- 1Animal Disease Prevention and Food Safety Key Laboratory of Sichuan Province, College of Life Sciences, Sichuan University, Chengdu, China
- 2College of Veterinary Medicine, Northwest A&F University, Yanglin, China
The appearance of transferable oxazolidinone resistance genes poses a major challenge to public health and environmental safety. These genes not only lead pathogenic bacteria to become resistant to linezolid but also reduce sensitivity to florfenicol, which is widely used in the veterinary field. To verify the dissemination of oxazolidinone resistance genes in enterococcal isolates from pigs at different production stages in a swine farm in China, we collected 355 enterococcal isolates that were resistant to florfenicol from 600 (150 per stage) fresh fecal swabs collected from a swine farm. Through initial PCR screening and whole-genome sequencing, 175 isolates harboring different oxazolidinone resistance genes were identified. All isolates carried the optrA gene. A total of 161 (92%, 161/175) isolates carried only the optrA gene. Three (1.71%, 3/175) isolates carried both the optrA and poxtA genes, and 11 (3.1%, 11/175) isolates contained the optrA gene and poxtA2 and cfr(D) variants. A total of 175 isolates that harbored oxazolidinone resistance genes included 161 E. faecalis, 6 E. faecium, and 8 E. hirae. By sequencing the whole genomes, we found that the 161 isolates of E. faecalis belonged to 28 different STs, including 8 new STs, and the 6 isolates of E. faecium belonged to four different STs, including one new ST. The phylogenetic tree based on SNPs of the core genome showed that both clonal spread and horizontal transfer mediated the diffusion of oxazolidone resistance genes in enterococcal isolates at specific stages in pig farms. Moreover, enterococcal isolates carrying oxazolidone resistance genes could spread from breeding pigs to fattening pigs, while transferable oxazolidone resistance genes in enterococcal isolates could persist on a pig farm throughout all production stages. Representative enterococcal isolates with different oxazolidinone resistance genes were further studied through Nanopore sequencing. We identified a novel plasmid, pM4-80 L4 (15,008 bp), carrying the poxtA2 and cfr(D) genes in enterococcal isolates at different stages. We also found three different plasmids harboring the poxtA gene with high genetic variation, and all poxtA genes were flanked by two copies of IS1216E elements. In addition, four genetically distinct plasmids carrying the optrA gene were identified, and Tn554 was found to mediate chromosome-localized optrA gene transfer. Our study highlighted that transferable oxazolidinone resistance genes in enterococcal isolates could persist throughout all production stages on a pig farm, and the prevalence and dissemination of oxazolidinone resistance genes in enterococcal isolates from animal farms should be continually monitored.
Introduction
Gram-positive enterococcal bacteria occur widely in the intestines of humans and animals. Enterococci in animals used for food production may contaminate food and the environment, creating a risk of human infection through anthropozoonosis (He et al., 2016). Enterococcus faecalis and Enterococcus faecium often cause urinary tract and soft tissue infections, and in severe situations, they may cause septicemia or meningitis (Yi et al., 2022). Notably, E. faecium and E. faecalis are important nosocomial pathogens worldwide (Bender et al., 2018). It has been reported that enterococci can not only exhibit inherent resistance to some antibiotics, but also easily acquire new resistance genes located on mobile genetic elements from other sources, including other bacteria. Thus, multidrug resistance can arise readily in enterococci, resulting in limited clinical treatment options (He et al., 2016). In addition, vancomycin-resistant E. faecium (VRE) has been listed by the World Health Organization as a pathogen requiring high vigilance since it was discovered at the end of the last century (WHO, 2017). Oxazolidinone antibiotics, mainly linezolid and tedizolid, have a bactericidal effect on various gram-positive bacteria (Wilson et al., 2008; Moellering, 2014). In 2000, the US FDA introduced linezolid for clinical treatment. Linezolid is usually used to treat severe clinical infections [Methicillin-resistant Staphylococcus aureus (MRSA) and Vancomycin-resistant Enterococcus faecium (VRE)] and is known as the last line of defense for the treatment of gram-positive bacteria (Bender et al., 2018). With the extensive clinical use of linezolid in the clinic, linezolid-resistant enterococci have gradually emerged. The emergence of linezolid-resistant enterococci poses a great challenge to human clinical treatment and public health (Schwarz et al., 2021).
Linezolid resistance mechanisms are associated with ribosomal mutations in the 23S rRNA and/or L3, L4 and L22 ribosomal proteins (Wang et al., 2020). However, the appearance of transferable oxazolidinone-resistant determinants in enterococci or other bacteria in many regions of the world should not be ignored (Ewa, 2018). Cfr is the first reported transferable oxazolidinone resistance gene and encodes a 23S rRNA methyltransferase. Bacteria carrying the cfr gene exhibit resistance to phenicols, oxazolidinones, lincosamides, pleuromutilins, and streptogramin A (PhLOPSA phenotype; Long et al., 2006; Shen et al., 2013). Since the cfr gene was originally discovered, four cfr-like genes have been reported worldwide, namely, cfr(B), cfr(C), cfr(D) and cfr (E) (Deshpande et al., 2015; Zhang et al., 2017; Stanley et al., 2019). OptrA is the second oxazolidinone resistance gene, reported in 2015, and encodes a ribosomal protection protein of the ABC-F family. Bacteria carrying the optrA gene exhibit resistance to oxazolidinones and phenicols (Wang et al., 2015). Since the initial description of the optrA gene, it and its many variants have been reported in several countries, revealing that it is more difficult to treat oxazolidinone-resistant bacteria (Schwarz et al., 2021). Unfortunately, another ribosomal protection protein of the ABC-F family, poxtA, was reported in MRSA of clinical origin in 2018 (Antonelli et al., 2018; Crowe-McAuliffe et al., 2022). The poxtA2 variant was also reported in 2021 (Baccani et al., 2021). Now, poxtA has been detected in clinical samples, animal samples and even marine plankton samples in many countries (Papagiannitsis et al., 2019; Dejoies et al., 2021). Although oxazolidinone antibiotics have never been approved for use in livestock breeding, we often detect the transferable determinants of oxazolidinone resistance in enterococci isolated from animals used in food production (Wang et al., 2020). Florfenicol is a phenicol compound that has broad spectrum antibacterial activity and few side effects and is used to treat respiratory and intestinal bacterial infections in animals used in food production (Crowe-McAuliffe et al., 2022). It was reported that the abundance of oxazolidinone resistance genes in livestock feces was related to florfenicol residue (Kim, 2021). Given that phenicol resistance could be caused by the extensive use of florfenicol, the widespread use of florfenicol in the veterinary field will not only promote the spread of antibiotic resistance genes of florfenicol but also promote the spread of oxazolidone resistance genes, which will cause great public health concerns.
Transferable oxazolidinone resistance genes are common in enterococci isolated from pig farms, as has been reported worldwide recently (Hao et al., 2019; Lei et al., 2019, 2021; Fioriti et al., 2021). However, there are few reports on the dissemination mode of transferable oxazolidinone resistance genes in pigs at different production stages in large-scale pig farms. In this study, we investigated the presence of oxazolidinone resistance genes in florfenicol-resistant enterococci isolated from a large-scale swine farm in China and determined the dissemination mechanisms of oxazolidinone resistance genes in enterococci isolated from healthy pigs in different production stages.
Materials and methods
Sample collection and florfenicol-resistant enterococcal isolation
The large-scale swine farm (approximately 1,000 breeding pigs and 10,000 fattening pigs) is located in Sichuan, one of China’s major pig raising provinces. In October and November 2021, Fresh fecal swabs were taken from 150 breeding pigs in the breeding pig section of the farm and from 450 fattening pigs aged between approximately 2, 4, and 6 months (150 fattening pigs each stage) in the fattening pig section of the farm. The swab samples were inoculated into 3 ml of Buffer Peptone Water (Oxoid, Basingstoke, United Kingdom) containing 8 mg/l florfenicol. The culture was incubated overnight at 37°C and 180 RPM, and following incubation, 0.1 ml aliquots of culture were streaked onto Pfizer Enterococcus Selective Agar (Qingdao Hope Bio-Technology, China) supplemented with florfenicol (8 mg/l), and the selective plate was incubated overnight at 37°C. If the plate had colonies, we randomly selected one enterococcal colony from the plate. Then, we used an automated system (BD Diagnostic Systems, Sparks, MD, United States) to identify the enterococcal isolates.
Detection of oxazolidinone resistance genes and antimicrobial susceptibility testing
Initial PCR screening was performed for all florfenicol-resistant enterococcal isolates using generic primers directed against the optrA, cfr and poxtA genes (Bender et al., 2019), and variants of optrA, cfr and poxtA were then identified by whole-genome sequencing. We sequenced the positive PCR products with Sanger (Chengdu Sangon, China). According to Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute, 2020), the minimum inhibitory concentrations (MICs) of linezolid, tedizolid, florfenicol, vancomycin, ampicillin, doxycycline, chloramphenicol and tetracycline were determined by the broth microdilution method. E. faecium ATCC29212 was used as a quality control strain for antimicrobial susceptibility testing.
Whole-genome sequencing and genomic analyses of enterococcal isolates that harbored oxazolidinone resistance genes
The MiniBEST bacterial genomic DNA Extraction Kit (Takara, Dalian, China) was used to extract the genomic DNA of enterococcal isolates containing oxazolidone resistance genes (optrA, poxtA and cfr). The entire genomes were sequenced using the Illumina HiSeq platform (150-bp paired-end reads, with an average coverage of approximately 200-fold). We used SPAdes_3.13.0 software to collect original sequencing data to map the genomes. Antimicrobial resistance genes were identified by using ResFinder 4.1.1 Linezolid resistance determinants were searched by LRE-Finder 1.0.2
To investigate the genetic environments of different oxazolidinone resistance genes, we selected 12 enterococcal isolates for further research, and the basis of our selection is described below. First, among the 11 enterococcal isolates carrying the optrA gene and the poxtA2 and cfr(D) variants, there were three ST-type isolates, and we selected one isolate for each ST (M2-9, M4-54, and M4-80). Second, we selected all enterococcal isolates carrying the optrA and poxtA genes, for a total of three isolates (B6, B54, and M2-95). Third, because there have been many reports on the genetic environment of optrA gene, and our manuscript mainly wanted to focus on the genetic environment of poxtA gene, poxtA2 and cfr(D) variants, we selected four representative enterococcal isolates out of 161 enterococcal isolates carrying only the optrA gene (M6-97, M6-130, B83, and B126). Finally, to study the phenomenon of heteroresistance, we selected two enterococcal isolates carrying only the optrA gene (M2-77 and M2-82). A Nanopore MinION Rapid Sequencing Kit was used to further sequence the genomes of representative enterococcal isolates with different oxazolidinone resistance genes. Using Nanopore sequencing data combined with Illumina sequencing data, the complete genome sequences were assembled by Unicycler. PCR linkage confirmed that there was a circular plasmid containing a transferable oxazolidinone resistance gene. Easyfig v2.2.23 was used for comparative analyses of the plasmids.
Molecular typing and phylogenetic analysis of enterococcal isolates that harbored oxazolidinone resistance genes
Sequence types (STs) were assigned on the basis of conventional MLST loci. STs of enterococcal isolates were determined by PubMLST,4 and the new STs were also assigned by PubMLST. MLST-based minimum spanning trees were created by GrapeTree v1.3.2.5 The core genome is present in all individuals of the same species, and the genes in the core genome are generally related to the stable biological function and phenotypic characteristics of the species, most of which are housekeeping genes (Tettelin et al., 2005; Yang and Gao, 2022). Using the core genomes of enterococci to construct phylogenetic trees can reduce the influence of variable genomes on genetic phylogenetic relationships. Sequences of isolates that harbored oxazolidinone resistance genes were annotated using Prokka v1.12 software, and the core genome was identified using Roary v3.11.2, then the SNPs of the core genome were extracted by Harvest tools (Seemann, 2014; Page et al., 2015; Dejoies et al., 2021). A phylogeny based on SNPs of the core genome was constructed by FastTree v2.1.11.6 The Newick file for the phylogenetic tree was modified in iTOL.
Transfer experiments
Using the criteria described previously in “Whole-genome sequencing and genomic analyses of enterococcal isolates that harbored oxazolidinone resistance genes,” eight isolates (M4-80, B6, B54, M2-95, M2-77, M2-82, M6-97, and M6-130) carrying different plasmids that were representative of the 175 enterococcal isolates carrying the oxazolidinone resistance gene in this study were selected as donors. The rifampicin-resistant E. faecalis JH2-2 was used as the recipient. Conjugal transfer was performed on a filter membrane as described previously (Brenciani et al., 2016). The donor and recipient bacteria were mixed in a 1:1 ratio on the filter membrane. Transconjugants were selected on Brain Heart Infusion Agar (Oxoid, Basingstoke, United Kingdom) plates containing 2 mg/l linezolid and 25 mg/l rifampicin. The transfer frequency was expressed as the ratio of the cell number (cfu/ml) of the transconjugant to that of the recipient.
Transconjugants were evaluated for their susceptibility to linezolid, tedizolid, chloramphenicol, florfenicol, tetracycline and doxycycline. Then, the detection of transferable oxazolidinone resistance genes, 16S rDNA sequencing and whole genome sequencing were also used to confirm transconjugants.
Nucleotide sequence accession numbers
The genomes of enterococcal isolates that harbored oxazolidinone resistance genes in this study have been deposited in the National Center for Biotechnology Information and registered BioProject number PRJNA856057. The complete nucleotide sequences of nine plasmids harboring different transferable oxazolidinone resistance genes have been deposited in GenBank and assigned accession numbers OP046170–OP046178.
Results
Prevalence of oxazolidinone resistance genes in florfenicol-resistant enterococcal isolates
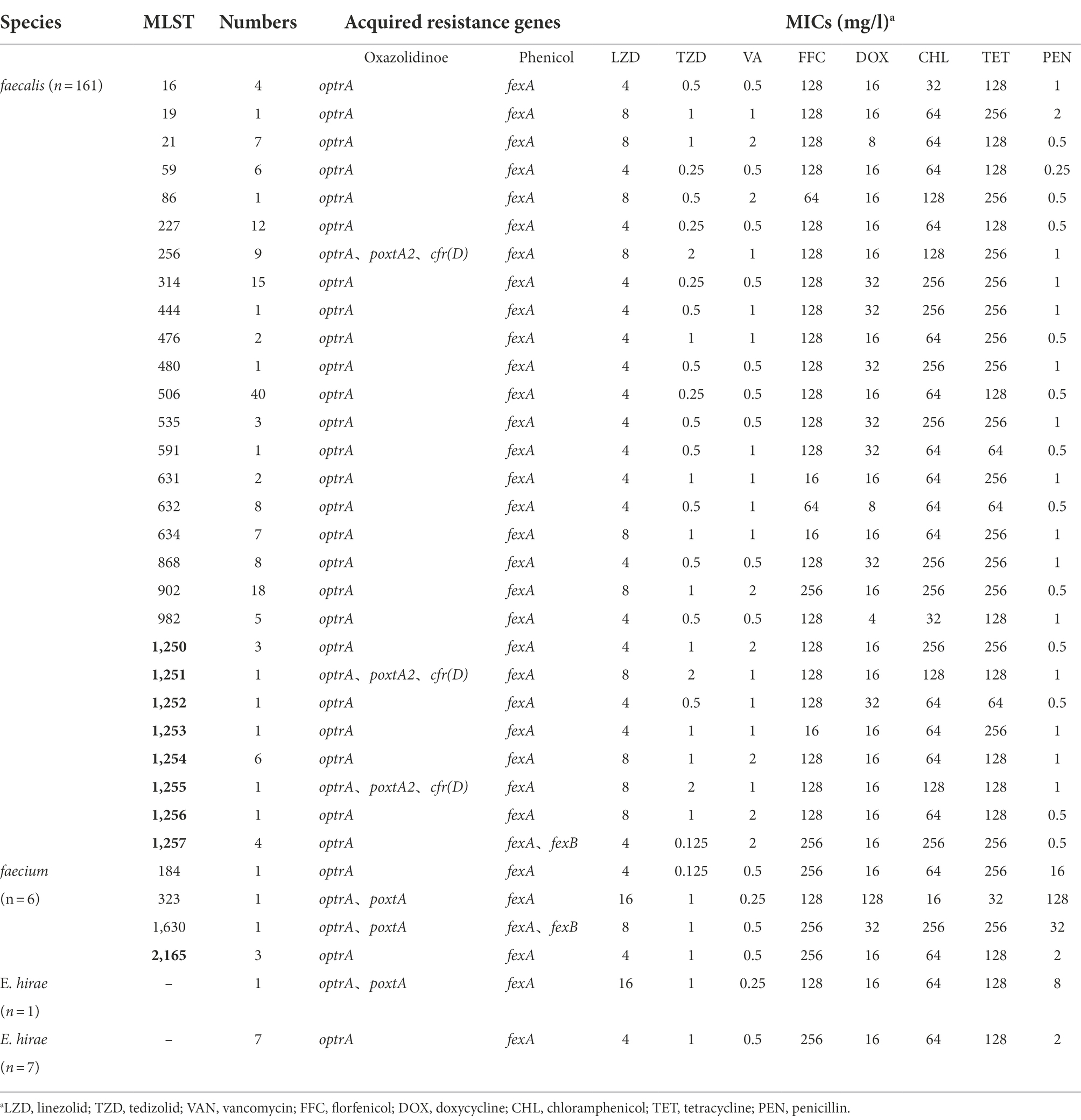
In this study, 355 isolates of florfenicol-resistant enterococcal isolates were collected from 600 fresh fecal swabs taken from pigs at different stages from breeding pig section and fattening pig section of the pig farm. Through initial PCR screening and whole-genome sequencing, 175 isolates harboring different oxazolidinone resistance genes were identified (Table 1). All isolates carried the optrA gene. A total of 161 (92%, 161/175) isolates carried only the optrA gene. Three (1.71%, 3/175) isolates carried both the optrA and poxtA genes, and eleven (3.1%, 11/175) isolates contained the optrA gene and poxtA2 and cfr(D) variants (Table 1). A total of 175 isolates that harbored oxazolidinone resistance genes included 161 E. faecalis, 6 E. faecium, and 8 E. hirae (Table 1). With respect to the isolation rate of enterococci carrying oxazolidinone resistance genes in each production stages, 49 (32.67%, 49/150) isolates came from the breeding pig section of the pig farm. The remaining isolates came from the fattening pig section of the pig farm, with 47 (31.33%, 47/150) from 2-month fattening pigs, 40 (26.67%, 40/150) from 4-month fattening pigs, and 37 (24.67%, 37/150) from 6-month fattening pigs.
Antimicrobial susceptibility testing indicated that the MIC values of linezolid against 161 enterococcal isolates that harbored only the optrA gene ranged from 4 to 8 mg/l, and the MIC values of tedizolid ranged from 0.125 to 1 mg/l (Table 1). Three isolates that carried the optrA and poxtA genes were resistant to linezolid and tedizolid. The MICs of the three isolates to linezolid and tedizolid were 8–16 mg/l and 1 mg/l, respectively (Table 1). In addition, 11 isolates carrying both the optrA gene and the poxtA2 and cfr(D) variants showed resistance to linezolid and tedizolid, and the MICs for these two antibiotics were 8 and 2 mg/l, respectively (Table 1). A total of 175 isolates exhibited a multidrug resistance phenotype and were resistant to chloramphenicol, florfenicol, tetracycline and doxycycline. Moreover, three isolates were resistant to penicillin (MICs ≥ 16 mg/l).
Genomic analyses of enterococcal isolates that carried oxazolidinone resistance genes
By sequencing the whole genomes of 175 isolates that carried oxazolidinone resistance genes, we found that the cfr and poxtA genes carried by 11 E. faecalis isolates were the cfr(D) and poxtA2 variants. The homology of the cfr(D) gene and wild type (accession number NG_067192) was 99.9%. The poxtA2 variant was recently detected in two E. faecalis isolates and one E. casseliflavus isolate from manure of a swine farm in Italy (Baccani et al., 2021). Unlike poxtA, the poxtA2 variant (accession number MZ171245) was not truncated by an IS1216 insertion at the 3′ end; thus, a new sequence consisting of eight amino acids (TPEEEQKY) replaced the six amino acids (GSVAKF) of the wild-type protein. Six different optrA variants were found by alignment of the optrA amino acid sequences with those found in E. faecalis E349 (Supplementary Table 1; Wang et al., 2015). Five of these variants have been reported previously (Cai et al., 2018). One variant, DKD (G40D, I287K, G393D), has not been reported previously (Supplementary Table 1). Different linezolid MICs were also present in isolates harboring different optrA variants (Supplementary Table 1).
In addition, all 175 isolates that contained the phenicol resistance gene fexA and five isolates also carried the fexB gene. All the isolates carried the tetracycline resistance genes tet(L) and/or tet(M) and macrolide resistance genes erm(A) and/or erm(B). Five isolates carried the macrolide resistance gene msr(C), and the M2-95 strain harbored both the mosaic tetracycline resistance gene tet(S/M) and the macrolide resistance gene msr(C). Four isolates of E. faecalis of ST1257 contained the mosaic tetracycline resistance gene tet(O/W/32/O) and chloramphenicol resistance gene cat. Thirteen to nineteen mutations in the pbp5 gene were found in six isolates of E. faecium. Two isolates carried the clpL gene.
Molecular typing and phylogenetic analysis of enterococcal isolates that harbored oxazolidinone resistance genes
MLST showed that the 161 isolates of E. faecalis that harbored oxazolidinone resistance genes belonged to 28 different STs, including ST21, ST59, ST256, ST314, ST506, ST632, ST634, ST868, ST902, and ST982, and eight new STs were assigned by PubMLST, including ST1250, ST1251, ST1252, ST1253, ST1254, ST1255, ST1256, and ST1257 (Table 1). Six E. faecium isolates that harbored oxazolidinone resistance genes belonged to 4 different STs, including ST184, ST323, ST1630 and one new ST, ST2165, assigned by PubMLST (Table 1). The 161 E. faecalis isolates were widely isolated from pigs in different production stages (Figure 1A). It is worth noting that isolates ST506 and ST632 were found in the samples from all four stages (Figure 1A). However, the distribution of the six E. faecium isolates was relatively homogeneous across different stages (Figure 1B). A phylogenetic tree based on SNPs of the core genome showed that the 175 enterococcal isolates carrying oxazolidone resistance genes were divided into 35 branches (Figure 2). The isolates with the same ST type were clustered into the same branch in the phylogenetic tree (Figure 2). For the isolates isolated from a certain stage, all ST902 isolates isolated from breeding pigs belonged to one branch, and the ST21 isolates isolated from 2-month fattening pigs also clustered together in the phylogenetic tree. Similar phenomena were found in 4-month fattening pigs and 6-month fattening pigs (Figure 2). For the isolates isolated from different stages, ST506 isolates carrying the optrA gene belonged to one clade and were isolated from samples in all four production stages (Figure 2). ST632 isolates carrying the optrA gene were clustered in the phylogenetic tree, and they were prevalent in all four production stages (Figure 2). The poxtA gene was carried by two isolates of E. faecium and one strain of E. hirae, and the poxtA2 and cfr(D) variants were carried by ST256, ST1251 and ST1255 isolates of E. faecalis (Figure 2).
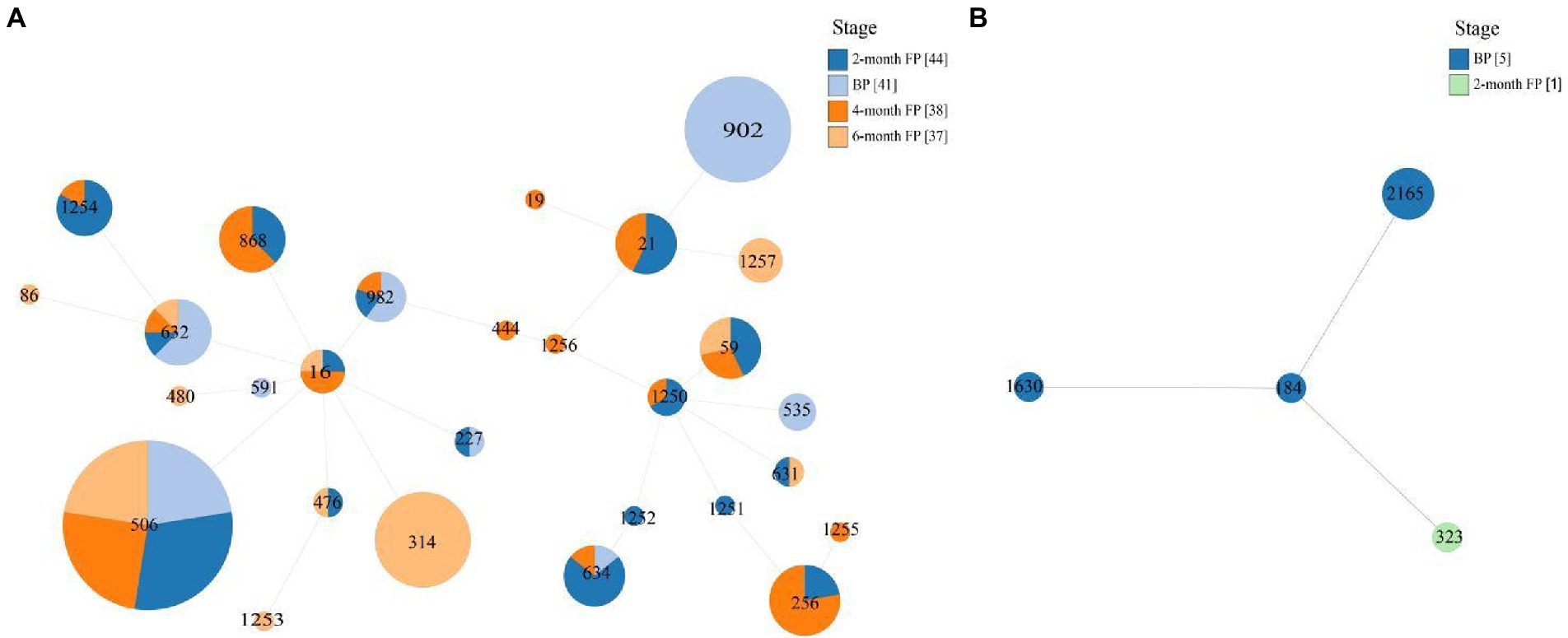
Figure 1. Multilocus Sequence Typing (MLST)-based minimum spanning trees of E. faecalis (A) isolates and E. faecium (B) isolates. The stage is indicated by different colors.
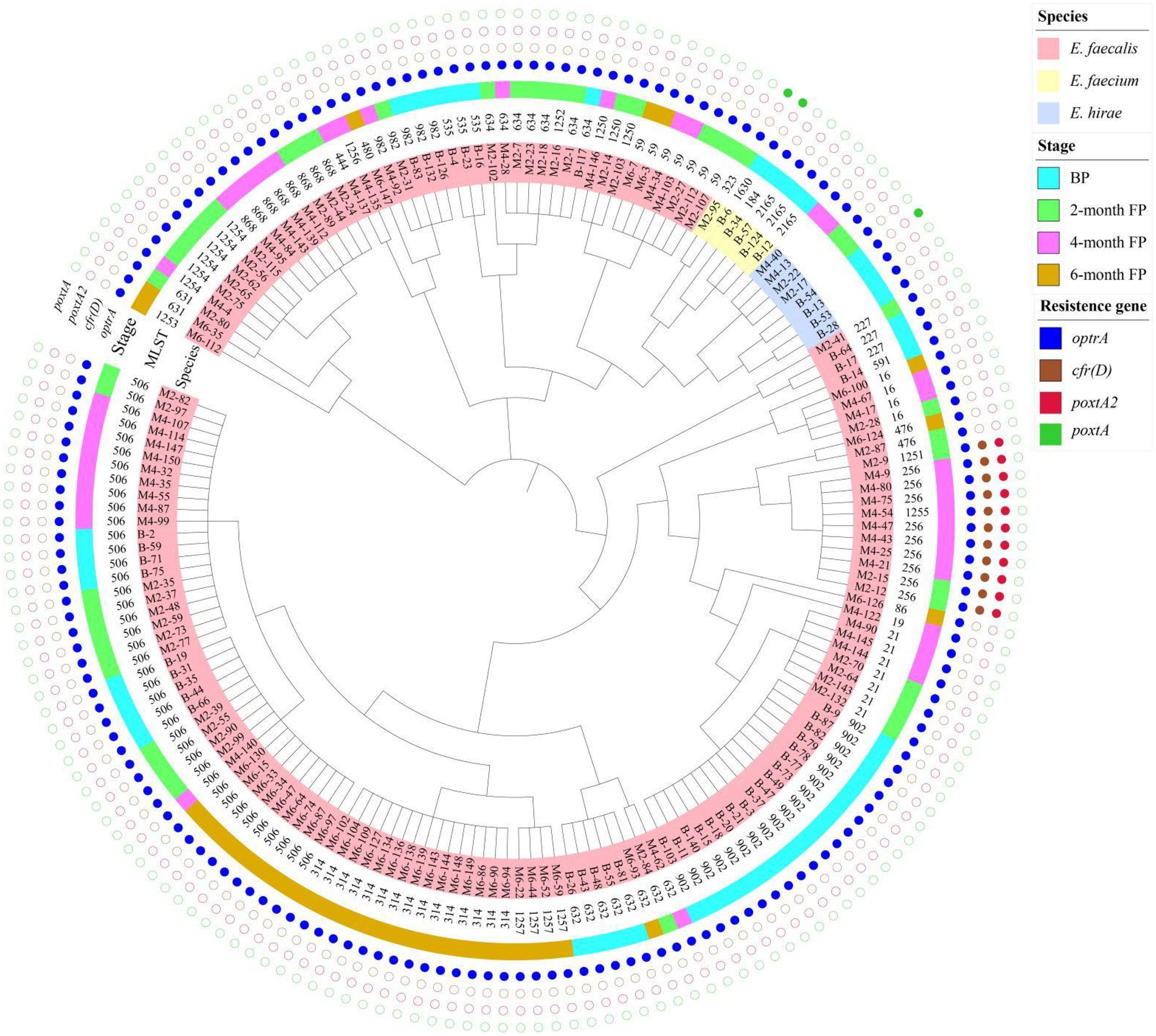
Figure 2. Phylogenetic tree based on SNPs of the core genome for 175 enterococcal isolates that harbored different oxazolidinone resistance genes. The species, stage, and oxazolidinone resistance genes are indicated by different colors.
The genetic environments of different oxazolidinone resistance genes
The genetic structures of plasmids containing oxazolidinone resistance genes and the genetic environments of different oxazolidinone resistance genes are shown in Figure 3. In three E. faecalis isolates (M2-9, M4-54 and M4-80) that carried the optrA gene and the poxtA2 and cfr(D) variants, bioinformatic data revealed that the optrA gene of three isolates was localized to the chromosome at Tn554, and a novel plasmid carrying poxtA2 and cfr(D) variants, named pM4-80 L4, was found concomitantly in these three isolates. This 150,008 bp plasmid (35.5% GC content) contained 17 ORFs. BLASTN analysis revealed that pM4-80 L4 was 98.15% identical (coverage 99%) to pV386 (33,48 kb in size; accession number MZ603802.1) carrying cfr(D) and poxtA2 variants. pV386 was recently found in an E. faecalis strain that was isolated from a swine environment in central Italy (Cinthi et al., 2022). Similar to pV386, pM4-80 L4 showed that the poxtA2 gene was closely associated with the cfr(D) gene (Figure 3A). Upstream of the poxtA2 gene, the fexA gene was found to be surrounded by two copies IS1216E in the same direction, and the rep1 gene (belonging to the Inc18 family) was also detected downstream of the cfr(D) gene (Figure 3A).
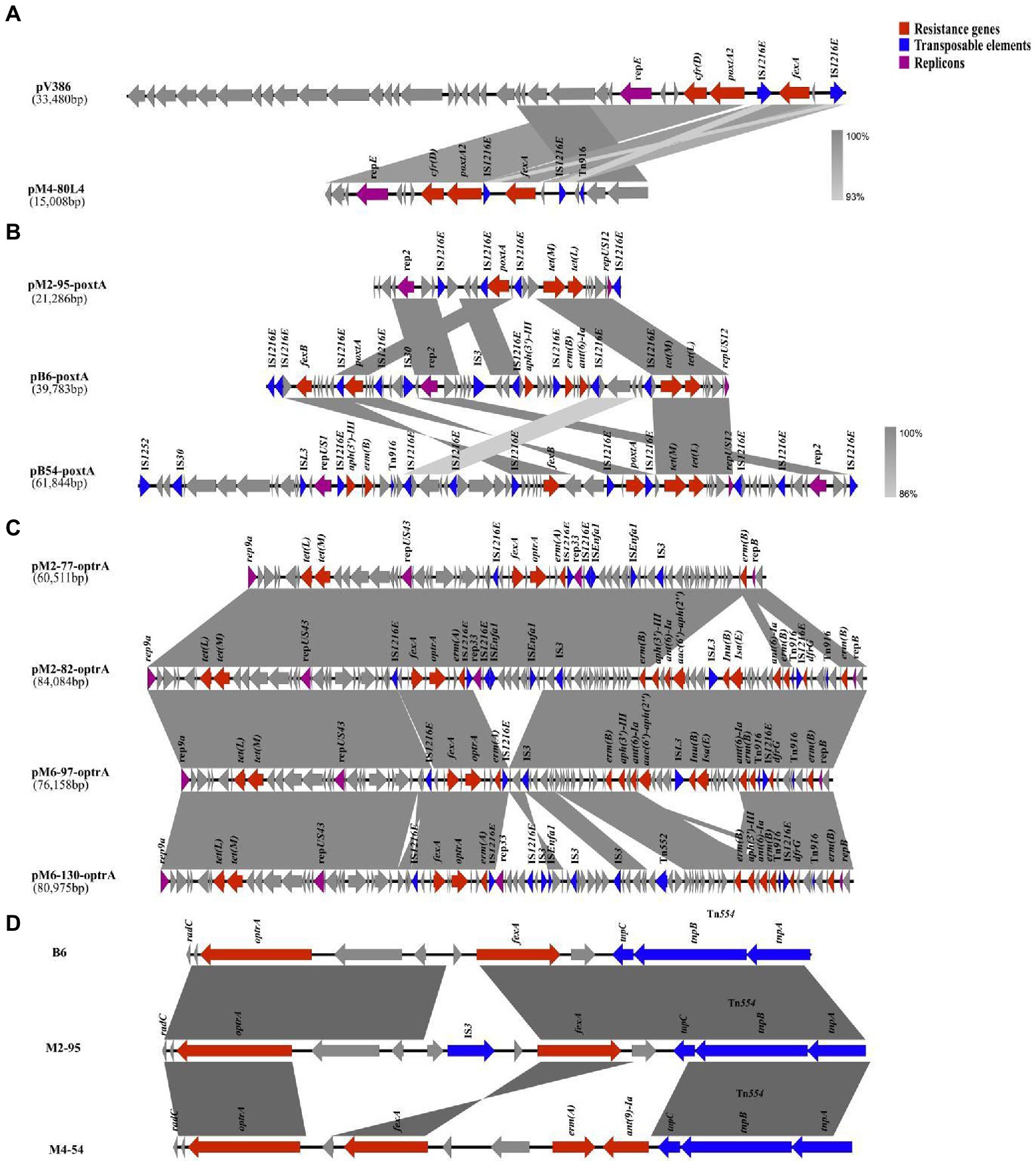
Figure 3. The genetic environments of different oxazolidinone resistance genes. Genes and ORFs are shown as arrows, and their transcription orientations are indicated by the arrowheads. (A) The genetic structure of pM4-80L4. (B) The genetic structures of three poxtA-carrying plasmids. (C) The genetic structures of four optrA-carrying plasmids. (D) The genetic structures of three different chromosomal optrA gene clusters.
Three isolates (B6, B54, and M2-95) harbored both the optrA and poxtA genes. In these three isolates, we found that the optrA gene was localized on the chromosome, whereas the poxtA gene was located in a plasmid in all isolates. Three different poxtA-carrying plasmids with sizes ranging from 21.2 to 61.8 kb were obtained after hybrid assembly (Table 2). Analysis of the poxtA nucleotide sequence location in each isolate showed that they exhibited 100% identity to that of S. aureus AOUC-0915, in which poxtA was initially described (Antonelli et al., 2018). Every poxtA gene was flanked by IS1216E at the left and/or right ends (Figure 3B). Two poxtA-carrying plasmids from two E. faecium isolates (B6 and M2-95) belonged to the same type that harbored rep2 and repUS12 replicons (Figure 3B). Two plasmids exhibited high genetic variation, and both of them carried the tetracycline resistance genes tet(L) and tet(M). pB6-poxtA also carried the phenicol resistance gene fexB, macrolide resistance gene erm(B) and aminoglycoside resistance gene aph(3′)-III; IS3 and IS30 were also found in pB6-poxtA (Figure 3B). The final poxtA-carrying plasmid, pB54-poxtA, from the E. hirae strain (B-54), carried three replicons (rep2, repUS12, and repUS1) and harbored the fexB, tet(L), tet(M), aph(3′)-III and erm(B) resistance genes, and IS1252, ISL3, IS30, and Tn916 were also identified in pB54-poxtA (Figure 3B).
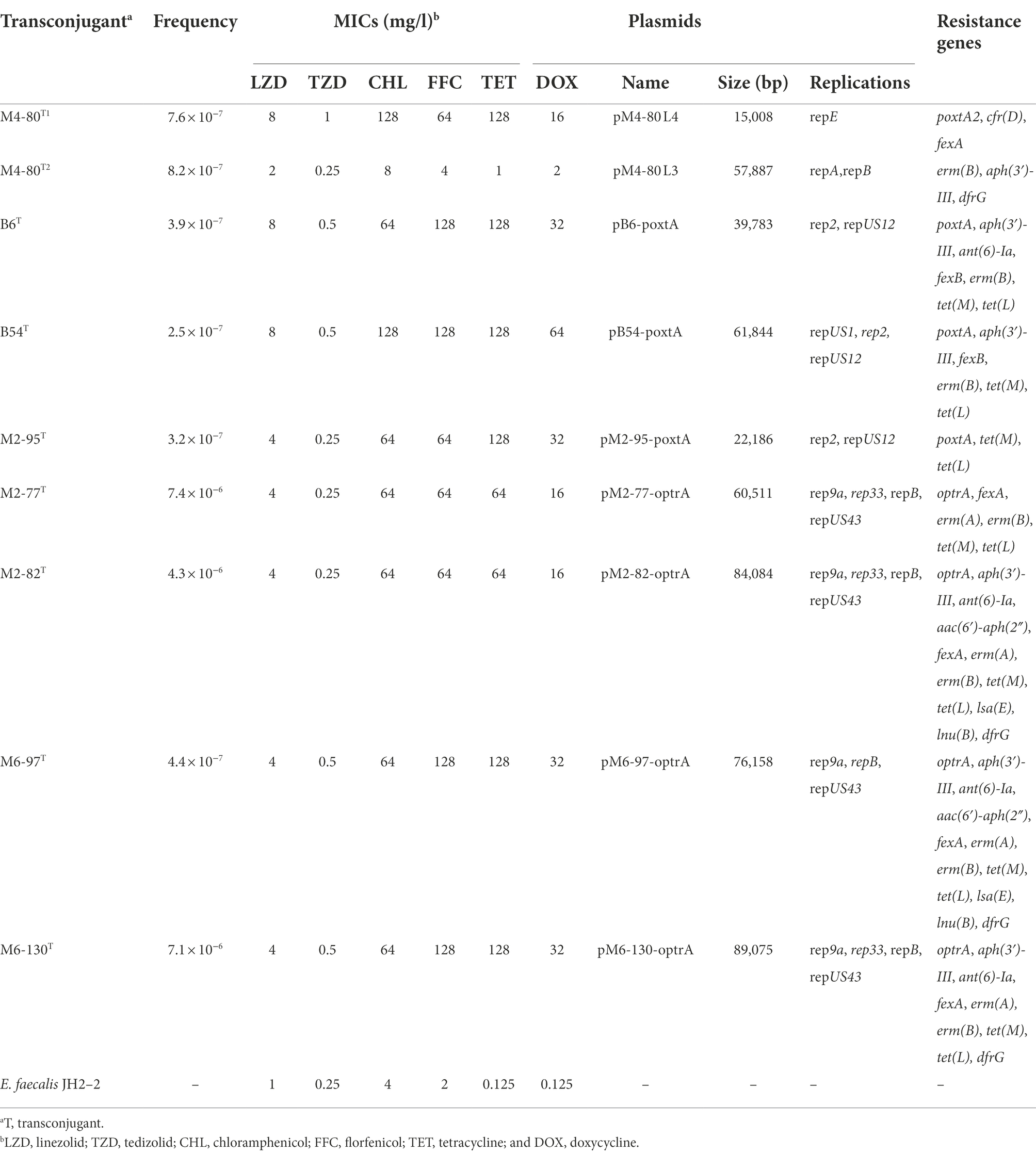
Table 2. Information of the different plasmids harboring transferable oxazolidinone resistance genes and antimicrobial susceptibilities of the transconjugants and recipient isolates.
Among the 161 enterococcal isolates that carried only the optrA gene, 27 isolates had the optrA gene localized to the chromosome at Tn554, while 134 isolates had the optrA gene localized to a plasmid. From the four optrA-carrying isolates that we selected (M2-77, M2-82, M6-97, and M6-130), four different optrA-carrying plasmids were identified, three of which belonged to the same type and carried rep9a, repUS43, rep33 and repB replicons (Table 2). The remaining plasmid carried the rep9a, repUS43 and repB replicons (Table 2). The genetic structure of the optrA-carrying plasmids is shown in Figure 3C. All optrA genes were associated with the phenicol resistance gene fexA and the macrolide resistance gene erm(A), and a similar genetic context IS1216E–fexA–optrA–erm(A)–IS1216E was found in four different optrA-carrying plasmids (Figure 3C). In the two optrA-carrying isolates we selected (B83 and B126) and the previous six isolates with optrA gene localization on chromosomes (M2-9, M4-54, M4-80, B6, B54, and M2-95), four different chromosomal optrA gene clusters were found. Three of these optrA genes were associated with Tn554 inserted into the radC gene, and the fexA—optrA segment was detected upstream of the Tn554 transposon (Figure 3D). The M4-54 isolate also carried the macrolide resistance gene erm(B) and the aminoglycoside resistance gene ant(9)-Ia upstream of the fexA-optrA segment, and IS3 inserted into the fexA-optrA segment was also found in M2-95 (Figure 3D). The genetic context IS1216E–fexA–optrA–erm(A)–IS1216E on the chromosome was identified in the remaining isolate (B54; Supplementary Figure 1).
Interestingly, the clonally related isolates had the heteroresistant genotype in ST506 and ST256, specifically in the ST506 isolates M2-77 and M2-82 (genotype M2-77 lacked aph(3′)-III, aac(6′)-aph(2″), ant(6)-Ia, lsa(E), lnu(B) and dfrG genes compared with M2-82) and the ST256 isolates M4-75 and M4-80 (genotype M4-75 lacked aph(3′)-III, erm(B) and dfrG genes compared with M4-80). Bioinformatics’ results revealed the genetic environments of differential resistance genes in clonally related isolates. Compared with M2-77, IS1216E and Tn916 might mediate the insertion of a 21,511 bp fragment that contained differential resistance genes into plasmids (pM2-82) of M2-82 (Figure 3C). However, compared with M4-75, M4-80 probably received a complete plasmid (pM4-80L3, 57.9 kb in size) that harbored differential resistance genes from the external environment (Supplementary Figure 1).
Transferability of plasmids containing different transferable oxazolidinone resistance genes
Using antimicrobial susceptibility testing, the detection of transferable oxazolidinone resistance genes, 16S rDNA sequencing and whole-genome sequencing, nine transconjugants containing transferable oxazolidinone resistance genes were obtained, with frequencies ranging from 7.1 × 10−6 to 2.5 × 10−7 (Table 2). The MICs of linezolid against those transconjugants varied from 4 to 8 mg/l, and the MICs of tedizolid varied from 0.25 to 1 mg/l (Table 2).
Discussion
Knowledge of the distribution of antimicrobial-resistant isolates in the food chain and edible animals is important in determining the potential risk of antimicrobial-resistant isolates to human health (Yoon et al., 2020). Although oxazolidinones have been approved for human use only, the cfr, optrA and poxtA genes have been detected in enterococcal isolates of animal and environmental origin and recently were even found in enterococci from coastal seawater samples (Freitas et al., 2021; Schwarz et al., 2021). Enterococci in edible animals that carry oxazolidinone resistance genes poses a threat to public health and the surrounding environment. In this study, we investigated the prevalence and genetic characteristics of oxazolidinone resistance genes in enterococcal isolates to better understand their resistance profiles and the dissemination of oxazolidinone resistance genes in enterococcal isolates obtained from pigs at different stages on a swine farm.
Since the three oxazolidinone resistance genes were discovered, these genes have been detected in enterococcal isolates from hospital patients, as well as livestock in the community and veterinary hospital (Carmen et al., 2018). In this study, we reported the presence of optrA, poxtA, poxtA2 and cfr(D) in 49.3% (175/355), 1.71% (3/175), 3.1% (11/355) and 3.1% (11/355) of florfenicol-resistant enterococcal isolates, respectively. The detection rate of the optrA gene was higher than that reported by Wang et al., who found that 24.8% (37/149) of enterococcal isolates of swine origin harbored the optrA gene (Wang et al., 2015). The prevalence of the poxtA gene was lower than that observed by Hao et al., who reported that the poxtA gene was present in 57.9% (66/114) of the florfenicol-resistant enterococcal isolates from two swine farms in Henan Province in China (Hao et al., 2019). The prevalence of the poxtA gene was close to that in florfenicol-resistant enterococci of swine origin in Italy (4.14%, 6/145; Fioriti et al., 2020). Moreover, Cinthi et al. reported that two E. faecalis isolates and one E. casseliflavus isolate from a pig farm environment carried poxtA2 and cfr(D) variants (Cinthi et al., 2022). These results indicated that the prevalence of three oxazolidinone resistance genes in enterococcal isolates from different swine farms exhibited huge differences, which might be associated with different veterinary antimicrobial agent usage schemes.
This study confirmed that WGS played an indispensable role in understanding the resistance profiles of oxazolidinone resistance genes and monitoring the dissemination of oxazolidinone resistance genes in enterococcal isolates. Searching for resistance genes among 355 florfenicol-resistant enterococcal isolates indicated no mutations in 23S rRNA (G2576T and G2505A) or in the L3, L4 and L22 ribosomal proteins in 175 enterococcal isolates carrying oxazolidinone resistance genes (Baccani et al., 2021); the remaining 180 florfenicol-resistant enterococcal isolates were not resistant to oxazolidinone, and no chromosomal mutations associated with oxazolidinone resistance were detected. In this study, three enterococcal isolates carried two oxazolidinone resistance genes, and 11 isolates carried three oxazolidinone resistance genes. Eleven E. faecalis harbored both the cfr-like variant cfr(D) and the poxtA2 variant. The poxtA2 variant was first identified from an isolate of E. gallinarum from a healthy child in Bolivia (Baccani et al., 2021). We also found six optrA variants, one of which has not been reported in E. faecalis. The presence of multiple oxazolidinone resistance genes in one strain might enhance resistance to oxazolidinones, and different variants of different oxazolidinone resistance genes and other factors in the different enterococcal species contribute to the level of linezolid resistance, which requires further investigation (Cai et al., 2018). We also found other antibiotic resistance genes in these 175 enterococcal isolates that harbored resistance genes for oxazolidines, such as tetracycline (tet(M), tet(S/M) and tet(O/W/32/O)), and macrolides (erm(A), erm(B) and msr(C)). Two isolates also carried the clpL gene, which encodes a chaperone family protein of HSP100/Clp (caseinolytic protease). It was found mainly in gram-positive bacteria, which might be associated with decreased penicillin susceptibility (Tran et al., 2011). Although we did not find vancomycin resistance genes, more attention should be given to monitoring the VRE that carries transferable oxazolidinone resistance genes (Yi et al. 2022). A total of 175 enterococcal isolates harbored multiple resistance genes, which indicated a broad antibiotic resistance spectrum of enterococcal isolates. This might make it difficult to treat antibiotic-resistant enterococcal isolates on swine farms.
On the basis of conventional MLST loci, we classified 175 enterococcal isolates harboring oxazolidinone resistance genes. A total of 161 E. faecalis isolates belonging to 28 different STs, including eight new STs, were identified. Of those clones, the most prevalent was ST506 (24.84%, 40/161). ST902 (11.18%, 18/161) was the second most prevalent clone. Moreover, four different STs and one new ST were found in six E. faecium isolates. Using WGS, MLST-based minimum spanning trees of E. faecalis isolates (161) and E. faecium isolates (6) were constructed. We found that the STs of E. faecalis were distributed across multiple production stages. In particular, ST506 and ST632 isolates were found in pigs at four production stages, indicating that these ST isolates may spread in pigs of different stages on swine farms. However, the distribution of E. faecium (6) isolates was simple because the number of E. faecium isolates was relatively small. A phylogenetic tree of the 175 enterococcal isolates was constructed based on SNPs of the core genome. The isolates with the same STs were clustered into the same branch on the phylogenetic tree, suggesting that there was a clonal correlation among the isolates. This result was consistent with the MLST results. For the isolates isolated from a certain stage, 18 isolates of ST902 from breeding pigs showed high similarity, and four isolates of ST21 from 2-month fattening pigs also showed high similarity. In contrast, different ST isolates isolated from the same production stage showed high diversity and did not cluster together in the phylogenetic tree. These results suggest that both clonal spread and horizontal transfer might mediate the diffusion of oxazolidone resistance genes in enterococcal isolates at specific stages in pig farms. For the isolates isolated from different production stages, we found that ST506 and ST632 isolates isolated from four stages showed high similarity, and the same ST isolates were also found to have clonal correlation between two adjacent stages (BP to 2M-FP, 2M-FP to 4M-FP, 4M-FP to 6M-FP). This meant that enterococcal isolates carrying oxazolidone resistance genes could spread from breeding pigs to fattening pigs, while transferable oxazolidone resistance genes in enterococcal isolates could persist throughout all production stages on a pig farm. Finally, the poxtA2 and cfr(D) variants were always carried by ST256 isolates isolated from the 2-month fattening pigs and 4-month fattening pigs. In addition, ST1251 and ST1255 isolates isolated from the 2-month fattening pigs and 4-month fattening pigs also carried the poxtA2 and cfr(D) variants, which were also close to ST256 isolates in the phylogenetic tree. This result indicated that ST1251 and ST1255 may have evolved from ST256.
Representative enterococcal isolates carrying different types of oxazolidinone resistance genes were further sequenced to investigate the locations and genetic environments of different oxazolidinone resistance genes. Mobile genetic elements contribute significantly to the transmission of resistance genes. For the 11 isolates carrying the optrA gene and the poxtA2 and cfr(D) variants, we identified a novel plasmid, named pM4-80 L4, that was prevalent in different STs of E. faecalis isolates from 2-month fattening pigs and 4-month fattening pigs. The plasmid harboring the poxtA2 and cfr(D) variants was similar to pV386 (accession number MZ603802.1), an isolate of swine origin from Italy, indicating that these two plasmids might have the same origin (Cinthi et al., 2022). The emergence of pM4-80 L4 harboring the poxtA2 and cfr(D) variants demonstrates that intense genetic exchanges between enterococcal isolates promoted the spread of oxazolidinone resistance determinants.
The poxtA gene shares 32% homology with the optrA gene (Dejoies et al., 2021). To date, there is no report on poxtA in enterococcal chromosomes. Among three enterococcal isolates carrying the optrA and poxtA genes, we identified three different plasmids carrying the poxtA gene. All poxtA genes were flanked by two IS1216E element copies in the same direction, which was consistent with other reports. The IS1216E—poxtA—IS1216E segment in our study is similar to that found in S. aureus AOUC-0915 and clinical E. faecium isolates from Italy and France, indicating that the genetic background of poxtA is relatively unique (Bender et al., 2018; Carmen et al., 2018; Dejoies et al., 2021; Coccitto et al., 2022). Moreover, a variety of antibiotic resistance genes were found on these plasmids, revealing co-transmission of antibiotic resistance genes. The main differences among insert sequences (ISs) are that they have different transposase properties and catalyze different chemical reactions (Wilson et al., 2008). The three different plasmids exhibited high genetic variation. Three to eight copies of IS1216E were found in the three plasmids, and we also identified other types of ISs. Different ISs might promote the diversity of poxtA-harboring plasmids. Early studies suggested that the transposition of ISs in the genome was random, while recent experiments confirmed that ISs are more inclined to insert into the plasmid in bacteria, which is conducive to the spread of ISs with plasmids as vectors (Siguier et al., 2006). Therefore, more attention should be given to bacterial antibiotic resistance mediated by ISs.
We also described the environment of optrA genes in different locations in this study. Because there have been many reports on the genetic environment of optrA gene, and our manuscript mainly wanted to focus on the genetic environment of poxtA gene, poxtA2 and cfr(D) variants, we selected four representative isolates from 161 isolates carried only optrA gene (M6-97 and M6-130 represent isolates with optrA gene localized to a plasmid, B83 and B126 represent isolates with optrA gene localized to the chromosome). We obtained two optrA-carrying plasmids by hybrid assembly in two isolates (M6-97 and M6-130). By comparing the sequences of the two optrA-carrying plasmids with the sequences of 134 isolates in which the optrA gene was localized to the plasmid, we found that regardless of the plasmid, its sequence coverage with the sequence of 134 isolates reached more than 85%. This showed that the optrA-carrying plasmids among 134 isolates were similar to these two plasmids obtained by hybrid assembly. In our study, we selected four isolates carrying only the optrA gene (M2-77, M2-82, M6-97, and M6-130), and four genetically distinct optrA-carrying plasmids were identified. A similar genetic context, IS1216E–fexA–optrA–erm(A)–IS1216E, was found in four plasmids, which was consistent with E. faecalis from human and animal origin (He et al., 2016). This indicates that IS1216E might promote the cotransfer of optrA, fexA and erm(A) among plasmids. In summary, IS1216E in Gram-positive bacteria (Enterococcus, Streptococcus suis and Listeria monocytogenes) belongs to the IS6/IS26 family of bacterial ISs and plays an important role in mobilizing antimicrobial resistance genes (Shan et al., 2020). Tn554 mediation of optrA gene transfer was identified in the chromosome carrying the optrA gene, consistent with other reports (He et al., 2016; Kang et al., 2019). The optrA gene was flanked by transposon (Tn554) or insertion sequences (IS1216E), indicating that the optrA gene can be transferred among different bacterial genera and species.
In addition, the phenomenon of heteroresistance complicates the analysis of antibiotic resistance in bacteria. We found heteroresistance of clonally related isolates in ST506 and ST256 by genomic analyses and phylogenetic tree construction. Then, we preliminarily clarified the mechanism of heteroresistance emergence, which was likely that mobile genetic elements mediated the insertion of DNA fragments or the acquisition of external plasmids. Last, but not least, these results suggest that caution must be taken to avoid the dissemination of oxazolidinone resistance genes in the environment as they reveal the genetic environment of different oxazolidinone resistance genes.
Conclusion
Our study highlighted that transferable oxazolidinone resistance genes in enterococcal isolates could persist throughout all production stages on a pig farm. Different mobile genetic elements, such as plasmids (pM4-80 L4), IS1216E and Tn554, mediated the dissemination of oxazolidinone resistance genes in enterococcal isolates in the swine farm. These results indicate that oxazolidinone resistance genes in enterococcal isolates had diverse dissemination characteristics in different production stages of large-scale pig farms. Although few data show that oxazolidinone-resistant enterococci could be directly transmitted from animals to humans, enterococci in animals used for food production could be an important repository of transferable oxazolidinone resistance genes. The prevalence and dissemination of oxazolidinone resistance genes in enterococcal isolates from animal farms should be continually monitored.
Data availability statement
The data presented in this study are deposited in the National Center for Biotechnology Information. The complete nucleotide sequences of nine plasmids harboring different transferable oxazolidinone resistance genes have been deposited in GenBank and assigned accession numbers OP046170–OP046178.
Author contributions
HW and ZH designed the study and supervised the work. YB, QW, XY, TZ, and XC participated, coordinated, and analyzed the data. ZH and YB wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. U21A20257).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1010513/full#supplementary-material
Footnotes
1. ^https://cge.cbs.dtu.dk/services/ResFinder/
2. ^https://cge.cbs.dtu.dk/services/LRE-finder/
3. ^http://mjsull.github.io/Easyfig/
References
Antonelli, A., D'Andrea, M. M., Brenciani, A., Galeotti, C. L., and Rossolini, G. M. (2018). Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 73, 1763–1769. doi: 10.1093/jac/dky088
Baccani, I., Antonelli, A., Di, P. V., Coppi, M., Di, M. T., Spinicci, M., et al. (2021). Detection of poxtA2, a presumptive ancestor, in a plasmid from a linezolid-resistant enterococcus gallinarum isolate. Antimicrob. Agents Chemother. 65:e695. doi: 10.1128/AAC.00695-21
Bender, J. K., Cattoir, V., Hegstad, K., Sadowy, E., Coque, T. M., Westh, H., et al. (2018). Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: towards a common nomenclature. Drug Resist. Updat. 40, 25–39. doi: 10.1016/j.drup.2018.10.002
Bender, J. K., Fleige, C., Klare, I., and Werner, G. (2019). Development of a multiplex-PCR to simultaneously detect acquired linezolid resistance genes cfr, optrA and poxtA in enterococci of clinical origin. J. Microbiol. Methods 160, 101–103. doi: 10.1016/j.mimet.2019.03.025
Brenciani, A., Morroni, G., Pollini, S., Tiberi, E., Mingoia, M., Varaldo, P. E., et al. (2016). Characterization of novel conjugative multiresistance plasmids carrying cfr from linezolid-resistant Staphylococcus epidermidis clinical isolates from Italy. J. Antimicrob. Chemother. 71, 307–313. doi: 10.1093/jac/dkv341
Cai, J., Schwarz, S., Chi, D., Wang, Z., and Wang, Y. (2018). Faecal carriage of optrA -positive enterococci in asymptomatic healthy humans in Hangzhou, China. Clin. Microbiol. Infect. 25, 630.e1–630.e6. doi: 10.1016/j.cmi.2018.07.025
Carmen, T., Carla, A. A., Laura, R., Ricardo, L., Rosa, D. C., and Teresa, M. C. (2018). Antimicrobial resistance in enterococcus spp. of animal origin. Microbiology. Spectrum 6. doi: 10.1128/microbiolspec.ARBA-0032-2018
Cinthi, M., Coccitto, S. N., Fioriti, S., Morroni, G., Simoni, S., Vignaroli, C., et al. (2022). Occurrence of a plasmid co-carrying cfr(D) and poxtA2 linezolid resistance genes in Enterococcus faecalis and Enterococcus casseliflavus from porcine manure, Italy. J. Antimicrob. Chemother. 77, 598–603. doi: 10.1093/jac/dkab456
Clinical and Laboratory Standards Institute (2020). Performance Standards for Antimicrobial Susceptibility Testing—30th Edn: M100. Wayne, PA.
Coccitto, S., Cinthi, M., Fioriti, S., Morroni, G., Simoni, S., Vignaroli, C., et al. (2022). Linezolid-resistant enterococcus gallinarum isolate of swine origin carrying cfr, optrA and poxtA genes. J. Antimicrob. Chemother. 77, 331–337. doi: 10.1093/jac/dkab408
Crowe-McAuliffe, C., Murina, V., Turnbull, K. J., Huch, S., Kasari, M., Takada, H., et al. (2022). Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics. Nat. Commun. 13:1860. doi: 10.1038/s41467-022-29274-9
Dejoies, L., Sassi, M., Schutz, S., Moreaux, J., Zouari, A., Potrel, S., et al. (2021). Genetic features of the poxtA linezolid resistance gene in human enterococci from France. J. Antimicrob. Chemother. 76, 1978–1985. doi: 10.1093/jac/dkab116
Deshpande, L. M., Ashcraft, D. S., Kahn, H. P., Pankey, G., and Mendes, R. E. (2015). Detection of a new cfr-like gene, cfr(B), in Enterococcus faecium isolates recovered from human specimens in the United States as part of the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 59, 6256–6261. doi: 10.1128/AAC.01473-15
Ewa, S. (2018). Linezolid resistance genes and genetic elements enhancing their dissemination in enterococci and streptococci. Plasmid 99, 89–98. doi: 10.1016/j.plasmid.2018.09.011
Finisterra, L., Duarte, B., Peixe, L., Novais, C., and Freitas, A. R. (2021). Industrial dog food is a vehicle of multidrug-resistant enterococci carrying virulence genes often linked to human infections. Int. J. Food Microbiol. 358:109284. doi: 10.1016/j.ijfoodmicro.2021.109284
Fioriti, S., Coccitto, S. N., Cedraro, N., Simoni, S., and Giovanetti, E. (2021). Linezolid resistance genes in enterococci isolated from sediment and zooplankton in two Italian coastal areas. Appl. Environ. Microbiol. 87:e2920. doi: 10.1128/AEM.02958-20
Fioriti, S., Morroni, G., Coccitto, S. N., Brenciani, A., Antonelli, A., Di Pilato, V., et al. (2020). Detection of Oxazolidinone resistance genes and characterization of genetic environments in enterococci of swine origin, Italy. Microorganisms 8:17. doi: 10.3390/microorganisms812201
Freitas, A., Finisterra, L., Tedim, A., Duarte, B., Novais, C., and Peixe, L. (2021). Linezolid- and multidrug-resistant enterococci in raw commercial dog food, Europe, 2019-2020. Emerg. Infect. Dis. 27, 2221–2224. doi: 10.3201/eid2708.204933
Hao, W., Shan, X., Li, D., Schwarz, S., and Zhang, S. (2019). Analysis of a poxtA- and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J. Antimicrob. Chemother. 74, 1771–1775. doi: 10.1093/jac/dkz109
He, T., Shen, Y., Schwarz, S., Cai, J., and Lv, Y. (2016). Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J. Antimicrob. Chemother. 71, 1466–1473. doi: 10.1093/jac/dkw016
Kang, Z. Z., Lei, C. W., Kong, L. H., Wang, Y. L., and Wang, H. N. (2019). Detection of transferable oxazolidinone resistance determinants in Enterococcus faecalis and Enterococcus faecium of swine origin in Sichuan Province, China. J. Global Antimicrob. Resist. 19, 333–337. doi: 10.1016/j.jgar.2019.05.021
Kim, H. Y. (2021). Prevalence and characteristics of phenicol-oxazolidinone resistance genes in Enterococcus faecalis and Enterococcus faecium isolated from food-producing animals and meat in Korea. Int. J. Mol. Sci. 22:11335. doi: 10.3390/ijms222111335
Lei, C. W., Chen, X., Liu, S. Y., Li, T. Y., Chen, Y., and Wang, H. N. (2021). Clonal spread and horizontal transfer mediate dissemination of phenicol-oxazolidinone-tetracycline resistance gene poxtA in enterococci isolates from a swine farm in China. Vet. Microbiol. 262:109219. doi: 10.1016/j.vetmic.2021.109219
Lei, C. W., Kang, Z. Z., Wu, S. K., Chen, Y. P., Kong, L. H., and Wang, H. N. (2019). Detection of the phenicol-oxazolidinone-tetracycline resistance gene poxtA in Enterococcus faecium and Enterococcus faecalis of food-producing animal origin in China. J. Antimicrob. Chemother. 74, 2459–2461. doi: 10.1093/jac/dkz198
Long, K. S., Pøhlsgaard, J., Kehrenberg, C., Schwarz, S., and Vester, B. (2006). The Cfr rRNA Methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin a antibiotics. Antimicrob. Agents Chemother. 50, 2500–2505. doi: 10.1128/AAC.00131-06
Moellering, R. (2014). Tedizolid: a novel oxazolidinone for gram-positive infections. Clin. Infect. Dis. 58, S1–S3. doi: 10.1007/s40121-015-0060-3
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Papagiannitsis, C. C., Tsilipounidaki, K., Malli, E., and Petinaki, E. (2019). Detection in Greece of a clinical enterococcus faecium isolate carrying the novel oxazolidinone resistance gene poxtA. J. Antimicrob. Chemother. 74, 2461–2462. doi: 10.1093/jac/dkz155
Schwarz, S., Zhang, W., Du, X. D., Krüger, H., Feler, A. T., Ma, S., et al. (2021). Mobile oxazolidinone resistance genes in gram-positive and gram-negative bacteria. Clin. Microbiol. Rev. 34:e18820. doi: 10.1128/CMR.00188-20
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shan, X., Li, X., Wang, N., Schwarz, S., Zhang, S., Li, D., et al. (2020). Studies on the role of IS1216E in the formation and dissemination of poxtA-carrying plasmids in an Enterococcus faecium clade A1 isolate. J. Antimicrob. Chemother. 75, 3126–3130. doi: 10.1093/jac/dkaa325
Shen, J., Yang, W., and Stefan, S. (2013). Presence and dissemination of the multiresistance gene cfr in gram-positive and gram-negative bacteria. J. Antimicrob. Chemother. 68, 1697–1706. doi: 10.1093/jac/dkt092
Siguier, P., Filée, J., and Chandler, M. (2006). Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 9, 526–531. doi: 10.1016/j.mib.2006.08.005
Stanley, P., Peter, B., Terence, L., Shane, G., Shu, J., Denise, D., et al. (2019). Linezolid-resistant ST872 Enteroccocus faecium harbouring optrA and cfr (D) oxazolidinone resistance genes. Int. J. Antimicrob. Agents 55:105831. doi: 10.1016/j.ijantimicag.2019.10.012
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc. Natl. Acad. Sci. U. S. A. 102, 13950–13955. doi: 10.1073/pnas.0506758102
Tran, D. H., Kwon, H. Y., Kim, E. H., Kim, K. W., Briles, D. E., Pyo, S., et al. (2011). Decrease in penicillin susceptibility due to heat shock protein ClpL in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 55, 2714–2728. doi: 10.1128/AAC.01383-10
Wang, Y., Li, X., Fu, Y., Chen, Y., Wang, Y., Ye, D., et al. (2020). Association of florfenicol residues with the abundance of oxazolidinone resistance genes in livestock manures. J. Hazard. Mater. 399:123059. doi: 10.1016/j.jhazmat.2020.123059
Wang, Y., Lv, Y., Cai, J., Stefan, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
WHO (2017). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18:318. doi: 10.1016/S1473-3099(17)30753-3
Wilson, D. N., Schluenzen, F., Harms, J. M., Starosta, A. L., Connell, S. R., and Fucini, P. (2008). The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. 105, 13339–13344. doi: 10.1073/pnas.0804276105
Yang, T., and Gao, F. (2022). High-quality pan-genome of Escherichia coli generated by excluding confounding and highly similar strains reveals an association between unique gene clusters and genomic islands. Brief. Bioinform. 23:6638794. doi: 10.1093/bib/bbac283
Yi, M., Zou, J., Zhao, J., Tang, Y., Yuan, Y., Yang, B., et al. (2022). Emergence of optrA-Mediated Linezolid Resistance in Enterococcus faecium: A Molecular Investigation in a Tertiary Hospital of Southwest China from 2014-2018. Infect. Drug. Resist. 15, 13–20. doi: 10.2147/IDR.S339761
Yoon, S., Son, S. H., Kim, Y. B., Seo, K. W., and Lee, Y. J. (2020). Molecular characteristics of optrA-carrying Enterococcus faecalis from chicken meat in South Korea. Poult. Sci. 99, 6990–6996. doi: 10.1016/j.psj.2020.08.062
Keywords: Enterococci, oxazolidinone, poxtA2-cfr(D) co-harboring, optrA, poxtA, genomic analyses, phylogenetic analysis, genetic environment
Citation: Huang Z, Bai Y, Wang Q, Yang X, Zhang T, Chen X and Wang H (2022) Persistence of transferable oxazolidinone resistance genes in enterococcal isolates from a swine farm in China. Front. Microbiol. 13:1010513. doi: 10.3389/fmicb.2022.1010513
Edited by:
David Christopher Coleman, Dublin Dental University Hospital, IrelandCopyright © 2022 Huang, Bai, Wang, Yang, Zhang, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongning Wang, whongning@163.com
†These authors have contributed equally to this work and share first authorship
 Zheren Huang
Zheren Huang Yilin Bai
Yilin Bai Qin Wang1
Qin Wang1 Xue Yang
Xue Yang Tiejun Zhang
Tiejun Zhang Hongning Wang
Hongning Wang