- 1Department of Biotechnology, Iranian Research Organization for Science and Technology (IROST), Tehran, Iran
- 2Department of Botany, Moravian Museum, Zelný trh 6, Brno, Czechia
- 3Department of Ecology, University of Kassel, Kassel, Germany
Agaric fungi are an important group of macromycetes with diverse ecological and functional properties, yet are poorly studied in many parts of the world. Here, we comprehensively analyzed 558 agaric species in Iran to reveal their resources of edible and poisonous species as well as their ecological guilds and luminescence potential. We also made a thorough survey of the antioxidant activity of the species. Phylogenetic relationships were reconstructed based on nuclear ribosomal LSU and ITS sequences. Our results reveal that agarics of Iran comprise about 189 edible, 128 poisonous, 254 soil saprotrophic, 172 ectomycorrhizal, 146 wood-inhabiting, 18 leaf/litter-inhabiting, 9 parasitic, and 19 luminescent species. Twenty percent of the Iranian agaric species possess antioxidant activity, phylogenetically distributed in four orders and 21 agaric families. About 5% of the antioxidant species can be considered strong antioxidants, many of which are also edible and could be utilized to develop functional foods. This is the first study combining phylogeny and antioxidant potential of agaric mushrooms in a large scale, and the obtained results would guide the selection of agaric taxa to be examined in the future for taxonomic revisions, biotechnological applications, and applied phylogeny studies.
Introduction
Agarics are mushroom-forming fungi also called euagarics and their hymenium is formed on gills. They belong to the subdivision Agaricomycotina, class Agaricomycetes (Moncalvo et al., 2002; Bauer et al., 2006). They produce important natural substances used in agriculture (e.g., strobilurines), medicine (e.g., pleuromutilines), and biotechnology (e.g., polysaccharides; Pointing et al., 2001; Webster and Weber, 2007; Kück et al., 2014; Hyde et al., 2019; Sandargo et al., 2019). Agaricales is the largest fungal order of agaric mushrooms comprising ca. 13,000 known species (Kirk et al., 2008). Thorough investigations of the agarics phylogeny have recently been provided by He et al. (2019) and Varga et al. (2019). Some agarics are important model organisms for research in genetics and basidiome development such as Coprinopsis cinerea and Cyclocybe cylindrica (Herzog et al., 2019). Among agarics, there are some of the most poisonous mushrooms such as Amanita phalloides, Cortinarius Subgen. Orellani, and Inosperma erubescens, frequently mixed up with edible mushrooms during culinary collecting and thus causing severe fatalities. Nevertheless, there is a large number of edible agaric mushrooms highly prized for culinary purposes such as Agaricus campestris, Coprinus comatus, Cyclocybe cylindrica, Macrolepiota procera, and the worldwide cultivated white button mushroom Agaricus bisporus. Several edible agaric species are saprotrophs and possible to cultivate, but there are also many edible species such as Russula spp. and Lactarius spp. which belong to the ectomycorrhizal ecological guild and thus not cultivable in artificial synthetic media. A number of species such as Lentinula edodes and Flammulina velutipes have culminated as functional mushrooms for developing mushroom-based functional foods and other valued mycochemicals (Chang, 1996; Cateni et al., 2022; Rodríguez-Seoane et al., 2022).
Numerous agarics have also been recognized as sources of antioxidant compounds (e.g., Ferreira et al., 2009; Asatiani et al., 2010; Guo et al., 2012; Wang and Xu, 2014; Sánchez, 2017; Islam et al., 2019; Thu et al., 2020). Antioxidant properties, or the ability to defend against and scavenge/reduce excess free radicals in biological systems, is among the important properties of living organisms and crucial for their survival (Xiao et al., 2020). Mushrooms as one of the most diverse natural antioxidant resources, have received attention in recent decades and are advantageous compared to plants because of their high diversity, fast growth, and culture possibilities (Gargano et al., 2017; Buswell, 2018).
A preliminary checklist of Iranian mushrooms appeared by Ghobad-Nejhad et al. (2020) listing 556 agaric and 29 bolete species. However, the species remain largely unexplored in terms of various important properties. Information about the edible, poisonous, and mycorrhizal agarics in Iran is principally lacking and currently, the antioxidant properties of Iranian agarics have remained largely unexplored.
Due to the lack of knowledge about the diversity of edible, poisonous, and mycorrhizal agarics in Iran, as well as their antioxidant properties, our study aimed to: (i) investigate Iranian agarics and reveal their resources of edible and poisonous species, (ii) present their ecological guilds and bioluminescence potential, and to (iii) explore the antioxidant properties of Iranian agarics and combine it with phylogenetic reconstructions. We believe our results would benefit a wide range of researchers involved in the study of agaric mushrooms.
Materials and methods
Sampling and molecular study
Taxon sampling for the molecular study was primarily done based on the list by Ghobad-Nejhad et al. (2020), supplemented by additional data in the present study. Species current names and species authorities follow Index Fungorum1 and MycoBank.2 Microscopy and morphological studies followed Ghobad-Nejhad et al. (2020). Sequences of the 28S rRNA (nLSU) and the ITS region (covering ITS1, 5.8, and ITS2) were carefully selected from GenBank, with special attention to the quality-controlled sequences (Nilsson et al., 2012) as well as to the authentic sequences obtained from Iranian specimens. For DNA extraction, we sampled more than 20 specimens and 12 samples were successfully sequenced and used in this paper. Genomic DNA was extracted from dried basidiomata using the DNA Extraction Mini Kit (FAVORGEN, Taiwan). The primers used for the amplification cycles were ITS1F/ITS4B or ITS1F/ITS4 (White et al., 1990; Gardes and Bruns, 1993) for the ITS region and LR0R/LR7 or LR0R/LR5 (Hopple and Vilgalys, 1999) for partial nLSU region. All sequences used in the phylogenetic analyses are listed in Table 1.

Table 1. Resources of agarics of Iran and their edibility (☺, edible; ☺, edible based on own observation in Iran; ☹, poisonous; ☹, poisonous based on own observation in Iran; ☺*, edible if well-cooked but poisonous if raw; X, inedible; ○, uncertain or unknown), ecological guild (♠, soil saprotroph; ☼, ectomycorrhizal; ▐, wood-inhabiting; ♣, leaf/litter-inhabiting; ◙, parasitic), luminescence, and antioxidant potential (S, strong; M, moderate; W, weak; ND, not determined; full details provided in the text).
Two concatenated datasets of nLSU + ITS were constructed, one representing the taxa belonging to the order Agaricales (dataset 1), and the other dataset for taxa of Cantharellales, Polyporales, and Russulales (dataset 2). Contumyces rosellus, the single Iranian agaric Hymenochaetales, was used as an outgroup for both datasets.
Sequences were aligned using MUSCLE (Madeira et al., 2019). To optimize the alignment, problematic columns were reduced with Noisy 1.5.12 (Dress et al., 2008) and were further identified and removed after careful visual inspection. Special attention was paid to excluding the poorly aligned columns of the ITS region and keeping the finely aligned parts. (Sequences of Amanita eliae, Mycena xantholeuca, Pluteus semibulbosus, and Tricholoma ustale were deleted from the final dataset due to poor alignment.)
Phylogenetic analyses
The sequence datasets were analyzed using Bayesian inference (BI) executed in MrBayes v. 3.2.7a (Ronquist et al., 2012). MrModeltest 2.3 was implemented to infer the best-fit model of nucleotide evolution for each alignment partition in each dataset (Nylander, 2004). Bayesian analyses were run for 40 (dataset 1) and 20 (dataset 2) million generations for four Markov chain Monte Carlo simulations, in two independent runs at the CIPRES Science Gateway (Miller et al., 2010), with the trees and parameters sampled every 5,000 generations, and the first 25% of the generations were discarded as burn-in. Posterior probabilities (PPs) were calculated from the posterior distribution of the retained trees. Maximum likelihood analyses were executed in raxmlGUI v.1.3 (Silvestro and Michalak, 2010) with the same parameters as used by Ghobad-Nejhad et al. (2021). The Bayesian phylograms were retained for tree visualizations and annotations.
Edibility, ecological guild and luminescence
The edibility rank of the species (edible, poisonous, inedible) and the ecological guilds (soil saprotrophic, ectomycorrhizal, leaf/litter-inhabiting, wood-inhabiting, parasitic) were assigned based on published literature as well as authors’ knowledge. The edibility of many species is highly subjective and evaluated differently in various countries. Here, the majority of our data are based on central and southern European literature, but even this literature was not necessarily confirmative. Therefore, for some species, more than one rank assignment was inevitably used. Besides the categories “edible” or “poisonous,” category “inedible” was also recognize (noted with symbol X in Table 1) for the species with an unpleasant taste, very small and tiny basidiomata and not usually collected for culinary purposes. Luminescence (bio/chemiluminescent) data were extracted from published literature as mentioned in Table 1 for each species.
Antioxidant properties
Antioxidant properties of the species were obtained via published references as well as own experiments performed in the present study (Tables 1, 2; Supplementary Table 1). A thorough literature survey was performed to extract and summarize the available data on the antioxidant properties of the agaric species. Published references were searched via Google Scholar, PubMed, and other standard repositories. Each literature was scrutinized carefully, avoiding poor quality and ambiguous data. Disqualified literature, unpublished data, and papers published in non-standard journals were removed from our analyses. In total, ca. 300 literature were surveyed and ca. 170 references were cited in this work and in Supplementary material. The majority of studies reported the antioxidant potential as EC50 values, i.e., half maximal effective concentration, based on DPPH (2,2-diphenyl-1-picrylhydrazyl) and ABTS (2, 2′-azino-bis-3-ethylbenzothiazoline-6-sulfonic acid) assays. To have an approximate comparison of the antioxidant potential of the species, we tentatively categorized the EC50 values as strong, moderate, and weak. For this, the EC50 values less than 1 mg/ml were considered as “strong” (S), EC50 values ranging from 1 to 10 mg/ml as “moderate” (M), and EC50 values more than 10 mg/ml were tentatively considered as “weak” (W) antioxidants (Table 1). For several species, we found different EC50 values reported in different studies. We preferred to keep the data as is for any future reference so that we assigned more than one code to classify the antioxidant potential of these species (e.g., SM standing for strong to moderate). (In a number of studies the antioxidant potential had been expressed only as radical scavenging activity% (RSA %). For these, the RSAs >80% were hesitantly considered as strong, RSA 50%–80% as moderate, and RSA < 50% were tentatively considered as weak, paying careful attention also to the values from the antioxidant standards; see Supplementary Table 1). In the cases where the antioxidant potential values were contrasting in different studies, we preferred to keep the data as is for any future reference, and therefore the antioxidant potential of the corresponding species are shown here with the combined codes SM, MW, and SMW, where applicable (Table 1).
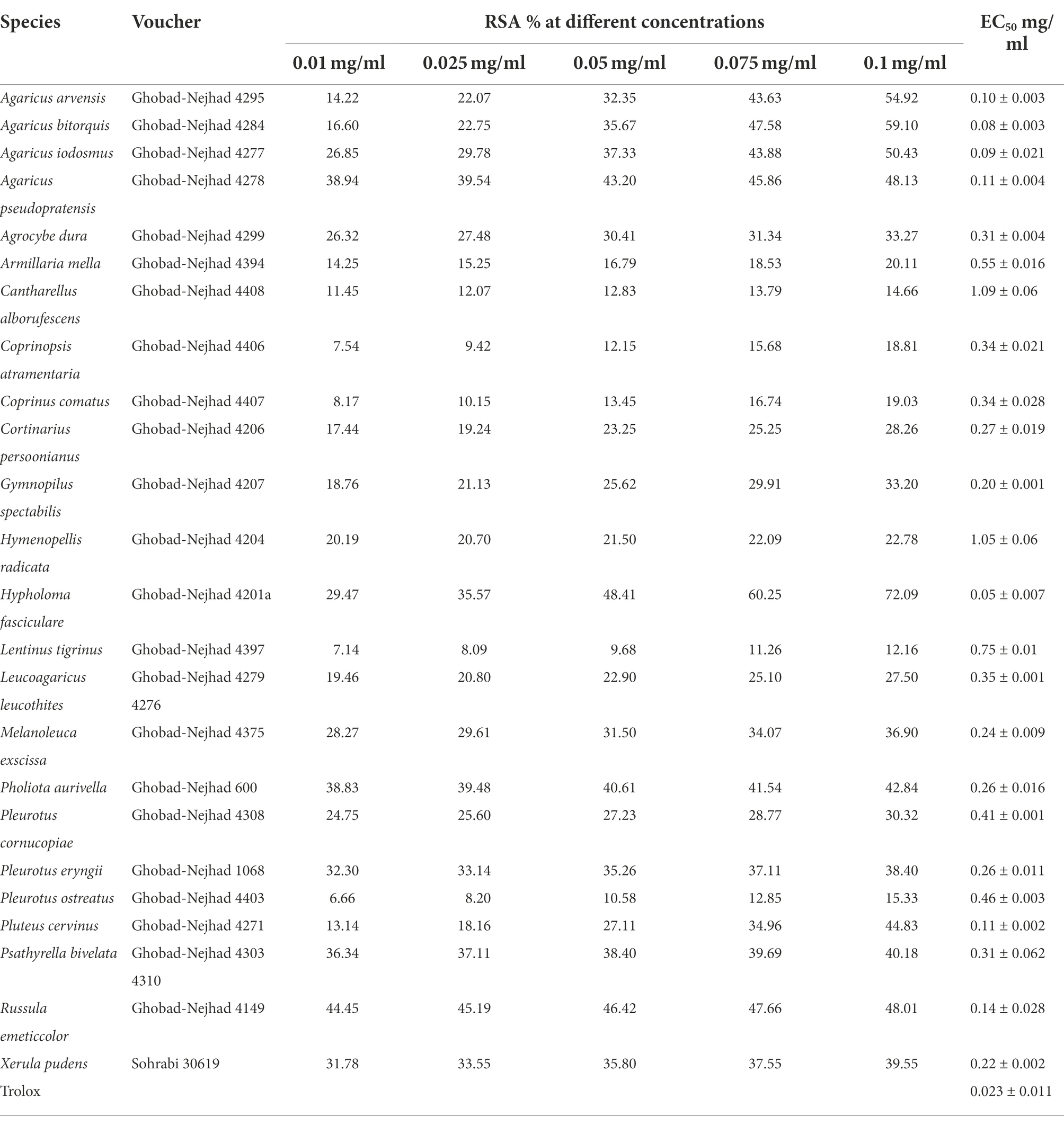
Table 2. The EC50 values and the percentage of radical scavenging activity (RSA) obtained by ABTS assays in this study.
Dried basidiomata from 24 species were sampled and examined for their antioxidant potential via ABTS assay following Re et al. (1999). Voucher samples were deposited at the Iranian Cryptogamic Herbarium (ICH) herbarium (acronym by Index Herbariorum) or at MG personal collection. The ABTS solution (7 mM) was prepared in 2.45 mM potassium sulfate and was kept at room temperature in the dark for 16 h. The mixture was then mixed with phosphate-buffered saline (PBS) as a control, and the absorbance reached 0.7 ± 0.02 at 734 nm. The extract samples with final concentrations of 0.01, 0.025, 0.05, 0.075, and 0.1 mg/ml were mixed with 980 μl of ABTS solution. The absorbance at 734 nm was measured after 6 min. The percentage of radical scavenging activity was calculated by the following equation, where A stands for absorbance (Öztürk et al., 2011):
The EC50 values were obtained through interpolation from linear regression analysis (Supplementary Figure 1). Trolox was used as a positive control at different concentrations (0.005, 0.01, 0.015, 0.02, 0.025, and 0.03 mg/ml).
Results
The results of our survey on the resources of agaric species in Iran are summarized in Table 1. Altogether, 558 agaric species from five orders were surveyed for their resources of edible and poisonous species, their ecological guilds, bioluminescence, and antioxidant potential. The two species Conocybe olivaceopileata and Inocybe ionolepis were added here to the Iranian mycota (see Table 1).
Phylogeny
The Agaricales dataset consisted of 428 taxa and 1,341 characters of which, 243 characters were constant, 144 variable, and 954 characters were informative. The best-fit evolutionary model suggested by MrModeltest was GTR + I + G for each of the LSU and ITS partitions. The Agaricales phylogram is shown in Figure 1. Nineteen families were phylogenetically retrieved with moderate to good posterior probabilities (PPs) and were shown in colored boxes, while the rest of the taxa were incertae sedis or received low to moderate branch support.
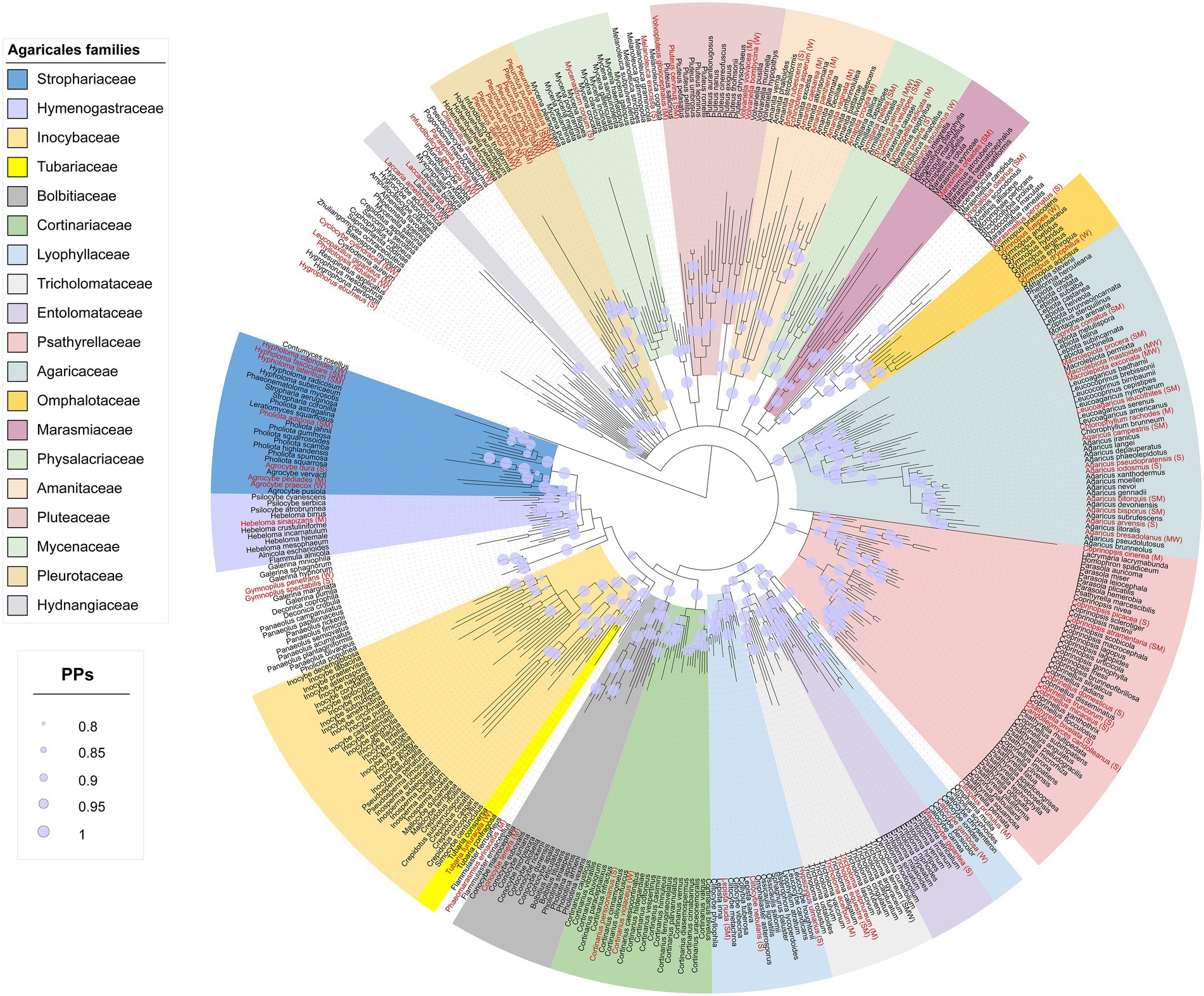
Figure 1. Phylogram from the combined nLSU + ITS sequence dataset representing the phylogenetic relationships of Iranian Agaricales. Posterior probabilities (PPs) ≥ 0.8 are shown as light lilac dots on the nodes. Terminals in red are species with antioxidant activity and the letters inside brackets are the tentative antioxidant codes: S, strong; M, moderate; W, weak (see the text for full details).
The species with antioxidant data were distributed in all the families shown in colored boxes except for the two families Entolomataceae and Inocybaceae (Figure 1). For some families such as Bolbitiaceae, Marasmiaceae, and Tubariaceae, there was only a single species with antioxidant activity, while other families such as Agaricaceae, Psathyrellaceae, and Pleurotaceae contained several antioxidant species.
Dataset 2 (Cantharellales, Polyporales, Russulales) consisted of 71 taxa and 1,528 characters of which, 279 characters were constant, 347 variable, and 902 characters were informative. The best-fit evolutionary model as suggested by MrModeltest was GTR + G for each of the LSU and ITS partitions. The phylogram obtained from the analyses of dataset 2 is presented in Figure 2. The orders Polyporales, Russulales, and Cantharellales were retrieved as moderate to well-supported monophyletic clades (PPs 0.75, 0.94, and 1.00, respectively). (For Polyporales, the two families Panaceae and Polyporaceae were not retrieved. Moreover, Panellus stipticus found a position close to the outgroup Contumyces rosellus, and we could not solve this.) The species with antioxidant data were distributed within the three orders in the phylogram (Figure 2). Out of six Cantharellales agaric members in Iran (Ghobad-Nejhad et al., 2020), four species have antioxidant data (Figure 2; Craterellus cinereus had no good LSU/ITS, so is missing in the phylogeny here). Polyporales has nine agaric species in Iran, four of which possess antioxidant activity (Figure 2). Russulales has 60 agaric species in Iran (Ghobad-Nejhad et al., 2020) from which, 16 species have antioxidant data (Figure 2; Table 1). Contumyces rosellus is the only Hymenochaetales agaric in Iran and has no antioxidant data.

Figure 2. Phylogram from the combined nLSU + ITS sequence dataset representing phylogenetic relationships of the Iranian Cantharellaeles, Russulales, and Polyporales. Posterior probabilities are shown below branches. Terminals in red are species with antioxidant activity and the letters inside brackets are the tentative antioxidant codes: S, strong; M, moderate; W, weak (see the text for full details).
Altogether, there were 50 agaric species lacking both ITS and LSU sequences and so did not appear in the phylogenetic analyses (Table 1); these species also lacked antioxidant data, except for Russula nigricans which was scored as a “strong” antioxidant species (Table 1).
Edibility, ecological guild, and luminescence
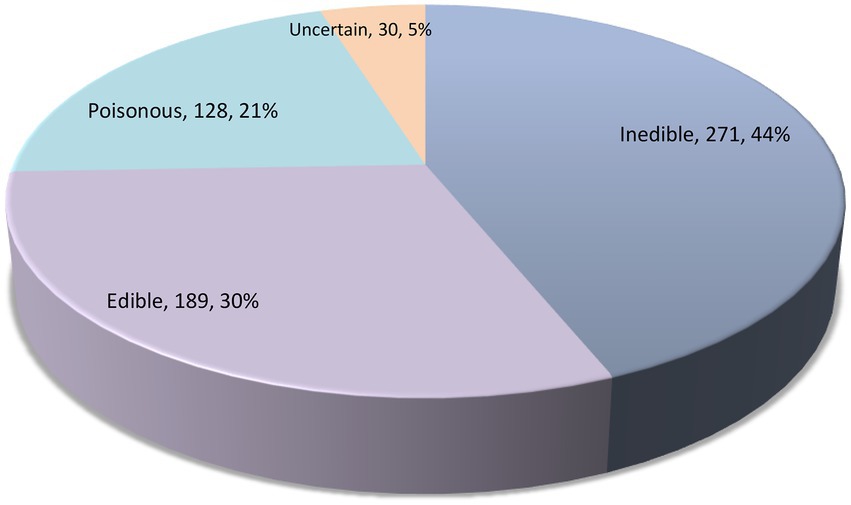
Results of the survey on edibility, ecological guilds, and luminescence of agaric species occurring in Iran are shown in Table 1 and Figures 3, 4. It is revealed that about 189 species of agarics in Iran can be classified as edible, 128 species as poisonous, and 271 species as inedible (Table 1; Figure 3). Moreover, 10 species can be assigned as edible only if well-cooked, whereas the edibility of 30 species is uncertain or unknown.
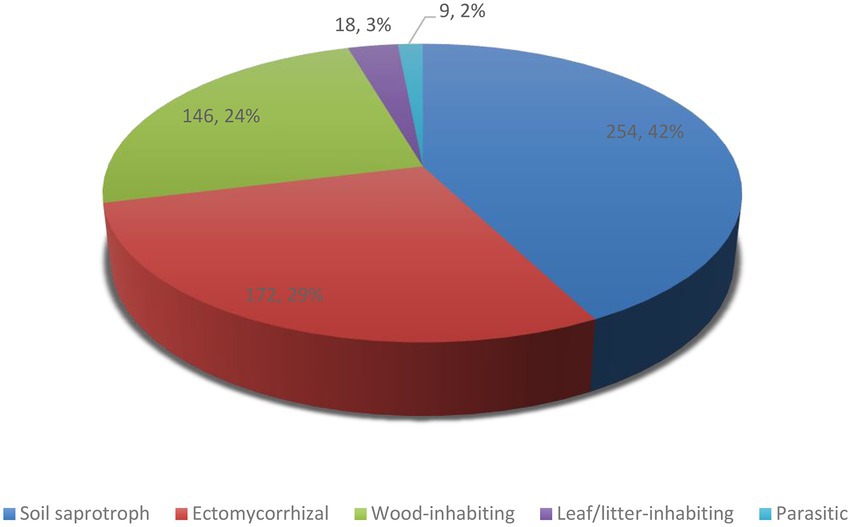
Concerning ecological guilds, our results show that about 254 species of agarics in Iran are soil saprotrophic, 172 species ectomycorrhizal, 146 species wood-inhabiting, 18 species leaf/litter-inhabiting, and nine species are parasitic (Table 1; Figure 4). Parasitic species include Armillaria borealis, A. cepistipes, A. gallica, A. mellea, Collybia tuberosa, Pleurotus eryngii, P. nebrodensis which are sapro-parasitic, Gymnopus fusipes which is wood-inhabiting parasitic, and Asterophora lycoperdoides which grows on basidiomata of Lactarius and Russula species (Table 1).
Among 558 agaric species in Iran, 19 species are categorized as luminescent (Table 1). These include Armillaria (four spp.), Collybia tuberosa, Flammulina velutipes, Mycena (six spp.), Omphalotus olearius, Panellus stipticus, and Russula (five spp.). The six species with chemiluminescence include Russula anthracina, R. cyanoxantha, R. delica, R. foetens, R. ochroleuca, as well as Panellus stipticus.
Antioxidant potential
Results of our survey on the antioxidant potential of agaric species occurring in Iran are shown in Table 1 and Figure 5 (see also Supplementary Table 1 for details on the antioxidant potential of the species and the corresponding references). According to the results, antioxidant activity data is available for 113 species phylogenetically distributed in four orders (Agaricales, Cantharellales, Russulales, Polyporales) and 21 agaric families including 17 families in the Agaricales (Strophariaceae, Hymenogastraceae, Tubariaceae, Bolbitiaceae, Cortinariaceae, Lyophyllaceae, Tricholmataceae, Psathyrellaceae, Agaricaceae, Omphalotaceae, Marasmiaceae, Physalacriaceae, Amanitaceae, Pluteaceae, Mycenaceae, Pleurotaceae, Hydnangiaceae; Figure 1), as well as Hydnaceae (=Cantharellaceae), Russulaceae, Polyporaceae, and Panaceae (Figure 2, families not shown on the tree). However, 445 species still lack information on their antioxidant potential (for a handful of species, the available antioxidant values in the literature had been expressed only by other methods such as TEAC and FRAP; as far as these cases were very few, they were not taken into account here, to keep the rest of the data comparable). The antioxidant potential of 24 species was assayed in this study and their EC50 values are reported in Table 2. Species assayed for the first time in this study included: Agaricus iodosmus, A. pseudopratensis, Agrocybe dura, Cantharellus alborufescens, Cortinarius persoonianus, Hymenopellis radicata, Melanoleuca exscissa, Psathyrella bivelata, Russula emeticolor, and Xerula pudens (Table 2; Supplementary Table 1). In general, the EC50 values of the agaric species ranged between 0.0015 mg/ml (for Psathyrella candolleana) up to 31.42 mg/ml (for Tricholoma terreum).
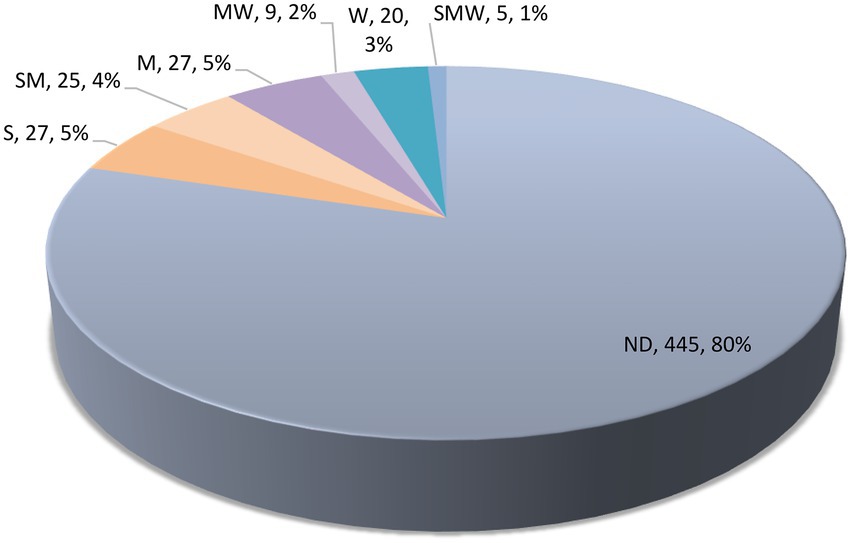
Figure 5. Categorization of antioxidant potential of Iranian agaric species. S, strong; M, moderate; W, weak; ND, not determined. See the text for full details.
Among the 113 species having antioxidant data, 27 species could roughly be classified as “strong,” 25 species as “strong to moderate,” 27 species as “moderate,” nine species as “moderate to weak,” and 20 species could be tentatively regarded as “weak” antioxidants (Figure 5). Some of the species in the S category are Agaricus arvensis, Agrocybe dura, Amanita rubescens, Candolleomyces candolleanus, Clitocybe nebularis, Coprinellus micaceus, Coprinopsis picacea, Craterellus tubaeformis, Gymnopilus spectabilis, Hygrophorus eburneus, Hypsizygus ulmarius, Macrocybe gigantean, Marasmiellus peronatus, Mycenastrum corium, Russula anthracina, Russula cyanoxantha, Russula emeticolor, Russula nigricans, Russula rosea, and Xerula pudens (Table 1).
The overall phylogenetic distribution of the agaric species with antioxidant data is shown in Figures 1, 2. Russula nigricans was the only species in our dataset with antioxidant data but lacked LSU/ITS DNA sequences in GenBank, so could not be used in our phylogenetic analyses.
Discussion
In this study, we comprehensively investigated the resources of agarics in Iran. Indeed, no published data have yet been available on number of recorded edible, poisonous, and other agarics in Iran, so the present work fills in these gaps. It is shown that there are currently about 189 edible, 128 poisonous, 254 soil saprotrophic, 172 ectomycorrhizal, 146 wood-inhabiting, 18 leaf/litter-inhabiting, 9 parasitic, and 19 luminescent agaric species in the country. The two species Conocybe olivaceopileata and Inocybe ionolepis were newly added to the Iranian mycota, new DNA sequences were obtained from Iranian samples, and the first phylogenetic reconstruction was provided for agarics of Iran. Evidently, this work is not final and therefore further studies of Iranian fungal diversity would add new species to the list presented here. About 500 agaric species belonging to the five orders Agaricales, Cantharellales, Polyporales, Russulales, and Hymenochaetales were phylogenetically analyzed based on nLSU + ITS sequence datasets. Thorough analyses with additional gene regions and vouchered samples must be utilized in the future to resolve the phylogenetic relationships of Iranian agarics. Yet, the preliminary phylogenetic analysis of agaric species presented here would help to inspire the investigation of many taxa in need of taxonomic revision. Phylogeny backbones can be used for visualization of the phylogenetic distribution of species possessing particular characteristics, herein, antioxidant potential, but also other features in the future. For instance, phylogenetic assessments have been used to screen the pleuromutilin-producing basidiomycete species (Hartley et al., 2009), fungal strains capable of degrading industrial compounds (Navarro et al., 2021), or other natural products (Adamek et al., 2019).
For a few species, the edibility assignment was based on own observation in Iran, but as stated earlier, most of the species were categorized based on available knowledge on central and southern European species. (It might be relevant to note that a number of previous studies have shown a high similarity of the Iranian mycota to that of Europe, e.g., Ghobad-Nejhad et al., 2012; Ghobad-Nejhad and Bernicchia, 2019.) Basically, edibility assignments should always be regarded with caution and it is generally recommended to avoid consuming raw or insufficiently identified mushrooms. There are still noticeable gaps in the knowledge of edible/poisonous mushrooms identification in Iran and the level of education, public awareness, and citizen science is far from medium standards. Concerning usage of edible fungi in Iran, published references are lacking, and our available data is fragmentary. In the reports and statistics on mushroom poisoning in Iran, there is no proper documentation of the species involved or at best, the species are only ambiguously characterized (Kiarsi et al., 2019).
The present work calculated as many as about 172 ectomycorrhizal agaric species for Iran. Ectomycorrhizal fungi are essential components of forest ecosystems to supply the symbiont trees with water and nutrients such as phosphorus and nitrogen, and therefore are highly important in forest sustainability (Varma and Hock, 2013). A large number of ectomycorrhizal agarics are also edible and may be harvested in the wild for culinary use, so they are in need of immediate conservation actions (Vaario and Matsushita, 2021); this is the case, especially with the Cantharellus species in northern Iran (Parad et al., 2018, 2020).
In this study, we listed 146 wood-inhabiting agaric species for Iran. There have been several studies on the diversity and taxonomy of wood-inhabiting aphyllophoroid fungi in Iran (e.g., Hallenberg, 1981; Ghobad-Nejhad and Hallenberg 2012; Amoopour et al., 2016; Ghobad-Nejhad and Langer, 2017; Nazari Mahroo et al., 2018; Ghobad-Nejhad and Bernicchia, 2019) but agarics growing on wood in Iran have not been studied systematically. Wood rotting fungi play a key role in terrestrial carbon cycling and have high potential in biotechnology, enzyme industry, biorefinery, and bioremediation of waste material and recalcitrant compounds (Gadd, 2001; Nguyen et al., 2018; Mäkelä et al., 2021). While Polyporales members are best known for their wood decomposition ability, genomic studies have revealed that several Agaricales taxa have evolved the enzymatic machinery comparable to the white-rot Polyporales (Floudas et al., 2020; Ruiz-Dueñas et al., 2020).
Another aspect surveyed in this study for the Iranian agaric species was bioluminescence. Bioluminescence, i.e., the ability of organisms to emit visible light, has been developed independently in the evolution of different organisms. Concerning fungi, 109 fungal taxa are known to exhibit bioluminescence all of which (except one Xylariales) are white-spored saprotrophic Basidiomycota distinguished in four phylogenetic lineages (Chew et al., 2015; Ke and Tsai, 2022) all sharing the same type of luciferin and luciferase (Oliveira et al., 2012). Interestingly, it has been shown that luminescence could be linked to the antioxidant/radical scavenging defense mechanism against some environmental stress factors (Vydryakova and Bissett, 2016; Oba et al., 2017). Moreover, the fungal bioluminescence capacity can be used in environmental biomonitoring of metals or organic compounds and to develop toxicity tests (Ke and Tsai, 2022).
In this work, a thorough survey was done to reveal the antioxidant potential of 558 agaric species and a new approach was used to combine antioxidant data with phylogeny of the species. Ten species were subjected to antioxidant analyses for the first time, belonging to the genera Agaricus, Agrocybe, Cantharellus, Cortinarius, Hymenopellis, Melanoleuca, Psathyrella, Russula, and Xerula. ABTS assay is one of the most frequently used method for quantification of antioxidant activity of mushrooms. Numerous antioxidant assays have been introduced which are usually classified into two groups based on the mechanism of action: single electron transfer and hydrogen atom transfer (Tan and Lim, 2015; Xiao et al., 2020). Compared to other methods, ABTS has the advantage of involving more or less both mechanisms (Prior et al., 2005). Yet, more examinations are required to fully investigate the antioxidant capacity of the species studied here, and to quantify and characterize the underlying bioactive compounds. Here, we could resume antioxidant data for 20% of agaric species (113 spp.), but noted that 80% of the species (445 spp.) have no antioxidant data. This is noteworthy compared to the fact that antioxidant tests are among the most popular bioactivity assays and it may show that macrofungi have remained little studied in this regard. The highest antioxidant capacities (the lowest EC50 values) were shown by the species categorized as S (27 spp.) and then as SM (25 spp.; Table 1; Figure 5). As noted earliers, in several cases, various EC50 values had been reported in different studies for some species, so that we assigned more than one code for them. We emphasize that such classification is approximate and for detailed comparisons, more precise methods are recommended to be applied. For five species, the EC50 measures ranged significantly, in a way that the code assignment could only be expressed as SMW: Pleurotus djamor, P. eryngii, P. ostreatus, P. pulmonarius, and Russula virescens. Of course, differences in the solvents, standards, modifications in the assays procedures, and even identification issues can account for the different measures under the same species name. Ideally, the identity of the voucher specimens should be fully characterized and the species should be assayed with exactly the same procedure so as to be able to have the best quality comparisons. In general, for the studies where both ABTS and DPPH assays had been conducted, ABTS values seemed to slightly outperform the DPPH values, showing lower EC50 measures. Many of the species in the S or “strong” antioxidant category are edible: Agaricus arvensis, Agrocybe dura, Amanita rubescens, Candolleomyces candolleanus, Clitocybe nebularis, Craterellus tubaeformis, Hygrophorus eburneus, Hypsizygus ulmarius, Macrocybe gigantea, Russula anthracina, R. cyanoxantha, R. emeticolor, R. nigricans, R. rosea, and Xerula pudens (Table 1). Oxidative stress is the root of a cascade of numerous acute and chronic human diseases (Kosanic et al., 2013). Diets rich in natural antioxidants enforce the native defense system and protect against oxidative damage (Ferreira et al., 2009). Mushroom species that are edible and possess high level of biological activities with perspectives on promoting human health are considered noteworthy candidates for developing functional foods and nutra-pharmaceutical products (Kozarski et al., 2015; Lu et al., 2020; Niego et al., 2021; Shaffique et al., 2021; El Sheikha, 2022). It is evident that thorough analyses are needed to fully characterize the mycochemical constitutes of such species and their various bioactivities.
Our results pave the avenue for advanced studies on edible, poisonous, saprotrophic, ectomycorrhizal, wood-inhabiting, parasitic, luminescent, and antioxidant species of agarics of Iran. Twenty percent of the Iranian agaric species possess antioxidant activity, phylogenetically distributed in four orders and 21 agaric families. About 5% of the antioxidant species can be considered strong antioxidants, many of which are also edible and could be utilized for the development of functional foods. Various edible agaric species are grown commercially in the world, while only 1–2 are commonly grown in Iran (personal comm.). Ectomycorrhizal and wood-inhabiting species are important components of forest sustainability. Forests in Iran are very scanty, comprising less than 10% of the total country area, and are on the verge of severe depletion due to numerous anthropological and environmental threats. Yet, Iranian old-growth forests, categorized as part of the northern hemisphere glacial refugia (Ghobad-Nejhad et al., 2012, 2020), harbor a rich reservoir of agaric fungi with diverse characteristics and beneficial aspects. Resources of Iranian agarics provide valuable opportunities for biotechnology and mycochemistry, and should be regarded for preservation and habitat conservation. Our preliminary phylogenetic trees would guide the selection of agaric taxa to be examined in the future for taxonomic revisions, biotechnological applications, and applied phylogeny studies. The thorough survey of antioxidant data of 558 agaric species would provide the state of the knowledge on agarics examined so far and the remaining gaps to be filled in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
MG-N conceptualized and designed the study, performed the molecular study and provided the first draft. VA contributed to the trait assignments. MG-N, VA, and EL wrote the manuscript. MG-N and MM performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Center for International Scientific Studies & Collaboration (CISSC), Ministry of Science, Research, and Technology of Iran. The studies of VA were enabled by the support provided to the Moravian Museum by the Ministry of Culture of the Czech Republic as part of its long-term conceptual development program for research institutions (DKRVO, ref. MK000094862).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1015440/full#supplementary-material
Footnotes
References
Adamek, M., Alanjary, M., and Ziemert, N. (2019). Applied evolution: phylogeny-based approaches in natural products research. Nat. Prod. Rep. 36, 1295–1312. doi: 10.1039/C9NP00027E
Amoopour, M., Ghobad-Nejhad, M., and Khodaparast, S. A. (2016). New records of polypores from Iran, with a checklist of polypores for Gilan Province. Czech Mycol. 68, 139–148. doi: 10.33585/cmy.68203
Antonín, V., Ďuriška, O., Jančovičová, S., Para, R., Kudláček, T., and Tomšovský, M. (2022). Multilocus phylogeny and taxonomy of European Melanoleuca subgenus Melanoleuca. Mycologia 114, 114–143. doi: 10.1080/00275514.2021.1966246
Asatiani, M. D., Elisashvili, V., Songulashvili, G., Reznick, A. Z., and Wasser, S. P. (2010). “Higher basidiomycetes mushrooms as a source of antioxidants” in Progress in Mycology. eds. M. Rai and G. Kövics (Springer: Dordrecht), 311–326.
Aslim, B., and Ozturk, S. (2011). Phenolic composition and antimicrobial and antioxidant activities of Leucoagaricus leucothites (Vittad.). Wasser. J. Med. Food 14, 1419–1424. doi: 10.1089/jmf.2010.0259
Bauer, R., Begerow, D., Sampaio, J. P., Weiss, M., and Oberwinkler, F. (2006). The simple-septate basidiomycetes: a synopsis. Mycol. Prog. 5, 41–66. doi: 10.1007/s11557-006-0502-0
Bermudes, D., Petersen, R. H., and Nealson, K. H. (1992). Low-level bioluminescence detected in Mycena haematopus basidiocarps. Mycologia 84, 799–802. doi: 10.2307/3760392
Bondar, V. S., Shimomura, O., and Gitelson, J. I. (2012). Luminescence of higher mushrooms. J. Siberian Univ. Biol. 5, 331–351. doi: 10.17516/1997-1389-0127
Buswell, J. A. (2018). “Mushroom-mediated protection from oxidative damage to DNA” in Biol. Macrofungi. eds. B. Singh, Lallawmsanga, and A. Passari (Cham: Springer), 115–127.
Cateni, F., Gargano, M. L., Procida, G., Venturella, G., Cirlincione, F., and Ferraro, V. (2022). Mycochemicals in wild and cultivated mushrooms: nutrition and health. Phytochem. Rev. 21, 339–383. doi: 10.1007/s11101-021-09748-2
Chang, R. (1996). Functional properties of edible mushrooms. Nutr. Rev. 54, S91–S93. doi: 10.1111/j.1753-4887.1996.tb03825.x
Chew, A. L. C., Desjardin, D. E., Tan, Y. S., Musa, M. Y., and Sabaratnam, V. (2015). Bioluminescent fungi from peninsular Malaysia – a taxonomic and phylogenetic overview. Fung. Divers. 70, 149–187. doi: 10.1007/s13225-014-0302-9
Crous, P. W., Cowan, D. A., Maggs-Kölling, G., Yilmaz, N., Larsson, E., Angelini, C., et al. (2020). Fungal planet description sheets: 1112–1181. Persoonia 45, 251–409. doi: 10.3767/persoonia.2020.45.10
Desjardin, D. E., Oliveira, A. G., and Stevani, C. V. (2008). Fungi bioluminescence revisited. Photochem. Photobiol. Sci. 7, 170–182. doi: 10.1039/b713328f
Dogan, H. H., and Aydin, S. (2013). Some biological activities of Lactarius vellereus (Fr.) Fr. In Turkey. Pakistan. J. Biol. Sci. 16, 1279–1286. doi: 10.3923/pjbs.2013.1279.1286
Dress, A. W., Flamm, C., Fritzsch, G., Grünewald, S., Kruspe, M., Prohaska, S. J., et al. (2008). Noisy: identification of problematic columns in multiple sequence alignments. Algorithms Mol. Biol. 3, 1–10. doi: 10.1186/1748-7188-3-7
El Sheikha, A. F. (2022). Nutritional profile and health benefits of Ganoderma lucidum “Lingzhi, Reishi, or Mannentake” as functional foods: current scenario and future perspectives. Foods 11:1030. doi: 10.3390/foods11071030
Ferreira, I. C., Barros, L., and Abreu, R. (2009). Antioxidants in wild mushrooms. Curr. Med. Chem. 16, 1543–1560. doi: 10.2174/092986709787909587
Floudas, D., Bentzer, J., Ahrén, D., Johansson, T., Persson, P., and Tunlid, A. (2020). Uncovering the hidden diversity of litter-decomposition mechanisms in mushroom-forming fungi. ISME J. 14, 2046–2059. doi: 10.1038/s41396-020-0667-6
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Molec. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Gargano, M. L., van Griensven, L. J., Isikhuemhen, O. S., Lindequist, U., Venturella, G., Wasser, S. P., et al. (2017). Medicinal mushrooms: valuable biological resources of high exploitation potential. Plant Biosyst. 151, 548–565. doi: 10.1080/11263504.2017.1301590
Ghobad-Nejhad, M., and Bernicchia, A. (2019). An outlook on the diversity of polypores shared between Iran and the Mediterranean area. Mycol. Iran. 6, 33–39. doi: 10.22043/MI.2020.121099
Ghobad-Nejhad, M., and Hallenberg, N. (2012). Checklist of Iranian non-gilled/non-gasteroid hymenomycetes (Agaricomycotina). Mycotaxon 119, 1–41. doi: 10.5248/119.493
Ghobad-Nejhad, M., Hallenberg, N., Hyvönen, J., and Yurchenko, E. (2012). The Caucasian corticioid fungi: level of endemism, similarity, and possible contribution to European fungal diversity. Fung. Divers. 52, 35–48. doi: 10.1007/s13225-011-0122-0
Ghobad-Nejhad, M., and Langer, E. (2017). First inventory of aphyllophoroid basidiomycetes of Zagros forests, W Iran. Plant Biosyst. 151, 844–854. doi: 10.1080/11263504.2016.1211199
Ghobad-Nejhad, M., Langer, E., Antonín, V., Gates, G., Noroozi, J., and Zare, R. (2020). The gilled fungi and boletes of Iran: diversity, systematics, and nrDNA data. Mycol. Iran. 7, 1–43. doi: 10.22043/MI.2021.123456
Ghobad-Nejhad, M., Langer, E., Nakasone, K., Diederich, P., Nilsson, R. H., Rajchenberg, M., et al. (2021). Digging up the roots: taxonomic and phylogenetic disentanglements in Corticiaceae s.s. (Corticiales, Basidiomycota) and evolution of nutritional modes. Front. Microbiol. 12:704802. doi: 10.3389/fmicb.2021.704802
Gitelson, J. I., Bondar, V. S., Medvedeva, S. E., Rodicheva, E. K., and Vydryakova, G. A. (2012). Chemiluminescent emission of tissues of fruit bodies of higher fungi. Dokl. Biochem. Biophys. 443, 105–108. doi: 10.1134/S1607672912020123
Guo, Y. J., Deng, G. F., Xu, X. R., Wu, S., Li, S., Xia, E. Q., et al. (2012). Antioxidant capacities, phenolic compounds, and polysaccharide contents of 49 edible macrofungi. Food Func. 3, 1195–1205. doi: 10.1039/c2fo30110e
Hallenberg, N. (1981). Synopsis of wood-inhabiting Aphyllophorales (basidiomycetes) and heterobasidiomycetes from N. Iran. Mycotaxon 12, 473–502.
Hartley, A. J., de Mattos-Shipley, K., Collins, C. M., Kilaru, S., Foster, G. D., and Bailey, A. M. (2009). Investigating pleuromutilin-producing Clitopilus species and related basidiomycetes. FEMS Microb. L. 297, 24–30. doi: 10.1111/j.1574-6968.2009.01656.x
He, M. Q., Zhao, R. L., Hyde, K. D., Begerow, D., Kemler, M., Yurkov, A., et al. (2019). Notes, outline and divergence times of Basidiomycota. Fung. Divers. 99, 105–367. doi: 10.1007/s13225-019-00435-4
Heleno, S. A., Barros, L., Martins, A., Queiroz, M. J., Santos-Buelga, C., and Ferreira, I. C. (2012). Phenolic, polysaccharidic, and lipidic fractions of mushrooms from northeastern Portugal: chemical compounds with antioxidant properties. J. Agric. Food Chem. 60, 4634–4640. doi: 10.1021/jf300739m
Herzog, R., Solovyeva, I., Bölker, M., Lugones, L. G., and Hennicke, F. (2019). Exploring molecular tools for transformation and gene expression in the cultivated edible mushroom Agrocybe aegerita. Mol. Gen. Genomics. 294, 663–677. doi: 10.1007/s00438-018-01528-6
Hopple, J. S. Jr., and Vilgalys, R. (1999). Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Molec. Phylogen. Evol. 13, 1–19. doi: 10.1006/mpev.1999.0634
Hyde, K. D., Xu, J., Rapior, S., Jeewon, R., Lumyong, S., Niego, A. G. T., et al. (2019). The amazing potential of fungi, 50 ways we can exploit fungi industrially. Fung. Divers. 97, 1–136. doi: 10.1007/s13225-019-00430-9
Islam, T., Ganesan, K., and Xu, B. B. (2019). New insight into mycochemical profiles and antioxidant potential of edible and medicinal mushrooms: a review. Int. J. Med. Mushrooms 21, 237–251. doi: 10.1615/IntJMedMushrooms.2019030079
Kaewnarin, K., Suwannarach, N., Kumla, J., and Lumyong, S. (2016). Phenolic profile of various wild edible mushroom extracts from Thailand and their antioxidant properties, anti-tyrosinase and hyperglycaemic inhibitory activities. J. Funct. Foods 27, 352–364. doi: 10.1016/j.jff.2016.09.008
Ke, H. M., and Tsai, I. J. (2022). Understanding and using fungal bioluminescence–recent progress and future perspectives. Curr. Opin. Green Sust. Chem. 33:100570. doi: 10.1016/j.cogsc.2021.100570
Kiarsi, M., Ostadtaghizadeh, A., Aghababaeian, H., Khaleghy, M., and Araghi, L. (2019). Mushroom poisoning of 1151 people in Iran, the lessons learnt: a brief report of cases and the literature review. Iran Red Crescent Med J 21:e91977. doi: 10.5812/ircmj.91977
Kirk, P. M., Cannon, P. F., Minter, D. W., and Stalpers, J. A. (2008). Dictionary of the Fungi, 10th ed.; CABI: Wallingford, p. 12.
Kosanic, M., Rankovic, B., and Dasic, M. (2013). Antioxidant and antimicrobial properties of mushrooms. Bulg. J. Agric. Sci. 19, 1040–1046.
Kotlobay, A. A., Sarkisyan, K. S., Mokrushina, Y. A., Marcet-Houben, M., Serebrovskaya, E. O., and Markina, N. M. (2018). Genetically encodable bioluminescent system from fungi. Proc. Natl. Acad. Sci. 115, 12728–12732. doi: 10.1073/pnas.1803615115
Kozarski, M., Klaus, A., Jakovljevic, D., Todorovic, N., Vunduk, J., Petrović, P., et al. (2015). Antioxidants of edible mushrooms. Molecules 20, 19489–19525. doi: 10.3390/molecules201019489
Kück, U., Bloemendal, S., and Teichert, I. (2014). Putting fungi to work: harvesting a cornucopia of drugs, toxins, and antibiotics. PLoS Pathog. 10:e1003950. doi: 10.1371/journal.ppat.1003950
Lu, H., Lou, H., Hu, J., Liu, Z., and Chen, Q. (2020). Macrofungi: a review of cultivation strategies, bioactivity, and application of mushrooms. Compr. Rev. Food Sci. Food Saf. 19, 2333–2356. doi: 10.1111/1541-4337.12602
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Mäkelä, M. R., Hildén, K. S., and Kuuskeri, J. (2021). Fungal lignin-modifying peroxidases and H2O2-producing enzymes. Encycl. Mycol. 2, 247–259. doi: 10.1016/B978-0-12-809633-8.21127-8
Malakauskienė, A. (2018). Reported and potential bioluminescent species in Lithuania. Biologija 64, 181–190. doi: 10.6001/biologija.v64i3.3823
Mihail, J. D. (2015). Bioluminescence patterns among north American Armillaria species. Fung. Biol. 119, 528–537. doi: 10.1016/j.funbio.2015.02.004
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, LA, 1–8.
Moncalvo, J. M., Vilgalys, R., Redhead, S. A., Johnson, J. E., James, T. Y., Catherine Aime, M., et al. (2002). One hundred and seventeen clades of euagarics. Mol. Phyl. Evol. 23, 357–400. doi: 10.1016/S1055-7903(02)00027
Navarro, D., Chaduli, D., Taussac, S., Lesage-Meessen, L., Grisel, S., Haon, M., et al. (2021). Large-scale phenotyping of 1,000 fungal strains for the degradation of non-natural, industrial compounds. Comm. Biol. 4, 1–10. doi: 10.1038/s42003-021-02401-w
Nazari Mahroo, S., Ghobad-Nejhad, M., and Khodaparast, S. A. (2018). A survey on Peniophora (Russulales, Basidiomycota) species in Iran. Nova Hedwig. 107, 257–270. doi: 10.1127/nova_hedwigia/2018/0468
Nguyen, K. A., Wikee, S., and Lumyong, S. (2018). Brief review: lignocellulolytic enzymes from polypores for efficient utilization of biomass. Mycosphere 9, 1073–1088. doi: 10.5943/mycosphere/9/6/2
Niego, A. G., Rapior, S., Thongklang, N., Raspé, O., Jaidee, W., Lumyong, S., et al. (2021). Macrofungi as a nutraceutical source: promising bioactive compounds and market value. J. Fungi 7:397. doi: 10.3390/jof7050397
Nilsson, R. H., Tedersoo, L., Abarenkov, K., Ryberg, M., Kristiansson, E., and Hartmann, M. (2012). Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. MycoKeys 4, 37–63. doi: 10.3897/mycokeys.4.3606
Nylander, J. A. A. (2004). MrModeltest v2. Program Distributed by the Author Sweden: Evolutionary Biology Centre, Uppsala University.
Oba, Y., Suzuki, Y., Martins, G. N., Carvalho, R. P., Pereira, T. A., Waldenmaier, H. E., et al. (2017). Identification of hispidin as a bioluminescent active compound and its recycling biosynthesis in the luminous fungal fruiting body. Photochem. Photobiol. Sci. 16, 1435–1440. doi: 10.1039/C7PP00216E
Oliveira, A. G., Desjardin, D. E., Perry, B. A., and Stevani, C. V. (2012). Evidence that a single bioluminescent system is shared by all known bioluminescent fungal lineages. Photochem. Photobiol. Sci. 11, 848–852. doi: 10.1039/c2pp25032b
Öztürk, M., Duru, M. E., Kivrak, Ş., Mercan-Doğan, N., Türkoglu, A., and Özler, M. A. (2011). In vitro antioxidant, anticholinesterase and antimicrobial activity studies on three Agaricus species with fatty acid compositions and iron contents: a comparative study on the three most edible mushrooms. Food Chem. Toxic. 49, 1353–1360. doi: 10.1016/j.fct.2011.03.019
Parad, G. A., Ghobad-Nejhad, M., Tabari, M., Yousefzadeh, H., Esmaeilzadeh, O., Tedersoo, L., et al. (2018). Cantharellus alborufescens and C. ferruginascens (Cantharellaceae, Basidiomycota) new to Iran. Cryptogam. Mycol. 39, 299–310. doi: 10.7872/crym/v39.iss3.2018.299
Parad, G. A., Tabari, M., Ghobad-Nejhad, M., Esmailzadeh, O., and Yousefzadeh, H. (2020). Environmental factors affecting the presence of edible Zarde-Kija mushroom (Cantharellus alborufescens) in plain forest of Noor (Mazandaran). Iran. J. Forest 12, 1–15.
Pointing, S. B., Pelling, A. L., Smith, G. J. D., Hyde, K. D., and Reddy, C. A. (2001). Screening of basidiomycetes and xylariaceous fungi for lignin peroxidase and laccase gene-specific sequences. Mycol. Res. 109, 115–124. doi: 10.1017/S0953756204001376
Prior, R. L., Wu, X., and Schaich, K. (2005). Standardized methods for the determination of antioxidant capacity and phenolics in foods and dietary supplements. J. Agric. Food Chem. 53, 4290–4302. doi: 10.1021/jf0502698
Re, R., Pellegrini, N., Proteggente, A., Pannala, A., Yang, M., and Rice-Evans, C. (1999). Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 26, 1231–1237. doi: 10.1016/S0891-5849(98)00315-3
Rodríguez-Seoane, P., Torres Perez, M. D., Fernández de Ana, C., Sinde-Stompel, E., and Domínguez, H. (2022). Antiradical and functional properties of subcritical water extracts from edible mushrooms and from commercial counterparts. Int. J. Food Sci. Technol. 57, 1420–1428. doi: 10.1111/ijfs.15383
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Ruiz-Dueñas, F. J., Barrasa, J. M., Sánchez-García, M., Camarero, S., Miyauchi, S., Serrano, A., et al. (2020). Genomic analysis enlightens Agaricales lifestyle evolution and increasing peroxidase diversity. Mol. Biol. Evol. 38, 1428–1446. doi: 10.1093/molbev/msaa301
Sánchez, C. (2017). Reactive oxygen species and antioxidant properties from mushrooms. Synth. Syst. Biotechnol. 2, 13–22. doi: 10.1016/j.synbio.2016.12.001
Sandargo, B., Chepkirui, C., Cheng, T., Chaverra-Muñoz, L., Thongbai, B., Stadler, M., et al. (2019). Biological and chemical diversity go hand in hand: basidiomycota as source of new pharmaceuticals and agrochemicals. Biotechnol. Adv. 37:107344. doi: 10.1016/j.biotechadv.2019.01.011
Shaffique, S., Kang, S. M., Kim, A. Y., Imran, M., Aaqil Khan, M., and Lee, I. J. (2021). Current knowledge of medicinal mushrooms related to antioxidant properties. Sustainability 13:7948. doi: 10.3390/su13147948
Shimomura, O. (1991). Superoxide-triggered chemiluminescence of extract of luminous mushroom Panellus stipticus after treatment with methylamine. J. Exp. Bot. 42, 555–560. doi: 10.1093/jxb/42.4.555
Silvestro, D., and Michalak, I. (2010). raxmlGUI: a graphical front-end for RAxML. Available at: http://sourceforge.net/projects/raxmlgui/ (Accessed July 15, 2022).
Tan, J. B., and Lim, Y. Y. (2015). Critical analysis of current methods for assessing the in vitro antioxidant and antibacterial activity of plant extracts. Food Chem. 172, 814–822. doi: 10.1016/j.foodchem.2014.09.141
Thu, Z. M., Myo, K. K., Aung, H. T., Clericuzio, M., Armijos, C., and Vidari, G. (2020). Bioactive phytochemical constituents of wild edible mushrooms from Southeast Asia. Molecules 25:1972. doi: 10.3390/molecules25081972
Treu, R., and Agerer, R. (1990). Culture characteristics of some Mycena species. Mycotaxon 38, 279–309.
Vaario, L. M., and Matsushita, N. (2021). Conservation of edible ectomycorrhizal mushrooms: understanding of the ECM fungi mediated carbon and nitrogen movement within forest ecosystems. Nitrogen Agric.-Physiol. Agric. Ecol. Aspects. doi: 10.5772/intechopen.95399
Varga, T., Krizsán, K., Földi, C., Dima, B., Sánchez-García, M., Sánchez-Ramírez, S., et al. (2019). Megaphylogeny resolves global patterns of mushroom evolution. Nat. Ecol. Evol. 3, 668–678. doi: 10.1038/s41559-019-0834-1
Varma, A., and Hock, B. (2013). Mycorrhiza: Structure, Function, Molecular Biology and Biotechnology. Germany: Springer Science & Business Media.
Vydryakova, G. A., and Bissett, J. (2016). Differential regulation of proteins and a possible role for manganese superoxide dismutase in bioluminescence of Panellus stipticus revealed by suppression subtractive hybridization. Adv. Microbiol. 06, 613–626. doi: 10.4236/aim.2016.69061
Wang, Y., and Xu, B. (2014). Distribution of antioxidant activities and total phenolic contents in acetone, ethanol, water and hot water extracts from 20 edible mushrooms via sequential extraction. Austin J. Nutr. Food Sci. 2:5.
Webster, J., and Weber, R. W. S. (2007). Introduction to Fungi ; Cambridge University Press: Cambridge.
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics” in PCR Protocols: A Guide to Methods and Applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, NY: Academic Press), 315–322.
Wu, F., Zhou, L. W., Yang, Z. L., Bau, T., Li, T. H., and Dai, Y. C. (2019). Resource diversity of Chinese macrofungi: edible, medicinal and poisonous species. Fung. Divers. 98, 1–76. doi: 10.1007/s13225-019-00432-7
Keywords: basidiomycetes, diversity, gilled mushrooms, ABTS assay, phylogeny
Citation: Ghobad-Nejhad M, Antonín V, Moghaddam M and Langer E (2022) Resources of Iranian agarics (Basidiomycota) with an outlook on their antioxidant potential. Front. Microbiol. 13:1015440. doi: 10.3389/fmicb.2022.1015440
Edited by:
Ludmila Chistoserdova, University of Washington, United StatesReviewed by:
Vidushi Neergheen, University of Mauritius, MauritiusIvan Milenkovic, University of Belgrade, Serbia
Copyright © 2022 Ghobad-Nejhad, Antonín, Moghaddam and Langer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masoomeh Ghobad-Nejhad, Z2hvYmFkbmVqaGFkQGdtYWlsLmNvbQ==; Z2hvYmFkbmVqaGFkQGlyb3N0Lmly; Ewald Langer, ZWxhbmdlckB1bmkta2Fzc2VsLmRl
†ORCID: Masoomeh Ghobad-Nejhad http://orcid.org/0000-0002-7807-4187
Vladimír Antonín http://orcid.org/0000-0002-6000-7285
Mohaddeseh Moghaddam http://orcid.org/0000-0002-7382-6044
Ewald Langer http://orcid.org/0000-0003-3480-2936
 Masoomeh Ghobad-Nejhad
Masoomeh Ghobad-Nejhad Vladimír Antonín2†
Vladimír Antonín2†