
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 26 September 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1019069
This article is part of the Research TopicApoplastic Effectors ― What Roles Do They Play In Plant-Pathogen Interactions?View all 7 articles
Plants are constantly exposed to diverse microbes and thus develop a sophisticated perceive system to distinguish non-self from self and identify non-self as friends or foes. Plants can detect microbes in apoplast via recognition of microbe-associated molecular patterns (MAMPs) by pattern recognition receptors (PRRs) on the cell surface to activate appropriate signaling in response to microbes. MAMPs are highly conserved but essential molecules of microbes and often buried in microbes’ complex structure. Mature MAMPs are released from microbes by invasion-induced hydrolytic enzymes in apoplast and accumulate in proximity of plasma membrane-localized PRRs to be perceived as ligands to activate downstream signaling. In response, microbes developed strategies to counteract these processing. Here, we review how the form, the concentration, and the size of mature MAMPs affect the PRR-mediated immune signaling. In particular, we describe some potential applications and explore potential open questions in the fields.
One of the most important evolutionary events in the history of life on earth is advent of the earliest land plants around 480 million years ago (Gehrig et al., 1996). During their establishment in terrestrial ecosystems, land plants have to adapt to an environment that houses a large variety of microbes such as fungi, oomycetes, bacteria, and viruses. From then on, if not earlier, plants and microbes have had continual interactions, influencing the evolution of both plants and microbes (Wang et al., 2010). To respond appropriately to such diverse microbes, plants have evolved abilities to distinguish self from non-self (Medzhitov and JanewayJr., 2002) and further identify non-self as friends or foes (Antolin-Llovera et al., 2014) through a sophisticated immune system. With these distinctions and identifications, plants can adapt to their environment by either activating immune responses to defend against pathogens or initiating symbiosis signaling to facilitate the accommodation of symbionts (Antolin-Llovera et al., 2014).
When microbes enter the apoplast through nature entries or wounded sites of plants, plants can detect microbes in apoplast via the recognition of Microbe/Pathogen-Associated Molecular Patterns (MAMPs/PAMPs; hereafter, referred to as MAMPs) by plant cell-surface localized Pattern Recognition Receptors (PRRs) (Macho and Zipfel, 2014) and activate appropriate downstream responses. The major classes of MAMPs include proteins, carbohydrates, lipids and nucleic acids, with common features: highly conserved structures, important functions for microbe survival and absence from the host plants (Nurnberger et al., 2004; Boutrot and Zipfel, 2017; Buscaill and van der Hoorn, 2021).
Plant PRRs are receptor-like kinases (RLKs) or receptor-like proteins (RLPs) that carry the extracellular leucine-rich repeat (LRR) or lysine motif (LysM) domain to confer the recognition of MAMPs (Buscaill and van der Hoorn, 2021). These MAMPs serve as the ligands for PRRs, while the binding specificity of ligand-receptor pairs is determined by both ligands (Felix et al., 1999) and PRR receptors (Gomez-Gomez and Boller, 2000). Such binding often induces the oligomerization of receptors (Liu et al., 2012) and their interaction with co-receptors, which is often required for the subsequent signal transduction (Shan et al., 2008; Liebrand et al., 2013). In addition to these typical MAMPs, some MAMPs (such as cerebroside) do not need a short ligand for recognizing (Koga et al., 1998). In general, effective physiological concentrations of ligands are required for their biological activities in plants (Albert et al., 2015).
The recognition of MAMPs by PRRs depends on both sides of MAMPs and PRRs; here we only focus on the MAMPs side to discuss how the form, the concentrations and the size of the MAMPs affect their recognitions and thus subsequent immune responses in host plants.
In higher plants, nearly all cells are connected directly or indirectly by plasmodesmata into a single “organism” named as symplast and the space outside this symplast is known as apoplast, including the cell wall and the aqueous intercellular space (Erickson, 1986).
In the scenario of plant-microbe interactions, the apoplast is the extracellular space in plant tissue that constitutes a source of nutrients and shelter for microbial inhabitants (Sattelmacher, 2001). Meanwhile, the apoplast is a hostile environment that contains hydrolytic enzymes and toxins for microbes (Sanchez-Vallet et al., 2015). Thus, the apoplast is defined as compartments of intracellular space beyond the plant plasma membrane, including the cell wall and structures that are formed during plant-microbe interactions (Figure 1). For example, fungal invasion forms a specialized structure known as appressorium. Further, invasive hyphae or haustoria are surrounded by a host-derived specialized membrane outside the invasive structure, known as the extrainvasive hyphal membrane (EIHM) (Kankanala et al., 2007), extrahaustorial membrane (EHM) (Karasov et al., 2017), or periarbuscular membrane (PAM) (Ivanov et al., 2019). The space between the microbe plasma membrane and the host extended membrane is also part of the apoplast (Wang et al., 2020; Figure 1).
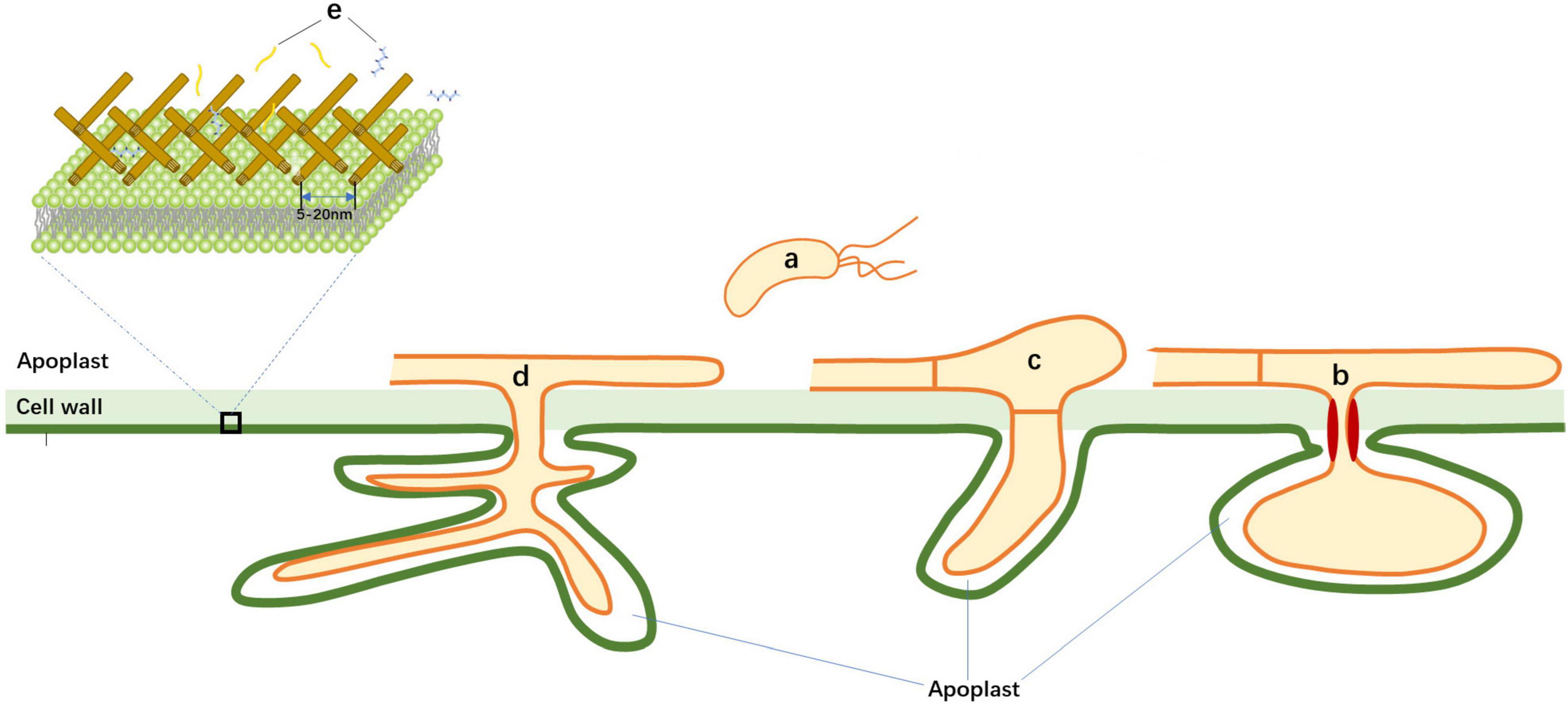
Figure 1. Schematic illustration of plant-microbe interface during invasion of pathogens. During invasion of microbes, bacterial, fungal, and oomycete pathogens colonize in apoplast space. Fungus/oomycete forms specialized structures known as the extrainvasive hyphal membrane (EIHM), the extrahaustorial membrane (EHM), or periarbuscular membrane (PAM). Mature microbe-associated molecular patterns (MAMPs) are derived from microbes and move through cell wall pores to be perceived by plasma-membrane localized Pattern Recognition Receptors (PRRs). (a) Bacterial pathogens; (b) Haustorial-forming fungal pathogens; (c) Non-haustorial-forming fungal pathogens; (d) Arbuscular mycorrhiza; and (e) Mature MAMPs.
Apoplast is a place where MAMPs matured from microbial complex. Apoplast constitutes a major battleground for plant-microbe interactions. Plants perceive microbial pattern via PRRs in apoplast to activate immune responses described as pattern-triggered immunity (PTI) (Saijo et al., 2018) while microbes develop extracellular strategies to avoid patterns recognition (Buscaill and van der Hoorn, 2021). Thus, the outcome of the apoplastic battle between plant and microbes will determine whether plants can stop invasion of microbes during early-stage infection.
Currently, the majority of known MAMPs are proteins, carbohydrates, lipids, and nucleic acids from bacteria, fungi, or oomycetes (see Table 1 and recent reviews for updated MAMPs and PRRs in Boutrot and Zipfel, 2017; Wang et al., 2020; Buscaill and van der Hoorn, 2021). Plant PRRs can perceive multiple MAMPs that are derived from flagellin, elongation factor Tu, peptidoglycans and lipopolysaccharides in bacteria, chitin in fungi, and Nep1-like Protein in oomycetes (Zipfel, 2014; Albert et al., 2015; Boutrot and Zipfel, 2017).
The right form of MAMPs is necessary for the recognition by PRRs and effective activation of immune responses in plants. The PRRs can recognize and bind with the small epitopes of MAMPs as ligands. For a certain host PRR, the specificity and affinity of ligand binding to the PRR depends on the proper form of MAMP, such as its sequence variants and modifications (Nurnberger et al., 1994; Chinchilla et al., 2006).
Microbe-associated molecular patterns are highly conserved patterns in microbes and maintain essential function for the microbe’s survival and therefore difficult to alter (Janeway, 1989; Medzhitov, 2007). Flg22 is one of the well-studied MAMPs derived from the bacterial flagellin (Felix et al., 1999; Zipfel et al., 2004). Flg22 is a highly conserved 22-amino acid of the N-terminal domain of bacterial flagellin (Table 1; Felix et al., 1999; Zipfel et al., 2004), and can be recognized by Flagellin Sensing 2 (FLS2) (Gomez-Gomez and Boller, 2000) to trigger immune responses.
However, some microbes can avoid the detection by PRRs through mutating sequence of MAMPs. Mutations in flg22 that are not recognized by FLS2 was reported in some flagellated bacteria (Cheng et al., 2021). An α-proteobacterium Agrobacterium carrying a flg22 variant does not trigger FLS2-mediated immune responses (Felix et al., 1999). With different versions of the flg22, some strains of the bacterial Ralstonia solanacearum and Xanthomonas campestris pv. campestris render them undetectable for FLS2-mediated immune system in host plants (Pfund et al., 2004; Sun et al., 2006). Likely, Xanthomonas oryzae pv. oryzae (Xoo) and pv. oryzicola (Xoc) can escape FLS2-mediated detection in rice due to substitutions in flg22 sequence (Wang et al., 2015). Further, X. arboricola pv. juglandis with the conserved flg22 sequence is non-pathogenic but strains with mutation within the flg22 sequence that can evade the recognition by FLS2 is pathogenic (Cesbron et al., 2015). The specific recognition of MAMPs relies on the interaction between the extracellular domain of PRRs and MAMPs, while the induction of immune responses in plant cells is determined by the activation of the intracellular domain of PRRs, the kinase domain. Interestingly, chimeric FLS2 receptors with partially swapped extracellular domains between the AtFLS2 and SlFLS2, could recognize both flg22 in Arabidopsis and flg15 in tomato (Mueller et al., 2012). However, whether a single native/chimeric PRR could recognize various MAMPs and mediate broad pathogen resistances in plants still requires further studies.
Besides flg22, elf18 is another well-studied MAMPs which contain the first 18 amino acids of N-terminus of the highly conserved bacterial protein Elongation factor (Table 1; Kunze et al., 2004), and can be recognized by the EF-Tu Receptor (EFR) to activate defense responses (Zipfel et al., 2006). Mutations in elf18 can also affect the immune activity in plants. The elf18 variants of X. campestris pv. campestris (Xcc) and Pseudomonas syringae pv. tomato DC3000 (PtoDC3000) trigger only 0.8–3.2% of the immune activity in comparison to that from Agrobacterium, Ralstonia, and other Xanthomonas and Pseudomonas strains (Lacombe et al., 2010).
Another example of MAMP with sequence variant is RaxX, a highly conserved protein in many pathogenic Xanthomonas species. RaxX is recognized by the rice receptor kinase XA21 to confer resistance to most strains of Xoo (Table 1; Wang et al., 1996; Pruitt et al., 2015; Luu et al., 2019). However, Xoo isolates with substitutions of the Y41 residue in RaxX can avoid XA21-mediated defense (Pruitt et al., 2015).
Modification of the MAMPs is another strategy to render them undetectable by plant PRRs and avoid host resistance.
Chitin is a structural unit of the fungal cell walls (Table 1) and chitin fragments are recognized as MAMPs by cell surface receptor Chitin Elicitor Receptor Kinase1 (CERK1) and Chitin Elicitor-Binding Protein (CEBiP) in plants (Kaku et al., 2006; Miya et al., 2007; Wan et al., 2008). Deacetylating chitin into chitosan prevents the recognition of chitin by receptors to evade host immunity (Vander et al., 1998). For example, fungal pathogens Verticillium dahliae and Fusarium oxysporum f. sp. vasinfectum secrete polysaccharide deacetylase1 (PDA1) to deacetylate chitin to prevent chitin-triggered immunity in cotton (Gao et al., 2019). The wheat stripe rust fungus Puccinia striiformis f. sp. tritici, the broad bean rust fungus Uromyces fabae, and the maize anthracnose fungus Colletotrichum graminicola also replace chitin with chitosan in their hyphae to avoid immunity (El Gueddari et al., 2002). In addition, it was reported that chitin deacetylases in the endophytic fungus Pestalotiopsis sp. are induced to avoid the recognition by plants (Cord-Landwehr et al., 2016).
Lipopolysaccharides (LPSs) are the major component of the outer membrane of Gram-negative bacteria (Table 1). LPS consists of lipid A and an oligosaccharide core that carries an O-polysaccharide (OPS) (Dow et al., 2000). LPS can be sensed by plant receptor-like kinase LORE and induce immune responses (Ranf et al., 2015). It was found that the pathogenic bacterium Xylella fastidiosa with an O-antigen chain delays the recognition by host plants while mutants of O-antigens strains induce faster immune responses, suggesting that glycan modification of LPS may affect plant immunogenic recognition (Rapicavoli et al., 2018).
There is an ongoing arm race between the immunorecognition by PRRs in host plants and the evasion of immunorecognition by pathogens. The race drives adaptive evolution of MAMPs in form to evade immunorecognition, though MAMPs are highly conserved patterns which are essential for microbes. Adaptive evolutions of MAMPs in sequences and modifications render them undetectable by PRRs and evade immunorecognition; nine different extracellular strategies have been recently summarized on how microbes avoid recognition by the host (Buscaill and van der Hoorn, 2021). In response to this, plants also evolve new versions of PRRs which can recognize these new forms of the MAMPs (Mueller et al., 2012).
In summary, immunogenic recognition of MAMPs by plant PRRs, which is determined by the specificity and affinity of ligand binding to PRRs, requires the proper form of MAMPs with right sequences and modifications.
Plants recognize MAMPs in a dose-dependent manner (Albert et al., 2015), and thus the effective activation of immune responses requires enough physiological concentration of the proper MAMPs at the infection site. The exact physiological concentration of MAMPs is difficult to measure due to technique limits. However, in vitro assays showed that the activity of MAMPs ranges from pM to nM (Table 1). We reason the exact physiological concentration of MAMPs on site should be lower or equal to pM level since not 100% of exogenous MAMPs could reach to the place where PRRs are. Upon infection, the concentration of MAMPs on site is dynamic and depends mainly on the production of MAMPs. It is generally known MAMPs are produced through two ways: biosynthesis and host hydrolyzation.
During infection, large amounts of peptidoglycan (PGN) building blocks are biosynthesized and some of these blocks are steadily shed into extracytoplasmic space. For instance, Bacillus subtilis released about 50% of its PGN in one generation during growth (Goodell and Schwarz, 1985). It has also been found that flagellin monomers are released into the supernatant of Pseudomonas aeruginosa cultures (Bardoel et al., 2011). Likewise, LPSs dropped into liquid culture when Escherichia coli grows in vitro (Mackowiak, 1984). Upon infection, these building blocks fallen into space surrounding cells and can serve as MAMPs for the activation of PTI signaling. For instance, culture supernatants from Bacillus sp. were shown to cause immune responses through the activation of nucleotide-binding oligomerization domain-containing protein (NOD), a signaling cascade in response to mostly either meso-diaminopimelic acid (mDAP) containing cell wall peptides (Fujimoto and Fukase, 2011). Pseudomonas aeruginosa aprA mutants induced an activation of TLR5 signaling via shedding monomeric flagellin into their environment (Bardoel et al., 2011). Similarly, growing fungal cells also shed chitin into the environment (Bueter et al., 2013).
To reduce the immune responses in plants, microbes downregulate the biosynthesis of MAMPs to reduce the concentrations of MAMPs on site. It was found that the biosynthesis of flagella in Pseudomonas is downregulated by the second messenger cyclic-di-GMP (cdG) (Hickman and Harwood, 2008). Increased cdG levels in the plant pathogen P. syringae, the plant opportunist P. aeruginosa and the plant commensal Pseudomonas fluorescens reduce flagellin levels, and thus reduce flg22 concentration on site, which contributes to the evasion of FLS2-mediated immune response in Nicotiana benthamiana and Arabidopsis (Pfeilmeier et al., 2016). Similarly, the maize fungal pathogen C. graminicola downregulates the expression of genes encoding KRE5 and KRE6, which are key enzymes for the biosynthesis of β-glucan (Oliveira-Garcia and Deising, 2016).
On the other hand, host hydrolytic activities establish decomposition of MAMPs precursors in apoplast to generate soluble ligands for PRRs. Plant apoplast contains hundreds of glycosidases, proteases, and other hydrolases (Grosse-Holz et al., 2018). In general, these hydrolases in the apoplast are not considered directly detrimental to pathogen growth, but are presumed to release pathogen MAMPs, which in turn trigger downstream defense responses. For example, fugal pathogens often induce plant chitinases to target the fungal cell walls, releasing chitin as MAMPs (Sanchez-Vallet et al., 2015). Upon bacterial infection, plants can produce a metazoan lysozyme-like hydrolase (LYS1), which releases soluble PGN from insoluble bacterial cell walls for triggering plant immunity (Liu et al., 2014). Similarly, immunogenic flagellin peptides are released from flagellin by host proteases with glycosidases to trigger immunity (Buscaill et al., 2019).
In response to host hydrolyzation, microbes can secret proteins or glycans to protect MAMPs precursors from hydrolases. One method is to secret proteins that cover the hydrolytic sites in MAMP precursors. Tomato leaf mold fungus produced Avr4 binds specifically to chitin in the fungal cell walls to protect it from plant chitinases (van den Burg et al., 2006). Similarly, xylem-invading fungus Verticillium nonalfalfae prevents chitin from hydrolysis by secreting VnaChtBP that binds chitin and suppresses chitin-induced immunity (Volk et al., 2019). Interestingly, the fungal vascular wilt pathogen V. dahliae strain VdLs17 could secret a lineage-specific LysM effector, Vd2LysM, which mimics the host PRR to bind chitin and suppresses chitin-induced immune responses (Akcapinar et al., 2015).
Glycosylation of MAMP precursors is another approach employed by microbes to evade host recognitions. For instance, glycosylation of bacterial flagellin and fungal chitin suppress the release of MAMPs. O-glycosylation of flagellin is observed in bacterial pathogens such as Xanthomonas, Pseudomonas, Burkholderia, Dickeya, Erwinia, Pantoea, and Pectobacterium (Taguchi et al., 2010; Ichinose et al., 2013; De Maayer and Cowan, 2016), presumably preventing the hydrolytic release of the flagellin MAMP (Buscaill and van der Hoorn, 2021). While mutants of the flagellin glycosyltransferase in P. syringae pv. tabaci 6605, P. syringae pv. glycinea race 4 and X. campestris pv. campestris Xca showed less virulent to their host plants (Takeuchi et al., 2003; Taguchi et al., 2006; Ichinose et al., 2013).
In bacterial pathogens Acidovorax avenae which cause rice leaf blight, flagellin from the N1141 strain but not K1 strain induces immune responses. The flagellin in two strains are identical in sequences but different in glycosylation pattern such as a 1,600 Da O-glucan for N1141 while a 1,600-Da one for K1 (Hirai et al., 2011).
Glycosylation of fungal cell walls could also prevent the release of chitin from cell wall. For instance, rice pathogens Magnaporthe oryzae, Cochliobolus miyabeanus, and Rhizoctonia solani accumulate α-1,3-glucan on the surface of infectious hyphae (Fujikawa et al., 2009, 2012). While fungal mutants with reduced α-1,3-glucan levels showed less virulence, suggesting that α-1,3-glucan may prevent chitin release from cell wall (Fujikawa et al., 2012).
In summary, the physiological concentration of the proper MAMPs, which is required for activation of effective immune responses at the infection site, is dependent on both biosynthesis of MAMPs by microbes and hydrolytic release of MAMPs by plants.
In apoplast, mature MAMPs accumulate in the proximity of the plasma membrane-localized PRRs through biosynthesis (Mackowiak, 1984; Goodell and Schwarz, 1985; Bardoel et al., 2011) or host hydrolytic enzymes (Guimil et al., 2005; Liu et al., 2014; Buscaill et al., 2019). The epitopes of MAMPs serve as ligands for PRR recognition and the immunogenic units of MAMPs are often buried in the MAMP precursors that are essential structures of microbes such as cell wall (Kaku et al., 2006; Miya et al., 2007) and flagella (Felix et al., 1999; Zipfel et al., 2004).
For the effective MAMP-PRR recognition, firstly MAMP molecules must be released as soluble fragments from precursors by host hydrolytic enzymes. Then these fragments need to move cross the host cell wall matrix before being recognized as ligands by plant plasma membrane-localized PRRs (Figure 1).
Plant cell walls compose of cross-linked polysaccharides with pores, ranged from 5 to 20 nm in size (Wang et al., 2016; Cunningham et al., 2018), suggesting that any MAMPs with size greater than these pores could be limited in their ability to pass through the cell wall matrix. Based on these criteria, the size of the mature MAMPs should be less than the pore size of cell wall matrix (5 nm). The immunogenic fragments of chitin, (NAG)7–8, can induce the highest immune activity in plants (Zipfel, 2008). Consistent with this, the length of (NAG)8 is around 4.14 and 4.08 nm (Figure 2), based on the crystal structures of the chitin receptors atCERK1 and OsCEBiP, respectively (Liu et al., 2012, 2016). Likewise, the length of flg22 from flagellin is 4.6 nm (Figure 2; Sun et al., 2013). Further, the length of atPep1 is 4.4 nm (Tang et al., 2015). As expected, these mature MAMPs have a size of less than 5 nm, which allows them to move through the cell wall matrix before specifical binding to their PM-localized PRRs.

Figure 2. The predicted size of the mature microbe-associated molecular patterns (MAMPs) from crystal structure of Pattern Recognition Receptor (PRR)-ligand pairs. (A) Chitin in atCERK1, (NAG)4 = 2.07 nm; (B) Chitin in OsCEBiP, (NAG)3 = 1.54 nm; (C) flg22, flg22 in Flagellin Sensing 2 (FLS2) length of flg22 = 4.06 nm; (D) atPEP1 in atPEPR, length of atPep1 = 4.4 nm.
However, extensive hydrolysis of the immunogenic MAMPs often leads to the loss of immunogenic activity (Liu et al., 2012, 2014). For instance, the heptamer and octamer of chitin fragments showed the highest immunogenic activity (Zipfel, 2008), but little (Petutschnig et al., 2010) or no activity (Zhang et al., 2002) was observed for tetramers and pentamers of chitin. Similarly, extensive digest of PGN into small fragments appears to abolish its immunogenic activity (Liu et al., 2014). Presumably, the minimum length of immunogenic fragments is required for the oligomerization of PRRs or interactions between PRRs and co-receptors, which is often necessary for effectively activating immune signaling (Liu et al., 2012). Furthermore, these small fragments by extensive hydrolysis of the immunogenic MAMPs may compete the immunogenic ligands for PRR binding, and therefore lead to the desensitization of the PRR-mediated perception (Felix et al., 1998).
In summary, the size of proper MAMPs with immunogenic activity falls in such a range that allow themselves to move through cell wall pores, as well as to bind with PRRs and initiate effective immune signaling.
During plant-microbe co-evolution, plant developed a sophisticated system to detect pathogens in apoplast and activate appropriate signaling in response to microbial invasions. Instead of direct perceiving entire microbes, plant only recognizes microbial signatures (MAMPs) by the cell surface receptors (PRRs) to initiate immune responses. Here, we discussed the form, the concentration, and the size of the mature MAMPs for effective activation of immune signaling in plants.
In plant apoplast, the cell wall matrix forms a barrier between apoplastic microbes and plant plasma membrane-localized PRRs (Figure 1). On one hand, the cell wall acts as a physical barrier to prevent microbes from contacting directly with plasma membrane. On the other hand, plant cell needs to overcome this distance barrier to directly detect the invasion of microbes to initiate immune responses. Investigation of the size range of immunogenic MAMPs will aid to understand the recognition and activation mechanisms of PTI, and develop new effective disease resistance strategies in plants. Furthermore, MAMPs can trigger plants to switch to a primed state of enhanced defense known as defense priming, which can also be induced by synthetic chemicals (Conrath et al., 2015). Knowledge of the size range of immunogenic MAMPs helps to design synthetic molecules as efficient priming inducers.
Recognition of MAMPs by PRRs triggers PTI which is locally and transiently, thus the fine-tuning signaling is required for plants in responds to such diverse microbes. We described that the PAMPs can be generated by both microbe biosynthesis and plant hydrolytic cleavage, while the concentration of PAMPs is temporally dynamic at the infection site of plant. In a temporal resolution, invasion of pathogens in apoplast initially generate low levels of MAMPs, which can be detected immediately by PRRs to induce signaling, including the upregulation of hydrolytic enzymes in apoplast (Liu et al., 2014). Consequently, increased levels of hydrolytic enzymes in apoplast release more MAMPs from pathogens and the elevated concentration of MAMPs then trigger stronger PTI. When pathogens are inhibited by immune responses and decreased in apoplast, MAMP precursors decrease subsequently. As a result, high levels of apoplastic hydrolytic enzymes over-digest the MAMPs into small fragments, leading to the low concentration of immunogenic MAMPs. Small fragments can further compete with immunogenic MAMPs for PRR binding and reduce PTI, which in turn prevents the upregulation of hydrolytic enzymes. Thus, the temporal dynamic of MAMPs enables plant to respond to the invasion of pathogens in an appropriate manner. However, it is unknown if there is a spatial regulation of MAMPs in apoplast since there is no any separated space in apoplast.
In conclusion, upstreaming events of recognition of MAMPs by PRRs are exciting research fields. However, there are still many holding secrets in how MAMPs mature from precursors, further understandings of these apoplastic events will ultimately lead to novel strategies to enhance pathogen resistances in plants.
XL: writing–original draft and supervision. PL, YL, XY, C-LS, and XL: conceptualization, writing–review and editing, and approved the submitted version.
This work was supported by Lushan Botanical Garden Start-Up Grant (2021ZWZX27) and Jiangxi Talent Project (20212BCJ25023).
Author C-LS was employed by ANGENOVO.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1019069/full#supplementary-material
Akcapinar, G. B., Kappel, L., Sezerman, O. U., and Seidl-Seiboth, V. (2015). Molecular diversity of LysM carbohydrate-binding motifs in fungi. Curr. Genet. 61, 103–113. doi: 10.1007/s00294-014-0471-9
Albert, I., Bohm, H., Albert, M., Feiler, C. E., Imkampe, J., Wallmeroth, N., et al. (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1:15140. doi: 10.1038/nplants.2015.140
Antolin-Llovera, M., Petutsching, E. K., Ried, M. K., Lipka, V., Nurnberger, T., Robatzek, S., et al. (2014). Knowing your friends and foes–plant receptor-like kinases as initiators of symbiosis or defence. New Phytol. 204, 791–802.
Bardoel, B. W., van der Ent, S., Pel, M. J., Tommassen, J., Pieterse, C. M., van Kessel, K. P., et al. (2011). Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS Pathog. 7:e1002206. doi: 10.1371/journal.ppat.1002206
Bohm, H., Albert, I., Oome, S., Raaymakers, T. M., Van den Ackerveken, G., and Nurnberger, T. (2014). A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog. 10:e1004491. doi: 10.1371/journal.ppat.1004491
Boutrot, F., and Zipfel, C. (2017). Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286. doi: 10.1146/annurev-phyto-080614-120106
Bueter, C. L., Specht, C. A., and Levitz, S. M. (2013). Innate sensing of chitin and chitosan. PLoS Pathog. 9:e1003080. doi: 10.1371/journal.ppat.1003080
Buscaill, P., and van der Hoorn, R. A. L. (2021). Defeated by the nines: Nine extracellular strategies to avoid microbe-associated molecular patterns recognition in plants. Plant Cell 33, 2116–2130. doi: 10.1093/plcell/koab109
Buscaill, P., Chandrasekar, B., Sanguankiattichai, N., Kourelis, J., Kaschani, F., Thomas, E. L., et al. (2019). Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 364:eaav0748.
Cesbron, S., Briand, M., Essakhi, S., Gironde, S., Boureau, T., Manceau, C., et al. (2015). Comparative genomics of pathogenic and nonpathogenic strains of Xanthomonas arboricola unveil molecular and evolutionary events linked to pathoadaptation. Front. Plant Sci. 6:1126. doi: 10.3389/fpls.2015.01126
Cheng, J. H. T., Bredow, M., Monaghan, J., and diCenzo, G. C. (2021). Proteobacteria contain diverse flg22 epitopes that elicit varying immune responses in Arabidopsis thaliana. Mol. Plant Microbe Interact. 34, 504–510. doi: 10.1094/MPMI-11-20-0314-SC
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476. doi: 10.1105/tpc.105.036574
Conrath, U., Beckers, G. J., Langenbach, C. J., and Jaskiewicz, M. R. (2015). Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119. doi: 10.1146/annurev-phyto-080614-120132
Cord-Landwehr, S., Melcher, R. L., Kolkenbrock, S., and Moerschbacher, B. M. (2016). A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 6:38018. doi: 10.1038/srep38018
Cunningham, F. J., Goh, N. S., Demirer, G. S., Matos, J. L., and Landry, M. P. (2018). Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 36, 882–897. doi: 10.1016/j.tibtech.2018.03.009
De Maayer, P., and Cowan, D. A. (2016). Comparative genomic analysis of the flagellin glycosylation island of the Gram-positive thermophile Geobacillus. BMC Genomics 17:913. doi: 10.1186/s12864-016-3273-2
Dow, M., Newman, M. A., and von Roepenack, E. (2000). The induction and modulation of plant defense responses by bacterial Lipopolysaccharides. Annu. Rev. Phytopathol. 38, 241–261. doi: 10.1146/annurev.phyto.38.1.241
El Gueddari, N. E., Rauchhaus, U., Moerschbacher, B. M., and Deising, H. B. (2002). Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 156, 103–112. doi: 10.1046/j.1469-8137.2002.00487.x
Erickson, R. O. (1986). Symplastic growth and symplasmic transport. Plant Physiol. 82:1153. doi: 10.1104/pp.82.4.1153
Felix, G., and Boller, T. (2003). Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 278, 6201–6208. doi: 10.1074/jbc.M209880200
Felix, G., Regenass, M., and Boller, T. (1993). Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: Induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4, 307–316. doi: 10.1046/j.1365-313X.1993.04020307.x
Felix, G., Baureithel, K., and Boller, T. (1998). Desensitization of the perception system for chitin fragments in tomato cells. Plant Physiol. 117, 643–650. doi: 10.1104/pp.117.2.643
Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. doi: 10.1046/j.1365-313x.1999.00265.x
Fujikawa, T., Kuga, Y., Yano, S., Yoshimi, A., Tachiki, T., Abe, K., et al. (2009). Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 73, 553–570. doi: 10.1111/j.1365-2958.2009.06786.x
Fujikawa, T., Sakaguchi, A., Nishizawa, Y., Kouzai, Y., Minami, E., Yano, S., et al. (2012). Surface alpha-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog. 8:e1002882. doi: 10.1371/journal.ppat.1002882
Fujimoto, Y., and Fukase, K. (2011). Structures, synthesis, and human Nod1 stimulation of immunostimulatory bacterial peptidoglycan fragments in the environment. J. Nat. Prod. 74, 518–525. doi: 10.1021/np100795d
Gao, F., Zhang, B. S., Zhao, J. H., Huang, J. F., Jia, P. S., Wang, S., et al. (2019). Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 5, 1167–1176. doi: 10.1038/s41477-019-0527-4
Gehrig, H., Schussler, A., and Kluge, M. (1996). Geosiphon pyriforme, a fungus forming endocytobiosis with Nostoc (cyanobacteria), is an ancestral member of the Glomales: Evidence by SSU rRNA analysis. J. Mol. Evol. 43, 71–81.
Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/s1097-2765(00)80265-8
Goodell, E. W., and Schwarz, U. (1985). Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J. Bacteriol. 162, 391–397. doi: 10.1128/jb.162.1.391-397.1985
Grosse-Holz, F., Kelly, S., Blaskowski, S., Kaschani, F., Kaiser, M., and van der Hoorn, R. A. L. (2018). The transcriptome, extracellular proteome and active secretome of agroinfiltrated Nicotiana benthamiana uncover a large, diverse protease repertoire. Plant Biotechnol. J. 16, 1068–1084. doi: 10.1111/pbi.12852
Guimil, S., Chang, H. S., Zhu, T., Sesma, A., Osbourn, A., Roux, C., et al. (2005). Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Natl. Acad. Sci. U.S.A. 102, 8066–8070. doi: 10.1073/pnas.0502999102
Gust, A. A., Biswas, R., Lenz, H. D., Rauhut, T., Ranf, S., Kemmerling, B., et al. (2007). Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 282, 32338–32348. doi: 10.1074/jbc.M704886200
Hickman, J. W., and Harwood, C. S. (2008). Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389. doi: 10.1111/j.1365-2958.2008.06281.x
Hirai, H., Takai, R., Iwano, M., Nakai, M., Kondo, M., Takayama, S., et al. (2011). Glycosylation regulates specific induction of rice immune responses by Acidovorax avenae flagellin. J. Biol. Chem. 286, 25519–25530. doi: 10.1074/jbc.M111.254029
Ichinose, Y., Taguchi, F., Yamamoto, M., Ohnishi-Kameyama, M., Atsumi, T., Iwaki, M., et al. (2013). Flagellin glycosylation is ubiquitous in a broad range of phytopathogenic bacteria. J. Gen. Plant Pathol. 79, 359–365. doi: 10.1007/s10327-013-0464-4
Ivanov, S., Austin, J. II, Berg, R. H., and Harrison, M. J. (2019). Extensive membrane systems at the host-arbuscular mycorrhizal fungus interface. Nat. Plants 5, 194–203. doi: 10.1038/s41477-019-0364-5
Janeway, C. A. Jr. (1989). Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54(Pt 1), 1–13. doi: 10.1101/sqb.1989.054.01.003
Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., Takio, K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091. doi: 10.1073/pnas.0508882103
Kankanala, P., Czymmek, K., and Valent, B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19, 706–724. doi: 10.1105/tpc.106.046300
Karasov, T. L., Chae, E., Herman, J. J., and Bergelson, J. (2017). Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 29, 666–680. doi: 10.1105/tpc.16.00931
Koga, J., Yamauchi, T., Shimura, M., Ogawa, N., Oshima, K., Umemura, K., et al. (1998). Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J. Biol. Chem. 273, 31985–31991. doi: 10.1074/jbc.273.48.31985
Kunze, G., Zipfel, C., Robatzek, S., Niehaus, K., Boller, T., and Felix, G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16, 3496–3507. doi: 10.1105/tpc.104.026765
Lacombe, S., Rougon-Cardoso, A., Sherwood, E., Peeters, N., Dahlbeck, D., van Esse, H. P., et al. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. doi: 10.1038/nbt.1613
Liebrand, T. W., van den Berg, G. C., Zhang, Z., Smit, P., Cordewener, J. H., America, A. H., et al. (2013). Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. U.S.A. 110, 10010–10015. doi: 10.1073/pnas.1220015110
Liu, S., Wang, J., Han, Z., Gong, X., Zhang, H., and Chai, J. (2016). Molecular mechanism for fungal cell wall recognition by rice chitin receptor OsCEBiP. Structure 24, 1192–1200. doi: 10.1016/j.str.2016.04.014
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. doi: 10.1126/science.1218867
Liu, X., Grabherr, H. M., Willmann, R., Kolb, D., Brunner, F., Bertsche, U., et al. (2014). Host-induced bacterial cell wall decomposition mediates pattern-triggered immunity in Arabidopsis. Elife 3:e01990.
Luu, D. D., Joe, A., Chen, Y., Parys, K., Bahar, O., Pruitt, R., et al. (2019). Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc. Natl. Acad. Sci. U.S.A. 116, 8525–8534. doi: 10.1073/pnas.1818275116
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Mackowiak, P. A. (1984). Relationship between growth temperature and shedding of lipopolysaccharides by gram-negative bacilli. Eur. J. Clin. Microbiol. 3, 406–410. doi: 10.1007/bf02017360
Medzhitov, R. (2007). Recognition of microorganisms and activation of the immune response. Nature 449, 819–826. doi: 10.1038/nature06246
Medzhitov, R., and Janeway, C. A. Jr. (2002). Decoding the patterns of self and nonself by the innate immune system. Science 296, 298–300. doi: 10.1126/science.1068883
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Mott, G. A., Thakur, S., Smakowska, E., Wang, P. W., Belkhadir, Y., Desveaux, D., et al. (2016). Genomic screens identify a new phytobacterial microbe-associated molecular pattern and the cognate Arabidopsis receptor-like kinase that mediates its immune elicitation. Genome Biol. 17:98. doi: 10.1186/s13059-016-0955-7
Mueller, K., Bittel, P., Chinchilla, D., Jehle, A. K., Albert, M., Boller, T., et al. (2012). Chimeric FLS2 receptors reveal the basis for differential flagellin perception in Arabidopsis and tomato. Plant Cell 24, 2213–2224. doi: 10.1105/tpc.112.096073
Nurnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198, 249–266. doi: 10.1111/j.0105-2896.2004.0119.x
Nurnberger, T., Nennstiel, D., Jabs, T., Sacks, W. R., Hahlbrock, K., and Scheel, D. (1994). High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 78, 449–460. doi: 10.1016/0092-8674(94)90423-5
Oliveira-Garcia, E., and Deising, H. B. (2016). Attenuation of PAMP-triggered immunity in maize requires down-regulation of the key beta-1,6-glucan synthesis genes KRE5 and KRE6 in biotrophic hyphae of Colletotrichum graminicola. Plant J. 87, 355–375. doi: 10.1111/tpj.13205
Petutschnig, E. K., Jones, A. M., Serazetdinova, L., Lipka, U., and Lipka, V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285, 28902–28911. doi: 10.1074/jbc.M110.116657
Pfeilmeier, S., Saur, I. M., Rathjen, J. P., Zipfel, C., and Malone, J. G. (2016). High levels of cyclic-di-GMP in plant-associated Pseudomonas correlate with evasion of plant immunity. Mol. Plant Pathol. 17, 521–531. doi: 10.1111/mpp.12297
Pfund, C., Tans-Kersten, J., Dunning, F. M., Alonso, J. M., Ecker, J. R., Allen, C., et al. (2004). Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol. Plant Microbe Interact. 17, 696–706. doi: 10.1094/MPMI.2004.17.6.696
Pruitt, R. N., Schwessinger, B., Joe, A., Thomas, N., Liu, F., Albert, M., et al. (2015). The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci. Adv. 1:e1500245. doi: 10.1126/sciadv.1500245
Ranf, S., Gisch, N., Schaffer, M., Illig, T., Westphal, L., Knirel, Y. A., et al. (2015). A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 16, 426–433. doi: 10.1038/ni.3124
Rapicavoli, J. N., Blanco-Ulate, B., Muszynski, A., Figueroa-Balderas, R., Morales-Cruz, A., Azadi, P., et al. (2018). Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat. Commun. 9:390. doi: 10.1038/s41467-018-02861-5
Saijo, Y., Loo, E. P., and Yasuda, S. (2018). Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 93, 592–613. doi: 10.1111/tpj.13808
Sanchez-Vallet, A., Mesters, J. R., and Thomma, B. P. (2015). The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 39, 171–183. doi: 10.1093/femsre/fuu003
Sattelmacher, B. (2001). The apoplast and its significance for plant mineral nutrition. New Phytol. 149, 167–192. doi: 10.1046/j.1469-8137.2001.00034.x
Shan, L., He, P., Li, J., Heese, A., Peck, S. C., Nurnberger, T., et al. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4, 17–27. doi: 10.1016/j.chom.2008.05.017
Sun, W., Dunning, F. M., Pfund, C., Weingarten, R., and Bent, A. F. (2006). Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18, 764–779. doi: 10.1105/tpc.105.037648
Sun, Y., Li, L., Macho, A. P., Han, Z., Hu, Z., Zipfel, C., et al. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628. doi: 10.1126/science.1243825
Taguchi, F., Takeuchi, K., Katoh, E., Murata, K., Suzuki, T., Marutani, M., et al. (2006). Identification of glycosylation genes and glycosylated amino acids of flagellin in Pseudomonas syringae pv. tabaci. Cell. Microbiol. 8, 923–938. doi: 10.1111/j.1462-5822.2005.00674.x
Taguchi, F., Yamamoto, M., Ohnishi-Kameyama, M., Iwaki, M., Yoshida, M., Ishii, T., et al. (2010). Defects in flagellin glycosylation affect the virulence of Pseudomonas syringae pv. tabaci 6605. Microbiology (Reading) 156, 72–80. doi: 10.1099/mic.0.030700-0
Takeuchi, K., Taguchi, F., Inagaki, Y., Toyoda, K., Shiraishi, T., and Ichinose, Y. (2003). Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. J. Bacteriol. 185, 6658–6665. doi: 10.1128/JB.185.22.6658-6665.2003
Tang, J., Han, Z., Sun, Y., Zhang, H., Gong, X., and Chai, J. (2015). Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 25, 110–120. doi: 10.1038/cr.2014.161
van den Burg, H. A., Harrison, S. J., Joosten, M. H., Vervoort, J., and de Wit, P. J. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 19, 1420–1430. doi: 10.1094/MPMI-19-1420
Vander, P., V rum, K. M., Domard, A., Eddine El Gueddari, N., and Moerschbacher, B. M. (1998). Comparison of the ability of partially N-acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol. 118, 1353–1359. doi: 10.1104/pp.118.4.1353
Volk, H., Marton, K., Flajsman, M., Radisek, S., Tian, H., Hein, I., et al. (2019). Chitin-Binding protein of Verticillium nonalfalfae disguises fungus from plant Chitinases and suppresses chitin-triggered host immunity. Mol. Plant Microbe Interact. 32, 1378–1390. doi: 10.1094/MPMI-03-19-0079-R
Wan, J., Zhang, X. C., Neece, D., Ramonell, K. M., Clough, S., Kim, S. Y., et al. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20, 471–481. doi: 10.1105/tpc.107.056754
Wang, B., Yeun, L. H., Xue, J. Y., Liu, Y., Ane, J. M., and Qiu, Y. L. (2010). Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 186, 514–525. doi: 10.1111/j.1469-8137.2009.03137.x
Wang, G. L., Song, W. Y., Ruan, D. L., Sideris, S., and Ronald, P. C. (1996). The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol. Plant Microbe Interact. 9, 850–855. doi: 10.1094/mpmi-9-0850
Wang, P., Lombi, E., Zhao, F. J., and Kopittke, P. M. (2016). Nanotechnology: A new opportunity in plant sciences. Trends Plant Sci. 21, 699–712. doi: 10.1016/j.tplants.2016.04.005
Wang, S., Sun, Z., Wang, H., Liu, L., Lu, F., Yang, J., et al. (2015). Rice OsFLS2-mediated perception of bacterial flagellins is evaded by Xanthomonas oryzae pvs. oryzae and oryzicola. Mol. Plant 8, 1024–1037. doi: 10.1016/j.molp.2015.01.012
Wang, Y., Wang, Y., and Wang, Y. (2020). Apoplastic proteases: Powerful weapons against pathogen infection in plants. Plant Commun. 1:100085. doi: 10.1016/j.xplc.2020.100085
Zhang, B., Ramonell, K., Somerville, S., and Stacey, G. (2002). Characterization of early, chitin-induced gene expression in Arabidopsis. Mol. Plant Microbe Interact. 15, 963–970. doi: 10.1094/MPMI.2002.15.9.963
Zipfel, C. (2008). Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20, 10–16. doi: 10.1016/j.coi.2007.11.003
Zipfel, C. (2014). Plant pattern-recognition receptors. Trends Immunol. 35, 345–351. doi: 10.1016/j.it.2014.05.004
Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J. D., Boller, T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760. doi: 10.1016/j.cell.2006.03.037
Keywords: apoplast, microbe-associated molecular patterns, pattern recognition receptors, ligands, receptor-like kinases, receptor-like proteins
Citation: Lü P, Liu Y, Yu X, Shi C-L and Liu X (2022) The right microbe-associated molecular patterns for effective recognition by plants. Front. Microbiol. 13:1019069. doi: 10.3389/fmicb.2022.1019069
Received: 14 August 2022; Accepted: 08 September 2022;
Published: 26 September 2022.
Edited by:
Xiao-Ren Chen, Yangzhou University, ChinaReviewed by:
Daolong Dou, Nanjing Agricultural University, ChinaCopyright © 2022 Lü, Liu, Yu, Shi and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaokun Liu, WGlhb2t1bi5saXVAbHNiZy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.