
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 03 November 2022
Sec. Microbial Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1009018
Oral microbes play a critical role in maintaining oral homeostasis. Microbial dysbiosis promotes disease pathogenesis through several mechanisms. Recent studies have revealed that microbial imbalance and sustained inflammation are involved in disease progression. The adverse interaction triggered by a host immune response to microorganisms can lead to oral and systemic diseases. Here, we reviewed how oral microbes communicate with hosts during the development of local and distant inflammation. Elucidation of these processes may reveal future directions in this field and the potential targets of novel biological therapies for oral and systemic diseases.
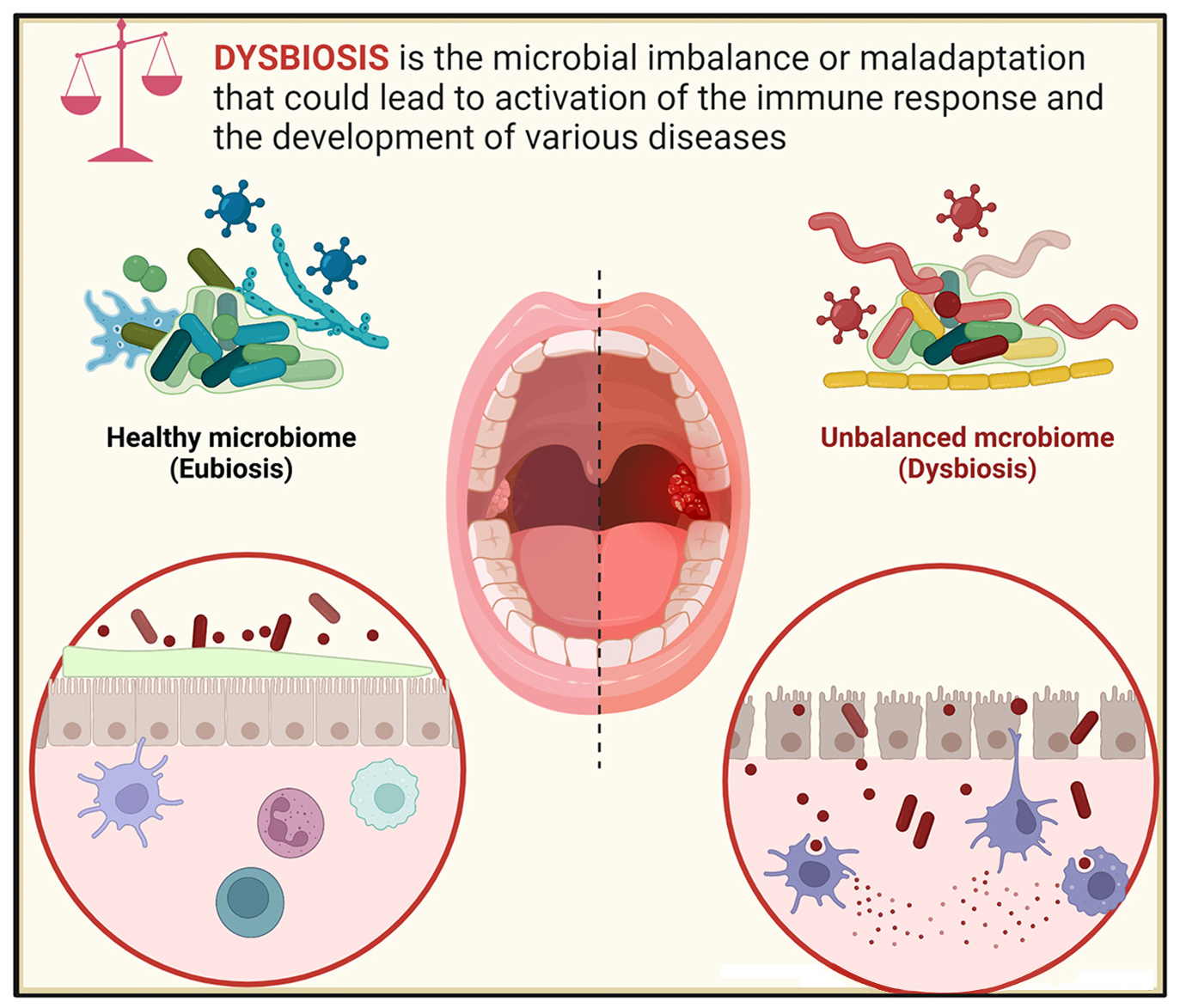
Graphical Abstract. The balance between microbial and immune activities maintains systemic homeostasis. Breaking this balance leads to oral and systemic diseases.
The human oral cavity contains diverse microhabitats. The healthy oral microbiota consists of rich communities of bacteria as well as fungi, viruses, and protozoa. Oral microbes and their interactions help protect the human body against invasion by undesirable organisms. When the microbial balance has been disturbed, inflammatory responses lead to tissue destruction, inducing an unremitting positive feedback loop of oral diseases, including dental caries, periodontal diseases, mucosal diseases (e.g., leukoplakia, lichen planus), and oral cancer, and even several chronic systemic disorders such as cardiovascular disease (CVD), diabetes, rheumatoid arthritis (RA), inflammatory bowel disease (IBD), Alzheimer’s disease (AD), and other cancers (Hajishengallis, 2015; Acharya et al., 2017; Lamont et al., 2018; Genco and Sanz, 2020).
In general, imbalanced microbes (dysbiosis) promote diseases through several mechanisms. First, increased microbial loads can induce an inflammatory response. For example, microbes in periodontal pockets are highly associated with periodontal disease (Diaz, 2012), with a 3-log increase in the microbiota load in periodontitis subjects.
High microbial biomass and community diversity are central features of periodontitis-associated oral microbes (Hajishengallis, 2014). In addition, damage may result from the direct action of microbial enzymes and toxins released by microbiota. Biofilm-forming microbes can destroy the collagen attachment between tooth and bone (You et al., 2017; Yousefi et al., 2020), ultimately causing infectious oral disease. In addition to the increases in microbial biomass and diversity, another mechanism is that periodontal pathogens or virulence factors released by oral microbes circulate through vessels and across the blood–brain barrier, which can induce distant disease (Han and Wang, 2013). For example, bacteremia induced by oral microbes increases the risk for adverse pregnancy outcomes (Saadaoui et al., 2021). Moreover, Porphyromonas gingivalis has been identified in the brains of AD patients (Dominy et al., 2019).
Accumulating evidence indicates that oral microbes are not simply a common risk factor, but multiple microorganism-induced immunological mechanisms are a key point to disease processes. Imbalanced interactions between the periodontal microbiota and the inflammatory response of the host are responsible for promoting some diseases via complex and diverse mechanisms.
Microbiota can induce the local or ectopic disease through activation of the host immune system. Several mechanisms have been proposed for this association. The local or ectopic microbiome can directly affect the proportion and composition of immune cells. For example, P. gingivalis and Lactobacillus rhamnosus GG regulate the balance of T-helper (Th)17 cells and T-cell regulators (Tregs) in colitis via Toll-like receptor (TLR)4 and TLR2 (Hao et al., 2015). Moreover, local or ectopic microbiota may release inflammatory cytokines or stimulate particular immune cells, leading to disease. For example, the oral microbiota promotes colitis via activation of the inflammasome in colonic mononuclear phagocytes and induction of migratory Th17 cells (Sun et al., 2020). Furthermore, tissue destruction in periodontal disease is caused by the host immune response rather than directly by microorganisms or their toxic products, as the host body prevents microorganisms from invading and damaging periodontal tissue. Thus, tissue destruction is an inevitable side effect of an exaggerated host inflammatory response (Wang K. et al., 2015). Microbial dysbiosis and local inflammation can enter a vicious cycle, each increasing the severity of the other.
In addition, recent studies have identified the reciprocal importance of periodontal disease in potentiating systemic disease states (Zheng et al., 2020). For example, periodontitis causes increased serum levels of interleukin (IL)-1β, IL-6, and acute-phase proteins, which might alter the ecological environment and thus support diabetes (Preshaw et al., 2012). Conversely, systemic infections can be detected in the bone marrow, where the process of increasing neutrophil and monocyte infiltration is initiated to fight oral infection (Yang et al., 2018; Cheng et al., 2021). Moreover, some systemic diseases cause the release of factors that worsen oral disease (Grassl et al., 2016). For example, diabetes enhances IL-17 levels and alters the oral microflora, increasing its pathogenicity.
The study of oral microbes and their relationship with the immune system is complex. Elucidating the interactions among oral microorganisms and their cooperation with the host is key to understanding disease progression. This review comprehensively presents an overview of oral and systemic diseases mediated by the interplay between dysbiosis of the oral microbiota and aberrant immune responses. This is important to provide novel insights to support effective prevention and meaningful guidance for future clinical applications.
The oral cavity is exposed to external microbiota, foreign antigens, and environmental threats starting at birth, leading to colonization of the oral mucosa by diverse microbes. Microbial colonization modulates the early development of the immune system in oral mucosal tissues, as well as the development of oral tolerance through the regulation of macrophages and innate lymphoid cells (Abusleme et al., 2013; Kitamoto et al., 2020). These early life events have critical impacts on later health and disease. However, the intricate mechanisms involved in host–microbiota interactions in the oral mucosa under normal conditions are just beginning to be revealed. The mammalian oral mucosa contains various epithelial and stromal cells, along with resident leukocytes, such as γδT cells, neutrophils, innate lymphoid cells (ILCs), and Langerhans cells (LCs) (Jepsen et al., 2018; Kirchner and leibundGut-Landmann, 2021). These innate lymphocytes are recruited before and after birth.
In mice and humans, γδT cells reside in the oral epithelium and gingiva (Figure 1A). They can be divided into Vγ6+, Vγ1+, and Vγ4+ subsets in mice. The Vγ6+ subset is located in the oral epithelium during embryogenesis and expands rapidly upon exposure to the microbiota, while the Vγ1+ and Vγ4+ subsets are recruited to the gingiva after birth (Xiao et al., 2017; Gaffen and Moutsopoulos, 2020). Vγ6+ γδT cells in both the neonatal epithelium and adult mice are the major source of IL-17, a cytokine that plays a key role in oral immunity (Wilharm et al., 2019; Koren et al., 2021). These Vγ6+ γδT17 cells mediate the recruitment of neutrophils to the neonatal oral epithelium in an IL-17-dependent manner (Wahaidi et al., 2011).
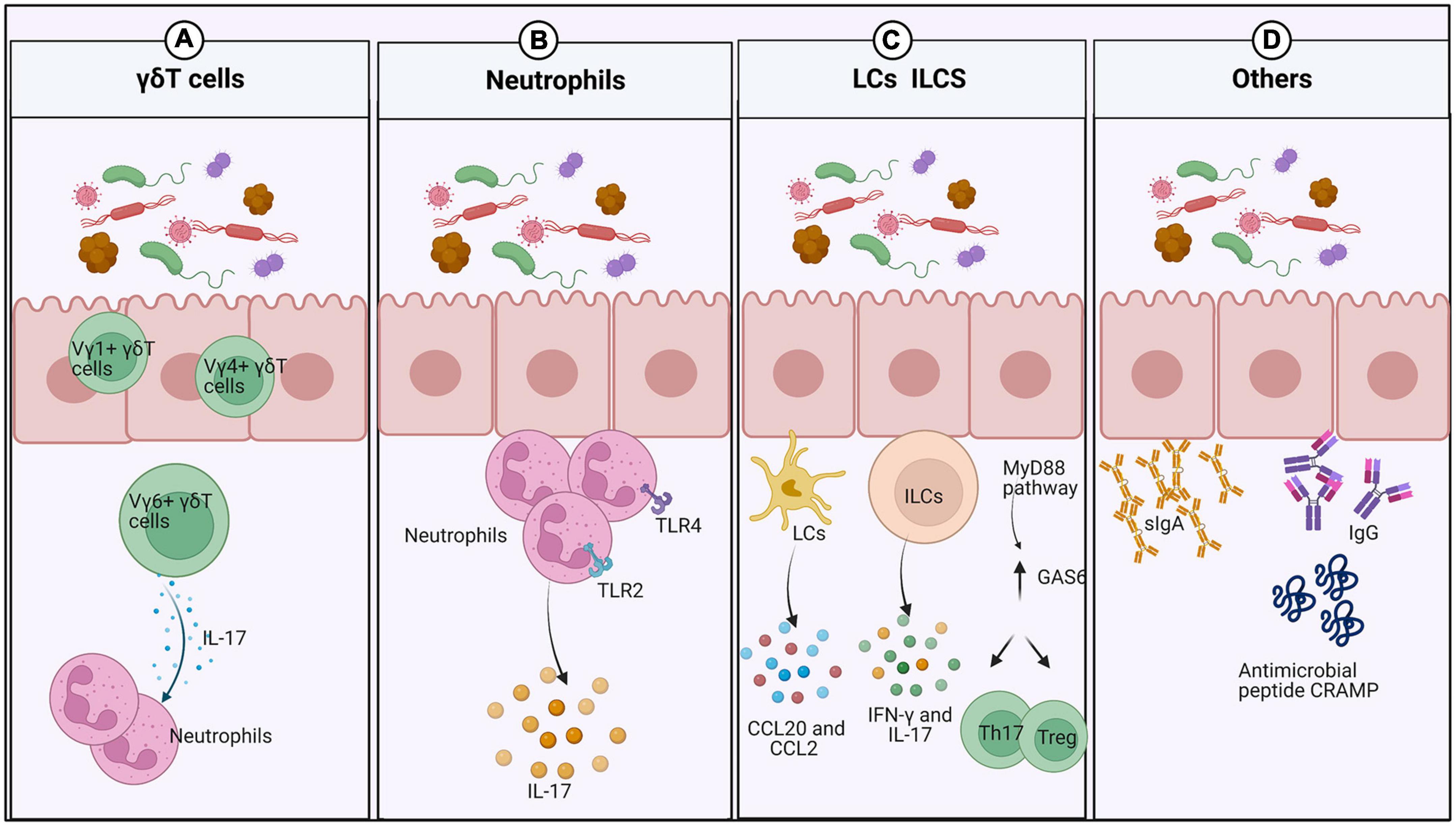
Figure 1. Interplay between the oral mucosal immune system and the microbiota under normal conditions. (A) After exposure to the external microbiota, numerous innate lymphocytes expand into the oral mucosa. γδdT cells increase rapidly in abundance and recruit neutrophils by producing IL-17. (B) Neutrophils increase in response to the local microbiota and IL-17 in the neonatal epithelium, but depend only on IL-17 in the gingiva. (C) Oral ILCs produce IFN-γ and IL-17 to protect the oral mucosa from viral and fungal infection. LCs are recruited by CCL20 and CCL2 to maintain mucosal homeostasis and prevent tissue destruction. In response to oral microbes, expression of GAS6 is upregulated in the gingival epithelium to maintain microbial homeostasis and downregulate gingival inflammation. (D) The oral microbiota promotes the induction of salivary antimicrobial components such as IgA and antimicrobial peptides.
Neutrophils are present prenatally in the oral epithelium, similar to γδT cells, and expand with exposure to the microbiota or transient recruitment by IL-17 (Figure 1B). However, they have also been detected in the gingiva of germ-free (GF) mice, but not in the buccal and gingival epithelium of IL-17–/– mice, suggesting that they are recruited to the gingiva in a manner dependent on IL-17 rather than microbes. Conversely, more neutrophils are present in the junctional epithelium of specific-pathogen-free (SPF) mice than in GF mice (Tordesillas and Berin, 2018; Koren et al., 2021). The contributions of TLR2 and TLR4 signaling in healthy periodontal tissue may account for that finding. Significant increases in the numbers of neutrophils in the junctional epithelium and significant changes in oral microbial composition have been observed in TLR2- and TLR4-deficient mice (Dutzan et al., 2016; Tordesillas and Berin, 2018).
Innate lymphoid cells are present in the oral mucosa of human infants (Williams et al., 2021). Furthermore, oral ILCs reside in the basal layers of the epithelium in mice and protect the uninfected epithelium of the oral mucosa against viral and fungal invasion (Krishnan et al., 2018; Wilharm et al., 2019; Figure 1C).
In mice, LCs, special antigen-presenting cells (APCs) of stratified squamous epithelia such as the skin and mucosa, are recruited by CCL20 and CCL2 to the epithelium postnatally depending on the local microbiota, where they maintain mucosal homeostasis and prevent tissue destruction (Jaitley and Saraswathi, 2012).
In addition to innate lymphocytes, GAS6 and a ligand in the TAM receptor family also play a fundamental role in homeostasis maintenance by the oral epithelium (Tsukamoto et al., 2012; Nassar et al., 2017). A few weeks after birth, oral microbes induce GAS6 expression in the gingival epithelium via a MyD88-dependent signaling pathway, while upregulated GAS6 expression maintains microbial homeostasis, as GAS6–/– mice exhibit elevated inflammation of the oral mucosa with an expansion of the oral microbiota. Moreover, GAS6 downregulates gingival inflammation, and a lack of GAS6 impairs the balance between Th17 cells and Tregs in the gingiva.
The oral microbial load is significantly higher in murine neonates than in adults. The decrease in oral microbial abundance is mediated by the upregulation of saliva production, which has been demonstrated in mice whose parotid salivary glands were surgically removed. However, the capacity to generate saliva flow is not affected by the microbiota, as the rates of saliva production are similar in adult GF and SPF mice (Wahaidi et al., 2011). The microbiota has a major impact on the induction of salivary antimicrobial components such as sIgA, IgG, and the antimicrobial peptide CRAMP (Koren et al., 2021; Figure 1D). Notably, sIgA plays a protective role in regulating and maintaining the normal oral microbiota to prevent microbial dysbiosis (Zenobia et al., 2013). Saliva can promote the attachment of certain bacteria and serve as a microbial nutrient source to affect the composition and activity of the oral microbiota (Chang et al., 2019). In addition, feeding methods also affect infant oral microbiota, and the microbial colonization patterns differ between breast-fed infants and formula-fed infants. There was a higher prevalence of Gemellaceae, Vogesella, Nocardioides (Duale et al., 2021), Actinobacteria, and Proteobacteria in breast-fed babies, while the phylum Bacteroidetes were more prevalent in formula-fed infants (Al-Shehri et al., 2016). Moreover, breast milk-derived cytokines and soluble receptors enhance oral tolerance development (Dawod and Marshall, 2019). It also remains to be seen if the higher relative abundance of some microbes in early life has oral health consequences in later life.
The relationship between oral bacterial colonization and the establishment of a healthy homeostatic mucosal immune system is not well understood. Recently, unique interactions between the epithelium and microbiota in the oral cavity of murine neonates have been studied, providing clues about the foundation of oral homeostasis (Simmerman et al., 2016). After the fragile balance between the host immune system and oral microbiota is established, it can be broken down by various endogenous and exogenous factors.
Orofacial myofunctional disorder (OMD) is a general term for a series of diseases that mainly affect the oral mucosa and soft tissues. It includes infectious diseases, mucosal spot disease, mucosal ulcer lesions, and oral cancer (Lin et al., 2021). Many factors are associated with the pathogenesis of OMD, including immune disorders, microbial infections, allergies, genetic susceptibility, hormonal imbalance, mechanical damage, and stress (Mock et al., 1983; Alrashdan et al., 2016; Vila et al., 2020). Recent studies have shown that imbalanced oral microbiota may lead to the development of oral mucosal diseases (Hu et al., 2019; Yu et al., 2020). Fluctuations in the mucosal microbiota may cause inflammation through interactions with both innate and adaptive immune responses (Baek and Choi, 2018; Pellon et al., 2020; Sami et al., 2020).
Herein, the host immune regulation mechanisms involved in the interactions between periodontal disease and host immune defenses are summarized to elucidate the immune mechanisms employed against the initiation and propagation of oral diseases.
Oral candidiasis (OC) is one of the most common fungal infections caused by Candida albicans, which is the most common Candida species (Akpan and Morgan, 2002) (Figure 2A). Oral epithelial cells (OECs) act as “first responders” to C. albicans infection and activate damage-associated immune effectors, including IL-1-family cytokines and β-defensins. C. albicans-induced IL-1α/β and IL-36 activate innate IL-17-producing cells (Verma et al., 2018). IL-17 binds to its receptor on OECs, inducing the release of antimicrobial effectors, including CXC chemokines and granulocyte colony-stimulating factor (G-CSF). These effectors promote a neutrophil response and direct fungicidal activity to maintain barrier integrity (Aggor et al., 2020). However, the Th17/IL-17 response is double-edged, as it also contributes to immunopathology and aberrant IL-17/Th17 responses, driving the pathogenesis of periodontitis and consequent bone and tooth loss (Bunte and Beikler, 2019). Moreover, the immune attack can be modified in various ways to destroy fungal pathogens. For example, immune cells secrete molecules such as antibodies and complementary compounds that bind to fungal wall components and enhance phagocytosis (Wich et al., 2021). Host-derived immune cells release enzymes to destroy the integrity of the fungal cell wall (Zhou et al., 2021). Thus, the interaction between mucosal immunity and C. albicans involves numerous interconnected host immune regulation mechanisms.
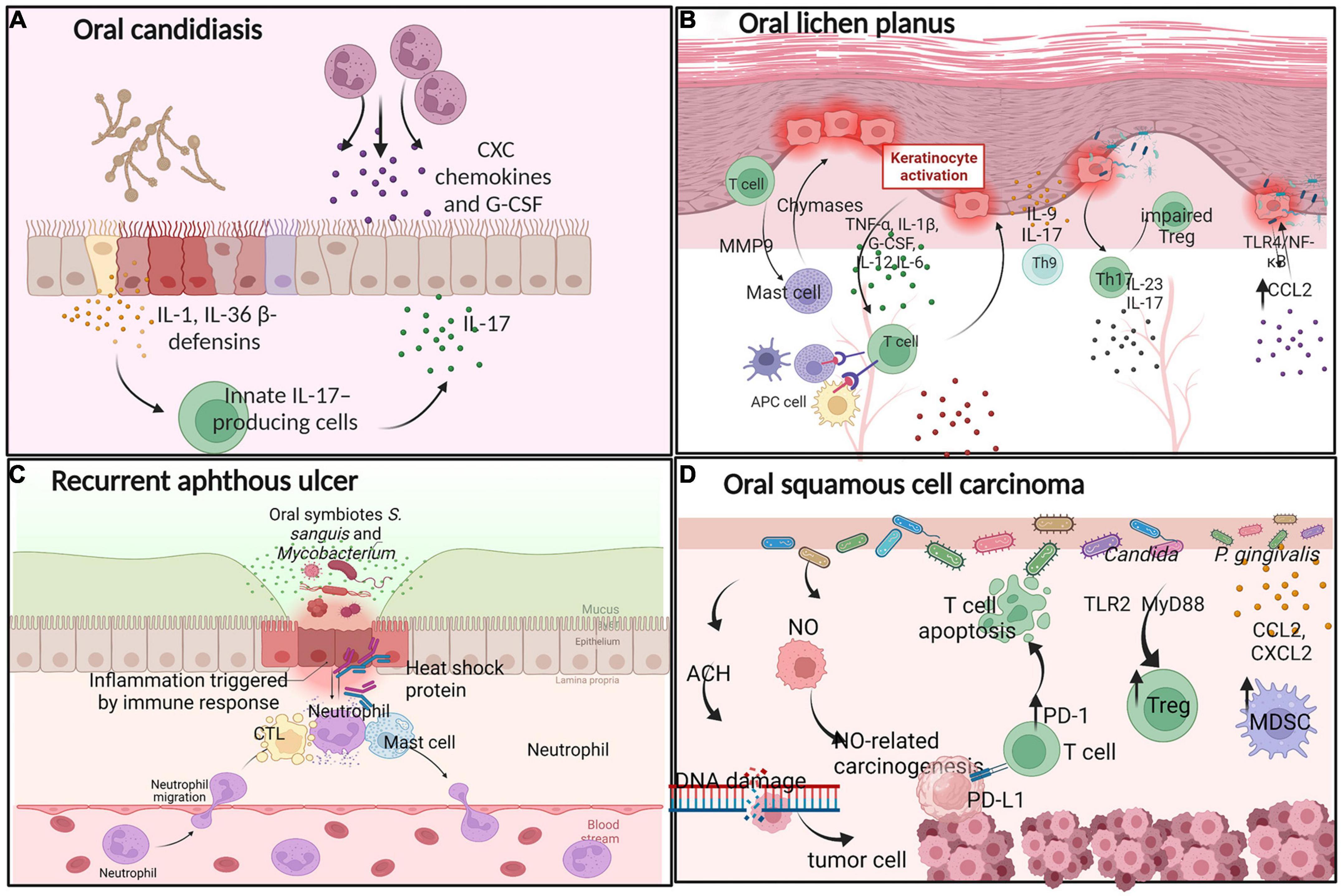
Figure 2. Interactions between oral microbes and immune cells in oral diseases. (A) Oral candidiasis: C. albicans-induced IL-1, IL-36, and β-defensins activate innate IL-17-producing cells to release CXC chemokines and G-CSF, promoting a neutrophil response. (B) Oral lichen planus: Oral keratinocytes release a multitude of proinflammatory cytokines, providing insights into the pathogenetic mechanism of OLP. (C) Recurrent aphthous ulcer: Driven by CTLs, neutrophils, and mast cells. (D) Oral squamous cell carcinoma: Some oral microbes produce ACH and NO, recruiting Treg and MDSCs to the tumor microenvironment to support oral cancer development.
Oral lichen planus (OLP) is a chronic autoimmune, T cell-mediated inflammatory disease of the oral mucosa (Nosratzehi, 2018; Figure 2B). When infection occurs in the oral cavity, OLP-mediating antigens recruit T cells to the target oral mucosa, where they direct an immune response that culminates in keratinocyte cell death and long-term disease. Oral keratinocytes release a multitude of proinflammatory cytokines, including TNF-α, IL-1β, G-CSF, IL-12, and IL-6, to recruit T cells. Activated APCs including dendritic cells (DCs), macrophages, mast cells, and activated oral keratinocytes activate T cells, produce the primary OLP immune response (El-Howati et al., 2022), in which cytotoxic CD8+ T cells drive keratinocyte apoptosis through the release of cytotoxins and CD4+ Th1 T lymphocytes maintain a pro-inflammatory state (Enomoto et al., 2018). Moreover, mast cells produce chymases, which damage the epithelial basement membrane directly or indirectly through the activation of MMP-9 (Wang et al., 2018). Some other Th subsets are involved in the development of OLP. Th17 cells secrete elevated levels of IL-17 and IL-23, while Th9 cells secrete IL-9 and IL-17 in OLP lesions, where these cells are thought to play key roles in oral immunity (Wang et al., 2017). Some evidence indicates that the proportion of peripheral Th17 cells and pro-inflammatory cytokine IL-17 levels are significantly elevated in patients with OLP (Xie et al., 2012; Wang et al., 2017). Pro-inflammatory cytokine IL-17 increases the proportion of functionally impaired CD4+CD25+ regulatory T cells in OLP patients, ultimately leading to OLP (Zhou et al., 2016).
Furthermore, oral bacteria within tissues provide insights into the pathogenetic mechanism of OLP. CCL2 is overproduced in oral keratinocytes and induced by lipopolysaccharide (LPS) via TLR4/NF-κB signaling; increased CCL2 expression is positively associated with the OLP disease score, and thus may sustain or exacerbate the chronic inflammation of OLP (Zeng et al., 2018; Osorio-Osorno et al., 2020). The above evidence suggest that the pathogenesis of OLP includes antigen-specific and non-specific mechanisms. Antigen-specific mechanisms involve T-cell activation following antigen presentation and apoptosis of oral keratinocytes. Non-specific mechanisms consist of the overproduction of CCL2, the activation of MMP-9, and mast cell degranulation. Thus, a complex immune cell relationship exists during the process of OLP.
Recurrent aphthous ulcer (RAU) is a chronic, inflammatory ulcerative disease of the oral mucosa, the pathogenesis of which remains unclear. Recent studies have provided compelling evidence that local immune dysfunction is closely associated with the occurrence of RAU (Saikaly et al., 2018; Figure 2C).
Recurrent aphthous ulcer is driven mainly by microbe-infected cytotoxic T lymphocytes (CTLs) destroying the mucosal epithelium, neutrophils, and mast cells. After ulceration, oral microbes penetrate the ulcerative oral mucosa to further trigger specific immune responses. Eventually, the inflammatory process results in the exudation of fibrinous and necrotic mucosal cells (Tanacan et al., 2022). In addition, metabolites produced by oral microbes damage the oral epithelial structure and may trigger the development of RAU (Hasan et al., 1995). A 65 kDa heat-shock protein produced by Streptococcus sanguinis and Mycobacterium tuberculosis can cross-react with peptides in OECs, producing anti-oral mucosa antibodies and promoting oral ulcers.
Hence, RAU may be driven by an imbalance of proinflammatory and anti-inflammatory cytokines, which leads to excessive immune activation of oral autoantigens and non-pathogenic microorganisms. Although the specific pathogenesis of RAU has been described, it requires further in-depth exploration.
Oral squamous cell carcinoma (OSCC) is the most prevalent and commonly studied malignant tumor of the oral cavity. Mounting evidence indicate that oral microbes are associated with the occurrence of OSCC (Figure 2D). Although smoking, drinking, and betel nut use are common risk factors for OSCC, the factors driving the malignant transformation process are poorly understood (Fantozzi et al., 2021). Emerging evidence suggest links between several types of metabolic damage and oral cancer. For example, some oral microbiota produces acetaldehyde (ACH), which causes DNA damage (Yokoyama et al., 2018). Bacteria release NO, contributing to NO-related carcinogenesis (Kakabadze et al., 2020). Previous research has revealed that the immunopathology of the oral microbiota is associated with the development of oral cancer. For example, Candida infection increases the proportion of Tregs through the TLR2/MyD88 pathway, increasing the fungal burden and disrupting mucous membrane immunity (Pandiyan et al., 2019). P. gingivalis expresses chemokines such as CCL2 and CXCL2 to recruit myeloid-derived suppressor cells (MDSCs) and promote tumor progression (Guo et al., 2021). Furthermore, P. gingivalis induces T cell inhibition via activation of PD-L1 binding to PD-1, which leads to apoptosis of T cells. Thus, P. gingivalis increases the invasiveness of oral cancer cells and their resistance to chemotherapeutic agents. Candida and Fusobacterium nucleatum have similar characteristics. Thus, the oral microbiota is involved in regulating the local growth and diffusion of cancer cells. As research progresses, oral squamous cell carcinoma will certainly become increasingly clear, improving treatment options for OSCC. Oral microbes have been proposed for use as a diagnostic indicator of oral cancer.
Altogether, the mechanisms of oral microbiota-induced carcinogenesis include microbial dysbiosis, colonization, and translocation of oral microbiota that lead to an excessive inflammatory reaction, immunosuppression of host, and promotion of malignant transformation. Here, we described the impact of oral microbial abnormalities on carcinogenesis and introduced new ideas for effective prevention of tumors.
A growing body of literature may become increasingly clear that localized inflammatory lesions may initiate secondary inflammatory infections or disease in distant organs through various modes of transmission. Oral microorganisms can ectopically colonize various tissues, leading to several conditions, including CVD, respiratory tract infection, IBD, AD, adverse pregnancy outcomes, and distant tumors. Here, we discuss how periodontal bacteria activate lymphocytes and cause comorbidities that disseminate to extraoral tissues.
An increasing number of studies suggest that periodontal inflammation and oral microorganisms are associated with elevated and accelerated risks of atherosclerosis and cardiovascular disease (CVD) (Figure 3A). The mechanism through which periodontal pathogens induce atherogenesis could be direct or indirect. The integrity of the oral mucosal basal layer is essential to ensuring that oral bacteria remain outside of the circulatory system (Xie et al., 2020). P. gingivalis is essential to ensure that it directly invades endothelial cells and stimulates numerous innate immune cell types through the TLR-NF-κB signaling pathway. Activated vascular endothelial cells recruit monocytes and elevate circulating lipids, thereby inducing atherogenesis. Furthermore, host genetics regulates lipid-related transcriptome affecting cardiometabolic diseases (CMDs) and controls the differentiation of innate immune cells activated in oral infections. Thus, genetics creates a complex relationship among oral infections (Janket et al., 2015).
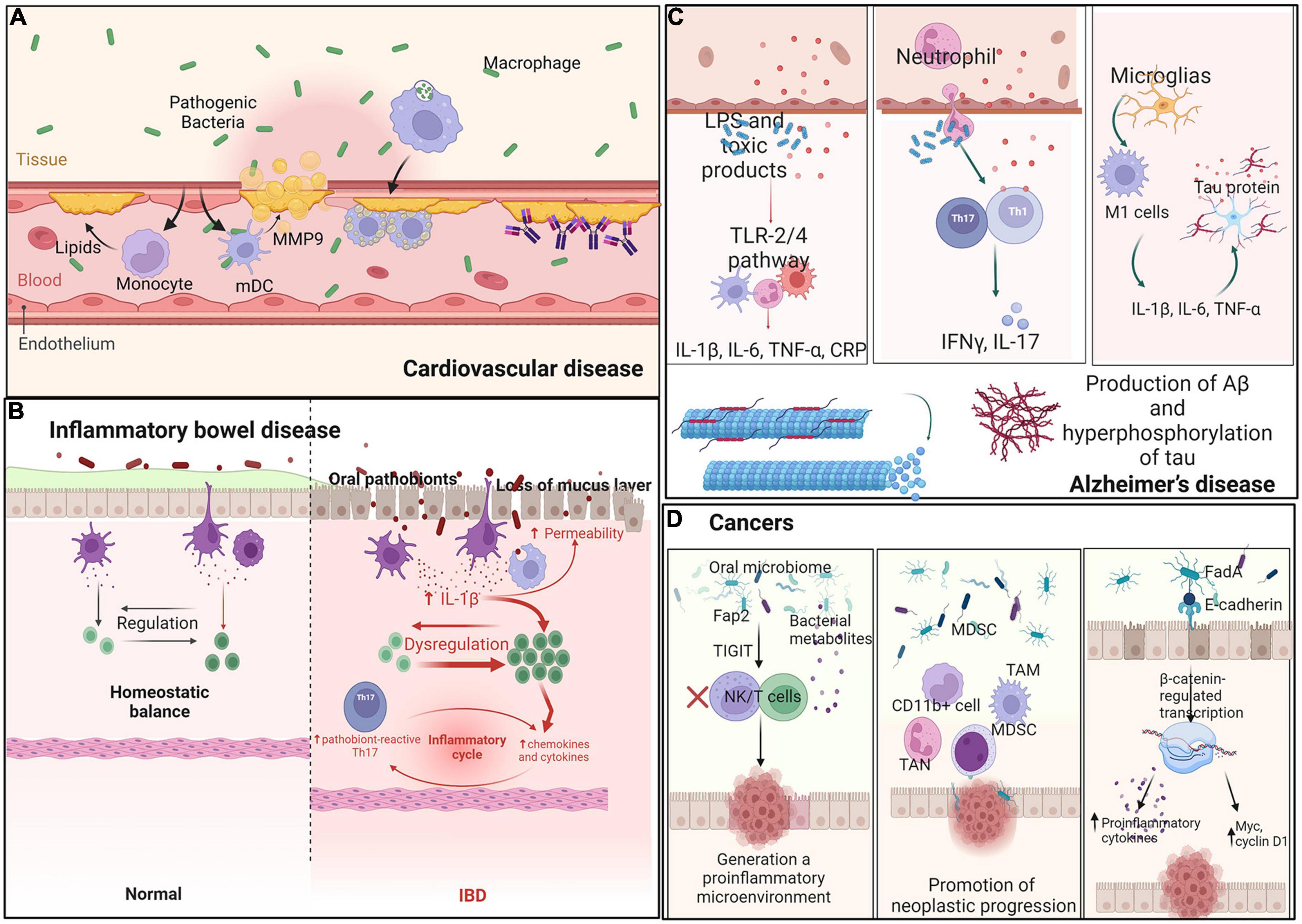
Figure 3. Interaction between oral microbes and immune cells in distant organs. Ectopic oral pathogens travel through other parts of the body, triggering inflammatory diseases. (A) Cardiovascular disease: Oral microbiota recruit monocytes and mDCs and secrete MMP-9 and lipids. Heat-shock proteins and antibodies against b2GP1 and oxLDL promote myocardial infarction. (B) Inflammatory bowel disease: Oral pathobionts translocate to the gut and promote colitis through induction of inflammasome-dependent IL-1β secretion and oral pathobiont-reactive Th17 cells. (C) Alzheimer’s disease: Oral pathobionts increase the production of Aβ and hyperphosphorylation of tau via the TLR-2 and TLR-4 signaling pathways, thereby increasing the levels of IL-17 and IFN-γ produced by Th-cells. (D) Cancer: Oral microbes engage the TIGIT receptor of immune cells, decreasing the abundances of MDSCs, TANs, and TAMs and increasing oncogene expression to generate a proinflammatory microenvironment.
Indirect interactions among periodontal pathogens in CVD include both innate and adaptive immune responses. For example, myeloid DCs (mDCs) infiltrate the oral submucosa via periodontitis and in atherosclerotic plaques. P. gingivalis enhances immature mDC differentiation and causes them to secrete high levels of MMP9 and other indicators of extracellular matrix destruction, plaque rupture, and myocardial infarction (Carrion et al., 2012). Oral gram-negative bacteria increase local levels of proinflammatory LPS molecules, thereby promoting inflammation-induced lipid accumulation in macrophages and ultimately leading to macrophage cholesterol accumulation and foam cell formation (Pietiainen et al., 2018). Another mechanism involves several antibodies recognizing host antigens and modulating their function (cross-reactivity). For example, antibodies are produced against the microbial heat-shock protein (Choi et al., 2021) or cardiolipin (targeting b2-glycoprotein 1, b2GP1) (Wang C.Y. et al., 2015). Saliva contains IgA and IgG antibodies binding to oxidized low-density lipoprotein (oxLDL), which show cross-reactive binding to P. gingivalis, and may participate in immune reactions involved in atherosclerosis and periodontal disease through molecular mimicry (Akhi et al., 2017). The third mechanism occurs with immune system recognition of periodontal bacteria as antigens. A strong combined IgG response to Aggregatibacter actinomycetemcomitans and P. gingivalis is associated with prevalent CVD (Choi et al., 2021). In summary, endotoxemia and systemic antibody responses are possible mediators linking oral dysbiosis to increased risk for CVD.
Oral microbiota can enter the blood circulation or colonize the whole respiratory system through the airways, which have long been recognized as a reservoir for infection of the whole respiratory system. Thus, oral microbes have easier access to the respiratory system than to other organ systems. Various lines of evidence draw links between oral microbes and pneumonia, chronic obstructive pulmonary diseases (COPD), lung cancer, and other respiratory diseases. On one hand, oral microbial dysbiosis leads to oral and respiratory diseases, while on the other hand, oral diseases and respiratory diseases worsen oral microbial dysbiosis.
Research has demonstrated that the same pathogens can be isolated from dental plaque, bronchoalveolar lavage fluid, and lung aspirates. In addition, patients tend to have a significantly lower risk for pneumonia when they receive periodontal treatment (Pathak et al., 2021). Moreover, oral and upper respiratory tract microbes have been detected in the lungs of COPD patients, likely due to the aspiration of oral secretions into the lungs. Periodontitis or caries can alter the oral microbiota and induce systemic inflammation, leading to asthma and bronchitis (Mammen et al., 2020). In turn, respiratory diseases cause oral microbial dysbiosis and oral diseases and can further exacerbate respiratory diseases in a vicious cycle. Future studies are needed to unravel the interactions between oral microorganisms and respiratory diseases.
The oral cavity is the entry point of the gastrointestinal tract, and oral microorganisms are essential drivers of the balance between health and disease. Large quantities of oral bacteria are constantly swallowed, affecting gut microbiota composition and leading to changes in gut epithelial permeability that can lead to bowel disease (Hajishengallis, 2015; Figure 3B). Messora et al. found that a ligature-induced periodontitis model showed changes in intestinal structure, such as defects of the villi, basal lamina degeneration, neutrophil infiltration into the small intestine, and direct damage to the intestinal barrier (Messora et al., 2013). Some IBD patients may have genetic factors leading to the weakening of the mucosal barrier, which increases susceptibility to disease (Merga et al., 2014). Moreover, in another study, the administration of P. gingivalis downregulated the levels of tight junction protein 1 and occludin in the small intestine (Nakajima et al., 2015), consistent with the ligature-induced periodontitis model.
Apart from direct damage to the bowel barrier, oral flora indirectly regulates the immune system to promote the development of bowel disease. Kitamoto et al. showed that amassed oral pathobionts are ingested and translocated to the gut, where they promote colitis through the induction of inflammasome-dependent IL-1β secretion in colonic mononuclear phagocytes. Moreover, oral microbes support the generation of oral pathobiont-reactive Th17 cells in the oral cavity, which then migrate to the inflamed gut. Translocated oral pathobionts, but not gut-resident microbes, activate Th17 cells of oral origin and initiate the development of colitis (Kitamoto et al., 2020). Atarashi et al. found that ectopic gut colonization by oral Klebsiella species increased the abundance of Th1 cells. The Klebsiella-induced Th1 cell response and secretion of IL-18 by TLR4-activated colonic epithelial cells appeared to amplify the Th1 cell response. Jia et al. explored the relationship between P. gingivalis and IBD, in which P. gingivalis exacerbated colitis by activating CD4+ T cells and upregulating the Th17/Treg ratio via the JAK-STAT signaling pathway (Jia et al., 2020), leading to bowel disease.
Taken together, these studies show that ectopic colonization by several oral bacteria may injure the intestinal epithelial barrier, disrupt the host immune system, and eventually alter gut microbiota composition, mainly characterized by decreased diversity along with disruption of immunoregulation and aggravation of chronic intestinal inflammation.
Alzheimer’s disease (AD) is a progressive neurodegenerative condition classically characterized by two hallmark pathologies: amyloid-β (Aβ) plaque deposition and neurofibrillary tangles of hyperphosphorylated tau. There is a strong correlation between AD and oral disease, particularly chronic periodontal diseases (Figure 3C). Dominy et al. reported the presence of P. gingivalis DNA and gingipain antigens in the brains of AD patients (Dominy et al., 2019). Moreover, several models explain the pathology of AD based on oral microbiota.
Periodontitis promotes AD through common pathophysiological pathways. Crossing of the blood–brain barrier, entry into brain tissue, and secretion of LPS by microbiota may occur in AD (Sansores-Espana et al., 2021). Moreover, organisms associated with the periodontal disease produce LPS and toxic products that spread from the ulcerated periodontal pocket into the systemic circulation, initiating classical innate immune signaling pathways via the TLR-2 and TLR-4 mechanisms. Then, the production of pro-inflammatory mediators in the brain, including IL-1β, IL-6, TNF-α, and C-reactive protein (CRP), ultimately increases the production of Aβ and hyperphosphorylation of tau, thus leading to neurodegeneration (Singhrao et al., 2015). Furthermore, peripheral neutrophils have been found in the brains of AD mouse models along with increased levels of IL-17 and interferon-gamma (IFN-γ) produced locally by Th cells (Sadrameli et al., 2020), supporting the hypothesis that chronic inflammation is a strong risk factor for the progressive development of neurodegenerative dementia.
Moreover, bacteria or their virulence factors have been suggested to spread from periodontal tissues to the brain through the trigeminal nerve. Such molecules could induce neuroinflammation mediated by the activated pro-inflammatory microglial phenotype. Under inflammatory conditions, microglia differentiate toward a modulator phenotype or M2, which is characterized by secretion of IL-10 and TGF-β1. When this inflammatory state becomes persistent, M2 shifts to the pro-inflammatory M1 phenotype. M1 cells increase the levels of IL-1β, IL-6, and TNF-α and modify the function of astrocytes, which increase their secretion of pro-inflammatory cytokines, generating Aβ and hyperphosphorylation of tau (Sureda et al., 2020). Moreover, LPS promotes the release of reactive oxygen and nitrogen species such as superoxide ions, iNOS, and NO. Then, vital neurons are destroyed and chronic inflammation occurs, with consequences for the development of disease.
A systematic review reported associations between periodontitis and adverse pregnancy outcomes (APOs). Translocation of bacterial pathogens to the placental barrier results in the restriction of fetal growth and APOs (Lin et al., 2007; Kaur et al., 2014). The most prevalent cultivable oral species associated with APOs is F. nucleatum, which has been detected in a wide variety of placental and fetal tissues. Other oral pathogens, such as P. gingivalis, can induce an approximately two-fold increase in the level of circulating proinflammatory cytokines. Thus, the hematogenous spread of periodontal pathogens, their byproducts (Cobb et al., 2017), and various mediators (Liu et al., 2007) of inflammation generated by the maternal or fetal immune responses cause decidual necrosis. Identification of the species or subspecies involved in APOs will improve early and accurate diagnosis and identification of individuals at risk.
Interactions between oral microbes and the immune system are thought to affect tumor development outside the oral cavity. Oral dysbiosis has been linked not only to head and neck cancer but also to the development of other tumors. Oral microbiota can induce carcinogenesis through the following three mechanisms: by promoting chronic inflammation, eventually leading to oncogene activation and angiogenesis; by affecting cell proliferation and inhibiting cellular apoptosis; and by producing carcinogenic substances. A published review summarized selected carcinogenesis mechanisms associated with oral microbes (Mascitti et al., 2019; Figure 3D).
Oral microbes are thought to influence the development of tumors in the gastrointestinal tract through various mechanisms. For example, LPS and cell extracts of F. nucleatum increase levels of inflammatory cytokines and chemokines, such as CCL20, generating a proinflammatory microenvironment. Fap2 protein engages the TIGIT receptor of immune cells, inhibiting NK and T cell activities and generating a proinflammatory microenvironment that supports colorectal carcinoma progression (Gur et al., 2015). F. nucleatum increases the abundance of CD11b+ myeloid cells, MDSCs, tumor-associated neutrophils (TANs), and tumor-associated macrophages (TAMs), promoting neoplastic progression (Gholizadeh et al., 2017). Moreover, binding of FadA to E-cadherin activates β-catenin-regulated transcription, which increases the expression of transcription factors to increase levels of proinflammatory cytokines and oncogenes, such as Myc and cyclin D1, and stimulates cell proliferation (Cuellar-Gomez et al., 2021). Activation of p38 leads to the secretion of MMP-9 and MMP-13, supporting cell invasion. Moreover, F. nucleatum attaches to host colorectal cancer cells and impairs intestinal barrier function, which might induce intestinal colonization by oral microbes (Wu et al., 2019).
The oral microbe has additionally been linked to pancreatic carcinogenesis. Elevated numbers of certain oral bacteria and the presence of serum antibodies against certain microbial species are associated with a two-fold higher risk for pancreatic cancer (Li et al., 2020). P. gingivalis triggers an innate immune response that involves recognition by TLR4, resulting in the release of pro-inflammatory cytokines. TLRs appear to play a role in pancreatic cancer development; their expression is explicitly higher in human pancreatic cancer (Kerr, 2015). Levels of oral bacteria are significantly elevated in the saliva of lung cancer patients. These mechanisms may provide novel targets and strategies for therapeutic intervention.
In summary, these results support the hypothesis that the interaction between the pathogenesis of periodontitis and disease in distant organs involves a complex interplay between immune-inflammatory responses and dysbiotic microbiota under the influence of environmental and genetic factors. Thus, clarification of the systemic effects of oral microorganisms would contribute to the goal of using the oral cavity to diagnose and treat non-oral diseases.
Evidence is emerging that oral organisms can increase the risk for or severity of autoimmune diseases. The relationship between periodontopathic microbiota and autoimmune diseases has been explored in recent years.
Rheumatoid arthritis (RA) is a systemic autoimmune disease that may reduce life expectancy. It is characterized by the presence of anti-citrullinated protein antibodies (ACPAs) and with clinical signs of enhanced inflammation and bone loss (Figure 4A). Evidence indicates that periodontitis can lead to RA through the production of P. gingivalis peptidyl-arginine-deiminase enzyme, in which the amino acid arginine is converted into citrulline, initiating an immune evasion response and the production of ACPAs (Picchianti-Diamanti et al., 2017). Recent studies have shown that periodontal pathogens directly promote experimentally induced autoimmune arthritis through induction of a TLR2- and IL-1-driven Th17 response, which leads to osteoclast differentiation and bone erosion (de Aquino et al., 2014). In turn, chronic systemic inflammation in RA may affect levels of inflammatory cytokines in the oral cavity (Graves et al., 2019), including TNF-α, IL-1, IL-6, and IL-17. Increased levels of inflammatory mediators in the periodontal tissues of subjects with RA and other diseases may alter local ecologic conditions to favor pathogenic microbial species and promote periodontitis.
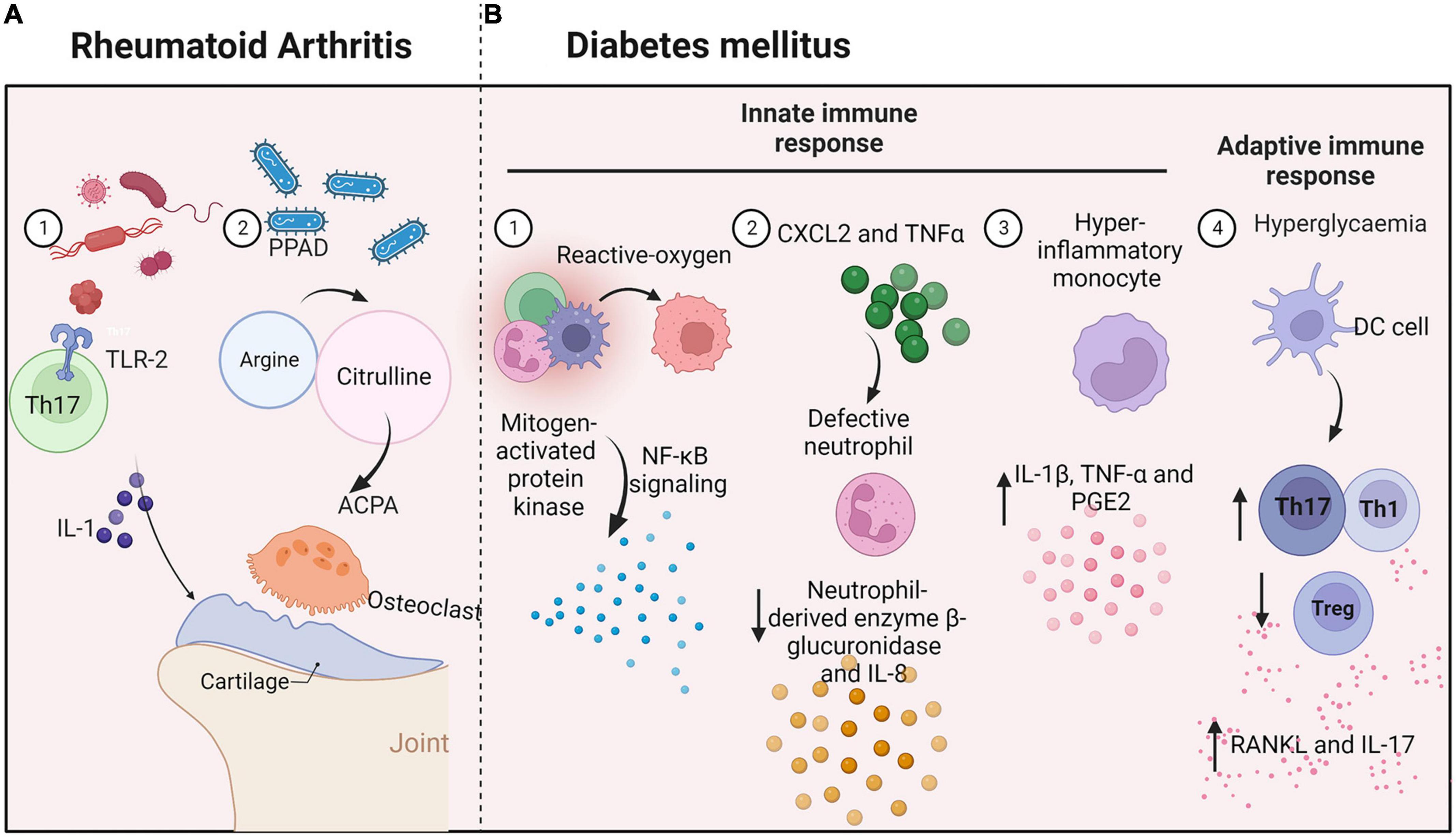
Figure 4. Interconnections between oral microbes and immune cells in systemic diseases. (A) Rheumatoid arthritis: Periodontal pathogens induce a TLR-2 and IL-1-driven Th17 response and increase inflammatory cytokine levels in oral tissues, promoting bone erosion. (B) Diabetes mellitus: Hyperglycemia produces reactive oxygen species and causes defective neutrophils to increase β-glucuronidase and IL-8 secretion. Monocytes with a hyper-inflammatory phenotype produce IL-1β, TNF-α, and PGE2. Moreover, high levels of glucose increase the generation of Th1 or Th17 lymphocytes and reduce the production of Treg cells.
Systemic lupus erythematosus (SLE) is a chronic autoimmune disorder characterized by the production of autoantibodies against nuclear and cytoplasmic antigens, affecting several organs. The diversity of microorganisms in periodontitis lesions is reduced in SLE patients (Aurlene et al., 2020), and microbial components differ between periodontitis and healthy periodontal tissue, indicating that SLE influences microbial diversity. Specifically, the proportion of anaerobic microorganisms (Correa et al., 2017), such as Prevotella nigrescens, Prevotella oulorum, Prevotella oris, and Selenomonas noxia, is elevated in the periodontal area of SLE patients. Meanwhile, the proportions of aerobic representatives of Prevotella, Selenomonas, and Treponema are lower. In turn, the influence of the subgingival microbiota in SLE patients aggravates the severity of periodontitis.
Systemic diseases have significant impacts on periodontitis, with diabetes mellitus having one of the strongest linkages. Diabetes is associated with increased prevalence, severity, and progression of periodontal diseases (Ohtsu et al., 2017; Figure 4B). For example, advanced glycation end-products increase IL-6 and TNF-α levels, leading to increased vascular permeability and the recruitment of inflammatory cells, thereby stimulating increased resorption. Moreover, high levels of glucose increase the levels of reactive oxygen species generated through both enzymatic and non-enzymatic pathways (Huang et al., 2020), leading to activation of mitogen-activated protein kinase and NF-κB signaling and the release of multiple inflammatory factors.
Hyperglycemia directly disrupts immune responses through impacts on immune cell function. For example, a neutrophil function is defective in diabetes, reducing levels of the neutrophil-derived enzyme β-glucuronidase and the chemokine IL-8 (Mutua and Gershwin, 2021). In turn, diabetes activates neutrophils and increases levels of reactive oxygen species, damaging the periodontium (Zheng et al., 2018). Diabetes increases NF-κB activity in periodontal ligament fibroblasts, increasing the expression of CXCL2 and TNFα, and leading to the recruitment of more neutrophils. A study demonstrated that monocytes from patients with type 1 diabetes mellitus have a hyper-inflammatory phenotype (Kraakman et al., 2014). The cells respond to LPS from periodontal bacteria by producing significantly higher levels of IL-1β, TNF-α, and PGE2, possibly prolonging the responses of neutrophils and monocytes/macrophages to plaque bacteria. Moreover, diabetes increases M1 macrophage polarization, increasing susceptibility to periodontal disease.
Diabetes also affects the adaptive immune response, leading to periodontitis. Diabetes can modulate dendritic cells, driving periodontal bone loss through increased abundances of Th1 or Th17 lymphocytes or a reduction of Treg cells (Graves et al., 2020). Th1 and Th17 cells regulate bone resorption and produce cytokines such as RANKL and IL-17, which are elevated in periodontitis patients with diabetes, increasing the pathogenicity of the oral flora, and leading to further recruitment of neutrophils. When IL-17 is inhibited, the microbial community shifts reflected in decreases in both pathogenicity and alveolar bone loss. Diabetes causes periodontal ligament cells, osteoblasts, and osteocytes to increase RANKL expression. Oral disease is associated with the development of systemic inflammation and the onset of insulin resistance in mice.
Although there are no known associations between the relative abundance of oral microbial species and asthma, allergic rhinitis, or atopic dermatitis, an imbalance of the oral flora is generally related to allergic diseases. For example, Proteobacteria is significantly enriched in the oral cavity of atopic dermatitis patients, while pathways for peptidoglycan biosynthesis, lysine biosynthesis, and galactose metabolism are decreased in abundance (Li et al., 2019). By contrast, the pathway for tryptophan metabolism is enriched in the oral cavity of allergic dermatitis patients. Interestingly, the microbes of the skin and oral cavity are relatively variable. A study that assessed the bacterial composition of saliva samples from children with allergic diseases (Dzidic et al., 2018), including eczema, gastrointestinal allergy, asthma, and allergic rhinoconjunctivitis found that Streptococcus parasanguinis and Gemella hemolysans were more prevalent in those children, while the genera Gemella (including G. sanguinis and G. hemolysans), Streptococcus (including S. mitis/dentisani, S. lactarius, and S. cristatus), and Alloprevotella were associated with allergy development (Matsui et al., 2019). In a murine food allergy model, the diversity of oral commensal bacteria was reduced, and the salivary IgA level was significantly elevated (Ho et al., 2021). Moreover, IL-4, IL-5, and IL-13 levels in saliva are elevated in subjects with peanut allergy, which may be associated with Veillonella species (Mariman et al., 2016).
The review has experienced a renewed interest with the recognition that other than traditional therapies which alter oral microbial composition to cure disease, such as the use of antibiotics, the perspectives on immune response modulation are present, which develop prevention and treatment strategies for improving the efficacy. These two effects combined may beneficially affect disease treatment and this combined therapeutic strategy requires intensive research. Moreover, baseline immune cells are a good predictor of clinical response in patients before and after treatment. Studies of the immunotherapy of oral microbiome’s effect on the immune response to oral and systematic diseases are worth pursuing.
This review provides an overview of the complex interactions triggered by the host immune response to the microorganisms in oral and systemic diseases (Supplementary Table). We note that microbial dysbiosis is not simply a common risk factor but triggers multiple microorganism-induced immunological mechanisms to promote disease onset and progression. Elucidation of the interactions will provide more meaningful guidance for future clinical applications. Research in this area is flourishing and in the coming years, more mechanisms and immunotherapies will be revealed.
YL, WQ, YM, and YZ respectively summarized part of this review. YL and CD finalized the manuscript. FC and MC provided professional instruction. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Science Foundation China (81991501), the open project of State Key Laboratory of Oral Diseases (SKLOD2021OF03), and the KCL and PKUHSC Joint Institute for Medical Research Fund (BMU2020KCL003). This work was also sponsored by the interdisciplinary medicine Seed Fund of Peking University (BMU2021MX021), National Key R&D Program of China (SQ2022YFE010688), and the Fundamental Research Funds for the Central Universities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1009018/full#supplementary-material
ACPAs, Anti-citrullinated protein antibodies; AD, Alzheimer’s disease; APCs, Antigen-presenting cells; APOs, Adverse pregnancy outcomes; b2GP1, B2-glycoprotein 1; COPD, Chronic obstructive pulmonary diseases; CTLs, Cytotoxic T lymphocytes; IBD, Inflammatory bowel disease; ILCs, Innate lymphoid cells; LCs, Langerhans cells; GAS6, Growth arrest specific 6; G-CSF, Granulocyte colony stimulating factor; GF, Germ-free; LPS, Lipopolysaccharide; mDCs, Myeloid DCs; MDSCs, Myeloid-derived suppressor cells; MyD88, Myeloid differentiation primary response gene 88; OC, Oral candidiasis; OECs, Oral epithelial cells; OLP, Oral lichen planus; OMD, Oral mucosal disease; OSCC, Oral squamous cell carcinoma; oxLDL, Oxidized-LDL; RAU, Recurrent aphthous ulcer; RA, Rheumatoid arthritis Receptor activator for nuclear factor- κ B ligand (RANKL); SLE, Systemic lupus erythematosus; sIgA, Secretory immunoglobulin A; TAM, TYRO3–AXL–MERTK; TAMs, Tumor-associated macrophages; TANs, Tumor-associated neutrophils; Th17, Type 17 T helper; tjp-1, Tight junction protein 1; TLR, Toll-like receptor; Tregs, T regulatory cells.
Abusleme, L., Dupuy, A. K., Dutzan, N., Silva, N., Burleson, J. A., Strausbaugh, L. D., et al. (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025. doi: 10.1038/ismej.2012.174
Acharya, C., Sahingur, S. E., and Bajaj, J. S. (2017). Microbiota, cirrhosis, and the emerging oral-gut-liver axis. JCI Insight 2:e94416. doi: 10.1172/jci.insight.94416
Aggor, F. E. Y., Break, T. J., Trevejo-Nunez, G., Whibley, N., Coleman, B. M., Bailey, R. D., et al. (2020). Oral epithelial IL-22/STAT3 signaling licenses IL-17-mediated immunity to oral mucosal candidiasis. Sci. Immunol. 5:eaba0570.
Akhi, R., Wang, C., Kyrklund, M., Kummu, O., Turunen, S. P., Hyvarinen, K., et al. (2017). Cross-reactive saliva IgA antibodies to oxidized LDL and periodontal pathogens in humans. J. Clin. Periodontol. 44, 682–691. doi: 10.1111/jcpe.12748
Akpan, A., and Morgan, R. (2002). Oral candidiasis. Postgrad. Med. J. 78, 455–459. doi: 10.1136/pmj.78.922.455
Alrashdan, M. S., Cirillo, N., and Mccullough, M. (2016). Oral lichen planus: A literature review and update. Arch. Dermatol. Res. 308, 539–551. doi: 10.1007/s00403-016-1667-2
Al-Shehri, S. S., Sweeney, E. L., Cowley, D. M., Liley, H. G., Ranasinghe, P. D., Charles, B. G., et al. (2016). Deep sequencing of the 16S ribosomal RNA of the neonatal oral microbiome: A comparison of breast-fed and formula-fed infants. Sci. Rep. 6:38309. doi: 10.1038/srep38309
Aurlene, N., Manipal, S., Prabu, D., and Rajmohan. (2020). Prevalence of oral mucosal lesions, dental caries, and periodontal disease among patients with systemic lupus erythematosus in a teaching hospital in Chennai, Tamil Nadu. J. Fam. Med. Prim. Care 9, 3374–3380. doi: 10.4103/jfmpc.jfmpc_1263_19
Baek, K., and Choi, Y. (2018). The microbiology of oral lichen planus: Is microbial infection the cause of oral lichen planus? Mol. Oral Microbiol. 33, 22–28. doi: 10.1111/omi.12197
Bunte, K., and Beikler, T. (2019). Th17 cells and the IL-23/IL-17 Axis in the pathogenesis of periodontitis and immune-mediated inflammatory diseases. Int. J. Mol. Sci. 20:3394. doi: 10.3390/ijms20143394
Carrion, J., Scisci, E., Miles, B., Sabino, G. J., Zeituni, A. E., Gu, Y., et al. (2012). Microbial carriage state of peripheral blood dendritic cells (DCs) in chronic periodontitis influences DC differentiation, atherogenic potential. J. Immunol. 189, 3178–3187. doi: 10.4049/jimmunol.1201053
Chang, A. M., Liu, Q., Hajjar, A. M., Greer, A., Mclean, J. S., and Darveau, R. P. (2019). Toll-like receptor-2 and -4 responses regulate neutrophil infiltration into the junctional epithelium and significantly contribute to the composition of the oral microbiota. J. Periodontol. 90, 1202–1212. doi: 10.1002/JPER.18-0719
Cheng, Z., Do, T., Mankia, K., Meade, J., Hunt, L., Clerehugh, V., et al. (2021). Dysbiosis in the oral microbiomes of anti-CCP positive individuals at risk of developing rheumatoid arthritis. Ann. Rheum. Dis. 80, 162–168. doi: 10.1136/annrheumdis-2020-216972
Choi, H., Dey, A. K., Priyamvara, A., Aksentijevich, M., Bandyopadhyay, D., Dey, D., et al. (2021). Role of periodontal infection, inflammation and immunity in atherosclerosis. Curr. Probl. Cardiol. 46:100638. doi: 10.1016/j.cpcardiol.2020.100638
Cobb, C. M., Kelly, P. J., Williams, K. B., Babbar, S., Angolkar, M., and Derman, R. J. (2017). The oral microbiome and adverse pregnancy outcomes. Int. J. Womens Health 9, 551–559. doi: 10.2147/IJWH.S142730
Correa, J. D., Calderaro, D. C., Ferreira, G. A., Mendonca, S. M., Fernandes, G. R., Xiao, E., et al. (2017). Subgingival microbiota dysbiosis in systemic lupus erythematosus: Association with periodontal status. Microbiome 5:34. doi: 10.1186/s40168-017-0252-z
Cuellar-Gomez, H., Ocharan-Hernandez, M. E., Calzada-Mendoza, C. C., and Comoto-Santacruz, D. A. (2021). Association of Fusobacterium nucleatum infection and colorectal cancer: A Mexican study. Rev. Gastroenterol. Mex. (Engl Ed). 87, 277–284. doi: 10.1016/j.rgmxen.2021.07.001
Dawod, B., and Marshall, J. S. (2019). Cytokines and soluble receptors in breast milk as enhancers of oral tolerance development. Front. Immunol. 10:16. doi: 10.3389/fimmu.2019.00016
de Aquino, S. G., Abdollahi-Roodsaz, S., Koenders, M. I., Van De Loo, F. A., Pruijn, G. J., Marijnissen, R. J., et al. (2014). Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2- and IL-1-driven Th17 response. J. Immunol. 192, 4103–4111. doi: 10.4049/jimmunol.1301970
Diaz, P. I. (2012). Microbial diversity and interactions in subgingival biofilm communities. Front. Oral Biol. 15:17–40. doi: 10.1159/000329669
Dominy, S. S., Lynch, C., Ermini, F., Benedyk, M., Marczyk, A., Konradi, A., et al. (2019). Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 5:eaau3333. doi: 10.1126/sciadv.aau3333
Duale, A., Singh, P., and Al Khodor, S. (2021). Breast milk: A meal worth having. Front. Nutr. 8:800927. doi: 10.3389/fnut.2021.800927
Dutzan, N., Konkel, J. E., Greenwell-Wild, T., and Moutsopoulos, N. M. (2016). Characterization of the human immune cell network at the gingival barrier. Mucosal Immunol. 9, 1163–1172. doi: 10.1038/mi.2015.136
Dzidic, M., Abrahamsson, T. R., Artacho, A., Collado, M. C., Mira, A., and Jenmalm, M. C. (2018). Oral microbiota maturation during the first 7 years of life in relation to allergy development. Allergy 73, 2000–2011. doi: 10.1111/all.13449
El-Howati, A., Thornhill, M. H., Colley, H. E., and Murdoch, C. (2022). Immune mechanisms in oral lichen planus. Oral Dis. 59, 174–179. doi: 10.1111/odi.14142
Enomoto, A., Sato, E., Yasuda, T., Isomura, T., Nagao, T., and Chikazu, D. (2018). Intraepithelial CD8+ lymphocytes as a predictive diagnostic biomarker for the remission of oral lichen planus. Hum. Pathol. 74, 43–53. doi: 10.1016/j.humpath.2017.12.008
Fantozzi, P. J., Bavarian, R., Tamayo, I., Bind, M. A., Woo, S. B., and Villa, A. (2021). The role of family history of cancer in oral cavity cancer. Head Face Med. 17:48. doi: 10.1186/s13005-021-00298-8
Gaffen, S. L., and Moutsopoulos, N. M. (2020). Regulation of host-microbe interactions at oral mucosal barriers by type 17 immunity. Sci. Immunol. 5:eaau4594. doi: 10.1126/sciimmunol.aau4594
Genco, R. J., and Sanz, M. (2020). Clinical and public health implications of periodontal and systemic diseases: An overview. Periodontol. 2000 83, 7–13. doi: 10.1111/prd.12344
Gholizadeh, P., Eslami, H., and Kafil, H. S. (2017). Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed. Pharmacother. 89, 918–925. doi: 10.1016/j.biopha.2017.02.102
Grassl, N., Kulak, N. A., Pichler, G., Geyer, P. E., Jung, J., Schubert, S., et al. (2016). Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome. Genome Med. 8:44. doi: 10.1186/s13073-016-0293-0
Graves, D. T., Correa, J. D., and Silva, T. A. (2019). The oral microbiota is modified by systemic diseases. J. Dent. Res. 98, 148–156. doi: 10.1177/0022034518805739
Graves, D. T., Ding, Z., and Yang, Y. (2020). The impact of diabetes on periodontal diseases. Periodontol. 2000 82, 214–224. doi: 10.1111/prd.12318
Guo, Z. C., Jumatai, S., Jing, S. L., Hu, L. L., Jia, X. Y., and Gong, Z. C. (2021). Bioinformatics and immunohistochemistry analyses of expression levels and clinical significance of CXCL2 and TANs in an oral squamous cell carcinoma tumor microenvironment of Prophyromonas gingivalis infection. Oncol. Lett. 21:189. doi: 10.3892/ol.2021.12450
Gur, C., Ibrahim, Y., Isaacson, B., Yamin, R., Abed, J., Gamliel, M., et al. (2015). Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42, 344–355. doi: 10.1016/j.immuni.2015.01.010
Hajishengallis, G. (2014). The inflammophilic character of the periodontitis-associated microbiota. Mol. Oral. Microbiol. 29, 248–257. doi: 10.1111/omi.12065
Hajishengallis, G. (2015). Periodontitis: From microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44. doi: 10.1038/nri3785
Han, Y. W., and Wang, X. (2013). Mobile microbiome: Oral bacteria in extra-oral infections and inflammation. J. Dent. Res. 92, 485–491. doi: 10.1177/0022034513487559
Hao, W., Xu, H., Chen, X., Zhou, Q., Zhang, P., Chen, F., et al. (2015). Changes in dental plaque microbial richness and oral behavioral habits during caries development in young Chinese children. Caries Res. 49, 116–123. doi: 10.1159/000366505
Hasan, A., Childerstone, A., Pervin, K., Shinnick, T., Mizushima, Y., Van Der Zee, R., et al. (1995). Recognition of a unique peptide epitope of the mycobacterial and human heat shock protein 65-60 antigen by T cells of patients with recurrent oral ulcers. Clin. Exp. Immunol. 99, 392–397. doi: 10.1111/j.1365-2249.1995.tb05563.x
Ho, H. E., Chun, Y., Jeong, S., Jumreornvong, O., Sicherer, S. H., and Bunyavanich, S. (2021). Multidimensional study of the oral microbiome, metabolite, and immunologic environment in peanut allergy. J. Allergy Clin. Immunol. 148, 627–632.e3. doi: 10.1016/j.jaci.2021.03.028
Hu, L., He, C., Zhao, C., Chen, X., Hua, H., and Yan, Z. (2019). Characterization of oral candidiasis and the Candida species profile in patients with oral mucosal diseases. Microb. Pathog. 134:103575. doi: 10.1016/j.micpath.2019.103575
Huang, C. T., Lue, J. H., Cheng, T. H., and Tsai, Y. J. (2020). Glycemic control with insulin attenuates sepsis-associated encephalopathy by inhibiting glial activation via the suppression of the nuclear factor kappa B and mitogen-activated protein kinase signaling pathways in septic rats. Brain Res. 1738:146822. doi: 10.1016/j.brainres.2020.146822
Jaitley, S., and Saraswathi, T. (2012). Pathophysiology of Langerhans cells. J. Oral Maxillofac. Pathol. 16, 239–244. doi: 10.4103/0973-029X.99077
Janket, S. J., Javaheri, H., Ackerson, L. K., Ayilavarapu, S., and Meurman, J. H. (2015). Oral infections, metabolic inflammation, genetics, and cardiometabolic diseases. J. Dent. Res. 94, 119S–127S. doi: 10.1177/0022034515580795
Jepsen, S., Caton, J. G., Albandar, J. M., Bissada, N. F., Bouchard, P., Cortellini, P., et al. (2018). Periodontal manifestations of systemic diseases and developmental and acquired conditions: Consensus report of workgroup 3 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J. Periodontol. 89 Suppl 1, S237–S248. doi: 10.1002/JPER.17-0733
Jia, L., Wu, R., Han, N., Fu, J., Luo, Z., Guo, L., et al. (2020). Porphyromonas gingivalis and Lactobacillus rhamnosus GG regulate the Th17/Treg balance in colitis via TLR4 and TLR2. Clin. Transl. Immunol. 9:e1213. doi: 10.1002/cti2.1213
Kakabadze, M. Z., Paresishvili, T., Karalashvili, L., Chakhunashvili, D., and Kakabadze, Z. (2020). Oral microbiota and oral cancer: Review. Oncol. Rev. 14:476. doi: 10.4081/oncol.2020.476
Kaur, M., Geisinger, M. L., Geurs, N. C., Griffin, R., Vassilopoulos, P. J., Vermeulen, L., et al. (2014). Effect of intensive oral hygiene regimen during pregnancy on periodontal health, cytokine levels, and pregnancy outcomes: A pilot study. J. Periodontol. 85, 1684–1692. doi: 10.1902/jop.2014.140248
Kirchner, F. R., and leibundGut-Landmann, S. (2021). Tissue-resident memory Th17 cells maintain stable fungal commensalism in the oral mucosa. Mucosal Immunol. 14, 455–467. doi: 10.1038/s41385-020-0327-1
Kitamoto, S., Nagao-Kitamoto, H., Jiao, Y., Gillilland, M. G. III, Hayashi, A., Imai, J., et al. (2020). The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell 182, 447–462.e14. doi: 10.1016/j.cell.2020.05.048
Koren, N., Zubeidat, K., Saba, Y., Horev, Y., Barel, O., Wilharm, A., et al. (2021). Maturation of the neonatal oral mucosa involves unique epithelium-microbiota interactions. Cell Host Microbe 29, 197–209.e5. doi: 10.1016/j.chom.2020.12.006
Kraakman, M. J., Murphy, A. J., Jandeleit-Dahm, K., and Kammoun, H. L. (2014). Macrophage polarization in obesity and type 2 diabetes: Weighing down our understanding of macrophage function? Front. Immunol. 5:470. doi: 10.3389/fimmu.2014.00470
Krishnan, S., Prise, I. E., Wemyss, K., Schenck, L. P., Bridgeman, H. M., Mcclure, F. A., et al. (2018). Amphiregulin-producing gammadelta T cells are vital for safeguarding oral barrier immune homeostasis. Proc. Natl. Acad. Sci. U.S.A. 115, 10738–10743. doi: 10.1073/pnas.1802320115
Lamont, R. J., Koo, H., and Hajishengallis, G. (2018). The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759. doi: 10.1038/s41579-018-0089-x
Li, P., Shu, Y., and Gu, Y. (2020). The potential role of bacteria in pancreatic cancer: A systematic review. Carcinogenesis 41, 397–404. doi: 10.1093/carcin/bgaa013
Li, W., Xu, X., Wen, H., Wang, Z., Ding, C., Liu, X., et al. (2019). Inverse association between the skin and oral microbiota in atopic dermatitis. J. Invest. Dermatol. 139, 1779–1787.e12. doi: 10.1016/j.jid.2019.02.009
Lin, D., Moss, K., Beck, J. D., Hefti, A., and Offenbacher, S. (2007). Persistently high levels of periodontal pathogens associated with preterm pregnancy outcome. J. Periodontol. 78, 833–841. doi: 10.1902/jop.2007.060201
Lin, D., Yang, L., Wen, L., Lu, H., Chen, Q., and Wang, Z. (2021). Crosstalk between the oral microbiota, mucosal immunity, and the epithelial barrier regulates oral mucosal disease pathogenesis. Mucosal Immunol. 14, 1247–1258. doi: 10.1038/s41385-021-00413-7
Liu, H., Redline, R. W., and Han, Y. W. (2007). Fusobacterium nucleatum induces fetal death in mice via stimulation of TLR4-mediated placental inflammatory response. J. Immunol. 179, 2501–2508. doi: 10.4049/jimmunol.179.4.2501
Mammen, M. J., Scannapieco, F. A., and Sethi, S. (2020). Oral-lung microbiome interactions in lung diseases. Periodontol. 2000 83, 234–241. doi: 10.1111/prd.12301
Mariman, R., Reefman, E., Tielen, F., Persoon-Deen, C., Van De Mark, K., Worms, N., et al. (2016). Lactobacillus plantarum NCIMB8826 ameliorates inflammation of colon and skin in human APOC1 transgenic mice. Benef Microbes 7, 215–225. doi: 10.3920/BM2015.0074
Mascitti, M., Togni, L., Troiano, G., Caponio, V. C. A., Gissi, D. B., Montebugnoli, L., et al. (2019). Beyond head and neck cancer: The relationship between oral microbiota and tumour development in distant organs. Front. Cell Infect. Microbiol. 9:232. doi: 10.3389/fcimb.2019.00232
Matsui, S., Kataoka, H., Tanaka, J. I., Kikuchi, M., Fukamachi, H., Morisaki, H., et al. (2019). Dysregulation of intestinal microbiota elicited by food allergy induces IgA-mediated oral dysbiosis. Infect. Immun. 88, e741–e719.
Merga, Y., Campbell, B. J., and Rhodes, J. M. (2014). Mucosal barrier, bacteria and inflammatory bowel disease: Possibilities for therapy. Dig Dis 32, 475–483. doi: 10.1159/000358156
Messora, M. R., Oliveira, L. F., Foureaux, R. C., Taba, M. Jr., Zangeronimo, M. G., Furlaneto, F. A., et al. (2013). Probiotic therapy reduces periodontal tissue destruction and improves the intestinal morphology in rats with ligature-induced periodontitis. J. Periodontol. 84, 1818–1826. doi: 10.1902/jop.2013.120644
Mock, D. M., Scott, M., Mock, N. I., and Thaler, M. M. (1983). The proportion of mitochondrial isoenzyme of aspartate aminotransferase is not elevated in Reye’s syndrome. Pediatr. Res. 17, 884–888. doi: 10.1203/00006450-198311000-00009
Mutua, V., and Gershwin, L. J. (2021). A review of neutrophil extracellular traps (NETs) in disease: Potential Anti-NETs therapeutics. Clin. Rev. Allergy Immunol. 61, 194–211. doi: 10.1007/s12016-020-08804-7
Nakajima, M., Arimatsu, K., Kato, T., Matsuda, Y., Minagawa, T., Takahashi, N., et al. (2015). Oral Administration of P. gingivalis Induces dysbiosis of gut microbiota and impaired barrier function leading to dissemination of enterobacteria to the liver. PLoS One 10:e0134234. doi: 10.1371/journal.pone.0134234
Nassar, M., Tabib, Y., Capucha, T., Mizraji, G., Nir, T., Pevsner-Fischer, M., et al. (2017). GAS6 is a key homeostatic immunological regulator of host-commensal interactions in the oral mucosa. Proc. Natl. Acad. Sci. U.S.A. 114, E337–E346. doi: 10.1073/pnas.1614926114
Nosratzehi, T. (2018). Oral lichen planus: An overview of potential risk factors, biomarkers and treatments. Asian Pac. J. Cancer Prev. 19, 1161–1167. doi: 10.22034/APJCP.2018.19.5.1161
Ohtsu, A., Shibutani, Y., Seno, K., Iwata, H., Kuwayama, T., and Shirasuna, K. (2017). Advanced glycation end products and lipopolysaccharides stimulate interleukin-6 secretion via the RAGE/TLR4-NF-kappaB-ROS pathways and resveratrol attenuates these inflammatory responses in mouse macrophages. Exp. Ther. Med. 14, 4363–4370. doi: 10.3892/etm.2017.5045
Osorio-Osorno, Y. A., Parada-Sanchez, M. T., Arango, J. C., and Arboleda Toro, D. (2020). Oral lichen planus: A chronic inflammatory model to study the regulation of the Toll-like receptor signaling in oral keratinocytes. J. Oral Biosci. 62, 115–122. doi: 10.1016/j.job.2020.05.003
Pandiyan, P., Bhaskaran, N., Zou, M., Schneider, E., Jayaraman, S., and Huehn, J. (2019). Microbiome dependent regulation of tregs and th17 cells in mucosa. Front. Immunol. 10:426. doi: 10.3389/fimmu.2019.00426
Pathak, J. L., Yan, Y., Zhang, Q., Wang, L., and Ge, L. (2021). The role of oral microbiome in respiratory health and diseases. Respir. Med. 185:106475. doi: 10.1016/j.rmed.2021.106475
Pellon, A., Sadeghi Nasab, S. D., and Moyes, D. L. (2020). New insights in Candida albicans innate immunity at the mucosa: Toxins, epithelium, metabolism, and beyond. Front. Cell Infect. Microbiol. 10:81. doi: 10.3389/fcimb.2020.00081
Picchianti-Diamanti, A., Rosado, M. M., and D’amelio, R. (2017). Infectious agents and inflammation: The Role of microbiota in autoimmune arthritis. Front. Microbiol. 8:2696. doi: 10.3389/fmicb.2017.02696
Pietiainen, M., Liljestrand, J. M., Kopra, E., and Pussinen, P. J. (2018). Mediators between oral dysbiosis and cardiovascular diseases. Eur. J. Oral. Sci. 126 Suppl 1, 26–36. doi: 10.1111/eos.12423
Preshaw, P. M., Alba, A. L., Herrera, D., Jepsen, S., Konstantinidis, A., Makrilakis, K., et al. (2012). Periodontitis and diabetes: A two-way relationship. Diabetologia 55, 21–31. doi: 10.1007/s00125-011-2342-y
Saadaoui, M., Singh, P., and Al Khodor, S. (2021). Oral microbiome and pregnancy: A bidirectional relationship. J. Reprod. Immunol. 145:103293. doi: 10.1016/j.jri.2021.103293
Sadrameli, M., Bathini, P., and Alberi, L. (2020). Linking mechanisms of periodontitis to Alzheimer’s disease. Curr. Opin. Neurol. 33, 230–238. doi: 10.1097/WCO.0000000000000797
Saikaly, S. K., Saikaly, T. S., and Saikaly, L. E. (2018). Recurrent aphthous ulceration: A review of potential causes and novel treatments. J. Dermatolog. Treat. 29, 542–552. doi: 10.1080/09546634.2017.1422079
Sami, A., Elimairi, I., Stanton, C., Ross, R. P., and Ryan, C. A. (2020). The role of the microbiome in oral squamous cell carcinoma with insight into the microbiome-treatment axis. Int. J. Mol. Sci. 21:8061. doi: 10.3390/ijms21218061
Sansores-Espana, D., Carrillo-Avila, A., Melgar-Rodriguez, S., Diaz-Zuniga, J., and Martinez-Aguilar, V. (2021). Periodontitis and Alzheimer s disease. Med. Oral. Patol. Oral. Cir. Bucal. 26, e43–e48. doi: 10.4317/medoral.23940
Simmerman, E., Qin, X., Marshall, B., Perry, L., Cai, L., Wang, T., et al. (2016). Innate lymphoid cells: A paradigm for low SSI in cleft lip repair. J. Surg. Res. 205, 312–317. doi: 10.1016/j.jss.2016.06.081
Singhrao, S. K., Harding, A., Poole, S., Kesavalu, L., and Crean, S. (2015). Porphyromonas gingivalis Periodontal Infection and Its Putative Links with Alzheimer’s Disease. Mediators Inflamm. 2015:137357. doi: 10.1155/2015/137357
Sun, J., Tang, Q., Yu, S., Xie, M., Xie, Y., Chen, G., et al. (2020). Role of the oral microbiota in cancer evolution and progression. Cancer Med. 9, 6306–6321. doi: 10.1002/cam4.3206
Sureda, A., Daglia, M., Arguelles Castilla, S., Sanadgol, N., Fazel Nabavi, S., Khan, H., et al. (2020). Oral microbiota and Alzheimer’s disease: Do all roads lead to Rome? Pharmacol. Res. 151:104582. doi: 10.1016/j.phrs.2019.104582
Tanacan, E., Dincer Rota, D., Oktem, R., and Erdogan, F. G. (2022). The correlation of systemic immune-inflammation index, neutrophil-to-lymphocyte ratio, derived neutrophil-to-lymphocyte ratio, and platelet-to-lymphocyte ratio with disease severity in recurrent aphthous stomatitis. J. Cosmet. Dermatol. doi: 10.1111/jocd.14838
Tordesillas, L., and Berin, M. C. (2018). Mechanisms of oral tolerance. Clin. Rev. Allergy Immunol. 55, 107–117. doi: 10.1007/s12016-018-8680-5
Tsukamoto, Y., Usui, M., Yamamoto, G., Takagi, Y., Tachikawa, T., Yamamoto, M., et al. (2012). Role of the junctional epithelium in periodontal innate defense and homeostasis. J. Periodontal. Res. 47, 750–757. doi: 10.1111/j.1600-0765.2012.01490.x
Verma, A. H., Zafar, H., Ponde, N. O., Hepworth, O. W., Sihra, D., Aggor, F. E. Y., et al. (2018). IL-36 and IL-1/IL-17 drive immunity to oral candidiasis via parallel mechanisms. J. Immunol. 201, 627–634. doi: 10.4049/jimmunol.1800515
Vila, T., Sultan, A. S., Montelongo-Jauregui, D., and Jabra-Rizk, M. A. (2020). Oral Candidiasis: A disease of opportunity. J. Fungi (Basel) 6:15. doi: 10.3390/jof6010015
Wahaidi, V. Y., Kowolik, M. J., Eckert, G. J., and Galli, D. M. (2011). Endotoxemia and the host systemic response during experimental gingivitis. J. Clin. Periodontol. 38, 412–417. doi: 10.1111/j.1600-051X.2011.01710.x
Wang, K., Miao, T., Lu, W., He, J., Cui, B., Li, J., et al. (2015). Analysis of oral microbial community and Th17-associated cytokines in saliva of patients with oral lichen planus. Microbiol. Immunol. 59, 105–113. doi: 10.1111/1348-0421.12232
Wang, C. Y., Chyuan, I. T., Wang, Y. L., Kuo, M. Y., Chang, C. W., Wu, K. J., et al. (2015). beta2-Glycoprotein I-dependent anti-cardiolipin antibodies associated with periodontitis in patients with systemic lupus erythematosus. J. Periodontol. 86, 995–1004. doi: 10.1902/jop.2015.140664
Wang, H., Bai, J., Luo, Z., Fu, J., Wang, H., and Sun, Z. (2017). Overexpression and varied clinical significance of Th9 versus Th17 cells in distinct subtypes of oral lichen planus. Arch. Oral Biol. 80, 110–116. doi: 10.1016/j.archoralbio.2017.04.003
Wang, H., Guan, X., Luo, Z., Liu, Y., Ren, Q., and Zhao, X. (2018). The association and potentially destructive role of Th9/IL-9 is synergistic with Th17 cells by elevating MMP9 production in local lesions of oral lichen planus. J. Oral Pathol. Med. 47, 425–433. doi: 10.1111/jop.12690
Wich, M., Greim, S., Ferreira-Gomes, M., Kruger, T., Kniemeyer, O., Brakhage, A. A., et al. (2021). Functionality of the human antibody response to Candida albicans. Virulence 12, 3137–3148. doi: 10.1080/21505594.2021.2015116
Wilharm, A., Tabib, Y., Nassar, M., Reinhardt, A., Mizraji, G., Sandrock, I., et al. (2019). Mutual interplay between IL-17-producing gammadeltaT cells and microbiota orchestrates oral mucosal homeostasis. Proc. Natl. Acad. Sci. U.S.A. 116, 2652–2661. doi: 10.1073/pnas.1818812116
Williams, D. W., Greenwell-Wild, T., Brenchley, L., Dutzan, N., Overmiller, A., Sawaya, A. P., et al. (2021). Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell 184, 4090.e–4104.e. doi: 10.1016/j.cell.2021.05.013
Wu, J., Li, Q., and Fu, X. (2019). Fusobacterium nucleatum contributes to the carcinogenesis of colorectal cancer by inducing inflammation and suppressing host immunity. Transl. Oncol. 12, 846–851. doi: 10.1016/j.tranon.2019.03.003
Xiao, E., Mattos, M., Vieira, G. H. A., Chen, S., Correa, J. D., Wu, Y., et al. (2017). Diabetes enhances IL-17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe 22, 120.e–128.e. doi: 10.1016/j.chom.2017.06.014
Xie, M., Tang, Q., Yu, S., Sun, J., Mei, F., Zhao, J., et al. (2020). Porphyromonas gingivalis disrupts vascular endothelial homeostasis in a TLR-NF-kappaB axis dependent manner. Int. J. Oral. Sci. 12:28. doi: 10.1038/s41368-020-00096-z
Xie, S., Ding, L., Xiong, Z., and Zhu, S. (2012). Implications of Th1 and Th17 cells in pathogenesis of oral lichen planus. J. Huazhong Univ. Sci. Technol. Med. Sci. 32, 451–457. doi: 10.1007/s11596-012-0078-7
Yang, L., Wang, J., Xiao, Y., Wang, X., Sun, Q., Shang, J., et al. (2018). Saliva dysfunction and oral microbial changes among systemic lupus erythematosus patients with dental caries. Biomed Res. Int. 2018:8364042. doi: 10.1155/2018/8364042
Yokoyama, S., Takeuchi, K., Shibata, Y., Kageyama, S., Matsumi, R., Takeshita, T., et al. (2018). Characterization of oral microbiota and acetaldehyde production. J. Oral. Microbiol. 10:1492316. doi: 10.1080/20002297.2018.1492316
You, M., Chan, Y., Lacap-Bugler, D. C., Huo, Y. B., Gao, W., Leung, W. K., et al. (2017). Oral treponeme major surface protein: Sequence diversity and distributions within periodontal niches. Mol. Oral Microbiol. 32, 455–474. doi: 10.1111/omi.12185
Yousefi, L., Leylabadlo, H. E., Pourlak, T., Eslami, H., Taghizadeh, S., Ganbarov, K., et al. (2020). Oral spirochetes: Pathogenic mechanisms in periodontal disease. Microb. Pathog. 144:104193. doi: 10.1016/j.micpath.2020.104193
Yu, F. Y., Wang, Q. Q., Li, M., Cheng, Y. H., Cheng, Y. L., Zhou, Y., et al. (2020). Dysbiosis of saliva microbiome in patients with oral lichen planus. BMC Microbiol. 20:75. doi: 10.1186/s12866-020-01733-7
Zeng, Q., Yang, X., Chen, X., Xia, J., Cheng, B., and Tao, X. (2018). Porphyromonas gingivalis lipopolysaccharide induces over production of CC chemokine ligand 2 via toll-like receptor-4 in oral lichen planus. J. Oral Pathol. Med. 47, 166–172. doi: 10.1111/jop.12655
Zenobia, C., Luo, X. L., Hashim, A., Abe, T., Jin, L., Chang, Y., et al. (2013). Commensal bacteria-dependent select expression of CXCL2 contributes to periodontal tissue homeostasis. Cell Microbiol. 15, 1419–1426. doi: 10.1111/cmi.12127
Zheng, D. W., Li, R. Q., An, J. X., Xie, T. Q., Han, Z. Y., Xu, R., et al. (2020). Prebiotics-encapsulated probiotic spores regulate gut microbiota and suppress colon cancer. Adv. Mater. 32:e2004529. doi: 10.1002/adma.202004529
Zheng, J., Chen, S., Albiero, M. L., Vieira, G. H. A., Wang, J., Feng, J. Q., et al. (2018). Diabetes activates periodontal ligament fibroblasts via NF-kappaB In Vivo. J. Dent. Res. 97, 580–588. doi: 10.1177/0022034518755697
Zhou, L., Cao, T., Wang, Y., Yao, H., Du, G., Chen, G., et al. (2016). Frequently increased but functionally impaired CD4+CD25+ Regulatory T cells in patients with oral lichen planus. Inflammation 39, 1205–1215. doi: 10.1007/s10753-016-0356-9
Keywords: immune system, oral microbiota, oral diseases, systematic diseases, human health
Citation: Liu Y, Qv W, Ma Y, Zhang Y, Ding C, Chu M and Chen F (2022) The interplay between oral microbes and immune responses. Front. Microbiol. 13:1009018. doi: 10.3389/fmicb.2022.1009018
Received: 06 August 2022; Accepted: 30 September 2022;
Published: 03 November 2022.
Edited by:
Aleksander M. Grabiec, Jagiellonian University, PolandReviewed by:
Anu Goenka, University of Bristol, United KingdomCopyright © 2022 Liu, Qv, Ma, Zhang, Ding, Chu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Chu, ZmFtb3VzQGJqbXUuZWR1LmNu; Feng Chen, Y2hlbmZlbmcyMDExQGhzYy5wa3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.