- 1College of Agronomy and Biosciences, Dali University, Dali, Yunnan, China
- 2Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 3Department of Entomology and Plant Pathology, Faculty of Agriculture, Chiang Mai University, Chiang Mai, Thailand
- 4School of Science, Mae Fah Luang University, Chiang Rai, Thailand
Rhododendron is an essential ornamental plant that is abundant in Yunnan province. In Cangshan Mountain, Yunnan, China, 61 species of Rhododendron have been reported. Endophytic fungi are internal plant tissue inhabitants that do not harm the host. It has emerged as an exciting research topic as they have the potential to provide numerous secondary metabolites. This study is focused on taxonomic novelties and new host records of endophytic fungi associated with Rhododendron plants collected from Cangshan Mountain in Yunnan Province, China. Pestalotiopsis fungi are associated with a vast array of plant species worldwide. In this study, fresh leaves of Rhododendron cyanocarpum, Rhododendron decorum, and Rhododendron delavayi were collected from Cangshan Mountain, Yunnan Province, China. Endophytic Pestalotiopsis fungi associated with Rhododendron were characterized based on phylogenetic analyses of combined ITS, TEF1-α, and TUB genes along with morphological characteristics. Six new species (Pestalotiopsis appendiculata, Pestalotiopsis cangshanensis, Pestalotiopsis daliensis, Pestalotiopsis fusoidea, Pestalotiopsis rosarioides, and Pestalotiopsis suae) and a new host record (Pestalotiopsis trachicarpicola) are described. Detailed descriptions and color photo plates of these species are provided. It is the first time that the endophytic fungi of Rhododendron plants in Cangshan Mountain have been studied.
Introduction
Rhododendron is the largest genus of woody plants in the Northern Hemisphere and the largest genus in Ericaceae (Fang et al., 2005; Shrestha et al., 2018). Rhododendron is an important component of montane ecosystems, with more than 1,025 species and approximately 581 species in China (Cai et al., 2016; Ma et al., 2017; Cao et al., 2022). Moreover, Yunnan province in southwest China is a center of diversity for Rhododendron (Ma et al., 2021). Rhododendron is a popular landscape plant and a food source (Negi et al., 2013; Lin et al., 2021). Due to the economic significance of this plant, it is essential to evaluate the fungi associated with it (Chaiwan et al., 2022). Pathogenic fungi, mycorrhizal fungi, and endophytic fungi have been isolated from Rhododendron in previous research (Zhang et al., 2019); however, there is no record of endophytic fungi associated with Rhododendron in Cangshan Mountain, Yunnan, China.
Endophytic fungi or endophytes exist widely inside the healthy tissues of living plants and are important components of plant micro-ecosystems (Jia et al., 2016). Endophytic fungi benefit their host plants by increasing their drought resistance, disease resistance, and growth-promoting properties (Rodriguez et al., 2009; De Silva et al., 2019; Rashmi et al., 2019). Endophytic fungi have the potential to produce metabolites with a wide range of biological activities, making them an appealing research topic (Huang et al., 2009; De Silva et al., 2019; Rashmi et al., 2019). More than 800 endophytic fungal genera have been reported worldwide, most speciose genera (>50 species) are Penicillium (103), Colletotrichum (78), Alternaria (61), Fusarium (59), Pestalotiopsis (53), and Aspergillus (52) (Rashmi et al., 2019). Amongst the different substrates, leaf endophytes have been studied and analyzed in more detail when compared to other parts (Rashmi et al., 2019).
Steyaert (1949) introduced Pestalotiopsis to accommodate species with fusiform conidia with three colored median cells and two colorless end cells, as well as two or more apical appendages. Traditionally, taxonomy and identification of Pestalotiopsis and allied genera were based mainly on conidial characters (Steyaert, 1949; Guba, 1961) and conidiogenesis (Sutton, 1980). Hu et al. (2007) reported that conidial characteristics, such as conidial length, median cell length, conidial width, and median cell color, were insignificantly different within Pestalotiopsis. Maharachchikumbura et al. (2014) selected internal transcribed spacer (ITS), partial β-tubulin (TUB), and partial translation elongation factor 1-alpha (TEF1-α), along with morphological characters to resolve the identification of Pestalotiopsis. Up to now, in a total of 92 Pestalotiopsis species have been introduced based on morphological and phylogenetic evidence (Maharachchikumbura et al., 2014; Liu et al., 2019; Rashmi et al., 2019; Shu et al., 2020; Monteiro et al., 2022). Pestalotiopsis is ordinarily isolable as endophytes in plants (Aly et al., 2010; Watanabe et al., 2010; Maharachchikumbura et al., 2012). However, there is only one Pestalotiopsis species viz., Pestalotiopsis baarnensis associated with Rhododendron (Rashmi et al., 2019).
As a part of the investigation on endophytic fungal diversity associated with Rhododendron plants in Cangshan Mountain, Yunnan province, China, we revealed seven Pestalotiopsis-like taxa from fresh leaves of Rhododendron. Their taxonomic positions were established based on morphological descriptions and multi-locus phylogenetic analyses. The endophytic fungal strain resources were stored for future study on their secondary metabolites.
Materials and methods
Isolation and morphology
Fresh Rhododendron (Ericaceae) leaves were obtained from Cangshan Mountain, Yunnan Province, China. The gathered leaves were placed in a sterile polyethylene bag and stored at 4°C. The symptomless leaves of each Rhododendron spp. were treated with gently running tap water to remove the surface debris. They were surface-sterilized by using 75% ethanol for 1 min, 0.1% HgCl2 for 3 min, and washed five times using sterile distilled water, finally dried on sterile filter paper (Tao et al., 2013). The 5-mm diameter leaf discs treated as above were placed on potato dextrose agar (PDA) plates without antibiotics. The PDA plates were incubated in ambient light at 25°C. When colonies appeared, they were transferred onto new PDA plates and further incubated in ambient light at 25°C for morphological examination. Sporulation was induced on pine needle medium (“pine needle” and 1/10-strength PDA). Macromorphological characters of conidiomata on PDA were observed using an Optec SZ 760 compound stereomicroscope. Temporarily prepared microscope slides were placed under a Nikon ECLIPSE Ni-U compound stereomicroscope for observation and micro-morphological photography. Part of the pure culture that produced spores was removed and put into a water–agar medium (WA) with glycerol and air-dried at room temperature (De Silva et al., 2019). All endophytic isolates are stored at the Culture Collection of Kunming Institute of Botany, the Chinese Academy of Sciences (KUNCC), and the China General Microbiological Culture Collection Center (CGMCC). The Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (Herb. KUN-HKAS) housed the herbarium specimens. The MycoBank1 number was registered (Crous et al., 2004).
DNA extraction, PCR amplification, and sequencing
Genomic DNA extraction was carried out from fresh mycelium growing on PDA at 25°C using the Trelief™ Plant Genomic DNA Kit according to the manufacturer’s instructions. The primer pairs ITS5/ITS4, EF1-728F/EF2, and Bt2a/Bt2b were used to amplify the ITS, TEF1-α, and TUB gene regions, respectively. The amplification was performed in a 25 μL reaction volume containing 12.5 μL of Master Mix (Tsingke Biotech, Yunnan, China), 1 μL of each primer (10 μm), 1 μL of template DNA, and 9.5 μL of deionized water. The PCR thermal cycles for three genes were performed under the following reaction conditions: an initial denaturing step for 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 45 s for ITS and TEF1-α, and 56°C for 60 s for TUB, elongation at 72°C for 1 min, and a final extension at 72°C for 10 min. PCR products were verified on 1% agarose electrophoresis gels stained with ethidium bromide. Sequencing was carried out by Tsingke Biological Engineering Technology and Services Co., Ltd. (Yunnan, China).
Molecular phylogenetic analyses
Sequence alignment
Sequences with high similarity indices were assembled in BioEdit, and those with low similarity indices were identified through a BLAST search for the closest matches with Pestalotiopsis taxa and from recently published data (Li et al., 2021). All consensus and reference sequences were automatically aligned with MAFFT v.7 using the Auto strategy (Katoh and Standley, 2013). The aligned sequences from each gene region (ITS, TEF1-α, and TUB) were combined and manually improved using Sequence Matrix (Hall, 1999). Uncertain regions were omitted from the alignment, and gaps were treated as data that was missing. Maximum likelihood (ML) and Bayesian inference were used to conduct phylogenetic analyses.
Phylogenetic analyses
Maximum likelihood analysis was performed at the CIPRES Science Gateway v.3.3 (Miller et al., 2010) using RAxML v.8.2.8 as part of the “RAxML-HPC2 on XSEDE” tool (Stamatakis, 2006; Stamatakis et al., 2008). The optimal ML tree search was conducted with 1,000 separate runs using the default algorithm of the programme from a random starting tree for each run. The final tree was selected amongst suboptimal trees from each run by comparing the likelihood scores using the GTR+GAMMA substitution model. Maximum likelihood bootstrap values equal to or greater than 60% were given as the first set of numbers above the nodes in the resulting ML tree.
Bayesian analysis was performed with MrBayes v.3.1.2 (Ronquist and Huelsenbeck, 2003) to evaluate posterior probabilities (Rannala and Yang, 1996) using Markov Chain Monte Carlo sampling (MCMC). The best-fit model of evolution was estimated using MrModeltest v.2.2 (Nylander, 2004). For Bayesian analysis, the best-fitting model of ITS, TEF1-α, and TUB was the GTR+I+G model. Posterior probabilities (PPs) (Rannala and Yang, 1996) were performed using Markov chain Monte Carlo sampling (BMCMC) in MrBayes v.3.1.2 (Liu et al., 2012). Six concurrent Markov chains were executed for 50 million generations, and samples of trees were taken every 5,000 generations (resulting in 10,000 trees). The initial 2,000 trees representing the burn-in phase of the analyses were discarded, while the remaining 8,000 trees were used to calculate PP in the majority rule consensus tree (Cai et al., 2006; Liu et al., 2012).
Phylogenetic trees were displayed in FigTree v. 1.4.4 (Rambaut, 2014) and edited in Adobe Illustrator CS5 (Adobe Systems, San Jose, CA, USA). Newly generated sequences were deposited in GenBank (Table 1).
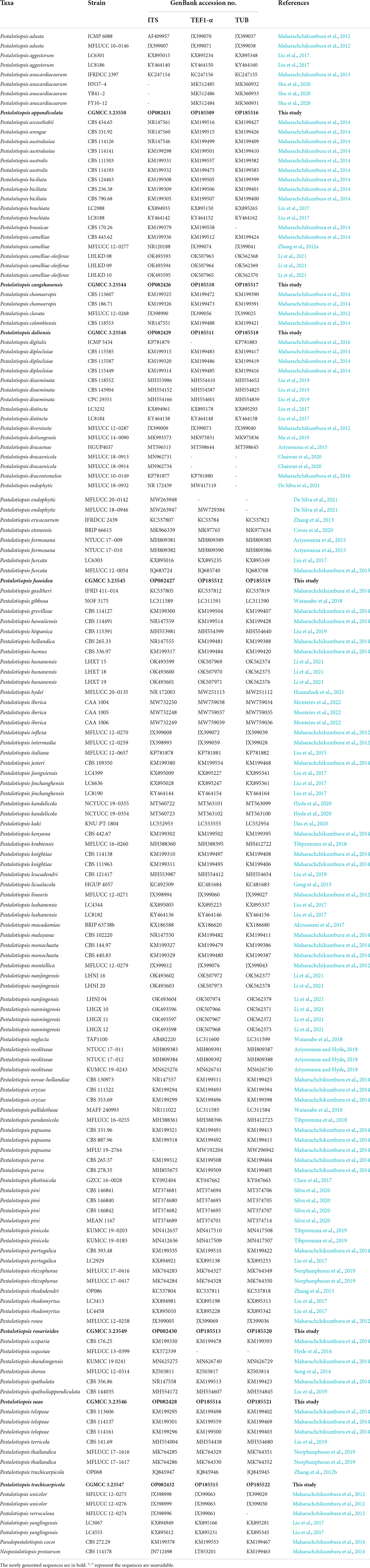
Table 1. GenBank numbers and culture collection accession numbers of species included in the phylogenetic study.
Results
Phylogenetic analyses
The combined ITS, TEF1-α, and TUB sequence dataset included 154 ingroup taxa and two outgroup taxa (Neopestalotiopsis protearum and Pseudopestalotiopsis cocos) with 2,160 characters (ITS: 1–538 bp; TEF: 539–1477 bp; TUB: 1478–2160 bp) overall post-alignment, including the gaps. The RAxML and Bayesian analyses of the combined dataset resulted in phylogenetic reconstructions with largely identical topologies and a ML analysis with a final likelihood value of −17482.622268, as shown in Figure 1. The matrix exhibited 993 distinct alignment patterns, with 24.14% undetermined characters or gaps. The estimated base frequencies were as follows: A = 0.237874, C = 0.294954, G = 0.216783, T = 0.250389; substitution rates AC = 1.059763, AG = 3.258532, AT = 1.260093, CG = 0.980806, CT = 4.659318, GT = 1.000000; gamma distribution shape parameter α = 0.309168. The bootstrap support values for RAxML greater than 60% and the Bayesian posterior probabilities greater than 0.95 are given at each node (Figure 1).
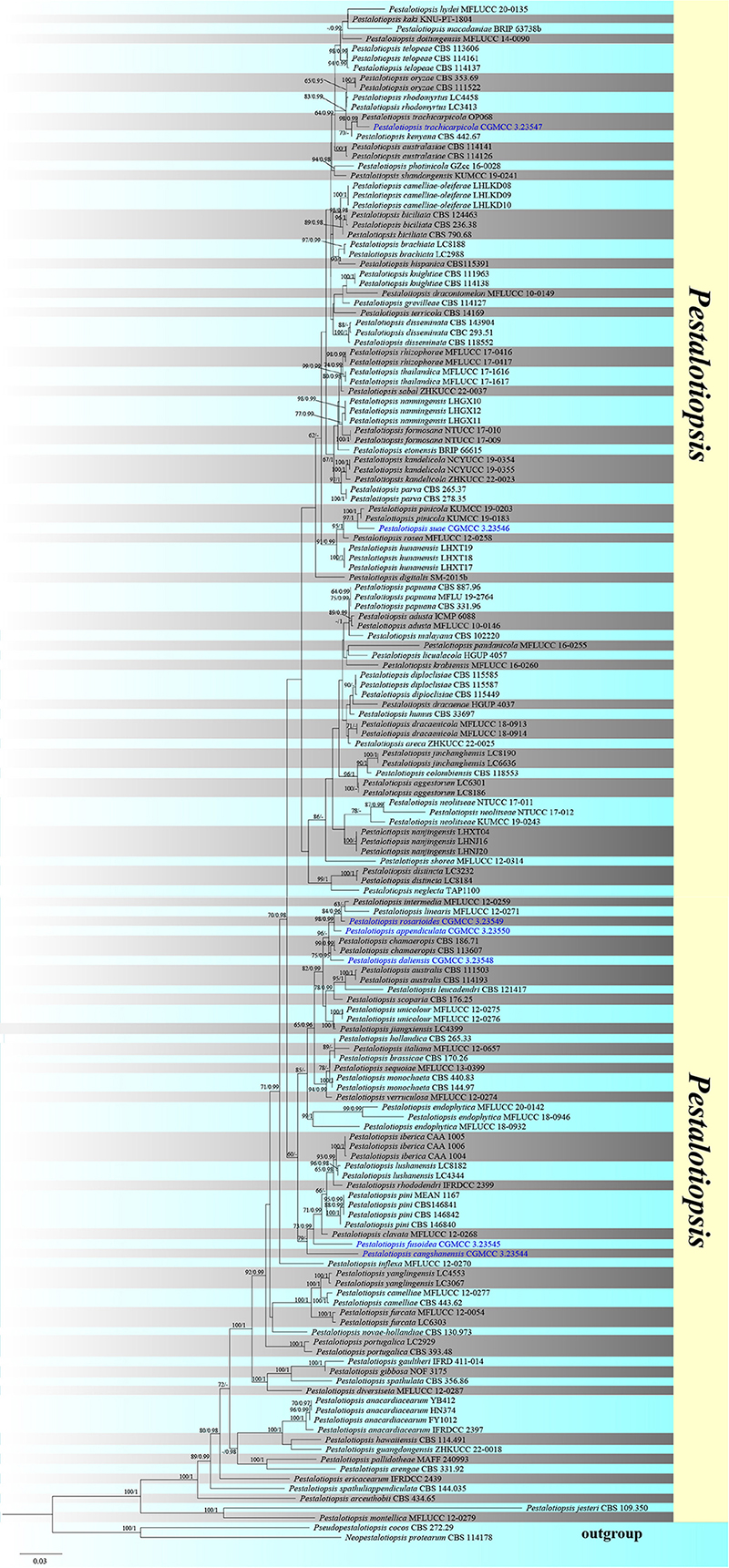
Figure 1. Phylogenetic tree based on RAxML analyses of a combined internal transcribed spacer (ITS), translation elongation factor 1-alpha (TEF1-α), and β-tubulin (TUB) dataset. Bootstrap support values for maximum likelihood ≥60% and Bayesian posterior probabilities ≥0.95 are indicated above the nodes as Maximum likelihood/Posterior probabilitie (ML/PP). The tree is rooted in Neopestalotiopsis protearum (CBS 114.178) and Pseudopestalotiopsis cocos (CBS 272.29). The new isolates are in blue.
In the phylogenetic analyses, all new strains were grouped with members of Pestalotiopsis. Pestalotiopsis rosarioides, Pestalotiopsis intermedia, and Pestalotiopsis linearis were grouped together; however, P. rosarioides has a separate branch with 84% ML and 0.96 BYPP support. Pestalotiopsis appendiculata established a distinct lineage with 98% ML and 0.99 BYPP bootstrap support. Pestalotiopsis suae was clustered as a sister taxon to Pestalotiopsis pinicola with a significant support (97% ML and 1 BYPP). Pestalotiopsis daliensis was clustered as a sister to Pestalotiopsis chamaeropis with significant support (75% ML and 0.95 BYPP). Pestalotiopsis fusoidea, Pestalotiopsis cangshanensis, Pestalotiopsis pini, Pestalotiopsis lushanensis, Pestalotiopsis rhododendri, and Pestalotiopsis clavate were grouped together in an independent clade within Pestalotiopsis, while P. fusoidea and P. cangshanensis formed distinct branches. Pestalotiopsis trachicarpicola clustered with the ex-type of P. trachicarpicola with strong support (98% ML and 0.99 BYPP).
Pestalotiopsis appendiculata D.F. Bao, R. Gu and Z.L. Luo, sp. nov.
MycoBank number: 845187, Figure 2.

Figure 2. Pestalotiopsis appendiculata (KUN-HKAS 124571, holotype). (A) Leaves of Rhododendron decorum. (B,C) Culture on potato dextrose agar (PDA) (upper and lower view). (D,E) Conidiomata on PDA. (F,G) Conidiophores, conidiogenous cells, and conidia. (H–O) Conidia. Scale bars: (F) 15 μm, (G) 10 μm, (H) 15 μm, (I–O) 10 μm.
Holotype–KUN-HKAS 124571
Etymology–“appendiculata,” denoting the fungus conidial appendages.
Endophytic in fresh Rhododendron decorum leaves. Sexual morph: Undetermined. Asexual morph: Conidiomata pycnidial in PDA culture, globose or clavate, aggregated or scattered, semi-immersed to erumpent, gray. There is no evidence of a conidiogenous cell. Conidia 19–24 × 5–6 μm ( = 21 × 5 μm, n = 30), fusoid, ellipsoid, straight to slightly curved, four-septate, slightly constricted at septa. Basal cell 2–4 μm long, conic to obconic with a truncate base, hyaline, verrucose, and thin-walled. Three-median cells doliiform, plicated, 13–15 μm ( = 14 μm, n = 30). Thin-walled, with a uniform light color on the third cell and the fourth cell relatively darker, the septa darker than the rest of the cells (second cell from the base, 4–6 μm long; third cell 5–6 μm long; fourth cell 4–6 μm long). Apical cell 2–4 μm long, hyaline, subcylindrical, or obconic with a truncated base, thin-walled, slightly rugose. With 2–3 tubular apical appendages arising from the apical crest, unbranched, filiform, 8–15 μm ( = 12 μm, n = 30). Basal appendage single, tubular, centric, or uncentred, 3–5 μm long.
Material examined: China, Yunnan Province, Dali City, Cangshan Mountain, isolated from healthy leaves of R. decorum (2489 m, 24.3240°N, 101.0140°E), April 2021, Z.Q. Zhang, D-60 (KUN-HKAS 124571, holotype), ex-type culture, CGMCC 3.23550 = KUNCC 22-12405.
Notes: P. appendiculata, P. intermedia, P. linearis, and P. rosarioides were grouped in the same clade in the phylogenetic analysis. Comparisons of ITS, TEF-1α, and TUB gene regions of P. appendiculata with P. linearis (2/538 in ITS, 12/398 in TEF-1α, and 23/450 in TUB), P. intermedia (3/537 in ITS, 17/398 in TEF-1α, and 13/479 in TUB), and P. rosarioides (3/553 in ITS, 22/553 in TEF-1α, and 22/458 in TUB) indicated significant differences. However, P. appendiculata can be distinguished from P. linearis and P. intermedia with its smaller conidia (Table 2). Moreover, the three-median cells of conidia in P. appendiculata are light gray; in contrast, they are brown in P. linearis, P. intermedia, and P. rosarioides. Based on the combined ITS, TEF-1α, and TUB sequence data, our phylogenetic analysis revealed that they are clearly distinct in the phylogram. P. appendiculata formed a separate branch with strong support values (98 ML/0.99 PP, Figure 1). Therefore, based on phylogenetic analysis and its morphological characteristics, P. appendiculata is introduced as a new species.
Pestalotiopsis cangshanensis H.W. Shen, R. Gu and Z.L. Luo, sp. nov.
MycoBank number: 845188, Figure 3.

Figure 3. Pestalotiopsis cangshanensis (KUN-HKAS 124573, holotype). (A) Leaves of Rhododendron delavayi. (B,C) Culture on potato dextrose agar (PDA) (upper and lower view). (D,E) Conidiomata on PDA. (F) Conidiophores, conidiogenous cells, and conidia. (G–O) Conidia with appendages. Scale bars: (F–H) 10 μm, (I–M) 15 μm, (N,O) 10 μm.
Holotype–KUN-HKAS 124573
Etymology–“cangshanensis,” referring to the Cangshan Mountain, where the species was obtained.
Endophytic in fresh Rhododendron delavayi leaves. Sexual morph: Undetermined. Asexual morph: Conidiomata pycnidial on PDA, dark brown to black conidial masses, globose, ink-shaped. Conidiophores indistinct and typically reduced to conidiogenous cells. Conidiogenous cells discrete, subcylindrical to ampulliform, hyaline, sometimes slightly wide at the base, truncated at the apex, 4–24 × 2–4 μm. Conidia 18–23 × 6–8 μm ( = 19 × 2 μm, n = 30), fusoid, straight to slightly curved, four-septate; three-median cells light or dark brown, 10–14 μm long ( = 12 μm, n = 30; second cell from the base pale-light brown 3–5 μm long; third cell 3–5 μm long; fourth cell 3–5 μm long), wall verruculose, concolourous. Base cell inverted funnel-shaped with a truncated base, 3–4 μm long ( = 4 μm), hyaline, thin-walled. Apical cell 4–5 μm long ( = 4 μm), hyaline, cylindrical to subcylindrical, thin, and smooth-walled. With three tubular apical appendages 9–19 μm long ( = 15 μm, n = 30) arising from the apical crest, filiform, unbranched. Basal appendage single, tubular, unbranched, centric, 5–8 μm long ( = 7 μm, n = 30).
Material examined: China, Yunnan Province, Dali City, Cangshan Mountain, isolated from healthy leaves of R. delavayi (2489 m, 25.4724°N, 99.5949°E), March 2021, Z.Q. Zhang, D-6 (KUN-HKAS 124573, holotype), ex-type culture, CGMCC 3.23544.
Notes: P. cangshanensis, P. clavate, P. lushanensis, P. rhododendri, and P. pini were grouped together. Comparisons of ITS, TEF-1α, and TUB gene regions of P. cangshanensis with P. lushanensis (2/505 in ITS, 16/932 in TEF-1α, and 12/789 in TUB), P. pini (2/605 in ITS, 17/474 in TEF-1α, and 11/792 in TUB), P. rhododendri (2/538 in ITS, 17/941 in TEF-1α, and 11/458 in TUB), and P. clavate (1/539 in ITS, 10/947 in TEF-1α, and 19/457 in TUB) exhibited significant differences. Morphologically, P. cangshanensis has smaller conidia than P. pini, P. clavata, P. rhododendri, and P. lushanensis (Table 3). Moreover, P. cangshanensis has shorter apical appendages than those of P. rhododendri (21–28 vs. 9–19 μm) and P. lushanensis (17–26 vs. 9–19 μm). Based on combined ITS, TEF1-α, and TUB sequence data, P. cangshanensis is clearly separated in the phylogram as it forms an independent clade. It indicates that P. cangshanensis can be introduced as a new species.
Pestalotiopsis daliensis H.W. Shen, R. Gu and Z.L. Luo, sp. nov.
MycoBank number: 845189, Figure 4.

Figure 4. Pestalotiopsis daliensis (KUN-HKAS 124576, holotype). (A) Leaves of Rhododendron decorum. (B,C) Culture on potato dextrose agar (PDA) (upper and lower view). (D,E) Conidiomata on PDA. (F) Conidiophores, conidiogenous cells, and conidia. (G–O) Conidia. Scale bars: (F) 25 μm, (G) 15 μm, and (H–O) 10 μm.
Holotype–KUN-HKAS 124576
Etymology–“daliensis,” referring to Dali City, where the species was obtained.
Endophytic in fresh R. decorum leaves. Sexual morph: Undetermined. Asexual morph: Conidiomata (on PDA) pycnidial, globose to clavate, solitary, exuding globose, dark-brown to black conidial masses. Conidiophores are usually indistinct and reduced to conidiogenous cells. Conidiogenous cells discrete, subcylindrical to ampulliform, hyaline, sometimes slightly wide at the base, truncated at the apex, 5–19 × 1–3 μm. Conidia 23–26 × 4–5 μm ( = 25 × 5 μm, n = 30), fusoid, ellipsoid, straight to slightly curved, four-septate; basal cell conic with a truncated base, hyaline or light-brown and thin-walled, 4–6 μm long ( = 5 μm, n = 30). Three-median cells dark, 13–16 μm long ( = 15 μm, n = 30), wall smooth, concolourous, septa darker than the rest of the cells (second cell from the base pale brown, 4–5 μm long; third cell, 4–5 μm long; fourth cell, 4–6 μm long). Apical cell 4–6 μm long ( = 5 μm, n = 30), hyaline, subcylindrical, thin-walled, and smooth-walled. With 2–3 tubular apical appendages 13–22 μm long ( = 18 μm, n = 30), arising from the apical crest, unbranched, filiform. Basal appendage 10–16 μm long ( = 13 μm, n = 30), single, tubular, unbranched, centric, straight, or slightly curved.
Material examined: China, Yunnan Province, Dali City, Cangshan Mountain, isolated from healthy leaves of R. decorum (2470 m, 25.5044°N, 100.0542°E), March 2021, Z.Q. Zhang, D-33 (KUN-HKAS 124576, holotype), ex-type culture, CGMCC 3.23548 = KUNCC 22-12403.
Notes: In the phylogenetic analysis, P. chamaeropis and P. daliensis are closely associated. Comparisons of ITS, TEF-1α, and TUB gene regions of P. daliensis with P. chamaeropis (2/599 in ITS, 6/478 in TEF-1α, and 8/774 in TUB) exhibited significant differences. However, the conidia of P. daliensis are relatively narrower than P. chamaeropis (4–5 vs. 7–9 μm). Moreover, P. daliensis has a much longer conidial basal appendage (10–16 vs. 4–9 μm). Hence, P. daliensis is introduced as a new species.
Pestalotiopsis fusoidea D.F. Bao, R. Gu and Z.L. Luo, sp. nov.
MycoBank number: 845190, Figure 5.

Figure 5. Pestalotiopsis fusoidea (KUN-HKAS 124579, holotype). (A) Leaves of Rhododendron delavayi. (B,C) Culture on potato dextrose agar (PDA) (upper and lower view). (D,E) Conidiomata on PDA. (F–H) Conidiophores, conidiogenous cells, and conidia. (I–N) Conidia. Scale bars: (F–H) 20 μm, (I–K) 15 μm, and (L–N) 10 μm.
Holotype–KUN-HKAS 124579
Etymology–“fusoidea,” referring to the fusoid conidia of this fungus.
Endophytic in fresh R. delavayi leaves. Sexual morph: Undetermined. Asexual morph: Colonies on PDA attaining 15–20 mm in diameter after 7 days at 25°C. Smooth edge, whitish, gregarious. Conidiomata formation black droplets, gregarious, reverse pale yellow. Conidia aggregate in culture to form black-droplet conidia masses. Conidiophores indistinct, typically reduced to conidiogenous cells. Conidiogenous cells discrete, subcylindrical to ampulliform, hyaline, and sometimes slightly wide at the base, 5–29 × 2–4 μm. Conidia 23–26 × 6–7 ( = 25 × 7 μm, n = 30), fusoid, four-septate, lightly, curved. Three-median cells 13–18 μm long ( = 16 μm, n = 30), brown or olive. Some of the third cells are the darkest, second cell 5–6 μm long ( = 5 μm, n = 30), third cell 4–6 μm long ( = 5 μm, n = 30), fourth cell 4–6 μm long ( = 5 μm, n = 30), apical cell 3–4 μm long ( = 4 μm, n = 30), hyaline, cylindrical to subcylindrical, with 2–4 (or mostly 3) tubular apical appendages 7–11 μm long ( = 8 μm, n = 30) long arising from the apical crest, filiform. The base cell is an inverted triangle 4–6 μm long ( = 4 μm, n = 30), with light brown or almost transparent hyaline. Basal appendage single, tubular, unbranched, centric, vertical, or curved, 4–6 μm long ( = 6 μm, n = 30).
Material examined: China, Yunnan Province, Dali City, Cangshan Mountain, isolated from healthy leaves of R. delavayi (2716 m, 25.5032°N, 100.4265°E), March 2021, Z.Q. Zhang, D-8 (KUN-HKAS 124579, holotype), ex-type culture CGMCC 3.23545 = KUNCC 22-12401.
Notes: Phylogenetically, P. fusoidea has a close with P. clavata, P. lushanensis, P. rhododendri, and P. pini. Comparisons of ITS, TEF-1α, and TUB gene regions of P. fusoidea with P. lushanensis (2/505 in ITS, 16/932 in TEF-1α, and 12/789 in TUB), P. rhododendri (2/538 in ITS, 13/941 in TEF-1α, and 11/458 in TUB), P. clavate (9/539 in ITS, 14/947 in TEF-1α, and 11/457 in TUB), P. pini (2/571 in ITS, 17/512 in TEF-1α, and 11/514 in TUB) exhibited significant differences. However, P. fusoidea has shorter apical appendages than P. pini (7–11 vs. 18–20 μm), P. clavate (7–11 vs. 20–25 μm), or P. rhododendri (7–11 vs. 21–29 μm). P. fusoidea has smaller conidia than P. lushanensis (23–26 × 6–7 vs. 18–23 × 6–8 μm). Based on combined ITS, TEF1-α, and TUB sequence data, P. fusoidea are apparently separated in the phylogram, forming a separate clade. It indicates that P. fusoidea can be introduced as a new species.
Pestalotiopsis rosarioides H.W. Shen, R. Gu and Z.L. Luo, sp. nov.
MycoBank number: 845191, Figure 6.

Figure 6. Pestalotiopsis rosarioides (KUN-HKAS 124574, holotype). (A) Leaves of Rhododendron decorum. (B,C) Culture on potato dextrose agar (PDA) (upper and lower view). (D,E) Conidiomata on PDA. (F,G) Conidiophores, conidiogenous cells, and conidia. (H–O) Conidia. Scale bars: (F–O) 15 μm.
Holotype–KUN-HKAS 124574
Etymology–“rosarioides,” referring to the rosary-like enlargement of the second and fourth cells of this fungus.
Endophytic in fresh R. decorum leaves. Sexual morph: Undetermined. Asexual morph: Conidiomata (on PDA) pycnidial, globose to clavate, solitary, exuding globose, dark-brown to black conidial masses. Conidiogenous cells are not obvious. Conidia 22–25 × 6–7 μm ( = 23 × 7 μm, n = 30), fusoid, ellipsoid, rosary, straight to slightly curved, four-septate. Basal cell conic with a truncated base, hyaline or light brown, and thin-walled, 4–5 μm long ( = 5 μm, n = 30). Three-median cells dark, 16–18 μm long ( = 17 μm, n = 30), smooth wall, concolourous, septa darker than the rest of the cells (second cell from the base pale brown and enlarged, 4–5 μm long; third cell 4–5 μm long; fourth cell expands to 3–6 μm long). Apical cell 4–7 ( = 5 μm, n = 30) long, hyaline, subcylindrical, smooth-walled. With 1–3 tubular apical appendages 5–9 μm long ( = 7 μm, n = 30) arising from the apical crest, unbranched, filiform. Basal appendage 4–5 μm long ( = 4 μm, n = 30), single, tubular, unbranched, centric.
Material examined: China, Yunnan Province, Dali City, Cangshan Mountain, isolated from healthy leaves of R. decorum (2784 m, 25.9454°N, 99.9915°E), July 2021, Z.Q. Zhang, D-46 (KUN-HKAS 124574, holotype), ex-type culture, CGMCC 3.23549 = KUNCC 22-12404.
Notes: From the phylogenetic analysis, P. intermedia, P. linearis, and P. rosarioides clustered within the same clade. Comparisons of ITS, TEF-1α, and TUB gene regions of P. rosarioides with P. intermedia (2/537 in ITS, 2/946 in TEF-1α, and 9/479 in TUB), and P. linearis (2/538 in ITS, 4/946 in TEF-1α, and 12/450 in TUB) exhibited significant differences. However, the second and fourth conidial cells of P. rosarioides are enlarged, which is distinct from other species in the genus. Moreover, P. rosarioides has much shorter apical appendages than P. linearis and P. intermedia (5–9 μm in P. rosarioides vs. 10–20 μm in P. linearis and 10–28 μm in P. intermedia). Furthermore, phylogenetic analysis indicates that P. rosarioides forms a distinct lineage within the clade (Figure 1), supporting it as a new species.
Pestalotiopsis suae H.W. Shen, R. Gu and Z.L. Luo, sp. nov.
MycoBank number: 845192, Figure 7.
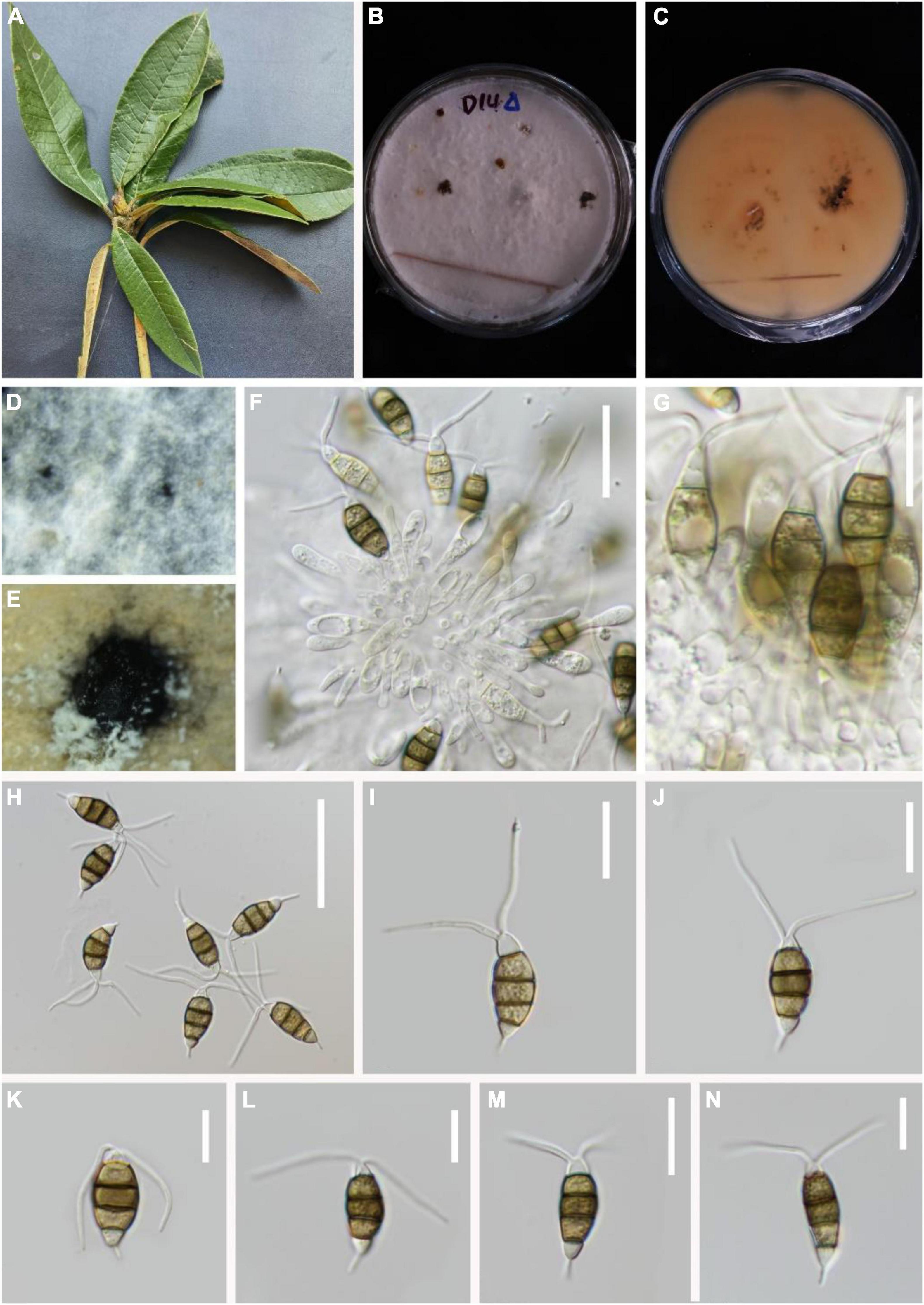
Figure 7. Pestalotiopsis suae (KUN-HKAS 124578, holotype). (A) Leaves of Rhododendron delavayi. (B,C) Culture on potato dextrose agar (PDA) (upper and lower view). (D,E) Conidiomata on PDA. (F,G) Conidiophores, conidiogenous cells, and conidia. (H–N) Conidia with appendages. Scale bars: (F) 20 μm, (G–J) 15 μm, (K–N) 10 μm.
Holotype–KUN-HKAS 124578
Etymology–“suae” in memory of the Chinese mycologist Prof. Hong-Yan Su, who kindly helped the authors in many ways and sadly passed away on 3 May 2022 during the preparation of the current article.
Endophytic in fresh R. delavayi leaves. Sexual morph: Undetermined. Asexual morph: Conidia aggregate in culture to form black-droplet conidia masses. Conidiophores indistinct and typically reduced to conidiogenous cells. Conidiogenous cells discrete, subcylindrical to ampulliform, hyaline, sometimes slightly wide at the base 5–19 × 1–3 μm. Conidia 17–24 × 4–8 μm ( = 23 × 7 μm, n = 30), fusoid, four-septate. A distinct dark separation exists between each cell, lightly curved, including three-median cells 7–16 μm long ( = 17 μm, n = 30), brown or olive, with the third cell darker. Apical cell 3–4 μm long ( = 4 μm, n = 30), hyaline, cylindrical to subcylindrical, with 2–3 tubular apical appendages (mostly 2), 7–11 μm long ( = 8 μm, n = 30), arising from the apical crest, filiform. Second cell 5–6 μm long ( = 5 μm, n = 30); third cell 4–6 μm long ( = 5 μm, n = 30); fourth cell 4–6 μm long ( = 5 μm, n = 30). Base cell is an inverted triangle, 4–6 μm long ( = 4 μm, n = 30), light brown or almost transparent hyaline. Basal appendage single, tubular, unbranched, centric, vertical, or curved, 4–6 μm long ( = 6 μm, n = 30).
Material examined: China, Yunnan Province, Dali City, Cangshan Mountain, isolated from healthy leaves of R. delavayi (2489 m, 25.4659°N, 99.5829°E), March 2021, Z.Q. Zhang, D-14 (KUN-HKAS 124578, holotype), ex-type culture, CGMCC 3.23546 = KUNCC 22-12402.
Notes: Based on phylogenetic analysis, the newly discovered P. suae is closely related to P. rosea and P. pinicola. Comparisons of ITS, TEF-1α, and TUB gene regions of P. suae with P. rosea (3/539 in ITS, 13/943 in TEF-1α, and 9/453 in TUB), and P. pinicola (10/608 in ITS, 9/467 in TEF-1α, and 5/409 in TUB) exhibited significant differences. However, P. suae is different from P. rosea due to its brown conidia, while the conidia of P. rosea are slightly red. P. suae can be distinguished from P. pinicola due to its size of apical and basal appendages; P. suae has shorter apical appendages (5–17 vs. 7–11 μm) and longer basal appendages (2–7 vs. 4–6 μm).
Pestalotiopsis trachicarpicola Y.M. Zhang and K.D. Hyde, Cryptog. Mycol. 33(3):311–318 (2012). Figure 8.
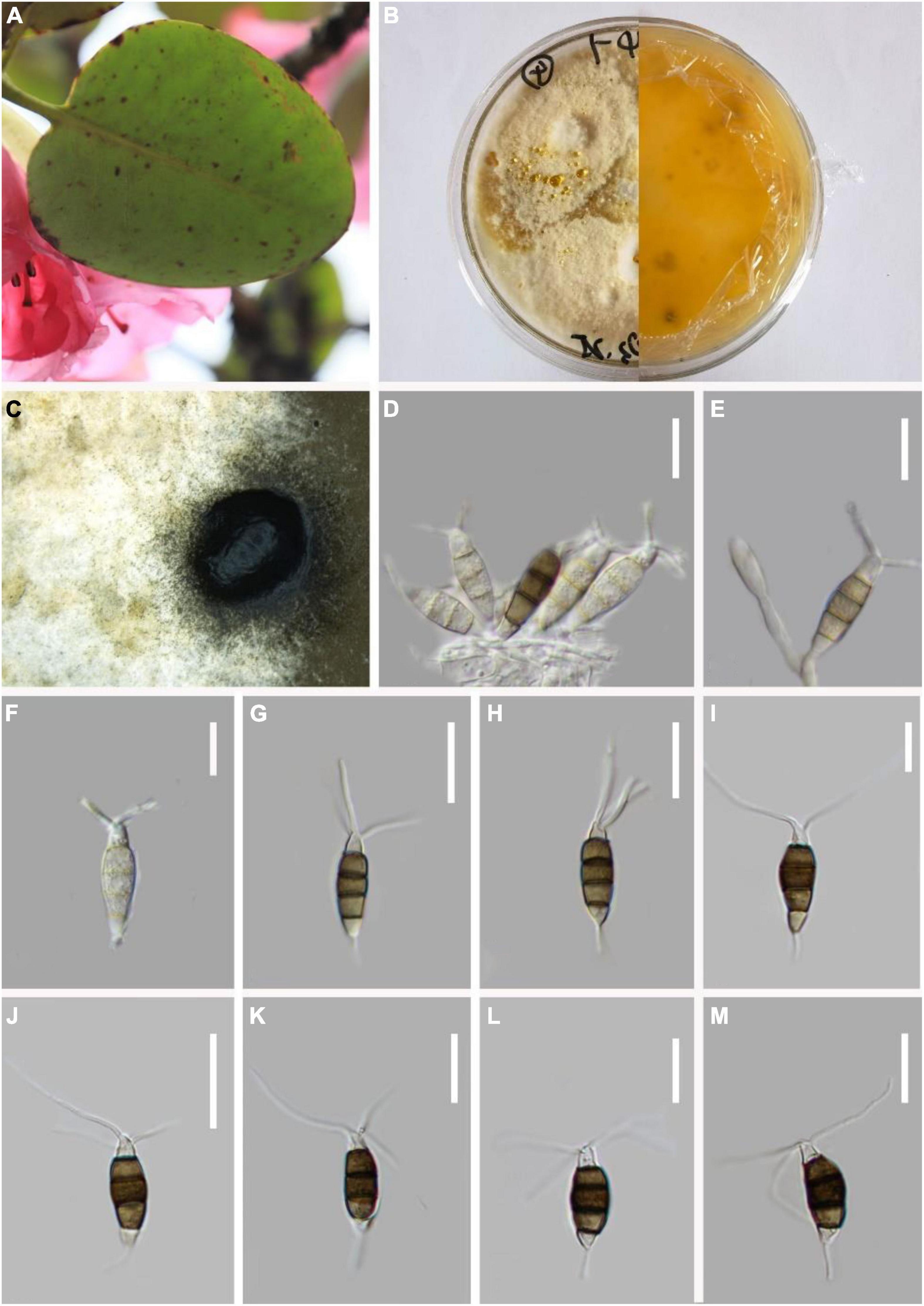
Figure 8. Pestalotiopsis trachicarpicola (KUN-HKAS 124577). (A) Leaves of Rhododendron cyanocarpum. (B) Culture on potato dextrose agar (PDA) (upper and lower view). (C) Conidiomata on PDA. (D,E) Conidiophores, conidiogenous cells, and conidia. (F–M) Conidia. Scale bars: (D–M) 10 μm.
Endophytic in fresh Rhododendron cyanocarpum leaves. Sexual morph: Undetermined. Asexual morph: Conidiomata pycnidial in culture on PDA, globose or clavate, aggregated or scattered, semi-immersed to erumpent, dark-brown to black. Conidiogenous cell is not obvious. Conidiophores are usually indistinct and reduced to conidiogenous cells. Conidia 19–23 × 5–6 μm ( = 21 × 5 μm, n = 30), fusoid, ellipsoid, straight to slightly curved, four-septate, slightly constricted at the septa. Basal cell conic to obconic with a truncated base, hyaline, verruculose, and thin-walled, 2–4 μm long. Three median cells doliiform, 11–14 μm ( = 12 μm, n = 30). Wall thin, color uniform (light or dark brown), septa darker than the rest of the cells, and the conidium constructed at the septum (second cell from the base, 4–5 μm long; third cell, 5–6 μm long; fourth cell, 4–6 μm long). Apical cell 2–4 μm long, hyaline, subcylindrical, or obconic with a truncated base, thin-walled, slightly rugose. With 2–3 tubular apical appendages arising from the apical crest, unbranched, filiform, 13–23 μm ( = 18 μm, n = 30). Basal appendage single, tubular, centric, or uncentred, 4–8 μm long.
Material examined: China, Yunnan Province, Dali City, Cangshan Mountain, isolated from healthy leaves of R. cyanocarpum, March 2021, Z.Q. Zhang, D-20 (KUN-HKAS 124577), living culture, CGMCC 3.23547.
Notes: Based on the phylogenetic analysis, P. trachicarpicola can be grouped with P. trachicarpicola (OP068) with strong support (98% ML and 0.99 BYPP). The morphologies of the two species are identical. For the first time, P. trachicarpicola is isolated from Rhododendron.
Discussion
Many fungal groups, such as Aspergillus, Ceratobasidium, Fusarium, Neocosmospora, Pestalotiopsis, Pyrenochaeta, Russula, Serendipita, Thanatephorus, and Trichoderma have been reported as endophytic fungi (Fu et al., 2022). As an ornamental plant, Rhododendron has achieved worldwide recognition (Cox and Cox, 1997). Recent research has isolated fungi from the leaf spots, mycorrhizae, and rhizosphere of Rhododendron (Medeiros et al., 2022). However, few studies have been conducted on the endophytic fungi of Rhododendron. Yunnan Province is one of the world’s most significant distribution centers for Rhododendron (Tian et al., 2011). There are 61 species of Rhododendron in Cangshan Mountain, Yunnan Province, China (Zhang et al., 2021). Our investigation indicates high diversity of Pestalotiopsis species in Rhododendron. However, the current study collected the leaves of three Rhododendron species only. In future research, it is possible to expand the survey area and collect additional Rhododendron leaves to obtain more endophytic fungal resources.
Previous studies mentioned that the color intensities of the median conidial cell, differences in the size variation of conidia, and the presence or absence of basal appendages can be used as additional taxonomic characteristics for distinguishing Pestalotiopsis species (Jeewon et al., 2003; Liu et al., 2017). Apical appendage characteristics, such as branching pattern, number, and attachment position, are also useful at the species level, but not at the generic level of a generic character (Crous et al., 2012). Therefore, it was proposed to implement additional morphological characteristics for species identification. ITS, TUB, and TEF1-α gene sequences can be combined to provide greater resolution for Pestalotiopsis (Li et al., 2021). In our phylogenetic tree, branch lengths of Pestalotiopsis cangshanensis, P. fusoidea, and some other species in this genus were notably short and the support values were relatively low. Further studies of Pestalotiopsis are, therefore, required to reveal whether the less informative loci lead to the poorly resolved phylogram.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/ (OP082426, OP185510, OP185517, OP082429, OP185511, OP185518, OP082431, OP185509, OP185516, OP082427, OP185512, OP185519, OP082430, OP185513, OP185520, OP082428, OP185514, OP185521, OP082432, OP185515, and OP185522).
Author contributions
RG conducted the experiments, analyzed the data, and wrote the manuscript. D-FB, Z-LL, and H-WS revised the manuscript. H-WS planned the experiments and analyzed the data. Z-LL and X-JS planned and funded the experiments. X-JS and Y-XL helped the experiments. All authors revised the manuscript.
Funding
This work was mainly supported by the National Natural Science Foundation of China (Project ID: 32060005 and 31900020) and the Yunnan Fundamental Research Project (grant nos. 202101AU070137 and 202201AW070001).
Acknowledgments
RG thanks Jie Gao and Zheng-Quan Zhang for help in collecting samples. We are grateful to Jun Lin, Wen-Juan Sun, and Jie Liu for their help in isolation, DNA extraction, and PCR amplification. Many thanks to Sajeewa S. N. Maharachchikumbura for his valuable comments and suggestions on this study. Also thanks to Shaun Pennycook for checking species names.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer KC declared a shared affiliation with the authors D-FB and H-WS to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Akinsanmi, O. A., Nisa, S., Jeff-Ego, O. S., Shivas, R. G., and Drenth, A. (2017). Dry flower disease of macadamia in Australia caused by Neopestalotiopsis macadamiae sp. nov. and Pestalotiopsis macadamiae sp. nov. Plant Dis. 101, 45–53. doi: 10.1094/PDIS-05-16-0630-RE
Aly, A. H., Debbab, A., Kjer, J., and Proksch, P. (2010). Fungal endophytes from higher plants: A prolific source of phytochemicals and other bioactive natural products. Fungal Divers. 41, 1–16. doi: 10.1016/j.micpath.2015.04.001
Ariyawansa, H. A., and Hyde, K. D. (2018). Additions to Pestalotiopsis in Taiwan. Mycosphere. 9, 999–1013. doi: 10.5943/mycosphere/9/5/4
Ariyawansa, H. A., Hyde, K. D., Jayasiri, S. C., Buyck, B., Chethana, K. W., Dai, D. Q., et al. (2015). Fungal diversity notes 111–252-taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 75, 27–274. doi: 10.1007/s12155-012-9194-2
Cai, L., Jeewon, R., and Hyde, K. D. (2006). Molecular systematics of Sordariaceae based on multiple gene sequences and morphology. Mycol. Res. 110, 137–150. doi: 10.1016/j.mycres.2005.09.014
Cai, L., Neilsen, J., Dao, Z. L., and Ma, Y. P. (2016). Rhododendron longipedicellatum (Ericaceae), a new species from South-eastern Yunnan, China. Phytotaxa 282, 296–300. doi: 10.11646/phytotaxa.282.4.7
Cao, Y., Ma, Y., Li, Z., Liu, X., Liu, D., Qu, S., et al. (2022). Genetic diversity and population structure of Rhododendron longipedicellatum, an endangered species. Trop. Conserv. Sci. 15, 1–10.
Chaiwan, N., Jeewon, R., Pem, D., Jayawardena, R. S., Nazurally, N., Mapook, A., et al. (2022). New species of Discosia rhododendricola, Neopestalotiopsis rhododendricola and new geographical record of Diaporthe nobilis from Rhododendron sp. J. Fungi. 8:907. doi: 10.3390/jof8090907
Chaiwan, N., Wanasinghe, D. N., Mapook, A., Jayawardena, R. S., Norphanphoun, C., and Hyde, K. D. (2020). Novel species of Pestalotiopsis fungi on dracaena from Thailand. Mycology 11, 306–315. doi: 10.1080/21501203.2020.1801873
Chen, Y. Y., Maharachchikumbura, S. S. N., Liu, J. K., Hyde, K. D., Nanayakkara, R. R., Zhu, G. S., et al. (2017). Fungi from Asian Karst formations I. Pestalotiopsis photinicola sp. nov., causing leaf spots of Photinia serrulata. Mycosphere 8, 103–110. doi: 10.5943/mycosphere/8/1/9
Cox, P. A., and Cox, K. N. E. (1997). The encyclopedia of Rhododendron species. Perth: Glendoick Publishing.
Crous, P. W., Gams, W., Stalpers, J. A., Robert, V., and Stegehuis, G. (2004). MycoBank: An online initiative to launch mycology into the 21st century. Stud. Mycol. 50, 19–22. doi: 10.1023/B:MYCO.0000012225.79969.29
Crous, P. W., Verkley, G., Christensen, M., Castañeda-Ruiz, R. F., and Groenewald, J. Z. (2012). How important are conidial appendages? Persoonia 28, 126–137. doi: 10.3767/003158512X652624
Crous, P. W., Wingfield, M. J., Chooi, Y. H., Gilchrist, C. L., Lacey, E., Pitt, J. I., et al. (2020). Fungal planet description sheets: 1042–1111. Persoonia 44, 301. doi: 10.3767/persoonia.2020.44.11
Das, K., Lee, S. Y., and Jung, H. Y. (2020). Pestalotiopsis kaki sp. nov., a novel species isolated from persimmon tree (Diospyros kaki) bark in Korea. Mycobiology 49, 54–60. doi: 10.1080/12298093.2020.1852703
De Silva, N. I., Brooks, S., Lumyong, S., and Hyde, K. D. (2019). Use of endophytes as biocontrol agents. Fungal Biol. Rev. 33, 133–148. doi: 10.1016/j.fbr.2018.10.001
De Silva, N. I., Maharachchikumbura, S., Thambugala, K., and Bhat, D. J. (2021). Morphomolecular taxonomic studies reveal a high number of endophytic fungi from Magnolia candolli and M. garrettii in China and Thailand. Mycosphere 12, 163–237. doi: 10.5943/mycosphere/12/1/3
Fang, M. Y., Fang, R. C., He, M. Y., Hu, L. Z., Yang, H. B., and David, F. C. (2005). Rhododendron. Flora China 14, 260–455.
Fu, H., Song, Z., Li, S. M., Lan, S. R., Zeng, X. H., and Huang, W. C. (2022). Effects of bamboo forest type and density on the growth of Bletilla striata and root endophytic fungi. Diversity 14, 391–405. doi: 10.3390/d14050391
Geng, K., Zhang, B., Song, Y. U., Hyde, K. D., Kang, J. C., and Wang, Y. (2013). A new species of Pestalotiopsis from leaf spots of Licuala grandis from Hainan, China. Phytotaxa 88, 49–54. doi: 10.11646/PHYTOTAXA.88.3.2
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hu, H. L., Jeewon, R., Zhou, D., Zhou, T. X., and Hyde, K. D. (2007). Phylogenetic diversity of endophytic Pestalotiopsis species in Pinus armandii and Ribes sppestalotiopsis: Evidence from rDNA and β-tubulin gene phylogenies. Fungal Divers. 24, 1–22 doi: 10.1055/s-0029-1185951
Huanaluek, N., Jayawardena, R. S., Maharachchikumbura, S., and Harishchandra, D. L. (2021). Additions to pestalotioid fungi in Thailand: Neopestalotiopsis hydeana sp. nov. and Pestalotiopsis hydei sp. nov. Phytotaxa 479, 23–43. doi: 10.11646/phytotaxa.479.1.2
Huang, W. Y., Cai, Y., Surveswaran, S., Hyde, K. D., Corke, H., and Sun, M. (2009). Molecular phylogenetic identification of endophytic fungi isolated from three Artemisia species. Fungal Divers. 36, 69–88. doi: 10.1016/j.funeco.2013.01.005
Hyde, K. D., Hongsanan, S., Jeewon, R., Bhat, D. J., McKenzie, E. H. C., Jones, E. B. G., et al. (2016). Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 80, 1–270. doi: 10.1007/s13225-016-0373-x
Hyde, K. D., Jeewon, R., Chen, Y. J., Bhunjun, C. S., Calabon, M. S., Jiang, H. B., et al. (2020). The numbers of fungi: Is the descriptive curve flattening. Fungal Divers. 103, 219–271. doi: 10.1007/s13225-020-00458-2
Jeewon, R., Liew, E. C. Y., Simpson, J. A., Hodgkiss, I. J., and Hyde, K. D. (2003). Phylogenetic significance of morphological characters in the taxonomy of Pestalotiopsis species. Mol. Phylogenet. Evol. 27, 372–383. doi: 10.1016/s1055-7903(03)00010-1
Jia, M., Chen, L., Xin, H. L., Zheng, C. J., Rahman, K., Han, T., et al. (2016). A Friendly relationship between endophytic fungi and medicinal plants: A systematic review. Front. Microbiol. 7:906. doi: 10.3389/fmicb.2016.00906
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Li, L. L., Yang, Q., and Li, H. (2021). Morphology, phylogeny, and pathogenicity of pestalotioid species on Camellia oleifera in China. J. Fungi. 7, 1080–1108. doi: 10.3390/jof7121080
Lin, L. C., Lin, C. Y., Lin, W. R., Tung, Y. T., and Wu, J. H. (2021). Effects of ericoid Mycorrhizal fungi or dark septate endophytic fungi on the secondary metabolite of Rhododendron pseudochrysanthum (R. Morii) seedlings. Appl. Ecol. Environ. Res. 19, 1221–1232. doi: 10.15666/aeer/1902_12211232
Liu, F., Bonthond, G., Groenewald, J. Z., Cai, L., and Crous, P. W. (2019). Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Stu. Mycol. 92, 287–415. doi: 10.1016/j.simyco.2018.11.001
Liu, F., Hou, L., Raza, M., and Cai, L. (2017). Pestalotiopsis and allied genera from Camellia, with description of 11 new species from China. Sci. Rep. 7, 1–19. doi: 10.1038/s41598-017-00972-5
Liu, J. K., Hyde, K. D., Jones, E. B., Ariyawansa, H. A., Bhat, D. J., Boonmee, S., et al. (2015). Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 72, 1–197.
Liu, J. K., Phookamsak, R., Doilom, M., Wikee, S., Li, Y. M., Ariyawansha, H., et al. (2012). Towards a natural classification of Botryosphaeriales. Fungal Divers. 57, 149–210. doi: 10.1007/s13225-012-0207-4
Ma, H., Li, T. Q., Liu, X. F., Wan, Y. M., Li, Y. Y., Liu, X. X., et al. (2017). Research progress in conservation biology of 8 tropical conservation science Rhododendron. World For. Res. 30, 13–17.
Ma, H., Liu, Y., Liu, D., Sun, W., Liu, X., Wan, Y., et al. (2021). Chromosome-level genome assembly and population genetic analysis of a critically endangered rhododendron provide insights into its conservation. Plant J. 107, 1533–1545. doi: 10.1111/tpj.15399
Ma, X. Y., Maharachchikumbura, S. S. N., Chen, B. W., Hyde, K. D., Mckenzie, E. H. C., Chomnunti, P., et al. (2019). Endophytic pestalotiod taxa in Dendrobium orchids. Phytotaxa 419, 268–286. doi: 10.11646/phytotaxa.419.3.2
Maharachchikumbura, S. S. N., Chukeatirote, E., Guo, L. D., Crous, P. W., McKenzie, E. H. C., and Hyde, K. D. (2013). Pestalotiopsis species associated with Camellia sinensis (tea). Mycotaxon 123, 47–61. doi: 10.5248/123.47
Maharachchikumbura, S. S. N., Guo, L. D., Cai, L., Chukeatirote, E., Wu, W. P., Sun, W. X., et al. (2012). A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Divers. 56, 95–129. doi: 10.1007/s13225-012-0198-1
Maharachchikumbura, S. S. N., Hyde, K. D., Groenewald, J. Z., Xu, J., and Crous, W. (2014). Pestalotiopsis revisited. Stud. Mycol. 79, 121–186. doi: 10.1016/j.simyco.2014.09.005
Maharachchikumbura, S. S. N., Larignon, P., Hyde, K. D., Abdullah, M. A., and Liu, Z. Y. (2016). Characterization of Neopestalotiopsis, Pestalotiopsis and Truncatella species associated with grapevine trunk diseases in France. Phytopathol. Mediterr. 55, 380–390. doi: 10.14601/Phytopathol_Mediterr-18298
Medeiros, J. S., Mann, M. A., Burns, J. H., Kyker, S., and Burke, D. (2022). Host ancestry and morphology differentially influence bacterial and fungal community structure of Rhododendron leaves, roots, and soil. Botany 100, 449–460. doi: 10.1139/cjb-2021-0104
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, 1–8.
Monteiro, P., Gonçalves, M. F., Pinto, G., Silva, B., Martín-García, J., Diez, J. J., et al. (2022). Three novel species of fungi associated with pine species showing needle blight-like disease symptoms. Eur. J. Plant Pathol. 162, 183–202. doi: 10.1007/s10658-021-02395-5
Negi, V. S., Maikhuri, R. K., Rawat, L. S., and Chandra, A. (2013). Bioprospecting of Rhododendron arboreum for livelihood enhancement in central Himalaya. J. Sci Technol. 8, 61–70. doi: 10.13140/2.1.1002.9762
Norphanphoun, C., Jayawardena, R. S., Chen, Y., Wen, T. C., Meepol, W., and Hyde, K. D. (2019). Morphological and phylogenetic characterization of novel pestalotioid species associated with mangroves in Thailand. Mycosphere 10, 531–578. doi: 10.5943/mycosphere/10/1/9
Nylander, J. A. A. (2004). MrModeltest v2 Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.
Rambaut, A. (2014). FigTree 1.4.2. Available online at: http://tree.bio.ed.ac.uk/software/figtree (accessed July 9, 2014).
Rannala, B., and Yang, Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Rashmi, M., Kushveer, J. S., and Sarma, V. V. (2019). A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 10, 798–1079. doi: 10.5943/mycosphere/10/1/19
Rodriguez, R. J., White, J. F., Arnold, A. E., and Redman, A. R. A. (2009). Fungal endophytes: diversity and functional roles. New phy. 182, 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Shrestha, N., Wang, Z., Su, X., Xu, X., Lyu, L., Liu, Y., et al. (2018). Global patterns of Rhododendron diversity: The role of evolutionary time and diversification rates. Glob. Ecol. Biogeogr. 27, 913–924. doi: 10.1111/geb.12750
Shu, J., Yu, Z., Sun, W., Zhao, J., Li, Q., Tang, L., et al. (2020). Identification and characterization of pestalotioid fungi causing leaf spots on mango in Southern China. Plant Dis. 104, 1207–1213. doi: 10.1094/PDIS-03-19-0438-RE
Silva, A. C., Diogo, E., Henriques, J., Ramos, A. P., Marcelo, S. D., Crous, P. W., et al. (2020). Pestalotiopsis pini sp. nov., an emerging pathogen on stone pine (Pinus pinea L.). Forests 11, 805–823. doi: 10.3390/f11080805
Song, Y., Tangthirasunun, N., Maharachchikumbura, S. S. N., Jiang, Y. L., Xu, J. J., Hyde, K. D., et al. (2014). Novel Pestalotiopsis species from Thailand point to the rich undiscovered diversity of this chemically creative genus. Cryptogamie Mycolo. 35, 139–149. doi: 10.7872/crym.v35.iss2.2014.139
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 75, 758–771. doi: 10.1080/10635150802429642
Steyaert, R. L. (1949). Contribution à l’étude monographique de Pestalotia de Not. et Monochaetia Sacc. (Truncatella gen. nov. et Pestalotiopsis gen. nov.). Bull. Jard. Bot. Brux. 19, 285–354. doi: 10.2307/3666710
Sutton, B. C. (1980). Fungi imperfecti with pycnidia, acervuli and stromata: colletotrichum. Coelomycetes 1980, 523–537. doi: 10.1016/s0007-1536(81)80170-2
Tao, G., Liu, Z. Y., Liu, F., Gao, Y. H., and Cai, L. (2013). Endophytic Colletotrichum species from Bletilla ochracea (Orchidaceae), with descriptions of seven new speices. Fungal Divers. 61, 139–164. doi: 10.1007/s13225-013-0254-5
Tian, W., Zhang, C. Q., Qiao, P., and Milne, R. (2011). Diversity of culturable ericoid mycorrhizal fungi of Rhododendron decorum in Yunnan, China. Mycologia 103, 703–709. doi: 10.3852/10-296
Tibpromma, S., Hyde, K. D., McKenzie, E. H., Bhat, D. J., Phillips, A. J., Wanasinghe, D. N., et al. (2018). Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 93, 1–160. doi: 10.1007/978-94-007-4447-9_4
Tibpromma, S., Mortimer, P. E., Karunarathna, S. C., Zhan, F., Xu, J., Promputtha, I., et al. (2019). Morphology and multi-gene phylogeny reveal Pestalotiopsis pinicola sp. nov. and a new host record of Cladosporium anthropophilum from edible pine (Pinus armandii) seeds in Yunnan province, China. Pathogens 8, 285–303. doi: 10.3390/pathogens8040285
Watanabe, K., Motohashi, K., and Ono, Y. (2010). Description of Pestalotiopsis pallidotheae: A new species from Japan. Mycoscience 51, 182–188. doi: 10.1007/S10267-009-0025-Z
Watanabe, K., Nozawa, S., Hsiang, T., and Callan, B. (2018). The cup fungus Pestalopezia brunneopruinosa is Pestalotiopsis gibbosa and belongs to sordariomycetes. PloS One 13:e0197025. doi: 10.1371/journal.pone.0197025
Zhang, Y., Bakenov, Z., Zhao, Y., Konarov, A., Malik, M., Paron, T., et al. (2012b). One-step synthesis of branched sulfur polypyrrole nanocomposite cathode for lithium rechargeable batteries. J. Power Sour. 208, 1–8. doi: 10.1016/j.jpowsour.2012.02.006
Zhang, Y., Maharachchikumbura, S. S. N., McKenzie, E. H. C., and Hyde, K. D. (2012a). A novel species of pestalotiopsis causing leaf spots of Trachycarpus fortunei. Cryptogam. Mycol. 33, 311–318. doi: 10.7872/crym.v33.iss3.2012.311
Zhang, Y. M., Maharachchikumbura, S. S. N., Tian, Q., and Hyde, K. D. (2013). Pestalotiopsis species on ornamental plants in Yunnan Province, China. Sydowia 65, 113–128.
Zhang, Y., Tang, F., Ni, J., Dong, L., and Sun, L. (2019). Diversity of root-associated fungi of Rhododendron simsii in subtropical forests: Fungal communities with high resistance to anthropogenic disturbances. J. For. Res. 30, 2321–2330. doi: 10.1007/s11676-019-01050-4
Keywords: six new species, endophytic fungi, Sordariomycetes, morphology, phylogeny
Citation: Gu R, Bao D-F, Shen H-W, Su X-J, Li Y-X and Luo Z-L (2022) Endophytic Pestalotiopsis species associated with Rhododendron in Cangshan Mountain, Yunnan Province, China. Front. Microbiol. 13:1016782. doi: 10.3389/fmicb.2022.1016782
Received: 11 August 2022; Accepted: 26 September 2022;
Published: 20 October 2022.
Edited by:
Samantha Chandranath Karunarathna, Qujing Normal University, ChinaReviewed by:
Kasun Thambugala, University of Sri Jayewardenepura, Sri LankaChayanard Phukhamsakda, Jilin Agriculture University, China
K. W. Thilini Chethana, Mae Fah Luang University, Thailand
Copyright © 2022 Gu, Bao, Shen, Su, Li and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zong-Long Luo, bHVvem9uZ2xvbmdmdW5naUAxNjMuY29t
 Rui Gu
Rui Gu Dan-Feng Bao1,2,3
Dan-Feng Bao1,2,3 Zong-Long Luo
Zong-Long Luo
