- 1Center for Electromicrobiology, Department of Biology, Aarhus University, Aarhus, Denmark
- 2Department of Oceanography, University of Concepción, Concepción, Chile
The sediments under the Oxygen Minimum Zone of the Eastern Boundary Current System (EBCS) along Central-South Peru and North-Central Chile, known as Humboldt Sulfuretum (HS), is an organic-matter-rich benthic habitat, where bacteria process a variety of sulfur compounds under low dissolved-oxygen concentrations, and high sulfide and nitrate levels. This study addressed the structure, diversity and spatial distribution patterns of the HS bacterial community along Northern and South-Central Chile using 16S rRNA gene amplicon sequencing. The results show that during the field study period, the community was dominated by sulfur-associated bacteria. Indeed, the most abundant phylum was Desulfobacterota, while Sva0081 sedimentary group, of the family Desulfosarcinaceae (the most abundant family), which includes sulfate-reducer and H2 scavenger bacteria, was the most abundant genus. Furthermore, a spatial pattern was unveiled along the study area to which the family Desulfobulbaceae contributed the most to the spatial variance, which encompasses 42 uncharacterized amplicon sequence variants (ASVs), three assigned to Ca. Electrothrix and two to Desulfobulbus. Moreover, a very high microdiversity was found, since only 3.7% of the ASVs were shared among localities, reflecting a highly diverse and mature community.
Introduction
In the Eastern Boundary Current System (EBCS) off the coasts of Chile and Peru, one of the world’s largest subsurface sulfidic benthic system occurs: the “Humboldt Sulfuretum”’ (HS) (Gallardo et al., 2016). While likely quite widespread in primeval oceans, ca. 3.5 Ga ago (Van Kranendonk et al., 2003; Ueno et al., 2008; Wacey et al., 2011; Schopf et al., 2017), these systems are rare and of limited extent in today’s Earth oceans. However, notwithstanding their reduced extension, ca. 1.15 million km2 (Helly and Levin, 2004), their contribution to the productivity of the world’s oceans is disproportionately high. In this thus rather rare, nonetheless significant biome, benthic bacteria process a variety of sulfur compounds under the absence or very low levels of dissolved oxygen, but rather high levels of reduced sulfur compounds, nitrate, and phosphate (Fossing et al., 1995; Høgslund et al., 2008; Gallardo et al., 2013a).
In similar settings, i.e., sulfide-rich/oxygen-deficient seafloor and subsea sediments, bacterial communities are characterized by anaerobic bacteria, such as sulfate-reducing anaerobic forms and fermenters of Chloroflexi, Proteobacteria, Firmicutes, or Candidatus Atribacteria as well as methanogenic, and methanotrophic archaea (Orsi, 2018). These communities are thus well-differentiated from those living in oxic sediments (Zinger et al., 2011; Bienhold et al., 2016). Under sulfidic conditions, the availability of electron donors and acceptors, geochemical and sedimentological properties control the structure of bacterial communities (Fry et al., 2008; Hoshino et al., 2020). Furthermore, lithotrophic, and mostly autotrophic, sulfur-oxidizing bacteria (SOB) are conspicuous inhabitants of these environments, i.e., the “megabacteria” Cand. Marithioploca araucae (Salman et al., 2011), and the “macrobacteria” Cand. Venteria Ishoeyi (Fonseca et al., 2017), which generate extensive mats in the HS (Gallardo, 1977; Maier and Gallardo, 1984; Schulz et al., 1996; Ferdelman et al., 1997).
Previous work in the HS, using 16S rRNA amplicon sequencing, focused on the diversity of the bacterial community in selected areas of this system. Thereby, following a combination of the principles that sustain the Thienemann-Sanders time-stability hypothesis, Gallardo et al. (2016) suggested that the HS sublitoral bacterial community inhabiting the Central-South distribution area (∼36°s), corresponds to an evolutionarily mature, “biologically accommodated” community, a condition that is characterized by the lack of statistically dominant species (OTUs). Moreover, recent work in the Northern Mejillones Bay, located in the permanent mid-water oxygen-deficient upwelling system off Northern Chile, reports that members of Anaerolineaceae, Thiotrichaceae, Desulfobulbaceae, Desulfarculaceae, and Bacteroidales play an important role in the differentiation of sampling points in this area (Zárate et al., 2021).
The present work is a contribution to the knowledge on the structure and diversity of the HS bacterial community through sediment samples obtained off the coast of Chile from five localities distributed between ca. −20 and −36.10′, using 16S rRNA gene amplicon sequencing.
Materials and methods
Sediment core samples
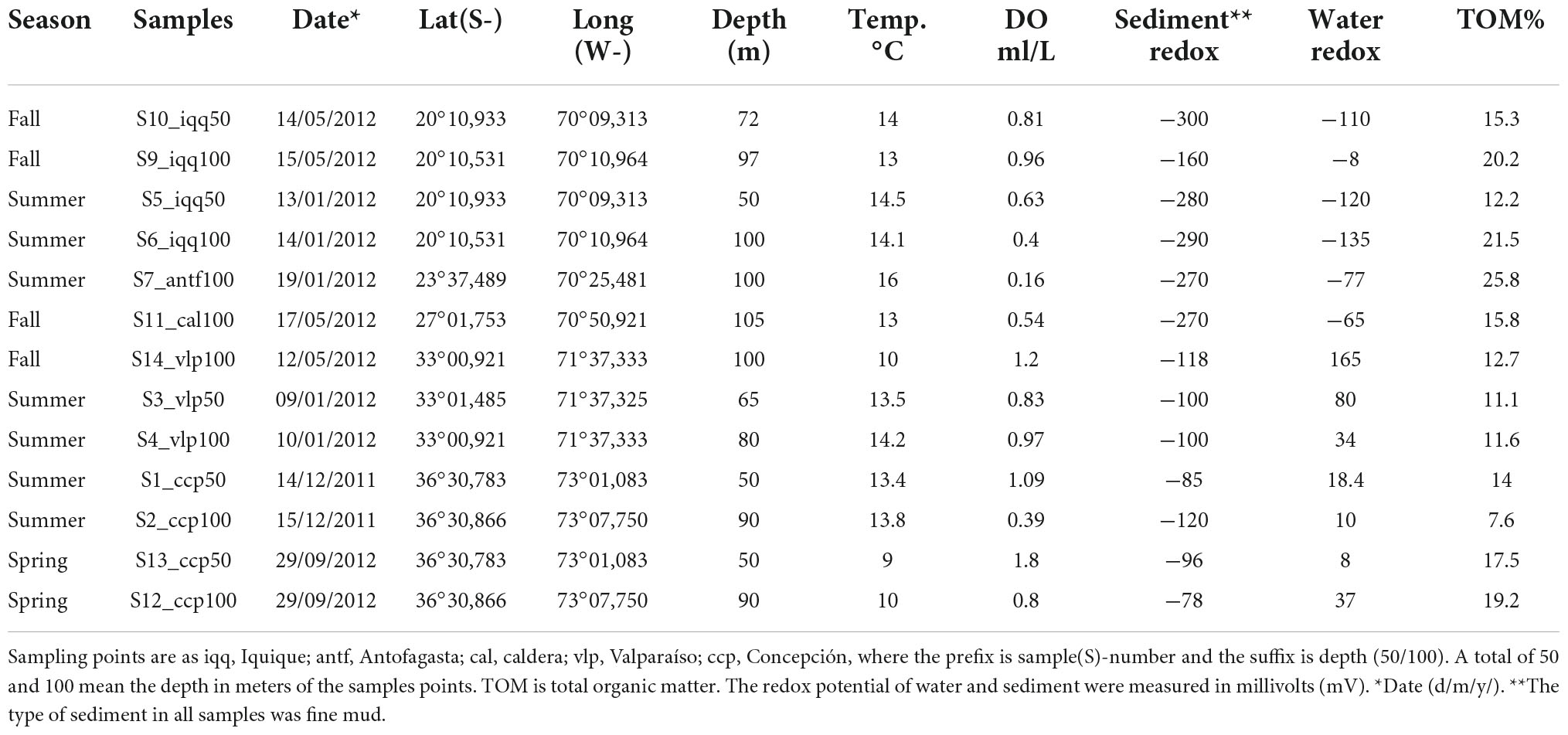
Off the coast of Chile sediment core samples were collected at approximately 50 and 100 m depth at five latitudes from −20 to −36 (Table 1), namely off Iquique (IQQ), Antofagasta (ANTF), Caldera (CAL), Valparaíso (VLP), and Concepcion (CCP) (Figure 1A). During samplings, the Northern IQQ, CAL, ANTF, and VLP substrates were evidently reduced and devoid of macrofauna, while at the Southern CCP station the scattered presence of polychetes and bivalves was noticeable.
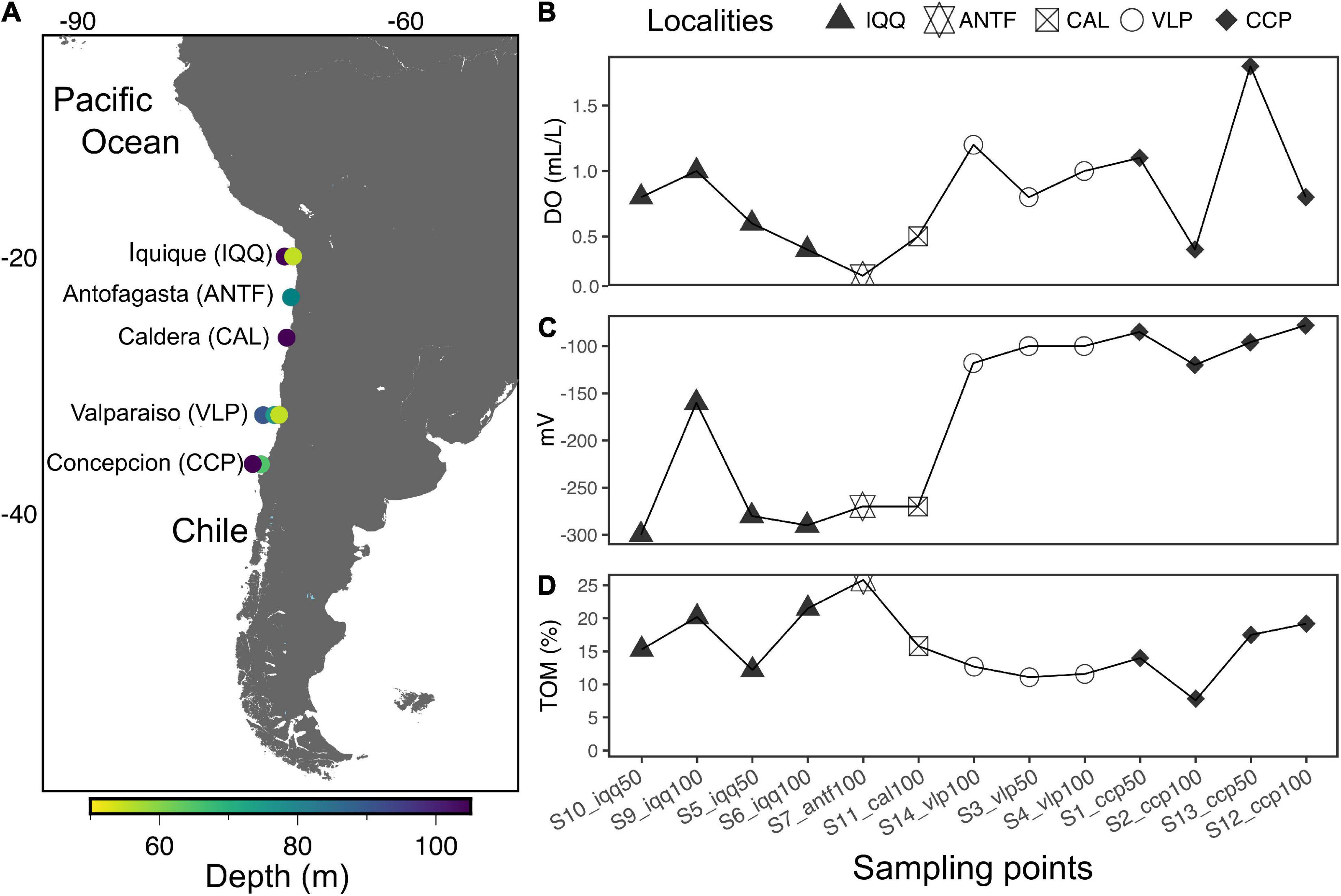
Figure 1. (A) Map of sampling points. (B) Dissolved oxygen (DO) of bottom water in ml/L. (C) Average redox potential of the first 5 cm of sediment, measured in millivolts (mV). (D) Average total organic matter (TOM) in percentage of the first 5 cm of sediment.
Sampling was carried out from the small motorboats “Otilia” (CCP), “Antares” (IQQ), and various fishing boats (ANTF, CAL, and VLP), with a small handy gravity mono-corer. Samples were sub-sampled on board with the upper 5 cm of sediment transferred to sterile tubes and kept at 4°C until DNA extraction (<12 h later). In addition, dissolved oxygen, from overlying water, sediment redox potential (Figures 1B,C), overlying water redox potential (using Ag/AgCl as reference electrode), and temperature, were immediately measured with an Oakton P650 multi-parameter instrument. Porosity and total organic matter (TOM) (Figure 1D) were determined by the ignition method (Mook and Hoskin, 1982).
DNA extraction and sequencing
DNA extraction was carried out (in triplicate) from 0.5 g of the upper 5 cm of sediments using the Fast DNA Spin Kit for Soil (MP laboratories) and quantified using a Nanodrop ND1000. Next, at the Marine Biological Laboratory (BPC-MBL), Woods Hole, MA, USA, the v4v6 region of the 16S rRNA gene was amplified and sequenced using a 454 GS FLX Titanium sequencing platform, Details of sequencing technology are described in Huber et al. (2007) and Huse et al. (2008). Data and experiments are available under project code PRJNA251688 in the NCBI database. Reads were demultiplexed and barcodes were removed for submission.
Quality control and filtering reads
After sequencing, the single-end raw reads were assessed for quality control using the FastQC package version 0.11.9,1 while the filtering and trimming were performed through the package PRINSEQ-lite version 0.20.4 (Schmieder and Edwards, 2011). Next, reads were subjected to denoising/chimera removal and clustering into amplicon sequence variants (ASVs) using the denoise-single method of the DADA2 package, version 1.14 (Callahan et al., 2016), as a plugin through the bioinformatics platform QIIME2, version 2019.10 (Bolyen et al., 2019). The ASV taxonomy assignment was performed through the feature-classifier classify-consensus-vsearch method in QIIME2, using the SILVA database 138.1 as reference (Quast et al., 2012). Singletons were removed from the final ASV table.
Data analyses
The main data analysis was performed using the R software environment version 3.6.3. Thereby, the bacteria abundance table was rarefied to the smallest sample size (8,000 reads per sample), to perform diversity and ordering analysis, using the function rarefy_even_depth of the Phyloseq package (R package), version 1.30.0 (McMurdie and Holmes, 2013). This function uses the standard R sample function to resample from the abundance values in the ASVs table so that all samples have the same size.
The NMDS ordination analysis was performed using the function ordinate of the package Phyloseq version 1.30.0, using the unifrac and Bray-Curtis distance indices on the ASV abundance table. In particular, unifrac accesses the abundance table, but also to a phylogenetic tree, previously built using the representative reads through QIIME2 and the de novo phylogenetic tree creation method, phylogeny fasttree.
The Shannon diversity index and observed ASVs were obtained using the R package Vegan version 2.5-7 (Oksanen et al., 2013). The Analysis of Similarity (ANOSIM) test was used to validate the ordination results using the Vegan function anosim, which statistically tests whether there is a significant difference between two or more groups of sampling units. The statistical comparisons between factors and groups were performed through the Vegan function Adonis statistical test (Permutational Multivariate Analysis of Variance) and the R package ALDEx2 version 1.22.0 (Fernandes et al., 2014). In addition, the correlation and statistical significance between the environmental variables and the Bray-Curtis dissimilarity matrix of the abundance table were performed using a combination of the bioEnv function and the Vegan package’s Mantel test. To evaluate the average percentage contribution of individual bacteria to the dissimilarity between groups in the Bray-Curtis dissimilarity matrices, the Vegan method of similarity percentage decomposition (SIMPER) was performed. The graphical representation of the data was achieved using ggplot2 version 3.3.3 (R package), Circos version 0.69-8 (Krzywinski et al., 2009), library Matplotlib from Python version 3.6, and the R package Metacoder version 0.3.4 (Foster et al., 2017).
Results
Community structure and diversity
The thirteen preprocessed libraries rendered 202,779 reads and 3,614 ASVs. The ASVs were distributed among 55 phyla, where the phylum Desulfobacterota (formerly the class Deltaproteobacteria within the phylum Proteobacteria) dominates with 22.7% of the total abundance, including several sulfur-associated bacteria. Next come the phyla Proteobacteria (18.4%), Bacteroidota (formerly the class Bacteroidetes), (15.8%), and Chloroflexi (9.4%) (Figure 2A). In this connection, there were no statistically significant differences between the most abundant phyla (>2% in abundance) and localities (p-value > 0.05), evaluated through the ALDEx2 statistical package.
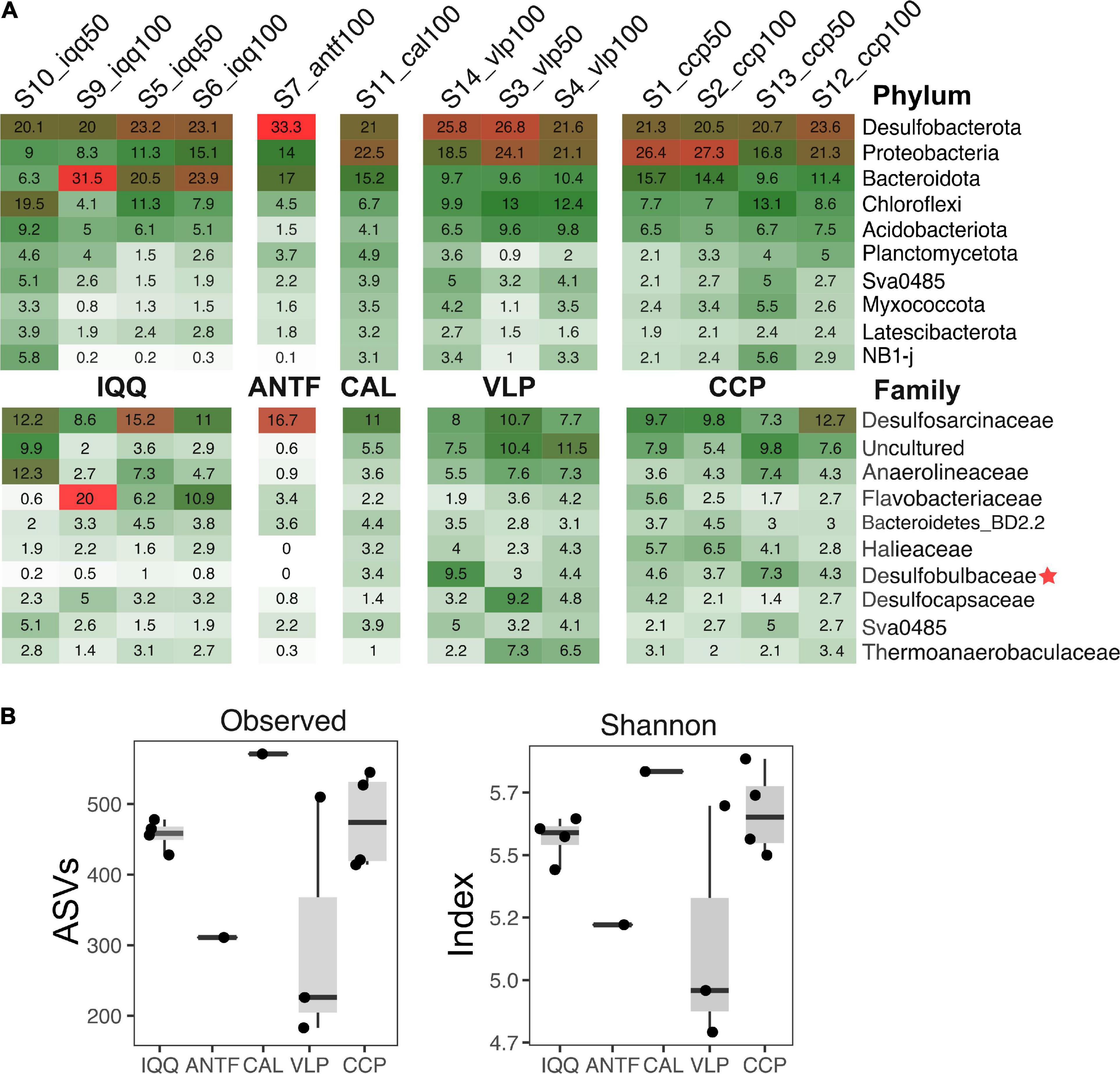
Figure 2. (A) Abundance of the ten most abundant phyla and families. The numbers in tiles represent the relative abundance in percent. The red star highlights taxa with statistically significant differences in abundance according to Aldex 2. (B) Number of observed ASVs and Shannon diversity index.
A total of 328 families were identified, where Desulfosarcinaceae (11%), in the phylum Desulfobacterota, is the most abundant; followed by an uncultured-unlabeled cluster (6.1%), and Anaerolineaceae (5.4%), of the phylum Chloroflexi (Figure 2A). According to ALDEx2, the family Desulfobulbaceae (3.2%) presents statistically significant differences in the abundances between IQQ and VLP-CCP localities (p-value < 0.05). On the other hand, although there were not statistically significant differences, the family Flavobacteriaceae (5.3%) presented greater abundance in IQQ than in VLP and CCP, while the Halieaceae family abundance was greater in VLP and CCP than in IQQ (see Supplementary Figures 2A,B for the ranking of the most abundant classes and orders). It is worth mentioning that although the Thiotrichaceae family is not among the ten most abundant families, it had 1.4% of the total abundance, ranking among the 20 most abundant and appearing with an average of 1.9% in CCP. This family harbors some of the most conspicuous large filamentous sulfur bacteria in the area, such as Ca. Marithioploca araucae, of the Gammaproteobacteria class.
Continuing with the taxonomic assignment, 397 different genera were retrieved (including one cluster for the uncultured and another for the unassigned). The most abundant genus was the Sva0081 sediment group (4.2%), in the family Desulfosarcinaceae (Supplementary Figure 2C). In this regard, it should be mentioned that after the Sva0081 sediment group, the genera SEEP-SRB1 (2.25%), LCP-80 (1.19%) and Desulfosarcina (0.86%) were identified in the family Desulfosarcinaceae (Supplementary Figure 3). While in the family Anaerolineaceae (5.4%), six genera were identified, of which Pelolinea (3.1%) was the most abundant and cosmopolitan. Furthermore, IQQ presented the largest diversity and abundance within that family (Supplementary Figure 3). Moreover, going down to the ASV ranking, the most frequent ASV was assigned to the genus Lutimonas (1.5%) in the phylum Bacteroidota, and the second in the frequency ranking (1.4%), to the order Syntrophobacterales, phylum Desulfobacterota (the graphical distribution of the most frequent thousand ASVs is in Supplementary Figure 1).
Compared with the rest of the sampling stations ANTF presented some different taxa with opposite abundances. Thus, the maximum abundance of Desulfobacterota and its family Desulfosarcinaceae was found in ANTF, while the abundance of families such as Anaerolineaceae (5.4%) was very low (0.9%), and others, such as Halieaceae and Desulfobulbaceae (which were present in IQQ, CAL, VLP, and CCP), were absent.
Regarding diversity indexes (Figure 2B), no statistically significant differences were found between the sampling stations (p-value > 0.05), although VLP had the lowest number of ASVs and Shannon diversity index, in which the sample s14_vlp100 was notably higher than s3_vlp100 and s4_vlp50, increasing the variance in the VLP locality. Furthermore, in terms of numbers of ASVs and Shannon diversity index, the localities of IQQ, CAL, and CCP, were very similar.
Spatial pattern
The NMDS analysis of the unifrac and Bray-Curtis distance matrix revealed a spatial pattern, in which the sampling points of IQQ, VLP, and CCP generated separate clusters (Figure 3). Although only localities with replicate samples (IQQ, VLP, and CCP) were considered valid for post-hoc analysis, it should be noted that the ANTF sampling point appeared distant from the complementary sampling points, while CAL appeared close to CCP. The post-hoc ANOSIM test confirmed that the clusters observed in the NMDS were statistically significant (R = 0.56, p-value = 0.0017), validating the spatial pattern.
Figure 3. Non-metric multidimensional scaling (NMDS) plot of the Bray-Curtis distance of the bacterial community with environmental parameters added using envfit. The NMDS was constructed over the Bray-Curtis dissimilarity matrix. The data was rarefied at 8,000 sequences per sample. The black arrows represent a statistically significant relationship (p < 0.05) measured through the Mantel or envfit test. The added parameters were the redox potential of sediment and water, the percentage of total organic matter (TOM), depth, temperature (Temp), and porosity (Por).
Regarding the influence of environmental measures on the Bray-Curtis dissimilarity of the microbial community, the percentage of TOM appeared as the most relevant variable (Supplementary Table 1), as is shown in the adjusted NMDS ordination analysis, built through the envfit method and the metaNMDS function from R (Figure 3). Furthermore, the bioEnv and Mantel test confirmed that TOM has a statistically significant positive correlation with bacterial abundance (r = 0.36, p-value = 0.02) as well as redox potential (r = 0.27, p-value; p = 0.02) (Supplementary Table 2).
To assess how taxa and, in particular, how families influenced the spatial pattern, the ten most abundant families (48% accumulated abundance) were evaluated through the Adonis statistical test, including the IQQ, VLP, and CCP localities as factors. The result showed that the family Desulfobulbaceae was the most important explanatory variable (Supplementary Table 3) for spatial pattern between the localities (R2 = 0.61; p-value < 0.01). Moreover, the second most important explanatory variable was Bacteroidetes BD2-2 (R2 = 0.58; p-value < 0.01), followed by Thermoanaerobaculaceae (R2 = 0.57; p-value < 0.01), and Desulfocapsaceae (R2 = 0.55; p-value < 0.01). Besides, at the ASV level, the SIMPER analysis showed that the greatest contribution to the dissimilarity pattern between IQQ and CCP was generated by a couple of ASVs assigned to the genus Lutimonas, one ASV assigned to Syntrophobacterales, one to the genera Desulfonema and Sulfurovum, which made 13.6% of the total contribution to dissimilarity. Differentially, between IQQ-VLP and CCP-VLP the greatest contribution was made by an ASV assigned to the genera B2M28 (class Gammaproteobacteria) (Supplementary Table 4).
The family Desulfobulbaceae
Since the family Desulfobulbaceae appeared as the most important taxonomic explanatory variable for the spatial pattern between localities, we investigated the distribution of the representatives of this family identified throughout the localities under study.
With 3.2% of the total abundance and 47 ASVs, the family Desulfobulbaceae contained three ASVs assigned to the genus Ca. Electrothrix, in the cable bacteria functional group, and two others, to the genus Desulfobulbus; the complement of 43 ASVs are uncharacterized forms (uncultured). In this regard, the phylogenetic relationship based on the ASV sequences showed four uncultured ASVs in the Ca. Electrothrix clade, seven in the Desulfobulbus clade, and the rest in a separate clade (Figure 4A). The Ca. Electrothrix-assigned ASVs were present in four samples from IQQ, CAL, and CCP, while Desulfobulbus was present in eleven samples from IQQ, VLP, and CCP, thus more ubiquitous and abundant than Ca. Electrothrix (Figure 4B). The uncultured genus ASVs were present in the CAL, VLP, and CCP sampling sites, and remarkably, IQQ was characterized by the almost exclusive presence of Desulfobulbus. Thus, the distribution pattern of Desulfobulbaceae emphasizes the general spatial pattern while further separating the IQQ cluster (Figure 4C). Sampling locality differences become more obvious when the numbers of shared and unique ASVs are considered, since only one Desulfobulbus assigned ASV (2.1%), is shared among IQQ, VLP, and CCP (Supplementary Figure 4). Concerning the influence of environmental variables on the abundance pattern of Desulfobulbaceae, the redox potential resulted in a statistically significant positive correlation (r = 0.41; p-value = 0.001).
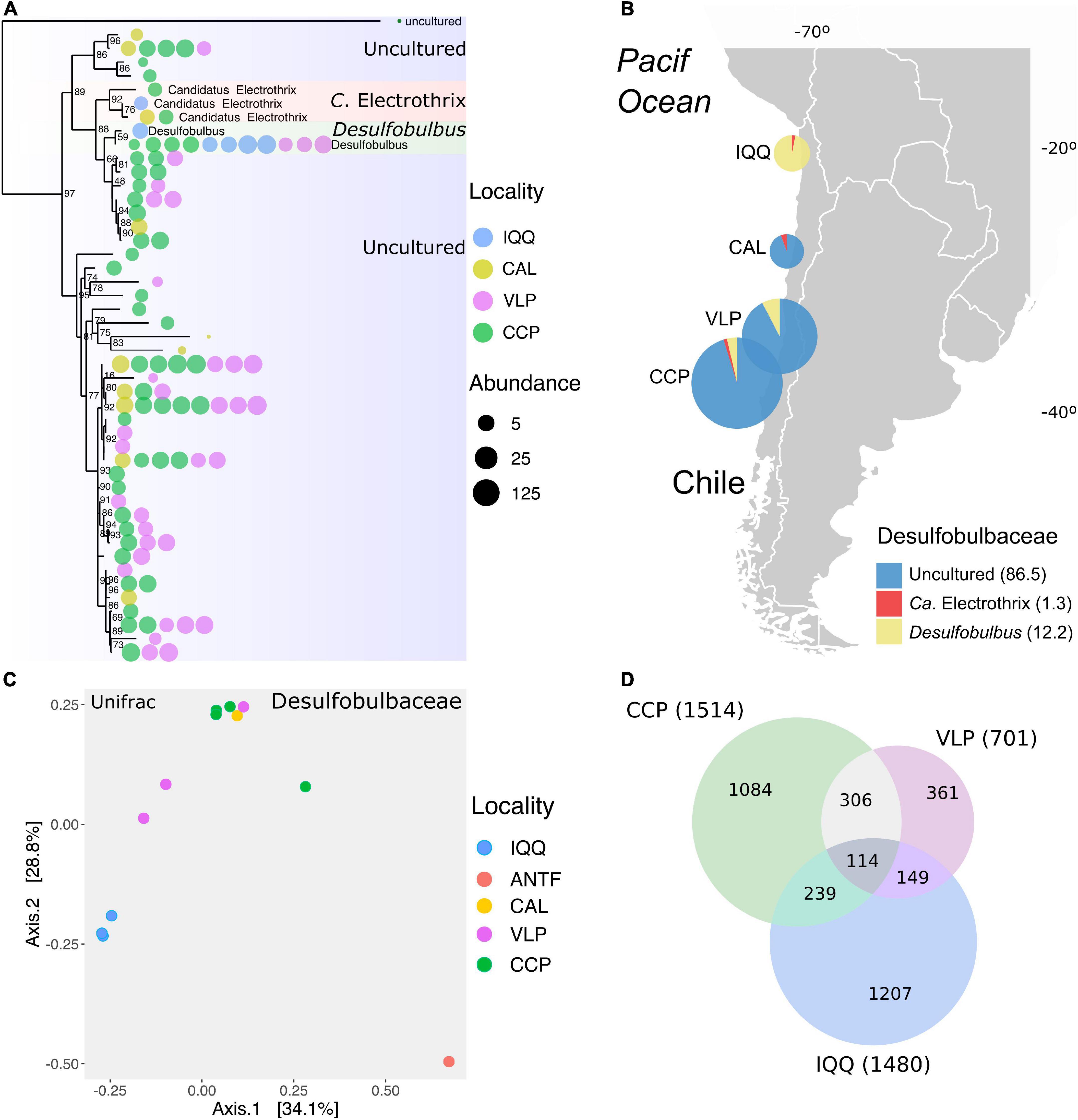
Figure 4. (A) Phylogenetic tree of the representatives of the family Desulfobulbaceae identified across the data. The color and size of the bubbles indicate the presence and abundance of taxa by locality. (B) Distribution of genera of the family Desulfobulbaceae by locality in percentage (C) Non-metric multidimensional scaling (NMDS) based on the unifrac matrix distance of the abundance in the Desulfobulbaceae family (Stress = 0.0005). (D) Venn chart of shared and unique ASVs across all data by locality. The number in parentheses is the total ASVs.
Shared and unique amplicon sequence variants
The latitudinal spatial pattern observed at the ASV level in the Desulfobulbaceae family, especially in IQQ, led to evaluating the shared and unique ASVs among IQQ, VLP, and CCP across the whole data. Removing the ANT and CAL data the ASVs shared among IQQ, VLP, and CCP were only 113 out of the total of 3,017 ASVs, which represent only 3.7% of the total ASVs (Figure 4D), and 26% of the total reads. Taxonomically, the phylum Desulfobacterota (23.5%) dominated among shared ASVs, followed by Bacteroidota (19.4%) and Proteobacteria (16.2%). Moreover, at the individual level, the first and third most abundant shared ASVs were assigned to the genus Lutimonas of the phylum Bacteroidota, and the second, to the phylum Desulfobacterota (order Syntrophobacterales).
From the uniqueness perspective, IQQ had the highest proportion of unique ASVs (Figure 5) (40%), closely followed by CCP with 36% and, both distant from VLP with 12%. Taxonomically, the phylum Desulfobacterota (20.8%) contained the larger number of unique ASVs in IQQ, while in CCP was Proteobacteria (21%), and in VLP Chloroflexi (16.6%).
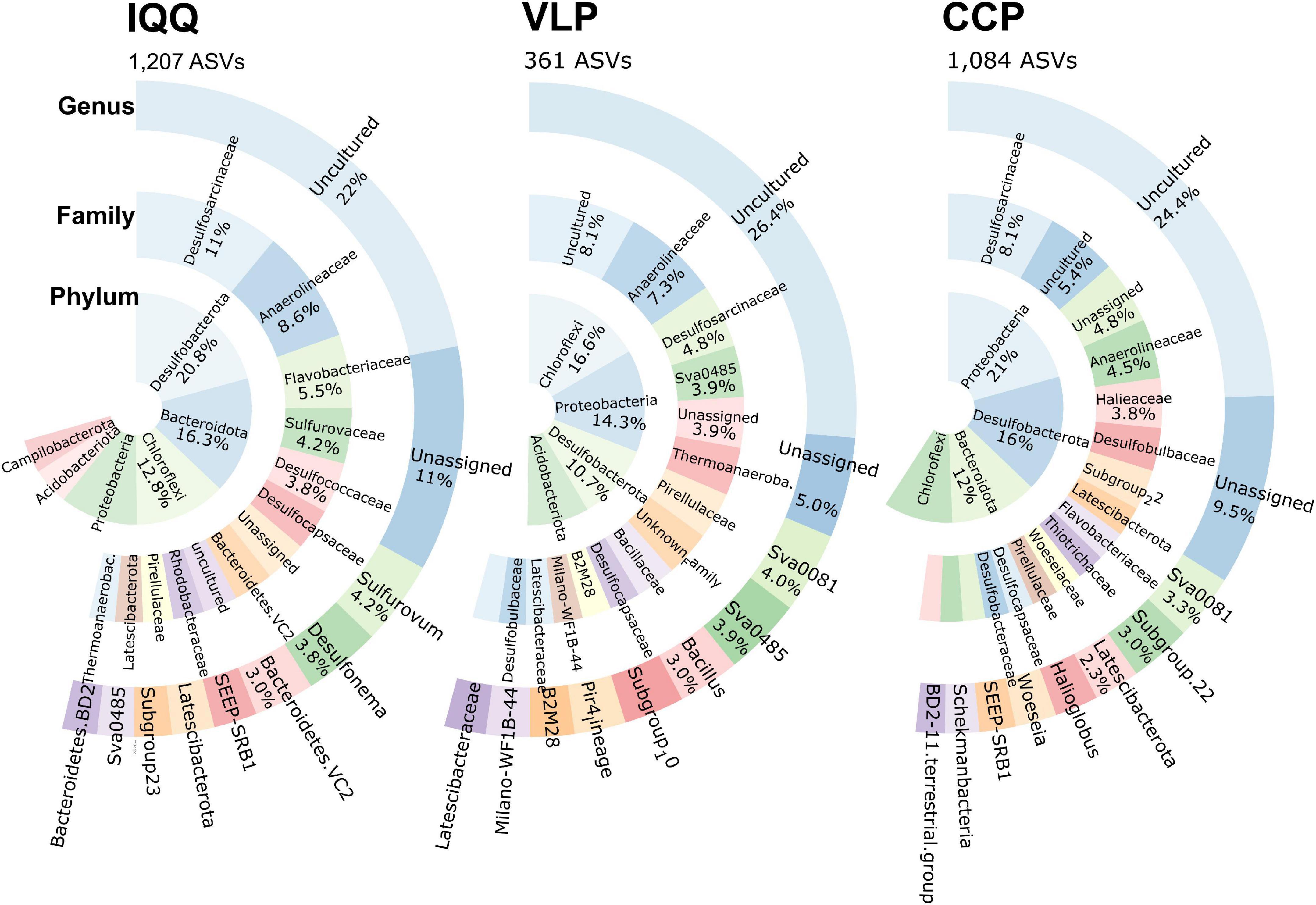
Figure 5. Relative abundance of the most frequent unique ASVs and its taxonomic assignment at the phyla, family, and genus levels by locality.
At the family level, and without considering the uncultured and the unassigned, the Desulfosarcinaceae and Anaerolineaceae contained the larger number of unique ASVs. After them, the family Flavobacteriaceae (5.5%) in IQQ; Halieaceae (3.8%) in CCP, and Sva0485 (formerly Deltaproteobacteria Candidate Sva0485 clade) in VLP appeared as the most abundant.
At the genus level and on the basis of unique ASVs, samples from IQQ, VLP, and CCP have a prominent sulfidic profile, with the presence of Sulfurovum (4.2%) (SOB) and Desulfonema (3.8%) (SRB) at the top of the abundance in IQQ, while, VLP and CCP coincide with the genus Sva0081 sediment group (SRB) as top taxa (after the unassigned and uncultured) (Figure 5). Conversely, in IQQ the genus Sva008 sediment group presented low abundance. In this sense, uncultured and unassigned assignments, ranging 31–35% of the accumulated abundance, leaves 65–69% of the sequences with known taxonomic affiliation at the genus level.
Discussion
Community structure and diversity
This study shows that the best represented bacteria phyla were Desulfobacterota (23%) (formerly the class Deltaproteobacteria), Proteobacteria (18%), Bacteroidota (16%) (the former phylum Bacteroidetes), and Chloroflexi (9%), a distribution that differs with descriptions of similar environments, such as the superficial anoxic OMZ sediments of the Arabian Sea and the Bay of Bengal (Lincy and Manohar, 2020), where Firmicutes (33.08%), Proteobacteria (32.59%), Bacteroidetes (17.48%), and Chloroflexi (5.52%) were the most abundant phyla. The Firmicutes phylum appears with only 1.5% of the total abundance in the present study, which may be attributable to patchy distribution or, as Cupit et al. (2019) posits, the fact that a wide range of Firmicutes species are not captured by conventional culture-independent DNA extraction methods, primarily due to the non-germinative state of Firmicutes endospores.
Global studies on the deeper anoxic layers, i.e., the subseafloor (Hoshino et al., 2020) have reported Chloroflexi, Planctomycetes, and Ca. Atribacteria (Ca. Caldatribacteriota or OP9/JS1) as the most frequent phyla. Indeed, in anoxic subseafloor sediments, Ca. Atribacteria is described as one of the most abundant bacterial taxa (Orsi, 2018; Hoshino et al., 2020), although it was scarcely present in the HS surface sediment samples. The most likely reason is that Ca. Atribacteria were distributed deeper in the sediments, whereas this study covers the first 5 cm of sediment.
In the HS the dominant classes were similar to those reported by Zinger et al. (2011) in their study of coastal sediments, where the most abundant classes were Gammaproteobacteria and Deltaproteobacteria, which are equivalent to the most abundant classes identified in the HS, taking into account that Desulfobacteria is the former Deltaproteobacteria class. Therefore, the HS community presents distinctive features at the phylum level with respect to similar environments, while at the class level it seems to agree with previous reports.
The two most abundant families in the HS, the Desulfosarcinaceae (10.9%) and Anaerolineaceae (5.4%) appear to thrive relatively well in this primeval sulfidic (anoxic) type of environment, a factor that supports the micropaleontological claim relating to the findings of filamentous microfossils in the Hadean (3,235 and 3,465 Ma ago) (Rasmussen, 2000; Schopf et al., 2018). The novel family Desulfosarcinaceae includes genera such as Desulfosarcina, Desulfatitalea, SEEP-SRB1, and Sva0081 sediment group, all of them sulfate-reducing bacteria (SRB). Further, the Sva0081 sediment group was the HS’s most abundant genus. According to previous studies, this uncultured genus is an important sink for acetate (Dyksma et al., 2018a) and for H2 in coastal marine sediments (Dyksma et al., 2018b). It proliferates fast in microaerobic conditions using the H2 that diffuses from fermentation deeper in the sediment (Dyksma et al., 2018a). Therefore, the dominant presence of Sva0081 sediment group in the HS suggests that H2 consumption would be an important metabolic pathway for energy, supporting populations like the Sva0081 sediment group.
As mentioned above, in the HS, the family Anaerolineaceae (5.4%) in the phylum Chloroflexi, was the second most abundant family after the Desulfosarcinaceae, with six genera. Among these, Pelolinea (3.1%), an anaerobic, cosmopolitan, filamentous genus (Imachi et al., 2014), was the most common. Most of the Anaerolineaceae reads (90.3%) are unknown and are assigned as uncultured bacteria. In general terms, this family is defined as consisting of strictly anaerobic and chemoheterotrophic multicellular filamentous bacteria, mostly using sugars and protein for fermentation (Yamada and Sekiguchi, 2018). In this connection, Anaerolineaceae could be playing an important role in the HS by generating organic acids, such as acetate (Liang et al., 2015). The latter molecule has been indicated as one of the most important energy substrates for sulfate-reducing bacteria in marine sediments (e.g., Parkes et al., 1989).
At the ASV level, the most frequent in the HS was assigned to the genus Lutimonas, which is an interesting coincidence with the Black Sea sediment bacterial community where it has been reported as the most abundant OTU (Jessen et al., 2017). Moreover, the genus Lutimonas is defined as symbiont, first isolated from the polychete Periserrula leucophryna (Yang et al., 2007), with variable capacity for nitrate reduction (Kim et al., 2014), and presumably also, nitrogen fixation (Petersen et al., 2016). Although the dominant presence of the symbiont genus Lutimonas in different sediments has been reported (Quero et al., 2015), its high abundance in the HS suggests that this local taxon is a free-living bacterium, not yet characterized.
This study further shows that the diversity and species richness in both the IQQ and CCP sampling stations are similar, while the VLP station presented the lowest diversity and relative abundances. This feature might be explained by the narrow shelf and thus of the sublittoral zone at this point, as compared with that off Concepcion and off Valparaiso (Völker et al., 2012) which in turn may be related to the quality and levels of available TOM. As suggested by Orsi (2018), these variables generally correlate with the number of cells and diversity in the sediments which depends on the lower or greater availability of electron donors in higher organic carbon fluxes.
Geographic patterns
The NMDS and ANOSIM analyses showed a spatial pattern that separated the sampling localities. Thereby, IQQ and CCP on one side and VLP on the other. In this regard, TOM and redox potential were statistically significantly correlated with bacterial abundance, in this case favoring the former two locations which shared productivity-enhancing factors, i.e., primary productivity-enhancing upwelling in IQQ. In addition to this factor, the nutrient input from the regional rivers (most important, the Bio-Bio and Itata rivers) may play a key factor at Concepcion. It is argued that productivity regimes, especially the content of organic matter in sediments, as well as the presence of oxygen, are key in shaping the structure and diversity of bacterial communities (Bienhold et al., 2016; Hoshino et al., 2020), changing spatially, even at microscales as a result of aggregation, creating very complex communities (Zorz et al., 2019).
Regarding spatial distribution patterns and taxa, the family Desulfobulbaceae turned out to be the most influential taxon in explaining the spatial pattern of the HS bacterial community, while it is the seventh most abundant family (3.2%). Indeed, this family presented statistically significant differences in abundance, where its presence is scarce in the locality of IQQ compared to VLP and CCP. In this regard, it has been shown that, as expected, the family Desulfobulbaceae tend to enhance their relative abundance toward anoxic conditions, especially those groups affiliated with sulfate-reducing bacteria, since the reduction of sulfates dominated the remineralization of organic matter with increasing hypoxia (Thamdrup and Canfield, 1996; Jessen et al., 2017). It is noteworthy mentioned that the family Thermoanaerobaculaceae, containing thermophilic bacteria, is present in all the sampling points of this study, and is the third most influential taxon in explaining the spatial pattern. In this regard, three uncultured genera were identified across the data; Subgroup 10, Subgroup 23 and TPD-58. According to Dedysh and Yilmaz (2020), sequences that share similarities with the 16S rRNA gene sequence of Thermoanaerobaculum aquaticum MP-01T, the only described member of the family, have been recovered from a variety of habitats including hot springs, saturated soils of water and sediment. The family Thermoanaerobaculaceae appears as ubiquitous in all localities and represent an interesting future focus of study in this environment.
The family Desulfobulbaceae
The family Desulfobulbaceae, in the phylum Desulfobacterota, includes SRBs and SOBs, and it has recently been suggested that it maintains important H2 scavengers subgroups (Dyksma et al., 2018b). Members of this family are commonly found in marine sediments at different latitudes, i.e., the Baltic Sea (Marzocchi et al., 2014; Marshall et al., 2019; Dam et al., 2021), and off the coast of Chile, e.g., in the coastal ecosystem off the Atacama desert, where between some points the Desulfobulbaceae is one of the dominant and key families (Zárate et al., 2021).
The data corresponding to Desulfobulbaceae contains ASVs assigned to the genera Desulfobulbus, Ca. Electrothrix (Trojan et al., 2016), and uncultured forms. The genus Desulfobulbus is strictly anaerobic, having both a respiratory and fermentative metabolism, with several ways to get energy, i.e., chemoorganoheterotrophic or chemolithoheterotrophic growth. Moreover, sulfate, sulfite, and often thiosulfate serve as terminal electron acceptors and are reduced to H2S (Galushko and Kuever, 2019). On the other hand, the genus Ca. Electrothrix corresponds to filamentous SOB, within the cable bacteria functional group, capable of conducting electrons over centimeter-long distances, coupling sediment electrogenic sulfur oxidation (e-SOx) to dissolved oxygen (Nielsen et al., 2010; Pfeffer et al., 2012), nitrate and nitrite reductions (Marzocchi et al., 2014). Cable bacteria have been found in different oceanographic settings and climate zones, and, in several different coastal habitats (Burdorf et al., 2017), and are a topic of increasing interest. Its presence in the HS system suggests that e-SOx could be occurring in this system, mostly sustained by high levels of nitrate (>20 μM) in both sediments Ahumada et al., 1983), and water column (see also Annexes in Gallardo et al., 2013b).
The relative abundance of Desulfobulbaceae in IQQ is low compared to those in VLP and CCP localities. Since Desulfobulbus represents 95% of the abundance of Desulfobulbaceae, along with a small abundance of Ca. Electrothrix (1 ASV; 5% of the abundance) in IQQ. Furthermore, unknown ASVs, assigned as uncultured Desulfobulbaceae, were not present in IQQ. Beyond that fact, the uncultured Desulfobulbaceae groups represent a “black box,” probably new species, close to Desulfobulbus and Ca. Electrothrix (Figure 4A).
The spatial pattern that separates IQQ, VLP, and CCP is clear in the family Desulfobulbaceae, where the redox potential appeared positively correlated. In fact, the redox potential is more negative in the northernmost locations (IQQ and ANTF) than in the Central-Southern locations. Therefore, the representatives of the Desulfobulbaceae family thrived better in more oxidized sediments in the HS.
The degree of shared Desulfobulbaceae ASVs was very low, thus only one ASV (Desulfobulbus) is shared between IQQ, VLP, and CCP localities, representing a large latitudinal fragmentation of the group among sites.
Sharing and uniqueness in the community
The spatial fragmentation of the community is clearer at the finer grain phylogenetic level as represented by the ASV clusters, which theoretically span species and subspecies layers. Therefore, the ANOSIM value for the spatial pattern is maximum at the ASV level (Supplementary Figure 5), supporting the idea that the spatial pattern is a better fit at this deep phylogenetic level.
This fine spatial pattern, at the ASV level, is probably an extended and general rule for sediment microbial communities, better reflecting environmental changes in microscales. In the Baltic Sea, for example, each site contained distinct ASV signatures of unique sulfate-reducing microorganisms in each family (Marshall et al., 2019). Along the same lines, the percentage of shared ASVs among IQQ, VLP, and CCP is very low, i.e., 3.7% (26% of the reads). This level is even lower than the shared ASVs (16.8%), between anaerobic and aerobic bacterial communities in subseafloor sediments (Hoshino et al., 2020), and those for the Bay of Bengal OMZ and Arabian Sea OMZ sediment bacterial community, with 30% shared OTUs (Lincy and Manohar, 2020). Thus, this result suggests the HS contains a high level of microdiversity, consistent with the Thienemann-Sander temporal stability hypothesis (Gallardo et al., 2016), but with microscale spatial changes, which drive the high microdiversity in a general context of stability, characterized by high abundance of sulfur species, nitrate and very low or no oxygen concentration.
The unique-shared perspective allows distinguishing those ASVs under the same taxonomic assignment, but present and absent in specific locations, revealing intraspecies variation, reflecting high microdiversity across the community. In this regard, IQQ was the locality with the highest proportion of unique ASVs (40%) and, although in a different order, they appeared predominantly assigned to Desulfobacterota, Proteobacteria, and Chloroflexi, and further, the most frequent families were Desulfosarcinaceae, and Anaerolineaceae (in the phylum Chloroflexi). Thus, at each site, there is a strong selection for taxonomically lower-level bacteria into the same groups (micro niches). Moreover, at the genus level, in the IQQ site, the most abundant unique ASVs were assigned to the genera Sulfurovum and Desulfonema, a facultative anaerobic SOB (hydrogen-oxidizing, thiosulfate-reducing), and SRB, respectively. In the VLP sampling site, the anaerobic SRB genera Sva0081 sediment group and Sva0485 (Candidatus Acidulodesulfobacterales), a facultatively anaerobic, sulfate/iron reducer, probably a sulfur oxidant under aerobic respiration (Tan et al., 2019), dominated the unique ASVs. In the CCP site, the Sva0081 sediment group was the most frequent, while poorly represented in IQQ suggesting different biogeochemical processes than those at VLP and CCP. Thus, according to these observations, both sulfur-reducing and sulfide-oxidzing processes occur simultaneously.
Conclusion
According to this study Desulfobacterota and Desulfosarcinaceae dominate in the HS, especially by SRB, reflected by the most abundant genus: the uncultivated genus Sva0081 sediment group. The revealed spatial pattern along the Chilean coast indicates changes at the bacteria community level, positively correlated with the percentage of TOM and redox potential.
Despite the fact that the Desulfobulbaceae species are still not properly characterized, they appeared as a key taxon with significant differences in their distribution influencing the general spatial pattern. Moreover, the spatial pattern is influenced by a large microdiversity, since a very small number of ASVs are shared (3.4%) among the localities included in the comparison, reflecting a diverse, mature and stable HS bacterial community.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA251688, PRJNA251688.
Author contributions
AF, VAG, and IPGM: conceptualization. AF: formal analysis. VAG, CE, and LN: resources. LPN, IPGM, and VAG: funding acquisition and supervision. AF, CE, and VAG: experimental studies and writing—original draft. AF, VAG, IPGM, and LPN: wrote and finalized the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This study was financed by the National Research and Development Agency (ANID) of Chile, in particular through the Conicyt Project 1110786 and a doctoral grant to AF. Besides, the Center for Electromicrobiology (CEM), Department of Biology at Aarhus University.
Acknowledgments
We sincerely appreciate the support of the National Agency for Research and Development of Chile (ANID–former Conicyt). In addition, we thank the Department of Marine Sciences, Universidad Arturo Prat, Iquique, Chile, the crew of the R/V “Kay Kay,” the Department of Oceanography, University of Concepción, Chile, and the Census of Marine Life (CoML) project with its program “International Census of Marine Microbes” (ICoML).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1016418/full#supplementary-material
Footnotes
References
Ahumada, R., Anny, R. G., and Victorino, M. M. (1983). Circulation and fertility of waters in Concepción Bay. Estuar. Coast. Shelf Sci. 16, 95–105. doi: 10.1016/0272-7714(83)90096-3
Bienhold, C., Zinger, L., Boetius, A., and Ramette, A. (2016). Diversity and biogeography of bathyal and abyssal seafloor bacteria. PLoS One 11:e0148016. doi: 10.1371/journal.pone.0148016
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Burdorf, L. D., Tramper, A., Seitaj, D., Meire, L., Hidalgo-Martinez, S., Zetsche, E. M., et al. (2017). Long-distance electron transport occurs globally in marine sediments. Biogeosciences 14, 683–701. doi: 10.5194/bg-14-683-2017
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cupit, C., Lomstein, B. A., and Kjeldsen, K. U. (2019). Contrasting community composition of endospores and vegetative firmicutes in a marine sediment suggests both endogenous and exogenous sources of endospore accumulation. Environ. Microbiol. Rep. 11, 352–360. doi: 10.1111/1758-2229.12679
Dam, A. S., Marshall, I. P., Risgaard-Petersen, N., Burdorf, L. D., and Marzocchi, U. (2021). Effect of salinity on cable bacteria species composition and diversity. Environ. Microbiol. 23, 2605–2616. doi: 10.1111/1462-2920.15484
Dedysh, S. N., and Yilmaz, P. (2020). Thermoanaerobaculaceae. Bergey’s manual of systematics of archaea and bacteria. Hoboken, NJ: John Wiley & Sons. doi: 10.1002/9781118960608.fbm00358
Dyksma, S., Lenk, S., Sawicka, J. E., and Mußmann, M. (2018a). Uncultured gammaproteobacteria and desulfobacteraceae account for major acetate assimilation in a coastal marine sediment. Front. Microbiol. 9:3124. doi: 10.3389/fmicb.2018.03124
Dyksma, S., Pjevac, P., Ovanesov, K., and Mussmann, M. (2018b). Evidence for H2 consumption by uncultured desulfobacterales in coastal sediments. Environ. Microbiol. 20, 450–461. doi: 10.1111/1462-2920.13880
Ferdelman, T. G., Lee, C., Pantoja, S., Harder, J., Bebout, B. M., and Fossing, H. (1997). Sulfate reduction and methanogenesis in a Thioploca-dominated sediment off the coast of Chile. Geochim. Cosmochim. Acta 61, 3065–3079. doi: 10.1016/S0016-7037(97)00158-0
Fernandes, A. D., Reid, J. N., Macklaim, J. M., McMurrough, T. A., Edgell, D. R., and Gloor, G. B. (2014). Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2:15. doi: 10.1186/2049-2618-2-15
Fonseca, A., Ishoey, T., Espinoza, C., Perez-Pantoja, D., Manghisi, A., Morabito, M., et al. (2017). Genomic features of “candidatus venteria ishoeyi”, a new sulfur-oxidizing macrobacterium from the humboldt sulfuretum off Chile. PLoS One 12:e0188371. doi: 10.1371/journal.pone.0188371
Fossing, H., Gallardo, V. A., Jørgensen, B. B., Hüttel, M., Nielsen, L. P., Schulz, H., et al. (1995). Concentration and transport of nitrate by the mat-forming sulphur bacterium Thioploca. Nature 374, 713–715. doi: 10.1038/374713a0
Foster, Z. S., Sharpton, T. J., and Grünwald, N. J. (2017). Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 13:e1005404. doi: 10.1371/journal.pcbi.1005404
Fry, J. C., Parkes, R. J., Cragg, B. A., Weightman, A. J., and Webster, G. (2008). Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol. Ecol. 66, 181–196. doi: 10.1111/j.1574-6941.2008.00566.x
Gallardo, V. A. (1977). Large benthic microbial communities in sulphide biota under Peru–Chile subsurface countercurrent. Nature 268, 331–332. doi: 10.1038/268331a0
Gallardo, V. A., Espinoza, C., Fonseca, A., and Musleh, S. (2013a). Las grandes bacterias del sulfureto de humboldt. Gayana (Concepción) 77, 136–170. doi: 10.4067/S0717-65382013000200008
Gallardo, V. A., Fonseca, A., Espinoza, C., Ruiz-Tagle, N., and Musleh, S. (2016). Bacteria of the humboldt sulfuretum comply with unifying macroecological principles. Mar. Biodivers. 46, 399–406. doi: 10.1007/s12526-015-0377-x
Gallardo, V. A., Fonseca, A., Musleh, S. S., and Espinoza, C. (2013b). Extrapolations of standing-stocks of big bacteria in humboldt eastern boundary current ecosystem (HEBCE). Oceanography 1:110. doi: 10.4172/2332-2632.1000110
Galushko, A., and Kuever, J. (2019). Desulfobulbus. Bergey’s manual of systematics of archaea and bacteria. Hoboken, NJ: John Wiley and Sons. doi: 10.1002/9781118960608.gbm01023.pub2
Helly, J. J., and Levin, L. A. (2004). Global distribution of naturally occurring marine hypoxia on continental margins. Deep Sea Res. Part I Oceanogr. Res. Pap. 51, 1159–1168. doi: 10.1016/j.dsr.2004.03.009
Høgslund, S., Revsbech, N. P., Cedhagen, T., Nielsen, L. P., and Gallardo, V. A. (2008). Denitrification, nitrate turnover, and aerobic respiration by benthic foraminiferans in the oxygen minimum zone off Chile. J. Exp. Mar. Biol. Ecol. 359, 85–91. doi: 10.1016/j.jembe.2008.02.015
Hoshino, T., Doi, H., Uramoto, G. I., Wörmer, L., Adhikari, R. R., Xiao, N., et al. (2020). Global diversity of microbial communities in marine sediment. Proc. Natl. Acad. Sci. U.S.A. 117, 27587–27597. doi: 10.1073/pnas.1919139117
Huber, J. A., Mark Welch, D. B., Morrison, H. G., Huse, S. M., Neal, P. R., Butterfield, D. A., et al. (2007). Microbial population structures in the deep marine biosphere. Science 318, 97–100. doi: 10.1126/science.1146689
Huse, S. M., Dethlefsen, L., Huber, J. A., Welch, D. M., Relman, D. A., and Sogin, M. L. (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. doi: 10.1371/journal.pgen.1000255
Imachi, H., Sakai, S., Lipp, J. S., Miyazaki, M., Saito, Y., Yamanaka, Y., et al. (2014). Pelolinea submarina gen. nov., sp. nov., an anaerobic, filamentous bacterium of the phylum Chloroflexi isolated from subseafloor sediment. Int. J. Syst. Evol. Microbiol. 64, 812–818. doi: 10.1099/ijs.0.057547-0
Jessen, G. L., Lichtschlag, A., Ramette, A., Pantoja, S., Rossel, P. E., Schubert, C. J., et al. (2017). Hypoxia causes preservation of labile organic matter and changes seafloor microbial community composition (Black Sea). Sci. Adv. 3:e1601897. doi: 10.1126/sciadv.1601897
Kim, E., Lin, Y., Kerney, R., Blumenberg, L., and Bishop, C. (2014). Phylogenetic analysis of algal symbionts associated with four North American amphibian egg masses. PLoS One 9:e108915. doi: 10.1371/journal.pone.0108915
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Liang, B., Wang, L. Y., Mbadinga, S. M., Liu, J. F., Yang, S. Z., Gu, J. D., et al. (2015). Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. AMB Express 5:117. doi: 10.1186/s13568-015-0117-4
Lincy, J., and Manohar, C. S. (2020). A comparison of bacterial communities from OMZ sediments in the Arabian Sea and the Bay of Bengal reveals major differences in nitrogen turnover and carbon recycling potential. Mar. Biol. Res. 16, 656–673. doi: 10.1080/17451000.2020.1840593
Maier, S., and Gallardo, V. A. (1984). Thioploca araucae sp. nov. and Thioploca chileae sp. nov. Int. J. Syst. Evol. Microbiol. 34, 414–418. doi: 10.1099/00207713-34-4-414
Marshall, I. P., Ren, G., Jaussi, M., Lomstein, B. A., Jørgensen, B. B., Røy, H., et al. (2019). Environmental filtering determines family-level structure of sulfate-reducing microbial communities in subsurface marine sediments. ISME J. 13, 1920–1932. doi: 10.1038/s41396-019-0387-y
Marzocchi, U., Trojan, D., Larsen, S., Louise Meyer, R., Peter Revsbech, N., Schramm, A., et al. (2014). Electric coupling between distant nitrate reduction and sulfide oxidation in marine sediment. ISME J. 8, 1682–1690. doi: 10.1038/ismej.2014.19
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Mook, D. H., and Hoskin, C. M. (1982). Organic determinations by ignition: Caution advised. Estuar. Coast. Shelf Sci. 15, 697–699. doi: 10.1016/0272-7714(82)90080-4
Nielsen, L. P., Risgaard-Petersen, N., Fossing, H., Christensen, P. B., and Sayama, M. (2010). Electric currents couple spatially separated biogeochemical processes in marine sediment. Nature 463, 1071–1074. doi: 10.1038/nature08790
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R., et al. (2013). Package ‘vegan’. Community ecology package, version. 2.
Orsi, W. D. (2018). Ecology and evolution of seafloor and subseafloor microbial communities. Nat. Rev. Microbiol. 16, 671–683. doi: 10.1038/s41579-018-0046-8
Parkes, R. J., Gibson, G. R., Mueller-Harvey, I., Buckingham, W. J., and Herbert, R. A. (1989). Determination of the substrates for sulphate-reducing bacteria within marine and esturaine sediments with different rates of sulphate reduction. Microbiology 135, 175–187. doi: 10.1099/00221287-135-1-175
Petersen, J. M., Kemper, A., Gruber-Vodicka, H., Cardini, U., Van Der Geest, M., Kleiner, M., et al. (2016). Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat. Microbiol. 2:16195. doi: 10.1038/nmicrobiol.2016.195
Pfeffer, C., Larsen, S., Song, J., Dong, M., Besenbacher, F., Meyer, R. L., et al. (2012). Filamentous bacteria transport electrons over centimetre distances. Nature 491, 218–221. doi: 10.1038/nature11586
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Quero, G. M., Cassin, D., Botter, M., Perini, L., and Luna, G. M. (2015). Patterns of benthic bacterial diversity in coastal areas contaminated by heavy metals, polycyclic aromatic hydrocarbons (PAHs) and polychlorinated biphenyls (PCBs). Front. Microbiol. 6:1053. doi: 10.3389/fmicb.2015.01053
Rasmussen, B. (2000). Filamentous microfossils in a 3,235-million-year-old volcanogenic massive sulphide deposit. Nature 405, 676–679. doi: 10.1038/35015063
Salman, V., Amann, R., Girnth, A. C., Polerecky, L., Bailey, J. V., Høgslund, S., et al. (2011). A single-cell sequencing approach to the classification of large, vacuolated sulfur bacteria. Syst. Appl. Microbiol. 34, 243–259. doi: 10.1016/j.syapm.2011.02.001
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Schopf, J. W., Kitajima, K., Spicuzza, M. J., Kudryavtsev, A. B., and Valley, J. W. (2018). SIMS analyses of the oldest known assemblage of microfossils document their taxon-correlated carbon isotope compositions. Proc. Natl. Acad. Sci. U.S.A. 115, 53–58. doi: 10.1073/pnas.1718063115
Schopf, J. W., Kudryavtsev, A. B., Osterhout, J. T., Williford, K. H., Kitajima, K., Valley, J. W., et al. (2017). An anaerobic ca. 3400 Ma shallow-water microbial consortium: Presumptive evidence of Earth’s Paleoarchean anoxic atmosphere. Precambrian Res. 299, 309–318. doi: 10.1016/j.precamres.2017.07.021
Schulz, H. N., Jorgensen, B. B., Fossing, H. A., and Ramsing, N. B. (1996). Community structure of filamentous, sheath-building sulfur bacteria, Thioploca spp., off the coast of Chile. Appl. Environ. Microbiol. 62, 1855–1862. doi: 10.1128/aem.62.6.1855-1862.1996
Tan, S., Liu, J., Fang, Y., Hedlund, B. P., Lian, Z. H., Huang, L. Y., et al. (2019). Insights into ecological role of a new deltaproteobacterial order Candidatus Acidulodesulfobacterales by metagenomics and metatranscriptomics. ISME J. 13, 2044–2057. doi: 10.1038/s41396-019-0415-y
Thamdrup, B. O., and Canfield, D. E. (1996). Pathways of carbon oxidation in continental margin sediments off central Chile. Limnol. Oceanogr. 41, 1629–1650. doi: 10.4319/lo.1996.41.8.1629
Trojan, D., Schreiber, L., Bjerg, J. T., Bøggild, A., Yang, T., Kjeldsen, K. U., et al. (2016). A taxonomic framework for cable bacteria and proposal of the candidate genera Electrothrix and Electronema. Syst. Appl. Microbiol. 39, 297–306. doi: 10.1016/j.syapm.2016.05.006
Ueno, Y., Ono, S., Rumble, D., and Maruyama, S. (2008). Quadruple sulfur isotope analysis of ca. 3.5 Ga dresser formation: New evidence for microbial sulfate reduction in the early Archean. Geochim. Cosmochim. Acta 72, 5675–5691. doi: 10.1016/j.gca.2008.08.026
Van Kranendonk, M. J., Webb, G. E., and Kamber, B. S. (2003). Geological and trace element evidence for a marine sedimentary environment of deposition and biogenicity of 3.45 Ga stromatolitic carbonates in the Pilbara Craton, and support for a reducing Archaean ocean. Geobiology 1, 91–108. doi: 10.1046/j.1472-4669.2003.00014.x
Völker, D., Geersen, J., Behrmann, J. H., and Weinrebe, W. R. (2012). “Submarine mass wasting off Southern Central Chile: Distribution and possible mechanisms of slope failure at an active continental margin,” in Submarine mass movements and their consequences, eds Y. Yamada et al. (Dordrecht, NL: Springer), 379–389. doi: 10.1007/978-94-007-2162-3_34
Wacey, D., Kilburn, M. R., Saunders, M., Cliff, J., and Brasier, M. D. (2011). Microfossils of sulphur-metabolizing cells in 3.4-billion-year-old rocks of Western Australia. Nat. Geosci. 4, 698–702. doi: 10.1038/ngeo1238
Yamada, T., and Sekiguchi, Y. (2018). “Anaerolineae,” in Bergey’s manual of systematics of archaea and bacteria, ed. W. B. Whitman (Hoboken, NJ: John Wiley & Sons). doi: 10.1002/9781118960608.fbm00301
Yang, S. J., Choo, Y. J., and Cho, J. C. (2007). Lutimonas vermicola gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from the marine polychaete Periserrula leucophryna. Int. J. Syst. Evol. Microbiol. 57, 1679–1684. doi: 10.1099/ijs.0.65060-0
Zárate, A., Dorador, C., Valdés, J., Molina, V., Icaza, G., Pacheco, A. S., et al. (2021). Benthic microbial diversity trends in response to heavy metals in an oxygen-deficient eutrophic bay of the Humboldt current system offshore the Atacama Desert. Environ. Pollut. 286:117281. doi: 10.1016/j.envpol.2021.117281
Zinger, L., Amaral-Zettler, L. A., Fuhrman, J. A., Horner-Devine, M. C., Huse, S. M., Welch, D. B. M., et al. (2011). Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS One 6:e24570. doi: 10.1371/journal.pone.0024570
Keywords: sulfur bacteria, sulfuretum, OMZ, anoxic sediments, microdiversity, bacterial community, Desulfobulbaceae, amplicon sequencing
Citation: Fonseca A, Espinoza C, Nielsen LP, Marshall IPG and Gallardo VA (2022) Bacterial community of sediments under the Eastern Boundary Current System shows high microdiversity and a latitudinal spatial pattern. Front. Microbiol. 13:1016418. doi: 10.3389/fmicb.2022.1016418
Received: 10 August 2022; Accepted: 16 September 2022;
Published: 30 September 2022.
Edited by:
Shan He, Ningbo University, ChinaReviewed by:
Songcan Chen, University of Vienna, AustriaAlexander Galushko, Agrophysical Research Institute, Russia
Copyright © 2022 Fonseca, Espinoza, Nielsen, Marshall and Gallardo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexis Fonseca, YWZvbnNlY2FAYmlvLmF1LmRr; Victor A. Gallardo, dmFnYWxsYXJAdWRlYy5jbA==
 Alexis Fonseca
Alexis Fonseca Carola Espinoza2
Carola Espinoza2 Lars Peter Nielsen
Lars Peter Nielsen Ian P. G. Marshall
Ian P. G. Marshall