- Guangdong Provincial Key Laboratory of Protein Function and Regulation in Agricultural Organisms, College of Life Sciences, South China Agricultural University, Guangzhou, Guangdong, China
The fatty acid synthesis (FAS) pathway is essential for bacterial survival. Acyl carrier proteins (ACPs), donors of acyl moieties, play a central role in FAS and are considered potential targets for the development of antibacterial agents. Ralstonia solanacearum, a primary phytopathogenic bacterium, causes bacterial wilt in more than 200 plant species. The genome of R. solanacearum contains five annotated acp genes, acpP1, acpP2, acpP3, acpP4, and acpP5. In this study, we characterized the five putative ACPs and confirmed that only AcpP1 is involved in FAS and is necessary for the growth of R. solanacearum. We also found that AcpP2 and AcpP4 participate in the polyketide synthesis pathway. Unexpectedly, the disruption of four acp genes (acpP2, acpP3, acpP4, and acpP5) allowed the mutant strain to grow as well as the wild-type strain, but attenuated the bacterium’s pathogenicity in the host plant tomato, suggesting that these four ACPs contribute to the virulence of R. solanacearum through mechanisms other than the FAS pathway.
Introduction
The acyl carrier proteins (ACPs) are universal, small, but flexible proteins (Prescott and Vagelos, 1972; White et al., 2005). The translation product of the acp gene is the inactive apo-form ACP. Apo-ACP must be converted to the active holo-form by 4′-phosphopantetheinyl transferase (PPTase), which transfers the 4′-phosphopantetheine (Ppant) group from coenzyme A (CoA) to a conserved serine residue in ACP with phosphoester bonding (Figure 1A; Elovson and Vagelos, 1968; Cronan and Thomas, 2009; Beld et al., 2014). Correspondingly, holo-ACP can be converted to apo-ACP by the cleavage of the Ppant arm by ACP phosphodiesterase (AcpH; Fischl and Kennedy, 1990; Thomas and Cronan, 2005). ACP belongs to a broad family of strongly related proteins with a conserved signature motif (Asp–Ser–Leu, DSL), and its structure typically consists of four α-helices (White et al., 2005). ACPs function as the carriers of acyl intermediates and are the essential cofactors of primary and secondary metabolic pathways, including those of fatty acid synthesis (FAS), polyketide synthesis (PKS), and the biosynthesis of phospholipids, glycolipids, and endotoxins (Byers and Gong, 2007). Because it plays a central role in FAS, which is essential for bacterial survival, ACP has become a target of interest for new antibacterial drugs (Sílvia et al., 2008).
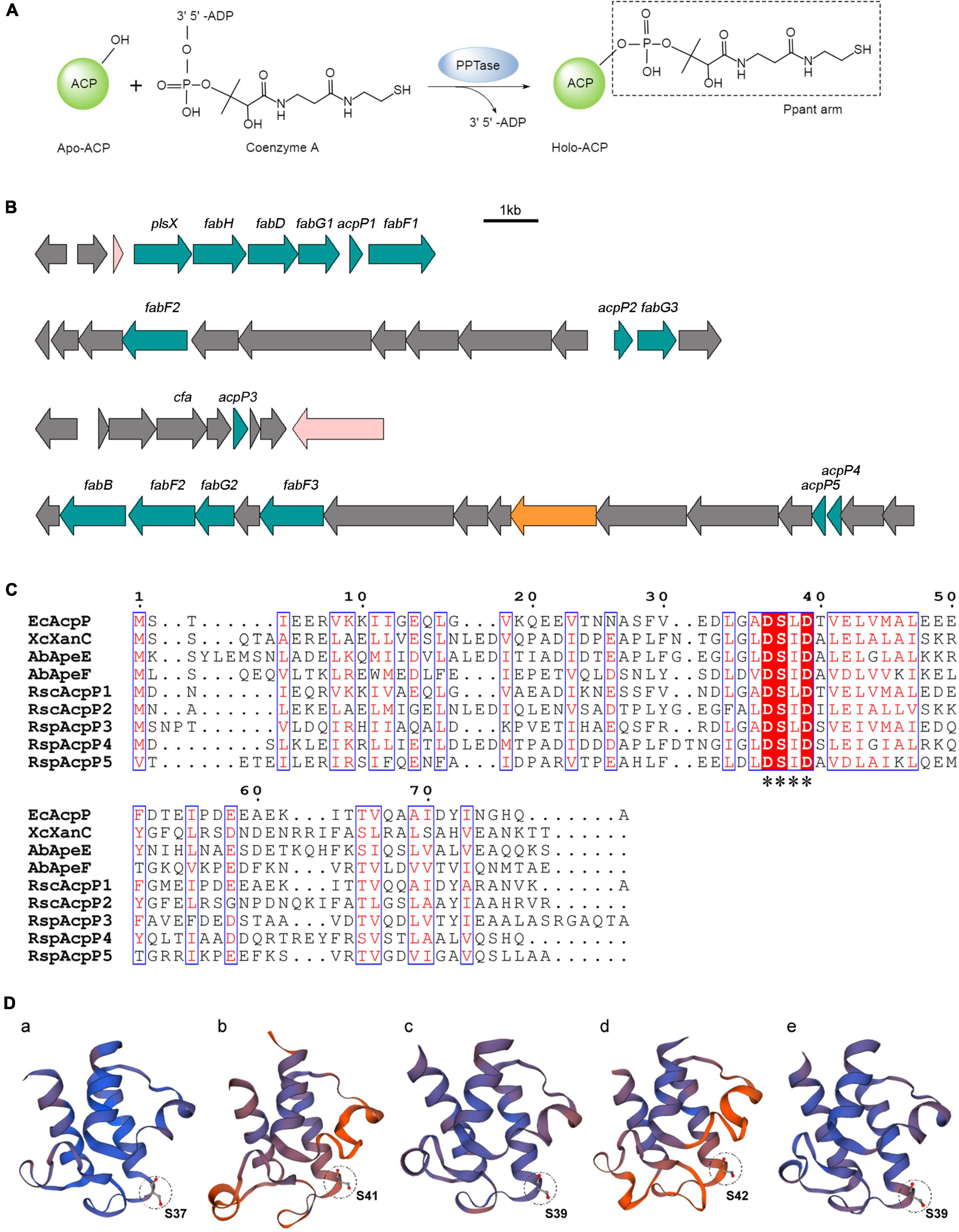
Figure 1. In silico analysis of R. solanacearum ACPs. (A) Posttranslational modification of acyl carrier proteins (ACPs). Apo-ACP was activated posttranslationally by the attachment of a Ppant arm derived from coenzyme A. The reaction was catalyzed by the 4′-phosphopantetheinyl transferase (PPTase). (B) Genetic organization of five acp genes in the R. solanacearum GMI1000 genome. (C) Sequence alignments of ACPs. The alignments were performed with T-Coffee (http://tcoffee.crg.cat/) and ESPript (https://espript.ibcp.fr/ESPript/ESPript/). White characters with red backgrounds are 100% identical, red characters with blue frames are similar. The conserved motif is labeled with asterisks. (D) Three-dimensional structure prediction of RsACPs using SWISS-MODEL (https://swissmodel.expasy.org/). The circle patterns show the conserved serine active site. (a) AcpP1 (template 4ihf.1.F), (b) AcpP2 (template 2l4b.1.A), (c) AcpP3 (template 3gzm.1.A), (d) AcpP4 (template 2 × 2b.1.A), and (e) AcpP5 (template 1 × 3o.1.A).
Escherichia coli encodes only a single and essential ACP, called AcpP, which acts in the synthesis of fatty acids, phospholipids, and lipid A (Cronan and Thomas, 2009). As well as interacting with the enzymes of lipid metabolism, E. coli AcpP also acts as a partner of various kinds of proteins, including MdoH, IscS, SpoT, and YchM (Therisod and Kennedy, 1987; Gully et al., 2003; Battesti and Bouveret, 2006; Babu et al., 2010). It suggests that E. coli AcpP is also involved in membrane-derived oligosaccharide (MDO) synthesis, cysteine thiolysis, (p)ppGpp metabolism, and bicarbonate transport.
However, most bacteria have more than one ACP, and some ACPs may not affect bacterial survival. Pseudomonas aeruginosa has three ACPs, of which AcpP1 functions as the essential ACP in fatty acid biosynthesis and the production of N-acyl homoserine lactones (AHLs), and AcpP3 is involved in the oxidative stress response (Ma et al., 2017; Chen et al., 2018). Enterococcus faecalis contains two ACPs involved in fatty acid metabolism: AcpA is mainly responsible for FAS, whereas AcpB may not play a role in FAS but channels exogenous acyl groups and forms the acyl–AcpB–FabT complex, which regulates the fatty acid biosynthesis (fab) operon (Zhu et al., 2019). In Xanthomonas campestris, there is one ACP, XanC, involved in PKS and the production of the yellow pigment xanthomonadin, and the typical AcpP also plays a role in FAS (Cao et al., 2018; Chen et al., 2022). The two ACPs of Myxococcus xanthus and Acinetobacter baumannii are reportedly involved in the secondary metabolism: MxTaB and MxTaE are involved in the antibiotic TA biosynthesis, and AbApeE and AbApeF are involved in the biosynthesis of aryl polyene (APE; Paitan et al., 1999; Grammbitter et al., 2019; Lee et al., 2021). Sinorhizobium meliloti expresses six ACPs, of which the canonical AcpP is responsible for general FAS and three ACPs are associated with fatty acids that form lipopolysaccharides (López-Lara and Geiger, 2000). NodF is involved in the synthesis of the C16 polyunsaturated fatty acid chains in the nodulation factor mono-N-acylated chitooligosaccharide; AcpXL in the transfer of long hydroxylated fatty acids to lipid A in lipopolysaccharides; and RkpF in the synthesis of polysaccharides that are rich in 3-deoxy-D-manno-2-octulosonic acid (Demont et al., 1993; Petrovics et al., 1993; Brozek et al., 1996; Epple et al., 1998). However, the functions of the other two ACPs are still unclear (Ramos-Vega et al., 2009; Davila-Martinez et al., 2010).
Ralstonia solanacearum is one of the most serious soil-borne phytopathogenic bacteria (Hayward, 1991; Mansfield et al., 2012; Peeters et al., 2013). It naturally invades plants through the root tips and the cracks caused by the secondary root emergence or elongation, and subsequently reaches and fills the xylem vessels, spreading systematically and forming vascular obstructions. This clogging causes wilting symptoms in the stem and leaves, and ultimately the death of the entire plant (Vasse et al., 1995; Lowe-Power et al., 2018). During this process, a bacterial quorum-sensing (QS) signal molecule [methyl 3-hydroxypalmitate (3-OH-PAME) or methyl 3-hydroxymyristate (3-OH-MAME)] autoregulates the production of various pathogenicity related factors and secondary metabolites (Flavier et al., 1997; Kai et al., 2015).
The R. solanacearum GMI1000 genome encodes five putative ACPs (Figure 1B; Salanoubat et al., 2002). AcpP1 is encoded by the gene RSc1053 (now named acpP1), which occurs in the fatty acid biosynthesis gene (fab) cluster. AcpP2 is encoded by RSc0434 (now named acpP2), also located on the chromosome. AcpP3, AcpP4, and AcpP5 are encoded by RSp1659, RSp0370, and RSp0369, respectively (now named acpP3, acpP4, and acpP5, respectively), and these genes are encoded on the megaplasmid. However, the function of none of these ACPs has been reported.
In this study, we characterized these five putative ACPs and confirmed that only AcpP1 is involved in FAS and is necessary for the survival of R. solanacearum. We also presented evidence that AcpP2 and AcpP4 can be involved in the PKS pathway, but their specific functions in R. solanacearum require further investigation. We also found that each acp gene product affected the bacterial capacity for swimming motility, and the disruption of four acp genes (acpP2, acpP3, acpP4, and acpP5) attenuated its pathogenicity against the host plant tomato.
Materials and methods
Materials
Coenzyme A, fatty acids, and antibiotics were obtained from Sigma-Aldrich (St. Louis, MO, USA). Sodium [1-14C] acetate was provided by American Radiolabeled Chemicals, Inc. (St. Louis, MO, USA). Ni-NTA agarose was provided by Invitrogen (Shanghai, China). Takara Biotechnology (Dalian, China) provided molecular biology reagents. All other reagents were of the highest available quality. Oligonucleotide primers and sequencing were provided by Sangon Biotech Co. (Shanghai, China).
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table 1. BG medium (bacto peptone 10 g/L, glucose 5 g/L, casamino acids 1 g/L, and yeast extract 1 g/L) and NYG medium (Cao et al., 2018) were used for the growth of the R. solanacearum and X. campestris strains, respectively. Luria-Bertani (LB) broth was used for the growth of E. coli, P. aeruginosa, and Agrobacterium tumefaciens. RB medium (Ulrich et al., 1983) was used to test the growth of E. coli acpP mutant strain CY1877. Antibiotics were used at the following concentrations: ampicillin 100 μg/ml, kanamycin 30 μg/ml, chloramphenicol 30 μg/ml, and gentamicin 10 μg/ml for R. solanacearum or 30 μg/ml for E. coli. Isopropyl-β-D-thiogalactoside (IPTG) and L-arabinose were used at final concentrations of 1 mM, and 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-Gal) was used at a concentration of 0.1 mM.
Recombinant DNA techniques and construction of plasmids
To clone the R. solanacearum acp genes, genomic DNA was extracted from R. solanacearum GMI1000 with the E.Z.N.A.® Bacterial DNA Kit (Omega Bio-Tek, Inc., Norcross, GA, USA). The PCR products amplified from strain GMI1000 genomic DNA with Pfu DNA polymerase and the primers listed in Supplementary Table 2 were digested with NdeI and HindIII and then inserted between the corresponding sites of pET-28b, pSRK-Km, or pSRK-Gm. To generate pTac85-derived plasmids, the PCR fragments were amplified from pSRK-Gm-derived plasmids and then digested with NcoI and SalI, the restriction sites for which had been designed into the primer sequences. After purification, the fragments were ligated into the vector pTac85. We used a similar process to construct the plasmids expressing site-directed mutant genes, except that the fragments were amplified by dividing the gene into two parts with primers carrying the mutation site. The two fragments of the gene were then purified and used for overlapping PCR. The plasmids used in this study are listed in Supplementary Table 1. The DNA fragments were confirmed with DNA sequencing by Shanghai Sangon, Inc. (Shanghai, China).
Generation of acp mutant strains
To mutate the R. solanacearum acp genes, suicide plasmids were constructed with three strategies (Supplementary Figure 1). To create unmarked deletion mutants of the acp genes, DNA fragments (of about 600 bp) flanking the acp genes were amplified with the primers listed in Supplementary Table 2. The PCR products were purified and used for overlapping PCR. The resulting 1.2-kb DNA fragments were digested with HindIII and XbaI or BamHI and inserted between the corresponding sites in pK18mobscaB. Using similar procedures, the EcacpP fragment and the fragments flanking acpP1 were ligated with overlapping PCR and cloned into plasmid pK18mobsacB to construct the suicide plasmids for the acpP1 replacement mutant. To create site-directed mutants, DNA fragments (of about 700 bp) flanking the acpP1 mutant sites were amplified with the primers listed in Supplementary Table 2. The PCR products were purified, ligated with overlapping PCR, and cloned into pK18mobsacB.
The unmarked acp mutant strains were constructed with allelic exchange using suicide vector pK18mobscaB (Supplementary Figure 1) (Schäfer et al., 1994), with the procedure described by Mao et al. (2015). Briefly, all pK18mobscaB-derived plasmids were introduced into R. solanacearum by conjugal transfer from E. coli S17-1. Single-crossover integrants into the strain GMI1000 chromosome were selected based on chloramphenicol and kanamycin resistance. Cultures grown from the integrants were plated onto BG plates containing 12% sucrose to select for the loss of the vector sequences from the GMI1000 chromosome. The successful construction of the designed mutants was evaluated with PCR and the primers listed in Supplementary Table 2, and was confirmed with DNA sequencing by Shanghai Sangon, Inc. (Shanghai, China). Using similar procedures, a two gene-mutated strain was generated in the background of the single gene-mutated strain, and was then used to generate the three gene-mutated strain, which was used, in turn, to generate the four gene-mutated strain.
Purification of Ralstonia solanacearum acyl carrier proteins
The pET-28b-derived plasmids were introduced into E. coli BL21(DE3) host cells. The ACPs with N-terminal 6 × His-tagged were expressed after the cells were induced with IPTG, and purified as described previously (Chen et al., 2022). To obtain apo-form ACPs, E. coli strain YY121 expressing E. coli AcpH was also cultured and the protein expressed in LB broth. The cells expressing ACPs and AcpH were harvested and lysed by sonication in AcpH reaction buffer [50 mM Tris–HCl (pH 8.8), 25 mM MgCl2, 1 mM dithiothreitol (DTT), 0.2 mM MnCl2; Cronan and Thomas, 2009]. Samples of holo-ACPs were converted to apo-ACPs with AcpH supernatant (the volumetric ratio of ACP solution to AcpH solution was 2:1). After incubation at 37°C for 4 h, the mixture was applied to Ni NTA Beads 6FF (Smart-Lifesciences Inc., China), according to the general protocol of the manufacturer. The eluted ACP was precipitated by the addition of trichloroacetic acid at a final concentration of 8%, and was then redissolved and dialyzed twice in AcpS reaction buffer [50 mM Tris-HCl (pH 8.8), 10 mM MgCl2, 5 mM DTT; Cronan and Thomas, 2009].
The plasmids pET-acpS, pET-Sfp, and pYFJ84 were introduced into E. coli BL21(DE3) host cells for the expression of E. coli AcpS, Bacillus subtilis PPTase Sfp, and Vibrio harveyi acyl-ACP synthetase (AasS) proteins, respectively. These proteins were also purified as described previously (Jiang et al., 2006; Chen et al., 2022).
Phosphopantetheinylatation and acylation of acyl carrier proteins in vitro
The phosphopantetheinylatation and acylation assays were adapted from those of Ma et al. (2017). Briefly, the reaction mixture for phosphopantetheinylatation contained 50 mM Tris–HCl (pH 8.8), 10 mM MgCl2, 1 mM DTT, 1 mM CoA, 50 μM apo-ACP, and 1 μM AcpS or Sfp, and was incubated at 37°C for 1 h. The reaction products were resolved with conformation-sensitive gel electrophoresis on 17.5% polyacrylamide gels containing 2.5 M urea, optimized for separation.
Vibrio harveyi AasS was used to acylate holo-ACP (Jiang et al., 2006). The reaction mixtures for acylation contained 100 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 10 mM ATP, 5 mM DTT, 0.5 mM hexanoic acid or dodecanoic acid, 25 μM holo-ACP (apo-ACP was incubated with PPTase), and 2 μM AasS, and were incubated at 37°C for 1 h. The reaction products were resolved on 17.5% polyacrylamide gels containing 4 M urea.
Analysis of phospholipid fatty acids
The R. solanacearum strains were cultured at 30°C in BG medium overnight, transferred to 5 ml of BG medium, and grown from an optical density at a wavelength of 600 nm (OD600) of 0.2 to an OD600 of 0.7–1.0 with or without additional sodium [1-14C] acetate (final concentration, 1 μCi/ml). To assay de novo FAS, the sodium [1-14C] acetate-labeled cells were pelleted and lysed with methanol-chloroform (2:1) solution, and the fatty acid methyl esters were extracted as previously described (Zhu et al., 2019). The labeled fatty acids were analyzed with thin-layer chromatography (TLC) and quantified with phosphorimaging. To assay the phospholipid compositions, fatty acid methyl esters were synthesized with sodium methoxide and extracted with petroleum ether, as described previously (Zhu et al., 2010), and were analyzed with GC–MS.
Assay of quorum-sensing signal molecule
An assay of the QS signal molecule was performed as described previously (Guo et al., 2018). P. aeruginosa strains were grown to an OD600 of 1.0, and the cells were removed by centrifugation at 4°C. The QS signal molecule HSL was twice extracted from 10 ml of supernatant from each sample with an equal volume of ethyl acetate. The organic extract was concentrated to dryness with a nitrogen bubbler, and the residue was resuspended in 100 μL of ethyl acetate. The biosensor A. tumefaciens NT1 (traR, tra::lacZ749) was grown in LB broth to an OD600 of 1.0 and then diluted to an OD600 of 0.1 to cover the ME medium plate (0.2 g/L MgSO4⋅7H2O, 10 g/L K2HPO4, 2 g/L C6H8O7⋅H2O, 3.5 g/L NaNH4HPO4⋅4H2O, pH 7.0) supplemented with X-Gal and IPTG. The extracts of the P. aeruginosa strains were dotted onto the ME plate described above. After overnight incubation at 30°C, a blue halo was observed when N-(3-oxo-dodecanoyl)-L-homoserine lactone (3-oxo-C12-HSL) was produced (Piper et al., 1993).
Extraction and quantification of xanthomonadins
The pigments were extracted from X. campestris with a previously described procedure (Cao et al., 2018). Briefly, X. campestris was grown to stationary phase (OD600 = 1.0) in NYG medium at 30°C. The cells from 10 ml of each culture were collected by centrifugation. The pigments were extracted by shaking with 1 ml of methanol for 5 min. The amounts of xanthomonadin pigment produced were expressed as the absorbance (OD445) of the crude pigment extracts.
RNA extraction, reverse transcription PCR, and quantitative real-time PCR
Ralstonia solanacearum strain GMI1000 was grown in BG medium at 30°C. The total RNA was extracted from the cells in 0.5 ml of culture from each growth phase with a E.Z.N.A.® Bacterial RNA Kit (Omega Bio-Tek, Inc.). Once extracted, 1 μg of total RNA was reverse transcribed with the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara Bio Inc.). Quantitative real-time PCR (qPCR) was performed with the 2 × RealStar Green Fast Mixture (GenStar). Relative quantitation was done by the comparative cycle threshold (ΔΔCT) method using the endogenous internal control 16S rDNA and gryB (DNA gyrase subunit B; Castillo and Greenberg, 2007) for sample normalization. The primers used in reverse transcription PCR (RT-PCR) and qPCR are listed in Supplementary Table 2.
Swimming motility assay
The motility of R. solanacearum was assayed on semisolid motility medium containing 1% (wt/vol) bacterial peptone and 0.3% (wt/vol) agar (BD). A 2 μL aliquot of cell suspension containing 1.0 × 108 CFU/ml was added to the center of the plate and cultured at 30°C. The diameters of migration were measured at 72 h postinoculation.
Pathogenicity tests
The bacterial virulence experiment using drenching infection assays was adapted from Morel et al. (2018). The roots of each 4-week-old tomato plant (Lycopersicon esculentum cv. Moneymaker) were partly cut, and were inoculated by pouring 20 ml of fresh bacterial suspension containing 5 × 109CFU of R. solanacearum onto the soil surrounding the plants. Once infected, the plants were incubated in a growth chamber (28°C day/27°C night, 85% relative humidity, 12 h light), and wilting symptoms were recorded daily on a severity rating visual scale, from 0 (no wilt) to 4 (death). Each strain tested was assessed for wilting in three independent experiments (15 plants each). The severity of the infection was expressed with the disease index (DI), which was calculated with the formula: DI = ∑(severity rating × number of plants in that rating)/(total number of plants × 4).
Data confirmation and statistical analysis
Experiments were repeated three times to confirm their reproducibility. All results were analyzed with GraphPad Prism 8 and are presented as means ± standard deviations (SD). The statistical significance of the difference between two measurements was determined with Student’s t-test. Results were considered statistically significant at P < 0.05.
Results
In silico analysis of Ralstonia solanacearum acyl carrier proteins
The multiple alignments of ACP amino acid sequences were performed with T-Coffee and ESPript (Figure 1C). AcpP1, encoded within a fab gene cluster, showed high identity with E. coli AcpP (68.35%), and AcpP3 and AcpP5 are also similar to E. coli AcpP (35.63% and 29.63% identity, respectively; Supplementary Table 3). The acpP2 gene is located in a cluster similar to the pig cluster, which is responsible for xanthomonadin biosynthesis in X. campestris (Cheng et al., 2012; Cao et al., 2018), and AcpP2 is 48.31% identical to XcXanC encoded by acpC in the pig cluster. The megaplasmid genes acpP4 and acpP5 are located in a cluster syntenic to the ape gene cluster, which is responsible for the biosynthesis of APE pigments in A. baumannii (Grammbitter et al., 2019; Lee et al., 2021). Two genes are located in the ape cluster, encoding AbApeE and AbApeF, and AcpP4 is 44.09% identical to AbApeE and AcpP5 is 41.46% identical to AbApeF. The alignment also showed that all ACPs contain the conserved serine catalytic triad Asp–Ser–Leu (DSL).
The prediction of the three-dimensional structures of the R. solanacearum ACPs with the SWISS-MODEL server showed that the topologies of the five ACPs are similar to that of E. coli AcpP (Figure 1D; White et al., 2005). They all have four helices (except AcpP2, which has no α3 helix) and the DSL motif at the end of helix α2 (Figure 1D and Supplementary Figure 2; Rock and Cronan, 1979; Roujeinikova et al., 2002; White et al., 2005). The results of a bioinformatic analysis imply that these five putative proteins in R. solanacearum are ACPs, among which AcpP1 and AcpP3 probably function in FAS and AcpP2, AcpP4, and AcpP5 in PKS.
Ralstonia solanacearum acyl carrier proteins are phosphopantetheinylated in vitro
In general, apo-ACP requires posttranslational modification for its activation (Figure 1A). To determine whether the five ACPs of R. solanacearum are functional, the apo-forms of AcpP1, AcpP2, AcpP3, and AcpP5 were isolated to examine their phosphopantetheinylatation. These apo-ACPs were purified as described in section “Materials and methods.” Curiously, attempts to isolate AcpP4 were unsuccessful, despite the optimization of its codons, varying the expression time (4–12 h) and temperature (18–37°C), its co-expression with the different protein partners (TrxA, MBP, and GST), and its fusion expression with different protein tags (MBP, GB1, Fh8, SUMO, and TrxA).
AcpS-type PPTases primarily act on the ACPs of the FAS pathway, whereas Sfp-type PPTases have a broader substrate range, and can modify the ACPs involved in the secondary or primary metabolism (Quadri et al., 1998; Flugel et al., 2000; Mootz et al., 2001; Beld et al., 2014). Therefore, E. coli AcpS and B. subtilis Sfp were used to examine the modification of the R. solanacearum ACPs. After incubation with AcpS or Sfp and CoA, the migration of AcpP1 and AcpP3 changed significantly, indicating that AcpP1 and AcpP3 are activated from apo-ACP to holo-ACP by these two types of PPTase (Figure 2). The migration of AcpP2 and AcpP5 changed in the presence of Sfp, but not in the presence of AcpS, indicating AcpP2 and AcpP5 are only activated by Sfp-type PPTases. These data show that these four R. solanacearum ACPs can be phosphopantetheinylated, and that AcpP1 and AcpP3 may be involved in FAS, whereas AcpP2 and AcpP5 may be involved in the secondary metabolism.
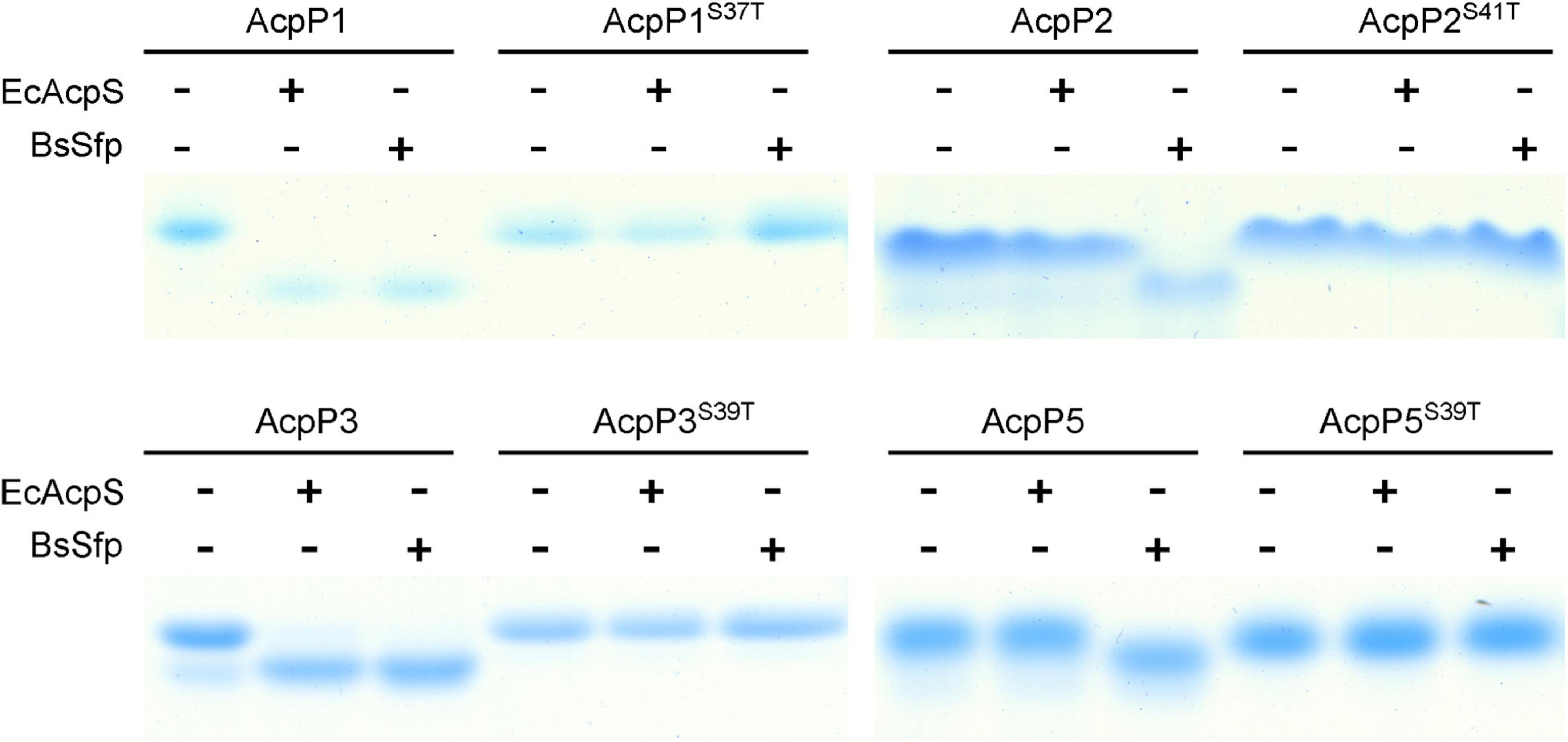
Figure 2. Four R. solanacearum ACPs were phosphopantetheinylated in vitro. The reaction mixture contained ACP, CoA, and E. coli AcpS or B. subtilis Sfp in AcpS reaction buffer. The apo-ACPs tested as the reaction substrate are given above each lane.
According to previous research, ACPs contain the conserved DSL motif, in which the serine is the active site through which the Ppant group is linked, and mutation at that residue causes protein loss of function (White et al., 2005; De Lay and Cronan, 2007). Therefore, the apo-forms of site-directed mutant proteins in which the Ser in DSL was replaced with Thr were also tested. After incubation with CoA, the migration of the four mutant proteins did not shift in the presence of either AcpS or Sfp, indicating that the mutant ACPs were not activated by these two types of PPTase (Figure 2), confirming that the Ser in the DSL motif is the key active site of the ACPs, as previously reported.
Only the acpP1 gene is necessary for Ralstonia solanacearum survival
To determine the function of each ACP, the acpP2, acpP3, acpP4, or acpP5 gene was deleted individually to generate mutant strain mRP2, mRP3, mRP4, or mRP5, respectively. A mutant lacking all four acp genes (mRP2345) was also generated. The growth of these mutants in the rich BG medium (Figure 3A) or minimal M63 medium (data not shown) was no different from that of the wild-type strain. In contrast, the acpP1-deletion mutant could not be isolated, suggesting that acpP1 is an important housekeeping gene in R. solanacearum.
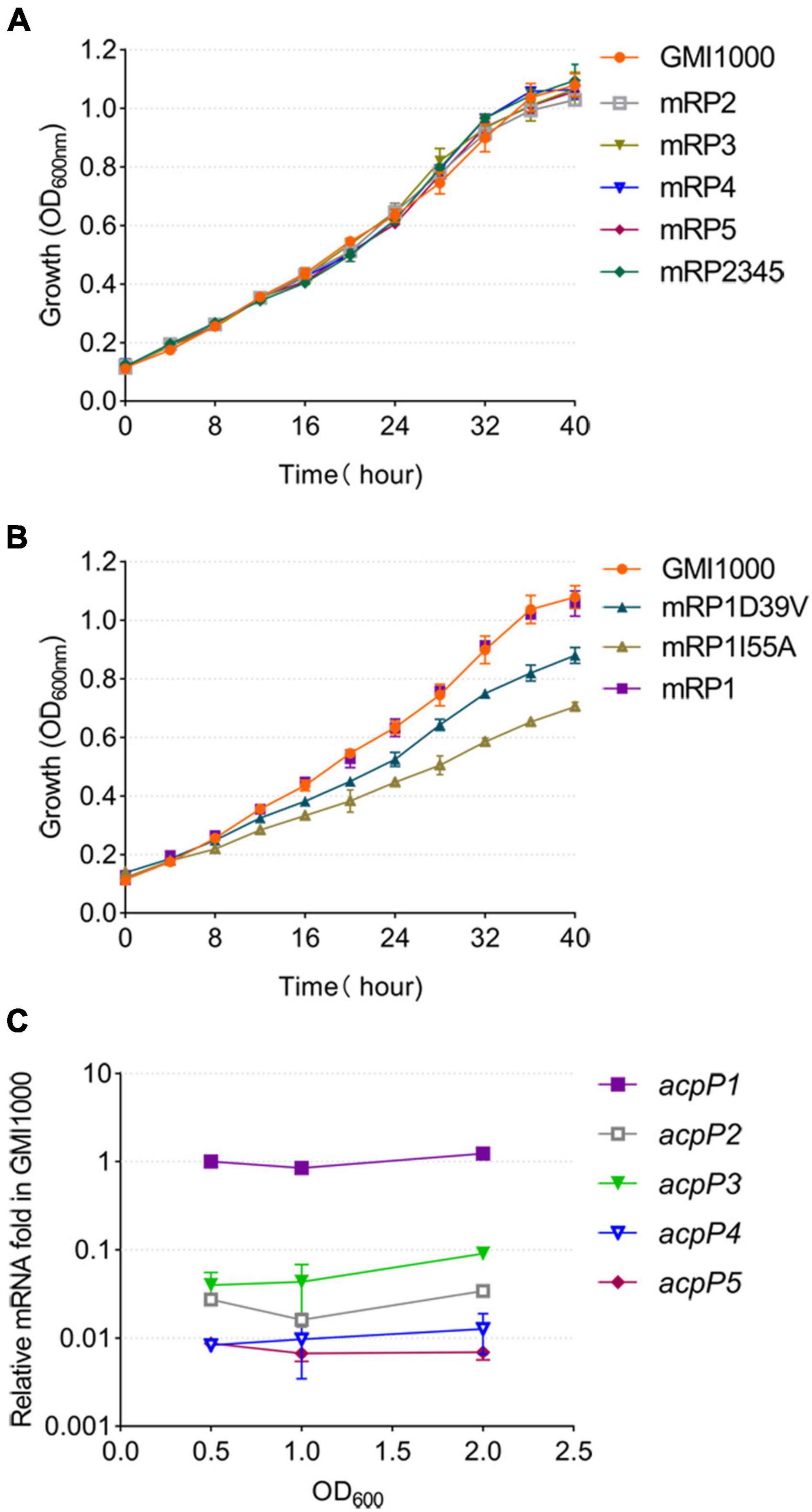
Figure 3. Only the AcpP1 affects the growth of R. solanacearum. (A,B) Growth of the strains in BG medium at 30°C. GMI1000, wild-type strain; mRP1D39V, mutant in which Asp39 of AcpP1 is altered to Val; mRP1I55A, mutant in which Ile55 of AcpP1 is altered to Ala; mRP1, mutant in which the acpP1 gene is replaced with E. coli acpP; mRP2, mutant lacking acpP2 gene; mRP3, mutant lacking acpP3 gene; mRP4, mutant lacking acpP4 gene; mRP5, mutant lacking acpP5 gene; mRP2345, mutant lacking acpP2, acpP3, acpP4, and acpP5 genes. (C) Expression of the five acp genes in R. solanacearum wild-type strain GMI1000. Expression level of the acpP1 gene in log phase (OD600 = 0.5) was set to 1.
Based on the importance of acpP1 and the similarity of the protein sequences encoded by acpP1 and E. coli acpP, the E. coli acpP gene was used to replace the acpP1 gene in R. solanacearum. The mutant strain in which the acpP1 gene was replaced with E. coli acpP (designated mRP1) was generated with this strategy. Previous research has shown that some single-site mutations of E. coli AcpP impair, but do not entirely destroy, the activity of the protein (De Lay and Cronan, 2007). Therefore, we attempted to site-specifically mutate the acpP1 gene with allelic replacement using the plasmid pK18mobscaB (Supplementary Figure 1). Two site-directed mutants were obtained: mRP1D39V (Asp39 mutated to Val) and mRP1I55A (Ile55 mutated to Ala). Although the replacement of the acpP1 gene with E. coli acpP did not affect the growth of the strain, the site-specific mutation of AcpP1 at Asp39 or Ile55 significantly impaired the growth of the strain (Figure 3B). All these data confirm that acpP1 is essential for R. solanacearum viability.
Using qPCR to quantify the transcription levels of all the acp genes in the R. solanacearum wild-type strain, we found that acpP1 is highly and stably expressed throughout its growth stages, whereas the expression of the other four acp genes was more than 10-fold lower than that of acpP1 (Figure 3C). These results indicate that acpP1 is highly expressed and required for the growth of R. solanacearum, and that mutations at certain sites result in significant growth retardation.
Only AcpP1 functions in fatty acid biosynthesis
In the genome of R. solanacearum GMI000, acpP1 is located in the canonical fab gene cluster, which includes plsX, fabH, fabD, fabG1, and fabF1 (Cheng et al., 2012; Feng et al., 2015; Mao et al., 2015). With a RT-PCR analysis of the intergenic regions, we showed that the acpP1 gene is cotranscribed with upstream and downstream genes and is located within a fab gene cluster (Figure 4A). This confirms that the expression of AcpP1 is closely related to the expression of FASII enzymes.
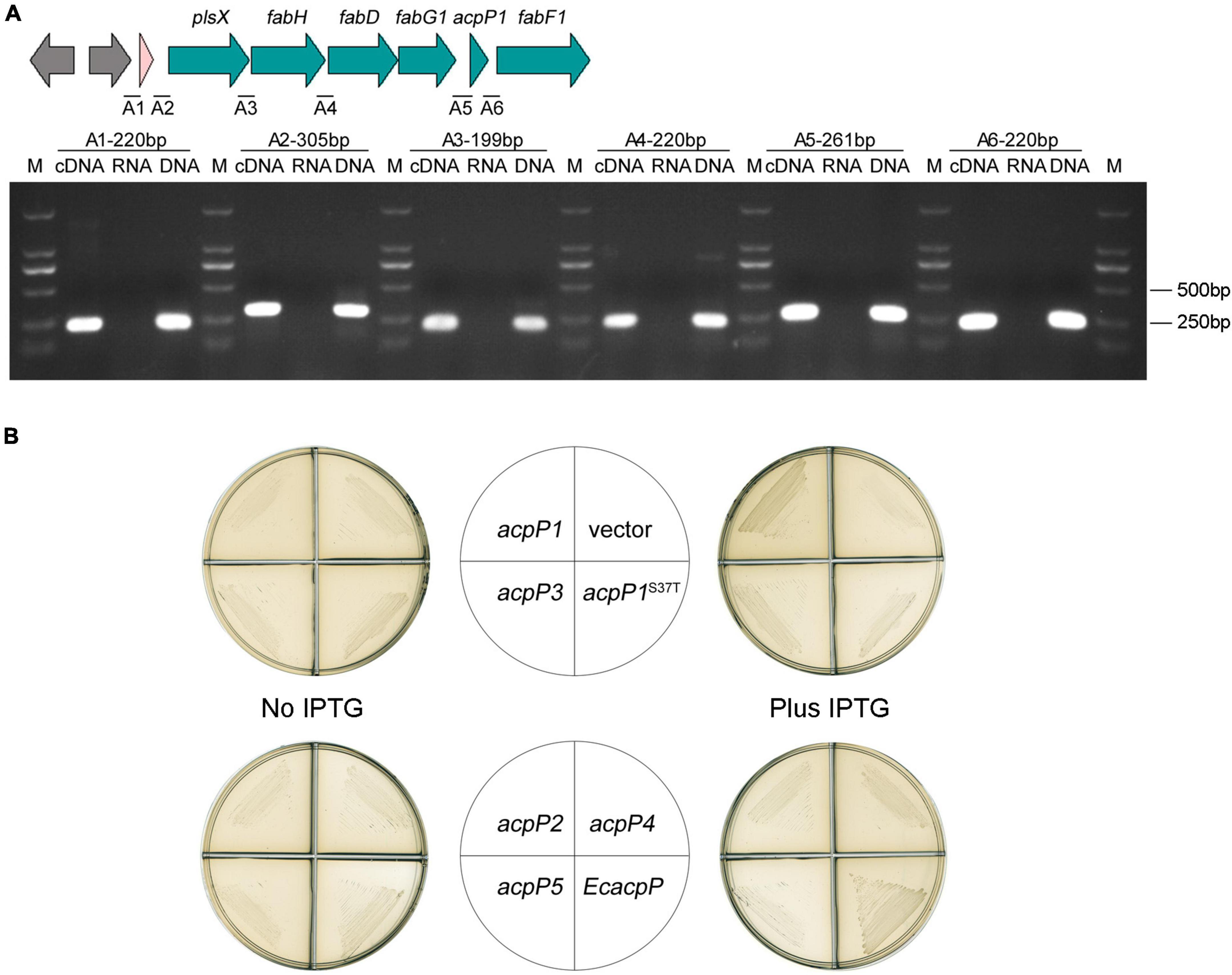
Figure 4. Only AcpP1 partially restored the growth of E. coli acpP-mutant strain. (A) RT-PCR of the intergenic regions. cDNA, RNA, or DNA from the GMI1000 strain was used as the template for PCR. The intergenic regions and lengths are marked above each lane. M: DNA marker DL2000. (B) Complementation of E. coli acpP mutant strain CY1877 with R. solanacearum acp genes. Strains CY1877 carrying IPTG-inducible expression vector pTac85 expressing an acp gene (as shown in the middle panels) were cultured on RB plates for 24 h. Left panels showed RB medium without the supplement of IPTG; right panels showed RB medium supplemented with IPTG; vector, pTac85.
Because E. coli AcpP acts in FASII, the E. coli acpP mutant strain CY1877, which requires arabinose to induce E. coli acpP expression for normal growth, was used to test the functions of the R. solanacearum acp-encoded proteins (Ma et al., 2017). The results showed that the overexpression of acpP1, induced with IPTG, partially restored CY1877 growth (Figure 4B), whereas the overexpression of the other acp genes failed to support the growth of the E. coli mutant, indicating that only R. solanacearum AcpP1 can functionally replace E. coli ACP.
Next, the R. solanacearum acp genes were overexpressed in P. aeruginosa mutant strain PA-A1, in which the chromosomal acpP1 gene was replaced with E. coli acpP, impairing QS signal production (Ma et al., 2017). The fatty-acid-derivative QS signal molecule, 3-oxo-C12-HSL, produced by P. aeruginosa PA-A1 was tested with the biosensor A. tumefaciens NT1 (traR, tra::lacZ749), which produces a blue halo in response to acyl homoserine lactones (acyl-HSLs; Piper et al., 1993). The growth of P. aeruginosa PA-A1 derivatives carrying the plasmid pSRK-Gm encoding the R. solanacearum acp genes did not differ from that of PA-A1, but the blue halo around PA-A1 carrying acpP1- or acpP3-encoded plasmids was larger than that around the PA-A1 strain carrying the empty vector (Figures 5A,B). These results suggest that AcpP1 functions in FAS, whereas AcpP3 seemed to be acylated. To confirm these findings, we tested the acylation of R. solanacearum ACPs using V. harveyi acyl-ACP synthetase (AasS) in vitro (Byers, 1989). After incubation of the R. solanacearum holo-ACPs with hexanoic acid or dodecanoic acid, ATP, and VhAasS, the migrations of holo-AcpP1 and holo-AcpP3 were altered, showing that these two holo-ACPs were acylated by VhAasS to carry a fatty acid chain (Figure 5C). In contrast, there was no change in the migrations of holo-AcpP2 and holo-AcpP5.
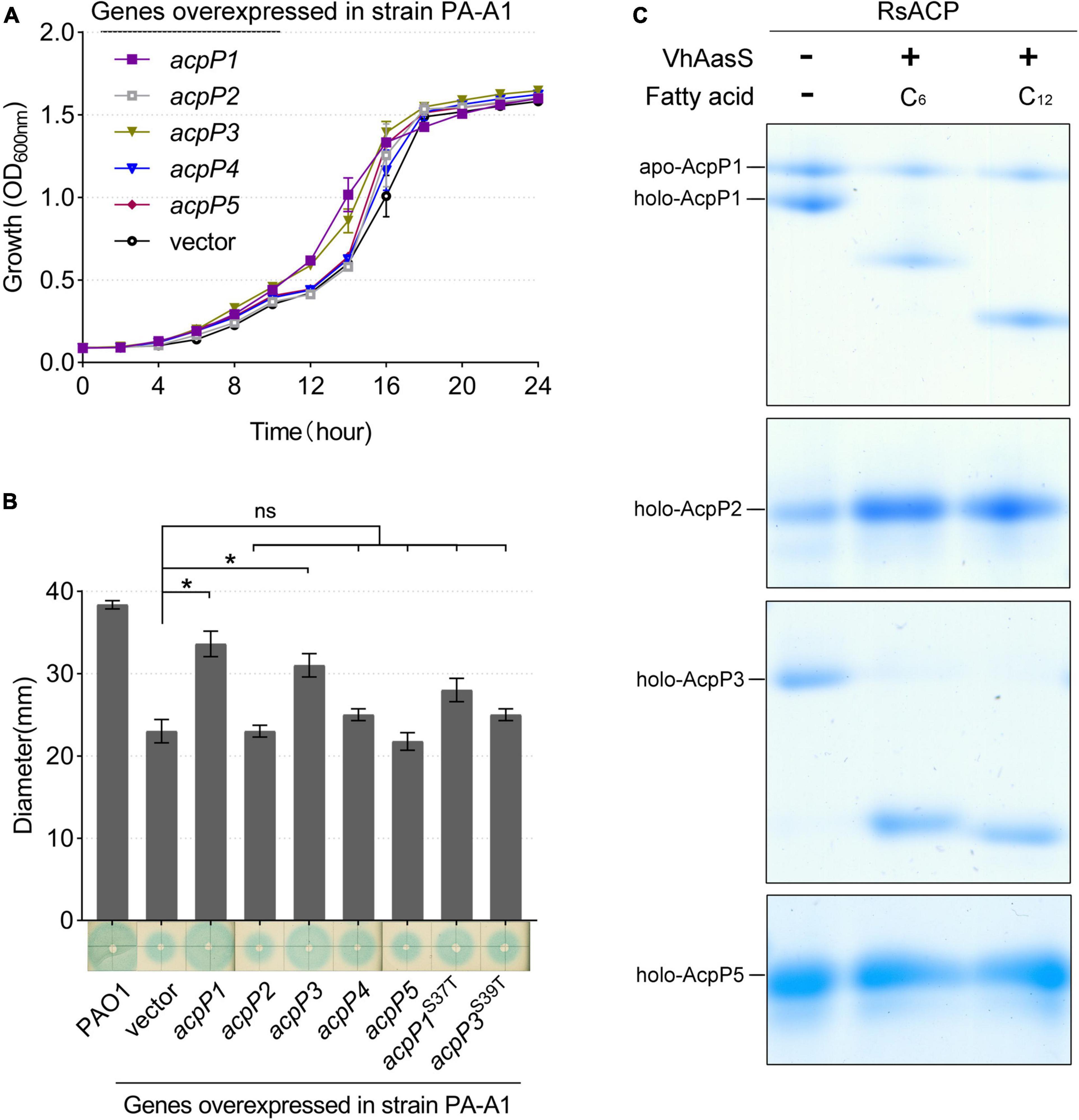
Figure 5. Identification of R. solanacearum AcpPs in vivo and in vitro. (A) Growth of P. aeruginosa acpP1-mutant strain PA-A1 harboring the R. solanacearum acp genes. Overexpression of R. solanacearum acp genes did not affect strain growth. (B) Detection of 3-oxo-C12-HSL signals produced by the complementation of strain PA-A1. The QS signal molecules were extracted as described previously and were dotted onto a ME medium plate that was supplemented with X-Gal and IPTG and covered with fresh A. tumefaciens NT1 (traR, tra::lacZ749; Guo et al., 2018). Student’s t-test was used to analyze the difference between PA-A1 carrying the empty vector and PA-A1 overexpressing the acp genes. *Indicates significant difference (P < 0.05). nsIndicates no significant difference (P > 0.05). (C) The R. solanacearum holo-AcpPs were acylated by VhAasS in vitro. The reaction mixtures contained holo-ACP, ATP, hexanoic acid or dodecanoic acid, and V. harveyi AasS.
To determine whether ACPs function in the FAS of R. solanacearum, the capacity of the acp mutants for FAS was determined by measuring their de novo FAS as the incorporation of [1-14C] acetate into their membrane phospholipids. The incorporation of [1-14C] acetate into the membrane phospholipids of the two acpP1 mutants, mRP1D39V and mRP1I55A, was twofold lower than that in the wild-type strain (Figure 6, lanes 2 and 4). However, neither single gene deletion mutants nor the quadruple gene deletion mutant of the other four acp genes affected the fatty acid biosynthesis of R. solanacearum (Figure 6, lanes 5–9). We also analyzed the fatty acid composition of the R. solanacearum strains with gas chromatography-mass spectrometry (GC–MS). All R. solanacearum strains had similar fatty acid profiles (Supplementary Table 4). These results confirm that only AcpP1 is involved in fatty acid biosynthesis.
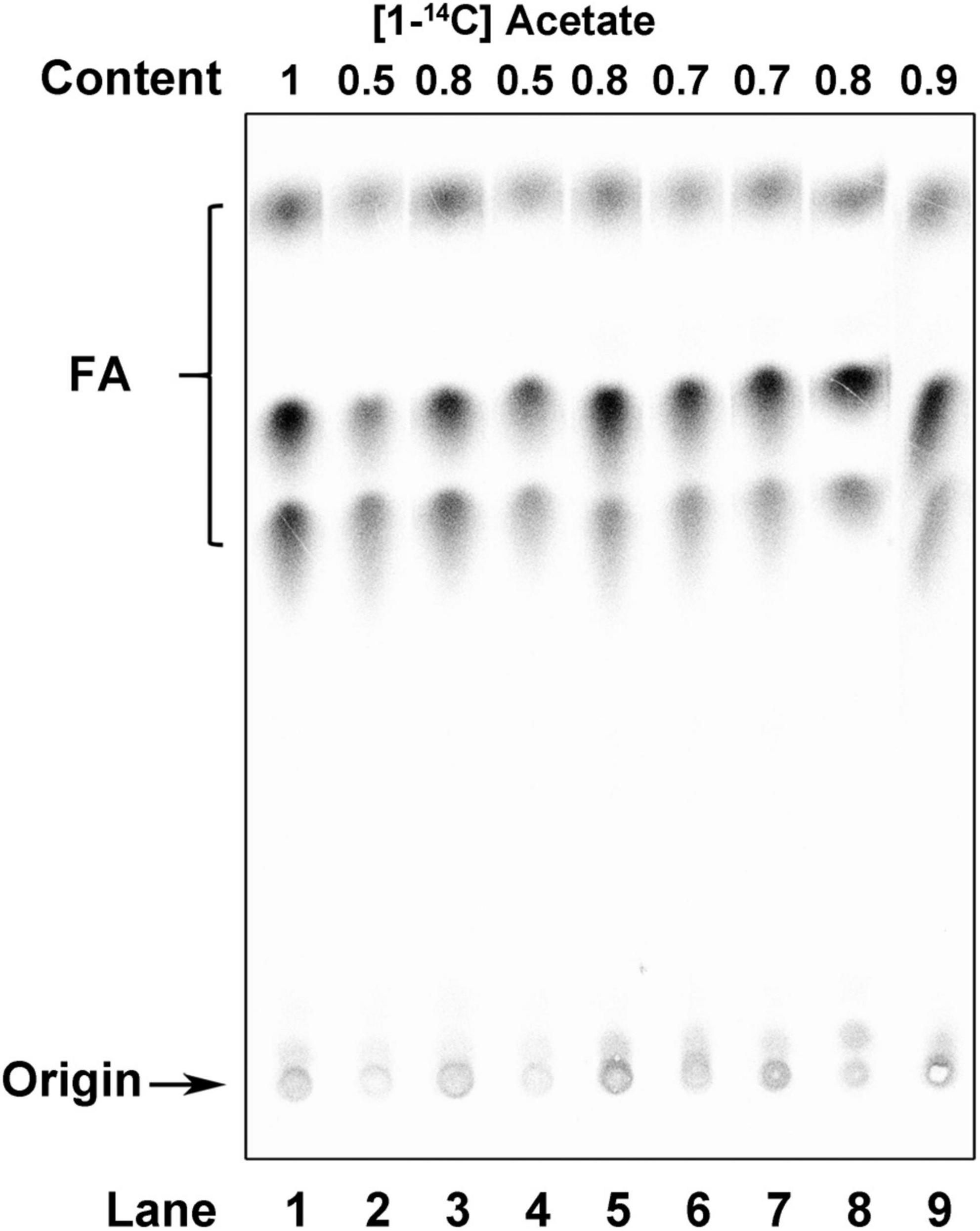
Figure 6. Analysis of de novo fatty acid synthesis in R. solanacearum strains. Argentation thin-layer chromatographic analysis of sodium [1-14C] acetate-labeled R. solanacearum strains, as described in section “Materials and methods.” Lane 1, wild-type strain; lane 2, mutant in which Asp39 of AcpP1 was mutated to Val; lane 3, mutant in which the acpP1 gene was replaced with E. coli acpP; lane 4, mutant in which Ile55 of AcpP1 was mutated to Ala; lane 5, mutant lacking genes acpP2, acpP3, acpP4, and acpP5; lane 6, mutant lacking gene acpP2; lane 7, mutant lacking gene acpP3; lane 8, mutant lacking gene acpP4; lane 9, mutant lacking gene acpP5. The numbers above the lanes give the relative incorporation value for each lane.
AcpP2 and AcpP4 participate in polyketide synthesis
According to the results described above, AcpP2, AcpP4, and AcpP5 are not involved in FAS, and genes acpP2, acpP4, and acpP5 are located in gene clusters related to the synthesis of APEs (Cao et al., 2018; Grammbitter et al., 2019). Therefore, we inferred that AcpP2, AcpP4, and AcpP5 are involved in PKS. In X. campestris, xanC encodes the ACP XanC, which is essential for the biosynthesis of xanthomonadin, a yellow APE pigment (Cao et al., 2018). Therefore, the X. campestris xanC-deletion mutant mXPC, which had lost the ability to synthesize yellow pigment, was constructed and used to examine whether R. solanacearum ACPs are involved in the PKS pathway. The overexpression of acpP2 or acpP4 from a plasmid pSRK-Km in the mutant mXPC restored xanthomonadin production, but the strain failed to produce xanthomonadin when the genes acpP1, acpP3, and acpP5 were overexpressed in the mXPC mutant (Figure 7A). These results show that AcpP2 and AcpP4 can act as ACPs in PKS and indicate that AcpP4 can be phosphopantetheinylated in vivo. Moreover, the strains expressing the site-specifically mutated genes acpP2S41T or acpP4S42T also did not produce yellow pigment, indicating that AcpP4, like AcpP2, has the typical active-site characteristics (Figure 7A).
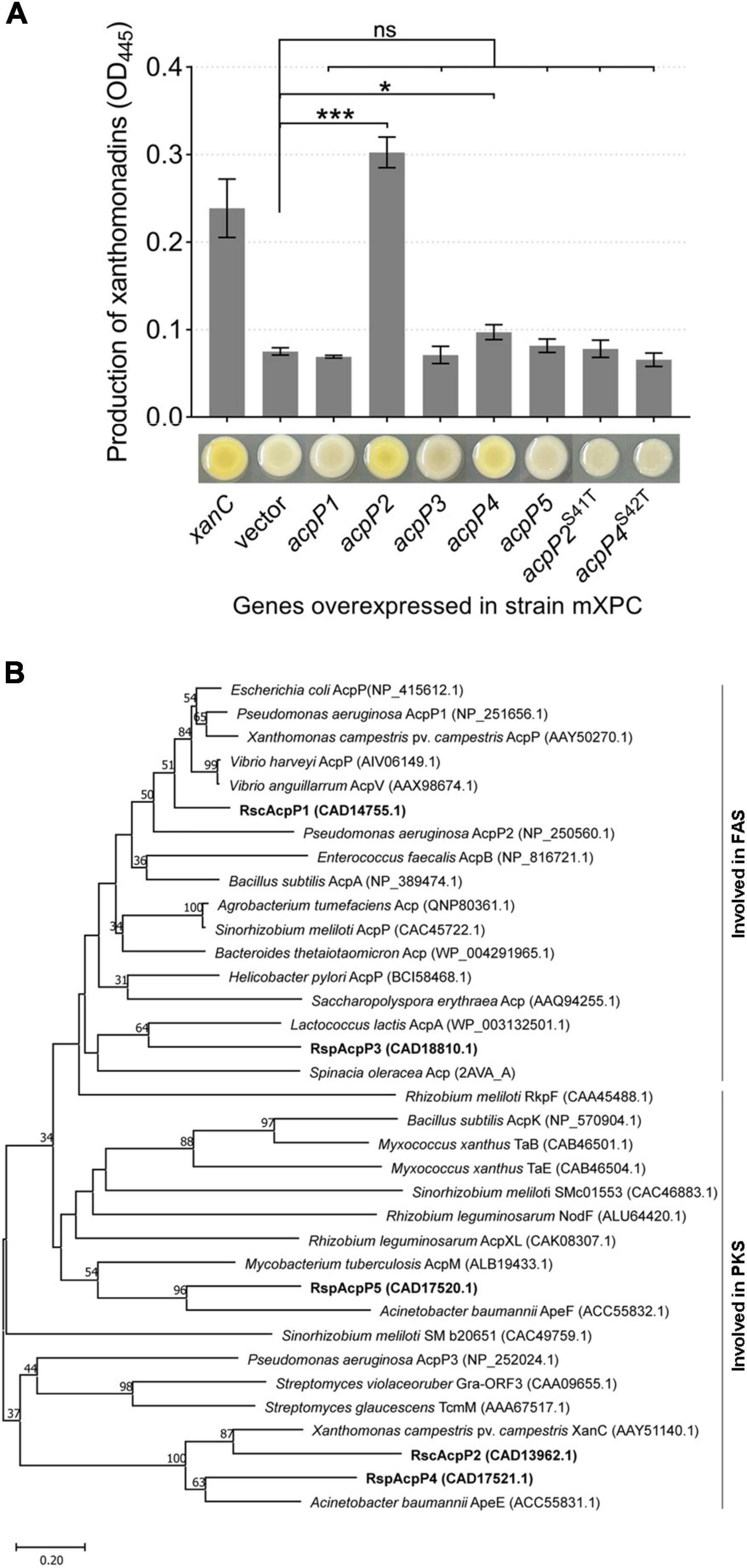
Figure 7. AcpP2 and AcpP4 participate in polyketide synthesis. (A) AcpP2 and AcpP4 restored the production of xanthomonadin in the X. campestris xanC mutant mXPC. Pigments were extracted with methanol from X. campestris using previously described procedures (Cao et al., 2018). Student’s t-test was used to compare mXPC carrying the empty vector and mXPC overexpressing the acp genes. Significant differences are indicated as ***P < 0.001 and *P < 0.05. (B) Neighbor-joining phylogenetic analysis of ACPs. The neighbor-joining tree was created with MEGA 11, with 1,000 bootstrap replicates. Proteins are shown with their NCBI (https://www.ncbi.nlm.nih.gov/) accession numbers in parenthesis. The RsACPs clustered into two regions, reflecting their involvement in fatty acid synthesis (FAS) or polyketide synthesis (PKS).
A phylogenetic analysis of the R. solanacearum ACPs and ACPs from different species, using the neighbor-joining algorithm in the molecular evolutionary genetics analysis software MEGA 11 (Tamura et al., 2021), was performed. On a phylogenetic tree constructed from the amino acid sequence alignment, these ACPs clustered in two major groups, involved in either FAS or PKS (Figure 7B). AcpP1 and AcpP3 clustered in the group involved in FAS, whereas AcpP2, AcpP4, and AcpP5 clustered with the other group. These data further support the inference that AcpP2 and AcpP4 are involved in the synthesis of an unknown secondary metabolite in R. solanacearum.
Ralstonia solanacearum mutant mRP2345 showed impaired virulence in tomato
To investigate the effects of these ACPs on the pathogenicity of R. solanacearum, tomato plants were infected by drenching the soil with the acp gene mutant strains. The strains caused plant wilting symptoms on days 4–6 after inoculation. By day 10, all strains caused wilting in more than 50% of the plants, except the four-gene deletion mutant mRP2345, which caused only 35.56% of the plants to develop the disease (Figure 8A). The disease severity index of mRP2345 (24.44%) was also significantly lower than that of the wild-type strain (44.44%; Figure 8B), indicating that the deletion of the four acp genes significantly reduced the virulence of R. solanacearum.
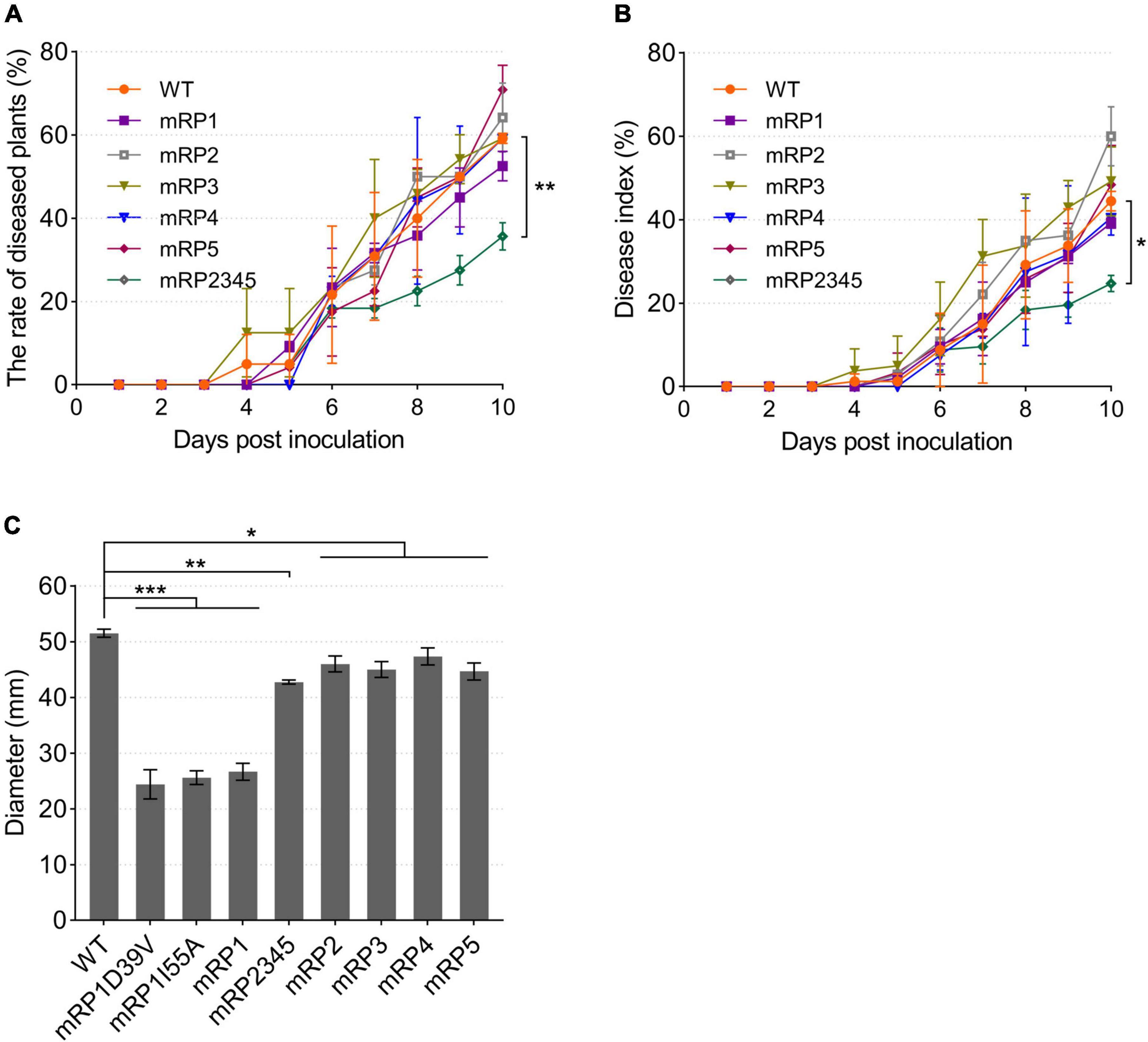
Figure 8. Pathogenicity and swimming motility of R. solanacearum strains. (A,B) Mutant mRP2345 showed significantly reduced virulence in tomato plants. Fresh cultures of the strains were used to infect tomato plants. Once infected, the numbers of diseased plants (left panel) and the disease severity rating for each plant (right panel) were recorded daily. The rate of diseased plants is the ratio of the number of diseased plants to the total number of plants. The disease index (DI), DI = ∑(severity rating × number of plants in that rating)/(total number of plants × 4). (C) The swimming motility patterns of the R. solanacearum strains were assessed on semisolid motility medium. WT, wild-type strain GMI1000; mRP1, mutant in which the acpP1 gene is replaced with E. coli acpP; mRP2, mutant lacking gene acpP2; mRP3, mutant lacking gene acpP3; mRP4, mutant lacking gene acpP4; mRP5, mutant lacking gene acpP5; mRP2345, mutant lacking genes acpP2, acpP3, acpP4, and acpP5; mRP1D39V, mutant in which Asp39 of AcpP1 is mutated to Val; mRP1I55A, mutant in which Ile55 of AcpP1 is mutated to Ala. Significant differences are indicated as ***P < 0.001, **P < 0.01, and *P < 0.05.
The pathogenicity related factors of the R. solanacearum acp mutant strains were also evaluated. First, semisolid plates were used to test the swimming motility of R. solanacearum. Compared with wild-type strain GMI1000, all the mutants formed smaller motility patterns, and the colonial patterns formed by the three acpP1 mutants were only half the size of the wild-type strain pattern (Figure 8C). The biofilms, extracellular cellulases, and extracellular polysaccharides produced by the R. solanacearum mutants were also tested (Supplementary Figure 3). However, these phenotypes were not significantly impaired in mutant mRP2345. Therefore, the mechanisms of action of AcpP2, AcpP3, AcpP4, and AcpP5 in the virulence of R. solanacearum remain to be clarified.
Discussion
The R. solanacearum GMI1000 genome contains five open reading frames, RSc1053, RSc0434, RSp1659, RSp0370, and RSp0369, that encode putative ACPs, AcpP1, AcpP2, AcpP3, AcpP4, and AcpP5, respectively. We confirmed that these ACPs are successfully phosphopantetheinylated and identified the active site at which the Ppant group is linked in these ACPs. Among the five acp genes, acpP1 is most strongly transcribed, suggesting that acpP1 is a housekeeping acp gene. acpP1 is also cotranscribed with the fab cluster and could not be deleted, demonstrated that AcpP1 is necessary for the survival of R. solanacearum. Therefore, like Burkholderia cenocepacia J2315 ACP, the mutation of which reduces its ability to colonize and kill nematodes, R. solanacearum acpP1 can also be considered a potential target for anti-infection drugs (Sílvia et al., 2008). As expectedly, two site-specific mutations in AcpP1 (D39V and I55A) impaired the growth and FAS of R. solanacearum. Asp39 is one of the conserved residues where AcpS binds, and previous research has shown that E. coli became temperature-sensitive when Asp39 of AcpP mutated to Val (De Lay and Cronan, 2007). Ile55 is also a key residue that interacts with the residues from the helices to provide space for the acyl chain and also interacts with FAS proteins (Roujeinikova et al., 2002, 2007). A previous study has shown that E. coli AcpPI54A can be phosphopantetheinylated efficiently (De Lay and Cronan, 2007). Therefore, we infer that the mutant mRP1I55A grew slowly because AcpP1I55A affected the space for the accepting acyl chain or impaired the interaction with R. solanacearum FAS proteins or acyl chains.
The expression of R. solanacearum acpP3 failed to restore the growth of the E. coli acpP-mutant strain CY1877, and the mutation of acpP3 did not affect fatty acid biosynthesis in R. solanacearum. Moreover, a mutant in which acpP3 functionally replaced acpP1 could not be generated. All these data indicate that AcpP3 is not involved in the FAS of R. solanacearum. Furthermore, AcpP3 does not affect the incorporation of exogenous fatty acids in R. solanacearum (data not shown), implying that AcpP3 differs from E. faecalis AcpB, which does not function in FAS but channels exogenous acyl groups (Zhu et al., 2019). However, like AcpP1, AcpP3 can be acylated by V. harveyi AasS, and AcpP3 restored the production of AHL in P. aeruginosa mutant strain PA-A1. Therefore, we infer that gene acpP3 was acquired by horizontal transfer, and AcpP3 cannot interact with FAS proteins of R. solanacearum and E. coli but of some species such as P. aeruginosa and V. harveyi. However, the functions of AcpP3 require further investigation.
Ralstonia solanacearum acpP2 restored the production of xanthomonadins to the X. campestris xanC mutant. acpP2 is located in a large gene cluster in which the genes are highly homologous to those encoded by the X. campestris pig gene cluster, although there is no homolog of the xanB2 gene in R. solanacearum (Cao et al., 2018). These data imply that the gene cluster in which acpP2 is located is involved in the synthesis of a secondary metabolite, similar to the xanthomonadins. acpP4 also restored the production of xanthomonadins to the xanC mutant but is located in another gene cluster, which is similar to the ape cluster in A. baumannii. In the production of APE, the AcpP4 homolog AbApeE acts as a starter, and the AcpP5 homolog AbApeF forms malonyl-ACP, which provides two carbon units for benzoyl-AbApeE (Grammbitter et al., 2019; Lee et al., 2021). However, in this study, we could not purify the AcpP4 protein or test the functions of AcpP4 and AcpP5 in vitro. Therefore, the functions of AcpP4 and AcpP5 require further investigation. Previous studies have shown that five protein residues of E. coli ACP, Phe28, Phe50, Ile54, Ala59, and Tyr71, affected the interaction of ACP and fatty acyl chain (Roujeinikova et al., 2007). In this study, AcpP2 and AcpP5 could not be acylation in vitro (Figure 5C), nor could AcpP2, AcpP4, and AcpP5 involve in FAS in vivo. Except because the possible inability of these three ACPs to interact with FAS proteins, it also may be due to these three ACPs cannot covalently attach to the fatty acyl chain, as most of the above five residues of these three ACPs are different from those of E. coli ACP (Figure 1C).
Single gene R. solanacearum mutants lacking acpP2, acpP3, acpP4, or acpP5 did not affect the pathogenicity of R. solanacearum. However, the pathogenicity of the mutant strain mRP2345, which lacked the acpP2, acpP3, acpP4, and acpP5 genes, was attenuated. This suggests that these acp genes encode redundant functions that allow R. solanacearum to invade its host or gain a competitive advantage in the soil. However, the functions of these ACPs in R. solanacearum require further research.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
YY constructed ACP deleted mutants, tested the pathogenicity of mutant strains, and carried out biochemical studies. RL cloned the ACP genes and constructed vectors. W-TL purified ACP proteins and tested the activity of ACP in vitro. W-BZ and ZH analyzed fatty acids composition of acpP strains. J-CM participated in the design of the study and helped to draft the manuscript. H-HW conceived of the study, and participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant numbers 31671987 and 31972232).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1014971/full#supplementary-material
References
Babu, M., Greenblatt, J. F., Emili, A., Strynadka, N. C., Reithmeier, R. A., and Moraes, T. F. (2010). Structure of a SLC26 anion transporter STAS domain in complex with acyl carrier protein: Implications for E. coli YchM in fatty acid metabolism. Structure 18, 1450–1462. doi: 10.1016/j.str.2010.08.015
Battesti, A., and Bouveret, E. (2006). Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62, 1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x
Beld, J., Sonnenschein, E. C., Vickery, C. R., Noel, J. P., and Burkart, M. D. (2014). The phosphopantetheinyl transferases: Catalysis of a post-translational modification crucial for life. Nat. Prod. Rep. 31, 61–108. doi: 10.1039/c3np70054b
Brozek, K. A., Carlson, R. W., and Raetz, C. R. (1996). A special acyl carrier proteinfor transferring long hydroxylated fatty acids to lipid A in Rhizobium. J. Biol. Chem. 271, 32126–32136. doi: 10.1074/jbc.271.50.32126
Byers, D. M. (1989). Elongation of exogenous fatty acids by the bioluminescent bacterium Vibrio harveyi. J. Bacteriol. 171, 59–64. doi: 10.1128/jb.171.1.59-64.1989
Byers, D. M., and Gong, H. S. (2007). Acyl carrier protein: Structure-function relationships in a conserved multifunctional protein family. Biochem. Cell Biol. 85, 649–662. doi: 10.1139/o07-109
Cao, X. Q., Wang, J. Y., Lian, Z., Bo, C., Yu, J., and He, Y. W. (2018). Biosynthesis of the yellow xanthomonadin pigments involves an ATP-dependent 3-hydroxybenzoic acid: Acyl carrier protein ligase and an unusual type II polyketide synthase pathway. Mol. Microbiol. 110, 16–32. doi: 10.1111/mmi.14064
Castillo, J. A., and Greenberg, J. T. (2007). Evolutionary dynamics of Ralstonia solanacearum. Appl. Environ. Microbiol. 73, 1225–1238. doi: 10.1128/AEM.01253-06
Chen, W., Wang, B., Gruber, J. D., Zhang, Y. M., and Davies, C. (2018). Acyl carrier protein 3 is involvedin oxidative stress responsein Pseudomonas aeruginosa. Front. Microbiol. 9:2244. doi: 10.3389/fmicb.2018.02244
Chen, Y. C., Hu, Z., Zhang, W. B., Yin, Y., Zhong, C. Y., Mo, W. Y., et al. (2022). HetI-like phosphopantetheinyl transferase posttranslationally modifies acyl carrier proteins in Xanthomonas. Mol. Plant Microbe Interact. 35, 323–335. doi: 10.1094/MPMI-10-21-0249-R
Cheng, J. L., Ma, J. C., Lin, J. S., Fan, Z. C., Cronan, J. E., and Wang, H. H. (2012). Only one of the five Ralstonia solanacearum long-chain 3-ketoacyl-acyl carrier protein synthase homologues functions in fatty acid synthesis. Appl. Environ. Microbiol. 78, 1563–1573. doi: 10.1128/AEM.07335-11
Cronan, J. E., and Thomas, J. (2009). Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 459, 395–433. doi: 10.1016/S0076-6879(09)04617-5
Davila-Martinez, Y., Ramos-Vega, A. L., Contreras-Martinez, S., Encarnacion, S., Geiger, O., and Lopez-Lara, I. M. (2010). SMc01553 is the sixth acyl carrier protein in Sinorhizobium meliloti 1021. Microbiology 156, 230–239. doi: 10.1099/mic.0.033480-0
De Lay, N. R., and Cronan, J. E. (2007). In Vivo functional analyses of the type II acyl carrier proteins of fatty acid biosynthesis. J. Biol. Chem. 282, 20319–20328. doi: 10.1074/jbc.M703789200
Demont, N., Debelle, F., Aurelle, H., Denarie, J., and Prome, J. C. (1993). Role of the Rhizobium meliloti nodF and nodE genes in the biosynthesis of lipo-oligosaccharidic nodulation factors. J. Biol. Chem. 268, 20134–20142. doi: 10.1016/S0021-9258(20)80704-2
Elovson, J., and Vagelos, P. R. (1968). Acyl carrier protein. X. Acyl carrier protein synthetase. J. Biol. Chem. 243, 3603–3611. doi: 10.1016/S0021-9258(19)34183-3
Epple, G., van der Drift, K. M., Thomas-Oates, J. E., and Geiger, O. (1998). Characterization of a novel acyl carrier protein, RkpF, encoded by an operon involved in capsular polysaccharide biosynthesis in Sinorhizobium meliloti. J. Bacteriol. 180, 4950–4954. doi: 10.1128/JB.180.18.4950-4954.1998
Feng, S. X., Ma, J. C., Yang, J., Hu, Z., Zhu, L., Bi, H. K., et al. (2015). Ralstonia solanacearum fatty acid composition is determined by interaction of two 3-ketoacyl-acyl carrier protein reductases encoded on separate replicons. BMC Microbiol. 15:223. doi: 10.1186/s12866-015-0554-x
Fischl, A. S., and Kennedy, E. P. (1990). Isolation and properties of acyl carrier protein phosphodiesterase of Escherichia coli. J. Bacteriol. 172, 5445–5449. doi: 10.1128/jb.172.9.5445-5449.1990
Flavier, A. B., Clough, S. J., Schell, M. A., and Denny, T. P. (1997). Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26, 251–259. doi: 10.1046/j.1365-2958.1997.5661945.x
Flugel, R. S., Hwangbo, Y., Lambalot, R. H., Cronan, J. E., and Walsh, C. T. (2000). Holo-(acyl carrier protein) synthase and phosphopantetheinyl transfer in Escherichia coli. J. Biol. Chem. 275, 959–968. doi: 10.1074/jbc.275.2.959
Grammbitter, G. L. C., Schmalhofer, M., Karimi, K., Shi, Y. M., Schöner, T. A., Tobias, N. J., et al. (2019). An uncommon type II PKS catalyzes biosynthesis of aryl polyene pigments. J. Am. Chem. Soc. 141, 16615–16623. doi: 10.1021/jacs.8b10776
Gully, D., Moinier, D., Loiseau, L., and Bouveret, E. (2003). New partners of acyl carrier protein detected in Escherichia coli by tandem affinity purification. FEBS Lett. 548, 90–96. doi: 10.1016/S0014-5793(03)00746-4
Guo, Q. Q., Zhang, W. B., Zhang, C., Song, Y. L., Liao, Y. L., Ma, J. C., et al. (2018). Characterization of 3-oxacyl-acyl carrier protein reductase homologue genes in Pseudomonas aeruginosa PAO1. Front. Microbiol. 10:028.
Hayward, A. C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29, 65–87.
Jiang, Y., Chan, C. H., and Cronan, J. E. (2006). The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry 45, 10008–10019. doi: 10.1021/bi060842w
Kai, K., Ohnishi, H., Shimatani, M., Ishikawa, S., Mori, Y., Kiba, A., et al. (2015). Methyl 3-hydroxymyristate, a diffusible signal mediating phc quorum sensing in Ralstonia solanacearum. Chem. Bio. Chem. 16, 2309–2318. doi: 10.1002/cbic.201500456
Lee, W. C., Choi, S., Jang, A., Son, K., and Kim, Y. (2021). Structural comparison of Acinetobacter baumannii β-ketoacyl-acyl carrier protein reductases in fatty acid and aryl polyene biosynthesis. Sci. Rep. 11:7945. doi: 10.1038/s41598-021-86997-3
López-Lara, I. M., and Geiger, O. (2000). Expression and purification offour different rhizobial acyl carrier proteins. Microbiology 146, 839–849. doi: 10.1099/00221287-146-4-839
Lowe-Power, T. M., Khokhani, D., and Allen, C. (2018). How Ralstonia solanacearum exploits and thrives in the flowing plant xylem environment. Trends Microbiol. 26, 929–942. doi: 10.1016/j.tim.2018.06.002
Ma, J. C., Wu, Y. Q., Cao, D., Zhang, W. B., and Wang, H. H. (2017). Only acyl carrier protein 1 (AcpP1) functions in Pseudomonas aeruginosa fatty acid synthesis. Front. Microbiol. 8:2186. doi: 10.3389/fmicb.2017.02186
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Mao, Y. H., Ma, J. C., Li, F., Hu, Z., and Wang, H. H. (2015). Ralstonia solanacearum RSp0194 encodes a novel 3-keto-acyl carrier protein synthase III. PLoS One 10:e0136261. doi: 10.1371/journal.pone.0136261
Mootz, H. D., Finking, R., and Marahiel, M. A. (2001). 4’-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J. Biol. Chem. 276, 37289–37298. doi: 10.1074/jbc.M103556200
Morel, A., Peeters, N., Vailleau, F., Barberis, P., Jiang, G., Berthomé, R., et al. (2018). “Plant pathogenicity phenotyping of Ralstonia solanacearum strains,” in Host-Pathogen Interactions. Methods in Molecular Biology, eds C. Medina and F. López-Baena (New York, NY: Humana Press). doi: 10.1007/978-1-4939-7604-1_18
Paitan, Y., Orr, E., Ron, E. Z., and Rosenberg, E. (1999). Genetic and functional analysis of genes required for the post-modification of the polyketide antibiotic TA of Myxococcus xanthus. Microbiology 145, 3059–3067. doi: 10.1099/00221287-145-11-3059
Peeters, N., Guidot, A., Vailleau, F., and Valls, M. (2013). Ralstonia solanacearum, a wide spread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 14, 651–662. doi: 10.1111/mpp.12038
Petrovics, G., Putnoky, P., Reuhs, B., Kim, J., Thorp, T. A., Noel, K. D., et al. (1993). The presence of a novel type surface polysaccharide in Rhizobium meliloti requires a new fatty acid synthase-like gene cluster involved in symbiotic nodule development. Mol. Microbiol. 8, 1083–1094. doi: 10.1111/j.1365-2958.1993.tb01653.x
Piper, K. P., Beck von Bodman, S., and Farrand, S. K. (1993). Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362, 448–450. doi: 10.1038/362448a0
Prescott, D. J., and Vagelos, P. R. (1972). Acyl carrier protein. Adv. Enzymol. Relat. Areas Mol. Biol. 36, 269–311. doi: 10.1002/9780470122815.ch8
Quadri, L. E., Weinreb, P. H., Lei, M., Nakano, M. M., Zuber, P., and Walsh, C. T. (1998). Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37, 1585–1595. doi: 10.1021/bi9719861
Ramos-Vega, A. L., Davila-Martinez, Y., Sohlenkamp, C., Contreras-Martinez, S., Encarnacion, S., Geiger, O., et al. (2009). SMb20651 is another acyl carrier protein from Sinorhizobium meliloti. Microbiology 155, 257–267. doi: 10.1099/mic.0.022079-0
Rock, C. O., and Cronan, J. E. (1979). Re-evaluation of the solution structure of acyl carrier protein. J. Biol. Chem. 254, 9778–9785. doi: 10.1016/S0021-9258(19)83584-6
Roujeinikova, A., Baldock, C., Simon, W. J., Gilroy, J., Baker, P. J., Stuitje, A. R., et al. (2002). X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site. Structure 10, 825–835. doi: 10.1016/S0969-2126(02)00775-X
Roujeinikova, A., Simon, W. J., Gilroy, J., Rice, D. W., Rafferty, J. B., and Slabas, A. R. (2007). Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J. Mol. Biol. 365, 135–145. doi: 10.1016/j.jmb.2006.09.049
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502. doi: 10.1038/415497a
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Pühler, A. (1994). Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Sílvia, A. S., Christian, G. R., Filipe, A., Luís, M., Julia, W., Stephan, S., et al. (2008). Burkholderia cenocepacia J2315 acyl carrier protein: A potential target for antimicrobials’ development? Microb. Pathog. 45, 331–336. doi: 10.1016/j.micpath.2008.08.002
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA 11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Therisod, H., and Kennedy, E. P. (1987). The function of acyl carrier proteinin the synthesis of membrane-derived oligosaccharides does not requireits phosphopantetheine prosthetic group. Proc. Natl. Acad. Sci. U.S.A. 84, 8235–8238. doi: 10.1073/pnas.84.23.8235
Thomas, J., and Cronan, J. E. (2005). The enigmatic acyl carrier protein phosphodiesterase of Escherichia coli: Genetic and enzymological characterization. J. Biol. Chem. 280, 34675–34683. doi: 10.1074/jbc.M505736200
Ulrich, A. K., de Mendoza, D., Garwin, J. L., and Cronan, J. E. (1983). Genetic and biochemical analyses of Escherichia coli mutants altered in the temperature-dependent regulation of membrane lipid composition. J. Bacteriol. 154, 221–230. doi: 10.1128/jb.154.1.221-230.1983
Vasse, J., Frey, P., and Trigalet, A. (1995). Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol. Plant Microbe Interact. 8, 241–251. doi: 10.1094/MPMI-8-0241
White, S. W., Zheng, J., Zhang, Y. M., and Rock, C. O. (2005). The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74, 791–831. doi: 10.1146/annurev.biochem.74.082803.133524
Zhu, L., Lin, J. S., Ma, J. C., Cronan, J. E., and Wang, H. H. (2010). Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob. Agents Chemother. 54, 689–698. doi: 10.1128/AAC.01152-09
Keywords: Ralstonia solanacearum, acyl carrier protein, fatty acid biosynthesis, pathogenicity, polyketide synthesis
Citation: Yin Y, Li R, Liang W-T, Zhang W-B, Hu Z, Ma J-C and Wang H-H (2022) Of its five acyl carrier proteins, only AcpP1 functions in Ralstonia solanacearum fatty acid synthesis. Front. Microbiol. 13:1014971. doi: 10.3389/fmicb.2022.1014971
Received: 09 August 2022; Accepted: 06 September 2022;
Published: 23 September 2022.
Edited by:
Huangen Ding, Louisiana State University, United StatesReviewed by:
Hongkai Bi, Nanjing Medical University, ChinaMaxuel Andrade, National Center for Research in Energy and Materials, Brazil
Copyright © 2022 Yin, Li, Liang, Zhang, Hu, Ma and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai-Hong Wang, d2FuZ2hoMzZAc2NhdS5lZHUuY24=; Jin-Cheng Ma, bWFqaW5jaGVuZ0BzY2F1LmVkdS5jbg==
 Yu Yin
Yu Yin Rui Li
Rui Li Wen-Bin Zhang
Wen-Bin Zhang Zhe Hu
Zhe Hu Jin-Cheng Ma
Jin-Cheng Ma Hai-Hong Wang
Hai-Hong Wang