- 1MOE Key Laboratory of Industrial Fermentation Microbiology, College of Biotechnology, Tianjin University of Science and Technology, Tianjin, China
- 2Tianjin Engineering Research Center of Microbial Metabolism and Fermentation Process Control, Tianjin University of Science and Technology, Tianjin, China
- 3National Technology Innovation Center of Synthetic Biology, Tianjin, China
Aspergillus niger is a major cell factory for citric acid production, and the process of citrate export from mitochondria to cytoplasm is predicted to be one of rate-limiting steps in citric acid accumulation. Currently, the mitochondrial citrate transporters (Ctps) in A. niger are not fully characterized. Here, six putative Ctp encoding genes (ctpA to ctpF) were identified based on their homology with a mitochondrial citrate transporter ScCtp1 from Saccharomyces cerevisiae. Disruption of individual ctpA to ctpF caused varying degrees of decline in citric acid accumulation at different fermentation stages, whereas a mutant strain S1696 with disruption of all six ctps showed complete loss of citiric acid production. S1696 also exhibited delayed growth, reduced conidia formation, and decreased pigmentogenesis. Exogenous addition of citrate partially restored the conidia formation and pigmentogenesis in S1696 mutant. Reintroduction of individual ctps (ctpA to ctpF) into S1696 at the amyA locus showed that ctpA, ctpB, and ctpD restored the citric acid titers to 88.5, 93.8, and 94.6% of the parent strain, respectively. Additionally, the formation of conidia and pigment production was partially restored after reintroduction of ctpA, ctpB, or ctpD. Overexpression of respective ctpA, ctpB, and ctpD in the parent strain resulted in increases in citric acid accumulation by 32.8, 19.3, and 24.2%, respectively. These results demonstrate that CtpA, CtpB, and CtpD play important roles in citric acid transport across the mitochondrial membrane and function in a redundant manner. Enhancement of citric acid transport process can serve as a target for boosting citric acid accumulation in A. niger.
Introduction
Citric acid is an intermediate metabolite of the tricarboxylic acid (TCA) cycle and has important commercial value. Due to its safety, good flavor, high solubility, metal-chelating, and buffering ability, citric acid is extensively used in diverse fields such as food, medicine, detergent, cosmetics (Soccol et al., 2006). The worldwide market of citric acid is estimated to reach USD3.6 billion by 2025 (Mores et al., 2021).
Although several bacteria and yeasts have been found to accumulate citric acid, the performance of most strains cannot meet the needs of commercial production. Currently, Aspergillus niger-based fermentation is the major process for industrial production of citric acid (Tong et al., 2019; Behera, 2020) because production strains of the fungus showed excellent yields with few by-products and the good biosafety, abundant extracellular hydrolase systems for the use of cheap industrial and agricultural raw materials. High levels of citric acid accumulation by A. niger depend on optimization of fermentation conditions including: (1) the carbon source must be quickly available and the concentration must be higher than 5%; (2) the nitrogen sources in the medium should be able to cause a pH decrease after utilization; (3) the phosphate concentrations should be suboptimal; and (4) trace-metal-ion (especially Mn2+) addition should be limited and pH needs to be maintained at < 3 (Karaffa and Kubicek, 2003; Behera, 2020).
Many studies have attempted to explain the molecular mechanism of high citric acid production by A. niger from different perspectives, especially the activity and regulation of various enzymes related to citric acid synthesis in the primary metabolism (Behera, 2020). Given that citric acid accumulation results from an interplay of multiple factors, further mining and analysis of the factors that promote citric acid accumulation could serve as a powerful tool for the metabolic engineering of more efficient production strains of A. niger. Through a quantitative model of carbohydrate degradation and oxalacetate formation, three pivotal steps for citric acid accumulation have been characterized, namely, sugar uptake and phosphorylation, citrate export from mitochondria to cytoplasm, and subsequent secretion outside the cell (Torres, 1994a, 1994b). Two kinds of glucose transporters, i.e., low- and high-affinity types, have been identified and their unique physiological roles in citric acid production have been elucidated in A. niger (Torres, 1994b; Torres et al., 1996; Sloothaak et al., 2015; Yin et al., 2017). Recently, through homology search and transcriptomic data analysis, a major citrate transporter, CexA, responsible for exporting citric acid from cytoplasm to extracellular medium has been identified by two independent research teams (Odoni et al., 2019; Steiger et al., 2019). Its impact on glucose transport, glycolysis, and citric acid accumulation was analyzed (Nakamura et al., 2020; Xu et al., 2020).
A mitochondrial citrate transporter CTP for the transport of citric acid from mitochondria to cytoplasm was identified in rat liver cells and found to be involved in the promiscuous antiport of citrate with other tricarboxylates, dicarboxylates, and phosphoenolpyruvates (Palmieri et al., 1972; Bisaccia et al., 1989; Palmieri, 1994). By contrast, the citrate transporter ScCtp1 identified in Saccharomyces cerevisiae showed a much higher substrate specificity than CTPs from higher eukaryotic organisms (Kaplan et al., 1995), and no obvious phenotype was found in ΔScCTP1 mutants, suggesting that other unidentified mitochondrial transporter protein may be involved in this process (Kaplan et al., 1996). During the shochu brewing process, citric acid production by Aspergillus luchuensis mut. Kawachii is a typical brewing characteristic. CtpA and YhmA in A. kawachii has been identified as mitochondrial citrate transporters with different counter substrates that participate in acetyl-CoA generation in the cytoplasm (Kadooka et al., 2019). In A. niger WU-2223 l, two putative Ctp encoding genes, ctpA and ctpB, were found, but no transcripts of ctpB were detected under all cultivation conditions examined. ctpA disruption was found to cause growth delay, low spore germination rates, and low citric acid accumulation only in the early-log phase, indicating that CtpA is not the sole mitochondrial citrate transporter and that other mitochondrial transporters may participate in citric acid accumulation (Kirimura et al., 2016).
The present study aimed to comprehensively characterize the putative genes of mitochondrial citrate transporters in A. niger and evaluate their roles in citric acid accumulation. Using yeast ScCtp1 as template, 6 possible genes encoding mitochondrial citrate transporters were identified in the genome databases of A. niger through homology search and domain analyses and their roles in citric acid accumulation were validated. Moreover, strains with elevated citric acid production were constructed by overexpression of Ctps in A. niger.
Materials and methods
Strains and culture conditions
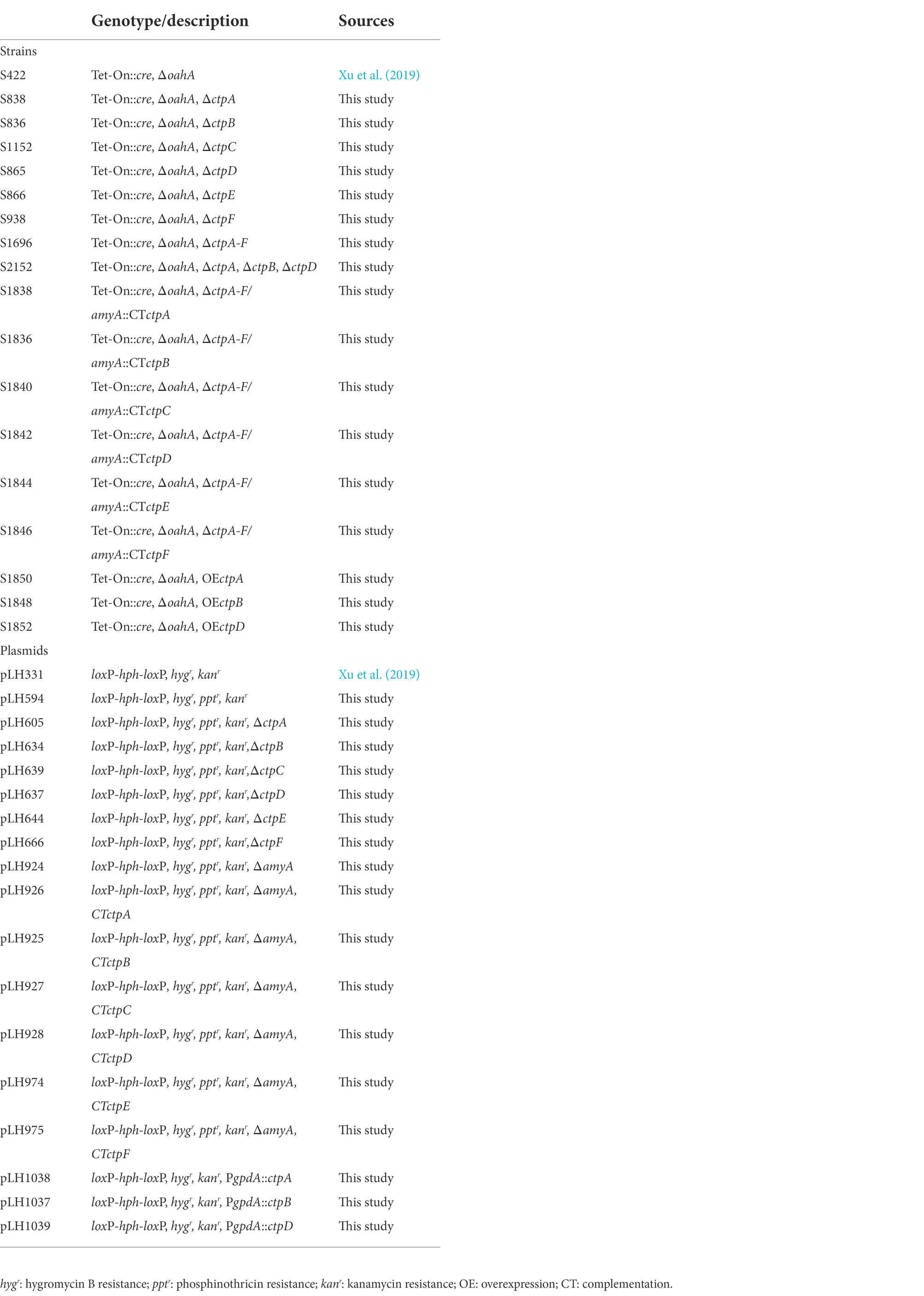
Aspergillus niger strain S422 (Table 1) derived from A. niger ATCC 1015 was used as parent strain in this study. Potato dextrose agar (PDA) was used for A. niger spore preparation, complete medium (CM) was used for transformant screening, and minimal medium (MM) was used for gene knock-out phenotype screening as previously described (Cao et al., 2020). Escherichia coli JM109 used as a host for plasmid construction, and Agrobacterium tumefaciens AGL-1 used for the genetic transformation of A. niger were cultivated in LB medium at 37 and 28°C, respectively (Xu et al., 2019). All A. niger strains used in this study are listed in Table 1.
Mining of putative mitochondrial citrate transporters (CTPs) in the Aspergillus niger genome
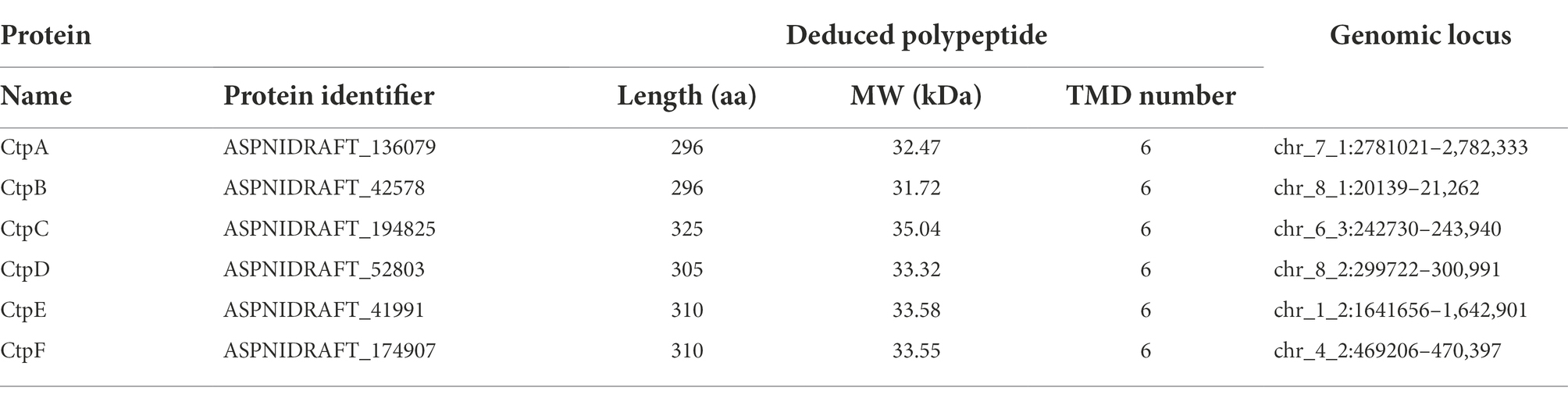
The A. niger 20131226 filtered model-protein database (A. niger ATCC 1015 v4.0)1 was searched with ScCtp1 protein sequence from S. cerevisiae S288c (accession no. NP_009850) through online blastp alignment program. With reference to genome annotation and the fact that shuttle transport of citric acid and dicarboxylic acid, sequences annotated as tri- and di-carboxylate transporter were both candidates for further analysis. Domain identification, annotation, and architectures of the obtained hit sequences were performed using the web resource SMART version 92 with the default parameters. Transmembrane domains were analyzed through the Phyre2 web portal.3 Proteins without the Mito_carr (PF00153) domain were eliminated (Table 2). Alignment of candidates was performed with DNAMAN software Version 10. A phylogenetic tree was also established with MEGA software Version 10.1.7 by neighbor-joining statistical method. Accession numbers of the sequences for phylogenetic-tree construction are listed in Supplementary Table 2.
Genomic DNA, total RNA isolation, and transcription analyses under citric acid accumulation condition
For genomic DNA isolation, 1 × 108 conidia were inoculated into 50 ml of potato dextrose broth for 24 h, and mycelia were harvested followed by washing with water twice. The washed mycelia were ground into powder in liquid nitrogen. The genomic DNA was extracted using DNA extraction buffer (100 mM NaCl, 50 mM EDTA, 50 mM Tris, and 1.0% SDS; pH 8.5). Proteins and RNA were eliminated with phenol buffer and RNaseA, respectively. For total RNA isolation, mycelia were collected under indicated conditions, and total RNA was isolated following a previously described method (Cao et al., 2020). cDNA was synthesized using PrimeScript™ II 1st-Strand cDNA Synthesis Kit (TaKaRa Biomedical Technology Co., Ltd., Beijing, China). Transcription levels of the putative ctpA to ctpF in A. niger were determined by RT-qPCR. Primers are listed in Supplementary Table 3.
Plasmid construction
Plasmids pLH605, pLH634, pLH639, pLH637, pLH644, pLH666, and pLH924 for ctpA to ctpF and amyA (ASPNIDRAFT_47911) disruptions were constructed as previously described (Cao et al., 2020). In a typical procedure, the dual flanked sequences of ctpA coding region were amplified with P3424/3425 and P3426/3427 followed by ligation into the upstream and downstream of the hygromycin resistance cassette (loxP-hph-loxP) in pLH594, respectively, to obtain the ctpA disruption plasmid pLH605. ctpB-ctpF and amyA disruption plasmids were constructed following the same strategy.
To construct the ctpA complementation plasmid integrated at the amyA locus, a ctpA expression cassette with its native promoter and terminator was obtained with P3407/3408. The resultant fragment was digested with BamH I/Sac I and ligated into pLH924 to obtain pLH926. The same strategy was used for the construction of ctpB to ctpF complement plasmids.
The plasmid pLH1038 for ctpA overexpression under the control of glyceraldehyde-3-phosphate dehydrogenase (gpdA) promoter was constructed as follows. P3619/3620 was used to amplify the cDNA of ctpA, and the obtained fragment was digested with BamH I/EcoR I, followed by ligation into pLH454 to obtain pLH1038. ctpB and ctpD gene overexpression plasmids were constructed following the same strategy. Primers used are listed in Supplementary Table 3.
Transformation of Aspergillus niger for disruption, individual reintroduction of ctpA to ctpF, and overexpression of ctpA, ctpB, and ctpD
Agrobacterium-mediated transformation was applied for A. niger genetic manipulation as previously described (Cao et al., 2020). ctpA disruptants were obtained by introducing pLH605 into A. niger S422. Transformants were selected on CM plates supplemented with cefotax thiazide sodium (0.10 g/l), hygromycin B (0.25 g/l), ampicillin (0.10 g/l), and streptomycin (0.10 g/l) at 28°C for 5 days, followed by screening on PDA plates with hygromycin B (0.25 g/l) and MM with glufosinate ammonium (1.00 g/l), respectively. According to the principle of homologous recombination, ΔctpA candidates that were hygromycin B resistant and glufosinate ammonium sensitive were identified by PCR analyses with P3416/P3417, P3416/P641, P642/P3419, P3418/P3419, and P3416/P3419. The resultant ctpA deficiency mutant was named S838 (Supplementary Figure 3A). In the same way, P1967/P1968, P1967/P641, P642/P1970, P1969/P1970, and P1967/P1970 were used for ΔctpB candidate identification; P2009/P2010, P2009/P641, P642/P2012, P2011/P2012, and P2009/P2012 were used for ΔctpC candidate identification; P2013/P2014, P2013P641, P642/P2016, P2015/P2016, and P2013/P2016 were used for ΔctpD candidate identification; P2017/P2018, P2017/P641, P642/P2020, P2019/P2020, and P2017/P2020 were used for ΔctpE candidate identification; and P2021/P2022, P2021/P641, P642/P2024, P2023/P2024, and P2021/P2024 were used for ΔctpF candidate identification, respectively. Individual deficiency mutants obtained for ctpB, ctpC, ctpD, ctpE, and ctpF were named S836, S1152, S865, S866, and S938 (Supplementary Figures 3B–F). Deletion mutant for all six ctps was also obtained by using the Cre-loxP-based genetic system (Xu et al., 2019) and named S1696 (ΔctpA-F). The primers used for PCR verification are listed in Supplementary Table 3.
To determine the role of CtpA in citric acid accumulation, pLH926 harboring ctpA expression cassette with its native promoter was reintroduced into the amyA locus in A. niger S1696 (ΔctpA-F). The obtained ctpA complementation strain was named S1838. Other ctps were also individually reintroduced into A. niger S1696 (ΔctpA-F), and the resultant strains were named S1836, S1840, S1842, S1844, and S1846, respectively.
Growth assay and conidia formation of Aspergillus niger parent and mutant strains
To investigate the growth differences, 5 × 104 conidia of A. niger parent and the indicated mutant strains in distilled water were inoculated on PDA plates at 28°C for 4 days. Morphology and conidia formation were photographed and analyzed. To establish the growth curve, 1 × 106 conidia of A. niger were inoculated into 20 ml of citric acid fermentation medium. At the indicated time, the culture broths were passed through qualitative filter paper (Whatman No.4), and trapped mycelia were completely dried at 105°C.
Shake-flask fermentation for assaying citric acid production
To assess the citric acid accumulations, 2 × 106 conidia/ml of A. niger mutants were inoculated into 50 ml of citric acid fermentation medium (10% sucrose, 0.25% NH4NO3, 0.1% MgSO4·7H2O, 0.1% KH2PO4, and yeast extract 0.05%; pH 2.5) in 250 ml Erlenmeyer flasks at 28°C and 200 rpm for 5 days. Fermentation broths were sampled at indicated time points for organic acid analyses as previously described (Cao et al., 2020).
Intracellular acetyl-CoA concentration measurement
A total of 2 × 106 conidia/mL for each A. niger strain were inoculated into 50 ml of citric acid fermentation medium in 250 ml Erlenmeyer flasks at 28°C, and 200 rpm for 3 days. The obtained mycelia were washed and ground into powder in the presence of liquid nitrogen. The acetyl-CoA concentration was measured with an Acetyl-CoA Assay Kit (Solarbio, BC0980), and the total protein concentration was determined using a BCA Protein Assay Kit (TaKaRa, T9300A) according to the manufacturer’s instructions. Optical density at 340 nm and 562 nm was measured for Acetyl-CoA and protein contents quantification, respectively, with a UV-3600i Plus system (Shimadzu, Japan).
Statistical analyses
All experiments were performed in triplicate, and the mean values were compared using two-tailed Student’s t-test. *p < 0.05, **p < 0.001, and ****p < 0.0001 were considered statistically significant.
Results
Identification of mitochondrial citrate transporters in Aspergillus niger (Ctps)
Previous studies have demonstrated that a protein CtpA was shown to play an important role in citrate transport across mitochondria membrane and citric acid production in A. niger WU-2223 l and A. kawachii SO2 (Kirimura et al., 2016; Kadooka et al., 2019). A Preliminary experiments showed that deletion of ctpA in A. niger strain S422 resulted in the production of citric acid decreased by 15.8 and 18.3% at 3 and 5 days, respectively (Supplementary Figure 1). This suggested that CtpA was indeed involved in the transport of citrate across the mitochondrial membrane, and other undiscovered Ctps might were also involved.
Here, ScCtp1 from S. cerevisiae is an ortholog of human SLC25A1 located in the inner mitochondrial membrane and is responsible for the citrate-malate shuttles (Kaplan et al., 1995; Gnoni et al., 2009; Palmieri, 2013). In the present study, using ScCtp1 as query sequence, 23 sequences were obtained (Supplementary Table 1). After sequence annotation screening and functional domain analysis, six putative Ctps were identified in A. niger ATCC 1015 model-protein database, namely CtpA to CtpF (Table 1). It should be noted that, based on the fact that the antiport of citrate and dicarboxylate (malate, oxoglutarate, oxaloacetate, etc) is the main manner of citrate transport from mitochondria to cytoplasm (Karaffa and Kubicek, 2003; Kirimura et al., 2016, 2019; Kadooka et al., 2019; Yang et al., 2019), the annotated putative dicarboxylate transporters were also named citrate transporters here. Among them, only CtpA has been investigated in A. niger WU-2223 l strain and A. luchuensis mut. Kawachii (Kirimura et al., 2016; Kadooka et al., 2019). Alignment with ScCtp1 showed that CtpA to CtpF exhibited 45.66, 42.9, 29.85, 23.89, 21.05, and 20.38% amino acid sequence identities, respectively. Six transmembrane domains (TMD I–VI; Figure 1A and Supplementary Figure 2) and three conserved motifs were found in each putative Ctp (Figure 1A), which were typical characteristics of all mitochondrial transporter family members (Palmieri, 2013). The predicted amino acid residues for dimer formation and citrate binding can also be found in most sequences and marked with different color symbols (Figure 1A; Kirimura et al., 2016; Kadooka et al., 2019). Functional domain analyses revealed that CtpA had two mito_carr domains, whereas other Ctps had three mito_carr domains in a tandem manner (Figure 1B). These analyses indicate that CtpA to CtpF might have citrate-transport activity. Phylogenetic analysis showed that CtpA, CtpB, AkCtpA, and ScCtp1 belong to one evolutionary branch, whereas CtpC, CtpD, CtpE, and CtpF are assigned to another common evolutionary branch (Figure 1C). It should be noted that although AkYhmA was reported to be a mitochondrial citrate transport protein (Kadooka et al., 2019), it had a relative distant evolutionary relationship with the putative Ctps (Figure 1C).
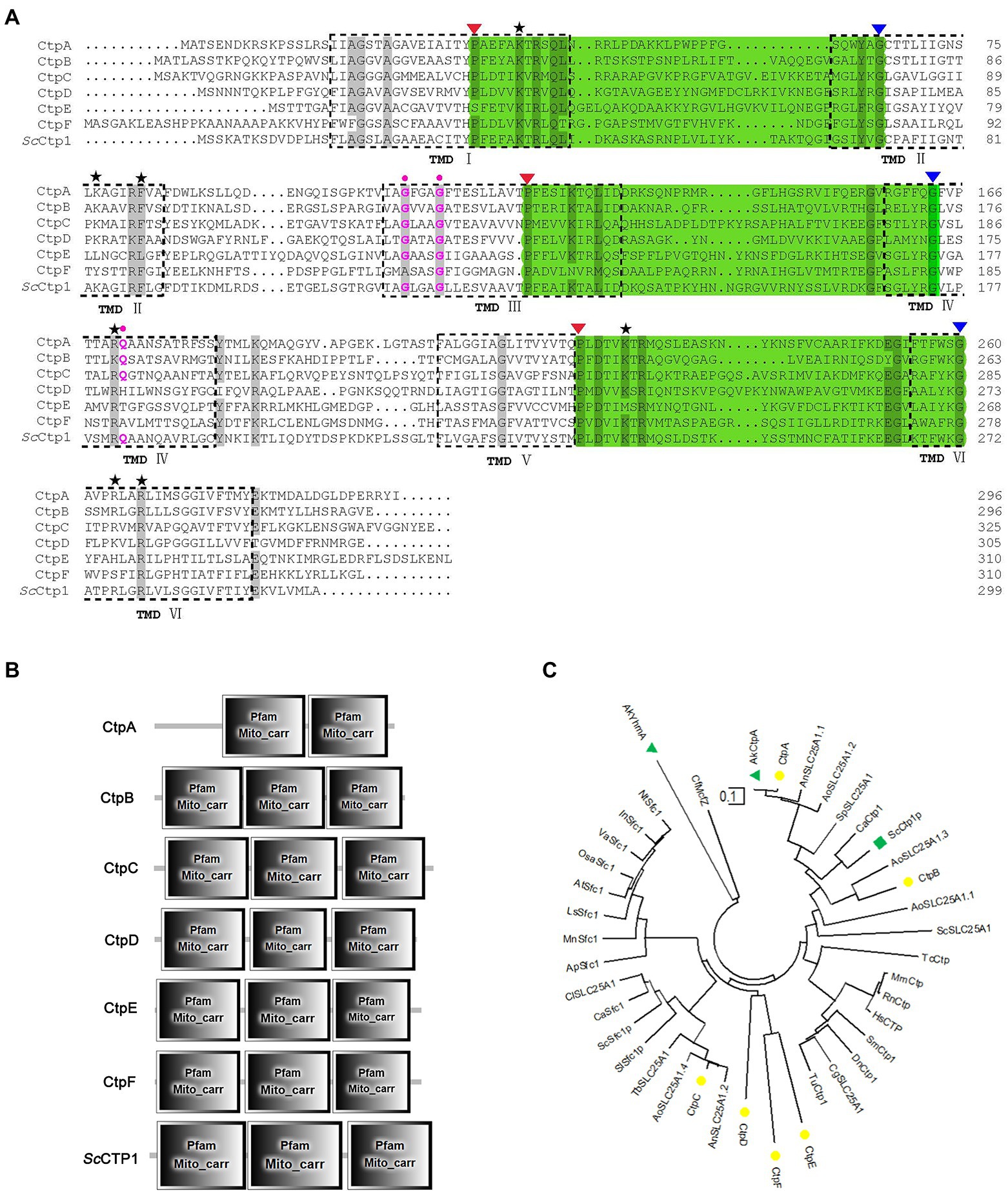
Figure 1. Sequence features and domain analyses of CtpA to CtpF. (A) Multiple sequence alignment of CtpA to CtpF amino acid sequence against their yeast ortholog ScCtp1. Six predicted transmembrane domains (TMD) based on ScCtp1 are marked as TMD I to VI. Note: three symbolic motifs PX[D/E]XX[K/R]X[K/R] (20–30 residues) [D/E]GXXXX[W/Y/F][K/R]G are in bottle-green background, and the conservative P and G are marked with red and blue inverse triangles, respectively; black Pentastar, citrate binding sites; pink dots, amino acid residues for dimer interface. (B) CtpA to CtpF and ScCtp1 domain architecture. (C) Neighbor-joining phylogenetic tree established using MEGA software Version 10.1.7. CtpA to CtpF are marked with yellow solid circle, ScCtp1 is marked with green solid triangle, and AkCtp and AkYhmA are marked with green square.
Transcription analyses of ctpA to ctpF
To determine the expression profiles of the putative ctpA to ctpF during citric acid accumulation, the total RNA of the parent strain A. niger S422 cultivated in citric acid fermentation medium for 3 and 5 d was, respectively, extracted for RT-qPCR analyses. As shown in Figure 2, except for ctpB that was transcribed only in the samples of 5-d fermentation, ctpA, ctpC, ctpD, ctpE, and ctpF were transcribed in the samples of both 3- and 5-day fermentation. Notably, in A. niger WU-2223 l, no transcription of ctpB was detected in SLZ medium (Kirimura et al., 2016). Citric acid fermentation medium in this study and SLZ medium are different in carbon source and mineral salt, which may lead to different expression profiles of ctpB.
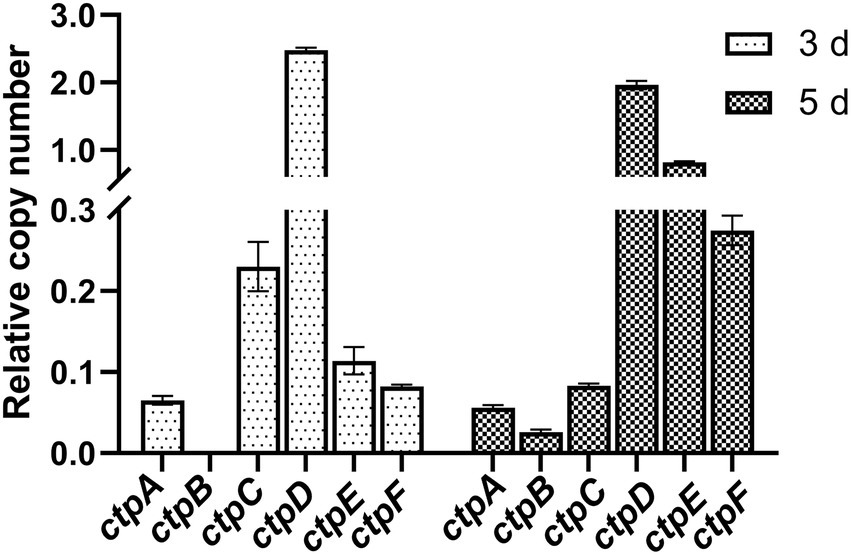
Figure 2. RT-qPCR analysis of ctpA to ctpF relative expression in S422 under citric acid fermentation conditions at 28°C, and 200 rpm for 3 days and 5 days, respectively. All expression values were normalized to the expression level of actA (ASPNIDRAFT_200483).
Phenotype analyses of A. niger mutants with individual ctp disruption or deletions of all six ctps
To investigate the physiological effects of CtpA to CtpF on A. niger growth and conidia formation, mutants with disruption of individual ctps or deletion of all six ctps were constructed (Supplementary Figures 3A–F). Compared with the parent strain S422 on PDA plates, all the individual ctp disruption mutants exhibited similar colonial morphologies, whereas S1696 (ΔctpA-F) colony became more fluffy and albino, and the conidia formation was significantly impaired (Figure 3A). Additionally, S1696 (ΔctpA-F) formed a smaller colony than the control strain S422 on MM plates (Figure 4B). Furthermore, the conidia formation of individual mutants was measured after their growth on PDA plates for 5 days and results showed that the number of conidia per square centimeter for individual ctp mutants decreased to varying degrees but was not statistically significant, whereas the number of conidia formed by S1696 (ΔctpA-F) was reduced to only 11.5% of that produced by the parent strain S422 (Figure 3B). Under citric acid fermentation condition, each ctp disruptant had a slightly slow growth rate, whereas S1696 (ΔctpA-F) exhibited a significantly reduced growth rate than S422 (Figure 3C). Similarly, previous studies also reported that ctpA disruption caused growth delay in A. niger strain WU-2223l and A. kawachii strain SO2 (Kirimura et al., 2016; Kadooka et al., 2019). Citrate transporters are considered to export citrate from mitochondria to cytoplasm and, in turn, citrate is cleaved to supply acetyl-CoA. In the cytoplasm, citrate participates in fatty acid and cholesterol biosynthesis and regulates acetyl-CoA carboxylase activity (Gnoni et al., 2009; Chen et al., 2014; Kadooka et al., 2019; Ruprecht and Kunji, 2020). Accordingly, the complete deficiency of ctps is predicted to result in the blockage of citrate transport, which, in turn, affected the supply of acetyl-CoA in the cytoplasm and caused growth defects. To test this hypothesis, the effects of exogenous addition of citrate or acetate on the growth defect of S1696 (ΔctpA-F) were investigated. As shown in Figure 3D, defects of pigment production and conidia formation in S1696 (ΔctpA-F) mutant were partially relieved with the addition of acetate or citrate. These results further demonstrated that disturbance of citrate transport caused by ctp deletion affected growth and conidia formation.
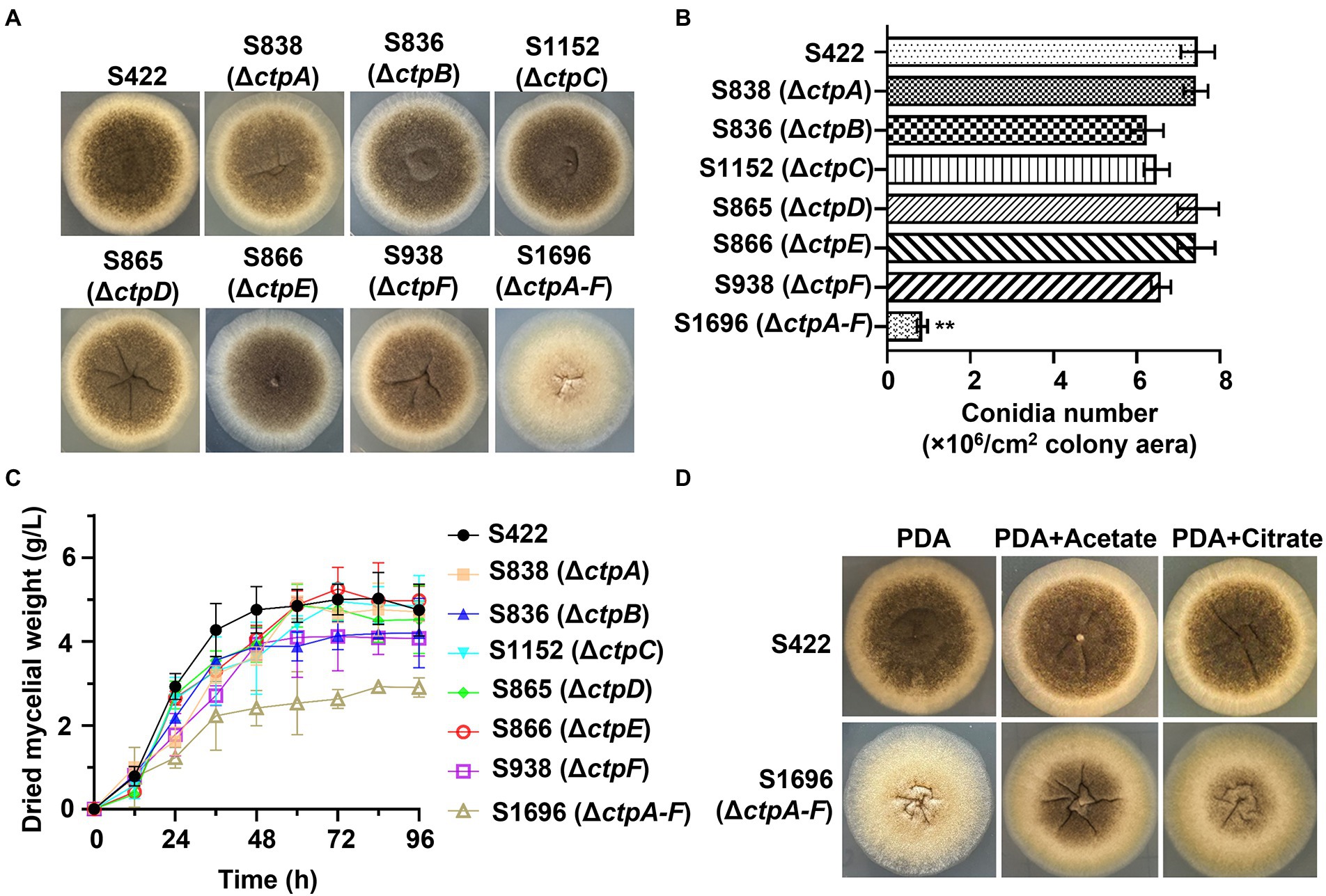
Figure 3. Phenotype analyses of ctp disruptions strains. (A) Morphology of A. niger S422, individual ctp disruption mutants, and ΔctpA-F mutant grown on PDA plate at 28°C for 4 d. (B) The number of conidia per square centimeter of the indicated strains grown on PDA plates at 28°C for 4 d. (C) Growth curves of A. niger S422 and the indicated mutants in 50 ml of citric acid fermentation medium at 28°C, and 200 rpm. (D) Morphology of A. niger S422 and S1696 (ΔctpA-F) grown on PDA plate or supplemented with 5 mM acetate or 10 mM citrate.
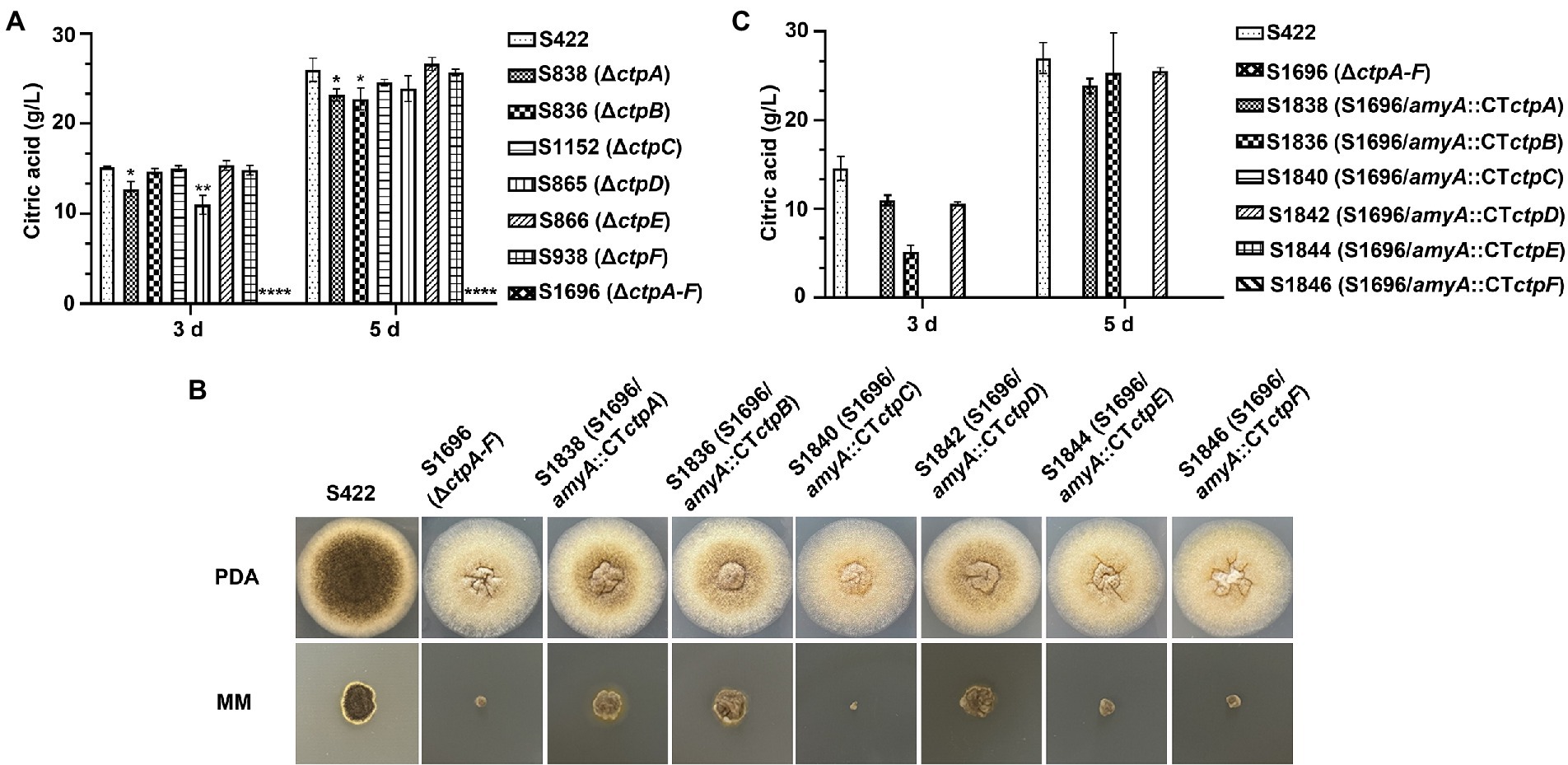
Figure 4. Citric acid production and morphology analyses of A. niger strains. (A) In a 250 ml shake flask, 1 × 108 conidia of each A. niger strain were inoculated in 50 ml of citric acid fermentation medium at 28°C for 3 and 5 days, respectively. Citric acid titers were determined by HPLC. (B) Morphology of A. niger S422 and S1696 (ΔctpA-F) and each Ctp reintroduction strain grown on PDA or MM agar plates at 28°C for 4 days. (C) Citric acid titers of each Ctp reintroduction A. niger strains were determined.
Effects of putative ctps on citric acid accumulation
To investigate the contribution of Ctp to citric acid production in A. niger, citric acid titers of individual ctp disruptants and S1696 (ΔctpA-F) were determined. As shown in Figure 4A, in the initial 3 days fermentation, S836 (ΔctpB), S1152 (ΔctpC), S866 (ΔctpE), and S938 (ΔctpF) strains had similar citric acid titers to the parent strain S422, whereas in S838 (ΔctpA) and S865 (ΔctpD) fermentation broth, the titers of citric acid decreased by 15.8 and 27.4%, respectively. On the 5 days of the late fermentation stage, S1152 (ΔctpC), S865 (ΔctpD), S866 (ΔctpE), and S938 (ΔctpF) strains had comparable levels of citric acid, whereas S838 (ΔctpA) and S836 (ΔctpB) exhibited a significant decrease in citric acid titers (18.3, 12.3%, respectively). To further investigate their roles in citric acid accumulation, mutants with double deletions of ctpA, ctpB, and ctpD were constructed (Supplementary Figures 3M–O). Although citric acid titers of mutants S2422 (ΔctpA, ΔctpB), S1920 (ΔctpA and ΔctpD), and S1995 (ΔctpB and ΔctpD) also declined by 16.6, 38.7 and 36.8% at 3 days, and 37.9, 21.1 and 16.9% at 5 days, respectively, S2152 (ΔctpA, ΔctpB, and ΔctpD) showed decreases of 89.9 and 91.0% on the 3 days and 5 days, respectively (Supplementary Figure 4). Similar to S2152 (ΔctpA, ΔctpB, and ΔctpD), deletions of all the six putative Ctps in S1696 resulted in no detectable levels of citric acid accumulation in the fermentation broth on the 3 days or 5 days of fermentation (Figure 4A). These results indicate that CtpA, CtpB, and CtpD are responsible for citrate export from mitochondria to cytoplasm at different fermentation stages in A. niger. In other words, CtpB may contribute in the late stage of fermentation, CtpD may primarily function in the early stage, and CtpA may play an important role throughout the entire fermentation stage. The effects of CtpA, CtpB, and CtpD on citric acid accumulation were also consistent with their expression profiles during the fermentation process (Figure 2).
To further determine whether ctpA to ctpF can restore citric acid production in S1696 (ΔctpA-F), ctpA to ctpF expression cassettes were individually introduced with their respective native promoters at the amyA locus (Supplementary Figures 3G–I) and all obtained strains were grown on PDA and MM agar plates, respectively. As shown in Figure 4B, defects of conidia formation, pigment production, and growth were partially suppressed by the reintroduction of ctpA, ctpB, or ctpD. The shake-flask fermentation test also showed that the citric acid titers of S1838 (S1696/amyA::ctpA), S1836 (S1696/amyA::ctpB), and S1842 (S1696/amyA::ctpD) recovered to the levels of 75.3, 35.4, and 72.6% at 3 days fermentation, as well as 93.8, 88.5, and 94.6% at 5 days, respectively, of the parent strain S422. These results indicate that CtpA, CtpB, and CtpD participate in citrate transport across the mitochondrial membrane in a redundant manner.
Effects of ctpA, ctpB, or ctpD reintroduction on intracellular acetyl-CoA levels
Citrate in the cytoplasm is cleaved by ATP-citrate lyase to supply acetyl-CoA for energy and biomolecule biogenesis, as well as protein acetylation. Thus, its sufficient supply is essential for normal growth and development (Chypre et al., 2012; Chen et al., 2014). Meanwhile, the addition of acetate and citrate partially recovered the conidia formation (Figure 3D) which may be due to the production of acetyl-CoA from acetate or citrate supplement. Based on the above results and prediction, the intracellular variations of acetyl-CoA with ctpA, ctpB, or ctpD reintroduction were further examined. As illustrated in Figure 5, loss of all ctps resulted in decrease in intracellular acetyl-CoA concentrations by 73.4%, whereas ctpA reintroduction into S1696 (ΔctpA-F) restored the acetyl-CoA concentrations from 3.11 nmol/mg to 8.26 nmol/mg, and ctpB or ctpD reintroduction elevated the acetyl-CoA concentrations to 7.06 nmol/mg and 8.14 nmol/mg, respectively, than that in S1696 (ΔctpA-F). These findings suggested that CtpA, CtpB, and CtpD were involved in citrate transport from mitochondria to cytoplasm and in turn affected acetyl-CoA biosynthesis. Moreover, the complete blockage of this process led to growth and development defects as displayed by strain S1696 (ΔctpA-F).
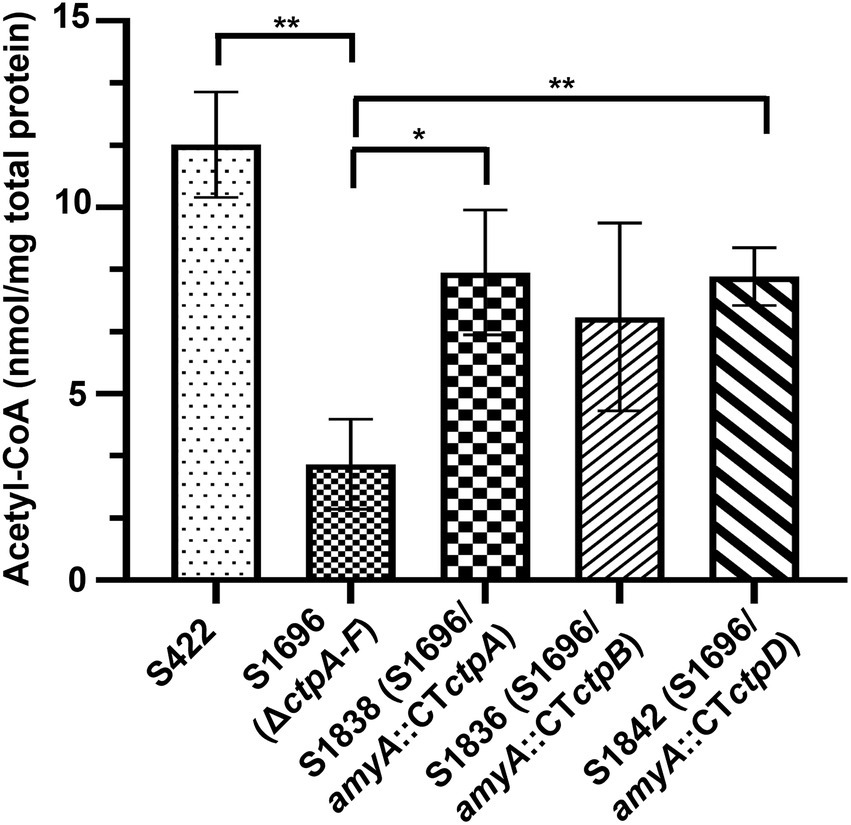
Figure 5. Intracellular acetyl-CoA concentrations of the indicated A. niger strains. About 1 × 108 conidia of each indicated A. niger strain were inoculated in 50 ml of citric acid fermentation medium at 28°C for 3 days, and the intracellular acetyl-CoA concentrations were determined. *p < 0.05, **p < 0.001, and ****p < 0.0001 were considered statistically significant. This meaning are described in “Statistical analyses” section.
Overexpression of respective ctpA, ctpB, or ctpD elevated citric acid accumulation
Given that CtpA, CtpB, and CtpD were shown to participate in citric acid transport across the mitochondrial inner membrane and affected citric acid accumulation, they were reasonable targets for engineering more efficient strains for citric acid production. Therefore, ctpA, ctpB, or ctpD overexpression cassettes driven by the constitutive promoter PgpdA were individually introduced into the parent strain S422. The overexpression transformants of ctpA, ctpB, or ctpD, namely, S1850 (S422/OEctpA), S1848 (S422/OEctpB) and S1852 (S422/OEctpD), were obtained and applied for shake-flask citrate fermentation. As shown in Figure 6, compared with S422, the respective overexpressions of ctpA, ctpB, and ctpD caused 32.8, 19.3, and 24.2% increase in citric acid accumulation at 5 days, respectively. The process of citrate export from mitochondria to cytoplasm has been predicted to be one of the bottlenecks in citrate production (Torres, 1994a,b), and enhancement of this process by the overexpression of these newly identified Ctps in the present study confirmed this prediction. Therefore, multiple Ctps are involved in citrate export from mitochondria to cytoplasm and enhancement of this process boosts extracellular citric acid accumulation.
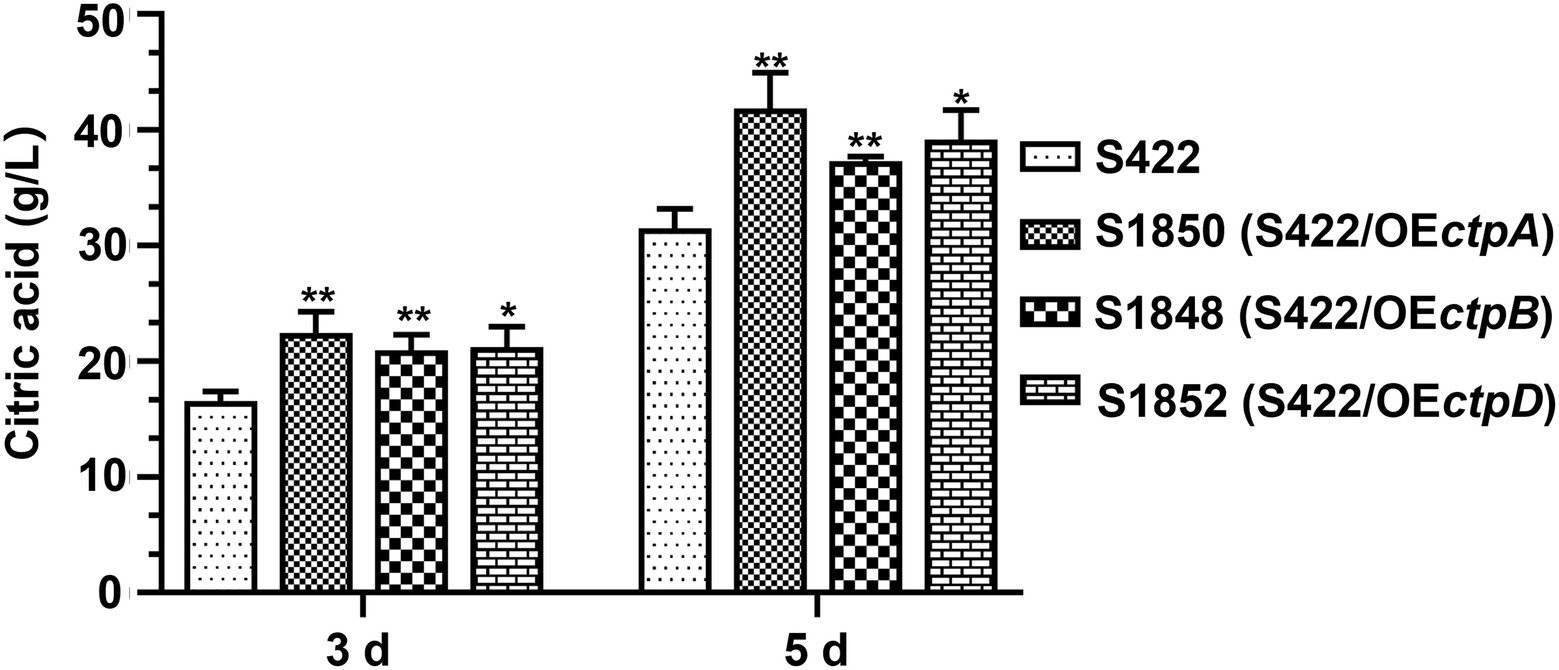
Figure 6. Citric acid production by A. niger S422 and S1848 (S422/OEctpB), S1850 (S422/OEctpA), and S1852 (S422/OEctpD). About 1 × 108 conidia of A. niger were inoculated in 50 ml of citric acid fermentation medium at 28°C for 3 and 5 days, respectively. Citric acid titers were determined by HPLC. *p < 0.05, **p < 0.001, and ****p < 0.0001 were considered statistically significant. This meaning are described in “Statistical analyses” section.
Discussion
Citric acid, primarily produced by the submerged aerobic fermentation of A. niger, is the largest consumed bulk organic acid and is extensively used in food, medicine, detergent, and cosmetics (Behera, 2020). Studies on improving the production process of citric acid fermentation are extensive. As the basis of fermentation, improvements in the citric acid-producing A. niger strain has important practical significance for promoting the development of the citric acid industry. In particular, most studies focus on engineering glycolysis and the TCA pathway for enhanced citric acid biosynthesis, elimination of inhibitor effects on citric acid biosynthesis and by-product formation, and enhancement in the supplement pathway (Yin et al., 2015). Additionally, citric acid transport processes are potential modification targets for enhancing its accumulation (Karaffa and Kubicek, 2003; Kadooka et al., 2019; Odoni et al., 2019; Steiger et al., 2019; Nakamura et al., 2020).
Here, the genes encoding mitochondrial citrate transporters responsible for citrate transport from mitochondria to cytoplasm were systematically mined and identified in A. niger genome. Six candidates, namely, CtpA to CtpF, were identified based on sequence homology with ScCtp1 from S. cerevisiae. All six Ctps exhibited similar domain-organization patterns and conserved functional amino acid residues but belong to different evolutionary branches (Figure 1C). A previous study on A. niger WU-2223l has shown that ctpB is not expressed under the conditions examined (Kirimura et al., 2016). However, under our citric acid fermentation conditions, ctpB transcripts were detected on the 5 days of fermentation (Figure 2), indicating that CtpB may be an important contributor of citric acid transport across the mitochondrial inner membrane in the late stage of citric acid fermentation. Other ctps were expressed during citric acid fermentation, suggesting that they may play different roles in citrate export in different fermentation stages. Although individual ctp disruptants exhibited similar growth and conidia formation to the parent strain, disruption of all six ctps led to growth defects, diminished pigmentogenesis, and diminished conidia formation (Figures 3A,B, 4B). Citrate supplement or introduction of respective CtpA/CtpB/CtpD also partially relieved the growth and development defects (Figures 3D, 4B). Additionally, no changes in temperature-dependent growth were observed, as previously reported in A. niger WU-2223 l and A. kawachii (data not shown; Kirimura et al., 2016; Kadooka et al., 2019). Ctps were also found to be involved in acetyl-CoA synthesis regulation (Figure 5). Ctps are considered to be responsible for the supply of cytosolic citrate to generate acetyl-CoA for cell growth and development (Mysyakina and Feofilova, 2011; Palmieri, 2013; Chen et al., 2014; Kadooka et al., 2019). The defects in growth and development in S1696 (ΔctpA-F) may be partially caused by the insufficient supply of citrate in the cytosol.
A previous study on A. niger WU-2223l found that CtpA participates in citric acid accumulation in the early-log phase of fermentation. In addition to CtpA, other Ctps were suggested to be involved in citric acid production in A. niger (Kirimura et al., 2016). In A. kawachii, two mitochondrial citrate transporters, AkCtpA (AKAW_03754) and AkYhmA (AKAW_06280), were shown to be involved in citric acid accumulation and required for acetyl-CoA biosynthesis (Kadooka et al., 2019). In the present study, disruption of individual ctpA, ctpB, or ctpD led to significantly decreased accumulation of extracellular citric acid (Figure 4A), and their respective reintroduction to S1696 (ΔctpA-F) partially restored the citric acid accumulation (Figure 4C). Importantly, overexpression of individual ctpA, ctpB, or ctpD under the constitutive promoter PgpdA caused significant increases in citric acid titers (Figure 6). These findings confirmed that CtpA, CtpB, and CtpD are major contributors to citrate transport in A. niger and function in a redundant manner. Although through blastp alignment program, one sequence ASPNIDRAFT_136079 (AnYhmA) was found to be a homolog of AkYhmA in A. niger, CtpA and AnYhmA exhibited only 18.87% sequence identity. Based on the results that citric acid accumulation was almost abolished when ctpA, ctpB, and ctpD were co-deleted (Supplementary Figure 4), AnYhmA might not participate in citrate transport across mitochondria in A. niger S422. Mitochondrial citrate transporters are considered to participate in citrate export, and Ctps such as ScCtp1, RnCtp, and AkCtpA were previously shown to exhibit different substrate specificities by reconstitution using liposomes. However, the molecular details of Ctps in A. niger are not completely understood and require further studies. Notably, although CtpE and CtpF share an evolutionary branch with the previously reported AkCtpA and AkYhmA, our data failed to support their independent involvement in extracelluar citric acid accumulation, similar to the case of CtpC. Further studies are needed to clarify the gene functions of ctpC, ctpE, and ctpF.
In summary, three mitochondrial citrate transporters, namely, CtpA, CtpB, and CtpD, were identified to be involved in citrate export from mitochondria to cytoplasm, thus contributing to extracellular citric acid accumulation. Additionally, given that citrate in the cytoplasm was the main substrate for acetyl-CoA synthesis, it is no surprising that CtpA, CtpB, and CtpD play important roles in strain growth, pigmentogenesis, and conidia formation. All these findings increased the understanding of the mechanisms of citrate transport and extracellular accumulation, thus providing fundamental knowledge for engineering more efficient commercial strains of A. niger for citrate production.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
WC analyzed the data and wrote the manuscript. LZ and LW generated all genetic constructs. MZ and JL performed the shake-flask fermentation experiments. JL and ZX contributed to scientific discussions and commented on the manuscript. HL supervised the work and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Key Research and Development Program of China (2021YFC2100700) and the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-006).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1009491/full#supplementary-material
Footnotes
1. ^https://mycocosm.jgi.doe.gov/pages/blast-query.jsf?db=Aspni7
2. ^http://smart.embl-heidelberg.de/
3. ^http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index
References
Behera, B. C. (2020). Citric acid from Aspergillus Niger: a comprehensive overview. Crit. Rev. Microbiol. 46, 727–749. doi: 10.1080/1040841X.2020.1828815
Bisaccia, F., De Palma, A., and Palmieri, F. (1989). Identification and purification of the tricarboxylate carrier from rat liver mitochondria. Biochim. Biophys. Acta (BBA)-Bioenergetics 977, 171–176. doi: 10.1016/S0005-2728(89)80068-4
Cao, W., Yan, L., Li, M., Liu, X., Xu, Y., Xie, Z., et al. (2020). Identification and engineering a C4-dicarboxylate transporter for improvement of malic acid production in Aspergillus Niger. Appl. Microbiol. Biotechnol. 104, 9773–9783. doi: 10.1007/s00253-020-10932-1
Chen, H., He, X., Geng, H., and Liu, H. (2014). Physiological characterization of ATP-citrate lyase in Aspergillus Niger. J. Ind. Microbiol. Biotechnol. 41, 721–731. doi: 10.1007/s10295-014-1418-3
Chypre, M., Zaidi, N., and Smans, K. (2012). ATP-citrate lyase: a mini-review. Biochem. Biophys. Res. Commun. 422, 1–4. doi: 10.1016/j.bbrc.2012.04.144
Gnoni, G., Priore, P., Geelen, M. J., and Siculella, L. (2009). The mitochondrial citrate carrier: metabolic role and regulation of its activity and expression. IUBMB Life 61, 987–994. doi: 10.1002/iub.249
Kadooka, C., Izumitsu, K., Onoue, M., Okutsu, K., Yoshizaki, Y., Takamine, K., et al. (2019). Mitochondrial citrate transporters CtpA and YhmA are required for extracellular citric acid accumulation and contribute to cytosolic acetyl coenzyme A generation in Aspergillus luchuensis. Appl. Environ. Microbiol. 85, e03136–e03118. doi: 10.1128/AEM.03136-18
Kaplan, R. S., Mayor, J., Gremse, D., and Wood, D. O. (1995). High level expression and characterization of the mitochondrial citrate transport protein from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270, 4108–4114. doi: 10.1074/JBC.270.8.4108
Kaplan, R. S., Mayor, J. A., Kakhniashvili, D., Gremse, D. A., Wood, D. O., and Nelson, D. R. (1996). Deletion of the nuclear gene encoding the mitochondrial citrate transport protein from Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 226, 657–662. doi: 10.1006/bbrc.1996.1411
Karaffa, L., and Kubicek, C. P. (2003). Aspergillus Niger citric acid accumulation: do we understand this well working black box? Appl. Microbiol. Biotechnol. 61, 189–196. doi: 10.1007/s00253-002-1201-7
Kirimura, K., Kobayashi, K., Ueda, Y., and Hattori, T. (2016). Phenotypes of gene disruptants in relation to a putative mitochondrial malate–citrate shuttle protein in citric acid-producing Aspergillus Niger. Biosci. Biotechnol. Biochem. 80, 1737–1746. doi: 10.1080/09168451.2016.1164583
Kirimura, K., Kobayashi, K., and Yoshioka, I. (2019). Decrease of citric acid produced by Aspergillus Niger through disruption of the gene encoding a putative mitochondrial citrate-oxoglutarate shuttle protein. Biosci. Biotechnol. Biochem. 83, 1538–1546. doi: 10.1080/09168451.2019.1574205
Mores, S., Vandenberghe, L. P. D. S., Magalhães Júnior, A. I., De Carvalho, J. C., De Mello, A. F. M., Pandey, A., et al. (2021). Citric acid bioproduction and downstream processing: status, opportunities, and challenges. Bioresour. Technol. 320:124426. doi: 10.1016/j.biortech.2020.124426
Mysyakina, I. S., and Feofilova, E. P. (2011). The role of lipids in the morphogenetic processes of mycelial fungi. Microbiology 80, 297–306. doi: 10.1134/S0026261711030155
Nakamura, E., Kadooka, C., Okutsu, K., Yoshizaki, Y., Takamine, K., Goto, M., et al. (2020). Citrate exporter enhances both extracellular and intracellular citric acid accumulation in the koji fungi Aspergillus luchuensis Mut. Kawachii and Aspergillus oryzae. J. Biosci. Bioeng. 131, 68–76. doi: 10.1016/j.jbiosc.2020.09.002
Odoni, D. I., Vázquez-Vilar, M., van Gaal, M. P., Schonewille, T., dos Santos, V. M. D. M., Tamayo-Ramos, J. A., et al. (2019). Aspergillus Niger citrate exporter revealed by comparison of two alternative citrate producing conditions. FEMS Microbiol. Lett. 366:fnz071. doi: 10.1093/femsle/fnz071
Palmieri, F. (1994). Mitochondrial carrier proteins. FEBS Lett. 346, 48–54. doi: 10.1016/0014-5793(94)00329-7
Palmieri, F. (2013). The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol. Asp. Med. 34, 465–484. doi: 10.1016/j.mam.2012.05.005
Palmieri, F., Stipani, I., Quagliariello, E., and Klingenberg, M. (1972). Kinetic study of the tricarboxylate carrier in rat liver mitochondria. FEBS J. 26, 587–594. doi: 10.1111/j.1432-1033.1972.tb01801.x
Ruprecht, J. J., and Kunji, E. R. S. (2020). The SLC25 mitochondrial carrier family: structure and mechanism. Trends Biochem. Sci. 45, 244–258. doi: 10.1016/j.tibs.2019.11.001
Sloothaak, J., Odoni, D. I., de Graaff, L. H., Martins dos Santos, V. A. P., Schaap, P. J., and Tamayo-Ramos, J. A. (2015). Aspergillus Niger membrane-associated proteome analysis for the identification of glucose transporters. Biotechnol. Biofuels 8:150. doi: 10.1186/s13068-015-0317-9
Soccol, C. R., Vandenberghe, L. P. S., Rodrigues, C., and Pandey, A. (2006). New perspectives for citric acid production and application. Food Technol. Biotechnol. 44, 141–149.
Steiger, M. G., Rassinger, A., Mattanovich, D., and Sauer, M. (2019). Engineering of the citrate exporter protein enables high citric acid production in Aspergillus Niger. Metab. Eng. 52, 224–231. doi: 10.1016/j.ymben.2018.12.004
Tong, Z., Zheng, X., Tong, Y., Shi, Y.-C., and Sun, J. (2019). Systems metabolic engineering for citric acid production by Aspergillus Niger in the post-genomic era. Microb. Cell Factories 18:28. doi: 10.1186/s12934-019-1064-6
Torres, N. V. (1994a). Modeling approach to control of carbohydrate metabolism during citric acid accumulation by Aspergillus Niger: I. model definition and stability of the steady state. Biotechnol. Bioeng. 44, 104–111. doi: 10.1002/bit.260440115
Torres, N. V. (1994b). Modeling approach to control of carbohydrate metabolism during citric acid accumulation by Aspergillus Niger: II. Sensitivity Analysis. Biotechnol. Bioeng. 44, 112–118. doi: 10.1002/bit.260440116
Torres, N. V., Riol-Cimas, J. M., Wolschek, M., and Kubicek, C. P. (1996). Glucose transport by Aspergillus Niger: the low-affinity carrier is only formed during growth on high glucose concentrations. Appl. Microbiol. Biotechnol. 44, 790–794. doi: 10.1007/BF00178620
Xu, Y., Shan, L., Zhou, Y., Xie, Z., Ball, A. S., Cao, W., et al. (2019). Development of a Cre-loxP-based genetic system in Aspergillus Niger ATCC 1015 and its application to construction of efficient organic acid-producing cell factories. Appl. Microbiol. Biotechnol. 103, 8105–8114. doi: 10.1007/s00253-019-10054-3
Xu, Y., Zhou, Y., Cao, W., and Liu, H. (2020). Improved production of malic acid in Aspergillus Niger by abolishing citric acid accumulation and enhancing glycolytic flux. ACS Synth. Biol. 9, 1418–1425. doi: 10.1021/acssynbio.0c00096
Yang, L., Linde, T., Hossain, A. H., Lübeck, M., Punt, P. J., and Lübeck, P. S. (2019). Disruption of a putative mitochondrial oxaloacetate shuttle protein in Aspergillus carbonarius results in secretion of malic acid at the expense of citric acid production. BMC Biotechnol. 19:72. doi: 10.1186/s12896-019-0572-0
Yin, X., Li, J., Shin, H., Du, G., Liu, L., and Chen, J. (2015). Metabolic engineering in the biotechnological production of organic acids in the tricarboxylic acid cycle of microorganisms: advances and prospects. Biotechnol. Adv. 33, 830–841. doi: 10.1016/j.biotechadv.2015.04.006
Keywords: citric acid, mitochondrial citrate transporter, transport, genetic characterization, Aspergillus niger
Citation: Cao W, Zhang L, Wu L, Zhang M, Liu J, Xie Z and Liu H (2022) Identification and genetic characterization of mitochondrial citrate transporters in Aspergillus niger. Front. Microbiol. 13:1009491. doi: 10.3389/fmicb.2022.1009491
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaReviewed by:
Ping Song, Nanjing Normal University, ChinaHu-Hu Liu, Hunan Agricultural University, ChinaCopyright © 2022 Cao, Zhang, Wu, Zhang, Liu, Xie and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Liu, bGl1aGFvQHR1c3QuZWR1LmNu
 Wei Cao
Wei Cao Licheng Zhang
Licheng Zhang Liu Wu
Liu Wu Mingyi Zhang
Mingyi Zhang Jiao Liu
Jiao Liu Zhoujie Xie
Zhoujie Xie Hao Liu
Hao Liu