
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 October 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1008110
This article is part of the Research Topic New Infectious Agents in Arthropod Vectors View all 9 articles
Rickettsia, Anaplasma, and Ehrlichia belonging to the order Rickettsiales are causative agents of tick-borne diseases in humans. During 2021, 434 ticks including Rhipicephalus microplus and R. haemaphysaloides were collected from three sampling sites in Yunnan Province, Southwest China, and analyzed for the presence of these bacteria. Nine bacterial species were identified, including two Rickettsia spp., three Anaplasma spp., and four Ehrlichia spp., some of which are potential human pathogens. Genetic and phylogenetic analysis on 16S rRNA, gltA, groEL, ompA, ompB, and sca4 genes indicated the presence of a novel spotted fever group Rickettsia (SFGR) named “Candidatus Rickettsia shennongii” in six of the 38 R. haemaphysaloides ticks from two locations, Dehong Autonomous Prefecture and Honghe City. Another SFGR species, Candidatus Rickettsia jingxinensis was detected in ticks from all three sites, with an overall positive rate of 62.67%. Three other human pathogenic species, Anaplasma ovis (1.38%, 6/434), Ehrlichia canis (16.36%, 71/434), and E. chaffeensis (0.23%, 1/434) were detected in these ticks and characterized. Moreover, Ehrlichia sp. (4.84%, 21/434), E. minasensis (7.37%, 32/434), A. marginale (6.91%, 30/434), and Cadidatus Anaplasma boleense (1.15%, 5/434) were detected in R. microplus ticks, for which pathogenicity to humans remains to be determined. The results reveal the remarkable diversity of Rickettsiales bacteria in ticks from Yunnan Province, Southwest China. The high infection rate of some human pathogenic bacteria in ticks may indicate potential infection risk in humans, and it highlights the need for surveillance in local populations.
Anaplasma spp. and Ehrlichia spp. belonging to order Rickettsiales are tick-borne intracellular bacteria, many of which are important pathogens of livestock. Anaplasma spp., Ehrlichia spp., as well as Rickettsia spp. bacteria also occasionally infect humans. Therefore, these bacteria are important for both veterinary and human public health (Gondard et al., 2017). SFGR represents a large group within the genus Rickettsia including Rickettsia rickettsii, R. conorii, R. australis, R. honei, R. japonica, R. africae, R. sibirica, etc. (Stenos et al., 2005). They are widely distributed and many of them are etiological agents of known human diseases, such as Rocky Mountain spotted fever (RMSF), Astrakhan fever, Mediterranean spotted fever, Indian tick typhus, Queensland tick typhus, Flinders Island spotted fever, Japanese spotted fever, African tick bite fever, and North Asian tick-typhus, etc. (Sentausa et al., 2012; Graham et al., 2017; Rudakov et al., 2019; Zazueta et al., 2021). Of those, R. rickettsii mainly distributes in North and South America and was reported to infect more than 4,000 patients during 2009–2019 at Mexico-United States Border (Zazueta et al., 2021). From 1997 to 2004, 415 children cases of Mediterranean spotted fever caused by R. conorii were recorded in Sicily, Italy (Colomba et al., 2006). Meanwhile, only sporadic infection cases were reported for R. australis, R. honei (in Australia), R. africae (in Africa), R. japonica (Asia), and other SFG members (Parola et al., 2013). For the genus Anaplasma, Anaplasma marginale, A. centrale, and A. bovis are common pathogens causing severe or mild anaplasmosis in ruminants, especially cattle (Rjeibi et al., 2018). Meanwhile, A. phagocytophilum, A. bovis, and A. capra are confirmed human pathogens (Li H. et al., 2015; Ismail and McBride, 2017; Lu et al., 2019). In the past decades, a total of 110 infection cases of A. phagocytophilum were reported (Dumic et al., 2022). Among Ehrlichia species, E. ruminantium and E. minasensis are known to infect cattle, causing severe fever, anemia, and thrombocytopenia (Peter et al., 2020). Ehrlichia chaffeensis, E. ewingii, E. muris, E. muris. Subsp. eauclairensis, and E. canis are reported to infect humans, with syndromes ranging from febrile to multiple organ failure (Saito and Walker, 2016; Pritt et al., 2017). From 2012–2016, 6,786 cases of E. chaffeensis infection were reported in the United States (Mogg et al., 2020).
Located in southwest China, Yunnan Province covers a vast area with diverse climates and unique biodiversity resources. The extraordinary biological and ecological diversity of this area makes the extensive diversity of Rickettsiales bacteria possible. In recent decades, much attention has been paid to Rickettsiales bacteria circulating in this area. Numerous studies have been performed and many bacterial species belonging to the order Rickettsiales have been characterized in Yunnan Province (Liang et al., 2012; Liu et al., 2020; Jiao et al., 2021). One study revealed high positive rates of SFGR in domestic animals (goats, dogs, and cattle), and proved the existence of R. heilongjiangensis and a distinct Rickettsia (Liang et al., 2012). Another study reported the presence of R. raoultii and Ca. R. jingxinensis in ticks from Yunnan Province (Liu et al., 2020). In Jiao et al. (2021) detected multiple tick-borne pathogens circulating in Rhipicephalus microplus ticks from Yunnan, including Ca. R. jingxinensis, A. marginale, and Coxiella burnetii. Furthermore, A. capra, A. phagocytophilum, and Candidatus Neoehrlichia mikurensis were also detected in R. microplus ticks from Yunnan Province (Jiao et al., 2021). Although many studies on tick-borne Rickettsiales bacteria have been performed in Yunnan Province, further exploration is still needed. To improve our knowledge on the biodiversity and epidemiology of Rickettsiales bacteria, we collected ticks from goats and cattle in three locations of Yunnan Province, and characterized Rickettsia, Anaplasma, and Ehrlichia in them.
During 2021, ticks were collected from three locations in Yunnan Province: Ruili county-level city of Dehong Dai-Jingpo Autonomous Prefecture (97.85°E, 24.01°N), Zhaoyang District of Zhaotong city (103.71°E, 24.32°N), and Shiping county of Honghe city (102.49°O, 23.71°N; Figure 1). Ticks were collected from cattle and goats, then brought to China CDC alive. Tick species were morphologically characterized by a sophisticated arthropod taxonomist based on the characteristics of the capitula, body, legs, anal groove, and caudal appendage (Namgyal et al., 2021). For further confirmation, randomly selected ticks were undergone molecular analysis by sequencing the mitochondrial cytochrome oxidase I (COI) gene sequences (Lu et al., 2013).
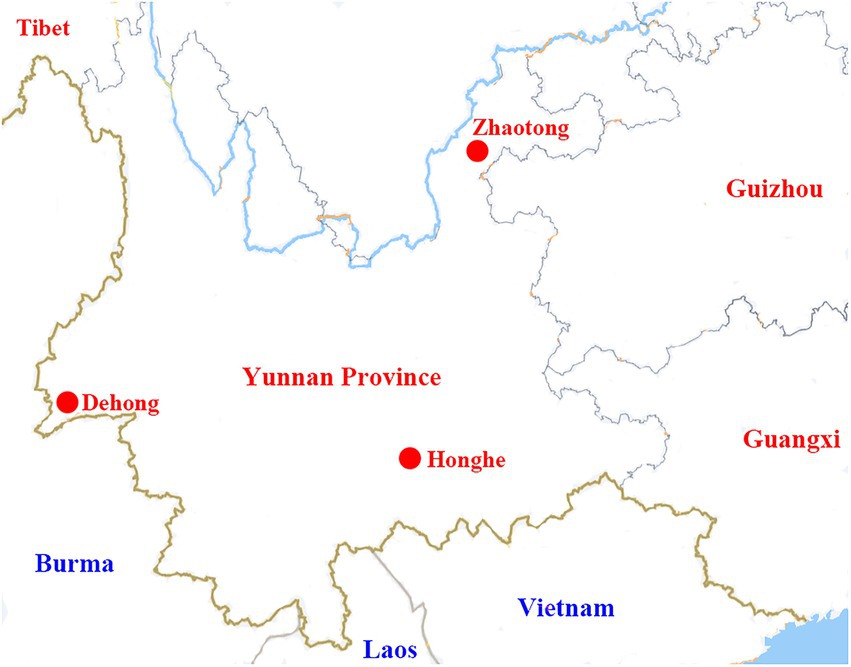
Figure 1. A map showing the locations of Dehong, Honghe, and Zhaotong in Yunnan Province, where the tick samples were collected.
After washing three times with sterile phosphate-buffered saline (PBS), ticks were individually homogenized in 500 μl of PBS using a Mixer Mill MM 400 (Retsch, Hann, Germany). The total DNA of each tick was extracted in 80 μl of eluate using a Mollusk DNA Extraction Kit (Omega Bio-Tek, United States) according to the manufacturer’s instructions.
Each tick DNA sample was screened by PCR for the presence of bacterial DNA including Rickettsia spp., Anaplasma spp., and Ehrlichia spp. Rickettsial DNA was detected by nested PCR as described previously using primers targeting the outer membrane protein A (ompA) gene, resulting in amplification of a 755-bp fragment (Lu et al., 2017). The DNA samples were also screened for Anaplasma and Ehrlichia DNA as described previously, amplifying a 550–850 bp fragment of the 16S rRNA gene (Guo et al., 2016). Negative control with distilled water in the PCR master mixture and positive control (DNA of R. felis, A. marginale and E. canis, respectively) were included in each test. After 1% agarose gel electrophoresis, amplification products were subjected to sequencing.
For further characterization and phylogenetic analysis of the detected bacterial strains, the partial sequences of the gltA gene (citrate synthase), the groEL gene (heat shock protein), and a long fragment of the 16S rRNA gene were obtained for representative Rickettsia, Anaplasma, and Ehrlichia positive samples. Additionally, nearly complete sequences of ompB and sca4 genes were obtained for the putative novel Rickettsia species. The primers used were described previously (Roux and Raoult, 2000; Sekeyova et al., 2001; Kang et al., 2014; Guo et al., 2016; Lu et al., 2017) and they are listed in Supplementary Table S1. All recovered sequences have been deposited in the GenBank database (GenBank numbers listed in Supplementary Table S2).
Sequences obtained in this study were analyzed by BLASTn1 and Clustal W within Molecular Evolutionary Genetics Analysis (MEGA) software, version 7.0 (Kumar et al., 2016). Phylogenetic analysis was also performed using PhyML 3.0. Confidence values for each branch of the phylogenetic trees were determined by bootstrap analysis with 1,000 replicates. Sequences recovered in this study were aligned with the reference sequences retrieved from the GenBank database.
During March to July 2021, 434 ticks (397 R. microplus and 37 R. haemaphysaloides) were collected from three locations in Yunnan Province. The three sampling locations (Ruili County-level City of Dehong Dai-Jingpo Autonomous Prefecture, Zhaoyang District of Zhaotong City, and Shiping County of Honghe City) were shown in Figure 1. Tick species were initially determined by morphological examination and then further determined by amplification and analysis of the COI gene (Lu et al., 2013). Except for four nymphs from Zhaotong, all ticks were adult, and most were fully or partially engorged. The COI sequences of ticks have been submitted to GenBank. The Accession numbers are OM959242-OM959313, OM959315-OM959321, and OM977035-OM977043. The tick species, quantity, and vertebrate hosts in different sampling sites were shown in Supplementary Table S3.
Nested PCR targeting the conserved domain of the ompA gene was performed to screen for Rickettsia in the collected tick samples. Based on agarose gel electrophoresis, DNA sequencing, and comparison with BLASTn, two Rickettsia species were initially identified: Ca. R. jingxinensis and a putative novel species. For further characterization, the 16S rRNA gene (1,172 bp), as well as the gltA (1,004 bp) and groEL (1038–1,042 bp) genes were successfully amplified from the total tick DNA samples.
DNA of Ca. R. jingxinensis was detected in ticks from all three sites, with strikingly high positive rates varying from 37.50 to 84.69% (48 of 128 ticks from Dehong, 166 of 196 ticks from Zhaotong, and 70 of 110 ticks from Honghe; Table 1). All Ca. R. jingxinensis strains were detected in R. microplus ticks except for one from R. haemaphysaloides. All the ompA sequences shared 100% identity with Ca. R. jingxinensis isolate Meixian-Hl-107 (MH932061.1) and Xian-Hl-79 (MH932069.1) reported in Haemaphysalis longicornis from ShaanXi Province, Northern China (Guo et al., 2019). The 16S rRNA, gltA and groEL genes shared 99.90–100% identity with each other in reference to homologous genes. The 16S rRNA and gltA gene sequences were 100 and 99.90% identical to other Ca. R. jingxinensis strains from China. Meanwhile, the groEL sequences shared highest 99.90% identity with Uncultured Rickettsia sp. clone tick28 (ON409665) and Uncultured Rickettsia sp. clone tick26 (ON409664) we previously identified in Ngawa, Sichuan Province, which actually also represent Ca. R. jingxinensis strains.
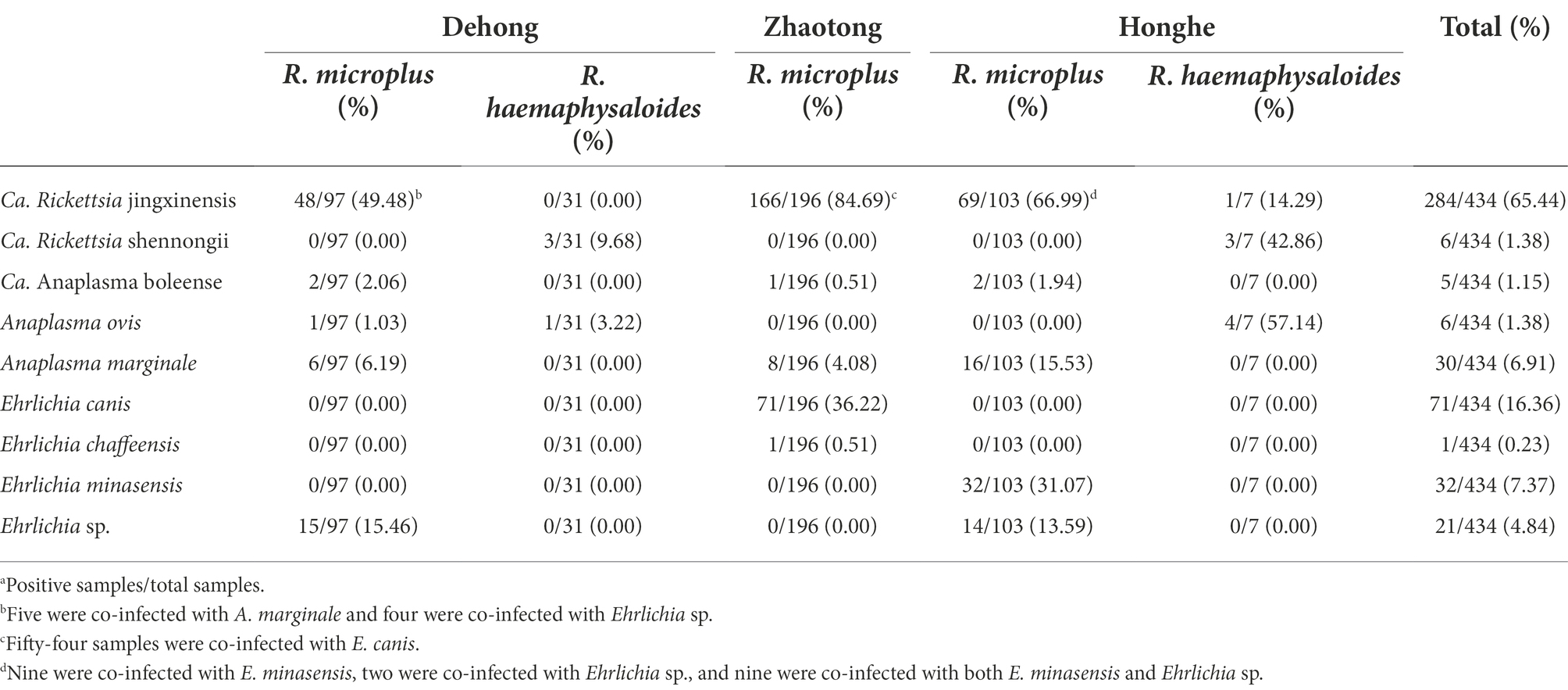
Table 1. Prevalence of Rickettsiales bacteria in ticks in Dehong, Zhaotong, and Honghe cities of Yunnan Province, Southwest China.
Notably, a putative novel SFGR species was identified in R. haemaphysaloides ticks from Dehong (3/31, 9.68%) and Honghe (3/7, 42.86%; Table 1). The ompA sequences of these were 100% identical to each other, and they shared 98.31% identity with R. rhipicephali str. 3-7-female6-CWPP (CP003342.1) and 98.17% with R. massiliae MTU5 (CP000683.1). The 16S rRNA (1,172 bp) gene sequences of all strains from Dehong and Honghe were identical, despite the geographic separation, and they shared 100.0% identity with Rickettsia sp. strain HB-9543 N2 (MT434770), 99.91% identity with R. raoultii isolate Tomsk (MK304546) and R. conorii str. Malish 7 (AE006914.1). The gltA (1,004 bp) gene sequences were found to be 99.70–99.80% identical to Rickettsia sp. strain HB-9543 N2 (MT434984), 99.30–99.40% to R. massiliae MTU5 (CP000683.1) and R. rhipicephali str. HJ#5 (CP013133). Regarding the groEL (1,042 bp) gene, all sequences shared high similarity with R. rhipicephali str. HJ#5 (CP013133, 99.42%) and R. rhipicephali str. 3-7-female6-CWPP (CP003342; 99.33%). For further confirmation, the ompB (4704–4,707 bp) and sca4 (2560–2,566 bp) genes were successfully recovered. Similar to other genes, the ompB gene shared 99.68–99.77% similarity to Rickettsia sp. strain HB-9543 N2 (MT434988) and 98.14–98.22% similarity with R. rhipicephali str. 3-7-female6-CWPP (CP003342.1), while the sca4 gene shared 99.73–99.96% identity with Rickettsia sp. strain HB-9543 N2 (MT434990) and 98.10–98.37% identity to R. rhipicephali str. 3-7-female6-CWPP (CP003342). Notably, the sca4 gene of all strains from Honghe City has an additional six-nucleotide insertion (AAGAAA). In the phylogenetic trees, all six genes of this Rickettsia formed a distinct clade closely related to R. rhipicephali and R. massiliae (Figure 2).
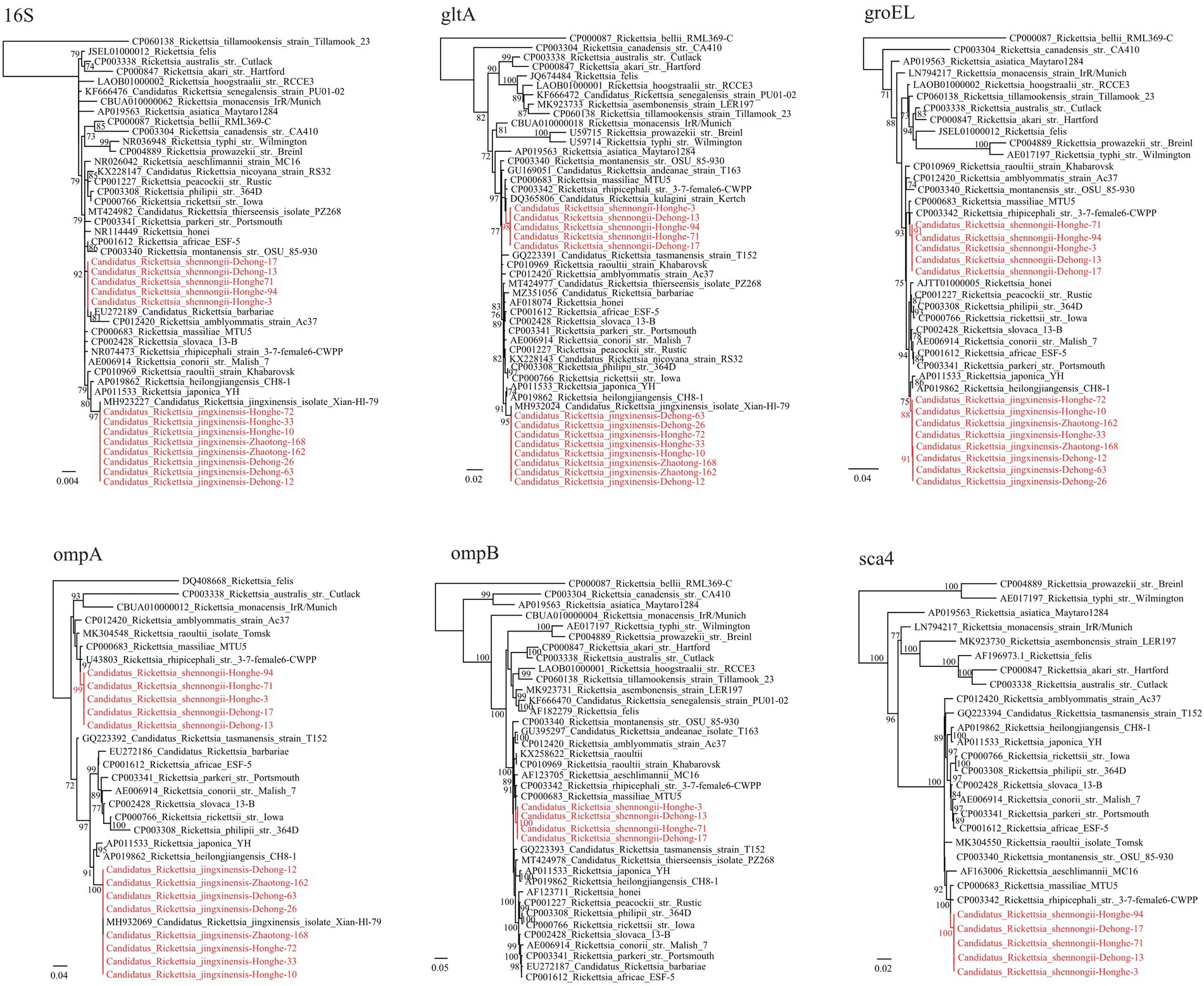
Figure 2. Phylogenetic trees constructed by the PhyML 3.0 software based on the nucleotide sequences of 16S rRNA (1,172 bp), groEL (1,038–1,042 bp), gltA (1,004 bp), ompA (664–712 bp), ompB (4,704–4,707 bp) and sca4 (2,560–2,566 bp) genes of Rickettsia strains. The bootstrap values were shown on the nodes. Red: the strains identified in this study.
According to the gene sequence-based criteria for the identification of a new Rickettsia species, a novel species should exhibit at most one of the following degrees of sequence similarity when comparing to validated Rickettsia species: ≥99.8 for the 16S, ≥99.9% for the gltA genes, and, when available, ≥98.8, ≥99.2, and ≥ 99.3% for the ompA and ompB and sca4 genes, respectively (Fournier et al., 2003). As indicated above, BLASTn shows that the 16S, gltA, ompA, ompB, and sca4 sequences of this Rickettsia have highest 99.91%, 99.30–99.40, 99.42, 98.31%, 98.14–98.22%, and 98.10–98.37% similarity to validated Rickettsia species, respectively (Supplementary Table S4). All the sequences match this criterion except the 16S gene has a similarity higher than 99.8%. This result clearly supports that they represent a novel Rickettsia species. Herein we name it “Candidatus Rickettsia shennongii” to memorize Shennong, a great master of agronomy and herbology in Chinese myth.
Three Anaplasma species were identified; Candidatus Anaplasma boleense, A. ovis, and A. marginale. DNA of Ca. A. boleense was detected in R. microplus ticks from Dehong, Zhaotong, and Honghe, with positive rates of 2.06% (2/97), 0.51% (1/196), and 1.94% (2/103), respectively (Table 1). The 16S rRNA (855 bp), gltA (672 bp), and groEL (857 bp) genes share 99.65–100, 99.11%, and 98.95–100% similarity with Ca. A. boleense strains reported in other locations of China. Anaplasma ovis was detected in ticks from Dehong and Honghe, mostly from R. haemaphysaloides. All the three genes (16S rRNA, 854–1,206 bp; gltA, 539 bp; groEL, 846 bp) are 100% identical to previously reported A. ovis sequences, and they are closely clustered with other A. ovis strains in the phylogenetic trees (Figure 3). As a widely distributed animal pathogen, A. marginale was detected in R. microplus ticks from all three locations, with prevalence rates from 4.08 to 15.53% (Table 1). All three genes (16S rRNA: 854–1,206 bp, gltA: 857 bp, groEL: 926 bp) share 100% identity with previously reported A. marginale sequences. To be noticed, it has been observed that the detection rates of Anaplasma spp. in ticks from the environment and those removed from livestock are different (von Fricken et al., 2021). Given all ticks in this study were removed from cattle and goats, the detection rates might be influenced.
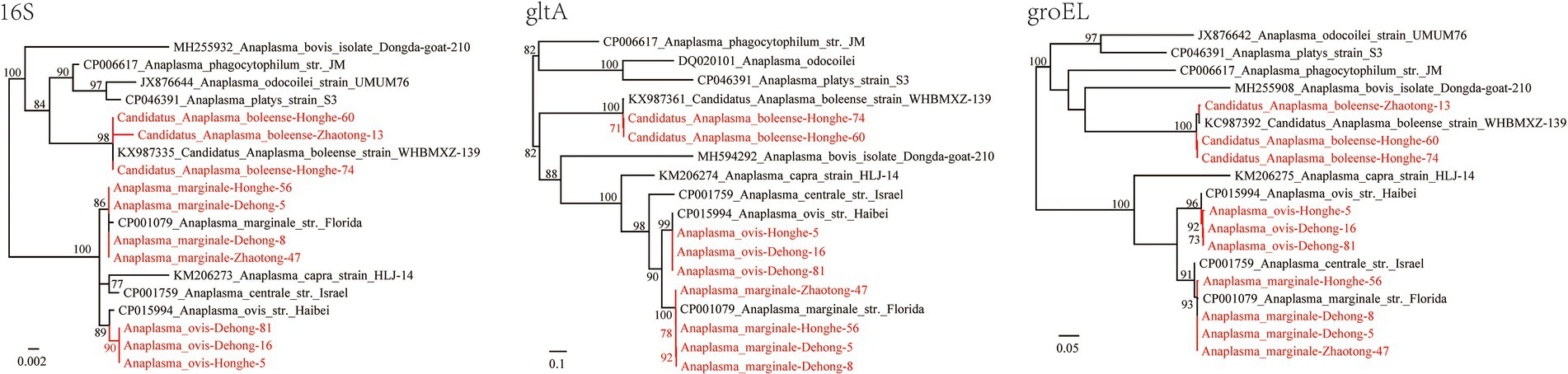
Figure 3. Phylogenetic trees constructed by the PhyML 3.0 software based on the nucleotide sequences of 16S rRNA (854–1,206 bp), gltA (539 bp), and groEL (846 bp) genes of Anaplasma strains. The bootstrap values were shown on the nodes. Red: the strains identified in this study.
Of the 434 ticks screened from three locations, 125 (28.80%) were positive for Ehrlichia including four species: E. minasensis, E. canis, E. chaffeensis, and Ehrlichia sp. (Table 1). In R. microplus ticks from both Dehong and Honghe city, an Ehrlichia sp. species closely related to Ehrlichia sp. strain WHBMXZ-43 was identified, which was first identified in R. microplus ticks from Wuhan city, South-Central China. The 16S rRNA genes of the detected strains share 99.84–99.91% identity with this Ehrlichia, while both gltA and groEL genes share 100% identity with this strain.
In R. microplus ticks from Honghe city, E. minasensis was detected, with a positive rate of 31.07% (32/103). The 16S rRNA, gltA, and groEL gene sequences all share 100% identity with E. minasensis strain B1 isolated from cattle in Brazil. In the phylogenetic trees based on these three genes, the two E. minasensis strains (Honghe-25 and Honghe-42) were both located in the same clade with E. minasensis strain B11 (Figure 4). This result indicated the prevalence and the high genetic conservation of E. minasensis in Yunnan Province.
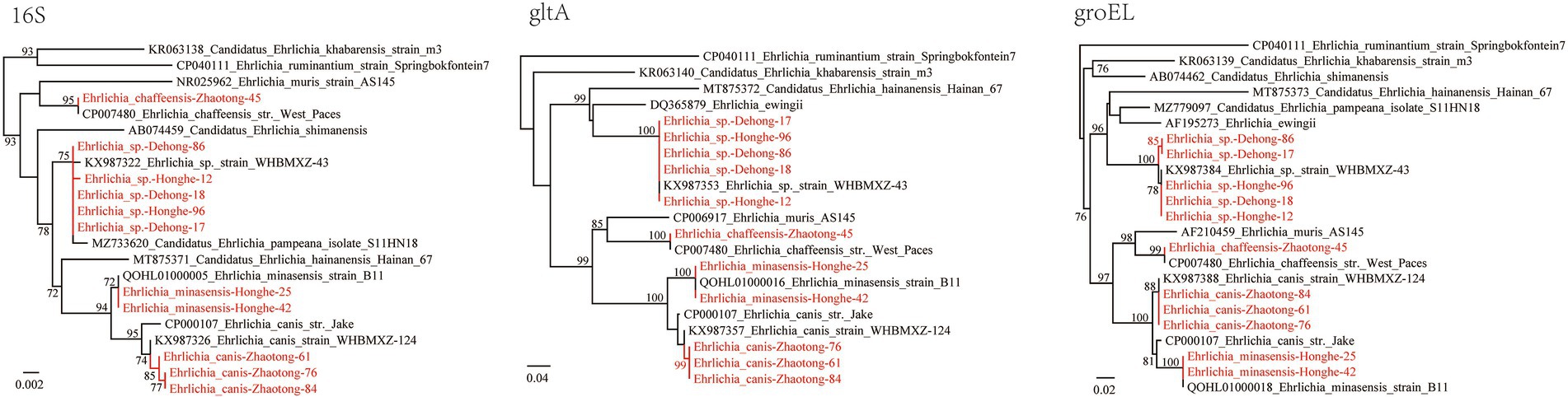
Figure 4. Phylogenetic trees constructed by the PhyML 3.0 software based on the nucleotide sequences of 16S rRNA (1,198–1,406 bp), gltA (864–973 bp), and groEL (1,115 bp) genes of Ehrlichia strains. The bootstrap values were shown on the nodes. Red: the strains identified in this study.
Of the 196 R. microplus ticks tested from Zhaotong city, two Ehrlichia species were detected: E. canis (71/196, 36.22%) and E. chaffeensis (1/196, 0.51%; Table 1). The prevalence in male, female, and nymph ticks was shown in Supplementary Table S5. The 16S rRNA gene sequences of detected E. canis strains (1,406 bp) share 99.57% similarity with E. canis clone CuD125 (MK507008.1) and E. canis strain YZ-1 (CP025749.1), etc. The gltA and groEL sequences are 100% identical between the detected strains. The gltA (973 bp) sequences are 97.84% identical to E. canis strain YZ-1 (CP025749.1) and E. canis isolate T85D7 (MW382940.1), while the groEL sequences share 98.66% identity with E. canis strain Mossesane (MG953295.1). Phylogenetic analysis clearly revealed that all strains are assembled with other E. canis strains (Figure 4). Additionally, an E. chaffeensis strain was detected in a single R. microplus tick. All 16S rRNA, gltA, and groEL genes share 100% similarity with E. chaffeensis str. West Paces, E. chaffeensis Arkansas, and other previously reported E. chaffeensis strains. These appear to be the only gltA and groEL sequences of E. chaffeensis from China submitted to GenBank.
Yunnan Province is a widely recognized biodiversity hotspot in China and worldwide (Li R. et al., 2015). In the present study, we revealed the extensive diversity and high positive rate of Rickettsiales bacteria in ticks from Yunnan Province. Nine bacterial species belonging to the genus Rickettsia, Anaplasma, and Ehrlichia were detected, including a novel SFGR species. Our result may contribute to the current knowledge of the biodiversity of Rickettsiales bacteria circulating in this area.
Rickettsia spp. have been recognized as pathogens of potential public health importance. Herein, the DNA of Ca. R. jingxinensis was detected in all three locations with extremely high positive rates varying from 37.50 to 84.69%. Cadidatus Rickettsia jingxinensis is a SFGR first identified in Ha. longicornis from northeast China in 2016 (Liu et al., 2016). It has since been detected in ticks from multiple provinces in China, including Shaanxi, Guangxi, Sichuan, and Yunnan (Guo et al., 2019; Liu et al., 2020; Jiao et al., 2021; Lu et al., 2022). Furthermore, it was also reported in Korea, Thailand, and India, spanning from East Asia to South Asia (Takhampunya et al., 2019; Bang et al., 2021) (GenBank No. MN463681-MN463688). Previous studies revealed strikingly high positive rates of this Rickettsia in certain areas (for instance, 44.4–69.7% in H. longicornis from Shaanxi and 24.61%in R. microplus from Yunnan; Guo et al., 2019; Jiao et al., 2021). Our findings confirmed the widespread circulation of Ca. R. jingxinensis in China, as well as its high positive rate in ticks removed from livestock. Because most ticks were fully or partially engorged in the present work, it is not clear whether the Rickettsia DNA was from the blood meal or the tick itself. In another word, the possibility remains that the livestock in these locations may be infected by Ca. R. jingxinensis. These results remind us that the potential risk of Ca. R. jingxinensis infecting animals should be considered, and more attention should be paid to its role in human/animal diseases.
In R. haemaphysaloides ticks from Dehong and Honghe, a novel SFGR species named Ca. R. shennongii was identified. According to the gene sequence-based criteria for the identification of a new Rickettsia species (Fournier et al., 2003), genetic analysis of key genes clearly indicated that these strains represent a novel Rickettsia species. Sequences from all strains were almost identical, and they formed distinct clades in the phylogenetic trees. Genetically, this Rickettsia is closely related to R. raoultii, R. massiliae, and R. conorii (16S rRNA gene shares 99.91% identity with R. raoultii and R. conorii, while groEL gene shares 99.33% identity with R. massiliae), all of which are recognized human pathogens causing spotted fever. Therefore, the pathogenicity of this Rickettsia is a concern. Interestingly, this Rickettsia was only detected in R. haemaphysaloides, a widely distributed three-host tick in China and South Asian countries (Zhou et al., 2006). This species is known to harbor pathogens including A. phagocytophilum, A. ovis, R. rhipicephali, R. slovaca, R. massiliae, and Babesia microti (Kuo et al., 2018; Li et al., 2018; Ghafar et al., 2020; Ali et al., 2021). Our results may suggest the possibility of R. haemaphysaloides ticks harboring Ca. R. shennongii.
Herein, the DNA of three Anaplasma species was detected in all three locations including Candidatus Anaplasma boleense, A. ovis, and A. marginale. Anaplasma ovis is a widely-distributed pathogen affecting sheep, goats, and wild ruminants. As the etiological agent of ovine anaplasmosis first reported in 1956, the pathogenicity of A. ovis has been well studied since. Anaplasma ovis infection in sheep is usually subclinical and mostly manifested as hemolytic anemia (Dahmani et al., 2019). Other main clinical manifestations include extreme weakness, anorexia, and weight loss, but these manifestations mostly occur under poor health conditions (Bauer et al., 2021). In 2010, a human anaplasmosis case caused by an A. ovis variant was reported in Cyprus, with clinical symptoms of fever, hepatosplenomegaly, and enlarged lymph nodes (Chochlakis et al., 2010), providing evidence that A. ovis might be a potential zoonotic pathogen that may occasionally infect humans. In the present study, ticks from Dehong and Honghe were both tested positive for A. ovis, suggesting that A. ovis circulation is common in Yunnan Province, and surveillance in local populations is needed.
In this study, Ca. A. boleense was detected in ticks from all three sites of Yunnan Province. Candidatus Anaplasma boleense was first identified in ticks from Bole City in the Xinjiang Uygur Autonomous Region, Northwest China (Kang et al., 2014). In recent years, it has been detected in mosquitoes and rodents (FJ182047) in multiple provinces of China (Guo et al., 2016). In 2018, Ca. A. boleense was found to infect deer, boars, buffalos, cows, and bats in Peninsular Malaysia (Koh et al., 2018). In 2020, Ca. A. boleense was reported to infect marsh deer in Argentina, South America (Orozco et al., 2020). Furthermore, some sequences submitted to the GenBank database suggest that this Anaplasma exists in Australia (MH500004) and South Africa (MK814450), suggesting that it is distributed worldwide. However, few studies have been performed on the pathogenicity of Ca. A. boleense toward humans or other animals. The wide host range, as well as its wide geographical distribution, suggest that this Anaplasma species merits further investigation.
Ehrlichia is a genus closely related to human diseases. In this study, the DNA of 4 Ehrlichia species were identified, including E. canis, E. chaffeensis, E. minasensis, and Ehrlichia sp., of which E. canis and E. chaffeensis are recognized human pathogens (Perez et al., 1996; Bouza-Mora et al., 2017). The most prevalent species was E. canis (36.22% positive rate in ticks from Zhaotong), the causative agent of canine monocytic ehrlichiosis. In China, E. canis has been detected in various hosts including ticks, goats, dogs, and deer in multiple locations (Xu et al., 2015; Li et al., 2016; Qiu et al., 2016; Yu et al., 2016; Lu et al., 2017; Zhang et al., 2017). Notably, E. canis is considered a potential agent of human disease (Perez et al., 1996; Bouza-Mora et al., 2017), although human infection cases have never been reported in China. As shown in Figure 2, the strains we detected are most closely related to E. canis strain WHBMXZ-124, a strain identified in R. microplus from Wuhan, China. However, considerable genetic distance exists between them, suggesting that they might represent a variant circulating in China. As a recently recognized species closely related to E. canis, E. minasensis is believed to have evolved from highly variable strains of E. canis, and it has been discovered in Canada, Brazil, France, Pakistan, Ethiopia, etc. (Li et al., 2019). Other than ticks, it also infects mammals including cattle and cervid. In China, there is only one report of E. minasensis in Hainan Province, South China (Li et al., 2019). In our current study, E. minasensis was detected in R. microplus ticks from Honghe city, with a high positive rate of 31.07%. To the best of our knowledge, this is only the second report of E. minasensis circulating in China. Due to its pathogenicity to mammals, the close relationship to human pathogenic species E. canis, and the high infection rate in ticks, its pathogenicity to humans and animals should be further explored.
In summary, this study revealed substantial diversity of Rickettsia, Ehrlichia, and Anaplasma in ticks from Yunnan Province, including a novel Rickettsia species. Notably, some of these bacteria are human pathogens (E. canis, E. chaffeensis, A. ovis, etc) and the positive rates are high. Considering the frequent contact between humans and tick hosts (goats and cattle), these results indicate the potential risk of zoonosis transmitted from ticks to humans. Additionally, the human pathogenicity of Ca. R. shennongii should be further studied and surveillance in these areas is clearly needed.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
This study was approved by the Ethics Committee of the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention.
KL conceived the study. KL and ML designed the experiments. KL, JT, and JH collected the samples. ML and WW performed the experiments. HZ, KL, and WG performed data analysis. HZ, HJ, and KL wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the National Natural Science Foundation of China (grant no. 82102390), the Key Supporting Scientific Research Projects of Beijing Road Medical Sector, General Hospital of Xinjiang Military Region (2022jzbjl16), the National Key Research and Development Program of China (grant nos. 2020YFA0907101, 2021YFC2301200, and 2021YFC2301202), and the Medical youth top talent project of Hubei.
We sincerely thank Wenjie Wang, Hejie Yang, Hongyu Ren, and Xiaojing Jin for their warmhearted help.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1008110/full#supplementary-material
Ali, A., Zahid, H., Zeb, I., Tufail, M., Khan, S., Haroon, M., et al. (2021). Risk factors associated with tick infestations on equids in Khyber Pakhtunkhwa, Pakistan, with notes on Rickettsia massiliae detection. Parasite Vector. 14:363. doi: 10.1186/s13071-021-04836-w
Bang, M. S., Kim, C. M., Pyun, S. H., Kim, D. M., and Yun, N. R. (2021). Molecular investigation of tick-borne pathogens in ticks removed from tick-bitten humans in the southwestern region of the Republic of Korea. PLoS One 16:e0252992. doi: 10.1371/journal.pone.0252992
Bauer, B. U., Răileanu, C., Tauchmann, O., Fischer, S., Ambros, C., Silaghi, C., et al. (2021). Anaplasma phagocytophilum and Anaplasma ovis-emerging pathogens in the German sheep population. Pathogens. 10:1298. doi: 10.3390/pathogens10101298
Bouza-Mora, L., Dolz, G., Solórzano-Morales, A., Romero-Zuñiga, J. J., Salazar-Sánchez, L., Labruna, M. B., et al. (2017). Novel genotype of Ehrlichia canis detected in samples of human blood bank donors in Costa Rica. Ticks Tick Borne Dis. 8, 36–40. doi: 10.1016/j.ttbdis.2016.09.012
Chochlakis, D., Ioannou, I., Tselentis, Y., and Psaroulaki, A. (2010). Human anaplasmosis and Anaplasma ovis variant. Emerg. Infect. Dis. 16, 1031–1032. doi: 10.3201/eid1606.090175
Colomba, C., Saporito, L., Polara, V. F., Rubino, R., and Titone, L. (2006). Mediterranean spotted fever: clinical and laboratory characteristics of 415 Sicilian children. BMC Infect. Dis. 6:60. doi: 10.1186/1471-2334-6-60
Dahmani, M., Davoust, B., Sambou, M., Bassene, H., Scandola, P., Ameur, T., et al. (2019). Molecular investigation and phylogeny of species of the Anaplasmataceae infecting animals and ticks in Senegal. Parasite Vector. 12:495. doi: 10.1186/s13071-019-3742-y
Dumic, I., Jevtic, D., Veselinovic, M., Nordstrom, C. W., Jovanovic, M., Mogulla, V., et al. (2022). Human granulocytic anaplasmosis - a systematic review of published cases. Microorganisms. 10:1433. doi: 10.3390/microorganisms10071433
Fournier, P. E., Dumler, J. S., Greub, G., Zhang, J., Wu, Y., and Raoult, D. (2003). Gene sequence-based criteria for identification of new Rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J. Clin. Microbiol. 41, 5456–5465. doi: 10.1128/JCM.41.12.5456-5465.2003
Ghafar, A., Khan, A., Cabezas-Cruz, A., Gauci, C. G., Niaz, S., Ayaz, S., et al. (2020). An assessment of the molecular diversity of ticks and tick-borne microorganisms of small ruminants in Pakistan. Microorganisms 8:1428. doi: 10.3390/microorganisms8091428
Gondard, M., Cabezas-Cruz, A., Charles, R. A., Vayssier-Taussat, M., Albina, E., and Moutailler, S. (2017). Ticks and tick-borne pathogens of the Caribbean: current understanding and future directions for more comprehensive surveillance. Front. Cell. Infect. Microbiol. 7:490. doi: 10.3389/fcimb.2017.00490
Graham, R. M. A., Donohue, S., McMahon, J., and Jennison, A. V. (2017). Detection of spotted fever group Rickettsia DNA by deep sequencing. Emerg. Infect. Dis. 23, 1911–1913. doi: 10.3201/eid2311.170474
Guo, W. P., Tian, J. H., Lin, X. D., Ni, X. B., Chen, X. P., Liao, Y., et al. (2016). Extensive genetic diversity of Rickettsiales bacteria in multiple mosquito species. Sci. Rep. 6:38770. doi: 10.1038/srep38770
Guo, W. P., Wang, Y. H., Lu, Q., Xu, G., Luo, Y., Ni, X., et al. (2019). Molecular detection of spotted fever group Rickettsiae in hard ticks, northern China. Transbound. Emerg. Dis. 66, 1587–1596. doi: 10.1111/tbed.13184
Ismail, N., and McBride, J. W. (2017). Tick-dorne emerging infections: ehrlichiosis and anaplasmosis. Clin. Lab. Med. 37, 317–340. doi: 10.1016/j.cll.2017.01.006
Jiao, J., Zhang, J., He, P., OuYang, X., Yu, Y., Wen, B., et al. (2021). Identification of tick-borne pathogens and genotyping of Coxiella burnetii in Rhipicephalus microplus in Yunnan Province, China. Front Microbiol. 12:736484. doi: 10.3389/fmicb.2021.736484
Kang, Y. J., Diao, X. N., Zhao, G. Y., Chen, M. H., Xiong, Y., Shi, M., et al. (2014). Extensive diversity of Rickettsiales bacteria in two species of ticks from China and the evolution of the Rickettsiales. BMC Evol. Biol. 14:167. doi: 10.1186/s12862-014-0167-2
Koh, F. X., Panchadcharam, C., Sitam, F. T., and Tay, S. T. (2018). Molecular investigation of Anaplasma spp. in domestic and wildlife animals in peninsular Malaysia. Vet. Parasitol. Reg. Stud. Rep. 13, 141–147. doi: 10.1016/j.vprsr.2018.05.006
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kuo, C. C., Huang, J. L., Chien, C. H., Shih, H. C., and Wang, H. C. (2018). First molecular detection of Anaplasma phagocytophilum in the hard tick Rhipicephalus haemaphysaloides in Taiwan. Exp. Appl. Acarol. 75, 437–443. doi: 10.1007/s10493-018-0283-6
Li, Y., Chen, Z., Liu, Z., Liu, J., Yang, J., Li, Q., et al. (2016). Molecular survey of Anaplasma and Ehrlichia of red deer and Sika deer in Gansu, China in 2013. Transbound. Emerg. Dis. 63, e228–e236. doi: 10.1111/tbed.12335
Li, R., Kraft, N. J., Yang, J., and Wang, Y. (2015). A phylogenetically informed delineation of floristic regions within a biodiversity hotspot in Yunnan, China. Sci Rep. 5:9396. doi: 10.1038/srep09396
Li, J., Liu, X., Mu, J., Yu, X., Fei, Y., Chang, J., et al. (2019). Emergence of a novel Ehrlichia minasensis strain, harboring the major immunogenic glycoprotein trp 36 with unique tandem repeat and C-terminal region sequences, in Haemaphysalis hystricis ticks removed from free-ranging sheep in Hainan Province, China. Microorganisms 7:369. doi: 10.3390/microorganisms7090369
Li, L. H., Zhang, Y., Zhu, D., and Zhou, X. N. (2018). Endosymbionts alter larva-to-nymph transstadial transmission of Babesia microti in Rhipicephalus haemaphysaloides ticks. Front. Microbiol. 9:1415. doi: 10.3389/fmicb.2018.01415
Li, H., Zheng, Y. C., Ma, L., Jia, N., Jiang, B. G., Jiang, R. R., et al. (2015). Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect. Dis. 15, 663–670. doi: 10.1016/S1473-3099(15)70051-4
Liang, C. W., Zhao, J. B., Li, J., Chang, L. T., Yu, H. L., Zhang, L. X., et al. (2012). Spotted fever group Rickettsia in Yunnan Province, China. Vector Borne Zoonotic Dis. 12, 281–286. doi: 10.1089/vbz.2011.0835
Liu, H., Li, Q., Zhang, X., Li, Z., Wang, Z., Song, M., et al. (2016). Characterization of Rickettsiae in ticks in northeastern China. Parasite Vector. 9:498. doi: 10.1186/s13071-016-1764-2
Liu, H., Liang, X., Wang, H., Sun, X., Bai, X., Hu, B., et al. (2020). Molecular evidence of the spotted fever group Rickettsiae in ticks from Yunnan Province, Southwest China. Exp. Appl. Acarol. 80, 339–348. doi: 10.1007/s10493-020-00467-5
Lu, M., Li, F., Liao, Y., Shen, J. J., Xu, J. M., Chen, Y. Z., et al. (2019). Epidemiology and diversity of Rickettsiales bacteria in humans and animals in Jiangsu and Jiangxi provinces, China. Sci Rep. 9:13176. doi: 10.1038/s41598-019-49059-3
Lu, X., Lin, X. D., Wang, J. B., Qin, X. C., Tian, J. H., Guo, W. P., et al. (2013). Molecular survey of hard ticks in endemic areas of tick-borne diseases in China. Ticks Tick Borne Dis. 4, 288–296. doi: 10.1016/j.ttbdis.2013.01.003
Lu, M., Tian, J. H., Pan, X. L., Qin, X. C., Wang, W., Chen, J. T., et al. (2022). Identification of Rickettsia spp., Anaplasma spp., and an Ehrlichia canis-like agent in Rhipicephalus microplus from southwest and south-Central China. Ticks Tick Borne Dis. 13:101884. doi: 10.1016/j.ttbdis.2021.101884
Lu, M., Tian, J. H., Yu, B., Guo, W. P., Holmes, E. C., and Zhang, Y. Z. (2017). Extensive diversity of rickettsiales bacteria in ticks from Wuhan, China. Ticks Tick Borne Dis. 8, 574–580. doi: 10.1016/j.ttbdis.2017.03.006
Mogg, M., Wang, H. H., Baker, A., Derouen, Z., Borski, J., and Grant, W. E. (2020). Increased incidence of Ehrlichia chaffeensis infections in the United States, 2012 through 2016. Vector Borne Zoonotic Dis. 20, 547–550. doi: 10.1089/vbz.2019.2595
Namgyal, J., Lysyk, T. J., Couloigner, I., Checkley, S., Gurung, R. B., Tenzin, T., et al. (2021). Identification, distribution, and habitat suitability models of Ixodid tick species in cattle in eastern Bhutan. Trop. Med. Infect. Dis. 6:27. doi: 10.3390/tropicalmed6010027
Orozco, M. M., Argibay, H. D., Minatel, L., Guillemi, E. C., Berra, Y., Schapira, A., et al. (2020). A participatory surveillance of marsh deer (Blastocerus dichotomus) morbidity and mortality in Argentina: first results. BMC Vet. Res. 16:321. doi: 10.1186/s12917-020-02533-x
Parola, P., Paddock, C. D., Socolovschi, C., Labruna, M. B., Mediannikov, O., Kernif, T., et al. (2013). Update on tick-borne rickettsioses around the world: a geographic approach. Clin. Microbiol. Rev. 26, 657–702. doi: 10.1128/CMR.00032-13
Perez, M., Rikihisa, Y., and Wen, B. (1996). Ehrlichia canis-like agent isolated from a man in Venezuela: antigenic and genetic characterization. J. Clin. Microbiol. 34, 2133–2139. doi: 10.1128/jcm.34.9.2133-2139.1996
Peter, S. G., Aboge, G. O., Kariuki, H. W., Kanduma, E. G., Gakuya, D. W., Maingi, N., et al. (2020). Molecular prevalence of emerging Anaplasma and Ehrlichia pathogens in apparently healthy dairy cattle in peri-urban Nairobi, Kenya. BMC Vet. Res. 16:364. doi: 10.1186/s12917-020-02584-0
Pritt, B. S., Allerdice, M. E. J., Sloan, L. M., Paddock, C. D., Munderloh, U. G., Rikihisa, Y., et al. (2017). Proposal to reclassify Ehrlichia muris as Ehrlichia muris subsp. muris subsp. nov. and description of Ehrlichia muris subsp. eauclairensis subsp. nov., a newly recognized tick-borne pathogen of humans. Int. J. Syst. Evol. Microbiol. 67, 2121–2126. doi: 10.1099/ijsem.0.001896
Qiu, H., Kelly, P. J., Zhang, J., Luo, Q., Yang, Y., Mao, Y., et al. (2016). Molecular detection of Anaplasma spp. and Ehrlichia spp. in ruminants from twelve provinces of China. Can. J. Infect. Dis. Med. 2016:9183861. doi: 10.1155/2016/9183861
Rjeibi, M. R., Ayadi, O., Rekik, M., and Gharbi, M. (2018). Molecular survey and genetic characterization of Anaplasma Centrale, A. marginale and A. bovis in cattle from Algeria. Transbound. Emerg. Dis. 65, 456–464. doi: 10.1111/tbed.12725
Roux, V., and Raoult, D. (2000). Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int. J. Syst. Evol. Microbiol. 50, 1449–1455. doi: 10.1099/00207713-50-4-1449
Rudakov, N., Samoylenko, I., Shtrek, S., Igolkina, Y., Rar, V., Zhirakovskaia, E., et al. (2019). A fatal case of tick-borne rickettsiosis caused by mixed Rickettsia sibirica subsp. sibirica and "Candidatus Rickettsia tarasevichiae" infection in Russia. Ticks Tick Borne Dis. 10:101278. doi: 10.1016/j.ttbdis.2019.101278
Saito, T. B., and Walker, D. H. (2016). Ehrlichioses: an important one health opportunity. Vet. Sci. 3:20. doi: 10.3390/vetsci3030020
Sekeyova, Z., Roux, V., and Raoult, D. (2001). Phylogeny of Rickettsia spp. inferred by comparing sequences of 'gene D', which encodes an intracytoplasmic protein. Int. J. Syst. Evol. Microbiol. 51, 1353–1360. doi: 10.1099/00207713-51-4-1353
Sentausa, E., El Karkouri, K., Robert, C., Raoult, D., and Fournier, P. E. (2012). Genome sequence of Rickettsia conorii subsp. caspia, the agent of astrakhan fever. J. Bacteriol. 194, 4763–4764. doi: 10.1128/JB.00992-12
Stenos, J., Graves, S. R., and Unsworth, N. B. (2005). A highly sensitive and specific real-time PCR assay for the detection of spotted fever and typhus group Rickettsiae. Am. J. Trop. Med. Hyg. 73, 1083–1085. doi: 10.4269/ajtmh.2005.73.1083
Takhampunya, R., Korkusol, A., Pongpichit, C., Yodin, K., Rungrojn, A., Chanarat, N., et al. (2019). Metagenomic approach to characterizing disease epidemiology in a disease-endemic environment in northern Thailand. Front. Microbiol. 10:319. doi: 10.3389/fmicb.2019.00319
von Fricken, M. E., Qurollo, B. A., Boldbaatar, B., Wang, Y. W., Jiang, R. R., Lkhagvatseren, S., et al. (2021). Genetic diversity of Anaplasma and Ehrlichia bacteria found in Dermacentor and Ixodes ticks in Mongolia. Ticks Tick Borne Dis. 11:101316. doi: 10.1016/j.ttbdis.2019.101316
Xu, D., Zhang, J., Shi, Z., Song, C., Zheng, X., Zhang, Y., et al. (2015). Molecular detection of vector-borne agents in dogs from ten provinces of China. Parasite Vector. 8:501. doi: 10.1186/s13071-015-1120-y
Yu, P. F., Niu, Q. L., Liu, Z. J., Yang, J. F., Chen, Z., Guan, G. Q., et al. (2016). Molecular epidemiological surveillance to assess emergence and re-emergence of tick-borne infections in tick samples from China evaluated by nested PCRs. Acta Trop. 158, 181–188. doi: 10.1016/j.actatropica.2016.02.027
Zazueta, O. E., Armstrong, P. A., Márquez-Elguea, A., Hernández Milán, N. S., Peterson, A. E., Ovalle-Marroquín, D. F., et al. (2021). Rocky Mountain spotted fever in a large metropolitan center, Mexico-United States border, 2009-2019. Emerg. Infect. Dis. 27, 1567–1576. doi: 10.3201/eid2706.191662
Zhang, J., Liu, Q., Wang, D., Li, W., Beugnet, F., and Zhou, J. (2017). Epidemiological survey of ticks and tick-borne pathogens in pet dogs in South-Eastern China. Parasite 24:35. doi: 10.1051/parasite/2017036
Keywords: Rickettsia, anaplasma, ehrlichia, Yunnan Province, Candidatus Rickettsia shennongii
Citation: Lu M, Tian J, Wang W, Zhao H, Jiang H, Han J, Guo W and Li K (2022) High diversity of Rickettsia spp., Anaplasma spp., and Ehrlichia spp. in ticks from Yunnan Province, Southwest China. Front. Microbiol. 13:1008110. doi: 10.3389/fmicb.2022.1008110
Received: 15 August 2022; Accepted: 20 September 2022;
Published: 13 October 2022.
Edited by:
Michael E. von Fricken, George Mason University, United StatesReviewed by:
Zhijun Hou, Northeast Forestry University, ChinaCopyright © 2022 Lu, Tian, Wang, Zhao, Jiang, Han, Guo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun Li, bGlrdW5AaWNkYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.