mutation analysis
Mutational profiles of the nucleotide or amino acid sequences were determined by comparing each gene (P, S, C, and X) before and during treatment with the corresponding reference sequence using Mega 7.026 (Kumar et al., 2016). Mutations’ impacts on treatment, immune response, and liver disease progression were analyzed based on the literature.
Results
HBV whole genome assembly
Whole genome sequences were obtained before and during treatment for 3 patients (1, 2, and 3). For the two remaining patients (4 and 5) we succeeded to obtain the whole genome before treatment. During treatment, the obtained sequence of patient 4 was lacking 408 bp (from nucleotide 45 to nt 453) and for patient 5 we could not be able to amplify the HBV genome which could be due to the low viral load.
HBV genotyping and subtyping
Phylogenetic analysis (Figure 2) showed that all the sequences belong to genotype D. Subgenotyping showed that patients 1 and 4 were infected with subgenotype D7; patients 2 and 3 with D1 and patient 5 with D8, supported by high bootstrap values: 100, 100, and 85, respectively.
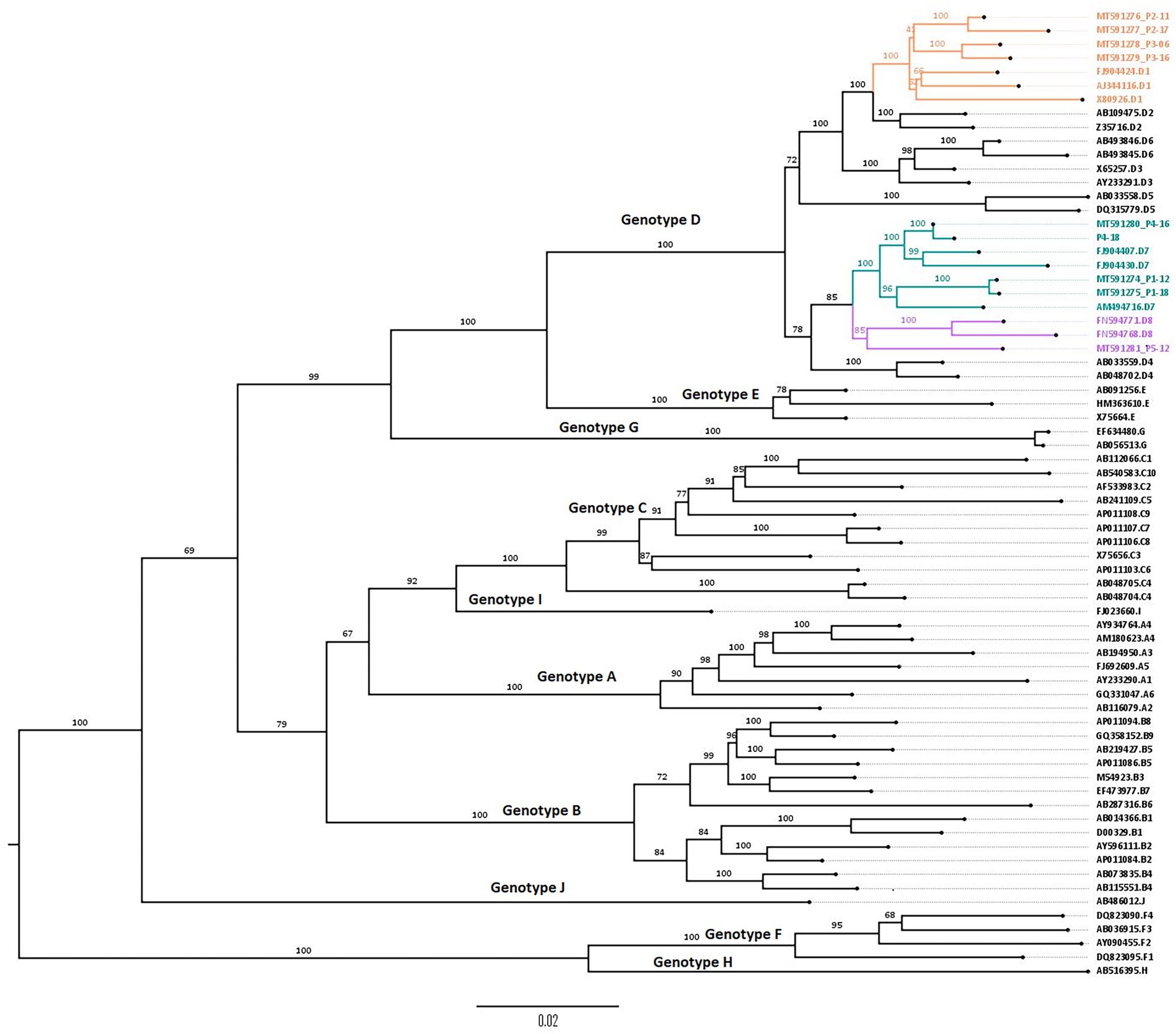
Figure 2. Phylogenetic tree constructed using the maximum likelihood method, showing D1, D7, and D8 subgenotypes from 5 Tunisian HBV patients. Phylogenetic tree of obtained genomic sequences with 58 reference sequences representing the 10 HBV genotypes (A–J) and their corresponding subgenotypes. The tree was constructed using the maximum likelihood method using the IQ tree web server and visualized by FigTree. Topology was supported by 1,000 bootstrap replicates. The tree was rooted using the midpoint rooting method.
Subtyping showed that the studied HBV strains belonged to the ayw2 subtype based on Arg122, Lys160, Pro127, Tyr140, and Gly159 positions.
Genetic variability in the P, S, C, and X genes
Mutational profile of the RT region in the polymerase gene
The mutational analysis of the RT region revealed a total of 28 aa substitutions ranging between 7 and 12 per patient, among them, several potential resistance-related mutations were detected. Primary mutations (category 1), rtS202G and rtM204V, occurred in patients 2 and 3 during treatment. Secondary/compensatory mutations (category 2), were found in 8 aa replacements; 5 were detected before treatment (rtL91I and rtT128N in patient 2; rtQ149K and rtP237T in patients 1, 4, and 5; rtQ267H in patients 4 and 5) and 3 changes emerged during treatment (rtL180M in patients 2 and 3; rtQ215S and rtF221Y in patient 2).
Three putative antiviral resistance mutations (category 3) were detected: rtR153W in treatment-naïve patients 1, 4, and 5 as well as rtD134E and rtC256S during treatment in patients 3 and 1, respectively. Three pre-treatment mutations (category 4) were also found: rtR110G and rtI266R in patient 1 and rtD263E in patient 5.
Mutations that did not fit categories 1 to 4 were classified into “novel amino acid substitutions” and were observed in 12 aa positions. Six variations namely rtE11D, rtH54Y, rtW257Y, rtD263E, rtQ267Y, and rtE271D were found in treatment-naïve patients and six variations namely rtL145M, rtL260F, rtQ267R, rtK270R, rtM309K, and rtN337T occurred during treatment. The aa changes detected in the RT region of the P gene are mentioned in Table 2; Figure 3.
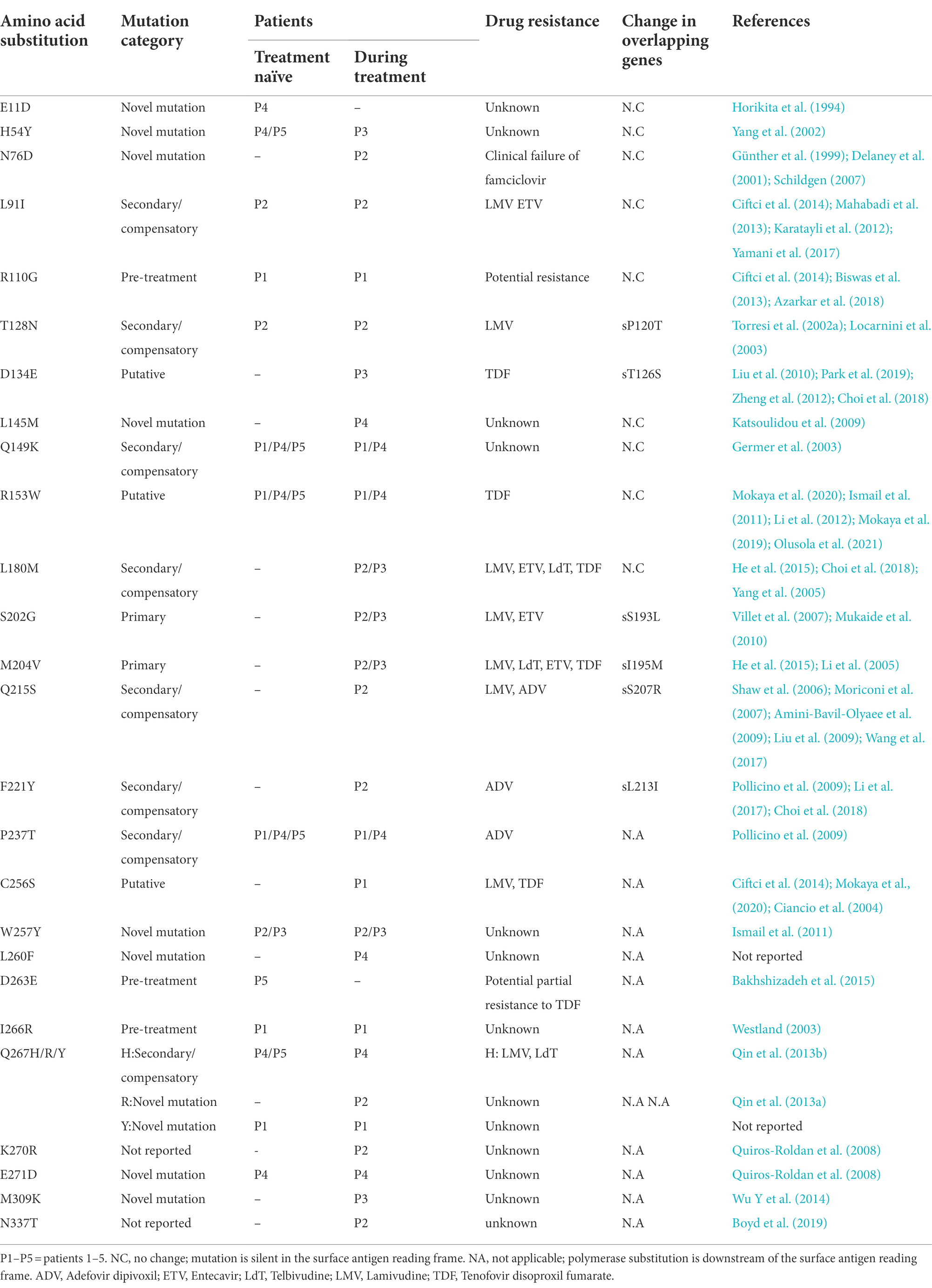
Table 2. Amino acid substitutions detected within the RT region sequences of the five HBV Chronic infected patients with their reported antiviral resistance.
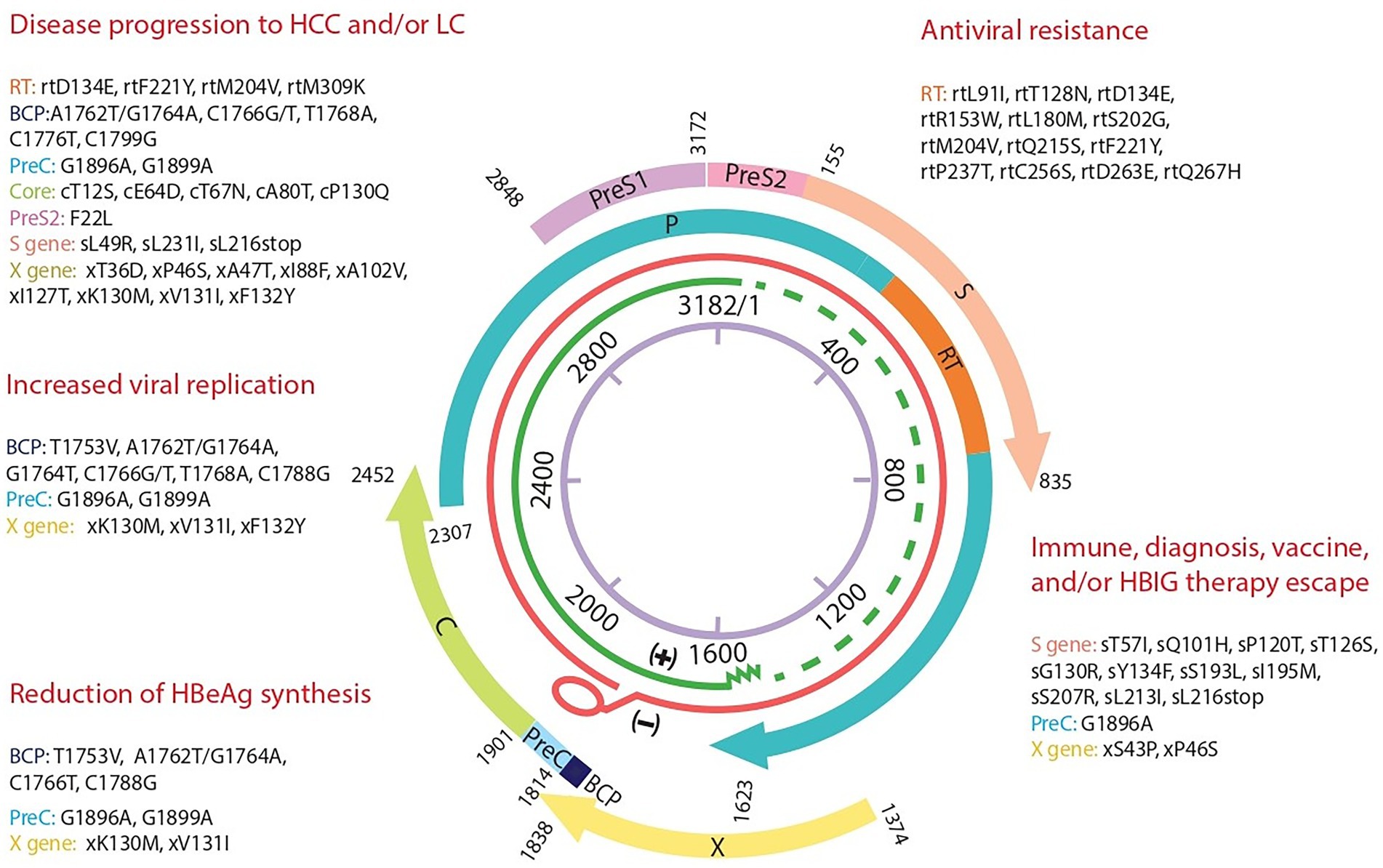
Figure 3. Hepatitis B genome map with summary of the most significant mutations and their clinical impact detected in 5 Tunisian HBV patients. A representation of the four overlapping genes encoding the polymerase P, the surface gene S, the capsid gene C, and X gene. The detected significant mutations are grouped depending on their clinical impact based on the literature. BCP, Basal core Promotor; C, core; PreC, precore; S, surface; RT, reverse transcriptase; HBIG, Immunoglobulin.
Mutational analysis of the pre-S/S coding regions
A total of 35 aa substitutions were observed in the whole S gene ranging between 3 and 18 mutations per patient, most of them (n = 16) were located in the S region. In the pre-S1 and pre-S2 regions, n = 9 and n = 10 substitutions were observed, respectively. Mutations detected in the S gene are summarized in Table 3; Figure 3. Out of the 16 aa changes in the S region, 10 were clustered in HBsAg epitopes including B-cells epitopes (aa100-160) as follows: 6 (sN3T, sL42R, sL49R, sT57I, sC76Y, and sQ101H) in HBs1 (upstream of aa120); 1 (sP120T) in HBs2 (aa120–123) and 3 (sT126S, sG130R and sY134F) in HBs3 (aa124–137). Furthermore, the major hydrophilic region (MHR, aa99–169) of the S region had accumulated 5 aa variations of which three were within the HBsAg “a” determinant region (aa124–147).
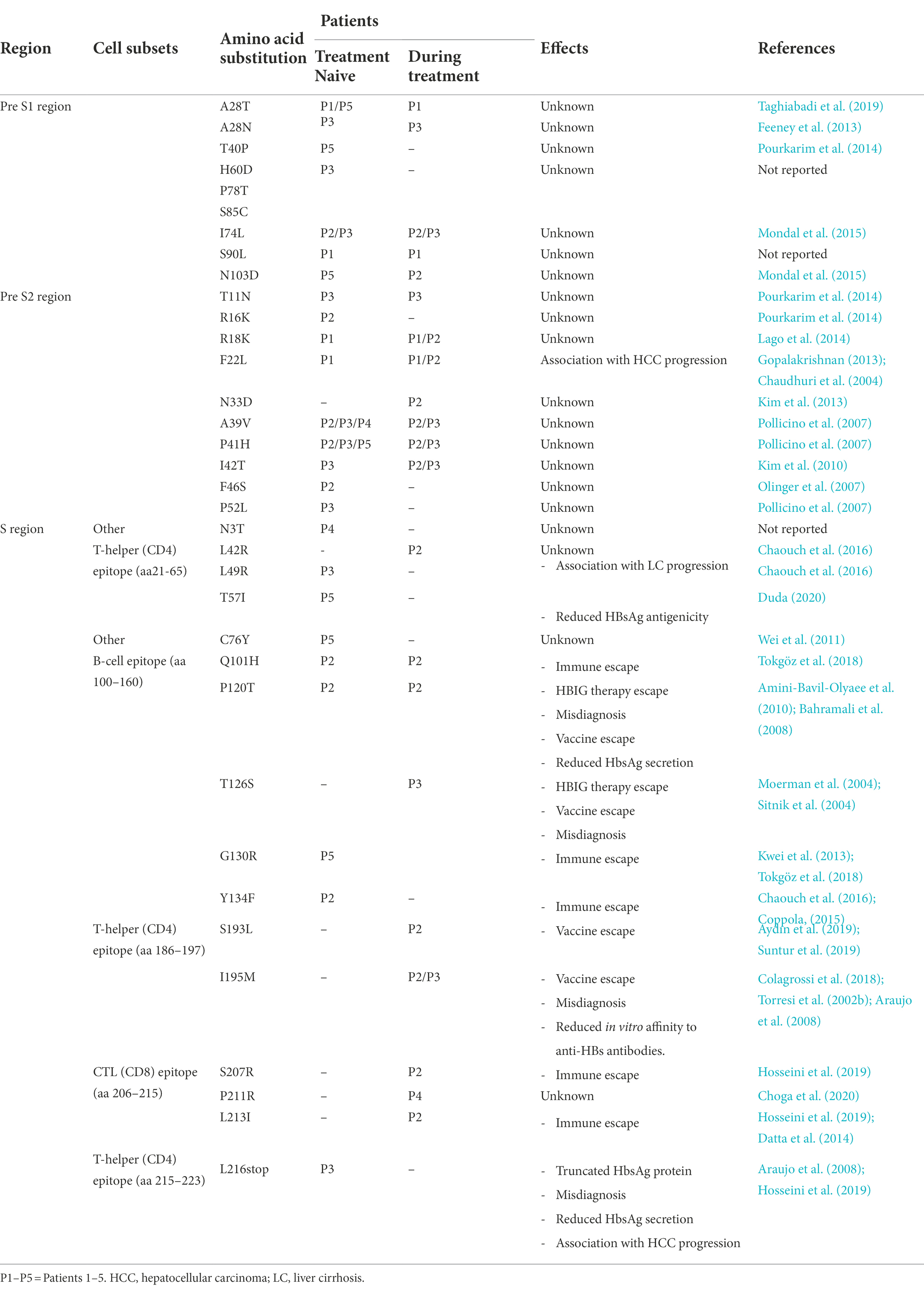
Table 3. Amino acid substitutions within the HBV surface gene sequences from the studied patients with their impact.
In addition, nine mutations out of the 16 substitutions in the S gene occurred in different CD4 and CD8 recognition epitopes with the following distribution: 6 aa changes (sL42R, sL49R, sT57I, sS193L, sI195M, and sL216*) in T-helper CD4 epitopes (aa21–65/aa186-197/aa215–223) and 3 (sS207R, sP211R, sL213I) within cytotoxic T lymphocyte CD8 epitopes (aa206-215).
Immune escape mutations (sQ101H, sG130R, sY134F, sS207R, and sL213I) were detected in patients 2 and 5, HBsAg vaccine escape mutations (sP120T, sT126S, sS193L, and sI195M) were found in patients 2 and/or 3, HBIG immunotherapy escape mutations (sP120T and sT126S) were detected in patients 2 and 3, respectively, and misdiagnosis mutations (sP120T, sT126S, sI195M, and sL216stop) were observed in patients 2 and/or 3. Other mutations such as sN3T, sL42R, sC76Y, and sP211R are either not reported or with unknown impacts are also detected in our study.
Mutational analysis of the basal core promotor BCP, precore, and core coding regions
The analysis of the 9 HBV genomic sequences bearing the BCP, pre-C, and Core regions is summarized in Table 4. In the BCP region, 14 different aa mutations were identified over 10 sites ranging between 2 and 8 per patient. Nucleotide mutation G1757A was detected in patients 2, 3, 4, and 5; A1762T, G1764A, and T1753V were detected in patients 1, 3, and 5; C1773T in patients 2 and 3, G1764T/C1766G in patient 2 and C1766T/T1768A in patient 3. The double mutation G1764A/A1762T was found in patients 1, 3, and 5.
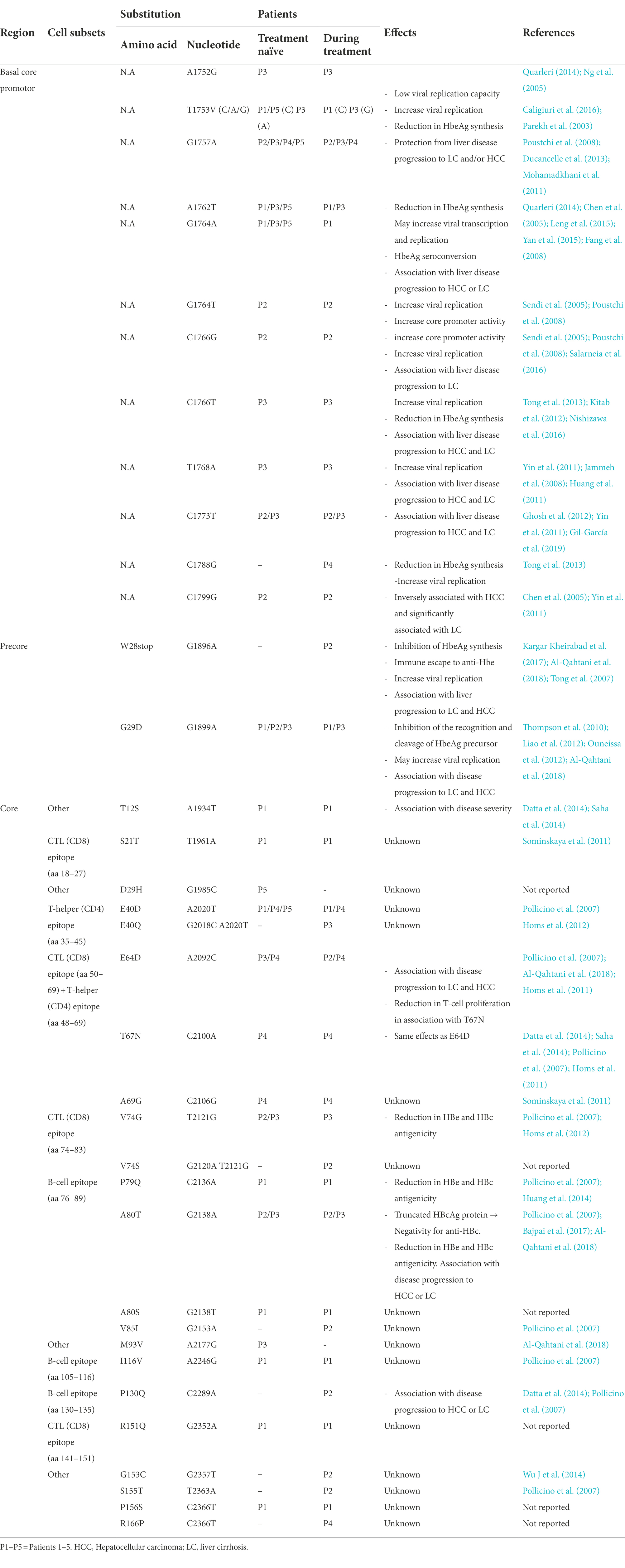
Table 4. Amino acid/nucleotide substitutions detected within the BCP, recure, and core sequences of the five chronic HBV infected patients with their impact.
In the precore region, 2 mutations were identified: G1896A (patient 2) and G1899A (patients 1, 2, and 3). Whereas, 22 mutations were observed in the core region ranging between 2 and 8 per patient. Among them, several amino acid substitutions were found within different antigen immunogenic epitopes. In particular, 4 aa changes were detected in the T-helper CD4 epitopes (aa 35–45 and 48–69; cE40D/Q, cE64D, cT67N, cA69G), 5 in the B-cell epitopes (aa76–89, 105–116, 130–135; cP79Q, cA80T/S, cV85I, cI116V and cP130Q) and 7 in the CTL CD8 epitopes (aa 18–27, 50–69, 74–83, 141–151; cS21T, cE64D, cT67N, cA69G, cV74S/G, cA80T/S, and cR151Q). The detected nucleotide/aa mutations found in the C gene are presented in Table 4; Figure 3.
Mutational analysis in the X coding region
In total, 30 aa substitutions were found in the X gene region ranging between 4 and 14 aa substitutions per patient of which 29 were before treatment beginning and only 1 was during it. Among them 10 variations were detected in the B-cell epitope (aa 29–48) namely: xL34I, xT36D/G, xS38P, xS39P, xP40S, xS41P, xL42P, xS43P, xP46S and xA47T. Four mutations; xK95N, xL98I, xA102, and xT105M; were detected within the T-helper CD4 epitope (aa 91–105) and 2 substitutions (xD119N and xL123W) were detected in CTL CD8 epitope (aa115–123).
As BCP overlaps partially with the HBx coding sequence, mutations at nucleotide positions T1753C/A/G, T1762T, G1764A/T, and C1788G; induce amino acid changes xI127T/D/G, xK130M, xV131I/L and xH139D near the C-terminus of the HBx protein, respectively.
The amino acid substitutions detected in the HBV X gene and their impact are summarized in Table 5; Figure 3.
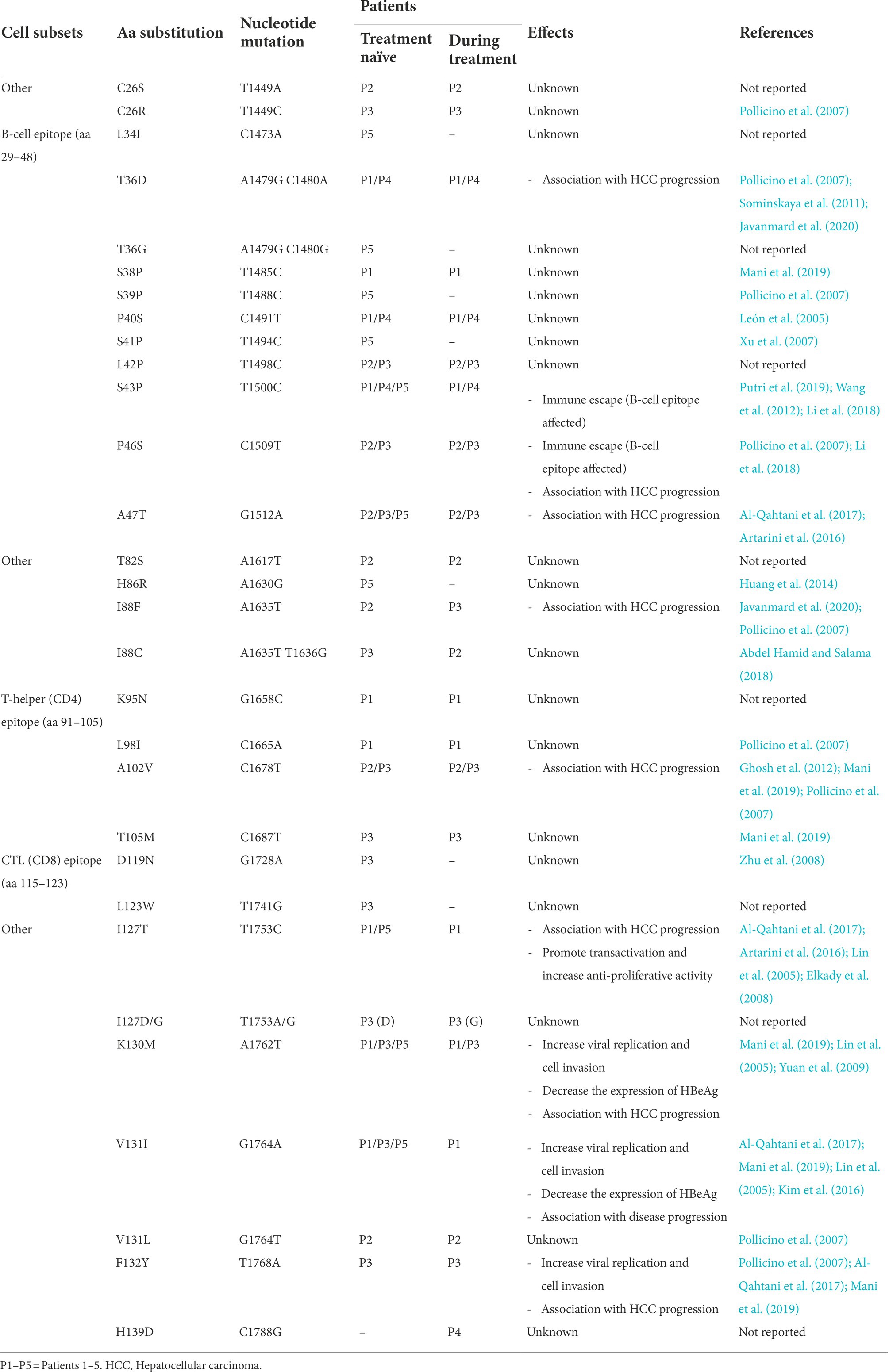
Table 5. Amino acid/nucleotide substitutions detected within the X gene sequences of the patients with their reported effects.
Discussion
In the present study, we have succeeded to generate the whole HBV genome by amplifying 3 overlapping PCR products covering the entire genome (3.2 kb) using Sanger technology. This technology remains of great importance despite the transition of most laboratories to Next generation sequencing (NGS) technologies. In fact, for small genomes, such as HBV, the Sanger technology is cost effective and more efficient for low viral loads < 103 IU/ml.
The whole genome was assembled for the 5 patients before the treatment and for 4 patients during the treatment. HBV genome was used for genotyping as well as to study the mutational profile in all the genes (S, P, X, and C) in order to give scientific proof of antiviral treatment resistance.
Genotyping showed that genotype D was detected in the 5 studied patients. This genotype was previously described as a predominant HBV genotype in Tunisia and the Maghreb region as well as in the Middle East with a low co-circulation rate of genotype E (Ayed et al., 2007; Ezzikouri et al., 2008; Ouneissa et al., 2013).
Subgenotypes D1 and D7, found in the present study, were previously described as the most prevalent subgenotypes circulating in Tunisia (Meldal et al., 2009). However, subgenotype D8 is to our knowledge detected for the first time in Tunisia. This subgenotype has been firstly detected in Niger and has been described as a recombinant strain between genotypes D and E (Chekaraou et al., 2010). The recombination analysis of the detected D8 strain, using the NCBI viral genotyping tool, was in line with the previous findings. Further studies are needed on larger population size to estimate the prevalence of this subgenotype in Tunisia.
In the second part of the present study, we have analyzed the mutational profile of all HBV genes P, S, C, and X.
The mutational profile of the RT region in the P gene showed high genetic variability with 28 different mutations. Before the treatment, 14 aa mutations were detected of which patient 2 had already 2 secondary/compensatory substitutions: rtL91I and T128N, described to be a resistance mutation to ETV and/or to LMV, respectively (Torresi et al., 2002a; Mahabadi et al., 2013; Ziaee et al., 2016). For the remaining patients, four mutations were detected and reported to be resistant to at least one of the following antivirals: rtQ267H in patients 4 and 5 to LMV and LdT; rtP237T in patients 1, 4, and 5 to ADV; rtR153W in patients 1, 4, and 5 in addition to rtD263E in patient 5 potentially to TDF (Pollicino et al., 2009; Qin et al., 2013b; Bakhshizadeh et al., 2015; Mokaya et al., 2020). The eight remaining substitutions were not previously described to have an impact on antiviral treatment.
During the treatment, 14 additional aa substitutions occurred. The most significant ones were rtM204V, rtL180M, and rtS202G detected in patients 2 and 3. Indeed, it has been described that the rtM204V substitution is usually associated with the compensatory mutation rtL180M, which restores the replication capacity of rtM204V mutants (Tenney et al., 2004). Thus, the pattern rtL180M, rtS202G, and rtM204V act synergistically not only to increase viral load but also to reduce treatment susceptibility and confer cross-resistance to ETV, TDF, LMV, and LdT (Kamiya, 2003; Li et al., 2005; He et al., 2015; Mokaya et al., 2020). Other emerged aa variations have been detected in our patients and previously described as resistance mutations that reduce the affinity and susceptibility to antiviral drugs namely rtQ215S and rtC256S to LMV; rtQ215S and rtF221Y to ADV; rtD134E, rtQ215S, and rtC256S to TDF (Moriconi et al., 2007; Amini-Bavil-Olyaee et al., 2009; Liu et al., 2009; Pollicino et al., 2009; Ciftci et al., 2014; Park et al., 2019; Mokaya et al., 2020).
Thus, our results support the need to introduce HBV genome sequencing as a pre-treatment diagnosis to predict potential resistance to available antiviral molecules, as well as to monitor the evolution of treatment response.
In addition, we have studied the mutational profile in the preS1, preS2, and S genes. As the coding sequence of the HBsAg is completely overlapped with the RT domain of the HBV polymerase, some mutations occurring in the RT region may lead to the emergence of escape mutants in the S region and vice versa. Thus, rtT128N, rtD134E, rtS202G, rtM204V, rtQ215S and rtF221Y substitutions observed in the RT region result in sP120T, sT126S, sS193L sI195M, sS207R and sL213I in the HBsAg gene, respectively. These mutations in addition to sT57I, sQ101H, sG130R, sY134F, and sL216stop could alter the antigenicity of HBsAg and reduce its expression and/or recognition by antibodies. Therefore, they could induce immune, vaccine, HBIG therapy, and/or diagnosis escape as well as influence HBsAg expression and treatment efficacy (Moerman et al., 2004; Sitnik et al., 2004; Bahramali et al., 2008; Amini-Bavil-Olyaee et al., 2010; Coppola, 2015; Ziaee et al., 2016; Rendon et al., 2017; Tokgöz et al., 2018; Aydın et al., 2019; Hosseini et al., 2019; Duda, 2020).
Regarding the mutational profile of BCP (nt 1,742–1,849), precore (nt 1,814–1,900), and core regions that code HBeAg and HBcAg proteins, the double mutants A1762T/G1764A, G1764T/C1766G and C1766T/T1768A, as well as the single mutations A1752G, T1753V (C/A/G), C1766T and C1788G, detected in BCP region, have been reported to enhance viral replication and/or reduce HBeAg synthesis by suppressing the transcription of the pre-C region (Parekh et al., 2003; Sendi et al., 2005; Poustchi et al., 2008; Tong et al., 2013; Caligiuri et al., 2016; Lazarevic et al., 2019). The single nucleotide mutations G1896A and G1899A in the precore region have been suggested to be mutational hotspots occurring most frequently in genotype D and were previously reported in Tunisian studies with an occurrence alone or in association (Triki et al., 2000; Bahri et al., 2006; Ayed et al., 2007; Poustchi et al., 2008; Ouneissa et al., 2012). These mutants result in a stop codon at position W28* and a substitution at position G29D, respectively, leading to the production of a truncated precore protein and then the abolition of HBeAg expression (Kobayashi et al., 2003; Thompson et al., 2010; Ducancelle et al., 2016). These variations are the major immune escape mutants of HBV as HBeAg is the main target for both cellular and humoral immune responses leading to a higher risk of liver HCC and LC progression (Tong et al., 2005; Liao et al., 2012; Suppiah et al., 2015; Pahal et al., 2016). In addition, precore mutants impose serious consequences on the treatment and enhance viral replication (Ouneissa et al., 2012; Kargar Kheirabad et al., 2017; Boyd et al., 2018).
Concerning the core mutations, cT67N within the T-helper CD4 epitope might be able to escape the host immune response (Datta et al., 2014; Saha et al., 2014). Moreover, cV74G, cP79Q, and cA80T mutations are known to reduce both HBe and HBc antigenicity (Pollicino et al., 2007; Huang et al., 2014). In addition, cA80T has resulted in the production of altered and truncated HBcAg protein leading potentially to abnormal immune reaction and negativity of anti-HBc (Bajpai et al., 2017).
In the last part of this study, we studied the mutational profile in the X gene. Substitutions xS43P and xP46S located in the B-cell epitope were detected in our study and have been suggested to be related with immune escape (Putri et al., 2019). Mutations xP46S, xA47T, xI88F, xA102V, xI127T, xK130M, xV131I, and xF132Y, were previously reported as significant HCC-related HBx mutants alone or combined such as (I127T + K130M + V131I) in patients 1, 3 and 5 and (xK130M + xV131I + xF132Y) in patient 3 (Pollicino et al., 2007; Ghosh et al., 2012; Ali et al., 2014; Al-Qahtani et al., 2017). Moreover, the double mutant xK130M + xV131I has been suggested to exacerbate the host’s immune response, increase viral replication, and lead to a truncated HBx protein (Wungu et al., 2019). In addition, it is associated with the activation of proto-oncogenes and inactivation of the tumor suppressor gene leading to a rapid progression of liver cirrhosis and/or HCC cell invasion and metastasis (Wang et al., 2016).
Several mutations previously reported to be significantly associated with an increased risk of severe liver disease progression to HCC and/or LC progression were also detected in other genes namely (rtD134E/rtF221Y/rtM204V/rtM309k) in the RT region; (sF22L) in the preS2 region; (sL49R, sL213I and sL216*) in the S region; (C1766T/T1768A double mutant, C1773T, C1799G, and C1766G) in the BCP region; and (cT12S/cE64D/cT67N/cA80T/cP130Q) in the core region of the C gene. These HCC-related mutations could be used as markers of HCC evolution in particular rtF221Y mutant which has been indicated as an independent risk factor for poor overall survival (Jammeh et al., 2008; Yin et al., 2011; Kitab et al., 2012; Zheng et al., 2012; Gopalakrishnan, 2013; Tong et al., 2013; Datta et al., 2014; Chaouch et al., 2016; Nishizawa et al., 2016; Kim et al., 2017; Li et al., 2017; Al-Qahtani et al., 2018; Choi et al., 2018; Hosseini et al., 2019). In contrast, the early development of G1757A in the BCP reduces the oncogenic potential of HBV suggesting that it might be a protective biomarker in chronic hepatitis B (Poustchi et al., 2008; Mohamadkhani et al., 2011; Ducancelle et al., 2013).
In addition to the commonly mentioned substitutions in all genes (P, S, C, and X), several nucleotide/amino acid substitutions have been detected in our patients (see Tables 2–5) but have never been reported previously or have been reported with unknown impact. Therefore, further studies are necessary to better understand and elucidate the effect of these mutations on HBV treatment, antigenicity, and disease evolution.
Conclusion
In conclusion, we would propose the whole genome sequencing as a pre-treatment diagnosis to predict potential resistance to available antiviral molecules, as well as to monitor the evolution of treatment response and prevent progression to cirrhosis or hepatocellular carcinoma. Thus, this could contribute to guiding national efforts to optimize relevant HBV treatment management in order to achieve the global hepatitis elimination goal by 2030.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
ZB, AC, HTr, MG, SA, LH, and MM: conceptualization. ZB, HTo, AS, WH, WK, and LY: methodology. ZB, AC, and HTr: validation. ZB and AC: formal analysis. ZB, AC, HTo, AS, WH, WK, and LY: investigation. ZB, AC, MG, SA, LH, and MM: data curation. ZB and KA: writing—original draft preparation. AC and HTr: editing and reviewing. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Tunisian Ministry of Higher Education [Programme d’Encouragement des Jeunes Chercheurs PEJC, 1ère Edition (2018; project code: 18PJEC07-09)], the Research Laboratory LR20IPT02: “Virus, Vectors and Hosts: One Health approach and technological innovation for a better health,” and the Clinical Investigation Center (CIC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ADV, Adefovir; aa, Amino acid; DNA, Deoxyribonucleic acid; ETV, Entecavir; HBcAg, hepatitis B core antigen; HBeAg, hepatitis B e antigen; HBIG, Hepatitis B immunoglobulin; HBsAg, Hepatitis B surface antigen; HBV, Hepatitis B virus; HCC, Hepatocellular carcinoma; LdT, Telbivudine; LMV, Lamivudine; MHR, Major hydrophilic region; NCBI, National Center for Biotechnology Information; NGS, Next Generation Sequencing; ORF, open reading frame; PCR, Polymerase chain reaction; RT, reverse transcriptase; TDF, tenofovir disoproxil fumarate.
Footnotes
1. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi.
2. ^https://mafft.cbrc.jp/alignment/server/
3. ^http://iqtree.cibiv.univie.ac.at
4. ^http://tree.bio.ed.ac.uk/software/figtree/
5. ^https://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi
References
Abdel Hamid, M., and Salama, A. (2018). X Gene variability and genotyping of hepatitis B virus (HBV) in sudanese patients with liver diseases, Khartoum, Sudan. Available at: https://www.researchgate.net/publication/325477946_X_GENE_VARIABILITY_AND_GENOTYPING_OF_HEPATITIS_B_VIRUS_HBV_IN_SUDANESE_PATIENTS_WITH_LIVER_DISEASES_KHARTOUM_SUDAN (Accessed October 22, 2020).
Al-Qahtani, A. A., Al-Anazi, M. R., Nazir, N., Ghai, R., Abdo, A. A., Sanai, F. M., et al. (2017). Hepatitis B virus (HBV) X gene mutations and their association with liver disease progression in HBV-infected patients. Oncotarget 8, 105115–105125.
Ali, A., Abdel-Hafiz, H., Suhail, M., Al-Mars, A., Zakaria, M. K., Fatima, K., et al. (2014). Hepatitis B virus, HBx mutants and their role in hepatocellular carcinoma. World J. Gastroenterol. 20, 10238–10248. doi: 10.3748/wjg.v20.i30.10238
Al-Qahtani, A. A., Al-Anazi, M. R., Nazir, N., Abdo, A. A., Sanai, F. M., Al-Hamoudi, W. K., et al. (2018). The correlation between Hepatitis B virus Precore/Core mutations and the progression of severe liver disease. Front. Cell. Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00355
Amini-Bavil-Olyaee, S., Herbers, U., Mohebbi, S. R., Sabahi, F., Zali, M. R., Luedde, T., et al. (2009). Prevalence, viral replication efficiency and antiviral drug susceptibility of rtQ215 polymerase mutations within the hepatitis B virus genome. J. Hepatol. 51, 647–654. doi: 10.1016/j.jhep.2009.04.022
Amini-Bavil-Olyaee, S., Vucur, M., Luedde, T., Trautwein, C., and Tacke, F. (2010). Differential Impact of Immune Escape Mutations G145R and P120T on the Replication of Lamivudine-Resistant Hepatitis B Virus e Antigen-Positive and -Negative Strains. J. Virol. 84, 1026–1033. doi: 10.1128/JVI.01796-09
Araujo, N. M., Branco-Vieira, M., Silva, A. C. M., Pilotto, J. H., Grinsztejn, B., de Almeida, A. J., et al. (2008). Occult hepatitis B virus infection in HIV-infected patients: evaluation of biochemical, virological and molecular parameters. Hepatol. Res. 38, 1194–1203. doi: 10.1111/j.1872-034X.2008.00392.x
Artarini, A., Geby Jessica, H., Rini Kartikasari, R., Riani, C., and Soefie Retnoningrum, D. (2016). Detection of Hepatitis B virus X gene mutation from local clinical samples. Microbiol Indones. 10, 9–14. doi: 10.5454/mi.10.1.2
Aydın, M., Tekin, S., Sayan, M., and Akhan, S. (2019). Molecular characterization of Hepatitis B virus strains isolated from chronic Hepatitis B patients in southeastern region of Turkey. Viral Hepat. J. 25, 40–44. doi: 10.4274/vhd.galenos.2019.2019.0015
Ayed, K., Gorgi, Y., Ayed-Jendoubi, S., Aouadi, H., Sfar, I., Najjar, T., et al. (2007). Hepatitis B virus genotypes and precore/core-promoter mutations in Tunisian patients with chronic hepatitis B virus infection. J. Infect. 54, 291–297. doi: 10.1016/j.jinf.2006.05.013
Azarkar, Z., Ziaee, M., Ebrahimzadeh, A., Sharifzadeh, G., and Javanmard, D. (2018). Epidemiology, risk factors, and molecular characterization of occult hepatitis B infection among anti-hepatitis B core antigen alone subjects. J. Med. Virol. 91.
Bahramali, G., Sadeghizadeh, M., Amini-Bavil-Olyaee, S., Alvaian, S. M., Behzad-Behbahani, A., Adeli, A., et al. (2008). Clinical, virologic and phylogenetic features of hepatitis B infection in Iranian patients. World J. Gastroenterol. 14, 5448–5453. doi: 10.3748/wjg.14.5448
Bahri, O., Cheikh, I., Hajji, N., Djebbi, A., Maamouri, N., Sadraoui, A., et al. (2006). Hepatitis B genotypes, precore and core promoter mutants circulating in Tunisia. J. Med. Virol. 78, 353–357. doi: 10.1002/jmv.20554
Bajpai, V., Gupta, E., Kundu, N., Sharma, S., and Shashtry, S. M. (2017). Hepatitis B core antibody negativity in a chronic hepatitis b infected patient: report of an unusual serological pattern. J. Clin. Diagn. Res. 11, DD04–DD06. doi: 10.7860/JCDR/2017/26821.10498
Bakhshizadeh, F., Hekmat, S., Keshvari, M., Alavian, S. M., Mostafavi, E., Keivani, H., et al. (2015). Efficacy of tenofovir disoproxil fumarate therapy in nucleoside-analogue naive Iranian patients treated for chronic hepatitis B. Hepat. Mon. 15. doi: 10.5812/hepatmon.15(5)2015.25749
Biswas, A., Panigrahi, R., Chandra, P. K., Banerjee, A., Datta, S., Pal, M., et al. (2013). Characterization of the occult hepatitis B virus variants circulating among the blood donors from eastern India. Sci. World J. doi: 10.1155/2013/212704
Boyd, A., Kouamé, M. G., Houghtaling, L., Moh, R., Gabillard, D., Maylin, S., et al. (2019). Hepatitis B virus activity in untreated hepatitis B e antigen-negative human immunodeficiency virus-hepatitis B virus co-infected patients from sub-Saharan Africa. Trans. R. Soc. Trop. Med. Hyg. 113, 437–445.
Boyd, A., Moh, R., Maylin, S., Abdou Chekaraou, M., Mahjoub, N., Gabillard, D., et al. (2018). Precore G1896A mutation is associated with reduced rates of HBsAg seroclearance in treated HIV hepatitis B virus co-infected patients from Western Africa. J. Viral Hepat. 25, 1121–1131. doi: 10.1111/jvh.12914
Caligiuri, P., Cerruti, R., Icardi, G., and Bruzzone, B. (2016). Overview of hepatitis B virus mutations and their implications in the management of infection. World J. Gastroenterol. 22, 145–154. doi: 10.3748/wjg.v22.i1.145
Chaouch, H., Taffon, S., Villano, U., Equestre, M., Bruni, R., Belhadj, M., et al. (2016). Naturally occurring surface antigen variants of Hepatitis B virus in Tunisian patients. Intervirology 59, 36–47. doi: 10.1159/000445894
Chaudhuri, V., Tayal, R., Nayak, B., Acharya, S. K., and Panda, S. K. (2004). Occult hepatitis B virus infection in chronic liver disease: full-length genome and analysis of mutant surface promoter. Gastroenterology 127, 1356–1371. doi: 10.1053/j.gastro.2004.08.003
Chekaraou, M. A., Brichler, S., Mansour, W., Gal, F., Garba, A., Dény, P., et al. (2010). A novel hepatitis B virus (HBV) subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J. Gen. Virol. 91, 1609–1620. doi: 10.1099/vir.0.018127-0
Chen, C. H., Lee, C. M., Lu, S. N., Changchien, C. S., Eng, H. L., Huang, C. M., et al. (2005). Clinical significance of hepatitis B virus (HBV) genotypes and precore and core promoter mutations affecting HBV e antigen expression in Taiwan. J. Clin. Microbiol. 43, 6000–6006. doi: 10.1128/JCM.43.12.6000-6006.2005
Choga, W. T., Anderson, M., Zumbika, E., Phinius, B. B., Mbangiwa, T., Bhebhe, L. N., et al. (2020). In Silico prediction of human leukocytes antigen (HLA) class II binding Hepatitis B virus (HBV) peptides in Botswana. Viruses 12:731. doi: 10.3390/v12070731
Choi, Y. M., Lee, S. Y., and Kim, B. J. (2018). Naturally occurring hepatitis B virus reverse transcriptase mutations related to potential antiviral drug resistance and liver disease progression. World J. Gastroenterol. 24, 1708–1724. doi: 10.3748/wjg.v24.i16.1708
Ciancio, A., Smedile, A., Rizzetto, M., Lagget, M., Gerin, J., and Korba, B. (2004). Identification of HBV DNA Sequences That Are Predictive of Response to Lamivudine Therapy. Hepatology 39, 64–73. doi: 10.1002/hep.20019
Ciftci, S., Keskin, F., Cakiris, A., Akyuz, F., Pinarbasi, B., Abaci, N., et al. (2014). Analysis of potential antiviral resistance mutation profiles within the HBV reverse transcriptase in untreated chronic hepatitis B patients using an ultra-deep pyrosequencing method. Diagn. Microbiol. Infect. Dis. 79, 25–30. doi: 10.1016/j.diagmicrobio.2014.01.005
Colagrossi, L., Hermans, L. E., Salpini, R., Di Carlo, D., Pas, S. D., Alvarez, M., et al. (2018). Immune-escape mutations and stop-codons in HBsAg develop in a large proportion of patients with chronic HBV infection exposed to anti-HBV drugs in Europe. BMC Infect. Dis. 18:251. doi: 10.1186/s12879-018-3161-2
Coppola, N. (2015). Clinical significance of hepatitis B surface antigen mutants. World J. Hepatol. 7:2729. doi: 10.4254/wjh.v7.i27.2729
Datta, S., Ghosh, A., Dasgupta, D., Ghosh, A., Roychoudhury, S., Roy, G., et al. (2014). Novel point and combo-mutations in the genome of hepatitis B virus-genotype D: characterization and impact on liver disease progression to hepatocellular carcinoma. PLoS One 9. doi: 10.1371/journal.pone.0110012
Delaney, W. E., Locarnini, S., and Shaw, T. (2001). Resistance of hepatitis B virus to antiviral drugs: Current aspects and directions for future investigation. Antivir. Chem. Chemother. 12, 1–35. doi: 10.1177/095632020101200101
Ducancelle, A., Abgueguen, P., Birguel, J., Mansour, W., Pivert, A., le Guillou-Guillemette, H., et al. (2013). High Endemicity and Low Molecular Diversity of Hepatitis B Virus Infections in Pregnant Women in a Rural District of North Cameroon. PLoS One 8:80346
Ducancelle, A., Pivert, A., Bertrais, S., Boursier, J., Balan, V., Veillon, P., et al. (2016). Different precore/core mutations of hepatitis B interact with, limit, or favor liver fibrosis severity. J. Gastroenterol. Hepatol. 31, 1750–1756. doi: 10.1111/jgh.13338
Duda, A. (2020). Influence de la variabilité des glycoprotéines d’enveloppe du virus de l’hépatite B sur la clairance de l’Ag HBs chez des patients co-infectés par le VIH suivis au CHU de Nancy. Available at: http://www.culture.gouv.fr/culture/infos-pratiques/droits/protection.htm (Accessed September 29, 2020).
Elkady, A., Tanaka, Y., Kurbanov, F., Oynsuren, T., and Mizokami, M. (2008). Virological and clinical implication of core promoter C1752/V1753 and T1764/G1766 mutations in hepatitis B virus genotype D infection in Mongolia. J. Gastroenterol. Hepatol. 23, 474–481. doi: 10.1111/j.1440-1746.2008.05321.x
Ezzikouri, S., Chemin, I., Chafik, A., Wakrim, L., Nourlil, J., Malki, A., et al. (2008). Genotype determination in Moroccan hepatitis B chronic carriers. Infect. Genet. Evol. 8, 306–312. doi: 10.1016/j.meegid.2008.01.010
Fang, Z. L., Sabin, C. A., Dong, B. Q., Ge, L. Y., Wei, S. C., Chen, Q. Y., et al. (2008). HBV A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAG carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am. J. Gastroenterol. 103, 2254–2262. doi: 10.1111/j.1572-0241.2008.01974.x
Feeney, S. A., Mccaughey, C., Watt, A. P., Agnaf, M. R. E., Mcdougall, N., Wend, U. C., et al. (2013). Reactivation of occult hepatitis B virus infection following cytotoxic lymphoma therapy in an anti-HBc negative patient. J. Med. Virol. 85, 597–601. doi: 10.1002/jmv.23513
Germer, J. J., Charlton, M. R., Ishitani, M., Forehand, C. D., and Patel, R. (2003). Characterization of Hepatitis B Virus Surface Antigen and Polymerase Mutations in Liver Transplant Recipients Pre- and Post-Transplant. Am. J. Transplant. 3, 743–753. doi: 10.1034/j.1600-6143.2003.00149.x
Ghosh, S., Mondal, R. K., Banerjee, P., Nandi, M., Sarkar, S., Das, K., et al. (2012). Tracking the naturally occurring mutations across the full-length genome of hepatitis B virus of genotype D in different phases of chronic e-antigen-negative infection. Clin. Microbiol. Infect. 18, E412–E418. doi: 10.1111/j.1469-0691.2012.03975.x
Gil-García, A. I., Madejón, A., Francisco-Recuero, I., López-López, A., Villafranca, E., Romero, M., et al. (2019). Prevalence of hepatocarcinoma-related hepatitis B virus mutants in patients in grey zone of treatment. World J. Gastroenterol. 25, 5883–5896. doi: 10.3748/wjg.v25.i38.5883
Gopalakrishnan, D. (2013). Hepatitis B virus subgenotype A1 predominates in liver disease patients from Kerala, India. World J. Gastroenterol. 19:9294. doi: 10.3748/wjg.v19.i48.9294
Günther, S., von Breunig, F., Santantonio, T., Jung, M. C., Gaeta, G. B., Fischer, L., et al. (1999). Absence of mutations in the YMDD motif/B region of the hepatitis B virus polymerase in famciclovir therapy failure. J. Hepatol. 30, 749–754. doi: 10.1016/S0168-8278(99)80124-X
He, X., Wang, F., Huang, B., Chen, P., and Zhong, L. (2015). Detection and analysis of resistance mutations of hepatitis B virus. Int. J. Clin. Exp. Med. 8, 9630–9639.
Homs, M., Buti, M., Tabernero, D., Quer, J., Sanchez, A., Corral, N., et al. (2012). Quasispecies dynamics in main core epitopes of hepatitis B virus by ultra-deep-pyrosequencing. World J. Gastroenterol. 18, 6096–6105. doi: 10.3748/wjg.v18.i42.6096
Homs, M., Jardi, R., Buti, M., Schaper, M., Tabernero, D., Fernandez-Fernandez, P., et al. (2011). HBV core region variability: effect of antiviral treatments on main epitopic regions. Antivir. Ther. 16, 37–49. doi: 10.3851/IMP1701
Horikita, M., Itoh, S., Yamamoto, K., Shibayama, T., Tsuda, F., and Okamoto, H. (1994). Differences in the entire nucleotide sequence between hepatitis B virus genomes from carriers positive for antibody to hepatitis B e antigen with and without active disease. J. Med. Virol. 44, 96–103. doi: 10.1002/jmv.1890440118
Hosseini, S. Y., Sanaei, N., Fattahi, M. R., Malek-Hosseini, S. A., and Sarvari, J. (2019). Association of HBsAg mutation patterns with hepatitis B infection outcome: Asymptomatic carriers versus HCC/cirrhotic patients. Ann. Hepatol. 18, 640–645. doi: 10.1016/j.aohep.2018.12.006
Huang, Y., Tong, S., Tai, A. W., Hussain, M., and Lok, A. S. F. (2011). Hepatitis B virus core promoter mutations contribute to hepatocarcinogenesis by deregulating SKP2 and its target, p21. Gastroenterology 141, 1412–1421.e5. doi: 10.1053/j.gastro.2011.06.048
Huang, F. Y., Wong, D. K. H., Seto, W. K., Zhang, A. Y., Lee, C. K., Lin, C. K., et al. (2014). Sequence variations of full-length hepatitis B virus genomes in Chinese patients with HBsAg-negative hepatitis B infection. PLoS One 9. doi: 10.1371/journal.pone.0115743
Ismail, A. M., Samuel, P., Eapen, C. E., Kannangai, R., and Abraham, P. (2011). Antiviral resistance mutations and genotype-associated amino acid substitutions in treatment-Naïve Hepatitis B virus-infected individuals from the Indian subcontinent. Intervirology 55, 36–44.
Jammeh, S., Tavner, F., Watson, R., Thomas, H. C., and Karayiannis, P. (2008). Effect of basal core promoter and pre-core mutations on hepatitis B virus replication. J. Gen. Virol. 89, 901–909. doi: 10.1099/vir.0.83468-0
Javanmard, D., Niya, M. H. K., Kalafkhany, D., Najafi, M., Ziaee, M., Babaei, M. R., et al. (2020). Downregulation of gsk3β and upregulation of urg7 in hepatitis b-related hepatocellular carcinoma. Hepat. Mon. 20, 1–11.
Kamiya, N. (2003). The mechanisms of action of antivirals against hepatitis B virus infection. J. Antimicrob. Chemother. 51, 1085–1089. doi: 10.1093/jac/dkg236
Karatayli, E., Karatayli, S. C., Cinar, K., Gokahmetoglu, S., Güven, K., Idilman, R., et al. (2012). Molecular characterization of a novel entecavir mutation pattern isolated from a multi-drug refractory patient with chronic hepatitis B infection. J. Clin. Virol. 53, 130–134. doi: 10.1016/j.jcv.2011.10.011
Kargar Kheirabad, A., Farshidfar, G., Nasrollaheian, S., and Gouklani, H. (2017). Prevalence and Characteristics of Precore Mutation in Iran and Its Correlation with Genotypes of Hepatitis B. Electron. Physician 9, 4114–4123. doi: 10.19082/4114
Katsoulidou, A., Paraskevis, D., Magiorkinis, E., Moschidis, Z., Haida, C., Hatzitheodorou, E., et al. (2009). Molecular characterization of occult hepatitis B cases in Greek blood donors. J. Med. Virol. 81, 815–825. doi: 10.1002/jmv.21499
Kim, J. H., Jung, Y. K., Joo, M. K., Kim, J. H., Yim, H. J., Park, J. J., et al. (2010). Hepatitis B viral surface mutations in patients with adefovir resistant chronic hepatitis B with A181T/V polymerase mutations. J. Korean Med. Sci. 25, 257–264. doi: 10.3346/jkms.2010.25.2.257
Kim, H., Lee, S. A., and Kim, B. J. (2016). X region mutations of hepatitis B virus related to clinical severity. World J. Gastroenterol. 22, 5467–5478. doi: 10.3748/wjg.v22.i24.5467
Kim, J. E., Lee, S. Y., Kim, H., Kim, K. J., Choe, W. H., and Kim, B. J. (2017). Naturally occurring mutations in the reverse transcriptase region of hepatitis B virus polymerase from treatment-naïve Korean patients infected with genotype C2. World J. Gastroenterol. 23, 4222–4232. doi: 10.3748/wjg.v23.i23.4222
Kim, H., Lee, S. A., Kim, D. W., Lee, S. H., and Kim, B. J. (2013). Naturally occurring mutations in large surface genes related to occult Infection of Hepatitis B virus Genotype C. Blackard J, editor. PLoS One 8:e54486. doi: 10.1371/journal.pone.0084194
Kitab, B., Essaid El Feydi, A., Afifi, R., Trepo, C., Benazzouz, M., Essamri, W., et al. Variability in the Precore and Core Promoter Regions of HBV Strains in Morocco: Characterization and Impact on Liver Disease Progression. R. Ray editor. PLoS One. (2012). 7:e42891, doi: 10.1371/journal.pone.0042891
Kobayashi, M., Arase, Y., Ikeda, K., Tsubota, A., Suzuki, Y., Saitoh, S., et al. (2003). Precore wild-type hepatitis B virus with G1896 in the resolution of persistent hepatitis B virus infection. Intervirology 46, 157–163. doi: 10.1159/000071456
Kumar, S., Stecher, G., Tamura, K., and Dudley, J. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets Downloaded from. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kwei, K., Tang, X., Lok, A. S., Sureau, C., Garcia, T., Li, J., et al. (2013). Impaired Virion secretion by Hepatitis B virus immune escape mutants and its rescue by wild-type envelope proteins or a second-site mutation. J. Virol. 87, 2352–2357. doi: 10.1128/JVI.02701-12
Lago, B. V., Mello, F. C., Ribas, F. S., Valente, F., Soares, C. C., Niel, C., et al. Analysis of Complete Nucleotide Sequences of Angolan Hepatitis B Virus Isolates Reveals the Existence of a Separate Lineage within Genotype E. I. A. Chemin, editor. PLoS One. (2014). 9:e92223, doi: 10.1371/journal.pone.0092223
Lazarevic, I., Banko, A., Miljanovic, D., and Cupic, M. (2019). Immune-escape hepatitis B virus mutations associated with viral reactivation upon immunosuppression. Viruses 11:778. doi: 10.3390/v11090778
Leng, X. H., Chen, E. Q., Du, L. Y., Bai, L., Gong, D. Y., Cheng, X., et al. (2015). Biological characteristics of the A1762T/G1764A mutant strain of hepatitis B virus in vivo. Mol. Med. Rep. 12, 5141–5148. doi: 10.3892/mmr.2015.4072
León, B., Taylor, L., Vargas, M., Luftig, R. B., Albertazzi, F., Herrero, L., et al. (2005). HBx M130K and V131I (T-A) mutations in HBV genotype F during a follow-up study in chronic carriers. Virol. J. 4:60
Li, M. W., Hou, W., Wo, J. E., and Liu, K. Z. (2005). Character of HBV (hepatitis B virus) polymerase gene rtM204V/I and rtL180M mutation in patients with lamivudine resistance. J. Zhejiang Univ. Sci. 6, 664–667. doi: 10.1631/jzus.2005.B0664
Li, H., Jia, J., Wang, M., Wang, H., Gu, X., Fang, M., et al. (2017). F221Y mutation in hepatitis B virus reverse transcriptase is associated with hepatocellular carcinoma prognosis following liver resection. Mol. Med. Rep. 15, 3292–3300. doi: 10.3892/mmr.2017.6362
Li, X. Y., Liang, C. H., Parkman, V., and Lv, Z. T. (2018). The association between TNF-a 238A/G and 308A/G polymorphisms and juvenile idiopathic arthritis: An updated PRISMA-compliant meta-analysis. Medicine 97. doi: 10.1097/MD.0000000000013964
Li, X. G., Liu, B. M., Xu, J., Liu, X. E., Ding, H., and Li, T. (2012). Discrepancy of potential antiviral resistance mutation profiles within the HBV reverse transcriptase between nucleos(t)ide analogue-untreated and -treated patients with chronic hepatitis B in a hospital in China. J. Med. Virol. 84, 207–216. doi: 10.1002/jmv.23182
Liao, Y., Hu, X., Chen, J., Cai, B., Tang, J., Ying, B., et al. (2012). Precore mutation of hepatitis B virus may contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. PLoS One 7. doi: 10.1371/journal.pone.0038394
Lin, X., Xu, X., Huang, Q. L., Liu, Y. Q., Zheng, D. L., Chen, W. N., et al. (2005). Biological impacts of “hot-spot” mutations of hepatitis B virus X proteins are genotype B and C differentiated. World J. Gastroenterol. 11, 4703–4708. doi: 10.3748/wjg.v11.i30.4703
Liu, B. M., Li, T., Xu, J., Li, X. G., Dong, J. P., Yan, P., et al. (2010). Characterization of potential antiviral resistance mutations in hepatitis B virus reverse transcriptase sequences in treatment-naïve Chinese patients. Antiviral Res. 85, 512–519. doi: 10.1016/j.antiviral.2009.12.006
Liu, S., Zhang, H., Gu, C., Yin, J., He, Y., Xie, J., et al. (2009). Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J. Natl. Cancer Inst. 101, 1066–1082.
Locarnini, S., and Mason, W. S. (2006). Cellular and virological mechanisms of HBV drug resistance. J. Hepatol. 44, 422–431. doi: 10.1016/j.jhep.2005.11.036
Locarnini, S., McMillan, J., and Bartholomeusz, A. (2003). The hepatitis B virus and common mutants. Semin. Liver Dis. 23, 5–20.
Mahabadi, M., Norouzi, M., Alavian, S. M., Samimirad, K., Azad, T. M., Saberfar, E., et al. (2013). Drug-related mutational patterns in hepatitis B virus (HBV) reverse transcriptase proteins from Iranian treatment-naïve chronic HBV patients. Hepat. Mon. 13. doi: 10.5812/hepatmon.6712
Mani, M., Vijayaraghavan, S., Sarangan, G., Barani, R., Abraham, P., and Srikanth, P. (2019). Hepatitis B virus X protein: the X factor in chronic hepatitis B virus disease progression. Indian J. Med. Microbiol. doi: 10.4103/ijmm.IJMM_19_421
Meldal, B. H. M., Moula, N. M., Barnes, I. H. A., Boukef, K., and Allain, J. P. (2009). A novel hepatitis B virus subgenotype, D7, in Tunisian blood donors. J. Gen. Virol. 90, 1622–1628. doi: 10.1099/vir.0.009738-0
Moerman, B., Moons, V., Sommer, H., Schmitt, Y., and Stetter, M. (2004). Evaluation of sensitivity for wild type and mutant forms of hepatitis B surface antigen by four commercial HBsAg assays. Clin. Lab. 50, 159–162.
Mohamadkhani, A., Montazeri, G., and Poustchi, H. (2011). The importance of hepatitis B virus genome diversity in basal core promoter region. Middle East J. Dig. Dis. 3, 13–19.
Mokaya, J., Maponga, T. G., McNaughton, A. L., van Schalkwyk, M., Hugo, S., Singer, J. B., et al. (2020). Evidence of tenofovir resistance in chronic hepatitis B virus (HBV) infection: An observational case series of South African adults. J. Clin. Virol. 129. doi: 10.1016/j.jcv.2020.104548
Mokaya, J., McNaughton, A., Bester, P., Goedhals, D., Barnes, E., Marsden, B., et al. (2019). Hepatitis B virus resistance to tenofovir: fact or fiction? A synthesis of the evidence to date. medRxiv [Preprint].
Mokaya, J., Mcnaughton, A. L., Bester, P. A., Goedhals, D., Barnes, E., Marsden, B. D., et al. (2020). Hepatitis B virus resistance to tenofovir: fact or fiction? A systematic literature review and structural analysis of drug resistance mechanisms [version 1; peer review: awaiting peer review].
Mondal, R. K., Khatun, M., Ghosh, S., Banerjee, P., Datta, S., Sarkar, S., et al. (2015). Immune-driven adaptation of hepatitis B virus genotype D involves preferential alteration in B-cellepitopes and replicative attenuation-an insight from human immunodeficiency virus/hepatitis B virus coinfection. Clin. Microbiol. Infect. 21, 710.e11–710.e20. doi: 10.1016/j.cmi.2015.03.004
Moriconi, F., Colombatto, P., Coco, B., Ciccorossi, P., Oliveri, F., Flichman, D., et al. (2007). Emergence of hepatitis B virus quasispecies with lower susceptibility to nucleos(t)ide analogues during lamivudine treatment. J. Antimicrob. Chemother. 60, 341–349. doi: 10.1093/jac/dkm187
Mukaide, M., Tanaka, Y., Shin-I, T., Yuen, M. F., Kurbanov, F., Yokosuka, O., et al. (2010). Mechanism of entecavir resistance of hepatitis B virus with viral breakthrough as determined by long-term clinical assessment and molecular docking simulation. Antimicrob. Agents Chemother. 54, 882–889. doi: 10.1128/AAC.01061-09
Ng, L. F. P., Chan, M., Chan, S. H., Cheng, P. C. P., Leung, E. H. C., Chen, W. N., et al. (2005). Host heterogeneous ribonucleoprotein K (hnRNP K) as a potential target to suppress hepatitis B virus replication. PLoS Med. 2, 0673–0683.
Nishizawa, T., Hoshino, T., Naganuma, A., Kobayashi, T., Nagashima, S., Takahashi, M., et al. (2016). Enhanced pregenomic RNA levels and lowered precore mRNA transcription efficiency in a genotype A hepatitis B virus genome with C1766T and T1768A mutations obtained from a fulminant hepatitis patient. J. Gen. Virol. 97, 2643–2656. doi: 10.1099/jgv.0.000566
Olinger, C. M., Weber, B., Otegbayo, J. A., Ammerlaan, W., Van Der Taelem-Brulé, N., and Muller, C. P. (2007). Hepatitis B virus genotype E surface antigen detection with different immunoassays and diagnostic impact of mutations in the preS/S gene. Med. Microbiol. Immunol. 196, 247–252. doi: 10.1007/s00430-007-0050-5
Olusola, B. A., Faneye, A. O., Oluwasemowo, O. O., Motayo, B. O., Adebayo, S., Oludiran-Ayoade, A. E., et al. (2021). Profiles of mutations in hepatitis B virus surface and polymerase genes isolated from treatment-naïve Nigerians infected with genotype E. J. Med. Microbiol. 70. doi: 10.1099/jmm.0.001338
Ouneissa, R., Bahri, O., Alaya-Bouafif, N., Ben Chouaieb, S., Yahia, A., Ben Sadraoui, A., et al. (2012). Frequency and clinical significance of core promoter and precore region mutations in Tunisian patients infected chronically with hepatitis B. J. Med. Virol. 84, 1719–1726. doi: 10.1002/jmv.23394
Ouneissa, R., Bahri, O., Ben Yahia, A., Touzi, H., Msaddak Azouz, M., Ben Mami, N., et al. (2013). Evaluation of PCR-RFLP in the Pre-S Region as Molecular Method for Hepatitis B Virus Genotyping. Hepat. Mon. 13. doi: 10.5812/hepatmon.11781
Pahal, V., Singh, J., and Dadhich, K. S. (2016). Hepatitis B virus core promoter and precore mutations and their relatedness to genotypes and disease pathogenesis. Pelagia Res. Lib. Adv. Appl. Sci. Res. 7, 70–80.
Parekh, S., Zoulim, F., Ahn, S. H., Tsai, A., Li, J., Kawai, S., et al. (2003). Genome replication, Virion secretion, and e antigen expression of naturally occurring Hepatitis B virus Core promoter mutants. J. Virol. 77, 6601–6612. doi: 10.1128/JVI.77.12.6601-6612.2003
Park, E. S., Lee, A. R., Kim, D. H., Lee, J. H., Yoo, J. J., Ahn, S. H., et al. (2019). Identification of a quadruple mutation that confers tenofovir resistance in chronic hepatitis B patients. J. Hepatol. 70, 1093–1102. doi: 10.1016/j.jhep.2019.02.006
Pollicino, T., Isgrò, G., di Stefano, R., Ferraro, D., Maimone, S., Brancatelli, S., et al. (2009). Variability of reverse transcriptase and overlapping S gene in hepatitis B virus isolates from untreated and lamivudine-resistant chronic hepatitis B patients. Antivir. Ther. 14, 649–654. doi: 10.1177/135965350901400504
Pollicino, T., Raffa, G., Costantino, L., Lisa, A., Campello, C., Squadrito, G., et al. (2007). Molecular and functional analysis of occult hepatitis B virus isolates from patients with hepatocellular carcinoma. Hepatology 45, 277–285. doi: 10.1002/hep.21529
Pourkarim, M. R., Vergote, V., Amini-Bavil-Olyaee, S., Sharifi, Z., Sijmons, S., Lemey, P., et al. (2014). Molecular characterization of hepatitis B virus (HBV) strains circulating in the northern coast of the Persian Gulf and its comparison with worldwide distribution of HBV subgenotype D1. J. Med. Virol. 86, 745–757. doi: 10.1002/jmv.23864
Poustchi, H., Mohamadkhani, A., Bowden, S., Montazeri, G., Ayres, A., Revill, P., et al. (2008). Clinical significance of precore and core promoter mutations in genotype D hepatitis B-related chronic liver disease. J. Viral Hepat. 15, 753–760. doi: 10.1111/j.1365-2893.2008.00998.x
Purdy, M. A., Talekar, G., Swenson, P., Araujo, A., and Fields, H. (2006). A new algorithm for deduction of hepatitis B surface antigen subtype determinants from the amino acid sequence. Intervirology 50, 45–51.
Putri, W. A., Yano, Y., Yamani, L. N., Lusida, M. I., Soetjipto, L. Y., et al. (2019). Association between HBx variations and development of severe liver disease among indonesian patients. Kobe J. Med. Sci. 65, 28–35.
Qin, B., Pei, R. J., He, T. T., Huang, Z. H., Pan, G. S., Tu, C. Y., et al. (2013a). Polymerase mutations rtN238R, rtT240Y and rtN248H of hepatitis B virus decrease susceptibility to adefovir. Chin. Sci. Bull. 58, 1760–1766. doi: 10.1007/s11434-013-5770-x
Qin, B., Zhang, B., Zhang, X., He, T., Xu, W., Fu, L., et al. (2013b). Substitution Rtq267h of hepatitis B virus increases the weight of replication and lamivudine resistance. Hepat. Mon. 13:12160. doi: 10.5812/hepatmon.12160
Quarleri, J. (2014). Core promoter: a critical region where the hepatitis B virus makes decisions. World J. Gastroenterol. 20, 425–435. doi: 10.3748/wjg.v20.i2.425
Quiros-Roldan, E., Calabresi, A., Lapadula, G., Tirelli, V., Costarelli, S., Cologni, G., et al. (2008). Evidence of long-term suppression of hepatitis B virus DNA by tenofovir as rescue treatment in patients coinfected by HIV. Antivir. Ther. 13, 341–348. doi: 10.1177/135965350801300315
Rajoriya, N., Combet, C., Zoulim, F., and Janssen, H. L. A. (2017). How viral genetic variants and genotypes influence disease and treatment outcome of chronic hepatitis B. Time for an individualised approach? J. Hepatol. 67, 1281–1297. doi: 10.1016/j.jhep.2017.07.011
Rendon, J. C., Cortes-Mancera, F., Restrepo-Gutierrez, J. C., and Hoyos, S. (2017). Navas MC. Molecular characterization of occult hepatitis B virus infection in patients with end-stage liver disease in Colombia. PLoS One 12:e0180447. doi: 10.1371/journal.pone.0180447
Saha, D., Pal, A., Biswas, A., Panigrahi, R., Sarkar, N., Das, D., et al. (2014). Molecular characterization of HBV strains circulating among the treatment-naive HIV/HBV co-infected patients of eastern India. PLoS One 9. doi: 10.1371/journal.pone.0090432
Salarneia, F., Zhand, S., Khodabakhshi, B., Tabarraei, A., Vakili, M. A., Javid, N., et al. (2016). Mutations at nucleotide 1762, 1764 and 1766 of Hepatitis B virus X gene in patients with chronic Hepatitis B and Hepatitis B-related cirrhosis. Med. Lab. J 10, 31–35.
Schildgen, O. (2007). Novel lamivudine resistance [3]. Antimicrob. Agents Chemother. 51:4533. doi: 10.1128/AAC.00840-07
Sendi, H., Mehrab-Mohseni, M., Zali, M. R., Norder, H., and Magnius, L. O. (2005). T1764G1766 core promoter double mutants are restricted to Hepatitis B virus strains with an A1757 and are common in genotype D. J. Gen. Virol. 86, 2451–2458. doi: 10.1099/vir.0.81023-0
Shaw, T., Bartholomeusz, A., and Locarnini, S. (2006). HBV drug resistance: mechanisms, detection and interpretation. J. Hepatol. 44, 593–606. doi: 10.1016/j.jhep.2006.01.001
Sheldon, J. (2008). Soriano V. Hepatitis B virus escape mutants induced by antiviral therapy. J. Antimicrob. Chemother. 61, 766–768. doi: 10.1093/jac/dkn014
Sheldon, J., Rodès, B., Zoulim, F., Bartholomeusz, A., and Soriano, V. (2006). Mutations affecting the replication capacity of the hepatitis B virus [Internet]. J. Viral Hepat. 13, 427–434. doi: 10.1111/j.1365-2893.2005.00713.x
Sitnik, R., Pinho, J. R. R., Bertolini, D. A., Bernardini, A. P., da Silva, L. C., and Carrilho, F. J. (2004). Hepatitis B virus genotypes and precore and core mutants in Brazilian patients. J. Clin. Microbiol. 42, 2455–2460. doi: 10.1128/JCM.42.6.2455-2460.2004
Sominskaya, I., Mihailova, M., Jansons, J., Legzdina, D., Sudmale, G., Pumpens, P., et al. (2011). Hepatitis B virus genotypes in Latvia. Open Hepatol. J. 3. doi: 10.2174/1876517301103010007
Sunbul, M. (2014). Hepatitis B virus genotypes: global distribution and clinical importance. World J. Gastroenterol. 20:5427. doi: 10.3748/wjg.v20.i18.5427
Suntur, B. M., Sayan, M., Kaya, H., and Ünal, N. (2019). Overlapping Pol/S gene analysis in chronic hepatitis B patients with coexisting Hbsag and Anti-HBs. Acta Medica Mediterranea. 35:3113
Suppiah, J., Zain, R. M., Bahari, N., Nawi, S. H., and Saat, Z. (2015). G1896A precore mutation and association with HBeAG status, genotype and clinical status in patients with chronic hepatitis B. Hepat. Mon. 15. doi: 10.5812/hepatmon.31490
Taghiabadi, M., Hosseini, S. Y., Gorzin, A. A., Taghavi, S. A., Monavari, S. H. R., and Sarvari, J. (2019). Comparison of pre-S1/S2 variations of hepatitis B virus between asymptomatic carriers and cirrhotic/hepatocellular carcinoma-affected individuals. Clin Exp Hepatol. 5, 161–168. doi: 10.5114/ceh.2019.84781
Tenney, D. J., Levine, S. M., Rose, R. E., Walsh, A. W., Weinheimer, S. P., Discotto, L., et al. (2004). Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to lamivudine. Antimicrob. Agents Chemother. 48, 3498–3507. doi: 10.1128/AAC.48.9.3498-3507.2004
Thompson, A. J. V., Nguyen, T., Iser, D., Ayres, A., Jackson, K., Littlejohn, M., et al. (2010). Serum hepatitis B surface antigen and hepatitis B e antigen titers: Disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology 51, 1933–1944. doi: 10.1002/hep.23571
Tokgöz, Y., Terlemez, S., Sayan, M., and Kırdar, S. (2018). Investigation of antiviral resistance and escape mutations in children with naive chronic hepatitis B patients and their parents. Turk. J. Pediatr. 60, 514–519. doi: 10.24953/turkjped.2018.05.007
Tong, M. J., Blatt, L. M., Kao, J. H., Cheng, J. T., and Corey, W. G. (2007). Basal core promoter T1762/A1764 and precore A1896 gene mutations in hepatitis B surface antigen-positive hepatocellular carcinoma: a comparison with chronic carriers. Liver Int. 27, 1356–1363. doi: 10.1111/j.1478-3231.2007.01585.x
Tong, S., Kim, K. H., Chante, C., Wands, J., and Li, J. (2005). Hepatitis B virus e antigen variants. Int. J. Med. Sci. 2, 2–7. doi: 10.7150/ijms.2.2
Tong, S., Li, J., Wands, J. R., and Wen, Y. (2013). Hepatitis B virus genetic variants: biological properties and clinical implications. Emerg. Microbes Infect. 2, 1–11.
Torresi, J., Earnest-Silveira, L., Civitico, G., Walters, T. E., Lewin, S. R., Fyfe, J., et al. (2002a). Restoration of replication phenotype of lamivudine-resistant hepatitis B virus mutants by compensatory changes in the “fingers” subdomain of the viral polymerase selected as a consequence of mutations in the overlapping S gene. Virology 299, 88–99. doi: 10.1006/viro.2002.1448
Torresi, J., Earnest-Silveira, L., Deliyannis, G., Edgtton, K., Zhuang, H., Locarnini, S. A., et al. (2002b). Reduced antigenicity of the Hepatitis B virus HBsAg protein arising as a consequence of sequence changes in the overlapping polymerase gene that are selected by lamivudine therapy. Virology 293, 305–313. doi: 10.1006/viro.2001.1246
Triki, H., Ben Slimane, S., Ben Mami, N., Sakka, T., Ben Ammar, A., and Dellagi, K. (2000). High circulation of hepatitis B virus (HBV) precore mutants in Tunisia, North Africa. Epidemiol. Infect. 125, 169–174. doi: 10.1017/S0950268899003921
Villet, S., Ollivet, A., Pichoud, C., Barraud, L., Villeneuve, J. P., Trépo, C., et al. (2007). Stepwise process for the development of entecavir resistance in a chronic hepatitis B virus infected patient. J. Hepatol. 46, 531–538. doi: 10.1016/j.jhep.2006.11.016
Wang, L., Han, F., Duan, H., Ji, F., Yan, X., Fan, Y., et al. (2017). Hepatitis B virus pre-existing drug resistant mutation is related to the genotype and disease progression. J. Infect. Dev. Ctries. 11, 727–732. doi: 10.3855/jidc.9021
Wang, Y., Zeng, L., and Chen, W. (2016). HBV X gene point mutations are associated with the risk of hepatocellular carcinoma: A systematic review and meta-analysis. Mol. Clin. Oncol. 4, 1045–1051. doi: 10.3892/mco.2016.847
Wang, Q., Zhang, T., Ye, L., Wang, W., and Zhang, X. (2012). Analysis of hepatitis B virus X gene (HBx) mutants in tissues of patients suffered from hepatocellular carcinoma in China. Cancer Epidemiol. 36, 369–374. doi: 10.1016/j.canep.2011.11.006
Wei, C., Yu-tian, C., Ji-zhi, W., Yong-wei, L., and Gang, L. (2011). Characterization of hepatitis virus B isolated from a multi-drug refractory patient. Virus Res. 155, 254–258. doi: 10.1016/j.virusres.2010.10.018
Westland, C. (2003). Week 48 resistance surveillance in two phase 3 clinical studies of adefovir dipivoxil for chronic hepatitis B. Hepatology 38, 96–103. doi: 10.1053/jhep.2003.50288
WHO (2021). Hepatitis B WHO guidlines. Available at: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (Accessed April 25, 2022).
Wu, Y., Gan, Y., Gao, F., Zhao, Z., Jin, Y., Zhu, Y., et al. (2014). Novel natural mutations in the Hepatitis B virus reverse transcriptase domain associated with hepatocellular carcinoma. PLoS One 9. doi: 10.1371/journal.pone.0115141
Wu, J. F., Ni, Y. H., Chen, H. L., Hsu, H. Y., and Chang, M. H. (2014). The impact of hepatitis B virus precore/core gene carboxyl terminal mutations on viral biosynthesis and the host immune response. J Infect Dis 209, 1374–1381. doi: 10.1093/infdis/jit638
Wungu, C. D. K., Amin, M., Ruslan, S. E. N., Purwono, P. B., Kholili, U., Maimunah, U., et al. (2019). Association between host TNF-α, TGF-β1, p53 polymorphisms, hbv x gene mutation, hbv viral load and the progression of hbv-associated chronic liver disease in indonesian patients. Biomed. Rep. 11, 145–153. doi: 10.3892/br.2019.1239
Xu, R., Zhang, X., Zhang, W., Fang, Y., Zheng, S., and Yu, X. F. (2007). Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology 46, 1810–1820. doi: 10.1002/hep.21893
Yamani, L. N., Yano, Y., Utsumi, T., Wasityastuti, W., Rinonce, H. T., Widasari, D. I., et al. (2017). Profile of mutations in the reverse transcriptase and overlapping surface genes of hepatitis B virus (HBV) in treatment-naïve indonesian HBV carriers. Jpn. J. Infect. Dis. 70, 647–655. doi: 10.7883/yoken.JJID.2017.078
Yan, L., Zhang, H., Ma, H., Liu, D., Li, W., Kang, Y., et al. (2015). Deep sequencing of hepatitis B virus basal core promoter and precore mutants in HBeAg-positive chronic hepatitis B patients. Sci. Rep. 9:5
Yang, H., Qi, X., Sabogal, A., Miller, M., Xiong, S., and Delaney, W. E. (2005). Cross-resistance testing of next-generation nucleoside and nucleotide analogues against lamivudine-resistant HBV. Antivir. Ther. 1359
Yang, H., Westland, C. E., Delaney, W. E., Heathcote, E. J., Ho, V., Fry, J., et al. (2002). Resistance surveillance in chronic hepatitis B patients treated with adefovir dipivoxil for up to 60 weeks. Hepatology 36, 464–473. doi: 10.1053/jhep.2002.34740
Yin, J., Xie, J., Liu, S., Zhang, H., Han, L., Lu, W., et al. (2011). Association between the various mutations in viral core promoter region to different stages of hepatitis B, ranging of asymptomatic carrier state to hepatocellular carcinoma. Am. J. Gastroenterol. 106, 81–92. doi: 10.1038/ajg.2010.399
Yuan, J. M., Ambinder, A., Fan, Y., Gao, Y. T., Yu, M. C., and Groopman, J. D. (2009). Prospective evaluation of hepatitis B 1762 T/1764 a mutations on hepatocellular carcinoma development in Shanghai, China. Cancer Epidemiol. Biomarkers Prev. 18, 590–594. doi: 10.1158/1055-9965.EPI-08-0966
Zheng, J., Zeng, Z., Zhang, D., Yu, Y., Wang, F., and Pan, C. Q. (2012). Prevalence and significance of Hepatitis B reverse transcriptase mutants in different disease stages of untreated patients. Liver Int. 32, 1535–1542. doi: 10.1111/j.1478-3231.2012.02859.x
Zhu, R., Zhang, H. P., Yu, H., Li, H., Ling, Y. Q., Hu, X. Q., et al. (2008). Hepatitis B virus mutations associated with in situ expression of hepatitis B core antigen, viral load and prognosis in chronic hepatitis B patients. Pathol. Res. Pract. 204, 731–742. doi: 10.1016/j.prp.2008.05.001
Keywords: HBV, antiviral resistance, liver cirrhosis, PCR, whole genome, Sanger sequencing, hepatocellular carcinoma
Citation: Belaiba Z, Ayouni K, Gdoura M, Kammoun Rebai W, Touzi H, Sadraoui A, Hammemi W, Yacoubi L, Abdelati S, Hamzaoui L, Msaddak Azzouz M, Chouikha A and Triki H (2022) Whole genome analysis of hepatitis B virus before and during long-term therapy in chronic infected patients: Molecular characterization, impact on treatment and liver disease progression. Front. Microbiol. 13:1020147. doi: 10.3389/fmicb.2022.1020147
Edited by:
Antoinette Van Der Kuyl, University of Amsterdam, NetherlandsReviewed by:
Jose A. Usme-Ciro, Universidad Cooperativa de Colombia, ColombiaLimin Chen, Chinese Academy of Medical Sciences and Peking Union Medical College, China
Copyright © 2022 Belaiba, Ayouni, Gdoura, Kammoun Rebai, Touzi, Sadraoui, Hammemi, Yacoubi, Abdelati, Hamzaoui, Msaddak Azzouz, Chouikha and Triki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zeineb Belaiba, emVpbmViLmJlbGFpYmFAcGFzdGV1ci51dG0udG4=; Anissa Chouikha, Y2hvdWlraGFhbmlzc2FAZ21haWwuY29t
†ORCID: Zeineb Belaiba, https://orcid.org/0000-0001-5187-0813
Anissa Chouikha, https://orcid.org/0000-0001-5616-0204
 Zeineb Belaiba
Zeineb Belaiba Kaouther Ayouni
Kaouther Ayouni Mariem Gdoura
Mariem Gdoura Wafa Kammoun Rebai
Wafa Kammoun Rebai Henda Touzi1
Henda Touzi1 Anissa Chouikha
Anissa Chouikha