
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 November 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1010584
Avibacterium paragallinarum-associated infectious coryza (IC) is an important threat in commercial poultry. Previous studies about the characteristics of A. paragallinarum are succeeded in revealing the course of IC disease, but whether and how resident microbes contribute to the infection remains unclear. To understand the role of commensal bacteria, we isolated 467 commensal bacteria, including 38 A. paragallinarum, from the respiratory tract of IC chicken. The predominant commensal isolates were Gram-positive bacteria belonging to Staphylococcus spp. [33.19%, 95% confidence interval (CI): 28.93–37.66%], Enterococcus spp. (16.49%, 95% CI: 13.23–20.17%), and Bacillus spp. (16.27%, 95% CI: 13.04–19.94%). These isolates were closely correlated with the survival of A. paragallinarum. We examined and found that commensal bacteria aggravate A. paragallinarum-associated infections because certain commensal species (28.57%, 95% CI: 15.72–44.58%) induced hemolysis and promoted the growth of A. paragallinarum in vitro. Notably, A. paragallinarum showed high resistance to routine antibiotics such as erythromycin (84.21%, 95% CI: 68.75–93.98%), tetracycline (73.68%, 95% CI: 56.90–86.60%) and carried diverse mobile resistance gene clusters. Overall, we found commensal bacteria especially Gram-positive bacteria facilitate the survival of multidrug-resistant A. paragallinarum to exacerbate infections, suggesting that novel strategies may diminish A. paragallinarum-associated infections by modulating the population dynamics of commensal bacteria.
HIGHLIGHTS
- Commensal bacteria isolated from respiratory tracts in IC chicken show correlative to the presence of A. paragallinarum and facilitate the survival.
- The symbiotic relationship between commensals and A. paragallinarum commonly present in respiratory tracts of IC chicken.
- Mobile resistance gene clusters are distributed globally in A. paragallinarum isolates, against frequently-used antimicrobial agents in commercial poultry.
Infectious coryza (IC) is an important threat to global animal health and causes huge economic losses, resulting in seriously decreased egg production and increased mortality in laying hens (Alvarez et al., 2020; Nouri et al., 2021). The distinguishing clinical symptoms are anorexia, facial swelling, nasal discharge, lacrimation, and even mortality (Xu et al., 2019). IC is an acute upper respiratory tract (URT) inflammation in chickens caused by a specific opportunistic pathogen, Avibacterium paragallinarum, historically named Haemophilus paragallinarum (Guo et al., 2022). Such bacterial pathogens are nutrient-dependent symbiotic bacteria, especially requiring sufficient accessory growth factors [X, hemin and V, nicotinamide adenine dinucleotide (NAD)] to proliferate in vivo (Mouahid et al., 1992). In homeostasis, commensal bacteria exclude exogenous microorganisms and directly inhibit the growth of pathogens (Clark, 2020) resulting in the limited establishment of opportunistic pathogens such as A. paragallinarum in respiratory tract (Long et al., 2012). When host immunity is compromised, A. paragallinarum initiates colonization and invasion, which is facilitated by commensals like Gallibacterium anatis (Paudel et al., 2017), Ornithobacterium rhinotracheale (Morales-Erasto et al., 2016), and Staphylococcus chromogenes (Wu et al., 2021). Resident species constitute a new niche to allow A. paragallinarum colonization and virulence by providing public goods and disrupting host barriers (Hoare et al., 2021; Wu et al., 2021). Additionally, protection mediated by indigenous microbial communities including extensive horizontal gene transfer (McInnes et al., 2020) and antibiotic inactivation (Gjonbalaj et al., 2020) increases the antimicrobial tolerance and resistance of pathogens. The benefit of commensal protection and facilitation is not confined to early growth, rather it is continuous during the infection. As a result, antimicrobials might not perform well in IC disease because of the combined effect within bacterial communities.
As is found that symbiotic interaction is critical for the survival of A. paragallinarum, multiple resistance phenotypes emerging in A. paragallinarum also contribute to the establishment in vivo and limit the efficacy of antimicrobials. To date, vaccination is still the most important implementation to provide effective protection against infection (Van den Biggelaar et al., 2020). However, this intervention is not sufficient, especially for infecting with high virulence and resistance strains (Trujillo-Ruiz et al., 2016). Once the vaccination is no longer effective, antimicrobial is considered as a complementary alternative to alleviate the symptom of the disease. However, irrational use of antibiotics accelerates the development of multidrug resistance and virulence in both pathogens like A. paragallinarum and indigenous microbiota (Nhung et al., 2017; Wongsuvan et al., 2018; Juricova et al., 2021). Further, published data about the genetic environment and transferability of virulence genes (VGs) and antimicrobial resistance genes (ARGs) in A. paragallinarum are limited. Therefore, characterization of resistant traits of A. paragallinarum is urgently needed for the appropriate use of antibiotics.
Although the causative agent of IC disease was defined, further efforts are still required to uncover the correlation between commensals and A. paragallinarum. Besides, the resistance genotypes and phenotypes of A. paragallinarum are needed to characterize for development of more effective treatment. In this study, we aimed to determine the promotion effect contributed by commensals through find out the association between A. paragallinarum and commensal bacteria in URT from IC chickens and subsequently analyzed the characteristics of antimicrobial resistance and virulence in all A. paragallinarum isolates to comprehensively understand the traits of resistant A. paragallinarum.
Layer chickens with symptom of IC (n = 38) and healthy chickens (n = 9) were collected from commercial layer farms in Tianjin, Hebei, and Guangxi in China from 2019 to 2022. All samples were vaccinated against IC and with no antibiotic treatment after infection. The heads of infected and non-infected birds were disinfected with 75% ethyl alcohol before bacterial isolation. Swab samples collected from nasal cavity and infraorbital sinuses were cultured on blood agar [trypticase soya agar (TSA) supplemented with 5% filter-sterilized sheep] and TSA plus agar (supplemented with 20 μg/ml NAD and 5% fetal bovine serum), respectively. TSA plus agar plates were used to isolate the A. paragallinarum and incubated aerobically at 5% CO2 and 37°C for 24–48 h. The colony morphology consisting with translucent light blue was selected as A. paragallinarum for further study. Commensals were cultured in blood agar plates at 37°C for 24 h and further purified in TSA agar. Afterward, all isolates were identified based on MALDI-TOF-MS identification and 16S rRNA gene sequence analysis. After confirming the taxonomic classification of these isolates, A. paragallinarum was freeze-dried with 10% skim milk and commensals were maintained in nutrient broth supplemented with 20% glycerol and stored at −80°C.
The growth-promotion effect contributed by commensal was determined through the observation of satellitic growth of A. paragallinarum. All commensal bacteria were cultured overnight and then diluted to 108 CFUs/ml. Meanwhile, A. paragallinarum was cultured in TBS plus broth for 18 h at 37°C, and then diluted to 106 CFUs/ml. Subsequently, 100 μl of diluted bacteria suspension was spread on the TSA blood agar, and then spotted 2 μl of the commensal bacteria solution on the center of the blood plate incubating in carbon dioxide incubator for 24–48 h. The experiment was repeated twice and the radius of growing circle of A. paragallinarum was recorded. The calculation equation of promotion rate = growth radius of A. paragallinarum − growth radius of commensal.
According to the previous research (Wu et al., 2021), the relationship between A. paragallinarum and commensals was defined as three synergistic indexes by the following formulae: (1) Isolation rate (%) = the number of isolates of a specific genus or species/total number of isolates; (2) Satellitism rate (%) = the number of specific species or genus of bacteria that facilitate the growth of A. paragallinarum/total number of commensal isolates that facilitate the growth of A. paragallinarum; (3) Colocalization rate (%) = the number of chickens infected with a specific species or genus of bacteria and A. paragallinarum/the number of chickens infected with A. paragallinarum.
To evaluate the hemolytic activity of commensal bacteria, isolates were spot on 5% sheep blood cells (RBCs) TSA agar after diluted to 106 CFUs/ml, and then recorded the transparent hemolysis radius after cultured for 12, 24, 36, 48, 60, and 72 h at 37°C. Meanwhile, hemolysis of supernatant from commensals was also determined according to the method described previously (Deng et al., 2021) with minor modifications. In brief, isolates were cultured in TSB for 24, 48, and 72 h to harvest the bacterial supernatant. RBCs were harvested by centrifugation (1,000 rpm/min, 10 min) and rinsed three times in phosphate buffered saline (PBS) solution. After RBCs was diluted to 8% (vol/vol), 100 μl bacterial supernatant was mixed with an equal volume of 8% RBCs and incubated at 37°C for 1 h, and then centrifuged at 1,000 rpm for 10 min at 4°C. The 4% RBCs suspension in PBS and Triton X-100 (0.1%) with RBCs suspension were used as negative control and positive control, respectively. The hemolytic activity was assessed by measuring the optical absorbance of the supernatant at OD576. The calculation equation of hemolysis: Hemolysis rate (%) = (ODSample − ODNegative)/(ODPositive − ODNegative) × 100%.
The broth microdilution method, as described by the Clinical and Laboratory Standards Institute guidelines (Clinical and Laboratory Standards Institute [CLSI], 2020), was used to determine the susceptibility of 38 A. paragallinarum isolates to 13 antimicrobial agents (the categories of antimicrobial agents were shown in Supplementary Table 2). The medium used in the susceptibility test was supplemented with 10% chicken serum and 0.0025% NAD (CAMHB plus broth) as described previously (Heuvelink et al., 2018). The MIC (minimum inhibitory concentration) ranges and breakpoints for ampicillin, erythromycin, and tetracycline were found via referral to the previous research (Blackall, 1988). For other antimicrobial agents (penicillin and meropenem), these criteria were referred to CLSI documents (Clinical and Laboratory Standards Institute [CLSI], 2016). Escherichia coli ATCC 25922 served as the quality control strain to validate the results obtained.
Genomic DNA of A. paragallinarum was prepared by using the TIANamp Bacterial DNA Kit (Tiangen Biotech, Beijing, China). DNA was then fragmented to prepare the library and was sequenced using Illumina NovaSeq 6,000 (Illumina, United States) in pair-end model. High-quality reads were de novo assembled by using SPAdes v3.12.0 (Bankevich et al., 2012) and annotated by Prokka v1.14.6 (Seemann, 2014). Phylogenetic relationships between the isolates were determined by bacterial core genome using bcgTree (Ankenbrand and Keller, 2016), and SNP analysis was conducted by Harvest v1.1.2 (Treangen et al., 2014). Mobile gene elements (MGEs) were screened including insertion sequences, prophage elements, integrative and conjugative elements (ICEs), and plasmid sequences. ISfinder (Siguier et al., 2006) and ICEberg (Liu et al., 2019) databases were used to identify complete or partial elements via BLASTn with detailed parameters (identity ≥70% and coverage ≥50%). Prophages and prophage-like elements were analyzed by Phigaro v2.3.0 (Starikova et al., 2020). The virulence factors and ARGs of A. paragallinarum were identified based on VFDB (Chen et al., 2005), ResFinder (Bortolaia et al., 2020), and the Comprehensive Antibiotic Resistance (CARD) (Alcock et al., 2020) database using BLASTn with a cut-off of 60% of coverage and 80% of identity. Finally, the genetic environment of VGs and ARGs gene clusters was further inspected by using CD-search and ORFFinder (Sayers et al., 2022), and collinearity analysis using clinker and clustermap.js (Gilchrist and Chooi, 2021) to compare the similarity of gene clusters with representative gene clusters previously reported.
The data was statistically analyzed by using GraphPad Prism 9. Tests of normal distribution were performed (Shapiro–Wilk test) and data were analyzed using one-way non-parametric (Kruskal–Wallis test) or one-way ANOVA with Tukey’s post hoc tests where applicable for continuous variables. LM was performed in the R package (version 4.1.3) to examine the correlation.
A total of 554 culturable bacterial isolates were identified from the respiratory tracts of healthy chickens and IC chickens (Figure 1). The URT bacterial composition in either IC chickens or healthy chickens was generally dominated by bacteria from two different phyla: Firmicutes and Proteobacteria (Supplementary Tables 1, 2). In healthy chickens, Proteobacteria was the predominant phylum, accounting for ∼60% of total isolates. By contrast, the diversity of culturable microbiome was significantly changed when the chicken was infected with A. paragallinarum, as Firmicutes were the most prominent phylum. In URT of IC chickens, a total of 467 commensal and pathogenic bacteria were to be isolated, including the causative agents such as A. paragallinarum, G. anatis, Pseudomonas aeruginosa, and Rothia nasimurium (Supplementary Table 1). Although A. paragallinarum is the primary etiologic agent in IC, most of the isolates cannot survive after being isolated from the host, resulting in a low isolation rate (57.89%, 22/38). In contrast to the healthy group, URT microbiota in infected chickens was mainly composed of Gram-positive bacteria, which was dominated by three most prominent genera: Staphylococcus (33.19%), Enterococcus (16.49%), and Bacillus (16.27%) (Figure 1). Each abundant genera had the predominant species, such as S. chromogenes, E. faecalis, and B. subtilis, with isolation rates of 19.7, 13.06, and 8.14%, respectively (Supplementary Table 1). Interestingly, the bacterial community in URT comprised large amounts of Bacillus spp. such as B. subtilis, B. haynesii, B. amyloliquefaciens, and B. velezensis (Supplementary Table 1), which were rarely reported in previous studies. Some oral and respiratory tract flora were also identified including Rothia spp., Acinetobacter spp., Corynebacterium spp., and G. anatis, with isolation rates of 5.57% (26/467), 4.91% (23/467), 4.28% (20/467), and 3.43% (16/467), respectively. The abundance of minor bacterial genera (less than 2% isolation rate) ranging from 1.71 to 0.21% contained respiratory tract pathogens such as Streptococcus pluranimalium and P. aeruginosa, implying that these isolates might not be the causative agents of IC.

Figure 1. Bacterial composition in upper respiratory tract from healthy chickens and IC chickens. In infection group, the bacterial genera with relative abundance of less than 2% were categorized into “Others” group, including Aerococcus spp. (0.21%), Carnobacterium spp. (0.21%), Enterobacter spp. (0.64%), Escherichia spp. (0.21%), Lysinibacillus spp. (1.71%), Paenibacillus spp. (0.21%), Streptococcus spp. (1.07%), Stenotrophomonas spp. (0.43%), Vibrio spp. (0.21%), and Kocuria spp. (1.71%).
The symbiotic growth of A. paragallinarum was associated with a part of commensals. To reveal the potential contribution of commensal to A. paragallinarum-associated infections, we determined the growth-promoting ability of commensal bacteria by measuring the radius of satellitic zone (Figure 2A). Based on the radius of satellitism, the strengths of promotion effect were categorized into three degrees: weak (<4 mm), medium (4–8 mm), and strong (>8 mm) (Figure 2B). In URT of IC chickens, the majority of bacterial members (31/42) showed growth-promoting effects on A. paragallinarum among which four species exhibited strong promotion, including B. safensis, B. wiedmanii, C. jeikeium, and S. epidermidis. The ability and strength of growth promotion showed species specificity in commensal bacteria. For instance, C. jeikeium was the particular species facilitating the growth of A. paragallinarum in genus of Corynebacterium spp. E. faecalis and K. kristinae displayed weak capability of promotion, and E. faecalis was the unique species that could interact with A. paragallinarum among enterococci. Similarly, in relatively low abundant genera (Rothia, Kocuria, and Paenibacillus), the promotion effect was restricted within certain species. In this case, the ability of growth promotion was not shared by closely related species. Conversely, in two abundant genera (Bacillus and Staphylococcus), all isolates showed promotion capacity with average satellitic radii ranging from 5 to 9 mm (Figure 2B), suggesting that at least some genera might have common capability that facilitated the survival of A. paragallinarum. Importantly, opportunistic pathogens in respiratory tract such as S. pluranimalium and Stenotrophomonas maltophilia were positive for the result of satellitism, implying that such pathogens may increase the possibility of co-infection in IC chicken.
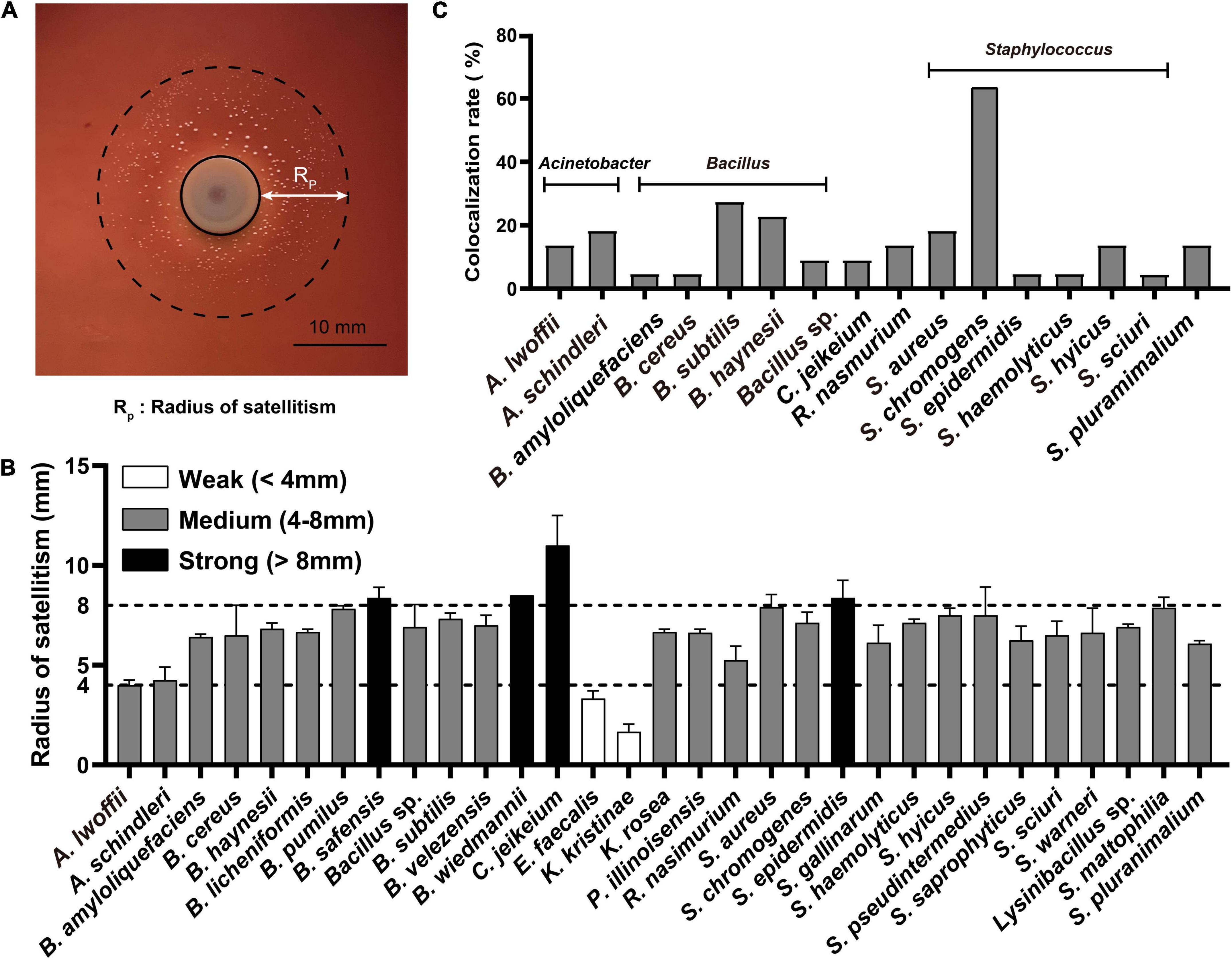
Figure 2. Commensals facilitated the growth of A. paragallinarum. (A) Representative image of the satellitism that commensal bacterium was surrounded by A. paragallinarum. (B) The satellitism profiles of commensals from IC chicken. (C) The colocalization profiles of commensals with strong growth-promoting effect on A. paragallinarum. The full names of genera were replaced by the abbreviation in this figure, and the full name of genera was listed in Supplementary Table 1.
The interaction between microbes is spatial short-range in communities (Dal Co et al., 2020), thus colocalization is a prerequisite synergistic index that commensals could contribute to the growth of A. paragallinarum. A total of 16 species were selected as candidates for further study as they showed strong or medium growth promotion effects and colocalized with A. paragallinarum (Figure 2C). It was notable that the genera of Bacillus and Staphylococcus were the prominent members co-isolating with A. paragallinarum and facilitating the survival of A. paragallinarum in vitro. In those candidate species, S. chromogenes showed extremely colocalized with A. paragallinarum. Interestingly, Gram-positive bacteria accounted for nearly 92% of all growth-promotion commensals, suggesting that Gram-positive commensals are primary microorganisms in feeding Gram-negative A. paragallinarum.
Given that interspecific interplays are common in commensal microbiota, it may contribute to flourishment of specialized kinds of commensals in IC chicken, thus we performed correlation analysis using linear regression model to investigate the relationship among bacterial isolation, colocalization, and satellitism. The p-value of correlation between isolation and colocalization at genus-level is less than 0.05, with Pearson’s correlation coefficient of 0.8156, indicating that positive correlation between colocalization and isolation irrespective of bacterial taxonomy (Figure 3A). Hence, we hypothesized that isolation and colocalization are closely associated with satellitism in IC chicken. To verify this hypothesis, we calculated the satellite rate in different genus isolates and examined the correlation between satellitism and isolation or colocalization by linear model. The results indicated that satellitism was significant positively correlated with isolation (p = 6.334e−08, R = 0.847) and colocalization (p = 0.001711, R = 0.4691) (Figures 3B,C). Of three predominant genera (Bacillus, Enterococcus, and Staphylococcus), the same positive correlations were observed as most of these isolates could support the growth of A. paragallinarum (Supplementary Figure 1), but such growth-promoting characteristics were slightly different in Enterococcus because the promotion effect was restricted in the specific species E. faecalis (Supplementary Table 1). The most noteworthy feature of these correlations was that abundant commensals in URT commonly colocalized with A. paragallinarum and could facilitate the survival of A. paragallinarum (Supplementary Table 1). In conclusion, the growth-promoting commensals showed positive interaction between A. paragallinarum, resulting in richness of promoting commensal bacteria.
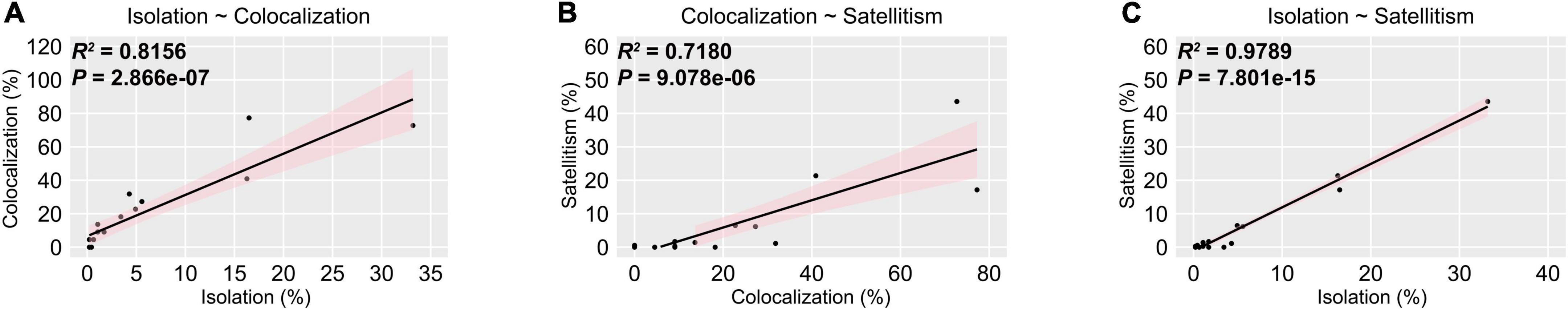
Figure 3. Isolation is positively correlated with co-colonization and satellitism at genus-level. The pink shaded areas represented the 95% confidence intervals. (A) Isolation is positively correlated with colocalization; (B) Colocalization is positively correlated with satellitism; (C) Isolation is positively correlated with satellitism.
To obtain nutrients from host, many respiratory tract microbes evolve the ability to disrupt the host barrier, particularly through hemolysis. Notably, the growth factors required by A. paragallinarum are available after microbiota-mediated RBCs lysis (Demarest et al., 2019). Thus, we screened the hemolytic activity of candidate commensals under different cultural conditions, to emphasize host-commensal interaction that benefits the A. paragallinarum survival. Five of these commensal species exhibited increasing hemolytic activity when cultured in blood agar (Figure 4A). Compared to the solid culture, more commensal species showed hemolysis to various extents in broth medium, such as the complete hemolysis in B. cereus and C. jeikeium, 50–90% of hemolysis rate in Bacillus sp., S. chromogens, and S. haemolyticus (Figure 4B and Supplementary Figure 2). These tendencies of hemolysis changes are different among colocalized commensals. Certain species (37.5%, 6/16) showed the highest hemolysis rate at 48 h, while 25% (4/16) and 18.75% (3/16) of tested species exhibited the strongest hemolysis at 24 and 72 h, respectively (Supplementary Figure 2). In contrast to static solid cultivation, more species showed hemolysis in liquid shake culture. Delving into more detail, seven species, including B. amyloliquefaciens, B. haynesii, B. subtilis, R. nasimurium, S. epidermidis, S. chromogenes, and S. haemolyticus, were positive for hemolysis in liquid cultures rather than in solid cultures, suggesting commensal-mediated hemolysis were prone to potentiate in liquid environment of host cell.
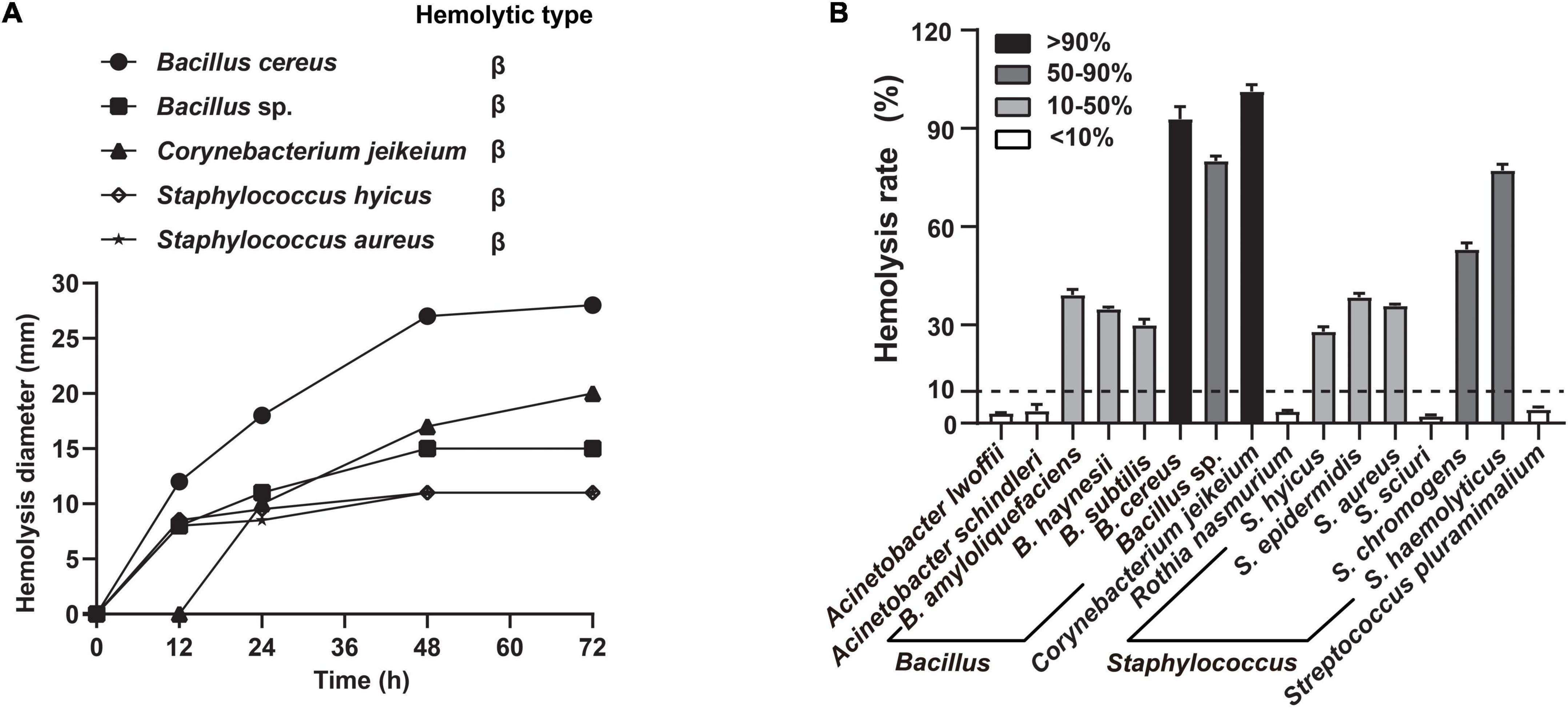
Figure 4. Commensals exhibited extensive hemolytic activity. The selected commensal bacteria were both exhibiting strong or medium growth-promoting effects and colocalizing with A. paragallinarum. (A) Five representative species were selected to determine the change of hemolysis on blood agar at 12, 24, 48, and 72 h; (B) distribution of hemolytic activity among 16 species commensals in supernatant test at 48 h. Hemolysis rate greater than 10% is considered positive hemolysis in this assay.
In the context of the commensal microbiota, resistant A. paragallinarum was promoted to colonize in vivo. The resistance characteristics in A. paragallinarum isolates further contribute to survival under antibiotic treatment. We found that A. paragallinarum isolates showed high MICs values to routine used antibiotics in poultry (Supplementary Table 2). Majority of isolates (73.68%) were resistant to tetracycline, consisting with the presence of tetracycline resistance gene tetB. The tetB resistance gene was located in Tn10 transposons and surrounded by mobile elements (Figure 5A). The Tn10 transposons in A. paragallinarum isolates were categorized into five representative sequence types on the basis of genetic environment. We noticed that the novel tetB hybrid cluster (Type E) was composed of multiple ARGs such as cat, aph, and sul, counting as Tn10 variant (Figure 5A). Except for tetracyclines, three kinds of antimicrobials are usually used in the treatment of IC, including macrolides, fluoroquinolones, and β-lactams. In terms of macrolides, 84.21% (32/38) of isolates were resistant to erythromycin and 31.58% (12/38) of isolates showed high MICs value of ≥ 64 μg/ml for tylosin, whereas only two isolates carried the macrolide resistance gene mef(B), implying that novel determinants could play a part in such resistance. As for antimicrobials of fluoroquinolones and β-lactams, we found two isolates showed extremely high MIC values of >16 μg/ml for enrofloxacin, and 28.94% (11/38) of isolates are ampicillin resistance while eight isolates (21.05%) were positive for β-lactam resistance genes including blaCTX–M–14, blaOXA–1, and blaROB–1 (Supplementary Table 2 and Supplementary Figure 3). The genetic environment of blaROB–1 in A. paragallinarum showed similarity to the plasmid pB1000 found in Glaesserella parasuis. Besides, compared with the breakpoint of streptomycin in A. paragallinarum, seven isolates (18.42%) showed resistance to gentamicin with a MIC value of >16 μg/ml. The genetic environment of type II gene cluster composed of several aminoglycoside resistance genes (ant, aac, and aph) shared high sequence similarity for the multi-drug resistance plasmid pYMH5, which was homologous to broad host-range plasmid pLS88 from H. ducreyi. Notably, 13 of 38 A. paragallinarum isolates exhibited potentially resistant to colistin with MICs value of 32 μg/ml or greater, but colistin resistance genes were not found, indicating a novel mechanism contributed to this phenotype.
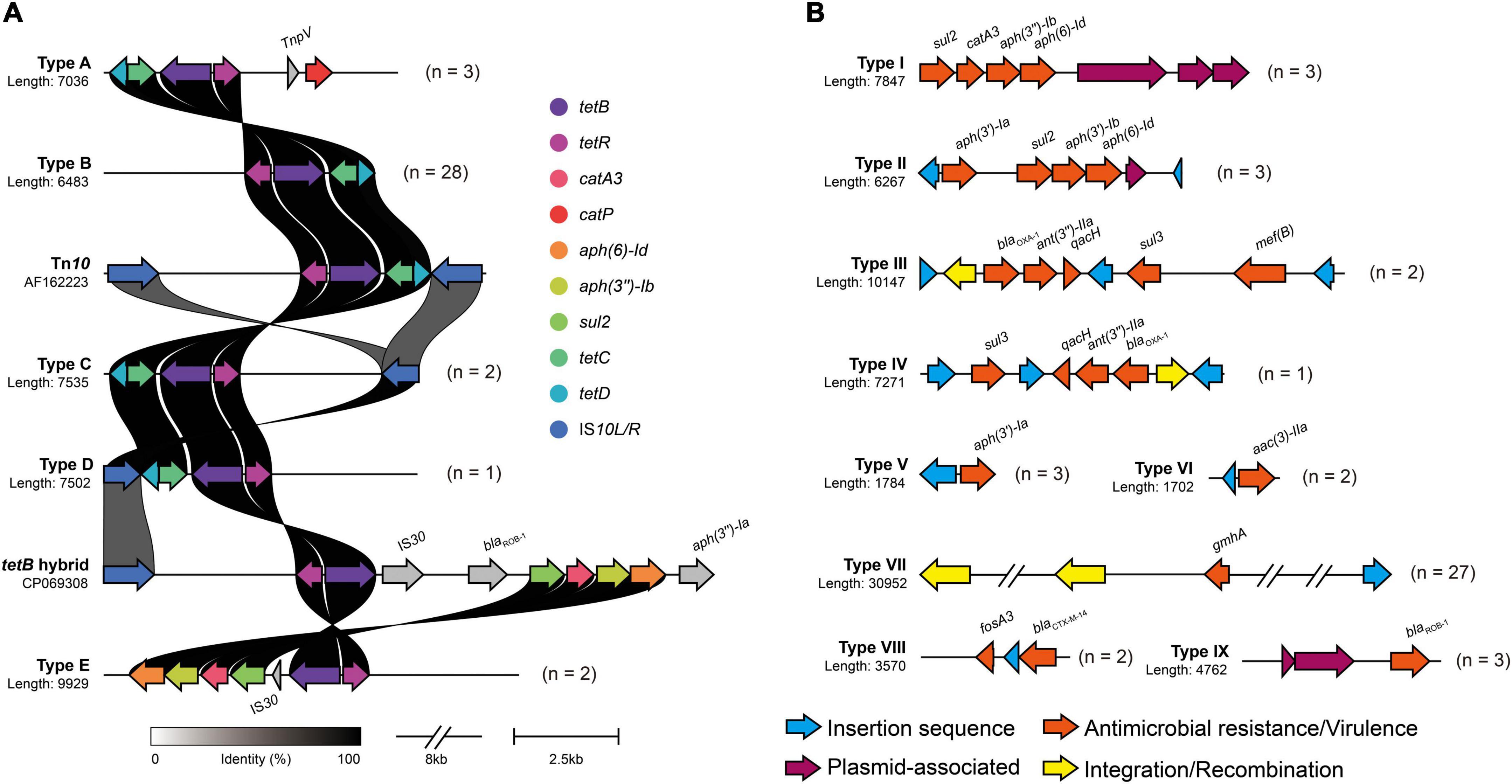
Figure 5. Genetic environment of mobile virulence and antimicrobial resistance gene in A. paragallinarum. Different types of antimicrobial resistance gene clusters were shown in above figure, and numbers followed by genetic profile represented the total numbers of similar gene clusters in all A. paragallinarum. (A) Profiles of tetB cluster in A. paragallinarum. The major functional proteins in Tn10 transposon, including TetB (tetracycline efflux protein), TetR (TetR family transcriptional regulator), TetC (transposon Tn10 TetC protein), TetD (transposon Tn10 TetD protein), and two insertion sequence IS10L and IS10R. Tn10 transposon in Shigella flexneri (accession: AF162223) and tetB hybrid sequence in Glaesserella parasuis (accession: CP069308) served as reference sequences for the comparison of tetracycline resistance gene cluster in A. paragallinarum. The gray shading genes indicated that these determinants were dissimilarity among other sequences. (B) Diversity of mobile gene cluster in A. paragallinarum. Mobile gene clusters excluding tetB clusters were categorized into nine sequence types.
Strikingly, resistance genes were almost flanked by mobile genetic elements. Several resistance genes associated with MGEs combined into a large gene cluster were globally distributed in this study as shown in Figure 5. Additionally, we also depicted the VGs and their genetic environment in A. paragallinarum. All of A. paragallinarum were carried VGs lpxC, manB/yhxB, and gmhA/lpcA, which involved in lipooligosaccharide (LOS) and exopolysaccharide biosynthesis. Endotoxin biosynthesis genes kdsA were restricted to group B A. paragallinarum, suggesting that kdsA acted as a marker gene in comparison to group A A. paragallinarum (Supplementary Figure 3). Notably, 71.05% of phosphohexose isomerase genes (gmhA/lpcA) were flanked by integrases and insertion sequences (Figure 5B). The positive virulence and antibiotic resistance genes were list in Supplementary Table 3.
The resident microbiota plays a crucial role in eradicating the colonization of pathogens and rebuilding microbial defense systems to maintain homeostasis (Li et al., 2019). However, certain commensal bacteria have been found to be positively correlated with the growth of pathobionts during infections (Stacy et al., 2014). The symbiotic relationship between A. paragallinarum and commensal microbiota may compromise therapeutic efficacies and aggravate persistent infections (Rawson et al., 2020; Westblade et al., 2021). Besides, the species of commensal bacteria that contribute to such infection remains elusive. Here, we find that several species of commensal bacteria promote the growth of A. paragallinarum, including Staphylococcus, Enterococcus, and Bacillus. These commensals are mainly composed of Gram-positive bacteria, consisting with previous observations that Gram-positive commensals create suitable niches for Gram-negative pathogens under resource-limited conditions (Piccardi et al., 2019). Hence, commensals are important for the survival of A. paragallinarum, especially for these co-existed Gram-positive bacteria in the respiratory tract.
The hemolytic activity may play a crucial role in interspecies interactions. The lysis of RBCs by commensal subsequently releases intracellular contents particularly NAD+ (Mouahid et al., 1992) to favor the survival of opportunistic pathogens. The capability of hemolysis in B. cereus, C. jeikeium, S. hyicus, S. aureus, and S. haemolyticus, can disrupt host barriers to accelerate the replications of A. paragallinarum in respiratory tract. For example, B. cereus is capable to produce diverse enterotoxins like Nhe and Hbl (Cui et al., 2016; Zhu et al., 2016), to promote the survival and growth of A. paragallinarum. Similarly, toxins secreted by Staphylococcus spp. potentiate infections as well (Cohen et al., 2016). Therefore, commensal-mediated hemolysis probably contributes to the increased abundance of A. paragallinarum.
The antimicrobial resistant A. paragallinarum aggravate infections. A. paragallinarum show increasing resistance to antimicrobial agents that are recommended for the treatment of IC such as ampicillin and tetracycline (Nhung et al., 2017; Fauziah et al., 2021). We find that 28.94% (11/38) of A. paragallinarum isolates show resistance to ampicillin. Nevertheless, Heuvelink et al. (2018) test 44 field isolates originating from 25 outbreaks and find all isolates are sensitive to ampicillin. Besides, tetB is present in all A. paragallinarum isolates and locates in the Tn10 transposon. The high level of tetracycline may promote the acquisition of mobile tetB gene in sensitive strains. Additionally, the genetic environment of Tn10 is similar to the ICEs in G. parasuis (Figure 2), suggesting that the transferable interspecies gene tetB is undergoing within Haemophilus (Sun et al., 2020). Notably, several strains show high MIC values (≥32 μg/ml) of colistin, whereas no mcr genes are detected in A. paragallinarum, indicating that additional undetermined resistant elements may contribute to such phenotype. Conclusively, the resistance characteristics emerging in A. paragallinarum enable them to better survival in vivo.
The composition and diversity of URT microbiota shift by the colonization of A. paragallinarum. Commensal bacteria particularly Gram-positive commensals show closely related to the presence of resistant A. paragallinarum and benefit the survival thereof. The commensal-mediated hemolysis further promotes the growth of A. paragallinarum and probably aggravates the infection. Therefore, targeting the population dynamics of commensal bacteria may be a promising therapeutic approach to treat IC.
The draft whole genome sequence assemblies in the study are deposited in the GenBank repository, accession number PRJNA836796 can found in the article/Supplementary material.
This animal study was reviewed and approved by the China Agricultural University. Written informed consent was obtained from the owners for the participation of their animals in this study.
JJZ: conceptualization and writing—review and editing. YSC: methodology, validation, formal analysis, investigation, and writing—review and editing. YFW: methodology, validation, and formal analysis. KZ and YQW: conceptualization, methodology, and funding acquisition. All authors contributed to the article and approved the submitted version.
This work was funded by grants from the Laboratory of Lingnan Modern Agriculture Project (NT2021006) and the National Natural Science Foundation of China (31922083).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1010584/full#supplementary-material
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Alvarez, K. L. F., Poma-Acevedo, A., and Fernandez-Diaz, M. (2020). A transient increase in MHC-II(low) monocytes after experimental infection with Avibacterium paragallinarum (serovar B-1) in SPF chickens. Vet. Res. 51:123. doi: 10.1186/s13567-020-00840-7
Ankenbrand, M. J., and Keller, A. (2016). bcgTree: Automatized phylogenetic tree building from bacterial core genomes. Genome 59, 783–791. doi: 10.1139/gen-2015-0175
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Blackall, P. (1988). Antimicrobial drug resistance and the occurrence of plasmids in Haemophilus paragallinarum. Avian Dis. 32, 742–747.
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
Clark, S. E. (2020). Commensal bacteria in the upper respiratory tract regulate susceptibility to infection. Curr. Opin. Immunol. 66, 42–49. doi: 10.1016/j.coi.2020.03.010
Clinical and Laboratory Standards Institute [CLSI] (2016). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, M45, 3rd Edn. Wayne, PA: CLSI.
Clinical and Laboratory Standards Institute [CLSI] (2020). Performance Standards for Antimicrobial Susceptibility Testing, M100, 30 Edn. Wayne, PA: CLSI.
Cohen, T., Hilliard, J., Jones-Nelson, O., Keller, A., Oday, T., Tkaczyk, C., et al. (2016). Staphylococcus aureus α toxin potentiates opportunistic bacterial lung infections. Sci. Transl. Med. 8:329ra31. doi: 10.1126/scitranslmed.aad9922
Cui, Y., Liu, Y., Liu, X., Xia, X., Ding, S., and Zhu, K. (2016). Evaluation of the toxicity and toxicokinetics of cereulide from an emetic Bacillus cereus strain of milk origin. Toxins 8:156. doi: 10.3390/toxins8060156
Dal Co, A., van Vliet, S., Kiviet, D. J., Schlegel, S., and Ackermann, M. (2020). Short-range interactions govern the dynamics and functions of microbial communities. Nat. Ecol. Evol. 4, 366–375. doi: 10.1038/s41559-019-1080-2
Demarest, T. G., Truong, G. T. D., Lovett, J., Mohanty, J. G., Mattison, J. A., Mattson, M. P., et al. (2019). Assessment of NAD(+)metabolism in human cell cultures, erythrocytes, cerebrospinal fluid and primate skeletal muscle. Anal. Biochem. 572, 1–8. doi: 10.1016/j.ab.2019.02.019
Deng, F., Chen, Y., Sun, T., Wu, Y., Su, Y., Liu, C., et al. (2021). Antimicrobial resistance, virulence characteristics and genotypes of Bacillus spp. from probiotic products of diverse origins. Food Res. Int. 139:109949. doi: 10.1016/j.foodres.2020.109949
Fauziah, I., Asmara, W., and Wahyuni, A. (2021). Antimicrobial sensitivity of Avibacterium paragallinarum isolates from layers in the special region of Yogyakarta, Indonesia. Vet. World 14, 1124–1127. doi: 10.14202/vetworld.2021.1124-1127
Gilchrist, C. L. M., and Chooi, Y. H. (2021). Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics [Epub ahead of print]. doi: 10.1093/bioinformatics/btab007
Gjonbalaj, M., Keith, J. W., Do, M. H., Hohl, T. M., Pamer, E. G., and Becattini, S. (2020). Antibiotic degradation by commensal microbes shields pathogens. Infect. Immun. 88, e00012–20. doi: 10.1128/IAI.00012-20
Guo, M., Liu, D., Chen, X., Wu, Y., and Zhang, X. (2022). Pathogenicity and innate response to Avibacterium paragallinarum in chickens. Poult. Sci. 101:101523. doi: 10.1016/j.psj.2021.101523
Heuvelink, A., Wiegel, J., Kehrenberg, C., Dijkman, R., Soriano-Vargas, E., and Feberwee, A. (2018). Antimicrobial susceptibility of Avibacterium paragallinarum isolates from outbreaks of infectious coryza in Dutch commercial poultry flocks, 2008-2017. Vet. Microbiol. 217, 135–143. doi: 10.1016/j.vetmic.2018.03.008
Hoare, A., Wang, H., Meethil, A., Abusleme, L., Hong, B. Y., Moutsopoulos, N. M., et al. (2021). A cross-species interaction with a symbiotic commensal enables cell-density-dependent growth and in vivo virulence of an oral pathogen. ISME J. 15, 1490–1504. doi: 10.1038/s41396-020-00865-y
Juricova, H., Matiasovicova, J., Kubasova, T., Cejkova, D., and Rychlik, I. (2021). The distribution of antibiotic resistance genes in chicken gut microbiota commensals. Sci. Rep. 11:3290. doi: 10.1038/s41598-021-82640-3
Li, K. J., Chen, Z. L., Huang, Y., Zhang, R., Luan, X. Q., Lei, T. T., et al. (2019). Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir. Res. 20:272. doi: 10.1186/s12931-019-1246-0
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., et al. (2019). ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, D660–D665. doi: 10.1093/nar/gky1123
Long, S. S., Pickering, L., and Prober, C. G. (2012). Principles and Practice of Pediatric Infectious Diseases, 4th Edn. Edinburg: Elsevier Saunders.
McInnes, R. S., Mccallum, G. E., Lamberte, L. E., and Van schaik, W. (2020). Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 53, 35–43. doi: 10.1016/j.mib.2020.02.002
Morales-Erasto, V., Falconi-Agapito, F., Luna-Galaz, G. A., Saravia, L. E., Montalvan-Avalos, A., Soriano-Vargas, E. E., et al. (2016). Coinfection of Avibacterium paragallinarum and Ornithobacterium rhinotracheale in chickens from peru. Avian Dis. 60, 75–78. doi: 10.1637/11265-082015-ResNote.1
Mouahid, M., Bisgaard, M., Morley, A. J., Mutters, R., and Mannheim, W. (1992). Occurrence of V-factor (n.d.) independent strains of Haemophilus paragallinarum. Vet. Microbiol. 31, 363–368. doi: 10.1016/0378-1135(92)90128-g
Nhung, N. T., Chansiripornchai, N., and Carrique-Mas, J. J. (2017). Antimicrobial resistance in bacterial poultry pathogens: A review. Front. Vet. Sci. 4:126. doi: 10.3389/fvets.2017.00126
Nouri, A., Bashashati, M., Mirzaie, S. G., Shoshtari, A., and Banani, M. (2021). Isolation, identification and antimicrobial susceptibility of Avibacterium Paragallinarum from backyard chicken in retail markets of Karaj and Tehran Cities, Iran. Arch. Razi Inst. 76, 1047–1053. doi: 10.22092/ari.2020.343173.1502
Paudel, S., Ruhnau, D., Wernsdorf, P., Liebhart, D., Hess, M., and Hess, C. (2017). Presence of Avibacterium paragallinarum and histopathologic lesions corresponds with clinical signs in a co-infection model with Gallibacterium anatis. Avian Dis. 61, 335–340. doi: 10.1637/11609-021317-RegR
Piccardi, P., Vessman, B., and Mitri, S. (2019). Toxicity drives facilitation between 4 bacterial species. Proc. Natl. Acad. Sci. U.S.A. 116, 15979–15984. doi: 10.1073/pnas.1906172116
Rawson, T. M., Moore, L. S. P., Zhu, N., Ranganathan, N., Skolimowska, K., Gilchrist, M., et al. (2020). Bacterial and fungal coinfection in individuals with coronavirus: A rapid review to support COVID-19 antimicrobial prescribing. Clin. Infect. Dis. 71, 2459–2468. doi: 10.1093/cid/ciaa530
Sayers, E. W., Bolton, E. E., Brister, J. R., Canese, K., Chan, J., Comeau, D. C., et al. (2022). Database resources of the national center for biotechnology information. Nucleic Acids Res. 50, D20–D26. doi: 10.1093/nar/gkab1112
Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Stacy, A., Everett, J., Jorth, P., Trivedi, U., Rumbaugh, K. P., and Whiteley, M. (2014). Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl. Acad. Sci. U.S.A. 111, 7819–7824. doi: 10.1073/pnas.1400586111
Starikova, E. V., Tikhonova, P. O., Prianichnikov, N. A., Rands, C. M., Zdobnov, E. M., Ilina, E. N., et al. (2020). Phigaro: High-throughput prophage sequence annotation. Bioinformatics 36, 3882–3884. doi: 10.1093/bioinformatics/btaa250
Sun, H. R., Cui, X. D., Liu, X. K., Li, S. H., Yi, K. F., Pan, Y. S., et al. (2020). Molecular characterization of a novel integrative conjugative element ICEHpa1 in Haemophilus parasuis. Front. Microbiol. 11:1884. doi: 10.3389/fmicb.2020.01884
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Trujillo-Ruiz, H. H., Shivaprasad, H. L., Morales-Erasto, V., Talavera-Rojas, M., Salgado-Miranda, C., Salazar-Garcia, F., et al. (2016). Virulence of serovar C-1 Strains of Avibacterium paragallinarum. Avian Dis. 60, 837–840. doi: 10.1637/11421-040716-ResNote
Van den Biggelaar, R., Van Eden, W., Rutten, V., and Jansen, C. A. (2020). Macrophage activation assays to evaluate the immunostimulatory capacity of Avibacterium paragallinarum in A multivalent poultry vaccine. Vaccines 8:671. doi: 10.3390/vaccines8040671
Westblade, L. F., Simon, M. S., and Satlin, M. J. (2021). Bacterial coinfections in coronavirus disease 2019. Trends Microbiol. 29, 930–941. doi: 10.1016/j.tim.2021.03.018
Wongsuvan, G., Wuthiekanun, V., Hinjoy, S., Day, N. P., and Limmathurotsakul, D. (2018). Antibiotic use in poultry: A survey of eight farms in Thailand. Bull. World Health Organ. 96, 94–100. doi: 10.2471/BLT.17.195834
Wu, Y., Wang, Y., Yang, H., Li, Q., Gong, X., Zhang, G., et al. (2021). Resident bacteria contribute to opportunistic infections of the respiratory tract. PLoS Pathog. 17:e1009436. doi: 10.1371/journal.ppat.1009436
Xu, Y., Cheng, J., Huang, X., Xu, M., Feng, J., Liu, C., et al. (2019). Characterization of emergent Avibacterium paragallinarum strains and the protection conferred by infectious coryza vaccines against them in China. Poult. Sci. 98, 6463–6471. doi: 10.3382/ps/pez531
Keywords: Avibacterium paragallinarum, commensal bacteria, satellitism, antimicrobial resistance, symbiosis
Citation: Zhu J, Chen Y, Wu Y, Wang Y and Zhu K (2022) Commensal bacteria contribute to the growth of multidrug-resistant Avibacterium paragallinarum in chickens. Front. Microbiol. 13:1010584. doi: 10.3389/fmicb.2022.1010584
Received: 03 August 2022; Accepted: 03 October 2022;
Published: 04 November 2022.
Edited by:
Neil Ross McEwan, Robert Gordon University, United KingdomReviewed by:
Ming-Kun Hsieh, National Chung Hsing University, TaiwanCopyright © 2022 Zhu, Chen, Wu, Wang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqiang Wang, dmV0d3lxQGNhdS5lZHUuY24=; Kui Zhu, emh1a0BjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.