- 1College of Forestry, Shenyang Agriculture University, Shenyang, China
- 2State Key Laboratory of Tree Genetics and Breeding, Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China
- 3Research Station of Liaohe-River Plain Forest Ecosystem, Chinese Forest Ecosystem Research Network (CFERN), Shenyang Agricultural University, Tieling, China
- 4Key Laboratory of Tree Breeding and Cultivation of State Forestry Administration, Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China
Decomposition of forest litter is an essential process for returning nutrients to the soil, which is crucial for preserving soil fertility and fostering the regular biological cycle and nutrient balance of the forest ecosystem. About 70% of the land-based forest litter is made up primarily of leaf litter. However, research on the complex effects and key determinants of leaf litter decomposition is still lacking. In this study, we examined the characteristics of nutrient release and microbial diversity structure during the decomposition of three types of litter in arid and semi-arid regions using 16S rRNA and ITS sequencing technology as well as nutrient content determination. It was revealed that the nutrient content and rate of decomposition of mixed litters were significantly different from those of single species. Following litter mixing, the richness and diversity of the microbial community on leaves significantly increased. It was determined that there was a significant correlation between bacterial diversity and content (Total N, Total P, N/P, and C/P). This study provided a theoretical framework for investigating the decomposition mechanism of mixed litters by revealing the microbial mechanism of mixed decomposition of litters from the microbial community and nutrient levels.
Introduction
Litter, also referred to as organic debris, is a general term for all organic matter in the ecosystem created by the withering of above-ground plant components and returned to the surface as a source of material and energy for decomposers to maintain ecosystem functions (Schlesinger and Lichter, 2001). Leaf litter makes up about 70% of the above-ground forest litter among them. Leaf litter currently dominates 72% of the research on litter in forest ecosystems (Jia B. R. et al., 2018; Wymore et al., 2018; Nicolas et al., 2019). The primary source of organic matter and nutrients in forest soils is litter decomposition, which also plays a crucial role in the energy flow and nutrient cycling in forest ecosystems (Swift et al., 1979). It also acts as a link between the two carbon pools found in soil and living things (Manzoni et al., 2010). It is the catalyst for biogeochemical cycles and significantly affects climate change (Nevins et al., 2018). The decomposition of leaf litter is a crucial indicator of the functioning of an ecosystem’s health because it can return plant carbon, nitrogen and phosphorus to the soil and the atmosphere, as well as other nutrients. This can have an impact on the global element balance, particularly the carbon balance (Cotrufo et al., 2015). Statistics show that every year, 68 GtC of carbon dioxide is released into the atmosphere by the decomposition of litter, making up roughly 70% of the annual carbon flux worldwide (Raich and Schlesinger, 1992). It also serves as a rich source of nutrients for microorganisms, and is crucial for maintaining soil fertility, promoting the regular biological cycle, and maintaining the nutrient balance in forest ecosystems (Manzoni et al., 2008; Jiang et al., 2013), all of which have long been of great concerned to researchers (Gui et al., 2017; Tan et al., 2020).
The nature of the litter, the composition and diversity of the decomposer community, soil conditions, and the climate all have an impact on how quickly litter decomposes (Handa et al., 2014; Stefano, 2017; Han et al., 2019; Sébastien et al., 2021). Climate has a significant impact on the rate at which litter decomposes both globally and locally. On a smaller scale, the type of the litter itself has the greatest impact on how quickly it decomposes. Litter leaf quality largely determines the decomposition of litter and the release of nutrients and minerals to the soil (Lucas-Borja et al., 2018), especially the carbon-nitrogen ratio of the litter (Bradford et al., 2016). The quality of this litter substrate will reduce its decomposition rate (Xiao et al., 2019), and higher C/N and C/P will inhibit the decomposition of leaf litter and its release of nutrients (Lucas-Borja et al., 2018). Coniferous litter typically has a higher C/N than broad-leaved litter. In view of this, understanding the nutrient cycle of leaf litter depends on understanding the stoichiometric characteristics of different shelterbelt species’ leaf litter.
The qualities of leaf litter (N and C/N) vary greatly between different plants, and the properties of leaf litter and the chemical structure of organic carbon are significant factors affecting the decomposition of the litter (Cotrufo et al., 2015). The primary source of leaf litter decomposition and the primary force driving the decomposition occurs when plant leaves fall to the ground (Davidson and Janssens, 2006; Cornelissen et al., 2007). The quality of the litter and its specialized microorganisms work together to cause litter to decompose. Uncertainty persists regarding the microbial community changes that occur as various plant leaf litter decomposes.
Leaf microbes are microorganisms that live as epiphytes or parasites on the leaves of plants (Jia T. et al., 2018; Yao et al., 2019). Phyllosphere microbial communities have different makeups depending on the type of plant and the environment it lives in. Although different species’ phyllosphere microbial communities differ significantly from one another (Bao et al., 2019), they all share a similar structure (Wallace et al., 2018). As plants progressively wither, variability of phyllosphere microbes gradually increases (Ferreira et al., 2016), and changes in phyllosphere microbial communities during decomposition (Whipps et al., 2010; Kembel et al., 2014) are influenced by leaves’ strong influence on physicochemical properties. The abundance of microbial communities significantly increased along with the degree of physical and biological fragmentation of litter (Guerreiro et al., 2017). Fungi are thought to play a significant role in the biogeochemical cycles and the decomposition of leaf litter (Osono et al., 2004). Saprophytic basidiomycetes are the primary decomposers of litter and have the ability to break down materials that are difficult to decompose, like the cellulose found in woody litter (Eichlerová et al., 2015; Urbanová et al., 2015; Baldrian, 2017). Bacteria, which can break down cellulose and hemicellulose, are also necessary for the decomposition of leaf litter. It is not yet clear, though, whether the succession of fungi during the decomposition of litter is consistent with bacterial succession. Additionally, and There is no information on the dynamic changes in phyllosphere microbial composition over time involving multiple decomposition stages, and most studies only concentrate on one stage of plant leaf decomposition or associated leaf litter surface microbes. A though understanding of the connection between the microbial community and litter decomposition over time can be gained by studying the phyllosphere’s microbial community.
Early research concentrated on the decomposition of a single leaf litter, but the majority of leaf litter in forest ecosystems was mixed, which had an impact on the nutrient release law during the decomposition process (Lu et al., 2020). Coniferous and broad-leaved litter’s physicochemical differences can have an impact on the structure and operation of the litter microbial community (Lin et al., 2021). In general, mixed litter has higher fungal and bacterial abundances and microbial community diversity than single litter, and they have significantly different microbial community compositions (Sun et al., 2018; Pereira et al., 2019; Zhang et al., 2019). This could cause variations in the metabolic functions of microbial communities (Li et al., 2015).
Microorganisms are the primary actors in the decomposition of litter, according to previous research (Muscolo et al., 2014; Lin et al., 2015; Scheibe et al., 2015), and they decompose more quickly in the decomposition of mixed litter than in the decomposition of asingle litter (Patoine et al., 2017; Tonin et al., 2017). According to some studies, the structure and operation of microbial communities can be affected in specific ways by the mixing of litter (Wang et al., 2019). This is due to the mixed litter’s higher levels of carbon sources and nutrients, which increase the nutrient supply to microorganisms and accelerate decomposition through complementary effects (Cline and Zak, 2015; Xiao et al., 2019). Evidently, there is currently a dearth of knowledge regarding the impact of microbial composition on litter decomposition. In addition, research on the structure of the microbial community involved in the decomposition of litter focuses less on that process and instead primarily on the structure of the microbial community in the soil (Sun et al., 2017). Therefore, it is useful to understand the mechanism of litter decomposition to study the changes in microbial community composition during decomposition.
“Three-North” Shelter Forest Program was launched by the Chinese government in 1978 in Northwest China, North China and Northeast China (collectively referred to as the “Three-North” Regions) in an effort to improve the ecological environment. Important tree species for afforestation in the “Three-North” Shelter Forest Program include Populus × canadensis Moench and Pinus sylvestris var. mongolica, both of which have straight trunks, a love of light, and a resistance to cold. In order to study the mixed decomposition characteristics of litter, Populus × canadensis Moench and Pinus sylvestris var. mongolica plantation litter in arid and semi-arid regions was chosen as the research object. The following hypotheses were put forth: (1) mixed litter decomposition altered the physicochemical characteristics of leaves in comparison to pure Pinus sylvestris var. mongolica forests; (2) conifer litter’s microbial community structure and diversity were affected by the addition of Populus × canadensis Moench litter; (3) the decomposition of Pinus sylvestris var. mongolica litter is accelerated by a mixture of withered leaves from coniferous and broad-leaved species. To provide a theoretical foundation for the decomposition of leaf litter, it’s necessary to reveal the interactions in mixed decomposition of Populus × canadensis Moench and Pinus sylvestris var. mongolica.
Materials and methods
Site description
The study site was established in the Liaohe Plain Forest Ecological Station of the National Forestry and Grassland Administration which is Fujia Forest Farm in Changtu County, Tieling City, Liaoning Province (43°21′143″–42°53′623″N, 123°53′623″–123°53′623″E). It is located in the southeastern part of Horqin Sandy Land, at the junction of Liaoning Province, Jilin Province, and Inner Mongolia Autonomous Region. The topography is flat, and there are a small number of sand dunes in the Liaohe throttle. It is a temperate semi-humid and semi-arid continental climate with an average annual precipitation of 400–550 mm, mainly concentrated in July and August, and evaporates 1,843 mm per year. The maximum temperature was 35.6°C, the lowest temperature was −31.5°C, and the average daily temperature was 6.3°C. The soil type contains low levels of organic substances and other nutrients.
Litter sampling
A litter collector of 1 m × 1 m was set up at the study site, and fresh litter samples of Populus × canadensis Moench and Pinus sylvestris var. mongolica were collected in October 2020. Litter samples were then mixed and stored in ice boxes for immediate shipment back to the lab. The leaves of Populus × canadensis Moench and Pinus sylvestris var. mongolica were air-dried and put into drop bags. The collected litter was packed into a nylon mesh decomposition bag with a size of 20 cm × 20 cm (the aperture was 1 mm × 1 mm, i.e., 16 mesh). The total weight of each bag was 8 g, and the mass ratios of litter leaves of Populus × canadensis Moench and Pinus sylvestris var. mongolica were 1:0, 1:1, and 0:1 (YS, YZ, and ZS in subsequent analysis). The litter bags were kept in a dehumidified, air-conditioned room. A decomposition test was set up under a Pinus sylvestris var. mongolica stand in April 2021. Eight litter decomposition bags of different proportions were set in each plot, and a total of four plots were repeated. In July 2021, bags’ surface waste was carefully removed, the litter bag retrieved and stored in an ice box for immediate transport back to the laboratory. Then 8 litter bags from the same plot were mixed, weighed and divided into 2 parts. A piece of the litter was dried at 65°C to a constant weight, then weighed to calculate its moisture content and total dry weight. The dried litter samples were crushed and pulverized through a 0.15 mm sieve (100 mesh). The chemical properties of dried litter samples were determined after crushing, including total carbon (total C), total nitrogen (total N), and total phosphorus (total P). The other part was put into a sterilization bag and stored in a − 80°C refrigerator for the determination of microbial diversity.
Determination of litter characteristics
An elemental analyzer (Elementar Vario EL III, Germany) was used to measure total C and total N in litter (Song et al., 2015). The total P in litter was determined by the molybdenum-antimony anti-spectrophotometric method (Ba et al., 2020). The weight loss rate of litter (Dw) was determined by the drying method.
In the formula: Dw is the weight loss rate of litter (%), Mo is the dry weight (g) of the sample in the decomposition bag at the time of delivery, and Mt. is dry weight (g) of litter after sample decomposition.
DNA extraction and amplification sequencing
Total DNA extraction was carried out according to the MoBio PowerSoil DNA Isolation Kit (MP Biomedicals, Santa Ana, CA, United States) Kit procedure of OMEGA, and each sample was weighed at about 0.5 g. A NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) determined the quantity and quality of the extracted DNA. Primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify the V3-V4 region of the bacterial 16S rRNA gene (Claesson et al., 2009). The fungal ITS region was amplified with primers ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′; White, 1994). The PCR amplification system has a total of 25 μl, including: DNA template 2 μl, each 1 μl upstream and downstream primers (10 μmol·L−1), buffer 5 μl, Q5 high-fidelity buffer 5 μl, 0.25 μl high-fidelity DNA polymerase (5 U·μl−1), dNTP (2.5 mmol·L−1) 2 μl, ultrapure water (dd H2O) 8.75 μl. PCR amplification conditions were first pre-denatured at 98°C for 2 min, then repeated 25 times in a cycle of 98°C for 15 s, 55°C for 30 s, and 72°C for 30 s, and finally extended at 72°C for 5 min. The PCR amplicons were purified by using Agincourt AM-Pure Beads (Beckman Coulter, Indianapolis, IN), and quantified by using the Pico Green dsDNA detection kit (Invitrogen, Carlsbad, CA, United States). The PCR products were sequenced by using the Illumina NovaSeq 6000 sequencing platform by Shanghai Personal Biotechnology Co., Ltd.
Microbial bioinformatics analysis
QIIME2 (2019.4) and UPARSE Pipeline were used to carry out correlation calculation and analysis on the original data sequences of high-throughput sequencing, and the sequence length was screened (Caporaso et al., 2010). QIIME2’s UCLUST, a sequence alignment tool, was used to merge the effective sequences with more than 97% similarity to OTU, and the sequence with the highest abundance of each OTU was selected as the representative sequence of OTU (Edgar, 2010), and compared with the template sequence of Silva database (Release123)1 to obtain the classification information of each OTU (Quast et al., 2012). Then, according to the number of sequences contained in each sample, a matrix file of OTU abundance in each sample is constructed, namely, OTU table. The richness indices of OTUs and diversity index, including Shannon index, Simpson index, and Chao1 index, were analyzed.
Statistical analysis
For data processing, Microsoft Excel (2019) was used, and the data in the table were all repeated Average ± Standard Error. SPSS 26.0 was used for statistical analysis. One-way analysis of variance was used to analyze the differences in physicochemical properties of different litter leaves, and two-way correlation analysis was used to compare the relationship between physicochemical properties of litter leaves and microbial diversity. Ecologists use alpha diversity and beta diversity indices to characterize the diversity of species within and between habitats, respectively, to comprehensively evaluate their overall diversity (Whittaker, 1960; Whittaker, 1972). In order to comprehensively assess the alpha diversity of microbial communities, Chao1 (Chao, 1984) and Observed species indices were used to characterize richness, and Shannon (Chiani, 1948; Shannon, 1948) and Simpson (Simpson, 1949) indices were used to characterize diversity. Evenness was characterized by Pielou’s evenness index (Pielou, 1966), and coverage by Good’s coverage index (Good, 1953). And the ggplot2 package of R (R v.3.4.4) was used to draw a boxplot. Principal coordinates analysis (PCoA) is one of the most classic unconstrained sorting (Classical Multidimensional Scaling, cMDScale) analysis methods (Alban, 2010). The differences in the β-diversity of litter leaves were analyzed and compared according to the OTU table and the ape package of R (R v.3.4.4). Among the samples, shared and unique OTUs of litter microbial communities were used in R (R v.3.4.4) and the “Venn Diagram” package to create Venn diagrams. Heatmaps for the top 50 taxonomic genera in each sample were constructed using R (R v.3.4.4) and the pheatmap package. The linear discriminant analysis (LDA) effect size (LEfSe) method was used to detect potentially biomarker-rich taxa based on a cross-group normalized relative abundance matrix using default parameters. The CoHeatmap analysis was carried out by the genescloud tool1 utilizing the Spearman rank correlation coefficient algorithm, R = 0.5. The matrix was constructed using Galaxy, an online interactive analysis of microbial community data.
Results
Characterization of physicochemical properties of different species of litter
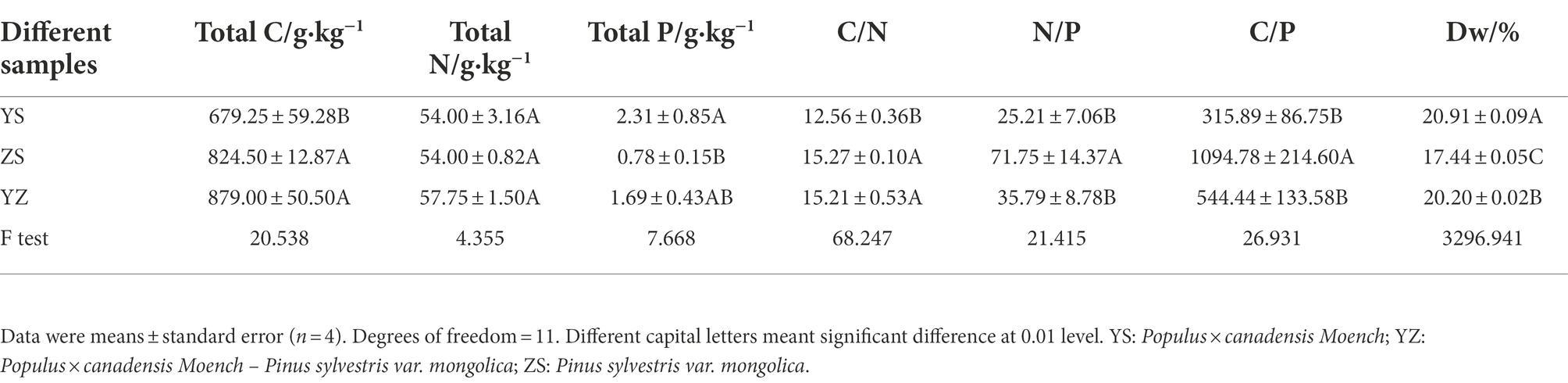
There were extremely significant differences in Dw (p < 0.01, F = 3296.941), total C (p < 0.01, F = 20.538), total P (p < 0.05, F = 7.668), C/N (p < 0.01, F = 68.247), N/P (p < 0.01, F = 21.415), and C/P (p < 0.01, F = 26.931) among the three types of leaf litter, but there was no significant difference in the content of total N (p > 0.05, F = 4.355), as shown in Table 1. The total C content of YS was 679.25 g·kg−1, which was significantly lower than that of YZ and ZS, while the total P content (2.31 g·kg−1) was significantly higher than that of YZ and ZS (p < 0.01). The total C, total N and total P contents of YZ were 879.00 g·kg−1, 57.75 g·kg−1 and 1.69 g·kg−1, respectively, which were higher than those of ZS. The significance rules of N/P and C/P of the three types of litter were consistent, and ZS was extremely significantly higher than YS and YZ (p < 0.01). The C/N of YZ was significantly different from that of YS (p < 0.01), but not significantly different from that of ZS (p > 0.05). Dw intuitively represents the rate at which litter decomposes. As can be seen from Table 1, YS had the highest decomposition rate, followed by YZ, and ZS was the lowest.
Characteristics of microbial community composition In different types of litter leaves
Venn plots were used to compare OTUs common to and unique to bacterial and fungal communities across all samples. At the bacterial level, a total of 24,192 OTUs were detected. Among them, the OTUs of YS, YZ and ZS were 10,311, 11,586, and 7,876, respectively. The total OTU of the three samples was 817. The OTUs specific to YS, YZ and ZS were 6,075, 6,999, and 6,372, respectively (Figure 1A). At the fungal level, however, the number of detected OTUs was only 1,314. Among them, the OTUs of YS, YZ and ZS are 608, 750, and 561. The common OTU among them was 152. The OTUs specific to YS, YZ, and ZS were 253, 332, and 276, respectively (Figure 1B). Principal coordinates analysis (PCoA) based on Bray-Curtis distance analysis of the different samples showed that the composition of bacterial and fungal communities between YS, YZ, and ZS were all different, forming three disjoint confidence ellipses, and all split along the PCo1 axis (Figure 2).
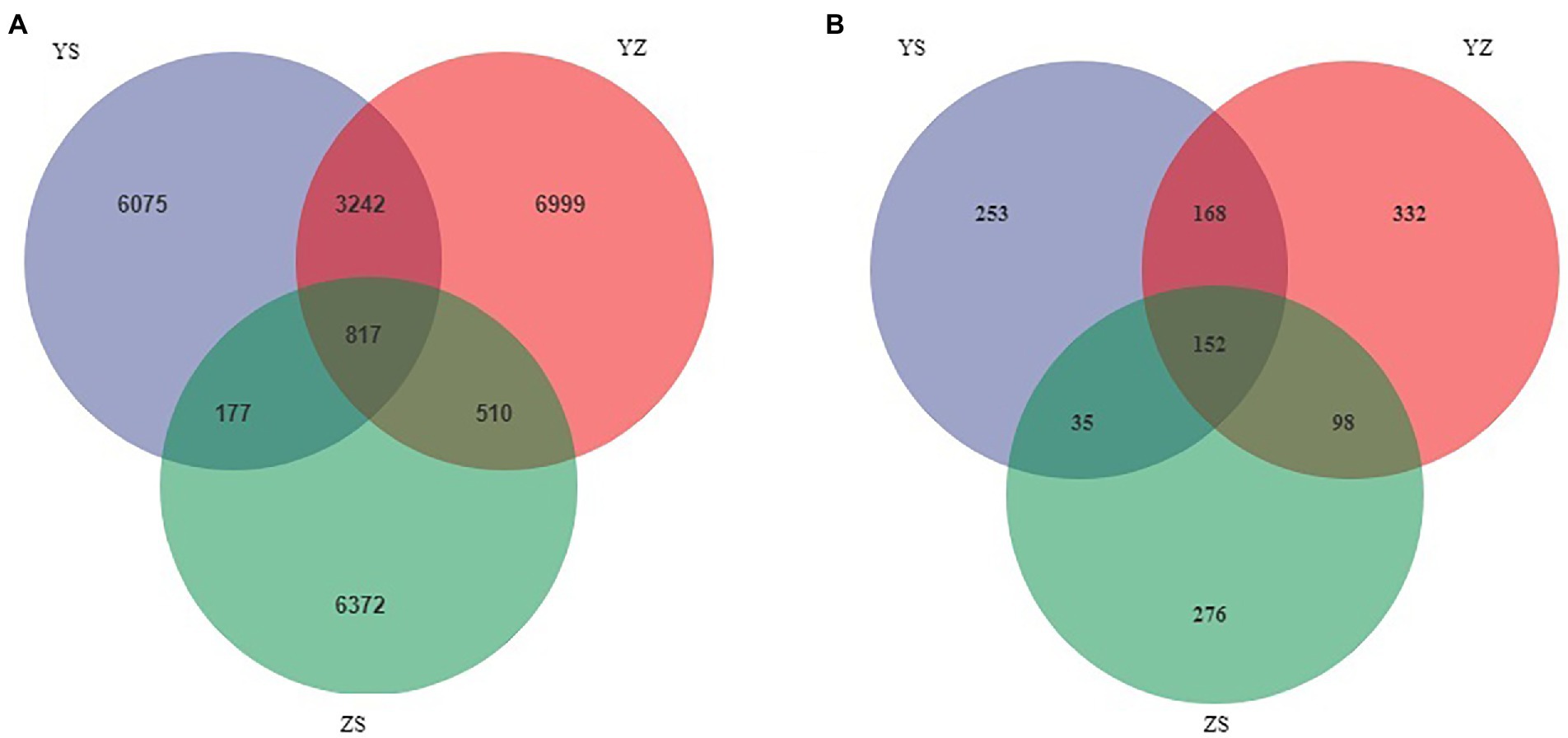
Figure 1. Venn diagrams showed the unique and shared OTU of litter microorganisms from three different samples. (A) unique and shared OTUs of leaf litter bacteria in three different samples; (B) unique and shared OTUs of leaf litter fungal in three different samples. YS: Populus × canadensis Moench; YZ: Populus × canadensis Moench-Pinus sylvestris var. mongolica; ZS: Pinus sylvestris var. mongolica.
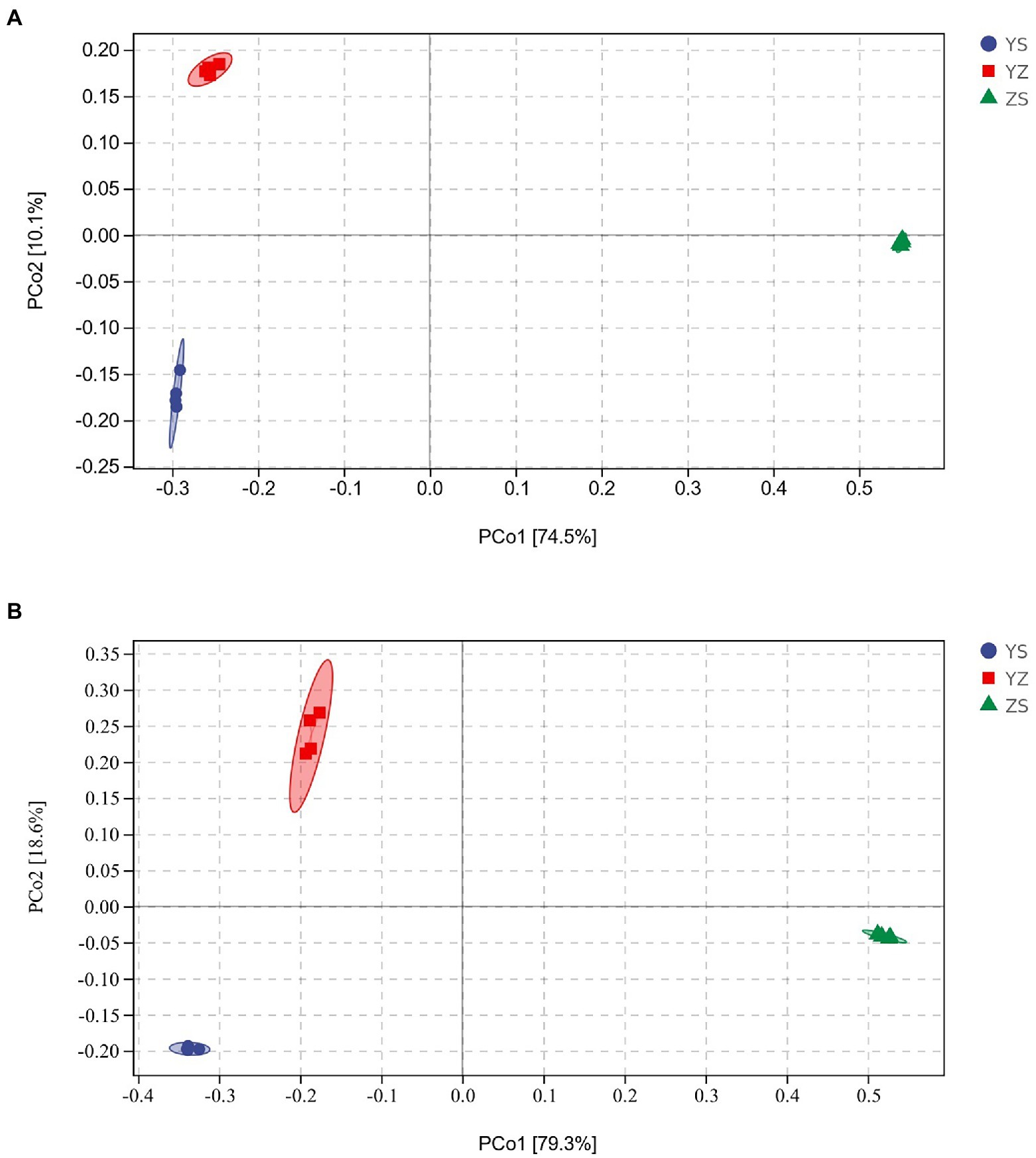
Figure 2. Priniciple coordinate analysis (PCoA) of Bray-Curtis’ diatance from all samples. (A) PCoA of bacteria communities in different samples; (B) PCoA of fungal communities in different samples. YS: Populus × canadensis Moench; YZ: Populus × canadensis Moench-Pinus sylvestris var. mongolica; ZS: Pinus sylvestris var. mongolica.
Alpha diversity refers to an index in ecology to estimate the richness, diversity and evenness of species and flora. Regarding the bacterial alpha-diversity, there was a distinction between the alpha diversity of the three litter types, including Chao1 index (F = 20.99; p = 0.0073), Goods_coverage (F = 22.77; p = 0.012), Shannon index (F = 25.98; p = 0.0073), Simpson index (F = 83.11; p = 0.0073), Pielou_e index (F = 43.12; p = 0.0073) and Observed_species (F = 14.18; p = 0.012). The six indexes of YZ were significantly different from those of ZS (p < 0.01), but the indexes of YZ and YS were not significantly different (p > 0.05). From Figure 3A, it could be seen intuitively that the Chao1 index, Pielou_e index, Shannon index, Simpson index and Observed_species index of YZ are the highest, followed by YS and ZS the lowest. And the Goods_coverage index showed the opposite law, namely ZS > YS > YZ. At the fungal level, only YS and YZ had significant differences between Pielou_e index (F = 97.84; p = 0.0073), Shannon index (F = 126.27; p = 0.0073), and Simpson index (F = 118.38; p = 0.0073), and there were no significant differences between the other indicators and samples (p > 0.05). The same as bacteria, YZ had the highest Chao1 index, Pielou_e index, Shannon index, Simpson index, and Observed_species index. In the above index, except that the Chao1 index of YS was higher than ZS, the rest were ZS higher than YS (Figure 3B).
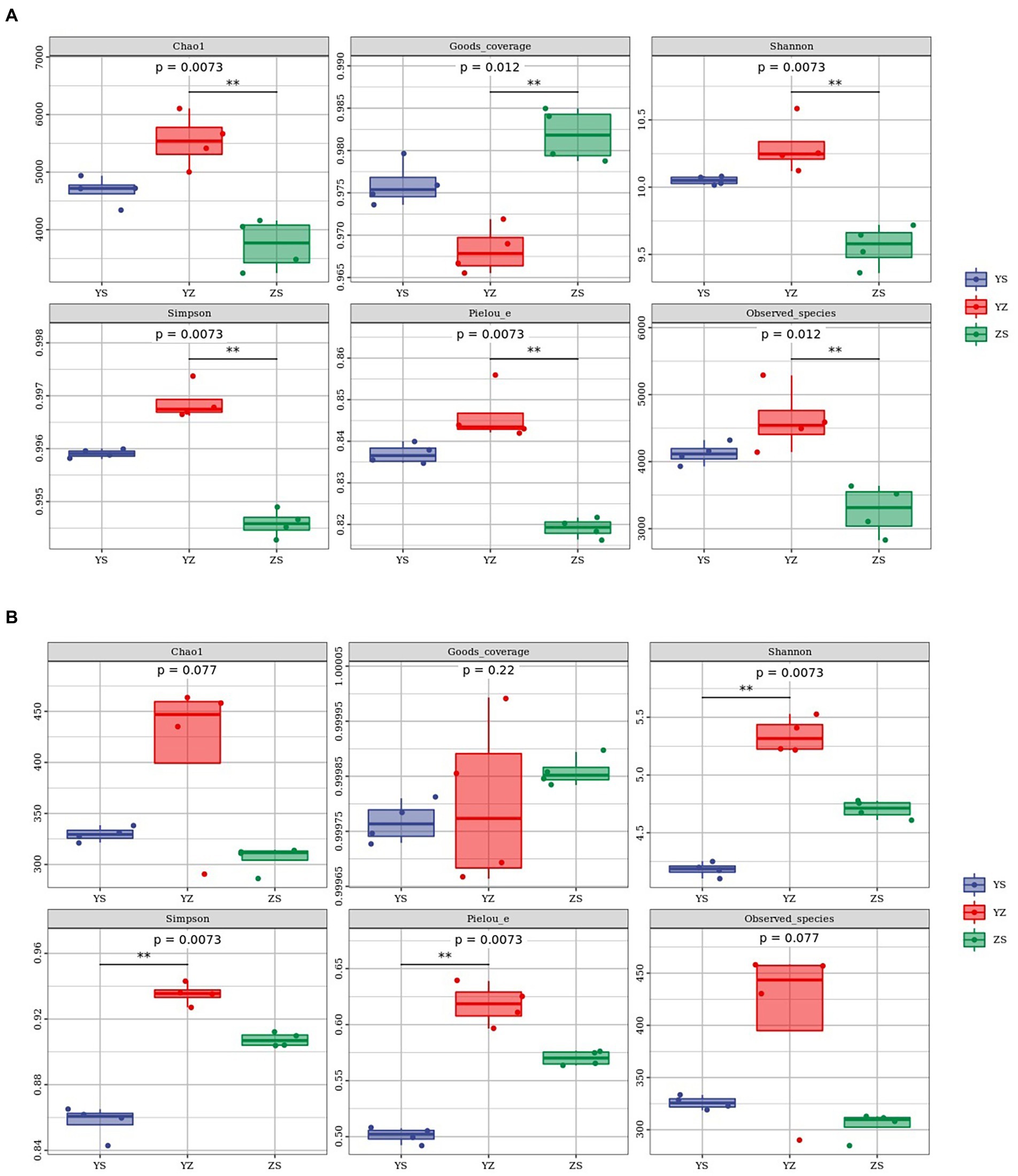
Figure 3. Litter microbial diversity index in YS, YZ and ZS. (A) Alpha diversity analysis of leaf litter bacterial community. (B) Alpha diversity analysis of leaf litter fungal community. YS: Populus × canadensis Moench; YZ: Populus × canadensis Moench-Pinus sylvestris var. mongolica; ZS: Pinus sylvestris var. mongolica. ** meant significant difference at 0.01 level.
The relative abundance of microorganisms in the samples of the three litter types was counted, and the top 10 relative abundances were drawn at the phylum level (others are shown) and genus level (others not shown), respectively, as shown in Figure 4. At the bacterial phylum level, the top 10 relative abundances were Proteobacteria, Actinobacteria, Bacteroidetes, Acidobacteria, Patescibacteria, Firmicutes, Chloroflexi, Cyanobacteria, Verrucomicrobia, Armatimonadetes. The relative abundance of Proteobacteria was the highest in ZS at 71.65%, followed by YZ (41.32%) and YS (41.00%). On the contrary, Actinobacteria had the highest relative abundance in YS at 47.6%, followed by YZ (46.23%) and ZS (19.05%). The relative abundance of Bacteroidetes was YZ > YS > ZS (Figure 4A). At the fungal phylum level, Ascomycota, Basidiomycota, Chytridiomycota, Mortierellomycota, Mucoromycota and Olpidiomycota were relatively abundant. Among them, only Ascomycota and Basidiomycota have relative abundance higher than 1%. The relative abundance of Ascomycota is the highest in YS at 82.75%, followed by YZ (70.70%) and the lowest in ZS (63.39%); the relative abundance of Basidiomycota is just the opposite, that is, ZS (33.29%) > YZ (3.38%) > YS (2.08%; Figure 4C).
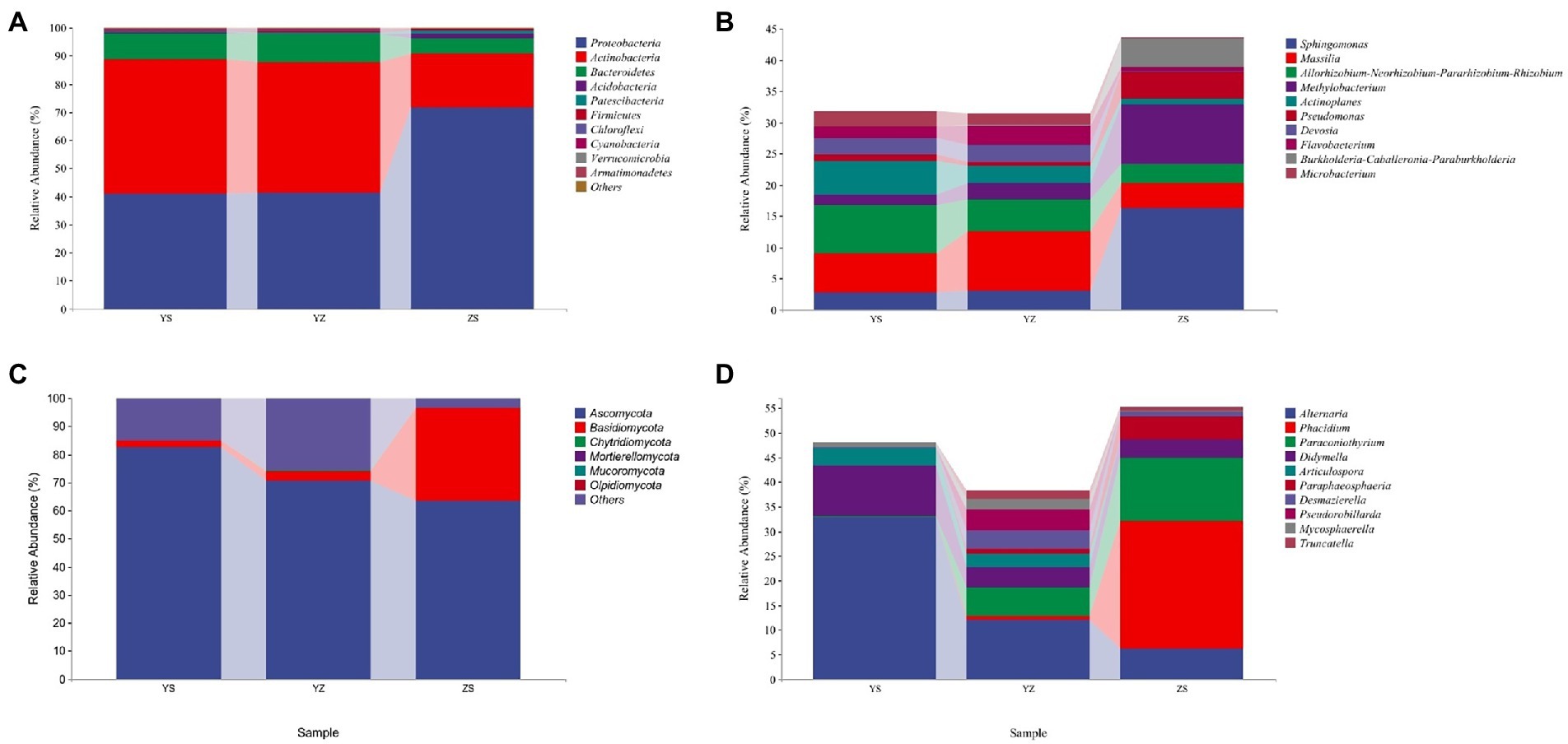
Figure 4. The relative abundance of microorganisms at pyhlum and genus levels in litter from different samples. (A) The relative abundance of dominant bacterial community at the phylum level; (B) The relative abundance of dominant bacterial community at the genus level (not showing others); (C) The relative abundance of dominant fungal community at the phylum level; (D) The relative abundance of dominant fungal community at the genus level (not showing others). YS: Populus × canadensis Moench; YZ: Populus × canadensis Moench-Pinus sylvestris var. mongolica; ZS: Pinus sylvestris var. mongolica.
At the bacterial genus level, the top 10 relative abundances were Sphingomonas, Massilia, Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Methylobacterium, Actinoplanes, Pseudomonas, Devosia, Flavobacterium, Burkholderia-Caballeronia-Paraburkholderia, and Microbacterium (Figure 4B). It is evident from Figure 5A that there was a correlation between the physicochemical characteristics of the litter and these 10 genera. Sphingomonas, Methylobacterium, Pseudomonas, Burkholderia-Caballeronia-Paraburkholderia were significantly positively correlated with litter N/P, C/P (p < 0.01), and negatively correlated with Dw (p < 0.01). Allorhizobium-Neorhizobium-Parararhizobium-Rhizobium, Actinoplanes, Devosia, Flavobacterium, and Microbacterium were significantly negatively correlated with litter N/P, C/P (Flavobacterium, p < 0.05; other else, p < 0.01) was significantly negatively correlated with N/P and positively correlated with TN, Dw (p < 0.05).
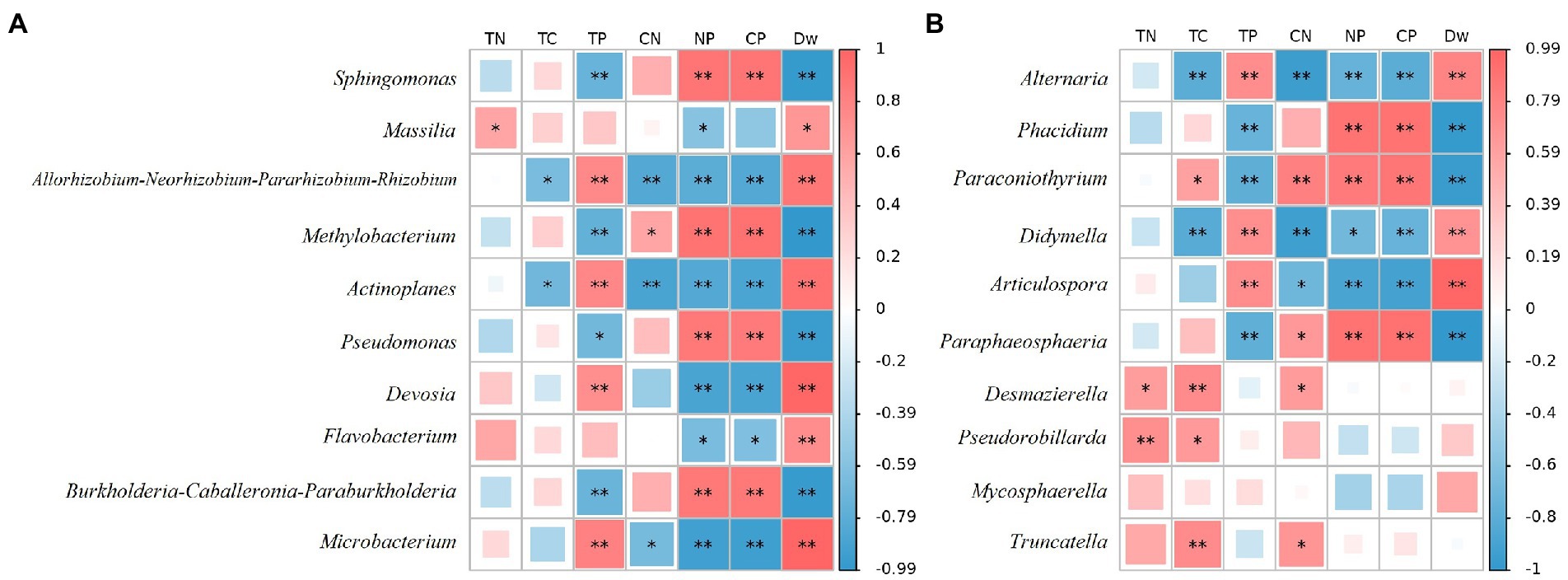
Figure 5. CoHeatmap shows the 10 genera relative abundance and chemical composition in various samples of litter. (A) Bacteria; (B) Fungi. The legend shows the correlation coefficient values, with red representing positive correlations and blue representing negative correlations. Shades of color indicate the strength of the correlation. *represents p < 0.05; **represents p < 0.01. TN: Total N; TC: Total C; TP: Total P; CN: C/N; NP: N/P; CP: C/P.
At the fungal genus level, the top 10 relative abundances were Alternaria, Phacidium, Paraconiothyrium, Didymella, Articulospora, Paraphaeosphaeria, Desmazierella, Pseudorobillarda, Mycosphaerella, and Truncatella (Figure 4D). Similarly, as can be seen from Figure 5B, Alternaria, Didymella, and Articulospora were significantly negatively correlated with litter N/P, C/P (Didymella, p < 0.05; other else, p < 0.01), and positively correlated with Dw (p < 0.01). Phacidium, Paraconiothyrium, and Paraphaeosphaeria were significantly positively correlated with litter N/P, C/P (p < 0.01), and negatively correlated with Dw (p < 0.01). Nevertheless, the relatively low levels of Desmazierella, Pseudorobillarda, Mycosphaerella, and Truncatella was not significantly correlated with Dw (p > 0.05).
Lefse is a recent analysis method based on Linear discriminant analysis (LDA) effect size. Its essence is to combine linear discriminant analysis with nonparametric Kruskal-Wallis and Wilcoxon rank sum tests to screen for key biomarkers (i.e., key community members; Segata et al., 2011). With LDA effect size scores >4.5, 16 bacterial taxa were significantly different across treatments (Figure 6A). When LDA effect size scores were > 5, 3 bacterial taxa were significantly different in litter from ZS and YS. Among them, at the bacterial phylum level, the main enriched bacterial taxa in YS leaf litter were Actinobacteria, ZS was mainly enriched by Proteobacteria, and YZ was Bacteroidetes (Figure 7A). As shown in Figure 6B, when the LDA effect size score was >4, the relative abundances of 19 fungal taxa were significantly different among different treatments (p < 0.05). Regarding the fungal alpha-diversity, Ascomycota was mainly enriched in YS litter, while Basidiomycota was mainly enriched in ZS (Figure 7B).
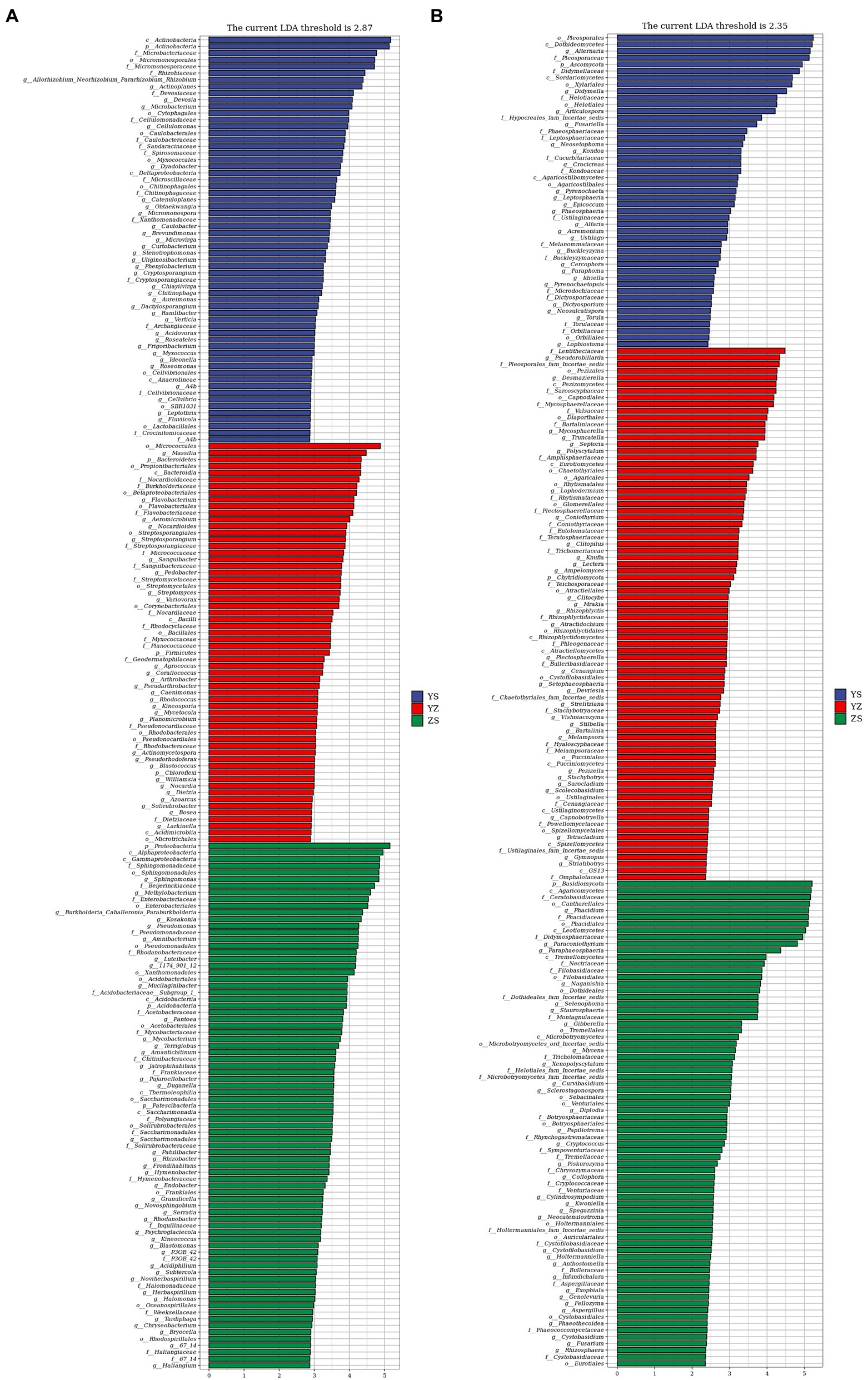
Figure 6. Microbial community of different leaf litter ratios with significantly different taxa. (A) litter bacterial communities; (B) litter fungal communities. YS: Populus × canadensis Moench; YZ: Populus × canadensis Moench-Pinus sylvestris var. mongolica; ZS: Pinus sylvestris var. mongolica. The longer the length, the more significant the difference between the taxon units, and the different color of the bar chart indicates the higher abundance sample group corresponding to the taxon.
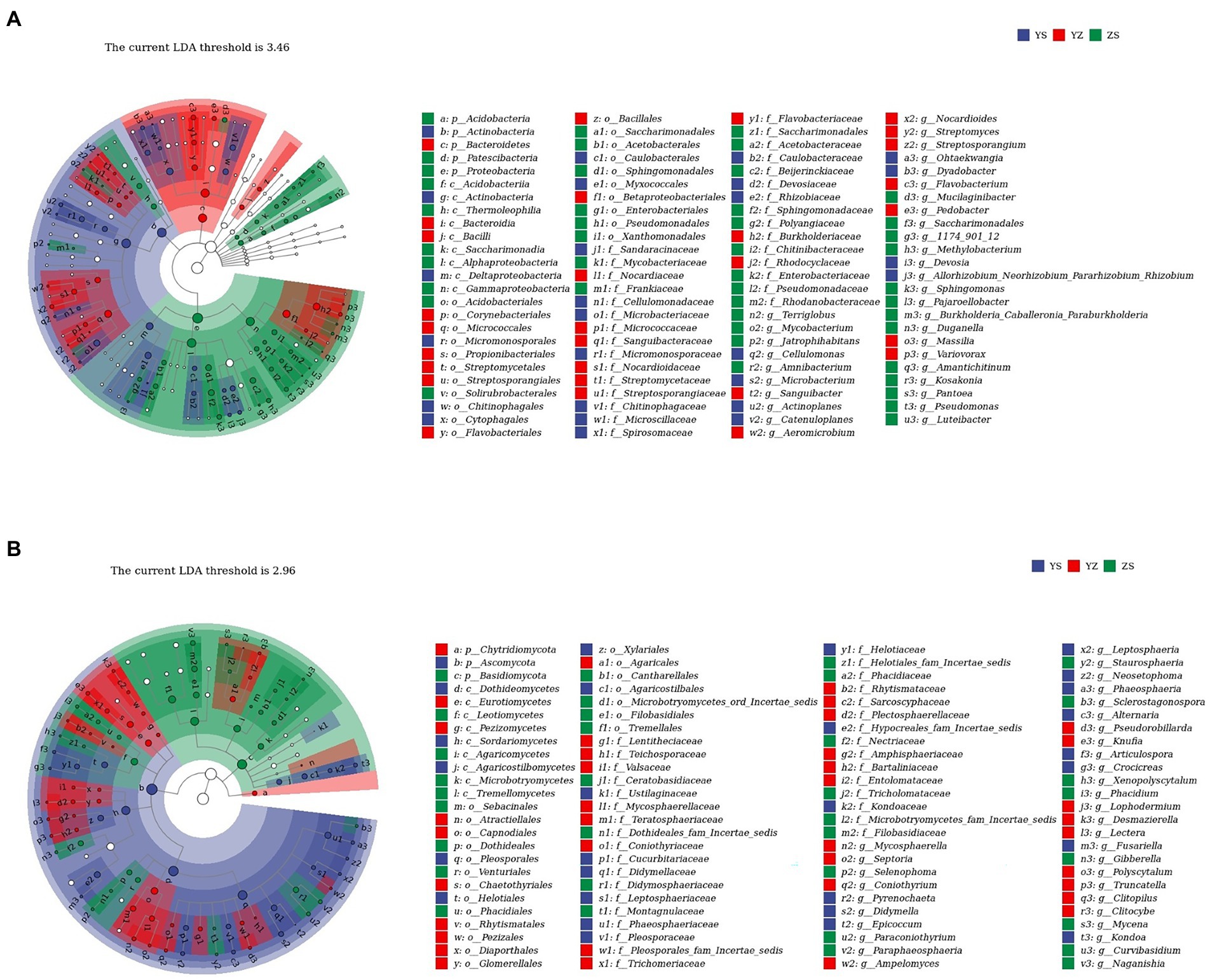
Figure 7. (A) Lefse with an LDA of 3.46 indicates that the 100 groups are significantly different between litter bacterial communities of YS, YZ, and ZS. (B) Lefse with an LDA of 2.96 indicates that the 100 groups are significantly different between litter fungal communities of YS, YZ, and ZS. YS: Populus × canadensis Moench; YZ: Populus × canadensis Moench-Pinus sylvestris var. mongolica; ZS: Pinus sylvestris var. mongolica. Circle radiation from inner to outer of evolutionary branch figure represents the classification of the level from phylum door genus; every small circle represents the level of a classification in different classification level. The diameter of the circle is prportional to the relative abundance.
The Bray-Curtis-based heatmap showed that the litter bacterial communities of YS and YZ were clustered together, indicating that the litter leaf communities from ZS were clearly distinct from those of YS and YZ (Supplementary Figure S1). The fungal community of the litter also showed the same conclusion (Supplementary Figure S2).
Correlation between physicochemical properties of litter and microbial community diversity in litter
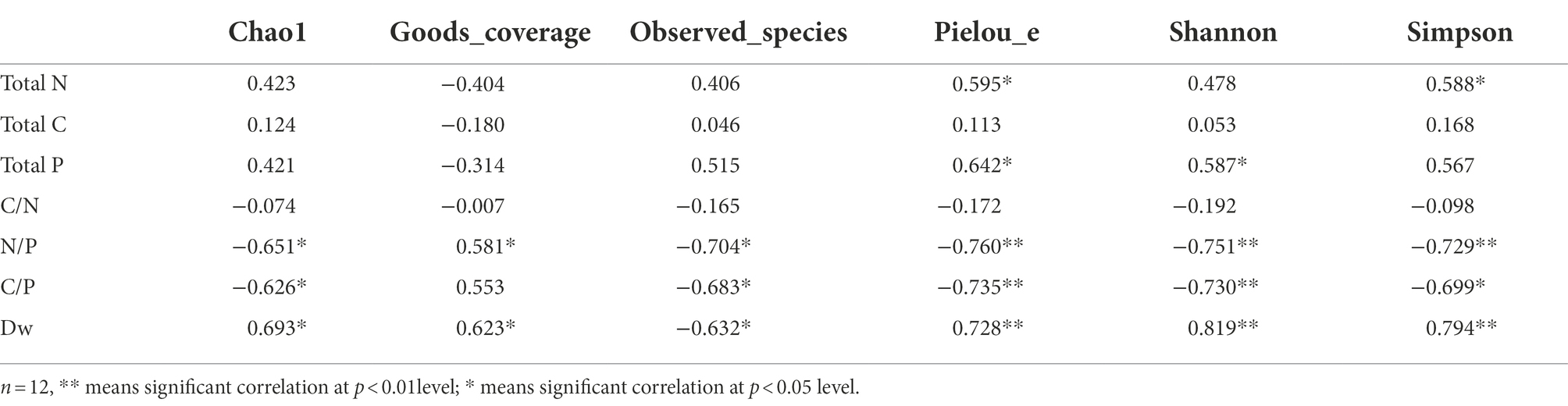
Regarding the bacterial alpha-diversity, total N of leaf litter was significantly positively correlated with Pielou_e (r = 0.595, p < 0.05) and Simpson index (r = 0.588, p < 0.05); total P was significantly positively correlated with Pielou_e (r = 0.642, p < 0.05) and Shannon index (r = 0.587, p < 0.05); N/P was significantly positively correlated with Goods_coverage (r = 0.581, p < 0.05); N/P and C/P were significantly negatively correlated with Chao 1 index (r = −0.651, p < 0.05; r = −0.626, p < 0.05), Observed_species index (r = −0.704, p < 0.05; r = −0.683, p < 0.05), Pielou_e index (r = −0.760, p < 0.01; r = −0.735, p < 0.01), Shannon index (r = −0.751, p < 0.01; r = −0.730, p < 0.01) and Simpson index (r = −0.729, p < 0.01; r = −0.699, p < 0.05). Dw was significantly positively correlated with Chao 1 index (r = 0.693, p < 0.05), Goods_coverage (r = 0.623, p < 0.05), Pielou_e index (r = 0.728, p < 0.01), Shannon index (r = 0.819, p < 0.01) and Simpson index (r = 0.794, p < 0.01). Dw was significantly negatively correlated with Observed_species index (r = −0.632, p < 0.05). There was no significant relationship between litter total C and bacterial community diversity index (Table 2).
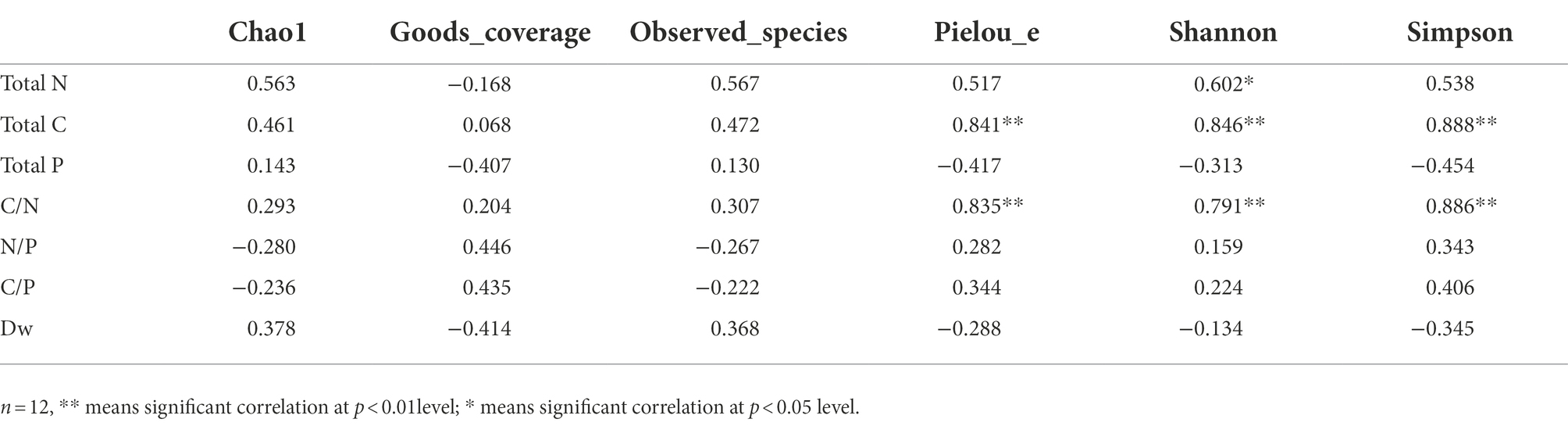
Regarding the fungal alpha-diversity, total N of leaf litter was significantly positively correlated with Shannon index (r = 0.602, p < 0.05); total C and C/N were significantly positively correlated with Pielou_e index (r = 0.841, p < 0.01; r = 0.835, p < 0.05), Shannon index (r = 0.846, p < 0.01; r = 0.791, p < 0.01) and Simpson index (r = 0.888, p < 0.01; r = 0.886, p < 0.01). There was no significant relationship between total P, N/P, C/P, Dw and microbial diversity index of litter (Table 3).
Discussions
“Litter mass,” according to Swift et al., (1989), refers to the physicochemical composition of litter. It is the nutrient composition and structure, which includes easily degradable parts (N, P) and resistant organic parts (lignin, cellulose, etc.). The primary controlling factor influencing the rate of litter decomposition is thought to be litter quality (Cornwell et al., 2008). The three elements C, N, and P are currently the primary physicochemical elements involved in research on litter decomposition in my country, and the majority of these studies concentrate on the quality of the litter matrix and the release dynamics of physicochemical elements (Jia et al., 2016). Given the strict stoichiometric requirements for microbial decomposer growth, it is possible that the scarcity of a particular nutrient will slow down the decomposition of litter (Ni et al., 2021). Numerous studies have discovered a strong correlation between the rate of litter decomposition and the initial N content of the litter as well as N-related substrate quality indicators. Indicators of C/N and C/P are frequently used to forecast the rate at which litter will decompose. The decomposition rate decreases as C/N and C/P values rise (Aber and Melillo, 1982; Magill et al., 2000; Sariyildiz and Anderson, 2003). It is consistent with the findings of this research (Table 1). The same pattern was shown in Figure 5. The decomposition rate of mixed litter was found in this study to be significantly higher than that of coniferous forest, but lower than that of pure broad–leaved forest. This is consistent with the findings of Pastor et al. (1984) who conducted experiments on nitrogen mineralization and litter decomposition in pine forests on Blackhawk Island in the United States. Sariyildiz also came to the same conclusion in his simulated experiments on beech and oak trees (Sariyildiz, 2015).
The primary factor influencing nutrient release is plant litter quality, and its C/N value is frequently regarded as an important attribute for measuring litter quality (Silver and Miya, 2001; Hu et al., 2006). For instance, Brady and Weil (1996) discovered that nitrogen release occurs when C/N values in the remaining litter are lower than 25 and nitrogen fixation occurs when C/N values are >25 (Killham, 1994). The rate of decomposition can be regulated by early litter decomposition C/N (Cotrufo and Ineson, 1995; Berg, 2000). Litter C/P has a threshold between 200 and 480, and this threshold affects how much litter P content is released. The litter P content is a net release when the litter C/P is <480 (Gosz et al., 1973; Dziadowiec, 1987; Manzoni et al., 2010). Furthermore, Liu’s research made the case that litter N/P can be used as a gauge for determining nutrient limitation. P limit how much litter can decompose if N/P is >25 (Liu et al., 2016). The results of this experiment demonstrated that the phenomenon that P limited the decomposition of coniferous species could very well be improved by the addition of broad–leaved species. In conclusion, adding broad–leaved litter can increase the rate at which coniferous litter decomposes, this is because mixed litter decomposes more quickly than litter from a single tree species.
Previous research has shown that single-species litter and mixed litter have significantly different community compositions and structures (Aneja et al., 2006; Chapman and Newman, 2010; Prescott and Grayston, 2013; Santonja et al., 2017). Microbial biomass and community composition can vary depending on the microenvironment or chemical makeup of the litter (Malosso et al., 2004; Das et al., 2007; Xu et al., 2013). The current study discovered differences between the alpha diversity of single-species litter and mixed litter microbial communities. Mixed litter made up of Populus × canadensis and Pinus sylvestris var. mongolica differed significantly from Pinus sylvestris var. mongolica litter in terms of bacterial richness, diversity, uniformity and coverage, but not from Populus × canadensis litter. However, the coverage and richness of mixed litter and Populus × canadensis litter were not significantly different, and the fungal community diversity and homogeneity of the Populus × canadensis – Pinus sylvestris var. mongolica litter were only significantly higher than those of Populus × canadensis. This has also been supported by earlier researches (Aneja et al., 2006; Chapman and Newman, 2010; Prescott and Grayston, 2013; Santonja et al., 2017). Proteobacteria and Ascomycetes are the most abundant taxa in the early stage of litter decomposition and are the main decomposers (Voříšková and Baldrian, 2012; Zhang et al., 2017). This study also found that Proteobacteria dominated the bacterial community and Ascomycota dominated the fungal community. According to research, fungi and bacteria work together to speed up the decomposition of litter (Wright and Covich, 2005). Fungi are primarily responsible for the decomposition of carbonaceous organic matter (Frey et al., 1999; Guggenberger et al., 1999; Pascoa and Cássio, 2004), while bacteria primarily consume nitrogenous organic matter (Six et al., 2002). Ascomycetes, the predominant decomposing bacteria in the three types of litter, are primarily in charge of breaking down cellulose and hemicellulose (Pointing and Hyde, 2000; Sánchez, 2009; Zhang et al., 2016), and Proteobacteria are in charge of breaking down proteins (Schweitzer et al., 2001; Kazakov et al., 2009).
In the breakdown of apoplast, various genera also play various roles. For instance, the top 10 species in terms of relative abundance in this study, Massilia, Allorhizobium-Neorhizobium-Parararhizobium-Rhizobium, Actinoplanes, Devosia, Flavobacterium, and Microbacterium, all have the job of breaking down organic acids, polyols, aromatic compounds, and other substances in the environment (Du et al., 2011; Feng et al., 2019; Menon et al., 2019). Both simple carbohydrates and refractory materials are thought to be capable of being broken down by Alternaria (Jatav et al., 2020). An endophytic fungus called Paraconiothyrium degrades lignin (Gao et al., 2011). They all play a role in the breakdown of apoplastic matter, which is consistent with the study’s findings.
The structure and operation of the litter microbial community are also impacted by the differences between coniferous and broad-leaved litter’s physical and chemical characteristics. Previous researches have also demonstrated a connection between the chemical properties of decomposed substrates and the diversity of the microbial community (Hawke and Vallance, 2015; Zeng et al., 2017). The two main microbial taxa involved in decomposition litter are fungi and bacteria, and depending on the ecosystem and type of litter, their functional characteristics and carbon requirements may vary (Lin et al., 2018). Although fungi are thought to decompose more quickly than bacteria, bacteria are more effective at using labile carbon compounds (Hunt et al., 1987). This may account for the stronger relationship between apomictic Dw and bacterial community alpha diversity as well as the lack of a relationship with fungal communities. When compared to coniferous pure forest litter, mixed forest litter has different carbon and mineral nutrient contents, offering a wider substrate for microbes that break down organic matter (Hooper and Vitousek, 1998; Hättenschwiler et al., 2011).
Conclusion
In this study, it was discovered that the decomposition effect of mixed litter was different from that of a species of tree by simulating the mixed decomposition of Populus × canadensis Moench and Pinus sylvestris var. mongolica litter. (1) Compared to leaf litter from a single tree species, mixed leaf litter had different nutrients. However, the contents of total P, C/N, N/P and C/P were as YZ was between YS and ZS. The total C and total N of YZ were higher than those of single tree species. (2) There were variations in the organization and composition of the microbial communities in mixed leaf litter compared to of single tree species, but the difference was not very significant. (3) The investigation revealed that leaf nutritional variables had an impact on the variety of the microbial community composition and organization in leaf litter. (4) The decomposition of Populus × canadensis Moench litter was significantly aided by the addition of Pinus sylvestris var. mongolica litter. N/P, C/P, total N, and total P had a substantial impact on the bacterial community, while C/N, total C, and total N had a large impact on the fungal community. Consequently, this research provided a theoretical framework for the investigation of the decomposition mechanism of mixed litter from the nutritional level and microbial community.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA832892.
Author contributions
WZu designed the research. JL, CD, and WZa performed the research. JL, CD, YW, and YZ analyzed the data. WZu, CD, and JL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (grant number 2021YFD2201205), the Basic Research Fund of RIF (grant no. CAFYBB2020SZ002), the National Natural Science Foundation of China (grant nos. 31870662 and 32271843), and Liaoning Province Scientific Research Funding Project (LSNQN202012).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1009091/full#supplementary-material
Footnotes
References
Aber, J. D., and Melillo, J. M. (1982). Nitrogen immobilization in decaying hardwood leaf litter as a function of initial nitrogen and lignin content. Can. J. Bot. 60, 2263–2269. doi: 10.1139/b82-277
Alban, R. (2010). Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62, 142–160. doi: 10.1111/j.1574-6941.2007.00375.x
Aneja, M. K., Sharma, S., Fleischmann, F., Stich, S., Heller, W., Bahnweg, G., et al. (2006). Microbial colonization of beech and spruce litter-influence of decomposition site and plant litter species on the diversity of microbial community. Microb. Ecol. 52, 127–135. doi: 10.1007/s00248-006-9006-3
Ba, H., Jiang, H., Shen, P., Cao, Y., and Li, L. (2020). Screening of phosphate-resolving bacteria in rhizosphere of cold sunflower and physiological and biochemical study. IOP Conf. Series Earth Environ. Sci. 526:012038. doi: 10.1088/1755-1315/526/1/012038
Baldrian, P. (2017). Forest microbiome: diversity, complexity and dynamics. FEMS Microbiol. Rev. 41, 109–130. doi: 10.1093/femsre/fuw040
Bao, L., Cai, W., Zhang, X., Liu, J., Chen, H., Wei, Y., et al. (2019). Distinct microbial community of phyllosphere associated with five tropical plants on Yongxing Island, South China Sea. Microorganisms 7:525. doi: 10.3390/microorganisms7110525
Berg, B. (2000). Litter decomposition and organic matter turnover in northern forest soils. Forest Ecol. Manag. 133, 13–22. doi: 10.1016/S0378-1127(99)00294-7
Bradford, M. A., Berg, B., Maynard, D. S., Wieder, W. R., and Wood, S. A. (2016). Understanding the dominant controls on litter decomposition. J. Ecol. 104, 229–238. doi: 10.1111/1365-2745.12507
Brady, N. C., and Weil, R. R.. (1996). The Nature and Properties of Soil. 11th ed. Englewood Cliffs: Prentice-Hall 740.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chao, A. (1984). Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270. doi: 10.2307/4615964
Chapman, S. K., and Newman, G. S. (2010). Biodiversity at the plant-soil interface: microbial abundance and community structure respond to litter mixing. Oecologia 162, 763–769. doi: 10.1007/s00442-009-1498-3
Chiani, M. (1948). A mathematical theory of communication. Bell Labs Tech. J. 27, 623–656. doi: 10.1002/j.1538-7305.1948.tb00917.x
Claesson, M. J., O'Sullivan, O., Wang, Q., Nikkilä, J., Marchesi, J. R., Smidt, H., et al. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. doi: 10.1371/journal.pone.0006669
Cline, L. C., and Zak, D. R. (2015). Initial colonization, community assembly and ecosystem function: fungal colonist traits and litter biochemistry mediate decay rate. Mol. Ecol. 24, 5045–5058. doi: 10.1111/mec.13361
Cornelissen, J. H., Van Bodegom, P. M., Aerts, R., Callaghan, T., Longtestijn, R., Alatalo, J., et al. (2007). Global negative vegetation feedback to climate warming responses of leaf litter decomposition rates in cold biomes. Ecol. Lett. 10, 619–627. doi: 10.1111/j.1461-0248.2007.01051.x
Cornwell, W. K., Cornelissen, J. H. C., Amatangelo, K., Dorrepaal, E., Eviner, V. T., Godoy, O., et al. (2008). Plant species traits are the predominant control on litter decomposition rates within biomes worldwide. Ecol. Lett. 11, 1065–1071. doi: 10.1111/j.1461-0248.2008.01219.x
Cotrufo, M. F., and Ineson, P. (1995). Effects of enhanced atmospheric CO2 and nutrient supply on the quality and subsequent decomposition of fine roots of Betula pendula Roth. And Picea sitchensis (bong.) Carr. Plant Soil 170, 267–277. doi: 10.1007/bf00010479
Cotrufo, M. F., Soong, J. L., Horton, A. J., Campbell, E. E., Haddix, M. L., Wall, D. H., et al. (2015). Formation of soil organic matter via biochemical and physical pathways of litter mass loss. Nat. Geosci. 8, 776–779. doi: 10.1038/ngeo2520
Das, M., Royer, T. V., and Leff, L. G. (2007). Diversity of fungi, bacteria, and actinomycetes on leaves decomposing in a stream. Appl. Environ. Microb. 73, 756–767. doi: 10.1128/AEM.01170-06
Davidson, E. A., and Janssens, I. A. (2006). Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440, 165–173. doi: 10.1038/nature04514
Du, Y., Yu, X., and Wang, G. (2011). Massilia tieshanensis sp. nov. isolated from mining soil. Int. J. Syst. Evol. Microbiol. 62, 2356–2362. doi: 10.1099/ijs.0.034306-0
Dziadowiec, H. (1987). The decomposition of plant litterfall in an oak-linden-hornbeam forest and an oak-pine mixed forest of the Bialoweza national park. Acta Soc. Bot. Pol. 56, 169–185. doi: 10.5586/asbp.1987.019
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eichlerová, I., Homolka, L., Žifčáková, L., Lisá, L., Dobiášová, P., and Baldrian, P. (2015). Enzymatic systems involved in decomposition reflects the ecology and taxonomy of saprotrophic fungi. Fungal Ecol. 13, 10–22. doi: 10.1016/j.funeco.2014.08.002
Feng, Y., Yuan, Q., Yang, Z., Zhang, T., Liu, X., Li, C., et al. (2019). Effect of Sphingomonas sp. strain on degradation of polyphenols in redried tobacco leaves. Acta Tabacaria Sinica 25, 19–24. doi: 10.16472/j.chinatobacco.2018.038
Ferreira, V., Raposeiro, P. M., Pereira, A., Cruz, A. M., Costa, A. C., Graca, M., et al. (2016). Leaf litter decomposition in remote oceanic island streams is driven by microbes and depends on litter quality and environmental conditions. Freshw. Biol. 61, 783–799. doi: 10.1111/fwb.12749
Frey, S. D., Elliott, E. T., and Paustian, K. (1999). Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biol. Biochem. 31, 573–585. doi: 10.1016/S0038-0717(98)00161-8
Gao, H., Wang, Y., Zhang, W., Wang, W., and Mu, Z. (2011). Isolation, identification and application in lignin degradation of an ascomycete GHJ-4. Afr. J. Biotechnol. 10, 4166–4174. doi: 10.4314/AJB.V10I20
Good, I. J. (1953). The population frequency of species and the estimation of the population parameters. Biometrics 40, 237–264. doi: 10.2307/2333344
Gosz, J. R., Likens, G. E., and Bormann, F. H. (1973). Nutrient release from decomposition leaf and branch litter in the Hubbard Brool Forest, New Hampshire. Ecolog. Monog. 43, 173–191. doi: 10.2307/1942193
Guerreiro, M. A., Brachmann, A., Begerow, D., and Peršoh, D. (2017). Transient leaf endophytes are the most active fungi in 1-year-old beech leaf litter. Fungal Divers. 89, 237–251. doi: 10.1007/s13225-017-0390-4
Guggenberger, G., Frey, S. D., Six, J., Paustian, K., and Elliott, E. T. (1999). Bacterial and fungal cell-wall residues in conventional and no-tillage agroecosystems. Soil Sci. Soc. Am. J. 63, 1188–1198. doi: 10.2136/sssaj1999.6351188x
Gui, H., Purahong, W., Hyde, K. D., Xu, J. C., and Mortimer, P. E. (2017). The arbuscular mycorrhizal fungus Funneliformis mosseae alters bacterial communities in subtropical forest soils during litter decomposition. Front. Microbiol. 8:1120. doi: 10.3389/fmicb.2017.01120
Han, C., Liu, T., Lu, X., Duan, L., Singh, V., and Ma, L. (2019). Effect of litter on soil respiration in a man-made Populus L. forest in a dune-meadow transitional region in China’s Horqin sandy land. Ecol. Eng. 127, 276–284. doi: 10.1016/j.ecoleng.2018.12.005
Handa, I. T., Aerts, R., Berendse, F., Berg, M. P., Bruder, A., Butenschoer, O., et al. (2014). Consequences of biodiversity loss for litter decomposition across biomes. Nature 509, 218–221. doi: 10.1038/nature13247
Hättenschwiler, S., Fromin, N., and Barantal, S. (2011). Functional diversity of terrestrial microbial decomposers and their substrates. C. R. Biol. 334, 393–402. doi: 10.1016/j.crvi.2011.03.001
Hawke, D. J., and Vallance, J. R. (2015). Microbial carbon concentration in samples of seabird and non-seabird forest soil: implications for leaf litter cycling. Pedobiologia 58, 33–39. doi: 10.1016/j.pedobi.2015.01.002
Hooper, D. U., and Vitousek, P. M. (1998). Effects of plant composition and diversity on nutrient cycling. Ecol. Monogr. 68, 121–149. doi: 10.2307/2657146
Hu, Y. L., Wang, S. L., and Zeng, D. H. (2006). Effects of single Chinese fir and mixed leaf litters on soil chemical, microbial properties and soil enzyme activities. Plant Soil 282, 379–386. doi: 10.1007/s11104-006-0004-5
Hunt, H. W., Coleman, D. C., and Ingham, E. R. (1987). The detrital food web in a short grass prairie. Biol. Fertil. Soils 3-3, 57–68. doi: 10.1007/BF00260580
Jatav, B. K., Sharma, T., and Dassani, S. (2020). Succession of microfungi on leaf litter of Acacia catechu in Datia, Madhya Pradesh, India. J. Pure Appl. Microbiol. 14, 581–590. doi: 10.22207/JPAM.14.1.60
Jia, T., Wang, R., Fan, X., and Chai, B. (2018). A comparative study of fungal community structure, diversity and richness between the soil and the Phyllosphere of native grass species in a copper tailings dam in Shanxi Province, China. Appl. Sci. 8:1297. doi: 10.3390/app8081297
Jia, B. R., Xu, Z. Z., Zhou, G. S., and Yin, X. J. (2018). Statistical characteristics of forest litter fall in China. Sci. China Life Sci. 61, 358–360. doi: 10.1007/s11427-016-9143-x
Jia, B., Zhou, G., and Xu, Z. (2016). Forest litterfall and its composition: a new data set of observational data from China. Ecology 97:1365. doi: 10.1890/15-1604.1
Jiang, Y. F., Yin, X. Q., and Wang, F. B. (2013). The influence of litter mixing on decomposition and soil fauna assemblages in a Pinus koraiensis mixed broad-leaved forest of the Changbai Mountains, China. Eur. J. Soil Biol. 55, 28–39. doi: 10.1016/j.ejsobi.2012.11.004
Kazakov, A. E., Rodionov, D. A., Alm, E., Arkin, A. P., Dubchak, I., and GelFaNd, M. S. (2009). Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in Proteobacteria. J. Bacteriol. 191, 52–64. doi: 10.1128/JB.01175-08
Kembel, S. W., O'Connor, T. K., Arnold, H. K., Hubbell, S. P., Wright, S. J., and Green, J. L. (2014). Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl. Acad. Sci. U. S. A. 111, 13715–13720. doi: 10.1073/pnas.1216057111
Li, T. Y., Kang, F. F., Han, H. R., Gao, J., Song, X., and Yu, S. (2015). Responses of soil microbial carbon metabolism to the leaf litter composition in Liaohe River nature reserve of northern Hebei Province, China. J. Appl. Ecol. 26, 715–722. doi: 10.13287/j.1001-9332.20150506.004
Lin, N., Bartsch, N., Heinrichs, S., and Vor, T. (2015). Long-term effects of canopy opening and liming on leaf litter production, and onleaf litter and fine-root decomposition in a European beech (Fagus sylvatica, L.) forest. Forest Ecol. Manag. 338, 183–190. doi: 10.1016/j.foreco.2014.11.029
Lin, H., Li, Y., Bruelheide, H., Zhang, S., Ren, H., Zhang, N., et al. (2021). What drives leaf litter decomposition and the decomposer community in subtropical forests – the richness of the above-ground tree community or that of the leaf litter? Soil Biol. Biochem. 160:108314. doi: 10.1016/j.soilbio.2021.108314
Lin, D., Pang, M., Fanin, N., Wang, H., Qian, S., Zhao, L., et al. (2018). Fungi participate in driving home-field advantage of litter decomposition in a subtropical forest. Plant Soil 434, 467–480. doi: 10.1007/s11104-018-3865-5
Liu, D., Keiblinger, K. M., Leitner, S., Mentler, A., and Zechmeister-Boltenstern, S. (2016). Is there a convergence of deciduous leaf litter stoichiometry, biochemistry and microbial population during decay? Geoderma 272, 93–100. doi: 10.1016/j.geoderma.2016.03.005
Lu, S., Xu, E., Wu, D., Lu, Y., Guo, J., and Yang, Y. (2020). Response of soil microbial community composition on litterfall input in a Castanopsis carlesii plantation. J. For. Environ. 40, 16–23. doi: 10.13324/j.cnki.jfcf.2020.01.00
Lucas-Borja, M. E., Hedod, S. J., Yang, Y., Shen, Y., and Candel-Pérez, D. (2018). Nutrient, metal contents and microbiological properties of litter and soil along a tree age gradient in Mediterranean forest ecosystems. Sci. Total Environ. 650, 749–758. doi: 10.1016/j.scitotenv.2018.09.079
Magill, A. H., Aber, J. D., Berntson, G. M., McDowell, W. H., Nadelhoffer, K. J., Melillo, J. M., et al. (2000). Long-term nitrogen additions and nitrogen saturation in two temperate forests. Ecosystems 3, 238–253. doi: 10.1007/s100210000023
Malosso, E., English, L., Hopkins, D. W., and O'Donnell, A. G. (2004). Use of 13 C-labelled plant materials and ergosterol, PLFA and NLFA analyses to investigate organic matter decomposition in Antarctic soil. Soil Biol. Biochem. 36, 165–175. doi: 10.1016/j.soilbio.2003.09.004
Manzoni, S., Jackson, R. B., Trofymow, J. A., and Porporato, A. (2008). The global stoichiometry of litter nitrogen mineralization. Science 321, 684–686. doi: 10.1126/science.1159792
Manzoni, S., Trofymow, J. A., Jackson, R. B., and Porporato, A. (2010). Stoichiometric controls on carbon, nitrogen, and phosphorus dynamics in decomposing litter. Ecol. Monogr. 80, 89–106. doi: 10.1890/09-0179.1
Menon, R. R., Kumari, S., Kumar, P., Verma, A., Krishnamurthi, S., and Rameshbumar, N. (2019). Sphingomonas pokkalii sp. nov., a novel plant associated rhizobacterium isolated from a saline tolerant pokkali rice and its draft genome analysis. Syst. Appl. Microbiol. 42, 334–342. doi: 10.1016/j.syapm.2019.02.003
Muscolo, A., Bagnato, S., Sidari, M., and Mercurio, R. (2014). A review of the roles of forest canopy gaps. J. Forestry Res. 25, 725–736. doi: 10.1007/s11676-014-0521-7
Nevins, C. J., Nakatsu, C., and Armstrong, S. (2018). Characterization of microbial community response to cover crop residue decomposition. Soil Biol. Biochem. 127, 39–49. doi: 10.1016/j.soilbio.2018.09.015
Ni, X., Lin, C., Chen, G., Xie, J., and Yang, Y. (2021). Decline in nutrient inputs from litterfall following forest plantation in subtropical China. Forest Ecol. Manag. 496:119445. doi: 10.1016/j.foreco.2021.119445
Nicolas, B., Alexandre, C., Jacinthe, R., Steven, W. K., and David, R. (2019). Microsite conditions influence leaf litter decomposition in sugar maple bioclimatic domain of Quebec. Biogeochemistry 145, 107–126. doi: 10.1007/s10533-019-00594-1
Osono, T., Bhatta, B. K., and Takeda, H. (2004). Phyllosphere fungi on living and decomposing leaves of giant dogwood. Mycoscience 45, 35–41. doi: 10.1007/S10267-003-0155-7
Pascoal, C., and Cássio, F. (2004). Contribution of fungi and bacteria to leaf litter decomposition in a polluted river. Appl. Environ. Microbiol. 70, 5266–5273. doi: 10.1128/AEM.70.9.5266-5273.2004
Pastor, J., Aber, J. D., Mc Claugherty, C. A., and Jerry, M. M. (1984). Aboveground production and N and P cycling along a nitrogen mineralization gradient on Blackhawk Island, Wisconsin. Ecology 65, 256–268. doi: 10.2307/1939478
Patoine, G., Thakur, M. P., Friese, J., Nock, C., Honig, L., Haase, J., et al. (2017). Plant litter functional diversity effects on litter mass loss depend on the macro-detritivore community. Pedobiologia 65, 29–42. doi: 10.1016/j.pedobi.2017.07.003
Pereira, A. P. A., Durrer, A., Gumiere, T., Robin, A., Wang, J., Verma, J. P., et al. (2019). Mixed eucalyptus plantations induce changes in microbial communities and increase biological functions in the soil and litter layers. Forest Ecol. Manag. 433, 332–342. doi: 10.1016/j.foreco.2018.11.018
Pielou, E. C. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Pointing, S. B., and Hyde, K. D. (2000). Lignocellulose-degrading marine fungi. Biofouling 15, 221–229. doi: 10.1080/08927010009386312
Prescott, C. E., and Grayston, S. J. (2013). Tree species influence on microbial communities in litter and soil: current knowledge and research needs. For. Ecol. Manag. 309, 19–27. doi: 10.1016/j.foreco.2013.02.034
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Raich, J. W., and Schlesinger, W. H. (1992). The global carbon dioxide flux in soil respiration and its relationship to vegetation and climate. Tellus 44, 81–99. doi: 10.1034/j.1600-0889.1992.t01-1-00001.x
Sánchez, C. (2009). Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol. Adv. 27, 185–194. doi: 10.1016/j.biotechadv.2008.11.001
Santonja, M., Rancon, A., Fromin, N., Baldy, V., Hättenschwilerc, S., Fernandez, C., et al. (2017). Plant litter diversity increases microbial abundance, fungal diversity, and carbon and nitrogen cycling in a Mediterranean shrubland. Soil Biol. Biochem. 111, 124–134. doi: 10.1016/j.soilbio.2017.04.006
Sariyildiz, T. (2015). Effects of tree species and topography on fine and small root decomposition rates of three common tree species (Alnus glutinosa, Picea orientalis, and Pinus sylvestris) in Turkey. Forest Ecol. Manag. 335, 71–86. doi: 10.1016/j.foreco.2014.09.030
Sariyildiz, T., and Anderson, J. M. (2003). Interactions between litter quality, decomposition and soil fertility: a laboratory study. Soil Biol. Biochem. 35, 391–399. doi: 10.1016/S0038-0717(02)00290-0
Scheibe, A., Steffens, C., Seven, J., Jacob, A., Hertel, D., Leuschner, C., et al. (2015). Effects of tree identity dominate over tree diversity on the soil microbial community structure. Soil Biol. Biochem. 81, 219–227. doi: 10.1016/j.soilbio.2014.11.020
Schlesinger, W. H., and Lichter, J. (2001). Limited carbon storage in soil and litter of experimental forest plots under increased atmospheric CO2. Nature 411, 466–469. doi: 10.1038/35078060
Schweitzer, B., Huber, I., Amann, R., Ludwig, W., and Simon, M. (2001). α-And β-Proteobacteria control the consumption and release of amino acids on lake snow aggregates. Appl. Environ. Microb. 67, 632–645. doi: 10.1128/AEM.67.2.632-645.2001
Sébastien, I., Brun, C., Millery, A., Piton, G., and Jean-Christophe, C. (2021). Litter and soil characteristics mediate the buffering effect of snow cover on litter decomposition. Plant Soil 460, 511–525. doi: 10.1007/s11104-020-04803-x
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shannon, C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 623–656. doi: 10.1002/j.1538-7305.1948.tb01338.x
Silver, W. L., and Miya, R. K. (2001). Global patterns in root decomposition: comparisons of climate and litter quality effects. Oecologia 129, 407–419. doi: 10.1007/s004420100740
Six, J., Feller, C., Denef, K., Ogle, S. M., and Albrecht, A. (2002). Soil organic matter, biota and aggregation in temperate and tropical soils-effects of no-tillage. Agronomie 22, 755–775. doi: 10.1051/agro:2002043
Song, P., Ren, H. B., Jia, Q., Guo, J. X., Zhang, N. L., and Ma, K. P. (2015). Effects of historical logging on soil microbial communities in a subtropical forest in southern China. Plant Soil 397, 115–126. doi: 10.1007/s11104-015-2553-y
Stefano, M. (2017). Flexible carbon-use efficiency across litter types and during decomposition partly compensates nutrient imbalances-results from analytical stoichiometric models. Front. Microbiol. 8:661. doi: 10.3389/fmicb.2017.00661
Sun, H., Wang, Q. X., Liu, N., Li, L., Zhang, C. G., Liu, Z. B., et al. (2017). Effects of different leaf litters on the physicochemical properties and bacterial communities in Panax ginseng-growing soil. Appl. Soil Ecol. 111, 17–24. doi: 10.1016/j.apsoil.2016.11.008
Sun, H., Wang, Q., Zhang, C., Li, L., Liu, Z., Liu, N., et al. (2018). Effects of different leaf litter on ginseng soil physicochemical properties and microbial community structure. Acta Ecol. Sin. 38, 3603–3615. doi: 10.5846/stxb201704180697
Swift, M. J., and Anderson, J. M. (1989). “Decomposition,” in Ecosystems of the World, 14B. Tropical Rain Forest Ecosystems: Biogeographical and Ecological Studies. eds. H. Lieth and M. J. A. Werger (Amsterdam: Elsevier), 547–569.
Swift, M. J., Heal, O. W., and Anderson, J. M.. (1979). Decomposition in Terrestrial Ecosystems. California: University of California Press.
Tan, X., Megan, B., Machmuller, M., Francesca, C., and Shen, W. (2020). Shifts in fungal biomass and activities of hydrolase and oxidative enzymes explain different responses of litter decomposition to nitrogen addition. Biol. Fert. Soils 56, 423–438. doi: 10.1007/s00374-020-01434-3
Tonin, A. M., Boyero, L., Monroy, S., Basaguren, A., Pérez, J., Pearson, R. G., et al. (2017). Stream nitrogen concentration, but not plant N-fixing capacity, modulates litter diversity effects on decomposition. Funct. Ecol. 31, 1471–1481. doi: 10.1111/1365-2435.12837
Urbanová, M., Šnajdr, J., and Baldrian, P. (2015). Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 84, 53–64. doi: 10.1016/j.soilbio.2015.02.011
Voříšková, J., and Baldrian, P. (2012). Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 7, 477–486. doi: 10.1038/ismej.2012.116
Wallace, J., Laforest-Lapointe, I., and Kembel, S. W. (2018). Variation in the leaf and root microbiome of sugar maple (Acer saccharum) at an elevational range limit. PeerJ 6:e5293. doi: 10.7717/peerj.5293
Wang, W., Chen, D., Sun, X., Zhang, Q., and Zhang, S. (2019). Impacts of mixed litter on the structure and functional pathway of microbial community in litter decomposition. Appl. Soil Ecol. 144, 72–82. doi: 10.1016/j.apsoil.2019.07.006
Whipps, J. M., Hand, P., Pink, D., and Bending, G. D. (2010). Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 105, 1744–1755. doi: 10.1111/j.1365-2672.2008.03906.x
White, T. J. (1994). Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics, Vol. 38. Pittsburgh, Pennsylvania, USA: Academic Press, 315–322.
Whittaker, R. H. (1960). Vegetation of the siskiyou mountains, Oregon and California. Ecol. Monogr. 30, 279–338. doi: 10.2307/1948435
Whittaker, R. H. (1972). Evolution and measurement of species diversity. Taxon 21, 213–251. doi: 10.2307/1218190
Wright, M. S., and Covich, A. P. (2005). Relative importance of bacteria and fungi in a tropical headwater stream: leaf decomposition and invertebrate feeding preference. Microb. Ecol. 49, 536–546. doi: 10.1007/s00248-004-0052-4
Wymore, A. S., Salpas, E., Casaburi, G., Liu, C. M., Price, L. B., Hungate, B. A., et al. (2018). Effects of plant species on stream bacterial communities via leachate from leaf litter. Hydrobiologia 807, 131–144. doi: 10.1007/s10750-017-3386-x
Xiao, W., Chen, H., Kumar, P., Chen, C., and Guan, Q. (2019). Multiple interactions between tree composition and diversity and microbial diversity underly litter decomposition. Geoderma 341, 161–171. doi: 10.1016/j.geoderma.2019.01.045
Xu, W., Shi, L., and Chan, O. (2013). Assessing the effect of litter species on the dynamic of bacteria and fungi communities during leaf decomposition in microcosm by molecular techniques. PLoS One 8:e84613. doi: 10.1371/journal.pone.0084613
Yao, H., Sun, X., He, C., Maitra, P., Li, X., and Guo, L. (2019). Phyllosphere epiphytic and endophytic fungal community and network structures differ in a tropical mangrove ecosystem. Microbiome 7:57. doi: 10.1186/s40168-019-0671-0
Zeng, Q., Yang, L., and An, S. (2017). Impact of litter quantity on the soil bacteria community during the decomposition of quercus wutaishanica litter. PeerJ 5:e3777. doi: 10.7717/peerj.3777
Zhang, L., Jia, Y., Zhang, X., Feng, X., Wu, J., Wang, L., et al. (2016). Wheat straw: an inefficient substrate for rapid natural lignocellulosic composting. Bioresour. Technol. 209, 402–406. doi: 10.1016/j.biortech.2016.03.004
Zhang, W., Lu, Z., Yang, K., and Zhu, J. (2017). Impacts of conversion from secondary forests to larch plantations on the structure and function of microbial communities. Appl. Soil Ecol. 111, 73–83. doi: 10.1016/j.apsoil.2016.11.019
Keywords: litter leaf, mixed decomposition, microbial community, Populus × canadensis Moench, Pinus sylvestris var. mongolica
Citation: Liu J, Ding C, Zhang W, Wei Y, Zhou Y and Zhu W (2022) Litter mixing promoted decomposition rate through increasing diversities of phyllosphere microbial communities. Front. Microbiol. 13:1009091. doi: 10.3389/fmicb.2022.1009091
Edited by:
Hui Li, Institute of Applied Ecology (CAS), ChinaReviewed by:
Xiyang Zhao, Jilin Agricultural University, ChinaBenye Xi, Beijing Forestry University, China
Copyright © 2022 Liu, Ding, Zhang, Wei, Zhou and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changjun Ding, changjund@126.com
 Jiaying Liu
Jiaying Liu