
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 September 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1014872
Leuconostoc species are important microorganisms in food fermentation but also cause food spoilage. Although these species are commercially important, their taxonomy is still based on inaccurate identification methods. Here, we used computational pangenome analysis to develop a real-time PCR-based method for identifying and differentiating the 12 major Leuconostoc species found in food. Analysis of pan and core-genome phylogenies showed clustering of strains into 12 distinct groups according to the species. Pangenome analysis of 130 Leuconostoc genomes from these 12 species enabled the identification of each species-specific gene. In silico testing of the species-specific genes against 143 publicly available Leuconostoc and 100 other lactic acid bacterial genomes showed that all the assays had 100% inclusivity/exclusivity. We also verified the specificity for each primer pair targeting each specific gene using 23 target and 124 non-target strains and found high specificity (100%). The sensitivity of the real-time PCR method was 102 colony forming units (CFUs)/ml in pure culture and spiked food samples. All standard curves showed good linear correlations, with an R2 value of ≥0.996, suggesting that screened targets have good specificity and strong anti-interference ability from food sample matrices and non-target strains. The real-time PCR method can be potentially used to determine the taxonomic status and identify the Leuconostoc species in foods.
The genus Leuconostoc belongs to the Lactobacillaceae family, also known as lactic acid bacteria. They inhabit several food sources, such as vegetables, silage, fruits, meat, fish, and dairy products (de Paula et al., 2015). Leuconostoc species can metabolize numerous sugars, alcohols, and carbohydrates and are used as a flavor starter in many fermented products (Guglielmotti et al., 2022). Specifically, they improve the physicochemical properties of fermented foods by producing organic acids, volatile compounds, and CO2, which contribute to the texture and flavor of dairy products (cheese, butter, and cream) (de Paula et al., 2015). Therefore, Leuconostoc species are important food microorganisms that positively influence food fermentation. However, certain species have detrimental effects and cause food spoilage (Hemme and Foucaud-Scheunemann, 2004). For example, some Leu. mesenteroides strains were considered opportunistic pathogens causing pulmonary infection and peritonitis and some Leu. gasicomitatum strains were identified as spoilage organism for broiler meat strips (Susiluoto et al., 2003; Menegueti et al., 2018). Leuconostoc species have traditionally been used as a probiotic candidates due to their ability to survive in the unfavorable conditions of the gastrointestinal tract (Björkroth et al., 2014). Although some Leuconostoc species are considered safe for use in the food industry and are known as “generally recognized as safe (GRAS)” organisms, they have been linked with diseases in immunocompromised patients (Kumar et al., 2022).
After undergoing several re-arrangements, the taxonomy of Leuconostoc currently includes 17 species,1 of which the following 12 were primarily isolated from food matrices: Leuconostoc (Leu.). carnosum, Leu. citreum, Leu. fallax, Leu. gasicomitatum, Leu. gelidum, Leu. holzapfelii, Leu. inhae, Leu. kimchii, Leu. lactis, Leu. mesenteroides, Leu. pseudomesenteroides, and Leu. suionicum (Hemme and Foucaud-Scheunemann, 2004; Padilla-Frausto et al., 2015). This genus has undergone several reclassifications. While some species that were originally classified under the genus Leuconostoc were reclassified as Oenococcus, Fructobacillus, and Weissella, others have been considered heterotypic synonyms (Björkroth et al., 2014; Bello et al., 2022). Most recently, the previously reported Leu. mesenteroides strain was reclassified as Leu. suionicum based on whole-genome-based sequence information (Kaushal and Singh, 2020). Another recent study suggested reclassifying Leu. gelidum subsp. gasicomitatum as Leu. gasicomitatum species (Wu and Gu, 2021). Leuconostoc species are often found in similar habitats as Lactococcus and Lactobacillus related species and were considered intermediates between Lactobacillus and Streptococcus (Kumar et al., 2022). Although Leuconostoc species are widely used along with Lactobacillus, its taxonomy is relatively less explored than Lactobacillus.
The selecting target genes and developing specific primer pairs are crucial factors for achieving accurate real-time PCR results (Martínez-Romero et al., 2018). Several previously described specific genes for Leuconostoc (16S rRNA gene, 23S rRNA gene, and 16S–23S intergenic region, hsp60, and rpoB genes) were considered while designing the specific primers (Kaur et al., 2017; Ricciardi et al., 2020; Guglielmotti et al., 2022). Several authors have found ribosomal genes (16S rRNA and 23S rRNA) problematic as they do not exhibit sufficient variability to allow differentiation between closely related species (Ricciardi et al., 2020; Guglielmotti et al., 2022). In the case of the genus Leuconostoc, a high degree of similarity (98.5 ~ 99.7%) was reported between the gene sequences for 16S rRNA (Wu and Gu, 2021). These authors also reported that some species, such as Leu. gelidum, Leu. gasicomitatum and Leu. inhae exhibit sequence similarities of 99% or higher (99.1–99.7%) between their 16S rRNA gene sequences. These facts highlight the need to find alternative specific genes to identify Leuconostoc species accurately.
Researchers have identified novel DNA markers to replace current, poorly specific markers using whole-genome sequencing-based methods. Whole-genome sequencing is widely used to determine the taxonomic position of microorganisms (Kumar et al., 2022). Recent comparative genomic studies have confirmed that the average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) might suffice to classify Leuconostoc species at the species or subspecies level (Wu and Gu, 2021; Bello et al., 2022). Also, more robust approaches (e.g., pan-and core-genome analysis) were used to account for strain diversity and classify closely related species or subspecies (Kim et al., 2021, 2022b,c). Although this method can provide the resolution necessary to identify bacterial genera or species within food samples, simpler methods such as real-time PCR provide higher resolution at lower cost and shorter testing time (Wang et al., 2022). Real-time PCR methods are advantageous for identification and differentiation compared with whole-genome sequencing as they are rapid, specific, and sensitive and do not require post-PCR processing. The food industry-accepted methods must be easy to use, affordable, and accurately provide species-level resolution.
Here, we developed an easy-to-use and accurate real-time PCR method based on novel marker genes obtained from computational pangenome analysis that can be used to identify the 12 Leuconostoc species that predominantly inhabit food matrices and are essential for food fermentation.
Whole-genomic sequences of 130 strains, including 13 Leu. carnosum, 10 Leu. citreum, 3 Leu. fallax, 14 Leu. gasicomitatum, 14 Leu. gelidum, 2 Leu. holzapfelii, 5 Leu. inhae, 2 Leu. kimchii, 12 Leu. lactis, 34 Leu. mesenteroides, 14 Leu. pseudomesenteroides, and 7 Leu. suionicum were retrieved from the National Center for Biotechnology Information (NCBI) database (last accessed on April 11, 2022) (Supplementary Table S1). The selection criteria for involving only 130 strain genome sequence data are as follows: the genome assembled at the complete level was preferentially used, and the species without complete genomes was used in order of scaffold and contig.
To ensure proper classification of all genomes, the pangenome and phylogenetic analysis were performed using the bacterial pangenome analysis (BPGA) tool version 1.3.0 (Chaudhari et al., 2016). Protein-coding gene sequences from each genome were used as input for the analysis. The identity cut-off was used as the default value (0.5) for the similarity calculation (Lim et al., 2021). MUSCLE (Multiple Sequence Comparison by Log-Expectation), a tool built into BPGA, was used for the phylogeny analysis. Core-genomic and pangenomic phylogenies were constructed based on linked core gene alignments and pan matrix (binary gene presence/absence matrix) and visualized using the interactive tree of life program version 6 (Letunic and Bork, 2021). The ANI was calculated using JSpeciesWS version 3.9.5 (Richter et al., 2016).
The species-specific genes were identified using a pangenome analysis pipeline. Briefly, assembled genomes were annotated using Prokka version 1.14.5 (Seemann, 2014), and these annotated assemblies, obtained in GFF3 format, were applied to calculate the pangenome analysis using Roary version 3.11.2 (Page et al., 2015). The pangenome was analyzed for each genome using a BLASTP identity cut-off of 95%. The presence/absence profiles of genes were converted into a 0/1 matrix using a script available with Roary. The matrix was employed to screen Leuconostoc species-specific genes according to the following criteria: 100% presence in target genomes and absence in non-target genomes. The identified specific genes were further selected from the whole-genome shotgun contigs databases using the nucleotide basic local alignment search tool (BLAST) version 2.13.0 with default parameters (word_size = 28, expect threshold = 0.05, and max matches in a query range = 0). The primers targeting each species-specific gene were designed using Primer Designer program (Scientific and Education Software, Durham, NC, United States).
We used 147 reference strains that were purchased from the Korean Collection for Type Cultures (KCTC, Daejeon, South Korea), the Korean Culture Center of Microorganisms (KCCM, Seoul, South Korea), the Korean Agricultural Culture Collection (KACC, Jeonju, South Korea), and the Korean Collection for Kimchi Microorganisms (KCKM, Gwangju, South Korea) (Supplementary Table S2).
The reference strains were cultured in MRS broth at 37°C for 48 h. The cells were collected by centrifugation at 13,000 × g for 5 min, and DNA was extracted using the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The concentration of the extracted DNA was estimated using the Maestro Nano-spectrophotometer (Maestrogen, Las Vegas, NV, United States). The genomic DNA was stored at −20°C until real-time PCR analysis.
A real-time PCR assay was performed in CFX96 Deep Well Real-Time System (Bio-Rad, Hercules, CA, United States) using a mixture consisting of 10 μl 2× A-Star Master Mix (BioFACT, Daejaon, South Korea), 1 μl of each primer pair (500 nM), 50 ng of template DNA, and distilled water up to a final volume of 20 μl. The real-time PCR conditions were initial denaturation at 95°C for 5 min, followed by 35 cycles at 95°C for 5 s and 60°C for 30 s. Melting curves were constructed by continuously increasing the temperature from 65°C to 95°C in 0.5°C increments, at 5 s per step. The real-time PCR assay was performed in triplicate.
We evaluated the specificity and amplification efficiency of the developed real-time PCR method, which helped us determine the lowest detectable DNA concentration.
The inclusivity/exclusivity of primer pairs was first assessed by in silico PCR2 analysis with genome sequence data of 133 target and 110 non-target strains obtained from GenBank. The specificity of the primer pairs was also evaluated using pure bacterial DNA. Genomic DNA was extracted from 147 lactic acid bacterial strains and used as a template.
Standard curves were established using serially diluted target bacterial strains ranging from 108 to 101 CFU/ml (Gómez-Rojo et al., 2015). For genomic DNA extraction, 1 ml of each dilution was taken and plated on lactobacilli MRS agar (Difco, Detroit, MI, United States) to determine the correlation between CFU/ml and the Ct value (Bustin et al., 2009). The amplification efficiency was calculated based on the formula: Efficiency = 101/slope−1 (Bustin et al., 2009).
Pork, lettuce, and pasteurized milk were purchased from the local markets in Korea. All samples were previously tested for the presence of 12 Leuconostoc species by real-time PCR. When not all Leuconostoc species were detected, these three samples were used for spiking with Leuconostoc species to prepare the contaminated samples. The cultured bacterial strains were diluted to concentrations from 108 to 101 CFU/ml with phosphate buffered saline (PBS). The cocktail of 12 Leuconostoc reference strains at concentration of 108 to 101 CFU/ml each was prepared. The bacterial cell number in the dilutions was confirmed by plate counting method. For pork meat and lettuce samples, 25 g of each sample was placed in stomacher bag and the cocktail was inoculated. After standing for 10 min, samples were homogenized with 225 ml PBS. Non-inoculated food samples were used as a negative control through the same procedure. The DNA extraction was conducted according to the method described in section “Bacterial strains” without additional incubation procedures and then analyzed using real-time PCR under the conditions described in section “Evaluation of real-time PCR assay.”
Bacterial pangenome analysis was used to construct phylogenetic tree based on pan and core-genome to confirm that the taxonomic labels of genomes used for analysis were correct. Roary was used to identify species-specific genes based on gene presence/absence matrix.
A total of 130 Leuconostoc genomes were obtained from the GenBank database, and the genomes were clustered using a phylogenetic tree based on pan and core genomes (Figure 1). Both methods subdivided the samples into 12 large clusters according to their species name Four genomes did not cluster with the same species, suggesting the species name was incorrect. These were Leu. inhae strains, which were Leu. gasicomitatum. The cluster containing Leu. gelidum group species was divided into Leu. gelidum, Leu. gasicomitatum, Leu. inhae, and Leu. kimchii strains. However, the Leu. inhae PB1a (GCA_900016185.1), KSL4-2 (GCA_900016165.1), PL111 (GCA_900016205.1), and C120c (GCA_900009505.1) strains clustered with Leu. gasicomitatum strains. In the phylogenetic tree based on pangenome, four Leu. inhae strains (PB1a, KSL4-2, PL111, and C120c) were clustered with Leu. gasicomitatum strains, whereas one Leu. inhae strain (DSM 15101) was clustered with Leu. kimchii. In the phylogenetic tree based on core-genome, four Leu. inhae strains (PB1a, KSL4-2, PL111, and C120c) were clustered with Leu. gasicomitatum strains, which was consistent with the pangenome tree, and Leu. inhae DSM 15101 existed independently between Leu. kimchii and Leu. gelidum. Therefore, a comparison of core and pan-based trees showed differences in order within species cluster but no differences in species assignment, which is consistent with previous study (Akwani et al., 2022).
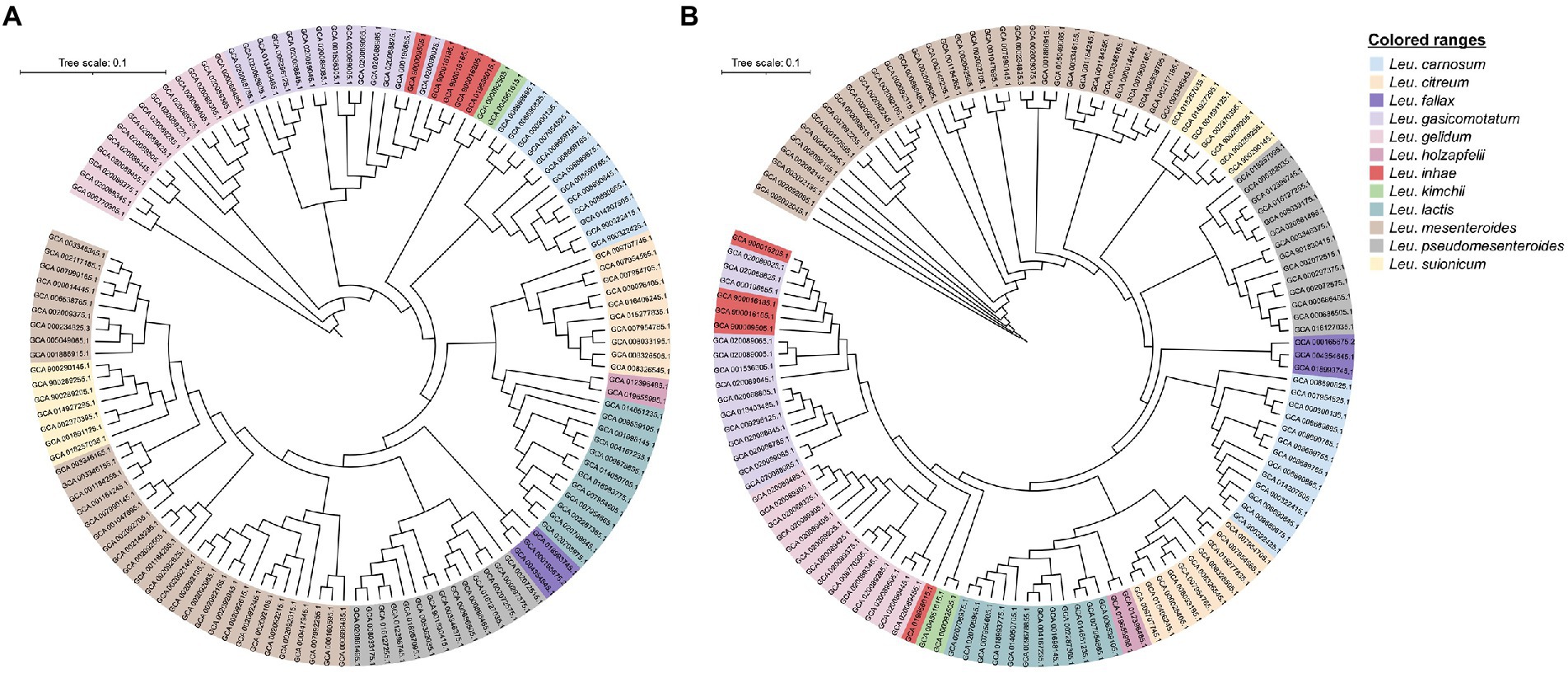
Figure 1. Phylogenetic relationship between 130 Leuconostoc strains. (A) Pangenome phylogenetic tree based on binary panmatrix (gene presence/absence (1/0) matrix). (B) Core-genome phylogenetic tree based on concatenated core gene alignment. Trees were visualized using Interactive Tree of Life software.
Average nucleotide identity analysis showed more than 95% identity among the same species, whereas less than 95% identity between different species (Supplementary Table S3). However, Leu. inhae PB1a, KSL4-2, PL111, and C120c strains had 86.94–87.1% identity with Leu. inhae type strain (DSM 15101T) while 99.04–99.62% identity with Leu. gasicomitatum type strain (LMG 18811T) (Figure 2). Also, Leu. inhae PB1a, KSL4-2, PL111, and C120c strains showed more similarity with other Leu. gasicomitatum genomes (97.72 to 99.96% identities) than Leu. inhae genome (86.94 to 87.1% identities).
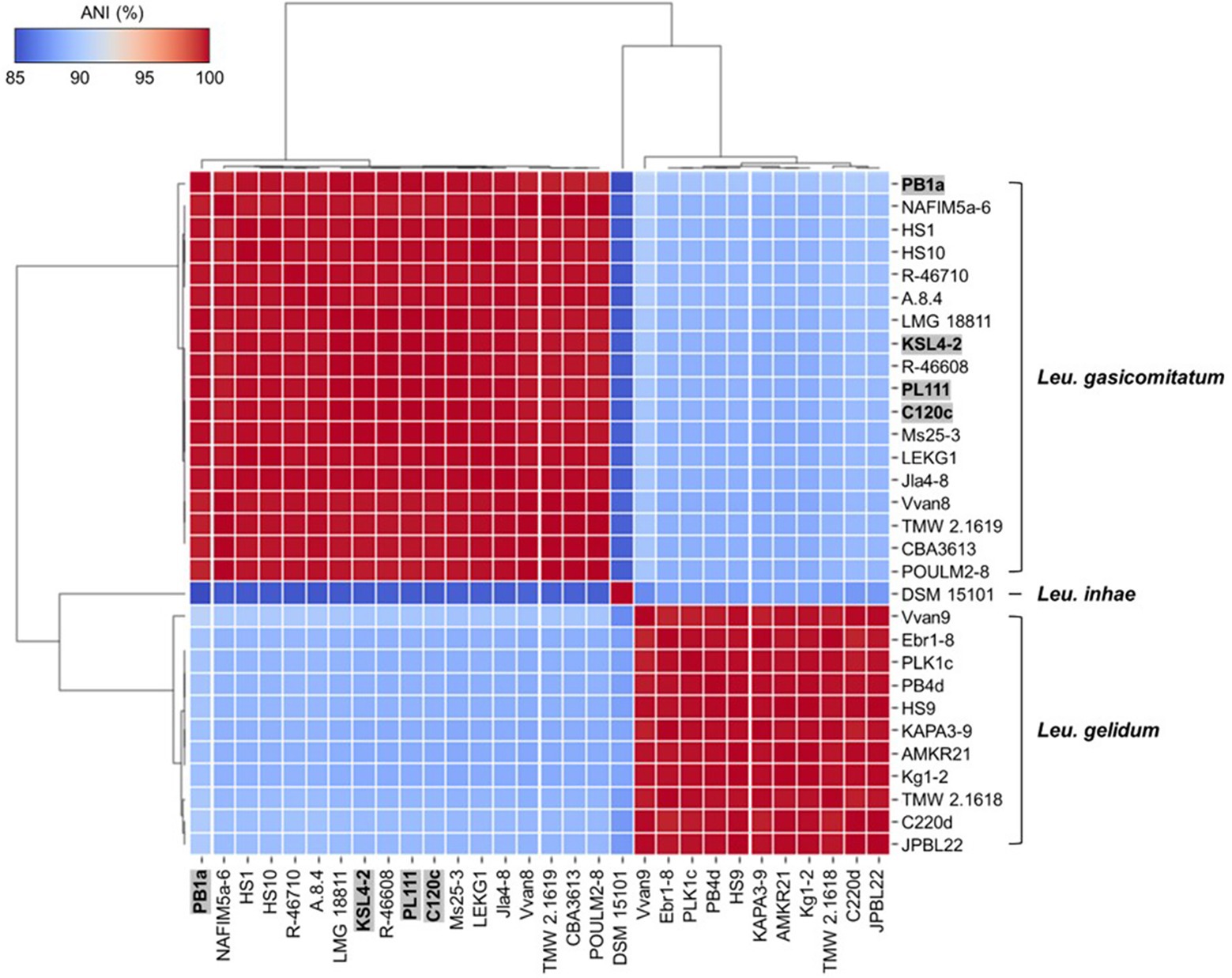
Figure 2. Heatmap of average nucleotide identity (ANI) values among Leu. gelidum, Leu. gasicomitatum, and Leu. inhae strains. Misclassified genomes are highlighted in grey box with bold letters. Strain names and accession numbers are listed in Supplementary Table S1.
Consistently, previous studies reported that incorrectly assigned taxonomic labels for bacterial species are prevalent with reported (Ghosh et al., 2019; Yang et al., 2021; Akwani et al., 2022). For example, Lacticaseibacillus paracasei was misclassified as Lacticaseibacillus casei and Enterococcus lactis as Enterococcus faecium, all of which are closely related species (Ghosh et al., 2019; Kim et al., 2022a). Critically, inaccurate genomic information might impede the development of methods distinguishing Leuconostoc species, so the information for the strains (Leu. inhae PB1a, KSL4-2, PL111, and C120c) should be corrected in the GenBank database to prevent further misidentification.
Recently, PCR methods targeting specific genes were developed from comparative genomics to accurately identify closely related species within genera. Pangenome analysis helps in finding more DNA markers for identifying closely related species (Belloso Daza et al., 2021). Highly specific genes identified via pangenome analysis are useful alternative genetic markers for differentiating closely related species. Thus, molecular assays targeting genetic markers achieve higher resolution within closely related species. The previous researchers have successfully identified for species, subspecies, or serovar-specific marker genes using the presence/absence matrix with a script built into Roary (Bannantine et al., 2021; Shang et al., 2021; Kim et al., 2022c). Here we selected novel species-specific genes that facilitate more accurate identification than the 16S rRNA gene and housekeeping genes using pangenome analysis.
A total of 130 genomes were used for the pangenome analysis and were clustered using gene presence in the accessory genome. The pangenome was screened for Leuconostoc species-specific genes using a gene presence/absence matrix, with species-specific genes defined as being present in 100% of the target species and absence in other species. The number of core genes in each species varied from 781 to 1,939 (Supplementary Table S4). We identified 783 species-specific genes, of which 42, 64, 305, 3, 3, 77, 106, 76, 18, 2, 68, and 19 were specific to Leu. carnosum, Leu. citreum, Leu. fallax, Leu. gasicomitatum, Leu. gelidum, Leu. holzapfelii, Leu. inhae, Leu. kimchii, Leu. lactis, Leu. mesenteroides, Leu. pseudomesenteroides, and Leu. Suionicum, respectively. These were further tested against 84,780,734 sequences using the BLAST nr/nt database and nine genomes representing phylogenetically related Leuconostoc species to target species (i.e., Leu. falkenbergense, Leu. rapi, Leu. litchii, Leu. miyukkimchii, and Leu. palmae). This reduced the number marker genes to 7 Leu. carnosum, 8 Leu. citreum, 34 Leu. fallax, 1 Leu. gasicomitatum, 1 Leu. gelidum, 8 Leu. holzapfelii, 40 Leu. inhae, 16 Leu. kimchii, 3 Leu. lactis, 1 Leu. mesenteroides, 10 Leu. pseudomesenteroides, and 1 Leu. suionicum-specific genes. Among these, we selected genes specific to each species based on their GC content and length (Table 1).
Primer pairs designed from species-specific genes are shown in Table 2. Twelve specific genes were confirmed using in silico PCR with the 143 Leuconostoc genomes and 100 other lactic acid bacteria genomes. For each species, we selected primer pairs representing specific genes of each species that showed 100% inclusivity and exclusivity in the in silico PCR (Supplementary Table S5). The amplicon sizes ranged from 100 to 211 bp. These primers were further tested using real-time PCR.
The specificity of primer pairs was tested using various reference strains available for this study. All target strains produced amplification curves for the corresponding primer pairs. Contrastingly, non-target strains did not produce any amplicons, indicating 100% specificity with no cross-reactivity (Figure 3). This validated the high specificity of the designed primer pairs. The Ct values of the amplification plot ranged from 9.86 to 12.5 (Supplementary Table S6). To calculate the efficiency of primer pairs, we generated standard curves using different concentrations of genomic DNA (108 to 101 CFU/ml) from 12 Leuconostoc species. Real-time PCR can detect up to 102 CFU/ml for all target species (Figure 4). Previous reports showed similar or lower sensitivities than our study for other lactic acid bacterial species, such as Lacticaseibacillus paracasei (102 CFU/ml) and Lactobacillus species (103 CFU/ml) (Jomehzadeh et al., 2020; Hu et al., 2021). To generate valid primer pairs, the slope and correlation coefficient (R2) for the standard curve should be −3.1 to −3.6 and ≥ 0.98, respectively (Broeders et al., 2014). The slops of the linear regression curves for the 12 Leuconostoc species ranged between −3.431 and −3.589, and the amplification efficiencies ranged from 90 to 95.6%, with an R2 value of ≥0.998. These results indicate that our real-time PCR method using species-specific primer pairs has high detection efficiency.
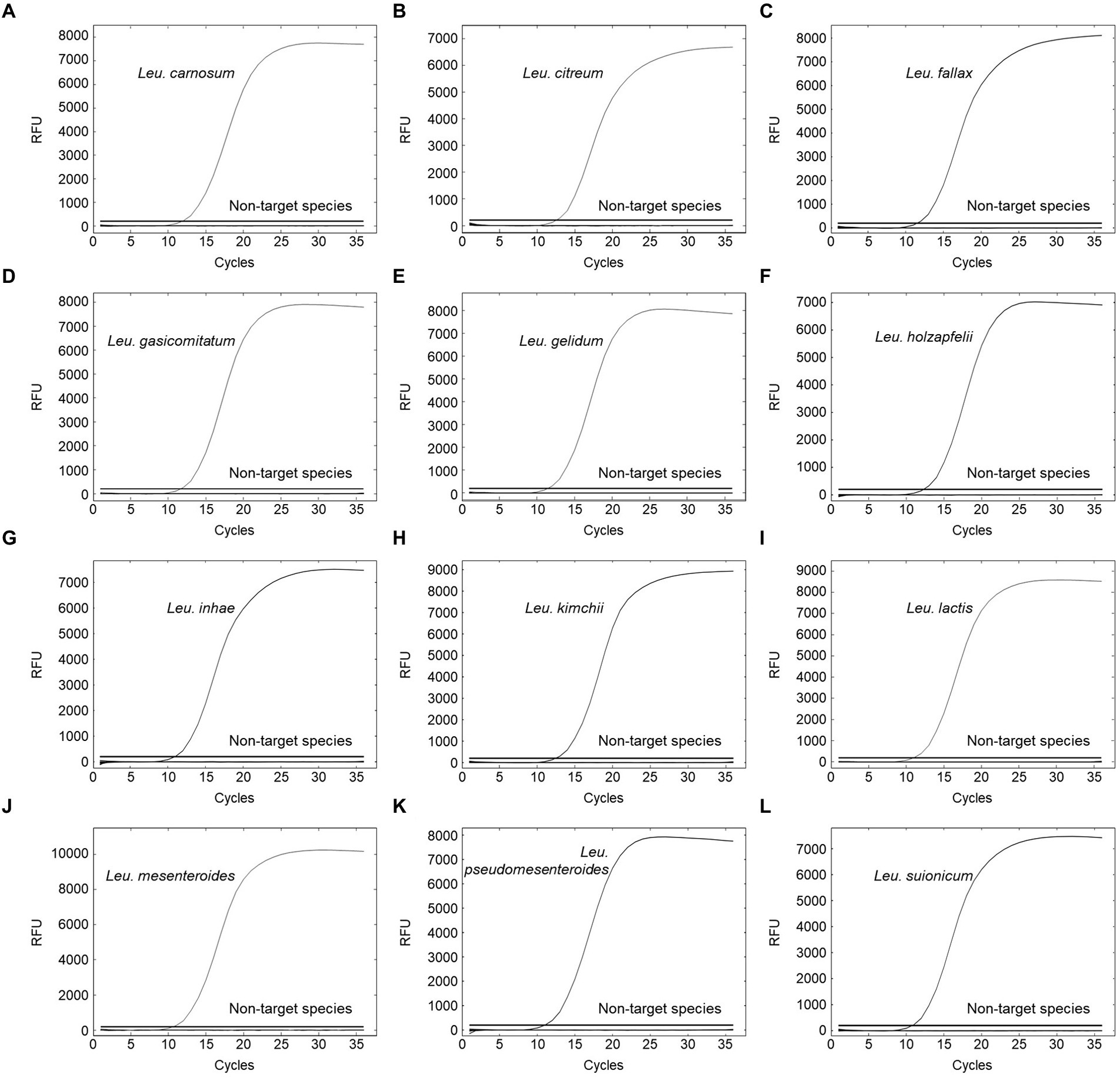
Figure 3. Specificity evaluation for (A) Leu. carnosum primer pair, (B) Leu. citreum primer pair, (C) Leu. fallax primer pair, (D) Leu. gasicomitatum primer pair, (E) Leu. gelidum primer pair, (F) Leu. holzapfelii primer pair, (G) Leu. inhae primer pair, (H) Leu. kimchii primer pair, (I) Leu. lactis primer pair, (J) Leu. mesenteroides primer pair, (K) Leu. pseudomesenteroides primer pair, and (L) Leu. suionicum primer pair. Test samples comprised 23 target strains of Leuconostoc species and 124 non-target strains.
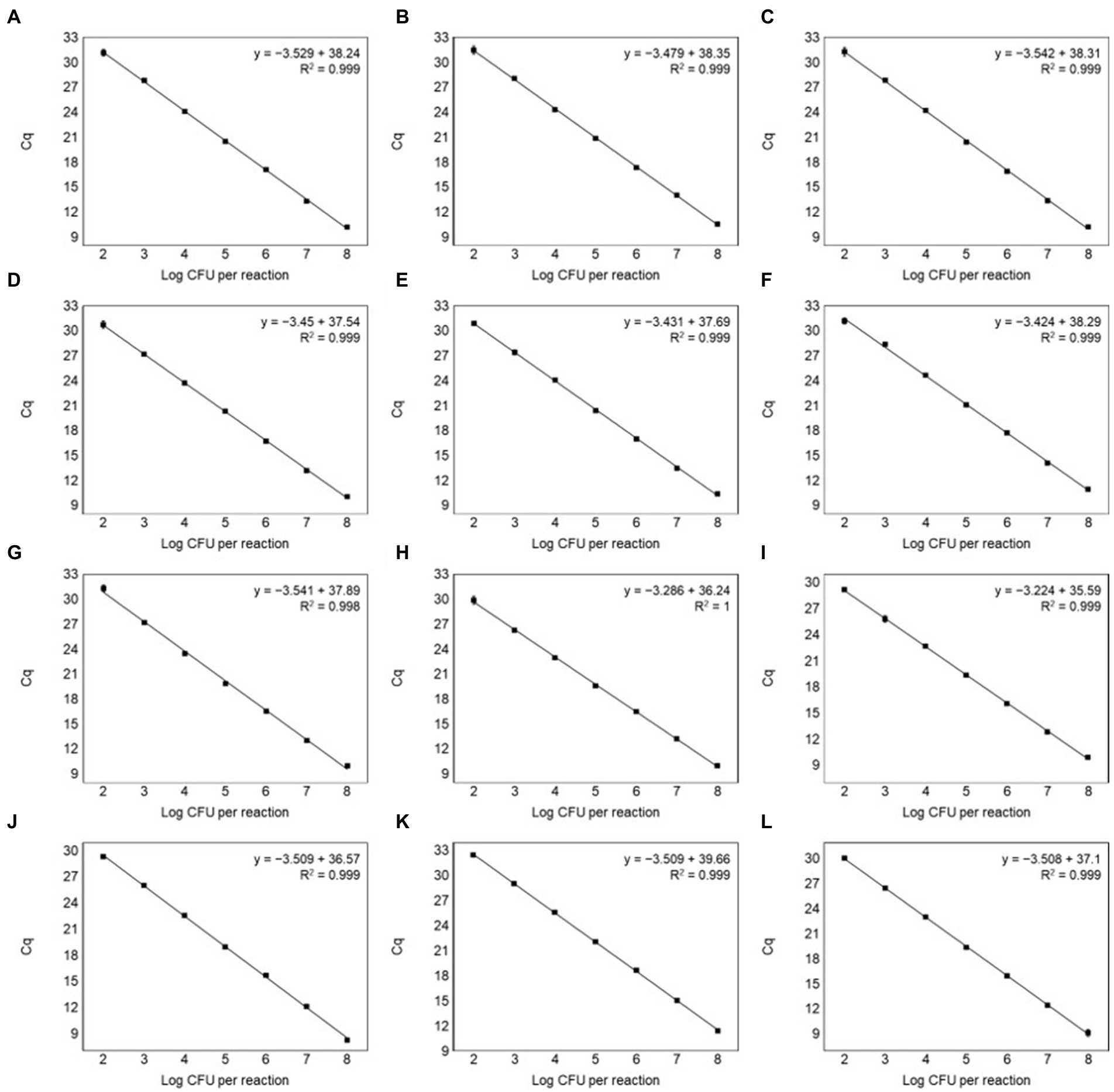
Figure 4. Standard curves by plotting Ct values against the log CFU/ml for (A) Leu. carnosum, (B) Leu. citreum, (C) Leu. fallax, (D) Leu. gasicomitatum, (E) Leu. gelidum, (F) Leu. holzapfelii, (G) Leu. inhae, (H) Leu. kimchii, (I) Leu. lactis, (J) Leu. mesenteroides, (K) Leu. pseudomesenteroides, and (L) Leu. suionicum in inhae pure culture. All samples were tested in triplicate.
To evaluate the suitability of real-time PCR, genomic DNA was extracted from pork, lettuce, and milk samples artificially contaminated with different concentrations of 12 Leuconostoc species. The sensitivity of the target genes in the artificially contaminated food samples was 102 CFU/g, equivalent to pure culture (Supplementary Table S7). Standard curves showed good linear correlations, with R2 values of ≥0.991 in all the samples (Table 3). The real-time PCR efficiencies ranged between 90.9 and 109.9%.
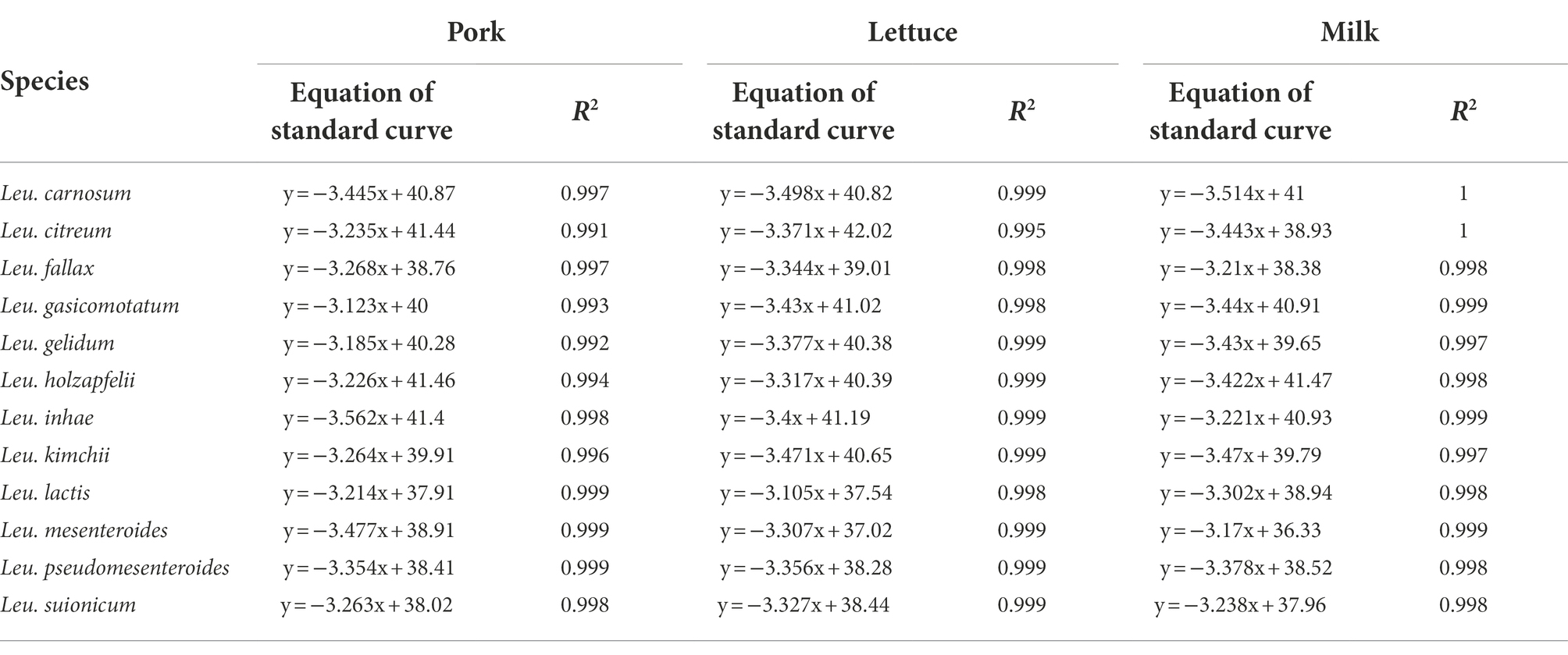
Table 3. The equation and coefficient of correlation values of standard curves in spiked pork, lettuce, and milk samples.
The sensitivity obtained for the food samples artificially inoculated with Leuconostoc species is greater than those reported for other bacterial species in various food samples, such as lactic acid bacteria from cold-smoked salmon (103 CFU/g), Pseudomonas aeruginosa in tomato (103 CFU/g), and Lactobacillus kefiri in kefir milk (103 CFU/g) (Kim et al., 2016; Jérôme et al., 2022; Wang et al., 2022). Contrastingly, we obtained sensitivity similar to that reported for the detection of Weissella viridescens in vacuum-packaged morcilla (102 CFU/ml) (Martins et al., 2020).
As meat, lettuce, and milk are complex foods containing starch, fat, and proteins, the possibility of finding contaminants in the extracted genomic DNA is higher. These components might affect the sensitivity and efficiency of the PCR reactions, as described in previous reports. Our real-time PCR method showed good linearity with regard to the standard curves for 12 Leuconostoc species inoculated into the three food types, suggesting that it is not affected by inhibitors (Zhou et al., 2022). This result indicates that the screened targets have good sensitivity and anti-interference ability for real-time PCR to identify Leuconostoc species rapidly and accurately in artificially spiked food samples.
To our knowledge, this is the first study demonstrating a novel real-time PCR-based method for identifying and discriminating between the major Leuconostoc species found in foods. Our real-time PCR method utilized newly discovered marker genes highly specific for identifying 12 Leuconostoc species, which displayed high specificity and good consistency for Leuconostoc species detection. Our qPCR method enabled rapid, specific, and sensitive Leuconostoc identification. It might be used as an alternative molecular method to identify these Leuconostoc species in food samples and possibly identify novel food-based Leuconostoc strains in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
EK and H-YK contributed to conception and design of this study. EK and S-MY performed pangenome analysis and unique gene extraction. I-SK and S-MY performed the experimental work. S-YL analyzed average nucleotide identity. H-YK supervised the work and reviewed and edited the manuscript. EK prepared a draft manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01662001)” Rural Development Administration, Republic of Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1014872/full#supplementary-material
Akwani, W. C., van Vliet, A. H. M., Joel, J. O., Andres, S., Diricks, M., Maurer, F. P., et al. (2022). The use of comparative genomic analysis for the development of subspecies-specific PCR assays for mycobacterium abscessus. Front. Cell. Infect. Microbiol. 12:816615. doi: 10.3389/fcimb.2022.816615
Bannantine, J. P., Stabel, J. R., Bayles, D. O., Conde, C., and Biet, F. (2021). Diagnostic sequences that distinguish M. avium subspecies strains. Front. Vet. Sci. 7:620094. doi: 10.3389/fvets.2020.620094
Bello, S., Rudra, B., and Gupta, R. S. (2022). Phylogenomic and comparative genomic analyses of Leuconostocaceae species: identification of molecular signatures specific for the genera Leuconostoc, Fructobacillus, and Oenococcus and proposal for a novel genus Periweissella gen. Nov. Int. J. Syst. Evol. Microbiol. 72:005284. doi: 10.1099/ijsem.0.005284
Belloso Daza, M. V., Cortimiglia, C., Bassi, D., and Cocconcelli, P. S. (2021). Genome-based studies indicate that the enterococcus faecium clade b strains belong to enterococcus lactis species and lack of the hospital infection associated markers. Int. J. Syst. Evol. Microbiol. 71:004948. doi: 10.1099/ijsem.0.004948
Björkroth, J., Dicks, L. M. T., Endo, A., and Holzapfel, W. H. (2014). “The genus Leuconostoc” in Lactic acid bacteria: biodiversity and taxonomy, eds. Holzapfel, W. H. and Wood, B. J. B. (Chichester, UK: John Wiley & Sons, Ltd), 391–404. doi: 10.1002/9781118655252.ch23
Broeders, S., Huber, I., Grohmann, L., Berben, G., Taverniers, I., Mazzara, M., et al. (2014). Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 37, 115–126. doi: 10.1016/j.tifs.2014.03.008
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
de Paula, A. T., Jeronymo-Ceneviva, A. B., Todorov, S. D., and Penna, A. L. B. (2015). The two faces of Leuconostoc mesenteroides in food systems. Food Rev. Int. 31, 147–171. doi: 10.1080/87559129.2014.981825
Ghosh, S., Sarangi, A. N., Mukherjee, M., Bhowmick, S., and Tripathy, S. (2019). Reanalysis of lactobacillus paracasei Lbs2 strain and large-scale comparative genomics places many strains into their correct taxonomic position. Microorganisms 7:487. doi: 10.3390/microorganisms7110487
Gómez-Rojo, E. M., Romero-Santacreu, L., Jaime, I., and Rovira, J. (2015). A novel real-time PCR assay for the specific identification and quantification of Weissella viridescens in blood sausages. Int. J. Food Microbiol. 215, 16–24. doi: 10.1016/j.ijfoodmicro.2015.08.002
Guglielmotti, D. M., Pujato, S. A., Quiberoni, A., and Suárez, V. B. (2022). Hsp60 gene as a reliable target for taxonomical identification and discrimination of Leuconostoc species of dairy origin. Int. Dairy J. 126:105227. doi: 10.1016/j.idairyj.2021.105227
Hemme, D., and Foucaud-Scheunemann, C. (2004). Leuconostoc, characteristics, use in dairy technology and prospects in functional foods. Int. Dairy J. 14, 467–494. doi: 10.1016/j.idairyj.2003.10.005
Hu, L., Xue, Y., Cui, L., Zhang, D., Feng, L., Zhang, W., et al. (2021). Detection of viable Lacticaseibacillus paracasei in fermented milk using propidium monoazide combined with quantitative loop-mediated isothermal amplification. FEMS Microbiol. Lett. 368:fnab148. doi: 10.1093/femsle/fnab148
Jérôme, M., Passerini, D., Chevalier, F., Marchand, L., Leroi, F., and Macé, S. (2022). Development of a rapid qPCR method to quantify lactic acid bacteria in cold-smoked salmon. Int. J. Food Microbiol. 363:109504. doi: 10.1016/j.ijfoodmicro.2021.109504
Jomehzadeh, N., Javaherizadeh, H., Amin, M., Rashno, M., and Teimoori, A. (2020). Quantification of intestinal lactobacillus species in children with functional constipation by quantitative real-time PCR. Clin. Exp. Gastroenterol. 13, 141–150. doi: 10.2147/CEG.S250755
Kaur, J., Lee, S., Park, Y. S., and Sharma, A. (2017). RAPD analysis of Leuconostoc mesenteroides strains associated with vegetables and food products from Korea. Food Sci. Technol. 77, 383–388. doi: 10.1016/j.lwt.2016.11.078
Kaushal, G., and Singh, S. P. (2020). Comparative genome analysis provides shreds of molecular evidence for reclassification of Leuconostoc mesenteroides MTCC 10508 as a strain of Leu. Suionicum. Genomics 112, 4023–4031. doi: 10.1016/j.ygeno.2020.06.040
Kim, D. H., Kang, I. B., Jeong, D., Kim, H., Kim, H. S., Lee, S. K., et al. (2016). Development of rapid and highly specific TaqMan probe-based real-time PCR assay for the identification and enumeration of lactobacillus kefiri in kefir milk. Int. Dairy J. 61, 18–21. doi: 10.1016/j.idairyj.2016.03.007
Kim, E., Kim, D.-S., Yang, S.-M., and Kim, H.-Y. (2022a). The accurate identification and quantification of six enterococcus species using quantitative polymerase chain reaction based novel DNA markers. LWT 166:113769. doi: 10.1016/j.lwt.2022.113769
Kim, E., Kim, D., Yang, S. M., and Kim, H. Y. (2022b). Validation of probiotic species or subspecies identity in commercial probiotic products using high-resolution PCR method based on large-scale genomic analysis. Food Res. Int. 154:111011. doi: 10.1016/j.foodres.2022.111011
Kim, E., Yang, S. M., Kim, D., and Kim, H. Y. (2021). Real-time PCR method for qualitative and quantitative detection of lactobacillus sakei group species targeting novel markers based on bioinformatics analysis. Int. J. Food Microbiol. 355:109335. doi: 10.1016/j.ijfoodmicro.2021.109335
Kim, E., Yang, S. M., Kim, I. S., and Kim, H. Y. (2022c). Identification of novel molecular targets for Weissella species-specific real-time PCR based on pangenome analysis. Appl. Microbiol. Biotechnol. 106, 4157–4168. doi: 10.1007/s00253-022-12003-z
Kumar, S., Bansal, K., and Sethi, S. K. (2022). Comparative genomics analysis of genus Leuconostoc resolves its taxonomy and elucidates its biotechnological importance. Food Microbiol. 106:104039. doi: 10.1016/j.fm.2022.104039
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Lim, J., Park, H. T., Ko, S., Park, H. E., Lee, G., Kim, S., et al. (2021). Genomic diversity of Mycobacterium avium subsp. paratuberculosis: pangenomic approach for highlighting unique genomic features with newly constructed complete genomes. Vet. Res. 52:46. doi: 10.1186/s13567-021-00905-1
Martínez-Romero, E., Rodríguez-Medina, N., Beltrán-Rojel, M., Silva-Sánchez, J., Barrios-Camacho, H., Pérez-Rueda, E., et al. (2018). Genome misclassification of Klebsiella variicola and Klebsiella quasipneumoniae isolated from plants, animals and humans. Salud Publica Mex. 60, 56–62. doi: 10.21149/8149
Martins, W. F., Longhi, D. A., de Aragão, G. M. F., Melero, B., Rovira, J., and Diez, A. M. (2020). A mathematical modeling approach to the quantification of lactic acid bacteria in vacuum-packaged samples of cooked meat: combining the TaqMan-based quantitative PCR method with the plate-count method. Int. J. Food Microbiol. 318:108466. doi: 10.1016/j.ijfoodmicro.2019.108466
Menegueti, M. G., Gaspar, G. G., Laus, A. M., Basile-Filho, A., Bellissimo-Rodrigues, F., and Auxiliadora-Martins, M. (2018). Bacteremia by Leuconostoc mesenteroides in an immunocompetent patient with chronic Chagas disease: a case report. BMC Infect. Dis. 18:547. doi: 10.1186/s12879-018-3452-7
Padilla-Frausto, J. J., Cepeda-Marquez, L. G., Salgado, L. M., Iturriaga, M. H., and Arvizu-Medrano, S. M. (2015). Detection and genotyping of Leuconostoc spp. in a sausage processing plant. J. Food Prot. 78, 2170–2176. doi: 10.4315/0362-028X.JFP-15-192
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Ricciardi, A., Storti, L. V., Zotta, T., Felis, G. E., and Parente, E. (2020). Analysis of rpoB polymorphism and PCR-based approaches for the identification of Leuconostoc mesenteroides at the species and subspecies level. Int. J. Food Microbiol. 318:108474. doi: 10.1016/j.ijfoodmicro.2019.108474
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shang, Y., Ye, Q., Wu, Q., Pang, R., Xiang, X., Wang, C., et al. (2021). PCR identification of salmonella serovars for the E serogroup based on novel specific targets obtained by pan-genome analysis. LWT 145:110535. doi: 10.1016/j.lwt.2020.110535
Susiluoto, T., Korkeala, H., and Björkroth, K. J. (2003). Leuconostoc gasicomitatum is the dominating lactic acid bacterium in retail modified-atmosphere-packaged marinated broiler meat strips on sell-by-day. Int. J. Food Microbiol. 80, 89–97. doi: 10.1016/S0168-1605(02)00123-X
Wang, C., Ye, Q., Jiang, A., Zhang, J., Shang, Y., Li, F., et al. (2022). Pseudomonas aeruginosa detection using conventional PCR and quantitative real-time PCR based on species-specific novel gene targets identified by pangenome analysis. Front. Microbiol. 13:820431. doi: 10.3389/fmicb.2022.820431
Wu, Y., and Gu, C. T. (2021). Rejection of the reclassification of Leuconostoc gasicomitatum as Leuconostoc gelidum subsp. gasicomitatum based on whole genome analysis. Int. J. Syst. Evol. Microbiol. 71:005027. doi: 10.1099/ijsem.0.005027
Yang, S. M., Kim, E., Kim, D., Kim, H. B., Baek, J., Ko, S., et al. (2021). Rapid real-time polymerase chain reaction for salmonella serotyping based on novel unique gene markers by pangenome analysis. Front. Microbiol. 12:750379. doi: 10.3389/fmicb.2021.750379
Keywords: computational pangenome analysis, identification, real-time PCR, species-specific genes, Leuconostoc
Citation: Kim E, Yang S-M, Kim I-S, Lee S-Y and Kim H-Y (2022) Identification of Leuconostoc species based on novel marker genes identified using real-time PCR via computational pangenome analysis. Front. Microbiol. 13:1014872. doi: 10.3389/fmicb.2022.1014872
Received: 09 August 2022; Accepted: 06 September 2022;
Published: 23 September 2022.
Edited by:
Si Hong Park, Oregon State University, United StatesReviewed by:
Hyunjin Yoon, Ajou University, South KoreaCopyright © 2022 Kim, Yang, Kim, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hae-Yeong Kim, aHlraW1Aa2h1LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.