
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 November 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1008905
This article is part of the Research Topic Global Dissemination and Evolution of Epidemic Multidrug-Resistant Gram-Negative Bacterial Pathogens: Surveillance, Diagnosis and Treatment Volume II View all 11 articles
 Zhu Zeng1†
Zhu Zeng1† Lei Lei2†
Lei Lei2† Linman Li3†
Linman Li3† Shengni Hua4
Shengni Hua4 Wenting Li5
Wenting Li5 Limei Zhang5
Limei Zhang5 Qiuping Lin5
Qiuping Lin5 Zhixiong Zheng5
Zhixiong Zheng5 Jing Yang6
Jing Yang6 Xiaohui Dou7*
Xiaohui Dou7* Luan Li8*
Luan Li8* Xiaobin Li5*
Xiaobin Li5*Klebsiella pneumoniae is a primary culprit of antibiotic-resistant nosocomial infections worldwide, and infections caused by NDM-producing strains are a major threat due to limited therapeutic options. The majority of blaNDM cases occur on plasmids; therefore, we explored the relationships between plasmids and blaNDM genes in K. pneumoniae by analyzing the variants of blaNDM, replicon types, conjugative transfer regions of 171 blaNDM-harboring plasmids from 4,451 K. pneumoniae plasmids. Of the nine identified blaNDM variants, blaNDM-1 (73.68%) and blaNDM-5 (16.37%) were the most dominant. Over half of the blaNDM-harboring plasmids of K. pneumoniae were classified into IncF plasmids. IncX3 single-replicon plasmids (46–57 kb) carried genes encoding relaxases of the MOBP family, T4CP genes of the VirD4/TraG subfamily, and VirB-like T4SS gene clusters, which were mainly geographically distributed in China. We found 10 blaNDM-harboring IncN plasmids (38.38–63.05 kb) carrying the NW-type origin of transfer (oriT) regions, genes coding for relaxases of MOBF family, genes encoding T4CPs of the TrwB/TraD subfamily, and Trw-like T4SS gene clusters, which were also mainly geographically distributed in China. Moreover, we identified 21 IncC plasmids carrying blaNDM-1 (140.1–329.2 kb), containing the A/C-type oriTs, genes encoding relaxases of MOBH family, genes encoding T4CPs belonging to TrwB/TraD subfamily, and Tra_F-like T4SS gene clusters. The blaNDM-harboring IncC plasmids were widely geographically distributed all over the world, mainly in the United States, China and Viet Nam. These findings enhance our understanding of the diversity of blaNDM-harboring plasmids in K. pneumoniae.
Klebsiella pneumoniae is a significant cause of nosocomial infections such as pneumonia, bloodstream infections, urinary tract infections, and septicemias (Pitout et al., 2015; Bengoechea and Sa Pessoa, 2019). Klebsiella pneumoniae represents one of the most concerning pathogens known for its high frequency and diversity of antimicrobial resistance (AMR) genes (Navon-Venezia et al., 2017; Wyres and Holt, 2018), and it has been classified as an ESKAPE organism (De Oliveira et al., 2020). The emergence and spread of carbapenem-resistant K. pneumoniae have become severe medical problems worldwide (Navon-Venezia et al., 2017). Resistance to carbapenems in K. pneumoniae involves diverse mechanisms, e.g., production of carbapenemases (e.g., KPC, NDM, and OXA-48-like), alterations in outer membrane permeability and the upregulation of efflux systems (Pitout et al., 2015).
New Delhi metallo-β-lactamase (NDM), belonging to Ambler class B β-lactamase, has the ability to hydrolyze all β-lactam antibiotics (including carbapenems) except the monobactam aztreonam (Nordmann et al., 2011). NDM-1 was first reported in a K. pneumoniae isolate recovered in a Swedish patient who traveled to New Delhi in 2008 (Yong et al., 2009). According to the records of the Beta-lactamase database (BLDB; Naas et al., 2017) on September 8th, 2022, more than 40 variants of NDM have been identified so far. A variety of infections caused by NDM-producing Enterobacterales strains are associated with inferior prognosis and high mortality, especially in high-risk immunocompromised patients (Guducuoglu et al., 2018). NDM-producing Enterobacterales clinical isolates, mainly K. pneumoniae and Escherichia coli, have been found worldwide, with a higher prevalence in the Indian subcontinent, the Balkans, and the Middle East (Albiger et al., 2015; Wu and Feng, 2019).
Antimicrobial resistance (AMR) in carbapenem-resistant Enterobacterales (CRE) strains is often encoded by the plasmid-borne genes (Rozwandowicz et al., 2018). Plasmids, especially conjugative plasmids, play an essential role in mediating horizontal gene transfer (HGT) and dissemination of AMR (Jiang et al., 2020). The conjugative transfer regions of conjugative plasmids typically comprise four key modules, including origin of transfer (oriT) region, gene encoding relaxase, gene encoding type IV coupling protein (T4CP), and gene cluster for the bacterial type IV secretion system (T4SS) apparatus (de la Cruz et al., 2010). The relaxase initiates the bacterial conjugation by recognizing and cleaving the oriT of the plasmid in a site-specific manner, forming a relaxosome (Llosa et al., 2002; Carballeira et al., 2014). Currently, nine types of plasmid-borne oriT1 and eight main relaxase families2 have been identified (Li et al., 2018). Conjugation requires a pilus, which is assembled by T4SS, to connect the donor and the recipient strains (de la Cruz et al., 2010). Currently, five main types of T4SS gene clusters are defined, including 18 different kinds of systems3 (Bi et al., 2013). The T4CP connects the relaxosome to T4SS, which is required for conjugation, and currently, two main subfamilies of T4CPs4 exist (Li et al., 2018).
Studies on the comprehensive analysis of blaNDM-harboring plasmids and their conjugative transfer regions in K. pneumoniae are scarce. In this work, we executed in silico typing and comparative analysis of blaNDM-harboring plasmids of K. pneumoniae using the bacterial plasmids available in the NCBI GenBank database. We systematically analyzed the variants of blaNDM, replicon types, phylogenetic patterns, and conjugative transfer regions of the blaNDM-positive plasmids of K. pneumoniae. This study provides deep insights into the characteristics and diversity of blaNDM-harboring plasmids in K. pneumoniae and further emphasizes their role in dissemination of resistance genes.
The GenBank Genome database (Benson et al., 2018) collect all the plasmids belonging to K. pneumoniae.5 A total of 4,451 plasmids (without duplicates) of K. pneumoniae (Supplementary Table S1) were downloaded on April 26th, 2022. Files in FASTA DNA format of the 4,451 plasmids were downloaded in batches into our Linux-based server.
The β-lactamase genes of the plasmids of K. pneumoniae were identified applying the ResFinder software, standalone version 4.1 (Bortolaia et al., 2020), with the minimum coverage of 60%, minimum identity of 90%, and species of “Klebsiella.” The term “blaNDM” was used to search in the “Resistance gene” list of the ResFinder results in order to judge the blaNDM-harboring plasmids of K. pneumoniae and identify the variants of the blaNDM genes. For some blaNDM-harboring plasmids, the variants of blaNDM were not determined by the ResFinder software; instead, they were submitted to the CARD database6 (Alcock et al., 2020) and the Beta-lactamase database (BLDB; Naas et al., 2017) for further analysis.
Replicon typing of the blaNDM-harboring plasmids was executed via the PlasmidFinder software (Carattoli and Hasman, 2020). Then, selecting the database “Enterobacteriales,” the FASTA-formatted DNA files were analyzed and classified in batches by using the PlasmidFinder tool version 2.0.1, with a minimum coverage cut-off of 60% and minimum identity cut-off of 95%. The database version was updated on November 29th, 2021.
The files of the blaNDM-harboring plasmids of K. pneumoniae, in GenBank format, were downloaded in batches using two Bioperl modules (Bio::SeqIO and Bio::DB::GenBank). Plasmid files containing protein sequences were compiled from the plasmid files in GenBank format through the Bioperl/Bio::SeqIO module. Phylogenetic cladogram based on the presence/absence of orthologous gene families of all the blaNDM-harboring plasmids of K. pneumoniae were constructed. First, a binary gene presence/absence matrix was built using OrthoFinder software (Emms and Kelly, 2019), and subsequently a hierarchical cluster result was generated by PAST3 (Hammer et al., 2001) and eventually displayed by iTOL (Letunic and Bork, 2016).
Information about geographic location of blaNDM-harboring plasmids and its host strains were extracted from the files of the blaNDM-harboring plasmids in GenBank format. Table containing the correspondence between strains and plasmids of K. pneumoniae were downloaded from the GenBank.7 The blaNDM-harboring plasmid-matched host K. pneumoniae strains were collected, and their DNA FASTA sequences were downloaded in batch using the Bioperl. The MLST software (Larsen et al., 2012) version 2.0.9 was downloaded from the website8 and installed on the Linux platform. The genomes of K. pneumoniae strains were analyzed in batch using MLST software.
Files in GenBank format of the blaNDM-harboring plasmids in K. pneumoniae were analyzed in batches using oriTfinder software (local version; Li et al., 2018) to identify the presence/absence of oriTs, relaxase-coding genes, T4CP-coding genes, and gene clusters for T4SS. Furthermore, the types of oriTs, relaxases, T4CPs, and T4SSs toward the plasmids were determined based on the exhibition of the oriTDB database9 (Li et al., 2018). In addition, the types of gene clusters for T4SS were classified based on the SecReT4 database10 (Bi et al., 2013).
The bipartite network was constructed based on all the blaNDM-harboring plasmids of K. pneumoniae using the AccNet software using default parameters (Lanza et al., 2017). The obtained network files including nodes, edges and clusters were then imported into the Cytoscape software (Shannon et al., 2003) for visualization. We displayed the relative genomic content of each plasmid by making the diameter of each node proportional to its degree.
Based on the results analyzed by ResFinder, 171 (3.84%) blaNDM-harboring plasmids (Supplementary Table S2) were identified from 4,451 plasmids of K. pneumoniae, which were downloaded from the GenBank Genome database. Among the 171 blaNDM-harboring plasmids of K. pneumoniae, nine different variants of blaNDM were identified (Figure 1A). Among the nine variants of blaNDM, blaNDM-1 was found to be the predominant variant, accounting for 73.68% (126 blaNDM-1-harboring plasmids), followed by blaNDM-5, accounting for 16.37% (28 blaNDM-5-harboring plasmids) (Figure 1A).
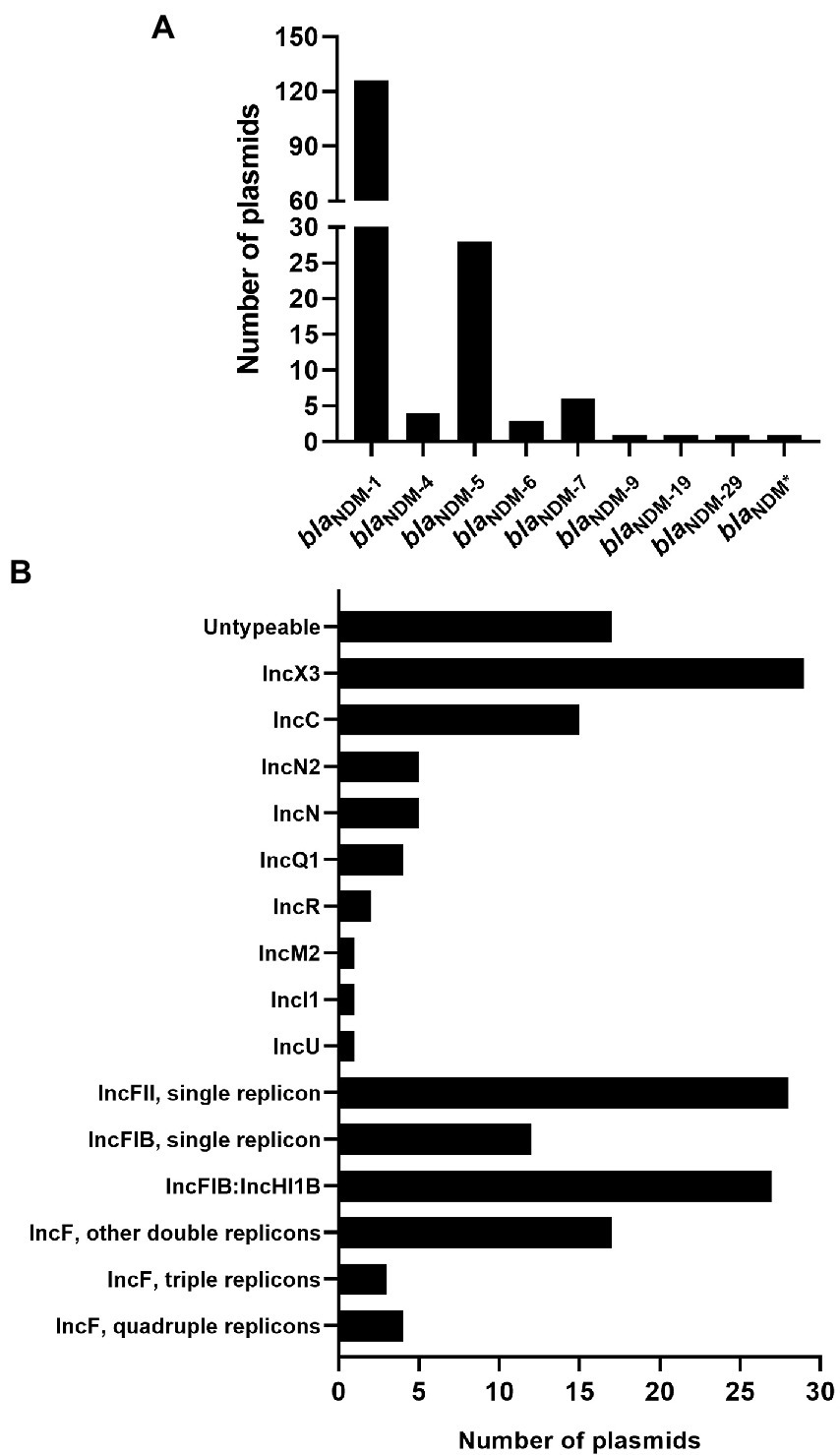
Figure 1. Characteristics of the 171 blaNDM-harboring plasmids of Klebsiella pneumoniae. (A) Histogram of number of variants of blaNDM genes among the 171 blaNDM-harboring plasmids of K. pneumoniae. blaNDM* representing an unknown variant of blaNDM. (B) An overview of the plasmid replicon types among the 171 blaNDM-harboring plasmids of K. pneumoniae analyzed using software PlasmidFinder.
Replicon typing of the 171 blaNDM-harboring plasmids of K. pneumoniae was executed using PlasmidFinder. Of the 171 plasmids, 154 were successfully identified with their replicon types, including 103 single-replicon plasmids and 51 multi-replicon plasmids (44 plasmids with two replicons, three plasmids with three replicons, and four plasmids with four replicons; Figure 1B; Supplementary Figure S1). For the 103 single-replicon plasmids harboring blaNDM in K. pneumoniae, the TOP5 prevalent replicons (in descending order) were IncX3 (29 plasmids), IncC (15 plasmids), IncFIB(pQil) (11 plasmids), IncFII (11 plasmids), and IncFII(Yp) (11 plasmids). Of the 44 blaNDM-harboring plasmids with two replicons, 25 contained replicons IncFIB(pNDM-Mar) and IncHI1B(pNDM-MAR), which were the most prevalent two-replicon plasmids harboring blaNDM in K. pneumoniae (Figure 1B; Supplementary Figure S1).
In summary, 21 of the 171 blaNDM-harboring plasmids of K. pneumoniae were found to carry the replicon of IncC, accounting for 12.28% of all the blaNDM-harboring plasmids of K. pneumoniae in this study (Figure 1B; Supplementary Figure S1). Notably, 91 of the 171 blaNDM-harboring plasmids in our study were found to be the IncF plasmids, including IncFI and IncFII plasmids, accounting for 53.22% of all the blaNDM-harboring plasmids of K. pneumoniae in this study (Figure 1B; Supplementary Figure S1).
To get the comprehensive overview of blaNDM-harboring plasmids in K. pneumoniae, we created a phylogenetic cladogram of the 171 blaNDM-harboring plasmids (Figure 2). Based on phylogenetic patterns of the 171 plasmids, combined with the replicon types, conjugative transfer regions, and genome sizes of the blaNDM-harboring plasmids, most of the 171 blaNDM-harboring plasmids were clustered into 10 main clades (clades I–X), representing 10 plasmid patterns carrying blaNDM genes in K. pneumoniae (Table 1).

Figure 2. Details of variants of blaNDM genes, replicon types, conjugative transfer regions, geographical distributions, host STs, and length distribution of the 171 blaNDM-harboring plasmids of K. pneumoniae. The four categories of information present in this figure include the phylogenetic cladogram, variants of blaNDM, replicon types, conjugative transfer regions (oriT, relaxase, T4CP, and T4SS), geographical distributions, ST types of host strains, and length distribution of the 171 blaNDM-harboring plasmids of K. pneumoniae. blaNDM* representing an unknown variant of blaNDM.
Clade I: A total of 29 IncX3 plasmids were found in the clade I cluster, mainly blaNDM-1 and blaNDM-5 (Figure 2). For the 29 IncX3 plasmids harboring blaNDM, their genome sizes varied from 45.1 to 159.3 kb (25th percentile = 46.2 kb; 75th percentile = 57.3 kb), with a median size of 53.1 kb (Supplementary Figure S2). For the conjugative transfer regions, all the plasmids belonging to clade I were found to carry genes encoding relaxases of the MOBP family characterized by the domain “Relaxase (Pfam: PF03432),” T4CPs of the VirD4/TraG subfamily characterized by the domain “T4SS-DNA_transf (Pfam: PF02534),” and VirB-like T4SS gene clusters (Figure 2; Supplementary Figure S3). Members of clade I were mainly geographically distributed in China (Figure 3; Table 1; Supplementary Table S3). No predominant ST types of isolates were found in the plasmids harboring blaNDM in K. pneumoniae (Table 1; Supplementary Table S3).
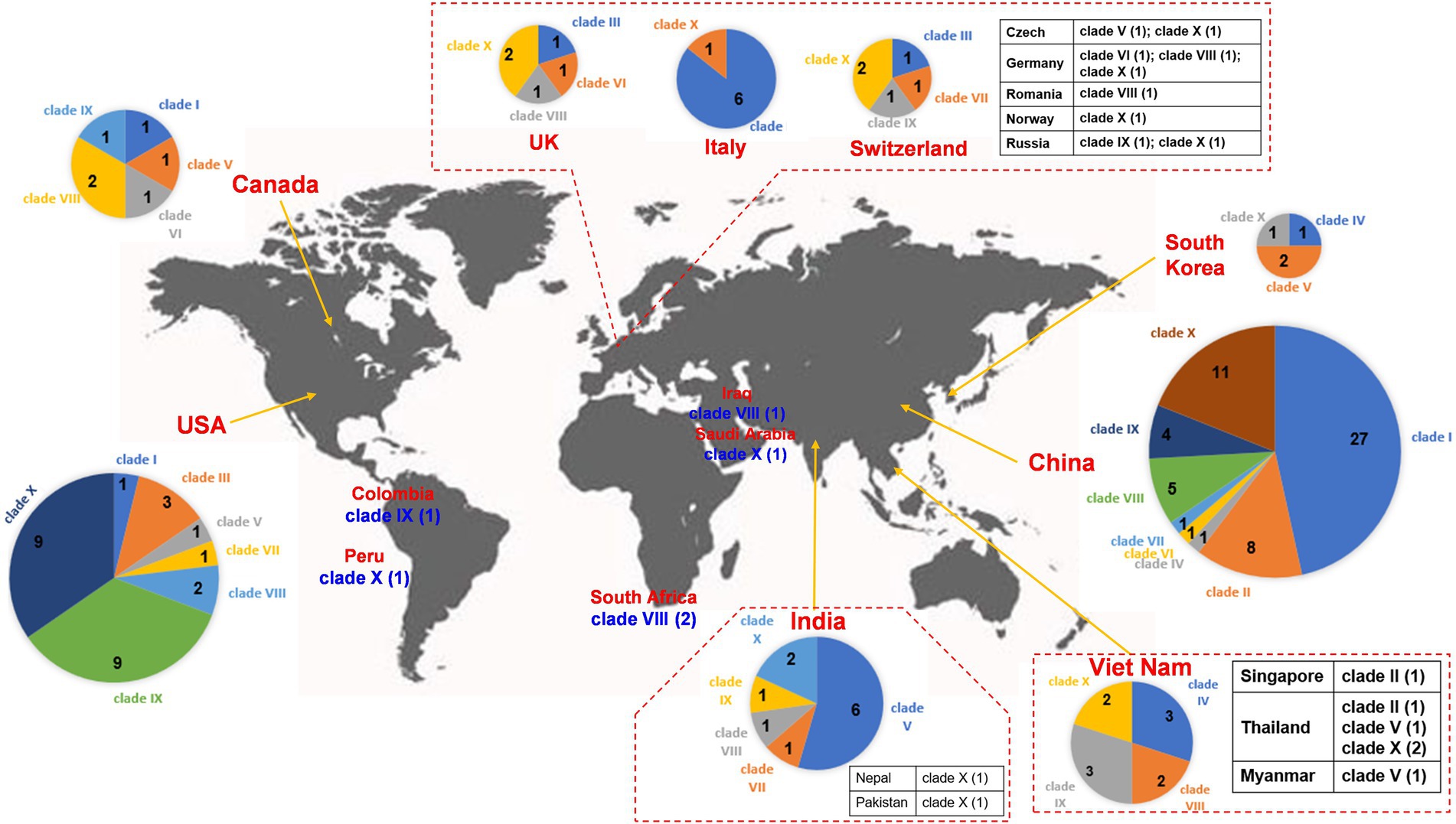
Figure 3. Worldwide distribution of blaNDM-harboring plasmids of K. pneumoniae. The geographical distribution of the 10 clades (Clade I–Clade X) from the blaNDM-positive plasmids of K. pneumoniae was calculated and displayed by pie chart. For the plasmids isolated in European countries, only those from the United Kingdom, Italy, and Switzerland were displayed by pie chart, others were displayed in the tabular form. For the plasmids isolated in Southeast Asia, only those from Viet Nam were displayed by pie chart, others were displayed in the tabular form. For the plasmids isolated in South Asia, only those from India were displayed by pie chart, others were displayed in the tabular form.
Clade II: Ten blaNDM-positive IncN plasmids were clustered into clade II, mainly carrying blaNDM-1 (Figure 2). The genome sizes of the 10 blaNDM-harboring IncN plasmids varied from 38.4 to 63.1 kb (25th percentile = 47.2 kb; 75th percentile = 59.8 kb), with a median size of 52.0 kb (Supplementary Figure S1). Almost all the IncN plasmids carried the NW-type oriTs and genes encoding relaxases of MOBF family characterized by the domain “TrwC (PF08751).” All the 10 blaNDM-positive IncN plasmids carried the genes encoding T4CPs of the TrwB/TraD subfamily characterized by the domain “TrwB_AAD_bind (PF10412)” and Trw-like T4SS gene clusters (Figure 2; Supplementary Figure S4). The members of clade II were mainly geographically distributed in China (Figure 3; Table 1; Supplementary Table S3). The 10 blaNDM-positive IncN plasmids were distributed in seven ST types of K. pneumoniae strains, and four plasmids were distributed in K. pneumoniae ST15 (Table 1; Supplementary Table S3).
Clade III: Eleven blaNDM-1-positive IncF plasmids with the IncFIB(pQil) replicon were grouped into clade III, and most were 54-kb plasmids (Figure 2). Moreover, no conjugative transfer regions were identified in the 11 plasmids of clade III, indicating that the 11 plasmids should be non-transferable. Plasmids belonging to clade III were mainly geographically distributed in Italy and United States (Figure 3; Table 1; Supplementary Table S3). All the members of clade III were harbored by the strains of K. pneumoniae ST147 (Table 1; Supplementary Table S3).
Clade IV: Five blaNDM-positive untypeable plasmids were clustered into clade IV, involving three blaNDM-1-positive plasmids and two blaNDM-4-positive plasmids (Figure 2). These five untyped plasmids, with lengths ranging from 75.3 to 86.0 kb, all carried the genes encoding relaxases of MOBF family, genes encoding T4CPs of TrwB/TraD subfamily, and Tra_F-like T4SS gene clusters (Figure 2; Supplementary Figure S5). For the five plasmids, three were found in Viet Nam, one was found in China, and one was found in South Korea (Figure 3; Table 1; Supplementary Table S3). The STs of K. pneumoniae host strains containing the clade IV plasmids were distributed into ST395 and ST16 (Table 1; Supplementary Table S3).
Clade V: Thirteen plasmids with the IncFII replicon, mainly carrying blaNDM-5, were classified into the clade V (Figure 2). For the 13 IncFII plasmids harboring blaNDM, genome sizes varied from 75.3 to 140.6 kb (25th percentile = 88.8 kb; 75th percentile = 101.4 kb), with a median size of 96.2 kb (Supplementary Figure S1). They all carried the F-type oriTs and Tra_F-like T4SS gene clusters (Figure 2; Supplementary Figure S6). Most of the plasmids clustered into clade V were found to carry genes encoding relaxases of the MOBF family and genes encoding T4CPs of the TrwB/TraD subfamily (Figure 2). The members of clade V were widely distributed in India, Southeast Asia, North America, East Asia, and Europe, with the highest prevalence in India (Figure 3; Table 1; Supplementary Table S3). The STs of K. pneumoniae host strains containing all Clade V plasmids were mainly distributed in ST16, ST147, and ST2096 (Table 1; Supplementary Table S3).
Clade VI: Four blaNDM-1-positive plasmids, including one IncM2 plasmid and three untyped plasmids, were classified into a small cluster named clade VI in our study (Figure 2). These four plasmids, with lengths ranging from 75.6 to 100.2 kb, all carried the L/M-type oriTs, genes encoding relaxases of MOBP family, genes encoding T4CPs of TrwB/TraD subfamily, and Tra_I-like T4SS gene clusters (Figure 2; Supplementary Figure S7). The four plasmids were sporadically discovered in Canada, Germany, United Kingdom, and China (Figure 3; Table 1; Supplementary Table S3). No prevalent STs of K. pneumoniae host strains containing all clade VI plasmids were found (Table 1; Supplementary Table S3).
Clade VII: Four blaNDM-1-positive plasmids with the IncFII(pKPX1) replicon were classified into clade VII (Figure 2). The genome sizes of the four IncFII(pKPX1) plasmids varied from 96.8 to 250.4 kb. Similar to the conjugative transfer regions of plasmids belonging to clade VI, they all carried the L/M-type oriTs, genes encoding relaxases of the MOBP family, genes encoding T4CPs of the TrwB/TraD subfamily, and Tra_I-like T4SS gene clusters (Figure 2; Supplementary Figure S8). The four plasmids were sporadically discovered in United States, India, Switzerland, and China (Figure 3; Table 1; Supplementary Table S3). No obvious common STs of strains were found (Table 1; Supplementary Table S3).
Clade VIII: Eighteen IncF plasmids, mainly carrying blaNDM-1, were grouped into the clade VIII cluster (Figure 2). Most of the IncF plasmids contained the IncFII(Yp) or IncFII(K) replicon in their genomes. For the 18 blaNDM-harboring plasmids of clade VIII, genome sizes varied from 94.4 to 316.2 kb (25th percentile = 106.8 kb, 75th percentile = 150.1 kb), with a median size of 110.6 kb (Supplementary Figure S1). Most of the plasmids of clade VIII were found to contain the F-type oriTs. They all carried the genes encoding relaxases of the MOBF family, genes encoding T4CPs of the TrwB/TraD subfamily, and Tra_F-like T4SS gene clusters (Figure 2; Supplementary Figure S9). Notably, K. pneumoniae strain JNQH116 plasmid pJNQH116-2 (NZ_CP070900), belonging to the clade VIII cluster, was found to contain both Tra_F-like and Trw-like T4SS gene clusters in its genome. For clade VIII, its members were widely geographically distributed all over the world, including China, India, Southeast Asia, Middle East, North America (Canada and United States), South Africa, and some European countries (e.g., Germany, Romania, and the United Kingdom; Figure 3; Table 1; Supplementary Table S3). No prevalent STs of K. pneumoniae host strains containing all clade VIII plasmids were found (Table 1; Supplementary Table S3).
Clade IX: A total of 21 IncC plasmids carrying blaNDM-1 were grouped into the clade IX cluster of the phylogenetic cladogram (Figure 2). Their genome sizes varied from 140.1 to 329.2 kb, with the 25th percentile, median size, and 75th percentile being 144.3, 147.9, and 178.7 kb, respectively (Supplementary Figure S1). For the conjugative transfer modules, all the plasmids belonging to clade IX carried the A/C-type oriTs, genes encoding relaxases of the MOBH family characterized by the domain “TraI_2 (Pfam: PF07514),” mostly genes encoding T4CPs of TrwB/TraD subfamily, and Tra_F-like T4SS gene clusters (Figure 2; Supplementary Figure S10). The IncC plasmids harboring blaNDM-1 were widely geographically distributed all over the world, mainly in the United States, Viet Nam, and China (Figure 3; Table 1; Supplementary Table S3). ST11 and ST1967 were the common STs strains containing blaNDM-1-harboring IncC plasmids (Table 1; Supplementary Table S3).
Clade X: A total of 40 mega plasmids, where the length range varied from 238.0 to 401.6 kb (25th percentile = 293.5 kb; median = 327.3 kb; 75th percentile = 355.1 kb), mainly carrying blaNDM-1, were grouped into a large cluster, named clade X in our study (Figure 2). Of the plasmids belonging to clade X, 27 (67.5%) were found to contain both replicons IncFIB(pNDM-Mar) and IncHI1B(pNDM-MAR), seven (17.5%) were unable to be typed, and four (10.0%) were IncQ1 plasmids. Moreover, all the plasmids of clade X carried genes encoding T4CPs of the TrwB/TraD subfamily and Tra_F-like T4SS gene clusters (Figure 2; Supplementary Figure S11). Most of the plasmids belonging to clade X were found to have no oriT and harbored the genes encoding relaxases of the MOBH family. For the clade with the largest number, clade X, its members were widely distributed all over the world, mainly in China and the United States (Figure 3; Table 1; Supplementary Table S3). ST14, ST11, and ST147 were the common STs strains containing the plasmids of clade X (Table 1; Supplementary Table S3).
We also perform a bipartite network analysis with the 171 blaNDM-harboring plasmids of K. pneumoniae. The bipartite network consisted of two classes of nodes: 171 plasmid units (PUs) and 2,502 homologous protein clusters (HPCs, protein families according to amino acid sequence identity, coverage, and E-value; Figure 4). Edges connected every PU with the HPC that it contained. The PUs of the bipartite network clearly showed distinct clustering phenomena. Overall, one homologous PU clusters (PUCs) contained the almost the same members of Clade X in the analysis above (Figure 4), which was clearly distinct from other PUs. One large region including clades III, VI – IX was also identified, which were mostly IncF and IncC plasmids. In addition, clades I, II, IV, and V were also found their corresponding PUCs in the PU-HPC bipartite network.
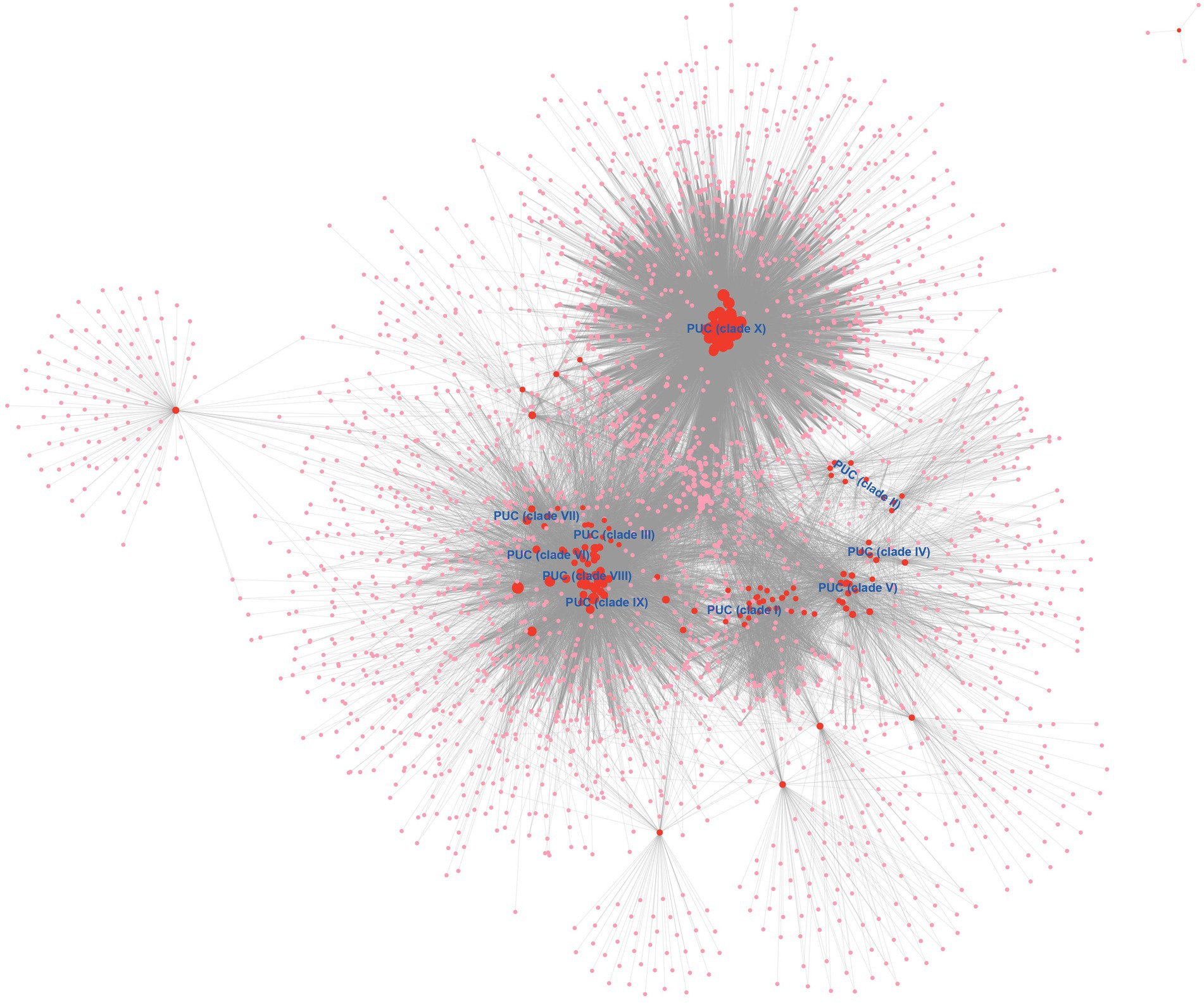
Figure 4. The PU-HPC bipartite network visualization of the blaNDM-harboring plasmids of K. pneumoniae. Plasmid units (PUs, 171) and homologous protein clusters (HPCs, 2,502) are represented as colored circles, with red for PUs and pink for HPCs. The size of a circle is ranked as the degree calculated with the Cytoscape’s built-in NetworkAnalyzer tool. The 10 clades (Clade I–Clade X) were labeled into the location of corresponding PUCs.
Global spread of the NDM-type carbapenemases can be partly attributed to the dissemination of various blaNDM-harboring plasmids (Lee et al., 2016; Dong et al., 2022). Therefore, to characterize plasmids harboring blaNDM in K. pneumoniae, we systematically analyzed the variants of blaNDM, replicon types, and conjugative transfer regions of 4,451 plasmids belonging to K. pneumoniae from the NCBI GenBank database. Overall, 171 blaNDM-harboring plasmids of K. pneumoniae were identified.
In our study, nine different variants of blaNDM were identified from the 171 blaNDM-harboring plasmids in K. pneumoniae, with blaNDM-1 and blaNDM-5 being highly prevalent; blaNDM-1-carrying plasmids were the most prevalent and accounted for 73.68% of the 171 blaNDM-harboring plasmids. NDM-1 was first reported in 2008 on a 180-kb plasmid of K. pneumoniae strain isolated from a Swedish patient hospitalized in New Delhi, India (Yong et al., 2009). After the first report, NDM-1 was reported in many clinical isolates, mainly K. pneumoniae and E. coli (Kumarasamy et al., 2010). In addition, blaNDM-5 was another common variant in our work, accounting for 16.37% of the 171 blaNDM-harboring plasmids. The variant NDM-5 was first reported on an IncF plasmid of E. coli EC405, isolated from a 41-year-old British patient who had a travel history to India (Hornsey et al., 2011). Notably, blaNDM-5 was reported to be the predominant variant in blaNDM-harboring plasmids of E. coli (Zhang et al., 2021).
Our results showed that IncX3 single-replicon plasmids were important carriers of blaNDM in K. pneumoniae, mainly blaNDM-1 and blaNDM-5. IncX3 plasmid is narrow-host range plasmids in Enterobacterales (Johnson et al., 2012), which has been reported to harbor diverse carbapenemase genes in CRE worldwide (Mouftah et al., 2019). Of the 29 blaNDM-harboring IncX3 plasmids grouped into clade I, most were relatively small with lengths of 46–57 kb (25th percentile = 46.2 kb; median size = 53.1 kb; 75th percentile = 57.3 kb). Based on the results analyzed by the oriTfinder software, all the 29 blaNDM-harboring IncX3 plasmids of clade I contained genes encoding for relaxases belonging to the MOBP family, with TraI protein encoded by the IncPα plasmid RP4 (Pansegrau et al., 1993) as a representative. T4CPs encoded by the 29 blaNDM-harboring IncX3 plasmids of clade I belonged to the VirD4/TraG subfamily, with the TraG protein of plasmid RP4 and the VirD4 protein of Ti plasmids as representatives (Gomis-Rüth et al., 2004). The blaNDM-harboring IncX3 plasmids classified into clade I contained VirB-like T4SS gene clusters, which are the best-characterized T4SS (Guglielmini et al., 2014). However, no known oriT site was found in most of the IncX3 plasmids harboring blaNDM belonging to clade I of the phylogenetic cladogram, indicating a new type of oriT site, different from the nine oriT families collected in the oriTDB database (Li et al., 2018).
We found 10 blaNDM-harboring IncN plasmids, with IncN or IncN2 replicons, clustered into clade II of the phylogenetic cladogram. They were also relatively small plasmids, with genome sizes varying from 38.38 to 63.05 kb. These blaNDM-harboring IncN plasmids carried the NW-type oriTs, which were characterized by the conserved nick region KGTST|ATAGC (“|” refers to the nic site of oriT), with oriT sites of IncN plasmid R46 (Hall and Vockler, 1987) and IncW plasmid R388 (Revilla et al., 2008) as representatives. Almost all the plasmids of clade II contained genes coding for relaxases of the MOBF family, which was characterized by the domain “TrwC (PF08751),” with R388 TrwC and F TraI as representatives (de la Cruz et al., 2010). The T4CPs encoded by the blaNDM-positive IncN plasmids belonged to the TrwB/TraD subfamily, which was characterized by the domain “TrwB_AAD_bind (PF10412),” with the TrwB encoded by plasmid R388 from E. coli as a representative (Gomis-Rüth et al., 2004). In addition, the blaNDM-positive IncN plasmids carried Trw-like T4SS gene clusters. The Trw T4SS clusters were regarded as the bacterial conjugation machines that mediate the spread of plasmids among bacterial populations (e.g., the trw locus of broad-host-range IncW plasmid R388; Seubert et al., 2003) while also mediating host-specific erythrocyte infection (e.g., the pathogenesis-related Trw system of Bartonella; Vayssier-Taussat et al., 2010).
Our work showed that 21 IncC plasmids carrying blaNDM-1, with genome sizes from 140.1 kb to 329.2 kb, were clustered into the clade IX of the phylogenetic cladogram constructed by the 171 blaNDM-harboring plasmids in K. pneumoniae. The broad-host-range IncC mega plasmids are essential contributors to the dissemination of antibiotic resistance genes, and more than 200 fully sequenced IncC plasmids have been reported (Ambrose et al., 2018). The blaNDM-1-harboring IncC plasmids of clade IX contained the A/C-type oriTs, with the oriT site of IncA/C conjugative pVCR94ΔX from Vibrio cholera as the prototype (Carraro et al., 2014). Furthermore, these blaNDM-1-harboring IncC plasmids carried genes encoding relaxases of the MOBH family, characterized by the domain “TraI_2 (Pfam: PF07514),” with TraI encoded by IncHI plasmid R27, TraI encoded by IncA/C plasmid pIP1202, TraI encoded by IncJ plasmid R391, and TraI encoded by IncT plasmid Rts1 as representatives (de la Cruz et al., 2010). In addition, most of the IncC plasmids clustered into clade IX contained genes encoding T4CPs of the TrwB/TraD subfamily and Tra_F-like T4SS gene clusters.
In our work, 53.22% (91 out of 171 plasmids) of the blaNDM-harboring plasmids of K. pneumoniae were found to be IncF plasmids, and most were multi-replicon IncF plasmids, especially IncFI-type plasmids. IncF plasmids are commonly low-copy-number plasmids, >100 kb in size (Villa et al., 2010); however, in our study, the blaNDM-harboring IncF plasmids in K. pneumoniae were heterogeneous in size. For example, the blaNDM-1-positive IncF plasmids, with the IncFIB(pQil) replicon, clustered into clade III were mostly 54-kb plasmids; the genome sizes of the IncFII plasmids grouped into clade V varied from 75.31 to 140.6 kb (25th percentile = 88.81 kb; 75th percentile = 101.4 kb); and the 27 plasmids with replicon IncFIB(pNDM-Mar) belonging to clade X were > 250 kb in size. The IncF plasmids comprise a diverse set of conjugative plasmids frequently found in Enterobacterales, which contribute to spreading AMR genes (Villa et al., 2010; Carattoli, 2011). The blaNDM-harboring IncF plasmids in K. pneumoniae were also heterogeneous in types of conjugative transfer regions. The IncFII-type plasmids, including clades V and VIII, carried F-type oriTs, genes encoding relaxases of the MOBF family, genes encoding T4CPs of the TrwB/TraD subfamily, and Tra_F-like T4SS gene clusters belonging to the classical F-like conjugative system (de la Cruz et al., 2010). Mega plasmids with replicons IncFIB(pNDM-Mar):IncHI1B(pNDM-MAR) belonging to clade X mostly harbored the genes encoding relaxases of the MOBH family. In our study, we found 11 blaNDM-1-positive IncFIB(pQil) plasmids classified into clade III without any classical conjugative transfer regions, which were predicted as non-transferable plasmids.
In this study, we analyzed the variants of blaNDM, replicon types, conjugative transfer regions, host STs, and geographical distributions of 171 blaNDM-harboring plasmids from 4,451 K. pneumoniae plasmids, which were downloaded from the GenBank database. Nine variants of blaNDM were found among the 171 blaNDM-positive plasmids, with blaNDM-1 (73.68%) and blaNDM-5 (16.37%) as the most dominant. Over half of the blaNDM-harboring plasmids of K. pneumoniae were classified into IncF plasmids. In addition, IncX3 single-replicon plasmids (46–57 kb), IncN plasmids (38.4–63.1 kb), IncC plasmids (140.1–329.2 kb) were also the common carriers of blaNDM in K. pneumoniae. The blaNDM-harboring IncX3 and IncN plasmids were mainly geographically distributed in China. The IncC plasmids harboring blaNDM-1 were widely geographically distributed all over the world, mainly in the United States, China, and Viet Nam. This study provides important insights into the diversity of blaNDM-harboring plasmids in K. pneumoniae and further addresses their role in the acquisition and spread of resistance genes. However, the genetic diversity and characteristics of blaNDM-harboring plasmids in other Gram-negative species need further study in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
XL, LuL, and XD: conceptualization. ZhuZ: methodology. LeL and LiL: software. SH and JY: validation. WL, LZ, QL, and ZhiZ: formal analysis. ZhuZ and XL: writing—original draft preparation. XL and LuL: writing—review and editing. XL: supervision. All authors contributed to the article and approved the submitted version.
This work was supported financially by the grants from the National Natural Science Foundation of China (grant nos. 81902460 and 82002170), the Xiangshan Talent Project of Zhuhai People’s Hospital (grant no. 2020XSYC-02), and the Cultivation Project of Zhuhai People’s Hospital (2019PY-19).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1008905/full#supplementary-material
1. ^https://bioinfo-mml.sjtu.edu.cn/oriTDB/browse_oriT_type_p.php
2. ^https://bioinfo-mml.sjtu.edu.cn/oriTDB/browse_relaxase.php
3. ^https://bioinfo-mml.sjtu.edu.cn/SecReT4/browse_type.php
4. ^https://bioinfo-mml.sjtu.edu.cn/oriTDB/browse_t4cp.php
5. ^https://www.ncbi.nlm.nih.gov/genome/browse/#!/plasmids/815/
7. ^https://www.ncbi.nlm.nih.gov/genome/browse/#!/prokaryotes/815/
8. ^https://cge.food.dtu.dk/services/MLST/
Albiger, B., Glasner, C., Struelens, M. J., Grundmann, H., and Monnet, D. L. (2015). Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro. Surveill. 20:30062. doi: 10.2807/1560-7917.es.2015.20.45.30062
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Ambrose, S. J., Harmer, C. J., and Hall, R. M. (2018). Evolution and typing of IncC plasmids contributing to antibiotic resistance in gram-negative bacteria. Plasmid 99, 40–55. doi: 10.1016/j.plasmid.2018.08.001
Bengoechea, J. A., and Sa Pessoa, J. (2019). Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol. Rev. 43, 123–144. doi: 10.1093/femsre/fuy043
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Ostell, J., Pruitt, K. D., et al. (2018). GenBank. Nucleic Acids Res. 46, D41–d47. doi: 10.1093/nar/gkx1094
Bi, D., Liu, L., Tai, C., Deng, Z., Rajakumar, K., and Ou, H. Y. (2013). SecReT4: a web-based bacterial type IV secretion system resource. Nucleic Acids Res. 41, D660–D665. doi: 10.1093/nar/gks1248
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Carattoli, A. (2011). Plasmids in gram negatives: molecular typing of resistance plasmids. Int. J. Med. Microbiol. 301, 654–658. doi: 10.1016/j.ijmm.2011.09.003
Carattoli, A., and Hasman, H. (2020). PlasmidFinder and in Silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol. Biol. 2075, 285–294. doi: 10.1007/978-1-4939-9877-7_20
Carballeira, J. D., González-Pérez, B., Moncalián, G., and la Cruz, F. D. (2014). A high security double lock and key mechanism in HUH relaxases controls oriT-processing for plasmid conjugation. Nucleic Acids Res. 42, 10632–10643. doi: 10.1093/nar/gku741
Carraro, N., Sauvé, M., Matteau, D., Lauzon, G., Rodrigue, S., and Burrus, V. (2014). Development of pVCR94ΔX from vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front. Microbiol. 5:44. doi: 10.3389/fmicb.2014.00044
de la Cruz, F., Frost, L. S., Meyer, R. J., and Zechner, E. L. (2010). Conjugative DNA metabolism in gram-negative bacteria. FEMS Microbiol. Rev. 34, 18–40. doi: 10.1111/j.1574-6976.2009.00195.x
De Oliveira, D. M., Forde, B. M., Kidd, T. J., Harris, P. N., Schembri, M. A., Beatson, S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33, e00181–e00119. doi: 10.1128/CMR.00181-19
Dong, H., Li, Y., Cheng, J., Xia, Z., Liu, W., Yan, T., et al. (2022). Genomic epidemiology insights on NDM-producing pathogens revealed the pivotal role of plasmids on blaNDM transmission. Microbiol. Spectrum 10, e02156–e02121. doi: 10.1128/spectrum.02156-21
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. doi: 10.1186/s13059-019-1832-y
Gomis-Rüth, F. X., Solà, M., de la Cruz, F., and Coll, M. (2004). Coupling factors in macromolecular type-IV secretion machineries. Curr. Pharm. Des. 10, 1551–1565. doi: 10.2174/1381612043384817
Guducuoglu, H., Gursoy, N. C., Yakupogullari, Y., Parlak, M., Karasin, G., Sunnetcioglu, M., et al. (2018). Hospital outbreak of a Colistin-resistant, NDM-1- and OXA-48-producing Klebsiella pneumoniae: high mortality from Pandrug resistance. Microb. Drug Resist. 24, 966–972. doi: 10.1089/mdr.2017.0173
Guglielmini, J., Néron, B., Abby, S. S., Garcillán-Barcia, M. P., de la Cruz, F., and Rocha, E. P. (2014). Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 42, 5715–5727. doi: 10.1093/nar/gku194
Hall, R. M., and Vockler, C. (1987). The region of the incN plasmid R46 coding for resistance to β-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasm ids and in Tn21-like transposons. Nucleic Acids Res. 15, 7491–7501. doi: 10.1093/nar/15.18.7491
Hammer, Ø., Harper, D. A., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9.
Hornsey, M., Phee, L., and Wareham, D. W. (2011). A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55, 5952–5954. doi: 10.1128/aac.05108-11
Jiang, Y., Wang, Y., Hua, X., Qu, Y., Peleg, A. Y., and Yu, Y. (2020). Pooled plasmid sequencing reveals the relationship between Mobile genetic elements and antimicrobial resistance genes in clinically isolated Klebsiella pneumoniae. Genomics Proteomics Bioinformatics 18, 539–548. doi: 10.1016/j.gpb.2020.12.002
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/s1473-3099(10)70143-2
Lanza, V. F., Baquero, F., de la Cruz, F., and Coque, T. M. J. B. (2017). AcCNET (accessory genome constellation network): comparative genomics software for accessory genome analysis using bipartite networks. Bioinformatics 33, 283–285. doi: 10.1093/bioinformatics/btw601
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/jcm.06094-11
Lee, C.-R., Lee, J. H., Park, K. S., Kim, Y. B., Jeong, B. C., and Lee, S. H. (2016). Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. doi: 10.3389/fmicb.2016.00895
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, X., Xie, Y., Liu, M., Tai, C., Sun, J., Deng, Z., et al. (2018). oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46, W229–w234. doi: 10.1093/nar/gky352
Llosa, M., Gomis-Rüth, F. X., Coll, M., and de la Cruz Fd, F. (2002). Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45, 1–8. doi: 10.1046/j.1365-2958.2002.03014.x
Mouftah, S. F., Pál, T., Darwish, D., Ghazawi, A., Villa, L., Carattoli, A., et al. (2019). Epidemic IncX3 plasmids spreading carbapenemase genes in the United Arab Emirates and worldwide. Infect. Drug. Resist. 12, 1729–1742. doi: 10.2147/idr.s210554
Naas, T., Oueslati, S., Bonnin, R. A., Dabos, M. L., Zavala, A., Dortet, L., et al. (2017). Beta-lactamase database (BLDB)–structure and function. J. Enzyme Inhib. Med. Chem. 32, 917–919. doi: 10.1080/14756366.2017.1344235
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Nordmann, P., Poirel, L., Walsh, T. R., and Livermore, D. M. (2011). The emerging NDM carbapenemases. Trends Microbiol. 19, 588–595. doi: 10.1016/j.tim.2011.09.005
Pansegrau, W., Schröder, W., and Lanka, E. (1993). Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc. Natl. Acad. Sci. U. S. A 90, 2925–2929. doi: 10.1073/pnas.90.7.2925
Pitout, J. D., Nordmann, P., and Poirel, L. (2015). Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59, 5873–5884. doi: 10.1128/AAC.01019-15
Revilla, C., Garcillán-Barcia, M. P., Fernández-López, R., Thomson, N. R., Sanders, M., Cheung, M., et al. (2008). Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob. Agents Chemother. 52, 1472–1480. doi: 10.1128/AAC.00982-07
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Seubert, A., Hiestand, R., De La Cruz, F., and Dehio, C. (2003). A bacterial conjugation machinery recruited for pathogenesis. Mol. Microbiol. 49, 1253–1266. doi: 10.1046/j.1365-2958.2003.03650.x
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Vayssier-Taussat, M., Le Rhun, D., Deng, H. K., Biville, F., Cescau, S., Danchin, A., et al. (2010). The Trw type IV secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog. 6:e1000946. doi: 10.1371/journal.ppat.1000946
Villa, L., García-Fernández, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Wu, W., and Feng, Y. (2019). NDM Metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32:e00115-18. doi: 10.1128/cmr.00115-18
Wyres, K. L., and Holt, K. E. (2018). Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 45, 131–139. doi: 10.1016/j.mib.2018.04.004
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, Bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/aac.00774-09
Keywords: Klebsiella pneumoniae, plasmid, blaNDM, replicon types, conjugative transfer region
Citation: Zeng Z, Lei L, Li L, Hua S, Li W, Zhang L, Lin Q, Zheng Z, Yang J, Dou X, Li L and Li X (2022) In silico characterization of blaNDM-harboring plasmids in Klebsiella pneumoniae. Front. Microbiol. 13:1008905. doi: 10.3389/fmicb.2022.1008905
Received: 01 August 2022; Accepted: 07 November 2022;
Published: 23 November 2022.
Edited by:
Fang He, Zhejiang Provincial People’s Hospital, ChinaReviewed by:
Deepjyoti Paul, Translational Health Science and Technology Institute (THSTI), IndiaCopyright © 2022 Zeng, Lei, Li, Hua, Li, Zhang, Lin, Zheng, Yang, Dou, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luan Li, bGlsdWFuODc5MUAxMjYuY29t; Xiaobin Li, bGkueGlhb2JpbjIwMDlAMTYzLmNvbQ==; Xiaohui Dou, MTM1MjcyNzY2NzFAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.