
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 October 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1018682
This article is part of the Research TopicAntimicrobial Resistance: Agriculture, Environment and Public Health within One Health FrameworkView all 17 articles
A correction has been applied to this article in:
Corrigendum: Antimicrobial resistance and genomic characterization of Escherichia coli from pigs and chickens in Zhejiang, China
 Wei Zhou1†
Wei Zhou1† Rumeng Lin2,3,4†
Rumeng Lin2,3,4† Zhijin Zhou1
Zhijin Zhou1 Jiangang Ma2
Jiangang Ma2 Hui Lin2,5
Hui Lin2,5 Xue Zheng2
Xue Zheng2 Jingge Wang2
Jingge Wang2 Jing Wu2
Jing Wu2 Yuzhi Dong2,4
Yuzhi Dong2,4 Han Jiang4
Han Jiang4 Hua Yang2
Hua Yang2 Zhangnv Yang6*
Zhangnv Yang6* Biao Tang2*
Biao Tang2* Min Yue7*
Min Yue7*Escherichia coli is considered an opportunistic pathogen and an indicator for antimicrobial resistance (AMR) monitoring. Despite many reports on its AMR monitoring, studies based on genome-based analysis of AMR genes are still insufficient. Here, 181 E. coli strains were isolated from anal swab samples collected from pigs and chickens of animal farms located in Eastern China and sequenced through the Illumina platform. The results showed that 87.85% (159/181) of the E. coli isolates were multidrug-resistant (MDR). Ampicillin (AMP)- spectinomycin (SPT)- tetracycline (TET)- florfenicol (FFC)- sulfisoxazole (SF)- trimethoprim/sulfamethoxazole (SXT) was the predominant AMR pattern. By whole-genome sequencing, we found that ST10 (10.49%, 19/181) and ST48 (7.18%, 13/181) were major sequence types. IncFIB and IncX1 were the most prevalent plasmid replicons. The AMR genes blaNDM-5 (1.10%, 2/181), mcr-1 (1.10%, 2/181), tet(X4) (1.10%, 2/181), and cfr (6.08%, 11/181) were also found in these isolates. In addition, among the 169 virulence genes detected, we identified astA (37.02%, 67/181), hlyA (1.66%, 3/181), hlyB (1.66%, 3/181) and hlyD (1.66%, 3/181), which were closely related to heat-stable enterotoxin 1 and α-hemolysin. In addition, there were 33 virulence genes associated with the iron uptake system, and 46 were adhesion-related genes. Our study highlighted the need for routine surveillance of AMR with advanced genomic approaches, providing up-to-date data on the prevalence of AMR for the development and execution of antimicrobial stewardship policy.
The recent emergence and rapid increase of multi-drug resistant (MDR, resistance to more than three kinds of antibiotics) bacteria have caused public concern, represented by Escherichia coli resistant to carbapenem, colistin and tigecycline which were recognized as the last line of resort (Li et al., 2017; He et al., 2019; Ma et al., 2022). Due to incorrect use and misuse of antibiotics, the spread of antimicrobial resistance (AMR) is accelerating (Wang et al., 2022; Tang et al., 2022a,b). The continuous spread of AMR not only increases the difficulty of preventing and controlling livestock and poultry diseases but also seriously threatens livestock products’ safety and endangers consumers’ health (Xu et al., 2022; Li et al., 2022a).
E. coli is a commonly used AMR indicator in human and food animal (Brisola et al., 2019; Ma et al., 2022). There have been many reports on the study in E. coli and monitoring of the spread of AMR. The emergence of plasmid-mediated carbapenem resistance genes, especially blaNDM, has seriously affected the efficacy of meropenem (0.4%, 1/219; Tang et al., 2019, 2021b). Colistin resistance mediated by a plasmid-encoded mcr-1 was first documented in China during routine surveillance of food animals (21%, 166/804; Liu et al., 2016). A retrospective survey showed that mcr-1 was first traced back to 1980 but was not prevalent among bacteria until 2009 (Shen et al., 2016). With the detection rate of mcr-1 in bacteria increasing year by year, a relatively high occurrence rate of the mcr-1 gene (1%) was detected from E. coli in human (Wang et al., 2017). Tigecycline resistance are mediated by two novel genes, tet(X3) and tet(X4), both of which can significantly reduce the efficacy of tigecycline (He et al., 2019; Sun et al., 2019; Guan et al., 2022). Additionally, bacterial strains carrying the cfr gene encoding 23S rRNA methylase, which is resistant to five classes of antimicrobials, including phenols, lincosamides, oxazolidinones, pleuromutilin, and streptomycin A,allowing bacteria to develop MDR (Deng et al., 2014; Tang et al., 2022c).
Whole genome sequencing (WGS) played a vital role in AMR study (Tang et al., 2020b; Peng X. et al., 2022; Li et al., 2022b). We can obtain the tested strains’ AMR and virulence genes by combining them with the antimicrobial sensitivity test (AST). It is helpful to better understand AMR’s development and transmission (Teng et al., 2022). WGS has become an indispensable and reliable tool for revealing the AMR mechanism in global pathogen surveillance (Boolchandani et al., 2019; Tang et al., 2021a; Yang et al., 2022).
In this study, we investigated the prevalence of AMR E. coli in pigs and chickens from animal farms in Eastern China and evaluated the AMR phenotypes, genotypes, virulence genes, and plasmids replicons. This study helps understand the AMR situation and provides a reference to formulate livestock AMR control policies to better protect food safety in China.
A total of 200 anal swab samples were collected from three cities (Lishui, Jinhua, and Quzhou) located in Zhejiang Province, Eastern China, from March to April 2021. The samples were randomly collected from 110 chickens and 90 pigs in 5 poultry farms and 4 swine farms, respectively (Table 1; Figure 1A). All experiment activities in this study were approved by the Institutional Review Board of Zhejiang Academy of Agricultural Sciences.
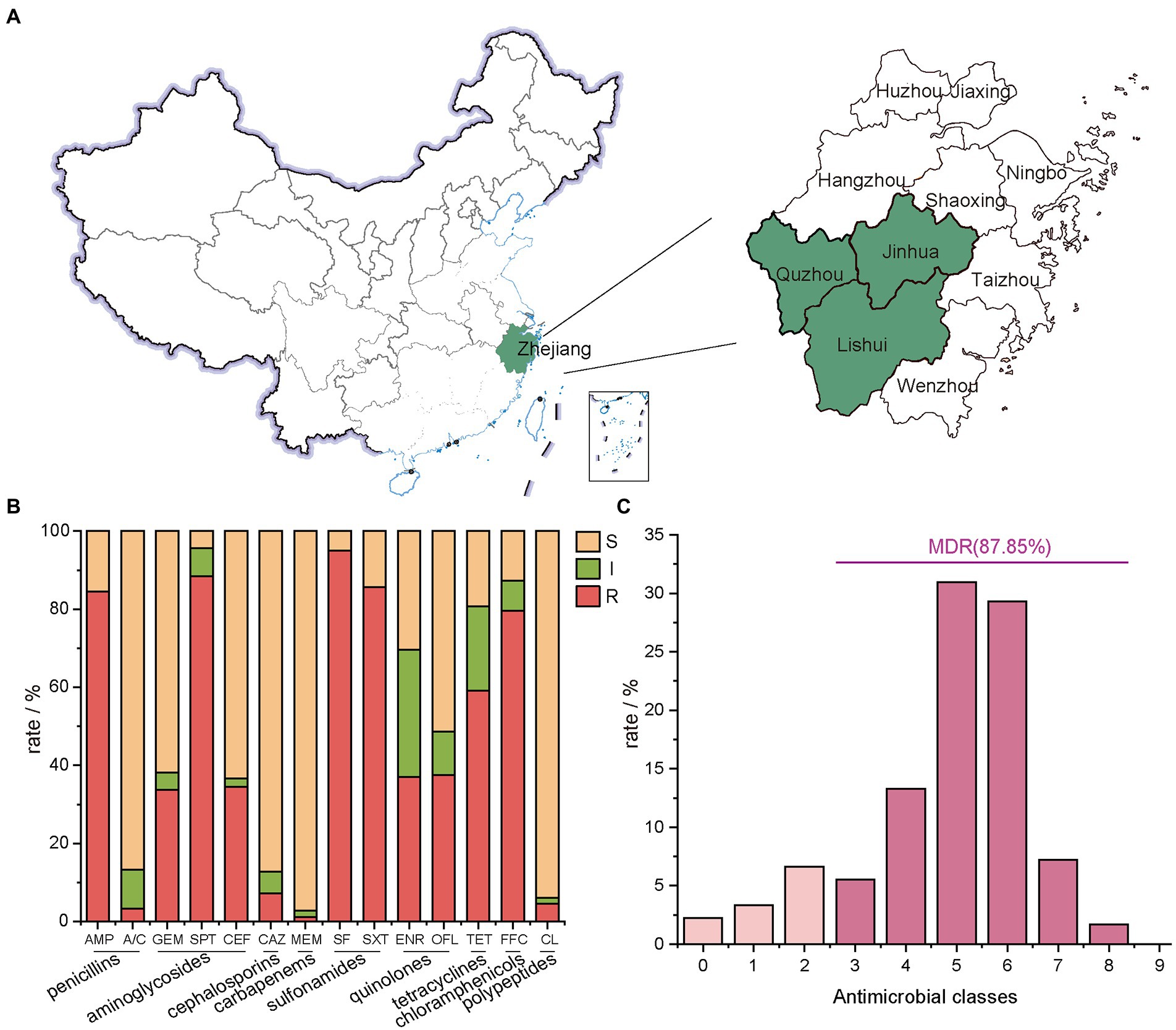
Figure 1. Geographical distribution of the sampling areas in Zhejiang Provinces, China, and AMR rates of E. coli isolates. (A) Sample sources of 181 strains of E. coli, Zhejiang Province in this study are shaded in green; (B) the number of E. coli isolates from farms in Zhejiang Province resistant to different antibiotics. In this experiment, 14 antibiotics with inflection points were divided into 9 categories; (C) the distribution of MDR strains.
Anal swab samples were enriched in 10 ml of buffered peptone solution (BPW, Land Bridge, Beijing, China). After initial pre-enrichment in BPW, 0.1 ml of enriched samples were streaked on MacConkey Agar (Land Bridge, Beijing, China) or Eosin Methylene Blue Agar (Land Bridge, Beijing, China) and incubated at 37°C for 24 h. Only one suspicious colony (round, moist, and show pink color on MacConkey Agar; purple-black color with green metallic sheen on EMB Agar) were selected and further cultured on Luria-Bertani (LB) Agar (Land Bridge, Beijing, China). Bacteria identification was carried out by MALDI-TOF MS. Confirmed isolates were stored at −80°C.
Micro-broth dilution method was used (Bio Fosun, Fosun Diagnostics, Shanghai, China) to determine the AMR profile of E. coli isolates (Tang et al., 2022b). The panel of antimicrobial compounds tested included ampicillin (AMP), augmentin (amoxicillin/clavulanic acid, A/C), gentamicin (GEM), tetracycline (TET), spectinomycin (SPT), florfenicol (FFC), sulfisoxazole (SF), trimethoprim/sulfamethoxazole (SXT), ceftiofur (CEF), ceftazidime (CAZ), enrofloxacin (ENR), ofloxacin (OFL), meropenem (MEM), ampicillin (APR), colistin (CL) and mequindox (MEQ). The 14 tested antibiotics are grouped into 9 classes (Figure 1B), including penicillins (AMP and A/C), aminoglycosides (GEM、SPT, and CEF), cephalosporins (CAZ), carbapenems (MEM), sulfonamides (SF and SXT), quinolones (ENR and OFL), tetracyclines (TET), chloramphenicols (FFC) and polypeptides (CL). The breakpoint for each antimicrobial was from the Clinical and Laboratory Standards Institute (CLSI, 2016:M100-S30). E. coli ATCC 25922 was used as quality control.
The genomic DNA extraction of E. coli was performed using a bacterial DNA extraction kit (Generay, Shanghai, China). The whole genome sequencing was performed on the Novaseq 6,000 (Illumina, SanDiego, CA, United States). Clean reads were assembled using SPAdesv3.12.0 (Bankevich et al., 2012). Genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline (Tatusova et al., 2016). ABRicate 1.0.1 tool1 and VFDB database were applied to predict the virulence genes and AMR genes. Replicons and sequence type (ST) were determined at the Center for Genomic Epidemiology (CGE).2 Phylogenetic analysis of the genome and plasmids was performed by kSNP 3.1 software based on the maximum-likelihood method (Gardner et al., 2015). Easyfig 2.2.5 was used for comparative analysis of the plasmids (Sullivan et al., 2011).
TBtools was used for clustering heat map analysis of AMR genes, AMR phenotypes, virulence genes, and plasmid replicons (Chen et al., 2020).
A total of 181 E. coli strains were isolated from all 200 anal swab samples with a detection rate of 90.50%. Among them, 96 isolates were from chickens, and 85 isolates were from pigs (Table 1).
E. coli isolates showed the lowest resistance rate to MEM at 2.76%, followed by A/C at 3.31%, CL at 5.52%, and CAZ at 7.18% (Figure 1B). A high resistance rate was shown for five antibiotics in descending order. They were SF (95.03%), SPT (88.40%), SXT (85.64%), AMP (84.53%) and FFC (79.56%). Except for TET with a resistance rate of 59.12%, the resistance rates of E. coli isolates to OFL, ENR, CEF and GEM ranged between 35.0% ~ 40.0% (Figure 2).
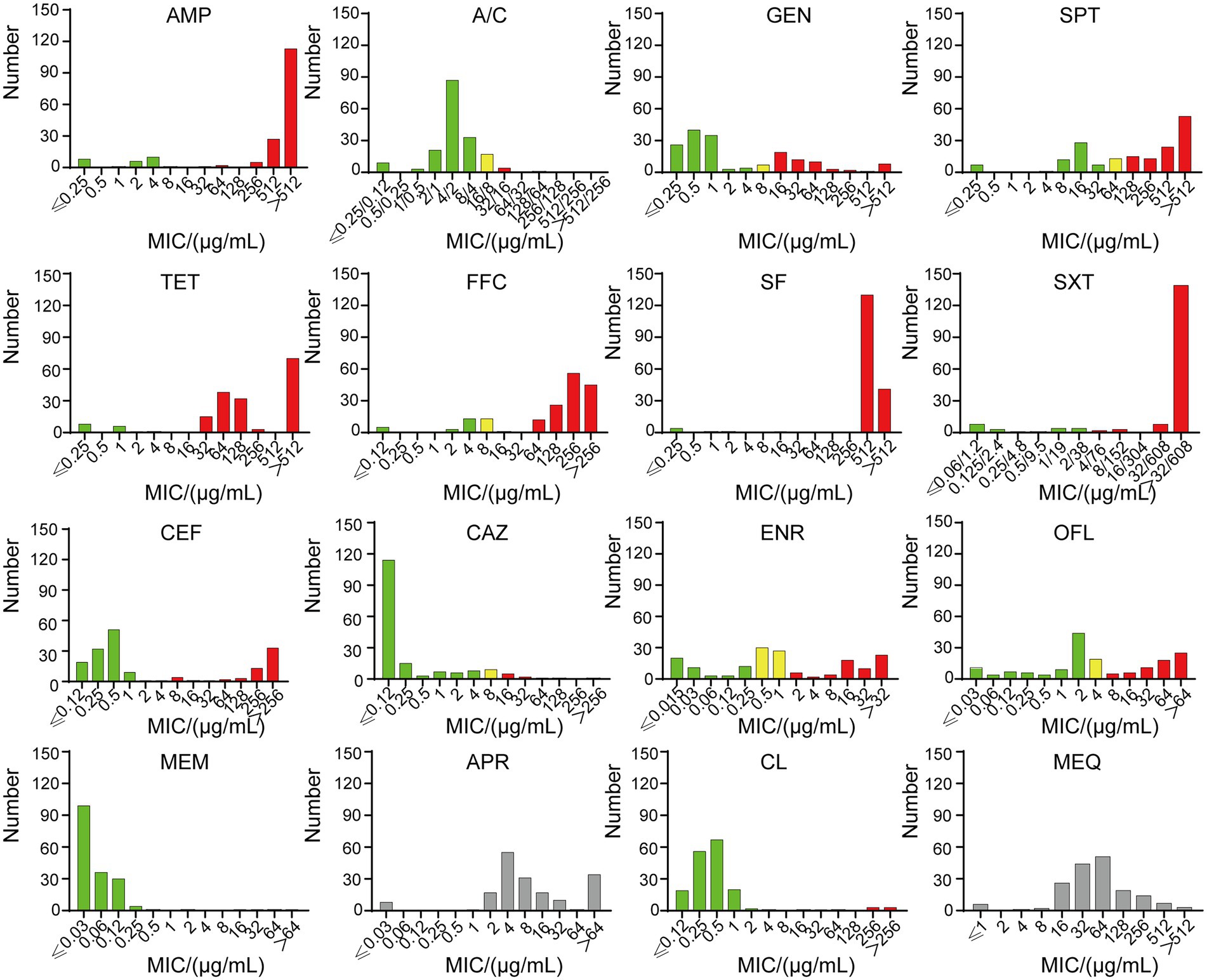
Figure 2. The MICs of 181 strains. Green indicates sensitivity, yellow indicates intermediary, and red indicates resistance. SF and SXT have no intermediate value, and APR and MEQ have no breakpoint. Ampicillin (AMP), amoxicillin/clavulanic acid (A/C), gentamicin (GEM), tetracycline (TET), spectinomycin (SPT), florfenicol (FFC), sulfisoxazole (SF), trimethoprim/sulfamethoxazole (SXT), ceftiofur (CEF), ceftazidime (CAZ), enrofloxacin (ENR), ofloxacin (OFL), meropenem (MEM), apramycin (APR), colistin (CL) and mequindox (MEQ).
The 14 tested antibiotics are grouped into 9 classes (Figure 1B), including penicillins (AMP and A/C), aminoglycosides (GEM、SPT, and CEF), cephalosporins (CAZ), carbapenems (MEM), sulfonamides (SF and SXT), quinolones (ENR and OFL), tetracyclines (TET), chloramphenicols (FFC) and polypeptides (CL). Carbapenems class of antibiotics, which in this study is MEM had the lowest resistance rate. Meanwhile, the sulfonamides class of antibiotics had the highest resistance rate with 181 resistant isolates, of which 172 were resistant to SF.
87.85% of isolates were MDR (Figure 1C; Supplementary Table S1), and the predominant MDR pattern (19.50%, 31/159) was resistance to AMP-SPT-TET-FFC-SF-SXT. Notably, three strains were determined to be resistant to 12 types of antibiotics. The patterns were AMP-A/C-GEM-SPT-TET-FFC-SF-SXT-CEF-CAZ-ENR-OFL and AMP-GEM-SPT-TET-FFC-SF-SXT-CEF-CAZ-ENR-OFL-CL, respectively.
Sixty-five different sequence types (STs) were generated in 181 E. coli isolates (Figure 3), which were further grouped into 16 clonal complexes (CCs) and 38 singletons. Among them, ST10 was most prevalent with 19 isolates (10.50%), followed by ST48 with 13 isolates (7.18%), ST58 and ST162 both with 9 isolates (4.97%).
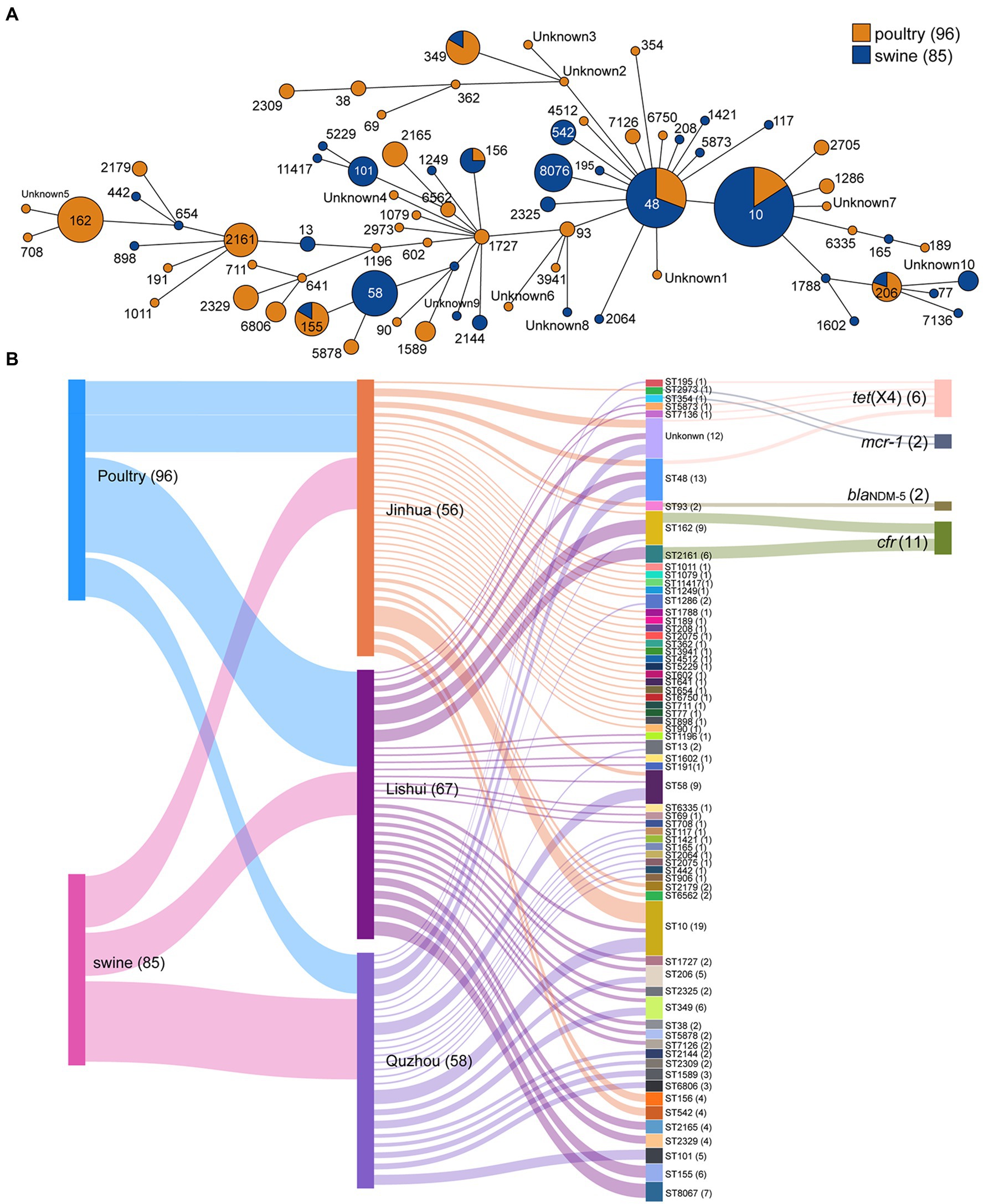
Figure 3. Minimum spanning tree of E. coli strains based on MLST and Sankey diagram combining the provinces, STs, farms, and sampling sources based on 181 E. coli isolates. (A) Each node represents a single ST. The size of the nodes is proportional to the number of isolates. The length of branches between each node is proportional to the number of different alleles that differ between two linked nodes; (B) the diameter of the line is directly proportional to the number of isolates, which is also marked with numbers. The lines are colored according to the city and the sampling source.
The plasmid replicon analysis (Figure 4) showed that a total of 40 types of plasmid replicons were detected in all 181 E. coli isolates, of which 107 (59.12%) carried the IncFIB (AP001918) replicon, followed by IncX1 replicon existing in 86 isolates (47.51%). 94.48% (171/181) of the isolates carried 2 ~ 6 replicons. Similar plasmid replicon types can be found in different cities and animals, suggesting that plasmids carrying AMR genes may be widely spread through horizontal gene transfer.
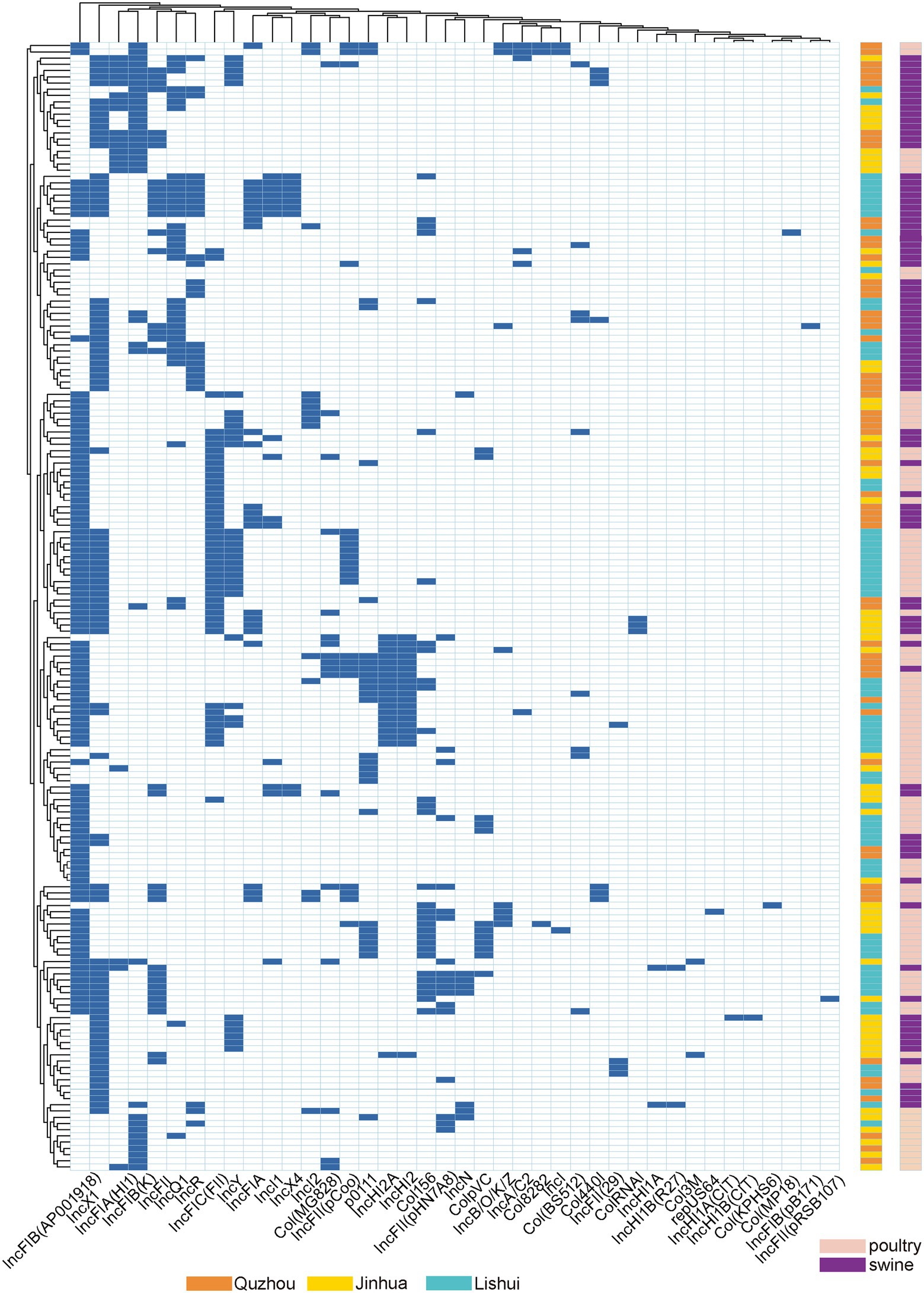
Figure 4. Dendrogram of hierarchical clustering heatmap of the 181 isolates and 40 plasmid replicons. Y-axis is the isolate number, and x-axis is the selected resistance genes identified. Non-expression of resistance is a white background and resistance phenotype is a blue background. Orange indicates isolates were isolated from Quzhou samples, aqua blue indicates isolates were isolated from Lishui, and bright yellow indicates isolates were isolated from Jinhua. Purple indicates that the strain is isolated from pig anal swab samples, and green indicates that the strain is isolated from bird anal swab samples.
As shown in Figure 5, 72 acquired AMR genes were detected in this study, among which mdf(A) was carried by all isolates. Notably, two isolates carried carbapenem resistance gene blaNDM-5, two isolates carried colistin resistance gene mcr-1, six isolates carried tigecycline resistance gene tet(X4), and eleven isolates carried linezolid resistance gene cfr.
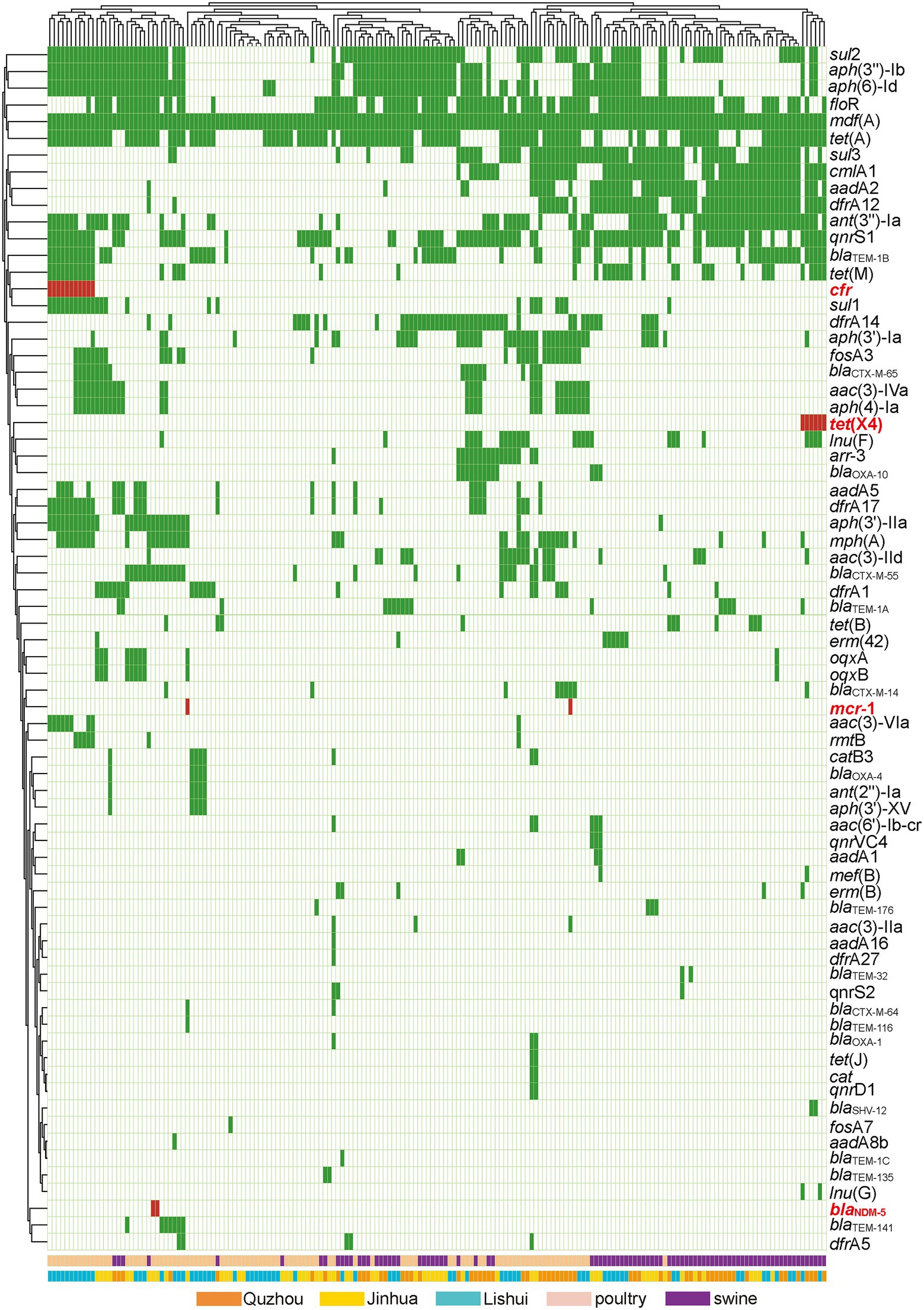
Figure 5. Antibiotic resistance patterns of E. coli isolates. Y-axis is the number of isolated strains, and x-axis is the selected AMR genes identified. Green indicates that the isolate is isolated from Quzhou sample, light blue indicates that it is isolated from Lishui, and light yellow indicates that it is isolated from Jinhua. Flesh color indicates that the isolate is isolated from poultry anal swab samples, and cinnabar indicates that it is isolated from pig anal swab samples. Dark green in the small grid indicates the presence of AMR genes, white indicates no, and red indicates the presence of risk AMR genes. The risk AMR genes mentioned here refer to the genes that have been focused on, such as tet(X4) (resistant to tigecycline), mcr-1 (resistant to colistin), blaNDM-5 (meropenem) or cfr (resistant to oxazolidinone, amphenicol, lincosamide), which pose a serious threat to public health and safety. Rifamycin (arr-3), Aminoglycoside (aac(3)-Iia, aac(3)-Iid, aac(3)-Iva, aac(3)-Via, aac(6′)-Ib-cr, aadA16, aadA1, aadA2, aadA5, aadA8b, ant(2″)-Ia, ant(3″)-Ia, aph(3″)-Ib, aph(3′)-Iia, aph(3′)-Ia, aph(3′)-XV, aph(4)-Ia, aph(6)-Id, rmtB), Beta-lactam (blaCTX-M-14, blaCTX-M-55, blaCTX-M-64, blaCTX-M-65, blaNDM-5, blaOXA-10, blaOXA-1, blaOXA-4, blaSHV-12, blaTEM-116, blaTEM-135, blaTEM-141, blaTEM-176, blaTEM-1A, blaTEM-1B, blaTEM-1C, blaTEM-32), Amphenicol (catB3, cat, cmlA1, floR, cfr), Folate pathway antagonist (dfrA12, dfrA14, dfrA17, dfrA1, dfrA27, dfrA5, sul1, sul2, sul3), Macrolide (erm(42), erm(B), mdf(A), mef(B), mph(A)), Fosfomycin (fosA3, fosA7), Lincosamide (lnu(F), lnu(G)), Polypeptides (mcr-1), Quinolone (oqxA, oqxB, qnrD1, qnrS1, qnrS2, qnrVC4), Tetracycline (tet(A), tet(B), tet(J), tet(M), tet(X)).
One hundred and sixty-nine virulence genes were found (Figure 6). Ten including entA, entB, entD, entE, entF, fepA, fepC, fepD, fes and ompA were detected in all isolates. The virulence genes carried by the other isolates ranged from 25 to 77. The encoding gene astA of heat-stable enterotoxin1 (East1) was detected in 37.02% (67/181) of the isolates. Three α-hemolysin encoding genes: hlyA, hlyB and hlyD, were present simultaneously in isolates ECLSZ21-06, ECQZZ21-39, and ECJHZ21-15. Thirty-three virulence genes detected here were related to the iron uptake system, and 46 were adhesion-associated genes.

Figure 6. Dendrogram of the hierarchical clustering heat map of isolates and virulence genes. The figure shows the predicted virulence genes factor profile of the studied isolates. Y-axis is the isolate number and x-axis is the selected virulence genes identified. The red color in the small cells indicates virulence genes associated with toxin production, the orange color indicates genes editing other virulence factors, and the white color indicates no virulence expression. Withered grass color indicates isolates isolated from Quzhou samples, yellow indicates isolates from Jinhua, and blue color indicates isolates from Lishui. Incarnadine pink indicates that the isolates were isolated from pig anal swab samples, and bright green indicates that they were isolated from birds.
In this study, we found that strains ECJHJ21-07 and ECJHJ21-14 carry the blaNDM-5 gene and have the same genetic context blaNDM-5-ble-trpF-dsbD. Strains ECLSZ21-04, ECLSZ21-15, ECQZZ21-02, ECQZZ21-05, ECQZZ21-04, and ECQZZ21-15 contain the tet(X4) gene, which is all adjacent to the estX gene. In the latter two strains, the tet(X4) gene is located in the gene arrangement estX-tet(X4)-ISVsa3 based on better sequencing quality. In addition, we found two strains carrying the mcr-1 gene, which have different genetic environments. The mcr-1-carrying plasmid in strain ECJHJ21-13 is homologous with plasmids pMCR4D31–3 and pHNSHP45 with an IncI2 type; the fragment harboring mcr-1 gene in strain ECQZJ21-13 is homologous with the IncHI2 plasmid (Figure 7).
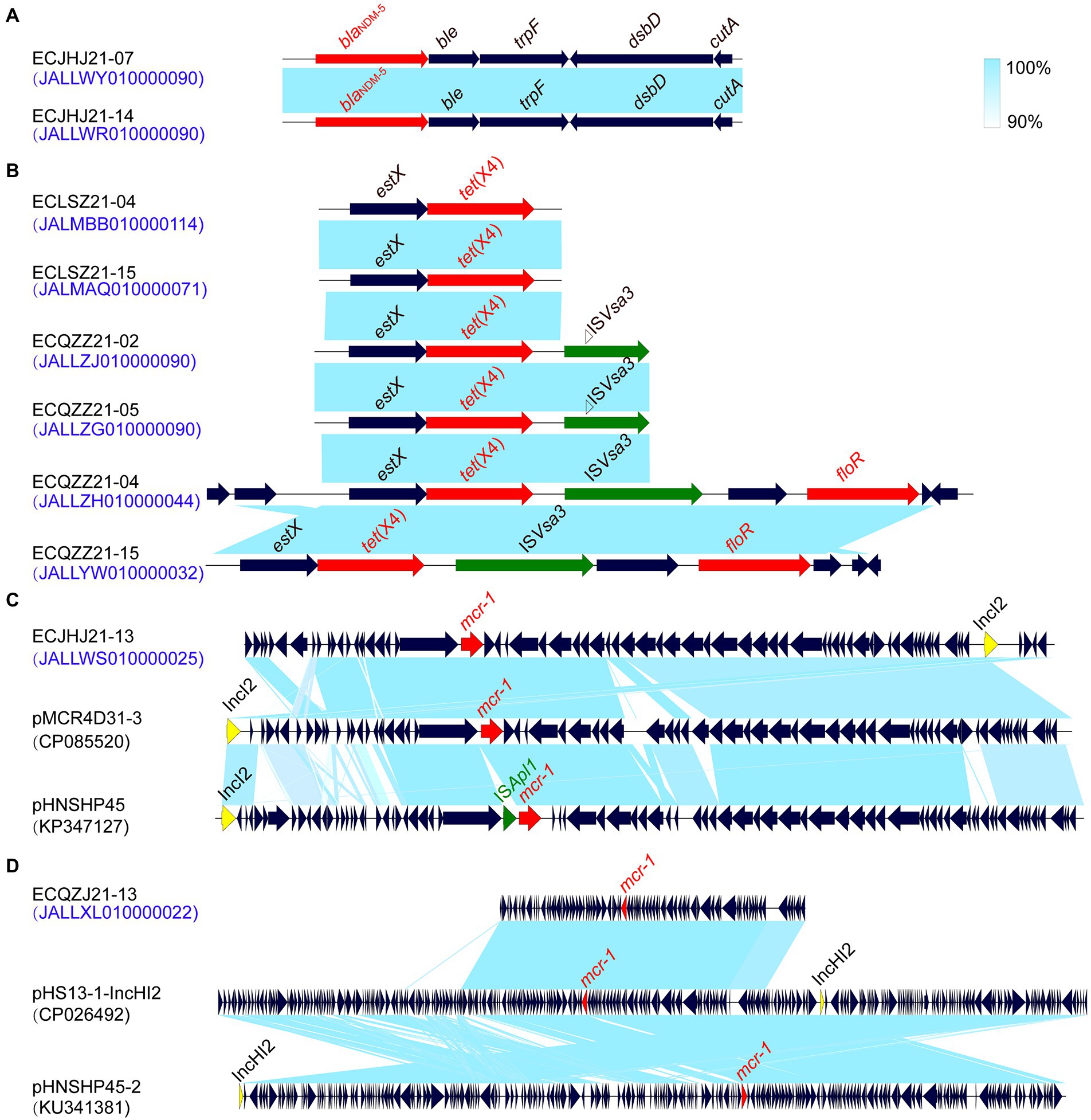
Figure 7. Comparison of the risk AMR gene environments in E. coli strains. (A) the blaNDM-5 gene environment. (B) the tet(X4) gene environment. (C) the mcr-1 gene environment of IncI2 plasmids. (D) the mcr-1 gene environment of IncHI2 plasmids.
Strains from different cities can gather on the same branch. As the last line of drugs, the AMR genes mcr-1, blaNDM, and tet(X4) are distributed in different branches, showing different genetic relationships (Figure 8). In the present study, these AMR genes are more likely to be horizontal gene transfer between E. coli strains. In addition, the cfr gene mainly exists in two branches, which may be the clonal transmission (Figure 8).
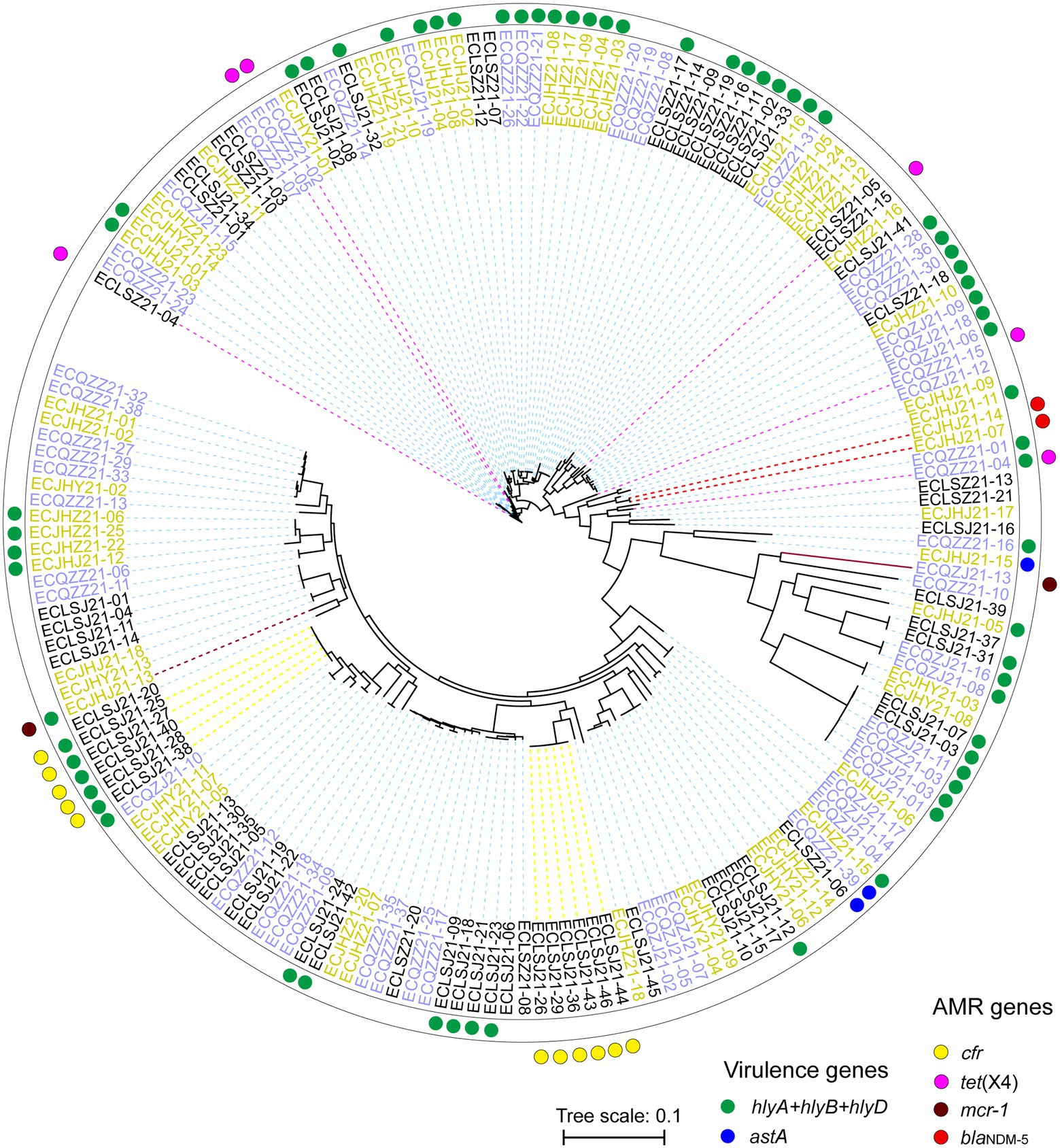
Figure 8. Phylogenetic SNP tree of E. coli strains with whole genome sequences. Red and blue dots indicate different virulence genes carrying strains, and yellow, pink, brick and red indicate different AMR genes carrying strains. The strains with different colors represent three different cities.
The highest concordance between genotype and phenotype was detected for carbapenems (100.00%), followed by polypeptides (93.92%), and tetracyclines (90.61%; Supplementary Table S2). In addition, two isolates carrying blaNDM-5 gene were found resistant to AMP, CEF, TET, FFC, SF, SXT, ENR, and OFL. All eleven isolates carrying the cfr gene showed resistance to AMP, SF, and SXT. Similarly, the isolates carrying mcr-1 showed resistance to AMP, TET, FFC, SF, CEF, ENR, and OFL, and the isolates carrying tet(X4) showed resistance to SPT, TET, FFC, SF, and SXT. These provide supporting evidence for AMR genes to explain drug resistance. The AMR gene cfr mainly existed in two STs, ST2161 and ST162. Two isolates carrying the blaNDM-5 were both assigned ST93. Isolates carrying mcr-1 belonged to ST2973 (ECJHJ21-13) and ST354 (ECQZJ21-13), respectively (Figure 3). Meanwhile, the isolates carrying tet(X4) demonstrated sequence type diversity, ST195, ST48, ST5873 and ST7136 were included.
The AMR of E. coli has become a worldwide public health problem (Rutuja et al., 2018). Pigs and poultry products may have a cross infection of AMR E.coli during processing and subsequent sale, which increases the risk of transmission and poses a significant threat to the sale of products and people’s health (Chang et al., 2020). To understand the current situation of AMR of E. coli, we analyzed AMR, MLST and virulence genes of E. coli isolated from anal swab samples of pigs and chickens in farms in Lishui, Quzhou and Jinhua, Zhejiang Province, Eastern China. In this study, relatively high AMR rates of FFC (79.56%), SXT (85.64%), SF (95.03%), SPT (88.40%) and AMP (84.53%) were detected that were consistent with the previously published results (Ma et al., 2022; Peng Z. et al., 2022). At the same time, the AMR rates of A/C (3.31%), MEM (2.76%), CL (5.52%) and CAZ (7.18%) were low. CL and MEM are recognized as the last line of defense against gram-negative bacteria (Lin et al., 2022; Wang et al., 2022) and low AMR rates reveal their resistance’s effectiveness is decreasing.
Genomic analysis revealed that the E. coli isolates harbored various AMR genes, which could be consistent with the AMR phenotype. All E. coli isolates contained the AMR gene mdf(A), and most of them also carried tet(A) (68.51%, 124/181), floR (62.98%, 114/181) and sul2 (56.35%, 102/181). These genes mediate resistance to tetracycline, chloramphenicol, and sulfonamide antibiotics. These critical AMR genes in E. coli isolates from food animals present a tremendous public health concern. It’s important to mention that the acquired AMR genes in bacterial genomes do not inevitably confer phenotypic resistance and vice versa (Boolchandani et al., 2019; Tang et al., 2022a). Other mechanisms such as SNPs and MDRtransporter also significantly contribute to the phenotypic resistance (Boolchandani et al., 2019). The phenotypic confirmation is still essential for validating of AMR profiles. As we found in this study, only the acquired genes of carbapenem, colistin and tetracycline resistance have the highest consistency with the AMR phenotypes. While other acquired AMR genes can not predict the resistance phenotype of the bacteria very well.
Sixty-five different STs were determined in all E. coli isolates. Among them, ST10 accounted for the largest proportion, up to 10.50% (19/181). Next was ST48, and 13 isolates (7.18%) were tested for this ST type. ST10 E. coli has been seen repeatedly in related pathogenicity studies, with multiple disease outbreaks associated with ST10 in a broiler production unit by testing over 18 months (Bojesen et al., 2022).
Nine of the ST10 E. coli isolates detected in our study carried the virulence gene astA, which plays an essential role in causing diarrhea and affecting the survival of animals (Meng et al., 2014; Paixão et al., 2016). In recent years, multiple researchers have found that some virulence genes can promote the AMR of bacteria strains (Fu et al., 2022). At the same time, some toxins will be enhanced under the pressure of antibiotics (Wangkheimayum et al., 2022). In our study, the prevalance of astA is up to 36.81%. The astA gene does not sufficiently lead to disease and is also found in healthy pigs, but when it cooperates with F pili or other virulence genes such as sta and stb, it will lead to diarrhea and pose a potential threat to food safety. Hemolysin is an important pathogenic factor of pathogenic E. coli. It not only plays a role in cracking cells but also promotes cell death by activating typical inflammatory bodies in cells. Hemolysin A (HlyA) toxin is important for triggering cell death in human macrophages(Verma et al., 2019; Gu et al., 2021). Our study obtained three isolates carrying encoding genes of three kinds of α-hemolysin, hlyA, hlyB, and hlyD. Here, we would also like to note the proportion of virulence genes associated with the iron uptake system (33/169, 19.53%) and adhesion (46/169, 27.22%). Adhesins are usually a class of glycoprotein or glycolipid biomolecules that are mainly distributed on the surface of bacteria (Berne et al., 2015). The adhesins identified in this study included F1C hairs, K88 hairs, type I hairs, and E. coli common hairs.
E. coli isolates harbored several kinds of plasmids, with the dominance of IncFIB (AP001918) followed by IncX1. These plasmids have been verified linked to resistance to several antimicrobial classes, including β-lactams, aminoglycosides, sulfonamides, tetracyclines, etc. Our research also showed that these plasmids were carried by isolates originating from chickens and pigs in Zhejiang provinces, indicating that these plasmids could disseminate widely among different hosts (Hu et al., 2016; Tang et al., 2019, 2020a).
In summary, a comprehensive study of AMR and genomic characterization of E. coli isolated from pigs and chickens anal swab samples in Eastern China was conducted. A high prevalence of MDR E. coli and many virulence determinants in the isolates posed a potential risk to food safety. This is a significant warning for public health safety in Zhejiang Province. It is essential to continue monitoring the MDR E. coli and implement antimicrobial stewardship programs for rational use of essential antimicrobials in animal farms to improve food safety and prevent the emergence of MDR bacteria.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
WZ, BT, ZY, and MY: conceptualization. HY, MY, and BT: funding acquisition. WZ, ZZ, HL, JWu, YD, HY, HJ, and BT: investigation. WZ, ZZ, and BT: methodology. ZY, BT, and MY: supervision. RL, JWa, XZ, and BT: visualization. RL and WZ: writing–original draft. All authors have read and agreed to the published version of the manuscript.
This work was supported by the Key Research and Development Program of Zhejiang Province (2020C02031), the Key Research and Development Program of Hangzhou (202203A08), the earmarked fund for China Agriculture Research System (CARS-42-27), the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products (2010DS700124-ZZ2102), and Collaborative Extension Plan of Major Agricultural Technologies in Zhejiang Province (2021XTTGXM03).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1018682/full#supplementary-material
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Berne, C., Ducret, A., Hardy, G. G., and Brun, Y. V. (2015). Adhesins involved in attachment to abiotic surfaces by gram-negative bacteria. Microbiol Spectr 3:10. doi: 10.1128/microbiolspec.MB-0018-2015
Bojesen, A. M., Ahmed, U., Skaarup, H., and Espinosa-Gongora, C. (2022). Recurring outbreaks by the same Escherichia coli ST10 clone in a broiler unit during 18 months. Vet. Res. 53:2. doi: 10.1186/s13567-021-01017-6
Boolchandani, M., D'Souza, A. W., and Dantas, G. (2019). Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 20, 356–370. doi: 10.1038/s41576-019-0108-4
Brisola, M. C., Crecencio, R. B., Bitner, D. S., Frigo, A., Rampazzo, L., Stefani, L. M., et al. (2019). Escherichia coli used as a biomarker of antimicrobial resistance in pig farms of southern Brazil. Sci. Total Environ. 647, 362–368. doi: 10.1016/j.scitotenv.2018.07.438
Chang, J., Tang, B., Chen, Y., Xia, X., Qian, M., and Yang, H. (2020). Two IncHI2 plasmid-mediated Colistin-resistant Escherichia coli strains from the broiler chicken supply chain in Zhejiang Province, China. J. Food Prot. 83, 1402–1410. doi: 10.4315/jfp-20-041
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Deng, H., Sun, J., Ma, J., Li, L., Fang, L. X., Zhang, Q., et al. (2014). Identification of the multi-resistance gene cfr in Escherichia coli isolates of animal origin. PLoS One 9:e102378. doi: 10.1371/journal.pone.0102378
Fu, D., Wu, J., Gu, Y., Li, Q., Shao, Y., Feng, H., et al. (2022). Corrigendum to the response regulator OmpR contributes to the pathogenicity of avian pathogenic Escherichia coli. Poult. Sci. 101:101876. doi: 10.1016/j.psj.2022.101876
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Gu, H., Cai, X., Zhang, X., Luo, J., Zhang, X., Hu, X., et al. (2021). A previously uncharacterized two-component signaling system in uropathogenic Escherichia coli coordinates protection against host-derived oxidative stress with activation of hemolysin-mediated host cell pyroptosis. PLoS Pathog. 17:e1010005. doi: 10.1371/journal.ppat.1010005
Guan, C., Tang, B., Yang, H., Ma, J., Huang, Y., and Liu, C. (2022). Emergence of plasmid-mediated tigecycline resistance gene, tet(X4), in Escherichia fergusonii from pigs. J Glob Antimicrob Resist 30, 249–251. doi: 10.1016/j.jgar.2022.06.029
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Hu, Y., Yang, X., Li, J., Lv, N., Liu, F., Wu, J., et al. (2016). The bacterial mobile resistome transfer network connecting the animal and human microbiomes. Appl. Environ. Microbiol. 82, 6672–6681. doi: 10.1128/aem.01802-16
Li, Y., Ed-Dra, A., Tang, B., Kang, X., Müller, A., Kehrenberg, C., et al. (2022a). Higher tolerance of predominant Salmonella serovars circulating in the antibiotic-free feed farms to environmental stresses. J. Hazard. Mater. 438:129476. doi: 10.1016/j.jhazmat.2022.129476
Li, Y., Kang, X., Ed-Dra, A., Zhou, X., Jia, C., Müller, A., et al. (2022b). Genome-based assessment of antimicrobial resistance and virulence potential of isolates of non-pullorum/gallinarum Salmonella serovars recovered from dead poultry in China. Microbiol Spectr 10:e0096522. doi: 10.1128/spectrum.00965-22
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72, 393–401. doi: 10.1093/jac/dkw411
Lin, J., Tang, B., Zheng, X., Chang, J., Ma, J., He, Y., et al. (2022). Emergence of Incl2 plasmid-mediated colistin resistance in avian Escherichia fergusonii. FEMS Microbiol. Lett. 369:fnac016. doi: 10.1093/femsle/fnac016
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Ma, J., Zhou, W., Wu, J., Liu, X., Lin, J., Ji, X., et al. (2022). Large-scale studies on antimicrobial resistance and molecular characterization of Escherichia coli from food animals in developed areas of eastern China. Microbiol Spectr 10:e0201522. doi: 10.1128/spectrum.02015-22
Meng, Q., Bai, X., Zhao, A., Lan, R., Du, H., Wang, T., et al. (2014). Characterization of Shiga toxin-producing Escherichia coli isolated from healthy pigs in China. BMC Microbiol. 14:5. doi: 10.1186/1471-2180-14-5
Paixão, A. C., Ferreira, A. C., Fontes, M., Themudo, P., Albuquerque, T., Soares, M. C., et al. (2016). Detection of virulence-associated genes in pathogenic and commensal avian Escherichia coli isolates. Poult. Sci. 95, 1646–1652. doi: 10.3382/ps/pew087
Peng, X., Ed-Dra, A., and Yue, M. (2022). Whole genome sequencing for the risk assessment of probiotic lactic acid bacteria. Crit. Rev. Food Sci. Nutr. 1–19. doi: 10.1080/10408398.2022.2087174
Peng, Z., Hu, Z., Li, Z., Zhang, X., Jia, C., Li, T., et al. (2022). Antimicrobial resistance and population genomics of multidrug-resistant Escherichia coli in pig farms in mainland China. Nat. Commun. 13:1116. doi: 10.1038/s41467-022-28750-6
Rutuja, D., Ragini, M., Dhananjaya, S., Kayzad, N., Appasaheb, G., and Tannaz, B. (2018). Antibiotic resistance characterization of Environmental E. coli isolated from river Mula-Mutha, Pune District, India. Int. J. Environ. Res. Public Health 15:1247. doi: 10.3390/ijerph15061247
Shen, Z., Wang, Y., Shen, Y., Shen, J., and Wu, C. (2016). Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 16:293. doi: 10.1016/s1473-3099(16)00061-x
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Chen, C., Cui, C. Y., Zhang, Y., Liu, X., Cui, Z. H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
Tang, B., Chang, J., Cao, L., Luo, Q., Xu, H., Lyu, W., et al. (2019). Characterization of an NDM-5 carbapenemase-producing Escherichia coli ST156 isolate from a poultry farm in Zhejiang, China. BMC Microbiol. 19:82. doi: 10.1186/s12866-019-1454-2
Tang, B., Chang, J., Chen, Y., Lin, J., Xiao, X., Xia, X., et al. (2022a). Escherichia fergusonii, an underrated repository for antimicrobial resistance in food animals. Microbiol Spectr 10:e0161721. doi: 10.1128/spectrum.01617-21
Tang, B., Chang, J., Luo, Y., Jiang, H., Liu, C., Xiao, X., et al. (2022b). Prevalence and characteristics of the mcr-1 gene in retail meat samples in Zhejiang Province, China. J. Microbiol. 60, 610–619. doi: 10.1007/s12275-022-1597-y
Tang, B., Chang, J., Zhang, L., Liu, L., Xia, X., Hassan, B. H., et al. (2020a). Carriage of distinct mcr-1-harboring plasmids by unusual serotypes of Salmonella. Adv. Biosyst. 4:e1900219. doi: 10.1002/adbi.201900219
Tang, B., Tang, Y., Zhang, L., Liu, X., Chang, J., Xia, X., et al. (2020b). Emergence of fexA in mediating resistance to Florfenicols in Campylobacter. Antimicrob. Agents Chemother. 64:e00260-20. doi: 10.1128/AAC.00260-20
Tang, B., Wang, Y., Luo, Y., Zheng, X., Qin, X., Yang, H., et al. (2021a). Coexistence of optrA and fexA in Campylobacter. mSphere 6:e00125-21. doi: 10.1128/mSphere.00125-21
Tang, B., Wang, J., Zheng, X., Chang, J., Ma, J., Wang, J., et al. (2022c). Antimicrobial resistance surveillance of Escherichia coli from chickens in the Qinghai plateau of China. Front. Microbiol. 13:885132. doi: 10.3389/fmicb.2022.885132
Tang, B., Yang, H., Jia, X., and Feng, Y. (2021b). Coexistence and characterization of Tet(X5) and NDM-3 in the MDR-Acinetobacter indicus of duck origin. Microb. Pathog. 150:104697. doi: 10.1016/j.micpath.2020.104697
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Teng, L., Liao, S., Zhou, X., Jia, C., Feng, M., Pan, H., et al. (2022). Prevalence and genomic investigation of multidrug-resistant Salmonella isolates from companion animals in Hangzhou, China. Antibiotics (Basel) 11:625. doi: 10.3390/antibiotics11050625
Verma, V., Gupta, S., Kumar, P., Rawat, A., Singh Dhanda, R., and Yadav, M. (2019). Efficient production of endotoxin depleted bioactive α-hemolysin of uropathogenic Escherichia coli. Prep. Biochem. Biotechnol. 49, 616–622. doi: 10.1080/10826068.2019.1591993
Wang, J., Tang, B., Lin, R., Zheng, X., Ma, J., Xiong, X., et al. (2022). Emergence of mcr-1- and blaNDM-5-harbouring IncHI2 plasmids in Escherichia coli strains isolated from meat in Zhejiang, China. J. Glob. Antimicrob. Resist. 30, 103–106. doi: 10.1016/j.jgar.2022.06.002
Wang, Y., Tian, G. B., Zhang, R., Shen, Y., Tyrrell, J. M., Huang, X., et al. (2017). Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 17, 390–399. doi: 10.1016/s1473-3099(16)30527-8
Wangkheimayum, J., Chanda, D. D., and Bhattacharjee, A. (2022). Expression of itaT toxin gene is enhanced under aminoglycoside stress in Escherichia coli harbouring aac(6′)-Ib. Gene Reports 26:101526. doi: 10.1016/j.genrep.2022.101526
Xu, L., Wan, F., Fu, H., Tang, B., Ruan, Z., Xiao, Y., et al. (2022). Emergence of Colistin resistance gene mcr-10 in Enterobacterales isolates recovered from fecal samples of chickens, slaughterhouse workers, and a nearby resident. Microbiol Spectr 10:e0041822. doi: 10.1128/spectrum.00418-22
Keywords: Escherichia coli, animal origin, antimicrobial resistance, genomic characterization, virulence genes
Citation: Zhou W, Lin R, Zhou Z, Ma J, Lin H, Zheng X, Wang J, Wu J, Dong Y, Jiang H, Yang H, Yang Z, Tang B and Yue M (2022) Antimicrobial resistance and genomic characterization of Escherichia coli from pigs and chickens in Zhejiang, China. Front. Microbiol. 13:1018682. doi: 10.3389/fmicb.2022.1018682
Received: 13 August 2022; Accepted: 06 October 2022;
Published: 24 October 2022.
Edited by:
Haihong Hao, Huazhong Agricultural University, ChinaReviewed by:
Tao He, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaCopyright © 2022 Zhou, Lin, Zhou, Ma, Lin, Zheng, Wang, Wu, Dong, Jiang, Yang, Yang, Tang and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhangnv Yang, em55YW5nQGNkYy56ai5jbg==; Biao Tang, dGFuZ2JpYW9AemFhcy5hYy5jbg==; Min Yue, bXl1ZUB6anUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.