
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 October 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1015588
Nitrogen (N) deposition is a worldwide issue caused by human activity. Long-term deposition of N strongly influences plant productivity and community composition. However, it is still unclear how the microbial community responds to long-term N addition in a desert ecosystem. Therefore, a long-term experiment was conducted in the Gurbantonggut Desert in northwestern China in 2015. Four N addition rates, 0 (CK), 5 (N1), 20 (N2), and 80 (N3) kg N ha−1 yr.−1, were tested and the soil was sampled after 6 years of N addition. High-throughput sequencing (HTS) was used to analyze the soil microbial composition. The HTS results showed that N addition had no significant effect on the bacterial α-diversity and β-diversity (p > 0.05) but significantly reduced the archaeal β-diversity (p < 0.05). The fungal Chao1 and ACE indexes in the N2 treatment increased by 24.10 and 26.07%, respectively. In addition, N addition affected the bacterial and fungal community structures. For example, compared to CK, the relative abundance of Actinobacteria increased by 17.80%, and the relative abundance of Bacteroidetes was reduced by 44.46% under N3 treatment. Additionally, N addition also changed the bacterial and fungal community functions. The N3 treatment showed increased relative abundance of nitrate-reducing bacteria (27.06% higher than CK). The relative abundance of symbiotrophic fungi was increased in the N1 treatment (253.11% higher than CK). SOC and NH4+-N could explain 62% of the changes in the fungal community function. N addition can directly affect the bacterial community function or indirectly through NO3−-N. These results suggest that different microbial groups may have various responses to N addition. Compared with bacteria and fungi, the effect of N addition was less on the archaeal community. Meanwhile, N-mediated changes of the soil properties play an essential role in changes in the microbial community. The results in the present study provided a reliable basis for an understanding of how the microbial community in a desert ecosystem adapts to long-term N deposition.
Human activity, such as the use of fossil fuels and N fertilizer, increases the N deposition rate (Norby, 1998; Decina et al., 2020). From 1980 to 2018, ammonia emissions from global agricultural production increased by 78% (Liu L. et al., 2022). In China, the average soil N increased by 8 kg N ha−1 yr.−1 between the 1980s and 2000s (Liu et al., 2013). An increase in N deposition dramatically impacts plants and microorganisms (Fisher et al., 1988; Bobbink et al., 2010; Freedman et al., 2015). Moderate N deposition can alleviate N-limitation in a terrestrial ecosystem and promote primary productivity (Tipping et al., 2019), while excess N deposition leads to adverse effects on the ecosystem, such as soil acidification and heavy metal toxicity (Aber et al., 1998; Tian and Niu, 2015).
The desert ecosystem is an essential part of the terrestrial ecosystem, covering about one-third of the total land area (Laity, 2009). Because desert ecosystems are typically N-deficient, N has been considered as the primary factor affecting the structure and function of desert ecosystems (Brooks, 2003). In a desert ecosystem, N usually penetrates the soil in the form of pulses, accompanied by precipitation (e.g., snow) and other forms, which can constitute an adequate short-term supply of nutrition (Austin et al., 2004). The critical N load of a desert ecosystem is usually lower than that in other ecosystem types (Fenn et al., 2010). On the one hand, the plant cover in a desert ecosystem is usually sparse and discontinuous; therefore, the natural nutrient pools in deserts are small (Zhou et al., 2018). On the other hand, N is relatively limited in the soil, making a desert ecosystem particularly vulnerable to anthropogenic N input (Ladwig et al., 2012). Recent intensification of urbanization around deserts has caused reactive N to enter adjacent deserts by diffusion due to human activity, which increases the N deposition rate (Fenn et al., 2003). However, research on N deposition has mainly focused on forest and grassland ecosystems and less on desert ecosystems (Gilliam, 2006; Stursova et al., 2006), which significantly limits our ability to consider and predict the effects of N deposition across different ecosystems.
The duration of N deposition is an essential factor affecting the structure and function of the desert ecosystem, primarily the long-term accumulation of N in the environment (Humbert et al., 2016). Long-term N addition affects the ecosystem more dramatically than short-term addition (Kivimäki et al., 2013). Han et al. (2019) found that long-term N addition negatively affected plant diversity, but short-term N addition did not. In addition, the impact of N addition on an ecosystem may exhibit a time lag, and short-term N addition does not accurately reflect the impact of N addition on the ecosystem (Zhou et al., 2018). Therefore, long-term research is necessary to fully reveal the ecological effects of N deposition.
Soil microorganisms play an essential role in soil organic matter decomposition, plant fitness, and nutrient cycling (Berg and Matzner, 1997; Lau and Lennon, 2012; Ford et al., 2016). N addition can affect the microbial community (Lu et al., 2011; Ying et al., 2017), but different responses are seen in different ecosystems. N addition for 2 years changed the fungal community in a forest ecosystem (He et al., 2021). However, it did not significantly affect the microbial community in a grassland ecosystem (McHugh et al., 2017). In addition, distinct microbial types may have different responses to N addition. For example, N addition significantly affected the bacterial community structure but had little effect on the fungal community structure (Mueller et al., 2015). Currently, the researches on effects of N deposition are mainly focused on the bacterial and fungal communities and relatively less on the archaeal community. Archaea are a class of unicellular organisms that have similar morphology as bacteria, but show a close relationship with the fungi (Zhao et al., 2022). It has been found in most ecosystems and are indispensable in many ecological and biogeochemical processes (Meng et al., 2014). Accordingly, the study of archaea when exploring the response patterns of various microbial types in a desert ecosystem is necessary.
A long-term in-situ experiment was conducted at the Fukang Desert Ecosystem Observation and Experimental Station in 2015 to understand the long-term effects of N addition on soil microbial communities and their underlying mechanisms in a desert ecosystem. Soil samples were taken in mid-May 2021, and HTS of 16S rRNA gene and ITS were performed to study the effect of N deposition on the bacterial, fungal, and archaeal community structure and function. The objectives of this study were (1) to explore the response of soil microbial diversity, community structure, and function to N addition, and (2) to elucidate the factors driving changes in the soil microbial community when N is added.
This study was conducted at the Fukang Desert Ecosystem Observation and Experimental Station, located in the southern periphery of the Gurbantonggut Desert, Northern Xinjiang, China (Supplementary Figures 1A,B). This area has a typical temperate continental climate. The annual mean temperature is 6.6°C, and the annual mean precipitation is 163 mm (Liu R. et al., 2022). The area has typical sandy soil with low nutrient status. The initial total N content of this soil was approximately 0.15 g kg−1 (Table 1), far lower than in a grassland or forest ecosystem (Zhang et al., 2014; Shi et al., 2018). The soil pH was 8–9 (Table 1), characterizing it as typical alkaline soil. The major plant community in this area includes shrubs and annual herbs, such as Haloxylon ammodendron, H. persicum, Nepeta micrantha, Erodium oxyrhinchum, and Meniocus linifolius (Huang et al., 2015; Yue et al., 2018).
The experimental plot was established in 2015 and adopted a completely randomized experimental design (Supplementary Figure 1C). The experimental area was maintained under natural conditions. Based on the actual atmospheric N deposition in this area (20–25 kg N ha−1 yr.−1, Zhang et al., 2012), four N treatments were established, including 0 (CK), 5 (N1), 20(N2), and 80 (N3) kg N ha−1 yr.−1. N2 treatment was approximately double the amount of atmospheric N deposition, while N1 and N3 treatments represented low and high N levels, respectively. Each treatment had three replicates. A total of twelve 2 m × 2 m quadrats were established and were bounded by a 0.5 m wide buffer strip. The form of N applied was NH4NO3 (35% pure N). The major plants in the sample plots were spring ephemerals that germinate in mid-late March and die in late May and early June (Huang et al., 2018). However, some Amaranthaceae plants continue their growth until October (Lu et al., 2013). Therefore, the added NH4NO3 was divided into six equal portions, and added to the plots during the plant growth season (i.e., April, May, and July to October) each year. The NH4NO3 was dissolved in 5 l of water and applied evenly with a sprayer. The same volume of water was supplied to the CK treatment to control for the effect of the water.
Soil samples were collected in the middle of May 2021. In this experiment, N addition continued until sampling. All plants within the sample plots were kept. The soil crust and plant litter were carefully removed from the soil surface during sampling. Soil samples were collected next to the plants at a depth of 0–5 cm using a three-point sampling method. Each soil sample was passed through a 2 mm mesh sieve, then divided into two parts, put into plastic self-sealing bags, packed into a refrigerated box, and transported to the laboratory. One subsample was used for measuring soil properties. The other subsample was stored at −20°C for the determination of the soil microbial community.
The soil pH was measured at a soil-to-water ratio of 1:5 with a pH meter (Seven Easy Plus, Shanghai, China; Rhoades, 1996). The soil organic carbon (SOC) was determined by the potassium dichromate external heating method (Ciavatta et al., 1991). The total nitrogen (TN) was determined by the Kjeldahl method (Bremner, 1965). The soil ammonium nitrogen (NH4+-N) and nitrate nitrogen (NO3−-N) were determined by indophenol blue colorimetry and ultraviolet spectrophotometry after extraction with 2 M KCl (Zhao et al., 2009). The available phosphorus (AP) was measured using the molybdenum antimony anti-colorimetry method (Zhang et al., 2015).
Microbial DNA from the soil samples was extracted by the CTAB method (Berry et al., 2005). The concentration and purity of the DNA sample were determined by electrophoresis in 1% (w/v) agarose gels. The DNA samples were subsequently diluted to 1 ng/uL using sterile water. Polymerase chain reaction (PCR) amplification was conducted with the 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) primers, which amplified the hypervariable regions V3–V4 of the bacterial 16S rRNA gene (Pichler et al., 2018). The primers ITS5-1737F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS2-2043R (5′-GCTGCGTTCTTCATCGATGC-3′) were used to amplify the fungal ITS1 region (Mukhtar et al., 2021). The hypervariable V4 regions of the archaeal 16S rRNA gene were amplified with the primer sets Arch519F (5′-CAGCCGCCGCGGTAA-3′) and Arch915R (5′-GTGCTCCCCCGCCAATTCCT-3′) (Li et al., 2022). All PCR reactions were carried out in a 30 μl reaction system with 15 μl of Phusion® High-Fidelity PCR Master Mix (BioLabs, Ipswich, MA, United States): approximately10 ng of template DNA, and 0.2 μM of forward and reverse primers. Thermal cycling conditions were as follows: initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s. Finally, 72°C for 5 min. The PCR products were visualized by agarose gel electrophoresis and were mixed in equidensity ratios. Then, the mixture of PCR products was purified with a Gene JET Gel Extraction Kit (Thermo Fisher Scientific). Sequencing libraries were generated using the Illumina TruSeq DNA PCR-Free Library Preparation Kit following the manufacturer’s recommendations, and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Fisher Scientific) and Agilent Bioanalyzer 2,100 system. Finally, the library was created by HTS on the Illumina NovaSeq 6,000 platform at Novogene (Beijing, China). All the sequences of bacterial and archaeal 16S rRNA, and of fungal ITS have been saved in the SRA of the National Center for Biotechnology Information under accession number PRJNA863934.
Paired-end reads from the original DNA fragments were spliced using the FLASH analysis tool (version 1.2.71; Magoč and Salzberg, 2011), and the obtained splicing sequence comprised the Raw tags. Raw tags obtained by splicing suffered strict filtering to produce high-quality Clean Tags. The QIIME software (version 1.9.12; Caporaso et al., 2010) was used to remove the chimeric sequence, and to obtain the Effective Tags. Using the Uparse algorithm (version 7.0.10013) to cluster all effective tags of all samples, sequences with ≥97% similarity were assigned to the same OTU. For bacteria and archaea, the Silva Database (version 138.14) based on the Mothur algorithm was used to annotate taxonomic information. Fungal species annotation was performed by using the blast algorithm in the QIIME software (version 1.9.15) and the Unite database (version 8.26). The Chao1 and ACE richness and the Shannon and Simpson diversity indices were calculated to measure microbial α-diversity using QIIME software (version 1.9.1). Bacterial and archaeal community functions in the ecological samples were predicted according to the FAPROTAX database (Sansupa et al., 2021), while the fungal community function was predicted according to the FUNGuild database (Nguyen et al., 2016).
One-way analysis of variance (ANOVA) with the least significant difference (LSD) was used to analyze the significant differences among N addition treatments for soil properties and the microbial community with the SPSS 22.0 software package (SPSS Inc., Chicago, IL, United States). Before the analysis, these data were transformed as necessary to meet the requirements of variance homogeneity and normal distribution. If the data were still not satisfied after conversion, the non-parametric test was carried out.
A Venn diagram was used to analyze the differences between shared and unique OTU numbers among the four N treatments. A principal co-ordinates analysis (PCoA) based on Bray–Curtis dissimilarity was constructed to study the differences between the four treatments using the vegan software package (version 2.15.3; Freedman and Zak, 2014). The microbial β-diversity index was calculated using the QIIME software (version 1.9.1), and pairwise differences were further compared based on the Wilcoxon rank sum test. A LEfSe analysis (LDA score ≥ 2.0) was used to determine the significant differences in microbial community from the phylum to the genus levels using the LEfSe software (version 1.0; Qin et al., 2021). The soil properties as explanatory variables and the dominant microbial phyla as species variables were used to perform a redundancy analysis (RDA) with the vegan packages in the R software (version 2.15.3). A Spearman correlation analysis was used to further explore the relationships between the soil properties and the dominant microbial phyla. A Mantel-test analysis based on the OTU level was used to describe the relationship between the soil properties and the microbial community structure. Structural equation modeling (SEM) was used to understand how the soil properties mediate the microbial community richness and function changes using AMOS 26.0. The microbial community richness was represented by the ACE index, which has a higher fitness than the Chao1 index. All functional groups of soil bacteria, fungi, and archaea were ranked using a principal component analysis (PCA), and the first principal component (PC1) was used in an SEM analysis. The fitness of the SEM was assessed using the Chi-square (χ2) test, the comparative fit index (CFI > 0.9), and the root mean square error of approximation (RMSEA <0.08; Zhao et al., 2020).
N addition did not significantly affect the pH, TN, and AP (p > 0.05) but significantly increased the content of SOC, NH4+-N, and NO3−-N (p < 0.05; Table 1). Compared with CK, the SOC content of the N1 treatment significantly increased by 64.63% (p < 0.05). The N3 treatment significantly increased the content of NH4+-N and NO3−-N by 124.77 and 615.87%, respectively (p < 0.001).
Among the four N treatments, bacteria, fungi, and archaea, respectively, shared 3,282, 304, and 24 OTUs (Figure 1). All OTUs were used in the subsequent statistical analyses. N addition did not significantly affect the bacterial and archaeal OTU and the α-diversity indexes (i.e., Shannon, Simpson, Chao1, and ACE), but the OTU, Chao1, and ACE indexes of the fungi were significantly increased (p < 0.05; Table 2). Compared to CK, the OTU and Chao1 index of fungi in the N2 treatment increased by 24.72 and 24.10%, respectively. The ACE index of fungi in the N1 and N2 treatments increased by 23.73 and 26.07%, respectively. A PCoA analysis based on the Bray–Curtis dissimilarity showed that the PC1 and PC2 dimensions explained 24.76 and 15.38% of the OTU information for the soil bacterial community (Figure 2A), 36.62 and 15.68% of the OTU information for the soil fungal community (Figure 2B), and 77.59 and 14.02% of the OTU information for the soil archaeal community (Figure 2C). The degree of dispersion between treatments in the PCoA analysis represents the difference between treatments, the more dispersed between treatments, the more significant differences in microbial communities. The PCoA analysis revealed that the N addition could affect the microbial community, but different microbial groups presented distinct responses (Figures 2A–C). Compared with fungi and archaea, the bacterial community was more sensitive to N addition since the bacterial communities in N2 and N3 treatments were separated from each other and from that in CK and N1 treatments. However, the communities of fungi and archaea in N2 and N3 treatments were separated from each other but not from that of CK and N1 treatments. Comparing the pairwise differences based on the Wilcoxon rank sum test showed that N addition did not significantly affect the bacterial and fungal β-diversity, but archaeal β-diversity was significantly reduced in the N3 treatment compared to CK (p < 0.05; Figures 2D–F).
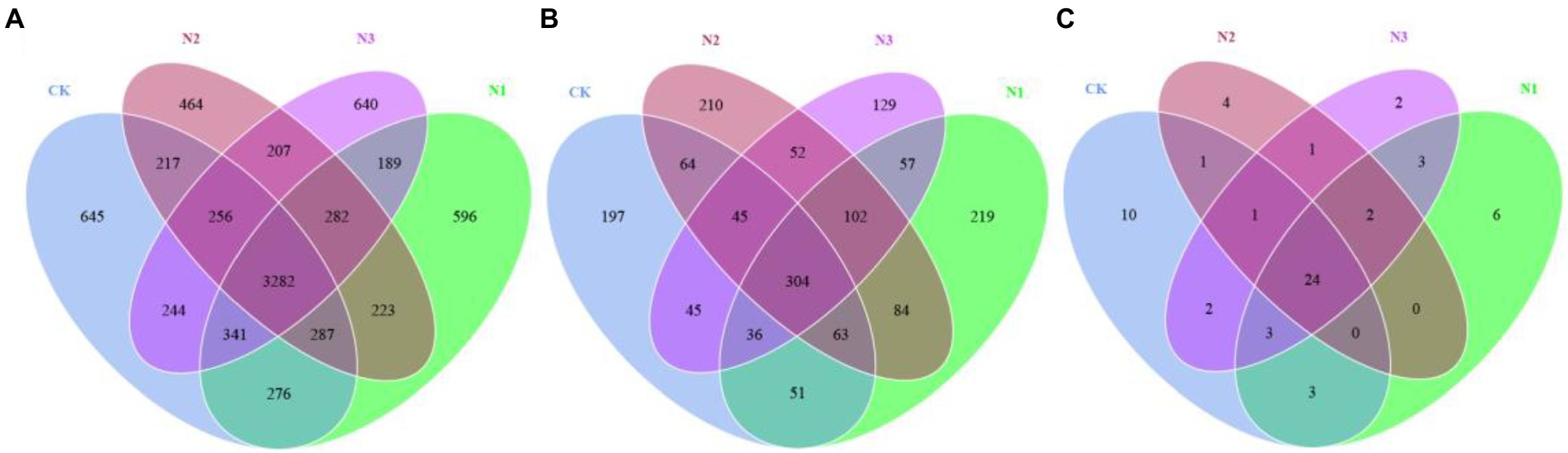
Figure 1. Venn analysis of the soil microbial community in different N deposition treatments based on OTU levels. (A) The number of bacterial OTU at different N treatments. (B) The number of fungal OTU at different N treatments. (C) The number of archaeal OTU at different N treatments. Each ellipse represents a treatment in the figure, and the number of shared OTUs between treatments is represented by the number of overlapping parts, while the number of non-overlapping parts represents the number of unique OTUs between treatments.
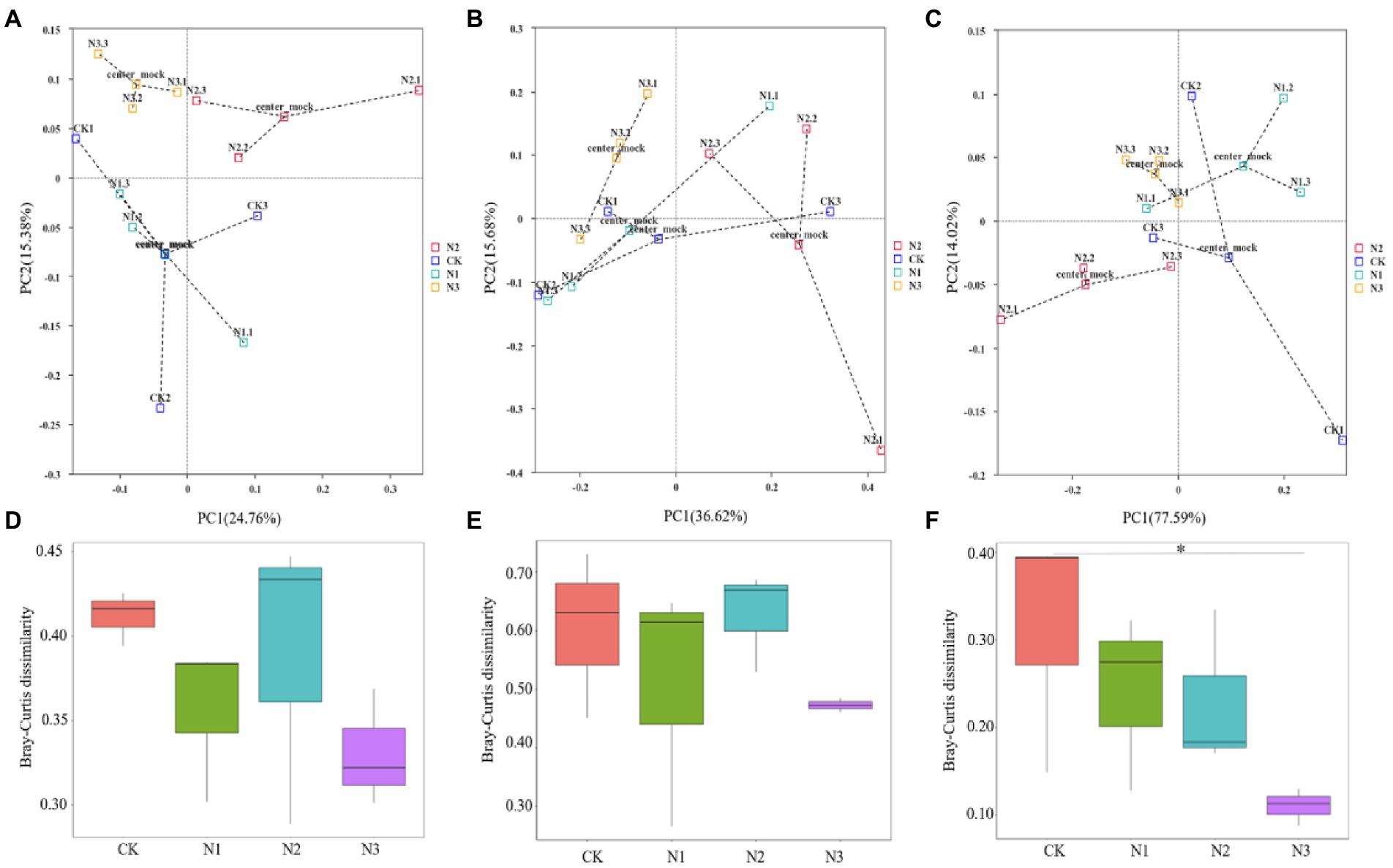
Figure 2. Effects of N addition on soil microbial community β-diversity revealed by PCoA analysis (A–C) and Wilcoxon rank sum test (D–F). (A,D) Bacterial community. (B,E) Fungal community. (C,F) Archaeal community. In (A–C), each point represents a sample, and points of the same color come from the same treatment. The closer the distance between two points is, the smaller the difference exists in community composition between them. The center mock represents the center point of the same sample confidence circle. * Indicates p < 0.05.
The relative abundance of the top 10 phyla of bacteria, fungi, and archaea was selected to evaluate the effect of N addition on soil microbial community structure (Figure 3). Actinobacteria (CK: 22.84%, N1: 22.02%, N2: 24.43%, N3: 26.91%), Bacteroidetes (CK: 17.05%, N1: 17.37%, N2: 16.22%, N3: 9.47%) and Proteobacteria (CK: 11.15%, N1: 11.84%, N2: 12.22%, N3: 11.28%) were the dominant bacterial phyla (Figure 3A). Compared with CK, the high N addition (N3 treatment) significantly increased the relative abundance of Actinobacteria by 17.80%, while significantly decreased the relative abundance of Bacteroidetes by 44.46% (p < 0.05). In addition, N addition presented a trend to increase the average relative abundance of Chloroflexi (CK: 5.39%, N1: 5.88%, N2: 6.28%, N3: 7.37%) and to decrease the average relative abundance of Verrucomicrobia (CK: 2.75%, N1: 2.15%, N2: 1.52%, N3: 1.63%). The relative abundance of Ascomycota was the highest among the fungal phyla (Figure 3B). Low level of N addition (N1 treatment) significantly increased the relative abundance of Basidiomycota, compared with CK, by 141.67% (p < 0.05). Moreover, N addition increased the average relative abundance of Mucoromycota (CK: 0.09%, N1: 0.43%, N2: 0.64%, N3: 1.19%), and decreased the average relative abundance of Chytridiomycota (CK: 2.74%, N1: 0.21%, N2: 0.62%, N3: 0.22%). A total of four archaeal phyla were detected in all the treatments (Figure 3C). The abundance of Crenarchaeota was the highest among the archaeal phyla (CK: 90.04%, N1: 93.87%, N2: 94.18%, N3: 95.01%). N addition increased the average relative abundance of Thermoplasmatota.

Figure 3. Effect of N addition on the soil microbial community (phylum level). (A) The relative abundance of the top 10 bacterial phyla. (B) The relative abundance of the top 10 fungal phyla. (C) The relative abundance of the archaeal phyla.
A LEfSE analysis was performed between treatments to identify the taxa (i.e., biomarkers) with significant differences in abundance (Figure 4). With an LDA threshold of 2, the LEfSe results showed 20 biomarkers in the bacterial community (Figure 4A): 12 in CK (including Thermophilales, Cyanobacteriaceae, Anaerococcus, and Fructobacillus), 5 in N1 treatment (including Muribaculaceae and Magnetospiraceae), 1 in N2 treatment (Helicobacter typhlonius), and 2 in N3 treatment (Solirubrobacter and Conexibacter). The LEfSe identified 40 biomarkers in the fungal community (Figure 4B): 9 in CK treatment, mainly including Orbiliaceae, Tremellaceae, Albatrellaceae, and Cryptococcus; 19 in N1 treatment (mainly the Basidiomycota taxa); 11 in N2 treatment, mainly including Leptosphaeriaceae and Psathyrellaceae; 1 in N3 treatment (Fusarium commune). No biomarker was found in the archaeal community in any treatment.
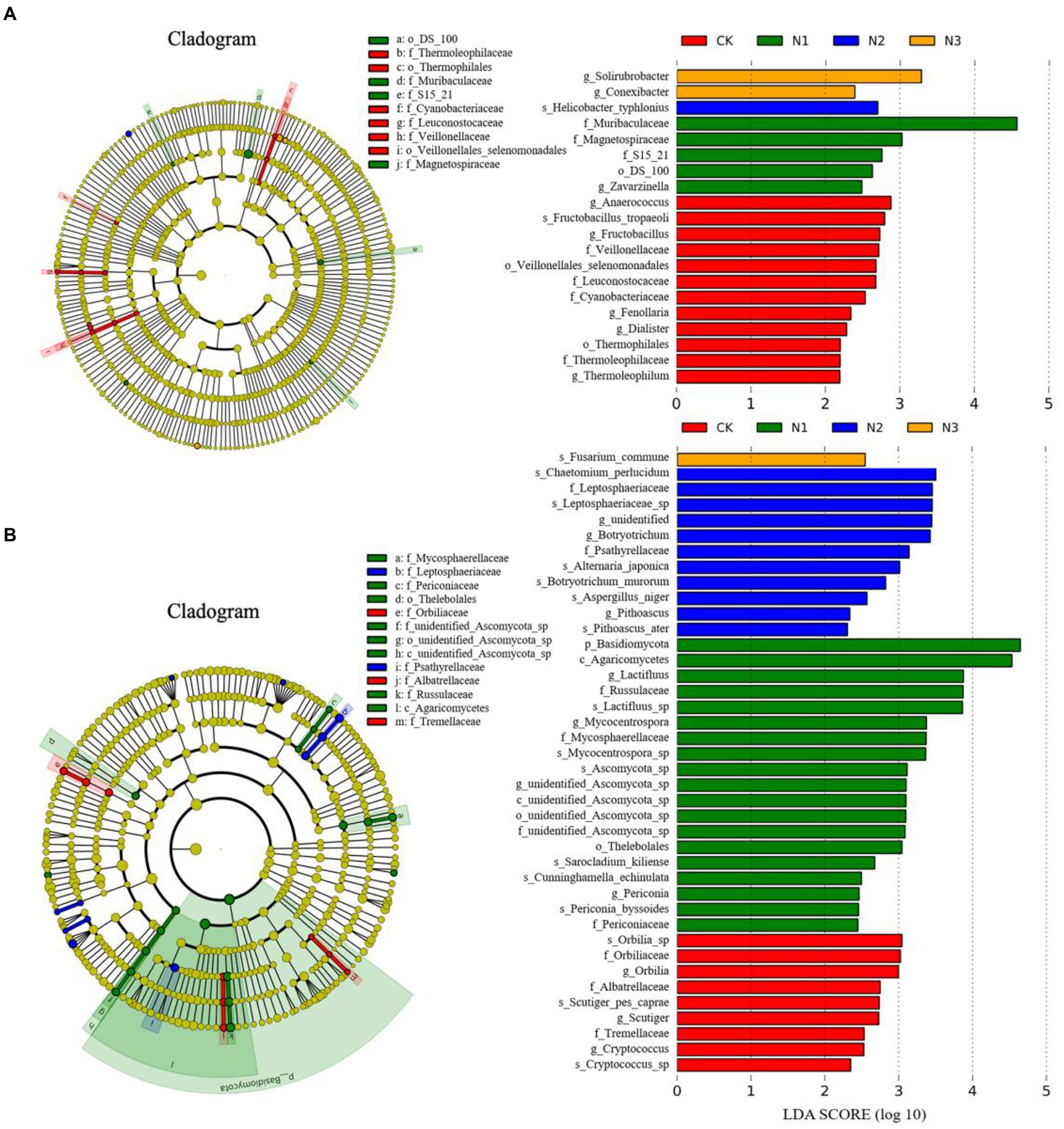
Figure 4. The LEfSe analysis of the soil microbial community structure regarding N addition (phylum-genus levels). (A) The LEfSe analysis of the bacterial community (LDA ≥ 2.0). (B) The LEfSe analysis of the fungal community (LDA ≥ 2.0).
According to the species classification results, 35 bacterial functions with a high relative abundance were selected for study (Figure 5A). The relative abundance of chemoheterotrophy (14.73–15.40%), aerobic chemoheterotrophy (13.09–14.33%), nitrate reduction (6.44–8.07%), and fermentation (1.64–2.83%) were high. The relative abundance of nitrate-reducing bacteria was significantly increased under N3 treatment (27.06%, p < 0.05) compared with that in CK. Additionally, the relative abundance of xylanolytic bacteria was significantly increased by 162.94% in N2 treatment (p < 0.05). Based on the annotated FUNGuild database, the fungal functional community showed 9 nutritional patterns, and pathotroph-saprotroph (26.96–56.87%) was the main fungal function (Figure 5B). The abundance of symbiotrophic fungi was significantly increased under N1 treatment (253.11%, p < 0.05) compared with that in CK. In addition, the average relative abundance of saprotrophic fungi was increased in N3 treatment. The archaeal functional community was annotated according to the FAPROTAX database. Two archaeal functions were detected, including aerobic ammonia oxidation and nitrification, and N addition did not affect their relative abundance (Figure 5C).
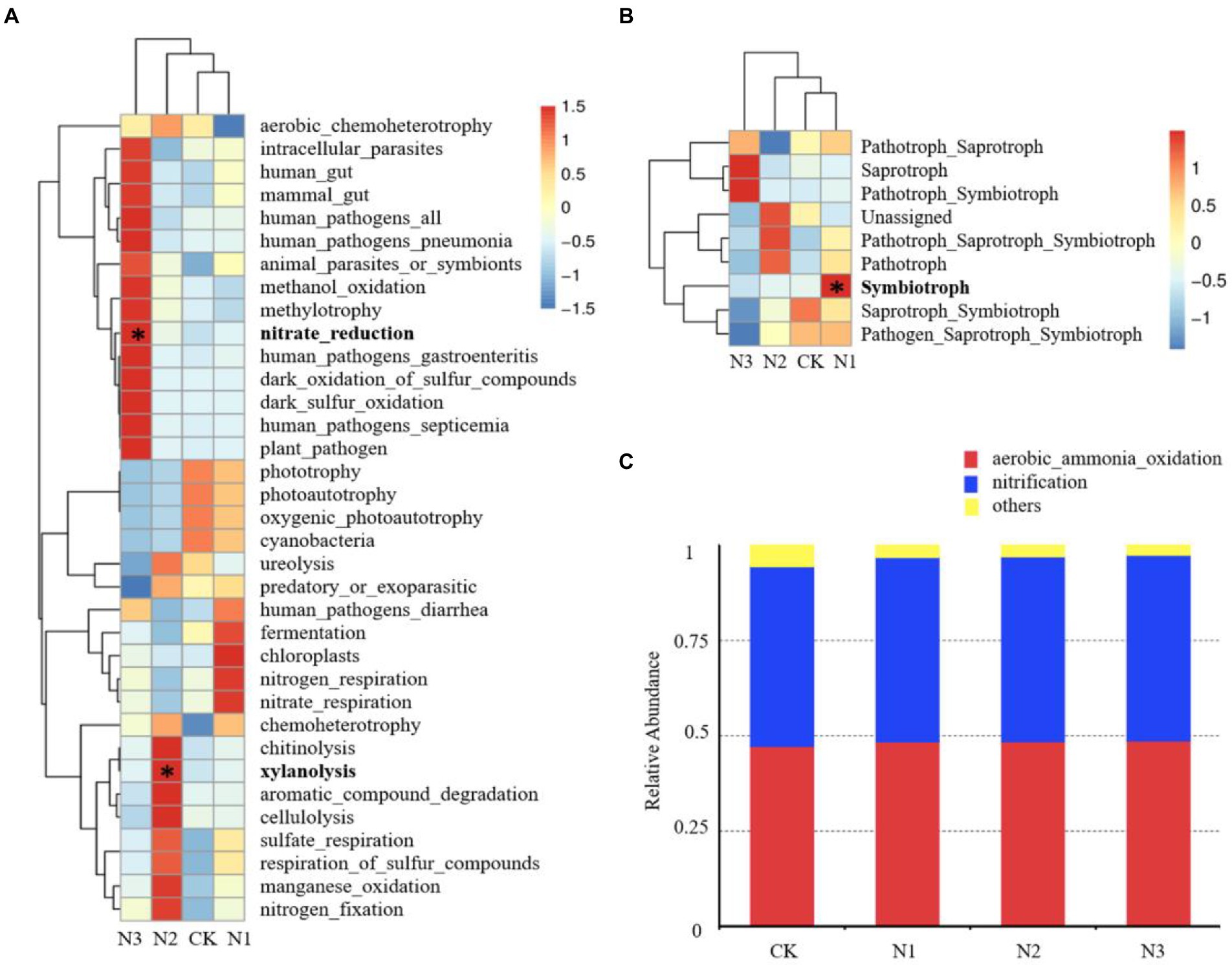
Figure 5. Effect of N addition on microbial community functions. (A) Effect of N addition on the relative abundance of the top 35 bacterial community functions. (B) Effect of N addition on the relative abundance of fungal community functions. (C) Effect of N addition on the relative abundance of archaeal community functions. * Indicates p < 0.05.
The relationship between the microbial community structure and soil properties at the phylum level was analyzed by RDA and Spearman analyses (Figure 6). The soil factors could explain 57.77% of the bacterial community variation; RDA1 explained 33.97% of the variation, and RDA2 explained 23.80% of the variation (Figure 6B). The concentrations of NH4+-N and NO3−-N were significantly positively correlated with Actinobacteria, unidentified bacteria, and Chloroflexi, but they were negatively correlated with Bacteroidetes. The pH was significantly positively correlated with Acidobacteria (p < 0.05; Figure 6A). The soil factors could explain 38.63% of the total variation of the fungal community; RDA1 explained 21.20% of the variation, and RDA2 explained 17.43% of the variation (Figure 6C). Glomeromycota was significantly positively correlated with NH4+-N (p < 0.01). NO3−-N was significantly positively correlated with Mucoromycota. AP was significantly positively correlated with Calcarisporiellomycota. TN was significantly negatively correlated with Rozellomycota (p < 0.05; Figure 6A). The soil factors could explain 30.79% of the total variation of the archaeal community; RDA1 explained 19.15% of the variation, and RDA2 explained 11.64% of the variation (Figure 6D). TN was significantly positively correlated with unidentified archaea (p < 0.01; Figure 6A).
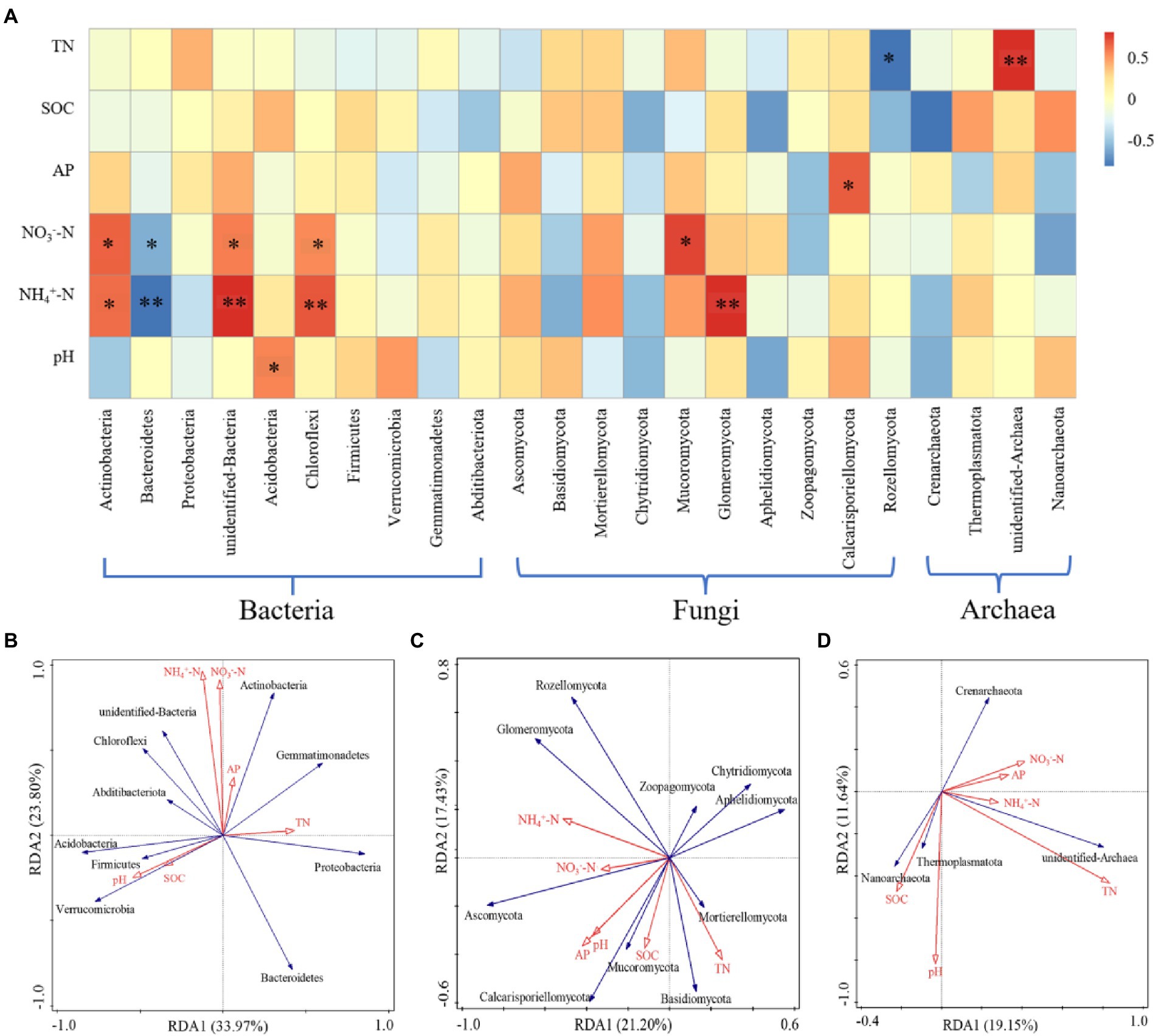
Figure 6. The analysis of the relationship between microbial community structure and soil properties (phylum level). (A) Spearman correlation analysis of microorganisms. (B) RDA analysis of bacteria. (C) RDA analysis of fungi. (D) RDA analysis of archaea. * Indicates p < 0.05, ** indicates p < 0.01.
In the present study, soil pH, TN content, and AP content were not significantly affected by the addition of N, meanwhile the contents of SOC, NH4+-N, and NO3−-N seemed N level dependent features, since the content of SOC was only significantly increased in the N1 treatment, and the contents of NH4+-N and NO3−-N were only significantly increased in the N3 treatment (Table 1). The increase of SOC might be related to the possibly enhanced input of plant residue (Sha et al., 2021), the relatively high abundance of photosynthetic bacteria, and the relatively low abundance of degradation bacteria and saprotrophic fungi in the N1 treatment (Figures 5A,B). The increase in contents of NH4+-N and NO3−-N by the high level of N addition (N3 treatment) was consistent with previous studies (Zheng et al., 2021). Although NH4+-N and NO3−-N were added at equimolar concentrations, their accumulation rates were different (NO3− > NH4+), similar with the observation of Huang et al. (2021) in the desert steppe. The NH4+-N and NO3−-N accumulation rates may be related to the alkaline soil (pH = 8.74; Table 1) in this study, since acidic soils favored the uptake of NO3−-N by plants, while alkaline soil favored the uptake of NH4+-N (Marcus-Wyner, 1983). The absence of change in TN content in treatments of N addition in this study might be explained by the assimilation of plants (Cui et al., 2019) and the emission of gases such as NH3 and N2O produced by nitrate respiration (Yue et al., 2019).
In this study, N addition did not affect the Shannon and Simpson indices of the soil microbes, which is consistent with previous studies (Yuan et al., 2020), but contrasted with the results obtained from a forest ecosystem (Liu Y. et al., 2022), in which 6 years of N addition reduced the microbial diversity. This might be related to the fact that the desert ecosystem differs from the forest ecosystem and their soil microorganisms have different sensitivity to N addition (Ramirez et al., 2010). N addition did not affect bacterial and archaeal richness but significantly increased fungal richness in the N2 treatment (Table 2), which might be dependent on the initial soil conditions (Wang et al., 2018). The fungal community richness increased with N at nutrient-poor sites but decreased at nutrient-rich sites (Moore et al., 2021). The SEM showed that SOC and NO3−-N could explain 20% of the changes in fungal richness (Figure 7). In addition, the variation among bacterial, fungal, and archaeal richness responses to N addition might be related to the demand and physiological structure of microorganisms themselves. Soil bacteria were mainly affected by predation rather than resource effectiveness (Wardle et al., 1995), while the fungal community was sensitive to changes in resource effectiveness associated with N deposition (Allison et al., 2007; Wang et al., 2021a). N addition did not affect bacterial and fungal β-diversity, which is consistent with Liu et al. (2021). In addition, N addition significantly reduced the archaeal β-diversity in the N3 treatment (Figure 2F), indicating that the N addition makes the archaeal community more convergent.
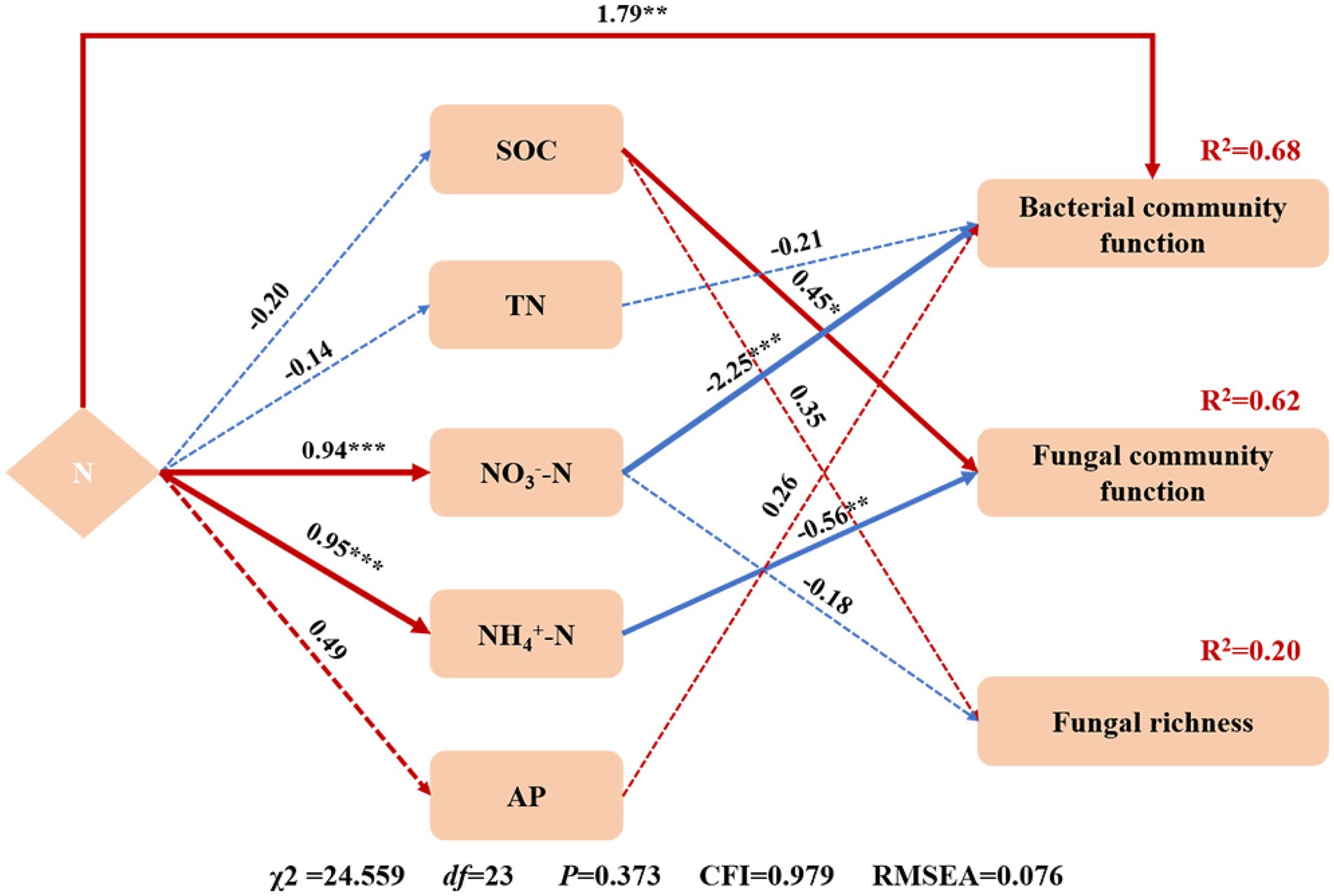
Figure 7. Structural equation model (SEM) of the effects of N addition on soil properties (SOC, TN, NO3−-N, NH4+-N, and AP) and microbial community richness and function. Arrows indicate the hypothesized direction of causation. The numbers next to the arrows are standardized path coefficients. The width of the arrows represents the strength of the relationships. Red arrows represent positive and blue arrows represent negative relationships. The solid arrows represent significant, and the dashed arrows represent non-significant relationships. To improve the adaptability of the model, delete the paths with less correlation. * Indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001.
N addition changed the bacterial and fungal community structures at the phylum level and had little effect on the archaeal community structure (Figure 3). A LEfSe analysis also confirmed this result (Figure 4). The relative abundances of Actinobacteria, Ascomycota, and Crenarchaeota were the highest among the phyla in bacteria, fungi, and archaea, respectively. N addition increased the relative abundances of the phyla Actinobacteria, Chloroflexi, and Mucoromycota, but decreased the relative abundances of Bacteroidetes and Verrucomicrobia. The co-trophic hypothesis could explain these results that nutrient-rich soil is conducive to the growth of copiotrophic taxa, with oligotrophic taxa exhibiting an opposite pattern (Fierer et al., 2012; Grant et al., 2022). In this study, N addition increased the relative abundance of copiotrophic taxa, such as Actinobacteria, and decreased the relative abundance of oligotrophic taxa, such as Verrucomicrobia.
Based upon the data in Supplementary Table 1, only the pH was significantly correlated with the community structure of fungi and archaea at the OTU level in the tested soil, while the other soil traits, including SOC, TN, NO3−-N, NH4+-N, and AP, had no significant effects on the community structure of bacteria, fungi, and archaea at the OTU level. This is consistent with previous studies that soil pH was the critical factor that affected the microbial community structure (Francioli et al., 2016; Ullah et al., 2019; Zheng et al., 2019).
Long-term N addition affected the bacterial and fungal community functions but did not affect the archaeal function (Figure 5). These effects were consistent with the response pattern of the microbial community structure. In this study, high level N addition (N3 treatment) significantly increased the relative abundance of the nitrate-reducing bacteria (Figure 5A), which are positively correlated with NO3−-N (Yang et al., 2017). The input of NH4NO3 increased the content of NO3−-N in soil (Table 1), thereby promoting the abundance of nitrate-reducing bacteria (Mohn et al., 2000). In addition, N2 treatment, but not N1 and N3 treatments, significantly increased the relative abundance of the xylanolytic bacteria (Figure 5A). It seems that this increase is not related to the SOC (Table 1), but possible related to the C/N ratio in soil. Xylan is the main component of hemicellulose, accounting for 20–30% of the total hemicellulose (Gírio et al., 2010). Cellulose and hemicellulose are the main components of the plant cell wall. The increased relative abundance of xylanolytic bacteria can promote the decomposition of plants. This result is consistent with the promotion of litter decomposition upon N addition (Hou et al., 2021). At the same time, N addition also increased the average relative abundance of saprotrophic fungi (Figure 5B). Saprotrophic fungi play an essential role in decomposition because they can attack the lignocellulose matrix in the litter (Adl, 2003). The relative abundance of saprotrophic fungi in the N3 treatment (81.17% higher than CK) further confirmed that N addition promoted plant litter decomposition. Mucoromycota is saprotrophic fungi, and can decompose organic matter (Rashmi et al., 2019). In this study, the average relative abundance increases of Mucoromycota may be an essential factor for the increase of the relative abundance of the saprotrophic fungi in general. The relative abundance of the symbiotrophic fungi increased significantly in the N1 treatment (Figure 5B), contrary to previous studies (van Diepen et al., 2010; Cheng et al., 2020). The relative abundance of symbiotrophic fungi was positively correlated with the carbon input (Högberg et al., 2010). The content of SOC increased significantly in the N1 treatment (Table 1), promoting symbiotrophic fungal growth. N addition had no effect on aerobic ammonia oxidizing archaea (Figure 5C), which is consistent with Long et al. (2012). N addition can change the abundance of aerobic ammonia oxidizing archaea in acidic soil (Isobe et al., 2012) but had no effect in alkaline soil (Shen et al., 2008). The experimental area soil is typically alkaline, with a pH of 8.74 (Table 1). Therefore, N addition did not affect the abundance of aerobic ammonia oxidizing archaea.
Soil properties are key factors affecting the microbial community. The SEM analysis explained the bacterial and fungal community function changes (Figure 7). N addition, TN, NO3−-N, and AP can explain 68% of the changes in the bacterial community function. N addition can directly affect bacterial community function. At the same time, it can also indirectly affect bacterial community function by changing the soil properties. In this study, NO3−-N is a vital soil factor affecting bacterial community function. The change in the fungal community function is mainly due to soil properties. SOC and NH4+-N can explain 62% of the changes in the fungal community function. SOC had a significant positive effect on the fungal community function, and NH4+-N significantly negatively impacted the fungal community function. The effect of SOC on the fungal community function may primarily promote the physiological metabolism of soil microorganisms through labile organic carbon (Cleveland et al., 2007; Wang et al., 2021b). N addition changed the content of SOC (Table 1), thus affecting the fungi community function.
In conclusion, N addition in the tested desert region presented different effects on the community structures and functions of bacteria, fungi, and archaea, respectively. Long-term (6 years) of N addition did not affect bacterial α-diversity and β-diversity. The fungal richness was significantly increased in the N2 (20 kg N ha−1 yr.−1) treatment. Although N addition did not affect the archaeal α-diversity, the β-diversity was significantly reduced in the N3 (80 kg N ha−1 yr.−1) treatment. In addition, N addition affects the bacterial and fungal community structures. Soil pH was significantly correlated with the fungal and archaeal community structures at the OTU level. N addition also changed bacterial and fungal community functions, directly or indirectly through NO3−-N for bacteria and via SOC and NH4+-N for fungi. Our study mainly focused on a desert ecosystem, but the effect of N deposition is highly environment-dependent and scale-dependent. To fully elucidate the effect of N deposition, further studies are necessary to carry out research on a regional and global scale.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA863934.
XZ and MW designed the research and wrote the manuscript. XZ and XS analyzed the data. XZ, HM, TW, and LH were responsible for performing the fieldwork. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (No. 31960260), Tianshan Youth Program of Xinjiang Uygur Autonomous Region (No. 2019Q078) and the Key Research and Development Program of the Xinjiang Uygur Autonomous Region (No. 2022B02003).
We thank Ping Yue for providing us with a field experiment site. We also greatly appreciate Junhui Cheng for their assistance in measuring soil properties. In addition, we are grateful to the reviewers for their insightful comments, which helped us improve this paper. We acknowledge Charlesworth (www.cwauthors.com) for linguistic assistance with the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1015588/full#supplementary-material
1. ^http://ccb.jhu.edu/software/FLASH/
2. ^http://qiime.org/scripts/split_libraries_fastq.html
3. ^http://www.drive5.com/uparse/
Aber, J., McDowell, W., Nadelhoffer, K., Magill, A., Berntson, G., Kamakea, M., et al. (1998). Nitrogen saturation in temperate forest ecosystems: hypotheses revisited. Bioscience 48, 921–934. doi: 10.2307/1313296
Adl, S. M. (2003). The ecology of soil decomposition. CABI. (Cambridge: CABI Publishing) doi: 10.1079/97808519966150000
Allison, V. J., Condron, L. M., Peltzer, D. A., Richardson, S. J., and Turner, B. L. (2007). Changes in enzyme activities and soil microbial community composition along carbon and nutrient gradients at the Franz Josef chronosequence New Zealand. Soil Biol. and Biochem. 39, 1770–1781. doi: 10.1016/j.soilbio.2007.02.006
Austin, A. T., Yahdjian, L., Stark, J. M., Belnap, J., Porporato, A., Norton, U., et al. (2004). Water pulses and biogeochemical cycles in arid and semiarid ecosystems. Oecologia 141, 221–235. doi: 10.1007/s00442-004-1519-1
Berg, B., and Matzner, E. (1997). Effect of N deposition on decomposition of plant litter and soil organic matter in forest systems. Environ. Rev. 5, 1–25. doi: 10.1139/a96-017
Berry, V., Gole, A., Kundu, S., Murphy, C. J., and Saraf, R. F. (2005). Deposition of CTAB-terminated nanorods on bacteria to form highly conducting hybrid systems. J. Am. Chem. Soc. 127, 17600–17601. doi: 10.1021/ja056428l
Bobbink, R., Hicks, K., Galloway, J., Spranger, T., Alkemade, R., Ashmore, M., et al. (2010). Global assessment of nitrogen deposition effects on terrestrial plant diversity: a synthesis. Ecol. Appl. 20, 30–59. doi: 10.1890/08-1140.1
Bremner, J. M. (1965). Total nitrogen. Methods Soil Anal. 9, 1149–1178. doi: 10.2134/agronmonogr9.2.c32
Brooks, M. L. (2003). Effects of increased soil nitrogen on the dominance of alien annual plants in the Mojave Desert. J. Appl. Ecol. 40, 344–353. doi: 10.1046/j.1365-2664.2003.00789.x
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cheng, Y., Wang, J., Wang, J., Wang, S., Chang, S. X., Cai, Z., et al. (2020). Nitrogen deposition differentially affects soil gross nitrogen transformations in organic and mineral horizons. Earth Sci. Rev. 201:103033. doi: 10.1016/j.earscirev.2019.103033
Ciavatta, C., Govi, M., Antisari, L. V., and Sequi, P. (1991). Determination of organic carbon in aqueous extracts of soils and fertilizers. Commun. Soil Sci. Plant Anal. 22, 795–807. doi: 10.1080/00103629109368455
Cleveland, C. C., Nemergut, D. R., Schmidt, S. K., and Townsend, A. R. (2007). Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry 82, 229–240. doi: 10.1007/s10533-006-9065-z
Cui, X., Yue, P., Wu, W., Gong, Y., Li, K., Misselbrook, T., et al. (2019). The growth and N retention of two annual desert plants varied under different nitrogen deposition rates. Front. Plant Sci. 10:356. doi: 10.3389/fpls.2019.00356
Decina, S. M., Hutyra, L. R., and Templer, P. H. (2020). Hotspots of nitrogen deposition in the world's urban areas: a global data synthesis. Front. Ecol. Environ. 18, 92–100. doi: 10.1002/fee.2143
Fenn, M. E., Allen, E. B., Weiss, S. B., Jovan, S., Geiser, L. H., Tonnesen, G. S., et al. (2010). Nitrogen critical loads and management alternatives for N-impacted ecosystems in California. J. Environ. Manag. 91, 2404–2423. doi: 10.1016/j.jenvman.2010.07.034
Fenn, M. E., Haeuber, R., Tonnesen, G. S., Baron, J. S., Grossman-Clarke, S., Hope, D., et al. (2003). Nitrogen emissions, deposition, and monitoring in the Western United States. Bioscience 53, 391–403. doi: 10.1641/0006-3568(2003)053[0391:nedami]2.0.co;2
Fierer, N., Lauber, C. L., Ramirez, K. S., Zaneveld, J., Bradford, M. A., and Knight, R. (2012). Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007–1017. doi: 10.1038/ismej.2011.159
Fisher, F. M., Zak, J. C., Cunningham, G. L., and Whitford, W. G. (1988). Water and nitrogen effects on growth and allocation patterns of creosotebush in the northern Chihuahuan Desert. J. Range Manag. 41, 387–391. doi: 10.2307/3899572
Ford, H., Roberts, A., and Jones, L. (2016). Nitrogen and phosphorus co-limitation and grazing moderate nitrogen impacts on plant growth and nutrient cycling in sand dune grassland. Sci. Total Environ. 542, 203–209. doi: 10.1016/j.scitotenv.2015.10.089
Francioli, D., Schulz, E., Lentendu, G., Wubet, T., Buscot, F., and Reitz, T. (2016). Mineral vs. organic amendments: microbial community structure, activity and abundance of agriculturally relevant microbes are driven by long-term fertilization strategies. Front. Microbiol. 7:1446. doi: 10.3389/fmicb.2016.01446
Freedman, Z. B., Romanowicz, K. J., Upchurch, R. A., and Zak, D. R. (2015). Differential responses of total and active soil microbial communities to long-term experimental N deposition. Soil Biol. Biochem. 90, 275–282. doi: 10.1016/j.soilbio.2015.08.014
Freedman, Z., and Zak, D. R. (2014). Atmospheric N deposition increases bacterial laccase-like multicopper oxidases: implications for organic matter decay. Appl. Environ. Microbiol. 80, 4460–4468. doi: 10.1128/aem.01224-14
Gilliam, F. S. (2006). Response of the herbaceous layer of forest ecosystems to excess nitrogen deposition. J. Ecol. 94, 1176–1191. doi: 10.1111/j.1365-2745.2006.01155.x
Gírio, F. M., Fonseca, C., Carvalheiro, F., Duarte, L. C., Marques, S., and Bogel-Łukasik, R. (2010). Hemicelluloses for fuel ethanol: a review. Bioresour. Technol. 101, 4775–4800. doi: 10.1016/j.biortech.2010.01.088
Grant, T., Sethuraman, A., Escobar, M. A., and Vourlitis, G. L. (2022). Chronic dry nitrogen inputs alter soil microbial community composition in Southern California semi-arid shrublands. Appl. Soil Ecol. 176:104496. doi: 10.1016/j.apsoil.2022.104496
Han, W. J., Cao, J. Y., Liu, J. L., Jiang, J., and Ni, J. (2019). Impacts of nitrogen deposition on terrestrial plant diversity: a meta-analysis in China. J. Plant Ecol. 12, 1025–1033. doi: 10.1093/jpe/rtz036
He, J., Jiao, S., Tan, X., Wei, H., Ma, X., Nie, Y., et al. (2021). Adaptation of soil fungal community structure and assembly to long-versus short-term nitrogen addition in a tropical forest. Front. Microbiol. 12:689674. doi: 10.3389/fmicb.2021.689674
Högberg, M. N., Briones, M. J., Keel, S. G., Metcalfe, D. B., Campbell, C., Midwood, A. J., et al. (2010). Quantification of effects of season and nitrogen supply on tree below-ground carbon transfer to ectomycorrhizal fungi and other soil organisms in a boreal pine forest. New Phytol. 187, 485–493. doi: 10.1111/j.1469-8137.2010.03274.x
Hou, S. L., Hättenschwiler, S., Yang, J. J., Sistla, S., Wei, H. W., Zhang, Z. W., et al. (2021). Increasing rates of long-term nitrogen deposition consistently increased litter decomposition in a semi-arid grassland. New Phytol. 229, 296–307. doi: 10.1111/nph.16854
Huang, G., Li, C. H., and Li, Y. (2018). Phenological responses to nitrogen and water addition are linked to plant growth patterns in a desert herbaceous community. Ecol. Evol. 8, 5139–5152. doi: 10.1002/ece3.4001
Huang, G., Li, Y., and Su, Y. G. (2015). Divergent responses of soil microbial communities to water and nitrogen addition in a temperate desert. Geoderma 251-252, 55–64. doi: 10.1016/j.geoderma.2015.03.018
Huang, J., Xu, Y., Yu, H., Zhu, W., Wang, P., Wang, B., et al. (2021). Soil prokaryotic community shows no response to 2 years of simulated nitrogen deposition in an arid ecosystem in northwestern China. Environ. Microbiol. 23, 1222–1237. doi: 10.1111/1462-2920.15364
Humbert, J. Y., Dwyer, J. M., Andrey, A., and Arlettaz, R. (2016). Impacts of nitrogen addition on plant biodiversity in mountain grasslands depend on dose, application duration and climate: a systematic review. Glob. Chang. Biol. 22, 110–120. doi: 10.1111/gcb.12986
Isobe, K., Koba, K., Suwa, Y., Ikutani, J., Fang, Y., Yoh, M., et al. (2012). High abundance of ammonia-oxidizing archaea in acidified subtropical forest soils in southern China after long-term N deposition. FEMS Microbiol. Ecol. 80, 193–203. doi: 10.1111/j.1574-6941.2011.01294.x
Kivimäki, S. K., Sheppard, L. J., Leith, I. D., and Grace, J. (2013). Long-term enhanced nitrogen deposition increases ecosystem respiration and carbon loss from a sphagnum bog in the Scottish Borders. Environ. Exp. Bot. 90, 53–61. doi: 10.1016/j.envexpbot.2012.09.003
Ladwig, L. M., Collins, S. L., Swann, A. L., Xia, Y., Allen, M. F., and Allen, E. B. (2012). Above-and belowground responses to nitrogen addition in a Chihuahuan Desert grassland. Oecologia 169, 177–185. doi: 10.1007/s00442-011-2173-z
Laity, J. J. (2009). Deserts and Desert Environments. Vol. 3. ed. J. Wiley Sons (Chichester, UK: Wiley-Blackwell).
Lau, J. A., and Lennon, J. T. (2012). Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. 109, 14058–14062. doi: 10.1073/pnas.1202319109
Li, Y., Liu, Y., Wang, X., Luo, S., Su, D., Jiang, H., et al. (2022). Biomethanation of syngas at high CO concentration in a continuous mode. Bioresour. Technol. 346:126407. doi: 10.1016/j.biortech.2021.126407
Liu, R., Feng, X., Li, C., Ma, J., Wang, Y., and Li, Y. (2022). The importance of stem photosynthesis for two desert shrubs across different groundwater depths. Front. Plant Sci. 13:804786. doi: 10.3389/fpls.2022.804786
Liu, W., Liu, L., Yang, X., Deng, M., Wang, Z., Wang, P., et al. (2021). Long-term nitrogen input alters plant and soil bacterial, but not fungal beta diversity in a semiarid grassland. Glob. Chang. Biol. 27, 3939–3950. doi: 10.1111/gcb.15681
Liu, Y., Tan, X., Fu, S., and Shen, W. (2022). Canopy and understory nitrogen addition alters organic soil bacterial communities but not fungal communities in a temperate Forest. Front. Microbiol. 13:888121. doi: 10.3389/fmicb.2022.888121
Liu, L., Xu, W., Lu, X., Zhong, B., Guo, Y., Lu, X., et al. (2022). Exploring global changes in agricultural ammonia emissions and their contribution to nitrogen deposition since 1980. Proc. Natl. Acad. Sci. 119:e2121998119. doi: 10.1073/pnas.2121998119
Liu, X., Zhang, Y., Han, W., Tang, A., Shen, J., Cui, Z., et al. (2013). Enhanced nitrogen deposition over China. Nature 494, 459–462. doi: 10.1038/nature11917
Long, X., Chen, C., Xu, Z., Oren, R., and He, J. Z. (2012). Abundance and community structure of ammonia-oxidizing bacteria and archaea in a temperate forest ecosystem under ten-years elevated CO2. Soil Biol. Biochem. 46, 163–171. doi: 10.1016/j.soilbio.2011.12.013
Lu, J. J., Tan, D. Y., Baskin, J. M., and Baskin, C. C. (2013). Trade-offs between seed dispersal and dormancy in an amphi-basicarpic cold desert annual. Ann. Bot. 112, 1815–1827. doi: 10.1093/aob/mct240
Lu, M., Yang, Y., Luo, Y., Fang, C., Zhou, X., Chen, J., et al. (2011). Responses of ecosystem nitrogen cycle to nitrogen addition: a meta-analysis. New Phytol. 189, 1040–1050. doi: 10.1111/j.1469-8137.2010.03563.x
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Marcus-Wyner, L. (1983). Influence of ambient acidity on the absorption of NO3− and NH4+ by tomato punts. J. Plant Nutr. 6, 657–666. doi: 10.1080/01904168309363133
McHugh, T. A., Morrissey, E. M., Mueller, R. C., Gallegos-Graves, L. V., Kuske, C. R., and Reed, S. C. (2017). Bacterial, fungal, and plant communities exhibit no biomass or compositional response to two years of simulated nitrogen deposition in a semiarid grassland. Environ. Microbiol. 19, 1600–1611. doi: 10.1111/1462-2920.13678
Meng, J., Xu, J., Qin, D., He, Y., Xiao, X., and Wang, F. (2014). Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J. 8, 650–659. doi: 10.1038/ismej.2013.174
Mohn, J., Schürmann, A., Hagedorn, F., Schleppi, P., and Bachofen, R. (2000). Increased rates of denitrification in nitrogen-treated forest soils. For. Ecol. Manag. 137, 113–119. doi: 10.1016/s0378-1127(99)00320-5
Moore, J. A., Anthony, M. A., Pec, G. J., Trocha, L. K., Trzebny, A., Geyer, K. M., et al. (2021). Fungal community structure and function shifts with atmospheric nitrogen deposition. Glob. Chang. Biol. 27, 1349–1364. doi: 10.1111/gcb.15444
Mueller, R. C., Belnap, J., and Kuske, C. R. (2015). Soil bacterial and fungal community responses to nitrogen addition across soil depth and microhabitat in an arid shrubland. Front. Microbiol. 6:891. doi: 10.3389/fmicb.2015.00891
Mukhtar, H., Lin, C. M., Wunderlich, R. F., Cheng, L. C., Ko, M. C., and Lin, Y. P. (2021). Climate and land cover shape the fungal community structure in topsoil. Sci. Total Environ. 751, 141721–141729. doi: 10.1016/j.scitotenv.2020.141721
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Norby, R. J. (1998). Nitrogen deposition: a component of global change analyses. New Phytol. 139, 189–200. doi: 10.1046/j.1469-8137.1998.00183.x
Pichler, M., Coskun, Ö. K., Ortega-Arbulú, A. S., Conci, N., Wörheide, G., Vargas, S., et al. (2018). A 16S rRNA gene sequencing and analysis protocol for the Illumina MiniSeq platform. Microbiology 7:e00611. doi: 10.1002/mbo3.611
Qin, J., Li, M., Zhang, H., Liu, H., Zhao, J., and Yang, D. (2021). Nitrogen deposition reduces the diversity and abundance of cbbL gene-containing CO2-fixing microorganisms in the soil of the Stipa baicalensis steppe. Front. Microbiol. 12:570908. doi: 10.3389/fmicb.2021.570908
Ramirez, K. S., Lauber, C. L., Knight, R., Bradford, M. A., and Fierer, N. (2010). Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Ecology 91, 3463–3470. doi: 10.1890/10-0426.1
Rashmi, M., Kushveer, J. S., and Sarma, V. V. (2019). A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 10, 798–1079. doi: 10.5943/mycosphere/10/1/19
Rhoades, J. D. (1996). Salinity: electrical conductivity and total dissolved solids. Methods Soil Anal. 5, 417–435. doi: 10.2136/sssabookser5.3.c14
Sansupa, C., Wahdan, S. F. M., Hossen, S., Disayathanoowat, T., Wubet, T., and Purahong, W. (2021). Can we use functional annotation of prokaryotic taxa (FAPROTAX) to assign the ecological functions of soil bacteria? Appl. Sci. 11, 688–706. doi: 10.3390/app11020688
Sha, M., Xu, J., Zheng, Z., and Fa, K. (2021). Enhanced atmospheric nitrogen deposition triggered little change in soil microbial diversity and structure in a desert ecosystem. Glob. Ecol. Conserv. 31:e01879. doi: 10.1016/j.gecco.2021.e01879
Shen, J. P., Zhang, L. M., Zhu, Y. G., Zhang, J. B., and He, J. Z. (2008). Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ. Microbiol. 10, 1601–1611. doi: 10.1111/j.1462-2920.2008.01578.x
Shi, X., Hu, H. W., Wang, J., He, J. Z., Zheng, C., Wan, X., et al. (2018). Niche separation of comammox nitrospira and canonical ammonia oxidizers in an acidic subtropical forest soil under long-term nitrogen deposition. Soil Biol. Biochem. 126, 114–122. doi: 10.1016/j.soilbio.2018.09
Stursova, M., Crenshaw, C. L., and Sinsabaugh, R. L. (2006). Microbial responses to long-term N deposition in a semiarid grassland. Microb. Ecol. 51, 90–98. doi: 10.1007/s00248-005-5156-y
Tian, D., and Niu, S. (2015). A global analysis of soil acidification caused by nitrogen addition. Environ. Res. Lett. 10:024019. doi: 10.1088/1748-9326/10/2/024019
Tipping, E., Davies, J. A., Henrys, P. A., Jarvis, S. G., Rowe, E. C., and Smart, S. M. (2019). Measured estimates of semi-natural terrestrial NPP in Great Britain: comparison with modelled values, and dependence on atmospheric nitrogen deposition. Biogeochemistry 144, 215–227. doi: 10.1007/s10533-019-00582-5
Ullah, S., Ai, C., Ding, W., Jiang, R., Zhao, S., and Zhang, J. (2019). The response of soil fungal diversity and community composition to long-term fertilization. Appl. Soil Ecol. 140, 35–41. doi: 10.1007/s10533-019-00582-5
van Diepen, L. T., Lilleskov, E. A., Pregitzer, K. S., and Miller, R. M. (2010). Simulated nitrogen deposition causes a decline of intra-and extraradical abundance of arbuscular mycorrhizal fungi and changes in microbial community structure in northern hardwood forests. Ecosystems 13, 683–695. doi: 10.1007/s10021-010-9347-0
Wang, J., He, N., Wang, Y., Li, J., and Li, M. (2021b). Divergent drivers determine soil bacterial β-diversity of forest and grassland ecosystems in Northwest China. Glob. Ecol. Conserv. 28:e01622. doi: 10.1016/j.gecco.2021.e01622
Wang, C., Liu, D., and Bai, E. (2018). Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 120, 126–133. doi: 10.1016/j.soilbio.2018.02.003
Wang, J., Shi, X., Zheng, C., Suter, H., and Huang, Z. (2021a). Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci. Total Environ. 755:142449. doi: 10.1016/j.scitotenv.2020.1
Wardle, D. A., Yeates, G. W., Watson, R. N., and Nicholson, K. S. (1995). Development of the decomposer food-web, trophic relationships, and ecosystem properties during a three-year primary succession in sawdust. Oikos 73:155. doi: 10.2307/3545904
Yang, W. H., Ryals, R. A., Cusack, D. F., and Silver, W. L. (2017). Cross-biome assessment of gross soil nitrogen cycling in California ecosystems. Soil Biol. Biochem. 107, 144–155. doi: 10.1016/j.soilbio.2017.01.004
Ying, J., Li, X., Wang, N., Lan, Z., He, J., and Bai, Y. (2017). Contrasting effects of nitrogen forms and soil pH on ammonia oxidizing microorganisms and their responses to long-term nitrogen fertilization in a typical steppe ecosystem. Soil Biol. Biochem. 107, 10–18. doi: 10.1016/j.soilbio.2016.12.023
Yuan, X., Niu, D., Weber-Grullon, L., and Fu, H. (2020). Nitrogen deposition enhances plant-microbe interactions in a semiarid grassland: the role of soil physicochemical properties. Geoderma 373, 114446–114458. doi: 10.1016/j.geoderma.2020.114446
Yue, P., Cui, X., Gong, Y., Li, K., Goulding, K., and Liu, X. (2018). Impact of elevated precipitation, nitrogen deposition and warming on soil respiration in a temperate desert. Biogeosciences 15, 2007–2019. doi: 10.5194/bg-15-2007-2018
Yue, P., Cui, X., Gong, Y., Li, K., Goulding, K., and Liu, X. (2019). Fluxes of N2O, CH4 and soil respiration as affected by water and nitrogen addition in a temperate desert. Geoderma 337, 770–772. doi: 10.1016/j.geoderma.2018.10.020
Zhang, G., Chen, Z., Zhang, A., Chen, L., and Wu, Z. (2014). Influence of climate warming and nitrogen deposition on soil phosphorus composition and phosphorus availability in a temperate grassland China. J. Arid Land 6, 156–163. doi: 10.1007/s40333-013-0241-4
Zhang, Y., Song, L., Liu, X. J., Li, W. Q., Lü, S. H., Zheng, L. X., et al. (2012). Atmospheric organic nitrogen deposition in China. Atmos. Environ. 46, 195–204. doi: 10.1016/j.atmosenv.2011.09.080
Zhang, W., Zhao, J., Pan, F., Li, D., Chen, H., and Wang, K. (2015). Changes in nitrogen and phosphorus limitation during secondary succession in a karst region in Southwest China. Plant Soil 391, 77–91. doi: 10.1007/s11104-015-2406-8
Zhao, S., Banerjee, S., White, J. F., Liu, J. J., Zhou, N., and Tian, C. Y. (2022). High salt stress increases archaeal abundance and network connectivity in saline agricultural soils. Catena 217:106520. doi: 10.1016/j.catena.2022.106520
Zhao, X., Li, Y., Xie, Z., and Li, P. (2020). Effects of nitrogen deposition and plant litter alteration on soil respiration in a semiarid grassland. Sci. Total Environ. 740:139634. doi: 10.1016/j.scitotenv.2020.139634
Zhao, X., Yan, X., Xiong, Z., Xie, Y., Xing, G., and Shi, S. (2009). Spatial and temporal variation of inorganic nitrogen wet deposition to the Yangtze River Delta region, China. Water Air Soil Pollut. 203, 277–289. doi: 10.1007/s42832-020-0026-6
Zheng, Q., Hu, Y., Zhang, S., Noll, L., Böckle, T., Dietrich, M., et al. (2019). Soil multifunctionality is affected by the soil environment and by microbial community composition and diversity. Soil Biol. Biochem. 136:107521. doi: 10.1016/j.soilbio.2019.107521
Zheng, X., Liu, Q., Ji, X., Cao, M., Zhang, Y., and Jiang, J. (2021). How do natural soil NH4+, NO3− and N2O interact in response to nitrogen input in different climatic zones? A global meta-analysis. Eur. J. Soil Sci. 72, 2231–2245. doi: 10.1111/ejss.13131
Keywords: nitrogen deposition, soil microbial community, desert ecosystem, bacteria, fungi, archaea
Citation: Zhang X, Song X, Wang T, Huang L, Ma H, Wang M and Tan D (2022) The responses to long-term nitrogen addition of soil bacterial, fungal, and archaeal communities in a desert ecosystem. Front. Microbiol. 13:1015588. doi: 10.3389/fmicb.2022.1015588
Received: 09 August 2022; Accepted: 26 September 2022;
Published: 13 October 2022.
Edited by:
Entao Wang, Instituto Politécnico Nacional (IPN), MexicoReviewed by:
Seán Storey, University College Dublin, IrelandCopyright © 2022 Zhang, Song, Wang, Huang, Ma, Wang and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mao Wang, d2FuZ21hbzg4QDEyNi5jb20=; Dunyan Tan, dGFuZHVueWFuQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.