- 1Co-Innovation Center of Jiangsu Marine Bio-industry Technology, Jiangsu Key Laboratory of Marine Bioresources and Environment, Jiangsu Ocean University, Lianyungang, China
- 2School of Food Science and Engineering, South China University of Technology, Guangzhou, China
- 3Jiangsu Institute of Marine Resources Development, Jiangsu Ocean University, Lianyungang, China
- 4School of Marine Science and Fisheries, Jiangsu Ocean University, Lianyungang, China
The cold-adapted and/or salt-tolerant enzymes from marine microorganisms were confirmed to be meritorious tools to enhance the efficiency of biocatalysis in industrial biotechnology. We purified and characterized a dextranase CeDex from the marine bacterium Cellulosimicrobium sp. THN1. CeDex acted in alkaline pHs (7.5–8.5) and a broad temperature range (10–50°C) with sufficient pH stability and thermostability. Remarkably, CeDex retained approximately 40% of its maximal activities at 4°C and increased its activity to 150% in 4 M NaCl, displaying prominently cold adaptation and salt tolerance. Moreover, CeDex was greatly stimulated by Mg2+, Na+, Ba2+, Ca2+ and Sr2+, and sugarcane juice always contains K+, Ca2+, Mg2+ and Na+, so CeDex will be suitable for removing dextran in the sugar industry. The main hydrolysate of CeDex was isomaltotriose, accompanied by isomaltotetraose, long-chain IOMs, and a small amount of isomaltose. The amino acid sequence of CeDex was identified from the THN1 genomic sequence by Nano LC–MS/MS and classified into the GH49 family. Notably, CeDex could prevent the formation of Streptococcus mutans biofilm and disassemble existing biofilms at 10 U/ml concentration and would have great potential to defeat biofilm-related dental caries.
Introduction
Dextran is a water-soluble glucan (105–107 Da) with mainly α-1,6-glycosidic linkages, generally synthesized from sucrose by dental caries pathogens and microorganisms polluting sugarcane and beet juice (Bashari, 2019; Martínez et al., 2021; Tanoue Batista et al., 2021). Dextranase (E.C.3.2.1.11) specially hydrolyzes the α-1,6-glucosidic linkages in dextran and generates low-molecular-weight (LMW) dextran, isomalto-oligosaccharide (IMO) and/or glucose (Khalikova et al., 2005). LMW dextran with antithrombotic activity is a blood substitute in emergencies and can be used to synthesize iron dextran to treat severe anemia (Khalikova et al., 2005). As one of the promising prebiotic candidates, IMOs modulate the composition of the intestinal microbiome and mediate health-promoting benefits (Sorndech, 2018; Huang et al., 2020; Park et al., 2021). In addition, dextranase can effectively restrain and eliminate dental plaque biofilm, which is greatly significant for the inhibition and treatment of dental caries and the exploitation of the dental caries vaccine (Walker et al., 2016; Pleszczyńska et al., 2017).
Dextranases were from plentiful microbes, such as Penicillium, Aspergillus, Chatomium, Lipomyces, Sporothrix, Streptococcus, Bifidobacterium, Arthrobacter, Pseudomonas, and others (Khalikova et al., 2005; Ren et al., 2018). Dextranases are classified into glycoside hydrolase (GH) 49, 66, 27 and 31 families based on protein structures and sequence similarity (Okazawa et al., 2015; Ren et al., 2019; Tsutsumi et al., 2019). Fungal dextranases often had high activity (generally more than 100 U/mg), but they possessed poor thermal stability and were also unstable under alkaline conditions (Huang et al., 2019). Their optimistic temperature was frequently higher than 50°C, and the optimistic pH was lower than 7 (Khalikova et al., 2005; Zhao et al., 2020; Pittrof et al., 2021). The marine dextranases with cold adaptation, thermostability and/or high catalytic activity in alkaline conditions had been reported and they had an advantage in removing plaque and preventing dental (Ren et al., 2018; Lai et al., 2019; Deng et al., 2020). The dextranases from marine bacteria, such as Cellulosimicrobium sp. PX02, Arthrobacter oxydans KQ11, Catenovulum agarivorans MNH15 and Bacillus aquimaris S5, have the characteristics of alkali resistance but did not possess cold-adaptation and salt tolerance (Lai et al., 2019; Liu et al., 2019; Ning et al., 2021). The concentration of PX02 dextranase needed to reach 15 U/ml to remove and inhibit more than 90% of the S. mutans biofilms (Ning et al., 2021). The dextranase Aodex from Arthrobacter oxydans KQ11 was a mesophilic enzyme (only reacted well at 45–60°C; Liu et al., 2019). When the concentration of Aodex was 6 U/ml, the biofilm inhibition ratio was 90% and the reduction ratio was only 65% (Wang et al., 2016). The dextranase of Catenovulum agarivorans MNH15 had an efficient effect on inhibiting the biofilm formed by S. mutans, but the SDS and lanric acid, the common reagents in oral hygiene products, had negative effects on enzyme activity (Lai et al., 2019). The dextranase from Catenovulum sp. DP03 was cold-adapted and alkali-resistant, some mouthwash ingredients could enhance its activity, and yet its biofilm inhibitory effect was inferior to dextranases from Arthrobacter oxydans KQ11 and Catenovulum agarivorans MNH15 (Ren et al., 2018; Deng et al., 2020). Many elements influence the activity and stability of dextranase, such as temperature, acid, base, metal ion, surfactant and other components of oral care products (Juntarachot et al., 2020a). It is still challenging to find a suitable and effective dextranase used in oral products because of their poor stability and low activity at medium and low temperatures (Qiu et al., 2016). Therefore, it is highly significant to find a dextranase with high catalytic efficiency at medium and low temperatures and high stability under different types of salts and chemical reagents.
There are a wide variety of microorganisms and other biological resources in marine environments (Kennedy et al., 2011). Because of the low temperature, saline and oligotrophic features of marine environment, marine bacteria evolved several physiological and molecular strategies to adapt to the suffered multiple stresses (Jin et al., 2019). For instance, a unique composition of the cytomembrane decreasing the membrane fluidity and producing compatible solutes (e.g., osmolytes; De Maayer et al., 2014; Collins and Margesin, 2019), ice-binding proteins (Bar Dolev et al., 2016; Vance et al., 2019) and cold-active enzymes (Santiago et al., 2016) all contribute to marine bacteria’s survival in low-temperature condition in the ocean (De Maayer et al., 2014; Mangiagalli and Lotti, 2021). Marine highly saline conditions, especially deep sea, exist salt-tolerant and/or halophiles bacteria (2–5 M NaCl or KCl; Jin et al., 2019). These bacteria balanced their osmotic pressure by importing inorganic ions (K+, Na+, Cl−) or synthesizing abundant specific organic osmoprotectants, and produced salt-active or salt-tolerant enzymes to maintain the basal metabolic rate (Tadeo et al., 2009; Ma et al., 2010; Yin et al., 2015; Jin et al., 2019). Therefore, more and more extremozymes with salt- and pH-tolerance, cold-adaption and thermostability were found in marine bacteria and have potential applications in the extreme conditions of industrial processes (Raddadi et al., 2015; Theerachat et al., 2018; Yang et al., 2018; Karan et al., 2020; Wang et al., 2021).
It is of great importance to explore and utilize marine cold-adapted, salt-tolerant and more stable marine dextranase resources for the dental health industry. This study was pursued to isolate marine dextranase-producing bacteria and obtained new marine dextranases with special characteristics. A cold-adapted and salt-tolerant dextranase from a marine bacterium was identified and characterized to reveal its enzymatic properties, hydrolysis characteristics, protein sequence and 3D structure and its effect on suppressing and removing dental plaque. It provides a new enzymatic tool for plaque removal oral products and will promote the commercial application of marine dextranase.
Materials and methods
Materials and chemicals
Blue Dextran 2000 and HiTrap Q FF were purchased from GE Healthcare (Uppsala, Sweden). Different molecular weight dextran and Coomassie brilliant blue G250 were obtained from Solarbio (Beijing, China). Oligosaccharide standards were available from Glycarbo (Osaka, Japan). The protein marker was from Sangon Biotech (Shanghai, China). The other reagents were of analytical grade and purchased from Sinopharm Chemical Reagent Corporation (Shanghai, China).
Screening and identification of strains
The sea mud samples were collected from Haizhou Bay of Lianyungang to screen dextranase-producing strains by the dilution separation method and the formation of clear zones on the blue dextran plate (Lai et al., 2019). The methods of observing colony morphology and phylogenetic analysis based on 16S rDNA were as previously described (Lai et al., 2019). The 16S rDNA gene sequence of strain THN1 was submitted to NCBI with a GenBank number ON815376.
Production and purification of dextranase
The strain THN1 was cultured in the medium containing dextran T20 at 5 g, tryptone at 15 g, NaCl at 5 g, bran at 8 g, and aged seawater at 1 l with pH 8.2, and incubated at 30°C for 24 h. The crude enzyme was the supernatant of fermented broth obtained by centrifugation (6,000 rpm, 4°C).
The supernatant was concentrated by the 30,000 NMWC centrifugal filter units (Millipore, Billerica, United States). The saturated ammonium sulfate was added to the concentrated enzyme solution slowly at 4°C. The protein precipitation of different concentrations of ammonium sulfate (20–90%) was collected by centrifugation, dissolved and dialyzed in 20 mM Tris–HCl buffer (pH 7.5) overnight at 4°C. Then the dextranase was purified by HiTrap Q Sepharose FastFlow on AKTA Pure Chromatography system (GE Healthcare, Uppsala, Sweden). The detailed operation referred to Liu et al. (2022). The fractions possessing high dextranase activity were gathered and analyzed by SDS-PAGE (8%).
Enzymatic characterization
Enzyme assay
The dextranase activity was determined by measuring the reduced sugar with a 3,5-dinitrosalicylic acid assay (DNS), with glucose as a standard. The enzyme activity (U) was defined as the amount of enzyme that produces 1 μmol reduced sugars per min under specific conditions. The protein concentration was measured by the BCA protein assay kit (Solarbio, Beijing, China). Unless stated otherwise, the reaction mixture contained 20 mM sodium phosphate buffer (pH 8.0) and 1% (w/w) dextran T20.
Effects of temperature and pH on dextranase activity and stability
The optimal temperature of the dextranase was measured at pH 8.0 in temperatures between 4 and 60°C. Thermostability was evaluated by testing the residual enzyme activity after the pre-incubation of the purified CeDex in 20 mM sodium phosphate buffer (pH 8.0) at 30, 40 and 50°C for 1–5 h. The optimal pH was tested at 40°C in 50 mM buffers with different pHs (3.0–9.0), which was determined in four different pH buffer systems. The pH stability was evaluated by measuring the residual activity under the optimal reaction condition after pre-incubation of CeDex in 50 mM buffers in the different pH buffers (3.0–9.0) at 4°C for 16 h.
Effects of NaCl on dextranase activity and stability
Determination of the effects of different NaCl concentrations on CeDex: the reaction mixtures with varying concentrations of NaCl (0–5 M) were prepared and reacted under the optimum condition. Determination of NaCl stability of CeDex: the purified CeDex solution with different concentrations of NaCl (0–5 M) was incubated at 4°C for 24 h, and then measured the residual enzyme activity under the optimum condition. The control group was under the same conditions without NaCl treatment.
Effects of metal ions and other compounds on dextranase activity
The effects of metal ions and other reagents were valued by testing the enzyme activity of different the CeDex reaction systems with various metal ions and reagents. The reagents included xylitol, sodium fluoride, sodium benzoate, ethanol, sodium dodecyl sulfonate, and lauric acid. The enzyme activity without metal ions or reagents was taken as the control.
Dextranase kinetics
The initial reaction velocities (v) of different substrate concentrations (0.1–2.5 mg/ mL) and different Mw dextran (T10, T20, and T70) were calculated under the optimum condition. The kinetic constants Km and Vmax were measured by Lineweaver-Burk plot (Lineweaver and Burk, 1934).
Hydrolysate analysis
Purified CeDex was incubated with 1% dextran T10, T20, T40, and T70 under the optimum condition for different times (4–24 h). The hydrolysis products were analyzed by -layer chromatography (TLC; aluminum sheet silica gel 60, F254; Merck, Darmstadt, Germany) using the n-propanol/ethyl acetate/acetonitrile/acetic acid/water (5,2,8,1,4.5 by vol.) solvent solution, then the plate was sprayed with the chromogenic reagent (0.2% (w/v) orcinol, 10% (v/v) H2SO4, and 70% (v/v) ethyl alcohol) followed with incubation at 95°C for 5 min. The standard sugars were glucose, isomaltose and isomaltotriose.
The hydrolysis products of CeDex on the substrate of dextran T20 were identified and quantified by Waters 600 with Waters Sugar-Pak1 (6.5 × 300 mm; Waters, Milford, MA, United States) and a differential refraction detector. The concrete process was performed as previously described (Lai et al., 2019). The peak area was quantified using Empower GPC (Gel Permeation Chromatography) software.
Amino acid sequence analysis and structure prediction of purified CeDex
The purified CeDex was sequenced by nano LC–MS/MS in Shanghai Applied Protein Technology Co. Ltd. (Shanghai, China) and searched in the whole-genome protein sequences of THN1. The protein sequence was blasted in NCBI, compared with other dextranases previously reported, and researched its conserved domains in the Conserved Domain Database1 on June 30, 2021. SignalP-5.0 Server2 on March 29, 2021, and ExPASy3 (2021.03.29) predicted the signal peptides, isoelectric points, and protein molecules of CeDex. The 3D structure of the protein was predicted by I-TASSER4 server on June 28, 2022. The TM-score valuing the quality of the predicted protein structure was calculated by ResQ server2 on July 23, 2022 (Yang et al., 2016). The Phylogenetic tree was established by the MEGA version 5.0 with the neighbor-joining algorithm.
Effect of CeDex on biofilm
Biofilm mass assay was performed according to the protocol of Ren et al. (2018). The minimum biofilm inhibition and reduction concentrations (MBIC and MBRC) of Streptococcus mutans ATCC 25175 (American Type Culture Collection (ATCC), Manassas, VA, United States) were measured.
The activated S. mutans ATCC 25175 was cultured anaerobically in BHI (1% sucrose) with different dextranase activities (0, 2, 4, 6, 8, 10 U) at 37°C for 24 h. After the aspiration of the excess solution, the formed biofilms were immobilized in methanol for 15 min and dried naturally at room temperature. MBIC of S. mutans ATCC 25175 was measured based on the biofilm mass assay. The MBRC of S. mutans ATCC 25175 was valued the same way, except that the CaDex was added after the biofilm of S. mutans ATCC 25175 was formed.
The experimental procedures for studying the effect of dextranase on S. mutans biofilm referred to Ning et al. (2021). The detailed procedures were as follows: the seed solution was inoculated to BHI medium (900 μl, 1% sucrose) added with diverse concentrations of dextranase (0, 2, 4, 6, 8, and 10 U/ml) in a 24-well plate containing a sterile coverslip. After cultivating at 37°C for 24 h, the slides were gently washed with double distilled water until the nonadhesive biofilms were eliminated. The adhesive biofilms were fixed with 2.5% glutaraldehyde at 4°C for 4 h and dehydrated with 50, 70, 80, 90, and 100% alcohol gradients, each for 30 min. Finally, the samples were sprayed with gold powder and observed under SEM (Model JFC-1600, JSM-6390LA; JEOL, Tokyo, Japan).
Results
Isolation and identification of the dextranase-producing strain
The strain THN1 (Figure 1A) was screened and showed dextranase-producing ability according to the formation of a clear zone on blue-dextran agar. THN1 was a yellow and rod-shaped bacterium by ager media and scanning electron micrograph (SEM; Figures 1A,B). Based on the alignment of 16S rDNA sequences and the results of phylogenetic analysis (Figure 1C), the THN1 strain was identified as Cellulosimicrobium sp. (CCTCC accession number: M2020430; China Center for Type Culture Collection, Wuhan, China).
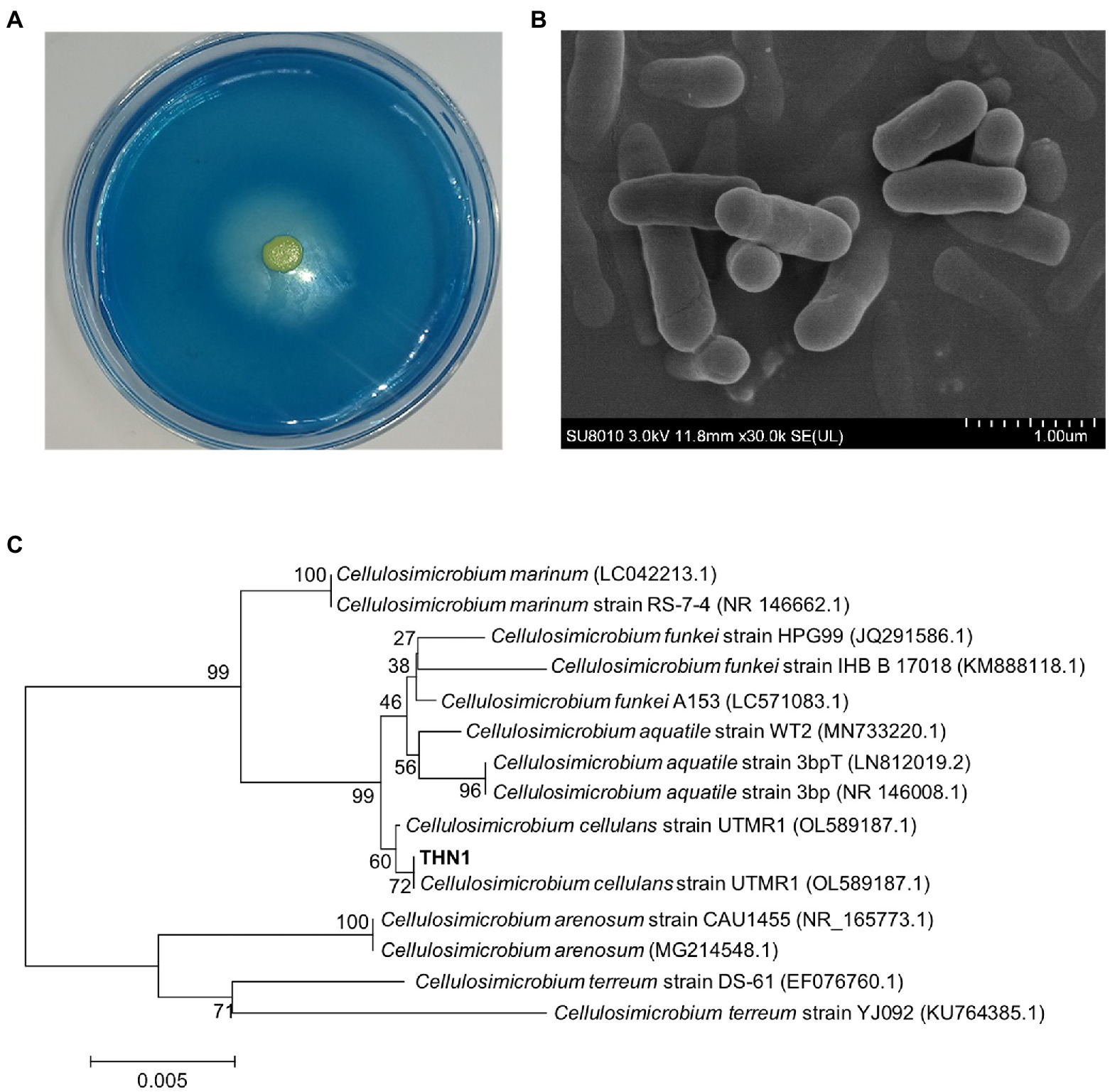
Figure 1. Morphological observation on the blue-dextran plate (A) and scanning electron micrograph (B). Phylogenetic tree of strain THN1 (C).
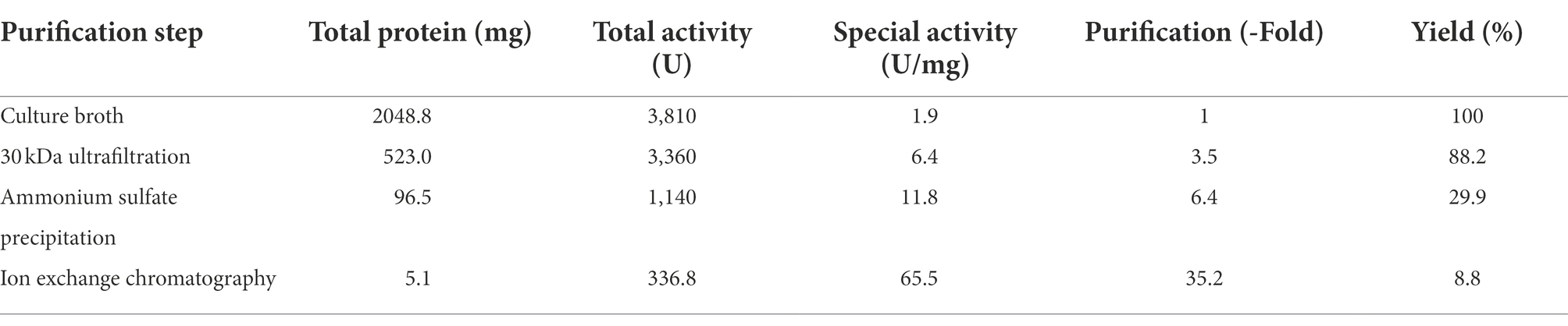
Purification of dextranase
By ultrafiltration, ammonium sulfate deposition, and ion exchange chromatography, the dextranase was purified to 65.5 U/mg (Table 1). During the step of ammonium sulfate precipitation, a large proportion of dextranase was precipitated and collected at 50–80% saturation. Anion exchange chromatography (sepharose Q FF) was used to optimize the purification. The eluates from tubes 19 to 23 with high dextranase activity were collected, merged and analyzed by SDS-PAGE. The result showed the purified dextranase (CeDex) was about 71 kDa (Figures 2A,B).
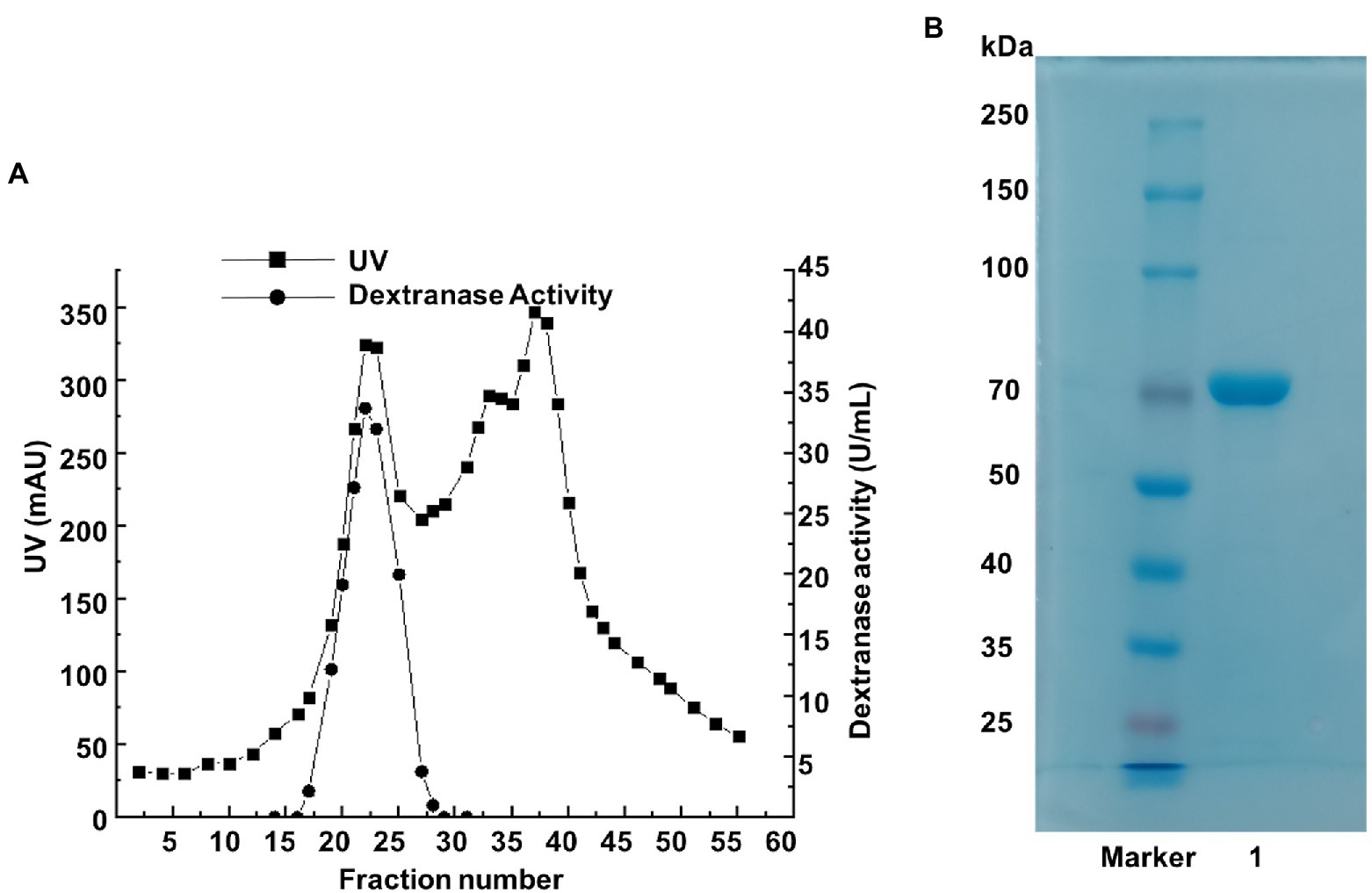
Figure 2. (A) The elution profile of the dextranase on a sepharose Q FF column. (B) SDS-PAGE analysis of tubes 19–23 collected from the ion-exchange chromatography. Lane Maker: protein marker. Lane 1: the purified dextranase.
Enzymatic characteristics of dextranase
Effects of temperature and pH on dextranase activity and stability
CeDex was tested at a temperature range of 4–60°C to get its apparent temperature dependence. The optimal reaction temperature was 40°C (Figure 3A). CeDex retained about 90% of the maximal activity at 20–35°C, 58% at 10°C, and 40% even at 4°C. So CeDex was a novel cold-adapted enzyme with stability in a wide range of moderate and low temperatures. Nevertheless, the CeDex activity was greatly reduced at 50°C and entirely deactivated at 60°C. The activity of CeDex at 30°C and 40°C was stabilized within 5 h (retained >80% activity); while reduced to 23% at 50°C after 1 h (Figure 3B). In addition, CeDex had the highest catalytic performance in the alkaline pH range (7–9) (Figure 3C) and excellent pH stability in pHs (3.0 to 11.0; Figure 3D).
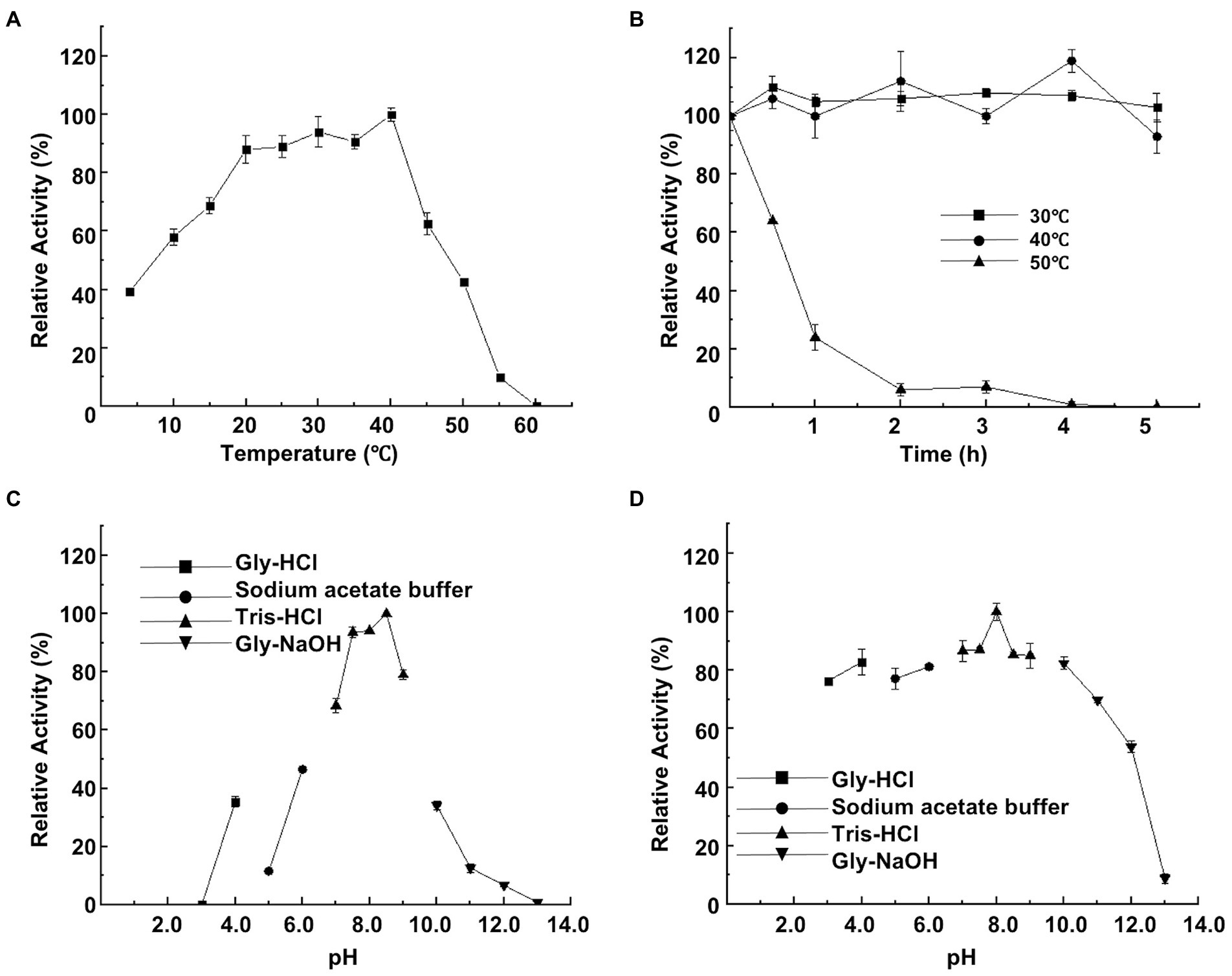
Figure 3. Effects of temperature and pH on CeDex activity. (A) optimal temperature; (B) thermal stability; (C) optimal pH; (D) pH stability. Error bars represent the mean ± SD (n = 3).
Effects of metal ions, high salt, and other compounds on dextranase activity
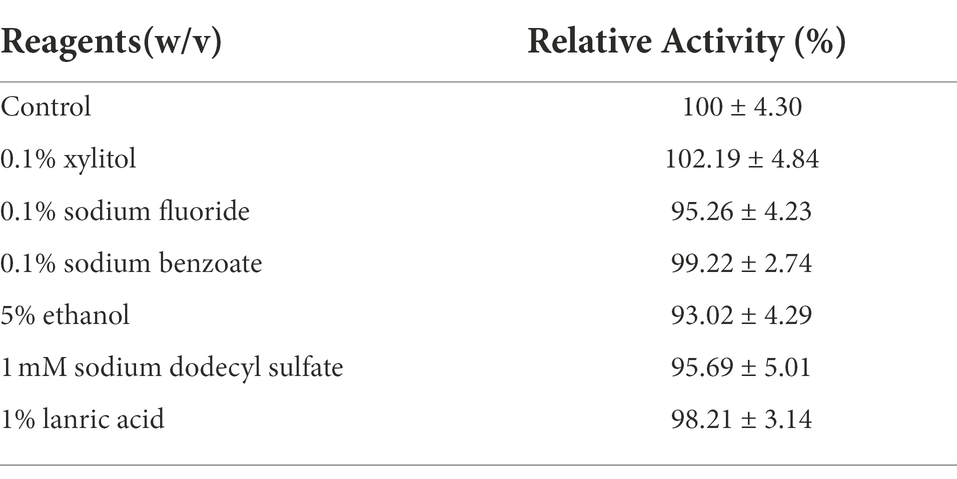
The effects of various metal ions on CeDex were researched in the optimal reaction condition (40°C, pH 8.5). Mg2+, Ba2+, Ca2+, Na+ and Sr2+ enhanced the CeDex activity by 1.1–2.5-fold (Figure 4A). Additionally, the relative activity of CeDex was the highest (160.5%) in 4 M NaCl solution and increased to 140–160% when the NaCl concentration was from 1 to 5 M (Figure 4B). Meanwhile, CeDex still retained 60% of its maximal activity after incubating in the 5 M NaCl solution (Figure 4C). So CeDex had excellent salt tolerance. Furthermore, the main compounds (xylitol, sodium benzoate, sodium fluoride, ethanol, sodium dodecyl sulfate, and lauric acid) in dental care products did not affect the CeDex activity significantly (Table 2).
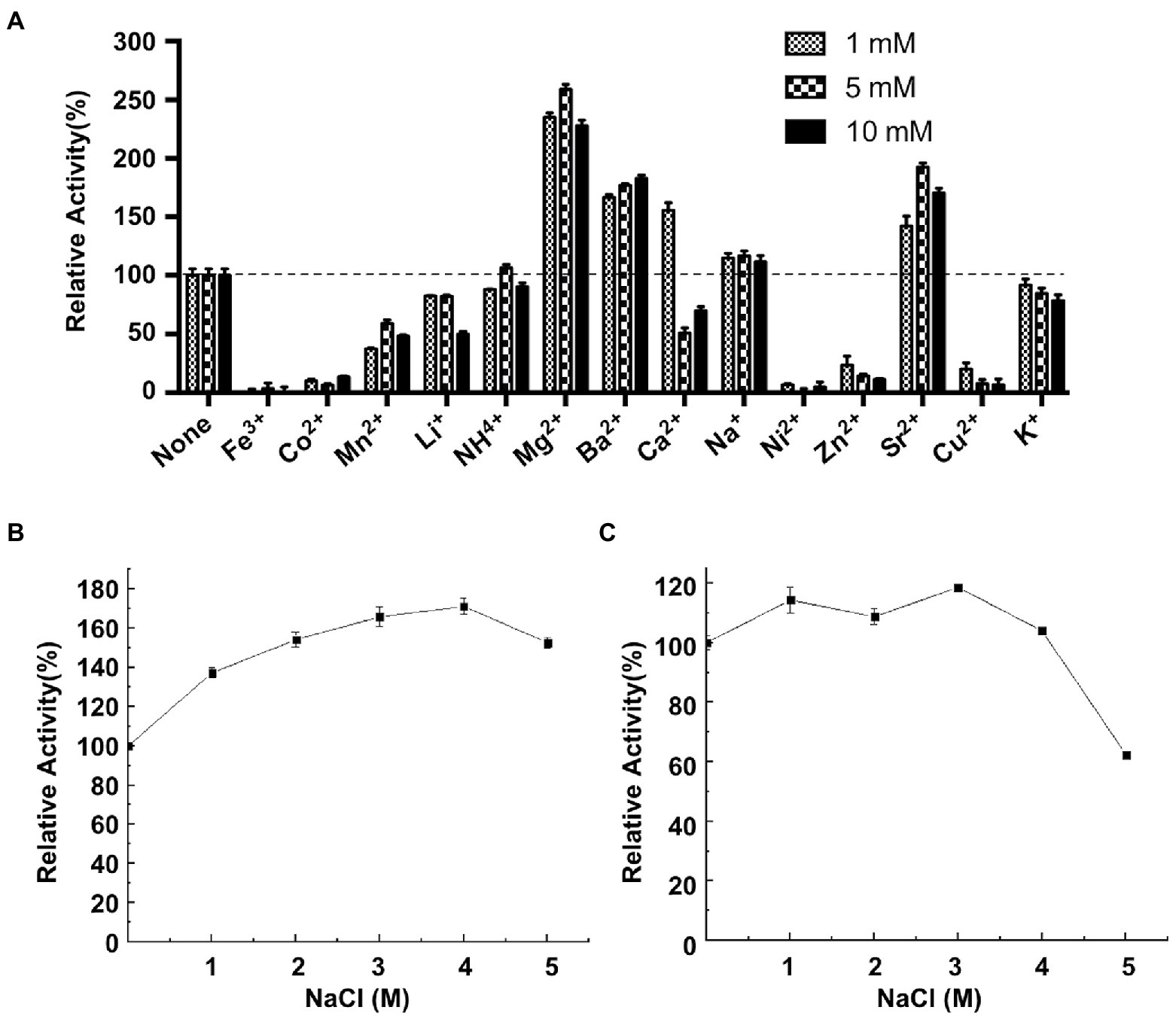
Figure 4. Effects of different metal ions and NaCl on CeDex activity. (A) metal ions; (B) NaCl; (C) NaCl stability. Error bars represent the mean ± SD (n = 3).
Substrate specificity and kinetic parameters of dextranase
The substrate specificity study showed CeDex mainly hydrolyzed the dextran with α-1,6 glycosidic bonds in its main chains (Supplementary Table 1), and was less effective for soluble starch, though the starch had a small amount of α-1,6 glycosidic bonds. No activity was detected when the substrates were pullulan and chitosan, mainly constituting α-1,4 glycosidic linkages and β-1,4 glycosidic linkages, respectively. Therefore, CeDex displayed high specificity on dextran.
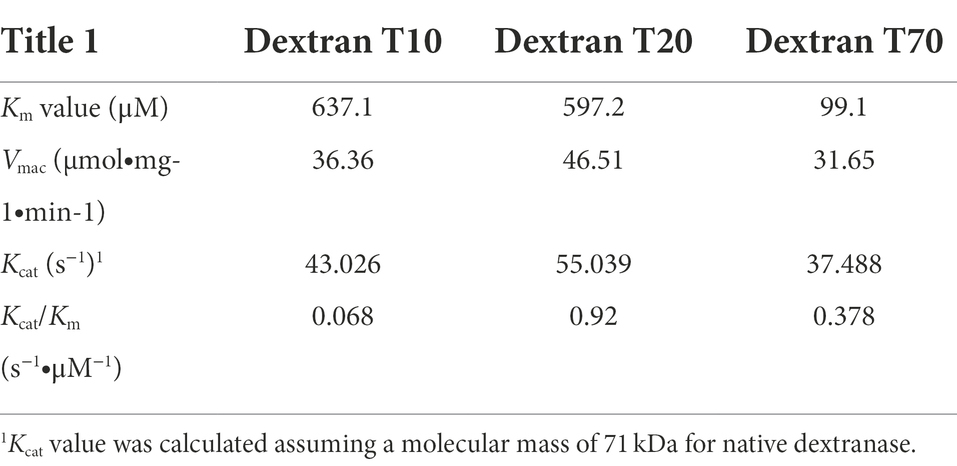
Table 3 displayed the enzyme kinetics of CeDex against dextran T10, T20, and T70. The Km of CeDex for dextran T70 was 0.0991 mM, less than those of dextranases from Arthrobacter oxydans (4.73 mM), Aspergillus allahabadii X26 (14.29 mM), Penicillium cyclopium CICC-4022 (2.61 mM) and Streptomyces sp. NK458 (94.30 mM; Huang et al., 2019), demonstrating the stronger affinity of CeDex for dextran. The Kcat and Kcat/Km values of CeDex for dextran T20 were the highest, indicating that CeDex had higher catalytic efficiency for dextran T20 than T10 and T70.
Analysis of hydrolysis products
The hydrolysates distribution of CeDex on different Mw dextrans (T10, T20, T40, T70) was revealed. The isomaltotetraose (G3) was the main product and higher molecular weight IMOs also existed (Figure 5). The hydrolysate distributions did not change significantly with the extension of the reaction time. The amounts of IMOs with different degrees of polymerization (DPs) in the hydrolysis products were measured by HPLC, in which isomaltotriose (G3) and DP ≥4 IMOs accounted for 58.1 and 40.7%, respectively, after 4 h of reaction. Moreover, the amounts of IMOs (DP 2–4) were increased as hydrolysis time extension with no glucose (Supplementary Table 2). Isomaltooligosaccharides are one kind of non-digestible oligosaccharides with low calorific value and can promote the growth of Bifidobacteria and Lactobacillus. Numerous isomaltooligosaccharides were produced by endodextranases and possessed broad applications in the food industry as dietary fibers and prebiotics (Sorndech, 2018). Intriguingly, the effective oligosaccharides with DPs 2–10 foreboded more applications and enormous business benefits in the food and health products (Seo et al., 2007; Liu et al., 2022).
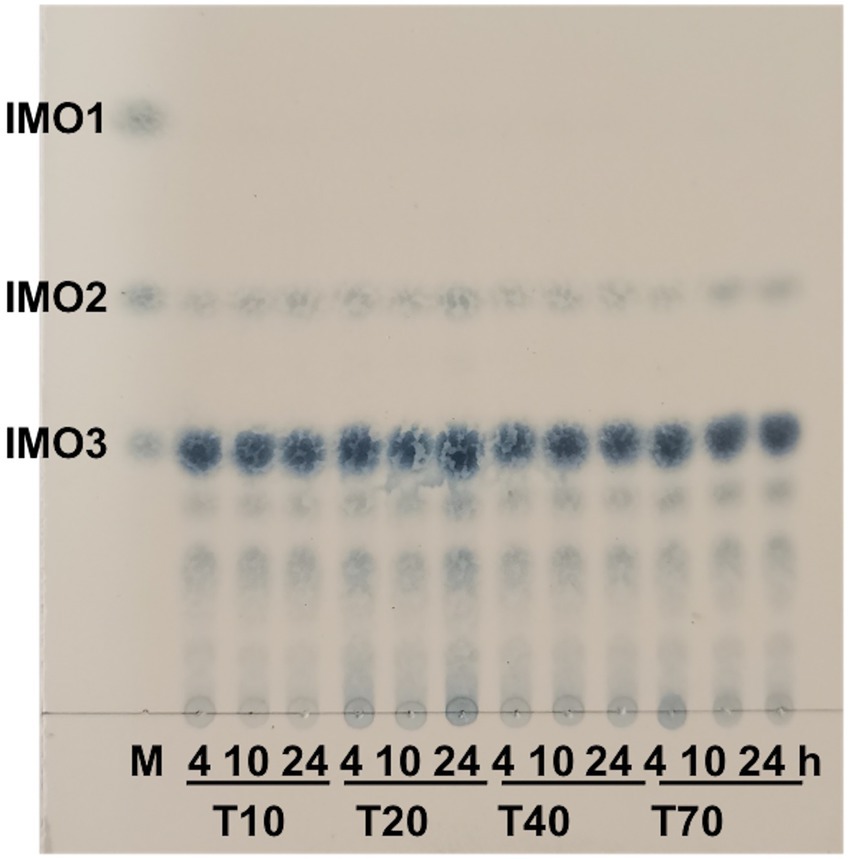
Figure 5. Products analysis of CeDex hydrolyzing the different molecular masses of dextran with various incubation times by TLC. M, the standard glucose (IMO1), isomaltose (IMO2) and isomaltotriose (IMO3).
Nano LC–MS/MS and structure analysis of purified CeDex
The peptide fragments (Supplementary Figure 1) of CeDex were obtained from nano LC–MS/MS followed by search analysis with Proteome Discoverer 1.4. The search database was the whole-genome protein sequence of THN1. The result showed the number of unique peptides of the orf2365 protein from Cellulosimicrobium sp. THN1 was the highest (35) and the coverage was 41.31% (Supplementary Figure 1). The orf2365 protein (CeDex2365, Genbank accession number ON856679) comprises signal peptides (M1-A23) and three primary domains -Domain of Glycosyl hydrolase family 49 (GH49, Thr42 to Ala627), Domain B of Glucodextranase (Pro638 to Thr721) and DOMON-like ligand-binding domain (Gly724 to Val965) and is identified as a multi-domain dextranase of GH49 family (Figure 6A). The peptides identified as parts of CeDex2365 by Nano LC–MS/MS were nearly all located in the GH49 domain (Supplementary Figure 1). CeDex2365 with 1,065 amino acids (AAs) has a predicted Mw of 114.49 kDa, which was utterly inconsistent with the result of the SDS-PAGE of CeDex. The N-terminal GH49 domain (A24-A637) with a predicted Mw of 68.2 kDa was close to the purified dextranase CeDex. Therefore, we deduced that CeDex was the GH49 domain of CeDex2365 (CeDex2365-49), and CeDex2365 was proteolytically cleaved after protein translation.
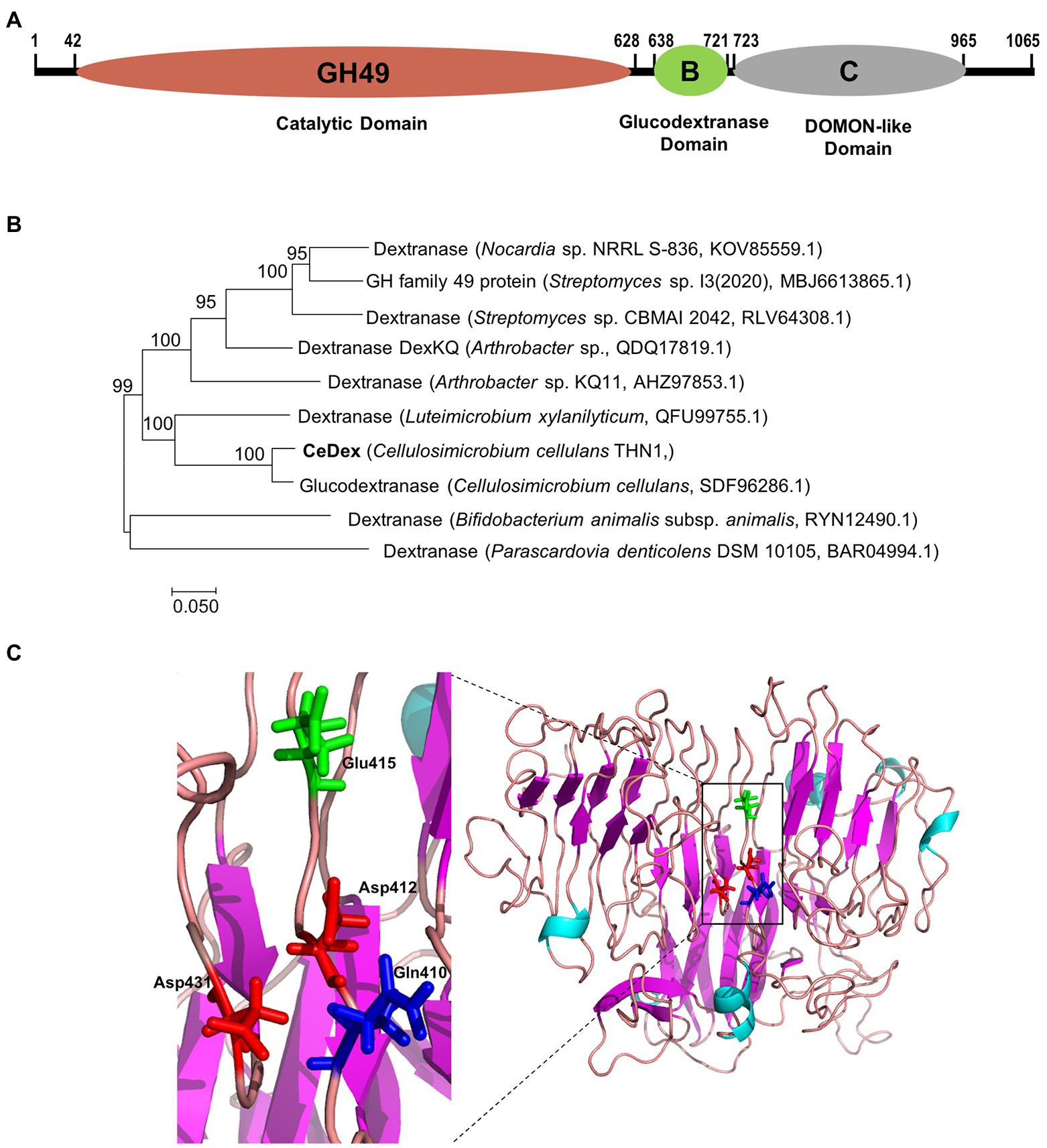
Figure 6. Sequence and three-dimensional structure analysis of CeDex2365. (A) Conserved domains analysis using the Conserved Domain Database (Lu et al., 2020). (B) Phylogenetic analysis of the GH49 domain of CeDex2365. (C) 3D structure of the GH49 domain of CeDex2365. Amino acid residues corresponding to catalysis are presented in the structure.
The phylogenetic analysis manifested CeDex2365 is closely connected with the putative glucodextranase from Cellulosimicrobium cellulans (Figure 6B). By submitting the sequence to I-TASSER5 server, the 3D structure of CeDex2365-49 was predicted (Figure 6C) using the crystal structure of Aodex from Arthrobacter oxydans KQ11 as a template (PDB entry: 6nzs.1. A; Ren et al., 2019). The TM-score of the predicted structure was the highest fitted score (0.61 ± 0.14; A TM-score > 0.5 indicates a model of correct topology; Zheng et al., 2021). The CeDex2365-49 consisted of a β-sandwich domain at N-terminal and a right-handed parallel β-helix domain at C-terminal (Larsson et al., 2003; Supplementary Figure 2). The signal peptide analysis by the SignalP 5.0 server showed CeDex was an extracellular protein with a signal sequence (1–23 AA of the N-terminus region; Supplementary Figure 1). Multiple sequence alignment suggested that the four predicted catalytic key residues were Asp412, Asp431, Glu415 and Gln410 (marked with stars in Supplementary Figure 2; Liu et al., 2019; Ren et al., 2019). Protein sequence alignment between CeDex and its homologs in the NCBI database indicated that CeDex exhibited high identities (92.8–99.6%) with the family 49 glycosyl hydrolases of Cellulosimicrobium genus (WP_257417428.1, WP_144683787.1, WP_194226287.1), which had not yet been structurally and functionally characterized. Among the reported GH49 dextranases, the catalytic GH49 domain exhibited the highest sequence identity (61.7%) with the dextranase AoDex from Arthrobacter oxydans KQ11 (GenBank accession number KJ571608). Then, the catalytic GH49 domain of CeDex shared 38.2 and 28.2% sequence identities with that of Catenovulum sp. DP03 cold-adapted dextranase (Deng et al., 2020) and Penicillium funiculosum dextranase (MH581385), respectively. The Gln418-Asp440 of catalytic center in CeDex was highly conserved and had high similarity with its homologs (Supplementary Figure 2).
According to recent research, one of the characteristic features of the cold-adapted and salt-tolerant enzymes was that the ratio of acidic (glutamic acid and aspartic acid) to basic (histidine, lysine, and arginine) amino acids was higher than that of thermophilic and non-salt tolerant enzymes (Ding et al., 2021). The amino acid composition of the different domains in CeDex2365 was analyzed and compared with other GH49 dextranases (Table 4). The proportion of acidic residues and basic residues in each domain of CeDex2365 was close to the cold-adapted dextranase CaDex. The net charge densities of CeDex2365 and CaDex were higher than that of AoDex and PcDex49, which did not possess cold-adaptation and salt tolerance. The high proportion of acidic amino acid in CeDex was in accord with that of the cold-adapted and salt-tolerant enzymes.
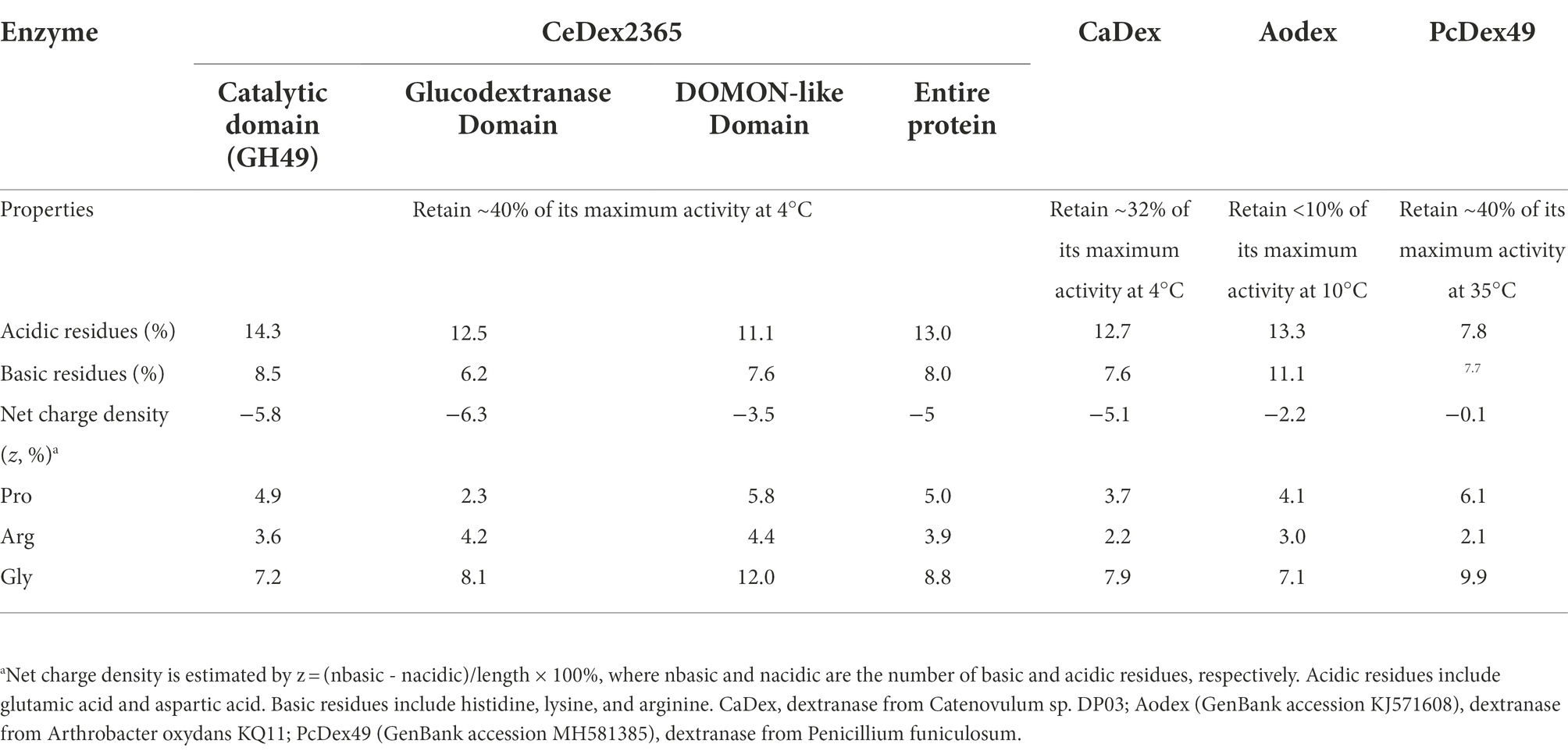
Table 4. Comparison of the properties and frequencies of charged amino acid residues for different domains in CeDex2365 and other dextranases.
Effects of CeDex on biofilm
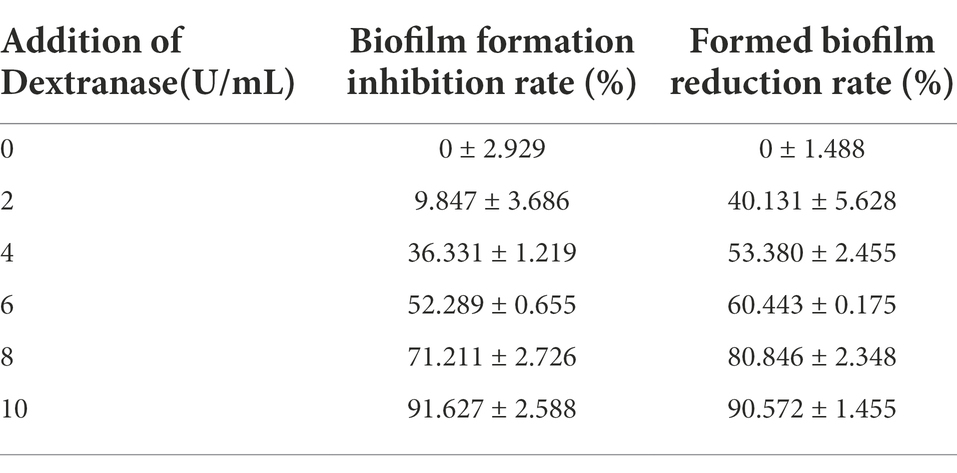
Dextran is the essential ingredient in dental plaque and protects oral bacteria within the biofilm from antimicrobials and harmful agents (Pleszczyńska et al., 2017). S. mutans is the primary bacteria forming dental plaque and causing dental decay (Juntarachot et al., 2020b). The minimum biofilm inhibition and reduction concentrations (MBIC and MBRC) of S. mutans ATCC 25175 were used to assess the effects of CeDex on biofilm (Table 5). The results showed that CeDex could inhibit biofilm formation and reduce the formed biofilm. As the concentration of CeDex increased, the inhibition and reduction rate increased. When adding 6 U/ml and 10 U/ml, CeDex could inhibit 52.3 and 91.6% of the plaque, respectively. The lowest CeDex concentration reducing biofilm by 50% was 4 U/ml, and when CeDex reached 10 U/ml, the reduction rate was up to 90.6%.
The morphology and biofilm of S. mutans were observed by scanning electron microscopy (SEM; Figure 7). The biofilm structure was tight and thick and had many channels formed by exopolysaccharides and bacteria. As the concentration of CeDex enhanced, the adhesion of S. mutans reduced, and the biofilm was gradually thinner and looser. The microplate crystal violet staining combined with SEM indicated that CeDex could significantly inhibit and reduce the plaque formed by S. mutans.
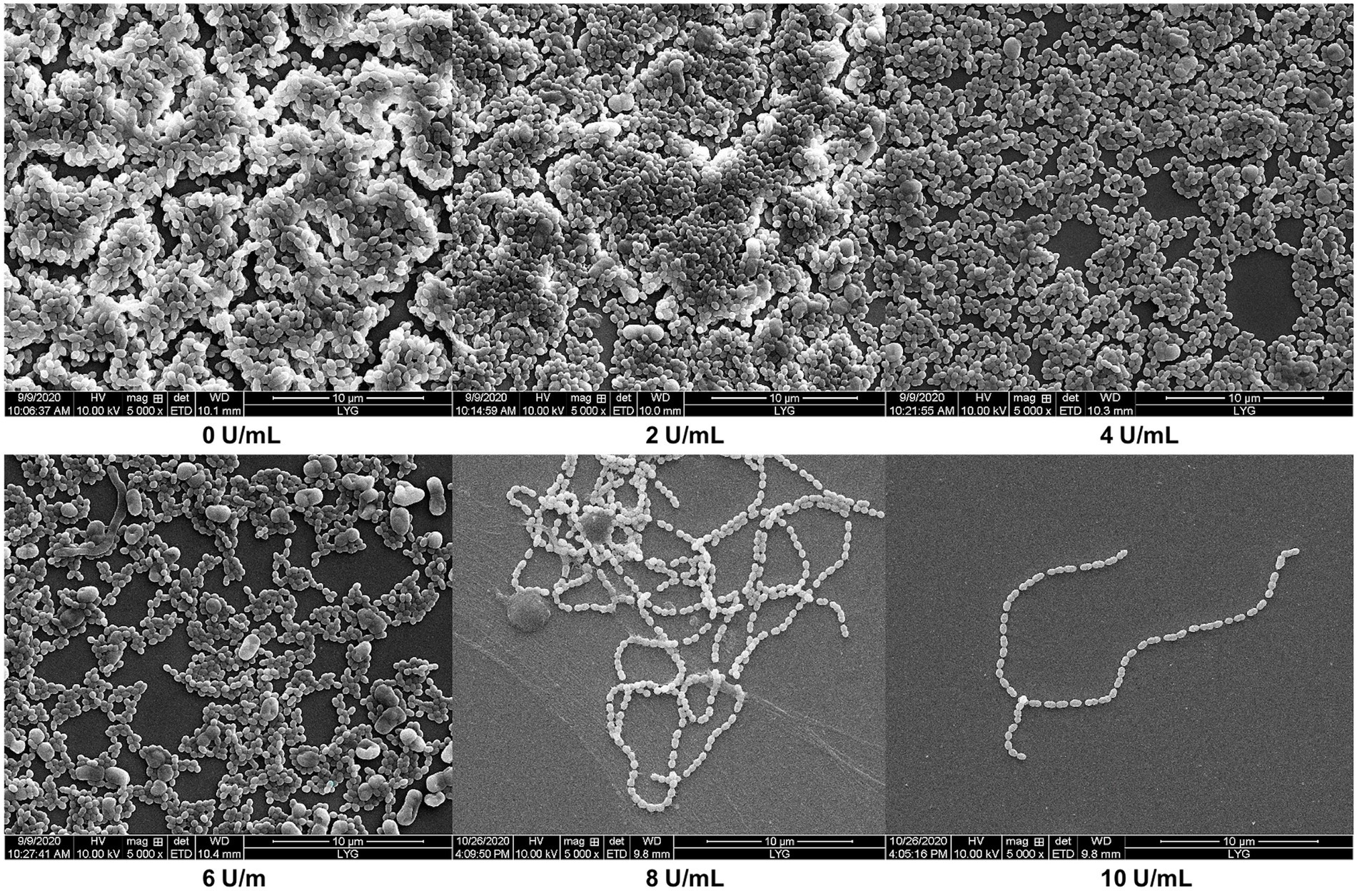
Figure 7. Effects of different CeDex concentrations on the biofilm formed by S. mutans on sterile coverslips (5000×). The presence of dextranase at the concentration of 0, 2, 4, 6, 8, and 10 U/ml. 0 U/ml presents a control with an equal amount of sterile deionized water instead of dextranase.
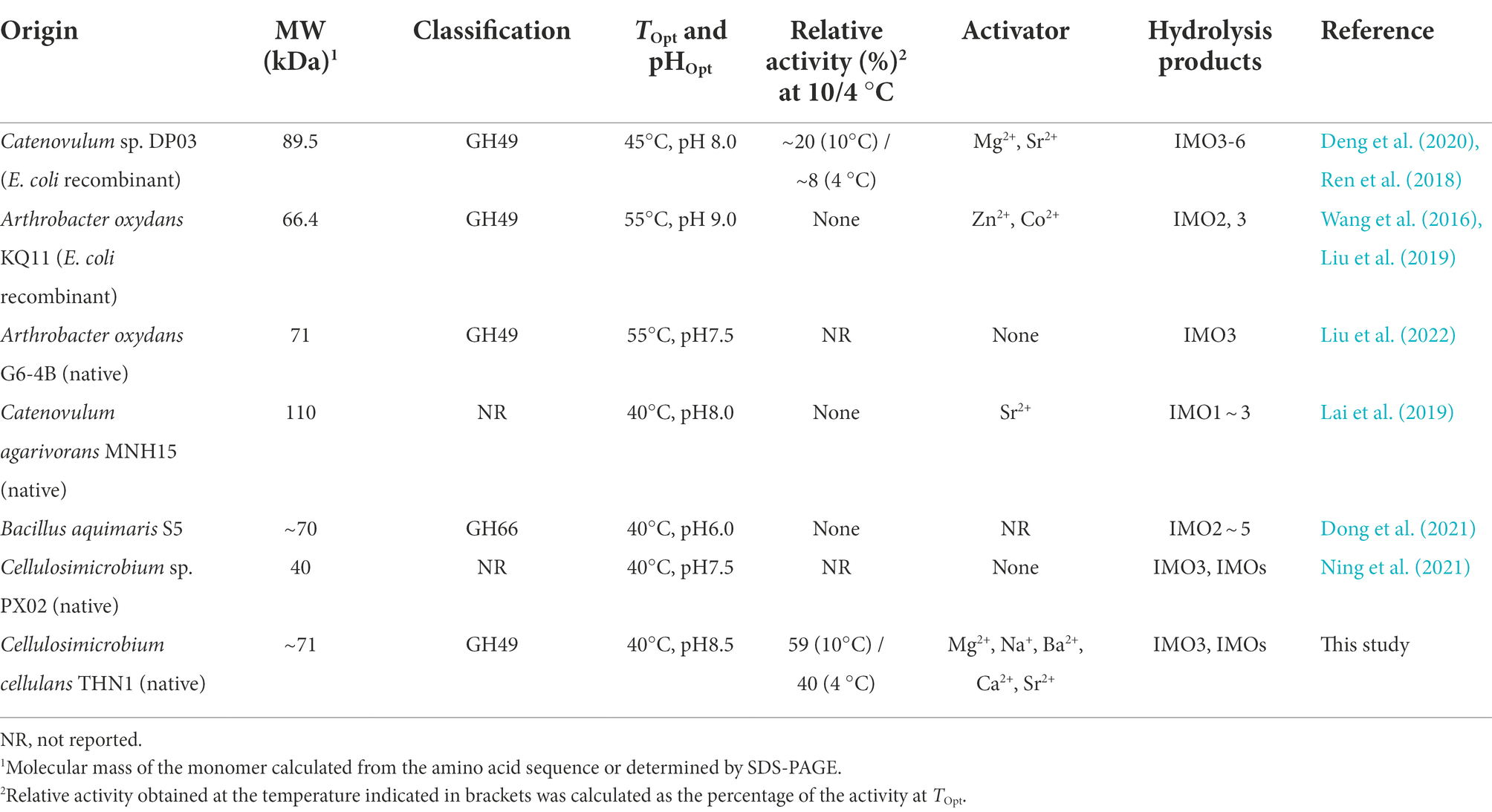
Discussion
In this study, we purified and characterized a dextranase CeDex from a marine bacterium Cellulosimicrobium sp. THN1. Previous research has reported several marine strains produced dextranases, such as Catenovulum sp. DP03, A. oxydans KQ11, Cellulosimicrobium sp. PX02, Bacillus aquimaris S5 and Catenovulum agarivorans MNH15 (Table 6). The Mws of dextranases from marine bacteria generally range from 40 to 110 kDa and therein lie the GH49 dextranases. The GH49 conserved domain (65–71 kDa) encompasses a β sandwich composed of 190 AAs at the N-terminal and a right-handed parallel β-helix consisting of three parallel sheets at the C-terminal (Ren et al., 2018, 2019; Lai et al., 2019; Deng et al., 2020; Liu et al., 2022). The theoretical Mw of orf2365 protein was larger than CeDex. According to the results of Nano LC–MS/MS and the Mw decided by SDS-PAGE, we estimated the extracellular dextranase CeDex was the GH49 domain in the N-terminal of protein orf2365. Proteolytic cleavage of proteins was a common and irreversible post-translational modification in bacteria and was executed by endoproteases with breakups at specific sites and exoproteases with cleaving the N- and C- termini. (Forrest and Welch, 2020). Therefore, we speculated the protein orf2365 was proteolytically cleaved at C-terminal after protein translation to generate the maturation protein CeDex, and the proteolytic site needs further verification.
To adapt to marine extreme environments, marine microorganisms produce cold-adapted and/or salt-tolerant enzymes, which have a promising prospect in applied biotechnology and basic research (Ding et al., 2021). The dextranase (CeDex) from the marine bacterium, Cellulosimicrobium sp. THN1, exhibited cold-adaptation, salt tolerance, excellent pH stability and sufficient thermostability. To the best of our knowledge, the dextranase CaDex from Catenovulum sp. DP03 was the only cold-adapted one and no salt-dextranase had been reported. CeDex kept more than 90% of the highest activities in a broad temperature range (20–40°C) and more than 58% at 10–20°C, which was distinguished from the conventional fungal dextranases (Wang et al., 2022). CaDex from Catenovulum sp. DP03 remained more than 70% of the highest activity at 30–50°C, about 40% at 20°C and 20% at 10°C (Table 6). In short, the CeDex activity at low temperatures (4–20°C) was higher than CaDex from DP03, and the cold-adapted property of CeDex was superior to other marine dextranases. Even though the fungal dextranases had higher enzymatic activity than bacterial dextranases, nearly all fungi dextranases were acidic and sensitive to alkaline in varying degrees (Khalikova et al., 2005). Previous research had demonstrated that alkaline dextranases had more advantages in the prevention and treatment of dental caries than acid dextranases because the alkaline mouthwash was more protective than enamel against acidic (Ren et al., 2018; Lai et al., 2019). Meanwhile, alkali production of oral bacteria plays an important part in oral ecology, pH homeostasis, and inhibiting the formation of dental caries (Huang et al., 2017). CeDex had enzyme activities in neutral and alkaline conditions (pH 7–9) and maintained good stability. So, CeDex was also an alkaline dextranase and has the potential to be a dental caries reagent. In recent research, marine dextranases were reported to effectively sweep away dental plaque, for example, dextranases from Cellulosimicrobium sp. PX02, Arthrobacter oxydans KQ11, Catenovulum agarivorans MNH15, Catenovulum sp. DP03 and Bacillus aquimaris S5 (Wang et al., 2016; Lai et al., 2019; Deng et al., 2020, 2021). In order for the inhibition rate of dental plaque biofilms to be 90%, the concentration of PX02 dextranase was at least 15 U/ml (Ning et al., 2021), AoDex and MNH15 dextranase needed to be 6–7 U/ml (Wang et al., 2016; Lai et al., 2019), the dextranase from Catenovulum sp. DP03 should be 8 U/ml (Dong et al., 2021). When the dextranase of Bacillus aquimaris S5 was 8 U/ml, the inhibition rate was 80% (Dong et al., 2021). Though AoDex had a highly efficient effect on inhibiting the biofilm formed by S. mutans, the biofilm reduction ratio was only 65% (6 U/ml; Wang et al., 2016). In addition, SDS had an effective inhibiting effect on the enzymatic activity of MNH15 dextranase and AoDex (Ren et al., 2018; Lai et al., 2019). Intriguingly, SDS and other compounds in dental care products did not suppress CeDex activity distinctly. Therefore, in consideration of the good preventing and treating effects on the biofilm of S. mutans (MBRC90 and MBIC90 were 10 U/ml) and excellent stability, the cold and alkalic CeDex will be more suitable for the exploitation of novel therapeutic agents for dental caries.
There has not been a clear definition of salt-tolerant enzymes until now. According to published research, they were a kind of enzyme with activity and/or stability under high NaCl concentrations. Different types of salt-tolerant enzymes had different tolerance ranges, for example, the NaCl concentration of salt-tolerant xylanase was 0.5–6.0 M (Cao et al., 2021), the NaCl concentration of halotolerant protease was 4–5 M (Mokashe et al., 2018), and the highest NaCl concentration of salt-tolerant esterase was 5 M (Wu et al., 2015; Jin et al., 2019). To our knowledge, no salt-tolerant dextranase has been reported to date. The highest NaCl concentration of retaining CeDex’s catalytic activities was 4 M and CeDex also had enzymatic activity in low NaCl conditions (~0.2 M; Figure 4B), which was different from most halophilic enzymes relying on high concentrations of NaCl to maintain their activities stably (Zhang et al., 2018). In addition, CeDex exhibited favorable stability in a high NaCl concentration (1–4 M NaCl). Salt-tolerant enzymes have great significance in biotechnology and research on the extreme environmental adaptation of microorganisms. Furthermore, halophilic enzymes with stable or stimulative activity in high-salt conditions could be available for the high-salt process (Schreck and Grunden, 2014; Yang et al., 2020). In the sugarcane industry, dextranase is mainly used to remove dextran, which pollutes the sugar juice and reduces the yield of sucrose (Bashari et al., 2019). Sugarcane juice contains cations like K+, Ca2+, Mg2+ and anions like Cl−, PO43− and SO32−, which inhibited dextranase activity (Purushe et al., 2012; Wang et al., 2016; Ren et al., 2018; Lai et al., 2019). The CeDex activity was significantly enhanced by Na+, Mg2+, Ba2+, Ca2+, and Sr2+ (1 mM) and K+, Li+, and NH4+ did not reduce the CeDex activity. In short, the excellent thermostability, salt tolerance, and metal ions stability made CeDex adaptable for the sugarcane industry (Purushe et al., 2012; Liu et al., 2022).
Conclusion
Our work revealed a new endo-type dextranase belonging to the GH49 family from a marine bacterium Cellulosimicrobium sp. THN1. CeDex was a cold-adapted dextranase with high activities at medium and low temperatures (10–40°C) and had excellent thermal and pH stability. Notably, CeDex was the first dextranase with activity and/or stability under a NaCl concentration of 1–4 M. CeDex was stimulated by Mg2+, Ca2+, and K+ ions in sugarcane juice, and effectively inhibit and/or eliminate the S. mutans biofilm unimpeded by some reagents of dental care products. Therefore, CeDex has great potential applications in the sugarcane industry and in manufacturing oral hygiene products to remove dental plaque.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
LX and YZ designed the experiments. YZ performed the experiments. LX, NL, ZWe, and ZWa analyzed the data. LX wrote the manuscript. SW and YW reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by research grants from the National Natural Science Foundation of China (grant no. 32172154), the Jiangsu Planned Projects for Postdoctoral Research Funds (2019 K215), the Research Start-up Fund of Jiangsu Ocean University (KQ17019), the Natural Science Foundation of Jiangsu Higher Education Institutions of China (grants no. 22KJB180015), the Natural Science Foundation of Jiangsu Province (grants no. BK20201028), the Postdoctoral Research Funds of Lianyungang.
Acknowledgments
We would like to thank the Analyses and Testing Center of Jiangsu Ocean University for its technical support and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1012957/full#supplementary-material
Footnotes
1. ^https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
2. ^http://www.cbs.dtu.dk/services/SignalP/
3. ^https://prosite.expasy.org/
References
Bar Dolev, M., Braslavsky, I., and Davies, P. L. (2016). Ice-binding proteins and their function. Annu. Rev. Biochem. 85, 515–542. doi: 10.1146/annurev-biochem-060815-014546
Bashari, M. (2019). Impact of dextran biodegradation catalyzed by dextranase enzyme on the crystallization rate of sucrose during sugar manufacturing. J. Food Nutr. Res. 7, 402–408. doi: 10.12691/jfnr-7-5-10
Cao, L., Zhang, R., Zhou, J., and Huang, Z. (2021). Biotechnological aspects of salt-tolerant xylanases: a review. J. Agric. Food Chem. 69, 8610–8624. doi: 10.1021/acs.jafc.1c03192
Collins, T., and Margesin, R. (2019). Psychrophilic lifestyles: mechanisms of adaptation and biotechnological tools. Appl. Microbiol. Biotechnol. 103, 2857–2871. doi: 10.1007/s00253-019-09659-5
De Maayer, P., Anderson, D., Cary, C., and Cowan, D. A. (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517. doi: 10.1002/embr.201338170
Deng, T., Feng, Y., Xu, L., Tian, X., Lai, X., Lyu, M., et al. (2020). Expression, purification and characterization of a cold-adapted dextranase from marine bacteria and its ability to remove dental plaque. Protein Expr. Purif. 174:105678. doi: 10.1016/j.pep.2020.105678
Ding, N., Zhao, B., Ban, X., Li, C., Venkataram Prasad, B. V., Gu, Z., et al. (2021). Carbohydrate-binding module and linker allow cold adaptation and salt tolerance of maltopentaose-forming amylase from marine bacterium Saccharophagus degradans 2-40 T. Front. Microbiol. 12:708480. doi: 10.3389/fmicb.2021.708480
Dong, D., Wang, X., Deng, T., Ning, Z., Tian, X., Zu, H., et al. (2021). A novel dextranase gene from the marine bacterium bacillus aquimaris S5 and its expression and characteristics. FEMS Microbiol. Lett. 368:fnab007. doi: 10.1093/femsle/fnab007
Forrest, S., and Welch, M. (2020). Arming the troops: post-translational modification of extracellular bacterial proteins. Sci. Prog. 103:36850420964317. doi: 10.1177/0036850420964317
Huang, S. X., Hou, D. Z., Qi, P. X., Wang, Q., Chen, H. L., Ci, L. Y., et al. (2020). Enzymatic synthesis of non-digestible oligosaccharide catalyzed by dextransucrase and dextranase from maltose acceptor reaction. Biochem. Biophys. Res. Commun. 523, 651–657. doi: 10.1016/j.bbrc.2019.12.010
Huang, X., Zhang, K., Deng, M., Ram, E., Liu, C., Zhou, X., et al. (2017). Effect of arginine on the growth and biofilm formation of oral bacteria. Arch. Oral Biol. 82:256. doi: 10.1016/j.archoralbio.2017.06.026
Huang, R., Zhong, L., Xie, F., Wei, L., Gan, L., Wang, X., et al. (2019). Purification, characterization and degradation performance of a novel dextranase from Penicillium cyclopium CICC-4022. Int. J. Mol. Sci. 20:1360. doi: 10.3390/ijms20061360
Jin, M., Gai, Y., Guo, X., Hou, Y., and Zeng, R. (2019). Properties and applications of extremozymes from deep-sea extremophilic microorganisms: a mini review. Mar. Drugs 17:656. doi: 10.3390/md17120656
Juntarachot, N., Kantachote, D., Peerajan, S., Sirilun, S., and Chaiyasut, C. (2020a). Optimization of fungal dextranase production and its antibiofilm activity, encapsulation and stability in toothpaste. Molecules 25:4784. doi: 10.3390/molecules25204784
Juntarachot, N., Sirilun, S., Kantachote, D., Sittiprapaporn, P., Tongpong, P., Peerajan, S., et al. (2020b). Anti-Streptococcus mutans and anti-biofilm activities of dextranase and its encapsulation in alginate beads for application in toothpaste. PeerJ. 8:e10165. doi: 10.7717/peerj.10165
Karan, R., Mathew, S., Muhammad, R., Bautista, D. B., Vogler, M., Eppinger, J., et al. (2020). Understanding high-salt and cold adaptation of a polyextremophilic enzyme. Microorganisms 8:1594. doi: 10.3390/microorganisms8101594
Kennedy, J., O’Leary, N. D., Kiran, G. S., Morrissey, J. P., O’Gara, F., Selvin, J., et al. (2011). Functional metagenomic strategies for the discovery of novel enzymes and biosurfactants with biotechnological applications from marine ecosystems. J. Appl. Microbiol. 111, 787–799. doi: 10.1111/j.1365-2672.2011.05106.x
Khalikova, E., Susi, P., and Korpela, T. (2005). Microbial dextran-hydrolyzing enzymes: fundamentals and applications. Microbiol. Mol. Biol. Rev. 69, 306–325. doi: 10.1128/MMBR.69.2.306-325.2005
Lai, X., Liu, X., Liu, X., Deng, T., Feng, Y., Tian, X., et al. (2019). The marine Catenovulum agarivorans MNH15 and dextranase: removing dental plaque. Mar. Drugs 17:592. doi: 10.3390/md17100592
Larsson, A. M., Andersson, R., Ståhlberg, J., Kenne, L., and Jones, T. A. (2003). Dextranase from Penicillium Minioluteum. Structure 11, 1111–1121. doi: 10.1016/s0969-2126(03)00147-3
Lineweaver, H., and Burk, D. (1934). The determination of enzyme dissociation constants. J. Am. Chem. Soc. 56Placeholder Text, 658–666.
Liu, N., Li, P., Dong, X., Lan, Y., Xu, L., Wei, Z., et al. (2022). Purification, characterization, and hydrolysate analysis of dextranase from Arthrobacter oxydans G6-4B. Front. Bioeng. Biotechnol. 9:813079. doi: 10.3389/fbioe.2021.813079
Liu, H., Ren, W., Ly, M., Li, H., and Wang, S. (2019). Characterization of an alkaline GH49 dextranase from marine bacterium Arthrobacter oxydans KQ11 and its application in the preparation of isomalto-oligosaccharide. Mar. Drugs 17:479. doi: 10.3390/md17080479
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Ma, Y., Galinski, E. A., Grant, W. D., Oren, A., and Ventosa, A. (2010). Halophiles 2010: life in saline environments. Appl. Environ. Microbiol. 76, 6971–6981. doi: 10.1128/AEM.01868-10
Mangiagalli, M., and Lotti, M. (2021). Cold-active β-galactosidases: insight into cold adaption mechanisms and biotechnological exploitation. Mar. Drugs 19:43. doi: 10.3390/md19010043
Martínez, D., Menéndez, C., Chacón, O., Fuentes, A. D., Borges, D., Sobrino, A., et al. (2021). Removal of bacterial dextran in sugarcane juice by Talaromyces minioluteus dextranase expressed constitutively in Pichia Pastoris. J. Biotechnol. 333, 10–20. doi: 10.1016/j.jbiotec
Mokashe, N., Chaudhari, B., and Patil, U. (2018). Operative utility of salt-stable proteases of halophilic and halotolerant bacteria in the biotechnology sector. Int. J. Biol. Macromol. 117, 493–522. doi: 10.1016/j.ijbiomac.2018.05.217
Ning, Z., Dong, D., Tian, X., Zu, H., Tian, X., Lyu, M., et al. (2021). Alkalic dextranase produced by marine bacterium Cellulosimicrobium sp. PX02 and its application. J. Basic Microbiol. 61, 1002–1015. doi: 10.1002/jobm.202100310
Okazawa, Y., Miyazaki, T., Yokoi, G., Ishizaki, Y., Nishikawa, A., and Tonozuka, T. (2015). Crystal structure and mutational analysis of isomalto-dextranase, a member of glycoside hydrolase family 27. J. Biol. Chem. 290, 26339–26349. doi: 10.1074/jbc.M115.680942
Park, B. R., Park, J. Y., Lee, S. H., Hong, S. J., Jeong, J. H., Choi, J. H., et al. (2021). Synthesis of improved long-chain isomaltooligosaccharide, using a novel glucosyltransferase derived from Thermoanaerobacter thermocopriae, with maltodextrin. Enzym. Microb. Technol. 147:109788. doi: 10.1016/j.enzmictec.2021.109788
Pittrof, S. L., Kaufhold, L., Fischer, A., and Wefers, D. (2021). Products released from structurally different dextrans by bacterial and fungal dextranases. Foods 10:244. doi: 10.3390/foods10020244
Pleszczyńska, M., Wiater, A., Bachanek, T., and Szczodrak, J. (2017). Enzymes in therapy of biofilm-related oral diseases. Biotechnol. Appl. Biochem. 64, 337–346. doi: 10.1002/bab.1490
Purushe, S., Prakash, D., Nawani, N. N., Dhakephalkar, P., and Kapadnis, B. (2012). Biocatalytic potential of an alkalophilic and thermophilic dextranase as a remedial measure for dextran removal during sugar manufacture. Bioresour. Technol. 115, 2–7. doi: 10.1016/j.biortech.2012.01.002
Qiu, Y. X., Mao, M. Y., Jang, D., Hong, X., Yang, Y. M., and Hu, T. (2016). Co-operative effect of exogenous dextranase and sodium fluoride on multispecies biofilms. J. Dent. Sci. 11, 41–47. doi: 10.1016/j.jds.2015.08.001
Raddadi, N., Cherif, A., Daffonchio, D., Neifar, M., and Fava, F. (2015). Biotechnological applications of extremophiles, extremozymes and extremolytes. Appl. Microbiol. Biotechnol. 99, 7907–7913. doi: 10.1007/s00253-015-6874-9
Ren, W., Cai, R., Yan, W., Lyu, M., Fang, Y., and Wang, S. (2018). Purification and characterization of a biofilm-degradable dextranase from a marine bacterium. Mar. Drugs 16:51. doi: 10.3390/md16020051
Ren, W., Liu, L., Gu, L., Yan, W., Feng, Y. L., Dong, D., et al. (2019). Crystal structure of GH49 dextranase from Arthrobacter oxidans KQ11: identification of catalytic base and improvement of thermostability using semirational design based on B-factors. J. Agric. Food Chem. 67, 4355–4366. doi: 10.1021/acs.jafc.9b01290
Santiago, M., Ramírez-Sarmiento, C. A., Zamora, R. A., and Parra, L. P. (2016). Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front. Microbiol. 7:1408. doi: 10.3389/fmicb.2016.01408
Schreck, S. D., and Grunden, A. M. (2014). Biotechnological applications of halophilic lipases and thioesterases. Appl. Microbiol. Biotechnol. 98, 1011–1021. doi: 10.1007/s00253-013-5417-5
Seo, E.-S., Nam, S.-H., Kang, H.-K., Cho, J.-Y., Lee, H.-S., Ryu, H.-W., et al. (2007). Synthesis of thermo- and acid-stable novel oligosaccharides by using dextransucrase with high concentration of sucrose. Enzyme Microb.Technol. 40, 1117–1123. doi: 10.1016/j.enzmictec
Sorndech, M. (2018). Isomalto-oligosaccharides: recent insights in production technology and their use for food and medical applications. LWT Food Sci. Technol. 95, 135–142. doi: 10.1016/j.lwt.2018.04.098
Tadeo, X., López-Méndez, B., Trigueros, T., Laín, A., Castaño, D., and Millet, O. (2009). Structural basis for the amino acid composition of proteins from halophilic archea. PLoS Biol. 12:e1000257. doi: 10.1371/journal.pbio.1000257
Tanoue Batista, M. C., Soccol, C. R., Spier, M. R., Libardi Junior, N., de Souza, P., and Vandenberghe, L. (2021). Potential application of dextranase produced by Penicillium aculeatum in solid-state fermentation from brewer’s spent grain in sugarcane process factories. Biocatal. Agric. Biotechnol. 35:102086. doi: 10.1016/j.bcab.2021.102086
Theerachat, M., Guieysse, D., Morel, S., Remaud-Siméon, M., and Chulalaksananukul, W. (2018). Laccases from marine organisms and their applications in the biodegradation of toxic and environmental pollutants: a review. Appl. Biochem. Biotechnol. 187, 583–611. doi: 10.1007/s12010-018-2829-9
Tsutsumi, K., Gozu, Y., Nishikawa, A., and Tonozuka, T. (2019). Structural insights into polysaccharide recognition by Flavobacterium johnsoniae dextranase, a member of glycoside hydrolase family 31. FEBS J. 287, 1195–1207. doi: 10.1111/febs.15074
Vance, T. D. R., Bayer-Giraldi, M., Davies, P. L., and Mangiagalli, M. (2019). Ice-binding proteins and the 'domain of unknown function' 3494 family. FEBS J. 286, 855–873. doi: 10.1111/febs.14764
Walker, G. V., Heng, N. C. K., Carne, A., Tagg, J. R., and Wescombe, P. A. (2016). Salivaricin E and abundant dextranase activity may contribute to the anti-cariogenic potential of the probiotic candidate streptococcus salivarius JH. Microbiology (Reading) 162, 476–486. doi: 10.1099/mic.0.000237
Wang, X., Cheng, H., Lu, M., Fang, Y., Jiao, Y., Li, W., et al. (2016). Dextranase from Arthrobacter oxydans KQ11-1 inhibits biofilm formation by polysaccharide hydrolysis. Biofouling 32, 1223–1233. doi: 10.1080/08927014.2016.1239722
Wang, W., Wang, J., Yan, R., Zeng, R., Zuo, Y., Wang, D., et al. (2021). Expression and characterization of a novel cold-adapted and stable β-agarase gene agaW1540 from the deep-sea bacterium Shewanella sp. WPAGA9. Mar. Drugs 19:431. doi: 10.3390/md19080431
Wang, X., Zhang, Y., Li, M., Qin, Q., and Xie, T. (2022). Purification and characterization of dextranase from Penicillium cyclopium CICC-4022 and its degradation of dextran. Int. J. Biol. Macromol. 204, 627–634. doi: 10.1016/j.ijbiomac.2022.01.196
Wu, G., Zhang, X., Wei, L., Wu, G., Kumar, A., Mao, T., et al. (2015). A cold-adapted, solvent and salt tolerant esterase from marine bacterium Psychrobacter pacificensis. Int. J. Biol. Macromol. 81, 180–187. doi: 10.1016/j.ijbiomac.2015.07.045
Yang, J., Wang, Y., and Zhang, Y. (2016). ResQ: an approach to unified estimation of B-factor and residue-specific error in protein structure prediction. J. Mol. Biol. 428, 693–701. doi: 10.1016/j.jmb.2015.09.024
Yang, Y., Yang, Y., Fan, Q., Huang, Z., Li, J., Wu, Q., et al. (2020). Molecular and biochemical characterization of salt-tolerant trehalose-6-phosphate hydrolases identified by screening and sequencing salt-tolerant clones from the metagenomic library of the gastrointestinal tract. Front. Microbiol. 11:1466. doi: 10.3389/fmicb.2020.01466
Yang, Q., Zhang, M., Zhang, M., Wang, C., Liu, Y., Fan, X., et al. (2018). Characterization of a novel, cold-adapted, and thermostable laccase-like enzyme with high tolerance for organic solvents and salt and potent dye decolorization ability, derived from a marine metagenomic library. Front. Microbiol. 9:2998. doi: 10.3389/fmicb.2018.02998
Yin, J., Chen, J. C., Wu, Q., and Chen, G. Q. (2015). Halophiles, coming stars for industrial biotechnology. Biotechnol. Adv. 33, 1433–1442. doi: 10.1016/j.biotechadv.2014.10.008
Zhang, W., Xu, H., Wu, Y., Zeng, J., Guo, Z., Wang, L., et al. (2018). A new cold-adapted, alkali-stable and highly salt-tolerant esterase from bacillus licheniformis. Int. J. Biol. Macromol. 111, 1183–1193. doi: 10.1016/j.ijbiomac.2018.01.152
Zhao, J., Wang, L., Wei, X., Li, K., and Liu, J. (2020). Food-grade expression and characterization of a dextranase from Chaetomium gracile suitable for sugarcane juice clarification. Chem. Biodivers. 17:e2000797. doi: 10.1002/cbdv.202000797
Keywords: salt-tolerant, cold-adapted, marine dextranase, dental plaque, isomalto-oligosaccharides5
Citation: Xu L, Zhang Y, Liu N, Wei Z, Wang Z, Wang Y and Wang S (2022) Purification and characterization of cold-adapted and salt-tolerant dextranase from Cellulosimicrobium sp. THN1 and its potential application for treatment of dental plaque. Front. Microbiol. 13:1012957. doi: 10.3389/fmicb.2022.1012957
Edited by:
Jinwei Zhang, University of Exeter, United KingdomReviewed by:
Kui Wang, Guangzhou University of Chinese Medicine, ChinaWei Ren, Hainan University, China
Copyright © 2022 Xu, Zhang, Liu, Wei, Wang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linxiang Xu, eHVsaW54aWFuZzE5ODhAMTYzLmNvbQ==; Yonghua Wang, eW9uZ2h3QHNjdXQuZWR1LmNu; Shujun Wang, c2p3YW5nQGpvdS5lZHUuY24=
 Linxiang Xu
Linxiang Xu Yan Zhang4
Yan Zhang4 Nannan Liu
Nannan Liu Zhen Wei
Zhen Wei Yonghua Wang
Yonghua Wang